Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections. C. trachomatis inorganic pyrophosphatase (CtPPase) hydrolyzes inorganic pyrophosphate during metabolism. A 2.2 Å resolution X-ray structure of CtPPase reveals shared structural features that may facilitate the repurposing of inhibitors identified for bacterial inorganic pyrophosphatases as starting points for new therapeutics.
Keywords: Chlamydia trachomatis, inorganic pyrophosphatase, infectious diseases, undergraduate education and training, Seattle Structural Genomics Center for Infectious Disease
Abstract
Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections globally and is one of the most commonly reported infections in the United States. There is a need to develop new therapeutics due to drug resistance and the failure of current treatments to clear persistent infections. Structures of potential C. trachomatis rational drug-discovery targets, including C. trachomatis inorganic pyrophosphatase (CtPPase), have been determined by the Seattle Structural Genomics Center for Infectious Disease. Inorganic pyrophosphatase hydrolyzes inorganic pyrophosphate during metabolism. Furthermore, bacterial inorganic pyrophosphatases have shown promise for therapeutic discovery. Here, a 2.2 Å resolution X-ray structure of CtPPase is reported. The crystal structure of CtPPase reveals shared structural features that may facilitate the repurposing of inhibitors identified for bacterial inorganic pyrophosphatases as starting points for new therapeutics for C. trachomatis.
1. Introduction
Chlamydiae are obligate intracellular bacteria that infect a wide range of eukaryotes, including humans, animals, insects and free-living amoebae. Chlamydia trachomatis is a Gram-negative coccus that causes a commonly known sexually transmitted infection often called chlamydia. Chronic chlamydia infection often leads to genital, ocular and respiratory disease (Lorenzini et al., 2010 ▸). The Chlamydia genus is phylogenetically distant from other bacteria, and 30% of its proteins are referred to as hypothetical proteins (Barta et al., 2013 ▸). Genital chlamydia is a major public health concern, with over 1.8 million cases reported to the US Centers for Disease Control and Prevention (CDC) in 2019. Furthermore, chlamydia is the most common bacterial sexually transmitted infection globally and is a leading cause of infertility (van Bergen et al., 2021 ▸; Dombrowski, 2021 ▸). The CDC recommends treating chlamydia in adults and adolescents with 100 mg doxycycline orally twice a day for seven days. Alternatively, a single 1 g oral dose of azithromycin or 500 mg levofloxacin can be administered. However, reinfection is common with all antibiotics, and compliance is low for doxycycline (Centers for Disease Control and Prevention, 2021 ▸). Efforts to identify new treatment strategies for chlamydia at the Seattle Structural Genomics Center for Infectious Disease (SSGCID) include structural studies of C. trachomatis proteins as the first steps towards rational drug discovery. C. trachomatis inorganic pyrophosphatase (CtPPase) was one of the investigated proteins because inorganic pyrophosphatases from other bacteria have shown promise as potentially selective targets (Pang et al., 2016 ▸; Lv et al., 2014 ▸). The production, crystallization and high-resolution structure of CtPPase are presented here.
2. Materials and methods
2.1. Macromolecule production
Cloning, expression and purification were conducted as part of the Seattle Structural Genomics Center for Infectious Disease (SSGCID) following standard protocols described previously (Bryan et al., 2011 ▸; Choi et al., 2011 ▸; Serbzhinskiy et al., 2015 ▸). The full-length gene for inorganic pyrophosphatase from C. trachomatis (CtPPase; UniProt O84777) encoding amino acids 1–209 was PCR-amplified from gDNA using the primers shown in Table 1 ▸. The gene was cloned into the ligation-independent cloning (LIC) expression vector pBG1861 encoding a noncleavable hexahistidine tag (Aslanidis & de Jong, 1990 ▸; Choi et al., 2011 ▸). Plasmid DNA was transformed into chemically competent Escherichia coli BL21(DE3)R3 Rosetta cells. The plasmid containing hexahistidine-tagged C. trachomatis inorganic pyrophosphatase (His-CtPPase) was expression-tested and 2 l of culture were grown using auto-induction medium (Studier, 2005 ▸). The expression clone ChtrB.01427.a.B1.GE42413 is available at https://www.ssgcid.org/available-materials/expression-clones/.
Table 1. Macromolecule-production information.
| Source organism | Chlamydia trachomatis (strain D/UW-3/Cx) |
| DNA source | Dr Kevin Hybiske (University of Washington, USA) |
| Forward primer | 5′-CTCACCACCACCACCACCATATGTCTAAAACACCATTATCCATAGC-3′ |
| Reverse primer | 5′-ATCCTATCTTACTCACTTACATAAAAAGATTGCAATAGTCTTCGT-3′ |
| Expression vector | pBG1861 |
| Expression host | E. coli BL21(DE3)R3 Rosetta cells |
| Complete amino-acid sequence of the construct produced | MAHHHHHHMSKTPLSIAHPWHGPVLTRDDYESLCCYIEITPADSVKFELDKETGILKVDRPQKFSNFCPCLYGLLPKTYCGDLSGEYSGQQSNRENIKGDGDPLDICVLTEKNITQGNILLQARPIGGIRILDSEEADDKIIAVLEDDLVYGNIEDISECPGTVLDMIQHYFLTYKATPESLIQAKPAKIEIVGLYGKKEAQKVIRLAHEDYCNLFM |
His-CtPPase was purified in a two-step protocol consisting of an immobilized metal-affinity chromatography (IMAC) step and size-exclusion chromatography (SEC). All chromatography runs were performed on an ÄKTApurifier 10 (GE Healthcare) using automated IMAC and SEC programs according to previously described procedures (Bryan et al., 2011 ▸). Thawed bacterial pellets were lysed by sonication in 200 ml buffer consisting of 25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 0.5% CHAPS, 30 mM imidazole, 10 mM MgCl2, 1 mM TCEP, 250 µg ml−1 AEBSF, 0.025% azide. After sonication, the crude lysate was clarified with 20 µl (25 U µl−1) Benzonase and incubated while mixing at room temperature for 45 min. The lysate was then clarified by centrifugation at 10 000 rev min−1 for 1 h using a Sorvall centrifuge (Thermo Scientific). The clarified supernatant was then passed over an Ni–NTA HisTrap FF 5 ml column (GE Healthcare) which was pre-equilibrated with loading buffer consisting of 25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 30 mM imidazole, 1 mM TCEP, 0.025% azide. The column was washed with 20 column volumes (CV) of loading buffer and was eluted with loading buffer plus 250 mM imidazole in a linear gradient over 7 CV. Peak fractions, as determined by the UV absorbance at 280 nm, were pooled and concentrated to 5 ml. A Superdex 75 SEC column (GE Healthcare) was equilibrated with running buffer consisting of 25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 2 mM DTT, 0.025% azide. The peak fractions were collected and analyzed for CtPPase using SDS–PAGE. The SEC peak fractions eluted as a single large peak at a molecular mass of ∼80 kDa, suggesting a trimeric enzyme. Peak fractions were pooled and concentrated to 62 mg ml−1 using an Amicon purification system (Millipore). Aliquots of 200 µl were flash-frozen in liquid nitrogen and stored at −80°C until use for crystallization.
2.2. Crystallization
Purified His-CtPPase was screened for crystallization in 96-well sitting-drop plates against the JCSG+ HTS (Rigaku Reagents) and MCSG1 (Anatrace) crystal screens. Equal volumes of protein solution (0.4 µl) and precipitant solution were set up at 287 K against a 80 µl reservoir in sitting-drop vapor-diffusion format. 3 mM inorganic pyrophosphate was added to the protein solution before crystallization experiments. Crystals were obtained using high sodium chloride and polyethylene glycol 3350 conditions (Table 2 ▸). A crystal was cryoprotected by exchange into precipitant supplemented with 15%(v/v) ethylene glycol and vitrified directly in liquid nitrogen.
Table 2. Crystallization.
| Method | Vapor diffusion, sitting drop |
| Plate type | 96-well Compact 300, Rigaku |
| Temperature (K) | 287 |
| Protein concentration (mg ml−1) | 31 |
| Buffer composition of protein solution | 3 mM inorganic pyrophosphate, 25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 2 mM DTT, 0.025% azide |
| Composition of reservoir solution | 2 M NaCl, 0.1 M Tris pH 8.5, 25%(v/v) PEG 3350 |
| Volume and ratio of drop | 0.4 µl protein plus 0.4 µl reservoir |
| Volume of reservoir (µl) | 80 |
| Composition of cryoprotectant solution | 2 M NaCl, 0.1 M Tris pH 8.5, 25%(v/v) PEG 3350, 15%(v/v) ethylene glycol |
2.3. Data collection and processing
Data were collected at 100 K on beamline 21-ID-F at the Advanced Photon Source, Argonne National Laboratory (see Table 3 ▸). Diffraction data (Table 3 ▸) were integrated using XDS and were reduced using XSCALE (Kabsch, 2010 ▸). Raw X-ray diffraction images are available at the Integrated Resource for Reproducibility in Macromolecular Crystallography at https://www.proteindiffraction.org.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Beamline 21-ID-F, APS |
| Wavelength (Å) | 0.97872 |
| Temperature (K) | 100 |
| Detector | RayoniX MX300HE CCD |
| Crystal-to-detector distance (mm) | 260 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 200 |
| Space group | C2221 |
| a, b, c (Å) | 77.16, 121.19, 124.50 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.24 |
| Resolution range (Å) | 44.99–2.25 (2.31–2.25) |
| Total No. of reflections | 226034 (16920) |
| No. of unique reflections | 27864 (2010) |
| Completeness (%) | 99.3 (99.2) |
| Multiplicity | 8.1 (8.4) |
| 〈I/σ(I)〉 | 25.24 (3.14) |
| R r.i.m. † | 0.046 (0.661) |
| Overall B factor from Wilson plot (Å2) | 56.57 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
The structure was solved by molecular replacement with Phaser (McCoy et al., 2007 ▸) from the CCP4 suite of programs (Collaborative Computational Project, Number 4, 1994 ▸; Krissinel et al., 2004 ▸; Winn et al., 2011 ▸) using PDB entry 5ls0 (Grzechowiak et al., 2019 ▸) as the search model. The structure was refined using iterative cycles of Phenix (Liebschner et al., 2019 ▸) followed by manual rebuilding of the structure using Coot (Emsley & Cowtan, 2004 ▸; Emsley et al., 2010 ▸). The quality of the structure was checked using MolProbity (Williams et al., 2018 ▸). All data-reduction and refinement statistics are shown in Table 4 ▸. The structure was refined to a resolution of 2.25 Å. Coordinates and structure factors have been deposited in the Protein Data Bank (https://www.rcsb.org) with accession code 6we5.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 44.99–2.25 (2.31–2.25) |
| Completeness (%) | 99.2 |
| σ Cutoff | F > 1.34σ(F) |
| No. of reflections, working set | 27834 (1787) |
| No. of reflections, test set | 2028 (156) |
| Final R cryst | 0.181 (0.275) |
| Final R free | 0.228 (0.374) |
| No. of non-H atoms | |
| Protein | 4694 |
| Ion | 3 |
| Ligand | 0 |
| Water | 65 |
| Total | 4762 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Angles (°) | 0.683 |
| Average B factors (Å2) | |
| Protein | 61.8 |
| Ion | 69.5 |
| Ligand | 0.0 |
| Water | 53.0 |
| Ramachandran plot | |
| Most favored (%) | 98.04 |
| Allowed (%) | 1.96 |
3. Results and discussion
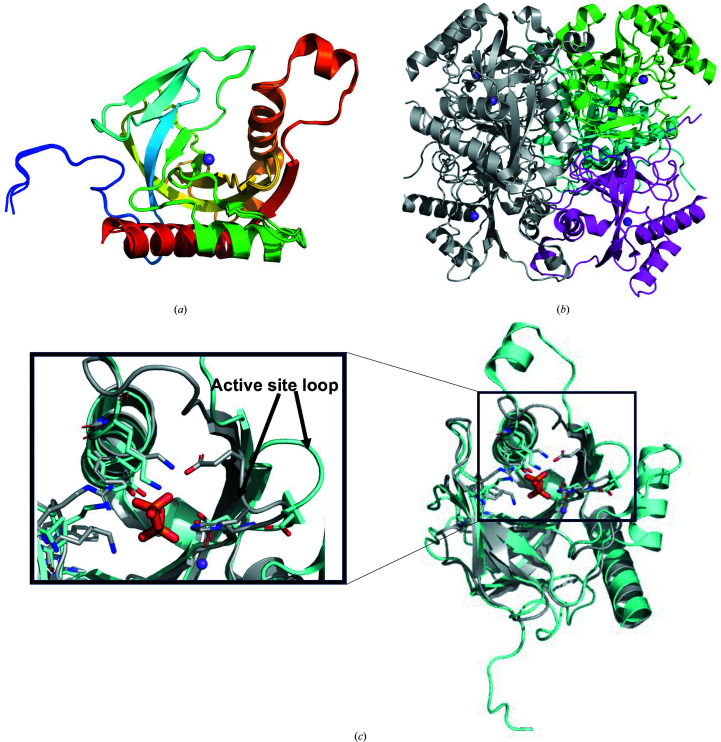
CtPPase is a small β-strand protein containing a core five-stranded oligonucleotide/oligosaccharide-binding (OB) fold. CtPPase has the prototypical family I pyrophosphatase (PPase) topology. Family 1 PPases are ubiquitous in all kingdoms of life (Kajander et al., 2013 ▸). The overall topology of CtPPase resembles an open fist (or baseball mitt) with the substrate-binding cavity sitting in the palm, while β-strands form finger-like structures surrounding the active site (Fig. 1 ▸ a).
Figure 1.
CtPPase structure. (a) The superposed CtPPase monomers are almost identical, with r.m.s.d.s of ∼0.3 Å for all atoms and ∼0.17 Å for Cα atoms. The monomers are colored from blue (N-terminus) to red (C-terminus). (b) A prototypical family I PPase hexamer was generated from the asymmetric unit trimer (monomers colored green, cyan and magenta) and a symmetry mate (shown in gray). The sodium ion bound in the active site of each monomer is shown as a purple sphere. (c) The CtPPase active-site loop (cyan) is in the open conformation compared with the closed conformation of M. tuberculosis PPase (MtPPase). The pyrophosphate (orange sticks) in the active site is from MtPPase (PDB entry 5kde), while the sodium ion (purple sphere) is from CtPPase (PDB entry 6we5).
The CtPPase structure was refined to 2.25 Å resolution in space group C2221 with three molecules in the asymmetric unit. Surface-area calculations by PISA (Krissinel, 2015 ▸) suggest a hexamer as the most likely biological assembly (Fig. 1 ▸ b). Hexamers were previously observed as the biological assembly in other well studied family I PPases, notably E. coli PPases (Cooperman et al., 1992 ▸). The CtPPase hexamer is similar to those of the well studied family I PPases. Electron density modeled as an Na atom was observed in the active site of each monomer. The active site is where the hydrolysis of pyrophosphate into two phosphate ions occurs. Despite the addition of pyrophosphate to the crystallization buffer, no density was observed for pyrophosphate or phosphate ions. Additionally, the flexible active-site loop is in the open conformation indicative of an apo structure without any substrate or product in the active site of CtPPase (Fig. 1 ▸ c). Future studies will include investigating whether the presence of the N-terminal hexahistidine tag renders CtPPase inactive and unable to hydrolyze pyrophosphate or form the biological hexamer in solution, or whether additional ions or cofactors need to be added to the enzyme before crystallization to generate the structure of the complex with pyrophosphate or phosphate.
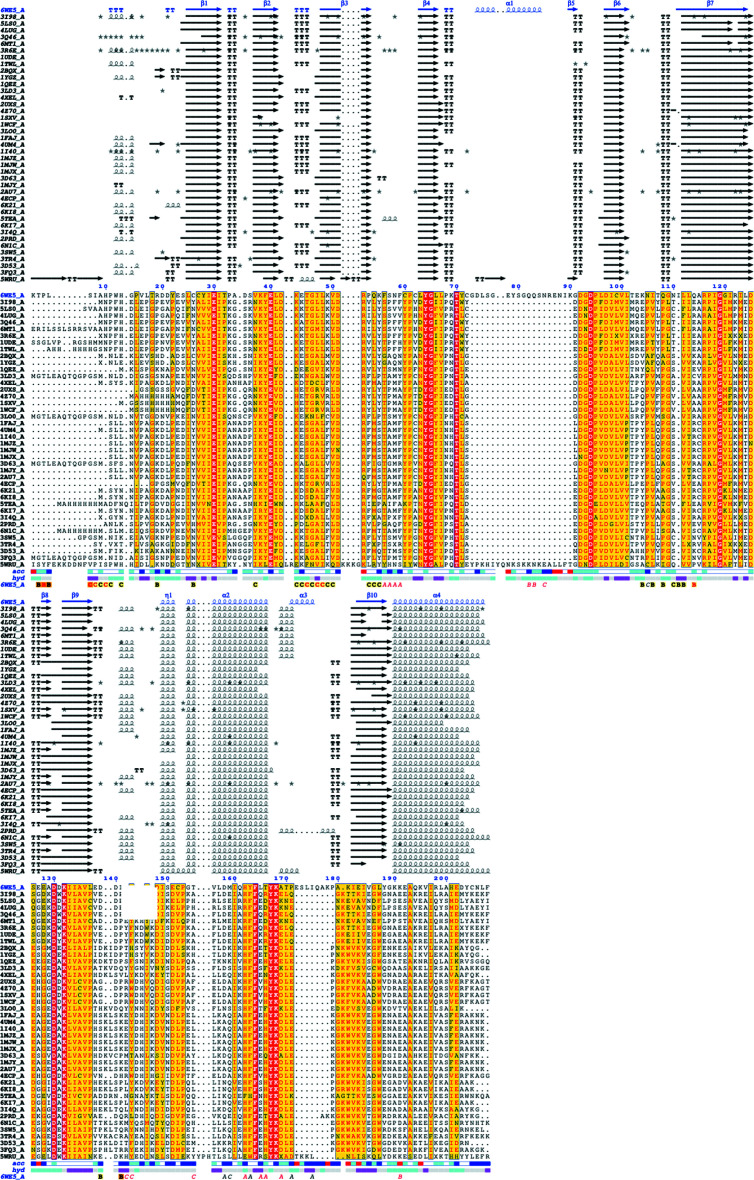
Since bacterial inorganic pyrophosphatases have shown promise as potentially selective targets (Pang et al., 2016 ▸; Lv et al., 2014 ▸), CtPPase was compared with other structures to determine whether it could be a viable drug target. PDBeFold analysis (http://www.ebi.ac.uk/msd-srv/ssm/; Krissinel & Henrick, 2004 ▸), the DALI server (http://ekhidna2.biocenter.helsinki.fi/dali/; Holm, 2020 ▸) and ENDscript analysis (Gouet et al., 2003 ▸; Robert & Gouet, 2014 ▸) were used to identify the closest structural neighbors of CtPPase. These analyses revealed that despite <37% sequence similarity, CtPPase shares significant secondary-structural similarity with several family I PPases, including some that have shown promise as drug targets (see supporting information and Fig. 2 ▸). The supporting information includes detailed results of the DALI (Supplementary Fig. S1) and PDBeFold (Supplementary Table S1) analyses. The overall core structure of CtPPase is highly similar to other bacterial PPases except for two major insertions (residues 71–86 and residues 170–180; Figs. 2 ▸ and 3 ▸). These insertions are on the exterior surface of the hexamer and do not participate in the formation of the hexamer or interact with the active site (Fig. 1 ▸ c).
Figure 2.
An ENDscript alignment identifies conserved residues in CtPPase and PPases. Multi-sequence alignment of CtPPase with 41 closest PPases obtained by a BLAST search against the PDBAA database. Identical and conserved residues are highlighted in red and yellow, respectively. Alternate residues are highlighted with gray stars. The different secondary-structure elements shown are α-helices (α), 310-helices (η), β-strands (β) and β-turns (TT).
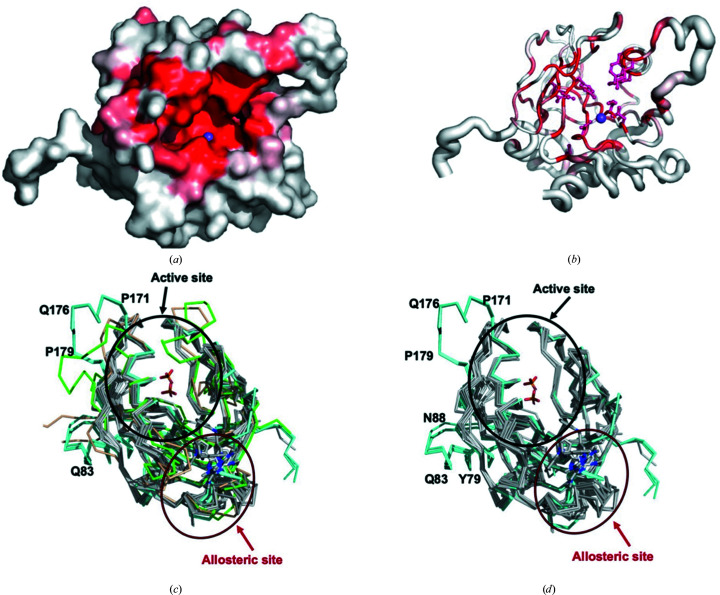
Figure 3.
Structural comparison of CtPPase with other PPases. (a) Solvent-accessible surface area colored by sequence conservation. Residues clustered in the active-site cleft are identified by the sodium ion present in the crystal structure of CtPPase (magenta sphere). (b) Coil diagram calculated by ENDscript. The circumference of the ribbon (sausage) represents the relative structural conservation compared with 41 other PPase structures (the same structures as indicated in Fig. 2 ▸). Thinner ribbons represent more conserved regions, while thicker ribbons represent less conserved regions. The ten identical residues cluster within or in proximity to the active site. Identical residues are indicated by red regions on the surface and a red ball-and-stick representation in ribbon diagrams. The sodium ion bound in the active site of each monomer is shown as a purple sphere. (c) Comparison of the active and allosteric sites of CtPPase (cyan) with bacterial PPases (gray) and eukaryotic PPases (Homo sapiens PPase, PDB entry 7btn, green; Plasmodium falciparum PPase, PDB entry 5wru, brown). (d) The same view of the structures without the eukaryotic PPases. The top ten unique bacterial PPases were selected from the ENDscript alignment. All three monomer chains of CtPPase are shown.
A comparison of CtPPase with 41 other PPases deposited in the Protein Data Bank using ENDscript identified 19 identical residues which cluster in the active-site pocket (Figs. 2 ▸ and 3 ▸). The active-site region contains a D-(S/G/N)-D-P-ali-D-ali-ali motif, where ali is C/I/L/M/V (Kankare et al., 1994 ▸). PDBeFold analysis also revealed that, as expected, bacterial PPases were structurally most similar to CtPPase (Supplementary Table S1). The most similar structure was from Thermococcus thioreducens, followed by Acinetobacter baumannii, with root-mean-square differences (r.m.s.d.s) of 1.18 and 1.33 Å over 196 and 162 residues, respectively. The top PPases that showed structural similarity are listed in Supplementary Table S1. Our preliminary analysis revealed structural differences between bacterial and eukaryotic PPases that may possibly be exploited for inhibitor design (Fig. 3 ▸ c).
A manual search of the entire PDB for structures of PPases from other organisms identified 80 different ligand-bound PPase structures. The majority of ligands were metals, ions, substrate or substrate mimics. Most ligands were bound in the active site. However, there were four structures with ligands bound outside the active site: two structures of Mycobacterium tuberculosis PPase (MtPPase), PDB entries 5kde and 5kd7 (Pang et al., 2016 ▸), and two structures of Burkholderia pseudomallei PPase (BpPPase), PDB entries 3ej2 and 3ej0 (Van Voorhis et al., 2009 ▸). The MtPPase ligands are low-micromolar IC50 allosteric inhibitors (Pang et al., 2016 ▸). The BpPPase ligands were discovered from fragment-based screens at the Seattle Structural Genomics Center for Infectious Disease. Superposition of the C. trachomatis (CtPPase) structures with MtPPase and BpPPase revealed that the ligands are small organic compounds that are located in a surface binding pocket on the opposite side to the pyrophosphate binding pocket (Fig. 3 ▸ c).
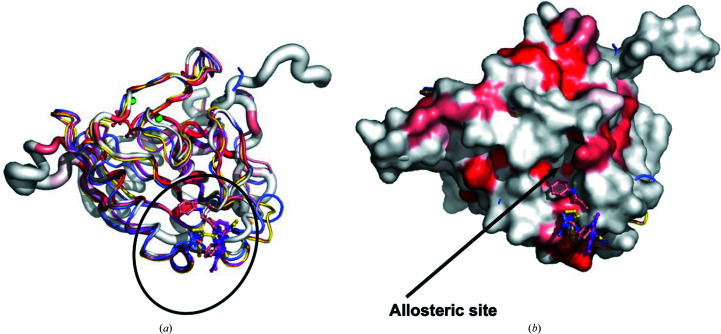
The two previous studies on MtPPase and BpPPase suggest the possibility of an allosteric binding site that small-molecule inhibitors of bacterial PPases could target. Comparison of the CtPPase structure with those of MtPPase and BpPPase shows that the loop adjacent to the putative allosteric binding site has moved into the pocket compared with MtPPase and BpPPase, closing off this site. Inspection of the solvent-accessible surface of CtPPase reveals a medium-sized cleft that partially occupies the fragment-binding site of BpPPase (Fig. 4 ▸). Future fragment-based screening targeting this cleft may generate allosteric inhibitors of CtPPase.
Figure 4.
The bacterial PPase allosteric binding site. The putative allosteric binding site of CtPPase identified from superposition of CtPPase (PDB entry 6we5) with MtPPase (PDB entries 5kde and 5kdf) and BpPPase (PDB entries 3ej0 and 3ej2). (a) Coil diagram of CtPPase (red and white) superimposed on the MtPPase structures with allosteric inhibitors (PDB entries 5kde, yellow, and 5kdf, magenta) and BpPPase bound with fragment compounds (PDB entries 3ej0, wheat, and 3ej2, blue). The location of the compounds is indicated with a black oval. The circumference of the coil represents the relative structural conservation compared with 41 other PPase structures (the same structures as indicated in Fig. 2 ▸). (b) A solvent-accessible surface diagram of CtPPase calculated with ENDscript reveals a potential binding pocket labeled the allosteric site on the CtPPase surface in proximity to the compounds.
4. Conclusion
We have determined the structure of an inorganic pyrophosphatase (PPase) from C. trachomatis. The overall structure is a prototypical bacterial PPase with additional amino acids inserted beyond the conserved active site. CtPPase has a pocket in proximity to the previously identified bacterial allosteric binding sites, suggesting the possibility of developing allosteric inhibitors of CtPPase. While the preliminary structural studies are promising, future studies include validating the enzymatic activity of CtPPase and probing the active and allosteric sites of CtPPase with substrates and potential inhibitors.
Supplementary Material
PDB reference: inorganic pyrophosphatase from Chlamydia trachomatis D/UW-3/Cx, 6we5
Supplementary Figure and Table. DOI: 10.1107/S2053230X22002138/tb5175sup1.pdf
Acknowledgments
This manuscript was generated as an educational collaboration between Hampton University (a Historically Black College and University) and the SSGCID. The SSGCID consortium is directed by Dr Peter Myler (Principal Investigator) and comprises many different scientists working at multiple centers towards determining the three-dimensional structures of proteins from biodefense organisms and emerging infectious diseases. In particular, we would like to thank the SSGCID cloning, protein production and X-ray crystallography groups at the Center for Global Infectious Disease Research, the University of Washington and UCB.
Funding Statement
This work was funded by National Institute of General Medical Sciences grants 1U01GM138433 and T34GM136489 to Oluwatoyin A. Asojo and Oluwatoyin A. Asojo; National Institute of Allergy and Infectious Diseases grants HHSN272201700059C, HHSN272201200025C, and HHSN272200700057C to Peter J. Myler.
References
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Barta, M. L., Hickey, J., Kemege, K. E., Lovell, S., Battaile, K. P. & Hefty, P. S. (2013). Acta Cryst. F69, 1196–1201. [DOI] [PMC free article] [PubMed]
- Bergen, J. E. A. M. van, Hoenderboom, B. M., David, S., Deug, F., Heijne, J. C. M., van Aar, F., Hoebe, C., Bos, H., Dukers-Muijrers, N., Götz, H. M., Low, N., Morré, S. A., Herrmann, B., van der Sande, M. A. B., de Vries, H. J. C., Ward, H. & van Benthem, B. H. B. (2021). Sex. Transm. Infect. 97, 501–506. [DOI] [PMC free article] [PubMed]
- Bryan, C. M., Bhandari, J., Napuli, A. J., Leibly, D. J., Choi, R., Kelley, A., Van Voorhis, W. C., Edwards, T. E. & Stewart, L. J. (2011). Acta Cryst. F67, 1010–1014. [DOI] [PMC free article] [PubMed]
- Centers for Disease Control and Prevention (2021). Sexually Transmitted Infections Treatment Guidelines, 2021. https://www.cdc.gov/std/treatment-guidelines/default.htm.
- Choi, R., Kelley, A., Leibly, D., Nakazawa Hewitt, S., Napuli, A. & Van Voorhis, W. (2011). Acta Cryst. F67, 998–1005. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cooperman, B. S., Baykov, A. A. & Lahti, R. (1992). Trends Biochem. Sci. 17, 262–266. [DOI] [PubMed]
- Dombrowski, J. C. (2021). Ann. Intern. Med. 174, ITC145–ITC160. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Grzechowiak, M., Ruszkowski, M., Sliwiak, J., Szpotkowski, K., Sikorski, M. & Jaskolski, M. (2019). Biochem. J. 476, 2297–2319. [DOI] [PubMed]
- Holm, L. (2020). Protein Sci. 29, 128–140. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kajander, T., Kellosalo, J. & Goldman, A. (2013). FEBS Lett. 587, 1863–1869. [DOI] [PubMed]
- Kankare, J., Neal, G. S., Salminen, T., Glumoff, T., Cooperman, B. S., Lahti, R. & Goldman, A. (1994). Protein Eng. 7, 823–830. [DOI] [PubMed]
- Krissinel, E. (2015). Nucleic Acids Res. 43, W314–W319. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krissinel, E. B., Winn, M. D., Ballard, C. C., Ashton, A. W., Patel, P., Potterton, E. A., McNicholas, S. J., Cowtan, K. D. & Emsley, P. (2004). Acta Cryst. D60, 2250–2255. [DOI] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Lorenzini, E., Singer, A., Singh, B., Lam, R., Skarina, T., Chirgadze, N. Y., Savchenko, A. & Gupta, R. S. (2010). J. Bacteriol. 192, 2746–2756. [DOI] [PMC free article] [PubMed]
- Lv, W., Banerjee, B., Molland, K. L., Seleem, M. N., Ghafoor, A., Hamed, M. I., Wan, B., Franzblau, S. G., Mesecar, A. D. & Cushman, M. (2014). Bioorg. Med. Chem. 22, 406–418. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Pang, A. H., Garzan, A., Larsen, M. J., McQuade, T. J., Garneau-Tsodikova, S. & Tsodikov, O. V. (2016). ACS Chem. Biol. 11, 3084–3092. [DOI] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Serbzhinskiy, D. A., Clifton, M. C., Sankaran, B., Staker, B. L., Edwards, T. E. & Myler, P. J. (2015). Acta Cryst. F71, 594–599. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Van Voorhis, W. C., Hol, W. G. J., Myler, P. J. & Stewart, L. J. (2009). PLoS Comput. Biol. 5, e1000530. [DOI] [PMC free article] [PubMed]
- Williams, C. J., Headd, J. J., Moriarty, N. W., Prisant, M. G., Videau, L. L., Deis, L. N., Verma, V., Keedy, D. A., Hintze, B. J., Chen, V. B., Jain, S., Lewis, S. M., Arendall, W. B., Snoeyink, J., Adams, P. D., Lovell, S. C., Richardson, J. S. & Richardson, J. S. (2018). Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: inorganic pyrophosphatase from Chlamydia trachomatis D/UW-3/Cx, 6we5
Supplementary Figure and Table. DOI: 10.1107/S2053230X22002138/tb5175sup1.pdf