Abstract
Evolutionary mutations in primate-specific genes drove primate cortex expansion. However, whether conserved genes with previously unidentified functions also play a key role in primate brain expansion remains unknown. Here, we focus on BRN2 (POU3F2), a gene encoding a neural transcription factor commonly expressed in both primates and mice. Compared to the limited effects on mouse brain development, BRN2 biallelic knockout in cynomolgus monkeys (Macaca fascicularis) is lethal before midgestation. Histology analysis and single-cell transcriptome show that BRN2 deficiency decreases RGC expansion, induces precocious differentiation, and alters the trajectory of neurogenesis in the telencephalon. BRN2, serving as an upstream factor, controls specification and differentiation of ganglionic eminences. In addition, we identified the conserved function of BRN2 in cynomolgus monkeys to human RGCs. BRN2 may function by directly regulating SOX2 and STAT3 and maintaining HOPX. Our findings reveal a previously unknown mechanism that BRN2, a conserved gene, drives early primate telencephalon development by gaining novel mechanistic functions.
A conserved gene, BRN2, drives brain development in primates, revealing a new mechanism on the evolutionary expansion of cortex.
INTRODUCTION
A challenge in brain development is to understand how extant mammals, such as monkeys and mice, use a similar number of genes yet have obviously more complex brain structure and functions (1), especially in the cortex. Cortical expansion in primate is characterized by a prolonged and increased proliferation of radial glial cells (RGCs) in the ventricular zone (VZ) of the telencephalon, with RGCs then expanding into the subventricular zone (SVZ) (2–5). In primates, RGCs are more abundant than that in mice and form a more extended SVZ, or the outer SVZ, which is driven by “basal” RGCs (bRGCs) (6). Nevertheless, the molecular mechanisms responsible for such expansion of primate RGCs remain largely unknown. One theory suggests that these adaptations evolved through gene expression changes (7–9). Identifying the genetic elements that endow RGCs with proliferation-promoting properties unique to primates is integral to elucidating primate-specific aspects of cortical expansion. One approach requires identifying which genes, genomic regulatory sequences, or signaling pathways exhibit primate-specific expression patterns with unique functions to RGCs.
Comparative analyses of mammalian genomes or gene expressions led to the identification of some primate-specific signatures of divergence that critically underlies the cortical expansion in primates. One driver of phenotypic evolution relates to changes in gene expression patterns, such as Osteocrin (1); extracellular matrix including collagens, laminins, proteoglycans, and integrins (10); ASPM (11); and platelet-derived growth factor D (PDGFD)–PDGF receptor β (PDGFRβ) signaling pathways (12). The second evolutionary driver is the emergence of primate-specific or novel genes, such as specific long noncoding RNA (13) and microRNA (14), TMEM14B (15), TBC1D3 (16), NOTCH2NL (17, 18), and ARHGAP11B (19, 20). Functional acquisitions of the two types of genes are mainly caused by evolutionary mutations of existing genes, including the acquisition of DNA regulatory elements, amino acid substitutions, and gene duplication. The third possible evolutionary driver implicates conserved genes among nonprimate and primate species that gain novel functions in primates. Identifying the third category of genes poses challenges, since the functions of these conserved genes are generally inaccessible for evaluation in rodent models.
In our preliminary study on the histological changes in cynomolgus monkey telencephalic development, we unexpectedly found that BRN2, a conserved and genuine proneural transcriptional factor (TF), was strongly expressed in the early RGCs of VZ at embryonic day 36 (E36) (fig. S1) but marks late RGC and upper-layer cortical neurons in mice (21–23). These results indicate that the expression of BRN2 is substantial earlier during the telencephalic development of monkeys than that in mice. Hence, we hypothesized that BRN2 plays a critical role during telencephalon development in monkeys, in contrast to its limited functions during the early development of mice. To test this hypothesis, we generated BRN2-knockout monkeys. Using histological analysis and single-cell sequencing, we investigated the following questions: (i) What are the spatiotemporal distribution differences of BRN2 between mouse and monkey cortices, (ii) what is the function of BRN2 on early primate telencephalon development, and (iii) what is the possible mechanism of BRN2 regulating primate telencephalon development.
RESULTS
BRN2 marks early RGCs in the developing cynomolgus monkey telencephalon
To screen key genes that exist in nonprimate species but trigger novel functions in primates, we investigated cell developmental axis and expression patterns of genes specific for cynomolgus monkey (Macaca fascicularis) embryonic brains. To do this, we first analyzed neural progenitor–specific marker expressions over cortical development by immunostaining. Human neuroepithelium cells (NEPCs) display specific molecular characteristics, such as high positivity of PAX6 and delayed expression of SOX1 (24). We observed that monkey neural cells at E29 were constituted by NEPCs with multiple pseudostratified layers (fig. S1A) like mouse E9.5 NEPCs (24). Specifically, we found ubiquitous expression of PAX6 but the absence of SOX1, EOMES [an intermediate progenitor (IP) marker], and NEUROD2 (a neuron marker), and asymmetrical distribution of ZO-1 (a marker of NEPCs for N-Cad–mediated adherens junctions) in the apical side (fig. S1A) (14). In contrast, the expression of SOX1, EOMES, and NEUROD2 at E36 indicated that the NEPCs have transitioned into RGCs (fig. S1B), equivalent to RGCs in the developing E10.5 mouse cortex (24). The gene expression patterns at E49 and E72 (fig. S1, C and D) remained in line with developmental cortices in E12.5 and E14.5 mouse (fig. S1, F and G), respectively, as summarized by Smart et al. (25) and Jiang and Nardelli (26).
Next, we examined the expression of general TFs specific for corticogenesis, including EOMES (an IP marker), NEUROD2, SATB2 (a cortical neuron marker), FOXP2 (a deep-layer neuron marker), and CUX1 and BRN2 (upper-layer neuron markers) (fig. S1, A to G). Obvious productions of SATB2+ cortical neurons and EOMES+ IPs were initiated from E36 SVZ cortical plates (CPs) (fig. S1B) and FOXP2+ deep-layer neurons from E49 CPs. CUX1+ upper-layer neurons were detected in E72 CPs (fig. S1D). Unexpectedly, BRN2 as an upper-layer neuron marker was strongly expressed not only in the E72 CPs but also in the RGCs of VZ and SVZ beginning from E30 (Fig. 1, A and B, and fig. S1, B to D). By comparing mouse cortical neurogenesis (26), we concluded that the cortical neurogenesis stages and expression patterns of genes, except BRN2 in E29 to E72 monkey cortices, resemble those in E9.5 to E14.5 mouse cortices (Fig. 1A to C and fig. S1, A to G) (25).
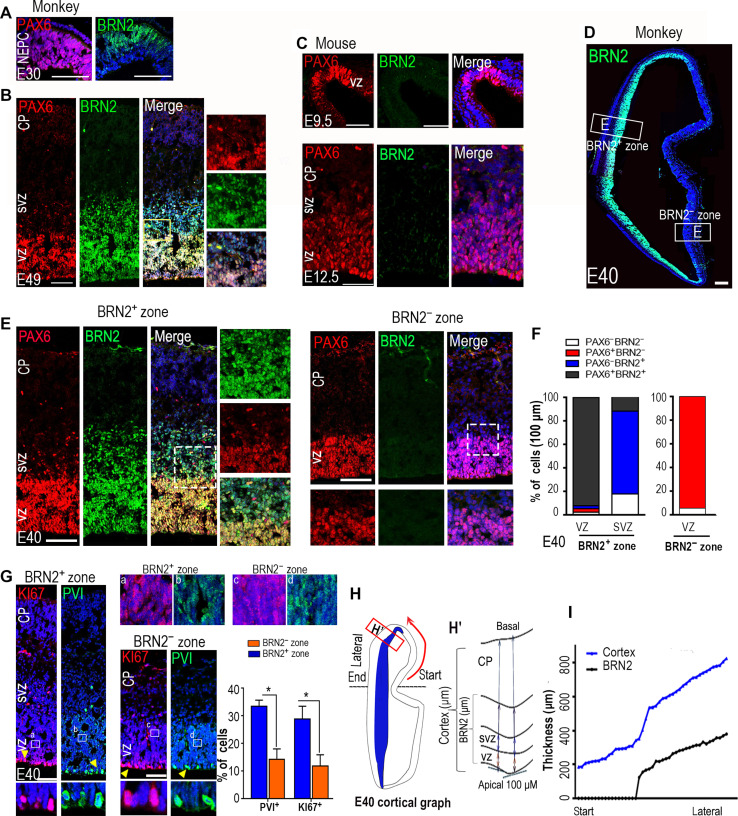
Fig. 1. The spatiotemporal distribution of BRN2 coordinates with RGC expansion in the developing cynomolgus monkey cortex.
(A and B) Dynamic expressions of PAX6 and BRN2 during the monkey cortical expansion through brain coronal sections of the primary visual cortex. BRN2 was initially expressed in NEPCs at embryonic day 30 (E30) (A) and gradually extended along the VZ spread to SVZ at E49 (B). Cortical layers were identified by staining of 4′,6-diamidino-2-phenylindole (DAPI) and PAX6. (C) Immunohistochemical analyses for Brn2 and Pax6 were performed in E9.5 (top) and E12.5 (bottom) mouse neocortices. (D) Images showing the expression patterns of BRN2 in E40 monkey coronal brain sections of the primary visual cortex. (E) Double staining of BRN2 and PAX6 in the E40 VZ and SVZ of BRN2-expressing (BRN2+) or BRN2-absent (BRN2−) zones, respectively. An obvious SVZ was formed in the BRN2+ zone but not in the BRN2− zone. (F) Quantification of BRN2 and PAX6 expression in the BRN2+ and BRN2− zones, respectively. Data are presented as means ± SEM (n = 3 slices); >300 cells per group were counted. (G) Expressions of p-VIMENTIN (PVI) and KI67 in the E40 VZ and SVZ of BRN2+ or BRN2− zones, respectively. The quantification data were only from the cells in the apical regions where cell division occurs. Data are presented as means ± SEM, n = 3 slices; >400 cells per group were counted. Arrowheads indicate positive cells. (a to d) Areas in rectangles are shown with higher magnification. (H) The left panel is a cortical graph at E40, and the right panel is the schematic diagram for measuring the thicknesses of whole-cortex and BRN2-expressing cells in monkey fetal brain. (I) Coordinating BRN2 distribution with the thicknesses of the cortex in E40 monkey neocortex, relative to (H). Start and lateral sites in (H) correspond to those in (I). Blue, DAPI, nuclear staining. Scale bars, 100 μm.
Spatiotemporal distribution differences of BRN2 in mouse and monkey cortices
BRN2 expression in early RGCs (fig. S1B) revealed that BRN2 may contribute to the unique aspects of primate cortical development at the early stage, prompting us to further examine its spatiotemporal distributions in monkey embryonic cortices from early (E29) to midgestation (E72) (25). Because of high conservation and mutual compensatory effects between BRN2 and BRN1 in mouse (22), we used a BRN2 monoclonal antibody that specifically recognizes the N-terminal peptide of BRN2 that is absent in BRN1 according to the manufacturer. Immunostaining showed that BRN2 was initially expressed in lateral NEPCs at E30, gradually extending along the VZ (PAX6-expressing cells) in the lateral-to-medial and ventral-to-dorsal directions by E40 (Fig. 1, A and B, and figs. S1, A to D, and S2, A to C). This distribution encompassed the entire cortical VZ/SVZ by E49 and spread across the entire cortex, including cortical upper layers, by E72 (fig. S2, B and C). Compared to PAX6+ cells specially locating in VZ/SVZ, BRN2+ cells gradually migrated toward the cortex over development and reached the CP at E72 (Fig. 1, A and B, and figs. S1, A to D, and S2, A to C). These results demonstrated that BRN2+ cortical neurons are produced at E72. In contrast, we did not observe Brn2 expression in the mouse E9.5 cortex (fig. S1E). We first detected it in the E12.5 ganglionic eminence (GE) but not in the cortical VZ region (Fig. 1C and fig. S2D). BRN2 was widely expressed in the E14.5 telencephalon, including the VZ and CP (fig. S2D). The expression patterns of BRN2 are in line with previous findings (21–23). These data showed that BRN2 expression during development begins earlier in the monkey cortex than in the mouse cortex.
To further validate the expression differences of BRN2 in primate and nonprimate species, we induced mouse and monkey embryonic stem cells (ESCs) into the neuroectoderm using a reported differentiation system [basic fibroblast growth factor (bFGF) induction] that enabled ESCs to spontaneously differentiate into general neuroepithelial stem cells (NESCs) (fig. S2E) (27). We found that BRN2 expression was initially detected in monkey-differentiated NESCs at day 6 postdifferentiation (pdD6) before SOX1 and SOX1 expression (fig. S2, F and G), whereas Brn2 first appeared in mouse pdD15 ESC-derived cells that lacked the expression of Sox1 (fig. S2, H and I). Thus, BRN2 expression pattern during in vitro neural differentiation is consistent with that in vivo.
Next, we examined the relationship of BRN2 expression with RGC proliferation and VZ expansion. By selecting the E40 cortex with asymmetrical distributions of BRN2 along the lateral-medial/dorsal VZ regions (Fig. 1D), we could evaluate the correlation of RGC expansion with BRN2 expression. Compared to the zone that is not expressed by BRN2 (BRN2− zone), we observed an increase at the VZ/SVZ in the BRN2-expressing zone (BRN2+ zone) (Fig. 1E). Double staining and quantification of BRN2 and PAX6 in the BRN2− and BRN2+ zones also supported this result (Fig. 1F). In addition, quantification of proliferation ratios in apical surface RGCs by P-Vimentin (PVI) and KI67 immunostaining showed an increase in the frequency of proliferative cells in the BRN2+ zone (Fig. 1G), implying that BRN2 expression correlates with RGC proliferation. The cell layer thickness showing BRN2 positivity is highly correlated with cortical thickness (Fig. 1, H and I). This result suggests that the onset and course of BRN2 expression are timed in precise concordance with the expansion of the VZ and cortex. Together, the spatial and temporal distribution differences in BRN2 between rodents and primates raise an intriguing possibility that BRN2 may play a critical role in primate RGC proliferation, SVZ expansion, and telencephalon development.
BRN2-knockout monkey fetuses are lethal before midgestation stage
Brn2 mutation fails to induce mouse neocortical deficits due to the compensatory effects of Brn1 (22, 28, 29). There are no significant differences in the number of RGCs before E14.5 and in the generation and migration of cortical interneurons in the Brn1/Brn2 double-mutant telencephalon (22). To investigate the functions of BRN2 in developing primate brains, we generated a biallelic BRN2-null cynomolgus monkey model using a CRISPR-Cas9 system (Fig. 2A) (30). We screened out two pairs of single-guide RNAs (sgRNAs) (type 1: sgRNA-A+B, 1-kb deletion; type 2: sgRNA-A+C, 0.5-kb deletion) that target the exon of the monkey BRN2 gene (Fig. 2B). Since using 0.5- and 1-kb deletions may be more beneficial to a functional knockout than using an sgRNA, we injected type 1 or 2 with sgRNA-Cas9 nuclease complex [ribonucleoprotein (RNP)] into 19 and 12 cynomolgus embryos, respectively. Polymerase chain reaction (PCR) and sequencing showed that 95% (19 of 20) of type 1–treated embryos were edited successfully at the BRN2 locus, among which 70% (14 of 20) of embryos displayed a 1-kb large-fragment deletion (fig. S3, A and B). All type 2–treated embryos (12 of 12) were successfully edited with 66.67% (8 of 12) 0.5-kb fragment deletion (fig. S3, A and B). Hence, we transferred 11 type 1–treated embryos and 67 type 2–treated embryos into 6 and 23 surrogate recipients, respectively. Eight embryos (10.25%, 1 and 7 for type 1– and type 2–treated embryos, respectively) were successfully implanted, but 2 embryos miscarried at 4 to 5 weeks (Fig. 2C), although the implantation and development efficiency of the control embryos was 26.92% (7 of 26 embryos) within this period. These results imply that the BRN2 mutation could affect embryo survival.
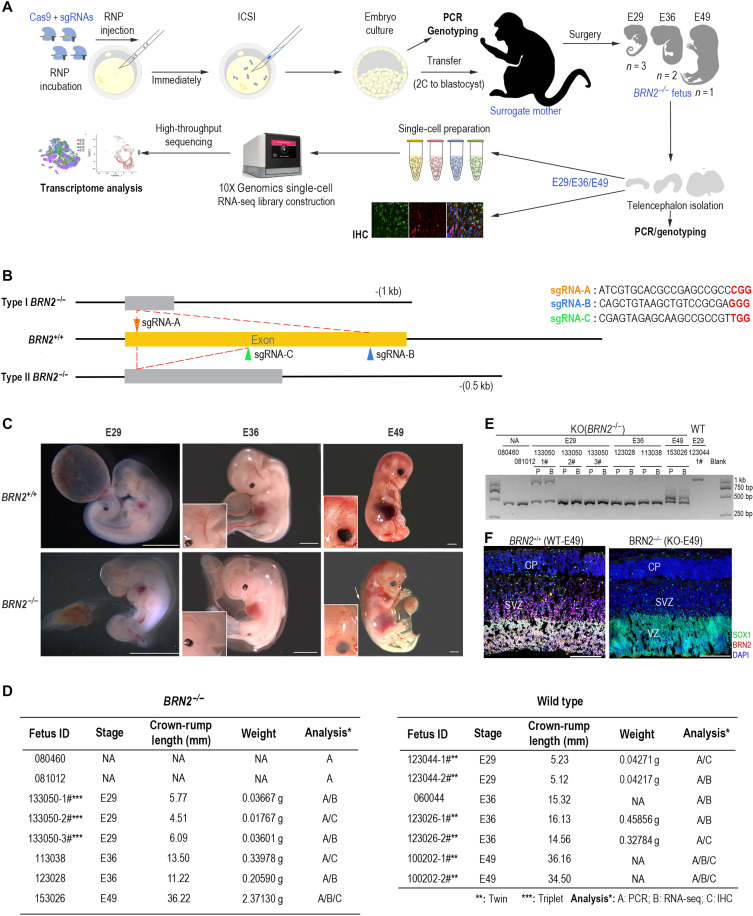
Fig. 2. Generation of BRN2-knockout (BRN2−/−) monkeys using a CRISPR-Cas9 system.
(A) Workflow of generation and analysis of BRN2-KO monkeys. RNP, gRNA-Cas9 nuclease complex. (B) Schematic diagram of target gene editing. Triangles with different colors show the different locations where the sgRNAs target. Two pairs of sgRNA (A+B or A+C) in principle induced type I [with about 1091–base pair (bp) deletion] and type II (with near-488-bp deletion) gene mutations, respectively. (C) Detailed information of all fetuses in this study. Two fetuses (080460 and 081012) were miscarried during gestation, and we failed to collect their tissues for further analysis. (D) Representative images of wild-type (WT; BRN2+/+) and BRN2-knockout (KO, BRN2−/−) fetus at E29, E36, and E49. Inserts are magnifications of monkey brains, showing abnormal cerebrovascular development in the BRN2−/− E36 and E49 fetuses. Scale bars, 3.7 mm. NA, non-examination. (E) Agarose gel electrophoresis of the PCR products from all knockout monkey samples. BRN2+/+ sample (123044-1#) is shown as a negative control of the target gene editing. P, placenta; B, brain cells. (F) Immunostainings of BRN2 and SOX1 in the BRN2+/+ and BRN2−/− monkey fetal cortex, confirming BRN2 deletion in the BRN2−/− cortex. Blue, DAPI, nuclear staining. Scale bars, 100 μm. IHC, immunohistochemistry.
On the basis of the development axis of monkey brain and temporal expression of BRN2 (fig. S1), we first collected E29 and E36 fetuses by caesarean for genotyping, transcription, and histology analysis (Fig. 2, C to F). No BRN2-knockout monkey fetus developed up to midgestation stage, equivalent to beyond the E12 stage in mouse. We detected a severe edema at E49 by ultrasonography and terminated the pregnancy. We observed clear abnormities of cerebrovascular development in E36 and E49 BRN2−/− fetuses (Fig. 2D). Together, these results showed that BRN2 knockout is lethal to monkey fetuses before midgestation. PCR and Sanger sequencing results showed that all gene-edited fetuses were successfully edited and carried the biallelic mutation (Fig. 2E and fig. S3C), which was further verified by BRN2 protein immunostaining (Fig. 2F).
Using two different guide RNA pairs guards against the possibility that the observed phenotypes arise from off-target mutations. Next, we further evaluated whether the CRISPR-Cas9 system caused other mutagenesis in the gene-modified monkeys by performing genomic DNA PCR and sequencing. Twenty high-potential off-target sites were analyzed (table S1). DNA sequencing confirmed the precise targeting of the BRN2 locus in all monkey fetuses without detectable off-target effects (table S1). On the basis of these results, we concluded that the BRN2 gene was successfully knocked out in these monkeys.
BRN2 deletion induces abnormal development of monkey cortices
Given that BRN2 is expressed in early RGCs, we evaluated the phenotypes of RGCs using RGC markers. We observed normal telencephalic development in the BRN2−/− E36 fetus, suggesting that BRN2 deletion did not affect the initial formation of telencephalon (fig. S4A). Immunostaining of E36 fetus showed that SOX2 loss was obviously detected in BRN2−/− GEs but not in the cortex, although there were no obvious differences about PAX6 expression and RGC proliferation between the BRN2−/− and BRN2+/+ (wild-type) telencephalons (fig. S4A). The SOX2 loss in BRN2−/− GEs concords with the initial expression of BRN2 in the GE region of early telencephalon (fig. S2, B and C). SOX2 expression was completely lost in the BRN2−/− whole-telencephalon RGCs at E49 (Fig. 3A), showing that the roles of BRN2 deletion on RGCs became obvious over development. SOX2 requires the coordination of BRN2 as a partner factor to function in neural specification and NP maintenance (31), implying that BRN2 functions in primate RGCs may specifically contribute to maintain SOX2. During early human brain development (before gestation week 10), most RGC divisions are vertically oriented (32, 33), so altered regulation of mitotic spindle orientation changes RGC cell numbers and neuronal numbers (34). Compared to the BRN2+/+ cortex, RGCs in BRN2−/− cortical VZ decreased the vertical RGC divisions with a coordinated increase in oblique divisions by KI67 and PVI staining (Fig. 3, B and B′) (34). The results indicate that BRN2 deletion decreases proliferation of RGCs by changing mitotic spindle orientation. In addition, activation of the neuron marker TUJ1 and NEURORD2 in VZ/SVZ RGCs in situ and increased production of SATB2+ neurons (Fig. 3C and fig. S4B) showed the neuron precocious differentiation of RGCs in BRN2−/− cortex. These data, together with no change in IP marker EOMES (fig. S4B), indicated that this precocious neuron production resulted from the direct neurogenesis of RGCs and not through IP. Since BRN2 expression correlated with the formation and expansion of the cortex (Fig. 1, H and I), we examined the expression of HOPX, an important transcription factor that reflects the abundance of bRGC abundance during primate oSVZ expansion (14, 35, 36). HOPX expression in primates occurred early in fetal VZ/SVZ but predominated expression in bRGCs of oSVZ at middle stages of corticogenesis (fig. S4C). BRN2 deficiency resulted in HOPX loss in VZ/SVZ (Fig. 3, D and D′). This result implies that BRN2 may contribute to RGC migration by activating HOPX. Overall, these findings revealed that BRN2 is crucial for RGC maintenance in primates, which differs from those in mouse. In mouse, knockout of both Brn1 and Brn2 inhibits neurogenesis and migration from the VZ at E12.5, while early overexpression of Brn2 promotes aberrant fate switching of RGC to upper-layer neurons in situ (21).
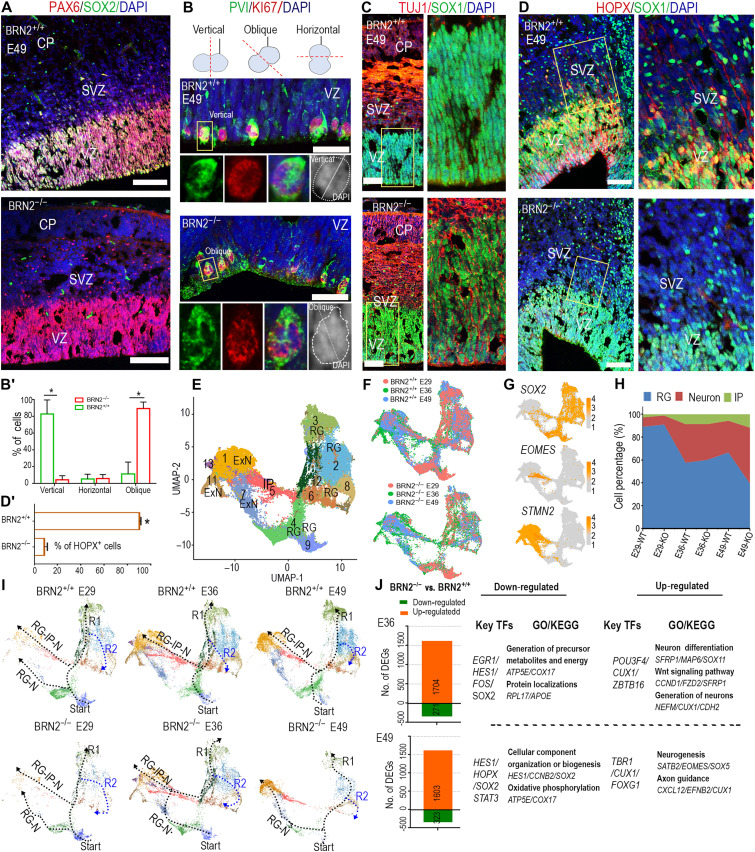
Fig. 3. BRN2 deletion results in abnormal development of monkey cortex.
(A to D) The coronal sections from the same region of primary somatosensory cortices were used to perform comparison analysis between E49 BRN2+/+ and BRN2−/− telencephalon (see Materials and Methods). (A) SOX2 and PAX6 expressions in the BRN2−/− and BRN2+/+ cortex, respectively. (B and B′) Representative staining images (B) and quantification (B′) of PVI, KI67, and DAPI in BRN2−/− and BRN2+/+ cortical cells. DAPI staining was used to identify cell division orientation. Quantification data of division orientations of RGCs are presented as means ± SEM (n = 6 representative images; >100 cells per group were counted, *P < 0.01). (C) Representative staining images of TUJ1 and SOX1 in BRN2−/− and BRN2+/+ cortical cells, respectively. (D and D′) HOPX and SOX1 expressions in the BRN2−/− and BRN2+/+ cortex, respectively. Right images are higher magnifications of left images. (D′) Quantification of HOPX cell migration into the SVZ region from VZ region (means ± SEM, n = 4 representative images; >500 cells per group were counted, *P < 0.01). Scale bars, 100 μm. (E and F) Visualization of major classes of neural cells from BRN2+/+ and BRN2−/− single cells across three different stages by Uniform Manifold Approximation and Projection (UMAP) analysis. (G) Plots of classical markers representing RGC (SOX2), intermediate progenitor (IP) (EOMES), and excitatory neuron (ExN) (STMN2), respectively. (H) Proportion dynamic of each cell type over cortical development. (I) Developmental trajectory of cortical cells across three different stages. RG-IP-N (RGC-IP-to-neuron) is the indirect way, while RG-N (RGC-to-neuron) is the direct way. The dotted lines show developmental trajectories of RGCs. The arrowheads show the directions of these trajectories. (J) DEGs between the BRN2−/− and BRN2+/+ cortex (see table S3). Representative transcription factors (TFs), gene ontology (GO) terms, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways are shown. Blue, DAPI, nuclear staining.
To further delineate the effects of BRN2 on cortical cell type composition and transcriptional profiles, we generated single-cell atlases for BRN2+/+ and BRN2−/− telencephalon at E29, E36, and E49 stages using a droplet-forming microfluidic device (10X Genomics platform) (Fig. 2A and fig. S2A). After quality control and filtration, we obtained 93,018 final cells for subsequent analyses (fig. S4D). Given the limited monkey resources to procure additional BRN2−/− fetuses, we first analyzed two E29 brains and observed a similar cell type composition between the two samples (fig. S4, E and F, and table S2). These results imply that the high-throughput technology is reproducible for constructing telencephalon atlases. Next, we integrated BRN2−/− and BRN2+/+ cells together across the three different stage samples (E29, E36, and E49) and annotated all cell types based on the expression levels of canonical cell type–specific markers (fig. S4, G to I, and table S2).
The mammalian neocortex contains two main neuron subpopulations: excitatory neurons (ExNs) and inhibitory interneurons. ExNs constitute 80% of neurons and originate from RGCs or IPs in the cortical VZ/SVZ. Thus, we first selected and integrated all RGC, IP, and ExN single cells sampled from BRN2+/+ and BRN2−/− cells across three stages to trace their development trajectories (Fig. 3, E and F). On the basis of classical markers and differentially expressed genes (DEGs), we categorized these cell populations into RGC (eight clusters), IP (one cluster), and ExN (four clusters) (Fig. 3, E to G; fig. S5, A and B; and table S3). By comparison, we did not observe obvious differences in cell type and proportion between BRN2+/+ and BRN2−/− cortices at E29 or E36, respectively (Fig. 3H). However, compared to the BRN2+/+ monkey, the RGC proportion exhibited a remarkable decrease in conjunction with an obvious increase in IPs and ExNs in E49 BRN2−/− cortex (Fig. 3H and table S3), which is consistent with our histology analysis.
By analyzing cell trajectory and pseudotime, we found that BRN2 deficiency changed the trajectory and direction of RGC development and neurogenesis (Fig. 3I and fig. S5C). In the BRN2+/+ cortex, RGC development followed two trajectories (R1 or R2 direction) from the start point independent of development stages (Fig. 3I and fig. S5C). However, the developmental trajectory for both the E36 and E49 BRN2−/− cortex was broken due to the loss of one population in cluster 4 (Fig. 3I and fig. S5C). In the BRN2+/+ cortex, neurogenesis followed the direct path [RGC-to-neuron (RG-N)] and indirect path [RGC-IP-to-neuron (RG-IP-N)] at E29, but the indirect path primarily predominated at E36 and E49 (Fig. 3I). As expected, since BRN2 is initially expressed after E29, BRN2−/− and BRN2+/+ cortical cells at E29 shared a similar trajectory of RGC development and neurogenesis (Fig. 3I). However, the direct path mainly contributed to neurogenesis of BRN2−/− cortical cells at E36 and dominated the process at E49 (Fig. 3I). Furthermore, pseudotime results confirmed the early initiation of the differentiation program for BRN2-deficient RGCs (fig. S5C). These results together with the histology analysis (Fig. 3, A to C, and fig. S4B) demonstrated the precocious and in situ neuron differentiation of RGCs. From E36 to E49, BRN2 deletion gradually induced abnormal development of the monkey cortex, in accord with BRN2 temporal expression patterns in the cortex. These results further demonstrated that BRN2 deletion drove these RGC phenotypes.
To characterize the molecular mechanisms and downstream genes including TFs of BRN2, we analyzed DEGs between BRN2+/+ and BRN2−/− cortex (Fig. 3J and table S3). The down-regulated transcription factors were related to neural progenitors, including HES1, HOPX, and SOX2, whereas up-regulated transcription factors mainly associated with neurogenic factors in BRN2−/− cells, such as TBR1 and SATB2, consistent with our immunohistochemical observations (Fig. 3J and fig. S5, D to H). In particular, HES1, an important effector of the Notch pathway that promotes cortical progenitor self-renewal (17, 37), showed a marked reduction in the E49 BRN2−/− cortex, especially in cluster 4 (fig. S5E). This result is contrary to the role of Brn2 in mice, which has minimal effect on HES1 (21). Consistent with the staining results (Fig. 3, A and D, and fig. S4A), HOPX and SOX2 expression patterns exhibited a marked decline in the E36 BRN2−/− cortex and differed significantly at E49 in the BRN2−/− cortex (fig. S5, D and F). Together, our results revealed that BRN2 regulates RGC maintenance and SVZ formation through multiple mechanisms.
Abnormal interneuron development in the BRN2−/− monkey
Cortical inhibitory interneurons have crucial roles in cortical development and function. Primate interneurons mostly originate from three distinct molecular and morphological regions: the medial, lateral, and caudal GEs (MGE, LGE, and CGE, respectively) (38). We observed strong expression of BRN2 in the whole developing monkey GEs (Fig. 4A and fig. S6A), which led us to hypothesize that BRN2 may function in interneuron development. We used COUP-TFII (NR2F2, a marker of CGE or LGE) and NKX2-1 [a marker of MGE (38)] to distinguish LGE/CGE and MGE by immunostaining (Fig. 4B and fig. S6D). Compared to the clear boundary of LGE and MGE in BRN2+/+ telencephalon, hematoxylin and eosin staining indicated that the BRN2−/− LGE and MGE were difficult to distinguish at E49 because of the absence of a distinct boundary between them (fig. S6, B and C). In contrast to the special distribution in BRN2+/+ CGE or LGE, COUP-TFII and NKX2.1 were widely activated in the entire BRN2−/− GEs, including MGE (Fig. 4B and fig. S6D). The data showed that the separation of MGE from CGE/LGE was impaired in the BRN2−/− GEs. To reveal the potential phenotypes and molecular characteristics, we analyzed the single-cell RNA sequencing (scRNA-seq) transcriptome of BRN2+/+ and BRN2−/− interneurons. We categorized these interneurons into 10 clusters, which highlighted interneuron progenitors (iNPs; clusters 3, 5, and 8) and differentiated interneurons (other 7 clusters) based on DEGs and classical markers (Fig. 4C; fig. S6, E and F; and table S4). As expected, the proportion of iNPs in the BRN2−/− GEs compared to that in BRN2+/+ GEs markedly decreased along with a distinct increase of differentiated interneurons (fig. S6G). Cell trajectory and pseudotime analysis showed that the BRN2 mutation changed the interneuron differentiation trajectory and maturation pathway (Fig. 4D and fig. S6H).
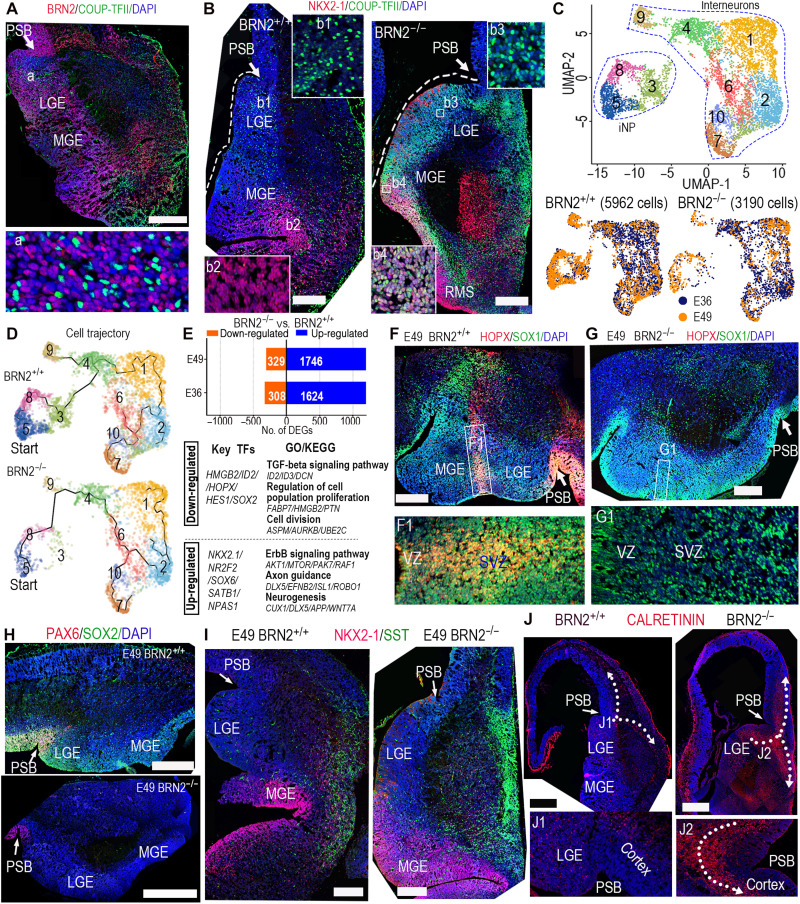
Fig. 4. BRN2 loss produced abnormities of monkey interneuron development.
(A) BRN2 and COUP-TFII staining showed the wide distribution of BRN2 in the telencephalic ganglionic eminences (GEs). PSB, pallial-subpallial boundary. (B) NKX2-1 and COUP-TFII expression in BRN2+/+ and BRN2−/− GEs, showing that COUP-TFII was specifically expressed in BRN2+/+ CGE/LGE, whereas it was obviously activated in the whole BRN2−/− GEs (including both LGE/CGE and MGE). The sections of BRN2+/+ and BRN2−/− were from equivalent coronal levels (details are in Materials and Methods). (C) Visualization of major classes of interneuron and interneuron progenitor (iNP) by UMAP analysis. (D) Developmental trajectory of interneurons and iNPs showing that BRN2 loss changed interneuron development trajectory. Cells in cluster 3 were markedly decreased in BRN2−/− monkeys. (E) DEGs between BRN2+/+ and BRN2−/− interneurons and iNPs (table S4). Representative transcription factors (TFs), GO terms, and KEGG pathways are shown. (F and G) The sections of BRN2+/+ and BRN2−/− telencephalon were from equivalent coronal levels. (F and G) Representative staining images of HOPX and SOX1 in BRN2+/+ (F) and BRN2−/− (G) GEs. F1 and G1 are magnifications of the squares in (F) and (G). (H) PAX6 and SOX2 expression in BRN2+/+ and BRN2−/− GEs. (I) SST and NKX2-1 expression in BRN2+/+ and BRN2−/− GEs, respectively. (J) CALRETININ (CR) expression in BRN2+/+ and BRN2−/− telencephalon. Arrows indicate the migration orientations of CR interneurons. All images of immunofluorescence were from coronal sections spanning the rostral-caudal extent of the telencephalon. Scale bars, 500 μm. Blue, DAPI, nuclear staining.
By comparative analysis, we identified DEGs and transcription factors, including HOPX, SOX2, and HES1, which were significantly down-regulated in the BRN2−/− GEs (Fig. 4E and table S4). Significantly down-regulated genes in BRN2−/− interneurons aligned with cell division and proliferation, whereas activated genes were enriched in axon guidance and neurogenesis (Fig. 4E). To further understand the phenotype, we examined cluster change and observed that one cell population [cluster 3 (C3)] was markedly absent in BRN2−/− interneurons (Fig. 4C and fig. S6G). Notably, some genes, such as HOPX and HES1, were specifically expressed in the C3 population of BRN2+/+ cells at E49 (Fig. 4E and fig. S6I), which implied that BRN2 could regulate these genes. Immunostaining on GEs showed that HOPX was expressed in an interneuron migration stream of BRN2+/+ MGE (Fig. 4F), whereas BRN2 null resulted in HOPX loss in the region (Fig. 4G). These data showed that C3 arises from these HOPX+ cells in the migration stream. Similar to that in the cortex, SOX2 expression in GEs was also markedly inhibited by BRN2 deficiency (Fig. 4H and fig. S6I). Together, these results showed that BRN2 is required for iNP maintenance, whereas its deficiency promotes the precocious production of interneurons.
Given that most primate neocortical gamma-aminobutyric acid-ergic (GABAergic) interneurons originate from MGE or LGE/CGE (38), we examined the interneuron genesis of the two regions. In the MGE, two main pathways control general aspects in interneuron development (39). NR2F1, an MGE marker, was markedly activated in BRN2−/− cells (fig. S7A), which prompted further evaluation of the changes in the two pathways. The first pathway was a NKX2-1/LHX6-SOX6/SATB1 transcriptional cascade that is crucial for MGE interneuron production (40). As expected, these gene expression patterns exhibited a marked up-regulation in BRN2−/− cells (fig. S7B), which implied pathway activation. The other pathway was the DLX family of homeodomain TFs that is regarded as the core on the gene regulatory network of interneuron development (40). We observed that BRN2 deficiency clearly activated the DLX gene network and promoted interneuron differentiation including somatostatin (SST) (Fig. 4I and fig. S7C).
Next, we investigated interneuron genesis in LGE/CGE. In primates, most CR (calretinin) neurons originated from the LGE and CGE, while LGE- and CGE-derived interneurons preferentially occupied the superficial cortical layers (38). In the LGE, BRN2 and NR2F2 (COUP-TFII) showed exclusive expression (Fig. 4A), while the expression of COUP-TFII markedly increased in BRN2−/− cortex, suggesting that BRN2 may restrict NR2F2 expression by maintaining RGCs. BRN2 deficiency activated CGE markers, including NR2F2 (41), NFIB, and APIS2 (42) (Fig. 4B and fig. S7D), and subsequently promoted differentiation, migration toward superficial cortical layers, and neocortical distribution of CR interneurons (Fig. 4J and fig. S7E). Furthermore, interneuron development abnormality caused by BRN2 deficiency was increasingly obvious over telencephalon development. Thus, BRN2 as an upstream factor controls interneuron genesis throughout the primate GEs.
BRN2 exhibits a conserved function for human RGCs
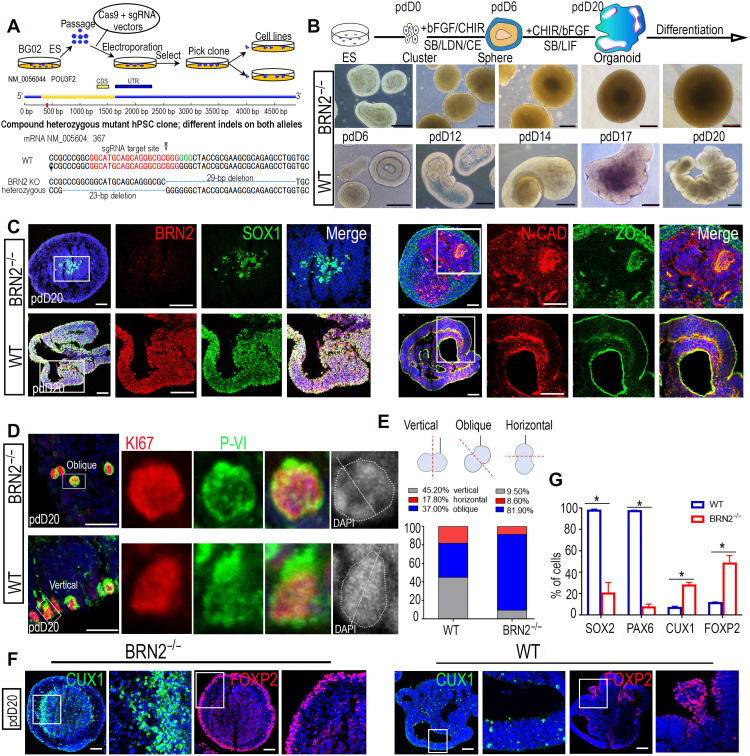
To investigate whether BRN2 function is conserved for human RGCs, we analyzed the temporal expression of BRN2 during human neural induction. As expected, BRN2 expressed in the human early NESCs and its expression pattern was highly consistent with that of primates (fig. S8A). We then generated one BRN2−/− human ESC derived from two different colonies by gene editing (Fig. 5A) and differentiated these cells into NESCs following the protocol (43) for generating cortical organoids (Fig. 5B). BRN2 deficiency failed to affect the generation of human NESCs and the formation of the typical two-layer neuroectoderm structures (43) (Fig. 5B and fig. S8, B to D), implying that BRN2 is not required for human neuroectoderm production, consistent with the function of BRN2 in monkey. After 20 days of culture and induction, BRN2+/+ NESCs further self-organized into cortical organoids, 94% of which exhibited a VZ-like structure, expressing typical NP-specific markers and forming NESC polarity in accord with the polarized distribution of N-CADHERIN and ZO-1 (Fig. 5, B and C, and fig. S8, B to D) (43). In contrast, almost all organoids derived from BRN2−/− NESCs lost NESC polarity and identity, and subsequently failed to generate cortical organoids (Fig. 5, B, C, and G, and fig. S8D).
Fig. 5. BRN2 function on monkey RGCs is conserved for human RGCs.
(A) Schematic of generating BRN2-knockout (BRN2−/−) human embryonic stem cells (ESCs) by CRISPR-Cas9 gene editing. (B) Generation of cortical organoids using human wild-type (BRN2+/+) and BRN2−/− ESCs, respectively, showing that BRN2−/− ESCs lost the ability to generate cortical organoids. The top panel shows the diagram of cortical organoid generation. (C) Expression of representative neuroepithelium markers in cortical organoids from BRN2+/+ and BRN2−/− ESCs. (D) Comparison of proliferation markers KI67 and PVI in BRN2+/+ or BRN2−/− cortical organoids. DAPI staining was used to identify cell division orientation. (E) Quantifications of cell division orientation in BRN2+/+ and BRN2−/− cells of cortical organoids (means ± SEM, n = 4 independent experiments; >200 cells per group were counted), respectively. (F) Comparison of CUX1 (an upper-layer neuron marker) and FOXP2 (a deep-layer neuron marker) expressions between BRN2+/+ and BRN2−/− cortical organoids, respectively. (G) Quantification of SOX2-, PAX6-, CUX1-, and FOXP2-positive cells in BRN2+/+ and BRN2−/− cortical organoids, respectively. Data are presented as means ± SEM, n = 3 slices; >300 cells per group were counted. *P < 0.05. Scale bars, 100 μm.
We also observed that BRN2 loss induced a reduction in RGC proliferation and decreased the vertical RGC divisions with a coordinated increase in oblique divisions by KI67 and PVI staining (Fig. 5, D and E) (34). Differentiation assays showed that BRN2 deletion promoted early generation of deep-layer neurons (Fig. 5, F and G, and fig. S8D). Thus, BRN2 is required for human RGC maintenance.
Mechanism of BRN2 regulating primate telencephalon development
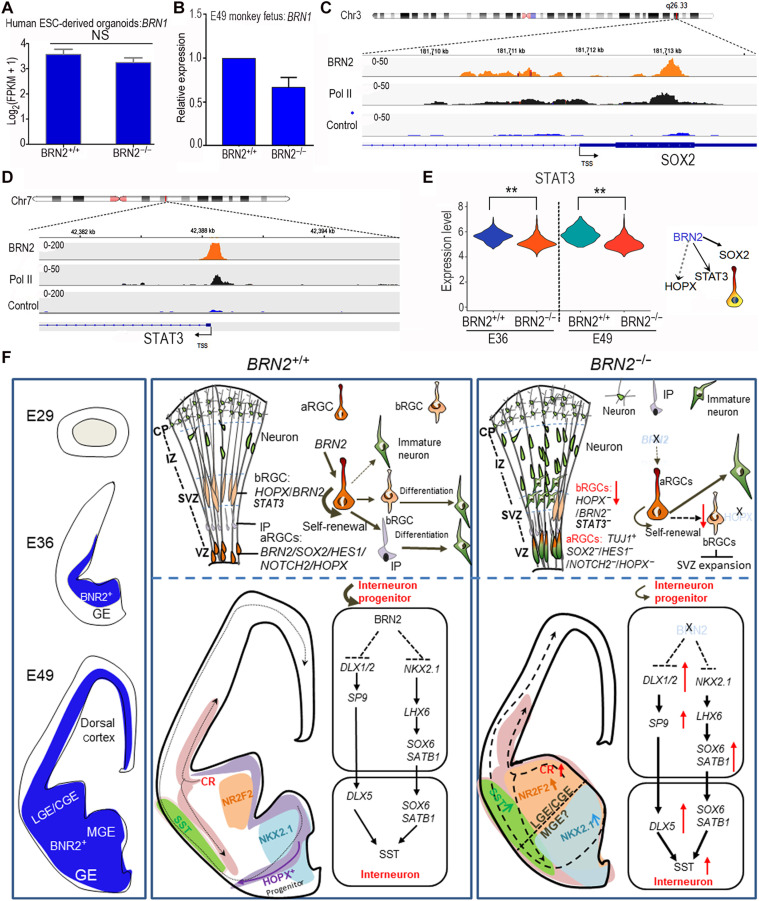
Since Brn1 can compensate for Brn2 functions in mouse neocortex when Brn2 is deleted (22, 28, 29), we analyzed BRN1 expression and did not observe any up-regulation of BRN1 in BRN2−/− human ESC–derived cortical organoids or monkey fetal cortical cells (Fig. 6, A and B). These results indicate that BRN2 deletions were unable to change BRN1 expression in primates and humans. These results revealed that a notable decrease of SOX2 was first detected in the E36 BRN2−/− monkey cortex (fig. S4A). Thus, we speculated that BRN2 could regulate the RGC expansion by directly activating SOX2. Using genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) of developing human cortex and human ESC–derived NESCs (43, 44), we identified direct targets of BRN2 (fig. S9A). We found a significant enrichment of BRN2 in the promoter region of SOX2 (Fig. 6C, fig. S9B, and table S5), a crucial gene that maintains RGC identity. This result suggests that BRN2 may maintain RGC identity through direct regulation of SOX2 expression. In addition, HOPX, a key marker of bRGCs (14, 35, 36), was completely lost in the BRN2-knockout monkey telencephalon. However, we did not detect direct binding of BRN2 on HOPX (table S5), implying that BRN2 may regulate HOPX expression indirectly. We noted that BRN2 can bind to the promoter region of STAT3 (Fig. 6D and fig. S9C). STAT3 is crucial for bRGC maintenance in the human brain (35, 45) and may be an upstream factor regulating HOPX (35, 45, 46). Furthermore, single-cell data showed that STAT3 expression was significantly inhibited in the BRN2−/− cortex (Fig. 6E). In conclusion, BRN2 may function as a key factor of early stages of neural development in primates through multiple potential mechanisms (Fig. 6F).
Fig. 6. The regulatory mechanisms of BRN2 on primate telencephalon development.
(A and B) Comparison of BRN1 expressions between BRN2+/+ and BRN2−/− human ESC–derived cortical organoids (A) and between BRN2+/+ and BRN2−/− monkey cortical cells (B). Data are presented as means ± SEM (n = 3). NS, no significance, P > 0.05. (C) Gene plots show densities of BRN2, polymerase II (Pol II), and control (IgG) at the SOX2 promoter in human developing cortex, indicating that BRN2 binds to the promoter region of SOX2. (D) Gene plots show densities of BRN2, Pol II, and control (IgG) at the STAT3 promoter in human developing cortex, indicating that BRN2 regulates STAT3 expression by directly binding to the promoter region of STAT3. (E) Comparison of STAT3 expressions in the BRN2+/+ and BRN2−/− cortex, which exhibited a significant down-regulation in the BRN2−/− cortex. **P < 0.01. (F) Summary of BRN2 regulatory mechanisms on monkey telencephalon development. aRGC, apical RGC. The thickness and fineness of the line represent the strong and weak activity, respectively. Arrows indicate the activation, and trident lines indicate the repression.
DISCUSSION
Using monkey knockout models and human organoid assays, the spatiotemporal expression and function of BRN2 during telencephalon development exhibit obvious differences between primate and mouse. BRN2 is expressed by RGCs of the VZ/SVZ in early fetal monkey neocortex and GEs, earlier than the RGCs in mouse (Fig. 1, A to C, and figs. S1, A to G, and S2, A to D) (21). Mutated BRN2 is lethal in monkey fetuses before midgestation, causes abnormal development of neocortex and GEs by decreasing RGC proliferation, and promotes precocious genesis of ExNs and interneurons. In contrast, Brn2 knockout mice do not induce lethality of mouse fetuses or neuronal loss in the cortex before birth (22, 29). Furthermore, we uncovered the predominant underlying molecular and cellular mechanisms by which BRN2 regulates primate neocortical development (Fig. 6). Overall, we demonstrated that BRN2, a conserved transcription factor, gains novel and more critical functions in monkeys compared to mice. Our study found previously unidentified critical functions of BRN2 for the developing primate telencephalon, which likely reveals a previously unknown mechanism of primate early telencephalon development.
Although BRN2 contributes to neural development in both primates and mice, its functions are markedly different between the species. In mice, Brn2 deficiency shows abnormalities only in limited brain regions, primarily in the hypothalamic supraoptic and paraventricular nuclei (22, 29), while BRN2 deficiency in primates induces abnormalities of the cortex and GEs. In Brn2-knockout mice, cells in the hypothalamic supraoptic and paraventricular nuclei also fail to differentiate into mature neurons, which leads to mortality within 10 days after birth (47). In contrast, mutated BRN2 causes lethality of monkey fetuses before midgestation, causes early abnormal development of neocortex and GEs by decreasing RGC proliferation, and promotes precocious genesis of ExNs and interneurons.
Elucidating the cellular and molecular architecture of the primate cerebral cortex is central to understanding our cognitive abilities and susceptibility to disease. Despite some success, we are only beginning to elucidate the genetic changes that occurred during evolution that define the primate brain (7). The evolutionary changes that expanded the neocortex likely used multiple processes at different levels (48), including changes in gene regulatory elements and changes in the genome that ranged from point mutations to entire gene duplications. These changes incorporated alterations in gene expression, produced genes with novel functional roles (9), and led to the emergence of primate-specific genes (1, 15, 17–20). Relevant changes in evolutionary functions likely arose from acquiring genetic mutations that induce altered or novel functions in the same cells (9). In the study, we noted that the regulatory mechanism of BRN2 for primate brain development differs from these reported mechanisms. BRN2 protein is highly conserved across species (fig. S9D), which implies that the differences in the spatiotemporal distribution of BRN2 between mouse and monkey cortex do not arise from the evolution of BRN2 protein sequence. Our results support the idea that diverse spatiotemporal distributions of genes play important roles in driving the telencephalon expansion during primate evolution. We posit that such distributions enable the emergence of previously unknown gene functions. Thus, our findings reveal a previously unidentified evolutionary mechanism in primate telencephalon development. In addition, differences in BRN2 expression patterns and functions between mouse and primate imply that primate brain development is poorly recapitulated in the mouse. Our findings underscore using species-specific genetic models to understand primate brain development.
Cortical neurons are produced via direct or indirect neurogenesis, either from RGCs or through RGCs-IPs, respectively. In primate cortices, indirect neurogenesis (RGCs-IPs-neurons) contributes most of the neurons. We found that BRN2 deletion induced differentiation of VZ RGCs in situ and switched the indirect neurogenesis to the direct neurogenesis (RGCs-neurons). We speculate that several possible mechanisms regulate these phenotypes (Fig. 6F). First, interactions between SOX and POU factors are conserved in all metazoans and play key roles in embryonic development. In this study, we noted that SOX2, but not SOX1, was significantly inhibited in BRN2−/− monkey RGCs, and BRN2 directly binds SOX2 promoter (Fig. 6C and fig. S9B). These findings, together with BRN2 expression in RGCs after E30, imply that BRN2 acts partially by retaining SOX2 expression to determine monkey RGC fate. Second, HOPX, a key marker of primate bRGCs (35, 36, 49), is absent in the E49 BRN2−/− cortex. Our findings revealed that BRN2 deficiency–induced HOPX loss subsequently inhibited RGC detachment from the VZ. HOPX+ RGCs in primate neocortex are significantly more abundant compared to their low abundance in the developing mouse neocortex (36, 50). In addition, our ChIP-seq data from the developing human cortex and human NESCs showed that BRN2 binds STAT3. STAT3 is reportedly a key determinant for RGC expansion accompanied by increasing the number of RGCs in the human brain (35). However, these findings require additional detailed mechanistic insight.
Abnormal interneuron development was detected in BRN2−/− monkeys. Our knowledge of cortical interneuron development primarily derives from mouse studies. Interneuron development is remarkably similar between mouse and primate, but there are important differences in the morphology, gene expression, and proportions of specific interneuron subsets (32, 33). Cortical interneurons mainly originate from the MGE, LGE, and CGE, respectively. Although some developmental and functional diversification of cortical interneurons and downstream factors that mark the MGE and LGE/CGE regions exists in primates (32, 38), the upstream regulators controlling MGE and LGE/CGE specification and separation remain unknown. Our findings showed that BRN2 deficiency significantly up-regulated COUP-TFII expression in the total GEs including MGE, which makes LGE and MGE undistinguishable. This result indicates that BRN2 is required for MGE and LGE/CGE specification, in contrast to having no effect on GEs in mouse. To our knowledge, we now revealed that BRN2 acts as an upstream factor that controls MGE and LGE/CGE specification in primates. Thus, we provide an example to emphasize the importance of directly studying the primate brain to further elucidate primate cortical interneuron development.
The data presented here provide crucial insights into the evolution of BRN2-induced telencephalic cortical development. These studies lay the foundation to uncover the mechanisms governing primate RGC proliferation and interneuron development. However, the regulatory elements governing the spatiotemporal distribution or downstream signaling of BRN2 still require further study as we unfortunately failed to uncover BRN2 functions for monkey neural development after midgestation lethality. Future work should develop inducible BRN2 conditional-knockout monkeys to resolve these issues.
MATERIALS AND METHODS
Animal ethics statement
Female cynomolgus monkeys (M. fascicularis), ranging in age from 5 to 8 years with body weights of 4 to 6 kg, were used in this study. All animals were housed at Yunnan Key Laboratory of Primate Biomedical Research (LPBR) and individually bred in an American standard cage at a light/dark cycle of 12 hours/12 hours. All animal procedures were approved in advance by the Institutional Animal Care and Use Committee of Yunnan Key LPBR and were performed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International for the ethical treatment of primates.
Caesarian section and fetus collection
Fetuses from six prenatal developmental stages (gestation periods of 29, 30, 36, 49, 55, and 72 days) of cynomolgus monkeys were collected by caesarian section as previously described (51).
Tissue processing and immunohistochemistry
After fetuses were collected, brain tissues were fixed; sequentially infiltrated with a series of 10, 20, and 30% (w/v) sucrose solutions; embedded in OCT (optimal cutting temperature compound; Sakura, 4583); and sectioned at the thickness of 10 to 15 μm on a Leica cryostat. For samples of E29, E30, and E36, the forebrain prominence was isolated from embryonic monkeys and sectioned at 10 μm thickness spanning the rostral-caudal extent of the brain. For samples of E40, E49, E55, and E72, the right embryonic hemispheres were isolated and sectioned at 15 μm thickness spanning the rostral-caudal extent of the brain (fig. S2A). For immunohistochemistry, sectioned tissues on slides were washed twice in phosphate-buffered saline (PBS), treated with 0.2% Triton X-100 (Gibco, 85112) overnight, washed three times in PBS, and blocked for 1 hour at room temperature with 3% bovine serum albumin (BSA; Solarbio, A8020). Slides were incubated with primary antibodies overnight, washed in PBS, and incubated for 2 hours at room temperature in blocking buffer containing Alexa Fluor 488–, 594–, or 647–conjugated secondary antibodies (1:600; Invitrogen). The following primary antibodies were used at 1:200 to 1:2000: SOX1, goat (AF3369, R&D Systems); DCX, mouse (MABN707, Millipore); TBR1, rabbit (AB10554, Millipore); TUJ1, mouse (MAB1637, Millipore); NESTIN, mouse (MAB5326, Millipore); TBR2, rabbit (ab23345, Abcam); N-Cadherin, rabbit (GTX127345, GeneTex); PAX6, rabbit (901301, BioLegend Covance); IBAL1, rabbit (019-19741, Wako); COUP-TFII, mouse (PP-H7147-00, R&D Systems); HOPX, rabbit (HPA030180, Sigma-Aldrich); NeuroD2, rabbit (ab104430, Abcam); GFAP, rabbit (z033429-2, Dako); SST, mouse (sc-55565, Santa Cruz Biotechnology); GAD65, mouse (ab26113, Abcam); BRN2, rabbit (12137S, Cell Signaling Technology); NKX2.1, rabbit (AB76013, Abcam); Vimentin, rabbit (14-9897, eBioscience); ZO-1, mouse (01-107, Invitrogen); PVI, mouse (D076-3S, MBL International); CUX1, rabbit (SC13042, Santa Cruz Biotechnology); FOXP2, rabbit (ab16046, Abcam); SATB2, mouse (ab51502, Abcam); Calretinin, mouse (AB5054, Millipore); SOX2, rabbit (AB5603, Millipore); and KI-67, rabbit (PA5-19462, Invitrogen).
For immunohistochemistry, whole brain was used for 10- to 15-μm coronal serial sections. To ensure that the sections of BRN2+/+ and BRN2−/− were from equivalent coronal levels, we used the following method. First, sections of the whole brain were numbered in positional order and then evenly categorized into three parts: the anterior, the middle, and the caudal part according to their positions. Second, two slides in the median of each part were chosen for simultaneous staining. The staining of the middle part is shown in Fig. 4B and fig. S6 (B and C), while the caudal part staining is shown in fig. S6D.
Preparation of gRNA
sgRNAs that target the BRN2 exon were designed by the CHOPCHOP online software in the website https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design. The top five with high scores were selected (table S1), and sgRNA oligos were amplified and transcribed in vitro using the GeneArt Precision gRNA Synthesis Kit (Invitrogen, A29377). Efficiency of these five sgRNAs was first verified on fibroblasts by electrotransfection, and the three of them with high efficiency were chosen for embryo editing.
Oocyte collection and in vitro fertilization
Oocyte collection and fertilization were performed as previously described (52). In brief, 10 healthy female cynomolgus monkeys aged 5 to 8 years with regular menstrual cycles were selected as oocyte donors for superovulation, which were performed by intramuscular injection with rhFSH (recombinant human follitropin alpha, GONAL-F, Merck Serono) for 8 days and rhCG (recombinant human chorionic gonadotropin alpha, OVIDREL, Merck Serono) on day 9. Oocytes were collected by laparoscopic follicular aspiration 32 to 35 hours after rhCG administration. Follicular contents were placed in Hepes-buffered Tyrode’s albumin lactate pyruvate (TALP) medium (Caisson Labs, IVL01) containing 0.3% BSA at 37°C. Oocytes were stripped of cumulus cells by pipetting after brief exposure (<1 min) to hyaluronidase (0.5 mg/ml) in TALP-Hepes to allow visual selection of nuclear maturity metaphase II (MII; first polar body present) oocytes.
RNP injection, in vitro fertilization, embryo culture, and transfer
Mature oocytes were subjected to intracytoplasmic sperm injection (ICSI) immediately and then cultured in CMRL-1066 containing 10% fetal bovine serum (FBS; Invitrogen, 16140071) at 37°C in 5% CO2. Fertilization was confirmed by the presence of a second polar body and two pronuclei. To produce BRN2-knockout embryos, we injected a mixture of sgRNA pair (2500 ng/μl, 1250 ng/μl for each sgRNA) and Cas9 nuclease (5000 ng/μl; Invitrogen, A36499) with a total volume of 5 pl into each oocyte before ICSI. Microinjections were performed in the cytoplasm of oocytes using a microinjection system under standard conditions. Zygotes (wild-type and BRN2 RNP-treated) were then cultured in chemically defined hamster embryo culture medium-9 (HECM-9) containing 10% FBS (Gibco, 16140071) at 37°C in 5% CO2 to allow embryo development. The culture medium was replaced every other day until the blastocyst stage. The cleaved embryos with high quality at the two-cell to blastocyst stage were transferred into the oviduct of the matched recipient mother monkeys. Thirty-eight monkeys were used as surrogate recipients. The earliest pregnancy diagnosis was performed by ultrasonography about 20 days after the embryo transfer. Both clinical pregnancy and number of fetuses were confirmed by fetal cardiac activity and the presence of a yolk sac as detected by ultrasonography.
Genomic DNA extraction and genotyping
Genomic DNA from tissues of mutant and wild-type fetuses was extracted by using the Wizard Genomic DNA Purification Kit (Promega, #A1125) according to the manufacturer’s instructions. Embryo genomes were amplified by the REPLI-g Single Cell Kit (QIAGEN, 150345). PCR was performed at 95°C for 5 min; 35 cycles at 95°C for 30 s, 64°C (drop down to 0.2°C per cycle) for 30 s, and 72°C for 2.5 min; and 72°C for 10 min for type I samples or 95°C for 5 min; 35 cycles at 95°C for 30 s, 59.5°C for 30 s, and 72°C for 1 min, and 72°C for 10 min for type II samples. Primers were as follows (BRN2-L-F/R and BRN2-F/R for type I and II samples, respectively): BRN2-L-F, 5′-CAGCAGTAATAGCAGGAGC-3′; BRN2-L-R, 5′-TTGGATAAAGCGGGTAGA-3′; BRN2-F, 5′-TGGCGACCGCAGCGTCTAAC-3′; BRN2-R, 5′-TTGCCATACAGAGTGCCCAGAG-3′. PCR products were conducted with Sanger sequencing and then analyzed. To identify whether chimeras exist, PCR products were further cloned into the vector Topo-clone (TSINGKE, 007VS) before sequencing.
Off-target analysis
We searched potential off-target sites and sequences by the online tool Cas-OFFinder (53) [www.rgenome.net/cas-offinder/; Macaca fascicularis (5.0) was chosen as Organism Type and Genomes]. Under the condition that the mismatch number is equal to or less than 3 and the DNA/RNA bulge size is equal to or less than 2387 and 516, potential target sites were provided (table S1). Twenty sites with high potential of off-targeting were selected for PCR amplification (product code of PCR enzyme is TSE004 from TSINGKE; primers are detailed in table S1) and sequencing. Those sites were selected as the principles as follows. Related reports showed that (i) protospacer adjacent motif (PAM)-proximal mismatches are less tolerated than PAM-distal mismatches; (ii) the sequence located at +4 to +7 position upstream of PAM is highly sensitive to target mismatch; (iii) Cas9 tolerates DNA bulges in three target sites: seven bases from PAM, the 5′ end (PAM-distal), and the 3′ end (PAM-proximal); and (iv) sgRNA bulges close to the PAM tend to prohibit cleavage (54, 55).
Single-cell suspension preparation
Single-cell isolation was performed primarily as previously described (56). Briefly, the telencephalon from monkey fetus (the whole forebrain prominence was collected at E29 and E36, and the left hemispheres were collected at E49) was dissected in Hibernate E medium (Invitrogen, A1247601) with deoxyribonuclease I (10 U/μl) (10 mg/ml; Roche, 11284932001), collagenase IV (2 mg/ml; Gibco, 17104-019), and papain (10 mg/ml; Worthington, LS003118). The cell suspension was vortexed and kept at 37°C for 10 min and then centrifuged at 140g for 5 min to obtain a cell pellet. Last, the cell pellet was resuspended in 500 μl of PBS and kept on ice.
10X RNA library preparation for high-throughput sequencing
High-throughput scRNA-seq was conducted following the instruction manual of 10X Genomics. Single cells were resuspended to a concentration of 1 million cells/ml in PBS supplemented with 0.04% BSA, and 16.7 μl of the suspension was then loaded into each 10X Chromium lane. 10X Chromium lane v2 was used for WT-E36 and WT-E49, while 10X Chromium lane v3 was used for the other samples instead, because the kit of v2 was not available after the kit update. The complementary DNA (cDNA) library was checked with a bioanalyzer (Agilent 2100 Bioanalyzer) for quality control. Following the library preparation, the sample was sequenced by NovaSeq S6000. Libraries were sequenced to an average depth of 25,000 to 50,000 reads per cell.
Single-cell bioinformatic analysis
Sequencing reads were aligned and annotated with the M. fascicularis (version 5.0) reference genome by using Cell Ranger (version 3.0.2) (57) and count pipeline that was provided by10X Genomics with default parameters. Cells with fewer than 500 unique molecular identifiers (UMIs), more than 4500 UMIs, or expression mitochondria gene fraction of more than 5% were discarded in the following analysis. The UMI counts were normalized to 10,000 counts per cell and converted to log scale (Seurat function NormalizeData). The batch, condition, and personal identifying information were added as metadata to the final Seurat object; nUMI and batch were regressed out using the ScaleData function. Cell doublets were removed by DoubletFinder V2.0 (58) assuming 10% doublets in our data, and other parameters were principal components (PCs) = 1:30 and the parameter selection for numbers of artificial doublets (pN) = 0.25. The parameter of PC space neighborhood size (PK) value of each sample was estimated independently by find.pk function. Standard integration analysis was then performed by combining wild-type (57) and knockout brain samples in E29, E36, and E49, respectively, with dim = 1:30 and anchor.features set to 6000 in FindIntegrationAnchors function. Highly variable genes in the integrated dataset were computed and used as input for principal components analysis. Significant principal components were used for downstream graph-based, semi-unsupervised clustering into distinct populations (FindClusters function), and Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction was then used to project these populations in two dimensions. For clustering, the resolution parameter, which indirectly controls the number of clusters, was approximated on the basis of the number of cells according to Seurat guidelines; a vector of resolution parameters was passed to the FindClusters function, and the optimal resolution that established discernible clusters with distinct marker gene expression was selected (59). To identify marker genes, the clusters were compared pairwise for differential gene expression using the Wilcoxon rank sum test for single-cell gene expression (FindAllMarkers function, min.pct = 0.4, min.diff.pct = 0.3, and false discovery rate < 0.05). To assign identities to these subpopulations, we cross-referenced their marker genes with known brain cell subtype markers from the literature, PAX6 for neural progenitor cell, STMN2 for neuron, etc. The cluster data of the neurons/RG (radial glia), interneuron, and microglial/endothelium cells were picked out from each time point, and then data of the same cell types were integrated together for all time points. The clustering approach was then repeated for the remaining cells. Reclustering analysis was performed as described above. For the DEG analysis in BRN2-knockout groups, the FindMarkers function was used on both wild-type and BRN2-knockout cells with the following parameters: Wilcoxon rank sum test, min.pct = 0.25, and logFC.threshold = 1. An additional adjusted P value (Bonferroni correction) cutoff of <1 × 10−10 was used to identify DEGs.
Regarding the concern about the different kit for E36 and E49, V2 and V3 were used in the study because of product upgrade, and their differences caused by V2 and V3 kit can be removed by the improvement of our analysis method. To minimize the batch effects of different kits, we analyzed our single-cell expression data by using scRNA-seq integration method in Seurat. This pipeline could identify cross-dataset pairs of cells that are in a matched biological state (“anchors”) and can be used both to correct technical differences between datasets (i.e., batch effect correction) and to perform comparative scRNA-seq analysis across experimental conditions. We also analyzed the RGs and interneurons independently from the other cells to reduce the potential batch effects.
Cell trajectory analysis
Pseudotime and trajectory analyses were performed using the Monocle3-alpha package, as described in the tutorials (http://cole-trapnell-lab.github.io/monocle-release/monocle3). UMAP projection data from Seurat output were transferred to monocle for cell visualization.
Establishment of the BRN2 gene-knockout ES cell line
For BG02 ES BRN2 knockout, sgRNAs were designed to target BRN2 exon (Fig. 5A), with the design of sgRNA using the online CRISPR design tool (https://zlab.bio/guide-design-resources), and cloned into the Pgl3-U6-2sgRNA-ccdB-EF1a-Cas9ZF-IRES (Addgene, 115519) vector. The sgRNA sequences of F-5′-ACCGGGCATGCAGCAGGGCGCGGG and R-5′-AAACCCCGCGCCCTGCTGCATGCC were used for cloning into this vector. Then, the sgRNA vector was electroporated into BG02 ESCs. Twelve hours after electroporation, the media were removed, and fresh primate ES cell medium with 3 μM Y-27632 (Selleck, S1049) was added. Two days later, cells were incubated transiently with puromycin (1 μg/ml; Meilun, MB2005) for 48 hours. Surviving colonies were picked out 1 week after puromycin selection.
Neural induction and organoid generation
Primate ESCs (including human BG02 and monkey CES1) were cultured on CF-1 mouse embryonic fibroblasts (MEFs) in medium as previously described (60). Mouse cell line C57 ESC was maintained on MEFs with the medium containing knockout Dulbecco’s modified Eagle’s medium (DMEM; Gibco, 10829-018) with 15% FBS (Gibco), 1% nonessential amino acids (NEAA) (Sigma-Aldrich, 11140050), 1% Glutamax (Sigma-Aldrich, 25030081), LIF (1000 U/ml; PeproTech), and 0.01 ß-mercaptoethanol (Invitrogen, 21985023).
For neural induction, two systems were used in the study. First, we followed the previously established protocol (43). Briefly, ESCs were digested into small clumps, transferred into a low–cell adhesion six-well plate with the reported neural induction medium including Neurobasal (Gibco, 21103-049)/DMEM/F12 (Gibco, 10565-018) = 1:1, 1× B27 (Gibco, 17504-044), 1× NEAA, 1% Glutamax, 3 mM CHIR99021 (Selleck, S2924), 5 mM SB431542 (Cellagentec, C7243), vitamin C (50 mg/ml; Merck Serono, A8960), bFGF (10 ng/ml; Millipore, GF003AF), 0.2 mM compound E (Millipore, 565790), and 0.1 mM LDN193189 (Cellagen Technology, S2618). For the second system, neural induction of ESCs was performed according to the published protocol (27).
Cortical organoids were generated as follows. Human BG02 ESCs were dissociated into single cells with TrypLE (Gibco, 12604021) and then transferred into low–cell adhesion 96-well plates at a density of 5000 cells per well, with a total of 150 μl of neural induction medium from day 0 to day 6. These cells were immediately reaggregated and formed neurospheres. On day 6, the aggregates were transferred to ultralow-adhesion six-well plates with NSC culture medium [Neurobasal medium surplus with B27, N2, and NEAA; 1% Glutamax; 3 mM CHIR99021; 5 mM SB431542; bFGF (10 ng/ml); and hLIF (1000 U/ml)]. The medium was changed every 2 to 3 days. On day 20, differentiation media were replaced with Neurobasal medium supplemented with 1× N2, 1× B27, 1× NEAA, and 1% Glutamax, and the medium was changed every 3 days.
Cell immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 30 min, washed three times with PBS, then treated with 0.2% Triton X-100 for 20 min, washed three times with PBS again, and incubated in blocking buffer 3% BSA for 30 min at room temperature. The cells were incubated with a primary antibody overnight at 4°C. The next day, the cells were washed with PBS and incubated with Alexa Fluor 488–, 594–, or 647–conjugated secondary antibodies.
mRNA extraction and qRT-PCR
For quantitative reverse transcription PCR (qRT-PCR), total RNA was isolated from different samples using TRIzol (Invitrogen, 15596026), and the cDNA was reserved with a PrimeScript first strand cDNA Synthesis kit (Takara) according to the manufacturer’s protocol and prepared for later qRT-PCR (Bio-Rad, CFX Connect Real-Time System) using SYBR Green PCR Supermixes (Bio-Rad). The primers used for semiquantitative PCR are listed as follows. The housekeeping gene β-actin was amplified as an internal control in gene expression analysis. Gene primers are as follows: β-ACTIN, F-CTTAGTTGCGTTACACCCTTTC; R-ACCTTCACCGTTCCAGTTTT; BRN1, F-ACGTCTACTCGCAGGTGGG; R-CTGCGCCCTGGCTTCACT.
ChIP sequencing
BRN2 cDNA was amplified from monkey embryonic brain and then cloned into pMXs-IRES-GFP retroviral vectors (Addgene, 72876). The primers used in amplification are as follows: BRN2 F, 5′-CGCGGATCCATGGCGACCGC-3′; BRN2 R, 5′-CCGCTCGAGTCACTGGACGG-3′. The viruses including BRN2 were produced in 293T cells and infected NESCs derived from BG02 ESCs with the previously reported protocol (61).
ChIP-seq for NESCs and human developing cortex at gestational week 9 from aborted embryos was performed as described previously (62). Briefly, approximately 100,000 cells were collected and processed for library preparation by using TruePrep Index Kit V2 for Illumina (Vazyme, TD202) according to the manufacturer’s protocol.
ChIP-seq analysis
Galaxy web server was used to analyze our ChIP-seq data (62). Both BRN2 ChIP-seq and IgG control data were aligned to the built-in human hg38 genome using bowtie2 with default settings. After mapping, BRN2 binding peaks were called by MACS2 callpeak software. These peaks were annotated by PAVIS online server (63).
Statistical analysis
Quantification data were expressed as means ± SD using GraphPad Prism. Three independent experiments were used for statistics, and the details were performed as stated in the legends and Materials and Methods. The significant difference between two samples was evaluated by a t test. P < 0.05 was considered as the significantly statistical difference.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (2018YFA0108500, 2018YFA0107900, 2020YFA0112700, and 2018YFA0801403), the National Natural Science Foundation of China (31760272, 31760268, and 81870307), Major Basic Research Project of Science and Technology of Yunnan (2018FA020, 202001BC070001, and 2019FY002), Chinese Postdoctoral Science Foundation (181155), Yunnan Provincial Innovation Team (2018HC003), and The Education Revitalization Project of Anhui Province: Stem Cell and Translational Medicine (Y05201374).
Author contributions: T.L., Y.N., and W.J. initiated the project. T.L. designed the experiments, organized and supervised the entire project, and wrote the manuscript. T.L. supervised X.Z. and Y.G. to perform data analysis. X.Z., D.L., J.Y., X.D., J.L., Z.A., Q.Z., and S.Z. performed immunostaining, ESC culture and differentiation, monkey fetal analysis, and data collection. Y.N., C.C., C.S., and Y.K. made the knockout monkeys, monkey genotyping analysis, and 10X RNA library construction. Y.G., K.D., and Y.Y. analyzed RNA-seq data.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCE AND NOTES
- 1.Ataman B., Boulting G. L., Harmin D. A., Yang M. G., Baker-Salisbury M., Yap E. L., Malik A. N., Mei K., Rubin A. A., Spiegel I., Durresi E., Sharma N., Hu L. S., Pletikos M., Griffith E. C., Partlow J. N., Stevens C. R., Adli M., Chahrour M., Sestan N., Walsh C. A., Berezovskii V. K., Livingstone M. S., Greenberg M. E., Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 539, 242–247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Namba T., Huttner W. B., Neural progenitor cells and their role in the development and evolutionary expansion of the neocortex. Wiley Interdiscip. Rev. Dev. Biol. 6, e256 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Dehay C., Kennedy H., Kosik K. S., The outer subventricular zone and primate-specific cortical complexification. Neuron 85, 683–694 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Florio M., Huttner W. B., Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Lein E. S., Belgard T. G., Hawrylycz M., Molnar Z., Transcriptomic perspectives on neocortical structure, development, evolution, and disease. Annu. Rev. Neurosci. 40, 629–652 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Hansen D. V., Lui J. H., Parker P. R., Kriegstein A. R., Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Cardenas A., Borrell V., Molecular and cellular evolution of corticogenesis in amniotes. Cell. Mol. Life Sci. 77, 1435–1460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Florio M., Heide M., Pinson A., Brandl H., Albert M., Winkler S., Wimberger P., Huttner W. B., Hiller M., Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife 7, e32332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heide M., Long K. R., Huttner W. B., Novel gene function and regulation in neocortex expansion. Curr. Opin. Cell Biol. 49, 22–30 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Fietz S. A., Lachmann R., Brandl H., Kircher M., Samusik N., Schroder R., Lakshmanaperumal N., Henry I., Vogt J., Riehn A., Distler W., Nitsch R., Enard W., Paabo S., Huttner W. B., Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. U.S.A. 109, 11836–11841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson M. B., Sun X., Kodani A., Borges-Monroy R., Girskis K. M., Ryu S. C., Wang P. P., Patel K., Gonzalez D. M., Woo Y. M., Yan Z., Liang B., Smith R. S., Chatterjee M., Coman D., Papademetris X., Staib L. H., Hyder F., Mandeville J. B., Grant P. E., Im K., Kwak H., Engelhardt J. F., Walsh C. A., Bae B. I., Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature 556, 370–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lui J. H., Nowakowski T. J., Pollen A. A., Javaherian A., Kriegstein A. R., Oldham M. C., Radial glia require PDGFD–PDGFRβ signalling in human but not mouse neocortex. Nature 515, 264–268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rani N., Nowakowski T. J., Zhou H., Godshalk S. E., Lisi V., Kriegstein A. R., Kosik K. S., A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowakowski T. J., Pollen A. A., Sandoval-Espinosa C., Kriegstein A. R., Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 91, 1219–1227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Liu W., Yang L., Wu Q., Zhang H., Fang A., Li L., Xu X., Sun L., Zhang J., Tang F., Wang X., The primate-specific gene TMEM14B marks outer radial glia cells and promotes cortical expansion and folding. Cell Stem Cell 21, 635–649 e638 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Ju X. C., Hou Q. Q., Sheng A. L., Wu K. Y., Zhou Y., Jin Y., Wen T., Yang Z., Wang X., Luo Z. G., The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. eLife 5, e18197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki I. K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Cheron J., Polleux F., Detours V., Vanderhaeghen P., Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiddes I. T., Lodewijk G. A., Mooring M., Bosworth C. M., Ewing A. D., Mantalas G. L., Novak A. M., van den Bout A., Bishara A., Rosenkrantz J. L., Lorig-Roach R., Field A. R., Haeussler M., Russo L., Bhaduri A., Nowakowski T. J., Pollen A. A., Dougherty M. L., Nuttle X., Addor M.-C., Zwolinski S., Katzman S., Kriegstein A., Eichler E. E., Salama S. R., Jacobs F. M. J., Haussler D., Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florio M., Namba T., Paabo S., Hiller M., Huttner W. B., A single splice site mutation in human-specific ARHGAP11B causes basal progenitor amplification. Sci. Adv. 2, e1601941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Florio M., Albert M., Taverna E., Namba T., Brandl H., Lewitus E., Haffner C., Sykes A., Wong F. K., Peters J., Guhr E., Klemroth S., Prüfer K., Kelso J., Naumann R., Nüsslein I., Dahl A., Lachmann R., Pääbo S., Huttner W. B., Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Dominguez M. H., Ayoub A. E., Rakic P., POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb. Cortex 23, 2632–2643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugitani Y., Nakai S., Minowa O., Nishi M., Jishage K. I., Kawano H., Mori K., Ogawa M., Noda T., Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16, 1760–1765 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagino-Yamagishi K., Saijoh Y., Ikeda M., Ichikawa M., Minamikawa-Tachino R., Hamada H., Predominant expression of Brn-2 in the postmitotic neurons of the developing mouse neocortex. Brain Res. 752, 261–268 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Huang C. T., Chen J., Pankratz M. T., Xi J., Li J., Yang Y., LaVaute T. M., Li X. J., Ayala M., Bondarenko G. I., du Z. W., Jin Y., Golos T. G., Zhang S. C., Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smart I. H., Dehay C., Giroud P., Berland M., Kennedy H., Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex 12, 37–53 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X., Nardelli J., Cellular and molecular introduction to brain development. Neurobiol. Dis. 92, 3–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S. C., Wernig M., Duncan I. D., Brustle O., Thomson J. A., In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 19, 1129–1133 (2001). [DOI] [PubMed] [Google Scholar]
- 28.McEvilly R. J., de Diaz M. O., Schonemann M. D., Hooshmand F., Rosenfeld M. G., Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295, 1528–1532 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Nakai S., Kawano H., Yudate T., Nishi M., Kuno J., Nagata A., Jishage K., Hamada H., Fujii H., Kawamura K., The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 9, 3109–3121 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W., Xiang A. P., Zhou J., Guo X., Bi Y., Si C., Hu B., Dong G., Wang H., Zhou Z., Li T., Tan T., Pu X., Wang F., Ji S., Zhou Q., Huang X., Ji W., Sha J., Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Lodato M. A., Ng C. W., Wamstad J. A., Cheng A. W., Thai K. K., Fraenkel E., Jaenisch R., Boyer L. A., SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLOS Genet. 9, e1003288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boldog E., Bakken T. E., Hodge R. D., Novotny M., Aevermann B. D., Baka J., Bordé S., Close J. L., Diez-Fuertes F., Ding S. L., Faragó N., Kocsis Á. K., Kovács B., Maltzer Z., McCorrison J. M., Miller J. A., Molnár G., Oláh G., Ozsvár A., Rózsa M., Shehata S. I., Smith K. A., Sunkin S. M., Tran D. N., Venepally P., Wall A., Puskás L. G., Barzó P., Steemers F. J., Schork N. J., Scheuermann R. H., Lasken R. S., Lein E. S., Tamás G., Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 21, 1185–1195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodge R. D., Bakken T. E., Miller J. A., Smith K. A., Barkan E. R., Graybuck L. T., Close J. L., Long B., Johansen N., Penn O., Yao Z., Eggermont J., Höllt T., Levi B. P., Shehata S. I., Aevermann B., Beller A., Bertagnolli D., Brouner K., Casper T., Cobbs C., Dalley R., Dee N., Ding S. L., Ellenbogen R. G., Fong O., Garren E., Goldy J., Gwinn R. P., Hirschstein D., Keene C. D., Keshk M., Ko A. L., Lathia K., Mahfouz A., Maltzer Z., McGraw M., Nguyen T. N., Nyhus J., Ojemann J. G., Oldre A., Parry S., Reynolds S., Rimorin C., Shapovalova N. V., Somasundaram S., Szafer A., Thomsen E. R., Tieu M., Quon G., Scheuermann R. H., Yuste R., Sunkin S. M., Lelieveldt B., Feng D., Ng L., Bernard A., Hawrylycz M., Phillips J. W., Tasic B., Zeng H., Jones A. R., Koch C., Lein E. S., Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaMonica B. E., Lui J. H., Hansen D. V., Kriegstein A. R., Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat. Commun. 4, 1665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollen A. A., Nowakowski T. J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C. R., Shuga J., Liu S. J., Oldham M. C., Diaz A., Lim D. A., Leyrat A. A., West J. A., Kriegstein A. R., Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaid S., Camp J. G., Hersemann L., Oegema C. E., Heninger A.-K., Winkler S., Brandl H., Sarov M., Treutlein B., Huttner W. B., Namba T., A novel population of Hopx-dependent basal radial glial cells in the developing mouse neocortex. Development 145, dev169276 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Kageyama R., Ohtsuka T., Shimojo H., Imayoshi I., Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247–1251 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Ma T., Wang C., Wang L., Zhou X., Tian M., Zhang Q., Zhang Y., Li J., Liu Z., Cai Y., Liu F., You Y., Chen C., Campbell K., Song H., Ma L., Rubenstein J. L., Yang Z., Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 16, 1588–1597 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Lim L., Mi D., Llorca A., Marin O., Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyoshi G., Elucidating the developmental trajectories of GABAergic cortical interneuron subtypes. Neurosci. Res. 138, 26–32 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Sultan K. T., Shi S.-H., Generation of diverse cortical inhibitory interneurons. Wiley Interdiscip. Rev. Dev. Biol. 7, e306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer C., Hafemeister C., Bandler R. C., Machold R., Batista Brito R., Jaglin X., Allaway K., Butler A., Fishell G., Satija R., Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X., Li B., Ai Z., Xiang Z., Zhang K., Qiu X., Chen Y., Li Y., Rizak J. D., Niu Y., Hu X., Sun Y. E., Ji W., Li T., A robust single primate neuroepithelial cell clonal expansion system for neural tube development and disease studies. Stem Cell Rep. 6, 228–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X., Ai Z., Hu X., Li T., Efficient generation of corticofugal projection neurons from human embryonic stem cells. Sci. Rep. 6, 28572 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foshay K. M., Gallicano G. I., Regulation of Sox2 by STAT3 initiates commitment to the neural precursor cell fate. Stem Cells Dev. 17, 269–278 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Tripathi S. K., Chen Z., Larjo A., Kanduri K., Nousiainen K., Äijo T., Ricaño-Ponce I., Hrdlickova B., Tuomela S., Laajala E., Salo V., Kumar V., Wijmenga C., Lähdesmäki H., Lahesmaa R., Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 19, 1888–1901 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Schonemann M. D., Ryan A. K., McEvilly R. J., O’Connell S. M., Arias C. A., Kalla K. A., Li P., Sawchenko P. E., Rosenfeld M. G., Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9, 3122–3135 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Sousa A. M. M., Meyer K. A., Santpere G., Gulden F. O., Sestan N., Evolution of the human nervous system function, structure, and development. Cell 170, 226–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nowakowski T. J., Rani N., Golkaram M., Zhou H. R., Alvarado B., Huch K., West J. A., Leyrat A., Pollen A. A., Kriegstein A. R., Petzold L. R., Kosik K. S., Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat. Neurosci. 21, 1784–1792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X., Tsai J. W., LaMonica B., Kriegstein A. R., A new subtype of progenitor cell in the mouse embryonic neocortex. Nat. Neurosci. 14, 555–561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lukaszewicz A., Savatier P., Cortay V., Giroud P., Huissoud C., Berland M., Kennedy H., Dehay C., G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47, 353–364 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niu Y., Yu Y., Bernat A., Yang S., He X., Guo X., Chen D., Chen Y., Ji S., Si W., Lv Y., Tan T., Wei Q., Wang H., Shi L., Guan J., Zhu X., Afanassieff M., Savatier P., Zhang K., Zhou Q., Ji W., Transgenic rhesus monkeys produced by gene transfer into early-cleavage–stage embryos using a simian immunodeficiency virus-based vector. Proc. Natl. Acad. Sci. U.S.A. 107, 17663–17667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae S., Park J., Kim J.-S., Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Y., Cradick T. J., Brown M. T., Deshmukh H., Ranjan P., Sarode N., Wile B. M., Vertino P. M., Stewart F. J., Bao G., CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 42, 7473–7485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng T., Hou Y., Zhang P., Zhang Z., Xu Y., Zhang L., Niu L., Yang Y., Liang D., Yi F., Peng W., Feng W., Yang Y., Chen J., Zhu Y. Y., Zhang L. H., du Q., Profiling single-guide RNA specificity reveals a mismatch sensitive core sequence. Sci. Rep. 7, 40638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong S., Zhang S., Fan X., Wu Q., Yan L., Dong J., Zhang H., Li L., Sun L., Pan N., Xu X., Tang F., Zhang J., Qiao J., Wang X., A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGinnis C. S., Murrow L. M., Gartner Z. J., Gartner, DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Soysa T. Y., Ranade S. S., Okawa S., Ravichandran S., Huang Y., Salunga H. T., Schricker A., del Sol A., Gifford C. A., Srivastava D., Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 572, 120–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang B., Xiang Z., Ai Z., Wang H., Li Y., Ji W., Li T., Generation of cardiac spheres from primate pluripotent stem cells in a small molecule-based 3D system. Biomaterials 65, 103–114 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Zhu X., Zhou W., Jin H., Li T., Brn2 alone is sufficient to convert astrocytes into neural progenitors and neurons. Stem Cells Dev. 27, 736–744 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Kaya-Okur H. S., Wu S. J., Codomo C. A., Pledger E. S., Bryson T. D., Henikoff J. G., Ahmad K., Henikoff S., CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang W., Loganantharaj R., Schroeder B., Fargo D., Li L. J. B., Pavis: A tool for peak annotation and visualization. Bioinformatics 29, 3097–3099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S5