ABSTRACT
The maternal epigenome may be responsive to prenatal metals exposures. We tested whether metals are associated with concurrent differential maternal whole blood DNA methylation. In the Early Autism Risk Longitudinal Investigation cohort, we measured first or second trimester maternal blood metals concentrations (cadmium, lead, mercury, manganese, and selenium) using inductively coupled plasma mass spectrometry. DNA methylation in maternal whole blood was measured on the Illumina 450 K array. A subset sample of 97 women had both measures available for analysis, all of whom did not report smoking during pregnancy. Linear regression was used to test for site-specific associations between individual metals and DNA methylation, adjusting for cell type composition and confounding variables. Discovery gene ontology analysis was conducted on the top 1,000 sites associated with each metal. We observed hypermethylation at 11 DNA methylation sites associated with lead (FDR False Discovery Rate q-value <0.1), near the genes CYP24A1, ASCL2, FAT1, SNX31, NKX6-2, LRC4C, BMP7, HOXC11, PCDH7, ZSCAN18, and VIPR2. Lead-associated sites were enriched (FDR q-value <0.1) for the pathways cell adhesion, nervous system development, and calcium ion binding. Manganese was associated with hypermethylation at four DNA methylation sites (FDR q-value <0.1), one of which was near the gene ARID2. Manganese-associated sites were enriched for cellular metabolism pathways (FDR q-value<0.1). Effect estimates for DNA methylation sites associated (p < 0.05) with cadmium, lead, and manganese were highly correlated (Pearson ρ > 0.86). DNA methylation sites associated with lead and manganese may be potential biomarkers of exposure or implicate downstream gene pathways.
KEYWORDS: Maternal epigenome, DNA methylation, trace metals
Introduction
Human exposure to trace metals occurs through various pathways, including contamination of water, soil, and food sources, in addition to ambient air [1–5]. Prenatal exposures to toxic metals such as lead (Pb), cadmium (Cd), and mercury (Hg) have been associated with several pregnancy outcomes, including preeclampsia, preterm birth, newborn weight and size, and changes in child neurodevelopment [6–9]. Essential metals, such as manganese (Mn) and selenium (Se), have important contributions to human biology and are co-factors for physiological processes such as gene expression, enzymatic activity, and cellular respiration [10,11]. However, both deficiencies and excess concentrations of Mn and Se can cause imbalances in these physiological processes [9,11–13]. Furthermore, pregnant women and the developing foetus have increased vulnerability to the toxic effects of trace metals. Some of the overlapping toxicological mechanisms by which trace metals can damage target tissues include interfering with cellular redox conditions, altering gene expression, damaging DNA, and immune system modulation [14–18]. Improved knowledge of biomarkers of exposure and downstream cellular responses will significantly advance understanding of the consequences of trace metals exposure during pregnancy.
DNA methylation is an epigenetic mechanism that contributes to the regulation of gene expression, and changes in the maternal DNA methylome can provide insight on health conditions during pregnancy [19]. Upon exposure, trace metals can influence changes in DNA methylation within circulating immune cells by interfering with enzymes involved in DNA methylation such as DNA methyl transferase, or its substrate S-adenosylmethionine [20]. Additionally, trace metals can alter DNA methylation by interfering with cellular redox conditions and gene expression. Ultimately, these DNA methylation changes can serve as a biomarker of trace metals exposure and potentially implicate relevant downstream biological processes and diseases. This may be particularly useful and important for pregnancy exposures to metals and early life child development.
Altered DNA methylation has been observed in cord blood in association with Cd [8,21–23], Pb [24–26], and Hg [27–30]. DNA methylation changes have also been observed in placental tissue in association with each of these toxic metals, in addition to Mn [31–36]. Altogether, these studies have identified numerous individual CpG sites near various genes. A consistent observation is the presence of associations of these metals with CpG sites near genes relevant to neurodevelopmental processes and immune responses.
Pregnancy is a particularly vulnerable period for the mother as well as the developing foetus. Currently, no studies have tested associations between maternal exposure to the aforementioned trace metals and maternal whole blood DNA methylation patterns early in pregnancy. In the present study, we conducted an epigenome-wide association study (EWAS) with the main goal of estimating differences in DNA methylation patterns in maternal whole blood associated with trace metals exposure. Through an EWAS approach, we investigated multiple methylation sites across the genome and gathered not only site-specific information, but also aggregate information from multiple sites to elucidate biological pathways. Several animal, in vitro, and human observational studies have observed altered immune signalling molecules, such as cytokines and chemokines, in association with exposure to toxic and essential trace metals, and this process has been hypothesized to occur partly through oxidative stress [16]. Based on this evidence, an a priori hypothesis underlining this present study is that immune cell profiles and corresponding DNA methylation profiles can be shifted beyond baseline proportions in response to maternal exposures to toxic and essential trace metals. By estimating these associations, we can inform potential antecedent mechanisms to adverse pregnancy outcomes and impaired foetal development.
Methods
Study sample
The present study was conducted on a subsample of the Early Autism Risk Longitudinal Investigation (EARLI) pregnancy cohort. Pregnant women recruited into EARLI were over the age of 18 and had previously given birth to a child with a confirmed diagnosis of autism spectrum disorder. Additional inclusion criteria pertained to the ability to communicate in English and being within 28 weeks of gestation at the time of enrolment. Recruitment occurred at four different study sites in the U.S. (Philadelphia, Baltimore, San Francisco/Oakland, and Sacramento). The institutional review boards (IRB) at organizations in the four study sites (Drexel University, Johns Hopkins University, University of California, Davis, and Kaiser Permanente Research) approved the EARLI study. The University of Michigan IRB also approved additional sample analyses. At the initial study visit we collected demographic and health information.
Trace metals measurement
Participants provided biological samples at two study visits during pregnancy. Maternal venous blood samples were collected in trace metal-free EDTA tubes. All samples were transported to the Johns Hopkins Biological Repository for storage at −80°C, or in liquid nitrogen at −120°C. Whole blood samples (n = 215) collected at the first study visit (in the first or second trimester) were selected for analysis of the non-essential trace metals Cd, Pb, and total Hg, and the essential trace metals Mn and Se. Among the entire set of blood samples, 101 samples were ineligible for quantification due to blood micro-clotting. A set of 114 samples was available for analysis. Cd, Mn, Pb, and Se concentrations were quantified using inductively coupled dynamic reaction cell plasma mass spectrometry (ELAN DRC II, PerkinElmer Norwalk, CT) (method DLS 3016.8, Centers for Disease Control and Prevention [CDC]). Total Hg concentration was quantified using triple spike isotope dilution gas chromatography and inductively coupled plasma dynamic reaction cell mass spectrometry (ELAN DRC II, PerkinElmer Norwalk, CT) (method DLS 3020.8, CDC). Bench quality control (QC) materials were characterized by at least 20 analytical runs and used to determine appropriate QC parameters, including methodological imprecision. For subsequent sample analyses, bench QC materials are included in the beginning and end of each analytical run. Standard Reference materials are from the National Institute of Standards and Technology (NIST) (SRM 955 c Levels 1–4) and used to verify method accuracy. The test system is also calibrated as part of the protocol for each analytical run with NIST-traceable calibration standards. The limit of detection (LOD) for each quantified trace metal is equivalent to LODs reported in the National Health and Nutrition Examination Survey administered by the CDC: Cd (0.1 μg/L), Mn (0.99 μg/L), Pb (0.07 μg/dL), Se (24.5 μg/L), and total Hg (0.28 μg/L).
DNA methylation measurement
An additional maternal venous blood sample was collected in standard EDTA tubes at the same time as those assayed for trace metals. Maternal venous blood samples contain platelets, lymphocytes, and erythrocytes, though only lymphocytes contain DNA for DNA methylation measures. We used the DNA Blood Midi kit (Qiagen, Valencia, CA) to extract genomic DNA from whole blood. Extraction was performed on the QIAsymphony automated workstation using the Blood 1000 protocol. Upon extraction, DNA was quantified using the Nanodrop (ThermoFisher Waltham, MA), and normalized DNA aliquots were transferred to the Johns Hopkins SNP Center/Center for Inherited Disease Research (Johns Hopkins University). For each participant, we bisulphite-treated 1 μg of DNA samples and applied a cleaning step using the EZ DNA methylation gold kit (Zymo Resaerch, Irvine, CA) according to manufacturer’s instructions. We randomly plated DNA samples and assayed for methylation using the Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA) (Bibikova et al., 2011). In the assay, we also incorporated methylation control gradients and between-plate repeated tissue controls. DNA methylation was quantified into β-values, which represents the proportion of methylation at a given CpG site.
Bioinformatic and statistical approach
We used R statistical software (version 3.3) to conduct statistical analyses. Raw Illumina image files were background fluorescence corrected using the minfi (version 1.22.1) Bioconductor package [37]. The methylation matrix was further corrected using the normal-exponential out-of-band (noob) function (Triche et al., 2013). We removed probes with failed detection p-value (>0.01) in >10% of samples (n = 635 probes) and cross-reactive probes (n = 29,154 probes) [38]. We also checked for samples with low overall array intensity (<10.5 relative fluorescence units) or samples that had over 20% of probes with failed detection p-value, and there were no samples that fit these criteria. Because the EARLI cohort was initially developed to investigate DNA methylation in biological samples from mothers, fathers, and cord blood samples, array probes located on sex chromosomes were excluded from downstream analyses during initial data pre-processing steps (n = 7,895 probes). We then applied the gaphunter function from the minfi package to remove probes with a gap in beta signal ≥ 0.05 (n = 60,725 probes). To prioritize DNA methylation sites with the greatest biologically relevant variance in per cent methylation, probes in the bottom 10% of standard deviation of per cent methylation were removed (n = 38,665 probes), resulting in a final methylation matrix of 348,438 probes. From the maternal blood samples, we applied a prediction algorithm to estimate adult cell type proportions from the methylation matrix [39]. We estimated proportions of granulocytes, CD8+ T-cells, CD4+ T-cells, natural killer cells, B-cells, and monocytes. For downstream analyses, we constructed principal components of estimated cell type proportions.
We compared the distributions of metals concentrations in our study sample to those in NHANES wave 2015–2016. We estimated bivariate associations between individual trace metals and potential covariates using t-tests for binary covariates (foetal sex and hybridization round), analysis of variance for categorical and ordinal covariates (maternal ethnicity, maternal education, and household income), and simple linear regression for continuous covariates (maternal and paternal age). We used a biomarker of global DNA methylation, which is calculated by Illumina array-wide average DNA methylation per sample [40]. We tested for differences in average DNA methylation with metal concentrations using multiple linear regression. We performed similar analyses stratified by genome location (CpG island, shore, shelf, open sea, and enhancers). As expected, toxic metals (Cd, Pb, and total Hg) each exhibited log-normal, highly right skewed distributions in univariate assessments. Therefore, since Cd, Pb, and total Hg are predictors in our models, they were log-transformed, and model diagnostics indicated better fit for linear regression using log-transformations. The essential metals (Mn and Se) were normally distributed in univariate assessments; therefore, transformations were not required. We chose to standardize the reporting of effect estimates by the interquartile range of each metal because this allows for clearer and more intuitive interpretation of regression results. Last, we applied multiple linear regression to evaluate associations between individual trace metals and per cent DNA methylation at each site using estimated β-values. Based on a priori knowledge, we selected maternal age, foetal sex, hybridization round (the date at which arrays were processed in the laboratory), along with two principal components of adult cell type proportions, as covariates in each of these adjusted models for approximated global array-wide DNA methylation and single-site analyses. As a sensitivity analysis to compare to our covariate model, we conducted surrogate variable analysis (Leek and Storey, 2007) to capture the heterogeneity in the methylation matrix and adjusted for surrogate variables in replacement of measured covariates. We compared Q–Q plots and genomic inflation factor (the ratio of expected versus observed –log10(p-values)) of each surrogate variable model and covariate model.
From the results of the single-site analysis, we extracted the top ~1,000 sites associated with each trace metal. We used these sites to test for enrichment in gene ontology biological processes by applying the gometh function in the missMethyl package [41], which uses the Wallenius’ noncentral hypergeometric distribution. Redundant gene ontologies were removed using REVIGO [42]. All associations were adjusted for false discovery rate to account for multiple comparisons [43]. We also evaluated multiple sites simultaneously to identify differentially methylated regions associated with each metal using DMRcate (version 1.8.6).
Finally, we evaluated correlations in single-site associations with multiple independent samples to inform future exploratory replication across study samples and tissue types. To do this, we prioritized studies that used the Infinium HumanMethylation450 BeadChip for DNA methylation measurement. Unfortunately, independent study samples were not always available for maternal blood metals measures or DNA methylation. Samples using other tissue types were examined to assess potential replication, with the understanding that cross-tissue correlation may be low even when blood-based associations truly exist. Comparison studies also differed in their use of transformation approaches for DNA methylation matrices (e.g., calculation of β matrix or M-value matrix). β-Values represent the per cent methylation at a given CpG site and M-values are logit-transformations of the β-values [44]. Although directions of associations are comparable, transformation may affect effect estimates in regression results. Therefore, we transformed our data to match the prior choices in the comparison studies.
Correlations for Pb findings were estimated using published data from a study in Project Viva, where Pb was measured in maternal whole blood during pregnancy and DNA methylation was measured in cord blood (n = 268) [26]. For Cd results, the most appropriate study eligible for comparison was a study conducted on data combined from the New Hampshire Birth Cohort Study (NHBCS) (n = 343) and the Rhode Island Child Health Study (RICHS) (n = 141) [33]. This study assessed Cd exposure and DNA methylation profiles in placental samples. Correlation for Hg results were conducted in a study of the NHBCS (n = 138), where Hg was measured in infant toenail samples and DNA methylation was measured in cord blood [28]. We identified one study of Mn measured in infant toenail samples and DNA methylation measured in cord blood (n = 61); however, this study did not report effect estimates and we were unable to test for correlation across single CpG sites [35]. There were no available studies of Se and DNA methylation in pregnancy.
Results
Univariate and bivariate statistics
Study participants in this subset of EARLI were between 21 and 44 years of age and predominantly non-Hispanic White (58%) (Table 1). None of the participants reported smoking during pregnancy or had any record of smoking prior to pregnancy. A majority of participants had some level of higher education (87%), and approximately 58% of participants had a household income that exceeded 50 USDk (Table 1). We observed 100% detection of Mn, Pb, and Se in the whole blood samples included in this analysis (Supplemental Table S1). Cd and total Hg were detected in 91.7% and 86.6% of samples, respectively (Supplemental Table S1). We observed similar concentrations of trace metals in EARLI compared to women of childbearing age (15–44 years) in the 2015–16 cycle of the National Health and Nutrition Examination Study (Supplemental Table S1). Maternal characteristics between the primary EARLI analytical subset (n = 97) did not differ substantively from the EARLI participants with micro-clotting blood samples (n = 82) (Supplemental Table S2).
Table 1.
The Early Autism Risk Longitudinal Investigation (EARLI) study sample (n = 97) demographic characteristics and maternal blood trace metal concentrations
| Overall |
Cd (μg/L) |
Mn (μg/L) |
Pb (μg/.dL) |
Se (μg/L) |
Hg (μg/L) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | GM (GSD) | p-Value | Mean (SD) | p-Value | GM (GSD) | p-Value | Mean (SD) | p-Value | GM (GSD) | p-Value | |
| Infant sexa | 0.01 | 0.88 | 0.45 | 0.27 | 0.77 | ||||||
| Male | 47 (48%) | 0.17 (1.9) | 12.5 (4.1) | 0.43 (1.7) | 199 (26.8) | 0.63 (2.15) | |||||
| Female | 50 (52%) | 0.23 (1.6) | 12.3 (3.2) | 0.47 (1.5) | 205 (30) | 0.66 (2.17) | |||||
| Maternal ageb | 0.33 | 0.11 | 0.12 | 0.27 | 0.09 | ||||||
| [21,31) | 25 (26%) | 0.18 (1.5) | 12.7 (3.8) | 0.36 (1.5) | 197 (21) | 0.55 (2.32) | |||||
| [31,34] | 23 (24%) | 0.24 (1.7) | 13.8 (3.3) | 0.55 (1.8) | 209 (37.4) | 0.58 (1.75) | |||||
| [34,37] | 24 (25%) | 0.19 (1.7) | 12.5 (3.5) | 0.46 (1.6) | 201 (30.5) | 0.71 (2.01) | |||||
| [37,44] | 25 (26%) | 0.21 (2) | 10.8 (3.3) | 0.46 (1.3) | 203 (23.9) | 0.77 (2.47) | |||||
| Maternal ethnicityc | 0.41 | <0.001 | 0.83 | 0.44 | 0.09 | ||||||
| Non-Hispanic White | 56 (58%) | 0.18 (1.7) | 11.4 (2.9) | 0.42 (1.5) | 199 (25.2) | 0.55 (2.14) | |||||
| Non-Hispanic Black | 11 (11%) | 0.22 (1.6) | 11.6 (3.9) | 0.46 (1.6) | 209 (28.1) | 0.95 (2.46) | |||||
| Hispanic/Latino | 16 (16%) | 0.18 (1.4) | 15 (4) | 0.41 (1.5) | 205 (29.9) | 0.62 (1.57) | |||||
| Missing | 14 (14%) | ||||||||||
| Maternal educationc | 0.40 | 0.57 | 0.22 | 0.74 | 0.93 | ||||||
| High school or less | 13 (13%) | 0.22 (1.4) | 12.9 (3) | 0.46 (1.4) | 207 (34.3) | 0.57 (2.15) | |||||
| Some college or associate’s degree | 24 (25%) | 0.22 (1.7) | 12.6 (4.2) | 0.4 (1.5) | 205 (23.6) | 0.66 (2.39) | |||||
| Bachelor’s degree | 27 (28%) | 0.17 (1.8) | 12.8 (3.4) | 0.42 (1.5) | 201 (20.9) | 0.63 (2.11) | |||||
| Graduate degree | 32 (33%) | 0.21 (1.9) | 11.6 (3.6) | 0.52 (1.8) | 198 (35.1) | 0.67 (1.94) | |||||
| Missing | 1 (1%) | ||||||||||
| Paternal ageb | 0.80 | 0.20 | 0.09 | 0.43 | 0.19 | ||||||
| [22,32) | 20 (21%) | 0.19 (1.9) | 13.3 (4.9) | 0.38 (1.5) | 194 (32.2) | 0.45 (1.92) | |||||
| [32,36) | 25 (26%) | 0.2 (1.8) | 11.8 (2.3) | 0.5 (1.5) | 209 (29.1) | 0.77 (2.19) | |||||
| [36,40) | 26 (27%) | 0.21 (1.8) | 12.1 (3.7) | 0.42 (1.6) | 205 (24.2) | 0.63 (1.96) | |||||
| [40,51] | 23 (24%) | 0.19 (1.5) | 12.1 (3.2) | 0.5 (1.8) | 203 (29) | 0.73 (2.48) | |||||
| Missing | 3 (3%) | ||||||||||
| Household incomec | 0.75 | 0.32 | 0.35 | 0.93 | 0.003 | ||||||
| <$50,000 | 38 (39%) | 0.21 (1.6) | 12.3 (3.5) | 0.45 (1.4) | 202 (30) | 0.46 (2.02) | |||||
| $50,000– $99,999 | 34 (35%) | 0.21 (2) | 12.9 (4) | 0.49 (1.9) | 201 (28.3) | 0.76 (1.98) | |||||
| ≥ $100,000 | 22 (23%) | 0.19 (1.6) | 11.4 (2.8) | 0.41 (1.4) | 204 (27.9) | 0.83 (2.3) | |||||
| Missing | 3 (3%) | ||||||||||
| Rounda | 0.48 | 0.06 | 0.97 | 0.16 | 0.29 | ||||||
| 1 | 6 (6%) | 0.24 (1.8) | 10.7 (1.9) | 0.45 (1.6) | 192 (16) | 0.45 (2.18) | |||||
| 2 | 91 (94%) | 0.2 (1.7) | 12.5 (3.7) | 0.45 (1.6) | 203 (29.1) | 0.66 (2.15) | |||||
aTwo-sample tt-test to test for differences in metal concentrations between infant sex, and round.
bSimple linear regression to test for differences in metal concentrations across maternal and paternal age.
cAnalysis of variance test for differences in metal concentrations across categories of maternal ethnicity, maternal education, and household income.
Abbreviations: cadmium (Cd); manganese (Mn); lead (Pb); selenium (Se); total mercury (Hg); geometric mean (GM); geometric standard deviation (GSD); standard deviation (SD).
In bivariate analyses, we observed a higher geometric mean of Cd in women with a female foetus (0.23 μg/L) compared to those with a male foetus (0.17 μg/L) (Table 1). Mn concentrations were notably higher in Hispanic/Latino women (>3 μg/L) compared to other racial groups (Table 1). Women in the highest household income group (≥$100k) had the highest mean concentration of total Hg (0.83 μg/L) (Table 1). Between trace metals, we observed the highest correlation between Cd and Pb (Spearman ρ = 0.36, p-value<0.001) (Supplemental Figure S1). When we tested for associations between metals and cell type proportions, Cd was positively associated with B-cell proportions (β [standard error] = 0.7 [0.3], p-value = 0.04) (Supplemental Table S3). Trace metal concentrations were not appreciably different by technical covariates such as hybridization date (p-value >0.05).
Associations between trace metals and single-site DNA methylation
Q–Q plots of expected versus observed –log10(p-values) across the genome are located in Supplemental Figure S2. Crude models with only individual trace metals as a predictor had genomic inflation factors (λ-values) ranging from 0.88 to 1.51 (Supplemental Figure S2). Although the range in λ-values was similar in adjusted covariate models (0.89–1.52), we observed marked improvement in genomic inflation as evidenced by the normality of the Q–Q plot for measured covariate adjusted models (Supplemental Figure S2). Approximated array-wide global DNA methylation trends in hyper- and hypo-methylation were illustrated in Supplemental Figure S3. When we tested for differences in average DNA methylation using linear regression, we did not observe statistically significant associations (p > 0.05) between overall average DNA methylation for Cd and Mn (Supplemental Table S4).
The top 10 CpG sites associated with each trace metal are reported in Figure 1(a-b). Pb and Mn were the only trace metals associated with individual CpG sites after adjustment for multiple comparisons with all five trace metals. Pb was associated (FDR q-value <0.1) with hyper-methylation at 11 CpG sites, near the genes CYP24A1, ASCL2, FAT1, SNX31, NKX6-2, LRRC4C, BMP7, HOXC11, PCDH7, ZSCAN18, and VIPR2 (Figure 1(a)). Mn was associated (FDR q-value <0.1) with hyper-methylation at four CpG sites, one of which was near the gene ARID2 (Figure 1b). At a less significant threshold (FDR q-value <0.2), Cd was associated with hyper-methylation at three sites, two of which were near the genes ITPKB and PCDH15 (Figure 1(a)), and total Hg was associated with hyper-methylation at one CpG site, near the gene SPTBN2 (Figure 1(a)). There were no sites that reached either of these significance thresholds for analyses of Se. In alignment with data visualization approaches to explore distributions of DNA methylation [26,29,33], for each of the metals, we created scatter plots between per cent DNA methylation at the top two CpG sites nearest genes in Supplemental Figures S4A and 4B. Scatter plots largely demonstrated linear relationships for each of the metals. Although our study sample included all non-smokers, metals exposure may result from environmental tobacco exposure and historical smoking. We compared top sites that were robust to FDR adjustment, to evaluate if any overlapped with CpG sites associated with cigarette smoking [45]. Three of the Pb-associated CpG sites near the genes BMP7 (cg20340302), HOXC11 (cg03536474), and VIPR2 (cg20673829) were also associated with current smoking exposure.
Figure 1.
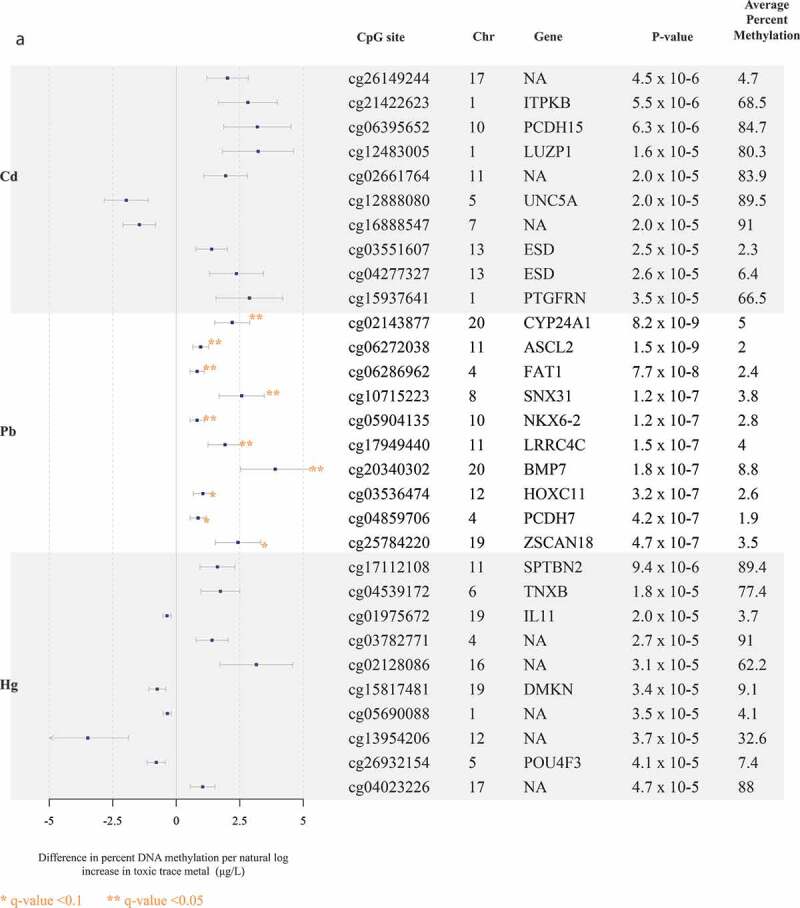
Top 10 CpG sites associated with blood trace metals from regression models adjusted for for cell type principal components 1 and 2, maternal age, foetal sex, and hybridization date (n = 97 samples). (Figure 1(a)) reports associations with cadmium (Cd), lead (Pb), and total mercury (Hg). (Figure 1(b)) reports associations with manganese (Mn) and selenium (Se).
Figure 1b.
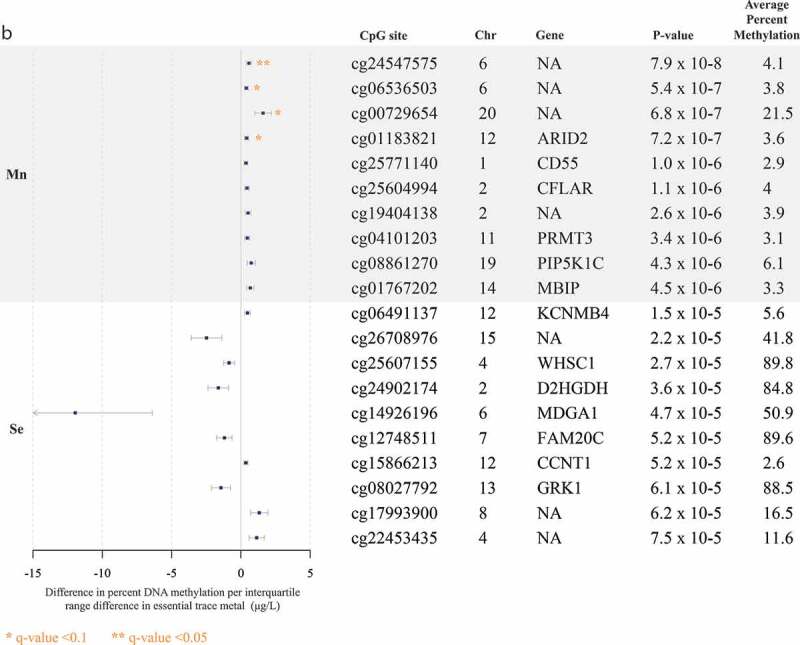
(Continued).
When we restricted to sites associated (p < 0.05) with Cd, Mn, and Pb (n = 96 probes), we observed very high correlations between Cd, Mn, and Pb single-site effect estimates (Pearson ρ range: 0.86–0.98) (Figure 2). Across all sites (n = 348,438 probes), there were lower magnitudes of correlations (Pearson ρ range: 0.03–0.3) (Supplemental Figure S5). Among the associated genes for each metal, the lowest p-value threshold where we observed three overlapping genes (SLC7A4, TFAP2A, NXN) for Cd, Mn, and Pb was at p = 1 × 10−3 (Supplemental Figure S6).
Figure 2.
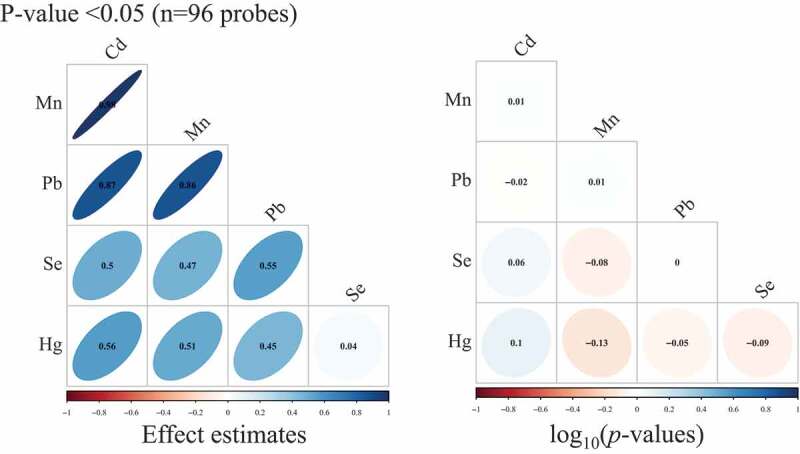
Pearson correlation matrices for effect estimates and Spearman correlation matrices for – log10(p-values) from single-site analyses of cadmium (Cd), manganese (Mn), lead (Pb), selenium (Se), and total mercury (Hg), where we restricted to only CpG sites that had p-value <0.05 in models of Cd, Mn, and Pb. (n = 97 samples).
Gene ontologies and differentially methylated regions
We observed several associations (FDR q-value <0.1) between gene ontologies and Pb, which included homophilic cell adhesion (p-value = 6.2 × 10−17), nervous system development (p-value = 1.2 × 10−9), biological adhesion (p-value = 3.5 × 10–8), developmental process (p-value = 1 × 10–8), calcium ion binding (p-value = 1.5 × 10–5), and cell fate commitment (p-value = 5.6 × 10–5) (Table 2). Mn was associated (FDR q-value <0.1) with cellular nitrogen metabolism (p-value = 1.4 × 10−6), cell cycle process (p-value = 1.5 × 10–5), nucleic acid metabolism (p-value = 5.3 × 10–5), nucleobase-containing compound metabolism (p-value = 5.7 × 10–5), and negative regulation of response to DNA damage stimulus (p-value = 7.1 × 10–5) (Table 2). We observed that total Hg was associated (FDR q-value <0.1) with organ morphogenesis (p-value = 2.2 × 10–7), tube development (p-value = 2.6 × 10−6), tissue development (p-value = 4.8 × 10−6), anterior/posterior pattern specification (p-value = 1.5 × 10–5), cartilage condensation (p-value = 2.8 × 10–5), cell aggregation (p-value = 3.2 × 10–5), cell–cell signalling (p-value = 3.7 × 10–5), and respiratory system development (p-value = 4.4 × 10–5) (Table 2). Finally, at an FDR q-value <0.1, Cd was associated with homophilic cell adhesion (p-value = 1 × 10–7) (Table 2). Assessment of differentially methylated regions indicated that among the metals Pb and Mn exhibited suggestive signals (Supplemental Table S5). For Pb, there were four suggestive differentially methylated regions; however, none of these was robust to false discovery adjustment: chromosomes 3 (adjusted p-value = 0.24), 4 (adjusted p-value = 0.13), 6 (adjusted p-value = 0.27), and 19 (adjusted p-value = 0.07). There was one differentially methylated region on chromosome 20 associated with Mn; however, that signal was not robust to false discovery adjustment (adjusted p-value = 0.73). Bump-hunting style analyses are not optimal for array data given the sparseness of CpGs in the array.
Table 2.
Gene ontology biological processes associated (~1,000 CpG sites ranked by minimum p-value) with single CpG sites and individual trace metals, using the covariate modela
| Rank |
GO ID |
GO term |
p-Value |
q-Value |
|---|---|---|---|---|
| Cd | ||||
| 1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1.0 × 10−7 | 0.002 |
| 2 | GO:0050851 | antigen receptor-mediated signaling pathway | 3.1 × 10−5 | 0.17 |
| 3 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic | 2.5 × 10−4 | 0.86 |
| 4 | GO:0005509 | calcium ion binding | 3.0 × 10−4 | 0.86 |
| 5 | GO:0002376 | immune system process | 1.2 × 10−3 | >0.9 |
| 6 | GO:0008142 | oxysterol binding | 1.4 × 10−3 | >0.9 |
| 7 | GO:0022410 | circadian sleep/wake cycle process | 1.6 × 10−3 | >0.9 |
| 8 | GO:2000107 | negative regulation of leukocyte apoptotic process | 1.7 × 10−3 | >0.9 |
| 9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity | 2.5 × 10−3 | >0.9 |
| 10 | GO:0042053 | regulation of dopamine metabolic process | 2.5 × 10−3 | >0.9 |
| Mn | ||||
| 1 | GO:0034641 | cellular nitrogen compound metabolic process | 1.4 × 10−6 | 0.002 |
| 2 | GO:0022402 | cell cycle process | 1.5 × 10−5 | 0.02 |
| 3 | GO:0090304 | nucleic acid metabolic process | 5.3 × 10−5 | 0.06 |
| 4 | GO:0006139 | nucleobase-containing compound metabolic process | 5.7 × 10−5 | 0.06 |
| 5 | GO:2001021 | negative regulation of response to DNA damage stimulus | 7.1 × 10−5 | 0.08 |
| 6 | GO:0006725 | cellular aromatic compound metabolic process | 1.1 × 10−4 | 0.1 |
| 7 | GO:0044237 | cellular metabolic process | 1.3 × 10−4 | 0.12 |
| 8 | GO:1901360 | organic cyclic compound metabolic process | 1.5 × 10−4 | 0.12 |
| 9 | GO:0046483 | heterocycle metabolic process | 1.5 × 10−4 | 0.12 |
| 10 | GO:0048534 | hematopoietic or lymphoid organ development | 2.9 × 10−4 | 0.21 |
| Pb | ||||
| 1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 6.2 × 10–17 | 1.4 × 10–12 |
| 2 | GO:0007399 | nervous system development | 1.2 × 10−9 | 6.6 × 10−6 |
| 3 | GO:0022610 | biological adhesion | 3.5 × 10−8 | 10.0× 10−5 |
| 4 | GO:0032502 | developmental process | 9.6 × 10−8 | 2.4 × 10−4 |
| 5 | GO:0005509 | calcium ion binding | 1.5 × 10−5 | 0.02 |
| 6 | GO:0045165 | cell fate commitment | 5.6 × 10−5 | 0.07 |
| 7 | GO:0051410 | detoxification of nitrogen compound | 1.6 × 10−4 | 0.2 |
| 8 | GO:0032501 | multicellular organismal process | 1.8 × 10−4 | 0.2 |
| 9 | GO:0001656 | metanephros development | 1.9 × 10−4 | 0.2 |
| 10 | GO:0022010 | central nervous system myelination | 1.9 × 10−4 | 0.2 |
| Se | ||||
| 1 | GO:0034062 | 5'-3' RNA polymerase activity | 1.1 × 10−4 | >0.9 |
| 2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity | 4.4 × 10−4 | >0.9 |
| 3 | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 5.2 × 10−4 | >0.9 |
| 4 | GO:1905214 | regulation of RNA binding | 6.5 × 10−4 | >0.9 |
| 5 | GO:0051276 | chromosome organization | 9.1 × 10−4 | >0.9 |
| 6 | GO:0001784 | phosphotyrosine residue binding | 1.2 × 10−3 | >0.9 |
| 7 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering | 1.3 × 10−3 | >0.9 |
| 8 | GO:0010836 | negative regulation of protein ADP-ribosylation | 1.8 × 10−3 | >0.9 |
| 9 | GO:0016233 | telomere capping | 2.1 × 10−3 | >0.9 |
| 10 | GO:0033044 | regulation of chromosome organization | 2.4 × 10−3 | >0.9 |
| Hg | ||||
| 1 | GO:0009887 | animal organ morphogenesis | 2.2 × 10−7 | 0.005 |
| 2 | GO:0035295 | tube development | 2.6 × 10−6 | 0.03 |
| 3 | GO:0009888 | tissue development | 4.8 × 10−6 | 0.04 |
| 4 | GO:0009952 | anterior/posterior pattern specification | 1.5 × 10−5 | 0.07 |
| 5 | GO:0001502 | cartilage condensation | 2.8 × 10−5 | 0.09 |
| 6 | GO:0098743 | cell aggregation | 3.2 × 10−5 | 0.09 |
| 7 | GO:0007267 | cell-cell signaling | 3.7 × 10−5 | 0.09 |
| 8 | GO:0060541 | respiratory system development | 4.4 ×10−5 | 0.09 |
| 9 | GO:0003401 | axis elongation | 6.4 × 10−5 | 0.11 |
| 10 | GO:0007389 | pattern specification process | 8.2 × 10−5 | 0.11 |
aCovariate model adjusted for cell type principal components 1 and 2, maternal age, foetal sex, and hybridization date.
Sensitivity analyses
The following number of surrogate variables were used to adjust for individual trace metals: Cd (n = 3), Mn (n = 5), Pb (n = 6), Se (n = 4), and Hg (n = 3). Q–Q plots for surrogate variables were comparable to adjusted covariate models (λ-values ranging from 0.99 to 1.03) (Supplemental Figure S2). Bivariate relationships between individual surrogate variables and known covariates are illustrated in Supplemental Figure S7. Overall, surrogate variables were associated with several known covariates, including cell type proportions, parental age, maternal ethnicity, education, and hybridization round (Supplemental Figure S7). Trends in the biomarker of global DNA methylation (array-wide average DNA methylation) were consistent between the known covariate adjusted models and the surrogate variable adjusted models (Figure 1 and Supplemental Figure S8). When we compared the single-site results between the known covariate adjusted model and the surrogate variable-adjusted model, we observed high correlations in effect estimates (Pearson ρ range: 0.75–0.94) and –log10(p-values) (Spearman ρ range: 0.5–0.85) (Supplemental Figure S8).
Correlation with independent study samples
Independent study samples were selected by exposure for most similar tissue and time period. We transformed our DNA methylation data to match the preprocessing that occurred in comparison studies. Among the correlations with independent samples, we observed the greatest positive correlations with Pb results for single CpG sites with p-values <0.05 (Spearman ρ range: 0.34–0.36) (Supplemental Figure S9). The correlation in findings for Pb did not differ substantially based on whether we used the β matrix or M-value matrix (Supplemental Figure S9). Notably, the independent study data we extracted for Pb involved Pb exposure assessment in maternal blood and DNA methylation measures in cord blood (Wu et al., 2017). The data from the Cd comparison study involved Cd exposure assessment and DNA methylation measures in placental tissue (Everson et al., 2018). Additionally, the data from the Hg comparison study involved Hg exposure assessment in infant toenail clippings and DNA methylation measures in cord blood (Cardenas et al., 2015). As such, correlation in results for Cd and Hg were not as consistent when comparing the EARLI study with the comparison studies: Cd (Spearman ρ range: −0.13–0.01) and Hg (Spearman ρ range: 0.09–0.15) (Supplemental Figure S9).
Discussion
To our knowledge this is the first study to report epigenome-wide associations between pregnancy exposures to multiple trace metals and maternal whole blood DNA methylation. After adjusting for multiple comparisons, Pb was associated (FDR q-value <0.1) with 11 individual CpG sites. Gene ontology analysis reported numerous biological processes associated with Pb, revealing linkage to neurodevelopment and immune perturbation pathways. Additionally, Mn was associated (FDR q-value <0.1) with four individual CpG sites. In gene ontology analysis of Mn, we observed pathways related to cellular metabolism. Across the metals, there was strong overlap and correlation in effect estimates for single-site findings with Pb, Cd, and Mn. Rigorous sensitivity analyses and correlations were estimated to independent samples across multiple tissues. Altogether, the CpG sites we reported are potential biomarkers of corresponding trace metals exposures and may implicate downstream toxicological mechanisms.
We observed the greatest number of absolute CpG sites and gene ontologies associated with Pb concentrations. Exposure to Pb results in deleterious effects on the nervous system, potentially through immunological mechanisms guided by epigenetic modifications [20,46]. The CpG site most associated with Pb was near the gene CYP24A1, which encodes for a member of the cytochrome p450 family of enzymes that is involved in vitamin D3 metabolism and cellular calcium homoeostasis[47]. Rat models have found vitamin D receptors abundantly expressed in astrocytes, and perturbations in vitamin D3 metabolism can alter nervous system maintenance [48]. Pb was also associated with the CpG site near the gene ASCL2, which encodes for achaete-scute homologue 2 – a basic helix–loop–helix transcription factor highly expressed in follicular T-helper cells – and regulates select chemokine receptors and influences T-cell migration [49]. Mice with ASCL2 knockouts or suppression exhibited reduced T-cell migration and follicular T-cell development, both of which are integral for resolving infection and protection against autoimmune disease [49]. In gene ontology analysis, we reported associations with nervous system development and precursors to immune cell migration, such as cell adhesion. Altogether, the CpG sites and gene ontologies associated with Pb are biologically consistent with the evidence supporting Pb as a neurotoxicant.
The comparison study we selected by Wu and colleagues (2017) found that prenatal Pb exposure was associated with altered DNA methylation in cord blood at sites near genes related to cell proliferation (CLEC11A) and vesicular transport in neurons (DNHD1). Although the top CpG sites in our study differed from those of the independent comparison study, we did observe moderate correlation in effect estimates across multiple sites associated at p < 0.05. What this suggests is that although specific sites may not be highly associated across studies, collectively we observe similar changes in per cent methylation across multiple sites. A separate study of women in Bangladesh observed associations between prenatal Pb and cord blood DNA methylation at site near genes involved in endothelial dysfunction (GP6) and immune modulation (HLA-DQ B1 and B2) [24]. Looking across generations, prenatal Pb has also been associated with altered DNA methylation near genes involved in nervous system development (NDRG4 and NINJ2) and immune system regulation (APOA5 and DOK3) in grandchildren of exposed mothers [50]. The combination of results from our study and previous studies indicates that Pb exposure in pregnancy is associated with DNA methylation signatures in multiple tissues.
In our analysis of Mn, we observed associations with multiple gene ontologies related to cellular metabolism. Interestingly, in vitro studies support these associations with findings that Mn exposure promotes sequestration of Mn into mitochondria and disrupts cellular respiration, intracellular calcium and redox homoeostasis, and metabolism [18,51]. Among the CpG sites nearest genes, we observed the greatest association between Mn and the CpG site near ARID2. The ARID2 gene encodes for AT-rich interactive domain 2 – a component of chromatin remodelling protein complexes – and essential for nucleotide excision repair of DNA damage sites [52]. The association between Mn and ARID2 is consistent with the toxicological evidence that exposure results in elevated reactive oxygen species, which could alter ARID2 function in circulating immune cells. Overall, these findings underline that future studies of Mn should consider exploring associations with biomarkers of altered cellular metabolism.
No studies were available to test for correlations with genome-wide single-site effect estimates for Mn. However, Maccani et al. (2015) reported associations between newborn toenail Mn and placental DNA methylation at CpG sites near genes associated with neurodevelopment (EMX2OS and ATAD2B). These two genes and their corresponding CpG sites were not among the top findings in our study; however, other CpG sites near these genes were associated (p-value <0.05) in our single-site analysis. The differences in exact CpG sites and associated genes are likely due to the fact that Mn was measured in infant toenail samples and DNA methylation was measured in placental tissues, which differed from the tissues used in EARLI (maternal blood).
Although single-site analysis did not reveal any single CpG site associated with Cd, Hg, or Se after adjustment of multiple comparisons, we did observe gene ontologies associated with Cd and Hg concentrations. Cd was associated (FDR q-value <0.05) with haemophilic cell adhesion. Albeit not within the FDR threshold of 0.05, we also observed immune and nervous system-related processes among the gene ontologies associated with Cd. We observed two CpG sites near the esterase D (ESD) gene may be associated with Cd exposure (however, the q-values were>0.1). Cd exposure is associated with increased ESD gene expression in human hepatocyte carcinoma cells [53] and decreased ESD gene expression in normal rat kidney epithelial cells [54]. We did not observe consistent site-specific correlation between the Cd findings in EARLI and the independent comparison study, which is likely due to the fact that the study measured Cd and DNA methylation in placental tissue [33]. However, Everson and colleagues (2018) did report associations with sites near genes involved in immune responses and cytokine production, as well as nervous system-related genes. Our participants were non-smokers and generally were exposed to low levels of Cd. These findings highlight the need to conduct additional replication studies of Cd and DNA methylation in maternal whole blood.
Exposure to Hg was associated (FDR q-value <0.05) with gene ontologies for organ morphogenesis, tube development (a precursor for neural tube development), and tissue development. We did not observe significant correlation in single-site results for Hg in EARLI compared to the independent comparison study [28]. Interestingly, this study reported relationships between Hg exposure and sites associated with tissue differentiation, which is broadly related to the biological pathways we observed [28]. The lack of consistency in single-site analysis between the studies may also be due to the fact that Cardenas and colleagues (2015) measured Hg in newborn toenail samples and DNA methylation was measured in cord blood.
Beyond the scope of describing biological pathways disrupted by prenatal metals exposures, the landscape of single-site DNA methylation profiles across the epigenome also reveals biomarker signals of metals exposures. Notably, across independent samples, replication approaches can be utilized to evaluate predictive capacity of a collection of DNA methylation sites to capture unique signatures of metals exposures. Upon establishing DNA methylation signatures for specific trace metals using replication samples, prediction algorithms can be constructed consisting of multiple DNA methylation sites. The ability to predict metals exposures using DNA methylation-derived prediction algorithms can have profound public health impacts. For example, existing birth cohort studies that have not previously measured metals exposures can leverage DNA methylation assays to reconstruct an individual’s exposure to metals for downstream inferential analysis of pregnancy and child health outcomes.
One of the major limitations of this study is the small sample size (n = 97), which reduced our power to assess single-site epigenome-wide associations for all of the trace metals. The independent comparison studies that we compared our results to had sample sizes ranging from 61 to 584 participants, and further, did not use the same tissue types. The cross-sectional study design is also another limitation, in that our findings are susceptible to reverse causation. For example, other factors aside from trace metals exposures – such as nutritional status and physical activity – may alter the maternal epigenome and alter gene expression in tissues that influence trace metal biotransformation and excretion. Cross-sectional studies are also unable to account for the temporal variability in environmental trace metals exposures. However, in a previous exposure assessment study, repeated measurements of blood metals in pregnancy showed that Pb exhibited high intra-class correlation coefficients (ICC = 0.78), indicating that a single time point measurement is substantively reliable, while other metals such as Cd, Hg, and Mn had moderate reliability (ICC ranging from 0.48 to 0.65) [55,56]. Another limitation of our study is that we did not have iron status available as a covariate, and due to overlapping reliance of divalent metal transporter 1 (DMT1), iron status may influence peripheral concentrations of Mn, Cd, and Pb [57–59]. These relationships may be more appropriately assessed by testing for statistical interactions or effect modification by iron status in future studies. Additionally, the profile of participants in EARLI does not represent the racial and socioeconomic diversity of U.S. residents, and therefore our study is limited in generalizability to the U.S. population.
Despite these limitations, our study was conducted in a well-characterized pregnancy cohort. Our study also implemented high-sensitivity exposure assessments of multiple trace metals early in pregnancy. Furthermore, the carefully applied protocols for whole blood processing and DNA methylation measurements yielded high proportions of viable CpG sites for single-site analysis. Our study is also unique in that it is the first to evaluate associations between five different trace metals and changes in maternal whole blood DNA methylation in early pregnancy. Another strength is that we applied rigorous sensitivity analysis by comparing surrogate variable analysis to standard adjustment of known covariates. Finally, we also evaluated our findings and their correlations across multiple single CpG sites in independent study samples.
In conclusion, during early pregnancy, maternal blood concentrations of Pb and Mn contributed the greatest associations with altered DNA methylation. The sites where we observed notable changes in DNA methylation were involved in immune responses and nervous system development and maintenance. Our findings are biologically consistent with experimental studies and previous epigenome-wide association studies of different tissues. Altered DNA methylation near the genes we reported may potentially contribute to adverse pregnancy outcomes, and future studies should test for potential mediation with the CpG sites we identified. Larger studies should seek to test for associations with the sites we identified with Pb and Mn.
Supplementary Material
Funding Statement
This work was supported by the National Institutes of Health [grants R01ES016443, R01ES017646, R01ES025531, R01ES025574, P30ES017885, R01AG055406]. Support for Max Aung was provided in part by a grant from the Robert Wood Johnson Foundation Health Policy Research Scholars programme;National Institute of Environmental Health Sciences [R01ES017646];National Institute of Environmental Health Sciences [R01ES025574];National Institute on Aging [R01AG055406];National Institute of Environmental Health Sciences (US) [P30ES017885];National Institute of Environmental Health Sciences (US) [R01ES016443];National Institute of Environmental Health Sciences (US) [R01ES025531];
Disclosure statement
The authors declare they have no conflict of interest.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].ATSDR. Cadmium Public Health Statement . 2013; 1–10. Available from: https://www.atsdr.cdc.gov/ToxProfiles/tp5-c1-b.pdf
- [2].ATSDR. Lead Public Health Statement . 2007; 1–13. Available from: https://www.atsdr.cdc.gov/ToxProfiles/tp13-c1-b.pdf
- [3].ATSDR. Manganese Public Health Statement . 2012; 1–10. Available from: https://www.atsdr.cdc.gov/ToxProfiles/tp151-c1-b.pdf
- [4].ATSDR. Mercury Public Health Statement . 1999; 1–20. Available from: https://www.atsdr.cdc.gov/ToxProfiles/tp46-c1-b.pdf
- [5].ATSDR. Selenium Public Health Statement . 2005; 1–8. Available from: https://www.atsdr.cdc.gov/ToxProfiles/tp92-c1-b.pdf
- [6].Poropat AE, Laidlaw MAS, Lanphear B, et al. Blood lead and preeclampsia_ A meta-analysis and review of implications. Environ Res. 2018;160:12–19. [DOI] [PubMed] [Google Scholar]
- [7].Rahman A, Kumarathasan P, Gomes J.. Infant and mother related outcomes from exposure to metals with endocrine disrupting properties during pregnancy. Sci total enviro. 2016;569–570: 1022–1031. [DOI] [PubMed] [Google Scholar]
- [8].Sanders AP, Desrosiers TA, Warren JL, et al. Association between arsenic, cadmium, manganese, and lead levels in private wells and birth defects prevalence in North Carolina: a semi-ecologic study. BMC Public Health. 2014;14(1):1322–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vrijheid M, Casas M, Gascon M, et al. Environmental pollutants and child health—A review of recent concerns. Int J Hyg Environ Health. 2016;219(4–5):331–342. [DOI] [PubMed] [Google Scholar]
- [10].Argüello JM, Raimunda D, González-Guerrero M. Metal transport across biomembranes: emerging models for a distinct chemistry. J Biol Chem. 2012;287(17):13510–13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lewicka I, Kocyłowski R, Grzesiak M, et al. Selected trace elements concentrations in pregnancy and their possible role — literature review. Ginekol Pol. 2017;88(9):509–514. [DOI] [PubMed] [Google Scholar]
- [12].Pieczyńska J, Grajeta H. The role of selenium in human conception and pregnancy. J Trace Elem Med Biol. 2015;29:31–38. [DOI] [PubMed] [Google Scholar]
- [13].Vinceti M, Filippini T, Wise LA. Environmental selenium and human health: an update. Curr Environ Health Rep. 2018;5(4):464–485. [DOI] [PubMed] [Google Scholar]
- [14].Li N, Liu X, Zhang P, et al. The effects of early life lead exposure on the expression of interleukin (IL) 1β, IL-6, and glial fibrillary acidic protein in the hippocampus of mouse pups. Hum Exp Toxicol. 2014;34(4):357–363. [DOI] [PubMed] [Google Scholar]
- [15].Hanson ML, Holásková I, Elliott M, et al. Prenatal cadmium exposure alters postnatal immune cell development and function. Toxicol Appl Pharmacol. 2012;261(2):196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Milnerowicz H, Ściskalska M, Dul M. Pro-inflammatory effects of metals in persons and animals exposed to tobacco smoke. J Trace Elem Med Biol. 2015;29:1–10. [DOI] [PubMed] [Google Scholar]
- [17].Pilones K, Tatum A, Gavalchin J. Gestational exposure to mercury leads to persistent changes in T-cell phenotype and function in adult DBF1 mice. J Immunotoxicol. 2009;6(3):161–170. [DOI] [PubMed] [Google Scholar]
- [18].Sarkar S, Malovic E, Harischandra DS, et al. Manganese exposure induces neuroinflammation by impairing mitochondrial dynamics in astrocytes. NeuroToxicol. 2018;64:204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bakulski KM, Fallin MD. Epigenetic epidemiology: promises for public health research. Environ Mol Mutagen. 2014;55(3):171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ruiz-Hernandez A, Kuo -C-C, Rentero-Garrido P, et al. Environmental chemicals and DNA methylation in adults: a systematic review of the epidemiologic evidence. Clin Epigenetics. 2015;7(1):9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cowley M, Skaar DA, Jima DD, et al. Effects of cadmium exposure on DNA methylation at imprinting control regions and genome-wide in mothers and newborn children. Environ Health Perspect [Internet]. 2018;126:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kippler M, Engström K, Mlakar SJ, et al. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics. 2014;8(5):494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vidal AC, Semenova V, Darrah T, Vengosh A, Huang Z, King K, Nye MD, Fry R, Skaar D, Maguire R, et al . Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharmacol Toxicol. 2015;16(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Engström K, Rydbeck F, Kippler M, et al. Prenatal lead exposure is associated with decreased cord blood DNA methylation of the glycoprotein VI gene involved in platelet activation and thrombus formation. Environ Epigenetics. 2015;1(1):dvv007–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pilsner JR, Hu H, Ettinger A, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117(9):1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu S, Hivert M-F, Cardenas A, et al. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in project viva: an epigenome-wide association study. Environ Health Perspect. 2017;125(8):087019–10. DOI: 10.1289/EHP1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bakulski KM, Lee H, Feinberg JI, et al. Prenatal mercury concentration is associated with changes in DNA methylation at TCEANC2 in newborns. Int J Epidemiol. 2015;44(4):1249–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cardenas A, Koestler DC, Houseman EA, et al. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics. 2015;10(6):508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cardenas A, Rifas-Shiman SL, Agha G, et al. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci Rep. 2017;7(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cardenas A, Rifas-Shiman SL, Godderis L, et al. Prenatal exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ Health Perspect. 2017;125(8):087022–10. DOI: 10.1289/EHP1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Appleton AA, Jackson BP, Karagas M, et al. Prenatal exposure to neurotoxic metals is associated with increased placental glucocorticoid receptor DNA methylation. Epigenetics. 2017;12(8):607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Everson TM, Armstrong DA, Jackson BP, et al. Maternal cadmium, placental PCDHAC1, and fetal development. Reprod Toxicol. 2016;65:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Everson TM, Punshon T, Jackson BP, et al. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two. U.S. Birth Cohorts Environ Health Perspect. 2018;126(1):017010–3. DOI: 10.1289/EHP2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Maccani JZJ, Koestler DC, Lester B, et al. Placental DNA methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environ Health Perspect. 2015;123(7):723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Maccani JZJ, Koestler DC, Houseman EA, et al. DNA methylation changes in the placenta are associated with fetal manganese exposure. Reprod Toxicol. 2015;57:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mohanty AF, Farin FM, Bammler TK, et al. Infant sex-specific placental cadmium and DNA methylation associations. Environ Res. 2015;138:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinfor (Oxford, England). 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen Y, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the illumina infinium humanmethylation450 microarray. Epigenetics. 2014;8(2):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bakulski KM, Dou JF, Feinberg JI, et al. Prenatal multivitamin use and MTHFR genotype are associated with newborn cord blood DNA methylation. Int j environ res public health. 2020;17(24):9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from illumina’s humanmethylation450 platform. Bioinf (Oxford, England). 2015;32(2):286–288. [DOI] [PubMed] [Google Scholar]
- [42].Supek F, Bošnjak M, Škunca N, et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B (Methodol). 1995;57(1):289–300. [Google Scholar]
- [44].Du P, Zhang X, Huang -C-C, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 2010;11(1):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Joehanes R, Just AC, Marioni RE, et al. Epigenetic signatures of cigarette smoking. Circ cardiovasc genet. 2016;9(5):436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chibowska K, Baranowska-Bosiacka I, Falkowska A, et al. Effect of lead (Pb) on inflammatory processes in the brain. Int J Mol Sci [Internet]. 2016;17(12):2140–17. DOI: 10.3390/ijms17122140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sun H, Jiang C, Cong L, et al. CYP24A1 inhibition facilitates the antiproliferative effect of 1,25(OH)2D3 through downregulation of the WNT/β-catenin pathway and methylation-mediated regulation of CYP24A1 in colorectal cancer cells. DNA Cell Biol [Internet]. 2018;37(9):742–749. [DOI] [PubMed] [Google Scholar]
- [48].Landel V, Stephan D, Cui X, et al. Differential expression of vitamin D-associated enzymes and receptors in brain cell subtypes. J Steroid Biochem Mol Biol. 2018;177:129–134. [DOI] [PubMed] [Google Scholar]
- [49].Liu X, Chen X, Zhong B, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Cell Mol Immunol. 2014;507:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sen A, Heredia N, Senut M-C, et al. Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Sci Rep. 2015 Sep 29;5(1):1-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Filipov NM, Seegal RF, Lawrence DA. Manganese potentiates in vitro production of proinflammatory cytokines and nitric oxide by microglia through a nuclear factor kappa B-dependent mechanism. Toxicol Sci. 2005;84(1):139–148. [DOI] [PubMed] [Google Scholar]
- [52].Oba A, Shimada S, Akiyama Y, et al. ARID2 modulates DNA damage response in human hepatocellular carcinoma cells. J Hepatol. 2017;66(5):942–951. [DOI] [PubMed] [Google Scholar]
- [53].Paesano L, Perotti A, Buschini A, et al. Data on HepG2 cells changes following exposure to cadmium sulphide quantum dots (CdS QDs). Data Brief. 2017;11:72–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yamanobe Y, Nagahara N, Matsukawa T, et al. Sex differences in shotgun proteome analyses for chronic oral intake of cadmium in mice. PLoS One. 2015;10(3):e0121819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ashrap P, Watkins DJ, Mukherjee B, et al. Predictors of urinary and blood Metal(loid) concentrations among pregnant women in Northern Puerto Rico. Environ Res. 2020;183:109178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rosner BBA. Fundamentals of biostatistics. Seventh ed. Boston: Brooks/ Cole, Cengage Learning; 2011. [Google Scholar]
- [57].Kim Y. Sex, pregnancy, and age-specific differences of blood manganese levels in relation to iron status; what does it mean? Toxicol Rep [Internet]. 2018;5: 28–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Illing AC, Shawki A, Cunningham CL, et al. Substrate profile and metal-ion selectivity of human divalent metal-ion transporter-1. J Biol Chem. 2012;287(36):30485–30496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Garrick MD, Dolan KG, Horbinski C, et al. DMT1: a mammalian transporter for multiple metals. Biometals. 2003;16(1):41–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


