Abstract
Background
Diarrhoea is the leading cause of morbidity and mortality in the world particularly in developing countries and among vulnerable groups of the population. Gram-negative enteric bacterial pathogens (GNEBPs) are a group of organisms that reside mainly in the intestine and induce diarrhoea. Antimicrobial agents are usually the part of their treatment regimen. The therapeutic effect of antimicrobials is hindered by the emergence and spread of drug-resistant strains. The information regarding the prevalence and antimicrobial resistance patterns of GNEBPs in Ethiopia is limited and found in a scattered form.
Objectives
This study was designed to determine the pooled prevalence and drug resistance patterns of GNEBPs by meta-analysis of data from diarrhoeic patients in Ethiopia.
Method
A comprehensive literature search was conducted through internet searches using Google Scholar, PubMed, Science Direct, HINARI databases, and reference lists of previous studies. Published articles were included in the study based on priorly set inclusion and exclusion criteria. Results were presented in the forest plot, tables, and figures with a 95% confidence interval (CI). The inconsistency index (I2) test statistics was used to assess heterogeneity across studies. The pooled prevalence estimate of GNEBPs and their drug resistance patterns were computed by a random-effects model. Software for Statistics and Data Science (STATA) version 14 statistical software was used for the analysis.
Result
After removing those articles which did not fulfil the inclusion criteria, 43 studies were included in the analysis. Studies were conducted in 8 regions of the country and most of the published articles were from the Amhara region (30.23%) followed by Oromia (18.60%) and Southern Nations, Nationalities, and Peoples’ region (SNNP) (18.60%). The pooled prevalence of GNEBPs was 15.81% (CI = 13.33–18.29). The funnel plot indicated the presence of publication bias. The pooled prevalence of GNEBPs in Addis Ababa, Amhara, SNNP, and Oromia regions were 20.08, 16.67, 12.12, and 11.61%, respectively. The pooled prevalence was 14.91, 18.03, and 13.46% among studies conducted from 2006–2010, 2011–2015, and 2016–2021, respectively and it was the highest (20.35%) in children having age less than or equal to 15 years. The pooled prevalence of Escherichia coli, Campylobacter spp., Shigella spp., and Salmonella enterica were 19.79, 10.76, 6.24, and 5.06%, respectively. Large proportions (60–90%) of the isolates were resistant to ampicillin, amoxicillin, tetracycline, and trimethoprim-sulphamethoxazole. The pooled prevalence of multidrug resistance (MDR) was 70.56% (CI = 64.56–76.77%) and MDR in Campylobacter spp., Shigella spp., E. coli, and S. enterica. were 80.78, 79.08, 78.20, and 59.46%, respectively.
Conclusion
The pooled estimate showed a high burden of GNEBPs infections and a high proportion of drug resistance characters to commonly used antimicrobial agents in Ethiopia. Therefore, performing drug susceptibility tests, establishing an antimicrobial surveillance system and confirmation by molecular techniques are needed.
Introduction
World Health Organization (WHO) defines diarrhoea as the passage of three or more loose or liquid stools per day (24 hours). During diarrhoea, the water content and volume of stool and defecation frequency will usually increase. The syndrome may be accompanied by other illnesses like vomiting, fever, dysentery, nausea, and abdominal cramps. It is the leading cause of morbidity and mortality in the world and contributes about 4% of all deaths and 5% of health loss to disability [1–3]. Diarrhoea is the fifth leading cause of death and it contributes to one in nine deaths among children younger than 5 years [4,5]. The problem is severe among the vulnerable population such as children, people with HIV, the elderly, and other individuals having weak immunity. Many factors contribute to diarrhoea; however, childhood wasting (low weight-for-height score), unsafe water, and unsafe sanitation are the leading risk factors [5]. The incidence of diarrhoea is different among the regions or continents of the world. It is highly prevalent in Sub-Saharan Africa and South Asia. The report from WHO showed that these countries account for about 78% of all diarrheal deaths among children in the developing world [2,4]. Ethiopia is one of the top three countries with very high child mortality due to diarrhoea in Africa [5–7].
Diarrhoea can be induced by a variety of causes. However, infectious agents like viruses and bacteria are among the leading causes. Bacteria, particularly Gram-negative enteric bacterial pathogens (GNEBPs) are the common causes of the syndrome. The group includes bacteria that reside mainly in the intestine. Genera such as Escherichia, Shigella, Campylobacter, Salmonella, Enterobacter, Klebsiella, Yersinia, Serratia, Proteus, and others are included in the group. However, the most common and significant pathogens are S. enterica, E. coli, Campylobacter, and Shigella spp. [8].
Antimicrobial agents are usually part of the treatment regimen, particularly on diarrhoea caused by bacteria. Due to the widespread and indiscriminate use of antimicrobials, several resistant strains are emerging which tend to spread globally [9,10]. Hence, antimicrobial resistance (AMR) is a global health threat and was recognized in the 2016 United Nations (UN) General Assembly [11]. It is one of the top challenges in achieving the 2030 UN sustainable development goals [10]. Infections caused by resistant organisms affect treatment outcomes, treatment costs, disease spread, and duration of illness, posing a challenge to the future of chemotherapy [12]. Some pathogenic strains are also developing resistance not only to one but to several agents, i.e., multidrug resistance [13,14].
Information regarding the prevalence and antimicrobial resistance patterns of GNEBPs in Ethiopia is limited, and available information is found in scattered forms. Hence, there is an interest to conduct a nationwide study. To fill this significant gap, this systemic review and meta-analysis was prepared. The review focused on the prevalence and antimicrobial resistance patterns of GNEBPs isolated from diarrheic patients in Ethiopia. The output of this systematic review and meta-analysis can be used by clinicians, policymakers, and researchers to make evidence-based decisions.
Methods
Literature search and selection
The published articles were searched based on preferred reporting items for systematic reviews and meta-analyses (PRISMA) guideline [15]. The search was performed from June to August 2021 using Google Scholar, PubMed, Science Direct, and HINARI databases. The search queries were set based on medical subject headlines (MESH) and Boolean logic. Relevant MeSH terms and keywords were used to retrieve all relevant articles from the databases listed above. The keywords and MeSH terms used were “enteric bacteria AND diarrhoea AND drug resistance AND Ethiopia”, “Salmonella AND diarrhoea, AND Ethiopia AND drug resistance”, “Shigella AND diarrhoea AND Ethiopia AND drug resistance”, “Escherichia coli AND diarrhoea AND Ethiopia AND drug resistance”, “Campylobacter AND diarrhoea AND Ethiopia AND drug resistance” “Yersinia AND diarrhoea AND Ethiopia AND drug resistance”. Each bacterial genus was searched separately, and a search was also conducted on reference lists of previous studies to increase the chance of getting more articles. Only those articles which fulfil the selection criteria were used to analyse the information.
Inclusion and exclusion criteria
Research conducted on GNEBPs from the diarrhoeic patient or their antimicrobial susceptibility in Ethiopia and full-length published articles in the English language were included in the analysis. To get updated information on the issue, articles published from 2010 to August 2021 were considered. Studies that did not focus on GNEBPs from the diarrhoeic patient or their antimicrobial susceptibility, anonymous reports, abstracts (incomplete information), and published articles before 2010 and unpublished information were not included in the study. Studies that were conducted to assess the knowledge, attitude, and practice (KAP) of the community or the professionals were not also included.
Data extraction
The selected articles were coded, and data were collected using a format prepared in Microsoft Excel. The format consists of the author’s name, study period, year of publication, study design, study region, study population, sample size, sample type, age, gender, isolated bacteria species, their prevalence, resistance patterns of the isolates, and prevalence of multidrug resistance. The extracted data were checked at least twice for their accuracy.
Quality control
The quality of eligible studies was checked using a set of criteria based on Joanna Briggs Institute critical appraisal tools including appropriateness of the research design to address the target population, adequate sample size, quality of paper, completeness of the information, and appropriateness of methods for isolation of the bacteria and appropriate statistical analysis [16]. The eligibility of selected articles was also assessed and approved by experts in the discipline.
Data analysis
The data were compiled in Excel 2010 (Microsoft, Redmond, WA, USA) spreadsheet and summarized by descriptive statistics. A random-effect model was used to determine the pooled prevalence and the 95% confidence interval (CI). All statistical analysis was achieved by using Software for Statistics and Data Science (STATA; https://www.stata.com/company/our-sites/) version 14. The data were described using forest plots, figures, and tables. The presence of publication bias was assessed by funnel plot. Sub-group analysis was performed based on the regions in the country, age group; (children, adults, and all age groups), and year of study (2006–2010, 2011–2015, and 2016–2021). Statistical heterogeneity was evaluated by the inconsistency index (I2) test. The I2 provides an estimate of the percentage of the variability in effect estimates that is due to heterogeneity rather than sampling error or chance differences. Hence, the I2 test measures the level of statistical heterogeneity among studies [17,18].
Results
Characteristics of published articles
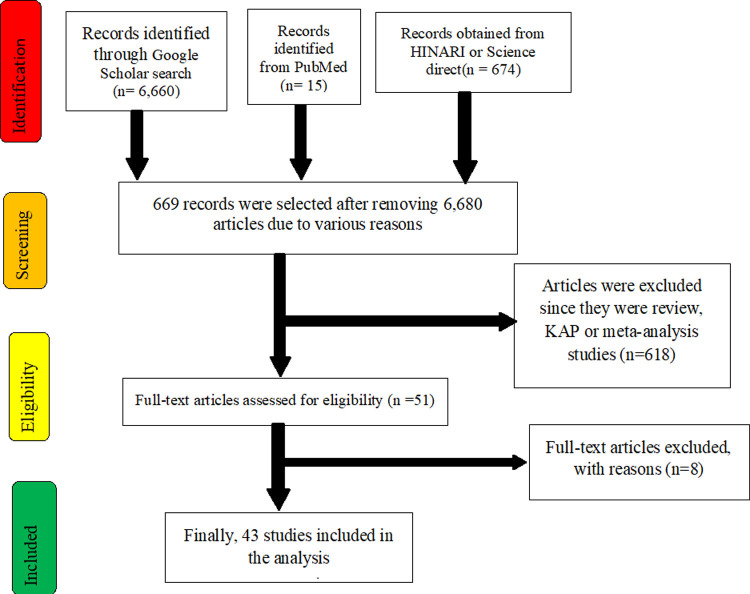
Of 7,349 identified studies, 6,680 articles were excluded upon reviewing the titles and abstracts because they were irrelevant (were not focusing on GNEBPs, diarrhoea, and drug resistance or were outside Ethiopia or duplicates). The remaining 669 articles were assessed for eligibility; of these, 626 articles were excluded since they were review, KAP, or meta-analysis studies. Finally, 43 studies meeting the inclusion criteria were included in this study. Selected articles were focusing on one or more GNEBPs. Fig 1 shows a flow diagram of the selection of articles for the analysis.
Fig 1. A flow diagram that shows the selection of articles for the analysis.
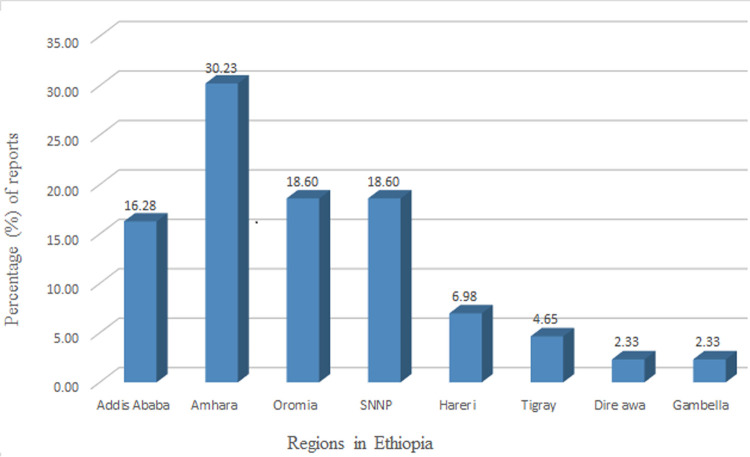
Table 1 shows the overall characteristics of articles included in the analysis, type and prevalence of Gram-negative isolates recovered from diarrheic patients. Studies were conducted in 8 regions of the country and most of the published articles were from the Amhara region (30.23%) followed by the Oromia region (18.60%) and South Nation and Nationalities Region (SNNP) (18.60%). Published articles were not found in other regions of the country (Afar, Somali, and Benishangul Gumuz regions) (Fig 2).
Table 1. Characteristics, quality, and the number of Gram-negative isolates recovered from diarrheic patients.
| References | Year of publication | Study period | Region | Study population | Age category | Gender | Number examined | No Positive | Prevalence | E. coli | Salmonella | Shigella | Klebsiella | Proteus | Enterobacter | Campylobacter | Citrobacter | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | |||||||||||||||||
| [19] | 2018 | Aug–Dec 2015 | Addis Ababa | Diarrheic patient | <15 yrs | 115 | 138 | 253 | 94 | 37.15 | 61 | 23 | 10 | |||||
| [20] | 2021 | Mar 2019 to Nov 2019 | SNNP | diarrheic patient | Adult > 15yrs | 151 | 127 | 278 | 24 | 8.63 | 15 | 9 | ||||||
| [21] | 2018 | Nov 2015 and Aug 2016 | Amhara | diarrheic patient | <15 yrs | 99 | 64 | 163 | 91 | 55.83 | 47 | 5 | 3 | 11 | 7 | 2 | ||
| [22] | 2020 | Jan to March 2018 | Amhara | Diarrheic patient, HIV + | all | 163 | 191 | 354 | 24 | 6.78 | 17 | 7 | ||||||
| [23] | 2020 | Jan to July 2014 | Oromia | diarrheic children | <15 yrs | 125 | 114 | 239 | 9 | 3.77 | 3 | 6 | ||||||
| [24] | 2018 | June to Sept 2017 | SNNP | diarrheic patient | <15 yrs | 101 | 103 | 204 | 19 | 9.31 | 3 | 17 | ||||||
| [25] | 2011 | Aug to Nov 2009 | Amhara | diarrheic patient | all | 125 | 90 | 215 | 32 | 14.88 | 32 | |||||||
| [26] | 2014 | Feb to May, 2014 | Amhara | diarrheic patient | all | 180 | 192 | 372 | 21 | 5.65 | 4 | 17 | ||||||
| [27] | 2018 | June to Oct, 2016 | Amhara | diarrheic patient | <15 yrs | 68 | 44 | 112 | 4 | 3.57 | 1 | 3 | ||||||
| [28] | 2014 | March to Nov 2012 | Oromia | diarrheic patient | <15 yrs | 114 | 146 | 260 | 22 | 8.46 | 16 | 6 | ||||||
| [29] | 2015 | March to May 2011 | Oromia | diarrheic patient | <15 yrs | 14 | 10 | 24 | 7 | 29.17 | 7 | |||||||
| [30] | 2020 | March and Aug 2019 | SNNP | HIV infected diarrheic | Adult >15 yrs | 84 | 96 | 180 | 15 | 8.33 | 5 | 2 | 8 | |||||
| [13] | 2011 | Jan to Aug 2006 | Addis Ababa | diarrheic patient | <15 yrs | 654 | 571 | 1225 | 126 | 10.29 | 65 | 61 | ||||||
| [31] | 2019 | Nov 2016 and May 2017 | Addis Ababa | diarrheic patient | <15 yrs | 155 | 135 | 290 | 42 | 14.48 | 13 | 7 | 22 | |||||
| [32] | 2013 | Oct 2011 to March 2012 | Amhara | diarrheic patient | <15 yrs | 144 | 141 | 285 | 44 | 15.44 | 44 | |||||||
| [33] | 2015 | Dec 2011 to Feb 2012 | Amhara | diarrheic patient | <15 yrs | 239 | 183 | 422 | 73 | 17.30 | 33 | 40 | ||||||
| [34] | 2014 | Feb to May 2011 | Harari | diarrheic patient | all | 193 | 191 | 384 | 56 | 14.58 | 56 | |||||||
| [35] | 2014 | June to Oct, 2011 | SNNP | diarrheic patient | <15 yrs | 81 | 77 | 158 | 35 | 22.15 | 4 | 11 | 20 | |||||
| [36] | 2014 | Dec 2011 to Feb 2012 | Amhara | diarrheic patient | <15 yrs | 422 | 33 | 7.82 | 33 | |||||||||
| [37] | 2019 | Feb 2017 to March 2017 | Oromia | diarrheic patient | all | 99 | 133 | 232 | 42 | 18.10 | 22 | 20 | ||||||
| [38] | 2020 | Nov 2016 to Jan 2017 | Amhara | diarrheic patient | all | 181 | 203 | 384 | 20 | 5.21 | 20 | |||||||
| [39] | 2015 | Dec 2011 to Feb 2012 | Amhara | diarrheic patient | <15 yrs | 183 | 239 | 422 | 204 | 48.34 | 204 | |||||||
| [40] | 2016 | Dec 2013 to Mar 2014 | Addis Ababa | diarrheic patient | <15 yrs | 125 | 131 | 256 | 78 | 30.47 | 78 | 7 | 33 | |||||
| [41] | 2019 | January to March 2018 | Amhara | diarrheic patient | <15 yrs | 151 | 121 | 272 | 29 | 10.66 | 29 | |||||||
| [42] | 2018 | Oct 2015 to Feb 2016 | Oromia | diarrheic patient | all | 223 | 199 | 422 | 39 | 9.24 | 30 | 9 | ||||||
| [43] | 2014 | July to Oct 2012 | Oromia | diarrheic patient | <15 yrs | 106 | 121 | 227 | 38 | 16.74 | 38 | |||||||
| [44] | 2018 | Mar to May, 2017 | SNNP | diarrheic patient | <15 yrs | 95 | 72 | 167 | 29 | 17.37 | 21 | 8 | ||||||
| [45] | 2019 | June to Dec 2017 | Gambella | diarrheic patient | <15 yrs | 74 | 60 | 134 | 55 | 41.04 | 4 | 14 | ||||||
| [46] | 2015 | August to Nov 2014 | Tigray | diarrheic patient | all | 109 | 107 | 216 | 15 | 6.94 | 15 | |||||||
| [47] | 2014 | Oct 2011 to June 2012 | SNNP | diarrheic patient | all | 221 | 161 | 382 | 57 | 14.92 | 40 | 17 | ||||||
| [48] | 2011 | Jan to Feb 2007 | Harari | diarrheic patient | all | 119 | 125 | 244 | 45 | 18.44 | 28 | 17 | ||||||
| [49] | 2019 | April to July 2016 | Oromia | diarrheic patient | <15 yrs | 179 | 243 | 422 | 47 | 11.14 | 29 | 18 | ||||||
| [50] | 2018 | Nov 2011 to March 2012 | Tigray | diarrheic patient | <15 yrs | 145 | 115 | 260 | 37 | 14.23 | 19 | 18 | ||||||
| [51] | 2011 | Oct 2006 to March 2007 | Amhara | diarrheic patient | All | 180 | 204 | 384 | 66 | 17.19 | 60 | 6 | ||||||
| [52] | 2021 | Apr to Aug 2019 | SNNPR | diarrheic patient | All | 130 | 133 | 263 | 21 | 7.98 | 1 | 20 | ||||||
| [53] | 2015 | May 2013 to Jan 2014 | Addis Ababa | Diarrheic patient | All | 425 | 532 | 957 | 59 | 6.17 | 59 | |||||||
| [54] | 2020 | January 2017 | Amhara | Diarrheic patient | <15 yrs | 152 | 192 | 344 | 45 | 13.08 | 35 | 6 | 4 | |||||
| [55] | 2016 | - | Oromia | Diarrheic patient | all | 84 | 92 | 176 | 21 | 11.93 | 19 | 2 | ||||||
| [56] | 2015 | Aug.-Dec. 2012 | Addis Ababa | diarrheic patient | <15 yrs | 115 | 138 | 253 | 104 | 41.11 | 61 | 10 | 23 | 10 | ||||
| [57] | 2017 | Feb to May, 2016 | SNNP | HIV-infected with Gastroenteritis | all | 103 | 112 | 215 | 27 | 12.56 | 2 | 11 | 3 | 13 | ||||
| [58] | 2018 | Dec. 2014 to March 2015 | Dire Dawa | Diarrheic patient | <15 yrs | 105 | 91 | 196 | 43 | 21.94 | 25 | 7 | 11 | |||||
| [59] | 2021 | - | Addis Ababa | diarrheic patient | All | 176 | 122 | 298 | 14 | 4.70 | 14 | |||||||
| [60] | 2014 | - | Harari | diarrheic patient | All | 208 | 176 | 384 | 56 | 14.58 | 56 | |||||||
yrs = years, SNNP = Southern Nations, Nationalities, and Peoples’ Region.
Fig 2. Percent of published articles in different regions of Ethiopia.
The country has 9 regions and two city administrations. Reports were from 8 of them which are indicated in the figure. Reports were not found from Afar, Somali, and Benishangul Gumuz regions during the period of data collection (SNNP = South Nation and Nationalities Region).
All studies were cross-sectional; conducted from 2006 to 2020 and published online from 2010 to August 2021. Almost all studies (97.67%) were institution-based (conducted on patients visiting health facilities). The stool samples were the specimen used to isolate the bacterial species. Isolates were characterized and their identities were confirmed by cultural and conventional biochemical tests, but molecular techniques were not used. Patients suffering from diarrhoea were used as the population of the studies and three studies were conducted on diarrhoeic patients with HIV. More than half of the studies (55.81%) were focusing on children less than equal to fifteen years of age. However, 39.53% of studies were considering all age groups (Fig 3 and Table 3).
Fig 3. Age categories and percentage of published articles.
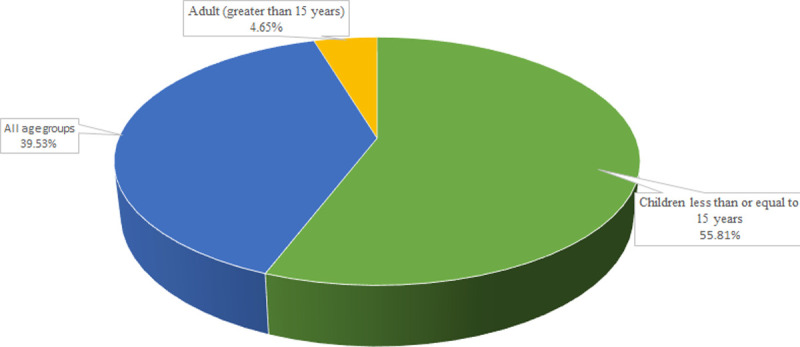
Table 3. Bacterial isolates reported by published articles from diarrheic patient.
| Bacterial isolates | No of Studies | Studies reporting the agent (%) | No of sample | No positive | Positives (%) | Prevalence (%) | ||
|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Pooled (95% CI) | ||||||
| Shigella spp. | 35 | 41.67 | 10,026 | 632 | 5.61 | 1.03 | 37.50 | 6.24 (4.66–7.81) |
| Salmonella enterica | 35 | 38.09 | 11,360 | 644 | 5.67 | 0.38 | 62.50 | 5.06 (4.04–6.09) |
| Escherichia coli | 9 | 7.1 | 2392 | 514 | 21.49 | 0.93 | 51.65 | 19.79 (10.48–29.09) |
| Campylobacter spp. | 5 | 6.0 | 1079 | 123 | 11.40 | 4.12 | 16.74 | 10.76 (5.62–15.91) |
| Citrobacter spp. | 1 | 1.2 | 253 | 10 | 3.95 | - | - | - |
| Enterobacter spp. | 1 | 1.2 | 163 | 2 | 1.23 | - | - | - |
| Klebsiella spp. | 1 | 1.2 | 163 | 11 | 6.75 | - | - | - |
| Proteus spp. | 1 | 1.2 | 163 | 7 | 4.29 | - | - | - |
| Total | 88* | |||||||
*An article may report one or more bacterial species from diarrheic patients; No = Number; % = Percent; CI = confident interval.
Prevalence of Gram-negative enteric bacterial pathogens
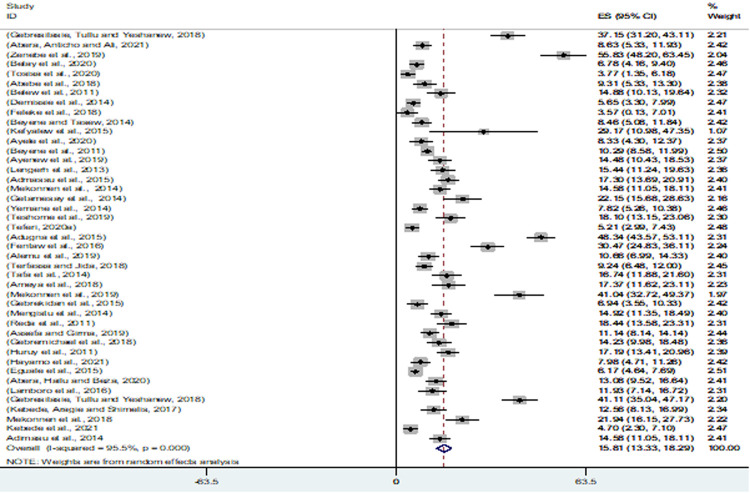
To isolate GNEBPs, in each study, 24 to 1,225 stool samples were collected. Totally, 13,350 stool samples were examined from 3,688 male and 3,822 female diarrheic patients and 1,962 (14.70%) samples were positive for GNEBPs. The minimum and maximum prevalence of GNEBPs in Ethiopia from diarrhoeic patients were 3.57% [27] and 55.83% [21]. The estimated pooled prevalence of GNEBPs in diarrheic patients from 43 studies was 15.81% (95% CI = 13.33–18.29) (Fig 4).
Fig 4. Forest plot of pooled prevalence estimates of Gram-negative enteric bacterial pathogens among diarrheic patients.
The middle solid vertical line represents the minimum possible prevalence value (0). The dashed line represents the mean pooled prevalence estimate. The black diamond at the centre of the grey box represents the point prevalence estimate of each study and the horizontal line indicates the 95% confidence interval of the estimates. The grey box shows the weight of each study contributing to the pooled prevalence estimate. The last row represents the overall pooled prevalence estimate with a 95% confidence interval.
The distribution of the studies using a funnel plot (Fig 5) showed the asymmetrical distribution of effect estimates; hence, there was a publication bias. To minimize the effect of the bias, subgroup analysis was used. Regionally, the pooled prevalence of GNEBPs from diarrheic patients in Addis Ababa, Amhara, SNNPs and Oromia were 20.08, 16.67, 12.12, and 11.61%, respectively. The pooled prevalence based on the study period was 14. 91, 18.03 and 13.46% among studies from 2006–2010, 2011–2015 and 2016–2021, respectively. The pooled prevalence was the highest (20.35%) in children having age less or equal to 15 years, followed by all age groups (10.83%) and adults greater than 15 years (8.51%) (Table 2).
Fig 5. Funnel plot for the prevalence of Gram-negative enteric bacterial pathogens among diarrheic patients.
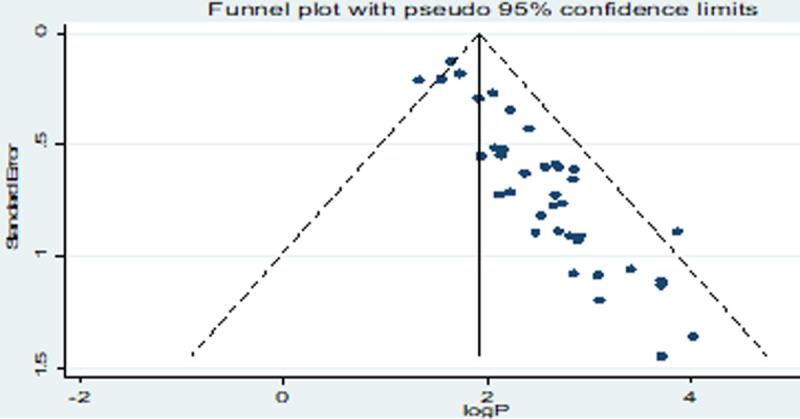
Table 2. The pooled prevalence of Gram-negative enterobacterial pathogens from diarrheic patients based on different subgrouping criteria.
| Subgrouping criteria | Categories | No of studies | Sample examined | No Positive | Pooled prevalence ((%), CI) | I2% (p-value) |
|---|---|---|---|---|---|---|
| Regions | Addis Ababa | 7 | 3532 | 517 | 20.08 (12.85–27.31) | 97.9 (0.00) |
| Amhara | 13 | 4151 | 686 | 16.67 (10.08–22.48) | 97.5 (0.00) | |
| SNNP | 8 | 1847 | 227 | 12.12 (9.15–15.08) | 75.7 (0.00) | |
| Oromia | 8 | 2002 | 225 | 11.61 (7.98–15.24) | 85.6 (0.00) | |
| Tigray | 2 | 476 | 52 | 10.47 (3.33–17.61) | 85.5 (0.00) | |
| Harari | 3 | 1012 | 157 | 15.39 (13.17–17.67) | 0.00 (0.38) | |
| Dire Dawa | 1 | 196 | 43 | 21.94 (16.15–27.73) | - | |
| Gambella | 1 | 134 | 55 | 41.04 (32.72–49.37) | - | |
| Study period | 2006–2010 | 4 | 2068 | 269 | 14.91 (10.44–19.39) | 84.3 (0.00) |
| 2011–2015 | 21 | 6548 | 1106 | 18.03 (13.70–22.36) | 96.8 (0.00) | |
| 2016–2021 | 18 | 4738 | 587 | 13.46 (10.11–16.80) | 93.8 (0.00) | |
| Age of the study subjects | Children <15 years | 24 | 7017 | 1308 | 20.35 (19.92–24.77) | 96.8 (0.00) |
| Adult > 15 years | 2 | 458 | 39 | 8.51 (5.96–11.07) | 0.00 (0.91) | |
| All age groups | 17 | 5882 | 615 | 10.83 (8.69–12.98) | 88.0 (0.00) |
No = number, % = percent, SNNP = Southern Nations, Nationalities, and Peoples’ Region, CI = confident interval, I = Inconsistency Index.
Table 3 shows the types of bacterial isolates reported by published articles from diarrheic patients. Shigella spp. were the most frequent isolate (41.67%), followed by S. enterica (38.09%). The pooled estimate of E. coli was the highest (19.79%) among enteric bacterial isolates. The pooled prevalence of Campylobacter spp., Shigella spp. and Salmonella enterica. were 10.76, 6.24 and 5.06%, respectively.
Escherichia coli was the most common (15.95%) isolate among children less than or equal to fifteen years of age whereas Salmonella enterica and Shigella spp. were common among studies that were focused on all age groups (Tables 1 and 4).
Table 4. Type bacterial isolates among different age groups.
| Age category (years) | Type of bacterial Isolates | No of studies | No of Sample | No Positive | Pooled prevalence ((95% CI) |
|---|---|---|---|---|---|
| < 15 | Campylobacter spp. | 3 | 670 | 102 | |
| E. coli | 8 | 2177 | 524 | 15.95 (14.52–17.38) | |
| Salmonella enterica | 19 | 5893 | 296 | 2.95 (2.52–3.37) | |
| Shigella spp. | 20 | 5767 | 344 | 3.95 (3.45–4.44) | |
| Adult >15 | Campylobacter spp. | 1 | 180 | 8 | |
| Salmonella spp. | 2 | 458 | 20 | ||
| Shigella spp. | 2 | 458 | 11 | ||
| All age groups | Campylobacter spp. | 1 | 215 | 13 | |
| E. coli | 1 | 215 | 2 | ||
| Salmonella enterica | 14 | 5067 | 381 | 3.21 (2.74–3.69) | |
| Shigella spp. | 13 | 3859 | 221 | 3.14 (2.59–3.68) |
No = Number; % = Percent; CI = confident interval.
Drug resistance patterns of Gram-negative enteric bacterial pathogens
Drug resistance in Shigella spp
Twenty-six antimicrobial panels were used to assess the drug resistance pattern of Shigella spp. The highest percentage of resistance was reported among members of the penicillin group such as ampicillin (85.01%) and amoxicillin (82.07%). Resistance against the tetracycline group was also very common (67.01%). A considerable proportion of resistance was also reported among cephalosporin groups, particularly on the first-generation agents and erythromycin. Resistance was not reported against carbapenems (Table 5).
Table 5. Prevalence of Shigella spp. resistance to different antimicrobial agents in Ethiopia.
| Antimicrobial Agents | The Main group of antimicrobial agents | No of studies | The total no of isolates tested | No of resistant isolate | Resistant isolate (%) | Pooled prevalence (%) (95% CI) | I2% (p-value) |
|---|---|---|---|---|---|---|---|
| Ampicillin | Penicillins | 30 | 519 | 452 | 87.09 | 85.01 (79.79–90.22) | 61.4 (0.00) |
| Amoxicillin | Penicillins | 14 | 175 | 153 | 87.43 | 82.07 (74.05–89.65) | 0.00 (0.47) |
| Augmentin (Amoxicillin and clavulanate potassium) | Penicillins and β-lactamase inhibitors | 10 | 196 | 91 | 46.43 | 43.87 (22.5–65.24) | 93.1 (0.00) |
| Tetracycline | Tetracyclines | 22 | 404 | 309 | 76.49 | 67.01 (55.83–78.36) | 89.9(0.00) |
| Doxycycline | Tetracyclines | 2 | 20 | 16 | 80.00 | - | - |
| Cephalothin | 1st G cephalosporins | 4 | 57 | 40 | 70.18 | 70.18 (60.56–100.65) | 100(-) |
| Cefoxitin | 2nd G cephalosporins | 1 | 20 | 6 | 30.00 | - | - |
| cefuroxime | 2nd G cephalosporins | 1 | 20 | 13 | 65.00 | - | - |
| Cefaclor | 2nd G cephalosporins | 1 | 17 | 11 | 64.71 | - | - |
| Ceftriaxone | 3rd G cephalosporins | 14 | 151 | 16 | 10.60 | 29.53 (7.80–51.25) | 79.5 (0.001) |
| ceftazidime | 3rd G cephalosporins | 6 | 62 | 7 | 11.29 | 8.24 (0.42–16.05) | 0.00 (0.46) |
| cefotaxime | 3rd G cephalosporins | 2 | 20 | 4 | 20.00 | - | - |
| Ceftizoxime | 3rd G cephalosporins | 1 | 40 | 11 | 27.5 | - | - |
| Chloramphenicol | Chloramphenicol | 30 | 508 | 220 | 43.31 | 36.95 (29.65–44.24) | 66.7 (0.00) |
| Kanamycin | Aminoglycosides | 2 | 34 | 7 | 20.59 | - | |
| Gentamicin | Aminoglycosides | 27 | 443 | 88 | 19. 86 | 25.38 (17.78–32.99) | 70.4 |
| Amikacin | Aminoglycosides | 3 | 22 | 2 | 9.09 | 14.29 (-4.04–32.62) | 100(-) |
| Streptomycin | Aminoglycosides | 1 | 32 | 31 | 96.87 | - | - |
| Nalidixic acid | Fluoroquinolones | 17 | 191 | 32 | 16.75 | 17.14 (11.34–22.94) | 0.00(0.69) |
| Norfloxacin | Fluoroquinolones | 14 | 229 | 14 | 6.11 | 8.34 (4.02–12.66) | 0.00 (0.976) |
| Ciprofloxacin | Fluoroquinolones | 28 | 468 | 32 | 6.84 | 11.86 (5.31–18.42) | 65.6 (0.001) |
| Trimethoprim-sulphamethoxazole | Folic acid metabolism inhibitors | 28 | 476 | 270 | 56.72 | 53.00 (44.34–61.67) | 72.90 (0.00) |
| Meropenem | Carbapenems | 2 | 11 | 0 | 0 | - | - |
| Erythromycin | Macrolides | 5 | 38 | 26 | 68.42 | 69.67(46.56–92.77) | 59.9 (0.058) |
| Azithromycin | Macrolides | 2 | 31 | 10 | 32.26 | - | - |
| Clindamycin | Macrolides | 1 | 8 | 4 | 50 | - | - |
No = Number; % = Percent; CI = confident interval, I = Inconsistency Index.
Drug resistance of Salmonella enterica
Twenty-five antimicrobial panels were used to assess the drug resistance patterns of S. enterica. The highest percentage of resistance was reported among members of the penicillin group such as ampicillin (64.98%) and amoxicillin (82.89%). Resistance against tetracycline and trimethoprim-sulphamethoxazole were also common. A considerable proportion of resistance was also reported against erythromycin and cephalosporin groups. Resistance was not reported against doxycycline, meropenem and azithromycin (Table 6).
Table 6. The pooled prevalence of Salmonella enterica resistance to different antimicrobial agents in Ethiopia.
| Antimicrobial Agents | The main group of antimicrobial agents | No of studies | The Total no of isolates tested | No of resistant isolate | Resistant isolate (%) | Pooled prevalence (%) (95% CI) | I2% (p-value) |
|---|---|---|---|---|---|---|---|
| Ampicillin | Penicillins | 29 | 588 | 447 | 76.02 | 64.98 (45.2–84.76) | 96.5 (0.00) |
| Amoxicillin | Penicillins | 12 | 255 | 223 | 87.45 | 82.89 (70.58–95.21) | 62.0 (0.072) |
| Augmentin (Amoxicillin and clavulanate potassium) | Penicillins and β-lactamase inhibitors | 8 | 185 | 75 | 40.54 | 45.34 (19.11–71.56) | 95.2 (0.00) |
| Tetracycline | Tetracycline | 24 | 555 | 287 | 51.71 | 54.59 (41.28–67.90) | 90.03 (0.00) |
| Doxycycline | Tetracycline | 2 | 34 | 0 | 0.00 | - | - |
| Cephalothin | 1st G cephalosporins | 2 | 9 | 1 | 11.11 | - | - |
| Cefaclor | 2nd G cephalosporins | 1 | 4 | 4 | 100.00 | - | - |
| Cefoxitin | 2nd G cephalosporins | 1 | 33 | 9 | 27.27 | - | - |
| Ceftizoxime | 2nd G cephalosporins | 1 | 21 | 5 | 23.81 | - | - |
| cefuroxime | 2nd G cephalosporins | 2 | 55 | 16 | 29.09 | - | - |
| Ceftriaxone | 3rd G cephalosporins | 16 | 384 | 109 | 28.39 | 33.1 (7.88–58.33) | 98.0 (0.00) |
| ceftazidime | 3rd G cephalosporins | 5 | 27 | 4 | 14.81 | 29.22 (22.42–8086) | 81.7 (0.02) |
| cefotaxime | 3rd G cephalosporins | 2 | 7 | 2 | 28.57 | - | - |
| Chloramphenicol | Chloramphenicol | 29 | 629 | 260 | 41.34 | 42.39 (27.39–57.38) | 96.1(0.00) |
| Kanamycin | Aminoglycosides | 3 | 73 | 24 | 32.88 | 33.7 (22.72–44.69) | 0.00 (0.67) |
| Gentamicin | Aminoglycosides | 24 | 491 | 121 | 24.64 | 17.36 (7.57–27.15) | 93.4 (0.00) |
| Amikacin | Aminoglycosides | 3 | 26 | 1 | 3.85 | - | - |
| Nalidixic acid | Fluoroquinolones | 19 | 462 | 66 | 14.29 | 14.60 (9.23–19.97) | 64.2 (0.00) |
| Norfloxacin | Fluoroquinolones | 10 | 200 | 9 | 4.50 | 5.17 (1.79–8.55) | 0.00(0.98) |
| Ciprofloxacin | Fluoroquinolones | 28 | 28 | 28 | 5.29 | 7.66 (2.67–12.66) | 74.8 (0.00) |
| Trimethoprim-sulphamethoxazole | Folic acid metabolism inhibitors | 25 | 464 | 218 | 46.98 | 46.72 (29.74–61.69) | 94.4 (0.00) |
| Meropenem | Carbapenems | 2 | 20 | 0 | 0.00 | - | - |
| Erythromycin | Macrolides | 5 | 45 | 25 | 55.56 | 52.97 (37.96–68.72) | 5.8 (0.364) |
| Azithromycin | Macrolides | 1 | 5 | 0 | 0.00 | - | - |
| Clindamycin | Macrolides | 1 | 113 | 1 | 0.88 | - | - |
No = Number; % = Percent; CI = confident interval, I = Inconsistency Index.
Drug resistance of Escherichia coli
Sixteen antimicrobial panels were used to assess the drug resistance pattern of E. coli. The highest percentage of resistance was reported on agents like ampicillin (77.97%). Resistance against tetracycline (76.87%) and trimethoprim-sulphamethoxazole (66.97%) were also common. A considerable proportion of resistance was also reported among cephalosporin groups. Resistance was not reported against meropenem (Table 7).
Table 7. The pooled prevalence of Escherichia coli resistance to different antimicrobial agents in Ethiopia.
| Antimicrobial Agents | The main group of antimicrobial agents | No of studies | The total no of isolates tested | No of resistant isolate | Resistant isolate (%) | Pooled prevalence (%) (95% CI) | I2 (p-value) |
|---|---|---|---|---|---|---|---|
| Ampicillin | Penicillins | 6 | 446 | 359 | 80.49 | 77.97 (70.17–85.76) | 71.4 (0.00) |
| Amoxicillin | Penicillins | 1 | 47 | 5 | 10.64 | - | - |
| Augmentin (Amoxicillin and clavulanate potassium) | Penicillins and B-lactamase inhibitors | 5 | 339 | 204 | 60.18 | 64.78 (42.00–87.57) | 95.0 (0.00) |
| Tetracycline | Tetracyclines | 3 | 253 | 194 | 76.68 | 76.87 (71.69–82.05) | 0.00 (0.563) |
| Ceftriaxone | 3rd G cephalosporins | 5 | 284 | 11 | 3.87 | 2.91 (0.74–5.08) | 11.5 (0.34) |
| Cephalothin | 1st G cephalosporins | 1 | 47 | 8 | 17.02 | - | - |
| cefotaxime | 3rd G cephalosporins | 1 | 204 | 50 | 24.51 | - | - |
| Gentamicin | Aminoglycosides | 6 | 388 | 102 | 26.29 | 19.72 (7.41–32.03) | 88.03 (0.00) |
| Amikacin | Aminoglycosides | 1 | 113 | 2 | 1.77 | - | - |
| Chloramphenicol | Chloramphenicol | 5 | 375 | 121 | 32.27 | 30.39 (20.95–39.84) | 67.0 (0.016) |
| Nalidixic acid | Fluoroquinolones | 5 | 224 | 38 | 16.96 | 16.71 (11.81–21.6) | 0.00 (0.713) |
| Norfloxacin | Fluoroquinolones | 2 | 206 | 20 | 9.71 | - | - |
| Ciprofloxacin | Fluoroquinolones | 7 | 439 | 27 | 6.15 | 5.57 (3.42–7.71) | 0.00 (0.063) |
| Trimethoprim-sulphamethoxazole | Folic acid metabolism inhibitors | 7 | 428 | 301 | 70.33 | 66.97 (56.21–77.71) | 80.7 (0.00) |
| Meropenem | Carbapenems | 1 | 133 | 0 | 0 | - | - |
| Erythromycin | Macrolides | 1 | 2 | 1 | 50 | - | - |
No = Number; % = Percent; CI = confident interval; I = Inconsistency Index.
Drug resistance of Campylobacter spp
Eighteen antimicrobial panels were used to assess the drug resistance patterns of Campylobacter spp. The highest percentage of resistance was reported for Cephalothin (81.52%). Resistance against ampicillin (65.61%) and trimethoprim-sulphamethoxazole (52.6%) were also common. Resistance was not reported against meropenem and azithromycin (Table 8).
Table 8. Pooled prevalence of Campylobacter spp. resistance to different antimicrobial agents in Ethiopia.
| Antimicrobial Agents | The main group of antimicrobial agents | No of studies | The total no of isolates tested | No of resistant isolate | Resistant isolate (%) | Pooled prevalence (%) (95% CI) | I2 (p-value) |
|---|---|---|---|---|---|---|---|
| Ampicillin | Penicillins | 4 | 110 | 72 | 65.45 | 65.61(44.90–86.32) | 83.3 (0.000 |
| Amoxicillin | Penicillins | 1 | 20 | 16 | 80.00 | - | - |
| Augmentin (Amoxicillin and clavulanate potassium) | Penicillins and B-lactamase inhibitors | 1 | 44 | 16 | 36.36 | - | - |
| Tetracycline | Tetracyclines | 5 | 123 | 60 | 48.78 | 42.17 (20.30–64.05) | 85.4 (0.00) |
| Doxycycline | Tetracyclines | 3 | 90 | 16 | 17.78 | 18.94 (10.5–27.38) | 0.00 (0.379) |
| Ceftriaxone | 3rd G cephalosporins | 4 | 85 | 15 | 17.65 | 39.43 (0.96–77.91) | 79.0 (0.029) |
| ceftazidime | 3rd G cephalosporins | 1 | 8 | 2 | 25.00 | - | - |
| Cephalothin | 1st G cephalosporins | 3 | 102 | 91 | 89.22 | 81.52 (63.78–99.27) | 63.2 (0.09) |
| Gentamycin | Aminoglycosides | 5 | 123 | 29 | 23.58 | 30.70 (8.04–53.36) | 88.0 (0.00) |
| Chloramphenicol | Chloramphenicol | 5 | 123 | 26 | 21.14 | 28.03 (10.33–45.85) | 74.4 (0.00) |
| Nalidixic acid | Fluoroquinolones | 4 | 115 | 13 | 11.30 | 10.45 (4.89–16.00) | 0.00 (0.711) |
| Norfloxacin | Fluoroquinolones | 3 | 95 | 10 | 10.53 | 12.02 (4.99–19.05) | 0.00 (0.665) |
| Ciprofloxacin | Fluoroquinolones | 5 | 123 | 19 | 15.45 | 13.9 (7.87–19.94) | 0.00 (0.528) |
| Trimethoprim-sulphamethoxazole | Folic acid metabolism inhibitors | 5 | 123 | 67 | 54.47 | 52.6 (33.76–71.43) | 78.4 (0.00) |
| Meropenem | Carbapenems | 1 | 8 | 0 | 0.00 | - | - |
| Erythromycin | Macrolides | 5 | 123 | 37 | 30.08 | 35.72 (18.34–35.10) | 78.4 (0.001) |
| Azithromycin | Macrolides | 1 | 8 | 0 | 0.00 | - | - |
| Clindamycin | Macrolides | 2 | 82 | 28 | 34.15 | - | - |
No = Number; % = Percent; CI = confident interval; I = Inconsistency Index.
Drug resistance of other enteric bacterial species
The number of published articles on other enteric bacterial species from diarrheic patients was very limited and impossible to summarize. However, one study conducted by Zenebe et al. [21] reported the presence of Klebsiella spp., Proteus spp., Enterobacter spp. in under-five children with diarrhoea. These bacteria were showing antimicrobial resistance character as indicated in Table 9.
Table 9. Pooled prevalence of other enteric bacterial resistance to different antimicrobial agents in Ethiopia.
| References | bacterial isolate | Ampicillin | Chloramphenicol | Gentamicin | Nalidixic acid | Tetracycline | Ciprofloxacin | Trimethoprim-sulphamethoxazole | Ceftriaxone | Amoxicillin | Cephalothin | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | No isolate tested | No resistance | % | ||
| [21] | Klebsiella | 11 | 3 | 27.27 | 11 | 2 | 18.18 | 11 | 4 | 36.36 | 11 | 1 | 9.09 | 11 | 9 | 81.82 | 11 | 0 | 0.00 | 11 | 8 | 72.73 | 11 | 0 | 0.00 | 11 | 4 | 36.36 | 11 | 2 | 18.18 |
| [21] | Proteus | 7 | 2 | 28.57 | 7 | 1 | 14.29 | 7 | 3 | 42.86 | 7 | 4 | 57.14 | 7 | 2 | 28.57 | 7 | 0 | 0.00 | 7 | 2 | 28.57 | 7 | 1 | 14.29 | 7 | 3 | 42.86 | 7 | 0 | 0.00 |
| [21] | Enterobacter | 2 | 1 | 50.0 | 2 | 0 | 0.00 | 2 | 1 | 50.0 | 2 | 1 | 50.0 | 2 | 0 | 0.0 | 2 | 0 | 0.0 | 2 | 0 | 0.0 | 2 | 0 | 0.0 | 2 | 0 | 0.0 | 2 | 0 | 0.00 |
Multidrug resistance
Out of 43 published articles on enteric bacterial pathogens, 32 (74.42%) reported multidrug-resistant (MDR) characters among the isolates. Among 1470 bacterial isolates, 1104 (75.52%) with a pooled prevalence of 70.56% (CI = 64.56–76.77%) were resistant to three or more antimicrobial agents (multidrug resistance). The report showed that the pooled prevalence of MDR in Campylobacter spp., Shigella spp., E. coli and S. enterica were 80.78, 79.08, 78.20 and 59.46%, respectively (Table 10).
Table 10. Multidrug resistance pattern of enteric bacteria pathogens from diarrheic patients.
| Type bacterial isolate | No of studies | The total no of isolates tested | No of multidrug-resistant isolates | Multidrug-resistant isolate (%) | Pooled prevalence (%) (95% CI) | I2 (p-value) |
|---|---|---|---|---|---|---|
| Shigella spp. | 24 | 443 | 363 | 81.94 | 79.08 (72.19–85.97) | 68.2 (0.00) |
| Salmonella enterica | 23 | 496 | 317 | 63.91 | 59.46 (46.13–72.79) | 91.1 (0.00) |
| Escherichia coli | 6 | 410 | 332 | 80.98 | 78.20 (67.46–88.93) | 84.7 (0.00) |
| Campylobacter spp. | 4 | 110 | 87 | 79.09 | 80.78 (65.04–96.52) | 94.6 (0.00) |
| Klebsiella spp. | 1 | 11 | 5 | 45.45 | - | - |
| 1470 | 1104 | 75.52 |
No = number, % = percent; CI = confidence interval, I = Inconsistency Index.
Discussion
Diarrhoea is a common health problem, causing mortality and morbidity for thousands of people around the globe. Both infectious and non-infectious agents can induce the problem, among the infectious agents, enteric bacterial pathogens like diarrheagenic E. coli, S. enterica, Shigella spp., and Campylobacter spp. play important roles in the induction or severity of diarrhoea [61]. Determining their burden in a given population is very essential to design strategies for the reduction of the incidence and influences of diarrhoea. The minimum and maximum prevalence of GNEBPs in Ethiopia from diarrhoeic patients were 3.57% [27] and 55.83% [21], respectively. The pooled prevalence of EBP isolates from the stool of diarrheic patients in Ethiopia was 15.81% (CI = 13.33–18.29). In line with this finding, Getie et al. [62] reported 13.2% prevalence of GNEBP in other groups of population (food handlers) in Gondar town, Northwest Ethiopia. On the other hand, Shah et al. [63] reported a 33.62% prevalence of GNEBP in Kenya. The difference may be due to the detection methods since they use molecular techniques in addition to the conventional culturing methods.
Almost all studies (97.67%) were institutional, and samples were collected from patients visiting health facilities. Focusing only on health facilities may not reflect the overall prevalence of GNEBPs and their drug resistance patterns in the country. Since some GNEBP infection cases may not arrive at health institutions and widespread use or misuse of antimicrobial drugs in the community may accelerate the occurrence of antimicrobial resistance [64,65].
Sub-grouping of the prevalence of GNEBPs based on the studies conducted in different regions of the country showed that the pooled prevalence was high in Addis Ababa (20.08%). In line with this, a spatial variation across the regions of Ethiopia was reported by Bogale et al. [66]. The difference based on the study period was not significant. However, a declining pattern of diarrhoea at the national level was reported by Bogale et al. [66]. Published articles were from 8 regions of Ethiopia, published articles were not found in Afar, Somali and Benishangul Gumuz regions of the country in this study. Hence, the pooled prevalence was calculated from 8 regions ignoring others. However, the scenario may not be equivalent to the region of the country having no reports.
Grouping of study participants based on their age showed that the pooled prevalence was the highest (20.35%) in children having age below or equal to 15 years which indicated that children are more exposed to the GNEBPs and express severe syndromes to visit health facilities. In line with this, Kotloff [4] and Havelaar et al. [67] reported that children contribute a huge proportion of diarrheal diseases in the world.
Diarrheagenic E. coli, Shigella spp., S. enterica and Campylobacter spp. were the most common isolates among GNEBPs. In line with this, Getie et al. [62] reported that Shigella spp. enterohemorrhagic E. coli (EHEC) and S. enterica were important isolates of GNEBPs among food handlers. In this study, the pooled estimate of E. coli was the highest (19.79%) among enteric bacterial pathogens. The result is very close to the report of Zenebe et al. [68] who reported that the pooled prevalence of E. coli was 25% in Ethiopia. A 33.8% pooled prevalence was reported by Oppong et al. [69] in Sub-Sharan Africa. Oppong et al. [69] also found that E. coli detection was the highest in the East African region and lowest in the middle part of Africa. The difference may be due to the number of studies in the summary and the targeted strain of E. coli. For example, in a single study in Niger, 11.1% of diarrhoeic children were positive for diarrheagenic E. coli [70]. Similarly, the Global enteric multicentre study on infants and children showed that diarrheagenic E. coli was among the four major pathogens responsible for diarrhoea in low-income and middle-income countries [71].
The pooled prevalence of Campylobacter spp. in this analysis was 10.76%. In line with this report, Kassie et al. [72] reported a prevalence of 10.5% among children in Denbia district, Ethiopia and 13.8% prevalence was also reported by Gedlu and Aseffa [73] among children in northwest Ethiopia. Oppong et al. [69] reported a 12.3% pooled prevalence of Campylobacter in East Africa. A high rate of Campylobacter infection among children was also reported in Kenya [63]. In contrary to the report of this study, Fletcher et al. [74] reported a pooled prevalence of 2.7% in Sub-Saharan countries. The difference may be related to the area coverage, disease prevention and control practices.
The pooled prevalence of Shigella spp. in this study was 6.24% which is equivalent to the report of Hussen et al. [75]. According to their report, the pooled prevalence of Shigella spp. in Ethiopia was 6.6%. Similarly, Oppong et al. [69] reported a pooled prevalence of 5.6% from children under five and Fletcher et al. [74] 4.3% of children aged less than 12 years in Sub-Saharan countries.
The pooled prevalence of S. enterica was 5.06% which is almost comparable with the reported pooled prevalence by Abate and Assefa [76], which was 4.8% among human stools and animal origin foods in Ethiopia.
Resistance to antimicrobial agents is a natural evolutionary process for the bacteria, however, the process is accelerated by human activities in terms of antimicrobial usage patterns and infection control or prevention practices. The risks are very high in developing countries like Ethiopia where there is a widespread use or misuse of antimicrobial agents with a high burden of infectious diseases [76–79].
In this meta-analysis, Shigella isolates were more resistant to the penicillin group of antimicrobial agents like ampicillin (85.01%) and amoxicillin (82.07%). A high percentage of resistance against ampicillin (83.1%) and amoxicillin (84.1%) were also reported by Hussen et al. [75]. There were also reports of drug resistance among the first-line drugs like ciprofloxacin (11.86%) and ceftriaxone (29.53%) for the treatment of Shigellosis. Resistance development against such types of antimicrobial agents was also reported by Hussen et al. [75]. They reported 8.9 and 9.3% resistance against ciprofloxacin and ceftriaxone, respectively.
In this analysis, high proportions of Salmonella isolates were also resistant against the penicillin group of antimicrobial agents like ampicillin (64.98%) and amoxicillin (82.89%). In line with this Tadesse [80] was also reported a high percentage (86.01%) of resistance of Salmonella against ampicillin and/or amoxicillin. Resistance of Salmonella isolates to fluoroquinolone like ciprofloxacin (2.27%) and third-generation cephalosporin (ceftriaxone 16.68%) were also reported. Similarly, according to Tadesse’s report, the pooled prevalence of ciprofloxacin resistance among Salmonella isolates was 3.61% [80] and a prevalence of 2.9% of resistance against ciprofloxacin was also reported in Iran [81].
Resistance was common among E. coli isolates in this analysis, particularly on antimicrobial agents like ampicillin (77.97%), tetracycline (76.87%) and trimethoprim-sulphamethoxazole (66.97%). Resistance of E. coli against a wide array of antimicrobials was also reported by Pormohammad et al. [82], Zenebe et al. [68] and Tuem et al. [83]. Resistance in non-pathogenic strains of E. coli may not have a direct effect on health, however, non-pathogenic resistant strains may acquire virulence genes and induce disease that may not be treated easily or non-pathogenic strains having resistant character may act as a reserve for the resistant character for other bacteria [82].
Among Campylobacter isolates in this analysis, the highest percentage of resistance was reported on antimicrobial agents like cephalothin (81.52%), ampicillin (65.61%) and trimethoprim-sulphamethoxazole (52.60%). A resistance pattern of 92.3–100% to erythromycin and the β—lactams, 61.5–86.7% to trimethoprim-sulfamethoxazole, 92.3–93.3% to tetracycline, 46.2–80% to chloramphenicol, 0–60% to aminoglycosides and 0% to imipenem were reported among Campylobacter spp. in Ghana [84].
In this study, among 1470 bacterial isolates 1104 (75.52%) with a pooled prevalence of 70.56%, were resistant to three or more antimicrobial agents (multidrug resistance) (MDR). In line with this finding, Alemayehu [85] reported that the pooled prevalence of multidrug resistance was 70.5% among bacterial isolates in Ethiopia. Another, meta-analysis study on multidrug resistance by Abayneh et al. [86] reported the pooled prevalence of 80.5% among Gram-negative bacteria. It was also very close to the reports from India (66.12%) [87] and Egypt (65.5%) [88,89]. In contrast to our finding in Ethiopia, a lower prevalence of MDR has been reported from Germany (60%) [90], Nepal (42.6%) [91], Australia (36%) [92], Indonesia (28.7%) [93], the USA (27%) [94], Spain (34.5%) [95] and France (11.6%) [96]. Several factors may play a role in the difference including the magnitude and style of antimicrobial use and infection prevention practices.
The MDR report among Campylobacter isolates in this analysis (80.78%) was higher than the report from Bangladesh (28.8%) [97] and Kenya 50% [98] but lower than the report from Ghana (97%) [84]. This may be due to differences in the use of antimicrobial agents, the area coverage, sample type and technique of detections. The MDR report of E. coli in this analysis (78.20) was not in line with other reports like 50% prevalence in Nigeria [99], 40% in Spain [95], 22% among human isolates in the world [82], 26% in China [100], 39.8% in Egypt isolates from animals [101], 28% in low and middle-income countries [102]. The MDR report of this meta-analysis among Shigella isolates (79.08) was almost similar to the report by Hussen et al. [75] (83.2%) but it was lower than other reports from Iran (89.4%) [103], and Bangladesh (94%) [104]. However, it was higher than other reports such as 53.8% among migrants in Europe [74], 60% in Kenya [98] and 19% in Somalia [105]. Factors like the magnitude and style of antimicrobial use and infection prevention practices may play roles in the differences.
The development of MDR character among S. enterica is also an important public health concern around the globe [106]. In this analysis, the prevalence of MDR character among Salmonella isolate was 59.46% which is comparable with the MDR report of Garedew et al. [107] (46.2%) among food handlers in Gondar. However, it was lower than the report from Dagnew et al. [108] (76%) and Admassu et al. [109] (100%). The difference may be due to the target population and study methods (single versus pooled meta-analysis reports).
Limitations of the study
Some of the studies included in the analysis were targeting the most common bacterial pathogens that did not rule out the absence of others. Therefore, for bacteria that are thought to be less frequent, the reported prevalence may not accurate. Publication bias and heterogenicity were observed in the analysis, but attempts were made to reduce their impact on the analysis by following the random effect model and subgrouping. However, these may not totally avoid their impact on the interpretation of the pooled results.
Conclusion
According to this analysis, the burden of Gram-negative enteric bacterial pathogens (GNEBPs) was high and may be considered a major cause of diarrhoea in Ethiopia. A significant proportion of the isolates exhibited resistance to the commonly used antibacterial agents which are expected to affect the treatment response and cost, morbidity and progression of the infection. Shigella spp., S. enterica, E. coli and Campylobacter spp. were the commonly isolated GNEPB from diarrheic patients in the country. The pooled estimate of Campylobacter spp. was the highest followed by E. coli, Shigella spp. and S. enterica. Resistance to antimicrobial agents was most common among the penicillin groups, followed by tetracycline, and trimethoprim-sulphamethoxazole. However, there were also resistant strains against very relevant drugs for the treatment of GNEBP such as fluoroquinolones and thirdgeneration cephalosporins. Almost all isolates were susceptible to meropenem.
Since the incidences of these bacterial diseases are related to hygiene, all activities that enhance hygienic practices (clean water and food, handwashing, proper use of latrine) must be advocated and implemented. Performing drug sensitivity tests for suspected diarrheagenic bacteria is extremely advantageous to select the appropriate antimicrobial drugs for the treatment. The antimicrobial resistance surveillance system must be established to understand the trend of resistance among pathogenic bacteria and to plan and implement mitigating strategies like proper control and prevention of infectious diseases and antimicrobial stewardship programs. Almost all studies were using conventional techniques for confirmation of the isolates, thus, adding molecular methods in the future will increase analysis precision.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank the researchers who publish their research work and make it available to the public through the internet.
Abbreviations
- GNEBPs
Gram-Negative Enteric Bacterial Pathogens
- HIV
Human Immunodeficiency Virus
- I2
Inconsistency Index
- KAP
Knowledge, Attitude and Practices
- MESH
Medical Subject headlines
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- SNNP
South Nation Nationalist People’sregion
- UN
United Nations
- USA
United State of America
- WHO
World Health Organization
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Caramia G, Silvi S, Verdenelli MC, Magdalena M. Treatment of acute diarrhoea: past and now. Int J Enteric Pathog. 2015; 3: e28612. doi: 10.17795/ijep28612 [DOI] [Google Scholar]
- 2.WHO. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007–2015. 2015. Available from: https://apps.who.int/iris/bitstream/handle/10665/199350/9789241565165_eng.pdf?sequence477= 1&isAllowed=y. [Google Scholar]
- 3.Girmay AM, Gari SR, Alemu BM, Martin R. Diarrheal disease and associated behavioural factors among food handlers in Addis Ababa, Ethiopia. AIMS Public Heal., 202; 7:100–113. doi: 10.3934/publichealth.2020010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotloff KL. The burden and etiology of diarrheal illness in developing countries. Pediatr. Clin North Am. 2017; 64:799–814. doi: 10.1016/j.pcl.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 5.Troeger C, Blacker BF, Khalil IA, Rao PC, Cao S, Zimsen SR, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the global burden of disease study 2016. Lancet Infect Dis. 2018; 18:1211–1228. doi: 10.1016/S1473-3099(18)30362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alebel A, Tesema C, Temesgen B, Gebrie A, Petrucka P, Kibret GD. Prevalence and determinants of diarrhoea among under-five children in ethiopia: A systematic review and meta-analysis. PLoS ONE. 2018: 13:e0199684. doi: 10.1371/journal.pone.0199684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ugboko HU, Nwinyi OC, Oranusi SU, Oyewale JO. Childhood diarrhoeal diseases in developing countries. Heliyon, 2020; 6:e03690. doi: 10.1016/j.heliyon.2020.e03690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks GF, Morse SA, Carroll KC, Mietzner TA, Butel JS. Jawetz, Melnick, & Adelberg’s Medical Microbiology. 26th ed. MC Graw Hill Medical, New York. 2013. [Google Scholar]
- 9.Lindahl JF, Grace D. The consequences of human actions on risks for infectious diseases: a Review. Infect Ecol Epidemiol. 2015; 5:30048. doi: 10.3402/iee.v5.30048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasovský D, Littmann J, Zorzet A, Cars O. Antimicrobial resistance: A threat to the world’s sustainable development. Ups J Med Sci. 2016; 121:159–164. doi: 10.1080/03009734.2016.1195900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.UN. United Nations general assembly. Draft resolution submitted by the president of the general assembly political declaration of the high-level meeting of the general assembly on antimicrobial resistance, Seventy-first session agenda item 127 Global health. 2016. [Google Scholar]
- 12.O’Neill J. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. 2014. https://iiif.wellcomecollection.org/file/b28552179_AMR%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations.pdf. [Google Scholar]
- 13.Beyene G, Nair S, Asrat D, Mengistu Y, Engers H, Wain J. Multidrug resistant Salmonella concord is a major cause of salmonellosis in children in Ethiopia. J Infect Dev Ctries. 2011; 5:023–033. doi: 10.3855/jidc.906 [DOI] [PubMed] [Google Scholar]
- 14.Teferi SC. Prevalence and antimicrobial resistance patterns of Shigella in Ethiopia from 2000 to 2018: A critical review. Chem Biomol Eng. 2020; 5:51–56. [Google Scholar]
- 15.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 Statement: An updated guideline for reporting systematic reviews. Syst Rev. 2021; 10:89. doi: 10.1186/s13643-021-01626-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.JBI. The joanna briggs institute critical appraisal tools for use in jbi systematic reviews; Checklist for systematic reviews and research syntheses. 2017. Available from: https://jbi.global/sites/default/files/2019-05/JBI-Critical-Appraisal-checklist-for-Systematic-Reviews2017-0.pdf. [Google Scholar]
- 17.Rücker G, Schwarzer G, Carpenter JR, Schumacher M. Undue reliance on I2 in assessing heterogeneity may mislead. BMC Med Res Methodol. 2008; 9:79. http://www.biomedcentral.com/1471-2288/8/79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haidich A. Meta-analysis in medical research. Hippokratia. 2010; 14:29–37. [PMC free article] [PubMed] [Google Scholar]
- 19.Gebresilasie YM, Tullu KD, Yeshanew AG. Resistance pattern and maternal knowledge, attitude and practices of suspected diarrheagenic Escherichia coli among children under 5 years of age in Addis Ababa, Ethiopia: A cross-sectional study. Antimicrob resist infect control. 2018; 7:110. doi: 10.1186/s13756-018-0402-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abera K, Anticho T L, Ali MM. Salmonella and Shigella and antimicrobial susceptibility profiles among adult patients with complaints of diarrhea at Hawassa comprehensive specialized. SAGE Open Med. 2021; 9:1–6. doi: 10.1177/20503121211000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zenebe T, Gebreyes D, Tesema A, Craddock H, Gishen N. Enteropathogens in under-five children with diarrhea in health facilities of Debre Berhan Town, North Shoa, Ethiopia. Ethiop J Heal Sci. 2019; 29:203. 10.4314/ejhs.v29i2.7, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belay A, Ashagrie M, Seyoum B, Alemu M, Tsegaye A. Prevalence of enteric pathogens, intestinal parasites and resistance profile of bacterial isolates among HIV infected and non-infected diarrheic patients in Dessie Town, Northeast. PLoS One, 2020; 15: e0243479. doi: 10.1371/journal.pone.0243479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tosisa W, Mihret A, Ararsa A, Eguale T, Abebe T. Prevalence and antimicrobial susceptibility of Salmonella and Shigella species isolated from diarrheic children in Ambo town. BMC Pediatr. 2020; 20:91. doi: 10.1186/s12887-020-1970-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abebe W, Earsido A, Taye S, Assefa M, Eyasu A, Godebo G. Prevalence and antibiotic susceptibility patterns of Shigella and Salmonella among children aged below five years with diarrhoea attending Nigist Eleni Mohammed memorial hospital, South Ethiopia. BMC Pediatr. 2018; 18:241. doi: 10.1186/s12887-018-1221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belew GD, Kibret M, Biadglegne F, Abera B. Prevalence and antimicrobial susceptibility patterns of Shigella species at Felege Hiwot Referral Hospital, Northwest Ethiopia. Ethiop Med J. 2011; 49:3. [PubMed] [Google Scholar]
- 26.Demissie TA, Wubie MT, Yehuala FM, Fetene DM, Gudeta GA. Prevalence and Antimicrobial susceptibility patterns of Shigella and Salmonella species among patients with diarrhea attending Gondar town health institutions, Northwest Ethiopia. Sci J Public Heal. 2014; 2:469–475. [Google Scholar]
- 27.Feleke H, Medhin G, Abebe A, Birhan HK, Asrat D. Enteric Pathogens and associated risk factors among under-five children with and without diarrhea in Wegera district, northwestern Ethiopia. Pan Afr Med J. 2018; 29:72. doi: 10.11604/pamj.2018.29.72.13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beyene G, Tasew H. Prevalence of intestinal parasite, Shigella and Salmonella species among diarrheal children in Jimma health center, Jimma southwest Ethiopia: A Cross Sectional Study. Ann Clin Microbiol Antimicrob. 2014; 13:1–7. doi: 10.1186/1476-0711-13-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kefyalew S, Kebede G, Keneni A. Prevalence of Shigella related diarrhea in Ambo town and antibiotic susceptibility of the isolated strains. Greener J Epidemiol Public Heal. 2015; 3:001–006. 10.15580/GJEPH.2015.1. [DOI] [Google Scholar]
- 30.Ayele AA, Tadesse D, Manilal A, Yohanes T, Seid M, Mekuria MS. Prevalence of enteric bacteria and enteroparasites in human immunodeficiency virus-infected individuals with diarrhoea attending antiretroviral treatment clinic, Arba Minch General Hospital, Southern. New Microbes New Infect. 2020; 38:100789. doi: 10.1016/j.nmni.2020.100789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayenew Z, Biazn H, Gebre-selassie S, Yeshitila B. Enteric pathogens and antimicrobial susceptibility profile among pediatric patients with diarrhea in Addis Ababa, Ethiopia. Ethiop Med J. 2019; 57–65. [Google Scholar]
- 32.Lengerh A, Moges F, Unakal C, Anagaw B. Prevalence, associated risk factors and antimicrobial susceptibility pattern of Campylobacter Species among under five diarrheic children at Gondar university hospital, northwest Ethiopia. BMC Pediatr. 2013;13: 82. doi: 10.1186/1471-2431-13-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Admassu M, Yemane G, Kibret M, Abera B, Nibret E, Adal M. Prevalence and antibiogram of Shigella and Salmonella spp. from under-five children with acute diarrhea in Bahir Dar town. Ethiop J Sci Technol. 2015; 8:27–35. [Google Scholar]
- 34.Mekonnen H, Kebede A, Menkir S. Isolation rate and drug resistance patterns of Shigella species among diarrheal patients attending at Hiwot Fana Hospital, Harar, Ethiopia. Ethiop J Sci Technol. 2014; 7:15–25. [Google Scholar]
- 35.Getamesay M, Getenet B, Ahmed Z. Prevalence of Salmonella and Campylobacter species and their susceptibility patters among under five children with diarrhea. Ethiop J Health Sci. 2014; 24:101–108.1. doi: 10.4314/ejhs.v24i2.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yemane G, Mulaw G, Gaim T. Prevalence and antimicrobial susceptibility of Salmonella species in diarrheal children under five-years in Bahir Dar. Int J Integr Sci Innov Technol. 2014; 3:2278–1145. [Google Scholar]
- 37.Teshome B, Teklemariam Z, Ayana DA, Marami D, Asaminew N. Salmonella and Shigella among patients with diarrhea at public health facilities in Adama, Ethiopia: Prevalence, antimicrobial susceptibility pattern, and associated factors. SAGE Open Med. 2019; 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teferi SC. Prevalence, Antibiotic Susceptibility Profile, and associated risk factors of Salmonella isolate among diarrheal patients visiting Dessie referral hospital, Northeast Ethiopia. Int J Microbiol., 2020; 2020:8834107. doi: 10.1155/2020/8834107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adugna A, Kibret M, Abera B, Nibret E, Adal M. Antibiogram of E. coli serotypes isolated from children aged under five with acute diarrhea in Bahir Dar town. Afr Health Sci. 2015; 15: 657–663. doi: 10.4314/ahs.v15i2.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fentaw S, Getahun M, Hussein M, Mamuye Y, Abebe A. Microbial aetiology of gastro-enteritis, antimicrobial resistance and associated factors among under-five children in Addis Ababa, Ethiopia. Ethiop J public Heal Nutr. 2016; 3:13–18. [Google Scholar]
- 41.Alemu A, Geta M, Taye S, Eshetie S, Engda T. Prevalence, associated risk factors and antimicrobial susceptibility patterns of Shigella infections among diarrheic pediatric population attending at Gondar town healthcare institutions, northwest Ethiopia. Trop Dis Travel Med. Vaccines: 2019: 5:7. 10.1186/s40794-019-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terfassa A, Jida M. Prevalence and antibiotics susceptibility pattern of Salmonella and Shigella species among diarrheal patients attending Nekemte referral hospital, Oromia, Ethiopia. Int J Microbiol. 2018; 2018:1–6. 10.1155/2018/9214689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tafa B, Sewunet T, Tassew H, Asrat D. Isolation and antimicrobial susceptibility patterns of Campylobacter species among diarrheic children at Jimma, Ethiopia. Int J Bacteriol., 2014; 1–7. doi: 10.1155/2014/560617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ameya G, Tsalla T, Getu F, Getu E. Antimicrobial susceptibility pattern, and associated factors of Salmonella and Shigella infections among under five children in Arba Minch, South Ethiopia. Ann Clin Microbiol Antimicrob. 2018; 17:1. doi: 10.1186/s12941-018-0253-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mekonnen GK, Mengistie B, Sahilu G, Kloos H, Mulat W. Etiologies of diarrhea and drug susceptibility patterns of bacterial isolates among under-five year children in refugee camps in Gambella Region, Ethiopia: A case control study. BMC Infect Dis. 2019; 19:1008. doi: 10.1186/s12879-019-4599-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gebrekidan A, Dejene TA, Kahsay G, Wasihun AG. Prevalence and antimicrobial susceptibility patterns of Shigella among acute diarrheal outpatients in Mekelle hospital. BMC Res Notes. 2015; 8: 611. doi: 10.1186/s13104-015-1606-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mengistu G, Mulugeta G, Lema T, Aseffa A. Microbial and biochemical technology prevalence and antimicrobial susceptibility patterns of Salmonella serovars and Shigella species. Microb Biochem Technol. 2014; S2. 10.4172/1948-5948.S2-006. [DOI] [Google Scholar]
- 48.Reda AA, Seyoum B, Yimam J, Andualem G, Fiseha S, Vandeweerd JM. Antibiotic susceptibility patterns of Salmonella and Shigella isolates in Harar, Eastern Ethiopia. J Infect Dis Immun. 2011; 3:134–139. [Google Scholar]
- 49.Assefa A. Girma M. Prevalence and antimicrobial susceptibility patterns of Salmonella and Shigella isolates among children aged below five years with diarrhea attending Robe general hospital and Goba referral hospital, South East. Trop Dis Travel Med Vaccines. 2019; 5:19. doi: 10.1186/s40794-019-0096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gebremichael G, Gebreegziabher G, Asrat D, Amanuel YW, Hagos T. Isolation and antimicrobial susceptibility profile of Shigella and Salmonella species from children with acute diarrhoea in Mekelle hospital and Semen health center, Ethiopia. Ethiop J Sci. 2018; 28: 197. 10.4314/ejhs.v28i2.11, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huruy K, Kassu A, Mulu A, Worku N, Fetene T, Gebretsadik S, et al. Intestinal parasitosis and shigellosis among diarrheal patients in Gondar teaching hospital, northwest Ethiopia. BMC Res Notes. 2011; 4:472. http://www.biomedcentral.com/1756-0500/4/472. doi: 10.1186/1756-0500-4-472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayamo M, Alemayehu T, Tadesse B, Mitiku E, Bedawi Z. Magnitude, risk factors and antimicrobial susceptibility pattern of Shigella and Salmonella among children with diarrhea in southern Ethiopia: A cross-sectional study. SAGE Open Med., 2021; 9:1–10. 2021. doi: 10.1177/20503121211009729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eguale T, Gebreyes WA, Asrat D, Alemayehu H, Gunn JS, Engidawork E. Non-typhoidal Salmonella serotypes, antimicrobial resistance and co-infection with parasites among patients with diarrhea and other gastrointestinal complaints in Addis Ababa, Ethiopia. BMC Infect Dis. 2015; 15:497. doi: 10.1186/s12879-015-1235-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abera B, Hailu T, Beza L. Aetiology of acute diarrhoea and antimicrobial usage among children aged under five years at health centres in Bahir Dar, Ethiopia. Trop Doct. 2020; l0:1–4. doi: 10.1177/0049475520912558 [DOI] [PubMed] [Google Scholar]
- 55.Lamboro T, Ketema T, Bacha K. Prevalence and antimicrobial resistance in Salmonella and Shigella species isolated from outpatients, Jimma University specialized hospital, southwest Ethiopia. Can J Infect Dis Med Microbiol. 2016. 10.1155/2016/4210760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mamuye Y, Metaferia G, Birhanu A, Desta K, Fantaw S. Isolation and antibiotic susceptibility patterns of Shigella and Salmonella among under 5 children with acute diarrhoea: A cross-sectional study at selected public health facilities in Addis Ababa, Ethiopia. Clin. Microbiol. Open Access: 2015; 04: 1–7. [Google Scholar]
- 57.Kebede A, Aragie S, Shimelis T. The common enteric bacterial pathogens and their antimicrobial susceptibility pattern among HIV-infected individuals attending the antiretroviral therapy clinic of Hawassa University Hospital, Southern Ethiopia. Antimicrob. Resist. Infect. Control. 2017; 6: 128. doi: 10.1186/s13756-017-0288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mekonnen M, Geda B, Teklemariam Z, Weldegebreal F, Balakrishnan S. Prevalence of childhood diarrhea and associated risk factors in Dire Dawa, Eastern Ethiopia. J of Public Heal From Theory to Pract. 2018; 26:29–37. [Google Scholar]
- 59.Kebede R, Alemayehu H, Medhin G. Eguale T. Nontyphoidal Salmonella and their antimicrobial susceptibility among diarrheic patients attending private hospitals in Addis Ababa, Ethiopia. Biomed Res Int. 2021: 6177741. doi: 10.1155/2021/6177741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adimasu DA, Kebede A, Menkir S. Prevalence of antibiotic resistant Salmonella isolates, Entermoeba histolytica and Giardia lamblia in Harar, Eastern Ethiopia. African J Microbiol Res. 8:2044–2053. [Google Scholar]
- 61.Walker CL, Sack D, Black RE. Etiology of Diarrhea in older children, adolescents and adults: A systematic review. PLoS Negl Trop Dis. 4:e768. doi: 10.1371/journal.pntd.0000768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Getie M, Abebe W, Tessema B. Prevalence of enteric bacteria and their antimicrobial susceptibility patterns among food handlers in Gondar town, northwest. Antimicrob. Resist Infect Control. 2019; 8:111. doi: 10.1186/s13756-019-0566-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shah M. Kathiiko C, Wada A, Odoyo E, Bundi M, Gabrie M, et al. Prevalence, seasonal variation, and antibiotic resistance pattern of enteric bacterial pathogens among hospitalized diarrheic children in suburban regions of central Kenya. Trop Med Health. 2016; 44:1–8. doi: 10.1186/s41182-016-0004-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lima AM, Oliveira DB, Quetz JS, HavtID A, Prata MG, Lima FN, et al. Etiology and severity of diarrheal diseases in infants at the semiarid region of Brazil: A case- control study. PLos Negelected Trop Dis. 2019;13:e0007154. doi: 10.1371/journal.pntd.0007154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giacomini E, Perrone V, Alessandrini Paoli DD, Luca CN, Esposti D. Evidence of antibiotic resistance from population-based studies: A Narrative Review. Infect Drug Resist. 2021; 14: 849–858. doi: 10.2147/IDR.S289741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bogale GG, Gelaye KA, Degefie DT, Gelaw YA. Spatial patterns of childhood diarrhea in ethiopia: data from ethiopian demographic and health surveys (2000, 2005 and 2011). BMC Infect Dis. 2017; 17:426. doi: 10.1186/s12879-017-2504-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med, 2015; 12:e1001923. doi: 10.1371/journal.pmed.1001923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zenebe T, Mitiku M, Alem Y. Prevalence of Escherichia coli in under-five children with diarrhea in Ethiopia: a systematic review and meta-analysis. Int J Microbiol. 2020; 2020:1–7. doi: 10.1155/2020/8844294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oppong B, Yang H, Amponsem-Boateng C, Kyere EK, Abdulai T, Duan G, Opolot G. Enteric pathogens associated with gastroenteritis among children under 5 years in Sub-Saharan Africa: A systematic review and met-analysis. Epidemiol Infect. 2020; 148:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langendorf C, Le Hello S, Moumouni A, Gouali M. Enteric Bacterial pathogens in children with diarrhea in niger: diversity and antimicrobial resistance. PLoS One, 2015; 10:e0120275. doi: 10.1371/journal.pone.0120275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kotloff KL, Nasrin D, Blackwelder WC, Wu Y, Farag T, Panchalingham S, et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the global enteric multicenter study. Lancet Glob Heal. 2019; 7:e568–e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kassie GM, Kassu A, Bayih AG. Campylobacter enteritis among children in Dembia District, Northwest. East Afr Med J. 2000; 77:654–657. [Google Scholar]
- 73.Gedlu E. Aseffa A. Campylobacter enteritis among children in north-west ethiopia a one-year prospective study. Ann Trop Paediatr. 1996; 16:207–212. doi: 10.1080/02724936.1996.11747828 [DOI] [PubMed] [Google Scholar]
- 74.Fletcher SM, Mclaws M, Ellis JT. Prevalence of gastrointestinal pathogens in developed and developing countries: Systematic review and meta-analysis. J Public Health Res. 2013; 2:e9. doi: 10.4081/jphr.2013.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hussen S, Mulatu G, Kassa ZY. Prevalence of Shigella species and its drug resistance pattern in ethiopia: a systematic review and meta‑analysis. Ann Clin Microbiol Antimicrob. 2019; 1:22. 10.1186/s12941-019-0321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abate D, Assefa N. Prevalence and antimicrobial resistance patterns of salmonella isolates in human stools and animal origin foods in Ethiopia: A Systematic review and meta-analysis. Int J Health Sci. 2021; 13: 43–55. [PMC free article] [PubMed] [Google Scholar]
- 77.Misganaw D, Abtew K. Evaluation of antibiotic utilization pattern during acute diarrheal disease at chefa-robit health. Drug Healthc Patient Saf, 2020; 12:169–175. doi: 10.2147/DHPS.S256330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moges F, Endris M, Mulu A, Tessema B, Belyhun Y, Shiferaw Y, et al. The growing challenges of antibacterial drug resistance in Ethiopia. J Glob Antimicrob Resist. 2014; 2: 148–154. doi: 10.1016/j.jgar.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 79.Muhie OA. Antibiotic use and resistance pattern in Ethiopia: Systematic review and meta-analysis. Int J Microbiol. 2019; 2019:2489063. doi: 10.1155/2019/2489063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tadesse G. A meta-analysis of the proportion of antimicrobial resistant human Salmonella isolates in Ethiopia. BMC Pharmacol Toxicol. 2014; 15:51. http://www.biomedcentral.com/2050-6511/15/51. doi: 10.1186/2050-6511-15-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khademi F, Vaez H, Ghanbari F, Arzanlou M, Sahebkar A. Prevalence of fluoroquinolone-resistant Salmonella serotypes in Iran: A meta-analysis. Pathog Glob Health. 2020; 114:16–29. doi: 10.1080/20477724.2020.1719701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pormohammad A, Nasiri MJ, Azimi T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: A systematic review and meta-analysis. Infect Drug Resist. 2019; 12:1181–1197. doi: 10.2147/IDR.S201324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tuem KB, Gebre AK, Atey TM, Bitew H, Yimer EM, Berhe DF. Drug resistance patterns of Escherichia coli in Ethiopia: A meta-analysis. Biomed Res Int. 2018; 2018:4536905. doi: 10.1155/2018/4536905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karikari AB, Obiri-danso K, Frimpong EH, Krogfelt KA. Antibiotic resistance in Campylobacter isolated from patients with gastroenteritis in a teaching hospital in Ghana. Open J Med Microbiol. 2017; 7:1–11. [Google Scholar]
- 85.Alemayehu T. Prevalence of multidrug-resistant bacteria in Ethiopia: A systematic review and meta-analysis. J Glob Antimicrob Resist. 2021; 26:133–139. doi: 10.1016/j.jgar.2021.05.017 [DOI] [PubMed] [Google Scholar]
- 86.Abayneh M, Hailemariam S, Asnake M. Bacterial profile and multi-drug resistance pattern of bacterial isolates among septicemia suspected cases: A meta-analysis report in Ethiopia. J Lab Med. 2021; 45:167–178. [Google Scholar]
- 87.Pattnaik D, Panda SS, Singh N, Sahoo S, Mohapatra I, Jena J. Multidrug resistant, extensively drug resistant and pan drug resistant gram negative bacteria at a tertiary care centre in Bhubaneswar. Int J Community Med Public Heal. 6: 567–572. [Google Scholar]
- 88.Elsalam MA, Gamal D, El Said M, Salem D, Aitta AA, El Gamal MS. Prevalence of plasmid-mediated quinolone resistance in multidrug-resistant gram negative bacilli in Egypt. Biomed Pharmacol J. 2018; 11:1927–1936. [Google Scholar]
- 89.Melake NA, Eissa NA, Keshk TF, Sleem AS. Prevalence of multidrug-resistant bacteria isolated from patients with burn infection. Menoufia Med J., 2015; 28:677–684. doi: 10.4103/1110-2098.167888 [DOI] [Google Scholar]
- 90.Lohr B, Pfeifer V, Heudorf U, Rangger C, Norris DE, Hunfeld KP. High prevalence of multidrug-resistant bacteria in Libyan war casualties admitted to a tertiary care hospital, Germany Microbial Drug Resistance. Microb Drug Resist, 2018; 24. 10.1089/mdr.2017.0141. [DOI] [PubMed] [Google Scholar]
- 91.Awasthi TR, Pant ND, Dahal PR, Prevalence of Multidrug resistant bacteria in causing community acquired urinary tract infection among the patients attending outpatient department of Seti zonal hospital, Dhangadi, Nepal. Nepal J Biotechnol, 2015; 3:55–59. [Google Scholar]
- 92.Lim CJ, Cheng AC, Kennon J, Spelman D, Hale D, Melican G et al. Prevalence of multidrug-resistant organisms and risk factors for carriage in long-term care facilities: a nested case–control study. J Antimicrob Chemother. 2014; 69:1972–1980. doi: 10.1093/jac/dku077 [DOI] [PubMed] [Google Scholar]
- 93.Adrizain R., Suryaningrat F, Alam A, Setiabudi D. Drug-resistant bacteria in children hospitalized at Dr. Hasan Sadikin general hospital Bandung Indonesia. IOP Conf Ser Earth Environ Sci. 2018; 125: 012077. doi: 10.1088/1755-1315/125/1/012077 [DOI] [Google Scholar]
- 94.Sainfer A, Smaldone A, Larson E. Prevalence of Multidrug-Resistant Gram-Negative Bacteria Among Nursing Home Residents- A Systematic Review and Meta-Analysis. Am. J. Infect. Control. 2017; 45:512–518. doi: 10.1016/j.ajic.2017.01.022 [DOI] [PubMed] [Google Scholar]
- 95.Rivera-izquierdo M, Benavente-Fernández A, López-Gómez J, Láinez-Ramos-Bossini AJ, Rodríguez-Camacho M, Valero-Ubierna MC, et al. Prevalence of multi-resistant microorganisms and antibiotic stewardship among hospitalized patients living in residential care homes in Spain. Antibiotics, 2020; 9:324. doi: 10.3390/antibiotics9060324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Buke C, Armand-Lefevre L, Laurence G, Isabelle D, Waafa R, et al. Epidemiology of multidrug-resistant bacteria in patients with long hospital stays infection control and hospital epidemiology Cambridge Core. Infect Control Hosp Epidemiol. 2007; 28: 1255–1260. doi: 10.1086/522678 [DOI] [PubMed] [Google Scholar]
- 97.Rahman A, Paul PR, Hoque N, Islam SS, Ziaul Haque AKM, Sikder MH, et al. Prevalence and antimicrobial resistance of Campylobacter species in diarrheal patients in Mymensingh, Bangladesh. Biomed Res Int. 2021; 2021: 9229485. doi: 10.1155/2021/9229485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zachariah OH, Lizzy MA, Rose K, Angela MM. Multiple drug resistance of Campylobacter jejuni and Shigella isolated from diarrhoeic children at Kapsabet County referral hospital, Kenya. BMC Infect Dis., 2021. 21: 109. doi: 10.1186/s12879-021-05788-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aworh MK, Adewusi OA, Mba N, Helwigh B, Hendriksen RS. Prevalence and risk factors for faecal carriage of multidrug-resistant Escherichia coli among slaughterhouse workers. Sci Rep. 2021; 11:133362. doi: 10.1038/s41598-021-92819-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu D, Ding Y, Yao K, Gao W, Wang Y. Antimicrobial resistance analysis of clinical Escherichia coli isolates in neonatal ward. Front Pediatr. 2021; 9:670470. doi: 10.3389/fped.2021.670470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suárez-Pérez A, Corbera JA, Tejedor-Junco MT, González-Martín M. Multidrug-resistant phenotypes of Escherichia coli isolates. Anim Artic. 2021; 11:1692. doi: 10.3390/ani11061692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nkansa-gyamfi NA, Kazibwe J, Nji E. Prevalence of multidrug, extensive drug, and pandrug-resistant commensal Escherichia coli isolated from healthy humans in community settings in low and middle-income countries: a systematic review and meta-analysis. glob health action. 12:1815272. doi: 10.1080/16549716.2020.1815272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abbasi E, Abtahi H, Van Belkum A. Multidrug-resistant Shigella infection in pediatric patients with diarrhea from central Iran. Infect Drug Resist. 2019; 12:1535–1544. doi: 10.2147/IDR.S203654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ud-din AI, Wahid UH, Latif HA, Shahnaij M, Akter M, Azmi IJ et al. Changing trends in the prevalence of Shigella species: Emergence of multi-drug resistant Shigella sonnei biotype—in Bangladesh. PLoS One, 2013; 8:e82601. doi: 10.1371/journal.pone.0082601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sheikh B, Nor A, Menza NC, Musyoki AM. Multidrug-resistant shigellosis among children aged below five years with diarrhea at Banadir hospital in Mogadishu, Somalia. Can J Infect Dis Med Microbiol. 2021; 2021:6630272. doi: 10.1155/2021/6630272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X, Biswas S, Paudyal N, Pan H, Li X. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front Microbiol. 2019; 10:985. doi: 10.3389/fmicb.2019.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garedew L, Wondafrash N, Feleke A. Identification of drug-resistant Salmonella from food handlers at the University of Gondar, Ethiopia. BMC Res Notes. 2014; 7:1–6. doi: 10.1186/1756-0500-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dagnew B, Alemayehu H, Medhin G, Eguale T. Prevalence and antimicrobial susceptibility of Salmonella in poultry farms and in-contact humans in Adama and Modjo Towns, Ethiopia. Microbiologyopen. 2020; 9: e1067. doi: 10.1002/mbo3.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Admassu D, Egata G, Teklemariam Z. Prevalence and antimicrobial susceptibility pattern of Salmonella enterica serovar Typhi and Salmonella enterica serovar Paratyphi among febrile patients at Karamara Hospital, Jigjiga, Eastern Ethiopia. SAGE Open Med. 2019; 7:1–7. doi: 10.1177/2050312119837854 [DOI] [PMC free article] [PubMed] [Google Scholar]