ABSTRACT
Applying antibiotics to susceptible bacterial cultures generates a minor population of persisters that remain susceptible to antibiotics but can endure them for extended periods. Recent reports suggest that antibiotic persisters (APs) of mycobacteria experience oxidative stress and develop resistance upon treatment with lethal doses of ciprofloxacin or rifampicin. However, the mechanisms driving the de novo emergence of resistance remained unclear. Here, we show that mycobacterial APs activate the SOS response, resulting in the upregulation of the error-prone DNA polymerase DnaE2. The sustained expression of dnaE2 in APs led to mutagenesis across the genome and resulted in the rapid evolution of resistance to antibiotics. Inhibition of RecA by suramin, an anti-Trypanosoma drug, reduced the rate of conversion of persisters to resistors in a diverse group of bacteria. Our study highlights suramin’s novel application as a broad-spectrum agent in combating the development of drug resistance.
KEYWORDS: antibiotic resistance, persistence, SOS response, mycobacteria, drug repurposing
INTRODUCTION
The remarkable success of antibiotics in treating infectious diseases has been overshadowed by the emergence of antibiotic resistance in bacteria. As a result, many antibiotics have been rendered ineffective, increasing the vulnerability of a large section of the population to infectious diseases. The WHO has recommended a global priority to tackle infections caused by 12 drug-resistant pathogens (WHO 2017), which reflects the grim situation caused by drug-resistant pathogens for public health. A general approach to counter antibiotic resistance is to introduce new molecules into the treatment pipeline to replace ineffective ones. However, the alarming rate of antibiotic resistance development in bacteria overwhelms the rate of discovery of new antibiotics.
The application of antibiotics to a bacterial culture results in a biphasic killing curve with the rapid killing of most of the bacterial population, leaving behind a subpopulation of survivors. These survivors, referred to as antibiotic persisters (APs), were first observed by Joseph W. Bigger when Staphylococcus was treated with a lethal dose of penicillin (1). Although APs were reported as early as 1944, the molecular and physiological causes of their occurrence remained elusive. The formation of APs is influenced by endogenous factors such as stochastic changes in the expression of toxin-antitoxin modules, the stringent response, and external stresses such as low pH, antibiotics, cell number, or a diauxic nutrient shift (2–8). The unique ability of APs to endure prolonged periods of antibiotic treatment with a limited loss of viability enables them to multiply and repopulate the niche upon antibiotic withdrawal. In a clinical setting, such an event would relapse into an infection (9, 10).
It has been demonstrated in Escherichia coli that APs can develop resistance when treated with different classes of antibiotics by mutagenesis caused by error-prone polymerases (11–13). The phenomenon of antibiotic-induced drug resistance (AIDR) in persister bacteria has also been observed in mycobacteria wherein antibiotic treatment resulted in three distinct phases: initial killing followed by the generation of persisters and a regrowth phase consisting of the resistant population (14, 15). In E. coli, AIDR has been reported by two different groups involving either DinB (polymerase IV [Pol IV]) or UmuD′2C (Pol V) (16, 17). Although mycobacterial APs were reported to experience high reactive oxygen species (ROS) levels, similar to their E. coli counterparts, the mechanistic details of AIDR remained unclear (14). There are significant differences in the effectors of the SOS response between E. coli and mycobacteria. The homolog of the E. coli translesion polymerase UmuD′2C, linked to AIDR in fluoroquinolone persisters, has not been identified in mycobacteria (18, 19). Moreover, the mycobacterial counterparts of E. coli Pol IV, Rv1537 and Rv3056 (MSMEG_3172 and MSMEG_2294 in Mycobacterium smegmatis), are dispensable for DNA damage tolerance (20, 21) and may not contribute to AIDR (16, 17). In Mycobacterium tuberculosis, SOS-induced mutagenesis is dependent on a C family polymerase, DnaE2, and two accessory proteins, ImuA′ and ImuB (22). The expression of DnaE2 has been linked to the emergence of drug resistance in mycobacteria upon induction by DNA damage stress. We hypothesized that DnaE2 is induced in APs and contributes to the emergence of antibiotic resistance in mycobacteria.
The prolonged duration of therapy and the use of multiple antibiotics to treat mycobacterial diseases warrant a detailed understanding of AIDR in this group. Here, we observed that high doses of ciprofloxacin (Cip) or rifampicin (Rif) resulted in redox imbalance leading to high ROS levels and the induction of the SOS response in the persisters. Our study reveals that the activation of SOS induces genome-wide mutagenesis through the DnaE2 error-prone polymerase. We further demonstrate that a WHO essential medicine, suramin (also called Germanin), which inhibits mycobacterial RecA (23), diminished the occurrence of drug-induced resistance in the persister bacteria of M. smegmatis and M. tuberculosis. Our findings highlight the potential of suramin to be repurposed for preventing the emergence of drug resistance from the persister population of bacteria. The WGS data is accessible at NCBI Genome Sequence Read Archive under the accession number PRJNA749550.
RESULTS
M. smegmatis antibiotic persisters display increased expression of dnaE2.
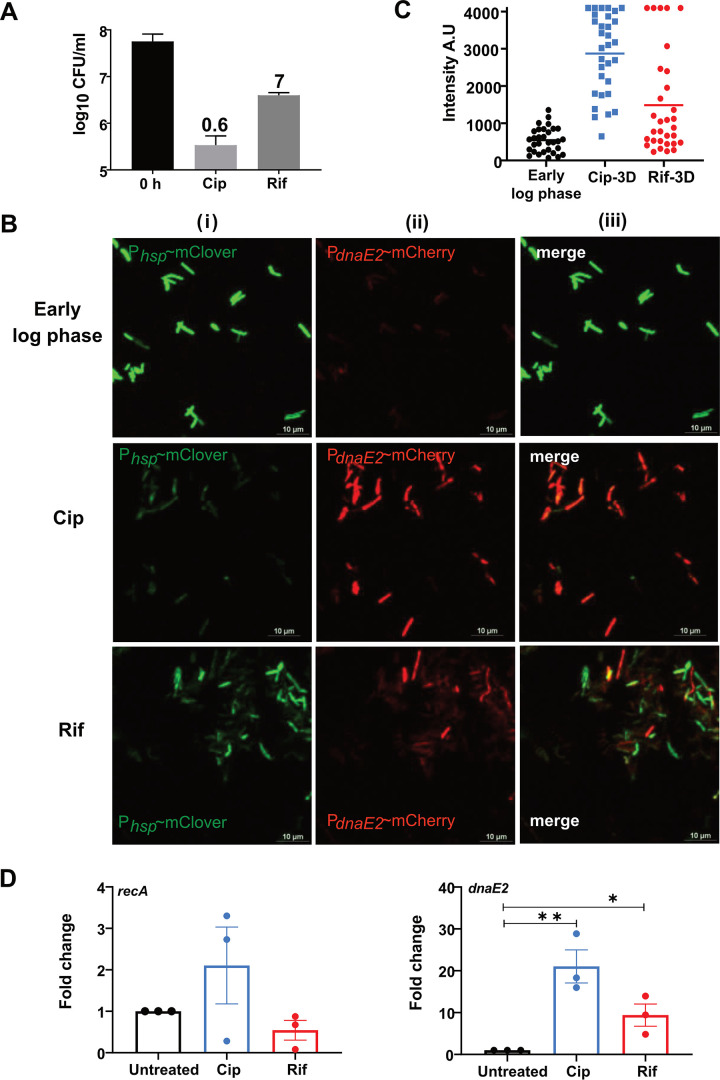
Treatment with a lethal dose of ciprofloxacin (Cip) (10 μg/mL) or rifampicin (Rif) (50 μg/mL) resulted in a significant loss of viability of M. smegmatis, leaving behind a minor population of APs (0.6% and 7%, respectively) (Fig. 1A). It has been reported that the AP population of bacteria could evolve into a resistant population through mutagenesis (12–14). In mycobacteria, the expression of the C family error-prone DNA polymerase DnaE2, along with additional accessory proteins, has been implicated in mutagenesis during the SOS response (22, 24). It has also been observed that dnaE2 was one of the highly expressed genes in the APs of M. tuberculosis treated with d-cycloserine (25). To further understand the expression pattern of dnaE2 in the persister population, we employed a dual-reporter strain in which destabilized versions of mClover and mCherry fluorescent proteins were expressed through a constitutive promoter (Phsp) and the dnaE2 promoter (PdnaE2), respectively. The dual-reporter strain was treated with Cip or Rif, and the reporters’ expression in APs was monitored at 24 h and 72 h. Seventy-two hours after antibiotic treatment, we observed a drastic reduction in the expression of Phsp∼mClover in the antibiotic-treated cells compared to the untreated early-log-phase cells, indicating that the cells were in a persistent state. Furthermore, within the same population, the expression level of the PdnaE2∼mCherry reporter was higher than the background level observed in the early-log-phase culture (Fig. 1B, panels i to iii). Quantitation of the mCherry signal (PdnaE2) indicated the increased expression of dnaE2 in antibiotic-treated cells, with Cip-treated cells showing higher expression levels than Rif-treated cells (Fig. 1C). Microscopic analysis of the dual-reporter strain indicated that dnaE2 was induced as early as 24 h in Cip treatment (see Fig. S1A at https://rgcb.res.in/suppl/Supplementary%20material.pdf). Additionally, quantitation of the dnaE2 mRNA level indicated that there was an ∼20-fold increase in the dnaE2 mRNA level in Cip-treated cells, compared to ∼9-fold in Rif-treated cells (Fig. 1D, right), supporting the microscopic data for higher expression levels of dnaE2 in Cip-treated cells. Since dnaE2 expression from the LexA-regulated promoter is RecA dependent (24, 26), we determined the expression of recA by real time quantitative reverse transcription PCR (qRT-PCR) in antibiotic-treated cells. We observed higher expression levels in Cip-treated cells (Fig. 1D, left). This observation was also reflected in AP cells using the PrecA∼mClover reporter strain, where we observed increased expression of recA (see Fig. S1B, panels ii and iii, at the URL mentioned above) 24 h after antibiotic treatment. Furthermore, a recA mutant strain failed to induce the expression of dnaE2 in APs (see Fig. S1C, panel ii, at the URL mentioned above), confirming the central role of RecA in inducing dnaE2 during the SOS response (24).
FIG 1.
Generation of persisters and analysis of dnaE2 expression in persister cells. (A) Survival of M. smegmatis cultures upon treatment with 10 μg/mL of ciprofloxacin (Cip) or 50 μg/mL of rifampicin (Rif) for 48 h. Viable counts were determined by serial dilution and plating. The mean survival rates ± standard deviations (SD) from five independent replicates were plotted using GraphPad Prism software. Percent survival is calculated as the ratio of viable cells at 48 h to those at 0 h and is depicted above each treatment. (B) M. smegmatis with a dual reporter was treated with Cip or Rif. After 72 h of incubation, an aliquot of the culture was subjected to confocal microscopy along with the untreated control. The experiment was repeated at least three times. (C) The intensity profile of dnaE2 expression was plotted by estimating the signal intensity of mCherry from ∼30 individual cells from 3 different fields. A.U, arbitrary units. (D) Triplicate cultures of wild-type M. smegmatis were treated with Cip and Rif for 6 h, along with the untreated control. Total RNA was isolated, followed by cDNA synthesis and qRT-PCR analysis of the recA and dnaE2 transcripts. Variations in gene expression in comparison with the untreated controls are represented as fold changes (2−ΔΔCT) on the graph (*, P value of <0.05; **, P value of <0.01).
The antibiotic persister population has high levels of ROS.
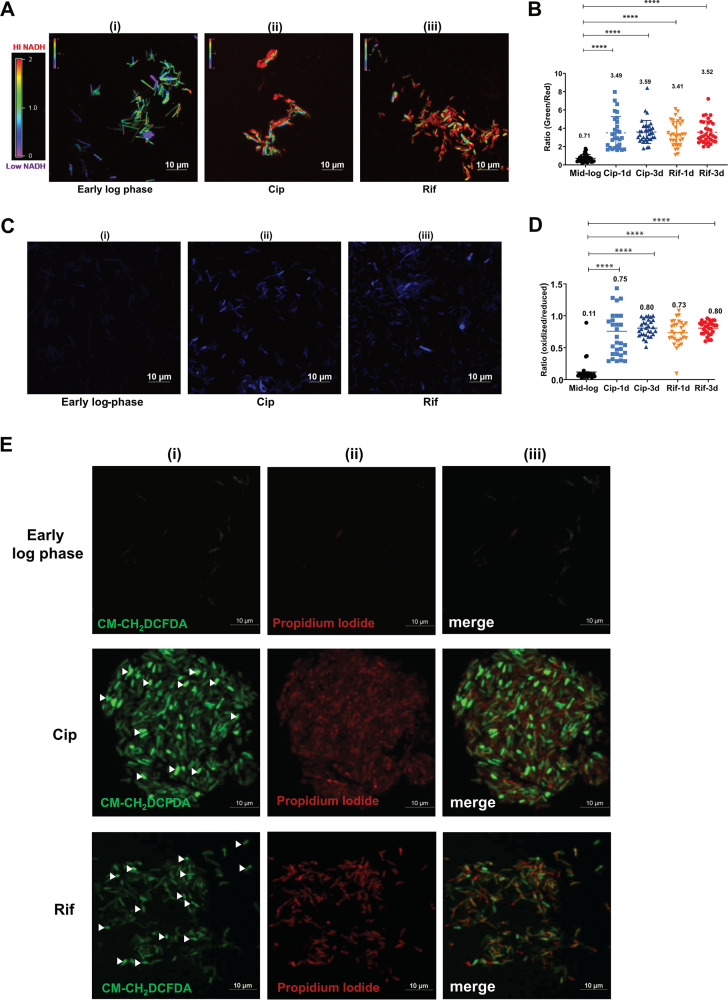
Metabolic perturbation in bacterial cells treated with sublethal doses of antibiotics has been associated with increased oxidative stress, resulting in DNA damage and the activation of RecA (17, 27, 28). We proposed a similar schema in the persister cells under a lethal dose of antibiotics. To probe the redox status of the persister cells, we employed a series of in vivo fluorescent biosensors that allowed us to assess the intracellular NADH/NAD+ ratio and oxidative stress in mycobacteria (29, 30). The M. smegmatis strain harboring the NADH reporter plasmid pMV762-Peredox-mCherry (29) was treated with Cip and Rif to select for APs, and the ratio of NADH/NAD+ was determined by fluorescence microscopy. We observed that by as early as 24 h after treatment, the APs accumulated 5-fold-higher levels of NADH under both Cip- and Rif-treated conditions (mean ratio values of 3.5 and 3.4, respectively) than the untreated early-log-phase culture (mean ratio value of 0.7) (Fig. 2B). NADH levels continued to be high in the APs at 72 h posttreatment (Fig. 2A and B). A high NADH/NAD+ ratio with limited ATP synthesis would generate superoxide by the direct transfer of electrons to O2, leading to high levels of ROS and oxidative stress (31, 32). We therefore studied the effect of high NADH levels on the redox status of persister bacteria using a highly sensitive MSH-dependent oxidoreductase (mycoredoxin-1; Mrx1) redox-sensitive GFP (Mrx1-roGFP2) reporter (30). We observed that persister bacteria experienced increased oxidative stress in both Cip and Rif treatments (Fig. 2C and D), consistent with previous reports (30). The results were further confirmed by staining antibiotic persisters with the ROS indicator dye CM-H2DCFDA (chloromethyl derivative of 2′,7′-dichlorodihydrofluorescein diacetate) (see Fig. S2, panels ii and iii, at https://rgcb.res.in/suppl/Supplementary%20material.pdf). The incorporation of an antioxidant, vitamin C, at 2.5 mM during Cip treatment reduced the ROS levels (see Fig. S2 at the URL mentioned above), supporting previous reports of better mycobacterial survival in the presence of vitamin C during antibiotic treatment. Dual staining of APs with CM-H2DCFDA (ROS) and propidium iodide (PI) (lethality) revealed that despite the lethal dose of Cip or Rif, a reasonable proportion of cells remained viable, sustaining high levels of ROS (Fig. 2E, panel i, cells indicated with arrowheads). In summary, APs of M. smegmatis subjected to lethal doses of Cip and Rif display NADH/NAD+ imbalance and high ROS levels.
FIG 2.
Analysis of redox imbalance and ROS levels in antibiotic persisters. (A) Ratiometric analysis of NADH/NAD+ levels in APs. After 72 h of treatment, the Cip and Rif APs of M. smegmatis harboring Peredox-mCherry were imaged along with an untreated early-mid-log-phase culture. The images were assigned a pseudocolor for the representation of the NADH:NAD+ levels in individual cells. (B) The observed ratios of green to red from ∼30 bacterial cells from 3 independent fields were determined. The data are presented as the means ± SD (****, P value of <0.0001). The experiment was repeated at least twice. (C) Determination of ROS levels in antibiotic persister cells using the Mrx1-roGFP sensor. Cip or Rif APs of M. smegmatis with the Mrx1-roGFP plasmid were analyzed after 72 h by confocal microscopy. The experiment was performed with two biological replicates, and a representative image is presented. (D) The ratio of blue to green was determined, and the values from individual cells were plotted. The data are presented as the means ± SD (****, P value of <0.0001). (E) The M. smegmatis wild type was treated with Cip and Rif and incubated for 72 h. After incubation, an aliquot was taken, stained with PI and CM-H2DCFDA (ROS indicator), and subjected to confocal laser scanning microscopy (CLSM). The fluorescence images were acquired, and a representative image is presented. Viable cells with high ROS levels are indicated with arrowheads.
Loss of dnaE2 results in UV sensitivity but does not enhance killing by antibiotics.
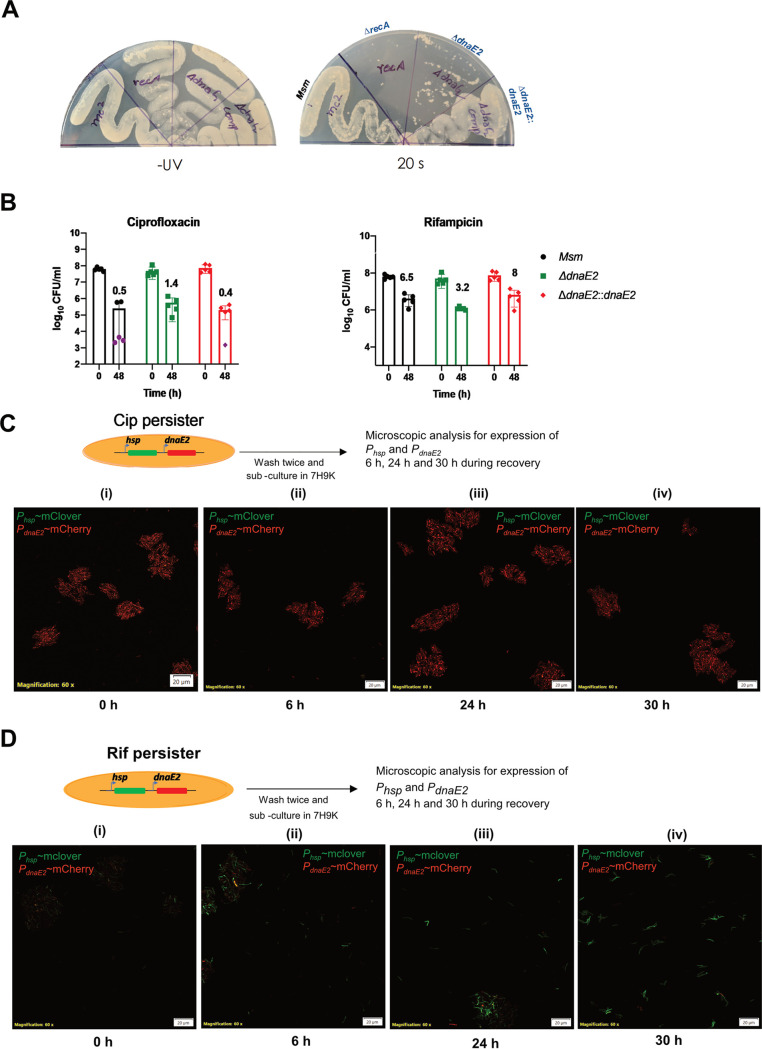
To understand the significance of the expression of DnaE2 in the persister population, we generated a gene replacement mutant of dnaE2 (see Fig. S3A and B at the URL mentioned above). Expectedly, the dnaE2 mutant strain showed increased sensitivity to UV compared to the parental strain but was less susceptible than the recA mutant (Fig. 3A, right). Genetic complementation of the dnaE2 mutant with a single copy of wild-type dnaE2, expressed from its native promoter, completely reversed its UV sensitivity phenotype (Fig. 3A, right). The MICs of Cip and Rif for the wild-type strain (M. smegmatis), the dnaE2 mutant (ΔdnaE2), and the dnaE2 mutant complemented with a wild-type copy of dnaE2 expressed under the control of its promoter (ΔdnaE2::dnaE2) were unchanged, suggesting that the dnaE2 mutation did not alter antibiotic susceptibility (see Fig. S3C and D at the URL mentioned above). We further sought to determine whether the loss of dnaE2 in the persister state resulted in differential killing rates for Cip and Rif. The M. smegmatis wild-type, ΔdnaE2, and ΔdnaE2::dnaE2 strains were treated with Cip and Rif for 48 h, and the CFU were enumerated in the surviving population by plating serially diluted cultures onto plain 7H10 agar plates. Rif and Cip treatments resulted in 1- and 3-log-scale killing, respectively, compared to the 0-h time point, and the corresponding surviving population accounted for ∼3 to 8% for Rif treatment and ∼0.4 to 1.4% in the case of Cip treatment (Fig. 3B). These observations indicated that the loss of dnaE2 did not significantly impact bacterial survival in an antibiotic-induced persister state.
FIG 3.
Characterization of the ΔdnaE2 strain and time course analysis of dnaE2 expression. (A) UV sensitivity analysis. Cultures of M. smegmatis wild-type (Msm), recA::kan, ΔdnaE2, and dnaE2::dnaE2 strains were streaked onto two 7H10 plates, one of which was exposed to UV for 20 s, while the other was left untreated. The plates were incubated at 37°C, and digital images of the plates were captured. (B) Evaluation of bacterial survivors upon antibiotic treatment. Five replicate cultures of M. smegmatis wild-type, ΔdnaE2, and ΔdnaE2::dnaE2 strains were treated with Cip or Rif for 48 h. Scatterplots of surviving bacteria from serial dilution and plating were plotted using GraphPad Prism software, with means ± SD. The average percent survival was determined and is represented above the corresponding bar for each antibiotic. (C and D) Time course expression analysis of dnaE2 was performed by recovering the M. smegmatis dual reporter from either Cip or Rif persister cells. The images were acquired at 0, 6, 24, and 30 h in the recovery stage. The images are representative of results from two independent replicates.
Expression of dnaE2 continues into the recovery phase of persister cells.
While dnaE2 was expressed in the persister stage, we were interested in determining if the expression continued if persisters were recovered in an antibiotic-free medium. We generated persisters in the dual-reporter strain by Cip and Rif treatments to further investigate this. We performed a time course microscopic analysis of PdnaE2 expression relative to Phsp during the recovery of APs in an antibiotic-free medium. The expression level of PdnaE2 was high in the Cip persisters when cultured in an antibiotic-free medium (Fig. 3C, panel i) and continued to remain high up to 30 h, while the expression of Phsp remained undetected or at a basal level (Fig. 3C, panels ii to iv). These observations indicated that the Cip persisters remained viable, as suggested by the high expression levels of destabilized mCherry from PdnaE2, but failed to resume active growth, demonstrated by the low expression levels of mClover from Phsp. A similar phenomenon has been reported in E. coli cells subjected to UV radiation, which causes extensive DNA damage and the activation of the SOS response (33). In contrast, the Rif persisters showed low expression levels of both PdnaE2 and Phsp at 0 h of recovery (Fig. 3D, panel i). Furthermore, there was considerable expression of Phsp (expression of mClover) as early as 6 h into the recovery phase that kept increasing at subsequent time points, indicating the resumption of active growth (Fig. 3D, panels ii to iv). These findings suggest that the level of dnaE2 expression in the recovery phase varies between Cip and Rif treatments.
Expression of dnaE2 results in the rapid emergence of antibiotic resistance in APs.
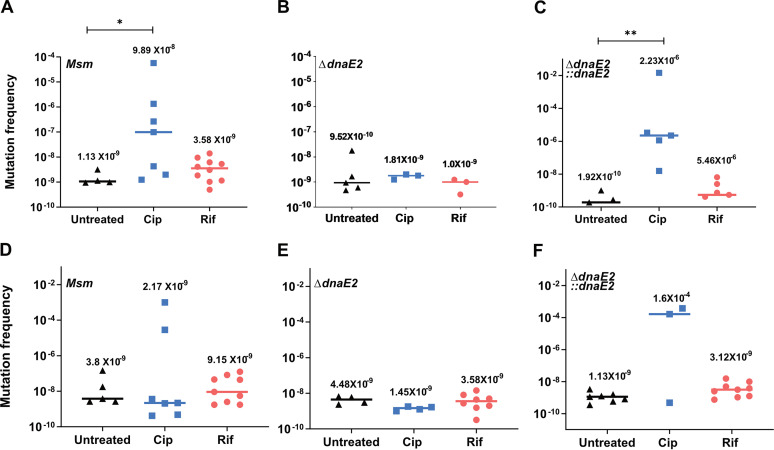
Since dnaE2 was expressed in both the persister and the recovery phases, we hypothesized that APs could display increased mutagenesis, resulting in antibiotic resistance development. We subjected 10 replicates of the actively growing cultures of M. smegmatis wild-type, ΔdnaE2, and ΔdnaE2::dnaE2 strains to Cip and Rif treatments separately for 2 days. Untreated controls were analyzed in parallel. Upon recovery in antibiotic-free medium and plating onto antibiotic plates, we observed a very high frequency (up to 5 orders of magnitude) of the conversion of Cip persisters to resistors to both Cip and streptomycin (Strep) (an antibiotic that the bacteria were never exposed to) (Fig. 4A and D, Msm). The median mutation frequency in the untreated case was 1.13 × 10−9, which increased to 9.89 × 10−8 upon Cip treatment. The Rif persisters also showed a modest increase in the extent of resistor formation, 3.58 × 10−9, compared to the untreated control (Fig. 4A, orange circles). The deletion of dnaE2 or the insertional inactivation of recA, necessary for the activation of the SOS response, reduced antibiotic-induced drug resistance (AIDR) to basal levels (Fig. 4B and E; see also Fig. S1D at the URL mentioned above). The reintroduction of dnaE2 into the knockout strain reproduced the AIDR phenomenon (Fig. 4C and F). These findings suggest that the mechanism underlying AIDR in M. smegmatis APs is similar to that reported in E. coli, wherein fluoroquinolone-induced persisters displayed enhanced resistance to multiple antibiotics, requiring SOS inducible error-prone polymerase activity (16).
FIG 4.
Expression of DnaE2 in persisters promotes enhanced rates of emergence of resistance to different antibiotics. Ten replicate cultures of M. smegmatis wild-type, ΔdnaE2, and ΔdnaE2::dnaE2 strains were subjected to either Cip or Rif to generate persisters along with untreated controls. The persisters from Cip and Rif were recovered in 7H9A medium, and the mutation frequency was determined by the ratio of the number of mutants to the viable count for each replicate. (A to C) Mutation frequencies for ciprofloxacin; (D to F) mutation frequencies for streptomycin. Under conditions where no mutant colonies were obtained, the frequency was calculated to be zero, and the values were not plotted. The individual values were plotted on a scatterplot using GraphPad Prism software v8, and the corresponding median value for each set is indicated above the plot (*, P value of <0.05; **, P value of <0.01).
Genome-wide mutagenesis in APs contributes to the emergence of drug resistance.
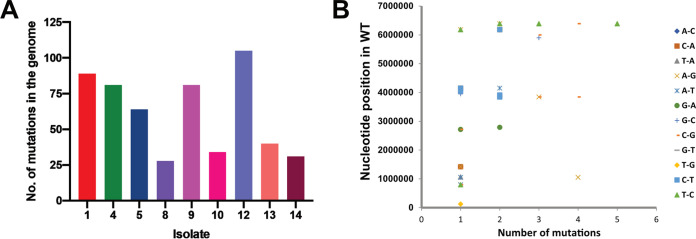
To further understand the magnitude of antibiotic-induced mutagenesis at the genomic scale, we subjected the M. smegmatis mc2155 strain to Cip and obtained mutants from persisters on both Cip and Strep plates as described above. A set of 9 isolates consisting of 3 Strep-resistant colonies and 6 Cip-resistant colonies was randomly selected and subjected to whole-genome sequencing along with the parental unstressed M. smegmatis mc2155 strain. Isolate 12 showed 106 mutations in the genome, compared to isolate 8, another ciprofloxacin-resistant isolate, which had 28 mutations in the genome (Fig. 5A). There was no specific pattern in the occurrence of mutations in the genome. We observed silent mutations that resulted in amino acid changes or affected the reading frame due to the insertion or deletion of nucleotides (Fig. 5B; see also Table S1 at the URL mentioned above). Interestingly, none of the sequenced Cip mutants had mutations mapping to gyrA (see Table S1 at the URL mentioned above) or the gyr locus in all the sequenced genomes. Generally, mutations that confer Cip resistance map to a 500-bp region within the DNA gyrase, referred to as the quinolone resistance-determining region. Nongyrase mutations that confer Cip resistance due to the expression of the proton antiporter efflux pump LfrA (MSMEG_6225) have been reported in M. smegmatis (34). Isolates 4, 8, 9, and 13 reported in this study contained a mutation in the gene MSMEG_6223 (LfrR), a TetR/AcrR family transcriptional regulator that is upstream of LfrA and has been implicated in downregulating the expression of the efflux pump (35). We believe that mutations that map to LfrR in these isolates render the protein ineffective, causing the derepression of LfrA and conferring Cip resistance.
FIG 5.
Genome-wide mutagenesis in antibiotic persisters. (A) Whole-genome sequencing analysis of resistant colonies from the persisters of M. smegmatis mc2155. The y axis corresponds to changes in the genome compared to the parental, untreated M. smegmatis mc2155 strain. Isolates 1 to 5 were obtained by streptomycin selection, while isolates 8 to 14 were obtained by ciprofloxacin selection. WT, wild type. (B) Base changes and corresponding positions in the genomes of all the resistant colonies.
All the AIDR isolates analyzed by whole-genome sequencing (WGS) displayed an increase in the MICs of Strep (isolates 1 to 5, MICs of >5 μg/mL, compared to 0.156 μg/mL for M. smegmatis mc2155) (Table 1) and Cip (isolates 8 to 14, MICs of 5 μg/mL, compared to 0.625 μg/mL for M. smegmatis mc2155) (Table 1). Isolate 1, which was resistant to streptomycin and was obtained in the Cip persister culture, showed the presence of K43T in the small ribosomal protein S12, which is known to confer resistance to Strep (36). Surprisingly, isolates 12 and 14 obtained on Cip plates also contained mutations in the S12 protein, and isolate 4 obtained on the streptomycin plate contained a mutation in the LfrR gene, indicating the evolution of multiple-drug resistance through AIDR (see Table S1 at the URL mentioned above). While isolate 14 had the less frequent P91H substitution, isolate 12 had the K43R substitution. The MIC analysis of the mutants further revealed that isolate 14 could sustain growth on 5 μg/mL of Strep, while isolate 12 failed to grow under similar conditions (Table 1). The replacement of lysine with arginine likely confers low-level Strep resistance. In the case of isolate 4, the presence of a mutation in LfrA failed to confer ciprofloxacin resistance, even though the strain displayed streptomycin resistance. Furthermore, isolates 4 and 5 did not contain mutations in S12, indicating that the mechanism conferring streptomycin resistance in these strains is different.
TABLE 1.
MICs of Cip and Strep for the 9 APs and wild-type M. smegmatis as determined by a REsazurin microtitre assay
| Strain | MIC (μg/mL) |
|
|---|---|---|
| Ciprofloxacin | Streptomycin | |
| M. smegmatis mc2155 | 0.625 | 0.156 |
| Isolate 1 | 0.625 | >5 |
| Isolate 4 | 0.3125 | >5 |
| Isolate 5 | 0.625 | >5 |
| Isolate 8 | 5 | 0.156 |
| Isolate 9 | 5 | 0.078 |
| Isolate 10 | 5 | 0.156 |
| Isolate 12 | 5 | 0.078 |
| Isolate 13 | 5 | 0.156 |
| Isolate 14 | 5 | >5 |
Inhibition of RecA through the administration of suramin eradicates antibiotic-induced drug resistance in persisters.
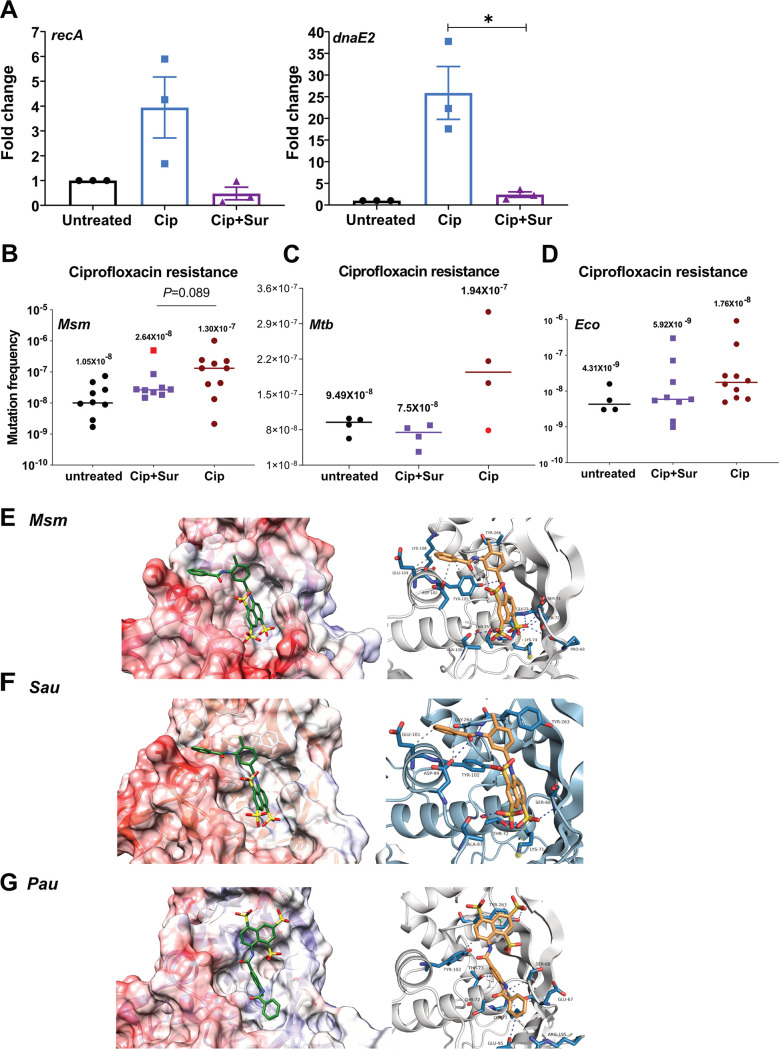
The Cip treatment that displayed robust DnaE2 expression and the AIDR phenotype induced DNA damage by (i) inhibition of gyrase activity and (ii) high levels of ROS. To better understand the contribution of each of these factors, we investigated the role of ROS in AIDR in the presence of 5 mM thiourea in Cip persisters. The presence of thiourea marginally reduced AIDR in Cip persisters, with two replicates showing reduced mutation frequencies compared to treatment with Cip alone (see Fig. S4A at the URL mentioned above), indicating a reasonable contribution of ROS to the mutational load in APs. A previous study by Barrett et al. (16) and our results present a strong case to target the error-prone polymerase to reduce AIDR. Although inhibition of ROS showed a reduction in AIDR, we considered an alternate approach of RecA inhibition by the antiprotozoal drug suramin to alleviate AIDR in persisters. Suramin has been used for almost a century to treat African sleeping disease caused by Trypanosoma brucei and has recently been used to treat several human diseases, including cancer (37). The molecule consists of polyunsaturated naphthyl urea. Due to its polyanionic nature, it interacts with mycobacterial RecA, inhibiting its ability to cleave LexA and thus preventing the induction of the SOS response (23, 38). We first monitored the effect of two different doses (0.2 mM and 0.5 mM) of suramin on the expression of dnaE2 in the Cip-treated persisters using the dual reporter. The concentrations of suramin used were ∼7-fold (0.2 mM) and 17-fold (0.5 mM) in excess of the molar concentration of Cip (10 μg/mL corresponds to 30 μM). We observed a dose-dependent decrease in the expression of dnaE2 in the persisters in the presence of suramin compared to the control cells grown in the absence of suramin (see Fig. S4B, panels iii and iv, at the URL mentioned above), in strong support of suramin’s ability to inhibit the SOS response in mycobacteria (23). Quantitation of the mRNA levels in Cip-treated and Cip-suramin-treated cells indicated a significant reduction in the expression of both recA and dnaE2 (Fig. 6A). Encouraged by this outcome, we tested the ability of suramin to reduce AIDR by adding it at a concentration of 0.2 mM to the culture during Cip treatment and during the recovery phase, followed by plating on Cip and Strep plates. In suramin’s presence, we observed a significant reduction in the Cip mutation frequency (Fig. 6B, purple squares), with a median value of 2.64 × 10−8, compared to the Cip-alone-treated cells, which showed higher rates of mutagenesis, with a median value of 1.30 × 10−7. When the individual values were analyzed, we observed a 0.5- to 50-fold decrease in the mutation frequency of Cip-suramin-cotreated cultures compared to the Cip-alone-treated cultures. The presence of suramin suppressed the emergence of drug resistance in Cip or isoniazid (Inh) persister cells of M. tuberculosis (Fig. 6C; see also Fig. S5B at the URL mentioned above), with a noticeable reduction in the Cip mutation frequency (median value of 7.5 × 10−8) compared with the Cip-alone-treated cells (1.94 × 10−7) (Fig. 6B). Similarly, in the case of Inh treatment, the presence of suramin reduced the mutation frequency by ∼2-fold (from 1.67 × 10−7 to 8.25 × 10−8) compared to Inh-only-treated cells (see Fig. S5B at the URL mentioned above).
FIG 6.
The presence of suramin (Sur) abrogates ciprofloxacin-mediated drug resistance. (A) Triplicate cultures of the M. smegmatis wild type were treated with Cip and Cip along with suramin (0.5 mM) for 6 h, along with the untreated control. The bacterial cultures treated with Cip along with suramin were washed and put for recovery with suramin for 15 h. Total RNA was isolated, which was followed by cDNA synthesis and qRT-PCR analysis of the recA and dnaE2 transcripts. Variations in gene expression in comparison to the untreated cells are represented as fold changes (2−ΔΔCT) on the graph. The experiment was repeated at least twice, with two technical replicates for each sample. (*, P value of <0.05). (B) Ten replicate cultures of the M. smegmatis wild-type generated persisters along with the untreated controls. The persisters from Cip treatment were recovered in 7H9A medium, while suramin-cotreated persisters were recovered in 7H9A medium with 0.2 mM suramin and plated onto ciprofloxacin. (C) Similarly, four replicate cultures of M. tuberculosis H37Rv (Mtb) generated persisters along with the untreated controls. The persisters from Cip treatment were recovered in 7H9A medium, while suramin-cotreated persisters were recovered in 7H9A medium with 0.2 mM suramin and plated onto ciprofloxacin. The outlier in the Cip- and suramin-treated sample is indicated by a red data point (*, P value of <0.05). (D) Ten replicate cultures of E. coli MG1655 (Eco) were treated with Cip or Cip and suramin to generate persisters along with the untreated controls. The persisters from Cip treatments and Cip-suramin cotreatments were recovered in LB medium with 0.5 mM suramin, and the mutation frequency was determined as described above. The median values under each condition are represented above the sample. (E to G) Molecular docking of suramin with RecA proteins of M. smegmatis (E), S. aureus (Sau) (F), and P. aeruginosa (Pau) (G).
In E. coli, AIDR is contributed by the RecA-dependent UmuD′2C and the σS pathways (16, 17). Since E. coli RecA (EcoRecA) is also susceptible to suramin (23), we examined if this molecule could mitigate AIDR albeit partially in E. coli. Treatment of E. coli cells with suramin in the medium during both the persister and the recovery stages resulted in a modest reduction of AIDR in E. coli (median value of 5.9 × 10−9) compared to the cells treated with Cip alone (median value of 1.76 × 10−8) (Fig. 6D). Interestingly, in the case of Strep mutation frequencies in M. smegmatis, the mutation frequency of the suramin-treated culture was noticeably lower (median value of 1.6 × 10−9) than that of the untreated culture (median value of 7.7 × 10−9) (see Fig. S5A at the URL mentioned above). Although we do not have a complete explanation for this observation, we speculate that suramin’s presence during the recovery of the APs may effectively reduce the mutagenesis caused by the spontaneous expression of DnaE2 in the untreated cells. Cotreatment with suramin had a positive impact on the growth of persisters during the recovery phase compared to Cip-alone-treated persisters (see Fig. S4B at the URL mentioned above). We believe that this could be due to the potential reduction of DnaE2 expression caused by the inhibition of RecA.
In silico binding studies implicate suramin’s potential broad-spectrum inhibition of RecA from diverse bacteria.
RecA is the master regulator of the SOS response, and it is conserved among bacteria, most notably those of the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) of pathogens. We modeled the structures of RecAs of Pseudomonas aeruginosa (PauRecA) and Staphylococcus aureus (SauRecA), which share nearly 71% sequence identity and 86% similarity with EcoRecA, and performed in silico docking of suramin on the modeled structures along with the available crystal structure of M. smegmatis RecA (MsmRecA). The docking studies indicated that suramin binds to the Walker A motif (typically 67 to 75 residues in the case of M. tuberculosis RecA [MtbRecA]), which interacts with beta and gamma phosphate oxygen atoms in the case of the Msm/MtbRecA crystal structures bound with ATPγS (PDB accession no. 1UBF and 1G18, respectively) (Fig. 6E; see also Fig. S5C at the URL mentioned above). The binding mode for suramin is similar to those for MtbRecA, EcoRecA, MsmRecA, and SauRecA; however, it is swapped in the case of PauRecA (Fig. 6E to G; see also Fig. S5C and D at the URL mentioned above). Interestingly, the binding of ATP is involved in the formation of RecA filaments. The binding of suramin at the site of ATPγS would destabilize the RecA filaments as previously reported by Nautiyal et al. (23). The occupancy of suramin at the ATPγS binding site could also reduce the protease activity of RecA essential for LexA cleavage, inhibiting the induction of the SOS response. Although the extent of AIDR reduction by suramin in P. aeruginosa or S. aureus warrants in vivo validation, our in silico modeling and docking studies indicate that this could be the most likely scenario.
DISCUSSION
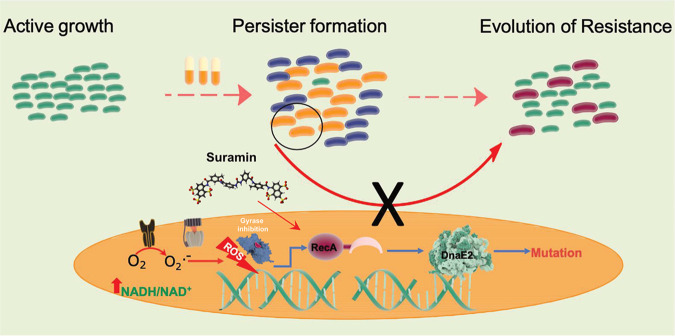
With the global consumption of antibiotics having doubled in the last decade (39), the incidences of APs becoming resistant further worsen the problem of antibiotic resistance. Resistance can be acquired from another resistant organism through horizontal gene transfer (HGT) or arise de novo by mutagenesis, often modifying the antibiotic target. In the genus Mycobacterium, HGT is not as widespread as in other bacteria, thus making a case for the de novo mode for the emergence of antibiotic resistance. It is worthwhile to mention that the course of treatment for pulmonary tuberculosis (TB) is a multidrug therapy for 6 to 9 months, implying that the occurrence of AIDR is highly probable in patients who are noncompliant in completing the entire duration of treatment. In the present study, we used M. smegmatis to understand the mechanism that facilitates the evolution of resistance in APs. Antibiotic treatment resulted in the extensive killing of the bacteria, leaving behind a minor population of persisters. Sublethal doses of antibiotics have been shown to affect bacterial metabolism, leading to an NADH/NAD+ imbalance in E. coli (31). A similar response was observed in M. tuberculosis within 24 h of exposure to antibiotics either in broth culture or within infected macrophages (29). In M. smegmatis, treatment with a very high dose of antibiotics resulted in NADH accumulation (Fig. 2A). Increased NADH levels with a concomitant reduction in ATP synthesis in the persisters would result in the generation of superoxide (O2·−), which is converted to hydrogen peroxide (H2O2) by superoxide dismutase (31). H2O2 reacts with Fe2+, generating OH·, thus increasing intracellular ROS levels (31, 40). Using a sensitive biological sensor, Mrx1-roGFP, we confirmed that the persisters had high ROS levels, corroborated by staining with the ROS indicator dye CM-H2DCFDA (Fig. 2C to E; see also Fig. S2 at https://rgcb.res.in/suppl/Supplementary%20material.pdf). High ROS levels inflict DNA damage by oxidizing nucleotides and causing strand breaks. Since the persisters experience sustained redox imbalance levels, they would experience a higher burden of DNA damage, activating the SOS response through RecA. DnaE2, an effector protein functioning downstream of RecA, is induced in the persisters (Fig. 1B), leading to genome-wide mutagenesis and resistance development (Fig. 7).
FIG 7.
Mechanism of antibiotic-induced drug resistance in M. smegmatis. A lethal dose kills the majority of the bacteria (blue cells), leaving behind a population of APs (orange cells). The APs display disturbed redox balance leading to the generation of ROS. The high levels of ROS in APs inflict DNA damage, activating RecA and triggering the SOS response, while Cip directly induces the SOS response by causing DNA strand breaks by the inhibition of DNA gyrase. DnaE2, an effector protein of the SOS response, induces mutagenesis, resulting in the emergence of drug resistance (red cells). The presence of suramin, a RecA inhibitor, mitigates antibiotic-induced drug resistance in persisters.
The expression analysis of dnaE2 in APs determined using the fluorescent reporters indicated a high degree of disparity between different antibiotic treatments. In our study, Cip treatment induced dnaE2 to the maximum level in a larger population of cells than with Rif treatment (Fig. 1B to D). Increased expression of dnaE2 in cip APs could result from the combination of DNA gyrase inhibition and ROS mediated DNA damage. In contrast, in the Rif-treated cells, despite having higher ROS levels (Fig. 2C to E), the inhibition of transcription would reduce global transcription, including that of recA (Fig. 1D, left). To rule out the possible effects of ciprofloxacin-associated DNA damage, an antibiotic that would affect the cell wall process would be most ideal. Interestingly, in M. tuberculosis, persisters generated by treatment with d-cycloserine (a cell wall inhibitor) displayed high levels of dnaE2 expression (25).
The expression of dnaE2 was not restricted to the persister state but was also observed during the recovery phase of Cip-treated cultures (Fig. 3C). Single-cell analysis of E. coli treated with a fluoroquinolone has also revealed SOS response induction during the recovery phase of ofloxacin persisters (41). Accordingly, the sustained expression of dnaE2 during the recovery state resulted in a mutation frequency that was higher by up to 4 to 5 orders of magnitude in APs (Fig. 4A and C). The difference in the magnitudes of resistors obtained by two different antibiotic treatments despite having comparable levels of redox stress (Fig. 2C to E) was surprising, especially as previous reports of M. tuberculosis persisters had reported an increase of nearly 3 to 6 orders of magnitude in the rate of resistance with Rif treatment (14). While a previous study by Swaminath et al. (15) detected mutations in the APs in the presence of the antibiotic, we detected resistors in APs that recovered in the absence of antibiotics. Despite having high dnaE2 expression levels in either the persister or recovery phase, the mutation frequencies in different replicates showed a distribution pattern similar to the ones seen for persister viability (Fig. 3B), with some replicates having lower mutation frequencies comparable to that of the untreated control. This high variation observed in our experiments could have arisen due to the method of detecting mutations, i.e., the occurrence of an antibiotic-resistant phenotype, while other mutations that do not confer resistance are missed in the analysis. An alternate explanation could be that the dnaE2 expression in the persister state may not account for the high rates of mutagenesis because the subsequent recovery phase that comprises a phase of DNA repair would reverse the mutations generated in the persister state. One of the earliest responses during spore germination or regrowth from the dormant state is DNA repair (42–44). Thus, the actual manifestation of mutants in the recovering population would depend on the stochastic variation of two opposing forces, promutagenesis influenced by the expression of dnaE2 and DNA repair in individual cells of the population. It is also worth mentioning that in the recovery phase of Rif persisters, the expression level of dnaE2 was much lower (Fig. 3D) than that in the Cip persisters (Fig. 3C). Accordingly, the magnitude of AIDR was modest (Fig. 4A and C, compare the values of Cip with those of Rif for the wild-type M. smegmatis and complemented strains).
While identifying new antibiotics is a possible solution to address the growing problem of antibiotic resistance, targeting the evolvability of antibiotic-susceptible bacteria to resistant strains is an emerging concept that is being considered (17, 45). Targeting DnaE2, the effector of mutagenesis, has been explored, and a purine analog that could potentially bind and inhibit DnaE2 was identified by in silico analysis (46). Since high ROS levels in the persisters are the root cause of mutagenesis through the DNA damage and SOS pathways, molecules that reduce ROS levels have also been tested to reduce AIDR. In E. coli, edaravone, an FDA-approved drug for lateral amyloid sclerosis that alleviates ROS levels, was shown to reduce AIDR (17). We explored if vitamin C, a widely recognized antioxidant with a history of being used in TB therapy, could alleviate antibiotic-mediated high ROS levels in APs. Although vitamin C showed a noticeable reduction of ROS in APs (see Fig. S2 at the URL mentioned above), it is also known to induce the hypoxia response in mycobacteria and to favor bacterial survival (47, 48) and therefore was not pursued further. However, the presence of thiourea during antibiotic treatment marginally reduced AIDR (see Fig. S4A at the URL mentioned above).
All the above-described strategies presented have some drawbacks: (i) targeting ROS may reduce the efficiency of the host immune response in eradicating pathogens, and (ii) inhibitors of error-prone polymerases will be bacterium specific and may not show broad-spectrum activity against a diverse group of bacterial pathogens. Therefore, we considered targeting RecA, the master regulator of the SOS response that is present across diverse groups of bacteria, as a potential target to mitigate AIDR. A small-molecule screen identified suramin and 17 other molecules that could inhibit RecA activity (38). Interestingly, suramin has been on the WHO’s list of essential drugs and is used to treat African sleeping sickness caused by T. brucei. The coadministration of suramin with Cip significantly reduced AIDR to both Cip and Strep in M. smegmatis (Fig. 6B; see also Fig. S5A at the URL mentioned above). Suramin’s effect on inhibiting AIDR was also observed in E. coli but to a lesser degree (Fig. 6D) than for M. smegmatis (Fig. 6B). This apparent difference could be because in E. coli, AIDR is also influenced by the RecA-independent σS pathway (17). Additionally, other factors, such as the biochemical properties of EcoRecA (49) and the differences in RecA expression levels and membrane permeability between the two organisms, could influence the outcome of suramin on AIDR. Low pH and starvation experienced by intracellular pathogens can trigger bacterial persistence and induce the SOS response (50). Based on the present study, it is essential to determine if other SOS-inducing conditions favor the development of drug resistance and to further test the ability of suramin as a widespread antievolution molecule against stress-induced mutagenesis. The high sequence and structural identities between MsmRecA and MtbRecA suggest that suramin’s benefits in reducing AIDR could be achieved by coadministering it during the prolonged course of TB therapy. Since eukaryotes encode multiple RAD51 family proteins (homologs of bacterial RecA), compared to a single RecA protein in bacteria (51), the potential adverse effects of suramin on recombination-related processes in humans could be minimal. Although repeated passaging of M. tuberculosis in THP-1 macrophage cell lines leads to mutagenesis (52), an in vivo model that reproduces our in vitro AIDR phenomenon is currently missing. Finally, the in silico docking studies suggest that suramin may exhibit anti-AIDR activity in other groups of bacteria such as Pseudomonas and Staphylococcus. Our findings have uncovered a novel broad-spectrum role of suramin in confronting the evolution of drug resistance.
MATERIALS AND METHODS
Lists of strains, plasmids, and oligonucleotides used in the study are presented in Tables S2, S3, and S4, respectively, at https://rgcb.res.in/suppl/Supplementary%20material.pdf.
Bacterial strains and culture conditions.
Glycerol stocks of strains of M. smegmatis were revived on Middlebrook 7H10 medium supplemented with 0.5% (vol/vol) glycerol and 0.05% (vol/vol) Tween 80 on plates containing the appropriate antibiotics. For culture under liquid conditions, Middlebrook 7H9 medium supplemented with 0.1% (vol/vol) glycerol and 0.05% Tween 80, along with the appropriate antibiotics, was used. When required, M. smegmatis cultures were selected by incorporating kanamycin (50 μg/mL), apramycin (25 μg/mL), and hygromycin (50 μg/mL). For cloning and plasmid DNA manipulation, E. coli TG1 or JM101 strains were used. E. coli strains were cultured in Luria-Bertani (LB) broth or LB agar at 37°C. For selection in E. coli, the antibiotics kanamycin (50 μg/mL), apramycin (50 μg/mL), hygromycin (150 μg/mL), and ampicillin (100 μg/mL) were used. All medium components were procured from Difco (MD, USA). Unless mentioned otherwise, all chemicals and reagents were procured from Sigma-Aldrich (MO, USA).
Time course analysis of dnaE2 expression during the recovery phase.
APs from the M. smegmatis dual-reporter strain (see Table S1 at the URL mentioned above) treated with Cip and Rif were generated as described above. After 48 h of treatment, the culture was harvested at 13,000 rpm for 1 min and washed twice with antibiotic-free 7H9 medium to remove traces of Cip. The washed bacterial pellet was resuspended in fresh 7H9K medium and incubated at 37°C at 175 rpm. An aliquot of this culture was concentrated, mounted onto a glass slide immediately with ProLong glass antifade mountant, and considered 0 h after recovery. Subsequently, a sample from the recovered APs was analyzed every 6 h for the expression of dual reporters by confocal microscopy. The images were acquired using an Olympus FluoView FV3000 instrument equipped with a 60× oil lens objective and a charge-coupled-device (CCD) camera connected to the FV3000 built-in software. Fields were selected randomly, and images were taken using Z-stacking with 60× oil immersion at a resolution of 512 by 512 pixels. The lens has a numerical aperture of 1.42. The images were captured by excitation with a 405-nm laser and emission using a 525/50-nm filter, and mCherry images were captured by excitation with a 561-nm laser and emission using a 625/50-nm filter. The postprocessing and analysis of the images were done using the Olympus cellSens software platform. In the cellSens software, the images that were acquired with 96 pixels per inch (ppi) were projected in a maximized view. The sharpness of the images was increased from the original 60 points to 65. The contrast optimization’s right overflow parameter was increased from 0 to 0.2 points (and the left overflow of the contrast optimization was kept unchanged). The images were saved as “.tiff” files.
Determination of AIDR in persisters.
The M. smegmatis wild-type, ΔdnaE2, and complemented strains were freshly revived on 7H10A medium. Due to the magnitude of the experiment, each strain was handled separately. For each strain, the antibiotics were freshly prepared. The strain was precultured in 20 mL 7H9A medium for 48 h. The culture was adjusted to an optical density at 600 nm (OD600) of 0.15 to 0.2 in antibiotic-free 7H9 medium, and 2 mL was distributed into 30 culture tubes. Ten replicates of these cultures were either treated with Cip or Rif or left untreated. The tubes were incubated for 48 h at 37°C at 175 rpm. After 48 h, the antibiotic-treated cultures were harvested at 13,000 rpm for 1 min at room temperature in a sterile 2-mL microcentrifuge tube, and the pellet was washed twice with 1 mL of antibiotic-free 7H9 medium to remove traces of Cip or Rif. The final pellet containing APs was resuspended in 0.2 mL of 7H9 medium, and each of the 10 replicates that were Cip or Rif treated was inoculated into 2 mL of fresh 7H9A medium. For the untreated control, which had attained saturation after 48 h, each replicate was serially diluted to a 10−4 dilution, and 0.2 mL of the diluted culture was inoculated into 2 mL of fresh 7H9A medium. This effectively resulted in an ∼10−5 dilution with the tube containing a few thousand bacteria. All 30 tubes were incubated at 37°C at 175 rpm until they attained saturation (2 to 6 days). From each replicate of the grown culture, an aliquot (50 μL) was used to make a 10-fold serial dilution to perform viable counts. The remaining culture was harvested in a sterile 2-mL microcentrifuge tube at 13,000 rpm for 1 min at room temperature. The supernatant was discarded, and the pellet was resuspended in 0.2 mL of 7H9 medium. This suspension was equally split (0.1 mL) and plated onto 7H10 plates containing 2.5 μg/mL of ciprofloxacin and 7H10 plates containing 50 μg/mL of streptomycin to enumerate the mutants in the culture. The plates were incubated for 6 days at 37°C, viable counts were recorded after 3 days, and mutants were recorded on the 6th day. The mutation frequency for each treatment was determined by calculating the number of mutants obtained on Cip and Strep plates with the corresponding viable counts and plotted using GraphPad Prism v8 software.
Effect of suramin on AIDR.
To determine the effect of suramin on AIDR, only M. smegmatis was used. The strain was revived, the preculture was adjusted to a final OD600 of ∼0.2, and 2 mL was aliquoted into 30 tubes. Ten replicates each were treated with Cip or Cip along with 0.2 mM suramin or left untreated. The tubes were incubated at 37°C for 48 h at 175 rpm. After 48 h, the tubes were processed as described above. While the untreated and Cip-alone-treated samples were recovered in 7H9A medium, the Cip- and suramin-cotreated cultures were recovered in 7H9A medium containing 0.2 mM suramin. The tubes were further incubated at 37°C with shaking until stationary phase (2 to 6 days), and the cultures from all 30 tubes were processed as described above to obtain mutants on Strep and Cip plates along with their corresponding viable counts. The data were analyzed, and graphs were plotted using GraphPad Prism software version 8.
Genomic DNA isolation and whole-genome sequence analysis of AIDR.
The M. smegmatis mc2155 strain was revived on 7H10 medium, and mutants from persisters were obtained on Cip and Strep plates as described in detail above. The antibiotic-resistant mutants of M. smegmatis from Cip plates (6 isolates) and Strep plates (3 isolates) and the parental wild-type strain M. smegmatis mc2155 were cultured in 10 mL plain 7H9 medium in a shaker incubator at 37°C at 175 rpm to stationary phase (OD600 of ∼1.5). The cultures were harvested at 10,000 rpm for 7 min in a sterile 50-mL Falcon tube and washed once with an equal volume of phosphate-buffered saline (PBS). The bacterial pellets were resuspended in 1 mL GTET buffer containing 25 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.1% Triton X-100, and 50 mM glucose, and 6 mg chicken egg white lysozyme was added. The suspension was incubated overnight at 37°C at 175 rpm in a shaking incubator. To this suspension, an SDS solution was added (final concentration of 1%) along with proteinase K (6 mg/mL). The suspension was incubated in a water bath at 60°C for 4 h with gentle intermittent mixing every 30 min. The bacterial lysate was extracted twice with a 1:1 volume of phenol containing chloroform and isoamyl alcohol (25:24:1, vol/vol). After the second extraction, the aqueous layer was mixed with ice-cold sodium acetate (0.3 M final concentration), and the genomic DNA (gDNA) was precipitated by the addition of a 1:1 volume of ice-cold isopropanol. The mixture was gently mixed and incubated on ice, and the precipitated gDNA was spooled, washed with 75% ice-cold ethanol twice, and air dried. The gDNA samples were dissolved in Tris-EDTA (TE) and stored at 4°C until further use. Genomic DNA samples (500 ng) were processed according to the manufacturer’s instruction to prepare libraries for the HiSeq platform (Illumina X ten). Sequencing was performed at 100× coverage with 150-bp paired-end (PE150) reads and a 1-GB output for every genome.
During assembly, raw paired reads were processed, and statistics were computed and visualized using FastQC v.0.11.3. Trim Galore v.6.4 was used for adaptor removal, quality trimming (Phred score of <20), and read length filtering (<50 bp) before assembly. The filtered reads were again processed, and statistics were computed using FastQC. The final high-quality short reads were used for de novo assembly using SPAdes v.3.11.1 with k-mers of a defined length (default settings). QUAST v.5.02 was used to generate metrics and to assess the quality of the assemblies. The resulting contigs were further filtered to remove those that were <1 kbp in length. Mauve (build 10, 26 February 2015) Contig Mover was utilized to arrange and orient the scaffolds against the reference sequence of M. smegmatis mc2155 (GenBank accession no. NC_008596). The arranged scaffolds were stitched together, removing all the gaps, resulting in a final no-gapped assembly for each genome.
To identify mutations in the AIDR colonies, the stitched assembly of the wild type was aligned to the reference genome (GenBank accession no. NC_008596) using BLASTN to map the gaps. A pairwise comparison of each test genome with the wild type was performed for finding single nucleotide polymorphisms (SNPs) and indels using Nucmer v.3.1. The mismatches were mapped to the annotated reference genome to identify the nature of the SNP.
Modeling and docking studies of suramin with RecA proteins.
Since the structures of RecA from S. aureus and P. aeruginosa have not been experimentally determined, they were determined by in silico modeling using the I-TASSER Web server of the University of Michigan (53). Briefly, the protein sequences of RecA from S. aureus (UniProt accession no. A5ISG9) and P. aeruginosa (UniProt accession no. P08280) were retrieved from UniProt and submitted to the I-TASSER Web server. The three-dimensional (3D) models with the highest template modeling (TM) and C score values among the predicted structures were used for binding studies. The TM and C scores for S. aureus and P. aeruginosa were 0.83 ± 0.08 and 0.83, respectively. In the case of M. tuberculosis, the available X-ray crystal structure (PDB accession no. 1G18) was used. To determine the molecular interaction between suramin and RecA, the suramin model from the crystal structure under PDB accession no. 3PP7 was obtained from the Protein Data Bank (www.rcsb.org) for docking analysis in Chimera 1.13.1 (54). AutoDock Vina (55) was used for the docking studies between the suramin molecule and the RecA proteins using Chimera as the front end. The result consisted of multiple possible dockings between the ligand and the protein with different docking scores. The interaction with the best docking score was chosen for analysis. The docking scores were −9.1 kcal/mol for M. tuberculosis, −8.9 kcal/mol for E. coli, −8.4 kcal/mol for M. smegmatis, −7.3 kcal/mol for P. aeruginosa, and −7.2 kcal/mol for S. aureus. The docking results were visualized and analyzed using UCSF Chimera 1.12, and the sites of interaction between RecA and suramin were visualized and analyzed using the Protein-Ligand Interaction Profiler (PLIP) (56), Chimera.
Statistical analysis.
The ratiometric data from the microscopic experiments were plotted using GraphPad Prism software, and statistical analysis was performed by one-way analysis of variance (ANOVA) using Dunnett’s test (****, P value of <0.0001). The effect of Cip treatment on AIDR in comparison to untreated samples was tested using Mann-Whitney’s test (*, P value of 0.05; **, P value of <0.01). The effect of suramin- and ciprofloxacin-cotreated samples compared with that of the ciprofloxacin-alone-treated sample was determined by Mann-Whitney’s test (**, P value of <0.01).
ACKNOWLEDGMENTS
We thank Umesh Varshney and Amit Singh, Indian Institute of Science; Ashwani Kumar, Institute of Microbial Technology, Chandigarh; Markus A. Seegur, University of Zurich; and Sadananda Singh, Indian Institute for Science Education and Research, Trivandrum, for sharing strains and plasmids used in the study. We thank G. Marcela Rodriguez, Rutgers University, and Pradeep Kumar, Rutgers University, for critical reading of the manuscript and their valuable inputs during the preparation of the manuscript. We acknowledge the Central Imaging facility, Rajiv Gandhi Centre for Biotechnology, and miBiomeTheraputics LLB (Mumbai) for the assistance in microscopy and whole-genome sequencing analysis, respectively. The schematics were generated using the Library of Science and Medical Illustrations available from somersault18:24.
K.K. acknowledges financial support from a Ramalingaswami Reentry fellowship from the Department of Biotechnology, Government of India; intramural funds from the Rajiv Gandhi Centre for Biotechnology; and an extramural grant from the Science and Engineering Board (CRG/2018/001209), Government of India.
We declare no conflicts of interest.
REFERENCES
- 1.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterlisation. Lancet 244:497–500. 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 2.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 3.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 4.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo J-M, Hardt W-D, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan M-W, Tenson T, Melderen LV, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brauner A, Shoresh N, Fridman O, Balaban NQ. 2017. An experimental framework for quantifying bacterial tolerance. Biophys J 112:2664–2671. 10.1016/j.bpj.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 7.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 9.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher RA, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 11.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 13.Windels E, Michiels J, Fauvart M, Wenseleers T, den Bergh B, Michiels J. 2019. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13:1239–1251. 10.1038/s41396-019-0344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebastian J, Swaminath S, Nair RR, Jakkala K, Pradhan A, Ajitkumar P. 2017. De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob Agents Chemother 61:e01343-16. 10.1128/AAC.01343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaminath S, Paul A, Pradhan A, Sebastian J, Nair RR, Ajitkumar P. 2020. Mycobacterium smegmatis moxifloxacin persister cells produce high levels of hydroxyl radical, generating genetic resisters selectable not only with moxifloxacin, but also with ethambutol and isoniazid. Microbiology (Reading) 166:180–198. 10.1099/mic.0.000874. [DOI] [PubMed] [Google Scholar]
- 16.Barrett TC, Mok WWK, Murawski AM, Brynildsen MP. 2019. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat Commun 10:1177. 10.1038/s41467-019-09058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pribis JP, García-Villada L, Zhai Y, Lewin-Epstein O, Wang AZ, Liu J, Xia J, Mei Q, Fitzgerald DM, Bos J, Austin RH, Herman C, Bates D, Hadany L, Hastings PJ, Rosenberg SM. 2019. Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol Cell 74:785–800.e7. 10.1016/j.molcel.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizrahi V, Andersen SJ. 1998. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol 29:1331–1339. 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 19.Ditse Z, Lamers MH, Warner DF. 2017. DNA replication in Mycobacterium tuberculosis. Microbiol Spectr 5:TBTB2-0027-2016. 10.1128/microbiolspec.TBTB2-0027-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kana BD, Abrahams GL, Sung N, Warner DF, Gordhan BG, Machowski EE, Tsenova L, Sacchettini JC, Stoker NG, Kaplan G, Mizrahi V. 2010. Role of the DinB homologs Rv1537 and Rv3056 in Mycobacterium tuberculosis. J Bacteriol 192:2220–2227. 10.1128/JB.01135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordonez H, Uson ML, Shuman S. 2014. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res 42:11056–11070. 10.1093/nar/gku752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warner DF, Ndwandwe DE, Abrahams GL, Kana BD, Machowski EE, Venclovas Č, Mizrahi V. 2010. Essential roles for imuA′- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA 107:13093–13098. 10.1073/pnas.1002614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nautiyal A, Patil KN, Muniyappa K. 2014. Suramin is a potent and selective inhibitor of Mycobacterium tuberculosis RecA protein and the SOS response: RecA as a potential target for antibacterial drug discovery. J Antimicrob Chemother 69:1834–1843. 10.1093/jac/dku080. [DOI] [PubMed] [Google Scholar]
- 24.Boshoff HI, Reed MB, Barry CE, III, Mizrahi V. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183–193. 10.1016/S0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 25.Keren I, Minami S, Rubin E, Lewis K. 2011. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2:e00100-11. 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wipperman MF, Heaton BE, Nautiyal A, Adefisayo O, Evans H, Gupta R, van Ditmarsch D, Soni R, Hendrickson R, Johnson J, Krogan N, Glickman MS. 2018. Mycobacterial mutagenesis and drug resistance are controlled by phosphorylation- and cardiolipin-mediated inhibition of the RecA coprotease. Mol Cell 72:152–161.e7. 10.1016/j.molcel.2018.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhat SA, Iqbal IK, Kumar A. 2016. Imaging the NADH:NAD+ homeostasis for understanding the metabolic response of Mycobacterium to physiologically relevant stresses. Front Cell Infect Microbiol 6:145. 10.3389/fcimb.2016.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, Bhave D, Kumar D, Carroll KS, Singh A. 2014. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog 10:e1003902. 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MP. 2009. How mitochondria produce reactive oxygen species. Biochem J 417:1–13. 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennington JM, Rosenberg SM. 2007. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet 39:797–802. 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takiff HE, Cimino M, Musso MC, Weisbrod T, Martinez R, Delgado MB, Salazar L, Bloom BR, Jacobs WR, Jr.. 1996. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc Natl Acad Sci USA 93:362–366. 10.1073/pnas.93.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X-Z, Zhang L, Nikaido H. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother 48:2415–2423. 10.1128/AAC.48.7.2415-2423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finken M, Kirschner P, Meier A, Wrede A, Böttger EC. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol 9:1239–1246. 10.1111/j.1365-2958.1993.tb01253.x. [DOI] [PubMed] [Google Scholar]
- 37.Wiedemar N, Hauser DA, Mäser P. 2020. 100 years of suramin. Antimicrob Agents Chemother 64:e01168-19. 10.1128/AAC.01168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wigle TJ, Singleton SF. 2007. Directed molecular screening for RecA ATPase inhibitors. Bioorg Med Chem Lett 17:3249–3253. 10.1016/j.bmcl.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein EY, Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R. 2018. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci USA 115:E3463–E3470. 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Zhao X. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother 53:1395–1402. 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goormaghtigh F, Melderen L. 2019. Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci Adv 5:eaav9462. 10.1126/sciadv.aav9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibarra JR, Orozco AD, Rojas JA, López K, Setlow P, Yasbin RE, Pedraza-Reyes M. 2008. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores. J Bacteriol 190:2031–2038. 10.1128/JB.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurthkoti K, Varshney U. 2010. Detrimental effects of hypoxia-specific expression of uracil DNA glycosylase (Ung) in Mycobacterium smegmatis. J Bacteriol 192:6439–6446. 10.1128/JB.00679-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinai L, Rosenberg A, Smith Y, Segev E, Ben-Yehuda S. 2015. The molecular timeline of a reviving bacterial spore. Mol Cell 57:695–707. 10.1016/j.molcel.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domenech A, Brochado AR, Sender V, Hentrich K, Henriques-Normark B, Typas A, Veening J-W. 2020. Proton motive force disruptors block bacterial competence and horizontal gene transfer. Cell Host Microbe 27:544–555.e3. 10.1016/j.chom.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Jadaun ADRS, Subbarao N, Dixit A. 2015. In silico screening for novel inhibitors of DNA polymerase III alpha subunit of Mycobacterium tuberculosis (MtbDnaE2, H37Rv). PLoS One 10:e0119760. 10.1371/journal.pone.0119760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taneja NK, Dhingra S, Mittal A, Naresh M, Tyagi JS. 2010. Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860. 10.1371/journal.pone.0010860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikri K, Duggal P, Kumar C, Batra SD, Vashist A, Bhaskar A, Tripathi K, Sethi T, Singh A, Tyagi JS. 2018. Multifaceted remodeling by vitamin C boosts sensitivity of Mycobacterium tuberculosis subpopulations to combination treatment by anti-tubercular drugs. Redox Biol 15:452–466. 10.1016/j.redox.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganesh N, Muniyappa K. 2003. Mycobacterium smegmatis RecA protein is structurally similar to but functionally distinct from Mycobacterium tuberculosis RecA. Proteins 53:6–17. 10.1002/prot.10433. [DOI] [PubMed] [Google Scholar]
- 50.Erill I, Campoy S, Barbé J. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 31:637–656. 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 51.Lin Z, Kong H, Nei M, Ma H. 2006. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA 103:10328–10333. 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guerrini V, Subbian S, Santucci P, Canaan S, Gennaro ML, Pozzi G. 2016. Experimental evolution of Mycobacterium tuberculosis in human macrophages results in low-frequency mutations not associated with selective advantage. PLoS One 11:e0167989. 10.1371/journal.pone.0167989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. 2015. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 43:W443–W447. 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]