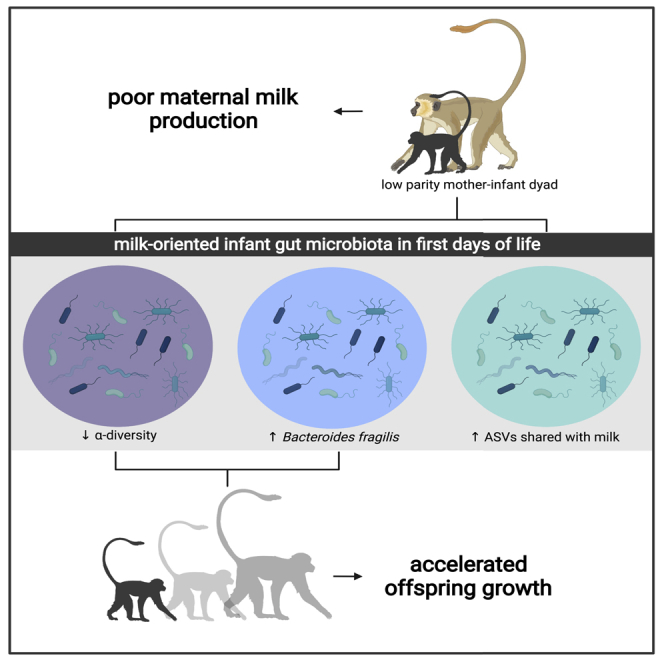
Summary
Maternal parity can impact offspring growth, but the mechanisms driving this effect are unclear. Here, we test the hypothesis that vertically transmitted microbiota may be one potential mechanism. We analyzed 118 fecal and milk samples from mother-offspring vervet monkey dyads across the first 6 months of life. Despite poorer milk production, offspring born to low parity females grew larger than their counterparts. These offspring exhibited reduced alpha diversity in the first days of life, stronger seeding of maternal milk microbiota, Bacteroides fragilis dominance, and a greater abundance of glycan utilization pathways. Moreover, the attainment of greater body mass by 6 months of age was mediated by reduced early life alpha diversity and B. fragilis dominance. This work demonstrates that the establishment of a specialized, milk-oriented gut microbiota promotes infant growth and suggests an evolutionarily conserved developmental role of B. fragilis in primates.
Subject areas: Microbiology, Microbiome
Graphical abstract
Highlights
-
•
Infants of low parity females grow fast despite poor maternal milk production
-
•
These infants have reduced gut microbiota diversity, more Bacteroides fragilis at 2–5 days old
-
•
The infant gut microbiota shares more ASVs with milk than with the maternal gut
-
•
A milk-oriented infant gut microbiota mediates faster growth to 6 months of age
Microbiology; Microbiome
Introduction
In both humans and nonhuman mammals, impaired postnatal growth during early life can result in delayed maturation, dysregulated metabolic function, and increased disease risk during later life (Barker, 2004; Festa-Bianchet et al., 2000; Wauters et al., 1993). The maternal environment is a particularly strong determinant of the pace of early growth: for example, mammalian infants born to reproductively inexperienced (i.e., low parity) mothers typically grow slower, are smaller in body mass for their age, and exhibit higher rates of mortality than infants born to multiparous females (Altmann and Alberts, 2005; Clutton-Brock et al., 1987; Clutton-Brock and Pemberton, 2004; Ibánez et al., 2013; Ruiz-López et al., 2010; Smuts and Nicolson, 1989). One potential explanation for these negative outcomes is that maternal differences in milk production result in poorer transfer of nutrients to fuel offspring growth in some mother-offspring pairs. Because milk volume increases with each successive reproduction, low parity mothers produce smaller quantities of milk than high parity mothers, resulting in fewer available nutrients for infants (Carnicella et al., 2008; Hinde et al., 2009; Lang et al., 2012; Roy et al., 2003; Sevi et al., 2000; Tanaka, 1997; Xiccato et al., 2004).
In captive nonhuman primates, however, infants of high and low parity mothers grow at similar rates despite poorer milk production in the latter group (Nuñez et al., 2015), indicating that milk volume alone may not fully explain parity effects on infant growth. Indeed, infant utilization of milk nutrients not only depends on milk quantity but also on the presence of milk-digesting host-associated microbiota in the infant gut (Marcobal and Sonnenburg, 2012; Zivkovic et al., 2013). In humans, gut microbes such as Bifidobacteria and Bacteroides encode genes responsible for breaking down milk glycans, otherwise indigestible compounds that regulate infant immunity and metabolism (Garrido et al., 2013; Marcobal et al., 2011). These microbes can affect infant growth directly by digesting milk oligosaccharides and glycosylated proteins (∼70% of human milk proteins, Kirmiz et al., 2018). For instance, in rodent transplant models of the human microbiome, pups with a greater relative abundance of gut Bacteroides fragilis grew faster, but only in the presence of milk glycans (Charbonneau et al., 2016).
Microbes like B. fragilis may also indirectly impact infant growth by modifying the resource landscape in the gut and reshaping microbiome community composition to prioritize somatic growth. Although a less diverse gut microbiota can indicate dysbiosis in adults, microbial homogeneity during early life may instead reflect a specialized microbiota canalized for nutrient assimilation. Studies in human infants have found mixed effects of alpha diversity on postnatal growth (Blanton et al., 2016; Gough et al., 2016), but studies in birds demonstrate a link between reduced early life diversity of the gut microbiota and faster later-life growth (Banerjee et al., 2018; Davidson et al., 2021). Alpha diversity may therefore approximate the microbial phenotype directly associated with early growth, reflecting taxonomic and functional homogeneity.
More broadly, the maternal environment may program the infant gut microbiota for enhanced milk utilization via vertical microbial transmission. Thus far, parity effects on maternal microbiota have been demonstrated in agricultural species (Berry et al., 2021; Bogado Pascottini et al., 2021), and more recently in humans (Kervinen et al., 2021; Lopez Leyva et al., 2021). However, in pigs, parity effects extend to the infant gut microbiota, suggesting that such effects may be vertically transmitted from maternal reservoirs (Berry et al., 2021). Large-scale investigations into maternal vertical transmission have identified the maternal gut as the largest source of colonizing bacteria to the infant gut (Bäckhed et al., 2015; Ferretti et al., 2018; Makino et al., 2013; Maqsood et al., 2019). Yet, milk remains notably absent from these studies, obscuring an understanding of its relative role in establishing the infant microbiota. Beyond nutrients and glycans, milk harbors an ephemeral yet diverse microbiota that colonizes the infant gut from birth through weaning (Fernández et al., 2013; Hunt et al., 2011; Meehan et al., 2018; Muletz-Wolz et al., 2019; Petrullo et al., 2019; Fehr et al., 2020). Microbial strains exclusively shared between milk and infant gut communities provide evidence of a unique milk-infant gut transmission pathway independent of other maternal reservoirs (Martín et al., 2012; Pannaraj et al., 2017).
Here, we investigate the relationship between parity, maternal and infant microbiota, and early growth in a nonhuman primate — the vervet monkey (Chlorocebus aethiops sabaeus). Vervet monkeys are established biomedical models for humans, and are particularly appropriate for investigating the relationship between maternal and infant microbiota and early growth. Like humans, vervets possess a milk microbiota that is highly diverse and abundant in growth-associated taxa such as B. fragilis (Petrullo et al., 2019). We characterize the milk and fecal microbiota of mother-infant vervet monkey dyads in a captive population with a high rate of growth-associated infant mortality (>30% in the first month of life, typically the smallest infants; Fairbanks and McGuire, 1984; Kavanagh et al., 2011). This high mortality rate offers a unique opportunity to investigate parity-dependent effects in a naturalistic system in which growth is under strong selective pressure and in which the environment is free of the confounds commonplace in human studies (e.g., socioeconomic status, birth mode).
We test the hypothesis that the infant microbiota mediates the effects of maternal parity on postnatal growth, and investigate if and how maternal microbiota contribute to these effects. We expect that maternal parity predicts the pace of infant growth, maternal and infant microbiota vary with parity, and the effects of maternal parity on postnatal infant growth occur via changes to the infant gut microbiota. We characterize infant gut microbiota in three ways, focusing on community diversity, temporal changes in the abundance of microbial taxa, and the sharing of microbiota with maternal reservoirs (i.e., vertical transmission). We further predict that parity-dependent variation in the infant gut microbiota originates in maternal microbial communities, and expect to find that these patterns are recapitulated in the maternal microbiota, particularly milk.
Results
Maternal parity predicts infant postnatal growth
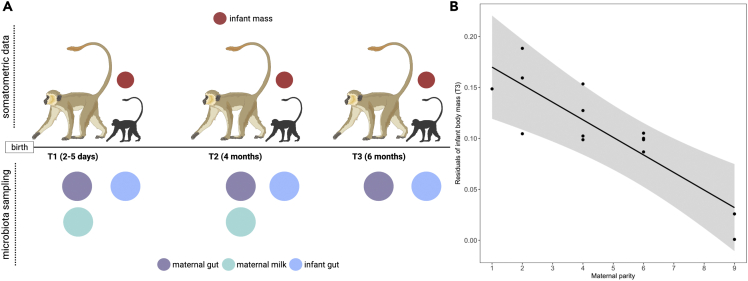
Mother-infant vervet monkey dyads were sampled at three time points across early postnatal life (T1 = 2–5 days old, T2 = 4 months, T3 = 6 months, Figure 1A). Despite poorer maternal milk production (Figure S1), infants born to low parity females were larger in body mass at T3 (β = −0.02 ± 0.004, t = −4.51 p < 0.01; Figure 1B). Each successive parity was associated with a 0.02 kg decrease in body size at both time points (mean infant body mass at T3 = 1.20 kg). There was no relationship between maternal parity and infant body mass in the first days of life (β = 0.02 ± 0.011, t = 1.42, p = 0.18), suggesting that parity-related differences in infant growth emerge primarily across the first 6 months of postnatal life and do not reflect variation in fetal growth or birth weight.
Figure 1.
Experimental design and relationship between maternal parity and offspring growth in the study population
(A) Sampling regimen and (B) relationship between infant body mass and prior maternal reproductive experience. Infants born to low parity females attain a significantly larger body mass by 6 months of age (T3) than infants born to high parity females. Plot shows partial residuals of infant body mass (kg) on the y axis and maternal parity (total number of pregnancies).
Parity shapes the infant and maternal microbiota
Alpha and beta diversity
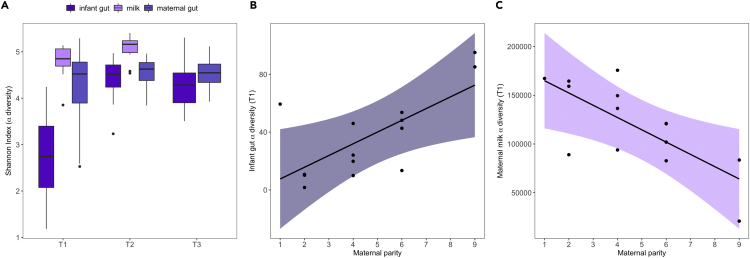
Amplicon sequencing of the 16S rRNA gene identified 1,956 unique amplicon sequence variants (ASVs) across all infant gut samples (mean ± SD = 276 ± 144 ASVs per sample, range = 45-542) compared to 2,019 unique ASVs across maternal gut samples (mean ± SD = 399 ± 94 ASVs per sample, range = 176-661) and 2,714 unique ASVs across maternal milk samples (mean ± SD = 484 ± 176 ASVs per sample, range = 152-742). Infant gut alpha-diversity (measured as Shannon Index) was low compared to maternal communities but increased sharply at T2 (β = 57.00 ± 7.63, t = 7.47, p < 0.0001; Figure 2A). Compositional differences between the gut microbiota of infants and their own mothers (i.e., dyadic beta diversity) were greater at T1 compared to T2 and T3 (β = −0.04 ± 0.007, t = −6.38, p < 0.0001; Figure S2), suggesting that infants achieve an adult-like gut microbiota by 4 months of age.
Figure 2.
Maternal parity predicts variation in alpha diversity of the infant gut and maternal milk microbiota
(A) The infant gut microbiome exhibits significantly lower Shannon Indices than maternal reservoirs at T1 (2–5 days old), but converges toward maternal levels by T2 (4 months old) and remain similarly high at T3 (6 months old).
(B) Shannon Indices (partial residuals) at T1 (2–5 days old) are lowest in low parity infants.
(C) The maternal milk microbiome of low parity females is more diverse (partial residuals of Shannon Indices) than high parity females.
Infants born to low parity females exhibited lower alpha-diversity than high parity infants at T1 (β = 6.13 ± 2.02, t = 3.04, p < 0.01; Figure 2B), but not at T2 or T3. Of the two maternal communities, only milk microbiota varied with maternal parity, and the pattern was opposite to the infant gut: low parity females exhibited more diverse milk microbiota than high parity females at T1 (β = −12,391 ± 3836, t = −3.23, p < 0.01; Figure 2C), and there was no relationship between maternal alpha diversity and parity at any other time point. There was also no relationship between maternal parity and beta diversity in the maternal or infant microbiota.
Differential abundance of taxa
ASVs in the infant gut resolved to 78 bacterial families, 42 of which were highly abundant (>1% of total community composition; Figure S3 and Table S1). Of these 42 families, 21 exhibited statistically significant changes in relative abundance from T1 to T2, and 11 exhibited changes from T2 to T3 (Benjamini-Hochberg adjusted p values: padj < 0.05; Table S2). At T1, the infant gut was dominated by a single ASV from the Bacteroidaceae family (assigned to B. fragilis; mean relative abundance 28.5%). By T2, infants had almost entirely lost Bacteroidaceae from the gut community, which instead became dominated by Prevotellaceae (36.0% relative abundance).
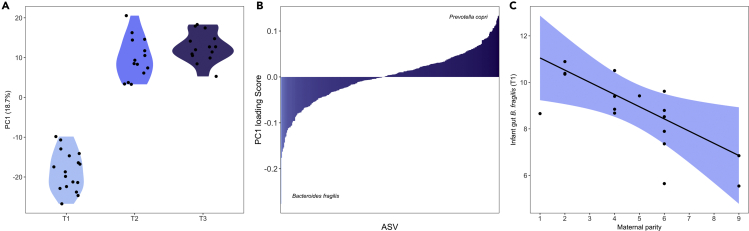
To identify which specific ASVs within these families contributed the most to the maturation of the infant gut microbiota, we used a Compositional Data Analysis (CoDA) framework to account for the compositional nature inherent to microbiome data (Gloor et al., 2017). PC1 explained 18.7% of the total variance in composition among infant samples and showed a clear distinction between early life (T1) samples and older samples (T2 and T3) (Figure 3A). Clustering on PC1 was most strongly explained by two ASVs, each with the highest absolute loading scores: B. fragilis, which dominated early life neonatal samples (T1), and Prevotella copri, which dominated older (T2 and T3) samples (Figure 3B and Table S2).
Figure 3.
Maturation of the infant gut microbiota is driven by a transition from Bacteroides fragilis dominance to Prevotella dominance
(A) Between-sample dissimilarity (Aitchison distance) on the first principal component (PC1) according to infant age.
(B) Loading scores of all infant gut ASVs on the first principal component, with the most influential ASVs depicted on the left for younger infants (B. fragilis) and on the right for older infants (P. copri).
(C) Partial residual plot demonstrating that infants born to low parity females exhibit significantly higher relative abundances of B. fragilis than high parity infants at T1 (2–5 days old).
Of these two taxa, only B. fragilis was associated with maternal parity. Infants born to low parity females exhibited stronger early life dominance of B. fragilis compared to high parity infants (β = −0.40 ± 0.14, t = −2.75, p < 0.01; Figure 3C). Moreover, a greater abundance of infant gut B. fragilis predicted lower alpha diversity at T1 (β = −1.15 ± 0.16, t = −7.25, p < 0.001), indicating that B. fragilis dominance drives homogeneity in the infant gut microbiota during early life.
Vertical transmission from maternal reservoirs
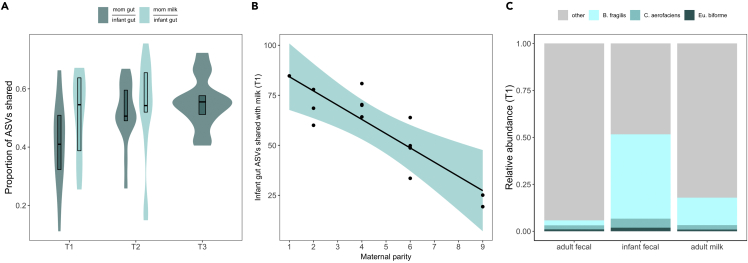
At T1, the vertical transmission of microbiota from milk to the infant gut was stronger than from the maternal gut (β = 9.50 ± 4.32, t = 2.20, p < 0.05; Figure 4A): infants shared 60 ASVs on average with their mother’s milk (51% transmission rate), compared to 41 ASVs with their mother’s gut (42% transmission rate). ASV sharing between the infant gut and maternal milk microbiota was greatest in low parity dyads (β = −7.51 ± 2.03, t = −3.69, p < 0.01; Figure 4B). There was no effect of maternal parity on infant gut ASVs shared with the maternal gut. By T2, the average transmission rates from milk and the maternal gut to the infant gut were not significantly different, and there was no longer an effect of maternal parity on transmission rates from milk, indicating a critical period of vertical transmission in the first days of life.
Figure 4.
Vertical transmission of milk microbiota varies with maternal parity
(A) Across all infants, the infant gut microbiome shares ∼10% more ASVs with milk than with the maternal gut at T1 (2-5 days old). Transmission rates do not differ at T2 (4 months old).
(B) Infants born to low parity females exhibit stronger ASV sharing with the milk microbiome at T1 (2–5 days old) compared to infants of high parity females.
(C) Comparison of the three high-frequency ASVs across microbial communities suggests that milk is the primary reservoir for B. fragilis to the infant gut.
To pinpoint which shared ASVs may be important for infant development, we identified ASVs that were both highly prevalent (i.e., present in all infant gut samples in our dataset) and shared at high frequency with the maternal milk and gut communities (i.e., present in matched mother-infant samples across ⪰90% of dyads). Three ASVs, B. fragilis, Collinsella aerofaciens, and Eubacteria biforme, were prevalent and shared at high frequencies. B. fragilis and Eu. biforme were shared exclusively between maternal milk and infant gut microbiota, whereas C. aerofaciens was shared universally across all three communities. The relative abundance of C. aerofaciens and Eu. biforme were similar across maternal and infant communities, but the infant gut housed the greatest proportion of B. fragilis (∼50% relative abundance, Figure 4C). Mothers housed ∼15x more B. fragilis in their milk compared to their gut (Figure 4C), providing evidence that infant gut B. fragilis may originate in milk.
There was no relationship between maternal parity and the relative abundance of C. aerofaciens or Eu. biforme in the infant gut, nor was there a relationship between maternal parity and these two microbial taxa in the maternal gut. However, low parity females housed more Eu. biforme (β = −0.09 ± 0.04, t = −2.47, p < 0.05) and B. fragilis (β = −0.09 ± 0.04, t = −2.01, p < 0.05) in their milk compared to high parity females.
Predicted functional capacity for glycan utilization
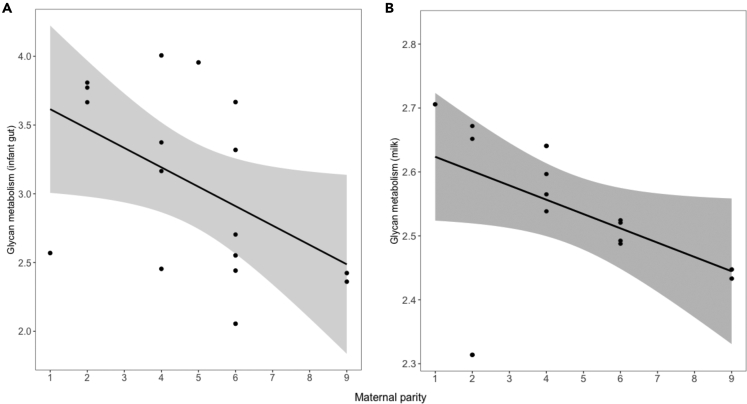
We generated data on predicted gene function for infant and maternal milk microbiota using the PICRUSt2 pipeline to determine whether the microbiota of low parity dyads was enriched in pathways for the utilization of milk glycans. Indeed, low parity infants exhibited a greater relative abundance of microbial gene functional pathways related to glycan biosynthesis and metabolism in their guts than high parity infants in the first days of life (T1) (β = −0.14 ± 0.05, t = −2.90, p = 0.006; Figure 5A). The milk microbiota of low parity mothers also housed a greater relative abundance of glycan utilization pathways than high parity mothers at this same timepoint (T1) (β = −0.01 ± 0.05, t = −2.90, p = 0.04; Figure 5B).
Figure 5.
Low parity dyads exhibit more pathways related to glycan utilization in their microbiota in the first days of life
Partial residual plots demonstrate that the relative abundance of predicted gene functional pathways related to glycan metabolism (y axis) is higher in the (A) infant gut (B) maternal milk microbiota of low parity dyads compared to high parity dyads at T1 (2–5 days old).
Infant gut microbiota mediate parity effects on postnatal growth
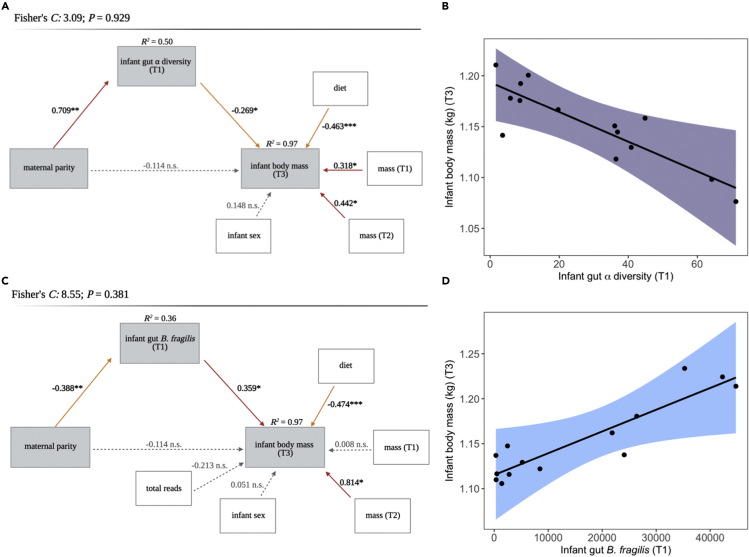
We constructed three path analyses corresponding to the three parity-dependent compositional measures of infant gut microbiota at T1 (alpha diversity, B. fragilis abundance, and ASVs shared with the maternal milk microbiota) and found that parity effects on infant body mass at T3 were mediated by the infant microbiota during early life. In the first model, the effect of maternal parity on neonatal infant gut alpha diversity was the strongest path present (β = 0.709, p < 0.01), and parity indirectly influenced infant mass via reduced infant gut alpha diversity (β = −0.269, p < 0.05) (Figures 6A and 6B). A reduction in each unit of early life Shannon Diversity was associated with a gain of approximately 0.3 kg in body mass. In the second model, maternal parity influenced infant mass via the relative abundance of B. fragilis in the infant gut (β = 0.359, Figures 6C and 6D). With each unit increase in the relative abundance of B. fragilis at T1, infants gained approximately 0.4 kg in body mass. The third model detected no mediation effect of the proportion of ASVs shared with milk on the rate of infant growth.
Figure 6.
A milk-oriented early life infant gut microbiome abundant in B. fragilis mediates the effects of maternal parity on infant growth
Path models demonstrate significant indirect effects of maternal parity on infant body mass at 6 months old (T3) mediated by infant gut microbiota (A) alpha diversity and (B) B. fragilis relative abundance in the first days of life (T1). Global goodness-of-fit was evaluated using Shipley’s test of d-separation (p ≥ 0.05 indicates a good fit) and is reported at the top of each path model. Variables of interest are housed in gray boxes, whereas control/confounding variables are in white boxes. Solid red arrows represent significant positive paths, solid orange arrows represent significant negative paths (p ≤ 0.05), and gray dashed arrows indicate nonsignificant paths. Asterisks denote significant paths (p ≤ 0.05 ∗, p ≤ 0.01 ∗∗, p ≤ 0.001 ∗∗∗). Coefficients of determination (R2) for each of the component models are given on top of the corresponding endogenous (dependent) variables. Partial residual plots of component models additionally highlight the mediation effects of infant gut alpha diversity (C) and B. fragilis relative abundance (D) on infant body mass, independent of maternal parity.
Discussion
To date, parity effects on maternal and infant microbiota have emerged primarily in agricultural animals, and have focused on microbial community changes without establishing a direct link to infant development. Here, we show that parity not only impacts the infant and maternal microbiota in a nonhuman primate, but also influences the pace of early growth via modeling of the infant gut microbiota in the first days of life. Low parity females produced less milk than high parity females, but infants born to low parity females housed a specialized, milk-oriented microbiota dominated by B. fragilis that promoted faster growth to 6 months of age. Patterns of B. fragilis were recapitulated only in the maternal milk microbiota, revealing this maternal reservoir to be the likely source of parity-dependent variation in the infant gut.
Consistent with previous studies in humans and rhesus macaques (Janiak et al., 2021; Thomson et al., 2018) but counter to a recent study in wild chimpanzees (Reese et al., 2020), alpha diversity of the vervet infant gut microbiota was lowest during early life (T1), increasing rapidly toward maternal levels by 4 months of age (T2). Changes in the infant gut microbiota with age were most strongly explained by B. fragilis and P. copri. A trade-off between Bacteroides and Prevotella has been well-described in microbiome research for over a decade as a reflection of dietary differences between human populations consuming diets high in protein and fat (characterized by more Bacteroides) and those consuming a fiber-rich diet (characterized by more Prevotella) (Ley, 2016; Lozupone et al., 2012). In infant vervet monkeys, a trade-off between these taxa may reflect adaptive plasticity of the gut microbiome as infants transition from the consumption of milk to solid foods. Unlike B. fragilis’s role in milk glycan consumption (Coyne et al., 2008; Marcobal et al., 2011), P. copri cannot digest glycans (Ley, 2016). Indeed, nursing piglets exhibit greater Bacteroidaceae, whereas weaned piglets exhibit greater Prevotellaceae (Frese et al., 2015).
We also found that low parity vervet infants exhibited less gut microbiota diversity than high parity infants, and low parity mothers harbored more diverse milk microbiota than high parity mothers. The opposing directionality of these relationships suggests that the reduced alpha diversity characteristic of the low parity infant gut microbiota is not simply a result of vertical transmission from a less diverse maternal milk microbiota. Instead, this pattern provides evidence that infants raised by reproductively inexperienced mothers may selectively seed taxa from a highly diverse milk microbiota (Coyte et al., 2015).
All infants, regardless of parity, selectively seeded B. fragilis in the first days of life (T1), but B. fragilis was most abundant among infants born to low parity mothers and drove homogeneity of the infant gut microbiota in early life. A greater abundance of B. fragilis at T1 predicted the overall lower gut microbiota alpha diversity at both T1 and T2. B. fragilis can modulate the biochemical makeup of the gut environment (Chatzidaki-Livanis et al., 2017; Coyne et al., 2008), in turn altering subsequent colonization by other taxa. Though largely absent from the infant gut microbiota at T2, B. fragilis dominance at T1 may therefore explain the persistence of reduced community diversity via regulation of immigration order (Coyne et al., 2008; Marcobal et al., 2011).
Infants shared ∼10% more ASVs with their mother’s milk microbiota than with her gut microbiota at T1 (i.e., a transmission rate from milk of ∼51 vs. ∼42% from the maternal gut), suggesting that vertical transmission from mothers to infants occurred predominantly via milk. These rates of transmission are higher than previously reported transmission rates in humans (milk: Pannaraj et al., 2017, maternal gut: Ferretti et al., 2018). Higher rates identified in our study may reflect stronger transmission via milk in nonhuman primates compared to humans, and contrast with previous studies in humans that suggest, despite the absence of milk samples, that the maternal gut is the largest source of taxa to the infant gut (Asnicar et al., 2017; Bäckhed et al., 2015; Ferretti et al., 2018). The incorporation of milk microbiota into the infant gut appears to be under strong evolutionary selection in primates, and may be otherwise obscured in human studies because of a historic focus on other maternal reservoirs.
Although there was no relationship between maternal parity and ASV sharing with the maternal gut microbiota at any age, infants of low parity females shared more ASVs with their mother’s milk microbiota than high parity infants at T1. There are a few potential explanations for this finding. First, the dispersal of milk microbiota may be enhanced in low parity females. The vervet monkey milk microbiota is highly individualized, with many ASVs unique to the host (Petrullo et al., 2019). Low parity females may possess more unique variants that are better adapted for dispersal to and survival within the infant gut (Duranti et al., 2017), an explanation also supported by our finding of greater taxonomic diversity in the milk of low parity females. Second, low parity females may exhibit variation in nonmicrobial components of milk (e.g., oligosaccharides, immunoglobulin A), which can modulate the infant gut environment in a manner that favors colonization by maternal milk ASVs over other taxa (Kirmiz et al., 2018; Rogier et al., 2014). Finally, low parity infants may selectively seed maternal milk microbiota more strongly than high parity infants via infant-specific mechanisms that promote colonization by maternal milk ASVs (e.g., phage activity, mucosal immunity, and pH) (Coyte et al., 2015).
High frequency sharing of particular ASVs in milk and infant gut communities provides evidence that the dyadic transmission and seeding of these taxa may be evolutionarily conserved (Korpela et al., 2018). At T1, there was a 15-fold higher abundance of B. fragilis in the milk microbiota compared to the maternal gut microbiota. Moreover, the relative abundance of B. fragilis was greater in the milk microbiota of low parity females compared to high parity females. Together, these results suggest that milk may be the primary reservoir of B. fragilis to the infant gut. Given that neither the maternal vaginal nor gut microbiota explain variation in infant Bacteroides colonization in prior studies in humans (Mitchell et al., 2020), our data suggest that milk may be the primary source of B. fragilis in primates.
Stronger vertical transmission of B. fragilis in low parity dyads may enhance host digestion of glycans from lower volumes of milk, resulting in a microbial community that is more efficient at assimilating milk components to fuel growth. Low parity females in this population can produce as much as 5x less milk than high parity females (Figure S1), necessitating a compensatory mechanism to mitigate the inequity in milk volume. We found that the maternal milk and infant gut microbiota of low parity dyads exhibited a greater abundance of gene pathways related to glycan utilization. Increased metabolism of milk glycans can provide the energetic resources to fuel host growth and development (Charbonneau et al., 2016). Thus, the B. fragilis dominance and enhanced glycan utilization characteristic of the low parity infant gut microbiota may reflect a compensatory strategy toward a homogeneous gut microbiota suited to assimilate energy from lower volumes of milk.
Finally, infants born to low parity females were larger in body mass at 6 months of age, and this effect was mediated by reduced alpha diversity at T1, in line with previous studies on birds (Banerjee et al., 2018; Davidson et al., 2021). Mass at 6 months was also mediated by B. fragilis dominance at T1, in line with experimental studies on the human microbiota (Blanton et al., 2016; Charbonneau et al., 2016). These findings support our hypothesis of a mediating role of microbiota on parity-dependent infant growth, but the faster, rather than slower, rates of growth among low parity infants are in contrast with prior work in wild mammals (e.g., Clutton-Brock and Pemberton, 2004; Ibánez et al., 2013; Ruiz-López et al., 2010). One potential explanation for this pattern is that the nutrient abundance present in captivity may exploit the compensatory mechanisms that allow low parity mothers to successfully rear infants despite poor milk production in the wild. Such exploitation, coupled with the “humanizing” effect of captivity on the nonhuman primate microbiota (Clayton et al., 2016; Houtz et al., 2021), may better approximate the microbial pathways that mediate maternal effects in modern human populations inhabiting calorie-rich environments. In addition, viability selection exerts substantial pressure on infant development in populations with high rates of early mortality (McAdam and Boutin, 2003; Mojica and Kelly, 2010). In our study population, infants face a growth-associated mortality rate of more than 30% (Fairbanks and McGuire, 1984) and individuals who die early in life are smaller in body size than their counterparts (Kavanagh et al., 2011), potentially amplifying growth-associated microbial effects. Our findings may thus extend to human populations experiencing stunted childhood growth and growth-associated early mortality by identifying B. fragilis as a key predictor of postnatal body mass.
Limitations of the study
This study was limited in a few ways. First, although maternal vaginal microbiota may contribute to or modify the above-described relationships, logistic constraints prevented vaginal microbiota sampling in this study. Despite this limitation, the vaginal microbiota is unlikely to be the primary contributor of maternal B. fragilis to the infant gut as Bacteroides are not common members of the human vaginal microbiota and even when present, do not appear to account for transmission to the infant gut (Mitchell et al., 2020). Given the importance of B. fragilis in mediating parity effects on infant growth in this population, our interpretation of milk as the potential source of B. fragilis is likely independent of vaginal microbiota’s effects. Second, the correlation between maternal parity and maternal age raises the question of whether the relationships described here may reflect age, rather than parity, effects. Because our hypotheses were centered on the effects of successive reproductions on maternal communities, and because prior work has shown that parity, not age, shapes the maternal microbiota (Berry et al., 2021), we did not include maternal age in our analyses to avoid issues of multicollinearity. Instead, we focused on parity because successive reproductions (independent of maternal age) shape the morphology of both the mammary gland (Rovai et al., 1999) and variation in milk production across mammalian species (e.g., Lang et al., 2012; Sevi et al., 2000), suggesting that similar effects may be found in maternal milk and infant gut microbiota.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Maternal (milk and fecal) and infant (fecal) samples used for microbiome analysis | Author's collection (Wake Forest School of Medicine Vervet Research Colony) | https://doi.org/10.6084/m9.figshare.14561925 |
| Deposited data | ||
| 16S rRNA amplicon sequences from 118 fecal and milk samples | This paper | www.ncbi.nlm.nih.gov/sra/PRJNA728247 (NCBI Sequence Read Archive) |
| Experimental models: Organisms/strains | ||
| 18 adult female and 18 mixed-sex infant vervet monkeys (Chlorocebus aethiops sabaeus) | Vervet Research Colony, Wake Forest University | N/A |
| Software and algorithms | ||
| QIIME2 | Hall and Beiko, 2018 | https://qiime2.org/ |
| DADA2 | Callahan et al., 2016 | https://benjjneb.github.io/dada2/ |
| MAFFT | Katoh et al., 2002 | https://mafft.cbrc.jp/alignment/software/ |
| PICRUSt2 | Douglas et al., 2020 | https://nephele.niaid.nih.gov/show_picrust2_details/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lauren Petrullo (petrullo@umich.edu).
Materials availability
This study did not generate new unique materials.
Experimental model and subject details
We studied 18 mother-infant captive vervet monkey (Chlorocebus aethiops sabaeus) dyads housed at the Vervet Research Colony (VRC) at the Wake Forest University School of Medicine in Winston-Salem, North Carolina, USA. Infant vervet monkeys <6 months of age of both sexes were included in this study. All animal use procedures and sample collection methods followed national and international guidelines and were approved by the Institutional Care and Use Committee (IACUC) at Wake Forest University School of Medicine. Vervets are highly social primates that breed once annually (Else et al., 1986). Mother-offspring dyads were housed at the VRC across 8 indoor-outdoor social groups organized by female kin. Thirteen of the dyads were provisioned daily with commercial monkey chow (Purina Monkey Chow, LabDiet 5038), while four dyads were fed a western-style lab diet (LabDiet 5L3K) as part of a different study. In addition, some dyads were fed ad libitum, while feeding was time-restricted for others. However, it is important to note that infants in this population do not appear to incorporate solid foods into their diets until about 4-6 months (Fairbanks, 1988), thus the effect of diet on early life microbiota at T1 in this study is expected to be minimal. Animals were supplemented with fresh fruits and vegetables daily. Infants included in this study were delivered via unassisted vaginal births between June 18, 2017 and August 27, 2017 after typical ∼5.5 month-long gestations.
Method details
We collected physiological data from dyads at three postnatal timepoints: T1 (2-5 days postpartum), T2 (4 months postpartum), and T3 (6 months postpartum). Two infants died within the first month of life as a probable consequence of poor maternal milk production: in one case, the mother was not producing any milk, and in the second case, the mother produced milk in only one mammary gland. Thus, both infants contributed fecal samples and somatometric data at T1 only. The T1 milk sample from the female with unilateral milk production was excluded from this study. Milk collection followed previously published protocols for the collection of milk from cercopithecine monkeys (Hinde et al., 2009; Petrullo et al., 2019). Both mammary glands were fully evacuated via manual expression into a single sample tube. Samples were placed immediately on ice, briefly vortexed, aliquoted into cryovials, and frozen at −80°C until shipment to the University of Washington. We collected maternal and infant fecal samples at all three sampling timepoints by briefly inserting standard (mothers) and pediatric (infants) flocked nylon swabs (FLOQSwabs, COPAN Diagnostics, Murrieta, CA) into the anal canal. Rectal swabs are reliable proxies for fecal samples when quantifying an animal’s gut microbiota, including among infants (Bassis et al., 2017; Reyman et al., 2019). Swabs were gently spun in the canal 2-3 times before removal and were snapped off into empty polypropylene tubes. Samples were immediately frozen at −80°C before being shipped to the University of Washington.
Quantification and statistical analysis
DNA extraction, amplification, and library preparation
We extracted microbial DNA from milk using the PowerFood Microbial kit (Qiagen) following the manufacturer’s kit protocols, with the addition of two front-end processing steps shown to increase overall yield from milk (Petrullo et al., 2019). For fecal swabs, microbial DNA was extracted using the Qiagen PowerLyzer PowerSoil kit. Samples were thawed to room temperature, swab tips were snapped into glass bead tubes (0.1 mm), and mechanically lysed (30 Hz for 2 min). Extractions then followed the manufacturer’s protocol.
We amplified the hypervariable V4 region of the 16S rRNA gene using PCR primer set 515F and 806R from The Human Microbiome Project (Gilbert et al., 2014; Yang et al., 2016), following previously published protocols (Petrullo et al., 2019). A fragment analyzer was used to confirm amplification of the V4 region prior to quantification via a qubit fluorometer.
Sequencing and bioinformatics
Amplicon libraries were balanced and pooled. Library complexity was increased by spiking libraries with PhiX prior to sequencing all libraries together on a single Illumina MiSeq flow cell using 301 bp paired-end sequences. This resulted in 1,456,972 reads for milk samples, 2,491,940 reads for adult fecal samples, and 2,147,458 reads for infant fecal samples. Sequences were analyzed using the QIIME2 platform (Caporaso et al., 2011; Hall and Beiko, 2018) and denoised with Divisive Amplicon Denoising Algorithm 2 (DADA2: Callahan et al., 2016) as a QIIME2 plug-in. In contrast to clustering sequencing reads based on a fixed dissimilarity threshold (e.g., the assignment of Operational Taxonomic Units [OTUs]), which can conflate sequencing errors with biological variation, DADA2 infers sequences exactly (resulting in amplicon sequence variants, hereafter referred to as ASVs), providing higher taxonomic resolution than OTUs (Callahan et al., 2016, 2017). This approach is crucial for assessing changes in microbial composition (Tikhonov et al., 2015) as well as community diversity (Rosen et al., 2015), particularly in microbial communities presumed to be low in diversity based on prior OTU clustering methods (e.g., the human vaginal microbiota, Callahan et al., 2017), as well as determining vertical transmission between maternal and infant communities. Forward and reverse reads were trimmed to 240 bases long to remove the low-quality portion of the sequences. Next, the forward and reverse reads were merged and chimeric sequences removed. After filtering, trimming, merging, and chimera removal, we retained 811,225 16S rRNA gene sequences from 29 milk samples (28,972 ± 17,165 reads per sample), 1,547,442 sequences from 45 adult female fecal samples (48,357 ± 15,100 reads per sample), and 2,147,458 sequences from 45 infant fecal samples (47,721 ± 12,580 reads per sample). Only fecal samples with >15,000 reads were retained for analyses, resulting in N=44 infant fecal samples. No other samples were removed at this stage as sequencing depths across samples were highly balanced. ASVs were aligned using MAFFT (Katoh et al., 2002) and a phylogenetic tree was constructed using fasttree2 (Price et al., 2010). Taxonomic assignment of ASVs was performed using the q2-feature-classifier in QIIME2 against the 13_8 version of the GreenGenes database (McDonald et al., 2012) based on 100% similarity to the reference sequence. One milk sample was excluded from the analysis as all of its 200,000+ reads were assigned to a single ASV.
Statistical analyses
All analyses for this study were performed in QIIME2 (Hall and Beiko, 2018) and R version 4.0.2 (R. Core Team, 2015). Taxonomy and count tables were imported from QIIME2 into R using the qiime2R package (Bisanz, 2018). As rarefaction of microbiota count data results in a loss of precision and variation (McMurdie and Holmes, 2014), we instead used a compositional data, or “CoDa”, approach when necessary (Gloor et al., 2017) and controlled for sequencing depth by including the log-transformed count of total reads in a sample as an offset in all statistical models. Where microbial taxa were converted to relative abundances, the taxonomy table was filtered to retain only those with > 1% relative abundance. Residuals of all linear models described below were found to be normally distributed following visual evaluation by Q-Q plots and Shapiro-Wilk tests. Because of the potential confounding effects of dietary composition and feeding schedule, diet was controlled for as a fixed factor in all analyses. Figures were created in R using ggplot2 (Wickham, 2011) and visreg for partial residual plots (Breheny and Burchett, 2017). The graphical abstract, Figures 1A, 6A, and 6C were created using biorender (www.biorender.com).
Milk yield
Linear models were used to test the effects of maternal parity on milk production (mL). Residuals for all models were normally distributed upon visual inspection and evaluation with a Shapiro-Wilk test, and therefore not transformed prior to analysis. We constructed models with milk yield (mL) as the dependent variable, with maternal parity as a fixed factor while controlling for offspring sex (Hinde, 2009; Hinde et al., 2009) and dietary composition.
Infant body mass
Linear models were used to test whether maternal parity predicted infant body mass at T3 (dependent variable), controlling for infant sex, approximate birth weight (body mass at T1), and dietary category. T3 body mass data were log-transformed prior to modelling to ensure normality of the model residuals.
Microbiome alpha & beta diversity
Microbiome samples were assessed at the community level using both alpha (within sample) and beta (between samples) diversity measures. Alpha diversity was calculated using the phyloseq package (McMurdie and Holmes, 2013) and quantified using the Shannon Index of alpha-diversity (Shannon, 1948; Spellerberg and Fedor, 2003), which takes into account both the richness and evenness of taxa. Shannon Indices were transformed by Tukey transformation to achieve normality of residuals, and linear mixed models were constructed to test associations between infant age (categorical fixed effect: T1, T2, T3) and gut microbiome diversity (Shannon Index, dependent variable), controlling for diet (fixed effect) and infant ID (random effect). Weighted UniFrac distance matrices were constructed in QIIME2 and were used to assess dissimilarity between the gut microbiome of infants and their own mothers (dyadic weighted UniFrac distance) as a measure of infant gut microbiome maturation. A linear mixed model was used to test whether infant age (fixed effect) predicted dyadic weighted UniFrac distance (dependent variable), controlling for diet (fixed effect) and infant ID (random effect).
Differential abundance testing
Differential abundance testing of taxa within the infant gut microbiome was performed in R using the NBZIMM package (Zhang and Yi, 2020). Negative binomial models (glmer.nb() function) were used test the effects of infant age (fixed effect) on the abundance of taxonomic families (dependent variable: raw counts), controlling for diet (fixed effect), sequencing depth fit as an offset (log transformation of total number of reads in the sample), and infant ID (random effect). Models that did not converge due to a high proportion of zeros were rerun using the same package as zero-inflated Gaussian mixed models (lme.zig() function). p-values were adjusted using the Benjamini-Hochberg FDR multiple-test correction (Benjamini and Hochberg, 1995) to account for multiple hypothesis testing and microbial taxa with adjusted p values < 0.05 were considered statistically significant.
Predicted functional profiles
To predict gene functional content from 16S rRNA data, we used the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) v.2.1.3-b software (Douglas et al., 2020) pipeline with default options (picrust2_pipeline.py). PICRUSt2 predicts gene family abundance by using the table of ASVs generated by QIIME2 and their representative sequences. We then inferred Kyoto KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathway abundances from the predicted KEGG ORTHOLOGY (KO) at Level 2 of the BRITE map (Kanehisa et al., 2014). Counts of functional pathways were transformed to relative abundance for each sample and a filtering threshold of 0.1% abundance was implemented to retain only the most abundant pathways. To estimate the accuracy of our PICRUSt2 predicted pathways, we calculated a weighted NSTI (Nearest Sequenced Taxon Index) score for each sample, which estimates the phylogenetic distance between OTUs and their sequenced reference genomes, to achieve an average NSTI score. The average NSTI score for all samples was 0.087 (SE ± 0.038, range: 0.011–0.28), reflecting high (∼91%) distinguishability.
Linear mixed-effect models were used to test the effects of maternal parity on glycan utilization and metabolism pathways in the infant gut microbiome (dependent variable) as a function of sampling timepoint, controlling for diet (fixed factors) and infant ID (random effect).
Principal Components Analysis & CoDa framework
A Compositional Data (CoDa) framework was used to investigate how specific ASVs contributed to the compositional patterns of infant gut microbiome maturation (Gloor et al., 2017). Raw count data of ASV reads were normalized using a centered log-ratio (clr) transformation and a pseudocount of 0.65 in place of zeroes (Aitchison, 1982). We then ordinated the samples based on their compositional dissimilarity using a Principal Components Analysis (PCA: Hotelling, 1933) using pairwise Euclidean distances between samples (i.e., Aitchison distance). Unlike Principal Coordinates Analysis (PCoA), which is more commonly used in microbiome data analysis, PCA is not driven by presence/absence data, is more reproducible, and is more robust against sparsity (Gloor et al., 2017). The PCA was used to visualize sample clustering and generate loading scores for each ASV on the first PCA axis (which captured 18.7% of variability and was highly predicted by infant age). The loading scores were then sorted by absolute value to determine the microbial taxa with the highest influence (e.g., absolute value loading score) on the observed clustering of the first PCA axis (i.e., the most predictive of infant age) (Martino et al., 2019). We then calculated a log-ratio of the relative abundance of the two most influential taxa (Bacteroides fragilis and Prevotella copri) and used Pearson’s correlation tests to determine the correlation coefficient between the log-ratios and samples’ coordinates on the first PCA axis. Subsequently, we used a linear mixed model to test whether the log-ratio of these taxa (fixed effect) predicted dyadic weighted UniFrac distances between infant and maternal gut microbiomes (dependent variable), controlling for infant age (fixed factor), diet (fixed factor), and dyad ID (random).
Vertical transmission
The proportion of ASVs shared between maternal and offspring microbiota is a commonly-used index of vertical transmission using 16S rRNA gene data (Björk et al., 2019; Korpela et al., 2018; Maqsood et al., 2019; Renelies-Hamilton et al., 2021). We calculated the proportion of shared ASVs as being the proportion of all ASVs present within the infant gut microbiome (denominator) that were also found in maternal milk and gut microbiomes (numerator). To test whether these proportions differed significantly between infant gut/maternal gut versus infant gut/maternal milk comparisons, we fit linear mixed models with proportion of ASVs shared at T1 and T2 as the dependent variable and comparison (milk vs. infant gut or maternal gut vs. infant gut) as a predictor variable, controlling for individual ID. To test whether maternal parity affected vertical transmission, generalized linear models were used to test whether maternal parity predicted the raw count of shared ASVs (dependent variable), controlling for diet and the total number of ASVs present within the infant gut.
Path analysis
To test for a mediation effect of the microbiome on infant growth, we performed path analyses by constructing three separate path models (one for each parity-dependent measure of infant gut microbiome composition: alpha diversity, B. fragilis abundance, and vertical transmission via the milk microbiome) using the piecewiseSEM() in R (v. 2.0.1) (Lefcheck, 2016). Unlike simple structural equation modeling, piecewiseSEM is robust to small sample sizes and can integrate mixed effects and generalized linear models (e.g., negative binomial models) (Lefcheck, 2016), making it more suitable for use with microbiome count data. Each path model was comprised of two recursive component models: 1) a model testing direct/indirect effects of both maternal parity and the relevant measure of infant gut microbiome composition at T1 on infant body mass at T3, controlling for confounding variables, and 2) a model testing the effect of maternal parity on the infant gut microbiome, controlling for confounding variables. Component models were integrated into a single causal network using the psem() function, which produced weighted coefficients and corresponding p-values to permit the assessment of the significance and relative strengths of each path within the overall network. The global goodness-of-fit was assessed for each model in piecewiseSEM() using Shipley’s test of d-separation (Shipley, 2013). All models had Fisher’s C statistics with p > 0.05, indicating that the models were suitable fits and no major paths were missing.
Acknowledgments
We would like to thank the veterinary and technical staff of the Vervet Research Colony, especially Edison Floyd and Chrissy Long, for their role in sample collections, and Katie Hinde for providing expertise and protocol guidance for milk sample collection. We would also like to thank Andreas Koenig, Carola Borries, and anonymous reviewers for comments on earlier versions of this manuscript. This research was funded by the National Institutes of Health CTSA pilot grant (UL1-TR001420, Donald Mcclain PI), a P40 grant (OD010965, Matthew Jorgensen PI), Stony Brook University, the University of Washington, and the Dr. W. Burghardt Turner Fellowship and AGEP-T FRAME Fellowship programs at Stony Brook University (to L.P.).
Author contributions
Concept, Design, and Interpretation of data, L.P. and A.L.; Data acquisition and Methodology, L.P., A.B., M.J.J., S.S., N.S.M., and A.L.; Writing – Original Draft, L.P. and A.L.; Review & Editing, A.B., M.J.J., S.S., and N.S.M.; Funding, L.P., M.J.J., N.S.M., and A.L.; Supervision, A.L. All authors have approved this version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103948.
Supplemental information
Negative loading scores correspond to younger infant samples, positive loading scores correspond to older infant samples.
Data and code availability
-
•
16S rRNA gene sequences are available under BioProject ID PRJNA728247 at the NCBI Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra/PRJNA728247) and asssociated metadata for this project are available at figshare: https://figshare.com/s/79f3cf6cf410f5a87d31.
-
•
Code for this project is available at figshare: https://figshare.com/s/79f3cf6cf410f5a87d31.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aitchison J. The statistical analysis of compositional data. J. R. Stat. Soc. Ser. B Stat. Methodol. 1982;44:139–160. [Google Scholar]
- Altmann J., Alberts S.C. Growth rates in a wild primate population: ecological influences and maternal effects. Behav. Ecol. Sociobiol. 2005;57:490–501. [Google Scholar]
- Asnicar F., Manara S., Zolfo M., Truong D.T., Scholz M., Armanini F., Ferretti P., Gorfer V., Pedrotti A., Tett A., Segata N. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. 2017;2:e00164-16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H., et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Sar A., Misra A., Pal S., Chakraborty A., Dam B. Increased productivity in poultry birds by sub-lethal dose of antibiotics is arbitrated by selective enrichment of gut microbiota, particularly short-chain fatty acid producers. Microbiology. 2018;164:142–153. doi: 10.1099/mic.0.000597. [DOI] [PubMed] [Google Scholar]
- Barker D.J.P. Developmental origins of adult health and disease. J. Epidemiol. Community Health. 2004;58:114–115. doi: 10.1136/jech.58.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis C.M., Moore N.M., Lolans K., Seekatz A.M., Weinstein R.A., Young V.B., Hayden M.K., CDC Prevention Epicenters Program Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017;17:78. doi: 10.1186/s12866-017-0983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Berry A.S.F., Pierdon M.K., Misic A.M., Sullivan M.C., O’Brien K., Chen Y., Murray S.J., Ramharack L.A., Baldassano R.N., Parsons T.D., Beiting D.P. Remodeling of the maternal gut microbiome during pregnancy is shaped by parity. Microbiome. 2021;9:146. doi: 10.1186/s40168-021-01089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisanz J.E. 2018. qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. Version 0. 99 13. [Google Scholar]
- Björk J.R., Díez-Vives C., Astudillo-García C., Archie E.A., Montoya J.M. Vertical transmission of sponge microbiota is inconsistent and unfaithful. Nat. Ecol. Evol. 2019;3:1172–1183. doi: 10.1101/425009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton L.V., Charbonneau M.R., Salih T., Barratt M.J., Venkatesh S., Ilkaveya O., Subramanian S., Manary M.J., Trehan I., Jorgensen J.M., et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogado Pascottini O., Spricigo J.F.W., Van Schyndel S.J., Mion B., Rousseau J., Weese J.S., LeBlanc S.J. Effects of parity, blood progesterone, and non-steroidal anti-inflammatory treatment on the dynamics of the uterine microbiota of healthy postpartum dairy cows. PLoS One. 2021;16:e0233943. doi: 10.1371/journal.pone.0233943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breheny P., Burchett W. Visualization of regression models using visreg. R. J. 2017;9:56. [Google Scholar]
- Callahan B.J., McMurdie P.J., Holmes S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U S A. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnicella D., Dario M., Ayres M.C.C., Laudadio V., Dario C. The effect of diet, parity, year and number of kids on milk yield and milk composition in Maltese goat. Small Rumin. Res. 2008;77:71–74. [Google Scholar]
- Charbonneau M.R., O’Donnell D., Blanton L.V., Totten S.M., Davis J.C.C., Barratt M.J., Cheng J., Guruge J., Talcott M., Bain J.R., et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 2016;164:859–871. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzidaki-Livanis M., Coyne M.J., Roelofs K.G., Gentyala R.R., Caldwell J.M., Comstock L.E. Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. mBio. 2017;28:e01902-17. doi: 10.1128/mbio.01902-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J.B., Vangay P., Huang H.U., Ward T., Hillman B.M., Al-Ghalith G.A., Travis D.A., Long H.T., Van Tuan B., Van Minh V., Cabana F., Nadler T., Toddes B., Murphy T., Glander K.E., Johnson T.J., Knights D. Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. U. S. A. 2016;113:10376–10381. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock T.H., Albon S.D., Guinness F.E. Interactions between population density and maternal characteristics affecting fecundity and juvenile survival in red deer. J. Anim. Ecol. 1987;56:857–871. [Google Scholar]
- Clutton-Brock T.H., Pemberton J.M. Cambridge University Press; 2004. Soay Sheep: Dynamics and Selection in an Island Population. [Google Scholar]
- Coyne M.J., Chatzidaki-Livanis M., Paoletti L.C., Comstock L.E. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U S A. 2008;105:13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte K.Z., Schluter J., Foster K.R. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- Davidson G.L., Somers S.E., Wiley N., Johnson C.N., Reichert M.S., Paul Ross R., Stanton C., Quinn J.L. A time-lagged association between the gut microbiome, nestling growth and nestling survival in wild great tits. J. Anim. Ecol. 2021;90:989–1003. doi: 10.1111/1365-2656.13428. [DOI] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotech. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranti S., Lugli G.A., Mancabelli L., Armanini F., Turroni F., James K., Ferretti P., Gorfer V., Ferrario C., Milani C., et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else J.G., Eley R.M., Wangula C., Worthman C., Lequin R.M. Reproduction in the vervet monkey (Cercopithecus aethiops): II. Annual menstrual patterns and seasonality. Am. J. Primatol. 1986;11:333–342. doi: 10.1002/ajp.1350110404. [DOI] [PubMed] [Google Scholar]
- Fairbanks L.A. Mother-infant behavior in vervet monkeys. Behav. Ecol. Sociobiol. 1988;23:157–165. [Google Scholar]
- Fairbanks L.A., McGuire M.T. Determinants of fecundity and reproductive success in captive vervet monkeys. Am. J. Primatol. 1984;7:27–38. doi: 10.1002/ajp.1350070106. [DOI] [PubMed] [Google Scholar]
- Fehr K., Moossavi S., Sbihi H., Boutin R.C.T., Bode L., Robertson B., Yonemitsu C., Field C.J., Becker A.B., Mandhane P.J., et al. Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers’ milk and the infant gut: the CHILD cohort study. Cell Host Microbe. 2020;28:285–297.e4. doi: 10.1016/j.chom.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Fernández L., Langa S., Martín V., Maldonado A., Jiménez E., Martín R., Rodríguez J.M. The human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 2013;69:1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Ferretti P., Pasolli E., Tett A., Asnicar F., Gorfer V., Fedi S., Armanini F., Truong D.T., Manara S., Zolfo M., et al. Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa-Bianchet M., Jorgenson J.T., Réale D. Early development, adult mass, and reproductive success in bighorn sheep. Behav. Ecol. 2000;11:633–639. [Google Scholar]
- Frese S.A., Parker K., Calvert C.C., Mills D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome. 2015;3:28. doi: 10.1186/s40168-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Dallas D.C., Mills D.A. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J.A., Jansson J.K., Knight R. The Earth Microbiome project: successes and aspirations. BMC Biol. 2014;12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J. Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough E.K., Stephens D.A., Moodie E.E.M., Prendergast A.J., Stoltzfus R.J., Humphrey J.H., Manges A.R. Linear growth faltering in infants is associated with Acidaminococcus sp. and community-level changes in the gut microbiota. Microbiome. 2016;4:5. doi: 10.1186/s40168-016-0149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M., Beiko R.G. 16S rRNA gene analysis with QIIME2. Methods Mol. Biol. 2018;1849:113–129. doi: 10.1007/978-1-4939-8728-3_8. [DOI] [PubMed] [Google Scholar]
- Hinde K. Richer milk for sons but more milk for daughters: sex-biased investment during lactation varies with maternal life history in rhesus macaques. Am. J. Hum. Biol. 2009;21:512–519. doi: 10.1002/ajhb.20917. [DOI] [PubMed] [Google Scholar]
- Hinde K., Power M.L., Oftedal O.T. Rhesus macaque milk: magnitude, sources, and consequences of individual variation over lactation. Am. J. Phys. Anthropol. 2009;138:148–157. doi: 10.1002/ajpa.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933;24:417–441. [Google Scholar]
- Houtz J.L., Sanders J.G., Denice A., Moeller A.H. Predictable and host-species specific humanization of the gut microbiota in captive primates. Molec. Ecol. 2021;30:3677–3687. doi: 10.1111/mec.15994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt K.M., Foster J.A., Forney L.J., Schütte U.M.E., Beck D.L., Abdo Z., Fox L.K., Williams J.E., McGuire M.K., McGuire M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6:e21313. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibánez B., Moreno E., Barbosa A. Parity, but not inbreeding, affects juvenile mortality in two captive endangered gazelles. Anim. Conserv. 2013;16:108–117. [Google Scholar]
- Janiak M.C., Montague M.J., Villamil C.I., Stock M.K., Trujillo A.E., DePasquale A.N., Orkin J.D., Bauman Surratt S.E., Gonzalez O., Platt M.L., et al. Age and sex-associated variation in the multi-site microbiome of an entire social group of free-ranging rhesus macaques. Microbiome. 2021;9:68. doi: 10.1186/s40168-021-01009-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nuc. Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K.-I., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K., Dozier B.L., Chavanne T.J., Fairbanks L.A., Jorgensen M.J., Kaplan J.R. Fetal and maternal factors associated with infant mortality in vervet monkeys. J. Med. Primatol. 2011;40:27–36. doi: 10.1111/j.1600-0684.2010.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervinen K., Holster T., Saqib S., Virtanen S., Stefanovic V., Rahkonen L., Nieminen P., Salonen A., Kalliala I. Parity and Gestational Age Are Associated with Vaginal Microbiota Composition in Term and Late Term Pregnancies. 2021. [DOI] [PMC free article] [PubMed]
- Kirmiz N., Robinson R.C., Shah I.M., Barile D., Mills D.A. Milk glycans and their interaction with the infant-gut microbiota. Annu. Rev. Food Sci. Technol. 2018;9:429–450. doi: 10.1146/annurev-food-030216-030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela K., Costea P., Coelho L.P., Kandels-Lewis S., Willemsen G., Boomsma D.I., Segata N., Bork P. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018;28:561–568. doi: 10.1101/gr.233940.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S.L.C., Iverson S.J., Bowen W.D. Primiparous and multiparous females differ in mammary gland alveolar development: implications for milk production. J. Exp. Biol. 2012;215:2904–2911. doi: 10.1242/jeb.067058. [DOI] [PubMed] [Google Scholar]
- Lefcheck J.S. piecewiseSEM: piecewise structural equation modelling in r for ecology, evolution, and systematics. Methods Ecol. Evol. 2016;7:573–579. [Google Scholar]
- Ley R.E. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016;13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- Lopez Leyva L., Gonzalez E., Li C., Ajeeb T., Solomons N.W., Agellon L.B., Scott M.E., Koski K.G. Human milk microbiota in an indigenous population is associated with maternal factors, stage of lactation, and breastfeeding practices. Curr. Dev. Nutr. 2021;5:nzab013. doi: 10.1093/cdn/nzab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H., Kushiro A., Ishikawa E., Kubota H., Gawad A., Sakai T., Oishi K., Martin R., Ben-Amor K., Knol J., Tanaka R. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One. 2013;8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqsood R., Rodgers R., Rodriguez C., Handley S.A., Ndao I.M., Tarr P.I., Warner B.B., Lim E.S., Holtz L.R. Discordant transmission of bacteria and viruses from mothers to babies at birth. Microbiome. 2019;7:156. doi: 10.1186/s40168-019-0766-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A., Barboza M., Sonnenburg E.D., Pudlo N., Martens E.C., Desai P., Lebrilla C.B., Weimer B.C., Mills D.A., German J.B., Sonnenburg J.L. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A., Sonnenburg J.L. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 2012;18:12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino C., Morton J.T., Marotz C.A., Thompson L.R., Tripathi A., Knight R., Zengler K. A novel sparse compositional technique reveals microbial perturbations. mSystems. 2019;4:e00016-19. doi: 10.1128/msystems.00016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Maldonado-Barragán A., Moles L., Rodriguez-Baños M., Campo R.D., Fernández L., Rodríguez J.M., Jiménez E. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 2012;28:36–44. doi: 10.1177/0890334411424729. [DOI] [PubMed] [Google Scholar]
- McAdam A.G., Boutin S. Variation in viability selection among cohorts of juvenile red squirrels (Tamiasciurus hudsonicus) Evolution. 2003;57:1689–1697. doi: 10.1111/j.0014-3820.2003.tb00374.x. [DOI] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. Package “phyloseq. Gan. 2013;2:7. [Google Scholar]
- Meehan C.L., Lackey K.A., Hagen E.H., Williams J.E., Roulette J., Helfrecht C., McGuire M.A., McGuire M.K. Social networks, cooperative breeding, and the human milk microbiome. Am. J. Hum. Biol. 2018;30:e23131. doi: 10.1002/ajhb.23131. [DOI] [PubMed] [Google Scholar]
- Mitchell C., Hogstrom L., Bryant A., Bergerat A., Cher A., Pochan S., Herman P., Carrigan M., Sharp K., Huttenhower C., et al. Delivery mode impacts newborn gut colonization efficiency. bioRxiv. 2020 doi: 10.1101/2020.01.29.919993. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica J.P., Kelly J.K. Viability selection prior to trait expression is an essential component of natural selection. Proc. Biol. Sci. 2010;277:2945–2950. doi: 10.1098/rspb.2010.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muletz-Wolz C.R., Kurata N.P., Himschoot E.A., Wenker E.S., Quinn E.A., Hinde K., Power M.L., Fleischer R.C. Diversity and temporal dynamics of primate milk microbiomes. Am. J. Primatol. 2019;81:164. doi: 10.1002/ajp.22994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez C.L., Grote M.N., Wechsler M., Allen-Blevins C.R., Hinde K. Offspring of primiparous mothers do not experience greater mortality or poorer growth: revisiting the conventional wisdom with archival records of Rhesus Macaques. Am. J. Primatol. 2015;77:963–973. doi: 10.1002/ajp.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannaraj P.S., Li F., Cerini C., Bender J.M., Yang S., Rollie A., Adisetiyo H., Zabih S., Lincez P.J., Bittinger K., et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrullo L., Jorgensen M.J., Snyder-Mackler N., Lu A. Composition and stability of the vervet monkey milk microbiome. Am. J. Primatol. 2019;81:e22982. doi: 10.1002/ajp.22982. [DOI] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R. Samurai Media Limited; 2015. An Introduction to R. [Google Scholar]
- Reese A.T., Phillips S.R., Owens L.A., Venable E.M., Langergraber K.E., Machanda Z.P., Mitani J.C., Muller M.N., Watts D.P., Wrangham R.W., et al. Age patterning in wild chimpanzee gut microbiota diversity reveals differences from humans in early life. Curr. Biol. 2020;31:613–620.e3. doi: 10.1016/j.cub.2020.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renelies-Hamilton J., Germer K., Sillam-Dussès D., Bodawatta K.H., Poulsen M. Disentangling the relative roles of vertical transmission, subsequent colonizations, and diet on cockroach microbiome assembly. mSphere. 2021;6:e01023-20. doi: 10.1128/mSphere.01023-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyman M., van Houten M.A., Arp K., Sanders E.A.M., Bogaert D. Rectal swabs are a reliable proxy for faecal samples in infant gut microbiota research based on 16S-rRNA sequencing. Sci. Rep. 2019;9:16072. doi: 10.1038/s41598-019-52549-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogier E.W., Frantz A.L., Bruno M.E.C., Wedlund L., Cohen D.A., Stromberg A.J., Kaetzel C.S. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen M.J., Davison M., Bhaya D., Fisher D.S. Microbial diversity. Fine-scale diversity and extensive recombination in a quasisexual bacterial population occupying a broad niche. Science. 2015;348:1019–1023. doi: 10.1126/science.aaa4456. [DOI] [PubMed] [Google Scholar]
- Rovai M., Such X., Piedrafita J., Caja G., Pujol M.R. Vol. 95. Publication-European Association for Animal Production; 1999. pp. 107–112. (Evolution of Mammary Morphology Traits during Lactation and its Relationship with Milk Yield of Manchega and Lacaune Dairy Sheep). [Google Scholar]
- Roy B., Mehla R.K., Sirohi S.K. Influence of milk yield, parity, stage of lactation and body weight on urea and protein concentration in milk of murrah buffaloes. Asian-australas. J. Anim. Sci. 2003;16:1285–1290. [Google Scholar]
- Ruiz-López M.J., Espeso G., Evenson D.P., Roldan E.R.S., Gomendio M. Paternal levels of DNA damage in spermatozoa and maternal parity influence offspring mortality in an endangered ungulate. Proc. Biol. Sci. 2010;277:2541–2546. doi: 10.1098/rspb.2010.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevi A., Taibi L., Albenzio M., Muscio A., Annicchiarico G. Effect of parity on milk yield, composition, somatic cell count, renneting parameters and bacteria counts of Comisana ewes. Small Rumin. Res. 2000;37:99–107. doi: 10.1016/s0921-4488(99)00133-9. [DOI] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. [Google Scholar]
- Shipley B. The AIC model selection method applied to path analytic models compared using a d-separation test. Ecology. 2013;94:560–564. doi: 10.1890/12-0976.1. [DOI] [PubMed] [Google Scholar]
- Smuts B., Nicolson N. Reproduction in wild female olive baboons. Am. J. Primatol. 1989;19:229–246. doi: 10.1002/ajp.1350190405. [DOI] [PubMed] [Google Scholar]
- Spellerberg I.F., Fedor P.J. A tribute to Claude Shannon (1916--2001) and a plea for more rigorous use of species richness, species diversity and the “Shannon--Wiener”Index. Glob. Ecol. Biogeogr. 2003;12:177–179. [Google Scholar]
- Tanaka I. Parity-related differences in suckling behavior and nipple preference among free-ranging Japanese macaques. Am. J. Primatol. 1997;42:331–339. doi: 10.1002/(SICI)1098-2345(1997)42:4<331::AID-AJP8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Thomson P., Medina D.A., Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol. 2018;75:37–46. doi: 10.1016/j.fm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- Tikhonov M., Leach R.W., Wingreen N.S. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015;9:68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters L., Bijnens L., Dhondt A.A. Body mass at weaning and juvenile recruitment in the red squirrel. J. Anim. Ecol. 1993;62:280–286. [Google Scholar]
- Wickham H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011;3:180–185. [Google Scholar]
- Xiccato G., Trocino A., Sartori A., Queaque P.I. Effect of parity order and litter weaning age on the performance and body energy balance of rabbit does. Livestock Prod. Sci. 2004;85:239–251. [Google Scholar]
- Yang B., Wang Y., Qian P.-Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics. 2016;17:135. doi: 10.1186/s12859-016-0992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yi N. NBZIMM: negative binomial and zero-inflated mixed models, with application to microbiome/metagenomics data analysis. BMC Bioinformatics. 2020;21:488. doi: 10.1186/s12859-020-03803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic A.M., Lewis Z.T., German J.B., Mills D.A. Establishment of a milk-oriented microbiota (MOM) in early life: how babies meet their MOMs. Funct. Food Rev. 2013;5:3–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Negative loading scores correspond to younger infant samples, positive loading scores correspond to older infant samples.
Data Availability Statement
-
•
16S rRNA gene sequences are available under BioProject ID PRJNA728247 at the NCBI Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra/PRJNA728247) and asssociated metadata for this project are available at figshare: https://figshare.com/s/79f3cf6cf410f5a87d31.
-
•
Code for this project is available at figshare: https://figshare.com/s/79f3cf6cf410f5a87d31.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.