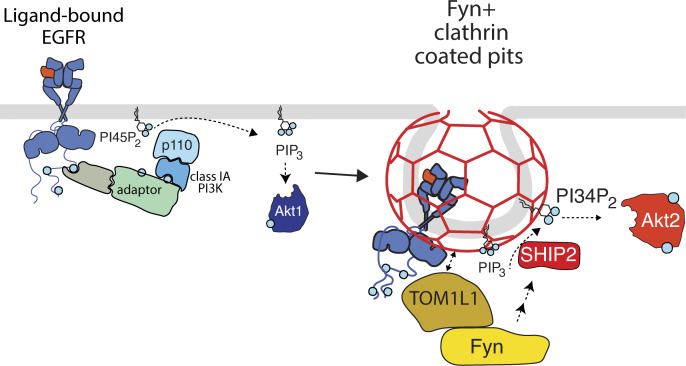
Ligand binding to EGF receptor (EGFR) triggers signaling and concomitant receptor recruitment to clathrin-coated pits. This study reveals that the signaling adaptor TOM1L1 recruits Fyn to a specialized subset of clathrin-coated pits and is required for SHIP2 recruitment and regulation of Akt signaling by EGFR.
Abstract
The epidermal growth factor (EGF) receptor (EGFR) controls many aspects of cell physiology. EGF binding to EGFR elicits the membrane recruitment and activation of phosphatidylinositol-3-kinase, leading to Akt phosphorylation and activation. Concomitantly, EGFR is recruited to clathrin-coated pits (CCPs), eventually leading to receptor endocytosis. Previous work uncovered that clathrin, but not receptor endocytosis, is required for EGF-stimulated Akt activation, and that some EGFR signals are enriched in CCPs. Here, we examine how CCPs control EGFR signaling. The signaling adaptor TOM1L1 and the Src-family kinase Fyn are enriched within a subset of CCPs with unique lifetimes and protein composition. Perturbation of TOM1L1 or Fyn impairs EGF-stimulated phosphorylation of Akt2 but not Akt1. EGF stimulation also triggered the TOM1L1- and Fyn-dependent recruitment of the phosphoinositide 5-phosphatase SHIP2 to CCPs. Thus, the recruitment of TOM1L1 and Fyn to a subset of CCPs underlies a role for these structures in the support of EGFR signaling leading to Akt activation.
Introduction
The epidermal growth factor (EGF) receptor (EGFR) is a receptor tyrosine kinase that controls many aspects of development and adult physiology, which at the cellular level involves enhancement of survival, growth, and proliferation (Avraham and Yarden, 2011; Pareja et al., 2015). EGFR must be tightly regulated, and dysregulation of this receptor has important roles in driving the growth and progression of certain cancers (Sigismund et al., 2018).
A major signaling intermediate phosphorylated and activated by EGFR to accomplish these outcomes is Akt (Vivanco and Sawyers, 2002; Cantley, 2002). Ligand binding to EGFR promotes EGFR dimerization and autophosphorylation, leading to activation of many intracellular signals (Lemmon et al., 2014; Freed et al., 2017; Bessman et al., 2014). Akt activation by EGFR requires binding of growth factor receptor-bound protein 2 (Grb2) to a motif containing phosphorylated Y1068 on EGFR, followed by recruitment of Grb2-associated binder 1 (Gab1; Lock et al., 2000). EGF-stimulated Gab1 phosphorylation (Kiyatkin et al., 2006) on a number of residues allows recruitment of several signals including class 1 phosphatidylinositol-3-kinase (PI3K), either directly (Holgado-Madruga et al., 1996; Mattoon et al., 2004) or via interaction with Gab1-bound SHP2 (Wu et al., 2001). PI3K recruitment to Gab1 results in production of phosphatidylinositol-3,4,5-trisphosphate (PIP3) from phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2; Rodrigues et al., 2000; Mattoon et al., 2004), which allows Akt membrane recruitment and phosphorylation on T308 and S473 by PDK1 and mTORC2, respectively, resulting in Akt activation (Matheny and Adamo, 2009).
There are three isoforms of Akt that exhibit similarities but that differ in their mechanism of activation by phosphoinositides (Liu et al., 2018). PIP3 directly recruits and activates Akt1 and Akt3, while Akt2 is recruited and activated by binding to phosphatidylinositol-3,4-bisphosphate (PtdIns(3,4)P2; Liu et al., 2018), which can be produced by the lipid phosphatase SHIP2 from PIP3 (Goulden et al., 2018; Liu et al., 2018). Hence, there are several critical control points for activation of Akt signaling, including the recruitment and activation of PI3K (Mattoon et al., 2004) as well as the possible regulation of SHIP2 by EGFR.
Concomitantly with activation of this and other signaling pathways, EGFR is recruited to clathrin-coated pits (CCPs), which in some cases leads to receptor internalization through clathrin-mediated endocytosis. CCPs are dynamic 50–200 nm structures that form on the inner leaflet of the plasma membrane by the assembly of clathrin, the adaptor protein AP2, and >50 other proteins (Mettlen et al., 2009; Schmid and McMahon, 2007; McMahon and Boucrot, 2011; Taylor et al., 2011). The formation of CCPs spans several stages, including nucleation, followed by CCP initiation, assembly, maturation, and eventual scission (Mettlen and Danuser, 2014; Kadlecova et al., 2017; Aguet et al., 2013; Delos Santos et al., 2017; McMahon and Boucrot, 2011).
In addition to the well-established role of CCPs in mediating internalization of various receptors, these structures may also modulate receptor signaling. Clathrin, but not EGFR endocytosis, regulates EGF-stimulated phosphorylation of Gab1 and Akt (Garay et al., 2015), and AP2 ablation impaired EGF-stimulated Akt phosphorylation (Pascolutti et al., 2019). Phosphorylated Gab1 (Lucarelli et al., 2016; Garay et al., 2015) and Akt (Rosselli-Murai et al., 2018) are enriched within CCPs, as is phosphatase and tensin homolog(PTEN) that elicits hydrolysis of PIP3 into PtdIns(4,5)P2 and thus negatively regulates EGFR signaling (Rosselli-Murai et al., 2018). SHIP2 is also localized to CCPs when overexpressed (Nakatsu et al., 2010), although it is not clear whether the endogenous protein is enriched in CCPs and whether EGFR signaling may control SHIP2 recruitment to CCPs. Hence, some CCPs may function as signaling nanodomains by dynamically scaffolding key signaling intermediates, leading to Akt activation. How CCPs may be uniquely equipped with specific proteins to promote activation of Akt signaling remains poorly understood.
Src family kinases regulate some aspects of EGFR signaling. While use of inhibitors PP1 or PP2 suggest a role for Src-family kinases in EGF-stimulated Gab1 phosphorylation (Furcht et al., 2015; Daub et al., 1997), PP2 impairs clathrin-mediated endocytosis in cells lacking the three ubiquitously expressed Src-family kinases c-Src, Fyn, and Yes (Sorkina et al., 2002). This suggests that PP2 and perhaps other Src-family kinase inhibitors may have effects on CCP dynamics independent of inhibition of Src family kinases.
That Src-family kinases may be candidates for clathrin-dependent signaling regulation suggests the need for adaptor proteins for enrichment of one or more of these kinases to clathrin structures. Target of Myb-1 like protein 1 (TOM1L1) is a signaling adaptor (Puertollano, 2005; Puertollano et al., 2001; Yamakami et al., 2003) that can directly bind clathrin heavy chain (Collin et al., 2007; Liu et al., 2009b) in addition to binding and activation of Src-family kinases, in particular Fyn (Seykora et al., 2002; Li et al., 2005). Phosphorylation of Y460 on TOM1L1 allows binding to Fyn via its SH2 domain (Seykora et al., 2002), leading to localized Fyn activation (Li et al., 2005). TOM1L1 directly interacts with a unique region within the C-terminus of clathrin heavy chain via a 447FDPL450 motif, such that mutation of this motif abolishes interaction with clathrin (Liu et al., 2009b). Whether TOM1L1 is recruited to CCPs at the plasma membrane and how it may regulate Fyn therein to participate in EGFR signaling remains poorly understood.
CCPs exhibit significant heterogeneity with respect to size, lifetime and protein composition (Liu et al., 2009a; Antonescu et al., 2010; Mettlen et al., 2010; Liu et al., 2010; Loerke et al., 2011; Nunez et al., 2011; Loerke et al., 2009; Antonescu et al., 2011; Puthenveedu and von Zastrow, 2006; Taylor et al., 2011; Mettlen et al., 2009; Aguet et al., 2013) in a manner related to the cargo receptor recruited therein (Mettlen et al., 2010; Puthenveedu and von Zastrow, 2006; Delos Santos et al., 2017). Molecularly distinct CCP subpopulations defined by the presence of AP2 or the endocytic accessory protein Epsin each have unique contributions to EGFR signaling (Pascolutti et al., 2019). This suggests that unique subpopulations of CCPs demarked by certain protein compositions may have specific functions, some of which may regulate specific aspects of EGFR signaling.
Here, we examine the recruitment of Fyn and TOM1L1 to plasma membrane clathrin structures, and how these proteins participate in EGFR signaling leading to the activation of specific Akt isoforms. We use perturbations of Fyn or TOM1L1, in conjunction with detection of the activation of EGFR signals and their localization vis-à-vis plasma membrane clathrin structures, to determine how TOM1L1 and Fyn participate in EGFR signaling. We also examine the properties of clathrin structures harboring Fyn or TOM1L1, finding that these signaling-capable clathrin structures have unique properties.
Results
Fyn and TOM1L1 regulate EGF-stimulated Akt phosphorylation
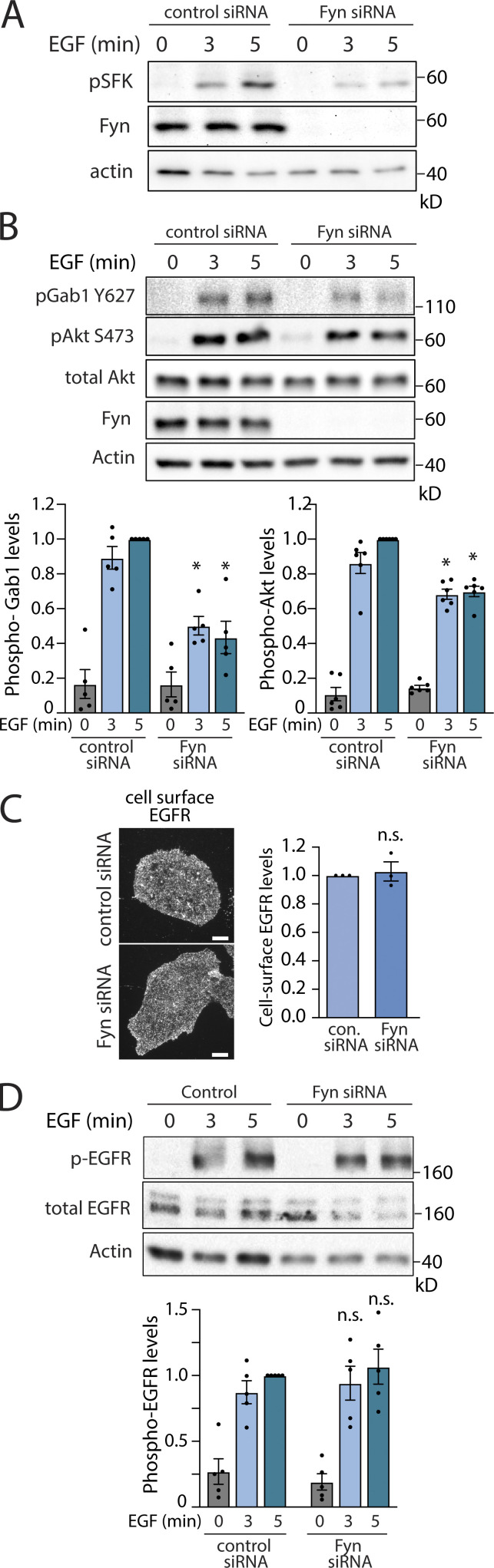
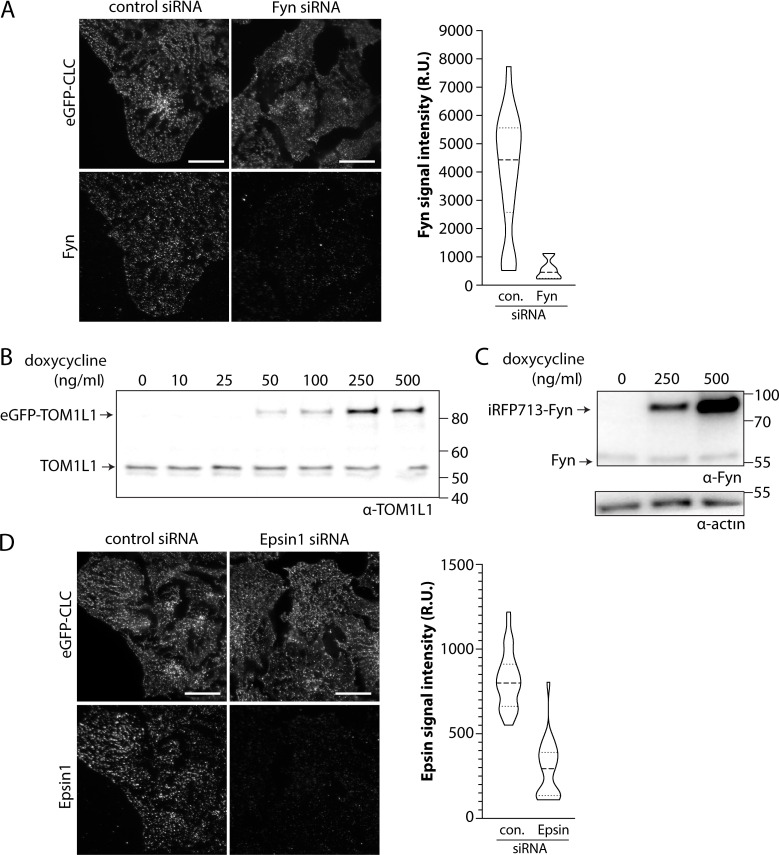
To determine how Fyn may contribute to EGF-stimulated Akt phosphorylation, we first detected Y420 phosphorylated Fyn (which corresponds to Y416 on c-Src). Unless otherwise indicated, we examine ARPE-19 cells, in which we previously found a role for clathrin, but not EGFR endocytosis, in EGF-stimulated Akt phosphorylation (Garay et al., 2015). The levels of the phosphorylated Src-family kinase were enhanced by EGF stimulation, which was impaired upon Fyn silencing (Fig. 1 A). This supports a previous study that found that this antibody preferentially detects phosphorylated Fyn (pY420; Githaka et al., 2016) and indicates that EGF stimulation leads to Fyn phosphorylation.
Figure 1.
Fyn regulates EGF-stimulated Gab1 and Akt phosphorylation. ARPE-19 cells were transfected with siRNA targeting Fyn or nontargeting siRNA (control), followed by stimulation with 5 ng/ml EGF for the time indicated. (A) Western blotting of whole-cell lysates probed with anti-phospho-Src-family kinase (corresponding to pY420 on Fyn). (B) Western blotting using anti-phospho-Gab1 (pY627) or anti-phospho-Akt (pY473, pan isoform). Also shown are the mean ± SEM phospho-Gab1 or phospho-Akt with points representing individual experiment measurements; n = 5–6; *, P < 0.05 relative to the control siRNA-treated EGF-stimulated conditions at each time point. (C) After siRNA transfection, intact cells were subjected to immunofluorescence microscopy with antibodies that selectively recognize the EGFR ectodomain (Alexa Fluor 488 labeling). Representative fluorescence microscopy micrographs of cell surface EGFR immunostaining are shown. Scale = 10 µm. Also shown are mean fluorescence intensity of cell-surface EGFR ± SEM with points representing individual experiment measurements; n = 3. (D) Western blotting using anti-phospho-EGFR (pY1068); also shown are the mean ± SEM phospho-EGFR with points representing individual experiment measurements; n = 5. Source data are available for this figure: SourceData F1.
To determine how Fyn may control EGFR signaling, we examined the effect of Fyn siRNA silencing. Fyn silencing substantially impaired EGF-stimulated Gab1 (Y627) and Akt (S473) phosphorylation (Fig. 1 B). In contrast, Fyn silencing was without effect on the cell surface levels of EGFR (Fig. 1 C) or EGF-stimulated EGFR phosphorylation (Fig. 1 D), indicating that defects in EGF-stimulated Gab1 and Akt phosphorylation upon Fyn silencing do not result from alterations in receptor availability for extracellular ligand and/or receptor activation. Hence, EGF stimulation results in Fyn phosphorylation, and Fyn contributes to EGF-stimulated Gab1 and Akt phosphorylation, but not EGFR phosphorylation.
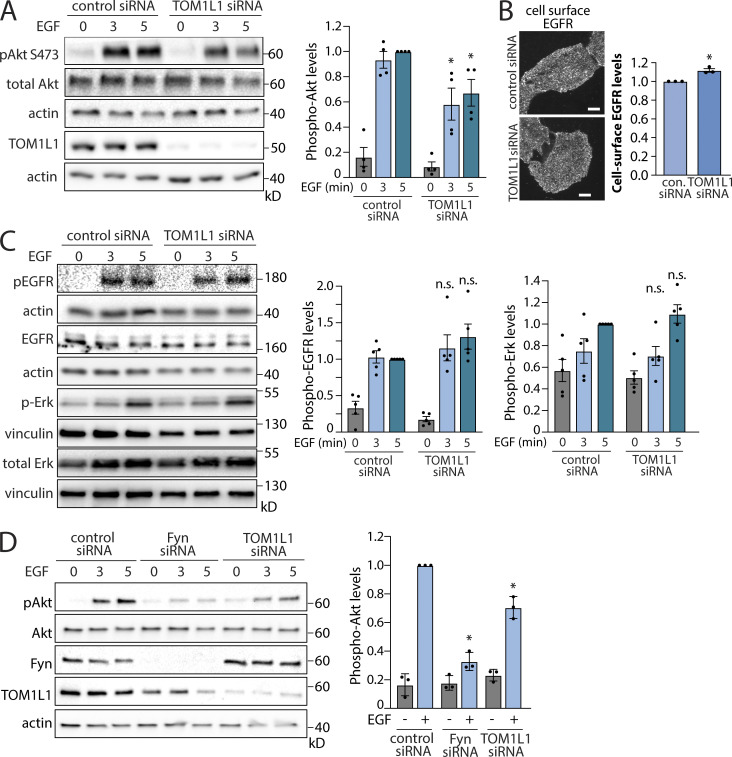
As Fyn silencing has a phenotype similar to that of clathrin with regard to EGFR signaling (Garay et al., 2015), and as TOM1L1 binds both clathrin and Fyn, we next sought to determine the contribution of TOM1L1 to EGFR signaling. Silencing of TOM1L1 significantly impaired EGF-stimulated Akt phosphorylation (Fig. 2 A), similarly to the perturbation of Fyn (Fig. 1 B) and clathrin (Garay et al., 2015). TOM1L1 silencing slightly but significantly increased cell surface EGFR levels (Fig. 2 B), and TOM1L1 silencing did not impair EGF-stimulated Gab1, EGFR, or Erk phosphorylation (Figs. S1 A and 2 C). This indicates that, similarly to Fyn silencing, TOM1L1 silencing selectively impairs EGF-stimulated Akt phosphorylation, without impairing EGFR ligand binding or receptor autophosphorylation. Gab1 phosphorylation on Y627 is distinct from that required for PI3K binding and activation (Rocchi et al., 1998). That silencing TOM1L1 did not impact EGF-stimulated Gab1 Y627 phosphorylation suggests that Fyn has additional functions in signaling, and that TOM1L1 and Fyn regulate EGF-stimulated Akt phosphorylation downstream of Gab1. Silencing TOM1L1 or Fyn also impaired EGF-stimulated Akt phosphorylation in MCF10A cells (Fig. 2 D), likewise mirroring the requirement for clathrin in these cells (Rosselli-Murai et al., 2018). Taken together, these data indicate that Fyn and TOM1L1 regulate EGF-stimulated Akt phosphorylation at a stage subsequent to EGF binding and EGFR autophosphorylation. Moreover, that Fyn or TOM1L1 perturbations are similar to those of clathrin suggests that these proteins may be working in concert within clathrin structures to control EGFR signaling to Akt.
Figure 2.
TOM1L1 regulates EGF-stimulated Akt phosphorylation. (A–D) ARPE-19 (A–C) or MCF10A (D) cells were transfected with siRNA targeting Fyn, TOM1L1, or nontargeting siRNA (control), as indicated, followed by stimulation with 5 ng/ml EGF for the time indicated. (A) Western blotting using anti-phospho-Akt (pY473, pan isoform). Also shown are the mean ± SEM phospho-Akt with points representing individual experiment measurements; n = 4; *, P < 0.05, relative to the control siRNA-treated EGF-stimulated condition at each time point. (B) After siRNA transfection, intact cells were subjected to immunofluorescence microscopy with antibodies that selectively recognize the EGFR ectodomain (Alexa Fluor 488 labeling). Representative fluorescence microscopy micrographs of cell surface EGFR immunostaining are shown. Scale = 10 µm. Also shown are mean fluorescence intensity of cell-surface EGFR ± SEM with points representing individual experiment measurements; n = 3. con., control. *, P < 0.05 relative to EGF-stimulated, control siRNA condition. (C) Western blotting using anti-phospho-EGFR (pY1068) or anti-phospho-Erk; also shown are the mean ± SEM phospho-EGFR or phospho-Erk levels with points representing individual experiment measurements; n = 5. (D) MCF10A cells: Western blotting using anti-phospho-Akt (pY473, pan isoform), as well as TOM1L1 or Fyn. Also shown are the mean ± SEM phospho-Akt at 3 min of EGF stimulation with points representing individual experiment measurements; n = 3; *, P < 0.05, relative to the control siRNA-treated EGF-stimulated conditions at each time point. Source data are available for this figure: SourceData F2.
Figure S1.
Additional information on TOM1L1 silencing and full-image panels of TIRFM and SIM micrographs shown in Fig. 3. (A) ARPE-19 cells were transfected with siRNA targeting Fyn, TOM1L1, or nontargeting siRNA (control), as indicated, followed by stimulation with 5 ng/ml EGF for the time indicated. Western blotting using anti-phospho-Gab1 (pY627) or actin. TOM1L1 siRNA silencing does not impair EGF-stimulated Gab1 phosphorylation (pY627). Molecular weight markers (kD) shown on the right. (B and C) Cell samples treated and prepared as in Fig. 3, A and B, were subjected to imaging by TIRFM. Shown here are the full-sized images of the representative images shown in Fig. 3, A and B, with a white box showing the area enlarged in Fig. 3, A and B. Scale bar = 20 μm. (D and E) Cell samples treated and prepared as in Fig. 3 C were subjected to SIM. (D) Shown here are the full-sized images of the representative images shown in Fig. 3 C, with a white box showing the area enlarged in Fig. 3 C. Scale bar = 10 µm. (E) Shown are linescan analyses of AP2 and Fyn-GFP in individual clathrin structures (bottom). Image inset width is 0.26 μm. Source data are available for this figure: SourceData FS1.
Fyn and TOM1L1 recruitment to plasma membrane clathrin structures is required for modulation of EGF-stimulated Akt phosphorylation
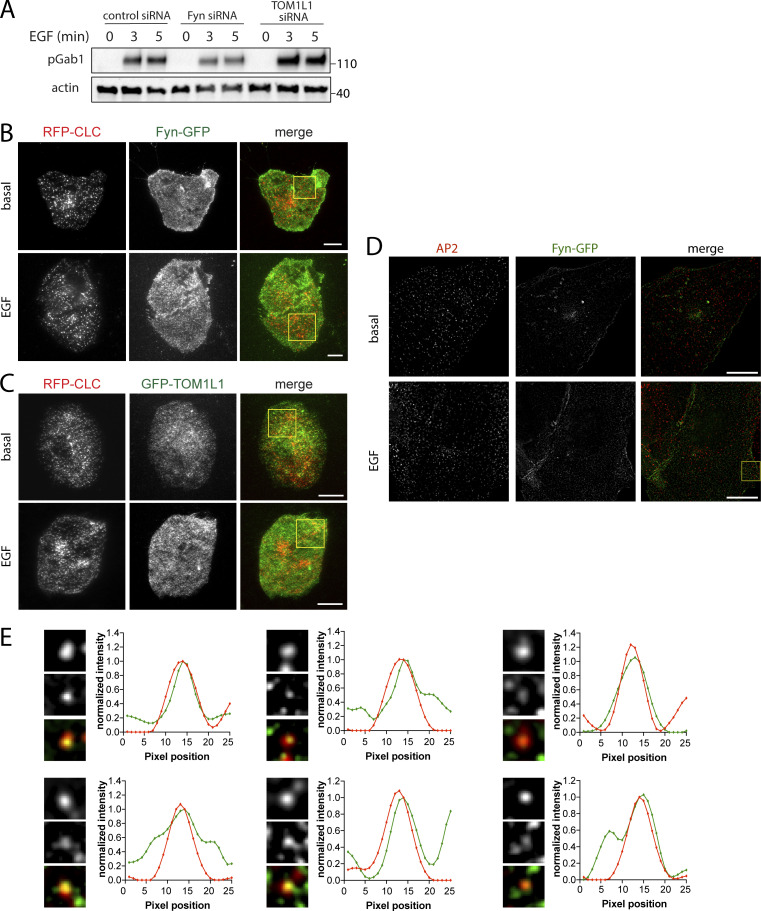
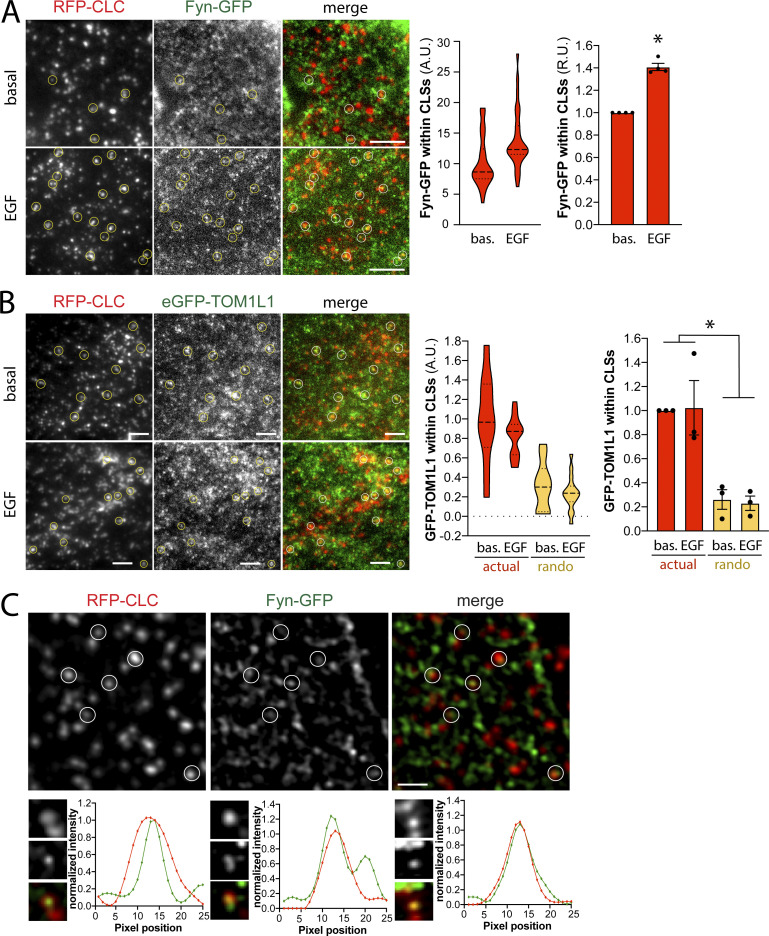
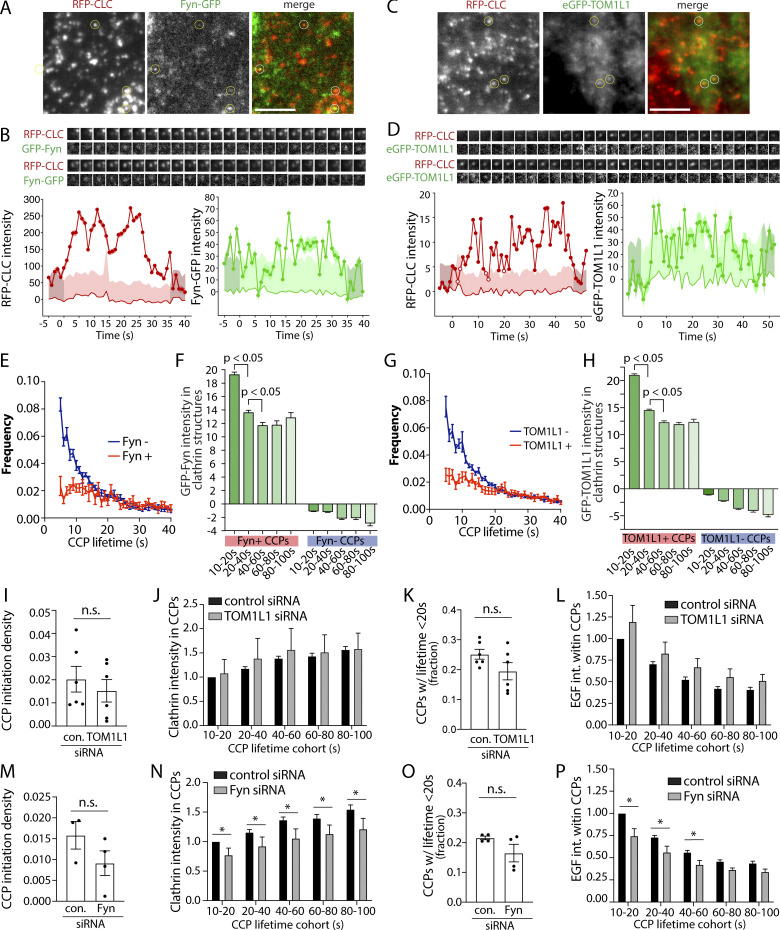
To determine whether Fyn is recruited to plasma membrane clathrin structures after EGF stimulation, we first used transfection of superfolder (sf)GFP-tagged Fyn (Fyn-GFP henceforth) at the lowest detectable levels in RPE cells stably expressing Tag-RFP-T–tagged clathrin light chain (RPE-RFP-CLC), followed by imaging using total internal reflection fluorescence microscopy (TIRFM) to selectively image the cell surface. Fyn-GFP exhibited little recruitment to cell surface clathrin structures in the basal state (Fig. 3 A). Notably, stimulation with 20 ng/ml EGF for 5 min caused a noticeable recruitment of Fyn-GFP within some clathrin structures, although Fyn-GFP could also be detected outside of clathrin structures. Automated detection and analysis of clathrin-labeled structures (CLSs) using a Gaussian-based modeling approach (Aguet et al., 2013), as we have used previously to detect specific proteins within CLSs (Delos Santos et al., 2017; Garay et al., 2015; Lucarelli et al., 2016), revealed a statistically significant increase in Fyn-GFP recruitment to CLSs upon EGF stimulation (Fig. 3 A, right). We use the term CLS to denote clathrin structures detected in fixed samples, since identification of bona fide CCPs from short-lived subthreshold clathrin structures requires live-cell analysis (Aguet et al., 2013; Kadlecova et al., 2017). The results of the measurement of Fyn-GFP within CLSs is shown as measurements of individual cells within a single experiment (Fig. 3 A, middle), as well as the results of the average of multiple independent experiments (Fig. 3 A, right).
Figure 3.
Fyn and TOM1L1 are recruited to a subset of plasma membrane clathrin structures. RPE cells stably expressing Tag-RFP-T-CLC were transfected with a plasmid encoding sfGFP fused to Fyn (Fyn-GFP) or eGFP fused to TOM1L1 (GFP-TOM1L1), as indicated. (A and B) Cells were stimulated with 20 ng/ml EGF for 5 min or left unstimulated (basal [bas.]), and then fixed and imaged using TIRFM. Shown (left) are representative micrographs depicting Fyn-positive clathrin structures (circles), identified manually. Scale = 5 µm. Full-size image panels obtained by TIRFM are available in Fig. S1. Micrographs obtained by TIRFM were subjected to automated detection and analysis of CLSs, allowing quantification of Fyn-GFP and RFP-CLC in each detected object. Shown (middle) are the measurements of Fyn-GFP or eGFP-TOM1L1 fluorescence intensity within CLSs, showing the distribution of the mean value of individual cells (each cell value determined from >300 CLSs) depicted as a violin plot, as well as median (long dashed line) and 25th/75th percentiles (short dashed line). Also shown (right) are the levels of eGFP-TOM1L1 or Fyn-GFP detected with CLSs in independent experiments (each experiment value determined from >15 cells per condition, depicted as points) as mean ± SEM; *, P < 0.05, relative to basal. In B, we also report measurements performed in the same image pairs in which one of the channels was rotated 180° to randomize the position of TOM1L1 structures relative to clathrin structures (rando). The number of CLSs and cells analyzed for each condition are as follows: Fyn-GFP expression, basal (44 cells, 6,641 CLSs) and EGF-stimulated (31 cells, 6,300 CLSs); eGFP-TOM1L1 expression, basal (31 cells, 6,574 CLSs) and EGF-stimulated (37 cells, 8,064 CLSs). (C) RPE cells transfected to express Fyn-GFP were subject to SIM. Shown are representative micrographs (top) and linescan analysis of AP2 and Fyn-GFP in individual clathrin structures (bottom). Scale = 1 μm (top); width of enlarged image (bottom) is 0.26 μm. Full-size image panels obtained by SIM are available in Fig. S1. A.U., arbitrary unit; R.U., relative unit.
To determine if TOM1L1 is also enriched within CLSs after EGF stimulation, we again used transfection to obtain the lowest detectable expression of eGFP-tagged TOM1L1 in RPE-RFP-CLC cells and subjected these cells to imaging by TIRFM followed by automated detection and analysis of CLSs (Garay et al., 2015). TOM1L1 could be observed and detected within plasma membrane CLSs in both the basal and EGF-stimulated conditions (Fig. 3 B, left, and red plots and bars). Scrambling the position of the eGFP-TOM1L1 fluorescence channel relative to the clathrin channel greatly reduced the eGFP-TOM1L1 detected within CLSs (Fig. 3 B, right), indicating that the recruitment of eGFP-TOM1L1 to CLSs was specific. This indicates that EGF stimulation elicits a gain in Fyn-GFP recruitment into CLS, while eGFP-TOM1L1 is present in some CLS before EGF stimulation.
To complement these experiments, we performed structured illumination microscopy (SIM) to observe the localization of Fyn-GFP and CLSs at higher resolution. We observed that clathrin structures again demonstrated partial overlap with Fyn-GFP (Figs. 3 C and S1 D). Notably, a linescan analysis of individual clathrin and Fyn structures revealed similar intensity profiles with similar centers (Figs. 3 C and S1 E), further supporting the specific recruitment of Fyn-GFP to some clathrin structures that we observed by systematic analysis of TIRFM images (Fig. 3 A).
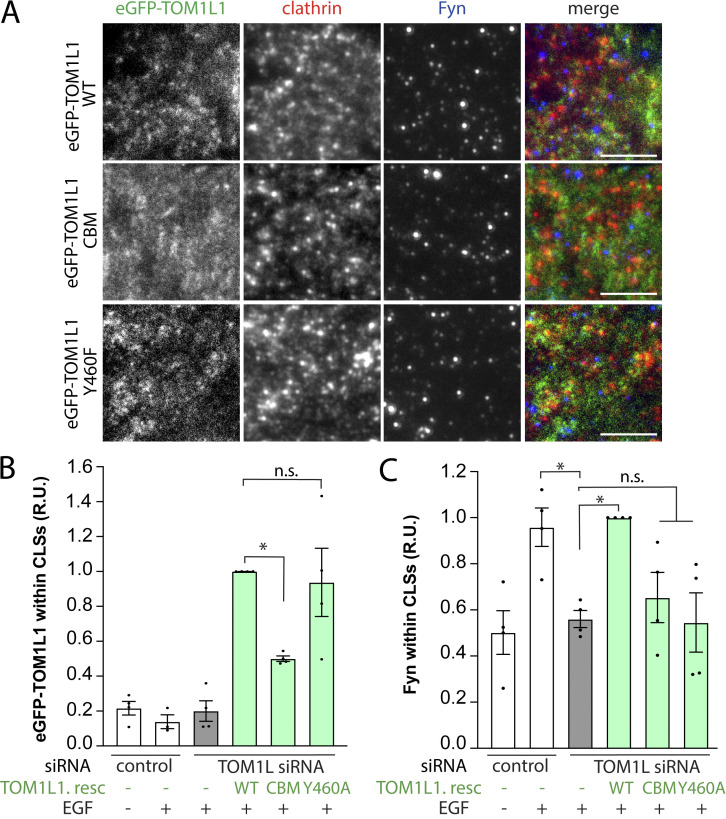
To determine the possible relationship between recruitment of TOM1L1 and that of Fyn to plasma membrane clathrin structures, we used a knockdown-rescue approach using siRNA-resistant mutants of TOM1L1 (fused to eGFP) that are defective in either clathrin-binding (clathrin-binding mutant, CBM: 447FDPL450 to 447AAAA450) or Fyn-binding (Y460F; Liu et al., 2009b). We expressed these eGFP-TOM1L1 constructs by generating ARPE-19 stable cells using the Sleeping Beauty transposon system (Kowarz et al., 2015); this allowed doxycycline-inducible expression of TOM1L1 at low, near-endogenous levels (Fig. S2 B). We first performed TIRFM on these knockdown-rescue cells after EGF stimulation and immunostaining of endogenous Fyn (Fig. S2 A) or clathrin. As expected, WT eGFP-TOM1L1 could be observed within CLSs, while eGFP-TOM1L1 CBM appeared less enriched in clathrin structures (Fig. 4 A). After automated detection of CLSs, eGFP-TOM1L1 CBM was significantly less detected in CLSs than WT eGFP-TOM1L1 or the Y460F mutant that retains an intact clathrin-binding motif (Fig. 4 B). These results show that eGFP-TOM1L1 is recruited to plasma membrane clathrin structures through the 447FDPL450 clathrin-binding motif.
Figure S2.
Optimization of Sleeping Beauty stable cell lines and antibody staining. (A and D) ARPE19 cells stably expressing eGFP-clathrin were treated with siRNA targeting Fyn, Epsin1, or nontargeting (control [con.]) siRNA. After transfection, cells were fixed and stained with antibodies recognizing Fyn (A) or Epsin (D), followed by imaging by TIRFM and quantification of signaling intensity per cell. Shown are representative TIRFM images (left) and the measurements of Fyn (A) or Epsin (D) signal intensity (right). Scale bar = 20 μm. Both Fyn and Epsin1 antibodies are highly specific. (B and C) Stable cell lines harboring a transgene for doxycycline-inducible expression of eGFP-TOM1L1 or Fyn-iRFP713 were treated with doxycycline as indicated for 24 h. Whole-cell lysates were resolved by Western blotting and probed with anti-TOM1L1 (B) or anti-Fyn antibodies (C). From these experiments, we optimized induction at 150 ng/ml doxycycline to obtain near-endogenous expression levels of eGFP-TOM1L1 and Fyn-iRFP713. Molecular weight markers (kD) are shown on the right. Source data are available for this figure: SourceData FS2. R.U., relative unit.
Figure 4.
TOM1L1 recruits Fyn to clathrin structures. ARPE-19 cells were engineered using the Sleeping Beauty transposon system to allow doxycycline-inducible expression of eGFP-TOM1L1 constructs as follows: WT, clathrin binding mutant (CBM: 447FDPL450 to 447AAAA450), or Fyn-binding mutant (Y460F). Each cell type was transfected with siRNA targeting TOM1L1 or nontargeting siRNA (control), followed by expression of eGFP-TOM1L1 using 150 ng/ml doxycycline, which resulted in levels of expression of eGFP-TOM1L1 comparable to endogenous; see Fig. S2 B. Cells were then stimulated with 5 ng/ml EGF for 5 min, fixed, subjected to antibody staining to fluorescently label clathrin (Alexa Fluor 647) and Fyn (Cy3), and imaged using TIRFM; see Fig. S2 A for specificity of Fyn antibody. (A) Shown are representative micrographs, scale bar = 5 µm. (B and C) Micrographs obtained by TIRFM were subjected to automated detection and analysis of clathrin structures, allowing quantification of eGFP-TOM1L1 (B) and endogenous Fyn (C) within RFP-CLC in each detected object. Shown are the levels of eGFP-TOM1L1 or Fyn-GFP detected with CLSs in individual experiments (each independent experiment value determined from >15 cells per condition, depicted as points) as mean ± SEM; *, P < 0.05. Gray bars represent cells subjected to TOM1L1 silencing with no rescue (resc.); green bars depict conditions in which siRNA silencing of endogenous TOM1L1 was rescued by doxycycline-induction of eGFP-TOM1L1 constructs, as indicated. The number of CLSs and cells analyzed in three independent experiments for each condition are as follows: control siRNA, basal (112 cells, 71,151 CLSs) and EGF-stimulated (102 cells, 59,551 CLSs); TOM1L1 siRNA, no rescue, EGF-stimulated (99 cells, 52,675 CLSs), TOM1L1 siRNA, eGFP-TOM1L1 WT rescue, EGF-stimulated (146 cells, 48,193 CLSs), TOM1L1 siRNA, eGFP-TOM1L1 CBM rescue, EGF-stimulated (113 cells, 44,141 CLSs), and TOM1L1 siRNA, eGFP-TOM1L1 Y460F rescue, EGF-stimulated (78 cells, 30,081 CLSs). R.U., relative unit.
We next examined the contribution of TOM1L1 to Fyn recruitment to CLSs in this knockdown-rescue experiment. First, in cells treated with control siRNA, we observed an increase in Fyn detected within CLSs upon EGF stimulation (Fig. 4 C, white bars). This shows that endogenous Fyn behaves similarly to Fyn-GFP expressed at low levels (Fig. 3 A). Silencing TOM1L1 in the absence of rescue significantly reduced Fyn detected within CLSs (Fig. 4 C, gray bars). Importantly, expression of the WT, but not the CBM or Fyn-binding mutant of eGFP-TOM1L1, rescued the loss of Fyn within CLSs in TOM1L1-silenced cells (Fig. 4 C, green bars). These experiments suggest that Fyn recruitment to CLSs is dependent on both the interaction of TOM1L1 with clathrin via its 447FDPL450 clathrin-binding motif, and the motif containing Y460 on TOM1L1 that is required for association with Fyn.
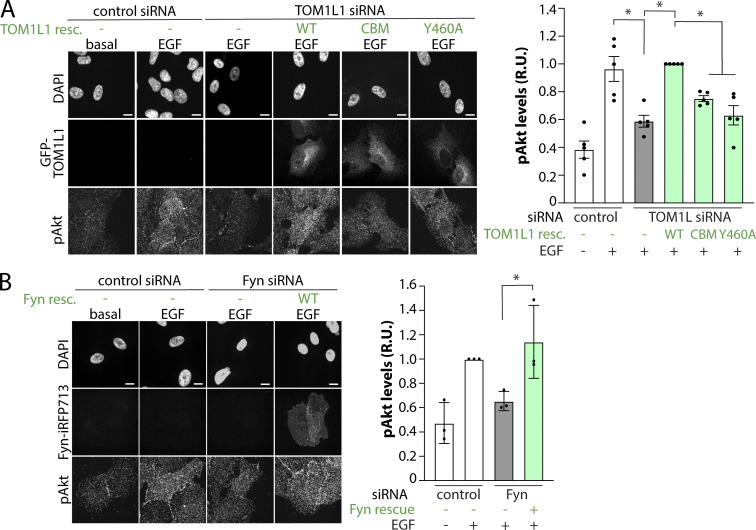
To determine if the recruitment of TOM1L1 and Fyn to CLSs regulates EGF-stimulated phosphorylation of Akt, we again used knockdown-rescue of TOM1L1. Because ∼50–60% of cells expressed detectable eGFP-TOM1L1, we detected Akt phosphorylation in individual cells where we could ascertain eGFP-TOM1L1 expression. As observed by Western blotting in Fig. 2, this immunofluorescence detection of pAkt revealed that in the absence of eGFP-TOM1L1 rescue, EGF stimulation triggered an increase in Akt phosphorylation, which was suppressed in TOM1L1-silenced cells (Fig. 5 A, white and gray bars). Expression of the WT, but not the CBM or Fyn-binding mutant of eGFP-TOM1L1, rescued the loss of Akt phosphorylation in EGF-stimulated cells observed upon silencing of endogenous TOM1L1 (Fig. 5 A, green bars). These results indicate that Akt phosphorylation triggered by EGF stimulation is regulated by the clathrin-interacting 447FDPL450 and Fyn-interacting Y460 motifs on TOM1L1. In turn, this suggests that recruitment of TOM1L1 and Fyn to clathrin structures at the plasma membrane contributes to EGFR signaling leading to Akt phosphorylation.
Figure 5.
Clathrin- and Fyn-interacting motifs within TOM1L1 are required for regulation of EGF-stimulated Akt phosphorylation. (A and B) ARPE-19 cells were engineered using the Sleeping Beauty transposon system to allow doxycycline-inducible expression of eGFP-TOM1L1 constructs as follows: WT, clathrin binding mutant (CBM: 447FDPL450 to 447AAAA450), or Fyn-binding mutant (Y460F; A) or Fyn-iRFP713 (WT; B). Cells were transfected with siRNA targeting TOM1L1, Fyn, or nontargeting siRNA (control), followed by expression of eGFP-TOM1L1 or Fyn-iRFP713 using 150 ng/ml doxycycline, which resulted in levels of expression of eGFP-TOM1L1 or Fyn comparable to endogenous; see Fig. S2, B and C. Cells were stimulated with 5 ng/ml EGF as indicated, and then fixed and subjected to detection of phospho-Akt (S473, pan-isoform, labeled with Cy3) by immunostaining. Shown (left) are representative images obtained by wide-field epifluorescence microscopy showing DAPI, eGFP-TOM1L1, Fyn-iRFP713, or pAkt, as indicated. Scale = 10 µm. Also shown (right) are the levels of pAkt detected per cell in individual experiments (each experiment value determined from >15 cells per condition, depicted as points) as mean ± SEM; n = 3–5 independent experiments; *, P < 0.05. Gray bars represent cells subjected to TOM1L1 or Fyn silencing with no rescue (resc); green bars depict conditions in which siRNA silencing of endogenous TOM1L1 or Fyn was rescued by doxycycline-induction of eGFP-TOM1L1 or Fyn-iRFP713 constructs, as indicated. R.U., relative unit.
Because expression of WT eGFP-TOM1L1 rescued the loss of EGF-stimulated Akt phosphorylation observed upon TOM1L1 knockdown, these experiments also indicate that the impairment of EGFR signaling by this knockdown (Fig. 2) was specific to loss of TOM1L1. We used a similar knockdown-rescue strategy for Fyn to determine if inducible expression of iRFP713-tagged Fyn (Fyn-iRFP713; Fig. S2 C) in stable cells could rescue the effect of Fyn silencing, which was indeed the case (Fig. 5 B). In the absence of inducible expression of Fyn-iRFP713, EGF stimulation led to an increase in Akt phosphorylation, which was suppressed in Fyn-silenced cells (Fig. 5 B, white and gray bars). Expression of Fyn-iRFP713 rescued the impairment of Akt phosphorylation in EGF-stimulated cells observed upon silencing of endogenous Fyn (Fig. 5 B, green bars).
These results indicate that Fyn recruitment to CLSs is dependent on TOM1L1 and that recruitment of TOM1L1 and Fyn to CLSs regulates EGF-simulated Akt phosphorylation.
Fyn is recruited to a distinct subset of plasma membrane clathrin structures
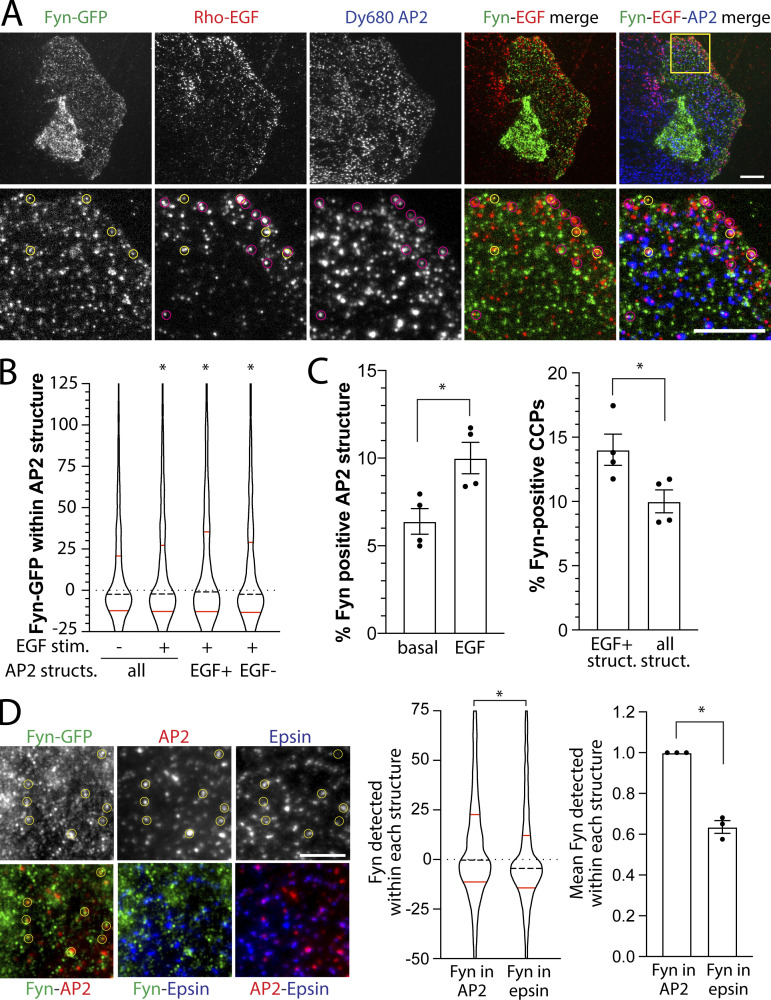
Fyn recruitment to CLSs may have some preference for CLSs with specific signaling receptors as cargo, such as EGFR, which are distinct from CLSs harboring transferrin receptor (Delos Santos et al., 2017). To examine this possibility, we used transient transfection of the lowest detectable levels of Fyn-GFP followed by stimulation with Rhodamine-conjugated EGF (Rho-EGF) to selectively label CLSs harboring ligand-bound EGFR, and finally detection of CLSs by immunofluorescence staining of AP2 (Fig. 6 A). When examining all CLSs regardless of EGFR recruitment, the distribution of Fyn-GFP intensities detected within CLSs shows that the majority of CLSs had little or no Fyn-GFP enrichment (seen by the large number of CLSs with near-zero levels of Fyn); however, a long “tail” of elevated Fyn-GFP intensities within CCPs shows that Fyn is detected in a subset of CLSs (Fig. 6 B). As the levels of Fyn-GFP detection within CLSs was determined by the amplitude of a Gaussian model at the position of detection of AP2 for each structure, negative values for Fyn-GFP localization within a CLS represents depletion of Fyn-GFP relative to local background in that structure. The distribution of Fyn-GFP detected within EGF+ CLSs appears elevated compared with EGF− CLSs (Fig. 6 B), suggesting that EGF stimulation triggers a recruitment of Fyn-GFP only to a subset of CLSs.
Figure 6.
Fyn is recruited to a distinct subset of clathrin structures at the plasma membrane. (A–D) ARPE-19 cells were transfected with a plasmid encoding Fyn-GFP, stimulated with 20 ng/ml Rho-EGF for 5 min, and then subjected to immunofluorescence staining to detect AP2 (A–C) or AP2 and Epsin1 (D), followed by imaging by TIRFM. (A) AP2 was labeled with DyLight 680. Representative TIRFM images, scale bar = 10 μm (top) or 5 μm (bottom). Images obtained by TIRFM were subjected to automated detection and analysis of AP2 structures, allowing quantification of Fyn-GFP or Rho-EGF in each detected object. (B) Shown is the distribution of Fyn-GFP in individual AP2 structures within an experiment. These data are shown for all detected AP2 structures or in AP2 structures (structs.) sorted by Rho-EGF content therein (EGF+ or EGF−; see Materials and methods), as indicated. *, P < 0.05, relative to all AP2 structures in non–EGF-stimulated (stim.) condition. (C) AP2-labeled structures were sorted by Fyn-GFP content and/or Rho-EGF levels therein (see Materials and methods). Shown are the measurements of the percentage of AP2 structures positive for Fyn-GFP (left) or AP2 structures positive for Fyn-GFP sorted by Rho-EGF content (right). In each case, the data shown represent the mean ± SEM of Fyn-GFP- or EGF-positive AP2 structures determined in individual experiments (each independent experiment value determined from >12 cells per condition, depicted as points). The total number of AP2-labeled structures and cells analyzed were 10,474 and 44 (total from three independent experiments). *, P < 0.05. (D) AP2 was labeled with Alexa Fluor 405 and Epsin1 with Cy3. Representative TIRFM images; scale bar = 5 μm. Images obtained by TIRFM were subjected to automated detection and analysis of AP2 or Epsin1 structures, allowing quantification of Fyn-GFP in each detected object. Shown (middle) is the distribution of Fyn-GFP in individual AP2 or Epin1 structures within an experiment. Also shown (right) is mean ± SEM of Fyn-GFP–positive AP2 or Epsin1 structures determined in individual experiments (each independent experiment value determined from >12 cells per condition, depicted as points). *, P < 0.05. The total number of structures and cells analyzed were 21,110 AP2-labeled structures and 40 cells in three independent experiments.
To determine whether Fyn-GFP recruitment occurs preferentially to the subset of CLSs that also contain EGFR, we sorted CLSs into Fyn+ and Fyn− cohorts based on an arbitrary but systematic threshold for Fyn-GFP, as we have done previously for other CLS components (Lucarelli et al., 2017; Delos Santos et al., 2017). As expected, we observed that EGF stimulation significantly increased the percentage of Fyn-GFP–positive CLSs (Fig. 6 C, left). Further, a significantly higher percentage of EGF+ CLSs were also Fyn+ compared with the percentage of all CLSs that were Fyn+ (not sorted by EGF status; Fig. 6 C, right), indicating that Fyn recruitment occurs preferentially, but not exclusively, to the subset of CLSs that also contain EGFR.
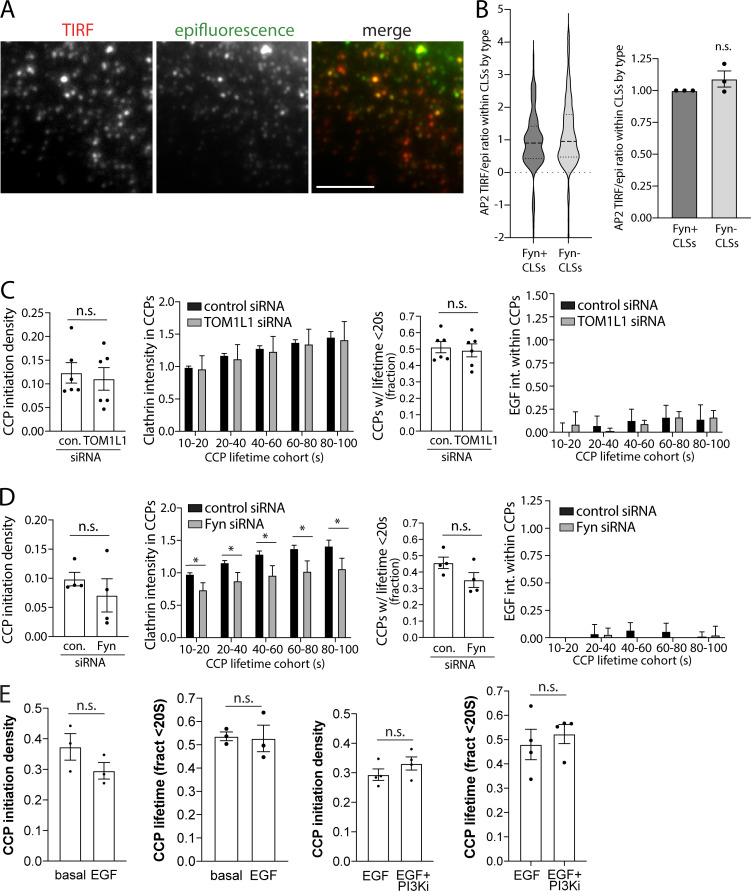
A recent study revealed the existence of AP2-independent CCPs, which were dependent instead on other clathrin adaptor proteins such as Epsin1 (Pascolutti et al., 2019). Interestingly, loss of AP2 but not that of Epsin proteins impaired EGF-stimulated Akt phosphorylation, suggesting the existence of subpopulations of CCPs defined by Epsin1 versus AP2 composition, with the latter contributing to regulation of EGFR signaling. To determine whether Fyn may be preferentially detected in Epsin1- versus AP2-positive plasma membrane structures, we examined the localization of endogenous Epsin1 (Fig. S2 D) and AP2 in Fyn-GFP–expressing cells (Fig. 6 D). Automated detection of AP2 or Epsin1 puncta, followed by measurement of Fyn-GFP recruitment in each, showed that Fyn-GFP is significantly more enriched in AP2 versus Epsin1 puncta, both at the level of individual structures (Fig. 6 D, middle) and when considering the mean level of Fyn-GFP in each type of structure obtained from independent experiments (Fig. 6 D, right). These results support the model that EGF stimulation triggers Fyn-GFP recruitment selectively to a unique subset of CCPs.
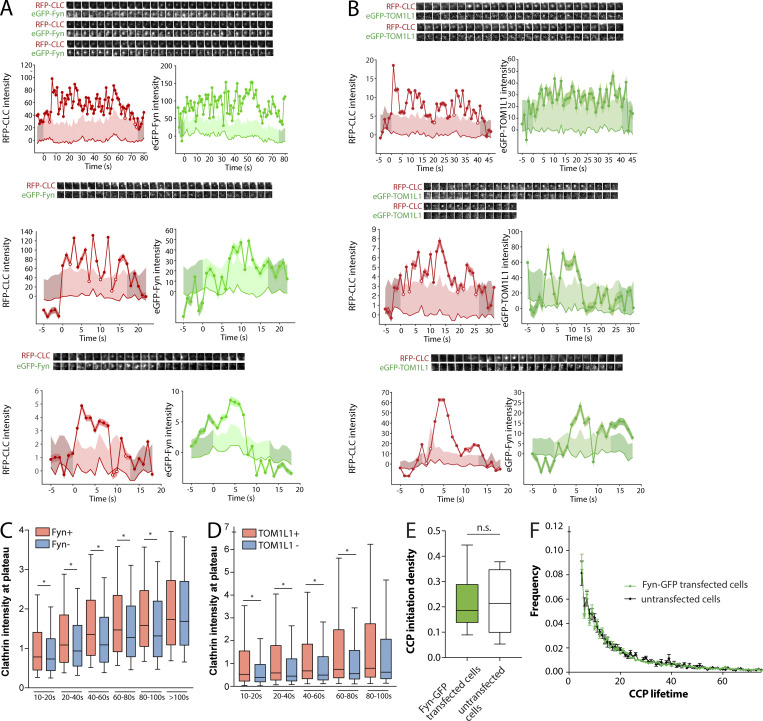
We next examined how TOM1L1 or Fyn was recruited to distinct CCP subpopulations based on CCP dynamics. We first used time-lapse imaging of EGF-stimulated RPE-RFP-CLC cells expressing the lowest detectable levels of Fyn-GFP. As in experiments in fixed cells, this strategy allowed ready observation of Fyn-GFP–positive CLSs (Fig. 7 A). These time-lapse image series were subjected to automated detection, tracking, and analysis, which allowed identification of Fyn+ and Fyn− CCPs (Aguet et al., 2013). This method was previously described and validated for classification of CCPs by recruitment of many different types of proteins including dynamin2 (Aguet et al., 2013) and receptor ligands transferrin and EGF (Delos Santos et al., 2017). Using this approach in cells also stimulated with unlabeled EGF, 15.6% ± 1.1% of CCPs were Fyn+, consistent with our analysis of fixed cells (Fig. 6 C). Individual CCP intensity traces revealed that many Fyn+ CCPs exhibited Fyn recruitment throughout the lifetime of the structure (Fig. 7 B), while others exhibited more transient detection of Fyn recruitment for a subset of the CCP lifetime (Fig. S3 A), although the apparent transient localization of Fyn to CCPs could be due to low signal of Fyn-GFP. Expression of Fyn-GFP at lowest detectable levels did not alter CCP dynamics relative to untransfected cells (Fig. S3, E and F). Using a similar live-cell imaging approach, we observed that eGFP-TOM1L1 was similarly recruited to a subset (8.4 ± 0.6%) of CCPs (Figs. 7, C and D; and Fig. S3 B).
Figure 7.
TOM1L1 and Fyn are recruited to a subset of CCPs with distinct lifetimes yet do not have a major contribution to clathrin assembly or CCP initiation or lifetime. (A–H) ARPE-19 cells stably expressing Tag-RFP-T-CLC were transfected with either Fyn-GFP (A, B, E, and F) or eGFP-TOM1L1 (C, D, G, and H) and then imaged using time-lapse TIRFM. (A and C) Representative single-image frames showing Fyn-GFP or eGFP-TOM1L1 and Tag-RFP-T-CLC taken from a sample time lapse. Scale bar = 5 μm. (B and D) Representative Fyn+ or TOM1L1+ CCP intensity traces, showing Fyn-GFP or eGFP-TOM1L1 and Tag-RFP-T clathrin signals (thick line), local background fluorescence (thin line), and 95% confidence interval of signal relative to background (shaded region). Also shown are fluorescence image time series (1.8-μm image width) corresponding to clathrin and TOM1L1/Fyn centered at the detected object. The representative image time-lapses show sequential frames obtained at intervals of 1 s and correspond to the timeline of the CCP plotted beneath. Additional traces of individual CCPs are shown in Fig. S3. (E–H) Time-lapse image series were subjected to automated detection, tracking, and analysis of CCPs, which allows sorting of CCPs by Fyn or TOM1L1 status, as described in Materials and methods. (E and G) Frequency distribution of CCP lifetimes by Fyn-GFP (E) or eGFP-TOM1L1 (G) status. (F and H) Mean intensity of Fyn-GFP (F) or eGFP-TOM1L1 (H) in each type of CCP (sorted by automated analysis first by Fyn/TOM1L1 status, then by CCP lifetime). The number of CCPs and cells analyzed, respectively, for each condition are as follows: Fyn-GFP expressing cells (18,383 CCPs, 17 cells); eGFP-TOM1L1–expressing cells (28,234 CCPs, 19 cells). (I and J) ARPE-19 cells stably expressing eGFP-clathrin were treated with siRNA targeting TOM1L1, Fyn, or nontargeting siRNA (control). Cells were then treated with 5 ng/ml Rho-EGF and imaged using time-lapse TIRFM, followed by automated detection, tracking, and analysis of CCPs, which allows sorting of CCPs by Rho-EGF status, as described in Materials and methods. (I–P) Shown selectively for EGF+ CCPs are the CCP initiation density (I and M); the intensity of eGFP-clathrin within CCPs, classified by CCP lifetime cohorts (J and N); the frequency of CCPs with lifetimes <20 s (K and O); and the Rho-EGF intensity within CCPs, classified by CCP lifetime cohorts (L and P). For I–P, shown are the average values per cell obtained from individual experiments (dots) and/or the mean (bars) ± SEM of these measurements. *, P < 0.05. The results of similar analysis of EGF− CCPs is shown in Fig. S4, C and D. For I–L, the number of CCPs and cells analyzed in six independent experiments for each condition are as follows: control siRNA, EGF (43 cells, 18,997 CCPs) and TOM1L1 siRNA, EGF (42 cells, 19,164 CCPs). For M–P: the number of CCPs and cells analyzed in four independent experiments for each condition are as follows: control siRNA, EGF (41 cells, 14,527 CCPs) and Fyn siRNA, EGF (37 cells, 9,692 CCPs).
Figure S3.
Additional information on live-cell imaging experiments shown in Fig. 7. (A and B) Cell samples were treated, prepared, and subjected to time-lapse TIRFM imaging as per Fig. 7, A–H. Shown are additional representative Fyn+ (A) or TOM1L1+ (B) CCPs showing fluorescence images (1.8-μm image width) corresponding to clathrin and Fyn/TOM1L1 centered at the detected object (top), as well as quantification of Fyn-GFP (A) or eGFP-TOM1L1 (B) and RFP-CLC within the object (bottom). (C–F) Expression of Fyn-GFP and eGFP-TOM1L1 results in recruitment of each to CCPs but does not alter CCP population dynamics under the conditions examined. Cell samples were treated, prepared, and subjected to time-lapse TIRFM imaging as per Fig. 7, A–H. (C and D) Shown are levels of Tag-RFP-T-clathrin in CCPs, sorted by detection of Fyn-GFP (C) or eGFP-TOM1L1 (D) as well as lifetime cohorts, as in Fig. 7, A–H; shown as median (line), 25th/75th percentile (boxes), and full range (whiskers). *, P < 0.05. eGFP-TOM1L1 and Fyn-GFP are preferentially recruited to larger CCPs. (E and F) The TIRFM time-lapse image series of cells transfected with Fyn-GFP (as in Fig. 7, A–H) was acquired alongside similar image series in RPE cells expressing only Tag-RFP-T-CLC (stably) but not Fyn-GFP. Each time-lapse TIRFM dataset of cells expressing Fyn-GFP or not transfected with Fyn-GFP (untransfected cells) was subjected to automated detection, tracking, and analysis of CCPs. (E) The CCP initiation density, depicting median (bar), interquartile range (boxes), and full range (whiskers). (F) The frequency distribution of CCP lifetimes. The expression of Fyn-GFP does not grossly alter the initiation density or lifetime distribution of the total cellular population of CCPs. The number of CCPs and cells analyzed, respectively, for each condition are as follows: Fyn-GFP–expressing cells (18,383 CCPs, 17 cells) and cells not expressing Fyn-GFP (16,412 CCPs, 6 cells).
To further understand how Fyn or TOM1L1 may be recruited to unique clathrin structures that may be specialized for signaling regulation, we first examined the lifetime and size of Fyn+ versus Fyn− structures. CCPs exhibit substantial heterogeneity in lifetimes and size (Aguet et al., 2013; Mettlen and Danuser, 2014; Kadlecova et al., 2017). Whereas Fyn was detected in CCPs over a broad range of lifetimes, the lifetime distribution of Fyn+ CCPs is skewed towards longer lifetimes compared with that of Fyn− CCPs (Fig. 7 E); a similar observation was made when examining TOM1L1+ CCPs versus those devoid of TOM1L1 (Fig. 7 G). CCPs positive for Fyn-GFP or eGFP-TOM1L1 also had significantly more RFP-clathrin intensity (Fig. S3, C and D). Fyn-positive CLSs had similar ratios of AP2 intensity in corresponding pairs of TIRFM and epifluorescence images (Fig. S4, A and B), indicating that the observed difference in clathrin intensity in CCPs by TIRF-M in Fyn+ CCPs was not due to differences in CCP curvature (Aguet et al., 2013), and instead was due to differences in CCP size. This is consistent with the preferential recruitment of Fyn-GFP to CCPs that also harbor EGFR, as EGFR+ CCPs exhibit longer lifetimes and larger size relative to all CCPs (Delos Santos et al., 2017). This further supports the specific recruitment of TOM1L1 and Fyn to unique subpopulations of CCPs, rather than broad recruitment to all CCPs.
Figure S4.
Additional information about TOM1L1 and Fyn recruitment to CCPs. (A and B) CLSs that recruit Fyn do not exhibit defects in curvature generation. RPE cells were transfected with Fyn-GFP, stimulated with (unlabeled) 20 ng/ml EGF for 5 min, fixed, and subjected to immunofluorescence staining of AP2. Scale bar = 5 µm. (A) Shown are representative images of AP2 staining detected by TIRFM and wide-field epifluorescence microscopy. Images were subjected to automated detection and analysis of AP2 structures, allowing quantification of each protein within each CLS (detected via AP2). (B) Shown (left) is the distribution of measurements of the ratio of AP2 intensity obtained by TIRFM to that obtained by epifluorescence microscopy within individual CLSs depicted as violin plot, as well as median (long dashed line) and 25th/75th percentiles (short dashed line). Also shown (right) are the mean AP2 TIRF/epifluorescence ratio of CLSs in individual experiments (each experiment value determined from >15 cells per condition, depicted as points) as mean ± SEM. The number of CLSs and cells analyzed, respectively, is 65,921 and 115, taken from three independent experiments. (C and D) Silencing of TOM1L1 or Fyn has minor effect on dynamics of EGFR-negative CCPs. Cell samples were treated, prepared, and subjected to time-lapse TIRFM imaging as per Fig. 7. ARPE-19 cells stably expressing eGFP-clathrin were treated with siRNA targeting TOM1L1, Fyn, or nontargeting siRNA (control [con.]). Cells were then treated with 5 ng/ml Rho-EGF and imaged using time-lapse TIRFM, followed by automated detection, tracking, and analysis of CCPs, which allows sorting of CCPs by Rho-EGF status, as described in Materials and methods. Shown selectively for EGF− CCPs are the CCP initiation density, intensity of eGFP-clathrin within CCPs, classified by CCP lifetime cohorts, frequency of CCPs with lifetimes <20 s, and Rho-EGF intensity within CCPs, classified by CCP lifetime cohorts. For measurements of CCP initiation density and lifetimes, shown are the average value per cell obtained from individual experiments (dots) and/or the mean (bars) ± SEM of these measurements. The results of similar analysis of EGF+ CCPs is shown in Fig. 7, I–P. Silencing of TOM1L1 has no effect on these aspects of CCP dynamics, while silencing Fyn has a modest effect on recruitment of clathrin to CCPs. *, P < 0.05. (E) Pictilisib treatment does not impact CCP initiation or lifetime distribution. Cell samples were treated, prepared, and subjected to time-lapse TIRFM imaging as per Fig. 9, A–E. ARPE-19 cells stably expressing eGFP-clathrin were engineered using the Sleeping Beauty transposon system to allow doxycycline-inducible expression of Fyn-iRFP713. Cells were treated with 10 μM Pictilisib (PI3Ki) for 30 min or 5 ng/ml EGF for 5 min, followed by time-lapse imaging using TIRFM. Time-lapse image series were subjected to automated detection, tracking, and analysis of CCPs, which allows sorting of CCPs by Fyn or TOM1L1 status, as described in Materials and methods. Shown is the mean CCP initiation density and the fraction of CCPs with lifetimes <20 s, comparing basal and EGF-treated cells or cells treated with both Pictilisib and EGF (EGF+PI3Ki) or EGF alone. Pictilisib treatment does not impact CCP initiation rate or lifetime distribution.
Previous studies found that while the overall population EGFR+ CCPs had longer lifetimes than EGFR− CCPs, the abundance of EGFR was higher in short-lived than long-lived CCPs (Delos Santos et al., 2017; Rosselli-Murai et al., 2018). The lipid phosphatase PTEN had similar enrichment in short-lived CCPs versus longer-lived CCPs (Rosselli-Murai et al., 2018). This suggests that while the abundance of CCP structural proteins such as AP2 and clathrin scale with a CCP lifetime (Loerke et al., 2011), some nonstructural regulators of CCP dynamics or receptor signaling may not, and are instead recruited to CCPs based on molecular determinants other than clathrin or AP2 abundance within CCPs. To determine whether TOM1L1 and Fyn recruitment to CCPs merely reflects CCP lifetime and/or clathrin content, we examined their levels in CCP lifetime cohorts. The mean level of GFP-Fyn detected over the entire lifetime of relatively short-lived (10–20 and 20–40 s) CCPs was significantly higher than that recruited to longer-lived CCPs (Fig. 7 F), a phenomenon also observed for eGFP-TOM1L1 (Fig. 7 H). Hence, the average Fyn or TOM1L1 recruitment level to CCPs does not scale with CCP lifetime and instead occurs to a higher extent in short-lived CCPs, a phenomenon similar to that observed for EGFR (Delos Santos et al., 2017; Rosselli-Murai et al., 2018). Of note is the efficient sorting of CCPs by Fyn or TOM1L1 composition, as determined by the mean Fyn-GFP (Fig. 7 F) or eGFP-TOM1L1 levels (Fig. 7 H) detected in each cohort of Fyn/TOM1L1-negative CCPs.
The increased abundance of Fyn-GFP and eGFP-TOM1L1 within short-lived versus longer-lived CCPs, together with the observation that overall Fyn+ or TOM1L1+ CCPs have longer lifetimes than Fyn− or TOM1L1− CCPs, respectively, suggests that the extent of TOM1L1 and Fyn recruitment may not impact CCP assembly or lifetime. Instead, these results suggest that TOM1L1 and Fyn are recruited as “passengers” from the perspective of regulation of clathrin assembly and CCP lifetime, such that they may instead only regulate signaling once recruited to CCPs. To probe this, we examined the effect of loss of Fyn or TOM1L1 on CCP dynamics. We used siRNA silencing of TOM1L1 or Fyn in cells expressing eGFP-clathrin and then performed live cell TIRFM in cells also stimulated with 5 ng/ml Rho-EGF. These time-lapse image series were subjected to automated detection, tracking and analysis, which allowed identification of EGF+ and EGF− CCP cohorts (Aguet et al., 2013; Delos Santos et al., 2017). When considering only EGF+ CCPs, TOM1L1 silencing did not significantly impact CCP initiation (Fig. 7 I), clathrin recruitment (Fig. 7 J), CCP lifetime (Fig. 7 K), or the intensity of Rho-EGF within CCPs (Fig. 7 L). While Fyn silencing also did not significantly impact CCP initiation (Fig. 7 M) or lifetime (Fig. 7 O), it did trigger a modest yet significant reduction of clathrin and Rho-EGF recruitment into CCPs, whether these were EGFR+ (Fig. 7, N–P) or EGFR− (Fig. S4, C and D). These results indicate that TOM1L1 and Fyn have limited roles in regulation of CCP initiation, clathrin assembly, lifetime, or EGFR recruitment. Instead, the localization of TOM1L1 and Fyn to a unique subset of CCPs allows regulation of EGFR signaling, perhaps by control of signaling intermediates within CCPs.
TOM1L1 and Fyn are selectively required for modulation of EGF-stimulated Akt2 phosphorylation and recruitment of SHIP2 to clathrin structures
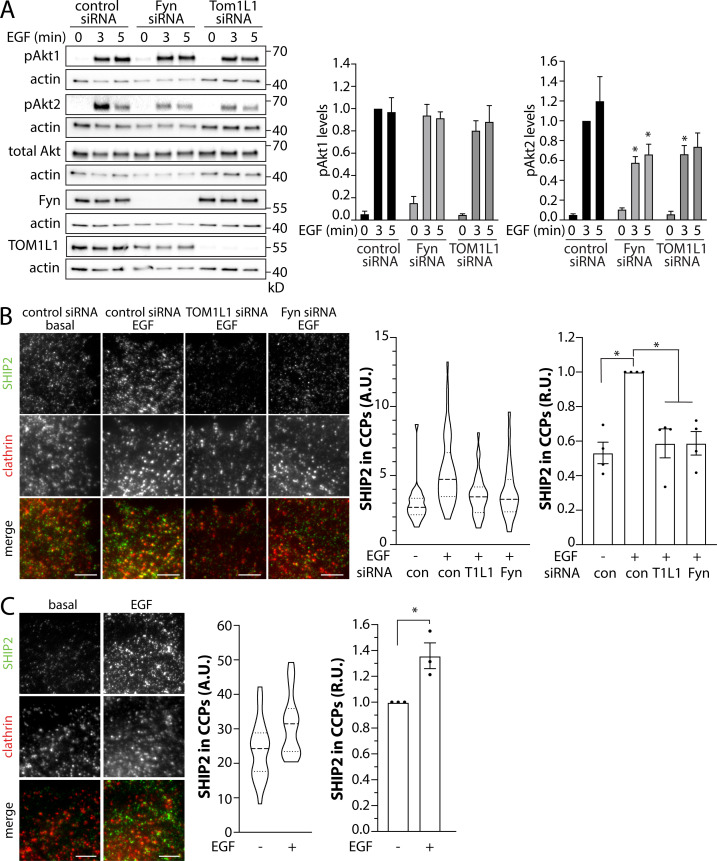
CCPs harbor many enzymes that regulate phosphoinositide dynamics (Sugiyama et al., 2019), including PTEN (Rosselli-Murai et al., 2018) and SHIP2 (Nakatsu et al., 2010). PTEN elicits the turnover of PIP3 to PtdIns(4,5)P2, which terminates PI3K signaling and Akt phosphorylation (Lee et al., 2018). In contrast, SHIP2 elicits turnover of PIP3 to PtdIns(3,4)P2 (Goulden et al., 2018; Liu et al., 2018), a lipid that selectively triggers phosphorylation of Akt2, but not Akt1 or Akt3 (Liu et al., 2018). SHIP2 inhibition slightly increased PDGF-stimulated Akt1 phosphorylation (S473), yet impaired Akt2 phosphorylation (S474; Liu et al., 2018). To resolve how TOM1L1 and Fyn may control phosphoinositide dynamics leading to regulation of Akt signaling by EGFR, we first examined the effect of TOM1L1 or Fyn silencing on EGF-stimulated phosphorylation of specific Akt isoforms. While silencing of TOM1L1 or Fyn did not impact the EGF-stimulated phosphorylation of Akt1, each led to a significant reduction in the phosphorylation of Akt2 (Fig. 8 A). This prompted us to consider that TOM1L1 and Fyn may control SHIP2, given the selective role of this lipid phosphatase in activation of Akt2 (Liu et al., 2018).
Figure 8.
TOM1L1 and Fyn are required for the selective modulation of EGF-stimulated Akt2 phosphorylation and SHIP2 recruitment to clathrin structures. (A–C) ARPE-19 cells (A and B) or MCF10A cells (C) were treated with siRNA targeting TOM1L1 or Fyn or with nontargeting control siRNA, as indicated, followed by stimulation with 5 ng/ml EGF for 5 min. (A) Immunoblotting of whole-cell lysates with antibodies that selectively recognize Akt1 or Akt2 when phosphorylated on S473/S474, total Akt, TOM1L1, Fyn, or actin (loading control). Also shown are the mean ± SEM phospho-Akt1 or Akt2; n = 5; *, P < 0.05, relative to the control siRNA-treated EGF-stimulated conditions at each time point. (B and C) After siRNA transfection, ARPE-19 cells stably expressing Tag-RFP-T clathrin were stained with antibodies recognizing endogenous SHIP2 (labeling with Alexa Fluor 488; B) or MCF10A cells were stained with antibodies to detect endogenous SHIP2 (labeling with Alexa Fluor 488) and clathrin (labeling with Alexa Fluor 647; C) and subjected to imaging using TIRFM. Shown are representative images; scale bar = 5 μm. Images obtained by TIRFM were subjected to automated detection and analysis of CLSs, allowing quantification of SHIP2 in each detected object. Shown (middle) are the measurements of SHIP2 fluorescence intensity within CLSs, showing the distribution of the mean value of individual cells (each cell value determined from >300 CLSs) depicted as a violin plot, as well as median (long dashed line) and 25th/75th percentiles (short dashed line). Also shown (right panels) are the levels of SHIP2 detected with CLSs in individual experiments (each experiment value determined from >15 cells per condition, depicted as points) as mean ± SEM; *, P < 0.05, relative to basal. For B, the number of CLSs and cells analyzed in four independent experiments for each condition are as follows: control siRNA, basal (73 cells, 48,307 CCPs), control siRNA, EGF (116 cells, 88,023 CCPs), TOM1L1 siRNA, EGF (88 cells, 56,918 CCPs); Fyn siRNA, EGF (99 cells, 73,347 CCPs). For C, the number of CLSs and cells analyzed in three independent experiments for each condition are as follows: basal (66 cells, 42,686 CCPs) and EGF-stimulated (57 cells, 35,445 CCPs). con, control. Source data are available for this figure: SourceData F8.
Thus, we next probed how TOM1L1 and Fyn may regulate SHIP2. We used immunofluorescence staining of endogenous SHIP2 in cells also expressing RFP-clathrin, imaging by TIRFM, and automated detection and analysis of CLSs (Fig. 8 B). EGF stimulation triggered a robust increase in SHIP2 recruitment within CLSs, as evident in the distribution of SHIP2 intensities within individual CLSs (Fig. 8 B, middle) as well as the results of the average of multiple independent experiments (Fig. 8 B, right). Importantly, silencing of TOM1L1 or Fyn impaired the EGF-stimulated gain in SHIP2 in CLSs. We observed a similar gain in SHIP2 recruitment to CLSs upon EGF stimulation in MCF10A cells (Fig. 8 C). Taken together, these results suggest that clathrin-localized TOM1L1 and Fyn may impact EGF-stimulated Akt2 phosphorylation via regulation of SHIP2 recruitment to CCPs.
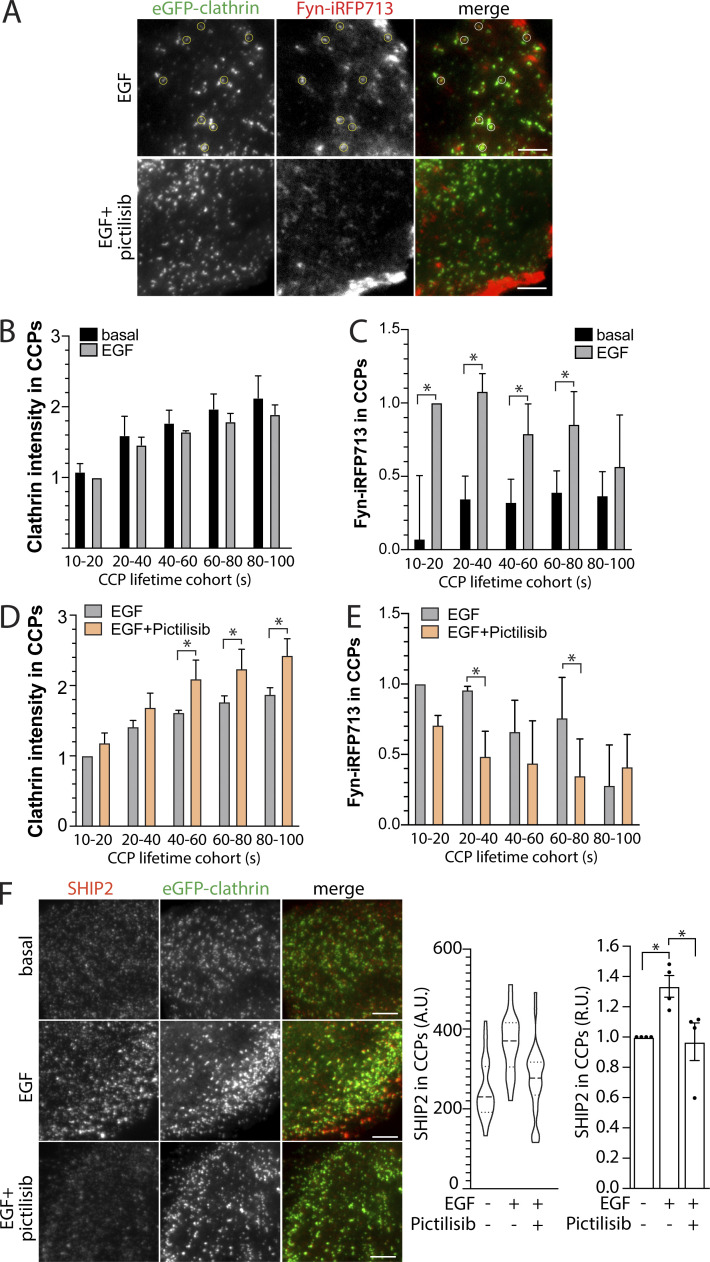
PIP3 has been proposed to regulate CCP dynamics and assembly (Rosselli-Murai et al., 2018; Dambournet et al., 2018), as have the downstream signals activated by PIP3 (Reis et al., 2015; Srinivasan et al., 2018). We next examined the possibility of regulation of Fyn and SHIP2 within CCPs by PI3K signaling. To do so, we used stable cells expressing eGFP-clathrin and induced to express Fyn-iRFP713 by doxycycline treatment (Fig. 5). Similarly to Fyn-GFP (Fig. 3) and endogenous Fyn (Fig. 4), in EGF-stimulated cells Fyn-iRFP713 could be observed within eGFP-clathrin puncta, which appeared disrupted in cells treated with the class I PI3K inhibitor Pictilisib (Fig. 9 A). These time-lapse image series were subjected to automated detection, tracking and analysis (Aguet et al., 2013). While EGF stimulation did not significantly alter the abundance of eGFP-clathrin within CCPs (Fig. 9 B), CCP initiation, or CCP lifetimes (Fig. S4 E), it did trigger an increase in Fyn-iRFP713 within CCPs (Fig. 9 C). Consistent with a role for PIP3 in regulation of CCP assembly, we observed a small but significant increase in CCP size in EGF-stimulated cells upon Pictilisib treatment (Fig. 9 D), although Pictilisib was without effect on CCP initiation or CCP lifetime (Fig. S4 E). Importantly, Pictilisib triggered a reduction of Fyn-iRFP713 within CCPs in EGF-stimulated cells (Fig. 9 E). This suggests that PI3K signaling contributes to regulation of Fyn recruitment to CCPs.
Figure 9.
Pictilisib suppresses EGF-stimulated recruitment of Fyn and SHIP2 to clathrin structures. (A–E) ARPE-19 cells stably expressing eGFP-clathrin were engineered using the Sleeping Beauty transposon system to allow doxycycline-inducible expression of Fyn-iRFP713. Cells were treated with 10 μM pictilisib for 30 min or 5 ng/ml EGF for 5 min, followed by time-lapse TIRFM. (A) Representative single-image frames showing eGFP-clathrin and Fyn-iRFP713. Scale bar = 5 μm. (B–E) Time-lapse image series were subjected to automated detection, tracking, and analysis of CCPs, which allows sorting of CCPs by Fyn or TOM1L1 status, as described in Materials and methods. Shown are the mean intensities of eGFP-clathrin (B and D) or Fyn-iRFP713 (C and E) within each type of CCP (sorted by CCP lifetime). Shown in B–E are the mean (bars) ± SEM of the average value per cell obtained from individual experiments. *, P < 0.05. (F) ARPE-19 cells stably expressing eGFP-clathrin were treated with 10 μM pictilisib for 30 min or 5 ng/ml EGF for 5 min, then stained with antibodies recognizing endogenous SHIP2 and subjected to imaging using TIRFM. Shown are representative images. Scale bar = 5 μm. Images obtained by TIRFM were subjected to automated detection and analysis of CLSs, allowing quantification of SHIP2 in each detected object. Shown (middle) are the measurements of SHIP2 fluorescence intensity within CLSs, showing the distribution of the mean value of individual cells (each cell value determined form >300 CLSs) depicted as violin plot, as well as median (long dashed line) and 25th/75th percentiles (short dashed line). Also shown (right) are the levels of SHIP2 detected with CLSs in individual experiments (each experiment value determined from >15 cells per condition, depicted as points) as mean ± SEM; *, P < 0.05, relative to basal. For B–E, the number of CCPs and cells analyzed in four independent experiments for each condition are as follows: basal, no drug (24 cells, 47,789 CCPs); EGF-stimulated, no drug (24 cells, 42,003 CCPs); and EGF-stimulated, Pictilisib-treated (24 cells, 45,456 CCPs). For F, the number of CLSs and cells analyzed in four independent experiments for each condition are as follows: basal, no drug (102 cells, 55,681 CCPs); EGF-stimulated, no drug (101 cells, 60,614 CCPs); and EGF-stimulated, Pictilisib-treated (93 cells, 55,715 CCPs).
Because Pictilisib suppressed Fyn-iRFP713 recruitment to CCPs, and because Fyn is required for the recruitment of SHIP2 to CCPs, we examined whether class I PI3K may contribute to SHIP2 recruitment to CCPs. EGF stimulation triggered an increase in SHIP2 recruitment to CCPs, which was blunted by treatment with Pictilisib (Fig. 9 F). This indicates that class I PI3K regulates the CCP recruitment of Fyn and SHIP2, and in doing so may promote Akt2 phosphorylation by more than merely supplying PIP3 substrate for SHIP2. Collectively, these experiments suggest that TOM1L1 and Fyn function to regulate SHIP2 recruitment to CCPs and thus regulate Akt2 activation.
Discussion
We previously observed that clathrin structures at the plasma membrane, but not EGFR endocytosis per se, are required for EGFR signaling to Gab1 and Akt (Garay et al., 2015; Lucarelli et al., 2017). Here, we uncover recruitment of TOM1L1 and Fyn to a subset of plasma membrane clathrin structures, and that both Fyn- and clathrin-binding by TOM1L1 are required for EGFR signaling leading to SHIP2 recruitment to CCPs, as well as the selective phosphorylation of Akt2 (Fig. S5). We also find that TOM1L1+ or Fyn+ clathrin structures have unique lifetimes, size, and composition. Therefore, we propose that TOM1L1 and Fyn define a unique subset of signaling-capable plasma membrane clathrin structures required for specific facets of EGFR signaling leading to Akt2 activation.
Figure S5.
Model of regulation of SHIP2 localization and Akt signaling by TOM1L1 and Fyn within specialized CCPs. EGFR signaling activation via engagement of clathrin-localized TOM1L1/Fyn, leading to SHIP2 recruitment to CCPs and Akt2 activation. Please refer to text for details.
Regulation of Akt signaling by clathrin structures
Fyn was required for EGF-stimulated phosphorylation of Y627 on Gab1 (Fig. 1). While Gab1 phosphorylation is required for recruitment and activation of class I PI3K (Mattoon et al., 2004), the recruitment of PI3K occurs to a motif on Gab1 harboring phosphorylated Y472 (Rocchi et al., 1998). Together with the lack of effect of Fyn and TOM1L1 silencing on phosphorylation of Akt1 (Fig. 8 A) that depends on PIP3 produced by class I PI3K (Liu et al., 2018), this suggests that Fyn may regulate Gab1 phosphorylation on Y627, but perhaps not phosphorylation on other sites of Gab1 leading to PI3K activation. Instead, that TOM1L1 and Fyn selectively regulate Akt2 phosphorylation and localization of SHIP2 to CCPs (Fig. 8 B) suggests their involvement in regulation of EGFR signaling subsequent to PI3K-dependent PIP3 production.
PI3K generates PIP3 at the plasma membrane, and this compartment is the major reservoir for its substrate PtdIns(4,5)P2 (Choy et al., 2017). Several lipid phosphatases and kinases act on internalized vesicles resulting in undetectable levels of PtdIns(4,5)P2 therein (He et al., 2017). In contrast to this model, PIP3 was detected on endosomes, in a manner that required the class I PI3K adaptor MAP4 (Thapa et al., 2020). Nonetheless, and consistent with activation of PI3K signaling at the plasma membrane, we previously reported that specific perturbations of dynamin2, which allow plasma membrane clathrin structure formation and receptor recruitment therein but impaired the formation of internalized vesicles, had no effect on Akt phosphorylation, while perturbation of clathrin impaired Akt phosphorylation (Garay et al., 2015). Furthermore, complete arrest of EGFR endocytosis in a cell lacking all three isoforms of dynamin did not impact Akt activation (Sousa et al., 2012). PtdIns(3,4)P2 was detected either exclusively at the plasma membrane (Goulden et al., 2018) or the plasma membrane and endosomes (Liu et al., 2018). These results suggest that PIP3 production, as well as that of subsequent SHIP2-generated PtdIns(3,4)P2, can occur at the plasma membrane, although some amount of these lipids can also be found on endosomes. Our results suggest that after synthesis by SHIP2 localized within clathrin structures (Fig. 8), PtsIns(3,4)P2 and/or associated Akt2 may rapidly diffuse within the plane of the membrane (He et al., 2017), may become immediately engaged with effector proteins, or may become internalized to vesicles or subsequent intracellular compartments (Liu et al., 2018). These findings contribute to the understanding of the close relationship of endocytic and signaling protein complexes and show that the control of SHIP2 localization to clathrin structures requires TOM1L1 and Fyn downstream of EGFR activation.
Consistent with the regulation of signaling by the endocytic machinery, phosphorylated Akt is detected within CCPs (Rosselli-Murai et al., 2018) and is recruited to a subset of early endosomes containing APPL1 and its interaction partner OCRL (Schenck et al., 2008; Erdmann et al., 2007). This suggests that subsequent to initial activation of Akt (and perhaps specifically Akt2) at the plasma membrane in a manner that does not require endocytosis per se, clathrin structures may further control Akt by vesicle traffic of phosphorylated Akt to specific endosomal compartments. Hence, TOM1L1 and Fyn may function within a subset of plasma membrane clathrin structures to regulate SHIP2 recruitment and the selective activation of Akt2, which may precede vesicle scission.
Control of Fyn within plasma membrane clathrin structures by TOM1L1
Many studies have examined the role of Src-family kinases in EGFR signaling, and c-Src, Fyn, and Yes can all be phosphorylated after EGF stimulation (Wilde et al., 1999). Src-family kinases can phosphorylate EGFR at Y845 to promote activation of receptor signaling, in particular for EGFR transactivation in the absence of ligand (Sato, 2013). Perturbations of TOM1L1 or Fyn had no effect on EGFR autophosphorylation and downstream signaling to Erk, thus indicating that the role of Fyn and TOM1L1 in EGFR signaling is unlikely to be due to control of EGF-stimulated EGFR phosphorylation. Instead, these results indicate that TOM1L1 and Fyn control EGFR signaling at a stage downstream of EGFR autophosphorylation, leading to regulation of Akt activation.
TOM1L1 is required for the recruitment of Fyn to plasma membrane clathrin structures (Fig. 4). Perturbation of the clathrin-binding (Liu et al., 2009b) or Fyn-binding (Seykora et al., 2002) abilities of TOM1L1 impairs EGF-stimulated Akt phosphorylation (Fig. 5 A). TOM1L1 interacts with clathrin via a motif distinct from the clathrin box that mediates interaction of many proteins with the N-terminal β-propeller structure of clathrin (Liu et al., 2009b). Instead, the 447FDPL450 motif on TOM1L1 interacts with the clathrin C-terminus (within residues 1,325–1,675; Collin et al., 2007; Liu et al., 2009b). An additional 401LQPVSL406 motif on TOM1L1 may also enhance binding to clathrin (Collin et al., 2007). Consistent with our observations (Fig. 3 B), TOM1L1 may thus be constitutively recruited to some plasma membrane clathrin structures, such that EGF-stimulated Y460 phosphorylation on TOM1L1 (Liu et al., 2009b) or some other cue allows stimulus-specific binding of Fyn to TOM1L1 and thus Fyn recruitment to CCPs.
In addition to recruitment of Fyn to a subset of clathrin structures, TOM1L1 binding enhances the activity of Fyn and Src (Li et al., 2005), suggesting that Fyn activation may be enhanced within these clathrin nanodomains. While we find that TOM1L1 is required to engage signaling to Akt by EGFR through its interactions with clathrin, TOM1L1 may also serve to negatively regulate signaling from platelet-derived growth factor receptor, by sequestration of Src family kinases from caveolae (Collin et al., 2007). Hence, TOM1L1 localization to clathrin structures may have other complex roles that distinctly impact signaling of different receptors. Moreover, while TOM1L1 is required for internalization of EGFR in A431 cells (Liu et al., 2009b) that have very high levels of EGFR expression, it is not required for internalization in pig aortic endothelial cells (Goh et al., 2010), engineered to express physiological EGFR levels similar to those in the ARPE-19 cells examined here. Hence, in addition to the key role in regulation of Akt activation by EGFR that we observe here, TOM1L1 may contribute to EGFR endocytosis when a high capacity of receptor internalization is required.
A subset of CCPs function as signaling platforms
We detected Fyn-GFP within a subset of plasma membrane clathrin structures (Figs. 3 A; 4; 7, A and B; and S3 A). This represented specific Fyn recruitment, since (a) silencing TOM1L1 abolished the EGF-stimulated gain in Fyn in clathrin structures (Fig. 4), (b) many Fyn-GFP and clathrin structures exhibited overlap when examined by the higher resolution afforded by SIM (Figs. 3 C and S1), (c) Fyn-positive clathrin structures have elevated lifetimes and size relative to Fyn-negative clathrin structures (Fig. 7), and (d) Fyn was more enriched in AP2-positive than Epsin-positive structures at the plasma membrane (Fig. 6). As such, we propose that a small subset of plasma membrane clathrin structures form with the ability to recruit Fyn, and thus represent specialized signaling-capable nanodomains.
The recruitment of Fyn to clathrin structures by TOM1L1 does not appear to play a major role in the regulation of clathrin assembly and CCP dynamics (Fig. 8), suggesting that TOM1L1 and Fyn are captured by some nascent CCPs to regulate signaling, but do not play a major role in regulating CCP dynamics. Src-family kinases can phosphorylate clathrin heavy chain, thus controlling clathrin structure dynamics (Wilde et al., 1999). Since we do not observe changes in CCP initiation or dynamics following TOM1L1 or Fyn silencing, this suggests that other Src-family kinases or other cellular contexts are required for regulation of clathrin dynamics involving clathrin phosphorylation.
The selective recruitment of Fyn to a subset of CCPs adds a new dimension to the heterogeneous composition and properties of these structures. For example, clathrin structures that contain EGFR and other specific signaling receptors are distinctly regulated by calcium signaling (Delos Santos et al., 2017; Reis et al., 2017), while the size and/or lifetimes of other subsets of CCPs harboring LDL receptor (Mettlen et al., 2010) or specific GPCRs (Puthenveedu and von Zastrow, 2006) are also uniquely regulated. Our work is also consistent with the formation of molecularly distinct subpopulations of CCPs based on AP2 recruitment (Pascolutti et al., 2019). Future work that uncovers the mechanisms that govern the distinct fates of different CCPs, for either specialized internalization of specific cargo or signaling phenomena, will thus be of great interest.
We also found that Pictilisib, an inhibitor of class I PI3K, had a modest effect on CCP assembly (increasing CCP size; Fig. 9 D), yet impaired the recruitment of Fyn-iRFP713 and SHIP2 to CCPs (Fig. 9, D and E). Other studies have reported more significant effects on CCP dynamics upon altering PIP3 dynamics by control of the 3-phosphatase PTEN or addition of exogenous PIP3 (Rosselli-Murai et al., 2018), or by treatment with the PI3K pan-class inhibitor LY2942002 (Dambournet et al., 2018). Pictilisib (GDC-0941) is specific for inhibition of class I PI3K (Folkes et al., 2008), so some of the effects of LY2942002 on CCP dynamics may be due to inhibition of class II or III PI3K. Further, as LY2942002 has distinct effects on CCP dynamics in specific cells (Dambournet et al., 2018) PI3K signaling may distinctly regulate CCP initiation, assembly, and turnover in a cell-dependent manner. Akt activated downstream of EGFR can in turn regulate CCP dynamics and EGFR signaling via glycogen synthase kinase 3β–dependent phosphorylation of dynamin1 (Reis et al., 2015; Srinivasan et al., 2018); however, we found that silencing dynamin1 did not impact EGFR endocytosis or signaling in ARPE-19 cells (Delos Santos et al., 2017), consistent with context-dependent regulation of CCP dynamics downstream of PI3K activation. Our results nonetheless indicate that some aspects of CCP formation, specifically, the EGF-stimulated recruitment of Fyn and SHIP2 may be class I PI3K-dependent. This further suggests that class I PI3K may be part of a feed-forward mechanism that promotes the formation of SHIP2-positive CCPs to control PtdIns(3,4)P2 dynamics and signaling downstream of PI3K activation.
The class II PI3K PI3KC2α is also recruited to CCPs, even in the absence of growth factor stimulation, and regulates CCP dynamics through the localized production of PtdIns(3,4)P2 (Posor et al., 2013; Wang et al., 2020) The levels of PtdIns(3,4)P2 produced by PI3KC2α locally within CCPs may be regulated to be just sufficient to recruit clathrin endocytic proteins such as SNX9 to CCPs (Schöneberg et al., 2017), suggesting that upon EGF stimulation, SHIP2 may enhance PtdIns(3,4)P2 production beyond that produced by PI3KC2α, thus supporting Akt2 phosphorylation.
The recruitment of Fyn to form signaling-capable clathrin structures to regulate EGF-stimulated Akt phosphorylation adds to the growing appreciation of the ability of clathrin structures to directly contribute to signal transduction. The PIP3 phosphatase PTEN is also recruited to a subset of clathrin structures and regulates the lifetime of clathrin coated pits (Rosselli-Murai et al., 2018). This suggests that clathrin structures may spatially link Fyn-driven signals that lead to SHIP2 recruitment and activation of Akt2 and the negative regulation of PIP3 signaling by PTEN. In addition to control of PI3K signaling by EGFR, clathrin structures regulate PI3K-Akt signaling by LPA1 stimulation (Leyton-Puig et al., 2017), as well as other signals by certain GPCRs (Eichel et al., 2016, 2018).
In summary, we identified that a subset of plasma membrane clathrin structures recruit TOM1L1 and Fyn and thus have unique properties and capabilities to regulate EGFR signaling. These signaling-capable clathrin structures may be unique nanodomains (Delos Santos et al., 2015; Lu and Fairn, 2018) that have an important role in control of phosphoinositide-regulating enzymes such as SHIP2, and in doing so, gate activation of Akt signaling before or concomitantly with receptor internalization.
Materials and methods
Materials
Antibodies recognizing specific proteins were as follows (with species and catalog numbers indicated for each): phospho-EGFR (pY1068, rabbit monoclonal, 3777), phospho-Gab1 (pY627, rabbit monoclonal, 3233), pAkt1 (S473, rabbit monoclonal, 9018), pAkt2 (S474, rabbit monoclonal, 8599), pAkt (S473, pan-isoform, for immunofluorescence, rabbit monoclonal, 4060), total Akt (pan isoform, mouse, 2920), SHIP2 (rabbit monoclonal, 2869), and Fyn (for IB, rabbit, 4023) obtained from Cell Signaling Technology; phospho-Akt (pS473, pan isoform for Western blotting, 44-621G) obtained from Life Technologies; AP2 (AP6, mouse monoclonal, ab2730), Fyn (for IF, rabbit monoclonal, ab125016), and Epsin1 (mouse monoclonal, ab75879) obtained from Abcam; and anti-actin and anti-clathrin heavy chain (TD.1) used for immunoblotting obtained from Santa Cruz Biotechnology. Fluorophore-conjugated or HRP secondary antibodies were from Jackson ImmunoResearch. Rho-EGF was generated in-house as previously described (Lucarelli et al., 2017).
WT human retinal pigment epithelial cells (ARPE-19; RPE cells herein) and ARPE-19 cells stably expressing CLC fused to enhanced GFP (RPE-GFP-CLC) or fused to TagRFP-T (RPE-RFP-CLC) were previously described (Bone et al., 2017; Aguet et al., 2013; Delos Santos et al., 2017). Cells were cultured in DMEM/F12 (Life Technologies) supplemented with 10% FBS (Life Technologies), 100 U/ml penicillin, and 100 µg/ml streptomycin (Life Technologies) at 37°C and 5% CO2.
MCF10A cells were cultured as previously described (Debnath et al., 2003). Briefly, MCF10A cells were cultured in DMEM/F12 medium supplemented with 5% horse serum (Life Technologies), 20 ng/ml EGF (Peprotech), 0.5 mg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml cholera toxin (Sigma-Aldrich), 10 μg/ml insulin (Sigma-Aldrich), and penicillin/streptomycin (Life Technologies) at 37°C and 5% CO2.
cDNA constructs for transient transfection
sfGFP-FYN-N-10 (sfGFP is superfolder GFP with enhanced stability and folding kinetics, derived from eGFP [Pédelacq et al., 2006]; herein we refer to this construct as Fyn-GFP) was a gift from Michael Davidson (Florida State University, Tallahassee, FL, plasmid 56294; Addgene). eGFP-tagged TOM1L1 (used in Figs. 3 B and 7) was generated by seamless cloning. Briefly, human TOM1L1 (pDNR-LIB, plasmid number 4612157, purchased from Dharmacon) was amplified by PCR using primers as follows: 5′-CGAGCTGTACAAGGGACTCAGATCTAGATCACAGATGGCGTTTGG-3′ and 5′-GCAGAATTCGAAGCTCTATTATTTACTTAAG-3′, and eGFP-C1 (Clontech) was amplified using primers as follows: 5′-CCAAACGCCATCTGTGATCTAGATCTGAGTCCCTTGTACAGCTCG-3′ and 5′-CTTAAGTAAATAATAGAGCTTCGAATTCTGC-3′. The resulting PCR products were mixed, subjected to digestion by Dpn1, and used to transform DH5α-competent Escherichia coli cells, followed by selection of resistant colonies and verification of GFP-TOM1L1 by sequencing.
Transient plasmid and siRNA transfections
cDNA transfections were performed using Lipofectamine 2000 (Life Technologies) as per manufacturer’s instructions and as previously described (Bone et al., 2017). Cells were washed in 1× PBS, and medium was replaced with Opti-MEM medium (Gibco, Life Technologies). Lipofectamine 2000 reagent (3 μl/well) and cDNA (1 µg/well) were each mixed separately in 50 μl OptiMem, combined in a 1:1 ratio, and incubated at room temperature for 10–15 min before pipetting the mixture dropwise onto plated cells in Opti-Mem. Experiments were performed 16–20 h after transfection.
siRNA transfections were performed using custom-synthesized siRNAs using RNAiMAX transfection reagent (Life Technologies) as per manufacturer’s instructions. Briefly, each siRNA was transfected at a concentration of 220 pmol/liter with transfection reagent in Opti-MEM Medium. Cells were incubated with the siRNA complexes for 4 h, after which cells were washed and replaced in regular growth medium. siRNA transfections were performed twice, 72 and 48 h before each experiment. Sequences used were as follows (sense): control (nontargeting), CGUACUGCUUGCGAUACGGUU; Fyn, AGGAAGAGCUCUGAAAUUAUU; TOM1L1, GUGAGAAACUGAAUGUAUUUU; and Epsin, ACUCAGAGGCGGAGAUCAA.
Stable transfections using Sleeping Beauty transposon system
pSBtet-BP was a gift from Eric Kowarz (Goethe-University of Frankfurt, Frankfurt, Germany, plasmid 60496; Addgene; http://n2t.net/addgene:60496; RRID:Addgene_60496; Kowarz et al., 2015). pCMV(CAT)T7-SB100 was a gift from Zsuzsanna Izsvak (Max Delbrück Center for Molecular Medicine, Berlin-Buch, Germany, plasmid 34879; http://n2t.net/addgene:34879; Addgene; RRID:Addgene_34879; Mátés et al., 2009). An oligonucleotide encoding eGFP fused to TOM1L1 were generated by BioBasic, using the ORF sequence of eGFP, followed by the sequence encoding a spacer peptide (5′-GGGGGGTCTGGTGGCAGTGGAGGGGGATCC-3′), followed by the ORF sequence of human TOM1L1, as per GenBank accession number NM_005486. This oligonucleotide sequence was subcloned into pSBtet-BP to generate pSBtet-BP-eGFP-TOM1L1 (WT). From this plasmid, Fyn-binding defective (Y460F) and clathrin-binding defective (447FDPL450AAAA) mutant TOM1L1 constructs were derived by BioBasic by site-directed mutagenesis. An oligonucleotide encoding Fyn-iRFP713 was synthesized using the sequence of human Fyn (GenBank accession number NM_002037), followed by the sequence encoding a spacer peptide (5′-GGGGGGTCTGGTGGCAGTGGAGGGGGATCC-3′), followed by the ORF sequence for iRFP713 (Shcherbakova and Verkhusha, 2013), and inserted into pSBtet-BP to generate pSBtet-BP-iRFP713-Fyn.
pSBtet-BP plasmids encoding various eGFP-TOM1L1 or Fyn-iRFP713 constructs alongside pCMV(CAT)T7-SB100 were cotransfected into ARPE-19 cells using FuGene HD transfection reagent, as per manufacturer’s protocol (Promega), followed by selection of stably engineered cells in media supplemented with 2 µg/ml puromycin for a period of 2–3 wk.
Cell and inhibitor treatments
All cells were serum deprived for 1 h before experimental assays unless otherwise stated. Cells were treated with 20 μm Pictilisib (unless otherwise indicated) or a corresponding volume of DMSO (vehicle control) for 20 min before stimulation with EGF (human; Life Technologies) or other experiments, as indicated. All inhibitor treatments were performed after 1-h serum deprivation.
Whole-cell lysates and Western blotting
After transfection, treatment with inhibitors, and/or stimulation with EGF, whole-cell lysates were prepared in Laemmli sample buffer (0.5 M Tris, pH 6.8, glycerol, 10% SDS, 10% β-mercaptoethanol, and 5% bromophenol blue; all from BioShop) supplemented with a protease and phosphatase inhibitor cocktail (1 mM sodium orthovanadate, 10 nM okadaic acid, and 20 nM protease inhibitor cocktail, each obtained from BioShop). Lysates were then heated at 65°C for 15 min and passed through a 27.5-gauge syringe. Proteins were resolved by glycine-Tris SDS-PAGE followed by transfer onto a polyvinylidene fluoride membrane; they were washed, blocked, and incubated with antibodies as previously described (Antonescu et al., 2011). Molecular weight markers used were Novex Sharp Pre-stained Protein Standard (LC5800) or PageRuler Prestained Protein Ladder (26617; Thermo Fisher Scientific). Signals to detect the intensity corresponding to phosphorylated proteins (e.g., pAkt) were obtained as previously described (Delos Santos et al., 2017). First, images were obtained using a Bio-Rad ChemiDoc Touch Imaging System upon soaking membranes in Luminata Crescendo HRP substrate (Millipore Sigma). Typical exposure times varied between 1 and 60 s and were selected to ensure that signal was not saturated at any pixel. Images were quantified using ImageJ software (National Institutes of Health; Schneider et al., 2012) by signal integration in an area corresponding to the appropriate lane and band for each condition. This measurement was then normalized to the loading control (e.g., actin) signal and then normalized to the total Akt signal, obtained after reblotting. In each experiment, the resulting normalized pAkt/total Akt signal in each condition was expressed as a fraction of the normalized pAkt/total Akt measurement in the control condition stimulated with EGF for 5 min. Statistical analysis was performed with ANOVA followed by Tukey posttest, with P < 0.05 used as a threshold for establishing differences between experimental conditions. Data distribution was assumed to be normal but was not formally tested.
Immunofluorescence staining
For detection of total cellular protein (all immunofluorescence experiments except Figs. 1 E and 2 C), after treatments as indicated cells were fixed 4% PFA for 30 min, followed by quenching of fixative in 100 mM glycine, cell permeabilization in 0.1% Triton X-100 (all solutions made in PBS), and blocking in Superblock Blocking Buffer (Thermo Fisher Scientific). Subsequently, cells were stained with primary and secondary antibodies (fluorochromes described for each experiment in corresponding figure legend) as indicated and mounted on glass slides in fluorescence mounting medium (Dako) or retained within aqueous medium for imaging by TIRFM.
Immunofluorescence staining of cell-surface EGFR (as in Figs. 1 C and 2 B) was performed as previously described (Garay et al., 2015), using anti-EGFR (mAb108, collected in-house from a hybridoma cell line from American Type Culture Collection). Briefly, after treatments, cells were rapidly washed with ice-cold PBS and incubated with primary antibody solution on ice, before cell permeabilization. After washing of unbound primary antibodies, cells were then fixed, permeabilized, and stained with secondary antibodies as described above.
Fluorescence microscopy
Wide-field epifluorescence microscopy experiments (Figs. 1 C and 2 B) were performed on an Olympus IX83 Inverted Microscope with a 100×/1.4-NA objective, coupled to a Hamamatsu ORCA-Flash4.0 digital camera using cellSens software (Olympus Canada). Imaging was performed in samples mounted in Dako at room temperature.
TIRFM experiments were performed on a Quorum Diskovery instrument, comprising a Leica DMi8 microscope equipped with a 63×/1.49-NA TIRF objective with a 1.8× camera relay (total magnification 108×). Imaging was done using 405-, 488-, 561-, or 637-nm laser illumination and 450/55, 525/50, 620/60, and 700/75 emission filters and acquired using a Zyla 4.2Plus sCMOS camera (Hamamatsu), with images acquired using MetaMorph (Molecular Devices). Fixed-cell TIRFM imaging was done at room temperature with samples mounted in PBS. For live-cell imaging experiments (Figs. 6 and 9), cells were maintained at constant 37°C during imaging, in phenol-free DMEM/F12 media (Gibco) supplemented with 20 mM Hepes and 20 ng/ml EGF. Some TIRFM experiments (Fig. S4, A and B) also involved acquisition of identical fields of view using wide-field epifluorescence microscopy.
SIM was performed using a Zeiss Elyra PS.1 superresolution inverted microscope using Zen software, as previously described (Bautista et al., 2018). Samples were imaged at an effective magnification of 101× (63×/1.4 objective + 1.6× optovar tube lens) on an oil-immersion objective. 488-, 561-, and 643-nm laser lines were directed into the microscope optical train via a multimode fiber coupler. The lasers were passed through a diffraction grating, and a series of diffraction orders (−1, 0, +1) were projected onto the back focal plane of the objective. These wavefronts were collimated in the objective to create a 3D sinusoidal illumination pattern on the sample. The diffraction grating was then rotated and translated throughout the acquisition to create patterned offset images containing encoded high-spatial-frequency information. Three lateral positions were acquired at each of five (72°) diffraction grating rotations for a total of 15 raw images. SIM imaging with all lasers was carried out at exposures varying from 50 to 250 ms, with laser power varying between 3% and 10%, and a gain level of <100. Imaging parameters were adjusted iteratively to achieve the best possible equalization of pixel intensity dynamic range across channels. Raw SIM image stacks were processed in Zen under the Structured Illumination toolbar. The noise filter for Wiener deconvolution was set to a value of 1.0 × 10–4.5 to maximize the recovery of high-spatial-frequency information while minimizing illumination pattern artifacts. Processed SIM images were then aligned via an affine transformation matrix of predefined values obtained using 100-nm multicolor Tetraspeck fluorescent microspheres (Thermo Fisher Scientific).
For all microscopy images, final image processing was limited to linear adjustments of brightness/contrast, which was applied identically for all images of the same channel in an experiment, using ImageJ (Schneider et al., 2012).
Analysis of fixed-cell fluorescence images
Fluorescence intensity of Akt phosphorylation
Fluorescence intensity of Akt phosphorylation (as shown in Fig. 5) was determined after antibody labeling of phosphorylated Akt, and wide-field fluorescence microscopy was determined using ImageJ (Schneider et al., 2012) as previously described (Antonescu et al., 2008; Garay et al., 2015). Briefly, a cell outline was determined manually, followed by measurement of the mean fluorescence intensity corresponding to pAkt in each cell. The nonspecific signal, determined similarly in cells subjected to immunofluorescence labeling without primary antibody, was subtracted from all values. Cells transfected for various TOM1L1 constructs were identified by significant fluorescence in the appropriate channel. Differences in the mean pAkt signal between different conditions were determined by one-way ANOVA with Tukey posttest, with a threshold of P < 0.05 for statistically significant difference between conditions. Data distribution was assumed to be normal but was not formally tested.
CLS detection and quantification of protein fluorescence intensity within CLSs
Systematic, unbiased detection and analysis of CLSs in fixed cells (Figs. 3, A and B; Figs. 4, 6, 8, and 9; and Fig. S4, A and B) was done as previously described (Delos Santos et al., 2017; Lucarelli et al., 2017), using custom software developed in Matlab (MathWorks Corp.), as described in Aguet et al. (2013) and Garay et al. (2015). Briefly, diffraction-limited clathrin structures were detected using a Gaussian-based model method to approximate the point-spread function of eGFP-CLCa, RFP-T-CLCa, epsin, or AP2 puncta (primary channel). The TIRFM intensity corresponding to various proteins in a secondary (or tertiary) channel (e.g., Fyn-GFP, eGFP-TOM1L1, Rho-EGF, epsin, or SHIP2) within CLSs was determined by the amplitude of the Gaussian model for the appropriate fluorescence channel for each CLS structure detected in the primary channel. As such, the measurements of fluorescently labeled proteins within CLSs represent their enrichment relative to the local background fluorescence in the immediate vicinity of the detected CLS. Similar measurements were done using wide-field epifluorescence microscopy images (as the secondary channel) after detection of CLSs in the corresponding TIRF channel (as the primary channel; Fig. S4, A and B). For experiments involving transfection of Fyn-GFP or eGFP-TOM1L1 (Fig. 3), the intensity of TOM1L1 or Fyn within CLSs is reported as the mean intensity within CLSs, normalized to the total intensity of Fyn-GFP or eGFP-TOM1L1 detected in the TIRF field (to normalize for variation in expression level), after selection of images that exhibited a restricted (low) level of Fyn-GFP or eGFP-TOM1L1 expression level.
Identification of EGF-positive or Fyn-GFP–positive CLSs
To identify a subpopulation of clathrin structures enriched in Rho-EGF or Fyn-GFP (Fig. 6), we established a threshold of the 85th percentile of either EGF or Fyn fluorescence intensity within CLSs in the control (no-inhibitor) condition in each experiment (Lucarelli et al., 2017; Delos Santos et al., 2017). Using this systematic threshold, we defined subsets of CLSs enriched in either fluorescent EGF or Fyn-GFP each condition as those with ligand fluorescence intensity above this threshold. For the experiments in Fig. 6 C (right), CLSs were first sorted by Fyn-GFP status, then by Rho-EGF status. The fraction of Fyn-GFP–positive CLSs in each EGF status cohort is also reported.
Measurements (mean levels of various proteins within specified CLS subset for each cell) were subjected to either two-sided Student’s t test or ANOVA followed by Tukey posttest, with a threshold of P < 0.05 for statistically significant differences between conditions. Data distribution was assumed to be normal but was not formally tested.
Analysis of CCPs time-lapse image series
Automated detection, tracking, and analysis of CCPs in time-lapse image series (as in Fig. 7; Fig. 9; Fig. S3, A and B; and Fig. S4) was performed as previously described (Aguet et al., 2013; Mettlen and Danuser, 2014; Kadlecova et al., 2017) after TIRF microscopy of RPE cells stably expressing Tag-RFP-T-CLCa and transfected with the lowest detectable levels of Fyn-GFP or eGFP-TOM1L1, or cells expressing eGFP-clathrin and Fyn-iRFP713. Unless otherwise indicated, cells were treated with 20 ng/ml (unlabeled) EGF at the time of imaging. Diffraction-limited clathrin structures were detected using a Gaussian-based model method to approximate the point-spread function (Aguet et al., 2013), and trajectories were determined from clathrin structure detections using the u-track software (Jaqaman et al., 2008). sCLSs were distinguished from bona fide CCPs as previously described, based on the quantitative and unbiased analysis of clathrin intensity progression in the early stages of structure formation (Aguet et al., 2013; Kadlecova et al., 2017). Both sCLSs and CCPs represent nucleation events, but only bona fide CCPs represent structures that undergo stabilization, maturation, and in some cases scission to produce intracellular vesicles (Aguet et al., 2013; Kadlecova et al., 2017). Here, we report only CCP data. CCPs were sorted into Fyn-GFP– or eGFP-TOM1L1–positive cohorts, using methods for sorting CCP subpopulation by composition of a secondary channel established previously (Aguet et al., 2013), which effectively sorts CCPs based on Fyn (Fig. 7 F) or TOM1L1 (Fig. 7 H) content. We report the distribution of CCP lifetimes (Fig. 7, E and G), as well as the plateau intensity of Fyn-GFP (Fig. 7 F), eGFP-TOM1L1 (Fig. 7 H), or RFP-CLC (Fig. S3, C and D). CCPs exhibit several phases that can be described as initiation, growth/assembly, plateau, and disassembly/scission (Loerke et al., 2011). Here we define the plateau intensity of each marker as the mean fluorescence of that protein within each clathrin structure, measured within time points corresponding to 30% and 70% of the total lifetime of that structure, during which time CCPs exhibit minimal growth or disassembly. Because CCPs are diffraction-limited objects, the amplitude of the Gaussian model of the fluorescence intensity of RFP-CLC informs about CCP size (Fig. 7, J and N). All measurements were subjected to ANOVA followed by Tukey posttest with a threshold of P < 0.05 for statistically significant differences between conditions. Data distribution was assumed to be normal but was not formally tested.
Online supplemental material
Fig. S1 contains additional information on TOM1L1 silencing and full image panels of TIRFM and SIM micrographs shown in Fig. 3. Fig. S2 shows optimization of Sleeping Beauty stable cell lines and antibody staining. Fig. S3 contains additional information on live-cell imaging experiments shown in Fig. 7. Fig. S4 contains additional information about TOM1L1 and Fyn recruitment to CCPs. Fig. S5 is a model of regulation of SHIP2 localization and Akt signaling by TOM1L1 and Fyn within specialized CCPs.
Supplementary Material
contains original blots for Fig. 1.
contains original blots for Fig. 2.
contains original blots for Fig. 8.
contains original blots for Fig. S1.
contains original blots for Fig. S2.
Acknowledgments
Funding for this research was provided by a Project Grant (PJT-156355) and a New Investigator Award from the Canadian Institutes of Health Research, as well as an Early Researcher Award (Ontario Ministry of Research, Innovation and Science) to C.N. Antonescu; an Ontario Graduate Scholarship to S. Lucarelli, J. Abousawan, and S. Rahmani; and a Canadian Institutes of Health Research Doctoral Research Award to R. Cabral-Dias. Contributions from R.J. Botelho were funded by a Discovery Grant (RGPIN-2020-04343) from the Natural Sciences and Engineering Council of Canada, the Canada Research Chairs Program (950-232333), Ryerson University, and the Canadian Foundation for Innovation (32957).
The authors declare no competing financial interests.
Author contributions: Conceptualization: R. Cabral-Dias, S. Lucarelli, W. Hong, G.D. Fairn, C.N. Antonescu. Funding Acquisition: R.J. Botelho, G.D. Fairn, C.N. Antonescu. Investigation: R. Cabral-Dias, S. Lucarelli, K. Zak, S. Rahmani, G. Judge, J. Abousawan, L.F. DiGiovanni, D. Vural, K.E. Anderson, M.G. Sugiyama, G. Genc. Formal Analysis: R. Cabral-Dias, S. Lucarelli, C.N. Antonescu. Supervision: P.K. Kim, R.J. Botelho, G.D. Fairn, C.N. Antonescu. Writing—original draft: S. Lucarelli, C.N. Antonescu. Writing—Review & Editing: R. Cabral-Dias, S. Lucarelli, K. Zak, S. Rahmani, J. Abousawan, L.F. DiGiovanni, D. Vural, K.E. Anderson, M.G. Sugiyama, G. Genc, W. Hong, R.J. Botelho, G.D. Fairn, P.K. Kim, C.N. Antonescu.
References
- Aguet, F., Antonescu C.N., Mettlen M., Schmid S.L., and Danuser G.. 2013. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 26:279–291. 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu, C.N., Aguet F., Danuser G., and Schmid S.L.. 2011. Phosphatidylinositol-(4,5)-bisphosphate regulates clathrin-coated pit initiation, stabilization, and size. Mol. Biol. Cell. 22:2588–2600. 10.1091/mbc.e11-04-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu, C.N., Danuser G., and Schmid S.L.. 2010. Phosphatidic acid plays a regulatory role in clathrin-mediated endocytosis. Mol. Biol. Cell. 21:2944–2952. 10.1091/mbc.e10-05-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu, C.N., Randhawa V.K., and Klip A.. 2008. Dissecting GLUT4 traffic components in L6 myocytes by fluorescence-based, single-cell assays. Methods Mol. Biol. 457:367–378. 10.1007/978-1-59745-261-8_27 [DOI] [PubMed] [Google Scholar]
- Avraham, R., and Yarden Y.. 2011. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 12:104–117. 10.1038/nrm3048 [DOI] [PubMed] [Google Scholar]
- Bautista, S.J., Boras I., Vissa A., Mecica N., Yip C.M., Kim P.K., and Antonescu C.N.. 2018. mTOR complex 1 controls the nuclear localization and function of glycogen synthase kinase 3β. J. Biol. Chem. 293:14723–14739. 10.1074/jbc.ra118.002800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman, N.J., Bagchi A., Ferguson K.M., and Lemmon M.A.. 2014. Complex relationship between ligand binding and dimerization in the epidermal growth factor receptor. Cell Rep. 9:1306–1317. 10.1016/j.celrep.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone, L.N., Dayam R.M., Lee M., Kono N., Fairn G.D., Arai H., Botelho R.J., and Antonescu C.N.. 2017. The acyltransferase LYCAT controls specific phosphoinositides and related membrane traffic. Mol. Biol. Cell. 28:161–172. 10.1091/mbc.e16-09-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley, L.C. 2002. The phosphoinositide 3-kinase pathway. Science. 296:1655–1657. 10.1126/science.296.5573.1655 [DOI] [PubMed] [Google Scholar]
- Choy, C.H., Han B.-K., and Botelho R.J.. 2017. Phosphoinositide diversity, distribution, and effector function: stepping out of the box. Bioessays. 39:1700121. 10.1002/bies.201700121 [DOI] [PubMed] [Google Scholar]
- Collin, G., Franco M., Simon V., Bénistant C., and Roche S.. 2007. The Tom1L1-clathrin heavy chain complex regulates membrane partitioning of the tyrosine kinase Src required for mitogenic and transforming activities. Mol. Cell. Biol. 27:7631–7640. 10.1128/mcb.00543-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambournet, D., Sochacki K., Cheng A., Akamatsu M., Taraska J., Hockemeyer D., and Drubin D.. 2018. Genome-edited human stem cells expressing fluorescently labeled endocytic markers allow quantitative analysis of clathrin-mediated endocytosis during differentiation. J. Cell Biol. 217:3301–3311. 10.1083/jcb.201710084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub, H., Wallasch C., Lankenau A., Herrlich A., and Ullrich A.. 1997. Signal characteristics of G protein-transactivated EGF receptor. EMBO J. 16:7032–7044. 10.1093/emboj/16.23.7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath, J., Muthuswamy S.K., and Brugge J.S.. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268. 10.1016/s1046-2023(03)00032-x [DOI] [PubMed] [Google Scholar]
- Delos Santos, R.C., Bautista S., Lucarelli S., Bone L.N., Dayam R.M., Abousawan J., Botelho R.J., and Antonescu C.N.. 2017. Selective control of clathrin- mediated endocytosis and clathrin-dependent signaling by phospholipase C and Ca2+ signals. Mol. Biol. Cell. 28:2802–2818. 10.1091/mbc.E16-12-0871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delos Santos, R.C., Garay C., and Antonescu C.N.. 2015. Charming neighborhoods on the cell surface: plasma membrane microdomains regulate receptor tyrosine kinase signaling. Cell. Signal. 27:1963–1976. 10.1016/j.cellsig.2015.07.004 [DOI] [PubMed] [Google Scholar]
- Eichel, K., Jullié D., Barsi-Rhyne B., Latorraca N.R., Masureel M., Sibarita J.B., Dror R.O., and Von Zastrow M.. 2018. Catalytic activation of β-Arrestin by GPCRs. Nature. 557:381–386. 10.1038/s41586-018-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichel, K., Jullié D., and von Zastrow M.. 2016. β-Arrestin drives MAP kinase signalling from clathrin-coated structures after GPCR dissociation. Nat. Cell Biol. 18:303–310. 10.1038/ncb3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, K.S., Mao Y., McCrea H.J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., and De Camilli P.. 2007. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell. 13:377–390. 10.1016/j.devcel.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkes, A.J., Ahmadi K., Alderton W.K., Alix S., Baker S.J., Box G., Chuckowree I.S., Clarke P.A., Depledge P., Eccles S.A., Friedman L.S., et al. 2008. The identification of 2-(1H-Indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J. Med. Chem. 51:5522–5532. 10.1021/jm800295d [DOI] [PubMed] [Google Scholar]
- Freed, D.M., Bessman N.J., Kiyatkin A., Salazar-Cavazos E., Byrne P.O., Moore J.O., Valley C.C., Ferguson K.M., Leahy D.J., Lidke D.S., and Lemmon M.A.. 2017. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 171:683–695.e18. 10.1016/j.cell.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furcht, C.M., Buonato J.M., and Lazzara M.J.. 2015. EGFR-activated Src family kinases maintain GAB1-SHP2 complexes distal from EGFR. Sci. Signal. 8:ra46. 10.1126/scisignal.2005697 [DOI] [PubMed] [Google Scholar]
- Garay, C., Judge G., Lucarelli S., Bautista S., Pandey R., Singh T., and Antonescu C.N.. 2015. Epidermal growth factor-stimulated Akt phosphorylation requires clathrin or ErbB2 but not receptor endocytosis. Mol. Biol. Cell. 26:3504–3519. 10.1091/mbc.e14-09-1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Githaka, J.M., Vega A.R., Baird M.A., Davidson M.W., Jaqaman K., and Touret N.. 2016. Ligand-induced growth and compaction of CD36 nanoclusters enriched in Fyn induces Fyn signaling. J. Cell Sci. 129:4175–4189. 10.1242/jcs.188946 [DOI] [PubMed] [Google Scholar]
- Goh, L.K., Huang F., Kim W., Gygi S., and Sorkin A.. 2010. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 189:871–883. 10.1083/jcb.201001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulden, B.D., Pacheco J., Dull A., Zewe J.P., Deiters A., and Hammond G.R.V.. 2018. A high-avidity biosensor reveals plasma membrane PI(3,4)P2 is predominantly a class I PI3K signaling product. J. Cell Biol. 218:1066–1079. 10.1083/jcb.201809026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Marsland R. III, Upadhyayula S., Song E., Dang S., Capraro B.R., Wang W., Skillern W., Gaudin R., Ma M., and Kirchhausen T.. 2017. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature. 552:410–414. 10.1038/nature25146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holgado-Madruga, M., Emlet D.R., Moscatello D.K., Godwin A.K., and Wong A.J.. 1996. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 379:560–564. 10.1038/379560a0 [DOI] [PubMed] [Google Scholar]
- Jaqaman, K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., and Danuser G.. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlecova, Z., Spielman S.J., Loerke D., Mohanakrishnan A., Reed D.K., and Schmid S.L.. 2017. Regulation of clathrin-mediated endocytosis by hierarchical allosteric activation of AP2. J. Cell Biol. 216:167–179. 10.1083/jcb.201608071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyatkin, A., Aksamitiene E., Markevich N.I., Borisov N.M., Hoek J.B., and Kholodenko B.N.. 2006. Scaffolding protein Grb2-associated binder 1 sustains epidermal growth factor-induced mitogenic and survival signaling by multiple positive feedback loops. J. Biol. Chem. 281:19925–19938. 10.1074/jbc.m600482200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarz, E., Löscher D., and Marschalek R.. 2015. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 10:647–653. 10.1002/biot.201400821 [DOI] [PubMed] [Google Scholar]
- Lee, Y.-R., Chen M., and Pandolfi P.P.. 2018. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 19:547–562. 10.1038/s41580-018-0015-0 [DOI] [PubMed] [Google Scholar]
- Lemmon, M.A., Schlessinger J., and Ferguson K.M.. 2014. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 6:a020768. 10.1101/cshperspect.a020768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton-Puig, D., Isogai T., Argenzio E., van den Broek B., Klarenbeek J., Janssen H., Jalink K., and Innocenti M.. 2017. Flat clathrin lattices are dynamic actin-controlled hubs for clathrin-mediated endocytosis and signalling of specific receptors. Nat. Commun. 8:16068. 10.1038/ncomms16068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Marshall C., Mei L., Dzubow L., Schmults C., Dans M., and Seykora J.. 2005. Srcasm modulates EGF and Src-kinase signaling in keratinocytes. J. Biol. Chem. 280:6036–6046. 10.1074/jbc.m406546200 [DOI] [PubMed] [Google Scholar]
- Liu, A.P., Aguet F., Danuser G., and Schmid S.L.. 2010. Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J. Cell Biol. 191:1381–1393. 10.1083/jcb.201008117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, A.P., Loerke D., Schmid S.L., and Danuser G.. 2009a. Global and local regulation of clathrin-coated pit dynamics detected on patterned substrates. Biophys. J. 97:1038–1047. 10.1016/j.bpj.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N.S., Loo L.S., Loh E., Seet L.-F., and Hong W.. 2009b. Participation of Tom1L1 in EGF-stimulated endocytosis of EGF receptor. EMBO J. 28:3485–3499. 10.1038/emboj.2009.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.-L., Wang Z.-G., Hu Y., Xin Y., Singaram I., Gorai S., Zhou X., Shim Y., Min J.-H., Gong L.-W., Hay N., et al. 2018. Quantitative lipid imaging reveals a new signaling function of phosphatidylinositol-3,4-bisphophate: isoform- and site-specific activation of Akt. Mol. Cell. 71:1092–1104.e5. 10.1016/j.molcel.2018.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock, L.S., Royal I., Naujokas M.A., and Park M.. 2000. Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J. Biol. Chem. 275:31536–31545. 10.1074/jbc.m003597200 [DOI] [PubMed] [Google Scholar]
- Loerke, D., Mettlen M., Schmid S.L., and Danuser G.. 2011. Measuring the hierarchy of molecular events during clathrin-mediated endocytosis. Traffic. 12:815–825. 10.1111/j.1600-0854.2011.01197.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke, D., Mettlen M., Yarar D., Jaqaman K., Jaqaman H., Danuser G., and Schmid S.L.. 2009. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7:e57. 10.1371/journal.pbio.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S.M., and Fairn G.D.. 2018. Mesoscale organization of domains in the plasma membrane – beyond the lipid raft. Crit. Rev. Biochem. Mol. Biol. 53:192–207. 10.1080/10409238.2018.1436515 [DOI] [PubMed] [Google Scholar]
- Lucarelli, S., Delos Santos R.C., and Antonescu C.N.. 2017. Measurement of epidermal growth factor receptor-derived signals within plasma membrane clathrin structures. Methods Mol. Biol. 1652:191–225. 10.1007/978-1-4939-7219-7_15 [DOI] [PubMed] [Google Scholar]
- Lucarelli, S., Pandey R., Judge G., and Antonescu C.N.. 2016. Similar requirement for clathrin in EGF- and HGF- stimulated Akt phosphorylation. Commun. Integr. Biol. 9:e1175696. 10.1080/19420889.2016.1175696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés, L., Chuah M.K.L., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., Ma L., et al. 2009. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41:753–761. 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- Matheny, R.W., and Adamo M.L.. 2009. Current perspectives on Akt Akt-ivation and Akt-ions. Exp. Biol. Med. 234:1264–1270. 10.3181/0904-mr-138 [DOI] [PubMed] [Google Scholar]
- Mattoon, D.R., Lamothe B., Lax I., and Schlessinger J.. 2004. The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC Biol. 2:24. 10.1186/1741-7007-2-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, H.T., and Boucrot E.. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12:517–533. 10.1038/nrm3151 [DOI] [PubMed] [Google Scholar]
- Mettlen, M., and Danuser G.. 2014. Imaging and modeling the dynamics of clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 6:a017038. 10.1101/cshperspect.a017038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen, M., Loerke D., Yarar D., Danuser G., and Schmid S.L.. 2010. Cargo- and adaptor-specific mechanisms regulate clathrin-mediated endocytosis. J. Cell Biol. 188:919–933. 10.1083/jcb.200908078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen, M., Stoeber M., Loerke D., Antonescu C.N., Danuser G., and Schmid S.L.. 2009. Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin-coated pits. Mol. Biol. Cell. 20:3251–3260. 10.1091/mbc.e09-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu, F., Perera R.M., Lucast L., Zoncu R., Domin J., Gertler F.B., Toomre D., and De Camilli P.. 2010. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J. Cell Biol. 190:307–315. 10.1083/jcb.201005018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez, D., Antonescu C., Mettlen M., Liu A., Schmid S.L., Loerke D., and Danuser G.. 2011. Hotspots organize clathrin-mediated endocytosis by efficient recruitment and retention of nucleating resources. Traffic. 12:1868–1878. 10.1111/j.1600-0854.2011.01273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareja, F., Pines G., and Yarden Y.. 2015. The EGFR/ERBB receptor family. In Receptor Tyrosine Kinases: Family and Subfamilies. Springer International Publishing, Cham. 107–164 pp. [Google Scholar]
- Pascolutti, R., Algisi V., Conte A., Raimondi A., Pasham M., Upadhyayula S., Gaudin R., Maritzen T., Barbieri E., Caldieri G., Tordonato C., et al. 2019. Molecularly distinct clathrin-coated pits differentially impact EGFR fate and signaling. Cell Rep. 27:3049–3061.e6. 10.1016/j.celrep.2019.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq, J.-D., Cabantous S., Tran T., Terwilliger T.C., and Waldo G.S.. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88. 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- Posor, Y., Eichhorn-Gruenig M., Puchkov D., Schöneberg J., Ullrich A., Lampe A., Müller R., Zarbakhsh S., Gulluni F., Hirsch E., Krauss M., et al. 2013. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 499:233–237. 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]
- Puertollano, R. 2005. Interactions of TOM1L1 with the multivesicular body sorting machinery. J. Biol. Chem. 280:9258–9264. 10.1074/jbc.m412481200 [DOI] [PubMed] [Google Scholar]
- Puertollano, R., Randazzo P.A., Presley J.F., Hartnell L.M., and Bonifacino J.S.. 2001. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 105:93–102. 10.1016/s0092-8674(01)00299-9 [DOI] [PubMed] [Google Scholar]
- Puthenveedu, M.A., and von Zastrow M.. 2006. Cargo regulates clathrin-coated pit dynamics. Cell. 127:113–124. 10.1016/j.cell.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Reis, C.R., Chen P.-H., Bendris N., and Schmid S.L.. 2017. TRAIL-death receptor endocytosis and apoptosis are selectively regulated by dynamin-1 activation. Proc. Natl. Acad. Sci. USA. 114:504–509. 10.1073/pnas.1615072114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, C.R., Chen P.-H., Srinivasan S., Aguet F., Mettlen M., and Schmid S.L.. 2015. Crosstalk between Akt/GSK3β signaling and dynamin-1 regulates clathrin-mediated endocytosis. EMBO J. 34:2132–2146. 10.15252/embj.201591518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi, S., Tartare-Deckert S., Murdaca J., Holgado-Madruga M., Wong A.J., and Van Obberghen E.. 1998. Determination of Gab1 (Grb2-associated binder-1) interaction with insulin receptor-signaling molecules. Mol. Endocrinol. 12:914–923. 10.1210/mend.12.7.0141 [DOI] [PubMed] [Google Scholar]
- Rodrigues, G.A., Falasca M., Zhang Z., Ong S.H., and Schlessinger J.. 2000. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol. Cell. Biol. 20:1448–1459. 10.1128/MCB.20.4.1448-1459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselli-Murai, L.K., Yates J.A., Yoshida S., Bourg J., Ho K.K.Y., White M., Prisby J., Tan X., Altemus M., Bao L., Wu Z.-F., et al. 2018. Loss of PTEN promotes formation of signaling-capable clathrin-coated pits. J. Cell Sci. 131:jcs208926. 10.1242/jcs.208926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K.-I. 2013. Cellular functions regulated by phosphorylation of EGFR on Tyr845. Int. J. Mol. Sci. 14:10761–10790. 10.3390/ijms140610761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck, A., Goto-Silva L., Collinet C., Rhinn M., Giner A., Habermann B., Brand M., and Zerial M.. 2008. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 133:486–497. 10.1016/j.cell.2008.02.044 [DOI] [PubMed] [Google Scholar]
- Schmid, E.M., and McMahon H.T.. 2007. Integrating molecular and network biology to decode endocytosis. Nature. 448:883–888. 10.1038/nature06031 [DOI] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg, J., Lehmann M., Ullrich A., Posor Y., Lo W.T., Lichtner G., Schmoranzer J., Haucke V., and Noé F.. 2017. Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat. Commun. 8:15873. 10.1038/ncomms15873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seykora, J.T., Mei L., Dotto G.P., and Stein P.L.. 2002. Srcasm: a novel Src activating and SignalingMolecule. J. Biol. Chem. 277:2812–2822. 10.1074/jbc.m106813200 [DOI] [PubMed] [Google Scholar]
- Shcherbakova, D.M., and Verkhusha V.V.. 2013. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 10:751–754. 10.1038/nmeth.2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund, S., Avanzato D., and Lanzetti L.. 2018. Emerging functions of the EGFR in cancer. Mol. Oncol. 12:3–20. 10.1002/1878-0261.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkina, T., Huang F., Beguinot L., and Sorkin A.. 2002. Effect of tyrosine kinase inhibitors on clathrin-coated pit recruitment and internalization of epidermal growth factor receptor. J. Biol. Chem. 277:27433–27441. 10.1074/jbc.m201595200 [DOI] [PubMed] [Google Scholar]
- Sousa, L.P., Lax I., Shen H., Ferguson S.M., De Camilli P., and Schlessinger J.. 2012. Suppression of EGFR endocytosis by dynamin depletion reveals that EGFR signaling occurs primarily at the plasma membrane. Proc. Natl. Acad. Sci. USA. 109:4419–4424. 10.1073/pnas.1200164109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S., Burckhardt C.J., Bhave M., Chen Z., Chen P.-H., Wang X., Danuser G., and Schmid S.L.. 2018. A noncanonical role for dynamin-1 in regulating early stages of clathrin-mediated endocytosis in non-neuronal cells. PLoS Biol. 16:e2005377. 10.1371/journal.pbio.2005377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, M.G., Fairn G.D., and Antonescu C.N.. 2019. Akt-ing up just about everywhere: compartment-specific Akt activation and function in receptor tyrosine kinase signaling. Front. Cell Dev. Biol. 7:70. 10.3389/fcell.2019.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M.J., Perrais D., and Merrifield C.J.. 2011. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9:e1000604. 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa, N., Chen M., Horn H., Choi S., Wen T., and Anderson R.. 2020. Phosphatidylinositol-3-OH kinase signalling is spatially organized at endosomal compartments by microtubule-associated protein 4. Nat. Cell Biol. 22:1357–1370. 10.1038/s41556-020-00596-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco, I., and Sawyers C.L.. 2002. The phosphatidylinositol 3-kinase–AKT pathway in human cancer. Nat. Rev. Cancer. 2:489–501. 10.1038/nrc839 [DOI] [PubMed] [Google Scholar]
- Wang, H., Loerke D., Bruns C., Müller R., Koch P.-A., Puchkov D., Schultz C., and Haucke V.. 2020. Phosphatidylinositol 3,4-bisphosphate synthesis and turnover are spatially segregated in the endocytic pathway. J. Biol. Chem. 295:1091–1104. 10.1074/jbc.RA119.011774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde, A., Beattie E.C., Lem L., Riethof D.A., Liu S.-H., Mobley W.C., Soriano P., and Brodsky F.M.. 1999. EGF receptor signaling stimulates SRC kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell. 96:677–687. 10.1016/s0092-8674(00)80578-4 [DOI] [PubMed] [Google Scholar]
- Wu, C.J., O’Rourke D.M., Feng G.S., Johnson G.R., Wang Q., and Greene M.I.. 2001. The tyrosine phosphatase SHP-2 is required for mediating phosphatidylinositol 3-kinase/Akt activation by growth factors. Oncogene. 20:6018–6025. 10.1038/sj.onc.1204699 [DOI] [PubMed] [Google Scholar]
- Yamakami, M., Yoshimori T., and Yokosawa H.. 2003. Tom1, a VHS domain-containing protein, interacts with tollip, ubiquitin, and clathrin. J. Biol. Chem. 278:52865–52872. 10.1074/jbc.m306740200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. 1.
contains original blots for Fig. 2.
contains original blots for Fig. 8.
contains original blots for Fig. S1.
contains original blots for Fig. S2.