Abstract
tRNA is the most extensively modified RNA in cells. On average a bacterial tRNA contains 8 and a eukaryotic tRNA contains 13 modifications per molecule. Recent studies reveal that tRNA modifications are highly dynamic and respond extensively to environmental conditions. Functions of tRNA modification dynamics include enhanced decoding of specific codons in response genes on demand and regulation of tRNA fragment biogenesis. This review summarizes recent advances in the studies of tRNA modification dynamics in biological processes, tRNA modification erasers, and in human associated bacteria. Furthermore, we use the term “metaepitranscriptomics” to describe the potential and approach of tRNA modification studies in the natural biological communities such as the microbiomes.
Short summary:
tRNA is highly modified in cells and tRNA modifications respond extensively to environmental conditions to enhance translation of specific genes and produce tRNA fragments on demand. We review recent advances in tRNA sequencing methods, tRNA modification dynamics in biological processes, and tRNA modification studies in the natural communities such as the microbiomes.
Introduction:
In all domains of cellular life, transfer RNAs (tRNAs) are the most extensively modified RNA family in cells. On average a bacterial tRNA contains 8 and eukaryotic tRNA contains 13 modifications per molecule, corresponding to ~1 in 10 to ~1 in 5 residues being modified with critical implications on cellular physiology. In broad categories, tRNA modifications in the anticodon loop are crucial to fine-tune mRNA decoding, whereas modifications outside of the anticodon loop play roles in tRNA stability, folding, localization, and quality control (Fig. 1).
Figure 1: Broad categories of tRNA modifications.
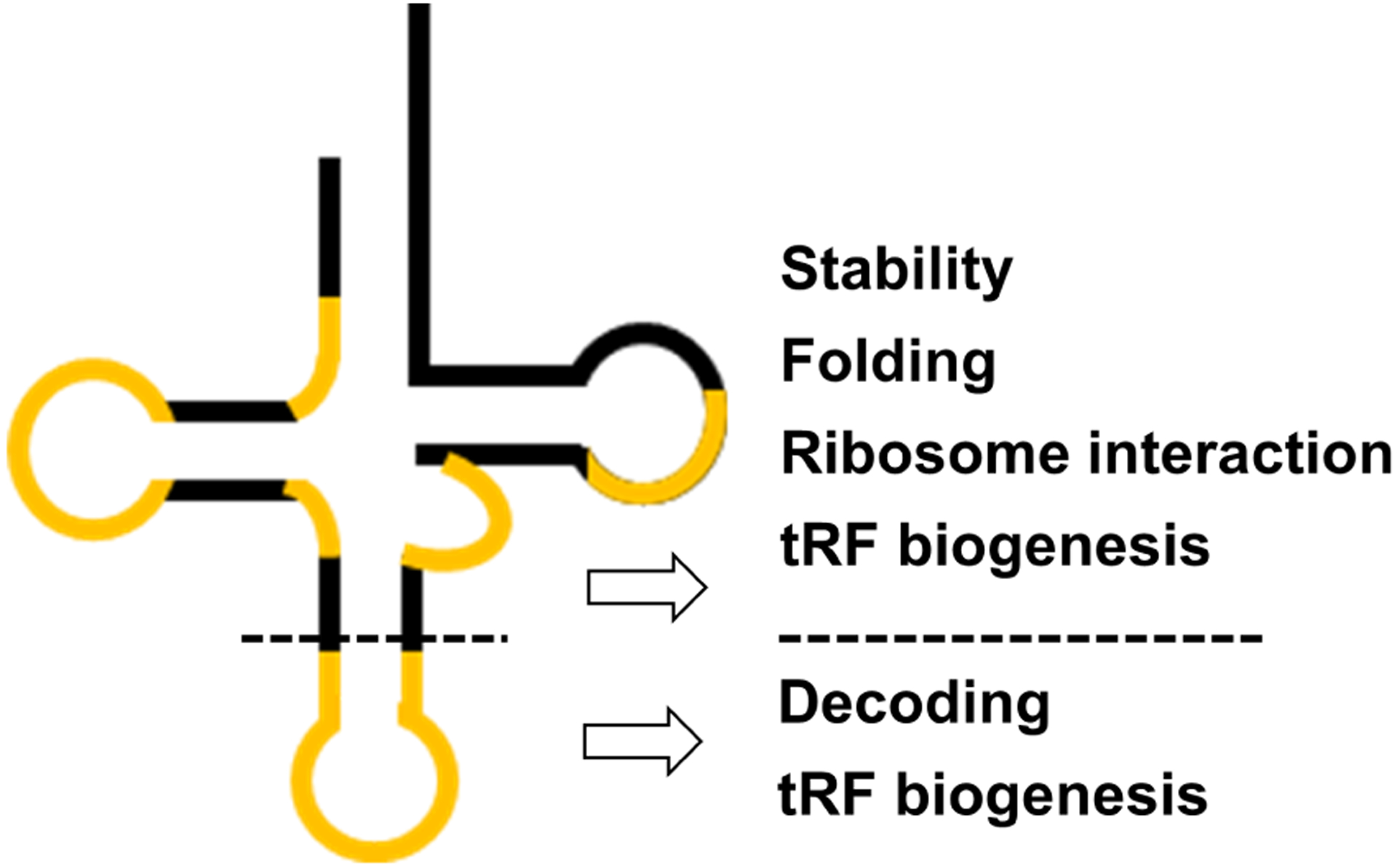
In the cloverleaf secondary structure of tRNA, the locations of the modified nucleotides are highlighted in orange. Dashed line separates the anticodon stem-loop from the rest of the tRNA. Broad categorization of tRNA modification function is on the right.
Chemically, the simplest tRNA modifications are base or 2’OH methylations. A human tRNA can contain up to 8 methylations including those ending in methyl groups and others followed by additional modifications (Boccaletto et al., 2018). Methylations are also primary markers for mRNA modification (e.g. N6-methyladenosine, m6A), chromosomal DNA modification (e.g. 5-methylcytosine, m5C), and proteins (e.g. N-methyl-lysine). On the other extreme, the wybutosine (W) modification in human tRNAPhe requires 7 enzymatic steps of de novo synthesis, and human tRNAAsp and tRNATyr are first modified to queuosine (Q), then glycosylated with mannose and galactose. This wide range of chemical complexity indicates that tRNA modifications are also part of the extensive cellular networks of metabolism.
Dynamic changes in the tRNA modification fraction at each site are expected among varying cellular activities and physiologies. Dynamic tRNA modification level changes can regulate translational efficiency and fidelity of specific genes that depend on the codon usage. Dynamic tRNA modification level changes can also regulate tRNA fragment biogenesis that in turn affects many cellular processes through their interactions with mRNAs and proteins. Many excellent reviews on tRNA modifications have been recently published that extensively describe our recent advances in the identification of tRNA modification enzymes (called writers), their functions in health and disease, the discovery and function of numerous new tRNA modifications, and the biology and mechanism of action of tRNA fragments (Boccaletto et al., 2018; de Crecy-Lagard et al., 2019; Huber et al., 2019; Kirchner and Ignatova, 2015; Schimmel, 2018; Suzuki, 2021).
This review focuses on the aspect of dynamic tRNA modifications: defined here as variations and changes in the modification fraction at specific sites that are dependent on cell type and cell state. We describe the recent technological advances in the studies of tRNA modification fraction, the biological investigations of tRNA modification response to stress and other cellular conditions, tRNA modification fraction response to physiological conditions and their potential functions, and finally an outlook on ‘metaepitranscriptomics’, an emerging area of study that provides a framework to study tRNA modifications in complex biological systems, such as the microbiome. The literatures in this area are increasing rapidly, we apologize to those we did not have the opportunity to cite in this review.
Sequencing methods of detection and quantitation of tRNA modifications
While liquid chromatography-mass spectrometry (LC/MS) continues to be a powerful approach to study tRNA modifications, recent advances in sequencing strategies have enabled comprehensive access to the modification dynamics with unprecedented throughput. Variations in the library preparation steps, that target the different chemical properties or response to enzyme treatments, provide much flexibility in either detecting a broad group of modifications or targeting a specific common modification. Leveraging specific properties of tRNAs and their modifications is a common theme in recently developed sequencing methods while others aim to alleviate common issues during library preparation steps through clever use of enzymes and specific adapters (Table 1). Several methods also examine tRNA dynamics on a broader, encompassing scale, such as within tissues or in naturally occurring, complex microbiomes. However, the existing sequencing methods target only a subset of tRNA modifications that are open to detection and quantitative assessment by reverse transcription (Fig. 2). There is much room for future development for more comprehensive and more precise measurements of tRNA modifications as current methods have many limitations (Table 1).
Table 1.
Summary of next-gen tRNA-seq methods
| Category | Method | Year Published | Description | Capabilities | Limitations | Reference |
|---|---|---|---|---|---|---|
| RT misincorporation signature | DM-tRNA-seq | 2015 |
|
|
|
Zheng et al., 2015 |
| ARM-seq | 2015 |
|
|
|
Cozen et al., 2015 | |
| mim-tRNA-seq | 2021 |
|
|
|
Behrens et al., 2021 | |
| Chemical treatment for specific modification | Ψ-seq; Pseudo-seq |
2014 2014 |
|
|
Schwartz et al., 2014; Carlile et al., 2014 | |
| RNA bisulfite sequencing | 2009 2012 2013 |
|
|
|
Schaefer et al., 2009; Squires et al., 2012; Edelheit et al., 2013 | |
| DM-Ψ-seq | 2020 |
|
|
|
Song et al., 2020 | |
| RBS-seq | 2019 |
|
|
|
Khoddami et al., 2019 | |
| AlkAniline-Seq TRAC-seq HAC-seq |
2018 2019 2021 |
|
|
|
Marchand et al., 2018; Lin et al., 2019; Cui et al., 2021 | |
| Library construction strategy | YAMAT-seq | 2017 |
|
|
|
Shigematsu et al., 2017 |
| Hydro-tRNAseq | 2017 |
|
|
|
Gogakos et al., 2017 | |
| LOTTE-seq | 2020 |
|
|
|
Erber et al., 2020 | |
| QuantM-tRNAseq | 2020 |
|
|
|
Pinkard et al., 2020 | |
| AQRNA-seq | 2021 |
|
|
|
Hu et al., 2021 |
TGIRT: Thermostable group II intron reverse transcriptase
RT: Reverse transcriptase.
CMC: 1-Cyclohexyl-(2-Morpholinoethyl)Carbodiimide.
Ψ: Pseudouridine.
Figure 2: Chemical structures of some tRNA modifications assessable by next-gen sequencing.
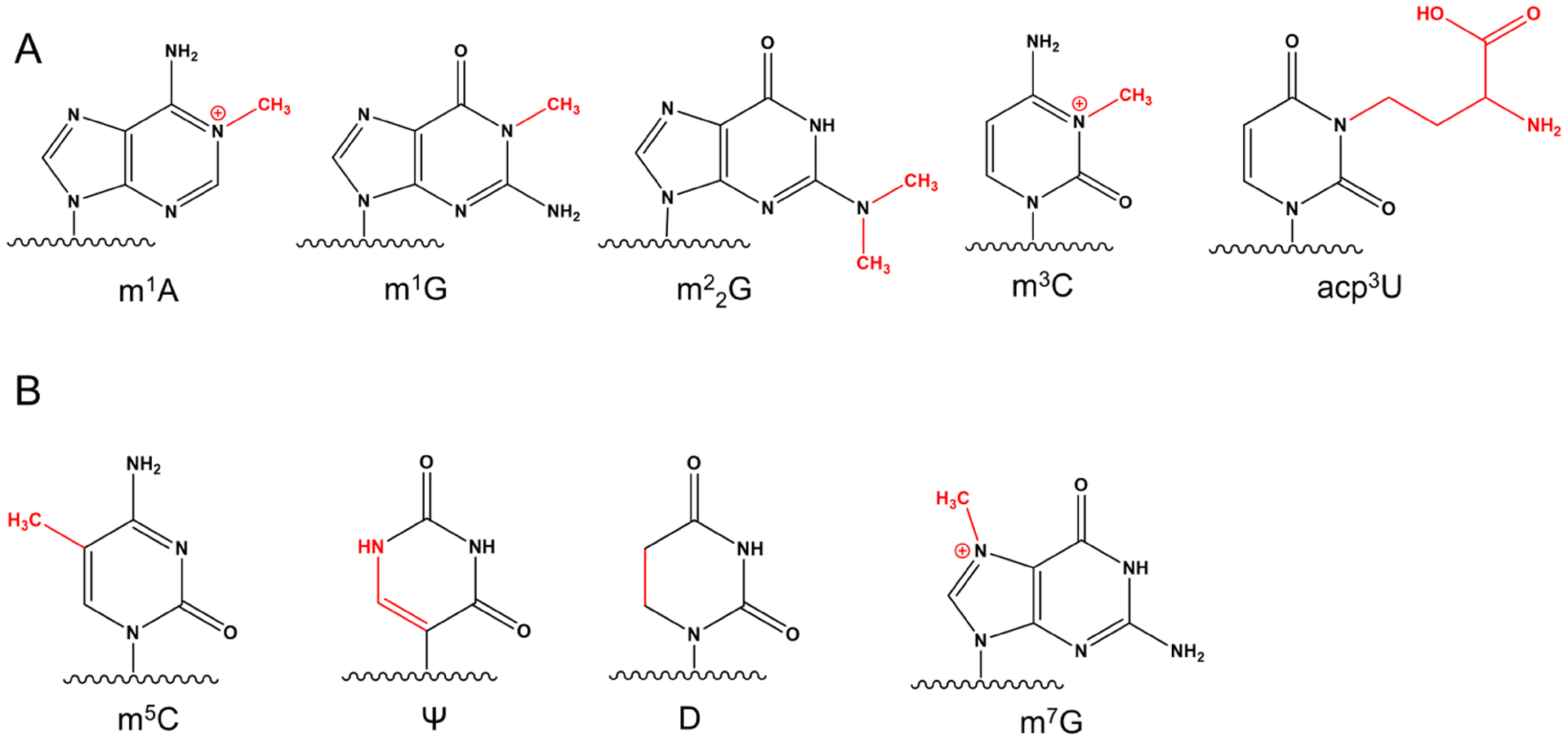
Modified chemical moieties are shown in red. (A) W-C face modifications: N1-methyladenosine (m1A), N1-methylguanosine (m1G), N2,2-dimethylguanosine (m22G), N3-methylcytidine (m3C), N3-(3-amino-3-carboxypropyl)uridine (acp3U). (B) After chemical treatment: 5-methylcytidine (m5C), pseudouridine (Ψ), dihydrouridine (D), N7-methylguanosine (m7G).
Reverse transcription misincorporation: m1A, m1G, m3C, m22G
Watson-Crick face methylations are abundant in tRNA; they introduce a major challenge in the reverse transcription step during cDNA synthesis. In eukaryotic tRNAs these include N1-methyladenosine (m1A), 1-methylguanosine (m1G), 3-methylcytidine (m3C), and N2,2-dimethylguanosine (m22G), which all cause some degree of misincorporation or truncated cDNA products during reverse transcription. However, several sequencing techniques take advantage of these observations of RT signatures derived from modifications. Ryvkin et al. developed the HAMR data analysis pipeline that identifies modifications through detection of nucleotides with significant sequencing error rates in Illumina sequencing results (Ryvkin et al., 2013). Detected modifications could be grouped into large modification families by similarities in their incorporation patterns.
The introduction of an enzymatic demethylation step in the sequencing library preparation allows for comparative analysis of treated and untreated samples to further pinpoint these modifications. ARM-seq (Cozen et al., 2015) and DM-tRNA-seq (Zheng et al., 2015) both take a similar approach in using the E. coli AlkB enzymes to demethylate RNA samples, and the latter also utilizes a highly processive thermostable group II intron reverse transcriptase (TGIRT) to increase read-through of these modifications in cDNA synthesis. These techniques solidify the characteristic properties of these methylations in producing specific RT signatures. Additional analysis of the DM-tRNA-seq results (Clark et al., 2016) introduces the modification index (MI) which combines the fraction of mutation and stop at each modification site to describe the semi-quantitative nature of measuring modification fractions.
Recently, mim-tRNAseq (Behrens et al., 2021) builds upon the analysis of modification-induced nucleotide misincorporation, with additional approaches to address cDNA synthesis issues and data analysis. mim-tRNAseq achieves a substantial increase in full length cDNA reads by using the TGIRT with DNA adapters at the 3’ end of tRNA and longer reaction time. Obtaining large portion of full-length tRNA reads is important in the precise mapping of mammalian tRNAs which are derived from many tRNA isodecoder genes with diverse sequences within the same anticodon family (Goodenbour and Pan, 2006).
Chemical treatment of specific modifications: Ψ, m5C, m7G, D, m3C.
Another fundamental theme in detecting and quantifying specific modifications by sequencing uses chemical treatments that cause targeted changes to select modification groups. Pseudouridine (Ψ) is a prevalent modification in all RNA families, and is also the most abundant modification in eukaryotic tRNA (Spenkuch et al., 2014). A common Ψ detection method uses the carbodiimide (CMC) reaction that specifically forms a chemical adduct for Ψ that can be detected by RT stops (Bakin and Ofengand, 1993). Ψ-seq (Schwartz et al., 2014) and Pseudo-seq (Carlile et al., 2014) use this CMC reaction approach to sequence Ψ in mRNAs and in tRNAs. DM-Ψ-seq (Song et al., 2020) combines demethylase treatment and CMC reaction which significantly enhances the data quality for Ψ analysis in tRNAs. Another Ψ detection method (RBS-seq) uses a bisulfite reaction that removes the Ψ base, thus producing a deletion signature in the RT reaction (Khoddami et al., 2019).
5-Methylcytosine (m5C) is another common modification in many RNA families. The most commonly used m5C detection method is the bisulfite treatment which converts unmodified C into U, but m5C remains as C in sequencing results. Several studies (Edelheit et al., 2013; Schaefer et al., 2009; Squires et al., 2012) apply this technique to study this modification in bacterial, archaeal and eukaryotic tRNAs.
Alkaline hydrolysis is not limited to specific modification changes, as shown by hydro-tRNAseq (Gogakos et al., 2017). Here, a limited alkaline hydrolysis step generates shorter, less structured RNA fragments containing fewer modifications, making them more amenable to cDNA library preparations. AlkAniline-Seq (Marchand et al., 2018) combines the alkaline hydrolysis step to generate abasic sites, followed by extensive 5’- and 3’-dephosphorylation and subsequent aniline cleavage to profile susceptible modifications of N7-methylguanosine (m7G), m3C and dihydrouridine (D).
Specific chemical reactions that target m7G (TRAC-seq) or m3C (HAC-seq) modifications result in highly efficient RT stops for their detection and quantitation (Cui et al., 2021; Lin et al., 2019).
A plethora of solutions to a common problem: Adapters, nanopore sequencing and new modification discovery
Several other approaches have been documented to overcome cDNA synthesis related issues to enrich for tRNA molecules, reduce truncated cDNA reads or increase full length tRNA reads, all resulting in higher quality sequencing results of tRNA. A common solution to these problems includes the construction of adapter molecules that select for tRNAs in the input sample, thus increasing the portion of sequencing data for tRNA in biological samples. Pang et al. (Pang et al., 2014) show the effects of stressors on tRNA levels in S. cerevisiae using two separate adapter ligation steps: the first adapter ligates selectively to the 3’ end of purified tRNA, while the second ligation to the 3’ end of the cDNA products enhances PCR amplification of truncated RT products. These two adapter ligation steps take into account modification induced falloffs during reverse transcription and increase read coverage. YAMAT-seq (Shigematsu et al., 2017) uses a Y-shaped adapter that specifically ligates to mature tRNA using T4 RNA Ligase 2. This high selectivity for full-length, mature tRNA is advantageous in comparing the tRNA expression levels in human cells using only a low amount of total RNA input. LOTTE-seq (Erber et al., 2020) addresses limitations of adapters that are selective for mature tRNA using an adapter capable of targeting both full length and prematurely terminated tRNA transcripts. This adapter is specifically ligated to the tRNA 3’-CCA end. Similarly, QuantM-tRNAseq (Pinkard et al., 2020) utilizes a split ligation strategy with complementary double-stranded adapters to monitor tRNA abundance and sequence variants in addition to tRNA modifications. AQRNA-seq (Hu et al., 2021) further addresses potential ligation biases towards specific RNA molecules in the library preparation steps. Using a two-step adapter ligation strategy with an optional AlkB treatment, Hu et al. compare their findings with commercially available small RNA sequencing kits and establishes an optimized pipeline with low level of length bias, as well as a potential direct correlation between sequencing read counts and RNA copy numbers.
These methods highlight the importance in examining and quantifying the potential biases and errors typical tRNA library preparation steps can introduce into the sequencing data, in which future methods and analysis pipelines should aim to further address, or at the very least, recognize (Table 1).
The emergence of nanopore sequencing technologies provides another framework in sequencing tRNA including the identification and quantification of modifications. One major benefit with nanopore sequencing is to bypass the issues associated with cDNA synthesis, as tRNAs can be sequenced directly. Nanopore also sequences single RNA molecules so that the coordination of multiple tRNA modifications can be studied simultaneously. However, the stable tRNA secondary structure can be problematic in translocating tRNA through the pores. Smith et al. (Smith et al., 2015) proposes possible methods to deal with linearizing tRNAs using specific adapters and polymerases to enhance processivity to avoid clogging the pores. Another limitation is that nanopore sequencing works poorly for short RNA, so that tRNA must be linked to long adaptor RNAs for sequencing runs. Thomas et al. (Thomas et al., 2021) sequence full length E. coli tRNAs by ligating double-stranded splint adapters to both ends of tRNA. The added bases to the 3’ and 5’ ends of tRNA enable efficient pore passthrough of both ends, which enables full-length tRNA to reach 76–92% of the aligned reads. In addition, systematic base miscalls indicate known tRNA modifications. With more nanopore tRNA sequencing methods currently in preprint, one can expect to see a boom of using nanopore to study tRNA modification dynamics.
With only a limited selection of complete tRNA modification profiles of mostly model organisms (Boccaletto et al., 2018), comparative analysis using these well described tRNA profiles is a useful approach for investigating the unknown tRNA profiles of other organisms. Kimura et al. (Kimura et al., 2020) leverages the known information on E. coli tRNA modifications to fully map and also identify novel V. cholerae specific tRNA modifications through combining tRNA sequencing and tRNA mass spectrometry.
tRNA modification dynamics in stress response and development
First global assessment
To investigate the dynamic modulation of tRNA modifications globally in yeast, Chan et al. developed a quantitative LC-MS/MS technique (Chan et al., 2010). Stress treatments cause a global tRNA modification profile reprogramming that is stress agent-specific and dose-dependent. Hydrogen peroxide, arsenite, methylmethane sulfonate (MMS), and hypochlorite induce distinct patterns of tRNA modification change. For example, the levels of 2’O-methyl-C (Cm), m5C, and m22G increase in response to H2O2 but decrease or are resistant to arsenite, MMS, and hypochlorite. As the concentration of H2O2 increases, the levels of m5C, Cm and m22G increase, while 5-methyl-U (m5U), m1G, m2G, 5-methoxycarbonylmethyl-2-thio-U (mcm5s2U), N6-isopentenyl-A (i6A), wybutosine (yW) and m1A decrease. MMS increases the level of m7G and decreases the level of m5C, mcm5s2U, Cm, i6A, and yW. tRNA 5-methoxycarbonylmethyl-U (mcm5U), m3C, m7G, mcm5s2U, i6A, yW, m5C, and Cm levels only decrease at highest concentration of NaAsO2 of 60 μM. NaOCl also shows dose-dependent effect on the increased levels of 2’O-methyl-A (Am) and 2’O-methyl-U (Um) and decreased levels of m5C. These changes in tRNA modification levels can be derived from the induction of new modification enzymes, altered activity of existing modification enzymes, or selective degradation of modified or unmodified tRNAs.
tRNA modifications in the anticodon loop affect decoding of stress-response genes
The m5C34 modification in tRNALeu(CAA) is the anticodon wobble nucleotide whose installation is catalyzed by TRM4 in yeast. H2O2 treatment increases its level (Chan et al., 2012). Luciferase reporter and proteomic analysis show that elevated level of m5C34 enhances selective translation of UUG codon enriched mRNA (e.g. the ribosomal protein RPL22A). Deletion of TRM4 or RPL22A results in cytotoxic hypersensitivity to oxidative stress. Thus, the wobble m5C modification and the m5C dependent upregulation of RPL22A is crucial for oxidative stress response in yeast.
The DNMT2 dependent tRNA m5C38 modification also plays an important role in stress response. m5C38 modulates the stability and fragmentation of substrate tRNAs in the bone marrow. Loss of m5C38 in tRNAAsp leads to misincorporation of near-cognate amino acids during translation of Asp codons in primary bone marrow cells (Tuorto et al., 2015). DNMT2 mutant fruit flies are more sensitive to oxidative stress. Under heat shock, DNMT2 re-localizes to stress granules in Drosophila ovaries (Schaefer et al., 2010).
The wobble anticodon uridines of tRNA are extensively modified. O2 of U34 can be modified to 2-thiouridine (s2U) by the URM1 pathway proteins (Chowdhury et al., 2012; Marelja et al., 2008; Noma et al., 2009; Schlieker et al., 2008; Termathe and Leidel, 2018). Modification at C5 of U34 generates 5-carboxymethyluridine (cm5U), 5-methoxycarbonylmethyluridine (mcm5U), 5-(carboxyhydroxymethyl)uridine methyl ester (mchm5U), and 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um) by the elongator complex consisting of ELP1-ELP6, ALKBH8 or TRMT9L (TRM9 in yeast) (Dauden et al., 2018; Fu et al., 2010a; Fu et al., 2010b; Songe-Moller et al., 2010). The URM1 and ELP pathway dependent mcm5U and mcm5s2U modifications increase in nutrient starvation and oxidative stress. Translation assays with tRNAs lacking mcm5U and mcm5s2U modifications show impaired translation of genes enriched with AAA, CAA, and GAA codons in S. cerevisiae and C. elegans (Deng et al., 2015; Nedialkova and Leidel, 2015; Rezgui et al., 2013). Both mcm5U and mcm5s2U modulate translation by promoting binding of tRNALys(UUU) to the ribosomal A-site. Loss of TRM9 causes decreased levels of wobble mcm5U and mcm5s2U modifications leading to decreased translation fidelity and increased levels of unfolded proteins which induces unfolded protein and heat shock responses (Nedialkova and Leidel, 2015; Patil et al., 2012a).
Hydroxyurea (HU)-induced S-phase cells have higher level of mcm5U which enables efficient translation of AGA codons and the RNR1 protein (Patil et al., 2012b). The level of mcm5U oscillates during the cell cycle. Loss of mcm5U modification causes a codon-specific downregulation of RNR1 and a delay in transition into S-phase after DNA damage (Patil et al., 2012b). Loss of mcm5s2U34 also induces multiple starvation responses in yeast (Bruch et al., 2020) and confers sensitivity to H2O2 stress (Garcia et al., 2016). Overexpression of tRNALys(UUU) largely suppresses ribosome pausing and the sensitivity to oxidative stress and simultaneously restores protein homeostasis, indicating that the stress sensitivity is related to the U34 modified tRNAs-mediated translation (Nedialkova and Leidel, 2015). Loss of s2U34 thiolation in tRNA leads to impaired global translation and confers resistance to tunicamycin. The level of s2U34 is reduced at elevated temperatures (Alings et al., 2015; Damon et al., 2015) which is due to the synthesis of newly unmodified tRNAs, not the degradation of s2U34 containing tRNAs nor the removal of s2U34 from tRNA (Alings et al., 2015). During nitrogen deprivation, mcm5U and mcm5s2U modification-dependent reciprocal regulation of TORC signaling and tRNA modification is activated (Candiracci et al., 2019).
Depletion of mcm5s2U34 installation enzymes in human breast cancer cell lines (MCF7 and MDA-MB231) leads to translational defects including ribosome pause and accumulation of ribosomes on XAA codons enriched transcripts (Rapino et al., 2021). The U34 wobble modification status regulates the translation of specific penta-hydrophilic amino acid motifs that determines the fates of the protein product such as aggregation, degradation, and decreased expression.
m3C modification is present at positions 32 in the anticodon loop of tRNASer/tRNAThr/tRNAArg (Clark et al., 2016). METTL2A/B and METTL6 catalyze the formation of m3C32 in cytosolic tRNAs, METTL2B may be responsible for the formation of m3C32 in mitochondrial tRNAs (de Crecy-Lagard et al., 2019; Ignatova et al., 2020; Xu et al., 2017). METTL6 methylates tRNASer in vitro and in human cells. METTL6 knockout leads to global changes in translation profile and ribosome occupancy, as well as loss of mouse stem cell pluripotency (Ignatova et al., 2020). Stress response studies of m3C tRNA modification in yeast reveal a stress-specific reprogramming of modifications and selective expression of different classes of stress response proteins mediated by codon-biased mRNAs translation (Chan et al., 2015). Stress-induced tRNA modification and mRNA expression patterns can distinguish different types of stresses in yeast exposed to multiple oxidants and alkylating agents. For example, SN2 alkylating agents cause increase of m3C32 in tRNAThr(IGU), which leads to selective expression of ACC/ACU codon enriched membrane protein genes (Chan et al., 2015).
N6-threonylcarbamoyladenosine (t6A) and 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A) are present at position 37 in the anticodon loop of tRNAs with NNU anticodons. t6A is catalyzed by a group of 6 proteins in the cytosol and YRDC and OSGEPL1 in mitochondria (de Crecy-Lagard et al., 2019; Thiaville et al., 2014; Zhou et al., 2020). CDKAL1 is responsible for the formation of ms2t6A37 in tRNALys(UUU) (Wei et al., 2011). The ms2t6A37 modification is crucial for maintaining translation fidelity of AAA and AAG codons. Depletion of ms2t6A37 in tRNALys(UUU) in mouse pancreatic β cell through CDKAL1 knockout causes impaired translation of proinsulin due to misincorporation of non-cognate amino acids at lysine codons (Wei et al., 2011). Loss of t6A modification in yeast confers sensitivity to stress treatments such as heat, ethanol, and salt and TOR pathway inhibitors, and leads to severe growth defects that cannot be rescued by the overexpression of any individual tRNA (Thiaville et al., 2016). t6A37 level is crucial for codon-biased mitochondrial translation under physiological conditions; it is also sensitive to environmental availability of CO2/bicarbonate which is rate-limiting for t6A37 formation in human mitochondria (Lin et al., 2018). t6A37 depletion by knockout of OSGEPL1, one of the enzymes required for t6A, leads to respiratory defect and reduced mitochondrial translation.
5-oxyacetyluridine (cmo5U) is present at the wobble anticodon position of four tRNA families. cmo5U is crucial for expanding the ability of wobble U to read G, A, U, and sometimes C (Nasvall et al., 2007; Weixlbaumer et al., 2007). CMOB is responsible for converting 5-hydroxyuridine (ho5U) to either 5-methoxyuridine (mo5U) or cmo5U based on the availability of S-carboxymethyl-S-adenosylmethionine (carboxy-SAM). CMOM further modifies cmo5U to mcmo5U (Kim et al., 2015; Sakai et al., 2016). Under constant hypoxic stress, M. bovis BCG shows distinct global change in 40 tRNA modifications in the three classical phases of hypoxia induced persistence of aerated growth and non-replicating persistence stages 1 and 2 (Chionh et al., 2016). Early hypoxia specifically increases the level of cmo5U and the abundance of tRNAThr(UGU) which leads to genome-wide codon-biased translation of mRNA enriched in ACG/ACA codons. The codon-mediated translation of DOSR as the master regulator of hypoxic bacteriostasis is confirmed and analyzed by codon re-engineering of DOSR.
5-taurinomethyluridine (τm5U) and 5-taurinomethyl-2-thiouridine (τm5s2U) are present at the wobble anticodon position of several mitochondrial tRNAs in mammals (Suzuki and Suzuki, 2014; Suzuki et al., 2002). MTO1 and GTPBP3 are responsible for the formation of the τm5U modification, while MTU1 is responsible for the formation of 2-thio group of τm5s2U (Asano et al., 2018; Suzuki and Suzuki, 2014; Umeda et al., 2005). 5,10-methylene-tetrahydrofolate and taurine are the metabolic sources of τm5U34 in mitochondrial tRNA (Asano et al., 2018). Cells salvage taurine from the medium (Suzuki et al., 2002) and the availability of taurine affects the τm5U34 modification level. Loss of τm5U34 by GTPBP3-knockout leads to respiratory defects and reduced mitochondrial translation. Taurine depletion decreases τm5U(s2)34 levels in HeLa cells and animal tissues, and leads to the replacement of τm5U34 modification by 5-carboxymethylaminomethyluridine (cmnm5U) where the taurine moiety is replaced by glycine in mitochondrial tRNAs (Asano et al., 2018).
Queuosine (Q) is present at the wobble anticodon position of tRNATyr/tRNAHis/tRNAAsn/tRNAAsp (Fergus et al., 2015). Q in tRNATyr and tRNAAsp is further glycosylated by galactose and mannose at the cis-diol group to generate galactosyl-Q (galQ) and mannosyl-Q (manQ) (Kasai et al., 1976; Kasai et al., 1975). QTRT1/QTRT2 heterodimer complex catalyzes the incorporation of queuine that is salvaged from diet and gut microbiome into tRNA (Boland et al., 2009). Q levels in tRNAs are controlled over development stages (White and Tener, 1973) and dynamically dependent on the availability of queuine in the diet (Reyniers et al., 1981) as well as the composition and functioning of the gut microbiome (Farkas, 1980). Q modification in tRNAHis enhances decoding of U-ending codons (Meier et al., 1985). In drosophila species, Q modification level changes correlate with genome-wide alteration of Tyr/His/Asn/Asp codons which is consistent with coordination of codon-mediated expression and translational fidelity (Zaborske et al., 2014). Queuine availability fine-tunes the translational speed and fidelity in S. pombe. Q modification in tRNAAsp and tRNAHis increases the translation speed at cognate C-ending codons while Q modification in tRNAAsn and tRNATyr decreases the translation speed at cognate U-ending codons (Muller et al., 2019). Q enhances the translation fidelity through inhibiting second-position misreading of the glycine codon (GGC) by tRNAAsp. The presence of Q modification moderately impairs mitochondrial functions under normal growth conditions by reducing the translation of mitochondrial proteins, while Q suppresses the inhibition effect on cell growth under CaCl2 induced stress (Muller et al., 2019).
Q levels regulate DNMT2-dependent m5C38 methylation of tRNAAsp. Q modification stimulates m5C38 installation in S. pombe and D. discoideum (Muller et al., 2015), in E. histolytica (Nagaraja et al., 2021), and in human cells (Tuorto et al., 2018).
Q modification increases the resistance of E. histolytica to oxidative stress and suppresses the downregulation of translation caused by oxidation stress through promoting the translation of oxidative stress response proteins, such as Hsp70, antioxidant enzymes, and DNA repair proteins (Nagaraja et al., 2021).
In human cells, queuine depletion causes reduced translation fidelity and formation of unfolded proteins that triggers endoplasmic reticulum stress and the unfolded protein response in HeLa and HCT116 cells and in germ-free mice (Tuorto et al., 2018). In HeLa cells, queuine depletion significantly impairs mitochondrial function, characterized by increased mitochondrial proton leak, decreased ATP synthesis, and reduction in cellular ATP levels. A breast tumor mouse model shows significant enrichment of microbes in the xenograft tumors in QTRT1 knockout tumor cells, indicating a possible role of Q modification in microbial recruitment to tumor sites (Zhang et al., 2020).
Inosine is present at positions 34 and 37 in the anticodon loop of eukaryotic tRNAs with ANN anticodons. ADAT2/ADAT3 are responsible for the formation of I at position 34 (Torres et al., 2014), and ADAT1 catalyzes the formation of I37 (Maas et al., 1999). In Arabidopsis, loss of the I37 modification due to the catalytically dead mutation of AtTAD1 shows no effect on tRNAAla(AGC) stability and no phenotype under normal conditions, but gains less biomass under cold and heat shock stress treatments (Zhou et al., 2013).
The I34 modification is conserved in eukaryotic tRNAs, it occurs in 8 human tRNA isoacceptors of all 3–4 codon boxes of Ala, Arg, Leu, Ile, Pro, Ser, Thr, Val with the exception of Gly (Novoa et al., 2012). Inosine expands the decoding potential in bacterial tRNAArg(ICG) to U/C/A ending codons. In human, I34 modification is catalyzed by ADAT2/ADAT3 heterodimer complex (hetADAT) which co-localizes in the nucleus. The level of I34 modification is dynamically controlled in response to the hetADAT protein levels (Torres et al., 2015). To our knowledge, dynamic changes of I34 modification in response to cellular conditions have not been characterized.
tRNA modifications outside of anticodon loop affect tRNA stability and localization
m5C is present at many positions in tRNA. NSUN2 is responsible for the formation of m5C at positions 40, 48–50 in the anticodon stem and T stem. (Blanco et al., 2014; Tuorto et al., 2012). NSUN6 catalyzes the formation of m5C at position 72 in the acceptor stem (Haag et al., 2015). m5C modification plays crucial roles in stress resistance and neurological development. Deletion of NSUN2 leads to the loss of m5C in specific tRNAs in mice and humans (Blanco et al., 2014). Loss of NSUN2-specific m5C modification does not reduce the abundance of specific tRNAs but causes an accumulation of 5’ tRNA-derived small RNA fragments (5’ tRF) by promoting angiogenin-mediated tRNA cleavage. During UV radiation and NaAsO2 treatment, NSUN2 re-localizes to nucleoplasm and cytoplasmic granules, leading to decreased methylation activity of NSUN2 on tRNA and increased 5’ tRF. 5’ tRF co-localizes with NSUN2 in cytoplasmic stress granules. NSUN2 depleted mouse and human cells are sensitive to UV radiation and NaAsO2 mainly due to the loss of NSUN2-mediated tRNA methylation. The accumulation of 5’ tRFs caused by NSUN2 loss inhibits translation, and this effect is sufficient to trigger cellular stress response pathways that lead to impaired survival of cortical, hippocampal and striatal neurons.
N2,N2-dimethylguanosine (m22G) is installed by TRMT1 and present at position 26 at the junction of anticodon and D stems in about half of cytosolic tRNAs and mitochondrial mt-tRNAIle (Clark et al., 2016; de Crecy-Lagard et al., 2019; Dewe et al., 2017). Loss of m22G modification due to TRMT1 knockout leads to impaired cell proliferation, disrupted global protein translation, and redox homeostasis perturbations, which leads to increased endogenous ROS levels and confers hypersensitivity to oxidative stress (Dewe et al., 2017).
3-(3-amino-3-carboxypropyl)uridine (acp3U) is present at position 20/20a in the D loop of human tRNAs, and position 47 in the variable loop of E. coli tRNA (Takakura et al., 2019). Acp3U47 in E. coli is installed by tapT (yfiP) and is crucial for tRNA thermal stability. Loss of acp3U47 in E. coli ΔtapT mutant also leads to genome instability under continuous heat shock stress. The human tapT homologues (DTWD1 and DTWD2) are shown to mediate the formation of acp3U in cytosolic tRNAs. DTWD1 and DTWD2 double knockout decreases cell growth (Takakura et al., 2019).
tRNA modifications and tRNA-derived fragments (tRFs)
tRNA fragments (tRF) are cleavage products derived from pre-tRNAs and mature tRNAs that have many biological functions such as regulating translation, enhancing cell survival, and mediating cell differentiation (Anderson and Ivanov, 2014). tRNA modifications play an important role in the regulation of tRNA cleavage and fragment generation. For example, DNMT2 dependent m5C38 modification (Blanco et al., 2014; de Crecy-Lagard et al., 2019; Schaefer et al., 2010; Tuorto et al., 2012) protects substrate tRNAs from angiogenin-mediated cleavage under oxidative stress and heat shock (Schaefer et al., 2010). The dynamic modulation of tRNA modifications by stress treatments leads to a dynamic pool of tRFs. The roles of tRNA modifications in tRF biogenesis and function have been summarized in several recent reviews (Durdevic and Schaefer, 2013; Fagan et al., 2021; Guzzi and Bellodi, 2020; Lyons et al., 2018). tRNA modifications such as m1A, Ψ, m5C, m3C, and Q can protect full-length tRNAs from stress-induced ribonucleases cleavage (Blanco et al., 2014; Chen et al., 2019; Schaefer et al., 2010; Wang et al., 2018). Other modifications such as mcm5s2U34 and Q may promote tRF generation (Donovan et al., 2017; Jablonowski et al., 2006). Besides, modifications in the tRNA fragments may be crucial for tRF stability and function. For example, Ψ is crucial for fine-tuning the activities of tRNA fragments in stem cells (Guzzi et al., 2018).
Erasing tRNA modifications
tRNA methylations are reversible and dynamically controlled through the action of tRNA methylases and demethylases. Currently, three tRNA demethylases, called erasers have been identified in humans: ALKBH1, ALKBH3, and FTO (Chen et al., 2019; Liu et al., 2016; Ueda et al., 2017; Wei et al., 2018). These erasers help control the dynamic level of tRNA methylations which fine-tune protein translation and tRNA fragment generation.
ALKBH1 demethylates m1A58 modification in ~10 tRNA isoacceptor families (Liu et al., 2016). M1A58 is located in the T loop of tRNA and adds a positive charge to the tRNA. ALKBH1 depletion leads to increased m1A58 levels in the target tRNA. ALKBH1 depletion also leads to an increase of tRNAiMet level. Coupled with the elevated protein translation and cell proliferation, these results suggest that m1A58 demethylation regulates translational initiation and elongation. In response to glucose deprivation, ALKBH1 protein level increases and the m1A level in target tRNA decreases which can explain the downregulation of translation under glucose starvation (Liu et al., 2016). ALKBH1-mediated tRNA demethylation also shows stress-specific patterns. Depletion of ALKBH1 improves both apoptosis and necrosis and reduced tRNA cleavage under arsenite stress. ALKBH1 depletion partially rescues tRNA fragment generation under arsenite and antimycin A stress, and conversely ALKBH1 overexpression enhances tRNA fragment generation through modulating m1A58 demethylation (Rashad et al., 2020).
In mitochondria, ALKBH1 can demethylate m1A in the bodies of tRNAArg and tRNALys (Kawarada et al., 2017), although the function of mt-tRNA m1A demethylation is unclear. ALKBH1 plays a dual role in tRNA modification, as it is also the writer of the cytosolic tRNALeu and mt-tRNAMet 5-formyl-C (f5C); depletion of ALKBH1 reduces mitochondrial translation and respiratory complex activities (Haag et al., 2016; Kawarada et al., 2017).
ALKBH3 demethylates m1A58 in 12 tRNA isoacceptor families and m3C in 4 tRNA families (Chen et al., 2019; Ueda et al., 2017). Unlike ALKBH1, ALKBH3 knockdown reduces cell proliferation through downregulation of translation (Chen et al., 2019; Ueda et al., 2017). ALKBH3 dependent tRNA demethylation also affects tRNA cleavage and fragment generation by angiogenin. These demethylation-dependent tRFs interact with the cytochrome c protein to reduce cell apoptosis and play a role in ALKBH3-induced cancer cell progression (Chen et al., 2019).
The third tRNA demethylase is FTO which also demethylates N6-methyladenosine (m6A) in mRNA. FTO demethylate m1A58 in many tRNA isoacceptor families, and codon-specific luciferase reporter assays indicate that FTO depletion leads to upregulation of translation (Wei et al., 2018).
These results from tRNA eraser studies suggest a role of the tRNA demethylation in the dynamic regulation of cellular functions in response to environmental stimuli. ALKBH1, ALKBH3, and FTO can all demethylate m1A58 in tRNAs. How these three demethylases interact and cooperate on m1A58 demethylation and cellular function needs further exploration.
Recently, ALKBH7 was shown to demethylate m22G in mitochondrial tRNAIle and m1A in mitochondrial tRNALeu1 pre-tRNA regions within nascent mitochondrial polycistronic RNA. Demethylation by ALKBH7 is crucial for normal processing of polycistronic mitochondrial RNA, which regulates steady-state mitochondrial RNA levels and downstream protein translation. Depletion of ALKBH7 leads to downregulation of mitochondrial proteins and decreases mitochondrial activity in human cancer cell lines and mouse tissues (Zhang et al., 2021).
tRNA modifications in the microbiome: a new world
A myriad of bacteria is present in a vast variety of environments ranging from the human gut to the bottom of the ocean, yet only a modest number of studies have been conducted beyond the laboratory cultures. Below we will briefly review some studies framed within the context of the interactions with human cells or in human health, highlighting another biological axis for future investigations.
Gut bacteria respond to food and antibiotic intake
The mammalian gut microbial communities live in multiple different environments, depending on the type of diet and metabolite intake by the host. Bacteria in the gut respond to these changes, either to utilize the influx of new nutrients, resist changes to the existing environment or respond in different manners to ensure survival.
One such example is the regulation of tRNA 2’-O-methylation (Nm) in E. coli tRNAs upon the onset of mild antibiotic stress and limited nutrient supply. The regulation of Nm modification does not significantly influence translation, but rather, the immune-stimulatory properties of the bacteria (Galvanin et al., 2020). tRNA 2’-O-methlyations, specifically at position G18 in the D loop, effectively suppress Toll-like receptors (TLR7) of the innate immune system in mammalian hosts (Gehrig et al., 2012). Under typical laboratory growth conditions, the lack of Gm18 minimally affects translation efficiency. However, Gm18 increases upon exposure to mild antibiotic conditions. E. coli tRNA isolated from mild antibiotic stress conditions causes lower activation of human plasmacytoid dendritic cells than those from conditions without antibiotic exposure. This tRNA modification dependent bacterial adaptation in response to antibiotics improves its survival within a human host.
Food intake also greatly influences gut bacterial physiology (Teng et al., 2018). The gut bacterium Lactobacillus rhanosus GG increases in abundance in the mouse gut in response to lemon-derived exosome-like nanoparticles (LELNs), likely through increased resistance to bile (Lei et al., 2021). The underlying mechanism may be specific tRNA decay, in which downregulation of proteins Msp1 and Msp3 leads to decreases in cell wall hydrolysis which reduces bile accessibility into cells. The minor change of Msp1 and Msp3 mRNA levels suggests a potential mechanism at the translational or post-translational level. Indeed, the tRNA processing enzyme RNase P seems to be involved in mediating tRNASer decay which decreases the expression of Msp1/3 proteins.
Pathogens utilize modifications to increase survival within hosts
Pathogenic bacteria also alter their tRNA dynamics to ensure survival during infection. On a global tRNA modification scale, high levels of tRNA methylations in E. coli are necessary in ensuring a general resistance against antibiotics (Masuda et al., 2019). In line with the use of modifications to control translation efficiency, the m1G37 modification status in the anticodon loop significantly affects the translation of several membrane proteins through proline decoding in their open reading frames (Masuda et al., 2019). The lack of the m1G37 writer impairs membrane structure of E. coli and Salmonella enterica, sensitizing them to antibiotics.
Upon onset of infection within a human host, reactive oxygen species production is a common general response in mitigating growth and spread of pathogenic bacteria. As a result, these pathogens have adapted specific methods to deal with this environmental stressor. Mycobacteria have been observed to change their tRNA patterns upon exposure to hypoxia, a commonly induced environment due to the inflammatory response by the immune system (Chionh et al., 2016). Each stage of hypoxia-induced change is associated with distinct patterns in tRNA modification. Specifically, the Doc regulon genes, which control the hypoxia response, have altered translation efficiency depending on their codon composition bias, particularly through decoding by tRNAThr(UGU). Pseudomonas aeruginosa modulates their expression of detoxifying genes such as KatA and KatB at the translational level upon exposure to hydrogen peroxide stress (Thongdee et al., 2019). Here, the m7G46 writer, TrmB is necessary to effectively translate Phe- and Asp-enriched mRNAs including the catalase enzymes KatA and KaB. Exposure to hydrogen peroxide stress is associated with increases in m7G levels, while the lack of TrmB leads to decreases in KatA and KatB expression and reduced resistance to hydrogen peroxide stress. These studies highlight specific mechanisms in which tRNA dynamic changes can be a main driver in mediating bacterial survival responses to human hosts defenses.
tRNA modification dynamics in gut microbiomes
All studies described above dealt with single organisms. However, complex biological communities also respond to the environment through tRNA modifications. Schwartz et al. (Schwartz et al., 2018) perform tRNA sequencing of the gut microbiome of mice fed with high-fat or low-fat diets to investigate how tRNA modifications respond to dietary conditions. In contrast to standard tRNA-seq where sequencing reads can be readily assigned to the tRNA genes in a reference genome, microbiomes do not have a readily available tRNA references for sequencing data alignment. To overcome this technical obstacle, for microbiome tRNA-seq analysis Schwartz et al. developed a pipeline that identifies each tRNA de novo using a defined signature of canonical tRNAs (Fig. 3A). Each tRNA is then assigned to a taxonomic group obtained from a database from >4,000 microbial genomes. This analysis also generates tRNA “seed” sequences that are most closely related to the tRNA genes in the microbial genomes. In the case of mouse gut microbiome on average ~10,000 seed sequences are obtained in each sample.
Figure 3: Gut microbiome tRNA-seq.
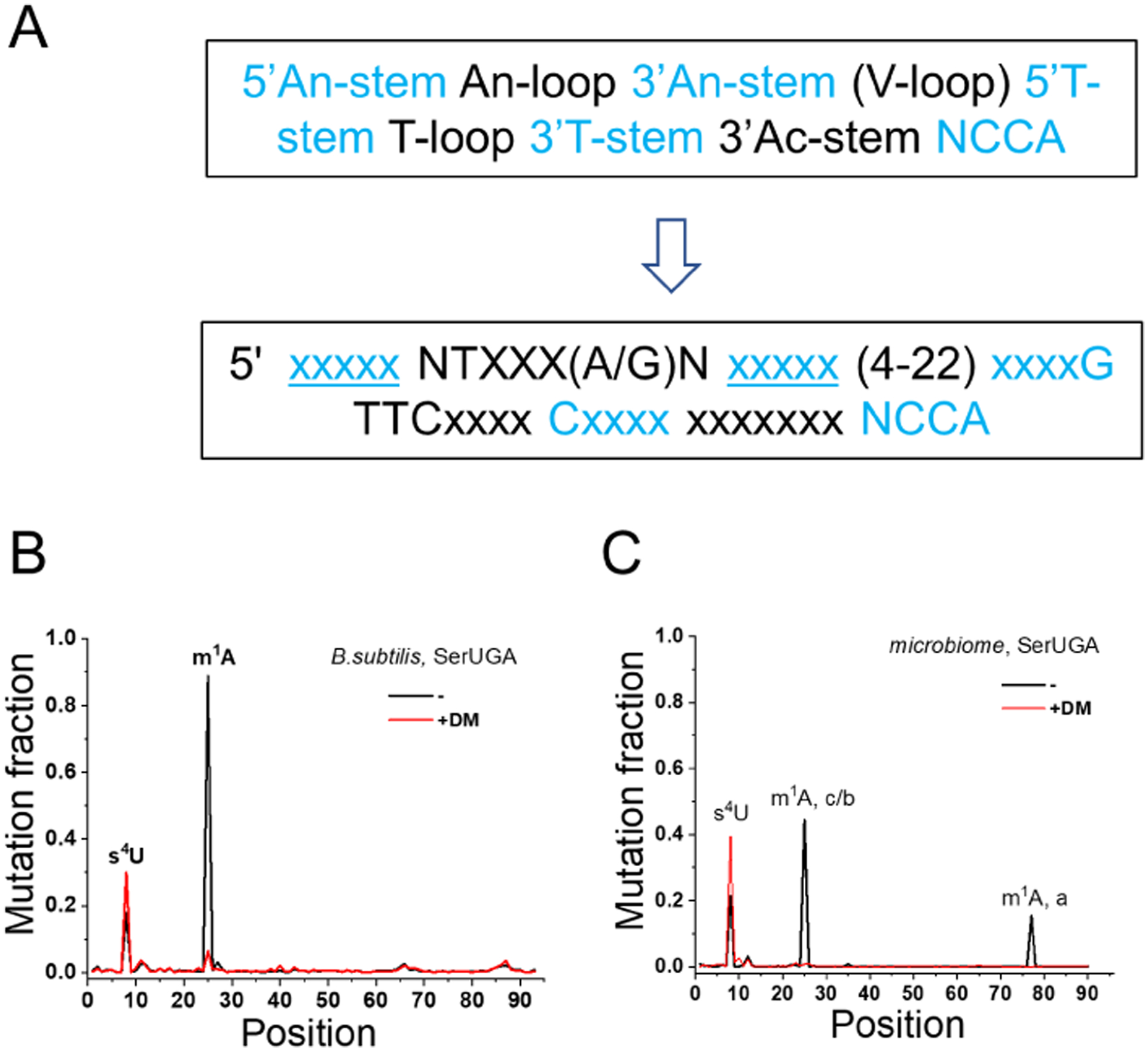
(A) Canonical tRNA signature used in de novo identification of tRNA reads. (B) tRNA modification identification by mutation in a bacterial culture. Demethylase (DM) treatment removes the mutation at nucleotide 22, assigning it to the known m1A modification. Position 8 mutation is in-sensitive to demethylase treatment; it corresponds to the well-characterization s4U modification in bacterial tRNA. (C) Same as panel B in a gut microbiome sample. The two demethylase removable peaks correspond to the m1A modification in clostridia and bacilli around nucleotide 22 (c/b), and in actinobacteria around nucleotide 77 in this type II tRNA.
tRNA modifications in the microbiome are identified by the misincorporation or “mutation” from the most closely related seed sequence. To distinguish mutation signature from actual tRNA modification versus those from single nucleotide polymorphism (SNP) in the microbial population, the same sample is sequenced with and without demethylase treatment using the DM-tRNA-seq procedure (Zheng et al., 2015). The most obviously verifiable tRNA modification is m1A which is “mutated” in high levels without and converts back to the seed sequence with demethylase treatment (Fig. 3B, 3C). Another modification identified through mutation signature is 4-thio-U (s4U) at the junction between the acceptor and D stems. Even though s4U does not respond to demethylase treatment, its position in tRNA is always U in canonical tRNA. Overall, among the 10 bacterial taxonomic classes found in the mouse gut microbiome in this study, m1A is present mostly in 3 and s4U present in 8 classes. Among the two most abundant bacterial classes in the gut microbiome, clostridia tRNAs have both m1A and s4U, bacteriodes tRNAs have neither. As measured by mutation fractions, the s4U levels are about the same in both high-fat and low-fat fed mice. A known function of s4U is response to UV radiation (Kramer et al., 1988). The gut microbiome samples are not exposed to light which can explain the low dependency of s4U level on diets.
In the gut microbiome, the tRNA m1A modifications respond strongly to the dietary conditions: higher in the same microbial tRNAs in the high-fat fed than the low-fat fed mice. In order to understand this result, Schwartz et al analyzed the published meta-proteome data from the high-fat and low-fat mice (Zhang et al., 2016). First, they found ~650 genes in the clostridia class that are differentially expressed. Performing the codon usage analysis of the protein group overexpressed in high-fat fed mice versus another protein group overexpressed in low-fat fed mice, they find that high-fat overexpressed proteins have strongly enriched Glu-Glu codon pair or other codon pairs that include Glu. Since tRNAGlu is one of the 4 tRNAs containing m1A and m1A level is higher in high-fat fed mice, this result is consistent with higher m1A level in tRNAGlu enhancing translation in a codon pair dependent manner. As for why only Glu codon, but not the codons read by other 3 tRNAs are affected, one possible explanation is that Glu codon response is specific for the dietary condition. One can hypothesize that codons read by the other 3 m1A-modified tRNAs, tRNACys, tRNAGln, and tRNASer may be preferred in translation under other conditions encountered by the gut microbiome such as inflammation or oxidative stress.
Metaepitranscriptomics
Applications of our evolving understanding of tRNA modifications to study microbial responses to environmental change within short timescales represents a new and exciting frontier. We live on a microbial planet (Flemming and Wuertz, 2019): thanks to their astonishing metabolic potential and diversity, microbes occupy all niches Earth has to offer, where they form complex and dense communities undertaking the recycling of critical ingredients of life through global biogeochemical cycles (Falkowski et al., 2008). Due to the profound impact of microbes on Earth’s habitability, understanding their ecology, evolution, and responses to environmental change has always been one of the most significant endeavors of the life sciences.
Emerging ‘omics approaches along with advances in sequencing strategies offer unprecedented insights into microbial ecosystems and lifestyles. For instance only during the past few years metagenomics, the study of the entire DNA content of a given environment, has enabled the discovery of new branches of life previously missed by culture-based studies (Castelle and Banfield, 2018), revealed new clades of human virome and their biogeography (Edwards et al., 2019), and resulted in key ecological (Delmont et al., 2018), evolutionary (Spang et al., 2015), and biotechnological (van Kessel et al., 2015) insights. However, as widely used high throughput ‘omics strategies typically target genetic information encoded in DNA or RNA molecules, studies that aim to understand environmental microbes lack insights into translation and its regulation. Only a small number of publications so far has ventured into this direction (Fremin et al., 2020; Schwartz et al., 2018). As a result, accents that fine-tune the complex metabolic orchestration of environmental bacteria beyond the information encoded in their genome are systematically absent in state-of-the-art analyses.
A bacterial cell has 30–45 different tRNA species, up to 100,000 tRNA transcripts, and 6–10 modifications per molecule. Based on the summary of bacterial tRNA modification types and locations (Fig. 4), the current strategies to sequence tRNA transcripts can reveal ~25% of sites on average of all bacterial tRNA modifications based on mutation signatures in cDNA synthesis. The dynamic range of tRNA modifications in complex environmental microbial populations is currently unknown. Yet, insights from the ‘epitranscriptomics’ of cells in culture which reveal responses to new environmental conditions within timescales of minutes, foreshadow not only the complexity of such signal in natural habitats but also the utility of ‘metaepitranscriptomics' of microbial communities to study temporal and spatial impact and magnitude of change. Since the codon composition of genomes and the tRNA genes they encode form a natural framework to study dynamics of tRNA transcripts, there is an inherent yet unexplored connection between popular ‘omics approaches and tRNA sequencing. Thus, the inclusion of metaepitranscriptomics into existing software platforms that currently offer data integration opportunities for popular ‘omics approaches (Eren et al., 2021) is critical to bring a currently missing perspective into microbial lifestyles by extending the computational tools available to environmental microbiologists and enable deeper insights into microbial life.
Figure 4. Bacterial tRNA modifications.
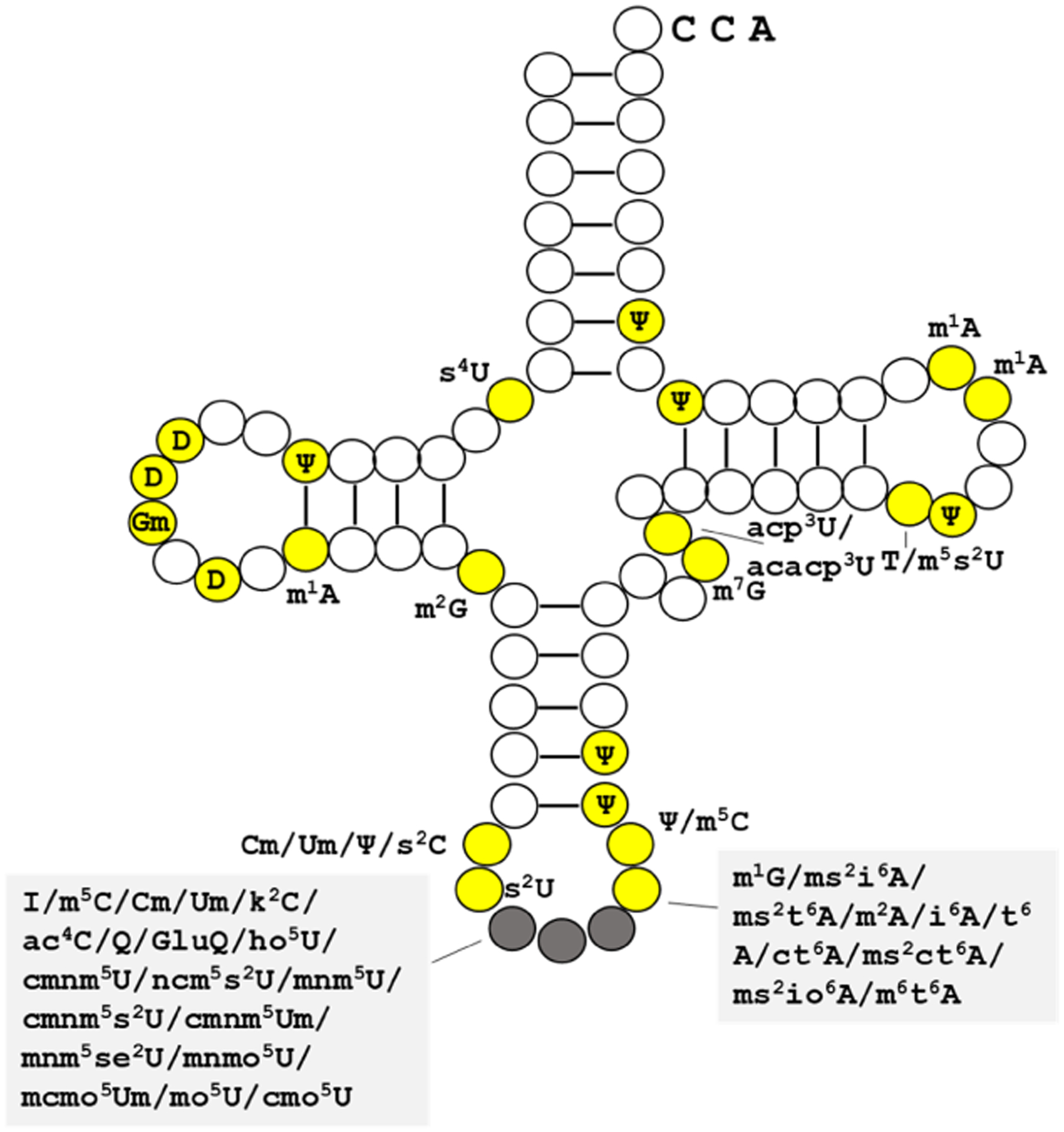
Modified positions are shown in yellow with the corresponding modification abbreviations. Anticodon nucleotides are in gray. Modification types and sites data from Modomics (Boccaletto et al. 2018).
In summary, current studies on dynamic tRNA modifications have already generated many exciting insights into how biological systems utilize them for their adaptive benefits. As an emerging area, future research shall reveal unprecedented and new insights not only for the biology of single organisms but also for the microbial and microbial-host communities.
Acknowledgments:
This work was supported by grants from the NIH (RM1HG008935, R01GM113194 to T.P., RC2 DK122394 to A.M.E.) and the Keck Foundation (to T.P. and A.M.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests:
Tao Pan is a co-founder of 4SR Biosciences Inc.
References
- Alings F, Sarin LP, Fufezan C, Drexler HC, and Leidel SA (2015). An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 21, 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Ivanov P (2014). tRNA fragments in human health and disease. FEBS Lett 588, 4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Suzuki T, Saito A, Wei FY, Ikeuchi Y, Numata T, Tanaka R, Yamane Y, Yamamoto T, Goto T, et al. (2018). Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res 46, 1565–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A, and Ofengand J (1993). Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762. [DOI] [PubMed] [Google Scholar]
- Behrens A, Rodschinka G, and Nedialkova DD (2021). High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell 81, 1802–1815 e1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. (2014). Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33, 2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland C, Hayes P, Santa-Maria I, Nishimura S, and Kelly VP (2009). Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J Biol Chem 284, 18218–18227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch A, Laguna T, Butter F, Schaffrath R, and Klassen R (2020). Misactivation of multiple starvation responses in yeast by loss of tRNA modifications. Nucleic Acids Res 48, 7307–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiracci J, Migeot V, Chionh YH, Bauer F, Brochier T, Russell B, Shiozaki K, Dedon P, and Hermand D (2019). Reciprocal regulation of TORC signaling and tRNA modifications by Elongator enforces nutrient-dependent cell fate. Sci Adv 5, eaav0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, and Gilbert WV (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, and Banfield JF (2018). Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life. Cell 172, 1181–1197. [DOI] [PubMed] [Google Scholar]
- Chan CT, Deng W, Li F, DeMott MS, Babu IR, Begley TJ, and Dedon PC (2015). Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes. Chem Res Toxicol 28, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, and Begley TJ (2010). A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6, e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, and Dedon PC (2012). Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, Lu Z, Zheng Z, Dai Q, and Wang H (2019). Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res 47, 2533–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chionh YH, McBee M, Babu IR, Hia F, Lin W, Zhao W, Cao J, Dziergowska A, Malkiewicz A, Begley TJ, et al. (2016). tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat Commun 7, 13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury MM, Dosche C, Lohmannsroben HG, and Leimkuhler S (2012). Dual role of the molybdenum cofactor biosynthesis protein MOCS3 in tRNA thiolation and molybdenum cofactor biosynthesis in humans. J Biol Chem 287, 17297–17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WC, Evans ME, Dominissini D, Zheng G, and Pan T (2016). tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22, 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, and Lowe TM (2015). ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12, 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Liu Q, Sendinc E, Shi Y, and Gregory RI (2021). Nucleotide resolution profiling of m3C RNA modification by HAC-seq. Nucleic Acids Res 49, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon JR, Pincus D, and Ploegh HL (2015). tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol Biol Cell 26, 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauden MI, Jaciuk M, Muller CW, and Glatt S (2018). Structural asymmetry in the eukaryotic Elongator complex. FEBS Lett 592, 502–515. [DOI] [PubMed] [Google Scholar]
- de Crecy-Lagard V, Boccaletto P, Mangleburg CG, Sharma P, Lowe TM, Leidel SA, and Bujnicki JM (2019). Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res 47, 2143–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Quince C, Shaiber A, Esen OC, Lee ST, Rappe MS, McLellan SL, Lucker S, and Eren AM (2018). Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol 3, 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Babu IR, Su D, Yin S, Begley TJ, and Dedon PC (2015). Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet 11, e1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe JM, Fuller BL, Lentini JM, Kellner SM, and Fu D (2017). TRMT1-Catalyzed tRNA Modifications Are Required for Redox Homeostasis To Ensure Proper Cellular Proliferation and Oxidative Stress Survival. Mol Cell Biol 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Rath S, Kolet-Mandrikov D, and Korennykh A (2017). Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durdevic Z, and Schaefer M (2013). tRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress? Bioessays 35, 323–327. [DOI] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, and Sorek R (2013). Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9, e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, Dinsdale EA, Cinek O, Aziz RK, McNair K, Barr JJ, et al. (2019). Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat Microbiol 4, 1727–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erber L, Hoffmann A, Fallmann J, Betat H, Stadler PF, and Morl M (2020). LOTTE-seq (Long hairpin oligonucleotide based tRNA high-throughput sequencing): specific selection of tRNAs with 3'-CCA end for high-throughput sequencing. RNA Biol 17, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, Fink I, Pan JN, Yousef M, Fogarty EC, et al. (2021). Community-led, integrated, reproducible multi-omics with anvi'o. Nat Microbiol 6, 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan SG, Helm M, and Prehn JHM (2021). tRNA-derived fragments: A new class of non-coding RNA with key roles in nervous system function and dysfunction. Prog Neurobiol, 102118. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Fenchel T, and Delong EF (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. [DOI] [PubMed] [Google Scholar]
- Farkas WR (1980). Effect of diet on the queuosine family of tRNAs of germ-free mice. J Biol Chem 255, 6832–6835. [PubMed] [Google Scholar]
- Fergus C, Barnes D, Alqasem MA, and Kelly VP (2015). The queuine micronutrient: charting a course from microbe to man. Nutrients 7, 2897–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, and Wuertz S (2019). Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol 17, 247–260. [DOI] [PubMed] [Google Scholar]
- Fremin BJ, Sberro H, and Bhatt AS (2020). MetaRibo-Seq measures translation in microbiomes. Nat Commun 11, 3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, and Samson LD (2010a). Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 30, 2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dai Q, Zhang W, Ren J, Pan T, and He C (2010b). The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl 49, 8885–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvanin A, Vogt LM, Grober A, Freund I, Ayadi L, Bourguignon-Igel V, Bessler L, Jacob D, Eigenbrod T, Marchand V, et al. (2020). Bacterial tRNA 2'-O-methylation is dynamically regulated under stress conditions and modulates innate immune response. Nucleic Acids Res 48, 12833–12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Encinar Del Dedo J, Ayte J, and Hidalgo E (2016). Genome-wide Screening of Regulators of Catalase Expression: ROLE OF A TRANSCRIPTION COMPLEX AND HISTONE AND tRNA MODIFICATION COMPLEXES ON ADAPTATION TO STRESS. The Journal of biological chemistry 291, 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M, et al. (2012). Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med 209, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogakos T, Brown M, Garzia A, Meyer C, Hafner M, and Tuschl T (2017). Characterizing Expression and Processing of Precursor and Mature Human tRNAs by Hydro-tRNAseq and PAR-CLIP. Cell Rep 20, 1463–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenbour JM, and Pan T (2006). Diversity of tRNA genes in eukaryotes. Nucleic Acids Res 34, 6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, and Bellodi C (2020). Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol 17, 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimkova K, Sommarin MNE, Munita R, Lubas M, et al. (2018). Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 173, 1204–1216 e1226. [DOI] [PubMed] [Google Scholar]
- Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hubner B, Seikowski J, Dennerlein S, Rehling P, et al. (2016). NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J 35, 2104–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag S, Warda AS, Kretschmer J, Gunnigmann MA, Hobartner C, and Bohnsack MT (2015). NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 21, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JF, Yim D, Ma D, Huber SM, Davis N, Bacusmo JM, Vermeulen S, Zhou J, Begley TJ, DeMott MS, et al. (2021). Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SM, Leonardi A, Dedon PC, and Begley TJ (2019). The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress. Toxics 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatova VV, Kaiser S, Ho JSY, Bing X, Stolz P, Tan YX, Lee CL, Gay FPH, Lastres PR, Gerlini R, et al. (2020). METTL6 is a tRNA m(3)C methyltransferase that regulates pluripotency and tumor cell growth. Sci Adv 6, eaaz4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D, Zink S, Mehlgarten C, Daum G, and Schaffrath R (2006). tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 59, 677–688. [DOI] [PubMed] [Google Scholar]
- Kasai H, Nakanishi K, Macfarlane RD, Torgerson DF, Ohashi Z, McCloskey JA, Gross HJ, and Nishimura S (1976). Letter: The structure of Q* nucleoside isolated from rabbit liver transfer ribonucleic acid. J Am Chem Soc 98, 5044–5046. [DOI] [PubMed] [Google Scholar]
- Kasai H, Oashi Z, Harada F, Nishimura S, Oppenheimer NJ, Crain PF, Liehr JG, von Minden DL, and McCloskey JA (1975). Structure of the modified nucleoside Q isolated from Escherichia coli transfer ribonucleic acid. 7-(4,5-cis-Dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanosine. Biochemistry 14, 4198–4208. [DOI] [PubMed] [Google Scholar]
- Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, and Suzuki T (2017). ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res 45, 7401–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, and Cairns BR (2019). Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc Natl Acad Sci U S A 116, 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Xiao H, Koh J, Wang Y, Bonanno JB, Thomas K, Babbitt PC, Brown S, Lee YS, and Almo SC (2015). Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification. Nucleic Acids Res 43, 4602–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Dedon PC, and Waldor MK (2020). Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat Chem Biol 16, 964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S, and Ignatova Z (2015). Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16, 98–112. [DOI] [PubMed] [Google Scholar]
- Kramer GF, Baker JC, and Ames BN (1988). Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and ApppGpp as components of an adaptive response. J Bacteriol 170, 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C, Teng Y, He L, Sayed M, Mu J, Xu F, Zhang X, Kumar A, Sundaram K, Sriwastva MK, et al. (2021). Lemon exosome-like nanoparticles enhance stress survival of gut bacteria by RNase P-mediated specific tRNA decay. iScience 24, 102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Miyauchi K, Harada T, Okita R, Takeshita E, Komaki H, Fujioka K, Yagasaki H, Goto YI, Yanaka K, et al. (2018). CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat Commun 9, 1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Liu Q, Jiang YZ, and Gregory RI (2019). Nucleotide resolution profiling of m(7)G tRNA modification by TRAC-Seq. Nat Protoc 14, 3220–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. (2016). ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 167, 816–828 e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Fay MM, and Ivanov P (2018). The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett 592, 2828–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Gerber AP, and Rich A (1999). Identification and characterization of a human tRNA-specific adenosine deaminase related to the ADAR family of pre-mRNA editing enzymes. Proc Natl Acad Sci U S A 96, 8895–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Ayadi L, Ernst FGM, Hertler J, Bourguignon-Igel V, Galvanin A, Kotter A, Helm M, Lafontaine DLJ, and Motorin Y (2018). AlkAniline-Seq: Profiling of m(7) G and m(3) C RNA Modifications at Single Nucleotide Resolution. Angew Chem Int Ed Engl 57, 16785–16790. [DOI] [PubMed] [Google Scholar]
- Marelja Z, Stocklein W, Nimtz M, and Leimkuhler S (2008). A novel role for human Nfs1 in the cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J Biol Chem 283, 25178–25185. [DOI] [PubMed] [Google Scholar]
- Masuda I, Matsubara R, Christian T, Rojas ER, Yadavalli SS, Zhang L, Goulian M, Foster LJ, Huang KC, and Hou YM (2019). tRNA Methylation Is a Global Determinant of Bacterial Multi-drug Resistance. Cell Syst 8, 302–314 e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Suter B, Grosjean H, Keith G, and Kubli E (1985). Queuosine modification of the wobble base in tRNAHis influences 'in vivo' decoding properties. The EMBO journal 4, 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Hartmann M, Schuster I, Bender S, Thuring KL, Helm M, Katze JR, Nellen W, Lyko F, and Ehrenhofer-Murray AE (2015). Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res 43, 10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Legrand C, Tuorto F, Kelly VP, Atlasi Y, Lyko F, and Ehrenhofer-Murray AE (2019). Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res 47, 3711–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja S, Cai MW, Sun J, Varet H, Sarid L, Trebicz-Geffen M, Shaulov Y, Mazumdar M, Legendre R, Coppee JY, et al. (2021). Queuine Is a Nutritional Regulator of Entamoeba histolytica Response to Oxidative Stress and a Virulence Attenuator. mBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasvall SJ, Chen P, and Bjork GR (2007). The wobble hypothesis revisited: uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA 13, 2151–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, and Leidel SA (2015). Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 161, 1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, and Suzuki T (2009). Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 37, 1335–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa EM, Pavon-Eternod M, Pan T, and Ribas de Pouplana L (2012). A role for tRNA modifications in genome structure and codon usage. Cell 149, 202–213. [DOI] [PubMed] [Google Scholar]
- Pang YL, Abo R, Levine SS, and Dedon PC (2014). Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res 42, e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Chan CT, Dyavaiah M, Rooney JP, Dedon PC, and Begley TJ (2012a). Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol 9, 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Dyavaiah M, Joseph F, Rooney JP, Chan CT, Dedon PC, and Begley TJ (2012b). Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell cycle 11, 3656–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkard O, McFarland S, Sweet T, and Coller J (2020). Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nature communications 11, 4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapino F, Zhou Z, Roncero Sanchez AM, Joiret M, Seca C, El Hachem N, Valenti G, Latini S, Shostak K, Geris L, et al. (2021). Wobble tRNA modification and hydrophilic amino acid patterns dictate protein fate. Nat Commun 12, 2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashad S, Han X, Sato K, Mishima E, Abe T, Tominaga T, and Niizuma K (2020). The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol 17, 1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyniers JP, Pleasants JR, Wostmann BS, Katze JR, and Farkas WR (1981). Administration of exogenous queuine is essential for the biosynthesis of the queuosine-containing transfer RNAs in the mouse. J Biol Chem 256, 11591–11594. [PubMed] [Google Scholar]
- Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, and Pedrioli PG (2013). tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci U S A 110, 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, Gregory BD, and Wang LS (2013). HAMR: high-throughput annotation of modified ribonucleotides. RNA 19, 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Miyauchi K, Kimura S, and Suzuki T (2016). Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res 44, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, and Lyko F (2009). RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, and Lyko F (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes & development 24, 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P (2018). The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nature reviews. Molecular cell biology 19, 45–58. [DOI] [PubMed] [Google Scholar]
- Schlieker CD, Van der Veen AG, Damon JR, Spooner E, and Ploegh HL (2008). A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A 105, 18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MH, Wang H, Pan JN, Clark WC, Cui S, Eckwahl MJ, Pan DW, Parisien M, Owens SM, Cheng BL, et al. (2018). Microbiome characterization by high-throughput transfer RNA sequencing and modification analysis. Nat Commun 9, 5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. (2014). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M, Honda S, Loher P, Telonis AG, Rigoutsos I, and Kirino Y (2017). YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucleic Acids Res 45, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Abu-Shumays R, Akeson M, and Bernick DL (2015). Capture, Unfolding, and Detection of Individual tRNA Molecules Using a Nanopore Device. Front Bioeng Biotechnol 3, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhuang Y, Zhu C, Meng H, Lu B, Xie B, Peng J, Li M, and Yi C (2020). Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol 16, 160–169. [DOI] [PubMed] [Google Scholar]
- Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, and Klungland A (2010). Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol 30, 1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jorgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, and Ettema TJG (2015). Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenkuch F, Motorin Y, and Helm M (2014). Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol 11, 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, and Preiss T (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T (2021). The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol 22, 375–392. [DOI] [PubMed] [Google Scholar]
- Suzuki T, and Suzuki T (2014). A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42, 7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T, Wada T, Saigo K, and Watanabe K (2002). Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J 21, 6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura M, Ishiguro K, Akichika S, Miyauchi K, and Suzuki T (2019). Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat Commun 10, 5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, Hutchins E, Mu J, Deng Z, Luo C, et al. (2018). Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 24, 637–652 e638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termathe M, and Leidel SA (2018). The Uba4 domain interplay is mediated via a thioester that is critical for tRNA thiolation through Urm1 thiocarboxylation. Nucleic Acids Res 46, 5171–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville PC, Iwata-Reuyl D, and de Crecy-Lagard V (2014). Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol 11, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville PC, Legendre R, Rojas-Benitez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, and de Crecy-Lagard V (2016). Global translational impacts of the loss of the tRNA modification t(6)A in yeast. Microb Cell 3, 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NK, Poodari VC, Jain M, Olsen HE, Akeson M, and Abu-Shumays RL (2021). Direct Nanopore Sequencing of Individual Full Length tRNA Strands. ACS nano 15, 16642–16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongdee N, Jaroensuk J, Atichartpongkul S, Chittrakanwong J, Chooyoung K, Srimahaeak T, Chaiyen P, Vattanaviboon P, Mongkolsuk S, and Fuangthong M (2019). TrmB, a tRNA m7G46 methyltransferase, plays a role in hydrogen peroxide resistance and positively modulates the translation of katA and katB mRNAs in Pseudomonas aeruginosa. Nucleic Acids Res 47, 9271–9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Pineyro D, Filonava L, Stracker TH, Batlle E, and Ribas de Pouplana L (2014). A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett 588, 4279–4286. [DOI] [PubMed] [Google Scholar]
- Torres AG, Pineyro D, Rodriguez-Escriba M, Camacho N, Reina O, Saint-Leger A, Filonava L, Batlle E, and Ribas de Pouplana L (2015). Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res 43, 5145–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Grone HJ, et al. (2015). The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 34, 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Legrand C, Cirzi C, Federico G, Liebers R, Muller M, Ehrenhofer-Murray AE, Dittmar G, Grone HJ, and Lyko F (2018). Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, and Lyko F (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nature structural & molecular biology 19, 900–905. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K, and Tsujikawa K (2017). AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep 7, 42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K, and Suzuki T (2005). Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J Biol Chem 280, 1613–1624. [DOI] [PubMed] [Google Scholar]
- van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, Jetten MS, and Lucker S (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Matuszek Z, Huang Y, Parisien M, Dai Q, Clark W, Schwartz MH, and Pan T (2018). Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, et al. (2011). Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 121, 3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. (2018). Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell 71, 973–985 e975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Murphy F.V.t., Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, and Ramakrishnan V (2007). Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nature structural & molecular biology 14, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BN, and Tener GM (1973). Activity of a transfer RNA modifying enzyme during the development of Drosophila and its relationship to the su(s) locus. J Mol Biol 74, 635–651. [DOI] [PubMed] [Google Scholar]
- Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, et al. (2017). Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem 292, 14695–14703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, and Drummond DA (2014). A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol 12, e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lu R, Zhang Y, Matuszek Z, Zhang W, Xia Y, Pan T, and Sun J (2020). tRNA Queuosine Modification Enzyme Modulates the Growth and Microbiome Recruitment to Breast Tumors. Cancers (Basel) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LS, Xiong QP, Pena Perez S, Liu C, Wei J, Le C, Zhang L, Harada BT, Dai Q, Feng X, et al. (2021). ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat Cell Biol 23, 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ning Z, Mayne J, Moore JI, Li J, Butcher J, Deeke SA, Chen R, Chiang CK, Wen M, et al. (2016). MetaPro-IQ: a universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, and Pan T (2015). Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12, 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JB, Wang Y, Zeng QY, Meng SX, Wang ED, and Zhou XL (2020). Molecular basis for t6A modification in human mitochondria. Nucleic Acids Res 48, 3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Karcher D, and Bock R (2013). Importance of adenosine-to-inosine editing adjacent to the anticodon in an Arabidopsis alanine tRNA under environmental stress. Nucleic Acids Res 41, 3362–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]


