Abstract
Craniofacial development is controlled by a large number of genes, which interact with one another to form a complex gene regulatory network (GRN). Key components of GRN are signaling molecules and transcription factors. Therefore, identifying targets of core transcription factors is an important part of the overall efforts toward building a comprehensive and accurate model of GRN. LHX6 and LHX8 are transcription factors expressed in the oral mesenchyme of the first pharyngeal arch (PA1), and they are crucial regulators of palate and tooth development. Previously, we performed genome-wide transcriptional profiling and chromatin immunoprecipitation to identify target genes of LHX6 and LHX8 in PA1, and described a set of genes repressed by LHX. However, there has not been any discussion of the genes positively regulated by LHX6 and LHX8. In this paper, we revisited the above datasets to identify candidate positive targets of LHX in PA1. Focusing on those with known connections to craniofacial development, we performed RNA in situ hybridization to confirm the changes in expression in Lhx6;Lhx8 mutant. We also confirmed the binding of LHX6 to several putative enhancers near the candidate target genes. Together, we have uncovered novel connections between Lhx and other important regulators of craniofacial development, including Eya1, Barx1, Rspo2, Rspo3, and Wnt11.
1. Introduction
In mammals, the face forms via embryonic intermediate structures called the frontonasal prominence and the first pharyngeal arch (PA1) (Marcucio et al., 2015; Sperber et al., 2010). The frontonasal prominence gives rise to the middle part of the face including the nose, while PA1 gives rise to the jaw and parts of the ear. PA1 is further divided into the maxillary arch (prospective upper jaw) and the mandibular arch (prospective lower jaw). PA1 contains the mesenchyme derived from the cranial neural crest and the mesoderm, which is covered by the ectoderm on the outside and the endoderm on the inside.
The neural crest-derived mesenchyme of PA1 contributes to diverse components of the jaw such as the odontoblasts of the teeth, the tendons and the ligaments, the dermis, and the bone. How these mesenchyme cells acquire positional identities and follow different fates has been studied extensively (Gou et al., 2015; Medeiros and Crump, 2012; Minoux and Rijli, 2010). Signaling by secreted proteins of WNT (wingless-type MMTV integration site), FGF (fibroblast growth factor), BMP (bone morphogenetic protein), TGF-β (transforming growth factor-β), endothelin, and hedgehog families sets up region-specific expression of a large number of transcription factors in PA1 mesenchyme, including members of DLX (distal-less homeobox), MSX (msh homeobox), PAX (paired box), HAND (heart and neural crest derivatives expressed), FOX (forkhead box), and LHX (LIM homeobox) families. In turn, these transcription factors regulate development of specific structures in each part of the face. Functions of individual signaling pathways and transcription factors have been investigated in detail over the past 30 years (Clouthier et al., 2010; Gou et al., 2015; Medeiros and Crump, 2012; Minoux and Rijli, 2010; Reynolds et al., 2019). More recently, large-scale transcriptomic and genomic studies have provided insights into the dynamics of gene expression and their regulation by epigenetic modifiers and cis-regulatory elements (Attanasio et al., 2013; Brinkley et al., 2016; Hooper et al., 2017; Wilderman et al., 2018).
To integrate the above information on craniofacial patterning into a comprehensive and coherent model of a gene regulatory network (GRN), it is important to identify targets of transcription factors that play key roles as nodes of the network. LHX6 and LHX8 are members of LIM-homeodomain transcription factors, and they have closely overlapping expression and functions in the oral mesenchyme of PA1 (Grigoriou et al., 1998). In mouse mutants lacking both LHX6 and LHX8, development of the molars and the secondary palate was arrested at an early stage (around embryonic day (E) 11.5), which indicated that the LHX transcription factors are crucial to development of oral structures from PA1 (Cesario et al., 2015; Denaxa et al., 2009). Previously, we combined genome-wide transcriptional profiling and chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) to identify transcriptional targets of LHX6 and LHX8 in the maxillary arch, and further described several genes repressed by LHX (negative targets) in relation to regulation of cell proliferation (Cesario et al., 2015). However, there has not been any follow-up report on target genes that are positively regulated by LHX in the embryonic face. In this study, we examined the list of candidate positive targets of LHX6 and LHX8, and provided verification for LHX-dependent expression and/or LHX binding for select genes with known connections to craniofacial development.
2. Results
2.1. Identification of candidate direct targets of LHX6 and LHX8 in the maxillary arch
To elucidate genetic pathways downstream of Lhx6 and Lhx8 in craniofacial development, we previously performed Affymetrix microarray-based transcriptional profiling from the maxillary arch of the mutants lacking both LHX6 and LHX8 (Lhx6PLAP/PLAP;Lhx8−/−) and control embryos at E10.5 (Cesario et al., 2015) (GEO accession: GSE 71285). This experiment identified 93 genes that were upregulated in the mutants and 119 genes downregulated in the mutants (= Lhx-regulated genes), with the criteria of a fold change>1.5, p<0.05, and an average probe signal in either genotype>100 (Cesario et al., 2015). In the same study, we mapped the binding of LHX6 in the genome through ChIP-seq from the maxillary arch of E11.5 wild type embryos (Cesario et al., 2015) (GEO accession: GSE71497). We chose E10.5 for transcriptional profiling because it was right before morphological phenotypes appeared in the mutants. However, we had to use E11.5 maxillary arch for ChIP-seq to obtain enough cells for this experiment. 6560 genomic regions showed enrichment of LHX6 binding (= LHX6 peaks), and each LHX6 peak was assigned to a gene whose transcription start site (TSS) was the nearest to the peak and less than 1000 kb away from the peak. We performed ChIP followed by quantitative real time PCR (ChIP-qPCR) for eight of the LHX6 peaks from ChIP-seq (three for Cesario et al., 2015, five for the current manuscript – see Sections 2.2 and 2.3), and we were able to confirm the ChIP-seq result for seven of them. One that failed validation was associated with Foxp1 (Cesario et al., 2015).
By intersecting the results of transcriptional profiling and ChIP-seq, we obtained a list of Lhx-regulated genes with one or more associated LHX6 peaks, which would be candidates for direct transcriptional targets of LHX in the maxillary arch. The list contained 43 genes that were negatively regulated by Lhx6 and Lhx8 (Table S1), and 49 genes that were positively regulated by the Lhx genes (Table S2). We had described before that LHX6 and LHX8 repressed Cdkn1c and several Fox genes to promote outgrowth of the palate (Cesario et al., 2015). Therefore, in the current study, we focused on the genes that were activated by LHX.
We prioritized the candidate positive targets of LHX based on known connections to craniofacial development. We first searched NCBI PubMed database (https://pubmed.ncbi.nlm.nih.gov/) for the gene symbol ‘AND craniofacial development’ for each of the 49 genes. 16 genes had at least 1 report, and 9 genes had evidence to suggest a role specifically in PA1 development (Eya1, Eya4, Dusp6, Msx1, Wnt11, Barx1, Rspo3, Rspo2, Shox2; see Section 3 for details). Interestingly, these 9 genes were mostly at the top of the list in which the genes were ranked by the number of associated LHX6 peaks (Table S2).
2.2. Transcription factors
Eya1 (EYA transcriptional coactivator and phosphatase 1) encodes a co-factor of a homeodomain transcription factor SIX1 (sine oculis-related homeobox 1) (Tadjuidje and Hegde, 2013). In our microarray data, the expression of Eya1 and its homolog Eya4 was reduced in Lhx6PLAP/PLAP;Lhx8−/− mutant maxillary arch by 1.5-fold and 1.6-fold, respectively. We confirmed the changes by RNA in situ hybridization on Lhx6CreER/CreER;Lhx8−/− mutants at E11.5. Lhx6CreER is functionally a null allele of Lhx6 just like Lhx6PLAP (see Section 4.1 and Figure S1 for details). At E11.5, the palate and tooth phenotypes of the Lhx mutants have just become noticeable morphologically, as reduced outgrowth of the palatal shelves and lack of transition from the dental lamina to the placode at the developing molars (Cesario et al., 2015; Denaxa et al., 2009). Eya1 was downregulated throughout the maxillary arch, while Eya4 was most affected in the medial and the ventral mesenchyme (Fig. 1D–G; Fig. S2). ChIP-seq found eight LHX6 peaks associated with Eya1 and 5 peaks associated with Eya4 (Fig. 1A,B). To assess the likelihood that these LHX6 peaks correspond to transcriptional enhancers, we compared the LHX6 ChIP-seq result with two datasets from ChIP-seq for markers of active enhancers from E11.5 maxillary arch. One dataset was from ChIP-seq for acetylation of lysine 27 of Histone H3 (H3K27ac), performed by our group using the same chromatin sample as LHX6 ChIP-seq (Landin Malt et al., 2014). The other dataset was from ChIP-seq for histone acetyltransferase p300, which was accessed through UCSC Genome browser FaceBase track (Brinkley et al., 2016; Pennacchio et al., 2017). All of the eight Eya1-associated LHX6 peaks and two of the Eya4-associated LHX6 peaks were enriched with H3K27ac (Fig. 1A,B). For Eya1, peaks #1, 4, and 8 were also enriched with p300 binding, making them strong candidates for enhancers (Table S3).
Figure 1. Lhx6 and Lhx8 positively regulate several transcription factors important for PA1 development.
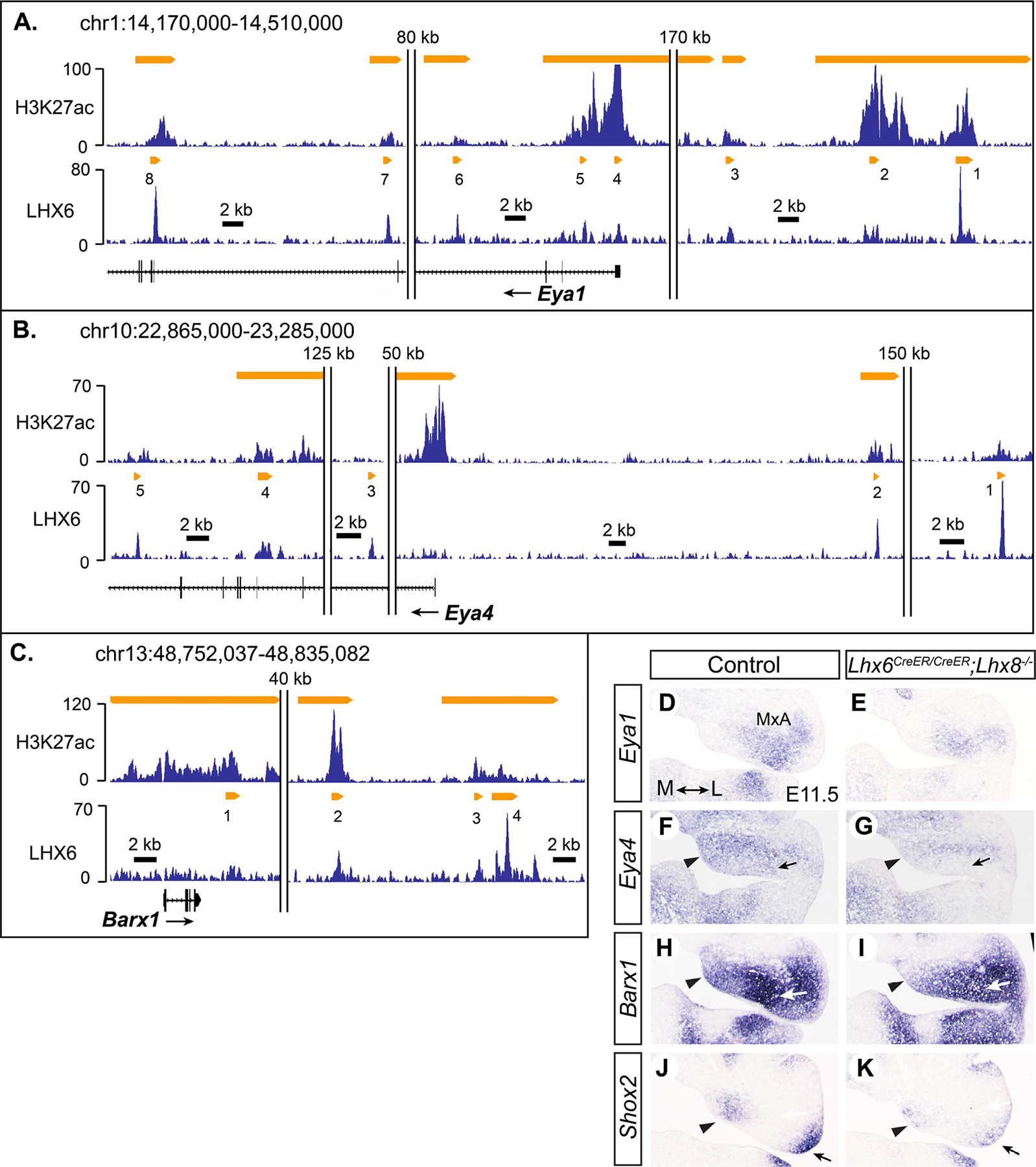
A-C) Results of H3K27ac ChIP-seq and LHX6 ChIP-seq visualized by Integrated Genome Browser (Freese et al., 2016). All genome coordinates in this paper are for NCBI37/mm9 assembly. The y-axis corresponds to fragment density derived from sequencing results. The orange bars indicate H3K27ac or LHX6 peaks determined by MACS peak-finding algorithm as described before (Cesario et al., 2015; Landin Malt et al., 2014). Double vertical lines indicate a gap in the genomic region presented in the figure. The size of the gap is indicated above the lines. D-K) RNA in situ hybridization on coronal sections of the head from E11.5 embryos. Medial-lateral axis is indicated in D. Arrowheads in F-K: reduced gene expression in the Lhx mutant in the medial mesenchyme of the maxillary arch (MxA). Arrows in F,G: decreased expression of Eya4 in the ventral mesenchyme of the mutant maxillary arch. Arrows in H,I: decreased expression of Barx1 in the upper molar mesenchyme in the mutant. Arrows in J,K: decreased expression of Shox2 in the lateral mesenchyme of the mutant maxillary arch.
Barx1 (BarH-like homeobox 1) and Shox2 (short stature homeobox 2) were downregulated by 3-fold and 3.8-fold, respectively, in Lhx6PLAP/PLAP;Lhx8−/− mutants at E10.5. At E11.5, both genes appeared only moderately affected in the Lhx mutants, with the most differences in the anterior part of the maxillary arch (Fig. 1H–K, Fig. S3). Barx1 expression was decreased in the medial mesenchyme and at the molar region. Shox2 expression was reduced in the medial end and the lateral end of the maxillary arch. There were four LHX6 peaks associated with Barx1, and two of them (#2, 4) were enriched with both H3K27ac and p300 (Fig. 1C, Table S3). Two LHX6 peaks were associated with Shox2, but they were low (Table S2, peak values 16 and 18, compared with an average of 25 for all 6560 LHX6 peaks), suggesting that LHX6 binding to these regions are marginal, if any.
A previous report showed that the expression of Msx1 (msh homeobox 1) and Pax9 (paired box 9) was affected in the molar mesenchyme of Lhx6;Lhx8 double knock mutants (Denaxa et al., 2009). The authors used different Lhx6 and Lhx8 mutant lines from our study, but we found the same result in Lhx6PLAP/PLAP;Lhx8−/− mutants. Msx1 expression was completely abolished in the mutant maxillary arch, while some Pax9 expression remained (Fig. 2D–G, Fig. S4). In our microarray experiment, Msx1 was identified as a gene downregulated in Lhx6PLAP/PLAP;Lhx8−/− maxillary arch (1.6-fold), but Pax9 did not show a significant difference between the mutant and control samples. This is most likely because robust expression of Pax9 in PA1 does not begin until E11 (Peters et al., 1998).
Figure 2. Msx1 and Pax9 are likely direct targets of LHX6 and LHX8.
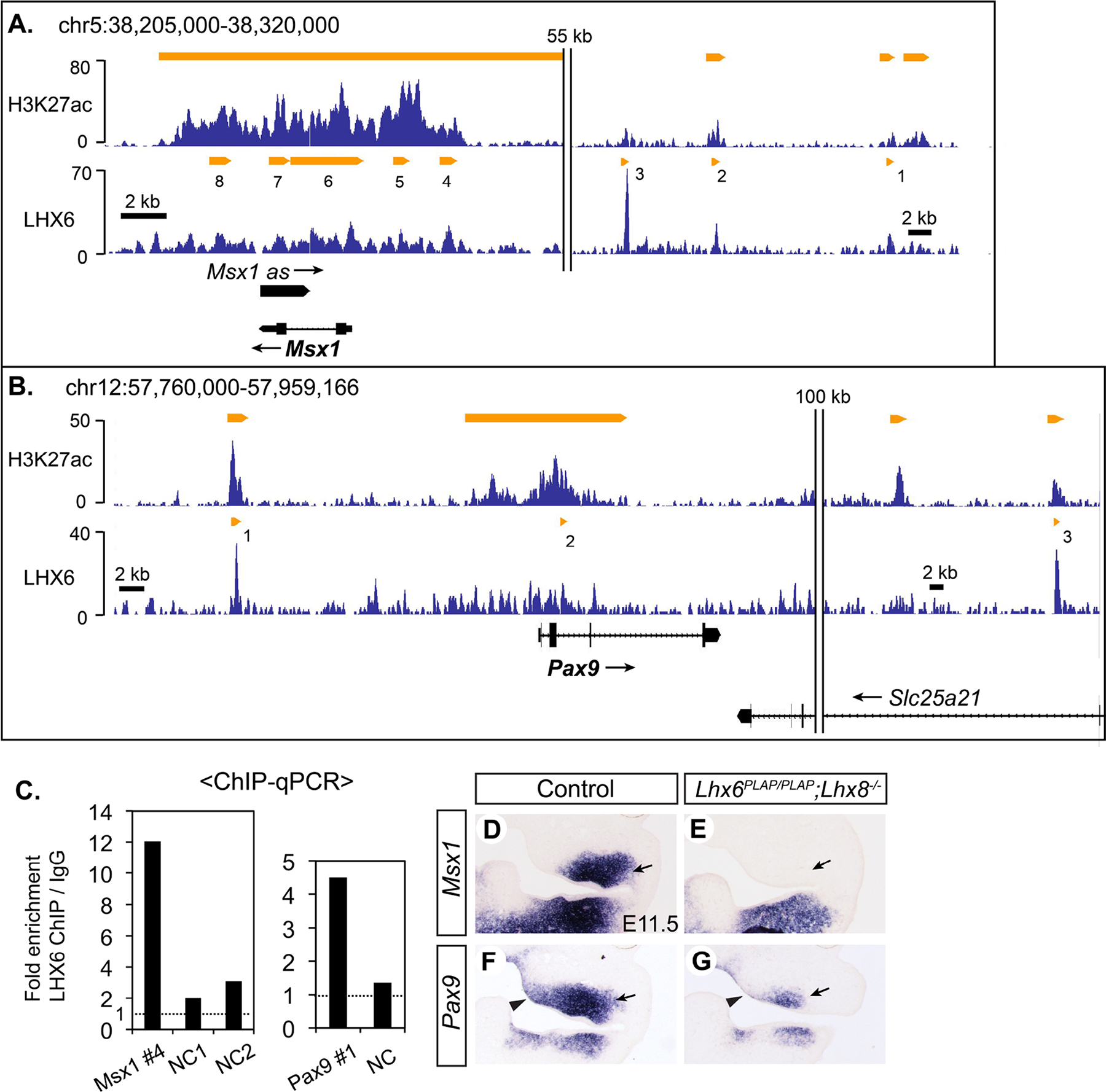
A,B) Results of H3K27ac ChIP-seq and LHX6 ChIP-seq around Msx1 and Pax9 visualized by Integrated Genome Browser. C) LHX6 ChIP-qPCR from E11.5 maxillary arch to confirm the result of ChIP-seq. NC: negative control sequence. D-K) RNA in situ hybridization on coronal sections of the head from E11.5 embryos. Arrows in D-G: decreased expression of Msx1 and Pax9 in the upper molar mesenchyme in the Lhx mutant. Arrowheads in F,G: decreased expression of Pax9 in the medial mesenchyme of the mutant maxillary arch.
8 LHX6 peaks were associated with Msx1, and 7 of them were also marked by H3K27ac and p300 (Fig. 2A, Table S3). Peaks #1, 2, 4, 5, and 8 had been tested by other researchers for an enhancer activity in transient transgenic mouse embryos (Attanasio et al., 2013; MacKenzie et al., 1997). Only peak #4 drove robust reporter expression in the maxillary arch (MacKenzie et al., 1997), and thus we performed LHX6 ChIP-qPCR validation for peak #4. We were able to confirm strong binding of LHX6 to this enhancer (Fig. 2C), establishing that Msx1 is a direct transcriptional target of LHX6. Pax9 had 3 associated LHX6 peaks, and all of them were marked by H3K27ac but not by p300 (Fig. 2B, Table S3). In an earlier study, Pax9 peak #1 showed an enhancer activity in a reporter assay in primary culture of maxillary arch cells, though this activity has yet to be confirmed in vivo (Landin Malt et al., 2014). We verified LHX6 binding to peak #1 by ChIP-qPCR (Fig. 2C).
2.3. Signaling molecules
R-spondins (RSPOs) are secreted glycoproteins that modulate WNT signaling. RSPOs usually augment the effects of WNT ligands for the canonical pathway involving β-catenin, but in some contexts, they appear to enhance non-canonical WNT signaling or inhibit the canonical signaling (Raslan and Yoon, 2019). Rspo2 and Rspo3 were downregulated in Lhx6PLAP/PLAP;Lhx8−/− mutant maxillary arch at E10.5 (1.9-fold and 3.1-fold, respectively). In addition, consistent with the function of RSPOs, our microarray data showed a modest decrease in the expression of Axin2 and Lef1, which are transcriptional targets of the canonical WNT pathway (Axin2: 1.4-fold, p=0.0057; Lef1: 1.3-fold, p=0.035) (Filali et al., 2002; Jho et al., 2002). From RNA in situ hybridization at E11.5, Rspo2 expression was most clearly affected in the Lhx mutants in the molar mesenchyme immediately underneath the dental lamina (Fig. 3D,E; Fig. S5). During normal development, Rspo3 was mainly expressed in the medial half of the maxillary arch, but this expression was significantly reduced in the Lhx mutants (Fig. 3F,G; Fig. S5). In contrast, Rspo3 expression appeared increased in the lateral part of the mutant maxillary arch, suggesting that Rspo3 may be repressed by LHX here. Still, the overall amount of Rspo3 mRNA was reduced in the Lhx mutants compared with controls.
Figure 3. Lhx6 and Lhx8 regulate Rspo2 and Rspo3 in the maxillary arch.
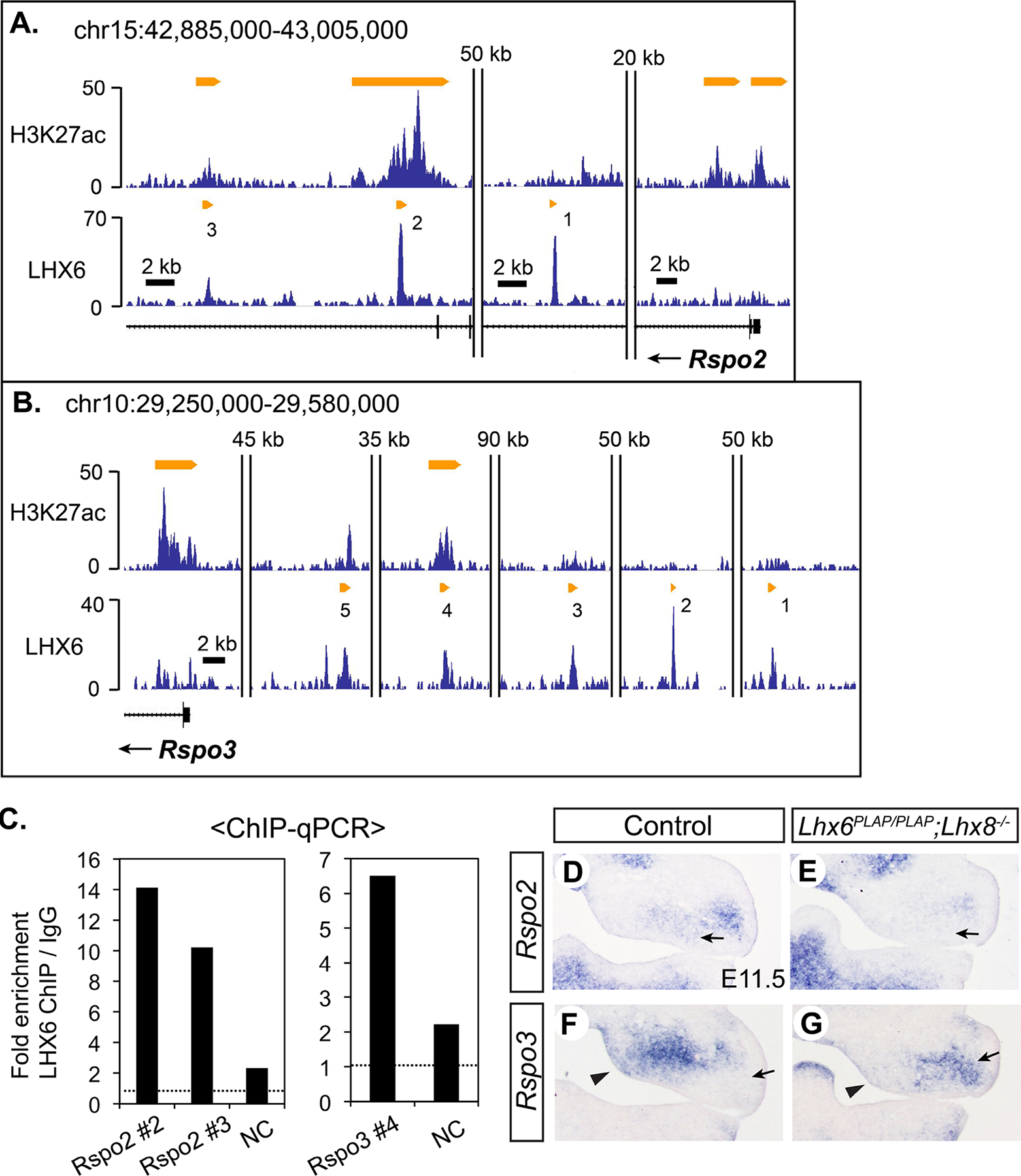
A,B) Results of H3K27ac ChIP-seq and LHX6 ChIP-seq around Rspo2 and Rspo3 visualized by Integrated Genome Browser. C) LHX6 ChIP-qPCR from E11.5 maxillary arch to confirm the result of ChIP-seq. Two independent rounds of ChIP-qPCR were performed, and the result from the second round is in Figure S5. D–G) RNA in situ hybridization on coronal sections of the head from E11.5 embryos. Arrows in D,E: decreased expression of Rspo2 in the upper molar mesenchyme in the Lhx mutant. Arrowheads in F,G: decreased expression of Rspo3 in the medial mesenchyme of the mutant maxillary arch. Arrows in F,G: ectopic expression of Rspo3 in the lateral mesenchyme of the mutant maxillary arch.
There were three LHX6 peaks within the introns of Rspo2 (Fig. 3A). Two of them (#2, #3) were marked by H3K27ac, and peak #2 was also occupied by p300 (Table S3). Rspo3 had five associated LHX6 peaks, which were spread over a large intergenic region upstream of Rspo3 (Fig. 3B). Only one peak (#4) was enriched with H3K27ac. Given the importance of the canonical WNT signaling in craniofacial development, we further confirmed LHX6 binding to the three putative enhancer regions (Rspo2 #2 and #3, Rspo3 #4) by ChIP-qPCR (Fig. 3C; Fig. S5).
Among 19 WNT 1igands in mammals, WNT11 is one of the few that primarily use non-canonical signaling pathways, such as Ca2+ pathway and planar cell polarity pathway (Uysal-Onganer and Kypta, 2012). Our transcriptional profiling found that Wnt11 expression was reduced in Lhx6PLAP/PLAP;Lhx8−/− mutant maxillary arch by 1.5-fold compared with controls at E10.5, and RNA in situ hybridization at E11.5 confirmed the difference in expression between the genotypes (Fig. 4C–F; Fig. S6). In the anterior part, Wnt11 was specifically downregulated in the medial corner of the maxillary arch in the mutants (Fig. 4C,D). More posteriorly, Wnt11 expression was decreased at the maxillary-mandibular junction in the mutants (Fig. 4E,F). Five LHX6 peaks were associated with Wnt11 in the genome, and all of them were enriched with H3K27ac but only one (#5) was enriched with p300 (Fig. 4A; Table S3). The most prominent LHX6 peak was in the first intron (#2), which was the third highest among all LHX6 peaks from this ChIP-seq.
Figure 4. Lhx6 and Lhx8 regulate Wnt11 and Dusp6 in the maxillary arch.
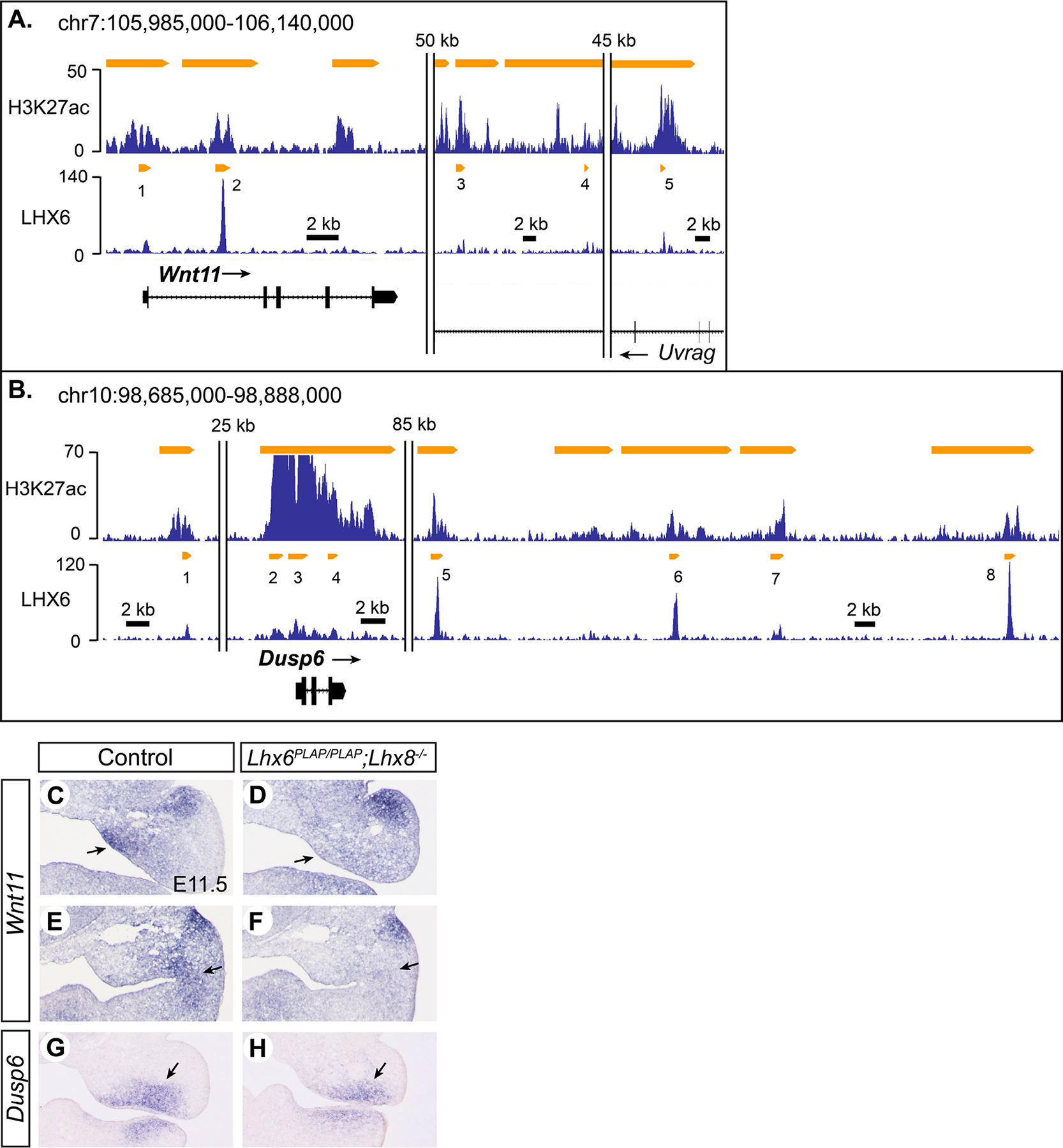
A,B) Results of H3K27ac ChIP-seq and LHX6 ChIP-seq around Wnt11 and Dusp6 visualized by Integrated Genome Browser. C-H) RNA in situ hybridization on coronal sections of the head from E11.5 embryos. Sections in E,F are posterior to those in C,D, from the same embryos. Arrows in C,D: decreased expression of Wnt11 in the medial mesenchyme of the mutant maxillary arch. Arrows in E,F: decreased expression of Wnt11 at the maxillary-mandibular junction in the mutant. Arrows in G,H: decreased expression of Dusp6 in the ventral mesenchyme of the mutant maxillary arch.
Dusp6 (dual specificity phosphatase 6) is another signaling molecule in the candidate positive target list of LHX6 and LHX8, and it encodes a negative feedback regulator of FGF-MAPK (mitogen-activated protein kinase) signaling pathway. Transcriptional profiling at E10.5 showed that Dusp6 was downregulated in Lhx6PLAP/PLAP;Lhx8−/− mutant maxillary arch (1.6-fold), and RNA in situ hybridization at E11.5 confirmed reduced expression in the mutants (Fig. 4G,H; Fig. S6). Because Dusp6 is an inhibitor as well as a readout of FGF-MAPK pathway (Eblaghie et al., 2003), we surveyed the microarray data for expression of several other genes known to be regulated by this pathway, namely, Spry1, Spry2, Spry4, Etv1, Etv4, and Etv5 (Mason et al., 2006; Munchberg and Steinbeisser, 1999; Raible and Brand, 2001). Only Etv1 was significantly different between the Lhx mutant and control groups (1.4-fold decrease in the mutant, p=0.0041), and so it appears unlikely that FGF-MAPK signaling in general was affected by inactivation of Lhx6 and Lhx8. From LHX6 ChIP-seq, we found 8 peaks associated with Dusp6 (Fig. 4B). All the peaks were marked with H3K27ac, and three of them (#3, #4, #5) were also bound by p300 (Table S3).
3. Discussion
In summary, by re-examining and validating previously published datasets, we identified putative target genes of a transcriptional activator function of LHX6 and LHX8. This study uncovered new regulatory relationships, for example, between Lhx and Eya genes. Also, while a couple of papers showed that Lhx6 and Lhx8 regulated the canonical WNT signaling in late stages of odontogenesis (He et al., 2021; Zhou et al., 2015), the connection to Rspo genes had been unknown. Figure 5 shows factors that are upstream or downstream of Lhx6 and Lhx8 within GRN of mammalian maxillary arch development. Even though Lhx6 and Lhx8 are expressed in closely overlapping patterns, their expression is under distinct regulations. Lhx6 expression in the maxillary arch was dependent on the functions of FGF8, DLX1, and DLX2 (Jeong et al., 2012; Trumpp et al., 1999), whereas Lhx8 expression was regulated by WNT/β-catenin signaling and a nuclear matrix protein SATB2 (special AT-rich sequence binding protein 2) (Dobreva et al., 2006; Landin Malt et al., 2014). For the genes downstream of Lhx6 and Lhx8, all the data were obtained from Lhx6;Lhx8 double mutants (this study, and Cesario et al., 2015; Denaxa et al., 2009), and thus it is impossible to distinguish contributions from each Lhx.
Figure 5. Lhx6 and Lhx8 within GRN of mammalian maxillary arch development.
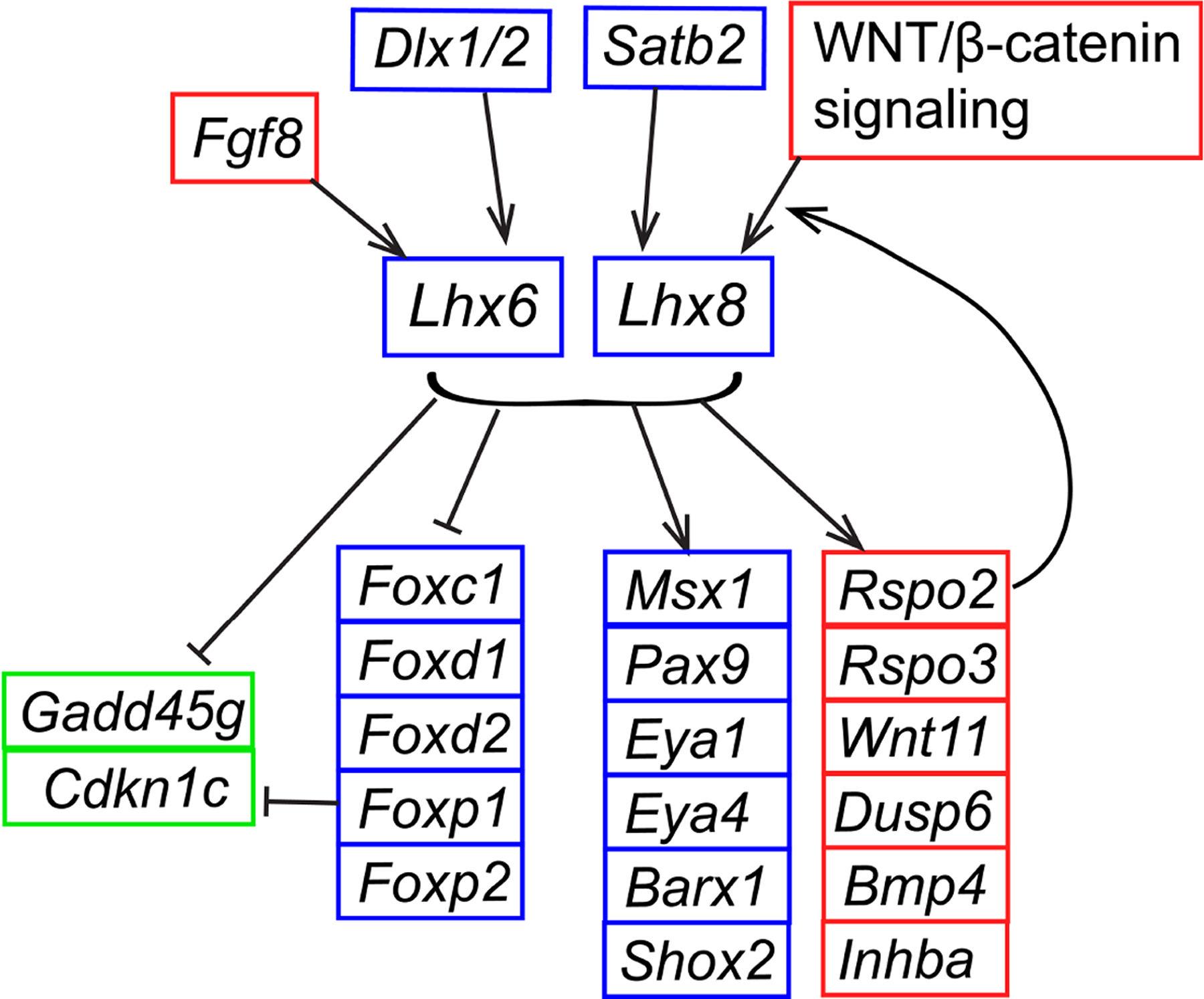
The schematic is based on data from this paper and previous reports (Cesario et al., 2015; Denaxa et al., 2009; Dobreva et al., 2006; Jeong et al., 2012; Landin Malt et al., 2014; Trumpp et al., 1999). It depicts the regulatory relationships demonstrated in the maxillary arch of mouse embryos at E9.5-E12.5.
Based on the current and our previous studies (Cesario et al., 2015), it appears that LHX6 and LHX8 can activate the expression of some genes while repressing others in the same tissue. The mechanistic basis of this dual function is unknown. One possibility is that transcriptional activation or repression by LHX is determined by an assortment of other transcription factors bound to the specific cis-regulatory element alongside LHX. These transcription factors may form a complex with LHX and alter the nature of interaction between LHX and the transcription machinery.
The genes examined in this paper were selected based on their known connections to craniofacial development. In humans, mutations of EYA1 underlie Branchio-otic syndrome (BOS) and Branchio-oto-renal syndrome (BOR), both of which include malformations of the ear. Similarly, deletion of Eya1 in mice resulted in hypoplasia of craniofacial skeleton and multiple defects in the ear (Xu et al., 1999). Eya4 has also been associated with craniofacial skeletogenesis because fusion of the palatal bone was delayed in Eya4 mutant mice (Depreux et al., 2008). Inactivation of Barx1 in mice caused cleft secondary palate and temporary stalling of molar development (Miletich et al., 2011). Shox2 mutation also caused cleft secondary palate, specifically in the anterior region (Yu et al., 2005). Msx1 and Pax9 encode key regulators of palatogenesis and odontogenesis (Lan et al., 2014; Lan et al., 2015), and loss of either gene led to cleft secondary palate and an arrest of tooth development at the bud stage (Peters et al., 1998; Satokata and Maas, 1994). Rspo2 mutant mice had partially penetrant cleft secondary palate, small lower jaw, and diastema teeth in the lower jaw (Jin et al., 2011; Yamada et al., 2009). Mutation of Rspo3 in the mouse craniofacial mesenchyme disrupted development of the lower incisors, while combined deletion of Rspo2 and Rspo3 resulted in severe hypoplasia of the face (Dasgupta et al., 2021). Knockout of Wnt11 in mice led to extensive embryonic lethality, precluding investigation of the craniofacial phenotype (Majumdar et al., 2003). However, knockdown of Wnt11 in mouse palate explant culture inhibited palatal shelf fusion (Lee et al., 2008), and a knockdown/overexpression study in chick showed that WNT11 regulates facial morphogenesis through the planar cell polarity pathway (Geetha-Loganathan et al., 2014). Mouse Dusp6 mutants were reported to have abnormal morphologies of the cranium and the middle ear (Li et al., 2007). In addition, a missense mutation in human DUSP6 was associated with malocclusion, a phenotype attributed to anomalies in growth and morphogenesis of the jaw (Nikopensius et al., 2013).
Clearly, a change in the expression of each target gene individually cannot fully recapitulate the profound craniofacial defects of Lhx6 and Lhx8 double mutants. Rather, the phenotype most likely arises from the combined effect of changes in multiple genes. It has been shown that mutations in Msx1 and Pax9 affected tooth development synergistically (Nakatomi et al., 2010), so did mutations in Barx1 and Msx1 (Miletich et al., 2011).
We acknowledge the limitations of this study in that unequivocally proving direct transcriptional regulation requires much more evidence than the data presented here. First, physical association between the promoter of a target gene and a putative LHX-regulated enhancer needs to be demonstrated by chromatin conformation capture (3C), which is technically challenging because 3C requires a large number of cells and the embryonic maxillary arch is small. Second, activity of the putative enhancer needs to be tested in vivo by transgenic reporter assays to confirm that it can drive gene expression in the maxillary arch in an LHX-dependent manner. However, generating transgenic animals takes a lot of resources, and thus it is beyond the scope of this paper. Nonetheless, this paper provides important new insights and guidance for future studies on GRN underlying development of the face.
4. Experimental Procedures
4.1. Animals
A mouse line for Lhx6 knockout with knockin of placental alkaline phosphatase (Lhx6PLAP, Mouse Genome Informatics (MGI) ID: 3584382) was generated by Regeneron using Velocigene technology (VG MAID #406) (Choi et al., 2005; Valenzuela et al., 2003). A mouse line for Lhx6 knockout with knockin of tamoxifen inducible Cre (Lhx6CreER, MGI ID: 4365737) was purchased from The Jackson Laboratory (Stock No: 010776) (Taniguchi et al., 2011). Lhx8 knockout line (Lhx8−, MGI ID: 2182594) was described before (Zhao et al., 1999). All the mutant lines were maintained in a mixed background that is predominantly CD1. Lhx6 and Lhx8 double knockout embryos were generated from an intercross of double heterozygote adults. Control embryos were selected from littermates that were wild type or a single heterozygote for Lhx6 or Lhx8. All the embryos were genotyped by PCR using DNA from the tail. Both Lhx6PLAP and Lhx6CreER lines were used in this study because while we initiated the project using Lhx6PLAP (Cesario et al., 2015), it was replaced with Lhx6CreER due to conditions associated with the material transfer agreement for Lhx6PLAP. We confirmed that Lhx6CreER/CreER;Lhx8−/− mutants had the same craniofacial phenotype as Lhx6PLAP/PLAP;Lhx8−/− mutants (Figure S1), which was also the same as the phenotype of another Lhx6 and Lhx8 double knockout mutants generated by other researchers (Denaxa et al., 2009).
All the animal experiments were performed in accordance with a protocol approved by Institutional Animal Care and Use Committee of New York University. The ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments) and National Institutes of Health Guide for the Care and Use of Laboratory Animals (8th edition) were followed.
4.2. ChIP-qPCR
ChIP-qPCR for LHX6 was performed from the maxillary arches of E11.5 wild type CD1 embryos as described before (Cesario et al., 2015). A negative control sequence for each LHX6 peak was selected from a nearby genomic region devoid of LHX6 binding based on the ChIP-seq profile (see Table S4 for genomic coordinates). ChIP-qPCR was an entirely separate experiment from the original ChIP-seq because fresh tissue was collected to prepare a new chromatin sample for each round of ChIP. The primers used for qPCR are listed in Table S4.
4.3. Cresyl violet staining and RNA in situ hybridization
Frozen sections of the head of an embryo were prepared and used for cresyl violet staining and RNA in situ hybridization as described before (Jeong et al., 2012). RNA in situ hybridization was performed on a series of coronal sections through the face that were separated by approximately 100 μm (see Fig. S2–S6). Each gene was examined in at least three pairs of the Lhx mutant and control littermates, and the results were consistent. The three pairs of embryos for each gene were from three different litters except for those used for Eya4, which were from two litters. Templates for the anti-sense RNA probes were obtained from other researchers (Barx1, Msx1, Pax9, Shox2, Wnt11), purchased from Open Biosystems as cDNA clones (Dusp6: GenBank BC003869, Rspo2: GenBank BC052844, Rspo3: GenBank BC103794), PCR-amplified from adult wild type CD1 mouse tail genomic DNA (Eya1: forward 5’-TGCATCATGCCTTGGAATTAGAG-3’, reverse with T3 polymerase site 5’-AATTAACCCTCACTAAAGGGACACGATTGTCTCAGTGATGTAC-3’), or PCR-amplified from E14.5 wild type CD1 head mesenchyme cDNA (Eya4: forward 5’-ACGCCTTACTCTTACCAAATGC-3’, reverse with T3 polymerase site 5’-AATTAACCCTCACTAAAGGGGCAAACAAAGGTTCGCACTACT-3’).
Supplementary Material
Highlights.
Candidate positive targets of LHX6 and LHX8 identified during upper jaw development
Expression of 10 genes examined in Lhx6;Lhx8 mutants by RNA in situ hybridization
Novel relationships found between Lhx and other key craniofacial regulators
Acknowledgement
We thank Krishnakali Dasgupta and Kesava Asam for assistance with dissecting and genotyping mouse embryos, and Dr Jean-Pierre Saint-Jeannet and his laboratory members for helpful discussions and sharing equipment. We thank Regeneron for allowing us to use Lhx6PLAP line.
Funding
This work was supported by NIH/NIDCR (R00 DE019486, R01 DE026798).
Footnotes
Supplementary Materials
Figures S1 – S6, Tables S1 – S4
Conflicts Of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, Phouanenavong S, Akiyama JA, Shoukry M, Afzal V, Rubin EM, FitzPatrick DR, Ren B, Hallgrimsson B, Pennacchio LA and Visel A, 2013. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 342, 1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley JF, Fisher S, Harris MP, Holmes G, Hooper JE, Jabs EW, Jones KL, Kesselman C, Klein OD, Maas RL, Marazita ML, Selleri L, Spritz RA, van Bakel H, Visel A, Williams TJ, Wysocka J, FaceBase C and Chai Y, 2016. The FaceBase Consortium: a comprehensive resource for craniofacial researchers. Development. 143, 2677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario JM, Landin Malt A, Deacon LJ, Sandberg M, Vogt D, Tang Z, Zhao Y, Brown S, Rubenstein JL and Jeong J, 2015. Lhx6 and Lhx8 promote palate development through negative regulation of a cell cycle inhibitor gene, p57Kip2. Hum Mol Genet. 24, 5024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Dong HW, Murphy AJ, Valenzuela DM, Yancopoulos GD, Swanson LW and Anderson DJ, 2005. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 46, 647–60. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Garcia E and Schilling TF, 2010. Regulation of facial morphogenesis by endothelin signaling: insights from mice and fish. Am J Med Genet A. 152A, 2962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta K, Cesario JM, Ha S, Asam K, Deacon LJ, Song AH, Kim J, Cobb J, Yoon JK and Jeong J, 2021. R-Spondin 3 Regulates Mammalian Dental and Craniofacial Development. Journal of Developmental Biology. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denaxa M, Sharpe PT and Pachnis V, 2009. The LIM homeodomain transcription factors Lhx6 and Lhx7 are key regulators of mammalian dentition. Dev Biol. 333, 324–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depreux FF, Darrow K, Conner DA, Eavey RD, Liberman MC, Seidman CE and Seidman JG, 2008. Eya4-deficient mice are a model for heritable otitis media. J Clin Invest. 118, 651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G and Grosschedl R, 2006. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 125, 971–986. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Lunn JS, Dickinson RJ, Munsterberg AE, Sanz-Ezquerro JJ, Farrell ER, Mathers J, Keyse SM, Storey K and Tickle C, 2003. Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr Biol. 13, 1009–18. [DOI] [PubMed] [Google Scholar]
- Filali M, Cheng N, Abbott D, Leontiev V and Engelhardt JF, 2002. Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem. 277, 33398–410. [DOI] [PubMed] [Google Scholar]
- Freese NH, Norris DC and Loraine AE, 2016. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 32, 2089–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Fu K and Richman JM, 2014. Avian facial morphogenesis is regulated by c-Jun N-terminal kinase/planar cell polarity (JNK/PCP) wingless-related (WNT) signaling. J Biol Chem. 289, 24153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou Y, Zhang T and Xu J, 2015. Transcription Factors in Craniofacial Development: From Receptor Signaling to Transcriptional and Epigenetic Regulation. Curr Top Dev Biol. 115, 377–410. [DOI] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT and Pachnis V, 1998. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 125, 2063–74. [DOI] [PubMed] [Google Scholar]
- He J, Jing J, Feng J, Han X, Yuan Y, Guo T, Pei F, Ma Y, Cho C, Ho TV and Chai Y, 2021. Lhx6 regulates canonical Wnt signaling to control the fate of mesenchymal progenitor cells during mouse molar root patterning. PLoS Genet. 17, e1009320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JE, Feng W, Li H, Leach SM, Phang T, Siska C, Jones KL, Spritz RA, Hunter LE and Williams T, 2017. Systems biology of facial development: contributions of ectoderm and mesenchyme. Dev Biol. 426, 97–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Cesario J, Zhao Y, Burns L, Westphal H and Rubenstein JL, 2012. Cleft palate defect of Dlx1/2−/− mutant mice is caused by lack of vertical outgrowth in the posterior palate. Dev Dyn. 241, 1757–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN and Costantini F, 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 22, 1172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YR, Turcotte TJ, Crocker AL, Han XH and Yoon JK, 2011. The canonical Wnt signaling activator, R-spondin2, regulates craniofacial patterning and morphogenesis within the branchial arch through ectodermal-mesenchymal interaction. Dev Biol. 352, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Jia S and Jiang R, 2014. Molecular patterning of the mammalian dentition. Semin Cell Dev Biol. 25–26, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Xu J and Jiang R, 2015. Cellular and Molecular Mechanisms of Palatogenesis. Curr Top Dev Biol. 115, 59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landin Malt A, Cesario JM, Tang Z, Brown S and Jeong J, 2014. Identification of a face enhancer reveals direct regulation of LIM homeobox 8 (Lhx8) by wingless-int (WNT)/beta-catenin signaling. J Biol Chem. 289, 30289–30301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Kim JY, Cho KW, Lee MJ, Cho SW, Kwak S, Cai J and Jung HS, 2008. Wnt11/Fgfr1b cross-talk modulates the fate of cells in palate development. Dev Biol. 314, 341–50. [DOI] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X and Mansour SL, 2007. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 134, 167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie A, Purdie L, Davidson D, Collinson M and Hill RE, 1997. Two enhancer domains control early aspects of the complex expression pattern of Msx1. Mech Dev. 62, 29–40. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Vainio S, Kispert A, McMahon J and McMahon AP, 2003. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 130, 3175–85. [DOI] [PubMed] [Google Scholar]
- Marcucio R, Hallgrimsson B and Young NM, 2015. Facial Morphogenesis: Physical and Molecular Interactions Between the Brain and the Face. Curr Top Dev Biol. 115, 299–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Basson MA and Licht JD, 2006. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 16, 45–54. [DOI] [PubMed] [Google Scholar]
- Medeiros DM and Crump JG, 2012. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev Biol. 371, 121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletich I, Yu WY, Zhang R, Yang K, Caixeta de Andrade S, Pereira SF, Ohazama A, Mock OB, Buchner G, Sealby J, Webster Z, Zhao M, Bei M and Sharpe PT, 2011. Developmental stalling and organ-autonomous regulation of morphogenesis. Proc Natl Acad Sci U S A. 108, 19270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M and Rijli FM, 2010. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 137, 2605–21. [DOI] [PubMed] [Google Scholar]
- Munchberg SR and Steinbeisser H, 1999. The Xenopus Ets transcription factor XER81 is a target of the FGF signaling pathway. Mech Dev. 80, 53–65. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Wang XP, Key D, Lund JJ, Turbe-Doan A, Kist R, Aw A, Chen Y, Maas RL and Peters H, 2010. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev Biol. 340, 438–49. [DOI] [PubMed] [Google Scholar]
- Nikopensius T, Saag M, Jagomagi T, Annilo T, Kals M, Kivistik PA, Milani L and Metspalu A, 2013. A missense mutation in DUSP6 is associated with Class III malocclusion. J Dent Res. 92, 893–8. [DOI] [PubMed] [Google Scholar]
- Pennacchio L, Lisgo S, Rubin E, FitzPatrick D, Yuzawa Y, Visel A and Chai Y, 2017. p300 ChIP-seq experiment on mouse embryonic craniofacial tissue at e11.5. FaceBase Consortium 10.25550/TYM [DOI] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K and Balling R, 1998. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12, 2735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F and Brand M, 2001. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev. 107, 105–17. [DOI] [PubMed] [Google Scholar]
- Raslan AA and Yoon JK, 2019. R-spondins: Multi-mode WNT signaling regulators in adult stem cells. Int J Biochem Cell Biol. 106, 26–34. [DOI] [PubMed] [Google Scholar]
- Reynolds K, Kumari P, Sepulveda Rincon L, Gu R, Ji Y, Kumar S and Zhou CJ, 2019. Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis Model Mech. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I and Maas R, 1994. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 6, 348–56. [DOI] [PubMed] [Google Scholar]
- Sperber GH, Sperber SM and Guttmann GD, 2010. Craniofacial Embryogenetics and development, 2nd ed. People’s Medical Pub. House USA, Shelton, CT. [Google Scholar]
- Tadjuidje E and Hegde RS, 2013. The Eyes Absent proteins in development and disease. Cell Mol Life Sci. 70, 1897–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB and Huang ZJ, 2011. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JLR, Bishop JM and Martin GR, 1999. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes & Development. 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal-Onganer P and Kypta RM, 2012. Wnt11 in 2011 - the regulation and function of a non-canonical Wnt. Acta Physiol (Oxf). 204, 52–64. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, Chernomorsky R, Boucher M, Elsasser AL, Esau L, Zheng J, Griffiths JA, Wang X, Su H, Xue Y, Dominguez MG, Noguera I, Torres R, Macdonald LE, Stewart AF, DeChiara TM and Yancopoulos GD, 2003. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 21, 652–9. [DOI] [PubMed] [Google Scholar]
- Wilderman A, VanOudenhove J, Kron J, Noonan JP and Cotney J, 2018. High-Resolution Epigenomic Atlas of Human Embryonic Craniofacial Development. Cell Rep. 23, 1581–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S and Maas R, 1999. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 23, 113–7. [DOI] [PubMed] [Google Scholar]
- Yamada W, Nagao K, Horikoshi K, Fujikura A, Ikeda E, Inagaki Y, Kakitani M, Tomizuka K, Miyazaki H, Suda T and Takubo K, 2009. Craniofacial malformation in R-spondin2 knockout mice. Biochem Biophys Res Commun. 381, 453–8. [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y and Chen Y, 2005. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 132, 4397–406. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Guo YJ, Tomac AC, Taylor NR, Grinberg A, Lee EJ, Huang S and Westphal H, 1999. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc Natl Acad Sci U S A. 96, 15002–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Yang G, Chen M, Wang C, He L, Xiang L, Chen D, Ling J and Mao JJ, 2015. Lhx8 mediated Wnt and TGFbeta pathways in tooth development and regeneration. Biomaterials. 63, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


