Abstract
The KEAP1/NRF2 pathway plays a physiologic protective role against xenobiotics and reactive oxygen species. However, activation of NRF2 provides a powerful selective advantage for tumors by rewiring metabolism to enhance proliferation, suppress various forms of stress, and promote immune evasion. Genetic, epigenetic and post-translational alterations that activate the KEAP1/NRF2 pathway are found in multiple solid tumors. Emerging clinical data highlights that alterations in this pathway results in resistance to multiple therapies. Here, we provide an overview of how dysregulation of the KEAP1/NRF2 pathway in cancer contributes to several hallmarks of cancer that promote tumorigenesis and leads to treatment resistance.
INTRODUCTION
The transcription factor nuclear factor erythroid 2-related factor 2 (NFE2L2, NRF2) plays a pivotal role in cellular physiology and tumorigenesis. The canonical role of NRF2 is to shift cellular metabolism to maintain redox balance. NRF2 protein levels are negatively regulated by the ubiquitin ligase scaffold protein Kelch-like-ECH associated protein 1 (KEAP1), which binds to NRF2 and facilitates its ubiquitination and proteasomal degradation (1,2). In the presence of cytotoxic oxidative stress, NRF2 accumulates and translocates to the nucleus where it regulates the transcription of a plethora of genes that promote antioxidant defenses (Figure 1) (2–7). In addition to its role in maintaining cellular redox homeostasis, NRF2 plays crucial roles in regulation of immune responses (8) and drug detoxification (9–11). The KEAP1/NRF2 pathway is genetically, epigenetically, and post-transcriptionally altered in multiple cancers including lung, breast, liver, esophageal, and pancreatic cancer (6,12–18). Mutually exclusive loss-of-function (LOF) mutations in KEAP1 or gain-of-function (GOF) mutations in NRF2, both of which result in stabilization of NRF2, have been identified to promote both tumorigenesis and resistance to multiple therapies (14,18–21). Alterations in this pathway also lead to metabolic vulnerabilities that can be exploited therapeutically. This review provides an overview of the KEAP1/NRF2 pathway in normal physiology, its importance in tumor development, cancer metabolism, mechanisms of treatment resistance, and novel therapeutic strategies to target tumors with NRF2 activation.
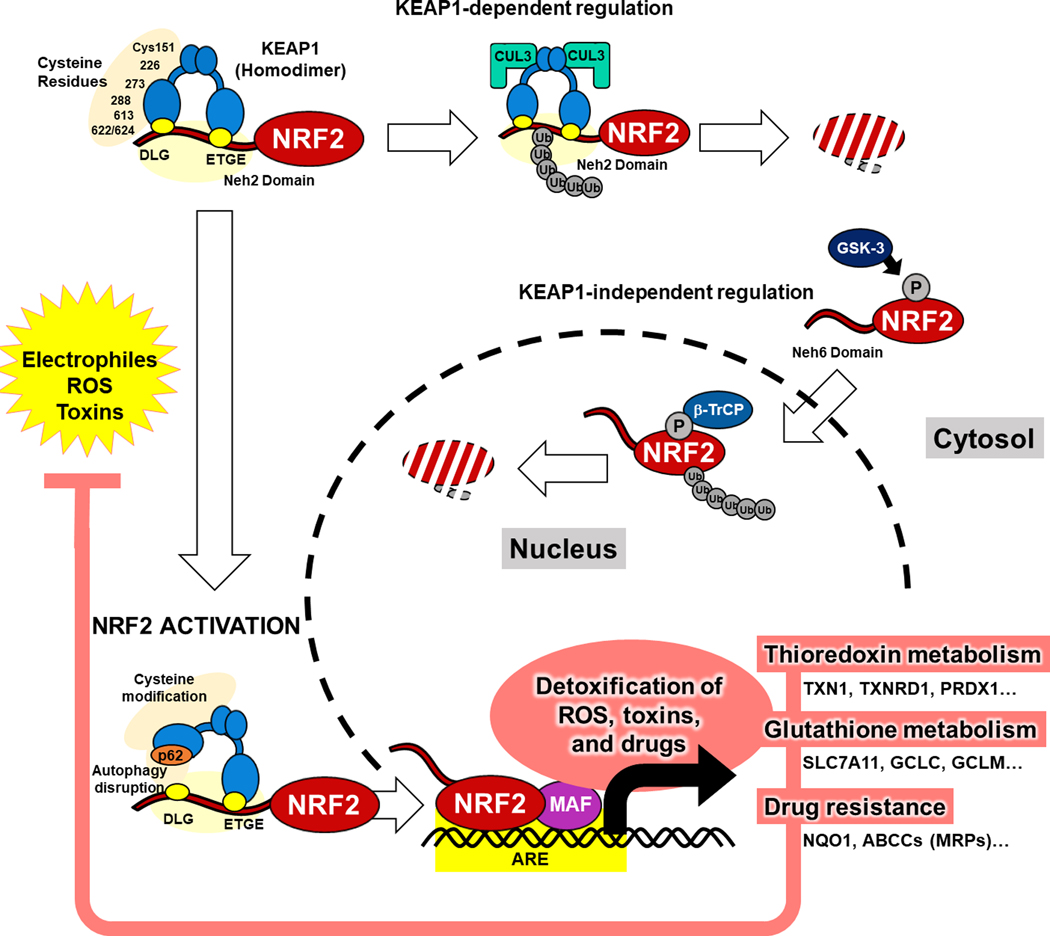
Figure 1: Physiological Activation and Regulation of NRF2.
Under basal conditions, NRF2 is bound by KEAP1 via the DLG and ETGE motifs in the Neh2 domain of NRF2 in cytosol and leads to binding of CUL3, poly-ubiquitination and proteasomal degradation. NRF2 is also regulated by KEAP1-indepent mechanisms via phosphorylation of the Neh6 domain by GSK-3 and proteasomal degradation by β-TrCP. Reactive oxygen species (ROS), drugs, and toxins react with cysteine residues on KEAP1 resulting in structural changes and the accumulation of NRF2 to translocate to the nucleus and function as a transcriptional factor. In the nucleus, NRF2 heterodimerizes with small MAF proteins and binds to antioxidant response elements to induce a series of target genes for detoxification of ROS, toxins, and drugs.
KEAP1, Kelch-like ECH-associated protein 1; NRF2, nuclear factor erythroid 2-related factor 2; β-TrCP, beta-transducin repeat containing protein; GSK-3, glycogen synthase kinase 3; Ub, ubiquitin; ARE, antioxidant response element; MAF, musculoaponeurotic fibrosarcoma; TXN, thioredoxin; TXNRD1, thioredoxin reductase 1; PRDX1, peroxiredoxin-1; GCLC, glutamate-cysteine ligase catalytic subunit; GCLM, glutamate-cysteine ligase modifier subunit; NQO1, NADPH-quinone Dehydrogenase 1; ABC, ATP-binding cassette
PHYSIOLOGIC IMPORTANCE OF THE KEAP1/NRF2 PATHWAY
The Yamamoto group was the first to describe the NRF2 transcriptional factor and its cytoprotective role through the regulation of a battery of genes that protect cells from toxins, drugs, and other toxic xenobiotics (4,22). Since then, multiple groups have evaluated the physiologic role of the KEAP1/NRF2 pathway using a series of genetically engineered mouse models (GEMMs). Although NRF2 loss has no gross impact on normal mouse development (23), the importance of NRF2 becomes apparent under various forms of environmental stress. Iizuka et al. used NRF2-null mice to demonstrate increased susceptibility to cigarette smoke-induced emphysema (24). Beyond cigarette exposure, NRF2-null mice and rats have increased susceptibility to multiple insults and the development of diverse pathologies (25–31).
To assess the impact of constitutive NRF2 activation, Wakabayashi et al. developed Keap1 knockout mice (32). At birth Keap1-null animals are normal in size but fail to survive past day 21 due to hyperkeratosis of the esophagus and forestomach that causes gastric obstruction and death (32). Critically, loss of Nrf2 in the context of Keap1 loss prevents the development of hyperkeratosis and death, demonstrating the epistatic relationship of KEAP1 and NRF2. To dissect the physiologic importance of NRF2 across tissues multiple studies have performed tissue-specific deletion of Nrf2 or achieved NRF2 activation through Keap1 deletion. Okawa et al. showed that hepatocyte-specific Keap1 loss was associated with increased tolerance to acetaminophen toxicity due to NRF2 activation and drug detoxification (33). NRF2 can suppress immune responses in multiple mouse models. In M1 macrophages, NRF2 suppresses proinflammatory cytokines by blocking transcription of Il6 and Il1β (8,34). The significance of tissue-specific deletion of Keap1 has been shown to regulate cell differentiation (35). Moreover, tissue-specific activation of NRF2 reduces tissue damage in sickle cell disease mouse models (36) and the development of type 1 diabetes in a non-obese diabetic mouse model (37). Although global loss of Nrf2 has minimal effects under homeostasis, in the context of cytotoxic stress, tissue-specific NRF2 activity and downstream biological consequences become evident.
PHYSIOLOGIC ACTIVATION OF NRF2 AND DYSREGULATION IN CANCER
Structural Features of KEAP1 and NRF2
Given the frequency of NRF2/KEAP1 pathway aberration in cancers (Figure 2A) the structural features of these proteins are important to comprehend how mutations disrupt interactions that lead to hyperactive NRF2 responses. Herein, we provide an overview of the structures of NRF2 and KEAP1, major domains, and the functions of these domains (Figure 2B and 2C). NRF2 is part of the Cap’ n’ collar (CNC) basic leucine zipper family of transcription factors. The CNC domain is a 43 amino acid sequence located at the N terminus of the DNA binding domain of this family which includes the transcription factors NRF1, NRF2, NRF3, BACH1, and BACH2 (38). The structure of NRF2 is broken down into 7 NRF2-ECH homology domains (Neh1-Neh7) (Figure 2C). The Neh1 domain mediates heterodimerization with the potein small musculoaponeurotic fibrosarcoma (sMAF) which together bind to DNA at designated Antioxidant Response Elements (ARE) with the sequence 5′-GTGACNNNGC-3′ (also called a CNC sMAF binding element – CsMBE) (39,40) (Figure 1). Neh1 also contains a nuclear localization signal while Neh5 has a redox sensitive nuclear export signal. The Neh2, 6, and 7 domains are involved in regulating NRF2 activity. Collectively, the Neh domains mediate specific protein-protein and protein-DNA interactions, resulting in the fine-tuning of the oxidative/xenobiotic stress signals governed by NRF2.The primary role of KEAP1 is to act as a substrate adaptor for the Cullin 3-RING E3 ubiquitin ligase complex. Each of the major domains of KEAP1 have an important role in this degradation process. KEAP1 contains five main domains (Figure 2B): The N-terminal region, Broad complex, Tramtrack and bric-à-brac (BTB) domain (41), intervening region (IVR), KELCH domain, and C-terminal region (Figure 2B). The KELCH domain contains six KELCH motif repeats that form six beta sheets (42). It is these KELCH repeats that bind to the Neh2 domain of NRF2 (43). Specifically, KEAP1 binds to two conserved degron motifs located in the Neh2 domain (44), ETGE and DLG (Figure 2C), with KEAP1 binding more strongly to the ETGE motif (45). The BTB domain facilitates the homodimerization of KEAP1 and binding to the Cullin 3-RING E3 ubiquitin ligase complex (46–49). This complex is formed by three components: Cullin 3 (CUL3), RING-box protein 1 (RBX-1), and E2. CUL3 binds a range of BTB-containing proteins including KEAP1 (Figure 1). These BTB proteins serve as substrate adaptors for the CUL3-RING E3 ligase complex. RBX-1 serves to bind E2 which has been conjugated to ubiquitin. Once the substrate is bound to this complex it is ubiquitinated and degraded. The IVR of KEAP1 plays an important role in redox homeostasis as this region contains key cysteine residues including C226, C273, and C288. These cysteine residues along with those in other domains seen in Figure 2B (C151, C613, and C622/624) are oxidized in the presence of electrophiles (50), reactive oxygen species (ROS) (51), and toxins (52). Modification of these cysteine residues results in a conformational change in KEAP1 impairing the degradation of NRF2 and enabling the accumulation of de novo NRF2 (53).
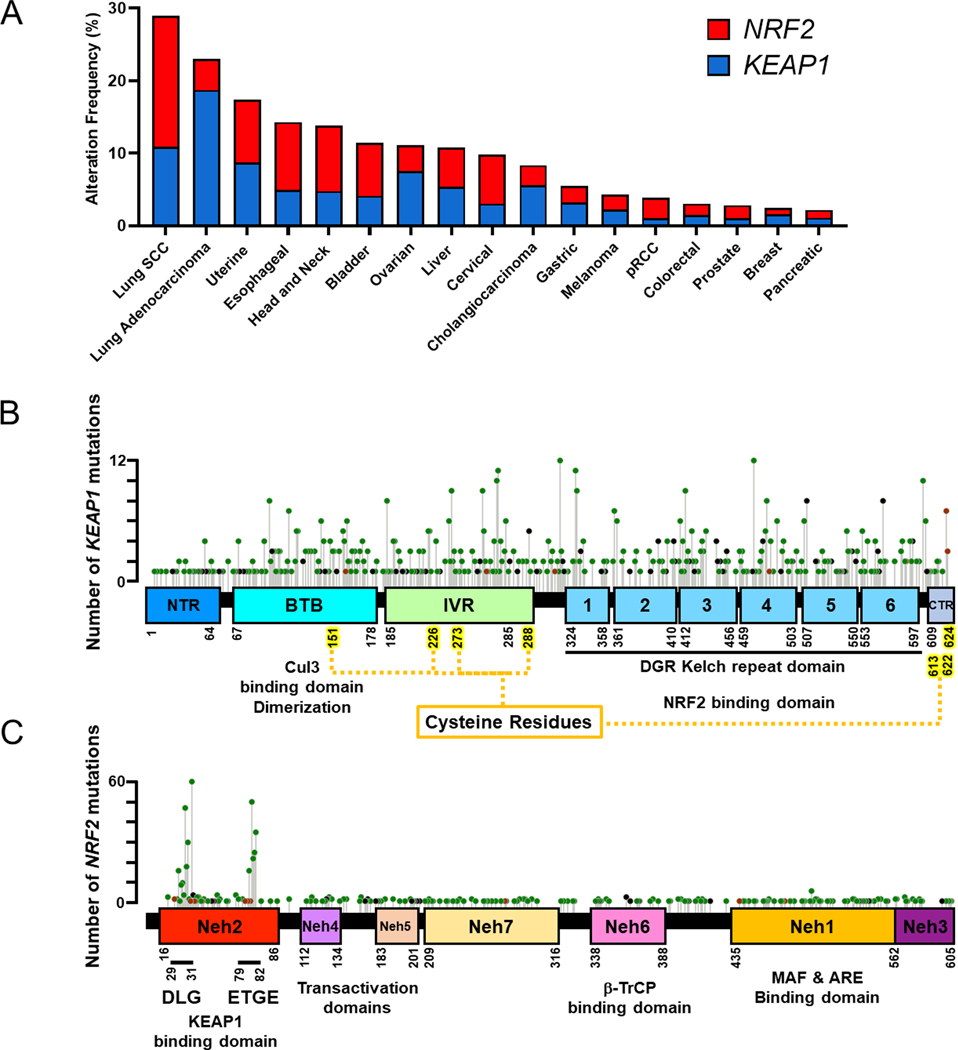
Figure 2: Mutation Spectrum in the KEAP1/NRF2 Pathway.
A) Frequency of mutations in NRF2 and KEAP1 in solid tumors generating using cBioportal TCGA data.
B) Map of KEAP1 with mutations based on cBioportal TCGA datasets. KEAP1 is divided up into the following domains: NTR, BTB, IVR, 6 Kelch domains, and CTR. Key cysteine residues for sensing ROS and toxins are indicated.
C) Map of NRF2 with 7 Neh domains and mutations labelled. The KEAP1 binding motifs, DLG and ETGE, are indicated in the Neh2 domain, while the loci of phosphorylation by β-TrCP is located in the Neh6 domain. Somatic mutations in NRF2 are highly concentrated in DLG and ETGE motifs.
SCC, squamous cell carcinoma; pRCC, papillary renal cell carcinoma; KEAP1, Kelch-like ECH-associated protein 1; NRF2, nuclear factor erythroid 2-related factor 2; NTR, N terminal region; BTB, Bric-a-brac; IVR, intervening region; DGR, double glycine repeat; CTR, C terminal region; Maf, musculoaponeurotic fibrosarcoma; ARE, antioxidant response element
Post-translational Regulation of NRF2
The sensory role of KEAP1 is enabled through cysteine residues that react with ROS (51) or electrophiles (50). Work by Wakabayashi et al. identified that reactive cysteine groups C273 and C278 respond to ROS and induce conformational changes in KEAP1 to promote NRF2 stabilization (54,55). In addition, there are a series of known electrophilic molecules capable of activating NRF2. Dinkova-Kostova et al. demonstrate that electrophiles such as sulforaphane react with cysteine to form disulfide links mainly at C257, C273, C288, and C297 (56). Other NRF2 inducers such as dexamethasone mesylate and tertiary butylhydroquinone (tBHQ) are known to react with cysteine residues on KEAP1 resulting in NRF2 stabilization (57,58). Similarly, McMahon et al. show that KEAP1 has three distinct sensor regions containing cysteines that react with Zn2+, nitrous oxide, or alkenals (59).
Multiple electrophilic metabolites, which can be aberrantly regulated in cancer, modify KEAP1 cysteine residues to alter KEAP1 conformation. This has been described especially in the context of the TCA (tricarboxylic acid) cycle–derived metabolites fumarate (60–63), itaconate (64) and glycolysis-derived metabolites such as methylglyoxal (57). Other metabolites such as polyunsaturated fatty acid alkenals can post-translationally stabilize NRF2. These findings suggest that KEAP1/NRF2 signaling respond not only to xenobiotic stressors and ROS but also to alteration in metabolites due to disruption of endogenous metabolism in cancer. The important role of metabolites in NRF2 activation and cancer risk is demonstrated by germline mutations of fumarate hydratase (FH) which results in the development of hereditary leiomyomatosis and renal cell carcinoma cancer syndrome (65). Inactivation of FH results in accumulation of the TCA cycle intermediate fumarate, which promotes NRF2 activation likely through succination of KEAP1 (61–63).
In addition to degradation through KEAP1-CUL3-ubiquitin ligase complex, NRF2 can also be degraded by the SKP1-CUL1 ubiquitin ligase complex. Chowdhry et al. show that NRF2 has two β-TrCP binding motifs –DSGIS and DSAPGS– in the Neh6 domain (66). Like KEAP1, β-TrCP acts as a substrate adaptor for CUL1 mediated degradation. Furthermore, binding at the DSGIS motif is enhanced by phosphorylation through glycogen synthase kinase-3 (GSK-3) (Figure 1). AKT inhibition results in an increase in GSK-3 activity, increased degradation of NRF2, and sensitivity to cisplatin in KEAP1-mutant A549 lung cancer cells (66). Activation of the PI3K/AKT mitogenic pathway mediates phosphorylation and inhibition of GSK-3 resulting in activation of NRF2. In breast cancer, where PI3K/AKT signaling is frequently dysregulated, NRF2 is post-translationally activated by PI3K/AKT to increase antioxidant capacity via the above mechanism (67).
Competitive Binding
The interaction between KEAP1 and NRF2 can be disrupted by proteins that compete with NRF2 from binding to KEAP1. For example, Ma et al. demonstrated that PALB2, which is known to bind and regulate the intranuclear localization and stability of BRCA2 (68), has an ETGE motif identical to NRF2 and competes for binding with KEAP1. Increased protein levels of PALB2 results in decreased KEAP1-dependent degradation of NRF2 (69). In a similar manner p62 (SQSTM1), an important component of autophagy and an NRF2 target, binds to KEAP1 through a KEAP1-interacting region (KIR) that competes with the ETGE motif of NRF2, resulting in inhibition of NRF2 degradation (70–72). Several other proteins are known to compete with NRF2 for binding to KEAP1, including BRCA1, p21, DPP3 (73,74). These competitive KEAP1 client proteins represent alternative non-oxidative stress related mechanisms by which NRF2 can be stabilized and thus feed multiple signaling inputs towards direct regulation of NRF2 and the downstream antioxidant response network.
Genetic Alterations in KEAP1/NRF2 pathway
Hyperactivation of NRF2 plays a critical role in multiple tumor types including lung, liver, gastric, ovarian, breast, and colorectal cancer (Figure 2A). Among them, activation of NRF2 is most extensively characterized in non-small cell lung cancer (NSCLC). Inactivating mutations in KEAP1 are located throughout the gene (Figure 2B). Hast et al. identified multiple classes of KEAP1 mutations that have differing mechanisms and degree of NRF2 activation (75). These somatic mutations result in KEAP1 LOF leading to stabilization and accumulation of NRF2 (76–81). Using an ARE-luciferase reporter, Hast demonstrated that these mutations enhance NRF2 activation. Interestingly, none of the mutations were in domains required for NRF2 binding (75). However, five of these mutations, including the more frequent G333C mutation, impaired NRF2 binding. Some of these mutations enhanced binding to NRF2, including R470C. Despite demonstrating increased NRF2 affinity, expression of KEAP1 super-binding mutants resulted in increased stability and nuclear localization of NRF2 with a concomitant increase in NRF2 transcriptional target genes. Furthermore, many KEAP1 mutations found in human lung tumors appear to be dominant negative mutations which establish varying degrees of NRF2 activation (44,82).
Kerins and Ooi provide a thorough review of key NRF2 mutations in cancer (83). Unlike KEAP1, activating gain-of-function mutations in NRF2 occur in specific hotspot regions corresponding to the ETGE and DLG domains (84) (Figure 2C). Mutations in NRF2 are more prevalent in lung squamous cell carcinoma while KEAP1 mutations are more frequent in lung adenocarcinoma (85–87). LOF mutations in KEAP1 and GOF mutations in NRF2 are generally mutually exclusive, suggesting that NRF2 activation is the main driver for selection of these mutations rather than other KEAP1-mediated effects. Work by Jamal-Hanjani et al. evaluated intra-tumoral heterogeneity and genomic evolution by analyzing multiple regions of lung tumors (88). They found that KEAP1 mutations are an early clonal driver mutation, emphasizing that mutations in KEAP1 are likely selected for during tumor initiation or progression.
Mutations in KEAP1 frequently co-occur with other mutations suggestive of cooperative events leading to selection. Notably, mutation in KRAS frequently co-occurs with KEAP1 mutation in lung adenocarcinoma (21). Furthermore, multiple patient cohorts have demonstrated that mutations in KEAP1 frequently co-occur with mutations in liver kinase B1 (LKB1/STK11) (21,89). Mutations in either KEAP1 or LKB1 are independently associated with unfavorable clinical outcomes (90). Interestingly, both genes are located on chromosome arm 19p along with SMARCA4, another frequently co-mutated gene with KEAP1 and LKB1 (90). While these mutations confer a survival advantage to tumors, they may provide the opportunity to therapeutically target genotype-specific vulnerabilities in these aggressive tumor subtypes.
Epigenetic Modification
KEAP1 can be epigenetically regulated, reflecting another mechanism to increase NRF2 levels. In vitro, Muscarella et al. found that 50% of NSCLC and 42% of small cell lung cancer cell lines had methylation in the promoter of KEAP1 and decreased expression (13). In their analysis of 47 patient NSCLC samples, methylation of KEAP1 was seen in 47% of cases (13). NRF2 activation as a result of KEAP1 promoter methylation has been association with poor patient outcomes in glioma, renal cell carcinoma, and colorectal cancer (12,91,92). An analysis of breast tumors found that KEAP1 promoter methylation was frequently observed and associated with ER positive HER2 negative tumors. Additionally, triple negative breast cancer patients with KEAP1 promoter hyper-methylation demonstrated a higher mortality (93).
Transcriptional Regulation
NRF2 can be regulated transcriptionally through multiple mechanisms. Interestingly, the promoter of Nrf2 contains an ARE sequence and therefore NRF2 can reinforce its own transcription (94). The aryl hydrocarbon receptor (AhR) has been shown to regulate Nrf2 by directly binding to xenobiotic response elements (XRE) found within the Nrf2 promoter (95). Wakabayashi et al. showed that Notch transcriptionally upregulates Nrf2. Notch-specific NRF2 regulation was shown to be important for liver development (96) as liver specific deletion of Nrf2 through Albumin-Cre results in reversal of both Notch induced hepatomegaly and increase in intrahepatic bile ducts (IHBD). Using mouse embryonic fibroblasts (MEFs), DeNicola et al. demonstrated that multiple oncogenes, including KrasG12D, BrafV619E, Myc transcriptionally upregulate Nrf2 to mitigate ROS (6). In breast cancer, BRCA1 is reported to regulate NRF2 through direct promoter binding (73,97) or via post-translational methods that lead to increased NRF2 expression (73).
Post-transcriptional Modifications
NRF2 levels can be regulated by various post-transcriptional mechanisms. Goldstein et al. identified a subset of tumors that had high expression of NRF2 and its transcriptional signatures without mutations in KEAP1 or NRF2 (98). They identified an NRF2 splice variant that skipped exon 2 which correspond to the Neh2 domain regulated by KEAP1 (98). In a series of in vitro experiments, they demonstrate that skipping of exon 2 resulted in NRF2 activation. This activity was not enhanced with knockdown of KEAP1 or impaired with KEAP1 overexpression, validating that the interaction between KEAP1 with NRF2 was lost through exon skipping.
Multiple microRNAs have also been found to regulate NRF2. Yang et al. identified that miR-28 negatively regulates Nrf2 transcripts (99). This same group also demonstrated that miR-200a negatively regulates Keap1 and thus stabilizes NRF2 in breast cancer cells (100,101). The microRNA miR-421 has been shown to reduce KEAP1 levels and is associated with resistance to paclitaxel in A549 lung cancer cells (102). Other microRNAs such as miR-155, miR-27a, miR142–5p, and miR144 have also been found to reduce NRF2 levels (103).
ROLE OF NRF2 IN TUMORIGENESIS
NRF2 has been shown to play a multi-faceted role at different stages of tumor development across several types of cancer. ROS accumulation promotes DNA damage leading to activation of oncogenes and inactivation of tumor suppressor genes, resulting in cellular transformation and tumor initiation. Many carcinogens used to initiate tumors in animals promote mutagenesis partly through buildup of ROS and DNA damage (104,105). Given the important role of NRF2 in suppressing ROS and drug detoxification, several studies have shown that NRF2 activation can suppress the formation of carcinogen-induced tumors. However, both cancer genomics and pre-clinical GEMM data suggest that activation of NRF2 through LOF mutations in KEAP1 or GOF mutations in NRF2 are associated with poor prognosis and more aggressive disease (79,106). Furthermore, there is evidence for tissue-specific protective roles of the KEAP1/NRF2 pathway in solid tumors driven by the same oncogenes. These observations suggest that NRF2 likely has a dual stage-specific pro-tumorigenic and anti-tumorigenic roles depending on the context. Here, we highlight the studies implicating the KEAP1/NRF2 pathway in tumorigenesis and discuss future work needed to clearly define the role of NRF2 in tumor initiation and progression across different malignancies.
To study carcinogenesis using mouse models, both chemical carcinogen-induced models or GEMMs have been employed. In some carcinogen-induced tumor models, NRF2 activation has been shown to suppress tumor initiation (31,107–109). Pharmacological activation of NRF2 by CDDO-Im, an NRF2 inducer currently in clinical trials for the treatment of diabetic kidney disease (110), suppressed tumorigenesis in a carcinogen-induced lung cancer model (111). In addition, germline Nrf2 knockout (KO) mice accelerated tumor formation in a vinyl carbamate-induced lung cancer model (112). Similarly, Nrf2 KO mice formed more lung tumors as compared to Nrf2 wild type mice upon exposure to urethane (105). Both the vinyl carbamate- and urethane-induced models acquire recurrent somatic mutations in known oncogenes, such as Hras and Kras, and Nrf2 activation in these models can suppress the initiation of carcinogen-induced tumors. Nrf2 KO mice also fail to develop tumors in carcinogen induced models of hepatocellular carcinoma and bladder cancer (113,114).
Based on genomic and clinical data of patients with lung cancer, it is clear that alterations in the KEAP1/NRF2 pathway are recurrent (21,76) and enriched in smokers and nearly absent in young never smokers (115,116). This correlation and the above pre-clinical findings have led to the hypothesis that smoking may induce NRF2 activation as a protective mechanism against damage induced by carcinogens to suppress tumor initiation. However, once an oncogenic driver mutation (e.g. KRAS) has occurred, high NRF2 expression may be selected for by protecting tumors from insults, supporting proliferative processes, and potentially suppressing anti-tumor immune responses.
DeNicola et al. were the first to demonstrate that Nrf2 is required for Kras-driven lung and pancreas tumorigenesis (6). They demonstrated that both Kras and Myc oncogenes constitutively increase Nrf2 transcription to elevate the basal activity of the antioxidant and cellular detoxification program required for tumorigenesis. Subsequent studies in Kras-driven pancreatic cancer models showed that Nrf2 KO repressed tumor formation (117). Using patient-derived organoid models, Chio et al. demonstrated that Nrf2 knockdown suppressed tumor initiation and maintenance (118). These studies support the idea that NRF2 is necessary for tumorigenesis in tumors with known oncogenic drivers such as Kras. Future studies using inducible approaches to suppress Nrf2 at initiation or progression will more precisely clarify the stage-specific requirement for Nrf2.
Genomic studies suggest that activation of the NRF2 pathway by genetic alterations in KEAP1/NRF2 play an important role in cancer development (14,78–80,106,119). The KEAP1/NRF2 axis in tumorigenesis has been most extensively studied in the setting of lung cancer. Romero et al. were the first to demonstrate the tumor suppressive role of KEAP1 in Kras-driven GEMMs of lung adenocarcinoma (120). Targeted somatic CRISPR/Cas9-based LOF of Keap1 resulted in increased tumor growth and histological grade (120). In addition, multiple studies have used conditional deletion of LOF point mutant (89,121–123) Keap1 alleles to demonstrate the tumors suppressive role of KEAP1 in Kras-driven lung adenocarcinoma. Furthermore, conditional Keap1 and Pten loss in the lung generated papillary lung adenomas, suggesting that PI3K/AKT pathway activation may cooperate with NRF2 activation in the absence of Kras alterations (124). Loss of Keap1 in basal cells also promotes the development of squamous cell carcinomas in the context of p53 loss (125).
A major caveat with some of the studies discussed above is that both the pharmacologic and genetic modulation of Nrf2 is systemic, which would also impact cells in the tumor microenvironment that play a role in tumor initiation and maintenance, including myeloid-derived cells whose differentiation and function is known to be regulated by NRF2 (8,28). Satoh et al. systematically tested the dual role of NRF2 activation in cancer development (126). Using a urethane-induced lung cancer model, they observed that Keap1 knockdown mice, with higher levels of NRF2, developed smaller tumors compared to wild-type mice. However, when tumors from these animals were transplanted into immunodeficient mice, Keap1 knockdown tumors grew faster than Keap1 wild-type tumors (126).
The role of NRF2 activation during tumorigenesis may also be tissue specific. Hamada et al. demonstrated that conditional loss of Keap1 in a Kras-driven pancreatic cancer GEMM leads to pancreatic atrophy and decreased tumor growth (127), while conditional loss of Keap1 in a Kras-driven lung cancer GEMM leads to more aggressive tumors (120). However, the dosage of NRF2 activation by Keap1 LOF likely also has a role in tumorigenesis (123). This is suggested given that human and mouse pancreatic tumors upregulate NRF2 in the setting of KRAS mutation but likely not to the extent seen in the setting of KEAP1 mutations (6).
Li et al. demonstrated that NRF2 activation not only leads to more aggressive tumors but may also play a role in driving histologic subtypes of NSCLC (128). Using a GEMM with conditional mutation of Kras and loss of Lkb1 that forms both adenocarcinoma and squamous lung carcinoma, they demonstrated that modulation of oxidative stress by overexpression of Nrf2 or treatment with N-acetyl cysteine (NAC) reduced the frequency of squamous tumors suggestive that ROS as a major driver of differentiation of tumors between lung adenocarcinomas vs squamous cell carcinomas (128). Interestingly, NRF2 is significantly upregulated in Type II endometrial cancer (serous and clear cell carcinomas) compared to Type I, suggesting that NRF2 is involved in driving histological subtypes of endometrial cancer (129).
In addition to the importance of the KEAP1/NRF2-pathway in tumor initiation and maintenance, NRF2 has been to shown to also promote metastases through its interaction with BACH1 (130). Accumulation of NRF2 in lung cancer causes the stabilization of BACH1 by induction of Heme Oxygenase 1 (HO-1), the enzyme that breaks down heme (131). Using a Kras-driven model, they demonstrate that loss of Keap1 or Fbxo22 induces metastasis in a BACH1-dependent manner. Furthermore, human metastatic lung cancers display higher levels of HO-1 and BACH1, which correlate with poor prognosis and increased incidence of metastasis in lung cancer patients. These findings were further supported by a study that demonstrated that dietary antioxidants lead to BACH1 stabilization and increased metastasis in the same Kras-driven adenocarcinoma model (132). Finally, dipeptidyl peptidase-4 inhibitors saxagliptin and sitagliptin can stabilize NRF2 resulting in an increased metastatic potential in xenograft models (133). This study also observed that increased NRF2 expression also correlated with increased metastasis in liver cancer. Overall, NRF2 activation by any number of mechanisms including somatic mutations, transcriptional regulation, or metabolic reprogramming-triggered modifications of KEAP1 promotes tumorigenesis.
NRF2 METABOLIC REWIRING
Redox Metabolism
NRF2 is traditionally known for its role in redox homeostasis through the regulation of glutathione and thioredoxin which are responsible for scavenging ROS, among other functions. Two genes critical for glutathione synthesis are transcriptionally regulated by NRF2: glutamate-cysteine ligase catalytic subunit (GCLC) and glutamate-cysteine ligase modifier subunit (GCLM) (134). GCLC catalyzes the reaction ligating glutamate and cysteine to form y-glutamyl cysteine while GCLM increases the affinity of GCLC for its substrates (Figure 3). Glutathione is then produced via glutathione synthetase using y-glutamyl cysteine and glycine as substrate and consuming ATP in the process. In addition, NRF2 transcriptionally regulates glutathione reductase (GSR) (135) and thioredoxin reductase 1 (TXNRD1) (136), both of which require NADPH generated by the pentose phosphate pathway (PPP), to reduce oxidized glutathione and thioredoxin (Figure 3).
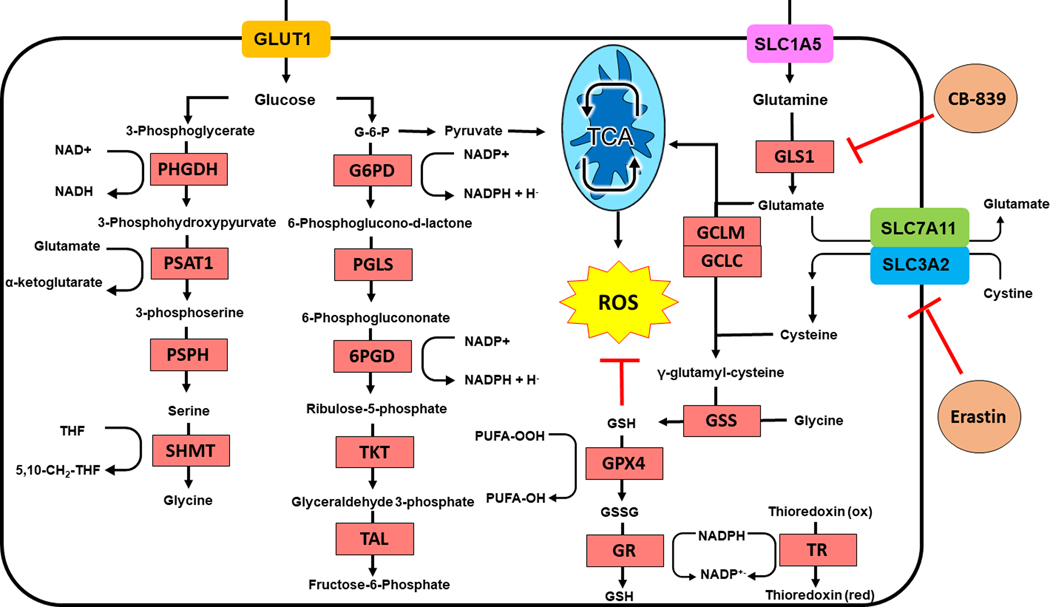
Figure 3: Metabolic Rewiring by NRF2.
Activation of NRF2 dramatically enhances generation of glutathione by increasing synthesis of glutathione from intracellular glutamate, cysteine, and glycine. Intracellular glutamate is derived from glutamine through GLS-1. Cystine is imported by the NRF2 target SLC7A11. Serine and glycine are synthesized via NRF2 dependent processes. NADPH is synthesized to support redox metabolism by the pentose phosphate pathway. GLS-1 and SLC7A11 function can be impaired by CB-839 and erastin respectively.
PHGD, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase; PSPH, phosphoserine phosphatase; SHMT, serine hydroxymethyltransferase; G6PD, glucose-6-phopshate dehydrogenase; PGLS, 6-phosphogluconolactonase; 6PGD, 6-phosphogluctonate dehydrogenase; TKT, transketolase; TAL, transaldolase; GPX4, glutathione peroxidase 4; GR, glutathione reductase; GLS1, glutaminase 1; GCLC, glutamate-cysteine ligase catalytic subunit; GCLM, glutamate-cysteine ligase modifier subunit; GSS, glutathione synthetase; TR, thioredoxin; NAD(P)H, nicotinic adenine dinucleotide (phosphate); PUFA, polyunsaturated fatty acid; THF, tetrahydrofolate
NADPH has two key roles in metabolism. First, it is used in the synthesis of multiple macromolecules including nucleotides (137), cholesterol (138), and fatty acids (139). Second, it is used to regenerate the reduced form of glutathione and thioredoxin described above. In addition to these well-established roles, it also acts as a cofactor for NAD(P)H Quinone Dehydrogenase 1 (NQO1), a target of NRF2 (140). NADPH can principally be generated from metabolism of TCA cycle intermediates, glycine/tetrahydrofolate (THF) metabolism, and by the PPP. The TCA cycle intermediates isocitrate and malate can be metabolized by isocitrate dehydrogenase and malic enzyme respectively to produce NADPH with both enzymes being regulated by NRF2 (7). The PPP enzymes involved in NADPH metabolism including G6PD, PGD, TKT and TALDO1 seem to be directly or indirectly regulated by NRF2 and are shown in Figure 3 (7). Overall NRF2 activation serves to increase the intracellular pool of NADPH by regulating enzymatic activity of key NADPH synthesis reactions.
Redox balance plays a critical role in cancer initiation and progression. Generation of ROS can induce tumor promoting mutations through DNA damage. However, contrary to common belief, use of antioxidants has been linked to cancer progression and poor outcomes in clinical trials. In a randomized clinical trial, Omenn et al. treated patients with high-risk exposure to smoking or asbestos with either a placebo or a combination of the antioxidants beta carotene and Vitamin A (141). The treatment arm had a significant increase in incidence of lung cancer and death. The study was stopped early because of these findings. In a more recent clinical trial, use of vitamin A, C, E, coenzyme Q10, and carotenoids was associated with increased risk of recurrence (142). Pre-clinical evidence also supports that supplementation with antioxidants promotes tumor progression. Utilizing KrasG12D- or BrafV600E-driven GEMMs of lung cancer, Sayin et al. showed that the addition of dietary antioxidants (NAC or Vitamin E) resulted in increased tumor burden and higher tumor grade (143). In a melanoma mouse model, treatment with NAC had no impact on primary tumor growth but increased circulating melanoma cells and metastases (35). Inhibition of NRF2 function results in reduced proliferation in vivo highlighting the importance of NRF2 activation in tumor progression (120,144). However, in addition to redox homeostasis, NRF2 regulates multiple other cellular pathways that also play a role in tumor progression.
Amino Acid Metabolism
While NRF2 is mainly known for regulating the cellular redox state, NRF2 activation has a key role in multiple pathways involving amino acid transport and metabolism (Figure 3). The synthesis of glutathione requires three amino acids: glutamate, cysteine, and glycine. NRF2 not only regulates the synthesis of glutathione, but also regulates the intracellular abundance of these amino acids. SLC7A11 is an NRF2 target that encodes for a protein that dimerizes with SLC3A2 to form the xc− antiporter system (xCT). xCT functions as a concentration-dependent antiporter which exports glutamate in exchange for cystine, the dimerized form of cysteine. This transporter serves to maintain intracellular stores of cysteine for glutathione synthesis. (7,120,145). Sayin et al. show that NRF2-mediated depletion of intracellular glutamate stores through export (xCT) and consumption (GSH synthesis) results in cancers that are dependent on extracellular glutamine (146). These tumors can be therapeutically targeted by inhibiting glutaminase, which ultimately catalyzes the conversion of glutamine to glutamate (Figure 3), and therefore depleting intracellular glutamate (57,64,146). Not only is glutamate a critical carbon source for many biosynthetic reactions but glutamate also serves as a nitrogen donor for the synthesis of non-essential amino acids including serine and glycine.
Under normal physiologic conditions, depletion of intracellular glutamate due to NRF2 activation likely has no impact on serine-dependent reactions, as serine and other non-essential amino acids can be imported from the microenvironment. However, LeBoeuf et al. have demonstrated that in serine deprived conditions, hyperactive NRF2 creates a metabolic vulnerability by depleting glutamate needed for serine synthesis (147). One central feature of NRF2 activation is the dependency on extracellular glutamine to replenish intracellular glutamate for numerous downstream pathways. Glutamine enters the cell through multiple transporters, including SLC1A5 (148) while GLS1 catalyzes the rate-limiting step in glutaminolysis to produce glutamate (149,150). This dependency on external glutamine can be exploited therapeutically.
Using a KRAS-driven both mouse and patient-derived xenograft (PDX) models, loss of KEAP1 sensitizes tumors to CB-839 (120,146). Several studies have demonstrated that inhibiting GLS1 with CB-839 or BPTES have been successful in impairing the growth of NRF2 addicted cancer cells (120,146,151,152). This has been further validated by Galan-Cobo who show that activation of NRF2 sensitizes human cell lines to glutaminase inhibition (153). Glutaminase inhibition sensitivity has been attributed to depletion of intracellular pools of glutamate which is exported by xCT in NRF2 active tumors as above. Sayin et al. showed that by blocking xCT using the small molecule erastin, and therefore blocking the export of glutamate, rescues sensitivity of Keap1 LOF or Nrf2 GOF mutant cell lines to CB-839 (146). This sensitivity is not specific to lung malignancies. Kras-mutant pancreatic cancers upregulate NRF2 activity in response to chemotherapy (154) or eIF4A inhibitors (155) and become sensitized to CB-839. In a HER2-driven breast cancer model, HER2 downregulation leads to ROS-mediated apoptosis. Resistant dormant cells escape apoptosis and lead to tumor recurrence by NRF2 driven activation of antioxidant pathways, which sensitizes them to glutaminase inhibition (156).
Another potential liability in NRF2 addicted tumors targets cysteine metabolism. Kang et al. identified that KEAP1-mutant tumors epigenetically silence the expression of cysteine dioxygenase 1 (CDO1), which metabolizes the entry of cysteine towards taurine biosynthesis (123). Overexpression of CDO1 in NRF2 hyperactive cells resulted in impaired proliferation suggesting a potential therapeutic vulnerability.
Autophagy
The KEAP1/NRF2 pathway interacts with the autophagy pathway through the adaptor p62. Normally p62 binds proteins (such as those misfolded under ER stress) and shuttles the protein to the autophagolysosome by interacting with LC3. As discussed above, p62 can bind KEAP1 but with lower affinity compared to NRF2. However, after phosphorylation at serine 349 by mTORC1, p62 binds KEAP1 with much higher affinity than NRF2 through competitive binding with the DLG motif (71,157). This interaction then leads to degradation of the p62-KEAP1 complex. Interestingly, NRF2 transcriptionally regulates p62 through an ARE and therefore forms a positive feedback loop (158).
The pathological role of the p62-KEAP1 interaction can be seen in a subset of bladder cancer patients with increased p62 expression. Overexpression of p62 in bladder cancer cell lines increases NRF2 levels and in vitro proliferation, whereas knockout of p62 impaired tumor growth in a xenograft model (159). Umemura et al. demonstrated, using multiple hepatocellular carcinoma models (carcinogen induced, constitutive mTORC1 activation, and a non-alcoholic steatohepatitis model), that hepatocyte specific loss of p62 reduced tumor initiation (160). They further showed that hepatocyte overexpression of p62 resulted in tumor formation and deletion of the KIR region of p62 (which interacts with KEAP1 and stabilizes NRF2) reversed the development of tumors. NRF2 stabilization by p62-KEAP1 binding has also been shown specifically in hepatitis C patients with hepatocellular carcinoma (161).
Using a CRISPR/Cas9-based screens, Romero et al. found that KEAP1 mutant cells are vulnerable to loss of Slc33a1, an endoplasmic reticulum associated protein implicated in ER homeostasis (122). Loss of this transporter results in an induction of autophagy markers and transcriptional signatures enriched in the unfolded protein response (UPR) pathway. The sensitivity of Keap1 mutant cells to loss of Slc33a1 was thought to be mediated by added UPR in the setting of increased glutathione synthesis. Targeting SLC33A1/UPR induction may be a promising therapeutic target in KEAP1 mutant lung adenocarcinoma.
Iron and Heme Metabolism
NRF2 transcriptionally induces a large transcriptional program that regulates heme and iron metabolism, which need to be tightly regulated and play a role in multiple physiological processes. Although heme is mostly known for its role in coordinating oxygen in hemoglobin, heme and its derivatives are important prosthetic groups for at least 100 different enzymes that catalyze multiple redox reactions including complex II, III, and IV of the electron transport chain (162,163) and lipid desaturation (164–166). Heme synthesis is a complex process that involves multiple reactions to synthesize the protoporphyrin IX (PPIX) to which ferrous iron (Fe2+) is bound in the mitochondria in order to generate heme. NRF2 induces the expression of the mitochondrial porphyrin transporter ATP binding cassette subfamily B member 6 (ABCB6), which imports porphyrin into the mitochondria, and ferrochelatase (FECH), which loads ferrous Fe2+ iron onto the PPIX ring (167). NRF2 also regulates heme degradation by inducing the expression of HO1 which degrades heme into ferrous iron and biliverdin (3). Regulation of heme plays a pivotal role in lung cancer metastasis. As discussed above, Lignitto et al. demonstrated that loss of KEAP1 leads to activation of NRF2, leading to an HO1-dependent heme degradation that then stabilizes the pro-metastatic transcription factor, BACH1 (130).
The iron atom in heme is a potential catalyst for Fenton reaction-mediated lipid oxidation through the production of superoxide that is scavenged by antioxidants (168). As a result, free heme levels must be tightly regulated in order to maintain redox homeostasis (131,168,169). The iron liberated from heme is recycled for heme synthesis, storage, or export from the cells. Therefore, NRF2 plays a critical role in regulating free iron to prevent Fenton reaction-mediated lipid peroxidation, which triggers a form of cell death called ferroptosis. These lipid peroxide radicals can be reduced to nontoxic lipids by glutathione in a reaction catalyzed by glutathione peroxidase 4 (GPX4) (Figure 3). Lipid radicals can accumulate in conjunction with free iron and in the absence of sufficient glutathione to trigger ferroptotic cell death. The KEAP1/NRF2 pathway likely plays a critical role in blunting ferroptotic cell death as several components of ferroptosis are regulated by NRF2. NRF2 prevents iron-dependent generation of hydroxyl radicals by promoting free iron sequestration through the transcriptional regulation of ferritin light (FTL) and heavy (FTH1) chains that together make up the 24 subunits that form ferritin (5). Export of free iron is solely facilitated by ferroportin 1 (FPN1) which has also been shown to be transcriptionally regulated by NRF2 in macrophages (170). In addition, NRF2 regulates glutathione synthesis at multiple levels including through regulation of xCT expression that facilitates cysteine availability for GSH synthesis. Reduction of lipid peroxides requires NADPH produced by NRF2 regulated enzymes in the PPP. There is a growing interest in exploiting ferroptosis to therapeutically target cancer cells. Despite this, there is limited data showing the importance of NRF2 activation suppressing ferroptosis in vivo. In vitro work has shown that NRF2 activation results in resistance to ferroptosis inducers, suggesting that these classes of drugs may have limited efficacy in NRF2 activated tumors (171). Recently, Takahashi et al. used 3D spheroid cultures that better mimic in vivo growth conditions, to demonstrate that the inner cells of tumor spheroids are exposed to higher ROS and lipid peroxidation and are susceptible to cell death after NRF2 knockdown (172). Furthermore, suppressing ferroptosis may be a key mechanism by which NRF2 active tumors resist radiation therapy. Lang et al. have shown in pre-clinical models that radiation therapy induced ferroptosis through downregulation of xCT (173). Therefore, upregulation of xCT may allow tumors to evade radiation induced ferroptosis. Kang et al. demonstrated that the NRF2 target GCLC suppresses ferroptosis by a GSH-independent mechanism (174). They show that GCLC activity serves as a glutamate sink through synthesis of γ-glutamyl dipeptides. This depletion of intracellular glutamate inhibited cystine starvation-dependent ferroptosis. Further work needs to be done to validate the role of NRF2 in ferroptosis in both humans and mouse models.
THE ROLE OF KEAP1/NRF2 IN THERAPY RESISTANCE
Chemotherapy and Radiotherapy Resistance
Disease recurrence and development of resistance to therapy pose a challenge to the clinical treatment of cancer. Hyperactivation of NRF2 in cancer is associated with chemotherapy and radiation therapy resistance and as a result poor prognosis (119,175,176). For example, NRF2-addicted cancer cell lines are less sensitive to the typical anti-cancer drugs such as cisplatin and doxorubicin (177). Platinum-based chemotherapy induces tumor cell death through two key mechanisms: the formation of DNA adducts that impair DNA replication and the induction of mitochondrial ROS (178,179). As previously described, in normal physiology NRF2 functions in a cyto-protective manner but also enables a mechanism for NRF2 mediated cisplatin resistance. Cancer cells with hyperactivation of NRF2 by somatic mutations in KEAP1 or NRF2 show increased expression of phase II detoxification enzymes like NQO1, GST, to conjugate reactive molecules like cisplatin with reduced form of glutathione, GSH. Once conjugated to GSH, cisplatin is excreted by phase III detoxification pumps MRP4 and MRP5 (180) (Figure 4). Other typical anti-cancer drugs, 5-FU, 6-TG, gemcitabine, cytarabine are also pumped out by this activation of ATP-binding cassette family transporters induced by NRF2. In addition, cisplatin is known to induce cell death through increased mitochondrial ROS which can be mitigated by the antioxidant pathways activated by NRF2. From a screening comparison of tumor suppressor vs. chemotherapy and targeted therapy, KEAP1 was detected as a marker of resistance across different organ backgrounds against arsenic trioxide (181). NRF2 hyperactivation can also sensitize to chemotherapy as NQO1 can activate mitomycin C, increasing its cytotoxicity (182,183). For similar reasons, hyperactive NRF2 tumors have been known to be resistant to radiation therapy likely mediated through regulation of radiation induced ROS (184). However, recent work has demonstrated that addition of the glutaminase inhibitor CB-839 can sensitize KEAP1-mutant cell lines to radiation therapy (19).
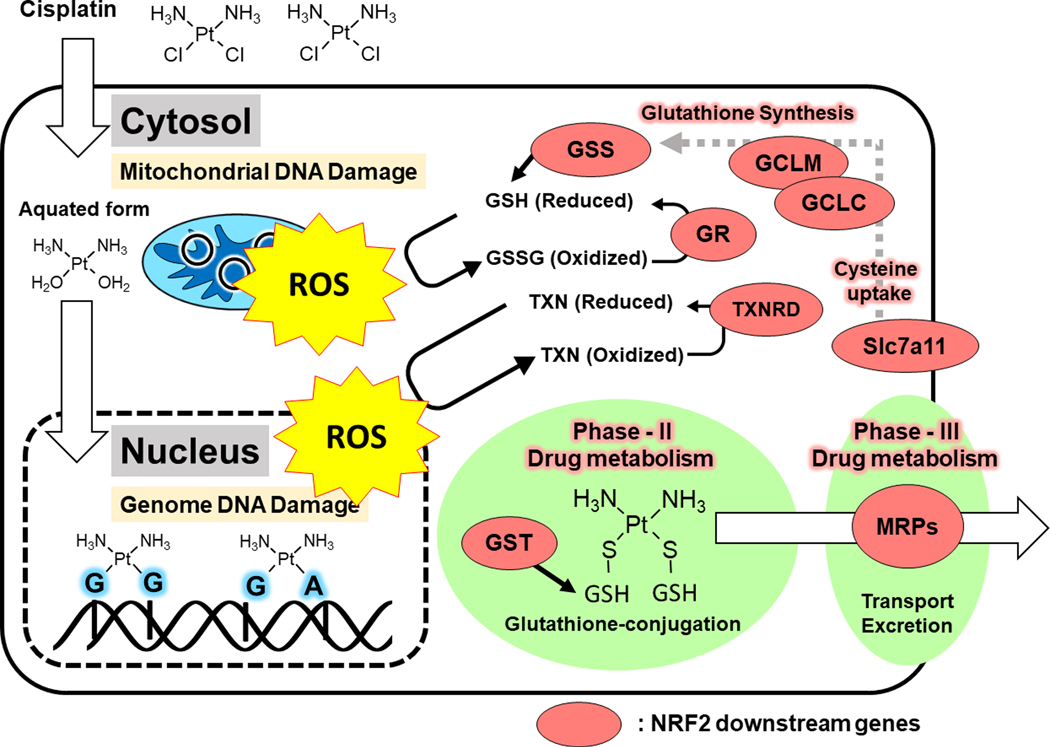
Figure 4: NRF2 Dependent Drug Detoxification.
Aquated form of cisplatin targets both mitochondrial and genomic DNA by anchoring to nucleotides at guanine-guanine or guanine-adenine bonds, which cause severe DNA damage and generates ROS. Hyperactivated NRF2 maintains high glutathione and thioredoxin levels, which scavenge ROS. Toxic aquated forms of cisplatin are conjugated with glutathione by a NRF2 induced transferase, GST, and then excreted by multi-drug resistant pumps, which are also transcriptional targets of NRF2.
ROS, reactive oxygen species, GSH, glutathione; GSSG, glutathione disulfide; TXN, thioredoxin; glutathione synthetase; GR, glutathione reductase; TXNRD1, thioredoxin reductase 1; GCLC, glutamate-cysteine ligase catalytic subunit; GCLM, glutamate-cysteine ligase modifier subunit; GST, glutathione s-transferase; MRP, multi-drug resistance protein
Targeted Therapy Resistance
Therapeutic targeting of mutations in EGFR, and ALK is a key component of treatment for patients with NSCLC with these specific driver mutations. Multiple lines of evidence suggest that LOF mutations in KEAP1 can reduce responses to targeted therapy. Using an in vitro CRISPR screen in human lung cancer cell lines, Krall et al. identified that KEAP1 loss results in resistance to the MEK inhibitor trametinib (185). Using a similar approach, it was demonstrated that KEAP1 mutations drive resistance to sorafenib in hepatocellular carcinoma cell lines (186). In addition, loss of KEAP1 in the BRAFV600E mutant lung cancer cell line HCC364 desensitizes cells to the BRAF inhibitor vemurafenib. The authors further verified targeted therapy resistance in EGFR and ALK mutant cells with KEAP1 mutations. Hellyer et al. validated these experimental findings in a cohort of 228 patients with EGFR mutations with or without KEAP1/NRF2/CUL3 mutations (187). Activation of NRF2 in these EGFR mutant tumors was associated with treatment failure. Other drugs including axitinib, a VEGF inhibitor, also appear to have reduced effectiveness in vitro in the context of elevated NRF2 activity in renal cell carcinoma (188). Recently in a phase II clinical trial using the KRASG12C inhibitor sotorasib there was a trend for reduced response rates in KEAP1 mutant tumors (189). The mechanism in which activation of NRF2 induces drug resistance in the context of targeted therapy is not clearly delineated but is likely related to already described mechanisms including drug detoxification and regulation of ROS.
Modulation of Anti-Tumor Immune Responses
Extensive clinical work has demonstrated that NRF2 activation in lung adenocarcinoma impairs anti-tumor immune response. In addition to reducing sensitivity to chemotherapy, both loss of function mutations in KEAP1 and activating mutations in NRF2 confer worse overall survival for patients treated with checkpoint blockade (21,190). These findings are not specific to lung as a pan-cancer analysis has shown similar findings across multiple tumor types (45,83). Surprisingly, mutations in KEAP1 were associated with high tumor mutational burden as well as increased PD-L1 expression which is strongly correlated with favorable responses to immune checkpoint blockade (191).
The mechanism of this resistance remains unclear and little work has been done to characterize the immune microenvironment of KEAP1-mutant tumors. Kadara et al. showed that in early stages, KEAP1-mutant tumors had higher peritumoral CD57+ and granzyme B+ cells suggestive of NK cells (192). Using a Keap1flox/flox; Ptenflox/flox (K1P) mouse model, Best et al. analyzed immune infiltration in tumor bearing and healthy mice. They found that lungs from tumor bearing mice had a reduced number of NK, B, and T cells. In addition, these studies showed that loss of Keap1 impairs the expansion of CD11c+ immune populations observed in the K1P mouse model (89).
The activation of NRF2 in specific immune populations can alter their function. Kobayashi et al. showed that NRF2 can inhibit LPS-induced expression of Il6 and Il1β in M1 macrophages (8). In a similar fashion, Thimmulappa et al. demonstrate Nrf2−/− peritoneal neutrophils produce less IL-6 and TNF-α in response to LPS stimulation (193). NRF2 can also alter cytokine expression in a tumor intrinsic context. The alarmin/cytokine IL-33 has been implicated in promoting tumor progression however the mechanism of IL-33 is not entirely clear. Using a skin squamous cell carcinoma it has been demonstrated that NRF2 promotes IL-33 release resulting in accumulation of pro-tumor macrophages (194). NRF2 also has been shown to regulate Il11. Using a mouse colorectal cell line, Nishina et al. induced Il11 transcription by treatment with the electrophile 1,2 naphthoquinone and further demonstrate that this upregulation was NRF2 dependent (195). Kitamura et al. demonstrated that in human breast cancer samples IL-11 protein levels correlated with NRF2 levels (196). They further showed that Keap1-null MEFs upregulate Il11 in 3D culture and that tumor engraftment was impaired by knockout of Il11. These studies demonstrate the importance of NRF2 activity in regulating cytokine production which may have an impact on tumorigenesis.
Hayashi and Kuga reported that Keap1-null and KrasG12D mutant lung tumors display infiltrating CD8+ T cells with higher expression of PD-L1 compared to Keap1 wild-type and KrasG12D-mutant tumor (197). In addition, they observed that NRF2 activation in the extra-tumoral area (described as a NRF2-charged microenvironment) resulted in reduction in area of Keap1-mutant KrasG12D-driven tumors. The NRF2-charged microenvironment also regulated the histopathology of tumors that developed. Tumors with a NRF2-charged microenvironment favored a more lepidic phenotype while in the non-charged microenvironment displayed a more aggressive papillary phenotype. Interestingly, Tnfα expression in CD8+ T cells was induced in the NRF2-charged microenvironment (197). Multiple other papers have shown that global NRF2 activation or NRF2 inactivation in immune populations can regulate tumor growth (126,198). These reports demonstrate the potential role for NRF2 modulation to suppress NRF2 hyperactivated cancers through enhancement of anti-cancer immunity. Extensive additional work is needed to characterize the alterations in the tumor immune microenvironment to determine the mechanism through which KEAP1-mutant tumors develop resistance to checkpoint blockade.
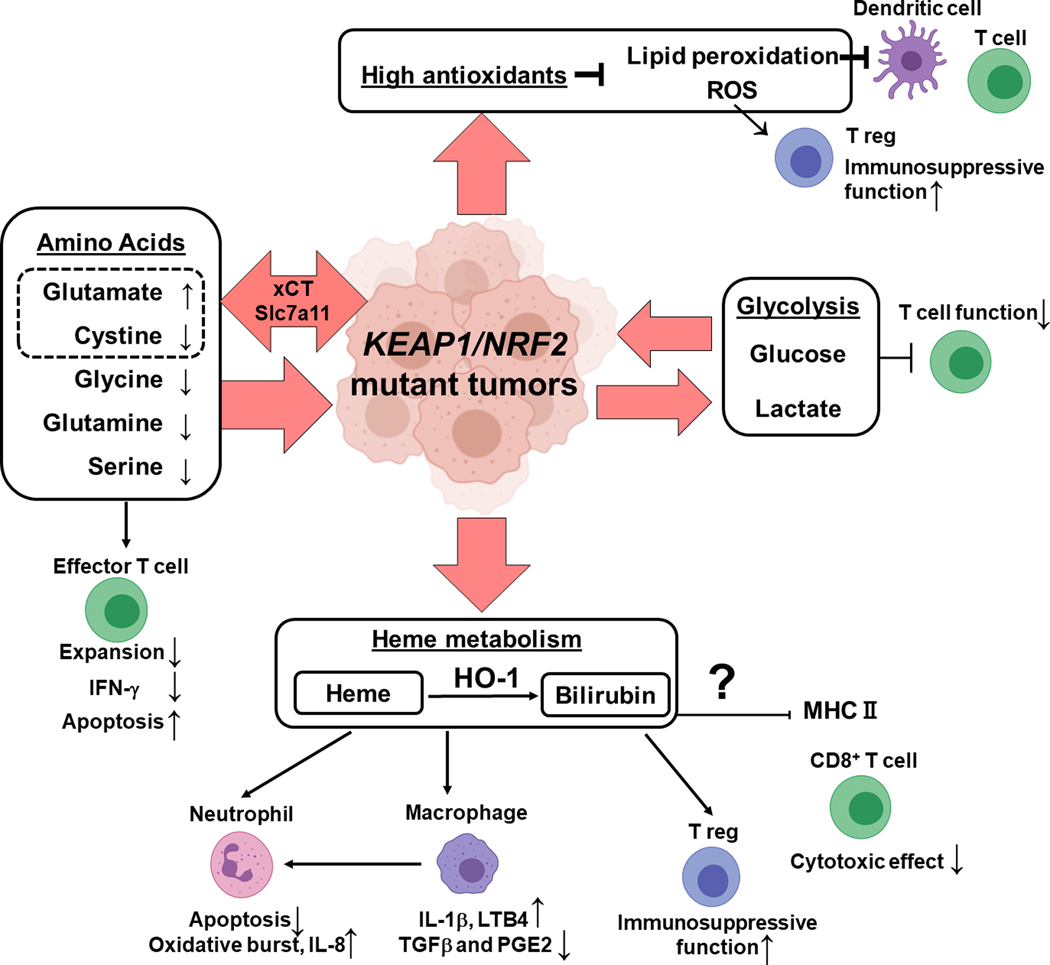
It is attractive to hypothesize that tumor cell metabolic rewiring driven by hyperactivation of NRF2 can alter the metabolic milieu of the tumor microenvironment and regulate immune cell effector functions (Figure 5). Nutrient competition is characterized by increased production of immunosuppressive metabolites such as lactate (199,200) and kynurenine (201) by the tumor cells but also increased consumption of metabolites necessary for immune cell activation such as glucose (199,202), glutamine (203–205), serine (206), and tryptophan (201,207). It is likely that the increased utilization of nutrients such as glucose, glutamine, alanine, glycine, and cystine (120,147) in Keap1-mutant tumors used to support anabolic metabolism may dampen immune responses via restricting effector immune cells of essential nutrients. Keap1 mutation could also be impacting immune responses via increased antioxidant production. This is an exciting line of investigation considering that immune populations have differential redox sensitivity. Lipid peroxidation products can inhibit dendritic cell function and subsequent T cell activation (208). Oxidative stress can increase the immunosuppressive ability of T regulatory cells (209). Interestingly, Wang et al. showed that interferon gamma released by CD8+ T cells downregulates the two subunits of the glutamate-cystine antiporter xCT promoting ferroptosis (SLC7A11, SLC3A2) (210). Increased SLC7A11 expression in the setting of KEAP1 LOF or NRF2 GOF may enable tumors to evade interferon gamma induced ferroptosis.
Figure 5: Impact of Keap1/Nrf2 Mutation on Tumor Microenvironment and Anti-tumor Immune Responses.
Keap1 LOF/ Nrf2 GOF mutation harboring tumors display increased uptake of non-essential amino acids such as glycine, serine, and glutamine. Cystine is imported through NRF2 regulated transporter xCT. Overall, in the microenvironment of Keap1/Nrf2 mutant tumors, glutamate is increased while cystine, glycine, glutamine, serine is depleted These metabolic changes can inhibit effector T cell function (expansion, production of IFN-γ) and induce apoptosis. Besides amino acids, Keap1/Nrf2 mutations result in increased glycolysis and thus increased glucose consumption and lactate secretion which can be deleterious for T cell function. NRF2 is a master regulator of antioxidants that decrease ROS and lipid peroxides which can impact dendritic cells, T cell effector cells and T regulatory cells. Hyperactivation of NRF2 results in altered heme metabolism and the byproducts of these pathways can impact neutrophils, macrophages, T regulatory cells and cytotoxic T lymphocytes.
We previously discussed that NRF2 plays a major role in regulating heme and iron metabolism. Lignitto et al. showed heme to be important for tumor growth and metastasis (130). NRF2 can induce multiple genes (ABCB6, FECH, HO1, BLVRB) central to heme regulation. Heme metabolism by-products as well as heme itself have been suggested to have immunomodulatory functions. In neutrophils, heme decreases apoptosis, promotes oxidative burst and IL-8 production (211). In macrophages, it promotes IL-1β production via inflammasome (NLRP3) activation, LTB4 production that drives neutrophil recruitment, and it also impairs production of anti-inflammatory molecules like TGFβ and PGE2 (212). Bilirubin which is produced by biliverdin reductase b (BLVRB), the last step of heme degradation, has been proposed to have potent immunosuppressive activity including the ability to downregulate MHCII expression (213,214), induce CD8+ T cell apoptosis (213), and promote T regulatory cell infiltration (215). Interestingly, carbon monoxide, a byproduct of HO1-catalyzed reactions, has been shown to suppress allograft rejection pointing to its immunosuppressive activity (216).
FUTURE THERAPEUTIC PERSPECTIVES
LOF KEAP1 mutations and GOF NRF2 mutations lead to more aggressive tumors through the mechanisms discussed above and serve as a prognostic marker associated with poor survival and poor multiple types of therapy (21,217,218). Furthermore, KEAP1/NRF2 mutations also lead to metabolic vulnerabilities in pre-clinical models, making these mutations predictive biomarkers of responsiveness to metabolic therapies (120). Currently there are four clinical trials specifically targeting KEAP1 mutations (summarized in Table 1). The most promising trial is the KEAPSAKE trial which combines the glutaminase inhibitor CB-839 (teleglenastat) with standard first line therapy (pembrolizumab and chemotherapy) in patients with tumors that carry LOF mutations in KEAP1 or activating mutations in NRF2. While inhibiting glutaminase may improve outcomes in patients with KEAP1 mutations by targeting tumor intrinsic vulnerabilities, CB-839 may have the added benefit of augmenting anti-tumor T cell responses. Leone et al. demonstrate that glutaminase inhibition can augment immune responses in pre-clinical mouse tumor models without hyperactivated NRF2 (219). They show that glutaminase inhibition has unique effects on T cell subsets including improved cytokine production, activation, and proliferation. The pro-drug DRP-104 (sirpiglenastat) is currently in phase 1 trials (Table 1, trial ID: NCT04471415). The active moiety of DRP-104, 6-Dizao-5-oxo-L-norluecine (DON), is a potent glutamine antagonist. While DON has previously been shown to reduce tumor growth, its high toxicity has prevented its therapeutic development (220). However, DRP-104 itself is inactive and was designed to limit systemic exposure to DON while targeting tumor cells.
Table 1:
Clinical Trials Targeting KEAP1/NRF2 Mutant Tumors
| Trial | Drug | Study Type | Inclusion | Intervention | Primary Outcomes |
|---|---|---|---|---|---|
|
KEAPSAKE
NCT04265534 |
CB-839 (Telaglenastat) | Phase 2 Randomized placebo controlled | Metastatic NSCLC with mutation in KEAP1, NRF2, or LKB1 | Pembrolizumab + Carboplatin + Pemetrexed +/− CB-839 | Progression Free Survival Safety and Tolerability Dosing |
|
BeGIN
NCT03872427 |
CB-839 | Phase 2 Open label single arm | Advanced tumor with mutation in NF1, KEAP1, or LKB1 | CB-839 | Best Overall Response Rate |
| NCT04471415 | DRP-104 (Sirpiglenastat) | Phase 1 and 2a Dose Escalation Dose expansion | Advanced NSCLC with mutation in KEAP1, NRF2, or LKB1 and already received 1st line therapy | DRP-104 + Atezolizumab | Maximum tolerated dose Area under plasma concentration Cmax of DRP-104 |
| NCT02417701 | Sapanisertib | Phase 2 Randomized open label | Stage IV or recurrent SCC with KEAP1 or NRF2 mutation | Docetaxel + Sapanisertib | Progression Free Survival |
While current clinical trials focus on targeting metabolic vulnerabilities in NRF2 addicted tumors, future trials are likely to involve targeting NRF2 signaling in itself. Several promising in vitro and in vivo studies have demonstrated the feasibility of inhibiting NRF2 to suppress tumor growth both as a single therapy or as a sensitizing agent. Tang et al. demonstrate that luteolin impairs the binding of NRF2 to AREs and promotes NRF2 degradation in A549 cells (221). They go on to show that luteolin sensitizes A549 derived cells to oxaliplatin, bleomycin, and doxorubicin. In subsequent work, the authors showed that combination luteolin and cisplatin treatment significantly impaired tumor growth compared to monotherapy in A549 xenograft models (222). Of note, luteolin has NRF2-independent effects including impairing EGFR signaling which may contribute to these results (223). In a similar set of experiments Ren et al. demonstrated that brusatol, a drug that inhibits NRF2 activity amongst other effects, sensitizes tumors to platinum chemotherapy in an A549 xenograft model (224). Inhibiting NRF2 activity has potential clinical implications, yet no clinical trials have used this approach to target tumors. The off-target effects of these drugs and the possibility of impairing physiologic antioxidant and immune responses may limit their use clinically.
CONCLUDING REMARKS
The KEAP1/NRF2 pathway has a key role in tumorigenesis. While NRF2 activation increases resistance to chemotherapy and radiation therapy, activation of these pathways leads to metabolic liabilities. Glutaminase inhibition has been extensively explored in pre-clinical models and has rapidly moved to clinical trials. But therapeutic vulnerabilities in other NRF2-dependent pathways including the PPP and heme synthesis need to be explored. In addition, while activation of NRF2 is clearly driving tumor intrinsic pro-tumor changes, there are NRF2 independent effects of KEAP1 mutations that should be explored and potentially exploited therapeutically. And finally, understanding additional co-occurring mutations in the context of KEAP1 mutation may lead to novel therapies in targeting not only KEAP1-mutant tumors but also other aggressive co-occurring mutations such as LKB1.
STATEMENT OF SIGNIFICANCE.
Alterations in the KEAP1/NRF2 pathway are found in multiple cancer types. Activation of NRF2 leads to metabolic rewiring of tumors that promote tumor initiation and progression. Here we present the known alterations that lead to NRF2 activation in cancer, the mechanisms in which NRF2 activation promotes tumors, and the therapeutic implications of NRF2 activation.
ACKNOWLEDGMENTS
R.P. is supported by the William Rom fellowship, the Stony Wold-Herbert Fund, and National Institutes of Health training grant T32 CA009161 (Levy).
Pharmaceuticals. MH is supported by the Uehara Memorial Foundation. T.P. is supported by NIH grants (R37CA222504 and R01CA227649) and an American Cancer Society Research Scholar Grant (RSG-17-200-01-TBE).
Authors’ Disclosures
T.P. has received Honoraria/Consulting fees from Calithera Biosciences, Vividion Therapeutics and research support from Bristol Myers Squibb, Dracen Pharmaceuticals, and Agios Pharmaceuticals.
Footnotes
R.P, M.H and A.M.Z. have no conflict of interests.
REFERENCES
- 1.Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 2004;24(16):7130–9 doi 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997;236(2):313–22 doi 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 3.Alam J, Stewart D, Touchard C, Boinapally S, Choi AMK, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. Journal of Biological Chemistry 1999;274(37):26071–8 doi 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 4.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes & Development 1999;13(1):76–86 doi 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietsch EC, Chan JY, Torti FM, Torti SV. Nrf2 Mediates the Induction of Ferritin H in Response to Xenobiotics and Cancer Chemopreventive Dithiolethiones. Journal of Biological Chemistry 2003;278(4):2361–9 doi 10.1074/jbc.M210664200. [DOI] [PubMed] [Google Scholar]
- 6.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011;475(7354):106–9 doi 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 2012;22(1):66–79 doi 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nature Communications 2016;7(1):11624 doi 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein1 in mouse embryo fibroblasts. Biochemical and Biophysical Research Communications 2003;310(3):824–9 doi 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 10.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/Small Maf Heterodimer Mediates the Induction of Phase II Detoxifying Enzyme Genes through Antioxidant Response Elements. Biochemical and Biophysical Research Communications 1997;236(2):313–22 doi 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 11.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, et al. The Cap ‘n’ Collar Basic Leucine Zipper Transcription Factor Nrf2 (NF-E2 p45-related Factor 2) Controls Both Constitutive and Inducible Expression of Intestinal Detoxification and Glutathione Biosynthetic Enzymes. Cancer Research 2001;61(8):3299. [PubMed] [Google Scholar]
- 12.Muscarella LA, Barbano R, D’Angelo V, Copetti M, Coco M, Balsamo T, et al. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics 2011;6(3):317–25 doi 10.4161/epi.6.3.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muscarella LA, Parrella P, D’Alessandro V, la Torre A, Barbano R, Fontana A, et al. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics 2011;6(6):710–9 doi 10.4161/epi.6.6.15773. [DOI] [PubMed] [Google Scholar]
- 14.Shibata T, Kokubu A, Saito S, Narisawa-Saito M, Sasaki H, Aoyagi K, et al. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia 2011;13(9):864–73 doi 10.1593/neo.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haque E, Karim MR, Salam Teeli A, Śmiech M, Leszczynski P, Winiarczyk D, et al. Molecular Mechanisms Underlying Hepatocellular Carcinoma Induction by Aberrant NRF2 Activation-Mediated Transcription Networks: Interaction of NRF2-KEAP1 Controls the Fate of Hepatocarcinogenesis. International journal of molecular sciences 2020;21(15):5378 doi 10.3390/ijms21155378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Wang Q, Yang Y, Lei C, Yang F, Liang L, et al. GSTZ1 deficiency promotes hepatocellular carcinoma proliferation via activation of the KEAP1/NRF2 pathway. Journal of Experimental & Clinical Cancer Research 2019;38(1):438 doi 10.1186/s13046-019-1459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Zhang C, Zhang L, Yang Q, Zhou S, Wen Q, et al. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer 2015;15(1):531 doi 10.1186/s12885-015-1541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic Alteration of Keap1 Confers Constitutive Nrf2 Activation and Resistance to Chemotherapy in Gallbladder Cancer. Gastroenterology 2008;135(4):1358–68.e4 doi 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 19.Binkley MS, Jeon Y-J, Nesselbush M, Moding EJ, Nabet BY, Almanza D, et al. KEAP1/NFE2L2 mutations predict lung cancer radiation resistance that can be targeted by glutaminase inhibition. Cancer Discovery 2020(10(12)):1826–41 doi 10.1158/2159-8290.CD-20-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, et al. Nrf2 Enhances Cell Proliferation and Resistance to Anticancer Drugs in Human Lung Cancer. Clinical Cancer Research 2009;15(10):3423 doi 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 21.Arbour KC, Jordan E, Kim HR, Dienstag J, Yu HA, Sanchez-Vega F, et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24(2):334–40 doi 10.1158/1078-0432.Ccr-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki T, Hidaka T, Kumagai Y, Yamamoto M. Environmental pollutants and the immune response. Nature Immunology 2020;21(12):1486–95 doi 10.1038/s41590-020-0802-6. [DOI] [PubMed] [Google Scholar]
- 23.Chan K, Lu R, Chang JC, Kan YW. NRF2, a Member of the NFE2 Family of Transcription Factors, is not Essential for Murine Erythropoiesis, Growth, and Development. Proceedings of the National Academy of Sciences of the United States of America 1996;93(24):13943–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iizuka T, Ishii Y, Itoh K, Kiwamoto T, Kimura T, Matsuno Y, et al. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes to Cells 2005;10(12):1113–25 doi 10.1111/j.1365-2443.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 25.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest 2006;116(4):984–95 doi 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi M, Takai J, Yu L, Motohashi H, Moriguchi T, Yamamoto M. Whole-Body In Vivo Monitoring of Inflammatory Diseases Exploiting Human Interleukin 6-Luciferase Transgenic Mice. Mol Cell Biol 2015;35(20):3590–601 doi 10.1128/MCB.00506-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taguchi K, Takaku M, Egner PA, Morita M, Kaneko T, Mashimo T, et al. Generation of a New Model Rat:Nrf2Knockout Rats Are Sensitive to Aflatoxin B1Toxicity. Toxicological Sciences 2016;152(1):40–52 doi 10.1093/toxsci/kfw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Q, Battelli L, Hubbs AF. Multiorgan autoimmune inflammation, enhanced lymphoproliferation, and impaired homeostasis of reactive oxygen species in mice lacking the antioxidant-activated transcription factor Nrf2. Am J Pathol 2006;168(6):1960–74 doi 10.2353/ajpath.2006.051113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoh K, Itoh K, Enomoto A, Hirayama A, Yamaguchi N, Kobayashi M, et al. Nrf2-deficient female mice develop lupus-like autoimmune nephritis11See Editorial by Byrd and Thomas, p. 1606. Kidney International 2001;60(4):1343–53 doi 10.1046/j.1523-1755.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 30.Hubbs AF, Benkovic SA, Miller DB, O’Callaghan JP, Battelli L, Schwegler-Berry D, et al. Vacuolar Leukoencephalopathy with Widespread Astrogliosis in Mice Lacking Transcription Factor Nrf2. The American Journal of Pathology 2007;170(6):2068–76 doi 10.2353/ajpath.2007.060898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proceedings of the National Academy of Sciences of the United States of America 2001;98(6):3410–5 doi 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet 2003;35(3):238–45 doi 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 33.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun 2006;339(1):79–88 doi 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki T, Murakami S, Biswal SS, Sakaguchi S, Harigae H, Yamamoto M, et al. Systemic Activation of NRF2 Alleviates Lethal Autoimmune Inflammation in Scurfy Mice. Mol Cell Biol 2017;37(15):e00063–17 doi 10.1128/MCB.00063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015;527(7577):186–91 doi 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keleku-Lukwete N, Suzuki M, Otsuki A, Tsuchida K, Katayama S, Hayashi M, et al. Amelioration of inflammation and tissue damage in sickle cell model mice by Nrf2 activation. Proceedings of the National Academy of Sciences 2015;112(39):12169–74 doi 10.1073/pnas.1509158112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagishita Y, Uruno A, Chartoumpekis DV, Kensler TW, Yamamoto M. Nrf2 represses the onset of type 1 diabetes in non-obese diabetic mice. Journal of Endocrinology 2019:403–16 doi 10.1530/joe-18-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Molecular and cellular biology 1996;16(11):6083–95 doi 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, et al. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic Biol Med 2016;91:45–57 doi 10.1016/j.freeradbiomed.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, et al. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 2012;40(20):10228–39 doi 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ 1995;6(9):1193–8. [PubMed] [Google Scholar]
- 42.Li X, Zhang D, Hannink M, Beamer LJ. Crystal Structure of the Kelch Domain of Human Keap1. Journal of Biological Chemistry 2004;279(52):54750–8 doi 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 43.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol 2006;26(8):2887–900 doi 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, et al. High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer cell 2016;30(2):214–28 doi 10.1016/j.ccell.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukutomi T, Takagi K, Mizushima T, Ohuchi N, Yamamoto M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol Cell Biol 2014;34(5):832–46 doi 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu L, Wei Y, Reboul J, Vaglio P, Shin T-H, Vidal M, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003;425(6955):316–21 doi 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 47.Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ Domain Proteins Are Putative Substrate Adaptors for Cullin 3 Ubiquitin Ligases. Molecular Cell 2003;12(3):783–90 doi 10.1016/S1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 48.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 2004;24(19):8477–86 doi 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ Dimerization Function Is Required to Sequester Nrf2 in Cytoplasm. Journal of Biological Chemistry 2002;277(39):36544–52 doi 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 50.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes to Cells 2003;8(4):379–91 doi 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki T, Muramatsu A, Saito R, Iso T, Shibata T, Kuwata K, et al. Molecular Mechanism of Cellular Oxidative Stress Sensing by Keap1. Cell Rep 2019;28(3):746–58 e4 doi 10.1016/j.celrep.2019.06.047. [DOI] [PubMed] [Google Scholar]
- 52.Saito R, Suzuki T, Hiramoto K, Asami S, Naganuma E, Suda H, et al. Characterizations of Three Major Cysteine Sensors of Keap1 in Stress Response. Mol Cell Biol 2016;36(2):271–84 doi 10.1128/MCB.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi A, Kang M-I, Watai Y, Tong KI, Shibata T, Uchida K, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 2006;26(1):221–9 doi 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang M-I, Kobayashi A, Yamamoto M, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America 2004;101(7):2040–5 doi 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 2004;24(16):7130–9 doi 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences of the United States of America 2002;99(18):11908–13 doi 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bollong MJ, Lee G, Coukos JS, Yun H, Zambaldo C, Chang JW, et al. A metabolite-derived protein modification integrates glycolysis with KEAP1–NRF2 signalling. Nature 2018;562(7728):600–4 doi 10.1038/s41586-018-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biagiotti S, Bianchi M, Rossi L, Chessa L, Magnani M. Activation of NRF2 by dexamethasone in ataxia telangiectasia cells involves KEAP1 inhibition but not the inhibition of p38. PLOS ONE 2019;14(5):e0216668 doi 10.1371/journal.pone.0216668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proceedings of the National Academy of Sciences 2010;107(44):18838 doi 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kinch L, Grishin NV, Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer cell 2011;20(4):418–20 doi 10.1016/j.ccr.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ooi A, Wong J-C, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, et al. An Antioxidant Response Phenotype Shared between Hereditary and Sporadic Type 2 Papillary Renal Cell Carcinoma. Cancer Cell 2011;20(4):511–23 doi 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 62.Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, et al. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol Cell 2013;51(2):236–48 doi 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer cell 2011;20(4):524–37 doi 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018;556(7699):113–7 doi 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 2016;374(2):135–45 doi 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013;32(32):3765–81 doi 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lien EC, Lyssiotis CA, Juvekar A, Hu H, Asara JM, Cantley LC, et al. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nature Cell Biology 2016;18(5):572–8 doi 10.1038/ncb3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nature Genetics 2007;39(2):159–61 doi 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 69.Ma J, Cai H, Wu T, Sobhian B, Huo Y, Alcivar A, et al. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Molecular and cellular biology 2012;32(8):1506–17 doi 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 2010;12(3):213–23 doi 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 71.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 2013;51(5):618–31 doi 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 72.Lau A, Wang X-J, Zhao F, Villeneuve NF, Wu T, Jiang T, et al. A Noncanonical Mechanism of Nrf2 Activation by Autophagy Deficiency: Direct Interaction between Keap1 and p62. Mol Cell Biol 2010;30(13):3275 doi 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorrini C, Baniasadi PS, Harris IS, Silvester J, Inoue S, Snow B, et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med 2013;210(8):1529–44 doi 10.1084/jem.20121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hast BE, Goldfarb D, Mulvaney KM, Hast MA, Siesser PF, Yan F, et al. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer research 2013;73(7):2199–210 doi 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hast BE, Cloer EW, Goldfarb D, Li H, Siesser PF, Yan F, et al. Cancer-Derived Mutations in KEAP1 Impair NRF2 Degradation but not Ubiquitination. Cancer Research 2014;74(3):808–17 doi 10.1158/0008-5472.CAN-13-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489(7417):519–25 doi 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 2013;27(20):2179–91 doi 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, et al. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res 2011;71(15):5081–9 doi 10.1158/0008-5472.CAN-10-4668. [DOI] [PubMed] [Google Scholar]
- 79.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 2008;135(4):1358–68, 68 e1–4 doi 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 80.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, et al. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol 2010;220(4):446–51 doi 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 81.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013;45(8):860–7 doi 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 82.Suzuki T, Maher J, Yamamoto M. Select Heterozygous Mutations Have a Dominant-Negative Effect on Wild-Type Keap1. Cancer Research 2011;71(5):1700 doi 10.1158/0008-5472.CAN-10-2939. [DOI] [PubMed] [Google Scholar]
- 83.Kerins MJ, Ooi A. A catalogue of somatic NRF2 gain-of-function mutations in cancer. Sci Rep 2018;8(1):12846- doi 10.1038/s41598-018-31281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taguchi K, Yamamoto M. The KEAP1–NRF2 System in Cancer. Frontiers in Oncology 2017;7(85):1–11 doi 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489(7417):519–25 doi 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cloer EW, Goldfarb D, Schrank TP, Weissman BE, Major MB. NRF2 Activation in Cancer: From DNA to Protein. Cancer Research 2019;79(5):889 doi 10.1158/0008-5472.CAN-18-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Imielinski M, Berger Alice H, Hammerman Peter S, Hernandez B, Pugh Trevor J, Hodis E, et al. Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell 2012;150(6):1107–20 doi 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. New England Journal of Medicine 2017;376(22):2109–21 doi 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 89.Best SA, Ding S, Kersbergen A, Dong X, Song JY, Xie Y, et al. Distinct initiating events underpin the immune and metabolic heterogeneity of KRAS-mutant lung adenocarcinoma. Nat Commun 2019;10(1):4190 doi 10.1038/s41467-019-12164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen R, Martin A, Ni A, Hellmann M, Arbour KC, Jordan E, et al. Harnessing Clinical Sequencing Data for Survival Stratification of Patients with Metastatic Lung Adenocarcinomas. JCO Precis Oncol 2019;3:10.1200/PO.18.00307 doi 10.1200/PO.18.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fabrizio FP, Costantini M, Copetti M, la Torre A, Sparaneo A, Fontana A, et al. Keap1/Nrf2 pathway in kidney cancer: frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget 2017;8(7):11187–98 doi 10.18632/oncotarget.14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 2012;12:66 doi 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barbano R, Muscarella LA, Pasculli B, Valori VM, Fontana A, Coco M, et al. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics 2013;8(1):105–12 doi 10.4161/epi.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kwak M-K, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol 2002;22(9):2883–92 doi 10.1128/mcb.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kensler TW, Wakabayashi N, Biswal S. Cell Survival Responses to Environmental Stresses Via the Keap1-Nrf2-ARE Pathway. Annual Review of Pharmacology and Toxicology 2007;47(1):89–116 doi 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 96.Wakabayashi N, Skoko JJ, Chartoumpekis DV, Kimura S, Slocum SL, Noda K, et al. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol 2014;34(4):653–63 doi 10.1128/MCB.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Regulation of Oxidative Stress Contributes to BRCA1 Tumor Suppression. Cancer Discovery 2013;3(9):964 doi 10.1158/2159-8290.CD-RW2013-161. [DOI] [Google Scholar]
- 98.Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, et al. Recurrent Loss of NFE2L2 Exon 2 Is a Mechanism for Nrf2 Pathway Activation in Human Cancers. Cell Rep 2016;16(10):2605–17 doi 10.1016/j.celrep.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 99.Yang M, Yao Y, Eades G, Zhang Y, Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Research and Treatment 2011;129(3):983–91 doi 10.1007/s10549-011-1604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. miR-200a Regulates Nrf2 Activation by Targeting Keap1 mRNA in Breast Cancer Cells. Journal of Biological Chemistry 2011;286(47):40725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discovery 2015;5(8):860 doi 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duan F-G, Wang M-F, Cao Y-B, Dan L, Li R-Z, Fan X-X, et al. MicroRNA-421 confers paclitaxel resistance by binding to the KEAP1 3′UTR and predicts poor survival in non-small cell lung cancer. Cell Death & Disease 2019;10(11):821 doi 10.1038/s41419-019-2031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Narasimhan M, Patel D, Vedpathak D, Rathinam M, Henderson G, Mahimainathan L. Identification of Novel microRNAs in Post-Transcriptional Control of Nrf2 Expression and Redox Homeostasis in Neuronal, SH-SY5Y Cells. PLOS ONE 2012;7(12):e51111 doi 10.1371/journal.pone.0051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 2015;517(7535):489–92 doi 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Satoh H, Moriguchi T, Taguchi K, Takai J, Maher JM, Suzuki T, et al. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis 2010;31(10):1833–43 doi 10.1093/carcin/bgq105. [DOI] [PubMed] [Google Scholar]
- 106.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, et al. Nrf2 and Keap1 Abnormalities in Non–Small Cell Lung Carcinoma and Association with Clinicopathologic Features. Clinical Cancer Research 2010;16(14):3743 doi 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tao S, Rojo de la Vega M, Chapman E, Ooi A, Zhang DD. The effects of NRF2 modulation on the initiation and progression of chemically and genetically induced lung cancer. Mol Carcinog 2018;57(2):182–92 doi 10.1002/mc.22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Long M, Tao S, Rojo de la Vega M, Jiang T, Wen Q, Park SL, et al. Nrf2-dependent suppression of azoxymethane/dextran sulfate sodium-induced colon carcinogenesis by the cinnamon-derived dietary factor cinnamaldehyde. Cancer Prev Res (Phila) 2015;8(5):444–54 doi 10.1158/1940-6207.CAPR-14-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bauer AK, Cho H-Y, Miller-Degraff L, Walker C, Helms K, Fostel J, et al. Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PloS one 2011;6(10):e26590-e doi 10.1371/journal.pone.0026590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kanda H, Yamawaki K. Bardoxolone methyl: drug development for diabetic kidney disease. Clin Exp Nephrol 2020;24(10):857–64 doi 10.1007/s10157-020-01917-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liby K, Royce DB, Williams CR, Risingsong R, Yore MM, Honda T, et al. The Synthetic Triterpenoids CDDO-Methyl Ester and CDDO-Ethyl Amide Prevent Lung Cancer Induced by Vinyl Carbamate in A/J Mice. Cancer Research 2007;67(6):2414 doi 10.1158/0008-5472.CAN-06-4534. [DOI] [PubMed] [Google Scholar]
- 112.Zhang D, Rennhack J, Andrechek ER, Rockwell CE, Liby KT. Identification of an Unfavorable Immune Signature in Advanced Lung Tumors from Nrf2-Deficient Mice. Antioxidants & Redox Signaling 2018;29(16):1535–52 doi 10.1089/ars.2017.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ngo HKC, Kim D-H, Cha Y-N, Na H-K, Surh Y-J. Nrf2 Mutagenic Activation Drives Hepatocarcinogenesis. Cancer Research 2017;77(18):4797 doi 10.1158/0008-5472.CAN-16-3538. [DOI] [PubMed] [Google Scholar]
- 114.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, et al. Nrf2 Is Essential for the Chemopreventive Efficacy of Oltipraz against Urinary Bladder Carcinogenesis. Cancer Research 2004;64(18):6424 doi 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 115.Frank R, Scheffler M, Merkelbach-Bruse S, Ihle MA, Kron A, Rauer M, et al. Clinical and Pathological Characteristics of KEAP1 and NFE2L2 Mutated Non–Small Cell Lung Carcinoma (NSCLC). Clinical Cancer Research 2018;24(13):3087–96 doi 10.1158/1078-0432.Ccr-17-3416. [DOI] [PubMed] [Google Scholar]
- 116.Luo W, Tian P, Wang Y, Xu H, Chen L, Tang C, et al. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. International Journal of Cancer 2018;143(7):1696–705 doi 10.1002/ijc.31542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hamada S, Taguchi K, Masamune A, Yamamoto M, Shimosegawa T. Nrf2 promotes mutant K-ras/p53-driven pancreatic carcinogenesis. Carcinogenesis 2017;38(6):661–70 doi 10.1093/carcin/bgx043. [DOI] [PubMed] [Google Scholar]
- 118.Chio IIC, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, et al. NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell 2016;166(4):963–76 doi 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 2010;9(2):336–46 doi 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 2017;23(11):1362–8 doi 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Singh A, Daemen A, Nickles D, Jeon S-M, Foreman O, Sudini K, et al. NRF2 Activation Promotes Aggressive Lung Cancer and Associates with Poor Clinical Outcomes. Clinical Cancer Research 2021;27(3):877–88 doi 10.1158/1078-0432.CCR-20-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Romero R, Sánchez-Rivera FJ, Westcott PMK, Mercer KL, Bhutkar A, Muir A, et al. Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1. Nature Cancer 2020;1(6):589–602 doi 10.1038/s43018-020-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kang YP, Torrente L, Falzone A, Elkins CM, Liu M, Asara JM, et al. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer. eLife 2019;8:e45572 doi 10.7554/eLife.45572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Best SA, De Souza DP, Kersbergen A, Policheni AN, Dayalan S, Tull D, et al. Synergy between the KEAP1/NRF2 and PI3K Pathways Drives Non-Small-Cell Lung Cancer with an Altered Immune Microenvironment. Cell Metabolism 2018;27(4):935–43.e4 doi 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 125.Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, et al. Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discovery 2017;7(1):86 doi 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Satoh H, Moriguchi T, Saigusa D, Baird L, Yu L, Rokutan H, et al. NRF2 Intensifies Host Defense Systems to Prevent Lung Carcinogenesis, but After Tumor Initiation Accelerates Malignant Cell Growth. Cancer Research 2016;76(10):3088 doi 10.1158/0008-5472.CAN-15-1584. [DOI] [PubMed] [Google Scholar]
- 127.Hamada S, Shimosegawa T, Taguchi K, Nabeshima T, Yamamoto M, Masamune A. Simultaneous K-ras activation and Keap1 deletion cause atrophy of pancreatic parenchyma. Am J Physiol Gastrointest Liver Physiol 2018;314(1):G65–g74 doi 10.1152/ajpgi.00228.2017. [DOI] [PubMed] [Google Scholar]
- 128.Li F, Han X, Li F, Wang R, Wang H, Gao Y, et al. LKB1 Inactivation Elicits a Redox Imbalance to Modulate Non-small Cell Lung Cancer Plasticity and Therapeutic Response. Cancer cell 2015;27(5):698–711 doi 10.1016/j.ccell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jiang T, Chen N, Zhao F, Wang X-J, Kong B, Zheng W, et al. High Levels of Nrf2 Determine Chemoresistance in Type II Endometrial Cancer. Cancer Research 2010;70(13):5486 doi 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell 2019;178(2):316–29.e18 doi 10.1016/j.cell.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kumar S, Bandyopadhyay U. Free heme toxicity and its detoxification systems in human. Toxicol Lett 2005;157(3):175–88 doi 10.1016/j.toxlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 132.Wiel C, Le Gal K, Ibrahim MX, Jahangir CA, Kashif M, Yao H, et al. BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell 2019;178(2):330–45 e22 doi 10.1016/j.cell.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 133.Wang H, Liu X, Long M, Huang Y, Zhang L, Zhang R, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Science Translational Medicine 2016;8(334):334ra51 doi 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- 134.Jyrkkanen HK, Kansanen E, Inkala M, Kivela AM, Hurttila H, Heinonen SE, et al. Nrf2 regulates antioxidant gene expression evoked by oxidized phospholipids in endothelial cells and murine arteries in vivo. Circ Res 2008;103(1):e1–9 doi 10.1161/CIRCRESAHA.108.176883. [DOI] [PubMed] [Google Scholar]
- 135.Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 2015;27(2):211–22 doi 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 136.Stafford WC, Peng X, Olofsson MH, Zhang X, Luci DK, Lu L, et al. Irreversible inhibition of cytosolic thioredoxin reductase 1 as a mechanistic basis for anticancer therapy. Sci Transl Med 2018;10(428) doi 10.1126/scitranslmed.aaf7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev 2009;19(1):32–7 doi 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Porter TD. Electron Transfer Pathways in Cholesterol Synthesis. Lipids 2015;50(10):927–36 doi 10.1007/s11745-015-4065-1. [DOI] [PubMed] [Google Scholar]
- 139.Wakil SJ. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 1989;28(11):4523–30 doi 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 140.Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Archives of biochemistry and biophysics 2010;501(1):116–23 doi 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Omenn GS, Goodman GE, Thornquist MD, Balmes J, Cullen MR, Glass A, et al. Effects of a Combination of Beta Carotene and Vitamin A on Lung Cancer and Cardiovascular Disease. New England Journal of Medicine 1996;334(18):1150–5 doi 10.1056/NEJM199605023341802. [DOI] [PubMed] [Google Scholar]
- 142.Ambrosone CB, Zirpoli GR, Hutson AD, McCann WE, McCann SE, Barlow WE, et al. Dietary Supplement Use During Chemotherapy and Survival Outcomes of Patients With Breast Cancer Enrolled in a Cooperative Group Clinical Trial (SWOG S0221). Journal of Clinical Oncology 2019;38(8):804–14 doi 10.1200/JCO.19.01203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Sci Transl Med 2014;6(221):221ra15 doi 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- 144.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011;475(7354):106–9 doi 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, et al. The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal 2013;18(5):522–55 doi 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BS, et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife 2017;6 doi 10.7554/eLife.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.LeBoeuf SE, Wu WL, Karakousi TR, Karadal B, Jackson SR, Davidson SM, et al. Activation of Oxidative Stress Response in Cancer Generates a Druggable Dependency on Exogenous Non-essential Amino Acids. Cell Metab 2020;31(2):339–50.e4 doi 10.1016/j.cmet.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen H, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res 2013;19(3):560–70 doi 10.1158/1078-0432.Ccr-12-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 2016;23(3):517–28 doi 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 2016;16(12):773 doi 10.1038/nrc.2016.131. [DOI] [PubMed] [Google Scholar]
- 151.Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J Clin Invest 2015;125(6):2293–306 doi 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cardnell RJG, Behrens C, Diao L, Fan Y, Tang X, Tong P, et al. An Integrated Molecular Analysis of Lung Adenocarcinomas Identifies Potential Therapeutic Targets among TTF1-Negative Tumors, Including DNA Repair Proteins and Nrf2. Clinical Cancer Research 2015;21(15):3480 doi 10.1158/1078-0432.CCR-14-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, et al. LKB1 and KEAP1/NRF2 Pathways Cooperatively Promote Metabolic Reprogramming with Enhanced Glutamine Dependence in <em>KRAS</em>-Mutant Lung Adenocarcinoma. Cancer Research 2019;79(13):3251 doi 10.1158/0008-5472.CAN-18-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mukhopadhyay S, Goswami D, Adiseshaiah PP, Burgan W, Yi M, Guerin TM, et al. Undermining Glutaminolysis Bolsters Chemotherapy While NRF2 Promotes Chemoresistance in KRAS-Driven Pancreatic Cancers. Cancer Research 2020;80(8):1630 doi 10.1158/0008-5472.CAN-19-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chan K, Robert F, Oertlin C, Kapeller-Libermann D, Avizonis D, Gutierrez J, et al. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat Commun 2019;10(1):5151 doi 10.1038/s41467-019-13086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fox DB, Garcia NMG, McKinney BJ, Lupo R, Noteware LC, Newcomb R, et al. NRF2 activation promotes the recurrence of dormant tumour cells through regulation of redox and nucleotide metabolism. Nat Metab 2020;2(4):318–34 doi 10.1038/s42255-020-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hancock R, Bertrand HC, Tsujita T, Naz S, El-Bakry A, Laoruchupong J, et al. Peptide inhibitors of the Keap1–Nrf2 protein–protein interaction. Free Radical Biology and Medicine 2012;52(2):444–51 doi 10.1016/j.freeradbiomed.2011.10.486. [DOI] [PubMed] [Google Scholar]
- 158.Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 2010;285(29):22576–91 doi 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li T, Jiang D, Wu K. p62 promotes bladder cancer cell growth by activating KEAP1/NRF2-dependent antioxidative response. Cancer science 2020;111(4):1156–64 doi 10.1111/cas.14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, et al. p62, Upregulated during Preneoplasia, Induces Hepatocellular Carcinogenesis by Maintaining Survival of Stressed HCC-Initiating Cells. Cancer Cell 2016;29(6):935–48 doi 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nature Communications 2016;7(1):12030 doi 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Atamna H, Liu J, Ames BN. Heme deficiency selectively interrupts assembly of mitochondrial complex IV in human fibroblasts: revelance to aging. J Biol Chem 2001;276(51):48410–6 doi 10.1074/jbc.M108362200. [DOI] [PubMed] [Google Scholar]
- 163.Lin KH, Xie A, Rutter JC, Ahn YR, Lloyd-Cowden JM, Nichols AG, et al. Systematic Dissection of the Metabolic-Apoptotic Interface in AML Reveals Heme Biosynthesis to Be a Regulator of Drug Sensitivity. Cell Metab 2019;29(5):1217–31 e7 doi 10.1016/j.cmet.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhu XG, Nicholson Puthenveedu S, Shen Y, La K, Ozlu C, Wang T, et al. CHP1 Regulates Compartmentalized Glycerolipid Synthesis by Activating GPAT4. Mol Cell 2019;74(1):45–58 e7 doi 10.1016/j.molcel.2019.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proceedings of the National Academy of Sciences 2003;100(6):3077–82 doi 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mitchell AG, Martin CE. A Novel Cytochrome b5-like Domain Is Linked to the Carboxyl Terminus of the Saccharomyces cerevisiae Δ−9 Fatty Acid Desaturase. Journal of Biological Chemistry 1995;270(50):29766–72 doi 10.1074/jbc.270.50.29766. [DOI] [PubMed] [Google Scholar]
- 167.Campbell MR, Karaca M, Adamski KN, Chorley BN, Wang X, Bell DA. Novel hematopoietic target genes in the NRF2-mediated transcriptional pathway. Oxidative medicine and cellular longevity 2013;2013:120305- doi 10.1155/2013/120305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Minotti G, Aust SD. The role of iron in oxygen radical mediated lipid peroxidation. Chem Biol Interact 1989;71(1):1–19. [DOI] [PubMed] [Google Scholar]
- 169.Sadrzadeh SM, Graf E, Panter SS, Hallaway PE, Eaton JW. Hemoglobin. A biologic fenton reagent. J Biol Chem 1984;259(23):14354–6. [PubMed] [Google Scholar]
- 170.Harada N, Kanayama M, Maruyama A, Yoshida A, Tazumi K, Hosoya T, et al. Nrf2 regulates ferroportin 1-mediated iron efflux and counteracts lipopolysaccharide-induced ferroportin 1 mRNA suppression in macrophages. Archives of Biochemistry and Biophysics 2011;508(1):101–9 doi 10.1016/j.abb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 171.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016;63(1):173–84 doi 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Takahashi N, Cho P, Selfors LM, Kuiken HJ, Kaul R, Fujiwara T, et al. 3D Culture Models with CRISPR Screens Reveal Hyperactive NRF2 as a Prerequisite for Spheroid Formation via Regulation of Proliferation and Ferroptosis. Molecular Cell 2020;80(5):828–44.e6 doi 10.1016/j.molcel.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lang X, Green MD, Wang W, Yu J, Choi JE, Jiang L, et al. Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11. Cancer discovery 2019;9(12):1673–85 doi 10.1158/2159-8290.CD-19-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kang YP, Mockabee-Macias A, Jiang C, Falzone A, Prieto-Farigua N, Stone E, et al. Non-canonical Glutamate-Cysteine Ligase Activity Protects against Ferroptosis. Cell Metabolism 2021;33(1):174–89.e7 doi 10.1016/j.cmet.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jeong Y, Hellyer JA, Stehr H, Hoang NT, Niu X, Das M, et al. Role of KEAP1/NFE2L2 Mutations in the Chemotherapeutic Response of Patients with Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2020;26(1):274–81 doi 10.1158/1078-0432.CCR-19-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yang H, Wang W, Zhang Y, Zhao J, Lin E, Gao J, et al. The Role of NF-E2-Related Factor 2 in Predicting Chemoresistance and Prognosis in Advanced Non-Small-Cell Lung Cancer. Clinical Lung Cancer 2011;12(3):166–71 doi 10.1016/j.cllc.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 177.Tsuchida K, Tsujita T, Hayashi M, Ojima A, Keleku-Lukwete N, Katsuoka F, et al. Halofuginone enhances the chemo-sensitivity of cancer cells by suppressing NRF2 accumulation. Free Radic Biol Med 2017;103:236–47 doi 10.1016/j.freeradbiomed.2016.12.041. [DOI] [PubMed] [Google Scholar]
- 178.Marullo R, Werner E, Degtyareva N, Moore B, Altavilla G, Ramalingam SS, et al. Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PloS one 2013;8(11):e81162-e doi 10.1371/journal.pone.0081162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pinto AL, Lippard SJ. Sequence-dependent termination of in vitro DNA synthesis by cis- and trans-diamminedichloroplatinum (II). Proceedings of the National Academy of Sciences of the United States of America 1985;82(14):4616–9 doi 10.1073/pnas.82.14.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ishikawa T, Ali-Osman F. Glutathione-associated cis-diamminedichloroplatinum(II) metabolism and ATP-dependent efflux from leukemia cells. Molecular characterization of glutathione-platinum complex and its biological significance. Journal of Biological Chemistry 1993;268(27):20116–25. [PubMed] [Google Scholar]
- 181.Ding H, Zhao J, Zhang Y, Yu J, Liu M, Li X, et al. Systematic Analysis of Drug Vulnerabilities Conferred by Tumor Suppressor Loss. Cell Reports 2019;27(11):3331–44.e6 doi 10.1016/j.celrep.2019.05.043. [DOI] [PubMed] [Google Scholar]
- 182.Begleiter A, Robotham E, Leith MK. Role of NAD(P)H:(quinone acceptor) oxidoreductase (DT-diaphorase) in activation of mitomycin C under hypoxia. Molecular Pharmacology 1992;41(4):677. [PubMed] [Google Scholar]
- 183.Siegel D, Gibson NW, Preusch PC, Ross D. Metabolism of mitomycin C by DT-diaphorase: role in mitomycin C-induced DNA damage and cytotoxicity in human colon carcinoma cells. Cancer Res 1990;50(23):7483–9. [PubMed] [Google Scholar]
- 184.Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, et al. Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discov 2017;7(1):86–101 doi 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ, et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. eLife 2017;6:e18970 doi 10.7554/eLife.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zheng A, Chevalier N, Calderoni M, Dubuis G, Dormond O, Ziros PG, et al. CRISPR/Cas9 genome-wide screening identifies KEAP1 as a sorafenib, lenvatinib, and regorafenib sensitivity gene in hepatocellular carcinoma. Oncotarget 2019;10(66):7058–70 doi 10.18632/oncotarget.27361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hellyer JA, Stehr H, Das M, Padda SK, Ramchandran K, Neal JW, et al. Impact of KEAP1/NFE2L2/CUL3 mutations on duration of response to EGFR tyrosine kinase inhibitors in EGFR mutated non-small cell lung cancer. Lung Cancer 2019;134:42–5 doi 10.1016/j.lungcan.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 188.Huang H, Wu Y, Fu W, Wang X, Zhou L, Xu X, et al. Downregulation of Keap1 contributes to poor prognosis and Axitinib resistance of renal cell carcinoma via upregulation of Nrf2 expression. Int J Mol Med 2019;43(5):2044–54 doi 10.3892/ijmm.2019.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. New England Journal of Medicine 2021;384(25):2371–81 doi 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33(5):843–52 e4 doi 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xu X, Yang Y, Liu X, Cao N, Zhang P, Zhao S, et al. NFE2L2/KEAP1 Mutations Correlate with Higher Tumor Mutational Burden Value/PD-L1 Expression and Potentiate Improved Clinical Outcome with Immunotherapy. Oncologist 2020;25(6):e955–e63 doi 10.1634/theoncologist.2019-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kadara H, Choi M, Zhang J, Parra ER, Rodriguez-Canales J, Gaffney SG, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol 2017;28(1):75–82 doi 10.1093/annonc/mdw436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, et al. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochemical and Biophysical Research Communications 2006;351(4):883–9 doi 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Taniguchi S, Elhance A, Van Duzer A, Kumar S, Leitenberger JJ, Oshimori N. Tumor-initiating cells establish an IL-33–TGF-β niche signaling loop to promote cancer progression. Science 2020;369(6501):eaay1813 doi 10.1126/science.aay1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nishina T, Deguchi Y, Miura R, Yamazaki S, Shinkai Y, Kojima Y, et al. Critical Contribution of Nuclear Factor Erythroid 2-related Factor 2 (NRF2) to Electrophile-induced Interleukin-11 Production. J Biol Chem 2017;292(1):205–16 doi 10.1074/jbc.M116.744755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kitamura H, Onodera Y, Murakami S, Suzuki T, Motohashi H. IL-11 contribution to tumorigenesis in an NRF2 addiction cancer model. Oncogene 2017;36(45):6315–24 doi 10.1038/onc.2017.236. [DOI] [PubMed] [Google Scholar]
- 197.Hayashi M, Kuga A, Suzuki M, Panda H, Kitamura H, Motohashi H, et al. Microenvironmental Activation of Nrf2 Restricts the Progression of Nrf2-Activated Malignant Tumors. Cancer Research 2020;80(16):3331–44 doi 10.1158/0008-5472.CAN-19-2888. [DOI] [PubMed] [Google Scholar]
- 198.Hiramoto K, Satoh H, Suzuki T, Moriguchi T, Pi J, Shimosegawa T, et al. Myeloid Lineage-Specific Deletion of Antioxidant System Enhances Tumor Metastasis. Cancer Prevention Research 2014;7(8):835–44 doi 10.1158/1940-6207.Capr-14-0094. [DOI] [PubMed] [Google Scholar]
- 199.Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015;162(6):1229–41 doi 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007;109(9):3812–9 doi 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 201.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med 1999;189(9):1363–72 doi 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chang CH, Curtis JD, Maggi LB Jr., Faubert B, Villarino AV, O’Sullivan D, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013;153(6):1239–51 doi 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 2014;40(5):692–705 doi 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol 2010;185(2):1037–44 doi 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chang W-K, Yang KD, Chuang H, Jan J-T, Shaio M-F. Glutamine Protects Activated Human T Cells from Apoptosis by Up-Regulating Glutathione and Bcl-2 Levels. Clinical Immunology 2002;104(2):151–60 doi 10.1006/clim.2002.5257. [DOI] [PubMed] [Google Scholar]
- 206.Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab 2017;25(2):345–57 doi 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 207.Mellor AL, Munn DH. Tryptophan Catabolism and Regulation of Adaptive Immunity. The Journal of Immunology 2003;170(12):5809 doi 10.4049/jimmunol.170.12.5809. [DOI] [PubMed] [Google Scholar]
- 208.Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, et al. ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell 2015;161(7):1527–38 doi 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Maj T, Wang W, Crespo J, Zhang H, Wang W, Wei S, et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat Immunol 2017;18(12):1332–41 doi 10.1038/ni.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569(7755):270–4 doi 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Graça-Souza AV, Arruda MA, de Freitas MS, Barja-Fidalgo C, Oliveira PL. Neutrophil activation by heme: implications for inflammatory processes. Blood 2002;99(11):4160–5 doi 10.1182/blood.v99.11.4160. [DOI] [PubMed] [Google Scholar]
- 212.Andrade BB, Araújo-Santos T, Luz NF, Khouri R, Bozza MT, Camargo LM, et al. Heme impairs prostaglandin E2 and TGF-beta production by human mononuclear cells via Cu/Zn superoxide dismutase: insight into the pathogenesis of severe malaria. J Immunol 2010;185(2):1196–204 doi 10.4049/jimmunol.0904179. [DOI] [PubMed] [Google Scholar]
- 213.Liu Y, Li P, Lu J, Xiong W, Oger J, Tetzlaff W, et al. Bilirubin possesses powerful immunomodulatory activity and suppresses experimental autoimmune encephalomyelitis. J Immunol 2008;181(3):1887–97 doi 10.4049/jimmunol.181.3.1887. [DOI] [PubMed] [Google Scholar]
- 214.Wu J, Ma J, Fan ST, Schlitt HJ, Tsui TY. Bilirubin derived from heme degradation suppresses MHC class II expression in endothelial cells. Biochem Biophys Res Commun 2005;338(2):890–6 doi 10.1016/j.bbrc.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 215.Lee SS, Gao W, Mazzola S, Thomas MN, Csizmadia E, Otterbein LE, et al. Heme oxygenase-1, carbon monoxide, and bilirubin induce tolerance in recipients toward islet allografts by modulating T regulatory cells. Faseb j 2007;21(13):3450–7 doi 10.1096/fj.07-8472com. [DOI] [PubMed] [Google Scholar]
- 216.Sato K, Balla J, Otterbein L, Smith RN, Brouard S, Lin Y, et al. Carbon monoxide generated by heme oxygenase-1 suppresses the rejection of mouse-to-rat cardiac transplants. J Immunol 2001;166(6):4185–94 doi 10.4049/jimmunol.166.6.4185. [DOI] [PubMed] [Google Scholar]
- 217.Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open 2020;5(2):e000706 doi 10.1136/esmoopen-2020-000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Di Federico A, De Giglio A, Parisi C, Gelsomino F. STK11/LKB1 and KEAP1 mutations in non-small cell lung cancer: Prognostic rather than predictive? European Journal of Cancer 2021;157:108–13 doi 10.1016/j.ejca.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 219.Leone RD, Zhao L, Englert JM, Sun I-M, Oh M-H, Sun I-H, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 2019;366(6468):1013–21 doi doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tenora L, Alt J, Dash RP, Gadiano AJ, Novotná K, Veeravalli V, et al. Tumor-Targeted Delivery of 6-Diazo-5-oxo-l-norleucine (DON) Using Substituted Acetylated Lysine Prodrugs. J Med Chem 2019;62(7):3524–38 doi 10.1021/acs.jmedchem.8b02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tang X, Wang H, Fan L, Wu X, Xin A, Ren H, et al. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radical Biology and Medicine 2011;50(11):1599–609 doi 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 222.Chian S, Thapa R, Chi Z, Wang XJ, Tang X. Luteolin inhibits the Nrf2 signaling pathway and tumor growth in vivo. Biochemical and Biophysical Research Communications 2014;447(4):602–8 doi 10.1016/j.bbrc.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 223.Lee E-J, Oh S-Y, Sung M-K. Luteolin exerts anti-tumor activity through the suppression of epidermal growth factor receptor-mediated pathway in MDA-MB-231 ER-negative breast cancer cells. Food and Chemical Toxicology 2012;50(11):4136–43 doi 10.1016/j.fct.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 224.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, et al. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proceedings of the National Academy of Sciences 2011;108(4):1433 doi 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]