Abstract
Protein biotinylation via chemical or enzymatic reactions is often coupled with streptavidin-based enrichment and on-bead digestion in numerous biological applications. However, the popular on-bead digestion method faces major challenges of streptavidin contamination, overwhelming signals from endogenous biotinylated proteins, the lost information on biotinylation sites, and limited sequence coverage of enriched proteins. Here, we explored thiol-cleavable biotin as an alternative approach to elute biotinylated proteins from streptavidin-coated beads for both chemical biotinylation and biotin ligase-based proximity labeling. All possible amino acid sites for biotinylation were thoroughly evaluated in addition to the primary lysine residue. We found that biotinylation at lysine residues notably reduces the trypsin digestion efficiency, which can be mitigated by the thiol-cleavable biotinylation method. We then evaluated the applicability of thiol-cleavable biotin as a substrate for proximity labeling in living cells, where TurboID biotin ligase was engineered onto the mitochondrial inner membrane facing the mitochondrial matrix. As a proof-of-principle study, thiol-cleavable biotin-assisted TurboID proteomics achieved remarkable intraorganelle spatial resolution with significantly enriched proteins localized in the mitochondrial inner membrane and mitochondrial matrix.
Keywords: cleavable biotin, proximity labeling, TurboID, mitochondrion, streptavidin
Graphical Abstract
INTRODUCTION
Biotin, also known as vitamin B7, is a cofactor for carboxylase enzymes required for fatty acid synthesis.1 Biotin can be covalently linked to proteins, peptides, and nucleic acids through chemical and enzymatic biotinylation. Because of the exceptionally high affinity between biotin and streptavidin (Kd ~ 10−14 M), biotin-streptavidin system has been widely applied in biotechnology for affinity purification, immobilization, imaging, and drug delivery.2–5 However, the extremely stable and strong biotin-streptavidin interaction is a double-edged sword. Biotinylated proteins can only be eluted from streptavidin under harsh denaturing conditions (e.g., boiling in denaturing buffer), on-bead digestion, or competitive elution with excess biotin in detergent.4,6,7 Elution with detergent containing buffer (e.g., SDS or Triton) is not compatible with mass spectrometry (MS) analysis and on-bead digestion often leads to overwhelming contamination signals of streptavidin peptides.7 The information on biotinylation sites is also lost from on-bead digestion since biotinylated peptides will remain attached to the beads. Alternative strategies have been developed in recent years that use structurally modified avidins with lower binding affinity (Kd ≈10−8 M), antibiotin antibodies, or biotin derivatives with a cleavable moiety that can be selectively released under mild elution conditions (e.g., UV radiation, chemical exposure, enzymes).8–12
A major class of applications of the biotin-streptavidin system involves attaching biotin to proteins of interest by chemical labeling or in situ enzymatic reactions. Recent advancements in enzymatic proximity labeling techniques have allowed both stable and transient protein networks and subcellular microenvironments to be captured in various living cells and organisms.13–15 Engineered peroxidase or biotin ligase can be tagged to the bait protein to biotinylate neighboring prey proteins upon activation, which can then be enriched by streptavidin-coated beads for MS-based proteomics.13 However, proximity labeling studies face the same challenges of using the biotin-streptavidin system. To address these challenges, antibiotin antibodies have been shown to sufficiently elute biotinylated peptides with excellent coverage of biotinylation sites,9,10 but antibiotin antibodies are much more expensive than streptavidin-coated beads and have potential issues of antibody nonspecific bindings. Cleavable biotin is another alternative but has rarely been used for proximity labeling studies.16 Therefore, on-bead digestion is still the most popular approach despite the lost information on biotinylation sites, streptavidin contamination, and limited sequence coverage of enriched proteins.
Here, we sought to evaluate cleavable biotin for both chemical biotinylation and enzymatic proximity labeling in living cells. Commercially available thiol-cleavable biotin (SS-biotin) was used which contains a disulfide bridge that can be cleaved with the addition of a reducing agent after capturing biotinylated proteins on the streptavidin-coated magnetic beads. We first chemically labeled protein standards and complex human cell lysate with amine-reactive NHS-SS-biotin which primarily modifies the lysine side chains of proteins. The amino acid sites of SS-biotinylation and protein digestion efficiency were investigated in comparison to NHS-biotin labeled and unlabeled proteins. We then applied the thiol-cleavable biotin method to proximity labeling in living cells. Recently developed TurboID biotin ligase was selected because of its highly efficient labeling activity compared to the BioID method to reduce the diffusion of the reactive biotin cloud.17 TurboID was genetically expressed onto the mitochondrial inner membrane, facing the mitochondrial matrix. Mitochondria are the powerhouses and the metabolic hubs of eukaryotic cells. The double-membraned structure of mitochondrion creates a unique compartment apart from the cytosolic environment. The inner mitochondrial membrane (IMM) is the major site for electron transport chain to produce ATP through aerobic respiration.18 In the presence of ATP and thiol-cleavable SS-biotin substrate, TurboID can catalyze the SS-biotinylation of nearby proteins within 10 nm labeling radius of the IMM, achieving remarkable intraorganelle spatial resolution for mitochondrial proteomics.
EXPERIMENTAL SECTION
In-vitro Protein Labeling with NHS-SS-biotin and NHS-biotin.
Amine-reactive NHS-SS-biotin and NHS-biotin reagents (APExBIO) were used to label BSA protein standard (Fisher Scientific) and whole-cell human protein extract (Promega). BSA protein standard and human cell lysate were prepared in phosphate buffer saline (PBS) at 2 mg/mL concentrations. Urea and Tris buffers contain primary amine groups and cannot be used for NHS labeling. NHS-SS-biotin and NHS-biotin reagents are moisture sensitive and were carefully prepared in DMSO (100 mM) to remain anhydrous in a desiccator at −30 °C. NHS reagent was mixed with each protein sample with a 50-fold molar excess for 30 min in a ThermoMixer at 25 °C. The labeling reaction was quenched with 10% hydroxylamine for 15 min, and labeled samples were stored at −80 °C.
To evaluate the labeling reaction, an aliquot of each labeled protein sample (40 μg) was transferred to a new tube to react with tris(2-carboxylethyl) phosphine (TCEP, 20 mM) in a ThermoMixer for 60 min at 37 °C to reduce/cleave the disulfide bonds. The free SH group was alkylated with 50 mM iodoacetamide (IAA) for 30 min, followed by an additional 20 mM TCEP treatment for 10 min to get rid of excessive IAA. Trypsin enzyme (Promega) was added (enzyme/protein = 1:30, w/w) for an in-solution digestion at 37 °C for 18 h. For the comparison of Trypsin, LysC, and Tryspin/LysC mix enzymes (Promega), 0.4 μg of enzymes were used to digest 40 μg of SS-biotin-labeled cell lysate in each replicate. Protein digestions were quenched with 10% trifluoroacetic acid (TFA) until pH < 3. Peptide samples were desalted on a C18 96-well μElution plate (Waters), dried under SpeedVac, and stored at −30 °C.
Cell Culture and Mitochondrial TurboID Stable Cell Line.
HEK293T and HeLa cells were cultured in DMEM medium supplemented with 10% FBS, 1 mM sodium pyruvate, 10 mM HEPES, nonessential amino acids, and GlutaMAX. All reagents were purchased from Gibco. TurboID was genetically expressed onto a mitochondrial inner membrane protein, stomatin-like protein 2 (STOML2), using the well-established TurboID method developed by the Ting group.19 pLVX-puro-STOML2-TurboID-HA plasmid was created by PCR amplification and subcloning into the EcoRI sites of pLVX-puro vector (Clontech). The sequence coding TurboID was amplified from TurboID-V5-KDEL-pDisplay (kindly provided by Dr. Alice Ting) with a sequence coding HA-tag. To generate a stably transfected cell line expressing STOML2-TurboID-HA, lentiviruses were packaged in HEK293T cells, and HeLa cells were transduced with these viruses with 10 μg/ mL polybrene (Sigma) and then optimized for protein expression via antibiotics selection. Wild-type HeLa cells without TurboID expression were used as the negative control group. Cells were cultured in 15 cm dishes until confluence. Both mitochondrial TurboID cells and control cells were incubated with 1 mM SS-biotin (Cayman) at 37 °C for 30 min, gently washed twice with PBS, and pelleted. Cell pellets were flash frozen in liquid nitrogen and stored at −80 °C.
Sample Preparation of Mitochondrial TurboID Cells.
Cell pellets were lysed in ice-cold lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 0.1% SDS, 1% Triton-X, Ultra mini protease inhibitor). Lysis buffer needs to avoid addition of reducing reagents such as TCEP or DTT. Cell lysates were homogenized, sonicated on ice for 15 min, and clarified by centrifugation at 16500 rpm for 15 min at 4 °C. The total protein concentrations of cell lysate samples were measured by a detergent-compatible colorimetric protein assay (DCA, BioRad). Streptavidin (SA) magnetic beads (Cytiva) were used to enrich biotinylated proteins with our previously developed enrichment and washing protocol.7 Briefly, after a 16 h incubation with the SA beads at 4 °C, supernatants were removed and the beads were washed twice sequentially with each wash buffer (buffer A: 2% SDS; buffer B: 50 mM Tris–HCl, 500 mM NaCl, 2% Triton-X; buffer C: 50 mM Tris–HCl, 250 mM NaCl, 0.5% SDS, 0.5% Triton-X, buffer D: 2 M Urea, 50 mM Tris-HCl). An additional two washes were performed with buffer D to ensure complete removal of detergent. Then, SS-biotin labeled proteins were cleaved off beads with elution buffer (20 mM TCEP, 0.1% Rapigest (Waters), 30 mM NaCl, and 50 mM Tris-HCl) in a ThermoMixer for 60 min at 37 °C. The beads were eluted again with 1% formic acid (FA) in 50 mM Tris-HCl for another 30 min to ensure complete protein elution. The eluents were combined for alkylation with IAA and quenched with TCEP as described above. Trypsin/Lys-C mix was used for protein digestion (1:30 ratio) for 16 h at 37 °C in a ThermoMixer. Protein digestion was quenched with 10% TFA until pH < 3, incubated at 37 °C for 45 min, and clarified by centrifugation at 13000 rpm for 10 min to remove Rapigest. Peptides were desalted, dried, and stored at -30 °C until LC–MS/MS analysis.
LC—MS/MS Analysis.
Peptides samples were reconstituted in 2% acetonitrile (ACN), 0.1% FA in LC-MS-grade water, briefly sonicated, and clarified by centrifugation at 16500 rpm for 10 min at 4 °C. A Dionex Ultimate 3000 RSLCnano system coupled with a Thermo Scientific Q-Exactive HFX mass spectrometer was used for LC-MS/MS analysis. The mobile phase A was 0.1% FA in water, and mobile phase B was 0.1% FA in ACN. Peptides were separated on an Easy-spray PepMap C18 column (2 μM, 100 Å, 75 μM × 75 cm) with a 2 h LC gradient and 55 °C column temperature. The flow rate was 0.2 μL/min. The quadrupole mass filtering was set from m/z 380 to 2000 with a resolving power of 120000 (at m/z 200FWHM). A top 30 data-dependent acquisition was used for MS/MS with a resolving power of 7500. Parent masses were isolated with a m/z 1.4 window and fragmented with higher-energy collision dissociation (HCD). The normalized collision energy was 30% and the dynamic exclusion time was 30 s. The maximum injection times were 30 ms for MS and 35 ms for MS/MS. The automatic gain control (AGC) was 1 × 106 for MS and 2 × 105 for MS/MS.
Data Analysis.
LC-MS/MS raw files were analyzed with Thermo Proteome Discoverer (2.4.1.15) software. The Uniprot BSA protein sequence was used to search labeled and unlabeled BSA sample. Swiss-Prot Homo sapiens database (reviewed) was used for human protein identification (1% false discovery rate cutoff). Common proteomics contamination database (from Max Planck Institute of Biochemistry) was included as the contamination marker. Trypsin was used as the enzyme with four maximum missed cleavages. Cysteine carbamidomethylation was included as a fixed modification. Methionine oxidation and acetylation of protein N-terminus were included as variable modification. Biotinylation (+226.0776 Da)or thiol-cleavable biotinylation (+145.0198 Da) was added as an additional variable modification at lysine residue for biotin and SS-biotin labeled samples, respectively. To evaluate potential labeling site, we also included other amino acid residues that could react with NHS group, including arginine, asparagine, glutamine, serine, threonine, tyrosine, and histidine. Chromatographic alignment was conducted with a maximum retention time shift of 2 min and a minimum signal-to-noise ratio of 5. Biotinylated proteins/peptides were also confirmed with Maxquant (1.6.17.0) software with the same parameters as the Proteome Discoverer software. For the analysis of mitochondrial TurboID samples, proteomics data was normalized to the endogenously biotinylated protein PCCA as described previously.7 Statistical analysis was conducted with a t test built in the Proteome Discoverer software. Protein Go-term analysis was conducted by the Enrichr online software platform.20 Raw LC–MS/MS data from this manuscript are available through the MassIVE repository (MSV000087256).
RESULTS AND DISCUSSION
Thiol-Cleavable Biotin for Chemical Labeling.
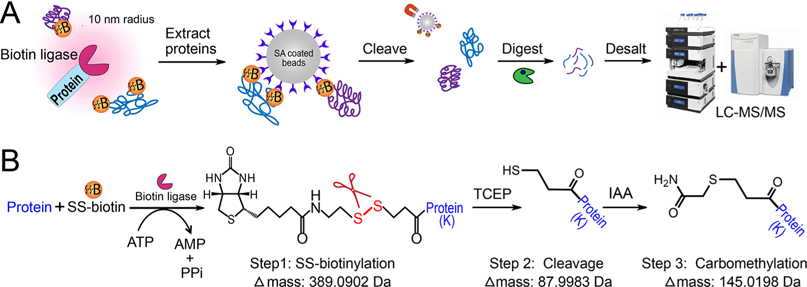
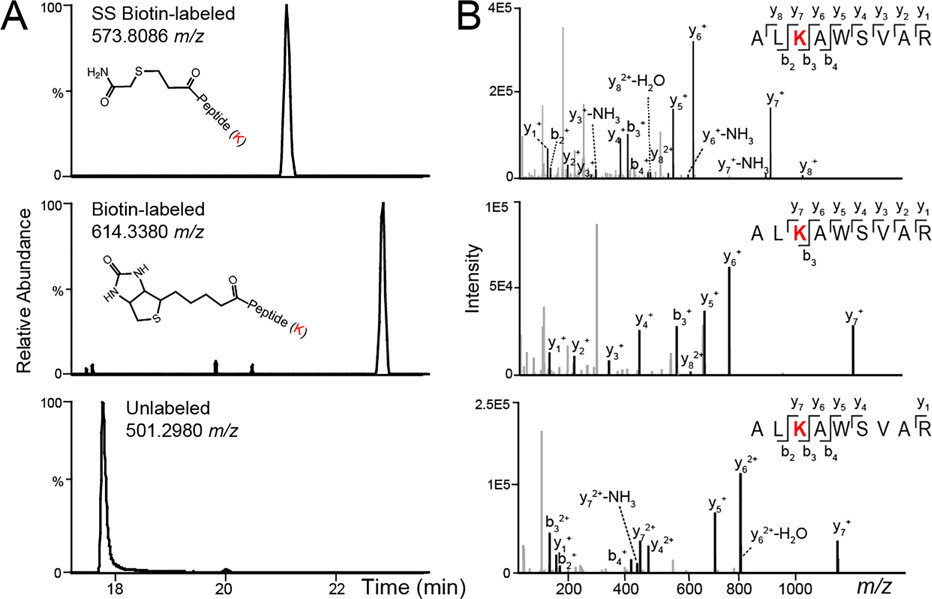
In proximity labeling, biotin ligases (e.g., BioID,21 TurboID17) catalyze the biotinylation of neighboring proteins in the presence of ATP and biotin substrate within a 10 nm labeling radius (Figure 1). Thiol-cleavable biotin can be a promising alternative to traditional biotin substrate, allowing the efficient and complete elution of biotinylated proteins from streptavidin beads while preserving the information on biotinylation sites. To imitate biotin ligase-based proximity labeling in cells, we first investigated the thiol-cleavable biotin labeling in vitro with the succinimidyl 2-(biotinamido)ethyl-1,3’-dithiopropinate (NHS-SS-biotin) reagent. NHS-SS-biotin is amine-reactive and labels the lysine side chain and N-terminus of proteins. To investigate chemical biotinylation, bovine serum albumin (BSA) standard and human whole cell lysate were labeled with NHS-SS-biotin reagent at the protein level. Biotinylated proteins can be enriched with streptavidin-coated magnetic beads. Reducing agent can then be added to cleave the disulfide bond and release the proteins from the beads to the supernatant. Then the eluted proteins can be digested in-solution for subsequent proteomics analysis. TCEP was chosen as the reducing agent because it is more stable, effective, and odor-less compared to DTT. IAA was added to the eluent to carbomethylate the free thiol group and prevent the reformation of disulfide bonds (Figure 1B). The concentration of TCEP and IAA was optimized to achieve nearly complete (99.5%) cleavage and carbomethylation reaction for human cell lysate samples (Table 1). For NHS-SS-biotin-labeled human cell lysate proteins, we identified a total of over 3000 modified peptides (Table S1). Additionally, to compare with the thiol-cleavable NHS-SS-biotin labeling, we also used the noncleavable biotin derivative, N-hydroxysuccinimidobiotin (NHS-biotin) to label BSA and human whole cell lysate. The complete protein IDs and quantification in three groups are provided in Supplemental Table S2. As shown in Figure 2 and Figure S1, peptides labeled with biotin at lysine residues have larger delta masses (+226.0776 Da) compared to the cleavable SS-biotin (+145.0198 Da). SS-biotin or biotin modification alters peptide hydrophobicity and size, causing delayed LC-MS retention times. Peptide backbone fragmentations are comparable if not better in SS-biotin-labeled peptides vs biotin-labeled and unlabeled peptides.
Figure 1.
Development of thiol-cleavable biotin-enabled proximity labeling. (A) General workflow for thiol-cleavable biotinylation and protein sample preparation. (B) Schematic of enzymatic reaction of SS-biotinylation, chemical cleavage of disulfide bonds to elute proteins off the streptavidin beads, and carbomethylation of the free thiol group to avoid reformation of disulfide bonds.
Table 1.
Reaction Efficiency of SS Cleavage and Carbomethylation from SS-Biotin-Labeled (K) Human Cell Lysate Proteins
| modification | Δ mass | biotinylated peptides (%) | avg percolator q-value | avg percolator PEP |
|---|---|---|---|---|
| cleaved, carbomethylateda | +145.0198 | 99.5 | 8.90 × 10−04 | 1.47 × 10−02 |
| cleaved, not carbomethylatedb | +87.9983 | 0.31 | 5.50 × 10−03 | 1.22 × 10−01 |
| SS-biotinylated, not cleavedb | +389.0902 | 0.20 | 2.50 × 10−04 | 8.46 × 10−04 |
Complete reaction of thiol-cleavable biotinylation.
Incomplete reaction of thiol-cleavable biotinylation.
Figure 2.
Example LC-MS chromatograms (A) and MS/MS spectra of peptide fragmentation (B) for SS-biotin-labeled, biotin-labeled, and unlabeled peptide (ALKAWSVAR) from bovine serum albumin (BSA) protein.
Thiol-Cleavable Biotin and Biotin Modify Proteins More Than the Lysine Residues.
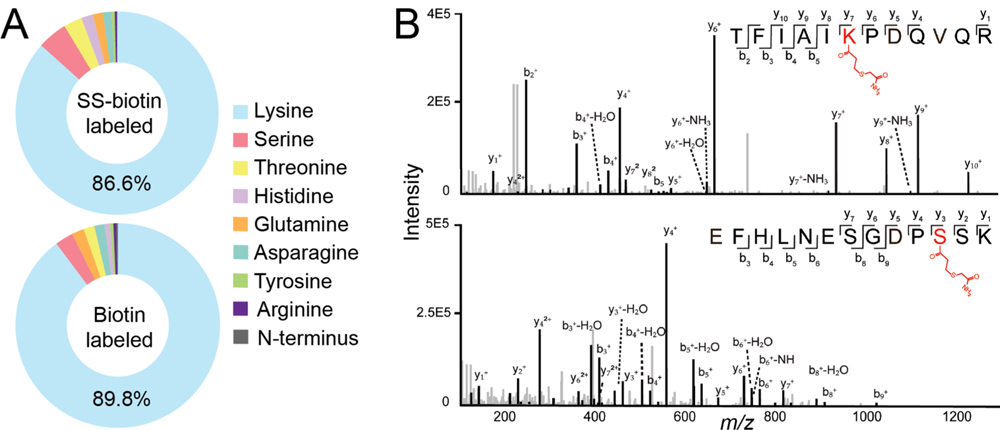
NHS-biotin reagents and biotin ligase-based proximity labeling primarily label proteins on the lysine side chain and protein N-terminus. NHS-biotin has also been shown to label serine and tyrosine residues that have hydroxyl groups.22 However, a systematic evaluation of all the possible biotinylation sites is still lacking. Here, we examined all amino acid residues that have a primary amine (lysine, arginine, asparagine, glutamine) or hydroxyl residue (serine, threonine, tyrosine) that could react with NHS group. We also included histidine residue, which contains a secondary amine group. We found that both SS-biotin and biotin primarily modify the lysine residues as expected (Figure 3). However, ~10–14% of proteins were also modified at other amino acid residues (e.g., serine, threonine, histidine) with confident peptide identification (percolator q-values <1 × 10−3) (Supplemental Table S3). Interestingly, only a few protein N-terminuses were labeled with biotin. This is probably due to the existing modifications at the protein N-terminus and the low percentage of N-terminus unique peptide identifications. Protein N-terminus have been shown to be co-or post-translational modified, such as acetylation, methylation, formylation, and methionine excision.23 Since biotinylation modifies more than the lysine residue, other amino acids should also be considered as variable modifications if researchers aim to obtain a comprehensive coverage of biotinylation sites. However, including more variable modifications requires exponentially increased data analysis time and computing power. Therefore, for studies that focus on overall protein quantifications, we recommend adding both lysine and serine as variable biotinylation sites for improved protein identification.
Figure 3.
Evaluation of protein biotinylation sites. (A) Percentage distribution of biotinylation sites from labeled human cell lysate. (B) Example fragmentation spectra of SS-biotin modified peptides.
Biotinylation at Lysine Reduces Trypsin Digestion Efficiency.
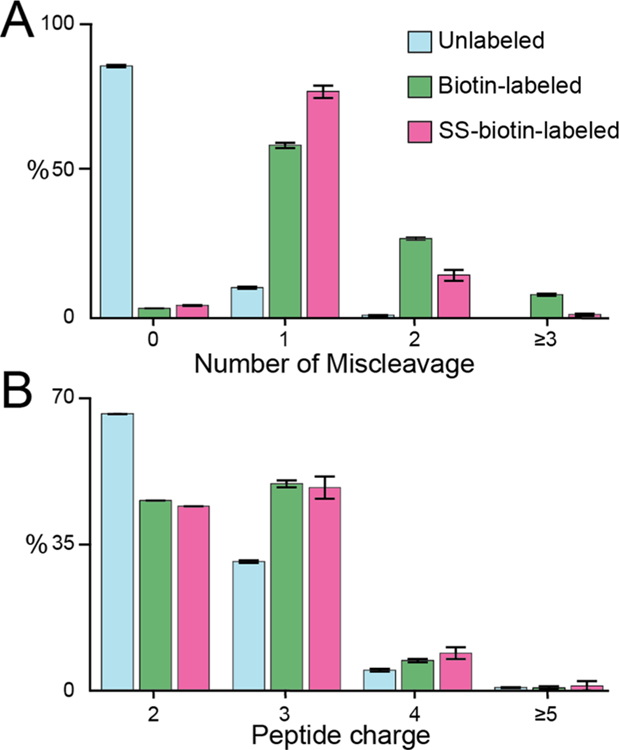
Trypsin is the most common enzyme for protein digestion that cleaves at the carboxyl side of lysine and arginine.24 Since biotinylation primarily modifies the lysine residue, we hypothesized that such modification may hinder trypsin digestion efficiency. Therefore, we directly compared the trypsin digestion efficiency for SS-biotin-labeled, biotin-labeled, and unlabeled human cell lysate. A total of over 5000 proteins were identified and quantified. As expected, unlabeled samples showed excellent cleavage efficiency, and the majority of unlabeled peptides have no miscleavages (Figure 4A). In contrast, more than 90% peptides modified with SS-biotin or biotin have at least one miscleavage. SS-biotin-modified peptides mitigated the problem compared to biotin modified peptides with more percentage of peptides with ≤1 miscleavages and less percentage of peptides with ≥2 miscleavages. The improved digestion efficiency is probably due to the excision of the biotin group from the protein/ peptide. We also compared the distribution of peptide charges and precursor masses to evaluate the influence of insufficient digestion. As illustrated in Figure 4B and Figure S2, SS-biotin and biotin-labeled peptides have significantly higher peptide charges and larger precursor masses compared to unlabeled samples. Since the addition of Lys-C enzyme has been shown to improve digestion efficiency at the lysine site for trypsin digestion, we directly compared the digestion efficiency of SS-biotin labeled cell lysate using trypsin, LysC, and Trysin/LysC mix enzymes.25 Although protein digestion with Trypsin/Lys-C mix provided the most protein/peptide identifications compared to trypsin or Lys-C alone, the majority of SS-biotin modified peptides still have at least one miscleavage (Figure S3). Therefore, we have proved that biotinylation at lysine reduces the protein digestion efficiency. This is critical for biotin ligase-based proximity labeling proteomics that routinely involves on-bead digestion to cleave the biotinylated proteins off the streptavidin beads. The miscleaved peptides will remain on the beads without sufficient elution and, therefore, sacrifice the protein/peptide identification and coverage. But peroxidase-based proximity labeling that biotinylates proteins at the electron-rich region (e.g., tyrosine residue) should not be influenced by this issue. In fact, we examined our previously developed LAMP1-APEX proximity labeling data and found that majority of peptides have no miscleavages (data not shown).7 For biotin ligase-based proximity labeling or chemical biotinylation at the lysine residue, enzymes that cleave at other residues, such as Glu-C, Asp-N, and Arg-C, may be used instead or in combination with trypsin digestion to improve protein coverage during on-bead digestion. Since our thiol-cleavable biotin method allows sufficient protein elution without the need for on-bead digestion, we can increase the maximum allowed miscleavages to three or four instead of routinely used two miscleavages for the proteomics data analysis.
Figure 4.
Evaluation of the trypsin digestion efficiency for unlabeled, biotin-labeled (K), and SS-biotin-labeled (K) peptides from human cell lysate samples. Histogram distributions of the number of miscleavages (A) and peptide charges (B).
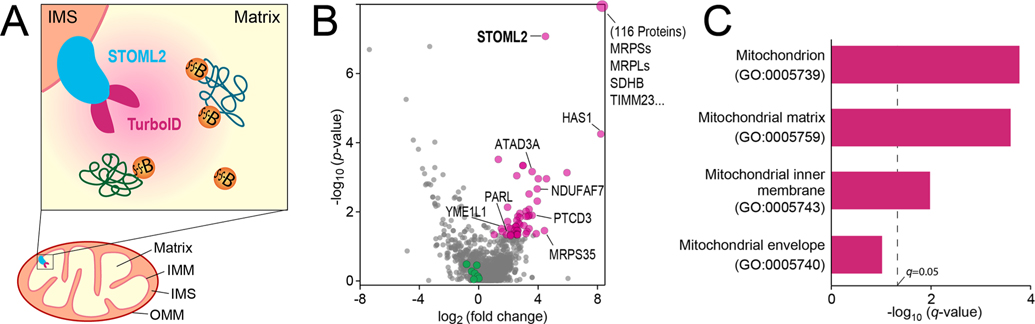
Application of Thiol-Cleavable Biotin to Mitochondrial TurboID Proximity Labeling.
We have demonstrated the thiol-cleavable protein biotinylation by chemical labeling in vitro. Here, we aim to evaluate its applicability to the enzymatic proximity labeling reaction in living cells. Using the well-established TurboID proximity labeling method, we genetically engineered TurboID onto the Stomatin-like protein 2 (STOML2), a mitochondrial protein belonging to SPFH (stomatin/prohibitin/flotillin/HfIKC) family. Members of SPFH family function as membrane organizers in various cellular membranes.26 STOML2 is peripherally associated with the matrix-side of the inner mitochondrial membrane and is suggested to act as membrane scaffolds of the inner mitochondrial membrane (Figure 5A).27 Thiol-cleavable biotin (SS-biotin) was added to the HeLa cells expressing STOML2-TurboID as well as the negative control group (N = 3). Through the proteomics workflow illustrated in Figure 1A, we identified and quantified a total of 1566 proteins (Supplemental Table S4). Statistical analysis revealed 173 significantly enriched proteins in the mitochondrial TurboID group vs control (p-value < 0.05, fold change > 2), of which 116 proteins were only identified and quantified in the TurboID group. These significantly enriched proteins are highlighted in red in the volcano plot, including the bait protein STOML2 and key mitochondrial proteins (Figure 5B). When compared to the routine on-bead digestion method, thiol-cleavable biotin method provided less nonspecific protein IDs (e.g., histones, actins) with greatly reduced streptavidin signals and enabled 305 biotinylate sites to be identified from ~200 modified peptides, mainly at K, but also at other amino acid residues like S, T, and H (Supplemental Table S5).
Figure 5.
Mitochondrial TurboID proteomics with thiol-cleavable biotin. (A) Schematic of STOML2-TurboID inside the mitochondrion using thiol-cleavable SS-biotin as the substrate. (B) Volcano plot of STOML2-TurboID proteomics vs negative control (N = 3). Significantly enriched proteins are highlighted in red. Endogenously biotinylated carboxylases were highlighted in green. (C) Go-term analysis of significantly enriched proteins (top 4 ranked Go-terms for cellular location). Mitochondrial structure: outer mitochondrial membrane (OMM), intermembrane space (IMS), inner mitochondrial membrane (IMM), and mitochondrial matrix.
In order to confirm that the biotinylated proteins were completely cleaved and eluted from the streptavidin beads, we also conducted on-bead digestion of the leftover beads after elution. Only 156 proteins were identified including streptavidin, trypsin, keratins, actins, histones, and other nonspecific binding proteins, proving that the cleavage/elution was complete (Supplemental Table S6). It is worth mentioning that highly abundant biotin-dependent carboxylases were identified in the leftover beads digest. In our previous study, we found that these endogenously biotinylated carboxylases were highly abundant in both lysosomal and cytosolic APEX proximity labeling data sets.7 Our thiol-cleavable mitochondrial TurboID method reduced but not removed these carboxylases, including PC, PCCA, PCCB, MCC1, MCC2, ACACA, and ACACB (highlighted in green in Figure 5B).28 These biotin-dependent carboxylases are probably partially biotinylated endogenously and partially SS-biotinylated due to the addition of SS-biotin during proximity labeling in living cells. Although these carboxylases cannot be removed, they serve as a quality control check point as they should remain unchanged among different groups and negative control that were all incubated with biotin substrate and, therefore, can be used to normalize the proximity labeling data set to reduce variation as we demonstrated previously.
Protein enrichment analysis was conducted for the significantly enriched proteins in mitochondrial TurboID vs control. Significantly enriched cellular location Go-terms include mitochondrion, mitochondrial matrix and mitochondrial inner membrane (IMM) (Figure 5C). TurboID labeling radius (~10 nm) is much smaller than the average diameter of the mitochondrion (~1 μm), achieving superspatial resolution of labeling activity inside the mitochondrion. Mitochondrial matrix and inner membrane proteins were much more highly enriched than other locations of the mitochondrion. For instance, many mitochondrial ribosomal proteins were enriched (MRPLs and MRPSs), which are localized to the matrix attaching to the IMM and are responsible for mitochondrial protein synthesis.29 Proteins related to the mitochondrial respiratory complexes localized at IMM were identified which are responsible for producing ATP.18,30 The known STOML2 interactor, YME1L and PARL were also enriched in our data set.31 Mitochondrial enzymes in the matrix were enriched such as ACOT2, HSD17B10, CHPF, and PPA2. Whereas only few mitochondrial outer membrane proteins were enriched. Traditionally, such spatial resolution inside the organelle can only be achieved by super resolution microscopy and electron microscopy, but one protein at a time. Mitochondrial APEX proximity labeling, originally developed in the Ting group, achieved remarkable spatial resolution and high throughput mitochondrial proteomic identification.32–34 As a proof-of-principle study, our thiol-cleavable biotin-assisted mitochondrial TurboID also demonstrated excellent intraorganelle resolution without the need for on-bead digestion and mitochondrial fractionation.
CONCLUSION
To summarize, we have developed both chemical and enzymatic biotinylation methods using thiol-cleavable biotin as the substrate. Through a comprehensive biotinylation site analysis, we demonstrated that NHS-reagent and biotin ligase-based biotinylation modify proteins more than the lysine side chain. We found that biotinylation at lysine residues reduces the trypsin digestion efficiency, resulting in the increased miscleavages and longer peptides with higher peptide charges. Our thiol-cleavable biotin labeling mitigated this issue compared to the traditional biotinylation, particularly with complete protein elution without the need for on-bead digestion. We also recommend setting the maximum allowed miscleavages to three or four for tryptic digested biotinylated proteins. Mitochondrial TurboID proteomics using the thiol-cleavable biotin method demonstrated intraorganelle spatial resolution of enriched mitochondrial proteins at mitochondrial inner membrane and matrix, providing a promising alternative method to study both chemical and enzymatic biotinylation of proteins in biological systems.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by National Institutes of Health grants R01NS121608 (L.H.) and the GW Faculty Startup Fund. L.H. acknowledges the ORAU Ralph E. Powe Junior Faculty Enhancement Award. A.M.F. and H.L. acknowledge the Bourdon F. Scribner Fellowship from GW Chemistry Department. We thank all the members in the Hao Lab for helpful discussions. S.S. is supported by the University of Pittsburgh, The Aging Institute Startup Seed, Samuel and Emma Winters Foundation, and UPMC Health System Competitive Medical Research Fund.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.1c00079.
Example LC–MS chromatograms (Figure S1), histogram distribution of precursor masses (M + H) of nonlabeled, biotin-labeled, and SS-biotin-labeled peptides from human cell lysate samples (Figure S2), and comparison of the protein digestion efficiency using Trypsin, LysC, and Trypsin/Lys-C mix for SS-biotin labeled human protein lysate (PDF)
Modified peptide sequences, protein IDs, average percolator q-values, SS-biotinylated peptides in STOML2-TurboID proteomics, Leftover bead digestion (Tables S1–S6) (XLSX)
Contributor Information
Haorong Li, Department of Chemistry, The George Washington University, Washington, DC 20052, United States.
Ashley M. Frankenfield, Department of Chemistry, The George Washington University, Washington, DC 20052, United States.
Ryan Houston, Aging Institute, Division of Cardiology, Department of Medicine, University of Pittsburgh, Pittsburgh, Pennsylvania 15219, United States.
Shiori Sekine, Aging Institute, Division of Cardiology, Department of Medicine, University of Pittsburgh, Pittsburgh, Pennsylvania 15219, United States.
Ling Hao, Department of Chemistry, The George Washington University, Washington, DC 20052, United States.
REFERENCES
- (1).Zempleni J; Wijeratne SSK; Hassan YI Biotin. BioFactors 2009, 35 (1), 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dundas CM; Demonte D; Park S Streptavidin-Biotin Technology: Improvements and Innovations in Chemical and Biological Applications. Appl. Microbiol. Biotechnol. 2013, 97 (21), 9343–9353. [DOI] [PubMed] [Google Scholar]
- (3).Sakahara H; Saga T Avidin-Biotin System for Delivery of Diagnostic Agents. Adv. Drug Delivery Rev. 1999, 37 (1–3), 89–101. [DOI] [PubMed] [Google Scholar]
- (4).Berg Luecke L; Gundry RL Assessment of Streptavidin Bead Binding Capacity to Improve Quality of Streptavidin-Based Enrichment Studies. J. Proteome Res. 2021, 20 (2), 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Guerrero C; Tagwerker C; Kaiser P; Huang L An Integrated Mass Spectrometry-Based Proteomic Approach: Quantitave Analysis of Tandem Affinity-Purified in Vivo Cross-Linked Protein Complexes (QTAX) to Decipher the 26 S Proteasome-Interacting Network. Mol. Cell. Proteomics 2006, 5 (2), 366–378. [DOI] [PubMed] [Google Scholar]
- (6).Cheah JS; Yamada S A Simple Elution Strategy for Biotinylated Proteins Bound to Streptavidin Conjugated Beads Using Excess Biotin and Heat. Biochem. Biophys. Res. Commun. 2017, 493 (4), 1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Frankenfield AM; Fernandopulle MS; Hasan S; Ward ME; Hao L Development and Comparative Evaluation of Endolysosomal Proximity Labeling-Based Proteomic Methods in Human IPSC-Derived Neurons. Anal. Chem. 2020, 92 (23), 15437–15444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).O’Sullivan VJ; Barrette-Ng I; Hommema E; Hermanson GT; Schofield M; Wu SC; Honetschlaeger C; Ng KKS; Wong SL Development of a Tetrameric Streptavidin Mutein with Reversible Biotin Binding Capability: Engineering a Mobile Loop as an Exit Door for Biotin. PLoS One 2012, 7 (4), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Udeshi ND; Pedram K; Svinkina T; Fereshetian S; Myers SA; Aygun O; Krug K; Clauser K; Ryan D; Ast T; Mootha VK; Ting AY; Carr SA Antibodies to Biotin Enable Large-Scale Detection of Biotinylation Sites on Proteins. Nat. Methods 2017, 14 (12), 1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kim DI; Cutler JA; Na CH; Reckel S; Renuse S; Madugundu AK; Tahir R; Goldschmidt HL; Reddy KL; Huganir RL; Wu X; Zachara NE; Hantschel O; Pandey A BioSITe: A Method for Direct Detection and Quantitation of Site-Specific Biotinylation. J. Proteome Res. 2018, 17 (2), 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Szychowski J; Mahdavi A; Hodas JJL; Bagert JD; Ngo JT; Landgraf P; Dieterich DC; Schuman EM; Tirrell DA Cleavable Biotin Probes for Labeling of Biomolecules via Azide-Alkyne Cycloaddition. J. Am. Chem. Soc. 2010, 132 (51), 18351–18360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Fonović M; Verhelst SHL; Sorum MT; Bogyo M Proteomics Evaluation of Chemically Cleavable Activity-Based Probes. Mol. Cell. Proteomics 2007, 6 (10), 1761–1770. [DOI] [PubMed] [Google Scholar]
- (13).Samavarchi-Tehrani P; Samson R; Gingras AC Proximity Dependent Biotinylation: Key Enzymes and Adaptation to Proteomics Approaches. Mol. Cell. Proteomics 2020, 19 (5), 757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Qin W; Cho KF; Cavanagh PE; Ting AY Deciphering Molecular Interactions by Proximity Labeling. Nat. Methods 2021, 18, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Richards AL; Eckhardt M; Krogan NJ Mass Spectrometry-based Protein-Protein Interaction Networks for the Study of Human Diseases. Mol. Syst. Biol. 2021, 17 (1), No. e8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rees JS; Li XW; Perrett S; Lilley KS; Jackson AP Selective Proteomic Proximity Labeling Assay Using Tyramide (SPPLAT): A Quantitative Method for the Proteomic Analysis of Localized Membrane-Bound Protein Clusters. Curr. Protoc. Protein Sci. 2015, 80 (1), 1–18. [DOI] [PubMed] [Google Scholar]
- (17).Branon TC; Bosch JA; Sanchez AD; Udeshi ND; Svinkina T; Carr SA; Feldman JL; Perrimon N; Ting AY Efficient Proximity Labeling in Living Cells and Organisms with TurboID. Nat. Biotechnol. 2018, 36 (9), 880–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sousa JS; D’Imprima E; Vonck J Mitochondrial Respiratory Chain Complexes. Subcell. Biochem. 2018, 87, 167–227. [DOI] [PubMed] [Google Scholar]
- (19).Cho KF; Branon TC; Udeshi ND; Myers SA; Carr SA; Ting AY Proximity Labeling in Mammalian Cells with TurboID and Split-TurboID. Nat. Protoc. 2020, 15 (12), 3971–3999. [DOI] [PubMed] [Google Scholar]
- (20).Kuleshov MV; Jones MR; Rouillard AD; Fernandez NF; Duan Q; Wang Z; Koplev S; Jenkins SL; Jagodnik KM; Lachmann A; McDermott MG; Monteiro CD; Gundersen GW; Ma’ayan A Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44 (W1), W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Roux KJ; Kim DI; Raida M; Burke B A Promiscuous Biotin Ligase Fusion Protein Identifies Proximal and Interacting Proteins in Mammalian Cells. J. Cell Biol. 2012, 196 (6), 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gabant G; Augier J; Armengaud J Assessment of Solvent Residues Accessibility Using Three Sulfo-NHS-Biotin Reagents in Parallel: Application to Footprint Changes of a Methyltransferase upon Binding Its Substrate. J. Mass Spectrom. 2008, 43 (3), 360–370. [DOI] [PubMed] [Google Scholar]
- (23).Giglione C; Fieulaine S; Meinnel T N-Terminal Protein Modifications: Bringing Back into Play the Ribosome. Biochimie 2015, 114, 134–146. [DOI] [PubMed] [Google Scholar]
- (24).Olsen JV; Ong SE; Mann M Trypsin Cleaves Exclusively C-Terminal to Arginine and Lysine Residues. Mol. Cell. Proteomics 2004, 3 (6), 608–614. [DOI] [PubMed] [Google Scholar]
- (25).Saveliev S; Bratz M; Zubarev R; Szapacs M; Budamgunta H; Urh M Trypsin/Lys-C Protease Mix for Enhanced Protein Mass Spectrometry Analysis. Nat. Methods 2013, 10 (11), i–ii. [Google Scholar]
- (26).Hinderhofer M; Walker CA; Friemel A; Stuermer CA; Möller HM; Reuter A Evolution of Prokaryotic SPFH Proteins. BMC Evol. Biol. 2009, 9 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mitsopoulos P; Chang Y-H; Wai T; König T; Dunn SD; Langer T; Madrenas J Stomatin-Like Protein 2 Is Required for In Vivo Mitochondrial Respiratory Chain Supercomplex Formation and Optimal Cell Function. Mol. Cell. Biol. 2015, 35 (10), 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tong L Structure and Function of Biotin-Dependent Carboxylases. Cell. Mol. Life Sci. 2013, 70 (5), 863–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Greber BJ; Ban N Structure and Function of the Mitochondrial Ribosome. Annu. Rev. Biochem. 2016, 85, 1–103. [DOI] [PubMed] [Google Scholar]
- (30).Kühlbrandt W Structure and Function of Mitochondrial Membrane Protein Complexes. BMC Biol. 2015, 13 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wai T; Saita S; Nolte H; Müller S; König T; Richter-Dennerlein R; Sprenger H; Madrenas J; Mühlmeister M; Brandt U; Krüger M; Langer T The Membrane Scaffold SLP2 Anchors a Proteolytic Hub in Mitochondria Containing PARL and the i -AAA Protease YME1L. EMBO Rep. 2016, 17 (12), 1844–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Rhee HW; Zou P; Udeshi ND; Martell JD; Mootha VK; Carr SA; Ting AY Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science (Washington, DC, U. S.) 2013, 339 (6125), 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Martell JD; Deerinck TJ; Sancak Y; Poulos TL; Mootha VK; Sosinsky GE; Ellisman MH; Ting AY Engineered Ascorbate Peroxidase as a Genetically Encoded Reporter for Electron Microscopy. Nat. Biotechnol. 2012, 30 (11), 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lee SY; Kang MG; Shin S; Kwak C; Kwon T; Seo JK; Kim JS; Rhee HW Architecture Mapping of the Inner Mitochondrial Membrane Proteome by Chemical Tools in Live Cells. J. Am. Chem. Soc. 2017, 139 (10), 3651–3662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.