Abstract
SNARE proteins and Sec1/Munc18 (SM) proteins constitute the core molecular engine that drives nearly all intracellular membrane fusion and exocytosis. While SNAREs are known to couple their folding and assembly to membrane fusion, the physiological pathways of SNARE assembly and the mechanistic roles of SM proteins have long been enigmatic. Here, we review recent advances in understanding the SNARE–SM fusion machinery with an emphasis on biochemical and biophysical studies of proteins that mediate synaptic vesicle fusion. We begin by discussing the energetics, pathways, and kinetics of SNARE folding and assembly in vitro. Then, we describe diverse interactions between SM and SNARE proteins and their potential impact on SNARE assembly in vivo. Recent work provides strong support for the idea that SM proteins function as chaperones, their essential role being to enable fast, accurate SNARE assembly. Finally, we review the evidence that SM proteins collaborate with other SNARE chaperones, especially Munc13-1, and briefly discuss some roles of SNARE and SM protein deficiencies in human disease.
Keywords: Membrane fusion, SNARE assembly, SM proteins, Munc18-1, template complex, optical tweezers
1. INTRODUCTION
Eukaryotic cells use vesicles and other membranous carriers to transport proteins and lipids among different cellular compartments. These vesicles bud from a donor membrane and fuse with a target membrane, delivering their cargo into the membrane or lumen of the corresponding compartment (1, 2). Membrane fusion is mediated by a conserved machinery comprising two key types of proteins: soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) and Sec1/Munc18 (SM)-family proteins (3). Additional proteins act on this machinery to precisely control the location, time, speed, and magnitude of membrane fusion. SNARE–SM-mediated fusion is ubiquitous in eukaryotes and underlies numerous physiological and pathological processes in humans. A prominent example is synaptic exocytosis, which is triggered by action potentials and forms the basis of all thoughts and actions. Vesicle–plasma membrane fusion also mediates insulin secretion for glucose homeostasis; antibody, cytokine, and cytotoxin secretion associated with immune responses; and the release of various hormones. Dysfunctions of the fusion machinery are linked to neurological disorders, diabetes, cancer, and immunological diseases (4-7).
SNARE proteins, most of which are integral membrane proteins with C-terminal anchors, contain characteristic SNARE motifs that are ~60 residues in length (Figure 1a) (8, 9). While most SNAREs contain one SNARE motif, a few instead consist of two SNARE motifs connected by an extended linker. Individual SNARE motifs are intrinsically disordered (10-13), whereas complementary sets of four SNARE motifs assemble to form parallel four-helix bundles (10, 12-15) (Figure 1b). The coupled folding and assembly of membrane-bridging (trans-) SNARE complexes draws two membranes into close proximity to produce the force required to overcome the energy barrier associated with membrane merger (16-21). In this manner, trans-SNARE complex assembly both induces membrane fusion and contributes to its specificity (1, 21). Fusion converts a trans-SNARE complex, in which the participating SNAREs are anchored in two different membranes, into a cis-SNARE complex, in which all of the SNAREs are embedded in the same fused membrane. These and other inactive SNARE complexes are then recycled—that is, forcibly disassembled—by the AAA+ ATPase NSF (22, 23). With the development of new technologies over the past decade, the assembly energetics, kinetics, and pathways of SNARE proteins in vitro have become relatively well characterized (17, 20, 24, 25). Much less is known about physiological SNARE assembly, guided by regulatory chaperones, in vivo (26). It is nonetheless clear that the physiological pathway(s) are different from the unsupervised pathways (27-29).
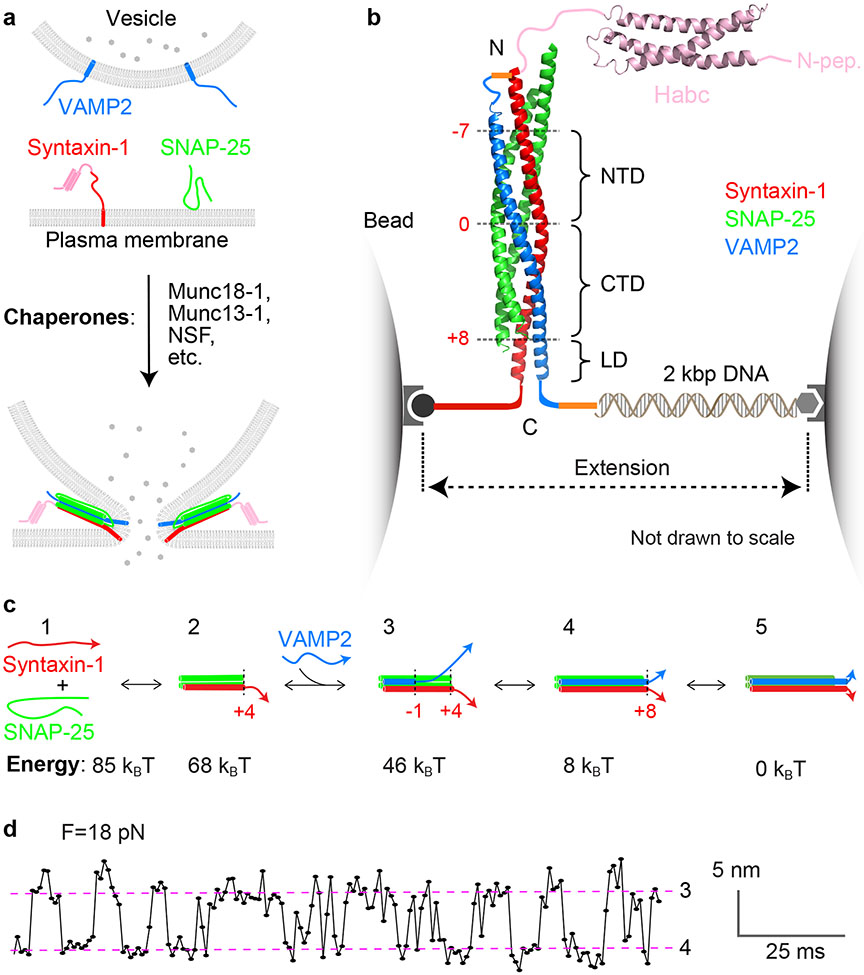
Figure 1.
Stepwise SNARE folding and assembly drives membrane fusion. (a) Schematic diagram of chaperoned SNARE assembly and membrane fusion. (b) Typical experimental setup to pull a single synaptic SNARE complex using dual-trap optical tweezers (17, 28). The cartoon of the fully assembled cytoplasmic SNARE complex incorporates the X-ray structure of the four-helix SNARE motif bundle (PDB ID: 3HD7) and the nuclear magnetic resonance structure of the syntaxin-1 Habc domain (PDB ID: 1BR0). A 2,260-base-pair DNA handle is used to attach a single SNARE complex to the surfaces of two polystyrene beads held in optical traps, whose separation is accurately controlled to change the pulling force applied to the SNARE complex. Red numbers indicate the hydrophobic layers and the ionic zero layer. (c) Energetics of uncatalyzed synaptic SNARE assembly as measured using the experimental setup in panel b. The free energies of states ❶–❹ relative to the fully assembled SNARE complex (state ❺) are shown below the state diagrams. (d) Representative time-dependent extension at constant force. The red dashed lines represent states ❸ and ❹ (as shown in panel c), and the transitions between them represent the rapid, reversible folding and unfolding of the SNARE CTD. Panel d modified [with permission] from Reference 17. Abbreviations: CTD, C-terminal domain; LD, juxtamembrane linker domain; Munc, mammalian homolog of unc; NTD, N-terminal domain; PDB ID, Protein Data Bank identifier; SNARE, soluble N-ethylmaleimide–sensitive factor attachment protein receptor.
Of fundamental importance for SNARE complex assembly in vivo are the SM proteins. One of the first SM proteins, UNC-18, was discovered by Brenner in 1974, via a genetic screen for Caenorhabditis elegans mutants that displayed uncoordinated movement (30). unc-18 mutants display a paralytic phenotype and accumulate the neurotransmitter acetylcholine, consistent with a defect in synaptic vesicle fusion (31). The yeast SM protein Sec1 was discovered five years later by Novick and Schekman, who reported the first of what would soon become a large collection of temperature-sensitive Saccharomyces cerevisiae secretion mutants (32, 33). At nonpermissive temperatures, sec1 yeast cells display a dramatic accumulation of secretory vesicles, suggesting a block in fusion at the plasma membrane. In 1993, the mammalian homolog of Unc18, Munc18-1, was isolated as a binding partner of the synaptic SNARE syntaxin-1 and was later found to be essential for neurotransmitter secretion (34, 35). Together, Sec1 and Munc18 gave rise to the name SM protein. Humans have seven SM proteins, including three—Munc18-1, Munc18-2, Munc18-3—that function in exocytosis (36, 37). Consistent with in vivo observations, SM proteins can promote SNARE-mediated fusion between reconstituted proteoliposomes (38). However, despite intensive studies over the past four decades, it is only recently that a detailed understanding has begun to emerge regarding how SM and SNARE proteins cooperate to drive membrane fusion.
This review focuses primarily on recent experimental approaches that have begun to elucidate the energetics, kinetics, and pathways of SNARE assembly in the absence and presence of SM proteins. These experiments suggest that SNAREs assemble stepwise with characteristic kinetics and that representative SM proteins chaperone SNARE assembly via conserved intermediates. We emphasize the well-studied synaptic SNARE–SM machinery, which mediates the fusion of synaptic vesicles with the plasma membrane to enable the release of neurotransmitters into the synaptic cleft (Figure 1a). This calcium-triggered process requires additional regulatory proteins, including Munc13-1, synaptotagmin, complexin, and NSF, whose specific functions have been recently reviewed (39-41). Of these proteins, we focus our discussion on Munc13-1, since recent experiments imply that it cooperates directly with Munc18-1 to chaperone SNARE assembly (42-47).
2. ENERGETICS, KINETICS, AND PATHWAYS OF SNARE ASSEMBLY
2.1. SNARE Domains and SNARE Complexes
The fusion of synaptic vesicles with the presynaptic plasma membrane requires three SNARE proteins: the vesicle (v-) SNARE VAMP2 and the target membrane (t-) SNAREs syntaxin-1 and SNAP-25 (48) (Figure 1a). Syntaxin-1 and VAMP2 contain C-terminal transmembrane anchors, whereas SNAP-25 associates with the plasma membrane by means of palmitoylated cysteine residues in the linker between its two SNARE motifs. Syntaxin-1, despite its relatively modest size (33 kDa), contains a surprisingly large number of functionally relevant regions. These include an N-terminal regulatory domain (NRD), a SNARE motif, a juxtamembrane linker domain (LD), and a C-terminal transmembrane domain (TMD). The NRD (pink in Figure 1b) consists of three parts: an N-terminal peptide (N-peptide, residues 1–26), a three-helical Habc domain (residues 27–146) (49), and a linker connecting the Habc domain to the SNARE motif (residues 147–187). The SNARE motif comprises N-terminal and C-terminal domains (NTD and CTD; Figure 1b), both in syntaxin-1 and in other SNAREs.
Crystal structures of fully assembled cis-SNARE complexes reveal parallel bundles of four α-helical SNARE motifs (14, 15, 50). The core of the bundle is composed of 16 layers, numbered from −7 to +8 (Figure 1b). Fifteen of these layers contain predominantly hydrophobic residues. The central zero layer, by contrast, comprises three glutamines and one arginine that interact with one another via charge-stabilized hydrogen bonds. These zero-layer residues, which are almost universally conserved among SNARE complexes, gave rise to a useful alternative nomenclature in which the glutamine-containing SNAREs are named the Qa-, Qb-, and Qc-SNAREs (with a, b, and c denoting their relative positions within the four-helix bundle), while the arginine-containing SNARE is an R-SNARE (8, 9). For the synaptic SNAREs, VAMP2 is the R-SNARE, syntaxin-1 is the Qa-SNARE, and SNAP-25—with two SNARE motifs separated by an extended linker—is termed a Qbc-SNARE. More typically, the Qb- and Qc-SNAREs are two separate proteins. In the most complete X-ray structure of a cis-SNARE complex, the α-helices adopted by the SNARE motifs of syntaxin-1 and VAMP2 extend through the juxtamembrane LDs and TMDs of both proteins (15).
2.2. SNARE Folding and Assembly
Diverse in vitro approaches have been used to characterize the folding rates and thermodynamic stabilities of cytoplasmic SNARE complexes (i.e., complexes among SNAREs lacking their TMDs) (Figure 1b) (25). Syntaxin-1 and SNAP-25 can form a binary 1:1 t-SNARE complex (or Qabc complex), which then binds VAMP2 to form a 1:1:1 ternary SNARE complex (10, 51) (Figure 1c). The binding of the t-SNARE complex to VAMP2 is relatively fast, with estimates of the rate constant ranging from 6 × 103 to 5 × 105 M−1s−1 (52, 53). When all three SNAREs are mixed simultaneously, however, assembly of the ternary complex is much slower—requiring from tens of minutes to hours (54)—presumably because of competing off-pathway reactions (see Section 2.3). Once assembled, the ternary SNARE complex is exceptionally stable, with a melting temperature over 90°C (15, 55).
The pronounced hysteresis of SNARE assembly and disassembly (55) has stymied efforts to measure the thermodynamic stability of the cytoplasmic SNARE complex using traditional bulk methods (25). A powerful alternative is single-molecule force spectroscopy (25). In a typical experiment, a single cytoplasmic SNARE complex is suspended between micron-sized polystyrene beads confined in dual optical traps (Figure 1b). This allows both the distance between the beads (the extension) and the force exerted by the SNARE complex on the beads to be measured simultaneously and with subnanometer and submillisecond resolution (17). Manipulating the beads applies a controlled pulling force to the C termini of syntaxin-1 and VAMP2, inducing SNARE unfolding, and the force is subsequently relaxed to allow SNARE refolding. The pulling force exerted by the optical traps mimics the membrane repulsive force on the trans-SNARE complex during SNARE assembly and membrane fusion. The resulting force-extension curves or extension-time trajectories reveal SNARE folding and unfolding transitions in real time (Figure 1d) (56). Unfolding energy is derived from the work (the force multiplied by the extension change) corresponding to each transition (56, 57). A similar experimental setup has been applied to characterize t-SNARE folding (58).
These measurements, interpreted in the light of other biophysical and structural studies, imply that the 1:1 t-SNARE complex (syntaxin-1–SNAP-25) is a parallel three-helix bundle with disordered C termini (layers +5 to +8) (Figure 1c, state ❷). The t-SNARE complex exhibits a thermodynamic stability of 17 kBT (1 kBT per molecule is equivalent to 0.6 kcal/mol) (58). VAMP2 binds to the t-SNARE complex in a stepwise manner: first, the VAMP2 NTD (from the −7 to −1 layers) (Figure 1c, state ❸), then the CTD (from the 0 to +8 layers) (Figure 1c, state ❹), and finally, the juxtamembrane LD (Figure 1c, state ❺) (17, 24). At each of these three steps, the corresponding region of VAMP2 folds into an α-helical conformation. The directional, stepwise folding and assembly of SNARE four-helix bundles is typically termed SNARE zippering. NTD zippering is slow, whereas CTD zippering is extremely fast without any energy barrier. NTD zippering stabilizes layers 0 to +4 of the t-SNARE complex (state ❸) (58, 59); CTD zippering then induces the folding of layers +5 to +8 of the t-SNAREs (state ❹) (24, 52, 58). Overall, assembly of a single synaptic SNARE complex outputs a total energy of approximately 85 kBT, including 68 kBT from zippering between the t- and v-SNAREs (24) and 17 kBT from the folding of the t-SNARE complex (58). The existence of intermediate states in the in vitro assembly of synaptic SNARE complexes has also been observed using a surface force apparatus (60), magnetic tweezers (61, 62), electrophysiological measurements (63, 64), and split VAMP2 peptides (52, 65). Several nonsynaptic SNARE complexes, when examined using optical tweezers, were found to zipper together in an analogous, stepwise manner (20).
In vivo, the different synaptic SNARE folding domains appear to have distinct functions (24, 66). NTD zippering is responsible for docking synaptic vesicles and priming the SNAREs for Ca2+-triggered fusion. CTD zippering is tightly coupled to membrane fusion and neurotransmitter release. Although both the LD and the TMD also contribute to the energy of SNARE zippering (15, 17, 20), their functions in membrane fusion are less clear. Nonetheless, both domains strongly interact with lipid bilayers, transducing the SNARE zippering force to the membranes (67, 68) and/or disrupting the membranes to facilitate fusion (69-74).
2.3. SNARE Misassembly
In the absence of chaperones, SNAREs misassemble into various off-pathway products that are incapable of mediating membrane fusion (75). For example, cytoplasmic syntaxin-1 forms homotetramers that are unable to bind SNAP-25 or VAMP2 (76, 77). In the presence of SNAP-25, syntaxin-1 forms off-pathway 2:1 complexes in addition to on-pathway 1:1 complexes (51, 53, 78, 79). The more stable 2:1 complexes, in which the second syntaxin-1 blocks VAMP2 binding, complicate bulk studies of SNARE assembly (51, 53). By contrast, optical tweezers make it possible to generate single 1:1 t-SNARE complexes by forcibly unzippering the VAMP2 molecule from a ternary SNARE complex. Upon relaxing the optical trap, the t-SNARE complex binds free VAMP2 present in the solution (58) with an apparent rate constant [2 × 106 M−1s−1 (28)] that is 4- to 300-fold higher than those inferred from bulk measurements (52, 53) (see Section 2.2). Thus, SNARE zippering is intrinsically rapid but is slowed down in vitro by off-pathway reactions.
Another type of SNARE misassembly involves the formation of complexes with antiparallel SNARE motifs (75, 80). Indeed, single-molecule fluorescence resonance energy transfer (smFRET) experiments showed that 40% or more of the complexes that formed upon mixing the three synaptic SNAREs contained antiparallel helices (43). In addition, it has long been known that cytoplasmic SNAREs can form cognate and noncognate complexes with similar efficiencies (81, 82). Overall, SNARE assembly in the absence of regulatory proteins is limited in both speed and accuracy.
3. SEC1/MUNC18–SNARE INTERACTIONS AND THEIR FUNCTIONAL IMPLICATIONS
3.1. Overview of Sec1/Munc18–SNARE Interactions
SM proteins interact with SNAREs and, like SNAREs, are essential for membrane fusion in vivo. Each SM protein interacts with one or more distinct sets of client SNAREs (36). X-ray structures of all four families of SM proteins have now been reported, and they are all very similar, the most notable feature being a large cleft created by a helical hairpin in domain 3a that extends, somewhat like a thumb, away from the globular body of the protein (27, 83-94) (Figure 2). The molecular mechanisms underlying SM function have been challenging to decipher. This is, at least in part, because the interactions between SM proteins and their cognate SNAREs are remarkably varied. For example, SM proteins can bind the extreme N terminus—the N-peptide—of Qa-SNAREs (85, 86, 95, 96), but the affinity of the interaction ranges from low nanomolar to almost undetectable (86, 87, 91, 92, 97, 98). More generally, it has been unclear whether there are universal principles that explain why SM proteins are essential for SNARE-mediated fusion. Recent developments have, however, seemed to move us closer to answering this question. Here, we describe representative SM–SNARE interactions and draw some conclusions about their likely roles in SNARE assembly and membrane fusion.
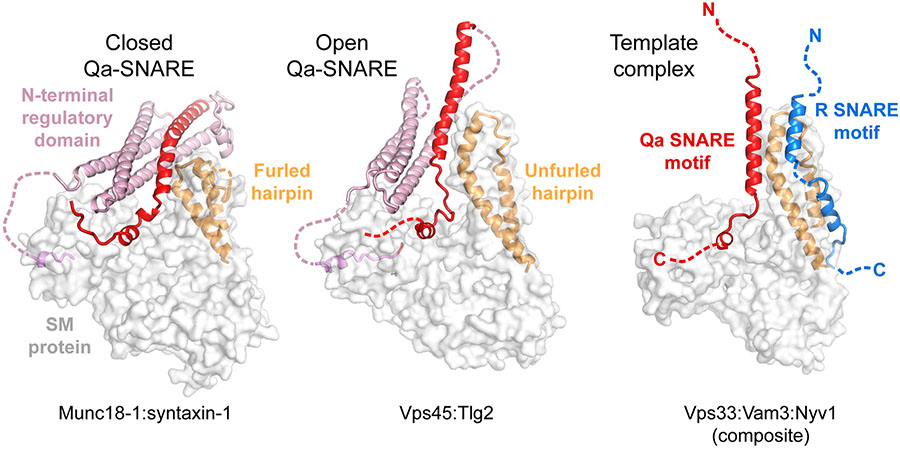
Figure 2.
Structures of SM–Qa-SNARE complexes. (a) The SM protein Munc18-1 bound to the Qa-SNARE syntaxin-1 (PDB ID: 3C98). Syntaxin-1 adopts a closed conformation in which the SNARE motif (red) interacts with the NRD (pink). The helical hairpin (gold) of Munc18-1 (gray surface) adopts a furled conformation, hiding the R-SNARE binding site. (b) The SM protein Vps45 bound to the Qa-SNARE Tlg2 (PDB ID: 6XM1). Tlg2 adopts an open conformation, with only minimal interactions between the SNARE motif and the NRD. In addition, the linker connecting the three-helical Habc domain (pink) to the SNARE motif (red) is disordered, unlike in the Munc18-1–syntaxin-1 structure. The helical hairpin is unfurled, exposing the presumptive R-SNARE binding site. (c) A composite model combining two SM–SNARE motif crystal structures: Vps33 with the Qa-SNARE Vam3 (PDB ID: 5BUZ) and Vps33 with the R-SNARE Nyv1 (PDB ID: 5BV0). In both the Munc18-1–syntaxin-1 and Vps45–Tlg2 structures, the N-peptide-binding site is at the left, behind the SM protein; Vps33 does not possess an N-peptide-binding site. Abbreviations: NRD, N-terminal regulatory domain; PDB ID, Protein Data Bank identifier; SM, Sec1/Munc18.
3.2. Sec1/Munc18–Qa-SNARE Complexes
Munc18-1 binds tightly to the Qa-SNARE syntaxin-1 with an affinity of approximately 1 nM (44, 86). Crystal structures revealed that Munc18-1 associates with the entire length of syntaxin-1’s cytoplasmic domain (86, 94) (Figure 2a). The bound SNARE is wedged into the cleft, with the Habc domain folded back on the N-terminal half of the SNARE motif to form a closed conformation. The SNARE motif, rather than being a single unbroken helix, as observed in the cis-SNARE complex, instead forms several shorter helices and is disordered at its C-terminal end. Most of its core residues are sequestered in interactions with the Habc domain (layers −7 to +1) or Munc18-1 (layers +2 and +3), preventing syntaxin-1 from associating with other SNAREs. Consequently, Munc18-1 inhibits SNARE assembly in vitro (86, 94). Nevertheless, the high-affinity Munc18-1–syntaxin-1 complex is important for the trafficking of syntaxin-1 to the presynaptic membrane (99) and appears to represent the physiologically relevant starting point for SNARE complex assembly (29)
The idea that SM proteins trap Qa-SNAREs in closed conformations has been highly influential, but is it the rule or the exception? Subsequent SM–Qa-SNARE structures, because they lacked major portions of the SNARE, left this question unresolved. One such structure, Sly1–Sed5, contained only the N-peptide of the Qa-SNARE, which binds with nanomolar affinity to the back side of the SM protein (85). Reexamination of the Munc18-1–syntaxin-1 structure (94) revealed that syntaxin-1’s N-peptide binds to an analogous site (86) (Figure 2a). Yet another SM–Qa-SNARE structure, of Vps33 bound to Vam3, contained only the SNARE motif of Vam3 (27). Very recently the structure of a second almost-complete Qa-SNARE (Tlg2) bound to an SM protein (Vps45) was reported (89) (Figure 2b). Strikingly, although Vps45 engages all of the same Qa-SNARE regions as Munc18-1—the N-peptide, the Habc domain, and the SNARE motif—the conformation of the bound SNARE is much more open. The SNARE motif makes the same interactions with the SM protein, centered around layers +2 and +3, but interacts only glancingly with the Habc domain. Interestingly, both syntaxin-1 and Tlg2 are prone to misassembling into tetramers, driven by their SNARE motif’s propensity to assemble into four-helix bundles (76, 77, 89). Both the closed complex formed by Munc18-1–syntaxin-1 and the open complex formed by Vps45–Tlg2 prevent this off-pathway association from occurring. Overall, it is now clear that SM-bound Qa-SNAREs can adopt at least two conformations, one closed and one (more) open. It is intriguing to speculate that the closed binding mode might be a specialization that evolved to provide stringent control of certain SNARE assembly reactions, such as those that prime synaptic vesicles for neurotransmitter release.
3.3. Sec1/Munc18–R-SNARE Complexes
Despite substantial evidence that SM proteins bind to R-SNAREs (44, 100-102), only one structure of an SM–R-SNARE complex has been reported: Vps33 bound to the SNARE motif of Nyv1 (27). The R-SNARE binding site, while distinct from the Qa-SNARE binding site, is formed almost entirely by the extended helical hairpin (Figure 2c). Once again, the bound SNARE motif forms several (in this case, two) shorter helices rather than one long one, with a break at the zero-layer arginine. Structure-guided mutations that weakened the interaction between Vps33 and Nyv1 compromised SNARE- and SM-mediated fusion, both in vitro and in vivo, supporting the functional significance of the SM–R-SNARE interaction (27).
Analogously, direct binding between Munc18-1 and VAMP2 was demonstrated in vitro using FRET assays, nuclear magnetic resonance, cross-linking mass spectrometry, and pull-down assays, with an estimated Kd in the range of 10–100 μM (43, 101, 103). As with Vps33–Nyv1, binding occurs on the helical hairpin of Munc18-1, and the Munc18-1 hairpin must be in its extended or unfurled conformation in order to bind VAMP2 (44, 101, 103, 104) (Figure 2b,c). Conversely the helical hairpin adopts a furled conformation in the Munc18-1–syntaxin-1 structure, with the tip of the hairpin folded over and a substantial portion of the R-SNARE binding site buried inside (Figure 2a). Mutations in the helical hairpin such as P335A and D326K, both of which appear to promote unfurling, enhance both VAMP2 binding and membrane fusion (44, 101). Although a high-resolution structure of the Munc18–VAMP2 complex has not been reported, the available data are consistent with a binding mode much like the one observed in the Vps33–Nyv1 complex (44, 105).
3.4. Sec1/Munc18–RQa Complexes (Template Complexes)
The crystal structures of the Vps33–Vam3 and Vps33–Nyv1 binary complexes demonstrated that the R-SNARE and the Qa-SNARE motifs bind to nonoverlapping sites, consistent with the observation by size-exclusion chromatography that all three proteins form a ternary complex (27). Combining the two crystal structures yielded a model for this ternary complex (Figure 2c) in which the NTDs (i.e., the N-terminal halves of the SNARE motifs) are helical and properly aligned for SNARE complex assembly. The CTDs, by contrast, diverge, with the R-SNARE following the groove in the helical hairpin, while the Qa-SNARE follows the same path as it does in the closed Munc18-1–syntaxin-1 structure. This model is evocative of a half-zippered SNARE complex. The SM protein functions as a template, nucleating SNARE assembly by holding the NTDs of the R- and Qa-SNARE motifs in alignment for binding to the NTDs of the Qb- and Qc-SNAREs.
Single-molecule force spectroscopy in conjunction with structural modeling and site-directed mutagenesis provided strong evidence that both Vps33 and Munc18-1 form structurally similar ternary template complexes with their cognate R- and Qa-SNAREs (28) (Figure 3). Importantly, these experiments allowed thermodynamic stabilities and folding/unfolding kinetics to be measured. The synaptic template complex was found to have an unfolding free energy of 5.2 kBT and a lifetime of 1.4 seconds. The stabilities of various mutant template complexes were measured to enable comparison with a rich set of existing data cataloging the impact of mutations on membrane fusion and/or neurotransmitter release. Mutations known to attenuate membrane fusion formed less stable template complexes whereas mutations that enhance membrane fusion formed superstable template complexes (28). This correlation suggests that formation of the template complex is a rate-limiting step for SNARE-mediated membrane fusion in vitro and in vivo.
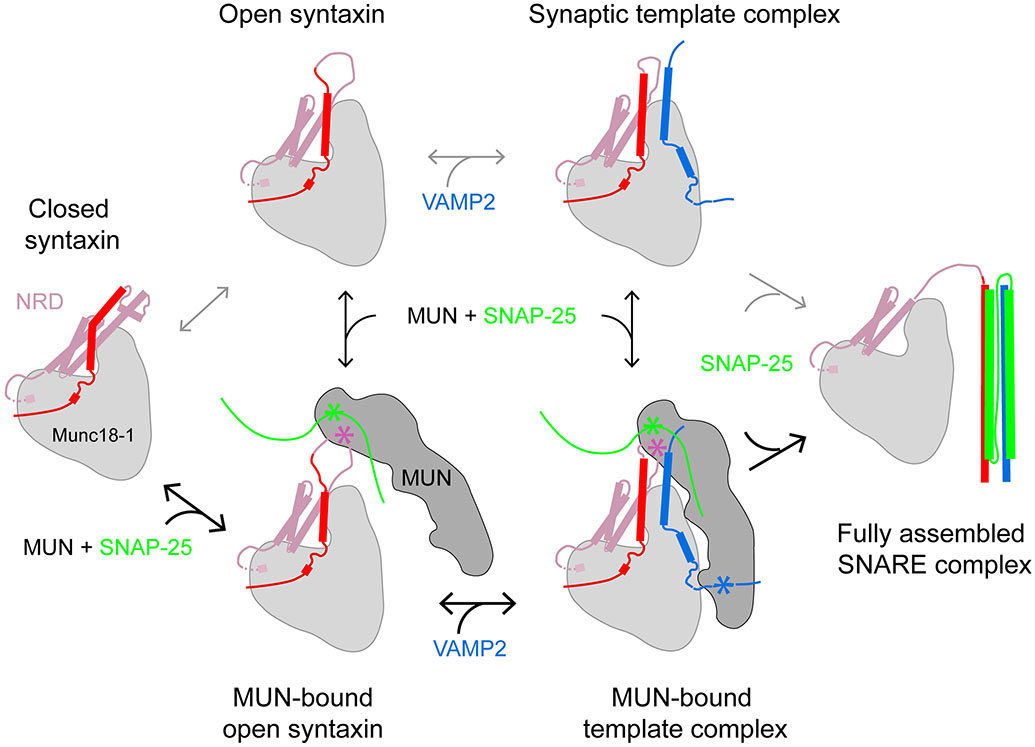
Figure 3.
Schematic diagrams of key intermediates and pathways of SNARE assembly chaperoned by Munc18-1 and the MUN domain of Munc13-1. Open syntaxin-1 can be generated by local unfolding of the syntaxin-1 linker region, through either thermal fluctuation, perturbation by mutation, or binding of Munc13-1. VAMP2 binding to the opened Munc18-1–syntaxin-1 complex forms the template complex, which in turn can be stabilized by the MUN domain of Munc13-1. SNAP-25 binding concludes SNARE assembly and displaces both chaperones from the four-helix bundle. Stars (outlined in yellow) indicate SNARE-binding sites on the MUN domain; the binding site for SNAP-25 has not been mapped and is therefore arbitrary. Abbreviation: NRD, N-terminal regulatory domain.
Single-molecule studies of Munc18-1 and SNARE mutant proteins also revealed new insights into the synaptic template complex (28) (Figure 3). Dissection of the syntaxin-1 NRD revealed that both the N-peptide and the Habc domain stabilize the template complex. The impact of the Habc domain was especially dramatic: When it was deleted, the template complex failed to form, but when it was added back as a separate molecule in trans, the template complex was once again observed. By contrast, removal of the linker region connecting the Habc domain to the SNARE motif had no apparent effect on the stability of the template complex (J. Yang & Y. Zhang, unpublished results). Thus, this linker region, which is well ordered in the Munc18-1–syntaxin-1 complex (Figure 2a), is likely to be unfolded in the template complex. Overall, single-molecule studies indicated that two regions of the NRD—the N-peptide and the Habc domain—stabilize the synaptic template complex and thereby promote membrane fusion. Nonetheless, unresolved discrepancies remain. Most notably, the roles of the N-peptide and Habc domain in liposome fusion and neurotransmitter release have been somewhat controversial, with some reports concluding that they are essential and others that they are dispensable (38, 106-110).
There are several limitations inherent in using the known structures of Vps33–Vam3 and Vps33–Nyv1 to model the structure of the synaptic template complex. First, the NRD of the Qa-SNARE is missing, so its role in stabilizing the template complex cannot be rationalized. Second, the R- and Qa-SNARE motifs, when present simultaneously, seem certain to influence one another’s conformations and perhaps also that of the helical hairpin. Indeed, single-molecule studies of mutant template complexes suggested that the VAMP2 NTD is helical all the way to the N-terminal −7 layer (28), whereas in the Vps33–Nyv1 structure, layers −7 to −4 are disordered (27). The −7 to −4 layers of VAMP2 presumably stabilize the template complex through interactions with the corresponding layers in syntaxin-1. For the CTDs, however, many aspects of the model based on Vps33–Vam3 and Vps33–Nyv1 are borne out by single-molecule studies. For example, replacing the highly conserved +6 layer phenylalanine of VAMP2 with alanine abolishes template-complex formation, consistent with the expectation that this residue inserts into a hydrophobic pocket on Munc18-1. Altogether, it appears that the majority of the R- and Qa-SNARE motifs contribute to the stability of the template complex—from the −7 to the +3 layer for syntaxin-1 and from the −7 to the +6 layer for VAMP2.
Four different template complexes, associated with various fusion machineries, have now been observed and characterized using optical tweezers. These include, in addition to Munc18-1–syntaxin-1–VAMP2 and Vps33–Nyv1–Vam3, Munc18-2–syntaxin-11–VAMP8 for cytotoxin release (J. Yang & Y. Zhang, unpublished results) and Munc18-3–syntaxin-4–VAMP2 for GLUT4 translocation (28). All four template complexes share similar stabilities and characteristic structural features. Formation of the three Munc18-associated template complexes requires the NRD of the Qa-SNARE. These findings, taken together, imply that the template complex may be a universally conserved intermediate for SM-chaperoned SNARE assembly.
3.5. Other Sec1/Munc18–SNARE Complexes
Munc18-1 binds to the ternary synaptic SNARE complex with modest affinity (Kd = 0.7 μM) (86, 111). Neither this SM–QabcR complex, nor any other, has been characterized in structural detail. Nonetheless, it is clear that Munc18-1 engages the SNARE complex predominantly by binding to the NRD of syntaxin-1. This conclusion follows from the observation that Munc18-1 binds with identical affinity to the isolated NRD and to the SNARE complex (86). The ability to bind to the NRD of syntaxin presumably gives Munc18-1 the capacity to engage other syntaxin-1-containing SNARE complexes as well, including 1:1 and 2:1 syntaxin-1–SNAP-25 complexes. The roles of these SM–Qabc complexes are still up for debate, but possibilities include rescuing syntaxin-1 from unproductive complexes and/or promoting SNARE assembly (79, 112). However, since the t-SNARE complex is subject to disassembly by NSF, it may not represent a functional intermediate in SNARE assembly in vivo (29). Likewise, the functional significance of Munc18-1 binding to the ternary SNARE complex is questionable, since point mutations that abrogate this interaction support normal synaptic transmission (108). Overall, the SM–SNARE complexes with the best-established functional roles in synaptic vesicle fusion are the Munc18-1–syntaxin-1 complex and the Munc-18–syntaxin-1–VAMP2 template complex.
4. SEC1/MUNC18 PROTEINS CHAPERONE SNARE ASSEMBLY
4.1. Sec1/Munc18 Proteins as SNARE Chaperones
It has long been suspected that SM proteins chaperone SNARE assembly. In 2004, for example, Peng et al. proposed that the yeast SM protein Sly1 promotes specific SNARE pairing by binding its cognate t- and v-SNAREs, thus proofreading SNARE assembly (113). In 2007, Shen et al. used a reconstituted proteoliposome fusion assay to demonstrate that Munc18-1 promotes fusion via a mechanism that depends on specific SM–Qa-SNARE and SM–R-SNARE interactions (38). This important study also showed that Munc18-1 enhances trans-SNARE complex assembly. In 2013, a landmark paper by Ma et al. introduced a reconstituted proteoliposome fusion assay that depends on both Munc18-1 and a second protein that is also essential for neurotransmitter release, Munc13-1 (29). This system differed from previous reconstitutions because it began with liposomes bearing Munc18-1–syntaxin-1 rather than t-SNARE complexes. Munc13-1 was included in this assay, at least in part, to open Munc18-bound closed syntaxin, thereby permitting Munc18-chaperoned SNARE assembly (114). In 2017, Lai et al. showed that Munc18-1 and Munc13-1 could cooperate to inhibit SNARE misassembly (43). These and many additional studies implied that SM proteins function as chaperones to enhance both the speed and the accuracy of SNARE assembly. However, while Munc18-1 is without doubt the most intensively studied SM protein, its ability to inhibit SNARE assembly (by forming a stable complex with syntaxin-1) made it difficult to study its ability to promote SNARE assembly (by functioning as a template).
4.2. The Template Hypothesis
The discovery that R- and Qa-SNAREs combine with SM proteins to form a conserved template complex provides an attractive mechanism for Munc18-catalyzed SNARE assembly (27, 28). In 2018, Jiao et al. demonstrated that Munc18-1 enhances the speed and accuracy of synaptic SNARE assembly in a template complex–dependent manner (28). Once the Munc18-1–syntaxin-1–VAMP2 template complex is formed, SNAP-25 rapidly binds to form the ternary SNARE complex (Figure 3). Upon the completion of SNARE assembly, Munc18-1 likely dissociates from the SNARE four-helix bundle (28, 103, 108).
The synaptic template complex is well suited to catalyzing SNARE assembly (27, 28). The syntaxin-1 and VAMP2 NTDs, aligned in a helical conformation on the Munc18-1 helical hairpin, nucleate assembly. The divergent CTD binding sites, meanwhile, ensure that CTD zippering weakens the SNARE–template interactions, promoting product release (Figure 3). Furthermore, the extensive interactions between SM and SNARE proteins, as well as interactions between the two templated SNAREs, confer specificity by helping to prevent noncognate or antiparallel SNAREs from assembling. To play these roles, a relatively weak template complex is necessary, because an overly stable template complex would act as a kinetic trap to slow down SNARE assembly. Nonetheless, within the range of stabilities of the wild type and mutant synaptic template complexes that have been tested, there is (as noted in Section 3.4) an excellent correlation between template-complex stability and function. The rate-determining role of the template complex enables the regulatory control of SNARE assembly by either SM or SNARE posttranslational modification or by other proteins that bind to the template complex.
4.3. Effects of Phosphorylation
Membrane fusion and exocytosis are regulated by the phosphorylation of SM and SNARE proteins (115). Munc18-1 is phosphorylated on Y473 by neuronal Src family kinases, abrogating Munc18-1-dependent membrane fusion in vitro and synaptic transmission in neurons (116). In contrast, Munc18-1 phosphorylation at S306 and S313 by protein kinase C boosts neurotransmitter release and contributes to short-term synaptic plasticity (117). In adipocytes, insulin triggers phosphorylation of Munc18-3 at Y219 and Y521 to activate fusion of GLUT4 storage vesicles for glucose uptake (118). In contrast, phosphorylation of VAMP8 at T47, T53, S54, or S61 inhibits SNARE zippering and mast cell exocytosis (119).
All of the sites of Munc18-1 phosphorylation mentioned above are located in the predicted SNARE-binding interfaces and presumably alter the stability of the template complex. Indeed, the Munc18-1 phosphomimetic mutation Y473D eliminates template-complex formation, while S306D and S313D stabilize the template complex, consistent with the observed effects of phosphorylation on exocytosis (28). Alternatively, phosphorylation could alter the energetics of CTD zippering. This Munc18-1-independent mechanism presumably explains the impact of VAMP8 phosphorylation on T47, S54, and S61, which are located at the +3, +5, and +7 layers of the SNARE motif. Munc18-1-dependent and independent contributions can be distinguished by using single-molecule strategies to measure the stabilities of the template complex and the SNARE complex, respectively.
5. COOPERATION WITH OTHER ESSENTIAL SNARE CHAPERONES
5.1. Munc13-1
Munc13-1 is a large (193 kDa) protein that is essential for synaptic vesicle fusion and neurotransmitter release (120). Munc13-1 facilitates the docking and priming of synaptic vesicles upstream of Ca2+-triggered fusion. It contains a C1 domain, three C2 domains (C2A, C2B, and C2C), and a MUN domain, arranged as shown in Figure 4. The MUN domain is homologous to the CATCHR (complexes associated with tethering containing helical rods) family of multisubunit tethering complexes (MTCs) (121). The largest fragment of Munc13-1 that has been structurally characterized, C1C2BMUN, revealed that the C1 and C2B domains pack against the N-terminal end of the rod-like MUN domain; the C2C domain, while not present in the crystallized construct, would presumably be located near the C-terminal end of the MUN domain (122). These structural features match up well with the critical role of Munc13-1 in tethering vesicles to the plasma membrane (123-125). Specifically, the C1 and C2B domains recognize the plasma membrane by binding diacyglycerol and negatively charged lipids, respectively, while the C2C domain binds to the synaptic vesicle membrane (123). In the absence of Ca2+, all of these interactions appear to be weak. Recent evidence suggests that at least six copies of Munc13-1 form a cluster that tethers a synaptic vesicle to the active zone (123, 126-128). The Munc13-1 membrane tether is further regulated by Ca2+ and by binding to RIM and other proteins that play important roles in short-term synaptic plasticity (129, 130).
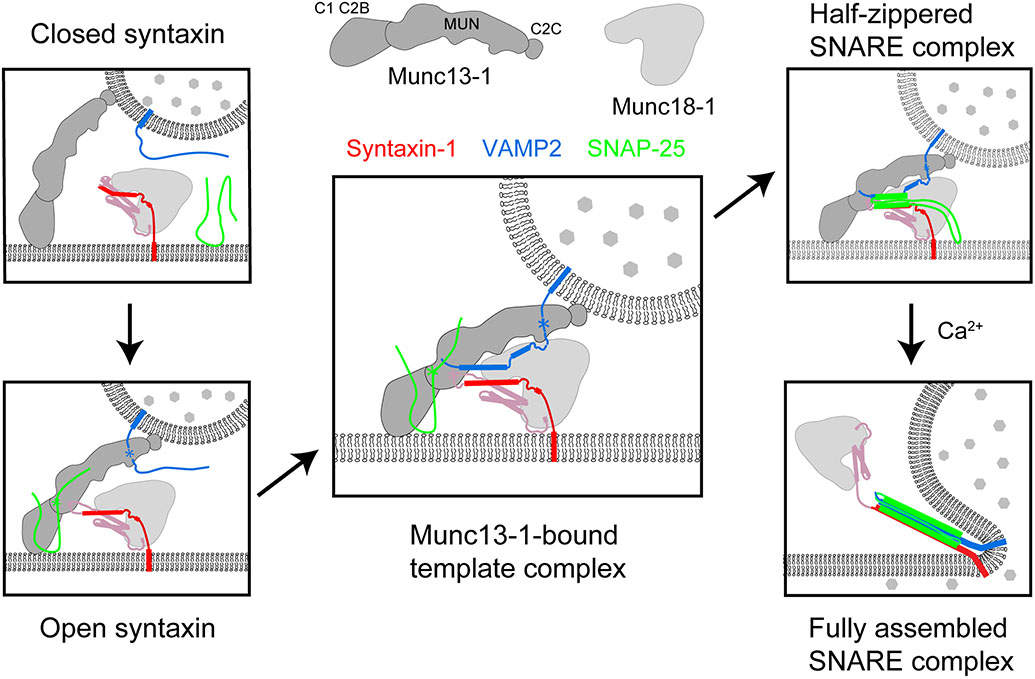
Figure 4.
Model for Munc18-1- and Munc13-1-chaperoned SNARE assembly and synaptic vesicle fusion. Munc13-1 first tethers the synaptic vesicle to the plasma membrane by simultaneously binding to both membranes through its terminal C2 and C1 domains and associates with VAMP2, SNAP-25, and Munc18-1-bound syntaxin. The latter association opens closed syntaxin. The open syntaxin then binds VAMP2 to form the template complex that is stabilized by Munc13-1. SNAP-25 binds to the templated SNAREs, generating a half-zippered SNARE complex in the presence of other SNARE chaperones like synaptotagmin and complexin. Finally, calcium triggers full SNARE assembly to induce membrane fusion and neurotransmitter release. (Top middle) The domain structure of Munc13-1 inferred from the crystal structure of the Munc13-1 fragment containing the C1, C2B, and MUN domains (122), with the N-terminal C2A domain omitted.
The MUN domain of Munc13-1 catalyzes SNARE assembly starting from Munc18-1–syntaxin-1 complexes (42, 114, 131). Catalysis requires that MUN bind to both syntaxin-1 and VAMP2: a central region of MUN binds to the linker region between the Habc domain and SNARE motif of syntaxin-1 with an affinity of ~40 μM (131), while a C-terminal region of MUN binds to the juxtamembrane LD of VAMP2 with an affinity of ~10 μM (46) (Figure 3). The ability of the MUN domain to interact with both syntaxin-1 and VAMP2 presumably underlies its ability to promote their proper orientation (i.e., parallel instead of antiparallel) in SNARE assembly experiments conducted in the absence of Munc18-1 (43). Nevertheless, the MUN domain alone fails to promote parallel pairing between syntaxin-1 and SNAP-25. It therefore seems likely that Munc13-1 cooperates with Munc18-1 in order to chaperone proper SNARE assembly.
5.2. Mechanism of Munc13-1 and Munc18-1 Cooperativity
Extensive studies have indicated that Munc13-1 and Munc18-1 cooperate to chaperone SNARE assembly by three mechanisms: opening closed syntaxin-1, recruiting SNAREs, and stabilizing the template complex (28, 29, 42, 45-47, 114, 131, 132) (Figure 3). Munc13-1 likely opens syntaxin-1 by binding to its linker region to induce its unfolding, exposing the syntaxin-1 SNARE motif for the initiation of SNARE assembly (131). Consistent with this view, a destabilizing mutation in the syntaxin-1 linker region [the so-called LE mutation (133)] enables Munc13-1-independent SNARE assembly (42). Single-molecule optical tweezers experiments revealed unfolding energies of 7.2 and 2.6 kBT for the Munc18-1-bound closed and open syntaxin-1, respectively (28), implying that the process of opening Munc18-1-bound syntaxin-1 requires an energy input of only 4.6 kBT. This is substantially less than the energy of ~22 kBT needed to wrest syntaxin-1 away from Munc18-1 (86). The structure of the opened syntaxin-1 is unknown but might, as noted in Section 3.2, resemble that of open Tlg2 bound to Vps45 (Figure 2). In summary, opening Munc18-1-bound syntaxin-1 involves a relatively small energy input, which may be provided by the weak binding of Munc13-1 to the syntaxin-1 linker region.
A recent study found that the MUN domain of Munc13-1, in addition to binding syntaxin-1 and VAMP2, also binds to the linker region of SNAP-25 with an affinity of ~30 μM (132, but see also 134) (Figure 3). It would be interesting to determine whether Munc13-1 can simultaneously bind to all three synaptic SNARE proteins in such a way as to enhance the rate of template-complex formation and SNARE assembly. Furthermore, single-molecule measurements suggest that the Munc13-1 MUN domain stabilizes the template complex, in part by binding to syntaxin-1 and VAMP2, and thereby supports efficient SNAP-25 binding (45). Together, these studies support a model in which the templating function of Munc18-1 joins with the multiple functions of Munc13-1 in syntaxin-1 opening, SNARE recruitment, and template-complex stabilization to cooperatively enhance SNAP-25 binding and SNARE zippering.
5.3. NSF/SNAP
SM proteins also cooperate with the SNARE disassembly machinery to promote proper SNARE assembly. Following membrane fusion, cis-SNARE complexes are disassembled by the AAA+ ATPase NSF and its adapter protein SNAP (no relation to SNAP-25) in an ATP-dependent manner (23). Other NSF/SNAP substrates include SNARE assembly intermediates—1:1 and 2:1 t-SNARE complexes, syntaxin-1 oligomers, and partially assembled trans-SNARE complexes—as well as various misassembled SNARE complexes (135-137). SM proteins appear to protect productive assembly intermediates from disassembly by NSF/SNAP, while permitting NSF/SNAP to recycle the off-pathway products of SNARE misassembly reactions. This role of SM proteins has been demonstrated for Munc18-1, Vps33, and Sly1 (136-139); however, its detailed molecular mechanism remains to be elucidated.
5.4. Multisubunit tethering complexes
The initial, reversible contact between a vesicle and its target membrane is thought to be mediated by MTCs (26, 140). For synaptic vesicles, Munc13-1 appears to play this role; in most other trafficking pathways, tethering requires an MTC from either the CATCHR or HOPS/CORVET family. There is abundant evidence that both families of MTCs, in addition to tethering the two membranes, promote SNARE assembly. The SNARE assembly activity of the HOPS/CORVET family relies in part on the SM protein Vps33, which is an integral subunit of the complex (27). There is also some evidence for direct interactions between CATCHR-family MTCs and SM proteins (141, 142). In addition, the MTCs appear to have SM-independent roles in SNARE assembly. For example, the CATCHR-family Dsl1 complex binds Qb- and Qc-SNAREs via their Habc domains, holding them in proximity and preventing them from forming closed conformations, while the CATCHR-family exocyst complex binds to the Qa-SNARE Sso2 such that it loosens its otherwise tightly closed conformation (143, 144). How MTCs and SM proteins collaborate to couple vesicle tethering to efficient SNARE assembly and membrane fusion is a fascinating area for future investigation.
6. THE SNARE–SEC1/MUNC18 FUSION MACHINERY AND HUMAN DISEASE
Mutations in SNAREs and SM proteins have been linked to diverse diseases and disorders. For example, hundreds of Munc18-1 heterozygous mutations have been identified in patients with epilepsy, intellectual disability, and movement disorders (4, 145, 146). The SNAP-25 substitution I67N causes myasthenia, cerebellar ataxia, cortical hyperexcitability, and intellectual disability, and bovine chromaffin cells expressing the mutant protein exhibited reduced evoked exocytosis (146). Defects in the SNARE–SM fusion machinery responsible for cytotoxin secretion in cytotoxic T lymphocytes and natural killer cells, including mutations in syntaxin-11, Munc18-2, and Munc13-4, are associated with the immune disorder hemophagocytic lymphohistiocytosis (7, 147-150). Mutations in human Vps33B cause defects in platelet α-granule biogenesis and are linked to arthrogryposis–renal dysfunction–cholestasis syndrome (151-153).
Defects in the SNARE–SM fusion machinery can cause disease via multiple mechanisms. First, SNARE mutations can directly alter the energetics of SNARE zippering. For example, the SNAP-25 mutation I67N, located in the +4 layer of the Qb-SNARE motif, lowers the CTD zippering energy by 14 kBT (5). Similarly, the SNAP-25 I67T mutation, identified in the blind-drunk mouse, reduces CTD zippering energy by 10 kBT. Second, SNARE or SM protein mutations can alter the stability of the template complex and thus the ability of the SM protein to chaperone SNARE assembly. The Munc18-1 disease mutations P335L and L341P (4), for example, destabilize and abolish the template complex, respectively, thereby compromising SNARE assembly (28). The effects of these mutations are not surprising, because all of them are located in the SM–SNARE binding interface. Third, many Munc18-1 mutants fail to fold properly and are degraded in the cell, causing Stxbp1 (another name for Munc18-1) haploinsufficiency (154, 155). Stxbp1 haploinsufficiency tends to reduce cortical inhibitory neurotransmission, causing seizures characteristic of epilepsies (154). Finally, many disease mutations are found on the surface of SM proteins outside the SNARE binding interface (4). These mutations are not expected to significantly destabilize the SM proteins nor to directly affect SNARE binding, suggesting mechanisms yet to be discovered.
Synaptic SNAREs are also chaperoned by other proteins, including two—α-synuclein and cysteine string protein-α (CSPα)—that are involved in neurodegeneration (156). Aggregation of α-synuclein in Lewy bodies is a defining feature of Parkinson’s disease (157). Interestingly, both α-synuclein and CSPα prevent SNAP-25 aggregation and degradation; α-synuclein additionally protects VAMP2 (158). The underlying molecular mechanisms, however, are not well understood.
7. CONCLUSIONS AND PERSPECTIVES
Current data support a working model for membrane fusion, using synaptic exocytosis as an example, that places essential SNARE chaperones in the context of the full fusion machinery (Figure 4). First, during vesicle docking, SNAREs and Munc18-1 are recruited to and thereby concentrated at the future site of membrane fusion, in part by tethering proteins including Munc13-1. Munc13-1 bridges vesicle and plasma membranes and catalyzes syntaxin-1 opening. Subsequently, Munc18-1 binds to VAMP2 to form the template complex, which is further stabilized by Munc13-1. Rapid binding of SNAP-25 generates a partially zippered SNARE complex. Synaptotagmin and complexin are likely to associate with the partially zippered SNARE complex, stabilizing it in an NSF/SNAP-resistant, primed state. Finally, calcium triggers fast zippering of the SNARE CTDs, inducing membrane fusion.
Fundamental questions remain to be addressed. First, how do SNAP-25 and other key regulatory proteins such as synaptotagmin and complexin interact with the synaptic template complex and potentially oligomerize to form a SNARE-chaperone supercomplex, as is outlined in the buttressed ring model (126)? Second, what is the precise structural nature of the template complex and subsequent SNARE assembly intermediates? Finally, how do the structures of the template complex and subsequent intermediates confer resistance to disassembly by NSF–SNAP and thereby allow productive SNARE assembly? These and other questions should be addressable through the synergistic application of structural biology, single-molecule methods, and other experimental approaches.
ACKNOWLEDGMENTS
We thank James Rothman, Jose Rizo, and Jingshi Shen for their critical reading of the manuscript. Work on SM–SNARE proteins in the authors’ labs is supported by NIH grants R35 GM131714 (to Y.Z.) and R01 GM071574 (to F.M.H.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Rothman JE. 2014. The principle of membrane fusion in the cell (Nobel lecture). Angew. Chem. Int. Ed 53:12676–94 [DOI] [PubMed] [Google Scholar]
- 2.Südhof TC. 2014. The molecular machinery of neurotransmitter release (Nobel lecture). Angew. Chem. Int. Ed 53:12696–717 [DOI] [PubMed] [Google Scholar]
- 3.Südhof TC, Rothman JE. 2009. Membrane fusion: grappling with SNARE and SM proteins. Science 323:474–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stamberger H, Nikanorova M, Willemsen MH, Accorsi P, Angriman M, et al. 2016. STXBP1 encephalopathy: a neurodevelopmental disorder including epilepsy. Neurology 86:954–62 [DOI] [PubMed] [Google Scholar]
- 5.Rebane AA, Wang B, Ma L, Qu H, Coleman J, et al. 2018. Two disease-causing SNAP-25B mutations selectively impair SNARE C-terminal assembly. J. Mol. Biol 430:479–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rorsman P, Ashcroft FM. 2018. Pancreatic β-cell electrical activity and insulin secretion: of mice and men. Physiol. Rev 98:117–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.zur Stadt U, Rohr J, Seifert W, Koch F, Grieve S, et al. 2009. Familial hemophagocytic lymphohistiocytosis type 5 (FHL-5) is caused by mutations in Munc18-2 and impaired binding to syntaxin 11. Am. J. Hum. Genet 85:482–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasshauer D, Sutton RB, Brunger AT, Jahn R. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. PNAS 95:15781–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloepper TH, Kienle CN, Fasshauer D. 2007. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18:3463–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasshauer D, Eliason WK, Brunger AT, Jahn R. 1998. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry 37:10354–62 [DOI] [PubMed] [Google Scholar]
- 11.Liang BY, Kiessling V, Tamm LK. 2013. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. PNAS 110: 19384–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lakomek NA, Yavuz H, Jahn R, Perez-Lara A. 2019. Structural dynamics and transient lipid binding of synaptobrevin-2 tune SNARE assembly and membrane fusion. PNAS 116:8699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Tu J, Zhang S, Cai B, Liu Z, et al. 2020. Different regions of synaptic vesicle membrane regulate VAMP2 conformation for the SNARE assembly. Nat. Commun 11:1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton RB, Fasshauer D, Jahn R, Brunger AT. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395:347–53 [DOI] [PubMed] [Google Scholar]
- 15.Stein A, Weber G, Wahl MC, Jahn R. 2009. Helical extension of the neuronal SNARE complex into the membrane. Nature 460:525–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. 1997. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90:523–35 [DOI] [PubMed] [Google Scholar]
- 17.Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, et al. 2012. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337:1340–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kesavan J, Borisovska M, Bruns D. 2007. v-SNARE actions during Ca2+-triggered exocytosis. Cell 131:351–63 [DOI] [PubMed] [Google Scholar]
- 19.Oelkers M, Witt H, Halder P, Jahn R, Janshoff A. 2016. SNARE-mediated membrane fusion trajectories derived from force-clamp experiments. PNAS 113:13051–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zorman S, Rebane AA, Ma L, Yang G, Molski MA, et al. 2014. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. eLife 3:e03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, et al. 1998. SNAREpins: minimal machinery for membrane fusion. Cell 92:759–72 [DOI] [PubMed] [Google Scholar]
- 22.Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. 1993. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75:409–18 [DOI] [PubMed] [Google Scholar]
- 23.Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, et al. 2015. Mechanistic insights into the recycling machine of the SNARE complex. Nature 518:61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Rebane AA, Yang G, Xi Z, Kang Y, et al. 2015. Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife 4:e09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y 2017. Energetics, kinetics, and pathway of SNARE folding and assembly revealed by optical tweezers. Protein Sci. 26:1252–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker RW, Hughson FM. 2016. Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol 17:465–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM. 2015. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349:1111–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao J, He M, Port SA, Baker RW, Xu Y, et al. 2018. Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife 7:e41771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma C, Su L, Seven AB, Xu Y, Rizo J. 2013. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339:421–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosono R, Hekimi S, Kamiya Y, Sassa T, Murakami S, et al. 1992. The unc-18 gene encodes a novel protein affecting the kinetics of acetylcholine metabolism in the nematode Caenorhabditis elegans. J. Neurochem 58:1517–25 [DOI] [PubMed] [Google Scholar]
- 32.Novick P, Field C, Schekman R. 1980. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21:205–15 [DOI] [PubMed] [Google Scholar]
- 33.Novick P, Schekman R. 1979. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. PNAS 76:1858–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hata Y, Slaughter CA, Südhof TC. 1993. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366:347–51 [DOI] [PubMed] [Google Scholar]
- 35.Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, et al. 2000. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287:864–69 [DOI] [PubMed] [Google Scholar]
- 36.Hong W 2005. SNAREs and traffic. Biochim. Biophys. Acta Mol. Cell Res 1744:120–44 [DOI] [PubMed] [Google Scholar]
- 37.Hong W, Lev S. 2014. Tethering the assembly of SNARE complexes. Trends Cell Biol. 24:35–43 [DOI] [PubMed] [Google Scholar]
- 38.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. 2007. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell 128:183–95 [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, Choi UB, Lai Y, Leitz J, White KI, Zhou Q. 2019. The pre-synaptic fusion machinery. Curr. Opin. Struct. Biol 54:179–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunger AT, Choi UB, Lai Y, Leitz J, Zhou Q. 2018. Molecular mechanisms of fast neurotransmitter release. Annu. Rev. Biophys 47:469–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizo J 2018. Mechanism of neurotransmitter release coming into focus. Protein Sci. 27:1364–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Wang S, Sheng Y, Zhang M, Zou W, et al. 2015. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat. Struct. Mol. Biol 22:547–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai Y, Choi UB, Leitz J, Rhee HJ, Lee C, et al. 2017. Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron 95:591–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitarska E, Xu J, Park S, Liu X, Quade B, et al. 2017. Autoinhibition of Munc18-1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. eLife 6:e24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu T, Jin H, Rothman JE, Zhang Y. 2020. Munc13-1 MUN domain and Munc18-1 cooperatively chaperone SNARE assembly through a tetrameric complex. PNAS 117:1036–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Li Y, Gong J, Ye S, Yang X, et al. 2019. Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly. Nat. Commun 10:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Gong J, Zhu L, Wang S, Yang X, et al. 2020. Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly. EMBO J. 39: e103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, et al. 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature 362:318–24 [DOI] [PubMed] [Google Scholar]
- 49.Fernandez I, Ubach J, Dulubova I, Zhang X, Südhof TC, Rizo J. 1998. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell 94:841–49 [DOI] [PubMed] [Google Scholar]
- 50.Antonin W, Fasshauer D, Becker S, Jahn R, Schneider TR. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol 9:107–11 [DOI] [PubMed] [Google Scholar]
- 51.Fasshauer D, Otto H, Eliason WK, Jahn R, Brünger AT. 1997. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem 272:28036–41 [DOI] [PubMed] [Google Scholar]
- 52.Li F, Tiwari N, Rothman JE, Pincet F. 2016. Kinetic barriers to SNAREpin assembly in the regulation of membrane docking/priming and fusion. PNAS 113:10536–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pobbati AV, Stein A, Fasshauer D. 2006. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science 313:673–76 [DOI] [PubMed] [Google Scholar]
- 54.Fasshauer D, Margittai M. 2004. A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J. Biol. Chem 279:7613–21 [DOI] [PubMed] [Google Scholar]
- 55.Fasshauer D, Antonin W, Subramaniam V, Jahn R. 2002. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat. Struct. Biol 9:144–51 [DOI] [PubMed] [Google Scholar]
- 56.Rebane AA, Ma L, Zhang YL. 2016. Structure-based derivation of protein folding intermediates and energies from optical tweezers. Biophys. J 110:441–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bustamante C, Alexander L, Maciuba K, Kaiser CM. 2020. Single-molecule studies of protein folding with optical tweezers. Annu. Rev. Biochem 89:443–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Rebane AA, Ma L, Li F, Jiao J, et al. 2016. Stability, folding dynamics, and long-range conformational transition of the synaptic t-SNARE complex. PNAS 113:E8031–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Q, Zhou P, Wang AL, Wu D, Zhao M, et al. 2017. The primed SNARE–complexin–synaptotagmin complex for neuronal exocytosis. Nature 548:420–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li F, Pincet F, Perez E, Eng WS, Melia TJ, et al. 2007.Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat. Struct. Mol. Biol 14:890–96 [DOI] [PubMed] [Google Scholar]
- 61.Min D, Kim K, Hyeon C, Cho YH, Shin YK, Yoon TY. 2013. Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism. Nat. Commun 4:1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shon MJ, Kim H, Yoon TY. 2018. Focused clamping of a single neuronal SNARE complex by complexin under high mechanical tension. Nat. Commun 9:3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu T, Rammner B, Margittai M, Artalejo AR, Neher E, Jahn R. 1999. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell 99:713–22 [DOI] [PubMed] [Google Scholar]
- 64.Das D, Bao H, Courtney KC, Wu LX, Chapman ER. 2020. Resolving kinetic intermediates during the regulated assembly and disassembly of fusion pores. Nat. Commun 11:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li F, Kummel D, Coleman J, Reinisch KM, Rothman JE, Pincet F. 2014. A half-zippered SNARE complex represents a functional intermediate in membrane fusion. J. Am. Chem. Soc 136:3456–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sorensen JB. 2010. Synaptobrevin N-terminally bound to syntaxin–SNAP-25 defines the primed vesicle state in regulated exocytosis. J. Cell Biol 188:401–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, et al. 2000. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol 150:105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou P, Bacaj T, Yang X, Pang ZP, Sudhof TC. 2013. Lipid-anchored snares lacking transmembrane regions fully support membrane fusion during neurotransmitter release. Neuron 80:470–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rathore SS, Liu YH, Yu HJ, Wan C, Lee M, et al. 2019. Intracellular vesicle fusion requires a membrane-destabilizing peptide located at the juxtamembrane region of the v-SNARE. Cell Rep. 29:4583–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dhara M, Martinez MM, Makke M, Schwarz Y, Mohrmann R, Bruns D. 2020. Synergistic actions of v-SNARE transmembrane domains and membrane-curvature modifying lipids in neurotransmitter release. eLife 9:e55152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang CW, Chiang CW, Gaffaney JD, Chapman ER, Jackson MB. 2016. Lipid-anchored synaptobrevin provides little or no support for exocytosis or liposome fusion. J. Biol. Chem 291:2848–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bao H, Goldschen-Ohm M, Jeggle P, Chanda B, Edwardson JM, Chapman ER. 2016. Exocytotic fusion pores are composed of both lipids and proteins. Nat. Struct. Mol. Biol 23:67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ngatchou AN, Kisler K, Fang QH, Walter AM, Zhao Y, et al. 2010. Role of the synaptobrevin C terminus in fusion pore formation. PNAS 107:18463–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han X, Wang CT, Bai JH, Chapman ER, Jackson MB. 2004. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science 304:289–92 [DOI] [PubMed] [Google Scholar]
- 75.Brunger AT. 2005. Structure and function of SNARE and SNARE-interacting proteins. Q. Rev. Biophys 38:1–47 [DOI] [PubMed] [Google Scholar]
- 76.Lerman JC, Robblee J, Fairman R, Hughson FM. 2000. Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry 39:8470–79 [DOI] [PubMed] [Google Scholar]
- 77.Misura KM, Scheller RH, Weis WI. 2001. Self-association of the H3 region of syntaxin 1A. Implications for intermediates in SNARE complex assembly. J. Biol. Chem 276:13273–82 [DOI] [PubMed] [Google Scholar]
- 78.Xiao W, Poirier MA, Bennett MK, Shin YK. 2001. The neuronal t-SNARE complex is a parallel four-helix bundle. Nat. Struct. Biol 8:308–11 [DOI] [PubMed] [Google Scholar]
- 79.Dawidowski D, Cafiso DS. 2016. Munc18-1 and the Syntaxin-1 N terminus regulate open-closed states in a t-SNARE complex. Structure 24:392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weninger K, Bowen ME, Chu S, Brunger AT. 2003. Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. PNAS 100:14800–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fasshauer D, Antonin W, Margittai M, Pabst S, Jahn R. 1999. Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J. Biol. Chem 274:15440–46 [DOI] [PubMed] [Google Scholar]
- 82.Yang B, Gonzalez L Jr, Prekeris R, Steegmaier M, Advani RJ, Scheller RH. 1999. SNARE interactions are not selective. Implications for membrane fusion specificity. J. Biol. Chem 274:5649–53 [DOI] [PubMed] [Google Scholar]
- 83.Baker RW, Jeffrey PD, Hughson FM. 2013. Crystal structures of the Sec1/Munc18 (SM) protein Vps33, alone and bound to the homotypic fusion and vacuolar protein sorting (HOPS) subunit Vps16. PLOS ONE 8:e67409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bracher A, Weissenhorn W. 2001. Crystal structures of neuronal squid Sec1 implicate inter-domain hinge movement in the release of t-SNAREs. J. Mol. Biol 306:7–13 [DOI] [PubMed] [Google Scholar]
- 85.Bracher A, Weissenhorn W. 2002. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 21:6114–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. 2008. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 27:923–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burkhardt P, Stegmann CM, Cooper B, Kloepper TH, Imig C, et al. 2011. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. PNAS 108:15264–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colbert KN, Hattendorf DA, Weiss TM, Burkhardt P, Fasshauer D, Weis WI. 2013. Syntaxin1a variants lacking an N-peptide or bearing the LE mutation bind to Munc18a in a closed conformation. PNAS 110:12637–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eisemann TJ, Allen F, Lau K, Shimamura GR, Jeffrey PD, Hughson FM. 2020. The Sec1/Munc18 protein Vps45 holds the Qa-SNARE Tlg2 in an open conformation. eLife 9:e60724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Graham SC, Wartosch L, Gray SR, Scourfield EJ, Deane JE, et al. 2013. Structural basis of Vps33A recruitment to the human HOPS complex by Vps16. PNAS 110:13345–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hackmann Y, Graham SC, Ehl S, Honing S, Lehmberg K, et al. 2013. Syntaxin binding mechanism and disease-causing mutations in Munc18-2. PNAS 110:E4482–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu SH, Christie MP, Saez NJ, Latham CF, Jarrott R, et al. 2011. Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. PNAS 108:1040–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu SH, Latham CF, Gee CL, James DE, Martin JL. 2007. Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. PNAS 104:8773–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Misura KM, Scheller RH, Weis WI. 2000. Three-dimensional structure of the neuronal-Sec1–syntaxin 1a complex. Nature 404:355–62 [DOI] [PubMed] [Google Scholar]
- 95.Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, et al. 2002. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 21:3620–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamaguchi T, Dulubova I, Min SW, Chen X, Rizo J, Südhof TC. 2002. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev. Cell 2:295–305 [DOI] [PubMed] [Google Scholar]
- 97.Demircioglu FE, Burkhardt P, Fasshauer D. 2014. The SM protein Sly1 accelerates assembly of the ER–Golgi SNARE complex. PNAS 111:13828–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morey C, Kienle CN, Klopper TH, Burkhardt P, Fasshauer D. 2017. Evidence for a conserved inhibitory binding mode between the membrane fusion assembly factors Munc18 and syntaxin in animals. J. Biol. Chem 292:20449–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Medine CN, Rickman C, Chamberlain LH, Duncan RR. 2007. Munc18-1 prevents the formation of ectopic SNARE complexes in living cells. J. Cell Sci 120:4407–15 [DOI] [PubMed] [Google Scholar]
- 100.Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. 2006. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J. Cell Biol 173:927–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parisotto D, Pfau M, Scheutzow A, Wild K, Mayer MP, et al. 2014. An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J. Biol. Chem 289:9639–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen C, Liu Y, Yu H, Gulbranson DR, Kogut I, et al. 2018. The N-peptide-binding mode is critical to Munc18-1 function in synaptic exocytosis. J. Biol. Chem 293:18309–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu Y, Su L, Rizo J. 2010. Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry 49:1568–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Munch AS, Kedar GH, van Weering JR, Vazquez-Sanchez S, He E, et al. 2016. Extension of helix 12 in Munc18-1 induces vesicle priming. J. Neurosci 36:6881–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andre T, Classen J, Brenner P, Betts MJ, Dorr B, et al. 2020. The interaction of Munc18-1 Helix 11 and 12 with the central region of the VAMP2 SNARE motif is essential for SNARE templating and synaptic transmission. eNeuro 7:ENEURO.0278-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rathore SS, Bend EG, Yu H, Hammarlund M, Jorgensen EM, Shen J. 2010. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. PNAS 107:22399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen J, Rathore SS, Khandan L, Rothman JE. 2010. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J. Cell Biol 190:55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meijer M, Burkhardt P, de Wit H, Toonen RF, Fasshauer D, Verhage M. 2012. Munc18-1 mutations that strongly impair SNARE-complex binding support normal synaptic transmission. EMBO J. 31:2156–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou P, Pang ZP, Yang X, Zhang Y, Rosenmund C, et al. 2013. Syntaxin-1 N-peptide and Habc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 32:159–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park S, Bin NR, Rajah MM, Kim B, Chou TC, et al. 2016. Conformational states of syntaxin-1 govern the necessity of N-peptide binding in exocytosis of PC12 cells and Caenorhabditis elegans. Mol. Biol. Cell 27:669–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dulubova I, Khvotchev M, Liu S, Huryeva I, Südhof TC, Rizo J. 2007. Munc18-1 binds directly to the neuronal SNARE complex. PNAS 104:2697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jakhanwal S, Lee CT, Urlaub H, Jahn R. 2017. An activated Q-SNARE/SM protein complex as a possible intermediate in SNARE assembly. EMBO J. 36:1788–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peng R, Gallwitz D. 2004. Multiple SNARE interactions of an SM protein: Sed5p/Sly1p binding is dispensable for transport. EMBO J. 23:3939–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma C, Li W, Xu Y, Rizo J. 2011. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat. Struct. Mol. Biol 18:542–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laidlaw KME, Livingstone R, Al-Tobi M, Bryant NJ, Gould GW. 2017. SNARE phosphorylation: a control mechanism for insulin-stimulated glucose transport and other regulated exocytic events. Biochem. Soc. Trans 45:1271–77 [DOI] [PubMed] [Google Scholar]
- 116.Meijer M, Dorr B, Lammertse HC, Blithikioti C, van Weering JR, et al. 2018. Tyrosine phosphorylation of Munc18-1 inhibits synaptic transmission by preventing SNARE assembly. EMBO J. 37:300–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Genc O, Kochubey O, Toonen RF, Verhage M, Schneggenburger R. 2014. Munc18-1 is a dynamically regulated PKC target during short-term enhancement of transmitter release. eLife 3:e01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jewell JL, Oh E, Ramalingam L, Kalwat MA, Tagliabracci VS, et al. 2011. Munc18c phosphorylation by the insulin receptor links cell signaling directly to SNARE exocytosis. J. Cell Biol 193:185–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Malmersjo S, Di Palma S, Diao J, Lai Y, Pfuetzner RA, et al. 2016. Phosphorylation of residues inside the SNARE complex suppresses secretory vesicle fusion. EMBO J. 35:1810–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Augustin I, Rosenmund C, Südhof TC, Brose N. 1999. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 400:457–61 [DOI] [PubMed] [Google Scholar]
- 121.Li W, Ma C, Guan R, Xu Y, Tomchick DR, Rizo J. 2011. The crystal structure of a Munc13 C-terminal module exhibits a remarkable similarity to vesicle tethering factors. Structure 19:1443–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu J, Camacho M, Xu Y, Esser V, Liu X, et al. 2017. Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. eLife 6:e22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Quade B, Camacho M, Zhao X, Orlando M, Trimbuch T, et al. 2019. Membrane bridging by Munc13-1 is crucial for neurotransmitter release. eLife 8:e42806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu XX, Seven AB, Camacho M, Esser V, Xu JJ, et al. 2016. Functional synergy between the Munc13 C-terminal C1 and C2 domains. eLife 5:e13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shin OH, Lu J, Rhee JS, Tomchick DR, Pang ZPP, et al. 2010. Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat. Struct. Mol. Biol 17:280–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rothman JE, Krishnakumar SS, Grushin K, Pincet F. 2017. Hypothesis—buttressed rings assemble, clamp, and release SNAREpins for synaptic transmission. FEBS Lett. 591:3459–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li F, Sundaram VK, Gatta AT, Coleman J, Krishnakumar S, et al. 2021. Vesicle capture by discrete self-assembled clusters of membrane-bound Munc13. bioRxiv 2020.08.17.254821. 10.1101/2020.08.17.254821 [DOI] [Google Scholar]
- 128.Sakamoto H, Ariyoshi T, Kimpara N, Sugao K, Taiko I, et al. 2018. Synaptic weight set by Munc13-1 supramolecular assemblies. Nat. Neurosci 21:41–49 [DOI] [PubMed] [Google Scholar]
- 129.Michelassi F, Liu H, Hu Z, Dittman JS. 2017. A C1–C2 module in munc13 inhibits calcium-dependent neurotransmitter release. Neuron 95:577–90.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Camacho M, Basu J, Trimbuch T, Chang SW, Pulido-Lozano C, et al. 2017. Heterodimerization of Munc13 C2A domain with RIM regulates synaptic vesicle docking and priming. Nat. Commun 8:15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang S, Choi UB, Gong J, Yang X, Li Y, et al. 2017. Conformational change of syntaxin linker region induced by Munc13s initiates SNARE complex formation in synaptic exocytosis. EMBO J. 36:816–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kalyana Sundaram RV, Jin H, Li F, Shu T, Coleman J, et al. 2020. Munc13 binds and recruits SNAP25 to chaperone SNARE complex assembly. FEBS Lett. 595:297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Richmond JE, Weimer RM, Jorgensen EM. 2001. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature 412:338–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Magdziarek M, Bolembach AA, Stepien KP, Quade B, Liu XX, Rizo J. 2020. Re-examining how Munc13-1 facilitates opening of syntaxin-1. Protein Sci. 29:1440–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Choi UB, Zhao ML, White KI, Pfuetzner RA, Esquivies L, et al. 2018. NSF-mediated disassembly of on- and off-pathway SNARE complexes and inhibition by complexin. eLife 7:e36497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Prinslow EA, Stepien KP, Pan YZ, Xu J, Rizo J. 2019. Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and αSNAP. eLife 8:e38880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stepien KP, Prinslow EA, Rizo J. 2019. Munc18-1 is crucial to overcome the inhibition of synaptic vesicle fusion by αSNAP. Nat. Commun 10:4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lobingier BT, Nickerson DP, Lo SY, Merz AJ. 2014. SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife 3:e02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu H, Jun Y, Thompson J, Yates J, Wickner W. 2010. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 29:1948–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ungermann C, Kummel D. 2019. Structure of membrane tethers and their role in fusion. Traffic 20:479–90 [DOI] [PubMed] [Google Scholar]
- 141.Laufman O, Kedan A, Hong W, Lev S. 2009. Direct interaction between the COG complex and the SM protein, Sly1, is required for Golgi SNARE pairing. EMBO J. 28:2006–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morgera F, Sallah MR, Dubuke ML, Gandhi P, Brewer DN, et al. 2012. Regulation of exocytosis by the exocyst subunit Sec6 and the SM protein Sec1. Mol. Biol. Cell 23:337–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Travis SM, DAmico K, Yu IM, McMahon C, Hamid S, et al. 2020. Structural basis for the binding of SNAREs to the multisubunit tethering complex Dsl1. J. Biol. Chem 295:10125–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yue P, Zhang Y, Mei K, Wang S, Lesigang J, et al. 2017. Sec3 promotes the initial binary t-SNARE complex assembly and membrane fusion. Nat. Commun 8:14236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Saitsu H, Kato M, Mizuguchi T, Hamada K, Osaka H, et al. 2008. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet 40:782–88 [DOI] [PubMed] [Google Scholar]
- 146.Shen XM, Selcen D, Brengman J, Engel AG. 2014. Mutant SNAP25B causes myasthenia, cortical hyperexcitability, ataxia, and intellectual disability. Neurology 83:2247–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bryceson YT, Rudd E, Zheng C, Edner J, Ma D, et al. 2007. Defective cytotoxic lymphocyte degranulation in syntaxin-11-deficient familial hemophagocytic lymphohistiocytosis 4 (FHL4) patients. Blood 110:1906–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cote M, Menager MM, Burgess A, Mahlaoui N, Picard C, et al. 2009. Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells. J. Clin. Investig 119:3765–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Al Hawas R, Ren Q, Ye S, Karim ZA, Filipovich AH, Whiteheart SW. 2012. Munc18b/STXBP2 is required for platelet secretion. Blood 120:2493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ye S, Karim ZA, Al Hawas R, Pessin JE, Filipovich AH, Whiteheart SW. 2012. Syntaxin-11, but not syntaxin-2 or syntaxin-4, is required for platelet secretion. Blood 120:2484–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gissen P, Johnson CA, Gentle D, Hurst LD, Doherty AJ, et al. 2005. Comparative evolutionary analysis of VPS33 homologues: genetic and functional insights. Hum. Mol. Genet 14:1261–70 [DOI] [PubMed] [Google Scholar]
- 152.Gissen P, Johnson CA, Morgan NV, Stapelbroek JM, Forshew T, et al. 2004. Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis–renal dysfunction–cholestasis (ARC) syndrome. Nat. Genet 36:400–4 [DOI] [PubMed] [Google Scholar]
- 153.Lo B, Li L, Gissen P, Christensen H, McKiernan PJ, et al. 2005. Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet α-granule biogenesis. Blood 106:4159–66 [DOI] [PubMed] [Google Scholar]
- 154.Chen W, Cai ZL, Chao ES, Chen H, Longley CM, et al. 2020. Stxbp1/Munc18-1 haploinsufficiency impairs inhibition and mediates key neurological features of STXBP1 encephalopathy. eLife 9:e48705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guiberson NGL, Pineda A, Abramov D, Kharel P, Carnazza KE, et al. 2018. Mechanism-based rescue of Munc18-1 dysfunction in varied encephalopathies by chemical chaperones. Nat. Commun 9:3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gorenberg EL, Chandra SS. 2017. The role of co-chaperones in synaptic proteostasis and neurodegenerative disease. Front. Neurosci 11:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hunn BH, Cragg SJ, Bolam JP, Spillantini MG, Wade-Martins R. 2015. Impaired intracellular trafficking defines early Parkinson’s disease. Trends Neurosci. 38:178–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Südhof TC. 2010. α-Synuclein promotes SNARE-complex. Science 329:1663–67 [DOI] [PMC free article] [PubMed] [Google Scholar]