Abstract
Objective
Antibiotic resistance by beta lactamase expression is a serious and growing threat. We aimed to determine whether beta-lactamase activity is detectable in urine specimens to enable faster identification of resistance.
Methods
Urine specimens from patients with extended spectrum beta lactamase (ESBL)-expressing urinary infections were incubated with beta lactam antibiotics. Beta lactam hydrolysis was determined by mass spectrometry methods.
Results
Ceftriaxone hydrolysis was observed in 45 of 45 ESBL-containing specimens from patients not treated with a beta lactamase inhibitor before specimen collection. Ceftriaxone hydrolysis was not observed in 108 of 108 non-ESBL-containing specimens. Spiking studies show that beta lactam hydrolysis can be observed within 30 minutes. Beta lactam hydrolysis is evidenced by mass spectrometry preceded by either liquid chromatography or matrix-assisted laser desorption ionization specimen processing methods.
Conclusion
Clinically significant beta lactamase activity is detectable directly from urine specimens. The described methods would enable the detection of beta lactam resistance 24 to 48 hours sooner than culture based methods.
Keywords: antibiotic resistance, beta lactamase, ESBL, beta lactam hydrolysis, mass-spectrometry, urinary tract infection, antibiotic resistance testing
The beta lactam class of antibiotics is a key component of the antimicrobial arsenal. The archetypal beta lactam is penicillin, which is naturally derived from penicillium mold and is synthetically manufactured for frequent human use.1 Since the discovery of penicillin, many other beta lactam antibiotics (all containing a beta lactam ring) have been developed to improve pharmacologic properties (eg, oral availability) and microbicidal properties (eg, targeting a broader spectrum of organisms).2 This class of antibiotics comprises 19 of 40 spots on the World Health Organization list of essential antibiotics and accounts for more than half of all antibiotics used globally.2-4
The beta lactam ring that defines the beta lactam class of drugs mimics the d-alanyl-d-alanine structure that nearly all bacterial species require for the composition of their cell wall.5 Beta lactam drugs irreversibly bind to cell-wall synthesis enzymes, leading to structural degradation and bacterial death. Resistance to beta lactams can be acquired by 3 mechanisms: (i) mutation of the beta lactam binding enzymes to prevent beta lactam binding, (ii) acquisition of a beta lactam efflux pump that prevents the drugs from entering the cell, and (iii) acquisition of a beta lactamase enzyme that hydrolyzes the beta lactam ring, rendering the molecule ineffective.6-8 The first 2 mechanisms of resistance are slow to be acquired and have a negative impact on the overall fitness of an organism.9 In contrast, the acquisition of a beta lactamase enzyme is fast because the gene encoding the enzyme can be transferred between bacteria (even those of different species) and production of the enzyme in sufficient quantities to confer resistance presents minimal metabolic cost to the organism.6,9
The evolution and spread of beta lactamase enzymes has led to a substantial health care burden. The rise and spread of extended spectrum beta lactamase (ESBL)-expressing Enterobacteriaceae have led to approximately 197,400 infections, 9,100 deaths, and $1.2 billion in estimated attributable health care costs annually in the United States.10,11 New beta lactamase enzymes are continuing to emerge and spread, presenting a serious public health threat.12,13
The typical workflow in a clinical microbiology laboratory leading to identification of antibiotic-resistant organisms consists of (1) specimen plating and mixed growth, (ii) pathogen isolation and growth, (iii) identification of the organism by biochemical tests or matrix-assisted laser desorption ionization-time of flight (MALDI-TOF), and (iv) observed growth in antibiotic-containing media to determine antibiotic susceptibility/resistance patterns.14-17 This workflow requires 36 to 72 hours, involves multiple manual steps, and may not detect inducible resistance.18 While awaiting results, most patients are empirically treated, and when susceptibility results are available the treatment strategy is adjusted.19 Inevitable delays in providing appropriate antibiotic treatment lead to higher patient morbidity and mortality and contribute to the spread of resistant organisms.20,21
Given the clinical challenges that beta lactamase expressing pathogens present, there is a clear need for faster identification to both enable effective treatment and enact isolation precautions preventing the further spread of resistant organisms. Resistant bacteria typically express high levels of beta lactamase to effectively counteract therapeutic doses of beta lactam antibiotics.22 We sought to determine whether beta lactamase activity could be sensitively and specifically detected in unprocessed clinical specimens to quickly (within hours) detect the presence of beta lactamase expressing organisms. Our approach used mass spectrometry to show beta lactamase activity directly in biological specimens by measuring the hydrolysis of target antibiotics through the disappearance of protonated molecular ions in the presence of resistant organisms.
Methods
Specimen Preparation
Residual urine specimens collected during routine clinical care were used for this study. The study was approved by the University of California, San Diego Institutional Review Board with a waiver of informed consent (protocol number 181656XL).
Urine specimens were collected in Becton Dickinson Biosciences vacutainer culture and susceptibility preservative tubes containing 2.63 mg/mL boric acid, 3.95 mg/mL sodium borate, and 1.65 mg/mL sodium formate to prevent bacterial overgrowth (Becton, Dickinson, and Company, Franklin Lakes, NJ) as part of routine clinical practice. All specimens tested were excess clinical specimens stored at 4°C for 7 days or less. Colony forming units (CFUs) were determined by colony counting after the application of specimens using a 0.001-mL loop. Specimens were centrifuged and reconstituted in drug-free urine (DFU) to remove preservatives, which inhibit hydrolysis activity. Next, 400 μL of the specimen was centrifuged at 21,000 g in a fixed-angle rotor for 10 minutes in 1.7 mL Eppendorf tubes. After centrifugation, the supernatant was aspirated off using an air-displacement pipette and discarded. The aspiration of supernatant was conducted in such a manner as to leave behind any pellet, regardless of whether it as visible to the naked eye. Next, 200 μL of certified DFU (UTAK Catalog #88121-CDF) containing 500 μg/mL of ceftriaxone was added to each specimen and mixed by vortexing for 1 minute. Specimens were then incubated at 37°C for at least 4 hours. After incubation, specimens were centrifuged at 13,000 g in a fixed-angle rotor for 5 minutes. In addition, 100 μL of the supernatant was diluted using 300 μL of 95% deionized H2O, 5% methanol containing internal standard (IS; 50 ng/mL amphetamine-d5). Isotopically labeled beta lactam was not used as an internal standard to prevent (i) ESBL-mediated hydrolysis of the internal standard or (ii) an internal standard acting as an inhibitor of ESBL-mediated hydrolysis. Finally, 20 μL of the diluted specimen was injected using a full-loop injection on a liquid chromatography (LC) quadrupole TOF mass spectrometer (MS) system (Waters ultra performance liquid chromatography, Waters Xevo quadrupole time-of-flight [QTOF]).
For multidrug analysis, specimen preparation was performed as above with the exception that specimens were incubated for 12 hours at 37°C and that 200 μL of certified DFU containing 500 μg/mL of each drug (ceftriaxone, cefazolin, and oxacillin or ceftriaxone, cefazolin, oxacillin, and meropenem) was added to each specimen. The kinetics of ceftriaxone hydrolysis and identification of its hydrolysis products were performed as stated above, with the exception that certified DFU containing 300 μg/mL of ceftriaxone was used. Incubation times were 5 minutes, 30 minutes, and 20 hours, with specimens being immediately injected into the LC-MS system after centrifugation and specimen dilution.
LC
For ceftriaxone analysis, LC was performed using a Waters BEH-C18 column (1.7 μm) on a Waters Acquity UPLC system. Mobile phase A was 5 mM ammonium formate, 0.1% formic acid, and 99.9% deionized H2O (buffer A). Mobile phase B was 0.1% formic acid and 99.9% acetonitrile (ACN). The LC method spanned a total of 8 minutes and is described in Supplemental Table 1. Specimens were injected using an autosampler with a full loop injection of 20 μL.
For multidrug analysis (ceftriaxone, cefazolin, oxacillin, meropenem), LC was performed with the same buffers, column, and injection parameters as above. The LC method spanned a total of 8 minutes and is described in Supplementary Table 1.
For ceftriaxone hydrolysis product analysis, LC was performed with the same buffers, column, and injection parameters as above. The LC method spanned a total of 15 minutes and is described in Supplementary Table 1.
QTOF-MS
Data for MS were acquired on a Waters Xevo G2 TOF MS. Mass spectra were acquired in a scanning mode across an m/z range (50–900 m/z) at a rate of 1 scan per 0.5 seconds in continuum mode. The total MS method spanned 6.5 minutes and was run in positive polarity with the analyzer in sensitivity mode. Electrospray ionization was used as the ionization method with a cone voltage of 25 volts and a fixed collision energy value of 6 volts.
Qualitative Matrix Interference Studies
Qualitative assessment of matrix interference was performed by injecting 5 unspiked ESBL-containing urine specimens and 5 unspiked ESBL-negative urine specimens while simultaneously infusing a solution containing 50 μg/mL of ceftriaxone reconstituted in 50% ACN:50% deionized H2O. Extracted ion chromatograms (EICs) of ceftriaxone were overlaid with the expected ceftriaxone peak for all 10 patient urine specimens.
MALDI-TOF
MALDI-TOF MS was performed on a Bruker Microflex MALDI-TOF mass spectrometer equipped with an N2 laser. Spectra were acquired in reflectron-positive mode, with 100 μm raster spot and roughly 80% laser power, 200 shots (random walk, 20 shots) detection gain 40×, and a range of 100–1000 m/z. Specimens were prepared as above and mixed 1:1 with 10 mg/mL α-cyano-4-hydroxycinnamic acid. Using the dried drop method, 2 μL of the mixture was spotted on a Bruker MSP 96 target ground steel plate. Calibration was performed using a ceftriaxone standard at 555.0533 m/z before data acquisition.
MS Data Processing
The LC-MS data were processed in MassLynx (Waters) using the integration parameters described in Supplemental Table 2. All traces were smoothed with 1 iteration using the mean of 2 adjacent scans. The MALDI-TOF data were processed using Flex Analysis 4.2 (Bruker Daltronics). Peak areas described throughout are measured in arbitrary units correlated to the quantity of ions colliding with the ion detector.
Data Analysis and Graphing
Data analysis was performed with R studio (rstudio.com). Graphs were created using the ggplot function within the tidyverse package (cran.r-project.org/web/packages/tidyverse).
Results
Hydrolysis of Ceftriaxone in Urine Specimens
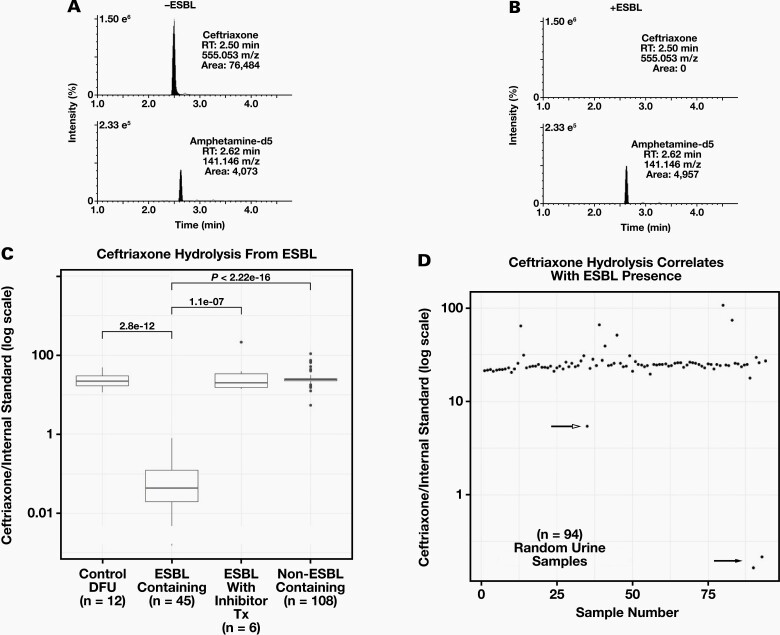
The ESBL hydrolysis of ceftriaxone was evaluated by examining EICs of ceftriaxone (500 μg/mL) and IS in DFU (control) after 4 hours of incubation at 37°C in the presence or absence of an ESBL-containing patient urine specimen (Figure 1). In DFU alone, peak areas of ceftriaxone and IS were 76,484 and 4,073, respectively. However, a discernible ceftriaxone peak was not observed after incubation with an ESBL, whereas a similar peak area for the IS was observed (4,957). It is clear when comparing Figure 1A with Figure 1B that ceftriaxone was hydrolyzed by the ESBL-containing specimen because the protonated molecular ion of ceftriaxone is not discernible in Figure 1B. The IS is easily seen in both Figure 1A and 1B and helps rule out a matrix effect that could potentially suppress the ceftriaxone signal in Figure 1B. Boxplots illustrating the median ratios of ceftriaxone area counts and IS area counts (CTX/IS ratio) after incubation with DFU, ESBL-containing patient specimens, ESBL-containing specimens from patients treated with a beta lactamase inhibitor (tazobactam or meropenem), and non-ESBL-containing patient specimens are shown in Figure 1C. Strikingly, there was at least a 10-fold difference in the median CTX/IS ratio between ESBL-containing specimens and specimens lacking ESBL activity. A cutoff of the CTX/IS of anywhere between 0.77 and 5.4 would allow perfect discrimination. To confirm the discriminatory utility of the hydrolysis assay, we tested 94 specimens in a blinded manner (Figure 1D). Two specimens showed clear evidence of hydrolysis (filled arrow), and these 2 specimens were confirmed to contain ESBLs.
Figure 1.
Evaluation of ESBL Hydrolysis of Ceftriaxone: (A) EICs of the protonated molecular ions (exact m/z) of ceftriaxone (top) and IS (bottom) after 4 hours of incubation with a sample not containing an extended spectrum beta lactamase (-ESBL) expressing pathogen. (B) EICs of the protonated molecular ions of ceftriaxone (top) and IS (bottom) after 4 hours of incubation with an ESBL containing specimen (+ESBL). Expected retention times for each compound are indicated to illustrate the loss of a peak. % ion intensity (indicated at top left of each EIC) is plotted on the Y-axis and Time (min) is plotted on the X-axis. (C) Boxplots of median ratios of ceftriaxone/IS after incubation with: Control DFU –drug free urine (n = 12), ESBL containing urine specimens (n = 45), ESBL containing urine specimens from patients receiving beta-lactamase inhibitor treatment (n = 6), and non-ESBL containing specimens (n = 108). Wilcoxon P-values are displayed. (D) Scatterplot illustrating ratios of ceftriaxone/IS plotted on log scale for 94 random urine culture specimens after incubation. Closed arrow indicates two samples with ESBL identified by urine culture and susceptibility assays. Open arrow indicates a non-ESBL containing specimen.
During a 12-month period from December 2019 to December 2020, the University of California, San Diego Clinical Microbiology Laboratory performed 37,731 urine cultures. Of these cultures, gram negative rods were observed in 11,560 (30.6%). In addition, ESBL-expressing organisms were observed in 1,283 cultures (3.4% of total cultures, 11.2% of gram negative rods). All ESBL-expressing organisms were gram negative rods: 987 (76.9%) were Escherichia coli, 280 (21.8%) were Klebsiella pneumoniae, and 16 (1.3%) were Klebsiella oxytoca. Thus, our finding that 2 out of 94 culture specimens contained ESBL organisms is consistent with the overall prevalence of ESBLs in our institution’s tested population.
Discrimination between ESBL- and non-ESBL-containing specimens was robust under conditions intended to simulate decreased analytical sensitivity or detect potential interferences. The robust identification of ESBL-containing specimens was also apparent when data were analyzed at unit resolution (Supplemental Figure 1), simulating the performance of an instrument with lower mass accuracy. To determine whether bacterial overgrowth or matrix effects could lead to false positives or false negatives, 5 specimens with ESBL and 5 specimens without ESBL, all with visible turbidity and/or discoloration, were analyzed on 5 nonconsecutive days in both a qualitative and a quantitative matrix effect study. The CTX/IS ratios were separated by at least an order of magnitude on all days, although some degradation of hydrolysis activity was observed (Supplemental Figure 2). An infusion of 50 μg/mL of ceftriaxone performed during an injection of urine specimens from the same set of 10 specimens revealed no significant ion suppression or enhancement across the ceftriaxone peak observed (Supplementary Figure 3).
Collective Hydrolysis of Ceftriaxone, Cefazolin, and Oxacillin in Urine Specimens
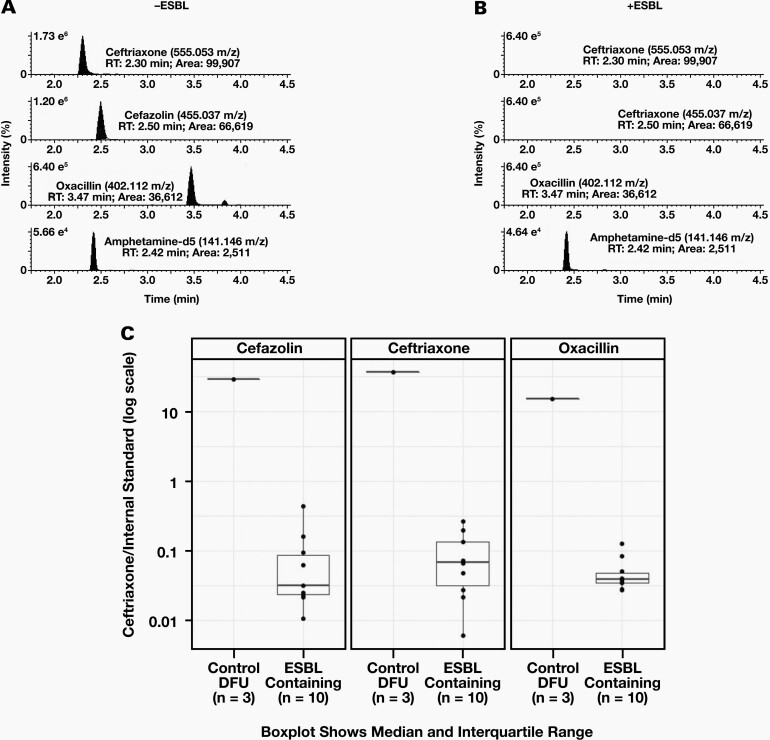
To show the generalizability of our approach, we looked for ESBL hydrolysis of a mixture of 3 beta lactam antibiotics (ceftriaxone, cefazolin, oxacillin [CCO]) by observing the EICs of ceftriaxone (500 μg/mL), cefazolin (500 μ g/mL), and oxacillin (500 μ g/mL), and the EIC of the IS in DFU were evaluated after 12 hours of incubation at 37°C, in the presence or absence of an ESBL (Figure 2). As expected, robust peak areas for the molecular ions corresponding to ceftriaxone (99,907), cefazolin (66,619), oxacillin (36,612), and IS (2,511) were observed in the absence of an ESBL (Figure 2A). Except for the IS, these peaks disappeared after incubation with an ESBL-containing specimen, indicating robust hydrolysis of all 3 beta lactam antibiotics (Figure 2B). Boxplots illustrating the median ratios of ceftriaxone/IS, cefazolin/IS, and oxacillin/IS after incubation with (n = 10) or without (n = 3) an ESBL-containing specimen are shown in Figure 2C. As seen with the hydrolysis of ceftriaxone alone, there was at least a 10-fold difference in the median ratios between ESBL-containing specimens and specimens lacking ESBL activity for the CCO mixture, with all beta lactam antibiotics being hydrolyzed in a similar fashion.
Figure 2.
Evaluation of ESBL Hydrolysis of CCO: (A) EICs of the protonated molecular ions of ceftriaxone, cefazolin, oxacillin, and IS are shown after incubation with a sample not containing extended spectrum beta lactamase (-ESBL) expressing pathogen. Exact m/z of the protonated molecular ion for each compound is indicated. (B) EICs of the protonated molecular ions of ceftriaxone, cefazolin, oxacillin, and IS are shown after incubation with a sample containing an ESBL containing specimen (-ESBL). Expected retention times of the protonated molecular ions for each compound are indicated to illustrate the loss of a peak. % ion intensity (indicated at top left of each EIC) is plotted on the Y-axis and Time (min) is plotted on the X-axis. (C) Boxplots plotted on a log scale of individual median ratios of CCO/IS after incubation with: Control DFU –drug free urine (n = 3) or ESBL containing urine specimens (n = 10). Dotted line indicates a ceftriaxone/IS ratio of 1.
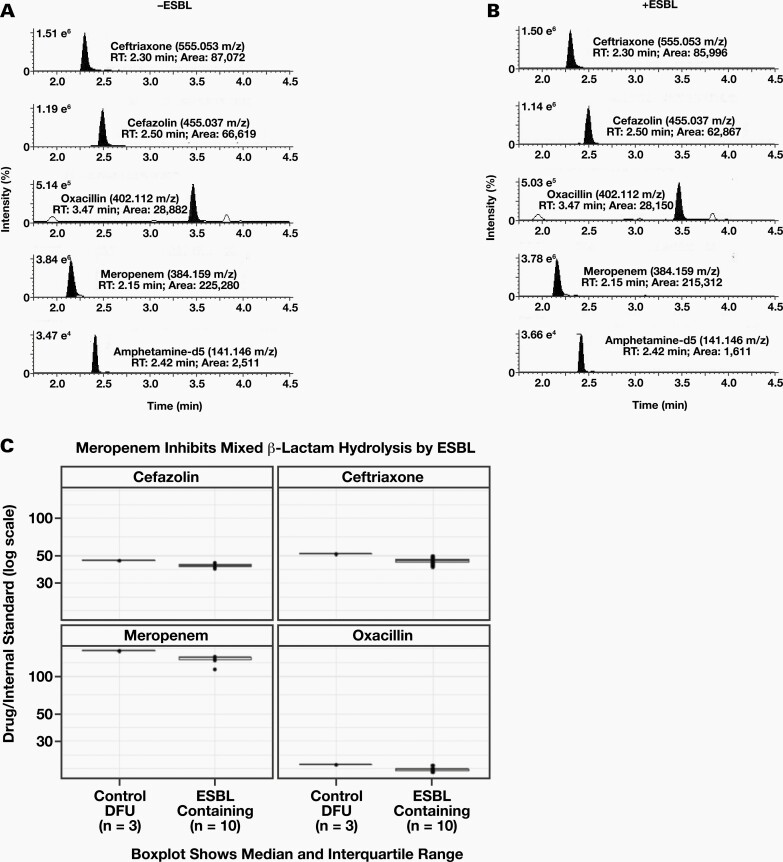
CCO Hydrolysis Inhibited by Meropenem in Noncultured Urine Specimens
Because a subpopulation of patients with suspected urinary tract infections (UTIs) take antibiotics or inhibitors that prevent ESBL activity, we monitored the hydrolysis of a CCO mixture in the presence of meropenem. The EICs for the protonated molecular ions of CCO, meropenem, and IS are shown in DFU after 12 hours of incubation at 37°C (Figure 3A). Peak areas of ceftriaxone (500 μg/mL), cefazolin (500 μ g/mL), oxacillin (500μg/mL), meropenem (500 μg/mL), and IS were 87,072; 66,190; 28,882; 225,280; and 1,524, respectively. As expected, these peak areas were essentially unchanged after a parallel incubation with an ESBL-containing specimen, indicating that meropenem inhibits the ESBL hydrolysis of CCO (Figure 3B). Boxplots illustrating the median ratios of ceftriaxone/IS, cefazolin/IS, oxacillin/IS, and meropenem/IS after incubation with (n = 10) or without (n = 3) an ESBL-containing specimen are shown in Figure 3C.
Figure 3.
Meropenem Inhibits ESBL CCO Hydrolysis: (A) EICs of the protonated molecular ions of ceftriaxone, cefazolin, oxacillin, meropenem and IS are shown after incubation with a sample not containing extended spectrum beta lactamase (-ESBL) expressing pathogen. Exact m/z of the protonated molecular ion for each compound is indicated. (B) EICs of the protonated molecular ions of ceftriaxone, cefazolin, oxacillin, meropenem, and IS are shown after incubation with a sample containing an ESBL containing specimen (-ESBL). Expected retention times of the protonated molecular ions for each compound are indicated to illustrate the loss of a peak. % ion intensity (indicated at top left of each EIC) is plotted on the Y-axis and Time (min) is plotted on the X-axis. (C) Boxplots plotted on a log scale of individual median ratios of CCO and meropenem/IS after incubation with: Control DFU –drug free urine (n = 3) or ESBL containing urine specimens (n = 10).
Kinetics of ESBL Ceftriaxone Hydrolysis and Identification of Hydrolysis Products
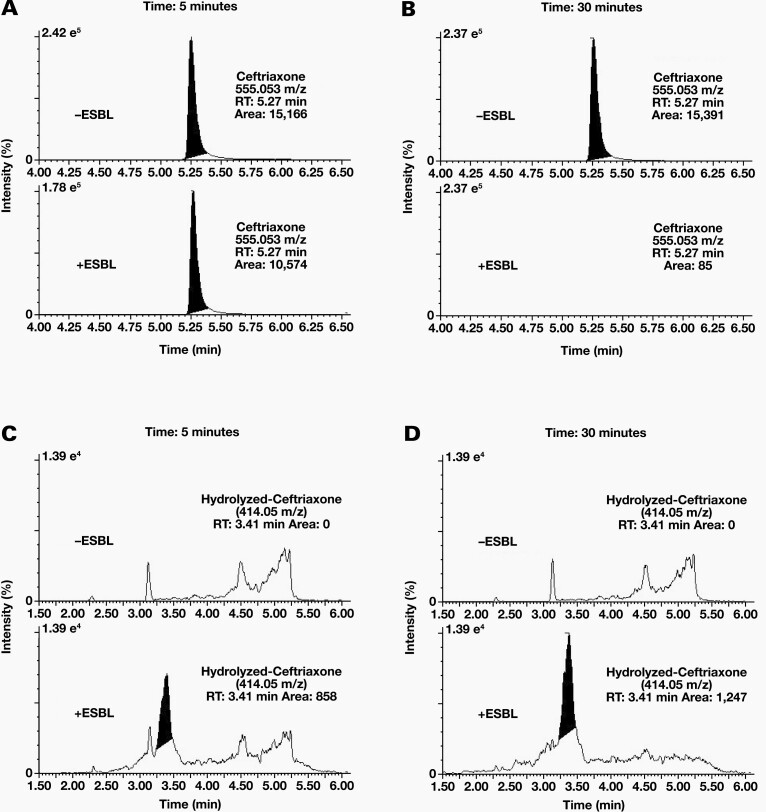
The kinetics of ceftriaxone hydrolysis by ESBL-containing specimens was evaluated by incubating ceftriaxone in the presence or absence of an ESBL-containing specimen for 5 minutes and 30 minutes at room temperature (RT). At the 5-minute time point, ceftriaxone peak areas were 15,166 in the absence of an ESBL and 10,574 in the presence of an ESBL (Figure 4A), an approximate 30% reduction in raw area counts. Hydrolysis was essentially complete by the 30-minute time point; the ceftriaxone peak area was 15,391 in the absence of an ESBL and was not observed in the presence of an ESBL (Figure 4B). The correlation of the CTX/IS ratios with the CFUs of the specimens showed no correlation (Supplemental Figure 4), suggesting that hydrolysis kinetics were substrate limited.
Figure 4.
Kinetics of Ceftriaxone Hydrolysis: Peak intensity of ceftriaxone after 5 minutes (A) and 30 minutes (B) of incubation with (+ESBL) or without (-ESBL) an ESBL containing specimen. The predicted hydrolysis product due to the loss of the ceftriaxone’s triazine-ylthiol group at 414.05 m/z (RT: 3.40 min) appears after 5 minutes (C) and 30 minutes (D) of incubation only in the presence of ESBL expressing bacteria. % ion intensity (indicated at top left of each EIC) is plotted on the Y-axis and Time (min) is plotted on the X-axis.
The presence of a ceftriaxone hydrolysis product was evaluated by monitoring the EICs of the protonated molecular ion of ceftriaxone after the loss of its triazine-ylthiol group (414.05 m/z).23 In the presence of an ESBL, a peak at 3.41 minutes was observed with an integrated area of 858. After 30 minutes, the peak at 3.41 minutes had an increased peak area of 1247 (Figure 4D). This peak was no longer observed after a 20-hour incubation at RT (Supplemental Figure 5), indicating that this hydrolysis product was likely unstable and unsuitable for end-point analysis.
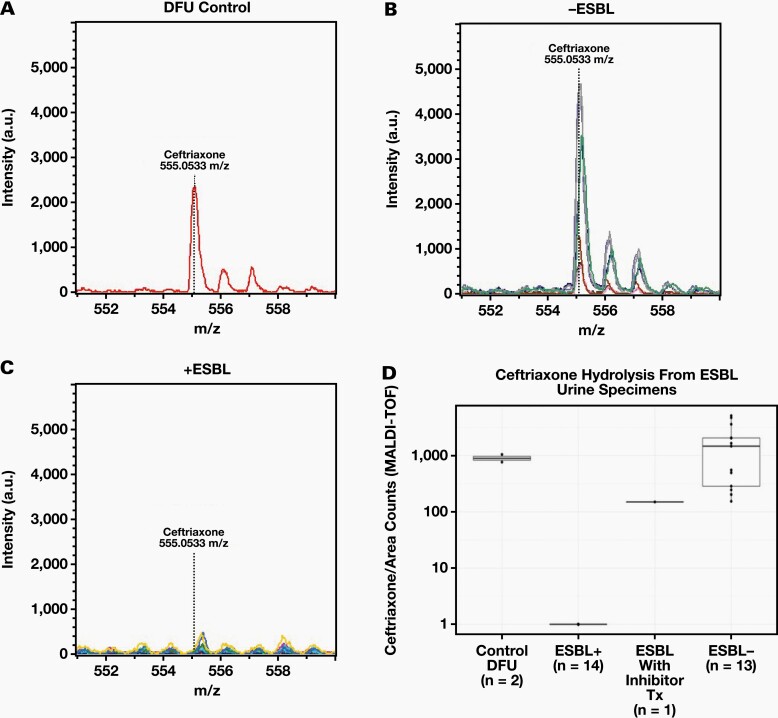
Monitoring of Ceftriaxone Hydrolysis by MALDI-TOF MS
Because many clinical microbiology laboratories are equipped with MALDI-TOF MS instruments, we evaluated ceftriaxone hydrolysis using a Bruker Microflex MALDI-TOF MS (Figure 5). Using the same specimen preparation used for LC-MS analysis, the protonated molecular ion of ceftriaxone was clearly observed at 555.0533 m/z in both the control and ESBL-negative specimens (Figure 5A and 5B). In contrast, this peak was absent in the ESBL-positive specimens, indicating ceftriaxone hydrolysis by ESBL (Figure 5C). Boxplots representing the median area counts for the protonated molecular ion of ceftriaxone are shown for controls (n = 2), ESBL-containing specimens (n = 14), ESBL-containing specimens from patients treated with a beta lactamase inhibitor (n = 1), and non-ESBL-containing specimens (n = 13; Figure 5D). The median ceftriaxone area counts for the controls, ESBL positives from patients treated with a beta lactamase inhibitor, and ESBL-negative specimens were at least 100-fold greater than the median area counts observed in ESBL-positive specimens.
Figure 5.
Evaluation of Ceftriaxone hydrolysis by MALDI-TOF MS. Protonated molecular ion of ceftriaxone after incubation with DFU (n = 2) (A), patient specimens (n = 8) without an ESBL (B), and patient specimens with an ESBL (n = 14) (C). The expected m/z of the protonated ceftriaxone molecular ion (555.0533 m/z) is indicated in each plot with a dotted line and each plot is shown using the same X and Y axis scale. (D) Box plots of the median area counts of ceftriaxone after incubation with DFU, ESBL +, ESBL + w/Inhibitor Treatment, and ESBL – specimens as measured by MALDI-TOF MS. Whiskers are up to but no greater than 1.5 times the inter-quartile range.
Discussion
The evolution and spread of antibiotic resistance among human pathogens represents a serious public health threat.11 Faster identification of the presence of antibiotic-resistant organisms is a key component in the effort to reduce the spread of antibiotic resistance, as evidenced by the inclusion of diagnostic development in the Centers for Disease Control and Prevention’s (CDC) national strategy to combat antibiotic resistance.24 Our findings indicate that beta lactamase activity from resistant organisms can be detected in clinical urine specimens. This finding enables a diagnostic strategy that could provide same-day detection of beta lactamase expressing organisms as compared with the several days required for traditional culture and susceptibility methods.
Same-day identification of beta lactamase expressing organisms could provide both clinical and infection control benefits. Timely treatment with effective antibiotics leads to improved clinical outcomes and decreased costs for individual patients.25 Infection control measures for patients with antibiotic-resistant organisms (eg, use of personal protective equipment) can prevent the nosocomial spread of resistant infections if implemented in a timely manner.26 In addition, our method may be useful for public health or agricultural monitoring for ESBL.27 Our data, and that from other centers, suggest that ESBL-expressing organisms account for 3% to 23% of all UTIs and are likely to increase as antibiotic resistance spreads.24,28-30 According to chart reviews of our small and randomly selected population of ESBL-expressing E. coli UTIs, 6 of 34 patients were empirically treated with ineffective cephalosporins (ceftriaxone or cefazolin). These patients may have had shorter lengths of infection had they been treated with effective antibiotics earlier. Furthermore, ESBL-expressing E. coli was discovered in one of these patients 48 hours after being discharged to a long-term care facility with an ineffective oral antibiotic regimen, a scenario associated with the continued spread of antibiotic-resistant organisms. Consistent with our findings, an observational study of hospitalized patients with ESBL pyelonephritis showed that ~65% of patients received ineffective antibiotic coverage.31 Direct detection of ESBL activity in urine specimens could be used to rule out empirical treatment for Gram-negative UTIs (eg, ceftriaxone or cefazolin).
Significant effort has gone into the detection of antibiotic-resistant organisms given the clinical need for faster identification. Primarily, breakthroughs have occurred in identifying the sequence of the genes responsible for resistance.32,33 Specifically, molecular identification of the mecA gene creating methicillin-resistant Staphylococcus aureus has shown clinical utility.34 However, the diversity of known beta lactamase genes (hundreds have been identified) and the emergence of novel genes limits the utility of this approach.32,33 Identification of ESBL enzyme activity from clinical specimens utilizing a chromogenic substrate has shown promise, but the reagents necessary are not readily available.35,36
Our findings show, for the first time, that the activity of ESBL enzyme present in a centrifuged urine specimen is sufficient for robust detection, using LC-MS, before the required culture and isolation of pathogenic organisms. Such direct detection of beta lactamase activity enables the fast detection of resistance in clinical scenarios involving other pathogens, clinical specimens, or beta lactam drugs. For example, detecting the presence of carbapenemase activity in sputum specimens from patients with suspected Acinetobacter infections could indicate the presence of carbapenemase expressing Acinetobacter infections, another class of antibiotic-resistant organisms classified as an urgent threat by the CDC.11 Additional studies need to be conducted to show that direct detection of beta lactamase activity could be used to differentiate a pathogenic infection from non-disease-causing colonization with a resistant organism, a common diagnostic challenge.37
Our data was primarily generated using LC-based separation methods followed by MS detection of parent beta lactam ions because of analytical sensitivity and ease of data interpretation. Because we did not rely on high-resolution data for identifying beta lactamase activity, this technique is generalizable to LC combined with most quadrupole mass spectrometers. In addition, we have shown that beta lactamase detection can be performed using MALDI techniques. The MALDI-TOF technology is utilized for organism identification in many clinical microbiology laboratories. Notably, the specific instrument type and matrix we used for our experiments is the same as those used for clinical specimens in our institution’s microbiology laboratory. The ready availability of both quadrupole and MALDI-TOF technology in clinical laboratories would enable the utilization of our described diagnostic approach with minimal additional instrumentation required.
Clinically administered beta lactamase inhibitors are a potential interferent of the described diagnostic strategy. As we observed, patients who received tazobactam or meropenem before specimen collection did not have appreciable beta lactamase activity in their urine. It should be noted that specimens intended for microbiology testing are ideally collected from patients before they receive treatment because antibiotics can also interfere with most culture, identification, and susceptibility testing methods. Thus, the potential for interference in the described method of beta lactamase activity assessment would not present an excessive burden compared to standard methods. Both tazobactam and meropenem have been shown to inhibit beta lactamase activity in vitro.38,39
Conclusion
Further development (eg, identifying optimal buffers and matrices, and the most stable hydrolysis products) of the described diagnostic strategy will be needed to improve turnaround time, which could be reduced to 30 minutes or less based on estimates of beta lactamase enzyme kinetics.40 Larger studies including unique clinical scenarios must be performed to understand sensitivity, specificity, optimal cutoffs, and concordance with traditional culture and susceptibility analysis for relevant pathogen, specimen, and antibiotic combinations.
Supplementary Material
Acknowledgments
We sincerely thank the staff of the clinical microbiology laboratory for assistance with data and specimen acquisition, especially Olympia Alverez and Janet Stagnaro.
This work was primarily funded using laboratory discretionary funds. A.M.C.R. and P.C.D. were supported by the National Science Foundation grant IOS-1656481 and National Institutes of Health grant number 1DP2GM137413-01. P.C.D. was supported by the Gordon and Betty Moore Foundation through grant GBMF7622 and the National Institutes of Health (P41 GM103484, R03 CA211211, R01 GM107550).
Raymond Suhandynata, Robert Fitzgerald, and Nicholas Bevins are listed as coinventors on a provisional patent application involving this work.
Glossary
Abbreviations
- ESBL
extended spectrum beta lactamase
- MALDI-TOF
matrix-assisted laser desorption ionization-time of flight
- CFU
colony forming unit
- DFU
drug-free urine
- IS
internal standard
- LC
liquid chromatography
- MS
mass spectrometry
- QTOF
quadrupole time of flight
- ACN
acetonitrile
- EIC
extracted ion chromatogram
- CTX-IS ratio
ceftriaxone-internal standard ratio
- CCO
ceftriaxone, cefazolin, oxacillin
- UTI
urinary tract infection
- RT
room temperature
- CDC
Centers for Disease Control and Prevention
References
- 1. Zaffiri L, Gardner J, Toledo-Pereyra LH. History of antibiotics. From salvarsan to cephalosporins. J Invest Surg. 2012;25(2):67–77. [DOI] [PubMed] [Google Scholar]
- 2. Bush K, Bradford PA. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med. 2016;6(8):a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elander RP. Industrial production of beta-lactam antibiotics. Appl Microbiol Biotechnol. 2003;61(5–6):385–392. [DOI] [PubMed] [Google Scholar]
- 4. Wirtz VJ, Hogerzeil HV, Gray AL, et al. Essential medicines for universal health coverage. Lancet. 2017;389(10067):403–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yocum RR, Rasmussen JR, Strominger JL. The mechanism of action of penicillin. Penicillin acylates the active site of Bacillus stearothermophilus D-alanine carboxypeptidase. J Biol Chem. 1980;255(9):3977–3986. [PubMed] [Google Scholar]
- 6. Essack SY. The development of beta-lactam antibiotics in response to the evolution of beta-lactamases. Pharm Res. 2001;18(10):1391–1399. [DOI] [PubMed] [Google Scholar]
- 7. Fani F, Leprohon P, Zhanel GG, Bergeron MG, Ouellette M. Genomic analyses of DNA transformation and penicillin resistance in Streptococcus pneumoniae clinical isolates. Antimicrob Agents Chemother. 2014;58(3):1397–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fuda CC, Fisher JF, Mobashery S. Beta-lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell Mol Life Sci. 2005;62(22):2617–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Autiero I, Costantini S, Colonna G. Modeling of the bacterial mechanism of methicillin-resistance by a systems biology approach. PLoS One. 2009;4(7):e6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jernigan JA, Hatfield KM, Wolford H, et al. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med. 2020;382:1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed July 21, 2021. [Google Scholar]
- 12. De Angelis G, Del Giacomo P, Posteraro B, Sanguinetti M, Tumbarello M. Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int J Mol Sci. 2020;21(14):5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1):e00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Processing, isolation, detection, and interpretation of aerobic bacteriology cultures. In: Leber AL, ed. Clinical Microbiology Procedures Handbook. Vol. 1–3, 4th ed. American Society for Microbiology; 2016: 3.3.1.1–3.3.2.15. [Google Scholar]
- 15. Beta-lactamase tests. In: Leber AL, ed. Clinical Microbiology Procedures Handbook. Vol. 1–3, 4th ed. American Society for Microbiology; 2016: 5.5.1–5.5.8. [Google Scholar]
- 16. Extended-spectrum beta-lactamase testing for Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Proteus mirabilis. In: Leber AL, ed. Clinical Microbiology Procedures Handbook. Vol. 1–3, 4th ed. American Society for Microbiology; 2016: 5.12.1–5.12.7. [Google Scholar]
- 17. Minimum bactericidal concentration testing. In: Leber AL, ed. Clinical Microbiology Procedures Handbook. Vol. 1–3, 4th ed. American Society for Microbiology; 2016: 5.14.1.1–5.14.3.6. [Google Scholar]
- 18. Harada S, Ishii Y, Yamaguchi K. Extended-spectrum β-lactamases: implications for the clinical laboratory and therapy. Korean J Lab Med. 2008;28(6):401–412. [DOI] [PubMed] [Google Scholar]
- 19. Leekha S, Terrell CL, Edson RS. General principles of antimicrobial therapy. Mayo Clin Proc. 2011;86(2):156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev. 2018;42(1):fux053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. San Millan A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 2018;26(12):978–985. [DOI] [PubMed] [Google Scholar]
- 22. Livermore DM. Beta-lactamases: quantity and resistance. Clin Microbiol Infect. 1997;3(Suppl 4):S10–S19. [PubMed] [Google Scholar]
- 23. Oviaño M, Gómara M, Barba MJ, Revillo MJ, Barbeyto LP, Bou G. Towards the early detection of β-lactamase-producing Enterobacteriaceae by MALDI-TOF MS analysis. J Antimicrob Chemother. 2017;72(8):2259–2262. [DOI] [PubMed] [Google Scholar]
- 24. Centers for Disease Control and Prevention. U.S. action to combat antibiotic resistance: a national priority.https://www.cdc.gov/drugresistance/us-activities.html. Accessed July 21, 2021.
- 25. Bonine NG, Berger A, Altincatal A, et al. Impact of delayed appropriate antibiotic therapy on patient outcomes by antibiotic resistance status from serious Gram-negative bacterial infections. Am J Med Sci. 2019;357(2):103–110. [DOI] [PubMed] [Google Scholar]
- 26. Kluytmans-van den Bergh MFQ, Bruijning-Verhagen PCJ, Vandenbroucke-Grauls CMJE, et al. ; SoM Study Group. Contact precautions in single-bed or multiple-bed rooms for patients with extended-spectrum β-lactamase-producing Enterobacteriaceae in Dutch hospitals: a cluster-randomised, crossover, non-inferiority study. Lancet Infect Dis. 2019;19(10):1069–1079. [DOI] [PubMed] [Google Scholar]
- 27. Kim YA, Kim H, Seo YH, Park GE, Lee H, Lee K. Prevalence and molecular epidemiology of extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli from multiple sectors of the swine industry in Korea: a Korean nationwide monitoring program for a one health approach to combat antimicrobial resistance. Ann Lab Med. 2021;41(3):285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flokas ME, Detsis M, Alevizakos M, Mylonakis E. Prevalence of ESBL-producing Enterobacteriaceae in paediatric urinary tract infections: a systematic review and meta-analysis. J Infect. 2016;73(6):547–557. [DOI] [PubMed] [Google Scholar]
- 29. Lee DS, Lee CB, Lee SJ. Prevalence and risk factors for extended spectrum beta-lactamase-producing uropathogens in patients with urinary tract infection. Korean J Urol. 2010;51(7):492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baek YJ, Kim YA, Kim D, et al. Risk factors for extended-spectrum-β-lactamase-producing Escherichia coli in community-onset bloodstream infection: impact on long-term care hospitals in Korea. Ann Lab Med. 2021;41(5):455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Talan DA, Takhar SS, Krishnadasan A, et al. Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli infections in patients with pyelonephritis, United States. Emerg Infect Dis. 2016;22(9):1594–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Angelis G, Grossi A, Menchinelli G, Boccia S, Sanguinetti M, Posteraro B. Rapid molecular tests for detection of antimicrobial resistance determinants in Gram-negative organisms from positive blood cultures: a systematic review and meta-analysis. Clin Microbiol Infect. 2020;26(3):271–280. [DOI] [PubMed] [Google Scholar]
- 33. Burnham CD, Leeds J, Nordmann P, O’Grady J, Patel J. Diagnosing antimicrobial resistance. Nat Rev Microbiol. 2017;15(11):697–703. [DOI] [PubMed] [Google Scholar]
- 34. Hiramatsu K, Ito T, Tsubakishita S, et al. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother. 2013;45(2):117–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallah S, Decré D, Genel N, Arlet G. The β-lacta test for direct detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae in urine. J Clin Microbiol. 2014;52(10): 3792–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nordmann P, Dortet L, Poirel L. Rapid detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2012;50(9):3016–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bajaj P, Singh NS, Virdi JS. Escherichia coli β-lactamases: what really matters. Front Microbiol. 2016;7:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bonomo RA, Rudin SA, Shlaes DM. Tazobactam is a potent inactivator of selected inhibitor-resistant class A beta-lactamases. FEMS Microbiol Lett. 1997;148(1):59–62. [DOI] [PubMed] [Google Scholar]
- 39. Nukaga M, Bethel CR, Thomson JM, et al. Inhibition of class A beta-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1. J Am Chem Soc. 2008;130(38):12656–12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Page MG. Extended-spectrum beta-lactamases: structure and kinetic mechanism. Clin Microbiol Infect. 2008;14(Suppl 1):63–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.