Abstract
The number and diversity of known CRISPR–Cas systems have substantially increased in recent years. Here, we provide an updated evolutionary classification of CRISPR–Cas systems and cas genes, with an emphasis on the major developments that have occurred since the publication of the latest classification, in 2015. The new classification includes 2 classes, 6 types and 33 subtypes, compared with 5 types and 16 subtypes in 2015. A key development is the ongoing discovery of multiple, novel class 2 CRISPR–Cas systems, which now include 3 types and 17 subtypes. A second major novelty is the discovery of numerous derived CRISPR–Cas variants, often associated with mobile genetic elements that lack the nucleases required for interference. Some of these variants are involved in RNA-guided transposition, whereas others are predicted to perform functions distinct from adaptive immunity that remain to be characterized experimentally. The third highlight is the discovery of numerous families of ancillary CRISPR-linked genes, often implicated in signal transduction. Together, these findings substantially clarify the functional diversity and evolutionary history of CRISPR–Cas.
CRISPR–Cas systems, which are best known as key components of a new generation of genome-engineering tools1,2, naturally function as adaptive immunity mechanisms in bacteria and archaea. The CRISPR–Cas immune response consists of three main stages: adaptation, expression and interference. At the adaptation stage, a distinct complex of Cas proteins binds to a target DNA, often after recognizing a distinct, short motif known as a protospacer-adjacent motif (PAM), and cleaves out a portion of the target DNA, the protospacer. After duplication of the repeat at the 5ʹ end of the CRISPR array, the adaptation complex inserts the protospacer DNA into the array, so that it becomes a spacer. Some CRISPR–Cas systems employ an alternative mechanism of adaptation — namely, spacer acquisition from RNA, via reverse transcription by a reverse transcriptase encoded at the CRISPR–cas locus.
At the expression stage, the CRISPR array is typically transcribed as a single transcript — the pre-CRISPR RNA (pre-crRNA) — that is processed into mature CRISPR RNAs (crRNAs), each containing the spacer sequence and parts of the flanking repeats. In different CRISPR–Cas variants, the pre-crRNA processing is mediated by a distinct subunit of a multiprotein Cas complex, by a single, multidomain Cas protein, or by non-Cas host RNases.
At the interference stage, the crRNA, which typically remains bound to the processing complex (protein), serves as a guide to recognize the protospacer (or a closely similar sequence) in the invading genome of a virus or plasmid, which is then cleaved and inactivated by a Cas nuclease (or nucleases) that either is part of the effector or is recruited at the interference stage. The above summary is a brief, oversimplified description of the CRISPR–Cas functionality that inevitably omits many details. These can be found in recent reviews on different aspects of CRISPR–Cas biology3–9.
Similar to other biological defence mechanisms, archaeal and bacterial CRISPR–Cas systems show a remarkable diversity of Cas protein sequences, gene compositions and architectures of the genomic loci3,5,10–15. Our knowledge of this diversity is continuously expanding through the screening of ever-growing genomic and metagenomic databases. To keep pace with such expansion, a robust classification of CRISPR–Cas systems based on their evolutionary relationships is essential for the progress of CRISPR research, but this presents formidable challenges, owing to the lack of universal markers and the fast evolution of the CRISPR–cas loci16. Therefore, the two previous CRISPR–Cas classifications, published in Nature Reviews Microbiology in 2011 and 2015, employed a multipronged approach that combined comparisons of the gene compositions of CRISPR–Cas systems and their loci architectures with sequence similarity-based clustering and phylogenetic analysis of conserved Cas proteins, such as Cas1 (REFS17,18). The 2015 classification included 5 types and 16 subtypes, as well as introducing the major division of CRISPR–Cas systems into two classes that radically differ with respect to the architectures of their effector modules involved in crRNA processing and interference. The class 1 systems have effector modules composed of multiple Cas proteins, some of which form crRNA-binding complexes (such as the Cascade complex in type I systems) that, with contributions from additional Cas proteins, mediate pre-crRNA processing and interference. By contrast, class 2 systems encompass a single, multidomain crRNA-binding protein (such as Cas9 in type II systems) that combines all activities required for interference and, in some variants, also those involved in pre-crRNA processing (BOX 1).
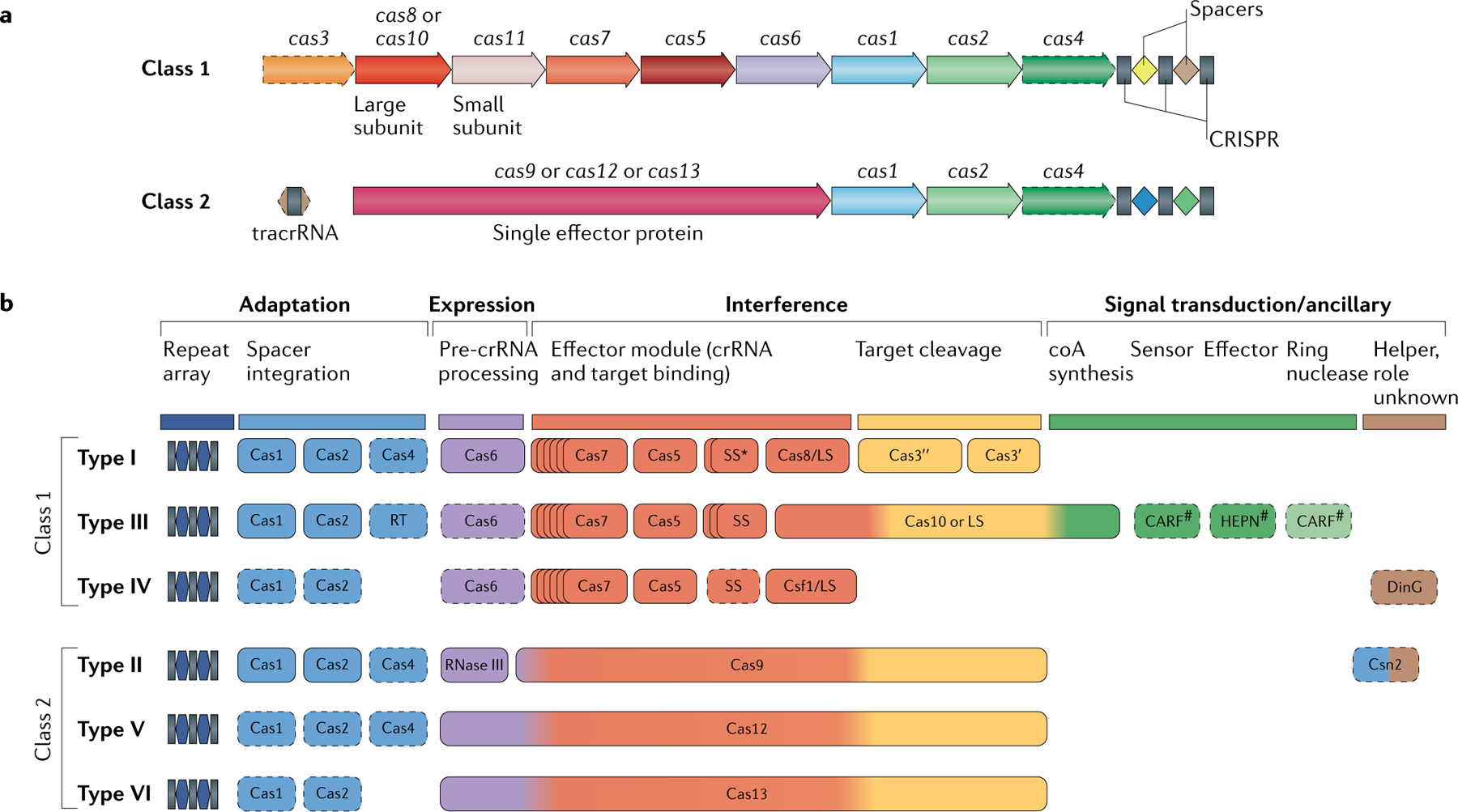
Box 1 |. The two classes of CRISPR–Cas systems and their modular organization.
Class 1 CRISPR–Cas systems have effector modules composed of multiple Cas proteins that form a crrNa-binding complex and function together in binding and processing of the target. Class 2 systems have a single, multidomain crRNA-binding protein that is functionally analogous to the entire effector complex of class 1. Part a of the figure illustrates the generic organizations of the class 1 and class 2 CRISPR–Cas loci. Part b of the figure shows the functional modules of CRISPR–Cas systems. The scheme shows the typical relationships between the genetic, structural and functional organizations of the six types of CRISPR–Cas systems. Protein names follow the current nomenclature. an asterisk indicates the putative small subunit that might be fused to the large subunit in several type I subtypes. The pound symbols (#) indicate that other unknown sensor, effector and ring nuclease protein families could be involved in the same signalling pathway. Dispensable (and/or missing, in some subtypes and variants) components are indicated by dashed outlines. Cas6 is shown with a thin solid outline for type I because it is dispensable in some, but not most, systems and with a dashed line for type III because most of these systems apparently use the Cas6 protein provided in trans by other CRISPR–cas loci. The three colours for Cas9, Cas10, Cas12 and Cas13 reflect the fact that these proteins contribute to different stages of the CRISPR–Cas response. The CRISPR-associated rossmann fold (CARF) and higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain proteins are the most common sensors and effectors, respectively, in the type III ancillary modules, but several alternative sensors and effectors have been identified, as well43. ring nucleases are a distinct variety of CARF domain proteins that cleave cyclic oligoa produced by Cas10 and thus control the indiscriminate RNase activity of the HEPN domain of Csx1 (REF.98). LS, large subunit; SS, small subunit; tracrrNa, transactivating CRISPR RNA. Figure modified from REF.18, Springer Nature limited.
Since the publication of the 2015 classification, there have been at least three major developments in the study of the diversity of CRISPR–Cas systems. First, driven partly by the interest in new tools for genome engineering, dedicated efforts have been undertaken to predict and experimentally validate additional class 2 systems19–28. As a result, the RNA-targeting type VI and multiple previously unknown subtypes of type V CRISPR–Cas systems have been discovered. Moreover, it has been shown that type V systems repeatedly evolved from transposon-encoded TnpB nucleases, yielding a large pool of type V variants, many of which can be expected to eventually become separate subtypes20,29,30. The second key development was the discovery of several class 1 and class 2 CRISPR–Cas variants that appear to lack targeted cleavage activity and thus likely perform functions distinct from adaptive immunity8,31,32. Such derived CRISPR–Cas systems include type IV, several variants of type I and at least one type V system variant, and these are often encoded within mobile genetic elements29,30,33. Recently, the involvement of two of these derived CRISPR–Cas variants, encoded by Tn7-like transposons, in crRNA-dependent DNA transposition has been demonstrated experimentally34,35. Although the origin of some of these derived forms from particular class 1 subtypes is readily identifiable, their placement in the CRISPR–Cas classification scheme remains problematic. The third important finding involves the identification of numerous gene families that are associated with specific variants of CRISPR–Cas systems, particularly of type III systems, and are implicated in signal transduction and regulatory roles8,31,32,36,37.
In this article, we reassess and update the classification of CRISPR–Cas systems, using the previously developed strategies along with analysis of the modular structure of bipartite networks of gene sharing. Special emphasis is put on classification of the quickly proliferating class 2 variants. The new class 2 classification now includes 3 types and 17 subtypes, compared with 2 types and 4 subtypes in the 2015 version, and opens the door for many more subtypes of type V systems to be identified. Although experimental study of the recently discovered class 2 variants is only in its initial phase, it is already clear that their properties are highly diverse and are difficult to predict from Cas protein sequences alone. Therefore, robust classification and systematic study of class 2 variants are essential for understanding their functionality in microorganisms and for the development of versatile genome-edit ing tools. In addition, several distinct class 1 variants, including three new subtypes, have been identified, bringing the total number of CRISPR–Cas subtypes to 33. We describe the current state and prospects of the classification and nomenclature of CRISPR–Cas systems and cas genes and, additionally, outline the emerging scenario of CRISPR–Cas evolution.
The classification approach
No genes are shared by all CRISPR–Cas systems, ruling out the possibility of a straightforward, comprehensive phylogenetic classification analogous to that employed for cellular life forms. Instead, a multipronged computational strategy has been adopted that includes the identification of signature genes for CRISPR–Cas types and subtypes, comparison of gene repertoires and genomic loci organizations, and sequence similarity-based clustering and phylogenetic analysis of the genes that are conserved in different subsets of CRISPR–Cas systems. Experimental data have also been taken into consideration, when available16–18,38 (BOX 2).
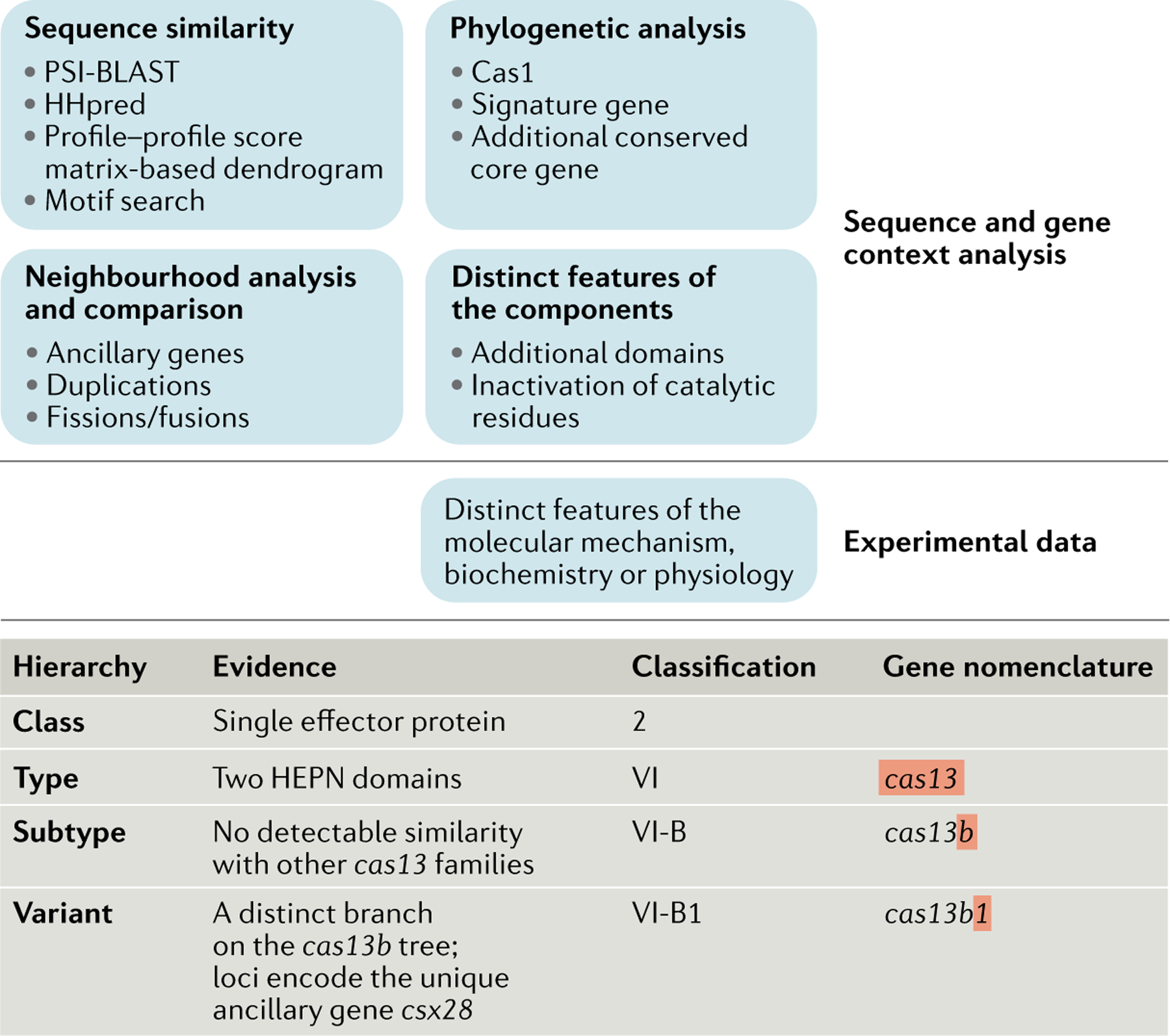
Box 2 |. Approaches for classification and nomenclature of CRISPR–Cas systems.
The top panel of the figure shows the hierarchy of the main sources of information that are used for the classification of CRISPR–Cas systems. Computational strategies exploit a combination of comparative genomic and experimental evidence, aiming to analyse the components of the cas loci, establish their organization and place them within the classification scheme. Given the fast evolution that has resulted in extensive sequence divergence of most Cas proteins, sensitive sequence similarity search and phylogenetic analysis methods are crucial for the correct assignment of the individual components; neighbourhood analysis is necessary for understanding the architecture of the specific variants of the system. experimental data are often essential to determining the distinct features of CRISPR–Cas systems and the molecular details of their mechanisms. experimental results guide additional computational analyses by providing information on functional similarity between the components of different CRISPR–Cas systems and on the contributions of different components to the system function. The bottom panel illustrates the three-level gene nomenclature scheme, and the evidence used for the classification of a variant of subtype VI-B is shown. Gene neighbourhood analysis allows for unambiguous classification of this system as class 2. motif search and profile–profile comparison of higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domains result in its classification as type VI. However, position-specific iterated BLAST searches do not detect sequence similarity to any of the previously identified type VI effector proteins. moreover, these loci encompass distinct ancillary genes, supporting their classification as a separate subtype (VI-B). The phylogenetic tree of Cas13b contains two strongly supported branches that are associated with distinct ancillary genes. accordingly, subtype VI-B is subdivided into two variants25.
Briefly, in this work, 566 amino acid sequence profiles (see Supplementary Methods), representing all variants of the 13 core cas genes, several still-uncharacterized components of their effector complexes, and reliably identified known ancillary genes, were compared to the protein sequences that are annotated in the 13,116 complete archaeal and bacterial genomes available at the NCBI as of March 1, 2019, using position-specific iterated BLAST39. This search, followed by extensive manual curation, resulted in the identification of 7,915 CRISPR–cas loci (Supplementary Dataset 1) that were fit into the previously developed classification, on the basis of the presence of the respective signature Cas proteins, sequence similarity between Cas proteins, the phylogenies of the most highly conserved Cas proteins (including Cas1 as well as the effector proteins for individual types and subtypes) and conservation of the locus organization. Loci that did not meet the criteria for inclusion in any of the previously identified subtypes were assigned to new subtypes (Supplementary Tables 1 and 2, Supplementary Dataset 1). The updated collection of Cas protein family profiles (Supplementary Dataset 2) is a resource for the identification of CRISPR–Cas systems in sequenced genomes and metagenomes.
Here, we additionally employed bipartite network analysis40,41 (Supplementary Dataset 3) for the identification of cohesive modules in the network that reflect both shared gene content and Cas protein sequence conservation, as well as for helping identify distinct CRISPR–Cas subgroups that might have subfunctionalized or neofunctionalized.
Functional modules and core genes
All cas genes can be subdivided into four distinct, although partially overlapping, functional modules (BOX 1)18,42. The adaptation module includes the gene encoding the key enzyme involved in spacer insertion (the Cas1 integrase) and the structural subunit of the adaptation complex Cas2, as well as the Cas4 nuclease in several CRISPR–Cas subtypes, the Csn2 protein in subtype II-A and reverse transcriptase in many type III systems. The expression processing module is responsible for pre-crRN A processing. In most class 1 systems, Cas6 is the enzyme that is directly responsible for processing. In type II systems, processing is catalysed by the bacterial RNase III (a non-Cas protein), whereas in many type V and apparently all type VI systems, the large effector Cas protein contains a distinct catalytic centre responsible for processing. The interference or effector module is involved in target recognition and nucleic acid cleavage. In class 1 CRISPR–Cas systems, the effector module consists of multiple Cas proteins — namely, Cas3 (sometimes fused to Cas2), Cas5–Cas8, Cas10 and Cas11, in different combinations, depending on the type and subtype (see BOX 1). By contrast, in class 2 systems, the effector module is represented by a single, large protein — Cas9, Cas12 or Cas13. The signal transduction or ancillary module is a diffuse collection of CRISPR-linked genes, most of which have roles in CRISPR–Cas systems that are, at best, tentatively predicted. However, for type III systems an essential signal transduction pathway has been characterized. This pathway involves activation of the Csm6 (or Csx1) higher eukaryotes and prokaryotes nucleotide-binding (HEPN) RNase by cyclic oligoA, which is synthesized by the Cas10 polymerase and binds the CRISPR-associated Rossmann fold (CARF) domain of Csm6 or Csx1 (REFS36,37,43).
Comparative genomic analyses have revealed partial independence of the adaptation and effector modules of CRISPR–Cas systems that, especially in the case of type III systems, appear to have recombined on many independent occasions44–46. As a result, the topology of the phylogenetic tree of Cas1 shows only limited agreement with the CRISPR–Cas classification (Supplementary Dataset 4).
The classification of CRISPR–Cas systems is based primarily on Cas protein composition differences and sequence divergence between the effector modules16,18. The class 1 effector complexes involved in pre-crRNA processing and target recognition have similar organizations between types I, III and IV, although the sequence conservation among these three types is minimal47,48. The backbone of the effector complexes in all three class 1 types is formed by the distantly related RNA recognition motif (RRM) domain-containing proteins of the repeat-associated mysterious proteins (RAMPs) Cas5 and Cas7, the latter typically present in multiple copies. In most class 1 CRISPR–Cas systems, the third RAMP, Cas6, is the dedicated RNase responsible for pre-crRNA processing and may or may not be physically associated with the effector complex. The RAMPs are characterized by extreme sequence divergence, so that the sequences of Cas5, Cas6 and Cas7 from different subtypes could be linked only by using the most sensitive methods for profile–profile sequence comparison or by direct comparison of protein structures. The large subunits of the effector complexes of type I and III systems, Cas8 and Cas10, respectively, occupy analogous positions in the complexes but show no sequence similarity and, at best, only remote structural similarity. Whether or not Cas8 and Cas10 are homologous remains an open question; if they are, the divergence is extreme, rendering any sequence of structural similarity effectively beyond detection49. Moreover, the Cas8 sequences show no detectable similarity even between some of the class 1 subtypes, such that the respective variants can serve as subtype signatures (Supplementary Table 2). The small subunit of the class 1 effector complexes (Cas11) shows no statistically significant sequence similarity between type I and III systems, but the structural similarity between the Cas11 proteins, as well as between Cas11 and the C-terminal α-helical domain of Cas10, strongly suggests that these are highly diverged homologues47,50. In type I systems, a key, standalone (although in some variants fused with Cas2) component of the effector module is Cas3, a large protein that typically consists of fused helicase and HD nuclease domains and is directly responsible for the target DNA cleavage. Type III systems differ fundamentally, with the HD nuclease being fused to Cas10, the large subunit of the complex involved in transcription-dependent cleavage of the target DNA.
Type IV CRISPR–Cas systems are highly derived variants that typically lack adaptation modules as well as the nucleases required for interference. Moreover, only Cas5 and Cas7 proteins are readily identifiable in type IV loci by sequence similarity with their counterparts in other types. Recent comparisons of the structures of the effector complexes of type IV and I systems have identified the type IV counterpart of the large subunit of the effector complex48, suggesting that type IV systems could be highly diverged type I or type III derivatives.
The class 2 effector modules are single large proteins, with their domain architectures clearly differentiating type II, V and VI systems (BOX 1; and see the discussion below)20. The types and subtypes within class 2 differ substantially with respect to the mechanisms of pre-crRNA processing51–54. In type VI and subtype V-A systems, the large effector protein also encompasses the pre-crRNA processing RNase activity55–57, whereas in type II and several type V subtypes, this processing activity is typically relegated to a non-Cas enzyme, RNase III. In the latter cases, the effector module includes an additional RNA molecule, the transactivating CRISPR (tracr) RNA, which forms stable duplexes with the partially complementary direct repeat of the pre-crRNA. After cleavage of the RNA duplex by RNase III, the mature guide RNA — that is, the crRNA–tracrRNA complex — remains stably bound to the effectors, allowing for specific DNA interference51–54.
The set of Cas1 to Cas13 proteins that comprise the adaptation and effector modules define the types and subtypes and thus represent the core of class 2 CRISPR–Cas systems. This core is accompanied by numerous ancillary proteins that are more loosely associated with CRISPR–Cas. The repertoire of the ancillary genes has recently expanded drastically, in large part through the use of dedicated computational protocols for the systematic detection of CRISPR-linked genes31,32. We discuss these ancillary genes in a later section, after describing the current state of the CRISPR–Cas classification.
In addition to the distinctions between the cas gene composition and the sequences and structures of Cas proteins, the types and subtypes of CRISPR–Cas systems can be, to some extent, differentiated by the distinct sequence and structural features of the repeats themselves58,59. However, the correspondence is incomplete, such that the branches in the cluster dendrogram of CRISPR–Cas collate multiple subtypes59.
CRISPR–Cas classification
Class 1 and its derivatives.
The classification of class 1 CRISPR–Cas systems, which include types I, III and IV, has remained relatively stable compared with the 2015 version18 (FIG. 1). The 2015 class 1 classification scheme included 12 subtypes that can be distinguished by sequence similarity clustering of effector proteins, as well as by comparison of loci organizations and the sequences of repeats. In the updated scheme, four subtypes are added — subtypes III-E, III-F, IV-B and IV-C. In addition, given the experimental demonstration of new spacer incorporation by subtype I-U sys tems60, this subtype was reclassified as subtype I-G.
Fig. 1 |. Updated classification of class 1 CRISPR–Cas systems.
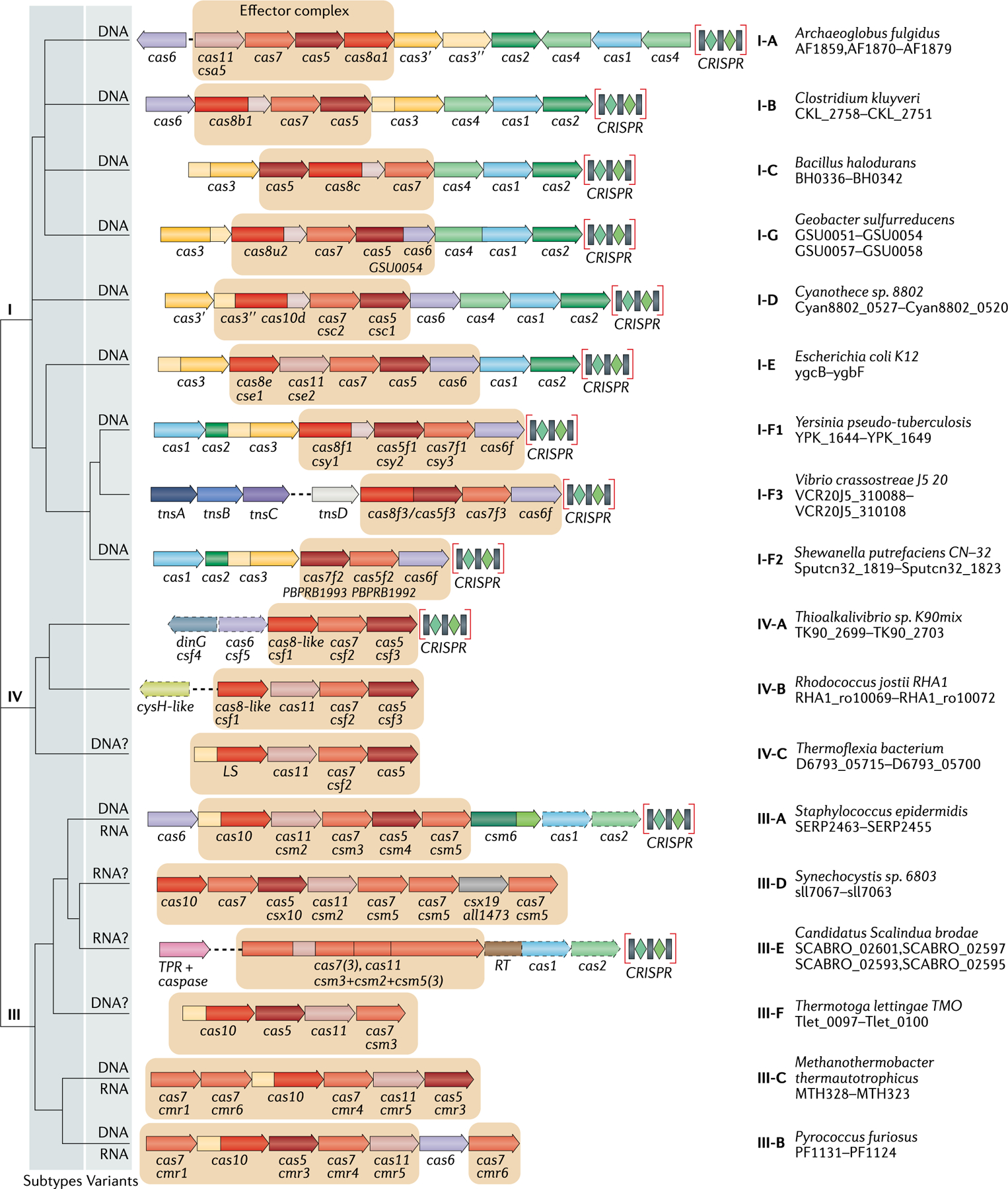
The figure schematically shows representative (typical) CRISPR–cas loci of each class 1 subtype and of selected distinct variants, with the dendrogram on the left showing the likely evolutionary relationships between the types and subtypes. The column on the right indicates the organism and the corresponding gene range. Homologous genes are colour-coded and identified by a family name. The gene names follow the previous classification18. Where both a systematic name and a legacy name are commonly used, the legacy name is given under the systematic name. The small subunit is encoded by csm2, cmr5, cse2, csa5 and several additional families of homologous genes that are collectively denoted cas11. The adaptation module genes cas1 and cas2 are dispensable in subtypes III-A and III-E (dashed lines). Gene regions coloured cream represent the HD nuclease domain; the HD domain in Cas10 is distinct from that in Cas3 and Cas3″. Functionally uncharacterized genes are shown in grey. The tan shading shows the effector module. The grey shading of different hues shows the two levels of classification: subtypes and variants. Most of the subtype III-B, III-C, III-E and III-F loci, as well as IV-B and IV-C loci, lack CRISPR arrays and are shown accordingly, although for each of the type III subtypes exceptions have been detected. CHAT, protease domain of the caspase family; RT, reverse transcriptase; TPR, tetratricopeptide repeat.
Subtype III-E, identified in 14 contigs from the NCBI non-redundant nucleotide sequence database that appear to come from 8 bacterial species (Supplementary Fig. 1, Supplementary Table 2, Supplementary Dataset 5), is characterized by a unique fusion of several Cas7 proteins and a putative Csm2-like small subunit (Cas11), such that the crRNA-binding part of the effector module is compressed within a single, large multidomain protein. In this respect, subtype III-E resembles class 2 CRISPR–Cas systems, although the domain composition and sequence analysis unequivocally place it within type III of class 1, and moreover, a specific relationship with subtype III-D could be traced (Supplementary Fig. 1). The multidomain subtype III-E effector is predicted to cleave pre-crRNA, given the conservation of aspartate residues that are known to be involved in RNA cleavage in the homologous Csm4 protein, and might also contribute to target RNA cleavage (Supplementary Fig. 2). The subtype III-E loci often include a putative ancillary gene encoding a large protein that contains a CHAT domain, a caspase family protease that is, typically, involved in programmed cell death61, fused to tetratricopeptide repeats (TPRs) (FIG. 1). The presence of this ancillary protein suggests subfunctionalization or neofunctionalization of subtype III-E systems and their potential involvement in complex defence pathways (FIG. 1, Supplementary Fig. 1).
The subtype III-F systems have been identified previously62 and are included in the 2015 CRISPR–Cas census18, but they were not classified as a distinct subtype, because their number was too small. Now that this variant has been found in 12 additional genomes, it has become apparent that they qualify as a separate subtype (Supplementary Fig. 3, Supplementary Table 2). The Cas7 and Cas5 subunits, as well as the large subunit of the subtype III-F effector complex, show a distant but substantial similarity with the corresponding components of other type III subtypes, whereas the putative small subunit does not show any similarity to Cas11. Unlike all other type III systems, subtype III-F contains only one Cas7-like protein. The HD domain fused to the Cas10-like large subunit retains all catalytic residues and therefore is predicted to cleave the target DNA. However, the cyclase or polymerase domain of the Cas10-like subunit is inactivated, as indicated by amino acid substitutions in the catalytic site, and furthermore, the subtype III-F loci lack any genes encoding CARF domain proteins. Thus, this type III subtype clearly does not function via cyclic oligoA signalling, as has been shown for subtype III-A and implied for the rest of the type III systems containing an active Cas10 polymerase36,37,43.
In the 2015 classification, subtype IV-B was reported as a variant that, unlike subtype IV-A systems, lacks the dinG gene but contains a distinct version of the predicted small subunit of the effector complex; furthermore, most of the subtype IV-B loci encompass the ancillary gene cysH32. These systems have been discovered on plasmids from numerous, diverse bacteria30, and accordingly, the variant was upgraded to a subtype. Subtype IV-C loci were also detected but not formally classified in the 2015 census. Now this type IV variant has been identified in nine contigs, mostly from thermophilic microorganisms (Supplementary Fig. 4, Supplementary Table 2), its classification as a distinct subtype also appears justified. The Cas7 and Cas5 homologues of the subtype IV-C systems show statistically significant similarity to the corresponding proteins of subtype IV-A and IV-B systems, whereas the putative large and small subunits of the effector complex lack any detectable similarity with their counterparts from any other CRISPR–Cas system. Notably, unlike in the other two type IV subtypes, the putative large subunit of subtype IV-C contains an HD nuclease domain, suggesting that it cleaves the target DNA. The order of the HD nuclease motifs is the same as in Cas3 in type I systems, but different from that in the HD nuclease domains fused to Cas10 in most of the type III systems, apparently as a result of a circular permutation occurring during the evolution of the CRISPR-associated HD nucleases.
Several additional, distinct variants of class 1 could become subtypes when more taxonomic diversity and/or more structural and experimental data become available. Among such cases, a distinct type III system variant found in archaea of the order Sulfolobales is represented by the loci YN1551_RS11700 to YN1551_RS11720 from Sulfolobus islandicus (Supplementary Dataset 1). This variant features extremely diverged Cas10 and Cas5 homologues and a unique, uncharacterized predicted component of the effector complex, Csx26. Another distinct type III system, so far found only in the archaeon Ignisphaera aggregans (loci Igag_0607 to Igag_0623), includes several proteins that are not similar to any known Cas or ancillary proteins.
A variety of derived, apparently defective variants of type I systems have been discovered, such as the ‘minimal’ subtype I-F and subtype I-B systems, which are encoded by distinct families of Tn7-like transposons30,33. These variants lack the helicase-nuclease Cas3 that is required for interference63 and therefore are predicted to perform functions distinct from adaptive immunity. A hypothesis has been proposed that these minimal type I variants mediate guide-RNA-dependent transposition30,33, and recently, such activity has been demonstrated experimentally35. Defective CRISPR–Cas systems have also been reported in preliminary studies to be encoded by some of the recently discovered giant phages, where their roles remain to be deciphered64. An analogous interference-deficient derivative of subtype I-E CRISPR–Cas systems was detected in the genomes of many bacteria of the genus Streptomyces32. This variant is not associated with any detectable mobile genetic elements but is tightly linked to a gene encoding a STAND superfamily NTPase65, suggesting involvement of these interference-deficient CRISPR–Cas systems in signal transduction and possibly in dormancy induction or programmed cell death. The differences in the Cas protein compositions between these minimal CRISPR–Cas variants and fully functional type I systems potentially could be used as an argument for classification of the defective variants into separate subtypes. However, Cas protein sequence comparison and phylogenetic analysis unequivocally demonstrate the origins of these variants in subtypes I-F, I-E and I-B, respectively30,32,33. Therefore, we propose to keep them within their respective subtypes as distinct variants — denoted, for example, I-F1, I-F2 and so forth (FIG. 1).
Apart from the newly identified subtype IV-C, most of the type IV systems are also defective CRISPR–Cas forms that lack the nucleases involved in target cleavage and thus resemble the transposon-encoded variants with respect to organization and, perhaps, functionality. Indeed, the distinctive biological features of type IV systems are their apparent (nearly) exclusive localization on plasmids, integrated conjugating elements and prophages30. Furthermore, preliminary data suggest that multiple spacers targeting heterologous plasmids have been detected in type IV CRISPR arrays, suggesting that one of the functions of type IV systems is inter-plasmid competition66.
Some derived variants are so distant from the canonical organization that their status as CRISPR–Cas systems appears questionable. A case in point is a recently described locus found in many Haloarchaea that only retain highly divergent forms of Cas5 and Cas7 (haloarchaeal RAMPs, or HRAMPs), along with an uncharacterized conserved protein and various nucleases67 (Supplementary Fig. 5A). The search of Asgard archaea genomes68 performed in the course of this work also revealed highly derived CRISPR–Cas variants that resemble HRAMPs in terms of their Cas protein composition and encompass an unusual large protein containing a diverged Cas1 domain, along with distinct variants of Cas5 (a fusion with an HD nuclease) and Cas7, as well as additional nucleases (Supplementary Fig. 5B). The functions of these extremely derived systems are unknown, and given the lack of adjacent CRISPR arrays, it is not even clear whether their activity is guide-RNA dependent. If these systems are shown to function via a CRISPR–Cas-like mechanism, they might qualify as distinct types, given the drastic reduction of the Cas protein repertoire.
Thus, the formation of derived variants that lack the interference capacity and are likely to perform functions distinct from adaptive immunity is a pervasive trend in the evolution of CRISPR–Cas. Additional highly divergent CRISPR–Cas derivatives are likely to be discovered, and their experimental characterization is likely to become a major research direction.
The expanding class 2.
Class 2 CRISPR–Cas systems include types II, V and VI. The distinguishing feature of these types is that their effector complexes consist of a single, large, multidomain protein, such as Cas9 in type II. Thanks to focused efforts on the computational discovery of new class 2 systems, partly in the quest for potential new genome-editing tools, this class has undergone a drastic expansion since the 2015 classification11,20–23. From 2 types and 4 subtypes in 2015, class 2 expanded to 3 types and 17 subtypes (FIG. 2). The new discoveries include multiple, diverse variants of type V as well as type VI systems, the first and so far the only variety of CRISPR–Cas systems that exclusively cleaves RNA.
Fig. 2 |. Updated classification of class 2 CRISPR–Cas systems.
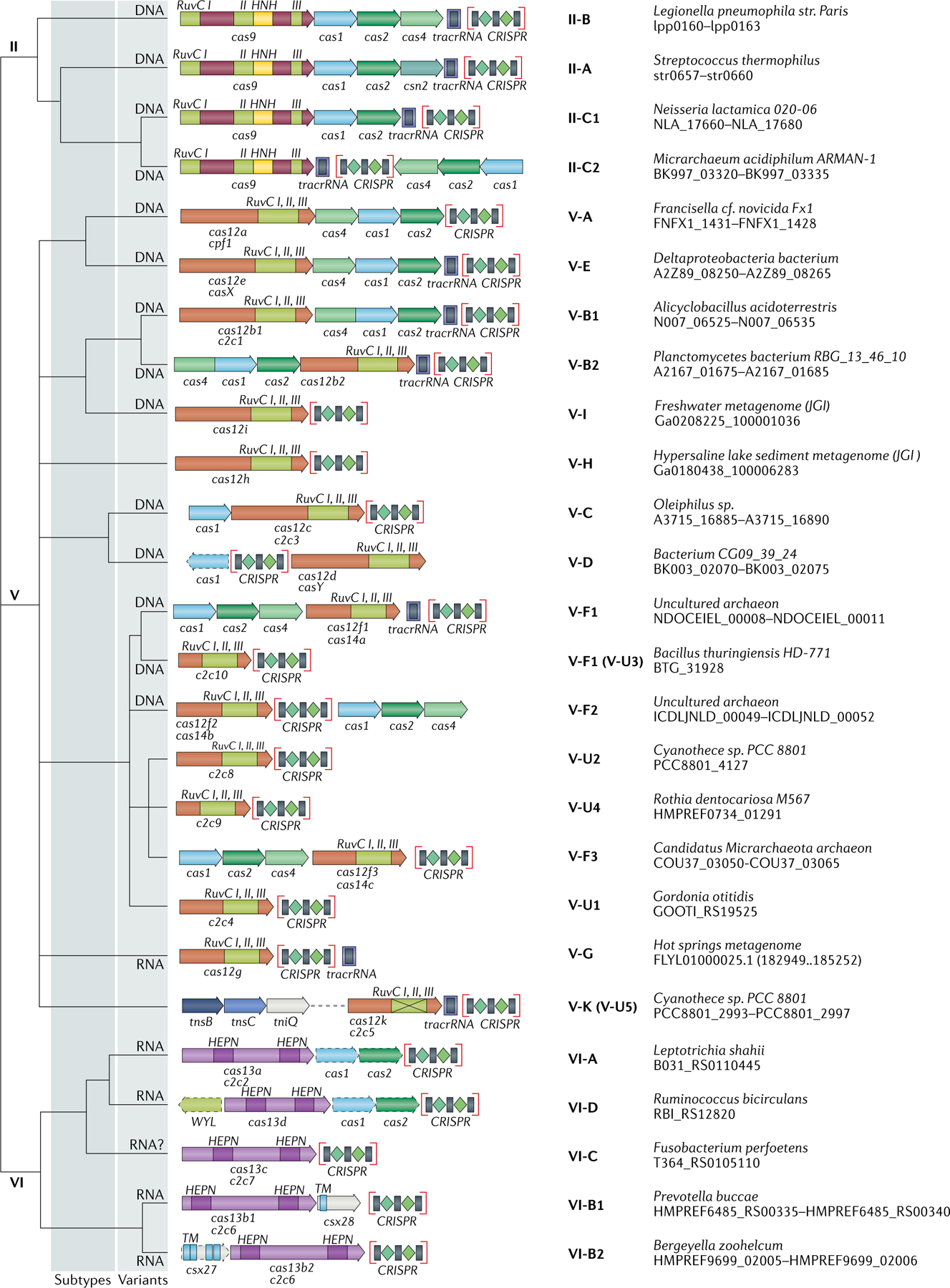
The figure schematically shows representative (typical) CRISPR–cas loci for each class 2 subtype and for selected distinct variants, with the dendrogram on the left showing the likely evolutionary relationships between the types and subtypes. The column on the right indicates the organism and the corresponding gene range. Homologous genes are colour coded and are identified by a family name following the previous classification18. Where both a systematic name and a legacy name are commonly used, the legacy name is given under the systematic name. The grey shading of different hues shows the two levels of classification: subtypes and variants. The adaptation module genes cas1 and cas2 are present in only a subset of the subtype V-D, VI-A and VI-D loci and are accordingly shown by dashed lines. The WYL-domain-encoding genes and csx27 genes are also dispensable and shown by dashed lines. Additional genes encoding components of the interference module, such as transactivating CRISPR RNA (tracrRNA), are shown. The domains of the effector proteins are colour-coded: RuvC-like nuclease, green; HNH nuclease, yellow; higher eukaryotes and prokaryotes nucleotide-binding (HEPN) RNase, purple; transmembrane domains, blue.
Type V systems fundamentally differ from type II by the domain architecture of their effector proteins. The type II effectors (Cas9) contain two nuclease domains that are each responsible for the cleavage of one strand of the target DNA, with the HNH nuclease inserted inside the RuvC-like nuclease domain sequence51. By contrast, the type V effectors (Cas12) only contain a RuvC-like domain that cleaves both strands69,70. Type VI effectors (Cas13) are unrelated to the effectors of type II and V systems, contain two HEPN domains and apparently target transcripts of invading DNA genomes. Cas13 proteins also display collateral, nonspecific RNase activity that is triggered by target recognition and induces dormancy in virus-infected bacteria71.
The assignment of subtypes within type II, V and VI systems is a challenge because of the uniform domain architecture of the respective effector proteins. The current practice (which, admittedly, involves a degree of arbitrariness) is to establish a new subtype for variants that do not show statistically significant sequence similarity to any of the already-established subtypes in BLAST searches39; the presence of additional accessory genes is also taken into consideration. This approach has so far resulted in the identification of 3 subtypes of type II systems, 10 subtypes of type V systems and 4 subtypes of type VI systems with typical, large effector proteins (FIG. 2).
In addition, a heterogeneous assemblage of putative type V variants with smaller RuvC-like domain-containing proteins, provisionally classified as subtype V-U, have been discovered20 (Supplementary Fig. 6). The putative subtype V-U effectors show high sequence similarity to TnpB proteins (predicted RuvC-like nucleases) encoded by IS605-like transposons and are thought to be intermediates on the evolutionary path from TnpB to fully fledged type V effectors. CRISPR–Cas systems evolved from different groups of TnpB on multiple, independent occasions, as has been shown by phylogenetic analysis of the TnpB family20. Recently, the interference activity of four subtype V-U effectors was validated experimentally, and as a result, one of these variants has been upgraded to a separate subtype, V-F 22,23. Notably, these newly characterized CRISPR–Cas variants show major differences in interference specificity compared with the previously characterized type V effectors and with one another. The subtype V-F effector, Cas12f (originally denoted Cas14), has been shown to cleave single-stranded DNA (ssDNA)22, although double-stranded DNA cleavage activity has subsequently been reported in a preliminary study as well72, whereas Cas12g is an RNA-guided RNase that also possesses collateral RNase and ssDNase activities23. These findings emphasize the remarkable functional diversity of CRISPR–Cas systems, which remains to be fully characterized through the discovery and study of new subtypes. Different variants within subtype V-F (currently, variants V-F1–V-F3) appear to originate from different groups of tnpB genes, as indicated by the phylogenetic analysis of the TnpB family20 (Supplementary Dataset 4). Nevertheless, given the highly significant sequence similarity between these effector proteins, they are all currently classified within a single subtype.
One of the former V-U variants, V-U5, contains an apparently inactivated RuvC-like nuclease domain, as indicated by the replacement of essential catalytic residues, and is encoded by cyanobacterial Tn7-like transposons30. The prediction that this variant evolved to function in transposons analogously to the defective type I systems — that is, by mediating guide RNA-dependent transposition — has recently been experimentally validated (and the subtype has accordingly been upgraded to subtype V-K) 34.
It is expected that the remaining subtype V-U variants will be classified into the already created or into additional subtypes as they are experimentally characterized. Furthermore, in all likelihood, multiple subtypes of type V systems that independently originated from TnpB nucleases remain to be discovered, and consequently, the number of recognized subtypes will grow further.
The origin of type VI systems is much less clear than the derivation of type V systems from TnpB. The HEPN RNase domain is widespread in various defence systems — in particular, as the toxin components of numerous toxin–antitoxin modules, which are likely to be the ultimate ancestors of CRISPR-associated HEPN domains7,73. Given that the presence of two HEPN domains is a unique signature of type VI effectors (Cas13), it is appealing to surmise that these effectors evolved from a common ancestor after duplication of the HEPN domain. However, the two HEPN domains in each of the Cas13 proteins are only distantly related to each other, and phylogenetic analysis results appear not to be compatible with the duplication scenario (Supplementary Fig. 7). In the phylogenetic tree of the HEPN family, the N-terminal and C-terminal HEPN domains form distinct branches, pointing to a common ancestor with two HEPN domains. This ancestral cas13 gene might have evolved by recombination between two genes encoding distinct HEPN-containing proteins and, possibly, a distinct family of toxin components of abortive infection modules73. Type VI systems appear to be far less diverse than type V systems, but the discovery of new subtypes remains possible. For example, we identified a distinct type VI system variant in Brachyspira species with a two-HEPN effector that shows no significant similarity to the Cas13 sequences from the four current subtypes (Supplementary Fig. 8). Presently, we refrain from calling it a new subtype because of its narrow spread in bacteria, but as the genomic database grows, this will be a strong candidate.
A bipartite gene-sharing network
In addition to the classification approaches outlined above, we performed a quantitative analysis of a bipartite network in which CRISPR–cas loci are connected through shared genes (Supplementary Fig. 9). To identify clusters of tightly connected loci that share overlapping gene sets, we applied a previously described consensus-clustering approach that combines bipartite modularity maximization and hierarchical clustering, followed by significance-based filtering of the results40. By highlighting distinct sets of genes and loci that are mutually associated, the identification of modules in the gene-sharing network could contribute to both CRISPR–Cas classification and functional prediction.
Altogether, 126 modules were identified in the CRISPR–Cas network, which can be roughly assigned to four categories: modules sharing distinct ancillary gene sets (category 1); derived variants characteristic of specific bacterial or archaeal lineages (category 2); mixed modules that apparently result from recombinational shuffling among CRISPR–cas loci that typically share closely related adaptation genes but have distinct effector genes (category 3); and modules that lack any of the above distinctive features but include highly diverged Cas proteins (category 4) (Supplementary Fig. 9, Supplementary Dataset 3). The recently characterized minimal variant of subtype I-F (I-F3) associated with Tn7-like transposons, a remarkable case of CRISPR–Cas neofunctionalization (module 16), is an example from category 1. Cyanobacteria-specific modules 65 and 98, which consist of distinct variants of subtype III-B, exemplify category 2. A case of previously described gene shuffling in Methanosarcina species62,74 is captured in module 84, which belongs in category 3. Most of the identified modules include CRISPR–cas loci that belong to the same subtype. The exceptions are modules that combine two or three subtypes of type I (modules 10 and 101) or type V (module 126) systems that share overlapping gene compositions. More notably, three modules (46, 93 and 108) join loci of types I and III systems, apparently reflecting recombinational events. Only a few relatively rare, low-abundance subtypes are represented by a single module. Most of the subtypes are divided into multiple modules, with subtypes I-E and I-B showing the highest heterogeneity (14 and 13 modules, respectively). This reflects the functional and evolutionary plasticity of these subtypes, which conceivably underlie their high abundance in current genomic databases (see below).
The fine-grained modules produced by bipartite network analysis could be useful for the identification of distinct functional variants of CRISPR–Cas systems that might be obscured by the conservative assignment of subtypes and variants. Moreover, this approach could provide a fast and straightforward way to assign new CRISPR–cas loci to predefined types and subtypes for which related loci have already been identified. In support of this possibility, the present bipartite network analysis was able to correctly assign most of the incomplete CRISPR–cas loci to the types and subtypes where they belong. To delineate coarse-grained modules that would facilitate the classification of novel CRISPR–Cas systems in an unsupervised way, more sophisticated multiresolution approaches will be required.
Distribution of CRISPR–Cas systems
The CRISPR–Cas systems are non-uniformly distributed among bacterial and archaeal phyla. We present a census of CRISPR–cas loci in the current collection of complete bacterial and archaeal genomes. Analysis of 13,116 complete genomes showed that CRISPR–cas loci are represented in a substantial majority of archaea (276 of 324 genomes (85.2%)), including almost all hyperthermophiles (89 of 92 genomes (96.7%)), but only in ~40% of bacteria (5,412 of 12,792 genomes (42.3%)) (FIG. 3, Supplementary Dataset 6). Clear trends are observed in the distributions of specific CRISPR–Cas classes, types and subtypes. In particular, class 2 remains nearly exclusive to bacteria. The absence of class 2 in archaea, at least in part, can be explained by the absence of RNase III, the pan-bacterial enzyme that is responsible for pre-crRNA processing in type II and some subtypes of type V systems — that is, in most of the class 2 systems46,75. By contrast, the genomes of Crenarchaeota are substantially enriched for type III systems of class 1. Overall, and in most groups of bacteria and archaea, class 1 is far more abundant than class 2. However, there are notable exceptions — for example, Tenericutes bacteria, in which only class 2 systems have been identified so far (FIG. 3). Some groups of bacteria, such as Chlamydia species (FIG. 3) or the recently discovered candidate phyla radiation, which appears to consist mostly of symbiotic microorganisms, are nearly devoid of CRISPR–Cas systems76–78. Conversely, the majority of type VI systems — and in particular, all instances of the most abundant sub type VI-B — have been identified in bacterial genomes of the phyla Bacteroidetes and Fusobacteria (FIG. 3).
Fig. 3 |. Distribution of the six types of CRISPR–Cas system in the major archaeal and bacterial phyla.
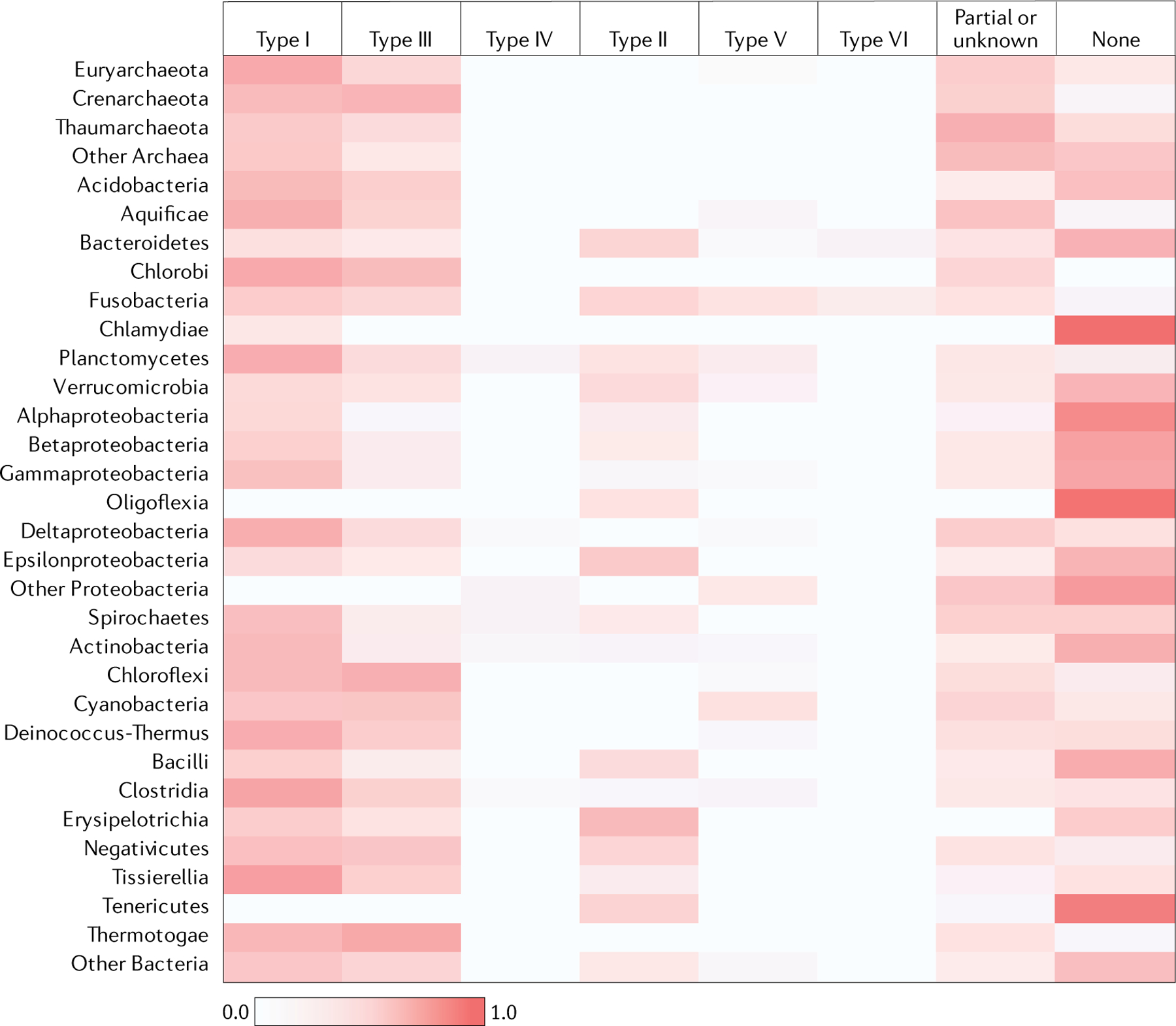
The heat map shows the weighted fraction (between 0 and 1.0) of the genomes in each of the major archaeal and bacterial phyla in which CRISPR–Cas systems of the respective type have been detected. Each CRISPR–cas locus of a given type within a taxon was assigned a weight equal to the weight of the respective genome (see the Supplementary Methods for details); additionally, the weights of the genomes that lack CRISPR–Cas loci were collected. The sum of the weights of the CRISPR–cas loci of each type was normalized by the sum total of the weights across the taxon. ‘Partial or unknown’ indicates CRISPR–cas loci that could not be assigned to any of the known types.
The biological underpinnings of the non-uniform phyletic spread of CRISPR–Cas systems remain to be elucidated. Considering the high horizontal mobility of CRISPR–cas loci, it appears likely that their loss or retention in prokaryotic genomes depends on the trade-off between the fitness cost, which is determined mostly by autoimmunity and the curtailment of horizontal gene transfer, and the benefits of defence conferred by adaptive immunity79–84. These benefits most likely depend on the abundance and diversity of viruses in specific habitats, as well as on the biology of host–parasite interactions in specific groups of microorganisms85,86. The evolutionary dynamics that determine the distribution of CRISPR–Cas among bacteria and archaea can be expected to become one of the major directions in CRISPR–Cas research in the next few years. In particular, these dynamics might depend, to a large extent, on the interactions between CRISPR–Cas and DNA repair mechanisms, such as the double-strand break repair systems87.
Core and ancillary cas genes
The components of the adaptation and effector modules comprise the suite of core Cas proteins. The core Cas proteins in the widespread CRISPR–Cas types and subtypes are well characterized, although the discovery of novel class 2 effector proteins continues to gradually expand the core gene repertoire. Furthermore, in the course of the systematic search for new CRISPR-linked proteins, many highly diverged variants of the core proteins have been identified32.
By contrast, the list of the (predicted) ancillary CRISPR-linked proteins has greatly expanded as a result of dedicated searches of CRISPR–Cas genomic neighbourhoods31,32. For the great majority of these proteins, no experimental data are available yet, but computational analysis of their domain architectures points to multiple connections to various signal transduction pathways, as well as membrane association or functional links to membrane transport processes for many CRISPR–Cas systems — particularly those of type III systems, which drastically stand out in the complexity of their gene repertoire among all CRISPR–Cas forms (FIG. 4). Several accessory proteins — for example, those in subtypes VI-B and VI-D — have been directly shown to modulate the activity of their respective effectors25,26. Furthermore, some of the genes that are currently classified as ancillary are actually represented in numerous CRISPR–Cas systems and could perform major roles in the immune response. The most obvious example is Csm6, a HEPN-domain RNase that is a component of the majority of subtype III-A CRISPR–Cas systems and is activated by the signal transduction pathway initiated by cyclic oligoA produced by the Cas10 polymerase30,31. Systematic experimental characterization of the roles of accessory proteins in CRISPR–Cas functions will undoubtedly be another key research area in the study of CRISPR–Cas biology for years to come.
Fig. 4 |. Ancillary genes in CRISPR–Cas systems.
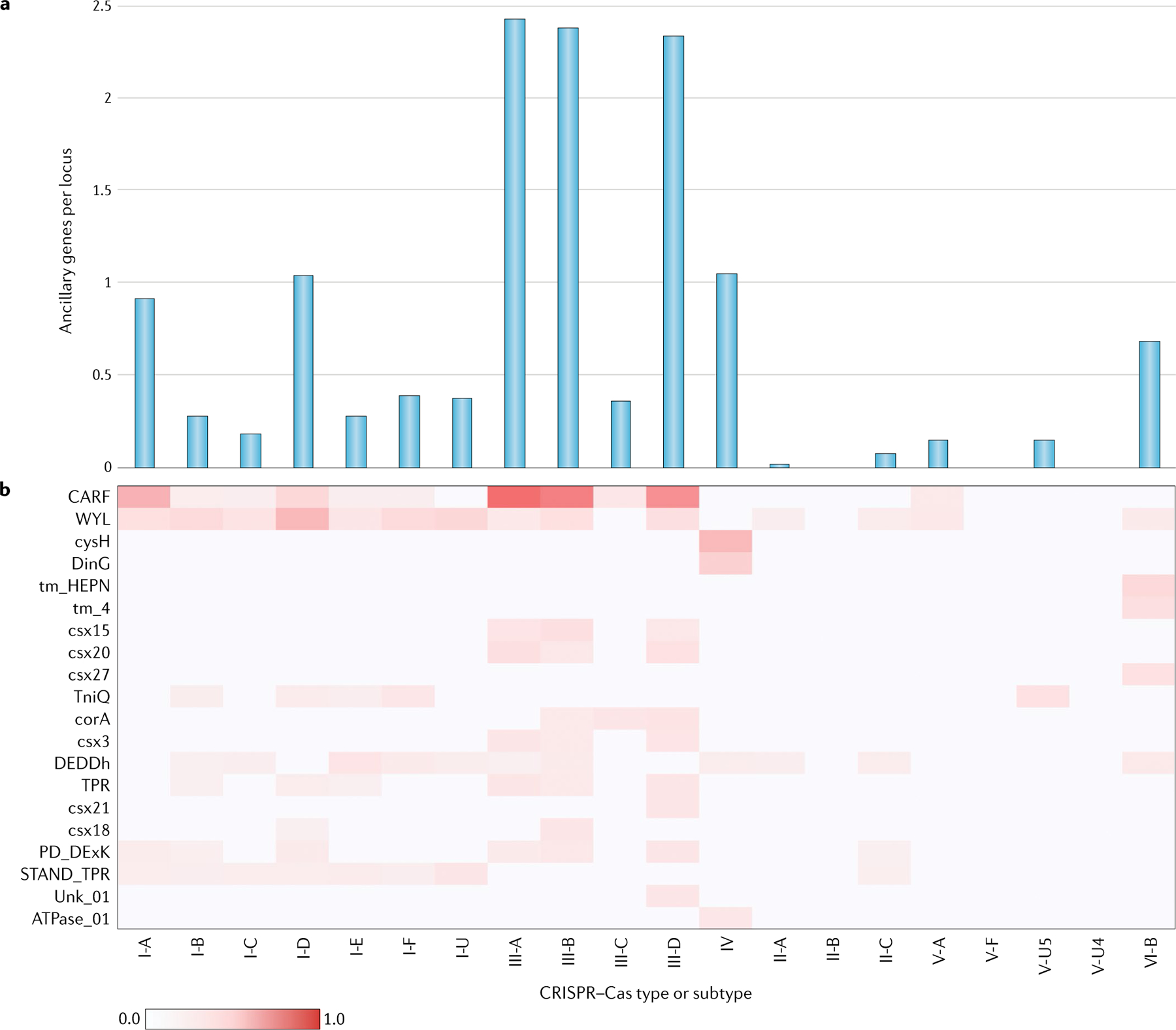
The basic molecular machinery of CRISPR–Cas systems consists of the cas core genes. The core genes are often accompanied by diverse ancillary genes that perform additional or regulatory functions. The ancillary genes are typically present only in subsets of the CRISPR–cas loci of the respective types and subtypes and often also occur in other, non-cas genomic contexts. Prediction of the ancillary genes was performed using the ‘CRISPRicity’ protocol, as we previously described32,99. Operationally, the list of ancillary genes includes families, labelled as ‘associated’ in the profFam.tab column in Supplementary Dataset 2. The numbers of occurrences (counts) of ancillary genes in each unambiguously classified CRISPR–cas locus were averaged across the system subtypes using genome weights, calculated as described in the Supplementary Methods. The occurrence of ancillary genes across the types and subtypes of CRISPR–Cas systems is shown (part a). The vertical axis shows the weighted mean numbers of ancillary genes per locus in different subtypes. The common ancillary genes and their distribution among CRISPR–Cas types and subtypes is also shown (part b). Gene families are denoted with the corresponding profile names (Supplementary Dataset 2). The weighted mean number of ancillary genes per locus in different subtypes is colour coded as per the scale shown at the bottom.
The discovery of new class 2 subtypes and numerous accessory proteins poses obvious problems for the systematic nomenclature of CRISPR-linked genes. So far, a conservative approach has been adopted, under which the cas designation is reserved for core genes, or more precisely, families of homologous core genes (Supplementary Table 1). The numbered cas gene names were originally assigned to the 11 most common genes among diverse CRISPR–Cas systems, and subsequently, cas12 and cas13 — the effectors of type V and type VI systems, respectively — have been added. Currently, the cas names are reserved for type-specific effector genes, whereas subtypes are specified by suffixes — for example, cas12a, cas12b, cas12c and so forth. The recent designation as Cas14 of small type V effector proteins related to those in subtype V-U systems22 does not conform with this criterion. We believe that the appropriate name for these proteins should be Cas12f 23, given that Cas12 is supposed to apply to all type V system effectors. Obviously, under this approach the number of cas genes cannot be expected to increase substantially, because both the discovery of new types and the identification of new core genes for already established types are rare. The ancillary genes continue to be known under their legacy names or as csx followed by a number, although a systematic nomenclature might be considered in the future.
Origins and evolution of CRISPR–Cas
Comparative analysis of CRISPR–Cas systems — in particular, the newly discovered class 2 subtypes — provides for the reconstruction, at least in outline, of a nearly complete scenario of CRISPR–Cas evolution (FIG. 5). A striking feature of the evolutionary history of CRISPR– Cas is the repeated recruitment of genes from different mobile genetic elements for various functions in adaptive immunity7,29. Thus, the adaptation module, along with the CRISPR repeats themselves, appears to originate from an immobilized transposon of the casposon family, so named because these elements employ a Cas1 homologue as the transposase88–90. The casposon could have contributed not only cas1 but also the cas4 gene, encoding another nuclease that is involved in PAM selection during adaptation in many CRISPR–Cas systems60,91–93, given that Cas4 homologues are among the cargo genes in some casposons.
Fig. 5 |. Outline of a complete scenario for the origins and evolution of CRISPR–Cas systems.
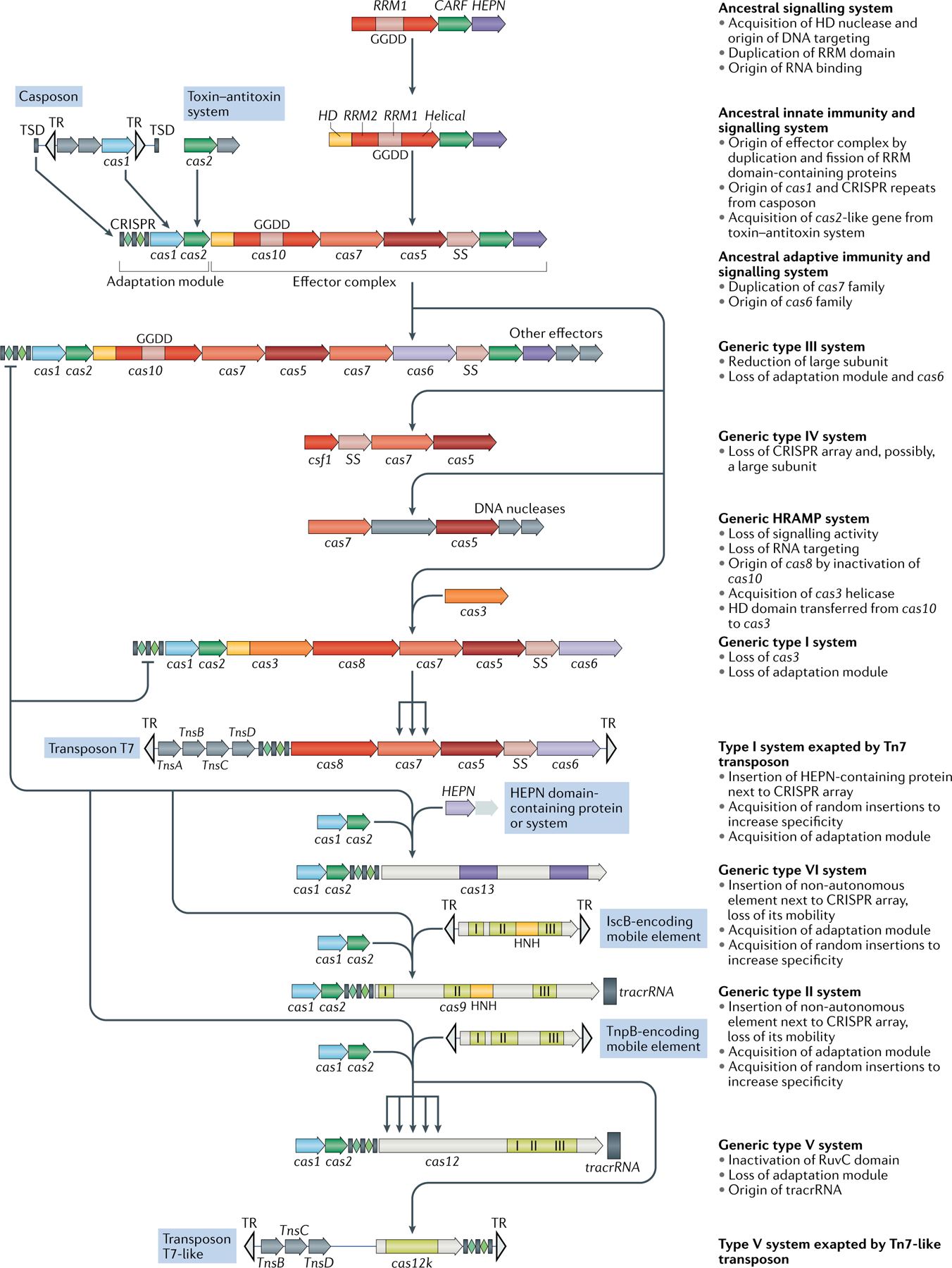
The figure depicts a hypothetical scenario of the origin of CRISPR–Cas systems from an ancestral signalling system (possibly an abortive infection defence system (Abi)). This putative ancestral Abi module shares a cyclic oligoA polymerase Palm domain (RNA recognition motif (RRM) fold) with Cas10 and is proposed to function analogously to type III CRISPR–Cas systems. Specifically, cyclic oligoA molecules that are synthesized in response to virus infection bind to the CRISPR-associated Rossmann fold (CARF) domain of the second protein in this system, resulting in activation of the RNase activity of the higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain, which induces dormancy through indiscriminate RNA cleavage. This putative ancestral Abi module would give rise to the type III-like CRISPR–Cas effector module via duplication of the RRM domain, with subsequent inactivation of one of the copies (the two RRM domains are denoted RRM1 and RRM2). The ancestral class 1 CRISPR–Cas system is inferred to have evolved through the merger of two modules: the adaptation module, including the CRISPR repeats, derived from a casposon, and the type III-like effector module, likely derived from the ancestral Abi system. The subsequent acquisition of the HD nuclease domain by the effector module provided for RNA-guided DNA cleavage. Inactivation of the oligoA polymerase domain in the effector complex, or possibly replacement of Cas10 by an unrelated protein and acquisition of the Cas3 helicase, led to the emergence of type I systems, which lack the cyclic oligoA-dependent signalling pathway and exclusively cleave double-stranded DNA. Class 2 systems of type II and different subtypes of type V appear to have evolved independently by the recruitment of distinct TnpB nucleases that are encoded by IS605-like transposable elements. Type VI likely originated from an RNA-cleaving, HEPN domain-containing abortive infection or toxin–antitoxin system. Some CRISPR–Cas systems, such as type IV and Tn7-linked systems I-F3 and V-K, were subsequently recruited by mobile genetic elements and lost their interference capacity along with the original defence function. The key evolutionary events are described to the right of the images. The typical CRISPR–cas operon organization is shown for each CRISPR–Cas subtype and for selected distinct variants. Homologous genes are colour-coded and identified by a family name following the previous classification18. The multiforking arrows denote events that have been inferred to have occurred on multiple, independent occasions during the evolution of CRISPR–Cas systems. GGDD, key catalytic motif of the cyclase or polymerase domain of Cas10 that is involved in the synthesis of cyclic oligoA signalling molecules; HRAMP, haloarchaeal repeat-associated mysterious proteins; TR, terminal repeats; tracrRNA, transactivating CRISPR RNA ; TSD, target site duplication, the likely source of ancestral repeats88.
The effector module of type III systems appears to be the best candidate for the ancestral state, given their widespread (especially in archaea) and complex gene composition, as well as the fact that, in most of the type III variants, the large subunit of the effector complex (Cas10) is an active enzyme, a cyclic oligoA polymerase7. The effector moiety of CRISPR–Cas could have started as a putative signalling system that has been identified in several bacteria and consists of a small-sized, ‘minimal’ Cas10 homologue and a homologue of Csm6 with fused CARF and HEPN domains7,94 (FIG. 5). This system is predicted to function analogously to the signal transduction pathway in type III CRISPR–Cas systems — namely, by synthesizing cyclic oligoA (most likely in response to stress) that is then bound by the CARF domain and allosterically activates the RNase activity of the HEPN domain36,37. Indiscriminate RNA cleavage by the HEPN domain would induce dormancy or programmed cell death. The putative ancestral system remains to be studied experimentally, but even without such validation, it resembles an abortive infection (Abi) module. Indeed, recently the HEPN-containing Csm6 protein of subtype III-A systems has been shown to act as a toxin causing growth arrest of the host cell95, which is compatible with the origin of the type III effector module from an Abi system. Similar to the known Abis10,96, the ancestor of the effector module is likely to be subject to extensive horizontal gene transfer and might, effectively, possess features of a mobile genetic element.
Thus, different types of mobile genetic elements seem to have given rise to both the adaptation and the effector parts of class 1 CRISPR–Cas systems. The subsequent evolution of the effector module would have involved serial duplication of the RRM domain of the Cas10 homologue and the capture of additional proteins — in particular, the target-cleaving HD nuclease7. The key event in the evolution of type I systems was the capture of the helicase-nuclease Cas3 and the replacement of the oligoA polymerase Cas10 with the enzymatically inactive Cas8 as the large subunit of the effector complex. Whether the latter event involved extreme divergence following the inactivation of Cas10 or the capture of an unrelated protein remains uncertain.
The origin of type IV systems remains uncertain, but the recent discovery of subtype IV-C systems, with the large subunit fused to an HD domain, together with the observation that both the Cas5 and Cas7 components of type IV systems share a greater sequence similarity with their counterparts from type III than with those from type I systems, suggests that type IV could have evolved from type III. These observations are compatible with the lack of association of the IV-C systems with any known mobile genetic elements. Similar lines of evidence could point to subtype I-D systems as a potential evolutionary intermediate between type III and type I systems. The structure of both the subtype I-D effector complex and the Cas10d protein should shed more light on the origin of type I systems. The origin of the HRAMP system, a highly derived CRISPR-less class 1 variant, is unclear as well, but both its Cas5 and Cas7 components are more similar to the respective proteins of type III than to those of type I systems, suggesting a route of evolution parallel to that of type IV systems67.
In class 2, the effectors of different subtypes of type V and, possibly, type II systems appear to have evolved, on multiple independent occasions, from TnpB nucleases encoded by yet another class of mobile genetic element, the IS605-like transposons20. Type II systems apparently evolved from a distinct variety of TnpB (denoted IscB) that contains an HNH nuclease domain inserted into the RuvC-like domain97. The type VI system effectors (Cas13) seem to originate from HEPN-containing components of an Abi module7,20 (FIG. 5). The functional analogy between Cas13a and Abi has been recently validated by experiments that have demonstrated growth arrest of phage-infected bacteria that is dependent on Cas13a activity71. A recurrent trend in the evolution of CRISPR–Cas effectors is the accretion of additional proteins (in class 1) or domains (in class 2), on top of the core nuclease domains, providing for the flexibility required to accommodate the crRNA and the target DNA or RNA7.
Another general trend in CRISPR–Cas evolution is the spawning of defective variants, many of which are appropriated by mobile genetic elements30,33. The defective forms of CRISPR–Cas systems are predicted to perform various functions that require target recognition but not cleavage. A striking case of such functionality is the crRNA-dependent, site-specific transposition that has recently been demonstrated for the transposon-encoded derived variants of subtype I-F and subtype V-K systems34,35.
Concluding remarks
Because the most abundant types and subtypes of CRISPR–Cas systems are now known, the overall structure of the current classification is likely to stand the test of time. However, the discovery of comparatively rare but functionally and evolutionarily interesting and informative variants has not stopped and, in all likelihood, will continue, especially as diverse environments are explored by methods of metagenomics and single-cell genomics. Some of these variants are distinct enough to become new subtypes, but so far, no new types have been identified after the discovery of type VI. According to the currently adopted criteria, to qualify as a new type, a CRISPR–Cas variant has to encompass an effector module unrelated (or extremely distantly related) to those of the known types. Other types might remain to be discovered, but it is becoming increasingly clear that, if such additional types exist, they are rare and/or highly specialized. Investigation of the numerous ancillary components of CRISPR–Cas is starting to uncover multiple connections between CRISPR–Cas and various functionally distinct systems of bacterial and archaeal cells, particularly those involved in different forms of signal transduction.
In summary, the diversity of the identified CRISPR– Cas systems has substantially increased over the last four years, thanks to a combination of computational and experimental approaches. Notably, the new varieties could be classified into distinct types and subtypes by using several criteria. Arguably, this granularity stems from punctuated evolution, whereby the diversification of emerging subtypes slows down after an initial period of rapid innovation. Notwithstanding the apparent distinctness of the subtypes, the increasing diversity of CRISPR–Cas creates further challenges to classification and nomenclature and calls for the development of robust classification criteria. The delineation of types and, to a large extent, subtypes will likely remain qualitative, given the paucity of shared components. However, the classification of variants within subtypes, some of which might qualify as separate subtypes, can be quantified — for example, by using bipartite network analysis, as shown here. On the whole, we believe that the classification of CRISPR–Cas systems has entered the era of consolidation and refinement. Experimental characterization of CRISPR–Cas functions still lags behind predictions produced by computational analysis. It is our hope that the updated classification will facilitate experimental studies and promote new directions.
Supplementary Material
CRISPR
Clustered regularly interspaced short palindromic repeats, present in most archaeal and many bacterial genomes.
Cas
CRISPR-associated (proteins).
Adaptation
First stage of the CRISPR–Cas response that involves spacer acquisition.
Interference
Final stage of the CRISPR–Cas response, which involves recognition and cleavage of the target DNA or RNA.
Protospacer-adjacent motif
(PAM). A short nucleotide sequence next to the protospacer that is required for target recognition by the crRNA effector.
Protospacer
Segment of DNA (typically, from a virus or plasmid) that is acquired by CRISPR–Cas systems via the activity of the adaptation complex.
CRISPR array
Genomic locus containing multiple, tandem CRISPR.
Spacer
Unique segment of DNA inserted between CRISPR units.
CRISPR–cas
Archaeal and bacterial system of adaptive immunity that consists of a CRISPR array and cas genes.
pre-crRNA
Long transcript of a CRISPR locus that is processed to yield the crRNA CRISPR–Cas system, where it is incorporated as a spacer.
crRNAs
Short RNA molecules containing the spacer sequence and parts of the CRISPR, used as the guide to target and cleave cognate foreign DNA or RNA.
Transposon
A mobile genetic element, typically flanked by inverted terminal repeats, that changes its location in the host genome by inserting into new sites with the help of a transposon-encoded enzyme known as transposase, integrase or recombinase.
Casposon
A member of a distinct class of transposons that employ a Cas1 homologue as the transposases and are thought to be the ancestors of CRISPR–Cas adaptation modules.
Acknowledgements
K.S.M., Y.I.W., J.I., S.A.S. and E.V.K. are supported through the Intramural Research Program of the US National Institutes of Health; F.J.M.M. was supported by grants BIO2014–53029-P (Ministerio de Ciencia, Innovación y Universidades, Spain), and 291815 Era-Net ANIHWA (7th Framework Programme, European Commission) and PROMETEO/2017/129 (Conselleria d’Educació, Investigació, Cultura i Esport, Generalitat Valenciana, Spain); S.A.S. was supported by RFBR (research project 18–34-00012) and a Systems Biology Fellowship from Philip Morris Sales and Marketing; S.M. was funded by funding from the Natural Sciences and Engineering Research Council of Canada (Discovery program) and holds a Tier 1 Canada Research Chair in Bacteriophages.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41579-019-0299-x.
References
- 1.Komor AC, Badran AH & Liu DR CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickar-Oliver A & Gersbach CA The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol 20, 490–507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohanraju P et al. Diverse evolutionary roots and mechanistic variations of the CRISPR–Cas systems. Science 353, aad5147 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Jackson SA et al. CRISPR–Cas: adapting to change. Science 356, eaal5056 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Barrangou R & Horvath P A decade of discovery: CRISPR functions and applications. Nat. Microbiol 2, 17092 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Jiang F & Doudna JA CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys 46, 505–529 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Koonin EV & Makarova KS Origins and evolution of CRISPR–Cas systems. Philos. Trans. R. Soc. Lond. B Biol. Sci 374, 20180087 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faure G, Makarova KS & Koonin EV CRISPR– Cas: complex functional networks and multiple roles beyond adaptive immunity. J. Mol. Biol 431, 3–20 (2019). [DOI] [PubMed] [Google Scholar]
- 9.McGinn J & Marraffini LA Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat. Rev. Microbiol 17, 7–12 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Koonin EV, Makarova KS & Wolf YI Evolutionary genomics of defense systems in archaea and bacteria. Annu. Rev. Microbiol 71, 233–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koonin EV, Makarova KS & Zhang F Diversity, classification and evolution of CRISPR–Cas systems. Curr. Opin. Microbiol 37, 67–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishino Y, Krupovic M & Forterre P History of CRISPR–Cas from encounter with a mysterious repeated sequence to genome editing technology. J. Bacteriol 200, e00580–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hille F & Charpentier E CRISPR–Cas: biology, mechanisms and relevance. Philos. Trans. R. Soc. Lond. B Biol. Sci 371, 20150496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright AV, Nunez JK & Doudna JA Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Klompe SE & Sternberg SH Harnessing ‘a billion years of experimentation’: the ongoing exploration and exploitation of CRISPR–Cas immune systems. CRISPR J 1, 141–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, Wolf YI & Koonin EV Classification and nomenclature of CRISPR–Cas systems: where from here? CRISPR J 1, 325–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova KS et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol 9, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova KS et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol 13, 722–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shmakov S et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shmakov S et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol 15, 169–182 (2017). This work demonstrates the relationships between the effectors of different types and subtypes of class 2 CRISPR–Cas systems and nucleases encoded by mobile genetic elements. On the basis of sequence comparison and phylogenetic analysis of Cas12 (type V effectors) and TnpB nucleases encoded by transposons, a scenario of independent recruitment of distinct TnpB variants, giving rise to different type V subtypes, is proposed.
- 21. Burstein D et al. New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237–241 (2017). This work describes the metagenomic discovery of two new subtypes of type V CRISPR–Cas systems and experimental validation of their activity.
- 22. Harrington LB et al. Programmed DNA destruction by miniature CRISPR–Cas14 enzymes. Science 362, 839–842 (2018). This work experimentally validates the enzymatic activity of small predicted effectors that have been assigned to subtype V-U by Shmakov et al. (2017) and are here reclassified as subtype V-F. It shows that these enzymes differ substantially from the previously characterized large type II and type V effectors and catalyse both crRNA-specific and non-specific cleavage of single-stranded DNA.
- 23. Yan WX et al. Functionally diverse type V CRISPR–Cas systems. Science 363, 88–91 (2019). This article reports the experimental characterization of CRISPR–Cas subtypes V-C, V-G, V-H and V-I. Whereas Cas12c, Cas12h and Cas12i proteins all demonstrate RNA-guided double-stranded DNA interference similar to that in previously described CRISPR–Cas effectors, Cas12g is shown to function as an RNase with collateral RNase and single-strand DNase activities.
- 24.Abudayyeh OO et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smargon AA et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yan WX et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339.e5 (2018). This study demonstrates RNA targeting by the smallest known type VI effector, Cas13d, and shows that the accessory WYL domain-containing protein stimulates this activity.
- 27.Murugan K, Babu K, Sundaresan R, Rajan R & Sashital DG The revolution continues: newly discovered systems expand the CRISPR–Cas toolkit. Mol. Cell 68, 15–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stella S, Alcon P & Montoya G Class 2 CRISPR–Cas RNA-guided endonucleases: Swiss army knives of genome editing. Nat. Struct. Mol. Biol 24, 882–892 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Koonin EV & Makarova KS Mobile genetic elements and evolution of CRISPR–Cas systems: all the way there and back. Genome Biol. Evol 9, 2812–2825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faure G et al. CRISPR–Cas in mobile genetic elements: counter-defense and beyond. Nat. Rev. Microbiol 17, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shah SA et al. Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families. RNA Biol 16, 530–542 (2019). Along with Shmakov et al. (2018), this study describes a computational approach to predict proteins that are functionally linked to CRISPR–Cas systems and applies this approach to type III systems.
- 32. Shmakov SA, Makarova KS, Wolf YI, Severinov KV & Koonin EV Systematic prediction of genes functionally linked to CRISPR–Cas systems by gene neighborhood analysis. Proc. Natl Acad. Sci. USA 115, E5307–E5316 (2018). Along with Shah et al. (2019), this article describes a computational approach for the systematic prediction of proteins that are functionally linked to CRISPR–Cas systems (‘CRISPRicity’ protocol) and applies that approach to all CRISPR–Cas types and subtypes.
- 33. Peters JE, Makarova KS, Shmakov S & Koonin EV Recruitment of CRISPR–Cas systems by Tn7-like transposons. Proc. Natl Acad. Sci. USA 114, E7358–E7366 (2017). This study describes, for the first time, defective CRISPR–Cas systems encoded in Tn7-like transposons and predicts their function in RNA-guided transposition.
- 34. Strecker J et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019). This work validates the prediction made in Shmakov et al. (2017), by showing that V-U5 variant effector proteins, which are inactivated TnpB homologues encoded in Tn7-like transposons, form a complex with the transposase subunit and enable crRNA-dependent transposition.
- 35. Klompe SE, Vo PLH, Halpin-Healy TS & Sternberg SH Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). This work complements Strecker et al. (2019) by experimentally validating the prediction made in Peters et al. (2017) that interference-deficient subtype I-F CRISPR–Cas systems encoded in Tn7-like transposons enable crRNA-dependent transposition.
- 36. Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G & Siksnys V A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609 (2017). Along with Niewoehner et al. (2017), this article describes the signalling pathway involved in the function of type III CRISPR–Cas systems, which involves the synthesis of cyclic oligoA molecules by Cas10, binding of these signalling molecules to the CARF domain of Csm6 and activation of the second domain of Casm6, the HEPN nuclease that catalyses promiscuous RNA cleavage.
- 37.Niewoehner O et al. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Makarova KS & Koonin EV Annotation and classification of CRISPR–Cas systems. Methods Mol. Biol 1311, 47–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iranzo J, Krupovic M & Koonin EV The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. MBio 7, e00978–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iranzo J, Martincorena I & Koonin EV Cancer-mutation network and the number and specificity of driver mutations. Proc. Natl Acad. Sci. USA 115, E6010–E6019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova KS, Wolf YI & Koonin EV The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans 41, 1392–1400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin EV & Makarova KS Discovery of oligonucleotide signaling mediated by CRISPR-associated polymerases solves two puzzles but leaves an enigma. ACS Chem. Biol 13, 309–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silas S et al. On the origin of reverse transcriptase-using CRISPR–Cas systems and their hyperdiverse, enigmatic spacer repertoires. MBio 8, e00897–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puigbo P, Makarova KS, Kristensen DM, Wolf YI & Koonin EV Reconstruction of the evolution of microbial defense systems. BMC Evol. Biol 17, 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrett RA, Vestergaard G & Shah SA Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol 19, 549–556 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Reeks J, Naismith JH & White MF CRISPR interference: a structural perspective. Biochem. J 453, 155–166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Özcan A et al. Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat. Microbiol 19, 89–96 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Makarova KS, Aravind L, Wolf YI & Koonin EV Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol. Direct 6, 38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venclovas C Structure of Csm2 elucidates the relationship between small subunits of CRISPR–Cas effector complexes. FEBS Lett 590, 1521–1529 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Chylinski K, Makarova KS, Charpentier E & Koonin EV Classification and evolution of type II CRISPR–Cas systems. Nucleic Acids Res 42, 6091–6105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briner AE & Barrangou R Guide RNAs: a glimpse at the sequences that drive CRISPR–Cas systems. Cold Spring Harb. Protoc 2016, pdb.top090902 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Faure G et al. Comparative genomics and evolution of trans-activating RNAs in Class 2 CRISPR–Cas systems. RNA Biol 16, 435–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chyou TY & Brown CM Prediction and diversity of tracrRNAs from type II CRISPR–Cas systems. RNA Biol 16, 423–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fonfara I, Richter H, Bratovic M, Le Rhun A & Charpentier E The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). [DOI] [PubMed] [Google Scholar]
- 56.East-Seletsky A et al. Two distinct RNase activities of CRISPR–C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168, 121–134 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Kunin V, Sorek R & Hugenholtz P Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8, R61 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lange SJ, Alkhnbashi OS, Rose D, Will S & Backofen R CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res 41, 8034–8044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almendros C, Nobrega FL, McKenzie RE & Brouns SJJ Cas4–Cas1 fusions drive efficient PAM selection and control CRISPR adaptation. Nucleic Acids Res 47, 5223–5230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koonin EV & Aravind L Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ 9, 394–404 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Vestergaard G, Garrett RA & Shah SA CRISPR adaptive immune systems of Archaea. RNA Biol 11, 156–167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinkunas T et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30, 1335–1342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Shayeb B et al. Clades of huge phage from across Earth’s ecosystems Preprint at 10.1101/572362 (2019). [DOI] [PMC free article] [PubMed]
- 65.Leipe DD, Koonin EV & Aravind L STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol 343, 1–28 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Newire E, Aydin A, Juma S, Enne V & Roberts AP Identification of a Type IV CRISPR–Cas system located exclusively on IncHI1B/IncFIB plasmids in Enterobacteriaceae Preprint at 10.1101/536375 (2019). [DOI] [PMC free article] [PubMed]
- 67.Makarova KS et al. Predicted highly derived class 1 CRISPR–Cas system in Haloarchaea containing diverged Cas5 and Cas7 homologs but no CRISPR array. FEMS Microbiol. Lett 366, fnz079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zaremba-Niedzwiedzka K et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Strecker J et al. Engineering of CRISPR–Cas12b for human genome editing. Nat. Commun 10, 212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swarts DC & Jinek M Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a. Mol. Cell 73, 589–600.e584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meeske AJ, Nakandakari-Higa S & Marraffini LA Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570, 241–245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karvelis T et al. PAM recognition by miniature CRISPR–Cas14 triggers programmable double-stranded DNA cleavage Preprint at 10.1101/654897 (2019). [DOI] [PMC free article] [PubMed]
- 73.Anantharaman V, Makarova KS, Burroughs AM, Koonin EV & Aravind L Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 8, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hudaiberdiev S et al. Phylogenomics of Cas4 family nucleases. BMC Evol. Biol 17, 232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Charpentier E, Richter H, van der Oost J & White MF Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol. Rev 39, 428–441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dudek NK et al. Novel microbial diversity and functional potential in the marine mammal oral microbiome. Curr. Biol 27, 3752–3762 e3756 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Castelle CJ et al. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat. Rev. Microbiol 16, 629–645 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Burstein D et al. Major bacterial lineages are essentially devoid of CRISPR–Cas viral defence systems. Nat. Commun 7, 10613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levin BR Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR-mediated adaptive immunity in bacteria. PLOS Genet 6, e1001171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iranzo J, Lobkovsky AE, Wolf YI & Koonin EV Evolutionary dynamics of the prokaryotic adaptive immunity system CRISPR–Cas in an explicit ecological context. J. Bacteriol 195, 3834–3844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iranzo J, Lobkovsky AE, Wolf YI & Koonin EV Immunity, suicide or both? Ecological determinants for the combined evolution of anti-pathogen defense systems. BMC Evol. Biol 15, 43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gurney J, Pleska M & Levin BR Why put up with immunity when there is resistance: an excursion into the population and evolutionary dynamics of restriction-modification and CRISPR–Cas. Philos. Trans. R. Soc. Lond. B Biol. Sci 374, 20180096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garcia-Martinez J, Maldonado RD, Guzman NM & Mojica FJM The CRISPR conundrum: evolve and maybe die, or survive and risk stagnation. Microb. Cell 5, 262–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Houte S et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS & Koonin EV Viral diversity threshold for adaptive immunity in prokaryotes. MBio 3, e00456–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Westra ER et al. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol 25, 1043–1049 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Bernheim A, Bikard D, Touchon M & Rocha EPC A matter of background: DNA repair pathways as a possible cause for the sparse distribution of CRISPR–Cas systems in bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci 374, 20180088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koonin EV & Krupovic M Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat. Rev. Genet 16, 184–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krupovic M, Beguin P & Koonin EV Casposons: mobile genetic elements that gave rise to the CRISPR–Cas adaptation machinery. Curr. Opin. Microbiol 38, 36–43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krupovic M, Makarova KS, Forterre P, Prangishvili D & Koonin EV Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR–Cas immunity. BMC Biol 12, 36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kieper SN et al. Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep 22, 3377–3384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee H, Zhou Y, Taylor DW & Sashital DG Cas4dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell 70, 48–59. e45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shiimori M, Garrett SC, Graveley BR & Terns MP Cas4 nucleases define the pam, length, and orientation of DNA fragments integrated at CRISPR loci. Mol. Cell 70, 814–824.e816 (2018). This work reveals the molecular details of the involvement of Cas4, an ancillary protein that cooperates with Cas1 and Cas2 in several CRISPR–Cas subtypes, in the process of adaptation.
- 94.Burroughs AM, Zhang D, Schaffer DE, Iyer LM & Aravind L Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res 43, 10633–10654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rostol JT & Marraffini LA Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat. Microbiol 4, 656–662 (2019). This work demonstrates that indiscriminate RNA cleavage by the HEPN RNase domain of the Csm6 protein of type III CRISPR–Cas systems induces growth arrest in the host bacteria, providing a backup defence mechanism.
- 96.Makarova KS, Wolf YI, Snir S & Koonin EV Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol 193, 6039–6056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kapitonov VV, Makarova KS & Koonin EV ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs. J. Bacteriol 198, 797–807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Athukoralage JS, Rouillon C, Graham S, Gruschow S & White MF Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562, 277–280 (2018). This works expands the characterization of the signalling pathway in type III CRISPR–Cas sequences by showing that a distinct variety of CARF domain cleaves the cyclic oligoA molecules produced by Cas10 and thus regulates the pathway.
- 99.Shmakov SA et al. Systematic prediction of functionally linked genes in bacterial and archaeal genomes. Nat. Protoc 14, 3013–3031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


