Significance
Dual-function RNAs base pair with target messenger RNAs as small regulatory RNAs and encode small protein regulators. However, only a limited number of these dual-function regulators have been identified. In this study, we show that a well-characterized base-pairing small RNA surprisingly also encodes a 15–amino acid protein. The very small protein binds the cyclic adenosine monophosphate receptor protein transcription factor to block activation of some promoters, raising the question of how many other transcription factors are modulated by unidentified small proteins.
Keywords: sRNA, dual-function RNA, small protein, carbon catabolite repression, SpfP
Abstract
The 109-nucleotide Spot 42 RNA whose transcription is repressed by the cyclic adenosine monophosphate receptor protein (CRP) is one of the best-characterized base-pairing small RNAs (sRNAs) in Escherichia coli. Consistent with high levels of Spot 42 in glucose-grown cells, the RNA blocks the expression of transporters and enzymes involved in the utilization of nonpreferred carbon, such as galactose. We now document that Spot 42 also encodes a 15–amino acid protein denoted SpfP. Previous studies showed that overexpression of Spot 42 reduces growth in galactose and other nonpreferred carbon sources. Overexpression of just the small protein from a Spot 42 derivative deficient in base-pairing activity also prevented growth on galactose, revealing that the sRNA and protein impact the same pathway. Copurification experiments showed that SpfP binds CRP. This binding blocks the ability of CRP to activate specific genes, such as the galETKM operon, impacting the kinetics of induction when cells are shifted from glucose to galactose medium. Thus, the small protein reinforces the feedforward loop regulated by the base-pairing activity of the Spot 42 RNA.
Carbon catabolite repression (CCR) is a mechanism used by bacteria to promote the use of a preferred carbon source in an environment where other substrates are also available (1). In gram-negative bacteria, like Escherichia coli, this is achieved by preventing expression of genes involved in the catabolism of nonpreferred carbon sources when glucose is present. In E. coli, the key components of the CCR pathway are the IIA component of the glucose-specific phosphotransferase system (PTS) (EIIAGlc; also called catabolite repression resistance [Crr] or EIIACrr), adenylate cyclase, cyclic AMP (cAMP), and the cAMP receptor protein (CRP; also denoted CAP) (2, 3). Under glucose-limiting conditions, the PTS proteins, including EIIAGlc, are predominantly phosphorylated. In this form, P-EIIAGlc activates adenylate cyclase, increasing cAMP synthesis. cAMP induces a conformational change in CRP that allows cAMP–CRP to bind the promoters of hundreds of catabolic genes, such as the galETKM operon for galactose metabolism and the malEFG operon for maltose metabolism (4). As a transcriptional activator, cAMP–CRP binds to sequences located upstream of (class I activation) or adjacent to (class II activation) promoters and participates in protein–protein interactions with RNA polymerase, leading to increased transcription initiation (5). cAMP–CRP also represses several genes by inhibiting transcription initiation (6, 7).
In addition to regulating the expression of protein-coding genes, E. coli CRP modulates the transcription of small regulatory RNAs (sRNAs), repressing Spot 42 sRNA expression and activating CyaR sRNA expression (8, 9). The 109-nucleotide (nt) Spot 42 sRNA encoded by the spf (Spot 42) gene is broadly found in γ-proteobacteria (10). Spot 42 is highly expressed when cells are grown with glucose, and its transcription is repressed by cAMP–CRP during growth in nonpreferred carbon sources (8). Many years after its first characterization (11–13), Spot 42 was shown to bind the RNA chaperone Hfq and base pair with messenger RNA (mRNA) targets to regulate their expression (14). The third gene in the galactose operon, galK encoding galactokinase (GalK), was the first identified Spot 42 target; down-regulation of this gene leads to discordant expression of the galactose operon (15). Later, Spot 42 was shown to base pair with and repress expression from a number of other mRNAs encoding proteins involved in the uptake and catabolism of nonpreferred carbon sources (16, 17). Consistent with this regulation, Spot 42 overexpression negatively impacts growth on a number of nonpreferred carbon sources (16).
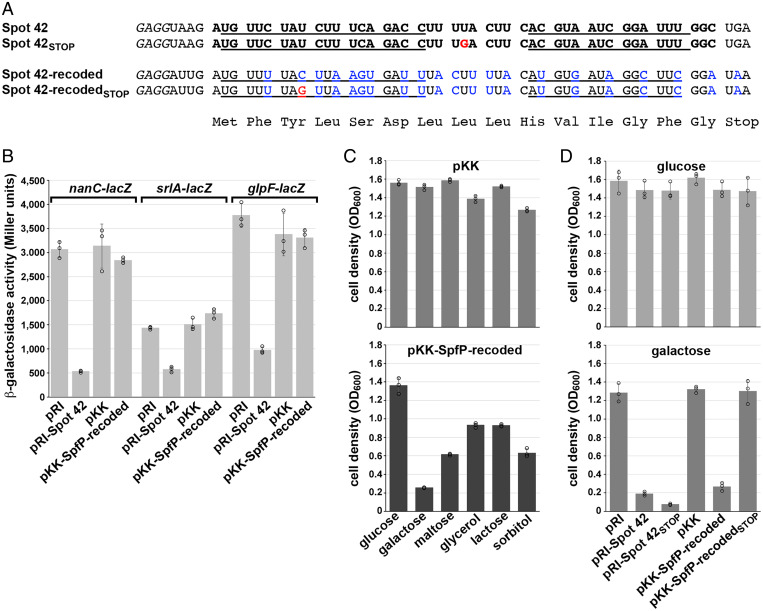
Apart from being an sRNA, Spot 42 RNA resembles a short mRNA. It contains a Shine–Dalgarno sequence followed by an AUG start codon, a 15–amino acid open reading frame (ORF), and a UGA stop codon (Fig. 1A) (18). However, an early study examining the affinity between Spot 42 and the 70S ribosome reported that although the RNA bound to ribosomes, it did so inefficiently and nonproductively (19). A fusion between the Spot 42 ORF to lacZ also did not support the synthesis of β-galactosidase. These studies led to the conclusion that Spot 42 does not function as an mRNA under the conditions tested (19).
Fig. 1.
Spot 42 is a dual-function sRNA. (A) Diagram of the Spot 42 RNA and sequence of the spf promoter and coding region. Boxes and text in light blue denote SpfP coding sequence, and the yellow box and highlighted text denote the region of Spot 42 base pairing with target mRNAs. The Spot 42 transcript is denoted by the gray box and is indicated in bold, with the +1 site of transcription (position 1988001 of the E. coli K-12 genome) in green font and the 3′-end of the transcript in red font. The SpfP ORF ribosome-binding site, start codon (ATG), and stop codon (TAA) are indicated by black boxes, the potential σ70 −10 sequence is underlined, and a predicted CRP-binding site (36) is highlighted in light gray on the sequence. (B) Diagram of Spot 42 secondary structure (taken from ref. 37) with the Shine–Dalgarno sequence, start codon, and stop codon boxed. Single-stranded regions involved in base pairing with mRNAs are highlighted in yellow. The Hfq-binding regions are indicated in blue, and double-stranded regions are indicated in red. (C) Expression of SpfP protein and Spot 42 RNA. Cells expressing SpfP–SPA (GSO1119) were grown to an OD600 of ∼0.4 at 30 °C in LB and then transferred to 30 °C, 42 °C, or 45 °C. Immunoblotting analysis was carried out on samples collected before transfer and 5 and 10 min after transfer. Anti-FLAG antibody was used to detect the SPA tag, and the Ponceau S stain documents approximately equal loading of the samples. The theoretical molecular weight of the 84–amino acid SPA-tagged SpfP protein is 9.8 kDa. For total RNA isolated for WT cells grown under the same conditions, the Spot 42 and 5S RNAs were detected with primers specific to each of these transcripts.
Here, we report that, in fact, Spot 42 encodes a functional small protein (SpfP) in addition to acting as a regulatory sRNA and thus should be classified as a dual-function RNA. This 15–amino acid protein, whose levels are elevated at higher temperature, exhibits the unique characteristic of binding the CRP transcription factor. High levels of SpfP block CRP-dependent activation of some targets. Thus, we propose that the SpfP interaction with CRP aids CCR by inhibiting the function of CRP in the presence of glucose and reinforcing the feedforward loop regulated by the base-pairing activity of Spot 42.
Results
Spot 42 Regulatory sRNA Encodes a 15–Amino Acid Protein.
Two recent studies (20, 21), in which ribosome profiling was conducted in the presence of translation inhibitors (Onc112 and retapamulin) that trap ribosomes on start codons, suggested the 15–amino acid ORF encoded by Spot 42 RNA might be translated (SI Appendix, Fig. S1A). To directly test whether the protein encoded by the predicted ORF is synthesized, we integrated the sequence for a sequential peptide affinity (SPA) tag onto the chromosome just upstream of the predicted stop codon. We then carried out immunoblotting analysis with anti-FLAG antibodies to detect the protein. Given that the potential ribosome-binding site and start codon could be sequestered by a stem-loop in the Spot 42 RNA structure, we thought higher temperatures might resolve the stem-loop and promote translation (Fig. 1B). Thus, cells with the chromosomal SPA fusion grown to exponential phase in lysogeny broth (LB) at 30 °C were shifted to 30 °C, 42 °C, or 45 °C. The tagged protein was detected at all temperatures (Fig. 1C), indicating that Spot 42 is also an mRNA encoding a 15–amino acid protein, hereafter referred to as SpfP. Interestingly, the levels of SpfP–SPA were higher at 42 °C and 45 °C than at 30 °C at the 5-min time point. The levels of wild-type (WT) Spot 42 RNA assayed for cells grown under the same conditions did not change significantly (Fig. 1C).
Spot 42 Base-Pairing and Protein-Coding Activities Can Be Genetically Separated.
After documenting SpfP expression, we wondered whether SpfP might be responsible for some of the previously observed phenotypes attributed to Spot 42 (16). To separate the protein-coding activity from the base-pairing activities of spf, we constructed an overexpression plasmid with the tac promoter on pKK177-3, driving expression of only the spfP ORF, in which the sequence of two of the Spot 42 base-pairing regions were recoded to eliminate base pairing potential while maintaining the amino acid sequence of SpfP (pKK–SpfP-recoded) (Fig. 2A).
Fig. 2.
SpfP expression impacts growth on galactose. (A) Sequence of region encoding 15–amino acid SpfP. Nucleotides changed in the recoded derivative are indicated in blue, and nucleotides changed in the STOP mutants are indicated in red. (B) β-galactosidase assay of Δspf cells with nanC-lacZ (GSO440), srlA-lacZ (GSO441), or glpF-lacZ (GSO519) transformed with pRI, pRI-Spot 42, pKK, or pKK–SpfP-recoded. The cells were grown to an OD600 of ∼1.0 in LB supplemented with 0.2% arabinose. (C) Growth assays of Δspf::kan cells (GSO433) transformed with pKK or pKK–SpfP-recoded. Strains were grown in M63 minimal medium supplemented with the indicated carbon sources all at 0.2% except for glycerol, which was at 0.4%. (D) Growth assays of Δspf::kan cells (GSO433) transformed with pRI, pRI-Spot42, pRI-Spot42STOP, pKK, pKK–SpfP-recoded, or pKK–SpfP-recoded STOP. Strains were grown in M63 minimal medium supplemented with either glucose (Top) or galactose (Bottom). For C and D, cells grown overnight in LB with ampicillin were diluted to an OD600 of ∼0.05 in M63 minimal medium with the indicated carbon sources and grown for 16 h, at which point the OD600 was measured. For B, C, and D, bars correspond to the average of three biological replicates with circles corresponding to individual data points and error bars representing 1 SD.
To check whether the recoded construct functioned as a regulatory sRNA, we assayed the effect of SpfP-recoded overexpression on lacZ fusions to three mRNAs known to be direct targets of Spot 42 (16). β-galactosidase activity was assayed in cells expressing nanC-lacZ, srlA-lacZ, and glpF-lacZ translational fusions transformed with vector controls or the corresponding plasmids overexpressing either Spot 42 (from pRI, a derivative of pKK177-3 without the ribosome-binding site) or SpfP-recoded (from pKK177-3). Regulation of the targets was only observed when the full-length Spot 42 transcript was overexpressed, suggesting that these targets are regulated by Spot 42 but not by the recoded construct (Fig. 2B). Thus, SpfP-recoded does not function as a base-pairing sRNA.
To test if SpfP contributes to Spot 42 phenotypes, we examined how expression of SpfP impacts growth on nonpreferred carbon sources as previously tested for Spot 42 (16). Δspf cells were transformed with the overexpression plasmids, and optical density at 600 nm (OD600) was measured after 16 h of growth in M63 minimal medium supplemented with either glucose or other nonpreferred carbon sources. As observed for Spot 42, overexpression of SpfP led to a growth defect in minimal medium with most of the nonpreferred carbon sources, particularly with galactose (Fig. 2C).
We also constructed stop codon mutants of Spot 42 and SpfP to further examine the effects of both the RNA and the small protein for growth on galactose. We observed that overexpression of full-length Spot 42 produces a growth defect in galactose that persists following introduction of a stop codon (Fig. 2D), showing that the sRNA itself confers a phenotype. In contrast, a stop codon mutation eliminated the growth phenotype associated with SpfP-recoded overexpression. Together, these data demonstrate that spf functions as both an sRNA and mRNA.
Histidine 10 Is Critical for SpfP Function.
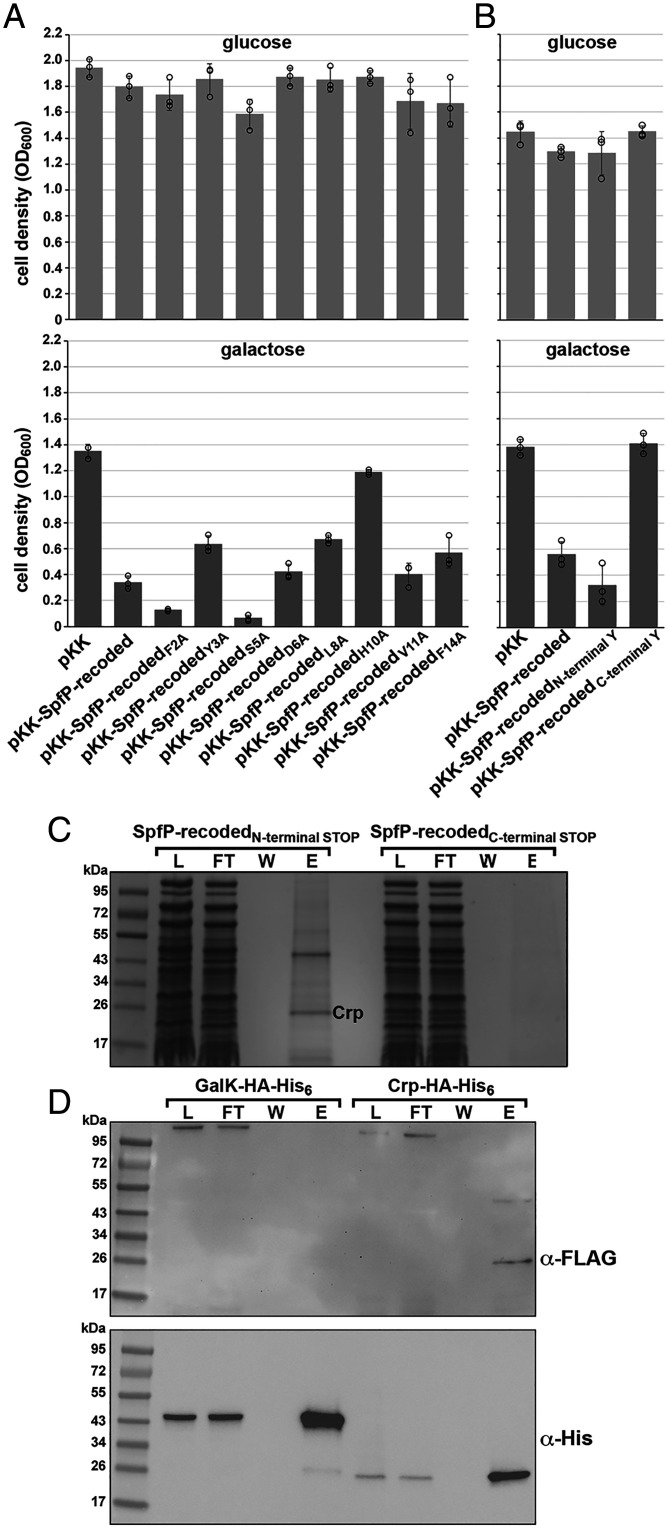
To determine which SpfP residues are important for observed growth defects in galactose medium, we carried out alanine scan mutagenesis of the SpfP coding sequence in the recoded sequence context. The mutants were overexpressed in a Δspf background, and growth in M63 galactose was measured after 16 h. As observed in the previous assay, cell density was ∼fourfold less following overexpression of the small protein (pKK–SpfP-recoded) than in the empty vector control. The F2A and S5A derivatives produced a more severe effect than pKK–SpfP-recoded (Fig. 3A). Conversely, Y3A, L8A, and F14A slightly relieved the growth defect, while H10A almost completely relieved the inhibitory effect of SpfP-recoded (Fig. 3A).
Fig. 3.
SpfP copurifies with CRP. (A) Growth assays of Δspf::kan cells (GSO433) expressing alanine substitution derivatives of pKK–SpfP-recoded. Strains were grown in M63 minimal medium supplemented with either glucose (Top) or galactose (Bottom). (B) Growth assays of Δspf::kan cells (GSO433) transformed with pKK–SpfP-recoded derivatives with either an additional N-terminal Y or C-terminal Y. Strains were grown in M63 minimal medium supplemented with either glucose (Top) or galactose (Bottom). For A and B, cells were assayed as in Fig. 2, and bars correspond to the average of three biological replicates with circles corresponding to individual data points and error bars representing 1 SD. (C) CRP copurifies with biotin-tagged SpfP. Δspf::kan cells (GSO433) with the SpfP-recodedN-terminal STOP or SpfP-recodedC-terminal STOP plasmids and the orthogonal tRNA and aminoacyl tRNA pair were grown in M63 glucose medium with 1 mM p-AzF to an OD600 of ∼0.5 and induced with 0.2% arabinose for 3 h. Cell lysates were treated with biotin–PEG-alkyne to biotinylate SpfP and passed over streptavidin beads. Fractions from the lysate (L), flow-through (FT), wash (W), and eluants (E) for each sample were subjected to SDS-PAGE followed by Coomassie blue staining. The unique ∼25-kDa band enriched in the eluant from the SpfP-expressing cells was excised from the gel and identified by mass spectrometry. (D) N-terminally FLAG-tagged SpfP copurifies with CRP–HA-His6 but not GalK–HA-His6. Δspf cells expressing CRP–HA-His6 or GalK–HA-His6 from the chromosome (GSO1061 and GSO1060, respectively) were transformed with pKK–SpfP-recodedN-FLAG and grown in LB until an OD600 of ∼0.5 was reached. The cell lysate was incubated overnight with 50 µL of Pierce anti-HA magnetic beads. The beads were collected using a magnet, and proteins were eluted with Laemmeli buffer. Immunoblots of fractions from the lysate (L), flow-through (FT), wash (W), and eluants (E) separated by SDS-PAGE were probed with anti-FLAG (Top) and anti-His (Bottom) antibodies. A cross-reacting band of high molecular weight is detected with anti-FLAG antibodies.
SpfP Associates With CRP.
To gain insight into the role of SpfP, we sought to identify coassociating proteins. As the SPA tag is almost three times the size of SpfP, we set out to generate an SpfP derivative with a nonnative amino acid, azido-modified phenylalanine amino acid (p-AzF), that is structurally similar to tyrosine and can be modified by biotin. We first tested the effect of introducing p-AzF on SpfP activity by inserting a tyrosine at either the N terminus of SpfP between the first and second amino acids or at the C terminus just upstream of the stop codon, again in the context of the recoded sequence. The growth assay indicated that the N-terminally tagged construct (pKK–SpfP-recodedN-terminal Y) has WT inhibitory activity for cells grown in M63 galactose, while the C-terminally tagged construct (pKK–SpfP-recodedC-terminal Y) does not and thus was used as a negative control in subsequent copurification experiments (Fig. 3B).
To purify SpfP, we introduced a stop codon between the first and second amino acids (SpfP-recodedN-terminal STOP) or just upstream of the stop codon (SpfP-recodedC-terminal STOP) as a control. Each of the stop codon derivatives of SpfP was then expressed under conditions where p-AzF was incorporated at the stop codon by using an engineered, orthogonal transfer RNA (tRNA) and aminoacyl-tRNA synthetase pair (pEVOL-p-AzF) (22). The SpfP derivatives bearing p-AzF were biotinylated and purified on streptavidin beads. The eluant from each column was separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). A prominent band at ∼25 kDa was observed for the tagged protein expressed from the SpfP-recodedN-terminal STOP construct (Fig. 3C and SI Appendix, Fig. S2A) but not from the SpfP-recodedC-terminal STOP construct (Fig. 3C). Mass spectrometric analysis revealed that CRP (23.6 kDa) was enriched in this ∼25-kDa band in two experiments. We noted that an additional ∼45-kDa band copurified with the protein expressed from the SpfP-recodedN-terminal STOP construct in both experiments but focused on CRP in this study given the connection to carbon metabolism.
As a further test for an interaction between SpfP and CRP, we carried out a reciprocal copurification experiment in which N-terminally FLAG-tagged SpfP was expressed in a Δspf background with chromosomally encoded CRP–hemagglutinin (HA)-His6 or GalK–HA-His6 as a cytosolic protein control. The inhibitory effect of SpfP on growth in minimal galactose medium was not altered in the CRP–HA-His6 strain (SI Appendix, Fig. S2B). The cells expressing the HA-His6–tagged proteins were grown in LB medium to an OD600 of ∼0.5, lysed, and incubated with anti-HA magnetic beads. The immunoblot of the fractions from these purifications probed with the anti-His antiserum showed that the tagged GalK and CRP derivatives were clearly enriched by incubation with the anti-HA beads (Fig. 3D). One prominent band of ∼25 kDa was observed when an immunoblot of the same fractions was probed with anti-FLAG antiserum to detect FLAG–SpfP. However, FLAG–SpfP should be ∼2 kDa. We hypothesized that the ∼25-kDa cross-reacting band was FLAG–SpfP associated with CRP. To test this hypothesis, we carried out a similar copurification with the chromosomally tagged CRP–HA-His6 compared to a Δcrp control strain (SI Appendix, Fig. S2C). Again FLAG-tagged SpfP migrated as a ∼25-kDa band, the same molecular weight as CRP–HA-His6, while the band was not detected with the Δcrp strain. We also mixed synthetic SpfP peptide biotinylated at the N terminus with extracts from CRP–HA-His6 Δspf and Δcrp Δspf cells and incubated the extracts with anti-HA magnetic beads to isolate CRP–HA-His6 together with associated proteins (SI Appendix, Fig. S2D). When we probed an immunoblot of these samples with streptavidin, we again observed a band that specifically comigrated with CRP–HA-His6. Together, these results indicate that SpfP associates with CRP, and this association is sufficiently tight that the 15–amino acid SpfP protein remains bound to CRP during SDS-PAGE.
SpfP Overexpression Blocks Activation of Some CRP-Dependent Operons.
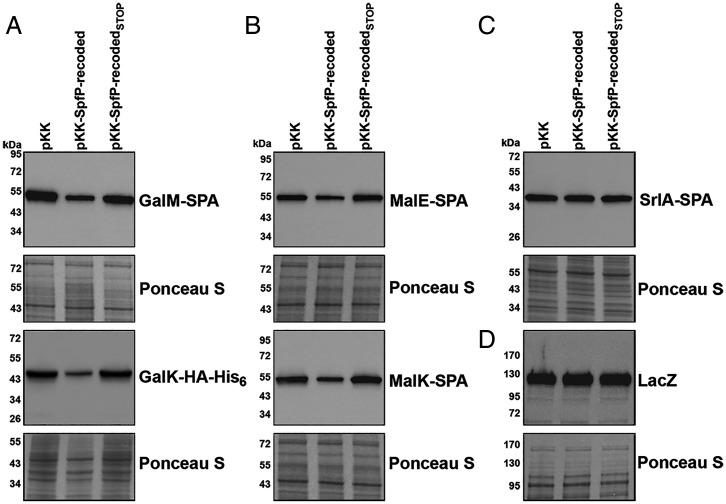
We wondered if SpfP binding to CRP could affect the activity of the transcription regulator. To test this possibility, we examined the effects of pKK–SpfP-recoded or pKK–SpfP-recodedSTOP, the stop mutant control, on the levels of proteins encoded by various CRP-activated or CRP-repressed operons. Strains with chromosomally tagged operons were grown with carbon sources that activated the expression of the specific operons. These experiments showed that pKK–SpfP-recoded, but not pKK nor pKK–SpfP-recodedSTOP, led to decreased levels of GalM–SPA and GalK–HA-His6 for cells grown in M63 with galactose (Fig. 4A) and decreased levels of MalE–SPA and MalK–SPA for cells grown in M63 with maltose (Fig. 4B). Consistent with the lack of a growth defect for the SpfP-recodedH10A mutant (Fig. 3A), this derivative did not reduce GalM–SPA and MalE–SPA levels (SI Appendix, Fig. S3A).
Fig. 4.
SpfP overexpression leads to down-regulation of CRP-activated genes. (A) Immunoblotting analysis of GalM–SPA and GalK–HA-His6 levels in Δspf strains with chromosomal galM-SPA::kan or galK-HA-His6::kan (GSO1066 and GSO106, respectively) transformed with pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP. Cells were grown in M63 galactose to an OD600 of ∼0.5. Anti-FLAG (Top) or anti-His antibody (Bottom) were used to detect GalM–SPA or GalK–HA-His6, respectively. (B) Immunoblotting analysis of MalE–SPA (Top) and MalK–SPA (Bottom) levels in Δspf strains with chromosomal malE-SPA::kan or malK-SPA::kan (GSO1067 and GSO1068, respectively) transformed with pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP. Cells were grown in M63 maltose to an OD600 of ∼0.5. Anti-FLAG antibodies were used to detect both proteins. (C) Immunoblotting analysis of SrlA–SPA levels in Δspf strains with chromosomal srlA-SPA::kan (GSO449) transformed with pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP. Cells were grown in M63 sorbitol to an OD600 of ∼0.5. Anti-FLAG antibody was used to detect the protein. (D) Immunoblotting analysis of LacZ levels in the Δspf strain (GSO1059) transformed with pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP. Cells were grown in M63 lactose to an OD600 of ∼0.5. Anti-β-galactosidase antibody was used to detect the protein. For A, B, C, and D, the Ponceau S–stained membrane documents approximately equal loading of the samples.
Interestingly, the levels of proteins encoded by other CRP-activated operons were not affected by overexpression of SpfP-recoded. Specifically, SrlA–SPA levels were unaffected by SpfP-recoded overexpression for cells grown in M63 medium with sorbitol (Fig. 4C), and SpfP-recoded overexpression did not lead to reduced β-galactosidase levels from lacZ for cells grown in M63 with lactose (Fig. 4D). We also did not detect differences in LB-grown cells in the levels of the AzuC–SPA small protein (SI Appendix, Fig. S3B), whose expression is repressed by CRP. The levels of CRP itself (SI Appendix, Fig. S3C), whose expression is both positively and negatively autoregulated, also did not change upon SpfP-recoded overexpression in cells grown in M63 medium with galactose. The levels of the Spot 42 RNA itself were slightly decreased by SpfP-recoded overexpression for cells grown in LB and slightly increased for cells grown in M63 medium with glucose (SI Appendix, Fig. S3D).
These assays showed that SpfP blocks the activation of some, but not all, CRP-dependent genes. We hypothesize that by binding CRP, SpfP directly or indirectly modulates the CRP interaction with RNA polymerase at affected promoters. However, given that malE and galE, class I and class II–activated genes, respectively, are both repressed by SpfP, while lacZ and srlA, also class I and class II–activated genes, respectively, are both unaffected by SpfP, the regulation is not based on the position of CRP binding relative to RNA polymerase.
SpfP Blocks CRP-Dependent Transcription from the galE P1 Promoter.
To test whether SpfP modulates CRP-mediated transcription activation, we assayed a transcriptional fusion of the P1 promoter of the galactose operon to lacZ (DM0021) (23) for cells carrying pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP grown in LB medium. The β-galactosidase activity assay showed that SpfP-recoded, but not SpfP-recodedSTOP, down-regulates transcription from the P1 promoter (SI Appendix, Fig. S3E). SpfP-recoded did not repress the levels of β-galactosidase activity for a galE P1-lacZ fusion with a C-to-G mutation at the −35 position in the CRP-binding site. However, as expected, β-galactosidase was significantly lower for the C35G derivative. These observations indicate that SpfP affects the ability of CRP to activate transcription of the galactose operon. As observed for GalM–SPA and MalE–SPA, the SpfP-recodedH10A mutant was defective at repressing the galE P1-lacZ fusion (SI Appendix, Fig. S3F).
Mutagenic Screen Reveals CRP Residues Important for Inhibition by SpfP.
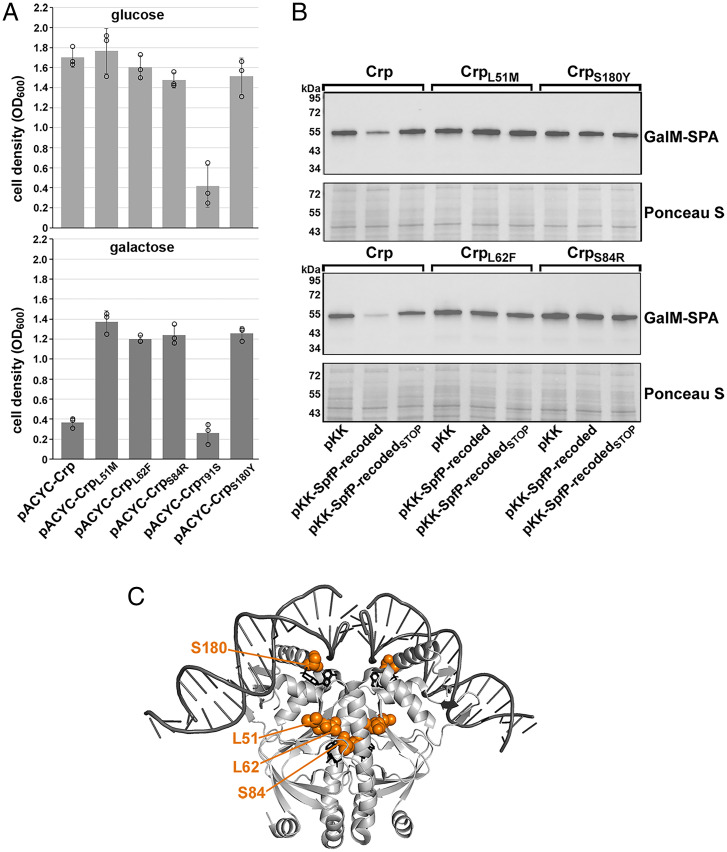
We next mutagenized crp and selected mutants that no longer had the galactose growth defect following overexpression of SpfP. A pACYC184 plasmid expressing CRP was treated with hydroxylamine and then transformed into a Δspf Δcrp strain carrying pKK–SpfP-recoded. Cells were plated on M63 medium supplemented with galactose, where growth is severely inhibited by overexpression of pKK–SpfP-recoded. Sequencing of the crp gene from the mutants that were able to grow revealed five different single mutations in crp: L51M, L62F, S84R, T91S, and S180Y. To verify that these mutants played a role in mitigating the effects of SpfP overexpression, we separately generated each mutation in pACYC-crp by site-directed mutagenesis. These plasmids were then transformed into the Δspf Δcrp strain with pKK–SpfP-recoded and grown for 16 h in glucose and galactose (Fig. 5A). The L51M, L62F, S84R, and S180Y mutations relieved the growth defect associated with SpfP overexpression for cells grown in galactose. In contrast, the T91S mutation produced a growth defect in both glucose and galactose.
Fig. 5.
Mutations that relieve the SpfP effect on CRP map to AR3. (A) Growth assays of Δspf Δcrp cells (GSO1133) expressing pKK–SpfP-recoded and pACYC-CRP derivatives with L51M, L62F, S84R, T91S, or S180Y mutations in M63 minimal medium supplemented with either glucose (Top) or galactose (Bottom). Bars correspond to the average of three biological replicates with circles corresponding to individual data points and error bars representing 1 SD. (B) Immunoblotting analysis of GalM–SPA levels in Δspf Δcrp strains with chromosomal galM-SPA::kan (GSO1134) transformed with pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP and pACYC-crpL51M, pACYC-crpL62F, pACYC-crpS84R, or pACYC-crpS180Y grown in M63 galactose to an OD600 of ∼0.5. Anti-FLAG antibody was used to detect GalM–SPA, and the Ponceau S–stained membrane documents approximately equal loading of the samples. (C) Structure of CRP bound to DNA (38) (Protein Data Bank [PDB] 1ZRF) depicted with PyMol with the L51, L62, S84, and S180 side chains highlighted in orange (numbering is shifted by one amino acid in PDB 1ZRF).
We also examined the ability of the L51M, L62F, S84R, and S180Y mutants to block the inhibitory effect of SpfP on GalM–SPA expression by transforming a galM-SPA Δcrp Δspf strain carrying pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP with the plasmids expressing crp or one of the four mutants (Fig. 5B). Consistent with the results of the genetic screen, SpfP overexpression no longer decreased GalM–SPA levels in the cells expressing mutant CRP. Mapping of these mutated residues on the structure of CRP showed that S180 is near one cAMP-binding site and the CRP DNA-binding domain, while L51, L62F, and S84 are clustered near a second cAMP-binding site and activating region 3 (AR3) implicated in interactions with RNA polymerase (Fig. 5C).
Relative Importance of Spot 42 Base-Pairing Activity and SpfP Protein Activity Varies with Temperature.
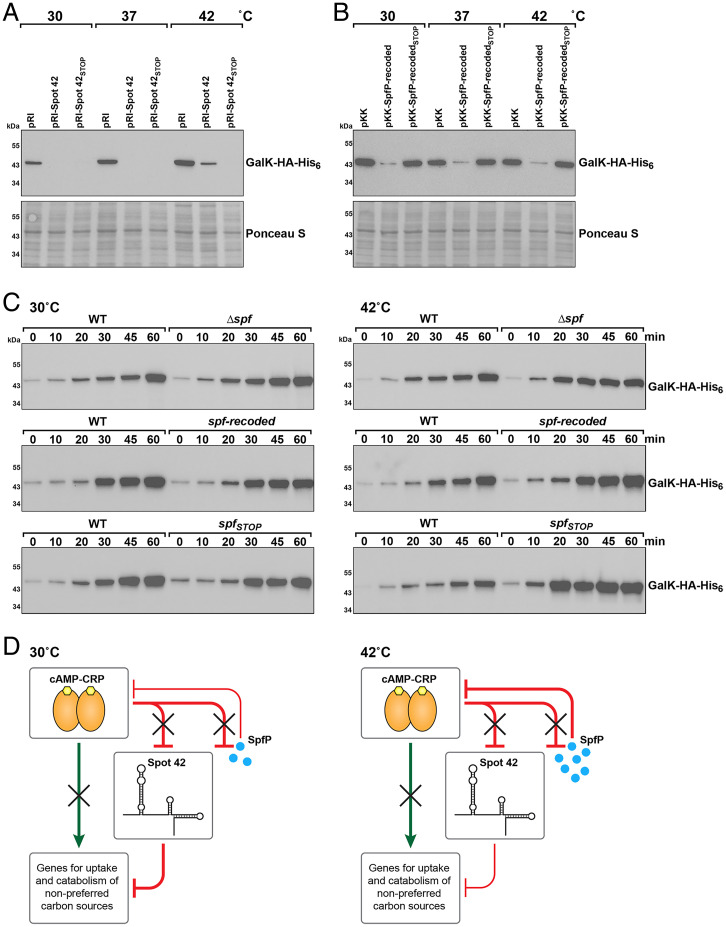
Given the somewhat higher levels of SpfP at 42 °C and 45 °C (Fig. 1C), we wondered whether the importance of the sRNA versus the mRNA activities of spf varied depending on the growth temperature. We repeated the β-galactosidase assay for RNA function by examining srlA-lacZ expression for cells grown at 30 °C, 37 °C, and 42 °C following Spot 42 and Spot 42STOP overexpression. WT Spot 42 overexpression resulted in less repression at 42 °C than at 30 °C and 37 °C (SI Appendix, Fig. S4). In contrast, Spot 42STOP overexpression led to the same repression at all temperatures. We also examined how the same constructs affected GalK–HA-His6 levels at the different temperatures. Consistent with the β-galactosidase activity assay, we observed that full-length Spot 42 was less effective at reducing GalK–HA-His6 levels at 42 °C, while Spot 42STOP overexpression led to strong repression at all temperatures (Fig. 6A). We interpret these results to mean that base pairing is more effective at repression at lower temperatures and that translation of Spot 42 can interfere with base-pairing activity at higher temperatures.
Fig. 6.
Spot 42 and SpfP have different effects at different temperatures. (A) Immunoblotting analysis of GalK–HA-His6 levels in a Δspf galK-HA-His6::kan strain (GSO1060) transformed with pRI, pRI-Spot 42, or pRI-Spot 42STOP and grown in LB at 30 °C, 37 °C, and 42 °C. (B) Immunoblotting analysis of GalK–HA-His6 levels in a Δspf galK-HA-His6::kan strain (GSO1060) transformed with pKK, pKK–SpfP-recoded, or pKK–SpfP-recodedSTOP and grown in LB at 30 °C, 37 °C, and 42 °C. For A and B, samples were collected at an OD600 of ∼0.5. Anti-His antibody was used to detect the HA-His6 tag. The Ponceau S–stained membrane documents approximately equal loading of the samples. (C) Immunoblotting analysis of GalK–HA-His6 levels in galK-HA-His6::kan (GSO1057), Δspf galK-HA-His6::kan (GSO1060), spf-recoded galK-HA-His6::kan (GSO1077), or spfSTOP::kan galK-HA-His6 (GSO1075) grown in M63 glucose to an OD600 of ∼0.4 at 30 °C (Left) or 42 °C (Right). Cells were collected and resuspended in M63 galactose at 30 °C or 42 °C, respectively, with samples collected at the indicated times. (D) The small protein SpfP reinforces the multioutput feedforward loop between CRP and Spot 42. For cells grown in the absence of glucose, CRP directly increases transcription of targets and represses Spot 42. When glucose is available (shown here), the Spot 42 RNA represses CRP-activated targets through base pairing, particularly at lower temperatures (Left), and the small protein SpfP blocks CRP-dependent activation, particularly at higher temperature (Right) (adapted from ref. 16).
Overexpression of SpfP-recoded led to similar, but slightly less, GalK–HA-His6 repression at all temperatures, while SpfP-recodedSTOP had no effect on GalK–HA-His6 levels (Fig. 6B). It is worth noting that, due to the recoded sequence, the spfP ribosome-binding site and start codon likely are no longer in a stem-loop structure, probably allowing equal translation at all temperatures.
Previous studies showed that Spot 42–repressed targets were induced more rapidly in Δspf cells than in WT cells upon a shift from glucose to a nonpreferred carbon source, allowing cells to be primed to utilize glucose for as long as possible (16). We repeated this experiment with a shift from glucose to galactose at 30 °C and 42 °C and similarly observed a more rapid induction of GalK–HA-His6 and GalE in the Δspf background when comparing the same time point between the WT and Δspf strain at both temperatures (Fig. 6C and SI Appendix, Fig. S5). A similar comparison between WT cells and cells in which chromosomal spf carried the recoded sequence (spf-recoded), and thus only expressed SpfP, showed little difference between the WT and spf-recoded strains at 30 °C and slightly stronger repression for the spf-recoded strain at 42 °C (Fig. 6C and SI Appendix, Fig. S5). These data indicate that the small protein is functioning to repress expression at both temperatures. When chromosomal spf had the same stop codon mutation as in Spot 42STOP, we also observed little difference between the WT and spfSTOP strains at 30 °C, probably due to effective Spot 42STOP function as an sRNA. However, we detected more rapid induction with the spfSTOP strain than with the WT strain at 42 °C (Fig. 6C and SI Appendix, Fig. S5). Thus, there is less repression at high temperature in the absence of small protein expression.
Discussion
When bacterial cells encounter environments with mixed nutrients, they selectively metabolize the most energetically favorable carbon sources to maximize growth. To accomplish this, bacteria can use transcription factors, RNA-binding proteins, regulatory sRNAs, and small proteins to specifically control gene expression to synthesize appropriate transporters and enzymes. In this study, we report that the base-pairing sRNA Spot 42 encodes a 15–amino acid protein, SpfP, that plays a role in catabolite repression. Previous work showed that Spot 42 RNA represses numerous targets, including galK, srlA, fucI, and nanC, encoding proteins involved in secondary metabolism and the uptake and use of nonpreferred carbon sources (16, 17). Through copurification analysis, we found that SpfP interacts with CRP (Fig. 3 C and D). We suggest that this interaction functions to block CRP-mediated activation of some target genes. Our discovery that Spot 42 can act as an mRNA demonstrates that this sRNA is a dual-function sRNA whose base-pairing activity and encoded small protein both impact carbon source utilization.
Impact of Dual-Function RNAs on Carbon Metabolism.
The findings reported here add another layer to the modulation of catabolite repression by Spot 42. When the preferred carbon source is available, Spot 42 coordinates with CRP in a multioutput feedforward loop to repress genes from auxiliary metabolic pathways to delay induction of these genes until glucose is consumed or repress expression as soon as glucose is available in order to prioritize the use of the preferred carbon source (16). As shown in Fig. 6D, we suggest that SpfP reinforces this regulatory loop by inhibiting CRP-mediated activation when cells are growing in glucose, particularly at high temperatures.
The regions of Spot 42 involved in base pairing overlap the SpfP coding sequence. This overlap raises the question of whether the sRNA or mRNA activity predominates under specific conditions. Given the potential for the predicted secondary structure overlapping the SpfP ribosome-binding site to be more open at higher temperatures and the slightly higher levels of SpfP at 42 °C and 45 °C, we hypothesized that SpfP-mediated effects may be more prominent at higher temperatures when base pairing may be less efficient. Consistent with this hypothesis, we found that Spot 42 repression of the srlA-lacZ translation fusion was less effective at higher temperatures (SI Appendix, Fig. S4). If Spot 42 base-pairing activity is decreased at temperatures where SpfP translation increases, perhaps SpfP extends the regulatory role of Spot 42 in carbon metabolism for the most critical genes when Spot 42 is no longer functioning as effectively as an sRNA.
Another factor to consider is the Hfq RNA chaperone protein, which is required for Spot 42 stability and base-pairing function (14). Additionally, the CsrA protein of the carbon storage regulator system binds the single GGA site within the SpfP coding sequence to also stabilize Spot 42 by protecting the RNA against RNase E–mediated cleavage (24). Whether the different factors, Hfq, CsrA, and ribosomes bind different pools of Spot 42 or whether the binding of one or the other predominates under specific conditions and how they each affect the base-pairing and protein-coding activities are interesting questions for future studies.
Given that only a handful of dual-function sRNAs have been characterized in bacteria, it is noteworthy that all dual-function sRNAs studied in E. coli thus far impact carbon utilization. The SgrST sRNA is specifically activated by the SgrR transcription factor when cells encounter glucose phosphate stress (25). As with Spot 42, both the base-pairing function and small protein target the same pathway. The sRNA and small protein encoded by SgrS both regulate glucose phosphate utilization metabolism by downregulating the levels and activity of the PtsG transporter (25, 26). In a parallel study, we characterized another dual-function sRNA with connections to carbon metabolism (27). This 164-nt RNA, AzuCR, whose transcription also is repressed by CRP, base pairs with galE, the first gene in the galETKM operon. The RNA is translated to give the 28–amino acid small protein AzuC, which binds to GlpD and leads to increased glycerol dehydrogenase activity. Thus, AzuCR modulates galactose and glycerol metabolism. While the small protein and base-pairing activities are encoded in separate parts of the 227-nt–long SgrST RNA, for both Spot 42 and AzuCR, the ORF overlaps the base-pairing region, leading to competition between the activities of these dual-function sRNAs. The perceived enrichment for dual-function sRNAs that modulate carbon metabolism may be a reflection of the fact that they are relatively abundant and thus readily detected. Alternatively, the strong selection imposed by metabolism may drive the evolution of dual-function sRNA regulators.
SpfP Interaction with CRP.
Another interesting question is the mechanism by which SpfP impacts CRP activity, a global transcription factor that regulates ∼300 genes (28). The functions of only a limited number of small proteins less than 50 amino acids have been elucidated in E. coli, the majority of which are localized to the inner membrane (29). At 15 amino acids, SpfP is among the smallest proteins to be characterized. The mutational studies indicate that the H10 residue of SpfP is needed for the activity of the small protein (SI Appendix, Fig. S3A), and mutations of the L51, L62, S84, and S180 residues of CRP abrogate the negative effect of SpfP overexpression (Fig. 5). Intriguingly, in a CRP–SpfP structure predicted with AlphaFold-Multimer (30), SpfP binds near the L51, L62, and S84 residues (SI Appendix, Fig. S6) in close proximity to the binding site for a second inhibitory cAMP molecule (31, 32) as well as AR3 of CRP (5). We suggest that the hydrophobic SpfP protein acts as a small molecule to affect cAMP binding to CRP or the ability of CRP to bind RNA polymerase, thereby blocking CRP-dependent transcription activation at specific promoters. Given how extensively CRP has been studied, it is intriguing that SpfP was not detected previously, although it is worth noting that a repressive, low-molecular–weight catabolite modulator factor was described decades ago (33). Fortunately, the existing tools and approaches to study CRP should facilitate further mechanistic studies.
SpfP is unique among the small proteins characterized in E. coli because it binds and modulates a transcription factor. A few other very small proteins or peptides have been found to regulate transcription factor activity in other bacteria. For example, the small 5- to 10-amino acid–long signaling peptides secreted by Bacillus and Streptococcus are recognized by sensory receptors from the RRNPP protein family (named for the representative proteins Rap, Rgg, NprR, PlcR, and PrgX) (34). Binding of these peptides to the RRNPP proteins induces conformational changes that favor oligomerization, DNA binding, and transcriptional activation of target genes. Similarly, the 6- to 10-amino acid–long arbitrium peptide released by phages modulates the lysis–lysogeny decision by interacting with the AimR transcription factor (35). We predict that other very small proteins will be found to target transcription factors and possibly other components of the transcriptional machinery. It is worth considering whether derivatives of these small proteins, which are easy to synthesize, could be exploited to control bacterial cell growth in specific ways.
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in Dataset S1. Details about strain and plasmid construction, including mutagenesis by hydroxylamine treatment, are provided in SI Appendix.
Bacterial Growth.
Bacterial growth was monitored in LB rich or M63 minimal medium supplemented with 0.001% vitamin B1 and glucose, galactose, or maltose at 0.2% and sorbitol at 0.4% and appropriate antibiotics, as described in SI Appendix.
Immunoblotting and Northern Analysis.
Specific tagged and untagged proteins were detected by immunoblotting analysis, and specific RNAs were detected by northern analysis, as described in detail in SI Appendix.
Protein Purification.
Proteins tagged with p-AzF or HA were purified based on their tags as described in detail in SI Appendix.
β-galactosidase Assays.
β-galactosidase activity assays were carried out as described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank S. Adhya and D. Lewis for advice and helpful discussions and anti-GalE antiserum, A. Buskirk for the spf browser image, and members of the G.S. group for comments on the manuscript. The work used the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov/). This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Reviewers: C.B., Helmholtz-Institut fur RNA-basierte Infektionsforschung; and S.B., University of Birmingham.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119866119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Brückner R., Titgemeyer F., Carbon catabolite repression in bacteria: Choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209, 141–148 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Görke B., Stülke J., Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Kolb A., Busby S., Buc H., Garges S., Adhya S., Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62, 749–795 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Zheng D., Constantinidou C., Hobman J. L., Minchin S. D., Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32, 5874–5893 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busby S., Ebright R. H., Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293, 199–213 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Aiba H., Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell 32, 141–149 (1983). [DOI] [PubMed] [Google Scholar]
- 7.Mallick U., Herrlich P., Regulation of synthesis of a major outer membrane protein: Cyclic AMP represses Escherichia coli protein III synthesis. Proc. Natl. Acad. Sci. U.S.A. 76, 5520–5523 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polayes D. A., Rice P. W., Garner M. M., Dahlberg J. E., Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J. Bacteriol. 170, 3110–3114 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Lay N., Gottesman S., The Crp-activated sRNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191, 461–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bækkedal C., Haugen P., The Spot 42 RNA: A regulatory small RNA with roles in the central metabolism. RNA Biol. 12, 1071–1077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikemura T., Dahlberg J. E., Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J. Biol. Chem. 248, 5033–5041 (1973). [PubMed] [Google Scholar]
- 12.Ikemura T., Dahlberg J. E., Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis. J. Biol. Chem. 248, 5024–5032 (1973). [PubMed] [Google Scholar]
- 13.Sahagan B. G., Dahlberg J. E., A small, unstable RNA molecule of Escherichia coli: Spot 42 RNA. II. Accumulation and distribution. J. Mol. Biol. 131, 593–605 (1979). [DOI] [PubMed] [Google Scholar]
- 14.Møller T., et al. , Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9, 23–30 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Møller T., Franch T., Udesen C., Gerdes K., Valentin-Hansen P., Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16, 1696–1706 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beisel C. L., Storz G., The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell 41, 286–297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beisel C. L., Updegrove T. B., Janson B. J., Storz G., Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 31, 1961–1974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahagan B. G., Dahlberg J. E., A small, unstable RNA molecule of Escherichia coli: Spot 42 RNA. I. Nucleotide sequence analysis. J. Mol. Biol. 131, 573–592 (1979). [DOI] [PubMed] [Google Scholar]
- 19.Rice P. W., Polayes D. A., Dahlberg J. E., Spot 42 RNA of Escherichia coli is not an mRNA. J. Bacteriol. 169, 3850–3852 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver J., Mohammad F., Buskirk A. R., Storz G., Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10, e02819-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meydan S., et al. , Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell 74, 481–493 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin J. W., et al. , Addition of p-azido-l-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 124, 9026–9027 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Geanacopoulos M., et al. , GalR mutants defective in repressosome formation. Genes Dev. 13, 1251–1262 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai Y.-J., et al. , CsrA regulation via binding to the base-pairing small RNA Spot 42. Mol. Microbiol. 117, 32–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderpool C. K., Gottesman S., Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54, 1076–1089 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Lloyd C. R., Park S., Fei J., Vanderpool C. K., The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J. Bacteriol. 199, e00869-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raina M., et al. , Dual-function AzuCR RNA modulates carbon metabolism. Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.2117930119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimada T., Fujita N., Yamamoto K., Ishihama A., Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6, e20081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemm M. R., Weaver J., Storz G., Escherichia coli small proteome. EcoSal Plus 9, 10.1128/ecosalplus.ESP-0031-2019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2021). 10.1101/2021.10.04.463034 (Accessed 13 December 2021). [DOI]
- 31.Passner J. M., Steitz T. A., The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc. Natl. Acad. Sci. U.S.A. 94, 2843–2847 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fic E., et al. , cAMP receptor protein from Escherichia coli as a model of signal transduction in proteins--a review. J. Mol. Microbiol. Biotechnol. 17, 1–11 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Ullmann A., Tillier F., Monod J., Catabolite modulator factor: A possible mediator of catabolite repression in bacteria. Proc. Natl. Acad. Sci. U.S.A. 73, 3476–3479 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neiditch M. B., Capodagli G. C., Prehna G., Federle M. J., Genetic and structural analyses of RRNPP intercellular peptide signaling of gram-positive bacteria. Annu. Rev. Genet. 51, 311–333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erez Z., et al. , Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keseler I. M., et al. , The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45, D543–D550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Updegrove T. B., Shabalina S. A., Storz G., How do base-pairing small RNAs evolve? FEMS Microbiol. Rev. 39, 379–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Napoli A. A., Lawson C. L., Ebright R. H., Berman H. M., Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: Recognition of pyrimidine-purine and purine-purine steps. J. Mol. Biol. 357, 173–183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.