Abstract
“Viable but non-culturable” (VBNC) states of bacteria pose challenges for environmental and clinical microbiology, but their biological mechanisms remain obscure. Mycobacterium tuberculosis (Mtb), the leading cause of death from infection until the coronavirus disease 2019 pandemic, affords a striking example of this phenotype. Mtb can enter into a “differentially detectable” (DD) state associated with phenotypic antimicrobial resistance. In this state, Mtb cells are viable but undetectable as colony-forming units. We found that Mtb cells enter the DD state when they undergo sublethal oxidative stress that damages their DNA, proteins, and lipids. Additionally, their replication process is delayed, allowing time for repair. Mycobacterium bovis and its derivative, BCG, fail to enter the DD state under similar conditions. These findings have implications for tuberculosis latency, detection, relapse, treatment monitoring, and development of regimens that overcome phenotypic antimicrobial resistance.
One Sentence Summary:
Mycobacterium tuberculosis that sustains moderate oxidative damage survives undetected on solid media, but growth delay restores replicative ability.
Introduction
In 2019, active tuberculosis (TB) sickened an estimated 10 million people and killed an estimated 1.4 million (1). Treatment usually requires at least 6 months of multi-drug chemotherapy to prevent relapse in a majority of those treated, even with regimens that effectively convert sputum cultures to negative at 2 months (2, 3). Better understanding of the survival strategies of Mycobacterium tuberculosis (Mtb), the causative agent of the vast majority of TB cases, could shorten treatment through improved targeting of slowly sterilized subpopulations.
Mtb can enter a state in which it is unable to grow on standard nutrient-rich solid media, and is therefore considered absent or dead, but is shown to be viable by other methods (4–6). For example, in the “Cornell model”, Mtb-infected mice were treated with TB drugs such that no Mtb could be cultured from any part of the carcass after treatment stopped, yet the disease reappeared later in some other members of the cohort and in almost all of the members of the cohort if they were immunosuppressed (4, 7–9). Limiting dilution assays have also revealed the existence of viable Mtb cells that do not form colonies on agar; that is, they do not represent colony-forming units (CFU). Enumeration of CFU has remained the gold standard assay for in vitro quantification of bacterial viability since Koch introduced the technique in the 1880’s, but this is undercut by these states of Mtb, interchangeably called “viable but non-culturable”, “differentially culturable” or “differentially detectable” (DD). DD Mtb have been found to outnumber CFU from three-fold up to multiple orders of magnitude in 21% to 86% of TB patients’ pre-treatment sputa, as well as in material from TB patients’ lymph nodes, pleural fluid, colon, and bone (5, 10–13). Moreover, previous studies have shown that the proportion of DD Mtb in the sputum rose within two weeks of antibiotic treatment as the total number of viable Mtb in the sputum declined (13). When generated in vitro, DD Mtb show marked phenotypic resistance to TB drugs (14, 15). Phenotypic resistance to antibiotics favors the emergence of genetic resistance (16, 17). Formation of DD Mtb might contribute to the requirement for prolonged therapy of TB, the variability in treatment outcomes, the emergence of genetic antimicrobial resistance, and the phenomenon of latent TB.
We recently developed an independently validated in vitro model of DD Mtb generation by starving Mtb of nutrients in phosphate-buffered saline (PBS) followed by exposure to high concentrations of rifampin (RIF), hereafter termed the PBS-RIF model. Worldwide, RIF is a backbone of the treatment of drug-susceptible and latent TB. In the PBS-RIF model, the starved cells, which are nonreplicating, are phenotypically resistant to conventional therapeutic doses of RIF. As judged by the reduction in CFU, RIF is required at concentrations 2 to 3 orders of magnitude above the conventional minimum inhibitory concentration (MIC) to kill 90 to 99% of the Mtb. In contrast, a liquid limiting dilution assay designed to avoid artefacts reveals a 1 to 2 log10 greater number of viable cells, based on the most probable number (MPN) calculation (14). In effect, RIF actually kills only some and often none of the starved cells, even at these high concentrations—a biologically striking and clinically worrisome example of phenotypic resistance (17).
Not accounting for DD Mtb may complicate the testing of TB drug regimens. It may confound a physician’s ability to determine when an individual patient can stop taking multiple potentially toxic drugs, something that is not reliably revealed by the negativity of sputum cultures in standard assays. However, the phenotypic limiting dilution/MPN assay is too cumbersome for routine clinical application. Therefore, a mechanistic understanding of DD Mtb is needed. This could provide insights that facilitate development of drugs that can kill DD Mtb, lay the groundwork for an improved clinical assay for DD Mtb, and reveal biology underlying TB transmission and latent TB infection.
Here we applied genetic and biochemical approaches to define the critical processes underlying DD Mtb formation. We found that DD Mtb arise when two conditions are met. First, Mtb cells experience an intermediate degree of oxidative damage such that they alter their growth phenotype. Second, delay of replication allows for recovery of replicative capacity. Limiting dilution allows for this delay and recovery, but is not the only such condition. Intriguingly, both Mycobacterium bovis BCG (BCG), the live strain used for TB vaccination worldwide, and M. bovis, the strain from which BCG was derived, experience greater oxidative stress than Mtb under the conditions tested and do not enter the DD state, apparently dying instead.
Results
DD Mtb undergo oxidative stress and partial loss of the oxidative stress response augments the DD phenotype.
Decades of research on other bacteria have implicated oxidative stress and damage in the formation of non-cultivable states (18, 19). Oxidative stress or oxidative damage (20) have been associated with both of the conditions in the PBS-RIF model (nutrient starvation and antibiotic exposure) that collectively lead to cultures composed of at least 90% DD Mtb as compared to vehicle control (PBS-dimethyl sulfoxide [DMSO]) (Fig. 1A). Recently, Hong et al. demonstrated that E. coli’s generation of reactive oxygen species (ROS) in response to antibiotics could prevent the cells from growing as CFU without killing them and ROS accumulation could continue after the antibiotics were removed (21). Thus, we hypothesized that increased oxidative stress and subsequent damage may occur during formation of DD Mtb.
Fig. 1. DD Mtb undergo oxidative stress.
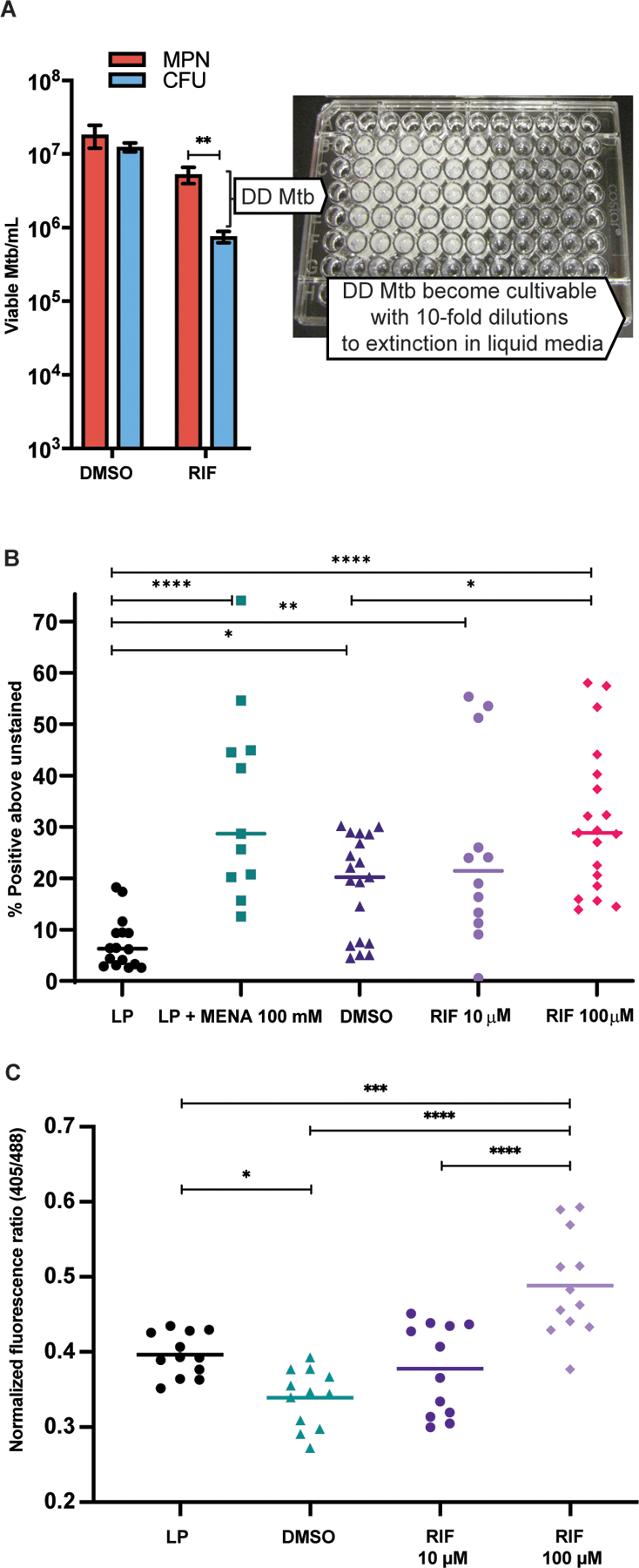
(A) Representative results of DD Mtb estimation in the PBS-RIF in vitro model are shown with photo of a representative liquid limiting dilution assay used for most probable number (MPN) calculations. Mtb cells were starved in PBS for at least 2 weeks, then exposed to either high dose RIF (10 to 100 µM) or DMSO (vehicle control). After washing out the RIF or DMSO, the culture undergoes 10-fold serial dilutions across 5 technical replicates, and the number of positive wells is inputted into an MPN calculator to estimate the original, undiluted concentration of viable cells. Data were analyzed using a mixed effects analysis with Sidak’s multiple comparisons test. (B) Flow cytometry analysis is shown of log phase (LP) Mtb, LP Mtb exposed to 100 mM menadione (LP + MENA), starved then DMSO-exposed Mtb (DMSO; vehicle control), and starved then RIF-exposed Mtb (RIF), after staining with CellROX Green. Data shown are representative of 7 experiments with 1 to 3 biological replicates each, except RIF 10 µM data which are from 4 experiments with 3 biological replicates each. Data were analyzed using a mixed effects analysis with Tukey’s multiple comparisons test. Horizontal bars indicate median. (C) Flow cytometry analysis of Mrx1-roGFP2 plasmid containing H37Rv (6 experiments, 12 biological replicates total) in LP, PBS starved then DMSO or RIF exposed Mtb. Statistics were calculated by one-way ANOVA with Tukey’s multiple comparisons test. Horizontal bars indicate mean. *adj p < 0.05, **adj p < 0.01, ***adj p < 0.001, ****adj p < 0.0001.
We first assessed this using CellROX green, a proprietary dye that fluoresces upon reaction with superoxide and subsequent binding to DNA. Based on CellROX fluorescence, McBee et al. reported that BCG produced superoxide during nutrient starvation or RIF treatment and did so to the highest degree after the combination of starvation and exposure to RIF (22). Similarly, we found that CellROX fluorescence increased significantly in nutrient starved Mtb and in Mtb in the PBS-RIF model as compared to logarithmically growing (log phase, LP) bacilli (P=0.01 for PBS-DMSO, P<0.0001 for PBS-RIF 100 µM, Fig. 1B), although baseline signal strength varied among experiments performed on separate days. After RIF was washed out and the cells were incubated a further week in PBS without RIF, the CellROX signal returned to the value seen in starved, non-RIF treated cells (fig. S1). The data are consistent with increased intrabacterial superoxide formation in both the PBS-DMSO and PBS-RIF models.
We then used an orthogonal assay to verify redox changes. The mycoredoxin-1-fused redox-sensitive green fluorescent protein (Mrx1-roGFP2) plasmid is a ratiometric fluorescence biosensor of the antioxidant glycopeptide mycothiol that can report the redox status of Mtb (23, 24). The biosensor indicated that there was only a slight increase in the reductive state in starved Mtb (PBS-DMSO) as compared to LP cells (Fig. 1C). However, in a dose dependent manner, subsequent RIF exposure led to oxidation of the biosensor (Fig. 1C). These results suggest that Mtb adapted to the increased superoxide formation we observed during starvation to preserve its redox balance, but that RIF interfered with this adaptation. Thus, the balance shifts toward oxidation in tandem with the vast majority of the culture entering the DD state. We hypothesized that the maintenance of redox balance, despite increased superoxide formation, renders starvation alone incapable of forming DD Mtb.
DD Mtb exhibit widespread damage to DNA, protein, and lipid components.
We next asked if the observed increase in oxidative stress during formation of DD Mtb impacted three classes of Mtb’s macromolecules: DNA, proteins, and lipids. We used terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) to fluorescently label single and double strand breaks in DNA. A protocol for TUNEL staining of eukaryotic cells was ineffective, likely due to inefficient permeabilization of the waxy mycobacterial cell wall and poor entry of terminal deoxynucleotidyl transferase (TdT). We adapted a protocol that uses organic solvent in reverse micelles to create Mtb cytoplasts (25) prior to fixation. This technique allowed discrimination between the signal seen before and after exposure of Mtb to the DNA-damaging agent bleomycin (fig. S2A and B). The modified TUNEL assay demonstrated a significantly higher percentage of positive cells in starved, RIF-exposed Mtb cells than in LP Mtb (P=0.0039, Fig. 2A), indicating an increased number of single- or double-strand DNA breaks in DD Mtb.
Fig. 2. DD Mtb exhibit widespread damage to DNA, proteins, and lipids.
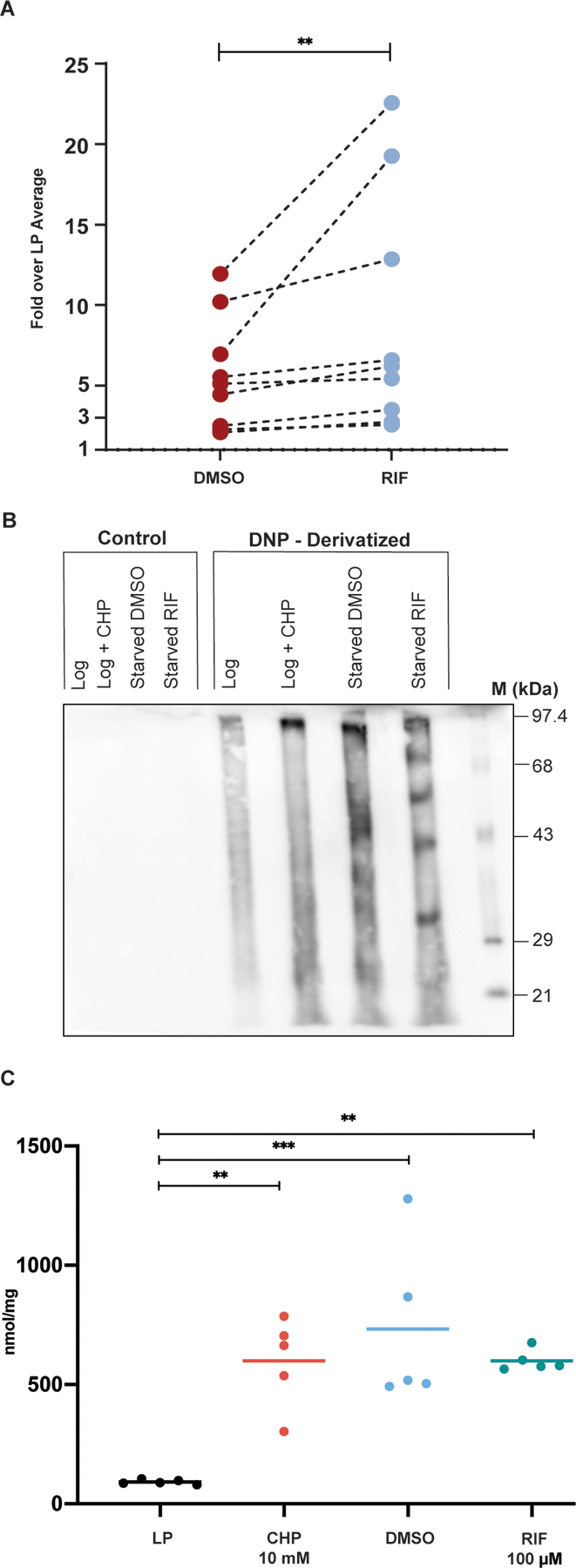
(A) TUNEL assay flow cytometry analysis of starved cells exposed to DMSO or RIF is shown normalized to the average LP percent positive. Data are representative of 3 experiments with 3 biological replicates each. Lines connecting DMSO and RIF indicate arms of the same biological replicate. Data were analyzed using a Wilcoxon matched-pairs signed rank test. **p < 0.01. (B) A representative western blot is shown of Oxyblot detection of carbonyl groups on proteins derivatized by 2,4-dinitrophenylhydrazine (DNPH) from LP cells, LP cells treated with 10 mM cumene hydroperoxide (CHP), and starved cells treated with DMSO or RIF. Control lanes were processed in the same way but lack DNPH. M: DNP-derivatized molecular weight protein standards. (C) Results of a Cayman LPO assay estimating lipid hydroperoxides is shown using lipid extracts prepared from LP cells, LP cells treated with 10 mM CHP, and PBS starved cells treated with DMSO or RIF. Data are representative of 3 experiments with 5 biological replicates total. Data were analyzed using a one-way ANOVA with Tukey’s multiple comparisons test. **adj p < 0.01, ***adj p < 0.001.
We used Oxyblot to detect carbonyl groups introduced into proteins by oxidative reactions. As compared with LP Mtb, starved cells exposed to either DMSO or RIF showed an increase in carbonylated proteins, as did the positive control, LP cells exposed to cumene hydroperoxide (Fig. 2B). Some protein species became carbonylated only in starved cells additionally exposed to RIF. This suggests that, although starved cells experienced oxidative stress, RIF either altered the pool of proteins available for oxidation or affected which proteins were oxidized. Experiments using coumarin hydrazine for fluorescent detection of carbonylation also showed an increase in carbonylation in starved, RIF-exposed Mtb as compared to LP cells (fig. S2C). The ability to form DD Mtb was not altered in strains of Mtb with disruption of individual genes important for certain forms of DNA repair (ΔuvrB, ΔrecAΔKu, and a recB transposon mutant) or protein homeostasis (ΔprcBA, ΔclpB) (fig. S3A to D). This could be due to their mutual redundancy or reliance of DD Mtb on different genes for protection or repair (26). Finally, starved cells exposed to either DMSO or RIF had significantly greater quantities of lipid hydroperoxides (oxidatively damaged lipids) than LP cells (P=0.005), although their quantities were equivalent regardless of the presence of DD Mtb (Fig. 2C).
Transcriptomic analysis of DD Mtb reveals dysregulation of central carbon metabolism and upregulation of genes related to redox status and damage repair.
With the above evidence of oxidative stress and damage, we turned our attention to mechanisms by which the bacilli react to and survive these insults as DD Mtb. We first used an unbiased approach comparing the transcriptome of Mtb in the PBS-RIF model to Mtb in PBS-DMSO controls. Because RIF inhibits RNA polymerase (RNAP) it might be expected almost to eliminate the transcriptome, as has been documented in exponentially growing Mtb exposed to RIF (27). Only a few transcripts in that study were elevated, thought to be secondary to differential mRNA stabilities (27). Strikingly, in starved Mtb, exposure to RIF led to nearly equivalent numbers of genes being upregulated and downregulated (data file S1 to S6; fig. S4A for principal component analysis). In one experiment, transcripts of 847 genes were upregulated (20.3% of the genome) and 897 downregulated with log2 fold change (FC) > 1.0. These unique alterations in relative abundance of transcripts may contribute to the DD phenotype.
As compared to treatment with DMSO alone, starved cells exposed to RIF displayed perturbations in the expression of genes encoding enzymes of the tricarboxylic acid cycle (Fig. 3A), most prominently in downregulation of icl-1, which encodes isocitrate lyase (ICL). Downregulation of icl1 leaves Mtb vulnerable to antibiotic-induced oxidative stress (28). Additionally, aceAa, icd1, and acn, all encoding enzymes related to isocitrate metabolism, were downregulated. However, there was relative upregulation of enzymes in the half of the cycle leading from α-ketoglutarate to oxaloacetate (korA, fumC, mdh). Shifts in metabolism linked to replication arrest and cell survival in the metabolome of Mtb during hypoxia similarly featured a central role of ICL (29). At the same time, starved Mtb exposed to RIF upregulated many genes encoding protein chaperones (grpE, clpB), detoxification processes (katG, trxA) and DNA repair (recA, ruvA) (Fig. 3A and fig. S4B). In all, the distinctive features of the transcriptome of a population enriched in DD Mtb portrayed cells that were combatting oxidative stress by enhancing mechanisms to protect themselves and repair damage.
Fig. 3. Characterization of DD Mtb by RNA-sequencing and Transposon-sequencing.
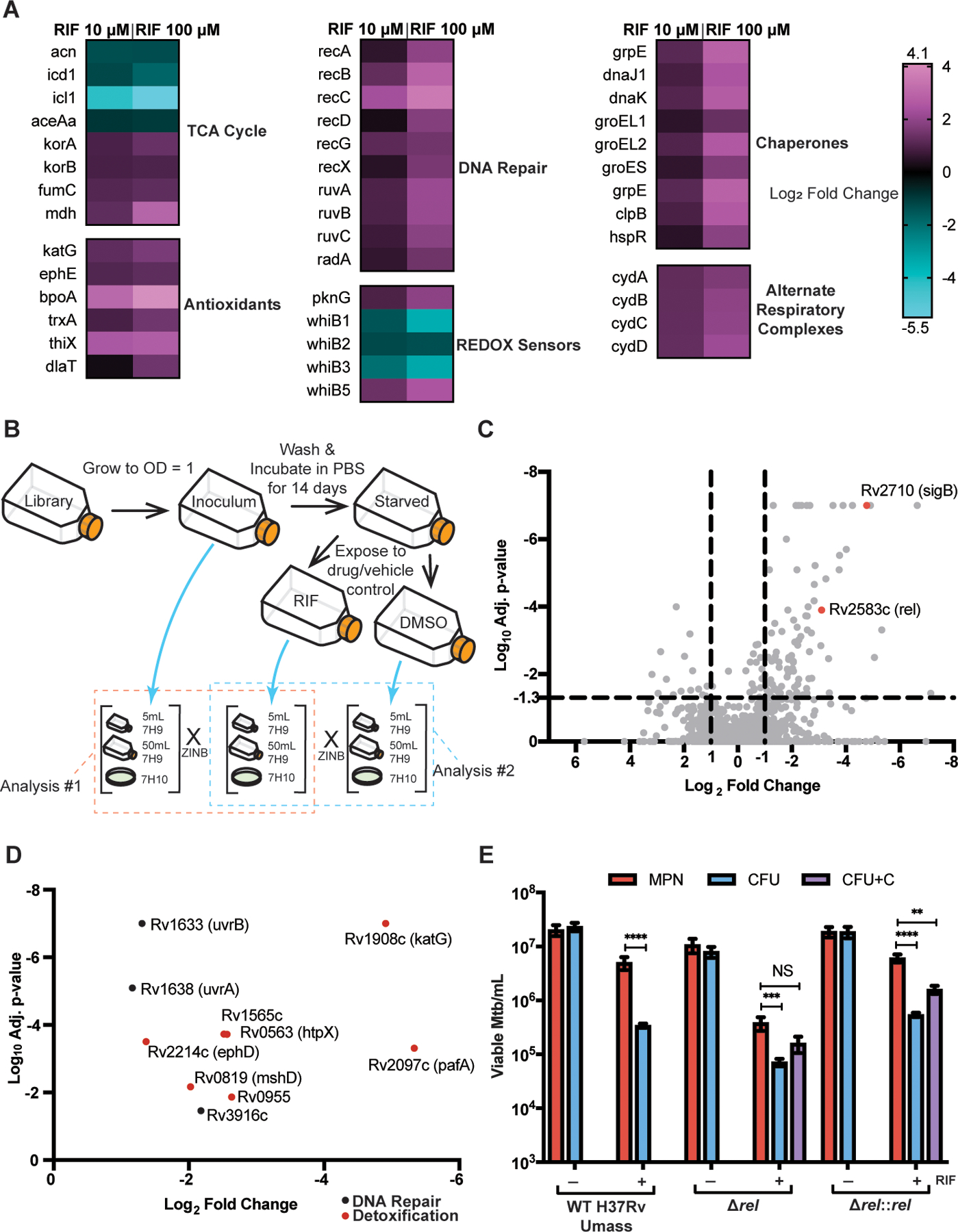
(A) Heat maps of selected significantly upregulated and downregulated genes as compared to vehicle control (DMSO) are shown. Data are representative of 2 experiments with 3 biological replicates each. (B) A schematic for TnSeq experiment and data analysis is shown. ZINB, zero-inflated negative binomial. (C) A volcano plot of transposon mutants depleted in starved, RIF exposed cells is shown as compared to input library. Data are representative of 2 experiments with 1 sample each. (D) Oxidative stress response genes and DNA damage repair genes from (C) are highlighted. (E) Viable cell counts are shown for wild type (WT) H37Rv UMass, ∆rel, and ∆rel::rel after starvation and 5 days of RIF or DMSO exposure. Data are representative of 2 experiments with 3 biological replicates each. CFU+C = CFU grown with charcoal supplementation. Data were analyzed using a mixed-effects analysis with Sidak’s multiple comparisons test. **adj p < 0.01, ***adj p < 0.001, ****adj p < 0.0001; NS, not significant.
Transposon sequencing (TnSeq) suggests that DD Mtb require oxidative stress responses, DNA repair and the stringent response
To complement RNA sequencing (RNAseq) with another untargeted method to characterize genetic determinants of entry into the DD state, we subjected a transposon (Tn) mutant library of Mtb to two weeks of starvation and 5 days of exposure to RIF. This strategy can identify genes that are non-essential for logarithmic phase growth in liquid media, but that become essential or important in the PBS-RIF model. Genes that are essential at baseline cannot be assessed with this technique. Because Tn library outgrowth on solid bacteriologic agar might artificially lower recovery, the Tn library was also grown at each time point in liquid 7H9 (5 mL and 50 mL) (Fig. 3B). In analysis #1, we used TRANSIT software and the ZINB method (30) to compare the input library to the final, RIF-exposed cultures in all three outgrowth formats. We found that 121 genes were differentially enriched with adj-p (q-value) < 0.05. Of these, 104 were more essential in RIF-exposed cultures, and 89 had a log2 FC < −1, suggesting their conditional essentiality for entering or maintaining the DD Mtb state (Fig. 3C and data file S6). Several of these genes were involved with oxidative stress and DNA repair (Fig. 3D), including katG, mshD, members of the nuo operon, uvrB, and uvrA. When comparing the DMSO and RIF arms for analysis #2, many of the same genes were conditionally essential, including mshD, nuo operon genes, uvrB, and uvrA (fig. S4C).
Comparisons between each of the liquid outgrowths and the 7H10 agar outgrowth for RIF- exposed cells revealed fewer changes, even when including genes conditionally essential in either outgrowth condition: 33 genes for 50 mL 7H9 versus agar and 21 genes for 5 mL 7H9 versus agar (data fileS7). Strikingly, part of a complex linking oxygen radical detoxification with thiol homeostasis, sseA (31), was conditionally essential for growth on agar after RIF exposure. This implies that this detoxification system is required to allow Mtb to grow as CFU in the PBS-RIF model.
The TnSeq results also suggested conditional essentiality of Rv2538c (relMtb), a mediator of the stringent response to amino acid starvation (32, 33). Consistent with this, ∆relMtb were unable to form a population of DD Mtb as compared to CFU growth on 7H10 plates supplemented with charcoal (Fig. 3E), and the ratio of MPN to CFU was significantly lower in the knockout strain as compared to the wild type in the PBS-RIF arm (P=0.0008, fig. S4D). Charcoal was added to mitigate RIF carry-over on cells; in our prior work, charcoal did not restore cultivability of Mtb CFU in the PBS-RIF model and was therefore omitted in those control arms (14, 34). The relative lack of a substantial DD population in ∆relMtb suggests that genetically encoded adaptation to stress is required for Mtb to exist in the DD state. The gene encoding sigma factor B, which is induced in multiple stress conditions (35–38) including nutrient starvation (39), also appeared to demonstrate conditional essentiality for DD Mtb. However, a ∆sigB strain demonstrated an overall loss in viability, explaining the positive hit in TnSeq, and the knockout strain did not show a difference in DD Mtb proportion after starvation and RIF exposure, suggesting that sigB does not play a specific role in entering the DD state (fig. S5A). Moreover, none of the single sigma factor knockouts produced a notable decrease in CFU after up to 14 days in starvation, and individual sigC, E, F, G, H, J, K, and M knockout strains also did not show alterations in proportions of DD Mtb after starvation and RIF exposure (fig. S5A and B). This approach, limited by use of single sigma factor knockouts, did not reveal a specific regulatory pathway. In sum, the above data suggest that DD Mtb formation is not reliant on a generic stress response.
Dysregulation of specific stress response genes influences the DD phenotype.
Using results of the unbiased screens, we next evaluated whether dysregulation of specific aspects of Mtb adaptation to oxidative stress could alter the DD phenotype. Mycothiol, the mycobacterial homolog of glutathione, buffers oxidative stress in Mtb (40–42). An enzyme involved in mycothiol synthesis, encoded by mshD, was found in the TnSeq to be conditionally essential in the PBS-RIF model. Mtb in which mshA, a gene essential for mycothiol synthesis, was disrupted formed an increased proportion of DD Mtb in the PBS-RIF model (Fig. 4A). Likewise, Mtb lacking icl1, one of the most downregulated genes after RIF exposure of starved Mtb, produced a higher proportion of DD upon RIF exposure than wild type Mtb and produced a significant amount of DD Mtb compared with starvation alone (P<0.0001, Fig. 4B). Ratios of MPN to CFU for both the mshA and icl1 knockout strains in the PBS-RIF condition were significantly higher than for their respective wild type strains (mshA, P< 0.0001; icl1 P<0.02, fig. S6A and B). In contrast, a strain lacking dlaT, which encodes one member of a four-protein peroxynitrite reductase/peroxidase (43) and which is highly sensitive to nitrosative stress (44), did not show an altered DD phenotype (fig. S6C). In addition, dlaT transcripts were only mildly increased in DD Mtb (average log2 FC 1.43). These results support the hypothesis that both oxidative stress and an altered cellular response to it are key to formation of DD Mtb, and that there is specificity in the type of stress or stress resistance involved in the formation of DD Mtb.
Fig. 4. Dysregulation of specific stress response genes influences the DD phenotype.
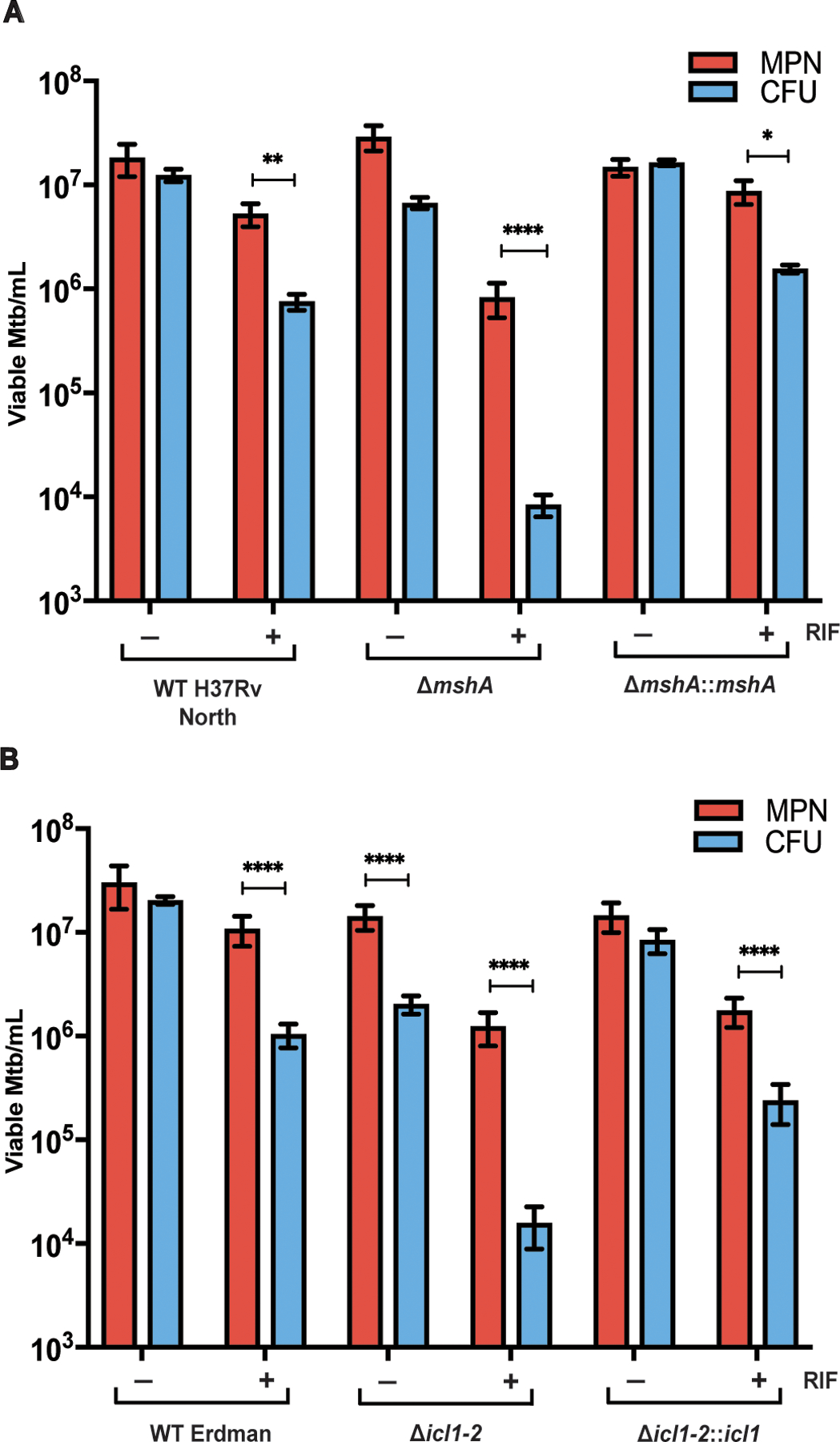
(A) Viable cell counts are shown for WT H37Rv, ∆mshA, and ∆mshA::mshA after starvation and 5 days of RIF or DMSO exposure. Data are representative of 2 experiments with 3 biological replicates each. Data were analyzed using a mixed-effects analysis with Sidak’s multiple comparisons test. (B) Viable cell counts are shown for parent wild type (WT) Erdman Mtb strain, ∆icl1–2, and ∆icl1–2::icl1 after starvation and 5 days of RIF or DMSO exposure. Data are representative of 3 experiments with 3 biological replicates each. Data were analyzed with a two-way ANOVA with Sidak’s multiple comparisons test. *adj p < 0.05, **adj p < 0.01, ***adj p < 0.001, ****adj p < 0.0001.
RNA polymerase inhibition of starved cells is not sufficient to produce DD Mtb.
The foregoing results with genetic knockouts suggested a specificity in the response to oxidative and starvation stress related to formation of DD Mtb. We then asked if there is also specificity in the way that RNAP is inhibited that affects DD Mtb formation. We previously demonstrated that DD Mtb formation in this model was seen only with rifamycins among compounds tested, and that mutations in RNAP that conferred resistance to RIF’s ability to kill replicating Mtb also blocked RIF from inducing DD Mtb in starved Mtb (14). To determine whether another RNAP inhibitor could replace a rifamycin, we exposed starved Mtb to Nα-aroyl-N-aryl-phenylalaninamides (AAP)—RNAP inhibitors whose mode of action is distinct from that of RIF (45). We used doses that reduced CFU by about 1-log10 so that any potential DD population could be recognized. Charcoal-supplemented 7H10 plates were also used for CFU assessment. No statistically significant proportion of DD Mtb was generated (P=0.932, Fig. 5A). This suggests that globally impaired transcription by itself is insufficient for DD Mtb formation, and that the way that RNAP is inhibited is critical. The mechanism of RNAP inhibition may influence which genes are affected and determine whether and to what extent a transcript is increased or reduced.
Fig. 5. DD formation is specific to rifamycins in the PBS-RIF model, and M. bovis is unable to enter the DD state in the PBS-RIF model.
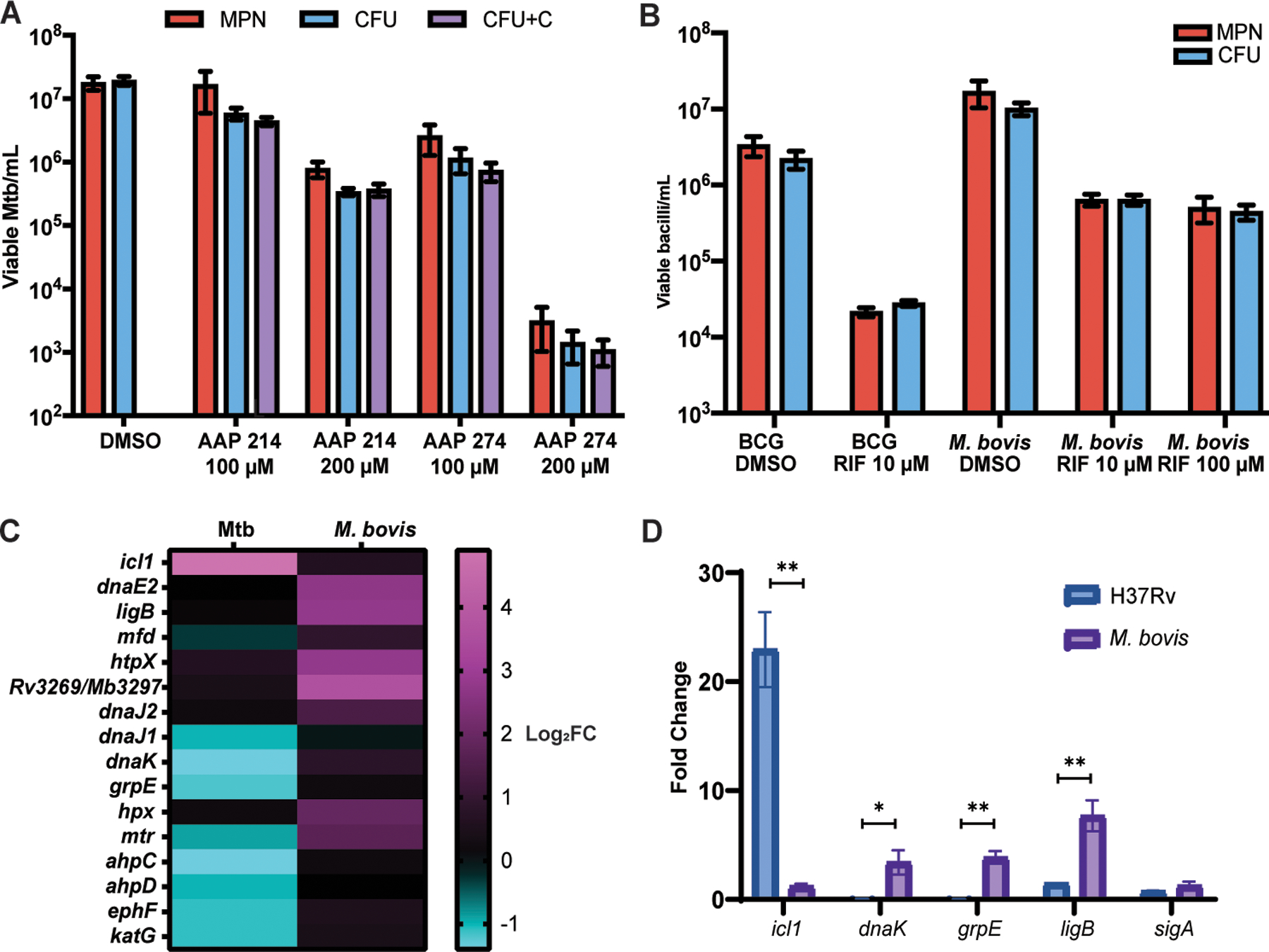
(A) Viable cell counts using CFU, CFU with charcoal (CFU+C), and MPN-limiting dilution assays are shown after starvation and 5 days of the exposure to Nα-aroyl-N-aryl-phenylalaninamide (AAP) direct RNA polymerase inhibitors (AAP 214 and 274) or DMSO exposure. Data are representative of 2 experiments with 3 biological replicates each. Data were analyzed using a mixed-effects analysis with Sidak’s multiple comparisons test. (B) Viable cell counts are shown for BCG Pasteur (2 experiments, 3 biological replicates each) and M. bovis (3 experiments, 3 biological replicates each, except R100, which is 2 experiments, 3 biological replicates each) after starvation and 5 days of RIF or DMSO exposure. Data were analyzed using a two-way ANOVA with Sidak’s multiple comparisons test. (C) A heat map of log2 fold change in transcripts is shown after PBS starvation without RIF exposure as compared to LP in Mtb and in M. bovis, according to RNAseq. (D) Fold change in icl1, dnaK, grpE, ligB, and sigA transcripts are shown as determined by qPCR after 4 weeks of PBS starvation and without RIF exposure as compared to LP cells, normalized to 16s rRNA, in H37Rv and M. bovis. Data were analyzed using multiple paired t-tests with Holm-Sidak correction for multiple comparisons. *adj p < 0.05, **adj p < 0.01.
M. bovis and BCG do not form DD under the same conditions as Mtb.
To test whether closely related mycobacteria could enter the DD state in the PBS-RIF model, we grew attenuated Mycobacterium bovis BCG Pasteur and virulent Mycobacterium bovis (American type culture collection (ATCC) 19210) to LP, starved them in PBS for at least 2 weeks and then exposed them to RIF. No DD populations formed, despite a decrease in the CFU (Fig. 5B). Nor did DD populations form when we used RIF at a 10-fold lower dosage (10 µM) to produce a decrease in CFU similar to that seen in Mtb in the PBS-RIF model. Screens aimed at reducing oxidative stress in BCG using 5 µM RIF after 4 weeks of starvation and using 10 µM RIF after only 1 week of starvation did not yield a DD population when comparing the limiting dilution assay to growth on charcoal supplemented 7H10 plates (fig. S7A and B).
The genomes of Mtb and BCG differ in 14 regions of the chromosome. An Mtb strain with deletions in both region of difference 1 (RD1) and panCD, each of which is required for virulence (46, 47), was able to form DD Mtb, suggesting that the critical genes for entering the DD state lie elsewhere (fig. S7C). The mce3 operon was upregulated in the RNAseq analysis and is absent from M. bovis (48), and we thus hypothesized that it might play a role in Mtb’s ability to enter the DD state. However, mce3A and mce3F transposon mutants in Mtb (49) did not demonstrate a difference in their DD phenotype compared to wild type Mtb (fig. S7D).
To further explore the difference between Mtb and M. bovis with respect to DD formation, we performed RNAseq in M. bovis in the PBS-RIF model. Results from one study with three biologic replicates were verified by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and compared directly to changes in Mtb. Striking differences between the species occurred during starvation, even prior to RIF exposure (Fig. 5C, data file S5, S8, and S9; table s1). icl1, one of the most upregulated genes in starved Mtb when compared to LP bacilli, was not upregulated during starvation in M. bovis (Fig. 5D). This suggests that alterations in icl1 during nutrient starvation are critical for subsequent survival as DD Mtb upon RIF exposure, and that M. bovis does not respond in the same way. Furthermore, damage response genes including dnaK and grpE, involved in proteostasis (50, 51), and ligB, involved in DNA repair (52, 53), were upregulated in M. bovis but not in Mtb during starvation alone (Fig. 5D). This implies that M. bovis is reacting to damaged macromolecules whereas Mtb survives without requiring this adaptation during nutrient starvation. Comparisons of macromolecular damage in PBS-RIF exposed M. bovis versus Mtb were confounded by lack of formation of DD M. bovis, whose surviving population consisted almost entirely of CFU, as opposed to Mtb, whose surviving population was highly enriched for DD cells. However, a comparison of oxyblot results during starvation alone, where both populations survived as CFU, showed a different pattern of protein expression and protein oxidation between the two species (fig. S7E and F).
Intermediate degrees of oxidative stress promote formation of DD Mtb.
The inability of M. bovis to enter a DD state and the above differences between the transcriptomes of M. bovis and Mtb led us to hypothesize that M. bovis may suffer greater oxidative stress than Mtb during the PBS-RIF model. M. bovis displayed significantly higher CellROX staining than Mtb (P<0.0001, Fig. 6A) during both starvation and RIF exposure. Likewise, exposure of starved Mtb to an Nα-aroyl-N-aryl-phenylalaninamide (AAP), which also did not lead to formation of DD Mtb, produced a higher degree of oxidation of the Mrx1-roGFP2 biosensor than exposure of starved Mtb to RIF (Fig. 6B). In both cases, lack of a DD population is indicative of more extensive bacterial death than experienced by Mtb in the PBS-RIF model. Streptomycin and fluoroquinolones, which did not generate DD Mtb when presented to starved Mtb (14), did not lead to an increase in oxidative stress (Fig. 6B). The rifamycins RIF and rifapentine (RPT) both cause moderate oxidative stress, and both can generate DD Mtb (14). Overall, these results suggest that: oxidative stress drives Mtb toward the DD state; Mtb must be able to mitigate oxidative stress to survive in the DD state; Mtb’s capacity to do so is limited, so that only intermediate degrees of oxidative stress lead to DD Mtb; and M. bovis strains are further limited in this capacity, such that they succumb under similar conditions.
Fig. 6. DD formation is correlated with intermediate degrees of oxidative stress.
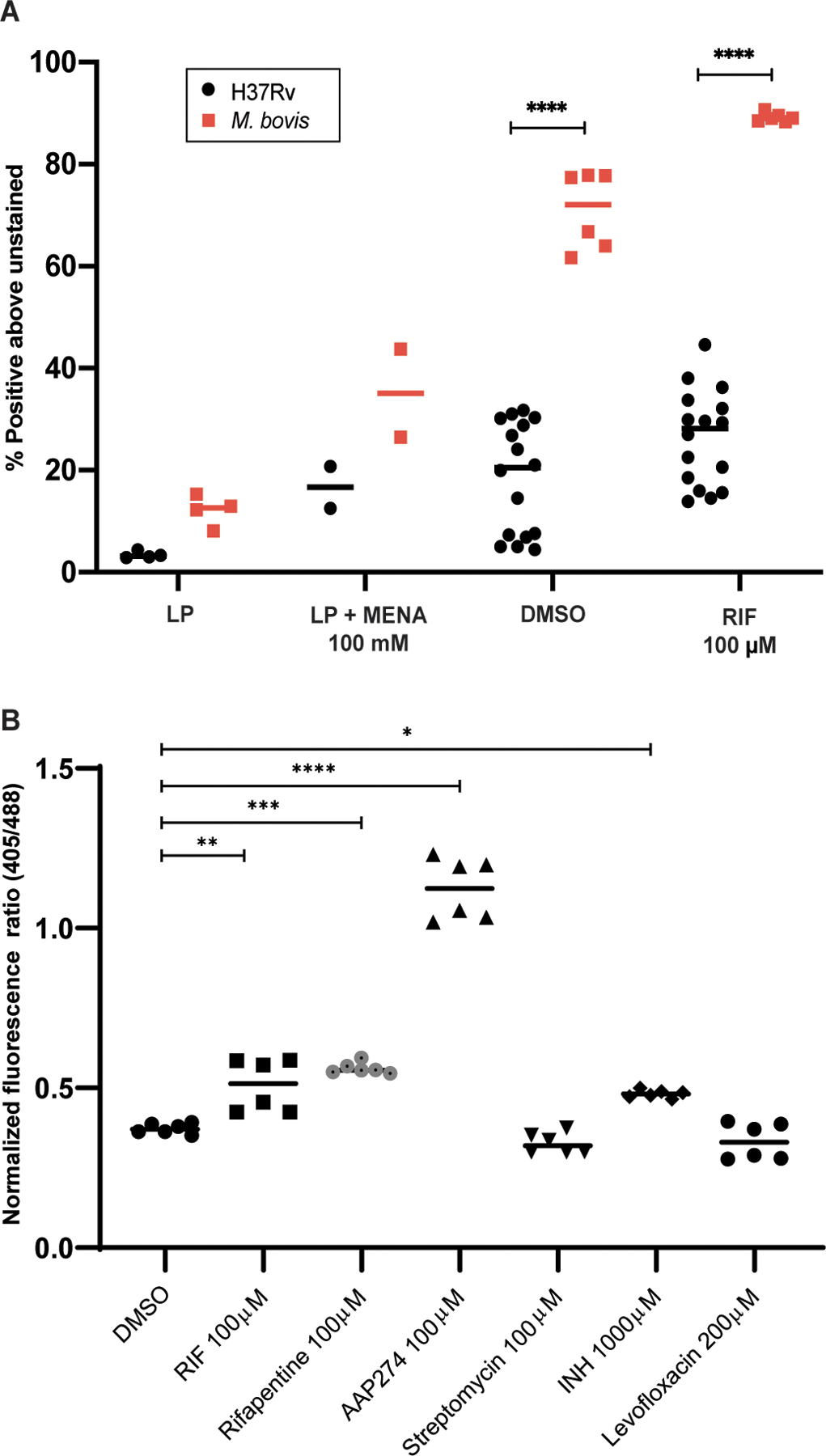
(A) Flow cytometry analysis is shown after staining M. bovis with CellROX Green as compared with Mtb. M. bovis data are from 2 experiments; Mtb data are as in Fig. 1B. Data were analyzed using a two-way ANOVA with Sidak’s multiple comparisons test. (B) Flow cytometry analysis is shown after starvation and exposure to RIF, rifapentine (RFP), isoniazid (INH), streptomycin (STR), levofloxacin (LVX), and AAP274 of H37Rv with the mrx1-roGFP2 plasmid. Data are representative of 2 experiments with 3 biological replicates each. Data were analyzed using a one-way ANOVA with Tukey’s multiple comparisons test. *adj p < 0.05, **adj p < 0.01, ***adj p < 0.001, ****adj p < 0.0001.
We hypothesized that mitigating oxidative stress after DD Mtb were plated on agar might allow a greater proportion to grow as CFU. However, there were no increases in starved, RIF-treated Mtb recovered on agar plates incubated at 1%, 5% or 21% oxygen, or on plates with the anti-oxidant N-acetylcysteine (NALC) (fig. S8A to C). Screens for improved CFU recovery on agar plates made with ultrapure agarose and a liquid/solid hybrid of filter paper floating on 7H9 also did not yield higher CFU counts (fig. S8D).
Growth delay is associated with recovery of DD Mtb’s ability to grow as CFU.
We next turned our attention to the question of why limiting dilution in nutrient-rich 7H9 medium allows oxidatively stressed Mtb to recover their ability to grow as CFU, when incubation on nutrient-rich agar without limiting dilution does not. Typically, when logarithmically replicating bacteria are diluted into fresh nutrient rich medium, they do not pause in their replication. In contrast, when stressed bacteria are re-suspended in fresh nutrient rich medium, there is a lag before they resume logarithmic replication. For Mtb, this lag has been described at the population level after exposure to RIF (54, 55) and at the single cell level after starvation in PBS (56). We hypothesized that a delay in replication may be a critical feature that allows DD Mtb to be detected by limiting dilution when the same cells plated directly on agar fail to grow. However, the reported studies of bacterial lag involved the transfer of stressed or starved cells to nutrient-rich liquid medium without limiting dilution. It is not clear whether limiting dilution itself imposes a lag in onset of bacterial replication. Therefore, we tested the effect of ten-fold serial dilutions on the number of CFU of Mtb collected in three conditions: during logarithmic replication, after starvation in PBS and exposure to DMSO vehicle alone (PBS-DMSO), or after being placed in the PBS-RIF model. The cells from each group were washed twice by centrifugation, re-suspended in 7H9, serially diluted, and sampled daily for up to 14 days for quantification of CFU.
Mtb collected from LP and subjected to limiting dilution continued logarithmic replication without a delay in onset (fig. S9A). In contrast, both PBS-DMSO and PBS-RIF cells displayed two different phases of growth: an initial slower growth phase, followed by a much faster phase. Starved cells exposed to DMSO alone displayed a 1- to 2-day delay in resumption of logarithmic growth, and this was unaltered by the extent of dilution (Fig. 7A). These cells continued this degree of growth until reaching stationary phase. RIF-exposed cells followed a more complex pattern that was influenced by the extent of dilution. These cells had the longest lag phase when undiluted or minimally (10x) diluted (Fig. 7B). We speculate this was due to residual cell-associated RIF. Samples diluted to an intermediate extent (100- to 1000-fold) grew more slowly for the first 6 days than they did thereafter but began replication earlier than other dilutions. Cells diluted even more extensively delayed their onset of rapid replication by as long as 15 days (Fig. 7B). The inset in Figure 7B displays the data with a linear y-axis to clearly delineate the differences in time of onset of faster replication. The results imply that in the limiting dilution assay, cells in the most dilute wells undergo a substantial replication delay that is independent of the RIF carry-over that may have affected the least diluted samples. In sum, three factors impose a delay in replication in the recovery phase in this model for DD Mtb generation: prior starvation, prior RIF exposure, and limiting dilution itself.
Fig. 7. Growth delay allows recovery of DD Mtb’s ability to grow as CFU.
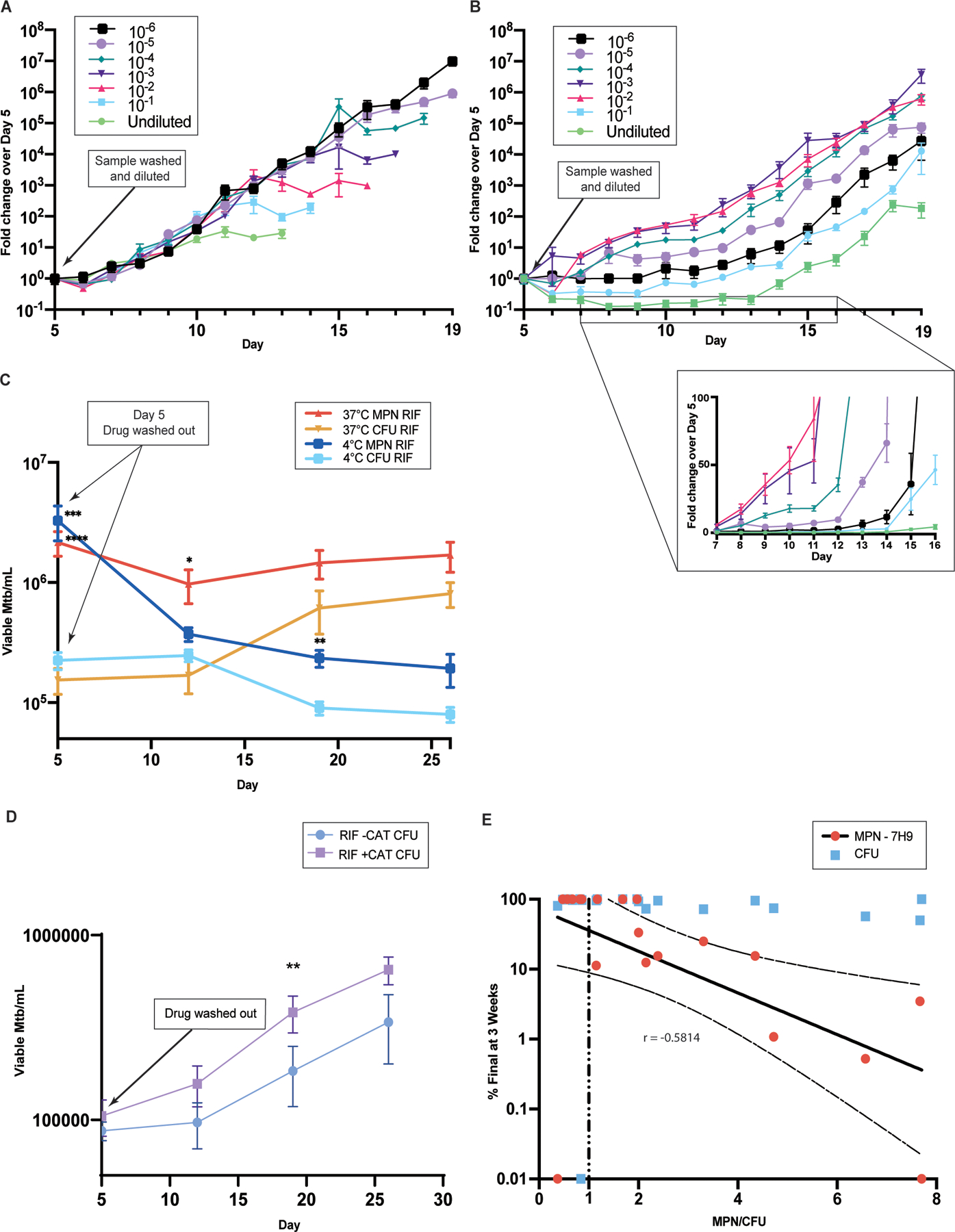
(A and B) Daily CFU counts of serially 10-fold diluted cultures in 7H9 complete are shown after 4 weeks of starvation and 5 further days of exposure to DMSO (1%) (A) or RIF (100 µM) (B), normalized to initial counts on the day of drug washout (day 5). Data are representative of 2 experiments with 3 biological replicates each. The inset in B shows days 7 to 16 with linear y-axis scale. (C) Serially sampled viable cell counts of DD Mtb re-suspended in PBS are shown after drug removal at 37 °C (data are representative of 7 experiments with 3 biological replicates each) and 4 °C (data are representative of 3 experiments with 3 biological replicates each). Data were analyzed using a mixed-effects analysis; only significant differences between MPN and CFU at the same temperature are shown; ***adj p < 0.001, 37 °C; ****adj p < 0.0001, 4 °C; *adj p < 0.05, only 37 °C; **adj p < 0.01, only 4 °C. (D) Serially sampled CFU counts are shown for Mtb subjected to starvation and exposure to RIF 10 µM and re-suspended in PBS after drug removal with and without 1400 units of human catalase (CAT) added one day prior to RIF exposure and again after drug removal. Data are representative of 2 experiments with 3 biological replicates each. Data were analyzed using multiple paired t-tests with Holm-Sidak correction for multiple comparisons. **adj p < 0.01 (E) The proportion of DD Mtb in patient sputa was plotted against the percentage of the final viable counts accounted for at 3 weeks of outgrowth, n=20. Shown with linear regression and 95% confidence intervals for MPN data after log transformation.
We next asked whether a growth delay unrelated to limiting dilution can suffice to restore CFU formation by stressed Mtb. DD Mtb formed in the PBS-RIF model were washed twice, re-suspended in PBS/0.02% tyloxapol (PBS-Tx) to the original volume and incubated at 37 °C. We sampled the cells weekly for both CFU and limiting dilution assays. The number of viable Mtb estimated by limiting dilution did not markedly change, but the number of Mtb recovered as CFU increased until, by 3 weeks, there was no longer a difference between estimations of viable Mtb derived by the limiting dilution/MPN and CFU assays (Fig. 7C). Thus DD Mtb recovered the ability to grow as CFU under a condition that precluded replication, without access to externally supplied nutrients and without dilution. In contrast, DD Mtb incubated in PBS at 4 °C could no longer be recovered by limiting dilution or CFU assays; they were presumably dead (Fig. 7C). Additionally, the increased TUNEL signal noted in PBS-RIF cells fell to match PBS-DMSO signal after 3 weeks of incubation in PBS-Tx, suggesting that the relative extent of DNA damage dropped to pre-drug exposure amounts (fig. S9B). We hypothesized that mitigating oxidative stress might alter CFU recovery, and found that DD Mtb cells recovered their ability to grow as CFU more quickly if we added the anti-oxidant catalase to the starved cells one day prior to RIF exposure and again after RIF was washed out (Fig. 7D). Although exogenous catalase did not prevent formation of DD Mtb, that catalase hastened the ability of DD Mtb to form CFU reinforces the relationship between oxidative stress and delayed return of replicative capacity.
DD Mtb from TB patients appear to grow more slowly ex vivo than CFU in the same samples.
Given the above in vitro evidence, we hypothesized that ex vivo DD Mtb recovered from patient sputa would also demonstrate a growth delay when recovering in the limiting dilution assay. To test this, we analyzed unpublished data compiled in an earlier study (13). In that work, 13 patients newly diagnosed with active TB in Haiti had Mtb quantified in sputum samples by both CFU and limiting dilution assays immediately prior to treatment and after 2 weeks of TB therapy with the standard 4-drug regimen for drug-sensitive TB, which includes RIF. CFU were counted and limiting dilution plates were read at 3, 5, 7, and 9 weeks, with termination of the assay any time after 5 weeks in which the counts were unchanged from the prior reading. For data points in which the 3-week read was available (11 patients, 20 samples), values of CFU and MPN from limiting dilution at 3 weeks were assessed as a percentage of the final values obtained from the same assay when scored after a longer incubation. Except for one outlier, all CFU reached at least 50% of their final value at 3 weeks, and 65% (13/20) reached at least 90% of their final value at 3 weeks. There was no significant correlation between this percentage for the CFU and the relative proportion of DD Mtb in the same sample (r = −0.3409, P=0.707 (Fig. 7E). In contrast, the percentage of final growth in the limiting dilution assay at 3 weeks was significantly negatively correlated (r = −0.5814, p<0.0036; Spearman correlation) with the ratio of MPN to CFU (Fig. 7E). Technical limitations associated with low bacillary load (<1000 per mL) and no evidence of growth at 3 weeks apply to two outliers with slow growth and without evidence of a DD population. In all, the results suggest that longer growth delay correlates with a higher proportionate recovery of DD Mtb in clinical samples, as it does for DD Mtb generated in vitro.
DD Mtb can form without starvation or antibiotics.
Clinical evidence suggests that entry into DD state can occur independent of rifamycin exposure, as DD Mtb have been recovered from sputa of patients who have not yet been treated (10–13). Given the highly specific requirements of the PBS-RIF model in forming DD Mtb, we next asked if antibiotic-independent methods of generating oxidative stress could produce DD Mtb. We discovered that LP cells incubated at 45 °C in nutrient rich medium for 24 hours displayed a marked decrease in CFU. Strikingly, however, there was no change in viable numbers of Mtb as assessed by limiting dilution (Fig. 8A). Within 2 days of being returned to 37 °C, the DD Mtb regained colony-forming capacity (Fig. 8A). The rate of recovery in numbers of CFU was faster than the initial remaining CFU population could have achieved by logarithmic replication at the rate seen in unstressed cells in the same medium. There was inter-experiment variability in the generation of DD Mtb by heat, especially after incubation at 50 °C (Fig. 8A and fig. S10). Nonetheless, heat-induced DD Mtb formation was positively correlated with a higher CellROX signal and a higher percent of TUNEL positivity (Fig. 8B and C) than in cells incubated at 37 °C.
Fig. 8. DD Mtb can form without starvation, antibiotics, or limiting dilution.
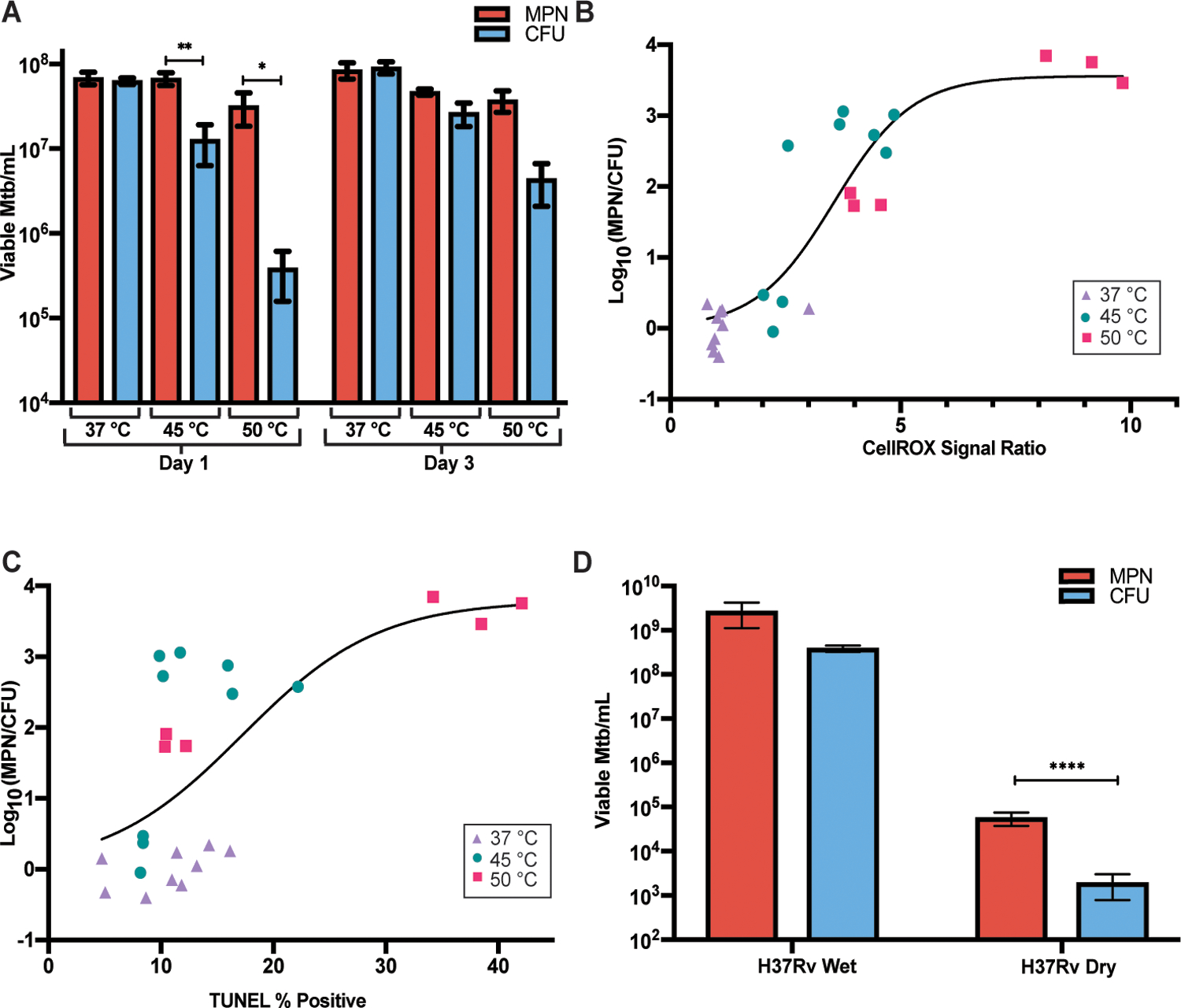
(A) Viable cell counts are shown for LP cells exposed for 24 hours to 37 °C, 45 °C and 50 °C immediately following heat exposure and after 48 hours at 37 °C. Data are representative of 3 experiments with 3 biological replicates each. Data were analyzed by a two-way ANOVA with Tukey’s multiple comparisons test. (B) CellROX % positive, normalized to average 37 °C CellROX % positive, is plotted versus proportion of DD Mtb after 1 day of indicated temperature exposures. Data are representative of 3 experiments with 3 biological replicates each and are shown with a nonlinear fit curve. (C) TUNEL positivity is plotted versus proportion of DD Mtb after 1 day of indicated temperature exposures. Data are representative of 3 experiments with 3 biological replicates each; one set of 50 °C was omitted due to extensive killing. Data are shown with a nonlinear fit curve. (D) Viable cell counts are shown for LP cells grown on filters and either desiccated or placed on a pool of saline for 7 days at 37 °C, 70% humidity, and room air CO2. Data are representative of 3 experiments with 2 to 3 biological replicates each. Data were analyzed by a two-way ANOVA with Sidak’s multiple comparisons test. All CFU were recorded and analyzed as the limit of detection, 100 CFU, if no growth was detected. *adj p < 0.05, **adj p < 0.01, ****adj p < 0.0001.
Transmission of TB occurs by aerosol, and aerosols are subject to evaporative desiccation. Yet it has been known for over a century that tubercle bacilli expectorated into the environment can maintain virulence for days or even months, as evidenced by the ability of recollected bacilli to cause TB in guinea pigs (57, 58). We hypothesized that desiccation stress might create a population of DD Mtb. To test this, Mtb cells were grown on filter paper and allowed to desiccate in room air for 7 days. As compared to Mtb on filter paper floating on saline, desiccation killed a large proportion of the population. The majority of survivors were DD Mtb (Fig. 8D). Thus, desiccation is another drug-independent in vitro model of DD Mtb formation, with implications for transmission biology.
Discussion
The remarkable resilience of oxidatively-damaged Mtb as it enters a DD state sheds light on fundamental questions in microbiology and introduces considerations for the clinical management of TB. Microbiologically, our findings demonstrate that oxidative stress drives the formation of DD Mtb; the transcriptome of these nonreplicating, phenotypically resistant cells is not “dormant” but is dramatically altered. Further, we show that survival of Mtb in the DD state requires specific genetic controls. The special ability of rifamycins to generate DD Mtb correlates with the intermediate degree of oxidative stress they generate in starved cells and may also reflect the specific identities of genes whose expression is enhanced or reduced by RIF. However, DD Mtb can form without antibiotic exposure (15, 59, 60), and there are other routes to imposition of intermediate amounts of oxidative stress in Mtb besides starvation and exposure to rifamycins, including exposure to the immune system (10–12) and, as shown here, heat. In the human host, it is possible that repeated exposure to febrile body temperatures might recapitulate what we saw in vitro with brief exposure to a supra-physiologic temperature, but this is speculative. DD Mtb created by heat stress shared the evidence of oxidative stress and DNA damage observed in the starvation-RIF model. Desiccation also drove Mtb into the DD state, which has implications for environmental biology and disease transmission.
This work adds to a rich literature in other bacteria relating changes in cultivability of viable bacteria to oxidative stress, nutrient availability, growth rate and the definition of viability itself (18, 20, 61, 62). Many bacterial species can enter a VBNC state (63), as first shown with chilled Vibrio and E. coli (64). In food safety literature, Staphylococcus aureus (65), Vibrio vulnificus (66) and Campylobacter jejuni (67) have all been shown to enter the VBNC state in association with oxidative stress and oxidative stress resistance. Similarly, creation of a VBNC subpopulation of E. coli by heat stress was associated with protein oxidation (68). Hong et al. (21) showed that, above a certain threshold of ROS, E. coli cells died, but below that threshold, provision of antioxidants revealed a cryptic population as CFU. Similarly, our study points to the ability of intermediate amounts of ROS to temporarily block the ability of an Mtb cell to be detected as a CFU.
Recovery of replication capacity on the part of oxidatively stressed Mtb was favored by a delay in replication, whether imposed by incubation without exogenous nutrients or by extensive dilution in nutritionally rich medium. Because the bulk of the Mtb populations in the DD models tested here were in the DD state, growth in the highest dilutions of the limiting dilution plates, dilutions at which no CFU remained, represented resuscitation of DD cells, as opposed to regrowth of a colony forming subpopulation—a distinction that must be considered in discussion of VBNC phenotypes (69). We hypothesize that recuperation requires repair, repair takes time, and replication before repair is suicidal. The notion of the lethality of premature replication is consistent with our observation of DNA strand breaks in DD Mtb. By the same reasoning, we take the eventual ability of the cells to proliferate as evidence that strand breaks have been repaired. The dependence of persistent cell survival on timing of DNA repair with respect to replication has been described in E. coli exposed to fluoroquinolones (70).
The discrepancy between Mtb and M. bovis/BCG in the ability to enter a DD state was surprising and reassuring. In many countries, viable BCG is given intradermally to newborns as a vaccine. BCG is also given intra-vesically to treat recurrent superficial bladder cancer. BCG appears to be cleared rapidly, except in some cancer patients and in immune-deficient infants, such as those with Severe Combined Immunodeficiency, Chronic Granulomatous Disease, AIDS (71), or the syndrome Mendelian Susceptibility to Mycobacterial Diseases, which involves deficient production or response to interferon-γ (72, 73). Recently, a live BCG vaccine was found to be more effective in macaques when administered intravenously than intradermally (74), an approach that favors the induction of “trained immunity” (75). Even after intravenous administration, BCG was apparently cleared (74). Virulent M. bovis can cause active TB that is clinically indistinguishable from the active disease caused by Mtb, but it remains unclear whether M. bovis routinely or readily enters a latent state of infection (76). Our studies support the expectation that clinically-administered BCG is unlikely to enter a DD state. Although stressed Mtb entered a state of reparable oxidative damage in vitro, BCG and M. bovis responded to the same conditions by accumulating even more ROS and did not recover. This differential response may be related to the equally striking differential regulation of icl1. A negative regulator of icl1, Rv0465 (Mtb)/Mb0474 (M. bovis), has a non-synonymous polymorphism in M. bovis (77). This might be related to the difference we observed in icl1 regulation between the two species.
RIF’s imposition of a severe and numerically symmetrical dysregulation of the transcriptome may be key to the special propensity of rifamycins to help form (13, 14) or select for (13) DD Mtb. Although starvation alone produced ROS in Mtb and damaged Mtb’s cellular components, starved Mtb showed no difficulty in growing on agar plates. Subsequent exposure to RIF appeared to disrupt cellular processes that detoxify ROS and control or repair the damage. This is in agreement with a study showing that RIF persisters undergo more oxidative stress than mid-LP, non-RIF exposed Mtb, and have a higher rate of de novo mutations during RIF exposure (78). Our observation that starved cells exposed to RIF upregulate expression of hundreds of genes is consistent with the report that Mtb cells phenotypically resistant to RIF incorporate [3H]uridine during RIF exposure (79). This work underscores the complexity of rifamycins’ mechanism of action by revealing the ability of cells seemingly killed by rifamycins to repair themselves, despite the post-antibiotic effect of RIF that has been studied for decades (50, 80, 81).
Clinically, DD Mtb are unaccounted for in the routine quantification of viable bacilli. The ability of DD Mtb to recover replicative capacity during a period of non-replication may complicate clinical management. Considering the varied environmental niches in different body compartments and the immunologic differences among patients and in a given patient over time, Mtb cells are likely to exist in the host at multiple degrees of oxidative stress and replicative capacity. Evidence of this has existed since the 1950s, when one study noted that samples taken from 44% of cavitary lesions not in communication with the airway required 3 to 10 months of incubation to display Mtb growth, and nothing grew from the remaining 56% of closed cavities. In contrast, nearly all ‘open’ cavitary lesions grew Mtb within 8 weeks of cultivation (82, 83). Efforts to combat the heterogeneity of Mtb in a host with active TB have led to the prolonged use of multiple antibiotics. Similarly, the months-long duration of prophylaxis and the associated drug toxicity have frustrated efforts to eliminate latent TB on a large scale in endemic areas. During active TB, drug regimens that afford the majority of patients a relapse-free cure can also result in over-treatment of many patients with often toxic drug combinations. People cured earlier in the treatment course cannot be reliably distinguished from those who require the full course of therapy. Taking DD Mtb into account may provide a new perspective on heterogeneity in tuberculosis infection and its outcomes. Indeed, a recent study found that, out of 41 TB patients that achieved clinical cure, 5 had viable, differentially culturable Mtb in their sputum or bronchoalveolar lavage fluid at the end of treatment, and 2 of these patients relapsed within one year from the treatment’s end (84).
The presence of DD Mtb also complicates current quantitative and semi-quantitative clinical diagnostic assays. For example, during RIF monotherapy and during standard four-drug therapy, discrepancies were identified between results of a liquid culture diagnostic assay, the mycobacterial growth indicator tube (MGIT), and CFU quantification (85). Our work suggests that the growth of stressed bacilli in liquid culture may be delayed in onset, leading to an inoculum-independent lengthening of time to positivity, the end-point in the MGIT assay. Moreover, tests of TB drugs typically involve collection of sputum overnight into a chilled container, and the specimens may be refrigerated for several days before the number of viable Mtb is determined. Our data show that oxidatively damaged Mtb are unable to recover in the cold, so that DD Mtb in sputum may die if collected in this manner, exaggerating the impact of the drug(s) under study. Thus, the ability to quantify DD Mtb in the clinic might improve diagnostics and treatment monitoring.
Our study has limitations. A minority CFU-capable population was present in every DD Mtb culture, complicating interpretation of RNA-seq and bulk measures of oxidative stress. To compensate, we relied on marked, quantifiable differences between limiting dilution and CFU assays and used orthogonal approaches, such as genetic knockouts and alternative models for generating DD Mtb. Moreover, DD Mtb that form in mice can escape detection by limiting dilution (86). This raises the specter that the DD Mtb detected in clinical specimens may under-report the proportion of DD Mtb in the human host.
Our future work will explore whether characteristics of the DD Mtb transcriptome can be leveraged to help identify DD Mtb in sputa without labor-intensive, months-long limiting dilution assays and potentially aid in assessing treatment response. If this could be tied to relapse-free outcome, then there is the potential to distinguish those cured with therapy shorter than the standard 6 months from those who require longer treatment. Screens for drugs effective against in vitro-generated DD Mtb have begun (14, 15) and might identify regimens that are more quickly and uniformly sterilizing.
Materials and Methods
Study Design
To explore the mechanisms of Mtb entry into the DD state, we used unbiased transcriptomic and transposon-sequencing techniques, and tested hypotheses using Mtb mutants with altered DD phenotypes and mycobacteria unable to enter the DD state in the same conditions. We used distinct reporters to quantify oxidative stress and oxidative damage, studied alternative methods of DD Mtb creation, and compared the growth rate and the replicative recovery of DD Mtb generated in vitro and those detected in an earlier study of patient sputa. “Biological replicates” refers to test samples initiated from the same LP culture but processed separately for exposure to an experimental stress, such as starvation. “Technical replicates,” such as those within an MPN plate, come from a single biological replicate, from which multiple samples were independently collected for quantification.
Statistical Analysis
Unless noted otherwise, statistical analyses were performed using Prism 8 (GraphPad) as detailed in the figure legends and results. Mixed-effects analyses were performed instead of two-way ANOVA when data were missing from contamination or other accidental loss of the sample. MPN estimations and CFU counts underwent log10 transformation prior to statistical analysis. Means ± SEM are shown when individual points are not plotted. Methods of statistical analysis are shown in the figure legends or the text. For RNA-Seq, differentially expressed genes were identified using the DESeq2 R package (87). Subsequent analysis of gene expression was performed in R (88, 89). For TnSeq, statistically significant differences in reads were identified using the Zero-Inflated Negative Binomial (ZINB) method by TRANSIT software v.3.0.1 (30). This allows simultaneous comparison of multiple variables, enabling appropriate q-value calculation. Data were normalized by trimmed total reads (TTR) normalization and adjusted for batch effects. Comparison between liquid and solid outgrowths for the RIF exposed cells was performed using a resampling method with TTR normalization and LOESS correction. Flow cytometry data were analyzed using FCS Express 7 Research Edition (De Novo Software). Gates were set two-dimensionally in unstained samples, using side scatter (SSC) versus either CellROX or TUNEL dot plots. To help ensure uniformity in analysis, gates were set to include the top 0.5 to 1.5% of unstained samples.
Supplementary Material
Acknowledgments
We thank Richard Ebright and his lab at Rutgers University and Fatouhi Nader of the TB Alliance for providing the Nα-aroyl-N-aryl-phenylalaninamides; William R Jacobs, Jr. and his lab at the Albert Einstein College of Medicine for providing the knockout strains for sigma factor, mshA, and RD1; Michael Glickman (Sloan Kettering Institute) for DNA repair mutants; Deborah Hung (Broad Institute) for transposon mutants; Amit Singh (Indian Institute of Science) for the Mrx1-roGFP2 plasmid; and Tanya Parish (Infectious Disease Research Institute) for a RIF-FITC conjugate molecular synthesis protocol. Creation of the transposon library was supported by the Broad Institute Tuberculosis donor group and the Pershing Square Foundation. We acknowledge Nitin Baliga, Eliza Peterson, and Mario Arrieta-Ortiz at the Institute for Systems Biology for their advice on RNAseq. We thank Jamie Bean (Sloan Kettering Institute) for his support in RNAseq data analysis. We thank Sabine Ehrt for assistance in transposon sequencing and providing knockout strains for prcBA; Kyu Rhee and his lab for sharing the technology of stripping outer lipids from the Mtb cell wall and advice regarding the desiccation model; Tania Lupoli for the clpB knockout strain; Kristin Burns-Huang, Sabine Ehrt, Carolina Trujillo, Selin Somerson-Karakaya, and Claire Frances Healy for useful discussions and Daniel Fitzgerald, Kathrine McAulay, and Les Centres GHESKIO for unpublished, de-identified data from a prior study. We acknowledge the help of the Weill Cornell Flow Cytometry core and the MSKCC Integrated Genomics Operation core and Peter Lopez, Michael Gregory, Kamilah Ryan, and Ludovic Desvignes for cell sorting technologies at NYU Langone’s Cytometry and Cell Sorting Laboratory, which is supported in part by grant P30CA016087 from the National Institutes of Health/National Cancer Institute. We thank the Jeffrey Aubé lab and Kelin Li at University of North Carolina for chemical synthesis support. We also thank Lee Riley (UC Berkeley School of Public Health) for sharing strains for which results were not included.
Funding
This work was supported by the Tri-Institutional TB Research Unit via National Institutes of Health grant U19 AI111143 (to CN), the Abby and Howard Milstein Program in Chemical Biology and Translational Medicine, National Institutes of Health grant T32 AI07613–19 (to CB), the Kellen Foundation Fund for the Future (to CB), and National Institutes of Health grant K08 AI139360 (to KS). The Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
Declaration of Interests
CN serves on advisory bodies for Tres Cantos Open Lab Foundation, Tri-Institutional Therapeutics Discovery Institute, Bridge Medicines, Pfizer External Sciences and Innovation, and Leap Therapeutics. All other authors declare no conflicts of interest related to this work.
Data Availability
All data associated with this study are in the paper or supplementary materials. Raw RNAseq and TnSeq data are available in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers: GSE180792 (TnSeq), GSE180658 (M. bovis RNAseq), and GSE180505 (Mtb RNAseq).
References
- 1.Global tuberculosis report 2020 (World Health Organization, Geneva, Switzerland, 2020). [Google Scholar]
- 2.Gillespie SH, Crook AM, McHugh TD, Mendel CM, Meredith SK, Murray SR, Pappas F, Phillips PPJ, Nunn AJ, Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. New England Journal of Medicine 371, 1577–1587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merle CS, Fielding K, Sow OB, Gninafon M, Lo MB, Mthiyane T, Odhiambo J, Amukoye E, Bah B, Kassa F, N’Diaye A, Rustomjee R, de Jong BC, Horton J, Perronne C, Sismanidis C, Lapujade O, Olliaro PL, Lienhardt C, A four-month gatifloxacin-containing regimen for treating tuberculosis. New England Journal of Medicine 371, 1588–1598 (2014). [DOI] [PubMed] [Google Scholar]
- 4.McCune RM, Feldmann FM, Lambert HP, McDermott W, Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J Exp Med 123, 445–468 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR, Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am J Respir Crit Care Med 181, 174–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shleeva MO, Bagramyan K, Telkov MV, Mukamolova GV, Young M, Kell DB, Kaprelyants AS, Formation and resuscitation of “non-culturable” cells of Rhodococcus rhodochrous and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology (Reading) 148, 1581–1591 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Scanga CA, Mohan VP, Joseph H, Yu K, Chan J, Flynn JL, Reactivation of latent tuberculosis: variations on the Cornell murine model. Infect Immun 67, 4531–4538 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pai SR, Actor JK, Sepulveda E, Hunter RL Jr., Jagannath C, Identification of viable and non-viable Mycobacterium tuberculosis in mouse organs by directed RT-PCR for antigen 85B mRNA. Microb Pathog 28, 335–342 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Nathan C, Taming tuberculosis: a challenge for science and society. Cell Host Microbe 5, 220–224 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Dhillon J, Fourie PB, Mitchison DA, Persister populations of Mycobacterium tuberculosis in sputum that grow in liquid but not on solid culture media. J Antimicrob Chemother 69, 437–440 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Chengalroyen MD, Beukes GM, Gordhan BG, Streicher EM, Churchyard G, Hafner R, Warren R, Otwombe K, Martinson N, Kana BD, Detection and quantification of differentially culturable tubercle bacteria in sputum from patients with tuberculosis. Am J Respir Crit Care Med 194, 1532–1540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosser A, Pareek M, Turapov O, Wiselka MJ, Mukamolova GV, Differentially culturable tubercule bacilli are generated during nonpulmonary tuberculosis infection. Am J Respir Crit Care Med 197, 818–821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAulay K, Saito K, Warrier T, Walsh KF, Mathurin LD, Royal-Mardi G, Lee MH, Ocheretina O, Pape JW, Fitzgerald DW, Nathan CF, Differentially detectable Mycobacterium tuberculosis cells in sputum from treatment-naive subjects in Haiti and their proportionate increase after initiation of treatment. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito K, Warrier T, Somersan-Karakaya S, Kaminski L, Mi J, Jiang X, Park S, Shigyo K, Gold B, Roberts J, Weber E, Jacobs WR Jr., Nathan CF, Rifamycin action on RNA polymerase in antibiotic-tolerant Mycobacterium tuberculosis results in differentially detectable populations. Proc Natl Acad Sci U S A 114, E4832–E4840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan SR, Venugopal U, Chandra G, Bharti S, Maurya RK, Krishnan MY, Effect of various drugs on differentially detectable persisters of Mycobacterium tuberculosis generated by long-term lipid diet. Tuberculosis (Edinb) 115, 89–95 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Gefen O, Ronin I, Bar-Meir M, Balaban NQ, Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Schrader SM, Vaubourgeix J, Nathan C, Biology of antimicrobial resistance and approaches to combat it. Sci Transl Med 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desnues B, Cuny C, Gregori G, Dukan S, Aguilaniu H, Nystrom T, Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep 4, 400–404 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arana I, Muela A, Iriberri J, Egea L, Barcina I, Role of hydrogen peroxide in loss of culturability mediated by visible light in Escherichia coli in a freshwater ecosystem. Appl Environ Microbiol 58, 3903–3907 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballesteros M, Fredriksson A, Henriksson J, Nystrom T, Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J 20, 5280–5289 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong Y, Zeng J, Wang X, Drlica K, Zhao X, Post-stress bacterial cell death mediated by reactive oxygen species. Proceedings of the National Academy of Sciences 116, 10064–10071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBee ME, Chionh YH, Sharaf ML, Ho P, Cai MW, Dedon PC, Production of superoxide in bacteria is stress- and cell state-dependent: a gating-optimized flow cytometry method that minimizes ROS measurement artifacts with fluorescent dyes. Front Microbiol 8, 459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, Bhave D, Kumar D, Carroll KS, Singh A, Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog 10, e1003902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra R, Kohli S, Malhotra N, Bandyopadhyay P, Mehta M, Munshi M, Adiga V, Ahuja VK, Shandil RK, Rajmani RS, Seshasayee ASN, Singh A, Targeting redox heterogeneity to counteract drug tolerance in replicating Mycobacterium tuberculosis. Sci Transl Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bansal-Mutalik R, Nikaido H, Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proceedings of the National Academy of Sciences 111, 4958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupuy P, Howlader M, Glickman MS, A multilayered repair system protects the mycobacterial chromosome from endogenous and antibiotic-induced oxidative damage. Proceedings of the National Academy of Sciences, 2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE 3rd, The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem 279, 40174–40184 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Nandakumar M, Nathan C, Rhee KY, Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nature Communications 5, 4306 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Eoh H, Rhee KY, Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America 110, 6554–6559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramaniyam S, Zaveri A, DeJesus MA, Smith C, Baker RE, Ehrt S, Schnappinger D, Sassetti CM, Ioerger TR, Statistical analysis of variability in TnSeq data across conditions using zero-inflated negative binomial regression. BMC Bioinformatics 20, 603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nambi S, Long JE, Mishra BB, Baker R, Murphy KC, Olive AJ, Nguyen HP, Shaffer SA, Sassetti CM, The oxidative stress network of Mycobacterium tuberculosis reveals coordination between radical detoxification systems. Cell Host Microbe 17, 829–837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE 3rd, The stringent response of Mycobacterium tuberculosis is required for long-term survival. Journal of bacteriology 182, 4889–4898 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss LA, Stallings CL, Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol 195, 5629–5638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gold B, Roberts J, Ling Y, Quezada LL, Glasheen J, Ballinger E, Somersan-Karakaya S, Warrier T, Warren JD, Nathan C, Rapid, semiquantitative assay to discriminate among compounds with activity against replicating or nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 59, 6521–6538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y, Coates AR, Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol 181, 469–476 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I, Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Molecular Microbiology 31, 715–724 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Lee JH, Karakousis PC, Bishai WR, Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J Bacteriol 190, 699–707 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontan PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I, The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol 191, 5628–5633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K, Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Molecular Microbiology 43, 717–731 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C, Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. Journal of Bacteriology 178, 1990–1995 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchmeier NA, Newton GL, Fahey RC, A mycothiol synthase mutant of Mycobacterium tuberculosis has an altered thiol-disulfide content and limited tolerance to stress. Journal of Bacteriology 188, 6245–6252 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchmeier NA, Newton GL, Koledin T, Fahey RC, Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Molecular Microbiology 47, 1723–1732 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C, Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295, 1073–1077 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Shi S, Ehrt S, Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infect Immun 74, 56–63 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin W, Mandal S, Degen D, Liu Y, Ebright YW, Li S, Feng Y, Zhang Y, Mandal S, Jiang Y, Liu S, Gigliotti M, Talaue M, Connell N, Das K, Arnold E, Ebright RH, Structural basis of Mycobacterium tuberculosis transcription and transcription inhibition. Mol Cell 66, 169–179 e168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR, Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis 187, 117–123 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR, Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Molecular Microbiology 51, 359–370 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zumarraga M, Bigi F, Alito A, Romano MI, Cataldi A, A 12.7 kb fragment of the Mycobacterium tuberculosis genome is not present in Mycobacterium bovis. Microbiology 145 ( Pt 4), 893–897 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Barczak AK, Avraham R, Singh S, Luo SS, Zhang WR, Bray MA, Hinman AE, Thompson M, Nietupski RM, Golas A, Montgomery P, Fitzgerald M, Smith RS, White DW, Tischler AD, Carpenter AE, Hung DT, Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog 13, e1006363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fay A, Glickman MS, An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genet 10, e1004516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lupoli TJ, Fay A, Adura C, Glickman MS, Nathan CF, Reconstitution of a Mycobacterium tuberculosis proteostasis network highlights essential cofactor interactions with chaperone DnaK. Proc Natl Acad Sci U S A 113, E7947–E7956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S, Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J Biol Chem 279, 20594–20606 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS, Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol 12, 304–312 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Dickinson JM, Jackett PS, Mitchison DA, The effect of pulsed exposures to rifampin on the uptake of uridine- 14 C by Mycobacterium tuberculosis. Am Rev Respir Dis 105, 519–527 (1972). [DOI] [PubMed] [Google Scholar]
- 55.Chan C-Y, Au-Yeang C, Yew W-W, Hui M, Cheng AFB, Postantibiotic effects of antituberculosis agents alone and in combination. Antimicrobial Agents and Chemotherapy 45, 3631 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manina G, Dhar N, McKinney JD, Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17, 32–46 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Caldwell ME, Viability of Mycobacterium tuberculosis in a semi-arid environment. The Journal of Infectious Diseases 37, 465–472 (1925). [Google Scholar]
- 58.Rogers JB, Studies on the viability of the tubercle bacillus. American Journal of Public Health 10, 345–347 (1920). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shleeva MO, Kudykina YK, Vostroknutova GN, Suzina NE, Mulyukin AL, Kaprelyants AS, Dormant ovoid cells of Mycobacterium tuberculosis are formed in response to gradual external acidification. Tuberculosis (Edinb) 91, 146–154 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Salina EG, Waddell SJ, Hoffmann N, Rosenkrands I, Butcher PD, Kaprelyants AS, Potassium availability triggers Mycobacterium tuberculosis transition to, and resuscitation from, non-culturable (dormant) states. Open Biol 4, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dukan S, Nystrom T, Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev 12, 3431–3441 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aldsworth TG, Sharman RL, Dodd CE, Stewart GS, A competitive microflora increases the resistance of Salmonella typhimurium to inimical processes: evidence for a suicide response. Appl Environ Microbiol 64, 1323–1327 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ayrapetyan M, Williams T, Oliver JD, Relationship between the viable but nonculturable state and antibiotic persister cells. J Bacteriol 200, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR, Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol 8, 313–323 (1982). [DOI] [PubMed] [Google Scholar]
- 65.Liao X, Liu D, Ding T, Nonthermal plasma induces the viable-but-nonculturable state in Staphylococcus aureus via metabolic suppression and the oxidative stress response. Appl Environ Microbiol 86, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abe A, Ohashi E, Ren H, Hayashi T, Endo H, Isolation of a viable but non-culturable suppression mutant of Vibrio vulnificus: role of antioxidant enzymes in surviving stationary phase and low temperatures. Fisheries Science 72, 656–664 (2006). [Google Scholar]
- 67.Oh E, McMullen L, Jeon B, Impact of oxidative stress defense on bacterial survival and morphological change in Campylobacter jejuni under aerobic conditions. Frontiers in Microbiology 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruhn-Olszewska B, Szczepaniak P, Matuszewska E, Kuczynska-Wisnik D, Stojowska-Swedrzynska K, Moruno Algara M, Laskowska E, Physiologically distinct subpopulations formed in Escherichia coli cultures in response to heat shock. Microbiol Res 209, 33–42 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Bogosian G, Bourneuf EV, A matter of bacterial life and death. EMBO Rep 2, 770–774 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mok WWK, Brynildsen MP, Timing of DNA damage responses impacts persistence to fluoroquinolones. Proc Natl Acad Sci U S A 115, E6301–E6309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casanova JL, Jouanguy E, Lamhamedi S, Blanche S, Fischer A, Immunological conditions of children with BCG disseminated infection. Lancet 346, 581 (1995). [DOI] [PubMed] [Google Scholar]
- 72.de Beaucoudrey L, Samarina A, Bustamante J, Cobat A, Boisson-Dupuis S, Feinberg J, Al-Muhsen S, Janniere L, Rose Y, de Suremain M, Kong XF, Filipe-Santos O, Chapgier A, Picard C, Fischer A, Dogu F, Ikinciogullari A, Tanir G, Al-Hajjar S, Al-Jumaah S, Frayha HH, AlSum Z, Al-Ajaji S, Alangari A, Al-Ghonaium A, Adimi P, Mansouri D, Ben-Mustapha I, Yancoski J, Garty BZ, Rodriguez-Gallego C, Caragol I, Kutukculer N, Kumararatne DS, Patel S, Doffinger R, Exley A, Jeppsson O, Reichenbach J, Nadal D, Boyko Y, Pietrucha B, Anderson S, Levin M, Schandene L, Schepers K, Efira A, Mascart F, Matsuoka M, Sakai T, Siegrist CA, Frecerova K, Bluetters-Sawatzki R, Bernhoft J, Freihorst J, Baumann U, Richter D, Haerynck F, De Baets F, Novelli V, Lammas D, Vermylen C, Tuerlinckx D, Nieuwhof C, Pac M, Haas WH, Muller-Fleckenstein I, Fleckenstein B, Levy J, Raj R, Cohen AC, Lewis DB, Holland SM, Yang KD, Wang X, Wang X, Jiang L, Yang X, Zhu C, Xie Y, Lee PP, Chan KW, Chen TX, Castro G, Natera I, Codoceo A, King A, Bezrodnik L, Di Giovani D, Gaillard MI, de Moraes-Vasconcelos D, Grumach AS, da Silva Duarte AJ, Aldana R, Espinosa-Rosales FJ, Bejaoui M, Bousfiha AA, Baghdadi JE, Ozbek N, Aksu G, Keser M, Somer A, Hatipoglu N, Aydogmus C, Asilsoy S, Camcioglu Y, Gulle S, Ozgur TT, Ozen M, Oleastro M, Bernasconi A, Mamishi S, Parvaneh N, Rosenzweig S, Barbouche R, Pedraza S, Lau YL, Ehlayel MS, Fieschi C, Abel L, Sanal O, Casanova JL, Revisiting human IL-12Rbeta1 deficiency: a survey of 141 patients from 30 countries. Medicine (Baltimore) 89, 381–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boisson-Dupuis S, The monogenic basis of human tuberculosis. Human Genetics, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darrah PA, Zeppa JJ, Maiello P, Hackney JA, Wadsworth MH 2nd, Hughes TK, Pokkali S, Swanson PA 2nd, Grant NL, Rodgers MA, Kamath M, Causgrove CM, Laddy DJ, Bonavia A, Casimiro D, Lin PL, Klein E, White AG, Scanga CA, Shalek AK, Roederer M, Flynn JL, Seder RA, Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature 577, 95–102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Netea MG, Giamarellos-Bourboulis EJ, Dominguez-Andres J, Curtis N, van Crevel R, van de Veerdonk FL, Bonten M, Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 Infection. Cell 181, 969–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sabio YGJ, Bigi MM, Klepp LI, Garcia EA, Blanco FC, Bigi F, Does Mycobacterium bovis persist in cattle in a non-replicative latent state as Mycobacterium tuberculosis in human beings? Vet Microbiol 247, 108758 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Bigi MM, Blanco FC, Araujo FR, Thacker TC, Zumarraga MJ, Cataldi AA, Soria MA, Bigi F, Polymorphisms of 20 regulatory proteins between Mycobacterium tuberculosis and Mycobacterium bovis. Microbiol Immunol 60, 552–560 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Sebastian J, Swaminath S, Nair RR, Jakkala K, Pradhan A, Ajitkumar P, De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob Agents Chemother 61, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Y, Mangan JA, Dhillon J, Sole KM, Mitchison DA, Butcher PD, Coates AR, Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriol 182, 6358–6365 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grumbach F, Canetti G, Le Lirzin M, Rifampicin in daily and intermittent treatment of experimental murine tuberculosis, with emphasis on late results. Tubercle 50, 280–293 (1969). [PubMed] [Google Scholar]
- 81.Dickinson JM, Mitchison DA, Suitability of rifampicin for intermittent administration in the treatment of tuberculosis. Tubercle 51, 82–94 (1970). [DOI] [PubMed] [Google Scholar]
- 82.Loring WW, Melvin I, Vandiviere HM, Willis HS, The death and resurrection of the tubercle bacillus. Trans Am Clin Climatol Assoc 67, 132–138 (1955). [PMC free article] [PubMed] [Google Scholar]
- 83.Bloom BR, McKinney JD, The death and resurrection of tuberculosis. Nature Medicine 5, 872–874 (1999). [DOI] [PubMed] [Google Scholar]
- 84.Beltran CGG, Heunis T, Gallant J, Venter R, du Plessis N, Loxton AG, Trost M, Winter J, Malherbe ST, Kana BD, Walzl G, Investigating non-sterilizing cure in TB patients at the end of successful anti-TB therapy. Front Cell Infect Microbiol 10, 443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bowness R, Boeree MJ, Aarnoutse R, Dawson R, Diacon A, Mangu C, Heinrich N, Ntinginya NE, Kohlenberg A, Mtafya B, Phillips PP, Rachow A, Plemper van Balen G, Gillespie SH, The relationship between Mycobacterium tuberculosis MGIT time to positivity and cfu in sputum samples demonstrates changing bacterial phenotypes potentially reflecting the impact of chemotherapy on critical sub-populations. J Antimicrob Chemother 70, 448–455 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Su H, Lin K, Tiwari D, Healy C, Trujillo C, Liu Y, Ioerger TR, Schnappinger D, Ehrt S, Genetic models of latent tuberculosis in mice reveal differential influence of adaptive immunity. JEM 218, e20210332 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.R: A language and environment for statistical computing (R Core Team, Vienna, Austria, 2016). [Google Scholar]
- 89.Kolde R, pheatmap: Pretty Heatmaps. R package version 1.0.8 (2015). [Google Scholar]
- 90.Andrews S, FastQC: a quality control tool for high throughput sequence data (2010). [Google Scholar]
- 91.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome S Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 94.Long JE, DeJesus M, Ward D, Baker RE, Ioerger T, Sassetti CM, Identifying essential genes in Mycobacterium tuberculosis by global phenotypic profiling. Methods Mol Biol 1279, 79–95 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Larsen MH, Biermann K, Tandberg S, Hsu T, Jacobs WR Jr, Genetic manipulation of Mycobacterium tuberculosis. Current protocols in microbiology 6, 10A. 12.11–10A. 12.21 (2007). [DOI] [PubMed] [Google Scholar]
- 96.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR, TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11, e1004401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or supplementary materials. Raw RNAseq and TnSeq data are available in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers: GSE180792 (TnSeq), GSE180658 (M. bovis RNAseq), and GSE180505 (Mtb RNAseq).


