Abstract
Since the late 19th century, the immune system has been known to play a role in cancer risk, initiation, and progression. Genome-wide association studies (GWAS) have identified hundreds of genetic risk loci for autoimmune and inflammatory diseases, yet the connection between human genetic variation and immune-mediated response to cancer treatments remains less well-explored. Understanding inherited genetic variation, with respect to germline genetic polymorphisms that affect immune system pathways, could lead to greater insights about how these processes may best be harnessed to successfully treat cancer. Our goal in this manuscript was to understand progress and challenges in assessing the role of inherited genetic variation in response to cancer treatments. Overall, the 39 studies reviewed here suggest that germline genetic variation in immune system related genes may potentially affect responses to cancer treatments. Although further research is needed, considering information on germline immune genetic variation may help, in some cases, to optimize cancer treatment.
Keywords: Germline, Genetic, Immunology, Cancer, Treatment, Outcomes
Introduction:
Over the past decade, several avenues of research have demonstrated the impact of the host, or germline genome, on cancer, from risk to progression to outcomes. Recent studies have shown that germline variants may be associated with increased risk for certain somatic mutations or may influence which mutations are selected for in the growing tumor (1,2). Furthermore, emerging evidence suggests germline variants can be used to predict the prognosis of cancer patients, tumor progression (3–7) and outcomes in response to cancer treatments (8–12). For example, germline variants in mismatch repair genes are associated with microsatellite instability and increased neoantigen production, rendering these tumors more susceptible to immune checkpoint blockade therapy (13,14). Additionally, recent studies identified variants that may affect genes with less obvious connections to cancer outcomes, such as genes that play roles in the vascular system (15–18).
The host immune system has been known to play an important role in cancer risk since the late 19th century, and growing evidence suggests that germline genetic variation influences host immunity both in general and in relation to cancer risk (19–22). Germline genetic variation in genes that code for immune modulatory proteins can significantly impact an individual’s immune function. Such inherited genetic variants may result in differences in immune system components that affect the abundance and activation states of immune cell types, the expression of immunomodulatory molecules, regulation of immune-related genes (23), and even the oncogenic mutations present in tumors (24,25). Hundreds of germline variants have been identified as causal variants and risk alleles in autoimmune diseases such as inflammatory bowel disease and rheumatoid arthritis (26). Certain human leukocyte antigen (HLA) variants are also associated with inflammatory conditions, autoimmune diseases, infection responses, and some cancers (20,21). Additionally, individuals with immune dysregulation, such as autoimmune diseases like rheumatoid arthritis, are at increased risk for developing certain types of cancer, demonstrating the link between immune function and cancer (22).
However, studies of germline variants in immune-related genes in association with outcomes in cancer patients are lacking. Given the emerging evidence that germline variation affects cancer outcomes (8,17), evidence that germline variation in immune related factors may affect the oncogenetic mutation (24,25) and immune composition of solid tumors (27), and recent progress in manipulating host immune components to treat cancer (28), we conducted an initial, broad review of published manuscripts to summarize information regarding types of cancers, outcomes, genes, and cancer treatments that were reported in epidemiologic studies examining germline immune genetic associations with cancer treatment outcomes. Our goal was to assess current progress in understanding whether germline genetic variation in immune system related genes could contribute to developing treatment strategies for a variety of cancers.
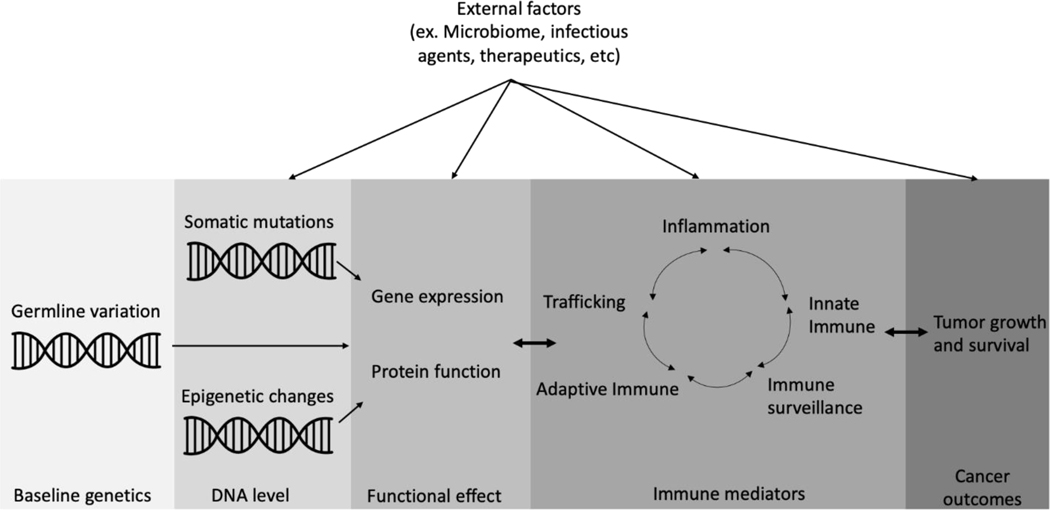
Overall, we identified 39 peer-reviewed epidemiologic studies published between 2010 and 2019 that found an association between germline genetic variants in immune-related genes and patient outcomes after cancer treatment. The genes that were analyzed in these studies encoded components of the innate and adaptive immune systems, inflammatory molecules, and immune surveillance components. We focused on studies published between 2010 and 2019, to coincide with the rise in use of immunotherapies for treating cancers (29), although we also included other types of cancer treatment. Identifying and characterizing germline genetic polymorphisms that affect host immune responses to tumors and treatments could lead to greater insights into how cancer treatments are mediated by the immune system and how these processes may best be harnessed to successfully treat cancer (Fig. 1).
Fig. 1.
Understanding genetic heterogeneity, with respect to germline genetic polymorphisms that affect host immune responses to tumors and treatments, could lead to greater insights about how cancer is mediated by the immune system and how these processes may best be harnessed to successfully treat cancer.
Methods
Study search
For this manuscript, we sought evidence of associations between immune germline genetics and cancer outcomes as reported in peer-reviewed epidemiologic research manuscripts. PubMed was searched for the period between January 1, 2010 through December 31, 2019 using the following search strategy: (Immunogenetics OR Neoplasms/immunology OR Genetics/immunology) AND (Neoplasms/drug therapy OR Neoplasms/therapy OR Neoplasms/therapeutic use OR Neoplasms/transplantation OR Neoplasms/complications OR Prognosis OR “disease progression”[mesh] OR immunotherapy) AND (Neoplasms OR Cancer OR malignancy OR tumor) AND (Genetic variation OR Mutation OR Polymorphism, genetic) AND (germline mutation/genetics OR germline OR GWA study OR “epidemiologic methods”[Mesh] OR neoplasms/epidemiology OR familial OR hereditary OR inborn) NOT (“Case Reports” [Publication Type:NoExp ] OR “Review” [Publication Type: NoExp] OR “Meta-Analysis” [Publication Type: NoExp]) and filters were applied (Species: Human; Languages: English; Publication dates: From 2010/01/01 to 2019/12/31). [11]
Study selection
Manuscripts identified by the literature search were excluded based on the following criteria: (i) studies not in humans with cancer; (ii) studies examining somatic mutations/tumor markers rather than germline immune genetics; (iii) articles with no original data (e.g. reviews, meta-analysis); (iv) studies that did not include human genetics; (v) case studies; (vi) studies that did not include cancer patient outcomes data such as recurrence, overall survival (OS), recurrence free survival (RFS), disease free survival (DFS), progression free survival (PFS), tumor shrinkage, response to treatment (complete response [CR], molecular response), relapse, event free survival (EFS), metastasis, survival time, or infections; (vii) studies in which germline genetics were not examined (e.g., the research analyzed only somatic mutations). The eligibility of each manuscript was assessed independently by two reviewers. A third reviewer resolved any disagreements.
Data abstraction
Data abstraction for the selected articles was primarily performed by one reviewer and cross-checked by at least one other reviewer. Data abstracted included: germline immune genes examined, cancer and treatment types studied, associations reported, outcomes examined, and prognosis/prediction goal of the article.
Results
Our PubMed search yielded 601 references, of which 454 were excluded after initial abstract review (Supplementary Fig. 1). Manuscripts were excluded primarily for lacking genetic data, including only tumor data (i.e., no germline data), not specifying treatment, and/or not examining cancer or cancer outcomes in humans. After abstract review and data abstraction, we excluded an additional 102 articles. The primary reasons for excluding articles at the data abstraction point were the article lacked cancer treatment information or did not test for associations between germline genetic variation in immune genes and cancer treatment outcomes. In total, 39 articles met the inclusion criteria for review.
Of the 39 studies we found (Tables 1–4), 25 different cancer types were assessed, including both solid tumors and blood cancers. Given that the role of the immune system in the formation and progression of hematologic malignancies is well recognized (30), it was not unexpected that the majority of papers focused on hematologic malignancies (18 papers). Also, as may have been expected, the second most studied cancer in these papers was melanoma (8 papers), which is recognized as one of the most immunogenic tumor types (31). Similarly, given the known role of immune cells in colorectal cancer formation and evolution (32), the third most common cancer in the examined studies was colorectal cancer (4 papers). Beyond hematologic malignancies, melanoma and colorectal cancer, the articles we found provided evidence that germline genetic variation in immune genes may influence treatment outcomes in a wide range of cancers. Such associations may even yield meaningful insight to treating rare cancers, such as bladder. For example, a study by Lim et al (33) found that lower expression of endoplasmic reticulum aminopeptidase 2 (ERAP2) was associated with better overall survival of bladder cancer patients (luminal subtype; HR=1.4, p=0.03) treated with the anti-PD1 drug atezolizumab. ERAP2 encodes a protein that is important in antigen presentation and processing. Genetic variants of ERAP2 that affect levels of expression could be further investigated as potential prognostic biomarkers and may be biologically meaningful by affecting possible interactions with HLA molecules.
Table 1.
Germline Immune Genetic Associations with Immunotherapy Outcomes
| Ref | Cancer | Treatment | Outcome | # subjects | Immune Genes | Immune gene function* | Main germline immune genetic associations found | Statistical significance of association |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| (44) | Bladder | BCG vaccine | Recurrence | 125 | TNFA, IL2RA, IL17A, IL17RA, IL18R1, ICAM-1, FASL, TRAILR1 | Cytokines – TNF family and Interleukins; Antimicrobial (regulates cell death); antigen processing and presentation | Patients with SNPs in immune and inflammatory genes had increased risk of recurrence after treatment. | HR values ranged between 1.70 and 5.19 |
| (33) | Bladder | Anti-PD-L1 (atezolizumab) | OS | 311 | TWAS identified variants associated with ERAP2 | Antigen processing and presentation | Lower ERAP2 expression levels were associated with improved response to atezolizumab (better OS) in luminal subtype bladder cancer patients. | p=0.03 |
| (42) | Ovarian | Farletuzumab (anti-folate receptor α) | PFS | 461 | FCGR2A, FCGR3A | Antigen-antibody - Immunoglobulin receptor on macrophages, neutrophils, Natural Killer cell | Enhanced clinical outcome was observed in patients with at least one high affinity allele of FCGR2A and FCGR3A (FCGR2A-131H and FCGR3A-158V alleles), with CA125<3xULN and received optimal farletuzumab. | HR=0.25, p=0.0202 |
| (43) | Renal cell carcinoma | High-dose aldesleukin (HD-IL2) | Tumor shrinkage | 106 | FCGR2A, FCGR3A, FCGR2C | Antigen-antibody - Immunoglobulin receptor on macrophages, neutrophils, Natural Killer cell | Higher affinity genotypes for FCGR2A, FCGR3A, FCGR2C together (i.e. favorable FCGR genotype group) were associated with increased tumor shrinkage. | p=0.03 |
| (46) | Melanoma | TAPcell vaccine | OS | 53 | TLR4 | Antimicrobial – Pathogen recognition and activation of innate immunity | Excluding patients with short post therapy response, patients bearing TLR4 896G allele had a significantly lower post therapy median survival than those with the normal allele (12 vs. 29 months). | p=0.026 |
| (35) | Melanoma | Anti-CTLA4 (ipilimumab, tremelimumab), anti-PD1 (nivolumab, pembrolizumab), or combined anti-CTLA4/anti-PD1 (ipilimumab/nivolumab) | Response to treatment | 436 | IL2, IL21 locus | Cytokine - Interleukin | rs17388568 (in IL2, IL21 locus) was associated with increased anti-PD1 response. | OR=0.26 (0.12–0.53), p=0.0002 |
| (58) | AML | Histamine dihydrochloride and low dose IL-2 immunotherapy | Leukemia free survival (LFS) and OS | 84 | HLA-B | Antigen processing and presentation | HLA-B-21M patients had improved LFS and OS compared to HLA-B-21T patients. | LFS (p=0.04; p=0.02); OS (p=0.007; p=0.003) |
| (59) | Melanoma and NSCLC | Anti–CTLA-4 or anti–PD-1 therapy | OS | 269 melanoma; 100 NSCLC | HLA-B44 | Antigen processing and presentation | Patients with B44 superfamily alleles had significantly better survival. | HR=0.61 (0.42–0.89), p=0.01 |
| (60) | Melanoma | Interferon | RFS | 286 | HLA genotypes | Antigen processing and presentation | HLA-Cw 06-positive patients had better RFS. | p=0.013 |
| (40) | Melanoma | Adoptive therapy (TILs) | Response to treatment | 140 | IRF5 genotype | Antimicrobial – virus mediated activation of interferon and modulation of cell growth | Lack of A allele in IRF5 genotype (rs10954213 G > A) was associated with non-response to TIL therapy. | p<0.005 |
| (49) | Melanoma | Adoptive therapy (TILs) | Response to treatment | 142 | CXCR3 and CCR5 genotypes | Cytokine – chemokine receptor | CXCR3 genotype and CCR5Δ32 deletion were associated with gene under expression and response to treatment. | OR=6.16 (complete response) and 2.32 (for overall response) |
| (61) | Melanoma | CTLA-4 blockade (ipilimumab or tremelimumab) | OS | 14 | Six CTLA4 polymorphisms (SNPs −1661A>G, −1577G>A, −658C>T, −319C>T, +49A>G, and CT60G>A) | T cell receptor signaling pathway | CTLA4−1577G/A and CT60G/A genotypes were significantly associated with improved overall survival. | p<0.006 |
Immune gene function: The immune system role of the protein encoded by the gene identified as being associated with cancer treatment outcome is indicated. The category designation was made as per Immport.org/shared/genelists and/or Genecards.org. Note that Cytokines can have various functions in mediating the immune system (such as inflammation, response to infection, etc.) through cell signaling; please refer to Genecards.org for a more comprehensive discussion of the gene functions.
Table 4.
Germline Immune Genetic Associations with Oncologic Surgery Outcomes
| Ref | Cancer | Treatment | Outcome | # subjects | Immune Genes | Immune gene function* | Main germline immune genetic associations found | Statistical significance of association |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| (70) | Colorectal | Surgery | RFS, OS | 668 | CCL2, PDCD1 | Cytokine – Chemokine/Antimicrobial (CCL2); T cell receptor signaling and antimicrobial (PDCD1) | CCL2 rs4586 TT genotype was strongly correlated with reduced OS. PDCD1 rs10204525 AA genotype was associated with shorter RFS and OS. |
CCL2: OS, HR=1.72 (1.14–2.61), p=0.010; PDCD1: OS, HR=1.48 (1.05–2.07), p=0.024; PDCD1: RFS, HR=1.66 (1.15–2.41), p=0.007 |
| (71) | Prostate | Surgery | Recurrence | 484 | IL10, CRP, IL1B, | Cytokine – Interleukin; Antimicrobial | SNPs in IL10, CRP and IL1B were associated with risk of recurrence independent of pathologic prognostic factors. | IL10 rs1800872 A allele: OR = 1.76 (1.00–3.10); IL10 rs1800896 G allele: OR=0.66 (0.48–0.91); CRP: OR= 0.65 (0.49–0.86); IL1B: OR=0.80 (0.60–1.06) |
| (72) | Lung NSCLC | Surgical resection | OS, RFS, metastasis | 385 | IL10 | Cytokine - Interleukin | Patients with IL10 non-ATA haplotypes had reduced OS and RFS. | OS: HR=1.43 (1.10–1.86), p=0.007; RFS: HR=1.56 (1.20–2.02), p<0.001 |
| (73) | Melanoma | Radiation, surgery, dacarbazine | Melanoma specific survival (MSS) | 241 | PDCD1 | T cell receptor signaling and antimicrobial | Poorer MSS was associated with the PD1.5 CC genotype. | HR=2.62 (1.16–5.92), p=0.02 |
| (74) | Lung NSCLC | Surgery | Survival time | 828 | CCL8 | Cytokine – Chemokine / antimicrobial | CCL8 rs3138035 was associated with improved survival time after surgery. | HR=0.47 (0.31–0.71) |
Immune gene function: The immune system role of the protein encoded by the gene identified as being associated with cancer treatment outcome is indicated. The category designation was made as per Immport.org/shared/genelists and/or Genecards.org. Note that Cytokines can have various functions in mediating the immune system (such as inflammation, response to infection, etc.) through cell signaling; please refer to Genecards.org for a more comprehensive discussion of the gene function.
Genes identified in these studies are involved in variety of roles in the immune system (see Tables 1–4). The majority of the genes could be identified as either encoding for cytokines (TNF, interleukins, chemokines) or involved with antigen processing and presentation (based on the broad categories from www.Immport.org/shared/genelists or www.genecards.org; see Tables 1–4). Germline variants in cytokine and cytokine receptor genes such as TNFA, IL2RA, IL17A, IL2, IL6, and IL10) may alter the immunomodulating functions of the encoded proteins, including regulating inflammation response and movement/expansion of immune cells, ultimately disrupting the balance of the immune system’s response to cancer treatments. For example, IL6 germline variants may increase serum levels of the encoded cytokine, which could increase inflammation and lead to poorer survival after HCT (34). As another example, the germline variant rs17388568 maps to the locus containing IL-2 and IL21 and is associated with increased anti-PD1 response (35). Variation at this locus may affect expression of these interleukin genes, leading to higher protein levels and increased stimulation of cytotoxic CD8+ T cells, thus improving the efficacy of anti-PD-1 treatment for melanoma. Germline variants in antigen processing and presentation genes (such as HLA, KIR, ICAM1, and ERAP2) may affect the efficient presentation of tumor specific antigens, which likely conduces to better outcomes in response to treatments such as immune checkpoint inhibitors (36). Inherited variants in KIR and HLA genes can alter the avidity of KIR-HLA interactions, which may then affect natural killer cell function and lead to better response to treatments, as was observed in studies of treatment of CML with tyrosine kinase inhibitors (37), metastatic colon cancer with FOLFIRI (38), and AML with HCT (39). Additionally, studies by Chat et al (35) and Uccellini et al (40) support a role for autoimmune susceptibility loci in modulating outcomes to cancer treatments (melanoma treated with anti-PD1 or tumor infiltrating lymphocytes), suggesting autoimmune risk loci could provide information on cancer treatment response.
Other genes identified in these studies function in immune processes such as antigen-antibody complex formation and elimination, antimicrobial activity or activation of innate immunity. Variants in genes such as FCGR2A, FCGR3A, and FCGR2C can affect how antigen-antibody complexes are formed and/or cleared, and were associated with enhanced response to trastuzumab and lapatinib for HER2+ breast cancer (41) and to immunotherapies for ovarian (42) and renal cell cancers (43) (farletuzumab and high-dose aldesleukin, respectively). Germline variants in genes involved with antimicrobial roles via activation of cell death pathways (e.g., FASL, TRAILR) may impair elimination of cancer cells in response to treatments, thereby affecting outcomes; for example, Lima et al (44) found variants in FASL and TRAILR were associated with increased risk of bladder cancer recurrence after treatment with BCG vaccine. Germline variants in genes involved with pathogen recognition and activation of innate immunity (e.g., TLR genes), were identified in association with worse outcomes in leukemia patients after HCT (45) and melanoma patients after TAPcell vaccine treatment (46). Variants in TLR genes may impact immune surveillance and inflammatory responses, leading to decreased response to cancer treatments that function by manipulating the host immune system, such as transplant and vaccines. The complement system is part of the innate immune system that is involved in opsonization and clearance of foreign materials, lysis of pathogens, and activation of inflammation. Charbonneau et al (47) found two germline variants of complement pathway genes that may augment host immune actions during treatment of follicular lymphoma patients with rituximab.
Several studies found similar associations among polymorphisms in genes in the same immunologic pathways, regardless of the type of cancer or treatment. For instance, variants in the toll-like receptors (TLRs) and associated genes were associated with poorer outcomes in melanoma (in response to TAPcell vaccine (46)), leukemia (in response to allogeneic HCT (45)), and multiple myeloma (in response to lenalidomide and bortezomib (48)) patients. The same CCR5 polymorphism was found to be associated with improved outcomes in both melanoma (in response to adoptive therapy (49)) and hematologic malignancies (in response to bone marrow transplant (50)) patients. Patients with weak or non-interacting KIR3DL1-HLAB alleles had better outcomes in response to allogeneic HCT for AML (39), to tyrosine kinase inhibitors for treatment of CML (37), and to anti-GD2 mAb for neuroblastoma (51). These results may suggest key immune system functions or players that are more likely to affect treatment outcomes.
The studies we included featured 28 different treatment regimens (i.e., drug types or combinations; see Tables 1–4). Treatments included those that directly modulate specific immune system components (such as anti-PD1 and anti-CTLA4) as well as broadly affect the immune system (such as hematopoietic stem cell and bone marrow transplants), non-immune system targeted therapies (such as tyrosine kinase inhibitors), chemotherapies (such as 5-fluorouraciol, leucovorin, irinotecan), and surgery. These findings imply value in studying germline immune gene variants in association with a variety of cancer treatments, and not just immune regulatory treatments.
Eighteen different outcome measures were used in the studies identified (see Tables 1–4). These included progression and survival metrics, complete response measures, as well as adverse effects such as fungal infection. It is difficult to compare effect sizes across the identified studies given the widely varying study designs and information reported within the manuscripts. However, several studies found significant differences in several outcome measures based on germline variation. Charbonneau et al (47) reported hazard ratios (HR) of 6.0 and 9.5 for two complement pathway germline variants (CFH rs376404 and CFHR5 rs6694672) in association with event free survival in 107 follicular lymphoma patients receiving rituximab. A study of melanoma and non-small cell lung cancer patients treated with anti-PD1 or anti-CTL4 found that the presence of certain HLAB44 superfamily alleles was associated with better survival (HR=0.61, p=0.01). Most of the studies were relatively small, with a median subject size of 243 (min = 14; max = 1328). The number of subjects in these studies is consistent with what was reported previously for an analysis of germline variants that associate with tumor progression and patient outcomes (8).
These studies all used a retrospective study design. The majority used a candidate gene approach in which germline variants in pre-selected immune genes were assessed and found that variation in 43 of these genes appeared to impact outcomes. Three others took pathway-based approaches that examined germline variants in several complement pathway genes (47), TLR genes (45), and HLA and KIR genes (37). However, three studies used more agnostic approaches. Pu et al (52) analyzed 11,930 inflammation-related SNPs and identified an HLA variant associated with decrease in median survival time and a KLRK1 rs2900420 variant that was associated with protective and prolonged overall survival in NSCLC patients following chemotherapy and radiation. Lim et al (33) used a TWAS approach and found ERAP2 expression levels were associated with improved response to atezolizumab (better OS) in luminal subtype bladder cancer patients. Lund et al (53) used a GWAS approach combined with a pathway analysis of variants in HLA-1 genes and found variants within that were associated with infections after induction therapy in childhood ALL patients.
Discussion
Although the studies we included are mainly of a preliminary or exploratory nature, they add to a growing body of evidence that germline genetic variation can be associated with differences in cancer outcomes (17). Collectively, these studies suggest that germline immune variants can affect and potentially provide information on how an individual will respond to cancer treatments. In general, the immune gene associations identified were not confined to a single immune system role for each cancer type and/or treatment. This suggests that study designs that examine immune pathways more comprehensively could yield useful insights for treatment strategies. This is supported by results from Shahamatdar et al (27) in which analyses at gene- and pathway-levels (rather than at the gene variant-level) may identify more biologically meaningful evidence of germline immune factors impacting outcomes. Although these associations need to be validated in other cohorts of patients and the impact of genetic variation on gene function examined further, they may present a starting point for unraveling the mechanisms that impact response to treatments and for identifying patients who could benefit most from a given treatment. Additionally, this broad review did not account for possible publication bias nor for study designs, sample size, or strength of associations. However, the results identified here can help inform the design of a focused, systematic review, or possibly even a meta-analysis focused on specific immune pathways, immune gene variants, treatments, outcomes and/or specific cancers.
The effects of immunogenetic variation on cancer outcomes may be specific to both cancer and treatment type. This is exemplified by the observation that three of the null results articles (Supplementary Table 1) failed to find an association between cancer outcomes and genes that were associated with treatment outcomes in other studies. For example, associations between CTLA4 and treatment were found for treatment of melanoma with ipilimumab (35) and lymphoma with HSCT (54), but not for interferon treatment of melanoma (30). Associations between treatment and HLA-G were found for treatment of colorectal cancer with primary surgery and then fluoropyrimidine or FL + oxaliplatin (55), but not for treatment of hematologic malignancies with bone marrow transplant; the authors speculate that because clinical factors (e.g., age, sex, time since diagnosis) have a significant effect on transplant outcomes, subtler genetic effects were not detectable in their study (31). They also demonstrate how germline genetic information will most likely be useful in the context of other patient characteristics, including the tumor microenvironment and the immune composition of tumors. Blank et al (56) have proposed a “cancer immunogram,” which is based on tumor characteristics and host immune status, that could help inform more personalized treatment; it is reasonable to expect that germline variation will affect aspects of host immune status.
Our review found several areas in which additional work is needed to fully realize the potential utility of germline variation information for cancer treatment. Most of the articles that we excluded lacked either information on germline variation or on treatment. To date, most clinical trials have not routinely collected germline genetic information; conversely, retrospective studies of outcomes may not have access to accurate and complete treatment data (17). Additionally, most studies of treatment mechanisms and efficacy have focused on tumor molecular characteristics.
Research analyzing adverse events in relation to germline immunogenetics is another area where further work is needed; most of the studies we identified focused on outcomes such as progression and survival. We found only two articles that focused on adverse events (infection in HCT recipients). Given that adverse events can affect the success of cancer treatment, work to determine whether variation in genes encoding immune system processes can distinguish among patients more or less likely to suffer an adverse event due to a particular treatment type could be beneficial for tailoring treatment. Research on ICI treatment has found that women are more likely to suffer adverse events than men, suggesting that host characteristics can influence the likelihood of adverse events (57).
The articles we found primarily used candidate gene approaches, in which known immune system genes were assessed for variation related to outcomes. Over the past decade, agnostic genome-wide approaches such as genome-wide association studies and whole exome and genome sequencing studies have led to new insights into disease etiology. These studies have identified novel roles for genes and pathways previously unknown to be involved in specific phenotypes. Although genome-wide approaches would be challenging in a treatment setting due to the need for discrete phenotypes and larger sample sizes, these approaches could identify novel genes and pathways that impact treatment outcomes. For example, in one of the more recent articles we found, Lim et al (33) conducted a TWAS to find common germline variants associated with gene expression and immune cell infiltration of the tumor. This led to the discovery that variation in the regulation of ERAP2, a protein involved in peptide presentation on MHC molecules, correlated with different treatment outcomes for bladder cancer patients receiving anti-PD1 therapy.
Another area where more work is needed is the collection of blood samples to provide germline DNA from patients, especially those participating in clinical trials. As exemplified by Lim et al (33), routine collection of samples from patients for future genomic interrogation can lead to new insights into how germline genetics could impact outcomes and help differentiate between patients likely to benefit from a treatment and those likely to see no benefit or even be harmed. The Society for Immunotherapy of Cancer (SITC) taskforce (19) suggested ways that this routine collection of samples could be made possible. Collaborative networks with harmonized collection of clinical and pathologic information are essential to establish large enough sample sizes for meaningful germline and somatic genetic studies. Ideally, oncology clinical trials could collect and analyze germline DNA to allow studies of the effects of germline variation on treatment outcomes. Additionally, epidemiology studies could collect detailed treatment, pathology, and outcomes data. Associations between germline variation and treatment outcomes could then be further explored to identify the underlying mechanisms through which variation impacts treatment. Work by population scientists and clinical epidemiologists could then determine the relevance of this information to the population and develop ways to apply this new knowledge in the clinic.
In addition, work to understand the functional effects of outcome-associated variants is greatly needed. The study by Lim et al (33) provides a strong example of how even variation in noncoding regions can be connected to biological mechanisms related to immune functions affecting outcome. Connecting coding region variation to drug effects may be slightly more straightforward, as demonstrated by work showing that certain FCGR3A polymorphisms associated with improved outcomes for ovarian cancer patients treated with farletuzumab had enhanced binding to the drug (42). Although many of the associations could plausibly have an effect on drug mechanisms, functional analyses such as those by Wang and Lim are needed to define the mechanisms linking genetic variation, treatment, and outcomes. Particularly in the case of GWAS, which often implicate non-coding variants, mechanistic work is needed.
Conclusion
Through this review, we identified the following areas in which further work is required in order to more fully realize the potential utility of germline immunogenetic variation for informing cancer outcomes: 1. Include treatment types and adverse events in epidemiologic studies; 2. Report null results; 3. Consider agnostic (genome-wide) approaches; 4. Implement routine collection of blood samples from patients for future genomic interrogation (especially those in clinical trials or cohorts); 4. Undertake mechanistic work to determine causality or functional effects of the variants; 5. Examine germline immune factors on the gene-level and pathway-level as well as at the genetic variant-level; and 6. Consider the micro-environmental conditions, tumor and patient specific characteristics, and progression stage of the disease.
Overall, these studies show that inherited genetic variation in a person’s immune system may affect responses to cancer treatments. By including the germline immune genetic contribution when assessing cancer, we can gain deeper insight into tumor biology and patient response. In combination with micro-environmental conditions, other cancer-associated genetic markers (such as DNA mismatch repair genes and KRAS), tumor characteristics (such as somatic mutations and tumor morphologic features), patient-specific characteristics, and stage of the disease, germline immune genetics may improve our ability to predict response to treatments and optimize treatment strategies. Including germline immune genetic information could also provide insight into the somatic mutation and immune cell landscape of tumors. Although validation by appropriately designed epidemiologic and clinical studies are needed, these studies support the need to consider information on germline immune genetic variants in cancer treatment optimization.
Supplementary Material
Table 2.
Germline Immune Genetic Associations with Transplant Outcomes
| Ref | Cancer | Treatment | Outcome | # subjects | Immune Genes | Immune gene function* | Main germline immune genetic associations found | Statistical significance of association |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| (62) | ALL, NHL, CLL, MM, or HL | Allogeneic HCT | OS | 186 | HLA-A, HLA-B, HLA-DRB1 | Antigen processing and presentation | Male patients with HLA-DRB1*04 had improved OS after allogenic-HSCT. | p=0.034, HR=0.35 (0.13–0.92) |
| (39) | AML | Allogeneic HCT | OS, relapse | 1328 | KIR3DL1, HLA-B | Antigen processing and presentation | Donors with weak or noninhibiting HLA-B/KIR3DL1 pairs had more favorable outcomes (lower relapse and overall mortality). | HR (relapse)=0.72 (0.58–0.90), p=0.004; HR (OS)=0.84 (0.72–0.98), p=0.03 |
| (63) | AML, ALL | Allogeneic HCT | DFS, relapse | 56 donor: patient pairs | HLA-E | Antigen processing and presentation | HLA-E*0103/0103 in recipient was associated with lower incidence of relapse and increased DFS period. | Relapse: p=0.02; DFS: p=0.001 |
| (64) | AML, CCL, CML, Lymphoma, MM | Bone marrow transplantation | OS | 652 | CXCL10 | Cytokine - Chemokine | CXCL10 rs3921 CG or GG genotype in transplant recipient was associated with improved OS. | p=0.02 |
| (50) | AML, ALL, MDS, ML, CML, MPN, myeloid malignancies, lymphoid malignancies | Bone marrow transplantation | DFS, OS | 329 | CCR5 | Cytokine – Chemokine Receptor | Recipients with CCR5 rs1800023-AA had better OS and DFS. | OS: p=0.028; DRS: p=0.015 |
| (34) | Acute leukemia, CML, lymphoma, other malignant haematologic disease | Allogeneic HCT | OS | 762 | IL-6, IL-10 | Cytokine - Interleukin | Donor IL-6 genotype was associated with reduced OS. Donor IL-10 genotype was protective for OS. | IL-6: p=0.007; IL-10: p=0.02 |
| (45) | AML, ALL, CML | Allogeneic HCT | OS, DFS | 816 | 29 SNPs in 10 Toll-like receptor genes | Antimicrobial – Pathogen recognition and activation of innate immunity | Minor allele of TLR8 rs3764879 in the donor was associated with reduced DFS and OS after allogeneic HCT. | OS: Male donor HR=1.41 (1.09–1.83), p=0.010; Female donor HR=2.78 (1.43–5.41), p=0.003. DFS: Male donor HR=1.45 (1.12–1.87), p=0.005; Female donor HR=2.34 (1.18–4.65), p=0.015 |
| (65) | AML | Allogeneic HCT | Relapse | 249 | IRF3 | Antimicrobial – innate immune response against DNA and RNA viruses | IRF3 rs2304205 AA in recipients and IRF3 rs7251 GG in donors were associated with a higher relapse incidence. | Both variants present: p=0.007 |
| (66) | AML, ALL, CML, MDS | Allogeneic HCT | OS | 186 | CASP8 | Caspase that activates inflammatory cytokines | Better OS was observed in patients receiving transplant from WT/WT donor compared with donors with a deletion. | RR=0.61 (0.38–0.98), p=0.04 |
| (54) | Acute leukemia, Chronic leukemia, lymphoma | Allogeneic HCT | OS, RFS | 164 | CTLA4 | T cell receptor signaling pathway | CTLA4 rs4553808 genotype in donors was associated with decreased RFS and OS. | RFS: HR=1.73 (1.10–2.71) p=0.017; OS: HR=1.84 (1.13–3.0) p=0.015 |
| (67) | AML, ALL, lymphoma, CLL | Autologous or allogeneic HSCT | Fungal infections | 198 | PTX3 | Inflammation and complement pathway | PTX3 polymorphisms were associated with the risk of invasive fungal infection. | rs2305619: OR=3.28 (1.24–8.69), p=0.02 rs1840680: OR=3.98 (1.52–10.4), p=0.0058 |
Immune gene function: The immune system role of the protein encoded by the gene identified as being associated with cancer treatment outcome is indicated. The category designation was made as per Immport.org/shared/genelists and/or Genecards.org. Note that Cytokines can have various functions in mediating the immune system (such as inflammation, response to infection, etc.) through cell signaling; please refer to Genecards.org for a more comprehensive discussion of the gene functions.
Table 3.
Germline Immune Genetic Associations with Targeted Therapy and Chemotherapy Outcomes
| Ref | Cancer | Treatment | Outcome | # subjects | Immune Genes | Immune gene function* | Main germline immune genetic associations found | Statistical significance of association |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| (41) | Breast | Preoperative chemotherapy plus trastuzumab (arm A), lapatinib (arm B) or both (arm C) | Response to treatment | 73 | FCGR3a | Antigen-antibody (Immunoglobulin receptor on macrophages, neutrophils) | FCGR3a V allele carriers had a significant improvement in pathological complete response rate with trastuzumab and lapatinib in HER2+ BC patients. | OR=9.4 (2.3–39.6), p=0.003 |
| (37) | CML | Tyrosine kinase inhibitors (imatinib, dasatinib, nilotinib, or bosutinib) | Complete molecular response (which was equivalent to undetectable BCR–ABL transcript expression) | 76 | KIR and HLA genes | Antigen processing and presentation | Weak interacting KIR3DL1*005 and HLA-B pairs were associated with improved molecular response to TKIs. | HR=14.22 (3.69–55.44), p<0.001 |
| (48) | Multiple myeloma | Lenalidomide and bortezomib | PFS, OS | 255 | TIRAP | Antimicrobial – Pathogen recognition and activation of innate immunity | TIRAP rs8177374 was associated with a decreased PFS and OS. | PFS: HR=1.74 (1.15–2.62), p=0.008; OS: HR=3.06 (1.74–5.4), p<0.001 |
| (47) | Follicular lymphoma; DLBCL | Rituximab | Event free survival | 107 FL; 82 DLBCL |
Several complement pathway genes | Complement pathway | CFH rs376404 and CFHR5 rs 6694672 variants were the most strongly associated with event free survival in follicular lymphoma patients receiving rituximab. |
CFH: HR=9.49 (2.59–505.6), p=0.0007 CFHR5: HR=6.00 (1.59–22.67), p=0.0083 |
| (51) | Neuroblastoma | Anti-GD2 mAb 3F8 | PFS, OS | 245 | KIR3DL1, HLA-B | Antigen processing and presentation | Noninteracting combinations had the most favorable PFS and OS outcomes. | HR (PFS)=0.43 (0.28–0.66), p<0.001; HR (OS)= 0.41 (0.25–0.65), p<0.001 |
| (38) | Colorectal | 5-fluorouracil, leucovorin, and irinotecan | CR (complete response) | 224 | KIR3DL1, HLA-Bw-I80 | Antigen processing and presentation | Presence of KIR3DL1/HLA-Bw4-I80 was associated with improved CR. | 1 Bw4-I80 allele: HR=2.7, p<0.001; 2 Bw4-I80 alleles: HR=1.8, p<0.006 |
| (55) | Colorectal | Primary surgery and then ADJ-CT based on fluoropyrimidine (FL) (i.e., 5-fluorouracil/folinic acid or capecitabine), or FL plus oxaliplatin (FL+OXA) | DFS, OS | 253 | HLA-G | Antigen processing and presentation | Presence of HLA-G +3035 C>T and +2960 14-bp INDEL were associated with improved DFS. Presence of HLA-G +3187 A>G was associated with worse DFS and OS. |
+2960: HR=0.60 (0.38–0.93), p-0.023; +3035: HR=0.51 (0.26–0.99), p=0.045; +3187: DFS, HR=2.46 (1.19–5.05), p-0.015; OS, HR=2.71 (1.16–6.63), p=0.022 |
| (52) | Lung NSCLC | First line Chemotherapy +/− radiotherapy | OS | 502 discovery; 355 internal validation |
11930 SNPs related to immune system | Antigen processing and presentation | HLA-DOB rs2071554 patients had decrease in median survival time. Variant was predicted to alter function. KLRK1 rs2900420 was protective and prolonged overall survival. |
HLA-DOB: HR=1.46 (1.02–2.09). KLRK1: HR=0.77 (0.61–0.99) |
| (68) | DLBCL | R-CHOP or R-CHOP-like treatment | OS, CR | 129 | C1qA | Complement pathway | C1qA 276 AA was associated with improved CR and OS. |
CR: p=0.0001
OR: p=0.023 |
| (69) | CLL | R-FC, R-CVP, chlorambucil, alemtuzumab | Response to treatment | 144 | CXCL12 | Cytokine -Chemokine | CXCL12 rs1801157 A allele was associated with poorer response to treatment (independent of the treatment type). | p<0.001 |
| (53) | Childhood ALL | Induction therapy (doxorubicin, vincristine, etc.) | Infections | 69 | GWAS - 34,000 SNPs in 2350 genes related to pharmacogenetics, immunogenetics, apoptosis, organ-specific toxicities, cell cycle control, and DNA repair and mitosis | Antigen processing and presentation | In pathway analyses, variants in Class I MHC-mediated antigen processing and presentation genes were predictive of infectious events. | AUC=0.83 |
Immune gene function: The immune system role of the protein encoded by the gene identified as being associated with cancer treatment outcome is indicated. The category designation was made as per Immport.org/shared/genelists and/or Genecards.org. Note that Cytokines can have various functions in mediating the immune system (such as inflammation, response to infection, etc.) through cell signaling; please refer to Genecards.org for a more comprehensive discussion of the gene functions.
Acknowledgements
The authors thank Drs. Mercy Prabhudas and Laura Amo Herrero from NIAID for critically reviewing the manuscript from the immunology perspective and Clare Rauch for her consultation on the literature review.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Carter H, Marty R, Hofree M, Gross AM, Jensen J, Fisch KM, et al. Interaction Landscape of Inherited Polymorphisms with Somatic Events in Cancer. Cancer Discov 2017;7(4):410–23 doi 10.1158/2159-8290.CD-16-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium ITP-CAoWG. Pan-cancer analysis of whole genomes. Nature 2020;578(7793):82–93 doi 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koessler T, Azzato EM, Perkins B, Macinnis RJ, Greenberg D, Easton DF, et al. Common germline variation in mismatch repair genes and survival after a diagnosis of colorectal cancer. Int J Cancer 2009;124(8):1887–91 doi 10.1002/ijc.24120. [DOI] [PubMed] [Google Scholar]
- 4.Shu X, Gu J, Huang M, Tannir NM, Matin SF, Karam JA, et al. Germline genetic variants in somatically significantly mutated genes in tumors are associated with renal cell carcinoma risk and outcome. Carcinogenesis 2018;39(6):752–7 doi 10.1093/carcin/bgy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stotz M, Herzog SA, Pichler M, Smolle M, Riedl J, Rossmann C, et al. Cancer Stem Cell Gene Variants in CD44 Predict Outcome in Stage II and Stage III Colon Cancer Patients. Anticancer Res 2017;37(4):2011–8 doi 10.21873/anticanres.11545. [DOI] [PubMed] [Google Scholar]
- 6.Summers MG, Maughan TS, Kaplan R, Law PJ, Houlston RS, Escott-Price V, et al. Comprehensive analysis of colorectal cancer-risk loci and survival outcome: A prognostic role for CDH1 variants. Eur J Cancer 2020;124:56–63 doi 10.1016/j.ejca.2019.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Świerniak M, Wójcicka A, Czetwertyńska M, Długosińska J, Stachlewska E, Gierlikowski W, et al. Association between GWAS-Derived rs966423 Genetic Variant and Overall Mortality in Patients with Differentiated Thyroid Cancer. Clin Cancer Res 2016;22(5):1111–9 doi 10.1158/1078-0432.Ccr-15-1746. [DOI] [PubMed] [Google Scholar]
- 8.Chatrath A, Ratan A, Dutta A. Germline Variants That Affect Tumor Progression. Trends Genet 2021;37(5):433–43 doi 10.1016/j.tig.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly AK. Pharmacogenetics: a general review on progress to date. Br Med Bull 2017;124(1):65–79 doi 10.1093/bmb/ldx035. [DOI] [PubMed] [Google Scholar]
- 10.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature 2015;526(7573):343–50 doi 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero Lagunes ML, Vera Badillo FE. Design and Implementing Pharmacogenomics Study in Cancer. Adv Exp Med Biol 2019;1168:43–77 doi 10.1007/978-3-030-24100-1_4. [DOI] [PubMed] [Google Scholar]
- 12.Weinshilboum RM, Wang L. Pharmacogenomics: Precision Medicine and Drug Response. Mayo Clin Proc 2017;92(11):1711–22 doi 10.1016/j.mayocp.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357(6349):409–13 doi 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372(26):2509–20 doi 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macauda A, Castelli E, Buda G, Pelosini M, Butrym A, Watek M, et al. Inherited variation in the xenobiotic transporter pathway and survival of multiple myeloma patients. Br J Haematol 2018;183(3):375–84 doi 10.1111/bjh.15521. [DOI] [PubMed] [Google Scholar]
- 16.Mosquera Orgueira A, Antelo Rodríguez B, Alonso Vence N, Díaz Arias J, Díaz Varela N, Pérez Encinas MM, et al. The association of germline variants with chronic lymphocytic leukemia outcome suggests the implication of novel genes and pathways in clinical evolution. BMC Cancer 2019;19(1):515 doi 10.1186/s12885-019-5628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatrath A, Przanowska R, Kiran S, Su Z, Saha S, Wilson B, et al. The pan-cancer landscape of prognostic germline variants in 10,582 patients. Genome Med 2020;12(1):15 doi 10.1186/s13073-020-0718-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatrath A, Kiran M, Kumar P, Ratan A, Dutta A. The Germline Variants rs61757955 and rs34988193 Are Predictive of Survival in Lower Grade Glioma Patients. Mol Cancer Res 2019;17(5):1075–86 doi 10.1158/1541-7786.Mcr-18-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedognetti D, Ceccarelli M, Galluzzi L, Lu R, Palucka K, Samayoa J, et al. Toward a comprehensive view of cancer immune responsiveness: a synopsis from the SITC workshop. J Immunother Cancer 2019;7(1):131 doi 10.1186/s40425-019-0602-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedognetti D, Tatari-Calderone Z, Marincola FM, Wang E. Intratumoral gene signatures and host genetic variations associated with immune responsiveness. In: Butterfield L, Kaufman L, Marincola FM, editors. SITC Cancer Immunotherapy Principle and Practice. 1st ed: Demos Medical; 2017. p 449–74. [Google Scholar]
- 21.Ferreiro-Iglesias A, Lesseur C, McKay J, Hung RJ, Han Y, Zong X, et al. Fine mapping of MHC region in lung cancer highlights independent susceptibility loci by ethnicity. Nat Commun 2018;9(1):3927 doi 10.1038/s41467-018-05890-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giat E, Ehrenfeld M, Shoenfeld Y. Cancer and autoimmune diseases. Autoimmun Rev 2017;16(10):1049–57 doi 10.1016/j.autrev.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Sayaman RW, Saad M, Thorsson V, Hendrickx W, Roelands J, Mokrab Y, et al. Germline genetic contribution to the immune landscape of cancer. Immunity 2021;54(2):367–86 e8 doi 10.1016/j.immuni.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marty Pyke R, Thompson WK, Salem RM, Font-Burgada J, Zanetti M, Carter H. Evolutionary Pressure against MHC Class II Binding Cancer Mutations. Cell 2018;175(2):416–28.e13 doi 10.1016/j.cell.2018.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marty R, Kaabinejadian S, Rossell D, Slifker MJ, van de Haar J, Engin HB, et al. MHC-I Genotype Restricts the Oncogenic Mutational Landscape. Cell 2017;171(6):1272–83.e15 doi 10.1016/j.cell.2017.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat Rev Genet 2016;17(3):160–74 doi 10.1038/nrg.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shahamatdar S, He MX, Reyna MA, Gusev A, AlDubayan SH, Van Allen EM, et al. Germline Features Associated with Immune Infiltration in Solid Tumors. Cell Rep 2020;30(9):2900–8.e4 doi 10.1016/j.celrep.2020.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hargadon KM, Johnson CE, Williams CJ. Immune checkpoint blockade therapy for cancer: An overview of FDA-approved immune checkpoint inhibitors. Int Immunopharmacol 2018;62:29–39 doi 10.1016/j.intimp.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Oiseth SJ, Aziz MS. 2017. Cancer immunotherapy: a brief review of the history, possibilities, and challenges ahead. In J Cancer Metastasis Treat. <https://jcmtjournal.com/article/view/2275>. Accessed 2020. [Google Scholar]
- 30.Gogas H, Dafni U, Koon H, Spyropoulou-Vlachou M, Metaxas Y, Buchbinder E, et al. Evaluation of six CTLA-4 polymorphisms in high-risk melanoma patients receiving adjuvant interferon therapy in the He13A/98 multicenter trial. J Transl Med 2010;8:108 doi 10.1186/1479-5876-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterhouse M, Duque-Afonso J, Wäsch R, Bertz H, Finke J. Soluble HLA-G molecules and HLA-G 14-base pair polymorphism after allogeneic hematopoietic cell transplantation. Transplant Proc 2013;45(1):397–401 doi 10.1016/j.transproceed.2012.05.073. [DOI] [PubMed] [Google Scholar]
- 32.Pearce KF, Lee SJ, Haagenson M, Petersdorf EW, Norden J, Collin MP, et al. Analysis of non-HLA genomic risk factors in HLA-matched unrelated donor hematopoietic cell transplantation for chronic myeloid leukemia. Haematologica 2012;97(7):1014–9 doi 10.3324/haematol.2011.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim YW, Chen-Harris H, Mayba O, Lianoglou S, Wuster A, Bhangale T, et al. Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc Natl Acad Sci U S A 2018;115(50):E11701-e10 doi 10.1073/pnas.1804506115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balavarca Y, Pearce K, Norden J, Collin M, Jackson G, Holler E, et al. Predicting survival using clinical risk scores and non-HLA immunogenetics. Bone Marrow Transplant 2015;50(11):1445–52 doi 10.1038/bmt.2015.173. [DOI] [PubMed] [Google Scholar]
- 35.Chat V, Ferguson R, Simpson D, Kazlow E, Lax R, Moran U, et al. Autoimmune genetic risk variants as germline biomarkers of response to melanoma immune-checkpoint inhibition. Cancer Immunol Immunother 2019;68(6):897–905 doi 10.1007/s00262-019-02318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mpakali A, Stratikos E. The Role of Antigen Processing and Presentation in Cancer and the Efficacy of Immune Checkpoint Inhibitor Immunotherapy. Cancers (Basel) 2021;13(1) doi 10.3390/cancers13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ureshino H, Shindo T, Kojima H, Kusunoki Y, Miyazaki Y, Tanaka H, et al. Allelic Polymorphisms of KIRs and HLAs Predict Favorable Responses to Tyrosine Kinase Inhibitors in CML. Cancer Immunol Res 2018;6(6):745–54 doi 10.1158/2326-6066.Cir-17-0462. [DOI] [PubMed] [Google Scholar]
- 38.De Re V, Caggiari L, De Zorzi M, Talamini R, Racanelli V, M DA, et al. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS One 2014;9(1):e84940 doi 10.1371/journal.pone.0084940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boudreau JE, Giglio F, Gooley TA, Stevenson PA, Le Luduec JB, Shaffer BC, et al. KIR3DL1/HLA-B Subtypes Govern Acute Myelogenous Leukemia Relapse After Hematopoietic Cell Transplantation. J Clin Oncol 2017;35(20):2268–78 doi 10.1200/jco.2016.70.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uccellini L, De Giorgi V, Zhao Y, Tumaini B, Erdenebileg N, Dudley ME, et al. IRF5 gene polymorphisms in melanoma. J Transl Med 2012;10:170 doi 10.1186/1479-5876-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musolino A, Naldi N, Dieci MV, Zanoni D, Rimanti A, Boggiani D, et al. Immunoglobulin G fragment C receptor polymorphisms and efficacy of preoperative chemotherapy plus trastuzumab and lapatinib in HER2-positive breast cancer. Pharmacogenomics J 2016;16(5):472–7 doi 10.1038/tpj.2016.51. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Somers EB, Ross EN, Kline JB, O’Shannessy DJ, Schweizer C, et al. FCGR2A and FCGR3A Genotypes Correlate with Farletuzumab Response in Patients with First-Relapsed Ovarian Cancer Exhibiting Low CA125. Cytogenet Genome Res 2017;152(4):169–79 doi 10.1159/000481213. [DOI] [PubMed] [Google Scholar]
- 43.Erbe AK, Wang W, Goldberg J, Gallenberger M, Kim K, Carmichael L, et al. FCGR Polymorphisms Influence Response to IL2 in Metastatic Renal Cell Carcinoma. Clin Cancer Res 2017;23(9):2159–68 doi 10.1158/1078-0432.Ccr-16-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lima L, Oliveira D, Ferreira JA, Tavares A, Cruz R, Medeiros R, et al. The role of functional polymorphisms in immune response genes as biomarkers of bacille Calmette-Guerin (BCG) immunotherapy outcome in bladder cancer: establishment of a predictive profile in a Southern Europe population. BJU Int 2015;116(5):753–63 doi 10.1111/bju.12844. [DOI] [PubMed] [Google Scholar]
- 45.Kornblit B, Enevold C, Wang T, Spellman S, Haagenson M, Lee SJ, et al. Toll-like receptor polymorphisms in allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant 2015;21(2):259–65 doi 10.1016/j.bbmt.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tittarelli A, González FE, Pereda C, Mora G, Muñoz L, Saffie C, et al. Toll-like receptor 4 gene polymorphism influences dendritic cell in vitro function and clinical outcomes in vaccinated melanoma patients. Cancer Immunol Immunother 2012;61(11):2067–77 doi 10.1007/s00262-012-1268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charbonneau B, Maurer MJ, Fredericksen ZS, Zent CS, Link BK, Novak AJ, et al. Germline variation in complement genes and event-free survival in follicular and diffuse large B-cell lymphoma. Am J Hematol 2012;87(9):880–5 doi 10.1002/ajh.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bagratuni T, Terpos E, Eleutherakis-Papaiakovou E, Kalapanida D, Gavriatopoulou M, Migkou M, et al. TLR4/TIRAP polymorphisms are associated with progression and survival of patients with symptomatic myeloma. Br J Haematol 2016;172(1):44–7 doi 10.1111/bjh.13786. [DOI] [PubMed] [Google Scholar]
- 49.Bedognetti D, Spivey TL, Zhao Y, Uccellini L, Tomei S, Dudley ME, et al. CXCR3/CCR5 pathways in metastatic melanoma patients treated with adoptive therapy and interleukin-2. Br J Cancer 2013;109(9):2412–23 doi 10.1038/bjc.2013.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horio T, Mizuno S, Uchino K, Mizutani M, Hanamura I, Espinoza JL, et al. The recipient CCR5 variation predicts survival outcomes after bone marrow transplantation. Transpl Immunol 2017;42:34–9 doi 10.1016/j.trim.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Forlenza CJ, Boudreau JE, Zheng J, Le Luduec JB, Chamberlain E, Heller G, et al. KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma. J Clin Oncol 2016;34(21):2443–51 doi 10.1200/JCO.2015.64.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pu X, Hildebrandt MA, Lu C, Roth JA, Stewart DJ, Zhao Y, et al. Inflammation-related genetic variations and survival in patients with advanced non-small cell lung cancer receiving first-line chemotherapy. Clin Pharmacol Ther 2014;96(3):360–9 doi 10.1038/clpt.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lund B, Wesolowska-Andersen A, Lausen B, Borst L, Rasmussen KK, Müller K, et al. Host genome variations and risk of infections during induction treatment for childhood acute lymphoblastic leukaemia. Eur J Haematol 2014;92(4):321–30 doi 10.1111/ejh.12243. [DOI] [PubMed] [Google Scholar]
- 54.Jagasia M, Clark WB, Brown-Gentry KD, Crawford DC, Fan KH, Chen H, et al. Genetic variation in donor CTLA-4 regulatory region is a strong predictor of outcome after allogeneic hematopoietic cell transplantation for hematologic malignancies. Biol Blood Marrow Transplant 2012;18(7):1069–75 doi 10.1016/j.bbmt.2011.12.518. [DOI] [PubMed] [Google Scholar]
- 55.Garziera M, Bidoli E, Cecchin E, Mini E, Nobili S, Lonardi S, et al. HLA-G 3’UTR Polymorphisms Impact the Prognosis of Stage II-III CRC Patients in Fluoropyrimidine-Based Treatment. PLoS One 2015;10(12):e0144000 doi 10.1371/journal.pone.0144000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science 2016;352(6286):658–60 doi 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 57.Jia XH, Geng LY, Jiang PP, Xu H, Nan KJ, Yao Y, et al. The biomarkers related to immune related adverse events caused by immune checkpoint inhibitors. J Exp Clin Cancer Res 2020;39(1):284 doi 10.1186/s13046-020-01749-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hallner A, Bernson E, Hussein BA, Ewald Sander F, Brune M, Aurelius J, et al. The HLA-B −21 dimorphism impacts on NK cell education and clinical outcome of immunotherapy in acute myeloid leukemia. Blood 2019;133(13):1479–88 doi 10.1182/blood-2018-09-874990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359(6375):582–7 doi 10.1126/science.aao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gogas H, Kirkwood JM, Falk CS, Dafni U, Sondak VK, Tsoutsos D, et al. Correlation of molecular human leukocyte antigen typing and outcome in high-risk melanoma patients receiving adjuvant interferon. Cancer 2010;116(18):4326–33 doi 10.1002/cncr.25211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Queirolo P, Morabito A, Laurent S, Lastraioli S, Piccioli P, Ascierto PA, et al. Association of CTLA-4 polymorphisms with improved overall survival in melanoma patients treated with CTLA-4 blockade: a pilot study. Cancer Invest 2013;31(5):336–45 doi 10.3109/07357907.2013.793699. [DOI] [PubMed] [Google Scholar]
- 62.Balassa K, Andrikovics H, Remenyi P, Batai A, Szilvasi A, Bors A, et al. Sex-specific survival difference in association with HLA-DRB1∗04 following allogeneic haematopoietic stem cell transplantation for lymphoid malignancies. Hum Immunol 2018;79(1):13–9 doi 10.1016/j.humimm.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Hosseini E, Schwarer AP, Jalali A, Ghasemzadeh M. The impact of HLA-E polymorphisms on relapse following allogeneic hematopoietic stem cell transplantation. Leuk Res 2013;37(5):516–9 doi 10.1016/j.leukres.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 64.Nakata K, Takami A, Espinoza JL, Matsuo K, Morishima Y, Onizuka M, et al. The recipient CXCL10 + 1642C>G variation predicts survival outcomes after HLA fully matched unrelated bone marrow transplantation. Clin Immunol 2013;146(2):104–11 doi 10.1016/j.clim.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 65.Martín-Antonio B, Suarez-Lledo M, Arroyes M, Fernández-Avilés F, Martínez C, Rovira M, et al. A variant in IRF3 impacts on the clinical outcome of AML patients submitted to Allo-SCT. Bone Marrow Transplant 2013;48(9):1205–11 doi 10.1038/bmt.2013.43. [DOI] [PubMed] [Google Scholar]
- 66.Shaw BE, Lee F, Krishnamurthy S, Byrne JL, Seedhouse C, Mayor NP, et al. Caspase-8 polymorphisms result in reduced Alemtuzumab-induced T-cell apoptosis and worse survival after transplantation. Bone Marrow Transplant 2015;50(2):237–43 doi 10.1038/bmt.2014.238. [DOI] [PubMed] [Google Scholar]
- 67.Herrero-Sánchez MC, Angomás EB, de Ramón C, Tellería JJ, Corchete LA, Alonso S, et al. Polymorphisms in Receptors Involved in Opsonic and Nonopsonic Phagocytosis, and Correlation with Risk of Infection in Oncohematology Patients. Infect Immun 2018;86(12) doi 10.1128/iai.00709-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin X, Ding H, Ding N, Fu Z, Song Y, Zhu J. Homozygous A polymorphism of the complement C1qA276 correlates with prolonged overall survival in patients with diffuse large B cell lymphoma treated with R-CHOP. J Hematol Oncol 2012;5:51 doi 10.1186/1756-8722-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butrym A, Gebura K, Iwaszko M, Kuliczkowski K, Bogunia-Kubik K, Mazur G. Dual role of the CXCL12 polymorphism in patients with chronic lymphocytic leukemia. Hla 2016;87(6):432–8 doi 10.1111/tan.12810. [DOI] [PubMed] [Google Scholar]
- 70.Yoon S, Kang BW, Park SY, Kim HJ, Park JS, Choi GS, et al. Prognostic relevance of genetic variants involved in immune checkpoints in patients with colorectal cancer. J Cancer Res Clin Oncol 2016;142(8):1775–80 doi 10.1007/s00432-016-2196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dluzniewski PJ, Wang MH, Zheng SL, De Marzo AM, Drake CG, Fedor HL, et al. Variation in IL10 and other genes involved in the immune response and in oxidation and prostate cancer recurrence. Cancer Epidemiol Biomarkers Prev 2012;21(10):1774–82 doi 10.1158/1055-9965.Epi-12-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang YC, Sung WW, Wu TC, Wang L, Chien WP, Cheng YW, et al. Interleukin-10 haplotype may predict survival and relapse in resected non-small cell lung cancer. PLoS One 2012;7(7):e39525 doi 10.1371/journal.pone.0039525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomez GVB, Rinck-Junior JA, Oliveira C, Silva DHL, Mamoni RL, Lourenço GJ, et al. PDCD1 gene polymorphisms as regulators of T-lymphocyte activity in cutaneous melanoma risk and prognosis. Pigment Cell Melanoma Res 2018;31(2):308–17 doi 10.1111/pcmr.12665. [DOI] [PubMed] [Google Scholar]
- 74.Ma H, Shu Y, Pan S, Chen J, Dai J, Jin G, et al. Polymorphisms of key chemokine genes and survival of non-small cell lung cancer in Chinese. Lung Cancer 2011;74(2):164–9 doi 10.1016/j.lungcan.2011.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.