Abstract
Immune evasion is key to cancer initiation and later at metastasis, but its dynamics at intermediate stages, where potential therapeutic interventions could be applied, is undefined. Here we show, using multi-dimensional analyses of resected tumours, their adjacent non-tumour tissues and peripheral blood, that extensive immune remodelling takes place in patients with stage I to III hepatocellular carcinoma (HCC). We demonstrate the depletion of anti-tumoural immune subsets and accumulation of immunosuppressive or exhausted subsets along with reduced tumour infiltration of CD8 T cells peaking at stage II tumours. Corresponding transcriptomic modification occur in the genes related to antigen presentation, immune responses, and chemotaxis. The progressive immune evasion is validated in a murine model of HCC. Our results show evidence of ongoing tumour-immune co-evolution during HCC progression and offer insights into potential interventions to reverse, prevent or limit the progression of the disease.
Subject terms: Hepatocellular carcinoma, Immunosurveillance, Functional clustering, Tumour heterogeneity
In order to design cancer immune therapies, it is important to understand how tumours evade the immune response that is mounted against them. Authors here analyse the distribution and properties of immune cells in hepatocellular carcinoma and describe a progressive tumour-immune co-evolution programme from early to late stage cancer.
Introduction
Hepatocellular carcinoma (HCC) remains a critical cancer burden ranking as the fourth most common cancer-related mortality worldwide1. The immune microenvironment plays a critical role in tumour development, particularly in HCC, where long-standing liver inflammation or chronic hepatitis infection often precedes carcinogenesis2–4. Moreover, the immune composition of the tumour microenvironment (TME) contributes to disease prognosis5–9, evidencing its importance in determining clinical outcomes of HCC patients. Indeed, tumour initiation and progression require escape from the immune system, representing an ongoing arms race between cancer and host. Studies have defined immune evasion as an early event in lung cancer10,11 or late event during metastasis in advanced colorectal cancer12. Thus both early and late phases of immune evasion could exist, with different characteristics and mechanisms to pave the way towards tumour progression. However, what happens in between these points throughout the intermediate stages of cancer is unknown.
By exploring the changing immune landscapes in patients with TNM (tumour size, lymph node involvement and metastasis) stage I–III HCC spanning across tumours, adjacent non-tumour tissues and blood, we show early and sustained immune escape. A second wave of immune evasion and concurrent tumour evolution peaks at stage II HCC. Our findings demonstrate continual immune-tumour co-evolution throughout tumour progression and identify intermediate tumour stage as a potential key interventional point for immunotherapy to prevent widespread HCC.
Results
Immune landscapes modification during HCC progression
To define the immune modifications along tumour progression, we first characterised the local and systemic immune landscapes in 38 treatment-naïve patients with TNM stage (S) I–III (S1-S3) HCC (n = 16 S1, n = 12 S2 and n = 10 S3), who underwent resection as first-line therapy (Supplementary Table 1). We collected two to five regions per tumour (T) samples (total T = 136) to account for intratumoural heterogeneity reported previously13,14, along with 38 matched adjacent non-tumour liver tissues (N) and peripheral blood (P) samples. All patients were clinically comparable, except for those parameters related to disease stage (i.e. tumour size and microvascular invasion) (Supplementary Table 1).
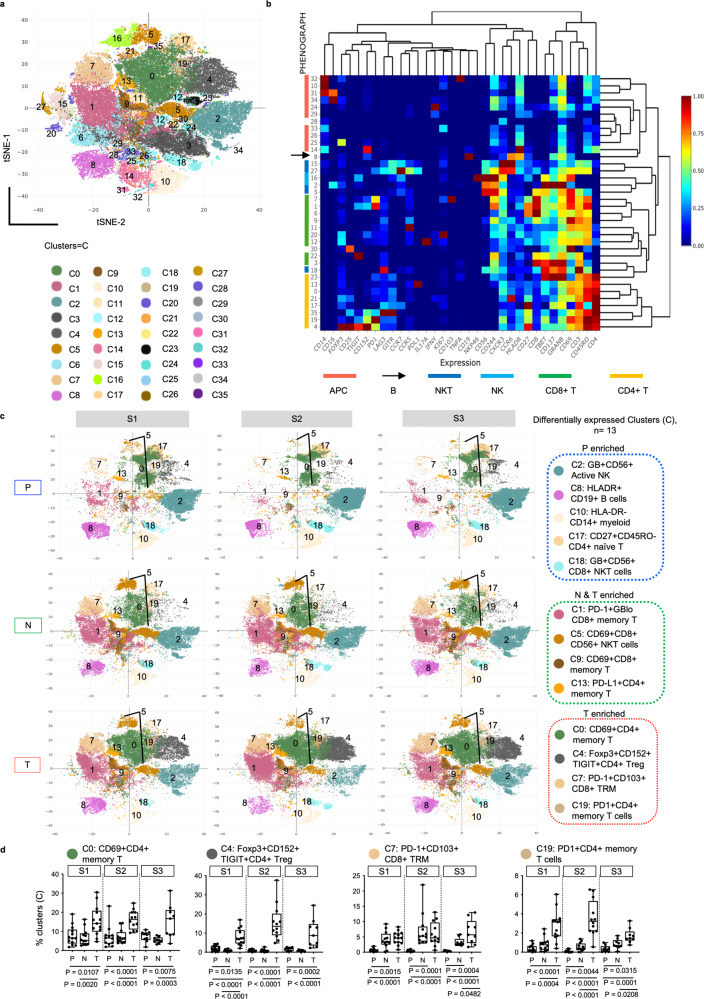
We first examined the phenotypes of immune cells in T, N and P compartments using mass cytometry by time-of-flight (CyTOF) to measure protein expression of 33 immune markers on or within individual single cells (Supplementary Table 2). Using Phenograph clustering15 and our in-house built web-based CyTOF analytics pipeline16, we identified 36 immune cell clusters (C0-C35) (Fig. 1a and Supplementary Fig. 1a) that were assigned to different immune subsets and major immune lineages according to their relative immune marker expressions (Fig. 1b and Supplementary Fig. 1b). UMAP dimensionality reduction plots showed the presence of more distinct immune clusters in T and to a lesser extent in N and P across all stages that were not confounded by patient-level differences (Supplementary Fig. 1c). Indeed, increased phenotypic heterogeneity or immune complexity was significant in tumours compared to adjacent non-tumour liver microenvironment or peripheral blood (Supplementary Fig. 1d).
Fig. 1. Diverse immune landscapes of HCC along tumour progression.
a Global tSNE plot showing 36 Phenograph immune clusters (C) from tumours (T), adjacent non-tumour liver tissues (N) and peripheral blood (P). Each cluster represented by one colour. b Heatmap representation of the same 36 immune clusters (rows) with normalised protein expression of 33 markers (columns) from all samples. The colour bars and arrow on the left indicate the major immune cell lineages shown below. c tSNE plots showing distributions and frequencies of 13 differentially expressed immune clusters across P, N or T immune compartments denoted with cluster colour code and their key phenotypes. d Graphs showing percentages of clusters (C) 0, 4, 7 and 19 with significant enrichment in T versus P and N at three stages of HCC. Clusters phenotypes are provided on top of each graph. Boxplots show median and the whiskers represent minimum and maximum values with the box edges showing the first and third quartiles. One-way ANOVA test with two-sided p-values by unpaired Mann–Whitney U (MWU) tests for two-group comparisons. a–d TNM stage I, II and III (S1, S2 and S3). nPS1 = 13, nPS2 = 12, nPS3 = 9, nNS1 = 12, nNS2 = 12, nNS3 = 9, nTS1 = 13, nTS2 = 12, nTS3 = 9. Source data are provided as a Source Data file.
From all three tumour stages, we identified the same 13 clusters that showed significant differences in their frequencies across tissue types: P, N and T using one-way ANOVA test (Fig. 1c and Supplementary Table 3). Among these, we observed PD-1+GBloCD45RO+CD8+ memory T (C1), CD69+CD8+ memory T cells (C9) and PD-L1+CD45RO+CD4+ memory T cells (C13) were enriched in both N and T compared to P (Supplementary Fig. 2a); while the antigen-presenting HLA-DR+CD19+ B cells (C8) and HLA-DR+CD14+ myeloid cells (C10), as well as CD27+CD45RO−CD4+ naïve T cells (C17) were significantly reduced in both N and T compared to P (Supplementary Fig. 2b). This shows a general accumulation of memory subsets and depletion of antigen-presenting cells in the N and T compartments as compared to P from all three tumour stages. More importantly, we observed enrichment of potentially exhausted and suppressive immune subsets in tumours: Foxp3+CD152+TIGIT+CD4+ regulatory T cells (Treg) (C4), PD1+CD103+CD45RO+CD8+ resident memory T cells (TRM) (C7) and PD1+ CD45RO+CD4+ memory T cells (C19) (Fig. 1d). Conversely, the frequencies of immunoactive subsets, including GB+CD56+ NK cells (C2) and GB+CD56+CD8+ NKT cells (C18) were significantly lower in T than N or P (Supplementary Fig. 2b).
Taken together, these data indicate that immune evasion is established early and maintained throughout the following stages of tumour development.
Progressive immune evasion peaking at stage II HCC
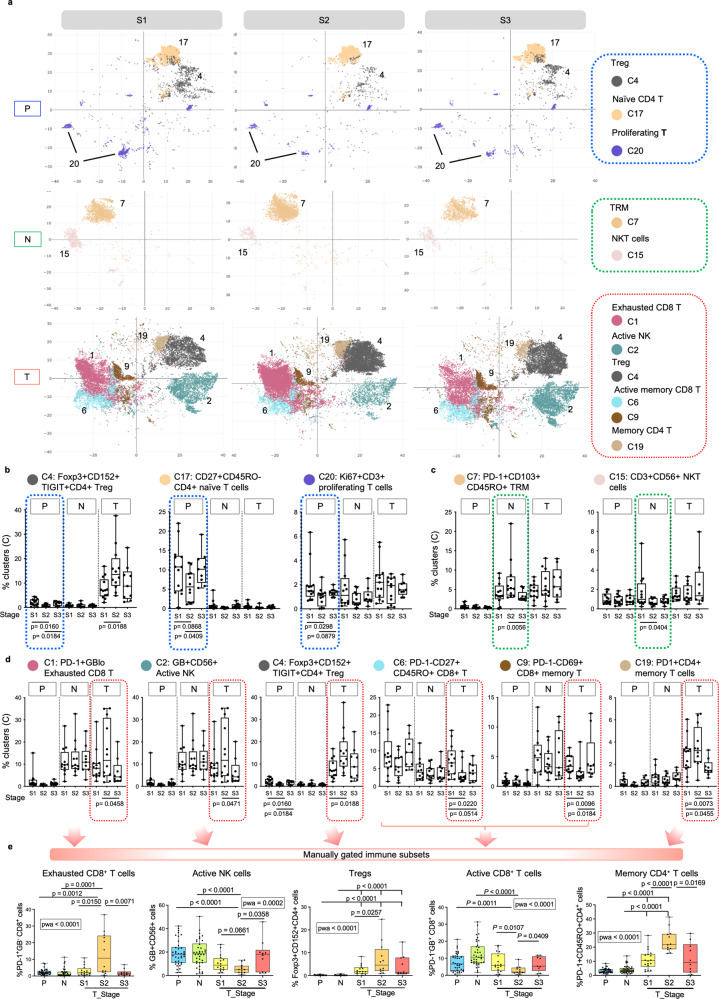
Next, to test if the immune landscapes within the same microenvironment differed among tumour stages, we compared the frequencies of the immune cell clusters among the three tumour stages within each tissue compartment using one-way ANOVA analysis (Supplementary Table 4). In P, Treg (C4), CD4+CD27+CD45RO− naïve T cells (C17) and CD3+Ki67+ proliferating T cells (C20) were less abundant in S2 compared to S1 or S3 (Fig. 2a, b). Whereas in N, increased CD8+PD-1+CD103+CD45RO+ TRM (C7) coupled with reduced CD3+CD56+ natural killer (NK) T cells (C15) were observed in S2 (Fig. 2a, c).
Fig. 2. Peak of immune evasion at stage II HCC.
a tSNE plots showing the distributions of selected immune clusters that are significantly different in abundance from PBMC (P), Non-tumour (N) or tumour (T) compartments across three TNM stages. b Graphs showing percentages of clusters with significant differences across TNM stages in P (blue boxes). c Graphs showing percentages of clusters with significant differences across TNM stages in N (green boxes). d Graphs showing percentages of clusters with significant differences across TNM stages in T (red boxes). e Graphs showing the percentages of manually gated immune populations from total live immune cells (by flowjo) in P, N and T across S1, S2 and S3 HCC. The bar and arrows indicate the major immune cell lineages shown below. a–d TNM stage I, II and III (S1, S2 and S3). nPS1 = 13, nPS2 = 12, nPS3 = 9, nNS1 = 12, nNS2 = 12, nNS3 = 9, nTS1 = 13, nTS2 = 12, nTS3 = 9. b–e Phenotypes are indicated on top. b–e Boxplots show median and the whiskers represent minimum and maximum values with the box edges showing the first and third quartiles. One-way ANOVA test with two-sided p-values by unpaired two-sided Mann–Whitney U (MWU) tests for two-group comparisons. e nPS1-S3 = 38, nNS1-S3 = 38, nTS1 = 16, nTS2 = 12, nTS3 = 10. Source data are provided as a Source Data file.
Tumours tissues exhibited the greatest inter-stage heterogeneity: six clusters showed significant difference in abundance between stages (Supplementary Table 4), particularly at S2 tumours (Fig. 2a). S2 tumours exhibited lower levels of pro-inflammatory or activated immune subsets: including GB+CD16+CD56+ active NK cells (C2, p = 0.0471, S2 vs S3), PD-1−GB+CD45RO+ (C6, p = 0.0220, S1 vs S2), and PD-1−CD69+CD45RO+ (C9, p = 0.009, S1 vs S2; p = 0.0184, S2 vs S3) active memory CD8+ T cells (Fig. 2d). Conversely, the frequencies of potentially exhausted and immunosuppressive clusters: PD-1+GBlowCD45RO+CD8+ exhausted T cells (C1, p = 0.0458, S2 vs S3), Treg (C4, p = 0.0188, S1 vs S2), and PD-1+GBlowCD45RO+CD4+ exhausted memory T cells (C19, p = 0.0073, S2 vs S3) were significantly higher in S2 tumours than in either S1 or S3 (Fig. 2d).
We also validated the above data using another clustering method, FlowSOM17 and found consistent trend of immune changes along tumour progression. Particularly, the immunosuppressive Treg and exhausted CD8+ T cells peaked in S2 tumours, while the active CD8+ T cells and active NK cells showed the lowest frequencies in S2 tumours (Supplementary Fig. 3a, b and Supplementary Table 5). Likewise, we validated the depletion of Treg in S2 PBMC and reduction of NKT cluster in S2 NILs (Supplementary Fig. 3c and Supplementary Table 5).
Manual gating (Supplementary Fig. 4a) validated that PD-1+GB−CD8+ exhausted T cells, Foxp3+CD152+CD4+ Treg, and PD-1+CD45RO+CD4+ memory T cells were enriched; while GB+CD56+ active NK cells and PD-1−GB+CD8+ activated T cells were depleted, specifically in S2 tumours compared to S1 or S3 tumours or both (Fig. 2e). Of note, the PD-1+CD8+ T cells expressed low level of granzyme B (Median 21.85% ± 23.05%) and co-expressed high level of CTLA-4 (Median 66.2% ± 21.71%), Tim3 (Median = 72.4% ± 17.61%) and to a lesser extent Lag3 (Median 36.3% ± 17.07%) (Supplementary Fig. 4b–d), supporting their exhausted phenotypes as also shown by our previous study of exhausted PD-1+CD8+ T cells in HCC18. These data indicate the apparent peak of immune evasion in S2 HCC tumours, followed by a potential partial immune recovery at S3.
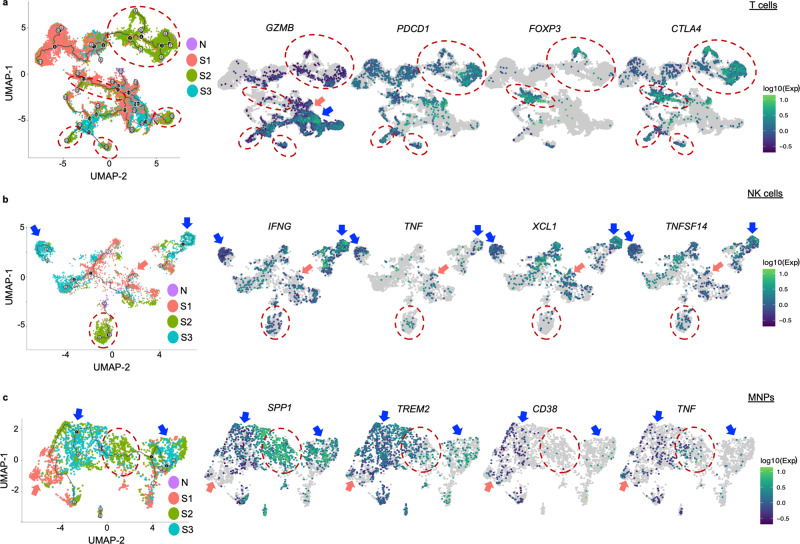
Single-cell immune trajectory along tumour progression
Next, we performed single-cell RNA sequencing on the tumour-infiltrating immune cells as previously described19 and analyzed their pseudotime trajectory along tumour progression using Monocle, an algorithm to learn the sequence of gene expression changes each cell must go through as part of a dynamic biological process20. Setting S1 tumours as the starting point, we first explored the T cells trajectory along tumour progression S1-S3. Consistently, we observed a more distinct immune phenotype at S2 tumours with reduced GZMB (granzyme B) and enriched PDCD1 (PD-1), FOXP3 and CTLA4 compared to N or S1 and S3 tumours (Fig. 3a). A consistent trend was also observed when analysed on CD4+ T and CD8+ T cells separately, where Treg markers and exhaustion markers were found to be enriched in S2 tumours respectively; while the proinflammatory gene GZMB was depleted in S2 tumours (Supplementary Fig. 5a).
Fig. 3. Single-cell pseudotime trajectory analysis of immune cells along HCC progression.
a Pseudotime trajectory ordering of tumour-infiltrating T cells along tumour progression. b Pseudotime trajectory ordering of tumour-infiltrating NK cells along tumour progression. c Pseudotime trajectory ordering of tumour-infiltrating mononuclear phagocytes (MNPs) along tumour progression. a–c Expression level of selected genes are shown. Trajectory is arranged according to the starting point of S1 tumours (n = 7), versus adjacent normal liver tissues (N) (n = 14), S2 tumours (n = 4) and S3 tumours (n = 3). S2 enriched areas are marked by red dotted line circles, and S1 or S3 enriched areas are marked by pink or blue arrows, respectively. .
From the NK cell subsets, proinflammatory cytokines such as IFNG and TNF; chemokine, XCL1, which could promote cDC1 recruitment critical for antitumor immunity;21 as well as TNFSF14, which is important for antitumour function of NK cells via DC maturation;22 were all depleted from S2 tumours but enriched in S1 or S3 tumours (Fig. 3b). In addition, for mononuclear phagocytes (MNPs) (Fig. 3c), we found several genes enriched in S2, such as SPP1, which is highly expressed in tumour-associated macrophages (TAMs) that promotes M2 polarization facilitating immune escape23 and TREM2, which marks the highly immunosuppressive TAMs24. In contrast, CD38, whose expression on myeloid cells predicts favourable prognosis hence its anti-tumoural activity in HCC25 and proinflammatory cytokine TNF, were both reduced in S2 and S3 tumours (Fig. 3c).
Taken together, these data further corroborate a peak of phenotypic immune evasion in S2 HCC tumours.
Tumour co-evolution alongside immune evasion
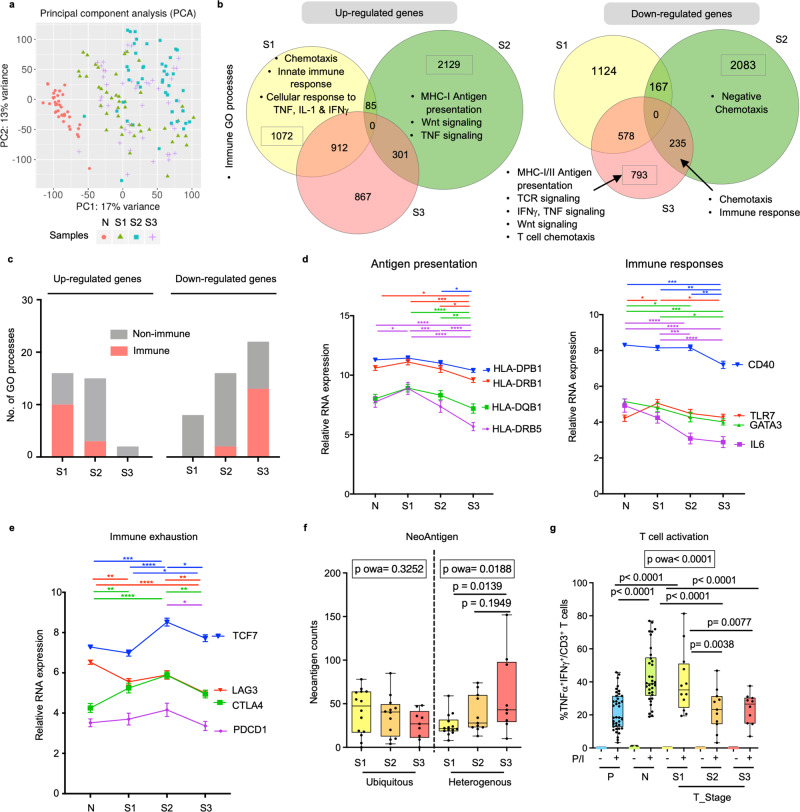
To better understand the onset of immunosuppression in the S2 TME, we performed transcriptomic analysis on parts of the same N and T tissues samples used for CyTOF. Principal component analysis (PCA) revealed that the greatest differences in gene expression were observed between N and T from any stage, while S2 tumours showed the greatest differences from N, S1 or S3 tumours (Fig. 4a), consistent with the most significant immune landscape changes at S2 tumours as shown above.
Fig. 4. Progressive tumour co-evolution along immune evasion at different HCC stages.
a Principal component analysis (PCA) of RNA sequencing profiles reveals sample distribution of HCC tumours from TNM stage I, II and III (S1, S2 and S3) and non-tumour tissues (N). b List of enriched immune-related Gene Ontology (GO) processes among tumours from three TMN stages. c Graphs compare the number of immune or non-immune related GO processes indicated by the upregulated or downregulated genes across different tumour stages (S1, S2 or S3). d Expression levels of genes involved in antigen-presentation (left) and immune responses (right) during tumour progression. e Expression levels of genes involved in immune exhaustion during tumour progression. d, e Two-way Anova test followed by respective unpaired Tukey’s pairwise comparison test colour coded by each gene; *, **, ***, **** denotes two-sided p-values <0.05, <0.01, <0.001 and <0.0001 respectively. Graphs show mean ± standard error of the mean. nN = 37, nS1 = 43, nS2 = 43 and nS3 = 45. f Changes of ubiquitous and heterogenous neoantigen counts across three tumour stages. nTS1 = 14, nTS2 = 12, nTS3 = 9. g Percentages of inflammatory TNFα+IFNγ+ active CD3+ T cells from peripheral blood (P) (n = 38), N (n = 34) and S1 (n = 12), S2 (n = 11) and S3 (n = 10) tumours (T_Stage), upon stimulation with PMA/Ionomycin (P/I) for 5 h. f, g Boxplots show median and the whiskers represent minimum and maximum values with the box edges showing the first and third quartiles. One-way ANOVA test (p owa) with two-sided p-values calculated by Tukey post-hoc multiple pairwise comparisons. Source data are provided as a Source Data file.
Next, we examined the differentially expressed genes (DEGs) in samples of the three tumour stages (Supplementary Fig. 6a). The highest number of upregulated immune-related pathways was found in S1 compared to S2 or S3 tumours (Fig. 4b, c). For instance, S1 tumours were enriched with genes involved in chemotaxis, innate immune response and cellular response to TNF, IL-1 and IFNγ; while S2 tumours were enriched with genes of the Wnt signalling pathway, which was shown to be involved in immune exclusion26 (Fig. 4b and Supplementary Table 6). Conversely, more immune-related pathways were downregulated in S3 tumours compared to S2 or S1 tumours (Fig. 4b, c); particularly those involved in antigen presentation, T cell receptor signalling and chemotaxis (Fig. 4b, c and Supplementary Table 7). We further examined the expression of specific genes involved in these key pathways and found that antigen presentation-related genes such as the HLAs genes and immune response-related genes including CD40, TLR7, GATA3 and IL6 showed progressively reduced expression in advanced compared to early stage tumours (Fig. 4d). Interestingly, we also observed a parabolic expression in genes involved in immune exhaustion (e.g. TCF7, LAG3, CTLA4 and PDCD1) (Fig. 4e), which is consistent with the peak of immune landscape remodelling at S2 tumours as shown by our data above. Of note, TCF7 despite being a key factor in establishing exhausted T cells27, could also present a stem-like memory T cell phenotype associated with response to immunotherapy28,29. We examined TCGA HCC cohort and concluded similar downtrends of these key genes involved in the immune processes along tumour progression (Supplementary Fig. 6b).
We hypothesized that the potential partial immune recovery in S3 tumours (Fig. 2d, e) could be induced by the emergence of neoantigens detected by the immune system11. We quantified neoantigens in tumour samples from each stage using whole-genome sequencing (Methods) and found significantly more heterogenous (occurring in at least one but not all tumour sectors), but not ubiquitous (occurring in all tumour sectors) neoantigens in S3 than S1 tumours (Fig. 4f). This late increase in heterogenous neoantigens, a hallmark of intratumoural heterogeneity, might induce a relative increase in activated immune cell subsets observed in S3 HCC. To understand the functional implications of this partial recovery in S3 HCC, we compared the ability of these immune cells to respond to stimulation in vitro following PMA/Ionomycin stimulation. Significantly lower percentages of CD3 + T cells expressing the inflammatory cytokines IFNγ and TNFα were detected in S2 and S3 tumours than S1 tumours (Fig. 4g and Supplementary Fig. 4e). This indicates that despite the apparent partial-recovery of immunoactive subsets and the increase in heterogenous neoantigen load in late HCC, the overall immunosuppressive TME in S2 tumours likely would render this recovery functionally ineffective and unable to prevent worsening of the disease. Taken together, we showed that the transcriptomic landscape of tumour co-evolved with a progressively immunosuppressive TME.
Immune exclusion with HCC progression
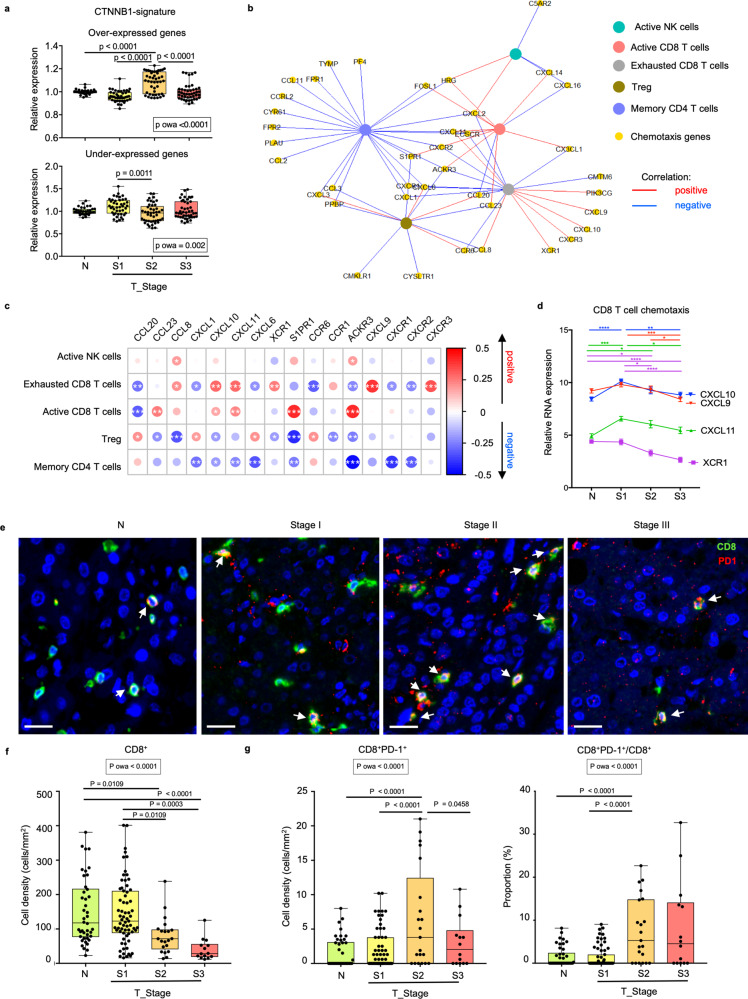
Another possible pathway for maintaining the suppressive TME despite the apparent recovery of active immune subsets seen in advanced tumours is immune exclusion, where the TME turns immunologically “cold”30. This is first implicated by transcriptomic upregulation of Wnt signalling pathway in S2 tumours (Fig. 4b and Supplementary Table 6) or activation of the CTNNB1 pathway that has been linked with immune exclusion in previous studies26,31,32. We next measured the tumour expression of genes that were over- or under-expressed in the previously reported CTNNB1-transcriptomic signature from HCC33 (Supplementary Fig. 7a). Indeed, we found significant regulation in genes involved in this CTNNB-1 signature, particularly in S2 tumours (Fig. 5a). Similar increase was also observed in some of the key Wnt pathway genes such as CTNNB1, AXIN1 and AXIN2 (Supplementary Fig. 7b) suggesting a peak of immune exclusion in S2 tumours.
Fig. 5. Immune exclusion from stage II HCC.
a Expression levels of over-expressed (top) or under-expressed (bottom) genes in beta-catenin (CTNNB1) signature in adjacent non-tumour liver tissues (N) (nN = 37) and tumours from different stages (nS1 = 43, nS2 = 45 and nS3 = 45). b Chemokine genes (yellow circles) versus immune subsets (multiple colours circles) network showing positive (red lines) and negative (blue lines) correlations (Spearman’s correlation test, rho ≥ 0.2, P < 0.05). c Heatmap shows positive (red circles) and negative (blue circled) correlations between 16 selected chemotactic genes (columns) and five immune subsets (rows). Circle sizes represent Spearman’s rho; *, **, ***, **** denotes p-values < 0.05, <0.01, <0.001 and <0.0001 respectively. d Expression levels of genes involved in CD8 T cell related chemotaxis. Two-way ANOVA test followed by respective unpaired Tukey’s pairwise comparison test colour coded by each gene; *, **, ***, **** denotes two-sided p-values < 0.05, <0.01, <0.001 and <0.0001, respectively. Graph shows mean ± standard error of the mean. nN = 37, nS1 = 43, nS2 = 43 and nS3 = 45. e Representative immunohistochemistry (IHC) images of CD8 (green) and PD-1 (red), respectively in N and S1-S3 HCC tumours from an independent validation cohort. CD8+PD-1+ populations are indicated by white arrows. Bar = 20 um. f Boxplots compare densities of CD8+ T cells and g densities of CD8+PD-1+ T cells (left) as well as proportions of PD-1+CD8+/CD8+ T cells (right) by IHC in N and tumours from S1-S3 stages. f, g nN = 42, nS1 = 67, nS2 = 21 and nS3 = 14. a, f and g Boxplots show median and the whiskers represent minimum and maximum values with the box edges showing the first and third quartiles. One-way ANOVA test (p owa) with two-sided p-values calculated by Tukey post-hoc multiple pairwise comparisons. Source data are provided as a Source Data file.
Next, as chemotaxis is known to dictate the recruitment of various immune subsets in the TME5,34, we further examined its relationship with the immune subsets in tumours. We selected the chemotaxis-related DEGs modified across S1–S3 tumours (Supplementary Table 6 and 7) and determined their correlations with the five key immune subsets altered within the tumours during HCC progression (Fig. 2e). In general, chemotactic genes that correlated positively or negatively with CD8+ T cells consistently showed the opposite correlation with Treg or CD4+ memory T cells, suggesting that distinct chemotactic pathways could control CD8 T cell vs immunosuppressive Treg tumour recruitment (Fig. 5b, c). Notably, the chemokines and their corresponding chemokine receptors: CCR1 (receptor for CCL23 and CCL8), CXCR2 (receptor for CXCL1 and CXCL6) and CXCR3 (receptor for CXCL9, CXCL10 and CXCL11) showed a consistent trend in correlation with the key immune subsets (Fig. 5c). More importantly, we observed a progressive downregulation of CD8+ T cell chemotactic genes such as CXCL9, CXCL10, CXCL11 and XCR1 (Fig. 5d), further indicating a progressively “cold” TME.
To validate the above observations with an actual immune exclusion, we performed immunohistochemistry (IHC) on FFPE tissues from an independent HCC cohort (n = 102 patient samples) to detect tumour infiltration of CD8+ or exhausted PD-1+CD8+ T cells. Indeed, we observed a progressive reduction in the density of total CD8+ T cells from S1 to S3 HCC tumours (Fig. 5e, f), consistent with progressive immune exclusion. Concurrently, both the density and proportion of PD-1+ exhausted CD8+ T cells were higher at S2 than S1 or N (Fig. 5g), consistent with the peak of immune exhaustion shown above (Fig. 2d, e). In summary, these data show that immune exhaustion and exclusion were established and peaked at S2 and persisted into S3 tumours promoting tumour progression.
Immune evasion trajectory validated in murine HCC model
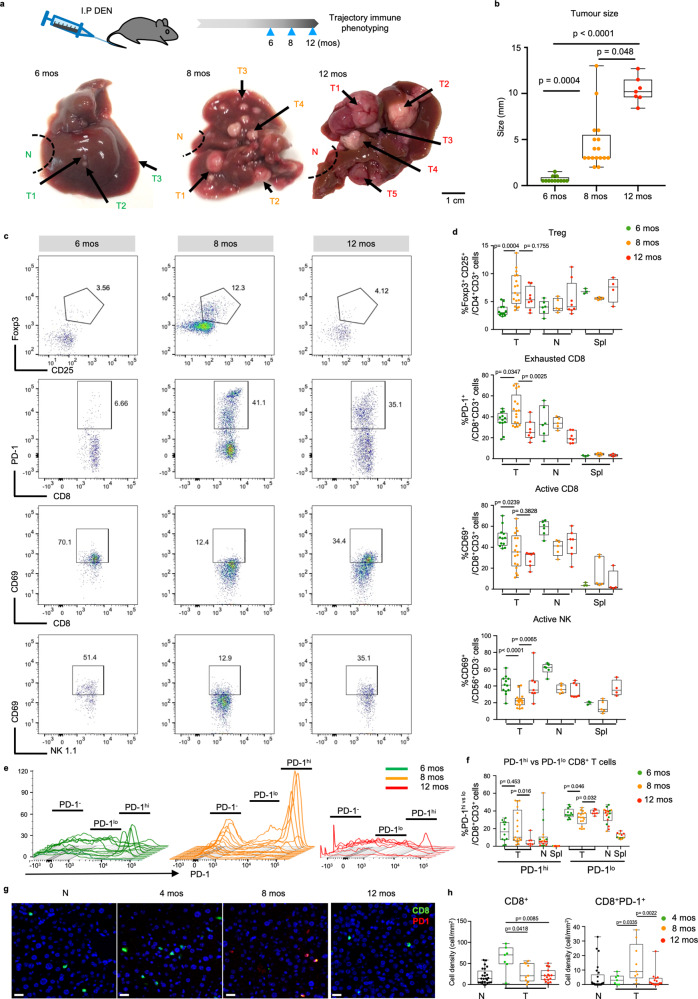
The human HCC data above demonstrated a dynamic pattern of co-evolution between tumour and immune compartments during HCC progression. To further validate the longitudinal process linking these stages, we employed the diethylnitrosamine (DEN)-induced HCC model, which as similar to the human condition, develops HCC from a background of chronic liver inflammation35,36. In these mice, early tumours at 6 months (mos) post DEN-treatment are generally small and monomorphic, while larger tumours emerge from eight to 12 mos post-injection37 (Fig. 6a, b), representing different stages of HCC development. When we examined the TILs from early (6 mos), intermediate (8 mos) and late (12 mos) stage post-DEN tumours from these mice by flow cytometry, we found significantly higher proportions of Foxp3+CD25+CD4+ Treg and PD-1+ exhausted CD8+ T cells, alongside significantly lower proportions of CD69+ active CD8+ T cells and NK 1.1+ NK cells at intermediate stage tumours at 8 mos post DEN induction (Fig. 6c, d and Supplementary Fig. 8a). This supports our human data, where tumours evaded the immune response progressively and peaked at the intermediate tumour stage.
Fig. 6. Validation of immune evasion during tumour progression in murine HCC model.
a Representative images of murine livers bearing multiple tumours (T) from 6, 8 or 12 months (mos) after DEN-induction. Bar = 1 cm. b Tumours from 6, 8 or 12 mos after DEN-induction showing varying tumour size. c Representative dot plots of Foxp3+CD25+ Treg, PD-1+ exhausted CD8 T cells, CD69+PD-1− active CD8 T cells and CD69+NK1.1+ active NK cells in 6, 8 or 12 mos post-DEN tumours. d Comparison of frequencies (%) of Treg, exhausted CD8 T cells, active CD8 T cells and active NK cells in 6, 8 or 12 mos T, adjacent non-tumour liver tissues (N) and spleen (spl). e Histograms showing relative expression of PD-1 further distributed to PD-1hi, PD-1lo and PD-1− CD8+ T cell populations from 6 mos (green lines), 8 mos (orange lines) and 12 mos (red lines) post-DEN induced murine HCC tumours. f Graph showing percentages of PD-1hi and PD-1lo CD8+ T cell populations across 6, 8 or 12 mos post-DEN T, N and spl. g Representative immunohistochemistry (IHC) images of CD8 (green) and PD-1 (red), respectively in N and murine HCC tumours from 4, 8 or 12 mos post-DEN induction. Bar = 20 um. h Boxplots compare densities of CD8+ T cells and CD8+PD-1+ T cells by IHC in N (n = 25) or T from 4 (n = 7), 8 (n = 9) or 12 (n = 15) mos post-DEN induction. b, d, f and h Boxplots show median and the whiskers represent minimum and maximum values with the box edges showing the first and third quartiles. Non-parametric one-way ANOVA Kruskal–Wallis test with two-sided p-values calculated by Dunn’s post-test between groups. b, d and f graphs show data from nT6mos = 12, nT8mos = 17, nT12mos = 7, nN6mos = 6, nN8mos2 = 5, nN12mos = 7, nspl6mos = 3, nspl8mos = 5, nspl12mos = 4 post-DEN murine samples. Source data are provided as a Source Data file.
We also detected PD-1hi, PD-1lo and PD-1− populations among CD8+ T cells with a significant accumulation of the PD-1hi and depletion of PD-1lo population within CD8+ T cells from intermediate 8 mos post-DEN HCC tumours compared to 6 or 12 mos post-DEN tumours (Fig. 6e, f). This again supports the immune evasion peaking at intermediate HCC. As PD-1hi CD8+ T cells are linked with immune exhaustion and are the prime target of immune-checkpoint blockade treatment for HCC38, these data support the potential benefit of immunotherapy in intermediate HCC.
For further validation of immune exclusion on these DEN-induced tumours, we performed IHC on tumour tissues from 4, 8 and 12 mos post-DEN induction and indeed found consistent trend of progressive immune exclusion with decreasing CD8+ T cell density from early to late stages tumours (Fig. 6g, h). Interestingly, the peak of density in CD8+PD-1+ populations occur consistently at intermediate 8 mos post-DEN tumours (Fig. 6h).
Taken together, tumours from this HCC murine model recapitulate our earlier findings in human HCC of an ongoing immune evasion peaking at intermediate tumour stage.
Discussion
In the current study, we have delineated the immune modifications across different tissue compartments at various stages of treatment naïve HCC, thereby uncovering strategic points for therapeutic intervention and advancing our understanding of the complex interplay between tumour cells and the immune system. In contrast to the current dogma that immune suppression/evasion occurs either early during carcinogenesis or late prior to metastasis, we found that in HCC, it is a progressive and continual process that peaks at intermediate stage II tumours. This peak is characterised by relatively higher frequencies of exhausted and suppressive immune subsets and reduced frequencies of activated immune subsets; concurrently, the immune landscape turns “cold”, with reduced numbers of infiltrating CD8+ T cells, rendering tumours refractory to immune-mediated suppression. This immune evasion could be due to the modifications in the tumour transcriptomic landscape, which concurrently mirrored the immune microenvironment, with lower expression of genes involved in several immune-related pathways as well as upregulation of CTNNB1-related genes associated to immune exclusion, both occurred at S2 HCC tumours.
Interestingly, we found potential evidence of partial immune recovery in stage III tumours corresponding with an increase in neoantigens, which have previously been shown to induce immune responses in the TME39. However, given the fact that most advanced HCC patients may not undergo resection as first-line therapy, these interesting findings will require careful interpretation against this inevitable confounding factor. Furthermore, the small recovery of immune cell subsets in stage III HCC tumours was coupled with overall lower immune activation status as well as immune exclusion: thus, the resurgence of immunosurveillance at this stage may not significantly impact disease progression. Nevertheless, these findings raise the possibility that immunotherapy, if timed carefully to exploit neoantigen exposure/immune resurgence in stage III tumours, could be a potentially powerful intervention in late-stage HCC, supported also by the efficacy of immune checkpoint blockade in advanced HCC40,41. The efficacy of immune checkpoint inhibitors, as well as Wnt inhibitors, remain an interesting area for further research.
In colorectal cancer, a similar inverse relationship between tumour-infiltrating CD8+ T cells and advanced tumours was observed42. In contrast, an in silico transcriptomic study in HCC failed to detect significant changes in immune composition across the various tumour stages43. The discrepancy in the later study might originate from technical differences between bulk transcriptomic deconvolution used in this previous study versus the comprehensive single-cell immunoprofiling performed in the current study. On the other hand, the concept of tumour evolution and the important role of immune contexture in immunosurveillance or immunoediting have been discussed elsewhere44. Immune evolution in HCC was proposed in our recent study to be manifested as immune intratumoural heterogeneity (immune-ITH)14. In addition, single-cell transcriptomic analysis of TILs in HCC also revealed immune evasion with T cell exhaustion45, myeloid/lymphoid cross-interaction46 and onco-foetal reprogramming of endothelial cells and macrophages19. In this study, we revealed an unprecedented progressive immune evolution alongside tumour progression peaking at the intermediate stage of HCC.
Longitudinal studies of tumour progression and its relationship to the immune landscape in human colorectal and lung cancer do suggest that evolution of these tumours is immune-related or immune-driven10,12, at least at the very early or very late disease stages captured in these studies. Similar to other human solid cancer studies, our current study has been limited to inferring the parallel cancer and immune evolution using specimens taken from different patients at various stages of tumour11,47,48. To demonstrate the trajectory of tumour landscapes evolution, both scRNA pseudotime trajectory prediction on tumour-infiltrating immune cells as well as early to late stages DEN-induced HCC model validated our discovery on a peak of phenotypic immune evasion at the intermediate tumour stage and a progressively immune excluded TME during tumour progression.
To conclude, our in-depth analysis of intratumoural immune dynamics during HCC progression has profound implications for our understanding of HCC evolution and will help guide the development of potential immunotherapeutic treatments.
Methods
Patient samples
The study was approved by the Central Institution Review Board (CIRB) of SingHealth, of which all National Cancer Centre Singapore, Singapore General Hospital and National University Hospital were constituent members (CIRB Ref: 2016/2626 and 2018/2112). Tumour (T), adjacent non-tumour liver tissue (N) and peripheral blood (P) were collected from 38 HCC patients, each provided written informed consent, with demographics and clinical characteristics as described in (Supplementary Table 1). For each patient, a single tumour slice was further dissected into 2 to 5 sectors (depending on the size of the tumour) with at least 1 cm separation between them. Adjacent non-tumour liver tissue from at least 2 cm away from the tumour was also harvested. Each tissue sector was divided for analysis by CyTOF, RNA and whole genome sequencing. Peripheral blood mononuclear cells (PBMC) (n = 38) were isolated by Ficoll-Paque Plus (GE Healthcare) density gradient centrifugation. Tumour-infiltrating leucocytes (TIL) (n = 135) and non-tumour tissue-infiltrating leucocytes (NIL) (n = 38) were isolated from each tissue separately using enzymatic digestion with 500 μg/mL collagenase IV (Thermo Fisher Scientific; Cat#: 17104019) and 50 μg/mL DNase I (Roche, Indianapolis, IN; Cat#: 4716728001) for 30 min in 37 °C. Cells were stored in liquid nitrogen with 10% DMSO in foetal bovine serum (FBS) until further analysis by CyTOF.
Cytometry by time of flight (CyTOF)
TILs, NILs and PBMCs were thawed and rested for at least 30 min in RPMI medium with 10% FBS and 1% penicillin/streptomycin. Only samples with >100,000 live cells after thawing will be subjected to downstream CyTOF analysis (note: only two tumour sectors with insufficient cell numbers were excluded from the entire cohort). Data was collected from at least two sectors of each tumour in this study. The cells were unstimulated or stimulated with 150 ng/mL PMA (phorbol myristate acetate; Sigma) and 100 ng/mL ionomycin (Sigma) and incubated for a total of 5 h in 37 °C and 5% CO2 and 3 μg/mL Brefeldin A (eBiosience) and 2 μM monesin (BioLegend) was added at the last 3.5 h of the incubation. The cells were then washed and stained with cisplatin-Live/Dead stain (Fludigm) before labelling with a combination of barcoding anti-CD45 antibody conjugated to three different metals, for simultaneous analysis of multiple samples49. Cells were then combined and labelled with metal-conjugated antibodies targeting surface markers before fixing with 1.6% paraformaldehyde, permeabilizing with 100% methanol and staining with metal-conjugated antibodies targeting intracellular molecules. All 33 antibodies and barcoding anti-CD45 antibodies were either conjugated in-house according to the manufacturer’s instructions (Fluidigm) or purchased pre-conjugated directly from Fluidigm (Supplementary Table 2). Finally, an iridium-containing DNA intercalator was added to identify single cells and the cells were washed and diluted with EQ Four Element Calibration Beads (Fluidigm) for signal normalization according to manufacturer’s instructions50, before data were acquired using Helios equipped with the CyTOF® 6.7 system control software (Fluidigm).
The generated files were analysed by FlowJo (v.10.2; FlowJo): live single-cells (cisplatin-negative and DNA-intercalator-positive) were debarcoded to each sample file based on their unique CD45 barcodes. Each file was then down-sampled to 10,000 cells and further analysed using our in-house Extended Polydimensional Immunome Characterization (EPIC) analysis pipeline with data visualization using the browser-based R shiny app ‘SciAtlasMiner’16. We also performed batch normalization, data visualization and statistical tests16. Clustering was performed using the Phenograph algorithm (v1.5.2)15 or FlowSOM algorithm17 and dimension reduction using tSNE. To compare the frequencies of cell populations between tissue types and clinical HCC stages, we applied one way ANOVA test with post-pairwise Mann–Whitney U-test. Of note, we averaged cluster frequencies from multiple tumour sectors to represent each patient for all statistical tests. Validation using manual gating was performed with FlowJo (v.10.2). Immune diversity was quantified by calculating the multivariate beta-dispersion of the Bray–Curtis distances between immune clusters for each tissue type using the ‘vegan’ R package v2.5. The beta-dispersion were compared among tissues and p-values were determined using Tukey’s HSD (honestly significant difference) test.
Single-cell RNA sequencing
Single cells were isolated from tumour and adjacent non-tumour tissues from 14 patients with S1 (n = 7), S2 (n = 4) and S3 (n = 3) HCC, followed the same enzymatic digestion as described previously19. Briefly, dead cells were removed using dead cell removal kit (Miltenyi, Cat#: 130-090-101). CD45+ cells were enriched using CD45 MicroBeads (Miltenyi, Cat#: 130-045-801) before CD45+ and CD45− cells were processed using the Chromium Single Cell 30 (v2 Chemistry) platform (10x Genomics, Pleasanton, CA) as previously described19. All data were then aggregated using cellranger aggr by normalizing all runs to the same sequencing depth. Downstream analysis was performed using Scanpy (version 1.4; all genes expressed by a minimum of 30 cells were considered and cells with <200 genes and >5% mitochondrial content were excluded from analysis). For clustering, best matched k-Nearest Neighbour is automatically weighted by the algorithm to compute the best UMAP topology (scanpy.api.tl.umap, minimum distance between 0.3 and 0.5). The Louvain method (scanpy.api.tl.louvain) is then used to detect a community of similar cells resolution parameter 0.6 to 1 as illustrated previously19. Pseudotime trajectory analysis of tumour-infiltrating immune cells were performed using the Monocle R package (version 3.0)20, on T cells (further sub-divided to CD3+CD8- and CD3+CD8+ T cells), NK cells and MNPs clusters. A Cell Data Set object was created according to Monocle’s pipeline. Log-normalisation and scaling was done using Monocle’s pre-processing function. Principle Component Analysis (PCA), dimension reduction and clustering were performed using the default parameters. Pseudotime trajectories were plotted for each partition using Monocle’s reversed graph embedding algorithm. For pseudotime ordering, cells from the Stage 1 (S1) tumour samples were taken as the starting point.
Immunohistochemistry (IHC) tissue staining
Formalin-fixed paraffin-embedded (FFPE) sections were from an independent cohort of 102 HCC patients who underwent curative resection from 1991 to 2009, and were obtained from the Department of Anatomical Pathology, Division of Pathology, Singapore General Hospital. Two cores of tumour and non-tumour FFPE tissues from each patient were stained with OpalTM Multiplex immunohistochemistry Detection Kit and images were acquired using Vectra 3.0 pathology imaging system microscope (Perkin Elmer) according to manufacturer’s instructions. We used antibodies: anti-CD8 (DAKO, Clone:C8/144B, 1:200) and anti-PD-1 (Abcam, Clone: NAT105, 1:100) and detection dye: Opal690 dye (CD8) and Opal650 dye (PD-1) with DAPI as nuclear counterstain. Quantification of stained cells was performed on the whole 1 mm core (Area = 0.785 mm2) and the average values of two cores from each patient sample was calculated and shown as the number of cells/mm2. Separately, IHC staining was done on FFPE tissues from DEN-induced mice (See description below). Adjacent non-tumour (N) and tumour samples from 4 mos (n = 7), 8 mos (n = 9) and 10 mos (n = 9) post-DEN mice were stained with Anti-CD8 (Abcam, Catalogue #ab203035; polyclonal; 1:500), anti-PD-1 (Abcam; clone EPR20665; 1:1000) and detection dye: Opal650 dye (CD8) and Opal620 dye (PD-1) with DAPI as nuclear counterstain. Quantification was performed and analyzed from at least 2–6 (limited by tumour size) random 0.6 × 0.5 mm2 regions.
Bulk RNA sequencing
Total RNAs from multiple tumour sectors and each non-tumour sector were isolated using Picopure RNA-Isolation kit (Arcuturus, Ambion), and cDNA was generated using the SMART-Seq® v4 UltraTM Low Input RNA Kit for Sequencing (Clontech, USA). Illumina indexed libraries were created using the Nextera XT DNA Library Prep Kit (Illumina, USA) and multiplexed for 2×101 bp-sequencing. RNA sequencing was performed on a HiSeq High output platform at the Genome Institute of Singapore (GIS).
The raw reads were aligned via STAR51 to the Human Reference Genome hg19, and the gene-level expected counts were calculated using RSEM52. Only protein-coding genes with at least one count per million reads in 5% or more of the samples were retained, and the data were normalized using DEseq253. Differential expression analysis was performed using R package Limma54 with the false discovery rate (FDR) adjusted for multiple testing using Benjamini–Hochberg procedure55. Differentially expressed genes (DEGs) were identified at an FDR < 0.05 with pairwise comparison. Pathway enrichment analyses were performed using integrated Differential Expression and Pathway analysis (iDEP)56 v0.91 and DAVID pathway analysis v.6.8, both with adjusted p-value < 0.01. For validation of selected genes of interest, the raw counts for the Liver HCC, The Cancer Genome Atlas (TCGA) dataset were downloaded from FireBrowse57. Only protein-coding genes with raw counts > 1 (TCGA dataset) in ≥5 samples were retained and data were normalized using DEseq2 (n = 297: S1, n = 151; S2, n = 75 and S3 n = 71).
Genes over- or under-expressed in CTNNB1-mutation signature were previously reported in HCC33, and the relative expression levels of these genes which were calculated for each tumour according to the median expression of each gene. The expression levels of genes of interest were compared among three tumour stages using one-way ANOVA test with post-hoc Tukey’s pairwise test. Correlation between genes and immune subsets were computed with Spearman’s rho and p-values and visualized using R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84) and “network”: Butts C (2020).
Whole genome sequencing and neoantigen prediction
DNA from tumour and adjacent non-tumour tissues was extracted using Qiagen AllPrep kit and sonicated into shorter fragments using the Covaris system. Quality check was performed using Agilent 2100 Bioanalyser. DNA fragments were end-repaired, ligated, amplified, and sequenced using the Illumina sequencing platform at GIS. Raw reads were mapped to the Human Reference Genome hg19 using the Burrows–Wheeler Aligner58. After removal of duplicated reads by PICARD (http://broadinstitute.github.io/picard/), base quality recalibration and realignment were performed using the Genome Analysis Tool Kit59. Comparison of tumour against non-tumour tissue was performed using Mutect (version 1.1.7)60 to call for somatic variants.
Neoantigen prediction was achieved using personalized Variant Antigens by Cancer Sequencing(pVacSeq)61, with variant calling information obtained from MuTect60. The variant calls were annotated using VEP: 8- to 11-mer epitopes with <500 nM predicted binding affinity to MHC-class 1. The ubiquitous (expressed by all tumour sectors) and heterogenous (expressed by at least one but not all sectors) neoantigens as defined previously62 were computed.
DEN-induced mouse model of HCC
The animal procedure was approved by and conducted in accordance with the regulations and guidelines of the Institutional Animal Care and Use Committee (IACUC) of the National University of Singapore and SingHealth Institute (IACUC reference number: R019-0666 and 2016/SHS/1228). HCC was induced by intraperitoneal injection of a single dose of diethylnitrosamine (DEN) (Sigma-Aldrich) at 25 mg per Kg body weight to 15 days old wild-type male C57BL/6 J mice, consistent with previously published method37. Mice were weaned at 21 days of age and maintained on the Chow diet and 12 h light/12 h dark cycles at 23 °C co-housed in pathogen-free animal facilities. Mice were euthanized by carbon dioxide-induced asphyxia. Tumours of various sizes corresponding to different stages of development were harvested at 4 or 6 mos (early stage), 8 mos (intermediate stage) and 12 mos (late stage) post DEN induction. Immune cells isolated from tumours, adjacent non-tumour liver tissues and spleens were analysed with flow cytometry (as detailed below). Additionally, FFPE tissues from adjacent non-tumour and tumour specimens were separately analysed with IHC (as detailed above). Tumours showed varying sizes with the median diameters as 3 mm.
Flow cytometry
Immune cells were isolated from mouse liver tumours or healthy livers by digestion with 500μg/mL collagenase IV (Thermo Fisher Scientific) and 50 μg/mL DNase I (Roche, Indianapolis, IN) and spleen from mechanical isolation. The isolated cells were analysed using antibodies listed in Supplementary Table 8. All flow cytometry analyses were done on a BD Fortessa flow cytometer equipped with five lasers (BD Biosciences, San Jose, CA), and the data were processed using FlowJo (v.10.2).
Statistics
Statistical comparisons of cell frequencies between groups were performed using the parametric one-way ANOVA test (OWA) with Tukey post-hoc multiple comparison test (human data) or unpaired two-sided Mann–Whitney U (MWU) tests for pairwise comparisons and non-parametric one-way ANOVA Kruskal–Wallis test with Dunn’s post-hoc multiple comparison test (murine data), both with two-sided p-values, in GraphPad Prism (V7.0d). The edgeR package63 was used for differential expression analysis of the RNA-seq data. Spearman’s correlation analysis was performed with the rho and p-values reported.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors would like to thank all members of TII, all participating patients, the clinical research coordinators from NCCS, SGH, NUHS for their contributions and Dr Lucy Robinson of Insight Editing London for scientific and language editing of this manuscript. This work was supported by the National Medical Research Council (NMRC), Singapore (ref numbers: NMRC/TCR/015-NCC/2016, NMRC/CIRG/1460/2016, NMRC/CSA-SI/0013/2017, NMRC/CSA-SI/0018/2017, NMRC/OFLCG/003/2018, NMRC/STaR/020/2013, NMRC/CG/M003/2017, LCG17MAY004 and NMRC/OFIRG/0064/2017) and National Research Foundation, Singapore (ref number: NRF-NRFF2015-04).
Source data
Author contributions
P.H.D.N. and M.W. obtained and analysed data and prepared the paper; C.T.T. performed the mouse experiments, analyzed the data and discussed the paper. C.J.L. processed the samples, performed flow cytometry and IHC experiments. H.H.L.L. C.Z.J.P. and S.M. analysed the genomic/transcriptomic data. J.J.W.S. and R.D. obtained and analyzed the scRNA seq data. J.Ya. performed the mouse experiments. S.D.S. processed the samples, performed flow cytometry and IHC experiments. W.L.T. provided genomic/transcriptomic analysis support. T.K.H.L., J.Ye., W.Q.L., Y.H.P., G. S., T.J.L. and W.K.W., prepared and provided tissue samples and discussed the data. C.Y.C., P.C.C., H.C.T., A.K., Y.Y.D. J.H.K., S.I., K.M., A.C., G.K.B. and B.K.P.G. recruited patients, provided samples and discussed the data. N.F. and V.C.Y. provided support for mouse model work. W.Z. designed and led the genomics analysis and discussed the data. S.A. designed the CyTOF pipeline and discussed the data. P.K.H.C. initiated the liver cancer programme, provided patient samples and discussed the data; V.C. designed and led the study, performed the analysis and prepared the paper.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The WGS and RNA-seq data generated in this study are deposited in European Genome-phenome Archive (EGA) under the accession code: EGAS00001003814. Single-cell RNA sequencing data were deposited under the accession code GSE156625. The remaining data are available within the Article, Supplementary Information or source data provided with this paper. Source data are provided with this paper.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Phuong H.D. Nguyen, Martin Wasser.
Contributor Information
Salvatore Albani, Email: salvo@duke-nus.edu.sg.
Pierce K. H. Chow, Email: pierce.chow@duke-nus.edu.sg
Valerie Chew, Email: valerie.chew@duke-nus.edu.sg.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-29122-w.
References
- 1.Yang JD, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J. Immunol. Res. 2014;2014:149185. doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galun E. Liver inflammation and cancer: the role of tissue microenvironment in generating the tumor-promoting niche (TPN) in the development of hepatocellular carcinoma. Hepatology. 2016;63:354–356. doi: 10.1002/hep.28344. [DOI] [PubMed] [Google Scholar]
- 4.Ray K. Liver cancer: a complex interplay between inflammation and immunity in liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2018;15:3. doi: 10.1038/nrgastro.2017.165. [DOI] [PubMed] [Google Scholar]
- 5.Chew V, et al. Chemokine-driven lymphocyte infiltration: an early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut. 2012;61:427–438. doi: 10.1136/gutjnl-2011-300509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chew V, et al. Inflammatory tumour microenvironment is associated with superior survival in hepatocellular carcinoma patients. J. Hepatol. 2010;52:370–379. doi: 10.1016/j.jhep.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Hou J, Zhang H, Sun B, Karin M. The immunobiology of hepatocellular carcinoma in humans and mice: Basic concepts and therapeutic implications. J. Hepatol. 2020;72:167–182. doi: 10.1016/j.jhep.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Garnelo M, et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66:342–351. doi: 10.1136/gutjnl-2015-310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J, et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology. 2007;132:2328–2339. doi: 10.1053/j.gastro.2007.03.102. [DOI] [PubMed] [Google Scholar]
- 10.Mascaux, C. et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature10.1038/s41586-019-1330-0 (2019). [DOI] [PubMed]
- 11.Rosenthal, R. et al. Neoantigen-directed immune escape in lung cancer evolution. Nature10.1038/s41586-019-1032-7 (2019). [DOI] [PMC free article] [PubMed]
- 12.Angelova M, et al. Evolution of metastases in space and time under immune selection. Cell. 2018;175:751–765 e716. doi: 10.1016/j.cell.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, L. Y. et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut10.1136/gutjnl-2019-318912 (2019). [DOI] [PMC free article] [PubMed]
- 14.Nguyen PHD, et al. Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. Nat. Commun. 2021;12:227. doi: 10.1038/s41467-020-20171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine JH, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeo, J. G. et al. The Extended Polydimensional Immunome Characterization (EPIC) web-based reference and discovery tool for cytometry data. Nat Biotechnol.10.1038/s41587-020-0532-1 (2020). [DOI] [PubMed]
- 17.Van Gassen S, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytom. A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 18.Chew V, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc. Natl Acad. Sci. USA. 2017;114:E5900–E5909. doi: 10.1073/pnas.1706559114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A, et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183:377–394 e321. doi: 10.1016/j.cell.2020.08.040. [DOI] [PubMed] [Google Scholar]
- 20.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bottcher JP, et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell. 2018;172:1022–1037 e1014. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes TD, et al. Licensed human natural killer cells aid dendritic cell maturation via TNFSF14/LIGHT. Proc. Natl. Acad. Sci. USA. 2014;111:E5688–E5696. doi: 10.1073/pnas.1411072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp. Cell Res. 2017;359:449–457. doi: 10.1016/j.yexcr.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 24.Katzenelenbogen Y, et al. Coupled scRNA-Seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell. 2020;182:872–885 e819. doi: 10.1016/j.cell.2020.06.032. [DOI] [PubMed] [Google Scholar]
- 25.Lam JH, et al. Expression of CD38 on macrophages predicts improved prognosis in hepatocellular carcinoma. Front. Immunol. 2019;10:2093. doi: 10.3389/fimmu.2019.02093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/beta-catenin pathway activation correlates with immune exclusion across human cancers. Clin. Cancer Res. 2019;25:3074–3083. doi: 10.1158/1078-0432.CCR-18-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, et al. TCF-1-Centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;51:840–855 e845. doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiqui I, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50:195–211 e110. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 29.Krishna S, et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science. 2020;370:1328–1334. doi: 10.1126/science.abb9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348:74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 31.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 32.Sia D, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology. 2017;153:812–826. doi: 10.1053/j.gastro.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Lachenmayer A, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin. Cancer Res. 2012;18:4997–5007. doi: 10.1158/1078-0432.CCR-11-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat. Rev. Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friemel J, et al. Characterization of HCC mouse models: towards an etiology-oriented subtyping approach. Mol. Cancer Res. 2019;17:1493–1502. doi: 10.1158/1541-7786.MCR-18-1045. [DOI] [PubMed] [Google Scholar]
- 36.Schneider C, et al. Adaptive immunity suppresses formation and progression of diethylnitrosamine-induced liver cancer. Gut. 2012;61:1733–1743. doi: 10.1136/gutjnl-2011-301116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naugler WE, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 38.Kim HD, et al. Association between expression level of PD1 by tumor-infiltrating CD8(+) T cells and features of hepatocellular carcinoma. Gastroenterology. 2018;155:1936–1950 e1917. doi: 10.1053/j.gastro.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 39.Yarchoan M, Johnson BA, 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer. 2017;17:209–222. doi: 10.1038/nrc.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yau T, et al. Nivolumab (NIVO) + ipilimumab (IPI) combination therapy in patients (pts) with advanced hepatocellular carcinoma (aHCC): results from CheckMate 040. J. Clin. Oncol. 2019;37:4012–4012. [Google Scholar]
- 41.Zhu AX, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940–952. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 42.Mlecnik B, et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J. Clin. Oncol. 2011;29:610–618. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 43.Foerster F, et al. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci. Rep. 2018;8:5351. doi: 10.1038/s41598-018-21937-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity. 2020;52:55–81. doi: 10.1016/j.immuni.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 45.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356 e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845 e820. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGranahan N, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271 e1211. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai L, Ong R, Li J, Albani S. A CD45-based barcoding approach to multiplex mass-cytometry (CyTOF) Cytom. A. 2015;87:369–374. doi: 10.1002/cyto.a.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytom. A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth, G. K. Limma: Linear Models for Microarray Data (Springer, 2005).
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995;57:289–300. [Google Scholar]
- 56.Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinforma. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cancer Genome Atlas Research N, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hundal J, et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WGS and RNA-seq data generated in this study are deposited in European Genome-phenome Archive (EGA) under the accession code: EGAS00001003814. Single-cell RNA sequencing data were deposited under the accession code GSE156625. The remaining data are available within the Article, Supplementary Information or source data provided with this paper. Source data are provided with this paper.