Significance
Biomolecular condensates are intracellular organelles that are not bounded by membranes and often show liquid-like, dynamic material properties. They typically contain various types of proteins and nucleic acids. How the interaction of proteins and nucleic acids finally results in dynamic condensates is not fully understood. Here we use optical tweezers and fluorescence microscopy to study how the prototypical prion-like protein Fused-in-Sarcoma (FUS) condenses with individual molecules of single- and double-stranded DNA. We find that FUS adsorbs on DNA in a monolayer and hence generates an effectively sticky FUS–DNA polymer that collapses and finally forms a dynamic, reversible FUS–DNA co-condensate. We speculate that protein monolayer-based protein–nucleic acid co-condensation is a general mechanism for forming intracellular membraneless organelles.
Keywords: optical tweezers, biomolecular condensates, co-condensation, FUS, DNA
Abstract
Biomolecular condensates provide distinct compartments that can localize and organize biochemistry inside cells. Recent evidence suggests that condensate formation is prevalent in the cell nucleus. To understand how different components of the nucleus interact during condensate formation is an important challenge. In particular, the physics of co-condensation of proteins together with nucleic acids remains elusive. Here we use optical tweezers to study how the prototypical prion-like protein Fused-in-Sarcoma (FUS) forms liquid-like assemblies in vitro, by co-condensing together with individual DNA molecules. Through progressive force-induced peeling of dsDNA, buffer exchange, and force measurements, we show that FUS adsorbing in a single layer on DNA effectively generates a sticky FUS–DNA polymer that can collapse to form a liquid-like FUS–DNA co-condensate. Condensation occurs at constant DNA tension for double-stranded DNA, which is a signature of phase separation. We suggest that co-condensation mediated by protein monolayer adsorption on nucleic acids is an important mechanism for intracellular compartmentalization.
Many cellular compartments that provide distinct biochemical environments are not separated by a lipid membrane. An important class of such membraneless compartments is formed by the condensation of proteins and other components in dynamic assemblies called biomolecular condensates (1–3). Biomolecular condensates increase the local concentration of their components, which can lead to substantially accelerated biochemical reactions (4, 5). Condensates that form beyond a saturation concentration can buffer the cellular concentration of molecules while at the same time clamping the concentration of phase-separated components inside (6). Biomolecular condensates could also localize reaction components, and by excluding molecules from condensates they can contribute to enhance specificity of biochemical processes. The formation of biomolecular condensates often relies on the existence of low-complexity domains (LCDs) (7–10). Condensates can show liquid-like material properties: they deform under shear stress, fuse, round up, and exchange their constituents with the environment (11–13).
Many condensed structures play essential roles in nuclear organization. For example, heterochromatin is a dense form of chromatin in which DNA co-condenses with specific factors as well as nucleosomes to form transcriptionally silent domains of chromatin (14–18). Furthermore, transcriptional hubs, or condensates, are dense and dynamic assemblies of transcription factors, associated proteins, DNA, and RNA. Such condensates have been suggested to play an important role in the generation of transcriptional hubs that could coordinate the expression of several genes and mediate enhancer function (19–23). Recently, it was shown that a pioneer transcription factor can form co-condensates together with DNA in vitro (24). Some membraneless compartments in the cell nucleus, such as the nucleolus, show all the features of liquid-like condensates (25, 26). However, for the majority of smaller nuclear compartments, the physical mechanisms by which they form remain controversial. In particular, the physicochemical mechanisms that drive co-condensation of proteins together with nucleic acids remain not well understood.
A prominent nuclear condensate is formed after DNA damage, where multiple proteins come together at the damage site to repair DNA (27, 28). Early components of the DNA damage condensate are members of the FET family such as the prion-like protein Fused-in-Sarcoma (FUS) (8, 29, 30). FUS has been shown to form liquid-like condensates in bulk solution at µM FUS concentrations (8, 31). However, its role in forming DNA repair compartments remains unknown.
FUS is a modular protein that consists of a nucleic acid binding domain (RNA Binding Domain [RBD]) containing various nucleic acid binding motifs such as an RNA recognition motif (RRM), a zinc finger motif, and two largely disordered R/G-rich regions implicated in nucleic acid binding (32). In addition, FUS contains an intrinsically disordered LCD that mediates FUS self-interaction (33, 34). FUS is involved in a multitude of physiological intracellular processes related to nucleic acid metabolism, for example, transcriptional regulation (35, 36), mRNA splicing (37), processing of noncoding RNA (38), DNA damage response (27–29, 39), ensuring mRNA stability (40), mRNA trafficking (41), and regulation of mRNA translation under stress conditions (42). FUS also forms higher-order aggregated and oligomeric assemblies in a set of neurodegenerative disorders (8, 29, 43).
While performing its physiological tasks, FUS typically acts in dynamic assemblies that are formed with or on nucleic acids or nucleic acid–like polymers. In the context of DNA damage, the formation and dissolution of FUS condensates depends on the presence or absence of poly(ADP ribose) (PAR), a DNA-like sugar polymer produced by PAR polymerases (8, 27, 29, 30, 39). Other examples for FUS-enriched condensates are stress granules, which are liquid-like, dynamic cytoplasmic hubs that form upon heat stress (8, 42, 44), or nuclear granules, which are associated with transcription and splicing (8, 45).
To investigate the physics underlying FUS–DNA condensate formation, we devised an in vitro assay based on optical tweezers combined with confocal microscopy. This allowed us to manipulate single DNA molecules in the presence of FUS protein in solution, image FUS proteins associating with the DNA molecule, and at the same time control and measure pN forces exerted on the DNA. Together, we reveal a mechanism of protein–nucleic acid co-condensation that is mediated by the adsorption of a protein monolayer on DNA.
Results
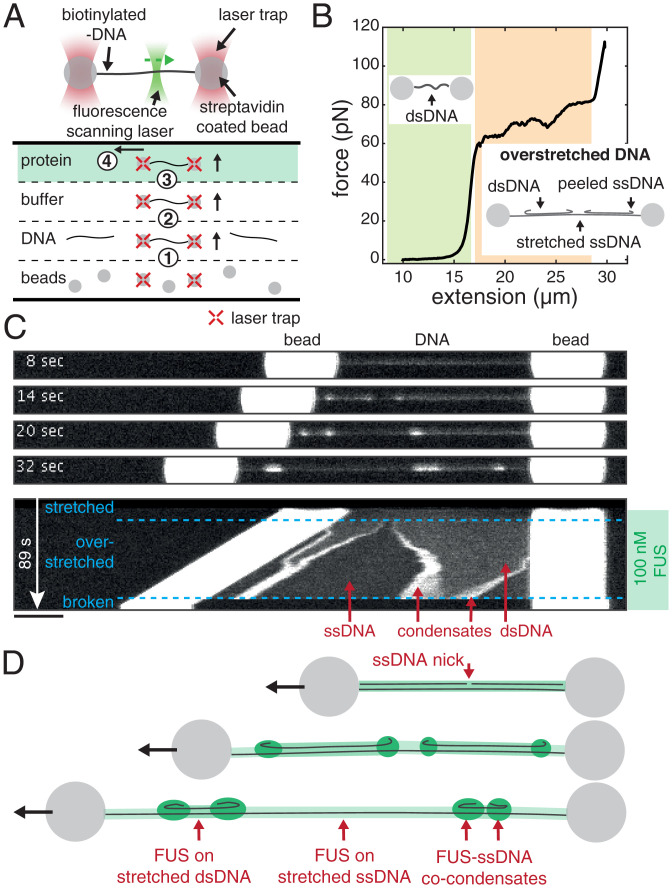
We established a biophysical assay based on optical tweezers and confocal microscopy to investigate collective interactions between FUS and DNA. For this, we exposed individual λ-DNA molecules stretched between two polystyrene beads each held in place in an optical trap to FUS-EGFP (from here on called “FUS”) inside a microfluidics flow chamber (Fig. 1A). Scanning confocal fluorescence microscopy was used to visualize the binding of FUS to DNA (49–51). We first trapped two streptavidin-coated polystyrene beads, which were then used to catch and stretch a lambda phage double-stranded DNA (dsDNA) molecule that was biotinylated at the two termini of only one of its two complementary strands. Next, we verified that indeed only a single DNA molecule was stretched by evaluating the mechanical properties of the connection and comparing it to the properties of a single λ-DNA molecule. Finally, we exposed the stretched DNA molecule to bulk FUS protein while imaging the system with a scanning confocal fluorescence microscope.
Fig. 1.
FUS forms co-condensates with ssDNA. (A) Schematics depicting the assembly and geometry of the optical tweezers-based assay. Steps for setting up the experiment in the flow chamber: 1) optical trapping of two Streptavidin-coated polystyrene beads, 2) catching of a λ-DNA molecule, 3) testing for a single DNA molecule, and 4) mechanical manipulation of the DNA in presence of FUS. (B) DNA mechanics and structure underlying our approach to study formation of FUS–ssDNA and FUS–dsDNA condensates. Investigation of FUS–dsDNA condensates is based on relaxed DNA. Investigation of FUS–ssDNA condensates is based on the gradual generation of peeled off ssDNA during DNA overstretching. See also Movie S1. (C) Snapshots and kymograph showing FUS–DNA interaction and FUS–ssDNA condensate formation during overstretching of DNA at 100 nM FUS. Note that the condensation propensity of FUS has been shown to be independent of the GFP tag (48–48). (Scale bar: 4 µm.) (D) Schematics depicting DNA overstretching in presence of FUS. FUS homogeneously coats stretched ssDNA and dsDNA and forms condensates with peeled relaxed ssDNA.
To study how FUS interacts with single-stranded DNA (ssDNA) and dsDNA, we exposed FUS to λ-DNA in different mechanical and structural states. The relationship between mechanical and structural properties of DNA is reflected in its force–extension curve (Fig. 1B) (51–54). At extensions (i.e., end-to-end distances) of up to about 0.9 times the contour length of the molecule (16.5 µm for λ-DNA) and at forces below ∼10 pN, DNA behaves as an entropic spring. We refer to this regime as relaxed. At higher forces and at extensions that are similar to the contour length, the DNA molecule behaves like a Hookian spring. At extensions significantly higher than the contour length, the DNA molecule is overstretched. In this regime, a progressive increase of the end-to-end distance of the molecule results in a progressive conversion of dsDNA to ssDNA while DNA tension remains constant at around 65 pN. In this process, ssDNA is peeled off, starting at free ssDNA ends. Free ends exist at nicks in the DNA backbone and at the ends of the dsDNA molecule. Note that the transition from B to S-DNA does not occur under the conditions used in this work (54). The overstretched DNA molecule then consists of three distinct structural types of DNA: sections of stretched dsDNA interspersed with sections of stretched ssDNA (both load-bearing and at tensions of ∼65 pN), with peeled and protruding ssDNA at the interfaces (Fig. 1 B, Insets). The ratio between dsDNA and ssDNA is defined by the end-to-end distance to which the DNA molecule is overstretched. In this work, we used relaxed dsDNA to study the formation of FUS–dsDNA co-condensates, and we made use of peeled ssDNA protruding from overstretched DNA to study the formation of FUS–ssDNA co-condensates.
FUS Forms Co-condensates with ssDNA.
To first investigate the interactions of FUS with ssDNA, we used optical traps to hold in place a single λ-DNA molecule extended to its contour length of 16.5 µm and transfer it into a microfluidics channel containing 100 nM FUS. Subsequently, we progressively increased its end-to-end distance to induce overstretching.
We observed that FUS attached to DNA in a spatially homogeneous manner upon transfer of the DNA molecule to the FUS channel (Fig. 1C and Movie S1). When the DNA end-to-end distance was increased to achieve overstretching, the originally homogenous coverage of DNA by FUS became interspaced by regions that exhibited lower fluorescence intensity. At the interface between regions of higher and lower FUS intensity, FUS puncta emerged. When we increased the DNA end-to-end distance further, the length of regions with higher intensity decreased while the length of lower-intensity regions increased. Concomitantly, the FUS puncta at the region interfaces grew in FUS intensity. Regions with high FUS intensity correspond to FUS unspecifically bound to stretched dsDNA (Fig. 1D). Regions with low intensity correspond to FUS bound to stretched ssDNA as these appear only during overstretching and grow with progressing overstretching (see SI Appendix, Fig. S2 for binding curves of FUS on stretched ssDNA and dsDNA). We interpret FUS puncta at interfaces between the low- and high-density FUS regions as co-condensates of FUS with ssDNA and provide evidence for condensation in the following sections. As the DNA is progressively overstretched, more and more peeled ssDNA is available, leading to growth of FUS–ssDNA co-condensates. We conclude that during overstretching, FUS binds to DNA in a manner that depends on the structural state of DNA: it homogeneously binds to dsDNA and ssDNA under tension and forms condensates together with peeled ssDNA that is not under tension.
FUS–ssDNA Co-condensate Formation Is Reversible.
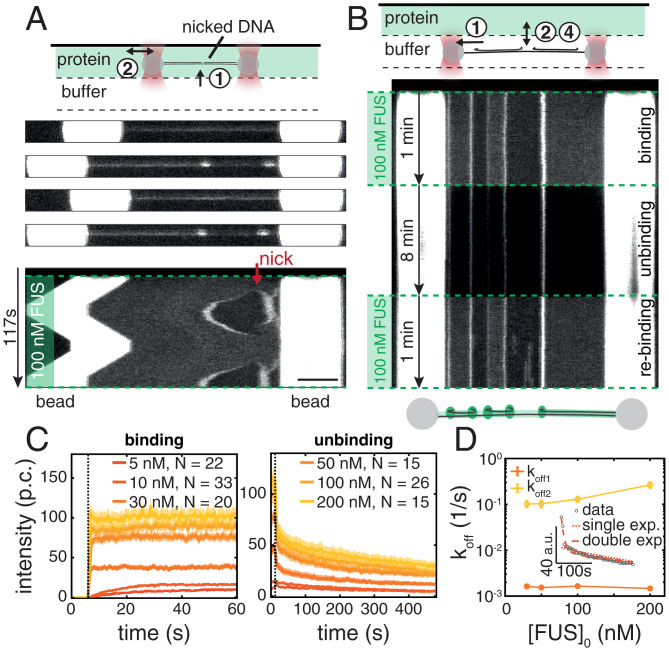
In what follows, we set out to study whether FUS–ssDNA co-condensates recapitulate typical dynamic properties of biomolecular condensates observed in vivo. We first investigated the reversibility of the formation of FUS–ssDNA co-condensates. To test if FUS–ssDNA co-condensates can be dissolved by the removal of ssDNA, we performed a repetitive stretch–relax experiment consisting of two subsequent overstretch–relaxation cycles. The approach was based on the rationale that overstretching progressively generates free and peeled ssDNA available for co-condensation, while relaxation progressively removes it. We first overstretched a DNA molecule in presence of 100 nM FUS, by increasing its extension from 17 to 21 µm at a speed of 0.1 µm/s. The molecule was then relaxed again, followed by a second overstretch cycle. We recorded the spatiotemporal distribution of FUS along the entire molecule throughout the process (Fig. 2A and Movie S2). In the example shown, we observed the formation of a condensate originating from a nick and a free terminal end on the right-hand side of the DNA molecule during the first overstretch. The size and brightness of condensates increased with progressive overstretching, in agreement with the findings presented in Fig. 1B. During the subsequent relaxation cycle, the size and brightness of condensates decreased progressively until they completely disappeared. Notably, condensates formed at precisely the same locations and with essentially the same dynamics during the second overstretching cycle as they did during the first one. We conclude that FUS–ssDNA co-condensates can be dissolved by removal of available ssDNA.
Fig. 2.
FUS–ssDNA co-condensate formation is reversible. (A) Snapshots and kymograph of repetitive overstretching experiments showing reversibility of condensate formation with respect to availability of an ssDNA scaffold. See also Movie S2. (Scale bar: 4 µm.) (B) Representative kymograph showing reversibility of FUS–ssDNA condensate formation with respect to availability of FUS tested in buffer exchange experiments. (C) Intensity–time traces of FUS–ssDNA condensates at different FUS concentrations during the binding and the unbinding step of the buffer exchange experiment. p.c., photon count. Mean ± STD. (D) Unbinding rates for condensates formed at different initial FUS concentrations. (Inset) Intensity–time traces fitted with single and double exponentials. Error bars show 95% confidence intervals.
To study if FUS–ssDNA co-condensates can be dissolved by the removal of free FUS from the environment, we performed binding–unbinding experiments by first overstretching a DNA molecule to 20 µm extension in absence of free FUS protein before moving it into (binding), out of (unbinding), and again into (rebinding) the FUS protein channel (Fig. 2B). We observed that in the binding process and upon entering the protein channel with 100 nM FUS, co-condensates rapidly formed, with a time scale that was below the temporal resolution of our imaging setup (0.5 s). Condensate formation was less rapid at lower concentrations of FUS (Fig. 2C). In the unbinding process and in absence of free FUS protein, the size and brightness of condensates decreased progressively. Notably, the intensity–time traces of condensate dissolution deviated from simple single-exponential behavior, indicating that multiple types of interaction might be involved in stabilization of FUS–ssDNA co-condensates (Fig. 2D). Interestingly, within 480 s of observation time, condensates did not disappear completely. It is possible that high local concentrations of FUS inside FUS–ssDNA condensates (∼5 µM; SI Appendix, Fig. S2B) enable long-lived interactions involving the RRM and LCD domains of FUS (8, 10, 55). We suspect that the associated aged or glassy state of FUS is responsible for slow unbinding time constants. Upon reexposure to free FUS protein during rebinding, condensates rapidly assumed the same size and intensity they had assumed in the initial binding step. Taken together, we conclude that FUS–ssDNA co-condensates dissolve when either ssDNA or free FUS is removed. FUS–ssDNA co-condensates form reversibly, which 1) is indicative of a significant amount of protein turnover in these condensates, 2) demonstrates that FUS–ssDNA interactions are key for co-condensation, and 3) demonstrates that FUS–FUS interactions, if they exist in these co-condensates, are not sufficient for maintaining a condensate in absence of ssDNA.
FUS–ssDNA Co-condensates Are Viscous Droplets with Liquid-Like Properties.
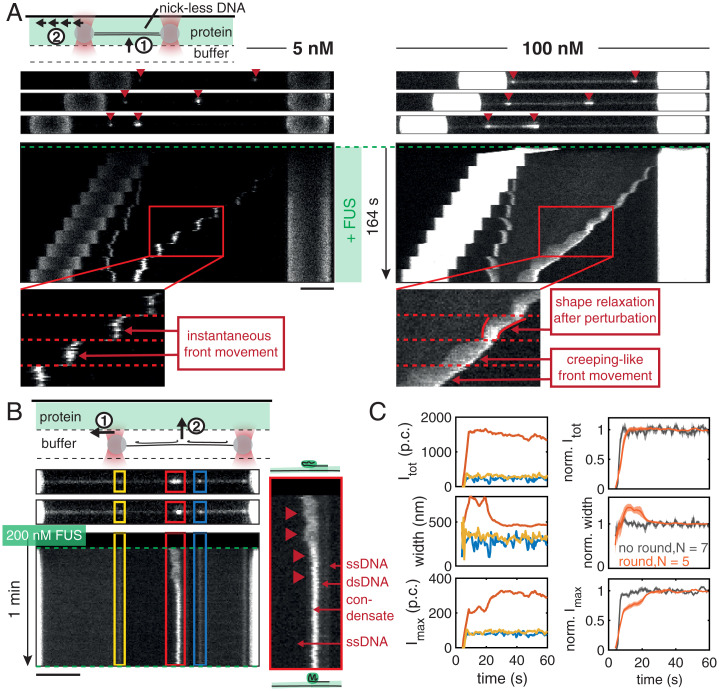
Biomolecular condensates often show properties of liquid-like droplets in vivo. They deform under shear stress and can exhibit shape relaxation driven by surface tension (11–13). We next investigated whether FUS–ssDNA co-condensates formed in vitro recapitulate this behavior. We first studied how these condensates react to the exertion of external mechanical perturbations. For that we increased the end-to-end distance of the DNA and hence the extent of overstretching in an abrupt and stepwise manner (steps every 10 s). This stepwise increase of the end-to-end distance within the overstretching regime instantaneously increases the amount of ssDNA substrate available for co-condensate formation and causes the condensates to move with the propagating peeling front.
At 5 nM FUS, small FUS–ssDNA co-condensates emerged from the ends of the DNA molecule, which appear to instantaneously follow the propagation of peeling fronts (Fig. 3 A, Left). When increasing the amount of overstretch in a stepwise manner, condensates also grew in a stepwise fashion. This indicates that relaxation times are fast and below the 1-s interval between confocal image recordings. However, at 100 nM FUS, we observed that FUS–ssDNA co-condensates followed the stepwise bead movement with time delay and in a smooth, creeping-like manner, reminiscent of viscous droplet being dragged along a string (Fig. 3 A, Right, and Movie S3). Leading and lagging edge of the condensates followed the bead movement on different response times, resulting in elongated condensate shapes. Elongated condensates relaxed toward more round shapes within the waiting time between steps (10 s). We conclude that FUS–ssDNA co-condensates formed at concentrations of ∼100 nM FUS display viscous material properties and exhibit viscoelastic shape relaxation.
Fig. 3.
FUS–ssDNA co-condensates are viscous droplets with liquid-like properties. (A) Representative snapshots and kymographs of FUS on DNA molecules overstretched in a stepwise manner in presence of different FUS concentrations. At 5 nM FUS, condensates had a point-like morphology and instantaneously grew and moved along the DNA when the DNA end-to-end distance was increased (Left, zoom). At 100 nM FUS, condensates grew and moved along the DNA in a creeping-like manner when the DNA end-to-end distance was increased. They elongated and showed shape relaxations on slow time scales compared to the fast-imposed external perturbations, reminiscent of viscous, liquid-like droplets (Right, zoom). (B) Representative snapshots and kymographs of a binding experiment performed at 200 nM FUS. Occasional shape changes from an initial elongated to a rounded morphology were observed (red condensate and zoom). (C) (Left) Quantification of shape changes of the example condensates. (Top) Total intensity, (Middle) width, and (Bottom) maximum intensity of individual condensates over time. While the total intensity of the red condensate remained constant over the course of the experiment, its width decreased while its maximum intensity increased until they leveled off. (Right) Quantification of shape changes of condensate ensemble. (Top) Normalized total intensity, (Middle) normalized width, and (Bottom) normalized maximum intensity of condensates over time. At 200 nM FUS, 5 out of 12 condensates showed rounding, decreasing their width to 70% of their initial width and increasing their maximum intensity to 140% of their initial intensity while keeping their total intensity constant within about 20 s. Traces show mean ± SEM, and p.c. denotes the photon count.
We next set out to find additional signatures for viscoelastic shape relaxation of FUS–ssDNA co-condensates. To this end we investigated condensate shape changes after their formation. Fig. 3B presents snapshots and the kymograph of a typical binding experiment performed at 200 nM FUS, showing how FUS assembles on the different segments of the overstretched DNA molecule upon exposure to FUS. In the representative example shown, while the two small condensates (marked in blue and yellow) did not change their shape after formation, the big condensate (marked in red) transitioned from an initially elongated toward a round shape within ∼120 s (Fig. 3C). The time constant of shape relaxation arises from co-condensate surface tension (see Fig. 6H), which co-condensates to adopt a spherical shape and co-condensate viscosity (11, 56). We conclude that FUS–ssDNA co-condensates display shape relaxations on a time scale of the order of 10 s, which together with our estimate of co-condensate surface tension (see Fig. 6H) reveals a co-condensate viscosity of ∼3 Pa⋅s (Materials and Methods) (56). We have thus revealed two types of shape relaxation of FUS–ssDNA co-condensates consistent with liquid-like behavior: they deform upon external mechanical perturbations, and they relax their shape after rapid formation.
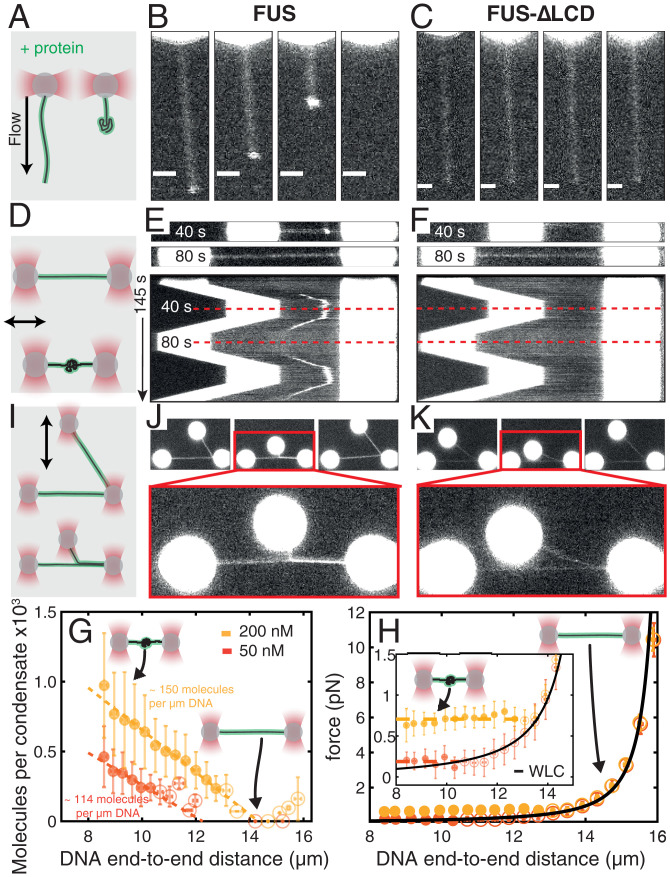
Fig. 6.
FUS monolayer adsorption on dsDNA and LCD mediated interactions lead to FUS–dsDNA co-condensate formation. (A–C) Individual dsDNA molecules attached via one end to beads are stretched by flow in presence of protein. (Scale bars: 2 µm.) (D–F) dsDNA attached to two beads was stretched and relaxed in presence of protein. (G) Number of FUS molecules in FUS–dsDNA condensates vs. DNA end-to-end distance estimated from condensate intensity (Methods and SI Appendix, Fig. S4), corresponding to one FUS molecule per 19.6 and 25.8 bp at 200 and 50 nM, respectively. (H) DNA tension vs. end-to-end distance in the presence of FUS. Black line, WLC. Dashed lines indicate linear fits. (I) Two dsDNA molecules were attached to three trapped beads in an L-like configuration allowing protein-mediated DNA zippering. (J) FUS mediates capillary-like forces between dsDNA strands. (K) DNA zippering is lost by deletion of the FUS LCD. In G and H, red indicates 50 nM FUS, n = 29 experiments; yellow indicates 200 nM FUS, n = 22 experiments. Filled circles indicate condensate; open circles indicate no condensate. Mean ± STD. Dashed lines indicate linear fits.
FUS Associating with ssDNA Generates a Sticky FUS–ssDNA Polymer.
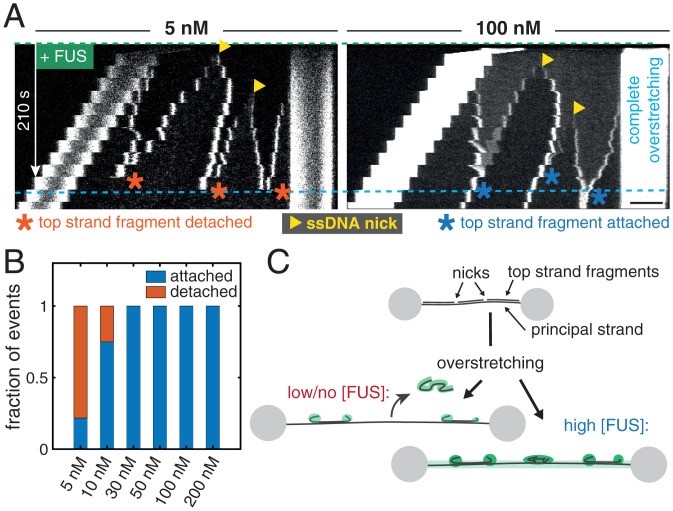
We speculate that FUS can form dynamic co-condensates with ssDNA because the association of FUS with ssDNA generates a self-interacting polymer which undergoes a globular collapse to form a liquid-like FUS–DNA co-condensate (57–59). Here FUS–FUS or additional FUS–DNA interactions could act like a molecular glue when two FUS-coated ssDNA fragments meet, which would prevent their dissociation. To test if FUS–DNA indeed behaves like a sticky polymer, we overstretched single DNA molecules whose top strands were by chance nicked at certain locations. We refer to the single strand of the dsDNA molecule that remains physically attached to the two polystyrene beads as the principal strand, while the complementary strand which becomes progressively peeled off during overstretching is referred to as the top strand. When a dsDNA molecule with a nicked top strand is overstretched to completeness (to 1.7 times its contour length), the peeled top strand fragments should dissociate and detach completely from the principal strand. We here tested if the interaction between FUS and ssDNA could interfere with this top strand detachment process.
Fig. 4 A, Left, shows the kymograph of a typical stepwise overstretching experiment performed at 5 nM FUS. We observe ssDNA peeled and condensation of ssDNA fragments with FUS, originating from the two terminal ends of the DNA molecule and from two nicks. When two peeling fronts met, they fused and subsequently disappeared from the field of view. This indicates that the corresponding ssDNA top strand fragment completely detached from the principal strand. Notably, all three ssDNA top strand fragments dissociated from the principal strand, but the principal strand was still intact after dissociation of the last top strand fragments. However, in the example kymograph for the experiment performed at 100 nM FUS (Fig. 4 A, Right), the top strand fragments did not fall off after peeling fronts of the individual fragments met in the course of overstretching. Rather, the top strand fragments remained attached to the principal strand. Taken together, our observations are consistent with the picture that FUS-coated ssDNA behaves like a sticky polymer, which serves to hold isolated fragments of ssDNA attached to regions of dissociation.
Fig. 4.
FUS associated with ssDNA generates a sticky FUS–ssDNA polymer. (A) Representative kymographs showing the influence of FUS–ssDNA interaction on the dissociation of DNA fragments when the DNA molecules are overstretched. The principal strand is the single strand of the dsDNA molecule attached to the beads. (Scale bar: 4 µm.) (B) Quantification of the fraction of fragments that detached from the principal strand vs. the fraction of fragments that stayed attached in stepwise overstretching experiments. Number of events: 5 nM, 23; 10 nM, 16; 30 nM, 14; 50 nM, 7; 100 nM, 23; and 200 nM, 18. (C) Illustration of the fragment detachment/attachment process.
If self-interactions of the FUS–ssDNA polymer arise from FUS–FUS interactions, or from FUS–ssDNA interactions that are in addition to the normal mode of association of FUS to ssDNA, we would expect that these self-interactions depend on FUS concentration. We analyzed peeling events from experiments performed in the concentration range between 5 and 200 nM FUS and classified them into detached (a top strand fragment disappeared from the principal strand when two corresponding peeling fronts met while the principal strand stayed intact) and attached (a top strand fragment remained attached to the principal strand when two corresponding peeling fronts met). We found that only for FUS concentrations below 30 nM, a considerable fraction of peeled top strand fragments detached from the principal strand (Fig. 4B), while they remained associated at higher concentrations. We conclude that self-interactions of the FUS–ssDNA polymer depend on FUS concentration. FUS–ssDNA co-condensates have liquid-like properties. Capillary forces are mechanical forces that are generated by a fluid when contacting a surface (24, 60). Given that self-interactions of the FUS–ssDNA polymer can generate a liquid phase, it is tempting to speculate that this can give rise to generalized capillary forces for liquid phases consisting of collapsed self-interacting polymers. For FUS, these could arise when the liquid phase contacts other FUS-coated DNA strands, resulting in the continued adhesion of condensates with principal strands in the experiments described in Fig. 4 and delaying force-induced disruption of dsDNA strands (SI Appendix, Fig. S3). It is tempting to speculate that these behaviors are related to the ability of FUS–dsDNA interactions to act as a molecular glue in the context of the DNA damage response. This is interesting as one might expect that an immediate response to DNA damage requires prevention of DNA fragments from leaving the damage site.
So far, we have shown that FUS forms dynamic co-condensates with ssDNA and that these condensates show various properties that are also typical for protein–nucleic acid-based organelles observed in vivo: their formation is reversible, they exchange constituents with the environment, and they show liquid-like material properties. Co-condensation also mediates stickiness and the adhesion of separate ssDNA strands. We next use the possibilities offered by our single molecule manipulation approach to reveal the physicochemical mechanisms underlying the formation of such FUS–ssDNA condensates.
FUS–ssDNA Co-condensation Is Based on FUS Adsorbing in a Single Layer on ssDNA.
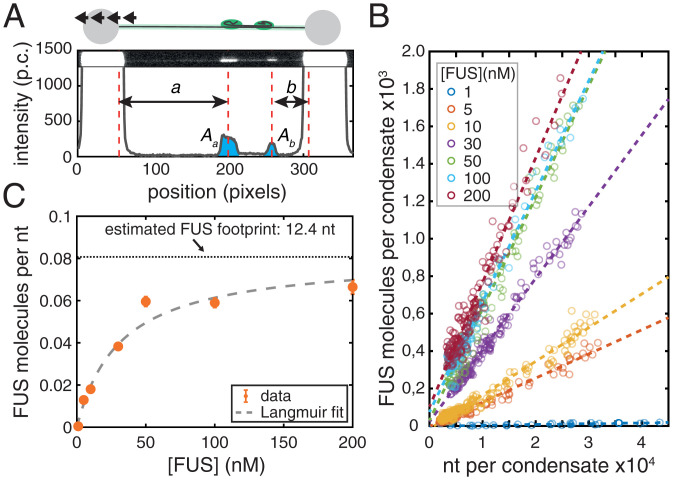
We were interested to understand whether ssDNA in FUS–ssDNA condensates is coated with a single adsorption layer of FUS with every FUS molecule directly binding to ssDNA, or if multiple layers of FUS are present with some FUS molecules not directly bound to ssDNA. We first investigated how the size of FUS–ssDNA co-condensates depends on the number of incorporated nucleotides. For this we utilized the stepwise overstretching assay introduced in Figs. 3 and 4. By controlling the end-to-end distance of the DNA molecule within the overstretching regime in a stepwise manner, we controlled the total number of peeled ssDNA nucleotides available for FUS–ssDNA co-condensate formation (Fig. 5A). By utilizing nick-free DNA molecules only, we ensured that ssDNA peeling during overstretching only occurred from the two ends of the DNA molecules. By measuring the distance between each of the two forming condensates and the respective beads, and taking into account the length of a single nucleotide under the applied tension of around 65 pN [0.58 nm (51–53)], we were able to determine the number of nucleotides available for incorporation into each of the two FUS–ssDNA co-condensates (see Analysis of FUS-ssDNA Co-condensate Composition for details). Further, we determined the integrated FUS fluorescence intensity associated with each condensate. Notably, we calibrated the FUS fluorescence intensity to arrive at a number of FUS-EGFP molecules in the condensate, using a calibration procedure that relied on individual dCas9-EGFP molecules tightly bound to λ-DNA molecules (Estimation of Number of FUS Molecules per Condensate and SI Appendix, Fig. S4) (61). We found that at all FUS concentrations investigated (between 1 and 200 nM FUS), the number of FUS molecules in a condensate was proportional to the number of incorporated nucleotides, with a slope that depends on the FUS concentration (Fig. 5B). This confirms that 1) the number of FUS molecules in a FUS–ssDNA co-condensate is determined by the amount of available ssDNA substrate and that 2) co-condensate stoichiometry (i.e., the ratio between number of proteins and number of nucleotides in a condensate) is independent of the size of the condensate, as is expected for co-condensation. More precisely, co-condensate stoichiometry is independent of the total number of ssDNA nucleotides in the co-condensate but depends on bulk FUS concentration (Fig. 5B). The ratio between the number of proteins and the number of nucleotides in a co-condensate (i.e., the slopes of the relations in Fig. 5B) informs about the degree of ssDNA substrate occupation by FUS. This ratio increased with increasing FUS concentrations between 1 and 50 nM and saturated at higher concentrations (Fig. 5C). Strikingly, this saturation curve was well described by a simple Langmuir adsorption model [Kd = 31.5 nM (11.3 to 51.8 nM) (numbers in parentheses indicate the lower and upper bounds of the 95% confidence interval, unless otherwise noted), saturation level p0 = 0.08 molecules/nt (0.06 to 0.10 molecules/nt) for FUS recruitment to ssDNA in FUS–ssDNA co-condensates]. The Langmuir adsorption model was originally used to describe the adsorption of gas molecule ligands on surfaces with negligible ligand–ligand interactions (56, 57). In our stepwise overstretching assay, the substrate for FUS binding is generated in a progressive manner, and co-condensate stoichiometry is set by FUS concentration. Notably, the adsorption curve saturates at a FUS density on ssDNA of approximately one FUS molecule every 12.4 nucleotides of ssDNA (Fig. 5C), consistent with the expected number of nucleotides that both the zinc finger and RRM domains would bind to (62). Taken together, our data suggest that FUS in FUS–ssDNA co-condensates forms a single adsorption layer on ssDNA, with every FUS molecule directly bound to ssDNA. Note that FUS–DNA co-condensates form in the absence of surface condensation via a prewetting transition (61). Binding occurs without detectable cooperativity despite the fact that FUS–FUS interactions within such a FUS–ssDNA co-condensate appear to collectively generate the capillary forces that drive co-condensation and condensate shape changes.
Fig. 5.
FUS–ssDNA co-condensation is based on FUS adsorbing in a single layer on ssDNA. (A) Intensity of FUS–ssDNA condensates extracted from stepwise overstretching experiments (p.c., photon count). A and A are integrated intensities of condensates, and a and b are the pieces of ssDNA incorporated in each of them. (B) Number of FUS molecules vs. number of nucleotides incorporated in each condensate. Number of events: 1 nM, 25; 5 nM, 72; 10 nM, 68; 30 nM, 69; 50 nM, 47; 100 nM, 38; and 200 nM, 59. An event is a single condensate observed during a single stretching step in a stepwise overstretching experiment. Dashed lines indicate linear fits to data points at the corresponding FUS concentration. Intensities were converted into numbers of FUS molecules by calibration with single dCas9-GFP molecules (SI Appendix, Fig. S4). (C) Number of FUS molecules per nucleotide in condensates vs. FUS concentration obtained from linear fitting in B. Data are fitted by a Langmuir binding isotherm. Dotted horizontal line indicates footprint of the FUS molecule inside FUS–ssDNA condensates. Orange shows the result of linear fitting in B within 95% confidence intervals. Gray dashed line shows the Langmuir fit.
FUS Monolayer Adsorption to dsDNA and LCD-Mediated Interactions Lead to FUS–dsDNA Co-condensate Formation.
Given that FUS has an affinity not only for ssDNA but also for dsDNA (Fig. 1C), we next investigated whether FUS can also form co-condensates with dsDNA. For this, we attached a single dsDNA molecule to a Streptavidin-coated bead held in an optical trap and applied an external buffer flow to stretch the DNA. We then moved the stretched bead–DNA construct to a channel containing 100 nM FUS while the flow was maintained (Fig. 6A). When moving the flow-stretched dsDNA molecule into the protein channel, we observed that 1) the dsDNA molecule became immediately coated with FUS (Fig. 6B and Movie S4) and 2) a co-condensate appeared to form at the free end of the dsDNA molecule, rapidly moving toward the bead and increasing in size with decreasing distance to the bead. Co-condensation was abolished when the LCD of FUS was not present (Fig. 6C), indicating that as expected, the LCD plays a role in mediating the FUS–FUS interactions necessary for co-condensation of FUS with dsDNA. Together, this provides evidence that the interaction of FUS with dsDNA leads to the formation of a FUS–dsDNA co-condensate even in presence of DNA tension.
To better investigate the co-condensation process, we next attached a dsDNA molecule to two beads held in place in optical traps and repeatedly relaxed and stretched the molecule between 8 and 16 µm end-to-end distance and thus to a length slightly below its contour length in a solution containing 200 nM FUS (Fig. 6D). Again, we observed that FUS assembled homogeneously on the stretched dsDNA molecule (Fig. 6E and Movie S5). Strikingly, a single FUS–DNA co-condensate emerged when the DNA was relaxed to an end-to-end distance below ∼14 µm, which grew in FUS amount with decreasing DNA end-to-end distance. The condensate dissolved again when the DNA was stretched beyond ∼14 µm, and it reformed with similar dynamics when the DNA was relaxed again, albeit at a slightly different position. Furthermore, the estimated FUS concentration in the co-condensate (∼5 µM) is above the reported saturation concentration for FUS under similar experimental conditions [∼2 µM (7)], as expected (SI Appendix, Fig. S2B). Again, condensate formation depended on the presence of the LCD of FUS (Fig. 6F). To conclude, FUS can form dynamic, reversible co-condensates with relaxed dsDNA.
We next asked whether these FUS–dsDNA co-condensates indeed form a separate physical phase. We draw an analogy to the phase transition between liquid water and vapor (63): when a pot of water is put onto a hot stove, the temperature of the water will not surpass 100 ∘C. Instead of increasing the temperature, energy input will cause water to transition from the liquid phase to the vapor phase while the temperature remains constant. This analogy is helpful for understanding the dissolution of FUS–dsDNA co-condensates by mechanically extracting FUS-coated dsDNA from the condensate. We predict two effects to occur when the end-to-end distance of an FUS-coated, condensed dsDNA molecule is increased. First, mass conservation implies that as FUS-coated DNA is progressively extracted from the condensate, the amount of material in the FUS–dsDNA co-condensate should decrease by corresponding amounts. Second, the dissolution of FUS–dsDNA co-condensates should occur at a constant DNA tension, similar to the constant temperature observed for the transition between liquid and gaseous water (24, 57).
To test the first prediction, we used the dual-trap experiment to form and dissolve FUS–dsDNA co-condensates at 50 and 200 nM FUS. Mass conservation implies that the number of FUS molecules inside a condensate increases proportionally with the amount of co-condensing dsDNA. In other words, the amount of FUS in the co-condensate should increase linearly with decreasing DNA end-to-end distance, which is what we observed (Fig. 6G). Furthermore, the absolute value of the slope of the linear relationship between number of FUS molecules in a co-condensate and the DNA end-to-end distance increased with increasing FUS concentration (Fig. 6G). At 50 nM FUS, ∼114 FUS molecules are bound per micrometer of DNA in a co-condensate (corresponding to a spacing of one FUS molecule every ∼26 bp), while at 200 nM FUS, ∼150 FUS molecules are bound per micrometer of DNA in a co-condensate (corresponding to a spacing of one FUS molecule every ∼20 bp) (also see SI Appendix, Fig. S2). This reveals that FUS adsorbs in a single layer on DNA at both concentrations investigated, with enough space between FUS molecules to allow each FUS molecule to directly bind to dsDNA. Note that the estimated FUS concentration in the co-condensate (∼5 µM) is above the reported saturation concentration for FUS under similar experimental conditions [∼2 µM (7)] (SI Appendix, Fig. S2B). Further, an analysis of the probability for co-condensate formation as a function of DNA end-to-end distance revealed a sharp transition at 10.5 µm (10.4 to 10.6 µm) at 50 nM FUS and 12.9 µm (12.7 to 13.1 µm) at 200 nM FUS (SI Appendix, Fig. S5D). This indicates that co-condensation occurs below a critical DNA end-to-end distance Lcrit that depends on FUS concentration. Taken together, we conclude that as FUS-coated DNA is progressively extracted from the FUS–dsDNA co-condensate, the amount of material in the FUS–dsDNA co-condensate decreases by corresponding amounts.
We next tested the second prediction and investigated the range of DNA tensions at which FUS–dsDNA co-condensates form (Fig. 6H and SI Appendix, Fig. S5G). Using the dual trap tweezer assay we found that as FUS-coated dsDNA is relaxed starting from an initially stretched configuration (16 µm end-to-end distance), the relation between force and DNA end-to-end distance follows the expected worm-like chain (WLC) behavior as long as its end-to-end distance is above Lcrit. Strikingly, when the end-to-end distance was reduced below Lcrit (and hence when a condensate forms), trap force remained constant [0.19 ± 0.05 pN at 50 nM FUS, 0.71 ± 0.05 pN at 200 nM FUS (mean ± STD)]. Furthermore, condensates of various sizes coexisted at essentially the same DNA tension (SI Appendix, Fig. S5G). Note also that in the region where the WLC transitions into the constant force regime a slight dip in force was observed, indicative of a small but finite surface tension of the condensate. A theoretical description of protein–DNA co-condensation in the optical trap suggests that this dip corresponds to a surface tension of the order of 0.15 pN/µm (SI Appendix, Fig. S5F and and Supplementary Methods). Together, this provides evidence that a first-order phase transition underlies the formation of FUS–dsDNA co-condensates.
We next set out to estimate the condensation free energy per FUS molecule (24). At DNA end-to-end distances far below the critical DNA length and in the case of low surface tension, the constant force generated by the co-condensate reeling in DNA is determined by the condensation free energy per volume μ and the DNA packing factor α. The packing factor is a measure for the scaling between length of condensed DNA and the volume of the condensate. We estimated α using the FUS concentration-dependent FUS coverage of dsDNA inside condensates (slope in Fig. 6G and SI Appendix, Fig. S5E) and the molecular volume of FUS inside condensates Vm (SI Appendix, Fig. S5F). We found that values of α (∼0.05 µ at 50 nM FUS and ∼0.06 µ at 200 nM FUS) were similar in magnitude to those reported for a DNA–protein phase containing the transcription factor FoxA1 (24). The condensation free energy per volume obtained using the packing factors and corresponding critical forces was ∼4.1 pN/µ at 50 nM FUS and ∼11.9 pN/µ at 200 nM FUS. With a FUS density inside condensates of about 2,500 molecules/µ (specified by the molecular volume), this provides an estimate of the condensation free energies of ∼0.4 kT/FUS at 50 nM FUS and ∼1.1 kT/FUS at 200 nM FUS. These values are consistent with previous reports of the interaction energies between Tyr–Tyr and Tyr–Arg residues in FUS (64), suggesting that interactions similar to those driving FUS liquid–liquid phase separation at higher concentrations (7) are present in co-condensates. Taken together, FUS adsorbing in a single layer on DNA effectively generates a sticky FUS–DNA polymer that can collapse to form a liquid-like FUS–DNA co-condensate. For dsDNA, this condensation occurs at constant DNA tension, which is a clear signature of a mesoscopic first-order phase transition.
We next set out to test if single-layer adsorbed FUS can mediate adhesion of separate dsDNA strands. For that we attached two FUS-coated dsDNA strands to three beads in an L-like configuration, with DNA strands held at tensions above the critical tension for FUS–dsDNA condensate formation (Fig. 6I). By moving one of the beads relative to the others, we were able to bring the two FUS-coated dsDNA molecules in close proximity in order to test if they adhere to each other. We found that the two FUS-coated dsDNA strands adhered to each other and zippered up at 100 nM FUS (Fig. 6 J and Movie S6). Zippering was reversed by pulling the DNA strands away from each other and reestablished by moving DNA strands closer. Furthermore, zippering depends on the presence of the LCD (Fig. 6H). Taken together, our data indicate that a single layer of FUS attached to DNA can mediate dynamic adhesion of separate DNA strands, opening up the possibility for this mechanism to be involved in long-range genome organization.
Discussion
The discovery that membraneless compartments can be formed by liquid-like biomolecular condensates and that phase separation can contribute to the spatiotemporal organization of intracellular biochemistry has opened up new perspectives in cell biology (1, 3). We show that FUS–ssDNA co-condensation is based on FUS adsorbing in a single layer on ssDNA well described by a Langmuir isotherm (Fig. 5 and SI Appendix, Fig. S2). Similarly, FUS monolayer adsorption to dsDNA together with LCD-mediated interactions lead to FUS–dsDNA co-condensate formation at low dsDNA tension (Figs. 6 and 7). Here changing dsDNA extension shifts the balance between the co-condensate and the FUS-coated dsDNA molecule and results in the co-condensate growing at the expense of stretched dsDNA. The process of co-condensation is a chemomechanical process that converts chemical potential changes to mechanical forces. These generalized capillary forces can exert tension on the DNA that remains outside the condensate. Growth of co-condensates occurs at constant DNA tension, consistent with a mesoscopic first-order phase transition as is expected for a physical condensation process. We find that the constant tension depends on the FUS concentration and is of the order of 1 pN. For comparison, forces required for unfolding individual proteins typically are higher and in the range of tens of pN (66–68). Also, the stall force of RNA Pol II is at least an order of magnitude higher, opening up the possibility that transcription can proceed essentially unhindered in the presence of such capillary forces (69). Together, this indicates that condensate formation by co-condensation can compete in force balance in the nucleus and with the formation of loops by the cohesin/condensin machinery (70) or with the association of DNA with lamins in the nuclear envelope (71). To conclude, the formation of a monolayer of FUS on both ssDNA and dsDNA effectively generates a sticky FUS–DNA polymer that subsequently condenses to form a dynamic, reversible FUS–DNA co-condensate.
Fig. 7.
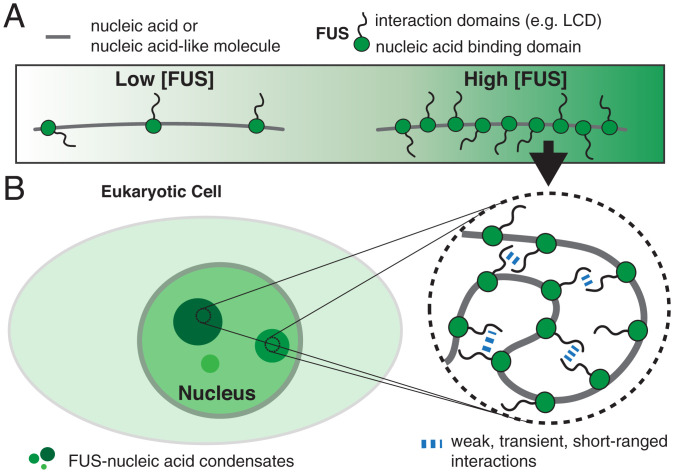
Biomolecular condensate formation based on monolayer protein recruitment to nucleic acids. (A) Nucleic acids or nucleic acid–like polymers recruit monolayers of proteins. (B) Protein adsorption on nucleic acids gives rise to an effective self-interacting protein–nucleic acid polymer. Collapse of this self-interacting polymer leads to the formation of protein–nucleic acid co-condensates reminiscent of biomolecular condensates observed in cell nuclei. This process is distinct from the globular collapse of a noninteracting freely jointed chain but shares similarities with the collapse of a polymer in poor–solvent-facilitating polymer self-interactions (65).
Protein–DNA co-condensation involves the collective binding of many proteins to a DNA substrate. Here we demonstrate that upon single-layer binding the FUS-coated DNA molecule undergoes co-condensation and maintains its DNA–protein stoichiometry as it grows. It is tempting to speculate that higher protein concentrations, closer to the saturation concentration for bulk phase separation, could lead to additional modes of interaction. These could involve multilayer protein adsorption on DNA (72) or the formation of surface condensates via a prewetting transition (61) (SI Appendix, Fig. S1), and quantitative methods for measuring protein–nucleic acid stoichiometry are required to distinguish between different co-condensation scenarios in vivo. Future work is necessary to characterize the full extent of collective interactions between FUS and DNA, to investigate a possible dependence on DNA sequence, and to reveal how differences in the mechanical properties of ssDNA and dsDNA affect material properties of FUS–DNA co-condensates.
We speculate that protein–DNA co-condensation plays an important role in biological processes such as heterochromatin formation driven by HP1α (14–18), the transcription-dependent organization of chromatin (19, 21, 22, 45) (SI Appendix, Fig. S6A), the formation of viral replication compartments (73–76) (SI Appendix, Fig. S6C), the compaction of viral ssRNA (77, 78), and the DNA damage response (27, 29, 30) (SI Appendix, Fig. S6B). With respect to the latter, we have shown that pairs of FUS-coated DNA can bind together and exert adhesion forces onto each other (Figs. 4 and 6). It is possible that inside the cell, such adhesion forces prevent DNA fragments from leaving damage sites during DNA repair. An interesting question for future research is to understand whether poly(ADP)ribose (PAR) triggers FUS–DNA co-condensation at the damage site, thereby preventing the escape of DNA damage fragments. Taken together, we suggest that co-condensation mediated by protein monolayer adsorption on nucleic acids constitutes a general mechanism for forming intracellular compartments.
Materials and Methods
Recombinant full-length FUS-EGFP and FUS-Δ LCD-EGFP were expressed and purified as described previously (8).
Optical Tweezers with Confocal Fluorescence Imaging.
Optical tweezer experiments were performed in a C-Trap G1 instrument which combines four optical traps with confocal fluorescence imaging and microfluidics (Lumicks). Experimental work flows were controlled using custom Python scripts through the inbuilt Bluelake software. Proteins were diluted in FUS buffer (70 mM KCl, 10 mM Tris, pH 7.4) to the final concentration. Biotinylated double-stranded λ-DNA (Lumicks) was diluted to ∼20 pg/µL in FUS buffer, and 4.4 µm Streptavidin-coated polystyrene beads (Spherotech) were diluted to 3% (w/v) in FUS buffer. DNA tethers were formed as described elsewhere (50). For fluorescence excitation, 2.14 µW of a 488-nm laser was used. Dwell time per pixel and pixel size were set to 0.05 ms and 100 nm × 100 nm. Buffer exchange experiments were recorded at 2 fps for 60 s (binding). For unbinding, DNA molecules were transferred back to the buffer channel while imaging at 0.25 fps during 480 s. Typically, an additional binding experiment (rebinding, same settings as initial binding) was performed. Stepwise overstretching experiments were performed in steps of 1 µm at 5 µm/s every 10 s and imaged at 1 fps. dsDNA flow-stretch experiments were imaged at 1 fps. All experiments were performed at 28 ∘C.
dsDNA Relaxation Cycle.
DNA molecules were relaxed and stretched between 16 and 8 µm at 0.5 µm/s with a waiting period of 20 s at each extreme position. After two rounds, the bead-to-bead distance was first increased to 31 µm to rupture the molecule and then again decreased to 8 µm to estimate the force base line. Imaging was performed at 2 fps.
dsDNA Zippering Experiments.
Three Streptavidin-coated polystyrene beads were trapped in three optical traps and moved to the DNA channel. In the presence of mild buffer flow, the two beads aligned in parallel to the flow direction (beads 1 and 2) captured a DNA molecule. The beads then were moved to the protein channel. Simultaneously, fluorescence imaging with a rate of approximately one frame every 5 s was started. Further, we approached bead 3 (already coated with DNA) toward the tether between beads 3 and 2 in order to enable contacting of the two FUS-coated DNA tethers.
Data Analysis.
Data analysis was performed using custom MATLAB (Mathworks) routines. Image representation was performed in FIJI v. 1.51h. For quantification of FUS intensities on DNA, images were background-subtracted by calculating the average background intensity away from the DNA for every frame and subtracting it from the whole image. Intensity profiles along the DNA direction were calculated by summing up background subtracted pixel intensities orthogonally to the DNA direction. Kymographs were generated from the time projection of these profiles.
Analysis of Buffer Exchange Experiments.
Kymographs were manually segmented into regions of stretched dsDNA, stretched ssDNA, and puncta (FUS–ssDNA condensates). Intensity–time traces of each DNA segment were calculated by averaging the intensities of all pixels in a segment for each frame. Average intensity–time traces of the different types of DNA (stretched ssDNA, stretched dsDNA, and puncta) for binding and unbinding experiments performed at different FUS concentrations were calculated by averaging the intensity–time traces of every segment obtained for the corresponding experiment type (binding or unbinding) at the corresponding FUS concentration.
To extract unbinding rates, the average intensity–time traces obtained from unbinding experiments were fitted using single or double exponential functions. Fitting was performed from the time point at which the background intensity dropped (indicating that the DNA had left the protein channel) to the last time point of the experiment (480 s). The quality of fitting (represented by the R2 value) drastically improved by using double exponentials instead of single exponentials, particularly at elevated FUS concentrations. Equilibrium intensities of FUS on stretched ssDNA and stretched dsDNA were calculated by averaging the intensity–time traces obtained from binding experiments performed at different FUS concentrations over the last 30 s (i.e., when the equilibrium was reached).
To study shape changes of FUS–ssDNA condensates over time, segments of puncta were obtained from kymographs of FUS binding experiments as described above. A custom peak finding algorithm was used to obtain the maximum intensity and the width of puncta in each frame of an experiment. The total intensity of a punctum was calculated as the product of maximum intensity and peak width.
For ensemble analysis, the individual time traces of maximum intensity, total intensity, and punctum width were normalized to their final value (last 10 s of the experiment). Puncta were classified according to whether they rounded up in the course of the binding experiment. A punctum was classified as rounded if the normalized final maximum intensity (last 10 s of the experiment) was at least higher than the normalized initial maximum intensity (first 10 s after punctum formation) plus four times the corresponding SD. Mean time traces of maximum intensity, total intensity, and punctum width were calculated according to this classification.
Analysis of FUS-Mediated ssDNA Adhesion.
Increasing the DNA end-to-end distance leads to progressive conversion of dsDNA to ssDNA via peeling from free ssDNA ends. Nicks in the ssDNA backbones of the dsDNA molecules define boundaries of potential ssDNA fragments. During overstretching, progressive peeling from the fragment boundaries will lead to dissociation of ssDNA fragments. For analysis, we evaluated events in which two peeling fronts propagated toward each other when the DNA end-to-end distance was increased. Events were classified into attached or detached based on the integrity of the tether. We only considered an event if the DNA tether remained intact for at least one more step of the stepwise increase of DNA end-to-end distance.
Analysis of FUS–ssDNA Co-condensate Composition.
The number of nucleotides peeled from each of the two ends of the overstretching fronts (for nicks-free molecules) and hence incorporated into each of the two condensates can be calculated from the distance between a condensate and the corresponding bead. This distance divided by the length of an ssDNA nucleotide at 65 pN (0.58 nm per nucleotide) yields the number of nucleotides in the corresponding condensate. For every step of a stepwise overstretching experiment, the intensity profile along the DNA molecule was calculated and the positions of the beads and the boundaries of the condensates selected. From this information we calculated the integrated intensity of a condensate and the corresponding number of incorporated nucleotides. For each FUS concentration we fitted the relation between condensate intensity and number of nucleotides with a linear function. The slope versus the corresponding FUS concentration was fitted to Langmuir isotherm of the form of to the data (qm being the saturation occupancy of nucleotides with FUS, Km being the FUS concentration at which the occupancy has reached half of its maximum value, and being the FUS concentration).
Estimation of the Viscosity of FUS–ssDNA Co-condensates.
The relation between viscosity and the shape relaxation time is given by (56) , where τ is the relaxation time, l is the characteristic size of droplets, η is the viscosity, and γ is the surface tension. Considering a relaxation time of ∼20 s, (Fig. 3C), a characteristic length of ∼1 µm (Fig. 3B), and a surface tension of 0.15 pN/µm (SI Appendix, Fig. S6F), we obtain a viscosity of η ∼3 Pa×s. This value is in good agreement with that reported for protein–ssDNA condensates in ref. 56 (5 to 6 Pa⋅s).
Analysis of FUS–dsDNA Co-condensates.
The free energy F of a FUS–dsDNA condensate containing the DNA length LD can be described using a volume contribution and a surface contribution (SI Appendix, Fig. S5A), building on a framework introduced in ref. 24:
μ is the condensation free energy per volume, α is the packing factor relating LD to the condensate volume, and γ is the surface tension. The force required to extract a piece of DNA from the condensate is
For small surface tension and high values of LD (corresponding to low DNA end-to-end distances), this expression approaches a constant DNA tension .
To analyze the mechanical properties and to finally estimate the condensation free energy per FUS molecule in FUS–dsDNA condensates, for each frame of a stack, we extracted the position, width (full width at half maximum [FWHM]) and maximum intensity of the condensate using a custom peak finding algorithm (SI Appendix, Fig. S5B). The total intensity of a detected condensate in each frame was calculated as the product of maximum intensity and peak width (SI Appendix, Fig. S5C). The baseline subtracted force–distance signal was synchronized with the fluorescence imaging data (raw force–distance signal [∼9 Hz] was down-sampled to 2 Hz). The intensity of tracked condensates for every frame in which a condensate was detected was correlated to the force at which it existed and to the corresponding DNA end-to-end distance. Downstream analysis was restricted to the part of the process where the initially stretched dsDNA molecule was relaxed from 16 to 8 µm end-to-end distance (unless indicated differently). Subsequent stretching and relaxation processes in presence of FUS appeared to alter the mechanical properties of the DNA. We first investigated at which DNA end-to-end distances FUS–dsDNA condensates exist (SI Appendix, Fig. S5D). For that, we analyzed the probability to find a condensate at the different DNA end-to-end distances between 16 and 8 µm during the initial relaxation process. The step-like shapes of the curves were fitted with error functions in the shape of
p is the probability to find a condensate, L is the DNA end-to-end distance, and Lcrit is the critical DNA end-to-end distance below which condensates typically form.
Packing factor α was obtained as the product of the negative slope of the linear relation between condensate intensity and DNA end-to-end distance α’ and the molecular volume VM of FUS in condensates (SI Appendix, Fig. S5E). VM was obtained from the expectation value of a Rayleigh distribution fit to the histogram of the ratio between volume and intensity of each detected condensate in each frame it was detected. For calculating the condensate volume, condensates were assumed to be spherical with a diameter equal to the FWHM obtained from tracking. α was obtained from a linear fit to the condensate intensity vs. DNA end-to-end distance at DNA end-to-end distances below Lcrit.
The number of FUS molecules bound per DNA length inside condensates was estimated using α and the intensity of a single GFP. This yielded ∼115 FUS molecules bound per micrometer of condensed DNA at 50 nM FUS (corresponding to a spacing of one FUS molecule every ∼26 bp) and ∼150 FUS molecules bound per micrometer of condensed DNA at 200 nM FUS (corresponding to a spacing of one FUS molecule every ∼20 bp).
The critical force f0 was finally obtained as the mean force exerted by the condensates below Lcrit (Fig. 6H). μ as energy per volume was obtained by dividing the critical force by the corresponding packing factor for 50 and 200 nM FUS. To estimate μ of a single FUS molecule, we approximated the number of FUS molecules per cubic micrometer using the molecular volume (in units of cubic micrometer per photon count) and the intensity of a single GFP molecule (in units of photon counts per molecule) extracted from SI Appendix, Fig. S4. To convert into units of kBT per FUS molecule we assumed a temperature of 303 K and hence a conversion relation of 1 kBT = 4.2e–3 pNµm.
Only events where a single condensate formed per single dsDNA molecule were analyzed. Imaging was performed at 2 fps.
Estimation of Number of FUS Molecules per Condensate.
dCas9-EGFP was incubated with four different types of guide RNA molecules that had sequences complementary to four adjacent sequences at about 1/3 of the length of λ-DNA (61). Individual preincubated λ-DNA molecules were imaged at the conditions used for FUS–DNA experiments for 360 s (pixel size 100 nm, pixel dwell time 0.05 ms, frame rate 1 fps). DNA molecules were held either at ∼60 pN or below 5 pN to allow for fluorescence calibration that could be used either for FUS–ssDNA condensates (formed on top of overstretched DNA tethers) or for FUS–dsDNA condensates (present at forces below 5 pN). Time traces of background subtracted sum intensities of puncta found in the DNA target regions were extracted (moving average over 30 frames). Multiple Gaussian distributions were fit to the probability density function of intensities. The position of the first peak (after the background peak) was used as an estimate of the intensity of a single EGFP.
Supplementary Material
Acknowledgments
We thank members of the S.W.G. laboratory for fruitful discussion. S.W.G. was supported by the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm 1782, Graduiertenschule 97, GR 3271/2, GR 3271/3, GR 3271/4) and the European Research Council (grant 742712). R.R. and A.H. acknowledge support by the NOMIS foundation. A.H. is supported by the Hermann und Lilly-Schilling Stiftung für medizinische Forschung im Stifterverband. F.J. acknowledges funding from the Volkswagen Foundation. We thank Stefan Golfier for contributing the sgRNA and dCas9-EGFP for single-molecule intensity quantification experiments. We further acknowledge K. M. Crell, S. Kaufmann, and F. Thonwart for providing reagents. We also thank J. Brugues, A. Gladfelter, A. Hyman, T. Mitchison, W. Snead, and T. Quail for discussions and critical comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107871119/-/DCSupplemental.
Data Availability
All data generated or analyzed in this study are available from Zenodo (https://doi.org/10.5281/zenodo.6241151). All materials will be made available upon request after completion of a Material Transfer Agreement. All other study data are included in the article and/or supporting information.
References
- 1.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A., Altmeyer M., Phase separation: Linking cellular compartmentalization to disease. Trends Cell Biol. 26, 547–558 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Hyman A. A., Weber C. A., Jülicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Hernández-Vega A., et al., Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 20, 2304–2312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li P., et al., Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klosin A., et al., Phase separation provides a mechanism to reduce noise in cells. Science 367, 464–468 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Wang J., et al., A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel A., et al., A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Kato M., et al., Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon I., et al., Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brangwynne C. P., et al., Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Jawerth L., et al., Protein condensates as aging Maxwell fluids. Science 370, 1317–1323 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Jawerth L. M., et al., Salt-dependent rheology and surface tension of protein condensates using optical traps. Phys. Rev. Lett. 121, 258101 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Larson A. G., Narlikar G. J., The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry 57, 2540–2548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson A. G., et al., Liquid droplet formation by HP1suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strom A. R., et al., Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keenen M. M., et al., HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife 10, e64563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanulli S., et al., HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henninger J. E., et al., RNA-mediated feedback control of transcriptional condensates. Cell 184, 207–225 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y. E., et al., Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabari B. R., et al., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho W. K., et al., Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A., A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quail T., et al., Force generation by protein–DNA co-condensation. Nat. Phys. 17, 1007–1012 (2021). [Google Scholar]
- 25.Brangwynne C. P., Mitchison T. J., Hyman A. A., Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 4334–4339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feric M., et al., Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aleksandrov R., et al., Protein dynamics in complex DNA lesions. Mol. Cell 69, 1046–1061 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Levone B. R., et al., FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J. Cell Biol. 220, e202008030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naumann M., et al., Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat. Commun. 9, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altmeyer M., et al., Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maharana S., et al., RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozdilek B. A., et al., Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 45, 7984–7996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Schwartz J. C., Cech T. R., Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 43, 7535–7543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz J. C., Wang X., Podell E. R., Cech T. R., RNA seeds higher-order assembly of FUS protein. Cell Rep. 5, 918–925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan A. Y., Riley T. R., Coady T., Bussemaker H. J., Manley J. L., TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl. Acad. Sci. U.S.A. 109, 6030–6035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L., Gal J., Chen J., Zhu H., Self-assembled FUS binds active chromatin and regulates gene transcription. Proc. Natl. Acad. Sci. U.S.A. 111, 17809–17814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogelj B., et al., Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci. Rep. 2, 603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shelkovnikova T. A., Robinson H. K., Troakes C., Ninkina N., Buchman V. L., Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum. Mol. Genet. 23, 2298–2312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singatulina A. S., et al., PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 27, 1809–1821 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Kapeli K., et al., Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nat. Commun. 7, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujii R., Takumi T., TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 118, 5755–5765 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Li Y. R., King O. D., Shorter J., Gitler A. D., Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alberti S., Dormann D., Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Protter D. S. W., Parker R., Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson V. F., et al., Transcription-dependent formation of nuclear granules containing FUS and RNA Pol II. Biochemistry 57, 7021–7032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogaert E., et al., Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 24, 529–537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burke K. A., Janke A. M., Rhine C. L., Fawzi N. L., Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA Polymerase II. Mol. Cell 60, 231–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monahan Z., et al., Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brouwer I., et al., Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature 535, 566–569 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Candelli A., et al., Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc. Natl. Acad. Sci. U.S.A. 111, 15090–15095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Mameren J., et al., Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl. Acad. Sci. U.S.A. 106, 18231–18236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross P., et al., Quantifying how DNA stretches, melts and changes twist under tension. Nat. Phys. 7, 731–736 (2011). [Google Scholar]
- 53.Smith S. B., Cui Y., Bustamante C., Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996). [DOI] [PubMed] [Google Scholar]
- 54.King G. A., et al., Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 110, 3859–3864 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee D. S. W., Wingreen N. S., Brangwynne C. P., Chromatin mechanics dictates subdiffusion and coarsening dynamics of embedded condensates. Nat. Phys., 17, 531–538 (2021). [Google Scholar]
- 56.Alshareedah I., Thurston G. M., Banerjee P. R., Quantifying viscosity and surface tension of multicomponent protein-nucleic acid condensates. Biophys. J. 120, 1161–1169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cristofalo M., et al., Cooperative effects on the compaction of DNA fragments by the nucleoid protein H-NS and the crowding agent PEG probed by magnetic tweezers. Biochim. Biophys. Acta, Gen. Subj. 1864, 129725 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Polotsky A. A., Daoud M., Borisov O. V., Birshtein T. M., A quantitative theory of mechanical unfolding of a homopolymer globule. Macromolecules 43, 1629–1643 (2010). [Google Scholar]
- 59.Halperin A., Goldbart P. M., Early stages of homopolymer collapse. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 61, 565–573 (2000). [DOI] [PubMed] [Google Scholar]
- 60.de Gennes P. G., Brochard-Wyart F., Quéré D., Capillarity and Wetting Phenomena (Springer, New York, 2004). [Google Scholar]
- 61.Morin J. A., et al., Sequence-dependent surface condensation of a pioneer transcription factor on DNA. Nat. Phys., 10.1038/s41567-021-01462-2 (2022). [Google Scholar]
- 62.Loughlin F. E., et al., The solution structure of FUS bound to RNA reveals a bipartite mode of RNA recognition with both sequence and shape specificity. Mol. Cell 73, 490–504 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Atkins P., de Paula J., Keeler J., Atkins’ Physical Chemistry 11th Edition: Volume 3: Molecular Thermodynamics and Kinetics (Oxford University Press, 2019). [Google Scholar]
- 64.Choi J. M., Hyman A. A., Pappu R. V., Generalized models for bond percolation transitions of associative polymers. Phys. Rev. E 102, 042403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Post C. B., Zimm B. H., Theory of DNA condensation: Collapse versus aggregation. Biopolymers 21, 2123–2137 (1982). [DOI] [PubMed] [Google Scholar]
- 66.Mojumdar S. Sen, et al., Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nat. Commun. 8, 1881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gupta A. N., et al., Pharmacological chaperone reshapes the energy landscape for folding and aggregation of the prion protein. Nat. Commun. 7, 12058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganim Z., Rief M., Mechanically switching single-molecule fluorescence of GFP by unfolding and refolding. Proc. Natl. Acad. Sci. U.S.A. 114, 11052–11056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yin H., et al., Transcription against an applied force. Science 270, 1653–1657 (1995). [DOI] [PubMed] [Google Scholar]
- 70.Golfier S., Quail T., Kimura H., Brugués J., Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. eLife 9, e53885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Schaik T., Vos M., Peric-Hupkes D., Hn Celie P., van Steensel B., Cell cycle dynamics of lamina-associated DNA. EMBO Rep. 21, e50636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitchison T. J., Beyond Langmuir: Surface-bound macromolecule condensates. Mol. Biol. Cell 31, 2502–2508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmid M., Speiseder T., Dobner T., Gonzalez R. A., DNA virus replication compartments. J. Virol. 88, 1404–1420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McSwiggen D. T., et al., Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife 8, e47098. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nevers Q., Albertini A. A., Lagaudrière-Gesbert C., Gaudin Y., Negri bodies and other virus membrane-less replication compartments. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heinrich B. S., Maliga Z., Stein D. A., Hyman A. A., Whelan S. P. J., Phase transitions drive the formation of vesicular stomatitis virus replication compartments. MBio 9, e02290–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cubuk J., et al., The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 12, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iserman C., et al., Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell 80, 1078–1091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in this study are available from Zenodo (https://doi.org/10.5281/zenodo.6241151). All materials will be made available upon request after completion of a Material Transfer Agreement. All other study data are included in the article and/or supporting information.