Abstract
Background: The presence of antimicrobial-resistant pathogens such as Klebsiella pneumoniae strains in the food supply is dangerous. The aim of this study was to assess the prevalence of Klebsiella pneumonia strains in Greek meat products and evaluate their phenotypes and genotypes. Methods: One hundred and ten meat specimens were cultured for the isolation of K. pneumoniae. In positive specimens, PCR (Polymerase Chain Reaction) analysis was performed to confirm the presence of K. pneumoniae. Genotypic and phenotypic evaluation of the isolated strains included multiplex immunoassay for the detection of carbapenemases, and PCR screening for the detection of resistance and virulence genes. Results: K. pneumoniae strains were recovered in 90 (81.8%) meat samples. The ecpA gene was identified in 30 (33.3%) isolates, while the fimH-1 and mrkA genes were present in 15 (16.7%) and 65 (72.2%) isolates, respectively. Sixty-five K. pneumoniae isolates (72.2%) were found to carry at least one resistance gene; of these, the blaNDM-like was the most commonly identified gene in 40 (61.5%) isolates, followed by the blaOXA-48 like gene in 20 isolates (30.8%). Conclusions: A high frequency of foodborne K. pneumoniae in Greece was found. Our results indicate that most strains carried resistance and virulence genes, indicating a high pathogenic potential and a significant risk to human health.
Keywords: Klebsiella pneumoniae, food, antimicrobial resistance, virulence
1. Introduction
Resistance to antibiotics is a global public health problem [1], hindering the treatment and extirpation of a growing number of bacterial, fungal and viral infections. Antibiotic-resistant bacteria are associated with high mortality in septic patients [2]. The rapid development of antibiotic-resistant strains is favored by the widespread and, in many cases, unjustified use of broad-spectrum antibiotics [3]. Resistance genes can also spread among different bacteria populations through the horizontal gene transfer (HGT) of mobile genetic elements (MGE) [4,5], facilitating the insurgence of multi-drug resistant strains [2,6,7].
K. pneumoniae is a common cause of community-acquired and nosocomial infections such as urinary tract infections, lower respiratory infections and liver abscesses. These pathogens can be found in various microbiological niches such as food and soil, but also in the skin, intestines and feces of mammals. Although K. pneumoniae it is not traditionally considered as a foodborne pathogen, there are reports of K. pneumoniae infections preceded by gut colonization, supporting the theory of food as a possible transmission vector for these pathogens [8,9,10,11]. Enterobacterales resistant to carbapenems and third-generation cephalosporin such as Klebsiella pneumoniae have been listed by the World Health Organization (WHO) as one of the top threats to public health worldwide, highlighting the need for the development of new treatment medications [6,7]. There are different resistance genes in K. pneumoniae which are located in transferable genetic elements and may be transferred to other bacteria. Unfortunately, antimicrobial-resistant K. pneumoniae strains have been detected in various food samples, such as raw meat, marketed fresh vegetables, ready-to-eat meals, and seafood [12,13,14,15,16,17]. Isolation of antimicrobial-resistant K. pneumoniae strains in the food supply is alarming, since they can become a large pool for resistance genes, carrying the risk for the emergence of new resistance mechanisms.
The aim of this study was to assess the contamination rate of Greek meat samples from different local markets with K. pneumoniae strains and evaluate the phenotypic and molecular characteristics of these strains. As there is a lack of data, limited information exists about the presence of highly resistant strains such as K. pneumoniae in meat products that are intended for consumption in market chains in Greece.
2. Materials and Methods
2.1. General Workflow
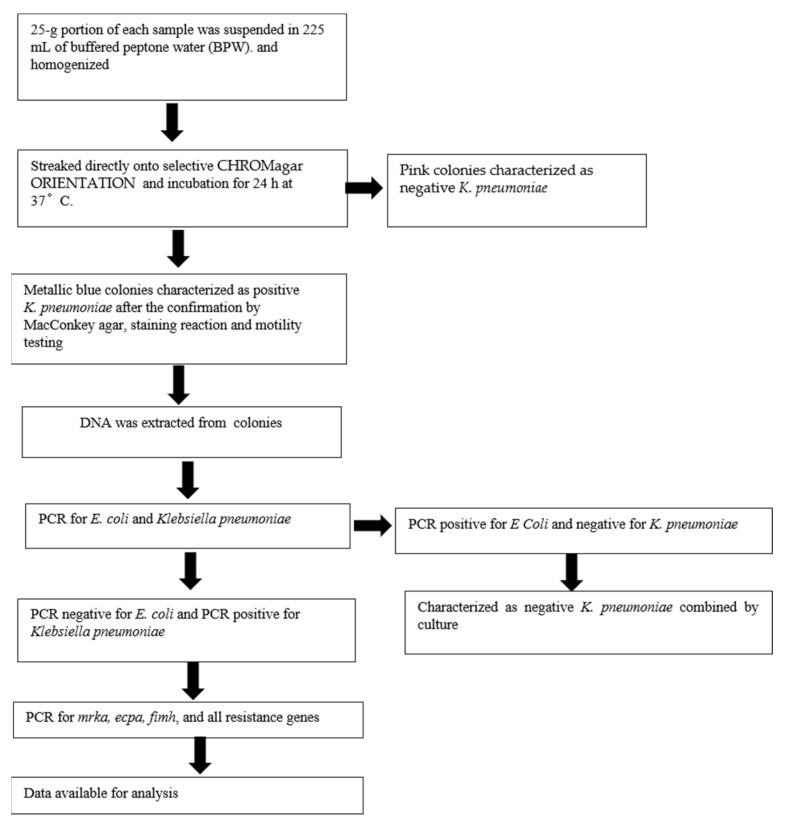
A general workflow was designed in order to understand the prevalence of Klebsiella pneumoniae in meat products. This study involved molecular screening of isolated Klebsiella pneumoniae. The general workflow of the study is provided in Figure 1.
Figure 1.
General Workflow. PCR: Polymerase Chain Reaction.
2.2. Sample Collection
A total of 110 meat samples were collected from different local markets in Athens. The samples included 40 (36.4%) meat specimens from pork, 35 (31.8%) from chicken and 35 (31.8%) from bovine.
2.3. Detection of K. pneumoniae Colonies
Twenty-five grams from each sample was collected in sterile bags and 225 mL of buffered peptone water (BPW) solution 1% was added and homogenized in a Stomacher (Laboratory Blender Stomacher 400, Seward Medical, London, UK). The sample suspensions were incubated overnight at 37 °C. A 100 μL aliquot of the pre-enrichment culture was streaked directly onto selective CHROMagar ORIENTATION (Bioprepare Microbiology, Athens, Greece) and incubated for 24 h at 37 °C. The specimens were inoculated and incubated on MacConkey agar. After 24–48 h of aerobic incubation at 36–37 °C, isolates with colonial appearance of Escherichia coli and Klebsiella pneumoniae characterized according to Gram staining reaction and motility testing method. Suspected isolates of Escherichia coli and Klebsiella species were confirmed. Colonies exhibiting red and metallic blue color from CHROMagar ORIENTATION were picked and regrown on tryptic soy agar (TSA; Oxoid) (Bioprepare Microbiology, Athens, Greece) for 24 h at 37 °C in order to have fresh cultures needed for further DNA isolation and confirmation by PCR.
2.4. DNA Isolation
Nucleic acids were extracted from the isolated colonies using the Zybio Nucleic Acids Isolation kit (Magnetic Beads Method) with an automatic extractor Zybio EXM 3000, following the manufacturer’s instructions. The DNA samples were separated into six aliquots for further analyses.
2.5. Confirmation of E. coli and K. pneumoniae by PCR
PCR assays were performed using GoTaq Hot Start Master Mix (Promega Gmbh, 68199, Mannheim, Germany), 0.2 mM of each gene’s specific primers (Table 1) [16] and 10 μL of eluted DNA and ddH2O (molecular grade) to bring the volume to 50 μL. PCR reactions were conducted in a thermal cycler (Applied Biosystems Athens, Greece). Amplification conditions consisted of an initial 5 min denaturation step at 95 °C, followed by 40 cycles of 35 s denaturation at 94 °C, 60 s annealing at 54 °C, and a 60 s extension step at 72 °C, and a final extension step at 72 °C for 10 min. PCR products were separated in a 2% agarose gel, stained with ethidium bromide (0.5 μg/mL). The genomic DNA of a clinical strain of Salmonella enteritis was used as a negative control.
Table 1.
Primers sequences.
| Target Genes | Primer Sequence | Ampillicon Size (bp) |
|---|---|---|
| blaIMP-1 | F: 5′-TGA GCA AGT TAT CTG TAT TC-3′ | 139 |
| R:5′-TTA GTT GCT TGG TTT TGA TG-3 | ||
| blaOXA-48 | F: 5′-TTG GTG GCA TCG ATT ATC GG-3′ | 281 |
| R: 5′-GAG CAC TTC TTT TGT GAT GGC-3′ | ||
| fimH | F: 5′-CGC CTG GTC CTT TGC CTG CA-3′ | 817 |
| R: 5′-CTG CAC GTT GCC GGC GGT AA-3′ | ||
| ecpA | F: 5′-AAT GGT TCA CCG GGA CAT CAT GTC C-3′ | 759 |
| R: 5′-AAG GAT GAA ATA TCG CCG ACA TCC-3′ | ||
| mrkA | F: 5′-GTT AAC GGC GGC CAG GGC AGC GA-3′ R: 5′-AGG TGA AAC GCG CGC CAT CA-3′ |
382 |
| blaVIM | F: 5′- GAT GGT GTT TGG TCG CAT A-3′ | 390 |
| R: 5′-CGA ATG CGC AGC ACC AG-3′ | ||
| blaNDM | F: 5′-GGT TTG GCG ATC TGG TTT TC-3′ | 521 |
| R: 5′- CGG AAT GGC TCA TCA CGA TC-3′ | ||
| blaKPC | F: 5′-ATG TCA CTG TAT CGC CGT CT-3′ | 538 |
| R: 5′-TTT TCA GAG CCT TAC TGC CC-3′ | ||
| E. coli | F: 5′-TGATTGAAGCAGAAGCCTGC R: 5′-CGCCAATCCACATCTGTGAA |
1.350 |
| Klebsiella pneumoniae | F 5′-ATTTGAAGAGGTTGCAAACGAT-3′ R 5′-TTCACTCTGAAGTTTTCTTGTGTTC-3′ |
130 |
2.6. Detection of the Virulence Genes ecpA, fimH, mrkA and the Resistance Genes blaKPC, blaNDM, blaVIM, bla IMP and blaOXA-48 by Multiplex PCR
We proceeded to conduct four multiplex assays to detect three combination sets of resistant and virulence genes. Set 1 referred to blaVIM and blaKPC genes, set 2 referred to blaNDM, Set 3 referred to blaIMP and blaOXA-48 genes and set 4 referred to ecpA, fimH, and mrkA genes. PCR assay were performed using GoTaq Hot Start Master Mix (Promega Gmbh, 68199, Mannheim, Germany), 0.2 mM of each gene’s specific primers (Table 1) [17,18,19], and 10 μL of eluted DNA and dH2O (molecular grade) to bring the volume up to 50 μL. PCR reactions were conducted in a thermal cycler (Veriti™ 96-Well Thermal Cycler, Applied Biosystems, Athens Greece). Amplification conditions consisted of an initial 5 min denaturation step at 95 °C, followed by 40 cycles of 35 s denaturation at 94 °C, 60 s annealing at 58 °C for set 4 and an annealing at 56 °C for the set 1, 2, 3, and a 60 s extension step at 72 °C, and a final extension step at 72 °C for 10 min. PCR products were separated in a 2% agarose gel, stained with ethidium bromide (0.5 μg/mL).
2.7. Phenotypic Analysis
Phenotypic analysis included an in vitro multiplex immunoassay for the detection and differentiation of five common carbapenemase families (KPC, OXA-48-like, VIM, IMP, and NDM) in the bacterial colonies, using the NG-Test Carba 5 (NG Biotech, Guipry, France) test. Following the manufacturer’s instructions, we inoculated 100 μL of the cultured suspension in the NG-Test Carba 5 kit using a transfer pipette that was included in the kit. The test was visually examined after 15 min for the presence or absence of the control and test lines, indicating the presence or absence of the respective carbapenemases in the tested specimen
2.8. Statistical Analysis
Statistical analysis included descriptive statistics of the tested samples. Categorical variables were compared using the chi square test. Statistical analysis was performed using the Stata 15.0 software (Stata Corp., College Station, TX, USA), and a p-value < 0.05 indicated statistical significance.
3. Results
Overall, K. pneumoniae strains were detected in 90 (81.8%) samples, whereas E. coli strains were detected in 20 (18.2%) samples. PCR analysis confirmed the presence of K. pneumoniae strains and E. coli strains in positive culture samples (Table 2). Among the 90 samples identified as K. pneumoniae-positive, 23 (25.6%) were from chicken, 32 (35.6%) from bovine samples and 35 (38.9%) from pork. Moreover, 23 of 35 (65.7%) chicken samples, 32 of 35 (91.4%) bovine samples, and 35 of 40 (87.5%) pork samples were positive for K. pneumoniae. The comparison of contamination rates among the different meat categories revealed that chicken samples were less contaminated than bovine (p = 0.008) and pork (p = 0.024) samples. However, the rates of K. pneumoniae contamination between pork and bovine samples did not differ (p = 0.58).
Table 2.
Klebsiella pneumoniae and Escherichia coli strains in meat samples.
| Strains | Overall (* n = 110) | Chicken (* n = 35) | Bovine (* n = 35) | Pork (* n = 40) |
|---|---|---|---|---|
| Klebsiella pneumoniae | 90 (81.8) | 23 (65.7) | 32 (91.4) | 35 (87.5) |
| Escherichia coli | 20 (18.2) | 7 (20.0) | 3 (8.5) | 10 (25.0) |
Data are presented as absolute values (percentages). * n refers to samples.
3.1. Phenotypic Analysis
Based on the in vitro multiplex immunoassay, 54 (60.0%) isolates were positive for the production of at least one carbapenemase. Specifically, 4 (7.4%) K. pneumoniae isolates were positive for KPC production, 5 (9.2%) K. pneumoniae isolates were positive for VIM production, 32 (59.2%) K. pneumoniae isolates were positive for NDM production, 5 (9.2%) K. pneumoniae isolates were positive for IMP production, and 17 (31.4%) K. pneumoniae isolates were positive for OXA-48 production (Table 3). Moreover, of the 54 isolates with at least one carbapenemase production, there were nine isolates with simultaneous production of two carpapenemases.
Table 3.
Carbapenemase production based on PCR and phenotypic assays.
| Carbapenemase | Positive Isolates Based on PCR (* n = 65) |
Positive Isolates Based on NG-Test Carba 5 (* n = 54) |
|---|---|---|
| KPC | 5 (7.6) | 4 (7.4) |
| OXA-48 | 20 (30.7) | 17 (31.4) |
| IMP | 5 (7.6) | 5 (9.2) |
| VIM | 5 (7.6) | 5 (9.2) |
| NDM | 40 (61.5) | 32 (59.2) |
Data are presented as absolute values (percentages). * n refers to samples.
3.2. Virulence and Resistance Genes
Regarding the virulence genes, 30 (33.3%) K. pneumoniae isolates amplified a 759-bp product corresponding to the molecular weight of the ecpA gene. The ecpA gene was detected in K. pneumoniae strains from 14 (46.7%) chicken and 16 (53.3%) bovine specimens. Moreover, the fimH-1 and mrkA genes, encoding type 1 and type 3 fimbrial adhesins, were present in 15 (16.7%) and 65 (72.2%) strains of K. pneumoniae isolates, respectively. Among the 15 samples, the fimH-1 gene was present in 33.3% of the three categories of meat products, as shown in Table 4. Among the 65 isolates, the mrkA gene was present in 50 of the chicken and bovine meat products and 15 of the pork meat products. Interestingly, 11.1% (10/90) of the strains possessed the cap+/fimH+/mrkA+ genotype, 0% (0/90) possessed the ecpA+/fimH+/mrkA− genotype, and 22.2% (20/90) of the strains had the cap+/fimH−/mrkA+ genotype.
Table 4.
Klebsiella pneumoniae strains carrying ecpA, fimH-1 and mrkA genes.
| Meat Specimens |
ecpA (n = 30) |
fimH-1 (n = 15) |
mrkA (n = 65) |
|---|---|---|---|
| Chicken | 14 (46.7) | 5 (33.3) | 25 (38.5) |
| Bovine | 16 (53.3) | 5 (33.3) | 25 (38.5) |
| Pork | 0 (0.0) | 5 (33.3) | 15 (23.1) |
Data are presented as absolute values (percentages).
Regarding the resistance genes, 65 (72.2%) K. pneumoniae isolates were found to carry at least one resistance gene. Of these, blaNDM-like was the most predominant gene identified in 40 (61.5%) isolates, followed by blaOXA-48-like in 20 (30.7%) isolates, while 5 (7.6%) isolates carried the blaIMP-type gene, 5 (7.6%) isolates carried the blaVIM-type gene, and 5 (7.6%) isolates carried the blaKPC-type gene (Table 3). The frequency of the blaNDM-like gene was significantly higher than those of the blaVIM-type, blaKPC-type, blaIMP-type, and blaOXA-48 genes (p < 0.001). Moreover, the frequency of the blaOXA-48 gene was significantly higher than those of the blaVIM-type, blaKPC-type and blaIMP-type genes (p < 0.001). Among the 65 isolates, 10 isolates had two resistance genes simultaneously, 5 had the blaOXA-48 and blaIMP genes, and 5 had the blaNDM and blaKPC genes.
4. Discussion
K. pneumoniae is not traditionally recognized as a common foodborne pathogen, and most studies investigating foodborne strains are focused on other common pathogens such as Escherichia coli, Salmonella and Shigella. Therefore, information regarding the contamination rate of retail food with K. pneumoniae strains and their characteristics such as antibiotic resistance and virulence profile that are necessary to evaluate their risk to public health is scarce [20,21,22,23]. In particular, in Greece, there are no reports regarding the prevalence and virulence of K. pneumoniae in food supply. It is of great importance to evaluate the genotypic and phenotypic characteristics of these foodborne pathogens and detect their antibiotic resistance since they can cause community and nosocomial outbreaks, similar to other common foodborne pathogens [24]. The evaluation of pathogens in food that harbor antibiotic resistance could allow public health authorities to implement more effective strategies to decrease the contamination rate and mitigate the impact of these pathogens on public health. This is even more prominent in certain areas such as Singapore, where most of the local food supply is imported, and therefore the first stages of production cannot be monitored and controlled for food contamination. Our findings raise concerns regarding the high contamination rate of meat products with K. pneumoniae in Greece (81.8%), while also most of the isolated pathogens carried at least one carbapenemase-resistant gene (72.2%), indicating a difficulty in the eradication of these pathogens. Our findings are in line with the reported high rates of antibiotic-resistant K. pneumoniae strains in various poultry and meat products, indicating that foodborne K. pneumoniae can become a potential pool for antibiotic resistance genes [25].
These pathogens constitute part of the normal human and animal flora, and therefore isolation of K. pneumoniae strains in food samples is not necessarily linked with infection transmission. However, their detection in the food chain can be indicative for non-hygienic practices of food preparation and handling, undercooking and suboptimal storage conditions, especially in cases of isolation in ready-to-eat cooked food. Although it has been also suggested that the incidence of liver abscesses due to K. pneumoniae worldwide is associated with the geographic distribution of certain virulent K. pneumoniae subtypes, the exact route of K. pneumoniae infections is not fully elucidated [10,26]. The association between foodborne K. pneumoniae and development of infections is not fully investigated, and there is a lack of justification for the incidence of K. pneumoniae as a foodborne pathogen. However, since the human gut microflora is closely related to the microbiome in consumed food, and in many cases colonization of the intestine with K. pneumoniae preceded infections, foodborne K. pneumoniae could be a potential source for the development of K. pneumoniae infections in the general population [16]. In line with this, Gorrie et al. found that half of the patients with K. pneumoniae infections in the intensive care unit tested positive for gastrointestinal carriage on admission, indicating that gut colonization is a likely entry source for K. pneumoniae infections [16].
The first finding emerging from the present study is the high presence of K. pneumoniae detected in meat. The prevalence of K. pneumoniae that was found in our study is significantly higher than those reported in other studies [20,23]. Guo et al. assessed the frequency of K. pneumoniae in food samples in Eastern China and reported a contamination rate of 9.9% (99 positive out of 998 tested samples) [23]. In another study, Hartantyo et al. evaluated the frequency of foodborne K. pneumoniae in Singapore, by screening 698 samples of raw and ready-to-eat retail food. The authors detected K. pneumoniae in 21% (147 out of 698) of samples tested, which is also significantly smaller than our contamination rate. The different contamination rates between our study and the other studies could be attributed to the heterogeneity in food sources among these studies, since many different categories of food samples such as vegetables, seafood, meat, raw and ready-to-eat food were analyzed those studies. The exact cause for the high frequency of K. pneumoniae observed in our study is not easily identifiable, since there are several stages from farmers’ production to consumer markets where infections could have been developed. Thus, further research is needed to clarify this. However, such prevalence of the detected pathogens, and especially in raw food samples, is of utmost importance, as it could be a serious “vector” for the development and the persistence of nosocomial infections. Upon the determination of the primary source responsible for the high contamination rate of meat samples, direct or indirect methods of protection and prevention are necessary.
Resistance to antibiotics is a multi-faceted and urgent global problem, which needs to be tackled under very different perspectives, involving simultaneously different stakeholders, such as pharmaceutical companies, medical doctors, pharmacies, policy makers, large inter-country health surveillance networks and initiatives, diagnostics companies as well as research institutes [19]. Drug resistance is said to be associated with the spread of transmissible plasmids that also carry virulence genes, and the acquisition of resistant determinants can contribute to the persistence of virulent microorganisms. Carbapenem resistance through carbapenemase production is an emerging threat to public health with a devastating socioecomonic and clinical impact worldwide. Our findings indicate that carbapenem-resistant genes are common among foodborne K. pneumoniae strains in Greece. Specifically, we found that 72.2% of the isolated K. pneumoniae carried at least one resistance gene for carbapenemase production. The blaNDM-l gene was the most predominant antibiotic resistance gene (61.5%), followed by the blaOXA-48 gene (30.7%). The rate and characteristics of carbapenemase resistance genes has not been evaluated in other studies investigating foodborne K. pneumoniae, and therefore we cannot draw any conclusions regarding the cause for the high rate of certain carbapenemase-production genes compared to some others, and we cannot compare our rates with the results of others studies. Guo et al. in his study isolated 99 K. pneumomiae strains from different food samples including cooked food, frozen raw food, and fresh raw meet samples and evaluated their resistance to antibiotics [23]. The authors conducted antimicrobial susceptibility testing for several antibiotics including carbapenems and performed PCR analysis for detection of several resistance genes. However, they did not evaluate carbapenemase resistance genes. As opposed to the results of our study, although many of the isolates in Guo et al.’s study showed resistance to other antibiotics such as ampicillin, none of them demonstrated resistance to carbapenems based on antimicrobial susceptibility testing. The authors reported that resistance to antibiotics was significantly higher in isolates from raw meat samples, and therefore the higher rate of resistance to carbapenems in our study may be attributed to the fact that all of our samples were raw meat products. In another study, Hartantyo et al. detected 147 K. pneumoniae isolates in raw and ready-to-eat retail food samples and evaluated their susceptibility to antibiotics in 97 of them [25]. Although antibiotic susceptibility testing based on the disk diffusion method was not performed for carbapenems, isolates were screened for the bla-KPC gene by PCR analysis. However, the authors did not report the rate of bla-KPC positive isolates in their study. Similar to the findings of Guo et al., the authors of this study also did not report on any positive isolate for the bla-KPC gene associated with resistance to carbapenems.
Another important finding of our study is the high rate of positive pathogens for certain virulence genes such as the ecpA gene (33.3%) the fimH-1 (16.7%) gene and the mrkA (72.2%) gene, indicating a high potential for biofilm formation. The fimH-1 and mrkA genes are encoding type 1 and type 3 fimbrial adhesins, while the ecpA gene encodes another pilus protein which is more common among E. coli species. These fimbrial structures constitute one of the main mechanisms for adherence to host cells. The multi-fimbrial nature of the isolated pathogens in our study highlights the high virulence potential of these strains, which in combination with the high prevalence of resistance genes can result in difficult-to-eradicate outbreaks.
There are some limitations of the study that must be addressed. First, the number of analyzed samples is too small to draw safe conclusions regarding the antimicrobial resistance of K. pneumoniae in meat products, and therefore large studies are needed to validate our results. Second, the evaluation of the genotype was limited, including only PCR for certain genes, while more advanced molecular techniques such as the whole-genome sequencing method and multi-locus sequence typing (MLST) to decipher the clonal relationship between the food isolates with other published isolated were not performed. Third, we did not document the association between a positive result for carbapenemase production based on the NG-Test Carba 5 and a positive result for carbapenemase resistance genes on PCR analysis, and therefore we cannot evaluate the agreement between these two methods. Last, the phenotype was assessed by only the immunoassay for the detection and differentiation of common carbapenemase enzymes, while further phenotypic characterization including antimicrobial susceptibility testing and the conjugation assay would be valuable.
5. Conclusions
In conclusion, we found that there is a high prevalence of K. pneumoniae in Greek meat products. Moreover, most of the isolated strains carried carbapenemase resistance genes and virulence genes, indicating a high pathogenic potential of these pathogens and a significant risk to human health. The data reported in this study will aid in our understanding of the potential risk of K. pneumoniae in retail food hygiene, food safety, and public health in Greece. However, more extensive research is needed in order to further characterize the frequency and resistance profile of foodborne K. pneumoniae strains.
Author Contributions
Conceptualization, D.H., A.G.T. and P.H.; methodology, N.A.T., I.B. and G.P.L.; software, H.S. and A.C.P.; validation, D.H., P.H. and A.G.T.; formal analysis, H.S., A.C.P. and G.P.L.; investigation, G.P.L., A.G.T., N.A.T. and I.B.; resources, All authors; data curation, D.H., A.G.T. and H.S.; writing—review and editing, D.H., A.G.T., P.H., N.A.T., I.B., A.C.P. and H.S.; visualization, D.H.; supervision, D.H.; project administration, D.H. and P.H.; funding acquisition, All authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Roca I., Akova M., Baquero F., Carlet J., Cavaleri M., Coenen S., Cohen J., Findlay D., Gyssens I., Heure O.E., et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer A., Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet. 2013;14:243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy S.B., Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004;10((Suppl. 12)):S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 4.Ochman H., Lawrence J.G., Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 5.Domingues S., da Silva G.J., Nielsen K.M. Integrons: Vehicles and pathways for horizontal dissemination in bacteria. Mob. Genet. Elem. 2012;2:211–223. doi: 10.4161/mge.22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Costa V.M., McGrann K.M., Hughes D.W., Wright G.D. Sampling the Antibiotic Resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 7.D’Costa V.M., King C.E., Kalan L., Morar M., Sung W.W.L., Schwarz C., Froese D., Zazula G., Calmels F., Debruyne R., et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 8.Karlowsky J.A., Jones M.E., Thornsberry C., Friedland I.R., Sahm D.F. Trends in antimicrobial susceptibilities among Enterobacteriaceae isolated from hospitalized patients in the United States from 1998 to 2001. Antimicrob. Agents Chemother. 2003;47:1672–1680. doi: 10.1128/AAC.47.5.1672-1680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siu L.K., Fung C.P., Chang F.Y., Lee N., Yeh K.M., Koh T.H., Ip M. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J. Clin. Microbiol. 2011;49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung C.P., Lin Y.T., Lin J.C., Chen T.L., Yeh K.M., Chang F.Y., Chuang H.C., Wu H.S., Tseng C.P., Siu L.K. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg. Infect. Dis. 2012;18:1322–1325. doi: 10.3201/eid1808.111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calbo E., Freixas N., Xercavins M., Riera M., Nicolás C., Monistrol O., Solé M.D.M., Sala M.R., Vila J., Garau J. Foodborne Nosocomial Outbreak of SHV1 and CTX-M-15-producing Klebsiella pneumoniae: Epidemiology and Control. Clin. Infect. Dis. 2011;52:743–749. doi: 10.1093/cid/ciq238. [DOI] [PubMed] [Google Scholar]
- 12.Falomir M.P., Rico H., Gozalbo D. Enterobacter and Klebsiella Species Isolated from Fresh Vegetables Marketed in Valencia (Spain) and Their Clinically Relevant Resistances to Chemotherapeutic Agents. Foodborne Pathog. Dis. 2013;10:1002–1007. doi: 10.1089/fpd.2013.1552. [DOI] [PubMed] [Google Scholar]
- 13.Nawaz M., Khan S., Tran Q., Sung K., Khan A., Adamu I., Steele R. Isolation and characterization of multidrug-resistant Klebsiella spp. isolated from shrimp imported from Thailand. Int. J. Food Microbiol. 2012;155:179–184. doi: 10.1016/j.ijfoodmicro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Wu H., Liu B.-G., Liu J.-H., Pan Y.-S., Yuan L., Hu G.-Z. Phenotypic and molecular characterization of CTX-M-14 extended-spectrum β-lactamase and plasmid-mediated ACT-like AmpC β-lactamase produced by Klebsiella pneumoniae isolates from chickens in Henan Province, China. Genet. Mol. Res. 2012;11:3357–3364. doi: 10.4238/2012.September.24.1. [DOI] [PubMed] [Google Scholar]
- 15.Gorrie C.L., Mirceta M., Wick R.R., Edwards D.J., Thomson N.R., Strugnell R.A., Pratt N.F., Garlick J.S., Watson K.M., Pilcher D.V., et al. Gastrointestinal car-riage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 2017;65:208–215. doi: 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Liu C., Zheng W., Zhang X., Yu J., Gao Q., Hou Y., Huang X. PCR detection of Klebsiella pneumoniae in infant formula based on 16S–23S internal transcribed spacer. Int. J. Food Microbiol. 2008;125:230–235. doi: 10.1016/j.ijfoodmicro.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Turton J., Perry C., Elgohari S., Hampton C.V. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 2010;59:541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 18.Ssekatawa K., Byarugaba D.K., Nakavuma J.L., Kato C.D., Ejobi F., Tweyongyere R., Eddie W.M. Prevalence of pathogenic Klebsiella pneumoniae based on PCR capsular typing harbouring carbapenemases encoding genes in Uganda tertiary hospitals. Antimicrob. Resist. Infect. Control. 2021;10:57. doi: 10.1186/s13756-021-00923-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement. CLSI Document M100-S24. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2014. [Google Scholar]
- 20.Barus T., Hanjaya I., Sadeli J., Lay B.W., Suwanto A., Yulandi A. Genetic Diversity of Klebsiella spp. Isolated from Tempe based on Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction (ERIC-PCR) Hayati J. Biosci. 2013;20:171–176. doi: 10.4308/hjb.20.4.171. [DOI] [Google Scholar]
- 21.Calhau V., Boaventura L., Ribeiro G., Mendonça N., da Silva G.J. Molecular characterization of Klebsiella pneumoniae isolated from renal transplanted patients: Virulence markers, extended-spectrum β-lactamases, and genetic relatedness. Diagn. Microbiol. Infect. Dis. 2014;79:393–395. doi: 10.1016/j.diagmicrobio.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Cao X., Xu X., Zhang Z., Shen H., Chen J., Zhang K. Molecular characterization of clinical multidrug-resistant Klebsiella pneumoniae isolates. Ann. Clin. Microbiol. Antimicrob. 2014;13:16. doi: 10.1186/1476-0711-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y., Zhou H., Qin L., Pang Z., Qin T., Ren H., Pan Z., Zhou J. Frequency, Antimicrobial Resistance and Genetic Diversity of Klebsiella pneumoniae in Food Samples. PLoS ONE. 2016;11:e0153561. doi: 10.1371/journal.pone.0153561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y.-T., Jeng Y.-Y., Chen T.-L., Fung C.-P. Bacteremic community-acquired pneumonia due to Klebsiella pneumoniae: Clinical and microbiological characteristics in Taiwan, 2001–2008. BMC Infect. Dis. 2010;10:307. doi: 10.1186/1471-2334-10-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartantyo S.H.P., Chau M.L., Koh T.H., Yap M., Yi T., Cao D.Y.H., Gutiérrez R.A., Ng L.C. Foodborne Klebsiella pneumoniae: Virulence Potential, Antibiotic Resistance, and Risks to Food Safety. J. Food Prot. 2020;83:1096–1103. doi: 10.4315/JFP-19-520. [DOI] [PubMed] [Google Scholar]
- 26.Puspanadan S., Afsah-Hejri L., Loo Y.Y., Nillian E., Kuan C.H., Goh S.G., Chang W.S., Lye Y.L., John Y.H.T., Rukayadi Y., et al. Detection of Klebsiella pneumoniae in raw vegetables using Most Probable Number-Polymerase Chain Reaction (MPN431 PCR) Int. Food Res. J. 2012;19:1757–1762. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is contained within the article.