Abstract
Eukaryotic cell organelles exert unique functions individually but also interact with each other for essential cellular functions. This physical interface between the organelles serves as an important platform for biomolecule trafficking and signaling. Mitochondria are a membrane-bound organelles and form a dynamic contact with other organelles. The interactions and communication between mitochondria and endoplasmic reticulum (ER) are facilitated by an ER specific domain, named mitochondria associated ER membrane (MAM). Due to its unique location, the MAM is a “hotspot” for important cell signaling and biochemical processes including calcium homeostasis, lipid synthesis/exchange, inflammasome and autophagosome formation, and mitochondria fission/fusion. Although techniques are available for isolation of organelle fractions including MAM, most utilize animal tissues and cell lines. Here we describe a protocol that is tailored to the isolation of highly purified MAM, mitochondria, ER and cytosol from human brain. In addition, we include a protocol for the isolation of total RNA and subsequent analysis of microRNAs from these highly purified organelle fractions. Finally, we include a panel of protein markers that are useful for validating the enrichment and purity of each subcellular fraction.
Keywords: mitochondria associated ER membrane (MAM), subcellular fractionation, neurodegenerative diseases, human brain, microRNA, RT-qPCR
1. Introduction
Subcellular fractionation is a process of separation and purification of cellular compartments using techniques based on size, density, shape, and surface charge [1]. The isolation of the subcellular constituents provides a valuable tool for studying localization, processing, trafficking, and signaling of biomolecules and is especially useful for studying functional contacts between organelles. One such specialized inter-organelle contact point is the mitochondria-associated endoplasmic reticulum (ER) membrane (MAM), which is a specific domain that functions as a physical conduit between the ER and mitochondria. MAM facilitates a number of crucial cellular activities including Ca2+ signaling, phospholipid exchange, inflammasome and autophagosome formation, and mitochondrial morphology and redox status [2, 3, 4]. Given this unique structural microdomain and functional properties, interest in MAM biology has broadened over the past decade to include examination of various tissue and cell types encompassing several pathological conditions. Procedures for the isolation of MAM from animal tissues and cells has been established and documented in several excellent reports [5, 6]. Here, we provide a protocol that is specifically tailored to isolate highly purified MAM, mitochondria, ER, and cytosol from human and rat brain tissues. Moreover, we describe a protocol for RNA isolation and microRNA (miRNA) analysis from these purified subcellular fractions. We reported previously that several inflammatory-related miRNAs are enriched in rat hippocampal mitochondria [7, 8] and now report that the MAM is also a subcellular localization site for miRNA, implicating a role for MAM in miRNA trafficking. Moreover, we found that several miRNAs including miR-146a and miR-107 were detected in the the MAM fraction and both miRNAs have been implicated in neurodegenerative diseases [9, 10] suggesting a potential cross-talk between miRNA and MAM. While MAM dysfunction has been implicated in multiple forms of neurodegenerative disease including Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis [11, 12, 13], the role of MAM in miRNA biology associated with neurodegenerative disease states remains elusive. To study this more closely, we adapted and developed a subcellular fractionation protocol that has been employed previously in our lab for mitochondria isolation. This protocol details the fractionation of mitochondria, MAM, ER, and cytosol and the isolation and analysis of miRNAs from these fractions. Based on our own practice, we provide important remarks at several critical steps. We also include a panel of protein markers for examining the enrichment and purity of these subcellular fractions. It is our opinion that this protocol would be useful for examining miRNA distribution patterns in human and experimental models of neurodegeneration, and the protocol can be easily modified to accommodate the fractionation of other tissues.
2. Materials
2.1. Brain tissues
Human brain tissue: human brain specimens should be obtained at a short postmortem interval (PMI) less than 6 hrs. (See Note 1). Human brain specimens were obtained from the University of Kentucky Alzheimer’s Disease Center (UK-ADC). The procedures and protocols related to procurement of the human brain specimens were approved by the University of Kentucky Institutional Review Board.
Rat brain tissue: use rat brain tissue as fresh as possible.
2.2. Subcellular Isolation:
2.2.1. Equipment and supplies:
Optima Xe-90 Ultracentrifuge (Beckman Coulter)
Table-top refrigerated centrifuge with speed up to 20,000 xg
SW 41 Ti rotor (Swinging bucket, 41,000 r.p.m., 288,000 xg)
SW 55 Ti rotor (Swinging bucket, 55,000 r.p.m., 285,000 xg)
Teflon glass homogenizer
Dounce glass tissue grinder with large clearance “A” pestle
Thinwall, Ultra-Clear™, 5 mL, 13 × 51 mm for SW55 Ti rotor
Thinwall, Ultra-Clear™, 13.2 mL, 14 × 89 mm for SW41 Ti rotor
1.7 ml and 5.0 mL microcentrifuge tubes
2.2.2. Chemicals and buffer preparation
All solutions should be prepared using deionized, nuclease-free water and buffers made fresh before beginning the subcellular fractionation procedure. The pH of the buffers (pH: 7.4) should be adjusted at 4°C. Keep the buffer on ice. (See Note 2)
Homogenization Buffer (HB): 30 mM Tris–HCl pH 7.4, 225 mM mannitol, 75 mM sucrose, 0.5 mM EGTA, protease inhibitor and 0.5 % BSA. (See Note 3)
Isolation Buffer A (IB-A): 30 mM Tris–HCl pH 7.4, 225 mM mannitol, 75 mM sucrose, and 0.25 % BSA. (See Note 3)
Isolation Buffer B (IB-B): 30 mM Tris–HCl pH 7.4, 225 mM mannitol, and 75 mM sucrose. (See Note 3)
Mitochondrial Resuspending Buffer (MRB): 5 mM HEPES, pH 7.4, 250 mM mannitol, and 0.5 mM EGTA (v/v). (See Note 3)
Percoll medium (PM): 25 mM HEPES, pH 7.4, 225 mM mannitol, 1 mM EGTA, 30 % Percoll (v/v). (See Note 4)
2.3. Protein isolation and Western blot analysis:
2.3.1. Equipment:
Table-top refrigerated centrifuge
Spectrophotometer for protein concentration determination
SDS-PAGE and Western blotting assembly
2.3.2. Reagents:
RIPA buffer
Protease inhibitors
Western blotting reagents
Primary and secondary antibodies (See Table 1)
Table 1:
Enrichment of protein markers in each fraction: Very high
| Protein Marker | Pure Cytosol | Pure Mitochondria | MAM | ER | Antibody Source | Dilution | Detection |
|---|---|---|---|---|---|---|---|
| PDZD8 | −− | −− | +++++ | ++ | Gifta | 1:500 | MAMb |
| FACL4 | −− | −− | ++++ | +++++ | Abgent (ap14406A) | 1:1000 | MAM/ERc |
| CALNEXIN | −− | −− | +++++ | +++++ | Abcam (ab22595) | 1:5000 | MAM/ERd |
| IP3R | −− | −− | + | +++++ | Santa Cruz (sc-377518) | 1:500 | MAM/ERe |
| GRP75 | −− | +++++ | ++++ | ++ | Santa Cruz (sc-133137) | 1:500 | Mito/MAMf |
| NDUAF9 | −− | +++++ | + | −− | ThermoFisher Scientific (459100) | 1:5000 | Mitog |
| VDAC1 | −− | +++++ | ++ | −− | Santa Cruz (sc-390996) | 1:500 | Mito/MAMh |
| TUBULIN | +++++ | −− | ++ | ++ | Abcam (ab6046) | 1:5000 | Cyto/MAM/ERi |
| Pro-CASPASE3 | +++++ | −− | ++ | ++ | CST (9662) | 1:1000 | Cyto/MAM/ERj |
| HSP90 | +++++ | −− | ++ | ++ | Santa Cruz (sc-7947) | 1:500 | Cyto/MAM/ERk |
Very high: +++++, high: ++++, medium: +++, low: ++, very low: +, not detected: −
The PDZD8 antibody was a generous gift from Dr. Joseph Sodroski [14]
PDZD8 (PDZ domain-containing protein 8) is a key protein tethering ER and mitochondria and regulates Ca2+ dynamics in neurons. PDZD8 is predominately enriched in MAM fractions and can be detected in ER
FASL4 (Fatty acid-CoA ligase 4 family protein) is a lipid metabolism enzyme, enriched in ER and MAM
Calnexin is an ER molecular chaperone. It can be detected in both ER and MAM fractions
IRP3 (trisphosphate receptor) is an ER Ca2+ channel protein. IRP3 is detected mostly in ER fractions
GRP75 (glucose-regulated protein 75) is a member of the mitochondrial HSP70 family and plays a key role in regulating Ca2+ transfer between ER and mitochondria. GRP75 can be detected in both mitochondria and MAM fractions
NDUFA9 (NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9) is a subunit of mitochondrial complex I located in the mitochondrial inner membrane. NDUFA9 is detected predominately in mitochondria fractions
VDAC1 (voltage-dependent anion channel 1) plays important role in exchange of ions and metabolites and is thought to be in direct contact with MAM. VDAC is located in the mitochondrial outer membrane and is often detected in both mitochondria and MAM
Tubulin is the major component of eukaryotic cytoskeleton and is widely used as cytosolic protein marker. However, given its role as a structural protein, low levels can be found in MAM and ER fractions
Pro-caspase 3 can be detected in both cytosol and MAM
HSP90 is a molecular chaperone expressed most abundantly in the cytoplasm. Trace amounts and can also be detected in MAM and ER
2.4. RNA isolation and miRNA Analysis:
2.4.1. Equipment:
Table-top refrigerated centrifuge
Nanodrop or equivalent equipment for quantifying total RNA levels following extraction
QuantStudio 7 Flex Real-Time PCR System or equivalent equipment
Biometra TAdvanced Thermocycler or equivalent with a fast ramping rate ≥ 5 °C/s
2.4.2. Reagents:
Trizole LS
20 μg/μl Glycogen
RNasin® Plus RNase Inhibitor or similar products
TaqMan™ MicroRNA Reverse Transcription Kit (See Note 5)
Megaplex™ RT Primers
TaqMan Custom microRNA Low Density Array Card
TaqMan™ MicroRNA Assay with RT/PCR primers
2✕ TaqMan® Universal Master Mix II, No AmpErase® UNG
2✕ TaqMan PreAmp Master Mix
25 mM MgCl2
Custom RT primer pool
3. Methods
3.1. Tissue Preparation
Human brain tissue: immerse tissue immediately in ice-cold IB-B with protease inhibitors and keep the tube containing specimen in ice. Remove meninges and visible blood vessels, and separate gray matter from most of white matter using a razor blade. (See Note 6)
Rat brain tissue: separate cortex from hippocampus and cerebellum and remove meninges and visible blood vessels. (See Note 7)
3.2. Homogenization and initial fractionations
Transfer 0.5 g of human brain tissue or one hemisphere rat cortex to a 10-ml Teflon glass homogenizer.
Homogenize tissue in 3 ml HB using a Teflon pestle for 8–10 strokes. (See Note 8)
Transfer homogenate to a 5 ml Eppendorf centrifuge tube.
Centrifuge homogenate at 630 xg for 5 min at 4 °C.
Collect supernatant in a new 5 ml tube. Resuspend pellet in 3 ml HB and repeat step 2–4.
Collect and combine supernatant from previous centrifugation (the volume of the two combined supernatants should be around 5 ml). Discard the pellet containing unbroken cell debris and nuclei.
Centrifuge the combined supernatant again at 630 xg for 5 min at 4 °C and transfer supernatant to a new tube.
Repeat centrifugation step as in step 7 and collect supernatant to a new tube. Save 350 μl supernatant into a 1.5 ml Eppendorf tube as nuclei-free lysate (Lysate).
Centrifuge the remaining nuclei-free lysate at 6300 xg for 10 min at 4 °C.
Save the supernatant as crude cytosol in a new 5 ml Eppendorf tube and keep on ice for further fractionation in section 3.2.4. (This crude cytosol contains ER and various multivesicular particles such as lysosome, microsomes and other cytosolic granules). The pellet contains crude mitochondria and MAM and proceed to Section 3.2.1 Mitochondria and MAM Separation for further fractionation.
3.2.1. Mitochondria and MAM Separation:
Add 2 ml ice-cold IB-A buffer to the crude mitochondria/MAM pellet (from step10 of Section 3.2 above) and gently transfer the pellet to a 7-ml glass Dounce tissue grinder using a 1.0 ml pipette tip that has been cut to obtain a larger opening. Apply 3–5 strokes using a loose, large clearance “A” pestle to resuspend the mitochondria/MAM pellet. (See Note 9)
Transfer the mitochondria/MAM resuspension to a fresh 5 ml Eppendorf tube and top the tube with 3 ml IB-A buffer.
Centrifuge the tube at 6300 xg for 8 min at 4 °C. Discard the supernatant after centrifugation.
Repeat step 1 to 3 using IB-B and gentle transfer of the pellet to the homogenizer.
Repeat Step 1 using 2 ml of MRB and keep resuspension on ice (Important Step: make sure the mitochondria/MAM pellet is well resuspended, if visible particles are seen, apply additional strokes)
Load 8 ml of PM into a 13.2-ml ultracentrifuge tube (See Note 10). Carefully layer the crude mitochondria/MAM suspended in MRB from Step 5 over PM solution without disturbing PM layer. (See Note 11)
Gently top the mitochondria/MAM layer with additional 1.5 ml MRB (Fig. 1). (Be sure to balance the tubes with MRB)
Centrifuge at 95,000 xg for 45 min at 4 °C using SW41 rotor (or similar rotor that is compatible with the tubes) in a ultracentrifuge. At the end of the centrifugation, two clear bands will be seen. The upper band is a wider, white to light gray band contains a mixture of MAM, fragmented mitochondria, and may also contain some larger multivascular bodies (MVBs). The lower band is close to the bottom of the percoll gradient which contains intact mitochondria (See Fig. 1).
Figure 1.
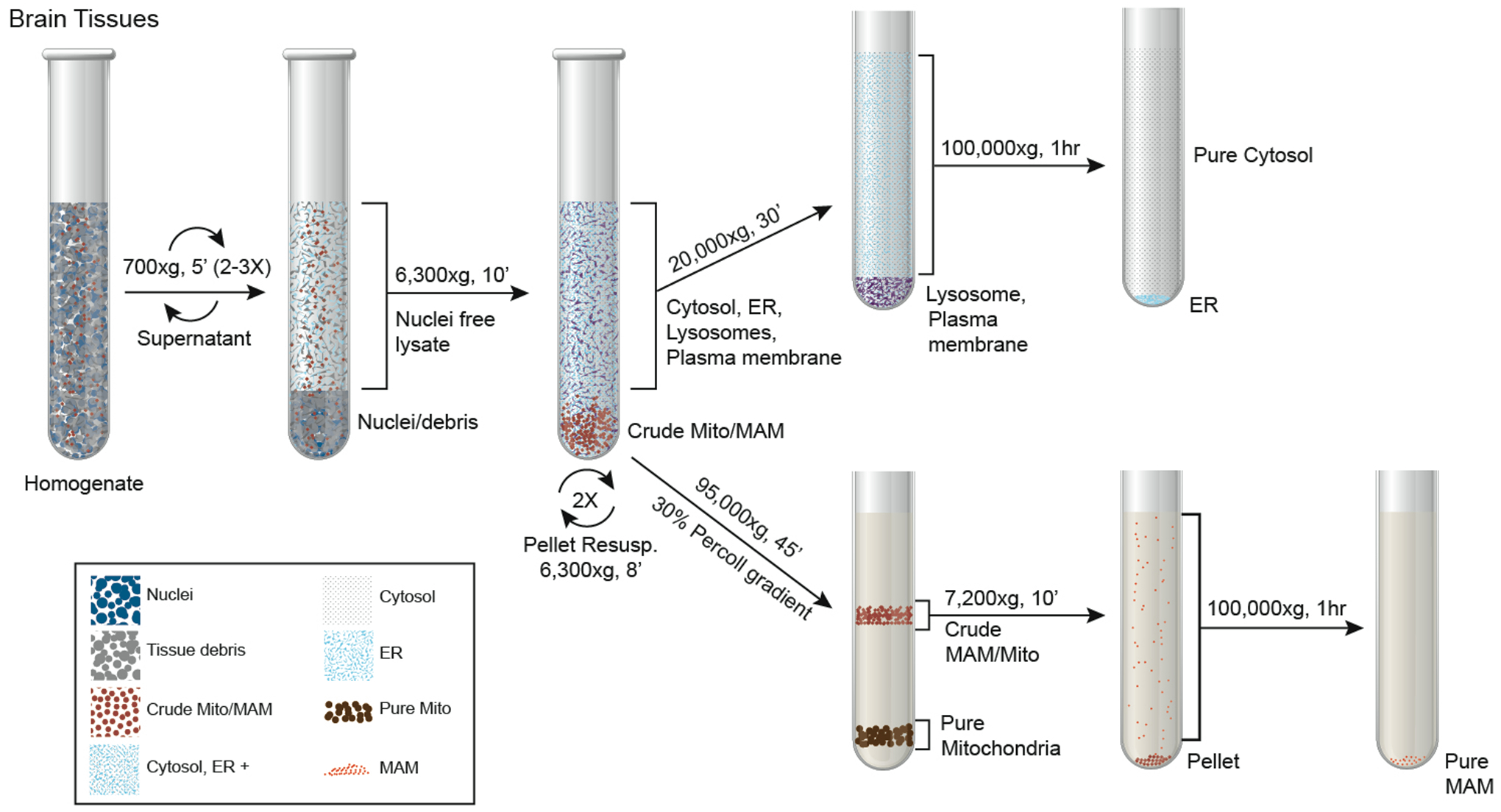
Schematic flowchart of subcellular fractionation procedure.
3.2.2. MAM Isolation:
Use a 1-ml pipette tip to collect the upper layer from step 8 in Section 3.2.1 (See Fig. 1) and dilute the collected solution 10 times with MRB in a 13.2-ml ultracentrifuge tube. (See Note 12)
Centrifuge the tube at 7200 xg for 10 min at 4 °C.
Transfer the supernatant (containing MAM fraction) into a new 13.2-ml ultracentrifuge and centrifuge at 100,000 xg for 60 min at 4 °C. After centrifugation, the purified MAM fraction can be seen as a floating sheet at the bottom of the tube.
Use a 1-ml pipette tip to carefully collect the MAM sheets and transfer to a 1.5-ml Eppendorf tube and top with MRB.
Centrifuge at top speed in a bench-top refrigerated centrifuge for 10 min at 4 °C.
Resuspend MAM pellet (MAM) in 200 μl RIPA lysis buffer. (See Note 13)
Use 100 μl of MAM lysis for RNA isolation and the remaining 100 μl for protein analysis.
3.2.3. Pure Mitochondria Isolation:
Collect the lower mitochondria band from step 8 in Section 3.2.1 (See Figure 1) and dilute the collected solution 10 times with MRB in a 13.2-ml ultracentrifuge tube. (See Note 12)
Centrifuge tube at 10,000 xg for 10 min at 4 °C.
Discard supernatant and resuspend the pellet containing purified mitochondria (pMito) in 200 μl RIPA lysis buffer. (See Note 13)
Use 100 μl of lysed mitochondria for RNA isolation and the remaining 100 μl for protein analysis.
3.2.4. ER Isolation:
Centrifuge crude cytosolic supernatant (from step 10 in Section 3.2) at 20,000 xg for 30 min at 4 °C in a bench-top refrigerated centrifuge.
Transfer supernatant into a new 5-ml ultracentrifuge tube and centrifuge at 100,000 xg for 60 min at 4 °C. This centrifugation step separates ER from cytosol.
Transfer supernatant into a new tube, this is the pure cytosolic fraction (pCyto). The resulting purified ER pellet (ER) is gently rinsed in-tube 3 times with MRB and then resuspended in 200 μl RIPA lysis buffer. (See Note 13)
Collect 275 μl pure cytosol for RNA isolation and 100 μl for protein analysis.
Collect 100 μl of ER lysis for RNA isolation and 100 μl for protein analysis. (See Note 13)
3.3. Protein isolation, quantification, and Western blot analysis:
Purified organelle fractions lysed in RIPA buffer (or other buffer solution) are incubated in ice for 20 min before subjected to centrifugation at 10,000 xg for 10 min at 4 °C.
Collect supernatants from each fraction.
Determine protein concentrations using BCA kit or other methodology of choice.
Add Laemmli sample buffer containing reducing agent (e.g. β–Mecaptoenothol or DTT) and boil the samples for 5 min. Aliquot and store the samples at −20 °C if not processed immediately.
To determine the enrichment/purity of each fraction, run Western blot analysis using antibodies against organelle selective protein markers suggested in Table 1. (Fig. 2a & b)
Figure 2. Western blot analysis of subcellular marker proteins in each organelle fraction.

Several conventionally recognized organelle marker proteins were used to determine the purity of the various subcellular fractions. A highly purified fraction is characterized by 1) enrichment of the corresponding organelle marker in that fraction and 2) the relative absence of markers associated with other organelles. The Western blots presented here represent a typical distribution of marker proteins in human brain cortical tissue (a) and rat brain cortical tissue (b).
3.4. RNA isolation and miRNA analysis (See Note 14)
3.4.1. RNA isolation
Add additional RIPA buffer to aliquots of purified MAM, mitochondria, ER, and cytosol to a final volume of 275 μl in 1.5-ml Eppendorf tube.
Add 750 μl TRIzol™ LS to the tube.
Cap the tube, mix the contents vigorously for 15 seconds and incubate for 5 min at room temperature.
After a quick spin (See Note 15), add 0.2 ml of chloroform, cap tube, and mix vigorously for 15 seconds followed by incubation for 5 min at room temperature.
Spin the tube at 10,000 xg for 30 sec at room temperature.
Transfer 750 μl of Trizol-sample mixture from the upper phase to a fresh tube avoiding the interface. (See Note 16)
Centrifuge the tube at 12,000 xg for 15 min at 4 °C
Collect 450 μl of the colorless upper aqueous phase into a new tube containing 2 μl glycogen (20 μg/μl). (See Note 17)
Vortex to mix followed by a quick spin, then add 450 μl isopropanol, mix well and leave overnight at −20°C. (See Note 18)
Centrifuge the tube at 12,000 xg for 15 min at 4 °C to pellet the precipitated RNA.
Carefully remove supernatant without disturbing RNA pellet.
Add 1 ml of 75% ethanol to rinse the RNA pellet.
Centrifuge tube at 10,000 xg for 5 min at 4 °C.
Decant off the supernatant and give the tube a quick spin.
Remove any remaining supernatant using a 200-μl pipette tip.
Briefly air-dry RNA pellet (See Note 19) and dissolve in 20–50 μl nuclease-free water (See Note 20) containing RNasin (0.5 Unit/μl). Set RNA in ice for several minute, then with brief vortex and short spin to obtain homogenous RNA solution.
Determine RNA concentration using NanoDrop Spectrophotometer (NanoDrop Technologies, Inc.) or any other method of choice.
Aliquot and store RNA samples at −80°C or proceed to next step.
3.4.2.
Single tube (3.5) or Low Density Array (3.6) reverse transcription (RT) for miRNAs using TaqMan® MicroRNA Reverse Transcription Kit.
3.5. Single tube RT
Normalize total RNA concentration to 10 ng/μl for all samples.
- Calculate and prepare RT master mixture for a 15-μl total reaction volume per sample (12 μl of reaction mixture + 3 μl RNA):
For 1 reaction: Nuclease-free water 6.16 μl 10✕ Reverse Transcription Buffer 1.50 μl RNase Inhibitor, 20 U/μl 0.19 μl 100mM dNTPs (with dTTP) 0.15 μl 5X RT Primer 3.00 μl MultiScribe™ Reverse Transcriptase, 50 U/μl 1.00 μl Aliquot 12 μl of reaction mixture to PCR reaction tubes or PCR plate.
Add 3 μl RNA to the reaction mixture.
Mix the content gently and briefly spin tube to bring down the solution to the bottom of the tube.
Incubate the tube in ice for 5 min.
- Perform RT reaction in a thermocycler using following program:
- Hold 30 minutes at 16 °C
- Hold 30 minutes at 42 °C
- Hold 5 minutes at 85 °C
- Hold at 4 °C or proceed to preamplification
3.6. TaqMan® Low Density Array RT using either Megaplex™ RT Primer pools or custom RT primer pools (See Note 21)
Normalize total RNA concentration to 10 ng/μl for all samples to be analyzed.
- Calculate and prepare RT master mixture for a 7.5-μl total reaction volume per sample (4.5 μl of reaction mixture + 3 μl RNA):
For 1 reaction: Nuclease-free water 0.20 μl 10✕ Reverse Transcription Buffer 0.80 μl RNase Inhibitor, 20 U/μl 0.10 μl 100mM dNTPs (with dTTP) 0.20 μl MgCl2 (25 mM) 0.90 μl Megaplex™ RT Primers (10✕) or custom RT primer pool 0.80 μl MultiScribe™ Reverse Transcriptase, 50 U/μl 1.50 μl Aliquot 4.5 μl reaction mixture to PCR reaction tubes or PCR plate.
Add 3 μl RNA to the reaction mixture.
Gently mix the contents and spin tube to bring down solution to the bottom of the tube.
Incubate the tube on ice for 5 min.
- Perform RT reaction in a thermocycler using following program:
- Hold 2 minutes at 16 °C
- Hold 2 minutes at 42 °C
-
Hold 1 second at 50 °C-Run 40 cycles of step 1–3
- Hold 5 minutes at 85 °C
- Hold at 4 °C or proceed to preamplification
Add 7.5 μl of nuclease-free water to 7.5 μl RT products (1:2 dilution) for preamplification or the sample can be further diluted for qPCR.
3.7. Pre-amplification
- Prepare preamplification master mixture for a 10-μl total reaction volume per sample (8 μl of reaction mixture + 2 μl RT product):
For 1 reaction: Nuclease-free water 1.50 μl 2✕ TaqMan PreAmp Master Mix 5.00 μl PreAmp Primers (See Note 21) 1.50 μl Aliquot 8 μl of reaction mixture to PCR reaction tubes or PCR plate.
Add 2 μl 2X diluted RT product to the reaction mixture.
Mix the contents and briefly spin the tube to bring down the solution to the bottom of the tube.
- Perform PreAmp reaction in a thermocycler using following program:
- Hold 10 minutes at 95 °C
- Hold 2 minutes at 55 °C
- Hold 2 minutes at 72 °C
- Hold 15 second at 95 °C
-
Hold 4 minutes at 60 °C-Run 12 cycles of step 4–5
- Hold 10 minutes at 99.9 °C
- Hold at 4 °C or proceed to qPCR
Add 30 μl of 0.1X TE buffer to 10 μl PreAmp products (1:4 dilution) for array cards. For single tube assays, PreAmp will be further diluted at least 10 times (final dilution for single tube assay is 1:40 or greater).
3.8. Real time PCR
3.8.1.1. Single tube TaqMan® assay
- Prepare PCR master mixture for a 10-μl total reaction volume per sample (7 μl of reaction mixture + 3 μl diluted PreAmp product or diluted RT product):
For 1 reaction: Nuclease-free water 2.50 μl 2✕ TaqMan® Universal Master Mix II, (No AmpErase® UNG) 5.00 μl 20✕ TaqMan® MicroRNA Assays 0.50 μl Aliquot 7 μl to PCR reaction plate or tubes.
Add 3 μl 40X diluted PreAmp product or diluted RT product to each designated well or tube.
Seal the plate with MicroAmp Optical Adhesive Film or cap the tubes.
Spin the plate or tube briefly.
- Perform qPCR reaction in QuantStudio 7 Flex Real-Time PCR System or equivalent equipment using following program (see Fig. 3 for representative amplification plot):
- Hold 10 minutes at 95 °C
- Hold 15 seconds at 95 °C
-
Hold 1 minutes at 60 °C-Run 40 cycles of step 2–3
- Hold at 4 °C
Figure 3. RT-qPCR detection of miRNAs in subcellular fractions.
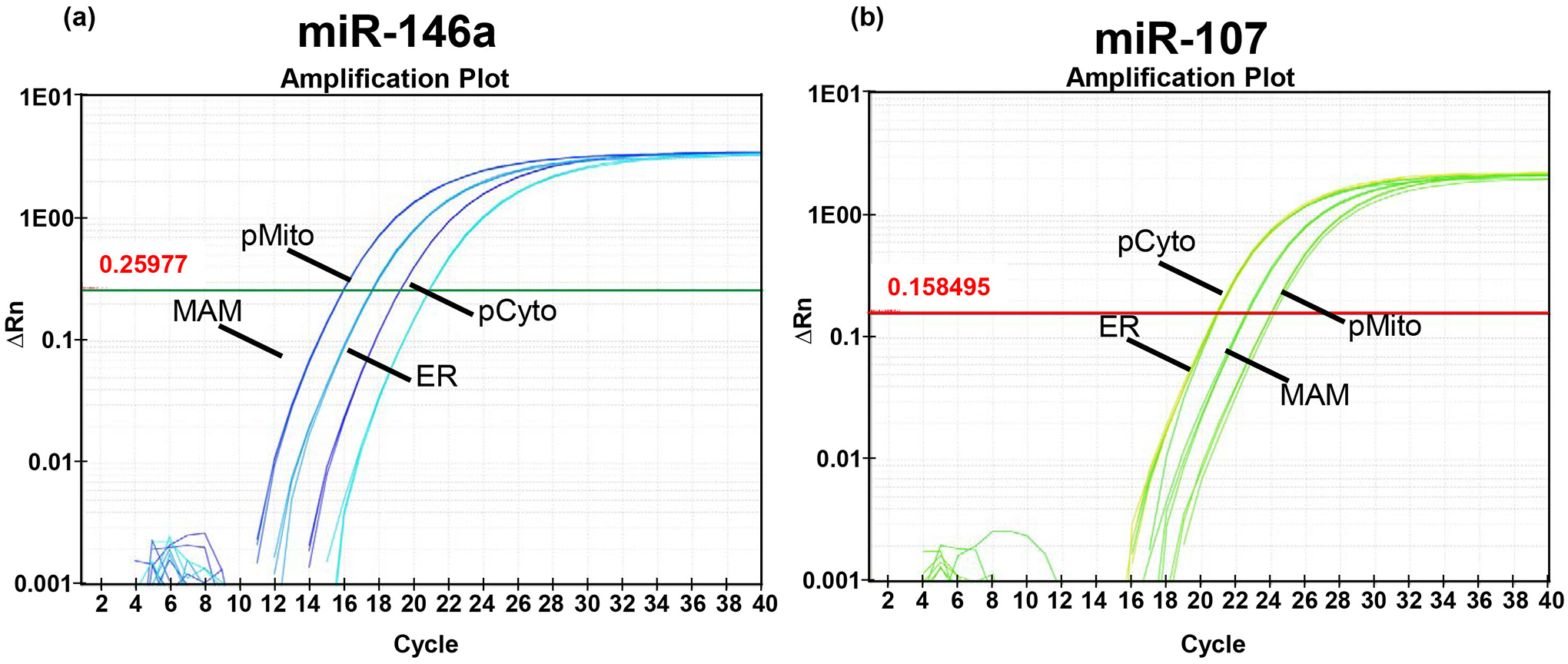
TaqMan® microRNA assay with preamplification is a highly sensitive method for detection of miRNAs from a relatively small quantity of starting total RNA (e.g. from mitochondria and MAM). The amplification plots presented in this figure demonstrate robust detection of miR-146a and miR-107 using 30 ng of total RNA isolated from purified mitochondria, MAM, ER, and cytosol fractions obtained from a 0.5 g human brain tissue sample.
3.8.2. TaqMan Custom microRNA Low Density Array:
- Prepare PCR master mixture for a 100-μl total reaction volume per sample (90 μl of reaction mixture + 10 μl diluted PreAmp):
For 1 reaction: Nuclease-free water 40 μl 2✕ TaqMan® Universal Master Mix II, (No AmpErase® UNG) 50 μl Aliquot 90 μl to reaction mixture to each tube.
Add 10 μl 4X diluted PreAmp product to each designated tube.
Cap the tube and mix by inversion several times, briefly spin tube.
Pipet 95 μl of the sample/reaction mixture to the port of the array card.
Centrifuge the array card and seal the plate.
- Perform qPCR reaction in QuantStudio 7 Flex Real-Time PCR System using following program:
- Hold 2 minutes at 50 °C
- Hold 10 minute at 95 °C
- Hold 15 seconds at 95 °C
-
Hold 1 minutes at 60 °C-Run 40 cycles of step 2–3
4. Notes
We observed that the fractionation yield is reduced when PMI is more than 8 hrs.
Prepare stock solutions of 0.5 M EGTA pH 7.4, 1 M Tris-HCl pH 7.4, 1M HEPES pH 7.4 ahead of time and stored at 4 °C.
This buffer should be prepared fresh before each experiment. The pH should be adjusted at 4 °C. Keep the buffer on ice.
Make PM immediately before the Percoll gradient step. Mix well before dispensing into the ultracentrifuge tubes to avoid forming regions with uneven PM concentrations.
We use TaqMan™ microRNA detection system, but it can be any other platform of analysis once total RNA is isolated.
Gray matter of human neocortex was used in this protocol.
Subcellular fractionation of rat cortex requires one hemisphere, while fractionation of rat hippocampus requires hippocampi from both hemispheres.
Pre-cool all glassware including homogenizer and pestle.
To avoid shearing, cut the tip of a 1.0 ml pipette tip (~0.5 cm from the tip) to transfer the mitochondria/MAM pellet into homogenizer. Thoroughly resuspend the pellet using a gentle motion with a loose fitting glass homogenizer/pestle.
Mix PM well before transfer into ultracentrifuge tube to avoid forming regions with uneven concentrations.
Avoid any movements that could disrupt layers.
A total of 1–1.2 ml may be collected.
Use of other buffers depends on the type of experiments and analysis to be conducted. For example, use buffers containing a less stringent detergent for the purpose of co-immunoprecipitation experiments.
Any RNA isolation method may be employed as long as good RNA quantity and quality are obtained. Likewise, other platforms can be used for miRNA analysis.
When using Trizol and chloroform, it is very important to minimize the carry-over of these organic solvents, which will interfere with downstream analysis. A quick spin can bring down the residue solution that may be on the cap and/or edges of the tube.
This 750 μl volume will contain most of aqueous phase with some lower pink Trizol solution.
Due to the small quantity of total RNA from purified mitochondria and MAM, it is essential to use a nucleic acid carrier (such as glycogen or bacteriophage MS2 RNA) to obtain a better total RNA yield. We use glycogen because it significantly enhances visibility of the RNA pellet.
In our hands, miRNAs are better precipitated with an overnight incubation.
The pellet usually dries within a couple minutes and care should be taken not to allow it to be over-dried.
In our experience, a volume of 20 μl is sufficient to dissolve pure mitochondria and MAM RNA pellets and 50 μl for ER RNA pellet obtained from 0.5 g of brain tissue.
Multiple miRNA RT and PreAmp primers can be pooled in a single RT reaction for a more efficient miRNA analysis procedure. For the protocol using custom pooled primers, see: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_094060.pdf
Acknowledgements
This work is supported by a grant from the Kentucky Spinal Cord and Head Injury Research Trust. The authors would like to thank Dr. Joseph Sodroski for the generous gift of the PDZD8 antibody.
References
- 1.Graham JM (2015) Fractionation of Subcellular Organelles. Curr Protoc Cell Biol 69:3 1 1–22 [DOI] [PubMed] [Google Scholar]
- 2.Janikiewicz J, Szymanski J, Malinska D et al. (2018) Mitochondria-associated membranes in aging and senescence: structure, function, and dynamics. Cell Death Dis 9:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchi S, Bittremieux M, Missiroli S et al. (2017) Endoplasmic Reticulum-Mitochondria Communication Through Ca(2+) Signaling: The Importance of Mitochondria-Associated Membranes (MAMs). Adv Exp Med Biol 997:49–67 [DOI] [PubMed] [Google Scholar]
- 4.Van Vliet AR, Verfaillie T, Agostinis P (2014) New functions of mitochondria associated membranes in cellular signaling. Biochim Biophys Acta 1843:2253–2262 [DOI] [PubMed] [Google Scholar]
- 5.Wieckowski MR, Giorgi C, Lebiedzinska M et al. (2009) Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc 4:1582–1590 [DOI] [PubMed] [Google Scholar]
- 6.Williamson CD, Wong DS, Bozidis P et al. (2015) Isolation of Endoplasmic Reticulum, Mitochondria, and Mitochondria-Associated Membrane and Detergent Resistant Membrane Fractions from Transfected Cells and from Human Cytomegalovirus-Infected Primary Fibroblasts. Curr Protoc Cell Biol 68:3 27 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang WX, Springer JE (2015) Role of mitochondria in regulating microRNA activity and its relevance to the central nervous system. Neural Regen Res 10:1026–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang WX, Visavadiya NP, Pandya JD et al. (2015) Mitochondria-associated microRNAs in rat hippocampus following traumatic brain injury. Exp Neurol 265:84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu NK, Xu XM (2011) MicroRNA in central nervous system trauma and degenerative disorders. Physiol Genomics 43:571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson PT, Wang WX, Rajeev BW (2008) MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol 18:130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paillusson S, Stoica R, Gomez-Suaga P et al. (2016) There’s Something Wrong with my MAM; the ER-Mitochondria Axis and Neurodegenerative Diseases. Trends Neurosci 39:146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Arribas M, Yakhine-Diop SMS, Pedro JMB et al. (2017) Mitochondria-Associated Membranes (MAMs): Overview and Its Role in Parkinson’s Disease. Mol Neurobiol 54:6287–6303 [DOI] [PubMed] [Google Scholar]
- 13.Vance JE (2014) MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta 1841:595–609 [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Sodroski J (2015) Efficient human immunodeficiency virus (HIV-1) infection of cells lacking PDZD8. Virology 481:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]


