Abstract
Zebrafish and their mutant lines have been extensively used in cardiovascular studies. In the current study, the novel system, Zebra II, is presented for prolonged electrocardiogram (ECG) acquisition and analysis for multiple zebrafish within controllable working environments. The Zebra II is composed of a perfusion system, apparatuses, sensors, and an in-house electronic system. First, the Zebra II is validated in comparison with a benchmark system, namely iWORX, through various experiments. The validation displayed comparable results in terms of data quality and ECG changes in response to drug treatment. The effects of anesthetic drugs and temperature variation on zebrafish ECG were subsequently investigated in experiments that need real-time data assessment. The Zebra II’s capability of continuous anesthetic administration enabled prolonged ECG acquisition up to 1 h compared to that of 5 min in existing systems. The novel, cloud-based, automated analysis with data obtained from four fish further provided a useful solution for combinatorial experiments and helped save significant time and effort. The system showed robust ECG acquisition and analytics for various applications including arrhythmia in sodium induced sinus arrest, temperature-induced heart rate variation, and drug-induced arrhythmia in Tg(SCN5A-D1275N) mutant and wildtype fish. The multiple channel acquisition also enabled the implementation of randomized controlled trials on zebrafish models. The developed ECG system holds promise and solves current drawbacks in order to greatly accelerate drug screening applications and other cardiovascular studies using zebrafish.
Keywords: Zebrafish, Electrocardiogram (ECG), Prolonged anesthesia, Cardiac disease, Drug screening
1. Introduction
Cardiovascular diseases (CVDs) are the leading cause of death worldwide. According to the 2020 AHA Annual Report, almost 860,000 people died of CVDs in the U.S. in 2017, and the overall financial burden from CVDs totaled $351.2 billion in 2014–2015, emphasizing the urgency to explicate the etiologies of CVDs (Virani et al., 2020). One such CVD is the sick sinus syndrome (SSS), a collection of progressive disorders marked by the heart’s inability to maintain a consistent rhythm of heart muscle contraction and relaxation (Monfredi and Boyett, 2015). The sick sinus syndrome is characterized by age-associated dysfunction of the sinoatrial node (SAN), with varying symptoms such as syncope, heart palpitations, and insomnia (Adán and Crown, 2003). The SSS has multiple manifestations on electrocardiogram (ECG) data, including sinus bradycardia, sinus arrest (SA), and sinoatrial block. The pathophysiology of SSS is not fully understood, but scientists have determined that it can be caused by numerous factors ranging from pharmacological medications and sleep disturbances to fibrosis and ion channel dysfunction (Semelka et al., 2013). Previous research has emphasized SSS-associated genetic pathways as potential avenues to a more permanent treatment for SSS (Bakker et al., 2012; Choudhury et al., 2018; Hu et al., 2014; Y et al., 2020).
The zebrafish serves as an ideal model for cardiovascular studies because of its similar homology to humans in both morphology, physiology, and genetics (Arnaout et al., 2007; Koopman et al., 2021). Despite having only two discernible chambers in the zebrafish heart compared to four in human hearts, the zebrafish heart possesses a similar contractile structure with an analogous conduction system (Genge et al., 2016; Nemtsas et al., 2010). Therefore, the zebrafish model is appropriate in the study of SSS and the correlation of related genetic pathways to the electrophysical phenotype via ECG. Currently, several research groups have developed systems to assess zebrafish ECG. Regarding sensor design, conventional needle electrodes are commonly used. Lin et al., (2018) designed and tested the needle electrode with different materials, including tungsten filament, stainless steel, and silver wire to investigate the recorded data quality. Along with a portable ECG kit, the authors aimed to provide a standard platform for research and teaching laboratories. The needle system was also deployed in other studies (Chaudhari et al., 2013; Lin et al., 2018; Liu et al., 2016; Stoyek et al., 2018) to conduct biological and/or drug-induced research. Although the system demonstrated promising results, the needles had to be gently inserted through the dermis of zebrafish in order to collect favorable signals. The sharpness of the needles could cause injury to the fish’s heart, thus possibly changing signal morphology (Liu et al., 2016). Moreover, it requires an intensive effort to precisely position the electrode on the tiny heart in order to achieve clear ECG acquisition. Therefore, several alternative probe systems have been developed, including the micro-electrode array (MEA) and the 3D-printed sensors. Our team and other research groups have demonstrated the use of MEA for acquisition and provided data with a favorable signal-to-noise ratio (SNR) and high spatial and temporal resolution (Cao et al., 2014; Lenning et al., 2018; Yu et al., 2012). For instance, we presented a MEA array covering the fish’s heart, which enabled site-specific ECG signals (Cao et al., 2014; Lenning et al., 2018). Cho et al., (2017) developed a MEA printed on a flexible printed circuit board (FPCB) based on a polyimide film for multiple electroencephalogram (EEG) acquisition for epilepsy studies. Although the MEA allowed multiple signal recordings, only one fish can be assessed at a time due to the limited number of electrode channels. To address this shortcoming, our group recently demonstrated a prolonged system for acquiring ECG from multiple fish simultaneously (Le et al., 2020). The MEA was replaced with two electrodes made of 125-μm thick polyimide with sputtered-Au electrodes embedded at the bottom of the housing for data acquisition. However, noise generated from a pump used for water circulation precluded high-SNR ECG signals. Furthermore, bulky and expensive acquisition tools were used to collect and transfer data through a cable to a computer. In the market, commercially available systems, such as the iWORX (Dover, NH), can provide improved system mobility with a compact amplifier. However, several challenges have not been resolved, such as i) the commercial systems only record for a short period of time (3–5 min), which is inadequate for experiments that need longer recording such as acute drug interactions; ii) the ECG acquisition requires anesthetized animals, rendering those systems stressful to the fish and inadequate to provide intrinsic cardiac electrophysiological data; iii) manual one-by-one measurement limits the capability of conducting studies necessitating a large number of fish; and iv) ECG data processing is carried out offline with exorbitant effort. Last but not least, no high throughput systems integrated with microelectronic systems have been reported for characterizing mutant phenotypes. Therefore, developing high throughput systems capable of prolonged ECG acquisition is an essential step for finding associations between arrhythmic phenotypes and mutant genotypes, identifying multiple arrhythmic phenotypes that are linked to a single mutant genotype, as well as elucidating cardiac drug efficacy using the zebrafish model.
In this work, the novel Zebra II system is introduced, which is capable of prolonged ECG acquisition from multiple fish simultaneously. An in-house electronic system was developed, leveraging the Internet of Thing (IoT) capability with wireless data transmission and data processing on a mobile application. The IoT capability enhanced the mobility and versatility of the system as well as supported distanced collaborations to conduct research on zebrafish models. The system was validated through numerous experiments, displaying its potential with 1) simultaneous ECG acquisition from 4 fish; 2) continuous ECG acquisition for up to 1 h compared to several minutes (min) of currently available systems; 3) reduction in confounding effects from anesthesia with the use of 50% lower Tricaine concentration. The system featured a robust capacity in prolonged ECG acquisition from different experiments including sodium-induced SA, temperature-induced heart rate variation, and drug-induced arrhythmia for Tg(SCN5A-D1275N) mutant and wildtype fish in standardized experimental conditions. The implementation of multiple electrode channels for prolonged ECG acquisition also enables the implementation of randomized controlled trials, with two fish per experimental group in identical experimental conditions. Finally, the developed ECG system holds promise and solves current drawbacks in order to greatly accelerate arrhythmic phenotype analysis and drug screening applications in zebrafish.
2. Materials and methods
2.1. Mutant Tg(SCN5A-D1275N) and zebrafish husbandry
Mutant Tg(SCN5A-D1275N), a transgenic zebrafish arrhythmia model bearing the pathogenic cardiac sodium channel mutation SCN5A-D1275N, was used to characterize and validate system performance, study sinus node dysfunction, and perform drug high throughput screening assays. Correlation between clinical phenotype and the mutant line has been reported for bradycardia, conduction-system abnormalities, episodes of SA (Yan et al., 2020).
Adult wild/mutant-type zebrafish with the age of 13–20 months (body lengths approximately 3–3.5 cm) were used in this study. Zebrafish were kept in a circulating system that was continuously filtered and aerated to maintain the water quality required for a healthy aquatic environment. The fish room was generally maintained between 26 and 28.5°C, and the lighting conditions were regulated within a 14:10 h light: dark cycle.
All animal protocols in this study were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) protocol (#AUP-18-115 at University of California, Irvine). All drug administration experiments (section 1–2, supplementary document), chemicals, reagents, and materials were performed in accordance with relevant guidelines and regulations.
2.2. Design and validation of the Zebra II system
The Zebra II system is composed of a perfusion system, an in-house electronic system, apparatuses, and sensors (Fig. 1a). The perfusion system comprised four syringes, four valves and tubing. The four syringes contained Tricaine solution with low concentration, which continuously fed to the fish through the tubing system. Tricaine (MS-222) was used as an anesthetic to reduce the fish’s aggressiveness and activity while maintaining their consciousness. The four valves were used to adjust the solution’s flow rate within a range of 5.5–6 ml/min, while housing apparatuses and sensors were improved from the previous work (Le et al., 2020). Specifically, multiple side-fitted housings were made of polydimethylsiloxane (PDMS), which provided comfort to the fish and minimized unwanted movements. Moreover, the top and bottom of the apparatus were designed to allow the fish to lay comfortably on electrodes within the curved bottom. The top was fitted with an additional part on the wall to keep the fish from escaping the apparatus. The zebrafish ECG system was placed within a home-made incubator (Fig. 1a). Temperature within the chamber ranged from 20 °C to 32 °C, as measured by a thermometer and controlled by a thermostat with an accuracy of ±1 °C.With the thermo box, a specific temperature was set by the thermostat control, and the light bulb was turned on so that the box’s temperature can be maintained at the setup temperature and vice versa. The electronic system and the mobile application are described in Fig. 1b–d and Sup. Fig. 1. The overall system electronic specifications are shown in Sup. Table 1.
Fig. 1.
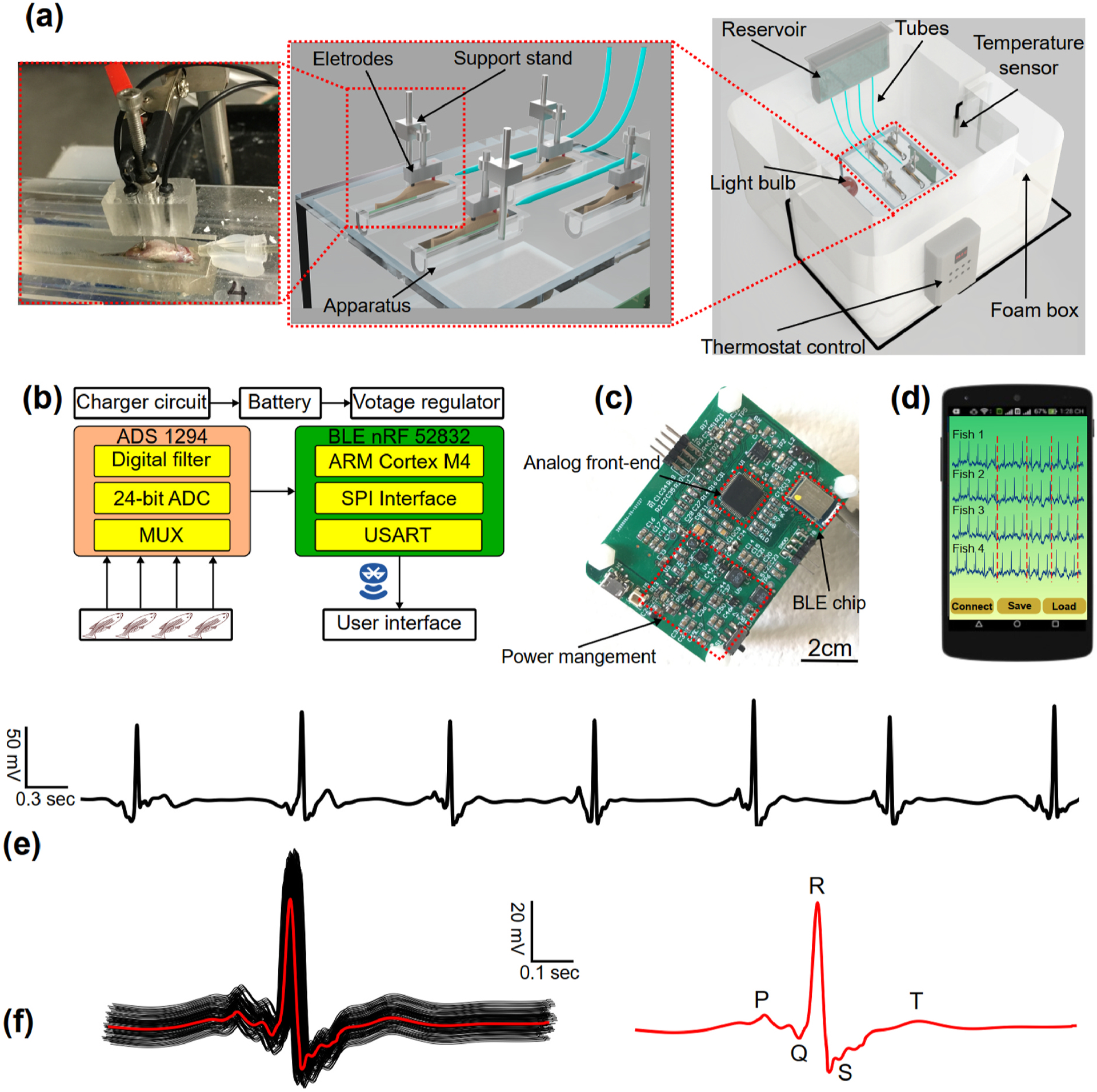
The prolonged ECG system for multiple adult zebrafish acquisition. (a) the prolonged ECG system design: the reservoir for containing Tricaine solution, the tube system for feeding the solution to the fish, the electrodes and support stand for acquiring ECG signals. (b) System-level block diagram showing analog front-end chip, signal transduction, wireless transmission from the ECG to user interface. (c) In-house electronic board having system-on-chip for wireless transmission, power management connecting to the electrode for ECG acquisition. Scale bar: 2 cm. (d) User interface of the mobile application receiving ECG data from multiple fish. (e) Representative ECG data collected by the system. (f) ECG data segments superimposed, and its average ECG segment in red with clear ECG waves (P wave, QRS complex and T wave).
The Zebra II wirelessly transmitted ECG signals to a smartphone, and the data were displayed on a mobile application as shown in Fig. 1d. To test the reproducibility, ECG signals were acquired from 8 wild type (WT) fish for 7 trials, and the variations in terms of signal to noise ratios (SNR), heart rate, QTc interval, and QRS interval were monitored. These results are further illustrated in Sup. Fig. 2. Furthermore, a robust and scalable real-time stream processing system leveraging the Google Cloud infrastructure was designed and implemented to facilitate remote monitoring and high-throughput ECG analysis. The processing system is illustrated in Sup. Fig. 3. The system was constructed to provide a collaborative platform for different research groups regardless of their geographical locations. With the capabilities of acquiring ECG from multiple fish simultaneously for up to 1 h, the proposed design can save time and efforts by nearly 40–50 fold compared with conventional approaches (Chaudhari et al., 2013; Lin et al., 2018; Yan et al., 2020).
Experiments were conducted to validate the performance of the developed system. First, zebrafish ECG signals were acquired simultaneously using the Zebra II and the commercial system developed by iWORX (Dover, NH) to assess the data quality from fish under Amiodarone treatment. Then, the optimal Tricaine concentration and temperature were determined for ECG acquisition (n = 64).
To further investigate this mutant line as a candidate for cardiac studies, we assessed the relationship between SCN5A and Meth (Tisdale et al., 2020)– a controlled substance. Several groups have studied its connection of using addictive drugs with sudden death. For instance, Nagasawa et al., screened several variations in the long QT syndrome-associated genes KCNQ1 (LQT1) and KCNH2 (LQT2), showing the increased risk of severe cardiac arrhythmia for addictive drug abusers (Nagasawa et al., 2018). However, they did not test for SCN5A variants, which was explored in the current study. Additionally, rescuing arrhythmic phenotypes induced by high sodium intake could provide insights into the nature of those arrhythmic phenotypes, as Meth has been previously demonstrated to increase HR after administration (Henry et al., 2012). Specifically, we treated two groups of zebrafish (labelled as “control” – WT fish and “mutant” – Tg(SCN5A-D1275N)) in 0.9‰ NaCl for 30 min before immersing the fish in 50 μM Meth for 30 min. As shown in Fig. 5, the HR, SDNN and QTc interval were compared between the two groups in the following three treatments: without drug treatment, with NaCl treatment, and with NaCl + Meth treatment (n = 12 WT fish and n = 8 Tg(SCN5A-D1275N fish)).
Fig. 5.
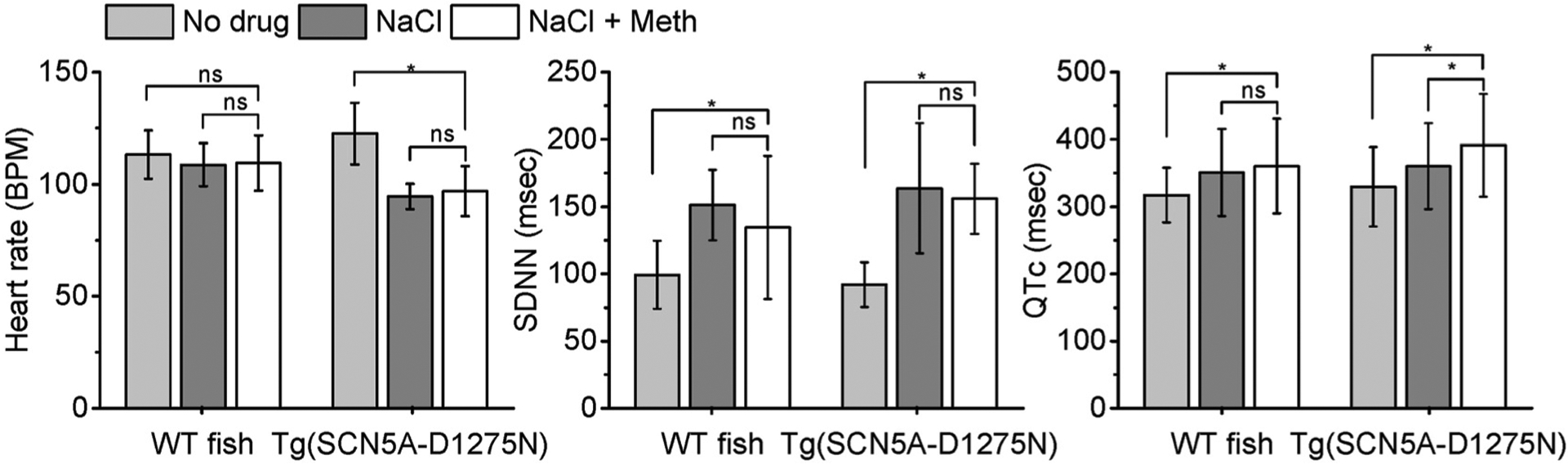
Investigation of methamphetamine (Meth)’s efficacy to rescue arrhythmic phenotypes after treatment with NaCl. The experiment analyzed and compared HR, QTc and SDNN among three treatments (e.g., no drug treatment, NaCl and, NaCl + Meth). The experiment was conducted in both WT fish and mutant fish Tg(SCN5A-D1275N). The average HR and SDNN for mutant fish treated with NaCl + Meth did not show significant difference from average HR and SDNN treated with NaCl only. *p < 0.05 (one-way analysis of variance with Turkey test). ns indicates not significant.
2.3. Signal processing and statistics
The recorded ECG data were analyzed, and several parameters were extracted, including heart rate (HR), QT, and QTc intervals (Le et al., 2019). The standard deviation of normal sinus beats (SDNN) was calculated based on the short-term, beat-to-beat variance of HR in each 5 min segment of the data, and the standard deviation of the average normal-to-normal (SDANN) intervals was calculated based on the variance of average HR from each 5 min segment. SDNN was used for short-term data analysis based on each 5 min segment, and SDANN was used for long-term data analysis for the whole 40 min segment (Shaffer and Ginsberg, 2017).
Statistical analysis was performed using the following tests with OriginLab 2019. Multiple comparisons were tested with one-way ANOVA, and significant results (P < 0.05) were analyzed with pairwise comparisons using Student’s t-test applying significance levels adjusted with the Bonferroni method. Significant P-values are indicated with asterisks (*) with *P < 0.05, **P < 0.01 and ***P < 0.001. Correlation analysis was performed using Pearson’s correlation.
3. Results and discussion
3.1. Multiple zebrafish ECG acquisition with Zebra II
The ECG data collected from Zebra II are shown in Fig. 1e. The data were pre-processed using the Wavelet technique (Le et al., 2019) to reduce various types of noise. Fig. 1f illustrates the ECG segments superimposed during the measurement, with each line representing one cardiac cycle. The full set of ECG waves symbolized with the P-wave, QRS-complex, and T-wave was present in the mean of ECG segments as highlighted in red, showing waveform reproducibility and stability during the recording period. Moreover, prior to the acquisition of ECG data, the impedance of the electrodes on zebrafish skin was verified (Sup. Fig. 4). The low standard deviation error indicated that ECG acquisition from the electrodes will remain stable for the duration of the measurement, ensuring that any variations in ECG acquisition were not caused by faulty electrodes. The relative standard deviation (RSD) of such parameters is presented in Sup. Table 2.
The acquired ECG data were then compared in both frequency domain and time domain (Sup. Fig. 5a and b). Specifically, the correlation coefficients were 98.78% and 96.54% in time domain and frequency domain, respectively. Moreover, the HR and QRS interval were also compared (Sup. Fig. 5c and d). As shown in the data, the Zebra II’s performance was comparable to that of the commercial iWORX system. Then, another experiment was performed on 36 wildtype (WT) zebrafish divided into the following 2 groups: 1) control (n = 20) and 2) Amiodarone treated (n = 16) fish. As shown in Sup. Fig. 6, HR, QRS duration and QTc interval were analyzed. With the control group, no significant difference (p-value > 0.05) between two systems in terms of QRS and QTc value was observed. Similarly, the HR value and QTc value showed no significant difference in the treated group. Furthermore, the Bland Altman analysis in Sup. Fig. 6b, d, f showed the agreement level between two systems, with most of HR values and QTc values located within the limit of agreement (LOA) region.
Electrode placement contributed significantly to the intensity of ECG data. With the use of the low Tricaine concentration to enable longer ECG acquisition, the fish tended to exhibit unexpected strong movement, thus leading to electrode dislocation. In the future, a mechanism may be utilized to automatically detect the impedance of electrode-tissue interface, which can be used to indicate the contact between electrode and fish’s chest. In terms of the current electronic system, the low-noise and high-resolution ADS chip provided favorable ECG data with additional data processing feature. However, scaling up to 8, 16 or even 24 channels to include more fish may be challenging at this point. For instance, it would require multiple ADS chips cascaded together, which would increase its computational power. However, wireless data transmission using BLE will no longer be compatible with this scenario due to the limit of bandwidth (Bulić et al., 2019). Another challenge was presented in terms of time delay. Since the recorded ECG data were wirelessly transmitted to the mobile app via BLE, the connection between the fish system and mobile phone needs to be established first. A set of connection parameters (e.g., connection interval, slave latency, and connection supervision timeout) was sent by the smartphone (BLE master device) to the fish system (BLE slave device), which takes about 2 s based on the observation during the experiment. Thus, establishing the initial connection contributed to the delay in wireless data transmission between the Zebra II and the phone. Additionally, with the sampling rate of 250 Hz, the operation time for ADC and BLE communication between the system (a slave device) and a phone was 4 msec. In regard to the cloud system, the time from acquiring ECG data to displaying the data on the cloud with Grafana was approximately 2 s. As a result, the total delay from the initial ECG acquisition to the presence of data on the cloud was 6 s. Thus, the entire operation can be considered as pseudo-real time.
3.2. Investigation of side effects of Tricaine and variable temperature cardiac rhythm
Tricaine (MS-222) and temperature have been shown to affect cardiac physiology of adult zebrafish and the HR of the treated subjects (Lin et al., 2018; Sun et al., 2009). At low temperatures (e.g., 18 – 20 °C), myocyte activity is reduced as a natural adaptive mechanism to aid survival during colder climates or seasons, which leads to a reduction in HR. At higher temperatures, increased HR facilitates greater cardiac output to support a higher metabolic activity/demand for oxygen consistent with normal physiological function (Maricondi-Massari et al., 1998). Therefore, with an optimal environment temperature, the prolonged ECG system introduced in this work will help lower the Tricaine concentration used in experiments, which can reduce the confounding effects of Tricaine in order to obtain intrinsic ECG data.
As shown in Fig. 2a, the ECG data from 8 fish per concentration group (75, 100, and 150 ppm) of Tricaine were obtained. The ECG data showed gill motion noise, manifested as low frequency, cyclic perturbations in the data. The noise interfered with the identification of ECG waves such as P waves, T waves and QRS complexes from fish treated with 75 ppm Tricaine, while the noise appeared to be more subdued in data obtained from fish treated with 100 ppm and 150 ppm. As a result, ECG data acquired from fish treated with 100 ppm and 150 ppm displayed clear ECG waves. After 40 min measurement, the recovery time and the survival rate after treatment were collected (Fig. 2b). It was found that fish treated with higher Tricaine concentrations necessitated longer recovery times. Specifically, fish treated with 150 ppm Tricaine took an average of 7 min to recover compared to the 3 min and 4.2 min for fish treated with 75 and 100 ppm, respectively. Furthermore, the survival rate of fish treated with 150 ppm Tricaine was about 75%, while other concentrations yielded survival rates above 90%. It reflected the effect of extremely high Tricaine concentration similar to that used for euthanasia (i.e., 168 ppm) (Matthews and Varga, 2012). Given the recovery time, the survival rate, and the acquired ECG data, the Tricaine concentration of 100 ppm was most optimal for ECG acquisition. Additionally, the variation of HR was also assessed in each 5-min segment during the 40-min measurement (Sup. Fig. 7). No significant difference was observed between the 5–min to 10–min, 10-min to 15–min, 15-min to 20–min, and 20-min to 25–min segments for ECG data from fish treated with either 75 ppm or 100 ppm Tricaine. The only difference occurred in the last 10 min of the 40-min measurement when the HR displayed more fluctuation with 75 ppm Tricaine (115.67 ± 25.29 BPM) than that with 100 ppm (115. 93 ± 11.28 BPM).
Fig. 2.
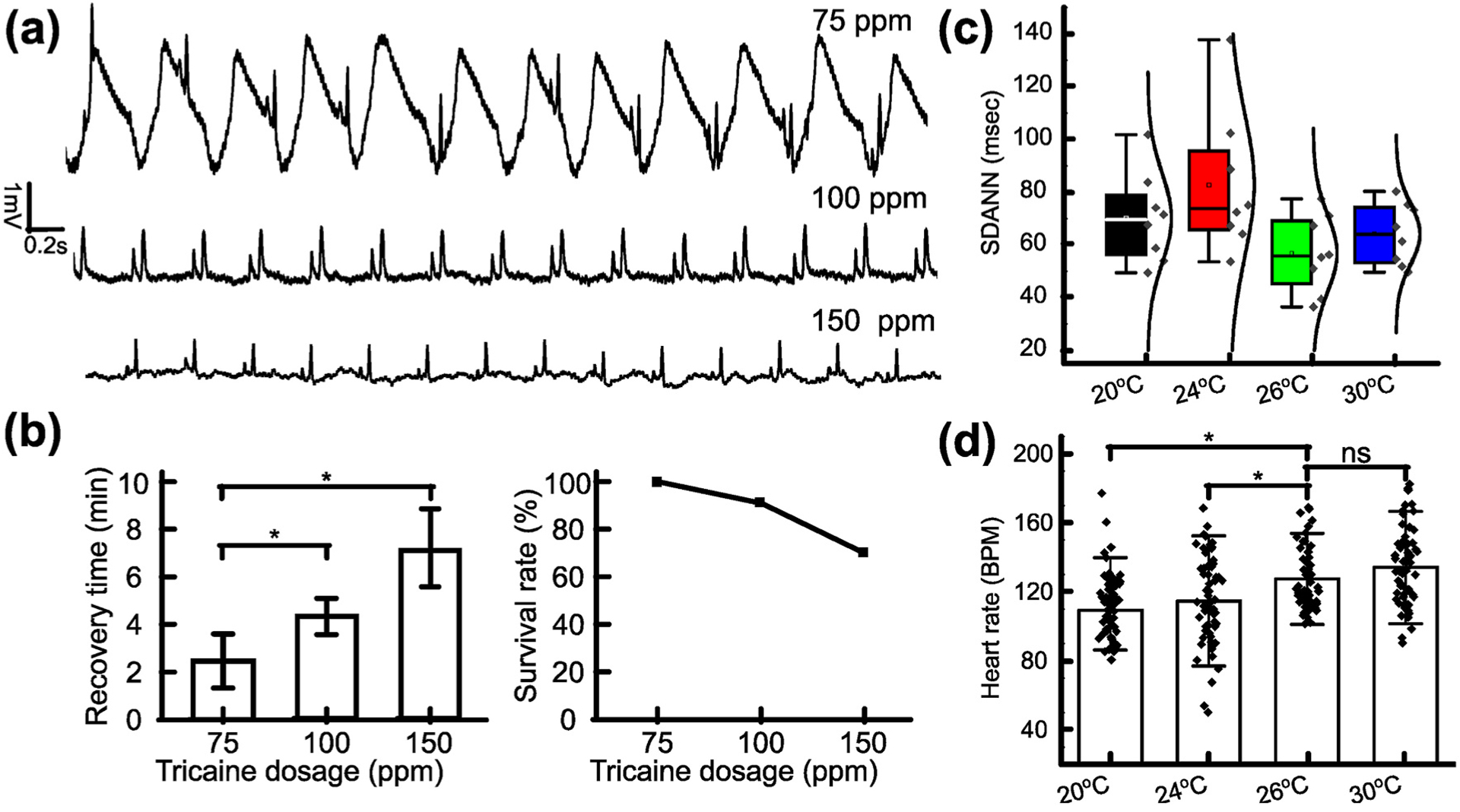
Investigation of Tricaine and temperature to reduce cardiac rhythm side effects. (a) Representative ECG data recorded from fish treated with different Tricaine concentrations. (b) Bar chart comparing recovery time needed after treatment for each Tricaine concentration. Line graph describing the survival rate of zebrafish treated by different Tricaine concentrations. (c) SDANN in WT fish with different temperatures. (d) HR in WT fish with different temperatures (N = 64, standard deviation (SD) of HR at 20 °C, 24 °C, 26 °C, 30 °C: 17.8 BPM, 25.2 BPM, 17.6 BPM and 21.7 BPM, respectively). *p < 0.05; **p < 0.01 (one-way analysis of variance). ns indicates not significant.
After determining the optimal Tricaine concentration, the effect of temperature on acquired ECG was investigated. As shown in Fig. 2c, SDANN at 26 °C had the lowest value with the range of 36 ms (msec) to 75 msec, while the SDANN at 24 °C is highest with the range of 50 msec–139 msec. Moreover, the data distribution from HR collected every 5 min at 26 °C was the most condensed (Fig. 2d). Thus, the HR obtained at 26 °C was the most stable compared to the values obtained at other temperatures.
3.3. Response analysis to drug treatment in real time with the Zebra II system
One of the key novelties of the Zebra II system is the capacity to test drugs with different concentrations on individual fish with a continuously prolonged assay. First, the effect of Amiodarone with different concentrations on zebrafish ECG was analyzed. Three doses of Amiodarone were consecutively filled in the reservoir to feed to the fish during ECG acquisition, and each dose lasted around 5 min. As shown in Fig. 3a, the changes in response to different dosages in all four fish were apparent. Zooming in on the data acquired from fish 1 at timepoints denoted from (1) to (4) corresponding to different Amiodarone concentrations revealed that the QTc interval displayed considerable changes. The QTc interval was 310 msec without drug treatment. Noticeable increases in QTc interval were observed after Amiodarone treatment. Specifically, the interval was 330 msec at 70 μM of Amiodarone, 476 msec at 100 μM, and 536 msec at 200 μM. Fig. 3b depicts the overall changes in terms of QTc interval, QRS interval, and HR in response to different Amiodarone concentrations. As Amiodarone concentration increased over the duration of the experiment, QTc interval and QRS interval increased, while the average HR decreased.
Fig. 3.
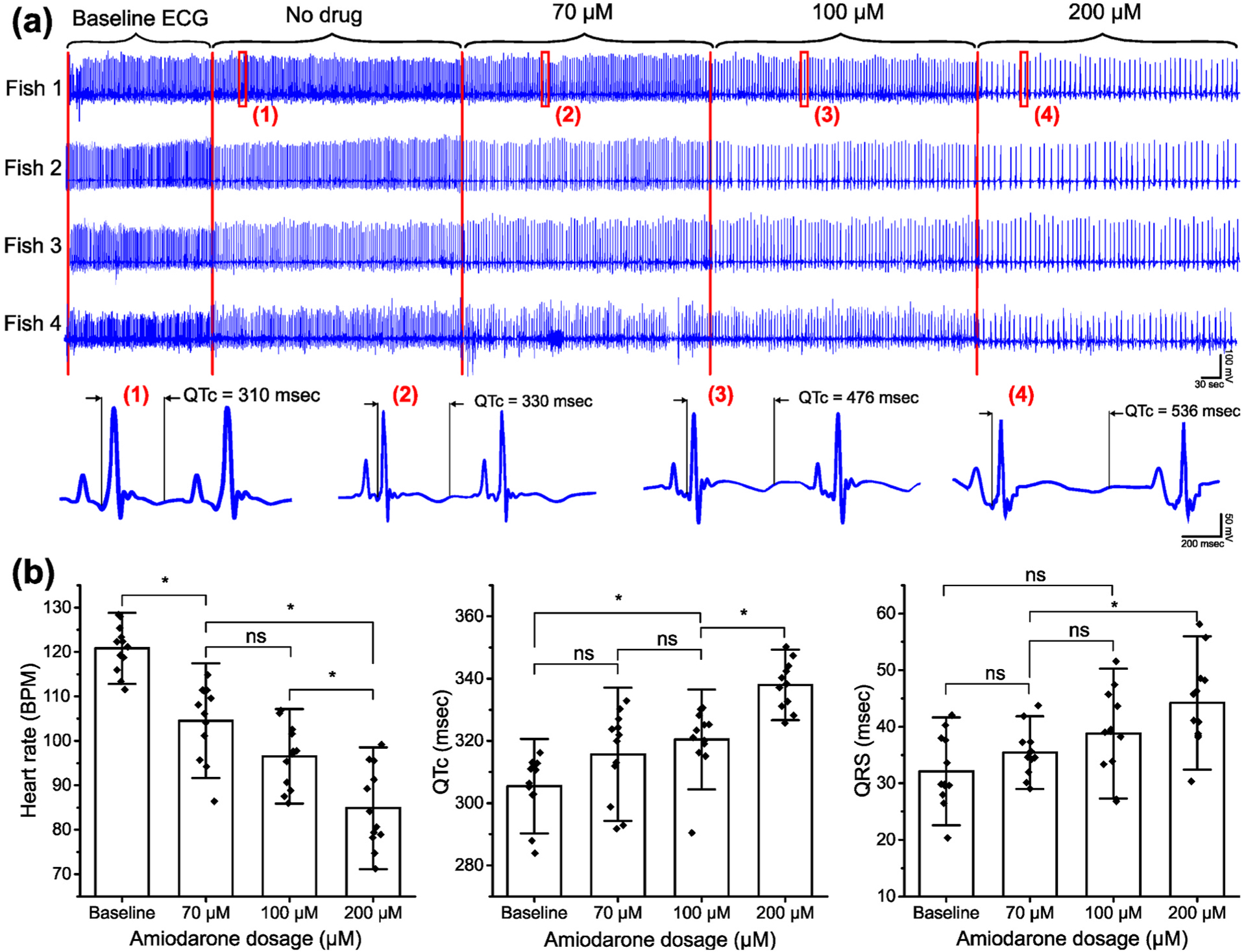
Demonstration of the prolonged system showing ECG changes in response to different Amiodarone concentrations. (a) Representative ECG data obtained by the developed system and its change in ECG parameters due to different Amiodarone concentrations. (b) Bar chart describing the discrepancy of HR, QTc interval and QRS interval in ECG data with different Amiodarone concentrations (n = 8 fish). Quantification of heart rate decreased in response to Amiodarone (120.8 ± 5.3 BPM without treatment, 84.9 ± 9.1 BPM with 200 μM Amiodarone) while the QTc and QRS interval tend to increase (337.9 ± 7.5 msec with no drug and 44.2 ± 7.9 msec with 200 μM Amiodarone). *p < 0.05 (one-way analysis of variance with Turkey test). ns indicates not significant.
In drug response studies, conducting real time or pseudo-real time measurements with the same fish is most ideal due to biological variability among subjects. The perfusion system is equipped with multiple chambers for multiple drug doses in order to provide seamless transitions of multiple drug treatments, decreasing potential noise and perturbations. The developed system was able to demonstrate that a longer acquisition enables the treatment of multiple drugs, as indicated by the successful demonstration of dose-response Amiodarone-associated ECG changes described earlier in this section, as well as sodium-associated ECG changes described later in section 3.4. The utility of the perfusion system can also be expanded to include the testing of multiple drugs simultaneously to assess ECG changes due to drug-drug interactions, a developing field of study (Zukkoor and Thohan, 2018). The chambers in the perfusion system can also contain multiple drugs for the precise modulation of zebrafish drug intake in order to accurately determine the effects of each tested drug as well as the onset of potential drug-drug interactions. While the subsequent ECG analysis of Tg (SCN5A-D1275N) indicates that Methamphetamine (Meth) did not improve SA frequency and HR, it demonstrated the assessment of effects of multiple drugs, as seen from the prolongation of the QTc interval after treatment of 50 μM of Meth (Fig. 5). In both groups, the QTc interval was longer (by 350 msec for WT and 385 msec for Tg(SCN5A-D1275N)) after Meth treatment. Thus, the robust performance of the system allowed incorporation of multiple drugs with different effects (e.g., antagonistic effects) in a single continuously prolonged assay to study the effect of drug-drug interactions on ECG changes, which was previously heavily performed on the short time course of other existing systems. Various ECG changes due to drug treatment were detected in prolonged ECG acquisition) to provide intuitive insights into drug-drug interaction effects, demonstrating the potential of the developed system to evaluate drug efficacy. As shown in Fig. 4d for the sodium sensitivity experiment, the average SDNN was 125 msec for WT fish but was 255 msec for Tg (SCN5A-D1275N), consistent with reduced conduction velocities due to sodium ion channel dysfunction (George, 2005).
Fig. 4.
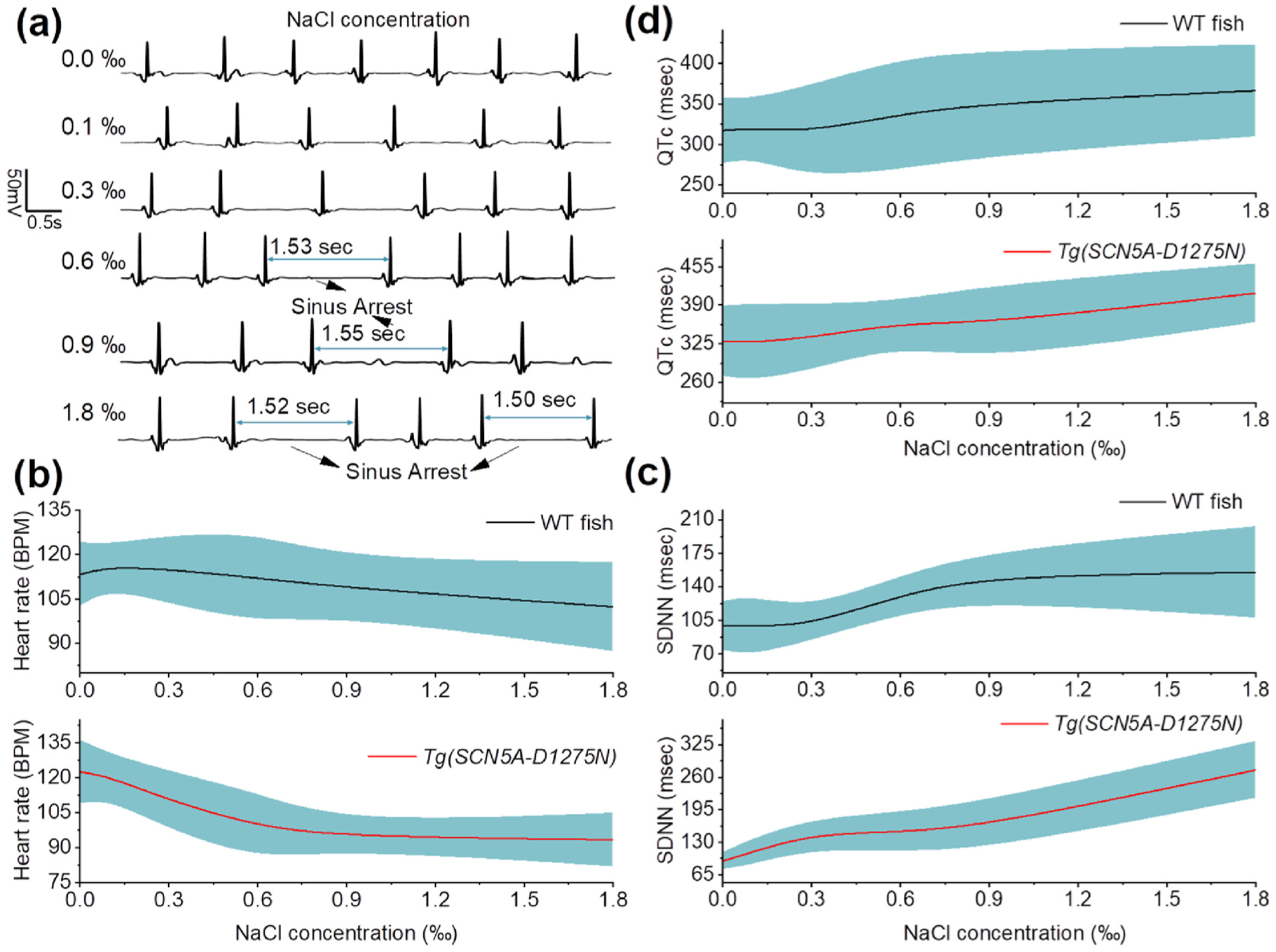
Evaluation of sodium sensitivity in the development of sinus arrest in Tg(SCN5A-D1275N). (a) Representative ECG data before and after NaCl treatment with different concentrations. SA appears more frequently in response to the increase of NaCl concentration. (b) The average HR of WT fish (n = 12), showing the slight decrease from 113.4 ± 10.8 BPM without treatment to 102.1 ± 15.0 BPM; the averaged HR of Tg(SCN5A-D1275N) mutant fish (n = 8), showing significant reduction from 122.6 ± 13.7 BPM without treatment to 93.3 ± 11.5 BPM after NaCl treatment. (c) SDNN of WT fish (n = 12) and mutant fish (n = 8), displaying similar variations in HR. (d) QTc values shows slight increase in both wild-type and mutant fish after NaCl treatment.
3.4. Evaluation of high sodium intake in the development of sinus arrest (SA) in Tg(SCN5A-D1275N)
No previous research studies investigated the role of high sodium intake on the development of SA for zebrafish with increased susceptibility to arrhythmia due to genetic causes. Here, the developed system demonstrated the role of excess sodium ions in inducing ECG changes in the Tg(SCN5A-D1275N) mutant, successfully characterizing the onset of arrhythmic phenotypes such as SA. Fig. 4a illustrates the ECG data obtained from Tg(SCN5A-D1275N) fish with different NaCl concentrations. Zebrafish started displaying a reduction in HR when treated with a low NaCl concentration of 0.1‰. More significant reductions were detected when the fish were treated with higher concentrations. According to the SA criteria (e.g., RR interval is greater than 1.5 s) determined in our previous work (Yan et al., 2020), SA appeared more frequently after treatment with 0.6‰ NaCl and above (Table 1). The result confirmed a strong association between high sodium intake and arrhythmic phenotypes, previously reported in hypertensive populations (Mente et al., 2016). High sodium intake is associated with alterations in various proteins responsible for transmembrane ion homeostasis and myocardial contractility. Recent studies provided important evidence that high sodium intake promotes structural and functional impairment of the heart, especially in populations bearing mutant phenotypes of the major cardiac sodium channels such as Nav1.5 and its corresponding gene SCN5A. However, there was a current lack of a functional prolonged ECG acquisition system to characterize arrhythmic phenotypes from Nav1.5 sodium channel mutants, including the functional response of Nav1.5 to initiate action potentials based on high sodium intake. SA induced by high sodium intake was observed in this study and may be associated with a rise in intracellular sodium concentration within cardiomyocytes due to the gain-of-function of Tg(SCN5A-D1275N) for sodium ions traveling into the cardiomyocyte. Detection of SA by the developed system implied that the Tg(SCN5A-D1275N) fish is susceptible to arrhythmic phenotypes after high sodium intake due to hastening epicardial repolarization and causing idiopathic ventricular conduction. These pathological changes were manifested as ECG changes and ventricular arrhythmias. ECG data acquired by the developed system were consistent with clinical reports, indicating that Brugada syndrome in human and animals resulted in ventricular conduction abnormalities due to high sodium intake (Antzelevitch, 2006; Mizusawa and Wilde, 2012). High sodium intake can cause destabilized closed-state inactivation gating of Nav1.5 that may attenuate the ventricular conduction delay as shown in the ECG data (Table 1).
Table 1.
Comparison of WT and Tg(SCN5A-D1275N) fish with NaCl treatment.
| Fish | WT | Tg(SCN5A-D1275N) | ||||||
|---|---|---|---|---|---|---|---|---|
| Drug | Average HR (BPM) | Percentage of fish with SA (No. of cycles) | SA frequency epm | QTc (msec) | Average HR (BPM) | Percentage of fish with SA (No. of cycles) | SA frequency epm | QTc (msec) |
| 0‰ NaCl | 113.3 ± 10.8 | 8.3(1) | 0.08 | 317.2 ± 40.7 | 122.6 ± 13.7 | 12.5 (1) | 0.125 | 329.8 ± 59.1 |
| 0.1‰ NaCl | 115.9 ± 7.5 | 8.3(1) | 0.08 | 319.7 ± 36.3 | 120.4 ± 9.9 | 12.5(1) | 0.125 | 327.2 ± 64.8 |
| 0.3‰ NaCl | 114.9 ± 11.5 | 8.3(1) | 0.08 | 316.1 ± 56.6 | 110.3 ± 12.6 | 37.5(7) | 0.875 | 335.7 ± 55.8 |
| 0.6‰ NaCl | 112.1 ± 15.2 | 16.6(2) | 0.16 | 337.2 ± 68.2 | 98.9 ± 14.4 | 75(8) | 1 | 359.9 ± 37.1 |
| 0.9‰ NaCl | 108.7 ± 9.5 | 25(3) | 0.25 | 350.7 ± 64.7 | 94.6 ± 5.7 | 75(15) | 1.875 | 360.1 ± 64.0 |
| 1.8‰ NaCl | 102.4 ± 15.0 | 50(7) | 0.42 | 366.1 ± 56.4 | 93.3 ± 11.5 | 87.5(17) | 2.125 | 410.5 ± 49.5 |
| 0.9‰ NaCl + 50 μM Meth | 109.4 ± 12.3 | 25(3) | 0.25 | 360.2 ± 70.3 | 96.95 ± 21.2 | 75(15) | 1.875 | 391.3 ± 76.1a |
Indicating the additive effect of Meth to QT prolongation.
As shown in Fig. 4b, mutant fish exhibited a significant decrease in HR after treatment of 0.6‰ NaCl. In contrast, NaCl treatment did not show a profound effect to the WT fish, as evidenced by the smaller decrease in HR after NaCl treatment. It was worth noting that these WT fish were at 1.5 years old, which could attribute to an increase of SA (Yan et al., 2020), and the slight reduction of HR in the experiment (Sup. Fig. 8). In terms of HRV, Tg(SCN5A-D1275N) fish showed a remarkable increase at high NaCl concentrations (0.9‰ and 1.8‰) compared with other concentrations. These results provided evidence that the Tg (SCN5A-D1275N) triggered more SA under NaCl treatment (Table 1). Moreover, the SDNN and QTc interval in response to NaCl treatment were also measured, exhibiting similar trends for both WT and mutant fish. (Fig. 4c and Sup. Fig. 9).
Notably, the results indicated that high sodium intake induced more drastic ECG changes in Tg(SCN5A-D1275N) fish. NaCl treatments at 0.6‰, 0.9‰, and 1.8‰ resulted in SA with durations of 1.53 s, 1.55 s and 1.52 s, respectively. Additionally, slower HR and prolonged QTc intervals were observed only in mutant fish. These results provided a significant association between the increased frequency of SA, slower HR, and prolonged QTc with increased sodium intake in mutants. According to previous reports (Darbar et al., 2008; Liu et al., 2014), Nav1.5 can disrupt the heart’s electrical activity and lead to a dramatic decrease of HR. The slow-conducting Tg(SCN5A-D1275N) mutant has been demonstrated previously by voltage-clamp measurement (McNair et al., 2011; Nguyen et al., 2008), which corroborated with the results in this study. The average QTc intervals were as high as 385 msec, indicating that the QTc intervals in mutant fish were generally more prolonged than wild type animals. Overall, high sodium intake led to various arrhythmic phenotypes, including slow HR, prolonged QTc, and increased SA frequency in Tg(SCN5A-D1275N) fish (Fig. 4d).
3.5. Rescue of arrhythmic phenotypes induced by high sodium intake in Tg(SCN5A-D1275N) fish
The average HR of mutant fish after the NaCl + Meth treatment (96.96 ± 7.61 BPM) was slightly higher than that of mutant fish solely treated with NaCl (94.59 ± 5.69 BPM) (Fig. 5); however, the difference was not significant (P > 0.05). Similarly, the SDNN value did not show any significant difference between mutant fish treated with NaCl + Meth and those solely treated with NaCl (Fig. 5). Meth was administered to the fish to determine if their cardiac systems could respond to the drug’s mechanism of inducing ECG changes (i.e., increased HR, QTc prolongation). However, the obtained data indicated that Meth treatment did not affect the HR and SDNN in both groups (Table 1), implying that NaCl administration resulted in some irreversible arrhythmic phenotypes (i. e., slow HR) that could not be easily rescued with other agents that increase HR. In contrast, a significant increase in QTc interval was detected in the mutant fish after Meth treatment (391.3 ± 76.1 msec vs. 360.1 ± 64.0 msec), indicating the additive effect of Meth to QTc interval prolongation (Table 1). Considering that no significant increase in QTc interval was seen in WT fish, the data indicated the mutant fish were more susceptible to agents causing QTc prolongation. Therefore, these results suggested different susceptibilities for arrhythmic phenotypes in the mutant Tg(SCN5A-D1275N).
The ECG data obtained from this study provided new evidence that high sodium intake increased the susceptibility of Tg(SCN5A-D1275N) fish to arrhythmic phenotypes. Results from mutant fish indicated the pathological slowing of HR, prolonged QTc interval, and higher frequency of SA. Although the developed system cannot provide other cardiac indices, such as ejection fraction and cardiac output, the system’s ability to detect arrhythmic phenotypes in real time is valuable for many applications such as drug screening and phenotype assessment.
4. Conclusion
The novelties of the developed Zebra II lie in the extended measurement for multi-step experiments (up to 1 h), high throughput screening with multiple zebrafish, controlled setting with minimal confounding effects, and automated cloud-based analytics. The system was successfully demonstrated to investigate the arrhythmic mutant line Tg(SCN5A-D1275N), revealing the effect of high sodium intake on the development of sinus arrest (SA), slow HR, and prolonged QTc. In the future, the Zebra II can be used for a host of cardiac disease studies, including phenotypic screening for genetic engineering studies and new drug screening applications.
Supplementary Material
Acknowledgement
The authors would like to acknowledge the financial support from the NSF CAREER Award #1917105 (H.C.), the NSF #1936519 (J.L. and H.C), the NIH #R44OD024874 (M.P.H.L and H.C.), the NIH HL107304 and HL081753 (X.X.). We thank Lauren Schmiess-Heine for managing the zebrafish facility; and staff members in the Xu lab at Mayo Clinic, MN, USA for maintaining and sharing the Tg(SCN5A-D1275N) fish.
Footnotes
CRediT authorship contribution statement
Tai Le: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Jimmy Zhang: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Anh H. Nguyen: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Ramses Seferino Trigo Torres: Methodology. Khuong Vo: Methodology, Writing – original draft, Writing – review & editing. Nikil Dutt: Writing – review & editing. Juhyun Lee: Writing – review & editing. Yonghe Ding: Writing – review & editing. Xiaolei Xu: Writing – review & editing. Michael P.H. Lau: Writing – review & editing. Hung Cao: Conceptualization, Investigation, Methodology, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113808.
References
- Adán V, Crown LA, 2003. Am. Fam. Physician 67 (8), 1725–1732. [PubMed] [Google Scholar]
- Antzelevitch C, 2006. Pacing Clin. Electrophysiol 29 (10), 1130–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R, Ferrer T, Huisken J, Spitzer K, Stainier DYR, Tristani-Firouzi M, Chi NC, 2007. Proc. Natl. Acad. Sci. Unit. States Am 104 (27), 11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, Tan HL, Moorman AF, t Hoen PA, Christoffels VM, 2012. Cardiovasc. Res 94 (3), 439–449. [DOI] [PubMed] [Google Scholar]
- Bulić P, Kojek G, Biasizzo A, 2019. Sensors 19 (17), 3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Yu F, Zhao Y, Zhang X, Tai J, Lee J, Darehzereshki A, Bersohn M, Lien C-L, Chi NC, Tai Y-C, Hsiai TK, 2014. Integr. Biol. : Quant.Biosci. from nano to macro 6 (8), 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari GH, Chennubhotla KS, Chatti K, Kulkarni P, 2013. J. Pharmacol. Toxicol. Methods 67 (2), 115–120. [DOI] [PubMed] [Google Scholar]
- Cho S-J, Byun D, Nam T-S, Choi S-Y, Lee B-G, Kim M-K, Kim S, 2017. Sci. Rep 7 (1), 3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury M, Black N, Alghamdi A, D’Souza A, Wang R, Yanni J, Dobrzynski H, Kingston PA, Zhang H, Boyett MR, Morris GM, 2018. J. Physiol 596 (24), 6141–6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL, Roden DM, 2008. Circulation 117 (15), 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genge CE, Lin E, Lee L, Sheng X, Rayani K, Gunawan M, Stevens CM, Li AY, Talab SS, Claydon TW, Hove-Madsen L, Tibbits GF, 2016. Reviews of Physiology, Biochemistry and Pharmacology, vol. 171. Springer International Publishing, Cham, pp. 99–136. [DOI] [PubMed] [Google Scholar]
- George AL Jr., 2005. J. Clin. Invest 115 (8), 1990–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry BL, Minassian A, Perry W, 2012. Addiction Biol 17 (3), 648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YF, Dawkins JF, Cho HC, Marbán E, Cingolani E, 2014. Sci. Transl. Med 6 (245), 245ra294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman CD, De Angelis J, Iyer SP, Verkerk AO, Da Silva J, Berecki G,Jeanes A, Baillie GJ, Paterson S, Uribe V, Ehrlich OV, Robinson SD, Garric L, Petrou S, Simons C, Vetter I, Hogan BM, de Boer TP, Bakkers J, Smith KA, 2021. Proc. Natl. Acad. Sci. Unit. States Am 118 (9), e2018220118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T, Lenning M, Clark I, Bhimani I, Fortunato J, Marsh P, Xu X, Cao H, 2019. IEEE Sensor. J 19 (11), 4283–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T, Zhang J, Xia X, Xu X, Clark I, Schmiess-Heine L, Nguyen AH, Lau MPH, Cao H, 2020. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society. EMBC, pp. 2610–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenning M, Fortunato J, Le T, Clark I, Sherpa A, Yi S, Hofsteen P, Thamilarasu G, Yang J, Xu X, Han H-D, Hsiai T, Cao H, 2018. Sensors 18 (1), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M-H, Chou H-C, Chen Y-F, Liu W, Lee C-C, Liu LY-M, Chuang Y-J, 2018. Sci. Rep 8 (1), 15986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Li L, Lam YW, Siu CW, Cheng SH, 2016. Sci. Rep 6, 25073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Yang K-C, Dudley SC, 2014. Nat. Rev. Cardiol 11 (10), 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricondi-Massari M, Kalinin AL, Glass ML, Rantin FT, 1998. J. Therm. Biol 23 (5), 283–290. [Google Scholar]
- Matthews M, Varga ZM, 2012. ILAR J 53 (2), 192–204. [DOI] [PubMed] [Google Scholar]
- McNair WP, Sinagra G, Taylor MRG, Di Lenarda A, Ferguson DA, Salcedo EE, Slavov D, Zhu X, Caldwell JH, Mestroni L, 2011. J. Am. Coll. Cardiol 57 (21), 2160–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mente A, O’Donnell M, Rangarajan S, Dagenais G, Lear S, McQueen M, Diaz R, Avezum A, Lopez-Jaramillo P, Lanas F, Li W, Lu Y, Yi S, Rensheng L, Iqbal R, Mony P, Yusuf R, Yusoff K, Szuba A, Oguz A, Rosengren A, Bahonar A, Yusufali A, Schutte AE, Chifamba J, Mann JFE, Anand SS, Teo K, Yusuf S, 2016. Lancet 388, 465–475, 10043. [DOI] [PubMed] [Google Scholar]
- Mizusawa Y, Wilde AAM, 2012. Circulation: Arrhythm. Electrophysiol 5 (3), 606–616. [DOI] [PubMed] [Google Scholar]
- Monfredi O, Boyett MR, 2015. J. Mol. Cell. Cardiol 83, 88–100. [DOI] [PubMed] [Google Scholar]
- Nagasawa S, Saitoh H, Kasahara S, Chiba F, Torimitsu S, Abe H, Yajima D, Iwase H, 2018. Sci. Rep 8 (1), 8443–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemtsas P, Wettwer E, Christ T, Weidinger G, Ravens U, 2010. J. Mol. Cell. Cardiol 48 (1), 161–171. [DOI] [PubMed] [Google Scholar]
- Nguyen TP, Wang DW, Rhodes TH, George AL, 2008. Circ. Res 102 (3), 364–371. [DOI] [PubMed] [Google Scholar]
- Semelka M, Gera J, Usman S, 2013. Am. Fam. Physician 87 (10), 691–696. [PubMed] [Google Scholar]
- Shaffer F, Ginsberg JP, 2017. Front. Public Health 5 (258). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyek MR, Rog-Zielinska EA, Quinn TA, 2018. Prog. Biophys. Mol. Biol 138, 91–104. [DOI] [PubMed] [Google Scholar]
- Sun P, Zhang Y, Yu F, Parks E, Lyman A, Wu Q, Ai L, Hu C-H, Zhou Q, Shung K, Lien C-L, Hsiai TK, 2009. Ann. Biomed. Eng 37 (5), 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale JE, Chung MK, Campbell KB, Hammadah M, Joglar JA, Leclerc J, Rajagopalan B, 2020. Circulation 142 (15), e214–e233. [DOI] [PubMed] [Google Scholar]
- Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Shay CM, Spartano NL, Stokes A, Tirschwell DL, VanWagner LB, Tsao CW, null n., 2020. Circulation 141 (9), e139–e596. [DOI] [PubMed] [Google Scholar]
- Y L, Y L, P Z, J Z, X W, 2020. J. Cardiol. Ther 7 (1), 913–921, 2020. [Google Scholar]
- Yan J, Li H, Bu H, Jiao K, Zhang AX, Le T, Cao H, Li Y, Ding Y, Xu X, 2020. PLoS One 15 (5), e0232457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Zhao Y, Gu J, Quigley KL, Chi NC, Tai Y-C, Hsiai TK, 2012. Biomed. Microdevices 14 (2), 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukkoor S, Thohan V, 2018. American College of Cardiology, pp. 653–655.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


