Abstract
The dental pulp has irreplaceable roles in maintaining healthy teeth and its regeneration is a primary aim of regenerative endodontics. This study aimed to replicate the characteristics of dental pulp tissue by using cranial neural crest (CNC)-like cells (CNCLCs); these cells were generated by modifying several steps of a previously established method for deriving NC-like cells from induced pluripotent stem cells (iPSCs). CNC is the anterior region of the neural crest in vertebrate embryos, which contains the primordium of dental pulp cells or odontoblasts. The produced CNCLCs showed approximately 2.5–12,000-fold upregulations of major CNC marker genes. Furthermore, the CNCLCs exhibited remarkable odontoblastic differentiation ability, especially when treated with a combination of the fibroblast growth factors (FGFs) FGF4 and FGF9. The FGFs induced odontoblast marker genes by 1.7–5.0-fold, as compared to bone morphogenetic protein 4 (BMP4) treatment. In a mouse subcutaneous implant model, the CNCLCs briefly fated with FGF4 + FGF9 replicated dental pulp tissue characteristics, such as harboring odontoblast-like cells, a dentin-like layer, and vast neovascularization, induced by the angiogenic self-assembling peptide hydrogel (SAPH), SLan. SLan acts as a versatile biocompatible scaffold in the canal space. This study demonstrated a successful collaboration between regenerative medicine and SAPH technology.
Keywords: Regenerative endodontics, iPSC, Cranial neural crest, Fibroblast growth factor, Odontoblastic differentiation, Angiogenic self-assembling peptide hydrogel
Graphical abstract
Highlights
-
•
Cranial neural crest like cells (CNCLCs) were generated by simplifying a previously established method for deriving neural crest-like cells from iPSCs.
-
•
The produced CNCLCs showed approximately ∼12,000-fold upregulations of major CNC marker genes.
-
•
The combination of fibroblast growth factors, FGF4 and FGF9, induced the CNCLCs toward odontoblastic differentiation more effectively than BMP4.
-
•
In a mice subcutaneous implant model, the CNCLCs replicated the characteristics of dental pulp harboring vast neovascularization with the aid of the angiogenic hydrogel, SLan.
1. Introduction
The dental pulp plays significant roles in sustaining healthy teeth. It nourishes dentin, senses external stimuli, accommodates resident immune cells, and harbors the precursors of odontoblast-like cells. These pulp activities can be easily abolished with inflammation or necrosis caused by dental caries or trauma. Regenerative endodontics aims to replace damaged dentin-pulp complexes with endogenous or exogenous cells to restore intact pulp functions. This new endodontic approach intends to embrace the concept of tissue engineering based on the triad, that is, stem cells to regenerate pulp tissue, biomimetic scaffolds, and bioactive growth factors [1]. Currently, two methodologies for cell-recovery in the debrided pulp space are being studied [2,3]—one utilizes stem-cells homing to the cleared pulp space from the patient's peri-apical region [4,5], while the other employs engrafting exogenous cell (also known as the ‘cell-based’ method) [6]. Both the methodologies have merits and demerits. Aged patients have difficulties with cell-homing approaches due to the insufficient regenerative capacity of their endogenous stem cells. In contrast, a variety of stem cell sources, such as dental pulp stem cells (DPSCs), stem cells from the apical papilla (SCAP), periodontal ligament stem cells (PDLSCs), and stem cells from human exfoliated deciduous teeth (SHED), have been studied in ‘cell-based’ methodology for the regeneration of pulp-dentin complex, (reviewed in Ref. [7]). These stem cells from patients can be cultured and propagated ex vivo to obtain sufficient number of cells required for transplantation.
Besides these autologous stem cells from the patient body, induced pluripotent stem cell (iPSC) is another attractive stem cell source for dentin-pulp regeneration [7,8]. iPSCs have the following important features: i) theoretically, iPSCs can be generated from any somatic cells of patients of any age [9], allowing treatment of elderly patients who lack enough stem cells mentioned above. ii) the tissue that originates from iPSCs potentially has the ability to bypass immunorejection in the recipient, enabling allografts of tissues produced from MHC-incompatible donors. Recently, three groups [[10], [11], [12]] demonstrated that iPSCs can differentiate into odontoblast-like cells via neural crest-like cells (NCLCs) as the precursor form. Neural crest is a multipotent cell population at the interface of the neuroepithelium and epidermis of a developing embryo, which can differentiate into numerous cell types, including dental mesenchymal cells [13,14].
Cranial neural crest (CNC) is one of the four distinct regions of the neural crest, which develops into craniofacial connective tissue, cranial neurons, and pharyngeal mesenchyme, including dental pulp and odontoblasts [[15], [16], [17], [18]]. In this study, we established a more facile method to induce cranial neural crest-like cells (CNCLCs) from iPSCs, compared to previous reports [[19], [20], [21]]. Furthermore, we showed that a combination of the fibroblast growth factors (FGFs) FGF4 and FGF9 can induce odontoblastic differentiation of CNCLCs with the highest efficiency. Lastly, it was demonstrated that our CNCLCs can replicate the characteristics of dental pulp tissue, by harnessing the angiogenic self-assembling peptide hydrogel (SAPH), SLan [22].
SLan is a SAPH containing the backbone sequence KSLSLSLSLSLSLK, which allows self-assembly into β-sheet-based nanofibrous ECM-mimetic structures [[23], [24], [25], [26]]. These synthetic peptide assemblies can be loaded with water-soluble drugs, biologics, and cells [25], enabling a suitable carrier for transplanted cells and required growth factors. When SLan is formulated in 1 × PBS and similar physiological/isotonic buffers, it forms a thixotropic gel—gel that becomes thin by shearing and regels instantaneously [23,[27], [28], [29], [30]]. The resultant in situ bolus demonstrates stable biocompatibility with animal models and degrades over week-long periods in vivo [25,27,31]. Notably, SLan has a key modification of the base peptide N-terminus with a vascular endothelial growth factor (VEGF) signaling domain, KLTWQELYQLKYKGI, derived from VEGF-165 [32], which allows mimicking the function of the original growth factor [33], inducing neovascularization into engrafted cell-SLan complexes. These key features of SLan make critical contributions to replicate the characteristics of dental pulp tissue in the present study, paving the way for material cell-based strategies in regenerative endodontics.
2. Materials and methods
2.1. Peptide synthesis and preparation
The angiogenic peptide SLan, K-(SL)6-K-G-KLTWQELYQLKYKGI, was synthesized following previously described standard synthetic procedures [27]. Briefly, peptides, resins, and coupling reagents were purchased from CEM corporation. Standard solid-phase peptide synthesis was performed using a CEM Liberty Blue microwave peptide synthesizer using standard Fmoc chemistry. All peptides were amidated at C-terminal and acetylated at N-terminal. The crude peptides were cleaved using 2.5% each of H2O, triisopropylsilane (TIS), and 3,6-dioxa-1,8-octanedithiol (DoDT), and 92.5% trifluoroacetic acid (TFA) (total volume of 10 mL for 0.1 mM scale synthesis) for 30 min at 37 °C. The cleaved peptides were crushed in cold (−20 °C) ether, centrifuged, ether decanted, and left to dry overnight. The resulting crude peptide pellets were dissolved in Milli-Q water at a concentration of ∼1 mg/mL, pH adjusted to 7.0, and dialyzed for 3 days using Spectra/Por S/P 7 RC dialysis tubing with a 2000 Da cut-off tubing against DI water. The peptides were subsequently lyophilized and >85% purity was confirmed using an Agilent 1100 series HPLC instrument with an Agilent (Santa Clara, CA) C3 reverse phase column. The molecular weights of the peptides were verified using an Orbitrap Q Exactive LC/MS instrument (Thermo Fisher Scientific, Waltham, MA, USA).
2.2. Cells and culture conditions
Human gingival fibroblasts were obtained by explanting gingival connective tissue attached to extracted third molars from healthy donors, according to the method described by Somerman et al. [34]. They were cultured in DMEM/F12 medium (Thermo Fisher Scientific) supplemented with 10% FBS and 1× Antibiotic-Antimycotic (Thermo Fisher Scientific). These protocols were approved by the Rutgers Institutional Review Board. Human dental pulp cells (DPCs) (PT-5025, Lonza, Switzerland), iPSCs, and CNCLCs were cultured in αMEM medium (Thermo Fisher Scientific) supplemented with 20% FBS (Thermo Fisher Scientific) and 1× Antibiotic-Antimycotic, Essential 8 medium (Thermo Fisher Scientific) supplemented with 10 μM Y-27632 (Tocris Bioscience, MN, USA), and DMEM/F12 medium supplemented with 10% FBS and 1× Antibiotic-Antimycotic, respectively. The osteo/odontogenic differentiation medium consisted of the above normal growth media, 50 μg/ml ascorbic acid, and 10 nM dexamethasone, and chased with 10 mM β-glycerophosphate for the last 5 days. For the odontoblastic differentiation assay, 1 × 105 CNCLCs were treated with osteo/odontogenic media in a 24-well plate for 3 weeks. Indicated FGFs were added to the medium at the concentration of 10 ng/ml. DSP expression was checked using anti-DSP (Santa Cruz Biotechnology, sc-73632) primary antibody and secondary antibody conjugated with Alexa Fluor 488 (Thermo Fisher Scientific). Unless otherwise noted, all the cells were cultured in a humidified 5% CO2 incubator at 37 °C.
2.3. RNA isolation and qPCR
Total RNA was isolated from approximately 2 × 105 cells using Direct-zol RNA Miniprep Plus (Zymo Research). cDNA was synthesized from at least 500 ng of isolated RNA using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), following the manufacturer's instructions. qPCR was performed using PowerUp™ SYBR® Green Master Mix (Thermo Fisher Scientific) and 200 pM gene-specific primers (Supplementary Table S1). Thermal cycler condition: 40 cycles of (95 °C for 15″, 60 °C for 30″ and 72 °C for 30″). All qPCR data shown in this study were obtained from the average of at least three independent samples. The error bars indicate the standard deviation. All statistical analyses in this study were performed using the Student's t-test assuming unpaired two-tailed data distribution.
2.4. Human iPSC production and validation
Human iPSCs were produced from human dermal fibroblasts or gingival fibroblasts using a CytoTune-iPS 2.0 Sendai Reprogramming kit (Thermo Fisher Scientific) with slight modifications to the protocol. Briefly, human fibroblasts cultured in DMDM/F12 medium (Thermo Fisher) on a 6-well plate at 2 × 105 cells/well were infected with Sendai virus with MOIs of 5, 5, and 3 for KOS, c-Myc, and Klf4, respectively. Following incubation for 7 days with daily media change, cells were lifted with TrypLE (Thermo Fisher Scientific) and transferred onto Matrigel (Corning) -coated 100 mm culture dish. The medium was changed to Essential 8 (Thermo Fisher Scientific) on the next day and was changed daily. Three weeks later, iPSC colonies were identified using the TRA-1-60 Alexa Fluor™ 488 Conjugate Kit (Thermo Fisher Scientific), picked up under a microscope using a 1 ml pipette and kept in a new plate containing Essential 8 medium and 10 μM Y-27632 dihydrochloride ROCK inhibitor (Tocris Bioscience).
The ability of iPSCs to differentiate into the three germ layer cells was validated using STEMdiff Trilineage Differentiation Kit (Stemcell Technologies, MA), following their protocol. Briefly, iPSCs pre-cultured with Essential 8 medium were plated in Matrigel-coated 24-well plates at a density of 4 × 105 cells/well (for ectoderm or endoderm differentiation), or 1 × 105 cells/well (for mesoderm differentiation). The next day, Essential 8 medium was changed to Trilineage Ectoderm, Mesoderm, or Endoderm Medium, inducing the differentiation of iPSCs into the three germ layer cells. After five days (mesoderm or endoderm differentiation) or seven days (ectoderm differentiation) with daily medium change, the differentiated cells were immuno-stained using FITC- or Alexa Fluor 488- conjugated antibodies for major markers of the three germ layer cells, followed by flow cytometry and fluorescence microscopy analyses. All other antibodies used in this section are listed in Supplementary Table S2.
Karyotyping of iPSCs with G-banding were performed by Institute of Genomic Medicine in Rutgers, The State University of New Jersey.
2.5. Human CNCLCs production
The protocol below is a modification of Lee's method [35]. Confluent iPSC cells cultured in Essential 8 (Thermo Fisher Scientific) media were lifted using TrypLE (Thermo Fisher Scientific) and thoroughly triturated to single cells using Pasteur pipette in fresh Essential 8 media. The cell suspension was filtered using a 40 μm pore size cell strainer to remove debris and cell clumps, followed by centrifugation for 5 min at 160×g and washing twice with Essential 8 medium. Resuspended cells in Essential 8 supplemented with 10 μM Y-27632 dihydrochloride ROCK inhibitor were spread on a Matrigel-coated 6-well plate at a density of 2 × 105 cells/well. After 15 min incubation at 37 °C, non-adherent cells were collected, centrifuged, washed as described above, resuspended in Essential 8 + 10 μM ROCK inhibitor, and spread onto Matrigel-coated 6-well plates at the density of 1 × 105 cells/well. When the cells reached 50%∼70% confluency with daily media changes, 10 μM SB431542 and 3 μM CHIR99021 were added to the media. This mixture of Essential 8, 10 μM SB431542, and 3 μM CHIR99021 was replaced daily for additional 2 days, followed by analysis with Anti-ETS1 (# MA5-15609, Thermo Fisher Scientific) and Alexa Fluor 488 donkey anti-mouse IgG (A21202, Thermo Fisher Scientific). All other antibodies used in this section are listed in Supplementary Table S2. Hereafter, CNCLCs were cultured in DMEM/F12 medium.
2.6. Preparation of root fragment from human third molar
All third molars used in this project were extracted from healthy donors. Approximately, 1 cm root fragments were cut out from the extracted third molar using a diamond disc (Brasseler USA). The root fragments were processed using endodontic rotary files to widen the apical foramen to approximately 1 mm, followed by removal of pulp tissue from the root canal using K-files. The hollow root fragments were antisepticized using 5.25% sodium hypochlorite and soaked in 17% EDTA to remove smear debris. After rinsing with PBS, the root fragments were subjected to a pulp regeneration assay. These protocols were approved by the Rutgers Institutional Review Board.
2.7. In vivo pulp regeneration assay
CNCLCs or DPCs cultured in a 24-well plate to 50% confluency were inoculated with lenti-tdTomato-puro vector (LV-CMV-tdTOMATO-Puro, Signagen) in serum-free DMEM/F12 + 10 μg/ml polybrene for 5 h. Following incubation of 48 h and media change to DMEM/F12 containing 10% serum, 0.5 μg/ml puromycin was added to select transduced cells. After 2 weeks of selection and passage, red fluorescence from the surviving cells was assessed microscopically.
The odontoblastic differentiation of the 5 × 105 labeled CNCLCs or DPCs was induced with osteo/odontogenic differentiation medium supplemented with 10 ng/ml each of FGF4 and FGF9 for 5 days. The 10–15 μl mixture of approximately 1 × 106 induced CNCLCs and 1% SLan were injected into the canal space of processed human root fragment, which was then capped on the crown side using glass ionomer and subcutaneously engrafted into the dorsa of 6-weeks-old NOD scid gamma mouse (NSG) mice (The Jackson Laboratory). These mice were housed in a single animal per cage in a facility with 12-h light—dark cycles and ad libitum food and water. The animal protocols were approved by the Rutgers University IACUC. The roots and surrounding tissues were collected 8 weeks later and subjected to histological analysis.
2.8. Histological analysis
The collected root fragments from the mice were fixed using 10% neutral buffered formalin and kept overnight at 4 °C. Following demineralization in 10% EDTA solution for 5–6 weeks at 4 °C, the samples were immersed in 10%, 20%, and 30% sucrose solutions. After embedding in Super Cryoembedding Medium (SCEM) (SECTION-LAB Co. Ltd. Hiroshima, Japan), the samples were frozen and sagittally sectioned, following the Kawamoto method (SECTION-LAB Co. Ltd. Hiroshima, Japan). The sections were stained with hematoxylin and eosin (H&E; Sigma-Aldrich, St. Louis, MO, USA). Immunohistochemical staining was performed using the EnVision system (Agilent), according to the manufacturer's instructions. The sections were incubated with anti-DSP (Santa Cruz Biotechnology, sc-73632) antibody at 4 °C, followed by labeling with HRP-conjugated anti-IgG antibody. Staining was completed after a 5 min incubation with 3,3-diaminobenzidine (DAB).
3. Results
3.1. CNCLC production and evaluation
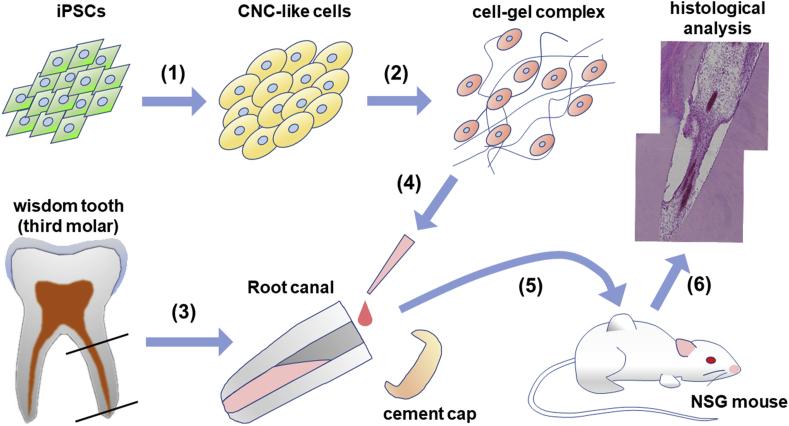
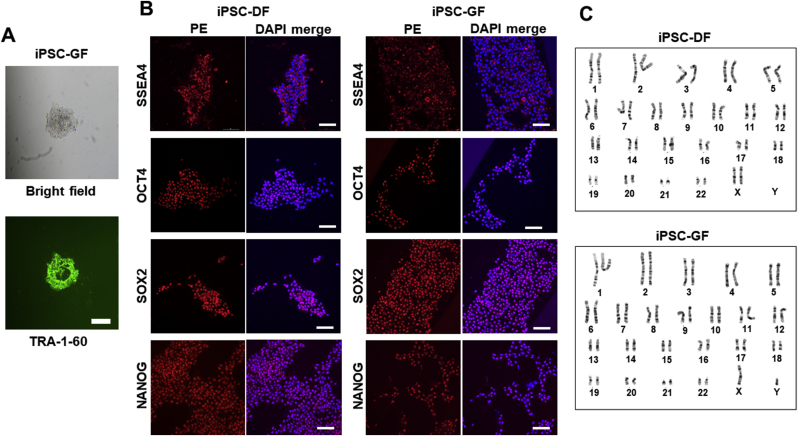
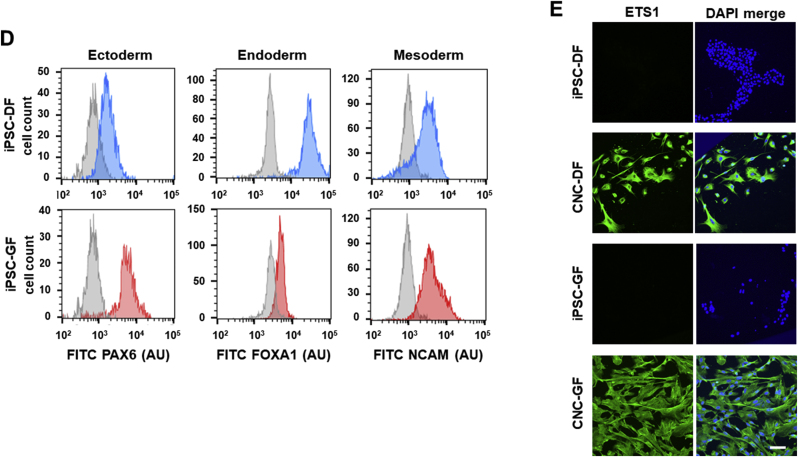
The overall strategy implemented in this study is illustrated in Fig. 1. First, CNCLCs were produced from iPSCs. To ensure the production of CNCLCs, iPSCs derived from two sources, human dermal fibroblasts (iPSC-DF) and human gingival fibroblasts (iPSC-GF), were used to produce two sets of CNCLCs (CNC-DF and CNC-GF). iPSC-DF was produced previously [36,37] and iPSC-GF was newly generated and isolated by colony-picking using TRA-1-60 live staining (Fig. 2A). The iPSCs obtained were validated by immunostaining for iPSC markers (Fig. 2B), by karyotyping with G-banding (Fig. 2C), and by assessing their ability to differentiate into the three germ layer cells using STEMdiff Trilineage Differentiation Kit, followed by immunostaining of ectoderm, endoderm, or mesoderm markers (Fig. 2D, Supplementary Fig. S1A–C). Both iPSC-DF and iPSC-GF showed the characteristics of authentic iPSCs and did not reveal any abnormalities in these analyses.
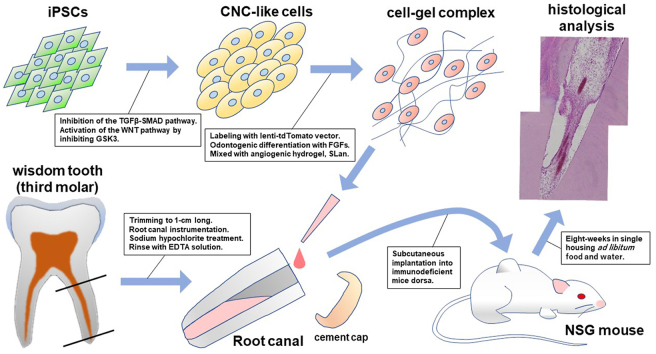
Fig. 1.
Illustrated flowchart of procedures taken in this study. (1) iPSCs differentiate into CNCLCs by the inhibition of TGFβ-SMAD pathway and the activation of WNT pathway by inhibiting GSK3, following the modified Lee's protocol [35]. (2) The CNCLCs were labeled with lenti-tdTomato vector and fated with FGFs and odontogenic differentiation media for 5 days. Mixing of cells with the angiogenic hydrogel immediately before injection to the scaffold. (3) Isolation of root parts of wisdom teeth (third molars) extracted from patients. The pulp cells were removed from them and antisepticized with sodium hypochlorite, followed by activation of the growth factors in the dentin with EDTA washing. (4) Injection of the cell-gel complex into the opened pulp space of the dentin scaffold. The apical foramen of the root was kept opened for nutrient supply to the internal CNCLCs from mouse body. The cell-gel complex does not leak from the apical foramen with the help of viscosity of the hydrogel. (5) After capping the crown side of the root with glass-ionomer cement, the dentin-scaffolds were engrafted into the dorsa of immunodeficient NSG mice subcutaneously. (6) Following eight-weeks in-vivo culture, the dentin-scaffolds were extracted and analyzed histologically.
Fig. 2.
Validation of the iPSCs produced in this study. (A) An iPSC-GF colony stained with anti-TRA1-Alexa Fluor 488 (green). Bright field (top) and TRA-1 (bottom). Scale bar = 200 μm. (B) iPSC markers detected with Immunostaining using PE-conjugated antibodies. Scale bar = 100 μm. (C) G-banding karyotype of iPSC-DF and iPSC-DF. (D) Validation of the iPSCs' ability to differentiate into the three germ layer cells. The differentiated cells and the progenitor iPSCs are immuno-stained with the FITC-conjugated antibodies for major markers of the three germ layer cells, followed by flowcytometry. Gray peaks and colored peaks correspond to untreated cells and cells treated with differentiation media, respectively. Y-axes correspond to cell number. AU stands for arbitrary unit. Scale bar = 100 μm. (E) The CNCLCs (CNC-DF and CNC-GF) and their progenitor cells (iPSC-DF and iPSC-GF) immuno-stained with anti-ETS1-Alexa 488 (green) and DAPI (blue). Scale bar = 100 μm.
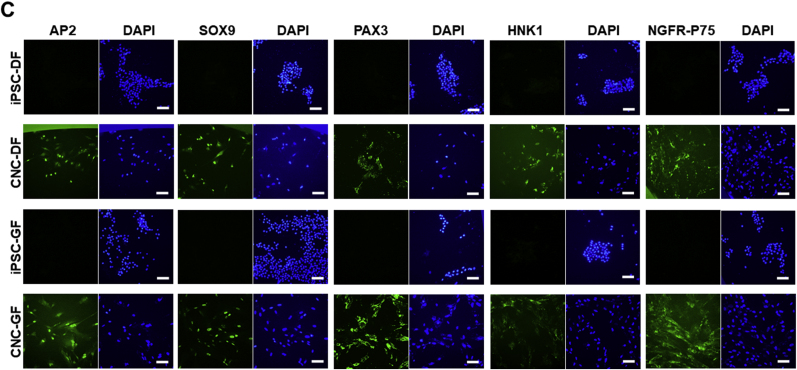
Presently, the most established strategy to generate neural crest (NC)-like cells from iPSCs is based on the activation of the canonical Wnt pathway by inhibiting GSK3 and the simultaneous inhibition of SMAD signaling [35,[38], [39], [40], [41]]. Meanwhile, Huang et al. reported that, in the absence of caudalization factors, such as retinoic acid, the NCs generated with this strategy show CNC-like features, such as high ETS proto-oncogene 1 transcription factor (ETS1) expression and low HOX genes expression [39]. Additionally, one of the established protocols using this WNT-SMAD strategy, published by Menendez et al., indicates that NCs are differentiated to peripheral neurons by N2-media (Thermo Fisher Scientific) treatment and to mesenchymal stem-like cells by FBS-containing media [38]. Considering these information and our final goal of generating dental pulp-like cells (belonging to mesenchymal lineage) from CNCLCs, we chose to eliminate the N2-media incubation steps from a facile but established protocol using the WNT-SMAD strategy, published by Lee et al. [35] and maintained the resulting CNCLCs in DMEM/F12 media supplemented with 10% FBS. With this modified method, both sets of CNCLCs (CNC-DF and CNC-GF) were generated and characterized by ETS1 expression via immunostaining. ETS1 is a transcription factor known to be specifically expressed in CNCs, but not in other portions of the neural crest [42,43]. ETS1 expression was clearly observed in the CNCLCs produced using our procedure (Fig. 2E), whereas it was absent in the undifferentiated iPSCs (Fig. 2E).
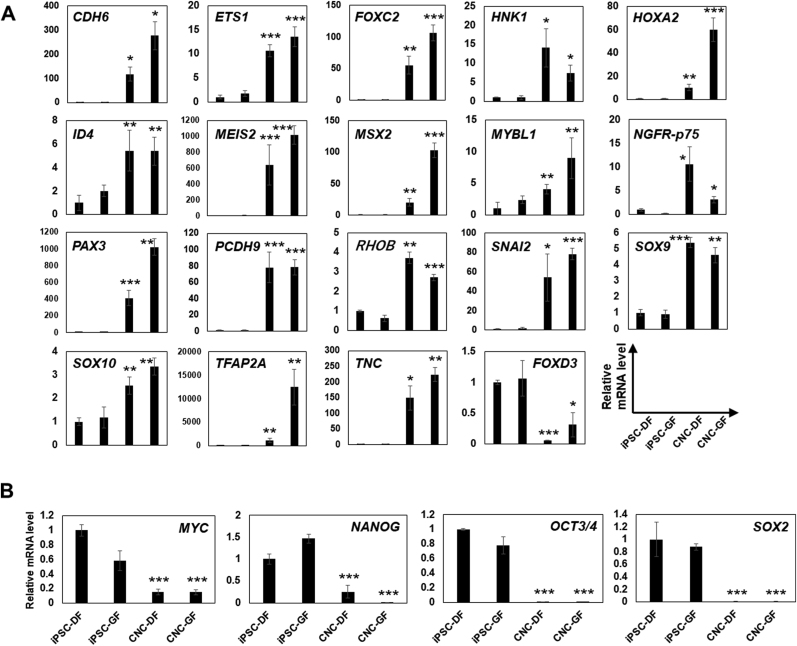
Next, the characteristics of the CNCLCs were evaluated by quantifying the expression levels of 19 published CNC marker genes using qPCR [21,44] (Fig. 3A and B). Interestingly, the expression of major CNC marker genes, such as ETS1, transcription factor AP-2 alpha (TFAP2A), 3-beta-glucuronosyltransferase 1 (B3GAT1 (HNK1)), low-affinity nerve growth factor receptor (NGFR (p75)), SRY-box transcription factor 9 (SOX9), 10 (SOX10), inhibitor of DNA binding 4 (ID4), Meis homeobox 2 (MEIS2), Msh homeobox 2 (MSX2), MYB Proto-Oncogene Like 1 (MYBL1), paired box gene 3 (PAX3), and snail family transcriptional repressor 2 (SNAI2), was upregulated in both CNC-DF and CNC-GF by approximately 2.5–12,000 fold compared to their parental cells (Fig. 3A). Furthermore, the expression of four pluripotent stem cell markers, c-Myc, octamer-binding transcription factor 3/4 (Oct3/4), homeobox protein NANOG, and SRY-box transcription factor 2 (SOX2), was significantly lower in the CNCLCs compared to their parental cells (Fig. 3B). These qPCR data were confirmed by immunostaining of the CNCLCs and the iPSCs with antibodies for the markers mentioned above (Fig. 3C, Supplementary Fig. S2) These data corroborated the validity of use of CNCLCs, including pulp-dentin complex regeneration, in further research. Subsequently, we decided to use CNC-GF for our model system since it has clearer CNC-like characteristics compared to CNC-DF (Fig. 3A and B). Especially, CNC-GF showed 5.1-fold higher MSX2 and 11.4-fold higher TFAP2A expression (Fig. 3A), while it expressed only 0.01-fold NANOG, an iPSC marker (Fig. 3B), compared to CNC-DF. Meanwhile, forkhead box D3 (FOXD3) expression in the CNCLCs was lower than that in the iPSC controls (Fig. 3A) (discussed later).
Fig. 3.
Relative expression levels of (A) indicated CNC marker genes and (B) pluri-potent stem cell marker genes (MYC, NANOG, OCT3/4, and SOX2) in iPSC-DF, iPSC-GF, CNC-DF, or CNC-GF. qPCR Ct-values were normalized with β-actin level and iPSC-DF values. Each column shows averaged value obtained from at least three independent samples. Error bars indicate standard deviations. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the parental iPSC group. (C) Immunostaining of major CNC markers on CNC-DF, CNC-GF, and their progenitor iPSCs with antibodies conjugated with FITC or non-conjugated antibodies detected by Alexa Fluor 488-conjugated secondary antibody (Supplementary Table S2). Scale bar = 100 μm.
3.2. With FGF4+9 stimulation, the CNCLCs effectively differentiate to odontoblast-like cells
One critical characteristic of dental pulp is the presence of odontoblast-like cell progenitors, which maturate in response to traumatic stimuli and produce reparative dentin [[45], [46], [47]]. Here, by investigating the ability of CNCLCs to differentiate into odontoblastic cells, we evaluated their potential to mimic dental pulp cells.
Odontoblast differentiation during tooth development is coordinately controlled by a multitude of signaling proteins, including fibroblast growth factors (FGFs), bone morphogenetic proteins (BMPs), Wnts, and sonic hedgehogs (SHHs) [48,49]. Recently, it was shown that BMP4 directly stimulates odontoblast differentiation of iPSCs derived from DPSCs [50]. However, when iPSCs are considered as a personalized regenerative medicine source, using the reported dose of BMP4 protein for differentiating each patient's iPSCs would be quite costly [51]. In addition, it is debatable to isolate DPSCs from a patient's healthy tooth as an iPSC source. These problems led us to test whether FGFs alone can induce odontoblastic differentiation of CNCLCs, since FGF proteins are commercially inexpensive and are known to contribute to odontoblast differentiation [48]. An additional rationale is that FGFs have previously induced a strong morphological change in CNCLCs in our laboratory (data not shown).
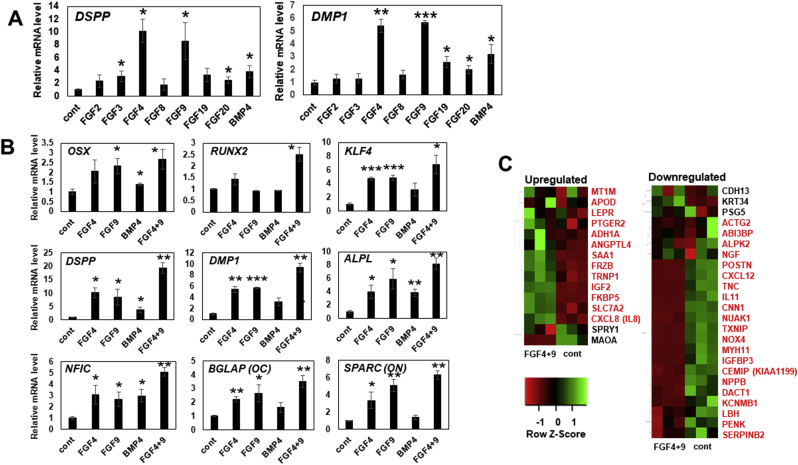
CNC-GFs were treated with odontogenic differentiation media supplemented with FGF2, 3, 4, 8, 9, 19, 20, or BMP4 for 14 days, and the expression of dentin sialophosphoprotein (DSPP) and dentin matrix acidic phosphoprotein 1 (DMP1) were compared (Fig. 4A). We chose these FGFs since they represent well other FGFs in the same subtypes to which they belong [52] and are involved in the odontogenesis process [48]. Interestingly, FGF4 and FGF9 induced both DSPP and DMP1 with the highest efficiency (Fig. 4A), whereas BMP4 showed a moderate effect. This suggested that BMP4 application for odontoblastic differentiation can be replaced with the combined use of FGF4 and 9.
Fig. 4.
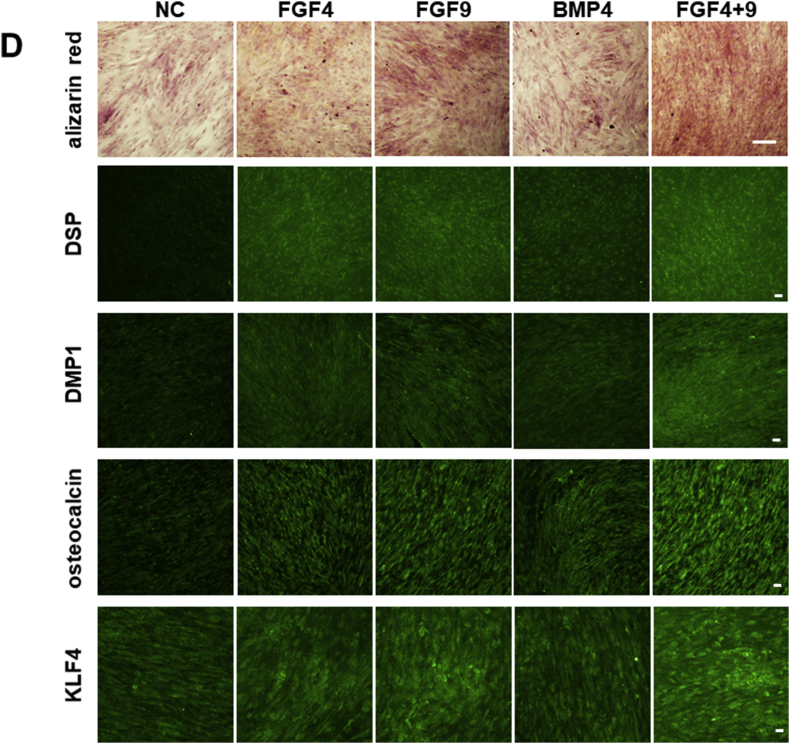
(A) Relative expression levels of odontoblast marker genes, DSPP and DMP1, in CNC-GF treated with 10 ng/ml of FGF2, FGF4, FGF8, FGF9, FGF19, FGF20, or 50 ng/ml BMP4, for 14 days. qPCR Ct-values were normalized with β-actin level and non-treated negative control values. Each column shows averaged value obtained from at least three independent samples. Error bars indicate standard deviations. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the control group. Cont: non-treated control. (B) Relative expression levels of genes upregulated through odontoblast differentiation (OSX, RUNX2, KLF4, DSPP, DMP1, ALPL, NFIC, OC, ON) in CNC-GF treated with FGF4, FGF9, BMP4, or FGF4+9 for 14 days. qPCR Ct-values were normalized with β-actin level and negative control values. Each column shows averaged value obtained from at least three independent samples. Error bars indicate standard deviations. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the control group. Cont: non-treated control. (C) Gene-expression-level heatmaps depicting odontogenic differentiation of CNC-GF treated with FGF4+9. Expression levels of the genes reported to be upregulated more than five folds (left column) and downregulated more than five folds (right column) in odontogenic differentiation process were assessed from the RNA-seq data of FGF4+9-treated CNC-GF or the non-treated negative control. RNA-seq data were taken from three independent samples for each condition. Z-score scale bar is shown in the left-bottom. Genes indicated in red behaved as published previously, while genes indicated in black did not. Cont: non-treated control. (D) CNC-GF treated with odontogenic media supplemented with indicated growth factor(s) for 14 days. NC stands for non-treated negative control. (First row) Alizarin Red staining. Calcium deposition forms red pigment with Alizarin Red S, visualizing odontoblast differentiation. Scale bar = 200 μm. (The second to fifth row) Immuno-staining using anti-DSP, anti-DMP1, anti-osteocalcin, or anti-KLF4 primary antibody and secondary antibody conjugated with Alexa fluor 488 (green). Scale bar = 100 μm.)
To evaluate the odontoblastic differentiation induced by FGF4 and 9, CNC-GFs were treated with FGF4, 9, 4 + 9, or BMP4 for 14 days, and the expression of nine genes known to characterize odontoblastic differentiation, which are osterix (OSX), runt-related transcription factor 2 (RUNX2), Kruppel-like factor 4 (KLF4), DSPP, DMP1, alkaline phosphatase, tissue-nonspecific isozyme (ALPL), nuclear factor 1 C-type (NFIC), osteocalcin (OC), and osteonectin (ON), was analyzed by qPCR (Fig. 4B). Interestingly, all the quantified genes were significantly upregulated in treated CNC-GFs. FGF4+9 combination showed the highest induction potency with 1.7–5.0 fold efficacy as compared to BMP4 (Fig. 4B). Similar results were obtained on immunostaining of CNC-GFs (treated as above) for major odontoblast markers using Alexa 488-conjugated antibodies (Fig. 4D, the second to fifth row). DSP, DMP1, osteocalcin, and KLF4 expression was highly augmented in cells treated with FGF4+9 (Fig. 4D).
To further evaluate the odontoblastic differentiation induced by FGF4+9, genome-wide gene expression of FGF4+9-treated CNC-GFs was analyzed using RNA-seq technology. From the RNA-seq data, we subsequently assessed expression levels of those genes that have been reported to be upregulated (UG) or downregulated (DG) more than five-fold in the odontoblastic differentiation of dental pulp cells [53] (Fig. 4C). Thirteen out of 16 UGs were upregulated, whereas protein sprouty homolog 1 (SPRY1) and monoamine oxidase A (MAOA) were downregulated and metallothionein-1G (MT1G) level was under the RNA-seq threshold (Fig. 4C, left). Out of 24 DGs, 21 were downregulated, whereas T-cadherin (CDH13), Keratin 34 (KRT34), and pregnancy-specific beta-1-glycoprotein 5 (PSG5) were upregulated (Fig. 4C, right). Additionally, to confirm the functionality of CNC-GF treated with FGF4+9, calcium deposition was assayed using alizarin red staining (Fig. 4D, top). After 14 days of the treatment, FGF4+9 induced the highest calcium deposition, whereas BMP4 showed moderate accumulation. These data indicated that CNC-GFs stimulated with FGF4+9 can differentiate into odontoblast-like cells, suggesting that CNC-GF is phenotypically potent to replicate the characteristics of dental pulp tissue.
3.3. The neovascularization induced by angiogenic hydrogel, SLan, aids CNC-GF replicate the characteristics of dental pulp tissue in vivo
To promote CNC-GF to replicate the characteristics of dental pulp tissue containing blood vessels, which is a key factor for regenerated tissue to survive in vivo, we introduced the cytocompatible pro-angiogenic hydrogel, SLan. SLan is a self-assembling peptide hydrogel (SAPH) that contains the functional mimetic of the VEGF-165 signaling domain, “QK” (seq. KLTWQELYQLKYKGI), at the peptide N-terminus [27,28,31]. Importantly, SLan has already shown its ability to induce angiogenesis in a dog model [22].
Prior to the in vivo assay, CNC-GFs were labeled with the lenti-tdTomato vector to be distinguished from the host cells. The odontoblastic differentiation of labeled cells was induced with FGF4+9 for a short period (five days), resuspended in SLan, and then injected into processed human tooth root fragments to mimic the real root canal environment [[54], [55], [56], [57]]. The roots were subcutaneously implanted into the dorsa of the NSG mice. Eight weeks later, the harvested cell-SLan complexes were analyzed histologically (Fig. 5).
Fig. 5.
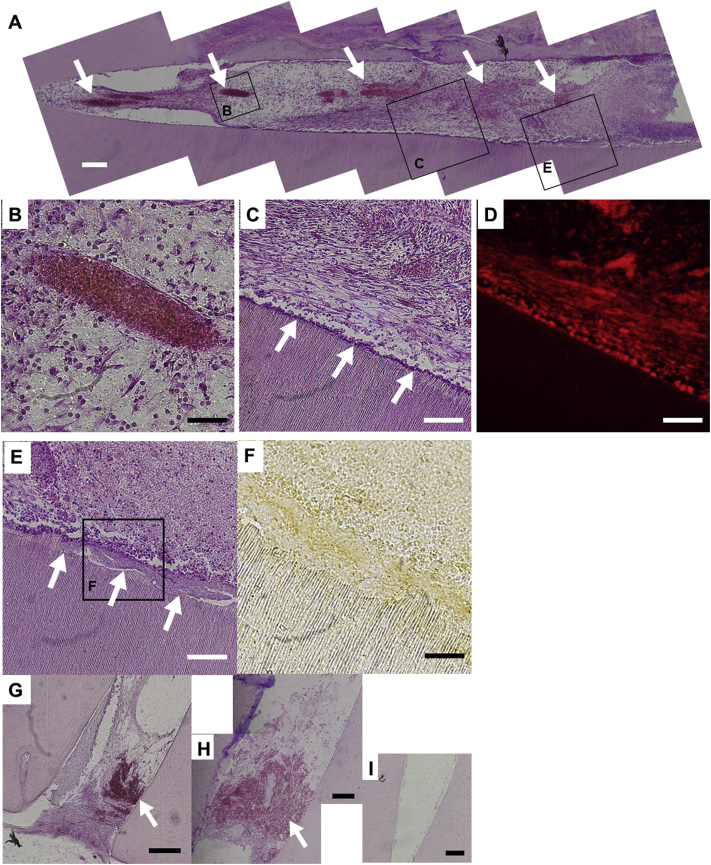
Histological analysis of the root containing the complex of SLan-hydrogel and CNC-GF pretreated with odontogenic medium including FGF4+9. Following in vivo culture for eight weeks, the engrafted scaffolds were extracted from mice dorsa, sliced with cryostat, and stained with H & E. (A) Full view of one of the scaffolds. The left side and right side correspond to apical- and crown-side of scaffold, respectively. Spindle-shaped red regions correspond to cross-sectioned neovascularized blood vessels (white arrows). Purple dots correspond to nuclei of CNC-GFs. Boxed areas are magnified and shown in indicated figures (B–E). Scale bar = 200 μm. (B) An example of neovascularization into the cell-SLan complex. Erythrocytes in the blood vessel are strongly red-stained with eosin. Scale bar = 50 μm. (C) The boundary between the cell-SLan complex (top) and dentin scaffold (bottom). CNC-GFs aligned on the surface of dentin scaffold (white arrows). Scale bar = 100 μm. (D) Another slice from region (C) showing red fluorescence. Red cells correspond to CNC-GFs. (E) Dentin-like layer formed in the space between CNC-GFs and dentin scaffold (white arrows). Scale bar = 100 μm. (F) Immunostaining of DSP in the cell-SLan complex with DAB, showing boxed area of image-(E). Regions with dense DSP are brown-stained. Scale bar = 50 μm. G, H, I denote the apical region of control root fragments, respectively; (G) DPCs and SLan, (H) SLan only, and (I) PBS only. Scale bar = 200 μm. Erythrocytes in blood vessels are red-stained (white arrows).
Interestingly, vast neovascularization was observed in the injected cell-SLan complex, which intruded into 82.7% of the cell-SLan complex length in an average of three independent tests (Fig. 5A and B). Neovascularization was present even in the control roots with DPC-SLan or SLan alone (Fig. 5G and H), but was absent in the control root fragment with PBS only (Fig. 5I). This suggests that SLan alone can effectively induce neovascularization in the root canal space. Moreover, aligned odontoblast-like cells (Fig. 5C) and dentin-like layers were detected by HE staining (Fig. 5E) and DSP immunostaining (Fig. 5F). These cells were confirmed to originate from CNC-GF due to their red fluorescence from tdTomato RFP (Fig. 5D). These data suggested that SLan induces intensive neovascularization, and CNC-GFs can differentiate into dental pulp-like tissue.
4. Discussion
In the present study, we replicated the characteristics of dental pulp tissues using CNCLCs derived from human iPSCs. Since theoretically iPSCs can be generated from somatic cells of any age group, it can be suitable for pulp regeneration in elderly patients, who lack a sufficient number of stem cells, such as DPSC, SHED, SCAP, or PDLSC. However, iPSC technology is reported to have practical obstacles to be forwarded to clinical trials, of which tumorigenicity is the most important concern [58]. iPSCs are at a risk of tumorigenesis when undifferentiated or immature iPSCs are contaminated in their target cell products, reprograming factors (especially cMyc) remain active even after their differentiation into the target cell type, or oncogenic mutations occur during the culture in vitro [58]. To circumvent these situations, not a few technologies have been developed or studied to date. Sawa and colleagues focused on CD30 epitope expressed by iPSCs and utilized brentuximab vedotin, an FDA-approved drug for CD30-positive lymphoma [59]. Brentuximab vedotin is an anti-CD30 antibody conjugated with the antimitotic agent, monomethyl auristatin E, which selectively bound to and eliminated undifferentiated iPSCs in their target cell product, cardiac myocytes, prior to engrafting. Hayashi et al. used a similar strategy to remove undifferentiated iPSCs from corneal epithelial cells utilizing an antibody for CD200, which is expressed by undifferentiated iPSCs but not by mature corneal epithelial cells [60]. The suicide gene strategy proposed by Kojima et al. could be effective in eliminating both undifferentiated iPSCs and tumor cells originating from them [61]. They introduced the herpes simplex virus type 1 thymidine kinase (HSVtk) gene into iPSCs. On treating iPSCs harboring HSVtk with ganciclovir (GCV), HSVtk converts the GCV to GCV-triphosphate, a DNA synthesis inhibitor that promotes apoptosis of dividing cells selectively due to cell cycle arrest [62]. These new technologies are based on the facile methods to distinguish iPSCs and their cancerous forms from the differentiated target cell type and on the existing FDA-approved drugs to eliminate them in vitro. Likewise, a facile method to distinguish iPSCs from dental pulp cells may be a key to forward iPSC technology for use in regenerative endodontics.
In the phenotypic evaluation of the CNCLCs, we found that CNC-DF and CNC-GF express 18 major CNC marker genes with high magnitude, while their expression levels of iPSC marker genes were considerably lower than those of iPSC-DF and iPSC-GF (Fig. 3A and B). This suggests that the CNCLCs we produced mimicked actual CNCs well. Meanwhile, the expression levels of FOXD3 in CNC-DF and CNC-GF were lower than those in their parental iPSCs. A similar phenomenon was reported previously [21], in which the mouse CNCLCs showed lower Foxd3 expression compared to the parental iPSCs. This phenomenon can be explained by the function of FOXD3 in pluripotent stem cells. FOXD3 is active in pluripotent stem cells to maintain their pluripotency [63] and also to promotes pluripotent stem cells to exit from their naïve state [64]. However, mesenchymal progenitors of NC do not express FOXD3 [65]. These observations suggest that iPSCs may express a high level of FOXD3 to maintain their pluripotency and that CNCLCs, which have undergone some level of differentiation, may not require FOXD3 expression at the level of iPSCs. In fact, the FOXD3 expression levels in CNC-DF and CNC-GF were not negligibly low in this study, reaching approximately 1/2–1/6 of TFAP2A, the most-upregulated CNC marker gene in this study relative to β-actin by qPCR, although these values are dependent on their PCR primer sensitivity.
One unexpected finding in this study is that FGF4 and FGF9 showed the highest inducibility for odontoblastic differentiation of CNCLCs. Both FGFs belong to the secreted FGF group, which signals to receptor tyrosine kinases (FGFRs) [52]. In humans, 18 secreted FGFs have been identified so far and classified into six subfamilies, that is, FGF1, FGF4, FGF7, FGF8, FGF9, and FGF15/19-subfamily. Each subfamily has a certain level of specificity for the partner FGFR [52]. Secreted FGFs play essential roles in the early stages of embryonic development and organogenesis, such as odontogenesis, including odontoblast differentiation. More specifically, during odontoblast development, the dental mesenchyme expresses FGFR1c, 2c, and 1b. Concurrently, pre-ameloblasts and ameloblasts secrete FGF3 (FGF7-subfamily, prefers FGFR2b), 4 (4-subfamily, 1c and 2c), 9 (9-subfamily, 3c), 15/19 (15/19-subfamily, weakly 1c, 2c. 3c and 4Δ), and 20 (9-subfamily, 3c) to stimulate odontoblast differentiation [48]. Considering the expression patterns of FGFs and FGFRs in odontoblast differentiation, FGF4 stimulating FGFR1c and 2c may be most effective for odontoblast differentiation. Meanwhile, in our assay assessing FGFs effect on CNC-GF's odontoblastic differentiation, FGF4 and, interestingly, FGF9 showed the highest efficacy (Fig. 4A). FGF9 has the highest affinity for FGFR3c, which has not been reported to be expressed in the dental mesenchyme, while FGF9 is secreted from enamel knot/ameloblasts during odontogenesis [48]. This discrepancy may not be due to the difference between CNC and dental mesenchyme, since FGF9 has been reported to induce odontoblastic differentiation in DPCs [66]. The weak affinity of FGF9 with FGFR2c [52] could be significant for the induction of odontoblastic differentiation of CNCLCs.
In the evaluation of phenotypic similarity to DPCs, CNC-GF differentiated into odontoblast-like cells with FGF4+9 stimulation. These odontoblast-like cells expressed the genes characteristic to the odontoblastic differentiation of DPCs (Fig. 4B and C), which proved that CNCLCs have DPC characteristics and that FGF4+9 can induce their odontoblastic differentiation. However, among the checked genes, SPRY1, MAOA, CDH13, KRT34, and PSG5 were not regulated, as previously reported [53] (Fig. 4C). This discrepancy could be explained by the following relationships between these genes and FGFs or NC/neural tissues: SPRY1 prevents excessive FGF signaling in multiple cell types throughout cerebellar development [67]. KRT34 and FGF9 are markedly upregulated in fibroblasts from patients with Dupuytren's disease, which is characterized by excessive local proliferation of fibroblasts and over-production of collagen and other components of the extracellular matrix (ECM) in the palmar fascia [68]. PSG5 expression is downregulated in human mesenchymal stem cells treated with FGF2 [69]. MAOA catalyzes the deamination of neurotransmitters, such as dopamine, norepinephrine, and serotonin. CDH13 expression alternates with migrating neural crest cells [70]. These findings indicate a relationship between the above five genes and FGFs or NC/neural tissues and suggest that the discrepancy in the regulation of the five genes could be due to the unique circumstances of this study using FGFs and CNCLC.
In this study, a self-assembling peptide hydrogel, SLan, significantly assisted CNC-GFs to replicate the characteristics of dental pulp tissue by inducing neovascularization. SLan recovers approximately 80% of the mechanical strength 1 min after shear-thinning, owing to its non-covalent self-assembling peptide backbone, K(SL)6K [23]. This characteristic enables SLan to be easily dispensed through a narrow bore syringe and near-instantaneously re-gel in situ. Furthermore, since SLan has a VEGF signaling domain in its own peptide, it can be tuned to undergo gradual degradation over 3–6 weeks in vivo [24], thereby extending the angiogenic induction period, as compared to classic hydrogels incorporated with rhVEGF (4–8 h) [32,33]. These two characteristics make SLan quite a suitable scaffold gel for pulp regeneration in situ.
The importance of angiogenesis in the survival of transplanted stem cells regenerating into dental pulp-like tissue has been well reported (reviewed in Ref. [71]). Since oxygen, which is required for cell survival, can diffuse only 150–200 μm deep from the surface of a cell clump [72,73], the long-term survival and function of transplanted 3-dimensional tissues depend on rapid angiogenesis, not only on the surface but also in the center of the tissue grafts. In fact, the induction of neovascularization is one of the major limitations in successful transplantation of tissue engineering products in clinical practice [74]. To achieve rapid angiogenesis, strategies involving incorporation of VEGF into hydrogels with controlled diffusion have been exploited [75]; however, many of them are challenged by the cost of rhVEGF and its rapid diffusion. Including the VEGF-motif in the hydrogel monomer sequence is a promising strategy [[26], [27], [28]]. Further work from our group has shown the facile modifiability of this platform to promote DPSC proliferation, neurogenesis, inflammation modulation, and others [[76], [77], [78], [79]]. Recently, stromal cell-derived factor 1 alpha (SDF-1α) was shown to enhance angiogenesis remarkably when overexpressed together with VEGF in DPSCs [80]. This finding could enhance our system utilizing CNCLCs and VEGF mimetics. SDF-1α can be incorporated into SLan hydrogel, which could augment the efficacy of pulp regeneration from CNCLCs. Furthermore, another group showed that VEGF- and BDNF-mimetic peptides have synergistic effects on peripheral nerve regeneration [81]. These synergistic effects from material combinations may be exploited to advance in-situ pulp regeneration toward practical treatment in the future.
5. Conclusions
We have shown that CNCLCs can be generated by simplifying the established method for deriving NCs from iPSCs. The produced CNCLCs showed upregulation of major CNC marker genes by several orders of magnitude. Furthermore, the combination of FGF4 and FGF9 induced the CNCLCs toward odontoblastic differentiation more effectively than BMP4. Moreover, by engrafting into mice dorsa, the CNCLCs briefly fated with FGF4 + FGF9 replicated the characteristics of dental pulp tissue harboring odontoblast-like cells, dentin-like layer, and vast neovascularization induced by the angiogenic SAPH, SLan. Thus, this study demonstrated a successful collaboration between regenerative medicine and SAPH technology.
Data availability statement
All data needed to evaluate the conclusions in the paper are presented in the paper and/or the Supplementary Materials. Additional data related to this study may be requested from the authors.
CRediT authorship contribution statement
Yoshifumi Kobayashi: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Julie Nouet: Investigation, Methodology, Visualization, Writing – review & editing. Erdenechimeg Baljinnyam: Investigation, Methodology, Writing – review & editing. Zain Siddiqui: Investigation, Visualization, Writing – review & editing. Daniel H. Fine: Resources, Supervision, Writing – review & editing. Diego Fraidenraich: Methodology, Supervision, Validation, Writing – review & editing. Vivek A. Kumar: Methodology, Resources, Supervision, Writing – review & editing. Emi Shimizu: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
VAK has equity interests in commercialization ventures aiming to translate these and related technologies.
Acknowledgements
This study was supported by NIH grants, R01DE025885 (E.S), R15EY029504 (VAK), and National Science Foundation NSF IIP 1903617 (VAK).
Footnotes
Headline: Angiogenic gel aids CNC-like cells to mimic dental pulp.
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.11.014.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Nakashima M., Akamine A. The application of tissue engineering to regeneration of pulp and dentin in endodontics. J. Endod. 2005;31(10):711–718. doi: 10.1097/01.don.0000164138.49923.e5. [DOI] [PubMed] [Google Scholar]
- 2.Huang G.T., Al-Habib M., Gauthier P. Challenges of stem cell-based pulp and dentin regeneration: a clinical perspective. Endod. Top. 2013;28(1):51–60. doi: 10.1111/etp.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin L.M., Huang G.T., Sigurdsson A., Kahler B. Clinical cell-based versus cell-free regenerative endodontics: clarification of concept and term. Int. Endod. J. 2021 doi: 10.1111/iej.13471. [DOI] [PubMed] [Google Scholar]
- 4.Lovelace T.W., Henry M.A., Hargreaves K.M., Diogenes A. Evaluation of the delivery of mesenchymal stem cells into the root canal space of necrotic immature teeth after clinical regenerative endodontic procedure. J. Endod. 2011;37(2):133–138. doi: 10.1016/j.joen.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Galler K.M., Eidt A., Schmalz G. Cell-free approaches for dental pulp tissue engineering. J. Endod. 2014;40(4 Suppl):S41–S45. doi: 10.1016/j.joen.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Cordeiro M.M., Dong Z., Kaneko T., Zhang Z., Miyazawa M., Shi S., Smith A.J., Nor J.E. Dental pulp tissue engineering with stem cells from exfoliated deciduous teeth. J. Endod. 2008;34(8):962–969. doi: 10.1016/j.joen.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Jung C., Kim S., Sun T., Cho Y.B., Song M. Pulp-dentin regeneration: current approaches and challenges. J. Tissue Eng. 2019;10 doi: 10.1177/2041731418819263. 2041731418819263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radwan I.A., Rady D., Abbass M.M.S., El Moshy S., AbuBakr N., Dorfer C.E., Fawzy El-Sayed K.M. Induced pluripotent stem cells in dental and nondental tissue regeneration: a review of an unexploited potential. Stem Cell. Int. 2020;2020:1941629. doi: 10.1155/2020/1941629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strassler E.T., Aalto-Setala K., Kiamehr M., Landmesser U., Krankel N. Age is relative-impact of donor age on induced pluripotent stem cell-derived cell functionality. Front. Cardiovasc. Med. 2018;5:4. doi: 10.3389/fcvm.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otsu K., Kishigami R., Oikawa-Sasaki A., Fukumoto S., Yamada A., Fujiwara N., Ishizeki K., Harada H. Differentiation of induced pluripotent stem cells into dental mesenchymal cells. Stem Cell. Dev. 2012;21(7):1156–1164. doi: 10.1089/scd.2011.0210. [DOI] [PubMed] [Google Scholar]
- 11.Seki D., Takeshita N., Oyanagi T., Sasaki S., Takano I., Hasegawa M., Takano-Yamamoto T. Differentiation of odontoblast-like cells from mouse induced pluripotent stem cells by Pax9 and Bmp4 transfection. Stem Cell Transl. Med. 2015;4(9):993–997. doi: 10.5966/sctm.2014-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M., Zhang X., Luo J., Yan R., Niibe K., Egusa H., Zhang Z., Xie M., Jiang X. Investigate the odontogenic differentiation and dentin-pulp tissue regeneration potential of neural crest cells. Front. Bioeng. Biotechnol. 2020;8:475. doi: 10.3389/fbioe.2020.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayor R., Theveneau E. The neural crest. Development. 2013;140(11):2247–2251. doi: 10.1242/dev.091751. [DOI] [PubMed] [Google Scholar]
- 14.Achilleos A., Trainor P.A. Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 2012;22(2):288–304. doi: 10.1038/cr.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santagati F., Rijli F.M. Cranial neural crest and the building of the vertebrate head. Nat. Rev. Neurosci. 2003;4(10):806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- 16.Cordero D.R., Brugmann S., Chu Y., Bajpai R., Jame M., Helms J.A. Cranial neural crest cells on the move: their roles in craniofacial development. Am. J. Med. Genet. 2011;155A(2):270–279. doi: 10.1002/ajmg.a.33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharpe P.T. Neural crest and tooth morphogenesis. Adv. Dent. Res. 2001;15:4–7. doi: 10.1177/08959374010150011001. [DOI] [PubMed] [Google Scholar]
- 18.Miletich I., Sharpe P.T. Neural crest contribution to mammalian tooth formation. Birth Defects Res C Embryo Today. 2004;72(2):200–212. doi: 10.1002/bdrc.20012. [DOI] [PubMed] [Google Scholar]
- 19.Mimura S., Suga M., Okada K., Kinehara M., Nikawa H., Furue M.K. Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells. Int. J. Dev. Biol. 2016;60(1–3):21–28. doi: 10.1387/ijdb.160040mk. [DOI] [PubMed] [Google Scholar]
- 20.Suga M., Hayashi Y., Furue M.K. In vitro models of cranial neural crest development toward toxicity tests: frog, mouse, and human. Oral Dis. 2017;23(5):559–565. doi: 10.1111/odi.12523. [DOI] [PubMed] [Google Scholar]
- 21.Odashima A., Onodera S., Saito A., Ogihara Y., Ichinohe T., Azuma T. Stage-dependent differential gene expression profiles of cranial neural crest-like cells derived from mouse-induced pluripotent stem cells. Med. Mol. Morphol. 2020;53(1):28–41. doi: 10.1007/s00795-019-00229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddiqui Z., Sarkar B., Kim K.K., Kadincesme N., Paul R., Kumar A., Kobayashi Y., Roy A., Choudhury M., Yang J., et al. Angiogenic hydrogels for dental pulp revascularization. Acta Biomater. 2021;126:109–118. doi: 10.1016/j.actbio.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aulisa L., Dong H., Hartgerink J.D. Self-assembly of multidomain peptides: sequence variation allows control over cross-linking and viscoelasticity. Biomacromolecules. 2009;10(9):2694–2698. doi: 10.1021/bm900634x. [DOI] [PubMed] [Google Scholar]
- 24.Moore A.N., Lopez Silva T.L., Carrejo N.C., Origel Marmolejo C.A., Li I.C., Hartgerink J.D. Nanofibrous peptide hydrogel elicits angiogenesis and neurogenesis without drugs, proteins, or cells. Biomaterials. 2018;161:154–163. doi: 10.1016/j.biomaterials.2018.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar V.A., Taylor N.L., Shi S., Wickremasinghe N.C., D'Souza R.N., Hartgerink J.D. Self-assembling multidomain peptides tailor biological responses through biphasic release. Biomaterials. 2015;52:71–78. doi: 10.1016/j.biomaterials.2015.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen P.K., Gao W., Patel S.D., Siddiqui Z., Weiner S., Shimizu E., Sarkar B., Kumar V.A. Self-assembly of a dentinogenic peptide hydrogel. ACS Omega. 2018;3(6):5980–5987. doi: 10.1021/acsomega.8b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar V.A., Taylor N.L., Shi S., Wang B.K., Jalan A.A., Kang M.K., Wickremasinghe N.C., Hartgerink J.D. Highly angiogenic peptide nanofibers. ACS Nano. 2015;9(1):860–868. doi: 10.1021/nn506544b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar V.A., Liu Q., Wickremasinghe N.C., Shi S., Cornwright T.T., Deng Y., Azares A., Moore A.N., Acevedo-Jake A.M., Agudo N.R., et al. Treatment of hind limb ischemia using angiogenic peptide nanofibers. Biomaterials. 2016;98:113–119. doi: 10.1016/j.biomaterials.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hitscherich P., Nguyen P.K., Kannan A., Chirayath A., Anur S., Sarkar B., Lee E.J., Kumar V.A. Injectable self-assembling peptide hydrogels for tissue writing and embryonic stem cell culture. J. Biomed. Nanotechnol. 2018;14(4):802–807. doi: 10.1166/jbn.2018.2583. [DOI] [PubMed] [Google Scholar]
- 30.Kim K.K., Siddiqui Z., Patel M., Sarkar B., Kumar V.A. A self-assembled peptide hydrogel for cytokine sequestration. J. Mater. Chem. B. 2020;8(5):945–950. doi: 10.1039/c9tb02250c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma X., Agas A., Siddiqui Z., Kim K., Iglesias-Montoro P., Kalluru J., Kumar V., Haorah J. Angiogenic peptide hydrogels for treatment of traumatic brain injury. Bioact. Mater. 2020;5(1):124–132. doi: 10.1016/j.bioactmat.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D'Andrea L.D., Iaccarino G., Fattorusso R., Sorriento D., Carannante C., Capasso D., Trimarco B., Pedone C. Targeting angiogenesis: structural characterization and biological properties of a de novo engineered VEGF mimicking peptide. Proc. Natl. Acad. Sci. U. S. A. 2005;102(40):14215–14220. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webber M.J., Tongers J., Newcomb C.J., Marquardt K.T., Bauersachs J., Losordo D.W., Stupp S.I. Supramolecular nanostructures that mimic VEGF as a strategy for ischemic tissue repair. Proc. Natl. Acad. Sci. U. S. A. 2011;108(33):13438–13443. doi: 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somerman M.J., Archer S.Y., Imm G.R., Foster R.A. A comparative study of human periodontal ligament cells and gingival fibroblasts in vitro. J. Dent. Res. 1988;67(1):66–70. doi: 10.1177/00220345880670011301. [DOI] [PubMed] [Google Scholar]
- 35.Lee G., Chambers S.M., Tomishima M.J., Studer L. Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 2010;5(4):688–701. doi: 10.1038/nprot.2010.35. [DOI] [PubMed] [Google Scholar]
- 36.Baljinnyam E., Venkatesh S., Gordan R., Mareedu S., Zhang J., Xie L.H., Azzam E.I., Suzuki C.K., Fraidenraich D. Effect of densely ionizing radiation on cardiomyocyte differentiation from human-induced pluripotent stem cells. Phys. Rep. 2017;5(15) doi: 10.14814/phy2.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkatesh S., Baljinnyam E., Tong M., Kashihara T., Yan L., Liu T., Li H., Xie L.H., Nakamura M., Oka S.I., et al. Proteomic analysis of mitochondrial biogenesis in cardiomyocytes differentiated from human induced pluripotent stem cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2020 doi: 10.1152/ajpregu.00207.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menendez L., Kulik M.J., Page A.T., Park S.S., Lauderdale J.D., Cunningham M.L., Dalton S. Directed differentiation of human pluripotent cells to neural crest stem cells. Nat. Protoc. 2013;8(1):203–212. doi: 10.1038/nprot.2012.156. [DOI] [PubMed] [Google Scholar]
- 39.Huang M., Miller M.L., McHenry L.K., Zheng T., Zhen Q., Ilkhanizadeh S., Conklin B.R., Bronner M.E., Weiss W.A. Generating trunk neural crest from human pluripotent stem cells. Sci. Rep. 2016;6:19727. doi: 10.1038/srep19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mica Y., Lee G., Chambers S.M., Tomishima M.J., Studer L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013;3(4):1140–1152. doi: 10.1016/j.celrep.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan A., Toh Y.C. Human pluripotent stem cell-derived neural crest cells for tissue regeneration and disease modeling. Front. Mol. Neurosci. 2019;12:39. doi: 10.3389/fnmol.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barembaum M., Bronner M.E. Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Dev. Biol. 2013;382(2):567–575. doi: 10.1016/j.ydbio.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tahtakran S.A., Selleck M.A. Ets-1 expression is associated with cranial neural crest migration and vasculogenesis in the chick embryo. Gene Expr. Patterns. 2003;3(4):455–458. doi: 10.1016/s1567-133x(03)00065-6. [DOI] [PubMed] [Google Scholar]
- 44.Simoes-Costa M., Tan-Cabugao J., Antoshechkin I., Sauka-Spengler T., Bronner M.E. Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome Res. 2014;24(2):281–290. doi: 10.1101/gr.161182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hargreaves K.M., Goodis H.E., Seltzer S. Quintessence Pub. Co.; Chicago: 2002. Seltzer and Bender's Dental Pulp. [Google Scholar]
- 46.Ogawa R., Saito C., Jung H.S., Ohshima H. Capacity of dental pulp differentiation after tooth transplantation. Cell Tissue Res. 2006;326(3):715–724. doi: 10.1007/s00441-006-0242-0. [DOI] [PubMed] [Google Scholar]
- 47.Morotomi T., Washio A., Kitamura C. Current and future options for dental pulp therapy. Jpn. Dent. Sci. Rev. 2019;55(1):5–11. doi: 10.1016/j.jdsr.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du W., Du W., Yu H. The role of fibroblast growth factors in tooth development and incisor renewal. Stem Cell. Int. 2018:7549160. doi: 10.1155/2018/7549160. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thesleff I., Mikkola M. The role of growth factors in tooth development. Int. Rev. Cytol. 2002;217:93–135. doi: 10.1016/s0074-7696(02)17013-6. [DOI] [PubMed] [Google Scholar]
- 50.Xie H., Dubey N., Shim W., Ramachandra C.J.A., Min K.S., Cao T., Rosa V. Functional odontoblastic-like cells derived from human iPSCs. J. Dent. Res. 2018;97(1):77–83. doi: 10.1177/0022034517730026. [DOI] [PubMed] [Google Scholar]
- 51.Garrison K.R., Donell S., Ryder J., Shemilt I., Mugford M., Harvey I., Song F. Clinical effectiveness and cost-effectiveness of bone morphogenetic proteins in the non-healing of fractures and spinal fusion: a systematic review. Health Technol. Assess. 2007;11(30):1–150. doi: 10.3310/hta11300. iii-iv. [DOI] [PubMed] [Google Scholar]
- 52.Ornitz D.M., Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015;4(3):215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo M.S., Hwang K.G., Kim H., Baek S.H. Analysis of gene expression during odontogenic differentiation of cultured human dental pulp cells. Restor. Dent. Endod. 2012;37(3):142–148. doi: 10.5395/rde.2012.37.3.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakai V.T., Zhang Z., Dong Z., Neiva K.G., Machado M.A., Shi S., Santos C.F., Nor J.E. SHED differentiate into functional odontoblasts and endothelium. J. Dent. Res. 2010;89(8):791–796. doi: 10.1177/0022034510368647. [DOI] [PubMed] [Google Scholar]
- 55.Galler K.M., D'Souza R.N., Federlin M., Cavender A.C., Hartgerink J.D., Hecker S., Schmalz G. Dentin conditioning codetermines cell fate in regenerative endodontics. J. Endod. 2011;37(11):1536–1541. doi: 10.1016/j.joen.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 56.Galler K.M., Hartgerink J.D., Cavender A.C., Schmalz G., D'Souza R.N. A customized self-assembling peptide hydrogel for dental pulp tissue engineering. Tissue Eng. 2012;18(1–2):176–184. doi: 10.1089/ten.TEA.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosa V., Zhang Z., Grande R.H., Nor J.E. Dental pulp tissue engineering in full-length human root canals. J. Dent. Res. 2013;92(11):970–975. doi: 10.1177/0022034513505772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamanaka S. Pluripotent stem cell-based cell therapy-promise and challenges. Cell Stem Cell. 2020;27(4):523–531. doi: 10.1016/j.stem.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 59.Sougawa N., Miyagawa S., Fukushima S., Kawamura A., Yokoyama J., Ito E., Harada A., Okimoto K., Mochizuki-Oda N., Saito A., et al. Immunologic targeting of CD30 eliminates tumourigenic human pluripotent stem cells, allowing safer clinical application of hiPSC-based cell therapy. Sci. Rep. 2018;8(1):3726. doi: 10.1038/s41598-018-21923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi R., Ishikawa Y., Katayama T., Quantock A.J., Nishida K. CD200 facilitates the isolation of corneal epithelial cells derived from human pluripotent stem cells. Sci. Rep. 2018;8(1):16550. doi: 10.1038/s41598-018-34845-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kojima K., Miyoshi H., Nagoshi N., Kohyama J., Itakura G., Kawabata S., Ozaki M., Iida T., Sugai K., Ito S., et al. Selective ablation of tumorigenic cells following human induced pluripotent stem cell-derived neural stem/progenitor cell transplantation in spinal cord injury. Stem Cell Transl. Med. 2019;8(3):260–270. doi: 10.1002/sctm.18-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zarogoulidis P., Darwiche K., Sakkas A., Yarmus L., Huang H., Li Q., Freitag L., Zarogoulidis K., Malecki M. Suicide gene therapy for cancer - current strategies. J. Genet. Syndr. Gene Ther. 2013;4 doi: 10.4172/2157-7412.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnakumar R., Chen A.F., Pantovich M.G., Danial M., Parchem R.J., Labosky P.A., Blelloch R. FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell. 2016;18(1):104–117. doi: 10.1016/j.stem.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Respuela P., Nikolic M., Tan M., Frommolt P., Zhao Y., Wysocka J., Rada-Iglesias A. Foxd3 promotes exit from naive pluripotency through enhancer decommissioning and inhibits germline specification. Cell Stem Cell. 2016;18(1):118–133. doi: 10.1016/j.stem.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mundell N.A., Labosky P.A. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011;138(4):641–652. doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong J., Tu X., Kong Y., Guo L., Li B., Zhong W., Cheng Y., Jiang Y., Jiang Q. LncRNA H19 promotes odontoblastic differentiation of human dental pulp stem cells by regulating miR-140-5p and BMP-2/FGF9. Stem Cell Res. Ther. 2020;11(1):202. doi: 10.1186/s13287-020-01698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu T., Yaguchi Y., Echevarria D., Martinez S., Basson M.A. Sprouty genes prevent excessive FGF signalling in multiple cell types throughout development of the cerebellum. Development. 2011;138(14):2957–2968. doi: 10.1242/dev.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forrester H.B., Temple-Smith P., Ham S., de Kretser D., Southwick G., Sprung C.N. Genome-wide analysis using exon arrays demonstrates an important role for expression of extra-cellular matrix, fibrotic control and tissue remodelling genes in Dupuytren's disease. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Solchaga L.A., Penick K., Goldberg V.M., Caplan A.I., Welter J.F. Fibroblast growth factor-2 enhances proliferation and delays loss of chondrogenic potential in human adult bone-marrow-derived mesenchymal stem cells. Tissue Eng. 2010;16(3):1009–1019. doi: 10.1089/ten.TEA.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ranscht B., Bronner-Fraser M. T-cadherin expression alternates with migrating neural crest cells in the trunk of the avian embryo. Development. 1991;111(1):15–22. doi: 10.1242/dev.111.1.15. [DOI] [PubMed] [Google Scholar]
- 71.Saghiri M.A., Asatourian A., Sorenson C.M., Sheibani N. Role of angiogenesis in endodontics: contributions of stem cells and proangiogenic and antiangiogenic factors to dental pulp regeneration. J. Endod. 2015;41(6):797–803. doi: 10.1016/j.joen.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Folkman J., Hochberg M. Self-regulation of growth in three dimensions. J. Exp. Med. 1973;138(4):745–753. doi: 10.1084/jem.138.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colton C.K. Implantable biohybrid artificial organs. Cell Transplant. 1995;4(4):415–436. doi: 10.1016/0963-6897(95)00025-s. [DOI] [PubMed] [Google Scholar]
- 74.Laschke M.W., Harder Y., Amon M., Martin I., Farhadi J., Ring A., Torio-Padron N., Schramm R., Rucker M., Junker D., et al. Angiogenesis in tissue engineering: breathing life into constructed tissue substitutes. Tissue Eng. 2006;12(8):2093–2104. doi: 10.1089/ten.2006.12.2093. [DOI] [PubMed] [Google Scholar]
- 75.Silva A.K., Richard C., Bessodes M., Scherman D., Merten O.W. Growth factor delivery approaches in hydrogels. Biomacromolecules. 2009;10(1):9–18. doi: 10.1021/bm801103c. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen P.K., Sarkar B., Siddiqui Z., McGowan M., Iglesias-Montoro P., Rachapudi S., Kim S., Gao W., Lee E.J., Kumar V.A. Self-assembly of an antiangiogenic nanofibrous peptide hydrogel. ACS Appl. Bio Mater. 2018;1(3):865–870. doi: 10.1021/acsabm.8b00283. [DOI] [PubMed] [Google Scholar]
- 77.Sarkar B., Siddiqui Z., Nguyen P.K., Dube N., Fu W., Park S., Jaisinghani S., Paul R., Kozuch S.D., Deng D., et al. Membrane-disrupting nanofibrous peptide hydrogels. ACS Biomater. Sci. Eng. 2019;5(9):4657–4670. doi: 10.1021/acsbiomaterials.9b00967. [DOI] [PubMed] [Google Scholar]
- 78.Harbour V., Casillas C., Siddiqui Z., Sarkar B., Sanyal S., Nguyen P., Kim K.K., Roy A., Iglesias-Montoro P., Patel S., et al. Regulation of lipoprotein homeostasis by self-assembling peptides. ACS Appl. Bio Mater. 2020;3(12):8978–8988. doi: 10.1021/acsabm.0c01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sarkar B., Siddiqui Z., Kim K.K., Nguyen P.K., Reyes X., McGill T.J., Kumar V.A. Implantable anti-angiogenic scaffolds for treatment of neovascular ocular pathologies. Drug Deliv. Transl. Res. 2020;10(5):1191–1202. doi: 10.1007/s13346-020-00753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu L., Dissanayaka W.L., Zhang C. Dental pulp stem cells overexpressing stromal-derived factor-1alpha and vascular endothelial growth factor in dental pulp regeneration. Clin. Oral Invest. 2019;23(5):2497–2509. doi: 10.1007/s00784-018-2699-0. [DOI] [PubMed] [Google Scholar]
- 81.Lu J., Yan X., Sun X., Shen X., Yin H., Wang C., Liu Y., Lu C., Fu H., Yang S., et al. Synergistic effects of dual-presenting VEGF- and BDNF-mimetic peptide epitopes from self-assembling peptide hydrogels on peripheral nerve regeneration. Nanoscale. 2019;11(42):19943–19958. doi: 10.1039/c9nr04521j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are presented in the paper and/or the Supplementary Materials. Additional data related to this study may be requested from the authors.