Abstract
The complex composition of bacterial membranes has a significant impact on the understanding of pathogen function and their development towards antibiotic resistance. In addition to the inherent complexity and biosafety risks of studying biological pathogen membranes, the continual rise of antibiotic resistance and its significant economical and clinical consequences has motivated the development of numerous in vitro model membrane systems with tuneable compositions, geometries, and sizes. Approaches discussed in this review include liposomes, solid-supported bilayers, and computational simulations which have been used to explore various processes including drug-membrane interactions, lipid-protein interactions, host–pathogen interactions, and structure-induced bacterial pathogenesis. The advantages, limitations, and applicable analytical tools of all architectures are summarised with a perspective for future research efforts in architectural improvement and elucidation of resistance development strategies and membrane-targeting antibiotic mechanisms.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12551-021-00913-7.
Keywords: Model membrane, Lipids, Membrane, Biophysics, Bacteria
Introduction
All organisms rely on the presence of biological membranes acting as barriers between the inside and outside cellular environments. The functionality of such membranes is dictated by the types of lipids and other molecules that make up their often highly complex structure (Watson 2015; Guidotti 1972).
The “ESKAPE” pathogens, a faction of Gram-negative (GN) and Gram-positive (GP) bacteria, are responsible for the majority of nosocomial infections and are deemed a great threat to global healthcare because of their multidrug resistance (MDR) (Boucher et al. 2009; Mar et al. 2017; Pendleton et al. 2013; Rice 2010; Santajit and Indrawattana 2016; Ventola 2015). MDR bacterial pathogens can overexpress intrinsic resistance markers via adaptive mutations and acquire various foreign resistance factors through gene transfer processes (Gould and Bal 2013; Ventola 2015; Chilambi et al. 2018; Fernández and Hancock 2012; Prestinaci et al. 2015; Jiang et al. 2019a). This makes them resistant to even the most effective antimicrobial medications, rendering once treatable infections untreatable (Mar et al. 2017; Renwick et al. 2016). Antimicrobial resistance has resulted in significant economic damage due to increased patient morbidity and mortality (Boucher et al. 2009; Ventola 2015; Renwick et al. 2016; Dutescu and Hillier 2021; D’Andrea et al. 2019; Tacconelli et al. 2018). Given the lack of success in marketing novel therapeutic antimicrobial agents including teixobactins, antimicrobial nanomaterials, and micro-engineered biomolecules (Mulani et al. 2019; Makabenta et al. 2021; Fatima et al. 2021; Mantravadi et al. 2019; Charbonneau et al. 2020; Hussein et al. 2020), current research has been devoted to sourcing natural antimicrobial products due to their chemical diversity and reported effectiveness as narrow- or broad-spectrum antibiotics (Hutchings et al. 2019; Quinto et al. 2019; Ghrairi et al. 2019). However, further research is required to ensure their clinical utility and to develop a better understanding of their mechanism of action. This highlights the critical requirement to understand the mechanisms behind pathogen resistance development and antimicrobial action.
The bacterial lipid membrane of MDR pathogens plays a significant part in the resistance development towards membrane-targeting antibiotics (polymyxins, β-lactams, glycopeptides, and lipopeptides), which typically penetrate the cell membrane to facilitate cellular entry of medication, or directly disrupt the cell membranes structural integrity to facilitate cell lysis (Kapoor et al. 2017; Epand et al. 2016; Tenover 2006; Dias and Rauter 2019). The membrane lipid profile can dictate the effectiveness of antibiotics and drug-efflux proteins that mediate the expulsion of antibiotics from the bacterium. Pathogen adaptation mechanisms alter the native lipid composition which facilitates structural modifications, including changes in membrane fluidity, organisation, and packing, that circumvents the effects of antibiotics and evades host immune attack (Jiang et al. 2019a, 2019b; Dadhich and Kapoor 2020; Han et al. 2018; Maifiah et al. 2016; Mishra et al. 2012). The unique structure of the membrane in GN bacteria is the primary reason for their rapid resistance development compared to GP bacteria (Breijyeh et al. 2020; Ghai and Ghai 2018). The lipid asymmetry, rigidity, and biochemistry of the LPS molecules in the membrane provide a considerable defensive barrier against numerous antibiotics (Breijyeh et al. 2020; Delcour 2009; Vasoo et al. 2015). Changes in the lipophilic composition and membrane structure can also influence various membrane-associated processes such as protein-lipid electrostatic interactions, ligand-binding, cell-to-cell communication, transport, and protein folding, translocation, and function (Corradi et al. 2019; Collinson 2019; Lin and Weibel 2016; Martens et al. 2019, 2016; Norimatsu et al. 2017; Du et al. 2018).
The bacterial lipid membrane is a viable target for novel antibiotic treatments as the lipophilic composition is crucial to antibiotic efficacy, and targeting the lipid membrane rather than biochemical pathways can prolong antibiotic resistance development (Dias and Rauter 2019; Lam et al. 2016). A better understanding of the bacterial lipid membrane and its interactions with antibiotics is thus imperative for subsequent antibiotic research and development efforts.
However, systematic studies of the bacterial cell membrane structure and its processes are difficult to perform when studying live bacterial cells due to the nanometre dimensions of their membranes as well as their high level of complexity (Behuria et al. 2020). Bacteria also possess a cell wall that requires removal prior to investigating membrane-mediated activities (Brown et al. 2010; Veron et al. 2008). The inherent complexity of biological bacterial cell membranes which contain numerous peptides, sugars, membrane proteins, lipids, and carbohydrates makes systematic investigations difficult (Andersson et al. 2018a; Castellana and Cremer 2006). Pathogenic bacteria especially pose unique investigatory challenges due to rigorous biosafety protocols (Behuria et al. 2020). An alternate method to analyse membrane-associated processes is to purify the bacterial membrane; however, the isolation process requires expensive instrumentation which is difficult to perform in common laboratories (Qing et al. 2019). Due to these limitations, progressions in the understanding of the organisation, structure, and processes that occur in biological bacterial membranes have been driven primarily through research on in vitro model membrane systems (Strahl and Errington 2017).
A variety of different model systems have been designed to mimic biological membranes in a controlled environment with only the most essential components (Salehi-Reyhani et al. 2017). Model membranes were developed as an accessible experimental platform to analyse membrane structure and function in an environment that replicates the fundamental environmental and physiochemical properties of biological membranes, whilst reducing their innate complexity (Andersson et al. 2018a, 2020, 2018b; Andersson and Köper 2016; Chan and Boxer 2007; Jackman et al. 2012; Siontorou et al. 2017). Model membrane systems are computationally modelled, free-standing, or solid-supported bilayer structures composed of various lipophilic compounds and proteins (Chan and Boxer 2007; Siontorou et al. 2017).
They enable the use of numerous microscopic, spectroscopic, electrochemical, reflectometric, and algorithmic analytical techniques often inaccessible when studying live cells (Wiebalck et al. 2016; Zieleniecki et al. 2016). The analytical techniques can, for example, reveal the mechanism of action surrounding membrane-targeting antibiotics (Peetla et al. 2009; Knobloch et al. 2015). Numerous model membrane systems have been designed to investigate membrane-drug interactions (Hollmann et al. 2018); however, few mimic bacterial membranes or the architecture of the ESKAPE pathogens.
Here, we provide an overview of the structure and lipophilic composition of GN and GP bacterial membranes and current membrane modelling systems for these structures, including liposomes, solid-supported bilayers, and computational simulations.
Bacterial membranes
Lipids in bacterial membranes serve as important structural and functional constituents and have important roles in membrane organisation, cell recognition, membrane fluidity, energy storage, direct modulation, membrane stability, cell signalling, and membrane formation (Solntceva et al. 2020; Carvalho and Caramujo 2018; Willdigg and Helmann 2021). To perform such complex and diverse functions, bacterial membranes are composed of approximately equivalent proportions of lipids and proteins and are complex structures with a high degree of organisation and variation between bacterial species and their GN and GP classifications (Strahl and Errington 2017; Epand and Epand 2009a; Sohlenkamp and Geiger 2016).
GN and GP bacterial lipid membranes are predominantly formed by phospholipids which are composed of a phosphate group, 2–4 hydrophobic fatty acid units, a variable hydrophilic head group, and a glycerol moiety (Sohlenkamp and Geiger 2016; Alagumuthu et al. 2019; Fahy et al. 2011). Phospholipids are organised in a classical bilayer described by the fluid-mosaic model (Singer and Nicolson 1972). The model has since been refined to accommodate the presence of lipid domains and cytoskeletal proteins that restrict and sectionalise lipid and protein diffusion (Strahl and Errington 2017; Meer et al. 2008; Barák and Muchová 2013). Both GN and GP bacteria contain a large variety of straight or branched, saturated, or unsaturated carboxylic acids with long aliphatic chains, known as fatty acids, that serve as essential building blocks for multiple lipophilic compounds (Carvalho and Caramujo 2018; Cronan and Thomas 2009). Numerous glycolipids, which are composed of a carbohydrate attached by a glycosidic bond containing 1–2 fatty acid units, are also typical constituents in the membranes of GN and GP bacteria (Bertani and Ruiz 2018; Reichmann and Gründling 2011). In addition to the aforementioned common lipid species, bacteria can also possess species-specific lipids (Solntceva et al. 2020).
Within bacterial species of different and the same Gram types, the lipid membrane contains a high degree of structural, chemical, and functional variability whereby numerous lipid molecular variants are present that differ in size, number, chemical composition, and isomeric form (Strahl and Errington 2017; Sohlenkamp and Geiger 2016; May and Grabowicz 2018; Rahman et al. 2000). Pathogens can also readily acquire multiple exogenous lipophilic bodies which generate substantial variation between pathogen strains and species (Jiang et al. 2019a; Jasim et al. 2018). The key lipid species present in the ESKAPE pathogens has been studied extensively (Table 1) (Sohlenkamp and Geiger 2016).
Table 1.
Diversity of membrane lipid species documented for the ESKAPE pathogens
| Bacterial species | Major membrane lipid species | References |
|---|---|---|
| E. faecium | PG, CL, Lysyl-PG, GP-DGDAG, Type I LTA, FA | Mishra et al. 2012; Theilacker et al. 2012) |
| S. aureus | PG, CL, Lyso-PG, GPL, Lysyl-PG, Type I LTA, FA | Epand and Epand 2009a; Song et al. 2020; Schneewind and Missiakas 2014; Kilelee et al. 2010; Malanovic and Lohner 2016; Oku et al. 2004; White and Frerman 1967) |
| K. pneumoniae | PG, PE, CL, SL, PC, Lysyl-PG, Lyso-PE, PI, PA, Lyso-PA, Lyso-PC, LPS, FA | Epand and Epand 2009a; Jasim et al. 2018; Vinogradov et al. 2002; Hobby et al. 2019) |
| A. baumannii | PE, PG, CL, Lyso-PE, Acyl-PG, PA, MLCL, PE-OH, CL-OH, MLCL-OH, LPS, FA | Jiang et al. 2019a; Unno et al. 2017; Jiang et al. 2020; Lopalco et al. 2017) |
| P. aeruginosa | PG, CL, PE, PC, OL, Alanyl-PG, RL, LPS, FA | Epand and Epand 2009a; Malanovic and Lohner 2016; Chao et al. 2010; Lam et al. 2011; Klein et al. 2009; Lewenza et al. 2011; Pramanik et al. 1990; Wilderman et al. 2002; Soberón-Chávez et al. 2005) |
| Enterobacter species† (E. cloacae, E. hormaechei, and E. aerogenes) | PG, PE, CL, LPS, FA | Epand and Epand 2009a; Bøse and Gjerde 1980; Gill and Suisted 1978; Kämpfer et al. 2015; Davin-Regli et al. 2019; Epand and Epand 2009b; Epand et al. 2010) |
†As there are 22 species found in the Enterobacter genus, only common species described in nosocomial infections were analysed and lipid compositions are assumed to be similar between each (same genus)(Davin-Regli et al. 2019; Epand et al. 2010; Villegas and Quinn 2002)
*See Supplementary Information (Sects. 1 and 2) for bacterial and lipid species acronym definitions, respectively
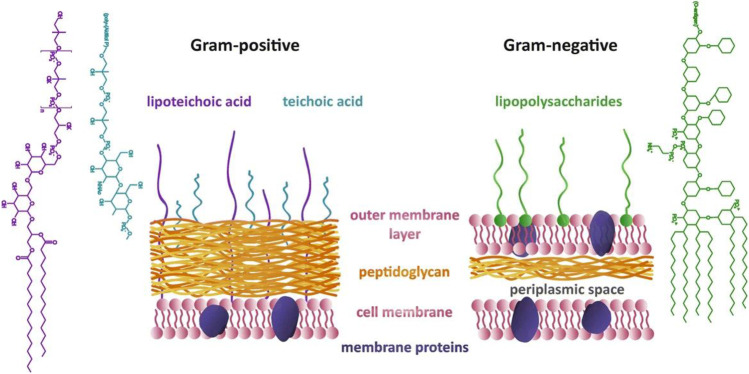
GN bacterial membranes consist of two lipid bilayers separated by a viscous, protein-enriched aqueous periplasmic space and a thin peptidoglycan (murein) wall (Fig. 1) (Kapoor et al. 2017; Barák and Muchová 2013; Silhavy et al. 2010). The inner membrane (IM) is comprised of an asymmetric phospholipid bilayer that encases the cytosol and harbours membrane proteins responsible for transport, energy production, protein secretion, and lipid biosynthesis (Silhavy et al. 2010; Bogdanov et al. 2020). The murein wall is responsible for protecting the bacterium against osmotic and mechanical stresses and maintaining bacterium shape (Kapoor et al. 2017; Silhavy et al. 2010). The outer membrane (OM) is attached to the murein wall via lipoproteins (Silhavy et al. 2010). The OM is an asymmetric lipid bilayer surrounding the periplasmic space (Kapoor et al. 2017; Paulowski et al. 2020). The proximal leaflet is comprised of phospholipids, whilst the distal leaflet is predominantly comprised of LPS which functions as a protective barrier (Silhavy et al. 2010; Cian et al. 2020). LPS is a glycolipid constructed of three distinct parts: lipid A (hydrophobic domain), the oligosaccharide core (hydrophilic domain), and the O-antigen (outmost polysaccharide domain) (Raetz and Whitfield 2002; Wang and Quinn 2010). The structure of LPS differs significantly between GN bacterial species due to survival adaptations in response to changes in environmental stimuli including pH, temperature, specific ion concentrations, osmolality, and toxins (including antibiotics) (Li et al. 2012; Needham and Trent 2013; Trent et al. 2006; Simpson and Trent 2019). Biochemical modifications to LPS domains or selective LPS production abandonment (specific to A. baumannii only) have been found to allow GN bacterial pathogens to evade host-immune attack, increase pathogenesis, and develop antimicrobial resistance (Needham and Trent 2013; Trent et al. 2006; Simpson and Trent 2019; Maldonado et al. 2016; Moffatt et al. 2010; Pelletier et al. 2013), for example, LPS modification adaptation strategies adopted by GN bacteria to protect themselves from cationic antimicrobials such as polymyxins include hydroxylation, dephosphorylation, palmitoylation, phosphatidylethanolamine addition, and 4-amino-4-deoxy-L-arabinose (L-Ara4N) addition to the lipid A portion (Dortet et al. 2020; Olaitan et al. 2014). The most common and effective modification to LPS in GN bacterial pathogens is the addition of L-Ara4N via cationic substitution of the 4’-phosphate group on the lipid A moiety (Olaitan et al. 2014; Nikaido 2003). This modification reduces the net charge of lipid A which, consequently, decreases the degree of electrostatic repulsion experienced between neighbouring LPS molecules. The incorporation of these cationic constituents results in a net positive charge of LPS upon biosynthesis which, inevitably, repulses cationic antimicrobials (Dortet et al. 2020; Olaitan et al. 2014). This repulsion results in antimicrobial resistance as the membrane has developed protection against OM disruption. In addition, murein lipoproteins and β-barrel proteins are present in the OM for murein wall anchoring and small (anions, maltodextrins, and maltose) and large molecule (antibiotics, vitamins and chelates) diffusion or transport (Silhavy et al. 2010).
Fig. 1.
Schematic depiction of the key structural differences in the cell walls of GN and GP bacteria (used with permission from (Pajerski et al. 2019))
The OM and LPS leaflets are absent in most GP bacteria which, in GN bacteria, are crucial in providing an additional stabilising layer around the bacterium and protect the bacterium from environmental hazards (Malanovic and Lohner 2016; Silhavy et al. 2010). To compensate for the OM deficit and withstand the osmotic and mechanical pressures exerted on the plasma membrane, GP bacteria are surrounded by a murein wall that is notably thicker (40–80 nm) in GP bacteria than those found in GN bacteria (7–8 nm) (Kapoor et al. 2017; Epand and Epand 2009a; Barák and Muchová 2013; Malanovic and Lohner 2016; Silhavy et al. 2010). Teichoic acids, including LTA, thread through the murein layers to anchor the murein wall to the membrane and regulate cell envelope function and structure (Malanovic and Lohner 2016; Silhavy et al. 2010). LTA is an alditol phosphate polymer linked by a glycolipid anchor that secures it to the lipid membrane (Solntceva et al. 2020; Percy and Gründling 2014). The structure of LTA varies significantly between GP bacterial species whereby there are five types of LTA (types I–V) that differ in core structure and glycolipid anchor (Percy and Gründling 2014; Shiraishi et al. 2013). Similarly to LPS in GN bacteria, biochemical modifications to the LTA backbone structure have been found to illicit antimicrobial resistance in GP bacterial pathogens (Percy and Gründling 2014; Gutmann et al. 1996; Saar-Dover et al. 2012). For example, the D-alanylation of LTA mediated by the dlt operon and/or incorporation of L-lysine in PG via the mprF gene can lead to an enhanced resistance against cationic antimicrobials (Percy and Gründling 2014; Saar-Dover et al. 2012; Abachin et al. 2002; Peschel et al. 1999; Reichmann et al. 2013). The modification increases the overall net positive surface charge of the membrane and reduces the binding affinity of cationic antimicrobials (Percy and Gründling 2014; Abachin et al. 2002; Peschel et al. 1999). However, other pathways may also be involved in resistance development. The addition of D-alanine, for example, also changes the conformation of LTA resulting in an increase in cell wall density and cell surface rigidity (Percy and Gründling 2014; Saar-Dover et al. 2012). This leads then to a reduction in the permeation of cationic antimicrobials through the cell. The membranes of GP bacteria are comprised of a single asymmetric phospholipid bilayer that encases the cytosol (Silhavy et al. 2010; Rosado et al. 2015; Jones et al. 2008). As there is no OM in GP bacteria to harbour extracellular proteins, GP bacteria are decorated with numerous proteins bound via peptide anchors, covalent interactions, lipid anchors, or non-covalent interactions to the membrane, murein wall, and/or teichoic acids that perform functions analogous to those found in GN bacteria (Malanovic and Lohner 2016; Silhavy et al. 2010; Scott and Barnett 2006).
Model membrane systems
Various model membrane systems have been established. Here, we focus on systems that specifically mimic microbial membranes.
Liposomes
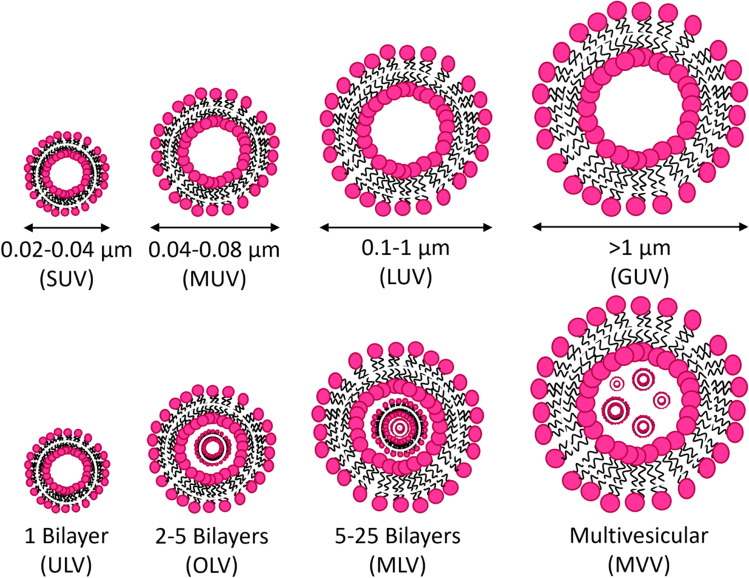
Liposomes are spherical-shaped vesicles ranging from nano- to micrometre diameters that are comprised of one or more phospholipid bilayers that encase an aqueous core (Siontorou et al. 2017; Akbarzadeh et al. 2013). Liposome structures are categorised according to their lamellar structure and vesicular size: unilamellar vesicles (ULV) can be small (SUV, 0.02–0.04 µm), medium (MUV, 0.04–0.08 µm), large (LUV, 0.1–1 µm), and giant (GUV, > 1 µm) (Siontorou et al. 2017; Akbarzadeh et al. 2013; Šturm and Poklar Ulrih 2021). Oligolamellar vesicles (OLV) are > 0.5 µm and can contain 2–5 concentrically arranged bilayers, multilamellar vesicles (MLV) are > 0.7 µm and can contain concentrically arranged 5–25 bilayers, and multivesicular vesicles (MVV) are 1–100 µm and can contain one or more non-concentrically arranged internal bilayers (Fig. 2) (Akbarzadeh et al. 2013; Navas et al. 2005; Giuliano et al. 2021; Mu et al. 2018). Liposomes are easily formed via numerous methods as reviewed elsewhere (Siontorou et al. 2017; Akbarzadeh et al. 2013; Šturm and Poklar Ulrih 2021). Liposome properties can differ depending on the method of preparation, size, lipophilic composition, surface charge, and functionalisation which allows for a considerable degree of customisation (Gabizon et al. 1998; Sherratt and Mason 2018; Fan et al. 2007; Bozzuto and Molinari 2015; Riaz et al. 2018; Sakai-Kato et al. 2019).
Fig. 2.
Schematic representation of different sizes (top) and lamellar structures (bottom) of liposomes
Liposomes have been constructed to mimic the OM, IM, and cytoplasmic space of various non-pathogenic and pathogenic bacteria (Table 2) (Behuria et al. 2020; Bogdanov et al. 2020; Paulowski et al. 2020; Tuerkova et al. 2020; Dombach et al. 2020; Jamasbi et al. 2014; Kumagai et al. 2019; Pérez-Peinado et al. 2018; Malishev et al. 2018; Kahveci et al. 2016; Lopes et al. 2012; Cheng et al. 2011; Marín-Menéndez et al. 2017; Fernandez et al. 2011; Domenech et al. 2009; Pinheiro et al. 2013; D’Errico et al. 2010; Furusato et al. 2018; Kiss et al. 2021; Jiménez et al. 2011; Sikder et al. 2019; Kubiak et al. 2011; Mohanan et al. 2020; Ruhr and Sahl 1985; Bharatiya et al. 2021).
Table 2.
Summary of cited liposome models, the lipid source, the lipid species utilised, and their corresponding research outcomes
| Model type | Reference | Lipid source | Lipid species | Research outcomes |
|---|---|---|---|---|
| GUV | Behuria et al. 2020) | E. coli polar lipid extract (DH5α) | PE, PG, CL | Development of a facile, inexpensive, and reproducible method for producing bacterial GUVs |
| Furusato et al. 2018) | Purchased synthesised lipids | POPC, POPG, Rhod-DOPE | Formation of membrane-associated proteins using a cell-free protein synthesis system inside GUVs | |
| Jiménez et al. 2011) | E. coli lipid extract (JM600) | Unspecified lipid content from the extracts | Incorporation of soluble proto-ring proteins into GUVs for probing of divisome component interactions | |
| Kubiak et al. 2011) | Purchased E. coli B (ATCC 11,303) polar lipid extracts, E. coli (O55:B5) LPS extracts, E. coli (EH-100) LPS extracts, E. coli (J5) LPS extracts, E. coli (F583) LPS, and lipid A extracts and synthesised lipids |
Extracted: PE, PG, CL, S-LPS, FITC-LPS, Ra-LPS, Rc-LPS, Rd-LPS, MPLA Synthesised: Rhod-DHPE |
Development of novel protocol for formation of GUVs composed of LPS species and E. coli extracts | |
| Mohanan et al. 2020) | E. coli B (ATCC 11,303) polar lipid extracts and purchased synthesised lipids |
Extracted: PE, PG, CL Synthesised: DOPG, Lysyl-PG, TOCL |
Development of GUV-based GN and GP bacterial membrane vesicles | |
| Saliba et al. 2014) | Purchased porcine brain extract, S. cerevisiae (yeast) extract, P. cirerri (yeast) extract, and synthesised lipids |
Extracted: PIP, SL (PHS including phosphate forms and phytocer) Synthesised: DAG, POPC, TOCL, DOPS, DOPG, POPE, DOPA, DOPI (including phosphate forms), SL (dihydrocer, cer (including phosphate and fluorescent forms), SO (including phosphate and florescent forms), DHS (including phosphate and fluorescent forms)), PEG350-PE, PEG2000-PE, ATTO647N-DMPE, NBD-PG, bodipy FL-PE Unknown source: ES |
Systematic characterisation of protein-lipid interactions using a microarray of liposomes | |
| Turner et al. 2015) | Purchased synthesised lipids | DOPE, DOPG, TOCL, Lysyl-PG | Analysis of C. botulinum toxin type A using culture and liposomal methods to assess loss of sterility | |
| Paulowski et al. 2020) | LPS extracts from P. mirabilis (R45). Purchased E. coli lipid extracts and synthesised lipids |
Extracted: PE, PG, R-LPS Synthesised: CL, Rhod-DHPE, NBD-PE, FITC-PE |
Demonstrate experimental methods to model the asymmetry of GN bacteria. The model’s usability was assessed for lipid domain analysis and peptide and protein interaction by characterising lipid flip-flop and phase behaviour | |
| LUV | Sikder et al. 2019) | Purchased synthesised lipids | DPPC, DPPG, DPPE | Programmable supramolecular assembly of π-amphiphile(s) for determination of interactions with bacteria and membrane mimicking liposomes |
| Som and Tew 2008) | Purchased E. coli B (ATCC 11,303) total lipid extracts and synthesised lipids |
Extracted: PE, PG, CL, unspecified lipid content, Egg-Lyso-PC Synthesised: DOPE, DOPC, DOPG, DOPS |
Use of a variety of lipid and lipid extract combinations to show that lipid structure and type could be more important than headgroup charge for determining membrane selectivity towards multiple antimicrobial oligomers | |
| Samuel and Gillmor 2016) | Purchased synthesised lipids | DOPG, DOPC, DPPC, DPPG | Examination of kinetics, behaviours and potential mechanisms of the NA-CATH peptide using SUVs | |
| Sborgi et al. 2016) | Purchased E. coli B (ATCC 11,303) polar lipid extract, porcine brain total lipid extract and synthesised lipids |
Extracted: PG, PE, CL, PA, PS, PI, PC, unspecified lipid content Synthesised: DMPC |
Determination that gasdermin D is the direct and final executor of pyroptotic cell death using liposome-inserted gasdermin D | |
| Carrasco-López et al. 2011) | Purchased E. coli B (ATCC 11,303) lipid extract (unspecified) | Polar (PE, PG, CL) or total (PE, PG, CL, unspecified lipid content) | Investigate the activation mechanism of AmpD peptidoglycan amidase to represent the regulatory processes that occur for other intracellular members of the amidase_2 family | |
| Sasaki et al. 2019) | Purchased E. coli B (ATCC 11,303) polar lipid extract and synthesised lipids |
Extracted: PG, PE, CL Synthesised: DAG |
Determination that YidC accelerates MPIase-dependent membrane protein integration | |
| Cheng et al. 2014) | Purchased synthesised lipids | POPC, POPG, POPE | Mechanistic contributions of membrane depolarisation in S. aureus towards the bactericidal activity of ramoplanin | |
| Lombardi et al. 2017) | Purchased bovine heart CL extract and synthesised lipids |
Extracted: CL Synthesised: DOPE, DOPG, DPPE, DPPG, NBD-PE, Rhod-PE, 5-SLPC, 14-SLPC |
Perturbation of lipid membranes by myxinidin mutant WMR due to anionic lipid segregation | |
| Zhang et al. 2014) | Purchased synthesised lipids | DMPC, DMPG, TOCL | Using cardiolipin in liposomes to show that changes in membrane lipid composition can allow bacteria to become resistant to daptomycin | |
| Domenech et al. 2009) | Purchased bovine heart CL extracts and synthesised lipids |
Extracted: CL Synthesised: POPC, DPPG, POPG, POPE |
Investigate the effect of vancomycin and oritavancin on the permeability and organisation of phospholipids in bacterial membrane models | |
| Fernandez et al. 2011) | Purchased synthesised lipids | DMPC, DMPG, d-DPMC, d-DMPG | Investigate the drug-membrane interactions between the synthetic antimicrobial peptide P5 and bacterial and human membrane models using solid-state NMR and circular dichroism | |
| Marín-Menéndez et al. 2017) | Purchased synthesised lipids | POPC, PG, CL | Develop bacterial model membranes to investigate the drug-membrane interactions and delivery mechanism of oligonucleotide therapeutics | |
| Lopes et al. 2012) | Purchased E. coli B (ATCC 11,303) total lipid extracts | PE, PG, CL, unspecified lipid content | Generate model membranes that represent Y. kristensenii and P. mirabilis to determine differences in lipid phase transitions with variations in lipid composition ratios | |
| Jamasbi et al. 2014) | Purchased synthesised lipids | POPE, POPG | Investigate and compare the cytosolic and antimicrobial mechanism of action of the lytic peptide, melittin, between prokaryotic and eukaryotic model membranes | |
| Tuerkova et al. 2020) | Purchased synthesised lipids | POPC, POPG | Investigate the mechanism of action regarding pore formation induced by kinked helical antimicrobial peptides via fluorescence leakage assays | |
| SUV | Kiss et al. 2021) | Purchased E. coli (EH100) LPS extracts and synthesised lipids |
Extracted: Ra-LPS Synthesised: DMPC |
Facile development of synthetic bacterial membrane models through the step-by-step construction of SUVs |
| Brian Chia et al. 2011) | Purchased E. coli B (ATCC 11,303) total lipid extract, bovine brain total lipid extract, and synthesised lipids |
Extracted: PG, PE, CL, unspecified lipid content Synthesised: DMPC, DMPG |
Investigation of peptide selectivity using vesicles to show that natural lipid extracts compare better to MIC values than synthetic lipids | |
| Bharatiya et al. 2021) | Purchased B. subtilis LTA extracts and synthesised lipids |
Extracted: LTA Synthesised: DPPG, DPPE, TMCL |
Investigate how different compositional variations of LTA alter the structural integrity and stability in model GP membranes | |
| Bogdanov et al. 2020) | Lipid extracts from E.coli strains W3110, W3899, EH150, UE54, BKT12, AL95, AT2033, and Y. pseudotuberculosis (O:1b IP32953). Purchased E. coli B (ATCC 11,303) polar lipid extracts and purchased plus in-house synthesised lipids |
Extracted: PE, PG, CL, PS, Lyso-PE, N-acyl-PE, PA, CDP-DAG Synthesised: DPPE, DPPS, TNP-PE, DNP-PE, TNP-LPE, TNP-LPS, TNP-PS, DFDNP-LPE, DFDNP-LPS, DFDNP-PE, DFDNP-PS |
To determine how phospholipids are distributed in the IM of GN bacteria and how different phospholipid species influences the distribution and regulation of phospholipid species across the leaflets. The phospholipid asymmetry is discussed in the context of bacterial growth, phospholipid synthesis and translocation, and adjustments in the physical and chemical properties of the membrane | |
| Cheng et al. 2011) | Purchased bovine heart CL extract and synthesised lipids |
Extracted: CL Synthesised: POPG, POPC, POPE |
Investigate how the lipid composition in GP and GN bacterial models influence the drug-membrane interactions between various cationic antimicrobial peptides | |
| MLV | D’Errico et al. 2010) | LPS extracts from B. cenocepacia ET-12 (LMG 16,656), B. multivorans (C1576), A. tumefaciens (TT111) and S. enterica (minnesota R595). Purchased synthesised lipids |
Extracted: R-LPS, S-LPS, Re-LPS Synthesised: DOPE |
Characterisation of liposome formation based on initial LPS molecular structure |
| Pinheiro et al. 2013) | Purchased synthesised lipids | DMPG, DPPE, DPPG | Investigate the drug-membrane interactions between Rifabutin and bacterial and human membrane models using wide- and small-angle X-ray scattering | |
| Kumagai et al. 2019) | Extracted LPS from P. aeruginosa (PA01) and purchased synthesised lipids |
Extracted: LPS Synthesised: POPE, POPG, TOCL, DOTAP |
Generate model GN and GP membranes to test the function of newly synthesised antimicrobial peptides. The antimicrobials were tested to assess the drug-membrane interactions and killing efficiency | |
| LUV and GUV | Kahveci et al. 2016) | Purchased bovine heart CL extract and synthesised lipids |
Extracted: CL Synthesised: DOPE, DOPG |
Analyse the interactions between mammalian and bacterial membrane models and conjugated fluorophores. The models were used to assess fluorophore-lipid binding affinity for the selective cell recognition |
| SUV and GUV | Malishev et al. 2018) | Purchased bovine heart CL extract and synthesised lipids |
Extracted: CL Synthesised: DOPE, DOPG |
Investigate the differences in protein-membrane interactions of amyloid protein, TasA, between mimic bacterial and eukaryotic cell membranes |
| SUV and LUV | Pérez-Peinado et al. 2018) | Purchased E. coli B (ATCC 11,303) polar lipid extract and synthesised lipids |
Extracted: PE, PG, CL Synthesised: POPC, POPG |
Determine the mechanism of action of the antimicrobial peptides, crotalicidin, and its fragment, on the Om of GN bacteria. Liposome models specifically were used to analyse preferential binding and the degree of membrane disruption |
| Unspecified liposome type | Su et al. 2011) | E. coli (WBB06) LPS extract and purchased synthesised lipids |
Extracted: Re-LPS Synthesised: POPE, POPG, DEPE |
Determination of Gram selectivity among β-hairpin AMPs using LPS-based model systems |
| Hancock and Nikaido 1978) | P. aeruginosa (PAO1) LPS and lipid extracts, and S. typhimurium (LT2M1) LPS and lipid extracts | Unspecified lipid content, R-LPS, S-LPS | Develop an improved method to separate the OM and IM of P. aeruginosa. Saccharide retention between liposomes and proteoliposomes was also investigated to compare exclusion limits between P. aeruginosa and enteric bacteria, S. enterica | |
| Ruhr and Sahl 1985) | S. cohnii (22), B. subtilis (W23), M. luteus (ATCC 4698) and soybean lipid extracts | Unspecified lipid content, Soy-PC | To determine the effect of the peptide antimicrobial, nisin, on the membrane potential and transport processes of GP bacteria | |
| Dombach et al. 2020) | Purchased E. coli B (ATCC 11,303) polar lipid extract | PE, PG, CL | Investigate the mechanism of action of a small molecule found in macrophages, JD1, that declines the survival and/or growth of GN bacteria |
*See Supplementary Information (Sect. 1 and 2) for bacterial and lipid species acronym definitions, respectively
Often GUVs or LUVs are used that contain either bacterial lipid extracts (> 4 lipid species), or synthetic lipids determined by the user (< 3 lipid species) asymmetrically arranged in a bilayer. Liposome formation using bacterial lipid extracts provide a more biologically attune system as various lipid species and their native molecular variants are inherently incorporated. Under an artificially user-defined composition, the inner and outer leaflets for GP liposome models commonly contain PG, lysyl-PG, and CL, whilst GN liposome models commonly contain PE, PG, and CL and uncommonly LPS. Liposome models have been utilised to investigate basic structural (lipid domain architecture, rigidity, diffusion, and lateral organisation) and rheological (constriction, shrinkage, and invagination) membrane properties. In addition, protein and peptide-lipid interactions (Saliba et al. 2014; Su et al. 2011), lipid composition-dependent uptake, release, and molecule function (i.e. membrane-targeting antibiotics) (Kilelee et al. 2010; Som and Tew 2008; Brian Chia et al. 2011), pore formation (Samuel and Gillmor 2016; Sborgi et al. 2016), and protein activity (Carrasco-López et al. 2011; Sasaki et al. 2019) have been explored.
Liposome models have been developed for the ESKAPE pathogens and have been used to investigate host–pathogen interactions, membrane permeability, and the effect of membrane composition on antimicrobial susceptibility (Turner et al. 2015; Cheng et al. 2014; Lombardi et al. 2017; Zhang et al. 2014; Hancock and Nikaido 1978; Ciesielski et al. 2013; Lee et al. 1992; Mitchell et al. 2016). Liposomes from synthetic PC and PG lipids and S. aureus lipid extracts were used to determine the effects of lipid acyl chain branching on antimicrobial peptide activity (Mitchell et al. 2016). This was achieved by measuring efflux kinetics of the encapsulated fluorescent dye carboxyfluorescein, mediated by the model peptide δ-lysin. Liposomes composed of anteiso-branched isomers were less susceptible to peptide-induced perturbations than liposomes containing iso-branched isomers. In addition, liposomes made from S. aureus extracts were more resistant to peptide-induced perturbation than liposomes composed of synthetic lipids, most likely due to the additional increased fraction of anteiso-branched fatty acids.
In a different approach, the association of LPS extracted from K. pneumoniae with eukaryotic lipids has been investigated with respect to host immunodetection strategies (Ciesielski et al. 2013). This was achieved by analysing liposome-liposome interactions between pathogen membrane model liposomes containing LPS and PC and host membrane model liposomes containing PC, SL, and cholesterol. LPS preferentially segregated in ordered SL/cholesterol rich domains which was linked to the evolutionary drive for eukaryotic cells to generate, within such domains, a sensory protein for bacterial detection. The permeability of various carbapenems via porins in proteoliposomes reconstituted from lipids extracted from the OM of susceptible and resistant strains E. cloacae has also been studied (Lee et al. 1992). Carbapenem permeability and efficacy was highly dependent on the lipophilic constitution of the OM and the amount and type of porins present.
While liposomes are very useful systems to study, they pose some challenges for detailed biophysical studies. Lipid composition is often difficult to control (Rideau et al. 2018; Weinberger et al. 2013). Methods to enhance compositional complexity have been developed (Göpfrich et al. 2019; Pautot et al. 2003); however, they can inhibit surface property analysis (Rideau et al. 2018). The metastable structure of liposomes and their susceptibility to lipophilic, oxidative, and hydrolytic degradation offers poor long-term stability (Akbarzadeh et al. 2013; Nkanga et al. 2019). Additionally, lipids often have relatively high phase transition temperatures which impede liposome formation (Eeman and Deleu 2010; Vestergaard et al, 2008). Finally, despite existing stabilisation methods (Schmid et al. 2015), protein reconstitution in liposomes still remains a challenge (Chan and Boxer 2007; Siontorou et al. 2017).
Solid-supported bilayers:
Solid-supported bilayer lipid membranes (sBLMs) consist of a lipid bilayer that is placed onto a solid substrate either via direct contact, via separation by a polymer cushion, or allowed to float directly above a covalently-bound self-assembled monolayer or a supported bilayer (Fig. 3) (Andersson and Köper 2016; Belegrinou et al. 2011; Sackmann 1996; Foglia et al. 2015). Tethered bilayer lipid membranes (tBLMs) are sBLMs with the proximal bilayer leaflet covalently linked to the substrate though thiolipid, oligopeptide, alkane- and aromatic-thiol, polymer, or protein anchors (Andersson and Köper 2016; Andersson et al. 2018b; Jackman et al. 2012; Li et al. 2015; Köper 2007). sBLMs and tBLMs have good electrical sealing properties, are air-stable, and can be formed via Langmuir transfer, vesicle fusion, or solvent-exchange techniques (Andersson et al. 2020; Jackman et al. 2012; Girard-Egrot and Maniti 2021; Kurniawan et al. 2018; Richter et al. 2003).
Fig. 3.
Schematic representation of various solid supported model membrane architectures. Please see text for details
Gold is the most commonly utilised substrate material for sBLMs and tBLMs due to its stability, facile functionalisation, and versatility in surface analysis techniques (Andersson and Köper 2016). However, other substrates including mercury, quartz, glass, aluminium oxide, indium tin oxide, silicon oxide, sapphire, mica, silver, and titanium oxide can also be utilised (Andersson et al. 2018b; Girard-Egrot and Maniti 2021; Clifton et al. 2020; Giess et al. 2004).
Surface sensitive techniques such as surface plasmon resonance, ellipsometry, neutron or X-ray reflectometry, atomic force microscopy, electrochemical impedance spectroscopy, quartz crystal microbalance with dissipation monitoring, and infrared reflection absorption spectroscopy are well-suited methods of surface analysis for these planar systems in aqueous solution (Ferhan et al. 2017; Wittenberg et al. 2014; Steltenkamp et al. 2006).
While these membrane systems commonly have simple lipid compositions, increased biological accuracy can be achieved in both sBLMs and tBLMs by customising the lipid composition to change membrane electrical sealing and structural properties (Andersson and Köper 2016; Andersson et al. 2018b; Girard-Egrot and Maniti 2021). tBLMs can also change the aforementioned membrane properties and facilitate protein incorporation by customising the tethering type, composition, and density. The OM and IM of various non-pathogenic and pathogenic bacteria have been modelled using both tBLMs and sBLMs (Table 3) (Paulowski et al. 2020; Pérez-Peinado et al. 2018; Weiss et al. 2010; Clifton et al. 2013; Paracini et al. 2018; Hughes et al. 2019; Dodd et al. 2008; Michel et al. 2017; Adhyapak et al. 2020; Nakatani et al. 2019; Hoiles and Krishnamurthy 2015; Schneck et al. 2009; Lee et al. 2020; Nedelkovski et al. 2013; Niu et al. 2017; Sharma et al. 2020; McGillivray et al. 2009).
Table 3.
Summary of cited solid supported bilayer models, the lipid source, the lipid species utilised and their corresponding research outcomes
| Model type | Reference | Lipid source | Lipid species | Research outcomes |
|---|---|---|---|---|
| tBLM | Andersson et al. 2018a) | Purchased synthesised lipids and E. coli (J5) LPS extracts |
Extracted: Rc-LPS Synthesised: DPhyPC, d-DPhyPC |
Generate a model membrane that mimics the OM of GN bacteria. Structural and electrical properties were investigated with respect to the influence of divalent ions and antibiotics |
| Weiss et al. 2010) |
Purchased E. coli B (ATCC 11,303) polar lipid extracts |
PE, PG, CL | Develop an assay to assess the activity of cytochrome bo3 in response to the substrate, ubiquinol-10, in the presence of multiple different inhibitors | |
| Nakatani et al. 2019) | Purchased E. coli B (ATCC 11,303) polar lipid extracts | PE, PG, CL | Develop a model bacterial architecture to analyse the catalytic behaviour of Type-II NADH:quinone oxidoreductase in the presence of various the substrates (quinone, quinone analogues and NADH) and inhibitors (phenothiazines) | |
| Hoiles and Krishnamurthy 2015) | Purchased synthesised lipids | POPG, Ether-DPhyPC, DPGE | Investigate pore formation dynamics and reaction-mechanism of the antimicrobial peptide, peptidyl-glycine leucine-carboxyamide, in archaebacterial model membranes | |
| Nedelkovski et al. 2013) | Purchased synthesised lipids | DPhyPC | Generate a biomimetic bacterial membrane architecture that produces enhanced infrared signals to better analyse the photoexcitation mechanism of photosynthetic reaction centres in R. sphaeroides | |
| Niu et al. 2017) | Purchased synthesised lipids and LPS extract from S. enterica (minnesota R595) |
Extracted: Lipid A Synthesised: DPhyPC, DPhyPG |
Investigate the molecular mechanism, interactions, and impact of the antimicrobial peptide, V4, on the electrical and mechanical properties of bacterial membrane models | |
| McGillivray et al. 2009) | Purchased synthesised lipids | DPhyPC | Develop a model bacterial membrane to analyse the structural and electrical properties and lipid-protein interactions of α-hemolysin channels derived from S. aureus | |
| Hsia et al. 2016) | Purchased synthesised lipids and E. coli (JC8031) lipid extracts |
Extracted: unspecified lipid content from the extracts Synthesised: DOPC, PEG5000-PE |
Develop a model membrane of the OM of GN bacteria. The formation of the membrane was characterised kinetically and acoustically to assess surface coverage, vesicle rupture and architecture mass. Properties including membrane diffusivity, mobility, viscoelasticity and lipid and protein symmetry were also investigated. Changes in membrane properties, mass and kinetics were also investigated in the presence of antibiotics | |
| Thomas et al. 1999) | Purchased synthesised lipids and E. coli (K12 D31m4) lipid A extracts |
Extracted: DPLA Synthesised: DMPC, Biotin-PE |
Investigate and identify the sequestering effectiveness and neutralisation mechanism between LPS and polymyxin B compared to polymyxin B synthetic peptide mimics | |
| Spencelayh et al. 2006) | E. coli JM1100 (pPER3) and purchased egg lipid extracts | Egg-PC, unspecified lipid content from the E. coli extracts | Generate a biomimetic bacterial membrane that facilitates the in vitro synthesis of peptidoglycan using native precursors. The binding behaviour between different antibiotics and the peptidoglycan precursors | |
| Mirandela et al. 2019) | Purchased synthesised lipids and E. coli B (ATCC 11,303) polar lipid extracts |
Extracted: PG, PE, CL Synthesised: POPC |
Investigate how the lipid-protein interaction between a mimetic GN lipid bilayer and an ammonium transporter protein native to E. coli affects transporter activity | |
| Maccarini et al. 2017) | Purchased synthesised lipids | DMPC, GDPE, DPEPC, DOPC, DOPE, DMPA, cholesterol | Develop a procedure to optimise the cell-free production of and incorporation of a porin from P. aeruginosa in a functional conformation | |
| Jeuken et al. 2006) | Purchased E. coli B (ATCC 11,303) polar lipid extracts | PG, PE, CL | Characterise the function and structure of redox-active enzyme, cytochrome bo3, derived from E. coli | |
| Jeuken et al. 2005) | Purchased E. coli B (ATCC 11,303) polar lipid extracts and egg lipid extracts. B. subtilis (3G18/pBSD1200) lipid extracts | PG, PE, CL, Lysine-Acyl-PG, egg-PC, unspecified lipid content from the B. subtilis extracts | Electrochemically characterise the function of redox-active membrane protein, succinate menaquinone oxidoreductase, native to B. subtilis | |
| Dupuy et al. 2018) | Purchased synthesised lipids and E. coli (O111:B4) LPS extracts |
Extracted: S-LPS Synthesised: POPE, POPG, TOCL, POPC, DOTAP, KDO2, DLPG |
Develop model GP and GN bacterial membranes to elude the biophysical interaction mechanism between the antimicrobial peptide Colistin and different lipid compositions | |
| Hughes et al. 2019) | Purchased synthesised lipids and E. coli (EH100) LPS extracts |
Extracted: Ra-LPS Synthesised: d-DPPC |
Collect biophysical information and investigate the physical properties of the OM of GN bacteria using model membranes and computational simulations | |
| Mohamed et al. 2021) | Lipid extracts from P. aeruginosa (PA14), A. baumannii (LAC-4) and E. cloacae (ATCC 13,407) and purchased synthesised lipids |
Extracted: unspecified lipid content but LPS was detected and quantified Synthesised: PEG5000-POPC, PEG5000-DHPE |
Generate OM model bilayers of three GN ESKAPE pathogens and investigate the model’s biophysical characteristics, and drug-membrane interactions with various antimicrobial compounds | |
| sBLM | Adhyapak et al. 2020) |
M. smegmatis (mc2155) lipid extracts |
PA, PE, PG and PI (including lyso forms); CL; DAG (including meromycolyl forms); SfL; DAT; GPepL; MA (including alpha and keto forms); PIM (including mono-acylated forms); TAT; MG; MPM; TDM; MB (including carboxy, cell-bound iron-loaded, monodeoxy, dideoxy and hybrid forms); MQ; PDIM; Ac2SGL; TG; DG; PCA (including hydroxy forms); CET; GPD; MCA; MPanA; MpenA; MSA; MCSA; L5P | Investigate the membrane lipid domain architecture, fluidity, packing, dynamics, synthesis regulation and lateral organisation in protein-free membrane models of mycobacteria |
| Schneck et al. 2009) | S. enterica (R60 and R595) lipid extracts | Lipid A, Ra-LPS-Ra, Re-LPS | Model the influences of different LPS mutations on the mechanical properties and intermembrane interactions in the presence and absence of divalent ions using GN bacterial OM models | |
| Lee et al. 2020) | E. coli BL21 (K-12 MG1655) total lipid extracts | PE, PG, PA | Investigate the impact of the antimicrobial peptide, maculatin 1.1, on the mechanical properties of lipid domains in bacterial membrane models simulating exponential and stationary growth phases | |
| Sharma et al. 2020) | Purchased E. coli B (ATCC 11,303) total lipid extracts and E. coli (O111:B4) LPS extracts and synthesised lipids |
Extracted: S-LPS, PE, CL, PG, unspecified lipid species Synthesised: POPE, ATTO488-DMPE, ATTO647N-DMPE |
Generate a model membrane that mimics the OM and IM of E. coli. Membrane lipid diffusiveness, fluidity, packing, and mobility was analysed with respect to the transport of the antimicrobial thymol | |
| Clifton et al. 2015) | Purchased E. coli (EH100) LPS extracts and synthesised lipids |
Extracted: Ra-LPS, Synthesised: DPPC, d-DPPC |
Generate an asymmetric model membrane that mimics the IM and OM of E. coli | |
| Li and Smith 2019) | Purchased synthesised lipids | POPG, DOTAP, TOCL, POPE, TopFluor-PE, TopFluor-TOCL | Develop model GP and GN asymmetric bacterial IMs. Lipid diffusion dynamics was investigated in the presence and absence of antimicrobial peptide binding | |
| Michel et al. 2017) | Purchased synthesised lipids and LPS extract from S. enterica (minnesota R595) |
Extracted: Re-LPS Synthesised: SOPE, SOPG, TOCL, d-POPG, d-POPE |
Develop and characterise a model GN asymmetrical bacterial IMs to antimicrobial plasticins | |
| Paulowski et al. 2020) | LPS extracts from P. mirabilis (R45). Purchased E. coli lipid extracts and synthesised lipids |
Extracted: PE, PG, R-LPS Synthesised: CL, Rhod-DHPE, NBD-PE, FITC-PE |
Demonstrate experimental methods to model the asymmetry of GN bacteria. The model’s usability was assessed for lipid domain analysis and peptide and protein interaction by characterising lipid flip-flop and phase behaviour | |
| Dodd et al. 2008) | Purchased synthesised lipids and E. coli (BL21(DE3)) lipid extracts |
Extracted: unspecified lipid content Synthesised: Egg-PC, TRF-DHPE, NBD-PC |
Generate sBLMs that contain mixtures of native E. coli lipids with Egg-PC with the intention of generating a simple model membrane for the study of drug-membrane interactions and numerous process that occur in bacterial membranes. The structural properties of the generated sBLMs were assessed using various surface sensitive analytical techniques | |
| Clifton et al. 2013) | Lipid A and LPS extracts from E. coli strains F583, EH100 and J5. Purchased synthesised lipids |
Extracted: lipid A, Ra-LPS, Rc-LPS Synthesised: DPPC, d-DPPC |
Develop a facile two-step approach to modelling the OM of GN bacteria. Via neutron reflectometry, the lipid distribution and coverage between leaflets, and membrane stability and structure were analysed | |
| Pérez-Peinado et al. 2018) | Purchased E. coli B (ATCC 11,303) polar lipid extract and synthesised lipids |
Extracted: PE, PG, CL Synthesised: POPC, POPG |
Determine the mechanism of action of the antimicrobial peptides, crotalicidin and its fragment, on the Om of GN bacteria. sBLM models specifically were used to analyse the membrane permeabilisation mechanism | |
| sBLM and tBLM | Chilambi et al. 2018) | Purchased synthesised lipids and E. faecalis OG1RF (wild type), EFC3C and EFC3Py (resistant strains) extracts |
Extracted: unspecified lipid species from extracts, various FAs Synthesised: DPDEPC, GPDE, DOPC, POPG |
Investigate the antimicrobial mechanism of antimicrobial conjugated oligoelectrolytes through changes in the fatty acid, genetic and uptake profiles between wild type and resistant strains of E. faecalis |
| Paracini et al. 2018) | Purchased synthesised lipids and LPS extract from E. coli (EH100) |
Extracted: Ra-LPS Synthesised: d-DPPC |
Investigate how the physical structure of the lipid OM of GN bacteria influences the drug-membrane interactions of polymyxin B |
*See Supplementary Information (Sect. 1 and 2) for bacterial and lipid species acronym definitions, respectively
These architectures often contain a limited number (1–4) of synthetic lipid species; however, they can also contain bacterial lipid extracts (> 4 lipid species) asymmetrically arranged in a bilayer. Unlike user-defined systems which are limited to the number and type of lipid species and their associated molecular variations incorporated, architectures formed from bacterial lipid extracts generate increasingly accurate biological models as various lipid species and their native molecular variants are inherently incorporated. Under user-defined compositions, the inner and outer leaflets of architectures modelling GN and GP bacteria commonly contain one molecular variation of PC. Few architectures have been developed where the inner and outer leaflets contain the most common lipid species or analogues thereof for GN (PE, PG and CL) and GP (PG, CL, and lysyl-PG) bacteria. For sBLM and tBLM systems, lysyl-PG is often substituted with DOTAP as it is more affordable for the increased quantities required to generate the architectures (Dupuy et al. 2018; Li and Smith 2019). Few architectures modelling the membrane of GN or GP bacteria have also been developed to contain LPS (Andersson et al. 2018a; Clifton et al. 2015; Hsia et al. 2016; Thomas et al. 1999) or murein (Spencelayh et al. 2006). The model architectures have been utilised to investigate general structural (thickness, roughness, and lipid density) and electrical membrane properties. In addition, the mechanism of interaction between antibiotic compounds and membrane constituents (Chilambi et al. 2018; Dupuy et al. 2018; Li and Smith 2019), lipid-protein interactions (Mirandela et al. 2019), ion transport (Maccarini et al. 2017), and redox-active enzyme function and characterisation (Jeuken et al. 2006, 2005) have been explored.
Limited architectures have been generated to model the ESKAPE pathogens and investigate electrochemical and structural changes with lipophilic composition (Jiang et al. 2019b; Mohamed et al. 2021; Zang et al. 2021). Recently, a tBLM for A. baumannii has been developed to model the OM in the presence and absence of exogenously incorporated omega-3 polyunsaturated fatty acid (PUFA) and docosahexaenoic acid (DHA) (Zang et al. 2021). Both tBLMs generated were asymmetrical and were constructed from lipid samples extracted from A. baumannii actively growing in the presence or absence of DHA. The tBLMs were used to determine whether DHA incorporation disrupted the function of efflux system AdeB due to impaired proton motive force retention from induced ion leakage. Both tBLM models were electrochemically similar therefore suggesting that AdeB dysfunction was not due to the membrane’s ability to maintain a proton motive force upon DHA incorporation. sBLM models for S. aureus have been developed to assess how upregulation in CL biosynthesis in daptomycin-resistant strains decreases antibiotic susceptibility (Jiang et al. 2019b). PG, lysyl-PG and CL in different concentration ratios were used to mimic resistant and susceptible strains. The daptomycin-resistant strain membrane was found to be thicker than the susceptible strain. The structural changes resulted in concentration-dependent changes in daptomycin interaction. At low daptomycin concentrations, the susceptible strain exhibited decreases in lipid volume whilst high concentrations induced considerable membrane penetration and disruption. In contrast, the resistant-strain exhibited only slight lipid volume reductions for all daptomycin concentrations analysed. This demonstrated that lipid-induced structural modifications can impair daptomycin efficacy.
Both sBLM and tBLM systems possess limitations unique to each architecture. sBLM systems can be unstable due to no linkage between the lipid bilayer and the substrate (Andersson and Köper 2016; Andersson et al. 2018b; Girard-Egrot and Maniti 2021). As a result, measurements requiring days or weeks are difficult to achieve. Direct bilayer-substrate contact can also create an insufficient amount of space for bilayer-spanning protein incorporation (Castellana and Cremer 2006; Andersson and Köper 2016; Alghalayini et al. 2019; Tamm and McConnell 1985). Protein-substrate contact induces denaturation or impaired function which hinders functional, electrical, or structural studies (Alghalayini et al. 2019; Tanaka and Sackmann 2005). Membrane structural and electrical properties are also subject to substrate topology, whereby any substrate imperfections will cause defects in the bilayer and hinder its resistance towards current transfer (Andersson and Köper 2016; Andersson et al. 2018b; Girard-Egrot and Maniti 2021). Using a polymer cushion to support the bilayer can partially reduce substrate topological effects, maintain bilayer fluidity, and prevent substrate-protein contact (Andersson and Köper 2016; Andersson et al. 2018b; Belegrinou et al. 2011). However, polymer cushion swelling behaviour, assembly, thickness, and morphology are difficult to control which dampens the electrical qualities of the lipid bilayer (Naumann et al. 2001, 2002). tBLMs were generated to circumvent all aforementioned limitations of sBLMs. However, the disadvantage of increased stability and electrical sealing in tBLM systems is decreased lateral lipid mobility (Andersson et al. 2018b). Depending upon the application, there are also disadvantages to using different types of tethers (Jackman et al. 2012). Similarly to liposomes, consideration of the lipid phase transition temperature can be crucial to successful lipid incorporation and architecture formation (Eeman and Deleu 2010; Vestergaard and d., Hamada, T., Takagi, M., 2008).
Computational modelling
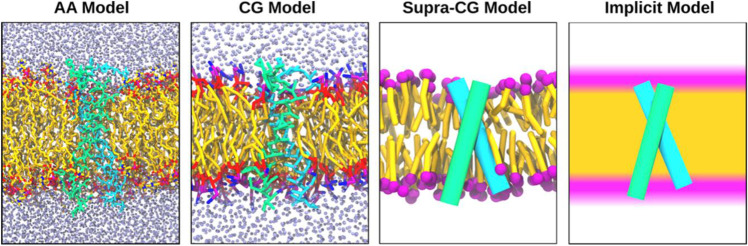
Despite the progress made in developing sophisticated experimental techniques that can directly investigate live bacterial cells and reveal complex lateral membrane organisation processes (Deleu et al. 2014; Lyman et al. 2018; Nickels et al. 2015), analysing the molecular details surrounding membrane organisation still proves difficult (Maity et al. 2015; Marrink et al. 2019). Molecular dynamics (MD) techniques can serve as a “computational microscope” whereby interactions between all constituents in the system can be analysed at an atomistic level (Marrink et al. 2019; Ingólfsson et al. 2016). The quality of the set of parameters that dictate particle interaction, known as the force field (FF), is crucial to the success of an MD simulation (MacKerell 2004). In biomolecular simulations, numerous FFs have been employed: implicit, supra-coarse-grain, coarse-grain, and all-atom (Marrink et al. 2019; Mori et al. 2016). All FFs are similar regarding their main approximations and function; however, the level of resolution between each is distinctive (Fig. 4) (MacKerell 2004). The highest level of resolution is full atomistic detail which is the most commonly utilised model for complex membrane systems. These include bacterial membranes, organelle membranes, plasma membranes and viral envelopes, protein folding, drug-membrane interactions, protein–ligand complex stability, protein–protein interaction modulators, lipid domain formation and behaviour, membrane curvature sensing and formation, membrane remodelling events, and lipid-protein binding site identification and binding strength (Matamoros-Recio et al. 2021; Bennett and Tieleman 2013; Chan et al. 2015; Kabedev et al. 2021; Khan et al. 2019; Lazim et al. 2020; Liu et al. 2021; Parkin et al. 2015; Reddy and Sansom 2016; Singharoy and Schulten 2017). Full atomistic detail significantly expands the predictive power of molecular dynamics simulations. To enhance the spatiotemporal range of MD simulations and decrease system complexity, the lower resolution level FFs can be utilised (Mori et al. 2016; Liu et al. 2021).
Fig. 4.
Schematic representation of different resolutions in molecular dynamics simulations of lipid membranes. All atom (AA) resolution explicitly considers all atoms. Coarse-grain (CG) resolution considers small atom groups and their associated hydrogens. Supra-CG resolution represents solvents implicitly and proteins and lipids as qualitative few-bead models. Implicit resolution further integrates out lipid molecules. (modified with permission from (Marrink et al. 2019))
Several MD models simulating the OM and IM of bacteria have been constructed at both the atomistic and coarse-grained levels of resolution (Table 4). (Bogdanov et al. 2020; Tuerkova et al. 2020; Hughes et al. 2019; Balusek and Gumbart 2016; Baltoumas et al. 2019; Gao et al. 2020; Kholina et al. 2020; Li and Guo 2013; Abellón-Ruiz et al. 2017; Berglund et al. 2015; Hsu et al. 2017a, 2017b; Ma et al. 2017a, 2017b, 2015; Mehmood et al. 2016; Orekhov et al. 2018; Shearer et al. 2019; Shearer and Khalid 2018; Rice and Wereszczynski 2018; Patel et al. 2016; Piggot et al. 2011; Carpenter et al. 2016; Fleming et al. 2016; Wu et al. 2013, 2014a; Duay et al. 2019; Khondker et al. 2019; Pandit and Klauda 2012; Pothula et al. 2016; Shahane et al. 2019).
Table 4.
Summary of cited computational models, the bacterial species modelled, and the lipid species utilised and their corresponding research outcomes
| Model type | Reference | Modelled bacterial species | Lipid species | Research outcomes |
|---|---|---|---|---|
| Atomistic (all-atom) | Balusek and Gumbart 2016) | _ | POPE, Ra-LPS modelled from E. coli strain K-12 | Investigate transport protein-LPS interactions and its effect on Ca2+ binding for vitamin transport in GN bacteria |
| Duay et al. 2019) | _ | POPE, POPG | Determine how Zn ions and pH affect the binding of the antimicrobial peptide, ClavA, to a membrane | |
| Khondker et al. 2019) | _ | POPC, POPS, DMPS | Investigate how the molecular density of a bilayer plays a significant role in the interactions of antimicrobial drugs with the membrane | |
| Pandit and Klauda 2012) | E. coli | POPE, POPG, PMPE, multiple molecular variations of PE and PG that mimic the main constituents of E. coli strains K12 LM 3118 and K12 NBRC 3301, PDSPE, PDSPC | Introduce cyclic moieties into the membrane to obtain a more realistic model | |
| Shahane et al. 2019) | _ | POPE, POPG | Determine how membrane composition influences the interaction with various antimicrobial peptides | |
| Khakbaz and Klauda 2015) | E. coli | POPE, PPPE, OSPE, PMPE, QMPE, PSPG, PMPG | Simulated parameters of complex membrane composition and compared how they differ significantly from simpler models | |
| Lim and Klauda 2011) | C. trachomatis | DPhyPC, 13-MpPPC, 14-MpPPC, DPPC, DMPE, DOPE, DOPG, SLPC, PPPE, DSPE, DLPE, POPE, cholesterol | Determine how increased lipid chain branching affects bilayer properties such as elastic modulus and chain order | |
| Jin et al. 2021) | _ | DOPC, POPE, POPG | Demonstrates the interaction of model membranes with various native and non-native small molecules used in quorum sensing | |
| Lee et al. 2017) | P. aeruginosa, E. coli | PPPE, PVPG, PVCL, R-LPS, S-LPS | Investigate how the composition of a membrane influences its interaction with an OM protein | |
| Ocampo-Ibáñez et al. 2020) | P. aeruginosa and K. pneumoniae | POPE, PMCL, POPG | Investigates how the interactions between the membrane and the cationic antimicrobial peptide, CecD, depends on the membrane composition | |
| Alkhalifa et al. 2020) | E. coli, S. aureus | POPC, DLPG, DLPE, TMCL | Determines how the membrane composition influences membrane interaction with various quaternary ammonium compounds | |
| Piggot et al. 2011) | E. coli, S. aureus | LPS, Lysyl-DPPG, POPE, POPG, DMPG, DPPE, CL | Demonstrates how membranes of various lipid composition show different electroporation properties | |
| Lins and Straatsma 2001) | P. aeruginosa (PAO1) | PE, R-LPS | Detailed description of the construction of an LPS membrane | |
| Yu and Klauda 2018) | P. aeruginosa (PAO1) | POPE, POPG, YOPE, PMSPG, PMSPE, DPPE, YOPG, DPPG | Description of a simulation using the CHARMM (Chemistry at Harvard Macromolecular Mechanics) FF to simulate in IM of P. aeruginosa | |
| Hwang et al. 2018) | E. coli | POPE, POPG, PMPE, QMPE, PMPG, PSPG, OSPE, Ra-LPS | Mechanical properties of the membrane are influenced by both the cell wall as well as the OM | |
| Bogdanov et al. 2020) | _ | DOPE, DOPG, TOCL, FDNB-PE | Elucidate the mechanism behind the inability of 1,5-difluoro-2,4-dinitobenzene to be able to cross-link PE based on phospholipid location in GN bacterial model membranes | |
| Piggot et al. 2013) | E. coli | POPC, PVPE, PVPG, PVCL, Rd-LPS | Model a transporter protein FecA, native to E. coli to identify various LPS-protein interactions and determine how it affects the conformational dynamics of FecA | |
| Kirschner et al. 2012) | P. aeruginosa (PAO1) | R-LPS, DPPE | Extend the GLYCAM06 FF to incorporate a new set of parameters that expands the number of monosaccharides that can be added to LPS and, consequently, improve the structure reproduction and membrane permeability for GN bacterial membrane models | |
| Wu et al. 2013) | E. coli | R-LPS, S-LPS | To build and model each LPS constituent based on chemical and spectroscopy investigations. Each consistent in LPS was used to gain insight on LPS properties, LPS molecule dynamics and LPS structure within an LPS bilayer. The addition of the O-antigen was also implemented to investigate how the O-antigen chain heterogeneity influenced membrane dynamins, structure, and properties. Simulations of the O-antigen were validated via NMR | |
| Dias et al. 2014) | P. aeruginosa | DPPE, R-LPS | Investigate how the chemical remodelling of LPS affects the electrostatic properties and structural dynamics of the OM of GN pathogen P. aeruginosa | |
| Wu et al. 2014a) | E. coli | PPPE, PVCL, PVPG, R-LPS | Investigate the structural properties the E. coli OM and any protein-lipid interactions experienced between the OM and phospholipase A | |
| Carpenter et al. 2016) | E. coli | Re-LPS, PE, PG, CL | Determine the free energy of permeation of ethane, benzene, hexane, ethanol, water, and acetic acid through an OM model of E. coli | |
| Fleming et al. 2016) | E. coli | R-LPS, PPPE, PVPG, PVCL | Investigate the conformation flexibility of transmembrane transporter protein, BamA, to determine how membrane interactions with the polypeptide transport-associated domain influence conformation dynamics | |
| Patel et al. 2016) | E. coli | PPPE, PVPG, PVCL, DMPC, R-LPS and S-LPS modelled from the LPS structure of E. coli strain K12 | Investigate the impact of how structural differences in various LPS molecules affect the function, dynamics, and structure of the transport protein OmpF. In addition, the importance of protein-LPS interactions was investigated to determine ion permeability and pore access behaviour in different LPS environments | |
| Rice and Wereszczynski 2018) | S. enterica | POPE, LPS (8 different variations both modified and unmodified) | Generate symmetric GN bacterial OMs to determine how the key lipid A differences in S. enterica alter bacterial virulence via changes in membrane properties | |
| Hughes et al. 2019) | _ | DPPC, R-LPS modelled from the LPS structure of E. coli strain K12 | Investigate the physical properties and biophysical behaviour of the GN bacterial OM including the lateral packing, lipid asymmetry, bilayer density and lipid profile. The results from the simulation were compared to experimental models to determine the degree of agreeability between the methods | |
| Li and Guo 2013) | _ | DOPE, DOPG | Investigate drug-membrane interactions to comprehend the mechanism of action of the antimicrobial EO-OPE-1 (C3) | |
| Gao et al. 2020) | _ | DPPG, PSPG, PVPG, R-LPS and S-LPS were modelled from the LPS structure of E. coli | Determine changes in membrane structural properties and lipid-membrane interactions upon the incorporation of enterobacterial common antigen glycoconjugates | |
| Course-grain | Ma et al. 2017b) | H. pylori, P. gingivalis, B. fragilis, B. pertussis, C. trachomatis, C. jejuni, N. meningitidis, and S. enterica | Lipid A variants from each species analysed, DPPE | To investigate how the molecular profile of lipid A significantly affects the biophysical properties of the membrane such as phase transition temperatures |
| Ma et al. 2015) | _ | R-LPS, S-LPS, DPPE, various polysaccharides | Simulate a full GN bacterial membrane with an OM, peptidoglycan later and an IM | |
| Oosten and Harroun 2016) | P. aeruginosa | R-LPS, POPE | An optimised simulation for a full LPS membrane | |
| Hsu et al. 2016) | _ | POPE, Re-LPS, Ra-LPS | Investigate how the interaction of fullerenes with membrane is dependent on the membrane composition, especially the LPS structure | |
| Shearer et al. 2020) | E. coli | POPE, POPG, CL, Re-LPS | To test numerous simulation methods to determine the best protocol for lipid convergence. This is tested by quantifying the potential of mean force for LPS and phospholipid extraction from model GN bacterial IM and OM bilayers, and lateral mixing of LPS and phospholipids within model GN bacterial IM and OM bilayers | |
| Shearer et al. 2019) | E. coli | Re-LPS, Ra-LPS, DPPC, POPG, POPE, S-LPS, S-LPS-PE | To investigate protein-lipid interactions influenced by the amount of LPS, lipid mobility and protein composition on the function of six native proteins in E. coli | |
| Berglund et al. 2015) | E. coli | Re-LPS, PVCL, PVPE, PVPG | Investigate the mechanisms of interaction between the antimicrobial peptide, polymyxin B1, with the OM and IM of E. coli | |
| Ma et al. 2017a) | E. coli | DPPE, POPG, CL, Lipid A alone, Lipid A attached to its core oligosaccharides | Determine the structural properties of Lipid A with and without its core oligosaccharides, and investigate the stepwise oligomerisation process of OmpF monomers into more complex dimer and trimer structures | |
| Hsu et al. 2017b) | _ | Ra-LPS, Re-LPS, POPG, POPE, CL | Generate a new feature for CHARMM-GUI Martini Maker via simulating micelle, nanodisc, vesicle, and bilayer systems in the absence and presence of membrane proteins to allow users to model complex bacterial OMs containing LPS | |
| Orekhov et al. 2018) | _ | DPPE, POPC, POPE, POPG, Ra-LPS modelled from P. aeruginosa strain PAO1 | Investigate the solvation behaviour of substituted polycationic metallophthalo-cyanines, which can result in photodynamic inactivation of GN and GP bacteria, in model bacterial membranes. The models were further utilised in investigating the molecular structure of substituted polycationic metallophthalo-cyanines, and their interactions with the membrane | |
| Mehmood et al. 2016) | E. coli | POPE, POPG, CL | Determine which phospholipids specifically bind to the ATP-binding cassette transporter McjD in different phospholipid membrane compositions, and investigate how they impact the function and stability of the transporter | |
| Shearer and Khalid 2018) | _ | POPE, POPG, CL, LPS | Investigate the differences in membrane dynamics and structure between symmetrical and asymmetrical GN bacterial membranes in the presence and absence of transmembrane proteins | |
| Hsu et al. 2017a) | E. coli (K12) | POPE, PVPG, CL, Re-LPS | Construct a model IM and OM of E. coli decorated with various native membrane proteins and connected by the transmembrane multi-drug efflux protein complex AcrBZ-ToIC. The model was used to investigate membrane curvature based, lipid diffusion, protein and lipid movement, lipid flow, lipid movement and protein-lipid interactions | |
| Kholina et al. 2020) | _ | POPG, POPE | Determine how various cationic antiseptics interact with model membranes by monitoring membrane structural changes | |
| Tuerkova et al. 2020) | _ | POPC, POPS, POPG | Determine how kinks in helical antimicrobial peptides affects membrane pore formation | |
| Atomistic (all-atom) and course-grain | Abellón-Ruiz et al. 2017) | _ | Re-LPS, POPE | Characterise and analyse the functional mechanism, structure, and lipid membrane interactions of the GN OM lipoprotein MlaA |
| Baltoumas et al. 2019) | _ | LPS (modelled from P. aeruginosa and E. coli), Lipid A (modelled from E. coli, P. aeruginosa, H. pylori, N. meningitidis), POPC, DOPC, DSPC, DPPC, DMPC, DLPC, POPE, DOPE, DSPE, DPPE, DMPE, DLPE, POPS, DOPS, DSPS, DPPS, DMPS, DLPS, POPG, DOPG, DSPG, DPPG, DMPG, DLPG, CL (both mono-and di-anionic forms) | Comparing the versatility and abilities of the program GNOMM (Gram-Negative Outer Membrane Modeler) in constructing and analysing the complex OM of GN bacteria across four different FFs |
*See Supplementary Information (Sect. 1 and 2) for bacterial and lipid species acronym definitions, respectively
These models often contain 2 or more different lipid species asymmetrically arranged in a bilayer, with the outer and inner leaflets composed primarily of LPS (restricted to the outer leaflet) and/or a mixture of PE, PG and sometimes CL. To compensate for the significant variation in the constituents of the phospholipids and LPS between bacterial strains and species, a range of different phospholipid and LPS fragments and variants have been parametrised for use in MD programs (Lee et al. 2018; Wu et al. 2014b). The models have been utilised to characterise and explore various membrane channels and bacterial membrane properties including divalent cation binding, density, diffusion, packing, rigidity, and average area per lipid. In addition, lipid changes between bacterial growth cycles (Khakbaz and Klauda 2015; Lim and Klauda 2011), effects of mechanical and oxidative stressors (Hwang et al. 2018), molecule permeation and partitioning (Jin et al. 2021; Hsu et al. 2016), and the lipophilic influence on membrane protein function and packing (Khalid et al. 2015; Patel et al. 2017) have also been explored.
Bacterial membranes modelling the ESKAPE pathogens have also been simulated to investigate drug-membrane interactions, lipid-protein interactions, and structural changes associated with bacterial pathogenesis (Zang et al. 2021; Piggot et al. 2011; Lee et al. 2017; Ocampo-Ibáñez et al. 2020; Alkhalifa et al. 2020; Lins and Straatsma 2001; Yu and Klauda 2018; Kirschner et al. 2012; Dias et al. 2014; Oosten and Harroun 2016; Chakraborty et al. 2020; Kim et al. 2016). Models for A. baumannii containing the OM/IM spanning AdeB RND drug-efflux complex in the presence and absence of incorporated host-derived PUFAs, arachidonic acid, and DHA have been developed within the coarse-grained FF to investigate PUFA-mediated antibiotic susceptibility (Zang et al. 2021). All three simulated membranes were asymmetrical, contained three different lipid species notably PG, CL, and PE and 2–7 molecular variations of each. PUFA incorporation was shown to morphologically disrupt AdeB, resulting in impaired efflux function and presented a potential weakness in A. baumannii’s MDR capacity. Chakraborty et al. (2020) also explored various drug-membrane-dependent interactions of two antimicrobial peptides, battacin analogues octapeptide 17 and pentapeptide 30, with the IM of S. aureus using an atomistic FF (Chakraborty et al. 2020). The IM was an asymmetric three-component mixture predominately of PG, lysine-PG, DPG, and CL. Kim et al. (2016) modelled homogenous bilayers from 12 pathogenic bacterial species, including A. baumannii, K. pneumoniae, and P. aeruginosa, using an atomistic FF to investigate atomistic-scale similarities and differences in membrane properties induced by the structural variations in LPS (Kim et al. 2016).
Molecular dynamic simulations can provide a detailed picture of membrane structure, yet they sometimes limited by the high complexity of biological membrane systems. For comprehensive reviews of the analytical limitations of MD simulations, see Marrink et al. (2019) (Marrink et al. 2019) and Goossens and Winter (2018). (Goossens and Winter 2018) Developments in the field are however very promising.
Outlook
The membrane models used to mimic pathogenic bacterial membranes and the techniques used to analyse them have provided useful information on the lateral organisation of these adaptable quasi two-dimensional architectures during resistance development. Each architecture possesses individual advantages and limitations when investigating drug-membrane interactions, lipid-protein interactions, host–pathogen interactions, and structure-induced bacterial pathogenesis. As in vitro modelling systems advance, the quest for increased realism has not ceased. Key challenges include observing and incorporating complex membrane proteins such as drug-efflux proteins, connecting theoretical and experimental results, and incorporating more complex lipophilic assemblies. Current model systems are created utilising well-defined lipid mixtures, and whilst simplification is necessary for specific membrane-mediated interaction analyses, oversimplification provides an insufficient understanding of complex bacterial membrane systems and processes. By incorporating more complex compositions (proteins and lipids), insights into essential pathogen resistance development processes, membrane-targeting antimicrobial mechanisms, and generating fully artificial architectures that safely captures numerous essential pathogenic biological features can be made to help combat the devastating consequences of antibiotic resistance.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge support from the Austrian Institute for Technology. A.B.C. acknowledges AINSE for an Honours scholarship.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Watson H (2015) Biological membranes. Essays Biochem 59:43–69. 10.1042/bse0590043 [DOI] [PMC free article] [PubMed]
- Guidotti G (1972) The composition of biological membranes. Arch Intern Med 129(2):194–201. 10.1001/archinte.1972.00320020038003 [PubMed]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48(1):1–12. 10.1086/595011 [DOI] [PubMed]
- Del Mar CB, Scott AM, Glasziou PP, Hoffmann T, van Driel ML, Beller E, Phillips SM, Dartnell J (2017) Reducing antibiotic prescribing in Australian general practice: time for a national strategy. Med J Aust 207(9):401–406. 10.5694/mja17.00574 [DOI] [PubMed]
- Pendleton JN, Gorman SP, Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11(3):297–308. 10.1586/eri.13.12 [DOI] [PubMed]
- Rice LB (2010) Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol 31(S1):S7–S10. 10.1086/655995 [DOI] [PubMed]
- Santajit, S.; Indrawattana, N., Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed research international2016,2016. 10.1155/2016/2475067 [DOI] [PMC free article] [PubMed]
- Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharmacy and Therapeutics. 2015;40(4):277. [PMC free article] [PubMed] [Google Scholar]
- Chilambi GS, Gao IH, Yoon BK, Park S, Kawakami LM, Ravikumar V, Chan-Park MB, Cho N-J, Bazan GC, Kline KA (2018) Membrane adaptation limitations in Enterococcus faecalis underlie sensitivity and the inability to develop significant resistance to conjugated oligoelectrolytes. RSC Adv 8(19):10284–10293. 10.1039/C7RA11823F [DOI] [PMC free article] [PubMed]
- Gould IM, Bal AM (2013) New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 4(2):185–191. 10.4161/viru.22507 [DOI] [PMC free article] [PubMed]
- Fernández L, Hancock RE (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25(4):661–681. 10.1128/cmr.00043-12 [DOI] [PMC free article] [PubMed]
- Prestinaci F, Pezzotti P, Pantosti A (2015) Antimicrobial resistance: a global multifaceted phenomenon. Pathogens and Global Health 109(7):309–318. 10.1179/2047773215Y.0000000030 [DOI] [PMC free article] [PubMed]
- Jiang, J.-H.; Hassan, K. A.; Begg, S. L.; Rupasinghe, T. W.; Naidu, V.; Pederick, V. G.; Khorvash, M.; Whittall, J. J.; Paton, J. C.; Paulsen, I. T., Identification of novel Acinetobacter baumannii host fatty acid stress adaptation strategies. Mbio2019,10 (1). 10.1128/mBio.02056-18 [DOI] [PMC free article] [PubMed]
- Renwick, M. J.; Simpkin, V.; Mossialos, E.; Organization, W. H., Targeting innovation in antibiotic drug discovery and development: The need for a One Health–One Europe–One World Framework. World Health Organization. Regional Office for Europe: 2016 [PubMed]
- Dutescu IA, Hillier SA (2021) Encouraging the Development of New Antibiotics: Are Financial Incentives the Right Way Forward? A Systematic Review and Case Study. Infect Drug Resist 14:415. 10.2147/IDR.S287792 [DOI] [PMC free article] [PubMed]
- D’Andrea, M. M.; Fraziano, M.; Thaller, M. C.; Rossolini, G. M., The urgent need for novel antimicrobial agents and strategies to fight antibiotic resistance. Multidisciplinary Digital Publishing Institute: 2019. 10.3390/antibiotics8040254 [DOI] [PMC free article] [PubMed]
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18(3):318–327. 10.1016/s1473-3099(17)30753-3 [DOI] [PubMed]
- Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR (2019) Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10:539. 10.3389/fmicb.2019.00539 [DOI] [PMC free article] [PubMed]
- Makabenta JMV, Nabawy A, Li C-H, Schmidt-Malan S, Patel R, Rotello VM (2021) Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat Rev Microbiol 19(1):23–36. 10.1038/s41579-020-0420-1 [DOI] [PMC free article] [PubMed]
- Fatima F, Siddiqui S, Khan WA (2021) Nanoparticles as novel emerging therapeutic antibacterial agents in the antibiotics resistant era. Biol Trace Elem Res 199(7):2552–2564. 10.1007/s12011-020-02394-3 [DOI] [PubMed]
- Mantravadi PK, Kalesh KA, Dobson RC, Hudson AO, Parthasarathy A (2019) The quest for novel antimicrobial compounds: emerging trends in research, development, and technologies. Antibiotics 8(1):8. 10.3390/antibiotics8010008 [DOI] [PMC free article] [PubMed]
- Charbonneau MR, Isabella VM, Li N, Kurtz CB (2020) Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun 11(1):1–11. 10.1038/s41467-020-15508-1 [DOI] [PMC free article] [PubMed]
- Hussein M, Karas JA, Schneider-Futschik EK, Chen F, Swarbrick J, Paulin OK, Hoyer D, Baker M, Zhu Y, Li J (2020) The killing mechanism of teixobactin against methicillin-resistant Staphylococcus aureus: an untargeted metabolomics study. Msystems 5(3):e00077-e120. 10.1128/mSystems.00077-20 [DOI] [PMC free article] [PubMed]
- Hutchings MI, Truman AW, Wilkinson B (2019) Antibiotics: past, present and future. Curr Opin Microbiol 51:72–80. 10.1016/j.mib.2019.10.008 [DOI] [PubMed]
- Quinto EJ, Caro I, Villalobos-Delgado LH, Mateo J, De-Mateo-Silleras B, Redondo-Del-Río MP (2019) Food Safety through Natural Antimicrobials. Antibiotics 8(4):208. 10.3390/antibiotics8040208 [DOI] [PMC free article] [PubMed]
- Ghrairi, T.; Jaraud, S.; Alves, A.; Fleury, Y.; El Salabi, A.; Chouchani, C., New insights into and updates on antimicrobial agents from natural products. Hindawi: 2019. 10.1155/2019/7079864 [DOI] [PMC free article] [PubMed]
- Kapoor G, Saigal S, Elongavan A (2017) Action and resistance mechanisms of antibiotics: A guide for clinicians. J Anaesthesiol Clin Pharmacol 33(3):300. 10.4103/joacp.JOACP_349_15 [DOI] [PMC free article] [PubMed]
- Epand RM, Walker C, Epand RF, Magarvey NA (2016) Molecular mechanisms of membrane targeting antibiotics. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(5):980–987. 10.1016/j.bbamem.2015.10.018 [DOI] [PubMed]
- Tenover FC (2006) Mechanisms of antimicrobial resistance in bacteria. Am J Med 119(6):S3–S10. 10.1016/j.amjmed.2006.03.011 [DOI] [PubMed]
- Dias C, Rauter AP (2019) Membrane-targeting antibiotics: recent developments outside the peptide space. Future Med Chem 11(3):211–228. 10.4155/fmc-2018-0254 [DOI] [PubMed]
- Dadhich R, Kapoor S (2020) Various Facets of Pathogenic Lipids in Infectious Diseases: Exploring Virulent Lipid-Host Interactome and Their Druggability. J Membr Biol 253(5):399–423. 10.1007/s00232-020-00135-0 [DOI] [PMC free article] [PubMed]
- Han, M.-L.; Zhu, Y.; Creek, D. J.; Lin, Y.-W.; Anderson, D.; Shen, H.-H.; Tsuji, B.; Gutu, A. D.; Moskowitz, S. M.; Velkov, T., Alterations of metabolic and lipid profiles in polymyxin-resistant Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy2018,62 (6). 10.1128/AAC.02656-17 [DOI] [PMC free article] [PubMed]
- Jiang J-H, Bhuiyan MS, Shen H-H, Cameron DR, Rupasinghe TW, Wu C-M, Le Brun AP, Kostoulias X, Domene C, Fulcher AJ (2019b) Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc Natl Acad Sci 116(9):3722–3727. 10.1073/pnas.1812066116 [DOI] [PMC free article] [PubMed]
- Maifiah MHM, Cheah S-E, Johnson MD, Han M-L, Boyce JD, Thamlikitkul V, Forrest A, Kaye KS, Hertzog P, Purcell AW (2016) Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci Rep 6(1):1–17. 10.1038/srep22287 [DOI] [PMC free article] [PubMed]
- Mishra NN, Bayer AS, Tran TT, Shamoo Y, Mileykovskaya E, Dowhan W, Guan Z, Arias CA (2012) Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PloS one 7(8):e43958. 10.1371/journal.pone.0043958 [DOI] [PMC free article] [PubMed]
- Breijyeh Z, Jubeh B, Karaman R (2020) Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 25(6):1340. 10.3390/molecules25061340 [DOI] [PMC free article] [PubMed]
- Ghai I, Ghai S (2018) Understanding antibiotic resistance via outer membrane permeability. Infect Drug Resist 11:523. 10.2147/idr.s156995 [DOI] [PMC free article] [PubMed]
- Delcour AH (2009) Outer membrane permeability and antibiotic resistance. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1794(5):808–816. 10.1016/j.bbapap.2008.11.005 [DOI] [PMC free article] [PubMed]
- Vasoo, S.; Barreto, J. N.; Tosh, P. K. In Emerging issues in gram-negative bacterial resistance: an update for the practicing clinician, Mayo Clinic Proceedings, Elsevier: 2015, 395–403. 10.1016/j.mayocp.2014.12.002 [DOI] [PubMed]
- Corradi V, Sejdiu BI, Mesa-Galloso H, Abdizadeh H, Noskov SY, Marrink SJ, Tieleman DP (2019) Emerging diversity in lipid–protein interactions. Chem Rev 119(9):5775–5848. 10.1021/acs.chemrev.8b00451 [DOI] [PMC free article] [PubMed]
- Collinson I (2019) The dynamic ATP-Driven mechanism of bacterial protein translocation and the critical role of phospholipids. Front Microbiol 10:1217. 10.3389/fmicb.2019.01217 [DOI] [PMC free article] [PubMed]
- Lin T-Y, Weibel DB (2016) Organization and function of anionic phospholipids in bacteria. Appl Microbiol Biotechnol 100(10):4255–4267. 10.1007/s00253-016-7468-x [DOI] [PubMed]
- Martens C, Shekhar M, Lau AM, Tajkhorshid E, Politis A (2019) Integrating hydrogen–deuterium exchange mass spectrometry with molecular dynamics simulations to probe lipid-modulated conformational changes in membrane proteins. Nat Protoc 14(11):3183–3204. 10.1038/s41596-019-0219-6 [DOI] [PMC free article] [PubMed]
- Martens C, Stein RA, Masureel M, Roth A, Mishra S, Dawaliby R, Konijnenberg A, Sobott F, Govaerts C, Mchaourab HS (2016) Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat Struct Mol Biol 23(8):744. 10.1038/nsmb.3262 [DOI] [PMC free article] [PubMed]
- Norimatsu Y, Hasegawa K, Shimizu N, Toyoshima C (2017) Protein–phospholipid interplay revealed with crystals of a calcium pump. Nature 545(7653):193–198. 10.1038/nature22357 [DOI] [PubMed]
- Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJ, Luisi BF (2018) Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16(9):523–539. 10.1038/s41579-018-0048-6 [DOI] [PubMed]
- Lam SJ, O’Brien-Simpson NM, Pantarat N, Sulistio A, Wong EH, Chen Y-Y, Lenzo JC, Holden JA, Blencowe A, Reynolds EC (2016) Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat Microbiol 1(11):1–11. 10.1038/nmicrobiol.2016.162 [DOI] [PubMed]
- Behuria, H.; Pal, N.; Munda, R.; Sahu, S., Preparation of Giant Unilamellar Vesicles (GUVS) from Bacterial Polar Lipid Extract: Developing a Prokaryotic Model Membrane System. In Biotechnology for Sustainable Utilization of Bioresources, Astral International Pvt. Ltd: New Delhi, 2020, 309-320
- Brown S, Meredith T, Swoboda J, Walker S (2010) Staphylococcus aureus and Bacillus subtilis W23 make polyribitol wall teichoic acids using different enzymatic pathways. Chem Biol 17(10):1101–1110. 10.1016/j.chembiol.2010.07.017 [DOI] [PMC free article] [PubMed]
- Veron W, Orange N, Feuilloley MG, Lesouhaitier O (2008) Natriuretic peptides modify Pseudomonas fluorescens cytotoxicity by regulating cyclic nucleotides and modifying LPS structure. BMC Microbiol 8(1):1–11. 10.1186/1471-2180-8-114 [DOI] [PMC free article] [PubMed]
- Andersson J, Fuller MA, Wood K, Holt SA, Köper I (2018a) A tethered bilayer lipid membrane that mimics microbial membranes. Phys Chem Chem Phys 20(18):12958–12969. 10.1039/C8CP01346B [DOI] [PubMed]
- Castellana ET, Cremer PS (2006) Solid supported lipid bilayers: From biophysical studies to sensor design. Surf Sci Rep 61(10):429–444. 10.1016/j.surfrep.2006.06.001 [DOI] [PMC free article] [PubMed]
- Qing G, Gong N, Chen X, Chen J, Zhang H, Wang Y, Wang R, Zhang S, Zhang Z, Zhao X (2019) Natural and engineered bacterial outer membrane vesicles. Biophysics Reports 5(4):184–198. 10.1007/s41048-019-00095-6
- Strahl H, Errington J (2017) Bacterial membranes: structure, domains, and function. Annu Rev Microbiol 71:519–538. 10.1146/annurev-micro-102215-095630 [DOI] [PubMed]
- Salehi-Reyhani A, Ces O, Elani Y (2017) Artificial cell mimics as simplified models for the study of cell biology. Exp Biol Med 242(13):1309–1317. 10.1177/1535370217711441 [DOI] [PMC free article] [PubMed]
- Andersson J, Bilotto P, Mears LL, Fossati S, Ramach U, Köper I, Valtiner M, Knoll W (2020) Solid-supported lipid bilayers–A versatile tool for the structural and functional characterization of membrane proteins. Methods 180:56–68. 10.1016/j.ymeth.2020.09.005 [DOI] [PubMed]
- Andersson J, Köper I (2016) Tethered and polymer supported bilayer lipid membranes: structure and function. Membranes 6(2):30. 10.3390/membranes6020030 [DOI] [PMC free article] [PubMed]
- Andersson J, Köper I, Knoll W (2018b) Tethered membrane architectures—design and applications. Front Mater 5:55. 10.3389/fmats.2018.00055
- Chan Y-HM, Boxer SG (2007) Model membrane systems and their applications. Curr Opin Chem Biol 11(6):581–587. 10.1016/j.cbpa.2007.09.020 [DOI] [PMC free article] [PubMed]
- Jackman JA, Knoll W, Cho N-J (2012) Biotechnology applications of tethered lipid bilayer membranes. Materials 5(12):2637–2657. 10.3390/ma5122637
- Siontorou CG, Nikoleli G-P, Nikolelis DP, Karapetis SK (2017) Artificial lipid membranes: Past, present, and future. Membranes 7(3):38. 10.3390/membranes7030038 [DOI] [PMC free article] [PubMed]
- Wiebalck S, Kozuch J, Forbrig E, Tzschucke CC, Jeuken LJ, Hildebrandt P (2016) Monitoring the transmembrane proton gradient generated by cytochrome bo 3 in tethered bilayer lipid membranes using SEIRA spectroscopy. J Phys Chem B 120(9):2249–2256. 10.1021/acs.jpcb.6b01435 [DOI] [PubMed]
- Zieleniecki JL, Nagarajan Y, Waters S, Rongala J, Thompson V, Hrmova M, Köper I (2016) Cell-free synthesis of a functional membrane transporter into a tethered bilayer lipid membrane. Langmuir 32(10):2445–2449. 10.1021/acs.langmuir.5b04059 [DOI] [PubMed]
- Peetla C, Stine A, Labhasetwar V (2009) Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery. Mol Pharm 6(5):1264–1276. 10.1021/mp9000662 [DOI] [PMC free article] [PubMed]
- Knobloch J, Suhendro DK, Zieleniecki JL, Shapter JG, Köper I (2015) Membrane–drug interactions studied using model membrane systems. Saudi J Biol Sci 22(6):714–718. 10.1016/j.sjbs.2015.03.007 [DOI] [PMC free article] [PubMed]
- Hollmann A, Martinez M, Maturana P, Semorile LC, Maffia PC (2018) Antimicrobial peptides: interaction with model and biological membranes and synergism with chemical antibiotics. Front Chem 6:204. 10.3389/fchem.2018.00204 [DOI] [PMC free article] [PubMed]
- Solntceva V, Kostrzewa M, Larrouy-Maumus G (2020) Detection of species-specific lipids by routine MALDI TOF mass spectrometry to unlock the challenges of microbial identification and antimicrobial susceptibility testing. Front Cell Infect Microbiol 10:914. 10.3389/fcimb.2020.621452 [DOI] [PMC free article] [PubMed]
- De Carvalho CC, Caramujo MJ (2018) The various roles of fatty acids. Molecules 23(10):2583. 10.3390/molecules23102583 [DOI] [PMC free article] [PubMed]
- Willdigg JR, Helmann JD (2021) Mini Review: Bacterial Membrane Composition and Its Modulation in Response to Stress. Front Mol Biosci 8:338. 10.3389/fmolb.2021.634438 [DOI] [PMC free article] [PubMed]
- Epand RM, Epand RF (2009) Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochimica et Biophysica Acta (BBA)-Biomembranes 1788(1):289–294. 10.1016/j.bbamem.2008.08.023 [DOI] [PubMed]
- Sohlenkamp C, Geiger O (2016) Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev 40(1):133–159. 10.1093/femsre/fuv008 [DOI] [PubMed]
- Alagumuthu M, Dahiya D, Nigam PS (2019) Phospholipid—the dynamic structure between living and non-living world; a much obligatory supramolecule for present and future [J]. AIMS Mol Sci 6(1):1–19. 10.3934/molsci.2019.1.1
- Fahy E, Cotter D, Sud M, Subramaniam S (2011) Lipid classification, structures and tools. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 1811(11):637–647. 10.1016/j.bbalip.2011.06.009 [DOI] [PMC free article] [PubMed]
- Singer SJ, Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science 175(4023):720–731. 10.1126/science.175.4023.720 [DOI] [PubMed]
- Van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9(2):112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed]
- Barák I, Muchová K (2013) The role of lipid domains in bacterial cell processes. Int J Mol Sci 14(2):4050–4065. 10.3390/ijms14024050 [DOI] [PMC free article] [PubMed]
- Cronan JE, Thomas J (2009) Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol 459:395–433. 10.1016/s0076-6879(09)04617-5 [DOI] [PMC free article] [PubMed]
- Bertani, B.; Ruiz, N., Function and biogenesis of lipopolysaccharides. EcoSal Plus2018,8 (1). 10.1128/ecosalplus.esp-0001-2018 [DOI] [PMC free article] [PubMed]
- Reichmann NT, Gründling A (2011) Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett 319(2):97–105. 10.1111/j.1574-6968.2011.02260.x [DOI] [PMC free article] [PubMed]
- May KL, Grabowicz M (2018) The bacterial outer membrane is an evolving antibiotic barrier. Proc Natl Acad Sci 115(36):8852–8854. 10.1073/pnas.1812779115 [DOI] [PMC free article] [PubMed]
- Rahman MM, Kolli VK, Kahler CM, Shih G, Stephens DS, Carlson RW (2000) The membrane phospholipids of Neisseria meningitidis and Neisseria gonorrhoeae as characterized by fast atom bombardment mass spectrometry. Microbiology 146(8):1901–1911. 10.1099/00221287-146-8-1901 [DOI] [PubMed]
- Jasim R, Han M-L, Zhu Y, Hu X, Hussein MH, Lin Y-W, Zhou QT, Dong CYD, Li J, Velkov T (2018) Lipidomic analysis of the outer membrane vesicles from paired polymyxin-susceptible and-resistant Klebsiella pneumoniae clinical isolates. Int J Mol Sci 19(8):2356. 10.3390/ijms19082356 [DOI] [PMC free article] [PubMed]
- Theilacker C, Kropec A, Hammer F, Sava I, Wobser D, Sakinc T, Codée JD, Hogendorf WF, van der Marel GA, Huebner J (2012) Protection against Staphylococcus aureus by antibody to the polyglycerolphosphate backbone of heterologous lipoteichoic acid. J Infect Dis 205(7):1076–1085. 10.1093/infdis/jis022 [DOI] [PubMed]
- Song H-S, Choi T-R, Han Y-H, Park Y-L, Park JY, Yang S-Y, Bhatia SK, Gurav R, Kim Y-G, Kim J-S (2020) Increased resistance of a methicillin-resistant Staphylococcus aureus Δ agr mutant with modified control in fatty acid metabolism. AMB Express 10(1):1–10. 10.1186/s13568-020-01000-y [DOI] [PMC free article] [PubMed]
- Schneewind O, Missiakas D (2014) Lipoteichoic acids, phosphate-containing polymers in the envelope of gram-positive bacteria. J Bacteriol 196(6):1133–1142. 10.1128/JB.01155-13 [DOI] [PMC free article] [PubMed]
- Kilelee E, Pokorny A, Yeaman MR, Bayer AS (2010) Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: implications for daptomycin resistance. Antimicrob Agents Chemother 54(10):4476–4479. 10.1128/AAC.00191-10 [DOI] [PMC free article] [PubMed]
- Malanovic N, Lohner K (2016) Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(5):936–946. 10.1016/j.bbamem.2015.11.004 [DOI] [PubMed]
- Oku Y, Kurokawa K, Ichihashi N, Sekimizu K (2004) Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150(1):45–51. 10.1099/mic.0.26706-0 [DOI] [PubMed]
- White DC, Frerman FE (1967) Extraction, characterization, and cellular localization of the lipids of Staphylococcus aureus. J Bacteriol 94(6):1854–1867. 10.1128/jb.94.6.1854-1867.1967 [DOI] [PMC free article] [PubMed]
- Vinogradov E, Frirdich E, MacLean LL, Perry MB, Petersen BO, Duus JØ, Whitfield C (2002) Structures of lipopolysaccharides from Klebsiella pneumoniae: Elucidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the Ochains. J Biol Chem 277(28):25070–25081. 10.1074/jbc.m202683200 [DOI] [PubMed]
- Hobby CR, Herndon JL, Morrow CA, Peters RE, Symes SJ, Giles DK (2019) Exogenous fatty acids alter phospholipid composition, membrane permeability, capacity for biofilm formation, and antimicrobial peptide susceptibility in Klebsiella pneumoniae. Microbiologyopen 8(2):e00635. 10.1002/mbo3.635 [DOI] [PMC free article] [PubMed]
- Unno, Y.; Sato, Y.; Nishida, S.; Nakano, A.; Nakano, R.; Ubagai, T.; Ono, Y., Acinetobacter baumannii Lipopolysaccharide Influences Adipokine Expression in 3T3-L1 Adipocytes. Mediators of inflammation2017,2017. 10.1155/2017/9039302 [DOI] [PMC free article] [PubMed]
- Jiang X, Yang K, Yuan B, Han M, Zhu Y, Roberts KD, Patil NA, Li J, Gong B, Hancock RE (2020) Molecular dynamics simulations informed by membrane lipidomics reveal the structure–interaction relationship of polymyxins with the lipid A-based outer membrane of Acinetobacter baumannii. J Antimicrob Chemother 75(12):3534–3543. 10.1093/jac/dkaa376 [DOI] [PMC free article] [PubMed]
- Lopalco P, Stahl J, Annese C, Averhoff B, Corcelli A (2017) Identification of unique cardiolipin and monolysocardiolipin species in Acinetobacter baumannii. Sci Rep 7(1):1–12. 10.1038/s41598-017-03214-w [DOI] [PMC free article] [PubMed]
- Chao J, Wolfaardt GM, Arts MT (2010) Characterization of Pseudomonas aeruginosa fatty acid profiles in biofilms and batch planktonic cultures. Can J Microbiol 56(12):1028–1039. 10.1139/w10-093 [DOI] [PubMed]
- Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D (2011) Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2:118. 10.3389/fmicb.2011.00118 [DOI] [PMC free article] [PubMed]
- Klein S, Lorenzo C, Hoffmann S, Walther JM, Storbeck S, Piekarski T, Tindall BJ, Wray V, Nimtz M, Moser J (2009) Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol Microbiol 71(3):551–565. 10.1111/j.1365-2958.2008.06562.x [DOI] [PubMed]
- Lewenza S, Falsafi R, Bains M, Rohs P, Stupak J, Sprott GD, Hancock RE (2011) The olsA gene mediates the synthesis of an ornithine lipid in Pseudomonas aeruginosa during growth under phosphate-limiting conditions, but is not involved in antimicrobial peptide susceptibility. FEMS Microbiol Lett 320(2):95–102. 10.1111/j.1574-6968.2011.02295.x [DOI] [PubMed]
- Pramanik B, Zechman J, Das P, Bartner P (1990) Bacterial phospholipid analysis by fast atom bombardment mass spectrometry. Biomed Environ Mass Spectrom 19(3):164–170. 10.1002/bms.1200190312 [DOI] [PubMed]
- Wilderman PJ, Vasil AI, Martin WE, Murphy RC, Vasil ML (2002) Pseudomonas aeruginosa synthesizes phosphatidylcholine by use of the phosphatidylcholine synthase pathway. J Bacteriol 184(17):4792–4799. 10.1128/jb.184.17.4792-4799.2002 [DOI] [PMC free article] [PubMed]
- Soberón-Chávez G, Lépine F, Déziel E (2005) Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 68(6):718–725. 10.1007/s00253-005-0150-3 [DOI] [PubMed]
- Bøse B, Gjerde J (1980) Fatty acid patterns in the classification of some representatives of the families Enterobacteriaceae and Vibrionaceae. Microbiology 116(1):41–49. 10.1099/00221287-116-1-41 [DOI] [PubMed]
- Gill C, Suisted J (1978) The effects of temperature and growth rate on the proportion of unsaturated fatty acids in bacterial lipids. Microbiology 104(1):31–36. 10.1099/00221287-104-1-31 [DOI] [PubMed]
- Kämpfer P, McInroy JA, Glaeser SP (2015) Enterobacter muelleri sp. nov., isolated from the rhizosphere of Zea mays. Int J Syst Evol Microbiol 65(Pt_11):4093–4099. 10.1099/ijsem.0.000547 [DOI] [PubMed]
- Davin-Regli A, Lavigne J-P, Pagès J-M (2019) Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev 32(4):e00002-19. 10.1128/cmr.00002-19 [DOI] [PMC free article] [PubMed]
- Epand RM, Epand RF (2009b) Domains in bacterial membranes and the action of antimicrobial agents. Mol BioSyst 5(6):580–587. 10.1016/j.bbamem.2008.08.023 [DOI] [PubMed]
- Epand RM, Epand RF, Arnusch CJ, Papahadjopoulos-Sternberg B, Wang G, Shai Y (2010) Lipid clustering by three homologous arginine-rich antimicrobial peptides is insensitive to amino acid arrangement and induced secondary structure. Biochimica et Biophysica Acta (BBA)-Biomembranes 1798(6):1272–1280. 10.1016/j.bbamem.2010.03.012 [DOI] [PubMed]
- Villegas, M. V.; Quinn, J. P., Enterobacter species. Antimicrobial therapy and vaccines. Maryland: Apple Trees Productions LLC2002, 255–63.
- Silhavy TJ, Kahne D, Walker S (2010) The bacterial cell envelope. Cold Spring Harbor Perspect Biol 2(5):a000414. 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed]
- Bogdanov M, Pyrshev K, Yesylevskyy S, Ryabichko S, Boiko V, Ivanchenko P, Kiyamova R, Guan Z, Ramseyer C, Dowhan W (2020) Phospholipid distribution in the cytoplasmic membrane of Gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent. Sci Adv 6(23):eaaz6333. 10.1126/sciadv.aaz6333 [DOI] [PMC free article] [PubMed]
- Paulowski L, Donoghue A, Nehls C, Groth S, Koistinen M, Hagge SO, Böhling A, Winterhalter M, Gutsmann T (2020) The beauty of asymmetric membranes: Reconstitution of the outer membrane of Gram-negative bacteria. Front Cell Dev Biol 8:586. 10.3389/fcell.2020.00586 [DOI] [PMC free article] [PubMed]
- Cian, M.; Giordano, N.; Mettlach, J.; Minor, K.; Dalebroux, Z., Separation of the Cell Envelope for Gram-negative Bacteria into Inner and Outer Membrane Fractions with Technical Adjustments for Acinetobacter baumannii. Journal of Visualized Experiments: Jove2020, (158). 10.3791/60517 [DOI] [PMC free article] [PubMed]
- Raetz CR, Whitfield C (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71(1):635–700. 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed]
- Wang, X.; Quinn, P. J., Endotoxins: lipopolysaccharides of gram-negative bacteria. In Endotoxins: structure, function and recognition, Springer: 2010; pp 3–25. 10.1007/978-90-481-9078-2_1 [DOI] [PubMed]
- Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD, Scott AJ, Masoudi A, Goodlett DR, Wang X (2012) LPS remodeling is an evolved survival strategy for bacteria. Proc Natl Acad Sci 109(22):8716–8721. 10.1073/pnas.1202908109 [DOI] [PMC free article] [PubMed]
- Needham BD, Trent MS (2013) Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11(7):467–481. 10.1038/nrmicro3047 [DOI] [PMC free article] [PubMed]
- Trent MS, Stead CM, Tran AX, Hankins JV (2006) Diversity of Endotoxin and Its Impact on Pathogenesis. J Endotoxin Res 12(4):205–223. 10.1179/096805106x118825 [DOI] [PubMed]
- Simpson BW, Trent MS (2019) Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17(7):403–416. 10.1038/s41579-019-0201-x [DOI] [PMC free article] [PubMed]
- Maldonado RF, Sá-Correia I, Valvano MA (2016) Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev 40(4):480–493. 10.1093/femsre/fuw007 [DOI] [PMC free article] [PubMed]
- Moffatt, J. H.; Harper, M.; Harrison, P.; Hale, J. D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St. Michael, F.; Cox, A. D., Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother2010,54 (12), 4971-4977. 10.1128/aac.00834-10 [DOI] [PMC free article] [PubMed]
- Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KR, Doi Y, Ernst RK (2013) Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57(10):4831–4840. 10.1128/aac.00865-13 [DOI] [PMC free article] [PubMed]
- Dortet L, Broda A, Bernabeu S, Glupczynski Y, Bogaerts P, Bonnin R, Naas T, Filloux A, Larrouy-Maumus G (2020) Optimization of the MALDIxin test for the rapid identification of colistin resistance in Klebsiella pneumoniae using MALDI-TOF MS. J Antimicrob Chemother 75(1):110–116. 10.1093/jac/dkz405 [DOI] [PMC free article] [PubMed]
- Olaitan AO, Morand S, Rolain J-M (2014) Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed]
- Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67(4):593–656. 10.1128/mmbr.67.4.593-656.2003 [DOI] [PMC free article] [PubMed]
- Pajerski W, Ochonska D, Brzychczy-Wloch M, Indyka P, Jarosz M, Golda-Cepa M, Sojka Z, Kotarba A (2019) Attachment efficiency of gold nanoparticles by Gram-positive and Gram-negative bacterial strains governed by surface charges. J Nanopart Res 21(8):1–12. 10.1007/s11051-019-4617-z
- Percy MG, Gründling A (2014) Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68:81–100. 10.1146/annurev-micro-091213-112949 [DOI] [PubMed]
- Shiraishi T, Yokota S-I, Morita N, Fukiya S, Tomita S, Tanaka N, Okada S, Yokota A (2013) Characterization of a Lactobacillus gasseri JCM 1131T lipoteichoic acid with a novel glycolipid anchor structure. Appl Environ Microbiol 79(10):3315–3318. 10.1128/AEM.00243-13 [DOI] [PMC free article] [PubMed]
- Gutmann L, Al-Obeid S, Billot-Klein D, Ebnet E, Fischer W (1996) Penicillin tolerance and modification of lipoteichoic acid associated with expression of vancomycin resistance in VanB-type Enterococcus faecium D366. Antimicrob Agents Chemother 40(1):257–259. 10.1128/AAC.40.1.257 [DOI] [PMC free article] [PubMed]
- Saar-Dover, R.; Bitler, A.; Nezer, R.; Shmuel-Galia, L.; Firon, A.; Shimoni, E.; Trieu-Cuot, P.; Shai, Y., D-alanylation of lipoteichoic acids confers resistance to cationic peptides in group B streptococcus by increasing the cell wall density. 2012. 10.1371/journal.ppat.1002891 [DOI] [PMC free article] [PubMed]
- Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P (2002) Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol 43(1):1–14. 10.1046/j.1365-2958.2002.02723.x [DOI] [PubMed]
- Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F (1999) Inactivation of the dlt Operon inStaphylococcus aureus Confers Sensitivity to Defensins, Protegrins, and Other Antimicrobial Peptides. J Biol Chem 274(13):8405–8410. 10.1074/jbc.274.13.8405 [DOI] [PubMed]
- Reichmann NT, Cassona CP, Gründling A (2013) Revised mechanism of D-alanine incorporation into cell wall polymers in Gram-positive bacteria. Microbiology 159(Pt 9):1868. 10.1099/mic.0.069898-0 [DOI] [PMC free article] [PubMed]
- Rosado H, Turner RD, Foster SJ, Taylor PW (2015) Impact of the β-lactam resistance modifier (−)-epicatechin gallate on the non-random distribution of phospholipids across the cytoplasmic membrane of Staphylococcus aureus. Int J Mol Sci 16(8):16710–16727. 10.3390/ijms160816710 [DOI] [PMC free article] [PubMed]
- Jones T, Yeaman MR, Sakoulas G, Yang S-J, Proctor RA, Sahl H-G, Schrenzel J, Xiong YQ, Bayer AS (2008) Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother 52(1):269–278. 10.1128/aac.00719-07 [DOI] [PMC free article] [PubMed]
- Scott JR, Barnett TC (2006) Surface proteins of gram-positive bacteria and how they get there. Annu Rev Microbiol 60:397–423. 10.1146/annurev.micro.60.080805.142256 [DOI] [PubMed]
- Akbarzadeh A, Rezaei-Sadabady R, Davaran S, Joo SW, Zarghami N, Hanifehpour Y, Samiei M, Kouhi M, Nejati-Koshki K (2013) Liposome: classification, preparation, and applications. Nanoscale Res Lett 8(1):1–9. 10.1186/1556-276X-8-102 [DOI] [PMC free article] [PubMed]
- Šturm L, Poklar Ulrih N (2021) Basic Methods for Preparation of Liposomes and Studying Their Interactions with Different Compounds, with the Emphasis on Polyphenols. Int J Mol Sci 22(12):6547. 10.3390/ijms22126547 [DOI] [PMC free article] [PubMed]
- Navas BP, Lohner K, Deutsch G, Sevcsik E, Riske K, Dimova R, Garidel P, Pabst G (2005) Composition dependence of vesicle morphology and mixing properties in a bacterial model membrane system. Biochimica et Biophysica Acta (BBA)-Biomembranes 1716(1):40–48. 10.1016/j.bbamem.2005.08.003 [DOI] [PubMed]
- Giuliano CB, Cvjetan N, Ayache J, Walde PJ (2021) Multivesicular Vesicles: Preparation and Applications. ChemSystemsChem 3(2):e2000049. 10.1002/syst.202000049
- Mu H, Wang Y, Chu Y, Jiang Y, Hua H, Chu L, Wang K, Wang A, Liu W, Li Y (2018) Multivesicular liposomes for sustained release of bevacizumab in treating laser-induced choroidal neovascularization. Drug Delivery 25(1):1372–1383. 10.1080/10717544.2018.1474967 [DOI] [PMC free article] [PubMed]
- Gabizon A, Goren D, Cohen R, Barenholz Y (1998) Development of liposomal anthracyclines: from basics to clinical applications. J Control Release 53(1–3):275–279. 10.1016/s0168-3659(97)00261-7 [DOI] [PubMed]
- Sherratt SC, Mason RP (2018) Eicosapentaenoic acid and docosahexaenoic acid have distinct membrane locations and lipid interactions as determined by X-ray diffraction. Chem Phys Lipid 212:73–79. 10.1016/j.chemphyslip.2018.01.002 [DOI] [PubMed]
- Fan M, Xu S, Xia S, Zhang X (2007) Effect of different preparation methods on physicochemical properties of salidroside liposomes. J Agric Food Chem 55(8):3089–3095. 10.1021/jf062935q [DOI] [PubMed]
- Bozzuto G, Molinari A (2015) Liposomes as nanomedical devices. Int J Nanomed 10:975. 10.2147/ijn.s68861 [DOI] [PMC free article] [PubMed]
- Riaz MK, Riaz MA, Zhang X, Lin C, Wong KH, Chen X, Zhang G, Lu A, Yang Z (2018) Surface functionalization and targeting strategies of liposomes in solid tumor therapy: A review. Int J Mol Sci 19(1):195. 10.3390/ijms19010195 [DOI] [PMC free article] [PubMed]
- Sakai-Kato K, Yoshida K, Izutsu K-I (2019) Effect of surface charge on the size-dependent cellular internalization of liposomes. Chem Phys Lipids 224:104726. 10.1016/j.chemphyslip.2019.01.004 [DOI] [PubMed]
- Tuerkova A, Kabelka I, Králová T, Sukeník L, Pokorná Š, Hof M, Vácha R (2020) Effect of helical kink in antimicrobial peptides on membrane pore formation. Elife 9:e47946. 10.7554/eLife.47946 [DOI] [PMC free article] [PubMed]
- Dombach JL, Quintana JL, Nagy TA, Wan C, Crooks AL, Yu H, Su C-C, Yu EW, Shen J, Detweiler CS (2020) A small molecule that mitigates bacterial infection disrupts Gram-negative cell membranes and is inhibited by cholesterol and neutral lipids. PLoS pathogens 16(12):e1009119. 10.1371/journal.ppat.1009119 [DOI] [PMC free article] [PubMed]
- Jamasbi E, Batinovic S, Sharples RA, Sani M-A, Robins-Browne RM, Wade JD, Separovic F, Hossain MA (2014) Melittin peptides exhibit different activity on different cells and model membranes. Amino Acids 46(12):2759–2766. 10.1007/s00726-014-1833-9 [DOI] [PubMed]
- Kumagai A, Dupuy FG, Arsov Z, Elhady Y, Moody D, Ernst RK, Deslouches B, Montelaro RC, Di YP, Tristram-Nagle S (2019) Elastic behavior of model membranes with antimicrobial peptides depends on lipid specificity and d-enantiomers. Soft Matter 15(8):1860–1868. 10.1039/c8sm02180e [DOI] [PMC free article] [PubMed]
- Pérez-Peinado C, Dias SA, Domingues MM, Benfield AH, Freire JM, Rádis-Baptista G, Gaspar D, Castanho MA, Craik DJ, Henriques ST (2018) Mechanisms of bacterial membrane permeabilization by crotalicidin (Ctn) and its fragment Ctn (15–34), antimicrobial peptides from rattlesnake venom. J Biol Chem 293(5):1536–1549. 10.1074/jbc.RA117.000125 [DOI] [PMC free article] [PubMed]
- Malishev R, Abbasi R, Jelinek R, Chai L (2018) Bacterial model membranes reshape fibrillation of a functional amyloid protein. Biochemistry 57(35):5230–5238. 10.1021/acs.biochem.8b00002 [DOI] [PubMed]
- Kahveci Z, Vázquez-Guilló R, Mira A, Martinez L, Falcó A, Mallavia R, Mateo CR (2016) Selective recognition and imaging of bacterial model membranes over mammalian ones by using cationic conjugated polyelectrolytes. Analyst 141(22):6287–6296. 10.1039/c6an01427e [DOI] [PubMed]
- Lopes SC, Neves CS, Eaton P, Gameiro P (2012) Improved model systems for bacterial membranes from differing species: the importance of varying composition in PE/PG/cardiolipin ternary mixtures. Mol Membr Biol 29(6):207–217. 152. 10.3109/09687688.2012.700491 [DOI] [PubMed]
- Cheng JT, Hale JD, Elliott M, Hancock RE, Straus SK (2011) The importance of bacterial membrane composition in the structure and function of aurein 2.2 and selected variants. Biochimica Et Biophysica Acta (BBA)-Biomembranes 1808(3):622–633. 10.1016/j.bbamem.2010.11.025 [DOI] [PubMed]
- Marín-Menéndez A, Montis C, Díaz-Calvo T, Carta D, Hatzixanthis K, Morris CJ, McArthur M, Berti D (2017) Antimicrobial nanoplexes meet model bacterial membranes: the key role of Cardiolipin. Sci Rep 7(1):1–13. 10.1038/srep41242 [DOI] [PMC free article] [PubMed]
- Fernandez DI, Sani M-A, Gehman JD, Hahm K-S, Separovic F (2011) Interactions of a synthetic Leu–Lys-rich antimicrobial peptide with phospholipid bilayers. Eur Biophys J 40(4):471–480. 10.1007/s00249-010-0660-5 [DOI] [PubMed]
- Domenech O, Francius G, Tulkens PM, Van Bambeke F, Dufrêne Y, Mingeot-Leclercq M-P (2009) Interactions of oritavancin, a new lipoglycopeptide derived from vancomycin, with phospholipid bilayers: effect on membrane permeability and nanoscale lipid membrane organization. Biochimica et Biophysica Acta (BBA)-Biomembranes 1788(9):1832–1840. 10.1016/j.bbamem.2009.05.003 [DOI] [PubMed]
- Pinheiro M, Nunes CU, Caio JM, Moiteiro C, Lúcio M, Brezesinski G, Reis S (2013) The influence of rifabutin on human and bacterial membrane models: Implications for its mechanism of action. J Phys Chem B 117(20):6187–6193. 10.1021/jp403073v [DOI] [PubMed]
- D’Errico G, Silipo A, Mangiapia G, Vitiello G, Radulescu A, Molinaro A, Lanzetta R, Paduano L (2010) Characterization of liposomes formed by lipopolysaccharides from Burkholderia cenocepacia, Burkholderia multivorans and Agrobacterium tumefaciens: from the molecular structure to the aggregate architecture. Phys Chem Chem Phys 12(41):13574–13585. 10.1039/C0CP00066C [DOI] [PubMed]
- Furusato T, Horie F, Matsubayashi HT, Amikura K, Kuruma Y, Ueda T (2018) De novo synthesis of basal bacterial cell division proteins FtsZ, FtsA, and ZipA inside giant vesicles. ACS Synth Biol 7(4):953–961. 10.1021/acssynbio.7b00350 [DOI] [PubMed]
- Kiss B, Bozó T, Mudra D, Tordai H, Herényi L, Kellermayer M (2021) Development, structure and mechanics of a synthetic E. coli outer membrane model. Nanoscale Adv 3(3):755–766. 10.1039/D0NA00977F [DOI] [PMC free article] [PubMed]
- Jiménez M, Martos A, Vicente M, Rivas G (2011) Reconstitution and organization of Escherichia coli proto-ring elements (FtsZ and FtsA) inside giant unilamellar vesicles obtained from bacterial inner membranes. J Biol Chem 286(13):11236–11241. 10.1074/jbc.m110.194365 [DOI] [PMC free article] [PubMed]
- Sikder A, Sarkar J, Barman R, Ghosh S (2019) Directional Supramolecular Assembly of π-Amphiphiles with Tunable Surface Functionality and Impact on the Antimicrobial Activity. J Phys Chem B 123(33):7169–7177. 10.1021/acs.jpcb.9b05193 [DOI] [PubMed]
- Kubiak J, Brewer J, Hansen S, Bagatolli LA (2011) Lipid lateral organization on giant unilamellar vesicles containing lipopolysaccharides. Biophys J 100(4):978–986. 10.1016/j.bpj.2011.01.012 [DOI] [PMC free article] [PubMed]
- Mohanan G, Nair KS, Nampoothiri KM, Bajaj H (2020) Engineering bio-mimicking functional vesicles with multiple compartments for quantifying molecular transport. Chem Sci 11(18):4669–4679. 10.1039/D0SC00084A [DOI] [PMC free article] [PubMed]
- Ruhr E, Sahl H-G (1985) Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob Agents Chemother 27(5):841–845. 10.1128/aac.27.5.841 [DOI] [PMC free article] [PubMed]
- Bharatiya B, Wang G, Rogers SE, Pedersen JS, Mann S, Briscoe WH (2021) Mixed liposomes containing gram-positive bacteria lipids: Lipoteichoic acid (LTA) induced structural changes. Colloids Surf B 199:111551. 10.1016/j.colsurfb.2020.111551 [DOI] [PubMed]
- Saliba A-E, Vonkova I, Ceschia S, Findlay GM, Maeda K, Tischer C, Deghou S, Van Noort V, Bork P, Pawson T (2014) A quantitative liposome microarray to systematically characterize protein-lipid interactions. Nat Methods 11(1):47–50. 10.1038/nmeth.2734 [DOI] [PubMed]
- Turner M, Singhrao SK, Dennison SR, Morton LHG, Crean S (2015) Challenging the Clostridium botulinum toxin type A (BoNT/A) with a selection of microorganisms by culture methods and extended storage of used vials to assess the loss of sterility. J Dent Appl 2(5):223–228
- Som A, Tew GN (2008) Influence of lipid composition on membrane activity of antimicrobial phenylene ethynylene oligomers. J Phys Chem B 112(11):3495–3502. 10.1021/jp077487j [DOI] [PMC free article] [PubMed]
- Samuel R, Gillmor S (2016) Membrane phase characteristics control NA-CATH activity. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(9):1974–1982. 10.1016/j.bbamem.2016.05.015 [DOI] [PubMed]
- Sborgi L, Rühl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Müller DJ, Broz P, Hiller S (2016) GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J 35(16):1766–1778. 10.15252/embj.201694696 [DOI] [PMC free article] [PubMed]
- Carrasco-López C, Rojas-Altuve A, Zhang W, Hesek D, Lee M, Barbe S, André I, Ferrer P, Silva-Martin N, Castro GR (2011) Crystal structures of bacterial peptidoglycan amidase AmpD and an unprecedented activation mechanism. J Biol Chem 286(36):31714–31722. 10.1074/jbc.M111.264366 [DOI] [PMC free article] [PubMed]
- Sasaki M, Nishikawa H, Suzuki S, Moser M, Huber M, Sawasato K, Matsubayashi HT, Kumazaki K, Tsukazaki T, Kuruma Y (2019) The bacterial protein YidC accelerates MPIase-dependent integration of membrane proteins. J Biol Chem 294(49):18898–18908. 10.1074/jbc.ra119.011248 [DOI] [PMC free article] [PubMed]
- Cheng M, Huang JX, Ramu S, Butler MS, Cooper MA (2014) Ramoplanin at bactericidal concentrations induces bacterial membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother 58(11):6819–6827. 10.1128/AAC.00061-14 [DOI] [PMC free article] [PubMed]
- Lombardi L, Stellato MI, Oliva R, Falanga A, Galdiero M, Petraccone L, D’Errico G, De Santis A, Galdiero S, Del Vecchio P (2017) Antimicrobial peptides at work: interaction of myxinidin and its mutant WMR with lipid bilayers mimicking the P. aeruginosa and E. coli membranes. Sci Rep 7(1):1–15. 10.1038/srep44425 [DOI] [PMC free article] [PubMed]
- Zhang T, Muraih JK, Tishbi N, Herskowitz J, Victor RL, Silverman J, Uwumarenogie S, Taylor SD, Palmer M, Mintzer E (2014) Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J Biol Chem 289(17):11584–11591. 10.1074/jbc.m114.554444 [DOI] [PMC free article] [PubMed]
- Brian Chia C, Gong Y, Bowie JH, Zuegg J, Cooper MA (2011) Membrane binding and perturbation studies of the antimicrobial peptides caerin, citropin, and maculatin. Pept Sci 96(2):147–157. 10.1002/bip.21438 [DOI] [PubMed]
- Su Y, Waring AJ, Ruchala P, Hong M (2011) Structures of β-hairpin antimicrobial protegrin peptides in lipopolysaccharide membranes: mechanism of gram selectivity obtained from solid-state nuclear magnetic resonance. Biochemistry 50(12):2072–2083. 10.1021/bi101975v [DOI] [PMC free article] [PubMed]
- Hancock R, Nikaido H (1978) Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol 136(1):381–390. 10.1128/jb.136.1.381-390.1978 [DOI] [PMC free article] [PubMed]
- Ciesielski F, Griffin DC, Rittig M, Moriyón I, Bonev BB (2013) Interactions of lipopolysaccharide with lipid membranes, raft models—A solid state NMR study. Biochimica et Biophysica Acta (BBA)-Biomembranes 1828(8):1731–1742. 10.1016/j.bbamem.2013.03.029 [DOI] [PubMed]
- Lee E-H, Collatz E, Trias J, Gutmann L (1992) Diffusion of β-lactam antibiotics into proteoliposomes reconstituted with outer membranes of isogenic imipenem-susceptible and-resistant strains of Enterobacter cloacae. Microbiology 138(11):2347–2351. 10.1099/00221287-138-11-2347 [DOI] [PubMed]
- Mitchell NJ, Seaton P, Pokorny A (2016) Branched phospholipids render lipid vesicles more susceptible to membrane-active peptides. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(5):988–994. 10.1016/j.bbamem.2015.10.014 [DOI] [PMC free article] [PubMed]
- Rideau E, Dimova R, Schwille P, Wurm FR, Landfester K (2018) Liposomes and polymersomes: a comparative review towards cell mimicking. Chem Soc Rev 47(23):8572–8610. 10.1039/C8CS00162F [DOI] [PubMed]
- Weinberger A, Tsai F-C, Koenderink GH, Schmidt TF, Itri R, Meier W, Schmatko T, Schröder A, Marques C (2013) Gel-assisted formation of giant unilamellar vesicles. Biophys J 105(1):154–164. 10.1016/j.bpj.2013.05.024 [DOI] [PMC free article] [PubMed]
- Göpfrich K, Haller B, Staufer O, Dreher Y, Mersdorf U, Platzman I, Spatz JP (2019) One-pot assembly of complex giant unilamellar vesicle-based synthetic cells. ACS Synth Biol 8(5):937–947. 10.1021/acssynbio.9b00034 [DOI] [PMC free article] [PubMed]
- Pautot S, Frisken BJ, Weitz D (2003) Engineering asymmetric vesicles. Proc Natl Acad Sci 100(19):10718–10721. 10.1073/pnas.1931005100 [DOI] [PMC free article] [PubMed]
- Nkanga, C. I.; Bapolisi, A. M.; Okafor, N. I.; Krause, R. W. M., General perception of liposomes: formation, manufacturing and applications. Liposomes-advances and perspectives2019. 10.5772/intechopen.84255
- Eeman M, Deleu M (2010) From biological membranes to biomimetic model membranes. Biotechnol Agron Soc Environ 14(4):719–736.
- Vestergaard MD, Hamada T, Takagi M (2008) Using model membranes for the study of amyloid beta: lipid interactions and neurotoxicity. Biotechnol Bioeng 99(4):753–763. 10.1002/bit.21731 [DOI] [PubMed]
- Schmid EM, Richmond DL, Fletcher DA (2015) Reconstitution of proteins on electroformed giant unilamellar vesicles. Methods Cell Biol 128:319–338. 10.1016/bs.mcb.2015.02.004 [DOI] [PMC free article] [PubMed]
- Belegrinou S, Menon S, Dobrunz D, Meier W (2011) Solid-supported polymeric membranes. Soft Matter 7(6):2202–2210. 10.1039/C0SM01163K
- Sackmann E (1996) Supported membranes: scientific and practical applications. Science 271(5245):43–48. 10.1126/science.271.5245.43 [DOI] [PubMed]
- Foglia F, Lawrence M, Barlow D (2015) Studies of model biological and bio-mimetic membrane structure: reflectivity vs diffraction, a critical comparison. Curr Opin Colloid Interface Sci 20(4):235–243. 10.1016/j.cocis.2015.08.001
- Li C, Wang M, Ferguson M, Zhan W (2015) Phospholipid/aromatic thiol hybrid bilayers. Langmuir 31(18):5228–5234. 10.1021/acs.langmuir.5b00476 [DOI] [PubMed]
- Köper I (2007) Insulating tethered bilayer lipid membranes to study membrane proteins. Mol BioSyst 3(10):651–657. 10.1039/B707168J [DOI] [PubMed]
- Girard-Egrot AP, Maniti O (2021) Why Do Tethered-Bilayer Lipid Membranes Suit for Functional Membrane Protein Reincorporation? Appl Sci 11(11):4876. 10.3390/app11114876
- Kurniawan J, de Ventrici Souza JOF, Dang AT, Liu GY, Kuhl TL (2018) Preparation and characterization of solid-supported lipid bilayers formed by Langmuir-Blodgett deposition: a tutorial. Langmuir 34(51):15622–15639. 10.1021/acs.langmuir.8b03504 [DOI] [PubMed]
- Richter RP, Him JLK, Brisson A (2003) Supported lipid membranes. Mater Today 6(11):32–37. 10.1016/S1369-7021(03)01129-5
- Clifton LA, Campbell RA, Sebastiani F, Campos-Terán J, Gonzalez-Martinez JF, Björklund S, Sotres J, Cárdenas M (2020) Design and use of model membranes to study biomolecular interactions using complementary surface-sensitive techniques. Adv Colloid Interface Sci 277:102118. 10.1016/j.cis.2020.102118 [DOI] [PubMed]
- Giess F, Friedrich MG, Heberle J, Naumann RL, Knoll W (2004) The protein-tethered lipid bilayer: A novel mimic of the biological membrane. Biophys J 87(5):3213–3220. 10.1529/biophysj.104.046169 [DOI] [PMC free article] [PubMed]
- Ferhan AR, Jackman JA, Cho N-J (2017) Probing Spatial Proximity of Supported Lipid Bilayers to Silica Surfaces by Localized Surface Plasmon Resonance Sensing. Anal Chem 89(7):4301–4308. 10.1021/acs.analchem.7b00370 [DOI] [PubMed]
- Wittenberg NJ, Wootla B, Jordan LR, Denic A, Warrington AE, Oh S-H, Rodriguez M (2014) Applications of SPR for the characterization of molecules important in the pathogenesis and treatment of neurodegenerative diseases. Expert Rev Neurother 14(4):449–463. 10.1586/14737175.2014.896199 [DOI] [PMC free article] [PubMed]
- Steltenkamp S, Müller MM, Deserno M, Hennesthal C, Steinem C, Janshoff A (2006) Mechanical Properties of Pore-Spanning Lipid Bilayers Probed by Atomic Force Microscopy. Biophys J 91(1):217–226. 10.1529/biophysj.106.081398 [DOI] [PMC free article] [PubMed]
- Weiss SA, Bushby RJ, Evans SD, Jeuken LJ (2010) A study of cytochrome bo3 in a tethered bilayer lipid membrane. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1797(12):1917–1923. 10.1016/j.bbabio.2010.01.012 [DOI] [PMC free article] [PubMed]
- Clifton LA, Skoda MW, Daulton EL, Hughes AV, Le Brun AP, Lakey JH, Holt SA (2013) Asymmetric phospholipid: lipopolysaccharide bilayers; a Gram-negative bacterial outer membrane mimic. J R Soc Interface 10(89):20130810. 10.1098/rsif.2013.0810 [DOI] [PMC free article] [PubMed]
- Paracini N, Clifton LA, Skoda MW, Lakey JH (2018) Liquid crystalline bacterial outer membranes are critical for antibiotic susceptibility. Proc Natl Acad Sci 115(32):E7587–E7594. 10.1073/pnas.1803975115 [DOI] [PMC free article] [PubMed]
- Hughes AV, Patel DS, Widmalm G, Klauda JB, Clifton LA, Im W (2019) Physical properties of bacterial outer membrane models: neutron reflectometry & molecular simulation. Biophys J 116(6):1095–1104. 10.1016/j.bpj.2019.02.001 [DOI] [PMC free article] [PubMed]
- Dodd CE, Johnson BR, Jeuken LJ, Bugg TD, Bushby RJ, Evans SD, Native E (2008) coli inner membrane incorporation in solid-supported lipid bilayer membranes. Biointerphases 3(2):FA59–FA67. 10.1116/1.2896113 [DOI] [PubMed]
- Michel J, Wang Y, Kiesel I, Gerelli Y, Rosilio V (2017) Disruption of asymmetric lipid bilayer models mimicking the outer membrane of gram-negative bacteria by an active plasticin. Langmuir 33(41):11028–11039. 10.1021/acs.langmuir.7b02864 [DOI] [PubMed]
- Adhyapak P, Srivatsav AT, Mishra M, Singh A, Narayan R, Kapoor S (2020) Dynamical organization of compositionally distinct inner and outer membrane lipids of mycobacteria. Biophys J 118(6):1279–1291. 10.1016/j.bpj.2020.01.027 [DOI] [PMC free article] [PubMed]
- Nakatani Y, Shimaki Y, Dutta D, Muench SP, Ireton K, Cook GM, Jeuken LJ (2019) Unprecedented properties of phenothiazines unraveled by a NDH-2 bioelectrochemical assay platform. J Am Chem Soc 142(3):1311–1320. 10.1021/jacs.9b10254 [DOI] [PubMed]
- Hoiles W, Krishnamurthy V (2015) Dynamic modeling of antimicrobial pore formation in engineered tethered membranes. IEEE Trans Mol Biol Multi-Scale Commun 1(3):265–276. 10.1109/TMBMC.2016.2537299
- Schneck E, Oliveira RG, Rehfeldt F, Demé B, Brandenburg K, Seydel U, Tanaka M (2009) Mechanical properties of interacting lipopolysaccharide membranes from bacteria mutants studied by specular and off-specular neutron scattering. Phys Rev E 80(4):041929. 10.1103/PhysRevE.80.041929 [DOI] [PubMed]
- Lee, T.-H.; Hofferek, V.; Sani, M.-a.; Separovic, F.; Reid, G.; Aguilar, M. I., The Impact of Antibacterial Peptides on Bacterial Lipid Membranes Depends on Stage of Growth. Faraday Discussions2020. 10.1039/D0FD00052C [DOI] [PubMed]
- Nedelkovski V, Schwaighofer A, Wraight CA, Nowak C, Naumann RL (2013) Surface-enhanced infrared absorption spectroscopy (SEIRAS) of light-activated photosynthetic reaction centers from Rhodobacter sphaeroides reconstituted in a biomimetic membrane system. J Phys Chem C 117(32):16357–16363. 10.1021/jp4056347
- Niu L, Wohland T, Knoll W, Köper I (2017) Interaction of a synthetic antimicrobial peptide with a model bilayer platform mimicking bacterial membranes. Biointerphases 12(4):04E404. 10.1116/1.5001020 [DOI] [PubMed]
- Sharma P, Parthasarathi S, Patil N, Waskar M, Raut JS, Puranik M, Ayappa KG, Basu JK (2020) Assessing barriers for antimicrobial penetration in complex asymmetric bacterial membranes: A case study with thymol. Langmuir 36(30):8800–8814. 10.1021/acs.langmuir.0c01124 [DOI] [PubMed]
- McGillivray DJ, Valincius G, Heinrich F, Robertson JW, Vanderah DJ, Febo-Ayala W, Ignatjev I, Lösche M, Kasianowicz JJ (2009) Structure of functional Staphylococcus aureus α-hemolysin channels in tethered bilayer lipid membranes. Biophys J 96(4):1547–1553. 10.1016/j.bpj.2008.11.020 [DOI] [PMC free article] [PubMed]
- Dupuy FG, Pagano I, Andenoro K, Peralta MF, Elhady Y, Heinrich F, Tristram-Nagle S (2018) Selective interaction of colistin with lipid model membranes. Biophys J 114(4):919–928. 10.1016/j.bpj.2017.12.027 [DOI] [PMC free article] [PubMed]
- Li X, Smith AW (2019) Quantifying Lipid Mobility and Peptide Binding for Gram-Negative and Gram-Positive Model Supported Lipid Bilayers. J Phys Chem B 123(49):10433–10440. 10.1021/acs.jpcb.9b09709 [DOI] [PubMed]
- Clifton LA, Holt SA, Hughes AV, Daulton EL, Arunmanee W, Heinrich F, Khalid S, Jefferies D, Charlton TR, Webster JR (2015) An accurate in vitro model of the E. coli envelope. Angew Chem Int Ed 54(41):11952–11955. 10.1002/anie.201504287 [DOI] [PMC free article] [PubMed]
- Hsia C-Y, Chen L, Singh RR, DeLisa MP, Daniel S (2016) A molecularly complete planar bacterial outer membrane platform. Sci Rep 6(1):1–14. 10.1038/srep32715 [DOI] [PMC free article] [PubMed]
- Thomas CJ, Surolia N, Surolia A (1999) Surface plasmon resonance studies resolve the enigmatic endotoxin neutralizing activity of polymyxin B. J Biol Chem 274(42):29624–29627. 10.1074/jbc.274.42.29624 [DOI] [PubMed]
- Spencelayh MJ, Cheng Y, Bushby RJ, Bugg TD, Li JJ, Henderson PJ, O’Reilly J, Evans SD (2006) Antibiotic action and peptidoglycan formation on tethered lipid bilayer membranes. Angewandte Chemie 118(13):2165–2170. 10.1002/ange.200504035 [DOI] [PubMed]
- Mirandela GD, Tamburrino G, Hoskisson PA, Zachariae U, Javelle A (2019) The lipid environment determines the activity of the Escherichia coli ammonium transporter AmtB. FASEB J 33(2):1989–1999. 10.1096/fj.201800782r [DOI] [PMC free article] [PubMed]
- Maccarini M, Gayet L, Alcaraz J-P, Liguori L, Stidder B, Watkins EB, Lenormand J-L, Martin DK (2017) Functional characterization of cell-free expressed OprF porin from Pseudomonas aeruginosa stably incorporated in tethered lipid bilayers. Langmuir 33(38):9988–9996. 10.1021/acs.langmuir.7b01731 [DOI] [PubMed]
- Jeuken LJ, Connell SD, Henderson PJ, Gennis RB, Evans SD, Bushby RJ (2006) Redox enzymes in tethered membranes. J Am Chem Soc 128(5):1711–1716. 10.1021/ja056972u [DOI] [PMC free article] [PubMed]
- Jeuken LJ, Connell SD, Nurnabi M, O’Reilly J, Henderson PJ, Evans SD, Bushby RJ (2005) Direct electrochemical interaction between a modified gold electrode and a bacterial membrane extract. Langmuir 21(4):1481–1488. 10.1021/la047732f [DOI] [PMC free article] [PubMed]
- Mohamed Z, Shin J-H, Ghosh S, Sharma AK, Pinnock F, Bint E Naser Farnush S, Dörr T, Daniel S (2021) Clinically relevant bacterial outer membrane models for antibiotic screening applications. ACS Infect Dis 7(9):2707–2722. 10.1021/acsinfecdis.1c00217 [DOI] [PubMed]
- Zang M, MacDermott-Opeskin H, Adams FG, Naidu V, Waters JK, Carey AB, Ashenden A, McLean KT, Brazel EB, Jiang J-H, Panizza A, Trappetti C, Paton JC, Peleg AY, Köper I, Paulsen IT, Hassan KA, O’Mara ML, Eijkelkamp BA (2021) The Membrane Composition Defines the Spatial Organization and Function of a Major Acinetobacter baumannii Drug Efflux System. mBio 12(3):1–6. 10.1128/mBio.01070-21 [DOI] [PMC free article] [PubMed]
- Alghalayini A, Garcia A, Berry T, Cranfield CG (2019) The use of tethered bilayer lipid membranes to identify the mechanisms of antimicrobial peptide interactions with lipid bilayers. Antibiotics 8(1):12. 10.3390/antibiotics8010012 [DOI] [PMC free article] [PubMed]
- Tamm LK, McConnell HM (1985) Supported phospholipid bilayers. Biophys J 47(1):105–113. 10.1016/s0006-3495(85)83882-0 [DOI] [PMC free article] [PubMed]
- Tanaka M, Sackmann E (2005) Polymer-supported membranes as models of the cell surface. Nature 437(7059):656–663. 10.1038/nature04164 [DOI] [PubMed]
- Naumann C, Knoll W, Frank C (2001) Hindered Diffusion in Polymer-Tethered Membranes: A Monolayer Study at the Air− Water Interface. Biomacromol 2(4):1097–1103. 10.1021/bm010022t [DOI] [PubMed]
- Naumann CA, Prucker O, Lehmann T, Rühe J, Knoll W, Frank CW (2002) The polymer-supported phospholipid bilayer: Tethering as a new approach to substrate− membrane stabilization. Biomacromol 3(1):27–35. 10.1021/bm0100211 [DOI] [PubMed]
- Deleu M, Crowet J-M, Nasir MN, Lins L (2014) Complementary biophysical tools to investigate lipid specificity in the interaction between bioactive molecules and the plasma membrane: A review. Biochimica et Biophysica Acta (BBA)-Biomembranes 1838(12):3171–3190. 10.1016/j.bbamem.2014.08.023 [DOI] [PubMed]
- Lyman E, Hsieh C-L, Eggeling C (2018) From dynamics to membrane organization: experimental breakthroughs occasion a “modeling manifesto.” Biophys J 115(4):595–604. 10.1016/j.bpj.2018.07.012 [DOI] [PMC free article] [PubMed]
- Nickels JD, Smith JC, Cheng X (2015) Lateral organization, bilayer asymmetry, and inter-leaflet coupling of biological membranes. Chem Phys Lipid 192:87–99. 10.1016/j.chemphyslip.2015.07.012 [DOI] [PubMed]
- Maity PC, Yang J, Klaesener K, Reth M (2015) The nanoscale organization of the B lymphocyte membrane. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1853(4):830–840. 10.1016/j.bbamcr.2014.11.010 [DOI] [PMC free article] [PubMed]
- Marrink SJ, Corradi V, Souza PC, Ingólfsson HI, Tieleman DP, Sansom MS (2019) Computational modeling of realistic cell membranes. Chem Rev 119(9):6184–6226. 10.1021/acs.chemrev.8b00460 [DOI] [PMC free article] [PubMed]
- Ingólfsson HI, Arnarez C, Periole X, Marrink SJ (2016) Computational ‘microscopy’of cellular membranes. J Cell Sci 129(2):257–268. 10.1242/jcs.176040 [DOI] [PubMed]
- MacKerell AD Jr (2004) Empirical force fields for biological macromolecules: overview and issues. J Comput Chem 25(13):1584–1604. 10.1002/jcc.20082 [DOI] [PubMed]
- Mori T, Miyashita N, Im W, Feig M, Sugita Y (2016) Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(7):1635–1651. 10.1016/j.bbamem.2015.12.032 [DOI] [PMC free article] [PubMed]
- Matamoros-Recio A, Franco-Gonzalez JF, Forgione RE, Torres-Mozas A, Silipo A, Martín-Santamaría S (2021) Understanding the Antibacterial Resistance: Computational Explorations in Bacterial Membranes. ACS Omega 6(9):6041–6054. 10.1021/acsomega.0c05590 [DOI] [PMC free article] [PubMed]
- Bennett WD, Tieleman DP (2013) Computer simulations of lipid membrane domains. Biochimica et Biophysica Acta (BBA)-Biomembranes 1828(8):1765–1776. 10.1016/j.bbamem.2013.03.004 [DOI] [PubMed]
- Chan C, Wen H, Lu L, Fan J (2015) Multiscale molecular dynamics simulations of membrane remodeling by Bin/Amphiphysin/Rvs family proteins. Chinese Physics B 25(1):018707. 10.1088/1674-1056/25/1/018707
- Kabedev A, Hossain S, Hubert M, Larsson P, Bergström CA (2021) Molecular dynamics simulations reveal membrane interactions for poorly water-soluble drugs: impact of bile solubilization and drug aggregation. J Pharm Sci 110(1):176–185. 10.1016/j.xphs.2020.10.061 [DOI] [PubMed]
- Khan SH, Prakash A, Pandey P, Lynn AM, Islam A, Hassan MI, Ahmad F (2019) Protein folding: Molecular dynamics simulations and in vitro studies for probing mechanism of urea-and guanidinium chloride-induced unfolding of horse cytochrome-c. Int J Biol Macromol 122:695–704. 10.1016/j.ijbiomac.2018.10.186 [DOI] [PubMed]
- Lazim R, Suh D, Choi S (2020) Advances in molecular dynamics simulations and enhanced sampling methods for the study of protein systems. Int J Mol Sci 21(17):6339. 10.3390/ijms21176339 [DOI] [PMC free article] [PubMed]
- Liu Y, de Vries AH, Pezeshkian W, Marrink SJ (2021) Capturing Membrane Phase Separation by Dual Resolution Molecular Dynamics Simulations. J Chem Theory Comput 17(9):5876–5884. 10.1021/acs.jctc.1c00151 [DOI] [PMC free article] [PubMed]
- Parkin J, Chavent M, Khalid S (2015) Molecular simulations of Gram-negative bacterial membranes: a vignette of some recent successes. Biophys J 109(3):461–468. 10.1016/j.bpj.2015.06.050 [DOI] [PMC free article] [PubMed]
- Reddy T, Sansom MS (2016) Computational virology: from the inside out. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(7):1610–1618. 10.1016/j.bbamem.2016.02.007 [DOI] [PMC free article] [PubMed]
- Singharoy A, Schulten K (2017) Atom-Resolved View of a Cell Organelle on a Computational Microscope. Biophys J 112(3):176a. 10.1016/j.bpj.2016.11.973
- Balusek C, Gumbart JC (2016) Role of the native outer-membrane environment on the transporter BtuB. Biophys J 111(7):1409–1417. 10.1016/j.bpj.2016.08.033 [DOI] [PMC free article] [PubMed]
- Baltoumas FA, Hamodrakas SJ, Iconomidou VA (2019) The gram-negative outer membrane modeler: Automated building of lipopolysaccharide-rich bacterial outer membranes in four force fields. J Comput Chem 40(18):1727–1734. 10.1002/jcc.25823 [DOI] [PubMed]
- Gao Y, Lee J, Widmalm G, Im W (2020) Modeling and Simulation of Bacterial Outer Membranes with Lipopolysaccharides and Enterobacterial Common Antigen. J Phys Chem B 124(28):5948–5956. 10.1021/acs.jpcb.0c03353 [DOI] [PubMed]
- Kholina EG, Kovalenko IB, Bozdaganyan ME, Strakhovskaya MG, Orekhov PS (2020) Cationic antiseptics facilitate pore formation in model bacterial membranes. J Phys Chem B 124(39):8593–8600. 10.1021/acs.jpcb.0c07212 [DOI] [PubMed]
- Li Y, Guo H (2013) Atomistic simulations of an antimicrobial molecule interacting with a model bacterial membrane. Theoret Chem Acc 132(1):1–8. 10.1007/s00214-012-1303-y
- Abellón-Ruiz J, Kaptan SS, Baslé A, Claudi B, Bumann D, Kleinekathöfer U, van den Berg B (2017) Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2(12):1616–1623. 10.1038/s41564-017-0046-x [DOI] [PubMed]
- Berglund NA, Piggot TJ, Jefferies D, Sessions RB, Bond PJ, Khalid S (2015) Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: a molecular dynamics study. PLoS Comput Biol 11(4):e1004180. 10.1371/journal.pcbi.1004180 [DOI] [PMC free article] [PubMed]
- Hsu P-C, Samsudin F, Shearer J, Khalid S (2017a) It is complicated: curvature, diffusion, and lipid sorting within the two membranes of Escherichia coli. J Phys Chem Lett 8(22):5513–5518. 10.1021/acs.jpclett.7b02432 [DOI] [PubMed]
- Hsu, P. C.; Bruininks, B. M.; Jefferies, D.; Cesar Telles de Souza, P.; Lee, J.; Patel, D. S.; Marrink, S. J.; Qi, Y.; Khalid, S.; Im, W., CHARMM‐GUI Martini Maker for modeling and simulation of complex bacterial membranes with lipopolysaccharides. Wiley Online Library: 2017. 10.1002/jcc.24895 [DOI] [PMC free article] [PubMed]
- Ma H, Khan A, Nangia S (2017a) Dynamics of OmpF trimer formation in the bacterial outer membrane of Escherichia coli. Langmuir 34(19):5623–5634. 10.1021/acs.langmuir.7b02653 [DOI] [PubMed]
- Mehmood S, Corradi V, Choudhury HG, Hussain R, Becker P, Axford D, Zirah S, Rebuffat S, Tieleman DP, Robinson CV (2016) Structural and functional basis for lipid synergy on the activity of the antibacterial peptide ABC transporter McjD. J Biol Chem 291(41):21656–21668. 10.1074/jbc.M116.732107 [DOI] [PMC free article] [PubMed]
- Orekhov PS, Kholina EG, Bozdaganyan ME, Nesterenko AM, Kovalenko IB, Strakhovskaya MG (2018) Molecular mechanism of uptake of cationic photoantimicrobial phthalocyanine across bacterial membranes revealed by molecular dynamics simulations. J Phys Chem B 122(14):3711–3722. 10.1021/acs.jpcb.7b11707 [DOI] [PubMed]
- Shearer J, Jefferies D, Khalid S (2019) Outer membrane proteins OmpA, FhuA, OmpF, EstA, BtuB, and OmpX have unique lipopolysaccharide fingerprints. J Chem Theory Comput 15(4):2608–2619. 10.1021/acs.jctc.8b01059 [DOI] [PubMed]
- Shearer J, Khalid S (2018) Communication between the leaflets of asymmetric membranes revealed from coarse-grain molecular dynamics simulations. Sci Rep 8(1):1–6. 10.1038/s41598-018-20227-1 [DOI] [PMC free article] [PubMed]
- Rice A, Wereszczynski J (2018) Atomistic scale effects of lipopolysaccharide modifications on bacterial outer membrane defenses. Biophys J 114(6):1389–1399. 10.1016/j.bpj.2018.02.006 [DOI] [PMC free article] [PubMed]
- Patel DS, Re S, Wu EL, Qi Y, Klebba PE, Widmalm G, Yeom MS, Sugita Y, Im W (2016) Dynamics and interactions of OmpF and LPS: influence on pore accessibility and ion permeability. Biophys J 110(4):930–938. 10.1016/j.bpj.2016.01.002 [DOI] [PMC free article] [PubMed]
- Piggot TJ, Holdbrook DA, Khalid S (2011) Electroporation of the E. coli and S. aureus membranes: molecular dynamics simulations of complex bacterial membranes. J Phys Chem B 115(45):13381–13388. 10.1021/jp207013v [DOI] [PubMed]
- Carpenter TS, Parkin J, Khalid S (2016) The free energy of small solute permeation through the Escherichia coli outer membrane has a distinctly asymmetric profile. J Phys Chem Lett 7(17):3446–3451. 10.1021/acs.jpclett.6b01399 [DOI] [PubMed]
- Fleming PJ, Patel DS, Wu EL, Qi Y, Yeom MS, Sousa MC, Fleming KG, Im W (2016) BamA POTRA domain interacts with a native lipid membrane surface. Biophys J 110(12):2698–2709. 10.1016/j.bpj.2016.05.010 [DOI] [PMC free article] [PubMed]
- Wu EL, Engström O, Jo S, Stuhlsatz D, Yeom MS, Klauda JB, Widmalm G, Im W (2013) Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys J 105(6):1444–1455. 10.1016/j.bpj.2013.08.002 [DOI] [PMC free article] [PubMed]
- Wu EL, Fleming PJ, Yeom MS, Widmalm G, Klauda JB, Fleming KG, Im W (2014) E. coli outer membrane and interactions with OmpLA. Biophys J 106(11):2493–2502. 10.1016/j.bpj.2014.04.024 [DOI] [PMC free article] [PubMed]
- Duay SS, Sharma G, Prabhakar R, Angeles-Boza AM, May ER (2019) Molecular dynamics investigation into the effect of zinc (II) on the structure and membrane interactions of the antimicrobial peptide Clavanin A. J Phys Chem B 123(15):3163–3176. 10.1021/acs.jpcb.8b11496 [DOI] [PMC free article] [PubMed]
- Khondker A, Dhaliwal AK, Saem S, Mahmood A, Fradin C, Moran-Mirabal J, Rheinstädter MC (2019) Membrane charge and lipid packing determine polymyxin-induced membrane damage. Commun Biol 2(1):1–11. 10.1038/s42003-019-0297-6 [DOI] [PMC free article] [PubMed]
- Ma H, Cummins DD, Edelstein NB, Gomez J, Khan A, Llewellyn MD, Picudella T, Willsey SR, Nangia S (2017b) Modeling diversity in structures of bacterial outer membrane lipids. J Chem Theory Comput 13(2):811–824. 10.1021/acs.jctc.6b00856 [DOI] [PubMed]
- Ma H, Irudayanathan FJ, Jiang W, Nangia S (2015) Simulating Gram-negative bacterial outer membrane: a coarse grain model. J Phys Chem B 119(46):14668–14682. 10.1021/acs.jpcb.5b07122 [DOI] [PubMed]
- Pandit KR, Klauda JB (2012) Membrane models of E. coli containing cyclic moieties in the aliphatic lipid chain. Biochimica et Biophysica Acta (BBA)-Biomembranes 1818(5):1205–1210. 10.1016/j.bbamem.2012.01.009 [DOI] [PubMed]
- Pothula KR, Solano CJ, Kleinekathöfer U (2016) Simulations of outer membrane channels and their permeability. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(7):1760–1771. 10.1016/j.bbamem.2015.12.020 [DOI] [PubMed]
- Shahane G, Ding W, Palaiokostas M, Azevedo HS, Orsi M (2019) Interaction of antimicrobial lipopeptides with bacterial lipid bilayers. J Membr Biol 252(4):317–329. 10.1007/s00232-019-00068-3 [DOI] [PMC free article] [PubMed]
- Khakbaz P, Klauda JB (2015) Probing the importance of lipid diversity in cell membranes via molecular simulation. Chem Phys Lipid 192:12–22. 10.1016/j.chemphyslip.2015.08.003 [DOI] [PubMed]
- Lim JB, Klauda JB (2011) Lipid chain branching at the iso-and anteiso-positions in complex chlamydia membranes: A molecular dynamics study. Biochimica et Biophysica Acta (BBA)-Biomembranes 1808(1):323–331. 10.1016/j.bbamem.2010.07.036 [DOI] [PubMed]
- Jin T, Patel SJ, Van Lehn RC (2021) Molecular simulations of lipid membrane partitioning and translocation by bacterial quorum sensing modulators. Plos one 16(2):e0246187. 10.1371/journal.pone.0246187 [DOI] [PMC free article] [PubMed]
- Lee J, Patel DS, Kucharska I, Tamm LK, Im W (2017) Refinement of OprH-LPS interactions by molecular simulations. Biophys J 112(2):346–355. 10.1016/j.bpj.2016.12.006 [DOI] [PMC free article] [PubMed]
- Ocampo-Ibáñez ID, Liscano Y, Rivera-Sánchez SP, Oñate-Garzón J, Lugo-Guevara AD, Flórez-Elvira LJ, Lesmes MC (2020) A Novel Cecropin D-Derived Short Cationic Antimicrobial Peptide Exhibits Antibacterial Activity Against Wild-Type and Multidrug-Resistant Strains of Klebsiella pneumoniae and Pseudomonas aeruginosa. Evol Bioinforma 16:1176934320936266. 10.1177/1176934320936266 [DOI] [PMC free article] [PubMed]
- Alkhalifa S, Jennings MC, Granata D, Klein M, Wuest WM, Minbiole KP, Carnevale V (2020) Analysis of the Destabilization of Bacterial Membranes by Quaternary Ammonium Compounds: A Combined Experimental and Computational Study. Chembiochem 21(10):1510. 10.1002/cbic.201900698 [DOI] [PMC free article] [PubMed]
- Lins RD, Straatsma T (2001) Computer simulation of the rough lipopolysaccharide membrane of Pseudomonas aeruginosa. Biophys J 81(2):1037–1046. 10.1016/S0006-3495(01)75761-X [DOI] [PMC free article] [PubMed]
- Yu Y, Klauda JB (2018) Modeling Pseudomonas aeruginosa inner plasma membrane in planktonic and biofilm modes. J Chem Phys 149(21):215102. 10.1063/1.5052629 [DOI] [PubMed]
- Hwang H, Paracini N, Parks JM, Lakey JH, Gumbart JC (2018) Distribution of mechanical stress in the Escherichia coli cell envelope. Biochimica et Biophysica Acta (BBA)-Biomembranes 1860(12):2566–2575. 10.1016/j.bbamem.2018.09.020 [DOI] [PMC free article] [PubMed]
- Piggot TJ, Holdbrook DA, Khalid S (2013) Conformational dynamics and membrane interactions of the E coli outer membrane protein FecA: a molecular dynamics simulation study. Biochimica et Biophysica Acta (BBA)-Biomembranes 1828(2):284–293. 10.1016/j.bbamem.2012.08.021 [DOI] [PubMed]
- Kirschner KN, Lins RD, Maass A, Soares TA (2012) A glycam-based force field for simulations of lipopolysaccharide membranes: parametrization and validation. J Chem Theory Comput 8(11):4719–4731. 10.1021/ct300534j [DOI] [PubMed]
- Dias RP, da Hora GC, Ramstedt M, Soares TA (2014) Outer membrane remodeling: the structural dynamics and electrostatics of rough lipopolysaccharide chemotypes. J Chem Theory Comput 10(6):2488–2497. 10.1021/ct500075h [DOI] [PubMed]
- Van Oosten B, Harroun TA (2016) A MARTINI extension for Pseudomonas aeruginosa PAO1 lipopolysaccharide. J Mol Graph Model 63:125–133. 10.1016/j.jmgm.2015.12.002 [DOI] [PubMed]
- Hsu P-C, Jefferies D, Khalid S (2016) Molecular dynamics simulations predict the pathways via which pristine fullerenes penetrate bacterial membranes. J Phys Chem B 120(43):11170–11179. 10.1021/acs.jpcb.6b06615 [DOI] [PubMed]
- Shearer J, Marzinek JK, Bond PJ, Khalid S (2020) Molecular dynamics simulations of bacterial outer membrane lipid extraction: Adequate sampling? J Chem Phys 153(4):044122. 10.1063/5.0017734 [DOI] [PubMed]
- Lee J, Patel DS, Ståhle J, Park S-J, Kern NR, Kim S, Lee J, Cheng X, Valvano MA, Holst O (2018) CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J Chem Theory Comput 15(1):775–786. 10.1021/acs.jctc.8b01066 [DOI] [PubMed]
- Wu, E. L.; Cheng, X.; Jo, S.; Rui, H.; Song, K. C.; Dávila‐Contreras, E. M.; Qi, Y.; Lee, J.; Monje‐Galvan, V.; Venable, R. M., CHARMM‐GUI membrane builder toward realistic biological membrane simulations. Wiley Online Library: 2014. 10.1002/jcc.23702 [DOI] [PMC free article] [PubMed]
- Khalid S, Berglund NA, Holdbrook DA, Leung YM, Parkin J (2015) The membranes of Gram-negative bacteria: progress in molecular modelling and simulation. Biochem Soc Trans 43(2):162–167. 10.1042/bst20140262 [DOI] [PubMed]
- Patel DS, Qi Y, Im W (2017) Modeling and simulation of bacterial outer membranes and interactions with membrane proteins. Curr Opin Struct Biol 43:131–140. 10.1016/j.sbi.2017.01.003 [DOI] [PubMed]
- Chakraborty, A.; Kobzev, E.; Chan, J.; de Zoysa, G. H.; Sarojini, V.; Piggot, T. J.; Allison, J. R., Molecular Dynamics Simulation of the Interaction of Two Linear Battacin Analogs with Model Gram-Positive and Gram-Negative Bacterial Cell Membranes. ACS Omega2020. 10.1021/acsomega.0c04752 [DOI] [PMC free article] [PubMed]
- Kim S, Patel DS, Park S, Slusky J, Klauda JB, Widmalm G, Im W (2016) Bilayer properties of lipid A from various Gram-negative bacteria. Biophys J 111(8):1750–1760. 10.1016/j.bpj.2016.09.001 [DOI] [PMC free article] [PubMed]
- Goossens K, De Winter H (2018) Molecular dynamics simulations of membrane proteins: An overview. J Chem Inf Model 58(11):2193–2202. 10.1021/acs.jcim.8b00639 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.