Abstract
Hepatitis C virus (HCV) is the major cause of non-A, non-B hepatitis worldwide. Current treatments are not curative for most infected individuals, and there is an urgent need for both novel therapeutic agents and small-animal models which can be used to evaluate candidate drugs. A small-animal model of HCV gene expression was developed with recombinant vaccinia virus vectors. VHCV-IRES (internal ribosome entry site) is a recombinant vaccinia viral vector containing the HCV 5′ nontranslated region (5′-NTR) and a portion of the HCV core coding region fused to the firefly luciferase gene. Intraperitoneal injection of VHCV-IRES produced high levels of luciferase activity in the livers of BALB/c mice. Antisense oligonucleotides complementary to the HCV 5′-NTR and translation initiation codon regions were then evaluated for their effects on the expression of these target HCV sequences in BALB/c mice infected with the vaccinia virus vector. Treatment of VHCV-IRES-infected mice with 20-base phosphorothioate oligonucleotides complementary to the sequence surrounding the HCV initiation codon (nucleotides 330 to 349) specifically reduced luciferase expression in the livers in a dose-dependent manner. Inhibition of HCV reporter gene expression in this small-animal model suggests that antisense oligonucleotides may provide a novel therapy for treatment of chronic HCV infection.
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus and a member of the family Flaviviridae. The 9.4-kb genomic RNA encodes a single polyprotein of approximately 3,010 amino acids (13, 42). The polyprotein is posttranslationally processed into at least 10 distinct structural and nonstructural proteins (6, 22, 29, 39). The genome contains a 5′ nontranslated region (5′-NTR) that is approximately 340 nucleotides in length (23) and that functions as an internal ribosome entry site (IRES) for the initiation of translation (38, 44, 46). The 3′-NTR consists of an internal poly(U/UC) tract followed by a highly conserved 98-base sequence. This 3′ structure is believed to be important in viral replication (28, 43, 48).
Infection with HCV has been identified as the major cause of posttransfusion non-A, non-B hepatitis. In the United States alone approximately 3.5 million people are believed to be infected. In selected populations in parts of Africa and the Middle East, the prevalence reaches 4 to 6% (40, 50). Most HCV infection leads to chronic liver disease in which liver cirrhosis often ensues through persistent viral replication, infection, and ensuing inflammatory activity. HCV also has a striking association with hepatocellular carcinoma (8, 37) and is a leading cause of end-stage liver disease requiring liver transplantation (40). Alpha interferon therapy is used to eradicate virus from chronically infected individuals, but long-term sustained responses are seen in only about 10 to 25% of patients after 6 months of therapy (17, 34a, 40). Vaccine production, meanwhile, has been hampered by the existence of quasispecies of the virus (10, 31) and the lack of a protective immune response (12, 47).
Antisense oligonucleotides are a very promising technology for use in the development of drugs with both high target specificity and reduced side effects (7, 15, 16, 18, 26, 32). Studies have reported antisense inhibition of viral gene expression in biochemical assays, in cultured cells, and in animal models (1–4, 35, 36, 45). Antisense oligonucleotides targeting human cytomegalovirus and human immunodeficiency virus are being evaluated in clinical trials.
Several cell culture systems (27, 41, 49) and mouse xenograft models with HCV-infected human liver fragment (21) have been reported to support HCV replication in vivo. However, their reliability and simplicity as models of HCV replication are still in question. Despite progress in molecular research, the inadequacies of cell culture replication systems and small-animal models of HCV infection continue to impede studies of viral pathogenesis and drug development.
In this work, a recombinant vaccinia virus vector containing a portion of the HCV genome was used to evaluate antisense oligonucleotide inhibition of HCV gene expression in vivo. Two phosphorothioate antisense oligonucleotides, ISIS 6547 and ISIS 14803, with sequences complementary to the sequence of the highly conserved region surrounding the HCV translation initiation codon were identified as having specific and dose-dependent inhibitory effects on HCV gene expression in the livers of HCV-vaccinia virus recombinant-infected mice.
MATERIALS AND METHODS
Antisense oligonucleotides.
All oligonucleotides used in this study were phosphorothioate oligodeoxyribonucleotides (ODNs). They were synthesized as described previously (33). Briefly, the synthesis was performed on a MilliGen/Biosearch 8800 DNA synthesizer. After thiation of phosphite linkages and deblocking, the ODNs were purified by high-pressure liquid chromatography with a Waters Prep LC 4000 pump system with a water-sodium acetate-methanol gradient. Detritylation was confirmed by capillary gel electrophoresis. The ODNs were further desalted by high-pressure liquid chromatography and assessed by spectrophotometry, polyacrylamide gel electrophoresis densitometry, electrospray-ionized mass spectrometry, and endotoxin assay. The final ODNs were >97.5% full length, had <1.5% phosphodiester linkages, and had <9 endotoxin units/mg. The sequences of oligonucleotides (5′ to 3′) were as follows: ISIS 6547, GTGCTCATGGTGCACGGTCT; ISIS 14803, GTGCmTCmATGGTGCmACmGGTCmT (where Cm represents 5-methylcytidine); and ISIS 1082, GCCGAGGTCCATGTCGTACGC.
DNA constructs.
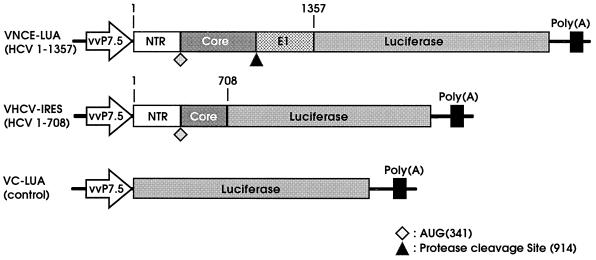
The vaccinia virus expression plasmid cassette pSC11 (11) uses the vaccinia virus early and late promoter vvP7.5 to express a foreign gene and the vaccinia virus late promoter vvP11 to express a lacZ gene. Portions of the vaccinia virus thymidine kinase (TK) sequence flank the expression cassette to facilitate homologous recombination into the vaccinia virus genome. The HCV sequence was derived from pHCV3, a cDNA clone obtained from a patient with HCV type H infection. Nucleotides 1 to 1357 of the HCV genome, including the HCV 5′-NTR, the entire core coding sequence, and part of the E1 coding sequence, were fused to the 5′ end of a luciferase gene containing simian virus 40 polyadenylation signal sequence (pGL-2 promoter vector; Promega). The fused DNA fragment was placed downstream of the vvP7.5 promoter in pSC11, and the resulting plasmid construct was designated pVNCE-LUA. A construct designated pVHCV-IRES was generated by restriction nuclease cleavage of pVNCE-LUA at nucleotides 709 to 1357 of the HCV genome and religation in the presence of a DNA oligomer connecting both sides of the deletion. pVC-LUA is a control virus construct in which the luciferase gene including the initiation codon and the polyadenylation signal was directly placed under control of the vvP7.5 promoter of pSC11. The constructs are depicted in Fig. 1.
FIG. 1.
Vaccinia virus-HCV recombinant constructs. HCV sequences were ligated in frame with the luciferase gene. The fused gene was cloned into pSC11 with the vaccinia virus promoter vvP7.5 to drive expression. Each clone was introduced into vaccinia virus WR via homologous recombination at the TK locus. Recombinant viruses that survived bromodeoxyuridine selection and that expressed both luciferase and β-galactosidase activities were isolated.
Recombinant virus.
The basic experimental procedures for the generation of recombinant vaccinia virus have been described previously (20). CV-1 cells were used for homologous recombination and viral plaque assays. Hu TK-negative (TK−) 143B cells for TK− selection were purchased from the American Type Culture Collection. BSC-40 cells for growth of virus were a gift from J. H. Strauss, California Institute of Technology. Vaccinia virus WR was a gift from B. Semler, University of California, Irvine. Plasmid DNA transfection was facilitated with lipofectin according to the recommendations of the supplier (GIBCO BRL). The selection of recombinant virus was made first by viral plaque formation in TK− 143B cells in the presence of bromodeoxyeridine and then by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining of plaques. Plaque-purified (three times) virus was used to prepare large quantities of virus stock. Virus-containing cells were harvested in Dulbecco modified Eagle medium with 0.5% fetal bovine serum followed by freezing-thawing three times to dissociate the virus. After centrifugation to remove the cellular debris, the supernatant was used for infection. Virus stock was ensured to be 100% pure by a plaque assay. The lowercase “p” in the designation of each plasmid DNA construct was removed from the designations of each recombinant virus recombined with the respective plasmid.
In vivo evaluation of oligonucleotides.
Six-week-old female BALB/c mice were purchased from Charles River Laboratories (Boston, Mass.) and housed at HTI Bio-service, Inc. (San Diego, Calif.). Randomized groups of 8 to 10 mice were subcutaneously pretreated with oligonucleotide once daily for 2 days before virus infection and were posttreated once at 4 h after infection. The infection was initiated by intraperitoneal injection of 108 PFU of virus in 0.5 ml of saline. At 24 h after infection the liver of each mouse was excised and was kept in dry ice. Hepatic tissue was homogenized by using the Tissue Tearor (Biospec Products Inc.) at 30,000 rpm for 30 s in 20 μl of luciferase reporter lysis buffer (Promega) per mg. Samples were then transferred to Eppendorf tubes, vortexed for 20 s, and then centrifuged at 10,000 rpm at 4°C for 3 min. A total of 20 μl of supernatant was transferred to each well of a 96-well microtiter plate (Dynatech Laboratories, Inc.), and 100 μl of Luciferase Assay Reagent (Promega) was added immediately prior to luminescence detection. The relative light units (RLUs) were measured with a luminometer (ML 1000; model 2.4; Dynatech Laboratories, Inc.).
RESULTS
Characterization of HCV-vaccinia virus recombinant.
In the vaccinia virus construct VHCV-IRES, the HCV target sequence (nucleotides 1 to 708) was fused to the luciferase gene and the fused product was placed downstream of the vaccinia promoter vvP7.5. This target-reporter construct uses the HCV initiation codon with the internal ribosomal entry initiation mechanism for translation (30, 38, 44). For the purpose of direct control for any other possible influence on luciferase gene readout except the expression of the HCV target sequence, pVC-LUA was engineered so that it had the same gene arrangements as VHCV-IRES except for the absence of the HCV target sequence. In pVC-LUA, luciferase gene expression is derived from its own initiation codon with a cap-dependent mechanism for translation (Fig. 1). VNCE-LUA contains additional HCV sequences (nucleotides 709 to 1357) fused to the luciferase gene.
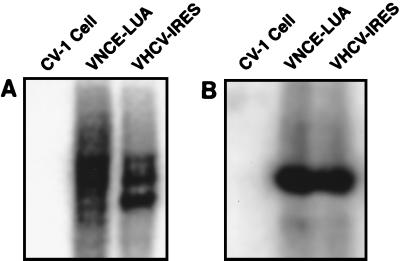
Both VHCV-IRES and VC-LUA were tested for the correct expression of specific RNAs. In infected CV-1 cells, target RNA expressed by VHCV-IRES was confirmed by HCV-specific Northern blotting and reverse transcription-PCR (RT-PCR) analysis. As shown in Fig. 2, VHCV-IRES produced shorter HCV-containing mRNA than VNCE-LUA due to the shorter HCV sequence in the DNA template. Both RNA species specifically hybridized to a radioactive HCV DNA probe covering the 5′ terminus. Two major HCV RNA species were observed for VHCV-IRES and VNCE-LUA. In each case the sizes of the slower-migrating RNAs correspond to the predicted sizes (2.9 kb for VNCE-LUA and 2.2 kb for VHCV-IRES). Multiple HCV RNA species expressed from cDNA containing the same HCV sequences as VNCE-LUA has previously been reported in transformed immortalized hepatocytes (24). The origin of the smaller RNA species expressed by these vectors has not been elucidated.
FIG. 2.
HCV-specific RNA in cells infected with the HCV-vaccinia virus recombinant. CV-1 cells were infected with virus recombinant at a multiplicity of infection of 5. Total RNA was isolated 24 h after infection and a Northern blot was prepared from a formaldehyde-agarose gel (A). (B) Southern blot of RT-PCR products from a reaction with HCV-specific primers. Both blots were hybridized with a 32P-radiolabeled probe of HCV (1 to 1,357 bp).
RNA from VHCV-IRES- or VNCE-LUA-infected CV-1 cells was reverse transcribed into DNA and amplified by PCR with primers bracketing nucleotides 93 to 314. The RT-PCR DNA fragments were of the predicted length and were also specifically recognized by the radioactive HCV DNA probe by Southern blot analysis.
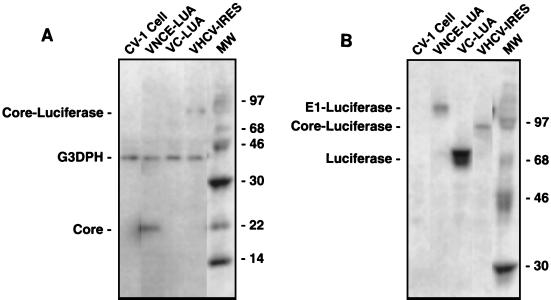
Protein production by the recombinant viruses was evaluated in infected CV-1 cells by Western blotting (Fig. 3). VHCV-IRES expressed a fusion protein that was identified with both a luciferase protein-specific antibody and an antibody specific for the HCV core protein. VC-LUA expressed only the full-length luciferase protein. VNCE-LUA expressed the HCV core protein and a fused E1-luciferase protein due to cleavage between the core protein and E1 by cellular signal peptidase. VHCV-IRES expressed less protein than VC-LUA, probably because of the lower efficiency of the HCV IRES translation mechanism. Accordingly, the luciferase activity produced by VHCV-IRES was less than that produced by VC-LUA in cell culture and mouse livers (data not shown).
FIG. 3.
Expression of proteins in cells infected with a vaccinia virus-HCV recombinant. CV-1 cells were infected with recombinant virus at a multiplicity of infection of 5. At 24 h following infection, protein extracts were prepared and proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After electrophoretic transfer to membranes, the HCV core protein was detected with a polyclonal HCV core protein-specific antibody (1:1,500) from an HCV-positive patient (A), and luciferase protein was detected with a polyclonal luciferase protein-specific antibody (1:5,000) from Promega (B). Immunoreactive proteins were visualized and quantitated with a phosphorimager following incubation with 125I-radiolabeled secondary antibody (1:3,000) from ICN. G3DPH, glyceraldehyde- 3-phosphate dehydrogenase; MW, molecular weight. Numbers on the right of each panel are molecular weights (in thousands).
Virulence and phenotypes of vaccinia virus-HCV recombinant.
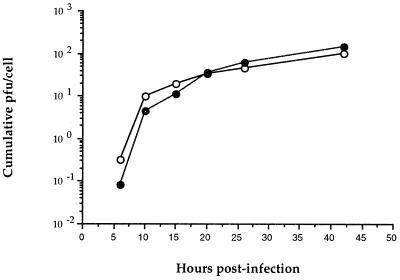
Genetic manipulation may produce phenotypic changes in a virus. To confirm that there was no difference in phenotype caused by the insertion of the HCV sequence, VC-LUA was compared with VNCE-LUA in a cumulative growth assay (Fig. 4). VNCE-LUA is the parental virus of VHCV-IRES and contains more HCV sequence (nucleotides 1 to 1357) than VHCV-IRES. Nevertheless, the growth rates of the two viruses were identical and their plaques were identical in size (data not shown).
FIG. 4.
Cumulative growth of VNCE-LUA (○) and VC-LUA (•). CV-1 cells were infected with vaccinia virus recombinant at a multiplicity of infection of 5. At the indicated times, cells were harvested and disrupted by freezing-thawing. Viral titers in supernatant were determined by viral plaque assay on a monolayer of CV-1 cells. Data represent the means from two independent sets of experiments.
In vivo luciferase activity.
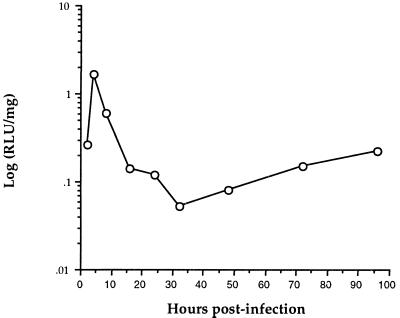
Luciferase was chosen as a reporter gene in this system because of the sensitivity of luciferase enzymatic assays, the lack of luciferase gene expression in mammals, and the wide quantitative response range of luminescence detection. The kinetics of luciferase protein expression in mouse liver after VHCV-IRES infection were evaluated (Fig. 5) in order to identify the optimum time after infection to determine expression levels. Luciferase activity peaks approximately 4 h after infection. This peak represents the initial wave of viral replication in the liver after intraperitoneal infection. Luciferase activity in the spleen displays a similar kinetic pattern (data not shown). From 16 to 96 h postinfection, luciferase activity stabilizes. The luciferase signal is only about 1/10 of its peak level at this time, but nonetheless, it is still enhanced several hundred-fold relative to the background levels. For the experiments reported on here, luciferase activities were determined 24 h after infection to minimize the variation introduced by the time needed for procedures with animals.
FIG. 5.
Kinetics of luciferase protein expression in livers (○) of mice infected with VHCV-IRES. BALB/c mice were infected with 108 PFU of VHCV-IRES by intraperitoneal injection. At different times following infection the mice were killed, their livers were removed, and hepatic luciferase activity was determined. Luciferase activity was expressed as RLUs per milligram of liver tissue (see Materials and Methods). Data represent the means for two mice at each time point.
The luciferase signal present in mouse liver is a direct result of infection, replication, and expression of recombinant virus. VHCV-IRES and VC-LUA recovered from infected mouse liver, spleen, and kidney 2 days after infection were capable of producing plaques on CV-1 cells. When VHCV-IRES was UV irradiated in the presence of psoralen prior to infection, the luciferase activity in liver was reduced to background level (data not shown), indicating the requirement for infectious virus.
Inhibitory effect of antisense phosphorothioate oligonucleotides on HCV gene expression.
Antisense inhibition of target gene expression requires specific sequence-dependent binding of oligonucleotides to complementary target RNA sequences. Although there is sequence diversity in the coding regions of HCV RNA from different strains of the virus, well-conserved sections of sequence found in the 5′-NTR provide suitable target sequences for antisense oligonucleotide inhibition. Previous biochemical experiments and cell culture assays identified ISIS 6547 as a potent inhibitor of HCV gene expression in vitro (24). This 20-base phosphorothioate oligonucleotide is complementary to sequences surrounding the HCV polyprotein initiation codon (nucleotides 330 to 349). In transformed human hepatocytes expressing the HCV 5′-NCR, the core protein, and part of E1 (HCV nucleotides 1 to 1357), ISIS 6547 demonstrated at least a 50% reduction of HCV RNA at a dose of 100 nM (24). This inhibitory effect was concentration dependent and sequence specific. HCV core protein expression in transformed hepatocytes was also inhibited by ISIS 6547.
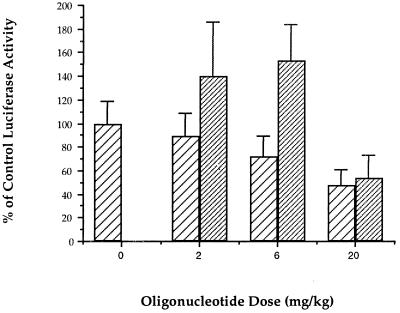
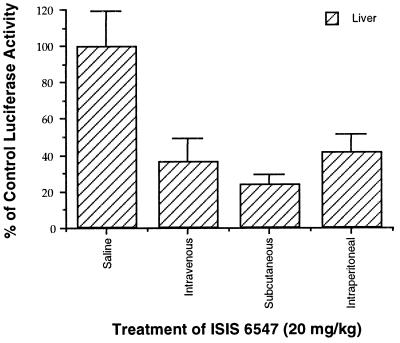
ISIS 6547 treatment was evaluated for its effects on HCV-luciferase expression in VHCV-IRES-infected mice. BALB/c mice were pretreated subcutaneously with oligonucleotide, infected intraperitoneally with VHCV-IRES, and posttreated subcutaneously with oligonucleotide. The effects of the oligonucleotides on HCV gene expression were measured by determining the luciferase activity in the mouse livers 24 h after infection. In a typical assay (Fig. 6), ISIS 6547 reduced the luciferase signal in a dose-dependent manner: 11% inhibition at 2 mg/kg of body weight, 28% inhibition at 6 mg/kg, and 52% inhibition at 20 mg/kg. A statistically significant reduction relative to that for the control group was achieved with the 20-mg/kg dose (P < 0.05). A control noncomplementary phosphorothioate oligonucleotide, ISIS 1082, exhibited no inhibitory activity at the two lower doses and appeared to stimulate expression at these doses. At a 6-mg/kg dose, the luciferase expression in ISIS 6547-treated mice was significantly less than the luciferase expression in ISIS 1082-treated mice (P < 0.05). At 20 mg/kg, ISIS 1082 nonspecifically inhibited the luciferase signal. Varying the route of antisense oligonucleotide injection (subcutaneous, intravenous, or intraperitoneal injection) did not make a significant difference in the inhibitory effect of ISIS 6547 (Fig. 7).
FIG. 6.
Inhibitory effects of HCV antisense oligonucleotide ISIS 6547 (▨) and a control oligonucleotide (ISIS 1082 [▨]) on luciferase expression in VHCV-IRES-infected mice. Members of a group of eight female BALB/c mice were treated subcutaneously with oligonucleotides at 48 and 24 h prior to infection and at 4 h after infection. Mice were infected by intraperitoneal infection of 108 PFU of VHCV-IRES. At 24 h following infection, the luciferase activity in the livers was determined. A control group of infected mice treated with saline instead of oligonucleotide was used to determine 100% control luciferase activity. Data represent the means ± standard errors of the means. ISIS 1082 is a control phosphorothioate oligonucleotide which is noncomplementary to HCV RNA. Luciferase expression levels in animals treated with ISIS 6547 or ISIS 1082 at 20 mg/kg were significantly different from those in control treated animals (P < 0.05), and luciferase expression in animals treated with 6 mg of ISIS 6547 per kg was significantly reduced relative to that in animals treated with ISIS 1082 (P < 0.05).
FIG. 7.
Inhibitory effects of HCV antisense oligonucleotide ISIS 6547 on luciferase expression in VHCV-IRES-infected mice by using alternative dosing routes of administration. Members of groups of eight female BALB/c mice were infected with VHCV-IRES and treated with ISIS 6547 as described in the legend to Fig. 6, except that ISIS 6547 was administrated by the intravenous, subcutaneous, and intraperitoneal routes of administration at a dose of 20 mg/kg. A control group of infected mice treated with saline instead of oligonucleotide via intravenous injection was used to determine 100% of luciferase activity. Data are presented as means ± standard errors of the means. For all three treatment groups the results were statistically significantly different from those for the saline-treated control group (P < 0.01).
To ensure that the oligonucleotides did not interfere with luciferase enzyme activity, ISIS 6547 was mixed with the liver extract from VHCV-IRES-infected mice treated with saline. ISIS 6547 at concentrations as high as 5 μM had no effect on luciferase enzyme activity. The luciferase activity of the mixture was measured under identical conditions, as described above for the animal studies. Therefore, ISIS 6547 does not directly interact with luciferase enzymatic activity.
Inhibitory effect of 5-methylcytidine-modified antisense oligonucleotides on HCV gene expression.
Replacement of the nucleoside cytidine with 5-methylcytidine in antisense oligonucleotides confers higher-affinity binding to target RNA while still permitting RNase H-mediated cleavage of hybrid message RNA (19). 5-Methylcytidine-modified oligonucleotides are also less immunostimulatory and can reduce host complement system activation and B-cell and natural killer cell proliferation (9, 25). ISIS 14803 is a 5-methylcytidine-modified oligonucleotide with the same sequence as ISIS 6547. When evaluated in the VHCV-IRES-infected mouse model, ISIS 14803 showed potency comparable to that of the parental oligonucleotide ISIS 6547 in the reduction of liver luciferase activity (Fig. 8).
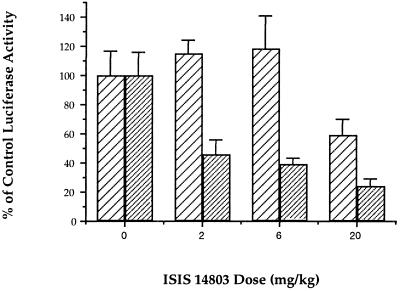
FIG. 8.
Comparison of inhibitory activities of ISIS 6547 (▨) and the 5′-methylcytidine-modified HCV antisense oligonucleotide ISIS 14803 ( ). Members of a group of eight female BALB/c mice were treated subcutaneously with oligonucleotide at 24 h and 2 h prior to infection and at 4 h after infection. Mice were infected by intraperitoneal injection of 108 PFU of VHCV-IRES. At 24 h following infection, the luciferase activity in the livers was determined. A control group of infected mice treated with saline instead of oligonucleotide was used to determine 100% control luciferase activity. Data are represented as means ± standard errors of the means.
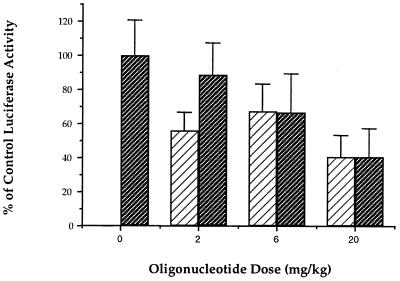
To ensure that this dose-dependent reduction was specific for HCV sequence and was not due to nonspecific effects on vaccinia virus replication, the levels of expression of luciferase reporter constructs VC-LUA and VHCV-IRES were compared in animals treated with ISIS 14803 (Fig. 9). ISIS 14803 did not inhibit luciferase expression from VC-LUA at doses of 2 and 6 mg/kg, while the dose-dependent inhibition of luciferase expression by VHCV-IRES was maintained. Differences in luciferase expression relative to that for the saline-treated control group were statistically significant for all groups treated with ISIS 14803 (P < 0.01). Luciferase expression levels in mice infected with VHCV-IRES and treated with ISIS 14803 were significantly lower than luciferase expression levels in mice infected with VC-LUA and treated with ISIS 14803 for all dose groups (P < 0.01).
FIG. 9.
Specific inhibitory effect of ISIS 14803 on luciferase expression in VHCV-IRES-infected mice. Members of groups of nine female BALB/c mice were infected and treated subcutaneously with oligonucleotide as described in the legend to Fig. 7. Control groups of mice infected with VHCV-IRES (▨) or VC-LUA (▨) but treated with saline instead of oligonucleotide were used to determine the respective values for 100% luciferase activity. Data are represented as means ± standard errors of the means. For all groups of mice infected with VHCV-IRES and treated with ISIS 14803 the results were statistically significantly different from those for the saline-treated control group infected with the same virus (P < 0.01). At each dose of oligonucleotide, the difference between the percentage of control expression for VHCV-IRES-infected mice and for VC-LUA-infected mice was statistically significant (P < 0.01).
At the high dose, 20 mg/kg, ISIS 14803 nonspecifically reduced the luciferase activity in animals infected with VC-LUA by 40.5% relative to that for the control group. Similar nonspecific activity was observed when mice infected with VHCV-IRES were treated with the control oligonucleotide ISIS 1082 (Fig. 6), indicating that at high doses phosphorothioate oligonucleotides have nonspecific effects on luciferase protein expression possibly resulting from nonspecific effects on vaccinia virus replication.
DISCUSSION
The in vivo results presented here demonstrate that antisense oligonucleotides specific for the HCV translation initiation region can specifically reduce HCV gene expression in the mouse liver. The sequence targeted by ISIS 6547 and ISIS 14803 is one of the most highly conserved regions among all different HCV strains and is therefore attractive from a drug development perspective.
Experimental evidence has shown that the inhibitory effects of antisense oligonucleotides can be achieved through several different mechanisms: inhibition of RNA splicing, inhibition of mRNA translation, or degradation of RNA (5, 14, 24, 33, 45). In transformed human hepatocytes expressing HCV target sequence, ISIS 6547 reduced target RNA levels by inducing cleavage within the oligonucleotide binding site (24), indicating that an RNase H-mediated mechanism was at least partly responsible. However, vaccinia virus and HCV both replicate in the cytoplasm of infected cells, an environment where RNase H levels may be reduced compared to the levels in the nucleus. In this in vivo model of HCV gene expression we have not attempted to discern the mechanism of inhibition. ISIS 6547 and ISIS 14803 could be exerting inhibitory activity through RNase H-mediated message degradation or translational arrest.
ISIS 6547 and ISIS 14803 treatment at moderate doses (2 and 6 mg/kg) specifically inhibited HCV-luciferase expression in the livers of recombinant vaccinia virus-infected mice. ISIS 1082, a sequence-irrelevant phosphorothioate oligonucleotide, inhibited HCV-luciferase expression only at the high dose (20 mg/kg). Similarly, ISIS 14803 treatment at a dose of 20 mg/kg reduced the level luciferase expression by the VC-LUA control virus. These results suggest that high doses of phosphorothioate oligonucleotides may exert nonspecific effects on vaccinia virus replication or gene expression. The mechanism of the observed nonspecific inhibition at high doses is unknown. However, phosphorothioate oligonucleotides have been reported to induce cytokine production and to exert proinflammatory effects which could affect vaccinia virus replication (34, 51). The apparently enhanced expression of luciferase in mice infected with VHCV-IRES and treated with the control oligonucleotide at 2 or 6 mg/kg also remains unexplained.
The 5-methylcytidine-modified anti-HCV oligonucleotide (ISIS 14803) and the unmodified anti-HCV oligonucleotide (ISIS 6547) showed comparable inhibitory activities in a head-to-head comparison (Fig. 8), despite the predicted enhanced hybridization affinity of the 5-methylcytidine-modified oligonucleotide. It is not likely that differences in the potencies of these compounds could be discerned given the variability inherent in the animal models used and the small increase in predicted affinity (the five substitutions would be expected to increase the melting temperature of ISIS 14803 and an RNA complement by approximately 2 to 3°C relative to that for ISIS 6547). It is encouraging nevertheless that in an independent experiment (Fig. 9) ISIS 14803 showed activity that was greater than the activity observed in any previous experiments with ISIS 6547.
The HCV-vaccinia virus recombinant model provides unique advantages for evaluation of inhibition of HCV gene expression. IRES-dependent expression of the luciferase reporter gene can easily be detected in the livers of infected mice. The liver is believed to be the primary site of HCV replication in patients. Furthermore, the vaccinia virus vector used for expression of the HCV luciferase reporter replicates in the cytoplasm of infected cells. Therefore, expression of the reporter gene is presumed to be cytoplasmic, as is expression for HCV.
However, there are limitations to the model as well. The vaccinia virus vector has a unique replication and expression system which, although cytoplasmic, is very different from that of HCV. Expression of the HCV-luciferase reporter gene from the vvP7.5 promoter is also likely to be much greater than that of HCV gene expression in infected hepatocytes. In addition, the mRNA produced by the vaccinia virus vector is likely to contain a 7-methylguanosine cap characteristic of transcripts for vaccinia virus. While IRES-dependent translation of the HCV-luciferase reporter is believed to predominate in this system since three out-of-frame initiation codons occur upstream of the authentic AUG in the HCV 5′-NTR, it is impossible to preclude the possibility of an altered translation mechanism in this system. The differences between the expression of HCV sequences in the HCV-vaccinia virus recombinant system and in HCV-infected hepatocytes are more likely to lead to quantitative rather than qualitative differences in the response to HCV antisense oligonucleotides. For example, the high levels of expression of the vaccinia virus system may be less sensitive to inhibition by antisense oligonucleotides than the low levels of expression likely to be encountered in HCV-infected hepatocytes.
Despite the imperfect nature of the HCV-vaccinia virus recombinant model, the results presented in this report demonstrate that antisense oligonucleotides directed to the IRES element of HCV can inhibit HCV gene expression in the livers of laboratory animals. Demonstration of the potential to inhibit HCV gene expression in vivo as well as in vitro suggests that antisense oligonucleotides may provide a novel approach to the control of HCV disease in patients.
ACKNOWLEDGMENTS
We thank C. Frank Bennett for stimulating suggestions and discussions throughout these studies and Cindy Vanderziel and Ray Ranken for excellent technical assistance. We are sincerely grateful to Kathleen Myers for critical reading of the manuscript.
REFERENCES
- 1.Agrawal S, Lisziewicz J. Potential for HIV-1 treatment with antisense oligonucleotides. J Biotechnol Healthcare. 1994;1:167–182. [Google Scholar]
- 2.Alt M, Renz R, Hofschneider P H, Paumgartner G, Caselmann W H. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligonucleotides. Hepatology. 1995;22:707–717. [PubMed] [Google Scholar]
- 3.Anderson K P, Fox M C, Brown-Driver V, Martin M J, Azad R F. Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrob Agents Chemother. 1996;40:2004–2011. doi: 10.1128/aac.40.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azad R F, Driver V B, Tanaka K, Crooke R M, Anderson K P. Antiviral activity of a phosphorothioate oligonucleotide complementary to RNA of the human cytomegalovirus major immediate-early region. Antimicrob Agents Chemother. 1993;37:1945–1954. doi: 10.1128/aac.37.9.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker B F, Lot S S, Condon T P, Cheng-Flournoy S, Lesnik E A, Sasmor H M, Bennett C F. 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM1 translation initiation complex in human umbilical vein endothelial cells. J Biol Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett C F, Kornbrust D, Henry S, Stecker K, Howard R, Cooper S, Dutson S, Hall W, Jacoby H I. An ICAM-1 antisense oligonucleotide prevents and reverses dextran sulfate sodium-induced colitis in mice. J Pharm Exp Ther. 1997;280:988–1000. [PubMed] [Google Scholar]
- 8.Bisceglie A M D, Simpson L H, Lotze M T, Hoofnagle J H. Development of hepatocellular carcinoma among patients with chronic liver disease due to hepatitis C viral infection. J Clin Gastroenterol. 1994;19:222–226. doi: 10.1097/00004836-199410000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Boggs R T, McGraw K, Condon T, Flournoy S, Villiet P, Bennett C F, Monia B P. Characterization of modulation of immune stimulation by modified oligonucleotides. Antisense Nucleic Acid Drug Dev. 1997;7:461–471. doi: 10.1089/oli.1.1997.7.461. [DOI] [PubMed] [Google Scholar]
- 10.Cabot B, Esteban J I, Martell M, Genesca J, Vargas V, Esteban R, Guardia J, Gomez J. Structure of replicating hepatitis C virus (HCV) quasispecies in the liver may not be reflected by analysis of circulating HCV viron. J Virol. 1997;71:1732–1734. doi: 10.1128/jvi.71.2.1732-1734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of β-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien D Y, Choo Q-L, Ralston R, Spaete R, Tong M, Houghton M, Kuo G. Persistence of HCV despite antibodies to both putative envelope glycoproteins. Lancet. 1993;342:933. doi: 10.1016/0140-6736(93)91983-s. [DOI] [PubMed] [Google Scholar]
- 13.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condon T P, Bennett C F. Altered mRNA splicing and inhibition of human E-selectin expression by an antisense oligonucleotide in human umbilical vein endothelial cells. J Biol Chem. 1996;271:30398–30403. doi: 10.1074/jbc.271.48.30398. [DOI] [PubMed] [Google Scholar]
- 15.Crooke S T. Therapeutic applications of oligonucleotides. Annu Rev Pharmcol Toxicol. 1992;32:329–376. doi: 10.1146/annurev.pa.32.040192.001553. [DOI] [PubMed] [Google Scholar]
- 16.Crooke S T. Oligonucleotide therapeutics. In: Wolff M E, editor. Burger’s medicinal chemistry and drug discovery. 5th ed. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 863–900. [Google Scholar]
- 17.Davis G L, Lau J N, Lim H L. Therapy for chronic hepatitis C. Viral Hepatitis. 1994;23:603–613. [PubMed] [Google Scholar]
- 18.Dean N, McKay R, Miraglia L, Howard R, Cooper S, Giddings J, Nicklin P, Meister L, Ziel R, Geiger T, Muller M, Fabbro D. Inhibition of growth of human tumor cell line in nude mice by an antisense oligonucleotide inhibitor of protein kinase C-α expression. Cancer Res. 1996;56:3499–3507. [PubMed] [Google Scholar]
- 19.Dean N M, Griffey R H. Identification and characterization of second-generation antisense oligonucleotides. Antisense Nucleic Acid Drug dev. 1997;7:229–233. doi: 10.1089/oli.1.1997.7.229. [DOI] [PubMed] [Google Scholar]
- 20.Earl P L, Moss B. Generation of recombinant vaccinia viruses. Curr Protocols Mol Biol. 1991;2:16.17.1–16.17.16. doi: 10.1002/cpmb.32. [DOI] [PubMed] [Google Scholar]
- 21.Galun E, Burakova T, Ketzinel M, Lubin I, Shezen E, Kahana Y, Eid A, Ilan Y, Rivkind A, Pizov G, Shouval D, Reisner Y. Hepatitis C virus viremia in SCID->BNX mouse chimera. J Infect Dis. 1995;172:25–30. doi: 10.1093/infdis/172.1.25. [DOI] [PubMed] [Google Scholar]
- 22.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Shyamala V, Richman K, Brauer B, Irvine B, Urdea M, Tekamp-Olson P, Kuo G, Choo Q-L, Houghton M. Characterization of terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanecak R, Brown-Driver V, Fox M C, Azad R F, Furusako S, Nozaki C, Ford C, Sasmor H, Anderson K P. Antisense oligonucleotide inhibition of hepatitis C virus gene expression in transformed hepatocytes. J Virol. 1996;70:5203–5212. doi: 10.1128/jvi.70.8.5203-5212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry S P, Monteith D, Bennett F, Levin A A. Toxicological and pharmacokinetic properties of chemically modified antisense oligonucleotide inhibitors of PKC-α and C-raf kinase. Anti-Cancer Drug Design. 1997;12:409–420. [PubMed] [Google Scholar]
- 26.Higgins K A, Perez J R, Coleman T A, Dorshkind K, McComas W, Sarmiento U M, Rosen C A, Narayanan R. Antisense inhibition of the p65 subunit of NF-κB blocks tumorigenicity and causes tumor regression. Proc Natl Acad Sci USA. 1993;90:9901–9905. doi: 10.1073/pnas.90.21.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito T, Mukaigawa J, Zhou J, Hirabayashi Y, Mitamura K, Yasui K. Cultivation of hepatitis C virus in primary hepatocyte culture from patients with chronic hepatitis C results in release of high titre infectious virus. J Gen Virol. 1996;77:1043–1054. doi: 10.1099/0022-1317-77-5-1043. [DOI] [PubMed] [Google Scholar]
- 28.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin C, Lindenbach B D, Prahai B M, McCourt D W, Rice C M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu H H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martell M, Esteban J I, Quer J, Vargas V, Esteban R, Guardia C, Gomez J. Dynamic behavior of hepatitis C quasispecies in patients undergoing orthotopic liver transplantation. J Virol. 1994;68:3425–3436. doi: 10.1128/jvi.68.5.3425-3436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monia B P, Johnston J F, Geiger T, Muller M, Fabbro D. Antitumor activity of a phosphorothioate antisense oligonucleotide targeted against C-raf kinase. Nat Med. 1996;2:668–675. doi: 10.1038/nm0696-668. [DOI] [PubMed] [Google Scholar]
- 33.Monia B P, Sasmor H, Johnston J F, Freier S M, Lesnik E A, Muller M, Geiger T, Altmann K-H, Moser H, Fabbro D. Sequence-specific antitumor activity of a phosphorothioate oligonucleotide targeted to human C-raf kinase supports an antisense mechanism of action in vivo. Proc Natl Acad Sci USA. 1996;93:15481–15484. doi: 10.1073/pnas.93.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monteith D K, Henry S P, Howard R B, Flournoy S, Levin A A, Bennett C F, Crooke S T. Immune stimulation—a class effect of phosphorothioate oligodeoxynucleotides in rodents. Anti-Cancer Drug Design. 1997;12:421–432. [PubMed] [Google Scholar]
- 34a.Nomura H, Kimmura Y, Tada H, Hisano C, Morita C, Okamoto O, Shiraishi G, Kashiwagi S. Predictive factors of a response to interferon therapy in chronic hepatitis C. J Clin Gastroenterol. 1996;23:185–190. doi: 10.1097/00004836-199610000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Offensperger W-B, Offensperger S, Walter E, Teubner K, Igloi G, Blum H E, Gerok W. In vivo inhibition of duck hepatitis B virus replication and gene expression by phosphorothioate modified antisense oligonucleotides. EMBO J. 1993;12:1257–1262. doi: 10.1002/j.1460-2075.1993.tb05767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raviprakash K, Liu K, Matteucci M, Wagner R, Riffenburgh R, Carl M. Inhibition of dengue virus by novel, modified antisense oligonucleotides. J Virol. 1995;69:69–74. doi: 10.1128/jvi.69.1.69-74.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Resnick R H, Koff R. Hepatitis C-related hepatocellular carcinoma: prevalence and significance. Arch Intern Med. 1993;153:1672–1677. [PubMed] [Google Scholar]
- 38.Rijnbrand R, Bredenbeek P, Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ nontranslated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 39.Santolini E, Migliaccio G, Monica N L. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharara A I, Hunt C M, Hamilton J D. Hepatitis C. Ann Intern Med. 1996;125:658–668. doi: 10.7326/0003-4819-125-8-199610150-00006. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu Y K, Iwamoto A, Hijikata M, Purcell R H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci USA. 1992;89:5477–5481. doi: 10.1073/pnas.89.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis c virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vidalin O, Major M E, Rayner B, Imbach J-L, Trepo C, Inchauspe G. In vitro inhibition of hepatitis C virus gene expression by chemically modified antisense oligodeoxynucleotides. Antimicrob Agents Chemother. 1996;40:2337–4804. doi: 10.1128/aac.40.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Le S-Y, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 47.Weiner A, Erickson A, Kansopon J, Crawford K, Muchmore E, Hughes A L, Houghton M, Walker C M. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci USA. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimomura H, Tsuji T, Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 49.Yoo B, Selby M, Choe J, Suh B, Choi S, Joh J, Nuovo G, Lee H-S, Houghton M, Han J. Transfection of a differentiated human hepatoma cell line (Huh7) with in vitro-transcribed hepatitis C virus (HCV) RNA and establishment of a long-term culture persistently infected with HCV. J Virol. 1995;69:32–38. doi: 10.1128/jvi.69.1.32-38.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zein N N, Rakela J, Krawitt E L, Reddy K R, Tominaga T, Persing D H, Group C S. Hepatitis C virus genotypes in the united states: epidemiology, pathogenicity, and response to interferon therapy. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Q, Temsamani J, Zhou R, Agrawal S. Pattern and kinetics of cytokine production following administration of phosphorothioate oligonucleotides in mice. Antisense Nucleic Acid Drug Dev. 1997;7:495–502. doi: 10.1089/oli.1.1997.7.495. [DOI] [PubMed] [Google Scholar]