Abstract
Aptamers have excellent specificity and affinity in targeting cell surface receptors, showing great potential in targeted delivery of drugs, siRNA, mRNA, and various nanomaterials with therapeutic function. A better insight of the receptor-mediated internalization process of aptameric conjugates could facilitate the design of new targeted drugs. In this paper, human transferrin receptor-targeted DNA aptamer (termed HG1-9)-fluorophore conjugates were synthesized to visualize the internalization, intracellular transport, and nano-environmental pH of aptameric conjugates. Unlike transferrin that showed high recycling rate and short duration time in cells, the synthetic aptameric conjugates continuously accumulated within cells at a relatively slower rate, besides recycling back to cell surface. After long incubation (≥2 h), only very small amounts of HG1-9 conjugates (approximately 5%) entered late endosomes or lysosomes, and more than 90% of internalized HG1-9 was retained in cellular vesicles (pH 6.0–6.8), escaping from degradation. And among the internalized HG1-9 conjugates, approximately 20% was dissociated from transferrin receptor. The lower recycling ratios of HG1-9 conjugates and their dissociation from receptors promote the accurate and efficient release of their loaded drugs. These results suggest that aptamer HG1-9 could be provided as a versatile tool for specific and effective delivery of diverse therapeutic payloads.
Keywords: aptamer, aptamer-drug conjugate, receptor-mediated internalization, dynamic imaging, transferrin receptor, endosomes, pH-sensitive ratiometric fluorophore, aptameric ligand
Graphical abstract
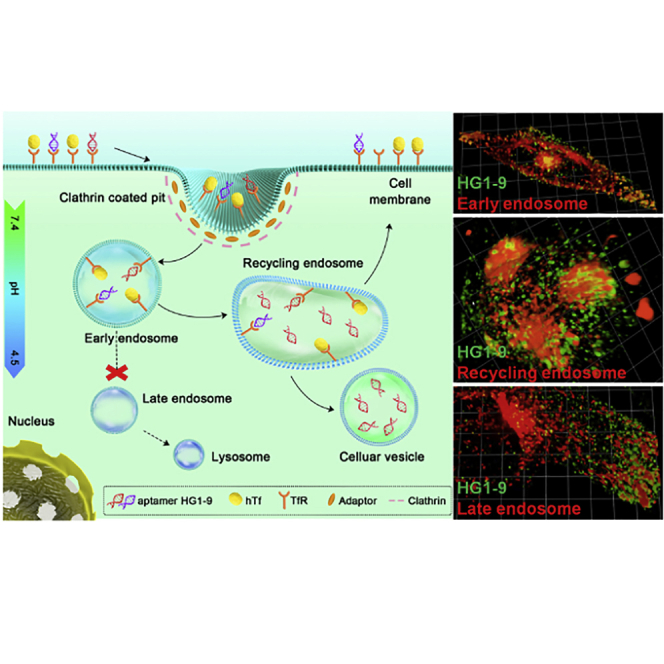
Fluorophore conjugates of human transferrin receptor-targeted aptamer (HG1-9) continuously enter cells at a slower rate than transferrin, stopping briefly in early endosomes. Then some of them enter recycling endosomes, and some dissociate from transferrin receptor and accumulate in cellular vesicles (pH 6.0–6.8). Few aptamer conjugates enter late endosomes or lysosomes
Introduction
Aptamers, also termed “chemical antibodies,” are single-stranded DNA or RNA oligonucleotides, which can specifically bind target molecules in the way similar to antibodies.1 Compared with antibodies, aptamers have more merits, such as high chemical stability, low immunogenicity, chemical synthesis, and ease in modification and linkage to various drugs without batch-to-batch variation.1,2 These advantages endow aptamers with great talent as targeting carriers for precise delivery of therapeutic agents.3 More recently, aptamer-drug conjugates (ApDCs) have been attracting much attention.4,5 Sullenger et al. reported an RNA aptamer that could specifically internalize into prostate cancer cells, and the aptamer-monomethyl auristatin (highly toxic drugs) conjugates could efficiently kills cancer cells without disturbing normal cells.6 Besides drugs, aptamers have also been investigated for targeted delivery of microRNAs, small interfering RNAs, and antisense oligonucleotides.7,8 However, compared with antibody-drug conjugates, which are already being used in clinics,9 there is still a long journey for ApDCs serving as clinical medicines. One of the roadblocks is that the internalization pathways of these aptameric conjugates are not quite clear, greatly constraining the design and application of ApDCs.
Oligonucleotides are often thought to enter cells with very low efficiency, due to the abundance of negative charge and relatively large molecular size.10 As an exception, aptamers that bind to cell surface receptors could be effectively transferred into cells, e.g., AS1411 (aptamer against nucleolin)11 and sgc8c (aptamer targeting protein tyrosine kinase 7, PTK7)12 were reported to be taken up within cells after binding their target proteins. Nevertheless, the fates of these aptamers, with degradation in lysosomes or not, are not clear yet.
Ligands of receptors on cell surfaces usually take receptor-mediated endocytic pathways. That is, ligand-receptor complexes on cell surfaces recruit a series of proteins to form clathrin-coated pits on the cell membrane, and they are transferred from the cell surface to early endosomes. Subsequently, different complexes are sorted into different intracellular vesicles for different destinations, such as recycling in Golgi apparatus/recycling endosomes or degradation in lysosomes.13 It is well-known that different receptors lead to different fates of ligands in cells; moreover, to one receptor, there are some different ligands that may initiate different endocytic pathways. For example, transferrin receptor (TfR, or CD71) can recognize multiple exogenous and endogenous ligands, such as transferrin (Tf), ferritin, hereditary hemochromatosis factor (HFE), and malaria parasite.14, 15, 16 For these ligands, their endocytic pathways are quite different, Tf-TfR complex is recycled back to cell surface, while HFE-TfR complex is destined for degradation in lysosomes; whereas, the endocytic pathways of other ligands have not been clarified yet.17,18 TfR has been widely used for targeted delivery of therapeutic agents against diseases, owing to the ubiquitous expression on cells and highly effective endocytosis.19, 20, 21 It is known that hTf can be rapidly internalized in early endosome and recycled back to the cell surface, where the high internalization and recycling rate of hTf leads to very short duration time in cells, which limits the release efficiency of hTf-loaded drugs. Thus, several ApDCs targeting TfR have been reported, such as a 2′-F modified RNA aptamer (aptamer Waz) for targeting delivery of siRNA-loaded liposomes22 and co-delivery of a chemotherapeutic drug (doxorubicin) and an inhibitor of a cell-survival factor23, and a DNA aptamer for targeting therapy of pancreatic cancer.24 However, comprehensive endocytic pathways of these ApDCs have seldomly been reported yet. Aptamer Waz has been creatively integrated with Broccoli aptamer to study targeted delivery of large RNA payload. This aptamer-RNA conjugate can maintain its functional form within cells.25 As unnatural ligands of receptors, aptameric conjugates may encounter altered fates, comparing to the endogenous ligands. Dynamic visualization of the internalization of these synthetic ligands will gain new insight into endocytosis of ApDCs and provide useful information for drug design.26, 27, 28
Herein, we report the detailed internalization and intracellular fate of a human TfR-targeted DNA aptamer, HG1-9. The aptamer-fluorophore conjugates are used to mimic ApDCs for investigation of the corresponding endocytosis behaviors and their fate within cells by dynamic imaging. In addition, the nano-environmental pH change of aptamer-containing vesicles along the endocytosis is also measured quantitatively by the conjugation of aptamer HG1-9 and a pH-sensitive ratiometric fluorophore-YNNA. Investigation of the internalization of these aptameric conjugates will provide important information for rational design and construction of aptamer-based drug delivery systems.
Results
Initial endocytosis of aptameric conjugates mediated by TfR
HG1-9, a new DNA aptamer (35 nt, 10.9 kDa), selected by cell-SELEX in our group was chosen for this investigation. HG1-9 was verified to bind human TfR with high specificity and affinity (Kd < 20 nM) and compete with transferrin to bind the same sites of human TfR.29,30 To monitor the internalization of HG1-9, enhanced green fluorescence protein (EGFP)-fused TfR (TfR-EGFP) was transfected into HeLa cells, and the transfected cells were incubated with Alexa Fluor 647 fluorophore (AF647) conjugated HG1-9 at 4°C and 37°C in binding buffer (PBS with 4.5 g/L glucose, 5 mM MgCl2, 1 mg/mL BSA, 0.1 mg/mL Herring sperm DNA), respectively. When incubated at 4°C, HG1-9 was observed to be located only on the cell membrane and overlaid well with TfR-EGFP; when incubated at 37°C, HG1-9 was located and well overlaid with TfR-EGFP both on cell membrane and in cytoplasm (Figure 1A). And the amount of HG1-9 in cytoplasm increased with the extension of incubation time (Figure S1), suggesting that HG1-9 conjugates could enter live cells through an active transport process mediated by TfR.
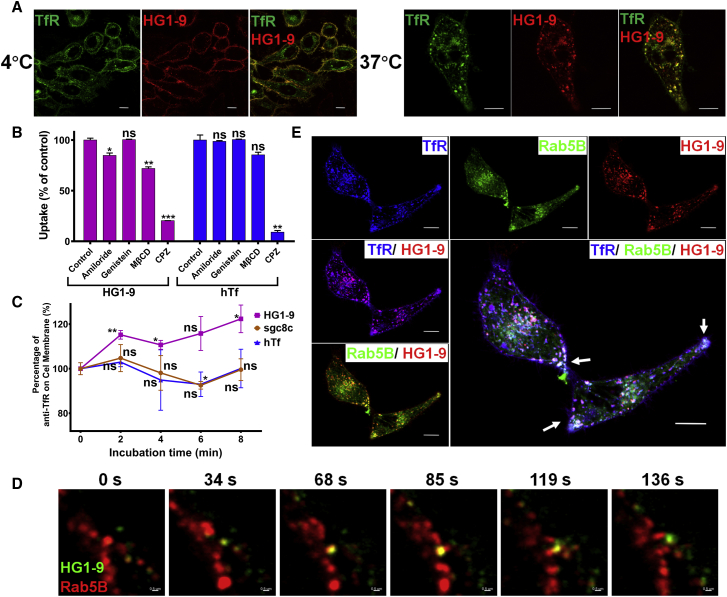
Figure 1.
Initial endocytosis of aptamer HG1-9 mediated by TfR
(A) Representative confocal images of the binding and distribution of aptamer HG1-9 (AF647 conjugated, 50 nM) to TfR-EGFP transfected HeLa cells at 4°C and 37°C for 30 min; scale, 10 μm. (B) The percentage of the uptake of aptamer HG1-9 and hTf by HeLa cells pre-treated with endocytosis inhibitors (50 μM amiloride, genistein, and chlorpromazine [CPZ] and 5 mM methyl-β-cyclodextrin [MβCD]) for 30 min at 37°C; the uptake of HG1-9 by untreated cells was 100%. (C) Flow cytometry analysis of the fluorescence of anti-TfR antibody on the surface of Jurkat cells (representing the content of TfR on cell surface) after incubation with unlabeled hTf (200 nM), HG1-9 (200 nM), and sgc8c (200 nM) at 37°C for a series of incubation times (0, 2, 4, 6, 8 min). The median fluorescence intensity of anti-TfR antibody on cell surface before incubation (0 min) was 100%, and the experiment was conducted three times independently, and each time point has three replicates. (D) Time-resolved spinning disk confocal images of HG1-9 (AF488 conjugated, 50 nM) in Rab5B-mCherry transfected HeLa cells for relative time from 0 s to 136 s; scale, 0.5 μm. (E) Representative confocal images of the binding and distribution of aptamer HG1-9 (AF647 conjugated, 50 nM) to TfR-EGFP and Rab5B-mCherry co-transfected HeLa cells after incubation for 60 min at 37°C; scale, 10 μm. The white arrows indicate the early endosomes containing HG1-9-TfR complex and Rab5B-mCherry near the cell periphery. t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005; ns, not significant.
It is well known that the uptake of hTf-TfR complex is clathrin dependent. To explore the endocytosis pathway of HG1-9 conjugates, endocytosis inhibitors (amiloride inhibits macropinocytosis; genistein interferes with caveolae-mediated endocytosis; methyl-β-cyclodextrin [MβCD] interferes with the lipid raft; and chlorpromazine [CPZ] inhibits the functions of clathrin) were used. After being pre-treated with inhibitors for 30 min respectively, HeLa cells were further incubated with HG1-9 conjugates for 1 h. The uptake of HG1-9 conjugates was not or slightly affected by amiloride, genistein, or MβCD, but significantly decreased (by 80%) in CPZ-treated cells. Similar results were also observed in the cellular uptake of hTf (Figures 1B and S2). These results suggest that the initial endocytosis of HG1-9 conjugates is clathrin dependent, and HG1-9 conjugate-TfR complexes could be internalized into cells through forming clathrin-coated pits on the cell membrane.
It has been reported that the ability of ligand-receptor complexes to stabilize the clathrin-coated pits on the cell surface strongly affects their internalization rates,31 so different ligand-TfR complexes may have different internalization rates. What is more, the evaluation of the initial internalization rate of the HG1-9 conjugates will help to estimate its potential as an effective drug carrier. The change of TfR amount on cell surface after incubation with HG1-9 or hTf could indirectly reflect the internalization rate of these ligands. Therefore, Jurkat cells were first pre-incubated with unconjugated HG1-9, hTf, or control aptamer sgc8c (aptamer against PTK7) at 37°C for different times (0, 2, 4, 6, 8 min) and quickly fixed by formaldehyde on ice to freeze the TfR on the cell surface. The amount of TfR on the cell surface was measured by flow cytometry after staining cells with anti-TfR antibody (the binding sites of antibody and HG1-9/hTf are different).30 After incubation with hTf at 37°C for different times, the change trend of TfR amount on cell surface was consistent with that of incubation with control aptamer sgc8c. However, the incubation with HG1-9 caused an increase of the TfR amount on the cell surface at the initial 8 min of incubation (Figure 1C). A similar trend was also observed in HeLa cells (Figure S3). This result suggests that HG1-9-TfR complex may retain longer on the cell surface than hTf-TfR complex, which means a relatively slower internalization rate of HG1-9-TfR complex. This relatively slower internalization rate of HG1-9-TfR complex may due to the electrostatic repulsion between the cell membrane and HG1-9 containing abundant negative charge, leading to less stable clathrin-coated pits.
After internalization, the clathrin-coated pits containing ligand-TfR complexes would form cellular vesicles, detach from the cell surface, and be transported to the first stop, early endosomes. Rab5B is a small GTPase and considered as the protein marker of early endosome.32 To indicate the early endosome, mCherry fluorescent protein-fused Rab5B (Rab5B-mCherry) expression plasmid was constructed and transfected in HeLa cells. To investigate the early endocytic pathway, dynamic behavior of internalized HG1-9 conjugates in Rab5B-mCherry expressed HeLa cell was visualized by time-resolved spinning disk confocal microscopy. The two-dimensional time-lapse imaging showed that HG1-9 co-located well with Rab5B near the cell periphery, where early endosomes are mainly located (Video S1). In single-particle tracking experiments, Rab5B-mCherry-fused early endosomes (red spots) quickly swallowed vesicles containing HG1-9 conjugates (green spots) in 68 s, and then the fission of this early endosome occurred in about 120 s (Figures 1D and Video S2). The steady colocalization of HG1-9/Rab5B and HG1-9/TfR after a long time of incubation was also analyzed in TfR-EGFP and Rab5B-mCherry co-transfected HeLa cells by the Mander's colocalization analysis, quantifying the colocalization by calculating directly as the fraction of one molecule that colocalizes with a second molecule.33 After incubation for 60 min, the Mander's colocalization coefficient (MCC) of HG1-9/Rab5B (the fraction of HG1-9 overlapping Rab5B) was calculated to be 0.636 ± 0.223 (n = 12 cells) and the MCC of HG1-9/TfR was 0.642 ± 0.125 (n = 9 cells); namely, more than half the amount of HG1-9 in cells located in Rab5B-fused endosomes, and more than half the amount of HG1-9 still associated with TfR in cells (Figures 1E, S4 and Table S2), suggesting that aptameric conjugates could continuously internalize into cells even after a long time of incubation.
Together, these observations suggest that at the early stage of endocytosis, the HG1-9 conjugates first bound TfR on the cell surface, then entered cells through a clathrin-dependent pathway with an uptake rate lower than that of hTf-TfR, and the continuously internalized HG1-9 conjugates stopped briefly in early endosomes before transferring to the next destination.
The fate of internalized aptameric conjugates within cells
In the hTf-TfR endocytic pathway, the hTf-TfR complex in early endosome is subsequently sorted into recycling endosome and recycled back to the cell surface instead of being transferred to the late endosome or lysosome for degradation. Thus, whether the fate of internalized HG1-9 conjugate is similar to that of hTf needs further investigation. The accumulation of Alexa Fluor 488 fluorophore (AF488)-conjugated HG1-9 and AF647-conjugated hTf within HeLa cells along with incubation time (5, 15, 30, 60, 90, 120 min) was first measured. The uptake ratios were acquired by dividing the fluorescence intensity of cells after incubation for a certain time by that after incubation for 5 min. The amount of hTf in cells was found to increase in 15 min, and then it stayed nearly unchanged up to 120 min, while the amount of HG1-9 in cells continuously increased (Figures 2A and S5). This result suggests that most hTf, accompanied with TfR, was recycled back to the cell surface, resulting in a quick balance of hTf in cells in 15 min, while at least a portion of HG1-9 conjugates were not shipped out of cells, resulting in accumulation in cells. Moreover, compared with another aptamer sgc8c, which has been frequently used in targeted delivery of drugs,34 aptamer HG1-9 showed much higher uptake efficiency than aptamer sgc8c (Figures S6 and S7). Furthermore, since the 5′-end conjugated HG1-9 showed a moderate nuclease resistance over the control sequence (Figure S8), the integrity of internalized HG1-9 conjugates was measured. After incubated with HeLa cells for 4 h at 37°C, the internalized HG1-9 was extracted and analyzed by gel electrophoresis (Figure 2B). The intact sequence of HG1-9 was found, suggesting that there are HG1-9 molecules retained intact instead of being degraded within cells. This is in agreement with the phenomenon of another reported TfR aptamer Waz within cells.25 To figure out the next stop of internalized HG1-9 conjugates after early endosome, the colocalization of HG1-9-fluorophore conjugates with Rab7A-mCherry (late endosome marker), LAMP1-mCherry (lysosome marker), and DND26 (dye for lysosomes) in HeLa cells was analyzed (Figures 2C and S9–S11). Confocal images showed that the fluorescence of HG1-9 poorly overlaid with that of Rab7A-mCherry (MCC of HG1-9/Rab7A: 0.211 ± 0.090, n = 6 cells), LAMP1-mCherry (MCC of HG1-9/LAMP-1: 0.075 ± 0.032, n = 5 cells), or DND26 (MCC of HG1-9/DND26: 0.128 ± 0.024, n = 16 cells), suggesting that HG1-9 conjugates seldom entered into late endosomes or lysosomes. Furthermore, the two-dimensional time-lapse images showed that vesicles containing HG1-9 conjugates (green spots) could contact with late endosomes containing Rab7A-mCherry (red spots), but they did not attempt to be assembled; in other words, they appeared to conduct a kiss-and-run fusion (Videos S3 and S4). This set of results indicates that a large proportion of internalized HG1-9 conjugates did not enter late endosomes and lysosomes in 2 h and could escape from degradation in lysosomes.
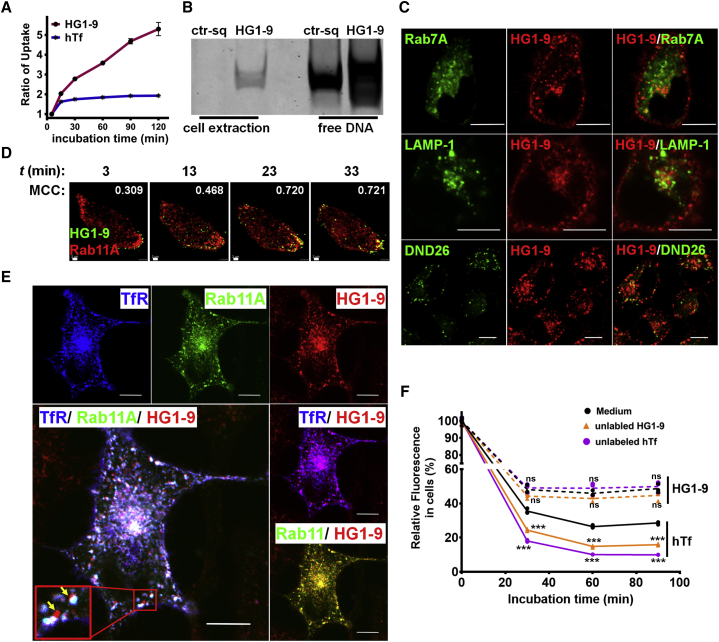
Figure 2.
The retention and recycling of HG1-9 conjugates within cells
(A) The accumulation of HG1-9 (AF488 conjugated, 50 nM) and hTf (AF647 conjugated, 50 nM) within HeLa cells along with incubation time (5, 15, 30, 60, 90, and 120 min) at 37°C; the fluorescence ratio of cells after incubation for 5 min was 1. (B) Gel analysis of the integrity of endocytic HG1-9 within cells, HG1-9 (TAMRA conjugated, 50 nM), or ctr-sq (TAMRA conjugated, 50 nM, negative control) was incubated with HeLa cells for 3.5 h; then cells were incubated in fresh medium for 0.5 h, where 20 pmol of free DNA was the reference. (C) Representative confocal images of the binding and distribution of aptamer HG1-9 within cells. Top row, HG1-9 (AF488 conjugated, 50 nM) was incubated with Rab7A-mCherry transfected cells for 60 min; middle row, HG1-9 (AF488 conjugated, 50 nM) incubated with LAMP-1-mCherry transfected cells for 120 min; bottom row, HG1-9 (TAMRA conjugated, 50 nM) and DND26 co-incubated with cells for 120 min; scale, 10 μm. (D) Time-resolved spinning disk confocal images of HG1-9 (AF488 conjugated, 50 nM) in Rab11A-mCherry transfected HeLa cells after incubation at 37°C for 3 to 33 min, and the corresponding MCC, the fractions of HG1-9 overlapping Rab11A are shown in each image; scale, 2 μm. (E) Representative confocal images of the distribution of HG1-9 (AF647 conjugated, 50 nM) in TfR-EGFP and Rab11A-mCherry co-transfected HeLa cells after incubation for 120 min at 37°C; scale, 10 μm. (F) The amount change of dye conjugated hTf and HG1-9 within cells after being recycled and competing by unlabeled ligands. HeLa cells were pre-incubated with AF647 conjugated-hTf (50 nM) and AF488 conjugated-HG1-9 (50 nM) for 15 min, respectively; then the cells were incubated with unlabeled HG1-9 (500 nM) or unlabeled hTf (500 nM) for different times (0, 30, 60, and 90 min). The fluorescence of cells before incubation with unlabeled ligand was 100%. The experiment was conducted at least twice independently, and each sample has three replicates. t test, ∗∗∗p < 0.005; ns, not significant.
To further investigate whether the internalized HG1-9 conjugates could also be sorted into recycling endosome, AF647-conjugated HG1-9 was incubated with Rab11A-mCherry (recycling endosome marker) transfected HeLa cells at 37°C. The two-dimensional time-lapse imaging showed that the fraction of HG1-9 overlapping Rab11A was increased from 0.309 to 0.721 with the incubation time from 3 to 33 min, suggesting the accumulation of HG1-9 in Rab11A-mCherry-fused recycling endosomes (Figure 2D and Video S5). Confocal imaging and the Mander's colocalization analysis showed that the fraction of HG1-9 overlapping Rab11A was 0.828 ± 0.023 (n = 10 cells) and the fraction of HG1-9 overlapping TfR was 0.812 ± 0.057 (n = 10 cells) in the Rab11A-mCherry and TfR-EGFP co-transfected HeLa cells after 120 min of incubation (Figures 2E, S12 and Table S2), suggesting most HG1-9 still associated with TfR and was enriched in recycling endosomes. Moreover, as shown in Figure 2E, some red dots (vesicles only containing HG1-9 conjugates) were observed (indicated by yellow arrows) without overlapping with TfR and Rab11A, suggesting that some of the HG1-9 conjugates might dissociate from TfR after entering cells and be restricted in cellular vesicles without TfR or Rab11A.
To study the possibility of HG1-9 conjugates recycled back to the cell surface, a competition experiment between fluorophore-conjugated HG1-9 and unlabeled HG1-9 was further conducted. If the fluorophore-conjugated HG1-9 in cells could be recycled back to the cell surface, the addition of a high concentration of unlabeled HG1-9 in bulk solution could gradually replace the recycled HG1-9 conjugates on the cell surface and inhibit its reinternalization, leading to the decrease of fluorescence in cells. To prove this, HeLa cells were incubated with fluorophore-conjugated HG1-9 or hTf at 37°C for 15 min; then, after incubated in fresh medium for another 15 min, the cells were incubated with the two unlabeled ligands (10 folds of conjugated ligands) or medium only for different times (30, 60, and 90 min). As shown in Figure 2F, all the fluorescence of fluorophore-conjugated HG1-9 or hTf in cells decreased in 30 min after incubation with unlabeled HG1-9, unlabeled hTf, and medium only, which may due to that internalized TfR ligands (HG1-9 and hTf) were transported out of cells through recycling endosomes or other pathways, e.g., extracellular vesicle secretion since Tf-TfR complexes are often found in exosomes.35 Nevertheless, there are some differences between the two ligands. For hTf, the addition of both unlabeled ligands (HG1-9 or hTf) could accelerate the fluorescence decrease of hTf within cells comparing to that in medium only, indicating that conjugated hTf could be continuously recycled back to the cell surface and replaced by unlabeled HG1-9 or hTf, which also verified the reliability of this experiment. Interestingly, for HG1-9, the addition of both unlabeled ligands (HG1-9 or hTf) did not cause significant difference of the fluorescence decrease curve of fluorophore-conjugated HG1-9 compared with that in medium only. At the first 30 min, the fluorescence decrease of HG1-9 conjugates in cells was significantly less than that of hTf conjugates, suggesting that some of the internalized aptameric conjugates are more likely to stay in cells instead of recycling back to the cell surface.
Together, these data suggest that HG1-9 conjugates could transfer into cells through the pathway mediated by TfR, and the fate of internalized aptameric conjugate is different from hTf conjugate. Some HG1-9 conjugates could be sorted in recycling endosomes in the form of HG1-9-TfR complex and recycled back to the cell surface just like hTf, but some internalized HG1-9 conjugates are prone to be retained in cells, which can explain the result in Figure 2A that HG1-9 conjugates could continuously accumulate within cells.
Ratiometric imaging of nano-environmental pH within cellular vesicles by YNNA-HG1-9
During the receptor-mediated endocytosis, the related vesicles gradually become acidic and mature, e.g., early endosomes (pH 6.0–6.5), late endosomes (pH 5–5.5), and lysosome (pH 4.5-5).36,37 The acidic pH gradient of these vesicles is one of the intrinsic properties of endocytosis.38 Thus, pH value can be another hallmark of endosomes.39 To determine the pH change along the internalization of this aptameric conjugate would provide further insights into the endocytic pathway of this synthetic ligand mediated by TfR. For monitoring the nano-environmental pH change of internalized aptamer HG1-9, YNNA, a pH-sensitive ratiometric fluorophore, was synthesized and conjugated to HG1-9 by azide-alkynyl click chemistry (Figures 3A, S13, and Scheme S1). YNNA was originated from a previously reported pH-sensitive fluorophore, ANNA, by changing the carboxyl group (ANNA) to an alkynyl group (YNNA). ANNA is a ratiometric fluorescent fluorophore reported by our group, which has been used to quantitively image intracellular and cancer microenvironmental pH in vitro and in vivo.40, 41, 42, 43
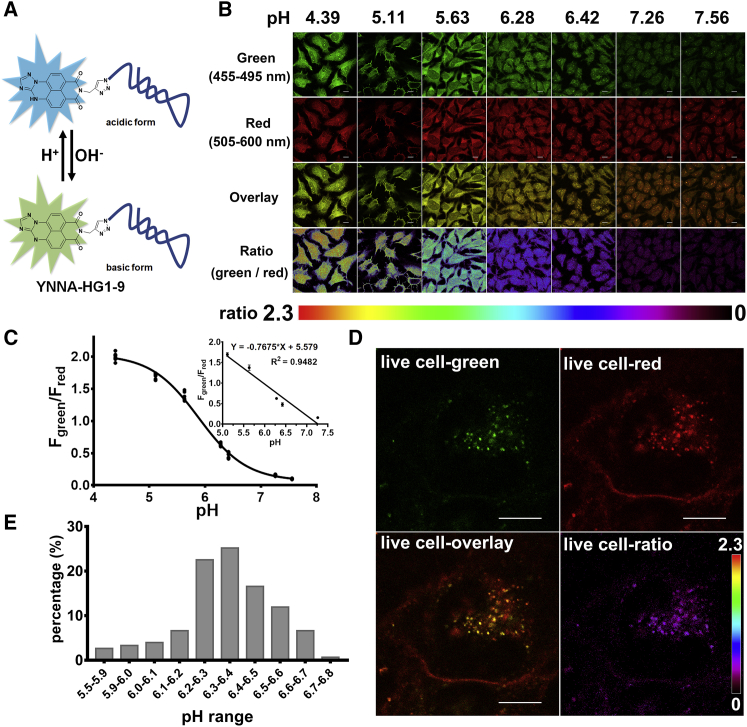
Figure 3.
The imaging and quantitative measurement of nano-environmental pH within vesicles containing YNNA-HG1-9
(A) Schematic illustration of pH response of YNNA-HG1-9. (B) Confocal images of methanol-fixed HeLa cells incubated with YNNA-HG1-9 in the HEPES buffers (125 mM KCl, 20 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, and 20 mM HEPES) with different pH values (4.39, 5.11, 5.63, 6.28, 6.42, 7.26, and 7.56), where the fluorescence was excited at 445 nm and collected in two channels (green, 455–495 nm; red 505–600 nm), and the pseudo-color ratiometric images (Fgreen/Fred) reflect the pH values. (C) The curve of fluorescence intensity ratios (Fgreen/Fred) versus pH values in fixed cells; linear range was inserted. (D) Representative confocal images of the distribution of internalized YNNA-HG1-9 within a live HeLa cell after incubation for 60 min at 37°C, and the pseudo-color ratiometric image reflected the pH of vesicles containing YNNA-HG1-9; scale, 10 μm. (E) The percentage and distribution of YNNA-HG1-9 contained vesicles with different pH (5.5–6.8); 151 regions of interest (ROIs) were randomly selected from six pseudo-color ratiometric images.
First, to investigate the effect of acidic pH on the binding ability of HG1-9 and hTf to TfR, the binding of pH-insensitive fluorophore-conjugated HG1-9 and hTf to fixed HeLa cells in binding buffers with different pH values was tested. The binding of hTf was nearly unchanged even when the pH was lowered to 5.34, while the binding of HG1-9 was slightly affected when the buffer became acidic (pH lower than 6.36); nevertheless, more than 75% of binding ability of HG1-9 remained when the pH was as low as 5.34, suggesting that some of aptameric conjugates dissociate with TfR, but the majority still bind TfR in relatively acidic conditions (Figures S14 and S15). Moreover, the binding ability of YNNA-HG1-9 to cells was verified by flow cytometry (Figure S16). The fluorescence spectra of YNNA and YNNA-HG1-9 in HEPES buffers with different pH values showed that YNNA-HG1-9 exhibited similar fluorescence spectra and pH response to those of YNNA in physiological pH range, and a linear pH response was found in the range of pH 4.42–7.32 (Figure S17). To image and quantitatively measure the nano-environmental pH along the internalization of YNNA-HG1-9 conjugates in live cells, the fluorescence ratio-pH curve in cells was acquired. The fixed HeLa cells were incubated with YNNA-HG1-9 in HEPES buffers with different pH values (4.39, 5.11, 5.63, 6.28, 6.42, 7.26, and 7.56). The fluorescence images were acquired in two channels: green (455–495 nm [Fgreen]) and red (505–600 nm [Fred]) under excitation of 445-nm laser. The pseudo-color ratiometric images showed that the fluorescence intensity ratios (Fgreen/Fred) varied along with the pH change (Figure 3B). The curve of fluorescence intensity ratios (Fgreen/Fred) versus pH values (ratio-pH curve) in cells was plotted. A linear relationship with pH values from pH 5.11 to pH 7.26 was obtained (Figure 3C).
The live cell imaging by YNNA-HG1-9 is shown in Figure 3D. After incubation with live cells for 60 min, YNNA-HG1-9 mainly distributed in two regions. One is near the cell membrane, which exhibited fluorescence only in the red channel; and another is in vesicles within cells, which exhibited fluorescence in both green and red channels, suggesting the relatively high pH around the cell membrane and relatively low pH in cellular vesicles. The pseudo-color ratiometric image showed that vesicles containing YNNA-HG1-9 exhibited a purple pseudo color, further indicating the relatively low pH of these cellular vesicles. To accurately analyze the nano-environmental pH of internalized YNNA-HG1-9, 151 regions of interest (ROIs) were randomly selected from six pseudo-color ratiometric images, and their pH values were calculated according to the fluorescence ratios (green/red) and the ratio-pH curve (Figures 3E and S18). The histogram of pH distribution of the ROIs showed that after incubation for 60 min, the pH values of the vesicles containing YNNA-HG1-9 were in the range of 5.5–6.8, and mainly (74.8%) distributed in the range of 6.0–6.5, which are consistent with the pH ranges of early endosomes (6.0–6.5) and endocytic recycling compartments (∼6.5).44 Moreover, there were approximately 19% of vesicles containing YNNA-HG1-9, which ranged in pH 6.5–6.8, which is slightly higher than the pH values of endosomes. These results were consistent with the conclusion earlier that HG1-9 conjugates first entered into early endosomes, then mainly were retained in recycling endosomes, and some could dissociate from TfR and be retained in cellular vesicles, while few HG1-9 conjugates entered into the lower pH vesicles, such as late endosomes and lysosomes. Given these observations, it has been evident that most of the internalized aptameric conjugates can escape from being degraded in lysosomes, which could also explain the finding of intact aptameric conjugates in cells.
Discussion
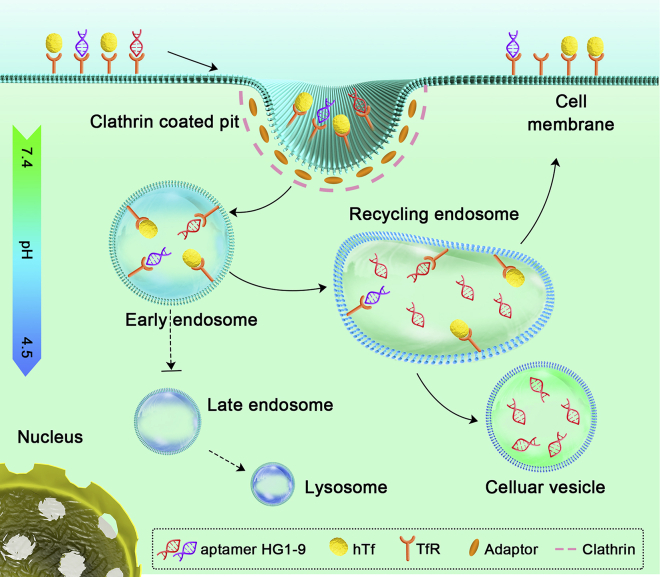
This work has comprehensively clarified the endocytic pathway of TfR-targeted aptamer HG1-9 conjugates (Figure 4): (1) The internalization of HG1-9 is first mediated by TfR through a clathrin-dependent pathway with a lower rate than that of hTf. (2) The internalized HG1-9 conjugates are then delivered into early endosomes and stay for a very short time. (3) A portion of internalized HG1-9 conjugates are transferred in recycling endosomes and recycled back to the cell surface, and some are dissociated from TfR and stay in the cellular vesicles without Rab11A or TfR. (4) After a long incubation (≥2 h), more than 90% of internalized HG1-9 was retained in cellular vesicles (pH 6.0–6.8), and only approximately 5% of HG1-9 conjugates entered acidic late endosomes or lysosomes, suggesting that HG1-9 conjugates could escape from degradation in lysosomes. Compared with hTf, HG1-9 possesses a lower internalization rate and a lower recycling ratio, and HG1-9 could be dissociated from TfR, which would promote the accuracy and efficiency of the loaded drugs release. Recently, messenger RNA (mRNA)-based vaccines have become a promising platform for infectious disease control and cancer immunotherapy.45 However, safe and efficient delivery of mRNA is now a major issue. HG1-9 may have the potential for efficient delivery and precise release of mRNA vaccines.
Figure 4.
The schematic illustration of endocytosis pathways of aptamer HG1-9 and human transferrin (hTf) within a cell
Materials and methods
Reagents and materials
The DNA sequences used in this work are listed in Table S1. Unless otherwise stated, the DNA sequences used were synthesized and purified by Sangon Biotech (Shanghai, China). Alexa Fluor 647 and Alexa Fluor 488 labeled DNA sequences were synthesized and purified by Thermo Fisher Scientific (Carlsbad, CA, USA). Unless otherwise stated, all the functional group were labeled at 5′-end of DNA sequences, and a thymic nucleotide was added between the fluorophore and the DNA sequence to avoid the fluorescent quenching by guanine. The stock solutions were prepared by dissolving DNA freeze-dried powders in phosphate buffer saline (PBS), and the concentration of solutions was measured by absorbance of 260 nm. For formation of stable structures, the dissolved DNA solutions were heated at 95°C for 5 min, cooled on ice for 10 min, and annealed to fold the stable structure at room temperature for at least 30 min. The annealed DNA stock solutions were stored at −20°C.
Anti-transferrin receptor antibody (3B8 2A1, sc-32272) and phycoerythrin (PE) labeled secondary antibody (m-IgGκ BP-PE, sc-516141) were purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). Human transferrin (unlabeled hTf) (T4132) was purchased from Sigma-Aldrich (St. Louis, MO). Alexa Fluor 647 and fluorescein (FITC) labeled human transferrin were purchased from Jackson ImmunoResearch (West Grove, PA, USA). Lipofectamin RNAiMAX Reagent and Lipofectamine 3000 were purchased from Invitrogen (Carlsbad, CA, USA). Amiloride, genistein, chlorpromazine, and methyl-β-cyclodextrin were purchased from J&K Scientific Ltd. (Beijing, China). Green Lysotracker probe (DND-26) was purchased from Beyotime Biotechnology (Jiangsu, China). Propargylamine was purchased from J&K Scientific (Beijing, China). 4-Bromo-1,8-naphthalic anhydride was purchased from Liangang Dye Chemical (Liaoning, China). 3-Amino-1,2,4-triazole was purchased from Alfa Aesar (Ward Hill, MA). TBTA (tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine) was purchased from Sigma-Aldrich. All the reagents were obtained from commercial suppliers and used without further purification.
Cell lines and cell culture
Jurkat E6-1 (Jurkat, human acute T lymphoblastic leukemia, identifier: 19375.09.3101HUMTCHU123) and HeLa (human cervical cancer, identifier: 19375.09.3101HUMTCHu187) cell lines were purchased from Typical Culture Preservation Commission Cell Bank, Chinese Academy of Sciences (Shanghai, China). All the cell lines were cultured in basic medium with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Corning) and routinely cultured at 37°C, in a humidified atmosphere with 5% CO2.
Flow cytometry analysis
Briefly, the flow cytometry analysis was conducted to determine the fluorescence intensity of aptamers or antibodies on/within cells with some certain treatment. Unless otherwise stated, HeLa cells that need flow cytometry analysis were prepared as monodispersed cell suspension after dissociated with PBS containing 0.2% EDTA. After being washed, the cell suspension was incubated with fluorophore-labeled aptamers or antibodies in binding buffer (PBS with 4.5 g/L glucose, 5 mM MgCl2, 1 mg/mL BSA, 0.1 mg/mL herring sperm DNA) at 4°C for 60 min. After incubation, the cells were washed by washing buffer (PBS with 4.5 g/L glucose and 5mM MgCl2) to remove nonspecific binding. For investigation of binding ability, cells were incubated with different concentrations of annealed fluorophore-labeled DNA sequences or transferrin protein for 30 min on ice in the dark. For competition binding, cells were simultaneously incubated with fluorophore-labeled DNA sequences and unlabeled transferrin for 30 min on ice in the dark. For antibody staining, cells were pre-incubated with 1% BSA and successively incubated with the monoclonal antibody (1:50) and PE labeled secondary antibody (1:100) for 30 min on ice in the dark. After incubation, the cells were carefully washed, resuspended, and filtered by a 400-mesh sieve. And the fluorescence intensity of cell samples was analyzed by a FACScalibur flow cytometer (Becton Dickinson, USA).
Live cell imaging by confocal laser scanning microscopy
For steady imaging of live cell, the cells were seeded at desired concentrations in the glass bottomed confocal dishes (NEST Biotechnology). After being cultured at least 24 h, the cells were washed and stained with dye-labeled aptamers, transferrin protein, or antibodies on ice or at 37°C. After incubation, cells were washed and imaged by confocal laser scanning microscopy using OLYMPUS FV3000-IX81 confocal microscope (Olympus Corporation, Japan) with 100× objective oil lens; and confocal images were acquired as 800× 800 pixels images and processed by Olympus FV31S-SW viewer software. For FITC, Alexa Fluor 488 (AF488) and EGFP, a 488-nm solid laser was used, and the emission was collected at 500–600 nm; for rhodamine (TAMRA), PE, and mCherry, a 561-nm solid laser was used, and the emission was collected at 570–620 nm, and Alexa Fluor 647 (AF647) was excited with a 640-nm solid laser and the emission was collected at 650–730 nm.
Plasmid transfection
To transfect the fluorescent protein fused proteins in HeLa cells, plasmid transfection was conducted. First, the total RNA of HeLa cells was extracted by an RNAeasy Animal Long RNA Isolation Kit with a Spin Column (Beyotime, China), and then the total RNA was used to reverse transcript the corresponding cDNA with ProtoScript First Strand cDNA synthesis kit (New England Biolabs [NEB], MA, USA). The Rab5B, Rab7A, LAMP1, Rab11A, and TfR genes cloned from the above cDNA using Q5 High-Fidelity 2X Master Mix (NEB) with forward and reverse primers (for Rab5B: forward: GCCGAATTCTATGACTAGCAGAAGCAC, reverse: GCCGGATCCTCAGTTGCTACAACACT; for Rab7A, forward: GCCGAATTCTATGACCTCTAGGAAGAAAGTGTTGC, reverse: GCCGGATCCTCAGCAACTGCAGCTTTCTG; for LAMP1, forward: GCCGAATTCTATGGCGGCCCCCG, reverse: GCCGGATCCCTAGATAGTCTGGTAGCCTGCGT; for Rab11A, forward: GCCGAATTCATGGGCACCCGCGAC, reverse: GCCGGATCCTTAGATGTTCTGACAGCACTGCACC; for TfR, forward: cgcGAATTCGCCACCATGATGGATCAAGCTAGATCAGCA, REVERSE: CGCGGATCCGCGCCGGTGGAGCCGGTGGAGCCGGTGGAGCCAAACTCATTGTCAATGTCCCAAA). Then, the Rab5B, Rab7A, LAMP1, and Rab11A genes were inserted into the pmCherry plasmids with restriction endonucleases (BamH1 and EcoR1) for mammalian expression plasmid pmCherry-Rab5B, pmCherry-Rab7A, pmCherry-LAMP1, and pmCherry-Rab11A construction. The TfR genes were inserted into pEGFP plasmid with restriction endonucleases (BamH1 and EcoR1, a gift from Professor Ming Wang) for mammalian expression plasmid pEGFP-TfR construction. For plasmids transfection, 2 × 105 HeLa cells were seeded in glass bottomed confocal dishes; after incubation for 24 h, the cells were transfected with plasmids using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s manual. After transfection for 6 h, the cells were grown for another 48 h in new, fresh media, and the cells were used for live cell imaging.
Comparison of uptake rate of aptamer HG1-9 and transferrin
To compare the speed of internalization of aptamer HG1-9 and transferrin mediated by TfR in HeLa cells, the amount of TfR on the cell surface at a series of internalization times was measured. 3 × 105 Jurkat or HeLa cells were respectively incubated with sgc8c (200 nM), transferrin (200 nM), or unlabeled HG1-9 (200 nM) at 37°C for a series of incubation times (0, 2, 4, 6, 8 min). After incubation, cells were immediately fixed by 4% formaldehyde at 4°C for 10 min, and fixation was terminated by glycine. After being carefully washed, the cells were blocked by 1% BSA, and then cells were subsequently incubated with anti-TfR antibody (1:50) for 30 min at 4°C and PE labeled secondary antibody (1:100) for 30 min at 4°C. After being labeled with antibody, cells were analyzed by flow cytometry.
Identification of internalization mechanism of aptamer HG1-9 by endocytosis inhibitors
To investigate the initial internalization mechanism, chemical inhibitors that can disturb endocytosis pathways were used to inhibit the cell uptake of HG1-9. HeLa cells were seeded in a 24-well plate with a concentration of 2 × 105 cells in each well for 24 h. After being pre-treated with 50 μM inhibitors (amiloride, genistein, and CPZ) and 5 mM MβCD for 30 min, the cells were respectively incubated with 200 nM AF647-HG1-9 and AF647-transferrin in the binding buffer containing inhibitors for 60 min at 37°C, and then the cells were monodispersed under the digestion by trypsin. After being washed, the cells were analyzed by flow cytometry. Each cell sample contained three repeats.
Dynamic cell imaging by spinning disk confocal microscopy
For spatiotemporal dynamic imaging of internalization of aptamer HG1-9 in live cells, two-dimensional time-lapse images were acquired by spinning disk confocal microscopy. HeLa cells that expressed mCherry fused protein Rab5B, Rab7A, or Rab11A were seeded in the glass bottomed confocal dishes. After being cultured for at least 24 h, the cells were washed and incubated in a CO2 chamber at 37°C, then the cells were stained with AF488-aptamers for 5 min in DMEM medium, and then the cells were imaged on a rotary disc confocal microscope system (UltraViewVox, Olympus IX83) with 100× objective oil lens. For AF488 dye, a 488-nm laser was used, and the emission was collected at 500–550 nm with an exposure time of 150 ms; for mCherry protein, a 561-nm laser was used and the emission was collected at 580–650 nm with an exposure time of 60 ms. For two-dimensional time-lapse imaging, the scan mode was set to high speed, and images of mCherry-Rab5B HeLa cells were acquired approximately every 16.8 s, images of mCherry-Rab7A HeLa cells were acquired approximately every 9.9 s, and images of mCherry-Rab11A HeLa cells were acquired approximately every 9.1 s. The videos made from collected images were processed by Imaris 9.3.1 software (Oxford Instruments, UK).
Investigation of integrity of endocytic HG1-9 by denatured-PAGE gel
To investigate the fate of aptamer HG1-9, the integrity of HG1-9 after being internalized within cells needs to be measured. HeLa cells were cultured in 100-mm dishes with a density of 1 × 106 cells/mL. After being cultured for 24 h, the cells were washed and incubated with 50 nM of TAMRA labeled HG1-9 or ctr-sq (control sequence) in binding buffer at 37°C for 3.5 h; then the cells were washed and further incubated with new, fresh medium at 37°C for 0.5 h. After incubation, the cells were carefully washed four times by PBS with 0.5% EDTA to remove the bound aptamer on the cell surface, and later the cells were dissociated. The endocytic HG1-9 was extracted according to proteinase-K-chloroform method: the cell pellets were lysed with lysis buffer (500 mM NaCl, 100 mM Tris-HCl, 50 mM EDTA, 0.5% SDS, 0.1mg/mL proteinase K, pH 8.0) at 50°C for 2 h. After lysis, the lysed cells were extracted with phenol-chloroform buffer, and supernatant containing total DNA was separated after centrifuge (4,000 rpm, 10 min). DNA was then acquired by ethanol precipitation. The endocytic HG1-9, ctr-sq, and their shortened sequences were captured by their corresponding complementary whole long sequences (c-HG1-9 and c-ctr-sq) that were labeled with biotin, which could be captured by streptavidin-coated magnetic beads. Then, the beads were washed and directly denatured by 7 M urea at 95°C for 10 min. 15% denatured-PAGE gel was used to analyze the endocytic HG1-9 and ctr-sq. And the gel was imaged by Typhoon Trio (GE Healthcare Life Sciences).
Determination of the recycling of hTf and HG1-9 within cells
HeLa cells were seeded in a 24-well plate with a concentration of 3 × 105 cells in each well. After culturing for 24 h, the cells were washed and incubated with 50 nM of AF488 labeled HG1-9 or AF647 labeled hTf in DMEM buffer (containing 10% heat-inactivated FBS and 5 mM MgCl2) at 37°C for 15 min. Then the cells were washed and further incubated with new, fresh medium at 37°C for 15 min, and the cells were subsequently incubated in medium only, or with unlabeled HG1-9 (500 nM) or hTf (500 nM) for 30, 60, and 90 min. After incubation, the cells were dissociated by trypsin and analyzed by flow cytometry.
Synthesis of YNNA and YNNA-HG1-9
YNNA (2-(prop-2-yn-1-yl)pyrido[3,4,5-gh][1,2,4]triazolo[1,5-a]perimidine-1,3(2H,6H)-dione) was synthesized in a way similar to our previously reported method.41 4-Bromo-1,8-naphthalic anhydride (300 mg, 1.08 mmoL) was added into ethanol and refluxed for 10min; then the temperature dropped down to 50°C, and propargylamine (150 μL, 2.3 mmoL) was slowly added, and the mixture was further refluxed for another 1 h. The product BNPY (6-bromo-2-(prop-2-yn-1-yl)-1H-benzo[de]isoquinoline-1,3(2H)-dione) was acquired as a grayish-white powder after being water washed and filtered. BNPY (90 mg, 0.29 mmoL), 3-amino-1,2,4-triazole (80 mg, 0.95 mmoL), and sodium hydroxide (28 mg, 0.7 mmoL) were dissolved in 2 mL of DMSO. The mixture was stirred at 135°C for 12 h under N2 atmosphere. Then hydrochloric acid was added into the mixture, and the solvent was evaporated under reduced pressure to acquire product YNNA as a reddish-brown powder. The crude product was dissolved in dichloromethane and purified by silica gel chromatography (CH2Cl2/MeOH, v/v, 40:1). Then, compound YNNA was confirmed by mass spectra (MS), MS(ESI) calculated for YNNA (C38H31N7O4): m/z 315.1, found, 314.1 (M-H)–.
To synthesize YNNA conjugated aptamer HG1-9, a click reaction was conducted. TBTA and CuSO4 and sodium ascorbate were dissolved in DMSO/water (70%/30%, v/v) and stirred for 10 min, then YNNA and azide-HG1-9 were added into the mixture and stirred at 40°C for 20 min to acquire YNNA-HG1-9. The crude product was purified by high-performance liquid chromatography (Shimadzu DGU-20A3R Prominence HPLC System, Kyoto, Japan) with C18 column: Agela, 5 μm, 100 Å, 4.6 × 250 mm; mobile phase A: 20 mM triethylamine acetate (pH 7.0), B: acetonitrile; the gradient program: 0–3 min, 5% B; 3–28 min, 5%–35% B; 28–33 min, 35%–60% B; 33–43 min 95% B, flow rate: 1 mL/min, UV detection at 260 nm and 455 nm, room temperature). YNNA-HG1-9 (93%, purity) was confirmed by MS. MS calculated for YNNA-HG1-9: m/z 11805.7, found, 11808.3 (M + H).
pH response of YNNA and YNNA-HG1-9 in buffer
YNNA was diluted in HEPES buffers (125 mM KCl, 20 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, and 20 mM HEPES) with different pH values (3.64, 4.04, 4.42, 4.85, 5.11 5.41 5.82, 6.52, 6.44, 6.77, 7.09, 7.32, 7.60, 7.99, and 9.09) to dilute a final concentration of 1 μM. YNNA-HG1-9 was diluted in HEPES buffers (125 mM KCl, 20 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, and 20 mM HEPES) with different pH values (3.92, 4.37, 4.68, 5.09, 5.46, 5.65, 6.34, 6.45, 6.64, 7.26, 8.12, 8.65, and 8.94) to dilute a final concentration of 200 nM. Then the fluorescence emission spectra were collected from 470 nm to 600 nm with excitation at 455 nm on a Hitachi F-4600 fluorescence spectrofluorometer. The curves of fluorescence intensity ratios (F480 nm/F510 nm) to corresponding pH values in buffers were plotted by Origin 8.5.
Ratiometric fluorescence imaging and measurement of nano-environmental pH along the endocytosis of HG1-9 in live cells
To realize ratiometric fluorescence imaging and quantitative measurement of nano-environmental pH along the endocytosis of HG1-9-YNNA conjugates in live cells, the fluorescence ratio-pH curve in cells needed to be acquired. HeLa cells were seeded in confocal dishes; after being cultured for 24 h, cells were fixed by cold methanol at −20°C overnight and treated with 0.05% Triton X-100; then the fixed cells were carefully washed by HEPES buffers with different pH values (4.39, 5.11, 5.63, 6.28, 6.42, 7.26, and 7.56) and incubated with YNNA-HG1-9 HEPES buffers with different pH values (4.39, 5.11, 5.63, 6.28, 6.42, 7.26, and 7.56) for 30 min. After incubation, the cells were imaged by CLSM. A 445-nm solid laser was used, and the fluorescence emission images were collected at two channels: 455–495 nm (green channel) and 505–600 nm (red channel). The pseudo-color ratiometric images (Fgreen/Fred) were processed by cellSense imaging software (Olympus, Japan), and fluorescence ratio values were calculated by dividing the fluorescence intensity of the green channel by that of the red channel after background subtraction, where at least seven ROIs in every image were chosen to acquire the average fluorescence ratio. The curve of fluorescence intensity ratios (Fgreen/Fred) to corresponding pH values in cells was plotted by Origin 8.5.
To quantitively measure the nano-environmental pH along the endocytosis of HG1-9 in live cells, the cultured HeLa cells were incubated with 500 nM YNNA-HG1-9 for 1 h at 37°C. After incubation, the live cells were carefully washed and imaged by confocal microscopy, and the data were analyzed by cellSense. The pH values in live cells were determined based on the fluorescence ratios of different ROIs and calculated according to the fluorescence ratio-pH curve. The total 151 ROIs were randomly selected from six images.
Statistical analysis
The data, needing to be quantified, were acquired from the experiments that were repeated at least two times and expressed as the means ± SE. Statistical analysis was performed by Origin 8.5.1. The statistical significance was estimated by Student's t test, and two-sided p values were calculated. For the colocalization analysis, the Mander's colocalization analysis was done by JACop plugin46 in NIH ImageJ software (http://rsbweb.nih.gov/ij).
Acknowledgments
This work was supported by National Natural Science Foundation of China (21705153, 21635008, 21874140, and 21877115). We thank Yanli Zhang from Tsinghua University for help on the use of the spinning disk confocal microscope.
Author contributions
N. Z. designed the study, conducted the experiments, analyzed the data, and wrote the manuscript. J.W. conducted the plasmid transfection. T.B. and X. L. provided experimental support. D. S. designed and coordinated the study, analyzed the data, revised the manuscript, and was responsible for the funding. All of the authors read and approved the final manuscript.
Declaration of interests
The authors declare no conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.02.006.
Supplemental information
References
- 1.Wang L., Bing T., Liu Y., Zhang N., Shen L., Liu X., Wang J., Shangguan D. Imaging of neurite network with an anti-L1CAM aptamer generated by neurite-SELEX. J. Am. Chem. Soc. 2018;140:18066–18073. doi: 10.1021/jacs.8b10783. [DOI] [PubMed] [Google Scholar]
- 2.Bing T., Shen L., Wang J., Wang L., Liu X., Zhang N., Xiao X., Shangguan D. Aptameric probe specifically binding protein heterodimer rather than monomers. Adv. Sci. 2019;6:1900143. doi: 10.1002/advs.201900143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon S., Rossi J.J. Aptamers: uptake mechanisms and intracellular applications. Adv. Drug Deliv. Rev. 2018;134:22–35. doi: 10.1016/j.addr.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K., Liu B., Yu B., Zhong W., Lu Y., Zhang J., Liao J., Liu J., Pu Y., Qiu L., et al. Advances in the development of aptamer drug conjugates for targeted drug delivery. Wiley Interdiscip. Rev. Nanomed Nanobiotechnol. 2017;9:e1438. doi: 10.1002/wnan.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xuan W.J., Peng Y.B., Deng Z.Y., Peng T.H., Kuai H.L., Li Y.Y., He J.X., Jin C., Liu Y.L., Wang R.W., et al. A basic insight into aptamer-drug conjugates (ApDCs) Biomaterials. 2018;182:216–226. doi: 10.1016/j.biomaterials.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 6.Powell Gray B., Kelly L., Ahrens D.P., Barry A.P., Kratschmer C., Levy M., Sullenger B.A. Tunable cytotoxic aptamer-drug conjugates for the treatment of prostate cancer. Proc. Natl. Acad. Sci. U S A. 2018;115:4761–4766. doi: 10.1073/pnas.1717705115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soldevilla M.M., Meraviglia-Crivelli de Caso D., Menon A.P., Pastor F. Aptamer-iRNAs as therapeutics for cancer treatment. Pharmaceuticals (Basel) 2018;11:108. doi: 10.3390/ph11040108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catuogno S., Esposito C.L., de Franciscis V. Aptamer-mediated targeted delivery of therapeutics: an update. Pharmaceuticals (Basel) 2016;9:69. doi: 10.3390/ph9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley A.M., Devine M., DeRemer D. Brentuximab vedotin: an anti-CD30 antibody-drug conjugate. Am. J. Health Syst. Pharm. 2013;70:589–597. doi: 10.2146/ajhp110608. [DOI] [PubMed] [Google Scholar]
- 10.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes-Reyes E.M., Teng Y., Bates P.J. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao Z.Y., Shangguan D.H., Cao Z.H., Fang X.H., Tan W.H. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry. 2008;14:1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 13.Kaksonen M., Roux A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 14.Chen A.C., Donovan A., Ned-Sykes R., Andrews N.C. Noncanonical role of transferrin receptor 1 is essential for intestinal homeostasis. Proc. Natl. Acad. Sci. U S A. 2015;112:11714–11719. doi: 10.1073/pnas.1511701112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giannetti A.M., Snow P.M., Zak O., Bjorkman P.J. Mechanism for multiple ligand recognition by the human transferrin receptor. PLoS Biol. 2003;1:E51. doi: 10.1371/journal.pbio.0000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies P.S., Zhang A.S., Anderson E.L., Roy C.N., Lampson M.A., McGraw T.E., Enns C.A. Evidence for the interaction of the hereditary haemochromatosis protein, HFE, with the transferrin receptor in endocytic compartments. Biochem. J. 2003;373:145–153. doi: 10.1042/BJ20030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Townsend A., Drakesmith H. Role of HFE in iron metabolism, hereditary haemochromatosis, anaemia of chronic disease, and secondary iron overload. Lancet. 2002;359:786–790. doi: 10.1016/S0140-6736(02)07885-6. [DOI] [PubMed] [Google Scholar]
- 18.Schmid S.L. A nostalgic look back 40 years after the discovery of receptor-mediated endocytosis. Mol. Biol. Cell. 2019;30:1–3. doi: 10.1091/mbc.E18-06-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniels T.R., Bernabeu E., Rodriguez J.A., Patel S., Kozman M., Chiappetta D.A., Holler E., Ljubimova J.Y., Helguera G., Penichet M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiley D.T., Webster P., Gale A., Davis M.E. Transcytosis and brain uptake of transferrin-containing nanoparticles by tuning avidity to transferrin receptor. Proc. Natl. Acad. Sci. U S A. 2013;110:8662–8667. doi: 10.1073/pnas.1307152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tortorella S., Karagiannis T.C. Transferrin receptor-mediated endocytosis: a useful target for cancer therapy. J. Membr. Biol. 2014;247:291–307. doi: 10.1007/s00232-014-9637-0. [DOI] [PubMed] [Google Scholar]
- 22.Wilner S.E., Wengerter B., Maier K., de Lourdes Borba Magalhaes M., Del Amo D.S., Pai S., Opazo F., Rizzoli S.O., Yan A., Levy M. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol. Ther. Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porciani D., Tedeschi L., Marchetti L., Citti L., Piazza V., Beltram F., Signore G. Aptamer-mediated codelivery of Doxorubicin and NF-kappaB Decoy enhances chemosensitivity of pancreatic tumor cells. Mol. Ther. Nucleic Acids. 2015;4:e235. doi: 10.1038/mtna.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X., Liu H., Han D., Peng B., Zhang H., Zhang L., Li J., Liu J., Cui C., Fang S., et al. Elucidation and structural modeling of CD71 as a molecular target for cell-specific aptamer binding. J. Am. Chem. Soc. 2019;141:10760–10769. doi: 10.1021/jacs.9b03720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porciani D., Cardwell L.N., Tawiah K.D., Alam K.K., Lange M.J., Daniels M.A., Burke D.H. Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines. Nat. Commun. 2018;9:2283. doi: 10.1038/s41467-018-04691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dean K.M., Palmer A.E. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem. Biol. 2014;10:512–523. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y., Li M., Qiu F., Zhang M., Zhang Y.H. Cell-permeable organic fluorescent probes for live-cell long-term super-resolution imaging reveal lysosome-mitochondrion interactions. Nat. Commun. 2017;8:1307. doi: 10.1038/s41467-017-01503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosendale M., Perrais D. Imaging in focus: imaging the dynamics of endocytosis. Int. J. Biochem. Cell Biol. 2017;93:41–45. doi: 10.1016/j.biocel.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Shangguan D., Zhang N., Bing T., Liu X., Shen L. 2016. A Transferrin Receptor (TfR) Targeting Aptamer Sequence and Application. China Patent. [Google Scholar]
- 30.Zhang N., Bing T., Shen L., Feng L., Liu X., Shangguan D. A DNA aptameric ligand of human transferrin receptor generated by cell-SELEX. Int. J. Mol. Sci. 2021;22:8923. doi: 10.3390/ijms22168923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M.L., Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Laifenfeld D., Patzek L.J., McPhie D.L., Chen Y., Levites Y., Cataldo A.M., Neve R.L. Rab5 mediates an amyloid precursor protein signaling pathway that leads to apoptosis. J. Neurosci. 2007;27:7141–7153. doi: 10.1523/JNEUROSCI.4599-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn K.W., Kamocka M.M., McDonald J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cansiz S., Zhang L.Q., Wu C.C., Wu Y., Teng I.T., Hou W.J., Wang Y.Y., Wan S.O., Cai R., Jin C., et al. DNA aptamer based nanodrugs: molecular engineering for efficiency. Chem. Asian J. 2015;10:2084–2094. doi: 10.1002/asia.201500434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang D., Oh S., Ahn S.M., Lee B.H., Moon M.H. Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation and nanoflow liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2008;7:3475–3480. doi: 10.1021/pr800225z. [DOI] [PubMed] [Google Scholar]
- 36.Ma X., Wang Y., Zhao T., Li Y., Su L.C., Wang Z., Huang G., Sumer B.D., Gao J. Ultra-pH-sensitive nanoprobe library with broad pH tunability and fluorescence emissions. J. Am. Chem. Soc. 2014;136:11085–11092. doi: 10.1021/ja5053158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., MacEwan S.R., Chilkoti A. Quantitative mapping of the spatial distribution of nanoparticles in endo-lysosomes by local pH. Nano Lett. 2017;17:1226–1232. doi: 10.1021/acs.nanolett.6b05041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grover A., Schmidt B.F., Salter R.D., Watkins S.C., Waggoner A.S., Bruchez M.P. Genetically encoded pH sensor for tracking surface proteins through endocytosis. Angew. Chem. Int. Ed. Engl. 2012;51:4838–4842. doi: 10.1002/anie.201108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Wang C., Li Y., Huang G., Zhao T., Ma X., Wang Z., Sumer B.D., White M.A., Gao J. Digitization of endocytic pH by hybrid ultra-pH-sensitive nanoprobes at single-organelle resolution. Adv. Mater. 2017;29:1603794. doi: 10.1002/adma.201603794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J., Fang C.L., Chang T.J., Liu X.J., Shangguan D. A pH sensitive ratiometric fluorophore and its application for monitoring the intracellular and extracellular pHs simultaneously. J. Mater. Chem. B. 2013;1:661–667. doi: 10.1039/c2tb00179a. [DOI] [PubMed] [Google Scholar]
- 41.Ma T.C., Hou Y., Zeng J.F., Liu C.Y., Zhang P.S., Jing L.H., Shangguan D., Gao M.Y. Dual-ratiometric target-triggered fluorescent probe for simultaneous quantitative visualization of tumor microenvironment protease activity and pH &ITin vivo&IT. J. Am. Chem. Soc. 2018;140:211–218. doi: 10.1021/jacs.7b08900. [DOI] [PubMed] [Google Scholar]
- 42.Hou Y., Zhou J., Gao Z.Y., Sun X.Y., Liu C.Y., Shangguan D.H., Yang W.S., Gao M.Y. Protease-activated ratio metric fluorescent probe for pH mapping of malignant tumors. ACS Nano. 2015;9:3199–3205. doi: 10.1021/acsnano.5b00276. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Zhang N., Lu S., Wang J., Bing T., Liu X., Chen C., Shangguan D. Dual-monitoring glycosylation and local pH in live cells by metabolic oligosaccharide engineering with a ratiometric fluorescent tag. Anal. Chem. 2019;91:13720–13728. doi: 10.1021/acs.analchem.9b03047. [DOI] [PubMed] [Google Scholar]
- 44.Jovic M., Sharma M., Rahajeng J., Caplan S. The early endosome: a busy sorting station for proteins at the crossroads. Histol. Histopathol. 2010;25:99–112. doi: 10.14670/hh-25.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung J.Y., Thone M.N., Kwon Y.J. COVID-19 vaccines: the status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2021;170:1–25. doi: 10.1016/j.addr.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolte S., Cordelieres F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.