ABSTRACT
The emergence of Klebsiella pneumoniae isolates carrying novel blaKPC variants conferring ceftazidime-avibactam (CAZ/AVI) resistance is being increasingly reported. We evaluated the accuracy of phenotypic methods commonly used in routine clinical laboratories in the detection of novel K. pneumoniae carbapenemase (KPC) enzymes. Additionally, we characterized by whole-genome sequencing (WGS) the KPC-ST307-K. pneumoniae isolates recovered in our hospital before and after CAZ/AVI therapy. Rectal colonization or infection by carbapenem-resistant KPC-3 K. pneumoniae isolates (imipenem MIC, 16 mg/L; meropenem MIC, 8 to >16 mg/L) and CAZ/AVI-susceptible isolates (CAZ/AVI MIC, 1 to 2 mg/L) were first detected in three intensive care unit (ICU) patients admitted between March 2020 and July 2020. KPC K. pneumoniae isolates with increased CAZ/AVI MICs (8 to 32 mg/L) and carbapenem susceptibility (imipenem and meropenem MIC, <1 mg/L) were recovered within 6 to 24 days after CAZ/AVI treatment. WGS confirmed that all KPC K. pneumoniae isolates belonged to the sequence type 307 (ST307) high-risk clone and carried identical antimicrobial resistance genes and virulence factors. The presence of the novel blaKPC-46, blaKPC-66, and blaKPC-92 genes was confirmed in the K. pneumoniae isolates with increased CAZ/AVI MICs and restored carbapenem activity. KPC production was confirmed by immunochromatography, the eazyplex Superbug CRE system, and the Xpert Carba-R assay in all KPC K. pneumoniae isolates, but not in any isolate using chromogenic agar plates for carbapenemase producers (ChromID-CARBA), the KPC/MBL/OXA-48 Confirm kit, and the β-CARBA test. Nevertheless, all grew in chromogenic agar plates for extended-spectrum β-lactamase (ESBL) producers (ChromID-ESBL). We report the failure of the most common phenotypic methods used for the detection of novel KPC carbapenemases but not of rapid molecular or immunochromatography assays, thus highlighting their relevance in microbiology laboratories.
KEYWORDS: novel blaKPC variants, carbapenemase phenotypic detection, KPC-3-ST307-K. pneumoniae high-risk clone, ceftazidime-avibactam susceptibility, novel blaKPC variants
INTRODUCTION
Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae isolates have been reported as a major pathogen causing human infections and nosocomial outbreaks (1, 2). KPC-type enzymes (class A β-lactamases) are able to hydrolyze all β-lactams and are poorly inhibited by traditional β-lactamase inhibitors (3). In KPC-producing K. pneumoniae isolates, carbapenem resistance is usually due to the KPC β-lactamase activity frequently combined with outer membrane permeability defects (4). The successful dissemination of KPC enzymes among high-risk clones of K. pneumoniae, such as sequence type 258 (ST258) and ST512, and more recently ST307, is one of the main mechanisms implicated in the global spread of these multidrug-resistant pathogens (5, 6). Currently, the high-risk clone ST307-K. pneumoniae is recognized as a new worldwide emerging sublineage with genetic features contributing to higher dissemination and persistence traits in the hospital setting (6, 7).
The high prevalence of infections caused by these microorganisms is of great concern in public health due to the limited therapeutic options and the high rate of associated morbidity and mortality (8). Ceftazidime-avibactam (CAZ/AVI) has been introduced in clinical practice to combat difficult-to-treat infections caused by carbapenem-resistant Enterobacterales isolates. Avibactam has the ability to restore the activity of ceftazidime against β-lactamases such as AmpC, extended-spectrum β-lactamase (ESBL), and serine carbapenemases, including KPCs and OXA-48-like enzymes (9). Although the clinical efficacy of CAZ/AVI in the treatment of infections caused by KPC-producing pathogens has been demonstrated, reduced activity has been reported due to some KPC variants, frequently emerging after CAZ/AVI exposure (with a frequency of ∼10−9) (10–15). Additionally, in some cases these clinical isolates expressing mutated KPC variants emerge with a phenotype that resembles an ESBL enzyme, thus making their detection difficult in routine clinical laboratories (16, 17).
In this work we evaluated the detection of KPC-producing K. pneumoniae ST307 isolates with increased CAZ/AVI susceptibility MIC values sequentially recovered from colonized or infected patients after CAZ/AVI therapy by using different phenotypic and molecular methods. Additionally, we used whole-genome sequencing to analyze the genetic characteristics of ST307-K. pneumoniae isolates recovered before and after treatment with CAZ/AVI.
MATERIALS AND METHODS
Isolates and bacterial identification.
Between March 2020 and July 2020, three patients admitted in the medical intensive care unit (ICU) of Ramón y Cajal University Hospital were infected or colonized by KPC-producing K. pneumoniae isolates with elevated CAZ/AVI MIC values after or during treatment with this antibiotic. All rectal colonization and clinical samples were prospectively collected. Rectal samples were directly seeded on ChromID-ESBL and ChromID CARBA SMART biplate selective chromogenic agar plates (bioMérieux, Marcy-l’Étoile, France) and incubated at 37°C for 24 or 48 h. Recovered clinical isolates were seeded and incubated in the chromogenic agar plates under the same conditions. Identification was performed by mass spectrometry by using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS; Bruker-Daltonics, Bremen, Germany).
Phenotypic detection of KPC carbapenemase.
Isolates were phenotypically confirmed as carbapenemase producers using the KPC/MBL/OXA-48 Confirm kit (Rosco Diagnostica, Taastrup, Denmark), the β-CARBA test (Bio-Rad, California, USA), the O.K.N.V.I. RESIST-5 (CORIS BioConcept, Gembloux, Belgium), and the NG-Test CARBA 5 (Hardy Diagnostics, California, USA) immunochromatography tests. Additionally, susceptibility testing by disk diffusion (DDST) and the ESBL/AmpC screen kit test (Rosco Diagnostica) were also performed. All tests were performed in triplicate.
Antimicrobial susceptibility testing and resistance genes characterization.
Antimicrobial susceptibility was studied by standard broth microdilution (lyophilized Sensititre panel; Thermo Fisher Scientific, formerly Trek Diagnostic Systems, Inc., Cleveland, OH). Ertapenem susceptibility was tested using MicroScan panels (Beckman, California, USA). The clinical breakpoint for cefepime-taniborbactam tentatively proposed by the manufacturer was susceptible ≤8/4 mg/L and resistant >8/4 mg/L (18). Carbapenemase and selected ESBL genes were detected with the eazyplex Superbug CRE system (Amplex-Biosystems, Delaware) and the Xpert Carba-R assay (Cepheid, Sunnyvale, CA) and confirmed by PCR and Sanger sequencing (19).
WGS and sequence processing.
For the subsequent genomic analysis, two KPC-K. pneumoniae isolates from each patient were selected (one isolate before treatment, resistant to carbapenems and susceptible to CAZ/AVI, and the first isolate after treatment, with restored carbapenem activity and decreased susceptibility to CAZ/AVI). Whole-genome sequencing (WGS) (Illumina NovaSeq 6000 platform; Oxford Genomics Centre [OGC], Oxford, UK) and sequence processing were carried out as previously described (16). Multilocus sequence type (MLST) calling was performed using MLST v2.19.0 (https://github.com/tseemann/mlst). Core genome alignment and variant calling of SNPs and small insertions/deletions (indels) were performed using the Snippy v4.6.0. program. K. pneumoniae serotypes based on the wzi gene and the K (capsule) and O (lipopolysaccharide [LPS]) antigens were predicted using Kleborate software v2.0.1. (20). Antimicrobial resistance and virulence genes were identified using ABRicate (ResFinder, ARG-ANNOT, and VFDB databases; threshold, 90% coverage; 98% identity). PointFinder software was used for the detection of chromosomal point mutations (21). K. pneumoniae integrative conjugative element (ICE K. pneumoniae)-associated virulence loci (yersiniabactin [ybt], colibactin [clb]) and virulence plasmid-associated loci (salmochelin [iro], aerobactin [iuc], hypermucoidy [rmpADC, rmpA2]) were investigated using Kleborate. The genetic environment and the mobile genetic elements implicated in the dissemination of the blaKPC genes were characterized using PlasmidFinder and BLAST (22).
Data availability.
The complete genome sequences of CAZ/AVI-susceptible and -resistant isolates were deposited at DDBJ/ENA/GenBank under accession numbers JAHUYV000000000 to JAHUZA000000000. The GenBank accession number of the novel blaKPC-92 gene is MZ461464. Information about genome characteristics and GenBank submission data are summarized in Table S1 in the supplemental material.
RESULTS
Case reports.
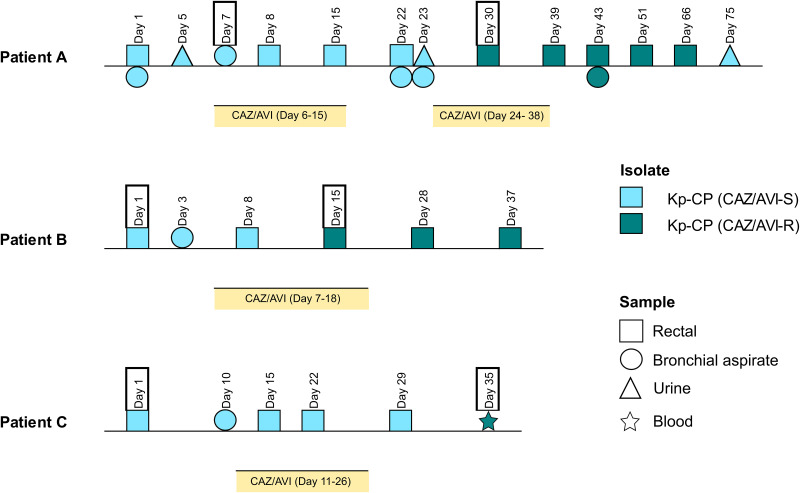
The timeline of events during the hospital admission and CAZ/AVI treatment received by each patient are represented in Fig. 1. Antibiotic susceptibility results of strains selected for the subsequent analysis are shown in Table 1.
FIG 1.
Timeline of events during the hospital admission and ceftazidime-avibactam treatment received by patients A, B, and C. Yellow bars represent duration of treatment with ceftazidime-avibactam. Samples with the framed dates are those that were subsequently tested for the screening of KPC-carbapenemase and analyzed by whole-genome sequencing. CAZ/AVI, ceftazidime-avibactam; CP, carbapenemase; ESBL, extended-spectrum β-lactamase.
TABLE 1.
Antibiotic susceptibility results in KPC-producing K. pneumoniae isolates
| Isolate | MIC (mg/L) for:a |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PTZ | AZT | CTX | CAZ | FEP | IMP | ERT | MER | C/T | FTB | CAZ/AVI | AZT/AVI | IMI/REL | MER/VAB | FDC | CIP | GEN | TOB | AMK | COL | FOS | TGC | |
| A-1 | >64/4 | >32 | >32 | >32 | 32 | 16 | >4 | 8 | 32 | 0.5 | 2 | 0.12 | 0.25 | ≤0.06 | 0.5 | >2 | >8 | >8 | ≤8 | >4 | 64 | 1 |
| A-2 | >64/4 | >32 | >32 | >32 | 32 | ≤1 | ≤0.5 | ≤ 0.12 | >32 | 2 | 32 | 0.06 | 0.25 | ≤0.06 | 1 | >2 | >8 | >8 | 32 | >4 | >64 | 1 |
| B-1 | >64/4 | >32 | >32 | >32 | >32 | 16 | >4 | >16 | >32 | 1 | 2 | 0.25 | 0.25 | ≤0.06 | 1 | >2 | >8 | >8 | 16 | >4 | 64 | 2 |
| B-2 | >64/4 | >32 | >32 | >32 | 32 | ≤1 | ≤0.5 | ≤ 0.12 | >32 | 1 | 8 | 0.25 | 0.25 | ≤0.06 | 2 | >2 | >8 | >8 | 16 | >4 | >64 | 2 |
| C-1 | >64/4 | >32 | >32 | >32 | >32 | 16 | >4 | >16 | >32 | 0.5 | 1 | 0.12 | 0.5 | ≤0.06 | 1 | >2 | >8 | >8 | ≤8 | ≤1 | >64 | 2 |
| C-2 | 32/4 | >32 | >32 | >32 | 16 | ≤1 | ≤0.5 | ≤ 0.12 | >32 | 0.5 | 8 | 0.06 | 0.25 | ≤0.06 | 2 | >2 | >8 | >8 | ≤8 | ≤1 | >64 | 2 |
PTZ, piperacillin-tazobactam; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; AZT, aztreonam; IMP, imipenem; ERT, ertapenem; MER, meropenem; CIP, ciprofloxacin; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; COL, colistin; FOS, fosfomycin; TIG, tigecycline; C/T, ceftolozane-tazobactam; CAZ/AVI, ceftazidime-avibactam; AZT/AVI, aztreonam-avibactam; MER/VAB, meropenem-vaborbactam; IMI/REL, imipenem-relebactam; FDC, cefiderocol; FTB, cefepime-taniborbactam.
In patient A (diagnosed with non-Hodgkin lymphoma), a K. pneumoniae isolate (A-1) resistant to meropenem (MICMEM, 8 mg/L), imipenem (MICIMP, 16 mg/L), and ertapenem (MICERT, >4 mg/L) and susceptible to CAZ/AVI (MICCAZ/AVI, 2 mg/L) was recovered from a bronchial aspiration sample on day 7 of admission. This patient was treated with two cycles of CAZ/AVI (2 g/0.5 g for 8 h intravenously [i.v.]) from day 6 to 15 and from day 24 to 38. On day 30, 6 days after the initiation of the second antibiotic cycle, a second K. pneumoniae isolate (A-2) with restored carbapenem susceptibility (MICMEM, ≤0.12 mg/L; MICIMP, ≤1 mg/L; MICERT, ≤0.5 mg/L) and resistance to CAZ/AVI (MICCAZ/AVI, 32 mg/L) was detected in a rectal sample.
In patient B (diagnosed with COVID-19), a K. pneumoniae isolate (B-1) compatible with carbapenemase production (MICMEM, >16 mg/L; MICIMP, >16 mg/L; MICERT, >4 mg/L) and susceptible to CAZ/AVI (MICCAZ/AVI, 2 mg/L) was recovered from a rectal sample. Two days later (on day 3), another K. pneumoniae isolate with the same phenotype was detected in a bronchial aspiration sample. CAZ/AVI therapy (2 g/0.5 g; 8 h i.v.) was implemented between day 7 and 18. On day 15, during the treatment with CAZ/AVI, a second K. pneumoniae isolate (B-2) with restored carbapenem susceptibility (MICMEM, ≤0.12 mg/L; MICIMP, ≤1 mg/L; MICERT, ≤0.5 mg/L) and a 4-fold increase in the CAZ/AVI MIC value (MICCAZ/AVI, 8 mg/L) was recovered from a rectal sample.
In patient C (diagnosed with non-Hodgkin lymphoma), a K. pneumoniae isolate (C-1) resistant to carbapenems (MICMEM, >16 mg/L; MICIMP, 16 mg/L; MICERT, >4 mg/L) and susceptible to CAZ/AVI (MICCAZ/AVI, 1 mg/L) was found in a rectal culture. On day 10, a K. pneumoniae isolate compatible with carbapenemase production was also recovered from a bronchial aspiration sample. CAZ/AVI treatment, adjusted to the renal function, was started on day 11 and continued until day 26 (2 g/0.5 g, 8 h i.v., between day 11 and 12; 1 g/0.25 g, 6 h i.v., from day 13 to 23; and 1 g/0.25 g, 8 h i.v., from day 24 to 26). On day 35, 24 days after initiation of treatment with CAZ/AVI, a second K. pneumoniae isolate (C-2) displaying susceptibility to carbapenems (MICMER, ≤0.12 mg/L; MICIMP, ≤1 mg/L; MICERT, ≤0.5 mg/L) and an increased CAZ/AVI MIC value (MICCAZ/AVI, 8 mg/L) was recovered from a blood sample.
All these K. pneumoniae isolates (A-1, A-2, B-1, B-2, C-1, and C-2) showed susceptible MIC values for cefepime-taniborbactam, aztreonam-avibactam, imipenem-relebactam, meropenem-vaborbactam, and cefiderocol (Table 1).
Detection of KPC carbapenemase.
All baseline K. pneumoniae isolates (A-1, B-1, and C-1) grew on both ChromID-ESBL and ChromIDR CARBA SMART agar plates and were phenotypically confirmed as carbapenemase producers using all phenotypic methods (Table 2). The presence of the blaKPC gene was confirmed in all these baseline K. pneumoniae isolates using the eazyplex SuperBug CRE system and the Xpert Carba-R assay. The K. pneumoniae isolates displaying restored carbapenem activity and elevated CAZ/AVI MIC values (A-2, B-2, and C-2) were only recovered from the ChromID-ESBL agar and were identified as non-carbapenemase producers using the KPC/MBL/OXA-48 Confirm kit and the β-CARBA test. The immunochromatography assays were positive for all K. pneumoniae isolates, including the A-2, B-2, and C-2 isolates (Table 2). In these three isolates (A-2, B-2, and C-2), production of ESBL was confirmed with the DDST and the ESBL/AmpC Screen kit test. The eazyplex SuperBug CRE system and the Xpert Carba-R test also demonstrated the presence of a blaKPC gene in all these isolates. CTX-M-1-group production was also verified in all K. pneumoniae isolates (A-1, A-2, B-1, B-2, C-1, and C-2) with the eazyplex SuperBug CRE system.
TABLE 2.
Patient data and carbapenem and ceftazidime-avibactam susceptibility of KPC-producing K. pneumoniae isolates and results of phenotypic methods for KPC detectiona
| Sample | Source | Date (day/mo/yr) | CAZ-AVI exposure at time of culture (days) | MIC values (mg/L) |
ChromID plates |
Phenotypic methods |
Rapid molecular methods |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MEM | CAZ/AVI | ESBL | CARBA SMART | IC |
β-CARBA test | Rosco Diagnostica |
DDST-ESBL | Eazyplex system | Xpert Carba-R | KPC variant | ||||||
| O.K.N.V.I. RESIST-5 | NG-test CARBA 5 | CP | ESBL/AmpC | ||||||||||||||
| A-1 | BAS | 02/03/2020 | +1 | 16 | 8 | 2 | + | + | + | + | + | + | −/– | − | + | + | KPC-3 |
| A-2 | Rectal | 25/03/2020 | +6 | ≤1 | ≤ 0.12 | 32 | + | − | + | + | − | − | +/– | + | + | + | KPC-46 |
| B-1 | Rectal | 02/06/2020 | 0 | 16 | >16 | 2 | + | + | + | + | + | + | −/– | − | + | + | KPC-3 |
| B-2 | Rectal | 16/06/2020 | +8 | ≤1 | ≤ 0.12 | 8 | + | − | + | + | − | − | +/– | + | + | + | KPC-66 |
| C-1 | Rectal | 09/06/2020 | 0 | 16 | >16 | 1 | + | + | + | + | + | + | −/– | − | + | + | KPC-3 |
| C-2 | Blood | 13/07/2020 | +24 | ≤1 | ≤ 0.12 | 8 | + | − | + | + | − | − | +/– | + | + | + | KPC-92 |
BAS, bronchial aspirate; IMP, imipenem; MEM, meropenem; CAZ/AVI, ceftazidime-avibactam; IC, immunochromatography test; CP, carbapenemases; DDST-ESBL, susceptibility testing by disk diffusion for ESBL detection; +, positive result for KPC carbapenemase detection, −, negative result for KPC carbapenemase detection.
Molecular typing.
All KPC-K. pneumoniae isolates belonged to ST307. The KL173-O2v2 serotype (associated with the wzi allele 173) was inferred in all ST307-KPC K. pneumoniae isolates. All core single nucleotide polymorphisms (SNPs) identified across the first and the second isolate of patients A, B, and C are shown in Table S2.
Resistome characterization.
The presence of the blaKPC-3 gene was confirmed by WGS in all ST307-KPC K. pneumoniae isolates displaying resistance to carbapenems and susceptibility to CAZ/AVI. Three variants of blaKPC-3 were identified by WGS in the isolates that showed elevated MIC values of CAZ/AVI and restored activity of carbapenems—patient A (isolate A-2, blaKPC-46, L168P), patient B (isolate B-2, blaKPC-66, E167_L168del), and patient C (isolate C-2, blaKPC-92, E167_N169delinsD). All substitutions and deletions/insertions occurred in the Ω-loop region (Arg164 to Asp179) of the KPC enzyme (Fig. S1). BlaKPC-3 and all novel blaKPC-46, blaKPC-66, and blaKPC-92 variants were encoded on a pKPQILtype plasmid (multireplicon IncFIIk-FIB plasmid) as a part of the 10-kb transposon, Tn4401a.
Screening for acquired resistance determinants revealed in all KPC ST307-K. pneumoniae isolates other resistance genes involving β-lactams (blaCTX-M-15, blaTEM-1, blaSHV-28, blaOXA-1, and blaOXA-9), aminoglycosides [aac(3)-IIa, aac(6′)-Ib-cr, strA, and strB], quinolones (oqxA5, oqxB19, and y qnrB1), sulfonamides (sul2), fosfomycin (fosA), and trimethoprim (dfrA14).
Investigation of major outer membrane porins showed an intact ompK35 gene in all KPC ST307-K. pneumoniae strains. However, identical known mutations associated with resistance to cephalosporins and/or carbapenems were found in OmpK36 (N49S, L59V, and T184P) and OmpK37 (I70M, I128M, and N230G) proteins in all strains susceptible and resistant to CAZ/AVI. The conserved ParC-80I and GyrA-83I fluoroquinolone resistance-associated mutations were also detected in all genomes.
Virulence determinants.
An identical virulence gene content was identified in all KPC ST307-K. pneumoniae strains—entA, entB, fepC, ompA, yagV/ecpE, yagW/ecpD, yagX/ecpC, yagY/ecpB, yagZ/ecpA, and ykgK/ecpR. On the other hand, the chromosomal siderophore yersiniabactin (ybt locus) and the genotoxin colibactin (clb locus) were not found, and an ICE K. pneumoniae lineage was not assigned. Horizontally acquired virulence factors encoding the siderophores aerobactin (iuc) and salmochelin (iro), the genotoxin colibactin (clb), and the hypermucoid factors (rmpA and rmpA2) were also not detected in these isolates.
DISCUSSION
In this study, we investigated the effect of novel blaKPC genes encoding enzymes that resemble those of ESBLs but confer reduced susceptibility to CAZ/AVI in the screening of KPC carbapenemases using the most commonly used phenotypic methods in routine clinical microbiology laboratories. Additionally, we used WGS to characterize the genetic features of KPC-producing K. pneumoniae ST307 strains recovered before and after treatment with CAZ/AVI.
ST307-K. pneumoniae is a successful high-risk clone, globally disseminated and largely associated with conserved IncF-type plasmids containing the blaCTX-M-15 gene (23). Currently, ST307 K. pneumoniae is emerging as an important lineage linked to the production of KPC enzymes and with a greater adaptation and persistence to hospital settings, replacing even the high-risk clones ST512 and ST258 in some areas of endemicity (6, 7). In our hospital, a high prevalence of the KPC-3-ST307-K. pneumoniae high-risk clone has been detected since 2018 in both colonized and infected patients (data not shown). The emergence of novel blaKPC-3 variants (KPC-49 and KPC-61) associated with increased CAZ/AVI MIC values and decreased carbapenem activity after treatment with CAZ/AVI has also been reported in our institution in patients infected both by the ST307-K. pneumoniae clone and by the ST131-Escherichia coli high-risk clone (16, 24). Nevertheless, during the follow-up period (March 2020 to July 2020) we only found these three patients infected or colonized with K. pneumoniae isolates with novel KPC variants. Overall, 30 patients received treatment with CAZ/AVI in the ICU in which they were admitted (data not shown), representing 10% emergence of CAZ/AVI resistance. This figure is similar to that previously reported (25).
Mutations in blaKPC usually occur following a prolonged exposure to CAZ/AVI and lead to treatment failure (10). In the present study, three mutants of the KPC-3 enzyme (KPC-46 and KPC-66, recently described, and KPC-92, first reported in this study) were detected in ST307-K. pneumoniae isolates colonizing or infecting patients during or after treatment with CAZ/AVI. Interestingly, all substitutions and deletions occurred in the Ω-loop region (Arg164 to Asp179) of the KPC-3 enzyme. Previous studies have demonstrated that KPC-3 variants carrying mutations in this region exhibit an enhanced affinity of ceftazidime that possibly reduces the binding of avibactam (12, 26–28). BlaKPC-3 and all variants blaKPC-46, blaKPC-66, blaKPC-92, were located on the conserved transposon Tn4401a as part of a conjugative pKpQIL-type plasmid. KPC-3-encoding pKpQIL plasmids are highly similar to those previously described in the ST258 and have also been related to the dissemination of other novel blaKPC genes (6, 29, 30). Some studies have reported that alterations affecting Ompk35 and Ompk36 porins along with the overexpression of KPC seem to be also involved in a reduced susceptibility to CAZ/AVI in K. pneumoniae isolates (30, 31). In our KPC-ST307 isolates, known mutations previously related to cephalosporin and/or carbapenem resistance were found in the OmpK36 and OmpK37 proteins, but differences between isolates displaying different carbapenems and CAZ/AVI MIC values were not found. According to these results and recent literature, the CAZ/AVI resistance phenotype detected in our isolates could be a consequence of the mutations in the blaKPC-3 gene following exposure to CAZ/AVI.
In addition to KPC genes, a large number of antibiotic resistance genes affecting different antimicrobial groups was detected in all ST307-K. pneumoniae isolates. Furthermore, although virulence loci associated with hypervirulent K. pneumoniae clones were not found, a large content of virulence determinants was detected in all strains. Accumulation of antibiotic resistance genes and virulence factors in the ST307-K. pneumoniae high-risk clone, added to the emergence of resistance to CAZ/AVI due to novel KPC enzymes, could lead to the establishment and persistence of this emerging lineage in the hospital setting causing infections leading to therapeutic failure.
Reliable identification of KPC carbapenemase production in clinically important clones is crucial to combat the nosocomial spread of these multidrug-resistant pathogens. The impact of novel KPC variants in the detection of KPC carbapenemase producers using phenotypic methods has been scarcely studied. In our work, meropenem, imipenem, and ertapenem susceptibility was fully restored in the isolates with clearly increased CAZ/AVI MIC values, resulting in KPC-producing K. pneumoniae isolates that are unable to grow on selective chromogenic medium for CPE and with a phenotype resembling that of ESBL producers. Additionally, the most common phenotypic methods in Europe used for carbapenemase screening, such as the KPC/MBL/OXA-48 Confirm kit and the β-CARBA test were not effective in the detection of novel KPC-46, KPC-66, and KPC-92 enzymes. In contrast, the immunochromatography tests were positive for detecting KPC-46, KPC-66, and KPC-92 enzymes. A recent study in Spain demonstrated that an immunochromatography assay detects as carbapenemases other novel KPC enzymes resembling an ESBL phenotype such as KPC-39, KPC-47, and KPC-48, but not KPC-31 (D179Y mutation) (32). In fact, a previous study reported that KPC-31 carbapenemase production is also not detected using colorimetric tests such as β-CARBA NP (33). It is important to consider that these novel KPC variants are potentially missing if ceftazidime-avibactam is not routinely tested on antimicrobial susceptibility testing (AST) and is only determined in isolates compatible with carbapenemase production.
The full or partial restoration of carbapenem susceptibility in isolates expressing mutated KPC enzymes that confer resistance to CAZ/AVI, a phenomenon known as “collateral sensitivity,” leaves carbapenems as a potential therapeutic option (10, 11, 34). Nevertheless, some studies have demonstrated that the carbapenem-resistant phenotype can be restored in isolates that still exhibit resistance to CAZ/AVI after the discontinuation of treatment with CAZ/AVI (35). The limitation of antimicrobial susceptibility methods in detecting isolates that carry KPC mutated enzymes with potential carbapenemase activity and that reverse their resistance phenotype after or during the antibiotic treatment represents a challenge for routine diagnostic laboratories. The emergence of these “silent” blaKPC genes in nosocomial pathogens causing both colonization and infection adds value to rapid molecular diagnostic tools and genomic techniques in the routine laboratories and highlights that further research in genotype-to-phenotype prediction using WGS is needed. Additionally, the spread of these “masked” KPC carbapenemases through epidemic K. pneumoniae clinical strains such as the ST307 high-risk clone could contribute to a higher dissemination of these enzymes in nosocomial settings and, as a consequence, increase difficult-to-treat infections.
In conclusion, we found K. pneumoniae isolates carrying KPC mutated enzymes conferring phenotypes resembling ESBLs which cannot be detected as KPC producers by the phenotypic methods most frequently used in the routine clinical laboratories. Additionally, the emergence of these novel KPC enzymes in the ST307-K. pneumoniae high-risk clone could lead to an increase in the epidemiological success of this lineage in the hospital setting. This finding should be a cause of great concern for public health.
ACKNOWLEDGMENTS
We thank Mary Harper for English correction of the manuscript.
M.H.-G. was supported by a contract from FIBIO (Fundación para la Investigación Biomédica del Instituto Ramón y Cajal de Investigación Sanitaria, IRYCIS, Madrid, Spain). We acknowledge financial support from Plan Nacional de I+D+I 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, the Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0011) cofinanced by the European Development Regional Fund “A Way to Achieve Europe” (ERDF), Operative Program Intelligent Growth 2014–2020, and CIBER de Enfermedades Infecciosas (CIBERINFEC) (CB21/13/00084), Instituto de Salud Carlos III, Madrid, Spain.
We declare no conflict of interest with the content of this article.
This study was approved by the Ramón y Cajal University Hospital Ethics Committee (reference 293/19).
Footnotes
Supplemental material is available online only.
Contributor Information
Rafael Cantón, Email: rafael.canton@salud.madrid.org.
Patricia J. Simner, Johns Hopkins
REFERENCES
- 1.Lerner A, Adler A, Abu-Hanna J, Cohen Percia S, Kazma Matalon M, Carmeli Y. 2015. Spread of KPC-producing carbapenem-resistant Enterobacteriaceae: the importance of super-spreaders and rectal KPC concentration. Clin Microbiol Infect 21:470.e1–470.e7. 10.1016/j.cmi.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulzova L, Navratilova L, Comor L. 2017. Alterations in outer membrane permeability favor drug-resistant phenotype of Klebsiella pneumoniae. Microb Drug Resist 23:413–420. 10.1089/mdr.2016.0017. [DOI] [PubMed] [Google Scholar]
- 5.Rojas LJ, Weinstock GM, de La Cadena E, Diaz L, Rios R, Hanson BM, Brown JS, Vats P, Phillips DS, Nguyen H, Hujer KM, Correa A, Adams MD, Perez F, Sodergren E, Narechania A, Planet PJ, Villegas M.v, Bonomo RA, Arias CA. 2017. An analysis of the epidemic of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae: convergence of two evolutionary mechanisms creates the “perfect storm”. J Infect Dis 217:82–92. 10.1093/infdis/jix524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, Endimiani A, Mammina C, Ocampo AM, Jimenez JN, Doumith M, Woodford N, Hopkins K, Carattoli A. 2017. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom 3:e000110. 10.1099/mgen.0.000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peirano G, Chen L, Kreiswirth BN, Pitout JDD. 2020. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob Agent Chemother 64:1–14. 10.1128/AAC.01148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porreca AM, Sullivan K.v, Gallagher JC, Gallagher JC. 2018. The epidemiology, evolution, and treatment of KPC-producing organisms. Curr Infect Dis Rep 20:13. 10.1007/s11908-018-0617-x. [DOI] [PubMed] [Google Scholar]
- 9.Pogue JM, Bonomo RA, Kaye KS. 2019. Ceftazidime/avibactam, meropenem/vaborbactam, or both? Clinical and formulary considerations. Clin Infect Dis 68:519–524. 10.1093/cid/ciy576. [DOI] [PubMed] [Google Scholar]
- 10.Shields RK, Chen L, Cheng S, Chavda KD, Press EG. 2016. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. 2017. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob Agents Chemother 61:e02534-16. 10.1128/AAC.02534-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobson CA, Bonacorsi S, Hocquet D, Baruchel A, Fahd M, Storme T, Tang R, Doit C, Tenaillon O, Birgy A. 2020. Impact of anticancer chemotherapy on the extension of beta-lactamase spectrum: an example with KPC-type carbapenemase activity towards ceftazidime-avibactam. Sci Rep 10:589. 10.1038/s41598-020-57505-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu G, Abraham T, Lee S. 2016. Ceftazidime-avibactam for treatment of carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 63:1147–1148. 10.1093/cid/ciw491. [DOI] [PubMed] [Google Scholar]
- 14.Gugliandolo A, Caio C, Mezzatesta ML, Rifici C, Bramanti P, Stefani S, Mazzon E. 2017. Successful ceftazidime-avibactam treatment of MDR-KPC-positive Klebsiella pneumoniae infection in a patient with traumatic brain injury. Medicine (Baltimore) 96:e7664. 10.1097/MD.0000000000007664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livermore DM, Warner M, Jamrozy D, Mushtaq S, Nichols WW, Mustafa N, Woodford N. 2015. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 59:5324–5330. 10.1128/AAC.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernández-García M, Sánchez-López J, Martínez-García L, Becerra-Aparicio F, Morosini MI, Ruiz-Garbajosa P, Cantón R. 2021. Emergence of the new KPC-49 variant conferring an ESBL phenotype with resistance to ceftazidime-avibactam in the ST131-H30R1 Escherichia coli high-risk clone. Pathogens 10:67. 10.3390/pathogens10010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobson CA, Bonacorsi S, Hervé J, Choudhury A, Magnan M, Cointe A, Bercot B, Tenaillon O, Birgy A. 2020. KPC beta-lactamases are permissive to insertions and deletions conferring substrate spectrum modifications and resistance to ceftazidime-avibactam. Antimicrob Agent Chemother 64:e01175-20. 10.1128/AAC.01175-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernández-García M, García-Castillo M, Ruiz-Garbajosa P, Siller-Ruiz M, Pitart C, Gracia-Ahufinger I, Mulet X, Tormo N, Cantón R. 2022. In vitro activity of cefepime-taniborbactam against carbapenemase producing Enterobacterales and Pseudomonas aeruginosa isolates recovered in Spain. Antimicrob Agent Chemother 10.1128/aac.02161-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Fernández S, Morosini MI, Marco F, Gijón D, Vergara A, Vila J, Ruiz-Garbajosa P, Cantón R. 2015. Evaluation of the eazyplex SuperBug CRE system for rapid detection of carbapenemases and ESBLs in clinical Enterobacteriaceae isolates recovered at two Spanish hospitals. J Antimicrob Chemother 70:1047–1050. 10.1093/jac/dku476. [DOI] [PubMed] [Google Scholar]
- 20.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carattoli A, Zankari E, Garciá-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyres KL, Hawkey J, Hetland MAK, Fostervold A, Wick RR, Judd LM, Hamidian M, Howden BP, Löhr IH, Holt KE. 2019. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother 74:577–581. 10.1093/jac/dky492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernández-García M, Sánchez-López J, Ponce-Alonso M, Morosini MI, Cantón R, Ruiz-Garbajosa P. 2019. Emergence of a new KPC-3 variant in the high-risk clone ST307- K. pneumoniae after ceftazidime-avibactam exposure in a tertiary hospital in Madrid. 12th International Meeting on Microbial Epidemiological Markers (IMMEM XII).
- 25.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. 2016. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis 63:1615–1618. 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen M-H, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Compain F, Arthur M. 2017. Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D179Y substitution in the KPC-2 β-lactamase. Antimicrob Agents Chemother 61:e00451-17. 10.1128/AAC.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel L, Vuillemin X, Juhas M, Masseron A, Bechtel-Grosch U, Tiziani S, Mancini S, Nordmann P. 2020. KPC-50 confers resistance to ceftazidime-avibactam associated with reduced carbapenemase activity. Antimicrob Agents Chemother 64:e00321-20. 10.1128/AAC.00321-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. 2010. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother 65:243–248. 10.1093/jac/dkp417. [DOI] [PubMed] [Google Scholar]
- 30.Galani I, Antoniadou A, Karaiskos I, Kontopoulou K, Giamarellou H, Souli M. 2019. Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin Microbiol Infect 25:763.e5–763.e8. 10.1016/j.cmi.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Humphries RM, Hemarajata P. 2017. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother 61:e00537-17. 10.1128/AAC.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cano Á, Guzmán-Puche J, García-Gutiérrez M, Castón JJ, Gracia-Ahufinger I, Pérez-Nadales E, Recio M, Natera AM, Marfil-Pérez E, Martínez-Martínez L, Torre-Cisneros J. 2020. Use of carbapenems in the combined treatment of emerging ceftazidime/avibactam-resistant and carbapenem-susceptible KPC-producing Klebsiella pneumoniae infections: report of a case and review of the literature. J Glob Antimicrob Resist 22:9–12. 10.1016/j.jgar.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Antonelli A, Giani T, di Pilato V, Riccobono E, Perriello G, Mencacci A, Rossolini GM. 2019. KPC-31 expressed in a ceftazidime/avibactam-resistant Klebsiella pneumoniae is associated with relevant detection issues. J Antimicrob Chemother 74:2464–2466. 10.1093/jac/dkz156. [DOI] [PubMed] [Google Scholar]
- 34.Shields RK, Nguyen MH, Press EG, Chen L, Kreiswirth BN, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance and restoration of carbapenem susceptibility in Klebsiella pneumoniae carbapenemase-producing K pneumoniae: a case report and review of literature. Open Forum Infect Dis 4:ofx101. 10.1093/ofid/ofx101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, Mehta M, Gomez-Simmonds A, Uhlemann A-C. 2018. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 62:e02101-17. 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download jcm.02245-21-s0001.pdf, PDF file, 0.2 MB (168.8KB, pdf)
Table S1. Download jcm.02245-21-s0002.xlsx, XLSX file, 0.01 MB (10.1KB, xlsx)
Table S2. Download jcm.02245-21-s0003.xlsx, XLSX file, 0.02 MB (18.1KB, xlsx)
Data Availability Statement
The complete genome sequences of CAZ/AVI-susceptible and -resistant isolates were deposited at DDBJ/ENA/GenBank under accession numbers JAHUYV000000000 to JAHUZA000000000. The GenBank accession number of the novel blaKPC-92 gene is MZ461464. Information about genome characteristics and GenBank submission data are summarized in Table S1 in the supplemental material.