Abstract
Transposable elements (TEs) are genomic parasites that can propagate throughout host genomes. Mammalian genomes are typically dominated by LINE retrotransposons and their associated SINEs, and germline mobilization is a challenge to genome integrity. There are defenses against TE proliferation and the PIWI/piRNA defense is among the most well understood. However, the PIWI/piRNA system has been investigated largely in animals with actively mobilizing TEs and it is unclear how the PIWI/piRNA system functions in the absence of mobilizing TEs. The 13-lined ground squirrel provides the opportunity to examine PIWI/piRNA and TE dynamics within the context of minimal, and possibly nonexistent, TE accumulation. To do so, we compared the PIWI/piRNA dynamics in squirrels to observations from the rabbit and mouse. Despite a lack of young insertions in squirrels, TEs were still actively transcribed at higher levels compared to mouse and rabbit. All three Piwi genes were not expressed, prior to P8 in squirrel testis, and there was little TE expression change with the onset of Piwi expression. We also demonstrated there was not a major expression change in the young squirrel LINE families in the transition from juvenile to adult testis in contrast to young mouse and rabbit LINE families. These observations lead us to conclude that PIWI suppression, was weaker for squirrel LINEs and SINEs and did not strongly reduce their transcription. We speculate that, although the PIWI/piRNA system is adaptable to novel TE threats, transcripts from TEs that are no longer threatening receive less attention from PIWI proteins.
Keywords: Argonautes, RNAi, nonmodel mammals, ping-pong cycle, retrotransposons
INTRODUCTION
Transposable elements (TEs) are genomic parasites that propagate by inserting copies of themselves into the genomes of their hosts. They account for up to 70% of mammalian genome content (de Koning et al. 2011). Because of their ability to mobilize, TEs are powerful mutagens, as novel TE insertions can disrupt exons, regulatory elements, and splice junctions, and facilitate nonhomologous recombination. As a result, TE insertions have been linked to genomic deletions, duplications, inversions, translocations, and chromosome breaks in a variety of genomes (Cheng et al. 2005; Franke et al. 2017; Platt et al. 2018). While some TE insertions have proven adaptive, TEs are generally considered a serious challenge to genome integrity.
Eukaryotic genomes have evolved mechanisms to restrict TE mobilization, especially in the germline. PIWI proteins and PIWI-interacting RNAs (piRNAs) have emerged as key components in protecting the genome against the proliferation of TEs, and probably evolved in response to the challenge presented by them (Aravin et al. 2007a, 2008; Brennecke et al. 2007; Malone and Hannon 2009; Siomi et al. 2011). piRNAs and PIWI proteins assemble into RNA-induced silencing complexes, which go on to neutralize TE-like targets by transcript cleavage or chromatin methylation (Aravin et al. 2007b; Carmell et al. 2007; Houwing et al. 2007; Molaro et al. 2014).
Piwi paralog counts vary among animals, Caenorhabditis elegans encodes two Piwis, Drosophila encode three Piwis, and mammals encode up to four (Reddien et al. 2005; Kerner et al. 2011; Lewis et al. 2016). Among vertebrates, the mouse (Mus musculus) is the most studied system (for review, see Ernst et al. 2017). Mouse piRNAs have been categorized into two major sets, pre-pachytene and pachytene, according to when their expression begins (Aravin et al. 2006; Girard et al. 2006). Pre-pachytene piRNAs are first expressed in the early stages of spermatogenesis, are enriched for TE-like sequences, and preferentially associate with the PIWIL2 and PIWIL4 proteins, and are most responsible for TE transcriptional silencing. In contrast, pachytene piRNAs begin their expression during the pachytene stage of meiosis I, are largely derived from intergenic regions, preferentially associate with PIWIL1, and current evidence suggests they play a role similar to microRNAs, silencing messenger RNAs in spermiogenesis (Li et al. 2013; Gou et al. 2014; Wu et al. 2020).
The mouse genome encodes three Piwi paralogs that are differentially expressed during development and associate preferentially with piRNAs of different sizes. PIWIL2 (MILI) preferentially binds piRNAs that are 26–27 nt long, is first expressed at embryonic day 12.5 (E12.5) in developing testes and is linked to the post-transcriptional silencing of TEs. PIWIL4 (MIWI2) preferentially binds to piRNAs that are ∼28 nt long, is expressed between E15.5 and postnatal day 3 (P3), and this period of time is linked to the de novo establishment of methylation marks in gonocytes (Carmell et al. 2007; Aravin et al. 2008; Molaro et al. 2014; Zoch et al. 2020). PIWIL2 is the primary driver of a feed-forward loop known as the ping-pong cycle that serves to reduce the abundance of TE transcripts through piRNA-guided cleavage. While PIWIL4 is active, PIWIL2 can also load secondary piRNAs onto PIWIL4, which then enters the nucleus to initiate methylation at TE loci (Aravin et al. 2007b, 2008; Kuramochi-Miyagawa et al. 2008; De Fazio et al. 2011; Manakov et al. 2015; Zoch et al. 2020). Defects in these paralogs lead to elevated TE activity and problems in the male germline (Carmell et al. 2007; Houwing et al. 2007; O'Donnell and Boeke 2007). PIWIL4 is also functional in undifferentiated spermatogonia in adult testis, although the link to TEs in these cell types is under investigation (Carrieri et al. 2017; Vasiliauskaite˙ et al. 2018). Finally, PIWIL1 (MIWI) becomes active at P14 during the pachytene stage of prophase I and remains active. This paralog preferentially binds to piRNAs ∼30 nt long derived from lncRNAs transcribed from intergenic space and is mostly linked to clearing mRNAs at the end of spermiogenesis (Li et al. 2013; Gou et al. 2014; Wu et al. 2020).
TE expression and accumulation rates vary among different mammals, both in terms of the type of TEs that are active and in the level of challenge presented by them (Pasquesi et al. 2020). However, piRNAs have only been described in a handful of vertebrates and a comparative framework is generally lacking (Lau et al. 2006; Liu et al. 2012; Chirn et al. 2015; Toombs et al. 2016; Vandewege et al. 2016; Sun et al. 2017). Further, whether the developmental changes in PIWI and piRNA expression are conserved, or how PIWIs/piRNAs repertoires respond to changing TE landscapes has not been addressed. In this regard, there is evidence that the PIWI/piRNA system is quickly adaptable to novel TE threats (Mourier 2011; Gainetdinov et al. 2017; Sun et al. 2017; Zhang et al. 2020), but it is not clear what happens when established TEs are no longer mobilizing or threatening the genome.
Comparisons among the 13-lined ground squirrel (Ictidomys tridecemlineatus), rabbit (Oryctolagus cuniculus), and mouse (Mus musculus) offer a natural experiment to test these questions. Genomic surveys indicate that the last LINE and SINE insertions in the 13-lined ground squirrel genome occurred between 4 and 5 million years ago and no retrotransposition-competent LINE loci have been found in its genome so far (Platt and Ray 2012). Thus, the squirrel offers a unique opportunity to study piRNA biogenesis, piRNA diversity, and the ping-pong cycle in a system where LINE and SINE mobilization appears to have ceased. In contrast, mouse and rabbit have typical mammalian genomic TE landscapes dominated by currently active retrotransposons, LINE-1 (L1), and associated SINEs, but have different patterns of TE expression and accumulation from one another. In the current study, we took advantage of the genomic resources available for these three species to (1) explore the relationship between TE abundance at the genome and transcript level in each species, (2) characterize developmental changes in the expression of PIWI paralogs and piRNAs, (3) measure the intensity of the ping-pong cycle, and (4) quantify TE expression changes in response to changing Piwi expression. We found that TE expression was much higher in the squirrel, despite the observed absence of recent TE insertions, but that the piRNA/PIWI response was reduced. Our results suggest that TEs that lack the capacity to generate de novo insertions elicit weak responses from the piRNA/PIWI pathway.
RESULTS
TE accumulation
We first characterized patterns of TE accumulation, diversity, and abundance in the mouse, rabbit, and squirrel genomes and found clear differences among them. We assessed the abundance and diversity of TEs present in each genome by measuring overall insertion numbers, young insertion numbers (<5% diverged from consensus), and median family distances. As in most mammals, LINEs, SINEs, and LTR retrotransposons account for the vast majority of TE insertions in these three species. Close to half of the TE-derived portion of the genome corresponds to LINEs, with a low number of insertions from DNA transposons in all three genomes (Fig. 1A). In contrast, there were differences in the relative contribution of SINEs and LTRs among them: SINEs accounted for close to half of the TE-derived portion of the genome in the rabbit, and LTR retrotransposons contributed a larger fraction in squirrel and mouse relative to the rabbit. These three species also differed in the historical patterns of TE accumulation. To reconstruct TE deposition history, we classified insertions by subfamilies and estimated the Kimura 2-parameter distance between consensus sequences of each TE family and individual insertions. According to the master gene model, TE insertions are driven by one master mobilizing element (Deininger et al. 1992; Cordaux et al. 2004), and younger insertions would more closely resemble the corresponding master mobilizing element than older insertions. Thus, distances to the corresponding consensus can be used to estimate the relative age of an insertion. In the case of the squirrel, these analyses confirmed a low number of young TE insertions, as previously reported, which would indicate a lack of mobilizing elements (Fig. 1B; Platt and Ray 2012).
FIGURE 1.
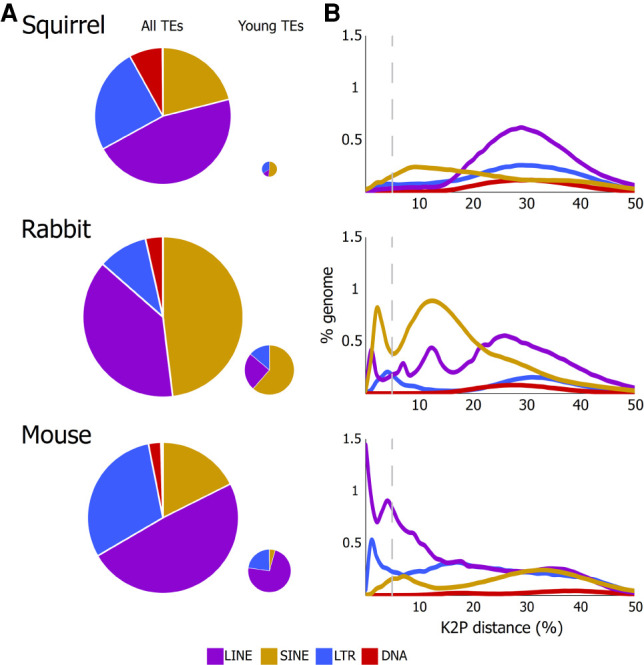
(A) Relative contribution of TEs to the corresponding genome. The small pie charts document the contribution of the more recent TE insertions, with the size of the pie chart proportional to the relative contribution of recent insertions to each genome. (B) Historical patterns of accumulation of the major TE categories inferred by calculating the Kimura 2-parameter distance between individual insertions and the corresponding consensus. Relatively young (<5% diverged) insertions to the left of the vertical line with lower genetic distances were deposited more recently. The insertions to the left of the dashed vertical lines were considered for the small pie charts.
PIWI expression and piRNAs
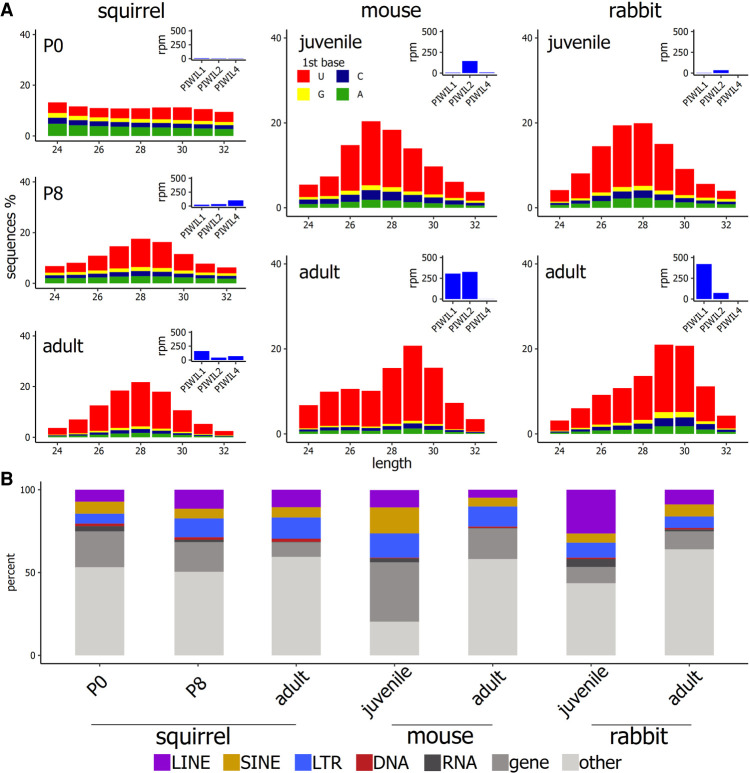
We then assessed changes in the expression of Piwi genes and associated piRNAs within developing testes in squirrel and rabbit and compared them to publicly available data from mouse. We conducted simultaneous RNA-seq and small RNA-seq experiments on a neonate (P0—0 d postbirth), postnatal juveniles (P2, P8, P10, P13), and one adult in the case of the squirrel, whereas in rabbits we sampled testes from a P21 juvenile and one adult (P > 90). As expected, we observed changes in the expression of the Piwi paralogs that corresponded with changes in piRNA repertoires. In squirrel, we did not detect the expression of any Piwi paralog in the P0 and P2 samples, and the small RNA libraries exhibited no clear hallmarks of PIWI processing that include a strong length bias between 24 and 32 bases and a U bias in the first 5′ base (Fig. 2A; Supplemental Fig. 1A). After P8, all three paralogs were expressed and the hallmarks of piRNA biogenesis became apparent (Fig 2A; Supplemental Fig. 1A). The robust presence of squirrel Piwil4 in adult testis was a little unique because Piwil4 expression was not measurably detected in our rabbit or mouse samples (Fig. 2A), but it is has been shown that Piwil4 is expressed in undifferentiated spermatogonia in mature testis (Carrieri et al. 2017; Vasiliauskaite˙ et al. 2018). In all three species, piRNAs became more abundant and diverse with the onset of Piwil1 expression in adult testis (Supplemental Table 1), a previously demonstrated phenomenon (Li et al. 2013).
FIGURE 2.
(A) Distribution length among piRNAs between 24 and 32 nt. Colors in the stacked bar plot represent the frequency of the first nucleotide. Corresponding expression of the PIWI proteins is measured as reads per million in each sample. (B) The proportion of piRNAs that mapped to discrete regions in the genome.
TEs are known to be a more common source of piRNAs in juvenile testis where pre-pachytene piRNAs are more common, whereas piRNAs are mostly derived from intergenic space in adult testis when PIWIL1 begins to process piRNAs from lncRNA loci (Li et al. 2013). This was observed in both the rabbit and mouse, but the change between juvenile and adult squirrel was not as extreme (Fig. 2B). In squirrel, TEs became a more common source of the small RNAs once Piwis were expressed, but TE-derived piRNAs were almost equally frequent in juvenile and adult testis (Fig. 2B).
TE expression
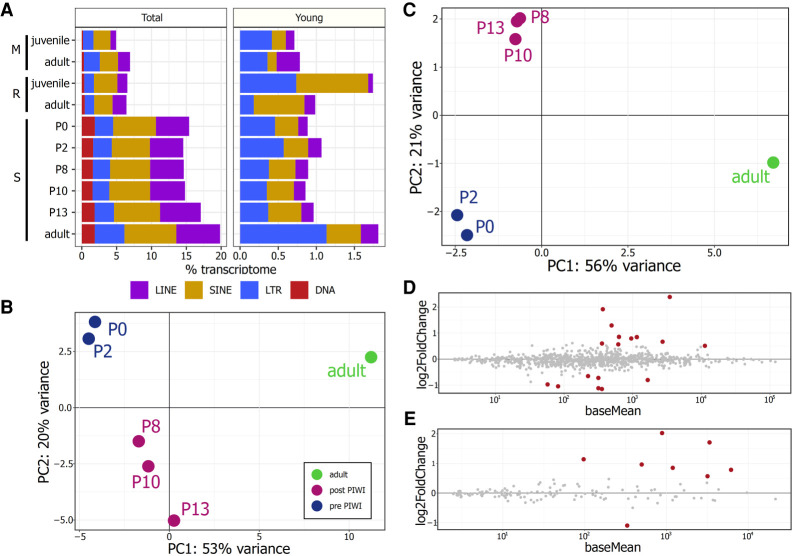
We mapped RNA-seq reads to corresponding genomes and measured TE family expression in units of reads per million (RPM). This metric can be expressed as a percent of the total RNA pool, which allows for comparisons across species. We found that squirrel TEs made up more than 15% of the transcriptome, almost doubling the relative amount of TE-derived RNA obtained from the corresponding mouse and rabbit samples (Fig. 3A). When we examined changes in TE expression over time, we noticed some general changes between the juvenile and adult testes of mouse and rabbit, but found that changes in TE expression during the development of squirrel testis were less dramatic. For example, LINE expression was lower in the testes of juvenile mouse and rabbit, compared to adult, but was roughly equivalent in all stages of squirrel samples examined (Supplemental Fig. 2A). Interestingly, the percent of the transcriptome made up of young TEs was similar across all species and developmental stages, with the exceptions of the juvenile rabbit and adult squirrel (Fig. 3A). These exceptions were largely caused by the high expression of a few LTR and SINE families (Supplemental Fig. 2B). The mouse genome contains numerous young LINE families that are actively mobilizing (Goodier et al. 2001; Sookdeo et al. 2013) and most of the young TEs from these families were expressed. In contrast, most young insertions in the rabbit and squirrel were generated by just two families that accounted for most of the RNA derived from young LINEs (Supplemental Fig. 2B).
FIGURE 3.
(A) PCA of squirrel testis derived from the expression of all TE insertions (B) and young insertions (C). Differentially expressed squirrel TEs between the pre-PIWI expressed samples (P0, P2) and post-PIWI expressed juvenile samples (P8, P10, P13) calculated from all TE insertions (D) and young insertions (E). Red circles reflect differentially expressed TEs with an adjusted P-value <0.1. Positive log2-fold change values reflect higher expression in pre-PIWI samples.
Since multiple life stages from the squirrel were sampled, we examined variation in TE expression among these developmental stages. We first conducted a principal component analysis (PCA) from the expression of all genes and TE insertions. We found that the first two principal components accounted for 90% of the variation and samples clustered into three groups that reflect the observed changes in PIWIL and piRNA expression. The P0 and P2 samples that lacked expressed PIWIs and piRNAs clustered in one group, the juvenile P8, P10, and P13 samples with expressed PIWIs and piRNAs clustered in another, and the adult sample was isolated (Supplemental Fig. 3A). When we excluded gene expression data, we found that samples grouped in a similar manner whether we considered expression from all TEs (Fig. 3B), or only young insertions (Fig. 3C), although samples were closer together in principle component space compared to the whole expression profile. Because the grouping of these clusters corresponded well with the observed changes in PIWI expression, we used these life stage replicates to test whether TE expression changed with the onset of PIWI paralog expression. We performed a differential expression analysis between the pre-PIWI and post-PIWI juvenile samples (P0 and P2 vs. P8, P10, and P13) to determine whether TEs were less expressed in post-PIWI samples—as the ping-pong model would predict. However, TE expression was similar among samples, and only 17 out of 794 families were differentially expressed. There was an almost equal mix of up- and down-regulated TE families: 10 families were more expressed in pre-PIWI samples, whereas seven were more expressed in the post-PIWI samples (Fig. 3D). Further, log2-fold expression was less than two in all but one family. Most of these 17 families had median K2P distances larger than 0.1 and were mostly LTRs (Supplemental Fig. 3B). We repeated this analysis for just the young insertions and found that eight out of 151 families with young insertions were differentially expressed, and that the log2-fold change was less than two in all cases (Fig. 3E). All differentially expressed families were LTRs and six were relatively young, with median K2P distances <0.1 (Supplemental Fig. 3C). Seven out of the eight families were up-regulated in pre-PIWI samples, suggesting PIWIs had a stronger effect on these young insertions compared to all insertions, as the ping-pong cycle would predict. However, very few families were differentially expressed, fold changes were relatively low, and only LTR insertions exhibited decreased expression with the onset of PIWI expression. These results suggest TE transcription is not heavily regulated in squirrel testis.
piRNAs and TE dynamics
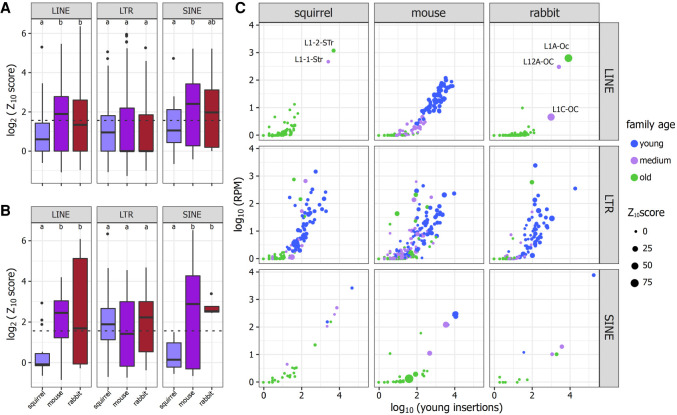
Our next step was to compare how PIWI and piRNA targeting among TE families varied among the three species. We estimated the strength of the piRNA/PIWI response to TEs by calculating Z10 scores from mapped piRNAs in LINEs, SINEs, and LTRs in all samples, for all insertions, and for only young insertions. The Z10 score measures the frequency of complementary piRNAs that overlap by 10 nt and reflects the strength of the ping-pong cycle. A Z10 score >1.96 reflects a nonrandom bias of complementary piRNAs overlapping by 10 bp and a P-value <0.05. When we compared Z10 scores among squirrel samples, our results were consistent with measures of PIWI expression. piRNA overlaps were largely random or nonexistent among piRNAs mapping to TE families in the P0 and P2 squirrel testes, and the ping-pong signature only became noticeable with the onset of PIWI expression in P8. However, the majority of TE families still lacked a strong ping-pong signature (Supplemental Fig. 4). To directly compare ping-pong cycle intensities across species, we averaged Z10 scores among samples within a species, but excluded the squirrel P0 and P2 samples because they lack PIWI proteins and piRNAs. An ANOVA revealed significant differences in Z10 score among species, so we conducted a post hoc Tukey's HSD test for pairwise comparisons and found that scores were significantly lower among squirrel LINEs than mouse and rabbit LINEs (P < 0.0001), Z10 scores were also significantly lower in squirrel SINEs compared to the mouse (P = 0.03), but not the rabbit (P = 0.45) (Fig. 4A). We then examined Z10 scores among young insertions, but only compared young TE Z10 scores from families that had >10 insertions <5% diverged from consensus and nonzero Z10 scores. The same overall pattern was present, except Z10 scores in squirrel SINEs were lower than both mouse and rabbit (P < 0.05) (Fig. 4B). The only TEs that had Z10 scores that reflected strong PIWI processing in the squirrel were the young LTRs (Fig. 4B), consistent with our differential expression results (Supplemental Fig. 3C).
FIGURE 4.
(A) Z10 score distribution among families among species. The dotted line reflects the position of a Z10 score = 1.96, P = 0.05. Significant differences among species means (as determined by Tukey's HSD test, P < 0.05) are indicated by different shared letters. (B) Z10 score distributions calculated from young insertions. (C) Number of young insertions plotted against the expression calculated from those insertions. Families are colored based on median K2P divergence, where young families are <10%, medium are between 10% and 20% diverged, and old families are >20%. Z10 scores calculated from young insertions are reflected by the size of the point.
We then compared average TE expression and Z10 scores to examine species-specific differences in TE and piRNA dynamics. First, we found that genome-wide TE expression strongly correlated with the total number of insertions, and that the age of the TE family was less relevant. In fact, the most expressed families were older than 20% diverged from consensus, and almost every family had some expressed insertions (Supplemental Fig. 5). Interestingly, Z10 scores did not strongly correlate with TE expression (Supplemental Fig. 6), suggesting that PIWI proteins are somewhat selective and do not merely target the families with the highest expression. For example, young mouse LINE families (<10% median divergence) elicited a stronger response from PIWIs than did older families, regardless of expression. When we examined expression values and Z10 scores from young insertions, there was a strong positive correlation between insertion number and expression with a family age component, that is, older families tended to have fewer young insertions that were less expressed (Fig. 4C). The mouse LINEs held most closely to this pattern, that is, young families with many young insertions were most expressed and exhibited more PIWI processing than insertions from medium-aged and old families. However, the two most active LINEs in squirrel and rabbit exhibited similar insertion number and expression values, but a dramatic difference in Z10 scores (Fig. 4C). Regardless of whether we examined all or young insertions, PIWI proteins were not targeting LINEs and SINEs in the squirrel as heavily as in mouse or rabbit, but it was apparent that squirrel LTRs were still mobilizing and cleaved by PIWI proteins.
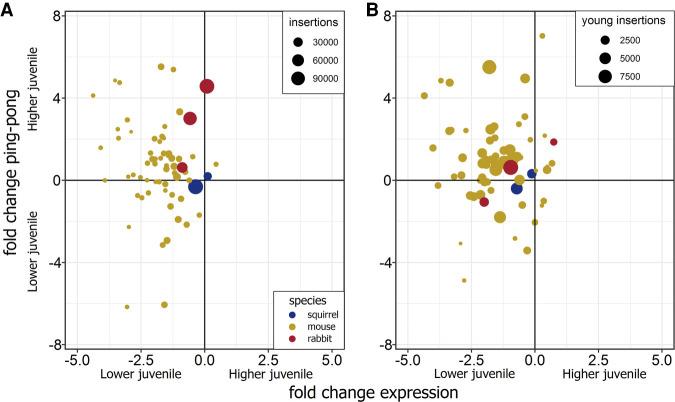
PIWI proteins can reduce the number of TE transcripts through direct cleavage and this targeting is reflected in the piRNA repertoire. piRNAs tend to derive from different TE sources in juvenile testis compared to adult testis, where LINEs are biased in juveniles and SINEs are biased in adults (Mourier 2011), suggesting a change in PIWI target preference. The ping-pong model predicts an inverse relationship between ping-pong cycle intensity and TE expression; that is, if a TE is heavily cleaved by PIWIs, piRNAs should be abundant and expression should be lower and vice versa. Therefore, we measured the log2-fold change in expression between juvenile (P13 of the squirrel) and adult testis, as well as the log2-fold change in Z10 scores in LINEs and tested this prediction in the context of species-specific differences. We were particularly interested to find out how much PIWI proteins reduced the expression of the two most recent squirrel LINE families in juvenile testis. We examined only families with more than 200 young insertions, as this excludes the older LINEs in squirrel and rabbit that lack Z10 scores but still provides 68 data points from mouse to produce a distribution for comparison. The overall prediction was accurate, as most LINEs in juvenile testis exhibited higher Z10 scores and lower expression (Fig. 5A). There was little expression or Z10 score change in the two squirrel LINEs, and we observed only a weak inverse relationship in one family when we examined these metrics from the youngest insertions (Fig. 5B). These analyses indicate that, while these two L1s have several thousand young insertions and have Z10 > 2.3 in all samples except P0 and P2, the strength of the ping-pong cycle in squirrel is not as intense as the cycle impacting active L1s in mouse or rabbit, and the PIWIs are not changing L1 expression.
FIGURE 5.
Changes in expression and the ping-pong cycle between a juvenile and adult for LINEs with more than 200 young insertions for (A) all insertions for that family and (B) just the young insertions. The juvenile squirrel is represented by the P13 sample.
PIWI evolution
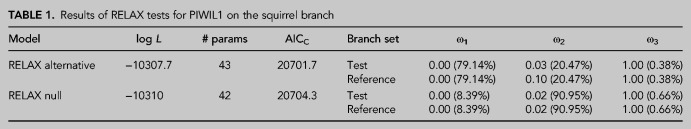
Finally, we tested whether reduced PIWI targeting could be attributed to the PIWI proteins themselves (i.e., is there evidence of reduced function in the squirrel PIWI proteins due to deleterious mutations?). To do so, we used selection tests that measure changes in dN/dS (ω). We specifically used the RELAX module in HyPhy that tests whether the strength of natural selection (positive or purifying) has been relaxed or intensified along a specified test branch. We constructed trees from the PIWI sequences of Glires (Supplemental Fig. 7) and used the squirrel branch as the test branch. This test was not significant for Piwil2 (K = 1.28, P = 0.321, likelihood ratio = 0.99) or Piwil4 (K = 0, P = 0.207, likelihood ratio = 1.59), but there was statistical support for the intensification of selection in Piwil1 (K = 1.49, P = 0.03, likelihood ratio = 4.69). However, ω was less than 1 among reference branches in the alternative model, and a K = 1.49 pushed ω in the test branch closer to 0, indicating an intensification of purifying selection (Table 1). This suggests that PIWIs have not experienced relaxed selection but have maintained their functional constraint such that the reduced targeting of squirrel LINEs and SINEs cannot be attributed to changes in PIWI function.
TABLE 1.
Results of RELAX tests for PIWIL1 on the squirrel branch
DISCUSSION
TE landscapes are variable among vertebrates and, given that there are several interactions between TEs and host defenses, what exactly causes this variability is still an unanswered question in genomics. Here we attempted to quantify the contribution of PIWI proteins to the dearth of novel TE insertions in the squirrel genome by comparing TE accumulation, TE expression, Piwi expression, and piRNA repertoires in relatively closely related species with a focus on the 13-lined ground squirrel that appears to lack mobilizing LINE and SINE families. We found that squirrel TEs were still highly expressed but lacked a strong PIWI response.
Although we did not treat small RNAs for 2′O-methylated 3′ ends, which would additionally validate piRNA presence, we are confident we accurately measured the behavior of the ping-pong cycle among species for the following reasons. We observed hallmark characteristics that suggested the majority of our small RNAs are bona fide piRNAs. piRNAs are the most abundant species of small RNA in mammalian testis (Czech and Hannon 2016) and most RNA between 24 and 36 nt long from the testis are going to be piRNAs. There was a strong 5′ U bias for small RNAs between 24 and 32 nt (Fig. 2A), which would not be expected from randomly degraded RNA fragments. Further, small RNAs treated for 2′O-methylated 3′ ends mirror the total population of small RNAs between 24 and 32 nt (Arensburger et al. 2011; Li et al. 2013; Praher et al. 2017). Evidence suggests treating for 2′O-methlayed 3′ ends is redundant when piRNA hallmarks are already present. Further, to ensure we examine small RNAs generated from the ping-pong cycle, we specifically studied complementary piRNAs that overlap and found 5′–5′ overlap distances between complementary piRNAs were biased for 10 nt, indicative PIWI processing, and the ping-pong cycle.
Piwi genes are expressed later in squirrels
Mouse Piwis, specifically Piwil2 and Piwil4, are initially expressed in prenatal testis; however, PIWIL4 ceases to be transcribed ∼3 d after birth but PIWIL2 is continuously expressed. Mice develop a little faster than squirrels, that is, 3- versus 4-wk pregnancy period, eyes open at P11–12 versus P21–24, and male mouse puberty occurs at 4 wk while squirrels go through a hibernation cycle and do not reach puberty until 11 mo after birth. Therefore, we wanted to address when PIWIs first become expressed in squirrel. We analyzed testis samples from squirrels between P0 and P13, and found that PIWIs are not expressed at least until P8. We acknowledge a 6-d gap between P2 and P8 when PIWIs could have become expressed, and we lack information on embryonic expression. Consistently, the hallmarks of piRNA biogenesis and TE processing were not observed until P8 in squirrel, but unlike adult rabbit and mouse, the distribution of piRNAs in adult squirrel testis stayed centered around 28 nt instead of 30 nt, confounding the difference between pre-pachytene and pachytene piRNAs in this species. It is possible that the squirrel's Piwil4 contributed to a higher abundance of short piRNAs, but this is currently unclear given a weak understanding of Piwil4 function in adult testis.
Since PIWIs are not expressed in P0 and P2, the small RNAs examined cannot be piRNAs, but once Piwis were expressed at P8, TEs became a more common source of these small RNAs (Fig. 2B). There were dramatic shifts in the sources of piRNAs between juvenile and adults of rabbits and mice, exhibiting differences in the sources of pre-pachytene and pachytene piRNAs, but this shift was not as apparent in squirrel. There could be two possible explanations for this: (1) because Piwil4 is so highly expressed in adult testis, TEs remain a common source of piRNAs, or (2) TE transcripts are not heavily targeted by PIWI proteins and do not make up a considerable proportion of pre-pachytene piRNAs. Perhaps some combination of both is also possible. Consistently among the species analyzed, when Piwil1 became expressed, diverse piRNAs synthesized from intergenic space became more abundant (Fig. 2B; Supplemental Table 1).
The ping-pong pathway weakly regulates TE expression in the squirrel
The piRNA pathway can silence TEs via methylation or cleavage. The ping-pong cycle regarding transcript cleavage only requires PIWIL2 and is constant once Piwil2 becomes expressed. Many TE loci are still expressed after methylation, and any fully intact expressed retrotransposon has the capacity to mobilize during spermatogenesis. The ping-pong cycle reduces the likelihood a retrotransposon will mobilize. We did not examine TE methylation since we were only able to collect tissues from birthed squirrels and rabbits, and little is known about methylation patterns and timing in nonmodel organisms and we specifically aimed to study the TE cleavage aspect of the ping-pong cycle.
We initially predicted that TE expression variation among species could be attributed to the number of insertions in the genome. While this appeared to be true within genomes (Supplemental Fig. 5), this was not the case among genomes (Fig. 3A). Given the small percentage of the squirrel genome derived from TEs, especially young TEs, we expected TEs would make up a small proportion of the squirrel transcriptome, but in fact TEs made up the highest percentage of the transcriptome among species. This example of a disconnect between TE insertions and their expression is consistent with the idea that transcription and successful reintegration are distinct phenomena; the first does not necessarily lead to the second, and insertion success may not be dependent on high levels of TE expression (Deininger et al. 2003; Kazazian 2004; Lu and Clark 2010). However, given the apparent absence of retrotransposition-competent L1 loci in the squirrel assembly (Platt and Ray 2012), the lack of new integrations while still observing transcription was unsurprising.
There was TE expression variation among the squirrel life stages revealed by PCA, where samples fell into three groups: a pre-PIWI group (P0 and P2), a post-PIWI juvenile group (P8, P10, and P13), and the adult group. Expression did not significantly change for 97.8% of TE families after Piwis became transcribed in P8. Further, among the 17 families that were differentially expressed, seven were expressed higher in samples with expressed Piwis. We yielded a similar result when we restricted analyses to young insertions, where 94.7% of families were not differentially expressed. However, of the eight differentially expressed families, seven experienced a reduction in transcription with the onset of Piwis. Almost all differentially expressed families were LTRs, and LTRs were the only group of elements with a strong ping-pong signature (Fig. 4). Unfortunately, we could not perform the same test in mouse and rabbit for comparison, but it has been reported that L1s are 5–10× more expressed in mouse Piwil2 knockouts (Aravin et al. 2007b). The absence of differentially expressed TEs could be explained by a lack of PIWI processing through the ping-pong cycle, particularly in LINEs and SINEs. The strength of the ping-pong cycle was significantly reduced among squirrel LINEs and SINEs relative to rabbit and mouse (Fig. 4A,B). We should note that the two most recently active L1s in squirrel had Z10 scores between 2.3 and 8.5, indicating an active ping-pong cycle, but substantially less so than the highest LINE ping-pong signatures in mouse and rabbit (max Z10 = 33.6 mouse, max Z10 = 81.3 rabbit). If a robust ping-pong cycle was absent in these species, our 5′–5′ 10 bp overlap Z10 score should be <1.96 (i.e., reads mapped randomly).
In a final approach to measure any LINE expression changes between squirrel juvenile and adult life stages, we took advantage of the fact that TE preference changes from LINEs to SINEs between juvenile testis and adult testis, respectively (Mourier 2011). We examined ping-pong and LINE expression–fold changes, and in our control samples we observed a pattern where the ping-pong cycle is stronger among LINEs and LINE expression is lower in juvenile testis compared to adults. In contrast, neither expression nor ping-pong intensity substantially changed in the most recent squirrel LINE families between juveniles and adults (Fig. 5). These results seem to imply that the ping-pong cycle is weakly regulating LINE expression in squirrel testes. This hypothesis could be further tested directly with the development of a PIWI knockout squirrel model.
One goal of ours was to test whether PIWIs were responsible for the “death” of the squirrel L1. If that were the case, we predicted that the hallmarks of PIWI suppression as evidenced by an intense ping-pong cycle and low expression of targeted TEs, would still be present. However, we observed the opposite relationship. If PIWIs were responsible for the immobility of LINEs, those hallmarks are gone and PIWIs respond to these L1s like an old, harmless family, so the question remains: how do TEs “die” and can PIWIs be responsible? Another unanswered question that remains is why TE expression is still so high in squirrels. One possible explanation could be due to the function of PIWI proteins. If there has been any functional relaxation in PIWIs, we should observe such a relaxation of functional constraint in the DNA sequences. To rule out this possibility, we collected PIWI paralog sequences from a range of Glires species and measured dN/dS changes along the squirrel lineage. We found no evidence of a relaxation effect, but indeed found an intensification of purifying selection on Piwil1. Further, there were still relatively strong ping-pong signatures among LTR families, indicating appropriate function. From these observations, we can rule out an explanation where PIWI proteins are at fault. Alternatively, Pasquesi et al. (2020) demonstrated that the majority of vertebrate TE transcripts are derived from older TE insertions and Molaro et al. (2014) determined that mouse L1 promoters least diverged from consensus were methylated more heavily than promoters from older L1s. Further, Byun et al. (2013) found older TEs in the human genome are less methylated at CpG cites than younger families. Therefore, since the squirrel genome is largely lacking young TEs, it is plausible these TEs are not as methylated and transcribed at higher levels. However, this does not necessarily explain why young squirrel L1 insertions lack a strong ping-pong signature. This leads to the last major question that has yet to be answered: Do PIWIs distinguish between an expressed TE and a mobilizing TE and, if so, how? The squirrel and rabbit genomes contain a similar number of young L1s that are nearly equivalently expressed, but demonstrate a difference in the strength of the ping-pong cycle (Fig. 4C). PIWIs are capable of targeting and cleaving TE transcripts from young families in a cluster-independent mechanism (Aravin et al. 2008; Gainetdinov et al. 2017), but these models do not explain how PIWIs are initially guided to the most threatening families, or how PIWIs differentiate between threatening and nonthreatening transcripts.
Conclusions
Taken together, our results raise interesting questions regarding the interplay between TEs, piRNAs, and Piwi paralogs. Our study indicates that TE transcript abundance is not a good predictor of TE accumulation. Contrary to our expectations, we found that TEs were actively transcribed in the squirrel even though most are not accumulating in the genome. The presence of squirrel Piwi transcripts largely did not have an effect on the expression of most TE families and, unlike recently active LINEs in the two other mammals we examined, the ping-pong cycle did not lead to the reduction of LINE transcripts. We hypothesize that this derives from the passive processing of TE-derived transcripts into piRNAs in the squirrel, which sharply contrasts with the active reduction of threatening TE transcripts in other mammals.
MATERIALS AND METHODS
Sample acquisition and RNA sequencing
All animal procedures conformed with federal and institutional guidelines for humane care and use and were preapproved by the respective Institutional Animal Care and Use Committees. 13-lined ground squirrels were reared in the captive breeding colony at the University of Wisconsin, Oshkosh (Merriman et al. 2012). For testis collection, euthanasia by cervical transection was conducted on neonates (P0), postnatal juveniles (P2, P8, P10, P13), and adults. We sampled one juvenile (P21) and one adult (P > 90) male rabbit. For each specimen, a cross section of testis was snap frozen in liquid nitrogen immediately following castration and stored at −80°C prior to RNA isolation. We isolated total RNA using TRIzol (Invitrogen) according to the manufacturer's specifications. Small RNA libraries were prepared using the Illumina TruSeq Small RNA kit and 1 × 50 bp reads were sequenced on the Illumina HiSeq 2000 platform. Directional RNA-seq libraries were prepped using the Illumina TruSeq v2 kit and 2 × 100 bp reads were also sequenced on a HiSeq 2000. All reads are available under the BioProject PRJNA528042. P7 mouse mRNA (SRR3659160), small RNA (SRR3659150), adult mRNA (SRR765631), and small RNA (SRR772033) libraries were collected from the NCBI short-read archive (SRA).
TE annotation
To visualize TE differences between the squirrel, rabbit, and mouse genomes, we masked the ground squirrel (SpeTri2.0), rabbit (OryCun2.0.73), and mouse (GRCm38) genome using RepeatMasker 4.0.5 “species Ictidomys,” “species Oryctolagus,” and “species Mus,” respectively. To estimate genetic distances, we used a modified calcDivergenceFromAlign.pl script included with RepeatMasker to calculate Kimura two-parameter (Kimura 1980) distances between each insertion and its respective consensus sequence. The option –noCpG was invoked to exclude highly mutable CpG sites from distance calculations. Redundant TE annotations were first identified using ClusterBed (Quinlan and Hall 2010), and the longest annotation with the lowest K2P was selected as the most probable annotation for that locus (script available at github.com/mike2vandy/squirrel_piRNA).
piRNA genomic mapping
Prior to small RNA mapping, we clipped barcodes, removed reads that had bases with Phred quality score <25, and removed identical reads using modules in the fastx toolkit. We also removed low complexity small RNA sequences by zipping the sequence and removing sequences that compressed by more than 75% (script available at github.com/mike2vandy/squirrel_ piRNA). We mapped piRNA sequences 24–32 nt long to the respective genomes using Bowtie 1.2 (Langmead et al. 2009) with the parameters –a –v 0 to allow reads to map to all perfect matches in the genome. We estimated the source (LINE, SINE, LTR, DNA, RNA, genic, or other) of piRNAs given the most frequently mapped annotation type per piRNA. Further, we examined the strength of the ping-pong cycle among TE families. We used intersectBed to intersect piRNAs with TE annotations. For each TE family, we calculated a 10 nt complementary overlap Z10 score where 1–9 and 11–20 nt overlaps were used as background following Han et al. (2015). We recorded the number of mapped positions for each small RNA and normalized piRNA abundances to all genome mapping reads as parts per million (ppm) so that our Z10 score takes into account piRNA source ambiguity and piRNA amplification. We repeated the same analysis for insertions that were <5% diverged from TE consensus sequences. Scripts available at github.com/mike2vandy/squirrel_piRNA.
TE expression
Prior to mapping, we removed adaptors and poor-quality sequences from RNA-seq data files using Trimmomatic v0.36 (Bolger et al. 2014). To estimate TE expression profiles through testis development, we mapped clean RNA-seq reads to the appropriate genome draft and Ensembl annotated gtf using STAR v2.7.1a (Dobin et al. 2013) with the parameters –outFilterMultimapNmax 100 and –winAnchorMultimapNmax 100 to allow reads to map up to 100 positions. We then used TEtranscripts v2.0.3 (Jin et al. 2015), specifically TEcount, to simultaneously estimate gene and genome-wide TE family expression from genome-mapped RNA-seq reads. TEcount requires two annotation files that specify gen and repeat coordinates. We used Ensembl annotations for genic coordinates and we modified our RepeatMasker output for compatibility with TEcount. Raw read counts were transformed into RPM counts using DESeq2 v1.26 (Love et al. 2014). We conducted a separate TE expression analysis to examine the expression patterns of just the “young” insertions. To do so, we filtered the TE gtf file for insertions that exhibited a Kimura 2-parameter distance <5% diverged from the consensus and reran TEcount.
To compare expression profiles among squirrel samples, we normalized read counts given all genes and TEs using estimateSizeFactors and estimateDispersions functions in DESeq2 after removing genes and TE families with less than 10 mapped reads across all samples. Patterns of expression variation were assessed by performing PCAs based on the blind variance stabilizing transformed data. We conducted PCAs with all genes and TEs, just TEs, and young TE insertions. We also performed a differential expression analysis between samples from juveniles that lacked expressed PIWI proteins (P0 and P2) and juveniles that expressed PIWI proteins (P8, P10, and P13) to identify TEs that were differentially expressed in the presence or absence of PIWI proteins. In these analyses, all TE families and genes were tested for differential expression and we determined statistical significance when the adjusted P-value was <0.1. We conducted these analyses for a data set that contained TE expression estimated from all TE insertions and a data set based on the expression of young TE insertions. We performed log fold shrinkage for lowly expressed TEs using apeglm when plotting fold change against expression (Zhu et al. 2019).
Selection tests
For each PIWI paralog, we collected orthologs from closely related rodent and lagomorph species from Ensembl (species and gene IDs are available in Supplemental Table 2). A codon alignment was made by translating coding sequences to amino acid, aligning amino acids using LINSI parameters in MAFFT (Katoh and Standley 2013), and using a custom script to reverse translate resulting alignments. Unrooted maximum likelihood trees were constructed using RAxML (Stamatakis 2014). We used the RELAX module (Wertheim et al. 2015) in HyPhy (Pond et al. 2005) to test for a relaxation or intensification of selection of the PIWI orthologs in squirrel. RELAX identifies shifts in the stringency of natural selection by performing a likelihood ratio test (LRT) between a null model that constrains a relation parameter K to 1 for all branches and an alternative model where K is a free parameter. K > 1 implies an intensification of selection and K < 1 suggests relaxation of selection along the test branch(es).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support from the National Science Foundation (DEB-1355176, RoL 1838283). Additional support was provided by the College of Agriculture and Life Sciences at Mississippi State University and the College of Arts and Sciences at Texas Tech University. In addition, we would like to thank the Texas Tech High-Performance Computing Center and the Mississippi State University Institute for Genomics Biocomputing and Biotechnology for providing the computational resources. Lastly, this work could not have been completed without the helpful assistance of Ildar Gainetdinov. The color palette was inspired by Tool's Lateralus (2001).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078862.121.
REFERENCES
- Aravin AA, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. 10.1038/nature04916 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. 2007a. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. 10.1126/science.1146484 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007b. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. 10.1126/science.1142612 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, Hice RH, Wright JA, Craig NL, Atkinson PW. 2011. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genomics 12: 606. 10.1186/1471-2164-12-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Byun HM, Motta V, Panni T, Bertazzi PA, Apostoli P, Hou L, Baccarelli AA. 2013. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol 10: 28. 10.1186/1743-8977-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Carrieri C, Comazzetto S, Grover A, Morgan M, Buness A, Nerlov C, O'Carroll D. 2017. A transit-amplifying population underpins the efficient regenerative capacity of the testis. J Exp Med 214: 1631–1641. 10.1084/jem.20161371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Pääbo S, et al. 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437: 88–93. 10.1038/nature04000 [DOI] [PubMed] [Google Scholar]
- Chirn G, Rahman R, Sytnikova YA, Matts JA, Zeng M, Gerlach D, Yu M, Berger B, Naramura M, Kile BT, et al. 2015. Conserved piRNA expression from a distinct set of piRNA cluster loci in eutherian mammals. PLoS Genet 11: e1005652. 10.1371/journal.pgen.1005652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Batzer MA. 2004. Retrotransposition of Alu elements: How many sources? Trends Genet 20: 464–467. 10.1016/j.tig.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 41: 324–337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263. 10.1038/nature10547 [DOI] [PubMed] [Google Scholar]
- de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA, Edgell MH. 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet 8: 307–311. 10.1016/0168-9525(92)90139-U [DOI] [PubMed] [Google Scholar]
- Deininger PL, Moran J V, Batzer MA, Kazazian HH. 2003. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 13: 651–658. 10.1016/j.gde.2003.10.013 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C, Odom DT, Kutter C. 2017. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat Commun 8: 1411. 10.1038/s41467-017-01049-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke V, Ganesh S, Karlic R, Malik R, Pasulka J, Horvat F, Kuzman M, Fulka H, Cernohorska M, Urbanova J, et al. 2017. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res 27: 1384–1394. 10.1101/gr.216150.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov I, Skvortsova Y, Kondratieva S, Funikov S, Azhikina T. 2017. Two modes of targeting transposable elements by piRNA pathway in human testis. RNA 23: 1614–1625. 10.1261/rna.060939.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. 10.1038/nature04917 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian J, Kazazian HH. 2001. A novel active L1 retrotransposon subfamily in the mouse. Genome Res 11: 1677–1685. 10.1101/gr.198301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou L-T, Dai P, Yang J-H, Xue Y, Hu Y-P, Zhou Y, Kang J-Y, Wang X, Li H, Hua M-M, et al. 2014. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 24: 680–700. 10.1038/cr.2014.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. PiRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. 10.1126/science.aaa1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82. 10.1016/j.cell.2007.03.026 [DOI] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M. 2015. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31: 3593–3599. 10.1093/bioinformatics/btv422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH. 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Kerner P, Degnan SM, Marchand L, Degnan BM, Vervoort M. 2011. Evolution of RNA-binding proteins in animals: insights from genome-wide analysis in the sponge Amphimedon queenslandica. Mol Biol Evol 28: 2289–2303. 10.1093/molbev/msr046 [DOI] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917. 10.1101/gad.1640708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367. 10.1126/science.1130164 [DOI] [PubMed] [Google Scholar]
- Lewis SH, Salmela H, Obbard DJ. 2016. Duplication and diversification of Dipteran Argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol Evol 8: 507–518. 10.1093/gbe/evw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZZ, Roy CKK, Dong X, Bolcun-Filas E, Wang J, Han BWW, Xu J, Moore MJJ, Schimenti JCC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81. 10.1016/j.molcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Lei B, Li Y, Tong K, Ding Y, Luo L, Xia X, Jiang S, Deng C, Xiong Y, et al. 2012. Discovery of potential piRNAs from next generation sequences of the sexually mature porcine testes. PLoS ONE 7: e34770. 10.1371/journal.pone.0034770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 1–21. 10.1186/gb-2014-15-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark AG. 2010. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res 20: 212–227. 10.1101/gr.095406.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. 2009. Small RNAs as guardians of the genome. Cell 136: 656–668. 10.1016/j.cell.2009.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. 2015. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep 12: 1234–1243. 10.1016/j.celrep.2015.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriman DK, Lahvis G, Jooss M, Gesicki JA, Schill K. 2012. Current practices in a captive breeding colony of 13-lined ground squirrels (Ictidomys tridecemlineatus). Lab Anim (NY) 41: 315–326. 10.1038/laban.150 [DOI] [PubMed] [Google Scholar]
- Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, Richard McCombie W, Smith AD, Hannon GJ. 2014. Two waves of de novo methylation during mouse germ cell development. Genes Dev 28: 1544–1549. 10.1101/gad.244350.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier T. 2011. Retrotransposon-centered analysis of piRNA targeting shows a shift from active to passive retrotransposon transcription in developing mouse testes. BMC Genomics 12: 440. 10.1186/1471-2164-12-440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD. 2007. Mighty Piwis defend the germline against genome intruders. Cell 129: 37–44. 10.1016/j.cell.2007.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquesi GIM, Perry BW, Vandewege MW, Ruggiero RP, Schield DR, Castoe TA. 2020. Vertebrate lineages exhibit diverse patterns of transposable element regulation and expression across tissues. Genome Biol. Evol 12: 506–521. 10.1093/gbe/evaa068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, Ray DA. 2012. A non-LTR retroelement extinction in Spermophilus tridecemlineatus. Gene 500: 47–53. 10.1016/j.gene.2012.03.051 [DOI] [PubMed] [Google Scholar]
- Platt RN, Vandewege MW, Ray DA. 2018. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res 26: 25–43. 10.1007/s10577-017-9570-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse S V. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679. 10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- Praher D, Zimmermann B, Genikhovich G, Columbus-Shenkar Y, Modepalli V, Ahoroni R, Moran Y, Technau U. 2017. Characterization of the piRNA pathway during development of the sea anemone Nematostella vectensis. RNA Biol 14: 1727–1741. 10.1080/15476286.2017.1349048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. 2005. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1327–1330. 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S. 2013. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4: 1–15. 10.1186/1759-8753-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YH, Xie LH, Zhuo X, Chen Q, Ghoneim D, Zhang B, Jagne J, Yang C, Li XZ. 2017. Domestic chickens activate a piRNA defense against avian leukosis virus. eLife 6: e24695. 10.7554/eLife.24695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, Sytnikova YA, Chirn G-W, Ang I, Lau NC, Blower MD. 2016. Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome. RNA 23: 504–520. 10.1261/rna.058859.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewege MW, Platt RN II, Ray DA, Hoffmann FG. 2016. Transposable element targeting by piRNAs in laurasiatherians with distinct transposable element histories. Genome Biol Evol 8: 1327–1337. 10.1093/gbe/evw078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskaite˙ L, Berrens R V, Ivanova I, Carrieri C, Reik W, Enright AJ, O'Carroll D. 2018. Defective germline reprogramming rewires the spermatogonial transcriptome. Nat Struct Mol Biol 25: 394. 10.1038/s41594-018-0058-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol 32: 820–832. 10.1093/molbev/msu400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PH, Fu Y, Cecchini K, Özata DM, Arif A, Yu T, Colpan C, Gainetdinov I, Weng Z, Zamore PD. 2020. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat Genet 52: 728–739. 10.1038/s41588-020-0657-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Pointer B, Kelleher ES. 2020. Rapid evolution of piRNA-mediated silencing of an invading transposable element was driven by abundant de novo mutations. Genome Res 30: 566–575. 10.1101/gr.251546.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Ibrahim JG, Love MI, Stegle O. 2019. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35: 2084–2092. 10.1093/bioinformatics/bty895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoch A, Auchynnikava T, Berrens R V, Kabayama Y, Schöpp T, Heep M, Vasiliauskaite˙ L, Pérez-Rico YA, Cook AG, Shkumatava A, et al. 2020. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature 584: 635–639. 10.1038/s41586-020-2557-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.