Abstract
Drug repositioning aims to reuse existing drugs, shelved drugs, or drug candidates that failed clinical trials for other medical indications. Its attraction is sprung from the reduction in risk associated with safety testing of new medications and the time to get a known drug into the clinics. Artificial Intelligence (AI) has been recently pursued to speed up drug repositioning and discovery. The essence of AI in drug repositioning is to unify the knowledge and actions, i.e. incorporating real-world and experimental data to map out the best way forward to identify effective therapeutics against a disease. In this review, we share positive expectations for the evolution of AI and drug repositioning and summarize the role of AI in several methods of drug repositioning.
Introduction
We reckon that Artificial Intelligence (AI) contributes to the ultimate unity of knowledge with actions, i.e. machine intelligence that perceives its environment and digests real-world data to take actions that maximize its chance of achieving goals. Long before AI became an active branch of computer science, it has been entertained by our ancestors and recorded by philosophers and litterateurs. Nevertheless, despite decades of developments, along with other aspects of scientific and technological advances, two of the three types of AI systems, artificial general intelligence or strong AI (e.g. the character Andrew Martin in the movie Bicentennial Man, who was able to deal with everything thrown at him by human life with human-like cognition and reasoning, and ultimately accepted as a member of human society) and artificial superintelligence (e.g. masterminds for robots uprising in dystopian science fictions that surpass the capacity of human intelligence and ability in everything we do) remain an illustration [1]. Meanwhile, there are great successes in realizing artificial narrow intelligence or weak AI, where goal-oriented systems were established to perform relatively singular tasks ranging from searching the internet, driving a car, or guarding a secured entry, to managing every connected part in a house, an assembly line, or a factory, to regulating certain aspects of daily life like traffic flow in the city and power grids in a country. Operating under established sets of constraints and limitations, ‘knowing’ through the ever-expanding technologies of data generation and storage, and reasoning through advanced machine learning and deep learning algorithms, weak AI systems are doing various jobs with accuracy and even assuming the role of trainer after beating the world’s best human players in games such as chess and Go [2,3].
Although full of promise and excitement since its first establishment as a scientific discipline in 1956, there have been several ‘AI winters’ featuring disappointments and loss of funding; yet through each ‘winter’, the AI community was able to establish new theories and approaches to overcome previous obstacles and attract renewed funding [4,5]. In the latest wave of AI, statistical deep learning methods took advantage of the advancement of computational power and availability of big real-world data to dominate AI research landscape. Given the widespread interests of applying AI to all aspects of lives and the chance of opening many new markets to grow business, industrial firms are joining academic institutes and government agencies as a major driving force for the development and deployment of AI. The combination of deep learning algorithms, high performance computing, and industrial powers has not produced successful transformation from weak to strong AI, yet the deep and wide incorporation of AI into the socioeconomic landscape created great opportunities for AI to evolve and become a driving force to unite ‘knowing’ and ‘doing’ in medicine.
Shortly after the end of the previous AI winter between late 1980s and early 1990s, the transformation from weak to strong AI was vividly depicted by the character of ‘The Doctor’ in the TV series of Star Trek: Voyager (1995–2001). As a holographic projected AI doctor who was designed as a short-term, task-oriented supplement to the medical staff during emergency situations, The Doctor was forced to step in as the medical chief for years and grew to be an integrated member of the USS Voyager. Given the unique characteristics of medical science, it is conceivable such a story about the growth and evolution of an AI system to be a trusted AI doctor. On one hand, dealing with the matters of life and death requires deep knowledge and proficient execution with accuracy, precision, and repeatability, which play into the strength of task-oriented AI and robotic systems. On the other hand, the efforts of respecting basic human rights are often hindered by so many disparities and socioeconomic challenges that a physician often requires exploring the most pragmatic therapeutic options acceptable by the patients and feasible to administer under the resource constraints. The exploration of pragmatic medical options starts as a compromise to limitations imposed by certain socioeconomic confinements, but when executed under rigorous scientific principles, such explorations may lead to new branches for medical science and optimized clinical practice. The development of bloodless surgery [6] provided a fine example for such pragmatic explorations. It was started as an effort to compromise with a group of patients who were candidates of heart transplantation but rejected blood transfusion due to religion belief. This ultimately prompted the surgeons to reconsider their over-reliance on blood transfusion during the transplantation and established novel technological standards and principles that eventually benefit many more heart transplant patients.
AI systems are being deployed in both healthcare and public health sectors as well as biomedical research. For example, smart wearable devices such as the Apple Watch are gaining momentum with FDA approval and double as readers for electrocardiography [7] and peripheral capillary oxygen saturation. Augmented intelligence decision supports and analytics are embedded with data sources big and small, ranging from personal wearable devices and electronic health record (EHR) to contact tracing systems covering the entire city, state, or country in order to provide timely reminders, flag adverse events, and request clinical assistance. A primary care physician longing for more time spent with the patients could equip the office with AI assisted scheduling and prescription management, natural language and voice processing enhanced access to EHR, and image diagnosis with deep learning tools. Yet when it comes to making treatment plans, the doctors tend to rely on their own judgment and medical knowledge. Although medical AI systems are sufficiently powered to find or recite optimal solutions to defined clinical diagnosis, those answers generated by the machine are often considered ‘optimal in vacuum’ [8,9]. Involving AI to generate hypotheses regarding patient care and training them to incorporate socioeconomic factors in due process during hypothesis generation would not only benefit the care providers, but also promote the evolution of AI to optimize patient care.
Drug repositioning is an attractive pursuit for AI to contribute and evolve. It aims to reuse existing drugs, shelved drugs, or drug candidates that failed clinical trials for different medical indications [10]. It could provide answers to diseases and conditions that attract limited attention or investments from pharmaceutical companies and thus would fill the gap of unmet medical needs. The repositioning of sildenafil citrate (brand name: Viagra) from a less-heralded hypertension drug to a world renowned therapy for erectile dysfunction [11] is one of the most well-known success stories of drug repositioning. Recent achievements do not even necessarily target a defined disease, but rather control side effects of therapies like chemotherapy induced diarrhea [12] and alleviating cancer drug toxicity [13] en route to improve patients’ quality of life and expand the patient population that can tolerate an effective treatment for devastating diseases. As showcased by the global frenzy of finding quick fixes in the beginning of the Covid-19 pandemic [14,15], drug repositioning can quickly become a hot topic when the standard drug discovery pipelines need much longer turn-around time to generate therapeutic options.
Unifying the knowledge and action thus forms the essence of AI in drug repositioning. Either taking advantage of the enriched data for existing drugs or shelved candidates and salvage them towards benefiting patient groups for less invested diseases and conditions [12,13,16,17], or providing the rationale for the best way forward when the world seems ready to rush into clinical trials based on high risk but far-from-perfect ideas [18–23], the application of AI in drug repositioning could promote constructive changes in both areas.
Advancements of drug repositioning studies require effective handling of multi-modality data
Drug repositioning studies converge multidisciplinary expertise from bioinformatics, cheminformatics, structural biology, medical informatics, systems biology, and drug screening and leverage multi-scale, multi-modality datasets from transcriptomic and proteomics profiles, electronical medical records and medical imaging, to social media and epidemiology surveillance to gain comprehensive understanding of disease mechanisms and drug effects.
Figure 1 illustrates the workflow of several types of drug repositioning methods [10] as organized into three modules, i.e. data and knowledge, modeling, and testing. Computational modeling methods (light green boxes) are shown to be the key to unite knowledge and actions in drug repositioning methods, that is, taking advantage of ever-expanding knowledge bases and data resources regarding targeted disease (light yellow boxes) and drugs (light blue boxes) to prioritize candidates for multi-layer testing and validations. Each drug repositioning method described in the workflow presents unique requests and challenges for computational modeling that can be addressed by AI.
Figure 1. The conceptual workflows for major types of drug repositioning methods.
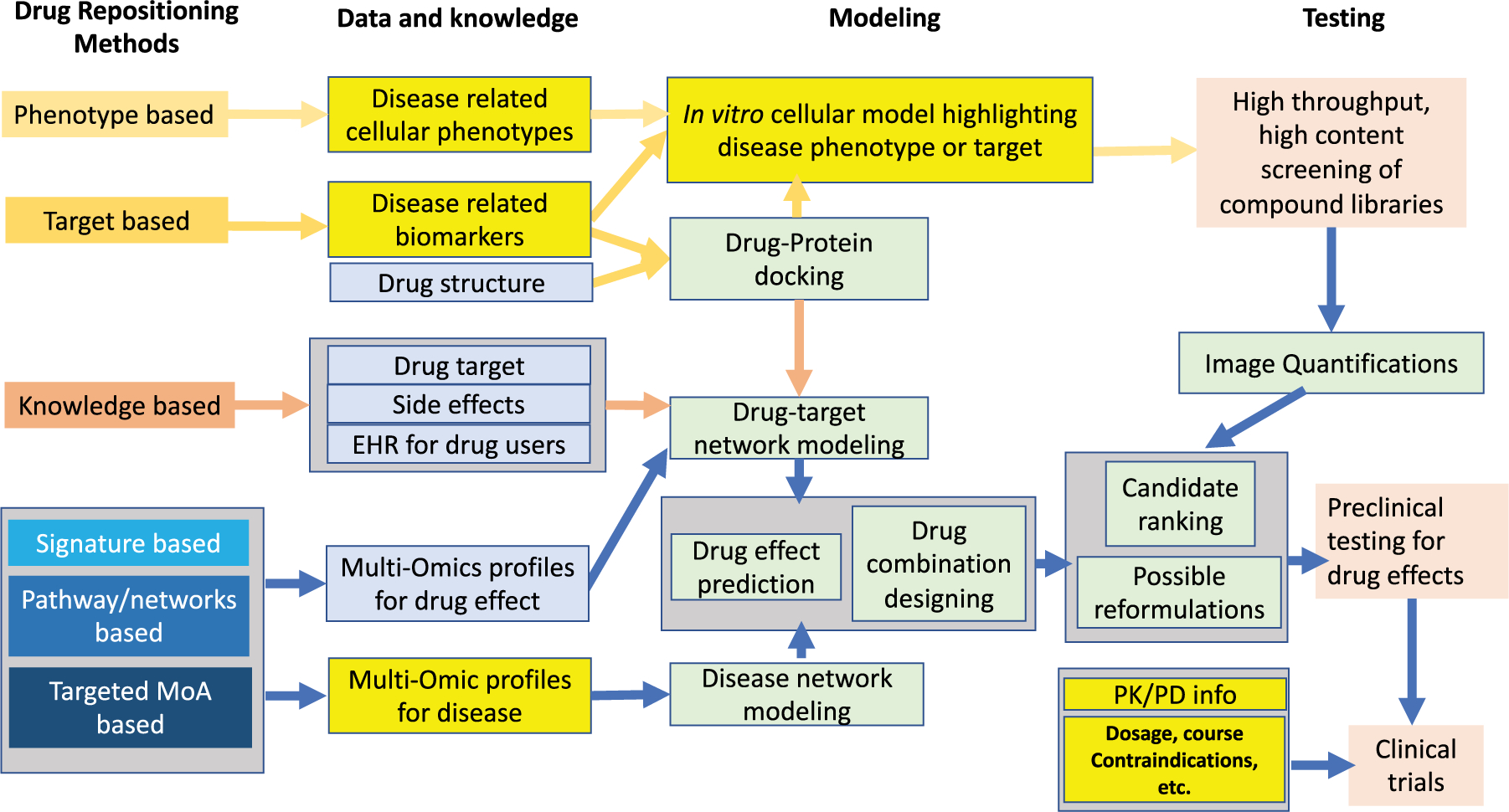
Each type of methods features unique combinations of disease (light yellow boxes) and drug (light blue) related data and knowledge, computational modeling methods (light green), and rigorous testing and validation.
Phenotype-based methods
Phenotype-based methods of drug repositioning are based on solid models of disease phenotyping and use high-throughput and/or high-content screening to obtain drug candidates that achieve desirable phenotypes in the model, e.g. compounds that can clear phosphorylated tau tangles in a multi-plate cell-based assay for Alzheimer’s disease drug screening [24,25]. Such methods do not require large amounts of pharmaceutical or biological information to start, but sophisticated biochemistry validations and meticulous follow-up studies are required to reveal mechanisms of action underlying the ability of the screening hits to achieve favorable phenotypes. Although often scolded as ‘blind’ or ‘brute-force,’ if applied to reliable disease models, phenotype-based methods can effectively cover large numbers of drug candidates and are regularly employed by large pharma companies and nascent biotech firms. The method is responsible for discovering a considerable portion of small molecules and biologics approved by FDA [26–28]. Off-label use of FDA approved drugs can be considered as a variant for phenotype-based methods as the underlying logic is to control the same biological process found in a different disease. For instance, Covid-19 pandemic has prompted a wide range of phenotype-based drug repositioning studies for candidates targeting different stages of virus infection [29–32].
It is worth noting that effective image quantification [33–36] is critical to the operation of phenotype-based methods, and the advancement of deep learning has drastically improved the performance of image quantification in high throughput and high content screening studies [37–40]. In addition, deep learning-based systems facilitate the incorporation of pharmaceutical, mechanical, and biological information, allow summarization of drug profile and toxicity [41], help identify novel mechanisms to achieve favorable phenotypes [42] en route to hit prediction, and enable smart in silico screening, which saves costs and time spent on in vitro screening.
Target-based methods
Target-based methods search for drugs that can impact known disease related target. It can be achieved through in vitro and in vivo (in animal models like zebra fish or drosophila) high-throughput/high-content screening of drugs targeting a protein or a biomarker of interest [43–45]. It is also feasible to carry out in silico screening through ligand-based screening or docking [46,47] to efficiently cover large searching space of compounds. Integration of target information into the drug repositioning process ensures a higher possibility of finding useful drugs. Drug chemical structure data was integrated with molecular activity and drug side effect data to check for drug similarity and predict drug-disease interactions [48]. An ‘expression profile’ was proposed by integrating 3-D drug chemical structure information, gene semantic similarity information, and drug-target interaction networks [49]. Free tools for compound-protein docking were being developed [50,51], and the results from multiple tools can be crosschecked and filtered [52], further facilitating the application of target-based drug repositioning.
Lack of reliable structure information for target proteins have long been the bottleneck for expanding the usage of in silico compound-protein docking. Accurate prediction of protein structures has been actively investigated by the deep learning community [53,54]. The recently reported high predictive accuracy achieved by AlphaFold [55] presents a great opportunity for pursuing compound-protein docking to a wider range of disease targets.
Knowledge-based methods
Knowledge-based methods apply bioinformatics or cheminformatics approaches to include the available information of drugs, drug-target networks [56–60], chemical structures [60], clinical trial information (adverse effects) [61,62], FDA approval labels [63], signaling or metabolic pathways [64] and drug effect profiles stored in EHR [65,66] into drug-repositioning studies. Knowledge-based methods incorporate known information into discovery of new disease mechanisms, including new targets for drugs, novel drug-drug similarities, and new diagnostic or prognostic biomarkers for diseases. While the discovery of artemisinin fifty years ago was inspired by a handful of records extracted from large amounts of traditional Chinese medicine literatures [67], today there are large amounts of drug effect profiles for patients stored electronically in EHR databases, providing opportunities for data mining or hypothesis generations on the beneficial [68,69] or adverse side effects [70] of drugs towards comorbidities. Natural language processing (NLP) is a branch of AI that has been used to extract, mine, and incorporate published information in medical science literature [71–73]. Its recent integration of deep learning further improves the efficacy and reliability of NLP programs–the quantity and quality of information used in the knowledge-based drug repositioning methods remains a critical condition for their success.
Signature-based methods
Signature-based methods analyze multiple types of -omics profiles, e.g. transcriptomics, proteomics, and meta-bolomics, of certain diseases and drug effects, use computational methods to extract target signatures, and search for candidate drugs with maximized impact on the target signatures. Whole-genome data allows for the discovery of off-target effects for drugs or unknown disease mechanisms, thus bringing a new dimension on the disease-drug relationships than the aforementioned methods of drug repositioning. Pioneered by the cMAP platform [74], where a database of drug effects on cancer cell lines allows the users to provide a customized target signature, there have been various public platforms with similar hypothesis generation functions, culminated by the Touchstone module in Clue.io [75,76], the direct heir of cMAP, where specific signatures were eliminated, and the similarity calculated across the whole transcriptomic profile was used to rank candidate compounds.
A major barrier for expanding signature-based methods using public data sources is the compatibility of drug effect profile to the biological questions at hand, as the drug effect profiles for Clue.io were generated by applying certain dosages of compounds for certain time periods on a small number of cancer cell lines. As non-cancer research communities speed up the efforts to build their own -omic databases for drug effects, it may be beneficial to carry out the signature-based drug repositioning in an iterative loop of prediction→validation→modeling, as shown in Figure 2. The key would be obtaining a list of successful vs. failed predictions after each round of validation, carrying out classifications between those transcriptomic profiles, and refining the target signature for a new round of prediction. While classification has been one of the most basic functions of AI, the ability of incorporating the validated and classified results to generate new target signatures with improved success rates for predicting hits would demand more sophisticated machine learning and decision-making algorithms.
Figure 2. An iterative loop of prediction→validation→modeling for improving the success rate of candidate predictions.
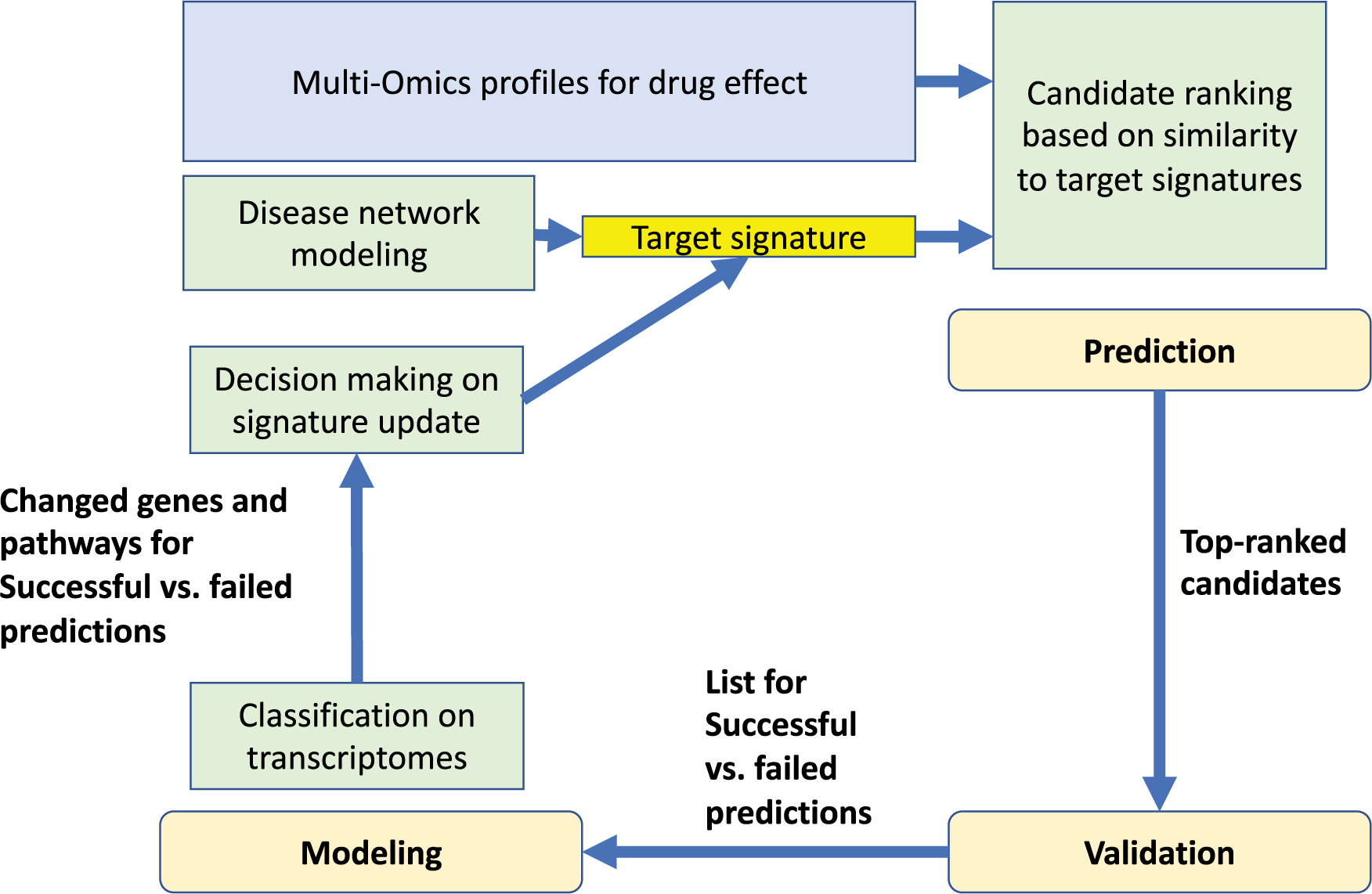
This iterative workflow converges disease (yellow box) and drug (blue box) related data and knowledge and uses computational modeling (light green) guided validation to compensate for the concerns of low predictive accuracy caused by factors such as imperfect drug effect profiles.
Pathway- or network-based methods
Pathway- or network-based methods use disease omics data, available signaling or metabolic pathways and protein interaction networks to reconstruct disease-specific pathways that provide the key targets for repositioned drugs [77,78]. Compared with the signature-based methods, this scheme requires in-depth modeling on the disease-specific omics data, incorporation of existing knowledge on pathways and functionally related gene sets, and the ability to prioritize significantly expressed targets. Due to the level of information redundancy within the pathway databases, a dataset on obesity-induced adipose inflammation reported that 12 of top 20 significant pathways center around the same group of genes [79,80]. Training AI programs to understand the hierarchical structure of the signaling networks is critical to improve the efficacy of this category of drug repositioning methods.
Targeted mechanism-based methods
Targeted mechanism-based methods are becoming highly popular in the era of precision medicine. Such methods integrate treatment omics data, available signaling pathway information, and protein interaction networks to delineate the unknown mechanisms of action of drugs [81–84]. The advancement of technologies like single cell RNA sequencing (scRNAseq) and spatial transcriptomics facilitates the in-depth modeling of disease phenotypes and allows novel types of drug targets, e.g. cross-talk between different cell types in a microenvironment or a signaling sub-network needing to be targeted by combinations of multiple drugs, to be defined. In a recent work, epithelial–stromal cross-talk signaling networks were found as potential drug target for chemo-resistant ovarian cancer [85]. Meanwhile, the systems biology community is busy creating novel methods to predict the synergy effects between drugs and create optimal combinations for network-level targets [16,86,87].
Requests on data sources and the strategies regarding imperfect datasets
Large amount of high-quality data is required in various steps within an Al-based drug repositioning pipeline. The most computational intensive, and thus data-hungry, steps are (1) generation of target signatures and (2) ranking of candidates based on the comparisons between the target signatures and drug effect profiles from the candidates. The quality of drug effect profiles has been a bottleneck in Al-based drug repositioning. On one hand, the popular choice of EHR data was originally designed for billing and financing purposes and often fail to document detailed drug effects on critical disease phenotypes, let alone molecular level disease biomarkers [88,89]. On the other hand, the most widely used transcriptomic dataset for drug effects, i.e. cMAP/Clue.io L1000 dataset, was generated mostly on cell lines related to solid tumors, covered very limited range of dosage and treatment time, and was a mixture of measurements of mRNA transcript for 987 landmark genes and mathematically inferenced expression profile for another 11 350 genes in the transcriptome [75]. Worse still, the reproducibility between data generated in different iterations of cMAP project is limited with respect to matching top-ranked drug candidates [90].
While the drug repositioning community eagerly awaiting the generation of public drug effect profiles applicable to various target diseases, AI combined with rigorous functional validations already showcased the ability of overcoming vague and noisy data resources and answering carefully defined drug repositioning questions. Scalable and accurate deep learning workflows are being developed [91,92], while cMAP/Clue.io L1000 dataset helped produce candidates for diseases far beyond solid tumor in adults, e.g. childhood cancer [16], leukemia [93] and SARS-Cov-2 infection [94], and also contributed in designing of drug combinations [87,95].
Several points can be made from those success stories regarding the preparation of data sources and dealing with imperfect datasets encountered in AI based drug repositioning.
In-house omics profiles for extracting disease-related target signatures
In-house omics profiles for extracting disease related target signatures are valuable resources. When planning the data generation, evaluate the ‘effect size’ achievable by the experimental design and technology in use, e.g. for comparing the difference of average through student’s t-test, the effect size can be evaluated by Cohen’s d [96]:
where
SDpool was obtained through investigation of the variance amongst samples. Different experimental designs require different strategies for evaluating desired effect sizes [97], and such evaluations are critical for obtaining enough samples and achieve sufficient statistical power. In addition, target-mechanism-based drug repositioning often requires multiple sets of data from each subject, e.g. transcriptome profiles from multiple cell types to model cell–cell communication [85,98], or matched multi-omics profiles to model the systems-level coordination [99]. The data generation procedures should be carefully planned to avoid batch effects.
Extracting target signatures from public resources
Extracting target signatures from public resources requires sufficient statistical power, yet the dataset also needs to be evaluated to select the samples that best fit the hypothesis of the specific drug repositioning applications. A highly re-used [100,101] single-cell RNAseq dataset from Alzheimer’s patients [102] contains patients with different levels of diseases, defined by both codex categories and image phenotypes. For larger data resources like TCGA, specialized tools were developed to summarize the metadata [103]. Researchers will benefit from an effective selection of outside dataset with matching disease subtypes and phenotypes that matches their specific applications.
Crosschecking the compound ranking results based on public drug effect data
Crosschecking the compound ranking results based on public drug effect data will also help improve the success rate of the AI-based drug repositioning. The touchstone tool of Clue.io provides the option of ranking the candidates based on all available or certain individual cell lines. We try to understand the genetic characteristics of the cell lines used in generating cMAP/Clue.io L1000 dataset and compare the candidate ranking results from all cell lines vs. certain subgroups. When possible, we will take positive control assays and generate in-house drug effect profiles to obtain reliable and concise target signature for the query with L1000 dataset. After the initial query, we will download the transcriptomic profiles for the top candidates and perform pathway overrepresentation analysis. This strategy evaluates whether the candidates are impacting the target signature in pathway level and compensates the bias of cMAP algorithm towards outliers.
Summary.
The field of drug repositioning has dealt with the coldest of cold cases over the years while also enjoying the status of the hottest of hot shots during the Covid-19 pandemic. Meanwhile, AI has been full of promise since its infancy, but experiencing several hype cycles, followed by disappointments, criticism, funding cuts, and then followed by renewed interest years later.
Recent enthusiasm and optimism on AI, especially the sub-field of machine learning, coupled with the widespread availability of big real-world data in healthcare and experimental data in disease biology and drug profiles led to a dramatic increase in funding and investment of drug repositioning and could bring cost-effective therapeutic solutions for various unmet medical needs quickly to the clinics.
While there are many types of AI methods, seemingly only a few of them, notably deep-learning and natural language processing, have been successfully incorporated into drug repositioning.
It is necessary to deploy the full repertoire of AI methods and establish effective feedback through testing and validation to train the AI models to trim existing information and generate meaningful hypotheses.
Search and optimization algorithms need to be updated to answer questions involved in drug combination design.
Logic programing and automatic reasoning are necessary to summarize validated results and create new target signatures.
Novel probabilistic methods are needed for reasoning involving incomplete or noise EHR datasets.
New graph convolutional neural network and machine learning constructs have to be built to provide more explainable processes and models that allow human users to comprehend and trust the results and output created for subsequent experimental validation and clinical studies.
Funding
This study is supported by Ting Tsung & Wei Fong Chao Foundation, John S Dunn Research Foundation, Cure Alzheimer’s Fund, NIA R01AG057635, NIA R01AG071496 and R01ES024165-S to S.T.C.W.
Abbreviations
- AI
artificial intelligence
- EHR
electronic health record
- NLP
natural language processing
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Fjelland R (2020) Why general artificial intelligence will not be realized. Humanit. Soc. Sci. Commun 7, 10 10.1057/S41599-020-0494-4 [DOI] [Google Scholar]
- 2.Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G et al. (2016) Mastering the game of Go with deep neural networks and tree search. Nature 529, 484–489 10.1038/nature16961 [DOI] [PubMed] [Google Scholar]
- 3.Schrittwieser J, Antonoglou I, Hubert T, Simonyan K, Sifre L, Schmitt S et al. (2020) Mastering Atari, Go, chess and shogi by planning with a learned model. Nature 588, 604–609 10.1038/s41586-020-03051-4 [DOI] [PubMed] [Google Scholar]
- 4.Toosi A, Bottino AG, Saboury B, Siegel E and Rahmim A (2021) A brief history of AI: how to prevent another winter (a critical review). PET Clin. 16, 449–469 https://doi.org/10.1016Zi.cpet.2021.07.001 [DOI] [PubMed] [Google Scholar]
- 5.Haenlein M and Kaplan A (2019) A brief history of artificial intelligence: on the past, present, and future of artificial intelligence. Calif. Manage Rev 61, 5–14 10.1177/0008125619864925 [DOI] [Google Scholar]
- 6.Resar LMS, Wick EC, Almasri TN, Dackiw EA, Ness PM and Frank SM (2016) Bloodless medicine: current strategies and emerging treatment paradigms. Transfusion (Paris) 56, 2637–2647 10.1111/trf.13736 [DOI] [PubMed] [Google Scholar]
- 7.Bayoumy K, Gaber M, Elshafeey A, Mhaimeed O, Dineen EH, Marvel FA et al. (2021) Smart wearable devices in cardiovascular care: where we are and how to move forward. Nat. Rev. Cardiol 18, 581–599 10.1038/s41569-021-00522-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lysaght T, Lim HY, Xafis V and Ngiam KY (2019) AI-Assisted Decision-making in healthcare. Asian Bioeth. Rev 11, 299–314 10.1007/s41649-019-00096-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shung DL and Sung JJY (2021) Challenges of developing artificial intelligence-assisted tools for clinical medicine. J. Gastroenterol. Hepatol 36, 295–298 10.1111/jgh.15378 [DOI] [PubMed] [Google Scholar]
- 10.Jin G and Wong STC (2013) Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines. Drug Discov. Today 19, 637–644 10.1016/j.drudis.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghofrani HA, Osterloh IH and Grimminger F (2006) Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nat. Rev. Drug Discov 5, 689–702 10.1038/nrd2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, Kong R, Cao H, Yin Z, Liu J, Nan X et al. (2018) Two birds, one stone: hesperetin alleviates chemotherapy-induced diarrhea and potentiates tumor inhibition. Oncotarget 9, 27958–27973 10.18632/oncotarget.24563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong R, Liu T, Zhu X, Ahmad S, Williams AL, Phan AT et al. (2014) Old drug new use-amoxapine and its metabolites as potent bacterial β-glucuronidase inhibitors for alleviating cancer drug toxicity. Clin. Cancer Res 20, 3521–3530 10.1158/1078-0432.CCR-14-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abubakar AR, Sani IH, Godman B, Kumar S, Islam S, Jahan I et al. (2020) Systematic review on the therapeutic options for COVID-19: clinical evidence of drug efficacy and implications. Infect. Drug Resist 13, 4673–4695 10.2147/IDR.S289037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali MJ, Hanif M, Haider MA, Ahmed MU, Sundas F, Hirani A et al. (2020) Treatment options for COVID-19: a review. Front. Med 7, 480 10.3389/fmed.2020.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Garrett Injac S, Cui K, Braun F, Lin Q, Du Y et al. (2018) Systems biology-based drug repositioning identifies digoxin as a potential therapy for groups 3 and 4 medulloblastoma. Sci. Transl. Med 10, eaat0150 10.1126/scitranslmed.aat0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan X, Wang J, Cheng H, Yin Z, Sheng J, Qiu B et al. (2019) Imatinib revives the therapeutic potential of metformin on ewing sarcoma by attenuating tumor hypoxic response and inhibiting convergent signaling pathways. Cancer Lett. 469, 195–206 10.1016/j.canlet.2019.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dotolo S, Marabotti A, Facchiano A and Tagliaferri R (2021) A review on drug repurposing applicable to COVID-19. Brief. Bioinform 22, 726–741 10.1093/bib/bbaa288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galindez G, Matschinske J, Rose TD, Sadegh S, Salgado-Albarrán M, Späth J et al. (2021) Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nat. Comput. Sci 1, 33–41 10.1038/s43588-020-00007-6 [DOI] [PubMed] [Google Scholar]
- 20.Sultana J, Crisafulli S, Gabbay F, Lynn E, Shakir S and Trifirò G (2020) Challenges for drug repurposing in the COVID-19 pandemic era. Front. Pharmacol 11, 588654 10.3389/fphar.2020.588654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X and Guan Y (2021) COVID-19 drug repurposing: A review of computational screening methods, clinical trials, and protein interaction assays. Med. Res. Rev 41, 5–28 10.1002/med.21728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Wang F, Tang J, Nussinov R and Cheng F (2020) Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health 2, e667–e676 10.1016/S2589-7500(20)30192-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand K, Niravath P, Patel T, Ensor J, Rodriguez A, Boone T et al. (2021) A phase II study of the efficacy and safety of chloroquine in combination with taxanes in the treatment of patients With advanced or metastatic anthracycline-refractory breast cancer. Clin. Breast Cancer 21, 199–204 https://doi.org/10.1016Zj.clbc.2020.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwak SS, Washicosky KJ, Brand E, von Maydell D, Aronson J, Kim S et al. (2020) Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer’s disease. Nat. Commun 11, 1377 10.1038/s41467-020-15120-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadimitriou C, Celikkaya H, Cosacak MI, Mashkaryan V, Bray L, Bhattarai P et al. (2018) 3D culture method for Alzheimer’s disease modeling reveals interleukin-4 rescues Aβ42-Induced loss of human neural stem cell plasticity. Dev. Cell 46, 85–101.e8 https://doi.org/10.10167i.devcel.2018.06.005 [DOI] [PubMed] [Google Scholar]
- 26.Pfister DG (2012) Off-Label Use of oncology drugs: The need for more data and then some. J. Clin. Oncol 30, 584–586 10.1200/JCO.2011.38.5567 [DOI] [PubMed] [Google Scholar]
- 27.Burstein HJ (2013) Off-Label Use of oncology drugs: too much, Too little, or just right? J. Natl Compr. Cancer Netw 11, 505–506 10.6004/jnccn.2013.0066 [DOI] [PubMed] [Google Scholar]
- 28.Stephen R, Knopf K, Reynolds M, Luo W and Fraeman K (2009) Poster session I. Value Health 12, A57 10.1016/S1098-3015(l0)73347-7 [DOI] [Google Scholar]
- 29.Mirabelli C, Wotring JW, Zhang CJ, McCarty SM, Fursmidt R, Pretto CD et al. (2021) Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19. Proc. Natl Acad. Sci. U.S.A 118, e2105815118 10.1073/pnas.2105815118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu T, Zheng W and Huang R (2021) High-throughput screening assays for SARS-CoV-2 drug development: current status and future directions. Drug Discov. Today 26, 2439–2444 10.1016/j.drudis.2021.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan W, Zhu S, Li S, Shang W, Zhang R, Li H et al. (2020) High-throughput screening of an FDA-approved drug library identifies inhibitors against arenaviruses and SARS-CoV-2. ACS Infect. Dis 7, 1409–1422 10.1021/acsinfecdis.0c00486 [DOI] [PubMed] [Google Scholar]
- 32.Zhu H, Chen CZ, Sakamuru S, Zhao J, Ngan DK, Simeonov A et al. (2021) Mining of high throughput screening database reveals AP-1 and autophagy pathways as potential targets for COVID-19 therapeutics. Sci. Rep 11, 6725 10.1038/s41598-021-86110-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin Z, Zhou X, Bakal C, Li F, Sun Y, Perrimon N et al. (2008) Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinformatics 9, 264 10.1186/1471-2105-9-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin Z, Zhou X, Sun Y and Wong STC (2009) Online phenotype discovery based on minimum classification error model. Pattern Recognit. Comput. Life Sci 42, 509–522 10.1016/j.patcog.2008.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Z, Sadok A, Sailem H, McCarthy A, Xia X, Li F et al. (2013) A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nat. Cell Biol 15, 860–871 10.1038/ncb2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin Z, Sailem H, Sero J, Ardy R, Wong STC and Bakal C (2014) How cells explore shape space: a quantitative statistical perspective of cellular morphogenesis. BioEssays 36, 1195–1203 10.1002/bies.201400011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang SJ, Lipnick SL, Makhortova NR, Venugopalan S, Fan M, Armstrong Z et al. (2019) Applying deep neural network analysis to high-content image-based assays. SLAS Discov. 24, 829–841 10.1177/2472555219857715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steigele S, Siegismund D, Fassler M, Kustec M, Kappler B, Hasaka T et al. (2020) Deep learning-based HCS image analysis for the enterprise. SLAS Discov. 25, 812–821 10.1177/2472555220918837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheeder C, Heigwer F and Boutros M (2018) Machine learning and image-based profiling in drug discovery. Curr. Opin Syst. Biol 10, 43–52 10.1016/j.coisb.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sommer C, Hoefler R, Samwer M and Gerlich DW (2017) A deep learning and novelty detection framework for rapid phenotyping in high-content screening. Mol. Biol. Cell 28, 3428–3436 10.1091/mbc.e17-05-0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green AJ, Mohlenkamp MJ, Das J, Chaudhari M, Truong L, Tanguay RL et al. (2021) Leveraging high-throughput screening data, deep neural networks, and conditional generative adversarial networks to advance predictive toxicology. PLoS Comput. Biol 17, e1009135 10.1371/journal.pcbi.1009135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham T-H, Qiu Y, Zeng J, Xie L and Zhang P (2021) A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nat. Mach. Intell 3, 247–257 10.1038/s42256-020-00285-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swamidass SJ (2011) Mining small-molecule screens to repurpose drugs. Brief. Bioinform 12, 327–335 10.1093/bib/bbr028 [DOI] [PubMed] [Google Scholar]
- 44.Crisman TJ, Parker CN, Jenkins JL, Scheiber J, Thoma M, Kang ZB et al. (2007) Understanding false positives in reporter gene assays: in silico chemogenomics approaches to prioritize cell-based HTS data. J. Chem. Inf. Model 47, 1319–1327 10.1021/ci6005504 [DOI] [PubMed] [Google Scholar]
- 45.Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK et al. (2007) A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem 50, 2385–2390 10.1021/jm061317y [DOI] [PubMed] [Google Scholar]
- 46.Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC et al. (2002) Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J. Med. Chem 45, 2213–2221 10.1021/jm010548w [DOI] [PubMed] [Google Scholar]
- 47.Kolb P, Ferreira RS, Irwin JJ and Sholchet BK (2009) Docking and chemolnformatlc screens for new ligands and targets. Curr. Opin. Biotechnol 20, 429–436 10.1016/j.copbio.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Chen S, Deng N and Wang Y (2013) Drug repositioning by kernel-based integration of molecular structure, molecular activity, and phenotype data. PLoS ONE 8, e78518 10.1371/journal.pone.0078518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan F, Yang R, Xu X, Chen X, Wang Y, Ma H et al. (2014) Drug repositioning by applying ‘expression profiles’ generated by integrating chemical structure similarity and gene semantic similarity. Mol. Biosyst 10, 1126–1138 10.1039/c3mb70554d [DOI] [PubMed] [Google Scholar]
- 50.Wang F, Wu F-X, Li C-Z, Jia C-Y, Su S-W, Hao G-F et al. (2019) ACID: a free tool for drug repurposing using consensus inverse docking strategy. J. Cheminformatics 11, 73 10.1186/s13321-019-0394-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pagadala NS, Syed K and Tuszynski J (2017) Software for molecular docking: a review. Biophys. Rev 9, 91–102 10.1007/s12551-016-0247-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ha EJ, Lwin CT and Durrant JD (2020) Liggrep: a tool for filtering docked poses to improve virtual-screening hit rates. J. Cheminformatics 12, 69 10.1186/s13321-020-00471-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torrisi M, Pollastri G and Le Q (2020) Deep learning methods in protein structure prediction. Comput. Struct. Biotechnol. J 18, 1301–1310 https://doi.org/10.1016Zi.csbj.2019.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T et al. (2020) Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 10.1038/s41586-019-1923-7 [DOI] [PubMed] [Google Scholar]
- 55.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O et al. (2021) Highly accurate protein structure prediction with alphaFold. Nature 596, 583–589 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildirim MA, Goh K-I, Cusick ME, Barabási A-L and Vidal M (2007) Drug—target network. Nat. Biotechnol 25, 1119–1126 10.1038/nbt1338 [DOI] [PubMed] [Google Scholar]
- 57.Zhao S and Li S (2010) Network-based relating pharmacological and genomic spaces for drug target identification. PLoS ONE 5, e11764 10.1371/journal.pone.0011764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G et al. (2012) Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol 8, e1002503 10.1371/journal.pcbi.1002503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alaimo S, Pulvirenti A, Giugno R and Ferro A (2013) Drug-target interaction prediction through domain-tuned network-based inference. Bioinformatics 29, 2004–2008 10.1093/bioinformatics/btt307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kinnings SL, Liu N, Buchmeier N, Tonge PJ, Xie L and Bourne PE (2009) Drug discovery using chemical systems biology: repositioning the safe medicine comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput. Biol 5, e1000423 10.1371/journal.pcbi.1000423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L and Agarwal P (2011) Systematic drug repositioning based on clinical side-effects. PLoS ONE 6, e28025 10.1371/journal.pone.0028025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campillos M, Kuhn M, Gavin AC, Jensen LJ and Bork P (2008) Drug target identification using side-effect similarity. Science 321, 263–266 10.1126/science.1158140 [DOI] [PubMed] [Google Scholar]
- 63.Bisgin H, Liu Z, Kelly R, Fang H, Xu X and Tong W (2012) Investigating drug repositioning opportunities in FDA drug labels through topic modeling. BMC Bioinformatics 13, S6 10.1186/1471-2105-13-S15-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.An SM, Ding QP and Li L (2013) Stem cell signaling as a target for novel drug discovery: recent progress in the WNT and Hedgehog pathways. Acta Pharmacol. Sin 34, 777–783 10.1038/aps.2013.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Y, Warner JL, Wang L, Jiang M, Xu J, Chen Q et al. (2019) Discovery of noncancer drug effects on survival in electronic health records of patients with cancer: a new paradigm for drug repurposing. JCO Clin. Cancer Inform 3, 1–9 10.1200/CCI.19.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu H, Aldrich MC, Chen Q, Liu H, Peterson NB, Dai Q et al. (2015) Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J. Am. Med. Inform. Assoc 22, 179–191 10.1136/amiajnl-2014-002649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, Xu C, Wong YK, Li Y, Liao F, Jiang T et al. (2019) Artemisinin, the magic drug discovered from traditional Chinese medicine. Engineering 5, 32–39 10.1016/j.eng.2018.11.011 [DOI] [Google Scholar]
- 68.Hilton CB, Milinovich A, Felix C, Vakharia N, Crone T, Donovan C et al. (2020) Personalized predictions of patient outcomes during and after hospitalization using artificial intelligence. NPJ Digit. Med 3, 51 10.1038/s41746-020-0249-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahmoudi E, Kamdar N, Kim N, Gonzales G, Singh K and Waljee AK (2020) Use of electronic medical records in development and validation of risk prediction models of hospital readmission: systematic review. BMJ 369, m958 10.1136/bmj.m958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choudhury O, Park Y, Salonidis T, Gkoulalas-Divanis A, Sylla I and Das AK (2020) Predicting adverse drug reactions on distributed health data using federated learning. AMIAAnnu. Symp. Proc 2019, 313–322 [PMC free article] [PubMed] [Google Scholar]
- 71.Öztürk H, Özgür A, Schwaller P, Laino T and Ozkirimli E (2020) Exploring chemical space using natural language processing methodologies for drug discovery. Drug Discov. Today 25, 689–705 10.1016/j.drudis.2020.01.020 [DOI] [PubMed] [Google Scholar]
- 72.Dinesh PM, Sujitha V, Salma C and Srijayapriya B (2021) A review on natural language processing: back to basics. In Innovative Data Communication Technologies and Application (Raj JS, Iliyasu AM, Bestak R and Baig ZA, eds), pp. 655–661, Springer Singapore, Singapore [Google Scholar]
- 73.Luo JW and Chong JJR (2020) Review of natural language processing in radiology. Neuroimaging Clin. N. Am. 30, 447–458 10.1016/j.nic.2020.08.001 [DOI] [PubMed] [Google Scholar]
- 74.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ et al. (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 10.1126/science.1132939 [DOI] [PubMed] [Google Scholar]
- 75.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X et al. (2017) A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17 10.1016/j.cell.2017.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Enache OM, Lahr DL, Natoli TE, Litichevskiy L, Wadden D, Flynn C et al. (2019) The GCTx format and cmap{Py, R, M, J} packages: resources for optimized storage and integrated traversal of annotated dense matrices. Bioinformatics 35, 1427–1429 10.1093/bioinformatics/bty784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK et al. (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292, 929–934 10.1126/science.292.5518.929 [DOI] [PubMed] [Google Scholar]
- 78.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D et al. (2003) Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet 34, 166–176 10.1038/ng1165 [DOI] [PubMed] [Google Scholar]
- 79.Deng T, Lyon CJ, Minze LJ, Lin J, Zou J, Liu JZ et al. (2013) Class II major histocompatibility complex plays an essential role in obesity-induced adipose inflammation. Cell Metab. 17, 411–422 https://doi.org/10.1016Zj.cmet.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin Z, Deng T, Peterson LE, Yu R, Lin J, Hamilton DJ et al. (2014) Transcriptome analysis of human adipocytes implicates the NOD-like receptor pathway in obesity-induced adipose inflammation. Mol. Cell. Endocrinol 394, 80–87 10.1016/j.mce.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao H, Jin G, Cui K, Ren D, Liu T, Chen P et al. (2013) Novel modeling of cancer cell signaling pathways enables systematic drug repositioning for distinct breast cancer metastases. Cancer Res. 73, 6149–6163 10.1158/0008-5472.CAN-12-4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin G, Fu C, Zhao H, Cui K, Chang J and Wong STC (2011) A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res. 72, 33–44 10.1158/0008-5472.CAN-11-2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin G, Zhao H, Zhou X and Wong STC (2011) An enhanced petri-net model to predict synergistic effects of pairwise drug combinations from gene microarray data. Bioinformatics 27, i310–i316 10.1093/bioinformatics/btr202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iskar M, Zeller G, Blattmann P, Campillos M, Kuhn M, Kaminska KH et al. (2013) Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Mol. Syst. Biol 9, 662–662 10.1038/msb.2013.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yeung T-L, Sheng J, Leung CS, Li F, Kim J, Ho SY et al. (2019) Systematic identification of druggable epithelial–stromal crosstalk signaling networks in ovarian cancer. JNCI J. Natl Cancer Inst 111, 272–282 10.1093/jnci/djy097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang L, Brunell D, Stephan C, Mancuso J, Yu X, He B et al. (2019) Driver network as a biomarker: systematic integration and network modeling of multi-omics data to derive driver signaling pathways for drug combination prediction. Bioinformatics 35, 3709–3717 10.1093/bioinformatics/btz109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang L, Li F, Sheng J, Xia X, Ma J, Zhan M et al. (2014) Drugcomboranker: drug combination discovery based on target network analysis. Bioinformatics 30, i228–i236 10.1093/bioinformatics/btu278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coleman N, Halas G, Peeler W, Casaclang N, Williamson T and Katz A (2015) From patient care to research: a validation study examining the factors contributing to data quality in a primary care electronic medical record database. BMC Fam. Pract 16, 11 10.1186/s12875-015-0223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shameer K, Glicksberg BS, Hodos R, Johnson KW, Badgeley MA, Readhead B et al. (2018) Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning. Brief. Bioinform 19, 656–678 10.1093/bib/bbw136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lim N and Pavlidis P (2021) Evaluation of connectivity map shows limited reproducibility in drug repositioning. Sci. Rep 11, 17624 10.1038/s41598-021-97005-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M et al. (2018) Scalable and accurate deep learning with electronic health records. NPJ Digit. Med 1, 18 10.1038/s41746-018-0029-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nelson CA, Butte AJ and Baranzini SE (2019) Integrating biomedical research and electronic health records to create knowledge-based biologically meaningful machine-readable embeddings. Nat. Commun 10, 3045 10.1038/s41467-019-11069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bonnet R, Nebout M, Brousse C, Reinier F, Imbert V, Rohrlich PS et al. (2020) New drug repositioning candidates for T-ALL identified via human/murine gene signature comparison. Front. Oncol 10, 2380 10.3389/fonc.2020.557643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mousavi SZ, Rahmanian M and Sami A (2020) A connectivity map-based drug repurposing study and integrative analysis of transcriptomic profiling of SARS-CoV-2 infection. Infect. Genet. Evol 86, 104610 10.1016/j.meegid.2020.104610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Regan-Fendt KE, Xu J, DiVincenzo M, Duggan MC, Shakya R, Na R et al. (2019) Synergy from gene expression and network mining (SynGeNet) method predicts synergistic drug combinations for diverse melanoma genomic subtypes. NPJ Syst. Biol. Appl 5, 6 10.1038/s41540-019-0085-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen J (1988) Statistical Power Analysis for the Behavioral Sciences, Routledge, 67 p [Google Scholar]
- 97.Berben L, Sereika SM and Engberg S (2012) Effect size estimation: methods and examples. Int. J. Nurs. Stud 49, 1039–1047 10.1016/j.ijnurstu.2012.01.015 [DOI] [PubMed] [Google Scholar]
- 98.Choi H, Sheng J, Gao D, Li F, Durrans A, Ryu S et al. (2015) Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell Rep. 10, 1187–1201 10.1016/j.celrep.2015.01.040 [DOI] [PubMed] [Google Scholar]
- 99.Auslander N, Yizhak K, Weinstock A, Budhu A, Tang W, Wang XW et al. (2016) A joint analysis of transcriptomic and metabolomic data uncovers enhanced enzyme-metabolite coupling in breast cancer. Sci. Rep 6, 29662 10.1038/srep29662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lagomarsino VN, Pearse RV, Liu L, Hsieh Y-C, Fernandez MA, Vinton EA et al. (2021) Stem cell-derived neurons reflect features of protein networks, neuropathology, and cognitive outcome of their aged human donors. Neuron 109, 3402–3420.e9 10.1016/j.neuron.2021.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang J, Wang C, Qi R, Fu H and Ma Q (2020) scREAD: a single-cell RNA-seq database for Alzheimer’s disease. iScience 23, 101769 10.1016/j.isci.2020.101769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ et al. (2019) Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 10.1038/s41586-019-1195-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chandran UR, Medvedeva OP, Barmada MM, Blood PD, Chakka A, Luthra S et al. (2016) TCGA expedition: a data acquisition and management system for TCGA data. PLoS ONE 11, e0165395 10.1371/journal.pone.0165395 [DOI] [PMC free article] [PubMed] [Google Scholar]


