Abstract
Respiratory viruses, especially coronaviruses, have resulted in worldwide pandemics in the past couple of decades. Saliva-based paper microfluidic assays represent an opportunity for noninvasive and rapid screening, yet both the sample matrix and test method come with unique challenges. In this work, we demonstrated the rapid and sensitive detection of SARS-CoV-2 from saliva samples, which could be simpler and more comfortable for patients than existing methods. Furthermore, we systematically investigated the components of saliva samples that affected assay performance. Using only a smartphone, an antibody-conjugated particle suspension, and a paper microfluidic chip, we made the assay user-friendly with minimal processing. Unlike the previously established flow rate assays that depended solely on the flow rate or distance, this unique assay analyzes the flow profile to determine infection status. Particle-target immunoagglutination changed the surface tension and subsequently the capillary flow velocity profile. A smartphone camera automatically measured the flow profile using a Python script, which was not affected by ambient light variations. The limit of detection (LOD) was 1 fg/μL SARS-CoV-2 from 1% saliva samples and 10 fg/μL from simulated saline gargle samples (15% saliva and 0.9% saline). This method was highly specific as demonstrated using influenza A/H1N1. The sample-to-answer assay time was <15 min, including <1-min capillary flow time. The overall accuracy was 89% with relatively clean clinical saline gargle samples. Despite some limitations with turbid clinical samples, this method presents a potential solution for rapid mass testing techniques during any infectious disease outbreak as soon as the antibodies become available.
Keywords: COVID-19, Respiratory virus, Smartphone-based biosensor, Capillary action, Particle immunoagglutination
1. Introduction
The emergence and re-emergence of infectious respiratory viral diseases, e.g., Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and the novel influenza A/H1N1 strain, has become a major global public health concern, and intense research has been in progress to cope with these diseases (Wu et al., 2020). The recent coronavirus disease 2019 (COVID-19) pandemic has taught us many important lessons regarding how to be better prepared to deal with any future infectious disease threats. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus causing the ongoing COVID-19 pandemic, was first reported in December 2019 and developed into a global pandemic within three months (Hoffman et al., 2020). It is highly contagious and has led to over 438 million confirmed cases globally as well as nearly 6 million deaths as of early March 2022 (retrieved from WHO's COVID-19 dashboard; https://covid19.who.int). The pandemic has become one of the major threats for people all over the world and has dramatically affected health, economics, and livelihood. COVID-19 can be easily transmitted via respiratory droplets from coughs, sneezes, speech, or breath. Moreover, it has been reported that some patients could be asymptomatic and still be able to transmit the virus (Byambasuren et al., 2020), which may be even riskier since the primary screening used in many countries (e.g., temperature measurement) would not be able to identify patients, and the infection could spiral out of control. This brings us to the need for more accurate diagnostic tools with high sensitivity and specificity for SARS-CoV-2 to manage the spread of the disease.
The current gold standard for COVID-19 diagnosis is reverse transcription-quantitative polymerase chain reaction (RT-qPCR), which is highly specific and sensitive (95% accuracy). However, false positive results can occur if the viral copies are too low (Healy et al., 2021), and this technique requires expensive laboratory equipment, sample pre-processing, a clean lab environment, and experienced operators. The overall process can take up to 1 day, or longer than weeks in an area with limited resources (Xu et al., 2020). Currently, biosensing technologies based on antibody-antigen binding (i.e., immunoassays) on platforms such as microfluidic chips or paper strips have gained increasing interest worldwide due to their robustness, low cost, and user-friendly features. They offer potential for a rapid point-of-care COVID-19 diagnosis (Choi, 2020). In particular, paper strip-based biosensors have been popularly utilized, especially in low-resource settings. These are commonly known as rapid antigen tests, as they detect the presence of virus antigens typically from nasopharyngeal or nasal swab samples. These tests are readily available in many forms, flexible, easy to modify, portable, and disposable. In addition, the cellulose structure provides filtration capabilities for filtering out unwanted large debris, and capillary action allows for spontaneous flow, reducing the need for additional pumps (Costa et al., 2014).
Many rapid antigen tests have been commercially available with emergency use authorization (EUA) approved by governmental agencies including the U.S. Food and Drug Administration (FDA). They have been used in many areas since they are inexpensive, offer rapid results (about 15–30 min), and allow on-site operation. Nonetheless, their accuracy (including sensitivity and specificity) are still very inferior, and they are not capable of detecting early phase or asymptomatic infections. Even the BinaxNOW rapid antigen test, which can be self-administered without a reader device and provide rapid at-home results, has been reported to have a sensitivity of only around 35.8%–64.2%, despite the high specificity of 99.8%–100% (Prince-Guerra et al., 2021).
To overcome these limitations and develop a rapid, highly sensitive, and highly specific COVID-19 diagnostic assay, we introduce a paper-based microfluidic biosensor chip assisted with a smartphone camera to detect the presence of SARS-CoV-2. Rather than detecting the signal in a colorimetric or fluorescent manner, the flow of the liquid is recorded in real-time with a smartphone camera. In this manner, we collect what we refer to as the “time to constant velocity” of the flow profile. This novel approach allows the data collection in a more consistent manner than simple flow rate or distance analysis (Klug et al., 2018). This represents the time it takes for samples and pre-loaded antibodies to interact before the sample flows at a constant velocity, indicating the presence or absence of SARS-CoV-2 in the sample. By simply capturing the flow rate over time, the assay can be conducted in ambient lighting conditions since neither magnification nor a dark environment is required. In addition, only a small amount of antibody-antigen binding is required to alter the interfacial tension, leading to the possibility of extremely sensitive detection and a very low LOD.
We utilize antibodies against SARS-CoV-2 nucleocapsid, which is the most abundant protein in coronaviruses and highly immunogenic. These antibodies are also reported to be more sensitive than the spike protein antibodies for early detection of COVID-19 (Oliveira et al., 2020; Burbelo et al., 2020). The antibodies against SARS-CoV-2 nucleocapsid are conjugated to submicron polystyrene particles. To start the assay, a 4 μL sample of human saliva (simulated or clinical) is loaded onto the loading area and the chip is allowed to dry for 10 min. Then, the user simply starts a video recording with a smartphone held above the chip, and 4 μL of the antibody-conjugated particle suspension (0.002–0.004% w/v) is loaded onto the same loading area of the chip and allowed to flow for about 2 min. To analyze the results, we developed an image processing Python code that automatically corrects the chip orientation and selects thresholding values from frame to frame to create a plot displaying the full flow rate profile.
The conventional method for collecting SARS-CoV-2 specimens is a nasopharyngeal (NP) swab. However, NP swab collection requires trained medical personnel with protective gear and may not be successful at the first attempt, causing discomfort to the patients and exposing staff to a high risk of infection (Czumbel et al., 2020). Other methods include a nasal swab, which can be accomplished by inserting 1 cm of the swab into the nostrils, which is less uncomfortable but markedly less sensitive compared to the NP swab (Teo et al., 2021). Saliva samples can be collected by sampling 5 mL of whole saliva, which is easy to collect without trained staff. However, these samples contain substantial amounts of mucous, requiring an extra dilution step (Goldfarb et al., 2021). Recent studies have shown a promising alternative: self-collected saline gargle samples (Worobey, 2020), which could simplify and accelerate COVID-19 diagnosis. Compared to NP swabs and saliva samples, saline gargle samples are more acceptable to the users and have sensitivity up to 98% (Worobey, 2020), while whole saliva sampling has only 79% sensitivity (Goldfarb et al., 2021; Ihling et al., 2020). Still, the complex matrix inherent to saline gargle samples presents unique challenges for the paper microfluidic and lateral flow assay platforms, such as salivary proteins and leftover food or toothpaste. Therefore, we used self-collected saline gargle samples, as well as simulated saline gargle samples spiked with a known concentration of SARS-CoV-2, as the sample collection method and systematically investigated the components of the clinical samples that affected assay performance. This approach is non-invasive, facilitates mass testing easily, and has high sensitivity. So far, saline gargle samples have only been successfully demonstrated with RT-qPCR, but not with a rapid antigen testing platform.
We aim to provide a low-cost point-of-care SARS-CoV-2 antigen test, facilitated with a smartphone camera and automatic flow profile detection. Advantages include easy sample collection and analysis, high specificity, high sensitivity, no sample pre-processing, no laboratory equipment, and no extensive training. This method has the potential to be used for detecting other respiratory viruses by changing the antibodies and optimizing experimental parameters.
2. Materials and methods
2.1. Paper chip design and fabrication
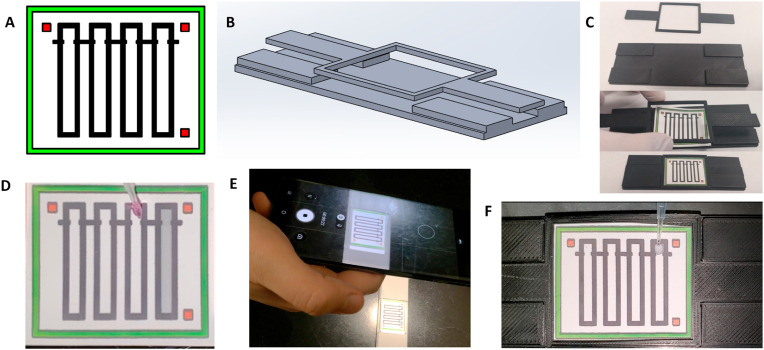
The microfluidic chip (shown in Fig. 1 A) was designed using SolidWorks 2020 software (Dassault Systèmes, Vélizy-Villacoublay, France). Each channel design consists of a 2.5 mm × 2.5 mm sample loading area, connected to a 2.5 mm × 18.5 mm straight channel. The loading area and the channel are separated by 0.5 mm × 1.5 mm indents extending from the channel edge, which are also used as reference lines. There are a total of 4 parallel channels on a single chip for high throughput analysis. An outer green box (28 mm × 34 mm) and three 1.5 mm × 1.5 mm red squares at three corners of the chip (top left, top right, and bottom right) allow for orientation recognition and locating the channel areas during automated flow measurement. The chip design was printed on Unisart® nitrocellulose membrane CN95 (Sartorius, Goettingen, Germany) using a wax printer (ColorQube 8550, Xerox, Norwalk, CT, USA). The printed wax was reheated using a hot plate set at 120 °C for 3 min to create hydrophobic barriers that penetrate the depth of the paper. The chip holder and chip lock (shown in Fig. 1B) are designed using SolidWorks 2020 and 3D-printed with a Creality Ender 3 printer (Creality, Shenzhen, China) using PETG filament (Overture, Wilmington, DE, USA).
Fig. 1.
Flow profile assay of SARS-CoV-2 on a paper-based microfluidic chip. (A) Paper-based microfluidic chip design containing green edge and three red squares for recognizing the chip area in automated flow distance measurement. (B) The chip holder and the chip lock. (C) A paper-based microfluidic chip was placed into a chip holder to flatten the chip. (D) 4 μL of sample was loaded directly onto the square inlet (top) of each channel and dried for 10 min. (E) A smartphone camera was held just above the chip to view the chip area and start recording the video. (F) 4 μL of Ab-particles were loaded, and the liquid flow on the paper microfluidic chip was recorded with the smartphone camera.
2.2. Antibody conjugated particles
Two types of antibodies were used in this study: rabbit monoclonal antibody to SARS-CoV-2 (2019-nCoV) nucleocapsid (Sino Biological US Inc., PA, USA) and rabbit polyclonal antibody to SARS-CoV/SARS-CoV-2 nucleocapsid (Sino Biological US Inc., PA, USA). Carboxylated, yellow-green, fluorescent polystyrene particles with 0.5 μm diameter (Magsphere Inc., Pasadena, CA, USA) were pre-washed with an activation buffer (2-(N-morpholino)ethanesulfonic (MES), pH 6.0) through centrifugation at 9,900×g for 10 min to remove surfactants from the stock solution. The fluorescence feature was not utilized for our lateral flow assays, except for confirming the particle immunoagglutination via fluorescence microscopic imaging during assay optimizations. For future applications of this technique, non-fluorescent particles can be used. Next, anti-SARS-CoV-2 antibodies were covalently conjugated to the pre-washed particles via EDAC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide] coupling reactions. Details on buffer preparations and conjugation protocols can be accessed via our laboratory's protocols.io site (Schackart, 2021). Particle concentration after conjugation was quantified using the spectrophotometer absorbance at 480 nm (USB4000 miniature spectrophotometer and Ocean View software; both from Ocean Insight, Inc, Orlando, FL, USA). Based on our previous work (Chung et al., 2021) and the results of optimization experiments, the final concentration of particles was adjusted to 0.02 μg/μL (0.002% w/v) for monoclonal antibody-conjugated particles (mAb-particles) and 0.04 μg/μL (0.004% w/v) for polyclonal antibody-conjugated particles (pAb-particles). Tween 20 surfactant was added to both mAb-particles and pAb-particles to improve their stability. 0.5% v/v Tween 20 was added to the particle suspension at 1:3 ratio (Tween 20: particle suspension) while maintaining the final concentrations of the particle suspensions as previously stated. After conjugation procedures, Ab-particles were mixed with DI water (negative control), and various concentrations of SARS-CoV-2 (0, 10, and 1000 fg/μL) in diluted saliva to validate the success of antibody conjugation on microparticles. The particle mixtures were pipetted onto a microscope glass slide, and fluorescence images were captured by a benchtop fluorescence microscope (Nikon Eclipse TS100, Minato, Tokyo, Japan) using ISCapture software with the blue excitation plus green emission wavelength filter attachment (A.G. Heinze, Lake Forest, CA, USA) as shown in Supplementary Fig. S1.
2.3. Simulated and clinical SARS-CoV-2 saline gargle samples
Simulated saline gargle samples consisted of 1% or 10% w/v human saliva from pooled donors (Lee Biosolutions, Inc., MO, USA), 0.9% w/v NaCl, and varying concentrations of heat-inactivated SARS-CoV-2 from virus culture diluted in DI water. SARS-CoV-2 viruses (USA-WA1/2020) were cultured on Vero cells (ATCC #CCL-81), and the cell lysates were collected and centrifuged. Solutions were heat-inactivated at 65 °C for 30 min. The final SARS-CoV-2 concentrations in the simulated samples were 0, 10, 100, and 1000 fg/μL, corresponding to approximately 0, 10, 100, and 1000 copies/μL. Virus cultures and heat inactivation were conducted in Dr. Janko Nikolich-Žugich's laboratory (Biosafety Level 3) at the University of Arizona, and the simulated sample preparations were conducted in Dr. Jeong-Yeol Yoon's laboratory (Biosafety Level 2) at the University of Arizona. All clinical samples tested in this publication were heat inactivated, and cultured virus for simulated samples was either UV or heat inactivated, so there was no risk of infection while conducting assays. In the potential translation of this technique to public use, strict protocols would be necessary to prevent any spread of disease while loading samples onto the chip. Additionally, sterilization of the chip holder may be advisable.
A saline gargle sample collection method has been successfully developed and implemented by one of our authors (Worobey, 2020). Clinical saline gargle samples were collected from student participants at the University of Arizona under the Test All Test Smart program. Participants were provided 5 mL of 0.9% w/v sterile saline (NaCl) and completed a 5 s swish followed by a 10 s gargle, all repeated 3 times, and the gargle sample was then deposited in the prepared container. Trained staff were not required to perform the sample collection. Then, samples were heat-inactivated at 65 °C for 30 min and tested for SARS-CoV-2 by using RT-qPCR with CDC RUO primers and protocol. There were 14 positive and 14 negative clinical samples used in this study. Flow assays were repeated 3 times for each clinical sample. Clinical samples were stored at 4 °C if not used immediately. Clinical sample collection protocol was approved by the Institutional Review Board of the University of Arizona (IRB number 2102491314), and the cycle threshold (Ct) values for the positive samples were also provided (Supplementary Table S1). The Ct value refers to the number of cycles run until amplification is observed during RT-qPCR. This can roughly be correlated to the amount of virus titer in each positive sample. A lower Ct value indicates that fewer cycles were required to see amplification of the target, in this case meaning a higher virus titer was initially present. Samples which were negative showed no amplification and therefore do not have an associated Ct value.
For clinical saline gargle samples, turbidity was observed by comparing the relative intensity of the solution in a tube against a matted black background, using a separate empty tube as a reference (Supplementary Fig. S2). ImageJ (U.S. National Institutes of Health; Bethesda, MD, USA) was used to find the intensity histogram, and the mode was derived. The derived values were normalized by dividing with the value of the empty tube, then a threshold was chosen by classifying relative intensity data into two groups separating turbid and non-turbid samples. Flow profiles were measured in a similar manner as the simulated mouth gargle samples.
For specificity testing, another simulated sample was prepared using purchased influenza A/H1N1 (NATFLUAH1-ERCM, ZeptoMetrix, NY, USA) which had a Ct value of around 25–28. This was mixed with the simulated saline gargle sample to 1% and 10% v/v and used to compare with 1 pg/μL SARS-CoV-2 simulated saline gargle sample to test for specificity.
In addition, to demonstrate how the turbidity could affect the flow behavior, an example of simulated turbid samples was made from mixing toothpaste with negative simulated mouth gargle samples to a concentration of 10 mg/mL w/v. This was compared to negative simulated saline gargle samples without toothpaste to determine the effect on the lateral flow assay. Surface tension analysis was also performed on the negative simulated sample and the simulated toothpaste sample for comparison.
2.4. Flow profile measurements
All experiments were conducted in a laboratory environment under consistent air flow, temperature, and humidity conditions. All sample loading was done by carefully suspending the droplet from the pipette, letting it hang from the pipette, then touching the suspended droplet to the loading area of the channel. This exact protocol was used to limit any variation between user performance in pressure or speed of loading the droplet. To run the experiment, the paper-based microfluidic chip was placed on the chip holder and flattened down with the chip lock (Fig. 1C). 4 μL of sample was pipetted onto the loading area of the channel (Fig. 1D). Liquid spontaneously flowed along the channel via capillary action (also known as wicking). The sample-loaded chip was allowed to dry at room temperature for about 10 min. Once the samples were completely dried, the smartphone camera was placed to capture the video of the entire chip (Fig. 1E). After starting the video recording, 4 μL of mAb- or pAb-particle suspension was loaded onto the loading area of each channel at the same locations that were preloaded with the (now dry) sample. The smartphone camera was used to capture a video of the capillary flow of Ab-particle suspension for about 2 min (Fig. 1F). Ab-particle suspension was prepared with and without 0.0125% v/v Tween 20 (final concentration in the suspension) and these were compared for Ab-particle optimization.
2.5. Video processing algorithm
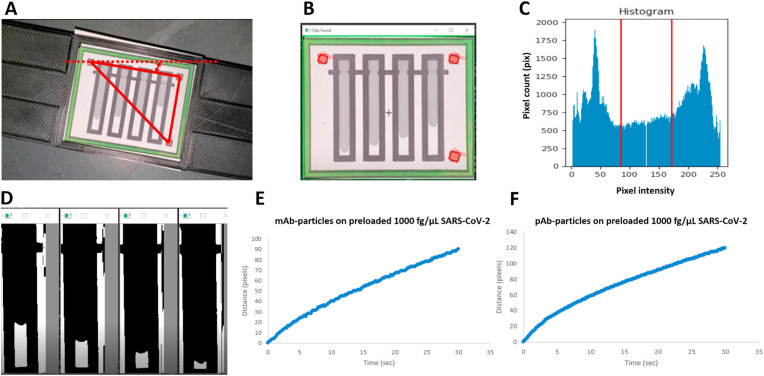
The flow profile was extracted from the recorded video files using our developed Python script provided under Supplementary Code S1. All necessary libraries (shown at the top of the Python script) should be downloaded before executing the code. Overall video processing is shown in Fig. 2 . The input video file is extracted into multiple image frames associated with time. Each image frame is scanned, and the centers of three red squares are detected for orientation correction by rotating the image until the chip aligns horizontally (Fig. 2A). The green box is recognized for the chip area (Fig. 2B), and the script proceeds to crop each flow channel to analyze separately. Each cropped channel undergoes pixel histogram analysis to automatically derive the threshold value using a multi-Otsu thresholding function that allows the flow to be easily distinguishable from the paper background (Fig. 2C). It is then converted to black and white using the derived threshold, and the Gaussian filtering function is used to remove noise. The flow is identified by black pixels increasing along the vertical centerline of each channel, where the white pixels represent empty paper (Fig. 2D). Flow tracking starts right after the flow passes the loading indents. With known chip dimensions, the flow distance in pixels can be converted to millimeters. Distance vs. time data and other flow characteristics can be derived for further analysis (Fig. 2E and F). The script uploads all data to an.xlsx file for storage. The script was run using Python 3.7.4 and 3.8.2 on a Windows OS using Visual Studio.
Fig. 2.
Video processing algorithm. Python code was developed to automatically obtain the flow distance over time. (A) The red squares were detected, and each frame is rotated for orientation correction. (B) The green edge was recognized and cropped. (C) The cropped area was analyzed to generate an intensity histogram plot. Using appropriate thresholding, the liquid flow was recognized on paper. (D) The flow on each channel was read separately by recognizing it as the black pixels increasing along the vertical centerline. (E) Flow distance vs. time profile of mAb-particles on the preloaded 1000 fg/μL SARS-CoV-2 spiked sample. (F) Flow distance vs. time profile of pAb-particles on the preloaded 1000 fg/μL SARS-CoV-2 spiked sample.
2.6. Pendant droplet experiment for surface tension analysis
Surface tension of the clinical saline gargle samples was found using the optical pendant droplet method (Daerr and Mogne, 2016), which was further described in Supplementary Method and Supplementary Fig. S3.
2.7. Bradford assay for total protein analysis
The Bradford Assay was used to determine the overall protein content of the clinical saline gargle samples, which was further described in Supplementary Method and Supplementary Fig. S4.
2.8. Statistical analysis
All data were derived from three replicates, each using a different paper chip (except for the specificity test with H1N1, in which 3 assays on the same chip were averaged). Statistical analyses were done by one-tailed Student's t-test in Microsoft Excel 365. Differences at p < 0.05 were considered statistically significant.
3. Results
3.1. Assay optimizations with simulated samples
Fluorescence images were captured for the antibody-conjugated particles mixed with DI water and viruses on a microscope glass slide (Supplementary Fig. S1A). There was no immunoagglutination present in the Ab-particle mixture in DI water. The extent of immunoagglutination increased as the virus concentration increased, indicating the successful conjugation of antibodies to the particle surfaces (Supplementary Fig. S1B).
After confirming successful antibody conjugation and subsequent immunoagglutination, the final concentrations of mAb-particles and pAb-particles were adjusted to 0.02 and 0.04 μg/μL (or 0.002% and 0.004% w/v). In order to optimize the particle concentration for each type of Ab-particles, flow assays were performed with both mAb-particles and pAb-particles at both concentrations (0.02 and 0.04 μg/μL) using the simulated SARS-CoV-2 samples with 1% saliva in DI water (no saline). The flow profile results (examples shown in Fig. 2E and F) showed a consistent relationship between distance and time for at least 30 s for both mAb-particles and pAb-particles. Therefore, the distance at 30 s was extracted from each flow profile to optimize various assay parameters.
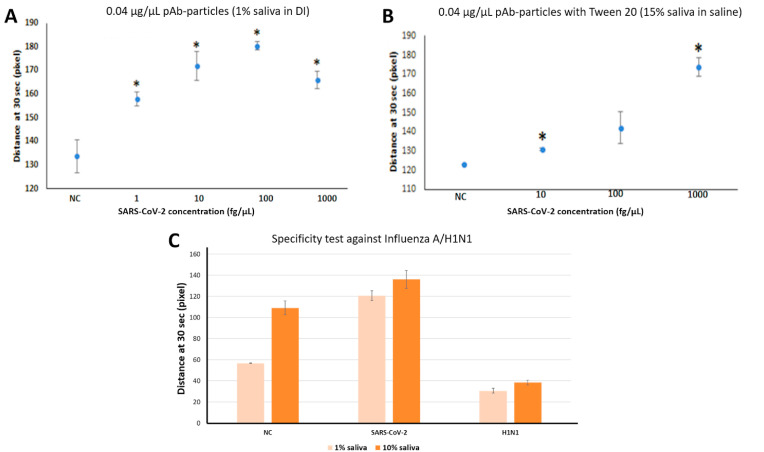
With pAb-particles and using SARS-CoV-2 spiked in 1% v/v human pooled saliva (Fig. 3 A), the characteristic bell-shaped curve was produced with an excellent LOD of 1 fg/μL or approximately 1 copy/μL. The curve started to decrease with very high SARS-CoV-2 concentration (1000 fg/μL), indicating the number of antibodies that could participate in immunoagglutination was smaller than the reaction-ready target antigens. These experiments were then repeated with the simulated saline gargle samples that are more similar to the clinical samples, i.e., with 15% saliva and 0.9% saline. To prevent particles from self-aggregating in the presence of saliva and saline, 0.5% w/v Tween 20 was added to the particle suspension (Cho et al., 2015; Chung et al., 2021). The LOD with pAb-particles was somewhat compromised at 10 fg/μL (or 10 copies/μL); however, this was the best LOD achieved in our optimization experiments using the simulated saline gargle samples. The decrease in high concentration was not observed, perhaps because there were fewer target antigens available due to their interactions with the higher saliva concentration or Tween 20. Therefore, pAb-particles at 0.04 μg/μL (0.004% w/v) with 0.5% w/v Tween 20 were used for the remainder of experiments. Results of experiments with mAb-particles and all other optimization experiments are shown in Supplementary Fig. S5.
Fig. 3.
Assay LOD and Specificity. NC indicates negative control and * shows p < 0.05 between sample and NC using one-tailed student's t-test with unequal variance. Error bars represent standard error. (A) Flow distances at 30 s on the paper microfluidic chips preloaded with SARS-CoV-2 spiked in 1% v/v human pooled saliva, using polyclonal antibody conjugated particles at 0.04 μg/μL (n = 3). (B) Flow distances at 30 s on the paper microfluidic chips preloaded with SARS-CoV-2 spiked in simulated saline gargle samples (∼15% v/v saliva and 0.9% saline), using polyclonal antibody conjugated particles at 0.04 μg/μL, with the addition of 0.5% w/v Tween 20 (n = 3). (C) Specificity test results with 1 pg/μL SARS-CoV-2 and influenza A/H1N1 (Ct values of 25–28) spiked in 1% v/v and 10% v/v saliva in 0.9% saline using the pAb-particles, shown together with the no target control samples (1% or 10% saliva in 0.9% saline) (n = 3).
Finally, specificity results with pAb-particles are shown in Fig. 3C. 10% v/v of the influenza A/H1N1 (Ct value = 25–28) was used as a specificity control. The flow distance results of the positive 1 pg/μL SARS-CoV-2 was significantly higher than the negative control (0 fg/μL) as well as the specificity control (H1N1).
3.2. Turbidity assessment of clinical saline gargle samples
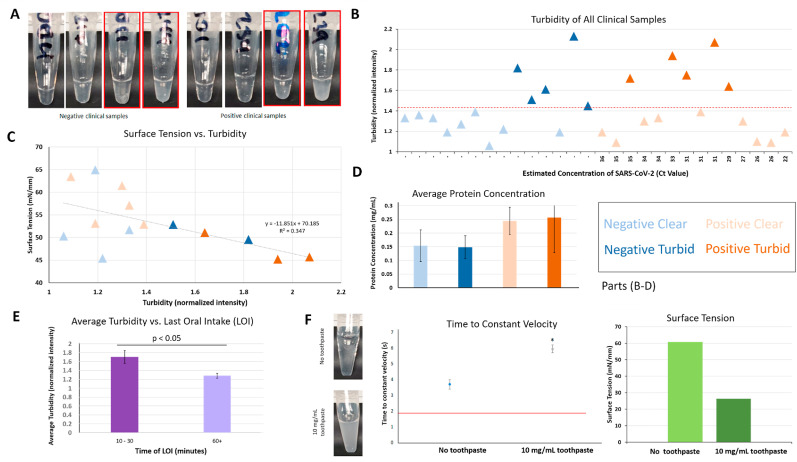
Turbidity may result from many components in saliva samples, so extensive testing was conducted. Example images of clinical saline gargle samples are shown in Fig. 4 A. There were 28 samples, with 14 negative and 14 positive samples as confirmed with RT-qPCR. Pixel intensities of the black background through the sample tube were obtained and divided by the value from the empty tube. We classified the samples into two categories: 1) turbid (normalized intensities >1.41) and 2) clear (<1.41) (Fig. 4B). There is some trend noticeable between turbidity and surface tension of the clinical samples (Fig. 4C), with turbid samples having lower surface tension than clear samples regardless of SARS-CoV-2 presence or absence. The Bradford assay did not show any difference between turbid and clear samples; however, it estimated a higher (not significant) protein concentration in positive than in negative samples (Fig. 4D). Some clinical samples had accompanying last oral intake (LOI) information, which represents the most recent time that the participant consumed food or beverage before providing their samples. There was a statistically significant (p < 0.05) difference using one-sided student's t-test in turbidity between the samples where patients had the LOI of 10–30 min (more turbid), versus those with the LOI of 60 or more minutes (clearer) (Fig. 4E). These results correspond well to the guidelines for these assays, which generally suggest that patients not consume food or beverage, use mouthwash, brush teeth, or smoke at least 30 min before testing. Finally, flow assays were conducted by adding toothpaste with the final concentration of 10 mg/mL to the NC simulated gargle samples (Fig. 4F). The time to reach constant velocity of the toothpaste-added NC sample was much higher than the clear NC sample. In addition, the surface tension of the 10 mg/mL toothpaste simulated sample was extremely low, compared to the 0 mg/mL toothpaste sample and even the turbid clinical samples (Fig. 4B). For the final assessment, 5 negative and 5 positive samples that were deemed turbid were excluded from the assay, leaving a total of 18 samples including 9 negative and 9 positive samples. Images of all clinical samples are shown in Supplementary Fig. S2.
Fig. 4.
Turbidity assessment of clinical saline gargle samples. Error bars represent standard error. (A) Photographs of negative and positive clinical saline gargle samples, obtained from human subjects. The normalized turbidity was determined by comparing the pixel intensities of the sample tubes against the black background. Red boxes indicate samples that were determined to be turbid using the procedure described in part B. (B) Using the normalized (to empty tube) turbidity, all clinical samples were classified into two categories, turbid and clear, using the threshold value of 1.41. Note: while all samples were classified in this manner, some could not undergo all subsequent testing due to low sample volume. (C) Surface tension measurements showed a decreasing trend with increased turbidity. (D) Total protein concentration of samples according to the Bradford assay. Turbid and clear samples showed no difference in total protein concentration, but SARS-CoV-2 positive samples had a higher (not significant) total protein concentration than negative samples. (n = 5 for negative clear, n = 3 for negative turbid, n = 6 for positive clear, and n = 4 for positive turbid). (E) Samples with a last oral intake (LOI) of 10–30 min prior to sample acquisition (n = 6) had higher turbidity than samples with a longer time since LOI (60+ min; n = 10), and the difference was statistically significant (p < 0.05). Average values are shown in the bar chart. (F) The time to constant velocity (n = 2) and surface tension of no toothpaste vs. toothpaste-added (10 mg/mL) NC samples, along with photos of the samples. Surface tension was measured at 0, 2, 4, 6, 8, and 10 s and the stabilized final value was chosen (hence no error bar). The accuracy of surface tension measurement is less than 1 mN/mm.
3.3. Clinical saline gargle sample assays
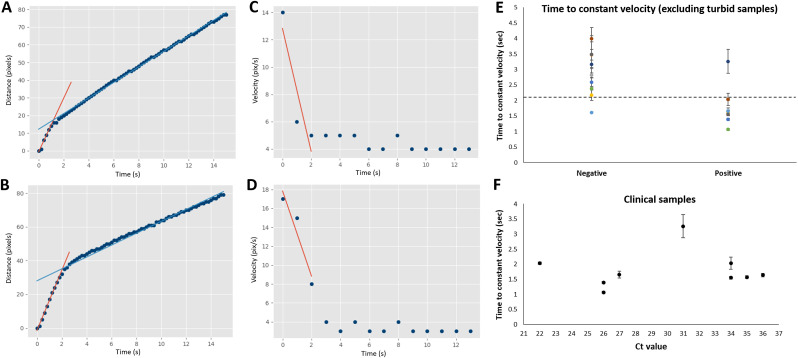
The clinical sample experiments were conducted in the same manner as the simulated samples, using 0.04 μg/μL pAb-particles and 0.5% w/v Tween 20. Fig. 5 A and B shows the raw flow distance profiles from a positive and negative clinical sample. Fig. 5C and D shows the numerically differentiated values, i.e., flow velocities over time. Both positive and negative clinical samples show constant flow velocities after 10 s and this trend can be observed with all clinical samples. Therefore, evaluating the parameters beyond this time point would not provide significantly different results between positive and negative samples. Two parameters were considered as potential criteria to make distinctions between positive and negative samples: the time to reach constant velocity, and the initial deceleration of flow. Fig. 5A and B shows the flow distance over time, overlaid with two linear trendlines for the initial (red line) and subsequent (blue line) flow velocities. The intersection of these lines represents the time at which the flow velocity decreased dramatically, which can also be seen in Fig. 5C and D as the time when velocity became nearly constant. Fig. 5C and D also show red lines which here represent the initial change in velocity over time (deceleration), which was calculated using the slope of the line. The time to reach constant velocity (where the lines in Fig. 5A and B intersect) clearly provided better distinction between positive (Fig. 5A) and negative (Fig. 5B) samples than the initial deceleration (the slopes of the lines in Fig. 5C and D), so this parameter was used. All time to reach constant velocity data are summarized in Supplementary Fig. S6.
Fig. 5.
Flow profile analysis of clinical saline gargle samples. (A–B) Flow distance profiles of representative positive and negative clinical samples, respectively. (C–D) Flow velocity profiles of the same, numerically differentiated from A and B. (E) The time to reach constant velocity (as demonstrated in A-B) for all clear negative and positive samples (n = 18). (F) Time to constant velocity for positive clear samples (n = 9) against the cycle threshold (Ct) values obtained with RT-qPCR. The lower the Ct value, the higher the virus titer in the sample.
Fig. 5E shows the time to reach constant velocity for all 18 clear clinical samples (9 negative and 9 positive samples) using the pAb-particles and Tween 20. The threshold to make the best separation between negative and positive samples was determined to be 2.1 s, giving one false-positive and one false-negative. Therefore, the overall accuracy, i.e., the number of true positive and true negative assessments divided by the total number of subjects, was [8 + 8]/18 = 89%. Sensitivity is the number of true positive assessments divided by the total number of all positive assessments, i.e., 8/9 = 89%. Specificity is the number of true negative assessments divided by the total number of all negative assessments, i.e., 8/9 = 89%. Fig. 5F shows the plot of the time to constant velocity data against the Ct values of all 9 positive clear samples, showing no obvious trend.
4. Discussion
We have demonstrated a very low LOD of 1 fg/μL (approximately 1 copy/μL) with 1% saliva samples and 10 fg/μL (approximately 10 copies/μL) with simulated mouth gargle samples (15% saliva and 0.9% saline). Considering the sample volume of 4 μL, the LOD of 10 copies/μL corresponds to 40 copies of virus. While this number may seem low, the number of nucleocapsid proteins that can bind to the antibody conjugated particles is still substantial, which explains how it is possible to achieve such a low LOD.
A good specificity was demonstrated using the influenza A/H1N1 spiked saliva samples. Interestingly, the difference in time to constant velocity between H1N1 and SARS-CoV-2 was much greater than that between the negative control and SARS-CoV-2, as shown in Fig. 3C. Perhaps the presence of a non-target may have eliminated any non-specific aggregation, leading to the maximum difference between H1N1 and SARS-CoV-2. Further investigation is necessary to draw a conclusion about this phenomenon, but the time to constant velocity is clearly different for H1N1 vs. SARS-CoV-2.
Assays were initially optimized to achieve the best detection limit for SARS-CoV-2 spiked into varying concentrations of saliva and saline, and flow distance at 30 s was used to distinguish positive and negative samples. Polyclonal antibodies at 0.04 μg/μL (0.004% w/v) showed the best assay performance (Fig. 3A) for SARS-CoV-2 spiked into 1% saliva with no saline. However, to better emulate clinical samples, saline and a higher concentration of saliva were then assayed. Tween 20 was added here to reduce potential instability that can occur due to hydrophobic interactions between styrene moieties of particles or via proteins and chemicals in the saliva (Kothekar et al., 2007). Addition of Tween 20 significantly attenuated the flow distances of negative samples but did not attenuate the signals of the positive samples as much, apparently resolving the self-aggregation issue and improving the sensitivity and LOD for clinically relevant samples with saline and high concentrations of saliva (Fig. 3B).
Previous studies have demonstrated that the Lucas-Washburn model shown below can be used to explain capillary flow by treating the porous structure of the paper as a bundle of capillary tubes (Camplisson et al., 2015; Klug et al., 2018):
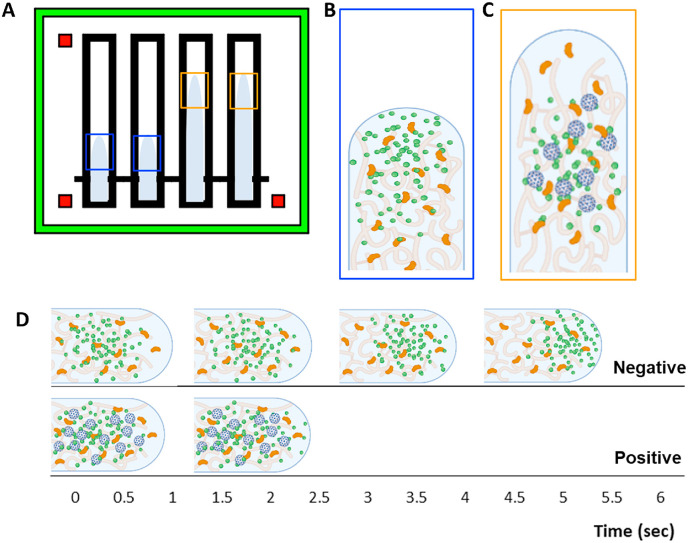
where l is the distance, t is time, R is the radius of the capillary (in this case the pore size of the paper), γ LG is the surface tension at the liquid-gas interface, θ is the water contact angle at the capillary wall (in this case at the paper fibers), and μ is the liquid viscosity. Capillary action is therefore the result of surface tension stemming from the intermolecular attraction at the liquid-air interface, which in our experiments is between the sample and paper chip. If R, γ LG, θ, and μ are constant throughout the assay, the flow distance profile l(t) should be a simple function of the square root of t. However, the flow distance profile does not fit such a simple function (Klug et al., 2018), indicating that the particle immunoagglutination occurs in a dynamic manner, changing the surface tension parameters (γ LG and θ) and viscosity (μ) in real-time. This also implies that particle immunoagglutination can change the flow behavior, providing a detection method for antibody-target immunoagglutination. Monodisperse particle solutions, such as negative samples with negligible immunoagglutination show rapid diffusion of particles to the wetting front. This significantly lowers surface tension and results in a slower flow rate as the particles at the wetting front decrease the hydrogen bond strength of the liquid. Polydisperse particle solutions, for example positive samples with a mixture of immunoagglutinated aggregates and singlets, show less diffusion of particles to the wetting front. This results in less change to the surface tension and a higher flow rate than monodisperse samples. This theory has previously been demonstrated and proven for E. coli and Zika virus by our group (Klug et al., 2018). Fig. 6 A represents the theory behind this phenomenon. With the negative samples, particles rarely aggregate, and the singlet Ab-particles diffuse to the wetting front (liquid-air interface) (Fig. 6B). These particles reduce the cohesive force between water molecules, lowering surface tension and causing the flow to be slower (following the Lucas-Washburn model). With the positive samples, the Ab-particles immunoagglutinate, forming larger and heavier particle clusters, which diffuse to the wetting front much more slowly. Bigger particle clusters are eventually trapped within the paper pores. This immunoagglutination leads to fewer singlet Ab-particles diffusing to the wetting front, leading to higher surface tension (than the negative samples) and faster flow (Fig. 6C). Increasing the SARS-CoV-2 concentration amplifies this phenomenon, leading to even faster flow, such as in Fig. 3B. For example images of the particle immunoagglutination, see Supplementary Fig. S7.
Fig. 6.
How particle immunoagglutination affects the flow distance and velocity profiles. (A) The flow distances at 30 s are longer with the positive samples (orange boxes) than the negative samples (blue boxes). (B) With no virus present, the singlet Ab-particles (green) quickly diffuse to the wetting front, lowering the surface tension and the flow velocity. Nitrocellulose fibers are colored in light orange and saliva proteins in dark orange. (C) With virus present (blue), immunoagglutination occurs, creating larger and heavier particle clusters, leaving very few singlet Ab-particles diffusing to the wetting front. (D) The Ab-particles on the negative sample are mostly in the singlet form and take more time to reach constant velocity (top), while fewer singlet Ab-particles on the positive sample are able to reach the flow interface due to immunoagglutination, so it takes less time to reach constant velocity (bottom).
With human clinical saline gargle samples, the flow distances at 30 s varied substantially depending on the quality of the clinical samples. This suggests that Ab-particles interacted with saline and saliva contents to some extent. Therefore, rather than measuring the flow distances at a fixed time point, we examined the flow velocity profiles through numerical differentiation of the flow distance profiles. For all samples, regardless of being positive or negative, the flow velocity approached a constant velocity (between 2.4 and 4 pixel/second). However, there was a clear difference between positive and negative samples in the time required to reach that plateau flow velocity. We defined this as “time to constant velocity,” as it is the time point where the flow distance profile changes its slope (i.e., velocity) to a constant value. Such time to constant velocity occurred in less than 5 s, i.e., substantially earlier than 30 s. With the positive samples, there were not many singlet Ab-particles left after immunoagglutination occurred, and constant velocity was achieved after a shorter time, while negative samples took a longer time to reach constant velocity, as shown in Fig. 6D. We have not seen analysis of this phenomenon using a flow rate profile in other publications.
We found that the clinical samples have a wide range of turbidity that could represent the results of food debris, toothpaste, or other individual conditions. The flow results of the positive and negative samples were obviously distinguishable when the samples were relatively clear (not overwhelmingly contaminated by, i.e., food debris). The turbidity that occurred due to the intentional mixture of toothpaste led to inconsistency of the flow profiles, and the flow rate results are also out of the expected range for negative samples. Because each clinical sample is derived from different individuals, it is difficult to determine how to set the criteria. Toothpaste, food debris, bacteria, sugar, fibers, oil, or small proteins can interfere with the interfacial tension and viscosity of the solution and ultimately affect the flow characteristics. Therefore, by setting aside samples that are significantly turbid, we were able to demonstrate 89% accuracy using relatively clear clinical samples. The high accuracy is attributed to this method's extremely low LOD, despite the low viral load in saliva samples.
5. Conclusions
In this study, we successfully demonstrate a low-cost, point-of-care COVID-19 test that works with clinical saline gargle samples, utilizing a smartphone camera and automatic flow profile detection. The saline gargle method may be considered simpler and more comfortable for patients compared to nasopharyngeal swabs. The LOD of our assay was as low as 1 fg/μL SARS-CoV-2 from 1% saliva samples and 10 fg/μL SARS-CoV-2 from simulated saline gargle samples (15% saliva and 0.9% saline), both using pAb-particles. This method also provided a high specificity as demonstrated against the influenza A/H1N1 virus. Our assay is easy to use and requires minimal sample processing and training, requiring only a smartphone, a paper microfluidic chip, and antibody-conjugated particles. The assay is not affected by ambient light variations. We have successfully developed a Python script that automatically searches for channels and provides results. The sample-to-answer assay time was less than 15 min, with capillary flow time of less than 1 min. This assay was also implemented with clinical samples and was able to achieve an accuracy of 89% for relatively clean samples. Further characterization of the clinical and simulated samples found that surface tension was somewhat correlated to turbid samples, and that the time of last oral intake was significantly related. Estimated total protein concentration was not related to turbidity, as positive SARS-CoV-2 samples had somewhat higher total protein concentration compared to SARS-CoV-2 negative samples, perhaps due to the presence or absence of antigen proteins. Despite the limitations encountered with these turbid samples, avoiding eating and brushing teeth at least 30 min before testing should improve clarity of samples for accurate testing, similar to existing recommendations for SARS-CoV-2 testing using nucleic acid amplification. Although there are some limitations when working with turbid clinical samples, this platform represents a potential solution for rapid mass testing during infectious disease outbreaks that can be further improved in the future.
CRediT authorship contribution statement
Patarajarin Akarapipad: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization. Kattika Kaarj: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – review & editing. Lane E. Breshears: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Investigation, Writing – review & editing. Katelyn Sosnowski: Methodology, Validation, Investigation, Formal analysis, Data curation, Writing – review & editing. Jacob Baker: Software, Validation, Investigation, Data curation. Brandon T. Nguyen: Validation, Investigation, Formal analysis. Ciara Eades: Data curation, Formal analysis. Jennifer L. Uhrlaub: Methodology, Resources. Grace Quirk: Methodology, Resources. Janko Nikolich-Žugich: Supervision, Project administration, Funding acquisition. Michael Worobey: Conceptualization, Methodology, Supervision, Project administration, Funding acquisition. Jeong-Yeol Yoon: Conceptualization, Methodology, Formal analysis, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project was funded by The University of Arizona's Test All Test Smart program. L.E.B. acknowledges the University of Arizona NASA Space Grant Fellowship. P.A. acknowledges the Royal Thai Government Scholarship from the Ministry of Higher Education, Science, Research and Innovation, Thailand. K.K. acknowledges the scholarship from the Development and Promotion of Science and Technology Talents Project (DPST) of Thailand. K.S. acknowledges the Computational and Mathematical Modeling of Biomedical Systems Training Grant from the National Institute of General Medical Sciences (NIGMS), grant number GM132008. J.B. acknowledges W. L. Gore & Associates, Inc. for the undergraduate research fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114192.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Cohen J.I. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.04.20.20071423. [DOI] [Google Scholar]
- Byambasuren O., Cardona M., Bell K., Clark J., McLaws M.-L., Glasziou P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: systematic review and meta-analysis. J. Assoc. Med. Microbiol. Infect. Dis. Can. 2020;5:223–234. doi: 10.3138/jammi-2020-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camplisson C.K., Schilling K.M., Pedrotti W.L., Stone H.A., Martinez A.W. Two-ply channels for faster wicking in paper-based microfluidic devices. Lab Chip. 2015;15:4461–4466. doi: 10.1039/C5LC01115A. [DOI] [PubMed] [Google Scholar]
- Cho S., Park T.S., Nahapetian T.G., Yoon J.-Y. Smartphone-based, sensitive μPAD detection of urinary tract infection and gonorrhea. Biosens. Bioelectron. 2015;74:601–611. doi: 10.1016/j.bios.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Choi J.R. Development of point-of-care biosensors for COVID-19. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Breshears L.E., Gonzales A., Jennings C.M., Morrison C.M., Betancourt W.Q., Reynolds K.A., Yoon J.-Y. Norovirus detection in water samples at the level of single virus copies per microliter using a smartphone-based fluorescence microscope. Nat. Protoc. 2021;16:1452–1475. doi: 10.1038/s41596-020-00460-7. [DOI] [PubMed] [Google Scholar]
- Costa M.N., Veigas B., Jacob J.M., Santos D.S., Gomes J., Baptista P.V., Martins R., Inácio J., Fortunato E. A low cost, safe, disposable, rapid and self-sustainable paper-based platform for diagnostic testing: lab-on-paper. Nanotechnology. 2014;25:94006. doi: 10.1088/0957-4484/25/9/094006. [DOI] [PubMed] [Google Scholar]
- Czumbel L.M., Kiss S., Farkas N., Mandel I., Hegyi A., Nagy Á., Lohinai Z., Szakács Z., Hegyi P., Steward M.C., Varga G. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front. Med. 2020;7 doi: 10.3389/fmed.2020.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daerr A., Mogne A. Pendent_Drop: an ImageJ plugin to measure the surface tension from an image of a pendent drop. J. Open Res. Software. 2016;4:e3. doi: 10.5334/jors.97. [DOI] [Google Scholar]
- Goldfarb D.M., Tilley P., Al-Rawahi G.N., Srigley J.A., Ford G., Pedersen H., Pabbi A., Hannam-Clark S., Charles M., Dittrick M., Gadkar V.J., Pernica J.M., Hoang L.M.N. Self-collected saline gargle samples as an alternative to health care worker-collected nasopharyngeal swabs for COVID-19 diagnosis in outpatients. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.02427-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy B., Khan A., Metezai H., Blyth I., Asad H. The impact of false positive COVID-19 results in an area of low prevalence. Clin. Med. 2021;21:e54–e56. doi: 10.7861/clinmed.2020-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman T., Nissen K., Krambrich J., Rönnberg B., Akaberi D., Esmaeilzadeh M., Salaneck E., Lindahl J., Lundkvist Å. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect. Ecol. Epidemiol. 2020;10 doi: 10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihling C., Tänzler D., Hagemann S., Kehlen A., Hüttelmaier S., Arlt C., Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J. Proteome Res. 2020;19:4389–4392. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- Klug K.E., Reynolds K.A., Yoon J.-Y. A capillary flow dynamics-based sensing modality for direct environmental pathogen monitoring. Chem. Eur J. 2018;24:6025–6029. doi: 10.1002/chem.201800085. [DOI] [PubMed] [Google Scholar]
- Kothekar S.C., Ware A.M., Waghmare J.T., Momin S.A. Comparative analysis of the properties of Tween‐20, Tween‐60, Tween‐80, Arlacel‐60, and Arlacel‐80. J. Dispersion Sci. Technol. 2007;28:477–484. doi: 10.1080/01932690601108045. [DOI] [Google Scholar]
- Oliveira S.C., de Magalhães M.T.Q., Homan E.J. Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince-Guerra J.L., Almendares O., Nolen L.D., Gunn J.K.L., Dale A.P., Buono S.A., Deutsch-Feldman M., Suppiah S., Hao L., Zeng Y., Stevens V.A., Knipe K., Pompey J., Atherstone C., Bui D.P., Powell T., Tamin A., Harcourt J.L., Shewmaker P.L., Medrzycki M., Wong P., Jain S., Tejada-Strop A., Rogers S., Emery B., Wang H., Petway M., Bohannon C., Folster J.M., MacNeil A., Salerno R., Kuhnert-Tallman W., Tate J.E., Thornburg N.J., Kirking H.L., Sheiban K., Kudrna J., Cullen T., Komatsu K.K., Villanueva J.M., Rose D.A., Neatherlin J.C., Anderson M., Rota P.A., Honein M.A., Bower W.A. Evaluation of abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection at two community-based testing sites — Pima county, Arizona, november 3-17, 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;70:100–105. doi: 10.15585/mmwr.mm7003e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackart K. 2021. Covalent Coupling Protein to Carboxylated Microparticles via EDAC. Protocols.Io. [DOI] [Google Scholar]
- Teo A.K.J., Choudhury Y., Tan I.B., Cher C.Y., Chew S.H., Wan Z.Y., Cheng L.T.E., Oon L.L.E., Tan M.H., Chan K.S., Hsu L.Y. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Sci. Rep. 2021;11:3134. doi: 10.1038/s41598-021-82787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M. Swish, gargle, repeat: UArizona researcher explores mouth rinse test as alternative to COVID-19 nasal swab. 2020. https://news.arizona.edu/story/swish-gargle-repeat-uarizona-researcher-explores-mouth-rinse-test-alternative-covid-19-nasal 6.20.21, URL.
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Hu Y., Song Z.-G., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. bioRxiv. 2020 doi: 10.1101/2020.01.24.919183. 2020.01.24.919183. [DOI] [Google Scholar]
- Xu M., Wang D., Wang H., Zhang X., Liang T., Dai J., Li M., Zhang J., Zhang K., Xu D., Yu X. COVID-19 diagnostic testing: Technology perspective. Clin. Transl. Med. 2020;10:e158. doi: 10.1002/ctm2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.