Summary
Understanding the molecular mechanisms of gene regulation is pivotal for understanding how cells establish and modify their identities and functions. Multiple transcription factors (TFs) coordinate to alter gene expression in cells; however, a method to quantitatively analyze the activity of each TF is lacking, particularly in vivo. Here, we introduce a viral-vector-based TF reporter battery that can be used to simultaneously analyze the activity of multiple TFs, visualized as the TF activity profile (TFAP) obtained by qPCR. We show that the cells possess distinct TFAPs that dynamically change according to experimental manipulation or physiological activity. We report a practical method to obtain the TFAP of a defined cell population and their experience-dependent changes in the mouse brain in vivo. The TFAP obtained by our method will help bridge the information gap between the genome and transcriptome and aid the multi-omics view of understanding the gene regulation system.
Subject areas: Biological sciences, Neuroscience, Cell biology, Biological sciences research methodologies, Biology experimental methods, Methodology in biological sciences
Graphical abstract

Highlights
-
•
A virus-based reporter battery for obtaining the TF activity profile of cells
-
•
TFAP analysis reveals the dynamic change of TF activity upon stimulation
-
•
Experience-dependent change of TFAP of the mouse brain in vivo
-
•
Neural and glial change of TF activity revealed by the cell-type-specific TFAP
Biological sciences; Neuroscience; Cell biology; Biological sciences research methodologies; Biology experimental methods; Methodology in biological sciences
Introduction
To better understand how organisms react and modify themselves according to environmental stimuli, biological research necessitates the investigation of how each cell changes its transcription. Recent advances in the DNA sequencing techniques have allowed us to have a deeper understanding of the genome, epigenome, and transcript profiles; however, combining such multi-omics data to obtain a comprehensive description of the molecular mechanisms of gene transcription is still challenging. To further advance our knowledge, it is necessary to quantitatively visualize the activity of transcription factors (TFs) that physically interact with genomic DNA and have a key role in converting the input of a cell to the transcriptional output (Vaquerizas et al., 2009). Several methods are currently used to directly or indirectly measure the activity of TFs in cells. An indirect way of predicting the activity of TFs is to analyze the gene regulatory elements of differentially expressed genes among the cells of distinct populations or conditions. This method can estimate the potential activities of TFs from the contents of the TF binding sequences (TFBS) in the promoter/enhancer region of differentially expressed gene sets (Essaghir et al., 2010); however, each gene is synergistically regulated by multiple TFs, and the presence of TFBS does not always reflect its interaction with TF, making results difficult to interpret. Physical associations of TFs with genomic DNA can be revealed by chromatin immunoprecipitation sequencing (ChIP-seq) analysis (Furey, 2012), but such interactions do not directly reflect the transcriptional activity of TFs (Sheng et al., 1988). For some TFs, the activity can be analyzed by measuring the posttranslational modification of the TF, such as phosphorylation or trans-localization of the TF to the nucleus; however, these modifications also do not directly indicate the gene transcription activity of the TF. In addition, such modifications can only be analyzed in certain TFs.
A widely accepted standard for measuring the transcriptional activity of TFs is the luciferase reporter assay (Alam and Cook, 1990). In this assay, in vitro cells are transfected with TF reporter plasmids bearing regulatory element sequences containing TFBSs of the target TF. The interaction between the TF and TFBS influences the transcription efficiency of the reporter gene, such as firefly luciferase. Thus, measuring the changes in reporter gene expression allows for direct measurement and comparison of the gene transcription activity of the targeted TF. Although this method is widely used in in vitro assays, measuring the TF activity in vivo remains challenging because the amount of reporter transcripts is strongly affected by the gene copy number, which is often difficult to control in in vivo. To overcome such difficulties, we have previously generated a lentivirus (LV)-based TF activity reporter (Abe et al., 2015). This system contains two independent promoters to compensate for the transfection efficiency. In the present study, we have expanded this reporter vector system to allow multiple TF analyses in the sample. This new TF reporter system allows us to directly measure the multiple TF activity as a TF activity profile in the cell.
Results
Developing the TF reporter virus battery
We previously developed a lentivirus-based reporter that allows quantification of endogenous gene transcription activity of the cAMP response element-binding protein (CREB) transcription factor (Abe et al., 2015). This construct expresses a reporter gene under a CREB-dependent promoter and a transfection reference gene (Ref-gene) under a constitutively active Pgk1 (phosphoglycerate kinase 1) promoter. By substituting the TFBS in the CREB-dependent promoter to other TFs, this construct can report the gene transcription activities of various TFs. We created reporter constructs for 56 distinct TFBSs of widely investigated TFs that were used in the conventional reporter assays, along with three control promoters (mini, Flt-pro, Arc-pro; Figure S1). Different sets of reporter and reference genes with different sequences were designed for each TF reporter, allowing quantification of the activity of multiple TFs in a single sample, to increase the throughput of analysis (Figure 1). Quantification of each TF activity was performed by using PCR primer sets specific to each of the Ref- and Rep-genes. By quantitatively analyzing the activity of multiple TFs in the sample, we obtain a TF activity profile (Romanov et al., 2008) (TFAP), which represents the endogenous activity of multiple TFs in the cell, indicating the transcriptional state of target cells (Figure 1).
Figure 1.
Schematics of TF activity profiling
(A) Image of lentiviral-vector-based TF activity reporter. This bipromoter vector expresses paired genes, a transfection reference gene (Ref) under the control of a constitutive Pgk promoter and TF activity reporter gene (Rep) under the TF activity-dependent promoter, which consists of a minimal promoter and TFBSs. FII, insulator sequence, LTR, long terminal repeat.
(B) Schematics of TF activity profiling procedures. The mRNAs from the cells transfected with multiple mixtures of the lentivirus-based TF activity reporter were collected and reverse transcribed using Rep-gene RT-primer and Ref-gene RT-primer, designed at the common sequence in all Rep-genes and Ref-genes. RT-PCR products were subjected to qPCR analysis using gene-specific primer pairs to each Ref and Rep gene. The activities of each TFs were calculated as Rep-gene expression value divided by each of the corresponding Ref-gene expression values.
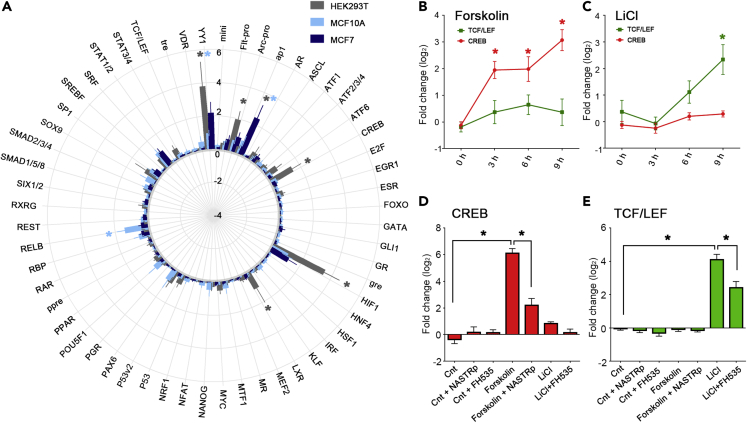
We obtained the TFAP of three cell lines (HEK293T, MCF7, and MCF10A) by transfecting cells with 56 TF reporter viruses and quantifying the activity of each TF (Figure 2A). The three cell lines had significantly different TFAPs representing the endogenous activities of 56 TFs (two-way ANOVA, F(2,931) = 4.223, p = 0.015). Post hoc analysis revealed significant changes in the activity of CREB, YY1, liver X receptor (LXR), hypoxia-inducible factor 1α (HIF1a), activator protein 1 (AP1), and a synthetic Arc/Arg3.1 promoter (Kawashima et al., 2009) in human embryonic kidney 293 (HEK293T) cells compared with MCF7 cells. Between MCF7 and MCF10A cells, we found a significant reduction of activity of YY1, AP1, and activation of RELB in MCF10A cells. These TFs may reflect the proliferation efficiency of HEK293T cells or the potential mechanism of tumor malignancy in MCF7 (Inglés-Esteve et al., 2012; Ludes-Meyers et al., 2001; Wan et al., 2012) and may contribute to the establishment of cell-line-specific transcriptomes (Uhlen et al., 2017; Yu et al., 2019).
Figure 2.
TF activity profile of cell culture
(A) Round bar plot showing the TF activity profile of HEK293T, MCF7, and MCF10A cells. TF activity as calculated by Rep expression divided by Ref expression value are shown. ∗p < 0.05, Dunnett’s test compared with MCF7, n = 3–7.
(B and C) Pharmacological effect on TF activity. HEK293T cells transfected with TCF/LEF (green) and CREB (red) reporter viruses were treated with 200 μM forskolin (B) or 20 mM LiCl (C) for 3, 6, or 9 h. Log2 fold change of TF activities compared with 0 h are shown. ∗p < 0.01, Dunnett’s test compared with 0 h, n = 12.
(D and E) Effect of inhibitors. Cells were pretreated with 10 μM NASTRp or 25 μM FH535 and stimulated with forskolin or LiCl for 9 h. Log2 fold change of TF activities compared with the control-treated culture (Cnt) are shown. ∗p < 0.01, Sidak’s test, n = 12. Mean ± SEM are shown.
The activity of TFs is dynamically regulated by cell signaling pathways. To understand how TFAP changes based on the state of the cells, HEK293T cells were treated with small-molecule reagents to manipulate cell signaling pathways. TFAP was obtained 0, 3, 6, and 9 h after drug initiation. We detected sustained upregulation of CREB beginning at 3 h after treating cells with forskolin, an adenylyl cyclase activator (Figure 2B). Treating cells with LiCl, a drug that inhibits glycogen synthase kinase (GSK) 3β and activates the β-catenin-TCF/LEF pathway, resulted in a significant upregulation of TCF/LEF activity after 9 h, without affecting the activity of CREB (Figure 2C). These activations were suppressed by the pretreatment of the cells with Naphthol AS-TR phosphate (Lee et al., 2015) (NASTRp) or FH535 (Handeli and Simon, 2008), specific inhibitors of CREB or TCF/LEF-dependent gene transcription (Figures 2D and 2E). Therefore, TF activity profiling allows a quantitative visualization of the physiological state of the cell and how it dynamically changes across stimulations.
Neural-activity-dependent TF activity profile changes
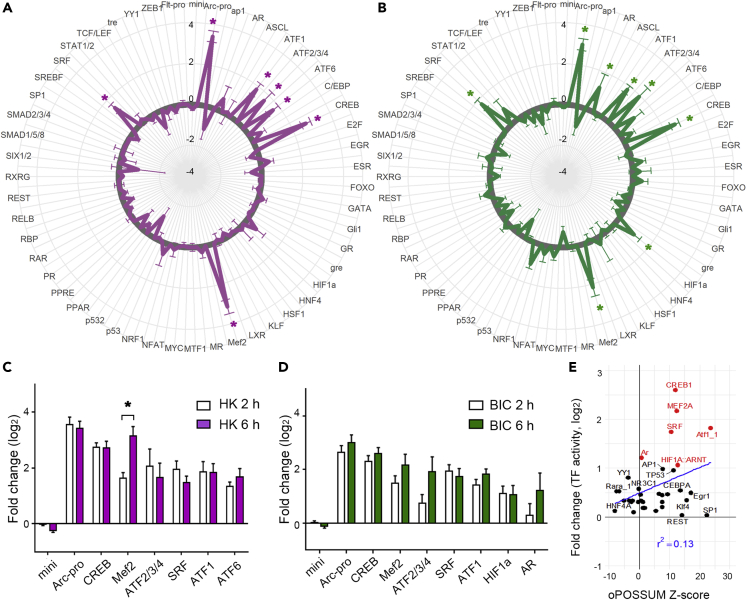
Several TFs in neurons are regulated by neural activity and are suggested to have important roles in establishing plastic changes in neural systems; however, how these TFs are coordinately activated is not well understood (Flavell and Greenberg, 2008). To further demonstrate the utility of our viral-vector-based TF activity profiling, we analyzed the changes in TFAP after neural stimulation. We analyzed the TF activity of mouse cortical primary cultures at 15 div (days in vitro) when neurons had functionally matured. We stimulated neurons with two different stimulation protocols: depolarization by high K+ buffer (Tao et al., 1998) (HK stim) and synaptic activity enhancement by inhibitory neurotransmitter antagonists (Hardingham et al., 2002; Kawashima et al., 2009) (bicuculine/4-AP stimuli, BIC stim). Both protocols sustainably increased action potential firing, and the activity of 49 TFs was compared with the nonstimulated control culture (Figures 3A and 3B). After stimulation for 6 h, a significant change of TFAP against that of no stimulated control was observed in both protocols (two-way ANOVA; HK stim, F(1, 853) = 13.37; BIC stim, F(1, 845) = 23.49; both p < 0.001). Notably, there was a high correlation between the activation pattern of the HK stim and BIC stim (r = 0.89, p < 0.001), indicating the robustness of our detection methods. Post hoc analysis demonstrated that compared with the control culture with no stimulation, significant upregulation of TF activity was observed in the reporter constructs for ATF1, ATF2/3/4, CREB, Mef2, and SRF in both protocols, ATF6 in HK stim and AR and HIF1a in BIC stim. Many of these TFs have been reported to be regulated by neural activity and have a role in learning and memory in rodents (Arias-Cavieres et al., 2020; Flavell and Greenberg, 2008; Lyons and West, 2011; Picot et al., 2016). A significant upregulation of activity in both stimulations was also observed in the synthetic promoter Arc-pro (Kawashima et al., 2009), which reflects the activity of the endogenous promoter of an activity-dependent gene, Arc/Arg3.1. Among the TFs with significantly enhanced activity after the 6 h stimulation, we also analyzed the activity after 2 h of stimulation (Figures 3C and 3D). Most TFs showed similar change in activity 2 h after stimulation, except for Mef2, which showed a significant increase in TF activity between 2 and 6 h after HK stimulation. These data show coordinated activation of TFs by neural activity, which may function during the activity-dependent gene expression in neurons.
Figure 3.
Profiling of the neural-activity-dependent change of TF activities.
(A and B) Radar graph showing the change of transcription factor activity change 6 h after stimulation with high K+ buffer (A) or bicuculine (B). Mouse cortical neuron culture at 15 div was stimulated and TFAP was obtained. For each TF, the change of TF activity against control stimulated culture is plotted as log2 fold change values. Values outside the gray circle indicate increased and values inside indicate decreased activity to control. Mean ± SEM are shown. ∗p < 0.05, Dunnett’s test, n = 4–33.
(C and D) TF activity change of neuron culture stimulated with high K+ buffer (HK, C) or bicuculine (BIC, D) for 2 and 6 h is shown as mean ± SEM ∗p < 0.05, Sidak’s test, n = 7, 8.
(E) Plot showing the absolute value of log2 fold change of TF activity after bicuculine treatment for 6 h to the enrichment of TFBSs in the promoter (oPOSSUM Z score) of selected 173 activity-dependent genes (Tyssowski et al., 2018). TFs with significant changes of activity by LV-based reporters are labeled in red.
Next, we compared the outcomes of our method with those of conventional methods. The activity of TFs involved in establishing the change in the transcriptome can be inferred by analyzing the differentially expressed gene sets. A commonly used approach is to investigate the enrichment of TFBS in the promoter region of those gene sets against those of the background genes. Using published gene set data (Tyssowski et al., 2018) and the oPOSSUM database (Ho Sui et al., 2005), we analyzed the enrichment of TFBS in differentially expressed genes after 6 h after bicuculine stimulation. We detected a proportional trend with the enrichment of TFBS, as revealed by the oPOSSUM Z score and the TF activity change demonstrated by our protocols (r = 0.359, p < 0.035; Figure 3E). In our protocols, compared with nonsignificant TFs, a significant increase in Z score was observed in the TFs, showing significant changes after stimulation (average of significant TFs = 11.99; nonsignificant TFs = 3.75; Welch’s t test, p = 0.04). These results demonstrate the relevance of our methods compared with previously existing methods to indirectly infer the activity of TFs.
TF activity profiling in vivo
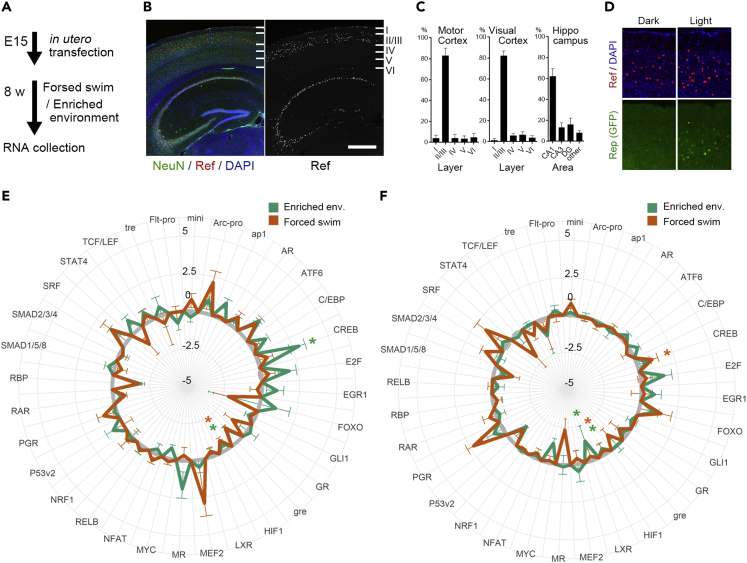
Studies quantifying TF activities were performed primarily in vitro using cultured cells. The TF reporter virus system developed in the present study allows for quantification of the TF activity in vivo (Abe et al., 2015). To obtain TFAP in vivo, it is necessary to restrict the expression of the reporter constructs to the restricted cell population, because lentiviruses transfect various cell populations that may have distinct TFAPs. To measure the TF activity in the adult mice in vivo, we injected the reporter viruses into the ventricle of the brain of embryonic day 15 (E15) embryos (Figure 4A). This procedure resulted in the expression of the transgene constructs in the defined cell populations in the adult brain. After the mice aged to 8 weeks, the expression was mainly detected in layer II/III of the cortex, CA1 pyramidal cell in the hippocampus (Figures 4B and 4C), and medium spiny neurons in the striatum. The expression of Ref and Rep gene can be observed in single-cell level (Figure 4D). The restricted expression of reporter constructs in a defined population allows us to reproductively quantify the activity of TFs in those cell populations after dissecting and collecting RNAs from these regions.
Figure 4.
TF activity profiling of neurons in vivo
(A) Experimental time course.
(B) Fluorescent image of a brain section of 8-week-old mouse, expressing the TF-reporter construct immunostained with antibodies recognizing neuronal nucleus (NeuN, green) and Ref gene (FLAG tag, red), shown with DAPI (blue). Scale bar, 500 μm.
(C) Classification of cells expressing Ref gene in M1 area of the motor cortex (left), V1 area of the visual cortex (middle), and the hippocampus (right), n = 6.
(D) Brain section of visual cortex immunostained with Ref gene (V5-tag, red) shown with DAPI (blue). The mouse expressing CREB-reporter construct was kept in the dark for 3 days and exposed to light for 4 h before fixation.
(E and F) TF-activity profiles of the cortex (E) and the hippocampus (F) from mice experienced enriched environment exploration (green) and forced swim (orange). For each TF, the change of TF activity against the home-caged mouse was calculated and plotted as log2 fold change. ∗p < 0.05, Dunnett’s test, n = 6–30. Mean ± SEM are shown.
To obtain TFAP in vivo and analyze how they change across stimulations, we quantified the activity of 30 TFs in the mouse brain 60 min after enriched environment exploration (Ramanan et al., 2005) or 120 min after the forced swim and compared them with home-caged controls (Figures 4E and 4F). We found a significant difference in the TF activity pattern among control, forced swim, and enriched mice (two-way ANOVA, F(2, 1159) = 4.639, p < 0.01). Post hoc analysis revealed significant upregulation of CREB activity in control versus enriched in the cortex, control versus forced swim in the hippocampus, downregulation of LXR in control versus enriched in the hippocampus, and HIF1a in control versus enriched and control versus forced swim mice in both cortex and hippocampus. Together, our method to quantify the TF activity in vivo offers an efficient analysis of the cell states and the experience-dependent change of them in the mouse brain.
Cell-type-specific TF activity profiling
Because each cell type has a different transcriptome, the transcription activity profile may differ between cell types (Zeisel et al., 2015). Consequently, to utilize our TF profiling, measurement of the TF profile of a specific cell type is required. To further obtain the TFAP of specific cell types, we utilized a Cre recombination system (Saunders et al., 2012) along with adeno-associated virus (AAV)-based TF reporter constructs (Figure 5A). Cre-dependent conversion of the direction of the transgene allowed the PCR-based detection of TF-activity-dependent expression of the reporter gene only in cells expressing Cre recombinase (Figure 5B). To analyze the cell-type-specific activity of TFs, we quantified the TF activities of neurons and glia in dissociated cultures of the mouse cortex. Cre recombinase was expressed specifically in neurons by the synapsin-1 promoter or glial cells by the glial fibrillary acidic protein (GFAP) promoter (Shigetomi et al., 2013) (Figure 5C). Among 47 TF reporters, we compared the endogenous activity between neurons and glial cells. We observed a significant difference in TFAP between neurons and glial cells (two-way ANOVA, F(1, 478) = 6.179, p = 0.0013), and the activity of the 14 reporters was increased or decreased more than 2-fold (Figure 5D). To further confirm the observed difference, we independently prepared neuronal and glial-cell-enriched cultures and obtained their TFAP using LV-based TF activity reporters (Figure 5E). Comparing the relative activity of 45 TFs in neuronal and glial cells, we observed a correlation between the AAV system and LV systems (r = 0.534, p < 0.0001; Figure 5F). In addition to restricted transfection of lentiviral vectors, the AAV system offers a cell-type-specific analysis of TFAP.
Figure 5.
AAV-based TF activity reporter for cell-type-specific TF activity profiling
(A) Schematics of the constructs. AAV for cell-specific expression of Cre recombinase (upper), Ref gene expression by a constitutive promoter (Pgk, middle), and TFBS-dependent Rep gene (bottom) are shown.
(B) Schematic of TF activity measurements. With the presence of Cre recombinase, the direction of the Rep gene flips, allowing the amplification of the cDNA by PCR primers.
(C) Cell-type-specific expression of GFP in 15 div cortical culture. Synapsin promoter (neurons) and GFAP promoter (glial cells) were used to express Cre in a specific cell type, GFP was used as the Ref gene. Cultures were also transfected with the synapsin-RFP virus. Scale bar, 50 μm.
(D) Log2 fold change of TF reporter activity in neuron/glia obtained using AAV-based system. n = 4–30.
(E) Log2 fold change of TF reporter activity in neuron/glia obtained using LV-based system. n = 4–7.
(F) Scatterplot showing the relation of relative TF activity of neuron and glia obtained from AAV-based and LV-based system.
Discussion
Regulation of gene expression is a highly complex process involving the joint activity of multiple layers of molecular factors, such as the genome, epigenome, and transcription factors. By direct measurement of the activity of each TFs in situ, our method provides an alternative way to the bioinformatical techniques to infer the TF activity and utilizing both data together will aid in bridging the gap between genome and transcriptome information. Our method is applicable to in vivo analyses and can aid in better understanding how the activity of multiple TFs simultaneously changes under physiological conditions or to drug treatments. Recent advances in transcriptome analysis have revealed that the expression of many genes change upon stimulation or in diseased conditions, but the methods to manipulate multiple genes are limited, hindering the research needed to understand the causal relationship between multiple gene regulation and the observed phenotype. Because TFs control the expression of multiple genes, analyzing and manipulating TF activity is promising for regulating various biological processes. TF profiling is also useful for high-throughput analysis of the effects and side effects of biochemicals (Medvedev et al., 2018); thus, our current method will be useful in assessing the effect of biochemicals in vivo. Because aberrant regulation of transcription factors is suggested to be involved in the pathophysiology of many diseases (Ebert and Greenberg, 2013; Lambert et al., 2018; Lee and Young, 2013; Maurano et al., 2012), analyzing the activity state of TFs will be helpful in developing effective treatments.
Gene expression is regulated not only by TF activity but also by epigenetic factors or enhancer-mediated structural modification of the genome, and each gene is regulated synergistically with multiple combinations of TFs (Lyons and West, 2011; Smith et al., 2013). In addition, the number and positional relation of TFBSs are known to influence the behavior of the regulatory region of the gene (Smith et al., 2013). TFAP revealed by our method should not be conceived as a direct reflection of the transcription mechanism of a genome locus in the endogenous chromatin contexts; instead, it should be considered as a method to assay the activity of each TF directly. After collecting the detailed activity of each TF, how multiple TFs and epigenetic regulation synergistically regulate the gene expression of a particular locus is an important future issue to be addressed. For example, combining this method with a massively parallel reporter assay (Smith et al., 2013) to analyze the activity of cis-regulatory sequences of endogenous chromatin contexts in vivo will increase the throughput of data acquisition and contribute to the complete understanding of transcription mechanisms in an organism.
Limitations of the study
Our current method offers a direct measurement of the transcriptional activity of each TF. Along with conventional methods such as transcriptome analysis, genome sequencing, ChIP-seq (Furey, 2012), ATAC-seq (Buenrostro et al., 2015), and Hi-C (Kempfer and Pombo, 2020), our method will aid research necessary to provide insights into the molecular mechanisms of gene regulation; however, because our methods largely rely on the designed TFBS of each reporter construct, we should be cautious about the interpretation of our present results. For example, because the optimal TFBS were overlapped in multiple TFs, and some TFs recognize multiple motifs, it is possible that our reporter constructs do not indicate the activity of a single TF (Inukai et al., 2017). It is also possible that the readout activity of TFs changes according to the TFBSs. Although additional confirmation to pinpoint the actual TF is required, our method offers a direct view of how the activities of TFs involved in the regulation of a particular gene regulatory sequence presented in the genome differentially changes upon stimulation.
We also do not know, at this moment, whether the TFAPs described in this study represent the TFAP of single cells shared within the cell population or rather an average of the defined cell population that have distinct TFAPs. Although our preliminary single-cell level analysis on the histological section suggests that each cell has different TF activity, more high-throughput measurements such as those using single-cell transcriptome sequencing will be necessary to address.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Flag tag | SIGMA | Cat#F7425; RRID: AB_439687 |
| Rabbit monoclonal anti-RFP | MBL | Cat#PM005; RRID: AB_591279 |
| Mouse monoclonal anti-EGFP | MBL | Cat#M048; RRID: AB_591822 |
| Mouse monoclonal anti-Fox3 | MERCK | Cat#MAB377; RRID: AB_2298772 |
| Mouse monoclonal anti-V5 tag | Thermo Fisher | Cat#R96025; RRID: AB_2556564 |
| Goat anti-Mouse IgG Alexa Fluor 488 | Thermo Fisher | Cat#A11001; RRID: AB_2534069 |
| Goat anti-Rabbit IgG Alexa Fluor 555 | Thermo Fisher | Cat#A21428; RRID: AB_2535849 |
| Bacterial and virus strains | ||
| AAV2/9-DIO-Rep | This Paper | N/A |
| AAV2/9-DIO-mCherry | This Paper | N/A |
| AAV2/9-hSynI-Cre | This Paper | N/A |
| AAV2/9-GFAP-Cre | This Paper | N/A |
| AAV2/9-hSynI-RFP | This Paper | N/A |
| AAV2/9-DIO-Pgk1-EGFP | This Paper | N/A |
| LV-Rep/Ref | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DL-amino-5-phosphonopentanoic acid | SIGMA | Cat#165304 |
| Tetrodotoxin | WAKO | Cat#206-11071 |
| 4-Aminopyridine | SIGMA | Cat#A78403 |
| Glycine | WAKO | Cat#073-00737 |
| Strychnine nitrate | WAKO | Cat#195-11151 |
| DAPI | Dojindo Molecular Technologies | Cat#D523 |
| EGF | Higeta Shoyu | Cat#REG100UG |
| Insulin | WAKO | Cat#093-06471 |
| B27 supplement | Thermo Fisher | Cat#17504044 |
| Cholera toxin | SIGMA | Cat#C9903 |
| Hydrocortisone | SIGMA | Cat#H0888 |
| Naphtol AS-TR phosphate | TCI | Cat#C2250 |
| FH535 | MedChemExpress | Cat#HY-15721 |
| Forskolin | WAKO | Cat#061-02191 |
| Lithium chloride | WAKO | Cat#123-01162 |
| Bicuculine methiodide | WAKO | Cat#023-16141 |
| DMEM high glucose | WAKO | Cat#043-30085 |
| Neurobasal Medium | Thermo Fisher | Cat#21103049 |
| Experimental models: Cell lines | ||
| HEK293T | TaKaRa | Cat#632180 |
| MCF7 | JCRB cell bank | JCRB0134 |
| MCF10A | ATCC | CRL-10317 |
| Experimental models: Organisms/strains | ||
| Mouse: Slc:ICR | SLC Japan | |
| Oligonucleotides | ||
| Rep-RT-primer 5′’-tggagggaagccgtgagaa-3’ | This paper | N/A |
| Ref-RT-primer 5’-ccacatagcgtaaaaggagcaac-3’ | This paper | N/A |
| Recombinant DNA | ||
| pAAV-DIO-Rep | This paper | N/A |
| pLV-Rep/Ref | This paper | N/A |
| pLV-TFRep1-creb | This paper | Addgene#182054 |
| pLV-TFRep2-creb | This paper | Addgene#182055 |
| pLV-TFRep3-creb | This paper | Addgene#182056 |
| pLV-TFRep4-creb | This paper | Addgene#182057 |
| pLV-TFRep5-creb | This paper | Addgene#182058 |
| pLV-TFRep6-creb | This paper | Addgene#182059 |
| pAAV-pgk-DIO-rvCRY-WPRE | This paper | Addgene#182060 |
| Software and algorithms | ||
| Adobe Photoshop CS6 | Adobe | N/A |
| GraphPad Prism 6 | GraphPad Software | Version 6.03 |
| Image J | NIH | Ver 1.53c |
| R | The R foundation | Ver 4.1.1 |
| BioRender | https://biorender.com/ | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact: Kentaro Abe (k.abe@tohoku.ac.jp).
Materials availability
Plasmids necessary to construct the plasmids and the viruses generated in this study have been deposited to Addgene, 182054, 182055, 182056, 182057, 182058, 182059, 182060.
Experimental model and subject details
Animals
The care and experimental manipulation of animals used in this study were reviewed and approved by the Institutional Animal Care and Use Committee at Kyoto University and Tohoku University. Male mice aged 8–12 week were used for the behaivoral analysis and TF activity profiling. Mice were purchased from Japan SLC and kept group housed on a 12-hour / 12-hour light cycle, water and food were given ad libitum.
Cell lines
HEK293T cells were purchased from TaKaRa (Lenti-X 293T cell line, 632180), MCF7 were obtained from JCRB cell bank (JCRB0134), MCF10A from ATCC (CRL-10317). HEK293T, MCF7, MCF10A are human derived cell line with the sex of female. HEK293T cells were grown at 37°C with Dulbecco's Modified Eagle Medium (DMEM high glucose, WAKO), 10% Fetal Calf Serum (Biowest) with penicillin and streptomycin (PS, WAKO). MCF7 cell were grown with Eagle's Minimum Essential Medium (E-MEM, Thermo Fisher) supplemented with non-essential amino acid (1×, Thermo Fisher), sodium pyruvate (1 mM, Thermo Fisher) and PS. MCF10A cell were grown with DMEM/F12 (WAKO), 5% Horse Serum (Thermo Fisher), EGF (20 ng/mL, Higeta shoyu), hydrocortisone (0.5 mg/mL, SIGMA), cholera toxin (100 ng/mL, SIGMA), insulin (10 μg/mL, WAKO) and PS. Cells were transfected with the reporter viruses and the total RNA was collected at 6-days after the transfection. In the drug treatment experiments, HEK293T cells transfected with lentiviruses were treated with Naphthol AS-TR phosphate (NASTPp; 10 μM, TCI), FH535 (20 μM, MedChemExpress), forskolin (200 μM, WAKO), or LiCl (20 mM, WAKO).
Primary cell culture
Cortical neuron culture was prepared from E15 embryo of ICR mouse (Slc: ICR) and maintained at 37°C in Neurobasal Medium (Thermo Fisher) supplemented with B27 supplement (Thermo Fisher), Gultamax-I (Thermo Fisher) and PS. To get cortical glial cell culture, the dissociated cortical cells were plated and maintained in DMEM, 10% Fetal Calf Serum with PS. Cells under passage 2–3 were used for analysis. Without the determination of the sex, embryos were mixed and used. Neurons were transfected with the viruses at 3 div. At 15 div, the cells were stimulated with a bath application of high K+ buffer (final concentration 55 mM), stimulated after overnight application of tetrodotoxin (1 μM, WAKO) and DL-amino-5-phosphonopentanoic acid, (DL-AP5; 100 μM, Tocris), or the inhibitory synaptic transmission inhibitors which consists from bicuculine methiodide (30 μM, WAKO), 4-aminopyridine (4-AP; 100 μM, SIGMA), glycine (100 μM, WAKO), strychnine nitrate (1 μM, WAKO) for either 2 h or 6 h. To analyze the difference of TFAP of neuronal and glial cells, glial culture was transfected with the same viruses on the same day of neural culture transfection (3 div), and both cells were lysed to collect total RNA at the same timing (15 div of neural culture) without stimulation.
Method details
TF reporter constructs
To construct lentivirus-based TF activity reporter constructs, we replaced the TFBS of LV-CREB-reporter (Abe et al., 2015) with TFBS of various TFs. Regulatory elements with TFBSs were selected from the literature (Figure S1). The TFBSs were synthesized and flanked with BamHI and HindIII sites to replace with CREB binding sequence of LV-CREB-reporter using BamHI and HindIII sites. For reporter gene, expressed by TF dependent promoter, the original reporter gene of LV-CREB-reporter, turboGFP, was replaced to GFPs with nucleotide sequence modifications, using AgeI and BsrGI sites. We used six reporter genes (Rep#1–6) having distinct sequences (Table S1). For reference gene, the original reference gene of LV-CREB-reporter, Histone-2B, was replaced with histone genes with nucleotide sequence modifications, using Aor51HI and NheI sites. We used six reference genes (Ref#1–6) each of which consisted of nucleotide sequence modified mouse Histone-2A or 2B tagged with distinct immunostaining tags (Table S1). This replacement result in the 6 constructs expressing different set of Rep and Ref genes (pLVTFrep#1 to pLVTFrep#6) for which TFBSs were inserted between the BamHI and HindIII site. A reporter-construct backbone without TFBS (mini, Promega pGL4.23), and a synthetic promotor element composed from human ferritin H promoter (Bevilacqua et al., 1992), or a synthetic Arc/Arg3.1 enhancer/promoter (Kawashima et al., 2009) were used as control promoters.
To construct AAV-based TF activity reporter constructs, we synthesized a double floxed cassette consisting of loxP and lox2272, as described before (Saunders et al., 2012), and cloned the Rep-genes in the reverse orientation using NheI and AscI sites. In the AAV system, all of the six Rep gene (Rep#1–6) and the five Ref gene (Ref#2–6) of LV system were used without distinction of the Rep gene and the Ref gene. The Ref#1 gene of the LV system and RFP (mCherry) gene were used as transfection reference gene expressed by Pgk1 promoter (AAV-DIO-Pgk1-Ref#1; DIO, double floxed inverted orientation (Saunders et al., 2012)). TFBS and the minimal-promoter region were obtained from the LV-based system and inserted into the constructs using BamHI and SalI sites. Each of AAV-based TF reporter constructs was created by subcloning the BamHI and SalI site from corresponding LV-based TF reporter constructs, and one of 11 reporter genes (Rep#1–6 or Ref#2–6 gene). Up to 11 distinct AAV vectors bearing Rep#1–6 or Ref#2–6, together with two reference-virus (AAV-DIO-Pgk1-Ref#1 and AAV-DIO-Pgk1-mCherry), and Cre expressing AAV vector (AAV-Syn1-Cre or AAV-GFAP-Cre) were mixed and used. AAV-GFAP-Cre was constructed from cloning the promoter region (GFAP-promoter) of pZac2.1-gfaABC1D-Lck-GCaMP3 (gift from Baljit Khakh, Addgene plasmid #44330). AAV-Syn1-Cre was constructed from cloning the promoter region (Syn1-promoter) of LSYN-EGFP-zCREB (Abe et al., 2015). AAV-DIO-Pgk1-EGFP was constructed from AAV-DIO-Pgk1-Ref by replacing Ref to EGFP sequence.
Virus preparation
Lentivirus based reporters were prepared as described before with a brief modification (Abe et al., 2015). Briefly, HEK293T cells were transfected with third-generation plasmids. 72 h after transfection, the medium was collected, then filtrated through a 0.45 μm pore size filter. The medium was mixed with PEG-8000 (polyethylene glycol 8000; final 8% w/v, Promega) overnight and centrifuged to sediment viral particles. Then, viruses were dissolved in phosphate-buffered saline (PBS) and further concentrated by ultrafiltration using VIVASPIN-500 (Sartorius). Lentiviruses with the titer of 2.0 × 1010 – 8.0 × 1012 infectious unit / mL were used.
For preparing AAV based reporter, HEK293T cells were transfected with pHelper, pAAV-RepCap, and pAAV ITR-expression vector (Stratagene). All the AAVs used in this study were made as AAV serotype 2/9. After 72 h of transfection, the medium was removed, cells were collected, and lysed by repetitive freeze and saw. The DNaseI resistant viral particles were purified by a chloroform PEG/aqueous two-phase partitioning purification (Guo et al., 2012) and further concentrated by ultrafiltration using VIVASPIN-500. Viruses with the titer of 1.0 × 1012 – 3.0 × 1014 DNaseI resisting viral particle / mL were used.
Sample preparation for TF activity profiling
For sample preparation for TF activity profiling in vitro, cell cultures were transfected with a mixture of reporter viruses. In using LV-based reporters, up to six viruses containing distinct TFBS and Rep-Ref pairs (Rep#1–6 / Ref#1–6) were mixed. In using AAV-based reporters, up to 14 viruses were mixed. Total RNA was obtained from the cell culture samples with homogenization and purification using MagExtractor-RNA (TOYOBO). Before processing further purification, each sample was frozen and stored at −80°C after the cell lysis.
For sample preparation for TF activity profiling in vivo, reporter LVs were injected into the ventricle of E15 embryo of ICR mouse (Slc: ICR). In using LV-based reporter, up to six viruses containing distinct TFBS and Rep-Ref pair (Rep#1–6 / Ref#1–6) were mixed, and the mixture solution up to 1 μL was injected into each embryo in utero. The profiling of TF activity was performed with 8–12-month mice that were transfected at E15 in utero. To obtain TFAP after experiencing an enriched environment, we kept mice singly for two days and then moved them into a novel cage (40 × 40 cm) about two times larger than their home cage, containing different textured beddings and enrichment toys. 60 min after the cage replacement, mice were euthanized, and their brain was rapidly collected. The control-treated mice were kept in their home cage. The forced swim was conducted at 8–12 months. For two consecutive days, mice were kept in the water of 25°C for 12 min each day, then dried and moved back to their home-cage. Mice showed a significant increase of immobility time during the 12-min-swim on the second day. 120 min after the second forced swim, mice were euthanized, and their brain was rapidly collected. The hippocampus and cortex of both hemispheres were dissected out from each mouse and subjected to total RNA purification. Total RNA was obtained from the tissue samples with homogenization and purification by guanidinium thiocyanate-phenol-chloroform extraction (Chomczynski and Sacchi, 1987). Before processing further purification, each sample was frozen and stored at −80°C after the tissue homogenization.
Obtaining TFAP
Purified RNAs from cell and tissue samples were reverse transcribed to obtain cDNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). Gene specific primers targeted to the common sequence of Rep-genes (5’-tggagggaagccgtgagaa-3’) and Ref-genes (5’-ccacatagcgtaaaaggagcaac-3’) were mixed and used for RTPCR. Then, the amount of each Rep-gene and Ref-gene were quantified using the real-time PCR system (Light Cycler 480, Roche; or CFX384, BioRad), using GoTaq qPCR Master Mix (Promega). The sequence of the primers used for the quantitative PCR are listed in Table S2. Each primer pairs were confirmed to amplify only the specific targeted genes among other Ref and Rep genes. To calculate the TF activity, the amount of reporter mRNA was normalized by the amount of reference mRNA for each TF construct. In the AAV system, each reporter expression was normalized with the reference AAV which contains Pgk1 promoter instead of the TF dependent promoter. For calculating TFAP of cultured neurons, log2 fold change against control-treated culture were calculated for each experiment and analyzed. For calculating TFAP in vivo, TF activity of each animal was stocked and log2 fold change against control-treated mouse was calculated and analyzed.
Analyzing TFAP data
In the neuron culture experiment, the published gene set of 173 genes which are differentially regulated in a similar experimental condition (Tyssowski et al., 2018) (mouse cortical neuron culture stimulated with bicuculine for 6 h) were analyzed as activity-dependent genes. This includes 19 rapid primary response genes (rPRG), 116 delayed primary response genes (dPRG), and 38 secondary response genes (SRG). Using the oPOSSUM (Ho Sui et al., 2005) ver3.0, the enrichment of TF regulatory motifs in the 5000 base pairs upstream and downstream of the transcription start site was queried against registered 29347 background genes using the single-site-analysis methods of default parameters. From the results, 35 TFs that our system covered in the bicuculine treatment experiment were further analyzed.
Immunostaining
Mice expressing LV-based TF reporters were euthanized by anesthetization and perfused with PBS, followed by 4% paraformaldehyde / PBS. The brains were collected and frozen at −80°C after cryoprotection with 30% sucrose / PBS, then sliced into 40 μm section using cryo-microtome (CM1850, Leica). Free-floating brain sections were permeabilized with 0.25% Triton X-100 / PBS and incubated with primary antibodies against Flag-tag (rabbit, 1:1000, SIGMA F7425) and NeuN/Fox3 (mouse, 1:750, Merck MAB377) or V5-tag (mouse, 1:2000, Thermo Fisher R96025). Brain-sections were washed and incubated with Alexa-488 or 555 conjugated secondary antibodies (goat, 1:500, Thermo Fisher A11001 and A21428) with 4',6-diamidino-2-phenylindole (DAPI; 1:4000, Dojindo Molecular Technologies). Brain sections were washed and mounted on a slide glass. Fluorescent images were obtained using tiling fluorescent microscopy (BZ9000, Keyence) using a 10 × objective lens. Cell layers and brain regions were determined using NeuN and DAPI stained signals, and cells expressing the Ref gene were counted using ImageJ (1.53c). For immunostaining of cultured cells, cells were transfected with the AAV virus at 3 div. A mixture of three viruses, AAV-hSyn1-RFP, AAV-DIO-Pgk1-EGFP together with either AAV-hSyn1-Cre or AAV-Gfap-Cre were used. At 15 div, the cells were fixed with 4% paraformaldehyde / PBS for 20 min and then permeabilized with 0.25% Triton X-100 / PBS. The culture was immunostained using anti-EGFP (mouse, 1:400, MBL M048) and anti-RFP (rabbit, 1:500, MBL PM005) antibodies followed by incubation with Alexa-488 or 555 conjugated secondary antibodies. Fluorescent images were obtained using fluorescent microscopy with a 20 × objective lens (Axiovert A1, Zeiss).
Quantifications and statistical analysis
All experiments were performed with a minimum of 3 independent experiments. The n values in this study represent biological replicates. All values were reported as mean± sem unless otherwise stated. Statistical analyses of multiple comparisons were performed using one-way or two-way ANOVA with post hoc analysis using Dunnett’s test or Sidak’s post hoc test. Two-tailed Welch’s t-test was used for analyses between two samples. The correlation of data was analyzed by Pearson’s correlation analysis. The significance level of P = 0.05 was used to reject the null hypothesis. Statistical analysis was performed using PRISM 6.03 software (Graph Pad).
Acknowledgments
We thank S. Matsui for assisting, D. Watanabe and the members of the Watanabe-lab at Kyoto University for their help in the initial phase of the research, and the members of Abe Lab at Tohoku University for help and fruitful suggestions. This research was supported by JSPS KAKENHI (21H05608, 19H04893, 19H03319, 16KT0067), Tohoku University Research Program “Frontier Research in Duo” (Grant No. 2101), and AMED under Grant Number JP18gm6110011 to KA.
Author contributions
KA conceived and initiated the project, performed all the experiment, data analysis, and wrote the manuscript. HA performed construct construction and in vitro analysis. All authors discussed and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103927.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abe K., Matsui S., Watanabe D. Transgenic songbirds with suppressed or enhanced activity of CREB transcription factor. PNAS. 2015;112:7599–7604. doi: 10.1073/pnas.1413484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam J., Cook J.L. Reporter genes: application to the study of mammalian gene transcription. Anal. Biochem. 1990;188:245–254. doi: 10.1016/0003-2697(90)90601-5. [DOI] [PubMed] [Google Scholar]
- Arias-Cavieres A., Khuu M.A., Nwakudu C.U., Barnard J.E., Dalgin G., Garcia A.J. A HIF1a-dependent pro-oxidant state disrupts synaptic plasticity and impairs spatial memory in response to intermittent hypoxia. ENeuro. 2020;7 doi: 10.1523/ENEURO.0024-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua M.A., Giordano M., D’Agostino P., Santoro C., Cimino F., Costanzo F. Promoter for the human ferritin heavy chain-encoding gene (FERH): structural and functional characterization. Gene. 1992;111:255–260. doi: 10.1016/0378-1119(92)90696-m. [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Ebert D.H., Greenberg M.E. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaghir A., Toffalini F., Knoops L., Kallin A., van Helden J., Demoulin J.-B. Transcription factor regulation can be accurately predicted from the presence of target gene signatures in microarray gene expression data. Nucleic Acids Res. 2010;38:e120. doi: 10.1093/nar/gkq149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S.W., Greenberg M.E. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey T.S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., El-Gohary Y., Prasadan K., Shiota C., Xiao X., Wiersch J., Paredes J., Tulachan S., Gittes G.K. Rapid and simplified purification of recombinant adeno-associated virus. J. Virol. Methods. 2012;183:139–146. doi: 10.1016/j.jviromet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S., Simon J.A. A small-molecule inhibitor of Tcf/β-catenin signaling down-regulates PPARγ and PPARδ activities. Mol. Cancer Ther. 2008;7:521–529. doi: 10.1158/1535-7163.MCT-07-2063. [DOI] [PubMed] [Google Scholar]
- Hardingham G.E., Fukunaga Y., Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat. Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Ho Sui S.J., Mortimer J.R., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglés-Esteve J., Morales M., Dalmases A., Garcia-Carbonell R., Jené-Sanz A., López-Bigas N., Iglesias M., Ruiz-Herguido C., Rovira A., Rojo F., et al. Inhibition of specific NF-κB activity contributes to the tumor suppressor function of 14-3-3σ in breast cancer. PLoS ONE. 2012;7:e38347. doi: 10.1371/journal.pone.0038347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai S., Kock K.H., Bulyk M.L. Transcription factor–DNA binding: beyond binding site motifs. Curr. Opin. Genet. Dev. 2017;43:110–119. doi: 10.1016/j.gde.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Okuno H., Nonaka M., Adachi-Morishima A., Kyo N., Okamura M., Takemoto-Kimura S., Worley P.F., Bito H. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. PNAS. 2009;106:316–321. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempfer R., Pombo A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 2020;21:207–226. doi: 10.1038/s41576-019-0195-2. [DOI] [PubMed] [Google Scholar]
- Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Park H.S., Park S.-A., Ryu S.-H., Meng W., Jürgensmeier J.M., Kurie J.M., Hong W.K., Boyer J.L., Herbst R.S., et al. A novel small-molecule inhibitor targeting CREB-CBP complex possesses anti-cancer effects along with cell cycle regulation, autophagy suppression and endoplasmic reticulum stress. PLoS ONE. 2015;10:e0122628. doi: 10.1371/journal.pone.0122628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludes-Meyers J.H., Liu Y., Muñoz-Medellin D., Hilsenbeck S.G., Brown P.H. AP-1 blockade inhibits the growth of normal and malignant breast cells. Oncogene. 2001;20:2771–2780. doi: 10.1038/sj.onc.1204377. [DOI] [PubMed] [Google Scholar]
- Lyons M.R., West A.E. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog. Neurobiol. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev A., Moeser M., Medvedeva L., Martsen E., Granick A., Raines L., Zeng M., Makarov S., Houck K.A., Makarov S.S. Evaluating biological activity of compounds by transcription factor activity profiling. Sci. Adv. 2018;4:eaar4666. doi: 10.1126/sciadv.aar4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot M., Billard J.-M., Dombret C., Albac C., Karameh N., Daumas S., Hardin-Pouzet H., Mhaouty-Kodja S. Neural androgen receptor deletion impairs the temporal processing of objects and hippocampal CA1-dependent mechanisms. PLOS ONE. 2016;11:e0148328. doi: 10.1371/journal.pone.0148328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan N., Shen Y., Sarsfield S., Lemberger T., Schütz G., Linden D.J., Ginty D.D. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- Romanov S., Medvedev A., Gambarian M., Poltoratskaya N., Moeser M., Medvedeva L., Gambarian M., Diatchenko L., Makarov S. Homogeneous reporter system enables quantitative functional assessment of multiple transcription factors. Nat. Methods. 2008;5:253–260. doi: 10.1038/nmeth.1186. [DOI] [PubMed] [Google Scholar]
- Saunders A., Johnson C., Sabatini B. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front. Neural Circuits. 2012;6:47. doi: 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M., Dougan S.T., McFadden G., Greenberg M.E. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol. Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E., Bushong E.A., Haustein M.D., Tong X., Jackson-Weaver O., Kracun S., Xu J., Sofroniew M.V., Ellisman M.H., Khakh B.S. Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J. Gen. Physiol. 2013;141:633–647. doi: 10.1085/jgp.201210949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.P., Taher L., Patwardhan R.P., Kim M.J., Inoue F., Shendure J., Ovcharenko I., Ahituv N. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat. Genet. 2013;45:1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Finkbeiner S., Arnold D.B., Shaywitz A.J., Greenberg M.E. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Tyssowski K.M., DeStefino N.R., Cho J.-H., Dunn C.J., Poston R.G., Carty C.E., Jones R.D., Chang S.M., Romeo P., Wurzelmann M.K., et al. Different neuronal activity patterns induce different gene expression programs. Neuron. 2018;98:530–546.e11. doi: 10.1016/j.neuron.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Wan M., Huang W., Kute T.E., Miller L.D., Zhang Q., Hatcher H., Wang J., Stovall D.B., Russell G.B., Cao P.D., et al. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am. J. Pathol. 2012;180:2120–2133. doi: 10.1016/j.ajpath.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D.M., van ’t Veer L.J., Butte A.J., Goldstein T., Sirota M. Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat. Commun. 2019;10:3574. doi: 10.1038/s41467-019-11415-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., Manno G.L., Juréus A., Marques S., Munguba H., He L., Betsholtz C., et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.