ABSTRACT
Assessment of T-cell responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigens may be of value to determine long-lasting protection to breakthrough infections or reinfections. Interferon gamma release assay is a validated method to test cellular immunity in mycobacterial infections and has been proposed for patients with SARS-CoV-2 infection or vaccination. Quantitative IgG to spike and qualitative IgG to nucleocapsid antigens were determined by chemiluminescence microparticle immunoassay using the Architect platform (Abbott), and interferon gamma release assays against two Qiagen proprietary mixes of SARS-CoV-2 spike protein (antigen 1 and antigen 2) were performed for a selected group of subjects. A total of 121 subjects in a cloistered institution after a COVID-19 outbreak was studied. IgG spike levels and interferon gamma concentrations were highest among subjects after two doses of vaccine, followed by patients with a longer history of past COVID-19 and no vaccination. The best cutoff for the interferon gamma assay was 25 IU/L for all subgroups of individuals and the two sets of SARS-CoV-2 antigens studied. Testing T-cell response may be of clinical utility to determine immunity after exposure to SARS-CoV-2 antigens, with the interferon gamma concentration of 25 IU/L as the best cutoff either after infection or vaccination.
KEYWORDS: IGRA, serology, SARS-CoV-2, vaccine, COVID-19, interferon gamma release assay
INTRODUCTION
After immunization, by either infection or vaccination, it is becoming more evident that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection will be frequent. Therefore, what can be the expected duration of immune protection from infection (1)? How should this protection be assessed (2)? And, more importantly, what level and type of residual immunity is needed to avoid severe disease after reinfection? These are some pending questions, along with the role of viral variants in immune escape that would finally help guide decisions as to when social restrictions may be lifted or the need of vaccine boosters. The validation of laboratory assays to determine protection, in which humoral and cellular immunity may play important roles, is an urgent task that may help answer some of these burning issues (3).
Testing humoral immunity is the common means of determination of past infection or vaccination, but it has some important caveats. Antibody response does not always serve as an indicator of prior coronavirus infection, particularly for milder disease, and is shorter-lived than T-cell responses (4). However, the neutralization activity of antibodies is related to disease severity and survival (5).
Cellular immunity, that based on T-lymphocyte responses, is a key element to sustain prolonged immunological response (6) and may be involved in SARS-CoV-2 clearance and protection from reinfection (7). In comparison with short-lived B cells, T-cell memory is also more enduring (4); in fact, the induction of sufficient T cells may be needed to maintain levels of anti-SARS-CoV-2 neutralizing antibodies (8), and this cellular response may be needed to prolong vaccine efficacy (9). This cellular response is mediated by both CD4+ and CD8+ T lymphocytes (10).
Herein, we report the results of a pilot study in a closed community with a single and recent exposure to SARS-CoV-2 that serves as validation of the interferon gamma (IFN-γ) release immune assay (IGRA) to test T-cell-based immunity, both mediated by CD4+ and CD8+ cells, after infection or vaccination.
MATERIALS AND METHODS
This is an immunological study of a cloistered community that in the beginning of May 2021 suffered for the first time an outbreak of COVID-19 that finally affected half of the members in the convent. Once the outbreak was over, and after some noninfected subjects had received either one or two doses of vaccine (BNT162b2; Pfizer-BioNTech), blood samples were extracted in one single round for immunological analyses.
At the time of the outbreak, with no intervention of the investigators and following local protocols, diagnosis of SARS-CoV-2 infection was initially done by antigen test (PanBio; Abbott). Nasal swabs achieve for this test a sensitivity of 91.1% and a specificity of 99.7%, with PCR nasopharyngeal swabs as the gold standard, according to package insert (11).
For subjects with positive antigen tests, confirmatory PCR in nasopharyngeal swabs was performed. The amplification technique used was reverse transcriptase PCR (RT-PCR) by Allplex 2019‐nCoV (Seegene Inc., Seoul, Republic of Korea), which targets E, N, and RdRP SARS-CoV-2 genes and has a limit of detection of 4,167 copies/mL (12). Sequencing analysis of PCR-positive samples was done with the MiSeq system (Illumina Inc., San Diego, CA).
The investigators carried out the immunological tests that specifically are part of this study. Humoral response was assessed by determination of IgG to nucleocapsid (IgG-N) and spike (IgG-S) proteins using the chemiluminescence microparticle immunoassay (CLIA) platform Architect (Abbott Inc., Abbott Park, IL) (13). Qualitative detection of IgG-N and the qualitative and semiquantitative detection of IgG-S were performed. The concentration of IgG-S was expressed in arbitrary units per milliliter (AU/mL), with a cutoff positivity of ≥50 AU/mL according to previous validation studies (14). For subjects with IgG-S concentrations above the upper limit of the analytical measuring interval (40,000 AU/mL), we considered the IgG-S concentration 2-fold above this level (namely, 80,000 AU/mL). For further analysis, the concentrations of IgG-S were transformed into a logarithmic distribution.
The SARS-CoV-2-specific T-cell responses were assessed in the Clinical Microbiology Laboratory, Hospital Enfermera Isabel Zendal in Madrid, by a whole-blood interferon gamma release immune assay (IGRA). The production of IFN-γ was measured using the sandwich CLIA platform approved for the determination of cellular immunity against Mycobacterium tuberculosis-specific antigens (QuantiFERON-TB Gold Plus, Liaison XL; DiaSorin, Saluggia, Italy) (15), but in this case, mycobacterial reactants were substituted with SARS-CoV-2 antigens (QuantiFERON SARS-CoV-2 Research Use Only; Qiagen, Hilden, Germany) to in vitro stimulation of lymphocytes. Briefly, venous whole-blood samples were collected directly into a core tube with lithium heparin and later transferred to the QuantiFERON tubes containing S peptides (antigen 1 [Ag1] and antigen 2 [Ag2]), as well as positive (mitogen) and negative (nil) controls. Whole blood was incubated at 37°C for 16 to 24 h and centrifuged to separate plasma. According to information provided in the package inserts, the SARS CoV-2 Ag1 tube contains CD4+ T-cell epitopes derived from the S1 subunit (receptor binging domain) of the spike protein, and the Ag2 tube contains CD4+ and CD8+ T-cell epitopes from the S1 and S2 subunits of the S protein (16). Ag1 and Ag2 at this time are in vitro diagnostic products labeled for research use only (RUO) and are not yet validated for clinical purposes. In conjunction with these tubes, blood containers that consist of nil and mitogen tests are used as negative and positive controls. Specimens were processed as per the manufacturer’s guidelines (17–19). The CLIA platform determines IFN-γ concentrations in international units per liter (IU/L), although the recommendation is that for clinical purposes a qualitative result is produced using a cutoff titer that is yet to be determined for SARS-CoV-2 infection. To calculate the final results per patient, the nil control test needs to be subtracted from mitogen, Ag1, and Ag2 results. The final IFN-γ concentration in the mitogen control test needs to be >500 IU/L to validate the final Ag1 and Ag2 results.
Statistical analyses were done using SPSS software, version 22 (IBM, Chicago, Ill). The Student's t test was used to compare normally distributed continuous variables. In the case of nonnormally distributed variables, Mann-Whitney's U test was applied. Comparison of proportions for categorical variables was done either by chi-square or Fisher’s exact test as required. Correlation tests were done using Spearman’s Rho test. The selection of the best cutoff for IFN-γ concentration with IGRA was done for both Ag1 and Ag2 using different clinical correlates in a dichotomic fashion (history of COVID-19, vaccination, exposure to SARS-CoV-2 S protein, serology to SARS-CoV-2) for the calculation of the area under the receiver operating characteristics curve (AUROC). After calculating the Youden index, the cutoff with the best sensitivity with a specificity above 90% was finally selected to ensure a minimal number of false positive results.
This pilot study was approved by the Ethics Committee of the Hospital Clínico San Carlos as part of the SeroVAC study (reference number 21/274-O_M_SP). Written informed consent was obtained from all participants.
RESULTS
On 5 May 2021, an outbreak of SARS-CoV-2 infection was declared in a cloistered religious community of 121 female members (median age, 39.0 years old; interquartile range [IQR], 15.5) with no history of known cases of COVID-19 nor of members with any type of immunosuppression. After two initial symptomatic cases presenting positive with a nasopharyngeal SARS-CoV-2 antigen (Ag) test, isolation measures were applied. Screening with Ag-tests of all members of the community detected 24 and 30 additional cases on 6 and 10 May, respectively. Subsequent tests detected two new cases on 13 May and 21 May; no more Ag tests were positive in subsequent rounds done every 3 to 5 days until 15 June when the outbreak was declared over. A total of 58 cases were finally detected, all with positive PCR in nasopharyngeal samples after the initial screening; most subjects were mildly symptomatic except for 3 cases that needed hospitalization, one of whom died. Sequencing analyses in PCR-positive samples revealed in all cases that the infecting SARS-CoV-2 variant was B.1.1.7 (Alpha variant), clade 20I/501Y.V1, which was then the most prevalent in Spain in that time.
The diagnosis of cases and screening of contacts was done by local medical services according to local protocols so that the investigations in this study were done with no opportunity to perform any direct diagnosis of SARS-CoV-2 infection. These clinical protocols contemplate the following: initial rapid Ag tests for screening of symptomatic cases, frequent rapid Ag tests for the screening of asymptomatic contacts, and PCR confirmation only for positive rapid Ag tests (20).
On 6 July 2021, blood samples were obtained for SARS-CoV-2 serology (IgG against nucleocapsid [N] and spike [S] antigens, Architect; Abbott) and interferon gamma (IFN-γ) release assay (IGRA).
A total of 117 subjects were assessed; 56 subjects had history of recent COVID-19 (mean [standard deviation (SD)] lag from diagnosis to test of 58.9 [2.8] days); 17 subjects had received one dose of mRNA vaccine after the outbreak (BNT162b2; Pfizer-BioNTech) (mean [SD] lag from vaccination to test of 19.1 [4.4] days); 15 subjects had received two doses of vaccine 3 weeks apart before or after the outbreak (mean [SD] lag from full vaccination to test of 55.0 [47.4] days); for 31 subjects there was no history of infection or vaccination. Only one subject in the one-dose and one subject in the two-dose vaccination groups had COVID-19 during the outbreak, so they were not considered for the immunological analyses. This left us with 115 valid samples for immunological analyses.
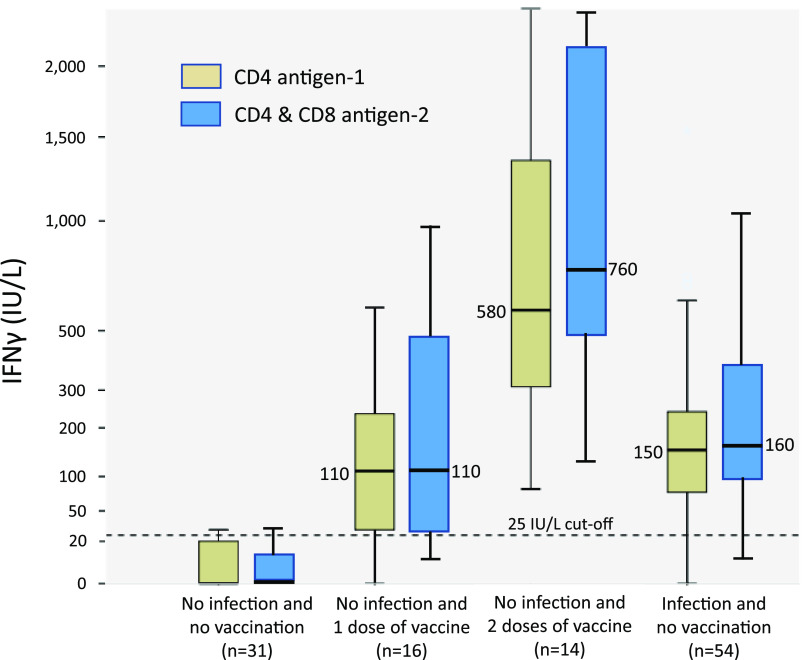
Table 1 shows the main immunological parameters of the different groups analyzed. We observed that history of exposure to SARS-CoV-2 antigens, by infection or vaccination, and having positive SARS-CoV-2 serology, either against S or N proteins, resulted in similar IFN-γ responses to Ag1 (median of 150 IU/L) or Ag2 (median of 170 IU/L). Administration of a second dose of vaccine significantly increased IFN-γ responses to Ag1 (median of 580 IU/L) or Ag2 (median of 760 IU/L).
TABLE 1.
Immunological assessment in groups of interest studied
| Group | No. of subjects | Mean (SD) lag from clinical event to test (days) | Positive IgG-spike (%); mean concn (SD) (logAU/mL)a | Positive IgG-nucleocapsid (%) | IFNγ (Ag1) concn median (IQR) (IU/L) | IFNγ (Ag2) concn median (IQR) (IU/L) |
|---|---|---|---|---|---|---|
| No infection and no vaccination | 31 | 2 (6.5); 0.23 (0.65) | 1 (3.2) | 0.0 (20.0) | 0.0 (10.0) | |
| No infection and 1 dose of vaccine | 16 | 19.1 (4.6) | 15 (93.8); 2.64 (0.80) | 0 (0) | 110 (210) | 110 (610) |
| No infection and 2 doses of vaccine | 14 | 51.4 (47.0) | 14 (100); 4.09 (0.51) | 0 (0) | 580 (1,195) | 760 (1,720) |
| Infection and no vaccination | 54 | 58.9 (2.8) | 54 (100); 2.67 (0.43) | 49 (90.7) | 150 (180) | 160 (290) |
| Exposure to SARS-CoV-2 antigens | 84 | 50.1 (24.3) | 83 (98.8); 2.90 (0.75) | 49 (58.3) | 150 (360) | 170 (530) |
| Negative SARS-CoV-2 serology (nucleocapsid or spike) | 30 | 0 (0) | 0 (0) | 0.0 (10.0) | 0.0 (10.0) | |
| Positive SARS-CoV-2 serology (nucleocapsid or spike) | 85 | 85 (100) | 50 (58.8) | 150 (365) | 170 (500) |
AU, arbitrary units; concn, concentration.
The correlation between IgG-S titers and IGRA against Ag1 and Ag2 was significant, particularly for the latter (Spearman’s rho, 0.56 and 0.83, respectively [P < 0.001]). Interestingly, IgG-S titers and IFN-γ responses run in parallel and were greater in subjects with two doses of vaccine than in subjects with partial vaccination or with recent COVID-19 (Table 1).
The 25 IU/L levels of IFN-γ were predictive of past COVID-19, SARS-CoV-2 vaccination, and positive SARS-CoV-2 serology with high sensitivity (80 to 93%) and better specificity (93 to 100%) after stimulation of T cells with any of the two sets of antigens used (Table 2). The concentration of IFN-γ after stimulation with Ag1 or Ag2 in the 4 groups studied is shown in Fig. 1. The IFN-γ 25 IU/L cutoff discriminated subjects with no exposure to SARS-CoV-2 from the other three groups (two of vaccinated and one of infected individuals).
TABLE 2.
Cutoff for IFN-γ release assay in subject after infection or vaccination immunity to SARS-CoV-2
| Group | IFN-γ to Ag1 ≥ 25 IU/L |
IFN-γ to Ag2 ≥ 25 IU/L |
||||
|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | AUROC (95% CI)a | Sensitivity (%) | Specificity (%) | AUROC (95% CI)a | |
| History of COVID-19 (yes vs no) | 89.4 | 93.3 | 0.96 (0.91–1.00) | 97.9 | 93.3 | 0.99 (0.97–1.00) |
| mRNA (BNT162b2) vaccination | ||||||
| yes vs no | 89.3 | 93.3 | 0.94 (0.86–1.00) | 89.3 | 93.3 | 0.96 (0.92–1.00) |
| complete vs no | 93.3 | 100 | 1.00 (1.00–1.00) | 93.3 | 100 | 1.00 (1.00–1.00) |
| partial vs no | 80.0 | 93.3 | 0.88 (0.75–1.00) | 80.0 | 93.3 | 0.93 (0.85–1.00) |
| Exposure to spike protein of SARS-CoV-2 (yes vs no) | 89.3 | 93.3 | 0.95 (0.91–0.99) | 94.7 | 93.3 | 0.98 (0.96–1.00) |
| Serology to SARS-CoV-2 (positive vs negative) | 88.3 | 96.4 | 0.95 (0.91–0.99) | 94.8 | 100 | 0.99 (0.97–1.00) |
P < 0.001. CI, confidence interval.
FIG 1.
Interferon gamma (IFN-γ) concentration in the different groups studied by IFN-γ release assay (IGRA).
DISCUSSION
According to our assessments, we propose the 25 IU/L cutoff for IGRA to determine T-cell response (CD4+ and CD8+) against SARS-CoV-2 S peptides, either after vaccination or infection. In the AUROC analysis, we selected the IFN-γ concentration with the best sensitivity and specificity always above 90%. Ensuring a reduced number of false positive results is key if this IGRA test is to be used to determine protection against SARS-CoV-2 infection or disease in, for example, patients with declining antibody levels as has been recently suggested (21). Two recent studies have found higher cutoffs (40 to 50 IU/L) for the same IGRA used in this study, in postinfection or postvaccination studies (22, 23), but showed a weaker correlation between humoral and cellular responses than in our study. Particularly, the time between infection and immune tests was much longer than in our study in one of the reports (22), making waning immunity or reinfections a possible source of variability. We feel that the homogeneity of our population, for which SARS-CoV-2 exposure was most likely restricted to a localized and recent outbreak or programmed vaccination, makes our cohort ideal for the purpose of validating the T-cell immune assay. Conversely, this same homogeneity in the selected sample may be viewed as a limitation of the study, as results may not be applicable to more diverse populations (i.e., males, older adults, children, reinfections, breakthrough infections, etc.). Further studies are needed to clarify this concern.
Our data show robust humoral and cellular responses after SARS-CoV-2 vaccination, already present after first dose and much improved after a second dose. These responses were better after vaccination than after natural infection, although these comparisons are not valid due to differences between these groups, as a greater age and a longer lag from SARS-CoV-2 exposure was observed among subjects infected than among subjects vaccinated. In fact, for the 14 patients with 2 doses of vaccine, mean IFN-γ concentration was thrice greater for those with the second dose given in the previous month than with those vaccinated greater than 1 month previously, suggesting a reduction of T-cell activity with time after exposure. A follow-up study is needed to determine immune response decay in infected versus vaccinated populations.
Given that Ag1 stimulates production of IFN-γ by CD4+ T cells and Ag2 stimulates both CD4+ and CD8+ T cells, there may be the possibility to discriminate cellular responses between these subpopulations. Still, it seems that production of IFN-γ by CD4+ T cells is more intense than that from CD8+ T cells, which may further simplify the IGRA to using just one single antigen (24).
It is yet to be established whether reduction in antibody titers against SARS-CoV-2 and/or selection of viral variants has correlates with the cases of reinfection or postvaccine infection (25). Previous studies suggest that T-cell immunity may be more durable, so that may help determine past COVID-19 in subjects with negative serology (26). Determination of T-lymphocyte response against SARS-CoV-2 may be studied as an additional marker of protection from COVID-19; as such, in immunodepressed subjects in whom antibody and T-cell responses are less robust (27), additional doses of vaccine are already being proposed. Finding that recent completion of vaccination provides greater levels of humoral and cellular immunity than recent COVID-19 itself is very encouraging and supports this strategy.
Some important limitations in this study need to be acknowledged. Firstly, the diagnosis of cases of SARS-CoV-2 infection was done according to medical protocols that call for an initial rapid Ag test and a confirmatory PCR if Ag positive. Although all positive Ag tests resulted in a positive PCR, we cannot exclude that, given the lower sensitivity of Ag tests, false negative results have missed cases of SARS-CoV-2 infection. Secondly, the validation of IGRA was done against the history of vaccination or infection and against humoral responses. We feel that definitive validation of IGRA before accepting its use on clinical grounds needs to be done against other tests that specifically assess cellular immunity (i.e., flow cytometry [28], major histocompatibility complex [MHC] tetramers [29], or activation-induced marker assay [21]). Also, internal control studies in pairs of samples from recruited patients were not done, which could have increased the information about the reliability of the technique.
Given the characteristics of our cohort, the IGRA used was only validated against the B.1.1.7 SARS-CoV-2 variant, which was at the time of the outbreak the most prevalent in Spain (86% reported frequency from 3 to 17 May 2021 [30]). Although T-cell responses have been shown to be less affected by variants of concern than humoral immune responses (31, 32), the IGRA probably needs validation in patients with known infection with other specific viral variants before being widely used.
As shown in a recent paper (33), up to 44% of subjects with negative SARS-CoV-2 serology may show a strong virus-specific T-cell response as assessed by IGRA. No such cases were detected in our cohort, as all 30 subjects with negative serology had negative IGRA, which indicates that false positive IGRA may be related with waning humoral immunity after old infection or because of cross-reaction with endemic coronavirus—both factors hypothetically affecting less our recently infected/vaccinated and cloistered cohort.
Important issues for further research are correlating clinical outcomes with the following: the risk of breakthrough infections; reinfection; severe COVID-19 according to IGRA results; or associated factors that modulate T-cell responses including age, severity of infection, or underlaying medical conditions. The closed setting where this study was done still offers a unique opportunity to shed light into these questions with the follow-up studies that are under way.
There is accumulating evidence that indicates that IGRA for assessing T-cell immunity may be a reliable test to be included in COVID-19 monitoring protocols, particularly after vaccination (23, 34–36). The clinical utility of IGRA is both because of the simplicity of the test compared with other alternatives (i.e., flow cytometry, MHC tetramers, or activation induced marker assay) and also the familiarity of most laboratories with this technique, which is already in use for the diagnosis of latent tuberculosis infection. In the context of waning humoral immunity, determination of T-cell responses may help identify subjects that after vaccination or infection remain protected against severe infection, while others may need primary vaccination or booster doses (37). This aspect may be of particular interest for patients with different types of B-cell defects—in whom humoral immunity is more affected than cellular immunity (38–40). Of note, in transplanted patients, both humoral and cellular immunity are similarly blunted (41). As T-cell responses can be detected early during COVID-19, earlier than the detection of antibodies, IGRA may also be considered for diagnostic purposes where molecular tests are not conclusive (42, 43). Other studies have shown that lower CD4+ and CD8+ T-cell responses are associated with longer duration of viral shedding, more severe cases, and increased mortality from COVID-19, so that prognostic information may also be provided (44) and set the indication of neutralizing antibodies (45).
In conclusion, recent vaccination is associated with robust humoral and cellular response to SARS-CoV-2 that are already present after one dose of vaccine and improved after a second dose. The levels of IFN-γ by IGRA may be a valid method to determine cellular responses after SARS-CoV-2 exposure. Production of ≥25 IU/L of IFN-γ by IGRA is strongly associated with evidence of T-cell activity after exposure to SARS-CoV-2 antigens.
ACKNOWLEDGMENTS
We thank all participants for their willingness to be a part of this study, which has been done, according to their words, for the interest of science and the benefit of other patients. We are also very grateful to Esperanza García-Caldevilla and Laura Barreiro for their aid in the logistics and to David Peck for the editing of the manuscript.
The study was funded by the Health Council, Community of Madrid. The costs of publication of this study were funded by Diasorin Iberia S.A.
We declare no conflict of interest.
Contributor Information
Pablo Barreiro, Email: pablo.barreiro@salud.madrid.org.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Cohen J, Burbelo P. 2021. Reinfection with SARS-CoV-2: implications for vaccines. Clin Infect Dis 73:e4223–e4228. 10.1093/cid/ciaa1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schäfer A, Muecksch F, Lorenzi JCC, Leist SR, Cipolla M, Bournazos S, Schmidt F, Maison RM, Gazumyan A, Martinez DR, Baric RS, Robbiani DF, Hatziioannou T, Ravetch JV, Bieniasz PD, Bowen RA, Nussenzweig MC, Sheahan TP. 2021. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J Exp Med 218:e20201993. 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Telenti A, Arvin A, Corey L, Corti D, Diamond MS, García-Sastre A, Garry RF, Holmes EC, Pang PS, Virgin HW. 2021. After the pandemic: perspectives on the future trajectory of COVID-19. Nature 596:495–504. 10.1038/s41586-021-03792-w. [DOI] [PubMed] [Google Scholar]
- 4.Hellerstein M. 2020. What are the roles of antibodies versus a durable, high quality T-cell response in protective immunity against SARS-CoV-2? Vaccine X 6:100076. 10.1016/j.jvacx.2020.100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, Hauser BM, Caradonna TM, Clayton KL, Nitido AD, Murali MR, Alter G, Charles RC, Dighe A, Branda JA, Lennerz JK, Lingwood D, Schmidt AG, Iafrate AJ, Balazs AB. 2021. COVID-19-neutralizing antibodies predict disease severity and survival. Cell 184:476–488. 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Li Z, Nampanya F, Patel S, Pessaint L, Van Ry A, Blade K, Yalley-Ogunro J, Cabus M, Brown R, Cook A, Teow E, Andersen H, Lewis MG, Lauffenburger DA, Alter G, Barouch DH. 2021. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590:630–634. 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephenson KE, Le Gars M, Sadoff J, de Groot AM, Heerwegh D, Truyers C, Atyeo C, Loos C, Chandrashekar A, McMahan K, Tostanoski LH, Yu J, Gebre MS, Jacob-Dolan C, Li Z, Patel S, Peter L, Liu J, Borducchi EN, Nkolola JP, Souza M, Tan CS, Zash R, Julg B, Nathavitharana RR, Shapiro RL, Azim AA, Alonso CD, Jaegle K, Ansel JL, Kanjilal DG, Guiney CJ, Bradshaw C, Tyler A, Makoni T, Yanosick KE, Seaman MS, Lauffenburger DA, Alter G, Struyf F, Douoguih M, Van Hoof J, Schuitemaker H, Barouch DH. 2021. Immunogenicity of the Ad26.COV2.S vaccine for COVID-19. JAMA 325:1535–1544. 10.1001/jama.2021.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens DS, McElrath MJ. 2020. COVID-19 and the path to immunity. JAMA 324:1279–1281. 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauer K, Harris T. 2020. An effective COVID-19 vaccine needs to engage T-cells. Front Immunol 11:581807. 10.3389/fimmu.2020.581807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aiello A, Najafi Fard S, Petruccioli E, Petrone L, Vanini V, Farroni C, Cuzzi G, Navarra A, Gualano G, Mosti S, Pierelli L, Nicastri E, Goletti D. 2021. Spike is the most recognized antigen in the whole-blood platform in both acute and convalescent COVID-19 patients. Int J Infect Dis 106:338–347. 10.1016/j.ijid.2021.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbott. 2022. PANBIO COVID-19 Ag rapid test device. Abbott, Abbott Park, IL. https://www.globalpointofcare.abbott/es/product-details/panbio-covid-19-ag-antigen-test.html. [Google Scholar]
- 12.Garg A, Ghoshal U, Patel SS, Singh DV, Arya AK, Vasanth S, Pandey A, Srivastava N. 2021. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol 93:2281–2286. 10.1002/jmv.26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maine GN, Lao KM, Krishnan SM, Afolayan-Oloye O, Fatemi S, Kumar S, VanHorn L, Hurand A, Sykes E, Sun Q. 2020. Longitudinal characterization of the IgM and IgG humoral response in symptomatic COVID-19 patients using the Abbott Architect. J Clin Virol 133:104663. 10.1016/j.jcv.2020.104663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung K, Shin S, Nam M, Hong YJ, Roh EY, Park KU, Song EY. 2021. Performance evaluation of three automated quantitative immunoassays and their correlation with a surrogate virus neutralization test in coronavirus disease 19 patients and pre-pandemic controls. J Clin Lab Anal 35:e23921. 10.1002/jcla.23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiaSorin. 2019. LIAISON QuantiFERON-TB Gold Plus (REF. 311010). DiaSorin, Stillwater, MN. https://www.accessdata.fda.gov/cdrh_docs/pdf18/P180047D.pdf. [Google Scholar]
- 16.Jaganathan S, Stieber F, Rao SN, Nikolayevskyy V, Manissero D, Allen N, Boyle J, Howard J. 2021. Preliminary evaluation of QuantiFERON SARS-CoV-2 and QIAreach anti-SARS-CoV-2 total test in recently vaccinated individuals. Infect Dis Ther 10:2765–2776. 10.1007/s40121-021-00521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiagen. 2021. QuantiFERON SARS-CoV-2 extended set blood collection tubes instructions for use (handbook). Qiagen, Hilden, Germany. https://www.qiagen.com/us/resources/resourcedetail?id=60729925-a7b2-4cf3-8d28-52e4106af16b&lang=en. [Google Scholar]
- 18.Qiagen. 2021. QuantiFERON control set blood collection tubes instructions for use (handbook). Qiagen, Hilden, Germany. https://www.qiagen.com/us/resources/resourcedetail?id=80996d9a-d50c-40b5-940f-b62959d54ab4&lang=en. [Google Scholar]
- 19.Qiagen. 2021. QuantiFERON SARSCoV-2 starter set blood collection tubes instructions for use (handbook). Qiagen, Hilden, Germany. https://www.qiagen.com/nl/resources/resourcedetail?id=2a36a234-bfde-45f4-ae08-7c28ef95df7b&lang=en. [Google Scholar]
- 20.Candel FJ, Barreiro P, San Román J, Abanades JC, Barba R, Barberán J, Bibiano C, Canora J, Cantón R, Calvo C, Carretero M, Cava F, Delgado R, García-Rodríguez J, González Del Castillo J, González de Villaumbrosia C, Hernández M, Losa JE, Martínez-Peromingo FJ, Molero JM, Muñoz P, Onecha E, Onoda M, Rodríguez J, Sánchez-Celaya M, Serra JA, Zapatero A. 2020. Recommendations for use of antigenic tests in the diagnosis of acute SARS-CoV-2 infection in the second pandemic wave: attitude in different clinical settings. Rev Esp Quimioter 33:466–484. 10.37201/req/120.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, Lundgreen KA, Reynaldi A, Khoury DS, Pattekar A, Gouma S, Kuri-Cervantes L, Hicks P, Dysinger S, Hicks A, Sharma H, Herring S, Korte S, Baxter AE, Oldridge DA, Giles JR, Weirick ME, McAllister CM, Awofolaju M, Tanenbaum N, Drapeau EM, Dougherty J, Long S, D’Andrea K, Hamilton JT, McLaughlin M, Williams JC, Adamski S, Kuthuru O, Frank I, Betts MR, Vella LA, Grifoni A, Weiskopf D, Sette A, Hensley SE, Davenport MP, Bates P, Luning Prak ET, Greenplate AR, Wherry EJ, The UPenn COVID Processing Unit. 2021. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 374:abm0829. 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand I, Gilberg L, Bruger J, Garí M, Wieser A, Eser TM, Frese J, Ahmed MIM, Rubio-Acero R, Guggenbuehl Noller JM, Castelletti N, Diekmannshemke J, Thiesbrummel S, Huynh D, Winter S, Kroidl I, Fuchs C, Hoelscher M, Roider J, Kobold S, Pritsch M, Geldmacher C. 2021. Broad T cell targeting of structural proteins after SARS-CoV-2 infection: high throughput assessment of T cell reactivity using an automated interferon gamma release assay. Front Immunol 12:688436. 10.3389/fimmu.2021.688436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-Gallo M, Esperalba-Esquerra J, Pujol-Borrell R, Sandá V, Arrese-Muñoz I, Fernández-Naval C, Antón-Pagarolas A, Cardona V, Labrador-Horrillo M, Pumarola-Suñé T, Hernandéz-González M. 2021. Commercialized kit to assess T-cell responses against SARS-CoV-2 S peptides. A pilot study in health care workers. medRxiv 10.1101/2021.03.31.21254472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murugesan K, Jagannathan P, Pham TD, Pandey S, Bonilla HF, Jacobson K, Parsonnet J, Andrews JR, Weiskopf D, Sette A, Pinsky BA, Singh U, Banaei N. 2020. Interferon-gamma release assay for accurate detection of SARS-CoV-2 T cell response. Clin Infect Dis 73:e3130–e3132. 10.1093/cid/ciaa1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown CM, Vostok J, Johnson H, Burns M, Gharpure R, Sami S, Sabo RT, Hall N, Foreman A, Schubert PL, Gallagher GR, Fink T, Madoff LC, Gabriel SB, MacInnis B, Park DJ, Siddle KJ, Harik V, Arvidson D, Brock-Fisher T, Dunn M, Kearns A, Laney AS. 2021. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings — Barnstable County, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep 70:1059–1062. 10.15585/mmwr.mm7031e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallais F, Velay A, Nazon C, Wendling M-J, Partisani M, Sibilia J, Candon S, Fafi-Kremer S. 2021. Intrafamilial exposure to SARS-CoV-2 associated with cellular immune response without seroconversion, France. Emerg Infect Dis 27:113–121. 10.3201/eid2701.203611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benotmane I, Gautier G, Perrin P, Olagne J, Cognard N, Fafi-Kremer S, Caillard S. 2021. Antibody response after a third dose of the mRNA-1273 SARS-CoV-2 vaccine in kidney transplant recipients with minimal serologic response to 2 doses. JAMA 326:1063–1065. 10.1001/jama.2021.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI-C, Wang L-F, Ooi EE, Kalimuddin S, Tambyah PA, Low JG-H, Tan Y-J, Bertoletti A. 2020. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584:457–462. 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 29.Quirós-Fernández I, Poorebrahim M, Fakhr E, Cid-Arregui A. 2021. Immunogenic T cell epitopes of SARS-CoV-2 are recognized by circulating memory and naïve CD8 T cells of unexposed individuals. EBioMedicine 72:103610. 10.1016/j.ebiom.2021.103610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodcroft E. 2021. Overview of variants in countries. CoVariants. https://covariants.org/per-country. [Google Scholar]
- 31.Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, Goodwin B, Rubiro P, Sutherland A, Wang E, Frazier A, Ramirez SI, Rawlings SA, Smith DM, da Silva Antunes R, Peters B, Scheuermann RH, Weiskopf D, Crotty S, Grifoni A, Sette A. 2021. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2:100355. 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geers D, Shamier M, Bogers S, den Hartog G, Gommers L, Nieuwkoop NN, Schmitz KS, Rijsbergen LC, van Osch JAT, Dijkhuizen E, Smits G, Comvalius A, van Mourik D, Caniels TG, van Gils MJ, Sanders RW, Oude Munnink BB, Molenkamp R, de Jager HJ, Haagmans BL, de Swart RL, Koopmans MPG, van Binnendijk RS, de Vries RD, Geurts van Kessel CH. 2021. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccine recipients. Sci Immunol 6:eabj1750. 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Echeverría G, Guevara Á, Coloma J, Ruiz AM, Vasquez MM, Tejera E, de Waard JH. 2021. Pre-existing T-cell immunity to SARS-CoV-2 in unexposed healthy controls in Ecuador, as detected with a COVID-19 interferon-gamma release assay. Int J Infect Dis 105:21–25. 10.1016/j.ijid.2021.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borobia AM, Carcas AJ, Pérez-Olmeda M, Castaño L, Bertran MJ, García-Pérez J, Campins M, Portolés A, González-Pérez M, García Morales MT, Arana-Arri E, Aldea M, Díez-Fuertes F, Fuentes I, Ascaso A, Lora D, Imaz-Ayo N, Barón-Mira LE, Agustí A, Pérez-Ingidua C, Gómez de la Cámara A, Arribas JR, Ochando J, Alcamí J, Belda-Iniesta C, Frías J, Martínez de Soto L, Rodríguez Mariblanca A, Díaz García L, Ramírez García E, Seco Meseguer E, Stewart Balbás SM, Marín Candón A, García García I, Urroz Elizalde M, Monserrat Villatoro J, de la Rosa P, Sanz García M, López Crespo C, Mauleón Martínez V, de Madariaga Castell R, Vitón Vara L, García Rodríguez J, Buño A, López Granados E, Cámara C, Rey Cuevas E, Ayllon García P, Jiménez González M, Hernández Rubio V, et al. 2021. Immunogenicity and reactogenicity of BNT162b2 booster in ChAdOx1-S-primed participants (CombiVacS): a multicentre, open-label, randomised, controlled, phase 2 trial. Lancet 398:121–130. 10.1016/S0140-6736(21)01420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Praet JT, Vandecasteele S, De Roo A, De Vriese AS, Reynders M. 2021. Humoral and cellular immunogenicity of the BNT162b2 messenger RNA coronavirus disease 2019 vaccine in nursing home residents. Clin Infect Dis 73:2145–2147. 10.1093/cid/ciab300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen RM, Tornby DS, Bistrup C, Johansen IS, Andersen TE, Justesen US. 2021. Negative SARS-CoV-2 antibodies, T-cell response and virus neutralization following full vaccination in a renal transplant recipient: a call for vigilance. Clin Microbiol Infect 27:1371–1373. 10.1016/j.cmi.2021.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noh JY, Jeong HW, Kim JH, Shin E-C. 2021. T cell-oriented strategies for controlling the COVID-19 pandemic. Nat Rev Immunol 21:687–688. 10.1038/s41577-021-00625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Abramo A, Vita S, Maffongelli G, Mariano A, Agrati C, Castilletti C, Goletti D, Ippolito G, Nicastri E, Spallanzani COVID-19 Case Investigation Team. 2021. Prolonged and severe SARS-CoV-2 infection in patients under B-cell-depleting drug successfully treated: a tailored approach. Int J Infect Dis 107:247–250. 10.1016/j.ijid.2021.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soresina A, Moratto D, Chiarini M, Paolillo C, Baresi G, Focà E, Bezzi M, Baronio B, Giacomelli M, Badolato R. 2020. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol 31:565–569. 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinti I, Lougaris V, Milito C, Cinetto F, Pecoraro A, Mezzaroma I, Mastroianni CM, Turriziani O, Bondioni MP, Filippini M, Soresina A, Spadaro G, Agostini C, Carsetti R, Plebani A. 2020. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol 146:211–213. 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crespo M, Barrilado‐Jackson A, Padilla E, Eguía J, Echeverria‐Esnal D, Cao H, Faura A, Folgueiras M, Solà‐Porta E, Pascual S, Barbosa F, Hurtado S, Ribera L, Río‐No L, Pérez‐Sáez MJ, Redondo‐Pachón D, Pascual J, Arias C, Buxeda A, Burballa C, Bach A, Pedreira G, Arenas MD, Collado S, Fernández M, Barbero E, Rodríguez E, Sans L, Márquez E, Vázquez S, Oliveras A, Galcerán B, Nuñez S, Ribas A, Iriarte M, Farrera J, Savall O, Causadias R, Muñoz J, Villegas E, Canal M, Grau S, Montero M, Villar J, Días P, for the Mariscovid Research Group. 22 September 2021. Negative immune responses to two-dose mRNA COVID-19 vaccines in renal allograft recipients assessed with simple antibody and interferon gamma release assay cellular monitoring. Am J Transplant 10.1111/ajt.16854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goletti D, Petrone L, Manissero D, Bertoletti A, Rao S, Ndunda N, Sette A, Nikolayevskyy V. 2021. The potential clinical utility of measuring severe acute respiratory syndrome coronavirus 2-specific T-cell responses. Clin Microbiol Infect 27:1784–1789. 10.1016/j.cmi.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrone L, Petruccioli E, Vanini V, Cuzzi G, Najafi Fard S, Alonzi T, Castilletti C, Palmieri F, Gualano G, Vittozzi P, Nicastri E, Lepore L, Antinori A, Vergori A, Caccamo N, Cantini F, Girardi E, Ippolito G, Grifoni A, Goletti D. 2021. A whole blood test to measure SARS-CoV-2-specific response in COVID-19 patients. Clin Microbiol Infect 27:286. 10.1016/j.cmi.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shrotri M, van Schalkwyk MCI, Post N, Eddy D, Huntley C, Leeman D, Rigby S, Williams SV, Bermingham WH, Kellam P, Maher J, Shields AM, Amirthalingam G, Peacock SJ, Ismail SA. 2021. T cell response to SARS-CoV-2 infection in humans: a systematic review. PLoS One 16:e0245532. 10.1371/journal.pone.0245532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta A, González-Rojas Y, Juárez E, Crespo Casal M, Moya J, Falci DR, Sarkis E, Solis J, Zheng H, Scott N, Cathcart AL, Hebner CM, Sager J, Mogalian E, Tipple C, Peppercorn A, Alexander E, Pang PS, Free A, Brinson C, Aldinger M, Shapiro AE, for the COMET-ICE Investigators. 2021. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N Engl J Med 385:1941–1950. 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]