Abstract
Owing to incurable castration-resistant prostate cancer (CRPC) ultimately developing after treating with androgen deprivation therapy (ADT), it is vital to devise new therapeutic strategies to treat CRPC. Treatments that target programmed cell death protein 1 (PD-1) and programmed death ligand-1 (PD-L1) have been approved for human cancers with clinical benefit. However, many patients, especially prostate cancer, fail to respond to anti-PD-1/PD-L1 treatment, so it is an urgent need to seek a support strategy for improving the traditional PD-1/PD-L1 targeting immunotherapy. In the present study, analyzing the data from our prostate cancer tissue microarray, we found that PD-L1 expression was positively correlated with the expression of heterogeneous nuclear ribonucleoprotein L (HnRNP L). Hence, we further investigated the potential role of HnRNP L on the PD-L1 expression, the sensitivity of cancer cells to T-cell killing and the synergistic effect with anti-PD-1 therapy in CRPC. Indeed, HnRNP L knockdown effectively decreased PD-L1 expression and recovered the sensitivity of cancer cells to T-cell killing in vitro and in vivo, on the contrary, HnRNP L overexpression led to the opposite effect in CRPC cells. In addition, consistent with the previous study, we revealed that ferroptosis played a critical role in T-cell-induced cancer cell death, and HnRNP L promoted the cancer immune escape partly through targeting YY1/PD-L1 axis and inhibiting ferroptosis in CRPC cells. Furthermore, HnRNP L knockdown enhanced antitumor immunity by recruiting infiltrating CD8+ T cells and synergized with anti-PD-1 therapy in CRPC tumors. This study provided biological evidence that HnRNP L knockdown might be a novel therapeutic agent in PD-L1/PD-1 blockade strategy that enhanced anti-tumor immune response in CRPC.
KEY WORDS: HnRNP L, PD-L1, YY1, Ferroptosis, Immune escape, Immune checkpoint blockade, Anti-PD-1 therapy, Castration-resistant prostate cancer
Abbreviations: ADT, androgen deprivation therapy; CRPC, castration-resistant prostate cancer; DMSO, dimethyl sulfoxide; ELISA, enzyme-linked immunosorbent assay; FBS, fetal bovine serum; Fer-1, ferrostatin-1; GSH, glutathione; HnRNP L, heterogeneous nuclear ribonucleoprotein L; IL, interleukin; INF-γ, interferon gamma; PD-1, programmed cell death protein 1; PD-L1, programmed death ligand1; qRT-PCR, quantitative reverse transcription polymerase chain reaction; ROS, reactive oxygen species; STAT, signal transducer and activator of transcription
Graphical abstract
Inhibition of HnRNP L diminishes PD-L1 expression and promotes antitumor immunity via destabilizing YY1 mRNA in CRPC and enhancing the T-cell-mediated cancer cell ferroptosis.
1. Introduction
Prostate cancer (PCa) is the second common malignant tumor among male cancer patients worldwide, which accounts for over 1414,259 new cases and approximately 375,304 mortalities in 20201. Although androgen deprivation therapy (ADT) is the standard treatment for early-stage PCa and advanced PCa, most of the patients respond resistant to this therapy ultimately, and it has become a major limitation for endocrine-based therapy2, 3, 4. Therefore, it is vital for the treatment of castration-resistant prostate cancer (CRPC) to identify new carcinogenic pathways, and more efficient targeted therapies are urgently needed. As immune escape is one of the major features of a variety of cancers5, 6, 7, 8, a better understanding of the biology of CRPC and its relationship with immune response would be important for the development of more effective therapeutic strategies.
Programmed death-ligand 1 (PD-L1), encoded by CD274 gene, is a transmembrane protein that induces immune suppression via binding to the inhibitory receptor programmed cell death protein 1 (PD-1) on T cells and eliciting T-cell anergy9. Many cancer cells, such as melanoma10, lung cancer11, bladder cancer12, breast cancer13 and prostate cancer14, upregulate the PD-L1 expression to escape immune surveillance. Targeting immune checkpoints such as the one mediated by PD-L1 and its receptor PD-1 has been approved for treating human cancers with appropriate clinical benefit15,16. However, many cancer patients, especially prostate cancer, fail to respond to the immune treatment with anti-PD-1 or anti-PD-L1 antibodies, and the underlying mechanism(s) is not well defined17, 18, 19. Recent advances in cancer immune therapies revealed that response to anti-PD-1/PD-L1 treatment might correlate with the PD-L1 expression levels in cancer cells20,21. Thus, it is crucial to explore the pathways controlling PD-L1 protein expression and stability, which is essential to help devise treatment strategies to strengthen prostate cancer immunotherapy.
Heterogeneous nuclear protein L (HnRNP L), a member of the HnRNP family, has been identified as a global splicing regulator that is able to regulate the transcription of precursor mRNAs and mature mRNAs22, 23, 24, 25. Recent studies have reported that aberrant expression of HnRNP L serves a critical role in regulating the tumorigenic capacity of a number of malignances, including lung cancer26, liver cancer27 and colorectal cancer28. Furthermore, in our previous study we found that HnRNP L is overexpressed in prostate cancer and exerts pro-proliferative and anti-apoptotic effects29. Interestingly, Fei et al.30 systematically identified HnRNP L to be a top essential RBP for prostate cancer progress by a genome-wide CRISPR screen. However, the specific role of HnRNP L in regulating the PD-L1 expression and mediating prostate cancer immune escape has not been elucidated. The aim of the present study therefore is to investigate the potential role and effects of HnRNP L in anti-tumor immunity of prostate cancer.
In this study, we show that HnRNP L is overexpressed in prostate cancer and positively correlates with the PD-L1 expression. Moreover, HnRNP L is responsible for YY1 mRNA stabilization and then promoting the transcription of PD-L1 in prostate cancer cell lines. Importantly, abrogation of HnRNP L sensitizes prostate cancer cells to activated Jurkat T cells mediated killing by downregulating the PD-L1 protein levels and promoting the activated Jurkat T cells-induced cancer cell ferroptosis, and enhances anti-PD-1 therapy efficacy by recruiting CD8+ T cells in vitro and in vivo. Our data shed a light on the therapeutic implications of HnRNP L inhibition in ameliorating the immunotherapy of CRPC.
2. Materials and methods
2.1. Reagents
The ferroptosis inhibitors S7243 (ferrostatin-1) was purchased from Selleck Chemicals (Houston, TX, USA) and dissolved in dimethyl sulfoxide (DMSO). Working solutions were prepared by diluting the stock solution with RPMI-1640. In all experiments, the final DMSO concentration was ≤0.1%.
2.2. Patients and clinical samples
Here, we detected the expression of HnRNP L protein using a tissue microarray (PRC481, Alenabio, Xi'an, China) containing several prostate cancer and normal prostate tissues by immunohistochemistry analysis. In addition, we analyzed the expression correlation between the HnRNP L and PD-L1 protein using a tissue microarray (HProA100PG01, Outdo Biotech, Shanghai, China) by immunofluorescence double staining. Four paired fresh tissues of patients were collected from radical prostatectomy performed at the Shenzhen Peking University Hospital between 2018 and 2020 and the HnRNP L protein expression levels in these tissues were measured by Western blotting. All of the patients in this study signed the informed consent following the ethical protocols of the Ethics Committee of Shenzhen Peking University Hospital, Peking University (Shenzhen, China).
2.3. Cell lines and culture
Two human CRPC cell lines (PC3 and DU145) and mouse prostate cancer RM-1 cells were obtained from American Type Culture Collection (ATCC). Cells were routinely cultured in complete medium (1640 medium [Gibco, China] was supplemented with 10% heat-inactivated fetal bovine serum [Sangon, China]), 1% penicillin/streptomycin (HyClone) and incubated at 37 °C in an atmosphere of 5% CO2.
2.4. Immunofluorescence assays
The transfected cells were cultured in 24-well plates for 24 h and fixed with 4% paraformaldehyde for 40 min. After washed for three times with PBS, cells were permeabilized with 0.3% Triton X-100 for 15 min and blocked with 5% BSA for 1 h at room temperature. Then, cells in 24-well plates were incubated with HnRNP L and PD-L1 primary antibodies overnight at 4 °C and incubated with appropriate secondary antibodies (ZSGB-Bio) for 1.5 h at room temperature. Finally, cells were counter-stained with DAPI for 5 min and detected under a confocal laser-scanning microscope.
2.5. RNAi and plasmid transfection
For RNA interference, the cells were transfected with 20 nmol/L small interfering RNA (siRNA) (GenePharma, Suzhou, China) targeting the HnRNP L: si#428: sence: CACUGGUGGAGUUUGAAGATT, antisence: UCUUCAAACUCCACCAGUGTT; si#613: sence: CCCAUUUAUUCGAUCACCATT, antisence: UGGUGAUCGAAUAAAUGGGTT; si#788: sence: GUUGCACUCUGAAGAUCGATT, antisence: UCGAUCUUCAGAGUGCAACTT, using Lipofectamine 3000 (L3000015, ThermoFisher, MA, USA) according to the manufacturer's instructions. For overexpression of HnRNP L in CRPC cells, the full-length homo sapiens HnRNP L was cloned into the pcDNA3.0 vector, producing pC3.0-HnRNP L plasmid. The transfected CRPC cells were harvested after 48 h later. The overexpression and the inhibition efficiency were determined by qRT-PCR and Western blotting.
2.6. Enzyme-linked immunosorbent assay (ELISA)
Levels of IL-2, PD-L1 and INF-γ in the supernatants were measured by ELISA (Elabscience, Wuhan, China) following its manufacturer's instructions. Briefly, 100 μL diluent buffer, 100 μL samples and 100 μL standard were added to the wells and incubated at 37 °C for 90 min and removed. And then, we added 100 μL biotinylated detection antibody into the wells at 37 °C for 1 h; immediately, we washed the samples for three times with 2 min each time. Next, 100 μL horseradish peroxidase conjugate was added to the wells and incubated the plates at 37 °C for 30 min. After washing five times, we added the substrate reagent and incubated them at 37 °C for 15 min in dark and then stopped the reaction. We used a microplate reader (Multiskan™ FC, ThermoFisher, MA, USA) to measure the absorbance at 450 nm.
2.7. Crystal violet staining assay
PC3 and DU145 cells that transfected with HnRNP L siRNA or plasmid were seeded into 12-well plates (1 × 105 cells per well) and incubated with Jurkat T cells (6 × 105 cells per well), which were activated by anti-CD3 plus anti-CD28 co-stimulation, for 2 days. After 2 days, the surviving cancer cells were washed with the cold PBS, fixed with 11% glutaraldehyde and stained with 0.1% crystal violet, and then measured by Microplate Reader (Multiskan™ FC, ThermoFisher).
2.8. Cell death analysis
After transfected with HnRNP L siRNA or plasmid for 24 h, human Jurkat T cells (activated by anti-CD3 plus anti-CD28 co-stimulation) were added at the ratio of 1:6 for another 48 h, and then harvested and stained with Annexin V-FITC and propidium iodide (PI) according to the kit's protocols. The cell death was validated by flow cytometry using a BD FACSCalibur system. All experiments were analyzed in triplicates.
2.9. Determination of lipid ROS generation
The transfected cells (2 × 105) in 6-well plates were co-cultured with the activated Jurkat T cells for 48 h and then change into 1 mL of fresh medium containing 5 μmol/L of BODIPY 581/591 C11 (GlpBio, Guangzhou, China) for 30 min at 37 °C. Cells were then washed twice for detecting under fluorescence microscope or trypsinized and resuspended in 0.5 mL of PBS for flow cytometry analysis. The wavelength of the oxidized fluorescent probe is in the green light band.
2.10. GSH assay
The Glutathione Assay Kit (Cayman Chemical, Michigan, USA) was used to measure the amount of glutathione (GSH) intracellularly. The transfected cells (2 × 105) were seeded in 6-well plates and co-cultured with the activated Jurkat T cells for 48 h, scraped into 500 μL of 10 mmol/L phosphate buffer containing 1 mmol/L EGTA, and then lysed by sonication. After centrifugation, supernatants were deproteinated by incubating with 500 μL of 10% metaphosphoric acid at room temperature for 5 min, and then centrifuged for 3 min at 4000 rpm (5425 R, Eppendorf, Germany). The resulted supernatants were mixed with 50 μL of 4 mol/L triethanolamine. 50 μL of each sample was then transferred to a 96-well plate, and incubated with 150 μL of Assay Cocktail containing the Ellman's reagent [5,5′-dithio-bis-2-(nitrobenzoic acid) or DTNB] at room temperature for 25 min. The absorbance at 405 nm was measured, and used to calculate the GSH amount in reference to a GSH standard curve.
2.11. Glutamate release assay
The Glutamate-Glo Assay kit (Promega, Madison, USA) was used to detect the content of glutamate released into condition medium. The transfected cells (2 × 105) in 6-well plates were co-cultured with the activated Jurkat T cells in glutamine-free medium for 48 h. To measure the glutamate level, 50 μL of condition medium was transferred to a 96-well plate, and mixed with 50 μL of a reaction mixture containing glutamate dehydrogenase, NAD, reductase, pro-luciferin, and luciferin detection solution following the manufacturer's protocols. The plate was shaken for 30–60 s and incubated at room temperature for 1 h, and luminescence was measured with SpectraMax L (Molecular Devices, Shanghai, China). The glutamate level was first calculated in reference to a glutamate standard curve, and then normalized to the total cell number determined by CCK-8 assays at the end of the experiment.
2.12. Establishment of stably transfected cell lines
The successful construction of lentivirus vector was finished in Shanghai Genechem Co., Ltd. Lentivirus-mediated HnRNP L overexpression and knockdown in RM-1 cell lines were achieved according to the manufacturer's instructions. The infection efficiency was validated by qRT-PCR and Western blotting.
2.13. In vivo tumor model
Twenty-four C57BL/6 or nude mice (3–4 weeks) were acquired from the Medical Experimental Animal Center of Guangdong Province (Guangzhou, China) and kept under specific pathogen-free conditions. All operations involving animals were conformed with the Chinese National Institute of Health Guide for the Care and Use of Laboratory Animals, and the study was approved by the Research Ethics Committee of Southern Medical University (Guangzhou, China).
We randomly divided these mice into four groups: the lentiviral-mediated HnRNP L stably overexpressed group (n = 6), the HnRNP L stable knockdown group (n = 6) and control groups for each (n = 6). 2 × 106 RM-1 cells were implanted subcutaneously into the right armpit and inguinal regions of each C57BL/6 or nude mouse. For the combination therapy C57BL/6 mice model, 100 μg of anti-PD1 (clone RMP1-14, BioXcell) antibody were injected on Days 7, 14, 21, and 28 after inoculating with HnRNP L knockdown or the control RM-1 cells. A total of 35 days for feeding, all mice were sacrificed and tumor weight and tumor volume were immediately measured respectively. Tumor volumes were calculated in accordance with Eq. (1):
| V = [Length (mm) × Width2 (mm)]/2 | (1) |
2.14. RNA-immunoprecipitation (RIP) assay
The binding of HnRNP L protein with YY1 mRNA was verified in this study by the RIP assay using the EZ-Magna RIP™ RIP Kit (Merck, Germany) following the manufacturer's instructions, in combination with qRT-PCR method. Briefly, after transfected with HnRNP L plasmid for 48 h, the CRPC (PC3 and DU145) cells were lysed in RIP lysis buffer, incubated with magnetic beads containing antibodies specifically recognizing HnRNP L proteins, and incubated with proteinase K. Magnetic beads conjugated with antibodies targeting human IgG were used as the negative control. After washing with the washing buffer, RNA samples bound in magnetic beads were eluted and used as the templates for RT. The relative contents of YY1 mRNA in elutes were analyzed by qRT-PCR method.
2.15. Chromatin immunoprecipitation (ChIP) assays
ChIP assay was performed using ChIP kit (Santa Cruz Biotechnology, CA, USA) according to the manufacturer's instructions. First, 1 × 108 CRPC (PC3 and DU145) cells were transfected with HnRNP L plasmid and co-cultured with the activated Jurkat T cells for 48 h. Then, cells were cross-linked in 1% formaldehyde for 15 min at room temperature and then lysed in SDS buffer. Sonication was used to fragment the DNA. ChIP for YY1 was performed using a YY1 primary antibody (CST, #63227S). Eluted DNA fragments were analyzed by qRT-PCR and agarose gel electrophoresis using the specific primers are listed as follows: PD-L1 (CD274) forward, TAGAAATACCATTTGACCCA; reverse, CCATTACTGGGTATATACCC.
2.16. Immunohistochemical analysis and evaluation
Tumor tissues were deparaffinized, rehydrated through incubation with xylene and ethanol and the endogenous antigen was restored by autoclave for 15 min in citric-acid buffer (10 mmol/L citrate buffer, pH 8.0). Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Then slides were incubated overnight with anti-HnRNP L, anti-PD-L1, anti-CD4, anti-CD8, anti-SLC7A11 and anti-GPX4 primary antibody at 4 °C and then HRP conjugated secondary antibody at 37 °C for 30 min. Signal was detected with DAB and nuclei were counterstained with hematoxylin. All antibodies were obtained from Abcam and diluted at the rate of 1:100. Intensity of stained cells was scored as follows: 0—lack of staining (−), 1—weak staining (+), 2—moderate staining (++) and 3—strong staining (+++). The percentage of stained cells was divided into five classes: 0 for negative cells, 1 for 1%–25%, 2 for 25%–50%, 3 for 50%–75%, and 4 for >75%. Total scores of stained cells ranged from 0 to 12. All sections were defined as having low expression—0 to 7 or high expression—8 or greater by semiquantitative score. Slides were assessed by 2 pathologists.
2.17. RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays
Total RNA was extracted from the cultured cells and human tissues using TRizol reagent (Takara Bio, Beijing, China), and first-strand cDNA was synthesized with the Prime Script RT Reagent Kit (Takara Bio, Beijing, China) following the manufacturer's protocols. Reverse transcription was performed with the Super Script First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) for qRT-PCR according to the manufacturer's instructions. qRT-PCR was performed using complementary DNA and SYBR-Green II (Takara Bio, Beijing, China). The relative levels of mRNAs in cells and tissues were normalized to the housekeeping gene GAPDH and calculated with the equation . Primers sequences were shown in Supporting Information Table S1.
2.18. Western blotting analysis
The treated cells were washed three times with cold PBS and lysed by RIPA buffer (KeyGEN, Jiangsu, China) containing protease and phosphatase inhibitors. The concentration of protein was detected by bicinchoninic acid method. For Western blotting analysis, proteins were separated by 10% polyacrylamide gel electrophoresis and then transferred onto 0.22 μm pore-size PVDF membrane (the PVDF membrane was activated by methanol). After blocking with 5% skim milk, the membrane was incubated with the appropriate primary antibodies at 4 °C overnight. Then, the membrane was washed 5 min for six times, followed by incubation with HRP conjugated secondary antibody for 2 h at room temperature. Finally, protein blots were visualized using ECL substrate reagents. Antibodies used in immunoblotting were as follows: mouse monoclonal antibody anti-HnRNP L (1:2000; Abcam #ab-6106), rabbit monoclonal antibody anti-PD-L1 (1:1000; Abcam #ab-213524), mouse monoclonal antibody anti-GAPDH (1:1000; Abcam #ab-8245), rabbit monoclonal antibody anti-SLC7A11 (1:1000; Abcam #ab-175186), rabbit monoclonal antibody anti-GPX4 (1:1000; Abcam #ab-125066), rabbit monoclonal antibody anti-YY1 (1:1000; CST#63227), rabbit monoclonal antibody anti-STAT1 (1:1000; Abcam#ab-109461).
2.19. Statistical analysis
Statistical analyses were performed using SPSS 20.0 (IBM Corp., Armonk, NY, USA). All assays were repeated at least three times and data are presented as the mean ± standard deviation (SD). Differences between groups were calculated utilizing ANOVA or a Student's t-test for continuous variables. The statistical significance of difference between groups was expressed by asterisks (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). P < 0.05 was considered statistically significant.
3. Results
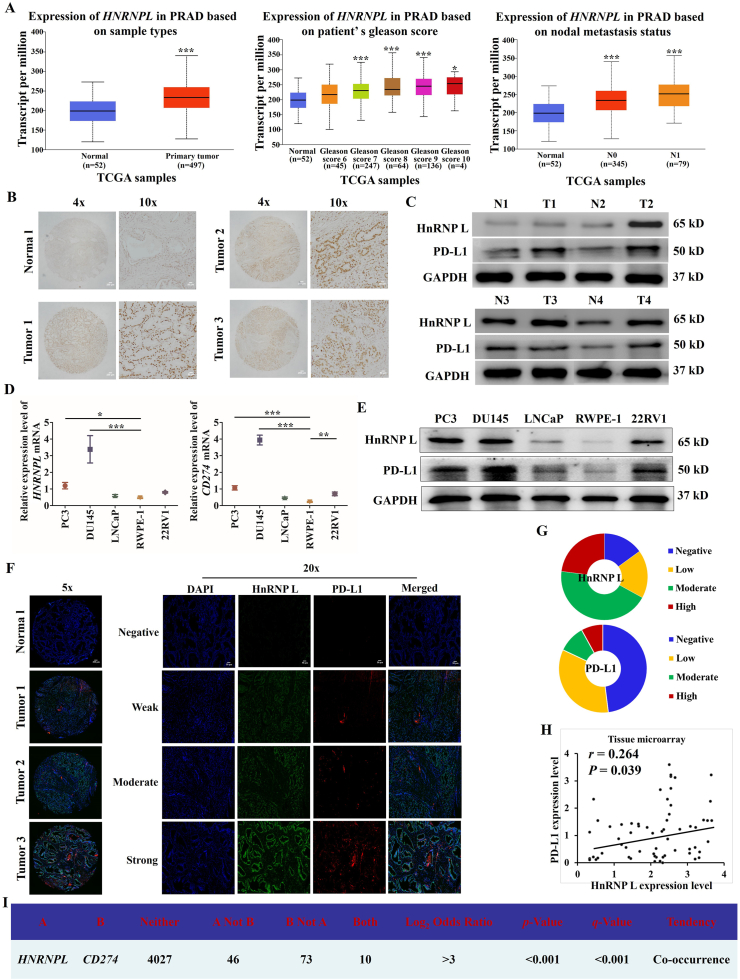
3.1. HnRNP L and PD-L1 are co-expressed in PCa cells and PCa tumors
We analyzed the expression of HNRNPL mRNA in 497 prostate cancer and 52 normal prostate tissues in TCGA database through TCGA data analysis. The results revealed significant upregulation of the expression of HNRNPL in the tumor tissues compared with the normal tissue (Fig. 1A). In addition, we also detected the protein expression of HnRNP L in 17 prostate cancer tissues and 7 non-prostate cancer tissues by IHC analysis and in 4 paired prostate cancer specimens by Western blotting analysis. We found that the HnRNP L protein levels in prostate cancer tissues were significantly higher than those in normal tissues (Fig. 1B and C). To test the potential role of HnRNP L in regulating PD-L1 expression, we first evaluated HnRNP L and PD-L1 proteins together with mRNA expression in control RWPE-1 cells and four different PCa cell lines by Western blotting and qRT-PCR. High expression of both HnRNP L and PD-L1 was detected in CRPC (PC3 and DU145) cells (Fig. 1D and E). Furthermore, IHC of 61 paraffin-embedded human PCa tissues and 3 normal prostate samples showed that expression of both HnRNP L and PD-L1 was dramatically increased in PCa tumors than the normal tissues (Fig. 1F and G). We further analyzed the expression correlation between these two proteins in our tissue microarray and the TCGA database and found a positive correlation between HnRNP L and PD-L1 both in mRNA (Fig. 1I) and protein level (r = 0.264, P = 0.039; Fig. 1H) in prostate carcinoma tissues. Collectively, these results imply that HnRNP L and PD-L1 are co-expressed in prostate cancer cells and tissues.
Figure 1.
HnRNP L and PD-L1 are co-expressed in PCa cells and PCa tumors. (A) The expression of HNRNPL in normal prostate tissues and prostate cancer tissues based on the data in TCGA database. (B) Expression of HnRNP L in a tissue microarray containing several prostate cancer and non-prostate cancer tissues (n = 24). Representative images of HnRNP L in Normal and Pca specimens examined by IHC. Four representative immunohistochemical staining images of HnRNP L in normal tissue (Normal) and tumor tissue samples (Tumor 1, Tumor 2 and Tumor 3) examined by IHC are shown as indicated. (C) Western blotting bands for HnRNP L and PD-L1 expression in each of the paired Pca tissue samples (T) and adjacent normal tissue samples (N) obtained from the same patients. The mRNA and protein expressions of HnRNP L and PD-L1 in the normal prostate epithelial cell line (RWPE-1) and four PCa cell lines were detected by qRT-PCR (D) and Western blotting (GAPDH was used as loading control) (E). (F) Representative images of IHC of anti-HnRNP L and anti-PD-L1 antibodies of PCa (n = 61) tissue sections. (G) Pie chart showing the staining index of HnRNP L and PD-L1 in PCa. (H) Correlation analysis of the staining index for expression of HnRNP L and PD-L1 in specimens of PCa patients (n = 61). (I) The co-expression data of HNRNPL and CD274 in prostate cancer through the TCGA database indicated their interconnections. Each bar represents the mean ± SD of three independent experiments. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.2. HnRNP L promotes the expression and secretion of PD-L1 in CRPC cells
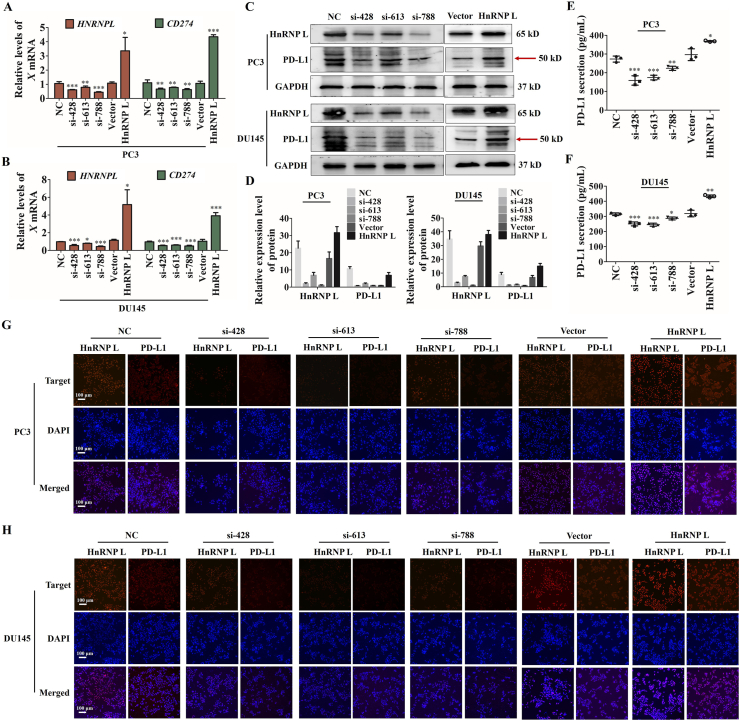
We sought to further validate the ability of HnRNP L-mediated PD-L1 expression in CRPC cells. As shown in Fig. 2A–D, qRT-PCR and immunoblotting revealed all HnRNP L siRNAs, 428, 613 and 788, decreased the abundance of CD274 (PD-L1) compared to the negative control, in contrast, overexpression of HnRNP L displayed the opposite effect. It has been reported that tumor cells could secrete PD-L1 into the extracellular fluid via vesicles. Therefore, we also detected the amount of PD-L1 in cell culture medium by ELISA and found that si-HnRNP L reduced the secretion of PD-L1 and HnRNP L overexpression increased the secretion (Fig. 2E and F). In addition, we performed the cellular immunofluorescence assay to further analyze the regulation of HnRNP L on PD-L1 and their location in CRPC cells and obtained the same effect (Fig. 2G and H). Since siRNA-428 and siRNA-613 was more effective than siRNA-788 in suppressing the secretion of HnRNP L, siRNA-428 and siRNA-613 was further used in the following experiment. Taken together, these results suggest that HnRNP L increased PD-L1 expression in CRPC cells.
Figure 2.
HnRNP L promotes the expression and secretion of PD-L1 in CRPC cells. The qRT-PCR analysis (A and B) and Western blotting analysis (C and D) of the expression of HnRNP L and CD274 (PD-L1) after HnRNP L inhibition or overexpression. Red arrow indicates the main bands for analysis. (E and F) PD-L1 in cell culture medium from the transfected CRPC cells (PC3 and DU145) was measured by enzyme-linked immunosorbent assay (ELISA). The immunofluorescence analysis of the expression of HnRNP L and CD274 (PD-L1) after HnRNP L inhibition or overexpression in PC3 (G) and DU145 (H) cells. Each bar represents the mean ± SD of three independent experiments. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
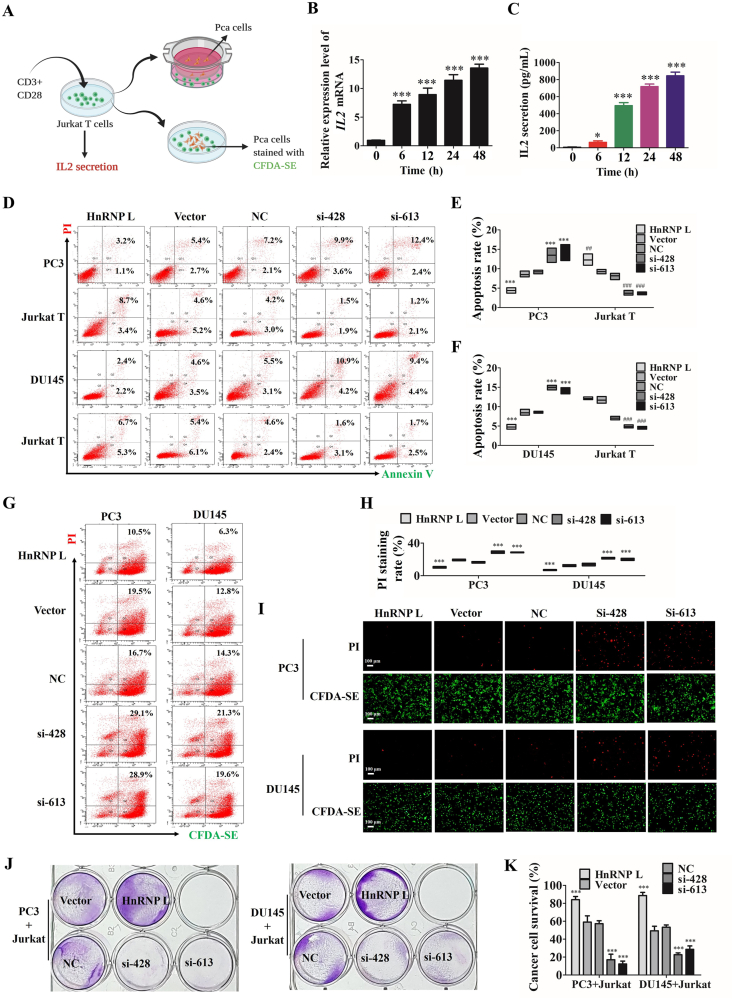
3.3. HnRNP L inhibits the killing activity of Jurkat T cells to CRPC cells
Because PD-L1 expressed on cancer cells binds to its homoreceptor PD-1 on infiltrating T cells and attenuates their antitumor activity31, we then detected whether HnRNP L affected the ability of CRPC cells bind to PD-1 and the killing activity of Jurkat T cells to CRPC cells. We used anti-CD3 plus anti-CD28 co-stimulation to activate the Jurkat T cells and found that the expression and secretion of IL-2, which reflected the activation of Jurkat T cells, was increased as the extend of the stimulating time (Fig. 3B and C). To validate the functional changes in HnRNP L inhibition mediated PD-L1 downregulation in CRPC cells, we co-cultured of PC3 or DU145 cells with the activated Jurkat T cells in a directly or indirectly manner (Fig. 3A) for 48 h and evaluated the effect of apoptosis in two human CRPC cells by flow cytometry analysis. Our results show increased apoptosis in CRPC cells treated with si-HnRNP L and decreased apoptosis in CRPC cells treated with HnRNP L overexpression after co-culture with the activated Jurkat T cells, while the opposite effect in co-cultured Jurkat T cells (Fig. 3D–F). For a direct-contact way in co-culture of CRPC cells with Jurkat T cells, we pre-stained the CRPC cells by CFDA-SE (Glpbio, Guangzhou, China) to distinguish the CRPC cells from the co-cultured Jurkat T cells and then analyzed the killing activity of Jurkat T cells to CRPC cells by flow cytometric (Fig. 3G and H) and fluorescence microscope (Fig. 3I) using CFDA-SE/PI staining. The results in a directly manner were consistent with the indirectly way above. The antitumor effect of HnRNP L inhibition was further estimated by detecting the surviving tumor cells using crystal violet staining after co-culturing CRPC cells with the activated Jurkat T cells. HnRNP L inhibition significantly reduced the surviving of CRPC cells than those with control, HnRNP L overexpression displayed the contrary effect (Fig. 3J and K).
Figure 3.
HnRNP L inhibits the killing activity of Jurkat T cells to CRPC cells. (A) Schematic diagram of the process of co-culture of PCa cells and Jurkat T cells. The qRT-PCR analysis (B) and ELISA analysis (C) of the expression and secretion of IL-2 from Jurkat T cells after activating by anti-CD3 plus anti-CD28 co-stimulation for 0, 6, 12, 24 and 48 h. Representative images of annexin-V/propidium iodide staining showing increased apoptosis in CRPC cells treated with si-HnRNP L and decreased apoptosis in CRPC cells treated with HnRNP L overexpression after co-culture with the activated Jurkat T cells, while the opposite effect in co-cultured Jurkat T cells (D). Statistical results are represented as mean ± SD of three independent experiments (E and F). HnRNP L inhibits the directly contacted Jurkat T cells induced-cell death of PC3 and DU145 cells (pre-stained by CFDA-SE), which were analyzed by flow cytometric (G and H) and fluorescence microscope (I) using CFDA-SE/PI staining. The data were analyzed by One-way ANOVA and post-hoc assays. (J) Jurkat T cells were activated by anti-CD3 plus anti-CD28 and co-cultured with the transfected CRPC cells (PC3 and DU145) in 12-well plates for 2 days and the surviving tumor cells were visualized by crystal violet staining. (K) Relative fold ratios of surviving cell intensity are shown (n = 3). The data are presented as mean ± SD. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
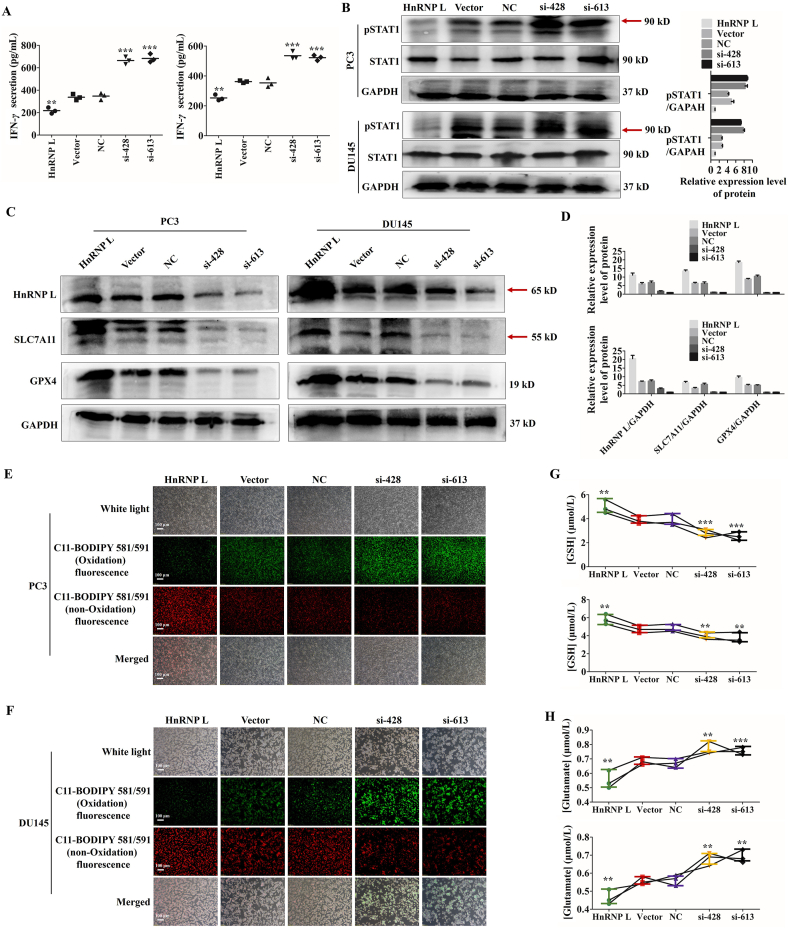
Interestingly, a recent study published in Nature revealed that CD8+ T cells played a pivotal role in cancer immunotherapy through generating IFN-γ, which activated STAT1 signaling, suppressed the SLC7A11 expression and induced the tumor cell ferroptosis32. In view of the above, we detected those events in our experimental models. As shown in Fig. 4A–D, HnRNP L inhibition in CRPC cells indeed induced massive release of IFN-γ from the co-cultured Jurkat T cells, upregulation of pSTAT1 and downregulation of SLC7A11 and GPX4. What's more, ferroptosis was an event characterized by lipid peroxidation and System Xc− obstacle33. As expected, we observed that HnRNP L inhibition induced the elevation of the lipid peroxide level (Fig. 4E and F) and an extensive amount of glutamate releasing into cultured media (Fig. 4G and H) in two CRPC cells after co-culturing with the activated Jurkat T cells. Similarly, HnRNP L overexpression displayed the opposite effect. All together, these results suggest that HnRNP L inhibition increases the cytotoxicity of T cells toward CRPC cells, appearing as high level of ferroptosis in cancer cells, by downregulating the PD-L1 expression in CRPC cells.
Figure 4.
HnRNP L inhibits the killing activity of Jurkat T cells to CRPC cells. (A) IFN-γ in cell culture medium from the activated Jurkat T cells after co-cultured with the transfected CRPC cells was measured by enzyme-linked immunosorbent assay (ELISA). (B) Western blotting analysis of the expression of p-STAT1 and STAT1 after co-culture with the activated Jurkat T cells in transfected CRPC cells. GAPDH was used as a loading control. Red arrow indicates the main bands for analysis. (C) and (D) Western blotting analysis of the expression of HnRNP L and the ferroptosis-related protein, SLC7A11 and GPX4 after co-culture with the activated Jurkat T cells in transfected CRPC cells (si-HnRNP L or HnRNP L overexpression). GAPDH was used as a loading control. Red arrow indicates the main bands for analysis. (E) and (F) The changes of lipid ROS accumulation in transfected PC3 and DU145 cells after co-culture with the activated Jurkat T cells were measured by fluorescence microscope. (G) and (H) The transfected PC3 and DU145 cells were co-cultured with the activated Jurkat T cells for 48 h, and the amount of glutamate released into culture medium was measured. Indicated cells co-cultured with the activated Jurkat T cells for 48 h were lysed, and the intracellular GSH level was measured. The data are presented as mean ± SD (n = 3). The data were analyzed by one-way ANOVA and post-hoc assays and student’ s t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, vs. NC or Vector control.
3.4. HnRNP L promotes tumor growth through immune escape in PCa tumors
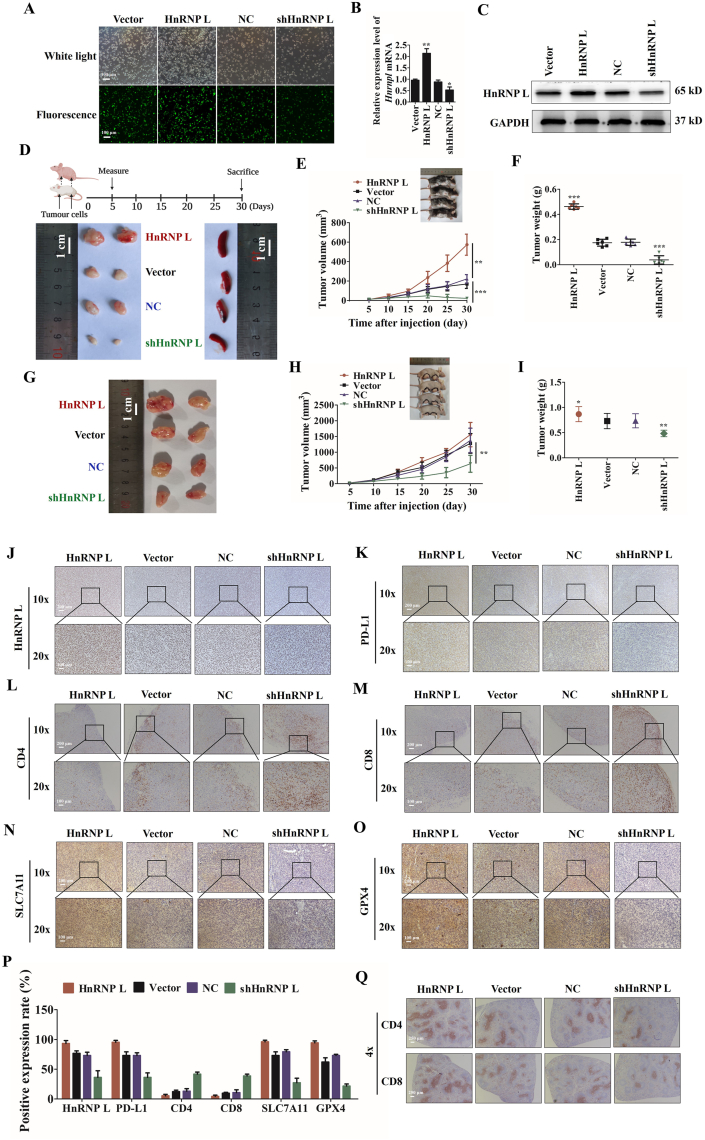
Based on the compelling in vitro evidence, we then evaluated the antitumor effect of HnRNP L inhibition in RM-1 xenograft models. Cell lines stably expressing HnRNPL were constructed by lentivirus vector transfection (Fig. 5A). Transfection efficiency was determined using qRT-PCR and Western blotting, and the results reveal that HnRNP L expression was significantly decreased in the HnRNP L knockdown group. Meanwhile, the expression of HnRNP L increased in the overexpression group compared with negative control cells, respectively (Fig. 5B and C). The transfected RM-1 cells (2 × 106) were implanted subcutaneously into the right armpit regions and right inguinal regions of each C57BL/6 mouse and each nude mouse. Interestingly, a knockdown of HnRNP L severely impaired the ability of the RM-1 cells to grow tumor in the immune competent C57BL/6 mice, while HnRNP L overexpression promoted the growth (Fig. 5D–F). In contrast, the same RM-1 cells grew faster when they were inoculated in the immunodeficient nude mice, while knocking down of HnRNP L only moderately retarded tumor growth in the nude mice (Fig. 5G–I). Further research on the mechanism of action of HnRNP L knockdown and overexpression on tumor growth was conducted by analyzing tumor protein expression, such as some immunological molecules and ferroptosis-related molecules, by immunohistochemistry. The results show that HnRNP L knockdown significantly reduced the expression of HnRNP L and PD-L1, which promoted the infiltration of CD4+ and CD8+ T cells, and decreased the SLC7A11 and GPX4 expression, which induced the ferroptosis. HnRNP L overexpression showed the opposite effect, which is good consistent with the in vitro results (Fig. 5J–Q). These data together suggest that HnRNP L knockdown in RM-1 cells could decrease the expression of PD-L1 in vivo, promote T cells mediated ferroptosis and thus inhibit tumor growth.
Figure 5.
HnRNP L promotes tumor growth through immune escape in PCa tumors. (A) The transfection efficiency of lentivirus vectors labeled with GFP. The ectopic expression of HnRNP L in RM-1 cells was analyzed using (B) qRT-PCR and (C) Western blotting. GAPDH was used as a loading control. Each bar represents the mean ± SD of three independent experiments. (D)–(F) 2 × 106 RM-1 cells (four groups: HnRNP L knockdown, NC, HnRNP L overexpression and Vector) were implanted subcutaneously into the right armpit regions and right inguinal regions of each C57BL/6 mouse. Tumor volumes were measured on the indicated days and drew into the tumor growth curves. (G)–(I) 2 × 106 RM-1 cells (four groups: HnRNP L knockdown, NC, HnRNP L overexpression and Vector) were implanted subcutaneously into the right armpit regions and right inguinal regions of each BALB/c nude mouse. Tumor size data (means ± SD, n = 6) were log-transformed before statistical analyses using generalized linear model. (J)–(O) Representative immunohistochemistry staining images of HnRNP L, PD-L1, CD4, CD8, SLC7A11, and GPX4 from tumor tissues (C57BL/6 mice) in each group. (P) Mean positive rates ± SD were shown (n = 6). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 (vs. NC or Vector Control). (Q) Representative immunohistochemistry staining images of CD4 and CD8 from spleen tissues (C57BL/6 mice) in each group.
3.5. HnRNP L regulates the expression of PD-L1 by stabilizing YY1 in CRPC cells
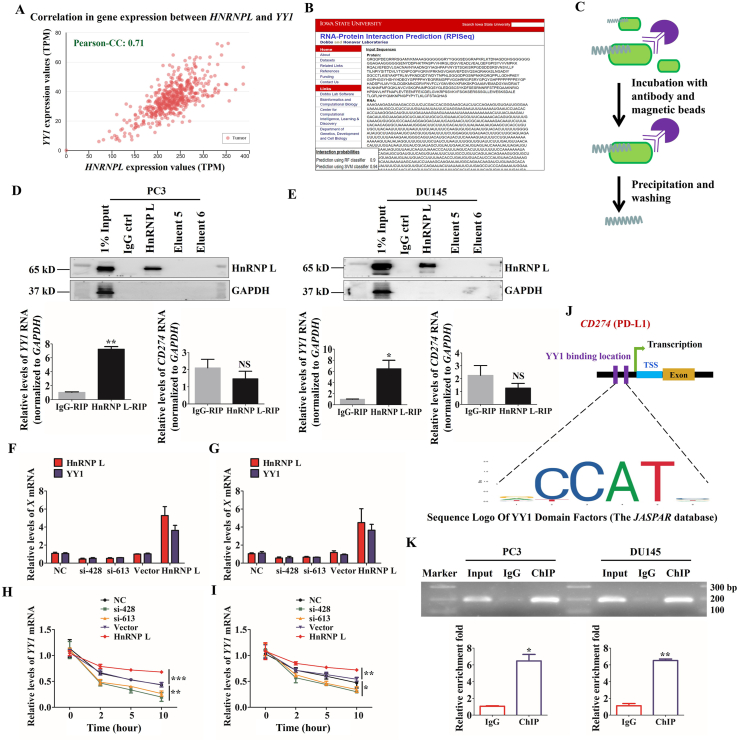
A recent review has demonstrated that PD-L1 is generally regulated by YY1, which was a transcriptional regulator participated in the life process of cells34. To clarify the possible contribution of YY1 to HnRNP L-mediated PD-L1 expression, we detected whether HnRNP L directly interacts with YY1. Through analyzing the TCGA database, we found that there was a high correlation between HNRNPL and YY1 (Fig. 6A). Besides, the data from RPISeq Software exhibited high probabilities of interaction between them (Fig. 6B). To this end, we performed RNA-binding protein immunoprecipitation (RIP) assay to confirm the interaction between HnRNP L and YY1 mRNA. Here we also tested the binding efficiency of HnRNP L antibody and found that there was little non-specific binding in our reaction system. The qRT-PCR results show that the ratio of co-purified YY1 mRNA in the HnRNP L antibody group to the IgG group was 6–8 in these two CRPC cells, while the difference for CD274 was not statistically significant (Fig. 6C–E). What's more, we found that HnRNP L inhibition significantly reduced the YY1 mRNA level; in contrast, overexpression of HnRNP L displayed the opposite effect (Fig. 6F and G). The qRT-PCR analysis of the expression of YY1 in 0, 2, 5, and 10 h after treating with actinomycin D in transfected CRPC cells (PC3 and DU145) revealed that HnRNP L possessed the potential enhancement for the stability of YY1 mRNA (Fig. 6H and I). In addition, we forecasted the binding site of YY1 on CD274 (PD-L1) gene through Jaspar Software (Fig. 6J). Furthermore, chromatin immunoprecipitation (ChIP) analysis of CRPC cells, which were transfected with HnRNP L plasmid and co-cultured with the activated Jurkat T cells revealed that endogenous YY1 occupied the promoter region of the CD274 gene (Fig. 6K). Taken together, these results suggest that HnRNP L upregulating the PD-L1 expression by directly binding to the YY1 mRNA.
Figure 6.
HnRNP L regulates the expression of PD-L1 by stabilizing YY1 in CRPC cells. (A) Correlation analysis of gene expression between HNRNPL and YY1 in specimens of PCa through TCGA database. (B) The interaction probabilities of HnRNP L and YY1 were predicted by RPISeq (RNA-Protein Interaction Prediction). (C) Schematic diagram of operation steps of RNA-immunoprecipitation (RIP) assay. (D) and (E) Adequate CRPC cell (PC3 and DU145) extracts were incubated with a monoclonal antibody for HnRNP L or IgG overnight at 4 °C before the purification of RNA and then Western blotting was performed to identify the efficiency of co-immunoprecipitation. IgG, the fifth and sixth eluent were served as negative control. YY1 mRNA identified as the co-purified mRNA with HnRNP L was proven by immunoprecipitation followed by qRT-PCR. (F) and (G) The qRT-PCR analysis of the expression of HnRNP L and YY1 after HnRNP L inhibition or overexpression in CRPC cells (PC3 and DU145). (H) and (I) The qRT-PCR analysis of the expression of YY1 in 0, 2, 5, and 10 h after treating with actinomycin D in transfected CRPC cells (PC3 and DU145). (J) Schematic diagram of YY1 binding location and sequence on human CD274 (PD-L1) gene. Identified YY1 binding sequence was compared with consensus sequence (CCAT sequence is essential for YY1 binding). TSS, transcription start site. (K) ChIP assay was carried out in PC3 and DU145 cells. Input served as a positive control for ChIP. IgG was used as a negative control for ChIP. The fold enrichment values of qRT-PCR were normalized to the positive control Input. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.6. HnRNP L inhibits Jurkat T cells-mediated ferroptosis of CRPC cells via the YY1/PD-L1 axis
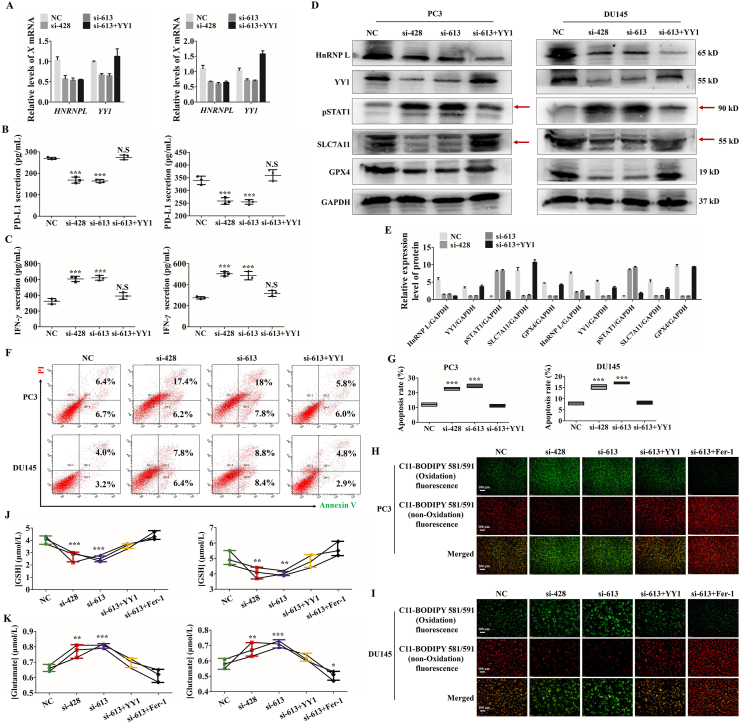
To detect the role of YY1 in HnRNP L-induced CRPC immune escape, YY1 was overexpressed in HnRNP L-inhibiting cells. The qRT-PCR, Western blotting and ELISA assays all demonstrated that YY1 overexpression attenuated si-HnRNP L-induced YY1 downregulation (Fig. 7A), the low release of PD-L1 from tumor cells (Fig. 7B) and the high release of IFN-γ from the activated Jurkat T cells (Fig. 7C), the activation of STAT1 signaling and the suppression of SLC7A11 and GPX4 (Fig. 7D and E). Consistent with the observations above, the cell death analysis by flow cytometry indicated that YY1 overexpression relieved the killing activity of Jurkat T cells to the HnRNP L-inhibiting cells (Fig. 7F and G). What's more, YY1 overexpression reversed HnRNP L inhibition induced the elevation of the lipid peroxide level (Fig. 7H and I) and an extensive amount of glutamate releasing into cultured media (Fig. 7J and K) in these two CRPC cells after co-culturing with the activated Jurkat T cells. To further identify the contribution of ferroptosis to HnRNP L-induced the cancer immune escape, we treated the si-HnRNP L transfected CRPC (PC3 and DU145) cells with ferrostatin-1 (Fer-1), which had been identified as a potent inhibitor of ferroptosis. As expected, we observed the similar effect with YY1 overexpression in PC3 and DU145 cells. All these results point out that HnRNP L promoted the PD-L1 expression and suppression of T cells activity by targeting YY1 in CRPC cells.
Figure 7.
HnRNP L inhibits Jurkat T cells-mediated ferroptosis of CRPC cells via the YY1/PD-L1 axis. (A) The qRT-PCR analysis of the expression of HnRNP L and YY1 in stable CRPC cells (PC3 and DU145) with or without HnRNP L inhibition and YY1 overexpression. (B) PD-L1 in cell culture medium from the stable CRPC cells (PC3 and DU145) with or without HnRNP L inhibition and YY1 overexpression was measured by enzyme-linked immunosorbent assay (ELISA). (C) IFN-γ in cell culture medium from the activated Jurkat T cells after co-cultured with the stable CRPC cells (PC3 and DU145) with or without HnRNP L inhibition and YY1 overexpression was measured by enzyme-linked immunosorbent assay (ELISA). (D) and (E) Western blotting analysis of the expression of HnRNP L, YY1, pSTAT1, SLC7A11 and GPX4 after co-culture with the activated Jurkat T cells in transfected CRPC cells (si-HnRNP L or/and YY1 overexpression). GAPDH was used as a loading control. Red arrow indicates the main bands for analysis. (F) and (G) Representative images of annexin-V/propidium iodide staining showing increased apoptosis in CRPC cells treated with si-HnRNP L and decreased apoptosis in CRPC cells treated with both si-HnRNP L and YY1 overexpression after co-culture with the activated Jurkat T cells (F). Statistical results were represented as mean ± SD of three independent experiments (G). (H) and (I) The changes of lipid ROS accumulation in transfected PC3 and DU145 (si-HnRNP L or/and YY1 overexpression or/and ferrostatin-1) cells after co-culture with the activated Jurkat T cells were measured by fluorescence microscope. (J) and (K) The stable CRPC cells (PC3 and DU145) with or without HnRNP L inhibition and YY1 overexpression or ferrostatin-1 were co-cultured with the activated Jurkat T cells for 48 h, and the amount of glutamate released into culture medium was measured. Indicated cells co-cultured with the activated Jurkat T cells for 48 h were lysed, and the intracellular GSH level was measured. The data are presented as mean ± SD (n = 3). The data were analyzed by one-way ANOVA and post-hoc assays and student’ s t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, vs. NC control.
3.7. Inhibition of HnRNP L enhances anti-PD-1 therapy efficacy by recruiting CD8+ T cells in PCa tumors
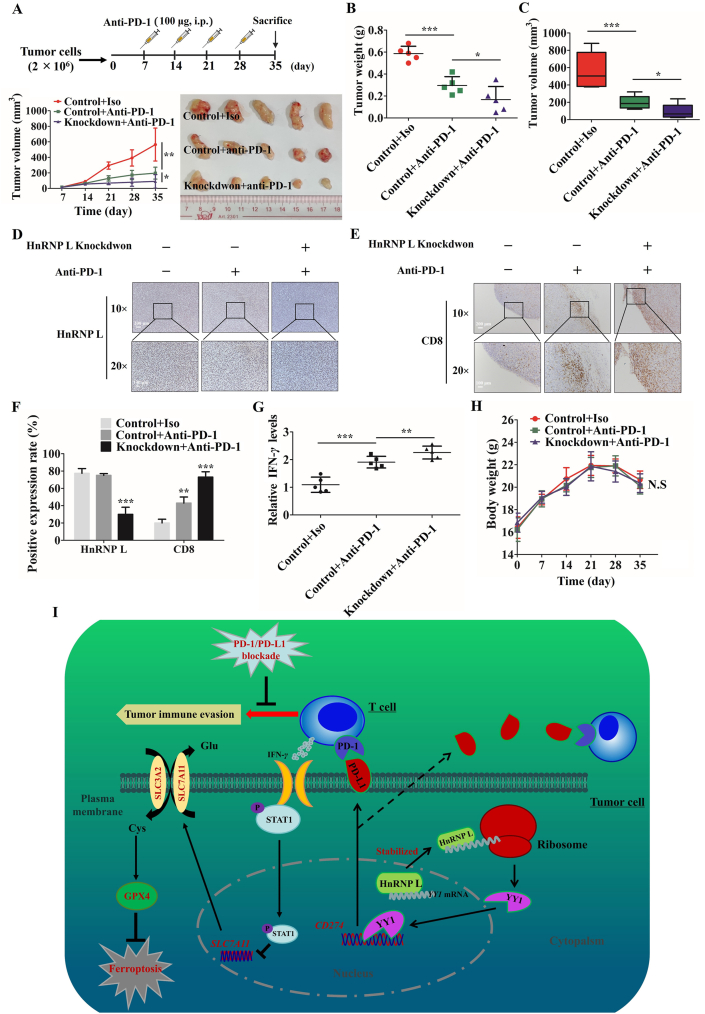
Thus, we have evaluated the functional importance of regulating PD-L1 expression by HnRNP L in tumor immune escape in vitro and in vivo. To further test the combination of HnRNP L knockdown with anti-PD-1 on CRPC immunotherapy, we carried out an in vivo experiment containing the control and HnRNP L-knockdown RM-1 tumour cells in C57BL/6 mice with or without anti-PD1 treating. The combination of HnRNP L knockdown with anti-PD-1 antibody therapy group showed a significant reduction in tumor burden than anti-PD-1 alone treatment group (Fig. 8A–C) with no clear evidence of cytotoxic activity as measured by the total body weight of the mice (Fig. 8H). Consistent with an enhanced anti-tumor effect, immune profiling of each cancer tissues indicated that more tumor infiltrating CD8+ T cells in mice receiving the combined HnRNP L knockdown and anti-PD-1 therapy (Fig. 8D–F). Importantly, the amount of interferon gamma (IFN-γ) was much more in HnRNP L knockdown and anti-PD-1 combination therapy group than other groups (Fig. 8G). All of the results demonstrated that HnRNP L knockdown enhanced the anti-PD-1 therapy in CRPC tumors by recruiting infiltrating CD8+ T cells.
Figure 8.
Inhibition of HnRNP L enhances anti-PD1 therapy efficacy by recruiting CD8+ T cells in PCa tumors. Tumor image (A), tumor weight (B) and tumor volume (C) from control and HnRNP L-knockdown RM-1 tumour cells in C57BL/6 mice with or without anti-PD-1 treating (n = 5 mice per group). (D) and (E) Representative immunohistochemistry staining images of HnRNP L and CD8 from tumor tissues (C57BL/6 mice) in each group. (F) Mean positive rates ± SD are shown (n = 5); ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. (G) IFN-γ in the mice tumors from these three groups was measured (n = 5). The collected tumors were homogenized and detected by using Quantikine ELISA (R&D Systems). (H) The body weight of each mouse was monitored during the experiment. N.S: P>0.05. (I) A consolidated model that illustrates a plausible sequence for the mechanism by which inhibition of HnRNP L downregulates PD-L1 and sensitizes castration-resistant prostate cancer cells to T cells killing.
4. Discussion
PCa is the most commonly diagnosed malignancy of the prostatic epithelium1. It can greatly shorten the lifespan and the life quality of patients. The current treatment for PCa includes surgical resection, chemotherapy, hormone therapy and radiation therapy, which have improved the survival rates. However, patients may inevitably develop severe complications, drug toxicity, therapy resistance and so on35,36. CRPC is considered the terminal stage of this disease37, and prolonging the occurrence of CRPC remains a therapeutic challenge. Immunotherapy with checkpoint inhibitors produced significant clinical responses in a subset of cancer patients who were resistant to prior therapies34. Thus, several efforts have been made to overcome or improve the resistance to checkpoint inhibitors, including regulating the expression of PD-L1 on cancer cells. To access the modulation of PD-L1 expression on prostate cancer cells, we have examined the role of HnRNP L in regulating tumor cell resistance to cytotoxic T-cell-mediated killing.
HnRNP L was originally identified as an RNA-binding protein that participates in the splicing, transportation and degradation of precursor mRNAs23,24,38. Several studies have reported that some splicing factors act as carcinogenic or anti-carcinogenic factors39, 40, 41. In addition, in our previous study, we found that HnRNP L could regulate the proliferation and apoptosis of prostate cancer cells29. However, the specific roles of HnRNP L in prostate cancer immune escape remain unclear. To address that issue, we first detected the co-expression of HnRNP L and PD-L1 in prostate cancer tissues and cells and found a significant positive correlation. Besides, consistent with this notion, we observed that HnRNP L regulated the expression and secretion of PD-L1 both in RNA and protein levels. The effects of altering HnRNP L expression provide further functional evidence of HnRNP L-mediated prostate cancer immune escape in vitro and in vivo. As we see, increased apoptosis in CRPC cells treated with si-HnRNP L and decreased apoptosis in CRPC cells treated with HnRNP L overexpression after co-culture with the activated Jurkat T cells, while the opposite effect in co-cultured Jurkat T cells. Knockdown of HnRNP L in RM-1 cells could decrease the expression of PD-L1 in vivo, promote immune response and thus inhibit tumor growth. Importantly, HnRNP L knockdown enhanced the anti-PD-1 therapy in CRPC tumors by recruiting infiltrating CD8+ T cells. The interesting research article published in Nature showed that CD8+ T cells played a pivotal role in cancer immunotherapy through ferroptosis32. Thus, we wondered that whether inhibition of HnRNP L enhanced the T-cell-mediated cancer cell killing partly via the increased levels of ferroptosis, which was mediated by CD8+ T cells, in castration-resistant prostate cancer.
Ferroptosis is a new cell death form distinct from apoptosis, necrosis, pyroptosis, and autophagy that can be driven by iron-dependent lipid ROS or small molecules42,43. Mechanistically, the ferroptosis is triggered by inhibition of the central regulating factor, SLC7A11, that indirectly inhibits GPX4 through hampering system Xc−, which is a cystine/glutamate antiporter that imports extracellular cystine in exchange for intracellular glutamate to generate glutathione33,44,45. Exploitation of ferroptosis for killing cancer cells in response to specific compounds are attracting more and more attention in cancer therapy46. Interestingly, the recent study by Wang et al.32 in Nature revealed that the activated T cells, especially CD8+ T cells, could induce ferroptosis in tumor cells through generating IFN-γ, which activated the Janus kinase (JAK) signal transducer and activator of the transcription 1 (STAT1) signaling pathway in tumor cells and then suppresses the transcriptional of SLC7A11 and contributed to lipid peroxidation and ferroptosis eventually. According to this, we show that HnRNP L knockdown inhibits the expression of PD-L1 and then relieve the suppression of cancer cells to T cells to a certain degree, which could produce more IFN-γ to induce the ferroptosis in CRPC cells via STAT1/SLC7A11/GPX4 signaling axis. What's more, an important finding in this study is that HnRNP L upregulate the expression of PD-L1 via strengthening the mRNA stability of YY1.
As a transcriptional regulator, YY1 has been identified that regulate many pathways participated in cell survival, growth, metabolism, epithelial to mesenchymal transition (EMT), resistance to chemotherapy and so on. Thus, many tumors have been shown to overexpress the YY1, which was correlate with poor outcomes47, 48, 49, 50. What's more, a review has provided systematic evidence demonstrating that YY1 could regulate the expression of PD-L1 via several indirect mechanisms, including P53, cytokines, growth factors and PTEN/PI3K/AKT/mTOR signaling pathways, etc.34. Clearly, the major mechanism by which YY1 regulates tumor resistance to cytotoxic immune functions is very likely through governing PD-L1 expression on tumor cells. In the present work, we identified that YY1 could directly bind to the promoter region of CD274 gene and boosted it transcription. In addition, overexpression of YY1 could attenuate the effect of HnRNP L knockdown on the expression of PD-L1 and the T cells-mediated cancer cells ferroptosis.
5. Conclusions
Taken together, results from our study reveal that inhibition of HnRNP L diminishes PD-L1 expression and promotes antitumor immunity partly via destabilizing YY1 mRNA in CRPC and enhancing the T-cell-mediated cancer cell ferroptosis (Fig. 8I). Besides, we also show that inhibition of HnRNP L could improve the anti-PD-1 therapy efficacy in vivo. Therefore, these data provide a strong evidence that HnRNP L inhibition functions as an immune-modulator by facilitating antitumor T-cell immunity and indicate that a potential utilization of HnRNP L inhibition in the immunotherapy of CRPC.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 81773277), Science and Technology Program of Guangzhou, China (Grant No. 201803010014), Guangdong Basic and Applied Basic Research Foundation (Grant Nos. 2020A1515110922 and 2019A1515110033, China), China Postdoctoral Science Foundation funded project (Grant Nos. 2018M643126 and 2019M662865), Distinguished Young Talents in Higher Education Foundation of Guangdong Province (Grant No. 2019KQNCX115, China) and Achievement Cultivation and Clinical Transformation Application Cultivation Projects of the First Affiliated Hospital of Guangzhou Medical University (Grant No. ZH201908, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.07.016.
Contributor Information
Chun Li, Email: 120601847@qq.com.
Bingkun Li, Email: 25840416@qq.com.
Xiangming Mao, Email: mxm@smu.edu.cn.
Author contributions
Xumin Zhou, Chun Li, Bingkun Li and Xiangming Mao designed research experiments; Xumin Zhou, Libin Zou and Hangyu Liao performed experiments; Xumin Zhou, Junqi Luo, Taowei Yang, Jun Wu, Wenbin Chen, Kaihui Wu, Shengren Cen, Daojun Lv, Fangpeng Shu and Yu Yang collected and analyzed data; Xumin Zhou, Chun Li, Binkun Li and Xiangming Mao prepared and edited the manuscript. All authors have given approval to the final version of the manuscript.
Conflicts of interest
All authors declared that they had no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Liu J.M., Yu C.P., Chuang H.C., Wu C.T., Hsu R.J. Androgen deprivation therapy for prostate cancer and the risk of autoimmune diseases. Prostate Cancer Prostatic Dis. 2019;22:475–482. doi: 10.1038/s41391-019-0130-9. [DOI] [PubMed] [Google Scholar]
- 3.Teo M.Y., Rathkopf D.E., Kantoff P. Treatment of advanced prostate cancer. Annu Rev Med. 2019;70:479–499. doi: 10.1146/annurev-med-051517-011947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson P.A., Arora V.K., Sawyers C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 6.O'Donnell J.S., Teng M., Smyth M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019;16:151–167. doi: 10.1038/s41571-018-0142-8. [DOI] [PubMed] [Google Scholar]
- 7.Feng M., Jiang W., Kim B., Zhang C.C., Fu Y.X., Weissman I.L. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer. 2019;19:568–586. doi: 10.1038/s41568-019-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 9.Cha J.H., Chan L.C., Li C.W., Hsu J.L., Hung M.C. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76:359–370. doi: 10.1016/j.molcel.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahmoud F., Shields B., Makhoul I., Avaritt N., Wong H.K., Hutchins L.F., et al. Immune surveillance in melanoma: from immune attack to melanoma escape and even counterattack. Cancer Biol Ther. 2017;18:451–469. doi: 10.1080/15384047.2017.1323596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du W., Zhu J., Zeng Y., Liu T., Zhang Y., Cai T., et al. KPNB1-mediated nuclear translocation of PD-L1 promotes non-small cell lung cancer cell proliferation via the Gas6/MerTK signaling pathway. Cell Death Differ. 2021;28:1284–1300. doi: 10.1038/s41418-020-00651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T.C., Sankin A.I., Porcelli S.A., Perlin D.S., Schoenberg M.P., Zang X. A review of the PD-1/PD-L1 checkpoint in bladder cancer: from mediator of immune escape to target for treatment. Urol Oncol. 2017;35:14–20. doi: 10.1016/j.urolonc.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Jing W., Guo X., Wang G., Bi Y., Han L., Zhu Q., et al. Breast cancer cells promote CD169+ macrophage-associated immunosuppression through JAK2-mediated PD-L1 upregulation on macrophages. Int Immunopharmacol. 2020;78:106012. doi: 10.1016/j.intimp.2019.106012. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Bu X., Wang H., Zhu Y., Geng Y., Nihira N.T., et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B., Song Y., Liu D. Recent development in clinical applications of PD-1 and PD-L1 antibodies for cancer immunotherapy. J Hematol Oncol. 2017;10:174. doi: 10.1186/s13045-017-0541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kline J., Gajewski T.F. Clinical development of mAbs to block the PD1 pathway as an immunotherapy for cancer. Curr Opin Investig Drugs. 2010;11:1354–1359. [PubMed] [Google Scholar]
- 17.O'Donnell J.S., Long G.V., Scolyer R.A., Teng M.W., Smyth M.J. Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treat Rev. 2017;52:71–81. doi: 10.1016/j.ctrv.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoffski P., Besse B., Gauler T., de Jonge M.J., Scambia G., Santoro A., et al. Efficacy and safety of biweekly i.v. administrations of the Aurora kinase inhibitor danusertib hydrochloride in independent cohorts of patients with advanced or metastatic breast, ovarian, colorectal, pancreatic, small-cell and non-small-cell lung cancer: a multi-tumour, multi-institutional phase II study. Ann Oncol. 2015;26:598–607. doi: 10.1093/annonc/mdu566. [DOI] [PubMed] [Google Scholar]
- 20.Brahmer J.R., Tykodi S.S., Chow L.Q., Hwu W.J., Topalian S.L., Hwu P., et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun C., Mezzadra R., Schumacher T.N. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinol-Roma S., Swanson M.S., Gall J.G., Dreyfuss G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol. 1989;109:2575–2587. doi: 10.1083/jcb.109.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shih S.C., Claffey K.P. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem. 1999;274:1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- 24.Hung L.H., Heiner M., Hui J., Schreiner S., Benes V., Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goehe R.W., Shultz J.C., Murudkar C., Usanovic S., Lamour N.F., Massey D.H., et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin Invest. 2010;120:3923–3939. doi: 10.1172/JCI43552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yau W.Y., Shih H.C., Tsai M.H., Sheu J.C., Chen C.H., Chow L.P. Autoantibody recognition of an N-terminal epitope of hnRNP L marks the risk for developing HBV-related hepatocellular carcinoma. J Proteomics. 2013;94:346–358. doi: 10.1016/j.jprot.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Hope N.R., Murray G.I. The expression profile of RNA-binding proteins in primary and metastatic colorectal cancer: relationship of heterogeneous nuclear ribonucleoproteins with prognosis. Hum Pathol. 2011;42:393–402. doi: 10.1016/j.humpath.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., Li Q., He J., Zhong L., Shu F., Xing R., et al. HnRNP-L promotes prostate cancer progression by enhancing cell cycling and inhibiting apoptosis. Oncotarget. 2017;8:19342–19353. doi: 10.18632/oncotarget.14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fei T., Chen Y., Xiao T., Li W., Cato L., Zhang P., et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 32.Wang W., Green M., Choi J.E., Gijon M., Kennedy P.D., Johnson J.K., et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockwell B.R., Friedmann A.J., Bayir H., Bush A.I., Conrad M., Dixon S.J., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hays E., Bonavida B. YY1 regulates cancer cell immune resistance by modulating PD-L1 expression. Drug Resist Updat. 2019;43:10–28. doi: 10.1016/j.drup.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Feldman B.J., Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 36.Sieber P.R. Emerging therapeutic for the treatment of skeletal-related events associated with metastatic castrate-resistant prostate cancer. Rev Urol. 2014;16:10–20. [PMC free article] [PubMed] [Google Scholar]
- 37.O'Reilly D., Johnson P., Buchanan P.J. Hypoxia induced cancer stem cell enrichment promotes resistance to androgen deprivation therapy in prostate cancer. Steroids. 2019;152:108497. doi: 10.1016/j.steroids.2019.108497. [DOI] [PubMed] [Google Scholar]
- 38.Guang S., Felthauser A.M., Mertz J.E. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol Cell Biol. 2005;25:6303–6313. doi: 10.1128/MCB.25.15.6303-6313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cappellari M., Bielli P., Paronetto M.P., Ciccosanti F., Fimia G.M., Saarikettu J., et al. The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene. 2014;33:3794–3802. doi: 10.1038/onc.2013.360. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Chen D., Qian H., Tsai Y.S., Shao S., Liu Q., et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26:374–389. doi: 10.1016/j.ccr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selvanathan S.P., Graham G.T., Erkizan H.V., Dirksen U., Natarajan T.G., Dakic A., et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc Natl Acad Sci U S A. 2015;112:E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao H., Bai Y., Jia Y., Zhao Y., Kang R., Tang D., et al. Ferroptosis is a lysosomal cell death process. Biochem Biophys Res Commun. 2018;503:1550–1556. doi: 10.1016/j.bbrc.2018.07.078. [DOI] [PubMed] [Google Scholar]
- 44.Xie Y., Hou W., Song X., Yu Y., Huang J., Sun X., et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen P., Wu Q., Feng J., Yan L., Sun Y., Liu S., et al. Erianin, a novel dibenzyl compound in Dendrobium extract, inhibits lung cancer cell growth and migration via calcium/calmodulin-dependent ferroptosis. Signal Transduct Target Ther. 2020;5:51. doi: 10.1038/s41392-020-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonavida B., Kaufhold S. Prognostic significance of YY1 protein expression and mRNA levels by bioinformatics analysis in human cancers: a therapeutic target. Pharmacol Ther. 2015;150:149–168. doi: 10.1016/j.pharmthera.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 48.Shi J., Hao A., Zhang Q., Sui G. The role of YY1 in oncogenesis and its potential as a drug target in cancer therapies. Curr Cancer Drug Targets. 2015;15:145–157. doi: 10.2174/1568009615666150131124200. [DOI] [PubMed] [Google Scholar]
- 49.Khachigian L.M. The Yin and Yang of YY1 in tumor growth and suppression. Int J Cancer. 2018;143:460–465. doi: 10.1002/ijc.31255. [DOI] [PubMed] [Google Scholar]
- 50.Cho A.A., Bonavida B. Targeting the overexpressed YY1 in cancer inhibits EMT and metastasis. Crit Rev Oncog. 2017;22:49–61. doi: 10.1615/CritRevOncog.2017020473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.