Abstract
Cellular redox homeostasis is precisely balanced by generation and elimination of reactive oxygen species (ROS). ROS are not only capable of causing oxidation of proteins, lipids and DNA to damage cells but can also act as signaling molecules to modulate transcription factors and epigenetic pathways that determine cell survival and death. Hsp70 proteins are central hubs for proteostasis and are important factors to ameliorate damage from different kinds of stress including oxidative stress. Hsp70 members often participate in different cellular signaling pathways via their clients and cochaperones. ROS can directly cause oxidative cysteine modifications of Hsp70 members to alter their structure and chaperone activity, resulting in changes in the interactions between Hsp70 and their clients or cochaperones, which can then transfer redox signals to Hsp70-related signaling pathways. On the other hand, ROS also activate some redox-related signaling pathways to indirectly modulate Hsp70 activity and expression. Post-translational modifications including phosphorylation together with elevated Hsp70 expression can expand the capacity of Hsp70 to deal with ROS-damaged proteins and support antioxidant enzymes. Knowledge about the response and role of Hsp70 in redox homeostasis will facilitate our understanding of the cellular knock-on effects of inhibitors targeting Hsp70 and the mechanisms of redox-related diseases and aging.
Keywords: redox homeostasis, oxidative stress, ROS, Hsp70, cysteine modifications, glutathionylation
1. Introduction
Cellular redox homeostasis relies on precise coordination between the generation and elimination of reactive oxygen species (ROS). ROS can act as signaling molecules to modulate transcription factors and epigenetic pathways controlling cell survival and death [1,2]. Cellular ROS levels undergo continuous fluctuations within the physiological redox range. The redox status of the cell is essential to regulate basic biological processes including cell proliferation/differentiation, metabolic homeostasis and immune responses [2,3]. Oxidative stress occurs when ROS generation exceeds ROS clearance and the redox status is outside the physiological redox range [4]. Continuous oxidative stress leads to cell, tissue and organ damage and is linked to pathogenesis in cancer, diabetes, cardiovascular disease, neurodegenerative diseases, reproductive system diseases and the process of aging [5].
ROS types can be divided into free radicals and nonradicals. The free radicals include nitric oxide (NO•), the superoxide radical anion (O2•−), the hydroxyl radical (OH•), the carbonate radical anion (CO3•−), nitrogen dioxide (NO2•) and alkoxyl/alkyl peroxyl (RO•/ROO•). Hydrogen peroxide (H2O2), peroxynitrite (ONOO−)/peroxynitrous acid (ONOOH) and hypochlorous acid (HOCl) are the major nonradicals. ROS generation includes exogenous input and endogenous production from the mitochondrial electric transport chain, as well as the catalytic products of the enzymes nitric oxide synthase (NOS), NADPH oxidase (NOX) and myeloperoxidase. ROS can be transformed from one type to another by a sequence of reactions. The clearance of ROS relies on both enzymatic and nonenzymatic antioxidants. Enzymatic antioxidants include superoxide dismutase (SOD), catalase (CAT), the glutathione peroxidase (GPX) system, the thioredoxin (TRX) system and peroxiredoxin (PRDX). Nonenzymatic antioxidants include glutathione (GSH), uric acid, Vitamin E, Vitamin C and bilirubin [3].
Oxidative stress can cause oxidation of proteins, lipids and DNA, potentially leading to cytotoxicity and/or cell death, but oxidative stress can also result in modification of protein cysteine residues, allowing the transfer of redox signals within different cellular signal transduction pathways [6,7,8,9,10]. Protein thiols can act as nucleophiles to undergo one- and two-electron oxidation reactions, forming thiol radicals or sulfenic acids for further reaction [11]. Reactions between ROS and protein thiols leads to multiple post-translational modifications (PTMs), including S-sulfenation (−SOH), S-sulfination (−SO2H), S-sulfonation (−SO3H), S-nitrosylation (−SNO), persulfidation (S-sulfhydration, −SSH), S-glutathionylation (−SSG) and disulfide bond formation (−S–S−) [12]. Modification of critical signaling molecules driven by ROS initiates signaling in a broad range of cellular processes [5,9,10,13] (Table 1).
Table 1.
Redox-regulated cellular signaling [5].
| Cellular Processes | Critical Signaling Molecules Modified by ROS |
|---|---|
| proliferation and survival | mitogen-activated protein kinases (MAPKs), phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K), phosphatase and tensin homolog deleted on chromosome ten (PTEN) and protein tyrosine phosphatases |
| redox homeostasis | thioredoxin, peroxiredoxin, redox factor-1 (Ref-1) and Kelch-like ECH-associated protein 1 (Keap1)/nuclear factor erythroid 2-related factor 2 (Nrf2) |
| mitochondrial oxidative stress and aging | p66Shc |
| iron homeostasis | iron response element–iron regulatory protein (IRE-IRP) containing iron–sulfur cluster |
| DNA damage response | ataxia telangiectasia mutated (ATM) |
Oxidative stress can also activate heat shock transcription factors (HSFs) and redox-sensitive transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) by multiple mechanisms to induce gene expression related to cytoprotection including heat shock proteins (HSPs) and antioxidant enzymes [14,15,16]. Oxidation of proteins by ROS often results in damage to protein structure and alteration of protein function, thus disturbing protein homeostasis (proteostasis) [17]. Hsp70 is the key chaperone in protein quality control and the central hub of the cellular proteostasis network, participating in numerous cellular processes by interacting with different clients [18]. Hsp70 is tightly related to redox homeostasis in several ways, including functional regulation of Hsp70 caused by PTMs (especially cysteine modifications), induced expression of Hsp70 caused by oxidative stress, Hsp70-dependent proteostasis under oxidative stress and redox-related signaling pathways involving Hsp70 (Figure 1).
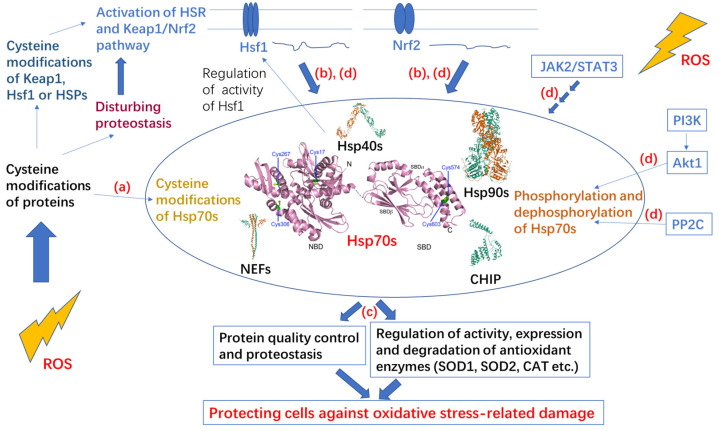
Figure 1.
Scheme of Hsp70 in redox homeostasis. Reactive oxygen species (ROS) often cause extensive cysteine modifications of proteins and disturb proteostasis, which can activate the heat shock response (HSR) and Kelch-like ECH-associated protein 1 (Keap1)/nuclear factor erythroid 2-related factor 2 (Nrf2) pathways by activation of heat shock transcription factor 1 (Hsf1) and redox-sensitive transcription factor Nrf2. ROS also widely modify different cellular signaling pathways. Hsp70s are involved in redox homeostasis in several aspects: (a) ROS can cause cysteine modifications of Hsp70s to modify their functions; (b) activation of Hsf1 and Nrf2 elevate expression of HSP70s; (c) Hsp70s work as central hubs in the protein quality control network to maintain proteostasis, including eliminating oxidized proteins and regulating activity, expression and degradation of antioxidant enzymes to contribute to redox homeostasis; and (d) Hsp70s are involved in redox-related signaling pathways, leading to phosphorylation or upregulated expression of Hsp70s.
2. Hsp70 System
Hsp70 members are highly conserved, ubiquitous and extremely important for maintenance of life in all organisms apart from some archaea [19,20]. Hsp70 members are multifunctional proteins and participate in multiple processes related to protein quality control, including de novo protein folding, refolding of misfolded proteins, protein transport, assembly and disassembly of protein complexes, prevention of protein aggregation, solubilizing aggregated proteins, translocation of proteins across membranes, regulating protein activity, cooperating with downstream chaperones such as Hsp90, which is responsible for the final folding and maturation of some substrate proteins, and cooperating with the cellular degradation machinery [21]. Hsp70 members not only have housekeeping functions to promote maturation of clients and adjust their activity in different cellular processes but also guard cells against disruption of protein homeostasis due to proteotoxic stresses, pathophysiological conditions and organismal aging [21,22,23]. From bacteria to humans, there are multiple Hsp70 members that exert overlapped and nonoverlapped functions, and more complicated regulation mechanisms of Hsp70 have evolved to adapt to the complex clients and cellular environment in higher organisms [24,25]. There are three Hsp70 members in Escherichia coli, 14 members in Saccharomyces cerevisiae and 17 members in Homo sapiens (including several Hsp110 proteins which share a similar structure with typical Hsp70s and generally act as nuclear exchange factors (NEFs) of typical Hsp70s) (Table 2). In eukaryotes, different Hsp70 members are often expressed in distinct cellular compartments (e.g., cytosol, nucleus, ER or mitochondria), and the expression levels are regulated as required under different conditions, such as growth, stress, disease, aging and tissue-specific activities (Table 2) [25].
Table 2.
Cysteine residues in bacteria, yeast and human Hsp70 homologs.
| Hsp70 Homolog | Cysteine Residues (Entry in UniProtKB) |
|---|---|
| Escherichia coli DnaK | Cys15 (P0A6Y8) |
| Escherichia coli HscA (Hsc66) | Cys315, Cys448 (P0A6Z1) |
| Escherichia coli HscC (Hsc62) | Cys236, Cys242, Cys261, Cys344, Cys360 (P77319) |
| Saccharomyces cerevisiae Ssa1 (cytosol) | Cys15, Cys264, Cys303 (P10591) |
| Saccharomyces cerevisiae Ssa2 (cytosol) | Cys15, Cys264, Cys303 (P10592) |
| Saccharomyces cerevisiae Ssa3 (cytosol) | Cys15, Cys304 (P09435) |
| Saccharomyces cerevisiae Ssa4 (cytosol) | Cys15, Cys304 (P22202) |
| Saccharomyces cerevisiae Ssb1 (cytosol) | Cys20, Cys435, Cys454 (P11484) |
| Saccharomyces cerevisiae Ssb2 (cytosol) | Cys20, Cys454 (P40150) |
| Saccharomyces cerevisiae Sse1 (cytosol Hsp110) | Cys142, Cys211, Cys228, Cys380, Cys484 (P32589) |
| Saccharomyces cerevisiae Sse2 (cytosol Hsp110) | Cys142, Cys211, Cys380, Cys484 (P32590) |
| Saccharomyces cerevisiae Ssz1 (cytosol) | Cys81, Cys86 (P38788) |
| Saccharomyces cerevisiae Ssc1 (mitochondria) | None (P0CS90) |
| Saccharomyces cerevisiae Ssc2 (Ssq1) (mitochondria) | Cys134 (Q05931) |
| Saccharomyces cerevisiae Ssc3 (Ecm10) (mitochondria) | None (P39987) |
| Saccharomyces cerevisiae Ssd1 (Kar2, BiP, Grp78) (ER) | Cys63 (P16474) |
| Saccharomyces cerevisiae Lhs1 (Grp170) (ER) | Cys520, Cys545, Cys547 (P36016) |
| Human HspA1A (Hsp72) (cytosol, nucleus, cell membrane, extracellular exosomes) | Cys17, Cys267, Cys306, Cys574, Cys603 (P0DMV8) |
| Human HspA1B (Hsp72) (cytosol, nucleus, extracellular exosomes) | Cys17, Cys267, Cys306, Cys574, Cys603 (P0DMV9) |
| Human HspA1L (cytosol, nucleus) | Cys19, Cys269, Cys308, Cys576, Cys605, Cys617, Cys622 (P34931) |
| Human HspA2 (cytosol, nucleus, cell membrane, extracellular exosomes) | Cys18, Cys191, Cys270, Cys577, Cys606 (P54652) |
| Human HspA4 (Apg2) (cytosol, extracellular exosome, mitochondrion, nucleus) | Cys13, Cys34, Cys38, Cys140, Cys146, Cys167, Cys213, Cys245, Cys270, Cys290, Cys310, Cys376, Cys380, Cys417, Cys779 (P34932) |
| Human HspA4L (Apg1, Osp94) (cytosol, nucleus) | Cys13, Cys34, Cys38, Cys140, Cys167, Cys213, Cys245, Cys270, Cys290, Cys310, Cys376, Cys380, Cys417, Cys421, Cys540, Cys589, Cys740, Cys782 (O95757) |
| Human HspA5 (BiP, Grp78) (ER, extracellular exosomes) | Cys41, Cys420 (P11021) |
| Human HspA6 (Hsp70B’) (cytosol, extracellular exosomes) | Cys19, Cys108, Cys269, Cys308, Cys387, Cys576, Cys605, Cys624 (P17066) |
| Human HspA7 (Hsp70B) (blood microparticles, extracellular exosomes) | Cys19, Cys108, Cys269, Cys308 (P48741) |
| human HspA8 (Hsc70, Hsc73) (cytosol, nucleus, cell membrane, extracellular exosomes) | Cys17, Cys267, Cys574, Cys603 (P11142) |
| Human HspA9 (Grp75, mt-Hsp70) (mitochondria, nucleus) | Cys66, Cys317, Cys366, Cys487, Cys608 (P38646) |
| Human HspA12A (extracellular exosomes, nucleus) | Cys80, Cys246, Cys502, Cys564, Cys621 (O43301) |
| Human HspA12B (endothelial cells, intracellular, blood plasma) | Cys36, Cys106, Cys250, Cys321, Cys365, Cys450, Cys570, Cys595, Cys610, Cys611, Cys626, Cys639 (Q96MM6) |
| Human HspA13 (Stch) (ER, extracellular exosomes, microsomes) | Cys43 (P48723) |
| Human HspA14 (Hsp60, Hsp70L1) (cytosol, membrane) | Cys10, Cys14, Cys89, Cys280, Cys293, Cys304, Cys311, Cys335, Cys394, Cys440, Cys492, Cys500 (Q0VDF9) |
| Human HspH1 (Hsp105, Hsp110) (microtubule, cytosol, extracellular region or secreted, nucleus) | Cys13, Cys34, Cys48, Cys140, Cys167, Cys213, Cys245, Cys270, Cys290, Cys310, Cys376, Cys380, Cys516, Cys650, Cys658, Cys796, Cys845 (Q92598) |
| Human Hyou1 (Grp170, Orp150) (ER, extracellular region or secreted) | Cys15, Cys240, Cys352, Cys805 (Q9Y4L1) |
Hsp70 members often share conserved sequence and structure, and the canonical Hsp70 structure is composed of an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD) joined by a flexible linker (Figure 2) [26]. The NBD has an actin-like fold containing two lobes (I and II), which can be further subdivided into four subdomains (IA, IB, IIA and IIB) (Figure 2) [27]. The deep cleft between the two lobes accommodates binding of ATP/ADP with the interaction with all four subdomains, controlling lobe movements [28]. The SBD consists of a β-sandwich substrate-binding subdomain (SBDβ) containing around eight β-strands and linking loops, an α-helical lid subdomain (SBDα) containing four to five α-helixes and a disordered and highly variable C-terminal tail (Figure 2) [29]. The SBDβ can accommodate exposed hydrophobic segments in nonnative polypeptides with its central hydrophobic cleft [29]. Mutations in the central hydrophobic cleft are often fatal for chaperone activity of Hsp70 [30]. Some mutations in the β6/β7 region of the Hsp70 substrate-binding domain are defective in prion propagation and heat-shock phenotypes in Saccharomyces cerevisiae [31,32]. In eukaryotic cytosolic and nuclear Hsp70 members, the C-terminal tail frequently ends with a conserved tetratricopeptide-repeat-domain-interaction EEVD motif, facilitating interactions with cochaperones [33,34]. In the E. coli Hsp70 homologue, DnaK, a ~15-residue motif in the C-terminal tail, can directly bind to misfolded protein substrates and enhance the protein refolding efficiency of DnaK [35]. In the Saccharomyces cerevisiae Hsp70 homolog, Ssa1, the 20-residue GGAP motif in the C-terminal tail, contributes to cell tolerance to temperature and cell-wall damage stress [36].
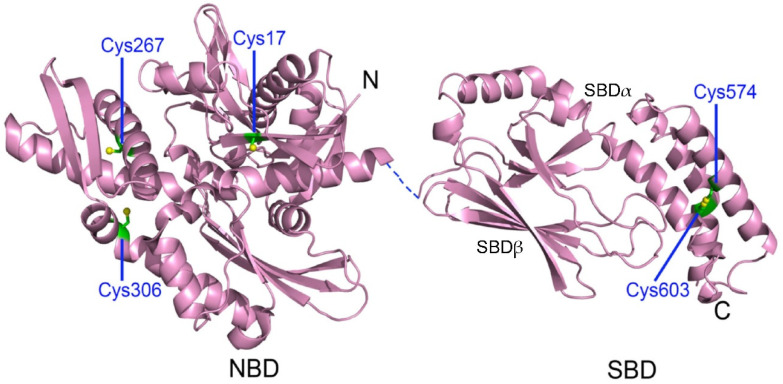
Figure 2.
Crystal structures of human HspA1A. The nucleotide-binding domain (NBD) in the ADP-bound state (PDB code 3AY9) and the substrate-binding domain (SBD, PDB code 4PO2) are shown. The dashed line represents the flexible linker between the NBD and SBD. The SBD contains a β-sandwich substrate-binding subdomain (SBDβ) and an α-helical lid subdomain (SBDα). The five Cys residues are labeled in green. Figure reproduced from ref. [52].
The conformation and function of Hsp70 are tightly related to allosteric mechanisms and binding of nucleotide and substrate [37]. In ADP-bound or nucleotide-free states, the two domains of Hsp70 remain separate as reflected by the NMR structure of DnaK [26]. ATP binding induces docking of the NBD and SBD with the association of the NBD and SBDꞵ, together with separation of the SBDα and SBDꞵ [38,39,40], so that the detached SBDα leans on the NBD [39,40]. The ATP-bound state also contains different functional states in equilibrium, such as a restrained state, which restricts ATP hydrolysis and blocks the client, and a stimulated state, which hydrolyzes ATP rapidly and binds the client with rapid on-and-off kinetics for DnaK [41]. The conformations between NBD and SBD show more obvious heterogeneity in human cytosolic stress-inducible Hsp70 (HspA1A); in the ADP-bound or nucleotide-free states, the NBD-SBD docked fraction is also observed in single-molecule fluorescence resonance energy transfer (smFRET) and NMR studies [38,42]. Generally, ATP binding and docking of the two domains tends to result in low substrate affinity and low ATPase activity [39]. Substrate binding to ATP-bound Hsp70 can change the equilibrium between the restrained state and the stimulated state and disrupt the interaction between the NBD and SBDꞵ, leading to undocking of the NBD and SBD and facilitating ATP hydrolysis [43]. ADP-bound Hsp70 has high affinity for substrates [43]. ATP-ADP exchange in the NBD restores Hsp70 to the ATP-bound state and facilitates the release of bound substrate [38]. ATP hydrolysis and ATP-ADP exchange in the NBD, and binding and release of substrates in the SBD, are coupled to form the functional cycle of Hsp70, which promotes protein folding and/or provides protection for clients by repeated temporary interactions. Hsp40 proteins trigger undocking of the NBD and SBDꞵ to stimulate ATP hydrolysis by Hsp70 and transfer substrates to Hsp70, and NEFs accelerate ATP-ADP exchange in the NBD by prompting ADP release [37,44,45,46]. Different Hsp40s and NEFs specifically tune this cycle, and the number of cochaperones has been observed to increase with evolution to guide Hsp70 to wider and more complex functions and regulatory mechanisms [20,21,47]. The functions of Hsp70 members are further extended by linking with other components of the proteostasis network such as other chaperones, including small heat shock proteins (sHSPs), Hsp60, Hsp90 and Hsp100, and protein degradation systems, including the ubiquitin–proteasome system (UPS) and chaperone-mediated autophagy (CMA) [21,48,49,50,51].
3. Post-Translational Modifications of Hsp70 under Oxidative Stress
ROS not only directly oxidize cysteine thiols in Hsp70 proteins but also cause covalent modification by production of lipoperoxides, altering the structure and chaperone activity of Hsp70 and affecting related cellular processes. Oxidation of extracellular Hsp70 alters its structure and signaling effects on macrophage function and viability, leading to lower proliferation, lower phagocytic activity and reduced TNF-α release [53]. These PTMs often inactivate the foldase activity of Hsp70 and change interaction with Hsp70 to modulate the activity of its clients and related signaling pathways. Thus, Hsp70 members can also act as redox sensors to transfer redox signals. For example, yeast Hsp70 Ssa1 can act as a sensor of thiol-reactive compounds, which probably involves modification of Cys264 and Cys303, activating Hsf1-mediated cytoprotection [54].
3.1. Cysteine Oxidation of Hsp70 and Redox Homeostasis
Most Hsp70 members contain at least one Cys residue, but different Hsp70 members vary in their distributions of Cys residues (Table 2). The number of Cys residues in Hsp70 increases with evolution, indicating the importance of cysteines for the function of Hsp70 to fit increasingly complex cellular environments (Table 2). These Cys residues often undergo different modifications under oxidative stress conditions to link Hsp70 with redox homeostasis (Table S1). Cys residues in Hsp70 generally show modest reactivity in global profiling of the cysteinome [55]. Over the past twenty years development of methods for enrichment of Cys-modified proteins and mass spectrometry (MS) detection has allowed extensive identification of cysteine modifications in Hsp70 members (Table S1), which have been summarized in databases for protein cysteine modifications [56,57,58,59,60]. Once cysteine modifications have been identified, the next goal is to identify the effects of cysteine modifications on the structure and function of Hsp70 and the physiological significance of these changes.
Cysteine modifications can interconvert, and different cysteine modifications are often detected at the same cysteine residue [61]. Glutathionylation, sulfenic modification, S-nitrosylation and S-sulfhydration of multiple Hsp70 members are frequently identified by MS in a variety of organisms (Table S1). Most human Hsp70 members containing reactive Cys residues have been detected as undergoing cysteine modifications, suggesting that they can sense redox change and participate in regulation of redox homeostasis (Table S1, Figure 3). Among these members, HspA1A/B, HspA4, HspA4L, HspA8 and HspH1 are the most frequently detected human Hsp70 homologues when screening for cysteine modifications (Table S1), possibly due to their higher cysteine reactivity or higher protein levels in cells.
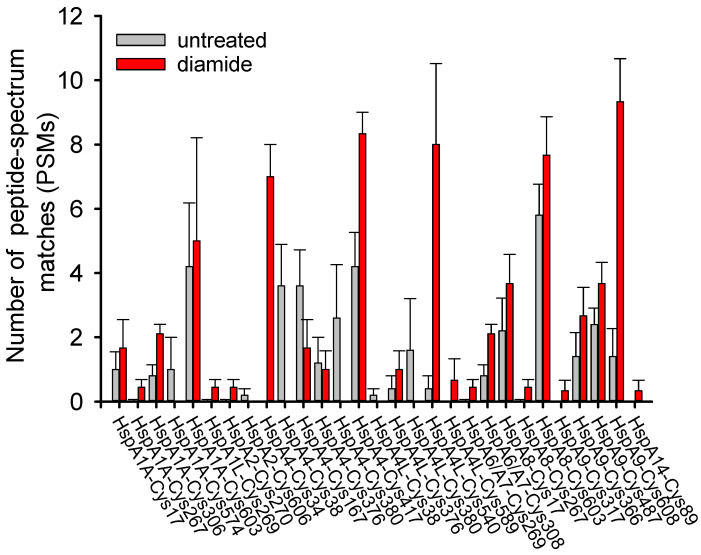
Figure 3.
Glutathionylation of the cysteine residues of Hsp70 family members in HeLa cells with or without diamide treatment detected by mass spectrometry. Figure adapted from ref. [52].
E. coli DnaK only has one Cys residue, Cys15, which lies on the β-sheet surface of the IA subdomain of the NBD. The cysteine at this position is highly conserved among Hsp70 family members, suggesting its functional importance (Figure 4, corresponding to Cys17 in Figure 2). Thiol reactivity of this cysteine is affected by nucleotides and substrate binding and thus is closely related with allostery of DnaK, and only the apo state of DnaK favors glutathionylation [62]. Glutathionylation has a dramatic effect on the structure and function of DnaK, resulting in altered allostery of DnaK including decreased ATPase activity and substrate-binding ability [62]. Glutathionylated DnaK also loses the ability to cooperate with its cochaperones DnaJ and GrpE to promote refolding of luciferase [62]. However, deglutathionylation can fully restore the structure and function of DnaK, and so the alteration caused by glutathionylation is fully reversible [62]. Thus, glutathionylation can turn off chaperone activity of DnaK under oxidative stress conditions, and this activity is recovered under reducing conditions. Chaperone activity of Hsp70 is related to suppression of activation of some proteins, such as heat shock response (HSR) transcription factors. In E. coli, binding of sigma32 to DnaK facilitates its degradation to repress the HSR under normal conditions and disabling of DnaK caused by glutathionylation can contribute to prolonging the lifetime of sigma32 and its activation of the HSR [63,64]. Thus, glutathionylation of DnaK not only protects DnaK from irreversible oxidative damage but also provides a link between oxidative stress and the HSR. Under oxidative heat treatment, H2O2 causes S-sulfenation, S-sulfination and S-sulfonation of a proportion of the cellular DnaK, resulting in its inactivation [65].
Figure 4.
The most conserved Cys residue in Hsp70 homologues. The conserved Cys is indicated by the green dashed box. In each line the last number indicates the position of the last residue in the corresponding Hsp70 homologue.
Human HspA1A contains five Cys residues: Cys17, Cys267 and Cys306, located in the NBD, and Cys574 and Cys603, located in the SBDα (Figure 2). All five Cys residues have the possibility to undergo glutathionylation in untreated or diamide-treated HeLa cells (Figure 3) [52]. Nucleotides inhibit modification of Cys residues in the NBD but not in the SBDα, and binding of peptide substrate has no effect on the activity of these cysteine residues of HspA1A [52]. Glutathionylation of Cys574 and Cys603, located in the SBDα, causes unfolding of the α helix bundle of the SBDα, and the unfolded region mimics the substrate to bind to and block the substrate-binding site in the SBDꞵ, resulting in promotion of intrinsic ATPase activity and decreased binding of external substrates, including heat shock transcription factor Hsf1 [52]. Similar to glutathionylation of DnaK, glutathionylation of HspA1A is fully reversible, allowing its structure and function to be fully restored [52]. Thus, glutathionylation and deglutathionylation of HspA1A caused by ROS followed by restoration of reducing conditions can act as a switch to turn the chaperone activity of HspA1A off and on and to transfer redox signals to its clients. The cysteine activities of the five Cys residues are different, for example Cys603 is more active than Cys574, and so can act as a redox sensor in HspA1A [52]. S-nitrosylation of Cys17, Cys306, Cys574 and Cys603 in HspA1A upon GSNO treatment or NO signaling and S-sulfhydration of Cys306 and Cys574 in HspA1A upon hydrogen sulfide (H2S) signaling have also been identified [66,67,68,69,70,71].
Human HspA8, which undertakes more housekeeping chaperone functions, including assisting de novo folding of newly synthesized peptides and CMA, shares 86% identity with stress-inducible HspA1A and has four Cys residues at the same positions as HspA1A but lacks Cys306. HspA1A and HspA8 are mainly located in the cytosol, nucleus, cell membrane and extracellular exosomes [25]. Expression of HSPA8 is constitutive and only slightly induced upon stress, while expression of HSPA1A varies in different tissues and is dramatically induced upon stress [25]. All four Cys residues in HspA8 have been detected as undergoing glutathionylation (Figure 3) [52,58]. However, HspA8 has much lower cysteine reactivity than HspA1A, indicating their different redox sensitivity and functional regulation [52]. The redox-active compound methylene blue (MB) cannot cause modification of Cys residues of HspA8 but can cause S-sulfenation of Cys306 in HspA1A, leading to exposure and oxidation of Cys267, resulting in decreased ATPase activity of HspA1A [72]. Thus, Cys306 distinguishes the different redox sensing of HspA1A and HspA8. The different sensitivity to redox and cysteine modifications of HspA1A and HspA8 may be related to their different functions under stress. S-sulfenation of Cys17, Cys574 and Cys603 in HspA8 and S-sulfinylation of Cys603 in HspA8 upon H2O2 treatment have also been detected by MS [73,74,75]. Expression of neuronal nitric oxide synthase (nNOS/NOS1) in SH-SY5Y cells releases nitric oxide (NO) and leads to S-nitrosylation of Cys17, Cys574 and Cys603 in HspA8 [76]. However, the regulation effect and physiological significance of S-nitrosylation of HspA8 is still unknown. Upon hydrogen sulfide (H2S) signaling, Cys574 and Cys603 in HspA8 also undergo S-sulfhydration [70,71].
In the ER, oxidative folding of proteins often generates ROS as a by-product. Yeast ER Hsp70 BiP (Kar2), like E. coli DnaK, has only one Cys residue, Cys63, which corresponds to the highly conserved Cys15 in DnaK (BiP has an extra N-terminal signal sequence) (Figure 4). Glutathionylation of yeast BiP is detected during ER oxidative stress and mediated by the cellular GSSG/GSH ratio, which is often measured to reflect the cellular redox status [77,78]. Glutathionylation decouples ATPase and peptide-binding activities of BiP, turning BiP from an ATP-dependent foldase into an ATP-independent holdase, which helps prevent protein aggregation [77]. The effect of glutathionylation on chaperone activity of BiP is also fully reversible upon removal of glutathionylation [77]. Glutathionylation promotes cell proliferation during oxidative stress, possibly due to the enhanced holdase activity of glutathionylated BiP. During oxidative stress, H2O2 also causes S-sulfenation of BiP, which leads to enhanced holdase activity of BiP to prevent protein aggregation, similar to the effects of glutathionylation. S-sulfenation of BiP can be converted into glutathionylation by reacting with GSH or can be converted into irreversible S-sulfination and/or S-sulfonation by further oxidation with H2O2 [77,79]. Thus, glutathionylation of BiP may also serve to prevent irreversible oxidation of BiP and preserve the capacity of BiP to undergo Cys reduction. Substituting the Cys residue in BiP with other amino acids to mimic unmodified or oxidized BiP results in decreased or increased cell viability in the presence of oxidants, respectively [80]. This indicates that levels of oxidized BiP can be tuned to maintain ER folding homeostasis under a range of redox conditions.
3.2. Covalent Modifications of Hsp70 and Redox Homeostasis
Oxidative stress often causes oxidation of lipids and overproduction of 4-hydroxynonenal (4-HNE), which is derived from peroxidation of polyunsaturated fatty acids [81]. 4-HNE is a highly reactive electrophile and can react with proteins, phospholipids and nucleic acids [81]. It can covalently modify His, Cys and Lys residues, causing functional impairment of proteins, leading to inhibition of enzyme activity and induction of apoptosis [82,83,84]. Thus, 4-HNE can act as a second messenger of free radicals and a growth regulatory factor [82,83,84]. The antibiotic plakortin can generate ROS and 4-HNE as products of lipoperoxides [85]. In plakortin-treated Plasmodium falciparum, 4-HNE-modified PfHsp70-1 and PfHsp70-2 (the BiP analogue) have been identified [85]. 4-HNE modification of Cys267 in rat Hsp72 was identified and found to decrease affinity of Hsp72 for ATP and reduce its luciferase refolding activity [86]. In a mouse model of alcoholic liver disease (ALD), Grp78 (a BiP analogue) undergoes modification by 4-HNE at some Lys and His residues within the NBD [87]. The 4-HNE modification decreases ATP binding and ATPase activity of Grp78 [87]. In human retinal pigment epithelial cells (ARPE-19), 4-HNE-modified Hsp70 cannot protect its client XIAP, resulting in promotion of XIAP degradation and apoptosis [88].
4. Protection Effect and Upregulated Expression of Hsp70 under Oxidative Stress
Hsp70 can counteract oxidative stress damage to proteins by inhibiting aggregation and/or facilitating degradation of oxidized proteins, as well as inducing the expression of key antioxidant enzymes such as SOD1 and CAT and adjusting the activity of antioxidant enzymes [15,89,90,91,92]. Overexpression of HSP70 in the muscle of mice antagonizes the aging-related increase in protein cysteine modifications including carbonylation, oxidation and formation of disulfide bonds [93]. Overexpression of mitochondrial HSP70/Hsp75 in rat brain protects mitochondria and results in the marked reduction in free radical generation [94]. Under oxidative stress, HspA1A and HspA8 can mediate the C-terminus of Hsp70-interacting protein (CHIP, also known as STUB1) to recognize and ubiquitinate ROS-stressed peroxisomes, resulting in their turnover by autophagy [95]. Overexpression of HSP70 suppresses ROS production from mitochondria in human lung microvascular endothelial cells (HLMVEC) exposed to bacterial toxins [96]. The HSPA1B rs1061581 polymorphism (1267 A/G), which is linked to the risk of developing multiple sclerosis, is related to ROS levels and has a role in the variation in HSPA1B expression levels under oxidative stimulus [97].
Oxidative stress can activate the HSR and Kelch-like ECH-associated protein 1 (Keap1)/Nrf2 pathway to upregulate expression of HSP70, and the increased protein level of Hsp70 protects cells against oxidative-stress-related damage [15,98,99]. However, different types of oxidative stress differ in their capacity to induce the HSR and elevate expression of HSP70, and the same oxidative stimulus may lead to distinct stress responses in different cells [100,101,102]. Oxidative-stress induced by 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) enhances HSP70 expression to exert anti-inflammatory effects [103]. Under mild oxidative stress conditions, expression of HSPA1A/B is upregulated in the mammalian neuronal cell line HT22 [90,104]. In the early phase after oxidative stress, HspA1A/B binds to partially unfolded oxidized proteins to prevent their aggregation [104]. Then, HspA1A/B facilitates the degradation of bound oxidized proteins by 20S proteasomes [104]. Expression of HspA8 is not affected by oxidative stress, and HspA8 has no effect on removal of proteins damaged by oxidation by the 20S proteasome [104]. Thus, HspA1A/B but not HspA8 mediates degradation of damaged proteins by the UPS under oxidative stress conditions [104], while HspA8-facilitated CMA is also activated by oxidative stress to facilitate degradation of oxidized proteins [105,106].
5. Hsp70 Participates in Multiple Redox-Related Signaling Pathways
Hsp70 and redox homeostasis are linked by multiple signaling pathways including inositol-4,5-bisphosphate 3-kinase (PI3K)/protein kinase B (PKB, Akt)/Hsp70, Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3)/Hsp70, and Keap1/Nrf2/Hsf1/Hsp70. Expression or activity of Hsp70 is often modulated by the related signaling pathways [107,108,109].
SOD2 protects cells against mitochondrial oxidative damage [110]. HspA1A can bind to SOD2 to prevent its aggregation and control its CHIP-mediated degradation or import it into mitochondria [107,111]. CHIP cooperates with HspA1A by binding to the C-terminus EEVD motif and the SBDα of HspA1A [112]. PI3K/Akt signaling is crucial for receiving input from the cell membrane and mitochondria [113]. Akt1 is the downstream kinase of PI3K and a client of HspA1A. Akt1 binds to the SBDα of HspA1A and phosphorylates the C-terminus Ser631 of HspA1A, resulting in decoupling of HspA1A and CHIP and inhibition of the degradation of SOD2 [107]. Then, the import of SOD2 into mitochondria is promoted. As a negative feedback loop, the resulting decrease in oxidative stress activation permits phosphatase 2C (PP2C) to dephosphorylate HspA1A to promote degradation of SOD2 [107]. Thus, Hsp70 phosphorylation involved in the PI3K/Akt/Hsp70 signaling pathway contributes to redox homeostasis.
The oncogenic signaling pathway JAK2/STAT3 plays crucial roles in regulating apoptosis, proliferation, differentiation, and the inflammatory response and participates in the occurrence and development of various tumors [114]. AG490 is an inhibitor of the JAK2/STAT3 signaling pathway [115]. It was found that H2O2-induced time-dependent increase in Hsp70 protein expression in vascular smooth muscle cells is inhibited by pretreatment with AG-490, suggesting that ROS can regulate HSP70 expression via the JAK2/STAT3 signaling pathway [116]. It was also found that both HSP70 RNAi and AG490 can increase ROS levels in Burkitt’s lymphoma Raji cells and reduce the activity of SOD and GPX, suggesting that inhibiting Hsp70 or the JAK2/STAT3 pathway may induce oxidative stress of Raji cells [108]. Therefore, the JAK2/STAT3 signaling pathway may contribute to redox homeostasis by modulating the expression of HSP70.
The Keap1/Nrf2 pathway is a thiol-based sensor–effector for maintaining redox homeostasis [16]. Keap1 acts as a cysteine-thiol-rich sensor of redox status, and Nrf2 is a transcription factor for some cytoprotective genes including some enzymatic antioxidants and Hsp70 [16]. The Keap1/Nrf2 complex facilitates degradation of Nrf2 and represses the activity of Nrf2 [16]. ROS or electrophiles can cause cysteine modification of Keap1, which contains multiple active thiols, to weaken the interaction between Keap1 and Nrf2 and promote the entrance of Nrf2 into the cell nucleus and activation of Nrf2 [16,117]. Hsf1 is an important transcription factor for genes related to HSR. Hsp90, Hsp70 and Hsp40 cooperate to inactivate Hsf1 by binding to it and preventing it from entering the cell nucleus, and they are also transcription targets of Hsf1, forming a negative feedback regulatory loop [118,119]. Although the mechanism by which Hsf1 is activated has not been fully elucidated, activation of Hsf1 is thought to be related to stress damage of proteins and modification of HSPs or Hsf1 [117,120]. Both Nrf2 and Hsf1 are critical for adaptation and survival, and there is crosstalk between the two pathways through overlapping transcriptional targets including HSP70 [99]. Some reports indicate that the Keap1/Nrf2 pathway is activated before the heat shock response upon oxidative or electrophilic stress [99,121,122]. When Hsf1 is mutated and therefore unable to activate the HSR, Nrf2 can be activated by heat shock to cause delayed upregulation of HSP70, suggesting the two pathways may compensate for each other [123]. Oxidative stress resulting from a lack of methionine induces HSPA1A expression through Nrf2 activation but not Hsf1 activation in HEK293 cells [109]. Both Hsf1 and Nrf2 pathways contribute to redox homeostasis and cooperate to promote a more reducing cellular environment.
6. Conclusions and Perspectives
Redox homeostasis relies on the balance between ROS generation and the antioxidation system. Hsp70 can buffer different kinds of stress including oxidative stress. Hsp70 members are important antioxidative components in eliminating damaged oxidized proteins and support antioxidative enzymes. Under oxidative stress, Hsp70 members will inevitably undergo oxidization, resulting in changes in their structure and chaperone activity. However, cysteine modifications often downregulate the chaperone activity of Hsp70, which is disadvantageous for dealing with protein damage due to oxidation. From this point of view, cysteine residues in Hsp70 are not conducive towards Hsp70 exerting its antioxidative functions. At the same time, only very few Hsp70 members do not contain any cysteine residues, and the number of cysteine residues in Hsp70 increases with evolution, suggesting important roles of cysteine residues in Hsp70. Hsp70 members can sense redox by cysteine modifications, then transfer redox signals by modulating the interaction between Hsp70 members and their clients. Hsp70 members are involved in cellular signaling due to the fact that their clients include key molecules in signaling pathways, including Hsf1 and Akt1. Cochaperones also guide Hsp70 members within different cellular processes, such as CHIP, acting as a link between Hsp70 and the UPS for degradation of Hsp70 clients. Thus, cysteine modifications of Hsp70 also change the fate of the clients by modifying the cooperation between Hsp70 and cochaperones. Therefore, cysteine modifications of Hsp70 may act to amplify the transfer of redox signaling to a broader range of proteins, achieving greater antioxidative potential in a shorter time and restoring the cellular environment by multiple pathways. As a consequence, Hsp70 proteins are expressed much more rapidly through different signaling pathways, and the newly synthesized Hsp70 proteins are not oxidized and have normal chaperone activity to deal with the damaged proteins that have accumulated during oxidative stress. Thus, combining an in-depth understanding of the basic principles of Hsp70 systems (including the relationship between the structure and function of Hsp70, the interaction between Hsp70 and its cochaperones, and the interaction between Hsp70 and its clients) and the extensive exploration of redox-related signaling pathways involving Hsp70 will lead to a fuller understanding of Hsp70 function in redox homeostasis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11050829/s1, Table S1: Cysteine modifications identified in Hsp70 members. References [124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163] were cited in the supplementary materials.
Author Contributions
H.Z. and S.P. conceived and wrote the review; all authors revised the review and approved the final version. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge support from the Chinese Ministry of Science and Technology (2017YFA0504000) and the National Natural Science Foundation of China (31920103011, 31770829, 32171443).
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holmström K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 2.Kong H., Chandel N.S. Regulation of redox balance in cancer and T cells. J. Biol. Chem. 2018;293:7499–7507. doi: 10.1074/jbc.TM117.000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Wang X., Cueto R., Effi C., Zhang Y., Tan H., Qin X., Ji Y., Yang X., Wang H. Biochemical basis and metabolic interplay of redox regulation. Redox Biol. 2019;26:101284. doi: 10.1016/j.redox.2019.101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sies H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ray P.D., Huang B.W., Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabak O., Gelisgen R., Erman H., Erdenen F., Muderrisoglu C., Aral H., Uzun H. Oxidative lipid, protein, and DNA damage as oxidative stress markers in vascular complications of diabetes mellitus. Clin. Investig. Med. 2011;34:E163–E171. doi: 10.25011/cim.v34i3.15189. [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Wang X., Shigenaga M.K., Yeo H.C., Mori A., Ames B.N. Immobilization stress causes oxidative damage to lipid, protein, and DNA in the brain of rats. Faseb J. 1996;10:1532–1538. doi: 10.1096/fasebj.10.13.8940299. [DOI] [PubMed] [Google Scholar]
- 8.Yang W.S., Stockwell B.R. Ferroptosis: Death by Lipid Peroxidation. Trends. Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forman H.J., Ursini F., Maiorino M. An overview of mechanisms of redox signaling. J. Mol. Cell Cardiol. 2014;73:2–9. doi: 10.1016/j.yjmcc.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forman H.J. Redox signaling: An evolution from free radicals to aging. Free Radic. Biol. Med. 2016;97:398–407. doi: 10.1016/j.freeradbiomed.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turell L., Zeida A., Trujillo M. Mechanisms and consequences of protein cysteine oxidation: The role of the initial short-lived intermediates. Essays Biochem. 2020;64:55–66. doi: 10.1042/EBC20190053. [DOI] [PubMed] [Google Scholar]
- 12.Yang J., Carroll K.S., Liebler D.C. The Expanding Landscape of the Thiol Redox Proteome. Mol. Cell Proteom. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szyller J., Bil-Lula I. Heat Shock Proteins in Oxidative Stress and Ischemia/Reperfusion Injury and Benefits from Physical Exercises: A Review to the Current Knowledge. Oxid. Med. Cell Longev. 2021;2021:6678457. doi: 10.1155/2021/6678457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalmar B., Greensmith L. Induction of heat shock proteins for protection against oxidative stress. Adv. Drug Deliv. Rev. 2009;61:310–318. doi: 10.1016/j.addr.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korovila I., Hugo M., Castro J.P., Weber D., Höhn A., Grune T., Jung T. Proteostasis, oxidative stress and aging. Redox Biol. 2017;13:550–567. doi: 10.1016/j.redox.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández-Fernández M.R., Valpuesta J.M. Hsp70 chaperone: A master player in protein homeostasis. F1000Research. 2018;7 doi: 10.12688/f1000research.15528.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Large A.T., Goldberg M.D., Lund P.A. Chaperones and protein folding in the archaea. Biochem. Soc. Trans. 2009;37:46–51. doi: 10.1042/BST0370046. [DOI] [PubMed] [Google Scholar]
- 20.Rebeaud M.E., Mallik S., Goloubinoff P., Tawfik D.S. On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life. Proc. Natl. Acad. Sci. USA. 2021;118:e2020885118. doi: 10.1073/pnas.2020885118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019;20:665–680. doi: 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- 22.Murphy M.E. The HSP70 family and cancer. Carcinogenesis. 2013;34:1181–1188. doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrestha L., Young J.C. Function and Chemotypes of Human Hsp70 Chaperones. Curr. Top. Med. Chem. 2016;16:2812–2828. doi: 10.2174/1568026616666160413142028. [DOI] [PubMed] [Google Scholar]
- 24.Kabani M., Martineau C.N. Multiple hsp70 isoforms in the eukaryotic cytosol: Mere redundancy or functional specificity? Curr. Genom. 2008;9:338–348. doi: 10.2174/138920208785133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radons J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertelsen E.B., Chang L., Gestwicki J.E., Zuiderweg E.R. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. USA. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuravleva A., Gierasch L.M. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc. Natl. Acad. Sci. USA. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Zuiderweg E.R. The 70-kDa heat shock protein chaperone nucleotide-binding domain in solution unveiled as a molecular machine that can reorient its functional subdomains. Proc. Natl. Acad. Sci. USA. 2004;101:10272–10277. doi: 10.1073/pnas.0401313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P., Leu J.I., Murphy M.E., George D.L., Marmorstein R. Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with peptide substrate. PLoS ONE. 2014;9:e103518. doi: 10.1371/journal.pone.0103518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuravleva A., Gierasch L.M. Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc. Natl. Acad. Sci. USA. 2015;112:E2865–E2873. doi: 10.1073/pnas.1506692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L., Gong W., Cusack S.A., Wu H., Loovers H.M., Zhang H., Perrett S., Jones G.W. The β6/β7 region of the Hsp70 substrate-binding domain mediates heat-shock response and prion propagation. Cell Mol. Life Sci. 2018;75:1445–1459. doi: 10.1007/s00018-017-2698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L., Zhang H., Cuskelly D.D., Doyle S., Perrett S., Jones G.W. Mutational analysis of the Hsp70 substrate-binding domain: Correlating molecular-level changes with in vivo function. Mol. Microbiol. 2021;115:1262–1276. doi: 10.1111/mmi.14671. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y., Rossi P., Kalodimos C.G. Structural basis for client recognition and activity of Hsp40 chaperones. Science. 2019;365:1313–1319. doi: 10.1126/science.aax1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumura Y., Sakai J., Skach W.R. Endoplasmic reticulum protein quality control is determined by cooperative interactions between Hsp/c70 protein and the CHIP E3 ligase. J. Biol. Chem. 2013;288:31069–31079. doi: 10.1074/jbc.M113.479345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smock R.G., Blackburn M.E., Gierasch L.M. Conserved, disordered C terminus of DnaK enhances cellular survival upon stress and DnaK in vitro chaperone activity. J. Biol. Chem. 2011;286:31821–31829. doi: 10.1074/jbc.M111.265835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong W., Hu W., Xu L., Wu H., Wu S., Zhang H., Wang J., Jones G.W., Perrett S. The C-terminal GGAP motif of Hsp70 mediates substrate recognition and stress response in yeast. J. Biol. Chem. 2018;293:17663–17675. doi: 10.1074/jbc.RA118.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuiderweg E.R., Bertelsen E.B., Rousaki A., Mayer M.P., Gestwicki J.E., Ahmad A. Allostery in the Hsp70 chaperone proteins. Top Curr. Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S., Hong L., Wang Y., Yu J., Yang J., Yang J., Zhang H., Perrett S. Kinetics of the conformational cycle of Hsp70 reveals the importance of the dynamic and heterogeneous nature of Hsp70 for its function. Proc. Natl. Acad. Sci. USA. 2020;117:7814–7823. doi: 10.1073/pnas.1914376117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kityk R., Kopp J., Sinning I., Mayer M.P. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Qi R., Sarbeng E.B., Liu Q., Le K.Q., Xu X., Xu H., Yang J., Wong J.L., Vorvis C., Hendrickson W.A., et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., Liu Q., Liu Q., Hendrickson W.A. Conformational equilibria in allosteric control of Hsp70 chaperones. Mol. Cell. 2021;81:3919–3933. doi: 10.1016/j.molcel.2021.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng W., Clerico E.M., McArthur N., Gierasch L.M. Allosteric landscapes of eukaryotic cytoplasmic Hsp70s are shaped by evolutionary tuning of key interfaces. Proc. Natl. Acad. Sci. USA. 2018;115:11970–11975. doi: 10.1073/pnas.1811105115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuravleva A., Clerico E.M., Gierasch L.M. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faust O., Abayev-Avraham M., Wentink A.S., Maurer M., Nillegoda N.B., London N., Bukau B., Rosenzweig R. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature. 2020;587:489–494. doi: 10.1038/s41586-020-2906-4. [DOI] [PubMed] [Google Scholar]
- 45.Polier S., Dragovic Z., Hartl F.U., Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Bracher A., Verghese J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2015;2:10. doi: 10.3389/fmolb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu X.B., Shao Y.M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kravats A.N., Hoskins J.R., Reidy M., Johnson J.L., Doyle S.M., Genest O., Masison D.C., Wickner S. Functional and physical interaction between yeast Hsp90 and Hsp70. Proc. Natl. Acad. Sci. USA. 2018;115:E2210–E2219. doi: 10.1073/pnas.1719969115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler J., Tyedmers J., Bukau B., Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stankiewicz M., Nikolay R., Rybin V., Mayer M.P. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. Febs J. 2010;277:3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- 51.Dice J.F. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 52.Yang J., Zhang H., Gong W., Liu Z., Wu H., Hu W., Chen X., Wang L., Wu S., Chen C., et al. S-Glutathionylation of human inducible Hsp70 reveals a regulatory mechanism involving the C-terminal α-helical lid. J. Biol. Chem. 2020;295:8302–8324. doi: 10.1074/jbc.RA119.012372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grunwald M.S., Pires A.S., Zanotto-Filho A., Gasparotto J., Gelain D.P., Demartini D.R., Schöler C.M., de Bittencourt P.I., Jr., Moreira J.C. The oxidation of HSP70 is associated with functional impairment and lack of stimulatory capacity. Cell Stress Chaperones. 2014;19:913–925. doi: 10.1007/s12192-014-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Gibney P.A., West J.D., Morano K.A. The yeast Hsp70 Ssa1 is a sensor for activation of the heat shock response by thiol-reactive compounds. Mol. Biol. Cell. 2012;23:3290–3298. doi: 10.1091/mbc.e12-06-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weerapana E., Wang C., Simon G.M., Richter F., Khare S., Dillon M.B., Bachovchin D.A., Mowen K., Baker D., Cravatt B.F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun M.A., Wang Y., Cheng H., Zhang Q., Ge W., Guo D. RedoxDB--a curated database for experimentally verified protein oxidative modification. Bioinformatics. 2012;28:2551–2552. doi: 10.1093/bioinformatics/bts468. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X., Huang B., Zhang L., Zhang Y., Zhao Y., Guo X., Qiao X., Chen C. SNObase, a database for S-nitrosation modification. Protein Cell. 2012;3:929–933. doi: 10.1007/s13238-012-2094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y.J., Lu C.T., Lee T.Y., Chen Y.J. dbGSH: A database of S-glutathionylation. Bioinformatics. 2014;30:2386–2388. doi: 10.1093/bioinformatics/btu301. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y.J., Lu C.T., Su M.G., Huang K.Y., Ching W.C., Yang H.H., Liao Y.C., Chen Y.J., Lee T.Y. dbSNO 2.0: A resource for exploring structural environment, functional and disease association and regulatory network of protein S-nitrosylation. Nucleic Acids Res. 2015;43:D503–D511. doi: 10.1093/nar/gku1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang P., Zhang Q., Li S., Cheng B., Xue H., Wei Z., Shao T., Liu Z.X., Cheng H., Wang Z. iCysMod: An integrative database for protein cysteine modifications in eukaryotes. Brief Bioinform. 2021;22:bbaa400. doi: 10.1093/bib/bbaa400. [DOI] [PubMed] [Google Scholar]
- 61.Fratelli M., Gianazza E., Ghezzi P. Redox proteomics: Identification and functional role of glutathionylated proteins. Expert Rev. Proteomics. 2004;1:365–376. doi: 10.1586/14789450.1.3.365. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H., Yang J., Wu S., Gong W., Chen C., Perrett S. Glutathionylation of the Bacterial Hsp70 Chaperone DnaK Provides a Link between Oxidative Stress and the Heat Shock Response. J. Biol. Chem. 2016;291:6967–6981. doi: 10.1074/jbc.M115.673608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gamer J., Multhaup G., Tomoyasu T., McCarty J.S., Rüdiger S., Schönfeld H.J., Schirra C., Bujard H., Bukau B. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. Embo J. 1996;15:607–617. doi: 10.1002/j.1460-2075.1996.tb00393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Müller A., Hoffmann J.H., Meyer H.E., Narberhaus F., Jakob U., Leichert L.I. Nonnative disulfide bond formation activates the σ32-dependent heat shock response in Escherichia coli. J. Bacteriol. 2013;195:2807–2816. doi: 10.1128/JB.00127-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winter J., Linke K., Jatzek A., Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol. Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 66.Chung H.S., Murray C.I., Venkatraman V., Crowgey E.L., Rainer P.P., Cole R.N., Bomgarden R.D., Rogers J.C., Balkan W., Hare J.M., et al. Dual Labeling Biotin Switch Assay to Reduce Bias Derived From Different Cysteine Subpopulations: A Method to Maximize S-Nitrosylation Detection. Circ. Res. 2015;117:846–857. doi: 10.1161/CIRCRESAHA.115.307336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mnatsakanyan R., Markoutsa S., Walbrunn K., Roos A., Verhelst S.H.L., Zahedi R.P. Proteome-wide detection of S-nitrosylation targets and motifs using bioorthogonal cleavable-linker-based enrichment and switch technique. Nat. Commun. 2019;10:2195. doi: 10.1038/s41467-019-10182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lefievre L., Chen Y., Conner S.J., Scott J.L., Publicover S.J., Ford W.C., Barratt C.L. Human spermatozoa contain multiple targets for protein S-nitrosylation: An alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics. 2007;7:3066–3084. doi: 10.1002/pmic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang B., Chen S.C., Wang D.L. Shear flow increases S-nitrosylation of proteins in endothelial cells. Cardiovasc. Res. 2009;83:536–546. doi: 10.1093/cvr/cvp154. [DOI] [PubMed] [Google Scholar]
- 70.Fu L., Liu K., He J., Tian C., Yu X., Yang J. Direct Proteomic Mapping of Cysteine Persulfidation. Antioxid. Redox Signal. 2020;33:1061–1076. doi: 10.1089/ars.2019.7777. [DOI] [PubMed] [Google Scholar]
- 71.Wu Q., Zhao B., Weng Y., Shan Y., Li X., Hu Y., Liang Z., Yuan H., Zhang L., Zhang Y. Site-Specific Quantification of Persulfidome by Combining an Isotope-Coded Affinity Tag with Strong Cation-Exchange-Based Fractionation. Anal. Chem. 2019;91:14860–14864. doi: 10.1021/acs.analchem.9b04112. [DOI] [PubMed] [Google Scholar]
- 72.Miyata Y., Rauch J.N., Jinwal U.K., Thompson A.D., Srinivasan S., Dickey C.A., Gestwicki J.E. Cysteine reactivity distinguishes redox sensing by the heat-inducible and constitutive forms of heat shock protein 70. Chem. Biol. 2012;19:1391–1399. doi: 10.1016/j.chembiol.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu L., Liu K., Ferreira R.B., Carroll K.S., Yang J. Proteome-Wide Analysis of Cysteine S-Sulfenylation Using a Benzothiazine-Based Probe. Curr. Protoc. Protein Sci. 2019;95:e76. doi: 10.1002/cpps.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang J., Gupta V., Carroll K.S., Liebler D.C. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akter S., Fu L., Jung Y., Conte M.L., Lawson J.R., Lowther W.T., Sun R., Liu K., Yang J., Carroll K.S. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 2018;14:995–1004. doi: 10.1038/s41589-018-0116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valek L., Heidler J., Scheving R., Wittig I., Tegeder I. Nitric oxide contributes to protein homeostasis by S-nitrosylations of the chaperone HSPA8 and the ubiquitin ligase UBE2D. Redox Biol. 2019;20:217–235. doi: 10.1016/j.redox.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J., Sevier C.S. Formation and Reversibility of BiP Protein Cysteine Oxidation Facilitate Cell Survival during and post Oxidative Stress. J. Biol. Chem. 2016;291:7541–7557. doi: 10.1074/jbc.M115.694810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flohé L. The fairytale of the GSSG/GSH redox potential. Biochim. Biophys. Acta. 2013;1830:3139–3142. doi: 10.1016/j.bbagen.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 79.Wang J., Pareja K.A., Kaiser C.A., Sevier C.S. Redox signaling via the molecular chaperone BiP protects cells against endoplasmic reticulum-derived oxidative stress. Elife. 2014;3:e03496. doi: 10.7554/eLife.03496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu M., Marsh H.M., Sevier C.S. A Conserved Cysteine within the ATPase Domain of the Endoplasmic Reticulum Chaperone BiP is Necessary for a Complete Complement of BiP Activities. J. Mol. Biol. 2016;428:4168–4184. doi: 10.1016/j.jmb.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Breitzig M., Bhimineni C., Lockey R., Kolliputi N. 4-Hydroxy-2-nonenal: A critical target in oxidative stress? Am. J. Physiol. Cell Physiol. 2016;311:C537–C543. doi: 10.1152/ajpcell.00101.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaudhary P., Sharma R., Sharma A., Vatsyayan R., Yadav S., Singhal S.S., Rauniyar N., Prokai L., Awasthi S., Awasthi Y.C. Mechanisms of 4-hydroxy-2-nonenal induced pro- and anti-apoptotic signaling. Biochemistry. 2010;49:6263–6275. doi: 10.1021/bi100517x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jaganjac M., Cindrić M., Jakovčević A., Žarković K., Žarković N. Lipid peroxidation in brain tumors. Neurochem. Int. 2021;149:105118. doi: 10.1016/j.neuint.2021.105118. [DOI] [PubMed] [Google Scholar]
- 84.Reyes-Jiménez E., Ramírez-Hernández A.A., Santos-Álvarez J.C., Velázquez-Enríquez J.M., Pina-Canseco S., Baltiérrez-Hoyos R., Vásquez-Garzón V.R. Involvement of 4-hydroxy-2-nonenal in the pathogenesis of pulmonary fibrosis. Mol. Cell Biochem. 2021;476:4405–4419. doi: 10.1007/s11010-021-04244-9. [DOI] [PubMed] [Google Scholar]
- 85.Skorokhod O.A., Davalos-Schafler D., Gallo V., Valente E., Ulliers D., Notarpietro A., Mandili G., Novelli F., Persico M., Taglialatela-Scafati O., et al. Oxidative stress-mediated antimalarial activity of plakortin, a natural endoperoxide from the tropical sponge Plakortis simplex. Free Radic. Biol. Med. 2015;89:624–637. doi: 10.1016/j.freeradbiomed.2015.10.399. [DOI] [PubMed] [Google Scholar]
- 86.Carbone D.L., Doorn J.A., Kiebler Z., Sampey B.P., Petersen D.R. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 2004;17:1459–1467. doi: 10.1021/tx049838g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Galligan J.J., Fritz K.S., Backos D.S., Shearn C.T., Smathers R.L., Jiang H., MacLean K.N., Reigan P.R., Petersen D.R. Oxidative stress-mediated aldehyde adduction of GRP78 in a mouse model of alcoholic liver disease: Functional independence of ATPase activity and chaperone function. Free Radic. Biol. Med. 2014;73:411–420. doi: 10.1016/j.freeradbiomed.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang L.L., Chen H., Wang J., Xia T., Sun H., Yuan C.H., Liu S.L., Chen J.B. 4-HNE Induces Apoptosis of Human Retinal Pigment Epithelial Cells by Modifying HSP70. Curr. Med. Sci. 2019;39:442–448. doi: 10.1007/s11596-019-2057-8. [DOI] [PubMed] [Google Scholar]
- 89.Polla B.S., Kantengwa S., François D., Salvioli S., Franceschi C., Marsac C., Cossarizza A. Mitochondria are selective targets for the protective effects of heat shock against oxidative injury. Proc. Natl. Acad. Sci. USA. 1996;93:6458–6463. doi: 10.1073/pnas.93.13.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khomenko I.P., Bakhtina L.Y., Zelenina O.M., Kruglov S.V., Manukhina E.B., Bayda L.A., Malyshev I.Y. Role of heat shock proteins HSP70 and HSP32 in the protective effect of adaptation of cultured HT22 hippocampal cells to oxidative stress. Bull. Exp. Biol. Med. 2007;144:174–177. doi: 10.1007/s10517-007-0282-9. [DOI] [PubMed] [Google Scholar]
- 91.Watanabe S., Ageta-Ishihara N., Nagatsu S., Takao K., Komine O., Endo F., Miyakawa T., Misawa H., Takahashi R., Kinoshita M., et al. SIRT1 overexpression ameliorates a mouse model of SOD1-linked amyotrophic lateral sclerosis via HSF1/HSP70i chaperone system. Mol. Brain. 2014;7:62. doi: 10.1186/s13041-014-0062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo S., Wharton W., Moseley P., Shi H. Heat shock protein 70 regulates cellular redox status by modulating glutathione-related enzyme activities. Cell Stress Chaperones. 2007;12:245–254. doi: 10.1379/CSC-265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Broome C.S., Kayani A.C., Palomero J., Dillmann W.H., Mestril R., Jackson M.J., McArdle A. Effect of lifelong overexpression of HSP70 in skeletal muscle on age-related oxidative stress and adaptation after nondamaging contractile activity. Faseb J. 2006;20:1549–1551. doi: 10.1096/fj.05-4935fje. [DOI] [PubMed] [Google Scholar]
- 94.Xu L., Voloboueva L.A., Ouyang Y., Emery J.F., Giffard R.G. Overexpression of mitochondrial Hsp70/Hsp75 in rat brain protects mitochondria, reduces oxidative stress, and protects from focal ischemia. J. Cereb. Blood Flow Metab. 2009;29:365–374. doi: 10.1038/jcbfm.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen B.H., Chang Y.J., Lin S., Yang W.Y. Hsc70/Stub1 promotes the removal of individual oxidatively stressed peroxisomes. Nat. Commun. 2020;11:5267. doi: 10.1038/s41467-020-18942-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li X., Yu Y., Gorshkov B., Haigh S., Bordan Z., Weintraub D., Rudic R.D., Chakraborty T., Barman S.A., Verin A.D., et al. Hsp70 Suppresses Mitochondrial Reactive Oxygen Species and Preserves Pulmonary Microvascular Barrier Integrity Following Exposure to Bacterial Toxins. Front. Immunol. 2018;9:1309. doi: 10.3389/fimmu.2018.01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pistono C., Monti M.C., Boiocchi C., Berzolari F.G., Osera C., Mallucci G., Cuccia M., Pascale A., Montomoli C., Bergamaschi R. Response to oxidative stress of peripheral blood mononuclear cells from multiple sclerosis patients and healthy controls. Cell Stress Chaperones. 2020;25:81–91. doi: 10.1007/s12192-019-01049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yan L.J., Christians E.S., Liu L., Xiao X., Sohal R.S., Benjamin I.J. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. Embo J. 2002;21:5164–5172. doi: 10.1093/emboj/cdf528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dayalan Naidu S., Kostov R.V., Dinkova-Kostova A.T. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends. Pharmacol. Sci. 2015;36:6–14. doi: 10.1016/j.tips.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 100.Girard P.M., Peynot N., Lelièvre J.M. Differential correlations between changes to glutathione redox state, protein ubiquitination, and stress-inducible HSPA chaperone expression after different types of oxidative stress. Cell Stress Chaperones. 2018;23:985–1002. doi: 10.1007/s12192-018-0909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Doulias P.T., Kotoglou P., Tenopoulou M., Keramisanou D., Tzavaras T., Brunk U., Galaris D., Angelidis C. Involvement of heat shock protein-70 in the mechanism of hydrogen peroxide-induced DNA damage: The role of lysosomes and iron. Free Radic. Biol. Med. 2007;42:567–577. doi: 10.1016/j.freeradbiomed.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 102.Adachi M., Liu Y., Fujii K., Calderwood S.K., Nakai A., Imai K., Shinomura Y. Oxidative stress impairs the heat stress response and delays unfolded protein recovery. PLoS ONE. 2009;4:e7719. doi: 10.1371/journal.pone.0007719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bianchi A., Moulin D., Hupont S., Koufany M., Netter P., Reboul P., Jouzeau J.Y. Oxidative stress-induced expression of HSP70 contributes to the inhibitory effect of 15d-PGJ2 on inducible prostaglandin pathway in chondrocytes. Free Radic. Biol. Med. 2014;76:114–126. doi: 10.1016/j.freeradbiomed.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 104.Reeg S., Jung T., Castro J.P., Davies K.J.A., Henze A., Grune T. The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic. Biol. Med. 2016;99:153–166. doi: 10.1016/j.freeradbiomed.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kiffin R., Christian C., Knecht E., Cuervo A.M. Activation of chaperone-mediated autophagy during oxidative stress. Mol. Biol. Cell. 2004;15:4829–4840. doi: 10.1091/mbc.e04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dodson M., Darley-Usmar V., Zhang J. Cellular metabolic and autophagic pathways: Traffic control by redox signaling. Free Radic. Biol. Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zemanovic S., Ivanov M.V., Ivanova L.V., Bhatnagar A., Michalkiewicz T., Teng R.J., Kumar S., Rathore R., Pritchard K.A., Jr., Konduri G.G., et al. Dynamic Phosphorylation of the C Terminus of Hsp70 Regulates the Mitochondrial Import of SOD2 and Redox Balance. Cell Rep. 2018;25:2605–2616. doi: 10.1016/j.celrep.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu N.W., Chen Y., Liu W., Chen Y.J., Fan Z.M., Liu M., Li L.J. Inhibition of JAK2/STAT3 Signaling Pathway Suppresses Proliferation of Burkitt’s Lymphoma Raji Cells via Cell Cycle Progression, Apoptosis, and Oxidative Stress by Modulating HSP70. Med. Sci. Monit. 2018;24:6255–6263. doi: 10.12659/MSM.910170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hensen S.M., Heldens L., van Enckevort C.M., van Genesen S.T., Pruijn G.J., Lubsen N.H. Activation of the antioxidant response in methionine deprived human cells results in an HSF1-independent increase in HSPA1A mRNA levels. Biochimie. 2013;95:1245–1251. doi: 10.1016/j.biochi.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 110.Smith M.R., Fernandes J., Go Y.M., Jones D.P. Redox dynamics of manganese as a mitochondrial life-death switch. Biochem. Biophys. Res. Commun. 2017;482:388–398. doi: 10.1016/j.bbrc.2016.10.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Afolayan A.J., Teng R.J., Eis A., Rana U., Broniowska K.A., Corbett J.A., Pritchard K., Konduri G.G. Inducible HSP70 regulates superoxide dismutase-2 and mitochondrial oxidative stress in the endothelial cells from developing lungs. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306:L351–L360. doi: 10.1152/ajplung.00264.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang H., Amick J., Chakravarti R., Santarriaga S., Schlanger S., McGlone C., Dare M., Nix J.C., Scaglione K.M., Stuehr D.J., et al. A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure. 2015;23:472–482. doi: 10.1016/j.str.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xie Y., Shi X., Sheng K., Han G., Li W., Zhao Q., Jiang B., Feng J., Li J., Gu Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review) Mol. Med. Rep. 2019;19:783–791. doi: 10.3892/mmr.2018.9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu R., Zhang Y.G., Sun J. STAT3 activation in infection and infection-associated cancer. Mol. Cell Endocrinol. 2017;451:80–87. doi: 10.1016/j.mce.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alas S., Bonavida B. Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin’s lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. Clin. Cancer Res. 2003;9:316–326. [PubMed] [Google Scholar]
- 116.Madamanchi N.R., Li S., Patterson C., Runge M.S. Reactive oxygen species regulate heat-shock protein 70 via the JAK/STAT pathway. Arterioscler. Thromb. Vasc. Biol. 2001;21:321–326. doi: 10.1161/01.ATV.21.3.321. [DOI] [PubMed] [Google Scholar]
- 117.Marinho H.S., Real C., Cyrne L., Soares H., Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ali A., Bharadwaj S., O’Carroll R., Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol. Cell Biol. 1998;18:4949–4960. doi: 10.1128/MCB.18.9.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shi Y., Mosser D.D., Morimoto R.I. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kmiecik S.W., Mayer M.P. Molecular mechanisms of heat shock factor 1 regulation. Trends. Biochem. Sci. 2021 doi: 10.1016/j.tibs.2021.10.004. [DOI] [PubMed] [Google Scholar]
- 121.Paul S., Ghosh S., Mandal S., Sau S., Pal M. NRF2 transcriptionally activates the heat shock factor 1 promoter under oxidative stress and affects survival and migration potential of MCF7 cells. J. Biol. Chem. 2018;293:19303–19316. doi: 10.1074/jbc.RA118.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Y., Ahn Y.H., Benjamin I.J., Honda T., Hicks R.J., Calabrese V., Cole P.A., Dinkova-Kostova A.T. HSF1-dependent upregulation of Hsp70 by sulfhydryl-reactive inducers of the KEAP1/NRF2/ARE pathway. Chem. Biol. 2011;18:1355–1361. doi: 10.1016/j.chembiol.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hensen S.M., Heldens L., van Genesen S.T., Pruijn G.J., Lubsen N.H. A delayed antioxidant response in heat-stressed cells expressing a non-DNA binding HSF1 mutant. Cell Stress Chaperones. 2013;18:455–473. doi: 10.1007/s12192-012-0400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wiktorowicz J.E., Stafford S., Rea H., Urvil P., Soman K., Kurosky A., Perez-Polo J.R., Savidge T.C. Quantification of Cysteinyl S-Nitrosylation by Fluorescence in Unbiased Proteomic Studies. Biochemistry. 2011;50:5601–5614. doi: 10.1021/bi200008b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ansong C., Wu S., Meng D., Liu X., Brewer H.M., Kaiser B.L.D., Nakayasu E., Cort J.R., Pevzner P., Smith R., et al. Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. USA. 2013;110:10153–10158. doi: 10.1073/pnas.1221210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Michelet L., Zaffagnini M., Vanacker H., Le Maréchal P., Marchand C., Schroda M., Lemaire S., Decottignies P. In Vivo Targets of S-Thiolation in Chlamydomonas reinhardtii. J. Biol. Chem. 2008;283:21571–21578. doi: 10.1074/jbc.M802331200. [DOI] [PubMed] [Google Scholar]
- 127.Huang J., Willems P., Wei B., Tian C., Ferreira R.B., Bodra N., Gache S.A.M., Wahni K., Liu K., Vertommen D., et al. Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. USA. 2019;116:21256–21261. doi: 10.1073/pnas.1906768116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hu J., Huang X., Chen L., Sun X., Lu C., Zhang L., Wang Y., Zuo J. Site-Specific Nitrosoproteomic Identification of Endogenously S-Nitrosylated Proteins in Arabidopsis. Plant Physiol. 2015;167:1731–1746. doi: 10.1104/pp.15.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chaki M., Shekariesfahlan A., Ageeva A., Mengel A., von Toerne C., Durner J., Lindermayr C. Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci. 2015;238:115–126. doi: 10.1016/j.plantsci.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 130.Fares A., Rossignol M., Peltier J.-B. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 2011;416:331–336. doi: 10.1016/j.bbrc.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 131.Aroca A., Benito J.M., Gotor C., Romero L.C. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 2017;68:4915–4927. doi: 10.1093/jxb/erx294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gong B., Shi Q. Identifying S-nitrosylated proteins and unraveling S-nitrosoglutathione reductase-modulated sodic alkaline stress tolerance in Solanum lycopersicum L. Plant Physiol. Biochem. 2019;142:84–93. doi: 10.1016/j.plaphy.2019.06.020. [DOI] [PubMed] [Google Scholar]
- 133.Gong B., Yan Y., Zhang L., Cheng F., Liu Z., Shi Q. Unravelling GSNOR-Mediated S-Nitrosylation and Multiple Developmental Programs in Tomato Plants. Plant Cell Physiol. 2019;60:2523–2537. doi: 10.1093/pcp/pcz143. [DOI] [PubMed] [Google Scholar]
- 134.Feng S., Chen Y., Yang F., Zhang L., Gong Y., Adilijiang G., Gao Y., Deng H. Development of a Clickable Probe for Profiling of Protein Glutathionylation in the Central Cellular Metabolism of E. coli and Drosophila. Chem. Biol. 2015;22:1461–1469. doi: 10.1016/j.chembiol.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 135.Su D., Gaffrey M.J., Guo J., Hatchell K.E., Chu R.K., Clauss T.R., Aldrich J.T., Wu S., Purvine S., Camp D.G., et al. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic. Biol. Med. 2013;67:460–470. doi: 10.1016/j.freeradbiomed.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Duan J., Kodali V.K., Gaffrey M.J., Guo J., Chu R.K., Camp D.G., Smith R.D., Thrall B.D., Qian W.-J. Quantitative Profiling of Protein S-Glutathionylation Reveals Redox-Dependent Regulation of Macrophage Function during Nanoparticle-Induced Oxidative Stress. ACS Nano. 2015;10:524–538. doi: 10.1021/acsnano.5b05524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fowler N.J., Blanford C.F., de Visser S., Warwicker J. Features of reactive cysteines discovered through computation: From kinase inhibition to enrichment around protein degrons. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-15997-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vanhecke G.C., Abeywardana M.Y., Huang B., Ahn Y., Young-Hoon A. Isotopically Labeled Clickable Glutathione to Quantify Protein S-Glutathionylation. ChemBioChem. 2019;21:853–859. doi: 10.1002/cbic.201900528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gould N.S., Evans P., Martínez-Acedo P., Marino S.M., Gladyshev V.N., Carroll K.S., Ischiropoulos H. Site-Specific Proteomic Mapping Identifies Selectively Modified Regulatory Cysteine Residues in Functionally Distinct Protein Networks. Chem. Biol. 2015;22:965–975. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Su D., Shukla A.K., Chen B., Kim J.-S., Nakayasu E., Qu Y., Aryal U., Weitz K., Clauss T.R., Monroe M.E., et al. Quantitative site-specific reactivity profiling of S-nitrosylation in mouse skeletal muscle using cysteinyl peptide enrichment coupled with mass spectrometry. Free Radic. Biol. Med. 2013;57:68–78. doi: 10.1016/j.freeradbiomed.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen Y.-J., Ku W.-C., Lin P.-Y., Chou H.-C., Khoo K.-H. S-Alkylating Labeling Strategy for Site-Specific Identification of theS-Nitrosoproteome. J. Proteome Res. 2010;9:6417–6439. doi: 10.1021/pr100680a. [DOI] [PubMed] [Google Scholar]
- 142.Lee T.-Y., Chen Y.-J., Lu C.-T., Ching W.-C., Teng Y.-C., Huang H.-D. dbSNO: A database of cysteine S-nitrosylation. Bioinformatics. 2012;28:2293–2295. doi: 10.1093/bioinformatics/bts436. [DOI] [PubMed] [Google Scholar]
- 143.Zaręba-Kozioł M., Szwajda A., Dadlez M., Wysłouch-Cieszyńska A., Lalowski M. Global Analysis of S-nitrosylation Sites in the Wild Type (APP) Transgenic Mouse Brain-Clues for Synaptic Pathology. Mol. Cell. Proteom. 2014;13:2288–2305. doi: 10.1074/mcp.M113.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ibáñez-Vea M., Huang H., De Morentin X.M., Pérez E., Gato M., Zuazo M., Arasanz H., Fernandez-Irigoyen J., Santamaría E., Fernández-Hinojal G., et al. Characterization of Macrophage Endogenous S-Nitrosoproteome Using a Cysteine-Specific Phosphonate Adaptable Tag in Combination with TiO2 Chromatography. J. Proteome Res. 2018;17:1172–1182. doi: 10.1021/acs.jproteome.7b00812. [DOI] [PubMed] [Google Scholar]
- 145.Kohr M.J., Aponte A.M., Sun J., Wang G., Murphy E., Gucek M., Steenbergen C. Characterization of potentialS-nitrosylation sites in the myocardium. Am. J. Physiol. Circ. Physiol. 2011;300:H1327–H13352011. doi: 10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Doulias P.-T., Tenopoulou M., Greene J.L., Raju K., Ischiropoulos H. Nitric Oxide Regulates Mitochondrial Fatty Acid Metabolism Through Reversible Protein S -Nitrosylation. Sci. Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kohr M., Aponte A.M., Sun J., Gucek M., Steenbergen C., Murphy E. Measurement of S -Nitrosylation Occupancy in the Myocardium With Cysteine-Reactive Tandem Mass Tags. Circ. Res. 2012;111:1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Qu Z., Meng F., Bomgarden R.D., Viner R.I., Li J., Rogers J.C., Cheng J., Greenlief C.M., Cui J., Lubahn D.B., et al. Proteomic Quantification and Site-Mapping of S-Nitrosylated Proteins Using Isobaric iodoTMT Reagents. J. Proteome Res. 2014;13:3200–3211. doi: 10.1021/pr401179v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gu L., Robinson R.A.S. High-throughput endogenous measurement of S-nitrosylation in Alzheimer's disease using oxidized cysteine-selective cPILOT. Anal. 2016;141:3904–3915. doi: 10.1039/C6AN00417B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Raju K., Doulias P.-T., Evans P., Krizman E.N., Jackson J.G., Horyn O., Daikhin Y., Nissim I., Yudkoff M., Nissim I., et al. Regulation of brain glutamate metabolism by nitric oxide and S-nitrosylation. Sci. Signal. 2015;8:ra68. doi: 10.1126/scisignal.aaa4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Smith J.G., Aldous S.G., Andreassi C., Cuda G., Gaspari M., Riccio A. Proteomic analysis of S-nitrosylated nuclear proteins in rat cortical neurons. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aar3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pan K.-T., Chen Y.-Y., Pu T.-H., Chao Y.-S., Yang C.-Y., Bomgarden R.D., Rogers J.C., Meng T.-C., Khoo K.-H. Mass Spectrometry-Based Quantitative Proteomics for Dissecting Multiplexed Redox Cysteine Modifications in Nitric Oxide-Protected Cardiomyocyte Under Hypoxia. Antioxidants Redox Signal. 2014;20:1365–1381. doi: 10.1089/ars.2013.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang H.-H., Lechuga T.J., Chen Y., Yang Y., Huang L., Chen D.-B. Quantitative Proteomics Analysis of VEGF-Responsive Endothelial Protein S-Nitrosylation Using Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and LC-MS/MS. Biol. Reprod. 2016;94:114. doi: 10.1095/biolreprod.116.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xie Y., Luo X., Li Y., Chen L., Ma W., Huang J., Cui J., Zhao Y., Xue Y., Zuo Z., et al. DeepNitro: Prediction of Protein Nitration and Nitrosylation Sites by Deep Learning. Genom. Proteom. Bioinform. 2018;16:294–306. doi: 10.1016/j.gpb.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chou C.-C., Chiang B.-Y., Lin J.C.-Y., Pan K.-T., Lin C.-H., Khoo K.-H. Characteristic Tandem Mass Spectral Features Under Various Collision Chemistries for Site-Specific Identification of Protein S-Glutathionylation. J. Am. Soc. Mass Spectrom. 2014;26:120–132. doi: 10.1007/s13361-014-1014-9. [DOI] [PubMed] [Google Scholar]
- 156.Konstantinidis D., Paletas K., Koliakos G., Kaloyianni M. The ambiguous role of the Na+–H+ exchanger isoform 1 (NHE1) in leptin-induced oxidative stress in human monocytes. Cell Stress Chaperon- 2009;14:591–601. doi: 10.1007/s12192-009-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hoppe G., Chai Y.-C., Crabb J.W., Sears J. Protein s-glutathionylation in retinal pigment epithelium converts heat shock protein 70 to an active chaperone. Exp. Eye Res. 2004;78:1085–1092. doi: 10.1016/j.exer.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 158.Majmudar J.D., Konopko A.M., Labby K.J., Tom C.T.M.B., Crellin J.E., Prakash A., Martin B.R. Harnessing Redox Cross-Reactivity To Profile Distinct Cysteine Modifications. J. Am. Chem. Soc. 2016;138:1852–1859. doi: 10.1021/jacs.5b06806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Forrester M.T., Thompson J.W., Foster M.W., Nogueira L., Moseley M.A., Stamler J.S. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xu Y., Ding J., Wu L.-Y., Chou K.-C. iSNO-PseAAC: Predict Cysteine S-Nitrosylation Sites in Proteins by Incorporating Position Specific Amino Acid Propensity into Pseudo Amino Acid Composition. PLoS ONE. 2013;8:e55844. doi: 10.1371/journal.pone.0055844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ben-Lulu S., Ziv T., Weisman-Shomer P., Benhar M. Nitrosothiol-Trapping-Based Proteomic Analysis of S-Nitrosylation in Human Lung Carcinoma Cells. PLoS ONE. 2017;12:e0169862. doi: 10.1371/journal.pone.0169862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu M., Hou J., Huang L., Huang X., Heibeck T.H., Zhao R., Pasa-Tolic L., Smith R.D., Li Y., Fu K., et al. Site-Specific Proteomics Approach for Study Protein S-Nitrosylation. Anal. Chem. 2010;82:7160–7168. doi: 10.1021/ac100569d. [DOI] [PubMed] [Google Scholar]
- 163.Tan C., Li Y., Huang X., Wei M., Huang Y., Tang Z., Huang H., Zhou W., Wang Y., Hu J. Extensive protein S-nitrosylation associated with human pancreatic ductal adenocarcinoma pathogenesis. Cell Death Dis. 2019;10:1–14. doi: 10.1038/s41419-019-2144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.