Abstract
Background
Kawasaki disease (KD) is a systemic vasculitis that is currently the most common cause of acquired heart disease in children. However, its etiology remains unknown. Long non-coding RNAs (lncRNAs) contribute to the pathophysiology of various diseases. Few studies have reported the role of lncRNAs in KD inflammation; thus, we investigated the role of lncRNA in KD inflammation.
Methods
A total of 50 patients with KD (median age, 19 months; 29 males and 21 females) were enrolled. We conducted cap analysis gene expression sequencing to determine differentially expressed genes in monocytes of the peripheral blood of the subjects.
Results
About 21 candidate lncRNA transcripts were identified. The analyses of transcriptome and gene ontology revealed that the immune system was involved in KD. Among these genes, G0/G1 switch gene 2 (G0S2) and its antisense lncRNA, HSD11B1-AS1, were upregulated during the acute phase of KD (P < 0.0001 and <0.0001, respectively). Moreover, G0S2 increased when lipopolysaccharides induced inflammation in THP-1 monocytes, and silencing of G0S2 suppressed the expression of HSD11B1-AS1 and tumor necrosis factor-α.
Conclusions
This study uncovered the crucial role of lncRNAs in innate immunity in acute KD. LncRNA may be a novel target for the diagnosis of KD.
Impact
This study revealed the whole aspect of the gene expression profile of monocytes of patients with Kawasaki disease (KD) using cap analysis gene expression sequencing and identified KD-specific molecules: G0/G1 switch gene 2 (G0S2) and long non-coding RNA (lncRNA) HSD11B1-AS1.
We demonstrated that G0S2 and its antisense HSD11B1-AS1 were associated with inflammation of innate immunity in KD.
lncRNA may be a novel key target for the diagnosis of patients with KD.
Introduction
Kawasaki disease (KD) is an acute inflammatory syndrome in the form of systemic vasculitis that occurs in children.1,2 It was first reported in 1967 by Tomisaku Kawasaki, a Japanese doctor. In Asia, especially in Japan, the number of patients with KD continues to increase. Currently, it is the most common cause of acquired cardiac disease in children in developed countries.3,4 Intriguingly, severe Kawasaki-like disease has been reported in pediatric patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a pandemic in 2020.5 KD is attracting attention worldwide and may be considered as a global disease.
Several studies have suggested that infectious agents, genetic factors, and/or environmental factors are involved in the pathogenesis of KD. Although the etiology of KD remains unknown, innate immunity plays a significant role in its pathogenesis according to the following two pieces of evidence.6 First, patients with KD have high recurrence rates within 1 year of onset.7 Second, the levels of damage-associated molecular patterns, which are induced in innate immunity, such as the S100A proteins, are elevated in the serum of patients with acute KD.8,9
Non-coding RNAs are non-protein-coding transcripts, and long non-coding RNAs (lncRNAs) are classified as those longer than 200 nucleotides.10 Recently, lncRNAs have been studied more for their potential role in the pathogenesis and progression of various diseases as they are involved in various biological processes during gene transcription regulation. Several recent studies have reported that lncRNAs play a significant role in cardiovascular diseases, including myocardial infarction, atherosclerosis, and cardiomyopathy. This suggests that they are therapeutic targets for these conditions.4,11–14
However, few studies have investigated the role of lncRNAs in KD. Particularly, no studies have focused on monocytes, although they significantly contribute to the inflammation of KD. Therefore, we hypothesized that lncRNAs significantly contribute to inflammation during the acute phase of KD. To test this hypothesis, we conducted the transcriptomic analysis using cap analysis gene expression sequencing (CAGE-seq) of monocytes from patients with KD to identify specific lncRNAs related to KD and elucidate their role during the acute phase of KD.
Methods
Enrollment of patients
A total of 50 patients with KD and 50 age-matched controls (25 febrile patients without KD and 25 healthy children) (KD vs. febrile control, P = 0.13; KD vs. healthy control: P = 0.16, respectively) were enrolled from August 2016 to March 2018 at the Toyama University Hospital and other affiliated hospitals. The study was approved by the ethics committee of the University of Toyama and conducted according to the Declaration of Helsinki. The patients and controls were enrolled after obtaining informed consent from their parents or legal guardians. KD was diagnosed using the previously defined clinical diagnostics.15 Patients with KD who responded to intravenous immunoglobulin (IVIG) treatment and those whose fever decreased within 48 h of treatment were designated as responders, whereas those with KD who did not respond to IVIG were designated as non-responders. Blood samples were collected at two time points for each patient: acute phase of KD before IVIG administration (pre-IVIG) and subacute phase approximately 7 days after IVIG injection (post-IVIG). The blood samples from healthy children were drawn at opportunities of usual examination, such as an allergy test.
Separation of monocytes and confirmation of accuracy by fluorescence-activated cell sorting (FACS)
Peripheral blood monocular cells were separated via centrifugation of leukocytes using polysucrose solution (Sigma-Aldrich, St. Louis, MO). The monocytes were separated and collected using magnetic beads carrying antibodies against the cluster of differentiation 14 (CD14) (Sigma-Aldrich, St. Louis, MO). The collected monocytes were subjected to FACS to ensure the purity of 98% using anti-CD14-FITC antibody (Beckman Coulter, Tokyo, Japan).
RNA isolation
Total RNA was extracted from the human monocyte samples using RNeasy Kits (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s protocols and then quantitated using a NanoVue (GE Health Care, Chicago, IL). The extracted RNA samples were stored at −150 °C until further processing.
CAGE-seq
We chose 3 IVIG responders, 3 IVIG non-responders, and 3 afebrile controls for CAGE-seq (Sup. Table 1). The sequenced read data were submitted to the DDBJ Sequence Read Archive (accession number DRA 013102). CAGE library preparation, sequencing, mapping, gene expression, and motif discovery analysis were conducted. In addition, total RNA quality was evaluated using a bioanalyzer (Agilent, Santa Clara, CA) to ensure that the RNA integrity number was >7.0; complementary DNAs (cDNAs) were also synthesized from total RNA using random primers. The ribose diols in the 5′ cap structures of RNAs were oxidized and then biotinylated. The biotinylated RNAs/cDNAs were captured using streptavidin magnetic beads (cap-trapping method). After RNA digestion using RNase ONE/H and adaptor ligation of both ends of the cDNA, double-stranded cDNA libraries (CAGE libraries) were constructed. The CAGE libraries were sequenced using single-end reads of 75 nt on a NextSeq 500 instrument (Illumina, San Diego, CA).
Differentially expressed gene (DEG) analysis
In CAGE-seq, DEG analysis was conducted as follows. After removing the reads derived from ribosomal RNA and reads containing non-A/T/G/C base, the remaining reads were mapped to the human reference genome (hg19) using BWA (ver. 0.7) and HISAT (ver.2.0.5).16,17 For tag clustering, the CAGEtag 5′ coordinates were entered for the REproducible CLUStering method (RECLU) clustering pipeline using the following parameters: maximum irreproducible discovery rate, 0.1; minimum count per million value, 0.1; and gene annotation (GENCODE human gene annotation release 19).18 The gene-level tag count outputs of the RECLU clustering were subjected to DEG analysis using R package edgeR (version 3.26).19 In the preprocessing, genes with low raw counts (<10) were removed according to the user manual, and counts were normalized using the relative log expression method. Then, the exact test in edgeR was conducted using the normalized counts as input to assess the differential expression of the genes for each comparison (healthy controls vs. KD pre-IVIG patients and IVIG non-responders vs. IVIG responders). Finally, genes with a false discovery rate (FDR) adjusted P value of <0.05 and an absolute log2 fold change value of ≥1 between groups were defined as DEGs. The volcano plot was drawn using the R package ggplot2 with the output of edgeR. The heatmap of the gene expression level of DEGs was drawn using the R software with the following parameters: clustering method (Ward) and distance measure (Euclidean).
Enrichment analysis
The DEGs were subjected to enrichment analysis using R package goseq (version 1.3).20 Gene ontology (GO) terms with P values <0.05 were defined as significantly enriched GO terms. Furthermore, significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were determined using the same criteria utilized in the GO enrichment analysis.
Real-time quantitative reverse transcription-polymerase chain reaction (qRT–PCR)
For real-time PCR (RT–PCR) analysis, RNA samples were reverse-transcribed using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Japan) according to the manufacturer’s protocol. RT–PCR was conducted using THUNDERBIRD SYBR qPCR Mix (Toyobo) on Thermal Cycler Dice Real-Time System II (Takara Bio, Kusatsu, Japan). Quantification was performed using the ΔΔCt method with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression as a reference gene. RT–PCR was conducted using human monocyte samples to validate the results of CAGE from 25 patients.
Cell culture
The human monocyte cell line, THP-1, was purchased from the JCRB Cell Bank (National Institute of Biomedical Innovation, Ibaraki, Japan). The cells were cultured with RPMI-1640 containing 10% heat-inactivated fetal calf serum, 1% penicillin G, and 0.1% gentamicin at 37 °C in a humidified atmosphere containing 5% CO2. To induce inflammation, 1.0 × 105 THP-1 cells were plated in 24-well plates containing 1-mL RPMI media; after 24 h, 1 μg/mL of lipopolysaccharide (LPS) (Sigma-Aldrich, St. Louis, MO) or 500 ng/mL of Pam3CSK4 (Pam) (Sigma-Aldrich, St. Louis, MO) was added to the media. RNA samples were collected 24 h after LPS or Pam stimulation.
Knockdown of G0S2 in THP-1 monocytes
The small interfering RNA (siRNA) delivery system (Dharmacon, Lafayette, CO) was utilized to knock down the expression of G0S2 (sense sequence: GCAuCCACCAAAGGAGuuuGGTT). This siRNA for G0S2 was designed on a gene corresponding to an intron region of HSD11b1-AS1 on the complementary strand to avoid the off-target effect for HSD11b1-AS1. siRNA sequence that had at least two mismatches with HSD11b1-AS1 were selected. In addition, we used BLAST to verify that other gene sequences were not affected. Control pools (Nippon Gene, Inc., Tokyo, Japan) containing non-targeting siRNAs were used as controls in all siRNA transfection experiments. THP-1 monocytes were plated in 24-well plates, cultured with growth media (RPMI medium supplemented with 10% FBS) for 24 h, and then transfected with siRNAs using Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol: a 10-μM siRNA solution was prepared by mixing the siRNA stock solution with Opti-MEM (Thermo Fisher Scientific, Waltham, MA) for 5 min; subsequently, 50 μL was added to the well containing 1 mL of media. The cells were cultured at 37 °C with 5% CO2 for 48 h and then treated with LPS or Pam, as described above.
Statistical analysis
Continuous variables were expressed as mean ± 1 standard deviation; categorical variables were expressed as numbers and percentages. Continuous variables were compared using the unpaired t test, nonparametric Mann–Whitney test, or one-way analysis of variance, as necessary. Conversely, categorical variables were compared using χ2 statistics or Fisher’s exact test as appropriate. P values <0.05 were considered statistically significant. Statistical analyses were conducted using JMP (version 13; SAS Institute, Cary, NC). Graphs were drawn using Prism 6.04 (GraphPad Software, Inc., La Jolla, CA).
Results
Clinical characteristics of patients with KD
Table 1 presents the specific demographic and clinical characteristics of the patients enrolled in this study. A total of 50 patients with KD (median age, 19 months; range, 4 months to 4 years; 29 males and 21 females), 25 febrile patients without KD, and 25 healthy volunteers were enrolled in this study. Among the patients with KD, 45 responded to IVIG treatment, and 5 showed no response. The data indicated that patients with KD had significantly longer fever duration, higher levels of C-reactive protein, and higher leukocyte and platelet counts than those in the febrile patient group. Furthermore, serum sodium was significantly lower in the KD group. Other items were not significantly different between the KD and febrile patient groups (Table 1).
Table 1.
Demographic and clinical characteristics of the enrolled patients.
| KD (n = 50) | Non-KD febrile (n = 25) | Healthy (n = 25) | P value (KD vs. febrile control) | ||
|---|---|---|---|---|---|
| Responder (n = 45) | Non-responder (n = 5) | ||||
| Age at onset, months, mean (range) | 18.2 (4–42) | 27 (12–55) | 14 (4–48) | 26.1 (12–72) | 0.13 |
| Male sex, n (%) | 25 (55) | 4 (80) | 11 (55.0) | 15 (60.0) | — |
| Fever duration, days, mean ± SD* | 5.5 (3–9) | 9.3 (8–11) | 3.5 (2–5) | — | <0.001 |
| CRP, mg/dL, mean ± SD* | 6.8 ± 5.7* | 4.6 ± 1.1 | 2.8 ± 1.0 | 0.2 ± 0.29 | 0.006 |
| WBC count, /mm3, mean ± SD* | 14,735 ± 3777 | 13300 ± 3874 | 10,835 ± 1521 | 8220 ± 836 | <0.001 |
| Neutrophils, %, mean ± SD | 65.8 ± 14.4 | 61.7 ± 19.8 | 65.9 ± 6.9 | 42.6 ± 5.5 | 0.871 |
| Platelet count, ×104/mm3, mean ± SD* | 37.5 ± 10.5 | 48.7 ± 16 | 26.2 ± 5.3 | 27.7 ± 4.7 | <0.001 |
| Na, mmol/L, mean ± SD* | 134.6 ± 2.4 | 135 ± 1.5 | 136.7 ± 3.0 | 137 ± 3.2 | 0.023 |
| AST, IU/L, mean ± SD | 90 (23–580) | 203 (23–549) | 33.6 ± 11.9 | 36.3 ± 8.4 | 0.095 |
| Maximum RCA (#1), mm, mean (range) | 1.92 (1.4–2.8) | 2.5 (1.8–3.4) | — | — | — |
| Maximum LCA (#5), mm, mean (range) | 2.1 (1.7–2.8) | 2.5 (2.2–4.0) | — | — | — |
KD Kawasaki disease, CRP C-reactive protein, WBC white blood cell, CA coronary artery, RCA right coronary artery, LCA left coronary artery.
*P < 0.05.
CAGE-seq identified differentially expressed lncRNAs in patients with acute KD
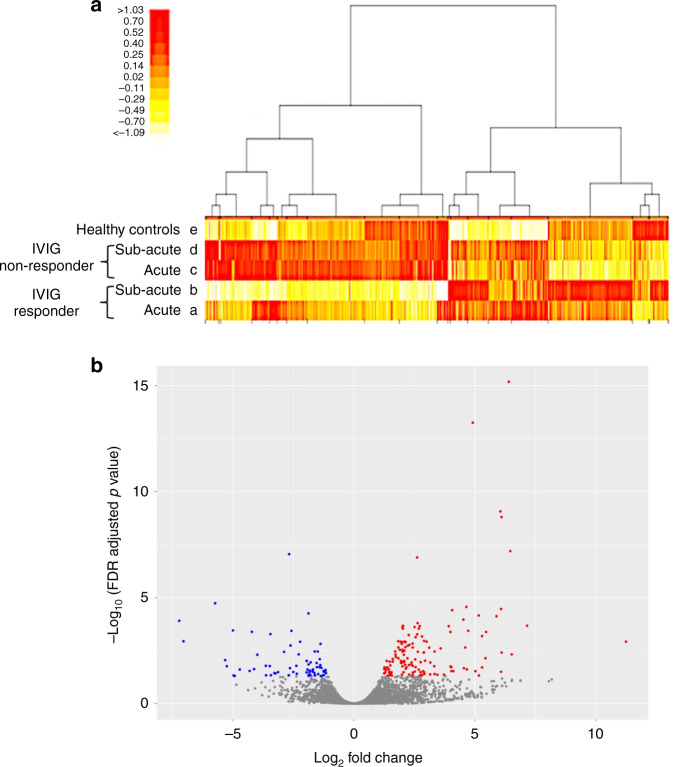
To identify widely affected expression profiles during the acute phase of KD, CAGE-seq analyses were conducted on peripheral blood monocyte RNAs (Fig. 1a). DEGs were detected with the following cutoff value: an expression difference of more than twofold (≥log2 value 1.00) and an FDR of <0.05. Comparing the pre-IVIG responders and non-responders, DEGs were not detected on lncRNAs. Then, we combined the pre-IVIG responders and non-responders as whole pre-IVIG KD and compared them with the healthy control group (Fig. 1b). Among all DEGs, 21 candidate lncRNA transcripts were identified; of these, 10 were elevated, and 11 were reduced (Table 2).
Fig. 1. Comprehensive gene expression analysis of monocytes by cap analysis gene expression sequencing (CAGE-seq).
a Heatmap of the expression levels of all transcription start points by CAGE-seq. Hierarchical clustering demonstrates the average change in the total transcript levels obtained from monocytes of patients with KD during the acute (a) and subacute phases (b) in the IVIG response group (IVIG responder) (n = 3), patients with KD during the acute (c) and subacute phases (d) in the IVIG refractory group (IVIG non-responder) (n = 3), and healthy controls (n = 3) (e). Regions indicated in red are genes with high expression levels, and those indicated in yellow are genes with low expression levels. The columns of the heatmap indicate transcripts and rows that present the participants. b Volcano plot comparing the gene expression of patients with KD pre-IVIG (responders and non-responders, n = 6) with that of healthy controls (n = 3). All the transcripts detected were depicted as dots, and DEGs were colored; genes with higher expression than healthy controls are indicated in red, and those with lower expression levels are indicated in blue.
Table 2.
The ranking of the gene expressions of 21 candidate lncRNA transcripts that were significantly expressed in the acute phase of KD.
| Gene | Log2 FC | Log2 CPM | FDR-adjusted P value |
|---|---|---|---|
| SNORD3C | −2.68 | 4.78 | 9.03E−08 |
| 7SK | −5.73 | 6.26 | 1.85E−05 |
| AC009950.2 | −4.99 | −1.32 | 3.56E−04 |
| SNORD3B-1 | −3.44 | 4.46 | 5.27E−04 |
| XIST | 11.24 | 3.46 | 1.20E−03 |
| HSD11B1-AS1 | 3.33 | 4.10 | 3.50E−03 |
| RP11-47I22.2 | 2.90 | 2.84 | 3.88E−03 |
| RP11-96C21.1 | 6.10 | 0.25 | 3.91E−03 |
| RP11-764K9.1 | −5.31 | 0.92 | 8.82E−03 |
| RP11-689B22.2 | 2.43 | 1.97 | 1.66E−02 |
| RP11-6N17.9 | −3.47 | −1.42 | 1.68E−02 |
| CTC-378H22.1 | −1.53 | 3.23 | 1.90E−02 |
| AC017002.2 | −2.64 | 3.03 | 2.12E−02 |
| MIR4435-1HG | −2.50 | 3.06 | 3.21E−02 |
| RP11-624L4.1 | 2.76 | 2.96 | 3.28E−02 |
| HLA-AS1 | 2.05 | 3.93 | 3.31E−02 |
| RP3-322G13.5 | 2.90 | 1.29 | 3.34E−02 |
| XXbac-BPG252P9.10 | 2.44 | 2.81 | 3.46E−02 |
| FAM157A | 3.15 | 0.04 | 4.22E−02 |
| AP003774.6 | −1.86 | 1.99 | 4.38E−02 |
| RP11-598F7.3 | −1.62 | 3.77 | 4.61E−02 |
FC fold change, CPM counts per million, FDR false discovery rate.
DEGs using CAGE-seq revealed the involvement of the immune system in acute KD inflammation
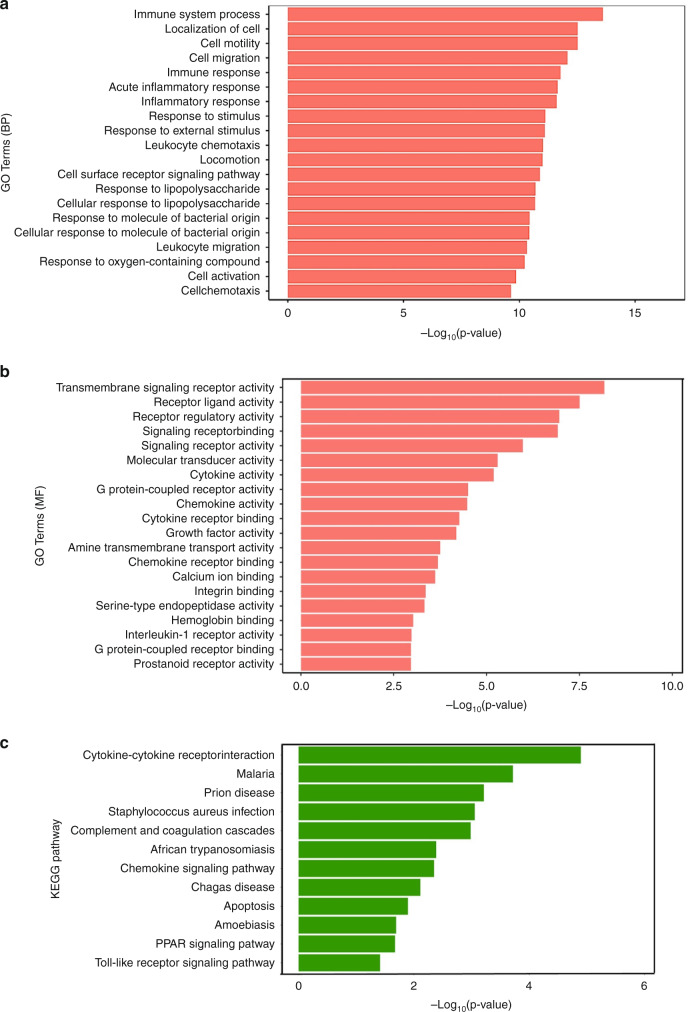
We conducted GO analysis of mRNAs that were differentially expressed during the acute phase of KD using the CAGE-seq results; the GO analysis identified several potential pathways involved in KD. The biological process revealed that the genes were involved in both the immune system process (−log P = 13.7) and the acute inflammatory response (−log P = 11.5) (Fig. 2a). The expressions of receptor ligands and receptor regulatory activities were increased in the molecular function analysis (Fig. 2b). In the KEGG pathway, numerous genes were actively involved in cytokine receptor interactions (−log P = 5) (Fig. 2c). Each gene in the GO analysis is described in Supplemental Table 2.
Fig. 2. Pathway analysis of differentially expressed genes (DEGs) by CAGE-seq in acute KD.
Gene ontology analysis of the biological process (BP) (a) and molecular function (MF) (b) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (c) for DEGs in the acute phase of KD (IVIG responder and non-responder) (n = 6) and healthy controls (n = 3) in CAGE-seq. Each gene in the GO analysis is described in Supplemental Table 2.
lncRNA HSD11B1-AS1 expression in monocytes was upregulated during the acute phase of KD and regulated by Toll-like receptor (TLR) ligands
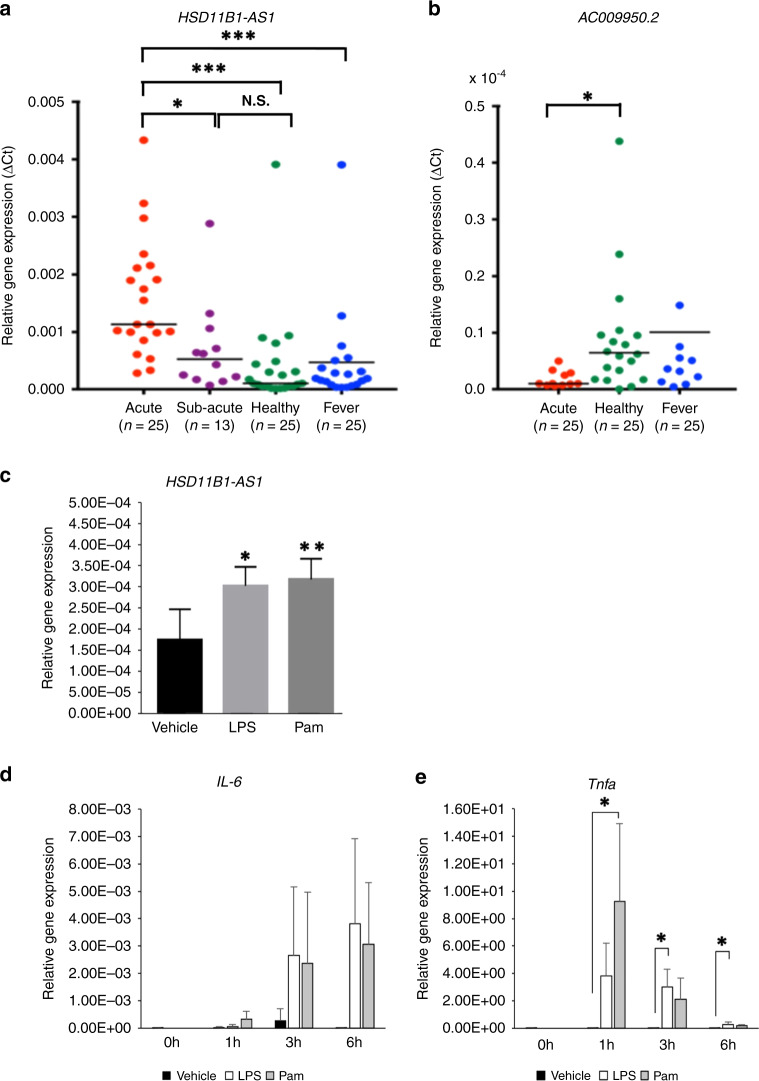
A total of 21 differentially expressed lncRNA transcripts were validated by qRT–PCR. Comparing the acute phase with the subacute phase, approximately 7 days after IVIG injection (post-IVIG), only the lncRNA HSD11B1-AS1 transcript exhibited a significant increase in the acute phase although 12 samples were missed due to early discharge (Fig. 3a) (acute KD vs. subacute KD: P < 0.004; acute KD vs. healthy controls: P < 0.0001; acute KD vs. fever controls: P < 0.0001). The lncRNA AC009950.2 transcript was significantly lower in patients with KD than in the healthy control group (P < 0.05) (Fig. 3b). The other 19 lncRNA transcripts did not show significant differences or did not express by qRT–PCR (Sup. Figure 1). To investigate whether HSD11B1-AS1 is associated with KD inflammation, THP-1 monocytes were stimulated with LPS, a TLR 4 ligand, and Pam, a TLR1/2 ligand. HSD11B1-AS1 significantly increased by 1.7 and 1.8 times 1 h following LPS (P = 0.02) and Pam (P = 0.01) stimulations, respectively (Fig. 3c). In addition, it was confirmed that the expression levels of interleukin (IL)-6 and tumor necrosis factor (TNF)-α increased by LPS and Pam stimulation (Fig. 3d, e).
Fig. 3. Expression levels of lncRNA transcripts during the acute phase of KD and changes in the expression levels of HSD11B1-AS1 stimulated with Toll-like receptor (TLR) ligand.
a The expression levels of HSD11B1-AS1 during the acute and subacute phases of KD and in the control group were measured via real-time PCR. ***P < 0.005. b The expression levels of AC009950.2 in patients with acute KD and the control group were measured via real-time PCR. *P < 0.05. c The relative gene expression of HSD11B1-AS1 in THP-1 monocytes was measured following a 1-h stimulation with lipopolysaccharide (LPS) or Pam3CSK4 (Pam) (n = 4). *P < 0.05, **P < 0.01. The relative gene expression of IL-6 (d) and TNF-α (e) in THP-1 monocytes was measured during a 6-h stimulation with LPS or Pam (n = 4). *P < 0.05. Results are expressed as relative units for each transcript compared with GAPDH.
G0S2 was associated with HSD11B1-AS1 and regulated by TLR ligands
The results of the GO analysis revealed that mRNAs were involved in the immune system process, acute inflammatory response, and cytokine receptor interaction. Therefore, network analysis was conducted to examine the association between mRNAs and HSD11B1-AS1 from the CAGE-seq results. Several mRNAs were identified using Pearson’s correlation coefficient (Tables 3 and 4). Tables 3 and 4 present the mRNAs with a high correlation coefficient (absolute value, >0.8) with HSD11B1-AS1. Among these mRNAs, G0S2 attracted the most attention owing to the following reasons. First, G0S2 had a high correlation coefficient (correlation coefficient, >0.8). Second, the location of G0S2 on the genome is on the opposite side of HSD11B1-AS1.
Table 3.
Ranking of positively correlated mRNAs with HSD11B1-AS1 during the acute phase of KD.
| Gene | Pearson’s correlation | P value | Q value |
|---|---|---|---|
| GPR97 | 0.950 | 0.0000851 | 0.00469779 |
| G0S2 | 0.942 | 0.000146 | 0.00566163 |
| IL8 | 0.935 | 0.000219 | 0.00635165 |
| PRKCH | 0.924 | 0.000363 | 0.00758993 |
| CD82 | 0.921 | 0.000426 | 0.00791498 |
| ADAM9 | 0.920 | 0.000439 | 0.00794197 |
| ALPL | 0.920 | 0.000446 | 0.00797516 |
| BCL2A1 | 0.920 | 0.000447 | 0.00797516 |
| CTD-3214H19.16 | 0.911 | 0.000631 | 0.00892573 |
| RP11-96C21.1 | 0.908 | 0.000719 | 0.00934358 |
| MAP3K8 | 0.907 | 0.000735 | 0.00939844 |
| LIN7A | 0.904 | 0.000831 | 0.00983774 |
| RP11-689B22.2 | 0.904 | 0.000835 | 0.00987318 |
| ANXA3 | 0.901 | 0.000920 | 0.01022796 |
| LRRC70 | 0.895 | 0.00110489 | 0.01096537 |
| PDE4D | 0.895 | 0.0011123 | 0.01098828 |
| SBNO2 | 0.895 | 0.00112411 | 0.01103843 |
| SLC25A37 | 0.894 | 0.00115259 | 0.01118092 |
| CXCL16 | 0.894 | 0.00115511 | 0.01118837 |
| ACTA2 | 0.893 | 0.00119349 | 0.01126866 |
| GRAP2 | 0.892 | 0.00122797 | 0.01139234 |
| CST7 | 0.888 | 0.00138299 | 0.01180303 |
| GZF1 | 0.886 | 0.00146193 | 0.01203551 |
| PPP1R3B | 0.885 | 0.00153032 | 0.01221561 |
| CXCR1 | 0.882 | 0.00163924 | 0.01253649 |
| ICAM1 | 0.882 | 0.00166005 | 0.0125918 |
| NAMPT | 0.880 | 0.00174838 | 0.01282802 |
| PPARG | 0.880 | 0.0017727 | 0.01292805 |
| RNF122 | 0.877 | 0.00190385 | 0.01322972 |
| LIMK2 | 0.876 | 0.0019362 | 0.01327902 |
| MCTP2 | 0.874 | 0.00208448 | 0.01363072 |
| MYL9 | 0.874 | 0.00208453 | 0.01363072 |
| RP11-47I22.2 | 0.869 | 0.002375 | 0.0143524 |
| KIAA0226L | 0.868 | 0.00241459 | 0.01443103 |
| MERTK | 0.864 | 0.00266999 | 0.01504887 |
| ABCA1 | 0.859 | 0.00300852 | 0.01574815 |
| SOD2 | 0.853 | 0.0034535 | 0.01664289 |
| HIF1A | 0.853 | 0.00347305 | 0.01666479 |
| PPBP | 0.851 | 0.00359032 | 0.01684495 |
| RP3-322G13.5 | 0.851 | 0.00360832 | 0.01688798 |
| PGS1 | 0.843 | 0.00434794 | 0.01812703 |
| SDCBP2 | 0.842 | 0.00441662 | 0.0182307 |
| RP11-624L4.1 | 0.841 | 0.00453802 | 0.01841602 |
| ARHGAP11B | 0.841 | 0.00454628 | 0.01841602 |
| LITAF | 0.834 | 0.00515066 | 0.01948279 |
| C10orf54 | 0.832 | 0.00543101 | 0.0199838 |
| WDR81 | 0.831 | 0.00546639 | 0.02003959 |
| CLU | 0.829 | 0.00569832 | 0.0204256 |
| AQP9 | 0.829 | 0.02045923 | 0.02045923 |
| AP001055.1 | 0.826 | 0.00605011 | 0.02095863 |
| CR1 | 0.825 | 0.00622001 | 0.02120348 |
| HAMP | 0.819 | 0.00688514 | 0.02219294 |
| SLC39A8 | 0.815 | 0.00739932 | 0.02296614 |
| NAMPTL | 0.812 | 0.0078275 | 0.02356404 |
| SLC22A16 | 0.810 | 0.00813223 | 0.02396141 |
| SLCO4A1 | 0.809 | 0.00831411 | 0.02417369 |
| FGF13 | 0.808 | 0.0083729 | 0.02425101 |
| TIMP1 | 0.808 | 0.00844191 | 0.02434853 |
Table 4.
Ranking of negatively correlated mRNAs with HSD11B1-AS1 during the acute phase of KD.
| Gene | Pearson’s correlation | P value | Q value |
|---|---|---|---|
| TRGV7 | −0.930 | 0.000274 | 0.00686965 |
| RASSF4 | −0.924 | 0.000375 | 0.00759138 |
| POLR1B | −0.918 | 0.000482 | 0.0081856 |
| MRPS33 | −0.914 | 0.000572 | 0.00866897 |
| XYLB | −0.905 | 0.000785 | 0.00961058 |
| GHRL | −0.881 | 0.00168009 | 0.01263453 |
| MRPS18B | −0.876 | 0.00195787 | 0.01333323 |
| TMA16 | −0.868 | 0.0024412 | 0.01449482 |
| C11orf21 | −0.862 | 0.00280742 | 0.01531237 |
| RABGAP1L | −0.834 | 0.00517758 | 0.01953136 |
| CTNND1 | −0.833 | 0.00526621 | 0.01967664 |
| MRPS25 | −0.821 | 0.00664845 | 0.02219294 |
| ARHGAP22 | −0.813 | 0.00769317 | 0.02334201 |
| HDAC9 | −0.806 | 0.00876556 | 0.02678924 |
| NAT10 | −0.801 | 0.00943516 | 0.02578498 |
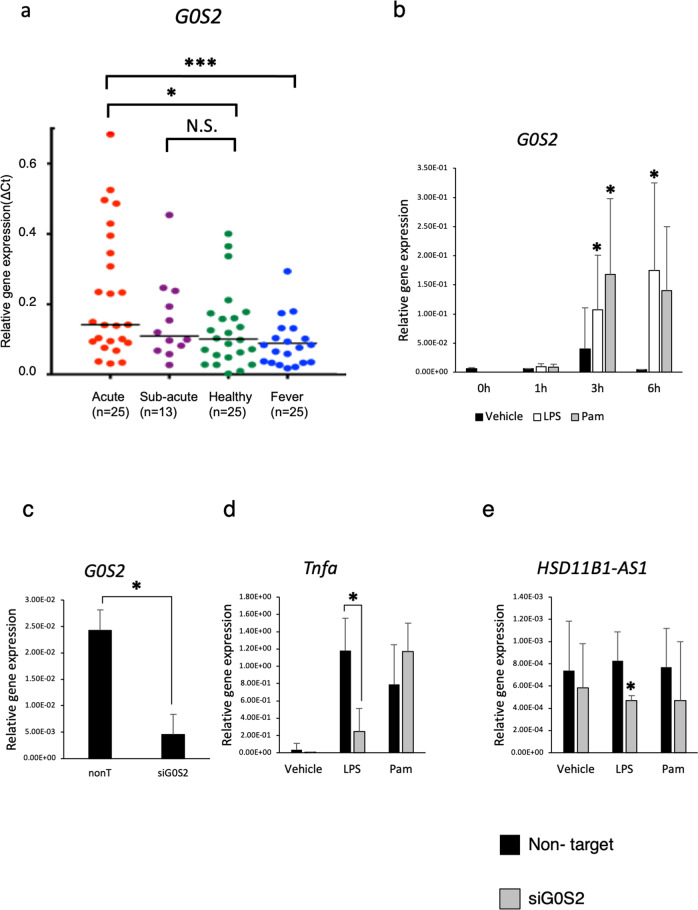
The expression of G0S2 significantly increased during the acute phase of KD compared with that in the febrile and healthy control groups and rapidly decreased in the subacute phase along with HSD11B1-AS1 (acute KD vs. healthy control: P = 0.02; acute KD vs. fever control: P = 0.002) (Fig. 4a).
Fig. 4. Expression levels of G0S2 in patients with KD and the potential role of G0S2 in the TLR pathway.
a The expression levels of G0S2 during the acute and convalescent phases of KD and in the control groups were measured via real-time PCR. *P < 0.05, ***P < 0.005. b The expression level of G0S2 in THP-1 monocytes was estimated along a time course until 6 h after stimulation with LPS or Pam (n = 4). *P < 0.05. c The relative gene expression of G0S2 in THP-1 monocytes treated with siG0S2 (n = 3). *P < 0.05. Relative gene expressions of TNF-α (d) and HSD11B1-AS1 (e) in THP-1 monocyte stimulated with LPS and Pam after treatment with siG0S2 (n = 3). *P < 0.05.
From these results, we hypothesized that HSD11B1-AS1 and G0S2 cooperated to contribute to the pathophysiology of KD inflammation. Therefore, we evaluated the mechanism underlying the effects of these two genes on the inflammatory response in monocytes during the acute phase of KD using THP-1 monocytes. The G0S2 expression was elevated 3 and 6 h after LPS and Pam stimulations (P < 0.05) (Fig. 4b).
To analyze the regulatory cluster of innate immunity between G0S2 and HSD11B1-AS1, the expression of G0S2 was knocked down using siRNA in THP-1 monocytes. Then, the cells were stimulated using these TLR ligands. The G0S2 expression was significantly downregulated after treatment with siRNA (P = 0.04) (Fig. 4c). Following LPS stimulation, the TNF-α expression was significantly suppressed in THP-1 monocytes, in which G0S2 was knocked down (P = 0.02), whereas TNF-α expression was not suppressed by Pam stimulation (Fig. 4d). Interestingly, HSD11B1-AS1 expression stimulated by LPS was also suppressed (P < 0.05) (Fig. 4e).
These data revealed that the expression of HSD11B1-AS1 was directly or indirectly regulated by G0S2 and that they regulate the inflammatory response in monocytes of patients with KD via an innate immunity pathway, particularly the TLR4 ligand.
Discussion
This is the first study on the comprehensive expression profiling of monocytes of patients with acute KD. We identified DEGs that included lncRNAs in the monocytes of patients with acute KD using CAGE-seq. Of these, the expression levels of lncRNA HSD11B1-AS1 and mRNA G0S2 were upregulated and rapidly decreased following the initial treatment with IVIG. G0S2 increased when LPS induced inflammation in THP-1 monocytes, and G0S2 silencing suppressed the expressions of HSD11B1-AS1 and TNFα.
These results indicated that HSD11B1-AS1 and G0S2 have strong interaction and may contribute to the entirety of processes controlling the innate monocytic response.
Originally, G0S2 was discovered as a gene involved in the cell cycle switch from G0 to G1 in cultured mononuclear cells.21 The G0S2 expression increased in the peripheral mononuclear cells of patients with small-sized vasculitis, such as Wegener’s granulomatosis, microscopic polyangiitis, and Churg–Strauss syndrome.22 A study has reported that monocytes or macrophages mediate inflammatory responses via adenosine triphosphate (ATP) signaling.23 In addition, G0S2 has been identified as a regulator of ATP production under cardiac ischemia.24 These pieces of evidence support our current findings and imply that G0S2 may be involved in the inflammatory response during acute KD as an ATP production regulator.
Studies have demonstrated that monocytes play an important role in the pathogenesis of KD. The number of monocytes secreting TNF-α increases in the peripheral blood during the acute phase of KD. Monocytes and macrophages infiltrate the coronary arteries of patients with KD.25–28 Administration of nucleotide-binding oligomerization domain-containing protein 1 ligand to severe combined immunodeficient mice could induce coronary artery vasculitis that resembles KD. This indicates that the innate immune system, including monocytes without T/B cells, was activated.29,30 These reports were consistent with our pathway analysis, indicating the activated immune system or acute inflammatory response of monocytes.
Our results revealed that TLR1/2 and TLR4 ligands may regulate both HSD11B1-AS1 and G0S2 as their expressions were upregulated when THP-1 monocytes were stimulated with LPS and Pam. Moreover, we found that the expression of HSD11B1-AS1 was suppressed when G0S2 was knocked down. Several studies have demonstrated that the TLR2 and TLR4 expressions were upregulated in monocytes during the acute phase of KD.31–33 The TLR4 pathway induces S100A8, A100A9, and S100A12 proteins in KD, and higher expression of nuclear factor kappa B (NF-κB) is exhibited in mononuclear cells.31,32,34 Our data suggested that HSD11B1-AS1 is regulated by G0S2 and that G0S2 and HSD11B1-AS1 mediate inflammation in the TLR pathway. Notably, HSD11B1-AS1 is located on the complementary strand of mRNA of G0S2 and is referred to as antisense lncRNA. In general, antisense lncRNAs exert the promotion or suppression of coding genes on the complementary strand.35 Thus, we hypothesized that G0S2 regulates the expression of the HSD11B1-AS1 gene. Although no studies have reported on the relationship between HSD11B1-AS1 and G0S2, our results indicated that HSD11B1-AS1 and G0S2 are directly or indirectly involved in KD inflammation via TLR signaling.
Recently, several studies have reported the association between KD and long non-coding RNAs. Using microarrays, Ko et al. have found that XLOC_00627, a lncRNA, was associated with coronary artery aneurysm in patients with KD.36 Li et al. have identified several lncRNAs that regulate the human macrophage response to an innate stimulus.37 Among them, lnc1992 was required for the expression of numerous immune-response genes, including post-transcriptional regulators of TNF-α expression, and its expression of the whole blood cells was upregulated in patients with KD using microarrays. Moreover, a recent study has revealed that the lncRNA SOCS2 and its antisense RNA AS1 were highly expressed in the serum of patients with KD, and they contributed to cell proliferation in the human umbilical vein endothelial cells of patients with KD by elevating CUEDC2 expression by sequestering miR-324-5p.38 Compared with those studies, this study adopted a different analysis method as CAGE-seq was employed to analyze gene expression. In addition, monocytes, which were key players of innate immunity, were analyzed because they initially appeared during the acute phase of KD. There are several advantages to employing CAGE-seq.39,40 Since the cap structure at the 5′ end of RNA is specifically biotinylated and collected (cap-trapping method), a full-length cDNA can be collected. With this process, the transcription start sites can be precisely detected in addition to the expression levels of mRNA and lncRNA.41 Moreover, since PCR amplification is not required during the library production process, CAGE-seq is considered a more qualitative and quantitative method compared with other methods.40,42 This methodology may allow researchers to conduct a precise and comprehensive analysis of their datasets.
Study limitations include a small sample size. It is also possible that the study lacked the power to comprehensively detect additional candidate lncRNAs. In addition, since the study excluded patients with coronary aneurysms, examining the relationship between patients and aneurysms was impossible. We might have lost the chance to capture genes with characteristic expression change taking place in the subacute phase because we did not parallelly compare gene expression between acute and subacute phases. Further studies such as overexpression of G0S2 are required to elucidate the role of HSD11B1-AS1 and G0S2 in KD.
In conclusion, our comprehensive analysis, including CAGE-seq, provided new insights into the immune system and the cytokine receptor interaction of lncRNA in KD inflammation. lncRNA HSD11B1-AS1 is regulated by G0S2 via the TLR pathways in monocytes and may control inflammation during the acute phase of KD.
CAGE-seq is an innovative methodology and may provide novel insights into the etiology of KD. This study revealed an important role of lncRNA in acute KD, and lncRNA may be a novel key target for the diagnosis of patients with KD.
Supplementary information
Acknowledgements
We are grateful to Professor Yuichi Adachi. We thank our colleagues and collaborating hospitals for always supporting us: Hitoshi Moriuchi, Haruna Hirai, Eriko Masuda, Miho Arai, Syokei Murakami, Shintaro Terashita, Yu Saito, Haruka Ushio, Taisuke Kato, Nao Sakata, Asami Takasaki, Osamu Higuchi, Tomoko Sakuma, Yoshie Okabe, Junko Yamaoto, Tatsuya Fuchizawa, Keiichiro Uese, Shinichi Tsubata, Itaru Yamaguchi, Yujiro Takegami, Ichiro Takasaki, Kesuke Okabe, and Koshi Kinoshita.
Author contributions
M.O. collected all data. M.O., K. Hirono, S.H., Y.H, M.H., and F.I. designed the study. S.T., N.M., H.N., K.I., S.O., K.W., I.H., H.T., and K. Hatasaki provided the specimens. M.O., S.H., Y.H., M.H., F.I., and K. Hirono analyzed the data. M.O., F.I., and K. Hirono wrote the manuscript. All authors reviewed the manuscript.
Funding information
This study was supported by Grant-in-Aid for Scientific Research 00831041 (to M.O.) from the Ministry of Education, Culture, Sports, Science and Technology, Grant-in-Aid from Japanese Kawasaki Disease Research Center (to M.O.), and a grant for Kawasaki Disease Research from Japan Blood Products Organization (to M.O.).
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the ethics committee of the University of Toyama and conducted according to the Declaration of Helsinki. Patients and controls were enrolled after obtaining informed consent from their parents or legal guardians.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41390-022-01999-9.
References
- 1.Newburger JW, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation. 2004;110:2747–2771. doi: 10.1161/01.CIR.0000145143.19711.78. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki T. Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children. Arerugi. 1967;16:178–222. [PubMed] [Google Scholar]
- 3.McCrindle BW, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation. 2017;135:e927–e999. doi: 10.1161/CIR.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 4.Dietz SM, et al. Dissecting Kawasaki disease: a state-of-the-art review. Eur. J. Pediatr. 2017;176:995–1009. doi: 10.1007/s00431-017-2937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdoni L, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara T, et al. Kawasaki disease: a matter of innate immunity. Clin. Exp. Immunol. 2016;186:134–143. doi: 10.1111/cei.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirata S, Nakamura Y, Yanagawa H. Incidence rate of recurrent Kawasaki disease and related risk factors: from the results of nationwide surveys of Kawasaki disease in Japan. Acta Paediatr. 2001;90:40–44. doi: 10.1111/j.1651-2227.2001.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 8.Ye F, et al. Neutrophil-derived S100A12 is profoundly upregulated in the early stage of acute Kawasaki disease. Am. J. Cardiol. 2004;94:840–844. doi: 10.1016/j.amjcard.2004.05.076. [DOI] [PubMed] [Google Scholar]
- 9.Ebihara T, et al. Differential gene expression of S100 protein family in leukocytes from patients with Kawasaki disease. Eur. J. Pediatr. 2005;164:427–431. doi: 10.1007/s00431-005-1664-5. [DOI] [PubMed] [Google Scholar]
- 10.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zangrando J, et al. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics. 2014;15:460. doi: 10.1186/1471-2164-15-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, et al. Early expressed circulating long noncoding RNA CHAST is associated with cardiac contractile function in patients with acute myocardial infarction. Int. J. Cardiol. 2020;302:15–20. doi: 10.1016/j.ijcard.2019.12.058. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, et al. Comprehensive analysis and co-expression network of mRNAs and lncRNAs in pressure overload-induced heart failure. Front. Genet. 2019;10:1271. doi: 10.3389/fgene.2019.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang F, et al. Long non-coding RNA RNF7 promotes the cardiac fibrosis in rat model via miR-543/THBS1 axis and TGFβ1 activation. Aging. 2020;12:996–1010. doi: 10.18632/aging.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayusawa M, et al. Revision of diagnostic guidelines for Kawasaki disease (the 5th revised edition) Pediatr. Int. 2005;47:232–234. doi: 10.1111/j.1442-200x.2005.02033.x. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohmiya H, et al. RECLU: a pipeline to discover reproducible transcriptional start sites and their alternative regulation using capped analysis of gene expression (CAGE) BMC Genomics. 2014;15:269. doi: 10.1186/1471-2164-15-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell L, Forsdyke DR. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol. 1991;10:581–591. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi S, et al. Expression profiling of PBMC-based diagnostic gene markers isolated from vasculitis patients. DNA Res. 2008;15:253–265. doi: 10.1093/dnares/dsn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakobsson PJ. Pain: how macrophages mediate inflammatory pain via ATP signaling. Nat. Rev. Rheumatol. 2010;6:679–681. doi: 10.1038/nrrheum.2010.175. [DOI] [PubMed] [Google Scholar]
- 24.Kioka H, et al. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proc. Natl Acad. Sci. USA. 2014;111:273–278. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa S, Matsubara T, Yabuta K. Mononuclear cell subsets and coronary artery lesions in Kawasaki disease. Arch. Dis. Child. 1992;67:706–708. doi: 10.1136/adc.67.6.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, et al. Up-regulation of miR-27a promotes monocyte-mediated inflammatory responses in Kawasaki disease by inhibiting function of B10 cells. J. Leukoc. Biol. 2020;107:133–144. doi: 10.1002/JLB.5A0919-075RR. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Oharaseki T, Yokouchi Y, Hiruta N, Naoe S. Kawasaki disease as a systemic vasculitis in childhood. Ann. Vasc. Dis. 2010;3:173–181. doi: 10.3400/avd.sasvp01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato N, Sagawa K, Sasaguri Y, Inoue O, Kato H. Immunopathology and cytokine detection in the skin lesions of patients with Kawasaki disease. J. Pediatr. 1993;122:198–203. doi: 10.1016/S0022-3476(06)80113-7. [DOI] [PubMed] [Google Scholar]
- 29.Nishio H, et al. Nod1 ligands induce site-specific vascular inflammation. Arterioscler. Thromb. Vasc. Biol. 2011;31:1093–1099. doi: 10.1161/ATVBAHA.110.216325. [DOI] [PubMed] [Google Scholar]
- 30.Motomura Y, et al. Identification of pathogenic cardiac CD11c+ macrophages in Nod1-mediated acute coronary arteritis. Arterioscler. Thromb. Vasc. Biol. 2015;35:1423–1433. doi: 10.1161/ATVBAHA.114.304846. [DOI] [PubMed] [Google Scholar]
- 31.Lin IC, et al. Augmented TLR2 expression on monocytes in both human Kawasaki disease and a mouse model of coronary arteritis. PLoS ONE. 2012;7:e38635. doi: 10.1371/journal.pone.0038635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armaroli G, et al. Monocyte-derived interleukin-1β as the driver of S100A12-induced sterile inflammatory activation of human coronary artery endothelial cells: implications for the pathogenesis of Kawasaki disease. Arthritis Rheumatol. 2019;71:792–804. doi: 10.1002/art.40784. [DOI] [PubMed] [Google Scholar]
- 33.Huang YH, et al. Identifying genetic hypomethylation and upregulation of toll-like receptors in Kawasaki disease. Oncotarget. 2017;8:11249–11258. doi: 10.18632/oncotarget.14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin W, et al. Expression of nuclear factor -κBp65 in mononuclear cells in Kawasaki disease and its relation to coronary artery lesions. Indian J. Pediatr. 2011;78:1378–1382. doi: 10.1007/s12098-011-0478-x. [DOI] [PubMed] [Google Scholar]
- 35.Carrieri C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 36.Ko TM, et al. Genome-wide transcriptome analysis to further understand neutrophil activation and lncRNA transcript profiles in Kawasaki disease. Sci. Rep. 2019;9:328. doi: 10.1038/s41598-018-36520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Zhou J, Huang K. Inhibition of the lncRNA Mirt1 attenuates acute myocardial infarction by suppressing NF-κB activation. Cell. Physiol. Biochem. 2017;42:1153–1164. doi: 10.1159/000478870. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J, Chen D. Kawasaki disease: SOCS2-AS1/miR-324-5p/CUEDC2 axis regulates the progression of human umbilical vein endothelial cells. Pediatr. Res. 2020;20:1–8. doi: 10.1038/s41390-020-1029-9. [DOI] [PubMed] [Google Scholar]
- 39.Arner E, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawaji H, et al. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res. 2014;24:708–717. doi: 10.1101/gr.156232.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S, et al. Annotation and cluster analysis of spatiotemporal- and sex-related lncRNA expression in rhesus macaque brain. Genome Res. 2017;27:1608–1620. doi: 10.1101/gr.217463.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Grady T, et al. Genome-wide transcript structure resolution reveals abundant alternate isoform usage from murine gammaherpesvirus 68. Cell Rep. 2019;27:3988.e5–4002.e5. doi: 10.1016/j.celrep.2019.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.