Abstract
Tanshinone ⅡA (TⅡA), a diterpene quinone with a furan ring, is a bioactive compound found in the medicinal herb redroot sage (Salvia miltiorrhiza Bunge), in which both furan and dihydrofuran analogs are present in abundance. Progress has been made recently in elucidating the tanshinone biosynthetic pathway, including heterocyclization of the dihydrofuran D-ring by cytochrome P450s; however, dehydrogenation of dihydrofuran to furan, a key step of furan ring formation, remains uncharacterized. Here, by differential transcriptome mining, we identified six 2-oxoglutarate-dependent dioxygenase (2-ODD) genes whose expressions corresponded to tanshinone biosynthesis. We showed that Sm2-ODD14 acts as a dehydrogenase catalyzing the furan ring aromatization. In vitro Sm2-ODD14 converted cryptotanshinone to TⅡA and thus was designated TⅡA synthase (SmTⅡAS). Furthermore, SmTⅡAS showed a strict substrate specificity, and repression of SmTⅡAS expression in hairy root by RNAi led to increased accumulation of total dihydrofuran-tanshinones and decreased production of furan-tanshinones. We conclude that SmTⅡAS controls the metabolite flux from dihydrofuran- to furan-tanshinones, which influences medicinal properties of S. miltiorrhiza.
A 2-oxoglutarate-dependent dioxygenase is responsible for tanshinone ⅡA biosynthesis in medicinal herb Salvia miltiorrhiza, which controls the metabolite flux from dihydrofuran- to furan-tanshinones.
Introduction
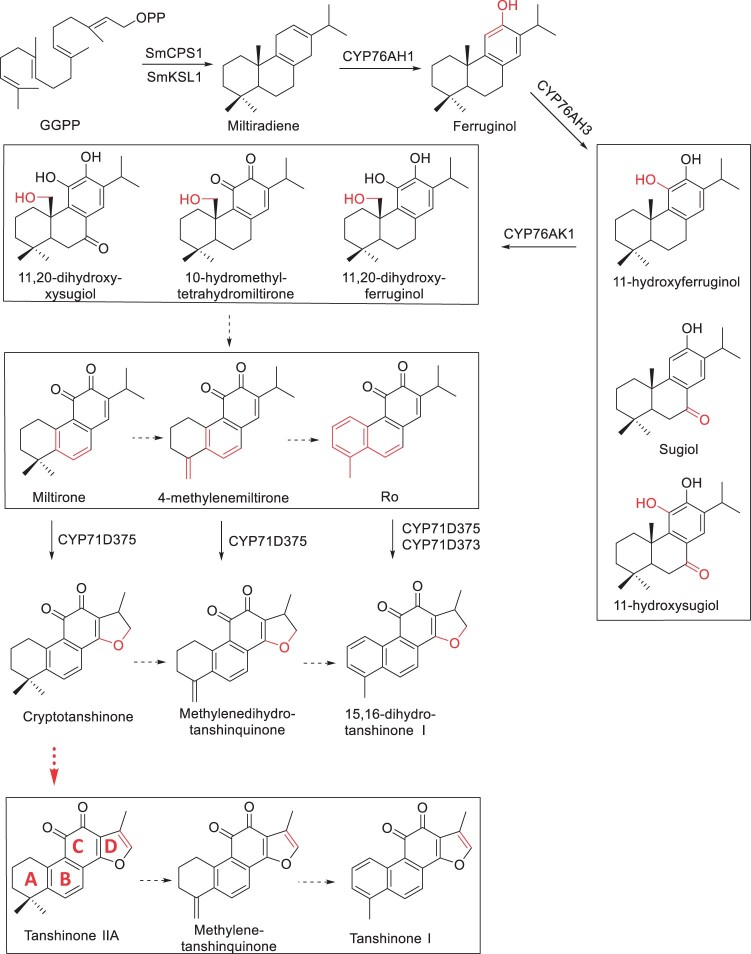
Plants of the family Lamiaceae produce a rich array of diterpenoids of varied skeletons (Johnson et al., 2019). Salvia miltiorrhiza Bunge, a medicinal species known as Danshen or red sage in China, accumulates in roots a group of abietane-type nor-diterpenes, collectively named tanshinones (Dong et al., 2011). To date, a total of 81 tanshinones were identified, which show therapeutically relevant biological activities including antibiotic, anti-inflammatory, and antioxidant properties (Wang et al., 2007). Due to their structural uniqueness and medicinal interests, the biosynthesis of tanshinones has been investigated intensively in last two decades (Figure 1). The diterpene synthases S. miltiorrhiza copalyl diphosphate synthase 1 and S. miltiorrhiza kaurene synthase-like 1 produce the tricyclic miltiradiene (Gao et al., 2009), then the CYP76AH subfamily enzymes hydroxylate and aromatize the C-ring, followed by C20 hydroxylation by CYP76AK1 (Guo et al., 2013, 2016). Very recently, two CYP71D enzymes (CYP71D375 and CYP71D373) were shown to catalyze the D-ring formation through C16 hydroxylation and 14,16-ether (hetero) cyclization, leading to the formation of dihydrofuran-tanshinones from their respective precursors (Ma et al., 2021).
Figure 1.
The biosynthetic pathway of tanshinones in S. miltiorrhiza. Solid arrows denote the known steps, dashed arrows denote the hypothetical steps, and the red arrow indicates the step will be elucidated in present study. The promiscuity of identified CYPs suggests a metabolic grid architecture for diterpenoid biosynthesis.
Among the tanshinones in S. miltiorrhiza, tanshinone ⅡA (TⅡA) is considered the major active ingredient (Fang et al., 2021). In 3T3-L1 cells and zebrafish TⅡA inhibited lipid accumulation (Park et al., 2017), and the derivate sodium TⅡA silate is used in China for alleviating cardiovascular and coronary heart diseases (Li et al., 2020b). Both T I and TⅡA, differing in A-ring aromatization, are typical tetracyclic tanshinones featuring an ortho-quinone C-ring and a furan D-ring (Figure 1), while cryptotanshinone (CT) and 15,16-dihydrotanshinone I (DTI) with dihydrofuran ring were believed to be their precursors, respectively (Ma et al., 2021). However, till now, aromatization of the D-ring to a furan in tanshinone biosynthesis remains uncharacterized.
Furan moieties occur in a wide variety of natural products, including many pharmaceuticals. Despite their importance, our understanding of the furan ring desaturation mechanism remains rudimentary. CYP71AJ1 and CYP71AJ4 in plants of the Apiaceae family catalyze the formation of linear and angular furanocoumarins, respectively, through concomitant carbon-chain cleavage and acetone releasing which, however, do not involve a separate desaturation step during furan formation (Larbat et al., 2007, 2009). In the biosynthesis of flavonoids, flavonol synthase and flavone synthase I, both belonging to the 2-oxoglutarate-dependent dioxygenase (2-ODD) family, catalyze C2–C3 desaturation to form the central pyran ring (Turnbull et al., 2004; Cheng et al., 2014). To date, the enzymes responsible for converting dihydrofuran or tetrahydrofuran to furan have not been identified.
In plants, the 2-ODD or 2-OGD superfamily comprises the second largest enzyme family besides the cytochrome P450s (CYPs). Based on similarity of the amino acid sequences, the plant 2-ODD family could be divided into DOXA, DOXB, and DOXC classes (Kawai et al., 2014). DOXA and DOXB function in nucleotide and protein modification, respectively; while DOXC class is involved in secondary metabolism of various phytochemicals including glucosinolates, flavonoids, and alkaloids (Araujo et al., 2014), by which the plant defend against biotic stresses (Ge et al., 2021). 2-ODD members facilitate numerous oxidative reactions, including hydroxylation, desaturation, demethylation, halogenation, epoxidation, and ring formation (Islam et al., 2018). Genome-wide analysis of 2-ODD superfamily has been reported in S. miltiorrhiza and, based on RNAi data obtained from hairy roots, 2OGD5 was found to affect the accumulation of CT, TⅡA, and miltirone (Xu and Song, 2017), but the biochemical evidence was lacking.
Here, we characterize a 2-ODD protein from S. miltiorrhiza, which catalyzes the desaturation of the dihydrofuran ring in CT and isoCT (iCT), and was designated S. miltiorrhiza TⅡA synthase (SmTⅡAS). In addition, among the 2-ODD candidates screened, SmTⅡAS is the only active dihydrofuran desaturase and exhibits high substrate selectivity. We propose that SmTⅡAS represents an example of furan synthase in general, in addition to be a key enzyme responsible for channeling dihydrofuran-tanshinones to the furan products.
Results
Mining 2-ODDs upregulated in root
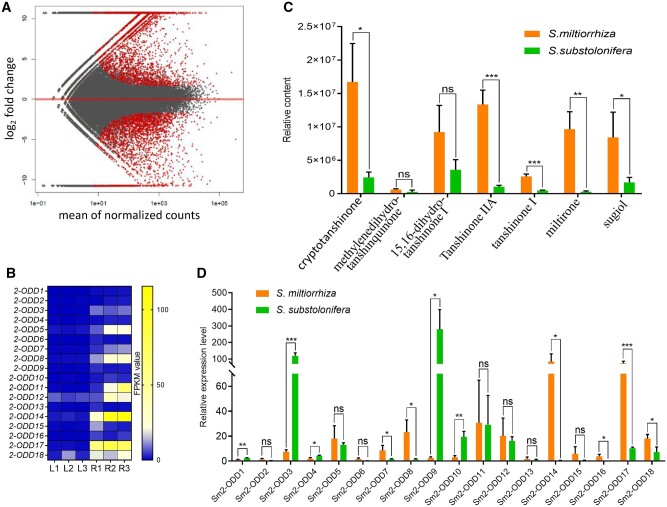
The biosynthesis and accumulation of secondary metabolites tend to be tissue-specific, and the enzymes involved are expected to share a similar gene expression pattern (Murata et al., 2008; Beaudoin and Facchini, 2014). Although trace amounts of tanshinones can be detected in the aerial tissues of S. miltiorrhiza, these diterpenoids are primarily synthesized and stored in root (Li et al., 2008a; Yang et al., 2013). To mine the candidate enzymes responsible for the desaturation of the dihydrofuran ring in tanshinones, we generated transcriptomes of root and leaf tissues of this species, respectively. The RNA-seq produced 112,846 unigenes, of which 44.41% were annotated in SWISSPROT database (Supplemental Table S1). After differential expression analysis (fold change > 2), we obtained 12,048 genes which were upregulated in root compared to leaf (Figure 2A), among which a total of 18 genes were annotated by scanning DIOX_N and 2-oxoglutaric acid (2OG)-Fell_Oxy domains to encode 2-ODDs, namely Sm2-ODD1 to 18 (Figure 2B;Supplemental Table S2). Except for Sm2-ODD11 and Sm2-ODD16, other 16 Sm2-ODDs have homologs (amino acid sequence identity ranged from 96.26% to 100%) to those previously reported by Xu and Song (2017) (Supplemental Table S2); however, none of Sm2-ODDs showed high identity to 2OGD5 (Xu and Song, 2017).
Figure 2.
The mining of candidate 2-ODDs involved in the tanshinone biosynthetic pathway in S. miltiorrhiza. A, Differentially expressed genes in leaf and root, the up- and down-regulated genes are indicated by red spots. B, The transcript levels of 18 2-ODD genes by FPKM in S. miltiorrhiza root (R) and leaf (L) transcriptomes. C, Accumulation of tanshinones in root of S. miltiorrhiza and S. substolonifera. D, Expression levels of the 18 candidate 2-ODDs by FPKM in roots of S. miltiorrhiza and S. substolonifera. For (C) and (D), Data are means ± sd (Standard Deviation) of three biological replicates. Statistical analysis was performed with Student’s t test. *P < 0.05; **P < 0.01, ***P < 0.001, and ns indicates no significance.
Another species of the genus, Salvia substolonifera, has a much lower relative content of tanshinones, such as CT, TⅡA, tanshinone I, and miltirone, in its root (Figure 2C;Supplemental Figure S1), which provided a good reference to select the genes of the tanshinone pathway. We thus compared the relative transcript levels of the 18 2-ODDs in roots of these two Salvia species. The FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) values from the Illumina-generated RNA-seq data narrowed the 18 2-ODD genes down to six, including Sm2-ODD7, Sm2-ODD8, Sm2-ODD14, Sm2-ODD16, Sm2-ODD17, and Sm2-ODD18, whose transcript levels were clearly higher in S. miltiorrhiza than in S. substolonifera (Figure 2D), suggesting that they could serve as oxidative enzymes in the biosynthesis of TⅡA, tanshinone I, or structurally related diterpenoids.
Functional characterization of Sm2-ODD14
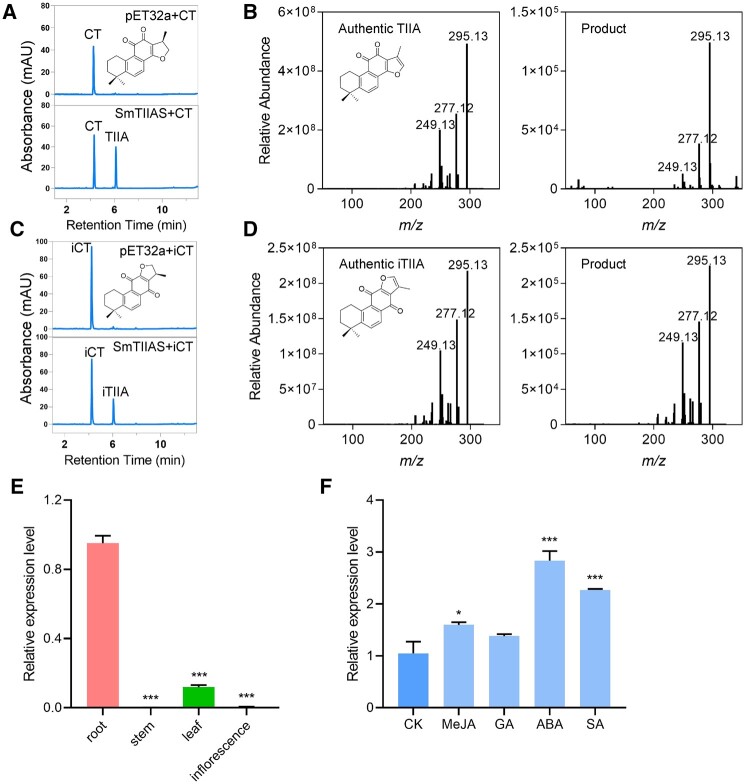
The six 2-ODDs, namely Sm2-ODD7, Sm2-ODD8, Sm2-ODD14, Sm2-ODD16, Sm2-ODD17, and Sm2-ODD18, were then expressed in Escherichia coli. In subsequent in vitro assays, only the recombinant protein of Sm2-ODD14 was active toward CT and converted it into a single product (Figure 3A), which was determined to be TⅡA by comparison to the authentic standard in Q Exactive (QE) plus mass analysis (Figure 3B); consequently, the enzyme was designated SmTⅡAS, because other products such as methylenetanshinquinone and tanshinone I derived from TⅡA were not detected in the enzymatic assay. To define the substrate spectrum of Sm2-ODD14, several structural analogs of CT, including methylenedihydrotanshinquinone, DTI, iCT, tetrahydrotanshinone I, and 1,2-didehydrocryptotanshinone, were tested. Sm2-ODD14 was found to be able to convert iCT to isotanshinone ⅡA (iTⅡA) (Figure 3, C and D), but inactive to other compounds (Supplemental Figure S2). Structurally, CT and iCT are different from other analogs in sharing a saturated A-ring with two methyl groups, which may be the key feature recognized by Sm2-ODD14. In conclusion, Sm2-ODD14 acted as a desaturase and introduced a double bond between C15 and C16 to complete the furan ring formation.
Figure 3.
Characterization of SmTⅡAS by in vitro assay and its relative expression. A, UPLC profile of the product generated by SmTⅡAS with CT as substrate. B, Authentic standard of TⅡA and the SmTⅡAS product, detected by QE. C, UPLC profile of the product of SmTⅡAS incubated with iCT as substrate. D, Authentic standard of iTⅡA and the SmTⅡAS product, detected by QE. The recombinant protein of SmTⅡAS were incubated with the indicated substrate at 20°C, for 7 min. E, Relative expression of SmTⅡAS in different tissues. F, Relative expression of SmTⅡAS after different phytohormones treatments, including MeJA, SA, ABA, GA, and with DMSO solution (5%) as control (CK). For (E) and (F), data are means ± sd of three biological replicates. Statistical analysis was performed with Student’s t test.
To optimize the reaction conditions, the activities of recombinant proteins were assessed with CT as substrate at different temperatures (20°C, 28°C, and 37°C) and pH values (6.5 and 7.4), and the optimal condition was fixed to be at 20°C and pH 6.5 (Supplemental Figure S3). To test the effects of cofactors on enzymatic activity, we first omitted 2OG in the assay, which abolished the SmTⅡAS activity. In addition, although not indispensable, the cofactors L-ascorbic acid (ASC) and Fe2+ facilitated the catalysis (Supplemental Figure S4a), likely through promoting the enzyme–substrate interaction. Kinetic analysis with CT and iCT in the presence of 2OG, ASC, and Fe2+ showed that the Km values of SmTⅡAS were 67.71 ± 7.30 and 29.71 ± 10.51 μM, the estimated kcat values were 0.60 ± 0.03 and 0.27 ± 0.09 s−1, and the kcat/Km were 8,843.45 ± 546.65 s−1M−1 and 9,200.85 ± 353.52 s−1M−1, respectively (Supplemental Figure S4, b and c). Although the enzyme showed higher affinity toward iCT, considering TⅡA being much more abundant than iTⅡA in S. miltiorrhiza, the function of SmTⅡAS in planta is mainly catalyzing the conversion of CT to TⅡA.
The tissue-specific expression pattern of SmTⅡAS were analyzed by using reverse transcription quantitative PCR (RT-qPCR) which showed the highest relative expression level in root and much lower levels in aerial tissues, especially in stem and inflorescence the expression levels were nearly undetectable (Figure 3E). This result is in agreement of the root-specific accumulation of tanshinones (Li et al., 2008a; Yang et al., 2013). Furthermore, the enhancement of the accumulation of tanshinones was reported to be triggered by treatment of several phytohormones including methyl jasmonate (MeJA), salicylic acid (SA), abscisic acid (ABA), and gibberellic acid (GA) (Pei et al., 2018). Accordingly, the expression levels of SmTⅡAS increased after treatment of the hairy roots culture of S. miltiorrhiza by ABA and SA and at a very limited level by MeJA (Figure 3F). These results further confirmed the function of SmTⅡAS in tanshinones biosynthesis.
Silencing SmTⅡAS in hairy root reduced the formation of furan-tanshinones
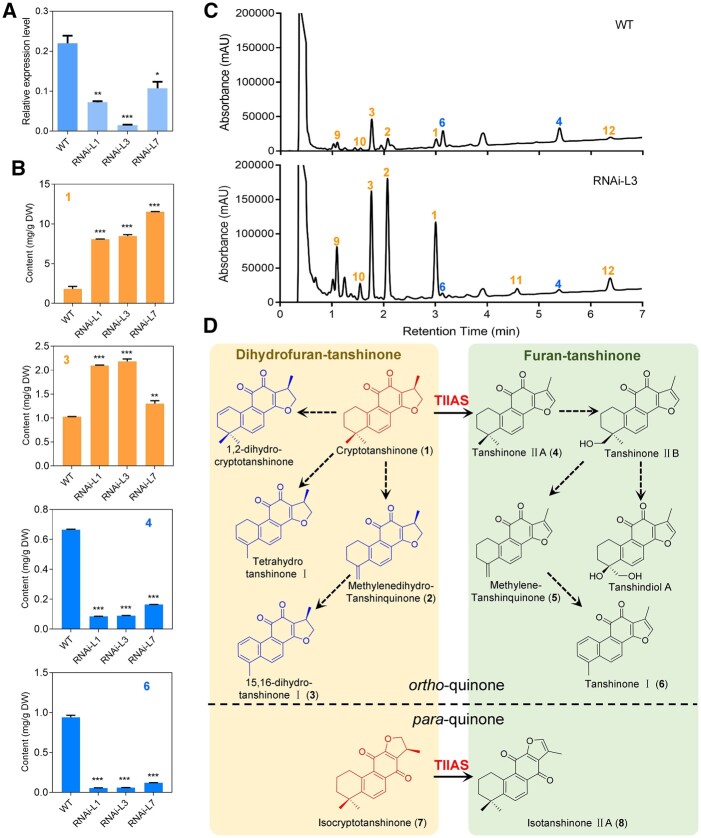
To prove the function of SmTⅡAS in vivo, we repressed the SmTⅡAS expression in S. miltiorrhiza hairy roots by RNA interference (RNAi), using a SmTⅡAS-specific region of 395 bp with no or low sequence identity to other 17 2-ODD sequences. Three RNAi hairy root lines (RNAi-L1, RNAi-L3, and RNAi-L7) were selected, in which the SmTⅡAS transcript level was decreased to 30%–50% of the WT level (Figure 4A). Analysis of the metabolites by UPLC (Ultra Performance Liquid Chromatography) showed that the level of TⅡA was decreased from 0.66 mg g−1 DW (dry weight) in the control to 0.08, 0.09, and 0.16 mg g−1 DW in the three RNAi hairy root lines, respectively; the SmTⅡAS substrate CT, in contrast, was over-accumulated in the RNAi hairy roots (Figure 4, B and C). Notably, the contents of methylenedihydrotanshinquinone and DTI also increased in the RNAi roots, while the tanshinone I level was reduced (Figure 4C;Supplemental Figure S5). Some tanshinones contents, such as methylenetanshinquinone, TⅡB, and tanshindiol A, were too low to be quantified both in the WT and the RNAi hairy root lines. Since SmTⅡAS could not accept methylenedihydrotanshinquinone or DTI as substrates, knocking-down the SmTⅡAS expression could not directly result in accumulation of these dihydrofuran-tanshinones. A more likely scenario is that the A-ring decorating enzymes do not distinguish between the dihydrofuran and furan D-ring, and the conversion from CT to TⅡA catalyzed by SmTⅡAS is the major path to the formation of furan-tanshinones, and the decreased transcript level of SmTⅡAS impeded the flux toward furan-tanshinones that resulted in accumulation of dihydrofuran-tanshinones, including CT, methylenedihydrotanshinquinone and DTI (Figure 4D).
Figure 4.
The effects of SmTⅡAS silencing by RNAi on the accumulation of tanshinones. A, Relative expression of SmTⅡAS in the RNAi hairy root lines detected by RT-qPCR (reverse transcription quantitative PCR). B, Contents of four tanshinones in the RNAi hairy roots (numbers shown in the panels correspond with the numbered compounds in (D). Data are means ± sd of three biological replicates. Statistical analysis was performed with Student’s t test. C, UPLC comparison of the WT and the RNAi hairy roots, line 3 (RNAi-L3). The mass spectra of the compounds were given in Supplementary Figure S5. Orange color and blue color indicate the increased and decreased content of compounds in the RNAi line, respectively. D, The catalytic role of SmTⅡAS in converting dihydrofuran-tanshinones to furan-tanshinones. The structures in red, but not those in blue, were accepted by SmTⅡAS. Peaks in the total ion chromatograms of (C) are numbered and assigned to corresponding structures (with identical numbers) in (D) while peaks marked with 9–12 could not be assigned structures, and the corresponding mass spectra were given in Supplemental Figure S5.
Lineage specificity of TⅡASs in Salvia species
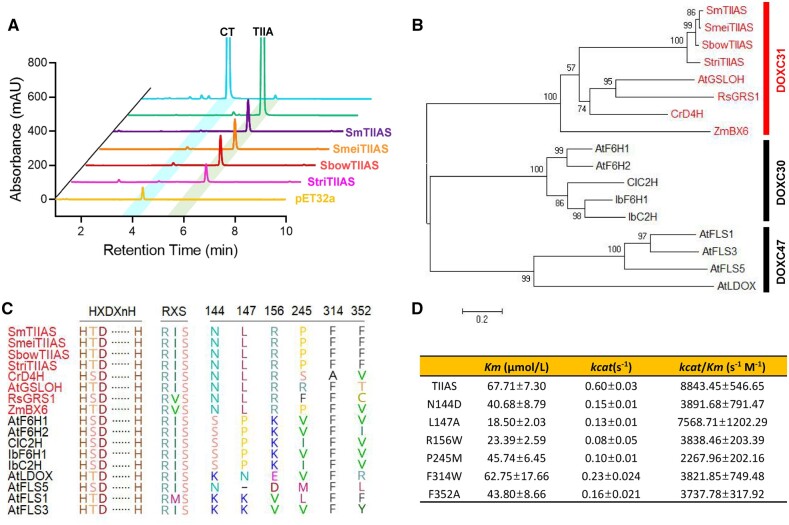
Salvia, the largest genus within Lamiaceae, is widely distributed throughout tropical and temperate regions of the world. Tanshinones are distributed in Salvia species in Himalayan and East Asia (Li et al., 2008b; Hu et al., 2018). Assay of SmTⅡAS orthologous proteins from three other tanshinone-producing Salvia species distributed in East Asia including S. meiliensis, S. bowleyana, and S. trijuga showed that they all have the desaturation activity against the dihydrofuran ring in CT (Figure 5A;Supplemental Figure S6).
Figure 5.
Evolution of TⅡASs. (A) Identification of TⅡASs from Salvia species. CT and TⅡA are the substrate and product, respectively. B Phylogenetic tree of TⅡASs with reported 2-ODD activities. The phylogenetic tree was constructed by the maximum likelihood method, and the branches indicate bootstrap values calculated by the 1,000 permutaion test. Scale bar indicates 0.2 substitutions per position in the sequence. The classification was according to Kawai et al. ( 2014) and red fonts indicate the DOXC D4H/BX6 clade. C, Multiple sequence alignment of TⅡASs and other 2-ODD proteins with the conserved and specified domains highlighted. D, Steady-state kinetic parameters of SmTIIAS and its substitution mutants by in vitro enzymatic assays. Assays were carried out in triplicate. Data represent the mean ± se of values.
2-ODDs of the DOXC class are classified into 57 phylogenetic clades (DOXC1-57) (Kawai et al., 2014). To discern the evolution of TⅡASs from ancestral 2-ODDs, we compared the TⅡASs with other DOXC 2-ODD proteins which have been characterized experimentally (Kawai et al., 2014; Caarls et al., 2017; Kakizaki et al., 2017; Nakayasu et al., 2017; Li et al., 2020a). In the phylogenetic tree constructed the SmTⅡAS and orthologous sequences from Salvia species were classified into the DOXC31 (namely D4H/BX6) clade (Figure 5B;Supplemental Figure S7), which in Arabidopsis (Arabidopsis thaliana) is the largest group (Kawai et al., 2014). Accordingly, the D4H/BX6 clade sequences were selected for multiple alignments. As a canonical 2-ODD, the 373-aa-long peptide of SmTⅡAS not only contains the C-terminal 2OG-FeⅡ_OXY domain with the dominate feature of the 2-HIS-1-carboxylate facial triad (residues His 242, Asp 244 and His 301) involved in iron binding, but also the key residues (Arg 308 and Ser 310) involved in the 2OG co-substrate binding. From the alignment, the residues Asn144, Leu147, and Arg156 are conserved in the D4H/BX6 clade, whereas Pro245 and Phe352 are specific to TⅡASs (Figure 5C). Data from substitution mutants supported the predicted residues to bind the substrate CT (Figure 5D).
Discussion
The 2-ODD superfamily enzymes catalyze various oxidative reactions including hydroxylation, demethylation, halogenation, desaturation, epoxidation, and ring formation (Islam et al., 2018). Furan moiety is present in several classes of natural products, such as furanoflavonoids, furanocoumarins, and terpenoids. Being a nonpolar aromatic component, furan derivatives have a unique place in the field of medicinal chemistry. For examples, rotundifuran, a labdane-type diterpene from the fruit of Beach vitex (Vitex rotundifolia), showed inhibitory effect on human myeloid leukemia HL-60 cell proliferation (Ko et al., 2001), and the furan-coumarin conjugate geiparvarin from leaves of Australian Willow (Geijera parviflora) has antitumor properties (Viola et al., 2004). Our finding reported herein provides a previously unknown mechanism for aromatization of furan structures.
The 2-ODDs are believed to originate from a common ancestor before land plant emerged, then underwent species-specific evolution under varied environments (Ge et al., 2021). Phylogenetic analysis showed that TⅡASs belong to the DOXC D4H/BX6 clade, which is the largest DOXC group in S. miltiorrhiza and may participate in the tanshinone biosynthesis based on genome-wide strategy (Xu and Song, 2017). This clade contains functionally diverse 2-ODDs among which most function as hydroxylase. For instance, ZmBX6 from maize (Zea mays), CrD4H from rosy periwinkle (Catharanthus roseus), and Arabidopsis AtGSLOH catalyze the hydroxylation of DIBOA glucoside, desacetoxyvindoline (monoterpenoid indole alkaloids), and 3-butenyl glucosinolate, respectively (Vazquez-Flota et al., 1997; Hansen et al., 2008; Jonczyk et al., 2008), whereas RsGRS1 from radish (Raphanus sativus) is responsible for the desaturation of the side chain of glucoerucin (Kakizaki et al. 2017). The phylogenetic analyses reveal that TⅡASs evolved in Salvia species distributed in the Himalayan and East Asian region, which served as a driving force of the furan-tanshinones innovation.
Both 2-ODDs and CYPs oxidize plant metabolites to create structural diversity. However, 2-ODDs invariably use hydrophilic substrates, often modified by CYPs, partially due to the cytosolic nature of the former (Kakizaki et al., 2017). Thus, it is suggested that 2-ODDs diversified following expansion of CYPs (Hedden and Thomas, 2012; Farrow and Facchini, 2014), which is also the case in Salvia. Notably, the CYPs in the Salvia diterpenoid pathway, such as CYP76AH3, CYP76AK1, and CYP71D375, generally show catalytic promiscuity (Figure 1), which create a metabolic grid for tanshinones biosynthesis. In contrast, SmTⅡAS is a rather specific enzyme that could not recognize the dihydrofuran-tanshinones with a modified A-ring (Figure 4D). Considering the promiscuity of the CYPs that act upstream to SmTⅡAS, the downstream CYPs may be similarly less stringent, and the metabolic grid also exists downstream of SmTⅡAS. Consistent with this hypothesis, silencing of SmTⅡAS shifted the diterpenoid flux toward the accumulation of not only CT but also other dihydrofuran-tanshinones (Figure 4, B and C). Our data suggest that SmTⅡAS functions at the bottleneck step to control the production of furan-tanshinones. Thus, a single enzyme SmTⅡAS extends the structural diversity of tanshinones in that quite a lot of tanshinones have either furan or dihydrofuran D-ring. Interestingly, in the genus Solanum, a short-chain dehydrogenase/reductase was found to be responsible for divergence of saturated/unsaturated steroidal glycoalkaloids (SGAs), two major group of SGAs (Sonawane et al., 2018). This coincidence implies presence or absence of double bonds in core scaffold of natural products may be a major source of structural diversity and the corresponding enzymes act on a key branch point in the biosynthesis pathways. Besides, except for SmTⅡAS identified here, there may be other 2-ODDs involved in tanshinone biosynthesis, such as 2OGD5, RNAi knockdown of which affects the accumulation of CT, TⅡA, and miltirone (Xu and Song, 2017).
Tanshinones are a group of abietane-type nor-diterpenes that present in plant in a mixture form. Biosynthetic studies revealed that these compounds are often biosynthetic intermediates from miltiradiene to tanshinone I, including CT, methylenedihydrotanshinquinone, DTI, and TⅡA. Thus, the biosynthesis of tanshinones is different from such pathways that accumulate one main end product such as gossypol in cotton (Gossypium spp.) or artemisinin in sweet Annie (Artemisia annua), in which the route is linear and the enzymes are more substrate-specific (Tian et al., 2018; Huang et al., 2021). Silencing of biosynthetic enzymes of these types led to reduction of end product and accumulation of corresponding intermediates that in some cases could be harmful to the host (Tian et al., 2019). Meanwhile, the biosynthesis of tanshinones involves metabolic grid and promiscuous enzymes, which release several intermediates with different pharmacological activities.
Among the mixture of diterpenoids in S. miltiorrhiza, TⅡA is the most abundant lipophilic constituent, and the furano-o-quinone is the core moiety unique to this group of nor-diterpenoids (Zhang et al., 2008; Ma et al., 2021). The presence of D-ring was reported to contribute substantially to anti-cancer activity of the tanshinones (Wang et al., 2014), and the furan oxygen plays a key role due to its involvement in the minor groove-binding of DNA (Zhang et al., 2008). Identification of the enzyme catalyzing the hetero-aromatization of dihydrofuran-tanshinones paves the way to design potential pharmaceutical resources.
Conclusion
Here we have demonstrated that one 2-ODD gene encodes SmTⅡAS in vitro and in planta. SmTⅡAS is the first enzyme characterized to dehydrogenate the dihydrofuran ring to a furan, a key step in furan ring formation. It shows a strict substrate specificity to CT and iCT and is a key enzyme responsible for channeling dihydrofuran- to the furan-tanshinones which, including TIIA, are considered the most effective ingredient in S. miltiorrhiza. Our finding is important not only to medicinal plants but also to plant secondary metabolism.
Materials and methods
Plant materials and chemicals
Plants of Salvia species were grown in nursery in Shanghai Chenshan Botanical Garden. Roots and leaves of the 1-year-old plants were sampled, three plants as triplicates, and desiccated in lyophilizer (Thermo) for subsequent analysis.
The standard compounds CT, DTI, TⅡA, and tanshinone I were purchased from Sigma-Aldrich Corp. (St Louis, USA), tetrahydrotanshinone I, 1,2-didehydrocryptotanshinone, iCT, and iTⅡA were purchased from Shanghai Yuanye Bio-Technology CO. Ltd., and methylenedihydrotanshinquinone was kindly provided by Prof. Juan Guo. MeJA was purchased from Sigma-Aldrich Corp., SA, ABA, and GA were purchased from Sangon Biotech (Shanghai) Co. Ltd.
RNA sequencing and candidate genes mining
The root and leaf RNA samples of Salvia species were profiled by HiSeq X Ten (Illumina San Diego, CA, USA) platform with paired-end method in which each sample contained three replicates, and the sequencing and data analysis were performed by Shanghai Oebiotech CO. Ltd. Annotation information was listed in Supplementary Table S1. Taking a significance level of P < 0.05 and Log2 fold-change > 2 as a threshold, we obtained 12,048 upregulated unigenes in root compared with leaf tissue of S. miltiorrhiza.
Based on the upregulated unigenes, Arabidopsis (A. thaliana) 2-ODD proteins (https://www.arabidopsis.org/) were used to local tblastn against the S. miltiorrhiza unigenes at a cut-off e-values of e−10. Then the newest HMM (hidden Markov model) profile of 2-ODD domain (PF03171 and PF14226) from Pfam database (http://pfam.xfam.org/) was used to search the Sm2-ODDs in protein database of S. miltiorrhiza on HMMER program (https://www.ebi.ac.uk/Tools/hmmer/) with a cut-off e-value of 10−4, which led to 18 common 2-ODDs candidate genes as listed in Supplementary Table S2.
Sequence alignment and phylogenetic analysis
The sequences for sequence alignment and phylogenetic analysis were obtained from National Center for Biotechnology Information (NCBI) database. Sequences alignments were performed using the MUSCLE algorithm in MEGA version 6 software package (Tamura et al., 2013). Phylogenetic tree was constructed by using the maximum-likelihood method on the LG model, with Gamma distributed rate variation among sites and a proportion of Invariant sites (G + I). Bootstrap statistics were calculated using 1,000 replicates. All phylogenetic analyses were conducted in MEGA version 6. The accession numbers of the proteins in the phylogenetic tree are listed in Supplemental Table S3.
cDNA cloning and heterogeneous expression
The RNA of S. miltiorrhiza root was extracted using cetyltrimethylammonium bromide (CTAB) solution (2% [w/v] CTAB, 2% [w/v] PVP, 100 mM Tris–HCl, 25 mM EDTA, 2M NaCl, and 2% β-mercaptoethanol) (Fang et al., 2017). Taking 1 μg of RNA of sample above performed reverse transcript following instruction of cDNA Synthesis Kit (TOYOBO, Osaka, Japan). The primers (Supplemental Table S4) were designed for amplifying 2-ODD sequences using KOD DNA polymerase (TOYOBO).
After digesting by Bam HI and Not I (Thermo Scientific, Waltham, MA, USA), the PCR amplicon of coding sequence was cloned into vector pET32a, and the pET32a-2ODD vectors were introduced into E. coli Rosetta 2 (DE3). The protein expression was induced by β-d-1-thiogalactopyranoside. After harvesting the cultures, crude protein lysate was centrifuged and purified with affinity chromatography with nickel nitrilotriacetic acid resin (Thermo Scientific), and the protein concentration was determined with bovine serum albumin standard.
Enzyme activity assays
In vitro activity assay was performed in a 200-μL reaction system consisting of 200 mM 2-(N-morpholino) ethanesulfonic acid (MES, pH 6.5), 200 μM 2OG, 200 μM L-ASC, 100 μM FeSO4, 200 μM adenosine triphosphate, the substrate (3.75–150 μM) and 100 μg purified recombinant protein. After shaking with sufficient air at 20°C for 7 min, the reaction was stopped by adding 1-mL ethyl acetate, extracting the product twice. The empty vector enzyme was used as negative control. The standard curves were plotted by authentic TⅡA and iTⅡA for quantification. Km and kcat were obtained by using GraphPad Prism version 5.0 (Motulsky, 2007), and the means ± se (standard error) were calculated from triplicates of assays.
The relative activities of SmTⅡAS and its variants were assayed in vitro as described above. Mutants of SmTⅡAS were generated by PCR using an overlap extension strategy with respective primers (Supplemental Table S4). The SmTⅡAS mutant sequences were inserted into pET32a and the proteins were produced as described above.
Hairy root and RNAi
MeJA, SA, ABA, and GA were dissolved in dimethyl sulfoxide (DMSO) and applied to the hairy roots culture at the final concentrations of 50 μM, 5 mM, 100 μM, and 50 μM, respectively, with addition of a DMSO solution (5%) as control. Hairy roots were harvested from the culture medium at 2 h after the treatments, then RNA was extracted. All treatments were performed in triplicate.
The region comprising nucleotides of 365–759 of SmTⅡAS open reading frame was selected for double-stranded RNA generation. To target SmTⅡAS precisely, this 395-bp fragment was compared with other 2-ODDs upregulated in S. miltiorrhiza root by blastn with cutoff of e-value ≤ 1e−10, which showed no substantial similarity. The fragment was cloned into gateway vector pDONR207, subsequently recombined into binary vector pK7GWIWG2R using a Gateway LR Clonase II enzyme mix (Invitrogen, Waltham, MA, USA). The disarmed Agrobacterium tumefaciens strain C58C1 containing pK7GWIWG2R-SmTⅡAS was applied to infect aseptic S. miltiorrhiza leaf tissue (0.5 × 0.5 cm) for 10 min, followed by transferring onto 6,7-V solid medium and co-cultured in the dark for 2 d, in presence of 400 mg/L carbenicillin to prevent Agrobacterium overgrowth. After 1 month, the hairy roots were transferred to liquid medium. The transgenic hairy roots were harvested for RNA isolation and metabolite extraction.
Quantitative reverse transcription PCR analysis
Hairy roots were collected and frozen in liquid nitrogen immediately, total RNA was isolated as described above and was reversed transcribed referring to the ReverTra Ace qPCR RT Kit (TOYOBO). RT-qPCR was performed with TB Green Premix Ex Taq (Takara, Shiga, Japan, RR420A). SmACTIN (Accession: HM231319) is used as internal reference gene, gene-specific primers as shown in Supplemental Table S4, the relative expression of SmTⅡAS was calculated following the formula of 2−ΔΔCt.
Metabolite extraction for UPLC and QE analysis
The plant tissues were dried by freeze dryer (Thermo), and 0.3 g each powder sample, was extracted with 5 mL of ethyl acetate, followed by ultrasonic crushing extraction for 1 h, drying ethyl acetate out by vacuum concentrator (Eppendorf, Hamburg, Germany ), and dissolving in methanol for detection of metabolites. Chromatographic separations were performed using the ACQUITY UPLC HSS C18 column (2.1 × 100 mm, 1.8 μm, Waters) based on Agilent 1260 infinity Ⅱ Prime LC system (Agilent, Santa Clara, CA, USA). A coupled Dionex UltiMate 3000 HPLC system and Q Exactive Plus Mass Spectrometer (Thermo Scientific) collected the MS (mass spectrum) data in positive-ion mode with the spray voltage 4 kV and capillary temperature 320°C, the stepped NCE (normalized collision energy) were set at 20, 40, and 70. Mobile phases with H2O consisting of 0.1% formic acid (A) versus acetonitrile containing 0.1% formic acid (B) were used. The gradient profile was performed as following: 0 min, 50% A/50% B; 8 min, 20% A/80% B; 8.5 min, 0% A/100% B; 11 min, 0% A/100% B; 11.5 min, 50% A/50% B, which was held on for 3 min for re-equilibration, giving a total run time of 14.5 min. And the temperature of the column was maintained at 40°C. The flow rate of mobile phase is 0.5 mL/min with the 270 nm of detection wavelength.
Accession numbers
The sequences of SmTⅡAS, SbowTⅡAS, SmeiTⅡAS, and StriTⅡAS isolated in this work were verified by complete gene sequencing and have been submitted to the NCBI database with the accession numbers of MW916096, MW928604, MW928605, and MW928606, respectively. The transcriptome sequence data were deposited to NCBI Sequence Read Archive (SRA) with the accession number of PRJNA771193 and PRJNA771195.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Aerial parts and roots of S. miltiorrhiza and S. substolonifera.
Supplemental Figure S2. UPLC profiles of enzyme activity of SmTⅡAS in vitro.
Supplemental Figure S3. Effect of temperature and pH on enzyme activities of SmTⅡAS using CT as substrate.
Supplemental Figure S4. Characterization of SmTⅡAS activities by in vitro assay.
Supplemental Figure S5. The metabolites altered in accumulation in SmTⅡAS-RNAi lines.
Supplemental Figure S6. The relative content of tanshinone IIA in S. miltiorrhiza, S. meiliensis, S. bowleyana and S. trijuga, respectively.
Supplemental Figure S7. Extended phylogenetic analysis of TⅡASs and Sm2-ODDs with other experimentally characterized 2-ODDs.
Supplemental Table S1. The annotation ratio statistics of S. miltiorrhiza and S. substolonifera transcriptome database.
Supplemental Table S2. The sequences of 18 Sm2-ODD candidate genes and the homologs from Xu and Song (2017).
Supplemental Table S3. Protein sequence information for the phylogenetic tree.
Supplemental Table S4. List of oligonucleotide primer sequences.
Supplementary Material
Acknowledgments
We thank Prof. Jian-Guo Cao (Shanghai Normal University) for supporting for this work. We also thank Dr Li-Jing Chang and Meng-Ying Cui for their generous help with this work.
Funding
This work was supported by the National Key R&D Program of China (Grant No. 2020YFA0907900), National Natural Science Foundation of China (Grant No. 32070338 and 31900255), the International Partnership Program of Chinese Academy of Sciences (153D31KYSB20160074), Strategic Biological Resources and Technology Supporting System from the Chinese Academy of Sciences (grant no. ZSZY-001), and the Special Fund for Shanghai Landscaping Administration Bureau Program (Grant No. G192419 and G222414).
Conflict of interest statement. The authors declare that there is no conflict of interest.
L.Y. and. X.-Y.C. designed and managed the project. J.-J.S. Y.L., and L.Y. isolated the genes and characterized the enzyme, X.F., C.-Y.L., S.W., Q.Z., and J.G. helped in the biosynthetic pathway analysis. J.-X.L., H.F., and J.-J.X. performed bioinformatics analysis. Y.-B.H. and Y.-K.W. collected and cultured plants. Y.K. assisted with LC-MS analysis. X.-Y.C. and Y.-H.H. analyzed and interpreted the data. L.Y., X.F., and X.-Y.C. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Lei Yang, leiyang@cemps.ac.cn
References
- Araujo WL, Martins AO, Fernie AR, Tohge T (2014) 2-Oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front Plant Sci 5: 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin GAW, Facchini PJ (2014) Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta 240: 19–32 [DOI] [PubMed] [Google Scholar]
- Caarls L, Elberse J, Awwanah M, Ludwig NR, de Vries M, Zeilmaker T, Van Wees SCM, Schuurink RC, Van den Ackerveken G (2017) Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc Natl Acad Sci USA 114: 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AX, Han XJ, Wu YF, Lou HX (2014) The function and catalysis of 2-oxoglutarate-dependent oxygenases involved in plant flavonoid biosynthesis. Int J Mol Sci 15: 1080–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Morris-Natschke SL, Lee KH (2011) Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat Prod Rep 28: 529–542 [DOI] [PubMed] [Google Scholar]
- Fang X, Li CY, Yang Y, Cui MY, Chen XY, Yang L (2017) Identification of a novel (-)-5-epieremophilene synthase from Salvia miltiorrhiza via transcriptome mining. Front Plant Sci 8: 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZY, Zhang M, Liu JN, Zhao X, Zhang YQ, Fang L (2021) Tanshinone IIA: a review of its anticancer effects. Front Pharmacol 11: 611087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow SC, Facchini PJ (2014) Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front Plant Sci 5: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Organic Lett 11: 5170–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Tang C, Zhu YX, Wang GF (2021) Genome-wide identification of the maize 2OGD superfamily genes and their response to Fusarium verticillioides and Fusarium graminearum. Gene 764: 145078. [DOI] [PubMed] [Google Scholar]
- Guo J, Ma X, Cai Y, Ma Y, Zhan Z, Zhou YJ, Liu W, Guan M, Yang J, Cui G, et al. (2016) Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol 210: 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, Zhang X, Liu W, Peters RJ, Chen X, et al. (2013) CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA 110: 12108–12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BG, Kerwin RE, Ober JA, Lambrix VM, Mitchell-Olds T, Gershenzon J, Halkier BA, Kliebenstein DJ (2008) A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol 148: 2096–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Hu GX, Takano A, Drew BT, Liu ED, Soltis DE, Soltis PS, Peng H, Xiang CL (2018) Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann Bot 122: 649–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JQ, Li DM, Tian X, Lin JL, Yang L, Xu JJ, Fang X (2021) Side products of recombinant amorpha-4,11-diene synthase and their effect on microbial artemisinin production. J Agric Food Chem 69: 2168–2178 [DOI] [PubMed] [Google Scholar]
- Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ (2018) 2-Oxoglutarate-dependent oxygenases. Annu Rev Biochem 87: 585–620 [DOI] [PubMed] [Google Scholar]
- Johnson SR, Bhat WW, Bibik J, Turmo A, Hamberger BEvolutionary Mint Genomics CHamberger B (2019) A database-driven approach identifies additional diterpene synthase activities in the mint family (Lamiaceae). J Biol Chem 294: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk R, Schmidt H, Osterrieder A, Fiesselmann A, Schullehner K, Haslbeck M, Sicker D, Hofmann D, Yalpani N, Simmons C, et al. (2008) Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol 146: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki T, Kitashiba H, Zou Z, Li F, Fukino N, Ohara T, Nishio T, Ishida M (2017) A 2-oxoglutarate-dependent dioxygenase mediates the biosynthesis of glucoraphasatin in radish. Plant Physiol 173: 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Ono E, Mizutani M (2014) Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J Cell Mol Biol 78: 328–343 [DOI] [PubMed] [Google Scholar]
- Ko WG, Kang TH, Lee SJ, Kim YC, Lee BH (2001) Rotundifuran, a labdane type diterpene from Vitex rotundifolia, induces apoptosis in human myeloid leukaemia cells. Phytother Res 15: 535–537 [DOI] [PubMed] [Google Scholar]
- Larbat R, Hehn A, Hans J, Schneider S, Jugde H, Schneider B, Matern U, Bourgaud F (2009) Isolation and functional characterization of CYP71AJ4 encoding for the first P450 monooxygenase of angular furanocoumarin biosynthesis. J Biol Chem 284: 4776–4785 [DOI] [PubMed] [Google Scholar]
- Larbat R, Kellner S, Specker S, Hehn A, Gontier E, Hans J, Bourgaud F, Matern U (2007) Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of furanocoumarin biosynthesis. J Biol Chem 282: 542–554 [DOI] [PubMed] [Google Scholar]
- Li DD, Ni R, Wang PP, Zhang XS, Wang PY, Zhu TT, Sun CJ, Liu CJ, Lou HX, Cheng AX (2020a) Molecular basis for chemical evolution of flavones to flavonols and anthocyanins in land plants. Plant Physiol 184: 1731–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JT, Dong JE, Liang ZS, Shu ZM, Wan GW (2008a) Distributional difference of fat-soluble compounds in the roots, stems and leaves of four Salvia plants. Fen zi xi bao sheng wu xue bao. J Mol Cell Biol 41: 44–52 [PubMed] [Google Scholar]
- Li MH, Chen JM, Peng Y, Wu Q, Xiao PG (2008b) Investigation of Danshen and related medicinal plants in China. J Ethnopharmacol 120: 419–426 [DOI] [PubMed] [Google Scholar]
- Li X, Luo D, Hou Y, Hou Y, Chen S, Zhan J, Luan J, Wang L, Lin D (2020b) Sodium tanshinone IIA silate exerts microcirculation protective effects against spinal cord injury in vitro and in vivo. Oxidat Med Cell Longev 2020: 3949575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Cui G, Chen T, Ma X, Wang R, Jin B, Yang J, Kang L, Tang J, Lai C, et al. (2021) Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza. Nat Commun 12: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky H (2007) Prism 5 statistics guide, 2007. GraphPad Softw 31: 39–42 [Google Scholar]
- Murata J, Roepke J, Gordon H, De Luca V (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu M, Umemoto N, Ohyama K, Fujimoto Y, Lee HJ, Watanabe B, Muranaka T, Saito K, Sugimoto Y, Mizutani M (2017) A dioxygenase catalyzes steroid 16alpha-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol 175: 120–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YK, Obiang-Obounou BW, Lee J, Lee TY, Bae MA, Hwang KS, Lee KB, Choi JS, Jang BC (2017) Anti-adipogenic effects on 3T3-L1 cells and Zebrafish by Tanshinone IIA. Int J Mol Sci 18: 2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei T, Ma P, Ding K, Liu S, Jia Y, Ru M, Dong J, Liang Z (2018) SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J Exp Bot 69: 1663–1678 [DOI] [PubMed] [Google Scholar]
- Sonawane PD, Heinig U, Panda S, Gilboa NS, Yona M, Kumar SP, Alkan N, Unger T, Bocobza S, Pliner M, et al. (2018) Short-chain dehydrogenase/reductase governs steroidal specialized metabolites structural diversity and toxicity in the genus Solanum. Proc Natl Acad Sci USA 115: E5419–E5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Fang X, Huang JQ, Wang LJ, Mao YB, Chen XY (2019) A gossypol biosynthetic intermediate disturbs plant defence response. Philos Trans R Soc Lond B Biol Sci 374: 20180319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Ruan JX, Huang JQ, Yang CQ, Fang X, Chen ZW, Hong H, Wang LJ, Mao YB, Lu S, et al. (2018) Characterization of gossypol biosynthetic pathway. Proc Natl Acad Sci USA 115: E5410–E5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima J, Welford RW, Yamazaki M, Saito K, Schofield CJ (2004) Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3beta-hydroxylase. J Biol Chem 279: 1206–1216 [DOI] [PubMed] [Google Scholar]
- Vazquez-Flota F, De Carolis E, Alarco AM, De Luca V (1997) Molecular cloning and characterization of desacetoxyvindoline-4-hydroxylase, a 2-oxoglutarate dependent-dioxygenase involved in the biosynthesis of vindoline in Catharanthus roseus (L.) G. Don. Plant Mol Biol 34: 935–948 [DOI] [PubMed] [Google Scholar]
- Viola G, Vedaldi D, dall’Acqua F, Basso G, Disaro S, Spinelli M, Cosimelli B, Boccalini M, Chimichi S (2004) Synthesis, cytotoxicity, and apoptosis induction in human tumor cells by geiparvarin analogues. Chem Biodivers 1: 1265–1280 [DOI] [PubMed] [Google Scholar]
- Wang L, Liu A, Zhang FL, Yeung JH, Li XQ, Cho CH (2014) Evaluation and SAR analysis of the cytotoxicity of tanshinones in colon cancer cells. Chin J Nat Med 12: 167–171 [DOI] [PubMed] [Google Scholar]
- Wang X, Morris-Natschke SL, Lee KH (2007) New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med Res Rev 27: 133–148 [DOI] [PubMed] [Google Scholar]
- Xu Z, Song J (2017) The 2-oxoglutarate-dependent dioxygenase superfamily participates in tanshinone production in Salvia miltiorrhiza. J Exp Bot 68: 2299–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ding G, Lin H, Cheng H, Kong Y, Wei Y, Fang X, Liu R, Wang L, Chen X, et al. (2013) Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS One 8: e80464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang J, Jin L, Song T, Wu G, Gao J (2008) Tanshinone IIA interacts with DNA by minor groove-binding. Biol Pharm Bull 31: 2342–2345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.