Summary
Proper segregation of chromosomes during mitosis depends on ‘amphitelic attachments’ – load-bearing connections of sister kinetochores to the opposite spindle poles via bundles of microtubules, termed the ‘K-fibers’. Current models of spindle assembly assume that K-fibers arise largely from stochastic capture of microtubules which occurs at random times and locations and independently at sister kinetochores. We test this assumption by following movements of all kinetochores in human cells and determine that most amphitelic attachments form synchronously at a specific stage of spindle assembly and within a spatially distinct domain. This biorientation domain is enriched in bundles of antiparallel microtubules and perturbation of microtubule bundling changes the temporal and spatial dynamics of amphitelic attachment formation. Structural analyses indicate that interactions of kinetochores with microtubule bundles are mediated by non-centrosomal short microtubules that emanate from most kinetochores during early prometaphase. Computational analyses suggest that momentous molecular motor-driven interactions with antiparallel bundles rapidly convert these short microtubules into nascent K-fibers. Thus, load-bearing connections to the opposite spindle poles form simultaneously on sister kinetochores. In contrast to the uncoordinated sequential attachments of sister kinetochores expected in stochastic models of spindle assembly, our model envisions formation of amphitelic attachments as a deterministic process in that the chromosomes connect with the spindle poles synchronously at a specific stage of spindle assembly and at a defined location determined by the spindle architecture. Experimental analyses of changes in the kinetochore behavior in cells with perturbed activity of molecular motors CenpE and dynein confirm predictive power of the model.
Graphical Abstract
In Brief
Based on analyses of chromosome behavior in human cells and computational modelling, Renda et al. propose a mechanism for synchronous attachment of sister kinetochores to the opposite spindle poles via dynamic interactions between short non-centrosomal microtubules at the kinetochore and bundles of antiparallel microtubules within the spindle.
Introduction
For proper segregation during mitosis, each chromosome must ‘biorient’ - physically connect with both poles of the mitotic ‘spindle’, a macromolecular machine that self assembles from microtubules (MTs). Load-bearing attachments of chromosomes to MTs are mediated by the ‘kinetochores’ (KTs), a pair of macromolecular complexes on the opposite sides of the chromosome’s centromere. The goal of spindle assembly is to attach sister KTs to the opposite spindle pole (‘amphitelic attachment’). Current models of spindle assembly stem from the ‘Search & Capture’ (S&C) hypothesis1 that envisions formation of amphitelic attachments via sequential capture of MTs emanating from the opposite spindle poles by the sister KTs. This stochastic process is facilitated by localized nucleation of MTs near chromosomes2–4, guidance of MT growth towards KTs5–8, stabilization of the initial connections9, and regulation of KT architecture10–13. Even with these facilitations, random discovery of sister KTs is expected to yield variable duration of spindle assembly and frequent errors arising from accidental capture of MTs produced by a ‘wrong’ spindle pole2,14–17. These expectations seem at odds with the rapid and robust cell division observed in chromosomally stable cells.
Here we analyze KT behavior and MT organization to determine when, where, and how amphitelic attachments form during mitosis in diploid human cells. We find that within a cell, chromosomes biorient synchronously at a defined stage of spindle elongation and within a spatially distinct ‘biorientation domain’ of the spindle. Computational analyses suggests that amphitelic attachments form in a single step via dynamic motor-mediated interactions between short MTs protruding from sister KTs and bundles of anti-parallel MT within the biorientation domain. Experimental perturbations of MT bundling or motor activities at the KTs change the dynamics of chromosome biorientation in a manner consistent with the model predictions. Thus, simultaneous connection of sister KTs to bundles of antiparallel MT is likely a major mechanism for chromosome biorientation in chromosomally stable human cells.
Results
Amphitelic attachments form predominantly at a specific stage of spindle elongation
We follow movements of KTs in chromosomally stable human RPE1 cells in 3-D at 5-s intervals. To minimize stress from fluorescence microscopy, we tag KTs and centrioles in the same color18,19 and discriminate these organelles by their behavior (Figure 1A, Video S1). Under these conditions, RPE1 cells initiate anaphase 23±3 min (N=17) after nuclear envelope breakdown (NEB) and show no chromosome mis-segregation as expected for normal mitosis10,19.
Figure 1. Amphitelic attachments form at a specific stage of spindle elongation.
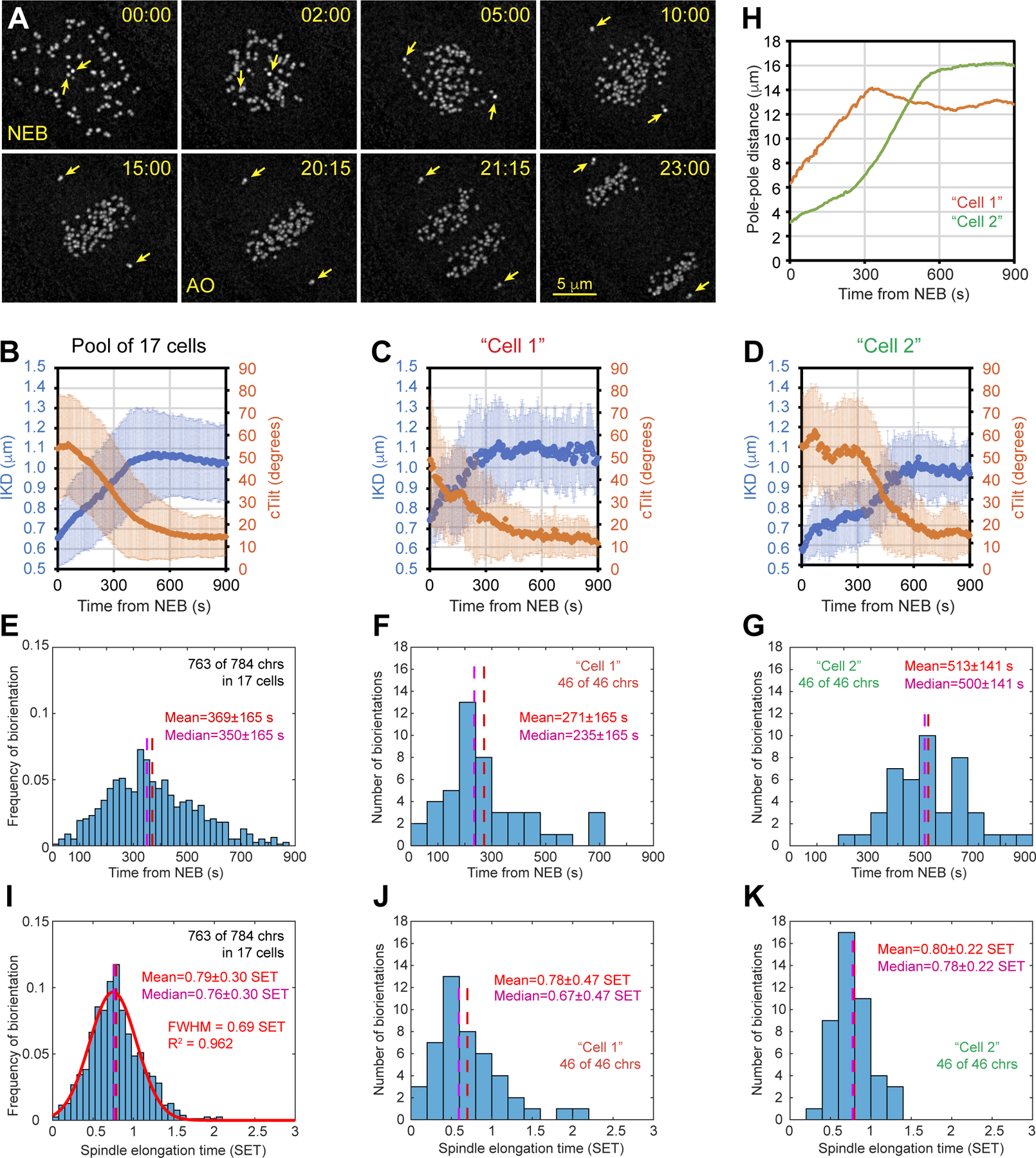
(A) Selected timepoints from a recording of mitosis in RPE1 cell at 5-s intervals. Frames are maximum-intensity projections of the entire cell. KTs and centrioles are tagged with CenpA-GFP and Centrin1-GFP. Arrows mark centrioles. Nuclear envelope breaks down at 00:00 (NEB) and anaphase onsets at 20:15 (AO). Scale bar, 5 μm. (B) Dynamics of the distance between sister KTs (IKD, blue) and angle between the centromere axis and spindle axis (cTilt, orange) for 784 KTs in 17 cells. (C-D) Similar to (B) but the plots present two selected cells. Images of Cell1 are shown in (A). (E) Temporal distribution of biorientation events in the population of 17 cells. (F-G) Similar to (E) but the plots present two selected cells. (H) Dynamics of spindle length in the two selected cells. (I) As in (E) but time is normalized by the duration of spindle elongation (SET) for each cell in the population. Notice that the distribution is nearly normal (red line). (J-K) As in (F) and (G) but time is expressed in SET. Error bars in (B)-(D) are standard deviation. See also Figure S1 and Video S1.
Two direct consequences of amphitelic attachment are a decrease in the angle between the line connecting sister KTs (i.e., centromere axis) and the line connecting spindle poles (i.e., spindle axis) as well as an increase in the distance between sister KTs (Figure S1A, cTilt and IKD). Consistent with previous reports19, we observe that the mean value of cTilt decreases while the mean IKD increases during the first 8 min of prometaphase in the population of 17 cells (Figure 1B, 784 chromosomes). However, significant variability exists in the dynamics of cTilt and IKD among individual cells. In some cells, these metrics change rapidly and plateau ~ 6 min after NEB (Figure 1C). In other cells, the changes are delayed for several minutes (Figure 1D).
Euploid cells remain in mitosis until all chromosomes become bioriented and therefore IKD and cTilt values observed just prior to anaphase onset (AO) characterize a pool of chromosomes with >99% amphitelic attachments. We reason that when both IKD and cTilt of a chromosome converge within one STD from the mean pre-AO values (Figure S1B), the chromosome has formed amphitelic attachments. Specifically, we probe trajectories of sister KTs for the timepoint when IKD exceeds 0.9 μm while cTilt remains below ~22.5° (π/8) for at least 30 seconds (Figure S1C). By these conservative criteria, 763 of 784 chromosomes (97.3%) in the 17 analyzed cells achieve biorientation <15 min after NEB with the maximal probability to form amphitelic attachments ~6 min after NEB (Figure 1E). However, temporal distributions of biorientation events vary significantly among individual cells. In some cells, most chromosomes biorient <4 min after NEB (Figure 1F, Median). In other cells, amphitelic attachments form en masse >8 min after NEB (Figure 1G). Biorientations occur earlier in cells where the spindle elongates to its full length rapidly (compare ‘Cell1’ and ‘Cell2’ in Figure 1C–D, F–G, and H). This observation prompted us to test whether the peak of biorientation events coincides with a specific stage of spindle elongation. For this purpose, we normalize progression through prometaphase by the duration of spindle elongation (0=NEB, 1=timepoint when spindle elongation stops; see Methods). On the ‘spindle elongation time’ (SET) scale, biorientations peaks coincide in various cells and the distribution of biorientation events in the population is narrow and nearly normal (Figure 1I–K). Thus, the majority of amphitelic attachments form during a short interval when the spindle reaches ~80% of its maximum length irrespective of when this stage of spindle assembly occurs in physical time. These data suggest that the state of the spindle determines when most chromosomes become bioriented.
Amphitelic attachments form rapidly near bundles of microtubules
To determine the trigger of amphitelic attachment formation we image SiR-Tubulin20 in cells with GFP-tagged KTs and centrioles. Spindle architecture and duration of mitosis are normal in cells followed at 30-s intervals (Video S2), which is sufficient for observing behavior of centromeres during formation of amphitelic attachments.
Recordings of 18 cells suggest that amphitelic attachments form when a KT encounters a bundle of MTs. Within a minute after the initial contact with a bundle, the centromere stretches to >0.9 μm and its axis aligns with the bundle (Figure 2A,B). To estimate the frequency of contacts between KTs and MT bundles we analyze 3-D distribution of spindle components in fixed prometaphase cells. MT bundles are detected via immunostaining for PRC1, a MT-associated protein known to bundle anti-parallel MTs21–24. In RPE1 cells, PRC1 decorates a subset of MTs throughout prometaphase (Figure S2A) and a similar pattern is observed in cells that express a full-length PRC1-GFP fusion (Figure S2B). In cells with ~12 μm spindles, which corresponds to ~80% of the full length and therefore to the stage of spindle elongation when most amphitelic attachments form (see Figure 1), PRC1-decorated bundles form a barrel around the spindle axis (Figure 2C) and the KTs are adjacent to the bundles (Figure 2D, mean distance 0.44±0.24 μm, 909 KTs in 11 cells).
Figure 2. Amphitelic attachments form near microtubule bundles.
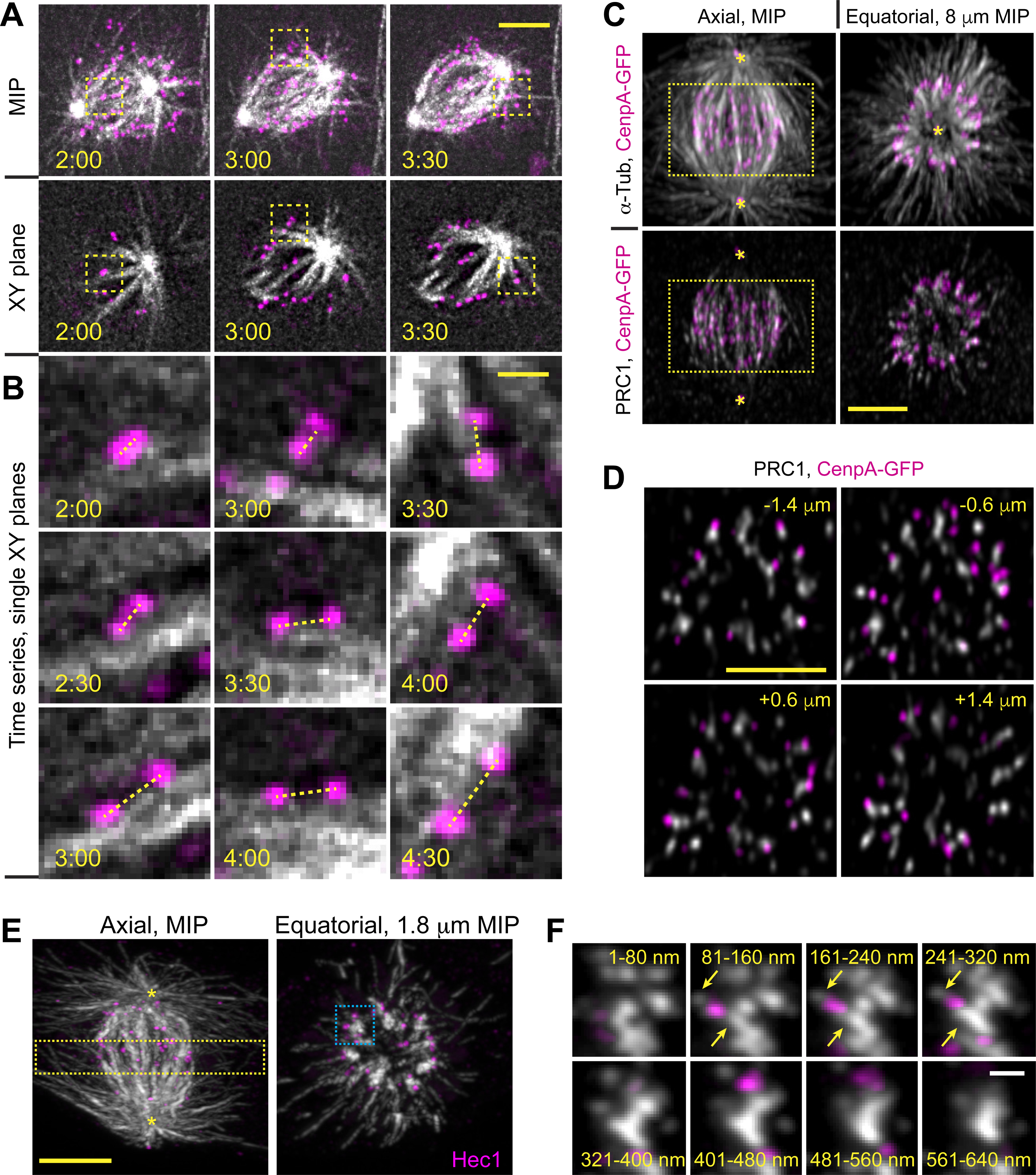
(A) Selected timepoints from a recording of RPE1 cell with GFP-tagged KTs and centrioles (shown in magenta) and SiR-Tubulin-labeled MTs (shown in grayscale). Maximum-intensity projections (top row) and selected single planes (bottom row) are shown for each timepoint. (B) Biorientation behavior of three KTs marked with boxes in (A). Notice that centromeres abruptly orient parallel to a MT bundle and stretch within 1 min after the initial contact with this bundle. (C) Spatial arrangement of MTs (α-Tubulin), MT bundles (PRC1), KTs (CenpA-GFP), and centrioles (Ctn1-GFP) in a prometaphase cell with ~12-μm long spindle. Axial view is a maximum-intensity projection of the entire spindle. Equatorial view presents a partial volume denoted by the box in Axial view. Asterisks denote centrioles. (D) Individual equatorial planes from the volume shown in (C). Distance from each plane to the spindle equator is shown. (E) Similar to (C) but this volume is constructed from a series of 80-nm sections (array tomography) and KTs are visualized via immunostaining for Hec1. (F) Sequential tomography slices detailing MT distribution near KTs marked with the blue box in (E). Arrows denote α-Tubulin spots between KTs and MT bundles. Scale bars, 5 μm in (A), (C), (D) and (E), 1 μm in (B), and 0.5 μm in (F). Asterisks mark positions of spindle poles. See also Figure S2 and Video S2.
To detail the interaction between KTs and MT bundles we employ array tomography25 (AT). A higher signal/noise ratio of AT reveals locations of short MTs that escape detection in conventional fluorescence microscopy26,27. Analysis of 5 prometaphase cells confirms the presence of MT bundles arranged in a ring and oriented roughly parallel to the spindle axis (Figure 2E). KTs reside near (~500 nm), yet not directly attached to these bundles. Instead, small tubulin spots bridge the Hec1-containing outer KT and the adjacent bundle (Figure 2F). Similar tubulin spots have been reported to contain variable numbers of short non-centrosomal MTs in correlative LM/EM analysis of early prometaphase RPE1 cells26.
Our observation that amphitelic attachments form rapidly near MT bundles prompted us to explore whether the time and place of amphitelic attachment formation change under conditions that perturb bundling of MTs within the spindle. Towards this goal we introduced an inducible shRNA construct against PRC1 to RPE1 cells with GFP-tagged KTs and centrioles (Methods). Consistent with previous reports, cells depleted of PRC1 progress through mitosis28,29; however, the central spindle that normally comprises MT bundles is not present during telophase (Figure S2C,D). For reproducibility, only cells that display this phenotype are included in our analyses (see Methods).
Depletion of PRC1 does not significantly change the shape and dimensions of the spindle (Figure 3A); however, MTs are distributed in a more homogeneous pattern within the spindle and the ring of MT bundles is not present during mid-prometaphase (Figure 3B, compare with Figure 2D).
Figure 3. Lack of microtubule bundles delays formation of amphitelic attachments.
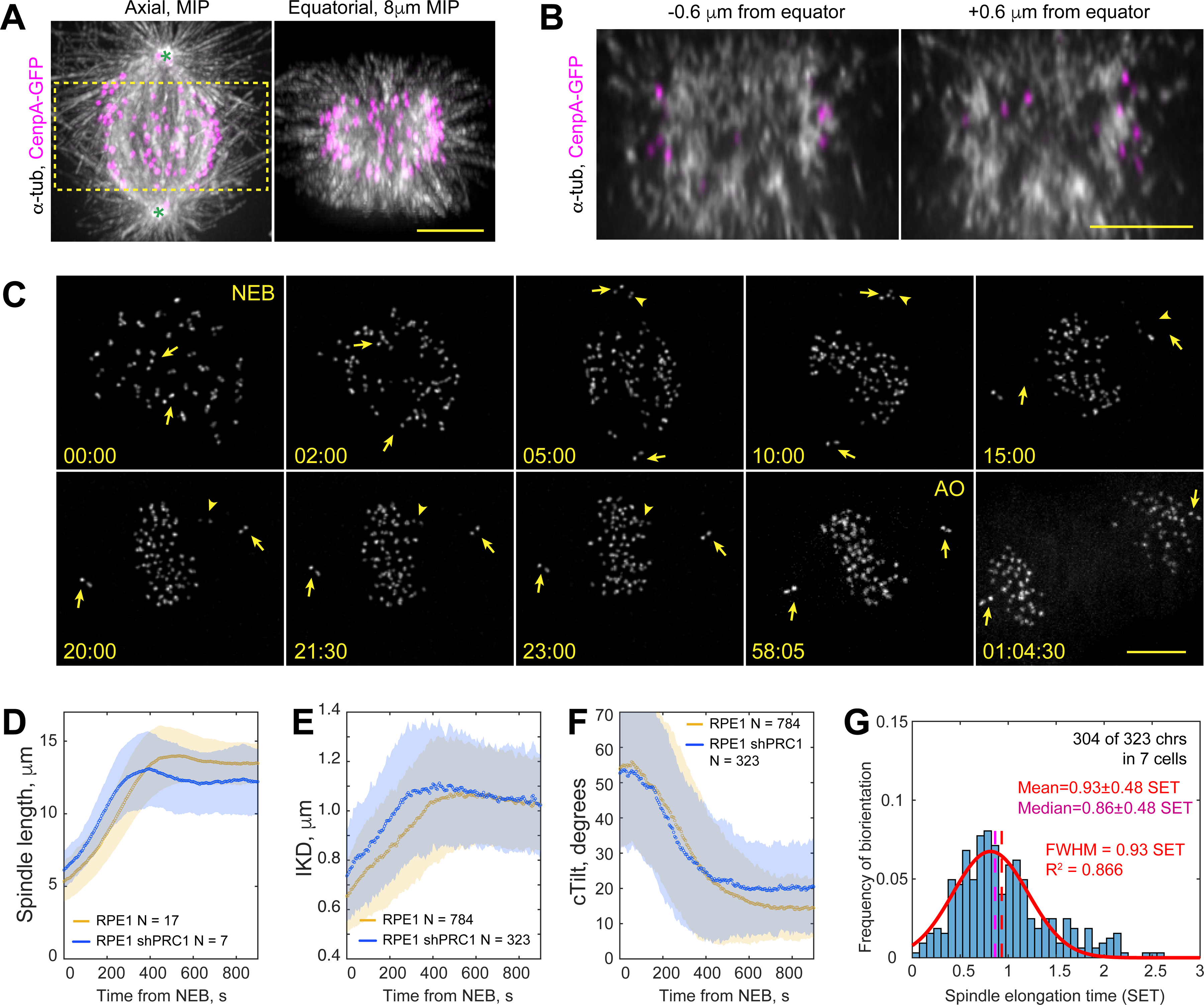
(A) Spatial arrangement of MTs (α-Tubulin), KTs (CenpA-GFP), and centrioles (Ctn1-GFP) in a prometaphase shRNA-depleted of PRC1. Axial view is a maximum-intensity projection of the entire spindle. Equatorial view presents a partial volume denoted by the box in Axial view. Asterisks denote centrioles (~12-μm spindle length). (B) Individual equatorial planes from the from the volume shown in (A). (C) Selected timepoints from a recording of PRC1-depleted RPE1 cell with GFP-tagged KTs and centrioles. Frames are maximum-intensity projections of the entire cell. Nuclear envelope breaks down at 00:00 (NEB) and anaphase onsets at 58:05 (AO). Arrows mark centrioles, arrowheads – a monooriented chromosome. (D-F) Dynamics of mean spindle length (D), distance between sister KTs (E, IKD), and the angle between the centromere and spindle axes (F, cTilt). Colored corridors are ±1 STD. (G) Temporal distribution of biorientation events in PRC1-depleted cells, normalized to spindle elongation time. Notice significant deviation from the normal distribution (red line). Scale bars, 5 μm in (A), (B) and (C). See also Figures S2, S3 and Video S3.
In ~75% of RPE1 cells, the centrosomes reside on the dorsal and ventral surfaces of the nucleus at NEB19,30. As the spindle elongates during prometaphase, its axis reorients from nearly orthogonal to nearly parallel to the coverslip surface (Figure 1A). This pattern as well as the rate of spindle elongation are similar in PRC1-depleted cells (Figure 3C,D, Video S3). The mean distance between sister KTs (IKD) in PRC1-depleted cells increases faster during early prometaphase yet it plateaus at the same level as in the wild-type (wt) RPE1 (Figure 3E). Orientation of centromeres (cTilt) improves similarly in the control vs. PRC1-depleted cells during earlier prometaphase. However, the mean value plateaus at a higher level and the STD is twice as large in the latter (Figure 3F, p<0.001 in Student’s T-test). The increased STD reflects instability in the orientation of individual centromeres that often ‘tumble’ repeatedly after a brief period of proper alignment. PRC1-depleted cells often display monooriented chromosomes that ultimately congress onto the metaphase plate prior to anaphase (Figure 3C). Consistent with the notion that monooriented chromosomes prevent mitotic exit31, both the mean and variability of mitotic duration increase significantly in PRC1 cells (37±10 min, N=30 vs. 23±3 min, N=17 in the wt RPE1, p < 0.001 in Student’s T-test).
Formation of amphitelic attachments in PRC1-depleted cells is delayed with both the mean and median values significantly larger than in wt RPE1 (Figure 3G, p < 0.0001 in Kruskal-Wallis test). Temporal distribution of biorientation events is positively skewed with many chromosomes achieving the amphitelic state during late prometaphase (Figure 3G, compare with Figure 1I). Thus, perturbation of MT bundling within the spindle impedes the normal dynamics of chromosome biorientation.
Microtubule bundles delineate a spatial domain that promotes chromosome biorientation
To determine where within the spindle the majority of amphitelic attachments form and how the chromosomes reach their biorientation locales, we analyzed centromere trajectories prior to and post biorientation of each chromosome. Extensive rotations and positional shifts of the spindle during prometaphase obscure centromere movements plotted in the conventional Cartesian coordinates linked to the microscope stage (Figure S3A). We overcome this problem by expressing positions in a spindle-centric cylindrical coordinate system (Figure S3A‘, see Methods), which allows us to view trajectories from fixed relative viewpoints, specifically in the Axial and Equatorial projections (Figure S3B,D).
Prior to biorientation, centromeres rapidly and linearly move toward the center of the spindle (Figure S3B). The linear inward movements stop abruptly when centromeres arrive 2.5–3.5 μm from the spindle axis and within ~3 μm from the equator (Figure S3B). Upon arrival to this part of the spindle, IKD and cTilt exceed the biorientation thresholds and the centromere begins to move roughly parallel to the spindle axis as expected for bioriented chromosomes (Figure S3B‘). The abrupt change in the motion pattern is consistent with the rapid formation of amphitelic attachments upon a contact with a MT bundle (Figure 2B). To assess the spatial distribution of biorientation events in multiple cells with variable dimensions of the spindle, we normalize distances by the maximal spindle length (MSL) achieved in each cell at the end of spindle elongation. This approach demonstrates that amphitelic attachments form predominantly within a doughnut (toroid) around the spindle axis (Figure 4A) with the mean equatorial radius of 0.23 MSL, thickness of the wall 0.19 MSL, and the axial length of 0.32 MSL (Figure S3C, 763 biorientations in 17 cells).
Figure 4. Amphitelic attachments form within a spindle domain enriched in microtubule bundles.
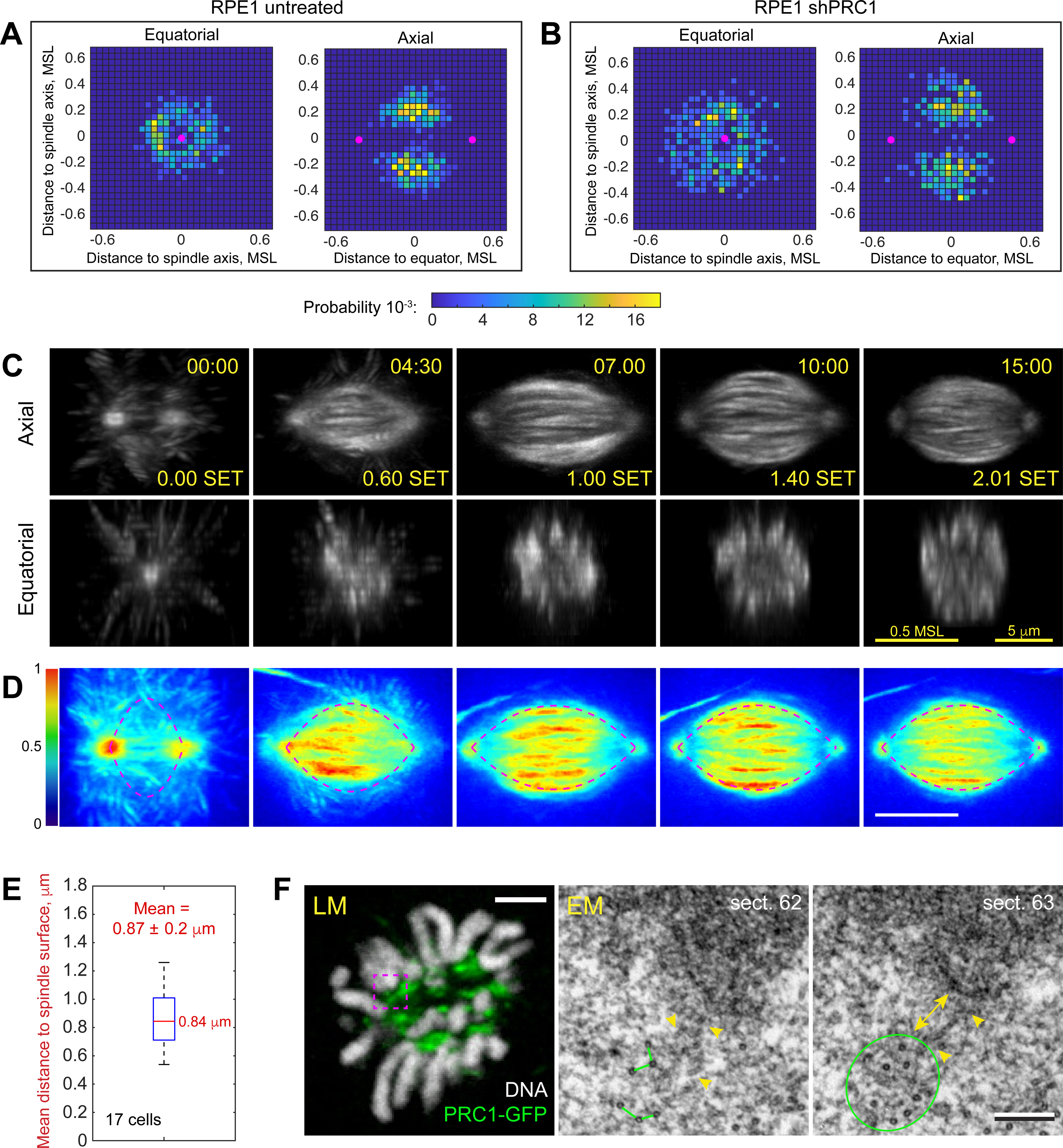
(A,B) Spatial distribution of biorientation events in the untreated (A) and PRC1-depleted (B) cells. 2D histograms in the Equatorial and Axial planes are shown. Distances are normalized to the Maximal Spindle Length in each cell. Magenta dots denote positions of spindle poles at the time with maximum probability of biorientation. (C) Selected timepoints from a timelapse recording of mitosis in RPE1 cells expressing PRC1-GFP. Axial and equatorial maximum-intensity projections of 3D volumes are shown. The volumes are aligned at each timepoint to stabilize the spindle position and orientation. Timestamps are in min:sec after NEB and in fraction of Spindle Elongation Time (SET). Scale bars are 5 μm and 0.5 of the Maximal Spindle Length (MSL) reached in this cell. (D) Average of 3-D time-lapse recordings aligned as in (A) and spatially normalized by the Maximal Spindle Length in each cell. Color map encodes intensity of PRC1-GFP in the averaged volume. Dashed lines approximate the edge of PRC1-enriched domain by an empirically constructed catenary function (see Methods). Timestamps are in SET. Scale bar is 0.5 MSL. (E) Tukey’s boxplot of Euclidian distances from centromeres to the catenary (edge of PRC1-enriched domain) at the time of amphitelic attachment formation. Mean value is reported with STD. (F) Typical arrangement of microtubules near kinetochores adjacent to PRC1-decorated bundles. LM – a single-plane image depicting PRC1-GFP (green) and chromosomes (Hoechst 33342, greyscale) in a fixed cell. EM – 80-nm serial sections through the area boxed in LM. Kinetochore plate is ~250 nm (yellow double arrow) from the edge of a bundle comprising 10 microtubules (green circle) with 50–70-nm spacing between individual microtubules (green lines). Short microtubules (arrowheads) bridge the bundle and the kinetochore plate. See also Video S4.
The rapid inward movement of centromeres during early prometaphase persists in cells depleted of PRC1. However, the centromeres do not display the abrupt change in the motion pattern typical for the wt RPE1. Instead, after the rapid delivery to within ~3.5 μm from the spindle axis, the centromeres drift in both axial and equatorial directions for variable times, which is manifested as jitter in the late segments of pre-biorientation trajectories (Figure S3D). These movements convert into more regular axial motion (Figure S3D‘) after the IKD and cTilt values exceed their biorientation thresholds. Formation of amphitelic attachments occurs within a large volume within the spindle (Figure 4B) and the distribution of biorientation events in the equatorial plane deviates from the normal distribution observed in the wt RPE1 (Figure S3E, compare with C). Thus, the sharply delineated barrel-shaped domain that promotes chromosome biorientation in the wt RPE1 cells (Figure 4A), disintegrates when MT bundling is perturbed via depletion of PRC1 (Figure 4B).
Dimensions of the spindle as well as its shape change as the cell progresses through prometaphase, thus the volume enriched in MT bundles is not constant. To delineate the shape of the biorientation domain at various stages of spindle assembly, we employ constitutive expression of a GFP-tagged full-length PRC1 in RPE1 cells. At a moderate expression level, localization of this construct (Figure S2B) is similar to the distribution of endogenous PRC1 (Figure S2A). Further, cells that express GFP-PRC1 progress through mitosis at a normal pace and segregate chromosomes properly (Video S4).
Live-cell recordings demonstrate that within the first 30 seconds of prometaphase PRC1-GFP is recruited to a subset of irregularly oriented MTs (Video S4). Within ~4 min as the spindle elongates to ~0.8 of its maximum length, these MTs organize into a hollow barrel-shaped array roughly parallel to the spindle axis (Figure 4C, Video S4). To reveal the typical shape of PRC1-GFP distribution, we averaged recordings of 12 cells with dimensions normalized by MSL. The edge of the PRC1-enriched domain (Figure 4D) resembles the shape of a chain hung from two posts, which prompted us to approximate this edge by a catenary function with coefficients proportional to the spindle length (see Methods). We find that over half of centromeres reside <0.85 μm from the catenary at the timepoint when IKD and cTilt exceed their biorientation thresholds irrespective of whether this occurs during earlier or later prometaphase (Figure 4E). Further, we find that ~94% of centromeres approach closer than 0.85 μm from the catenary prior to their biorientation.
To detail the interactions between MTs and KTs adjacent to MT bundles we employed correlative light/electron microscopy in RPE1 cells expressing PRC1-GFP. Analysis of two prometaphase cells with ~12-μm spindles (~0.8 MSL) demonstrates presence of short (300–600 nm) MTs that bridge the KT plates with bundles of 10–15 PRC1-decorated MTs (Figure 4F). These observations are consistent with the previous report that KTs residing on the spindle surface are end-on attached to numerous short non-centrosomal MTs that emanate from the KT and intermix with the spindle MTs26.
Computational model of biorientation on bundled antiparallel microtubules
Our observations suggest that during normal mitosis, amphitelic attachments form rapidly within a defined spatial domain where short MTs emanating from the KTs encounter MT bundles decorated with PRC1. Previous investigations identify multi-valent complexes of the minus-end directed molecular motor dynein and NuMA as the linker that connects the minus ends of MTs protruding from KTs to the adjacent MT bundles and forcefully pull KTs poleward27,32–34. These findings prompted us to computationally explore whether motor-mediated interactions between the minus ends of disorganized MTs emanating from the KT (Figure 5A) and bundles of antiparallel MT bundles within the spindle provide an efficient means for rapid biorientation. We developed a stochastic spatial mechanical model in which sister KTs are the ends of the centromeric spring (Figure 5B). Short MTs randomly pivot around their plus ends anchored at the sister KTs when dyneins at their minus ends are unbound from the long spindle MTs. Binding can occur when short MT minus ends are near a long spindle MT. While bound, dyneins pull the short MT minus ends toward the long MT minus ends. Between these kinetic events, the positions of the KTs evolve via forces mediated through bound short MTs (see Methods S1 for further details and mathematics of the model). Intuitively, stochastic dynein-mediated connections at the short MT ends emanating from the KTs in various directions (Figure 5A) would jerk the KTs around and lead to unstable cTilt and low IKD values. However, we establish numerically that a simple assumption leads to a different outcome: if the rate of the short-long MT unbinding is lower when short MTs are pulled in the directions that are more parallel to the centrosomal axis (Figure 5B), then the following geometric-mechanical positive feedback ensues. Even when the centromere axis is initially perpendicular (Figure 5C, t=0s) to the long MTs, the random forces from dynein tilt the axis (Figure 5C, t=10s). Then, the short MTs oriented more parallel to the centrosomal axis are bound stably, while the short MTs oriented more normal to the centrosomal axis unbind rapidly, swing and rebind, so ultimately most of the short MTs from each KT bind only to those long MTs that lead the short MT minus ends in the same direction to which the KT is tilted (Figure 5C, t=30s). This MT polarity sorting resulting from the dynein motors’ tug-of-war aligns the centromere with the spindle MTs (decreases cTilt). The improvement in centromere orientation further increases the disparity between the oppositely pulling short MTs, so the initially disorganized array of short MTs protruding from the KT transforms into a bundle of parallel MTs and the forces acting along these ‘nascent K-fibers’ stretch the centromere (increase IKD).
Figure 5. Computational model of chromosome biorientation.
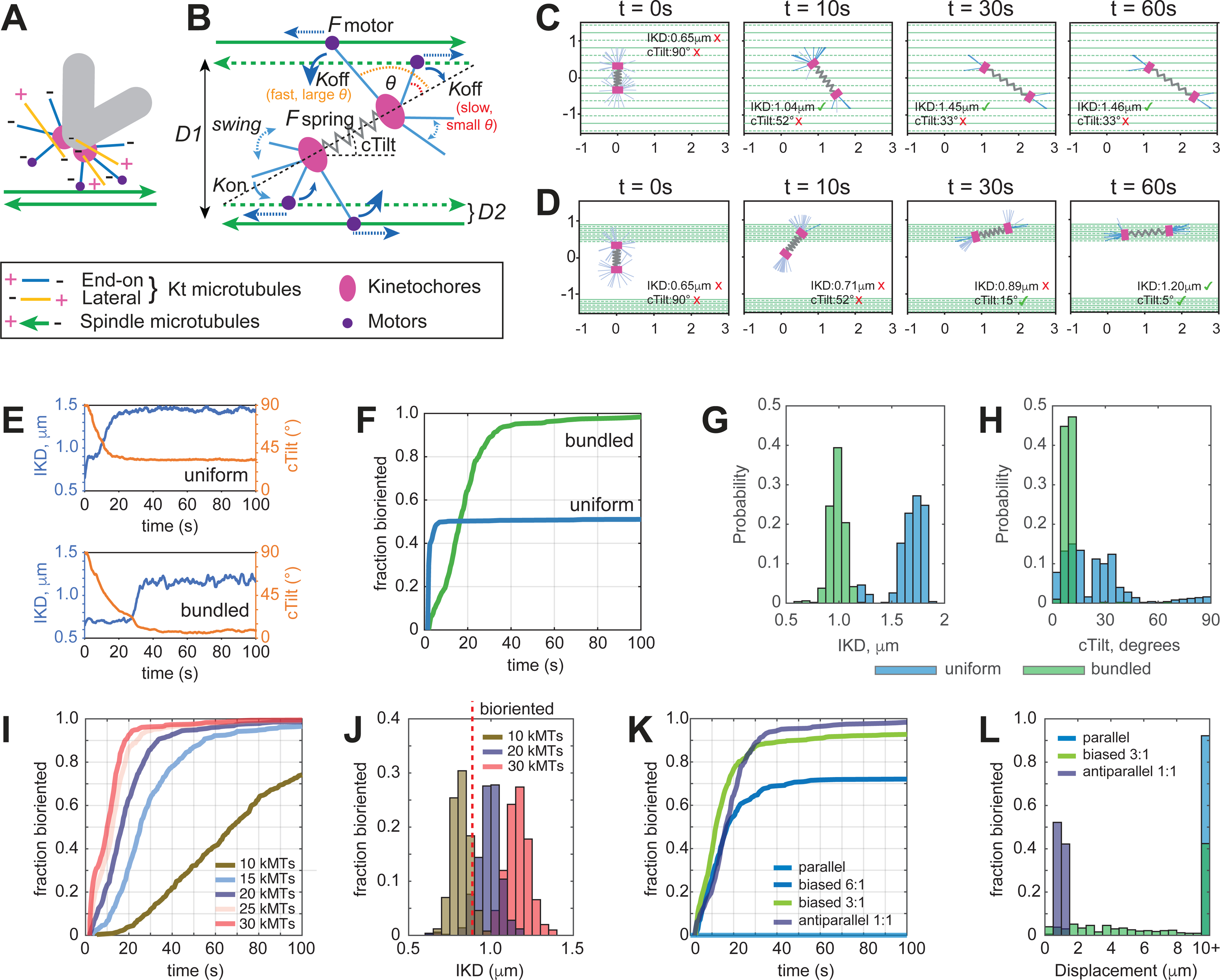
(A) MT arrangement at the KTs considered in the model. Only MTs with the plus end attached to the KT and the minus end protruding outwards (blue lines) contribute to the interaction with the spindle. This interaction is mediated by a minus end-directed motor (dynein, purple dots). (B) Principal framework of the model. Protruding KT MTs pivot around the KTs until their minus ends connect to the walls of reachable spindle MTs with rate defined by Kon when in proximity. Once connected, the minus end attempts to move along the spindle MT towards its minus end. Connections of MTs protruding from sister KTs to spindle MTs of opposite polarity stretches the centromere and increases longevity of the connection by decreasing the rate Koff via their spatial organization. Orientation of the spindle MTs is intermittent, and their organization is characterized by the distances D1 and D2. See Supplemental Text for details. (C-E) Examples of behavior predicted for centromeres (single simulation run). (C) On spindle surface comprising evenly spaced (D1=D2=200 nm distance) MTs of intermittent polarity, the centromere stretches but fails to orient along the spindle axis. (D) On spindle surface comprising MT bundles separated by D1=2 μm (10 MTs of intermittent polarity, D2=50 nm), the centromere orients and stretches to the level expected for bioriented chromosomes. (E) dynamics of IKD and cTilt for centromeres shown in (C) and (D). (F) Fraction of centromeres predicted to achieve biorientation at various times for the evaluated scenarios. (G,H) Predicted distributions of IKD and cTilt after 100 s of interaction with the spindle surface comprising evenly spaced individual MTs (blue) vs. MT bundles (green). (I) Fraction of bioriented centromeres for various numbers of MT minus ends protruding from the KT. (J) Distributions of IKD predicted for various numbers of MT minus ends after 100 s of interaction with MT bundles. (K) Fraction of bioriented centromeres for various ratios of MTs with the opposite polarity. Notice that biorientation fails on parallel MT bundles (light blue). (L) Predicted displacement from the point of initial contact towards the bundle terminus with the greater number of minus ends for bundles with various polarity bias (100 s of interaction, 20 MTs protruding from the kinetochore). See also Video S5.
Our assumption of differences in the stability of motor-mediated MT interactions is supported by the observations of increased detachment rates under higher angles between the pulling force vector and the track of molecular motors35,36. Further, the notion that pulling sister KTs towards the opposite spindle poles stabilize MT attachments is generally accepted37–41.
To test feasibility of rapid biorientation via interaction between the short MTs at KTs and the spindle we run a series of computational simulations based on the rules and forces presented in Figure 5B (see Methods S1 for parameters). First, we explore whether the proposed mechanism depends on the distribution of long MTs within the spindle and more specifically on whether the surface of the spindle comprises a uniform array of evenly spaced individual MTs vs. a series of MT bundles (Figure 5C–E). We find that centromeres interacting with a surface of evenly spaced antiparallel MTs stretch rapidly because short MT find many long MTs to attach to and pull along. However, these interactions fail to orient the centromere parallel to the spindle axis, because the randomly selected long MTs along which the short MTs pull, could be widely separated (Figure 5C, Video S5). In contrast, a centromere that interacts with a bundle comprising ~10 anti-parallel MTs at ~50 nm spacing, which resembles the configuration observed by correlative LM/EM (see Figure 4F), both stretches and orients parallel to the bundle (Figure 5D, Video S5). Within ~100 s from the onset of the interaction, virtually all modeled centromeres on a bundle reach values of IKD and cTilt expected for bioriented chromosomes. In contrast, only about 50% of centromeres on a uniform MT surface satisfy both biorientation criteria (Figure 5F). Interestingly, interactions with a surface of individual MTs are predicted to stretch the centromere to a greater extent than interactions with a bundle (Figure 5G). The model also predicts a greater variability in cTilt angles for centromeres that interact with individual MTs (Figure 5H). These predictions are consistent with the dynamics of IKD and cTilt values observed in the wt vs. PRC1-depleted cells that lack MT bundles (see Figure 3).
Exploration of the model by parameter sweeps identify two factors important for rapid and efficient biorientation. First, the process depends on the number of MT minus ends protruding from the KT when it encounters a bundle. While KTs with >20 attached MTs biorient efficiently, the time required for the formation of amphitelic attachments increases rapidly for <10 attached MTs (Figure 5I). The delay arises because KTs with a lower number of attached microtubules fail to stretch the centromere above the biorientation threshold after 100 s of interaction (Figure 5J). Conversely, centromeres with 30 or more attached MTs tend to over-stretch (Figure 5J). Thus, ~20 MT minus ends protruding from the KT are optimal for biorientation. This number is consistent with the number of short MTs detected in EM reconstructions of KTs positioned on the spindle surface in early-prometaphase RPE1 cells26. Second, an important determinant of biorientation efficiency is the ratio of MTs with opposite polarity within the bundle. When the polarity bias exceeds 3:1, many centromeres fail to form amphitelic attachments in a reasonable time (Figure 5K). Further, dynein-mediated interactions with a polarity-biased bundle are predicted to shift the centromere from the place of the initial encounter towards the terminus of the bundle with the higher number of MT minus ends. In the context of the spindle, this means that interactions with polarity-biased microtubules promote chromosome monoorientation (Figure 5L). Thus, rapid formation of amphitelic attachments is predicted to be most efficient near the spindle equator, where polarity bias within the bundles is expected to be minimal.
Changes in chromosome behavior upon inactivation of microtubule motors at the kinetochore are consistent with the model prediction
The model predicts that rapid formation of amphitelic attachments occurs when centromeres with an optimal number of short MTs attached to sister KTs promptly gather within the biorientation domain enriched in MT bundles. Thus, perturbation of MT bundling, delayed delivery of centromeres to the bundle-enriched domain, or an insufficient number of MT minus ends protruding from the KT would all affect the temporal and spatial distributions of biorientation events. Consistent with the model prediction, we observe a delayed and less synchronous formation of amphitelic attachments within a larger volume when MT bundling is inhibited via PRC1 depletion (Figure 4A,B). To test whether abnormal transport of centromeres to the biorientation domain or a lower number of MT minus ends protruding from sister KTs yield effects that are consistent with the model, we perturb the activities of molecular motors CenpE (kinesin 7) or cytoplasmic dynein at the KT. Chemical inhibition of CenpE has been shown to decrease the number of short MTs end-on attached to KTs during early prometaphase26, likely due to role of this motor in the conversion from lateral to end-on interactions with captured MTs42. Dynein has been implicated in the transport of chromosomes towards the spindle during prometaphase43–45. Thus, KTs lacking this motor are likely to encounter MT bundles at a later stage of spindle assembly.
A cell permeable inhibitor GSK923295 offers an efficient means for inhibiting CenpE activity46. To assess the role of dynein at the KT, we employ RPE1 cells with genetically ablated Rod, an adapter protein required for the recruitment of dynein to KTs12,47,48. Inhibition of CenpE or failure to recruit dynein to the KTs neither noticeably affect the spindle architecture (Figure S4A) nor it changes the pattern of spindle orientation and the rate of spindle elongation during prometaphase (Figure S4B). However, dynamics of chromosome biorientation change prominently in these cells as evident from changes in the dynamics of cTilt and IKD (Figure S4C,D). Monooriented chromosomes remaining near a spindle pole for an extended time are commonly observed (Figure 6A–B, Videos S6,S7). While many chromosomes reside closer to one spindle pole (<0.25 of the contemporary spindle length) at NEB, linear movements toward the spindle center (Figure S3B‘) rapidly decrease the number of these initially monooriented chromosomes in the untreated RPE1 (Figure 6C). In CenpE-inhibited cells, the number of monooriented chromosomes decreases similarly during the first 400 s of prometaphase but subsequently it increases (Figure 6C) as many chromosomes move towards the spindle center in early prometaphase and then migrate along the spindle axis towards a spindle pole (Figure 6C, S4E). In contrast, the number of monooriented chromosomes in RodΔ/Δ RPE1 declines slower although steadily throughout prometaphase in (Figure 6C). The slower decline correlates with the lack of rapid centripetal movements of centromeres during early prometaphase (Figure S4F). Peripheral chromosomes in RodΔ/Δ gradually migrate closer to the spindle axis and the equator via directionally unstable movements (Figure S4F).
Figure 6. Dynamics and localization of amphitelic attachment formation in cells lacking activities of CenpE or dynein at the kinetochore.
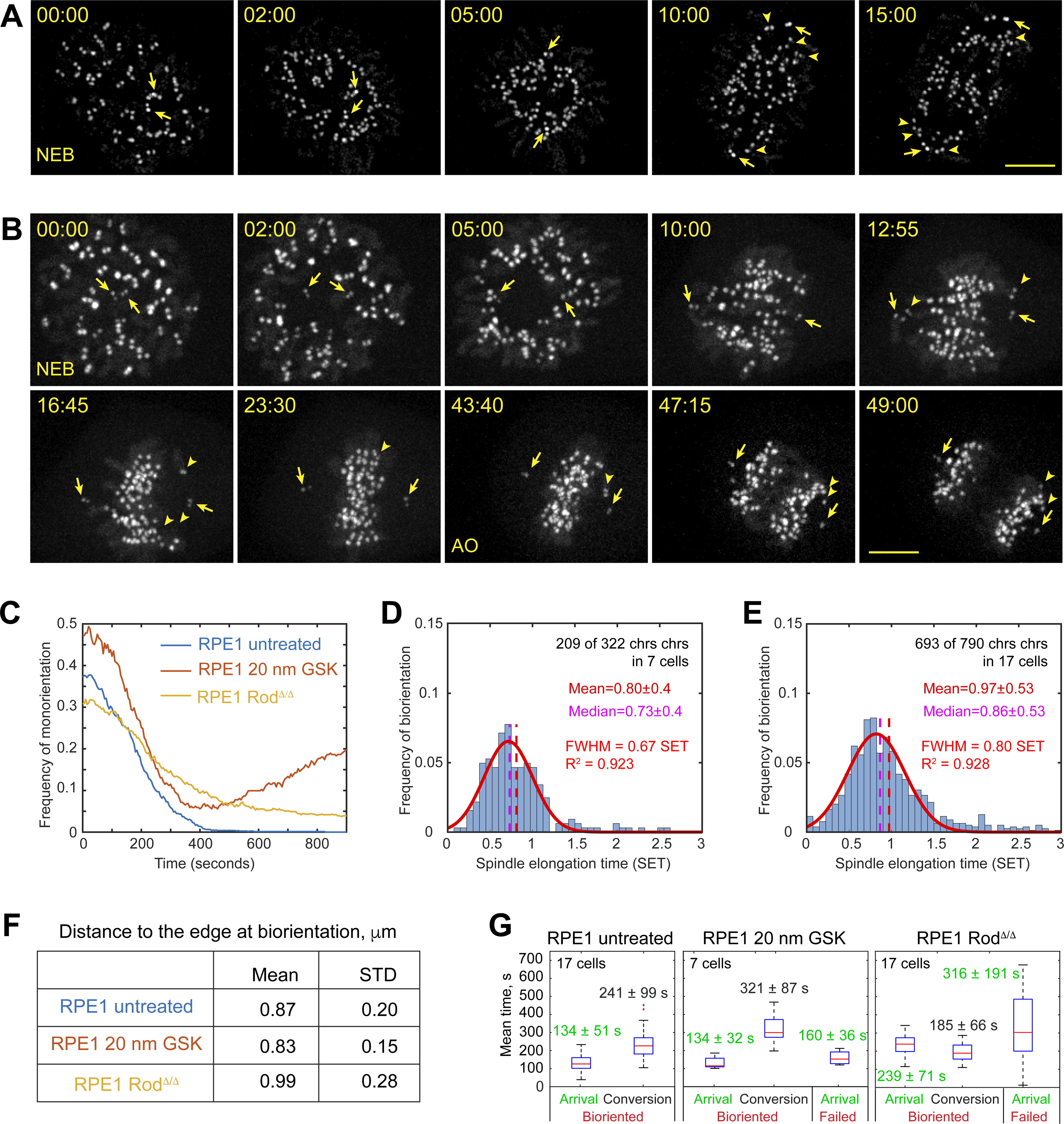
(A,B) Selected timepoints from recording of mitosis in the wt RPE1 cell treated with 20-nM GSK923295 (A) or RPE1 RodΔ/Δ (B). KTs and centrioles are tagged with CenpA-GFP and Centrin1-GFP. Arrows denote centrioles. Arrowheads point at KTs on monooriented chromosomes. Scale bar, 5 μm. (C) Fraction of chromosomes with centromeres residing <0.25 of the spindle length from a pole (i.e., monooriented). (D,E) Temporal distribution of biorientation events (normalized by spindle elongation time) in cells lacking activity of CenpE (D) or dynein at the KT (E). (F) Mean and STD values for distances from centromeres to the edge of the bundle-enriched spindle domain during formation of amphitelic attachments. (G) Tuckey’s box plots for times when centromeres arrive within 0.85 μm from the edge of the bundle-enriched domain (Arrival) and intervals from the arrival to the formation of amphitelic attachments (Conversion). Arrival times of centromeres that formed amphitelic attachments and centromeres that fail to biorient in the first 15 min of prometaphase are reported separately. Mean values are reported with STD. See also Figure S4, Videos S6 and S7.
Consistent with the observation of persistent monoorientation, ~37% (119/321) chromosomes in CenpE-inhibited and ~13% (97/790) chromosomes in RodΔ/Δ cells fail to form amphitelic attachments within 15 min after NEB. However, temporal dynamics of biorientation are markedly different in CenpE-inhibited vs. RodΔ/Δ cells (Figure 6D,E). In the former, although a lower number of chromosomes achieve biorientation, most amphitelic attachments form at the same stage of spindle elongation as in untreated RPE1 (0.80±0.40 SET vs. 0.79±0.30 SET, compare Figs. 6D and 1I, p=0.7179 in Kruskal-Wallis test). In RodΔ/Δ cells, formation of amphitelic attachments is delayed (0.97±0.53 SET, compare Figs. 6E and 1I, p < 0.0001 in Kruskal-Wallis test) and the distribution is skewed with many chromosomes achieving biorientation during late prometaphase (Figure 6E). Slower biorientation in RodΔ/Δ is consistent with longer and more variable duration of mitosis (41±15 min, N=17 vs. 23±3 min, N=17 in the wt RPE1, Student’s T-test p<0.001,).
As in untreated RPE1 cells, formation of amphitelic attachments in both CenpE-inhibited and RodΔ/Δ cells occurs predominantly within the spatial domain delineated by the same catenary function as in the wt RPE1 cells. Only insignificant differences are detected in the mean distance from the centromere to the catenary at the time of biorientation (Figure 6F). In contrast, the number of centromeres that do not enter the biorientation domain (remain >0.85 μm to the catenary throughout prometaphase) increases from ~6% in the wt (48/784) and CenpE-inhibited (21/321) cells to ~18% in RodΔ/Δ (140/790). Importantly, the number of chromosomes that fail to form amphitelic attachments is significantly higher among those that do not enter the biorientation domain (53%).
To assess the efficiency of amphitelic attachment formation near MT bundles, we analyze when centromeres enter the biorientation domain and the interval from their arrival to the formation of amphitelic attachment (Figure S4G). The mean arrival times in the untreated vs. CenpE-inhibited cells do not differ significantly irrespective of whether the chromosome subsequently forms amphitelic attachments (Figure 6G). However, the interval from the arrival to amphitelic attachment formation is significantly longer in CenpE-inhibited cells. In RodΔ/Δ cells, arrival to the biorientation domain is significantly delayed, particularly for the chromosomes that fail to form amphitelic attachments (Figure 6G). In contrast, the interval from the arrival to amphitelic attachments formation is shorter in RodΔ/Δ cells (Figure 6G). Thus, consistent with the model predictions, a lower number of MTs at the KT has no effect on the timely delivery of centromeres to the biorientation domain near the spindle equator; however, these centromeres often fail to form amphitelic attachments and subsequently shift poleward. In contrast, lack of dynein at the KT interferes with the delivery of centromeres to the biorientation domain but does not decrease the efficiency of amphitelic attachment formation on centromeres that encounter MT bundles.
Discussion
We propose a mechanism for synchronous formation of load-bearing connections of sister kinetochores to the opposite spindle poles (Figure 7). In contrast to the random sequential attachment of sister kinetochores envisioned in models based on the S&C hypothesis9,49–54, our model predicts almost instantaneous formation of amphitelic attachments on centromeres delivered to the biorientation domain of the spindle. Thus, proper architecture of the spindle determines where and when chromosomes achieve biorientation. Significant changes in the dynamics and spatial distribution of biorientation events in cells under conditions that interfere with various aspects of the proposed mechanism suggests that it is a major contributor during normal mitosis.
Figure 7. Centromere behaviors predicted in the model.
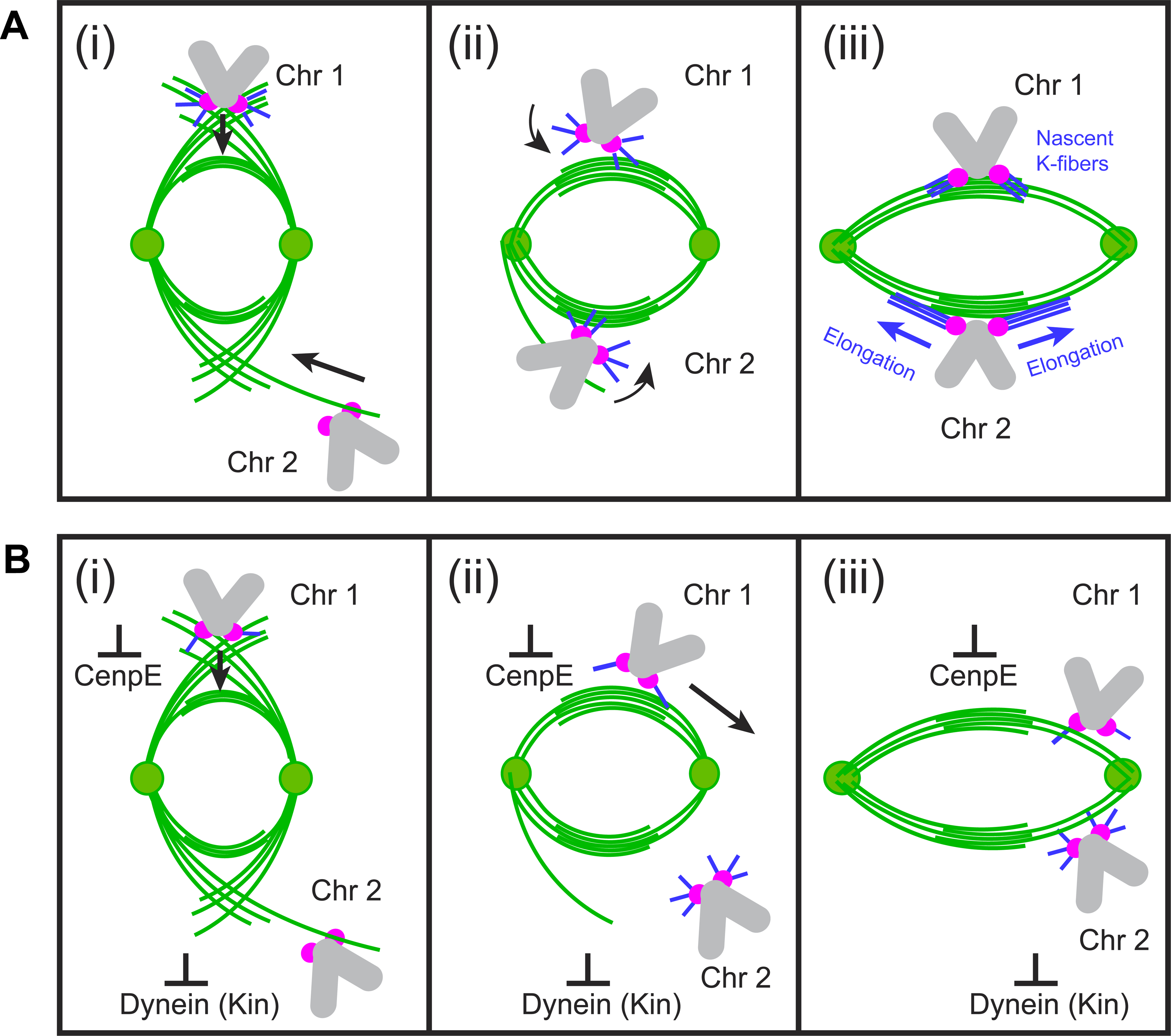
(A) Normal mitosis. At early stages of spindle elongation (i), interactions between short MTs at KTs and spindle MTs (Chr1) as well as direct interactions between KTs and astral MTs (Chr2) move centromeres towards the spindle domain enriched with bundles of antiparallel MTs. Both movements are driven by dynein that acts at the minus ends of short MTs (Chr1) or at the KT (Chr2). Near the bundles (ii), dynein-mediated interactions at the minus ends sort MTs protruding from the kinetochores into nascent K-fibers, that support load-bearing connections of sister kinetochores to the opposite spindle poles (iii, Chr1). Nascent K-fibers elongate (iii, Chr2) and their minus ends eventually reach spindle poles. (B) Effects of abnormal motor activities at KTs. Lower number of MT minus ends protruding from KTs in CenpE-inhibited cells does not significantly interfere with centripetal movement of centromeres on MT arrays with uniform polarity during early prometaphase (i, Chr1); however, sorting of short MTs into nascent K-fibers is impeded (ii, Chr2). As a result, chromosomes congress at the equator but many subsequently shift poleward and become monooriented (iii, Chr1). In contrast, absence of dynein at the KT interferes with prompt delivery of peripheral chromosomes to the equatorial zone where antiparallel bundles are numerous (I, Chr2). Encounters with the fully elongated spindle at later times increases probability of interactions with polarity biased MT arrays away from the equator, which promotes monoorientation
A key feature of our model is that load-bearing connections form by KTs that are already attached to plus end of short non-centrosomal MTs (Figure 7). Presence of these MTs at most KTs during earliest stages of spindle formation has been demonstrated26,55,56 and incorporation of MTs nucleated at the KT into K-fibers appears to continue throughout mitosis57. Live-cell microscopy demonstrates that MTs nucleated at KT develop into bundles that grow outwards and eventually connect to the spindle poles27,32,33,58,59. However, how the initial array of MTs at the KT converts into a K-fiber with proper polarity is unknown. Our model suggests that efficient sorting of MTs into two bundles oriented toward the opposite spindle poles arises from transient interactions between MTs protruding from the KTs and bundles of antiparallel MTs. A key prediction that a low number of minus ends protruding from the KT slows the conversion (Figure 5I) is consistent with the increased conversion time in CenpE-inhibited cells (Figure 6G) where the number of protruding MTs is lower26.
Consistent with the proposed model, amphitelic attachments form over a longer period and within a greater volume in cells depleted of PRC1, where bundles of anti-parallel MTs are scarce. However, all chromosomes in these cells eventually biorient and thus proximity to antiparallel bundles is not essential. Indeed, our model predicts that sorting of short MTs also occurs on the surface comprising antiparallel individual MTs, although the efficiency is reduced (Figure 5F). Interestingly, centromeres are overstretched during early prometaphase in PRC1-depleted cells (Figure 3E), consistent with the model predictions. Alternatively, spindle assembly in the absence of MT bundles may occur primarily via conventional S&C. Several features of mitosis in PRC1-depleted cells are consistent with this possibility. First, temporal distribution of biorientation is positively skewed with a tail indicating that a fraction of KTs is captured only after a very long and variable time as expected in stochastic S&C49. Second, centromeres in PRC1-depleted cells exhibit extended poleward movements (Figure S3D) and higher frequency of monoorientation as expected for uncoordinated attachments of sister kinetochores. Irrespective of the mechanism(s) that allow amphitelic attachments to form under abnormal conditions, our data indicate that most amphitelic attachments arise near antiparallel bundles when the bundles are accessible. This notion gains further support from the association of mature K-fibers with PRC1-decorated MT bundles of MTs that ‘bridge’ K-fibers of sister kinetochores in various cell types23,24,60–62.
Our current computational analyses quantitatively address only the mechanism of centromere biorientation upon arrival to the spindle surface. The preceding step, centripetal convergence of the peripheral chromosomes, requires additional exploration. In some cell types, the force that gathers peripheral chromosomes on the spindle arises from an actin cage63–65. This mechanism, that acts on the whole spindle would explain the synchrony with which initially scattered chromosomes initiate their movements and arrive at the spindle surface. However, suppression of the rapid inward movement of centromeres observed in RodΔ/Δ cells is more consistent with the notion of KTs gliding alongside of captured astral MTs. This movement is known to be driven by dynein bound directly to KTs43,47,66,67. Thus, conventional S&C may play an important role during initial stages by gathering chromosomes in the spindle compartment that supports nearly synchronous and rapid formation of amphitelic attachments.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alexey Khodjakov (alexey.khodjakov@health.ny.gov).
Material Availability
All unique reagents generated in this study are available from the Lead Contact without restrictions.
Data and code availability
Kinetochore tracking data have been deposited at Zenodo and are publicly available as of the date of publication. DOI is listed in the key resources table
Computer simulation code has been deposited at Zenodo and is publicly available as of the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE Antibodies | SOURCE | DENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat# T9026; RRID: AB_477593 |
| Rabbit polyclonal PRC1 | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.68 | N.C |
| Mouse monoclonal 9G3/Hec1 | Abcam | Cat# ab3613; RRID: AB_303949 |
| Mouse Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-11032; RRID: AB_2534091 |
| Goat anti-Mouse IgG Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21236; RRID: AB_2535805 |
| Goat anti-Rabbit Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-11012; RRID: AB_141359 |
| Goat anti-Mouse IgG1 (γ1) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21121; RRID: AB_2535764 |
| Goat anti-Mouse IgG2a Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-21135; RRID: AB_2535774 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| GSK-923295 | MedChemExpress | Cat# HY-10299 |
| PIPES | Sigma-Aldrich | E006757; CAS: 5625-37-6 |
| EGTA | Sigma-Aldrich | E4378; CAS: 67-42-5 |
| MgCl2 | Sigma-Aldrich | CAS 7791-18-6 |
| Triton X-100 | Sigma-Aldrich | X-100; CAS No: 9036-19-5 |
| Glutaraldehyde | Sigma-Aldrich | G5882; CAS: 111-30-8 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# 15714 |
| EDTA | Sigma-Aldrich | E-5134; CAS: 6381-92-6 |
| Phosphate-buffered saline, pH 7.2 | Thermo Fisher Scientific | Cat# 20012050 |
| Tween 20 | Sigma-Aldrich | P1379; CAS 9005-64-5 |
| Sodium Borohydride | Sigma-Aldrich | 452882; CAS: 16940-66-2 |
| Hoechst 33342 | Molecular Probes | Cat# H3570 |
| CaCl2 | Acros Organics | AC123350025; CAS 10035-04-8 |
| HEPES | Sigma-Aldrich | H4034; CAS 7365-45-9 |
| KCl | Fisher | Cat# P217-3; CAS 7447-40-7 |
| NaCl | Fisher | Cat# S640-3; CAS 7647-14-5 |
| Na2HPO4 | Sigma | Cat# S374-3; CAS 7558-79-4 |
| Dextrose | Fisher | Cat# BP350-1; CAS 50-99-7 |
| Polybrene | Sigma-Aldrich | Cat# H9268; CAS: 28728-55-4 |
| Blasticidin | InvivoGen | ant-bl-05 |
| Puromycin | Sigma-Aldrich | P7255; CAS 58-58-2 |
| Doxycycline Hyclate | Sigma-Aldrich | D9891; CAS: 24390-14-5 |
|
| ||
| Critical commercial assays | ||
|
| ||
| SiR-Tubulin Kit | Spirochrome AG | CY-SC002 |
| Lipofectamine 2000 | ThermoFisher | 11668027 |
|
| ||
| Deposited data | ||
|
| ||
| Kinetochore tracking data | This study | https://doi.org/10.5281/zenodo.5803448 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: hTERT-RPE-1(retinal pigmented epithelium, female) co-expressing CENP-A-GFP and centrin1-GFP | Laboratory of Alexey Khodjakov, Wadsworth Center, New York State Department of Health, Albany, NY.19 | N/A |
| Human: hTERT-RPE-1 expressing GFP-PRC1 | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.68 | N/A |
| Human: hTERT-RPE-1 expressing Sh-PRC1 RPE1 | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.29 | N/A |
| Human: hTERT RPE KNTC1-/- | Laboratory of Prasad V. Jallepalli, loan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY.12 | N/A |
| Human: hTERT-RPE-1 TetON Sh-PRC1 RPE1 | This study | N/A |
| Human: Ampho-293 (embryonic kidney, female) | Clontech | 631505 |
| Human: hTERT-RPE1 RodΔ/Δ co-expressing CENP-A-GFP and centrin1-GFP | This study | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| shRNA targeting sequence: PRC1; 5-GTGATTGAGGCAATTCGAG-3′ | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.29 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pMSCVblast vector69 | Addgene | Addgene # 7508 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ/Fiji | NIH | https://imagej.nih.gov/ij/ |
| MATLAB, R2021a | Mathworks | https://www.mathworks.com |
| Imaris | Oxford Instruments | https://imaris.oxinst.com |
| Adobe Suite CC 2021 | Adobe | https://www.adobe.com |
| SoftWoRx 5.0 | Applied Precision | http://www.api.com/softworx.asp |
| Simulation code | This study | https://doi.org/10.5281/zenodo.5804405 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and chemicals
Cell lines used in this study are listed in the Key Recourse Table. hTERT RPE1 (human retinal pigment epithelial, female) co-expressing CENP-A-GFP and centrin1-GFP19, hTERT RPE1 expressing GFP-PRC1 or Sh-PRC1, hTERT-RPE1 RodΔ/Δ co-expressing CENP-A-GFP and centrin1-GFP cells were maintained in antibiotic-free DMEM/F-12 medium supplemented with 10% fetal bovine serum (FBS, Gibco) at 37 °C, 5% CO2. Culture media for hTERT-RPE1 RodΔ/Δ were additionally supplemented with 1-mM sodium pyruvate (Gibco). Ampho-293 cells (human embryonic kidney, female) were grown in DMEM with 10% FBS and penicillin/streptomycin (Sigma). hTERT RPE1 cells expressing TetON Sh-PRC1 cells were cultured in DMEM with tetracycline-free FSB (Gibco). CenpE was inhibited by 20-nM GSK-923295 (MedChemExpress) added to the growth medium 0.5–2.5 h prior to initiation of live cell recordings of fixation.
METHOD DETAILS
Transfection
To generate hTERT-RPE1 RodΔ/Δ cell line with stable expression of CenpA-GFP and Centrin1-GFP, hTERT RPE KNTC1−/− cells12, a kind gift from Dr. Prasad V Jallepalli, (Memorial Sloan Kettering Cancer Center), were transfected with lentivirus constructs as previously described19.
Constitutive expression of GFP-PRC1 in hTERT RPE1 was achieved by retroviral transduction as previously described68. Cells with GFP expression were selected by flow cytometry on a BD FACS Aria system 2 (BD Biosciences) equipped with a 488 nm excitation line and a GFP emission filter.
Two approaches to PRC1 knockdown were utilized. In both, the target sequence 5′-GTGATTGAGGCAATTCGAG-3′ was used, as it had previously been shown to efficiently knock down PRC169. The shPRC1 construct was generated as previously described29 and transfected into hTERT RPE1 cells expressing CenpA-GFP and Centrin1-GFP with Lipofectamine 2000 (ThermoFisher). Live-cell recording of these cells were obtained 48–72 hrs after transfection. In the second approach, we generated cells with tetracycline-inducible expression of the same shRNA construct. The tetracycline repressor sequence was cloned into the pMSCVblast expression vector70 obtained from Addgene (hereafter ‘TetRpMSCVblast’). TetRpMSCVblast construct was first transfected into Ampho-293 cells for retrovirus production. Transfection was performed using the calcium phosphate transfection method. Briefly, a mixture of calcium chloride (CaCl2), TetRpMSCVblast (plasmid DNA) and MilliQ water is made to yield a final concentration of 0.25 M CaCl2 (Acros Organics) and 3 μg of plasmid DNA. A solution of 2X HBS (50mM HEPES (Sigma), 10 mM KCl (Fisher), 12 mM Dextrose (Fisher), 280mM NaCl (Fisher),1.5mM Na2HPO4 (Sigma), pH 7.0) is then added dropwise to the plasmid DNA mixture to yield a 1X HBS mixture, while expelling air from a 2 mL pipette. The final mixture of plasmid DNA and HBS is then added dropwise to ampho-293 cells and incubated overnight. After replacing the medium of transfected cells twice (~ 6 and 24 hr post transfection), the medium was harvested, passed through a 0.45 μm filter (PALL), and added directly to hTERT-RPE1 cells expressing CenpA-GFP and Centrin1-GFP in the presence of 4 μg/ml polybrene (Sigma). Stable inducible clones were selected with Blasticidin (InvivoGen). Next, the shRNA target sequence in PRC1 was transfected into the inducible clones via retroviral transduction. Clones that stably incorporates the construct were selected with Puromycin (Sigma). For induction of shPRC1, cells were incubated with 5 μg/mL Doxycycline in full growth media 48–72 hours prior live-cell recordings.
Live-cell microscopy
Cells were grown on #1.5 glass coverslips in Petri dishes for 48–72 hours. One day prior to the recording, regular culture media was replaced with phenol-red free mixture of DMEM/F-12 containing 10% FBS. Approximately 2 hours prior to the recording, coverslips were mounted on Rose chambers and placed on the microscope stage. The chambers were maintained at 37.0±0.3°C within a custom-built enclosure. Imaging was done with a spinning-disc confocal scanner (Yokogawa, X1) attached to a Nikon Ti2E microscope equipped with a λPlanApo 100×1.45 NA oil-immersion objective. 488-nm excitation light intensity was kept at ~10 nW/μm2 (~40 μW out of the lens). For tracking KT movements, Z-series of 17–20 sections were collected every 5 s at 100–150 ms exposures and 500–750 nm steps. For shPRC1 RPE1 that display higher variability of mitosis duration, recordings were done at 5-s intervals for the first 20–30 minutes of prometaphase and at 60-s intervals at later timepoints. The cells were fixed during telophase and immunostained for α-Tubulin. Only cells with no MT bundles and disorganized central spindle were included in the analyses of the KT movements. For recordings of MTs, cells were incubated with 75-nM SiR-Tubulin (Spirochrome, CY-SC002) and 10-μM Verapamil for 2–3 hours prior to imaging. 640-nm excitation light intensity was kept at ~10 nW/μm2 (40 μW out of the lens). All SiR-Tubulin fluorescence recording were done in combination with either CenpA-GFP+Centrin1-GFP or PRC1-GFP at 30-s intervals. All images were captured on a Photometrics 95B Prime camera at 110-nm XY pixel size. The system was controlled by NIS-Elements Imaging Software.
Fixed-cell immunofluorescence
For MT visualization, cells were pre-extracted in warm PEM buffer (100-mM PIPES, pH 6.9, 2.5-mM EGTA, 5-mM MgCl2) supplemented with 0.5% Triton X-100 for 1 min and fixed with 1% glutaraldehyde in PEM for 10 min. Cells were then stained a monoclonal antibody against α-Tubulin (T9026; Sigma-Aldrich) followed by a secondary antibody conjugated with Alexa Fluor 594 or 647 (Thermo Fisher Scientific).
For PRC1 visualization, cells were pre-extracted in warm PEM buffer (100-mM PIPES pH 7, 1-mM EDTA, 1-mM MgCl2) supplemented with 0.5%Triton X-100 for 30 seconds and fixed with 3.2% paraformaldehyde and 0.1% glutaraldehyde in warm PEM buffer. Cells were then stained with a rabbit polyclonal antibody29 at 1:1000 followed by a secondary antibody conjugated with Alexa Fluor 594 (Thermo Fisher Scientific). Staining for different antigens was done sequentially. Chromosomes were stained with Hoechst 33343 at 1 μg/ml.
Images of fixed cells were collected on the same microscope as live-cell recordings at 73 or 110-nm XY pixels and 200-nm Z-steps. All images were deconvolved with the SoftWoRx 5.0 (Applied Precision) and objective lens-specific point spread function. Precise Axial and Equatorial views of the spindle were generated by rotating the volume in 3-D to orient the spindle axis defined by the 3-D coordinates of both spindle poles.
Array Tomography
Array Tomography reconstruction were obtained as previously described26. KTs and MTs were visualized with monoclonal 9G3/Hec1 (Abcam ab3613) at 1:200 and DM1α antibody (Sigma T9026) antibodies followed by isotype-specific secondary antibodies against mouse γ1 (conjugated to Alexa Fluor 488, Thermo Fisher Scientific, A-21121) and γ2a (conjugated to Alexa Fluor 594, Thermo Fisher Scientific, A-21135). Precise Axial and Equatorial views of the spindle were generated by rotating the volume in 3-D to orient the spindle axis defined by the 3-D coordinates of both spindle poles.
Correlative Light Electron Microscopy
GFP-PRC1 RPE1 cells were fixed for 30 minutes in PBS containing 2.5% glutaraldehyde (Sigma-Aldrich). Chromosomes were stained with Hoechst 33342 at 1 μg/ml for 5 min. Complete Z-series were collected as in fixed-cell immunofluorescence preparations. EM embedding and serial sectioning were done as previously described71. 80-nm sections were imaged on a JEM 1400 microscope (JEOL) operated at 80 kV using a side-mounted 4.0-megapixel XR401 sCMOS AMT camera (AMT). Complete image series recorded at 8K magnification were used to reconstruct partial volumes containing PRC1 bundles. These volumes were aligned with the light microscopy images by matching positions of chromosome arms. Serial higher-magnification images (40K) were then collected to detail the distribution of the PRC1-decorated microtubule bundles near kinetochores.
QUANTIFICATION AND STATISTICAL ANALYSIS
Kinetochore tracking and analysis
KTs and centrioles were detected and tracked in Imaris (Bitplane). Due to a significant number of errors in tracking, particularly at the early stages of spindle assembly, each trajectory was verified and edited by a human operator. Verified trajectories were transferred to Matlab for visualization and analysis.
Temporal synchronization of various recordings was achieved by detecting when spindle elongation is completed in spindle pole trajectories smoothened with the Savitzky-Golay filter over 50 timepoints. The time corresponding to the end of spindle elongation was assigned the value of 1. Progression of time in individual recordings was then normalized to this value. For synchronization of spatial coordinates among multiple cells, all distances were normalized by assigning the value of 1 to the length of the spindle at the timepoint when spindle elongation was completed.
Centromere trajectories were constructed by calculating the center between sister KTs and analyzed in a spindle-centric cylindrical coordinate system, in which, at every time point, the spindle axis is a chosen reference z-axis of the cell 3D space, with the origin in the middle between the centrosomes. A KT position is given by three coordinates: distance along the axis z, radial distance ρ from the axis and angular direction φ around the axis. In this system, centrosomes simply segregate symmetrically along the straight axis, while movements of centromeres can be conveniently viewed by either projecting their trajectories onto the plane orthogonal to the spindle axis, where we can see ρ and φ coordinates but not z-coordinates (Figure S3B,D, Equatorial), or by plotting z and ρ coordinates (while ignoring φ coordinate) on any plane coming through the spindle axis (Figure S3B,D, Axial). In this view, z is the horizontal axis, and ρ is the coordinate in the vertical direction of the plane. For convenience, we randomly invert the sign of ρ coordinate for half of trajectories so that the appearance of the plot resembles a spindle.
Computational model
The computational model describes the dynamic mechanical interactions between short MTs, long MTs, and KTs in two spatial dimensions. At every time step, stochastic binding and unbinding events between short and long MTs are processed via the Gillespie algorithm, then mechanical forces are computed and used to update positions using a Euler-Maruyama integration scheme for stochastic movements. KTs are connected via chromatin, modeled as a Hookean spring force. Short MTs, modeled as stiff springs, emanate from each KT and angularly diffuse while unbound. When bound, molecular motors exert a constant force on minus ends of the short MTs and in the minus-end direction of the bound long MT. Binding occurs with a fixed probability per unit time when the minus-end tip of a short MT tip is in proximity to a long MT. Both ends of the short MTs can unbind. Unbinding from the KT of the plus-end of a short MT occurs at a rate depending on the angle formed with the KT-KT axis, and assuming rapid rebinding at the KT: the short MT is reattached to the KT at a random orientation, keeping the number of short MTs fixed. Unbinding at the minus-end of the short MT is assumed to occur at constant rate. The long MT configurations (geometries and polarities) are fixed in each simulation and modeled with infinite length. The output of the computational model is a time series corresponding to the KT positions, from which the IKD and cTilt angle can be computed and compared to experimentally observed values. Specific equations and computational details are described in Methods S1.
Characterization of the biorientation domain
To estimate the shape of biorientation domain, 12 recordings of RPE1 cells expressing PRC1-GFP were individually scaled to equalize the maximum length of the spindle among all cells. Each time point in every recording was then rotated to orient the spindle parallel to the abscissa and translated to place the center of the spindle at 0,0,0 coordinates. Maximum-intensity projections were then calculated for each recording and these projections were used to calculate a single average of all 12 recordings. The edge of the domain with high concentration of PRC1-GFP was then empirically matched to a catenary function y=k*cosh(x / 1.2); where x is spindle length at the timepoint and k=−1.8*x1.2.
Statistical methods
Mean values were compared in the two-tailed heteroscedastic Student’s T-test. Median values were compared in the Kruskal-Wallis test.
Supplementary Material
Methods S1. Description of the model and computational simulations. Related to STAR Methods. Assumptions, geometric constraints, parameters, and mathematical equations used in the construction of the model.
Video S1. Mitosis in a wild-type RPE1 cell. Example of recordings used for tracking chromosome movements. Related to Figure 1. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP; however, these organelles can be discriminated due to their different behavior. At the onset of spindle assembly (00:00, NEB), centriole pairs appear to be in a common complex (within the yellow circle); however, in 3D space they reside on the opposite (dorsal and ventral) sides of the nucleus. As the spindle elongates during early-mid prometaphase (00:00 – 05:50) it also rotates from nearly orthogonal to nearly parallel to the coverslip surface. Anaphase initiates at 20:15 (AO). Chromatids segregate into two groups and initiate formation of daughter nuclei. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected timepoints from this video are presented in Figure 1A.
Video S2. Mitosis in a wild-type RPE1 cell. Example of recordings used for visualization of centromere behavior during biorientation. Related to Figure 2. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP; microtubules are stained with 75-nM SiR-Tubulin. The video starts 1 min prior to NEB and the cell enters anaphase at 22:30. Notice that all 46 chromosomes segregate normally. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected frames from this video are presented in Figure 2A.
Video S3. Mitosis in RPE1 depleted of PRC1 cell. Example of recordings used for tracking chromosome movements. Related to Figure 3. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP. At the onset of spindle assembly (00:00, NEB), centriole pairs (yellow circles) reside on the dorsal and ventral sides of the nucleus. Notice increased intervals between frames from 30:05 to 58:05 to minimize phototoxicity. Selected frames from this video are presented in Figure 3C.
Video S4. Mitosis in an RPE1 cell expressing GFP-tagged PRC1. Related to Figure 4. Example of recordings used for delineation of the spindle domain with high concentration of microtubule bundles. Microtubules are stained with 75-nM SiR-Tubulin. The video starts 1 min prior to NEB and the cell enters anaphase at 19:30. Notice that at this level of expression, recruitment of PRC1-GFP to microtubule bundles during early prometaphase is apparent only with a stretched Look-Up Table (stretched LUT, right panel). At the normal LUT spanning from the dimmest to the brightest value of the entire recordings PRC1-GFP decoration of microtubule bundles is not detectable until metaphase. Time is shown in min:sec on the left and Spindle Elongation Time units (SET) on the right.
Video S5. Examples of simulated centromere behavior on a surface comprising evenly spaced antiparallel microtubules (top) or a bundle of antiparallel microtubules (bottom). Related to Figure 5. Predicted distances between sister kinetochores, and the angle between the centromere axis and spindle axis are shown for each timepoint (seconds, microns, and degrees). Minus ends of spindle microtubules are on the left for the yellow and on the right for the green microtubules. Selected frames from this video are shown in Figure 5 C,D.
Video S6. Mitosis in an RPE1 with inhibited CenpE. Example of recordings used for tracking chromosome movements. Related to Figure 6. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP. Imaging and culture conditions are similar to Video S1 but the media contains high concentration of CenpE inhibitor GSK923295 (20 nM). Inhibition of CenpE does not significantly affect centrosome positioning at NEB (yellow circle in 00:00 frame) or spindle elongation (00:00 – 11:30). Peripheral chromosomes exhibit rapid inward movements towards the spindle axis during early prometaphase (00:00 – 03:30). However, approximate half of chromosomes fail to establish amphitelic attachments and become monooriented towards one of the spindle poles. RPE1 cells with inactive CenpE remain arrested in mitosis for > 8 hrs. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected timepoints from this video are presented in Figure 6A.
Video S7. Mitosis in a RodΔ/Δ RPE1 cell. Example of recordings used for tracking chromosome movements. Related to Figure 6. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP. Imaging and culture conditions are similar to Video S1. Behavior of centrioles (yellow circle in 00:00 frame) and the pattern of spindle elongation (00:00 – 05:15) are similar to wild-type RPE1 cells although the spindle is shorter in most cells. Rapid inward movements towards the spindle axis are not apparent during early prometaphase (00:00 – 04:00). Several chromosomes achieve biorientation after a long delay and one chromosome remains monooriented through the onset of anaphase at 43:40. Both chromatids of this chromosome are incorporated into the same daughter nucleus. Multiple lagging chromatids are apparent during anaphase (43:50 – 48:30) although all ultimately segregate into appropriate daughter nuclei. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected timepoints from this video are presented in Figure 6B.
Highlights.
Amphitelic attachments form synchronously at a specific stage of spindle elongation
Amphitelic attachments form within a spatial domain defined by microtubule bundles
Bundles of antiparallel microtubules facilitate chromosome biorientation
CenpE and dynein at kinetochore affect the efficiency and rapidity of biorientation
Acknowledgements
This work was supported by the National Institutes of Health NIGMS grants GM130298 (A.K.) and GM130234 (TMK). LC was supported by the Swiss National Science Foundation grant P400PB 183828.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kirschner M, and Mitchison T (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell 45, 329–342. [DOI] [PubMed] [Google Scholar]
- 2.Heald R, and Khodjakov A (2015). Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J Cell Biol 211, 1103–1111. 10.1083/jcb.201510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petry S (2016). Mechanisms of Mitotic Spindle Assembly. Annu Rev Biochem 85, 659–683. 10.1146/annurev-biochem-060815-014528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prosser SL, and Pelletier L (2017). Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol 18, 187–201. 10.1038/nrm.2016.162. [DOI] [PubMed] [Google Scholar]
- 5.David AF, Roudot P, Legant WR, Betzig E, Danuser G, and Gerlich DW (2019). Augmin accumulation on long-lived microtubules drives amplification and kinetochore-directed growth. J Cell Biol 218, 2150–2168. 10.1083/jcb.201805044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalab P, Pralle A, Isacoff EY, Heald R, and Weis K (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701. [DOI] [PubMed] [Google Scholar]
- 7.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, and Mattaj IW (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83–93. [DOI] [PubMed] [Google Scholar]
- 8.Carazo-Salas RE, Gruss OJ, Mattaj IW, and Karsenti E (2001). Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nature Cell Biology 3, 228–234. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn J, and Dumont S (2017). Spindle assembly checkpoint satisfaction occurs via end-on but not lateral attachments under tension. J Cell Biol 216, 1533–1542. 10.1083/jcb.201611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, McEwen BF, Mogilner A, and Khodjakov A (2015). Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol 17, 1134–1144. 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacristan C, Ahmad MUD, Keller J, Fermie J, Groenewold V, Tromer E, Fish A, Melero R, Carazo JM, Klumperman J, et al. (2018). Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat Cell Biol. 10.1038/s41556-018-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Rodriguez JA, Lewis C, McKinley KL, Sikirzhytski V, Corona J, Maciejowski J, Khodjakov A, Cheeseman IM, and Jallepalli PV (2018). Distinct Roles of RZZ and Bub1-KNL1 in Mitotic Checkpoint Signaling and Kinetochore Expansion. Curr Biol 28, 3422–3429 e3425. 10.1016/j.cub.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira C, Reis RM, Gama JB, Celestino R, Cheerambathur DK, Carvalho AX, and Gassmann R (2018). Self-Assembly of the RZZ Complex into Filaments Drives Kinetochore Expansion in the Absence of Microtubule Attachment. Curr Biol 28, 3408–3421 e3408. 10.1016/j.cub.2018.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renda F, and Khodjakov A (2021). Role of spatial patterns and kinetochore architecture in spindle morphogenesis. Seminars in cell & developmental biology. 10.1016/j.semcdb.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walczak CE, Cai S, and Khodjakov A (2010). Mechanisms of chromosome behaviour during mitosis. Nature Reviews Molecular Cell Biology 11, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tolic IM, and Pavin N (2021). Mitotic spindle: lessons from theoretical modeling. Mol Biol Cell 32, 218–222. 10.1091/mbc.E20-05-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIntosh JR, Molodtsov MI, and Ataullakhanov FI (2012). Biophysics of mitosis. Q Rev Biophys 45, 147–207. 10.1017/S0033583512000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magidson V, and Khodjakov A (2013). Circumventing photodamage in live-cell microscopy. Methods Cell Biol 114, 545–560. 10.1016/B978-0-12-407761-4.00023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, and Khodjakov A (2011). The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukinavicius G, Reymond L, D’Este E, Masharina A, Gottfert F, Ta H, Guther A, Fournier M, Rizzo S, Waldmann H, et al. (2014). Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods. 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 21.Mollinari C, Kleman JP, Jiang W, Schoem G, Hunter T, and Margolis RL (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. The Journal of Cell Biology 157, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, and Kapoor TM (2010). Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell 142, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajtez J, Solomatina A, Novak M, Polak B, Vukusic K, Rudiger J, Cojoc G, Milas A, Sumanovac Sestak I, Risteski P, et al. (2016). Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat Commun 7, 10298. 10.1038/ncomms10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagric M, Risteski P, Martincic J, Milas A, and Tolic IM (2021). Optogenetic control of PRC1 reveals its role in chromosome alignment on the spindle by overlap length-dependent forces. eLife 10. 10.7554/eLife.61170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micheva KD, and Smith SJ (2007). Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 55, 25–36. 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikirzhytski V, Renda F, Tikhonenko I, Magidson V, McEwen BF, and Khodjakov A (2018). Microtubules assemble near most kinetochores during early prometaphase in human cells. J Cell Biol 217, 2647–2659. 10.1083/jcb.201710094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sikirzhytski V, Magidson V, Steinman JB, He J, Le Berre M, Tikhonenko I, Ault JG, McEwen BF, Chen JK, Sui H, et al. (2014). Direct kinetochore-spindle pole connections are not required for chromosome segregation. The Journal of Cell Biology 206, 231–243. 10.1083/jcb.201401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mollinari C, Kleman JP, Saoudi Y, Jablonski SA, Perard J, Yen TJ, and Margolis RL (2005). Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol Biol Cell 16, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pamula MC, Carlini L, Forth S, Verma P, Suresh S, Legant WR, Khodjakov A, Betzig E, and Kapoor TM (2019). High-resolution imaging reveals how the spindle midzone impacts chromosome movement. J Cell Biol 218, 2529–2544. 10.1083/jcb.201904169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunes V, Dantas M, Castro D, Vitiello E, Wang I, Carpi N, Balland M, Piel M, Aguiar P, Maiato H, and Ferreira JG (2020). Centrosome-nuclear axis repositioning drives the assembly of a bipolar spindle scaffold to ensure mitotic fidelity. Mol Biol Cell 31, 1675–1690. 10.1091/mbc.E20-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rieder CL, Schultz A, Cole R, and Sluder G (1994). Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. The Journal of Cell Biology 127, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiato H, Rieder CL, and Khodjakov A (2004). Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. The Journal of Cell Biology 167, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elting MW, Hueschen CL, Udy DB, and Dumont S (2014). Force on spindle microtubule minus ends moves chromosomes. The Journal of Cell Biology 206, 245–256. 10.1083/jcb.201401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rusan NM, Tulu US, Fagerstrom CJ, and Wadsworth P (2002). Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. The Journal of Cell Biology 158, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yildiz A, Tomishige M, Gennerich A, and Vale RD (2008). Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134, 1030–1041. 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khataee H, and Howard J (2019). Force Generated by Two Kinesin Motors Depends on the Load Direction and Intermolecular Coupling. Phys Rev Lett 122, 188101. 10.1103/PhysRevLett.122.188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicklas RB, and Koch CA (1969). Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. The Journal of Cell Biology 43, 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicklas RB, and Ward SC (1994). Elements of error correction in mitosis: microtubule capture, release, and tension. The Journal of Cell Biology 126, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, and Nasmyth K (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317–329. [DOI] [PubMed] [Google Scholar]
- 40.Lampson MA, and Cheeseman IM (2011). Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol 21, 133–140. 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicklas RB, Waters JC, Salmon ED, and Ward SC (2001). Checkpoint signals in grasshopper meiosis are sensitive to microtubule attachment, but tension is still essential. J Cell Sci 114, 4173–4183. [DOI] [PubMed] [Google Scholar]
- 42.Chakraborty M, Tarasovetc EV, Zaytsev AV, Godzi M, Figueiredo AC, Ataullakhanov FI, and Grishchuk EL (2019). Microtubule end conversion mediated by motors and diffusing proteins with no intrinsic microtubule end-binding activity. Nat Commun 10, 1673. 10.1038/s41467-019-09411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Tulu US, Wadsworth P, and Rieder CL (2007). Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol 17, 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheerambathur DK, Gassmann R, Cook B, Oegema K, and Desai A (2013). Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science 342, 1239–1242. 10.1126/science.1246232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barisic M, Aguiar P, Geley S, and Maiato H (2014). Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat Cell Biol 16, 1249–1256. 10.1038/ncb3060. [DOI] [PubMed] [Google Scholar]
- 46.Wood KW, Lad L, Luo L, Qian X, Knight SD, Nevins N, Brejc K, Sutton D, Gilmartin AG, Chua PR, et al. (2010). Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc Natl Acad Sci USA 107, 5839–5844. 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Yu W, Liang Y, and Zhu X (2007). Kinetochore dynein generates a poleward pulling force to facilitate congression and full chromosome alignment. Cell Res 17, 701–712. 10.1038/cr.2007.65. [DOI] [PubMed] [Google Scholar]
- 48.Karess R (2005). Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends in Cell Biology 15, 386–392. 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, and Mogilner A (2005). Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr Biol 15, 828–832. [DOI] [PubMed] [Google Scholar]
- 50.Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, and Mogilner D (2009). Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proceedings of National Academy of Sciences USA 106, 15708–15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalinina I, Nandi A, Delivani P, Chacon MR, Klemm AH, Ramunno-Johnson D, Krull A, Lindner B, Pavin N, and Tolic-Norrelykke IM (2013). Pivoting of microtubules around the spindle pole accelerates kinetochore capture. Nat Cell Biol 15, 82–87. 10.1038/ncb2640. [DOI] [PubMed] [Google Scholar]
- 52.Blackwell R, Sweezy-Schindler O, Edelmaier C, Gergely ZR, Flynn PJ, Montes S, Crapo A, Doostan A, McIntosh JR, Glaser MA, and Betterton MD (2017). Contributions of Microtubule Dynamic Instability and Rotational Diffusion to Kinetochore Capture. Biophys J 112, 552–563. 10.1016/j.bpj.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edelmaier C, Lamson AR, Gergely ZR, Ansari S, Blackwell R, McIntosh JR, Glaser MA, and Betterton MD (2020). Mechanisms of chromosome biorientation and bipolar spindle assembly analyzed by computational modeling. eLife 9. 10.7554/eLife.48787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pavin N, and Tolic-Norrelykke IM (2014). Swinging a sword: how microtubules search for their targets. Systems and synthetic biology 8, 179–186. 10.1007/s11693-014-9134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witt PL, Ris H, and Borisy GG (1980). Origin of kinetochore microtubules in Chinese hamster ovary cells. Chromosoma 81, 483–505. [DOI] [PubMed] [Google Scholar]
- 56.Snyder JA, and McIntosh JR (1975). Initiation and growth of microtubules from mitotic centers in lysed mammalian cells. The Journal of Cell Biology 67, 744–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiewisz R, Fabig G, Conway W, Needleman D, and Müller-Reichert T (2021). Three-dimensional structure of the kinetochore-fibers in human mitotic spindles. bioRxiv, 2021.2011.2013.468347. 10.1101/2021.11.13.468347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khodjakov A, Copenagle L, Gordon MB, Compton DA, and Kapoor TM (2003). Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. The Journal of Cell Biology 160, 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tulu US, Fagerstrom C, Ferenz NP, and Wadsworth P (2006). Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol 16, 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nixon FM, Honnor TR, Clarke NI, Starling GP, Beckett AJ, Johansen AM, Brettschneider JA, Prior IA, and Royle SJ (2017). Microtubule organization within mitotic spindles revealed by serial block face scanning electron microscopy and image analysis. J Cell Sci 130, 1845–1855. 10.1242/jcs.203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDonald KL, O’Toole ET, Mastronarde DN, and McIntosh JR (1992). Kinetochore microtubules in PTK cells. The Journal of Cell Biology 118, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Toole E, Morphew M, and McIntosh JR (2020). Electron tomography reveals aspects of spindle structure important for mechanical stability at metaphase. Mol Biol Cell 31, 184–195. 10.1091/mbc.E19-07-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mogessie B, and Schuh M (2017). Actin protects mammalian eggs against chromosome segregation errors. Science 357. 10.1126/science.aal1647. [DOI] [PubMed] [Google Scholar]
- 64.Booth AJ, Yue Z, Eykelenboom JK, Stiff T, Luxton GWG, Hochegger H, and Tanaka TU (2019). Contractile acto-myosin network on nuclear envelope remnants positions human chromosomes for mitosis. eLife 8. 10.7554/eLife.46902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lenart P, Daigle N, Hand AR, Eils R, Terasaki M, and Ellenberg J (2005). A contractile nuclear actin network drives chromosome congression in oocytes. Nature 436, 812–818. [DOI] [PubMed] [Google Scholar]
- 66.Vorozhko VV, Emanuele MJ, Kallio MJ, Stukenberg PT, and Gorbsky GJ (2008). Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma 117, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rieder CL, and Alexander SP (1990). Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. The Journal of Cell Biology 110, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian R, Ti SC, Tan L, Darst SA, and Kapoor TM (2013). Marking and Measuring Single Microtubules by PRC1 and Kinesin-4. Cell 154, 377–390. 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voets E, Marsman J, Demmers J, Beijersbergen R, and Wolthuis R (2015). The lethal response to Cdk1 inhibition depends on sister chromatid alignment errors generated by KIF4 and isoform 1 of PRC1. Sci Rep 5, 14798. 10.1038/srep14798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, and Mu D (2007). Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A 104, 16663–16668. 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rieder CL, and Cassels G (1999). Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods in Cell Biology 61, 297–315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods S1. Description of the model and computational simulations. Related to STAR Methods. Assumptions, geometric constraints, parameters, and mathematical equations used in the construction of the model.
Video S1. Mitosis in a wild-type RPE1 cell. Example of recordings used for tracking chromosome movements. Related to Figure 1. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP; however, these organelles can be discriminated due to their different behavior. At the onset of spindle assembly (00:00, NEB), centriole pairs appear to be in a common complex (within the yellow circle); however, in 3D space they reside on the opposite (dorsal and ventral) sides of the nucleus. As the spindle elongates during early-mid prometaphase (00:00 – 05:50) it also rotates from nearly orthogonal to nearly parallel to the coverslip surface. Anaphase initiates at 20:15 (AO). Chromatids segregate into two groups and initiate formation of daughter nuclei. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected timepoints from this video are presented in Figure 1A.
Video S2. Mitosis in a wild-type RPE1 cell. Example of recordings used for visualization of centromere behavior during biorientation. Related to Figure 2. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP; microtubules are stained with 75-nM SiR-Tubulin. The video starts 1 min prior to NEB and the cell enters anaphase at 22:30. Notice that all 46 chromosomes segregate normally. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected frames from this video are presented in Figure 2A.
Video S3. Mitosis in RPE1 depleted of PRC1 cell. Example of recordings used for tracking chromosome movements. Related to Figure 3. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP. At the onset of spindle assembly (00:00, NEB), centriole pairs (yellow circles) reside on the dorsal and ventral sides of the nucleus. Notice increased intervals between frames from 30:05 to 58:05 to minimize phototoxicity. Selected frames from this video are presented in Figure 3C.
Video S4. Mitosis in an RPE1 cell expressing GFP-tagged PRC1. Related to Figure 4. Example of recordings used for delineation of the spindle domain with high concentration of microtubule bundles. Microtubules are stained with 75-nM SiR-Tubulin. The video starts 1 min prior to NEB and the cell enters anaphase at 19:30. Notice that at this level of expression, recruitment of PRC1-GFP to microtubule bundles during early prometaphase is apparent only with a stretched Look-Up Table (stretched LUT, right panel). At the normal LUT spanning from the dimmest to the brightest value of the entire recordings PRC1-GFP decoration of microtubule bundles is not detectable until metaphase. Time is shown in min:sec on the left and Spindle Elongation Time units (SET) on the right.
Video S5. Examples of simulated centromere behavior on a surface comprising evenly spaced antiparallel microtubules (top) or a bundle of antiparallel microtubules (bottom). Related to Figure 5. Predicted distances between sister kinetochores, and the angle between the centromere axis and spindle axis are shown for each timepoint (seconds, microns, and degrees). Minus ends of spindle microtubules are on the left for the yellow and on the right for the green microtubules. Selected frames from this video are shown in Figure 5 C,D.
Video S6. Mitosis in an RPE1 with inhibited CenpE. Example of recordings used for tracking chromosome movements. Related to Figure 6. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP. Imaging and culture conditions are similar to Video S1 but the media contains high concentration of CenpE inhibitor GSK923295 (20 nM). Inhibition of CenpE does not significantly affect centrosome positioning at NEB (yellow circle in 00:00 frame) or spindle elongation (00:00 – 11:30). Peripheral chromosomes exhibit rapid inward movements towards the spindle axis during early prometaphase (00:00 – 03:30). However, approximate half of chromosomes fail to establish amphitelic attachments and become monooriented towards one of the spindle poles. RPE1 cells with inactive CenpE remain arrested in mitosis for > 8 hrs. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected timepoints from this video are presented in Figure 6A.
Video S7. Mitosis in a RodΔ/Δ RPE1 cell. Example of recordings used for tracking chromosome movements. Related to Figure 6. Kinetochores and centrioles are tagged with CenpA-GFP and Centrin1-GFP. Imaging and culture conditions are similar to Video S1. Behavior of centrioles (yellow circle in 00:00 frame) and the pattern of spindle elongation (00:00 – 05:15) are similar to wild-type RPE1 cells although the spindle is shorter in most cells. Rapid inward movements towards the spindle axis are not apparent during early prometaphase (00:00 – 04:00). Several chromosomes achieve biorientation after a long delay and one chromosome remains monooriented through the onset of anaphase at 43:40. Both chromatids of this chromosome are incorporated into the same daughter nucleus. Multiple lagging chromatids are apparent during anaphase (43:50 – 48:30) although all ultimately segregate into appropriate daughter nuclei. Each timepoint is shown as a maximum-intensity projection of the entire cell volume. Selected timepoints from this video are presented in Figure 6B.
Data Availability Statement
Kinetochore tracking data have been deposited at Zenodo and are publicly available as of the date of publication. DOI is listed in the key resources table
Computer simulation code has been deposited at Zenodo and is publicly available as of the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE Antibodies | SOURCE | DENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat# T9026; RRID: AB_477593 |
| Rabbit polyclonal PRC1 | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.68 | N.C |
| Mouse monoclonal 9G3/Hec1 | Abcam | Cat# ab3613; RRID: AB_303949 |
| Mouse Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-11032; RRID: AB_2534091 |
| Goat anti-Mouse IgG Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21236; RRID: AB_2535805 |
| Goat anti-Rabbit Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-11012; RRID: AB_141359 |
| Goat anti-Mouse IgG1 (γ1) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21121; RRID: AB_2535764 |
| Goat anti-Mouse IgG2a Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-21135; RRID: AB_2535774 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| GSK-923295 | MedChemExpress | Cat# HY-10299 |
| PIPES | Sigma-Aldrich | E006757; CAS: 5625-37-6 |
| EGTA | Sigma-Aldrich | E4378; CAS: 67-42-5 |
| MgCl2 | Sigma-Aldrich | CAS 7791-18-6 |
| Triton X-100 | Sigma-Aldrich | X-100; CAS No: 9036-19-5 |
| Glutaraldehyde | Sigma-Aldrich | G5882; CAS: 111-30-8 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# 15714 |
| EDTA | Sigma-Aldrich | E-5134; CAS: 6381-92-6 |
| Phosphate-buffered saline, pH 7.2 | Thermo Fisher Scientific | Cat# 20012050 |
| Tween 20 | Sigma-Aldrich | P1379; CAS 9005-64-5 |
| Sodium Borohydride | Sigma-Aldrich | 452882; CAS: 16940-66-2 |
| Hoechst 33342 | Molecular Probes | Cat# H3570 |
| CaCl2 | Acros Organics | AC123350025; CAS 10035-04-8 |
| HEPES | Sigma-Aldrich | H4034; CAS 7365-45-9 |
| KCl | Fisher | Cat# P217-3; CAS 7447-40-7 |
| NaCl | Fisher | Cat# S640-3; CAS 7647-14-5 |
| Na2HPO4 | Sigma | Cat# S374-3; CAS 7558-79-4 |
| Dextrose | Fisher | Cat# BP350-1; CAS 50-99-7 |
| Polybrene | Sigma-Aldrich | Cat# H9268; CAS: 28728-55-4 |
| Blasticidin | InvivoGen | ant-bl-05 |
| Puromycin | Sigma-Aldrich | P7255; CAS 58-58-2 |
| Doxycycline Hyclate | Sigma-Aldrich | D9891; CAS: 24390-14-5 |
|
| ||
| Critical commercial assays | ||
|
| ||
| SiR-Tubulin Kit | Spirochrome AG | CY-SC002 |
| Lipofectamine 2000 | ThermoFisher | 11668027 |
|
| ||
| Deposited data | ||
|
| ||
| Kinetochore tracking data | This study | https://doi.org/10.5281/zenodo.5803448 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: hTERT-RPE-1(retinal pigmented epithelium, female) co-expressing CENP-A-GFP and centrin1-GFP | Laboratory of Alexey Khodjakov, Wadsworth Center, New York State Department of Health, Albany, NY.19 | N/A |
| Human: hTERT-RPE-1 expressing GFP-PRC1 | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.68 | N/A |
| Human: hTERT-RPE-1 expressing Sh-PRC1 RPE1 | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.29 | N/A |
| Human: hTERT RPE KNTC1-/- | Laboratory of Prasad V. Jallepalli, loan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY.12 | N/A |
| Human: hTERT-RPE-1 TetON Sh-PRC1 RPE1 | This study | N/A |
| Human: Ampho-293 (embryonic kidney, female) | Clontech | 631505 |
| Human: hTERT-RPE1 RodΔ/Δ co-expressing CENP-A-GFP and centrin1-GFP | This study | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| shRNA targeting sequence: PRC1; 5-GTGATTGAGGCAATTCGAG-3′ | Laboratory of Tarun M Kapoor, The Rockefeller University, New York, NY.29 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pMSCVblast vector69 | Addgene | Addgene # 7508 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ/Fiji | NIH | https://imagej.nih.gov/ij/ |
| MATLAB, R2021a | Mathworks | https://www.mathworks.com |
| Imaris | Oxford Instruments | https://imaris.oxinst.com |
| Adobe Suite CC 2021 | Adobe | https://www.adobe.com |
| SoftWoRx 5.0 | Applied Precision | http://www.api.com/softworx.asp |
| Simulation code | This study | https://doi.org/10.5281/zenodo.5804405 |


