Abstract
Purpose
Previous studies have shown the role of ten-eleven translocation 2 (TET2) in CD4+ T cells. However, its function in CD4+ T cells under allergic inflammation is unclear. We aimed to investigate the epigenomic distribution of DNA 5-hydroxymethylcytosine (5hmC) and the role of TET2 in CD4+ T cells of allergic rhinitis (AR).
Methods
The hMeDIP-seq was performed to identify sequences with 5hmC deposition in CD4+ T cells of AR patients. Tet2-deficient or wild type mice were stimulated with ovalbumin (OVA) to develop an AR mouse model. The histopathology in nasal mucosae, Th1/Th2/Treg/Th17 cell percentage, concentrations of Th-related cytokines, expression of Tet and differential hydroxymethylated genes (DhMG), and the global deposition of 5hmC in sorted CD4+ T cells were detected.
Results
Epigenome-wide 5hmC landscape and DhMG in the CD4+ T cells of AR patients were identified. Tet2 depletion did not led to spontaneous inflammation. However, under the stimulation of allergen, OVA, loss of Tet2 resulted in the exacerbation of allergic inflammation, which was characterized by severer allergic symptoms, more inflammatory cells infiltrating the nasal lamina propria, sharper imbalances between Th1/Th2 and Treg/Th17 cells, and excessive secretion of OVA-specific IgE and Th2-related cytokines. Moreover, altered mRNA production of several DhMG and sharp decrease in 5hmC deposition were also observed in Tet2-deficient OVA-exposed mice.
Conclusions
TET2 may regulate DNA 5hmC, DhMG expressions, and CD4+ T cell balance in AR.
Keywords: Allergic rhinitis; 5-hydroxymethylcytosine; CD4-Positive T-Lymphocytes; Tet2 protein, mouse; ovalbumin; epigenomes; cytokines
INTRODUCTION
Allergic rhinitis (AR), characterized by dysfunction of gene expression regulation, is a heterogeneous immune disorder. Recently, AR incidence has dramatically increased,1,2 which cannot be fully explained by classic genetic mechanism. Epigenetic modifications provide insights to the mechanisms of allergic diseases that translate the influence of exposure into alterations in genes expression.3 In eukaryotic cells, DNA methylation is the most common epigenetic modification, which contributes to allergic inflammation and abnormal differentiation of T cells in AR.4,5 Epigenome-wide association studies (EWAS) have shown that there are distinct DNA methylation landscapes between immune dysregulation and degenerative disease, and between AR and asthma.6 DNA methylation signatures are associated with pollution or allergen exposure,7,8 treatment,9 and prognosis,10 suggesting their potential as clinical biomarkers.
Currently, EWAS for DNA methylation in allergic diseases mainly focus on 5-methylcytosine (5mC). However, 5-hydroxymethylcytosine (5hmC) has hardly been studied. The 5hmC is the oxidation product of 5mC, which is catalyzed by ten-eleven translocation (TET) enzymes—TET1, TET2, and TET3.11 The 5hmC can be successively oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), and ultimately transformed to unmodified cytosine, highlighting the DNA demethylation mechanism.12 In airway epithelial cells, exposure to house dust mites (HDMs) and diesel exhaust particles results in elevated TET1 expression; this was associated with global changes in 5mC and 5hmC expression.13,14 Furthermore, TET1 defect exacerbated allergic airway inflammation by regulating interferon and aryl hydrocarbon receptor signaling possibly via 5mC/5hmC in airway epithelial cells.15 TETs and global 5hmC levels of dendritic cells are higher in AR patients than in healthy controls.16 A recent study in nasal mucosa showed the epigenome-wide distribution profile of 5hmC in a murine model of AR.17 The loss of TET2 could lead to 5hmC alteration associated with specific immune pathways.17
CD4+ T cells undergo a series of transcriptional reprogramming and play a vital role in allergic reactions. TET-mediated transformation of 5mC to 5hmC contributes to the expression of lineage-specific signature genes and the differentiation of human and mouse CD4+ T cells.18,19 Hypermethylation occurs at high levels in DNA of naïve T cells.18 Increase in 5hmC can predict DNA demethylation and lineage-specific signature gene activation in differentiated T cells.18,19 Thus, TETs regulate the transformation from naïve T cells to T helper cells (Th)1, Th2, regulatory T cells (Treg cells), and Th17 cells.18,19,20 Our recent study showed that TET2 was related to the Foxp3 hypermethylation in Treg cells of AR.21 However, the epigenome landscape of 5hmC and its role in CD4+ T cells in AR are unknown.
In this study, we generated an epigenome-wide distribution profile of 5hmC in CD4+ T cells of AR patients and investigated the function of TET2 in gene expression and DNA hydroxymethylation in CD4+ T cells using ovalbumin (OVA)-exposed Tet2-deficient mice.
MATERIALS AND METHODS
Human subjects
All protocols for the human study were approved by the Ethics Committee of Renmin Hospital of Wuhan University (WDRY2018-K020). Fifteen healthy volunteers and 15 AR patients were enrolled from the Department of Otolaryngology-Head and Neck Surgery, Renmin Hospital of Wuhan University. The diagnosis of AR was established according to the Chinese Society of Allergy Guidelines for Diagnosis and Treatment of Allergic Rhinitis2: (1) clinical history of nasal symptoms, including sneezing, rhinorrhea, nasal itching, and nasal obstruction with or without ocular symptoms; (2) nasal examinations demonstrating pale and edematous nasal mucosa, and watery nasal discharge; and (3) positive results of serum specific IgE measurement or skin test.
Individuals who did not have any symptoms of allergic diseases (AR, asthma, atopic dermatitis, and food allergy) and who were not diagnosed with allergic diseases or chronic rhinitis were enrolled as healthy controls. Subjects were excluded if they: (1) were diagnosed with infectious diseases, autoimmune diseases, tumors, or severe systemic and psychiatric disorders; (2) were pregnant; (3) were being or had been treated with specific immunotherapy, or had received antihistamines, steroids, antibiotics, or immune drugs in recent 4 weeks; or (4) were ≤14 years old or ≥65 years old. Written informed consent was obtained from all subjects before their participation in the study. The visual analogue scores of symptoms and quality of life,22 clinical information and mucosal samples were obtained at the attack stage for each AR patient. Subject characteristics are presented in Supplementary Table S1.
Mice
All protocols for animal experiments were approved by the Laboratory Animal Welfare & Ethics Committee of Renmin Hospital of Wuhan University (WDRM20181109). Tet2-deficient B6(Cg)-Tet2tm1.2Rao /J mice were obtained from The Jackson Laboratory (West Grove, PA, USA). Wild type (WT) controls and homozygous (Tet2-knockout [KO]) mice were bred from heterozygous parents. All animals were kept in the Animal Experiment Center of Renmin Hospital of Wuhan University.
OVA-exposed mouse models of AR
A mouse model of AR was established by OVA in a specific pathogen-free biohazard containment facility as previously described.23 There were 5 female and 5 male mice (6- to 8-week-old on day 1) in each group. Mice were sensitized by intraperitoneal injection of 300 μL of phosphate-buffered saline (PBS) containing 50 μg of OVA (grade V; Sigma, St. Louis, MO, USA) and 1 mg of aluminum hydroxide on days 1, 8, and 15. From days 22 to 28, the sensitized mice were intranasally challenged with 20 μL of PBS (10 μL per nostril) containing 400 μg OVA. The control group received equivalent amounts of PBS. Symptoms in the mice were observed 5 minutes after the final challenge. Under the anesthesia of pentobarbital sodium, mice were sacrificed and dissected for testing 24 hours after the last challenge.
Isolation of CD4+ T cells
Fresh human lymphocytes were isolated from peripheral blood, and mouse lymphocytes were obtained from the spleens using the Human Blood Lymphocyte Separation Medium Kit and the Mouse Spleen Lymphocyte Separation Medium Kit, respectively (TBD Science, Tianjin, China). Then, human and mouse CD4+ T cells were sorted by positive selection using Anti-Human CD4 Magnetic Particles and Anti-Mouse CD4 Magnetic Particles (BD Biosciences, San Jose, CA, USA), respectively. The purity of CD4+ cells was generally higher than 95%. Genomic DNA and total RNA of the sorted CD4+ T cells were extracted using DNA/RNA isolation kits (Tiangen Biotech, Beijing, China).
Dot blot
Each DNA sample was diluted to 150 ng/µL and denatured at 99°C for 5 minutes. The denatured DNA (2 µL) was spotted on a nitrocellulose membrane (Biosharp, Hefei, China) and air dried. The membrane was blocked by soaking in 5% BSA in TBS-T at room temperature for 1 hour, following which it was immunoblotted with antibody against 5hmC (1:10,000; Active Motif, Carlsbad, CA, USA) and 5mC (1:2,000; Active Motif) at 4°C overnight. Later, the membrane was incubated with horse radish peroxidase-conjugated anti-rabbit IgG (1:40,000 for 5hmC, 1:2,000 for 5mC; Biosharp) at room temperature for 30 minutes. The membrane was then incubated with enhanced chemiluminescent substrate (Biosharp) for 1 minute. Images were taken using the ChemiDocTM Touch Imaging System (Bio-Rad, Hercules, CA, USA).
5hmC DNA immunoprecipitation coupled with next generation sequencing (hMeDIP-seq) and data analysis
The hMeDIP-seq was performed on Illumina HiSeq X ten (Illumina Inc., San Diego, CA, USA) using the hMeDIP Kit (Active Motif) to identify sequences with 5hmC deposition. MACS2 (version: 2.1.2) software was used to call 5hmC-occupied peaks representing enriched hydroxymethylation sites from a filtered alignment file with default parameters. Occupancy analysis and normalization of the peaks were conducted using the R package DiffBind (version: 2.10.0). DiffBind used edgeR to analyze differential hydroxymethylated peaks/regions (DhMR). A P value and false discovery rate (FDR) were assigned to each candidate peak indicating the significance of their being differentially hydroxymethylated. Only peaks with FDR ≤ 0.05 were identified as significantly differential peaks.
A promoter was defined as 2,000 bp upstream and 500 bp downstream of the transcription start site (TSS). CpG islands (CGIs) are the genomic regions where CpG sites occur with high frequency. There are different standards for the definition of CGI. The definition for CGI in this study is based on UCSC Genome Browse database,24 which states that CGI is a region with at least 200 bp, a GC percentage of more than 50%, and an observed-to-expected CpG ratio higher than 60%. The observed value in the “observed-to-expected CpG ratio” was calculated as the number of CpG, and the expected value was calculated as: (number of C × number of G)/length of sequence.25
Fisher's exact test and the Benjamini-Hochberg procedure were used for Gene Ontology (GO) enrichment analysis of differential hydroxymethylated genes (DhMG). The ontology contained 3 parts: biological process, cellular component, and molecular function.
Reverse real-time quantitative transcription polymerase chain reaction (RT-qPCR)
Complementary DNA was produced from the total RNA using the PrimeScript™ RT reagent kit with gDNA Eraser (TaKaRa Bio, Beijing, China). qRT-PCR reactions were performed on the CFX96 Real Time PCR Detection System (Bio-Rad) using TB Green™ Premix Ex Taq™ II (TaKaRa Bio). Relative mRNA levels were calculated using comparative Ct values after normalizing to β-actin (2−ΔΔCT method). The qPCR primer sequences are listed in Supplementary Table S2.
Western blot analysis
Protein was extracted from CD4+ T cells, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and detected with antibody to TET2 (Proteintech, Wuhan, China). Images were taken using the ChemiDocTM Touch Imaging System (Bio-Rad). Relative protein levels were quantified by band intensity after normalizing to glyceraldehyde 3-phosphate dehydrogenase.
Histopathological analysis of nasal mucosae
After mice were sacrificed, mouse noses were removed and fixed with 4% paraformaldehyde overnight, decalcified in an ethylenediaminetetraacetic acid decalcifying solution for 2 weeks, dehydrated in a series of increasing concentrations of ethanol, and embedded in paraffin. The paraffin-embedded tissues were sectioned coronally by continuous microtoming at a thickness of 4 μm. Histopathology of nasal mucosae of the mice in all groups was evaluated with hematoxylin and eosin staining.
Immunofluorescence
Human nasal mucosae were obtained from patients who had turbinate hypertrophy with or without AR during partial turbinectomy. After fixing and embedding, nasal mucosae were sectioned into 4-μm slices. Slices were stained with Anti-Human CD4 antibody and Anti-Human CD4 antibody (Proteintech). The immunofluorescence intensity of TET2 was calculated in manually selected CD4+ cells by ImageJ (National Institutes of Health, Bethesda, MD, USA). Briefly, after merging the channels of CD4 and DAPI, the CD4 and DAPI double-positive cells were manually selected. Then, the selected areas were projected onto TET2-labelled channel. The immunofluorescence intensity of each selected site in TET2 channel was calculated. Each slice was counted in 3 randomly selected high-power fields at 400× magnification.
OVA-specific immunoglobulin E (OVA-sIgE) and cytokine measurement
Serum and nasal lavage fluid (NLF) were obtained from each mouse. The concentrations of OVA-sIgE in serum were detected using the Anti-Ovalbumin IgE (mouse) ELISA Kit (Cayman Chemical, Ann Arbor, MI, USA) and an EnSight Multimode Plate Reader (PerkinElmer, Waltham, MS, USA). The concentrations of IFN-γ, IL-4, IL-5, IL-13, IL-10, and IL-17A were detected using the Bio-Plex suspension array system (Bio-Rad) according to the Bio-Plex Pro Mouse Cytokine Assays Quick Guide.
Flow cytometric analysis
Mouse lymphocytes were extracted from the fresh mouse spleens using the Mouse Spleen Lymphocyte Separation Medium Kit (TBD Science) and were stained with anti-CD4-FITC (BD Biosciences). To induce secretion, the lymphocytes were stimulated with Leukocyte Activation Cocktail (BD Biosciences) for 6 hours. After surface marker staining with anti-CD4-FITC (BD Biosciences), lymphocytes were permeabilized and incubated with anti-IFN-γ-APC, anti-IL-4-PerCP-Cy5.5, anti-FOXP3-APC, and anti-IL-17A-PerCP-Cy5.5 (BD Biosciences). Flow cytometry was performed on a BD FACSCalibur flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (version 10.0.7; Treestar, Ashland, KY, USA).
Statistical analysis
GraphPad Prism software (version 8.3; GraphPad Software, San Diego, CA, USA) and SPSS statistical software (version 22.0; IBM, Chicago, IL, USA) were used for data analysis. To compare differences between 2 groups, unpaired 2-tailed Student’s t test was used if data followed the Gaussian distribution. The Mann-Whitney test was used if the data did not follow the Gaussian distribution. Data are presented as mean ± standard error of mean. It was considered statistically significant if P < 0.05.
RESULTS
Global DNA 5hmC deposition and TET family expression in CD4+ T cells of AR patients
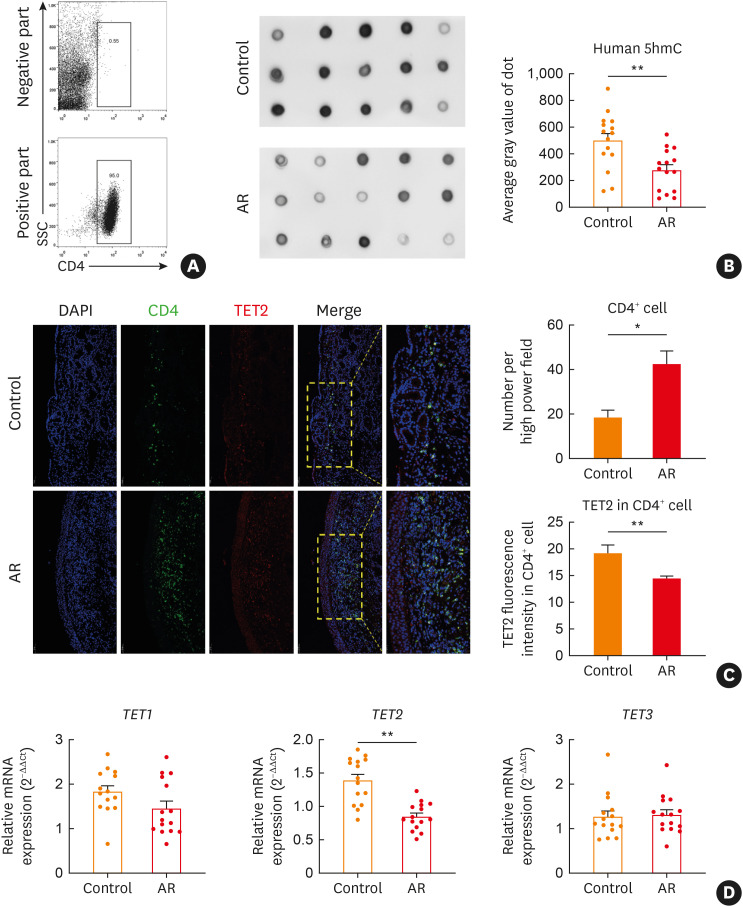
The global expression of DNA 5hmC was analyzed by dot blot analysis (Fig. 1A). Results showed that AR patients had distinctly lower global DNA 5hmC levels in CD4+ T cells than healthy volunteers (Fig. 1B). TET family proteins catalyze the generation of 5hmC. Immunofluorescence staining indicated that there were more CD4+cells in the nasal mucosae of AR patients than of healthy volunteers. While TET2 expression was lower in CD4+ cells of AR subjects (Fig. 1C and D), TET1 and TET3 mRNA levels were not different between the 2 groups (Fig. 1D). These results led us to hypothesize that TET2-mediated DNA hydroxymethylation could be a key player in CD4+ T cells of AR.
Fig. 1. Global DNA 5hmC deposition and TET family expression in CD4+ T cells of AR patients. (A) After positive selection using anti-human CD4 magnetic nanoparticles, the positive and negative fractions were verified by flow cytometry. (B) Global DNA 5hmC deposition in CD4+ T cells was detected by dot blot and quantified by average gray value of dot. (C) Expression of CD4 and TET2 in human nasal tissues was indicated by immunofluorescence. The immunofluorescence intensity of TET2 was calculated in manually selected CD4+cells. (D) Relative mRNA expression of the TET family (TET1, TET2, and TET3) in CD4+ T cells was measured by RT-qPCR and calculated using comparative Ct values after normalizing to β-actin. Bars indicate mean ± standard error of the mean.
Control, healthy volunteers; AR, allergic rhinitis; 5hmC, 5-hydroxymethylcytosine; TET, ten-eleven translocation; RT-qPCR, reverse real-time quantitative transcription polymerase chain reaction.
*P < 0.05, **P < 0.01.
Epigenome-wide 5hmC distribution in CD4+ T cells of AR patients
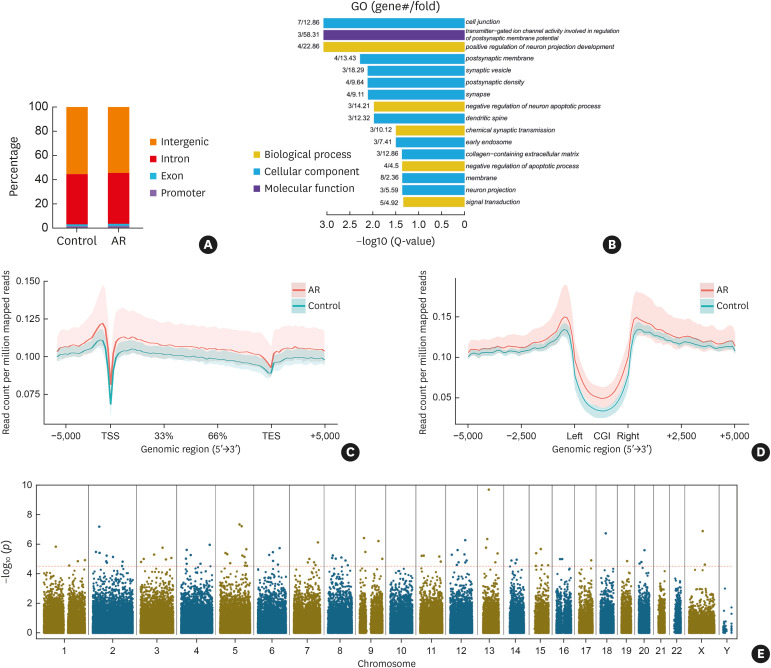
Noting the global alteration in 5hmC in CD4+ T cells of AR patients, we performed hMeDIP-seq to generate an epigenome-wide distribution profile of 5hmC in CD4+ T cells. Five AR patients and 5 healthy controls were included. We identified 81,768 ± 10,757 5hmC-occupied peaks in the AR group and 59,384 ± 9,328 in the control group. According to the annotation from the UCSC Genome Browse of ‘‘known genes,’’ the peak distribution was analyzed in 4 types of regions in the human genome: promoter, exon, intron, and intergenic regions.24,26 Results showed that the 5hmC distribution in the AR and control groups displayed similar patterns. Most of the 5hmC-occupied peaks were located in the intergenic regions, ranging from 53% to 55% in the AR group and 55% to 56% in the control group, followed by introns which ranged from 41% to 42% in both groups. In contrast, only 2%–3% of 5hmC-occupied peaks were located in promoters and 1%–2% in exons in both groups (Fig. 2A).
Fig. 2. Epigenome-wide 5hmC distribution and DhMR in CD4+ T cells of AR patients. (A) The percentage of 5hmC-occupied peaks in promoter, exon, intron, and intergenic regions. (B) DhMG were analyzed by GO and enriched in biological process, cellular component, and molecular function. The P value indicates the significance of DhMG enriched in GO terms. (C) The normalized read count for 5hmC across the gene body ± 5 kb flanking regions with 100 bp resolution is shown. (D) The normalized read count for 5hmC around ± 5 kb regions flanking CGI centers with 100 bp resolution is shown. The solid red and blue lines indicate the average count for the AR and control group, respectively, while the red and blue shadows indicate the range of counts for the AR and control groups, respectively. (E) The Manhattan plot shows the chromosome distribution and significance of the differences in all the 143,812 5hmC-occupied peaks between AR patients and healthy volunteers. The red dotted horizontal line represents the FDR corrected threshold (FDR < 0.05) of genome-wide significance.
Control, healthy volunteers; AR, allergic rhinitis; 5hmC, 5-hydroxymethylcytosine; DhMR, differentially hydroxymethylated regions; DhMG, differentially hydroxymethylated genes; GO, Gene Ontology; CGI, CpG island; FDR, false discovery rate.
We next analyzed the profile of 5hmC within the gene body and its 5,000 bp upstream and downstream. CGIs are genomic regions where CpG sites occur with high frequency. The profiles of 5hmC within CGI and its 5,000 bp upstream and downstream were also generated. As shown in Fig. 2C and E, the distribution of 5hmC around the gene body and CGI was similar in both groups. Moreover, our results revealed the depletion of 5hmC at the TSS and the center of CGI (Fig. 2D and E). In mammalian genomes, CGI are usually associated with promoters.27 This was consistent with our result that there were only a few 5hmC-occupied peaks located in promoters (Fig. 2A).
DhMR between CD4+ T cells of AR patients and healthy volunteers
Differential hydroxymethylation analysis was conducted among 143,812 5hmC-occupied peaks between AR patients and healthy controls. The chromosome distribution of all peaks is shown in a Manhattan plot (Fig. 2E). A total of 92 DhMR (the dots above the red line) were identified, containing 19 hyperhydroxymethylatied regions and 73 hypohydroxymethylatied regions. GO analysis showed that DhMG were significantly enriched in multiple terms such as cell junction, transmitter-gated ion channel activity, positive regulation of neuron projection development, membrane, and signal transduction (Fig. 2C).
Validating the expression of DhMG in CD4+ T cells of AR patients and healthy volunteers
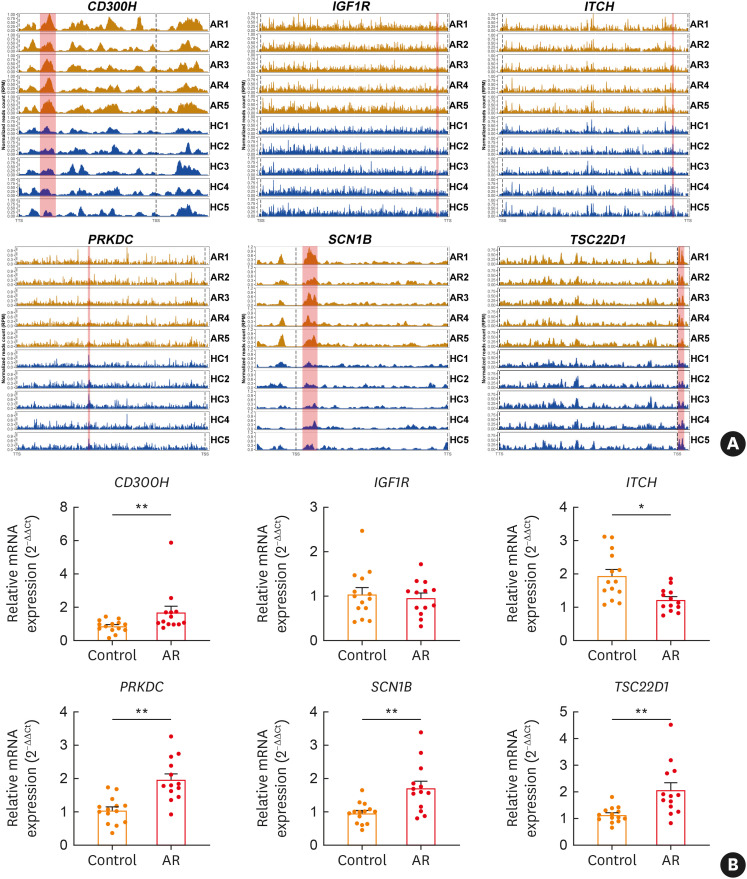
Previous studies reported that CD300H, insulin-like growth factor 1 receptor (IGF1R), itchy E3 ubiquitin protein ligase (ITCH), protein kinase, DNA-activated, catalytic subunit (PRKDC), sodium voltage-gated channel beta subunit 1 (SCN1B), and transforming growth factor β-stimulated clone 22 (TSC-22) domain family member 1 (TSC22D1) played an important role in the immune or allergic response.28,29,30,31,32,33 Some DhMR were located in the aforementioned genes (Fig. 3A). The mRNA expression of these genes was verified by RT-qPCR. Result showed that AR patients had lower ITCH mRNA production and more CD300H, PRKDC, SCN1B, and TSC22D1 mRNA production in CD4+ T cells than in healthy controls (Fig. 3B).
Fig. 3. Validating the expressions of DhMG in CD4+ T cells of AR patients and that of healthy volunteers. (A) The normalized read count for 5hmC around DhMG including CD300H, IGF1R, ITCH, PRKDC, SCN1B, and TSC22D1 is shown. The DhMR are highlighted in red. (B) Relative mRNA expressions of CD300H, IGF1R, ITCH, PRKDC, SCN1B, and TSC22D1 in CD4+ T cells were measured by RT-qPCR and calculated using comparative Ct values after normalizing to β-actin. Bars show mean ± standard error of the mean.
DhMG, differentially hydroxymethylated genes; Control, healthy volunteers; AR, allergic rhinitis; 5hmC, 5-hydroxymethylcytosine; IGF1R, insulin-like growth factor 1 receptor; ITCH, itchy E3 ubiquitin protein ligase; PRKDC, protein kinase, DNA-activated, catalytic subunit; SCN1B, sodium voltage-gated channel beta subunit 1; TSC22D1, transforming growth factor β-stimulated clone 22 domain family member 1; RT-qPCR, reverse real-time quantitative transcription polymerase chain reaction.
*P < 0.05, **P < 0.01.
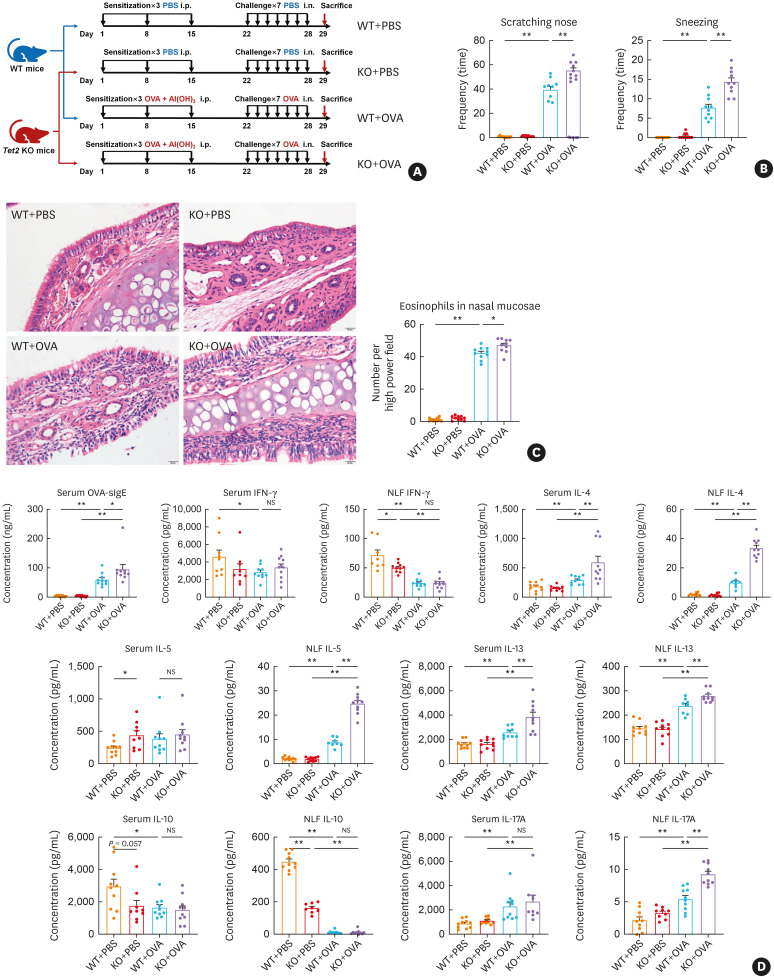
Tet2 deficiency exacerbated the allergic inflammation in OVA-exposed mice
To delineate the role of TET2 in DNA hydroxymethylation and gene expression in allergic inflammation, we used KO mouse and OVA-exposed AR mouse models (Fig. 4A). The main nasal symptoms of OVA-exposed mice were nose scratching and sneezing. The WT+OVA mice showed obvious nose scratching and sneezing than the WT+PBS mice. The KO+OVA mice sneezed and scratched the nose even more frequently than the WT+OVA mice (Fig. 4B). Hematoxylin and eosin staining (Fig. 4C) showed that the PBS-exposed KO mice (KO+PBS) did not suffer from noticeable infiltration of eosinophils or other inflammatory cells in the nasal lamina propria. The pseudostratified ciliated columnar epithelium was intact in this group. These were similar findings in the PBS-exposed WT mice (WT+PBS). However, under the stimulation of OVA, the KO mice (KO+OVA) exhibited higher levels of infiltration of eosinophils and other inflammatory cells in the nasal lamina propria than the WT mice (WT+OVA). Tissue edema and cilia exfoliation were also observed in both groups.
Fig. 4. Tet2 deficiency exacerbates allergic inflammation in OVA-exposed mice. (A) OVA-exposed mice models of AR were developed using WT and KO mice via intraperitoneal sensitization followed by intranasal administration of OVA. (B) Symptoms of nose scratching and sneezing were observed in 5 minutes after the final challenge. (C) Histopathology of nasal mucosae in mice was evaluated with hematoxylin and eosin staining. Eosinophils in nasal mucosae were counted in high-power fields at 400× magnification. (D) OVA-sIgE and Th-related cytokines in serum or NLF are shown.
Tet, ten-eleven translocation; OVA, ovalbumin; AR, allergic rhinitis; WT, wild type; KO, Tet2 knockout; PBS, phosphate-buffered saline; NLF, nasal lavage fluid; OVA-sIgE, ovalbumin-specific immunoglobulin E; NS, not significant.
*P < 0.05, **P < 0.01.
We also detected OVA-sIgE and Th-related cytokines in serum and NLF (Fig. 4D). Results showed that ablation of Tet2 had a minor effect on the secretion of OVA-sIgE and Th-related cytokines in the PBS-exposed mice, demonstrating decreases in IFN-γ in serum and IL-10 in NLF as well as increases in IL-5 in NLF. The WT+OVA mice showed a decrease in IFN-γ and IL-10 in serum and NLF as well as an increase in OVA-sIgE, IL-4, IL-5, IL-13, and IL-17A in the serum and/or NLF. Remarkably, Tet2 deficiency aggravated the excessive secretion of OVA-sIgE, IL-4, IL-5, IL-13, and IL-17A in OVA-exposed mice.
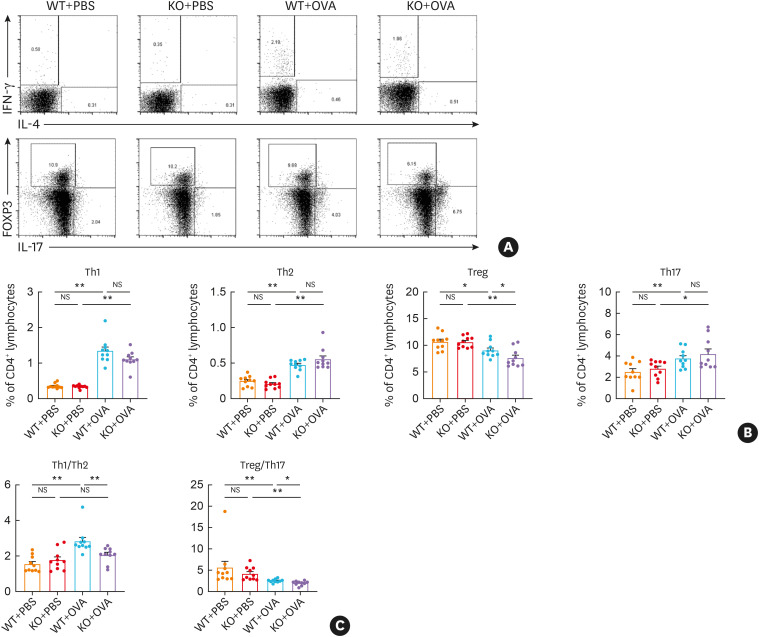
Flow cytometry showed the percentages of CD4+IFN-γ+Th1 cells, CD4+IL-4+Th2 cells, CD4+FOXP3+Treg cells, and CD4+IL-17+Th17 cells in CD4+ cells (Fig. 5A and B). Without allergen stimulation, the percentage of Th1, Th2, Treg, and Th17 cells in the KO+PBS mice less significantly changed compared with the WT+PBS mice. The ratio of Treg/Th17 cells and Th1/Th2 cells also remained unchanged in the KO+PBS mice. However, under the stimulation of OVA, the KO+OVA mice showed a lower Treg cell percentage than the WT+OVA mice. The ratio of Th1/Th2 to Treg/Th17 cells reduced more significantly in the KO+OVA group (Fig. 5).
Fig. 5. Tet2 deficiency aggravates the imbalance between Th1/Th2 and Treg/Th17 cells in OVA-exposed mice. (A) Lymphocytes were extracted from fresh the mouse spleens and the expressions of anti-IFN-γ, IL-4, FOXP3 and IL-17A in gated CD4+ T cells were analyzed. (B) The percentages of CD4+IFN-γ+Th1 cells, CD4+IL-4+Th2 cells, CD4+FOXP3+Treg cells, and CD4+IL-17+Th17 cells in CD4+ cells were calculated. (C) The ratio of Treg/Th17 cells to that of Th1/Th2 cells is shown.
Tet, ten-eleven translocation; Th, T helper; Treg, regulatory T; PBS, phosphate-buffered saline; OVA, ovalbumin; WT, wild type; KO, Tet2 knockout; NS, not significant.
*P < 0.05, **P < 0.01.
Tet2 modulates DhMG expression and DNA methylation in CD4+ T cells of OVA-exposed mice
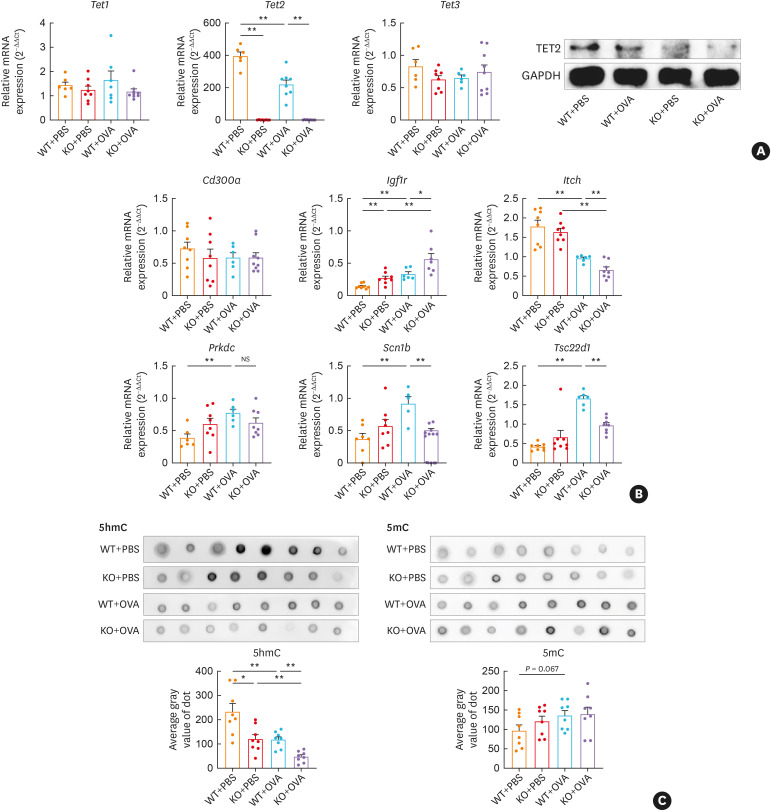
The 3 TET proteins show distinct and overlapping functions in differentiated cells, such as T cells and mast cells.34,35,36 We confirmed the expression of Tets in CD4+ T by RT-qPCR and western blot analysis. Data showed that there was no compensatory increase in Tet1 or Tet3 mRNA levels because of KO in CD4+ T cells (Fig. 6A).
Fig. 6. Tet2 modulates DhMG expressions and DNA methylation in CD4+ T cells of OVA-exposed mice. (A) Relative mRNA expressions of Tet1, Tet2, and Tet3 in CD4+ T cells were determined by RT-qPCR and calculated using comparative Ct values after normalizing to β-actin. The relative protein levels were quantified by western blotting after normalizing to GAPDH. (B) Relative mRNA expressions of Cd300a, Igf1r, Itch, Prkdc, Scn1b, and Tsc22d1 in CD4+ T cells were determined by RT-qPCR and calculated using comparative Ct values after normalizing to β-actin. (C, D) Global DNA 5hmC deposition in mice CD4+ T cells was detected using dot blot and quantified using the average gray value of dot.
Tet, ten-eleven translocation; DhMG, differentially hydroxymethylated genes; PBS, phosphate-buffered saline; OVA, ovalbumin; WT, wild type; KO, Tet2 knockout; RT-qPCR, reverse real-time quantitative transcription polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Igf1r, insulin-like growth factor 1 receptor; Itch, itchy E3 ubiquitin protein ligase; Prkdc, protein kinase, DNA-activated, catalytic subunit; Scn1b, sodium voltage-gated channel beta subunit 1; Tsc22d1, transforming growth factor β-stimulated clone 22 domain family member 1; NS, not significant.
*P < 0.05, **P < 0.01.
We next analyzed the influence of Tet2 on the aforementioned DhMG. The mRNA expressions of mouse homologous genes were analyzed in CD4+ T cells. Because the murine counterpart of CD300H has not been identified, we analyzed Cd300a, the homolog of CD300A, which had the closest relationship with CD300H in the phylogenetic tree.28 KO resulted in the up-regulation of Igf1r and the down-regulation of Itch, Scn1b, and Tsc22d1. However, Tet2 depletion had no influence on the expression of Cd300a mRNA (Fig. 6B).
Furthermore, the global 5hmC levels were considerably altered by OVA stimulation and KO. Significantly, OVA stimulation resulted in the decrease in 5hmC of CD4+ T cells, which was further aggravated when Tet2 was depleted. Mild up-regulation of 5mC was concurrently noticed after OVA stimulation and KO (Fig. 6C).
DISCUSSION
The production of TET-oxidized 5mC contained 5hmC, 5fC, and 5caC. The 5mC accounts for 1% of bases and 5% of cytosines. The 5hmC accounts for 0.1% of bases and 0.5% of cytosines.37 The 5fC and 5caC are even less abundant, regardless of the cell type.37,38 Most articles concerning these oxidization products focused on 5hmC, and there was little research into the function of 5fC and 5caC in immune cells.39,40 A recent study by Onodera et al.39 has showed that although loss of TET enzymes disrupted gene expression, loss of TDG enzymes resulted in the accumulation of 5fC/5caC with no effects. Changes in 5fC and 5caC made only a minor contribution to the differentiation and activation of Th2 cells and LPS-activated macrophages.39 Caldwell et al.40 identified 5fC/5caC, rather than 5hmC, which was the major driver of DNA demethylation during induced pluripotent stem cells reprogramming, and played important roles in gene regulation and chromatin reorganization. The roles of 5fC and 5caC in allergic diseases were unclear. While accumulating studies have shown that TETs and 5hmC were closely related with the antigen-induced immune response. In vitro, Tet2 was up-regulated in mouse CD4+ T cells by TCR signaling.41 In a zebrafish model, elevations in IL-4 and IL-13A were induced by soluble antigens (lipopolysaccharides and keyhole limpet hemocyanin), along with the up-regulation of Tet1 and Tet3 in the spleen, kidneys, and peripheral lymphocytes.42 Exposure to HDMs could result in elevations in TET1 and changes in 5mC and 5hmC in human airway epithelial cells.13 Chemical allergens induced 5mC and 5hmC changes in DNA from draining lymph nodes of mice.43 In this study, we reported that AR patients and OVA-exposed mice had less global DNA 5hmC deposition and TET2 expression in CD4+ T cells. These observations indicates that TETs and 5hmC may have essential functions in immune responses induced by antigens, especially allergens.
Emerging studies have highlighted the roles of TETs and 5hmC in allergic airway diseases. Increases in global DNA 5mC and 5hmC levels were detected in the lungs of HDM-induced mouse model of asthma.44 Hypomethylation of TET1 promoters and an increase in global 5hmC levels in human saliva, peripheral blood mononuclear cells (PBMCs), and bronchial epithelial cells were significantly associated with asthma in humans.14 PM2.5 particles aggravated allergic inflammation in the lungs of a murine asthma model and disturbed the balance of Treg/Th17 cells, probably by inhibiting TET2 activity.45 Our previous study showed that the decline in TET2 was related to hypermethylation of Foxp3 in Treg cells of AR subjects.21 It was reported that AR patients had higher TETs and global 5hmC levels in PBMCs and TET1 in dendritic cells.16 A recent study indicated an elevation in Tet2 mRNA and a decrease in genome-wide 5hmC in nasal mucosa obtained from a murine model of AR.17 We focused on CD4+ T cells. After observing the global alteration in 5hmC and TET2 expression in CD4+ T cells of AR, we generated an epigenome-wide distribution profile of 5hmC by hMeDIP-seq. The distribution characteristics of 5hmC, which was enriched in intergenic regions and introns as well as depleted at promoters, TSS, and the center of CGI, suggested that 5hmC may primarily function as enhancers, but not as promoters.19
We also identified DhMR and DhMG in AR. GO analysis showed that they were associated with cell junction, ion channel activity, membrane, and signal transduction. Expressions of DhMG, including CD300H, IGF1R, ITCH, PRKDC, SCN1B, and TSC22D1 mRNA, were verified in CD4+ T cells of AR subjects. Tet2 deficiency resulted in an increased production of Igf1r mRNA and a decreased production of Itch, Scn1b, and Tsc22d1 mRNA in CD4+ T cells of an OVA-exposed AR mouse model. CD300H belongs to the CD300 family, which in turn belongs to the immunoglobulin superfamily.27 The CD300 family is expressed on granulocytes, dendritic cells, monocytes-macrophages, mast cells, and lymphocytes, suggesting that they have vital functions in immune response and inflammation.46 IGF1R has tyrosine kinase activity. IGF-1 could suppress allergic contact dermatitis in a manner, depending on Treg cells.47 ITCH, as a member of the Nedd4 family of HECT domain E3 ubiquitin ligases, induced the up-regulation of Foxp3 and contributed to the suppression of airway inflammation in vivo. 30 Other studies also suggested the roles of ITCH in CD4+ T cells, follicular helper T cells, CD8+T cells, and B cells.48,49 The PRKDC encodes the catalytic subunit of the DNA-dependent protein kinase (DNA-PK). DNA-PK is involved in dendritic cell-induced airway inflammatory responses to HDMs.31 Furthermore, ablation of DNA-PK attenuated allergic inflammation in a murine asthma model.31,50 SCN1B modulates the kinetics of channel inactivation. Ion channels have key functions in the immune system as previously documented.32 TSC22D1 is a member of TSC-22 domain family of leucine zipper transcription factors. TSC-22 can bind to Smad3 and Smad4, and regulate their transcriptional activity, thereby enhancing TGF-β signaling.33 For the functions of these DhMG in immunity, TET2 may influence the allergic response in AR by mediating DNA hydroxymethylation and by regulating the expression of these genes.
The effects of TET gene deletions under certain inflammatory conditions have been studied in previous studies. TET2 knockout mice are viable and fertile with a mild hematopoietic phenotype.51 In a murine model of asthma, Tet1 deficiency aggravated asthma severity, including lung eosinophilia and airway hyperresponsiveness.15 Tet2 deficiency resulted in an increase in IL-6 and contributed to the inflammation of endotoxin shock and colitis.52 Tet1- and Tet2-deficient mice displayed gain of methylation in Foxp3 and impaired Treg cell function and autoimmune inflammation in the colon, lungs, and liver.53 Li et al.16 also observed that TET1 inhibition in dendritic cells dampened its function of activating Treg cells. They attributed this to the hypothesis that dendritic cells from AR patients who had higher TET1 expression were susceptible to TET1 decrease.16 Meng et al.17 reported that loss of Tet2 could significantly increase the severity of allergy symptoms and the production of IL-17. In our study, Tet2 depletion did not lead to detectable spontaneous inflammation. However, loss of Tet2 dampened immune homeostasis which was unfolded under the stimulation of allergen, OVA. Tet2 deficiency resulted in the exacerbation of OVA-induced allergic inflammation, characterized by a sharper imbalance between CD4+ Th cells and an excessive secretion of Th2-related cytokines. This result was consistent with the effects of TETs gene deletions in the above studies.
In particular, López-Moyado et al.54 found dual mechanisms of TET-mediated DNA methylation. TET knockout in diverse cell types (such as embryonic stem cells [ESCs], neural precursor cells differentiated from ESCs, hematopoietic stem/precursor cells, T cells, and pro-B cells) not only resulted in the expected DNA hypermethylation in euchromatin, but also unexpected hypomethylation in the heterochromatin. This could lead to the silencing of most active enhancers and most highly expressed genes in euchromatin and the reactivation of heterochromatin.54 Based on the above knowledge, we hypothesize that when TET2 is deficient, genes were repressed to various extents in heterochromatin are reactivated. This may lead to the dysfunction of immunity and may make the organism more susceptible to allergens.
There are several limitations in the present study. First, TET2 and 5hmC were altered in AR and OVA-exposed mice. However, how allergens induced the changes in Tet2 expression is still unknown. Secondly, we only investigated the results of KO. More research concerning the function of TET1 and TET3 in AR would be warranted. Thirdly, the roles of DhMG in AR need to be further clarified. Studies delineating the relationship among TET2, DhMG, and AR are warranted.
In conclusion, AR patients had less global DNA 5hmC deposition and TET2 expression in CD4+ T cells than healthy volunteers. The 5hmC was enriched in intergenic regions and introns as well as was depleted at the promoters, TSS, and the center of CGI. DhMG, including CD300H, IGF1R, ITCH, PRKDC, SCN1B, and TSC22D1, were identified. Tet2 depletion did not lead to detectable spontaneous inflammation. However, loss of Tet2 dampened immune homeostasis, which was unfolded under the stimulation of allergen, OVA. Tet2 deficiency led to the exacerbation of allergic inflammation, which was characterized by more severe allergy symptoms, more inflammatory cells infiltrating the nasal lamina propria, sharper imbalance between Th1/Th2 and Treg/Th17 cells, and more excessive secretion of OVA-sIgE, IL-4, IL-5, IL-13, and IL-17A. These may have been attributed to the abnormal deposition of DNA methylation, aberrant expression of DhMG, and dysfunction of CD4+ Th cells caused by Tet2 deficiency. For the first time, this study delineated the epigenome-wide 5hmC landscape in CD4+ T cells of AR patients. A Tet2-deficient mouse model of AR suggested that TET2 could regulate DNA 5hmC, DhMG expressions, and balance between CD4+ T cells in AR.
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (No. 81770986 and 82071017), National Natural Science Foundation of Hubei Province (No. 2021CFB125) and the Fundamental Research Funds for the Central Universities (No. 2042021kf0093).
Footnotes
Disclosure: There are no financial or other issues that might lead to conflict of interest.
SUPPLEMENTARY MATERIALS
Characteristics of the subjects
Primer sequences
References
- 1.Zhang Y, Zhang L. Increasing prevalence of allergic rhinitis in China. Allergy Asthma Immunol Res. 2019;11:156–169. doi: 10.4168/aair.2019.11.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng L, Chen J, Fu Q, He S, Li H, Liu Z, et al. Chinese Society of Allergy guidelines for diagnosis and treatment of allergic rhinitis. Allergy Asthma Immunol Res. 2018;10:300–353. doi: 10.4168/aair.2018.10.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long A, Bunning B, Sampath V, DeKruyff RH, Nadeau KC. Epigenetics and the environment in airway disease: asthma and allergic rhinitis. Adv Exp Med Biol. 2020;1253:153–181. doi: 10.1007/978-981-15-3449-2_6. [DOI] [PubMed] [Google Scholar]
- 4.Alashkar Alhamwe B, Alhamdan F, Ruhl A, Potaczek DP, Renz H. The role of epigenetics in allergy and asthma development. Curr Opin Allergy Clin Immunol. 2020;20:48–55. doi: 10.1097/ACI.0000000000000598. [DOI] [PubMed] [Google Scholar]
- 5.Tumes DJ, Papadopoulos M, Endo Y, Onodera A, Hirahara K, Nakayama T. Epigenetic regulation of T-helper cell differentiation, memory, and plasticity in allergic asthma. Immunol Rev. 2017;278:8–19. doi: 10.1111/imr.12560. [DOI] [PubMed] [Google Scholar]
- 6.Clark AD, Nair N, Anderson AE, Thalayasingam N, Naamane N, Skelton AJ, et al. Lymphocyte DNA methylation mediates genetic risk at shared immune-mediated disease loci. J Allergy Clin Immunol. 2020;145:1438–1451. doi: 10.1016/j.jaci.2019.12.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prunicki M, Stell L, Dinakarpandian D, de Planell-Saguer M, Lucas RW, Hammond SK, et al. Exposure to NO2, CO, and PM2.5 is linked to regional DNA methylation differences in asthma. Clin Epigenetics. 2018;10:2. doi: 10.1186/s13148-017-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung KH, Torrone D, Lovinsky-Desir S, Perzanowski M, Bautista J, Jezioro JR, et al. Short-term exposure to PM2.5 and vanadium and changes in asthma gene DNA methylation and lung function decrements among urban children. Respir Res. 2017;18:63. doi: 10.1186/s12931-017-0550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kere M, Gruzieva O, Ullemar V, Söderhäll C, Greco D, Kull I, et al. Effects of inhaled corticosteroids on DNA methylation in peripheral blood cells in children with asthma. Allergy. 2020;75:688–691. doi: 10.1111/all.14043. [DOI] [PubMed] [Google Scholar]
- 10.Vermeulen CJ, Xu CJ, Vonk JM, Ten Hacken NH, Timens W, Heijink IH, et al. Differential DNA methylation in bronchial biopsies between persistent asthma and asthma in remission. Eur Respir J. 2020;55:1901280. doi: 10.1183/13993003.01280-2019. [DOI] [PubMed] [Google Scholar]
- 11.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Chen X, Weirauch MT, Zhang X, Burleson JD, Brandt EB, et al. Diesel exhaust and house dust mite allergen lead to common changes in the airway methylome and hydroxymethylome. Environ Epigenet. 2018;4:dvy020. doi: 10.1093/eep/dvy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somineni HK, Zhang X, Biagini Myers JM, Kovacic MB, Ulm A, Jurcak N, et al. Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J Allergy Clin Immunol. 2016;137:797–805.e5. doi: 10.1016/j.jaci.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burleson JD, Siniard D, Yadagiri VK, Chen X, Weirauch MT, Ruff BP, et al. TET1 contributes to allergic airway inflammation and regulates interferon and aryl hydrocarbon receptor signaling pathways in bronchial epithelial cells. Sci Rep. 2019;9:7361. doi: 10.1038/s41598-019-43767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Lu T, Sun W, Ma R, Zhong H, Wei Y, et al. Ten-eleven translocation (TET) enzymes modulate the activation of dendritic cells in allergic rhinitis. Front Immunol. 2019;10:2271. doi: 10.3389/fimmu.2019.02271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng C, Gu L, Li Y, Li R, Cao Y, Li Z, et al. Ten-eleven translocation 2 modulates allergic inflammation by 5-hydroxymethylcytosine remodeling of immunologic pathways. Hum Mol Genet. 2021;30:1985–1995. doi: 10.1093/hmg/ddab167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nestor CE, Lentini A, Hägg Nilsson C, Gawel DR, Gustafsson M, Mattson L, et al. 5-hydroxymethylcytosine remodeling precedes lineage specification during differentiation of human CD4+ T cells. Cell Reports. 2016;16:559–570. doi: 10.1016/j.celrep.2016.05.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue X, Trifari S, Äijö T, Tsagaratou A, Pastor WA, Zepeda-Martínez JA, et al. Control of Foxp3 stability through modulation of TET activity. J Exp Med. 2016;213:377–397. doi: 10.1084/jem.20151438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan L, Qiu T, Xiang R, Cao C, Deng Y, Tao Z, et al. Down-regulation of Tet2 is associated with Foxp3 TSDR hypermethylation in regulatory T cell of allergic rhinitis. Life Sci. 2020;241:117101. doi: 10.1016/j.lfs.2019.117101. [DOI] [PubMed] [Google Scholar]
- 22.Pfaar O, Demoly P, Gerth van Wijk R, Bonini S, Bousquet J, Canonica GW, et al. Recommendations for the standardization of clinical outcomes used in allergen immunotherapy trials for allergic rhinoconjunctivitis: an EAACI Position Paper. Allergy. 2014;69:854–867. doi: 10.1111/all.12383. [DOI] [PubMed] [Google Scholar]
- 23.Kim EH, Kim JH, Samivel R, Bae JS, Chung YJ, Chung PS, et al. Intralymphatic treatment of flagellin-ovalbumin mixture reduced allergic inflammation in murine model of allergic rhinitis. Allergy. 2016;71:629–639. doi: 10.1111/all.12839. [DOI] [PubMed] [Google Scholar]
- 24.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47:D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 26.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, et al. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33:e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niizuma K, Tahara-Hanaoka S, Noguchi E, Shibuya A. Identification and characterization of CD300H, a new member of the human CD300 immunoreceptor family. J Biol Chem. 2015;290:22298–22308. doi: 10.1074/jbc.M115.643361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piñeiro-Hermida S, Gregory JA, López IP, Torrens R, Ruíz-Martínez C, Adner M, et al. Attenuated airway hyperresponsiveness and mucus secretion in HDM-exposed Igf1r-deficient mice. Allergy. 2017;72:1317–1326. doi: 10.1111/all.13142. [DOI] [PubMed] [Google Scholar]
- 30.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9:245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra A, Brown AL, Yao X, Yang S, Park SJ, Liu C, et al. Dendritic cells induce Th2-mediated airway inflammatory responses to house dust mite via DNA-dependent protein kinase. Nat Commun. 2015;6:6224. doi: 10.1038/ncomms7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaeth M, Feske S. Ion channelopathies of the immune system. Curr Opin Immunol. 2018;52:39–50. doi: 10.1016/j.coi.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi SJ, Moon JH, Ahn YW, Ahn JH, Kim DU, Han TH. Tsc-22 enhances TGF-beta signaling by associating with Smad4 and induces erythroid cell differentiation. Mol Cell Biochem. 2005;271:23–28. doi: 10.1007/s11010-005-3456-7. [DOI] [PubMed] [Google Scholar]
- 34.Montagner S, Leoni C, Emming S, Della Chiara G, Balestrieri C, Barozzi I, et al. TET2 regulates mast cell differentiation and proliferation through catalytic and non-catalytic activities. Cell Reports. 2016;15:1566–1579. doi: 10.1016/j.celrep.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lio CW, Zhang J, González-Avalos E, Hogan PG, Chang X, Rao A. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife. 2016;5:e18290. doi: 10.7554/eLife.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsagaratou A, Rao A. TET proteins and 5-methylcytosine oxidation in the immune system. Cold Spring Harb Symp Quant Biol. 2013;78:1–10. doi: 10.1101/sqb.2013.78.020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsagaratou A, Lio CJ, Yue X, Rao A. TET methylcytosine oxidases in T cell and B cell development and function. Front Immunol. 2017;8:220. doi: 10.3389/fimmu.2017.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onodera A, González-Avalos E, Lio CJ, Georges RO, Bellacosa A, Nakayama T, et al. Roles of TET and TDG in DNA demethylation in proliferating and non-proliferating immune cells. Genome Biol. 2021;22:186. doi: 10.1186/s13059-021-02384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caldwell BA, Liu MY, Prasasya RD, Wang T, DeNizio JE, Leu NA, et al. Functionally distinct roles for TET-oxidized 5-methylcytosine bases in somatic reprogramming to pluripotency. Mol Cell. 2021;81:859–869.e8. doi: 10.1016/j.molcel.2020.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair VS, Oh KI. Down-regulation of Tet2 prevents TSDR demethylation in IL2 deficient regulatory T cells. Biochem Biophys Res Commun. 2014;450:918–924. doi: 10.1016/j.bbrc.2014.06.110. [DOI] [PubMed] [Google Scholar]
- 42.Yang C, Li Z, Kang W, Tian Y, Yan Y, Chen W. TET1 and TET3 are essential in induction of Th2-type immunity partly through regulation of IL-4/13A expression in zebrafish model. Gene. 2016;591:201–208. doi: 10.1016/j.gene.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 43.Chapman VL, Terranova R, Moggs JG, Kimber I, Dearman RJ. Evaluation of 5-methylcytosine and 5-hydroxymethylcytosine as potential biomarkers for characterisation of chemical allergens. Toxicology. 2016;340:17–26. doi: 10.1016/j.tox.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Cheng RY, Shang Y, Limjunyawong N, Dao T, Das S, Rabold R, et al. Alterations of the lung methylome in allergic airway hyper-responsiveness. Environ Mol Mutagen. 2014;55:244–255. doi: 10.1002/em.21851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L, Fu J, Lin SH, Sun JL, Xia L, Lin CH, et al. Particulate matter of 2.5 μm or less in diameter disturbs the balance of TH17/regulatory T cells by targeting glutamate oxaloacetate transaminase 1 and hypoxia-inducible factor 1α in an asthma model. J Allergy Clin Immunol. 2020;145:402–414. doi: 10.1016/j.jaci.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Vitallé J, Terrén I, Orrantia A, Bilbao A, Gamboa PM, Borrego F, et al. The expression and function of CD300 molecules in the main players of allergic responses: mast cells, basophils and eosinophils. Int J Mol Sci. 2020;21:3173. doi: 10.3390/ijms21093173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johannesson B, Sattler S, Semenova E, Pastore S, Kennedy-Lydon TM, Sampson RD, et al. Insulin-like growth factor-1 induces regulatory T cell-mediated suppression of allergic contact dermatitis in mice. Dis Model Mech. 2014;7:977–985. doi: 10.1242/dmm.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Zhang Y, Wei Y, Wang Z, Zhu G, Fang Y, et al. The E3 ubiquitin ligase Itch is required for B-cell development. Sci Rep. 2019;9:421. doi: 10.1038/s41598-018-36844-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aki D, Li Q, Li H, Liu YC, Lee JH. Immune regulation by protein ubiquitination: roles of the E3 ligases VHL and Itch. Protein Cell. 2019;10:395–404. doi: 10.1007/s13238-018-0586-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghonim MA, Pyakurel K, Ju J, Rodriguez PC, Lammi MR, Davis C, et al. DNA-dependent protein kinase inhibition blocks asthma in mice and modulates human endothelial and CD4+ T-cell function without causing severe combined immunodeficiency. J Allergy Clin Immunol. 2015;135:425–440. doi: 10.1016/j.jaci.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang R, Qu C, Zhou Y, Konkel JE, Shi S, Liu Y, et al. Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity. 2015;43:251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López-Moyado IF, Tsagaratou A, Yuita H, Seo H, Delatte B, Heinz S, et al. Paradoxical association of TET loss of function with genome-wide DNA hypomethylation. Proc Natl Acad Sci U S A. 2019;116:16933–16942. doi: 10.1073/pnas.1903059116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of the subjects
Primer sequences