Novel models of kinase-rearranged sarcomas show that MAPK pathway feedback activation dampens responses to tyrosine kinase inhibitors, revealing the potential of combinatorial therapies to combat these tumors.
Abstract
Kinase fusions have been identified in a growing subset of sarcomas, but a lack of preclinical models has impeded their functional analysis as therapeutic targets in the sarcoma setting. In this study, we generated models of sarcomas bearing kinase fusions and assessed their response to molecularly targeted therapy. Immortalized, untransformed human mesenchymal stem cells (HMSC), a putative cell of origin of sarcomas, were modified using CRISPR-Cas9 to harbor a RET chromosomal translocation (HMSC-RET). In parallel, patient-derived models of RET- and NTRK-rearranged sarcomas were generated. Expression of a RET fusion activated common proliferation and survival pathways and transformed HMSC cells. The HMSC-RET models displayed similar behavior and response to therapy as the patient-derived counterparts in vitro and in vivo. Capicua (CIC)-mediated suppression of negative MAPK pathway regulators was identified as a potential mechanism by which these sarcomas compensate for RET or NTRK inhibition. This CIC-mediated feedback reactivation was blocked by coinhibition of the MAPK pathway and RET or NTRK in the respective models. Importantly, the combination of RET and ERK inhibitors was more effective than single agents at blocking tumor growth in vivo. This work offers new tools and insights to improve targeted therapy approaches in kinase-addicted sarcomas and supports upfront combination therapy to prolong responses.
Significance:
Novel models of kinase-rearranged sarcomas show that MAPK pathway feedback activation dampens responses to tyrosine kinase inhibitors, revealing the potential of combinatorial therapies to combat these tumors.
Introduction
The rapid development and routine clinical implementation of next-generation sequencing for molecular diagnostics has significantly increased the number of patients who can benefit from biomarker-directed therapy. While significant progress has been made in the field of carcinomas (e.g., lung, breast, colon), only a marginal benefit has been achieved in patients with soft-tissue sarcomas. This is partly due to the low prevalence of sarcomas (1% of malignancies), which limits the number of patients for clinical trials. In addition, sarcomas represent a highly heterogeneous group, with more than 100 different entities recognized to date (1, 2).
Most sarcomas have either molecular alterations directly affecting gene expression (e.g., transcription factor fusions or alterations in the epigenetic machinery) that are historically challenging to target (3), or a yet undefined driver. A small, but continuously growing proportion of sarcomas harbor actionable oncogenic drivers that act mainly via deregulation of RTK signaling. The successful targeting of KIT and PDGFRA mutants in advanced/metastatic gastrointestinal stromal tumors (GIST) with imatinib was the first demonstration of successful application of targeted therapy to sarcomas (4). More recently, the benefit of targeted therapy was observed in NF1-deficient and NTRK-rearranged soft-tissue tumors treated with MEK and NTRK inhibitors, respectively (5, 6).
The rarity and biological heterogeneity of soft-tissue sarcomas present challenges for molecular subtype-specific investigation of biology and therapy development. While murine models and ectopic expression of cDNAs of genes of interest are commonly used to model and study cancer biology, including sarcomagenesis, recent advances in genetic engineering and human stem cell technologies present new opportunities to improve sarcoma modeling. Our group and others have previously demonstrated the feasibility of using CRISPR-Cas9–mediated genomic editing in human cells to model chromosomal translocation (7–9).
RET fusions have recently been described in undifferentiated pediatric sarcomas (10, 11). While many of these fusions appear within the infantile fibrosarcoma spectrum (ETV6-NTRK3 fusions are more typically expressed in this histology), it is important to note that some cases occur in adults (10). The spectrum and prevalence of RET fusion–positive sarcomas in adults are yet to be defined well. Here, we used CRISPR-Cas9 and immortalized, untransformed human mesenchymal stem cells (HMSC) as putative cells-of-origin of soft-tissue sarcomas to model a RET oncogene fusion, SPECC1L-RET, which was first identified in a sarcoma. To complement this genomically engineered model, we generated the first patient-derived xenograft model and primary cancer cells from a sarcoma harboring this RET fusion. We used these novel preclinical sarcoma models to investigate oncogenic properties of RET fusion and explore different therapeutic strategies. We show that dynamic feedback regulation of the MAPK pathway via Capicua (CIC) impinges on growth and response to RET-targeted therapy. Importantly, we found that a combination of RET and ERK inhibitors is more effective in vivo at suppressing tumor growth than monotherapy. We extended these findings to an ETV6-NTRK3-rearranged infantile fibrosarcoma patient-derived model, supporting the broader relevance of our results.
Materials and Methods
Materials
All cell culture growth media, antibiotics, and PBS were prepared by the MSK Media Preparation Core Facility. FBS was procured from Atlanta Biologicals. Primocin was obtained from InvivoGen. Human mesenchymal stem cells (HMSC) were obtained from Lonza. HEK293T cells were purchased from ATCC (RRID:CVCL_0063). All other cell lines were generated as described below. Small-molecule inhibitors (except LOXO-292) were obtained from Selleckchem. Promega's Caspase Glo 3/7 activity kit, AlamarBlue viability dye, tissue culture plastic wares, and all Western blotting reagents were obtained from Thermo Fisher Scientific. Protease inhibitor cocktail, RIPA lysis buffer (10×), cloning discs and all other chemicals not listed above were purchased from EMD-Millipore Sigma. All oligonucleotides used for PCR assays were custom designed by the authors and obtained from Integrated DNA Technologies. The small-molecule RET inhibitor LOXO-292 (selpercatinib) was provided by LOXO Oncology. For Western blotting and IHC, the antibody clones, manufacturers, and dilution used are listed in Supplementary Table S1.
Growth and propagation of cell lines
All cell lines were maintained in a humidified incubator infused with 5% CO2 and subcultured when the stock flasks reached approximately 75% confluence at a 1:3 dilution. All cell lines were maintained in DMEM:F12 (high glucose) growth medium supplemented with 10% FBS and 100 μg/mL primocin (complete growth media). Cell lines were tested for Mycoplasma every four months (MycoAltert Kit, Lonza) with the most recent testing conducted three months prior to completion of the studies in this manuscript. A new vial of cells was thawed and used for up to 5–10 passages (every two months) and the known fusion oncogene was verified by RT-PCR each time.
Patient, patient-derived xenograft models, and cell lines
The patient was given selpercatinib on a single patient use protocol and the study was conducted in accordance with the Declaration of Helsinki as previously reported (12). Tissue samples were collected under an institutional IRB-approved biospecimen collection protocol and written informed consent was obtained from the patients. Animals were monitored daily and cared for in accordance with guidelines approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee and Research Animal Resource Center. The patient-derived xenograft (PDX) models SR-Sarc-0001pdx (infantile fibrosarcoma, resected brain tumor metastasis sample, SPECC1L-RET rearrangement) and the SR-Sarc-0002pdx (infantile fibrosarcoma, cerebellar tumor sample, ETV6-NTRK3F617I rearrangement) were generated by mincing the tumor specimens and then mixing with 50% (vol/vol) Matrigel and implanted into the subcutaneous flank of a 6-week-old female NOD/SCID gamma (NSG) mouse (The Jackson Laboratory). PDX tumors were transplanted for at least three serial passages before the model was considered established and used for efficacy studies. The SR-Sarc-0001 and SR-Sarc-0002 cell lines were generated from PDX tumor tissues. Briefly, fresh tumors were cut into small pieces and then digested in a cocktail of tumor dissociation enzymes (Miltenyi Biotec, 130–095–929) in 5 mL serum-free DME:F12 media for 30 minutes, 37°C, with vortexing every 5–10 minutes, according to manufacturer's instructions. Digested samples were resuspended in 45 mL complete growth media to inactivate the tissue digestion enzymes and then cells were pelleted by centrifugation. Finally, cells were plated in complete growth media and allowed to propagate over multiple generations, trypsinized when necessary to subculture, and eventually only single cells remained. Studies with the SR-Sarc-0001 cells were conducted during the first five passages.
The immortalized HMSC-TS cell line was generated by sequential lentiviral transfer of TERT (T) and SV40 large-T antigen cDNAs, into primary HMSC cells (Lonza) and then stable cells were selected with 500 μg/mL geneticin and 1 μg/mL puromycin, respectively. Throughout this article, the immortalized HMSC-TS cells are referred to as HMSC. To faithfully model a SPECC1L-RET fusion, the following guide RNAs (gRNA) were designed using benchling.com: RET intron 11 guide #1 TCACGCCACCATAAGCTGCG; RET intron 11 guide #2 CATCCGGAGCAGTCCCAAGT; SPECC1L intron 10 guide #1 GCAGTGTCAAGGTTTACACG; SPECC1L intron 10 guide #2 GAGAACCACTTTAACCCGGG. The placement of these gRNAs aimed to minimize splicing interference by being positioned at least 50 to 250 bp from the splice site. To form a complete ribonucleoprotein (RNP) complex, an sgRNA-specific Alt-R CRISPR-Cas9 crRNA, universal Alt-R CRISPR-Cas9 tracrRNA and Alt-R S.p. HiFi Cas9 Nuclease V3 were combined (Integrated DNA Technologies). The resulting RNP complex was introduced into HMSC cells by electroporation (Amaxa nucleofector II system, Lonza). The cells were seeded into 6-well plates thereafter. Seventy-two hours after nucleofection, the cells were harvested, RNA isolated, and the expression of SPECC1L-RET fusion in the pool of cells was confirmed using RT-PCR. The cells were then seeded for subclonal selection at a density of 5,000 cells per 10-cm dish. Single-cell colonies were isolated using cloning discs and propagated in 96-well plates. Expression of SPECC1L-RET fusion in the clones was tested using RT-PCR. A clone of SPECC1L-RET–positive immortalized HMSCs (HMSC-RET) was used in the subsequent experiments.
Xenograft formation of HMSC-RET cells
To test the tumorigenic potential of the isogenic cell lines, HMSC or HMSC-RET cells were mixed with 50% (vol/vol) Matrigel and implanted into a subcutaneous flank of female NSG mice (10 million cells per flank). Each cell line was implanted into four mice. Once tumors reached approximately 600 mm3, animals were sacrificed, and xenograft tumors were transplanted to generate tumor seeds for subsequent efficacy studies.
Efficacy studies
Fresh SR-Sarc-0001pdx samples or HMSC-RET xenograft tumor samples (15 mm3) were cleaned, minced, then mixed with Matrigel (50%) and implanted into a subcutaneous flank of 6-week-old female NSG mice to generate xenografts. After randomizing into groups of five, tumor-bearing animals were treated with vehicle, vandetanib (50 mg/kg, once daily) or selpercatinib (10 mg/kg, twice daily; 25 mg/kg, twice daily) when tumors reached approximately 120–130 mm3 volume. Treatment was initiated 13 days after implantation for both models in the monotherapy studies. This is indicated as day 0 for treatment time on the graphs. In the drug combination studies, treatment was initiated nine and 13 days after implantation of the HMSC-RET and SR-Sarc-0001pdx tumors, respectively. Selpercatinib was administered orally in 1% carboxymethyl-cellulose sodium and 0.5% Tween 80 in deionized water. Vandetanib was administered orally in 1% carboxymethyl-cellulose sodium in deionized water. Tumor size and body weight were measured twice weekly. Tumor volume was calculated using the formula: length × width2 × 0.52. One animal bearing HMSC-RET tumor in the selpercatinib group died during the first week of treatment of an unknown cause; therefore, this study arm had four mice at the completion of the experiment.
Immunofluorescence
Cells were seeded in 4-well Lab Tek II Chamber Slides (Thermo Fisher Scientific). The following day, cells were fixed for 15 minutes with 4% paraformaldehyde, washed, and incubated in blocking buffer for 1 hour (PBS with 5% FBS and 0.3% Triton X-100). Blocking buffer was aspirated and cells were incubated with primary antibody overnight in the dark at 4°C. The following day, cells were washed, incubated with fluorophore-conjugated secondary antibody for 1 hour at room temperature in the dark, washed with PBS containing Hoechst 33342 (1 μg/mL), then mounted using ProLong Gold Antifade reagent (Cell Signaling Technology).
Whole-transcriptome sequencing and analysis
Cells were seeded in 6-well plates at a density of 500,000 cells per well. The media were refreshed with DMEM:F12 media supplemented with 0.5% FBS the next day. Eighteen hours later, three wells of HMSC-RET and SR-Sarc-0001 cells were treated with 100 nmol/L selpercatinib in DMEM:F12 media supplemented with 0.5% for 6 hours. Cells were then lysed, and total RNA extracted using RNeasy Mini Kit (Qiagen). Whole-transcriptome sequencing was performed by Genewiz. FASTQ files were aligned against the human genome (assembly GRCh38) using RNASTAR aligner (13). Gene read counts were estimated using featureCounts program (RRID:SCR_012919) and GENCODE GRCh38 gene annotations (RRID:SCR_014966). Gene expression analysis and Gene Set Expression Analysis were performed using DEseq2 (RRID:SCR_015687) and clusterProfiler (RRID:SCR_016884), respectively, in R programming environment.
Histology
Tissue from HMSC-RET and SR-Sarc-0001 xenograft tumors was fixed in formalin, embedded in paraffin, mounted on glass slides and stained with hematoxylin and eosin.
Viability
For viability assays, cells were plated in clear-bottom, white 96-well plates at a density of 7,500 cells per well and incubated with compounds for 96 h as described previously (14). The relative number of viable cells was determined using alamarBlue viability dye, and fluorescence was measured using a Molecular Devices SpectraMax M2 multimodal plate reader (Ex: 485 nm, Em: 530 nm) (14). Data was analyzed by non-linear regression and curves fitted using GraphPad Prism software to generate IC50 values.
Apoptosis assay
For apoptosis assays, cells were plated directly into compounds at a density of 25,000 cells per well in white, clear-bottom 96-well plates. Each plate contained DMSO-treated wells as negative controls and 1 μmol/L carfilzomib-treated wells as positive controls. Caspase-3/7 enzymatic assay was performed 48 h after treatments using Caspase-Glo 3/7 Assay System (Promega). Luminescence was measured using a BioTek Synergy H1 microplate reader at 10-minute intervals (Agilent). Each condition was assayed in triplicate determinations in at least two independent experiments. Data were normalized to vehicle-treated wells and analyzed by non-linear regression using Graphpad Prism v8.0. The results of representative experiments are shown (mean ± SD).
Spheroids assay
For assessment of anchorage-independent growth, 3,000 cells were plated into Corning Costar ultra-low attachment 96-well plates (Thermo Fisher Scientific). Cells were observed visually and images captured every 24 hours. AlamarBlue assay was used to assess metabolic activity.
Synergy analysis
For synergy analysis, cells were plated into internal wells of clear-bottom, white 96-well plates at a density of 7,500 cells per well and incubated with compounds for 96 hours as described previously (14). Three to four plates were used for each combination of drugs for each cell line, representing 3–4 replicates. The relative number of viable cells was determined using AlamarBlue viability as described above. To determine synergy or antagonism, we applied Combenefit software tool using classical Loewe synergy model (15).
Preparation of whole-cell, cytoplasmic and nuclear extracts, and Western blotting
For Western blotting studies of whole-cell extracts, cells were grown in medium containing 10% FBS and then deprived of serum by growing in medium containing 0.5% FBS for 24 hours prior to inhibitor treatments and preparation of extracts. Protein levels and phosphorylation state were detected by Western blotting as previously described (16). Cells were lysed in 1× RIPA lysis buffer containing phosphatase and protease inhibitors. Cell lysis and extraction of separate cytoplasmic and nuclear protein fractions was performed using NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Phosphatase and protease inhibitors were added to the reagents. An approximate ratio of cytoplasmic to nuclear protein ratio of 4:1 yield was obtained. Whole-cell lysates and subcellular fractions were denatured in 2× sample buffer at 55°C for 15 minjutes. Fifteen and 3.75 μg protein were resolved on 3%–8% NuPAGE Tris-Acetate gels (Invitrogen) for cytoplasmic and nuclear samples, respectively. For whole-cell extracts, 20 μg total protein was resolved on 4%–12% NuPAGE or Bolt gels (Invitrogen). Proteins were transferred onto polyvinylidene fluoride (PVDF) membranes, which were then blocked in 3% (wt/vol) BSA in Tris-buffered saline supplemented with 0.1% Tween-20 (vol/vol) for 1 hour at room temperature, and probed with primary antibodies. Bound antibodies were detected with peroxidase-labeled goat antibody raised to mouse or rabbit IgG (R&D Systems) and imaged with enhanced chemiluminescence (ECL) Western blotting detection reagent (GE Healthcare). Images were captured on X-ray films. Western blotting was conducted at least two times from independently prepared samples.
Generation of lentiviruses and transduction
Lentiviral plasmids carrying a nontargeting shRNA (NTsh) or shRNAs targeting DUSP4 or DUSP6 were obtained from the MSK RNAi Core Facility (TRC clones). To generate lentiviruses, plasmids (packaging and transfer) were transfected into HEK293T cells using Fugene HD transfection reagent and viral supernatant was harvested at 48, 72, and 96 hours after transfection and combined. Transduction was performed in 6-well plates using 10 μg/mL polybrene transfection reagent (17).
RT-PCR experiments
For detection of the SPECC1L-RET fusion transcript in cell lines and PDX, RNA was extracted using RNeasy Mini Kit (Qiagen) and cDNAs were synthesized using SuperScript IV VILO (Thermo Fisher Scientific) according to the manufacturer's instructions. The SPECC1L-RET fusion was detected by RT-PCR using 5′-ACC AAA CCC TGC TGC AGC TGC A-3′ (forward, SPECC1L exon 10) and 5′-TGC TCT GCC TTT CAG ATG GAA GG-3′ (reverse, RET exon 12) primers.
Statistical analysis
For animal studies, AUC analysis was used to compare the average tumor volume between groups. Briefly, AUC values and their SEs were computed as an estimation of the surface area between baseline values (mean value of the tumor volumes at the beginning of the treatment) and growth curves for vehicle and each treatment conditions. Regression of tumor volume below the baseline value is indicated by negative AUC values. Treatment response was compared to the vehicle group using multiple Student t tests. All data was plotted and analyzed using GraphPad Prism v8 software (RRID:SCR_002798). IC50 values were compared using the 95% CI values for significance.
Data availability statement
Reagents created by our laboratory and used in this study can be obtained by contacting the corresponding author directly.
Results
Generation of isogenic HMSC cells with a SPECC1L-RET rearrangement using CRISPR-Cas9
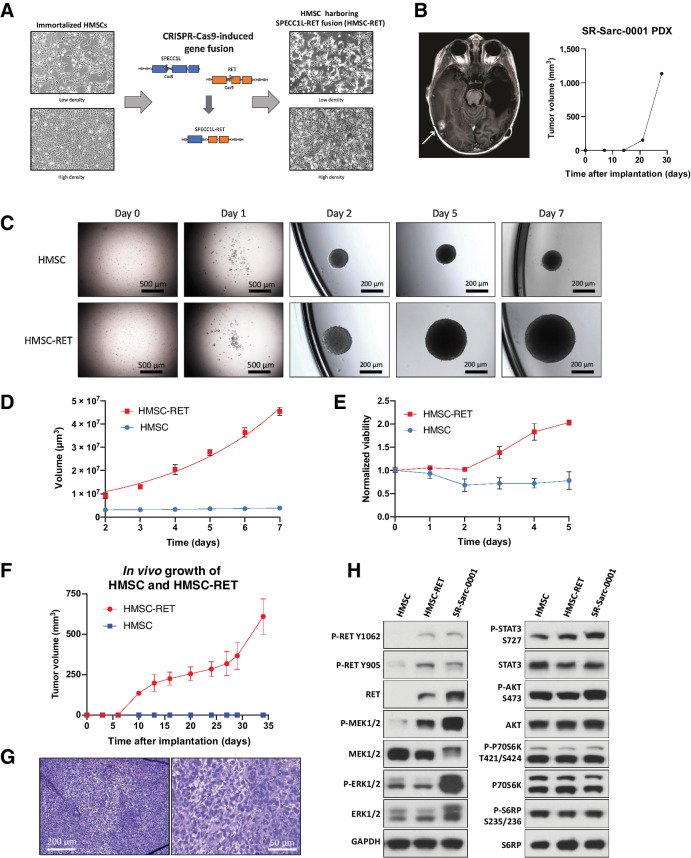
Modeling sarcomas to investigate biology and develop therapy has relied mainly on exogenous constructs in murine models or viral-mediated cDNA overexpression in immortalized untransformed cells, or cancer cells (18–20). Here we aimed to investigate the feasibility of using human cells to introduce genomic alterations using the CRISPR-Cas9 system to model sarcomas driven by a RET rearrangement. HMSCs are believed to be the ultimate precursors for many cell types that give rise to soft-tissues and are likely the putative cells-of-origin of soft-tissue sarcomas (21, 22). The HMSCs were immortalized by sequential expression of TERT and SV40 large T-antigen. We then used CRISPR-Cas9 to introduce an oncogenic SPECC1L-RET fusion by genomic alteration of the endogenous alleles of SPECC1L and RET in the immortalized HMSC cells. This fusion was previously identified in soft-tissue sarcomas of mainly children (10, 11). Guides targeting introns 10 and 11 of SPECC1L and RET, respectively, were used to enable aberrant recombination between these introns, resulting in a chimeric gene encoding a fusion of exon 10 of SPECC1L to exon 12 of RET. HMSC cells harboring the SPECC1L-RET rearrangement (HMSC-RET) demonstrated greater cellular atypia than parental HMSC cells (Fig. 1A).
Figure 1.
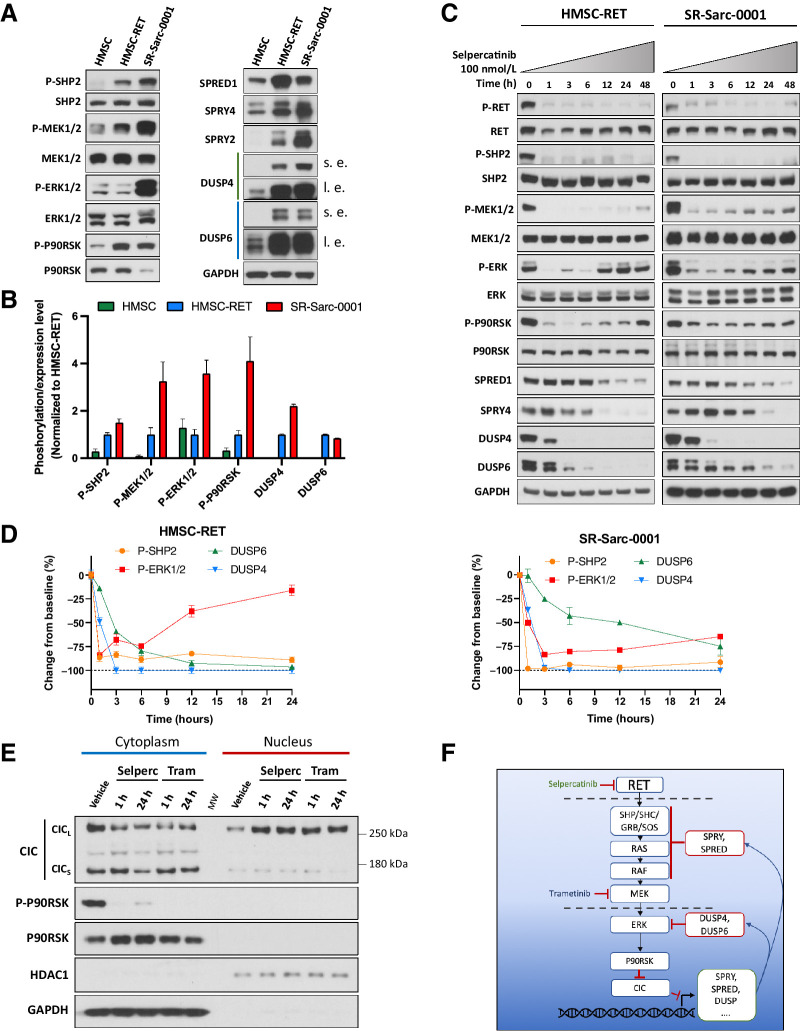
Establishment of novel SPECC1L-RET–rearranged sarcoma models. A, Generation of a SPECC1L-RET chromosomal rearrangement using CRISPR-Cas9–mediated genomic engineering in immortalized HMSC. B, Imaging of the brain metastasis of the SPECC1L-RET–rearranged sarcoma (left) that was used to establish SR-Sarc-0001 PDX (PDX growth in time is depicted in the right panel) C, HMSC and HMSC-RET cells were plated into ultra-low attachment plates to form spheroids and images were captured every 24 hours. Data represent the mean ± SD of 10 measurements. D, Spheroid volume. E, Metabolic activity of the spheroids was measured every 24 hours using alamarBlue cell viability dye. Resulting values were normalized to the initial values obtained at the first measurement (24 hours after seeding). Data represent mean ± SD of 12 measurements. F, HMSCs and HMSC-RET cells were implanted subcutaneously (10 million cells per implantation) to assess tumorigenesis. All HMSC-RET implantations resulted in tumors (100% penetrance), while HMSCs did not form tumors. Data represent the mean ± SD of 4 tumors. G, HMSC-RET xenografts were extracted, fixed in formalin, and embedded in paraffin. Hematoxylin and eosin staining of the obtained tissue slides is shown. H, HMSC, HMSC-RET, and SR-Sarc-0001 cells were serum-starved for 24 hours and then whole-cell extracts were prepared for Western blot profiling for the total and phosphorylated proteins shown.
To have a patient-derived counterpart for the HMSC-RET model, we generated PDX and cell line models (SR-Sarc-0001pdx and SR-Sarc-0001cl, respectively) from a sarcoma harboring SPECC1L-RET rearrangement (Fig. 1B). The initial clinical history of the patient from which this tumor sample was obtained was previously published (12) and here we report the follow-up clinical course (depicted schematically in Supplementary Fig. S1A). Given severe toxicities with conventional chemotherapy, at 21 months of age, the patient was treated with 48 mg selpercatinib, an FDA-approved RET inhibitor for lung and thyroid cancers (23). She had a partial response after the first two cycles of selpercatinib but then developed an isolated brain metastasis at the right posterior temporal occipital junction, which was resected. A PDX model was established from this resection (Fig. 1B). Histologic characterization of the PDX tumor and the corresponding patient sample is depicted in Supplementary Fig. S1B. The SPECC1L-RET fusion was confirmed by RT-PCR in the HMSC-RET cells and the patient-derived models (Supplementary Fig. S1C). The fusion point was determined by sequencing of the PCR amplicon (Supplementary Fig. S1D).
Following intracranial disease progression in the absence of an acquired resistance mutation, selpercatinib dose was increased to 100 mg, and the radiographic response in all target lesions improved to 66% (in aggregate) from baseline. The patient's functional status gradually improved from 30% to 70% on the Lansky scale with selpercatinib therapy. She remained on selpercatinib monotherapy until 42 months of age, when it became apparent that she had progression of multiple brain lesions but otherwise stable extracranial disease. The patient subsequently passed away.
Characterization of RET fusion–positive sarcoma cells
We characterized the sarcoma models with RET fusions by evaluating cellular signaling and tumorigenic properties. Anchorage-independent growth is one of the cornerstones of malignant transformation and is commonly tested to confirm oncogenic properties (24). To determine whether CRISPR-Cas9–induced SPECC1L-RET rearrangement conferred anchorage-independent growth, HMSC-RET and HMSC cells were seeded into ultra-low attachment plates and observed for spheroid formation, growth, and metabolic activity. Both cell lines formed spheroids by day two (Fig. 1C). However, only HMSC-RET spheroids showed growth over the seven days of observation (Fig. 1C), with a corresponding increase in volume of the spheroids (Fig. 1D). HMSC-RET spheroids also showed increased metabolic activity over time, consistent with elevated growth (Fig. 1E). In contrast, HMSC spheroids showed decreased metabolic activity by day two, followed by a plateau (Fig. 1E). To gain some insights into the difference in growth rates, we examined the expression level of proteins that regulate proliferation and survival. HMSC-RET cells had higher levels of the cell-cycle activator cyclin D1 and lower levels of both the cell-cycle inhibitor P27 and the proapoptotic marker cleaved PARP (Supplementary Fig. S2). These results suggest that the faster growth rate of the HMSC-RET cells was likely due to prominent activation of proliferation pathways and reduced cell death.
To determine whether the CRISPR-Cas9–induced SPECC1L-RET rearrangement can transform HMSCs, we implanted the immortalized control and RET-rearranged HMSC cells subcutaneously into immunodeficient mice. HMSC-RET cells formed palpable tumors as early as ten days after implantation and grew for more than three weeks, at which point, tumor-bearing animals were sacrificed (Fig. 1F). All implantations resulted in tumors. The control immortalized HMSCs did not form tumors (Fig. 1F). Histologic characterization of the resulting xenografts demonstrated disordered growth of undifferentiated pleomorphic cells (Fig. 1G).
We next characterized the isogenic and patient-derived RET fusion-positive cell line models by assessing expression and activation state of known effectors of growth-promoting pathways. As expected, only cells with RET fusion displayed RET protein expression (migrating in SDS-PAGE at approximately 150 kDa, consistent with a predicted translation of SPECC1L-RET transcript of 3861 bases encoding 1287 amino acids) and RET phosphorylation at two distinct sites, namely Y905 and Y1062 (Fig. 1H). Increased phosphorylation of MEK1/2 and STAT3 (S727) was also observed in both HMSC-RET and SR-Sarc-0001 cells (Fig. 1H). Interestingly, only SR-Sarc-0001 cells displayed elevated ERK1/2 phosphorylation, despite elevated phospho-MEK1/2 in both RET fusion–positive cell lines. There was no detectable increase in phosphorylation of AKT or elements of the mTOR pathway in cells with RET fusion compared with the control HMSCs.
To determine the subcellular localization of SPECC1L-RET, we performed immunofluorescence staining in the HMSCs, HMSC-RET, and SR-Sarc-0001 cells using anti-RET anti-sera (Supplementary Fig. S3). A primarily diffuse cytoplasmic signal was observed in the HMSC-RET and SR-Sarc-0001 cells, consistent with previously reported SPECC1L localization (25). Conversely, only nuclear staining was observed in the parental HMSC cells with the anti-RET antibody, presumably due to nonspecific antibody staining (no signal was observed in these cells by Western blotting analysis).
Expression of SPECC1L-RET sensitizes cells to RET inhibition
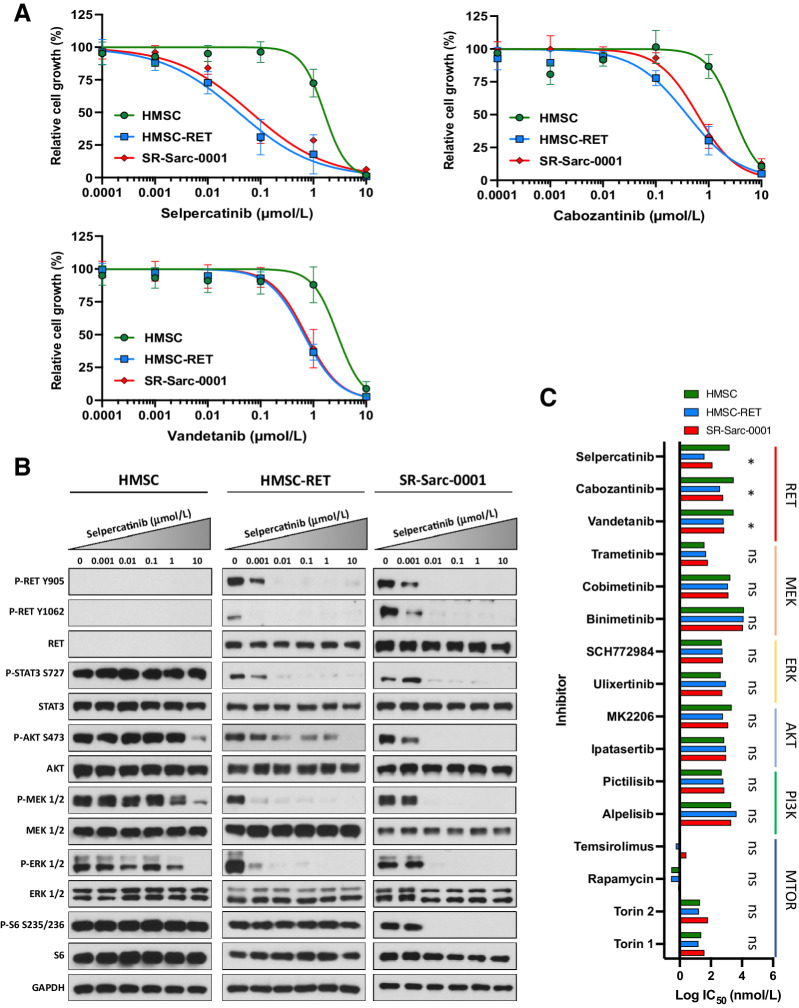
Having shown that cells with SPECC1L-RET fusion had higher levels of RET phosphorylation and activation of downstream signaling, we next sought to determine the efficacy of RET small-molecule inhibitors on growth and cell signaling in our models with RET fusion (Fig. 2A). Growth of HMSC-RET and SR-Sarc-0001 cells was inhibited by selpercatinib with IC50 values of 38 nmol/L (95% CI: 26–55 nmol/L) and 66 nmol/L (95% CI, 46–94), respectively, and at higher concentrations of cabozantinib [IC50: HMSC-RET 389 nmol/L (95% CI, 297–506); SR-Sarc-0001 617 nmol/L (95% CI: 504–754 nmol/L)] and vandetanib [IC50: HMSC-RET 662 nmol/L (95% CI, 581–748); SR-Sarc-0001 720 nmol/L (95% CI, 579–875)], demonstrating a potentially broad therapeutic window compared with HMSCs where the IC50 values for growth inhibition by selpercatinib, cabozantinib, and vandetanib were 1.6 μmol/L (95% CI, undetermined-2.1 μmol/L), 2.9 μmol/L (95% CI, 2.1–4.1 μmol/L), and 2.9 μmol/L (1.9–4.0 μmol/L), respectively. Treatment of HMSC-RET and SR-Sarc-0001 cells with selpercatinib achieved complete inhibition of RET phosphorylation (Y905 and Y1062) at 10 nmol/L and this was accompanied by loss of AKT, MEK1/2, ERK1/2, and STAT3 phosphorylation (Fig. 2B). Phosphorylation of S6 in SR-Sarc-0001 cells (inhibited with 10 nmol/L) was more sensitive to selpercatinib compared with HMSC-RET cells (inhibited at > 1 μmol/L; Fig. 2B).
Figure 2.
SPECC1L-RET–rearranged sarcoma models are sensitive to RET inhibition. A, Sensitivity of HMSC, HMSC-RET, and SR-Sarc-0001 cells to RET inhibitors was assessed using alamarBlue cell viability assay after 96-hour treatment. Data represent the mean ± SD of 6 measurements. Resulting IC50 values and 95% CIs are shown in the bottom right panel. B, Cells were treated with the indicated concentrations of selpercatinib for 90 minutes, then whole-cell extracts were prepared for Western blot profiling. C, Sensitivity of HMSC, HMSC-RET, and SR-Sarc-0001 cells to different RET, MEK, ERK, AKT, PI3K, or mTOR inhibitors was determined. IC50 values are shown as a bar plot. IC50 values and their 95% CIs of HMSC-RET and SR-Sarc-0001 cells were compared with the parental HMSCs to determine differential sensitivity. *, P < 0.05; ns, not significant.
Despite similar basal levels of phosphorylated ERK, AKT, and STAT3 in HMSC and HMSC-RET cells (implying that no additional activation was tolerated upon expression of the SPECC1L-RET oncoprotein), selpercatinib treatment resulted in reduced phosphorylation of these proteins, strongly suggesting that the RET fusion coopted them to drive proliferation. Phosphorylation of ERK, AKT, and STAT3 remained largely unaffected by selpercatinib treatment in HMSC cells (Fig. 2B). The lack of any further activation of ERK, AKT, and STAT in HMSC cells when SPECC1L-RET is expressed suggests that hyperactivation of these pathways may not be tolerated, and feedback mechanisms likely exist to keep them from being activated beyond a critical threshold.
We further aimed to establish if RET fusion–dependent growth was associated with increased sensitivity to inhibition of downstream signaling. We used small molecule inhibitors targeting MEK (trametinib, cobimetinib, binimetinib), ERK (ulixertinib, SCH772984), AKT (ipatasertib, MK2206), PI3K (alpelisib, pictilisib), and mTOR (rapamycin, temsirolimus, torin 1, torin 2). The RET fusion–positive cell lines did not show differential sensitivity to any of these agents compared to parental HMSC cells (Fig. 2C). These findings imply that inhibition of only one downstream pathway is insufficient to block growth of cells driven by a RET fusion.
Selpercatinib inhibits proliferation and induces apoptosis in RET-rearranged sarcoma models
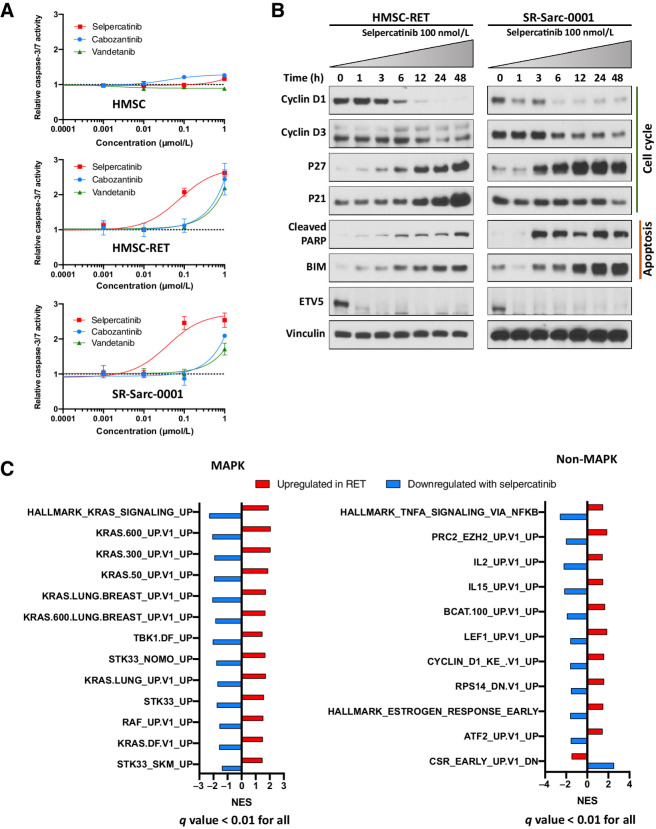
To elucidate potential mechanisms behind the reduced number of RET-rearranged cells after treatment with RET inhibitors, we investigated apoptosis by assessing caspase-3/7 enzymatic activity after exposure to the RET inhibitors selpercatinib, cabozantinib, and vandetanib. No increase in caspase-3/7 enzymatic activity was observed in the control HMSCs at doses tested (0.001– 1 μmol/L) of each drug (Fig. 3A). However, caspase-3/7 activation was observed following treatment with 1 μmol/L selpercatinib (HMSC-RET: 2.6 ± 0.07-fold, P < 0.0001; SR-Sarc-0001: 2.5 ± 0.17-fold, P < 0.0001) in HMSC-RET and SR-Sarc-0001 cells, with EC50 values for enzymatic activation overlapping with the IC50 values for growth inhibition (HMSC-RET, selpercatinib EC50 77 nmol/L, 95% CI, 47–126 nmol/L; SR-Sarc-0001 selpercatinib EC50 39 nmol/L, 95% CI, 19–79 nmol/L; Fig. 3A). Consistent with the viability assay results, selpercatinib outperformed vandetanib and cabozantinib.
Figure 3.
RET inhibition induces apoptosis and markers of cell-cycle arrest, and prominent MAPK-related transcriptomic deregulation in the RET-rearranged models. A, Cells were treated with the indicated doses of selpercatinib, cabozantinib, or vandetanib for 48 hours and then caspase-3/7 activity was determined. Each condition was assayed in triplicate determinations in two independent experiments. Results represent the mean ± SD of one representative experiment. B, Cells were treated with 100 nmol/L selpercatinib for the indicated times and then whole-cell extracts prepared for Western blotting analysis. One representative immunoblot from two independent experiments is shown for each protein. C, Transcriptomic analysis of the overlapping oncogenic and Hallmark signatures that were upregulated in HMSC-RET and SR-Sarc-0001 cells as compared with the parental HMSCs and downregulated in HMSC-RET and SR-Sarc-0001 upon treatment with selpercatinib (6 hours, 100 nmol/L) shows a prominent role for the MAPK pathway.
To better understand the signaling mechanisms that were impaired and likely resulted in loss of cell growth, we treated SR-Sarc-0001 and HMSC-RET cells with selpercatinib (100 nmol/L) for up to 48 hours and examined the level of several markers of cell cycle and apoptosis by immunoblotting. We observed downregulation of the cell cycle activators cyclin D1 and cyclin D3, and upregulation of the cell-cycle inhibitor p27 and the proapoptotic markers cleaved PARP and BIM in both cell lines (Fig. 3B). In HMSC-RET cells, induction of p21 was also detected (Fig. 3B). Finally, decreased expression of ETV5, an oncogenic transcription factor previously demonstrated to be part of RET signaling (26), was observed (Fig. 3B).
Transcriptomic profiling reveals prominent RET-dependent activation of the MAPK pathway
To investigate transcriptomic changes in our sarcoma models, we performed whole transcriptome sequencing of the HMSCs, HMSC-RET, and SR-Sarc-0001 cell lines, as well as selpercatinib-treated HMSC-RET and SR-Sarc-0001 cells (100 nmol/L, 6 hours). To rigorously detect mRNA changes associated with RET expression, we generated two orthogonal differential gene expression datasets: (i) genes differentially expressed in HMSC-RET and SR-Sarc-0001 cells compared with parental HMSC (Supplementary Table S2); (ii) genes differentially expressed in HMSC-RET and SR-Sarc-0001 cells after treatment with selpercatinib (Supplementary Table S3). Ranked lists of both datasets were submitted for Gene Set Enrichment Analysis (GSEA) using “Hallmark” and “Oncogenic signature” gene sets from the Molecular Signature Database (Supplementary Table S4 for gene sets upregulated in RET and Supplementary Table S5 for gene sets downregulated with selpercatinib treatment; ref. 27). We further overlapped the significant GSEA results (FDR q value < 0.01) from the two datasets to identify consistent changes (positively enriched in RET fusion–positive cells and negatively enriched with selpercatinib treatment or vice versa) that could be attributed to RET activity with high confidence. As a result, we identified 24 gene sets (Fig. 3C; Supplementary Table S6). A close review of these gene sets revealed that 13 of 24 are related to the MAPK pathway (e.g., KRAS- and RAF-related signatures, as well as TBK1 and STK33 signatures that were previously reported as important coactors in KRAS-dependent oncogenesis; refs. 28, 29), confirming a prominent role for this pathway in the oncogenesis and response to treatment of RET-fusion positive cells. A cyclin D1 signature was upregulated in RET fusion–positive cells and downregulated following selpercatinib treatment. Correspondingly, expression of CCND1 (which encodes cyclin D1) was upregulated in HSMC-RET and SR-Sarc-0001 cells (two-fold increase compared with parental HMSC cells, Padj < 0.001), and downregulated with selpercatinib treatment (0.23-fold, Padj < 0.0001), consistent with our results from immunoblotting analysis. No other pattern or pathway was prominently represented in this analysis.
In vivo treatment of RET-rearranged soft-tissue sarcoma models with selpercatinib induces a durable and complete response
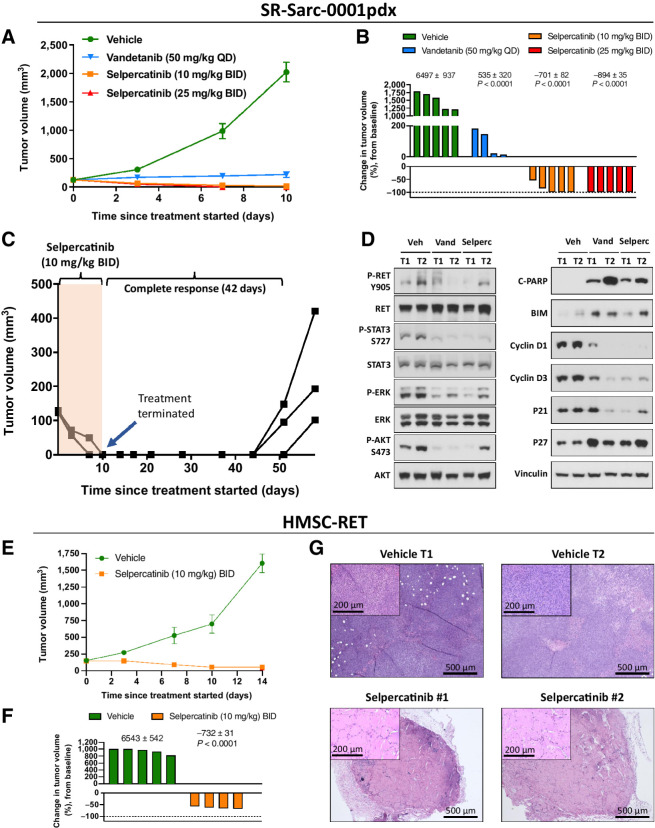
To extend our in vitro findings, we tested the effect of two RET inhibitors on growth of SR-Sarc-0001pdx tumors in vivo. The tumor volumes over the course of treatment are shown in Fig. 4A. Administration of selpercatinib resulted in no measurable disease in 3 of 5 mice with the 10 mg/kg twice daily dose and 5/5 in the 25 mg/kg twice daily group by the 10th day of treatment (Fig. 4A and B). Human equivalents of both doses are clinically achievable and well tolerated. Growth inhibition but no regression was observed in tumors treated with the RET multikinase inhibitor vandetanib. Treatment of animals bearing the three tumors that appear to regress completely in the selpercatinib 10 mg/kg twice-daily group was ceased to investigate the durability of the selpercatinib-induced responses. Remarkably, there was no palpable tumor for up to 42 days, demonstrating durability of response that persisted long after the expected drug clearance (Fig. 4C). Tumor regrowth, however, was evident after this stage.
Figure 4.
In vivo efficacy of RET inhibition in RET-rearranged sarcoma models. A, SR-Sarc-0001pdx tumors were implanted into the subcutaneous flank of NSG mice. Treatment commenced 13 days after implantation. Data represents mean ± SE of five tumors per group. While no tumor regression was observed with vandetanib, both selpercatinib 10 mg/kg twice daily and 25 mg/kg twice daily induced complete responses in most tumors. B, The percent change in tumor volume on day 10 when treatment was stopped. One tumor in the vandetanib group was too small to show up on the scale used. The numbers above the corresponding bar plots show the AUC values used to statistically compare selpercatinib and vandetanib treatments to the vehicle group. The resulting AUC values were used to calculate the P values displayed. C, Animals (three) in the selpercatinib 10 mg/kg twice daily group were observed for tumor recurrence after cessation of treatment. Complete responses were sustained for 42 days with no therapy. After this point, each line on the graph represents an individual tumor. D, Mice bearing SR-Sarc-0001pdx tumors were treated for two days with 10 mg/kg twice daily selpercatinib (three doses) or 50 mg/kg once daily vandetanib (two doses) and the tumors removed to profile cell signaling, cell cycle, and apoptosis proteins using Western blot analysis. Two tumors (T) are represented from each group. E, HMSC-RET xenograft tumors were treated with vehicle or 10 mg/kg twice daily selpercatinib 13 days after implantation. There were five animals per group. F, The percent change in tumor volume on day 10 when treatment was stopped. The numbers above the corresponding bar plots show the AUC values used to statistically compare the two conditions and calculate the P value for the 10 mg/kg twice daily selpercatinib group. G, Histology of HMSC-RET xenograft tumors isolated at the end of the study. The selpercatinib-treated tumors were composed of hypocellular stroma, consistent with necrotic scarring. BID, twice daily; QD, once daily.
To corroborate our in vitro findings, we treated animals bearing SR-Sarc-0001 PDX tumors for two days and then performed Western blot analysis to profile the activation state of signaling molecules, and expression of makers of cell cycle and apoptosis. There was a high level of correlation between in vivo and in vitro observations, namely inhibition of RET, STAT3, AKT, and ERK phosphorylation. Induction of proapoptotic markers (BIM and cleaved PARP) and cell-cycle inhibitor proteins, and downregulation of cell-cycle activators (cyclin D1 and D3) were also observed (Fig. 4D).
To support the above findings obtained in the patient-derived model, we tested the efficacy of selpercatinib on the growth of HMSC-RET xenograft tumors. Treatment with selpercatinib (10 mg/kg twice daily) achieved significant tumor control, resulting in 60%–70% reduction in tumor volume (Fig. 4E and F). After 14 days of treatment, tumors were excised to compare histology. Remarkably, while dense cellular content was observed in the vehicle-treated tumors, selpercatinib-treated tumors were composed of hypocellular stroma, which was consistent with necrotic scarring. It is thus likely that volumetric assessment underestimated the therapeutic efficacy of selpercatinib in these xenografts.
Negative MAPK pathway regulators dynamically regulate RET-induced activation and mediate restoration of MAPK activity upon treatment
The data presented above indicate that the MAPK pathway is critical in RET-induced oncogenesis and response to treatment. These results prompted us to further define the interplay between RET fusion and the MAPK pathway. First, we aimed to clarify the inconsistency between a clear dependence of ERK1/2 and MEK1/2 phosphorylation on RET kinase activity and lack of ERK1/2 hyper-phosphorylation in HMSCs when SPECC1L-RET was introduced. On the basis of our observation of increased MEK1/2 phosphorylation, we hypothesized that phosphorylation of ERK1/2 immediately downstream of MEK1/2 may be counterbalanced by ERK1/2 phosphatases, such as the dual-specificity protein phosphatases (DUSP). Indeed, inspection of the RNAseq dataset revealed overexpression of multiple members of the DUSP family such as DUSP4, DUSP5, DUSP6, DUSP9, DUSP15, DUSP10, and DUSP23 in RET fusion–positive cells (Padj < 0.05, Supplementary Table S2). Treatment with selpercatinib significantly suppressed expression of DUSP4, DUSP5, and DUSP6 in these cells (Padj < 0.05, Supplementary Table S3). The expression of DUSPs may play an important role in “titrating” MAPK activation in the setting of mutational activation of signaling: we and others have previously shown that, while DUSP6 is upregulated in KRAS-mutated lung cancers, DUSP4 is downregulated in EGFR-mutated lung cancers (30–32)
We further expanded our analysis to other negative regulators of the MAPK pathway, such as sprouty (SPRY) and sprouty-related (SPRED) proteins. SPRY1, SPRY2, SPRY4, SPRED1, SPRED2, and SPRED3 were significantly upregulated in RET fusion–positive cells (Padj < 0.05, Supplementary Table S2). At the same time, SPRY4 and SPRED3 were significantly downregulated with selpercatinib treatment (Padj < 0.05, Supplementary Table S3), demonstrating a tight dynamic regulation of MAPK pathway in these cells. We confirmed that the negative regulators of the MAPK pathway were upregulated in our RET fusion–positive models with immunoblotting for DUSP4, DUSP6, SPRY2, SPRY4, and SPRED1 (Fig. 5A).
Figure 5.
MAPK pathway is dynamically regulated in RET-rearranged cells and may impinge upon RET inhibition. A, HMSC, HMSC-RET, and SR-Sarc-0001 cells were serum-starved for 24 hours and then whole-cell extracts were prepared for Western blotting. s.e, shorter exposure; l.e., longer exposure. One representative immunoblot from two independent experiments is shown for each protein. B, Densitometry signals were quantified and normalized to corresponding loading controls (total SHP2, MEK1/2, ERK1/2, and P90RSK for corresponding phosphoproteins; GAPDH for DUSP4 and DUSP6). The resulting values were normalized to HMSC-RET. C, Cells were treated with 100 nmol/L selpercatinib for the indicated times and then whole-cell extracts prepared for Western blotting. One representative immunoblot from two independent experiments is shown for each protein. D, Densitometry signals were quantified and phosphoproteins were normalized to their corresponding totals while DUSP4 and DUSP6 were normalized to GAPDH. E, HMSC-RET cells were treated with 100 nmol/L selpercatinib or 100 nmol/L trametinib for 1 and 24 hours, and then cytoplasmic and nuclear extracts prepared for Western blotting. GAPDH represents a predominantly cytoplasmic protein control and HDAC1 represents a predominantly nuclear protein control. Selperc, selpercatinib 100 nmol/L; Tram, trametinib 100 nmol/L; CICs, short CIC isoform; CICL, long CIC isoform. F, Schematic illustration of the proposed model of RET–MAPK–CIC signaling.
Our observations suggest that RET fusions increase MAPK pathway output, which then activates mediators of a negative feedback loop, resulting in suppression of ERK phosphorylation. However, sustained expression of negative regulators in this scenario mandates that the MAPK pathway output is also maintained. To confirm this hypothesis, we immunoblotted for phospho-P90RSK, a substrate of ERK1/2. Indeed, we observed a significant increase in phospho-P90RSK in HMSC-RET cells when compared with parental HMSC cells, despite equal levels of phospho-ERK1/2 and P90RSK in the two cell lines (Fig. 5A, quantified in Fig. 5B). These findings suggest a new dynamic equilibrium of phospho-ERK1/2 is established in these cells when the activity of the MAPK pathway is increased. Of note, MEK phosphorylation was higher in SR-Sarc-0001 cells than in the HMSC-RET cells and this correlated with higher ERK phosphorylation despite the two cell lines having equivalent expression of the negative regulators of the MAPK pathway (Fig. 5A). These observations would suggest that direct phosphorylation of ERK by MEK takes precedence over downregulation by phosphatases. Nevertheless, SR-Sarc-0001 cells can still modulate the output from ERK as evidenced by the lower expression of P90RSK, which ultimately resulted in SR-Sarc-0001 and HMSC-RET having equivalent phosphorylation of P90RSK (Fig. 5A).
We next aimed to determine the consequence of MAPK pathway regulation on treatment with selpercatinib. To this end, we treated HMSC-RET and SR-Sarc-0001 cells with selpercatinib (100 nmol/L) for up to 48 hours and then examined the phosphorylation state and expression level of the MAPK pathway actors and negative regulators (Fig. 5C and D). Selpercatinib treatment induced sustained inhibition of RET and SHP2 phosphorylation. In agreement with transcriptomic findings, a decline in DUSP4 and DUSP6 (starting after 1 hour of treatment) and SPRED1 and SPRY4 (starting at 12–24 hours after treatment) protein levels were observed in our cells. This was accompanied by reactivation of MEK1/2, ERK1/2, and P90RSK. Restoration of phospho-ERK was close to the basal level (84% of basal value) following 24 hours of selpercatinib treatment in HMSC-RET cells. In SR-Sarc-0001, a significant but less prominent reactivation of ERK1/2 was also observed 24 hours after selpercatinib treatment (3.3-fold increase above the 3-hour time point (P = 0.003). To further understand the relationship between pERK and DUSP4/6, we performed shRNA-mediated knockdown of DUSP4 and DUSP6 and then examined pERK levels. shRNAs targeting DUSP4 and DUSP6 effectively reduced expression of the corresponding proteins and resulted in upregulation of pERK (Supplementary Fig. S4A and S4B). Of note, only the shRNA that caused complete loss of DUSP6 was effective at increasing pERK levels (Supplementary Fig. S4A and S4B).
To confirm that the changes in expression of the MAPK pathway regulators can be attributed to the MAPK pathway output, we treated HMSC-RET cells with a MEK inhibitor (trametinib, 100 nmol/L) and an ERK inhibitor (ulixertinib, 500 nmol/L) for 1, 12, and 24 hours. (Supplementary Fig. S5A). A rapid and profound decrease in DUSP4, DUSP6, SPRY2, SPRY4, and SPRED1 protein expression was observed following treatment with trametinib. These changes were less prominent with ulixertinib treatment, consistent with lower inhibition of phospho-P90RSK. Of note, while an increase in ERK1/2 phosphorylation was observed with ulixertinib treatment at all time points, only minor reactivation of phospho-ERK1/2 occurred at the 12 and 24 hours of trametinib treatment. This observation suggests that interplay between residual MEK activity and DUSP downregulation is responsible for ERK1/2 rebound in selpercatinib treatment.
Inhibition of RET leads to nuclear accumulation of capicua transcriptional repressor in RET fusion–positive cells
CIC is a transcriptional repressor functioning through a default repression mechanism. Unless deactivated, it is located in the nucleus and constitutively inhibits expression of multiple genes involved in RTK signaling including several that we showed above to be regulated by RET-induced MAPK signaling such as DUSP4, DUSP6, SPRY2, SPRY4, SPRED1, and others (33–35). RTKs, in turn, can deactivate CIC via the MAPK-P90RSK axis: P90RSK phosphorylates CIC, which promotes its interaction with 14–3-3 and nuclear export (36, 37). We therefore hypothesized that selpercatinib-induced downregulation of the MAPK pathway regulators is mediated by nuclear accumulation of CIC. To this end, we treated HMSC-RET cells for one or 24 hours with selpercatinib (100 nmol/L) or trametinib (100 nmol/L) to confirm participation of the MAPK pathway. We then isolated nuclear and cytoplasmic compartments to determine treatment-induced changes in subcellular localization of CIC. In agreement with our hypothesis, selpercatinib and trametinib induced a prominent increase in nuclear CIC, with a concomitant decrease in the cytoplasmic fraction (Fig. 5E, longer exposure in Supplementary Fig. S5B). Of the two known CIC isoforms, only the long isoform (CICL) showed clear accumulation in the nucleus. A proposed model is illustrated schematically in Fig. 5F.
Combination of RET and ERK inhibitors is synergistic in isogenic and patient-derived RET-rearranged sarcoma models
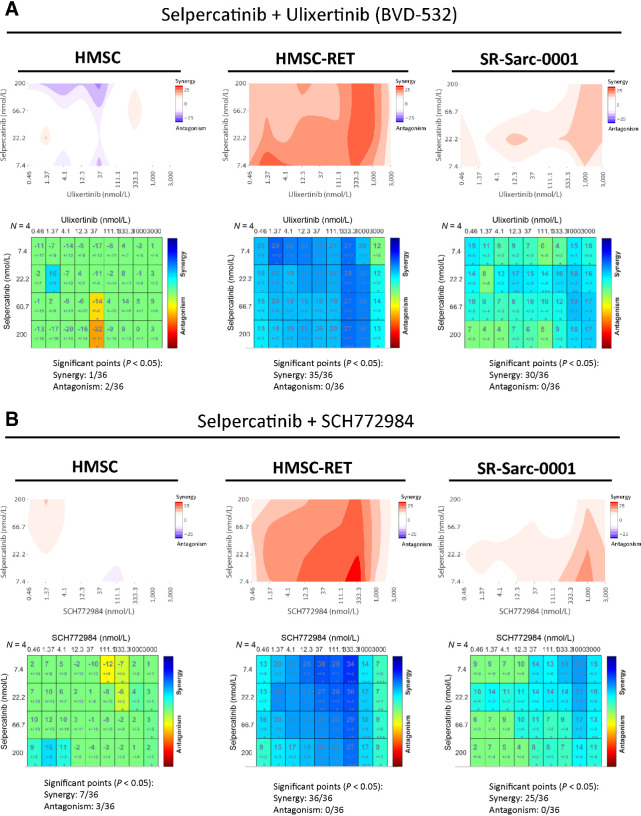
We hypothesized that reactivation of ERK in cells treated with selpercatinib results in lower therapeutic efficacy and that a combination of ERK and RET inhibitors would be synergistic. To this end, we combined the ERK inhibitors ulixertinib (38) and SCH772984 (39) with the RET inhibitor selpercatinib and applied Loewe synergy analysis (15) to the growth data. This experiment was performed in HMSC-RET and SR-Sarc-0001 cells, as well as parental HMSCs to confidently attribute our findings to the RET–MAPK interaction. Indeed, selpercatinib and ulixertinib were significantly synergistic in 35 of 36 combinations in HMSC-RET and 30/36 combinations in SR-Sarc-0001, while no prominent synergy was observed in the parental HMSC cells (1/36 synergistic, 2/36 antagonistic; Fig. 6A). Similarly, combination of selpercatinib and SCH772984 was synergistic in 36 of 36 combinations in HMSC-RET and 30/36 combinations in SR-Sarc-0001, while only 7 of 36 doses were synergistic and 3 of 36 antagonistic in HMSCs (Fig. 6B).
Figure 6.
Combination of RET and ERK inhibitors is synergistic in RET-rearranged cells. A and B, HMSC, HMSC-RET, and SR-Sarc-0001 cells were treated with the indicated combinations of selpercatinib and ulixertinib (A) or selpercatinib and SCH772984 (B) for 96 hours and then cell viability was determined. Loewe synergy analysis was applied to determine each combination synergy score and their statistical significance. Resulting scores are shown as a contour map (top) or a matrix table (bottom).
To gain some insights into possible mechanisms of how combined inhibition of ERK and RET could act synergistically, we looked at the effect of the drug combinations on expression of markers of apoptosis and proliferation. Treatment of HMSC-RET cells with a combination of selpercatinib and ulixertinib was more effective at inhibiting ERK pathway output, as measured by P90RSK phosphorylation, compared with selpercatinib alone (Supplementary Fig. S6A). Combined inhibition of RET and ERK was also more effective at inducing expression of the proapoptotic protein BIM and the cycle inhibitors p21 and p27 (Supplementary Fig. S6A). Immunoblots were quantitated and are shown in Supplementary Fig. S6B. These results suggest that the synergistic effect of combined RET and ERK inhibitors likely results from enhanced cell-cycle inhibition and apoptosis.
Combination of RET and MAPK pathway inhibitors is more effective than single agents in vivo
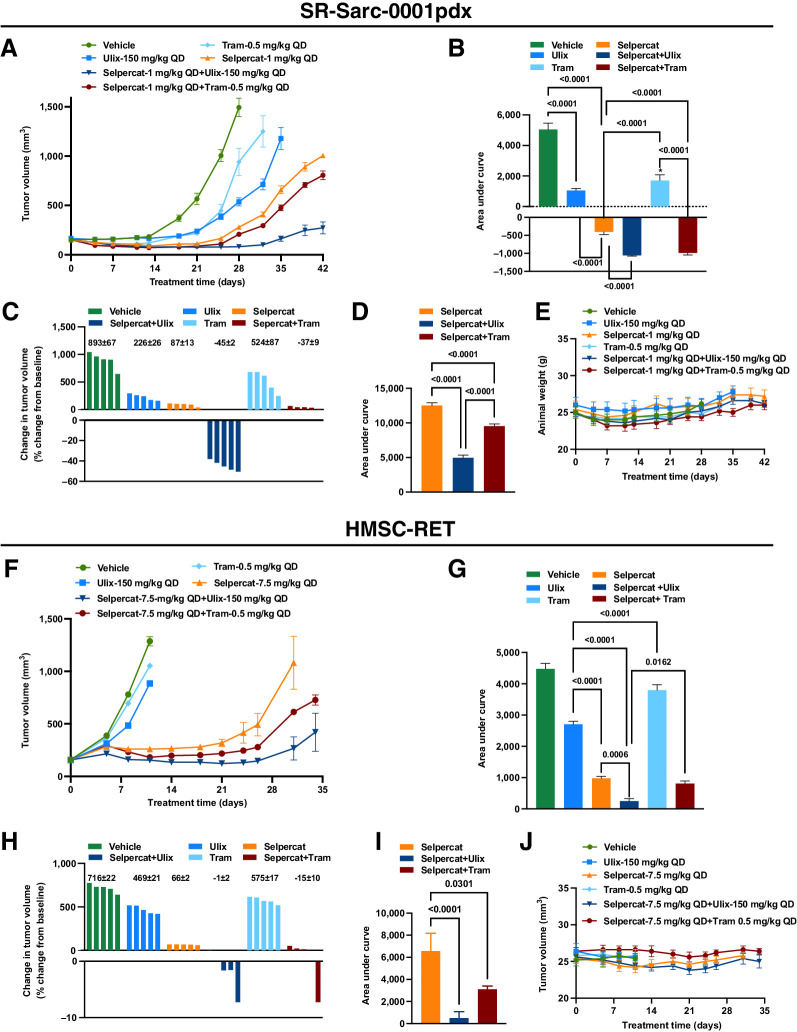
Given the synergy observed above between RET and ERK in the RET-rearranged sarcoma cell line models, we sought to examine the efficacy of combined RET and MAPK inhibitors in vivo. Animals bearing SR-Sarc-0001pdx or HMSC-RET xenograft tumors were treated with vehicle, selpercatinib, ulixertinib, or trametinib as single agents or in combination (Fig. 7). Inhibitor doses that would give less than maximal effect as single agents were chosen for these studies. The time course of treatment of the SR-Sarc-0001pdx model is given in (Fig. 7A). AUC analysis was performed on data up to day 28 (last day all animals were alive) and is illustrated in Fig. 7B, respectively. Treatment with all single agents caused a significant reduction in growth of SR-Sarc-0001pdx tumors (Fig. 7A), with selpercatinib administration resulting in the greatest reduction in tumor growth ((Fig. 7B). Combined inhibition of RET and ERK, or RET and MEK resulted in significant tumor shrinkage, which was superior to the response seen with any of the single agents (Fig. 7B). At the doses used, the combination of ulixertinib and selpercatinib was the most effective treatment, with all tumors in the group shrinking by >40% (Fig. 7C). At the end of the study, the two sets of combinations were still more effective than selpercatinib treatment alone (Fig. 7D). None of the single agents or combinations had any adverse effect on animal health or animal weight (Fig. 7E). These treatments and combinations were also evaluated in the HMSC-RET xenograft model, yielding qualitatively equivalent results (Fig. 7F–J) with the combination of ulixertinib and selpercatinib again the most active treatment strategy (Fig. 7H).
Figure 7.
Combine targeting of RET and MEK or ERK is more effective at inhibiting tumor growth than single agents. Mice bearing SR-Sarc-0001pdx or HMSC-RET xenograft tumors were treated with vehicle, selpercatinib (Selpercat), ulixertinib (Ulix), trametinib (Tram), or the indicated combinations. A, Volume of SR-Sarc-0001pdx tumors. Treatment began 13 days after implantation (day 0 on graph). B, AUC analysis of data up to day 28 of treatment (last day that all animals were alive). The mean tumor volume of each treatment group was significantly different from the vehicle group (P < 0.0001). C, AUC analysis of data up to day 42 of treatment (last measurement). D, Percent change in volume of individual tumors on day 28. The mean ± SEM values are shown above the bars. All data represent the mean tumor volume ± SEM of five tumors. E, Animal weight. No treatment caused a statistically significant change in animal weight or other aspects of health. F, Volume of HMSC-RET xenograft tumors. Treatment began nine days after implantation (day 0 on graph). G, AUC analysis of data up to day 11 of treatment (last day that all animals were alive). Tumor volume of all treatment groups was significantly different from the vehicle group (P < 0.0001). H, AUC analysis of data up to day 31 of treatment (last day all animals were alive in the three groups). I, Percent change in volume of individual tumors on day 11. The mean ± SEM values are shown above the bars. All data represent the mean tumor volume ± SEM of five tumors. J, Animal weight. No treatment caused a statistically significant change in animal weight or other aspects of health. BID, twice daily; QD, once daily.
NTRK3-rearranged sarcoma displays dynamic modulation of the MAPK pathway, providing a rationale for coinhibition of NTRK and ERK
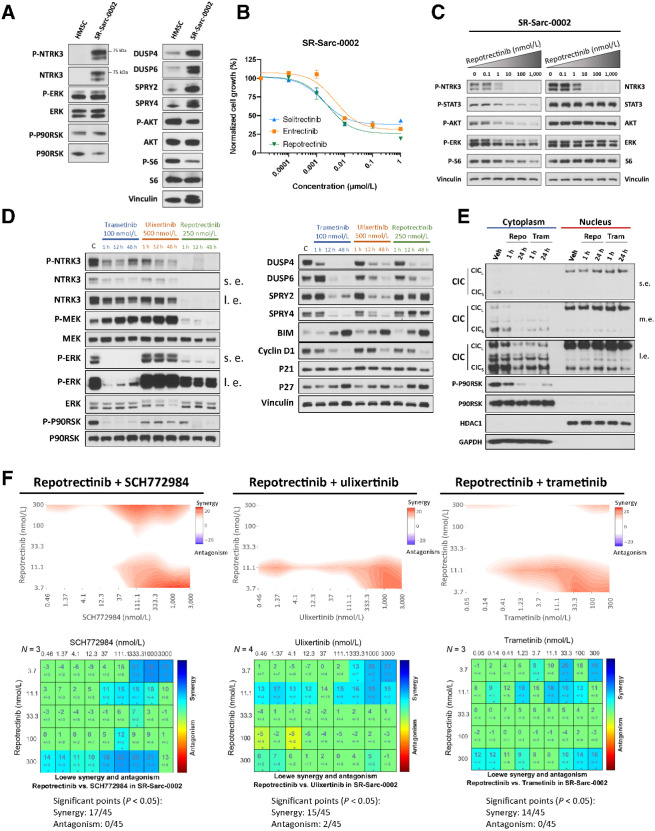
Finally, we wished to examine if other RTK fusion–driven sarcomas show a similar pattern of MAPK pathway activation and feedback regulation. NTRK fusions are also known oncogenic drivers in some sarcomas, with ETV6-NTRK3 being the most common RTK fusion occurring in infantile fibrosarcoma (IFS; ref. 40). Given the application of NTRK kinase inhibitors to treat sarcoma patients (5), we investigated whether feedback inhibition of the MAPK pathway may also act to dampen the therapeutic efficacy of NTRK kinase inhibitors as it does for RET inhibitors. To assess this, we used a patient-derived ETV6-NTRK3-rearranged IFS model (SR-Sarc-0002, immunoblotting characterization in Fig. 8A). Consistent with our findings in the RET models, SR-Sarc-0002 cells displayed overexpression of multiple negative regulators of the MAPK pathway, such as DUSP4, DUSP6, SPRY2, and SPRY4 compared with HMSCs (Fig. 8A). As observed in the RET-driven models, phospho-ERK levels in SR-Sarc-0002 cells were comparable with HMSCs, despite elevated levels of pNTRK3 (Fig. 8A).
Figure 8.
ETV6-NTRK3–rearranged sarcoma model confirms a broad role of MAPK regulation in RTK oncogenesis and response to therapy. A, HMSC and SR-Sarc-0002 (ETV6-NTRK3F617I–rearranged infantile fibrosarcoma cell line) cells were serum-starved for 24 hours and then whole-cell extracts were prepared for Western blot profiling. B, SR-Sarc-0002 cell were treated with the indicated doses of repotrectinib (TPX-0005), entrectinib, and selitrectinib (LOXO-195) for 96 hours and then cell viability was assessed using alamarBlue. Data represent the mean ± SD of three measurements. C, SR-Sarc-0002 cells were treated with the indicated doses of repotrectinib for 90 minutes and then whole-cell extracts were prepared for Western blot profiling. D, SR-Sarc-0002 cells were treated with trametinib (100 nmol/L), ulixertinib (500 nmol/L), or repotrectinib (250 nmol/L) for the indicated times and then whole-cell extracts prepared for Western blotting. s.e., shorter exposure; l.e., longer exposure. E, SR-Sarc-0002 cells were treated with repotrectinib (250 nmol/L) or trametinib (100 nmol/L) for 1 and 24 hours and nuclear and cytoplasmic extracts were prepared for Western blotting. Repo, repotrectinib 250 nmol/L; Tram, trametinib 100 nmol/L; m.e., medium exposure. F, SR-Sarc-0002 cells were treated with the indicated combinations of repotrectinib and SCH772984 (left), repotrectinib and ulixertinib (middle), and repotrectinib and trametinib (right) for 96 hours and then cell viability was determined. Loewe synergy analysis was applied to determine the synergy scores and their statistical significance for each combination. Resulting scores are shown as a contour map (top) or a matrix table (bottom). Representative immunoblots from two independent experiments are shown in A, C, D, E.
We next determined sensitivity of this model to different NTRK inhibitors [entrectinib, TPX-0005 (repotrectinib), LOXO-195 (selitrectinib); Fig. 8B]. All inhibitors demonstrated comparable efficiency with IC50 in the low nanomolar range. Treatment of SR-Sarc-0002 cells with repotrectinib resulted in inhibition of NTRK3, ERK1/2, AKT, STAT3, and S6 phosphorylation in a dose-dependent manner (Fig. 8C). We further treated cells with trametinib, ulixertinib, and repotrectinib for one, 12, and 48 hours and examined the phosphorylation state of MAPK pathway effectors (Fig. 8D). Consistent with our findings in the RET-rearranged models, every inhibitor induced rapid suppression of DUSP4 and DUSP6. Interestingly, expression of SPRY2 and SPRY4 was not prominently altered by repotrectinib, whereas in the same experiment, trametinib and ulixertinib showed strong suppression of SPRY2 and SPRY4 levels. All inhibitors increased the levels of BIM and p27 and decreased expression of cyclin D1.
To determine whether inhibition of NTRK3, or the MAPK pathway directly, may affect nuclear traffic of CIC in this model, we isolated nuclear and cytoplasmic fractions after treatment with repotrectinib (250 nmol/L) and trametinib (100 nmol/L) for 1 and 24 hours (Fig. 8E). Although there was no significant enrichment of nuclear CIC at 1 hour of repotrectinib treatment, increased nuclear:cytoplasmic ratio was observed at 24 hours of repotrectinib and at 1 and 24 hours of trametinib treatment (Supplementary Fig. S7A). The less prominent inhibition of P90RSK phosphorylation observed at one hour of repotrectinib treatment may be responsible for the delay in nuclear CIC accumulation (Fig. 8E; Supplementary Fig. S7B).
We further combined repotrectinib with ulixertinib, SCH772984, or trametinib and assessed the effects of these drug combinations on cell growth. These combinations demonstrated significant synergy with 17 of 45 significantly synergistic combinations in SCH772984, 15 of 45 in ulixertinib, and 14 of 45 in trametinib (Fig. 8F), albeit less so than in the RET-rearranged models.
Discussion
Here, we report the characterization of patient-derived and isogenic in vitro and in vivo models of RET fusion–driven soft-tissue sarcoma, to our knowledge the first such sarcoma models. While the patient-derived models closely recapitulate the characteristics of patient tumors, engineered isogenic models allow for comparative studies to provide novel insights into receptor tyrosine kinase (RTK)-driven sarcomagenesis. In addition, isogenic models can be invaluable in cases of rare entities, where a paucity of models hampers advancement in therapy development or understanding tumor biology. We employed immortalized untransformed HMSCs, which are considered a rational choice to approximate the putative tumor-initiating cells of soft-tissue sarcomas (41, 42) to demonstrate how these cells may be exploited to investigate sarcoma biology and therapy development, using RET fusion as proof-of-concept. Importantly, we opted for a lentiviral-free, CRISPR-Cas9–induced system to introduce the RET chromosomal translocation to more closely reproduce naturally occurring tumorigenesis since the endogenous alleles were altered, ensuring appropriate physiological oncogene expression level. Specifically, CRISPR-Cas9–generated fusions retain their native promoter, genomic location, and required splicing so that their regulation is more representative of the naturally occurring gene fusions driving human malignancies. Alternative methods, such as cDNA overexpression, while easier to execute, come at the cost of an ectopic genomic placement of a sole transcript isoform constitutively expressed from a strong promoter, often at supraphysiologic levels.
Introduction of the RET fusion induced anchorage-independent growth in HMSCs, thereby confirming its oncogenicity in vitro. In vivo, subcutaneous implantation of HMSC-RET, but not HMSC, leads to rapid and sustained growth in mice. The resulting HMSC-RET xenograft tumors resemble undifferentiated pleomorphic sarcomas. HMSC-RET models demonstrated highly consistent findings when compared with the patient-derived models. Growth of HMSC-RET and SR-Sarc-0001 cells was inhibited by RET inhibitors with similar IC50 values. The EC50 values of RET inhibitors for apoptosis induction were also similar in the two cell lines. Mechanistically, similar signaling pathways and markers of cell cycle and apoptosis were activated or inhibited when cells were treated with the RET inhibitor selpercatinib, and these changes followed coherent temporal dynamics. In vivo, significant reductions in tumor volume were achieved with selpercatinib in both models, with no measurable disease in most animals implanted with PDXs, and small fibrotic residues in HMSC-RET xenografts. These findings underscore the reliability of our approach, especially for studies evaluating oncogenic pathways and preclinical drug development.
One limitation of our study is a lack of a more comprehensive genomic comparison of the HMSC-RET models with the patient-derived SR-Sarc-0001 models. Although these studies have the potential to more clearly define the molecular properties of the two sets of disease models and perhaps provide a better understanding of the cell of origin of RET-mutated sarcomas, they are beyond the scope of the work described here. In general, for primitive sarcomas of uncertain lineage, we believe HMSCs represent a reasonable choice as the starting cell. The oncogenicity of chimeric transcription factors has shown considerably more cellular context-specificity than that of kinase fusions, which can transform a much broader spectrum of “cells-of-origin” (including many epithelial cells). Another limitation of our study is the use of only one clone of HMSC-RET cells and patient-derived models established from only one patient. However, due to the rarity of RET fusion–driven sarcomas, there are very few opportunities to obtain patient tumors to generate more patient-derived models. As for the CRISPR models, generation of the fusion events using CRISPR is extremely low-yield, and we have only managed to generate one clone. Nevertheless, most experiments conducted in the HMSC-RET model orthogonally confirmed the findings in the patient-derived model.
The small-molecule RET-specific inhibitor selpercatinib has been approved in patients with non–small cell lung cancer (NSCLC) and thyroid carcinomas, with 85% and 79% overall response rates (ORR) observed, respectively (23, 43). More recently, another small-molecule RET-specific inhibitor pralsetinib (BLU-667; ref. 44) was approved for NSCLC, with a 70% ORR. However, due to their rarity, little is known about the responses of RET fusion–positive tumors of other histologies to molecularly targeted drugs. Given the successful results of treating NTRK fusion–positive cancers (another RTK fusion occurring in tumors of multiple histologies), demonstration of RET inhibitors efficacy outside of the lung and thyroid tumor spectrum could effectively expand the number of patients who may benefit from this therapy in a tumor-type agnostic manner. To this end, our preclinical findings strongly support the clinical application of RET inhibitors in soft-tissue sarcomas with RET fusions.
Despite encouraging efficacy of RET inhibitors, the emergence of therapy resistance is the most common reason for discontinuation of the targeted agents, limiting clinical benefit. Broadly, resistance mechanisms can be divided into two groups: (i) “on-target” resistance, caused by a mutation in the target protein, hindering binding of an inhibitor; (ii) “off-target” resistance, commonly caused by activation of alternate signaling pathways, bypassing the original oncogenic pathway. The types of prevalent bypass mechanisms are often dependent on the primary driver [e.g., MET amplification in EGFR-mutated lung adenocarcinoma treated with osimertinib (45) and activating KRAS mutations in MET exon 14 skipping-mutated lung adenocarcinoma (46, 47)]. A body of work has shown that oncogenic dependency on, or “addiction” to, a given signaling axis is partly manifested by the emergence of multiple mechanisms to regain its activity when the primary driver is inhibited (48).
Here, we studied the role of the MAPK pathway in driving growth and modulating response to therapy in novel RET fusion–driven sarcoma models developed for this study. We extended our observations to a novel NTRK3-rearranged patient-derived model that was also developed for this study. Transcriptomic profiling of RET fusion–positive cells identified prominent expression of multiple MAPK pathway signatures, which were downregulated in cells treated with selpercatinib. Inhibition of RET or MEK leads to nuclear accumulation of the transcriptional repressor CIC. CIC is an HMG-box transcriptional repressor that exists in a complex with ATXN1 in the nucleus and regulates cell growth by controlling MAPK pathway signaling (49). Thus, CIC functions as a tumor suppressor and loss-of-function mutations have been identified in oligodendroglioma (50). CIC controls expression of the ERK phosphatases DUSP4 and DUSP6, as well as SPRY and SPRED family members, which are suppressors of MAPK signaling upstream of ERK (33–35). We observed that following RET or NTRK inhibition, CIC accumulates in the nucleus concomitantly with a loss of expression of several negative regulators of the MAPK pathway. In parallel, reactivation of MEK and ERK was detected, likely as a consequence of the loss of the negative regulators. These findings support a model whereby transformation by RET and NTRK is accompanied by tight regulation of MAPK signaling through changes in expression of negative regulators. Cells adapt to RET inhibition by downregulating the negative regulators of the MAPK pathway, thereby providing partial restoration of ERK signaling that is no longer dependent on the kinase fusion, and this may blunt response to therapy. Consistent with these findings, combination of RET or NTRK inhibitors with ERK or MEK inhibitors was synergistic in the in vitro models. Importantly, in the two RET fusion–driven xenograft models, combination of selpercatinib with the ERK inhibitor ulixertinib or the MEK inhibitor trametinib was more effective at reducing tumor growth and prolonging the therapeutic response compared with TKI monotherapy. These observations provide a preclinical rationale for the clinical evaluation of these TKI combinations in treating kinase fusion–driven sarcomas.
Authors' Disclosures
S. Kohsaka reports grants from Boehringer-ingelheim, Astrazeneca, Eisai, and grants from Chordia Therapeutics outside the submitted work. J.L. Glade Bender reports other support from Loxo-Oncology, other support from Lilly, and non-financial support from Helsinn during the conduct of the study; other support from Roche, Merck, Amgen, Bristol Meyers Squibb, Eisai, Novartis, Celgene, Ignyta, and other support from Bayer outside the submitted work; and serves as an uncompensated consultant for Springworks, Merck and Pfizer (DSMC) and BMS and Eisai (pediatric advisory boards). M. Ladanyi reports grants from LOXO oncology during the conduct of the study and personal fees from LOXO Oncology outside the submitted work. R. Somwar reports grants and non-financial support from Loxo Oncology during the conduct of the study; grants and non-financial support from Helsinn Health Care, Merus, and grants and non-financial support from Elevation Oncology outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank Drs. William Lockwood (University of British Columbia) and Monika Davare (Oregon Health and Science University) for valuable insights during this study. This work was supported by research grants from Cycle for Survival (to M. Ladanyi), LOXO Oncology (to M. Ladanyi and R. Somwar), and grants from the NIH (to E. de Stanchina; U54 0D020355), and a Cancer Center Support grant to Memorial Sloan Kettering Cancer Center (P30 CA008748).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
I. Odintsov: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M.V. Ortiz: Data curation, investigation, writing–review and editing. I. Khodos: Investigation. M.S. Mattar: Resources, formal analysis, investigation, writing–review and editing. A.J.W. Lui: Data curation, formal analysis, writing–review and editing. S. Kohsaka: Resources, investigation. E. de Stanchina: Resources, funding acquisition, project administration, writing–review and editing. J.L.G. Bender: Supervision, writing–original draft, project administration. M. Ladanyi: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. R. Somwar: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Sbaraglia M, Bellan E, Dei Tos AP. The 2020 WHO classification of soft tissue tumours: news and perspectives. Pathologica 2020;113:70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Board WCoTE. WHO classification of tumors: Soft tisse and bone tumors. 2020.
- 3. Knott MML, Holting TLB, Ohmura S, Kirchner T, Cidre-Aranaz F, Grunewald TGP. Targeting the undruggable: exploiting neomorphic features of fusion oncoproteins in childhood sarcomas for innovative therapies. Cancer Metastasis Rev 2019;38:625–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joensuu H, Roberts PJ, Sarlomo-Rikala M, Andersson LC, Tervahartiala P, Tuveson D, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med 2001;344:1052–6. [DOI] [PubMed] [Google Scholar]
- 5. Doebele RC, Davis LE, Vaishnavi A, Le AT, Estrada-Bernal A, Keysar S, et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov 2015;5:1049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, et al. Activity of Selumetinib in Neurofibromatosis type 1-related plexiform neurofibromas. N Engl J Med 2016;375:2550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spraggon L, Martelotto LG, Hmeljak J, Hitchman TD, Wang J, Wang L, et al. Generation of conditional oncogenic chromosomal translocations using CRISPR-Cas9 genomic editing and homology-directed repair. J Pathol 2017;242:102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun 2014;5:3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vanoli F, Tomishima M, Feng W, Lamribet K, Babin L, Brunet E, et al. CRISPR-Cas9-guided oncogenic chromosomal translocations with conditional fusion protein expression in human mesenchymal cells. Proc Natl Acad Sci U S A 2017;114:3696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Antonescu CR, Dickson BC, Swanson D, Zhang L, Sung YS, Kao YC, et al. Spindle cell tumors with RET gene fusions exhibit a morphologic spectrum Akin to tumors with NTRK gene fusions. Am J Surg Pathol 2019;43:1384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis JL, Vargas SO, Rudzinski ER, Lopez Marti JM, Janeway K, Forrest S, et al. Recurrent RET gene fusions in paediatric spindle mesenchymal neoplasms. Histopathology 2020;76:1032–41. [DOI] [PubMed] [Google Scholar]
- 12. Ortiz MV, Gerdemann U, Raju SG, Henry D, Smith S, Rothenberg SM, et al. Activity of the highly specific RET inhibitor selpercatinib (LOXO-292) in pediatric patients with tumors harboring RET gene alterations. JCO Precis Oncol 2020;4:PO.19.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Widmann J, Stombaugh J, McDonald D, Chocholousova J, Gardner P, Iyer MK, et al. RNASTAR: An RNA STructural alignment repository that provides insight into the evolution of natural and artificial RNAs. RNA 2012;18:1319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Somwar R, Shum D, Djaballah H, Varmus H. Identification and preliminary characterization of novel small molecules that inhibit growth of human lung adenocarcinoma cells. J Biomol Screen 2009;14:1176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32:2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Somwar R, Kim DY, Sweeney G, Huang C, Niu W, Lador C, et al. GLUT4 translocation precedes the stimulation of glucose uptake by insulin in muscle cells: potential activation of GLUT4 via p38 mitogen-activated protein kinase. Biochem J 2001;359:639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li GG, Somwar R, Joseph J, Smith RS, Hayashi T, Martin L, et al. Antitumor activity of RXDX-105 in multiple cancer types with RET rearrangements or mutations. Clin Cancer Res 2017;23:2981–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee SB, Kolquist KA, Nichols K, Englert C, Maheswaran S, Ladanyi M, et al. The EWS-WT1 translocation product induces PDGFA in desmoplastic small round-cell tumour. Nat Genet 1997;17:309–13. [DOI] [PubMed] [Google Scholar]
- 19. Banito A, Li X, Laporte AN, Roe JS, Sanchez-Vega F, Huang CH, et al. The SS18-SSX oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell 2018;33:527–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kobos R, Nagai M, Tsuda M, Merl MY, Saito T, Lae M, et al. Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein. J Pathol 2013;229:743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedenstein AJ, Gorskaja JF, Kulagina NN. Fibroblast precursors in normal and irradiated mouse hematopoietic organs. Exp Hematol 1976;4:267–74. [PubMed] [Google Scholar]
- 22. Ankrum JA, Ong JF, Karp JM. Mesenchymal stem cells: immune evasive, not immune privileged. Nat Biotechnol 2014;32:252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drilon A, Oxnard GR, Tan DSW, Loong HHF, Johnson M, Gainor J, et al. Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. N Engl J Med 2020;383:813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol 2004;5:816–26. [DOI] [PubMed] [Google Scholar]
- 25. Saadi I, Alkuraya FS, Gisselbrecht SS, Goessling W, Cavallesco R, Turbe-Doan A, et al. Deficiency of the cytoskeletal protein SPECC1L leads to oblique facial clefting. Am J Hum Genet 2011;89:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, et al. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet 2009;41:1295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 2009;137:821–34. [DOI] [PubMed] [Google Scholar]
- 29. Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chitale D, Gong Y, Taylor BS, Broderick S, Brennan C, Somwar R, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene 2009;28:2773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Unni AM, Harbourne B, Oh MH, Wild S, Ferrarone JR, Lockwood WW, et al. Hyperactivation of ERK by multiple mechanisms is toxic to RTK-RAS mutation-driven lung adenocarcinoma cells. Elife 2018;7:e33718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A 2009;106:4519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weissmann S, Cloos PA, Sidoli S, Jensen ON, Pollard S, Helin K. The tumor suppressor CIC directly regulates MAPK pathway genes via histone deacetylation. Cancer Res 2018;78:4114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. LeBlanc VG, Firme M, Song J, Chan SY, Lee MH, Yip S, et al. Comparative transcriptome analysis of isogenic cell line models and primary cancers links capicua (CIC) loss to activation of the MAPK signalling cascade. J Pathol 2017;242:206–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okimoto RA, Breitenbuecher F, Olivas VR, Wu W, Gini B, Hofree M, et al. Inactivation of Capicua drives cancer metastasis. Nat Genet 2017;49:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dissanayake K, Toth R, Blakey J, Olsson O, Campbell DG, Prescott AR, et al. ERK/p90(RSK)/14–3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicua. Biochem J 2011;433:515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ren Y, Ouyang Z, Hou Z, Yan Y, Zhi Z, Shi M, et al. CIC is a mediator of the ERK1/2-DUSP6 negative feedback loop. iScience 2020;23:101635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sullivan RJ, Infante JR, Janku F, Wong DJL, Sosman JA, Keedy V, et al. First-in-class ERK1/2 inhibitor Ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: Results of a phase I dose-escalation and expansion study. Cancer Discov 2018;8:184–95. [DOI] [PubMed] [Google Scholar]
- 39. Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 2013;3:742–50. [DOI] [PubMed] [Google Scholar]
- 40. Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet 1998;18:184–7. [DOI] [PubMed] [Google Scholar]
- 41. Tolar J, Nauta AJ, Osborn MJ, Panoskaltsis Mortari A, McElmurry RT, Bell S, et al. Sarcoma derived from cultured mesenchymal stem cells. Stem Cells 2007;25:371–9. [DOI] [PubMed] [Google Scholar]
- 42. Mohseny AB, Hogendoorn PC. Concise review: mesenchymal tumors: when stem cells go mad. Stem Cells 2011;29:397–403. [DOI] [PubMed] [Google Scholar]
- 43. Wirth LJ, Sherman E, Robinson B, Solomon B, Kang H, Lorch J, et al. Efficacy of selpercatinib in RET-altered thyroid cancers. N Engl J Med 2020;383:825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Subbiah V, Gainor JF, Rahal R, Brubaker JD, Kim JL, Maynard M, et al. Precision targeted therapy with BLU-667 for RET-driven cancers. Cancer Discov 2018;8:836–49. [DOI] [PubMed] [Google Scholar]
- 45. Schoenfeld AJ, Chan JM, Kubota D, Sato H, Rizvi H, Daneshbod Y, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 2020;26:2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suzawa K, Offin M, Lu D, Kurzatkowski C, Vojnic M, Smith RS, et al. Activation of KRAS mediates resistance to targeted therapy in MET Exon 14-mutant non-small cell lung cancer. Clin Cancer Res 2019;25:1248–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bahcall M, Awad MM, Sholl LM, Wilson FH, Xu M, Wang S, et al. Amplification of wild-type KRAS imparts resistance to crizotinib in MET Exon 14 mutant non-small cell lung cancer. Clin Cancer Res 2018;24:5963–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction–a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol 2006;3:448–57. [DOI] [PubMed] [Google Scholar]
- 49. Tanaka M, Yoshimoto T, Nakamura T. A double-edged sword: the world according to Capicua in cancer. Cancer Sci 2017;108:2319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011;333:1453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Reagents created by our laboratory and used in this study can be obtained by contacting the corresponding author directly.