Abstract
Purpose:
PARP inhibitors are approved for use in breast, ovarian, prostate and pancreatic cancer, which are the solid tumor types that most frequently have alterations in key homologous recombination (HR) genes, such as BRCA1/2. However, the frequency of HR deficiency in other solid tumor types, including bladder cancer, is less well characterized.
Experimental Design:
Specific DNA aberration profiles (mutational signatures) are induced by homologous recombination deficiency (HRD) and the presence of these “genomic scars” can be used to assess the presence or absence of HR deficiency in a given tumor biopsy even in the absence of an observed alteration of an HR gene. Using whole exome and whole genome data, we measured various HR deficiency-associated mutational signatures in bladder cancer.
Results:
We found that a subset of bladder tumors have evidence of HR deficiency. In addition to a small number of tumors with bi-allelic BRCA1/2 events, approximately 10% of bladder tumors had significant evidence of HR deficiency associated mutational signatures. Increased levels of HRD signatures were associated with promoter methylation of RBBP8 which encodes CtIP, a key protein involved in HR.
Conclusion:
A subset of bladder tumors have genomic features suggestive of HR deficiency and therefore may be more likely to benefit from therapies such as platinum agents and PARP inhibitors that target tumor HR deficiency.
Keywords: DNA repair, homologous recombination repair, homologous recombination deficiency, RBBP8/CtIP promoter methylation, cancer genomics, bladder cancer
Introduction
Cisplatin-based chemotherapy is a first-line treatment for muscle-invasive and metastatic urothelial cancer (UC). However, responses to platinum-based chemotherapy vary widely among patients, and de novo or acquired cisplatin resistance is common. Despite recent approvals of several anti-PD-1/PD-L1 and targeted agents, the median survival for cisplatin-resistant metastatic UC remains less than one year (1).
Platinum agents are highly mutagenic and create both intrastrand and interstrand DNA lesions. Intrastrand lesions are typically repaired by the nucleotide excision repair (NER) pathway, and urothelial tumor NER deficiency caused by somatic mutations in the NER gene, ERCC2, drives cisplatin sensitivity. In patients with muscle-invasive bladder cancer (MIBC) who received neoadjuvant cisplatin-based chemotherapy, ERCC2 mutations were associated with significantly higher rates of pathologic complete response and improved overall survival (2,3). However, less than 25% of bladder cancer cases are NER deficient as defined by an ERCC2 mutation (4) and/or by the presence of an ERCC2-associated mutational signature (5). Since approximately one-third of patients achieve a complete pathologic response to neoadjuvant cisplatin-based chemotherapy and approximately half of patients with metastatic disease have an initial response to platinum-based chemotherapy, other tumor features beyond NER deficiency may also drive cisplatin sensitivity.
The homologous recombination (HR) repair pathway repairs DNA double-strand breaks created by lesions such as platinum-induced interstrand crosslinks. Alterations in HR genes are associated with increased platinum sensitivity in multiple tumor types including breast and ovarian cancer (6,7). HR gene alterations are also present in a subset of urothelial tumors, including BRCA1 (1–2%) and BRCA2 (2–3%) (4,8). In several studies, mutation(s) in one or more DNA repair genes (including several HR genes) is associated with increased sensitivity to platinum-based chemotherapy in the neoadjuvant or metastatic UC settings, suggesting that HR deficiency may drive platinum sensitivity in a subset of urothelial tumors (8–11).
In addition to driving sensitivity to platinum-based agents, HR deficiency can also sensitize tumors to small molecule inhibitors of poly (ADP ribose)-polymerase (PARP), and PARP inhibitors are now FDA approved for HR-deficient breast, ovarian, prostate, and pancreatic tumors. Several PARP inhibitors trials in metastatic UC are underway. A preliminary report from the ATLAS trial of rucaparib in metastatic UC patients who had progressed on cisplatin and/or anti-PD-1/PD-L1 therapy showed no confirmed responses and a 28% stable disease rate among 97 evaluable patients (12). Targeted sequencing revealed mutations in HR genes (BRCA1/2, PALB2, RAD51) in 6 patients; however, the functional relevance of these mutations and whether they were accompanied by loss of heterozygosity (LOH) of the wild-type (WT) allele were not known. These data suggest that additional methods may be required to accurately identify HR-deficient urothelial tumors.
HR deficiency can be present in a tumor even in the absence of canonical HR gene mutations, and such cases can often be detected by the presence of HR deficiency-associated mutational patterns (signatures). HR deficiency induces multiple types of genetic alterations ranging from single nucleotide variations to large scale genomic (13) rearrangements. The presence and frequency of these events can be used to calculate clinically applicable composite mutational signatures, such as the HRD score (14) and HRDetect score (15). High levels of these HR deficiency indicators are associated with better response to PARP inhibitor- or platinum-based therapy in ovarian and breast cancer (16,17).
Here, we apply several validated genomic signatures of HR deficiency to characterize the landscape of HR deficiency in bladder cancer. We find that a small but significant fraction of bladder tumors have elevated levels of HR deficiency signatures and that the frequency of HR deficiency in urothelial cancer may be higher than predicted based on BRCA1/2 mutational status alone. We also propose that some of the cases with elevated HR deficiency associated signatures are caused by the suppression of RBBP8, which encodes CtIP, an exonuclease intimately involved in HR.
Materials and Methods
Patients and cohorts
In this study 533 whole genome (WGS) and whole exome sequenced (WES) pretreatment samples were analyzed from three urothelial bladder tumor cohorts (Supp. Table 1).
1. TCGA cohort
The WGS normal and tumor bam files were downloaded from the ICGC data portal (https://dcc.icgc.org/). The WES normal and tumor bam files, as well as the vcf files generated by MuTect2 were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/).
2. DFCI/MSKCC and Philadelphia cohorts
The normal and tumor bam files were downloaded from the database of Genotypes and Phenotypes (dbGaP) upon request (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000771.v2.p1) using the phs000771.v2.p1 accession code. The average coverage of the analyzed WGS and WES samples in each cohort is shown in Supp. Figures 1, 2.
Mutation, copy number, and structural variant calling
Germline variants were called with HaplotypeCaller in key DNA damage response (DDR) genes, while somatic point mutations and indels were called with MuTect2 (GATK, v3.8). The high fidelity of the reported variants was ensured by the application of additional hard filters in addition to the tools’ default filters (Supp. Tables 2, 3). Allele-specific copy number profiles were estimated using Sequenza (18) and FACETS (19) (Supp. Figures 3, 4). Germline and somatic mutations were annotated using InterVar (20) and the genotype of the samples was determined (Supp. Figures 5-7). The identified BRCA1/2-deficient samples and other HR-deficient samples are listed in Supp. Tables 4, 5. The frequency of pathogenic mutations in DNA damage checkpoint genes are shown in Supp. Table 6. Structural variants were detected by BRASS (v6.0.0. https://github.com/cancerit/BRASS). Further details are available in the Supplementary Methods.
Mutational signatures
Somatic single base substitution signatures were determined with the help of the deconstructSigs R package (21), using the COSMIC signatures as a mutational process matrix (https://cancer.sanger.ac.uk/signatures/). Doublet base substitutions and indels in each sample were classified into a 78-dimensional doublet base substitution and an 83-dimensional indel catalog, respectively, with the help of the ICAMS R package (22), and the previously described matrices of doublet base substitution and indel signatures were used (23) in a non-negative least-squares problem to estimate the matrix of exposures to mutational processes. In addition, deletions were classified into three groups: deletions with complete repetitions, microhomology-mediated deletions, and unique deletions; and the deletion profile of the samples was determined. The extraction of rearrangement signatures was executed as described previously (24). The extracted single base substitution signatures, doublet base substitution signatures, indel signatures, rearrangement signatures and deletion profiles are shown in Supp. Figures 8-15. Further details are provided in the Supplementary Methods.
HRD score and HRDetect score calculation
Homologous recombination deficiency (HRD)-associated genomic scar scores were calculated from the copy number profile of the samples using the scarHRD R package (25) (Supp. Figures 16, 17). HRDetect scores were calculated using the extracted signatures and deletion profiles as described in the Supplementary Methods.
DNA methylation and RNA expression analysis
DNA methylation data measured by the Illumina HumanMethylation450 platform and RNA expression data were downloaded from the Xena platform (https://xenabrowser.net/) (26). Adjacent CpG loci were divided into clusters with a default maximum gap of 500 bp and a maximum cluster gap of 1500 bp using the minfi R package (27). The mean β value of the loci within each cluster was calculated and used as a single methylation estimate per cluster. In the TCGA BLCA cohort, methylation clusters of RBBP8 showed high correlation (Supp. Fig. 18), thus the mean methylation of the clusters was used to determine the methylation status of the RBBP8 gene in each sample. The Fragments Per Kilobase of transcript per Million mapped reads (FPKM) normalization method was used and the data were then log-transformed. Additional details are described in the Supplementary Methods.
Survival analysis
Survival analysis is described in the Supplementary Methods and survival curves are shown in Supp. Figures 19-22.
Cell lines
The CCLE mutation and copy number (CN) calls (28) and ABSOLUTE CN analysis results (29) were downloaded from the DepMap data portal (https://depmap.org/portal/) and the scarHRD R package (25) was used to calculate the HRD scores of twenty-three BLCA cell lines (Supp. Fig. 23). The PharmacoGx R package (30) was used to download and analyze cell lines sensitivity data from the Cancer Therapeutics Response Portal version 2 (CTRPv2) (31) (Supp. Fig. 24). The CCLE RNAseq normalized and log-transformed expression data (29) were downloaded from the DepMap data portal (https://depmap.org/portal/) and is shown in Supp. Fig. 25A. DNA methylation raw data measured by the Illumina HumanMethylation450 platform (32) were downloaded from the GEO database using the GSE68379 accession number. The minfi R package (27) was used for the analysis as described above. The correlation between RBBP8 mRNA expression and RBBP8 promoter-associated methylation is shown in Supp. Fig 25B.
Code availability
There are no restrictions to access the custom code used for the analyses presented in this study. Information is available from the authors on request.
Results
Frequency of homologous recombination and DNA damage checkpoint gene mutations in MIBC whole exome sequencing cohorts
Whole genome sequencing (WGS) data contain more numerous and a wider variety of HR deficiency induced mutational events than WES data (33). While point mutations and short insertions/deletions (indels) can be identified from both WES and WGS data, only WGS data allow the detection of large-scale rearrangements. In addition, WGS data contain about 100-fold more mutational events than WES data (33). However, only 23 cases in the TCGA bladder cancer cohort had WGS data available, whereas three WES cohorts totaling 493 cases with high quality data were available. These included the TCGA cohort (4) (n=395) and two cohorts that contained information about response to platinum treatment (DFCI/MSKCC (2), n=50, and Philadelphia (3), n=48) (Supp. Table 1). Therefore, we began our analysis using the WES cohorts.
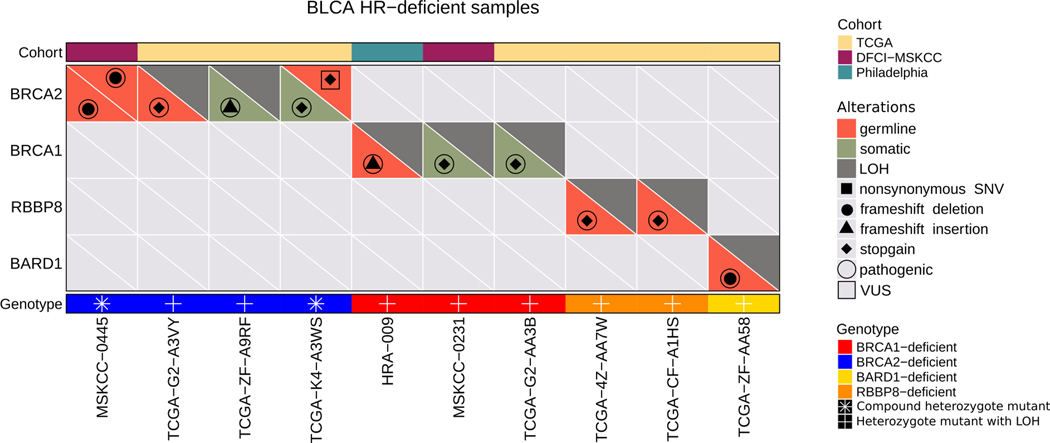
Mutations in the canonical HR genes BRCA1 and BRCA2 are present in 3–5% of urothelial tumors (34); however, only a fraction of these cases have both a predicted loss-of-function mutation as well as loss of heterozygosity (LOH) of the wild-type (WT) allele (Supp. Figures 5-7). Since LOH of the WT allele is typically required to confer HR deficiency (35) in the setting of a BRCA1/2 mutation, we considered only those BRCA1/2 mutant cases that had biallelic mutations or a mutation accompanied by LOH to be HR deficient. In the TCGA WES data set (n=395 cases), we identified two cases with a pathogenic BRCA2 mutation accompanied by LOH, one case with biallelic BRCA2 pathogenic mutations, and one case in which a pathogenic BRCA1 mutation was accompanied by LOH (Figure 1, Supp. Table 4). We similarly identified two of 50 cases in the DFCI/MSKCC cohort and one of 48 cases in the Philadelphia cohort with either a pathogenic BRCA1/2 mutation accompanied by LOH or biallelic BRCA1/2 pathogenic mutations (Figure 1, Supp. Table 4).
Figure 1:
Landscape of germline and somatic mutations and copy number alterations of HR genes across the three bladder cancer WES cohorts. We identified 4 BRCA2-deficient samples, 3 BRCA1-deficient samples, 2 RBBP8-deficient samples, and 1 BARD1-deficient sample in the three cohorts.
Loss of function of PALB2, RAD51C/D and other HR genes can also induce HR deficiency associated mutational signatures (36,37). Across the three cohorts (493 cases), we identified three such cases (one BARD1 and two RBBP8 mutations in the TCGA cohort) with a pathogenic mutation accompanied by LOH (Figure 1, Supp. Table 5). We also determined mutation and LOH status for DNA damage checkpoint genes such as ATM, ATR, CHK2, TP53, and RB1 (Supp. Figures 5-7, Supp. Table 6), although loss of function of these genes is usually not associated with an HR deficiency mutational signature (13). TP53 and RB1 were the most frequently altered DNA damage checkpoint genes in the cohorts (Supp. Table 6).
Genomic scar-based HRD scores in bladder cancer
The first scoring systems to quantify the degree of HRD were based on data obtained from hybridization microarrays such as single nucleotide polymorphism (SNP) arrays. Three such scoring systems have been widely used: (1) The HRD-LOH score is calculated by tallying the number of LOH regions exceeding 15 Mb in size but less than the whole chromosome (38); (2) the Large-scale State Transitions (LST) score (39) is defined as the number of chromosomal breaks between adjacent regions of at least 10 Mb, with a distance between them not larger than 3Mb; and (3) the number of Telomeric Allelic Imbalances (TAI) (40) is the number of AIs (unequal contribution of parental allele sequences) that extend to the telomeric end of a chromosome. These measures were later adapted to next generation sequencing and the sum of these scores is often referred to as the HRD score (14), which was recently approved by the FDA as a companion diagnostic for prioritizing patients with ovarian cancer for PARP inhibitor therapy (41).
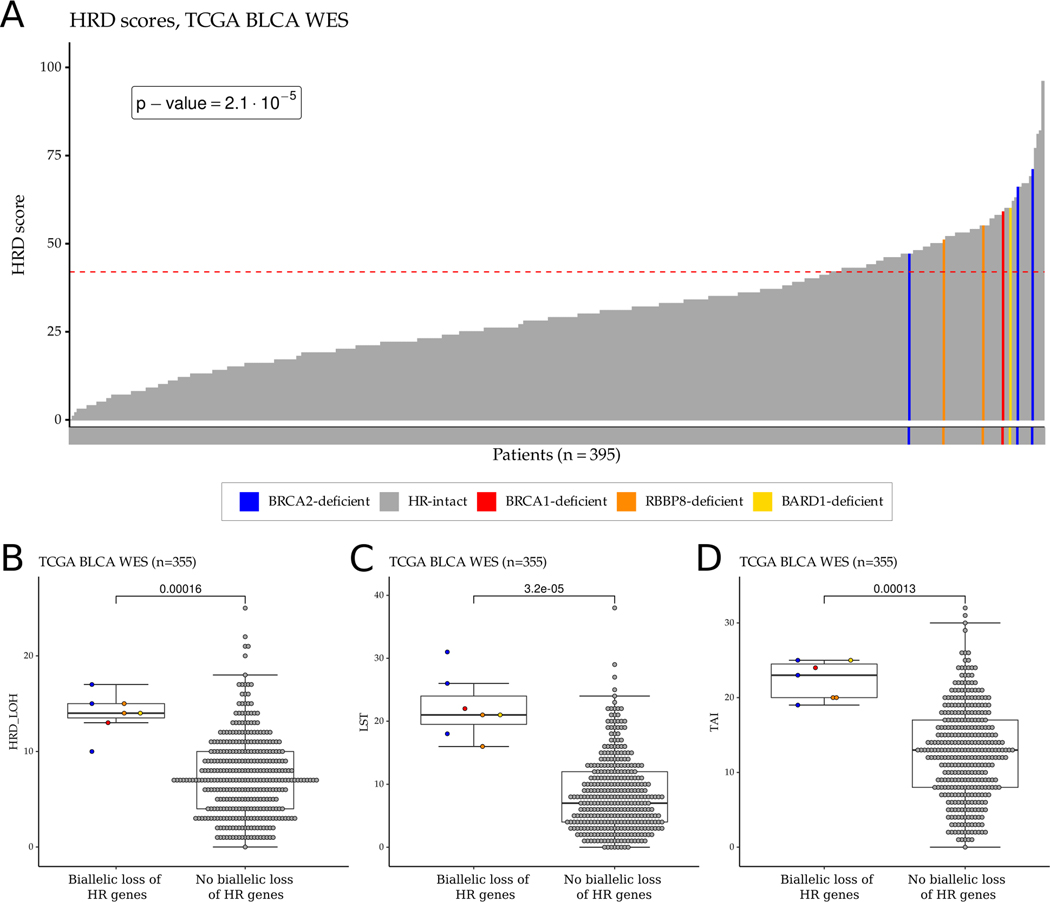
In the TCGA WES bladder cancer cohort, all four BRCA1/2 deficient as well as the one BARD1 and two RBBP8 deficient cases had an HRD score ≥42 (Figure 2, p-value = 2.1×10−5, Fisher exact test), which is the threshold for HR deficiency previously defined for ovarian cancer (14). However, there were an additional 80 of 388 cases without a BRCA1/2 or other HR gene mutation that also had an HRD score ≥42 (Figure 2A). Interestingly, four out of five tumors with a BRCA1/2 variant of uncertain significance (VUS) with accompanying LOH (Supp. Table 4) were also associated with high HRD score, suggesting that these VUS cases may also be pathogenic (Supp. Fig. 26).
Figure 2:
HRD score distribution and HRD score components in the TCGA bladder cancer WES cohort. A. HRD score distribution of the samples in the TCGA bladder cancer WES cohort. The dashed red line represents the cut-off value (≥42) for HR deficiency previously defined for ovarian cancer. B-D. Boxplots showing the distribution of the number of HR deficiency-associated loss of heterozygosity (HRD-LOH) (B), large-scale state transition (LST) (C), and telomeric allelic imbalance (TAI) (D) events in the TCGA bladder cancer WES cohort. For each boxplot, the dark horizontal line within each box represents the median, the edges of the box represent the lower and upper bounds of the interquartile range (IQR), the upper whisker is the min(max(x), Q3+1.5×IQR) and the lower whisker is the max(min(x), Q1–1.5×IQR). ERCC2 mutant samples or samples with high ERCC2mut score were excluded from the boxplots. P-values were calculated by the Wilcoxon rank-sum test and no mathematical correction was applied for multiple comparisons.
HRD scores derived from WES data can be impacted by the relatively low number of SNVs (33); therefore, we also calculated HRD scores from WGS data for the 23 cases that had both WGS and WES data available. Similar to other solid tumors, the WES and WGS derived HRD scores showed a strong correlation (0.93; p = 2.4×10−10, Supp. Fig. 27).
Patterns of HR deficiency associated mutational signatures and HRDetect scores in bladder cancer
Next generation sequencing allows the detection of mutations on a broader scale than is possible with hybridization microarrays, and mutations ranging from single base substitutions to megabase (Mb) deletions or large-scale rearrangements can be detected (Supp. Figures 8-15). The first catalogue of mutational signatures (COSMIC signatures v2) contained mainly single nucleotide substitution signatures (42). Recently, a more comprehensive list of mutational signatures was published that includes single nucleotide substitution, doublet base substitution and short insertion/deletion (indel) signatures (COSMIC signatures v3) (23).
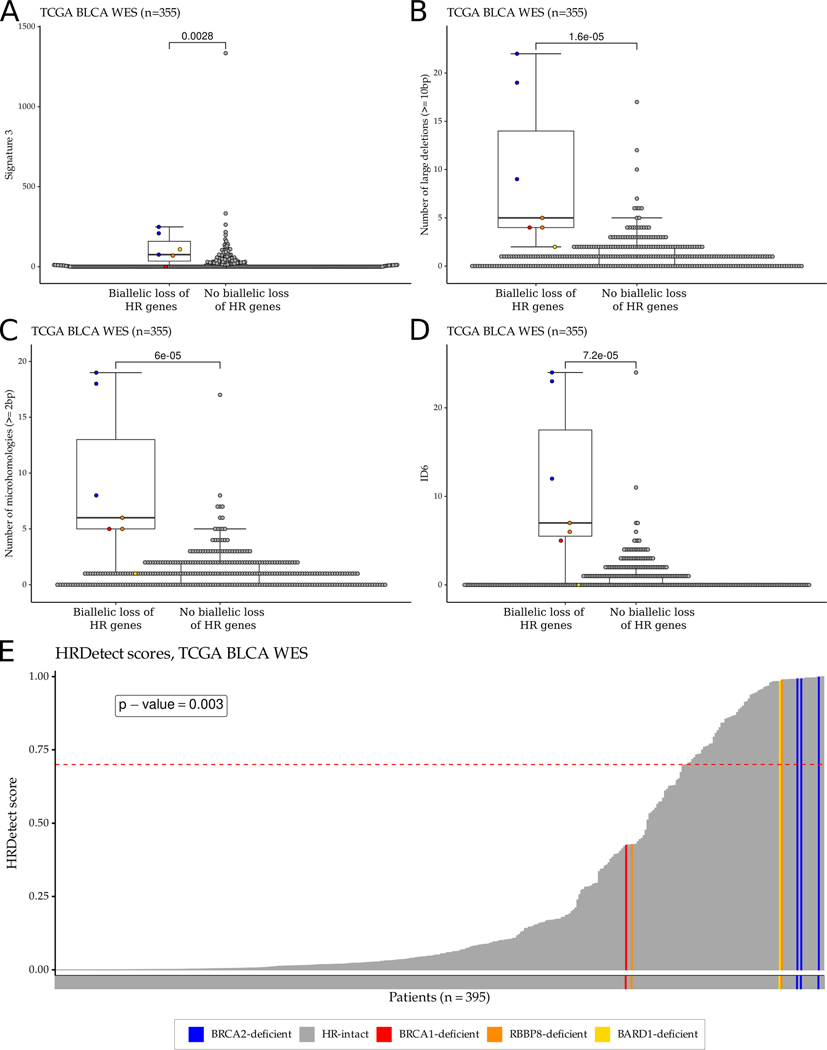
Loss of function of BRCA1 or BRCA2 is associated with distinct mutational patterns (signatures) that can be detected by WES analysis. Signature 3 (also known as the “BRCAness” signature or SBS3 in COSMIC signatures v3) (23,42) and an increased number of deletions with flanking microhomology (Signature ID6 in COSMIC signatures v3) are common features of defective homologous recombination (13). Consistent with this, we found that signature 3 (both COSMIC v2 and v3) was higher in the four BRCA1/2 deficient and the BARD1 and RBBP8 deficient TCGA bladder cancer cases than in the BRCA1/2 intact tumors (p=0.0028) (Figure 3A and Supp. Fig. 28). In addition, three different short indel based signatures often associated with loss of function of BRCA1/2 were also higher in the BRCA1/2 deficient and the BARD1 and RBBP8 deficient TCGA cases than in BRCA1/2 intact cases. These include: 1) the number of ≥10bp deletions (Figure 3B), 2) the number of deletions with flanking microhomology (Figure 3C), and 3) the ID6 signature from COSMIC signatures v3 (Figure 3D). Similar observations were made in the DFCI/MSKCC and Philadelphia cohorts, although the sample sizes were too small to reach statistical significance (Supp. Figures 29, 30). These results suggest that loss of function mutations in BRCA1 or BRCA2 are associated with the same mutational signatures in bladder cancer as are observed in ovarian, breast and prostate cancer (15,33).
Figure 3:
HR deficiency-associated mutational signatures and HRDetect score distribution in the TCGA bladder cancer WES cohort. A-D. Boxplots showing the distributions of HR deficiency-associated mutational signatures: signature 3 (COSMIC v2) (A), the number of large (≥10bp) deletions (B), the number of microhomology-mediated deletions (C), and ID6 (COSMIC v3) (D). For each boxplot, the dark horizontal line within each box represents the median, the edges of the box represent the lower and upper bounds of the IQR, the upper whisker is equal to min(max(x), Q3+1.5×IQR) and the lower whisker is equal to max(min(x), Q1–1.5×IQR). ERCC2 mutant samples or samples with high ERCC2mut score were excluded from the boxplots. P-values were calculated by the Wilcoxon rank-sum test and no mathematical correction was applied for multiple comparisons. E. HRDetect score distribution of the samples in the TCGA bladder cancer WES cohort. The dashed red line represents the threshold (≥0.7) for HR deficiency previously defined for breast cancer.
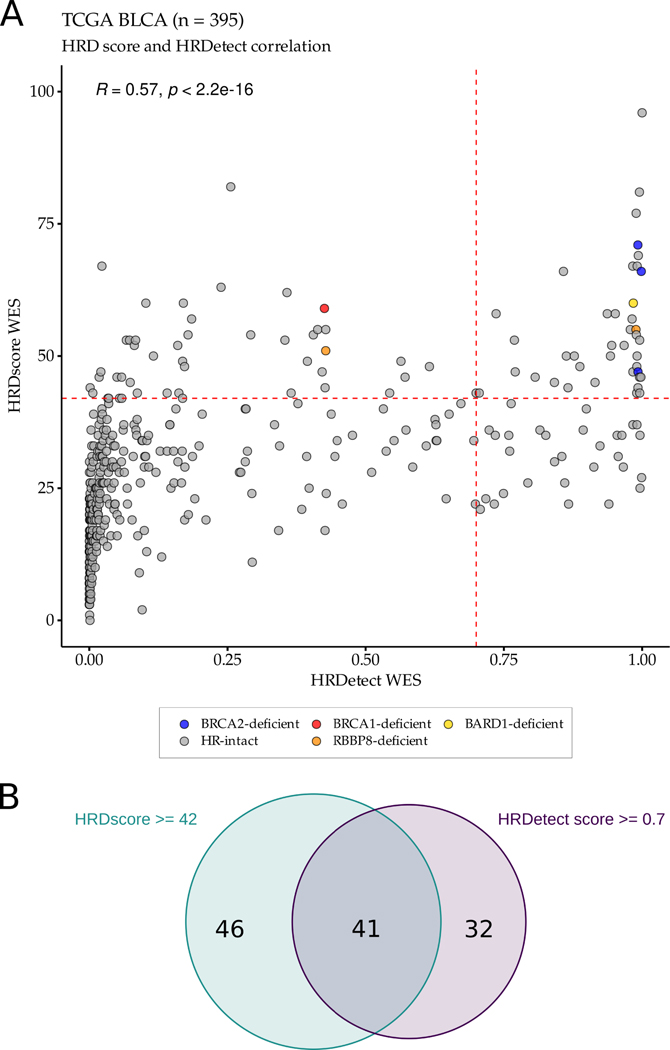
HRDetect is a composite mutational signature that combines features of the HRD score with HR deficiency-induced point mutation and short indel profiles (15). We calculated HRDetect scores using the bladder TCGA WES data and found that all three BRCA2 deficient cases, but not the BRCA1 deficient case, had an HRDetect score ≥0.7 (Figure 3E, p-value = 0.003, Fisher exact test), the threshold commonly used to detect HR deficiency in breast cancer data (15). The single BARD1 deficient case and one of the two RBBP8 deficient cases also had an HRDetect value of close to 1 (Figure 3E). When we compared the HRD scores to the HRDetect values, we observed a correlation of 0.57 (p < 2.2×10−16) (Figure 4A) and about half (41 of 87) of the cases with an HRD score ≥42 also had an HRDetect score ≥0.7 (Figure 4B).
Figure 4:
Correlation between HRD scores and HRDetect scores in the TCGA bladder cancer WES cohort. A. Correlation between HRD scores and HRDetect scores (r = 0.57, p-value < 2.2×10−16). B. Of the 395 samples, 87 samples had high HRD scores, 73 samples had high HRDetect scores, and 41 samples had high HRD and high HRDetect scores.
These results suggest that HR deficiency may occur in bladder tumors without observable LOF mutations in BRCA1 or BRCA2. The frequency, even when only those cases with both an HRD score of ≥42 and an HRDetect score of ≥0.7 are considered could be as high as 10% (41 out of 395 TCGA cases) of all bladder cancer cases.
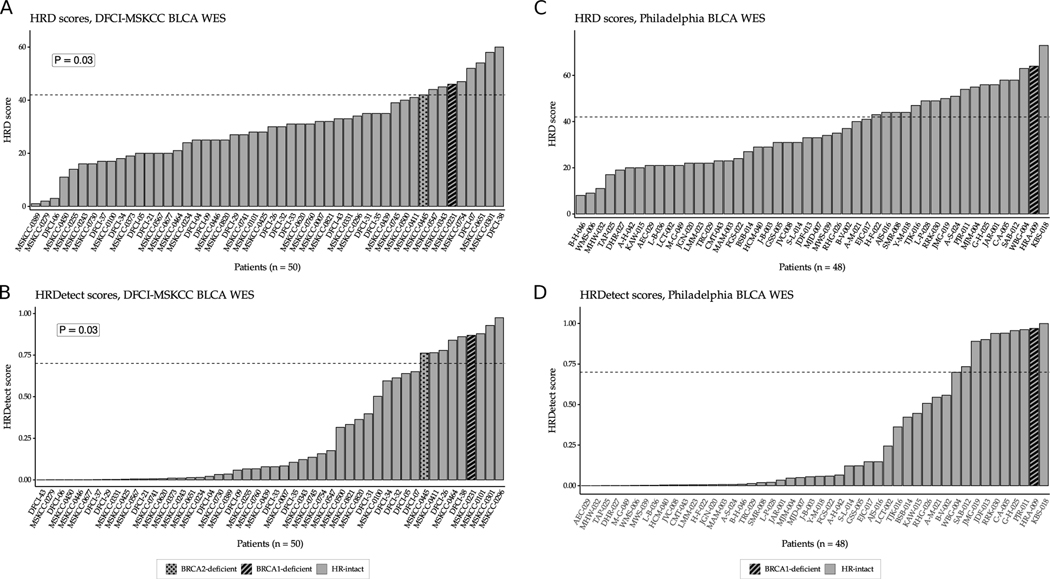
We also calculated the HRD score and HRDetect values for the DFCI/MSKCC and Philadelphia cohorts (2,3). In the DFCI/MSKCC cohort, nine of the 50 cases, including the two BRCA1/2 deficient cases, had an HRD score of 42 or higher (Figure 5A). Similarly, nine cases including the two BRCA1/2 deficient cases also had an HRDetect value ≥0.7 (Figure 5B). Four cases had both a high HRD score and high HRDetect value, including the two BRCA1/2 deficient cases (Supp. Fig. 31A).
Figure 5:
HRD score and HRDetect score distributions in the DFCI/MSKCC and Philadelphia bladder cancer WES cohorts. A. HRD score distribution of tumors in the DFCI/MSKCC bladder cancer WES cohort. The dashed red line represents the cut-off value (≥42) for HR deficiency previously defined for ovarian cancer. B. HRDetect score distribution of tumors in the DFCI/MSKCC bladder cancer WES cohort. The dashed red line represents the cut-off value (≥0.7) for HR deficiency previously defined for breast cancer. C. HRD score distribution of the samples in the Philadelphia bladder cancer WES cohort. The dashed red line represents the cut-off value (≥42) for HR deficiency previously defined for ovarian cancer. D. HRDetect score distribution of the samples in the Philadelphia bladder cancer WES cohort. The dashed red line represents the cut-off value (≥0.7) for HR deficiency previously defined for breast cancer. The BRCA1- and the BRCA2-deficient samples in both cohorts had high HRD and high HRDetect scores. P-values were calculated by the Fisher exact test.
In the Philadelphia cohort 18 of 48 cases had an HRD score ≥42, including the BRCA1 deficient case (Figure 5C). Nine cases, including the BRCA1 deficient case, had an HRDetect value ≥0.7 (Figure 5D) all of which also had an HRD score ≥42 as well (Supp. Fig. 31B).
The DFCI/MSKCC and Philadelphia cohorts are comprised of patients who received cisplatin-based chemotherapy followed by radical cystectomy. We previously showed that bladder tumors with an ERCC2 mutant specific mutational signature, with or without an actual mutation in ERCC2, have increased sensitivity to cisplatin-based therapy (5). After removing all cases with an ERCC2 mutation or an ERCC2 mutation specific signature, there were 32 cases remaining in the DFCI/MSKCC cohort, including 7 cases with an HRD score ≥42 or HRDetect score ≥0.7. Within this limited cohort we did not detect a significant correlation between elevated HRD or HRDetect scores and response to platinum treatment (3 of 7 putative HRD tumors responded [43%] versus 6 of 25 non-HRD tumors [24%]); however, both BRCA1/2-mutant cases were responders. Similarly, we did not detect a correlation between increased HRD or HRDetect scores and response to cisplatin in the subset of the Philadelphia cohort without ERCC2 mutations or ERCC2 mutation specific signature. However, the samples in this cohort were derived from FFPE tissues and the artifacts introduced during processing may have influenced the copy number analysis and the calculation of HRD score. The single BRCA1-deficient case was a responder.
Methylation driven suppression of RBBP8 is associated with increased HR deficiency associated mutational signatures in bladder tumor cases
Across the three analyzed cohorts only a minority of cases with high levels of HR deficiency associated mutational signatures harbored predicted loss of function in BRCA1/2 or other HR genes. However, suppression of HR gene function via other mechanisms such as promoter methylation - as occurs commonly for BRCA1 in breast cancer (43) - can also lead to HR deficiency. In a recent analysis of promoter methylation status and gene expression level of DNA damage genes across tumor types (44), RBBP8 promoter methylation and accompanying reduction in gene expression was present in 37% of bladder tumors but was rare in other tumor types. RBBP8 encodes the protein CtIP, which interacts directly with BRCA1 and plays a critical role in end resection at double strand DNA breaks prior to HR (45). Accordingly, suppression of RBBP8 has been linked to HR deficiency and increased PARP inhibitor sensitivity (46,47).
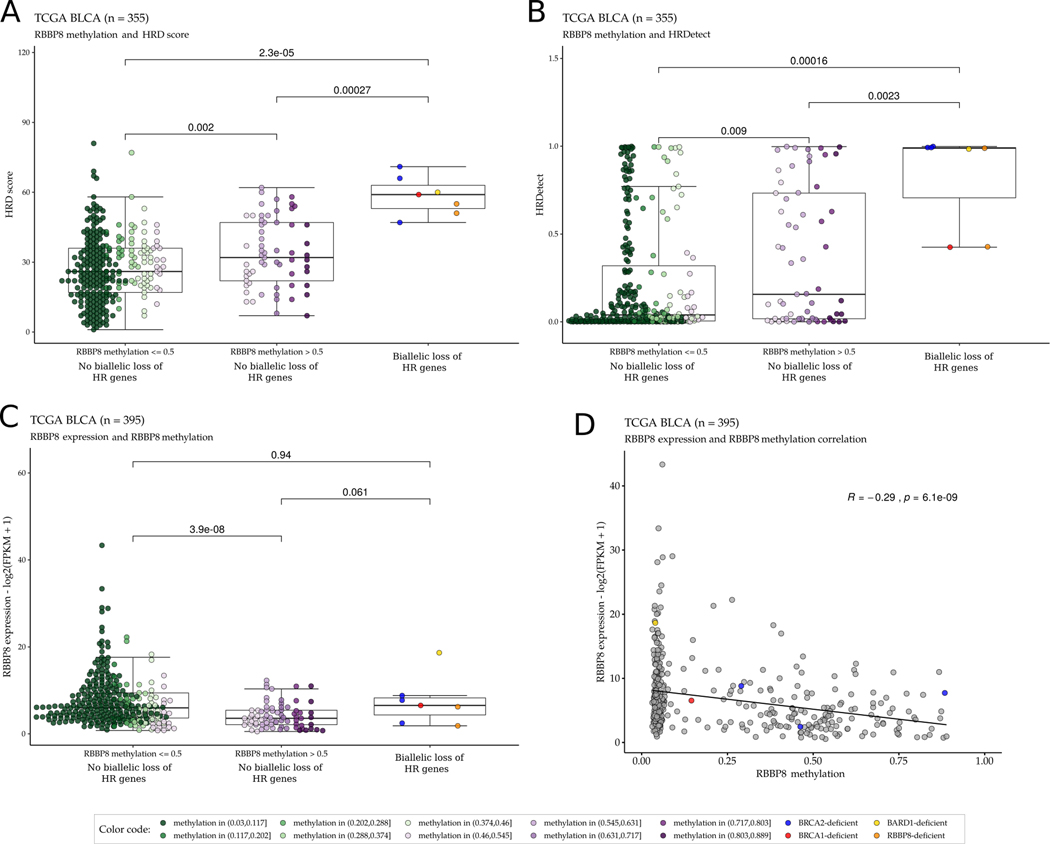
We found that promoter methylation of RBBP8 was significantly associated with higher HRD and HRDetect scores in the TCGA BLCA WES cohort (Figure 6A, B). Increased RBBP8 methylation was also strongly associated with decreased RBBP8 mRNA levels in WT BRCA1/2 cases (Figure 6C), and a significant negative correlation was observed between RBBP8 methylation and RBBP8 mRNA expression levels (Figure 6D). This defines a putative epigenetic mechanism for HR deficiency in bladder cancer that could occur in the absence of observed mutations in HR genes.
Figure 6:
Promoter methylation of RBBP8 is associated with higher HRD and HRDetect scores in the TCGA bladder cancer WES cohort. A-B: Tumors with no identified biallelic HR gene alterations and high RBBP8 methylation (β value >0.5) have significantly higher HRD scores (A) and HRDetect scores (B) than tumors with low RBBP8 methylation (β value ≤0.5) and with no biallelic HR gene alterations. C. RBBP8 expression was significantly lower in RBBP8 methylated (β value >0.5) tumors than in tumors with low RBBP8 methylation (β value ≤0.5) or tumors with biallelic HR gene alterations in the TCGA bladder cancer WES cohort. For each boxplot, the dark horizontal line is the median, the edges of the box represent the lower and upper bounds of the IQR, the upper whisker is the min(max(x), Q3+1.5×IQR) and the lower whisker is the max(min(x), Q1–1.5×IQR). P-values were calculated by the Wilcoxon rank-sum test. D. There is an inverse correlation between RBBP8 gene expression and RBBP8 DNA methylation (r = −0.29, p-value = 6.1×10−9) in the TCGA bladder cancer WES cohort. The binary logarithm of the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) normalized expression values are shown on the y axis, and the DNA methylation β values are shown on the x axis.
Increased HRD scores are associated with PARP inhibitor sensitivity and lower RBBP8 expression levels in bladder cancer cell lines
To further characterize the relationship among HRD score, PARP inhibitor sensitivity, and RBBP8 expression, we used publicly available WES data to calculate the HRD score of twenty-three bladder cancer cell lines. Six of the 23 cell lines had an HRD score ≥42 (Supp Fig. 23), and these six cell lines with HRD score ≥42 were significantly more sensitive to the PARP inhibitor olaparib (Supp. Fig. 24) and trended towards lower RBBP8 expression (Supp. Fig. 25) than cell lines with HRD score <42. These cell line data suggest that tumors with higher HRD scores are more likely to respond to PARP inhibitor therapy, and that decreased RBBP8 expression may contribute to HR deficiency in a subset of bladder tumors.
Discussion
Homologous recombination deficiency is often driven by loss of function of BRCA1 or BRCA2 in tumor cells, which is frequently the result of an inactivating mutation coupled with the loss of the accompanying wild type allele (loss of heterozygosity). Consequently, many HR deficient cases can be identified by sequencing BRCA1/2, and this approach led to initial approval of PARP inhibitors in tumor types including breast, ovarian, and prostate cancer. However, it is clear that mechanisms beyond BRCA1/2 loss may also drive tumor HR deficiency. These mechanisms include loss or mutation of other HR genes (such as PALB2, RAD51C, and others), suppression of expression of BRCA1 or other HR genes by methylation, and perhaps through other, as-yet uncharacterized mechanisms.
Mutational signature-based approaches have recently been applied to improve prediction of HR deficiency because they detect the consequences of HR deficiency rather than the underlying cause. The first diagnostic HR deficiency mutational signature (HRD score) was recently approved to direct PARP inhibitor therapy in ovarian cancer cases without BRCA1/2 mutations based on data showing that patients without mutations in the canonical HR genes (BRCA1, BRCA2, etc.) but with high HR deficiency associated mutational signatures benefitted from PARP inhibitor therapy (16). These findings raise the possibility that other tumors with high levels of HR deficiency associated mutational signatures in the absence of BRCA1/2 mutations may also be HR deficient and could benefit from PARP inhibitor therapy.
Bladder cancer presents a complex picture in terms of DNA repair pathway aberrations. Approximately 20% of bladder tumors have loss of nucleotide excision repair (NER), primarily caused by somatic mutations in the NER gene ERCC2 (5). In this work, we identify a separate subset of bladder tumors with evidence of HR deficiency as defined by high levels of HR deficiency-associated mutational signatures. We find that BRCA1/2 loss-of-function mutations – when present in both alleles or when coupled with LOH – are associated with the same HR deficiency associated mutational signatures as are present in BRCA1/2-mutant ovarian, breast, or prostate tumors. Therefore, BRCA1/2 mutant bladder tumors, although rare, appear to have bona fide HR deficiency. However, it is important to note that only bladder tumors with loss of function of both BRCA1/2 alleles (via predicted deleterious mutation(s) and/or LOH) would be expected to exhibit an HR deficient phenotype.
In addition to the small percentage of HR-deficient bladder tumors due to loss of BRCA1/2, we also found that a significant number of bladder tumors with WT BRCA1/2 showed levels of HRD associated mutational signatures as high as those observed in BRCA1/2 deficient cases. Although numerous mechanisms beyond HR gene mutations could drive HR deficiency, we identified methylation of RBBP8 as one potential non-canonical driver of HR deficiency in bladder cancer. RBBP8 encodes the protein CtIP, which plays a critical role in coordinating repair events at DNA double strand breaks and stalled replication forks (48). Germline RBBP8 mutations were recently found in patients with early-onset breast cancer (47) and RBBP8 suppression has been shown to confer HR deficiency and increased PARP inhibitor sensitivity in MCF7 and U2OS cells (46,47). Therefore, the increased activity of HR deficiency mutational signatures observed in bladder tumors with reduced RBBP8 expression suggests a novel mechanism of HR deficiency that may be operant in a subset of bladder tumors. However, additional work will be required to firmly establish a causative role for RBBP8 suppression as a driver of HRD in bladder cancer and to define parameters such as the extent of RBBP8 suppression required to drive clinically relevant HR deficiency. For example, the average HRD score in RBBP8 methylated cases was higher than in non-methylated cases but was lower than in the BRCA1/2 deficient cases. One possible explanation for this observation is that RBBP8 gene methylation may be a subclonal event in some cases. Clarifying this correlation will require additional approaches such as spatial transcriptomics or immunohistochemistry-based measurements of RBBP8 levels.
Reliable detection of HR-deficient bladder tumors may have important clinical implications. Patients with HR deficient tumors may be prioritized for cisplatin-based chemotherapy. Furthermore, although cisplatin resistance develops in the majority of patients with bladder cancer (49), platinum resistance may not confer cross-resistance to other HR-targeting therapies such as PARP inhibitors. For example, ovarian tumors that are resistant to platinum often respond to PARP inhibitors, and progression on platinum can be significantly delayed by PARP inhibitors in HR deficient cases (16,50). In addition, nearly half of all patients with urothelial cancer are ineligible for cisplatin-based chemotherapy due to medical comorbidities (51), but cisplatin-ineligible patients with an HR deficient tumor may be eligible to receive a PARP inhibitor.
HR deficiency associated mutational signatures were first identified in breast and ovarian cancer, which harbor the highest frequency of BRCA1/2 inactivating events. We now show that the same signatures may also be useful in identifying HR deficiency in bladder cancer, a tumor type with far less frequent alterations in BRCA1/2. These findings may have implications for PARP inhibitor clinical trials and suggest that mutational analysis of known HR genes such as BRCA1/2 could be combined with mutational signature approaches (such as the HRD score) to identify cases most likely to harbor HR deficiency. Additional studies will be required to define mechanisms of HR deficiency in bladder cancer, optimize threshold values of the HR deficiency mutational signatures for clinical application, and understand the therapeutic implications of HR deficiency in bladder cancer.
Supplementary Material
Translational relevance.
PARP inhibitors are an effective treatment for homologous recombination deficient (HRD) solid tumors, such as ovarian, breast, pancreatic, and prostate cancer. It is likely that subsets of other solid tumor types may also harbor homologous recombination deficiency. We applied mutational signature approaches to evaluate HRD in bladder cancer and found that BRCA1/2-deficient bladder tumors have similar HRD-associated mutational signatures as BRCA1/2-deficient breast and ovarian tumors. In addition, we found that nearly 10% of bladder tumors with wild-type BRCA1/2 also exhibit mutational signatures of HRD, suggesting that alternative mechanisms, such as the promoter methylation of RBBP8, may drive HRD in these cases. Bladder cancer cases with high levels of HRD associated mutational signatures may be candidates for agents such as PARP inhibitors that preferentially target HRD tumors.
Acknowledgments
This work was supported by the Research and Technology Innovation Fund (KTIA_NAP_13-2014-0021 and 2017-1.2.1-NKP-2017-00002 to Z.S.), Breast Cancer Research Foundation (BCRF-20-159 to Z.S.), Kræftens Bekæmpelse (R281-A16566 to Z.S.), the Novo Nordisk Foundation Interdisciplinary Synergy Programme Grant (NNF15OC0016584 to Z.S.), the Novo Nordisk Foundation Center for Protein Research core grant (NNF14CC0001 to S.B.), Department of Defense through the Prostate Cancer Research Program (W81XWH-18-2-0056 to Z.S.), Det Frie Forskningsråd Sundhed og Sygdom (7016-00345B to Z.S.), Burroughs-Wellcome Fund (to K.W.M.), the National Cancer Institute (5K08CA219504 to K.W.M), and the Velux Foundation (00018310 to Zs.S. and J.B.). Results shown here are based in part from data generated by the TCGA Research Network: http://cancergenome.nih.gov/ and the International Cancer Genome Consortium (ICGC): https://icgc.org/. Results presented in the current publication are based in part on the use of study data downloaded from the dbGaP website, under phs000771.v2.p1 accession code (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000771.v2.p1).
Footnotes
Conflict of interest
Z. Szallasi is an inventor on a patent used in the myChoice HRD assay.
References
- 1.Perez-Gracia JL, Loriot Y, Rosenberg JE, Powles T, Necchi A, Hussain SA, et al. Atezolizumab in Platinum-treated Locally Advanced or Metastatic Urothelial Carcinoma: Outcomes by Prior Number of Regimens. Eur Urol. 2018;73:462–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Allen EM, Mouw KW, Kim P, Iyer G, Wagle N, Al-Ahmadie H, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, Plimack ER, Hoffman-Censits J, Garraway LA, Bellmunt J, Van Allen E, et al. Clinical Validation of Chemotherapy Response Biomarker ERCC2 in Muscle-Invasive Urothelial Bladder Carcinoma. JAMA Oncol. 2016;2:1094–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Börcsök J, Sztupinszki Z, Bekele R, Gao SP, Diossy M, Samant AS, et al. Identification of a synthetic lethal relationship between nucleotide excision repair (NER) deficiency and irofulven sensitivity in urothelial cancer. Clin Cancer Res. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miron B, Hoffman-Censits JH, Anari F, O’Neill J, Geynisman DM, Zibelman MR, et al. Defects in DNA Repair Genes Confer Improved Long-term Survival after Cisplatin-based Neoadjuvant Chemotherapy for Muscle-invasive Bladder Cancer. Eur Urol Oncol. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teo MY, Bambury RM, Zabor EC, Jordan E, Al-Ahmadie H, Boyd ME, et al. DNA Damage Response and Repair Gene Alterations Are Associated with Improved Survival in Patients with Platinum-Treated Advanced Urothelial Carcinoma. Clin Cancer Res. 2017;23:3610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer G, Balar AV, Milowsky MI, Bochner BH, Dalbagni G, Donat SM, et al. Multicenter Prospective Phase II Trial of Neoadjuvant Dose-Dense Gemcitabine Plus Cisplatin in Patients With Muscle-Invasive Bladder Cancer. J Clin Oncol. 2018;36:1949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taber A, Christensen E, Lamy P, Nordentoft I, Prip F, Lindskrog SV, et al. Molecular correlates of cisplatin-based chemotherapy response in muscle invasive bladder cancer by integrated multi-omics analysis. Nat Commun. 2020;11:4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grivas P, Loriot Y, Feyerabend S, Morales-Barrera R, Teo MY, Vogelzang NJ, et al. Rucaparib for recurrent, locally advanced, or metastatic urothelial carcinoma (mUC): Results from ATLAS, a phase II open-label trial. Journal of Clinical Oncology [Internet]. American Society of Clinical Oncology; 2020. [cited 2020 Jul 17]; Available from: 10.1200/JCO.2020.38.6_suppl.440 [DOI] [Google Scholar]
- 13.Póti Á, Gyergyák H, Németh E, Rusz O, Tóth S, Kovácsházi C, et al. Correlation of homologous recombination deficiency induced mutational signatures with sensitivity to PARP inhibitors and cytotoxic agents. Genome Biol. 2019;20:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res. 2016;22:3764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N Engl J Med. 2019;381:2416–28. [DOI] [PubMed] [Google Scholar]
- 17.Zhao EY, Shen Y, Pleasance E, Kasaian K, Leelakumari S, Jones M, et al. Homologous Recombination Deficiency and Platinum-Based Therapy Outcomes in Advanced Breast Cancer. Clin Cancer Res. 2017;23:7521–30. [DOI] [PubMed] [Google Scholar]
- 18.Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am J Hum Genet. 2017;100:267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boot A, Huang MN, Ng AWT, Ho S-C, Lim JQ, Kawakami Y, et al. In-depth characterization of the cisplatin mutational signature in human cell lines and in esophageal and liver tumors. Genome Res. 2018;28:654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sztupinszki Z, Diossy M, Krzystanek M, Reiniger L, Csabai I, Favero F, et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer. 2018;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnov P, Safikhani Z, El-Hachem N, Wang D, She A, Olsen C, et al. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics. 2016;32:1244–6. [DOI] [PubMed] [Google Scholar]
- 31.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov. 2015;5:1210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sztupinszki Z, Diossy M, Krzystanek M, Borcsok J, Pomerantz MM, Tisza V, et al. Detection of Molecular Signatures of Homologous Recombination Deficiency in Prostate Cancer with or without BRCA1/2 Mutations. Clin Cancer Res. 2020;26:2673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009;69:6381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li A, Geyer FC, Blecua P, Lee JY, Selenica P, Brown DN, et al. Homologous recombination DNA repair defects in PALB2-associated breast cancers. NPJ Breast Cancer. 2019;5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mutter RW, Riaz N, Ng CK, Delsite R, Piscuoglio S, Edelweiss M, et al. Bi-allelic alterations in DNA repair genes underpin homologous recombination DNA repair defects in breast cancer. J Pathol. 2017;242:165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–62. [DOI] [PubMed] [Google Scholar]
- 40.Birkbak NJ, Wang ZC, Kim J-Y, Eklund AC, Li Q, Tian R, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Health C for D and R. Myriad myChoice CDx - P190014. FDA [Internet]. FDA; 2019. [cited 2020 Jul 30]; Available from: https://www.fda.gov/medical-devices/recently-approved-devices/myriad-mychoice-cdx-p190014 [Google Scholar]
- 42.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017;49:1476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mijnes J, Veeck J, Gaisa NT, Burghardt E, de Ruijter TC, Gostek S, et al. Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin Epigenetics. 2018;10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Ding Q, Fujimori H, Motegi A, Miki Y, Masutani M. Loss of CtIP disturbs homologous recombination repair and sensitizes breast cancer cells to PARP inhibitors. Oncotarget. 2016;7:7701–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarrizi R, Higgs MR, Voßgröne K, Rossing M, Bertelsen B, Bose M, et al. Germline RBBP8 variants associated with early-onset breast cancer compromise replication fork stability. J Clin Invest. 2020;130:4069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Przetocka S, Porro A, Bolck HA, Walker C, Lezaja A, Trenner A, et al. CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress. Mol Cell. 2018;72:568–582.e6. [DOI] [PubMed] [Google Scholar]
- 49.Hussain SA, James ND. The systemic treatment of advanced and metastatic bladder cancer. Lancet Oncol. 2003;4:489–97. [DOI] [PubMed] [Google Scholar]
- 50.Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med. 2016;375:2154–64. [DOI] [PubMed] [Google Scholar]
- 51.Sonpavde G, Watson D, Tourtellott M, Cowey CL, Hellerstedt B, Hutson TE, et al. Administration of cisplatin-based chemotherapy for advanced urothelial carcinoma in the community. Clin Genitourin Cancer. 2012;10:1–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.