Abstract
Follicle development beyond the preantral stage is dependent on gonadotropins. FSH signaling is crucial for the advancement of preantral follicles to the antral stage, and LH signaling is essential for further maturation of preovulatory follicles. Estrogen is intricately tied to gonadotropin signaling during the advanced stages of folliculogenesis. We observed that Erβnull ovarian follicles fail to develop beyond the antral stage, even after exogenous gonadotropin stimulation. As ERβ is primarily expressed in the granulosa cells (GCs), we explored the gonadotropin-regulated GC genes that induce maturation of antral follicles. Synchronized follicle development was induced by administration of exogenous gonadotropins to wildtype 4-wk-old female rats. The GC transcriptome was analyzed via RNA-sequencing before and after gonadotropin stimulation. An Erβnull mutant model that fails to show follicle maturation was also included in order to identify the ERβ-regulated genes involved at this step. We observed that specific groups of genes were differentially expressed in response to PMSG or hCG administration in wildtype rats. While some of the PMSG or hCG-induced genes showed a similar expression pattern in Erβnull GCs, a subset of PMSG- or hCG- induced genes showed a differential expression pattern in Erβnull GCs. These latter ERβ-regulated genes included previously known FSH or LH target genes including Lhcgr, Cyp11a1, Cyp19a1, Pgr, Runx2, Egfr, Kiss1, and Ptgs2, which are involved in follicle development, oocyte maturation, and ovulation. We also identified novel ERβ-regulated genes including Jaml, Galnt6, Znf750, Dusp9, Wnt16, and Mageb16 that failed to respond to gonadotropin stimulation in Erβnull GCs. Our findings indicate that the gonadotropin-induced spatiotemporal pattern of gene expression is essential for ovarian follicle maturation beyond the antral stage. However, expression of a subset of those gonadotropin-induced genes is dependent on transcriptional regulation by ERβ.
Keywords: Gonadotropins, ERβ, Follicle maturation, Granulosa cells, Transcriptome analyses
1. INTRODUCTION
Ovarian granulosa (GCs) and theca cells (TCs) play essential regulatory roles in follicle development and oocyte maturation (Eppig, 2001; Jones and Shikanov, 2019; Orisaka et al., 2009). While ovarian follicle activation is gonadotropin-independent, later stages of follicle development require follicle stimulating hormone (FSH) and luteinizing hormone (LH) to induce maturation and ovulation (Goldman and Mahesh, 1968; Goldman and Mahesh, 1969; Grimek et al., 1976; Schwartz, 1974). Gonadotropin signaling initiates steroidogenesis in TCs, which produce androgens (Fortune and Armstrong, 1977) that are converted into estrogens by the neighboring GCs (Erickson and Hsueh, 1978; Liu and Hsueh, 1986). Follicular estrogen in turn regulates the gonadotropin secretion from the H-P axis (Yasin et al., 1995). Gonadotropins also induce differentiation of GCs, which is essential for the expression of factors that regulate the final stages of follicle development, oocyte maturation, and ovulation (Greep et al., 1942; LOSTROH and JOHNSON, 1966). FSH acts through binding the FSH receptor (FSHR) expressed exclusively on GCs, whereas LH acts through binding the LH receptor (LHCGR) expressed on both TCs and GCs (Gromoll et al., 1996; Moudgal, 2012; RAJANIEMI et al., 1977; Simoni et al., 1997). The expression of FSHR on GCs is follicular-stage-specific (Oktay et al., 1997; Peters, 1979), however, follicular-stage-specific expression of LHCGR on GCs is induced by FSH signaling (Dierich et al., 1998; Zeleznik et al., 1974).
Estrogen signaling plays a dynamic role in ovarian folliculogenesis and steroidogenesis, as well as in the hypothalamic-pituitary-ovarian (H-P-O) axis regulation of ovarian functions (Richards, 1980; Shupnik, 2002; Wójtowicz et al., 2007). While TCs predominantly express estrogen receptor alpha (ERα), GCs primarily express estrogen receptor beta (ERβ). Mutant mouse and rat models homozygous for the Erβ null mutation suffer from defective follicle maturation and failure of ovulation (Couse et al., 2005a; Rumi et al., 2017). Although Erβ mutants exhibit an attenuated gonadotropin surge (Jayes et al., 2014; Rumi et al., 2017), administration of exogenous gonadotropins fails to compensate for the defective follicle maturation and induction of ovulation, which suggests a primary intraovarian defect(s) caused by the loss of ERβ (Rumi et al., 2017). Due to the predominant expression of ERβ in GCs, we suspect that dysregulation of ERβ-regulated genes in GCs is responsible for the failure of gonadotropin-induced follicle maturation.
Gonadotropin stimulation induces the differentiation of somatic cells in the ovary and regulates the expression of genes that impact follicle development and oocyte maturation (Dekel and Beers, 1978; Gómez et al., 1993). We observed that Erβnull ovarian follicles fail to develop beyond the antral stage. Previous studies have focused on the attenuated gonadotropin surge and defective expression of GC genes involved in the preovulatory process (Khristi et al., 2018; Rumi et al., 2017). Our previous study identified differential expression of several genes in Erβnull GCs 10h after hCG administration (Khristi et al., 2018). However, the loss of ERβ may interfere with the development of ovarian follicles at different stages before and after gonadotropin stimulation. Previous studies have shown that stage-specific changes in GC genes can play a crucial role in follicle development (Ota et al., 2006; Praveen Chakravarthi et al., 2015; Rice et al., 2007; Xu et al., 2002). Therefore, in this study, we have analyzed the transcriptomes of wildtype and Erβnull GCs before and after gonadotropin stimulation to identify the ERβ-regulated genes, which play important roles at different stages of follicle development. ERβ is the predominant estrogen receptor expressed in the ovary and expression is initiated during the early stages of embryogenesis (Byers et al., 1997b; Couse et al., 2004a; Pelletier and El-Alfy, 2000a; Słomczyńska et al., 2001a). Thus, in the absence of ERβ, ovarian follicle defects may develop due to alerted gene expression prior to gonadotropin stimulation. Therefore, in this study, we have analyzed the GC transcriptome in Erβnull rat ovaries before and after gonadotropin stimulation and compared them with those of wildtype rats. This systematic approach generated an invaluable source of data for understanding the molecular mechanisms involved in ERβ-regulated follicle maturation beyond the antral stage.
2. MATERIALS AND METHODS
2.1. Animal models
4-wk-old Erβnull and age-matched wildtype female Holtzman Sprague-Dawley (HSD) rats were used in this study. Erβnull mutant rats were generated by targeted deletion of exon 3 of the rat Erβ gene (Rumi et al., 2017). Experimental animals were generated by breeding heterozygous mutant male and female rats carrying the exon 3 deletion within Erβ gene. Presence of the Erβ gene mutation was screened for using PCR-based genotyping (Rumi et al., 2017). All animal experiments were performed in accordance with the protocols approved by the University of Kansas Medical Center (KUMC) Animal Care and Use Committee.
2.2. Induction of follicle maturation by gonadotropin administration
Synchronized follicular growth was initiated by administration of exogenous gonadotropins to 4-wk-old Erβnull and wildtype female rats as described previously by our laboratory (Chakravarthi et al., 2020a; Chakravarthi et al., 2018; Khristi et al., 2018; Rumi et al., 2017). Briefly, 30IU of pregnant mare’s serum gonadotropin (PMSG, BioVendor, MO) was injected i.p. into immature female rats, and 48h later some of PMSG treated females received an i.p. injection of 30IU of human chorionic gonadotropin (hCG, BioVendor, MO) (Fig. 2A).
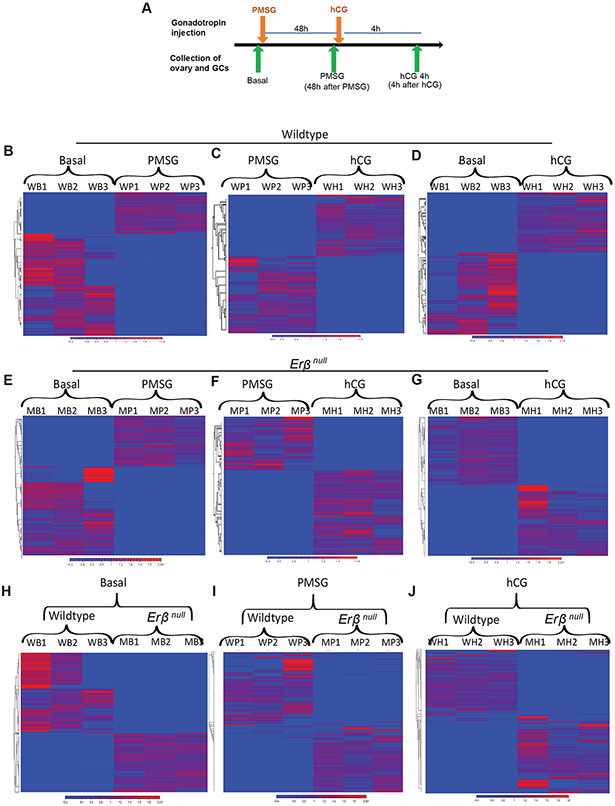
Figure 2. RNA-sequencing of granulosa cells collected from gonadotropin-treated wildtype and Erβnull rat ovaries.
4-wk-old wildtype and Erβnull rats were treated with gonadotropins, and ovaries were collected before and after treatment for isolation of granulosa cells (GCs): before treatment (Basal), 48 hours after PMSG injection (PMSG), and 4h after hCG injection (hCG 4h) (A). Sequencing was performed on total RNA extracted from GCs, and data were analyzed by using CLC Genomics Workbench. Hierarchical clustering was performed on the differentially expressed genes (DEGs) among replicates at different stages of gonadotropin treatment in wildtype (B-D) or Erβnull (E-G) GCs. In addition, clustering was performed on the DEGs between wildtype and Erβnull rat GCs in Basal (H), PMSG (I), and hCG 4h (J) groups.
2.3. Follicle counting in the serial sections of whole ovary
Ovarian tissues were collected in PMSG-treated rats 4h after the hCG injection and fixed in 4% formaldehyde overnight, processed, and embedded in paraffin following standard procedures (Chakravarthi et al., 2020b; Roby, 2001; Roby et al., 2005; Roby et al., 1999). Whole ovaries were serially sectioned at 6 μM thickness and stained with hematoxylin and eosin (H&E). Antral and large antral follicles were counted in every fifth section and was subsequently multiplied by five to estimate total follicle counts. To avoid counting the same follicle more than once, follicles containing nucleus will alone be counted (Chakravarthi et al., 2020b; Roby, 2001; Roby et al., 2005; Roby et al., 1999). Follicles of 900-1000μM diameter, were considered to be large antral (Graafian) follicles (Picut et al., 2015). Ovaries from three wildtype and three Erβnull rats were processed for follicle counting and the data are represented as mean ± SEM, *P ≤ 0.05.
2.4. Collection of GCs and RNA-sequencing
Ovariesarian tissues were collected prior to gonadotropin administration (Basal group), 48h after PMSG injection (PMSG group), and in PMSG-treated rats at 4h or 10h following hCG injection (hCG 4h and hCG 10h groups). Ovaries were collected immediately after sacrificing the rats. The GCs were isolated from the ovaries in M199 media by needle puncture under microscopic examination, followed by filtration with a 40μM cell strainer, centrifuged, and used for total RNA extraction. This whole process took about 15 min (Chakravarthi et al., 2018; Khristi et al., 2018). Total RNA was extracted from the GCs using TRI Reagent (Sigma-Aldrich, St. Louis, MO) following the manufacturer’s instructions. RNA quality was assessed on an Agilent Bioanalyzer, and samples with RIN values over 9 were selected for mRNA-seq library preparation. Each library was prepared using RNA samples from 3 individual rats and 3 independent libraries were made for each experimental time point. 500 ng of total RNA was used for each RNA-seq library preparation using a TruSeq Standard mRNA kit (Illumina, San Diego, CA) as described in our previous publications (Khristi et al., 2018; Khristi et al., 2019). The cDNA libraries were first evaluated for quality at the KUMC Genomics Core and then sequenced on an Illumina HiSeq X sequencer (Novogene Corporation, Sacramento, CA). All RNA-seq data have been submitted to the Sequencing Read Archive as shown in Supplementary Table 1. In addition, our previously published RNA-seq data of wildtype and Erβnull GCs collected 10h post-hCG were used for comparisons (Khristi et al., 2018).
2.5. Analyses of RNA-sequencing data
RNA-sequencing data were demultiplexed, trimmed, aligned, and analyzed using CLC Genomics Workbench 20 (Qiagen Bioinformatics, Germantown, MD) as described previously (Chakravarthi et al., 2020a; Chakravarthi et al., 2019; Khristi et al., 2019). Trimming was performed to remove low quality reads, and good quality reads were aligned with the Rattus norvegicus genome (Rnor_6.0) using the default parameters: (a) maximum number of allowable mismatches was 2; (b) minimum length and similarity fraction was set at 0.8; and (c) minimum number of hits per read was 10. Expression values were measured in TPM (transcripts per million). The threshold p-value was determined according to the false discovery rate (FDR). Differentially expressed genes (DEGs) were selected if the absolute fold change in expression was ≥ 2 with an FDR p-value of ≤0.05 as described previously (Chakravarthi et al., 2020a; Chakravarthi et al., 2019; Khristi et al., 2019).
2.6. Pathway analyses
In the first approach, DEGs were identified between the ‘Basal to PMSG’ and ‘PMSG to hCG 4h’ groups for either wildtype or Erβnull GCs. In the second approach, DEGs were identified in Basal, PMSG, or hCG 4h groups by comparing the wildtype and Erβnull data. These DEGs were sub-grouped into upregulated and downregulated genes. Using the Meta-Chart (https://www.meta-chart.com/) software, Venn diagrams were generated by overlapping the upregulated or downregulated DEGs in wildtype and Erβnull data as identified in the first approach. For further analyses, these Venn diagrams were overlapped with genes upregulated or downregulated in Erβnull GCs as identified in the second approach. PMSG- or hCG-regulated genes in wildtype GCs that showed a differential expression in Erβnull GCs were selected as ERβ-regulated genes and subjected to Ingenuity Pathway Analysis (IPA; Qiagen Bioinformatics) to determine their involvement in folliculogenesis.
2.7. Validation of RNA-sequencing data by RT-qPCR
For validation of RNA-seq data, selected DEGs were analyzed using RT-qPCR. cDNA was prepared from 1 μg of total RNA purified from the GCs collected at the indicated time points (Basal, PMSG, hCG 4h and hCG 10h) from both genotypes (Fig. 2A) using the High Capacity cDNA Reverse Transcription kit (ThermoFisher Scientific). qPCR was performed using Power SYBR Green Master Mix (ThermoFisher Scientific). A list of qPCR primer sequences used in this study is shown in Supplementary Table 2. The results of RT-qPCR were normalized to Rn18s expression and calculated by the comparative ΔΔCT method (Chakravarthi et al., 2020b; Khristi et al., 2018; Khristi et al., 2019).
2.8. Statistical analyses
Each experimental group consisted of a minimum of 6 rats, and all procedures were repeated for reproducibility (except for RNA-sequencing). The experimental results are presented as the mean ± standard error (SE). The results were analyzed for one-way ANOVA, and the significance of mean differences was determined by Duncan’s post hoc test, with p< 0.05. All the statistical calculations were done using SPSS 22 (IBM, Armonk, NY). Each RNA-seq library was prepared using pooled RNA samples from 3 individual wildtype or Erβnull rats. Each group of RNA-sequencing data consisted of three different libraries.
3. RESULTS
3.1. Defective follicle development in Erβnull rats
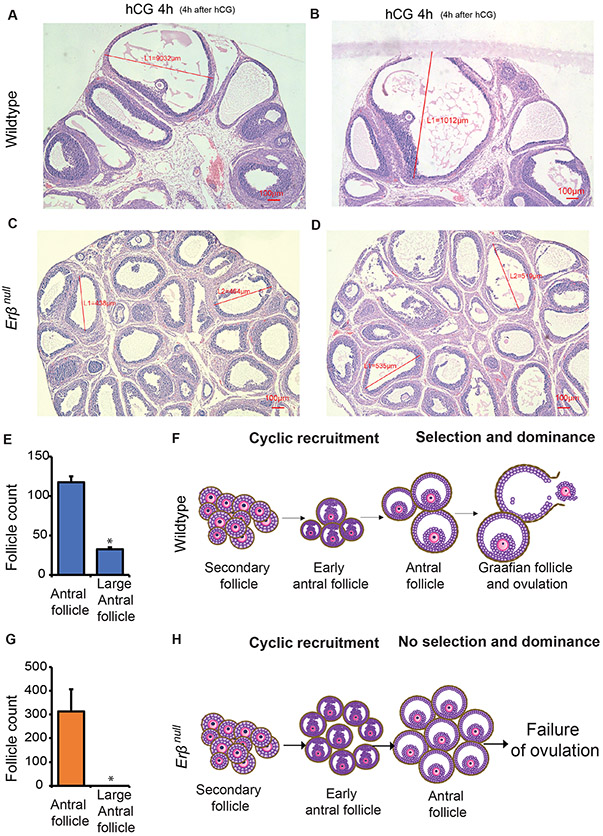
Wildtype rat ovaries responded to exogenous gonadotropins and follicles were observed to have varying stages of development including preantral, antral, and large antral (Graafian) follicles (Fig. 1A-B). In contrast, gonadotropin-stimulated Erβnull rat ovaries exhibited an abundance of antral follicles of similar size (Fig. 1C-D). Wildtype ovaries had antral follicles of 900-1000μM diameter, which are considered to be large antral follicles (Graafian follicles) (Picut et al., 2015). In Erβnull ovaries, follicles did not grow beyond 600μM, which indicates that Erβnull ovaries lack the ability for antral follicles to undergo normal selection and dominance which is evidenced by the follicle size and count (Fig.1 A-D, E, G). While the follicles in wildtype ovaries showed the classic signs of selection and dominance in response to gonadotropin stimulation (Fig. 1F), the antral follicles in Erβnull ovaries failed to undergo such preovulatory development to Graafian follicles (Fig. 1H).
Figure 1. Defective follicle maturation in Erβnull ovaries.
Hematoxylin and Eosin stained sections of ovaries collected from gonadotropin-treated 4-wk-old wildtype rats show different stages of follicles including antral, and preovulatory or Graafian follicles (A-B). In contrast, the sections of ovaries collected from Erβnull rats show numerous antral follicles of similar size (C-D).
Follicle counts were conducted to determine the numbers of antral and preovulatory follicle (Graafian follicle) in wildtype and in Erβnull ovaries (E and G). Ovaries from wildtype rats contain antral and graafian follicles, but Erβnull ovaries are devoid of preovulatory (Graafian) follicles (>600mm) and showing only numerous antral follicles (A-D, E and G). Erβnull follicles do not undergo selection and dominance in response to gonadotropin and lack preovulatory maturation, which is observed in wildtype ovaries following stimulation with PMSG and hCG (F and H). hCG 4h, 4h after hCG administration to PMSG treated rats. Follicle count data represented as mean ± SEM. n ≥ 3. *P ≤ 0.05.
3.2. GC-transcriptome during gonadotropin-induced follicle development
GC mRNA-seq data for Basal, PMSG, and 4h post-hCG groups were analyzed in two different approaches. In the first approach, DEGs were identified across the treatment groups from Basal to PMSG, and from PMSG to 4h post-hCG within the same genotype (i.e., wildtype or Erβnull; Fig. 2B and 2C). PMSG administration resulted in differential expression of 1720 genes in wildtype and 1085 genes in Erβnull GCs. At 4h post-hCG, 3061 genes were differentially expressed in wildtype and 1055 genes in Erβnull GCs. The PMSG and 4h post-hCG DEGs in wildtype and Erβnull GCs are presented in Supplementary Tables 3-6. In the second approach, DEGs were identified within the Basal, PMSG, or 4h post-hCG groups across the wildtype and Erβnull RNA-seq data (Fig. 2D-F). In in Erβnull GCs, 970 genes were differentially expressed at the Basal stage, 1162 genes after PMSG stimulation and 1526 genes at 4h post-hCG. A list of the DEGs in Erβnull GCs compared to wildtype at Basal, PMSG, and hCG 4h time points are presented in Supplementary Tables 7-9.
3.3. ERβ-regulated genes in GCs 48h post-PMSG
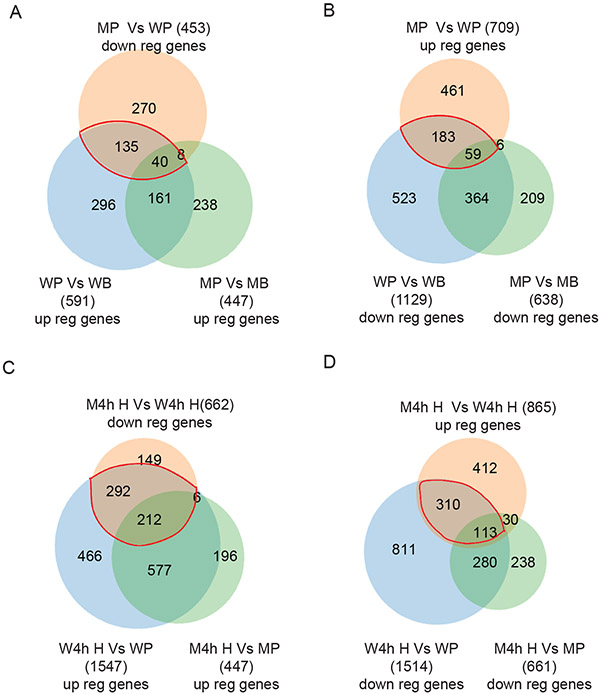
Comparison of the PMSG group to the unstimulated (Basal) group within the same genotype exhibited differential expression of 1720 (591 + 1129) genes in wildtype GCs, and 1085 (447 + 638) genes in Erβnull GCs. Direct comparison of PMSG-treated Erβnull GCs to PMSG-treated wildtype GCs identified a total of 1162 DEGs with 453 genes showing downregulation and 709 showing upregulation. These results show a wide divergence in the gene expression profiles in GCs from wildtype and Erβnull rat ovaries. We compared the PMSG-upregulated (from Basal) genes in wildtype GCs (total 591) and Erβnull GCs (total 447) to those DEGs (total 453) that were downregulated in PMSG-treated Erβnull GCs compared to wildtype GCs (Fig. 3A). In the side-by-side comparison of Erβnull and wildtype GCs, only 201 (161+40) genes showed a similar pattern of response to PMSG in both genotypes. In contrast, 431 (296+135) genes were upregulated in wildtype GCs only and 246 (238+8) genes in Erβnull GCs only. Remarkably, 175 (135+40) genes identified as being upregulated by PMSG in either wildtype or Erβnull GCs had a reduced level of induction/expression in Erβnull GCs as compared to wildtype PMSG-treated GCs (Fig. 3A).
Figure 3. Identification of PMSG- and hCG-regulated genes that are dependent on ERβ signaling.
Upregulated (up reg) genes from the Basal to PMSG treated group in wildtype (WP vs WB) granulosa cells (GCs) were compared with that of Erβnull GCs (MP vs MB). These two sets of DEGs were then compared with the genes that were differentially downregulated (down reg) in PMSG-treated Erβnull GCs compared to that of wildtype GCs (MP vs WP) to identify the potential PMSG-induced genes that are dependent on ERβ signaling (A). Downregulated genes from the Basal to PMSG group in wildtype GCs (WP vs WB) were compared with that of Erβnull GCs (MP vs MB). These two sets of DEGs were then compared with the genes that were upregulated in PMSG-treated Erβnull GCs (MP vs WP) to identify the PMSG-inhibited genes that are dependent on ERβ (B). Similarly, upregulated genes in PMSG to hCG 4h wildtype GCs (W4h H vs WP) were compared to that of Erβnull GCs (M4h H vs MP), and subsequently compared with the genes downregulated in Erβnull GCs 4h after hCG treatment (M4hH vs W4h H), to identify the hCG-induced genes that are dependent on ERβ (C). Finally, downregulated genes from PMSG to hCG 4h wildtype GCs (W4h H vs WP) were compared to that of Erβnull GCs (M4h H vs MP), and then with the differentially upregulated genes in Erβnull hCG 4h GCs (M4h H vs W4h H), to detect the hCG inhibited genes that are dependent on ERβ (D). WB, Wildtype Basal; WP, Wildtype PMSG; W4h H, Wildtype 4h post-hCG; MB, Erβnull Basal; MP, Erβnull PMSG; M4h H, Erβnull 4h post-hCG.
We next made a similar analysis, but focused on genes whose expression decreased following treatment with PMSG (Figure 3B). We compared the genes downregulated (from Basal) after PMSG treatment (1129 in wildtype and 638 in Erβnull GCs) to the 709 DEGs that were upregulated in PMSG-treated Erβnull GCs as compared to wildtype GCs. Again, the side by side comparison of Erβnull and wildtype GCs indicated that only a small proportion of 423 (59+364) genes showed a similar pattern in response to PMSG in both, while 706 (523+183) genes were downregulated in wildtype GCs only and 215 (209+6) genes in Erβnull GCs only (Fig. 3B). Remarkably, 242 (183+59) genes which were downregulated following stimulation with PMSG in wildtype and Erβnull GCs actually had a higher level of expression in Erβnull GCs when compared to wildtype PMSG-treated GCs (Fig. 3B).
3.4. ERβ-regulated genes in GCs 4h post-hCG
Comparative analyses were performed with the 4h post-hCG GCs (Fig 3C and D) and their respective PMSG controls as described above for the post-PMSG group. While hCG treatment altered the expression of 3061 genes (1547 upregulated and 1514 downregulated) in wildtype GCs, expression of only 1108 genes (447 upregulated and 661 downregulated) were significantly modulated in Erβnull GCs. Direct comparison of hCG-treated Erβnull GCs to hCG-treated wildtype GCs identified a total of 1527 DEGs, of those 662 genes were downregulated (Fig. 3C) and 865 were upregulated (Fig. 3D) in the Erβnull GCs. Of the total 1547 upregulated genes in wildtype hCG 4h GCs, 789 (577 +212) genes showed similar expression patterns in Erβnull hCG 4h GCs (Fig. 3C), while 758 (466 +292) upregulated genes were unique to wildtype GCs and 212 (196 + 6) to Erβnull GCs. Remarkably, 504 (292+212) genes that were upregulated in wildtype hCG 4h GCs had a significantly lower level of expression/induction in Erβnull hCG 4h GCs (Fig. 3C).
Of the total 1514 genes downregulated in wildtype hCG 4h GCs, 393 (280+113) genes were also downregulated in Erβnull hCG 4h GCs (Fig. 3D), while an additional 1121 (811+310) downregulated genes were unique to wildtype GCs and 268 (238 +30) unique to Erβnull GCs (Fig. 3D). Furthermore, we identified 423 (310 +113) genes that were downregulated in wildtype hCG 4h GCs that had a significantly higher level of expression/induction in ERβnull hCG 4h GCs (Fig. 3D).
3.5. ERβ-regulated FSH/FSHR target genes
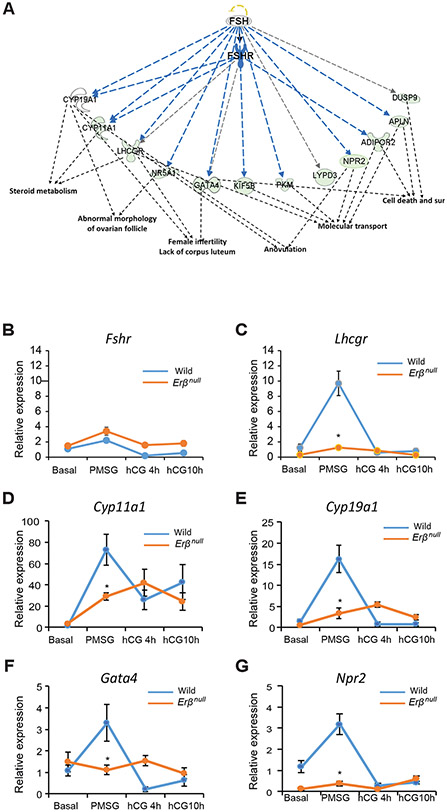
As PMSG predominantly acts on FSH receptors (FSHRs), we examined the PMSG-regulated genes using Ingenuity Pathway Analysis (IPA) specifically for FSH/FSHR signaling. IPA demonstrated that a large number of the 175 genes (Fig. 3A) that were upregulated by PMSG treatment (from Basal) in wildtype or Erβnull GCs, and whose expression differed between the genotypes following PMSG treatment, have roles in folliculogenesis (Fig. 4A). Further analyses determined that those genes identified as having a reduced level of induction following PMSG in Erβnull GCs are specifically involved in steroid metabolism, follicle development, molecular transport, cell survival, ovulation, and fertility (Fig. 4A).
Figure 4. PMSG-regulated genes that are ERβ dependent and involved in folliculogenesis.
PMSG and ERβ dependent target genes that were identified in the Fig. 3A were subjected to Ingenuity Pathway Analysis (IPA). IPA revealed a group of downregulated genes that have FSH/FSHR as an upstream regulator (A) and are involved in steroid metabolism, folliculogenesis, ovulation, molecular transport, cell death, and cell survival (sur). The expression of these genes was further validated by RT-qPCR analysis (B-G). We did not detect any significant difference in the expression levels or pattern of Fshr between wildtype and Erβnull GCs (B). In wildtype GCs, Lhcgr, Cyp11a1 Cyp19a1, Gata4 and Npr2 expression significantly increased from Basal to PMSG and then returned to basal levels at 4h after hCG and remained unchanged at 10h after hCG (C-G). In Erβnull GCs the pattern of expression of Lhcgr was similar to that of wildtype GCs, but following PMSG Lhcgr expression was significantly lower in Erβnull GCs compared to wildtype GCs (C). Cyp11a1 and Cyp19a1 expression showed an increasing pattern from Basal to hCG4h and remained unchanged at hCG10h, however, following PMSG Cyp11a1 and Cyp19a1 expression was significantly lower in Erβnull GCs compared to wildtype GCs (D, E). There was no significant change in the expression of Gata4 and Npr2 from Basal to 10h hCG. But following PMSG treatment, Gata4 and Npr2 expression was significantly lower in Erβnull GCs compared to wildtype GCs (F, G). RT-qPCR data are represented as mean ± SEM. n ≥ 6. *P ≤ 0.05.
We observed that the expression of Fshr was not decreased in PMSG-treated Erβnull GCs (Fig. 4B), but several other genes known to be upregulated by FSH/FSHR signaling failed to respond appropriately (Fig. 4). Interestingly, expression of Lhcgr, Cyp11a1, Cyp19a, Gata4, and Npr2 genes were quite low in both wildtype and Erβnull GCs at the Basal stage prior to gonadotropin administration, elevated at 48h after PMSG injection, and then declined back to Basal levels 4h after hCG injection (Fig. 4C-G). Even though Erβnull GCs showed similar pattern of Lhcgr expression, PMSG-induced upregulation of Lhcgr was significantly low in Erβnull GCs compared to wildtype GCs (Fig. 4C). The expression of Cyp11a1, Cyp19a1 in Erβnull GCs showed increasing pattern from basal to hCG4h and then decreased slightly which was not significant. There was a significant increase in the expression of Cyp11a1 in the Erβnull GCs after PMSG treatment, but it was remarkably lower than that of wildtype. PMSG-induced upregulation of other steroidogenesis-related genes including Cyp19a1 in the Erβnull GCs was also lower than that in wildtype GCs.” (Fig. 4D and E). There is no significant change in the expression of Gata4 and Npr2 in Erβnull GCs across different stages of gonadotropin stimulation from Basal to 10h after hCG. The expression of these transcriptional regulators (Gata4, and Npr2) failed to be induced by PMSG in Erβnull GCs (Fig. 4F and G).
3.6. ERβ-regulated LH/LHCGR target genes
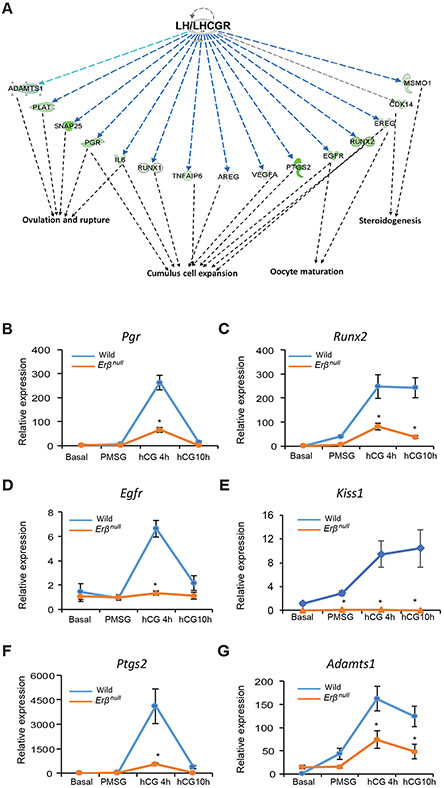
hCG is known to act on the LH receptors (LHCGRs) expressed on GCs, therefore, we have examined the hCG-regulated genes using IPA specifically focusing on LH/LHCGR signaling. To identify the hCG-induced genes that are regulated by ERβ, genes that were upregulated by hCG from the levels after PMSG treatment in both wildtype and Erβnull GCs were overlapped with the genes that were downregulated in Erβnull GCs by hCG treatment compared to wildtype (Fig. 3C). This comparative analysis identified 504 genes (292+212) (Fig. 3C) as hCG induced genes that need ERβ for their induction. These 504 genes were subjected to IPA analysis to determine the genes involved in the cumulus cell expansion, oocyte maturation, steroidogenesis, and ovulation (Fig. 5A).
Figure 5. hCG-regulated genes that are dependent on ERβ and involved in follicle maturation.
hCG-regulated genes that were identified in Fig. 3C were subjected to Ingenuity Pathway Analysis (IPA). IPA revealed a subset of LH/LHCGR-regulated genes involved in steroidogenesis, cumulus cell expansion, oocyte maturation, and ovulation (A). The expression of some of the key genes involved in follicle maturation and ovulation were further confirmed by RT-qPCR (B-G). The expression of Pgr, Egfr and Ptgs2 remained unchanged from Basal to PMSG but increased significantly at hCG 4h and then came to basal levels at 10h following hCG in wildtype GCs (B, D, F). The expression levels and pattern of Pgr, Egfr and Ptgs2 in Erβnull GCs were similar to that of wildtype GCs except at hCG4h, where the expression was significantly lower in Erβnull GCs compared to wildtype GCs (B, D, F). The expression of Runx2, Kiss1 and Adamts1 increased slightly from Basal to PMSG but increased rapidly at hCG 4h and then remained unchanged at hCG 10h in Erβnull GCs (C, E, G). The expression pattern of Runx2 and Adamts1 in Erβnull GCs was similar to that of wildtype GCs, but thereafter at hCG4h and hCG 10h expression was significantly lower in Erβnull GCs compared to wildtype GCs (C, G). The expression of Kiss1 remained unchanged from Basal to hCG 10h in Erβnull GCs. In contrast, Kiss1 expression level at PMSG, hCG4h and hCG 10h was significantly higher in wildtype GCs as compared to Erβnull GCs (E). RT-qPCR data are represented as mean ± SEM. n ≥ 6. *P ≤ 0.05.
In wildtype GCs expression levels of Pgr, Egfr, and Ptgs2 were low basally and after PMSG treatment, increased dramatically after hCG and them declined back to basal levels at 10h after hCG (Fig. 5B, D, F). In contrast, Runx2, Kiss1 and Adamts1 expression was elevated over basal levels following gonadotropin administration and was higher after hCG than that observed following PMSG treatment (Fig. 5C, E, G). In Erβnull GCs the expression pattern of Pgr, Egfr, and Ptgs2 was similar to that of wildtype GCs with the exception of that observed at 4hr post-hCG where expression was significantly lower than that overserved in wildtype GCs(Fig. 5B, D, F). The expression pattern for Runx2 and Adamts1 in Erβnull GCs was similar to that of wildtype GCs, but levels were significantly lower at 4h and 10h after hCG compared to wildtype GCs (Fig. 5C, G). In the case of Kiss1, expression remained unchanged from Basal to 10h after hCG and was significantly lower at all stages in Erβnull GCs compared to wildtype GCs. The hCG-induced upregulation of Kiss1 expression was completely absent in Erβnull GCs (Fig. 5E).
3.7. ERβ-regulated novel genes involved in follicle development
We also identified novel gonadotropin-stimulated genes that were differentially upregulated in wildtype GCs but failed to respond in Erβnull GCs. We speculate that these genes may contribute to gonadotropin-stimulated ovarian follicle development. However, IPA analyses did not link these genes to either gonadotropin or ERβ signaling. We have selected and validated the expression of these novel genes based on their higher TPM values and relative fold changes.
Among these genes, Jaml (Fig. 6A), Galnt6 (Fig. 6B), Znf750 (Fig. 6C) and Dusp9 (Fig. 6D) were high basally or upregulated by PMSG treatment and subsequently downregulated after hCG administration in wildtype GCs. Interestingly, the expression of Jaml (Fig. 6A), which is a cell adhesion molecule, was significantly higher at the Basal stage in wildtype GCs as compared to Erβnull GCs. Two other novel genes, Wnt16 (Fig. 6E) and Mageb16 (Fig. 6F), were upregulated in wildtype GCs only after hCG treatment. While the expression of Wnt16 (Fig. 6E) declined to Basal levels 10h after hCG treatment, Mageb16 (Fig. 6F) levels remained elevated. These genes exhibited minimal changes in Erβnull GCs after gonadotropin treatment.
Figure 6. Gonadotropin-induced and ERβ-regulated novel genes with potential roles in follicle development.
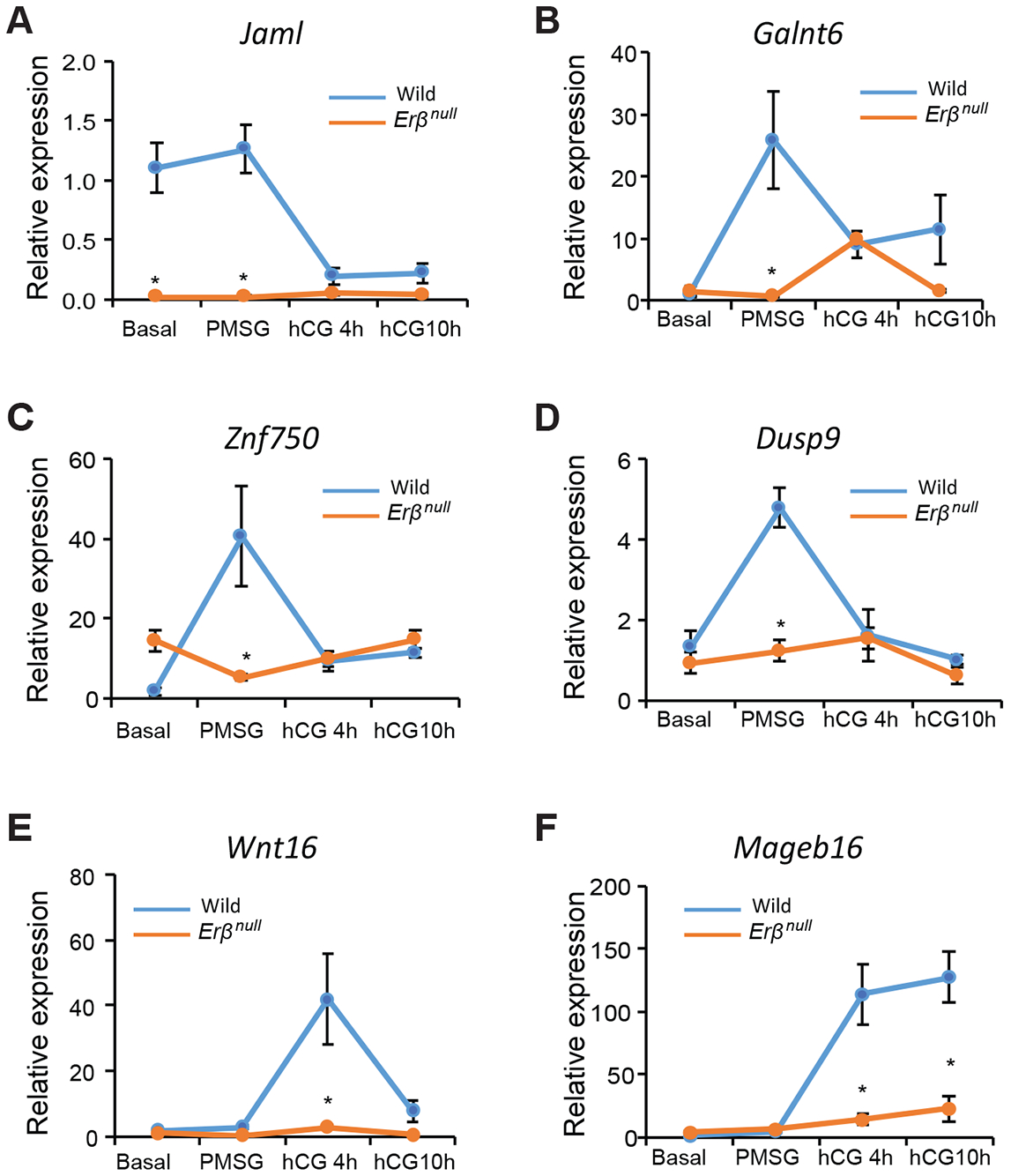
We identified several PMSG (A-D) and hCG (E, F) regulated genes, which were not previously shown to be regulated by ERβ or gonadotropins. We identified these genes based on their relative abundance (TPM values) and fold change in Erβnull GCs. PMSG induced genes included Jaml, Galnt6, Znf750, and Dusp9 (A-D). The expression of Jaml was higher basally and following PMSG in wildtype GCs as compared to Erβnull GCs and declined following stimulation with hCG (A) whereas Erβnull GCs the expression remained unchanged following gonadotropin stimulation (A). The expression of Galnt6, Znf750, and Dusp9 increased significantly from Basal to PMSG and declined after hCG stimulation in wildtype GCs (B-D). In Erβnull GCs, expression of Galnt6 remained unchanged from Basal to PMSG, increased slightly 4h after hCG, 4h and then decreased at hCG10h. The PMSG-induced increase in Galnt6 observed in wildtype GCs was completely absent in Erβnull GCs (B). The expression of Znf750 and Dusp9 in Erβnull GCs remained unchanged from Basal to hCG10h, which is in contrast to PMSG induction of expression in wildtype GCs (C, D). The expression of Wnt16 remained unchanged from Basal to PMSG, increased significantly from PMSG to hCG 4h and then declined to basal levels at hCG10h in wildtype GCs (E). The expression of Wnt16 remains unchanged from Basal to hCG10h in Erβnull GCs (E). Expression of Mageb16 was increased at 4 and 10h following hCG stimulation in wildtype GCs (F). Expression of Mageb16 was also increased by hCG in Erβnull GCs, but this induction was markedly lower than that achieved in wildtype GCs at hCG4h and hCG10h (F). RT-qPCR data are represented as mean ± SEM. n ≥ 6. *P ≤ 0.05.
4. DISCUSSION
Erβnull female rodents are infertile due to defective follicle development and failure of ovulation (Couse and Korach, 1999; Couse et al., 2003; Rumi et al., 2017). In order to elucidate the specific defects in follicle development, we studied exogenous-gonadotropin-induced synchronized follicle development using 4-wk-old female rats. We observed that gonadotropin stimulation fails to initiate the selection and dominance of antral follicles in Erβnull rats. The presence of many antral follicles of similar size and lack of preovulatory Graafian follicles suggest that ERβ is essential for follicle development beyond the antral stage.
ERβ is the major estrogen receptor in the ovary (Byers et al., 1997a; Couse et al., 2004b; Pelletier and El-Alfy, 2000b; Słomczyńska et al., 2001b), and GCs are the major cell-type in ovarian follicles that express ERβ (Cheng et al., 2002; Paterni et al., 2014). ERβ plays an important role in the transcriptional regulation of GC genes that are involved in the GC-oocyte interactions required for the development of ovarian follicles (Couse et al., 2005b; Khristi et al., 2018; Rumi et al., 2017). In this study, we analyzed the GC transcriptome before and after gonadotropin administration to identify the ERβ-regulated genes (direct or indirect) that are involved in follicle maturation beyond the antral stage. Using a systematic approach, we identified the DEGs that were induced by PMSG and hCG during the development of wildtype ovarian follicles We subsequently identified DEGs that showed a similar or opposite expression pattern in Erβnull GCs. This unique approach of differential transcriptome analysis enabled us to identify the ERβ-regulated genes that are dysregulated after PMSG or hCG administration in Erβnull rats. These ERβ-regulated genes were further subjected to IPA to identify key genes involved in steroidogenesis, follicle development, cumulus expansion, oocyte maturation, and ovulation.
It is noteworthy that, for the transcriptome and subsequent IPA analyses, we have considered PMSG treatment to be representative of FSH/FSHR activation and hCG treatment of LH/LHCGR activation. Despite a normal level of Fshr expression in Erβnull GCs, PMSG treatment failed to induce many of the FSH/FSHR target genes including Lhcgr, Cyp11a1, Cyp19a1, Gata4, and Npr2; however, expression of some other known targets like Star remained unaffected. This suggests that upregulation of a subset of PMSG-induced genes also requires the presence of ERβ in GCs. In wildtype GCs, a low level of gene expression before gonadotropin treatment (Basal stage) and marked upregulation after PMSG administration suggests that induction of these proteins is necessary during this stage of follicle development or to execute the responses of LH/LHCGR signaling during the subsequent stage. Moreover, a sharp decrease of selective transcript levels after hCG administration suggests that either LH/LHCGR signaling leads to an abrupt downregulation of transcription or degradation of those mRNAs. Such spatiotemporal patterns of expression in response to gonadotropins are characteristic of many of the GC genes involved in follicle maturation (Bédard et al., 2003; Chakravarthi et al., 2015; Chakravarthi et al., 2016; Chen et al., 2009; Lakshminarayana et al., 2014; Ronen-Fuhrmann et al., 1998). However, those reports were based on studying one or a few genes, while the current study demonstrates the spatiotemporal pattern of gene expression among the whole transcriptome in wildtype and Erβnull GCs.
PMSG stimulation failed to upregulate the expression of Lhcgr in Erβnull GCs, which is essential for follicle development beyond the antral stage (LaPOLT et al., 1990; Lu et al., 1993; Richards and Ascoli, 2018). Lhcgr knockout mice exhibited an ovarian phenotype with normal development of primary, secondary, and antral follicles but a lack of preovulatory Graafian follicles (Lei et al., 2001). Together these studies emphasize the importance of ERβ-regulated Lhcgr expression during the late stages of follicle development. Similarly, Cyp19a1 knockout mice also developed antral follicles that failed to ovulate (Fisher et al., 1998). It has been reported that decreased levels of Cyp11a1 or Cyp19a1 disrupt the development of antral and Graafian follicles (Gershon and Dekel, 2020). The reduced level of Gata4 expression in Erβnull GCs may contribute to reduced GC proliferation and theca cell recruitment, which would impair the maturation of follicles to the preovulatory stage (Bennett et al., 2013; Efimenko et al., 2013; LaVoie, 2014). Additionally, NPR2 plays an important role in GCs by inducing cGMP levels, which inhibits phosphodiesterase 3A, prevents cAMP hydrolysis, and releases oocytes from meiotic arrest (Arroyo et al., 2020; Potter et al., 2006; Zhang et al., 2010).
We identified that LH/LHCGR target genes, particularly the expression of genes involved in cumulus cell expansion, steroidogenesis, oocyte maturation, and ovulation, were affected in Erβnull GCs. This effect was in part due to the failure of PMSG-induced Lhcgr upregulation in Erβnull GCs. We found that hCG-induced upregulation of Pgr in GCs is also dependent on ERβ. Progesterone signaling is essential for follicle maturation and induction of ovulation (Jo et al., 2002; Kubota et al., 2016; Robker et al., 2009). Progesterone signaling regulates the expression of Egfr, Ptgs2, and Adamts1 (Robker et al., 2000), and their expression was disregulated in Erβnull GCs. Studies have shown that EGFR signaling can regulate the expression of Runx2 and Plat, induce production of steroids, and promote oocyte maturation (Jamnongjit et al., 2005). EGFR signaling also plays an essential role in the transition of antral follicles to the preovulatory stage (Panigone et al., 2008; Wu et al., 2019). RUNX2 regulates the expression of Ptgs2 and Runx1, impacting the ovulatory process (Lee-Thacker et al., 2018; Liu et al., 2009).
Expression of Ptgs2 and Runx2 showed significant increases in Erβnull GCs in response to hCG, but the level of induction was significantly lower as compared to wildtype GCs, which suggests that a relatively lower level of Runx2 may in turn affect the upregulation of Ptgs2. Importantly, PTGS2 is involved in prostaglandin synthesis, preovulatory follicle maturation, and ovulation (Laurincik et al., 1993; Siddappa et al., 2015). We previously reported that the expression of Kiss1 in GCs is induced by gonadotropins and dependent on ERβ signaling (Chakravarthi et al., 2018). Intraovarian administration of kisspeptin increased the number of preovulatory follicles and decreased the number of antral follicles, whereas treatment with kisspeptin antagonist KP234 had the opposite effect, suggesting a requirement for kisspeptin during preovulatory follicle maturation (Fernandois et al., 2016). Recently, we have also shown that GC-derived KISS1 plays a role in follicle development and oocyte maturation (Chakravarthi et al, 2020). In addition to Ptgs2, hCG-induced upregulation of Adamts1 in Erβnull GCs did not reach to the level that was observed in wildtype GCs It has been suggested that LH/LHCGR activated ERK1/2 induce the expression of ADAMTS1, a metalloprotease, which is also involved in cumulus expansion and induction of ovulation (Arroyo et al., 2020; Brown et al., 2010; Madan et al., 2003; Russell et al., 2003; Shozu et al., 2005; Yung et al., 2010).
We also identified a subset of Erβ-dependent, gonadotropin-induced genes in GCs, that were previously not known to be regulated by gonadotropin or ERβ signaling or to be involved in ovarian follicle development. Among these novel genes, JAML is a cell-cell adhesion protein and engaged in homophilic interactions (Moog-Lutz et al., 2003). Expression of Jaml was high in the Basal and PMSG groups and decreased after hCG stimulation in wildtype, while expression remained low at all stages in Erβnull GCs. The high level of expression in the Basal and PMSG stages suggests that JAML might have a role in preantral follicles, especially in GC-GC or GC-oocyte interactions. Induction by hCG may suppress Jaml expression, leading to a reduction in these interactions in preparation for ovulation. GALNT6 has been shown to be essential for the activation of EGFR by phosphorylation and by modification of O-glycosylation (Lin et al., 2017). It is well known that LH-induced EGFR signaling plays a critical role in preovulatory follicle development by activating MAPK pathways and inducing the steroidogenic enzymes (Jamnongjit et al., 2005; Panigone et al., 2008; Wu et al., 2019). Dual-specific phosphatases (DUSPs) dephosphorylate MAPKs and thereby inhibiting their activities (Chen et al., 2019). We speculate that PMSG-induced Dusp9 suppresses ERK activation to allow proper development of preantral follicles in wildtype ovaries. In contrast, in Erβnull ovaries, the lack of DUSP9 results in aberrant activation of ERK and leads to premature formation of many small antral follicles. ZNF750 is a cell cycle regulator that acts by inducing Klf4 gene expression (Boxer et al., 2014; Liu et al., 2019; Sen et al., 2012). KLF4 increases the susceptibility of GCs to apoptosis by downregulating Bcl2 expression and promotes LH induced luteal transition of GCs (Choi and Roh, 2018). In this study, we also observed a failure of PMSG- or hCG-stimulated induction of either Wnt16 or Mageb16. WNT16 is a positive regulator of the canonical WNT pathway and it plays a potential role in the dominant follicle selection (Gupta et al., 2014). Mage proteins interact with nucleic acids or target proteins and regulate different cell processes (Rajagopalan et al., 2011). Mageb16 was reported to be a testis-specific gene (Liu et al., 2014), and the expression of Mageb16 was found to be low in the ovary (Liu et al., 2014). Interestingly, we observed a dramatic increase in the expression of Mageb16 in wildtype GCs after gonadotropin injection, which implicates gonadotropin stimulation is being necessary for the expression of Mageb16 in ovaries. Based on the expression and its general function, Mageb16 may have a role in the later stages of follicle development and further studies are needed to confirm its role.
Ours results demonstrate the spatiotemporal pattern of gonadotropin-induced gene regulation in GCs that is essential for ovarian follicle maturation. These datasets may serve as the reference for understanding gonadotropin regulation of GC genes. Although a group of gonadotropin-regulated genes showed similar expression patterns in both wildtype and Erβnull GCs, others were unique to each genotype. We were particularly interested in a subset of gonadotropin-regulated genes characterized by a remarkably different expression pattern in Erβnull GCs. These included FSH signaling regulated genes like Lhcgr, Cyp11a1, Cyp19a1, Gata4, Npr2, Jaml, Galnt6, Znf750, and Dusp6 and LH regulated genes Pgr, Egfr, Adamts1, Ptgs2, Kiss1, Runx2, Wnt16, and Mageb16 (Fig.7). We also identified several novel genes including Jaml, Galnt6, Znf750, Dusp6, Wnt16, and Mageb16, which are gonadotropin-induced but dependent on Erβ. Reported functions for these novel genes suggest that they may have important roles in follicle development and ovulation however, further studies are required to confirm their involvement in the ovary.
Figure 7. ERβ-regulated granulosa cell genes that are crucial for gonadotropin induced follicle maturation.
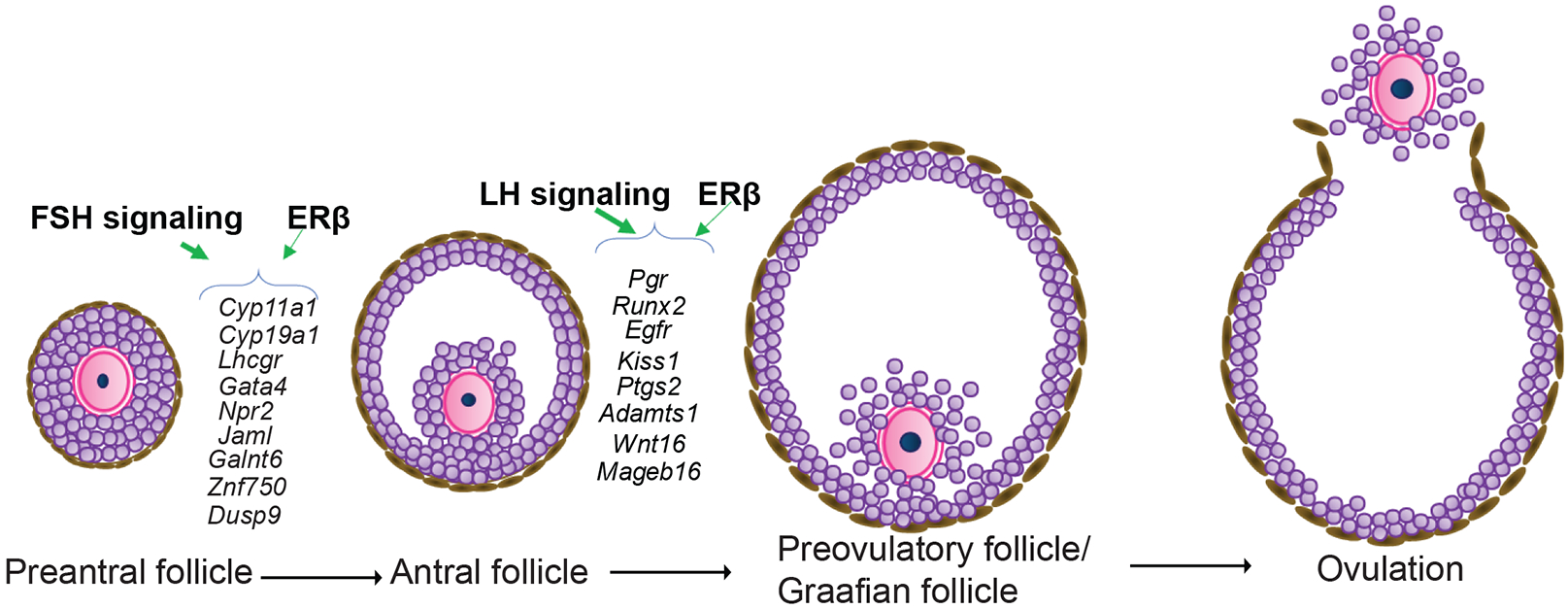
Expression of FSH/FSHR-induced genes including Cyp11a1, Cyp19a1, Lhcgr, Gata4, Npr2, Jaml, Galnt6, Znf750, and Dusp9 as well as expression of LH/LHCGR-induced genes including Pgr, Runx2, Egfr, Kiss1, Ptgs2, Adamts1, Wnt16, and Mageb16 are dependent on ERβ-signaling. These genes play a crucial role in the preovulatory maturation of ovarian follicles.
Supplementary Material
Acknowledgments
Generation of these datasets was supported by funding from the KUMC SOM, COBRE (P30 GM122731), and K-INBRE.
Footnotes
Disclosure: The authors do not have any conflicts of interest.
6. REFERENCES
- Arroyo A, Kim B, and Yeh J. 2020. Luteinizing Hormone Action in Human Oocyte Maturation and Quality: Signaling Pathways, Regulation, and Clinical Impact. Reproductive sciences (Thousand Oaks, Calif.). 27:1223–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bédard J, Brûlé S, Price CA, Silversides DW, and Lussier JG. 2003. Serine protease inhibitor-E2 (SERPINE2) is differentially expressed in granulosa cells of dominant follicle in cattle. Molecular reproduction and development. 64:152–165. [DOI] [PubMed] [Google Scholar]
- Bennett J, Baumgarten SC, and Stocco C. 2013. GATA4 and GATA6 silencing in ovarian granulosa cells affects levels of mRNAs involved in steroidogenesis, extracellular structure organization, IGF-I activity, and apoptosis. Endocrinology. 154:4845–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer LD, Barajas B, Tao S, Zhang J, and Khavari PA. 2014. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes & development. 28:2013–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, and Russell DL. 2010. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biology of reproduction. 83:549–557. [DOI] [PubMed] [Google Scholar]
- Byers M, Kuiper GG, Gustafsson J.-A.k., and Park-Sarge O-K. 1997a. Estrogen receptor-β mRNA expression in rat ovary: down-regulation by gonadotropins. Molecular Endocrinology. 11:172–182. [DOI] [PubMed] [Google Scholar]
- Byers M, Kuiper GG, Gustafsson JA, and Park-Sarge OK. 1997b. Estrogen receptor-beta mRNA expression in rat ovary: down-regulation by gonadotropins. Molecular endocrinology (Baltimore, Md.). 11:172–182. [DOI] [PubMed] [Google Scholar]
- Chakravarthi VP, Ghosh S, Dai E, Pathak D, and Rumi MAK. 2020a. Transcriptome datasets of ESR2-regulated genes in rat granulosa cells during gonadotropin-induced follicle maturation. Data in brief. 30:105405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthi VP, Ghosh S, Roby KF, Wolfe MW, and Rumi MAK. 2020b. A Gatekeeping Role of ESR2 to Maintain the Primordial Follicle Reserve. Endocrinology. 161. [DOI] [PubMed] [Google Scholar]
- Chakravarthi VP, Ghosh S, Roy R, Dai E, Pathak D, and Rumi MAK. 2019. Transcriptome datasets of gonadotropin-induced ESR2-regulated genes in rat oocytes. Data in brief. 27:104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthi VP, Khristi V, Ghosh S, Yerrathota S, Dai E, Roby KF, Wolfe MW, and Rumi MAK. 2018. ESR2 Is Essential for Gonadotropin-Induced Kiss1 Expression in Granulosa Cells. Endocrinology. 159:3860–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthi VP, Kona S, Kumar AS, Bhaskar M, and Rao V. 2015. Quantitative expression of antiapoptotic and proapoptotic genes in sheep ovarian follicles grown in vivo or cultured in vitro. Theriogenology. 83:590–595. [DOI] [PubMed] [Google Scholar]
- Chakravarthi VP, Kona S, Kumar AS, Bhaskar M, and Rao V. 2016. Quantitative patterns of expression of gap junction genes during in vivo or in vitro development of ovarian follicles in sheep. Small Ruminant Research. 143:35–42. [Google Scholar]
- Chen AQ, Wang ZG, Xu ZR, Yu SD, and Yang ZG. 2009. Analysis of gene expression in granulosa cells of ovine antral growing follicles using suppressive subtractive hybridization. Animal reproduction science. 115:39–48. [DOI] [PubMed] [Google Scholar]
- Chen HF, Chuang HC, and Tan TH. 2019. Regulation of Dual-Specificity Phosphatase (DUSP) Ubiquitination and Protein Stability. International journal of molecular sciences. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Weihua Z, Mäkinen S, Mäkelä S, Saji S, Warner M, Gustafsson J-A, and Hovatta O. 2002. A role for the androgen receptor in follicular atresia of estrogen receptor beta knockout mouse ovary. Biology of reproduction. 66:77–84. [DOI] [PubMed] [Google Scholar]
- Choi H, and Roh J. 2018. Role of Klf4 in the Regulation of Apoptosis and Cell Cycle in Rat Granulosa Cells during the Periovulatory Period. International journal of molecular sciences. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse JF, and Korach KS. 1999. Estrogen receptor null mice: what have we learned and where will they lead us? Endocrine reviews. 20:358–417. [DOI] [PubMed] [Google Scholar]
- Couse JF, Yates MM, Deroo BJ, and Korach KS. 2005a. Estrogen receptor-beta is critical to granulosa cell differentiation and the ovulatory response to gonadotropins. Endocrinology. 146:3247–3262. [DOI] [PubMed] [Google Scholar]
- Couse JF, Yates MM, Deroo BJ, and Korach KS. 2005b. Estrogen receptor-β is critical to granulosa cell differentiation and the ovulatory response to gonadotropins. Endocrinology. 146:3247–3262. [DOI] [PubMed] [Google Scholar]
- Couse JF, Yates MM, Sanford R, Nyska A, Nilson JH, and Korach KS. 2004a. Formation of cystic ovarian follicles associated with elevated luteinizing hormone requires estrogen receptor-beta. Endocrinology. 145:4693–4702. [DOI] [PubMed] [Google Scholar]
- Couse JF, Yates MM, Sanford R, Nyska A, Nilson JH, and Korach KS. 2004b. Formation of cystic ovarian follicles associated with elevated luteinizing hormone requires estrogen receptor-β. Endocrinology. 145:4693–4702. [DOI] [PubMed] [Google Scholar]
- Couse JF, Yates MM, Walker VR, and Korach KS. 2003. Characterization of the hypothalamic-pituitary-gonadal axis in estrogen receptor (ER) null mice reveals hypergonadism and endocrine sex reversal in females lacking ERα but not ERβ. Molecular Endocrinology. 17:1039–1053. [DOI] [PubMed] [Google Scholar]
- Dekel N, and Beers WH. 1978. Rat oocyte maturation in vitro: relief of cyclic AMP inhibition by gonadotropins. Proceedings of the National Academy of Sciences of the United States of America. 75:4369–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierich A, Sairam MR, Monaco L, Fimia GM, Gansmuller A, LeMeur M, and Sassone-Corsi P. 1998. Impairing follicle-stimulating hormone (FSH) signaling in vivo: targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proceedings of the National Academy of Sciences of the United States of America. 95:13612–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko E, Padua MB, Manuylov NL, Fox SC, Morse DA, and Tevosian SG. 2013. The transcription factor GATA4 is required for follicular development and normal ovarian function. Developmental biology. 381:144–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JJ 2001. Oocyte control of ovarian follicular development and function in mammals. Reproduction (Cambridge, England). 122:829–838. [DOI] [PubMed] [Google Scholar]
- Erickson GF, and Hsueh AJ. 1978. Stimulation of aromatase activity by follicle stimulating hormone in rat granulosa cells in vivo and in vitro. Endocrinology. 102:1275–1282. [DOI] [PubMed] [Google Scholar]
- Fernandois D, Na E, Cuevas F, Cruz G, Lara HE, and Paredes AH. 2016. Kisspeptin is involved in ovarian follicular development during aging in rats. The Journal of endocrinology. 228:161–170. [DOI] [PubMed] [Google Scholar]
- Fisher CR, Graves KH, Parlow AF, and Simpson ER. 1998. Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. Proceedings of the National Academy of Sciences of the United States of America. 95:6965–6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune JE, and Armstrong DT. 1977. Androgen production by theca and granulosa isolated from proestrous rat follicles. Endocrinology. 100:1341–1347. [DOI] [PubMed] [Google Scholar]
- Gershon E, and Dekel N. 2020. Newly Identified Regulators of Ovarian Folliculogenesis and Ovulation. International journal of molecular sciences. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman BD, and Mahesh VB. 1968. Fluctuations in pituitary FSH during the ovulatory cycle in the rat and a possible role of FSH in the induction of ovulation. Endocrinology. 83:97–106. [DOI] [PubMed] [Google Scholar]
- Goldman BD, and Mahesh VB. 1969. A possible role of acute FSH-release in ovulation in the hamster, as demonstrated by utilization of antibodies to LH and FSH. Endocrinology. 84:236–243. [DOI] [PubMed] [Google Scholar]
- Gómez E, Tarín JJ, and Pellicer A. 1993. Oocyte maturation in humans: the role of gonadotropins and growth factors. Fertility and sterility. 60:40–46. [PubMed] [Google Scholar]
- Greep RO, Van Dyke H, and CHOW BF. 1942. Gonadotropins of the swine pituitary I. Various biological effects of purified thylakentrin (FSH) and pure metakentrin (ICSH). Endocrinology. 30:635–649. [Google Scholar]
- Grimek HJ, Nuti LC, Nuti KM, and McShan WH. 1976. Effect of neuraminidase treatment on the biological activity of highly purified ovine FSH and LH in hypophysectomized immature male and female rats. Endocrinology. 98:105–110. [DOI] [PubMed] [Google Scholar]
- Gromoll J, Simoni M, Nordhoff V, Behre HM, De Geyter C, and Nieschlag E. 1996. Functional and clinical consequences of mutations in the FSH receptor. Molecular and cellular endocrinology. 125:177–182. [DOI] [PubMed] [Google Scholar]
- Gupta PS, Folger JK, Rajput SK, Lv L, Yao J, Ireland JJ, and Smith GW. 2014. Regulation and regulatory role of WNT signaling in potentiating FSH action during bovine dominant follicle selection. PloS one. 9:e100201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamnongjit M, Gill A, and Hammes SR. 2005. Epidermal growth factor receptor signaling is required for normal ovarian steroidogenesis and oocyte maturation. Proceedings of the National Academy of Sciences of the United States of America. 102:16257–16262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayes FL, Burns KA, Rodriguez KF, Kissling GE, and Korach KS. 2014. The naturally occurring luteinizing hormone surge is diminished in mice lacking estrogen receptor Beta in the ovary. Biology of reproduction. 90:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Komar CM, and Fortune JE. 2002. Gonadotropin surge induces two separate increases in messenger RNA for progesterone receptor in bovine preovulatory follicles. Biology of reproduction. 67:1981–1988. [DOI] [PubMed] [Google Scholar]
- Jones ASK, and Shikanov A. 2019. Follicle development as an orchestrated signaling network in a 3D organoid. Journal of biological engineering. 13:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khristi V, Chakravarthi VP, Singh P, Ghosh S, Pramanik A, Ratri A, Borosha S, Roby KF, Wolfe MW, and Rumi MAK. 2018. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Molecular and cellular endocrinology. 474:214–226. [DOI] [PubMed] [Google Scholar]
- Khristi V, Ratri A, Ghosh S, Pathak D, Borosha S, Dai E, Roy R, Chakravarthi VP, Wolfe MW, and Karim Rumi MA. 2019. Disruption of ESR1 alters the expression of genes regulating hepatic lipid and carbohydrate metabolism in male rats. Molecular and cellular endocrinology. 490:47–56. [DOI] [PubMed] [Google Scholar]
- Kubota K, Cui W, Dhakal P, Wolfe MW, Rumi MA, Vivian JL, Roby KF, and Soares MJ. 2016. Rethinking progesterone regulation of female reproductive cyclicity. Proceedings of the National Academy of Sciences of the United States of America. 113:4212–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayana B, Chakravarthi VP, Brahmaiah K, and Rao V. 2014. Quantification of P450 aromatase gene expression in cultured and in vivo grown ovarian follicles in sheep. Small Ruminant Research. 117:66–72. [Google Scholar]
- LaPOLT PS, OIKAWA M, JIA X-C, DARGAN C, and HSUEH AJ. 1990. Gonadotropin-induced up-and down-regulation of rat ovarian LH receptor message levels during follicular growth, ovulation and luteinization. Endocrinology. 126:3277–3279. [DOI] [PubMed] [Google Scholar]
- Laurincik J, Oberfranc M, Hyttel P, Grafenau P, Tomanek M, and Pivko J. 1993. Characterization of the periovulatory period in superovulated heifers. Theriogenology. 39:537–544. [DOI] [PubMed] [Google Scholar]
- LaVoie HA 2014. The GATA-keepers of ovarian development and folliculogenesis. Biology of reproduction. 91:38. [DOI] [PubMed] [Google Scholar]
- Lee-Thacker S, Choi Y, Taniuchi I, Takarada T, Yoneda Y, Ko C, and Jo M. 2018. Core Binding Factor β Expression in Ovarian Granulosa Cells Is Essential for Female Fertility. Endocrinology. 159:2094–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei ZM, Mishra S, Zou W, Xu B, Foltz M, Li X, and Rao CV. 2001. Targeted disruption of luteinizing hormone/human chorionic gonadotropin receptor gene. Molecular endocrinology (Baltimore, Md.). 15:184–200. [DOI] [PubMed] [Google Scholar]
- Lin TC, Chen ST, Huang MC, Huang J, Hsu CL, Juan HF, Lin HH, and Chen CH. 2017. GALNT6 expression enhances aggressive phenotypes of ovarian cancer cells by regulating EGFR activity. Oncotarget. 8:42588–42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Park ES, and Jo M. 2009. Runt-related transcription factor 1 regulates luteinized hormone-induced prostaglandin-endoperoxide synthase 2 expression in rat periovulatory granulosa cells. Endocrinology. 150:3291–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang Y, Xu C, Yang H, Chen S, and Chen H. 2019. RNA sequencing analysis of the CAL-27 cell response to over-expressed ZNF750 gene revealed an extensive regulation on cell cycle. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 118:109377. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang M, Jiang S, Lu Y, Tao D, Yang Y, Ma Y, and Zhang S. 2014. Demethylation of CpG islands in the 5' upstream regions mediates the expression of the human testis-specific gene MAGEB16 and its mouse homolog Mageb16. BMB reports. 47:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YX, and Hsueh AJ. 1986. Synergism between granulosa and theca-interstitial cells in estrogen biosynthesis by gonadotropin-treated rat ovaries: studies on the two-cell, two-gonadotropin hypothesis using steroid antisera. Biology of reproduction. 35:27–36. [DOI] [PubMed] [Google Scholar]
- LOSTROH AJ, and JOHNSON RE. 1966. Amounts of interstitial cell-stimulating hormone and follicle-stimulating hormone required for follicular development, uterine growth and ovulation in the hypophysectomized rat. Endocrinology. 79:991–996. [DOI] [PubMed] [Google Scholar]
- Lu D, Peegel H, Mosier SM, and Menon K. 1993. Loss of lutropin/human choriogonadotropin receptor messenger ribonucleic acid during ligand-induced down-regulation occurs post transcriptionally. Endocrinology. 132:235–240. [DOI] [PubMed] [Google Scholar]
- Madan P, Bridges PJ, Komar CM, Beristain AG, Rajamahendran R, Fortune JE, and MacCalman CD. 2003. Expression of messenger RNA for ADAMTS subtypes changes in the periovulatory follicle after the gonadotropin surge and during luteal development and regression in cattle. Biology of reproduction. 69:1506–1514. [DOI] [PubMed] [Google Scholar]
- Moog-Lutz C, Cavé-Riant F, Guibal FC, Breau MA, Di Gioia Y, Couraud PO, Cayre YE, Bourdoulous S, and Lutz PG. 2003. JAML, a novel protein with characteristics of a junctional adhesion molecule, is induced during differentiation of myeloid leukemia cells. Blood. 102:3371–3378. [DOI] [PubMed] [Google Scholar]
- Moudgal N 2012. Gonadotropins and gonadal function. Elsevier. [Google Scholar]
- Oktay K, Briggs D, and Gosden RG. 1997. Ontogeny of follicle-stimulating hormone receptor gene expression in isolated human ovarian follicles. The Journal of Clinical Endocrinology & Metabolism. 82:3748–3751. [DOI] [PubMed] [Google Scholar]
- Orisaka M, Tajima K, Tsang BK, and Kotsuji F. 2009. Oocyte-granulosa-theca cell interactions during preantral follicular development. Journal of ovarian research. 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Choi KB, Gilks CB, Leung PC, and Auersperg N. 2006. Cell type- and stage-specific changes in HOXA7 protein expression in human ovarian folliculogenesis: possible role of GDF-9. Differentiation. 74:1–10. [DOI] [PubMed] [Google Scholar]
- Panigone S, Hsieh M, Fu M, Persani L, and Conti M. 2008. Luteinizing hormone signaling in preovulatory follicles involves early activation of the epidermal growth factor receptor pathway. Molecular endocrinology (Baltimore, Md.). 22:924–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterni I, Granchi C, Katzenellenbogen JA, and Minutolo F. 2014. Estrogen receptors alpha (ERα) and beta (ERβ): subtype-selective ligands and clinical potential. Steroids. 90:13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G, and El-Alfy M. 2000a. Immunocytochemical localization of estrogen receptors alpha and beta in the human reproductive organs. The Journal of clinical endocrinology and metabolism. 85:4835–4840. [DOI] [PubMed] [Google Scholar]
- Pelletier G, and El-Alfy M. 2000b. Immunocytochemical localization of estrogen receptors α and β in the human reproductive organs. The Journal of Clinical Endocrinology & Metabolism. 85:4835–4840. [DOI] [PubMed] [Google Scholar]
- Peters H 1979. Some aspects of early follicular development. Ovarian follicular development and function:1–13. [Google Scholar]
- Picut CA, Dixon D, Simons ML, Stump DG, Parker GA, and Remick AK. 2015. Postnatal ovary development in the rat: morphologic study and correlation of morphology to neuroendocrine parameters. Toxicol Pathol. 43:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter LR, Abbey-Hosch S, and Dickey DM. 2006. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocrine reviews. 27:47–72. [DOI] [PubMed] [Google Scholar]
- Praveen Chakravarthi V, Kona SS, Siva Kumar AV, Bhaskar M, and Rao VH. 2015. Quantitative expression of antiapoptotic and proapoptotic genes in sheep ovarian follicles grown in vivo or cultured in vitro. Theriogenology. 83:590–595. [DOI] [PubMed] [Google Scholar]
- Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, and Kulkarni P. 2011. A majority of the cancer/testis antigens are intrinsically disordered proteins. Journal of cellular biochemistry. 112:3256–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAJANIEMI HJ, MIDGLEY JR AR, DUNCAN JA, and REICHERT JR LE. 1977. Gonadotropin receptors in rat ovarian tissue: III. Binding sites for luteinizing hormone and differentiation of granulosa cells to luteal cells. Endocrinology. 101:898–910. [DOI] [PubMed] [Google Scholar]
- Rice S, Ojha K, Whitehead S, and Mason H. 2007. Stage-specific expression of androgen receptor, follicle-stimulating hormone receptor, and anti-Müllerian hormone type II receptor in single, isolated, human preantral follicles: relevance to polycystic ovaries. The Journal of clinical endocrinology and metabolism. 92:1034–1040. [DOI] [PubMed] [Google Scholar]
- Richards JS 1980. Maturation of ovarian follicles: actions and interactions of pituitary and ovarian hormones on follicular cell differentiation. Physiological Reviews. 60:51–89. [DOI] [PubMed] [Google Scholar]
- Richards JS, and Ascoli M. 2018. Endocrine, paracrine, and autocrine signaling pathways that regulate ovulation. Trends in Endocrinology & Metabolism. 29:313–325. [DOI] [PubMed] [Google Scholar]
- Robker RL, Akison LK, and Russell DL. 2009. Control of oocyte release by progesterone receptor-regulated gene expression. Nuclear receptor signaling. 7:e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robker RL, Russell DL, Espey LL, Lydon JP, O'Malley BW, and Richards JS. 2000. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proceedings of the National Academy of Sciences of the United States of America. 97:4689–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby KF 2001. Alterations in follicle development, steroidogenesis, and gonadotropin receptor binding in a model of ovulatory blockade. Endocrinology. 142:2328–2335. [DOI] [PubMed] [Google Scholar]
- Roby KF, Son DS, Taylor CC, Montgomery-Rice V, Kirchoff J, Tang S, and Terranova PF. 2005. Alterations in reproductive function in SRC tyrosine kinase knockout mice. Endocrine. 26:169–176. [DOI] [PubMed] [Google Scholar]
- Roby KF, Son DS, and Terranova PF. 1999. Alterations of events related to ovarian function in tumor necrosis factor receptor type I knockout mice. Biology of reproduction. 61:1616–1621. [DOI] [PubMed] [Google Scholar]
- Ronen-Fuhrmann T, Timberg R, King SR, Hales KH, Hales DB, Stocco DM, and Orly J. 1998. Spatio-temporal expression patterns of steroidogenic acute regulatory protein (StAR) during follicular development in the rat ovary. Endocrinology. 139:303–315. [DOI] [PubMed] [Google Scholar]
- Rumi MAK, Singh P, Roby KF, Zhao X, Iqbal K, Ratri A, Lei T, Cui W, Borosha S, Dhakal P, Kubota K, Chakraborty D, Vivian JL, Wolfe MW, and Soares MJ. 2017. Defining the Role of Estrogen Receptor beta in the Regulation of Female Fertility. Endocrinology. 158:2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DL, Doyle KM, Ochsner SA, Sandy JD, and Richards JS. 2003. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. The Journal of biological chemistry. 278:42330–42339. [DOI] [PubMed] [Google Scholar]
- Schwartz NB 1974. The role of FSH and LH and of their antibodies on follicle growth and on ovulation. Biology of reproduction. 10:236–272. [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, and Khavari PA. 2012. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Developmental cell. 22:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shozu M, Minami N, Yokoyama H, Inoue M, Kurihara H, Matsushima K, and Kuno K. 2005. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. Journal of molecular endocrinology. 35:343–355. [DOI] [PubMed] [Google Scholar]
- Shupnik M 2002. Oestrogen receptors, receptor variants and oestrogen actions in the hypothalamic-pituitary axis. Journal of neuroendocrinology. 14:85–94. [DOI] [PubMed] [Google Scholar]
- Siddappa D, Beaulieu É, Gévry N, Roux PP, Bordignon V, and Duggavathi R. 2015. Effect of the transient pharmacological inhibition of Mapk3/1 pathway on ovulation in mice. PloS one. 10:e0119387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni M, Gromoll J, and Nieschlag E. 1997. The follicle-stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endocrine reviews. 18:739–773. [DOI] [PubMed] [Google Scholar]
- Słomczyńska M, Duda M, and Galas J. 2001a. Estrogen receptor alpha and beta expression in the porcine ovary. Folia histochemica et cytobiologica. 39:137–138. [PubMed] [Google Scholar]
- Słomczyńska M, Duda M, and Galas J. 2001b. Estrogen receptor alpha and beta expression in the porcine ovary. Folia histochemica et cytobiologica. 39:137. [PubMed] [Google Scholar]
- Wójtowicz A, Kajta M, and Gregoraszczuk E. 2007. DDT-and DDE-induced disruption of ovarian steroidogenesis in prepubertal porcine ovarian follicles: a possible interaction with the main steroidogenic enzymes and estrogen receptor beta. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society. 58:873–885. [PubMed] [Google Scholar]
- Wu Y, Xiao H, Pi J, Zhang H, Pan A, Pu Y, Liang Z, Shen J, and Du J. 2019. EGFR promotes the proliferation of quail follicular granulosa cells through the MAPK/extracellular signal-regulated kinase (ERK) signaling pathway. Cell cycle (Georgetown, Tex.). 18:2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Oakley J, and McGee EA. 2002. Stage-specific expression of Smad2 and Smad3 during folliculogenesis. Biology of reproduction. 66:1571–1578. [DOI] [PubMed] [Google Scholar]
- Yasin M, Dalkin AC, Haisenleder DJ, Kerrigan JR, and Marshall JC. 1995. Gonadotropin-releasing hormone (GnRH) pulse pattern regulates GnRH receptor gene expression: augmentation by estradiol. Endocrinology. 136:1559–1564. [DOI] [PubMed] [Google Scholar]
- Yung Y, Maman E, Konopnicki S, Cohen B, Brengauz M, Lojkin I, Dal Canto M, Fadini R, Dor J, and Hourvitz A. 2010. ADAMTS-1: a new human ovulatory gene and a cumulus marker for fertilization capacity. Molecular and cellular endocrinology. 328:104–108. [DOI] [PubMed] [Google Scholar]
- Zeleznik AJ, Midgley AR Jr., and Reichert LE Jr. 1974. Granulosa cell maturation in the rat: increased binding of human chorionic gonadotropin following treatment with follicle-stimulating hormone in vivo. Endocrinology. 95:818–825. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su YQ, Sugiura K, Xia G, and Eppig JJ. 2010. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science (New York, N.Y.). 330:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.