Abstract
Because of the evolutionary variants of SARS-CoV-2, development of broad-spectrum neutralizing antibodies resilient to virus escape is urgently needed. We identified a group of high-affinity nanobodies from camels immunized with receptor-binding domain (RBD) of SARS-CoV-2 spike protein and resolved the structures of two non-competing nanobodies (NB1A7 and NB1B11) in complex with RBD using X-ray crystallography. The structures show that NB1A7 targets the highly conserved cryptic epitope shared by SARS-CoV-2 variants and some other coronaviruses and blocks ACE2 receptor attachment of the spike protein, and NB1B11 epitope overlaps with the contacting surface of ACE2 and is different from the binding site of NB1A7. These two nanobodies were covalently linked into multivalent and bi-paratopic formats, which significantly improved the avidity and neutralization potency and may further inhibit viral escape. The results contribute to the structure-guided design of antibodies against future variants of SARS-CoV-2 virus to combat coronavirus epidemics and pandemics.
Keywords: SARS-CoV-2 virus, receptor-binding domain, nanobody, crystal structure, bi-paratopic nanobodies
Graphical abstract
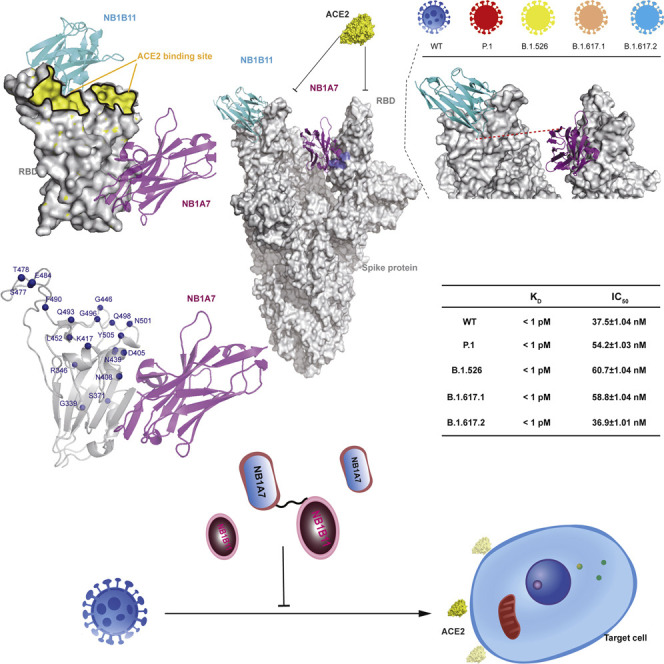
Shi et al. identify two neutralizing nanobodies against SARS-CoV-2. Crystal structures reveal how they prevent the virus and its variants from entering cells. The combination of two nanobodies significantly improves neutralization potency, which is a potential therapeutic candidate to reduce the probability of the emergence of escape variants of SARS-CoV-2.
Introduction
The COVID-19 pandemic was caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Currently, many COVID-19 vaccines have been proved to be effective, which brings hope for the gradual alleviation and containment of the pandemic (Brouwer et al., 2020; Cao et al., 2020; Hansen et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Wec et al., 2020; Wrapp et al., 2020). However, the scale of the pandemic and fast evolving antigenic variants leads to vaccine-escape mutants (Wang et al., 2021b, 2021d; Wu et al., 2021). Therefore, there is an urgent need to develop broad-spectrum neutralizing antibodies against emerging variants.
Camelid VHH single-domain antibody, also known as a nanobody, consists of the variable domain of heavy chain-only antibody without its light-chain counterpart, which is still able to selectively bind to a specific antigen (Muyldermans, 2013). As single chains, these nanobodies are easily bioengineered into bi-paratopic and multivalent molecules to enhance binding affinity and neutralization potency against the virus and suppress viral mutational escape (Bracken et al., 2021; Koenig et al., 2021; Schoof et al., 2020; Xiang et al., 2020b).
The spike (S) protein of SARS-CoV-2 guides the entry of the virion into the host cell (Ke et al., 2020; Turoňová et al., 2020). It is a homotrimeric glycoprotein that consists of S1 and S2 subunits. The receptor-binding domain (RBD) of S1 located at the top of S protein binds angiotensin-converting enzyme 2 (ACE2) as a canonical receptor on the host cell surface, whereas S2 subunit is responsible for the fusion of the viral and host cell membranes. The RBD in the metastable prefusion state undergoes hinge-like conformational movements between the inaccessible down conformation and the accessible up conformation. In the up position, ACE2 binding rearranges the conformation of the prefusion state of S protein, exposing the proteolytic site to the host proteases, shedding of the S1 subunit, and changing the S2 conformation to fuse with the host cell membrane (Cai et al., 2020; Wang et al., 2020; Yan et al., 2020). Thus, antibody interference with SARS-CoV-2 S protein function is an attractive target for the prevention and treatment of SARS-CoV-2 infection. In the past year, many neutralizing monoclonal antibodies from convalescent plasma have been developed for therapy (Chen et al., 2021; Cohen, 2021; Weinreich et al., 2021), but emerging escape mutants have eluded these neutralizing antibodies.
Since the start of the COVID-19 pandemic, the structural biology community has tried to understand the mechanism of SARS-CoV-2 host cell entry and antibody neutralization. A large number of structures of spike protein in complex with antibodies have been resolved, showing the diversity of neutralizing epitopes and mechanisms. Most antibodies or nanobodies bind close to the ACE2-interacting surface on the RBD to prevent ACE2 engagement, but some antibodies neutralize SARS-CoV-2 by destroying the spike protein function, by stabilizing the inactive spike form or premature induction of the post-fusion conformation (Chi et al., 2020; Du et al., 2020; Lv et al., 2020; Sun et al., 2020, 2021; Wang et al., 2021a; Yao et al., 2021; Koenig et al., 2021).
Here, we identified dozens of RBD-specific nanobodies from a phage display library generated from a camel immunized with recombinant RBD. Two non-competing nanobodies (NB1B11 and NB1A7) with high affinity were selected, and the crystal structure in complex with the RBD was resolved. The results revealed two distinct neutralizing epitopes and provided insights into their antiviral mechanisms. Interestingly, NB1A7 binds to a highly conserved cryptic epitope of RBD that could block ACE2 interaction with most RBD mutants of the emerging viral lineages. Moreover, NB1A7 binding epitope is shared with SARS-CoV-2 and SARS-CoV-1. Guided by the structure of nanobody and RBD complex, we designed different multivalent nanobodies to significantly increase the binding affinity and greatly improve the neutralization potency to SARS-CoV-2 virus. Furthermore, targeting different epitopes could suppress the emergence of escape mutants.
Results
Identification of camelid nanobodies blocking RBD interaction with ACE2
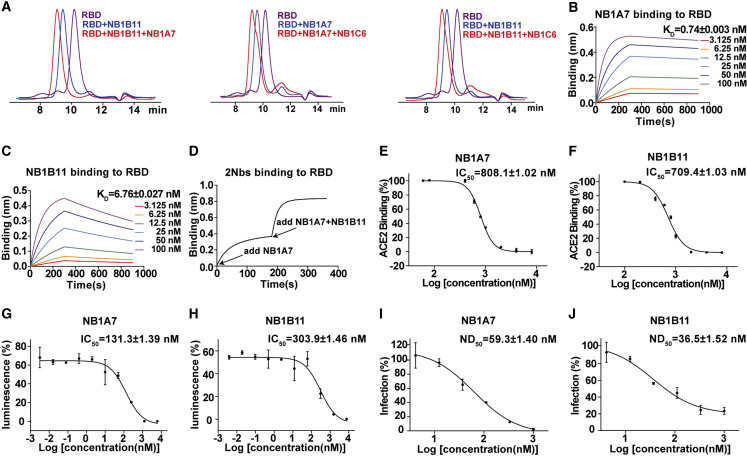
We immunized a camel with the recombinant RBD and generated nanobody phage display library. Subsequently, we performed two rounds of bio-panning on RBD and used ELISA to verify specific binding to RBD and identified 29 high-affinity nanobodies (Figure S1A). The panel of nanobodies was stratified into three groups on the basis of noncompetitive binding epitopes using size exclusion chromatography (SEC). According to SEC profile, NB1B11 and NB1A7, NB1B11 and NB1C6, and NB1A7 and NB1C6 can simultaneously bind to RBD, indicative of three non-overlapping epitopes (Figures 1A; Table 1 ). NB1B11 and its competing nanobodies were termed group I and NB1A7 and its overlapping nanobodies group II, while the binding epitope of NB1C6 was not shared with any other identified nanobodies but was termed group III. The binding kinetics of these nanobodies were measured using biolayer interferometry (BLI) on a ForteBio Octet Red96e system, all of them showing high affinities to RBD with K D values of 0.74 ± 0.003 nM, 6.76 ± 0.027 nM, and 5.07 ± 0.05 nM for NB1A7, NB1B11, and NB1C6 (Figures 1B, 1C, and S1B). Furthermore, epitope binding with BLI confirmed that NB1B11, NB1A7, and NB1C6 had non-competing epitopes on the RBD (Figures 1D and S1C). We then assessed whether these three nanobodies could block RBD interaction with ACE2 using competitive ELISA. NB1A7 and NB1B11 significantly inhibited ACE2 attachment to RBD with half-maximal inhibitory concentration (IC50) values of 808.1 ± 1.02 nM and 709.4 ± 1.03 nM, respectively (Figures 1E and 1F), while NB1C6 did not. Subsequently, we used a SARS-CoV-2-luciferase pseudovirus neutralization assay to test and characterize the antiviral activities of NB1B11 and NB1A7. NB1A7 and NB1B11 had inhibitory activity, with IC50 values of 131.3 ± 1.39 nM and 303.9 ± 1.46 nM, respectively (Figures 1G and 1H). Finally, we evaluated the neutralizing activity of these two nanobodies using a plaque reduction neutralization test (PRNT) using live SARS-CoV-2 virus infection of Vero E6 cells. NB1A7 and NB1B11 neutralized SARS-CoV-2 virus with 50% neutralizing doses (ND50) of 59.3 ± 1.40 nM and 36.5 ± 1.52 nM (Figures 1I, 1J, and S2). Taken together, NB1A7 and NB1B11 bound different antigenic sites, significantly inhibited the binding of ACE2 to RBD, and potently neutralized SARS-CoV-2 virus.
Figure 1.
Camelid nanobodies against two different epitopes on SARS-CoV-2 spike RBD neutralize virus infection
(A) Analysis of nanobody binding epitopes was performed using size exclusion chromatography (SEC).
(B and C) The binding affinity of NB1A7 or NB1B11 and RBD was measured using BLI. The process of association and dissociation was fitted by a 1:1 binding data fit model.
(D) The non-competing epitopes of NB1B11 and NB1A7 were confirmed using BLI.
(E and F) Competitive ELISA was used to assess whether NB1A7 or NB1B11 block RBD interaction with ACE2 in vitro. NB1A7 and NB1B11 showed IC50 values of 808.1 ± 1.02 nM and 709.4 ± 1.03 nM, respectively. The results are shown as mean ± SEM (n = 3).
(G and H) SARS-CoV-2-luciferase pseudovirus neutralization assay to test antiviral activities of NB1A7 (IC50 = 131.3 ± 1.39 nM) and NB1B11 (IC50 = 303.9 ± 1.46 nM). The results are shown as mean ± SEM (n = 2).
(I and J) SARS-CoV-2 plaque reduction neutralization test (PRNT) was performed to detect neutralizing activity of NB1A7 (ND50 = 59.3 ± 1.40 nM) and NB1B11 (ND50 = 36.5 ± 1.52 nM). The results are shown as mean ± SEM (n = 2).
See also Figures S1 and S2 and Table 1.
Table 1.
The amino acid sequences of NB1A7, NB1B11, and NB1C6
| Nanobody | Sequence |
|---|---|
| NB1A7 | QVQLQESGGGSVQAGGSLRLSCAASGYT FSSYCLGWFRQAPGKE |
| REGVAAIDSDGSTSYADSVKGRFTISRDN AKNTLYLQMNSLKPEDT | |
| AMYYCAAEGGPSLSYCTGGYGFLLSGL MYNSWGQGTQVTVSS | |
| NB1B11 | QVQLQESGGGSVQAGGSLRLSCAASGY TVSVGCMAWFRQAPGKE |
| REGVAGIDASGITKYSDSVKGRFTISKDN AKNALDLQMNGLKPEDT | |
| AMYHCAA GLVRGSCTDVLDHPSYLGVWGQGTQV TVSS | |
| NB1C6 | QVQLQESGGGSVQAGGRLRLSCAASG DTYSSYCMGWFRQAPGKE |
| REGVAAIYIGGDNTYYADSAKGRFTISQD YDKNTAYLQMNSLKSEDT | |
| AMYYCAAELFCPWPDIGTMSPAEYKY WGQGTQVTVSS |
Structure determination of the RBD-NB1A7
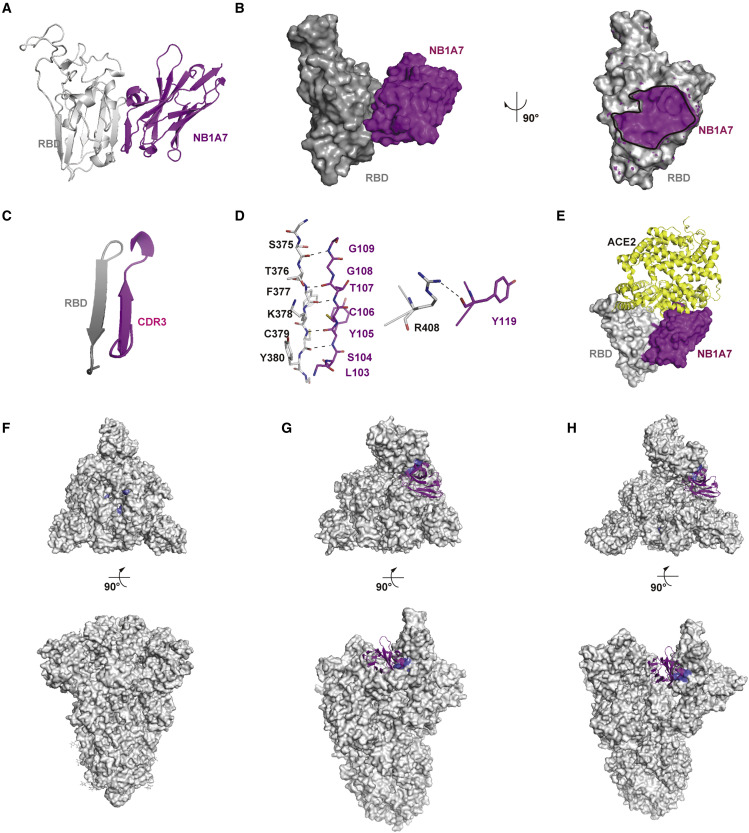
We solved the crystal structure of NB1A7 with the RBD complex at 2.00 Å resolution with R work of 18.1% and R free of 22.3% (Figure 2A; Table 2 ). NB1A7 binds to one side of RBD core, which consists of a twisted five-stranded antiparallel β sheet. The binding epitope is almost perpendicular to the receptor-binding motif (RBM) and does not overlap with receptor-binding region (Figure 2A). NB1A7 buries 1,665 Å2 solvent-accessible surface area on RBD (Figure 2B) and uses the CDR3 loop to interact with the β2 strand of RBD core, resulting in the residues S104-G109 from CDR3 of NB1A7, which form one β strand adhering to the antiparallel β sheet of RBD core. Such engagement interferes with the local conformation of RBD (Figures 2A and 2C), which extend the length of β2 strand of RBD from the segment T376-Y380 to the residues S375-G381 and make a minor conformational change of α2 helix and α3 helix of RBD. The main chain of residues S104-G109 from CDR3 of NB1A7 and main chain of the residues S375-G381 interact through the opposite amino-to-carboxyl hydrogen bonding patterns, which is the characteristic bond of the adjacent β strands in antiparallel β sheet structure (Figures 2C and 2D). Additionally, the electrostatic interaction from the main chain of Y119 of NB1A7 to the side chain of R408 from RBD is observed (Figure 2D).
Figure 2.
Crystal structure analysis of RBD-NB1A7 complex
(A) Cartoon presentation of NB1A7 (in purple) in complex with RBD (in gray).
(B) Surface presentation of NB1A7 (in purple) in complex with RBD (in gray).
(C and D) Detailed interaction between NB1A7 and RBD.
(E) Steric clash of NB1A7 and ACE2 (in yellow) on RBD on the basis of PDB: 6M0J.
(F and G) The alignments of RBD-NB1A7 complex and different conformations of trimeric SARS-CoV-2 spike protein (in gray). NB1A7 (in purple) cannot bind to the RBD in all down conformations (PDB: 6XLU) and one up conformation (PDB: 6XM4), because of steric hindrance.
(H) NB1A7 binds to spike conformation with 2-up RBDs (PDB:7A93).
See also Table 2.
Table 2.
X-ray data collection and refinement statistics
| RBD-NB1B11 | RBD-NB1A7 | |
|---|---|---|
| Wavelength | 0.9785 | 0.9791 |
| Resolution range | 49.37–2.08 (2.13–2.08) | 19.72–2.00 (2.07–2.00) |
| Space group | P 21 2 21 | C 2 2 2 |
| Cell parameter | ||
| a, b, c (Å) | 62.5, 88.8, 190.1 | 110.6, 183.8, 42.2 |
| α, β, γ(°) | 90, 90, 90 | 90, 90, 90 |
| Total reflections | 640,941 (44,904) | 381,050 (30,986) |
| Unique reflections | 64,558 (4,474) | 30,004 (2,951) |
| Multiplicity | 9.9 (10.0) | 12.7 (10.5) |
| Completeness (%) | 99.96 (100.00) | 100.00 (100.00) |
| Mean I/sigma (I) | 15.0 (1.5) | 11.21 (2.14) |
| Wilson B-factor (Å2) | 37.17 | 25.04 |
| Rmerge | 0.118 (1.763) | 0.220 (1.103) |
| Rmeas | 0.125 (1.855) | 0.229 (1.158) |
| Rpim | 0.039 (0.571) | 0.064 (0.347) |
| CC1/2 | 0.999 (0.587) | 0.960 (0.838) |
| Rwork | 0.197 | 0.181 |
| Rfree | 0.226 | 0.223 |
| Number of non-hydrogen atoms | 4,996 | 2,720 |
| Macromolecules | 4,661 | 2,443 |
| Ligands | 5 | 28 |
| Solvent | 330 | 249 |
| Protein residues | 614 | 320 |
| RMS (bonds) (Å) | 0.002 | 0.015 |
| RMS (angles) (°) | 0.60 | 1.33 |
| Ramachandran favored (%) | 97.01 | 96.82 |
| Ramachandran allowed (%) | 2.66 | 3.18 |
| Ramachandran outliers (%) | 0.33 | 0.00 |
| Rotamer outliers (%) | 0.00 | 0.00 |
| Average B-factor (Å2) | 42.49 | 33.18 |
| Macromolecules | 42.24 | 31.83 |
| Solvent | 45.84 | 39.46 |
Double mechanism of NB1A7 neutralizing SARS-CoV-2
Superposition of the RBD-NB1A7 complex with the RBD-ACE2 demonstrate that the NB1A7 binding epitope is distant from RBM, and it attaches to RBD without disturbing the conformation of the region recognized by the ACE2, while the CDR opposite apex of NB1A7 sterically clashes with zinc-containing subdomain and C terminus-containing subdomain of ACE2 (Figure 2E). Thus, the interaction between ACE2 and RBD can be effectively blocked.
We also analyzed the binding epitope within the S protein. The recently reported cryoelectron microscopy structures of the S protein demonstrated that the RBDs nested at the top of S protein underwent a hinge like movement, switching between “up” and “down” conformations (Koenig et al., 2021). The down conformation is an inactive state inaccessible to ACE2, while the up conformation is an active state that can be recognized by ACE2. We aligned the RBD-NB1A7 complex structure to the different conformations of the S protein. We found that part of the binding epitope was buried when RBD was in a “down” conformation (Figure 2F), which indicated that NB1A7 could not access to the close state of RBD within S protein. We superposed RBD-NB1A7 complex with the only up RBD state in context of S protein and found that NB1A7 still had steric hindrance with the adjacent clockwise RBD in down conformation (Figure 2G). However, the clash can be relieved when the targeted RBD and the clockwise adjacent RBD are both in the “up” conformation, suggesting that NB1A7 can bind to the “double-up” conformational RBDs in trimeric spike protein and sandwich between them (Figure 2H).
It was reported that ACE2 could only bind to the RBD in the open state, and its binding induced the conformational change, which leads to S protein proteolysis, and S2 fuse with host cell membrane (Koenig et al., 2021). NB1A7 binding could facilitate the RBD in up conformation to expose the proteolysis site and promote the shedding of S1, leading to premature activation from prefusion to post-fusion state to inactivate the fusion machinery. It is possible, as reported, that the neutralizing nanobodies stabilizing the RBD up conformation induced the fusion activity of spike (Koenig et al., 2021). As the above analysis indicates, dual neutralization mechanisms of NB1A7 against S protein can prevent the virus from entering the host cell.
NB1A7 binding epitope is highly conserved
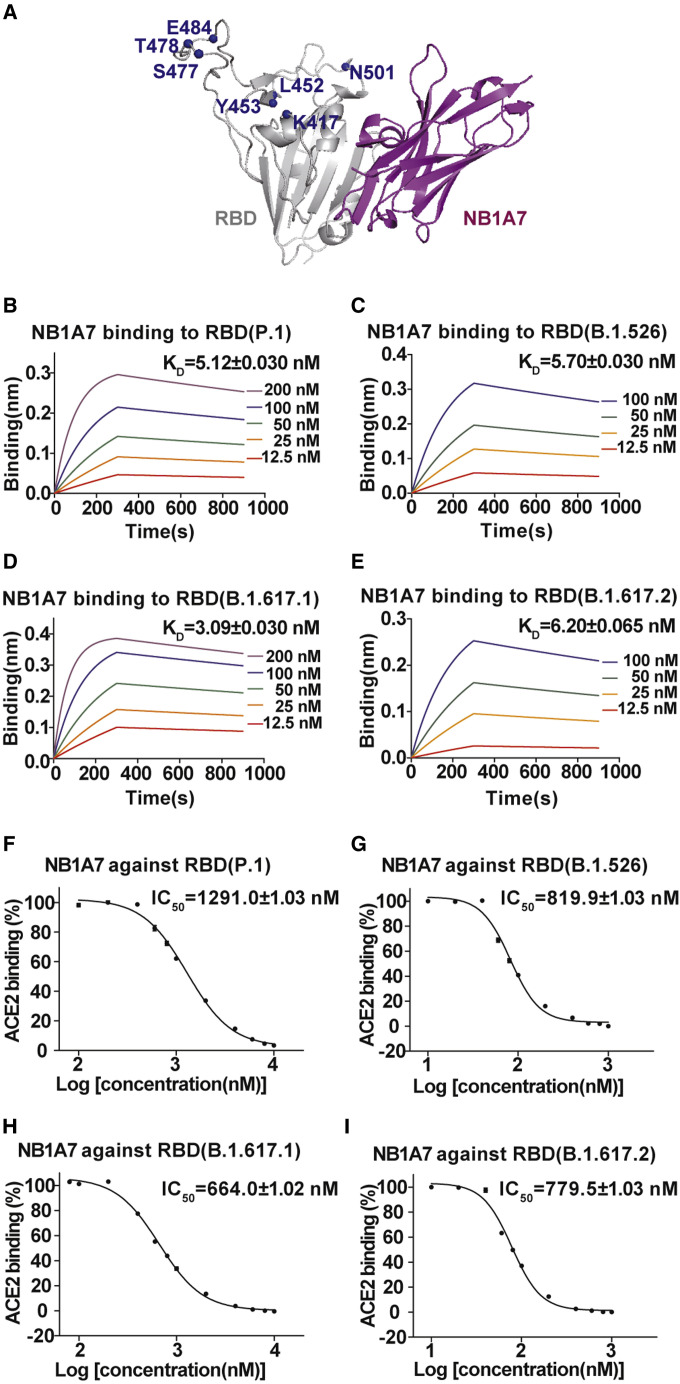
The serologic antibody response against SARS-CoV-2 in an infected individual leads to viral escape mutant. The region of RBD interacting with ACE2 receptor is a major target for the neutralizing human antibody, hence the RBM is the primary region where SARS-CoV-2 variants arise with mutations (Wang et al., 2021c, 2021d; Zhou et al., 2021; Cele et al., 2021; Davies et al., 2021; Wibmer et al., 2021; Yadav et al., 2021). Greaney et al. thoroughly summarized all mutations that escape binding by three major classes of antibodies against the SARS-CoV-2 RBD. They found that the most prominent emerging viral lineages include K417T/N, E484K/Q, L452R, N501Y, S477N, and T478K mutations (Figure 3 A) (Greaney et al., 2021). From sequence alignment and structural information, the neutralizing epitope of NB1A7 is a highly conserved cryptic region, and distant from the receptor binding site, so it could resist the plasma-escaping variants (Figures 3A and S3A). We further determined the affinities of NB1A7 to the RBD mutants (P.1 strain [K417T, E484K and N501Y], B.1.526 strain [E484K, S477N], B.1.617.1 strain [L452R, E484Q], and B.1.617.2 strain [L452R, T478K]) with K D values of 5.12 ± 0.03 nM, 5.70 ± 0.03 nM, 3.09 ± 0.03 nM, and 6.20 ± 0.065 nM (Figures 3B–3E and Table S1). Furthermore, competitive ELISA was performed to measure the inhibitory activity of NB1A7 to the mutants, with IC50 values of 1,291.0 ± 1.03 nM, 819.9 ± 1.03 nM, 664 ± 1.02 nM, and 779.5 ± 1.03 nM, respectively (Figures 3F–3I). Our data indicate that NB1A7 retains high binding affinity to RBD carrying most of mutations of the emerging viral lineages.
Figure 3.
The conservation of NB1A7
(A) Mapping mutations of circulating strains to RBD-NB1A7 complex. Mutations are indicated with ball in blue. The binding epitope of NB1A7 is distant from these mutations.
(B–E) The binding affinity of NB1A7 and RBD from P.1 strain, B.1.526 strain, B.1.617.1 strain, or B.1.617.2 strain was measured using BLI. The KD values are all at the nanomole level.
(F–I) Competitive ELISA was used to determine IC50 values of NB1A7 blocking ACE2 on RBD variants. The IC50 values of NB1A7 are 1291.0 ± 1.03 nM for P.1, 819.9 ± 1.03 nM for B.1.526, 664.0 ± 1.02 nM for B.1.617.1 and 779.5 ± 1.03 nM for B.1.617.2 strain, respectively.
See also Figures S3 and S4 and Tables S1 and S2.
However, the interface interactions between NB1A7 and RBD primarily include the hydrogen bond networks among the main chain of the residues of the β2 strand and the main chain of the residues of CDR3 of NB1A7. There is no side-chain interaction except R408 (Figure 2D). Therefore, the mutant interfering with the formation of the β2 strand of RBD will be the escape mutant for NB1A7. During the preparation of our manuscript, emerging viral lineages (BA.2 and omicron) were reported. We found that the mutation sites R408S, T376A, and S371F from the BA.2 lineage and the mutation sites S371L, S375F, and S73P from the emerging omicron variant are located in or related to the epitope of NB1A7. The mutation R408S directly disrupted the electrostatic interaction between Y119 and R 408, and the S (S371 or S373 or S375) with a small hydrophilic side chain is replaced by F or P or L with a bulky hydrophobic side chain, possibly interfering with the formation of the β2 strand of RBD, reducing the binding affinity of NB1A7 to RBD and escape neutralization (Figure S3).
The S protein RBDs of SARS-CoV-2 and SARS-CoV-1 are closely related in phylogeny and have 73% sequence identity. It was found that NB1A7 binding epitope of RBD was completely conserved in SARS-CoV-1 (Figure S4A). We assessed the binding affinity of NB1A7 against SARS-CoV-1 spike protein RBD, and the K D value was 471 nM (Figure S4G). Compared with SARS-CoV-2, the binding affinity of NB1A7 to SARS-CoV-1 significantly decreased, although the epitope of NB1A7 on the SARS-CoV-2 is identical to that of the SARS-CoV-2.
Superposition of the SARS-CoV-2 RBD and SARS-CoV-1 RBD showed that the segment from S375 to Y380 formed a ꞵ strand in the SARS-CoV-2 RBD, while the corresponding peptide from S362 to Y367 in the SARS-CoV-2 RBD is loosely organized, and the ꞵ strand of the SARS-CoV-1 become shorter than that of the SARS-CoV-2 RBD (Figures S4B–S4F). The binding affinity of NB1A7 to SARS-CoV-2 RBD is regulated primarily by interactions of β2 strand of RBD and the β strand of CDR3 from NB1 A7.
The alignment of two RBD sequences shows that the proline 384 of SARS-CoV-2 is replaced with alanine at corresponding site of SARS-CoV-1, which is close to the interface between NB1A7 and RBD (Figure S4A). The structure of SARS-CoV-2 RBD showed that the proline 384 residues of SARS-CoV-2 RBD assume the cis configuration, and there is a hydrogen bond between the main chain carbonyl group of the proline and the main chain amine group of lysine 386; as a result, the peptide from S383 to K386 forms a turn structure in SARS-CoV-2 RBD, and a hydrophobic core is formed by the P384, Y369 of α helix (Y365-N370), and F377 of the strand (S375-Y380) (Figures 2C and 2D). It is the hydrophobic interaction that stabilizes the integrity of the ꞵ strand from S375 to Y380 in the SARS-CoV-2 RBD. The replacement of proline with alanine results in the corresponding fragment with a wavy loop conformation, in which a hydrogen bond is formed between the carbonyl group of S370 and the hydroxy of Y356 of the α helix composed with a segment fromY352 to N357, and the hydrogen bond pulls down the α helix, which destroys the hydrophobic interaction between the Y356 of the α helix and the F364 in the SARS-CoV-1 RBD, so that the segment from S362 to Y367 is loosely organized and the binding with NB1A7 is not as well as that with SARS-CoV-2 RBD.
Therefore, the corresponding β2 strand of RBD was disrupted and shorter in the SARS-CoV-1 RBD, so that the binding affinity of NB1A7 to SARS-CoV-1 significantly decreased (Figure S4G).
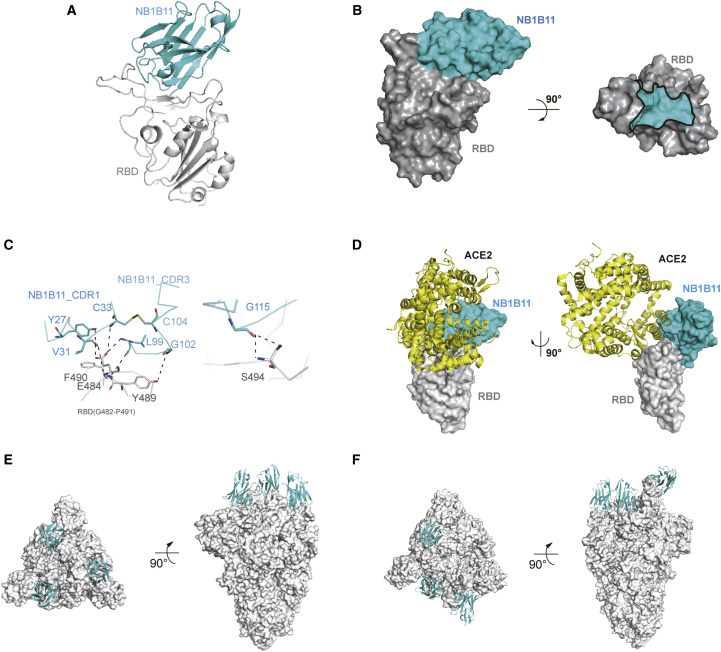
Structure determination of the NB1B11-RBD complex
To understand the neutralizing mechanism of NB1B11 against SARS-CoV-2, we determined the crystal structure of RBD-NB1B11 complex at a resolution of 2.08 Å with R work of 19.7% and R free of 22.6% (Figure 4 A; Table 2). In the structure, NB1B11 uses its CDR1 and CDR3 to bind to the distal end of the extended external loop of RBM and buries 1,710 Å2 solvent-accessible surface area on RBD (Figure 4B), which is the ACE2-binding site on the RBD. NB1B11 engages with RBD through shape complementarity, which has no disruption of the conformation of RBD (Figure 4A). The concave outer surface of the RBM accommodates the CDR3 of NB1B11, and the side chain of E484 inserts into the cleft composed of CDR1 and CDR3 from NB1B11, and its side chain carbonyl group forms double hydrogen bonds with the side chain of Y27 and the main chain of L99 from NB1B11, respectively. The main chain amine group of E484 forms the hydrogen bond with the main chain carbonyl group of V31 from NB1B11 (Figure 4C). In addition, the aromatic rings of the two adjacent residues (Y489 and F490 of RBD) extend in opposite directions, forming hydrophobic interaction with L99 and Y27 from NB1B11, respectively (Figure 4C). Moreover, the hydroxyl of Y489 side chain of RBD forms a hydrogen bond with G102 from CDR3 of NB1B11, and the hydrogen bond between F490 of RBD and L99 from NB1B11 stabilizes the two hydrophobic patches. In addition, the amine chain, and the hydroxyl of side chain of S494 from β5 of RBD and G115 of NB1B11 form double hydrogen bonds (Figure 4C).
Figure 4.
Crystal structure of RBD in complex with NB1B11
(A) Cartoon presentation of NB1B11 (in cyan) in complex with RBD (in gray).
(B) Surface presentation of NB1B11 (in cyan) in complex with RBD (in gray).
(C) The detailed interaction between NB1B11 and RBD.
(D) The binding site of NB1B11 on the RBD is overlapping with the contacting surface of ACE2 (in yellow) with RBD on the basis of PDB: 6M0J.
(E and F) The alignments of RBD-NB1B11 complex and different conformations of SARS-CoV-2 spike protein trimer (in gray). (E) NB1B11 not only can attach to all down conformation (PDB: 6XLU) (F) but can also bind to the RBD in its one-up conformation accessible to ACE2 (PDB: 6XM4).
See also Table 2.
Superposition of the RBD-NB1B11 complex on the RBD-ACE2 shows that NB1B11 clash with the bottom side of the small lobe of ACE2, and the epitope partially overlaps with the contacting surface of N-terminal helix of the ACE2 (Figure 4D), which was consistent with the biochemical and neutralization data that supported blockade of RBD/ACE2 attachment. We also aligned RBD-NB1B11 complex with the different conformation of RBD (up and down) in context of the S trimer and found that NB1B11 can bind to the RBD in its “up” conformation accessible to ACE2 and also attach to the “down” conformation that is inaccessible to ACE2 (Figures 4E and 4F). Thus, it has excellent antiviral efficacy.
Structure-based development of bi-paratopic and multivalent nanobodies
Superposing the structures of RBD-NB1A7 and RBD-NB1B11 complexes on the basis of RBD shows that NB1A7 and NB1B11 bind non-overlapping epitope of RBD. We designed two different constructs on the basis of the structural model of NB1A7 and NB1B11 simultaneously binding to the intra-RBD or the “double-up” conformational RBDs in trimeric spike protein of SARS-CoV-2. First, the binding sites of NB1A7 and NB1B11 on intra-RBD are in close proximity, and their C-terminal orientations are upward. Therefore, we generated bi-paratopic heterodimeric nanobodies (format I) through their C-term fused with Fc domain of IgG2B, respectively (Figure 5A). Second, the C-term of NB1A7 was linked to the N-term of NB1B11 by a linker followed by Fc to assemble format II, which bound to inter-RBDs (at less double-up conformation) (Figure 5B). Format II is a bi-paratopic and multivalent nanobody.
Figure 5.
Structure-based development of multivalent nanobodies against SARS-CoV-2
(A) Schematic of multivalent nanobody format I binding to one RBD. The C-term of NB1A7 and NB1B11 were fused with the Fc domain of mouse IgG2B to generate bi-paratopic heterodimeric nanobodies.
(B) Schematic of multivalent nanobody format II binding to inter-RBDs. The C-term of NB1A7 links the N-term of NB1B11 by a 20 amino acid (GGGGS)4 linker and then attached with Fc to generate bi-paratopic and multivalent nanobodies.
(C and D) Affinity binding curves of format I (C) and format II (D) with spike using BLI. The KD values are 0.31 ± 0.01 nM and less than 1 pM for format I and format II, respectively. The process of association and dissociation was fitted by a 1:1 binding data fit model.
(E and F) Competitive ELISA was used to assess whether format I or format II block spike interaction with ACE2 in vitro. Format I and format II blocked the ACE2 attachment on spike with IC50 values of 64.9 ± 1.01 nM and 20.8 ± 1.02 nM, respectively. The results are shown as mean ± SEM (n = 3).
(G and H) Neutralizing activity of format I or format II against SARS-CoV-2-luciferase pseudovirus with IC50 values of 2.8 ± 2.15 nM and 3.1± 1.50 nM, respectively. The results are shown as mean ± SEM (n = 2).
(I and J) Neutralizing activity of format I (NC50 = 1.0 ± 1.50 nM) or format II (NC50 = 0.6 ± 1.0 nM) against live SARS-CoV-2 virus. The results are shown as mean ± SEM (n = 2).
See also Figure S5.
As we expected, the binding experiment of spike protein by BLI showed that these two kinds of junctions significantly increased the avidity. Format I bound to the spike protein with a K D value of 0.31 ± 0.01 nM, while format II has binding affinity reaching the femtomole range (beyond the limit of instrument detection), at least 1,000-fold higher affinity to spike protein than each nanobody monomer (Figures 5C and 5D). After binding to the spike protein, no dissociation of format II was observed for 20 min, and both formats can block ACE2 attaching to the spike protein. Competitive ELISA experiments showed that format II inhibited ACE2 binding to spike protein with an IC50 of 20.8 ± 1.02 nM, while format I blocked the interaction of ACE2 and spike protein with an IC50 of 64.9 ± 1.01 nM (Figures 5E and 5F). We also used a SARS-CoV-2-luciferase pseudovirus neutralization assay to assess neutralizing potency of format I and format II. Both formats showed stronger neutralization activity than the stand-alone monomer, with IC50 values 2 orders of magnitude lower than that of isolated monomeric nanobody (2.8 ± 2.15 nM and 3.1 ± 1.50 nM for format I and format II, respectively) (Figures 5G and 5H). Finally, we used PRNT to determine the viral neutralization activity of each module reformats. Format I and II prevented the live SARS-CoV-2 virus infection of Vero E6 cells with IC50 values of 1.0 ± 1.5 nM and 0.6 ± 1.19 nM, respectively (Figures 5I, 5J, and S5). Collectively, the assembly of NB1A7 and NB1B11 significantly increased the binding affinity and substantially improved the neutralization potency to the live SARS-CoV-2 virus.
To assess the breadth of these nanobodies, we next measured the binding activity of formats I and II to RBD variants carrying the mutations of P.1 strain, B.1.526 strain, B.1.617.1 strain, and B.1.617.2 strain. Format I molecule bound to RBD variants with a K D value of about 1 nM (Figures S6A–S6E), while format II bound to RBD variants with K D values up to the femtomole range, which is comparable with that of its binding to wild-type (WT) RBD (Figures 6A–6E; Table S1). We also measured the inhibitory activity of multivalent nanobodies on ACE2 engagement to WT and mutants of RBD. Format I showed IC50 values between 47.5 ± 1.05 nM and 142.5 ± 1.04 nM for WT and mutate RBDs (Figures S6F–S6J). Compared with format I, format II has a slightly stronger inhibitory efficacy, with IC50 values between 37.5 ± 1.04 nM and 60.7 ± 1.04 nM, for WT RBD and other four RBD variants (Figures 6F–6J). Slight differences in the inhibitory efficacy of the multivalent nanobodies against different variants likely reflect the increased affinity of some these variants to ACE2.
Figure 6.
Format II against SARS-CoV-2 and its mutants
(A–E) Affinity binding curves of format II with RBD from WT virus or P.1, B.1.526, B.1.617.1, and B.1.617.2 lineage variants, with KD less than 1 pM.
(F–J) Competitive ELISA was used to assess the inhibitory activity of format II on ACE2 engagement to WT and mutants of RBD, and the IC50 values are 37.5 ± 1.04, 54.2 ± 1.03, 60.7 ± 1.04, 58.8 ± 1.04, and 36.9 ± 1.01 nM for RBD WT virus and P.1, B.1.526, B.1.617.1, and B.1.617.2 lineage variants, respectively. The results are shown as mean ± SEM (n = 3).
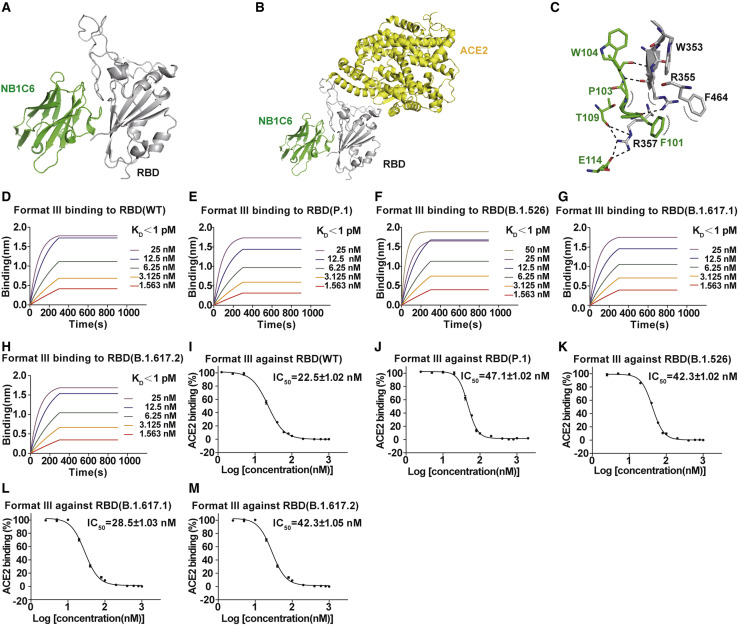
The residue E484 is located at the epitope of NB1B11 on RBD, and many emerging viral lineages contain mutations at this site. Thus, NB1A7 and NB1B11 are not the most ideal combination. Although we did not obtain the crystal structure of NB1C6-RBD complex, structural modeling of NB1C6 and RBD was conducted with HDOCK. The structural model showed that the epitope of NB1C6 on RBD is almost perpendicular to the receptor-binding motif, localized on the opposite side of NB1A7, and does not overlap with receptor-binding region (Figures 7A–7C). On the basis of the structural model, we constructed bispecific nanobodies using NB1C6 and NB1A7. The C-term of NB1A7 was linked to the N-term of NB1C6 by a (GGGGS)4 linker followed by Fc to assemble a bispecific nanobody (format III). The bispecific nanobody showed significantly higher binding affinity to the variants of SARS-CoV-2, with K D values less than 1 pM (Figures 7D–7H; Tables S1 and S2), and its inhibitory effect was slightly better than that of the bispecific nanobodies composed of NB1A7 and NB1B11 (Figures 7I–7M).
Figure 7.
Structure model of RBD-NB1C6 complex and the binding affinity and competitive capacity of format III
(A) The structural model of RBD-NB1C6 complex.
(B) The binding site of NB1C6 (green) on the RBD (gray) does not overlap with the contacting surface of ACE2 (yellow) with RBD on the basis of PDB: 6M0J.
(C) Details of interaction between NB1C6 (green) and RBD (gray).
(D–H) Binding kinetics of format III to RBD wild-type and mutants.
(I–M) The competitive capacity of format III with ACE2 binding RBD wild-type and mutants.
Discussion
To deal with the crisis brought on by the COVID-19 pandemic, the scientific community and pharmaceutical industry have used advanced strategies to develop vaccines and neutralizing antibodies. But the evolving variants of SARS-CoV-2 elude neutralizing antibodies generated from initial SARS-CoV-2 virus infection or substantially reduce the neutralizing potency of convalescent and vaccine-induced sera (Wang et al., 2021b, 2021d; Wu et al., 2021). Viral adaptation and antigenic drift resulted in emerging variants of SARS-CoV-2 that became the globally dominant circulating strains. Therefore, the development of a universal or broadly reactive effective antiviral therapeutic is urgently needed to prevent the emergence of vaccine resistant viral lineages and future variants.
We identified a panel of nanobodies with high affinity to RBD from a camel immunized with recombinant RBD, which adds to the growing reports of neutralizing nanobodies against SARS-CoV-2 as potential therapeutics for COVID-19 patients (Bracken et al., 2021; Huo et al., 2020; Koenig et al., 2021; Muyldermans, 2013; Pymm et al., 2021; Schoof et al., 2020; Wrapp et al., 2020; Xiang et al., 2020a; Xu et al., 2021). We also classified these nanobodies into three groups according to their antigenic sites and non-competing epitopes. We then selected one representative nanobody from each of the three clusters (NB1B11, NB1A7, and NB1C6) for further study and characterization, including binding strength with RBD, inhibition of ACE2 binding, and viral neutralization through the pseudovirus luciferase reporter assay or authentic SARS-CoV-2 virus PRNT assay. The results demonstrated that NB1B11 and NB1A7 have high neutralizing potency against SARS-CoV-2 virus. Indeed NB1A7 and NB1B11 have potential, but as shown in the Supplemental information, NB1C6 may, as part of a bi-head, become important as well to prevent the generation of escape mutants of SARS-CoV-2. To understand the potential neutralization mechanism of these nanobodies to SARS-CoV-2 virus, we determined the structure RBD-NB1A7. The structure of RBD-NB1A7 in the context of SARS-CoV-2 spike protein shows that NB1A7 binds at a highly conserved cryptic epitope in RBD and required the adjacent clockwise RBD in open conformation. Although its epitope does not overlap with the ACE2-contacting surface on the RBD, it sterically interferes with RBD attachment to ACE2. In addition, NB1A7 stabilizes the RBD in the active up conformation accessible to ACE2 receptor. It is possible that NB1A7 binding induces premature activation of the spike protein that without host cell membranes leads to irreversible conformational changes to a post-fusion state and disruption of the spike function. Therefore, NB1A7 has strong neutralizing effects on the SARS-CoV-2 virus by blocking the interaction between ACE2 and RBD and destroying its spike function. It is exciting that the NB1A7 binding epitope is shared between SARS-CoV-1 and SARS-CoV-2, which indicates NB1A7 has a potential cross-reactive nanobody that might work against SARS-CoV-1, SARS-CoV-2, and current or future variants. However, although the mutations do not directly interact with the NB1A7, nearby mutations can affect binding of NB1A7 to the emerging viral lineages (Tables S1 and S2). The binding affinity of NB1A7 to RBD is regulated through mechanisms involving both direct and long-range interactions, and some mutation sites, despite located outside the contact interface with NB1A7, may maintain the structural integrity of the epitope. As concluded by Starr et al., viral escape mutations generally occur in the antibody-RBD interface (Starr et al., 2021). Our data indicate that NB1A7 retains high binding affinity to RBD carrying most of the mutations present on emerging viral lineages (Figures 3B–3E). However, some mutations of the BA.2 lineage and the emerging omicron variant are located in or related with the epitope of NB1A7, which possibly reduces the binding affinity of NB1A7 to RBD and escape neutralization.
The structure of RBD-NB1B11 reveals that the NB1B11 binding epitope partially overlaps with the interface between ACE2 and RBD, which directly blocks ACE2 binding with RBD. Similarly, the epitopes of the neutralizing monoclonal antibodies currently approved or under clinical development also target the ACE2 receptor binding site (Wang et al., 2021a, 2021b, 2021c, 2021d). The recently reported neutralizing nanobody NB20 also binds this region, and it can bind both open and closed RBD within SARS-CoV-2 spike protein (Xiang et al., 2020a).
Compared with conventional antibodies, nanobodies are easy to engineer to form multivalent nanobodies for bioactivity enhancement. On the basis of the structure of both of RBD-NB1A7 and RBD-NB1B11, NB1A7 and NB1B11 have different antigenic sites on the RBD of the spike protein, so we designed two formats of bi-paratopic multivalent molecules composed of these two nanobodies. Format I is a heterodimer consisting of NB1A7-Fc-fusion and NB1B11-Fc-fusion that bind intra-RBD, while format II is a C terminus-NB1A7 and N terminus NB1B11 linked with a (GGGGS)4 linker and then fusion with Fc, which bind inter-RBD within spike protein. As expected, both formats dramatically increased the binding affinity and enhanced the neutralization activity against SARS-CoV-2 with synergistic effect. The binding affinity of formats II and III reached the femtomole range. The combined treatments have a very low tolerance for future variants and can protect against the predominant SARS-CoV-2 virus variants currently in circulation and future escape mutant virus. To the best of our knowledge, this would reflect the most potent neutralizing nanobody targeting RBD of SARS-CoV-2 spike protein reported to date. Furthermore, the high conserved epitope of NB1A7 combined with a bi--paratopic strategy should prevent the emergence of viral escape mutants. In summary, this study provides an excellent proof of concept for rational development of broadly reactive neutralizing nanobodies, and the candidates described here will be moved into animal experiments and clinical development.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L)(Goat monoclonal) | Yeasen | Cat# 33201ES60; RRID: AB_10015289 |
| HA-Tag, (Mouse monoclonal) | InvivoGen | Cat# ab-hatag; RRID: AB_391833 |
| Bacterial and virus strains | ||
| BL21(DE3) | New England Biolabs | Cat#: C2527I |
| TG1 | Lucigen | Cat#: 60502 |
| TOP10F' | Huayueyang Biotech | WXR15-100S |
| Chemicals, peptides, and recombinant proteins | ||
| Anti-FLAG Affinity Gel | Smart-Lifesciences | Cat#: SA042C |
| Flag peptide DYKDDDDK | Genscript | N/A |
| NB1A7 | This paper | N/A |
| NB1B11 | This paper | N/A |
| RBD-Flag | This paper | N/A |
| Deposited data | ||
| The binding affinity of nanobodies | This paper | https://data.mendeley.com/datasets/jjpdzrb5ht/draft?a=a4c83cb6-324c-493c-a6a7-24f5e6df40e5 |
| The block data of nanobodies | This paper | https://data.mendeley.com/datasets/jjpdzrb5ht/draft?a=a4c83cb6-324c-493c-a6a7-24f5e6df40e5 |
| PDB accession code of the RBD-NB1A7 crystal structure | This paper | PDB: 7FAT |
| PDB accession code of the RBD-NB1B11 crystal structure | This paper | PDB: 7FAU |
| Experimental models: Cell lines | ||
| FreeStyle™ 293-F cells |
Gibco |
11625019 |
| RRID: CVCL_6642 | ||
| Oligonucleotides | ||
| 5’-CGGAATTCGATATCGCAGGTGCAGCT GCAGGAGTC-3’ (NB1A7-Fc-F primer & NB1B11-Fc-F primer) |
This paper | N/A |
| 5’-CTAGCCATGGCGGAACCTCCA CCTCCTGAGGAGACGGTGACCTGGG-3’ (NB1A7-Fc-R primer & NB1B11-Fc-R primer) |
This paper | N/A |
| 5’-CGGAATTCGATATCGCAGGTGCAGCT GCAGGAGTC-3’ (NB1A7-NB1B11-Fc-F1) |
This paper | N/A |
| 5’-GCTCCCTCCGCCACCA GACC-3’ (NB1A7-NB1B11-Fc-R1) |
This paper | N/A |
| 5’-GGTCTGGTGGCGGAGGGAGC CAAGTACAATTACAAGAAAGCGGAG GAGGCTCGGTGCAGGC-3’ (NB1A7-NB1B11-Fc-F2) |
This paper | N/A |
| 5’-GCCCGCTGGGAGATCTAACCATGG-3’ (NB1A7-NB1B11-Fc-R2) | This paper | N/A |
| 5’-CGGAATTCGATATCG CAGGTGCAGCTGCAGGAGTC-3’ (NB1A7-NB1C6-Fc-F1) |
This paper | N/A |
| 5’-GACTCCTGCAGCTGCACC TGGCTCCCTCCGCCACCAGACC-3’ (NB1A7-NB1C6-Fc-R1) |
This paper | N/A |
| 5’-GGTCTGGTGGCGGAGG GAGCCAGGTGCAGCTGCAGG AGTC-3’ (Linker-NB1C6-F2) |
This paper | N/A |
| 5’-CATGCCATGGCTCCACCGCTG CCACCTGAGGAGACGGTGACC TGGG-3’ (NB1A7-NB1C6-Fc-R2) |
This paper | N/A |
| Remaining primers can be found in Table S3 | This paper | N/A |
| Recombinant DNA | ||
| pMECS nanobody | This paper | N/A |
| pCMV-RBD-Flag | This paper | N/A |
| Software and algorithms | ||
| PHENIX | http://www.phenix-online.org/ | Version 1.19.2 RRID:SCR_014224https://phenix-online.org/download |
| Coot | Coot (cam.ac.uk) | Version 0.9.4 RRID:SCR_014222https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/binaries/pre-releases/coot-0.9.4-pre-revision-10190-binary-Linux-x86_64-ubuntu-16.04.6-python-gtk2.tar.gz |
| MolProbity | https://doi.org/10.1107/S0907444909042073 | RRID:SCR_014226http://molprobity.biochem.duke.edu |
| PyMol | Schrodinger | Version 2.5 RRID:SCR_000305https://pymol.org/installers/PyMOL-2.5.2-Windows-x86_64.exe |
| GraphPad Prism 7 | GraphPad | RRID:SCR_002798https://www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Y. Geng (gengyong@simm.ac.cn).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Microbe strains
(HEK) 293F cells (human) were purchased from Gibco (Cat# R790-07, RRID: CVCL_6642), which were grown in SMM 293-TII medium (Sino Biological Inc.) supplemented with 2% (v/v) FBS (Gibco) and 8% CO2 for maintenance. E. coli BL21(DE3) was cultured at 37°C in LB medium supplemented with 100 mg/mL of ampicillin and induced with isopropyl β-D-1-thiogalactopyranoside (IPTG, final concentration 0.1 mM) at 16°C for 10 hours, when an OD600 of 0.6 was reached for recombinant proteins expression. All the cell lines tested negative for mycoplasma contamination.
Method details
Constructs
The gene encoding amino acids 333-523 of receptor binding domain (RBD) of SARS-CoV-2, its variants P.1 (K417T, E484K, N501Y), B.1.526 (E484K, S477N), B.1.617.1 (L452R, E484Q) and B.1.617.2 (L452R, T478K), followed by an Avi-tag and a Flag epitope tag at C-terminus was cloned into a modified pEG BacMam vector, respectively.
For the construct of SARS-CoV-2 Spike protein, the extracellular domain of S protein (residues 1-1211) was cloned into a modified pEG BacMam vector with proline substitutions at residues 986K and 987V, a “GSAS” substitution at residues 682 to 685(RRAR), a C-terminal T4 fibritin trimerization motif followed by a Flag epitope tag.
The gene encoding SARS-CoV RBD (306-527) followed by an Avi-tag and a Flag epitope tag at C-terminus was also cloned into a modified pEG BacMam vector.
Protein expression and purification
The recombinant SARS-CoV-2 RBD, RBD mutants, SARS-CoV RBD or SARS-CoV-2 S protein were overexpressed in the HEK293F cells (ATCC) at 37°C in a humidified 8% CO2 incubator (Eppendorf, Germany). For each protein, the plasmid was transiently transfected into the cells using polyethylenimines (PEIs) with the cell density at 2×106 cells per ml. 10 mM sodium butyrate was added 12 h post-transfection. Cells were then incubated at 30°C with gentle rotation and the supernatant was collected after 48 h transfection by centrifugation at 5000×g for 30 min. The protein was purified by anti-FLAG M2 affinity resin. After the protein binding to the M2 beads, the resin was washed with 20 mM Tris pH 8.0, 150 mM NaCl and subsequently eluted by the buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, 0.2 mg mL-1 flag peptide. The eluted protein was then injected on a High-load Superdex 200 16/600 GL column. The peak fractions of SARS-CoV-2 RBD were collected in the buffer containing 20 mM Tris pH 8.0, 150 mM NaCl and aliquot for camel immunization.
Nanobody library generation
Camel immunization and nanobody (Nb) library generation were performed as described previously. In brief, the camel was immunized with 1 mg RBD protein combined with Gerbu FAMA adjuvant once a week for 7 weeks. This animal work was approved and supervised by Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Permit Number: SYXK 2015-0027). The peripheral blood lymphocytes (PBLs) were isolated by Ficoll-Paque Plus (GE Healthcare, USA) for naïve library construction. Total RNA was extracted and the cDNA was synthesized by using a Super-ScriptTM III FIRST-Strand SUPERMIX Kit (Invitrogen, USA). The VHH genes were amplified by two-step nest-PCR and cloned into a phagemid pMECS vector as previously described. The primers (CALL-F and CALL-R) were used in the first PCR to amplify the fragments from the leader signal sequence of all V elements of family III to the second constant heavy-chain domain (CH2) that is conserved. The primers (VHH-F and VHH-R) were used to amplify the nanobody encoding genes via the second nested PCR. The phagemids were then transformed into the electro-competent E. coli TG1 cells. The size of constructed nanobody library was evaluated by the number of positive colonies. The cells were then scraped and stored at −80°C.
Screening and identification of Nbs by phage display technology
To screen and identify RBD specific Nbs, phages presenting RBD-VHHs were enriched 2 rounds of bio-panning, which was performed in the 96 wells microtiter plates (Thermo, USA). For each round of bio-panning, phages were dispensed into RBD coated plates and incubated for 2 h. The wells were then washed with PBST for 10 times and PBS for 5 times. The retained phages were trypsinized and collected for the next round of bio-panning. The collected phages infected E. coli TG1 cells to obtain the cells expressing VHH. Overall 950 clones were picked to confirm whether bound to RBD.
These clones were selected by RBD-coated ELISA plates, which were detected by HRP-conjugated anti-HA antibody. All positive clones were sequenced and grouped based on the homologues of complementary determining regions (CDRs). Finally, a total of 29 nanobodies with different sequence of CDR3 were identified.
Expression and purification of Nbs
The genes encoding RBD-specific Nbs were cloned into pET-28a (+) vector with N terminal 6×His tag, followed by SUMO fusion protein. It was expressed in the periplasm of E. coli BL21(DE3) and grown to OD600nm 0.6 at 37°C in 2×YT medium containing 100 μg mL−1 ampicillin, 0.1% (w/v) glucose and 1mM MgCl2. 1mM IPTG was added to induce the expression of Nbs at 28°C for 12 h. Cells were harvested by centrifugation at 5000 × g for 30 min and lysed in the buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1 mM PMSF) by sonication, then centrifuged to remove the cell debris. The supernatant was collected and applied to a nickel affinity chromatography column and the eluent was further purified by size-exclusion chromatography using a HiLoad 16/600 Superdex 75 column. The purified His tagged-SUMO-Nbs were incubated with ubiquitin-like-specific protease 1 (ULP1) (prepared in-house) to remove the His tag and SUMO fusion. After loaded onto the nickel affinity chromatography column, the flow through that contained the Nb was applied to the HiLoad 16/600 Superdex 75 column for further purification. Finally, the Nb was spin concentrated to 1 mg mL−1.
Determination of different epitopes of RBD protein
To initial screen the binding sites of Nbs to RBD, purified recombinant RBD and Nbs were mixed at a molar ratio of 1:1:1 and incubated for 30 mins before injected on a superdex 200 increase 5/150 GL. Protein signals were detected by UV280 nm. Finally, three groups were detected to bind at the different epitopes of RBD protein.
The biolayer Interferometry (BLI) was performed to further confirm these three binding epitopes. The biotinylated RBD was immobilized on the Streptavidin Dip and ReadTM Biosensors for 120 s until the instrument response was about 1 nm relative units (RUs). Baseline was measured in the kinetic buffer (PBS buffer and 0.02% Tween 20) and the sensors were exposed to 500 nM NB1A7 until the curve reached plateau phase. Thuen the sensors were further exposed to both 500 nM NB1A7 and NB1B11. Finally, the sensors were exposed to both NB1B11, NB1A7 and NB1C6 until the curve was stable at plateau phase.
Kinetic measurements by biolayer interferometry (BLI) binding assays
The biolayer Interferometry (BLI) was used to measure the binding affinity (KD) of Nbs. The biotinylated RBD was immobilized on the Streptavidin Dip and ReadTM Biosensors for 120 s until the instrument response was about 1 nm relative units (RUs). Baseline was measured in the kinetic buffer (PBS buffer and 0.02% Tween 20) and the sensors were exposed to nanobodies with twofold serial dilutions from 100 nM to 3.125 nM for 300 s. Then the dissociation was monitored in kinetic buffer for 600 s. All the curves were fitted by a 1:1 binding data fit model using the ForteBio Data Analysis Software to calculate the KD, which is the ratio of Koff/Kon.
Binding inhibition of ACE2
Streptavidin was coated onto the ELISA plates overnight at 4°C. Wells were washed 5 times in PBST and blocked with 2% BSA for 2 h. And then biotinylated RBD protein was added to wells. 0.1 μg mL−1 ACE2-Fc (prepared in house) supplement with Nbs in different concentrations from 900 nM to 0.0047 nM in threefold serial dilution were added to the wells to competitively bind RBD protein. After wash 5 times in PBST, goat anti-human IgG-Fc secondary antibody (HRP) was added and incubated for 1 h. TMB substrate (Thermo Fisher Scientific, USA) was added and the reactions were stopped by 2 M H2SO4. Then, the plates were measured at OD450nm.
Pseudotyped virus neutralization
Pseudotyped virus neutralization was performed in order to evaluate the neutralization activity of Nbs. In brief, HEK293T cells that stably expressed the ACE2 (HEK-ACE2 cells) were cultured in 96-well plates. When the cell density reached 3×104 cells per ml, NB1A7 or NB1B11 in different concentrations from 100 μg ml−1 to 0.0000512 μg ml−1 in fivefold serial dilution mixed with SARS-CoV-2-Luciferase pseudovirus were incubated with cells at 37°C, 5% CO2 for 72 hours. The luciferase activity was calculated by using the Luciferase Assay System (Promega, USA). The analysis was performed on Prism (GraphPad) to calculate half-maximal inhibitory concentration (IC50) values.
Authentic SARS-CoV-2 plaque reduction neutralization test (PRNT)
PRNT was performed to detect the Nbs neutralizing the SARS-CoV-2 virus. Vero E6 cells were seeded into 24-well culture plates at the density of 1.5×106 cells per well the day before infection. Next day, Nbs were prepared in a serial dilution and mixed with SARS-CoV-2 suspension containing 200 TCID50 in DMEM that supplemented with 2% FBS. After 1 h incubation at 37°C, the mixture was added into Vero E6 cells. The mixture was removed after 1 h incubation at 37°C and substituted with the medium containing DMEM, 2%FBS and 0.9% carboxymethyl cellulose (Promega). Plaques were stained by 0.5% crystal violet after 3 days incubation at 37°C, 5% CO2. Plaques were counted and the 50% neutralizing dose (ND50) were calculated.
Crystallization and data collection
RBD protein, NB1A7 and NB1B11 were prepared as described above. The peak fractions of RBD were incubated with a 1.2 molar excess of NB1A7 or NB1B11 before injected on the High-load Superdex 200 16/600 GL column. The fractions in buffer containing 20 mM Tris pH 8.0, 100 mM NaCl were collected and concentrated to 10 mg mL−1 for further crystallization.
The crystals of NB1A7 bound RBD and NB1B11 bound RBD were obtained using hanging drop vapor diffusion technique. The crystals of NB1A7 bound RBD were grown at 20°C in a reservoir solution containing 8% PEG6000, 0.1 M citric acid pH 3.5 and 2% ethylene glycol, while the crystals of NB1B11 bound RBD were formed at 20°C in a reservoir solution of 16% PEG3350 and 0.1 M Zinc acetate, dehydrate. All the crystals were flash-cooled with liquid nitrogen in the reservoir buffer supplemented with 20% glycerol.
Diffraction data were collected at 100K on the beamline BL19U1 at Shanghai Synchrotron Radiation Facility, China with a wave length of 0.979 Å. Data were processed using XDS and AIMLESS as a part of the CCP4 programs.
Structure determination and refinement
The structures of NB1A7 bound RBD and NB1B11 bound RBD were solved by molecular replacement using Phaser. The searching model include the RBD (PDB:7CH5) was manually fit into the density in COOT (Emsley and Cowtan, 2004). The refinement of the structures was performed by PHENIX suite (Adams et al., 2002). The final validation of models was done by MolProbity (Chen et al., 2010).
Expression and purification of multivalent Nbs
For bi-paratopic heterodimeric nanobodies (Format I), the gene encoding NB1A7 or NB1B11 was cloned into a pFUSE-mlgG2B-Fc2 vector, respectively. For bi-paratopic and multivalent nanobodies (Format II), the gene encoding NB1A7 and NB1B11 that linked by a 20 amino acid (GGGGS)4 linker, followed by a GGGGS linker at the C-terminus was cloned into the pFUSE-mlgG2B-Fc2 vector. For Format III, NB1B11 was replaced by NB1C6 compared with Format II. For the bi-paratopic and multivalent nanobodies (Format III), the gene encoding NB1A7 and NB1C6 that linked by a 20 amino acid (GGGGS)4 linker, followed by a GGGGS linker at the C-terminus was cloned into the pFUSE-mlgG2B-Fc2 vector.
For the expression of Format I, Format II and Format III, co-transient transfection of NB1A7-Fc and NB1B11-Fc or transient transfection of NB1A7-(GGGGS)4-NB1B11-Fc or NB1A7-(GGGGS)4-NB1C6-Fc incubated with PEIs was performed in HEK293F at the density of 2×106 cells per mL. 10 mM sodium butyrate was added 12 h post-transfection. Cells were then incubated at 30°C with gentle rotation and the supernatant was collected after 48 h transfection by centrifugation at 5000×g for 30 min. The proteins were purified by rProtein A Beads (SMART LIFESCIENCES, China) for initial purification. Then the proteins were washed by the buffer containing 20 mM Tris pH 7.0, 150 mM NaCl and eluted by 0.1 M Glycine buffer pH 3.0 and 1M Tris pH 8.5 was add to the eluted protein to adjust the pH value to the neutrality. The multivalent Nbs were concentrated and applied to the HiLoad 16/600 Superdex 200 column. The peak fractions were collected and stored at −80°C for further experiment.
Quantification and statistical analysis
The statistical analysis was performed on Prism (GraphPad) to calculate halfmaximal inhibitory concentration (IC50) values and halfmaximal neutralizing Dose (ND50).
Competitive ELISA was used to assess whether nanobodies block RBD interaction with ACE2 in vitro. The results shown are mean ± SEM (n = 3).
SARS-CoV-2-Luciferase pseudovirus neutralization assay to test antiviral activities. The results shown are mean ± SEM (n = 2).
SARS-CoV-2 plaque reduction neutralization test (PRNT) was performed to detect neutralizing activity. The results shown are mean ± SEM (n = 2).
Acknowledgments
We thank the staff of the BL19U1 beamline of the National Facility for Protein Science in Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility for assistance during data collection (Zhang et al., 2019). This work is supported by National Natural Science Foundation of China (grant 31670743), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA12040326), the Science and Technology Commission of Shanghai Municipality (grant 18JC1415400), the Joint Research Fund for Overseas, Hong Kong and Macao Scholars (grant 81628013), the Natural Science Foundation of Shanghai (grant 16ZR1442900), the National Science Foundation for Young Scholar projects (grant 81803599), grants from the Zhejiang University COVID-19 Special Project (grant 2020XGZX092) and the Shanghai Institute of Materia Medica, Chinese Academy of Sciences (grants CASIMM0120164013, SIMM1606YZZ-06, SIMM1601KF-06, 55201631121116101, 55201631121108000, 5112345601, and 2015123456005).
Author contributions
Y. Geng conceived and designed the study. Z.Z. Shi and X.Y. Li expressed and purified RBDs. Z.Z. Shi and X.Y. Li discovered nanobodies and analyzed their binding affinity and activity. H.W. Zhang performed plaque reduction neutralization testing, and R. Gong supervised the experiments. Z.C. Sun and X.Y. Li prepared crystals. Z.C. Sun and L. Wang collected crystal data. L. Wang built and refined models of the crystal structures of RBD-NB, and C.P. Fan supervised this process. Q.Q. Cui and X.C. Chen prepared spike protein. Y. Geng, L. Wang, X.C. Chen, X.Y. Li, and Z.Z. Shi wrote the manuscript. H.R. Qiao, Z.Y. Lan, X. Zhang, X.H. Li, and L.Y. Li purified nanobodies, and J.F. Xu guided this process. Y. Geng. supervised the project.
Declaration of interests
The authors declare no competing interests.
Published: March 10, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.str.2022.02.011.
Supplemental information
Data and code availability
The crystal structures of SARS-CoV-2 RBD in complex with NB1A7 are deposited in the Protein Data Bank (PDB) with accession codes PDB: 7FAT.
The crystal structures of SARS-CoV-2 RBD in complex with NB1B11 are deposited in the Protein Data Bank (PDB) with accession codes PDB: 7FAU.
Raw data from Figures 1, 3, 5, 6, and 7 were deposited on Mendeley at https://data.mendeley.com/datasets/jjpdzrb5ht/draft?a=a4c83cb6-324c-493c-a6a7-24f5e6df40e5.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bracken C.J., Lim S.A., Solomon P., Rettko N.J., Nguyen D.P., Zha B.S., Schaefer K., Byrnes J.R., Zhou J., Lui I., et al. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat. Chem. Biol. 2021;17:113–121. doi: 10.1038/s41589-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/s0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M.S. Monoclonal antibodies to disrupt progression of early Covid-19 infection. N. Engl. J. Med. 2021;384:289–291. doi: 10.1056/NEJMe2034495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected Hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P.A., Das H., Liu H., Kümmerer B.M., Gohr F.N., Jenster L.M., Schiffelers L.D.J., Tesfamariam Y.M., Uchima M., Wuerth J.D., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371 doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lv Z., Deng Y.Q., Ye Q., Cao L., Sun C.Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Pymm P., Adair A., Chan L.J., Cooney J.P., Mordant F.L., Allison C.C., Lopez E., Haycroft E.R., O'Neill M.T., Tan L.L., et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Kobe B., Qi J. Targeting multiple epitopes on the spike protein: a new hope for COVID-19 antibody therapy. Signal. Transduct. Target. Ther. 2020;5:208. doi: 10.1038/s41392-020-00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wang L., Feng R., Wang N., Wang Y., Zhu D., Xing X., Yang P., Zhang Y., Li W., Wang X. Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms. Cell Res. 2021;31:597–600. doi: 10.1038/s41422-021-00497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bülow S., Gecht M., Bagola K., Hörner C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Sun Y., Feng R., Wang Y., Guo Y., Zhang L., Deng Y.Q., Wang L., Cui Z., Cao L., et al. Structure-based development of human antibody cocktails against SARS-CoV-2. Cell Res. 2021;31:101–103. doi: 10.1038/s41422-020-00446-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.25.428137. Preprint at. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.01.25.427948. Preprint at. [DOI] [Google Scholar]
- Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile, multivalent nanobody cocktails efficiently neutralize SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.24.264333. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xu K., Jung S., Conte A., Lieberman J., Muecksch F., Lorenzi J.C.C., Park S., Schmidt F., Wang Z., et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 2021 doi: 10.1038/s41586-021-03676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P.D., Sapkal G.N., Abraham P., Ella R., Deshpande G., Patil D.Y., Nyayanit D.A., Gupta N., Sahay R.R., Shete A.M., et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab411. [DOI] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Sun Y., Deng Y.Q., Wang N., Tan Y., Zhang N.N., Li X.F., Kong C., Xu Y.P., Chen Q., et al. Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res. 2021;31:25–36. doi: 10.1038/s41422-020-00444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.-Z., Tang J.-C., Wang S.-S., Wang Z.-J., Qin W.-M., He J.-H. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 2019;30 doi: 10.1007/s41365-019-0683-2. [DOI] [Google Scholar]
- Zhou D., Chan J.F., Zhou B., Zhou R., Li S., Shan S., Liu L., Zhang A.J., Chen S.J., Chan C.C., et al. Robust SARS-CoV-2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe. 2021;29:551–563.e5. doi: 10.1016/j.chom.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structures of SARS-CoV-2 RBD in complex with NB1A7 are deposited in the Protein Data Bank (PDB) with accession codes PDB: 7FAT.
The crystal structures of SARS-CoV-2 RBD in complex with NB1B11 are deposited in the Protein Data Bank (PDB) with accession codes PDB: 7FAU.
Raw data from Figures 1, 3, 5, 6, and 7 were deposited on Mendeley at https://data.mendeley.com/datasets/jjpdzrb5ht/draft?a=a4c83cb6-324c-493c-a6a7-24f5e6df40e5.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.