Graphical abstract
Keywords: SARS-CoV-2 variant coinfection, Viral transmission simulation, Coinfection index, Heterozygous single-nucleotide polymorphisms
Abstract
SARS-CoV-2 is a single-stranded RNA betacoronavirus with a high mutation rate. The rapidly emerging SARS-CoV-2 variants could increase transmissibility and diminish vaccine protection. However, whether coinfection with multiple SARS-CoV-2 variants exists remains controversial. This study collected 12,986 and 4,113 SARS-CoV-2 genomes from the GISAID database on May 11, 2020 (GISAID20May11), and Apr 1, 2021 (GISAID21Apr1), respectively. With single-nucleotide variant (SNV) and network clique analyses, we constructed single-nucleotide polymorphism (SNP) coexistence networks and discovered maximal SNP cliques of sizes 16 and 34 in the GISAID20May11 and GISAID21Apr1 datasets, respectively. Simulating the transmission routes and SNV accumulations, we discovered a linear relationship between the size of the maximal clique and the number of coinfected variants. We deduced that the COVID-19 cases in GISAID20May11 and GISAID21Apr1 were coinfections with 3.20 and 3.42 variants on average, respectively. Additionally, we performed Nanopore sequencing on 42 COVID-19 patients and discovered recurrent heterozygous SNPs in twenty of the patients, including loci 8,782 and 28,144, which were crucial for SARS-CoV-2 lineage divergence. In conclusion, our findings reported SARS-CoV-2 variants coinfection in COVID-19 patients and demonstrated the increasing number of coinfected variants.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped and single-stranded RNA betacoronavirus of 29.9 k bases that belongs to the family Coronaviridae [1], [2]. Since 2000, we have witnessed and experienced two other outbreaks of highly widespread pathogenic coronaviruses in human populations: severe acute respiratory syndrome (SARS)-CoV in 2002–2003 and Middle East Respiratory Syndrome (MERS)-CoV in 2012 [3]. All three viruses can lead to acute respiratory distress syndrome (ARDS) in human hosts, which may cause pulmonary fibrosis and lead to permanent lung function reduction or death [4]. Although SARS-CoV and MERS-CoV have higher mortality rates than SARS-CoV-2, SARS-CoV-2 can invade a wide variety of host cells and cause rapid spread among people [5].
To address these challenges, researchers have conducted various studies to explore the genomic sequences of SARS-CoV-2 [6], [7], [8]. Qianqian Li et al. analysed 13,406 spike sequences of SARS-COV-2 variants in the GISAID database and divided the variants into seven evolutionary groups using neutralizing monoclonal antibodies [6]. Correspondingly, the Centers for Disease Control and Prevention also reported newly emerged SARS-CoV-2 variants that circulate globally, including the B.1.1.7 lineage in the United Kingdom, B.1.351 lineage in Nelson Mandela Bay and South Africa, P.1 lineage in Japan and Brazil, B.1.429 lineage in the United States, etc. [9]. From Pengfei Wang et al.’s study, we learned that extensive mutations in the spike protein of B.1.1.7 and B.1.351 variants could enhance their resistance to neutralization by convalescent and postvaccination sera [10]. These reports enforce the notion that the newly emerged SARS-CoV-2 variants would increase viral transmissibility and disease severity and reduce the protective ability of vaccines [10], [11], [12].
In addition to the rapidly emerging SARS-CoV-2 variants, previous studies also reported coinfection with SARS-CoV-2 and other respiratory pathogens [13], [14]. In their study, David Kim and his colleagues found that 116 COVID-19 patients were also positive for other microbial pathogens, such as influenza A/B, respiratory syncytial virus, human metapneumovirus, and Chlamydia pneumoniae [13]. Additionally, reinfection with different SARS-CoV-2 variants in a COVID-19 patient has been reported. Richard L Tillett et al. presented a COVID-19 patient who tested positive for SARS-CoV-2 in Apr 2020 and was reinfected by a different SARS-CoV-2 variant in June 2020 [15]. This was an astonishing discovery, and it was hard to explain why previous exposure to SARS-CoV-2 failed to provide immunity protection to the patient. Since coinfection is prevalent in viral infections [16], [17], [18], these studies have inspired us to explore whether coinfection with multiple SARS-CoV-2 variants exists in COVID-19 patients, providing clues for prolonged viral shedding time and severe symptoms [19].
To detect whether coinfection with multiple SARS-CoV-2 variants exists, we adopted graph theory in this study, and each clique represents a subnetwork composed of vertices and a set of edges. In previous reports, researchers normally focused on the maximal clique in a dataset to investigate the original network features, such as biomedical structure [20], [21], [22], protein–protein interactions [23], [24], [25], and disease-related gene detection [26], [27]. Moreover, single-nucleotide polymorphisms (SNPs) that construct cliques can be applied for bacterial horizontal gene cotransfer detection [28], covariate gene expression determination [29], and SNP-based bacterial genome recombination identification [30]. Hence, we planned to investigate the features of the maximal SNP cliques for the datasets and detect whether coinfection with multiple SARS-CoV-2 variants exists.
We collected 12,986 SARS-CoV-2 genomic sequences from the GISAID database on May 11, 2020, and noted them as GISAID20May11. After constructing the SNP coexistence network, we discovered that the maximal SNP clique was size 16. We proved that it is possible to achieve such a large SNP clique with coinfection. By simulating the transmission route and SNP accumulation in variant genomes, we discovered that coinfection with variants provided explanations of such a large clique and revealed a significant linear relationship between the size of the maximal clique and the average number of coinfected variants. According to the linear relationship obtained by the simulation, we discovered 3.20 averaged coinfected variants in the COVID-19 patients from the GISAID20May11 dataset. To validate the methods and results, we extracted 4,113 additional genomes from the GISAID database on Apr 1, 2021 (GISAID21Apr1) and discovered an increased coinfected variant number of 3.42. Then, we performed Nanopore sequencing on sputum samples from 42 COVID-19 patients and found recurrent heterozygous SNPs on some loci of the SARS-CoV-2 genome. In particular, loci 8,782 and 28,144, which are crucial for phylogenetic divergence, proved multiple variant coinfection. Hence, our study proposed a computational simulation method to detect the number of coinfected variants in COVID-19 patients and confirmed coinfection with multiple SARS-CoV-2 variants, suggesting an increase in coinfections with multiple variants throughout the epidemic.
2. Materials and methods
2.1. GISAID datasets and mutation detection
This study collected SARS-CoV-2 genomic sequences from the GISAID database (https://www.gisaid.org/) and divided them into two genomic datasets according to their releasing date: for the 12,986 SARS-CoV-2 genomic sequences published before May 11, 2020, we noted them as GISAID20May11 dataset; for the 4,113 SARS-CoV-2 genomic sequences posted on Apr 1, 2021, we noted them as GISAID21Apr1 dataset. All genomes in these two datasets were tagged as complete (>29,000 nt) and high coverage (<1% Ns with < 0.05% unique amino acid mutation) in GISAID. We adopted MUMmer (version 3.23) to obtain the SNVs of the SARS-CoV-2 genomes [31]. Each SARS-CoV-2 genome was aligned with the SARS-CoV-2 reference genome (MN908947.3) to obtain the homologous region using the nucmer function with the default parameters [31]. Then, we obtained the SNP matrix from the alignment results with the show-SNP function [31] and prepared for SNP clique analysis.
2.2. SNP coexistence network and clique analysis
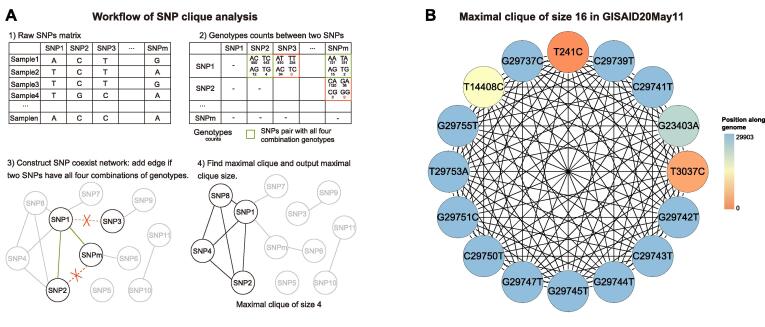
To evaluate the complexity of SNP cooccurrences within the GISAID dataset, we applied single-nucleotide variant (SNV) clique analysis by in-house scripts. After obtaining all SNPs, we checked the alleles at every locus of the SARS-CoV-2 genome. Over 92% of the SNP loci (5,671/6,178) had two alleles. Focusing on the loci with two alleles, we removed the SNP loci with three or four alleles. We labelled the major allele of the SNP locus as R and the minor allele as A. Thus, it had four possible genetic combinations for every pair of two SNP loci: RR, RA, AR, and AA. We recognized each SNP locus as a vertex and created an edge between a locus pair only if all four genetic combinations existed in at least one assembly genome within the GISAID dataset (Fig. 1A). We obtained the maximal clique from the network. Based on the cliques, we can tell whether SARS-CoV-2 coinfection exists since the existence of a large clique will be intractable to explain using phylogeny.
Fig. 1.
Workflow of SNP clique analysis and the maximal clique in the GISAID20May11 dataset. A. The workflow of SNP clique analysis. First, we constructed an SNP, single-nucleotide polymorphism, and coexisting network from the SNP matrix. Every SNP locus is a vertex, and we add an edge between a locus pair if they have all four major genotypes. We then extract the maximal clique from the network. B. The maximal 16-SNP clique was found in the GISAID20May11 dataset with 11,179 SARS-CoV-2 genomes.
2.3. Simulation of viral transmission route
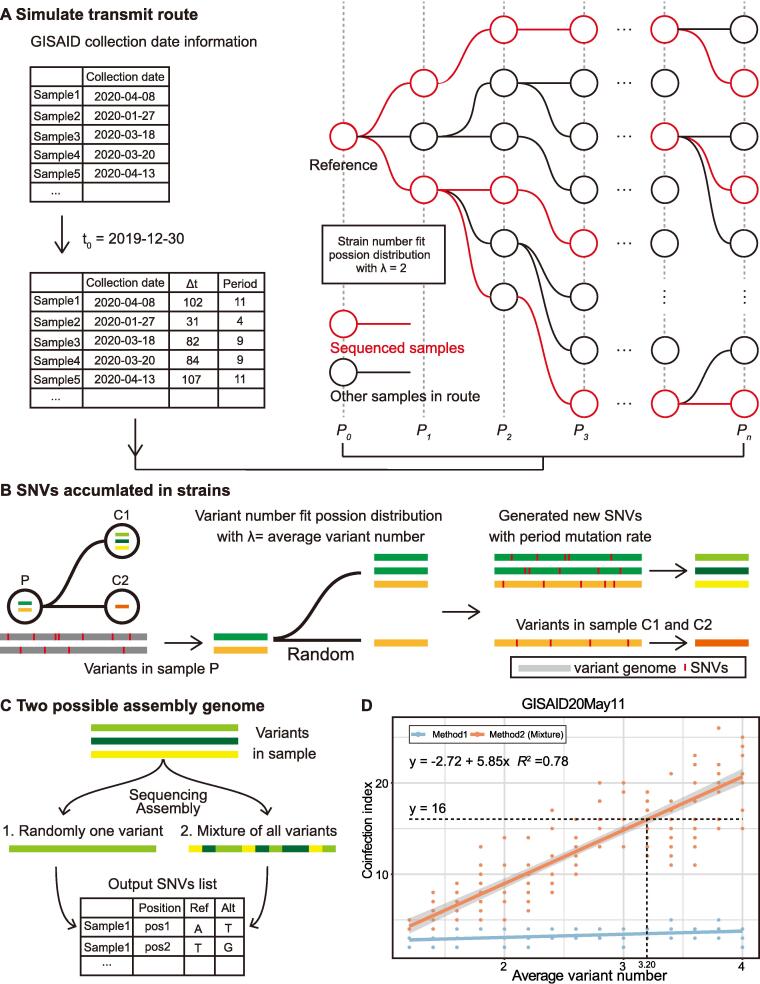
We simulated the virus transmission route based on the epidemiological information of SARS-CoV-2. The reproduction number (R0) represents the average number of people a COVID-19 patient can infect in the infectious period. During the epidemic, the R0 is constantly changing. Our simulations are executed assuming that R0 equals 2, which means that each patient could infect two people on average. The distribution of the number of people infected by the same patient conforms to the Poisson distribution.
In addition, we make other assumptions to clarify the transmission routes. We assume each patient’s infectious period lasts ten days, and the transmission ends after the infectious period. According to the sample collection time provided in GISAID, we can count the number of samples collected in each period and note the period with the maximum sample number as Pn. We note the collection time and the period of the reference genome as t0 and P0, respectively. Dividing the duration between the sample collection date and t0 to the infectious period, we obtained the period number for each sample (Fig. 3A).
Fig. 3.
The simulation flowchart of viral SNVs in samples and the regression of variant number and the coinfection index. A. We simulated the transmission route based on known epidemiological information of SARS-CoV-2 and constructed a transmission tree. Then, we selected the sequenced samples based on their release date in the GISAID database. B. Variant numbers in all samples fit the Poisson distribution with λ equal to the average variant number. In a single transmission branch, variants in child nodes are randomly inherited from the parent sample. In addition, new SNVs, single-nucleotide variants, would also emerge in the child nodes based on the given mutation rate. C. We simulated two possible methods to obtain assembled genomes in the datasets. In method 1, the assembled genome exhibited one of the variants. In method 2, the assembled genome represented the mixed genome of multiple variants. Then, we acquired the SNV list for all samples as the output. D. The distribution of the coinfection index with different average variant numbers in the GISAID20May11 dataset. The dash lines suggested the coinfected variant number corresponding to the 16-SNP clique with method 2.
We constructed a transmission route tree based on the above assumptions. For each sample in Pi (1 < i < n), the number of samples infected by it in Pi+1 is determined by a Poisson distribution (λ = R0), as we mentioned above. We will generate samples randomly if the total sample number is less than the actual sample number, ensuring sufficient samples for the selection in the following steps. Next, we randomly selected samples in each period to simulate random sample sequencing. The number of selected samples is the same as the confirmed sample number in the period. We retained all samples in the route from the reference to selected samples and constructed a subtree for further simulation experiments.
2.4. Simulation of SNVs accumulations in variants
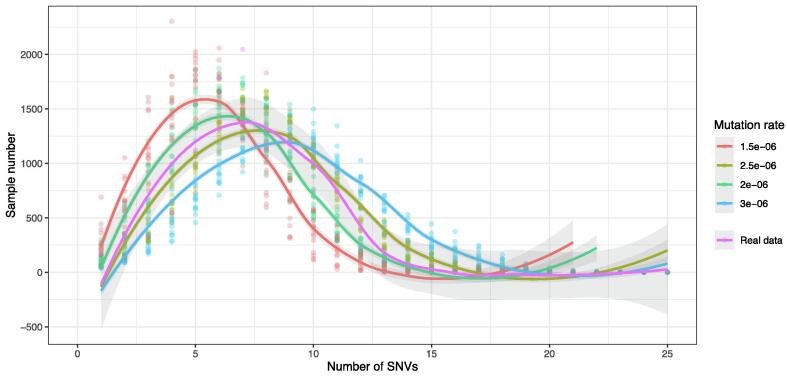
We simulate the SNV accumulations in variants with two variables: the mutation rate (r) and the average variant number (w). In a previous report, the estimated mutation rate of SARS-CoV-2 ranged from 2.88 × 10-6 to 3.45 × 10-6 substitutions per site per day [32], [33], [34]. However, in our preliminary test, the obtained SNV distribution curve does not fit the distribution curve from the real dataset (Fig. 4). In our simulations, the mutation rate has four possible values: 1.5 × 10-6, 2 × 10-6, 2.5 × 10-6, and 3 × 10-6. The average variant number in the simulation has fifteen possible values ranging from 1.2 to 4, with an interval of 0.2. The distribution of strain numbers in all samples conformed to a Poisson distribution.
Fig. 4.
The distribution of samples with different SNVs in the GISAID20May11 dataset and the simulation under different mutation rates. We had 15 possible average numbers of variants and ten duplicates for each pair of mutation rates and the average variant number. We plotted the sample number in all simulations and regressed the sample number vs the number of SNVs of all simulations with a specific mutation rate, and the 95% CI region is shown in grey.
We can obtain a period mutation rate from the infectious period and mutation rate, representing the mutation rate between two neighbouring periods. In a single transmission branch, variants in samples at child branches are random heritages from the variants in the parent branch. Meanwhile, new SNVs would also emerge based on the period mutation rate. Since reversible mutation occurred, we aligned the genomes of new variants to the reference genome to obtain the SNV list (Fig. 3B).
2.5. Simulation of SNVs in assembly genomes
We conducted two methods to simulate the actual SNVs on the assembled genome (Fig. 3C). In the first method, each sample has one randomly selected variant that is fully sequenced. In the second method, we hypothesized that the genomic sequence is an assembled mixture of genomes from all variants in the sample. For the second method, we set a window of 100 nt and slide it across the entire genomic sequence. In each window, its SNVs come from a randomly selected strain (Fig. 3C).
With four possible mutation rate values and fifteen possible average variant numbers, we conducted a total of 60 simulations and performed ten repetitions for each simulation. Finally, we performed SNP clique analysis with simulated SNVs in assembly genomes and recorded the maximum clique size for the GISAID20May11 and GISAID21Apr1 datasets.
2.6. Collection of sputum samples from COVID-19 patients
To confirm coinfection with SARS-CoV-2 variants, we performed multiplex PCR on sputum samples collected from COVID-19 patients. Forty-two patients were recruited from the First Affiliated Hospital of Guangzhou Medical University and Guangdong Second Provincial General Hospital, China, between Jan and Mar 2020 (Table S1). The sputum samples from the patients were inactivated at 56 °C for 30 min following WHO and Chinese guidelines [35], [36]. The specimens were stored at 4 °C until ready for shipment to the Guangdong Centers for Disease Control and Prevention.
2.7. Nanopore sequencing on the products of multiplex PCR
We extracted the total RNA from the samples according to the protocol of the RNA isolation kit (RNAqueous Total RNA isolation Kit, Invitrogen, China) and determined the RNA concentration by Qubit (Thermo Fisher Scientific, China). Based on two pools of primers (98 pairs of primers in total, Table S2), the entire genomic sequence of SARS-CoV-2 was amplified segmentally by reverse transcription. Then, libraries were built by adding the adapter and barcode to the amplified genomic fragments with a Nanopore library construction kit (EXP-FLP002-XL, Flow Cell Priming Kit XL, YILIMART, China). The samples were sequenced on the MinIon sequencing platform (Oxford Nanopore Technologies, UK).
2.8. Nanopore sequencing data filtration
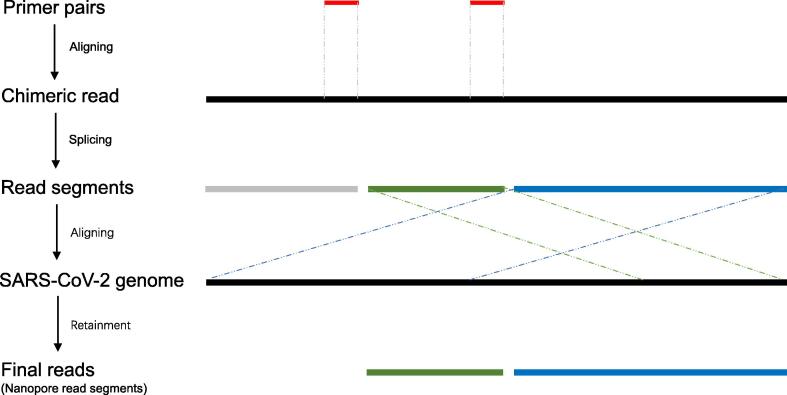
The MinIon sequencer generated Fast5 format data, converted into fastq format with guppy base caller (version 3.0.3). By applying NanoFilt (version 1.7.0) [37], we performed data filtration on the raw fastq data with the following criteria: the read lengths should be longer than 100 bp after removing the adapter sequences, and the overall quality of reads should be higher than 10. Furthermore, the chimeric reads should be processed to avoid false identification of virus recombination or host integration due to the random connection of multiplex PCR amplicons. Therefore, we positioned the primers on the sequencing reads to identify the chimeric reads, split the identified chimeric reads into segments corresponding to PCR amplicons, and retained the final reads by aligning the segments to the viral genome (Fig. 7). This method allowed us to salvage a massive amount of sequencing data, leading to more accurate alignment and higher coverage.
Fig. 7.
The procedure of chimeric read identification and read splicing. To remove the chimeric reads, we first aligned the multiplex PCR primer to the Nanopore sequencing reads and split the reads into segments according to the aligning results. Then, we aligned the read segments to the SARS-CoV-2 genome and discarded those segments that could not be aligned to the reference genome. Finally, we removed the original reads and kept the genome-aligned segments.
2.9. Mutation detection with Nanopore sequencing data
We aligned the filtered and segmented reads to the SARS-CoV-2 reference genome (MN908947.3) with Minimap2 by applying the default parameters for Oxford Nanopore reads [38]. The aligned PCR amplicons were separated according to the corresponding primer pool. With the separated alignment results, the genomic variations with average quality larger than ten were called with bcftools (version 1.8) [39]. Mutations with less than ten supported reads were filtered. We also filtered the variations within ten bp upstream or downstream of the primer region within the corresponding primer pool to reduce the PCR amplification effects. The filtered mutations for different primer pools were then merged as the final mutations. The final mutations were annotated by in-house software based on the gene information in the SARS-CoV-2 reference genome.
3. Results
3.1. The 16-SNP clique reveals the coinfection with multiple SARS-CoV-2 variants in the GISAID20May11 dataset
In this study, we tried SNP clique analyses to detect the maximal cliques for the SARS-CoV-2 genomic datasets heuristically. The GISAID20May11 dataset contains 12,986 SARS-CoV-2 genomes published between Dec 30, 2019, and May 11, 2020. After filtering 1,804 duplicated sequences, we aligned the remaining 11,182 viral genomes to the SARS-CoV-2 reference genome to obtain SNVs. Then, we removed three viral genomes with over 1,000 SNVs and obtained 11,179 genomes for the following-up analysis. With 57,548 SNVs on 6,178 SNP loci, we performed SNP clique analysis (Fig. 1A) and constructed SNP coexistence networks with 1,150 vertices and 8,003 edges. Among the networks, we discovered the maximal clique with 16 coexisting loci (Fig. 1B).
To better understand the existence of the 16-SNP clique in the GISAID20May11 dataset, we designed a formula to calculate the possibility to obtain such a big clique without considering coinfection or recombination. The formula calculates the probability of obtaining n-cliques from the sequenced genomes at time t:
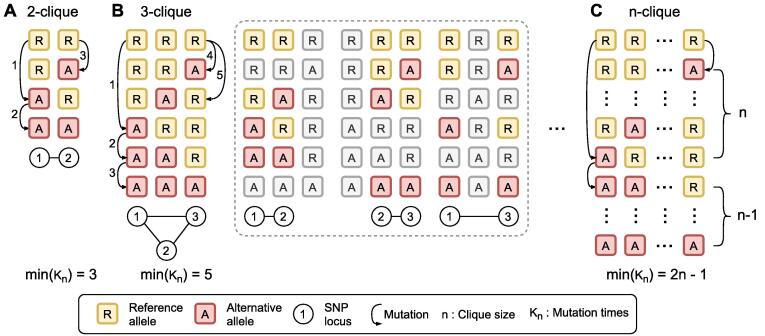
In the formula, R stands for the mutation rate of SARS-CoV-2 per substitute per day, t stands for the duration time from the collection date of the reference genome to the publishing date of the sequenced sample, Kn stands for the number of mutations required to obtain n-SNP clique from the reference genome, and Nt stands for the number of COVID-19 sequences at time t. According to previous reports, we set the highest mutation rate of 3.45 × 10-6 as R, and we can determine the minimal Kn by the clique size. The reference genome experienced at least three mutations, corresponding to three new haplotypes for the formation of a 2-SNP clique (Fig. 2A). The reference genome experienced at least five mutations, resulting in five new haplotypes to generate a 3-SNP clique (Fig. 2B). Hence, we summarized that the reference genome experienced at least 2n − 1 mutations to obtain an n-SNP clique, and the formula can be further changed into:
Fig. 2.
The minimum mutation frequency to access n-SNP clique from the reference genome. A, B, and C illustrated the formation of 2, 3, and n-SNP cliques with minimum mutation. Each block represents the alleles at the SNP loci. The yellow blocks (shown with “R”) and the red blocks (shown with “A”) represent major and minor alleles, respectively. The circles with numbers exhibit the loci of the alleles in the reference genome. n represents the size of the maximal clique of the SNP coexistence network. Kn stands for the number of mutations required to form n-clique from the reference genome. From the plot, we concluded that the reference genome experienced at least 2n − 1 mutations to obtain an n-SNP clique. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
While, in a specific database with known SNP loci, we can confirm that the n haplotypes with only one alternative mutation had already existed in the dataset (shown at the top of Fig. 2C). In that condition, min(Kn) should be (2n − 1) - n, which is n − 1. For the GISAID20May11 dataset (t = 134 and Nt = 11,179), the maximal probability of obtaining 16 cliques was 1.05 × 10-46. In addition, we discovered 130 new haplotypes in the dataset for the 16 loci, which is significantly larger than the lower bound, 2n − 1. If we set Kn as 130, the possibility of obtaining the number of haplotypes was even slighter, which nearly equalled 0. Although linkage exists between different loci, which affects the parameter n, it barely affects the final generation possibility of the clique or explains so many haplotypes. Therefore, the 16-SNP clique in the dataset is unlikely to be explained by single variant infection, except hypermutation and reversible mutation. With such inference, we deduced that the coinfection of multiple SARS-CoV-2 variants occurred in GISAID20May11 dataset and some assembly genomes were mixed sequences of multiple coinfected variants.
3.2. An average of three SARS-CoV-2 variants coexisted in samples from the GISAID20May11 dataset
To further confirm the coinfection of multiple SARS-CoV-2 variants, we selected the maximal clique from the SNP coexistence networks, noted its size as the coinfection index, and determined the average coinfected variant number with computational simulations. By simulating the transmission route tree of COVID-19, we traced the virus transmission among the infected individuals. Based on the publishing date of the sequences, we divided them into different transmission periods, simulated SNV accumulation in their genome sequences, and calculated the coinfection index using SNP clique analysis (Fig. 3A, see detailed method). Using different mutation rates and the average coinfected variant number in the simulation, we obtained a chart of the average variant number against the coinfection index under a specific mutation rate (Fig. 4). We find the simulation with the mutation rate of 2.5 × 10-6 has the smallest Mean squared error. The subsequent result is based on the mutation rate of 2.5 × 10-6. During transmission, the variants in a sample at the child node were randomly inherited from the sample at the parent node. At the same time, new SNVs would also emerge based on a given simulated mutation rate (Fig. 3B). In the simulation, we proposed two methods to decipher how the coinfected variants construct their assembled genome. The first method randomly selected a variant from the coinfected sample and detected its SNVs. The second method (the mixed method) generated an assembly genome, which was a mixture of all variants. We split the genome into windows with a fixed size of 100 nt for the second method, and each window comes from a randomly selected variant in the sample. Using these two methods, we obtained SNVs in the assembled genomes (Fig. 3C).
After plotting the coinfection index against the average variant number, we obtained two regression lines for the two aforementioned methods (Fig. 3D). With the results, we noticed that only the regression line based on the mixed method could achieve a coinfection index of 16 for the GISAID20May11 dataset. Then, we determined the averaged variant number in the GISAID20May11 dataset with the coinfection index line. We performed regression analysis between the averaged variant number and coinfection index and discovered a significant linear relationship between the two variables with method 2 (F-statistic P-value < 2.2 × 10-16, adjusted R-squared = 0.78, Fig. 3D).
According to the obtained fitting equation, we deduced that the corresponding average variant number was 3.20 when the coinfection index was 16 in GIDAID20May11 (Fig. 3D).
3.3. The coinfection index increased during the COVID-19 pandemic
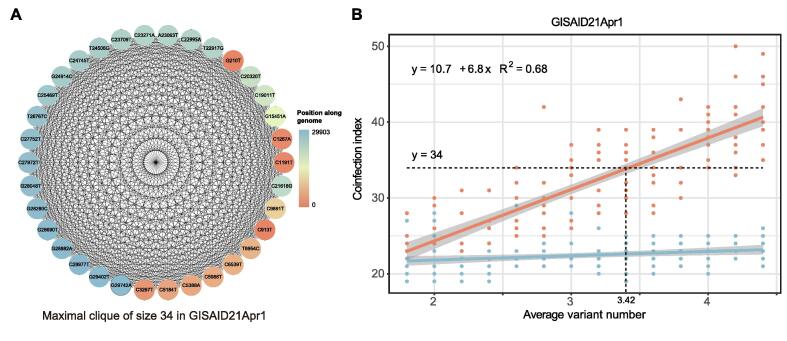
For the verification of SARS-CoV-2 variant coinfection and the simulation method, we collected 4,113 additional genomes from the GISAID database on Apr 1, 2021. The genomes of GISAID21Apr1 were sampled from five different continents. Europe supplied 3,023 samples; North America provided 1,047 samples; and Asia, South America, and Oceania had 27, 12, and 4 samples, respectively. We found 28 SNPs in over 3,000 samples, revealing that those samples should have the same or related ancestor. We selected samples from the same branch to ensure that those samples had the same ancestor to fit the SNV distribution in the GISAID21Apr1 dataset. With the GISAID21Apr1 dataset, we obtained a maximal clique with 34 coexisting SNPs from 140,348 SNVs on 6,415 genomic loci (Fig. 5A). Adopting the aforementioned formula, we discovered that the maximal probability of obtaining 34-SNP clique was 1.48 × 10-89 in the GISAID21Apr1 dataset (t = 458 and Nt = 4,113), and 107 haplotypes existed in this dataset. Then, we constructed the coinfection index curve with the dataset and determined the average variant number. The regressed linearity of the coinfection index and the average number of variants also showed a significant linear relationship (F-statistic P-value < 2.2 × 10-16, adjusted R-squared = 0.68, Fig. 5B). The fitting equation revealed an average variant number of 3.42 in the GISAID21Apr1 dataset. Hence, we deducted that the virus could transfer between continents in this pandemic and increased the coinfection risks of different variants.
Fig. 5.
The maximal SNP clique and the coinfection analysis in the GISAID21Apr1 dataset. A. The maximal SNP clique in the GISAID21Apr1 dataset. The 4113 SARS-CoV-2 genomes in the GISAID21Apr1 dataset contained the maximal SNP clique of size 34. B. The regression of variant number and the coinfection index in the GISAID21Apr1 dataset. The dash line exhibited that the averaged variant number of 3.42 corresponding to the coinfection index of 34 in the GISAID21Apr1 dataset.
3.4. Recurrent heterozygous SNPs implied coinfection with multiple SARS-CoV-2 variants
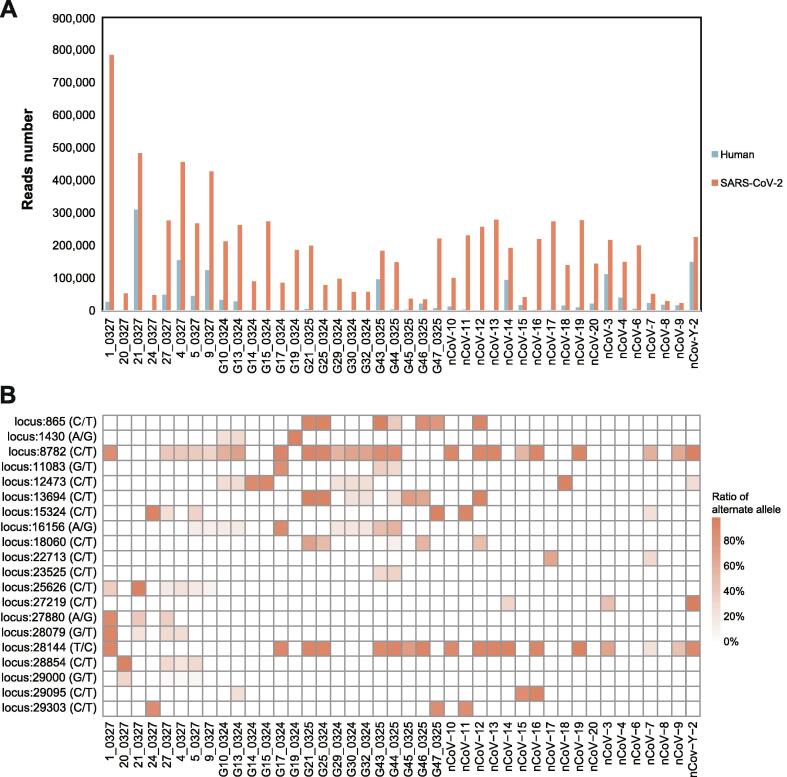
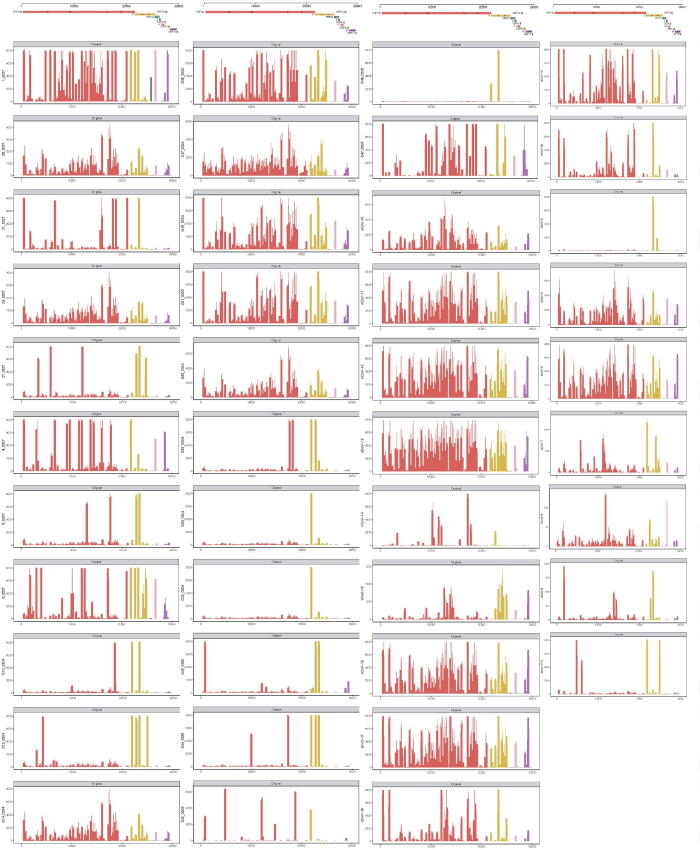
In addition, we performed Nanopore sequencing on sputum samples from 42 COVID-19 patients recruited from the First Affiliated Hospital of Guangzhou Medical University and Guangdong Second Provincial General Hospital for SARS-CoV-2 genome acquisition and mutation detection (Table S1). After sequencing the multiplex polymerase chain reaction (PCR) products (Table S2), we obtained a total of 7,877,736 clean reads, and each sample has 187,565 ± 143,719.55 (mean ± SD) reads on average (Fig. 6A). To eliminate the chimeric caused by the random connection of the PCR amplicons, we developed a software tool named CovProfile, performed data filtration, and detected the mutations in SARS-CoV-2 variants (Fig. 7). After the removal of the chimeric reads, we aligned the clean reads to the SARS-CoV-2 genome and human transcriptome and discovered that the ratio of aligned sequences ranged from 3.86% to 99.74% in the SARS-CoV-2 genome and from 0.13% to 70.5% in the human transcriptome database (Fig. 6A). Moreover, the SARS-CoV-2 genomic coverage reached over 99.7% with > 1,800 × depth in each sample, ensuring adequate data volume for SNP calling (Fig. 8). The raw Nanopore sequencing data can be accessed in the Genome Sequence Archive of the National Genomics Data Center (accession ID: CRA002522) [40], [41].
Fig. 6.
Statistics of Nanopore sequencing data and the distribution of recurrent SNPs for the 42 COVID-19 samples. A. After low-quality filtration, we aligned the Nanopore sequencing reads to the SARS-CoV-2 genome and human transcriptome. The histograms in red and blue exhibit the number of reads aligned to the SARS-CoV-2 genome and human transcriptome, respectively. B. The heatmap exhibited twenty recurrent SNPs for the 42 COVID-19 patients. The squares with deeper colour represent the higher ratios of the alternate allele in the sample. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Coverage of depth of aligned data in the 42 COVID-19 samples. The X coordinate represents the location of the SARS-CoV-2 genome, and the Y coordinate represents the sequencing depth. The red, yellow, green, pink, brown, light green, purple and dark brown bars represent the genetic regions of ORF1ab, S, ORF3a, M, ORF6, ORF8, N and ORF10, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
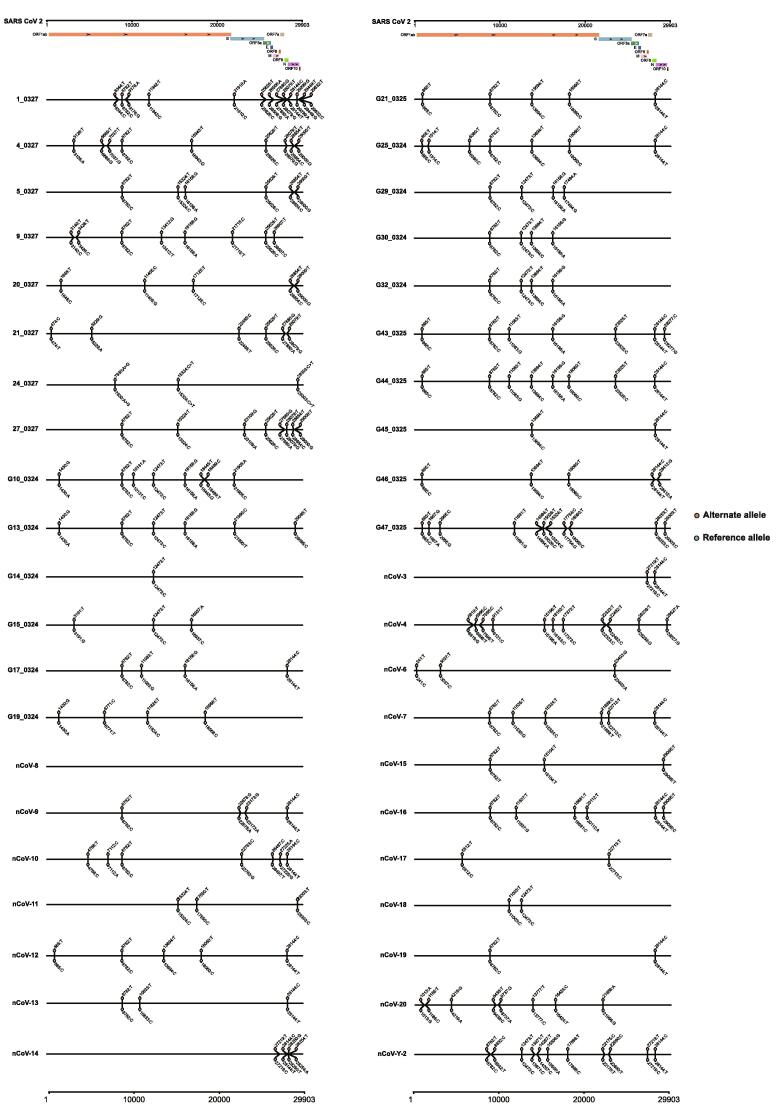
With the read alignment in the SARS-CoV-2 genome, we explored the distributions of SARS-CoV-2 mutations in the 42 samples. In all samples, we discovered a total of 115 SNPs, 108 of which were located in genetic regions, including the ORF1ab, S, ORF3a, N, M, ORF6, ORF8, and ORF10 genes. Furthermore, we discovered heterozygous SNPs in 41 of the enrolled samples (Table S3, Fig. 9). Although heterozygous SNPs can result from a mutation spectrum generated by a single infection [42], twenty heterozygous SNPs existed in over two samples, suggesting the probability of coinfected variants, such as C865T, A1430G, C8782T, G11038T, etc. (Fig. 6B). Notably, we discovered that 14 samples contained two genotyped SNPs on loci 8,782 and 28,144 simultaneously, which were significant loci for SARS-CoV-2 phylogenetic clade identification. Therefore, the sequencing data confirmed the multiple variants coinfection in COVID-19 patients.
Fig. 9.
Distributions of heterozygous SNPs in the 42 COVID-19 samples. After obtaining the SARS-CoV-2 genomic reads, we detected the SNPs for each sample and exhibited the genotypes for the reference and mutations. For each sample, the mutated and reference genotypes were marked with red and blue, respectively. At the top of the figure, we exhibited the locations of the genes in the SARS-CoV-2 genome and annotated them with different colours. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
SARS-CoV-2 poses a significant threat to human lives, and recent studies have reported rapidly emerging variants and their impact on clinical severity and vaccine protection [7], [9], [43], [44], [45], [46]. In this study, we aimed to detect whether coinfection with multiple SARS-CoV-2 variants existed in COVID-19 patients, which might be associated with frequent homologous recombination and greater clinical severity.
4.1. The existence of maximal SNP cliques encourages the coinfection with multiple SARS-CoV-2 variants
The study performed SNP coexistence network analysis to detect the “coinfection index” based on the maximal SNP cliques in the collected GISAID datasets. With the formula to calculate the probability of obtaining maximal cliques of single variant infection, we discovered that the probability of obtaining the maximal cliques for the GISAID20May11 and GISAID21Apr1 datasets were 1.05 × 10-46 and 1.48 × 10-89, respectively, which nearly equalled 0. After ruling out the single variant infection, we have three potential explanations for the maximal SNP cliques: the coexistence of multiple SARS-CoV-2 variants; the recombination of the SARS-CoV-2 variants, although the occurrence of recombination is also based on the premise of coinfection; and hypermutation and reversible mutation. Since the mutations are undirected and we observed two genotypes for the locus in most samples, hypermutation and reversible mutation could barely explain the situation. Hence, coinfection with multiple SARS-CoV-2 variants provides the most likely explanations for the maximal SNP cliques.
Then, we simulated the process of coinfection with multiple variants (Fig. 3) and deciphered the number of coinfected variants for SARS-CoV-2 in hosts with linear regression between the coinfection index and the average variant number. In the simulation of variant coinfection, the final assembly genome contained mixed SNVs from multiple variants. These variants may cotransmit to other patients at the same time. Hence, coinfection with multiple variants was the only way to explain the large SNP clique. With the GISAID20May11 and GISAID21Apr1 datasets, we discovered that the number of coinfected variants increased from 3.20 to 3.42 in COVID-19 patients. Considering the rapidly emerging SARS-CoV-2 variants worldwide, we hypothesized that the coinfected variants in hosts would aggravate the clinical severity, increase the change in viral recombination, and pose a greater threat to us [47], [48]. Although coinfection explained the large clique detected in the SNP coexistence networks in the datasets, the discoveries still need to be verified experimentally.
4.2. Heterozygous SNPs on the phylogenetically diverging loci reveal coinfection with multiple SARS-CoV-2 variants
To verify coinfection with multiple SARS-CoV-2 variants, we performed Nanopore sequencing on 42 COVID-19 patients and implemented CovProfile for sequencing data processing and genomic mutation detection. Our results confirmed the reliability of the multiplex PCR method in identifying SARS-CoV-2 and discovered recurrent heterozygous SNPs in 41 of 42 samples. Since heterozygous SNPs can be caused by a mutation spectrum generated by a single infection [42], [49] or the presence of sample bias [50], we further detected the existence of recurrent heterozygous SNPs and discovered two genotyped SNPs on loci 8,782 and 28,144 in fourteen patients. Since loci 8782 and 28,144 were important for identifying the L and S lineages of SARS-CoV-2 [51], [52], [53], the heterogeneous genotypes on these loci suggested coinfection in the patients. Corresponding to the simulation results, the discoveries of recurrent heterozygous SNPs, especially those crucial for phylogenetic clade identification, suggested multiple variant coinfection in COVID-19 patients.
The discovery of SARS-CoV-2 variant coinfection provided explanations for the severe clinical symptoms in some COVID-19 patients and significantly affected the application of vaccines [9], [54], [55]. Since vaccines were developed referencing a specific SARS-CoV-2 variant, the infection of variants limited the protection afforded by vaccines [9], [10]. For instance, the SARS-CoV-2B.1.351 variant, widespread in Nelson Mandela Bay and South Africa, can evade the immune response stimulated by vaccines and significantly reduce the vaccine's protective effect on the population [43], [56]. Moreover, Nicole Pedro et al. also discovered dual SARS-CoV-2 variant coinfection in a patient with severe COVID-19 in Portugal, which supported our discoveries [19]. Therefore, coinfection with multiple SARS-CoV-2 variants raised another challenge to which we need to stay alert in the battle against the COVID-19 epidemic.
4.3. Application and limitation of the coinfection simulation
The coinfection simulation fits in other epidemics. With such an approach, we could detect whether coinfection with multiple variants exists during viral transmission, decipher the average number of the coinfected variants in each host, and explore the trends of coinfected variant numbers in an epidemic. In addition, we could predict the epidemic development by combining the simulation algorithm with other epidemiological models [57]. Although the findings from algorithm derivation implied the coinfection with multiple SARS-CoV-2 variants in patients, this method still has several limitations. In the simulation, we assumed that the first submitted sequence was the source of all SARS-CoV-2 variants. In the pandemic, the first infective SARS-CoV-2 variant should have emerged long before being discovered [58], [59], [60]. The study by Giovanni Apolone et al. proposed that SARS-CoV-2 RBD-specific antibodies were detected in the serum samples of Italian cohorts collected as early as March 2019, indicating that the source variants of all currently sequenced variants should appear earlier [61]. Determining the virus's origin is difficult, so we chose an exact time point during the simulation, but it does not affect our conclusions on host coinfection with multiple variants. Moreover, there was no guarantee considering the quality of the viral variants submitted to GISAID, which might influence the accuracy and potential phylogenetic study. Finally, the discovered heterozygous SNPs need to be verified with biological duplication, and we should identify coinfected viral lineages in COVID-19 patients and their potential impacts on host health [62].
In conclusion, our study proposed a computational simulation approach to decipher the number of coinfected variants, declared coinfection with multiple SARS-CoV-2 variants in COVID-19 patients, and reported increased coinfected variants in the COVID-19 epidemic.
5. Ethics statement
The ethics committees at the First Affiliated Hospital of Guangzhou Medical University approved this study (Ethical number: 2020–36), and the study conformed to the principles expressed in the Declaration of Helsinki. All patients provided written informed consent and volunteered to receive investigation for scientific research.
6. Data availability
The Nanopore sequencing data in this paper have been deposited in the Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn/gsa), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (BioProject No: PRJCA002503, accession ID: CRA002522).
7. Code availability
The programs for the simulation of viral coinfection are available in the GitHub repository (https://github.com/deepomicslab/CoV_Simulation).
CRediT authorship contribution statement
Yinhu Li: Conceptualization, Methodology, Visualization, Data curation, Writing – original draft. Yiqi Jiang: Conceptualization, Methodology, Software, Visualization, Data curation, Writing – original draft. Zhengtu Li: Investigation, Resources, Writing – review & editing. Yonghan Yu: Methodology, Software, Visualization, Writing – original draft. Jiaxing Chen: Software, Investigation. Wenlong Jia: Software, Visualization. Yen Kaow Ng: Supervision. Feng Ye: Conceptualization, Resources, Writing – review & editing, Supervision. Shuai Cheng Li: Conceptualization, Methodology, Software, Writing – review & editing. Bairong Shen: Conceptualization, Methodology, Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by the Chengdu Science and Technology Project of China for COVID-19 prevention and control (Grant no. 2020-YF05-00281-SN); the Guangzhou Institute of Respiratory Health Open Project (Funds provided by China Evergrande Group) - Project No. (2020GIRHHMS14); the Guangzhou Municipal Science and Technology Bureau; and Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020003). We want to thank Beijing YuanShengKangTai (ProtoDNA) Genetech Co. Ltd. for their assistance with Nanopore sequencing. We would like to thank all the doctors at the First Affiliated Hospital of Guangzhou Medical University for their assistance with a specimen and clinical data collection. Furthermore, we acknowledge the AJE team for polishing the English language of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.03.011.
Contributor Information
Feng Ye, Email: tu276025@gird.cn.
Shuai Cheng Li, Email: shuaicli@cityu.edu.hk.
Bairong Shen, Email: bairong.shen@scu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picchianti Diamanti A., Rosado M.M., Pioli C., Sesti G., Lagana B. Cytokine Release Syndrome in COVID-19 Patients, A New Scenario for an Old Concern: The Fragile Balance between Infections and Autoimmunity. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21093330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cyranoski D. Profile of a killer: the complex biology powering the coronavirus pandemic. Nature. 2020;581:22–26. doi: 10.1038/d41586-020-01315-7. [DOI] [PubMed] [Google Scholar]
- 6.Li Q., Wu J., Nie J., Zhang L., Hao H., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182(1284–1294) doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J., Peng P., Wang K., Fang L., Luo F.Y., et al. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell Mol Immunol. 2021 doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hourdel V., Kwasiborski A., Baliere C., Matheus S., Batejat C.F., et al. Rapid Genomic Characterization of SARS-CoV-2 by Direct Amplicon-Based Sequencing Through Comparison of MinION and Illumina iSeq100(TM) System. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.571328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John P., Moore P.A.O. SARS-CoV-2 Vaccines and the Growing Threat of Viral Variants. JAMA. 2021 doi: 10.1001/jama.2021.1114. [DOI] [PubMed] [Google Scholar]
- 10.Wang P.F., Nair M.S., Liu L.H., Iketani S., Luo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 11.Burioni R., Topol E.J. Assessing the human immune response to SARS-CoV-2 variants. Nat Med. 2021;27:571–572. doi: 10.1038/s41591-021-01290-0. [DOI] [PubMed] [Google Scholar]
- 12.Bianchi M., Borsetti A., Ciccozzi M., Pascarella S. SARS-Cov-2 ORF3a: Mutability and function. Int J Biol Macromol. 2021;170:820–826. doi: 10.1016/j.ijbiomac.2020.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of Co-infection Between SARS-CoV-2 and Other Respiratory Pathogens. JAMA. 2020;323:2085–2086. doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo Y., Miyazaki S., Yamashita R., Ikeda T. Coinfection with SARS-CoV-2 and influenza A virus. BMJ Case Rep. 2020;13 doi: 10.1136/bcr-2020-236812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teweldemedhin M., Asres N., Gebreyesus H., Asgedom S.W. Tuberculosis-Human Immunodeficiency Virus (HIV) co-infection in Ethiopia: a systematic review and meta-analysis. BMC Infect Dis. 2018;18:676. doi: 10.1186/s12879-018-3604-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuya-Kanamori L., Liang S., Milinovich G., Soares Magalhaes R.J., Clements A.C., et al. Co-distribution and co-infection of chikungunya and dengue viruses. BMC Infect Dis. 2016;16:84. doi: 10.1186/s12879-016-1417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villamil-Gomez W.E., Gonzalez-Camargo O., Rodriguez-Ayubi J., Zapata-Serpa D., Rodriguez-Morales A.J. Dengue, chikungunya and Zika co-infection in a patient from Colombia. J Infect Public Health. 2016;9:684–686. doi: 10.1016/j.jiph.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Pedro N, Silva CN, Magalhaes AC, Cavadas B, Rocha AM, et al. (2021) Dynamics of a Dual SARS-CoV-2 Lineage Co-Infection on a Prolonged Viral Shedding COVID-19 Case: Insights into Clinical Severity and Disease Duration. Microorganisms 9. [DOI] [PMC free article] [PubMed]
- 20.Hattori M., Okuno Y., Goto S., Kanehisa M. Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J Am Chem Soc. 2003;125:11853–11865. doi: 10.1021/ja036030u. [DOI] [PubMed] [Google Scholar]
- 21.Fukagawa D., Tamura T., Takasu A., Tomita E., Akutsu T. A clique-based method for the edit distance between unordered trees and its application to analysis of glycan structures. BMC Bioinf. 2011;12(Suppl 1):S13. doi: 10.1186/1471-2105-12-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsunaga T., Yonemori C., Tomita E., Muramatsu M. Clique-based data mining for related genes in a biomedical database. BMC Bioinf. 2009;10:205. doi: 10.1186/1471-2105-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Wu M., Kwoh C.K., Ng S.K. Computational approaches for detecting protein complexes from protein interaction networks: a survey. BMC Genomics. 2010;11(Suppl 1):S3. doi: 10.1186/1471-2164-11-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohseni-Zadeh S., Brezellec P., Risler J. Cluster-C, an algorithm for the large-scale clustering of protein sequences based on the extraction of maximal cliques. Comput Biol Chem. 2004;28:211–218. doi: 10.1016/j.compbiolchem.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Zhang B., Park B.H., Karpinets T., Samatova N.F. From pull-down data to protein interaction networks and complexes with biological relevance. Bioinformatics. 2008;24:979–986. doi: 10.1093/bioinformatics/btn036. [DOI] [PubMed] [Google Scholar]
- 26.Li C., Xu J. Feature selection with the Fisher score followed by the Maximal Clique Centrality algorithm can accurately identify the hub genes of hepatocellular carcinoma. Sci Rep. 2019;9:17283. doi: 10.1038/s41598-019-53471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H., Wang Y., Zhang Z., Li H. Identification of KIF18B as a Hub Candidate Gene in the Metastasis of Clear Cell Renal Cell Carcinoma by Weighted Gene Co-expression Network Analysis. Front Genet. 2020;11:905. doi: 10.3389/fgene.2020.00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan Y., Wick R.R., Zobel J., Ingle D.J., Inouye M., et al. GeneMates: an R package for detecting horizontal gene co-transfer between bacteria using gene-gene associations controlled for population structure. BMC Genomics. 2020;21:658. doi: 10.1186/s12864-020-07019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curis E., Courtin C., Geoffroy P.A., Laplanche J.L., Saubamea B., et al. Determination of sets of covariating gene expression using graph analysis on pairwise expression ratios. Bioinformatics. 2019;35:258–265. doi: 10.1093/bioinformatics/bty629. [DOI] [PubMed] [Google Scholar]
- 30.Lai Y.P., Ioerger T.R. A statistical method to identify recombination in bacterial genomes based on SNP incompatibility. BMC Bioinf. 2018;19:450. doi: 10.1186/s12859-018-2456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delcher A.L., Phillippy A., Carlton J., Salzberg S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trevor Bedford RN, James Hadfield, Emma Hodcroft, Misja Ilcisin, Nicola Muller (2020) Genomic analysis of nCoV spread. Situation report 2020-01-23.: ResearchWorks Archive.
- 33.Baric R.S., Yount B., Hensley L., Peel S.A., Chen W. Episodic evolution mediates interspecies transfer of a murine coronavirus. J Virol. 1997;71:1946–1955. doi: 10.1128/jvi.71.3.1946-1955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johns Hopkins Center for Health Security. SARS-CoV-2 Genetics 2020.
- 35.Li T. Diagnosis and clinical management of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection: an operational recommendation of Peking Union Medical College Hospital (V2.0) Emerg Microbes Infect. 2020;9:582–585. doi: 10.1080/22221751.2020.1735265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prevention CCfDCa (2020) The guideline of diagnosis and treatment of COVID-19 (the seventh edition).
- 37.De Coster W., D'Hert S., Schultz D.T., Cruts M., Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T., Chen X., Zhang S., Zhu J., Tang B., et al. Genomics Proteomics Bioinformatics; 2021. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Members C.-N., Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021;49:18–28. [Google Scholar]
- 42.Teng X., Li Q., Li Z., Zhang Y., Niu G., et al. Compositional Variability and Mutation Spectra of Monophyletic SARS-CoV-2 Clades. Genomics Proteomics Bioinformatics. 2020;18:648–663. doi: 10.1016/j.gpb.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Chen X.-Y., Yang L., Yao Q., Chen K.P. Human SARS-CoV-2 has evolved to increase U content and reduce genome size. Int J Biol Macromol. 2022;204:356–363. doi: 10.1016/j.ijbiomac.2022.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boehm E., Kronig I., Neher R.A., Eckerle I., Vetter P., et al. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021;27:1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papanikolaou V., Chrysovergis A., Ragos V., Tsiambas E., Katsinis S., et al. From delta to Omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene. 2022;814 doi: 10.1016/j.gene.2021.146134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Y., He S., Tian W., Li D., An M., et al. First complete-genome documentation of HIV-1 intersubtype superinfection with transmissions of diverse recombinants over time to five recipients. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Escalera A., Gonzalez-Reiche A.S., Aslam S., Mena I., Laporte M., et al. Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Badua C.L.D.C., Baldo K.A.T., Medina P.M.B. Genomic and proteomic mutation landscapes of SARS-CoV-2. J Med Virol. 2021;93:1702–1721. doi: 10.1002/jmv.26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q., Zhao S., Shi C.M., Song S., Zhu S., et al. Population Genetics of SARS-CoV-2: Disentangling Effects of Sampling Bias and Infection Clusters. Genomics Proteomics Bioinformatics. 2020;18:640–647. doi: 10.1016/j.gpb.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiaolu Tang C.W., Li X., Song Y., Yao X., Xinkai W.u., Duan Y., et al. On the origin and continuing evolution of SARS-CoV-2. Natational Science Review. 2020;7:12. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife. 2021;10 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams TC, Burgers WA (2021) SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir Med. [DOI] [PMC free article] [PubMed]
- 55.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., et al. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q.Q., Nie J.H., Wu J.J., Zhang L., Ding R.X., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valba O.V., Avetisov V.A., Gorsky A.S., Nechaev S.K. The beacon: journal for studying ideologies and mental dimensions 3. 2020. Evaluating Ideologies of Coronacrisis-Related Self-Isolation and Frontiers Closing by SIR Compartmental Epidemiological Model. [Google Scholar]
- 58.Awadasseid A., Wu Y.L., Tanaka Y., Zhang W. SARS-CoV-2 variants evolved during the early stage of the pandemic and effects of mutations on adaptation in Wuhan populations. International Journal of Biological Sciences. 2021;17:97–106. doi: 10.7150/ijbs.47827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.59. Gerilovych AP, Rev. Fr, Stegniy, Borys T., Kornieikov, Oleksandr M., Muzyka, Denys V., Gerilovych, Iryna O., Bolotin, Vitaliy I., Kovalenko, Larysa V., Arefiev, Vasiliy L., Zlenko, Oksana B., & Kolchyk, Olena V. (2020) Coronavirus Infections of Animals and Humans: Ideological Use in Media vs Evidence-Based Scientific Approach. The Beacon: Journal for Studying Ideologies and Mental Dimensions 3.
- 60.Bloom J.D. Recovery of Deleted Deep Sequencing Data Sheds More Light on the Early Wuhan SARS-CoV-2 Epidemic. Mol Biol Evol. 2021;38:5211–5224. doi: 10.1093/molbev/msab246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Apolone G, Montomoli E, Manenti A, Boeri M, Sabia F, et al. (2020) Unexpected detection of SARS-CoV-2 antibodies in the prepandemic period in Italy. Tumori: 300891620974755 [DOI] [PMC free article] [PubMed]
- 62.Jogalekar M.P., Veerabathini A., Gangadaran P. SARS-CoV-2 variants: A double-edged sword? Exp Biol Med. 2021;246:1721–1726. doi: 10.1177/15353702211014146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Nanopore sequencing data in this paper have been deposited in the Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn/gsa), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (BioProject No: PRJCA002503, accession ID: CRA002522).