Abstract
The early-stage repair of bone injuries dominated by the inflammatory phase is significant for successful bone healing, and the phenotypic transition of macrophages in the inflammatory phase plays indispensable roles during the bone healing process. The goal of this paper is to design a microRNA delivery nanocarrier for strictly temporal guidance of the polarization of macrophages by the sequential delivery of different microRNAs. The results showed that microRNA nanocarriers, synthesized through free radical polymerization, could be internalized by macrophages with about a cellular uptake efficiency of 80%, and the sequential delivery of microRNA-155 nanocarriers and microRNA-21 nanocarriers proved, for the first time, that it could promote an efficient and timely switch from the M1 to the M2 phenotype along the time point of bone tissue repair. The strategy proposed in this paper holds potential for controlling sequential M1-to-M2 polarization of macrophages, which provides another perspective for the treatment of bone tissue regeneration.
1. Introduction
Bone healing is a complex process involving inflammation, callus formation, and remodeling.1,2 Macrophages play pivotal and dynamic roles in the inflammatory stage and mediate almost all phases of bone healing.3 Macrophages can be activated into M1 or M2 phenotypes in response to microenvironmental stimuli.4−6 Macrophages sequential polarization to M1 and M2 phenotypes have been recognized as the core event in the inflammatory phase, governing the fate of bone repair.7
M1 phenotype macrophages play a chemotactic role, participate in the recruitment of mesenchymal stem cells, and stimulate angiogenesis, which dominate in the first 3 days after injury.8−12 Subsequently, macrophages are transiently transformed into M2 phenotypes, which have been reported to inhibit inflammation and promote osteogenic differentiation of mesenchymal stem cells.13,14 However, if proinflammatory cytokines persist and M1 phenotype macrophages dominate for more than 4 days, they lead to chronic inflammation, destroy osteogenesis, cause the synthesis of a mineralized extracellular matrix, and finally delay bone repair or result in failed bone repair.15,16 Therefore, the intervention treatment of bone injuries should not only give full play to the role of M1 phenotype macrophages in promoting inflammation and triggering subsequent bone healing but also achieve the timely transformation of M1 phenotype macrophages to M2 phenotypes.17
At present, it has been reported that many cytokines can regulate the phenotypic transformation of macrophages. For example, Spiller et al. reported a decellularized bone scaffold for sequentially delivering interferon-γ (IFNγ) and IL-4 to regulate the sequential transformation of macrophage phenotype from M1 to M2, which promoted angiogenesis and healing.6 Jumana et al. designed a biomimetic calcium phosphate coating for the delivery of IFNγ followed by simvastatin, resulting in the effective M1-to-M2 phenotype transition of macrophages.18 Many reports have proved that timing regulation of macrophage phenotype switch has emerged as a potential strategy to promote bone tissue regeneration. Although cytokines have been proved to have a good effect on the regulation of macrophage phenotypes, their application dose often exceeds the physiological range, resulting in serious side effects.19,20
MicroRNAs are widely used in the treatment of various diseases owing to the advantage of multiple targets, which can avoid the above shortcomings of cytokines.21−24 Saleh et al. developed an adhesive hydrogel containing miR-223-5p-loaded nanoparticles to control macrophage polarization to M2 for promoting wound healing.25 Although a single microRNA molecule has been proved in other diseases that it can promote tissue repair by regulating macrophage polarization, the method of promoting any phenotypic polarization without considering the time node is not suitable for bone tissue.7,17 Therefore, complying with the natural process of bone repair, the sequential delivery of multiple genes to regulate macrophage polarization may be a forceful strategy to accelerate bone healing, and microRNAs have been reported to be differentially expressed in the activated macrophages, such as microRNA-127-3p, miR-181a, and miR-451.26−29 Among them, microRNA-155 is highly expressed in M1 macrophages and less expressed in M2 macrophages.30−32 The precise role of microRNA-21 on polarization of macrophage phenotypes is controversial,27,33,34 but our unpublished data have proved that the delivery of microRNA-21 can promote macrophage polarization toward M2 phenotypes. Therefore, this paper intends to use microRNA-155 and microRNA-21 to sequentially stimulate macrophages toward M1 and M2 phenotype polarization, which strictly follows the natural process of bone healing and will be a new strategy to promote bone repair.
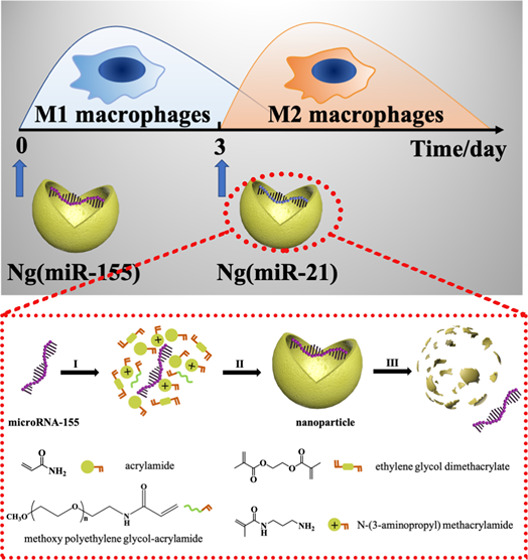
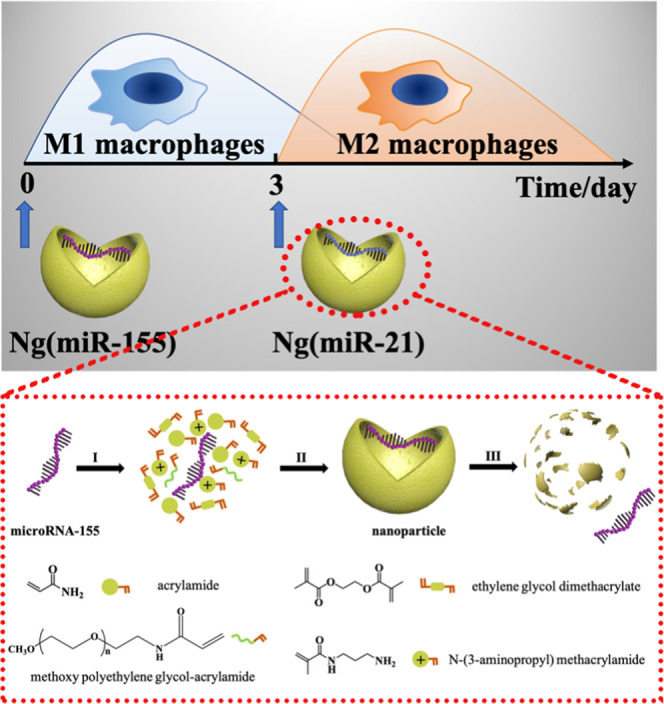
An important challenge of gene therapy is to develop safe and effective gene vectors because exposed microRNA molecules cannot easily pass through the negatively charged cell membrane and will be rapidly degraded by enzymes once they enter the circulatory system.35−37 In the present study, we designed a kind of positively charged nanocarrier-loaded microRNAs to guide the phenotypic transition of macrophages (Scheme 1). According to the physiological process of bone repair, microRNA-155 nanocarriers were first applied to accelerate the transition of macrophages to M1 phenotypes, and then microRNA-21 nanocarriers were used to transiently convert macrophages to M2 phenotypes at the appropriate time point (day 3). This strategy of sequential delivery and the appropriately selected microRNAs make sure that the phenotypic transformation of macrophages strictly follows the physiological process of bone repair, which is verified through in vitro experiments. The sequential delivery of two microRNAs conforming to the physiological process of bone tissue repair provides a new strategy for bone repair.
Scheme 1. Schematic Illustration of the Sequential Delivery of Different MicroRNAs for Modulating M1-to-M2 Macrophage Polarization.
2. Materials and Methods
2.1. Materials
The microRNA-21 mimic (miR-21), microRNA-155 mimic (miR-155), and 6-carboxy-fluorescein (FAM)-labeled microRNAs were synthesized by GenePharma (Shanghai, China). The sense strand sequence of miR-155 was 5′-UUAAUGCUAAUCGUGAUAGGGGUU-3′. The sense strand sequence of miR-21 was 5′-UAGCUUAUCAGACUGAUGUUGA-3′. All of the chemical reagents used in this paper were purchased from Sigma-Aldrich unless otherwise noted, and all reagents were analytical grade. Methoxy poly(ethylene glycol)-acrylamide (mPEG-AC, Mw 2000) was purchased from Huateng Pharma (Hunan, China). Culture medium, paraformaldehyde, and CCK-8 kit were supplied by Dalian Meilun Biotechnology Co., Ltd. (Dalian, China). 4′,6-Diamidino-2-phenylindole (DAPI) was purchased from Beyotime Biotechnology Co., Ltd. (Shanghai, China).
2.2. Preparation and Characterization of Nanocarriers
The miR-155 nanocarriers (Ng(miR-155)) and miR-21 nanocarriers (Ng(miR-21)) were synthesized using the same preparation method. Ng(miR-155) was taken as an example and a series of characterization techniques were carried out. Briefly, a neutral monomer (acrylamide), a positively charged monomer (N-(3-aminopropyl)-methacrylamide), a hydrophilic monomer (methoxy poly(ethylene glycol)-acrylamide, Mw 2000), and miR-155 were mixed in a ratio of 3500:350:150:1. Then, an acid-degraded cross-linker (ethylene glycol dimethacrylate) was added to the system, and the ratio of the monomers/cross-linker was 8:1. Subsequently, the polymerization was initiated by ammonium peroxydisulfate and N,N,N′,N′-tetramethylethylenediamine. Then, the reaction was carried out at 4 °C for 2 h, followed by dialysis using a 10 KDa dialysis bag against phosphate-buffered solution (PBS, pH = 7.4) to remove free miRNAs, unreacted monomers, and initiators.
The ζ potential and particle size distribution of Ng(miR-155) were measured by dynamic light scattering (DLS, BI-90Plus, Brookhaven Instruments Ltd.). Transmission electron microscopy (TEM, Jem-2100 f, JEOL, Japan) was used to observe the morphology of Ng(miR-155), which was stained with 2% phosphotungstic acid. The migration of miR-155 and Ng(miR-155) bands was observed by agarose gel electrophoresis containing ethidium bromide. Further, after freeze-drying, an appropriate amount of Ng(miR-155) was scanned by Fourier transform infrared spectroscopy (FTIR, JASCO FT/IR-420) for verifying the composition of polymer shells. The elements of polymer shells and naked miR-155 were analyzed by X-ray photoelectron spectroscopy (XPS) for proving that miR-155 was completely wrapped by the polymer shells.
2.3. Cell Viability Assays and Cellular Uptake Efficiency
RAW264.7 cells were used as a cellular model for evaluation of macrophage phenotypes and were maintained in 1640 culture medium containing 10% fetal bovine serum and 1% penicillin-streptomycin at 37 °C in 5% CO2 and 100% humidity. The cell viability was evaluated by the CCK-8 kit on day 1.38 RAW264.7 cells were seeded in 96-well plates at a density of 5 × 103 per well, and a certain concentration gradient of Ng(miR-155) was incubated with the cells for 24 h. Then, 10% CCK-8 was added to each well and reacted for 1 h. The absorbance value of solution at 450 nm wavelength was read out using a microplate reader (BioTek).
NIH 3T3 cells were used for evaluation of cytotoxicity. The NIH 3T3 cells were incubated at 37 °C in 5% CO2 and 100% humidity for 3 days after treatment with PBS or 50 nM Ng(miR-155). Then, the cells were stained with DAPI and FITC-phalloidin and observed by confocal laser scanning microscopy (A1R+, Nikon).
The endocytosis efficiency of Ng(miR-155) was quantified by detecting the intracellular fluorescence intensity.39 Ng(miR-155) was used in subsequent experiments with a concentration of 50 nM. RAW264.7 cells were seeded in six-well plates at a density of 1 × 105 per well for 24 h. After being incubated with miR-155 or Ng(miR-155) for another 4 h, the RAW264.7 cells were washed with PBS solution to remove the remaining molecules, followed by fixing with 4% paraformaldehyde for 10 min. Under dark conditions, the cytoskeleton and nuclei were labeled with phalloidin and DAPI, respectively. Finally, it was observed and photographed by confocal laser scanning microscopy, and the quantitively estimated cellular endocytosis efficiency was tested by flow cytometric analysis (BD Biosciences). The cells were collected and fixed, followed by washing with PBS solution. The percentage of fluorescence-positive RAW264.7 cells and mean fluorescence intensity (MFI) were evaluated. All data were analyzed using FlowJo software.
2.4. Flow Cytometric Analysis of Macrophage Phenotypes
The RAW264.7 cells were maintained in six-well plates at 1 × 105 cells per well and treated with PBS, Ng(NC), Ng(miR-155), or Ng(miR-21) for 24 h. Then, the cells were collected and incubated with PE-conjugated anti-mouse CCR7 (M1 marker, BioLegend) and APC-conjugated anti-mouse CD206 (M2 marker, BioLegend) at 37 °C for 1 h under dark conditions. Then, the cells were washed with PBS three times to remove excess antibodies, followed by resuspension with 500 μL of PBS. Finally, the fluorescence intensity of the samples was detected with a flow cytometer, and the effect of Ng(miR-21) on macrophage phenotypes was studied in detail in our other articles, but we verified it again in this paper.
2.5. Intracellular Reactive Oxygen Species Detection
Reactive oxygen species (ROS) could be produced in large quantities by M1 phenotype macrophages. 2′,7′-Dichlorodihydrofluorescein diacetate (DCFH-DA) could be hydrolyzed by esterase to produce fluorescence, which was used as a probe for detecting intracellular ROS. The RAW264.7 cells were treated with PBS or Ng(miR-155), and cultured with DCFH-DA (10 × 10–6 M) in PBS (37 °C, 30 min). Finally, fluorescence images were captured by confocal laser scanning microscopy.
2.6. Immunofluorescence Staining
The ratio of RAW264.7 cells positive for iNOS (M1 marker) and CD206 (M2 marker) was evaluated by immunofluorescence staining. After treatment with PBS, Ng(miR-155), or Ng(miR-21) for 24 h, the cells were fixed with 4% paraformaldehyde for 10 min, treated with 0.1% Triton X-100 for 5 min, and blocked in 1% BSA for 1 h. Subsequently, the cells were incubated with antibodies overnight at 4 °C, followed by washing three times. Then, the samples were incubated with secondary antibodies under dark conditions for 1 h at 37 °C, followed by treatment with DAPI for 10 min. The images were taken by confocal laser scanning microscopy. The semiquantitative analysis of fluorescence was evaluated by ImageJ.
2.7. Cytokine Secretion
The cell culture medium stimulated by the above PBS, Ng(miR-155), or Ng(miR-21) for 24 h was tested by an enzyme-linked immunosorbent assay (ELISA) kit for detection of the concentration of proinflammatory cytokine TNF-α and anti-inflammatory cytokine IL-10. The supernatants were centrifuged to remove cellular debris and measured according to the manufacturer’s instructions. The experiment was performed in triplicate.
For the sequential stimulation of macrophages, Ng(miR-155) was added to the culture medium. The culture medium was changed after 24 h followed by washing three times, and then Ng(miR-21) was added to stimulate it for 24 h. Subsequently, the secretion of cytokines was tested according to the above steps. Lipopolysaccharides (LPS) were used as a positive control group to stimulate the inflammatory conditions. RAW264.7 cells were pretreated with 100 ng/mL LPS for 24 h, and other steps were performed according to the above descriptions.
2.8. Total RNA isolation and qPCR analysis
To analyze the gene expression level of cytokines, a fluorescence quantitative polymerase chain reaction (qPCR) was used. The total RNA was isolated using a TRNzol Universal Reagent (Tiangen Biotech Co., Ltd.) according to the manufacturer’s instructions. cDNA was synthesized from 2000 ng of samples’ RNA using a FastQuant RT Kit (Tiangen Biotech Co., Ltd.). Then, a SuperReal PreMix Plus kit (Tiangen Biotech Co., Ltd.) was used for detection expression of mRNA by a Real-Time PCR Detection System (Bio-Rad). Each sample was repeated three times, and the comparative cycle threshold (CT) method (2–ΔΔCT) was used to calculate the fold changes of mRNAs (iNOS, iL-6, and iL-10). The cycling parameters were shown as follows: initial denaturation at 95 °C for 15 min, 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, and extension at 72 °C for 20 s. The sequences of primers were listed as follows: iNOS, forward CAGCTGGGCTGTACAAACCTT and reverse CATTGGAAGTGAAGCGTTTCG; iL-6, forward AGTTGCCTTCTTGGGACTGA and reverse TCCACGATTTCCCAGAGAAC; iL-10, forward ACTCTTCACCTGCTCCACTG and reverse GCTATGCTGCCTGCTCTTAC; GAPDH, forward ATCACTGCCACCCAGAAG and reverse TCCACGACGGACACATTG. GAPDH was used as a quantitative control for RNA levels.
2.9. Statistical Analysis
All data were shown as the mean ± standard deviation (SD), and the statistical analyses were carried out using Prism GraphPad software version 8. Values of p less than 0.05 were considered statistically significant.
3. Results and Discussion
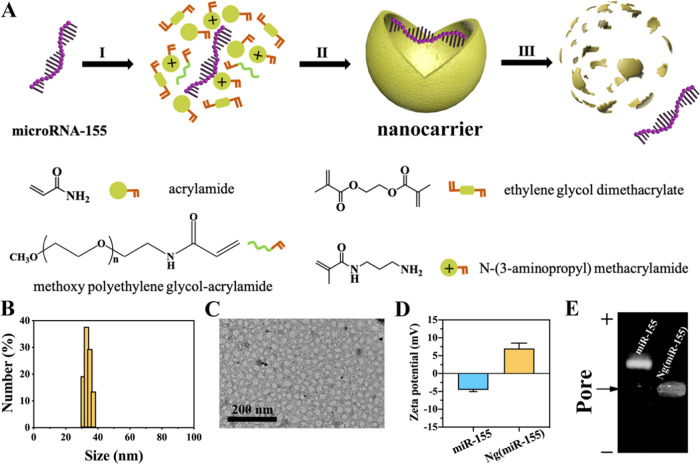
A nanocarrier for high-efficiency transfection of macrophages was designed for the delivery of microRNAs. The nanocarriers were synthesized by free radical polymerization, and the microRNAs were encapsulated within polymer shells (Figure 1A). An acid-degraded cross-linker, ethylene glycol dimethacrylate (EGDMA), was used to synthesize the microRNA-21 nanoparticles. The cross-linker molecule degraded within the acidic endosomes (pH = 5.4) followed by the release of microRNAs. Here, microRNA-155 was loaded in the nanocarrier for verifying the successful preparation of microRNA nanocarriers. The results of DLS showed that the particle size of Ng(miR-155) was evenly distributed at 30–40 nm (Figure 1B). The sphere-like structure of Ng(miR-155) could be more intuitively observed from the TEM images (Figure 1C). Figure 1D shows that microRNA-155 was negatively charged, and the ζ potential was about −4.57 mV. The Ng(miR-155) was positively charged due to the amino groups on the surface of nanocarriers, and the ζ potential was about 7.01 mV. Under the electric field, microRNA-155 migrated from the negative electrode to the positive electrode, and Ng(miR-155) remained in the pores (Figure 1E).
Figure 1.
Characterization of microRNA nanocarriers. (A) Schematic illustration of the synthesis of microRNA nanocarriers. (I) Enriching of the monomers and cross-linkers around microRNA molecules. (II) Formation of Ng(miR-155) by free radical polymerization. (III) Release of microRNA-155 from the nanocarrier upon degradation of the polymer shell. MicroRNA-155 was taken as an example. (B) Particle size distribution of nanocarriers by DLS. (C) TEM images of nanocarriers. (D) ζ potential of miRNA-155 and Ng(miR-155). (E) Agarose gel electrophoresis. Native miR-155 served as the control.
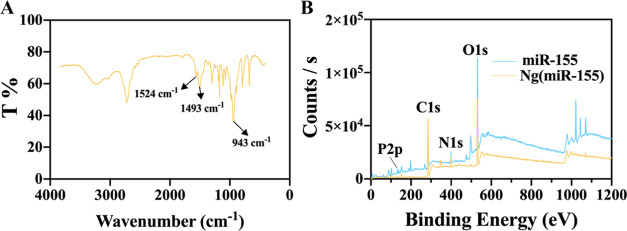
In addition, the characteristic groups of the polymer shell (1524 and 1493 cm–1, C=O; 943 cm–1, C–N) could also be seen from the FTIR spectrum, indicating that the microRNAs had indeed been successfully encapsulated in the nanocarriers (Figure 2A). XPS showed that the content of the P element decreased greatly in Ng(miR-155), which further indicated that the microRNAs were encapsulated inside the nanocarriers (Figure 2B). Therefore, the microRNA-loaded nanocarriers were successfully prepared with a weak positive charge and uniform small size.
Figure 2.
(A) FTIR spectrum of Ng(miR-155). (B) XPS images of miRNA-155 and Ng(miR-155).
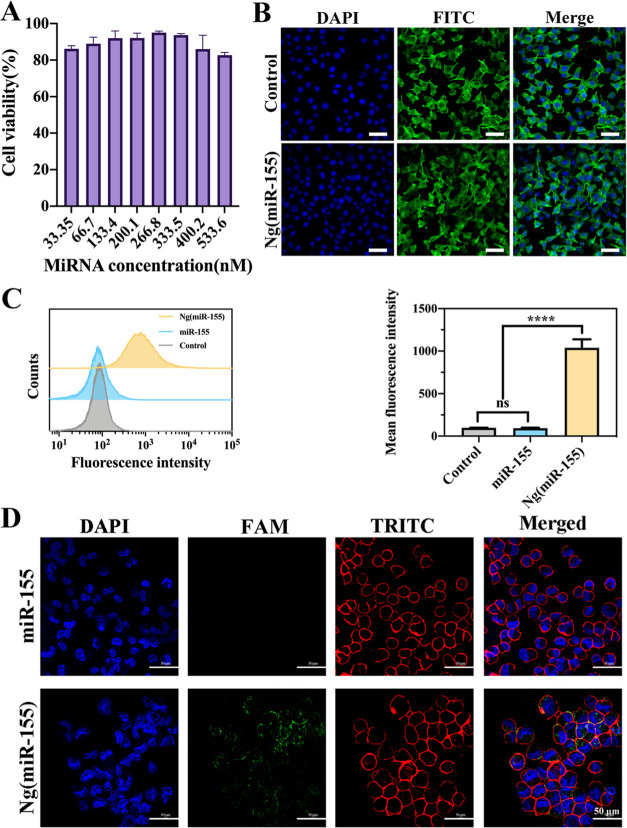
After that, the macrophage cell viability was verified by the CCK-8 kit. As shown in Figure 3A, after incubation with nanocarriers with all concentration gradients for 24 h, the cell viability of macrophages reached 85%. The effective concentration used in subsequent experiments in this study was 50 nM, and the cell viability was about 90%. Figure 3B shows that there was no difference in the NIH 3T3 cell morphology in control and Ng(miR-155) groups. The results indicated that Ng(miR-155) had perfect biocompatibility. After incubation with macrophages for 4 h, the uptake of Ng(miR-155) by macrophages was analyzed by flow cytometry (Figure 3C). The results showed that the curve of the Ng(miR-155) group significantly moved to the right compared with the free miRNA-155 group. The mean fluorescence intensity of cells treated with Ng(miR-155) was much higher than that of free miRNA-155. The results were further confirmed by confocal laser scanning microscopy images, which are shown in Figure 3D. There was almost no fluorescence in the free miRNA-155 group, indicating the poor uptake efficiency by macrophages. In contrast, Ng(miR-155) was concentrated around the cell nucleus and located in the cell cytoplasm. Therefore, the results intuitively showed the nanocarriers could be uptaken by macrophages with high endocytosis efficiency.
Figure 3.
(A) RAW264.7 cell viability in different concentration gradients of Ng(miR-155). (B) NIH 3T3 cell morphology after being treated by Ng(miR-155). Cells were counterstained with DAPI (nuclei) and FITC-labeled phalloidin (actin). (C) Flow cytometric analyses of RAW264.7 cells after incubation with free miR-155 or Ng(miR-155). Quantitative analyses of fluorescence were shown by mean fluorescence intensity. (D) Confocal laser scanning microscopy images of the RAW264.7 cells incubated with free miR-155 and Ng(miR-155). Cells were counterstained with DAPI (nuclei) and TRITC-labeled phalloidin (actin). Scale bars are 50 μm. ***P < 0.001, ns means no significant difference.
Although microRNA-based therapy has been applied to various diseases, there are few studies on the modulation of macrophage polarization by microRNAs, probably because the application of microRNAs is hindered by a lack of effective delivery systems for macrophages that are more difficult to transfect than general cell types.36 Compared with traditional transfection methods, such as electroporation, lipofectamine reagent, and plasmid,40,41 the nanocarriers reported in this paper have the advantages of low toxicity and high transfection efficiency. To the best of our knowledge, nanoparticles are seldom reported to deliver microRNAs to macrophages, and nanoparticles that can efficiently transfect other cells are not necessarily suitable for macrophages. We previously reported a nanoparticle that could be uptaken into mesenchymal stem cells,24 which was also based on free radical polymerization, and the physicochemical properties of nanoparticles such as particle size and electrical properties could be regulated by the ratio and type of monomers, which might be suitable for different kinds of cells. The transfection effect targeting macrophages was verified for the first time in this article. Therefore, the above results showed that the nanocarriers reported in this paper provided a feasible delivery system for gene therapy targeting macrophages.
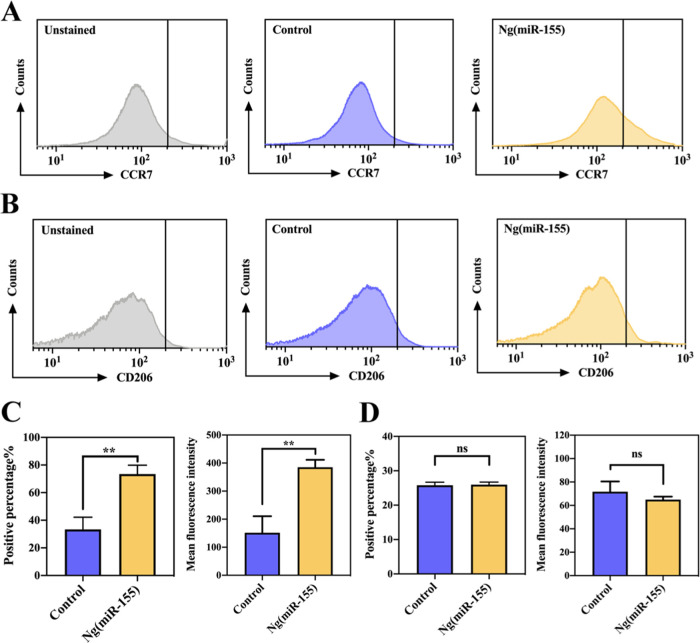
The results had shown that the designed nanocarriers could effectively deliver microRNAs into macrophages. Subsequently, we further studied the polarization of macrophages by Ng(miR-155). After incubation with Ng(miR-155) for 24 h, the expressions of CCR7 (M1 marker) and CD206 (M2 marker) of macrophages were detected by flow cytometry. Figure 4A shows the expression of the M1 marker, which indicated that after stimulation with Ng(miR-155), the curve significantly moved to the right (positive results). In addition, quantitative analysis showed that the positive percentage of the Ng(miR-155) group was about 31%, which was much higher than the control group (Figure 4C). To exclude the effect of the polymer shell on macrophage polarization, a disordered microRNA sequence (negative control for microRNA mimics) was used to synthesize NC nanocarriers. Figure S1 shows that the polymer shell did not affect the polarization of macrophages to M1 phenotypes. Therefore, Ng(miR-155) could promote the transformation of macrophages to M1 phenotypes owing to microRNA-155 molecules rather than the polymer shell. The trend of mean fluorescence intensity was consistent with the above results. Figure 4B,D shows the expression of the M2 marker, which indicated that irrespective of the qualitative curve or quantitative analysis, there was no significant difference between the control and Ng(miR-155) groups. The results of flow cytometry showed that microRNA-155 nanocarriers could promote the expression of the M1-related marker in macrophages and had no interference with the M2-related marker.
Figure 4.
Flow cytometric analyses of macrophage phenotypes treated with Ng(miR-155). (A) The M1 macrophage-related marker CCR7. (B) The M2 macrophage-related marker CD206. (C) Quantitative analyses of the fluorescence of A. (D) Quantitative analyses of the fluorescence of B. Scale bars are 50 μm. **P < 0.01; ns indicates that the groups are not significantly different from each other.
Macrophages are highly plastic with the stimulation of the microenvironment. The M1 phenotype macrophages present a round shape with a number of pseudopodia, while the M2 phenotypes are elongated spindle-shaped. The representative images of RAW264.7 cells are shown in Figure 5A. The macrophages were round with the treatment of PBS, while the macrophages treated with Ng(miR-155) were with a number of pseudopodia, which was consistent with the morphology of M1 macrophages. In addition, M1 phenotype macrophages could produce a high concentration of ROS, which acted to regulate biological processes. The level of ROS was evaluated via DCFH-DA, an ROS probe. The results showed that Ng(miR-155) exhibited a remarkable ROS improvement compared with the control group (Figure 5B). These results confirmed that microRNA-155 nanocarriers could promote macrophage polarization toward M1 phenotypes.
Figure 5.
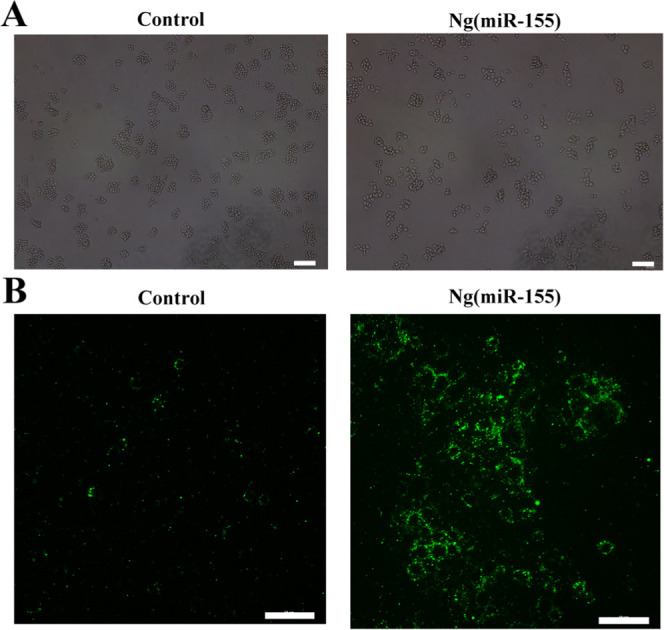
(A) Representative cell morphology of RAW264.7 cells. (B) Detection of ROS production. Scale bars are 50 μm.
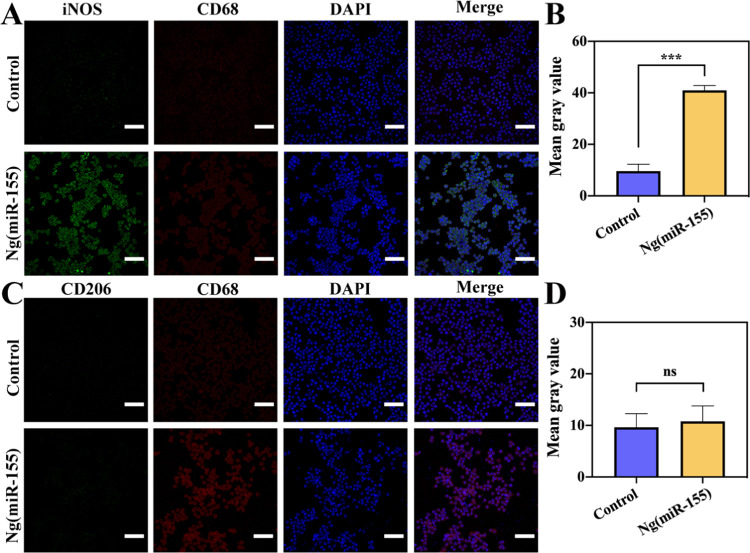
To further study the effect of microRNAs on phenotype transformation of macrophages, the immunofluorescence staining analysis of iNOS (M1 marker) and CD206 (M2 marker) was carried out. CD68 was used as a universal marker of macrophages. The images of macrophages treated by Ng(miR-155) showed that the fluorescence intensity of the iNOS marker was stronger than that of the control group (Figure 6A), and the expression of the CD206 marker did not show a significant difference (Figure 6C), which indicated that microRNA-155 nanocarriers could promote the transformation of macrophages to M1 phenotypes. This was also confirmed by the results of semiquantitative analyses (Figure 6B,D). The results also showed that after stimulation with Ng(miR-155), the ratio of M1/M2 was improved significantly, which could accelerate the process of inflammation. Many research studies had reported that M1 phenotype macrophages could recruit mesenchymal stem cells to the injury site and stimulate the initial formation of blood vessels by the secretion of TNF-α, IFN-γ, and IL-6,42−44 which showed that microRNA-155 nanocarriers could activate the follow-up process of bone repair.
Figure 6.
Immunofluorescence images of macrophages with the treatment of Ng(miR-155). (A) The expression of iNOS (M1 phenotype-related marker). (B) Quantitative analyses of the fluorescence of iNOS. (C) The expression of CD206 (M2 phenotype-related marker). (D) Quantitative analyses of the fluorescence of CD206. The nuclei were counterstained with DAPI. Scale bars are 100 μm. ***P < 0.001; ns indicates that the groups are not significantly different from each other.
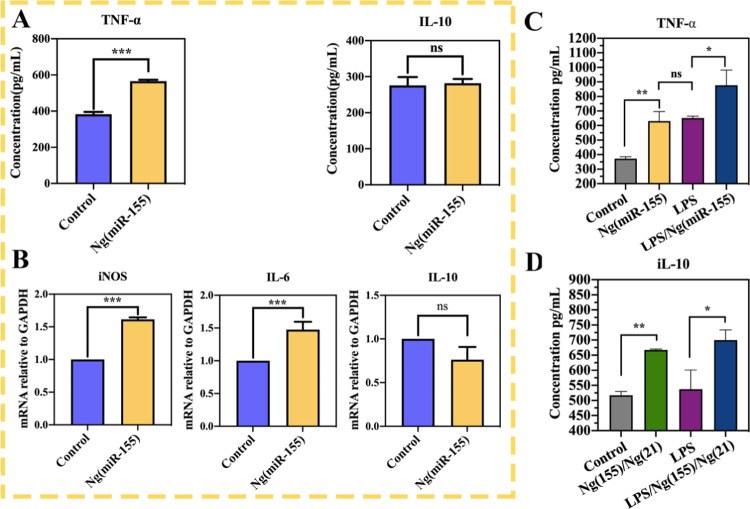
Macrophages regulate the process of inflammation by secreting cytokines, and macrophages with different phenotypes secrete different cytokines. Therefore, we can further prove the effect of microRNAs on macrophage polarization through the identification of cytokines. The proinflammatory cytokine TNF-α and anti-inflammatory cytokine IL-10 were detected in the supernatant of macrophages stimulated by Ng(miR-155) for 24 h (Figure 7A). The results showed that the level of TNF-α increased significantly in the Ng(miR-155) group, and the concentration increased from about 390 pg/mL to about 590 pg/mL. However, the concentration of anti-inflammatory cytokine IL-10 did not change significantly, which remained at about 260 pg/mL. Therefore, compared with the untreated control group, microRNA-155 nanocarriers significantly increased the secretion of proinflammatory cytokines, but had little effect on the expression of anti-inflammatory cytokines, which further showed that microRNA-155 nanocarriers could promote the macrophage polarization toward M1 phenotypes, not M2 phenotypes.
Figure 7.
(A) ELISA assay for TNF-α and iL-10 in the supernatant of RAW264.7 cells treated with Ng(miR-155). (B) Real-time PCR analysis of relative gene expression of the M1-related iNOS, IL-6, and M2-related IL-10. C and (D) Secretion of TNF-α and iL-10 after the sequential delivery of Ng(miR-155) and Ng(miR-21). *P < 0.05, **P < 0.01, and ***P < 0.001; ns indicates that the groups are not significantly different from each other.
The above data have proved that microRNA-155 nanocarriers could improve the secretion of proinflammatory cytokines. Then, we further detected the expression of genes related to macrophage phenotype by RT-qPCR. As shown in Figure 7B, Ng(miR-155) significantly upregulated the expression of proinflammatory M1 macrophage-related genes, such as iNOS and IL-6, but slightly decreased the mRNA expression of anti-inflammatory M2 macrophage-related genes, such as IL-10, with no significant difference compared with the control group. All of the above data showed that microRNA-155 nanocarriers could promote macrophage polarization to M1, which was consistent with ELISA analysis.
The above results showed that microRNA-155 nanocarriers upregulated the proinflammatory cytokines iNOS, TNF-α, and IL-6 by promoting the polarization of macrophages toward M1 phenotypes. Our another unpublished paper had demonstrated that microRNA-21 nanocarriers could regulate macrophage polarization to M2 phenotypes in vitro and in vivo. Here, considering the integrity of the paper, the phenotypic conversion effect of microRNA-21 nanocarriers on macrophages was reverified, and the relevant data were listed in the Supporting Information, which was consistent with our previous results (Figures S2–S4). It was proved again that microRNA-21 nanocarriers could promote macrophage transformation toward M2 phenotypes. Therefore, the sequential delivery of Ng(miR-155) and Ng(miR-21) could theoretically achieve M1 and M2 polarization of macrophage phenotypes in sequence.
To demonstrate that the phenotypes of macrophages can be switched sequentially, the secretion of TNF-α and iL-10 after the sequential delivery of Ng(miR-155) and Ng(miR-21) was detected by ELISA (Figure 7C,D). The results showed that under the stimulation of Ng(miR-155), the secretion of TNF-α increased significantly, with or without LPS, which proved the emergence of M1 phenotypes (Figure 7C). Subsequently, the intervention of Ng(miR-21) increased the secretion of IL-10, which demonstrated that macrophages were rapidly switched to M2 phenotypes. Therefore, the sequential delivery of microRNA-155 and microRNA-21 nanocarriers could regulate the M1-to-M2 macrophage phenotype switch.
There have been some reports on the systems for promoting sequential transformation of macrophage phenotypes. For example, Li et al. reported an IFNγ-loading calcium silicate/β-tricalcium phosphate scaffold for modulating M1-to-M2 macrophage transformation.45 However, IFNγ was sequentially released over 5 days, and the Si ions released from the scaffold reached 20 ppm in the first 3 days. Namely, this might lead to the persistence of M1 phenotype macrophages and the early emergence of M2 phenotype macrophages. Currently, it is recognized that the ideal strategy to promote bone healing is to repair it, complying with the natural healing process of bone tissue.46 If macrophages can be strictly regulated to express M1 phenotypes in the first 3 days of bone injury and then change to M2 phenotypes, it may be more conducive to bone tissue regeneration. Similar to the above reports, most of the reported release systems cannot accurately control the release of drugs,6,47 which cannot match the phenotypic conversion of macrophages during the natural healing of bone tissue. In comparison, the strategy of the sequential delivery of microRNA-155 and microRNA-21 nanocarriers can sequentially achieve M1 and M2 polarization of macrophage phenotypes at precise time points. Moreover, an enzyme-sensitive hydrogel system that achieved the accurate release of two cargoes was designed by us.48 By comparison, the method described in this paper can be completed only by injecting different nanoparticles at time points with a syringe, which greatly simplifies the operation steps. Therefore, the sequential delivery of two microRNA nanocarriers conforming to the time points of macrophage phenotype transformation during the physiological process provides an alternative strategy for bone repair.
4. Conclusions
Multiple gene therapy was first proposed to regulate the M1-to-M2 phenotype transition of macrophages according to the physiological process of bone tissue repair, and the sequential delivery of microRNA-155 and microRNA-21 nanocarriers could sequentially achieve M1 and M2 polarization of macrophage phenotypes. Our findings demonstrated that the designed nanocarriers could be effectively internalized by macrophages with about 80% of endocytosis efficiency within 4 h. MicroRNA-155 nanocarriers could instantaneously modulate macrophage polarization toward M1 phenotypes as well as improve M1 phenotype-related cytokines secretion. The positive percentage of M1-related markers in the microRNA-155 nanocarrier group reached about 31%. Subsequently, the intervention of microRNA-21 nanocarriers could rapidly reverse M1 phenotypes to M2 phenotypes at the appropriate time point. The therapy strategy of the sequential delivery of multiple gene targeting phenotype sequential transition of macrophages provides a new perspective for bone regeneration.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 51673144, 51871163, and 31600769).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c00297.
Flow cytometric analyses of macrophage phenotypes treated with NC nanocarriers miR-21 nanocarriers (Figures S1 and S2); immunofluorescence images of macrophages with the treatment of miR-21 nanocarriers (Figure S3); and ELISA assay and real-time PCR analysis for testing transition of macrophage phenotypes under the stimulation of miR-21 nanocarriers (Figure S4) (PDF)
Author Contributions
X.L.: conceptualization, investigation, methodology, formal analysis, data curation, and writing—original draft. S.X.: investigation, formal analysis, and data curation. Q.Z.: methodology, formal analysis, and data curation. X.S.: methodology, resources, and funding acquisition. N.C.: methodology and investigation. S.L.: writing—review and editing. J.Z.: conceptualization, validation, supervision, funding acquisition, project administration, and writing—review and editing. X.H.: project administration and writing—review and editing. X.Y.: conceptualization, funding acquisition, project administration, and writing—review and editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Claes L.; Recknagel S.; Ignatius A. Fracture healing under healthy and inflammatory conditions. Nat. Rev. Rheumatol. 2012, 8, 133–143. 10.1038/nrrheum.2012.1. [DOI] [PubMed] [Google Scholar]
- Bahney C. S.; Zondervan R. L.; Allison P.; Theologis A.; Ashley J. W.; Ahn J.; Miclau T.; Marcucio R. S.; Hankenson K. D. Cellular biology of fracture healing. J. Orthop. Res. 2019, 37, 35–50. 10.1002/jor.24170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss K. M.; Reddy G. B.; Hyzy S. L.; Schwartz Z.; Boyan B. D.; Olivares-Navarrete R. Titanium surface characteristics including topography and wettability alter macrophage activation. Acta Biomater. 2016, 31, 425–434. 10.1016/j.actbio.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggatt L. J.; Wullschleger M. E.; Alexander K. A.; Wu A. C. K.; Millard S. M.; Kaur S.; Maugham M. L.; Gregory L. S.; Steck R.; Pettit A. R. Fracture healing via periosteal callus formation requires macrophages for both initiation and progression of early endochondral ossification. Am. J. Pathol. 2014, 184, 3192–3204. 10.1016/j.ajpath.2014.08.017. [DOI] [PubMed] [Google Scholar]
- Gordon S.; Pluddemann A.; Martinez E. F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol. Rev. 2014, 262, 36–55. 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller K. L.; Nassiri S.; Witherel C. E.; Anfang R. R.; Ng J.; Nakazawa K. R.; Yu T.; Vunjak-Novakovic G. Sequential delivery of immunomodulatory cytokines to facilitate the M1-to-M2 transition of macrophages and enhance vascularization of bone scaffolds. Biomaterials 2015, 37, 194–207. 10.1016/j.biomaterials.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlundt C.; Khassawna T. El.; Serra A.; Dienelt A.; Wendler S.; Schell H.; Rooijen N.; Radbruch A.; Lucius R.; Hartmann S.; Duda G. N.; Schmidt-Bleek K. Macrophages in bone fracture healing: their essential role in endochondral ossification. Bone 2018, 106, 78–89. 10.1016/j.bone.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Yin X.; Li Y.; Yang C.; Weng J.; Wang J.; Zhou J.; Feng B. Alginate/chitosan multilayer films coated on IL-4-loaded TiO2 nanotubes for modulation of macrophage phenotype. Int. J. Biol. Macromol. 2019, 133, 503–513. 10.1016/j.ijbiomac.2019.04.028. [DOI] [PubMed] [Google Scholar]
- Lu L. Y.; Loi F.; Nathan K.; Lin T. H.; Pajarinen J.; Gibon E.; Nabeshima A.; Cordova L.; Jamsen E.; Yao Z.; Goodman S. B. Pro-inflammatory M1 macrophages promote osteogenesis by mesenchymal stem cells via the COX-2-prostaglandin E2 pathway. J. Orthop. Res. 2017, 35, 2378–2385. 10.1002/jor.23553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai-Aql Z. S.; Alagl A. S.; Graves D. T.; Gerstenfeld L. C.; Einhorn T. A. Molecular mechanisms controlling bone formation during fracture healing and distraction osteogenesis. J. Dent. Res. 2008, 87, 107–118. 10.1177/154405910808700215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser D. M.; Edwards J. P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller K. L.; Koh T. J. Macrophage-based therapeutic strategies in regenerative medicine. Adv. Drug Delivery Rev. 2017, 122, 74–83. 10.1016/j.addr.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasnik S.; Rundle C. H.; Baylink D. J.; Yazdi M. S.; Carreon E.; Xu Y.; Qin X. Z.; Lau K. H. W.; Tang X. L. 25-Dihydroxyvitamin D suppresses M1 macrophages and promotes M2 differentiation at bone injury sites. JCI Insight 2018, 3, 98773 10.1172/jci.insight.98773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L.; Zhao Y.; Zhang Y.; Ruan Z. The macrophage polarization regulates MSC osteoblast differentiation in vitro. Ann. Clin. Lab. Sci. 2016, 46, 65–71. [PubMed] [Google Scholar]
- Schmidt-Bleek K.; Schell H.; Schulz N.; Hoff P.; Perka C.; Buttgereit F.; Volk H. D.; Lienau J.; Duda G. N. Inflammatory phase of bone healing initiates the regenerative healing cascade. Cell Tissue Res. 2012, 347, 567–573. 10.1007/s00441-011-1205-7. [DOI] [PubMed] [Google Scholar]
- O’Brien E. M.; Risser G. E.; Spiller K. L. Sequential drug delivery to modulate macrophage behavior and enhance implant integration. Adv. Drug Delivery Rev. 2019, 149-150, 85–94. 10.1016/j.addr.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao W.; Xie H. Z.; Fang J. H.; Shen J.; Li W. T.; Shen D. N.; Wu J.; Wu S. L.; Liu X. Y.; Zheng Y. F.; Cheung K. M. C.; Yeung K. W. K. Sequential activation of heterogeneous macrophage phenotypes is essential for biomaterials-induced bone regeneration. Biomaterials 2021, 276, 121038 10.1016/j.biomaterials.2021.121038. [DOI] [PubMed] [Google Scholar]
- Alhamdi J. R.; Peng T.; Al-Naggar I. M.; Hawley K. L.; Spiller K. L.; Kuhn L. T. Controlled M1-to-M2 transition of aged macrophages by calcium phosphate coatings. Biomaterials 2019, 196, 90–99. 10.1016/j.biomaterials.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y.; Wang Z.; Shi Y.; Dong L.; Wang C. Modulating macrophage activities to promote endogenous bone regeneration: Biological mechanisms and engineering approaches. Bioact. Mater. 2021, 6, 244–261. 10.1016/j.bioactmat.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian V.; Onaca O.; Enea R.; Hughes D. W.; Palivan C. G. Protein delivery: from conventional drug delivery carriers to polymeric nanoreactors. Expert Opin. Drug Delivery 2010, 7, 63–78. 10.1517/17425240903394520. [DOI] [PubMed] [Google Scholar]
- Filipowicz W.; Bhattacharyya S. N.; Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?. Nat. Rev. Genet. 2008, 9, 102–114. 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Friedman R. C.; Farh K. K. H.; Burge C. B.; Bartel D. P. Most mammalian miRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H.; Shan P.; Wang Y.; Li P.; Wang K.; Yang L. Nanomedicines for the efficient treatment of intracellular bacteria: the “ART” principle. Front. Chem. 2021, 9, 924 10.3389/fchem.2021.775682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. P.; Li S. D.; Qi H. Z.; Han D. L.; Chen N.; Zhan Q.; Li Z. Y.; Zhao J.; Hou X.; Yuan X. B.; Yang X. J. Early healing of alveolar bone promoted by microRNA-21-loaded nanoparticles combined with Bio-Oss particles. Chem. Eng. J. 2020, 401, 126026 10.1016/j.cej.2020.126026. [DOI] [Google Scholar]
- Saleh B.; Dhaliwal H. K.; Portillo-Lara R.; Shirzaei S. E.; Abdi R.; Amiji M. M.; Annabi N. Local immunomodulation using an adhesive hydrogel loaded with miRNA-laden nanoparticles promotes wound healing. Small 2019, 15, 1902232 10.1002/smll.201902232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Zhang M.; Zhong M.; Suo Q.; Lv K. Expression profiles of miRNAs in polarized macrophages. International Journal of Molecular Medicine. Int. J. Mol. Med. 2013, 31, 797–802. 10.3892/ijmm.2013.1260. [DOI] [PubMed] [Google Scholar]
- Sen C. K.; Roy S. MicroRNA 21 in tissue injury and inflammation. Cardiovasc. Res. 2012, 96, 230–233. 10.1093/cvr/cvs222. [DOI] [Google Scholar]
- Yu J. H.; Long L.; Luo Z. X.; Li L. M.; You J. R. Anti-inflammatory role of microRNA let-7c in LPS treated alveolar macrophages by targeting STAT3. Asian Pac. J. Trop. Med. 2016, 9, 72–75. 10.1016/j.apjtm.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Lin J.; Mohamed I.; Lin P. H.; Shirahama H.; Milbreta U.; Sieow J. L.; Peng Y.; Bugiani M.; Wong S. C.; Levinson H.; Chew S. Y. Modulating macrophage phenotype by sustained microRNA delivery improves host-implant integration. Adv. Healthcare Mater. 2020, 9, 1901257 10.1002/adhm.201901257. [DOI] [PubMed] [Google Scholar]
- Cai X.; Yin Y.; Li N.; Zhu D.; Zhang J.; Zhang C. Y.; Zen K. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J. Mol. Cell Biol. 2012, 4, 341–343. 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- Jablonski K. A.; Gaudet A. D.; Amici S. A.; Popovich P. G.; Guerau-de-Arellano M. Control of the inflammatory macrophage transcriptional signature by miRNA-155. PLoS One 2016, 11, e0159724 10.1371/journal.pone.0159724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P.; Wang H.; Luo X.; Liu H.; Lu B.; Li T.; Yang S.; Gu Q.; Li B.; Wang F.; Sun X. MicroRNA-155 inhibits polarization of macrophages to M2-Type and suppresses choroidal neovascularization. Inflammation 2018, 41, 143–153. 10.1007/s10753-017-0672-8. [DOI] [PubMed] [Google Scholar]
- Sheedy F. J. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 2015, 6, 19 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoon L.; Millard S. M.; Raggatt L. J.; Pettit A. R. Osteomacs and bone regeneration. Curr. Osteoporos Rep. 2017, 15, 385–395. 10.1007/s11914-017-0384-x. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Li Y.; Chen Y. E.; Chen J.; Ma P. X. Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects. Nat Commun. 2016, 7, 10376 10.1038/ncomms10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. A.; Wyatt H.; Susser L.; Geoffrion M.; Rasheed A.; Duchez A. C.; Cottee M. L.; Afolayan E.; Farah E.; Kahiel Z.; Cote M.; Gadde S.; Rayner K. J. Delivery of microRNAs by chitosan nanoparticles to functionally alter macrophage cholesterol efflux in vitro and in vivo. ACS Nano 2019, 13, 6491–6505. 10.1021/acsnano.8b09679. [DOI] [PubMed] [Google Scholar]
- Condorelli G.; Latronico M. V.; Cavarretta E. MicroRNAs in cardiovascular diseases: current knowledge and the road ahead. J. Am. Coll. Cardiol. 2014, 63, 2177–2187. 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- Yang L. J.; Han D. L.; Zhan Q.; Li X. P.; Shan P. P.; Hu Y. J.; Ding H.; Wang Y.; Zhang L.; Zhang Y.; Xue S.; Zhao J.; Hou X.; Wang Y.; Li P. F.; Yuan X. B.; Qi H. Z. Blood TfR plus exosomes separated by a pH-responsive method deliver chemotherapeutics for tumor therapy. Theranostics 2019, 9, 7680–7696. 10.7150/thno.37220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long L. X.; Cheng L. J.; Hou J. J.; Wang L. M.; Wang X.; He L. G.; Li S. D.; Zhao J.; Hou X.; Kang C. S.; Yuan X. B. The effect of umbrella-type branching on the blood circulation and tumor targeting of star-branched PLA-PMPC copolymer micelles. Sci. China: Technol. Sci. 2021, 64, 71–82. 10.1007/s11431-020-1645-6. [DOI] [Google Scholar]
- Zhou W.; Su L.; Duan X. Y.; Chen X.; Hays A.; Upashyayula S.; Shivde J.; Wang H. Z.; Li Y.; Huang D. M.; Liang S. MicroRNA-21 down-regulates inflammation and inhibits periodontitis. Mol. Immunol. 2018, 101, 608–614. 10.1016/j.molimm.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhyastha R.; Madhyastha H.; Nurrahmah Q. I.; Purbasari B.; Maruyama M.; Nakajims Y. MicroRNA 21 elicits a pro-inflammatory response in macrophages, with exosomes functioning as delivery vehicles. Inflammation 2021, 44, 1274–1287. 10.1007/s10753-021-01415-0. [DOI] [PubMed] [Google Scholar]
- Schlundt C.; Fischer H.; Bucher C. H.; Rendenbach C.; Duda G. N.; Schmidt-Bleek K. The multifaceted roles of macrophages in bone regeneration: a story of polarization, activation and time. Acta Biomater. 2021, 133, 46–57. 10.1016/j.actbio.2021.04.052. [DOI] [PubMed] [Google Scholar]
- Stefanowski J.; Lang A.; Rauch A.; Aulich L.; Kohler M.; Fiedler A. F.; Buttgereit F.; Schmidt-Bleek K.; Duda G. N.; Gaber T.; Niesner R. A.; Hauser A. E. Spatial distribution of macrophages during callus formation and maturation reveals close crosstalk between macrophages and newly forming vessels. Front. Immunol. 2019, 10, 2588 10.3389/fimmu.2019.02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller K. L.; Anfang R. R.; Spiller K. J.; Ng J.; Nakazawa K. R.; Daulton J. W.; Vunjak-Novakovic G. The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials 2014, 35, 4477–4488. 10.1016/j.biomaterials.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Peng M. Z.; Yang Z. Z.; Zhou X. J.; Deng Y.; Jiang C.; Xiao M.; Wang J. W. 3D-printed IFN-γ-loading calcium silicate-β-tricalcium phosphate scaffold sequentially activates M1 and M2 polarization of macrophages to promote vascularization of tissue engineering bone. Acta Biomater. 2018, 71, 96–107. 10.1016/j.actbio.2018.03.012. [DOI] [PubMed] [Google Scholar]
- Chen Z. T.; Klein T.; Murray R. Z.; Crawford R.; Chang J.; Wu C. T.; Xiao Y. Osteoimmunomodulation for the development of advanced bone biomaterials. Mater. Today 2016, 19, 304–321. 10.1016/j.mattod.2015.11.004. [DOI] [Google Scholar]
- Gao L. L.; Li M. T.; Yin L.; Zhao C. J.; Chen J. H.; Zhou J.; Duan K.; Feng B. Dual-inflammatory cytokines on TiO2 nanotube-coated surfaces used for regulating macrophage polarization in bone implants. J. Biomed. Mater. Res. 2018, 106, 1878–1886. 10.1002/jbm.a.36391. [DOI] [PubMed] [Google Scholar]
- Xue S. L.; Li X. P.; Li S. D.; Chen N.; Zhan Q.; Long L. X.; Zhao J.; Hou X.; Yuan X. B. Bone fracture microenvironment responsive hydrogel for timing sequential release of cargoes. Colloids Surf., A 2021, 629, 127413 10.1016/j.colsurfa.2021.127413. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.