Abstract
Renal fibrosis is the common and final pathological process of kidney diseases. As a dynamic and reversible post-translational modification, SUMOylation and deSUMOylation of transcriptional factors and key mediators significantly affect the development of renal fibrosis. Recent advances suggest that SUMOylation functions as the promising intervening target against renal fibrosis, and natural products prevent renal fibrosis via modulating SUMOylation. Here, we introduce the mechanism of SUMOylation in renal fibrosis and therapeutic effects of natural products. This process starts by summarizing the key mediators and enzymes during SUMOylation and deSUMOylation and its regulation role in transcriptional factors and key mediators in renal fibrosis, then linking the mechanism findings of SUMOylation and natural products to develop novel therapeutic candidates for treating renal fibrosis, and concludes by commenting on promising therapeutic targets and candidate natural products in renal fibrosis via modulating SUMOylation, which highlights modulating SUMOylation as a promising strategy for natural products against renal fibrosis.
Keywords: renal fibrosis, sumoylation, SUMO-specific protease, natural products, NF-κB
1 Introduction
Epigenetic modification is dynamic and responses to environmental influences, including chromatin remodeling proteins and histone post-translational modifications (PTM). SUMOylation is a PTM process. SUMOylation shares similar reaction scheme and enzyme classes, but the conjugation involves in small ubiquitin-like modifiers (SUMOs) rather than ubiquitin. Emerging evidences support SUMOylation as a promising therapeutic target for renal fibrosis through modulating transcription factor and key mediator. NR5A2 is a key mediator on the transcriptional regulation of the calreticulin gene, and its SUMOylation exacerbates renal fibrosis in the unilateral ureteric obstruction model (Arvaniti et al., 2016). In addition, SUMOylation of Samd4 by SUMO2/3 activates TGF-β/Smad signaling and then upregulates the expression of fibronectin in high glucose induced renal mesangial cells (Zhou et al., 2014). Here, we introduce the mechanism of SUMOylation in renal fibrosis and therapeutic effects of natural products. This process starts by summarizing the key mediators and enzymes during SUMOylation and deSUMOylation and its regulation role in transcriptional factors and key mediators in renal fibrosis, then linking the mechanism findings of SUMOylation and natural products to develop novel therapeutic candidates for treating renal fibrosis, and concludes by commenting on promising therapeutic targets and candidate natural products in renal fibrosis via modulating SUMOylation.
2 SUMOylation
2.1 SUMO Proteins
SUMO proteins are small acidic proteins with distant homology to ubiquitin. At primary amino acid sequence, they are 10–20% identical to ubiquitin. On a structural level, and their relatedness is much more pronounced. As shown in Figure 1, ubiquitin and SUMO share the classical ubiquitin superfold. Characteristic for all members of the SUMO family and absent from ubiquitin or other ubiquitin-related proteins, is the N-terminal flexible extension of 10–30 amino acids. The function of this extension remains however currently unknown.
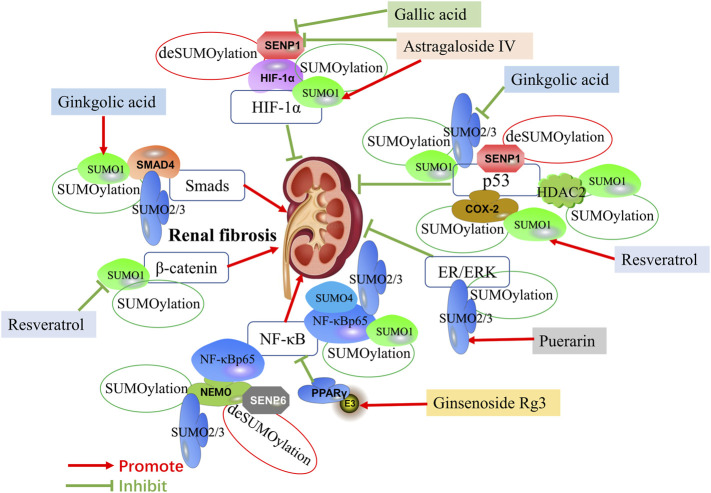
FIGURE 1.
Mechanisms of SUMOylation and deSUMOylation by transcription factors in renal fibrosis and therapeutic targets of natural products against renal fibrosis via SUMOylation. Mechanisms of SUMOylation and deSUMOylation by transcription factors in renal fibrosis are including TGF-β/Smad signaling, HIF-1α signaling, p53 pathway, ER/ERK pathway, NF-κB signaling, and β-catenin pathway. Smad4 is SUMOylated by SUMO1 and SUMO2/3. HIF-1α is SUMOylated by SUMO1. P53 is SUMOylated by SUMO1 and SUMO2/3, and also is upregulate by HDAC2 and COX-2 SUMOylated by SUMO1. ER/ERK is SUMOylated by SUMO2/3. NF-κBp65 is SUMOylated by SUMO1, SUMO2/3 and SUMO4, and also is upregulate by NEMO SUMOylated by SUMO2/3. PPARγ activated by SUMO ligase E3 could inhibit NF-κB signaling. β-catenin is SUMOylated by SUMO1. SNEP1 deSUMOylates HIF-1α and p53. Additionally, SNEP6 deSUMOylates NEMO. Ginkgolic acid promotes the expression of Smad4 by upregulating SUMO1, while inhibits the expression of p53 by downregulating SUMO2/3. Astragaloside IV promotes the expression of HIF-1α by upregulating SUMO1 and downregulating SENP1. Gallic acid also promotes the expression of HIF-1α by downregulating SENP1. Resveratrol induces SUMOylated COX-2 by SUMO1, thus enhancing the expression of p53. In addition, Resveratrol inhibits the expression of β-catenin by downregulating SUMO1. Puerarin activates ER/ERK pathway by upregulating SUMO2/3. Ginsenoside Rg3 inhibits NF-κB by upregulating PPARγ via activating E3.
As a type of PTMs, SUMOylation are widely existed in eukaryotes. SUMOs modification mediates the protein–protein interactions of target substrates, and affects their enzymatic function, subcellular localization, and stability (Kukkula et al., 2021). SUMO proteins covalently bind to their target proteins at lysine (k) residues. To date, five SUMO proteins (SUMO1–5) have been found in human genome harbors genes (Enserink, 2015). Due to the sequence of SUMO2 and SUMO3 is very similar, they are commonly known as SUMO2/3 (Saitoh and Hinchey, 2000). Although SUMO2/3 and SUMO1 share 50% sequence homology, significant differences have been shown in function and location. Interestingly, SUMO1 knockout mice are survivable because SUMO2/3 can compensate for most functions of SUMO1 (Zhang et al., 2008; Wang et al., 2014a). SUMO1 and SUMO2/3 are common, while SUMO4 is only expressed in few organs, such as spleen and kidney (Du et al., 2021). SUMO4 is associated with the pathogenesis of type I diabetes mellitus. SUMO5 is first discovered in promyelocytic leukemia, which mediates the growth and disruption of promyelocytic leukemia nuclear bodies (Liang et al., 2016). However, whether SUMO5 is expressed at the protein level remains controversial.
2.2 SUMOylation and deSUMOylation Process
The SUMOylation starts from precursor synthesis, hydrolytic activation to covalent binding to the substrate protein that relies on the synergistic effect of three enzymes: E1 (SUMO-activating enzyme); E2 (SUMO- transferring enzyme); and E3 (SUMO ligase) (Yeh, 2009). First, SUMOs are activated by E1, and the C-terminal glycine is connected to the cysteine residue of E1 through a thiolipid bond. Thus, its group is adenylated to provide ATP energy and completes the activation of SUMOs. Then, SUMOs are transferred to the cysteine residues of E2, which can directly recognize the substrate, and finally couple SUMO to the lysine of the substrate protein through an isopeptide bond residues. There are three main types of E3: activated STAT protein inhibits protein inhibitor of activated STAT (PIAS) family members, Pc2 and Ran BP2. It can enhance the efficiency and specificity of E2 transferring SUMOs to substrate proteins (K et al., 2021).
As a highly regulated and reversible process, SUMOylation is regulated by a series of proteases (Wilkinson and Henley, 2010). As one of the key proteases, SUMO-specific proteases (SENPs) contain seven types (SENP1–3 and SENP5–8). SENP1/2 prefers SUMO1–3 as the substrate (Wilkinson and Henley, 2010), while SENP3/5 and SENP6/7 have broad specificity for SUMO2/3 (Mukhopadhyay and Dasso, 2007), and SENP8 does not act on SUMO protein (Kunz et al., 2018). Under the action of SENPs, SUMOs are cleaved from the substrate protein, and the SUMOs precursor is processed into mature SUMOs re-enter the new SUMO cycle.
The abnormal SUMOylation usually leads to disorders of function of target proteins, and causes the damage of important physiological processes (Bertke et al., 2019). The balance of SUMOylation and deSUMOylation is crucial for embryonic development, since knockout of SENP1, SENP2, or SENP3 leads to embryonic lethality in mice (Chiu et al., 2008; Lao et al., 2018).
3 SUMOylation in Renal Fibrosis
SUMOs involve in many cellular processes including DNA damage repair, protein stability regulation, cell cycle progression and regulation of signal transduction (Eifler and Vertegaal, 2015; Zeng et al., 2020). Consequently, abnormal SUMO modification can also results in many diseases such as cancers, diabetes, liver diseases, and kidney diseases (Bettermann et al., 2012; Li et al., 2019; Yang et al., 2019; Xie et al., 2020; Zeng et al., 2020). Especially, SUMOylation is activated by some factors, such as hypoxia, metabolic stress, oxidation, genotoxicity, and osmosis (Sun et al., 2018; Appelman et al., 2021; Ma et al., 2021).
Notably, recent studies indicate that SUMOylation functions as a critical role in maintaining renal fibrosis process. As the final pathological process of kidney diseases, renal fibrosis is caused by a decrease in matrix degradation, inflammatory cell infiltration, dysregulation of cell-matrix interaction, and the accumulation of extracellular matrix (ECM) proteins, which mainly involves fibronectin, various collagen and laminin. Although many researches have described the pathogenesis of renal fibrosis, few studies have investigated the role and mechanism of protein SUMOylation during renal fibrosis. Under certain pathological conditions, the signal pathway regulated by SUMOylation is associated with the pathophysiological changes of renal fibrosis. The regulation of renal fibrosis processes involves multiple signaling pathways, therefore we will elaborate on the SUMOylation of transcriptional factors and key mediators in renal fibrosis (Table 1). The molecular mechanisms involved in SUMOs and renal fibrosis are present in Figure 1 and Figure 2.
TABLE 1.
The SUMOylation of transcription factors and key mediators in renal fibrosis.
| Model | SUMO proteins or enzymes | Target genes | References |
|---|---|---|---|
| Modulating transcription factors | |||
| High glucose induced renal mesangial cells | SUMO2/3 | Smad4 | Zhou et al. (2014) |
| Senp1(−/−) mice | SENP1 | HIF-1α | Xu et al. (2010) |
| Co-culture models of glomerular endothelial cells with podocytes | SENP1 | HIF-1α | Wang et al. (2015) |
| Puromycin aminonucleoside-induced podocyte | SENP1 | p53 | Wang et al. (2014) |
| Renal mesangial cells | SUMO1 | p53 | Wu et al. (2021) |
| Rat kidney proximal tubular cells | SUMO2/3 | p53 | Guo et al. (2015) |
| HEK 293T, HCT- 116 p53−/− and all RKO cells | SUMO1 | p53 | Wagner et al. (2015) |
| High glucose stimulated rat glomerular mesangial cells | SUMO1 and SUMO2/3 | NF-κB p65 | Huang et al. (2013) |
| High glucose stimulated rat glomerular mesangial cells | E3 | NF-κB p65 | Huang et al. (2017) |
| Lipopolysaccharide (LPS)-induced human renal proximal tubular cells | E3 | PPARγ | Lu et al. (2013) |
| diabetic GK rats | SUMO4 | NF-κB p65 | Chen et al. (2014) |
| C57BL/6 mice, HEK293T, MEF and RAW264.7 cells | SUMO2/3 | NEMO/IKKγ | Liu et al. (2013) |
| C57BL/6 mice, HEK293T, MEF and RAW264.7 cells | SENP6 | NEMO | Liu et al. (2013) |
| Modulating key mediators | |||
| C57/Bl6 mice and podocytes HEK-293T | SUMO1 and SUMO2/3 | Nephrin | Tossidou et al. (2014) |
| Podocytes | SUMO1 and SUMO2/3 | CIN85 | Tossidou et al. (2012) |
| HEK293 and HeLa cells | SENP3 | Drp1 | Guo et al. (2017) |
| COS-7 fibroblasts | SENP2 | OAT3 | Wang and You, (2019) |
| COS-7 fibroblasts | SUMO2/3 | OAT3 | Wang et al. (2019) |
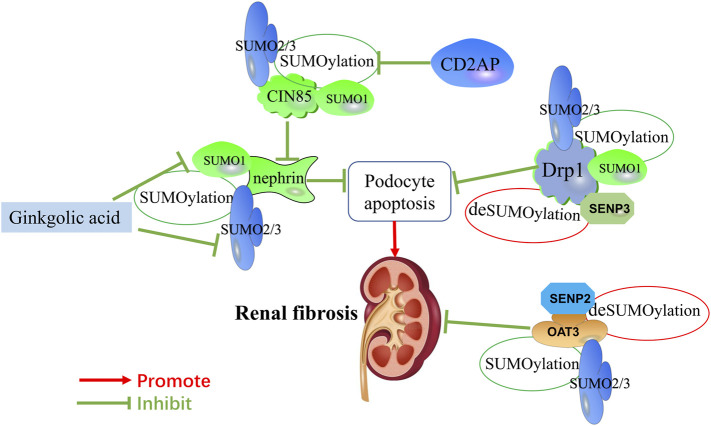
FIGURE 2.
Mechanisms of SUMOylation and deSUMOylation by key mediators in renal fibrosis and therapeutic targets of natural products against renal fibrosis via SUMOylation. Mechanisms of SUMOylation and deSUMOylation by key mediators in renal fibrosis are including podocyte apoptosis, and OAT3-mediated transfer channels. As one of component of the slit diaphragm, nephrin is SUMOylated by SUMO1 and SUMO2/3. CD2AP promotes the expression of nephrin by inhibiting the SUMOylation of CIN85. Drp1 is SUMOylated by SUMO1 and SUMO2/3. OAT3 is SUMOylated by SUMO2/3. SNEP3 deSUMOylates Drp1, and SNEP2 deSUMOylates OAT3. Ginkgolic acid inhibits the expression of nephrin by downregulating SUMO1 and SUMO2/3.
3.1 The SUMOylation of Transcription Factors in Renal Fibrosis
3.1.1 Smads
TGF-β/Smad signaling pathways play a critical role in renal fibrosis through Smad and non-Smad pathways (Meng et al., 2015; Frangogiannis, 2020). Sumoylation of type I TGF-β receptor increases its ligand recruitment ability by phosphorylation of Smad3 (Kang et al., 2008). Smad3 is involved in TGF-β-mediated signaling pathway. E3 ligase PIASy suppresses TGF-β signaling through SUMOylating Smad3 and Smad4 (Imoto et al., 2008; Lee et al., 2014). SUMOylated Smad4 increases the activation of TGF-β1/Smad signaling (Lee et al., 2003; Long et al., 2004; Wang Z. et al., 2018). In high glucose induced renal mesangial cells, SUMOylation of Samd4 by SUMO2/3 activates TGF-β/Smad signaling and then upregulates the expression of fibronectin (Zhou et al., 2014). In addition, some studies reveal that SUMO1 also enhances Smad4 SUMOylation (Ohshima and Shimotohno, 2003; Wang Z. et al., 2018; Liu et al., 2020). Ski and SnoN can inhibit TGF-β pathway through blocking its interaction with Smads. Arkadia (a RING domain E3 ubiquitin ligase) mediates noncovalent interaction with poly-SUMO2, and activates TGF-β/Smad signaling via degradation of Ski and SnoN (Erker et al., 2013). Taken together, targeting SUMOylation of TGF-β/Smad signaling might function as a promising therapeutic approach against renal fibrosis.
3.1.2 HIF-1α Pathways
Decreased oxygen significantly affects gene expression, metabolic changes and regeneration processes, including angiogenesis and stimulation of stem cell proliferation, differentiation, and migration (Hadanny and Efrati, 2020). Glomerular damage leads to a decrease in oxygen supply, and recent studies have emphasized the key role of hypoxia in the development of renal fibrosis (Bessho et al., 2019; Ishiuchi et al., 2020; Wakashima et al., 2020). The expression of SUMO proteins is increased under conditions of hypoxia in vivo and in vitro (Comerford et al., 2003; Shao et al., 2004).
The expression of hypoxia-inducible factor-1α (HIF-1α) in kidney is upregulated under conditions of hypoxia (Bessho et al., 2019). Meanwhile, the upregulation of HIF-1α expression can improve the viability and angiogenesis of bone marrow mesenchymal stem cells (Luo et al., 2019). Evidences show that vascular endothelial growth factor A (VEGFA) promotes the survival of bone marrow mesenchymal stem cells and angiogenesis, while HIF-1α can upregulate the expression of VEGFA (Utispan and Koontongkaew, 2021; Wang B. et al., 2021). Hypoxia enhances the SUMOylation of HIF-1α by SUMO1 and SUMO2/3 (Carbia-Nagashima et al., 2007); whereas SENP1 could deconjugate SUMOylated HIF-1α and reduce HIF-1α degradation during hypoxia (Cheng et al., 2007). SENP1 enhances HIF-1α deSUMOylation and increases VEGF production in endothelial cells following exposure to hypoxia (Xu et al., 2010; Cui et al., 2017). In SENP1(−/−) mice, the vascular endothelial cells in embryonic renal glomeruli and the VEGF production were significantly reduced (Xu et al., 2010). In co-culture models of glomerular endothelial cells with podocytes, the expression of SENP1 in podocytes increases under conditions of hypoxia, which induces endothelial cells survival via deSUMOylation of HIF-1α signaling (Wang et al., 2015). Therefore, SUMOylation of HIF-1α has the potential to be clinically developed as an antifibrotic target.
3.1.3 p53 Pathways
Podocyte apoptosis is a main cause of the decrease in the number of podocytes, which can lead to proteinuria, glomerulosclerosis and renal fibrosis. Recent studies found that the p53 protein participates in the pathogenesis of podocyte apoptosis (Choy et al., 2021; Liang et al., 2021; Yang et al., 2021). As a tumor suppressor, p53 could safeguard the genome and prevent malignant transformation by halting the cell cycle and promoting apoptosis (Jiang et al., 2021; Thoms and Stark, 2021). p53 is increased under stimulation, and then upregulates the expression of pro-apoptotic genes (Eliaš and Macnamara, 2021). SUMOylated p53 could bind to the anti-apoptotic factor Bax and Bcl-2, and lead to apoptosis (Nowakowski et al., 2021). SENP1 deficiency significantly increases podocyte apoptosis by increasing expression of the p53 target pro-apoptotic genes (Noxa, PUMA and BAX) and aggravating accumulation of SUMOylated p53 protein in puromycin aminonucleoside-induced podocyte injury (Wang et al., 2014b). Mesangial cell proliferation is a core pathological feature of many kidney diseases. In renal mesangial cells, Krϋppel-like factor 15 (KLF15) targeted SUMO1 to inhibit cell proliferation via enhancing the stability of p53 (Wu et al., 2021). However, ginkgolic acid (a pharmacological inhibitor of SUMOylation) suppresses p53 SUMOylation by SUMO2/3 and enhances apoptosis in rat kidney proximal tubular cells (Guo et al., 2015). SUMOylation of histone deacetylase 2 (HDAC2) by SUMO1 reduced DNA damage-induced apoptosis via deacetylation of p53 (Wagner et al., 2015).
3.1.4 NF-κB Signaling
As a nuclear transcription factor, nuclear factor κB (NF-κB) regulates the accumulation and release of cytokines and adhesion molecules by leukocytes (Wenzl et al., 2021). During the resting state, NF-κB binds to the inhibitor of κB (IκB) in the cytoplasm (Lee et al., 2021). SUMOylation of NF-κB plays a core role in the regulation of renal inflammation and fibrosis (Al Za’abi et al., 2021; You et al., 2021). With the modification by SUMO2/3, IκBα is separated from NF-κB, and enhance the activation of NF-κB (Chang and Abe, 2016). In high glucose stimulated rat glomerular mesangial cells, IκBα SUMOylation reduces the expression of monocyte chemotactic protein 1 (MCP-1), and ameliorates cellular inflammatory response through inhibiting NF-κB signaling (Huang et al., 2013). As one of E3 ligases, PIAS promotes SUMOylation of NF-κB via enhancing the expression of SUMO1 and SUMO2/3, and leads to the release of MCP-1 and IL-6 from glomerular mesangial cells (Huang et al., 2017).
The utilization of renal energy is partially determined by peroxisome proliferator activated receptors (PPARs) mediated fatty acid oxidation (Aranda-Rivera et al., 2021). PPARγ plays a key role in physiological and pathological processes of renal fibrosis. The reduction of PPARγ SUMOylation suppresses the activation of anti-apoptotic and anti-inflammatory cytokines through inhibiting the NF-κB activity (Xie et al., 2018; Zhao et al., 2021b). In lipopolysaccharide (LPS)-induced human renal proximal tubular cells, rosiglitazone (an agonist of PPARγ) decreases chemokines expression via inhibiting NF-κB activation by activating PPARγ SUMOylation (Lu et al., 2013). SUMO4 also plays a key role in regulating NF-κB signaling in glomerular cells (Chen et al., 2014). SUMOylation NEMO/IKKγ (NF-κB essential modifier) by SUMO2/3 enhanced the activity of NF-κB pathway, whereas SENP6 reverses this process by catalyzing the deSUMOylation of NEMO (Liu et al., 2013). These results indicate that the regulation of NF-κB SUMOylation has become a potential therapeutic strategy for renal fibrosis.
3.2 The SUMOylation of Key Mediators in Renal Fibrosis
3.2.1 Podocyte Apoptosis Pathway
Podocyte injury is the basic pathological feature of many kidney diseases, including decreased expression of the fissure membrane components and ultrastructural changes. Recently, SUMOylation has also been implicated in podocyte injury. As one of components of the slit diaphragm, nephrin is a target protein for SUMOylation by SUMO1 and SUMO2/3 (Tossidou et al., 2014). In podocytes, SUMOylation plays a key role in the tight orchestration of nephrin turnover at the slit diaphragm. The SUMOylation inhibitor ginkgolic acid decreases PI3K/AKT signaling and reduces membrane expression of nephrin (Tossidou et al., 2014). Besides nephrin, CD2-associated protein (CD2AP) also plays a crucial role in slit diaphragm (Chung et al., 2020; Basgen et al., 2021). Due to high sequence and structural similarities with CD2AP, Cbl-interacting protein of 85 kDa (CIN85) is a binding partner of nephrin and then increases endocytosis of nephrin (Tossidou et al., 2010). CD2AP reduces the binding of CIN85 to nephrin via CIN85 SUMOylation (Tossidou et al., 2012).
Mitochondrial dysfunction caused excessive mitochondrial fission, and then promoted podocyte apoptosis through the overproduction of reactive oxygen species (Su et al., 2021; Zhu et al., 2021). Moreover, mitochondrial dysfunction in damaged podocytes causes the development of marked glomerulosclerosis and albuminuria (Zhou et al., 2019). SUMOylation of dynamin-related protein (Drp) 1 activates mitochondrial autophagy and inhibits ROS production via regulating mitochondrial morphology and division (Din et al., 2013). SUMO1 SUMOylates Drp1, and then prevents the mitochondrial translocation (Jin et al., 2021). On the other hand, SENP2 deSUMOylates Drp1 by removing SUMO1, whereas SENP3 and SENP5 removes SUMO2/3 from deSUMOylation of Drp1 (Fu et al., 2014; Shimizu et al., 2016). DeSUMOylation of Drp1 by SENP3 promotes cell death by enhancing Drp1 partitioning to the mitochondrial outer membrane and improving cytochrome c release and apoptosis (Guo et al., 2017).
3.2.2 OAT3-Mediated Transfer Channels
Organic anion transporter 3 (OAT3) is an important transporter that located in the basolateral membrane of proximal renal tubules. OAT3 plays vital parts in removing a variety of drugs from the kidney, so as to avoid their possibly toxic side effects in the body (Fu et al., 2021). Activated OAT3 attenuates renal lipid accumulation and alleviates renal injury associated with renal inflammation and fibrosis in high-fat diet-induced obese rats (Pengrattanachot et al., 2020). In COS-7 fibroblasts, the downregulation of Senp2 results in an increased OAT3 SUMOylation via enhancing OAT3 expression and transport activity (Wang and You, 2019). In addition, protein kinase A (PKA) accelerates the OAT3 SUMOylation by SUMO2/3 (Wang et al., 2019).
4 Natural Products Against Renal Fibrosis via Regulating SUMOylation
4.1 Natural Products Against Renal Fibrosis via Regulating the SUMOylation of Transcription Factors
Numerous natural products have been widely applied to the treatment of fibrosis via inhibition of SUMOylation (Table 2). However, few studies have investigated the role and mechanism of natural products against renal fibrosis via modulating SUMOylation. Recent studies reveal that ginkgolic acid, a natural product from ginkgo, could regulate the SUMOylation of p53 to attenuate renal fibrosis (Tossidou et al., 2014; Guo et al., 2015).
TABLE 2.
Natural products and therapeutic targets against renal fibrosis via SUMOylation.
| Compounds | Resource | SUMO proteins or enzymes | Therapeutic target | References |
|---|---|---|---|---|
| Modulating transcription factors | ||||
| Ginkgolic acid | Ginkgo | SUMO2/3 | p53 | Guo et al. (2015) |
| SUMO1 | Smad4 | Liu et al. (2020) | ||
| Astragaloside IV | Astragalus membranaceus | SUMO1 | HIF-1α | Wang et al. (2021) |
| SENP1 | — | Liu et al. (2021) | ||
| SUMO1 | — | Wang et al. (2021a) | ||
| Resveratrol | Vitis L., Veratrum L., Arachis, Polygonum | SUMO1 | β-catenin | Wang et al. (2020) |
| SUMO1 | COX-2 | Cheng et al. (2018); Lin et al. (2011) | ||
| Puerarin | Radix Puerariae | SUMO2 | ER/ERK | Zhao et al. (2021) |
| Gallic acid | gallnut, sumac, tea leaves, and oak bark | SENP1 | HIF-1α | Taghvaei et al. (2021) |
| Ginsenoside Rg3 | Panax ginseng | E3 | NF-κB | Zou et al. (2018) |
| Modulating key mediators | ||||
| Ginkgolic acid | Ginkgo | SUMO1 and SUMO2/3 | nephrin | Tossidou et al. (2014) |
Astragaloside IV, isolated from Astragalus membranaceus, plays a key role in antioxidant, antifibrosis, antitumor and anti-diabetic effects (Zhang et al., 2020). Astragaloside IV improves angiogenesis in adverse hypoxic conditions through stabilizing the presence of HIF-1α protein via enhancing SUMO1 expression (Wang B. et al., 2021).
Additionally, natural products also play protective effects via SUMOylation in extrarenal tissue. Wnt/β-catenin signaling pathway plays a key role in promoting fibrosis (Li. S. S. et al., 2021). Resveratrol, a natural active antioxidant, alleviates inflammatory response and fibrosis by modulating SUMO1 through Wnt/β-catenin pathway in dextran sodium sulfate-induced inflammatory bowel disease mice (Wang et al., 2020). Resveratrol also induces SUMOylated cyclooxygenase (COX)-2 by SUMO1, thus enhancing the expression of pro-apoptotic genes via activating p53 in human prostate cancer LNCaP cells and human ovarian carcinoma (OVCAR-3) cells (Lin et al., 2011; Cheng et al., 2018). In heart failure mice, astragaloside IV reduces the levels of reactive oxygen species (ROS), and improves cardiac function by inhibiting the overexpression of SENP1 (Liu et al., 2021). Meanwhile, astragaloside IV promotes the proliferation and migration of vascular endothelial cells and antagonizes the adverse microenvironment of hypoxia and high glucose by enhancing SUMO1 expression in human umbilical vein endothelial cells (Wang B. S. et al., 2021). In cardiomyocytes, puerarin attenuates cellular inflammatory response and fibrosis through activating ER/ERK pathway by increasing the expression of SUMO2 (Zhao et al., 2021a). In oral squamous cell carcinoma cells, ginkgolic acid suppresses tumorigenicity and tumor progression through inhibition of the enhancement of SUMOylation of Smad4 (Liu et al., 2020). Ginsenoside Rg3, extracted from Panax ginseng, inhibits NF-κB signaling through increasing the phosphorylation of RanBP2 (E3 SUMO-protein ligase) in breast cancer cells (Zou et al., 2018).
Numerous studies have demonstrated that SENP1 could bind to HIF-1α for deSUMOylation (Bai et al., 2021; Taghvaei et al., 2021). Gallic acid plays an essential role in antioxidant, anti-inflammatory, antimutagenic and anticancer through reducing deSUMOylation via inhibiting SENP1 expression (Taghvaei et al., 2021). Natural products against renal fibrosis via regulating the SUMOylation of transcription factors are present in Figure 1.
4.2 Natural Products Against Renal Fibrosis via Regulating the SUMOylation of Key Mediators
Ginkgolic acid, extracted from Ginkgo biloba leaves and seed coat, have beneficial effects including anti-inflammatory (Li et al., 2018), antitumor (Zhao Q. et al., 2021), and anti-bacterial effects (Bhutta et al., 2021). Unfortunately, ginkgolic acid could aggravate kidney damage by enhancing apoptosis and downregulating the expression of nephrin. Ginkgolic acid enhances apoptosis through suppressing SUMOylation by SUMO2/3 and sensitizes renal tubular cells to apoptosis during cisplatin treatment of rat kidney proximal tubular cells (Guo et al., 2015). Additionally, ginkgolic acid reduces membrane expression of nephrin via suppressing SUMOylation by SUMO1 and SUMO2/3, leading to a significant proteinuria in mice (Tossidou et al., 2014). Natural products against renal fibrosis via regulating the SUMOylation of key mediators are present in Figure 2.
5 Discussion and Conclusion
In summary, the present study provides a systemic review of the role of SUMOylation in renal fibrosis, and the therapeutic effects of natural products through modulating transcription factors and key mediators. The pathological processes of renal fibrosis are regulated by complex signal pathways and factors, thus leading to a series of stress responses. The imbalance of protein SUMOylation is involved signal pathways and factors in renal fibrosis. SUMOylation significantly affects renal fibrosis through modulating transcription factors and key mediators, including Smads, HIF-1α, p53, NF-κB, podocyte apoptosis pathway, OAT3-mediated transfer channels, and Wnt/β-catenin pathway.
However, the effective treatment via SUMOylation remains a formidable challenge. The lack of effective treatment for renal fibrosis indicates that a deep understanding of the molecular mechanisms contributing to renal fibrosis remain urgent. Fortunately, researches of natural products in modulating SUMOylation have been carried out recently, which probably reveals the role of SUMOylation in the progression of renal fibrosis soon, including astragaloside IV, resveratrol, puerarin, gallic acid, and ginsenoside Rg3.
Additionally, the techniques that help to high-throughput screening of effective natural products targeting SUMOylation also facilitate to discover the candidate drug for treating renal fibrosis via modulating SUMOylation. It is worth noting that natural products mentioned above still needs more attention since the lack of underlying mechanism are far not enough for a candidate drug clinically. The detailed and in-depth basic research are urgent to investigate the underlying mechanisms of natural products against renal fibrosis, and the finding in reveal novel signaling pathways and treatment targets are necessary.
Author Contributions
PL, JZ, YW, CW, XQ, and D-QC mainly drafted the work critical for important intellectual content. PL and D-QC participated in the funding acquisition. All authors read and approved the final manuscript. PL, JZ, and YW contributed equally to this work.
Funding
This work was supported by supported by Research Projects of the National Natural Science Foundation of China (Nos 81904174, 82104511), China Postdoctoral Science Foundation (No. 2021M693579).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Al Za’abi M., Ali H., Ali B. H. (2021). Effect of Flaxseed on Systemic Inflammation and Oxidative Stress in Diabetic Rats with or without Chronic Kidney Disease. PLoS One 16 (10), e0258800. 10.1371/journal.pone.0258800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelman M. D., van der Veen S. W., van Mil S. W. C. (2021). Post-translational Modifications of FXR; Implications for Cholestasis and Obesity-Related Disorders. Front. Endocrinol. (Lausanne) 12, 729828. 10.3389/fendo.2021.729828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Rivera A. K., Cruz-Gregorio A., Aparicio-Trejo O. E., Pedraza-Chaverri J. (2021). Mitochondrial Redox Signaling and Oxidative Stress in Kidney Diseases. Biomolecules 11 (8), 1144. 10.3390/biom11081144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvaniti E., Vakrakou A., Kaltezioti V., Stergiopoulos A., Prakoura N., Politis P. K., et al. (2016). Nuclear Receptor NR5A2 Is Involved in the Calreticulin Gene Regulation during Renal Fibrosis. Biochim. Biophys. Acta 1862 (9), 1774–1785. 10.1016/j.bbadis.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Bai Y. T., Xiao F. J., Wang H., Ge R. L., Wang L. S. (2021). Hypoxia Protects H9c2 Cells against Ferroptosis through SENP1-Mediated Protein DeSUMOylation. Int. J. Med. Sci. 18 (7), 1618–1627. 10.7150/ijms.50804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basgen J. M., Wong J. S., Ray J., Nicholas S. B., Campbell K. N. (2021). Podocyte Foot Process Effacement Precedes Albuminuria and Glomerular Hypertrophy in CD2-Associated Protein Deficient Mice. Front. Med. (Lausanne) 8, 745319. 10.3389/fmed.2021.745319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertke M. M., Dubiak K. M., Cronin L., Zeng E., Huber P. W. (2019). A Deficiency in SUMOylation Activity Disrupts Multiple Pathways Leading to Neural Tube and Heart Defects in Xenopus Embryos. BMC Genomics 20 (1), 386. 10.1186/s12864-019-5773-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho R., Takiyama Y., Takiyama T., Kitsunai H., Takeda Y., Sakagami H., et al. (2019). Hypoxia-inducible Factor-1α Is the Therapeutic Target of the SGLT2 Inhibitor for Diabetic Nephropathy. Sci. Rep. 9 (1), 14754. 10.1038/s41598-019-51343-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettermann K., Benesch M., Weis S., Haybaeck J. (2012). SUMOylation in Carcinogenesis. Cancer Lett. 316 (2), 113–125. 10.1016/j.canlet.2011.10.036 [DOI] [PubMed] [Google Scholar]
- Bhutta M., Shechter O., Gallo E., Martin S., Jones E., Doncel G., et al. (2021). Ginkgolic Acid Inhibits Herpes Simplex Virus Type 1 Skin Infection and Prevents Zosteriform Spread in Mice. Viruses 13 (1), 86. 10.3390/v13010086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbia-Nagashima A., Gerez J., Perez-Castro C., Paez-Pereda M., Silberstein S., Stalla G. K., et al. (2007). RSUME, a Small RWD-Containing Protein, Enhances SUMO Conjugation and Stabilizes HIF-1alpha during Hypoxia. Cell 131 (2), 309–323. 10.1016/j.cell.2007.07.044 [DOI] [PubMed] [Google Scholar]
- Chang E., Abe J. I. (2016). Kinase-SUMO Networks in Diabetes-Mediated Cardiovascular Disease. Metabolism 65 (5), 623–633. 10.1016/j.metabol.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Yang T., Liu F., Li H., Guo Y., Yang H., et al. (2014). Inflammatory Factor-specific Sumoylation Regulates NF-Κb Signalling in Glomerular Cells from Diabetic Rats. Inflamm. Res. 63 (1), 23–31. 10.1007/s00011-013-0675-3 [DOI] [PubMed] [Google Scholar]
- Cheng J., Kang X., Zhang S., Yeh E. T. (2007). SUMO-specific Protease 1 Is Essential for Stabilization of HIF1alpha during Hypoxia. Cell 131 (3), 584–595. 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T. M., Chin Y. T., Ho Y., Chen Y. R., Yang Y. N., Yang Y. C., et al. (2018). Resveratrol Induces Sumoylated COX-2-dependent Anti-proliferation in Human Prostate Cancer LNCaP Cells. Food Chem. Toxicol. 112, 67–75. 10.1016/j.fct.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Chiu S. Y., Asai N., Costantini F., Hsu W. (2008). SUMO-specific Protease 2 Is Essential for Modulating P53-Mdm2 in Development of Trophoblast Stem Cell Niches and Lineages. Plos Biol. 6 (12), e310. 10.1371/journal.pbio.0060310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy J., Kan Y., Cifelli S., Johnson J., Chen M., Shiao L. L., et al. (2021). High-Throughput Screening to Identify Small Molecules that Selectively Inhibit APOL1 Protein Level in Podocytes. SLAS Discov. 26 (9), 1225–1237. 10.1177/24725552211026245 [DOI] [PubMed] [Google Scholar]
- Chung J. J., Goldstein L., Chen Y. J., Lee J., Webster J. D., Roose-Girma M., et al. (2020). Single-Cell Transcriptome Profiling of the Kidney Glomerulus Identifies Key Cell Types and Reactions to Injury. J. Am. Soc. Nephrol. 31 (10), 2341–2354. 10.1681/ASN.2020020220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford K. M., Leonard M. O., Karhausen J., Carey R., Colgan S. P., Taylor C. T. (2003). Small Ubiquitin-Related Modifier-1 Modification Mediates Resolution of CREB-dependent Responses to Hypoxia. Proc. Natl. Acad. Sci. U S A. 100 (3), 986–991. 10.1073/pnas.0337412100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C. P., Wong C. C., Kai A. K., Ho D. W., Lau E. Y., Tsui Y. M., et al. (2017). SENP1 Promotes Hypoxia-Induced Cancer Stemness by HIF-1α deSUMOylation and SENP1/HIF-1α Positive Feedback Loop. Gut 66 (12), 2149–2159. 10.1136/gutjnl-2016-313264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din S., Mason M., Völkers M., Johnson B., Cottage C. T., Wang Z., et al. (2013). Pim-1 Preserves Mitochondrial Morphology by Inhibiting Dynamin-Related Protein 1 Translocation. Proc. Natl. Acad. Sci. U S A. 110 (15), 5969–5974. 10.1073/pnas.1213294110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Chen X., Su Q., Lu W., Wang Q., Yuan H., et al. (2021). The Function of SUMOylation and its Critical Roles in Cardiovascular Diseases and Potential Clinical Implications. Int. J. Mol. Sci. 22 (19), 10618. 10.3390/ijms221910618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler K., Vertegaal A. C. O. (2015). SUMOylation-Mediated Regulation of Cell Cycle Progression and Cancer. Trends Biochem. Sci. 40 (12), 779–793. 10.1016/j.tibs.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliaš J., Macnamara C. K. (2021). Mathematical Modelling of P53 Signalling during DNA Damage Response: A Survey. Int. J. Mol. Sci. 22 (19), 10590. 10.3390/ijms221910590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M. (2015). Sumo and the Cellular Stress Response. Cell Div 10, 4. 10.1186/s13008-015-0010-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erker Y., Neyret-Kahn H., Seeler J. S., Dejean A., Atfi A., Levy L. (2013). Arkadia, a Novel SUMO-Targeted Ubiquitin Ligase Involved in PML Degradation. Mol. Cell. Biol. 33 (11), 2163–2177. 10.1128/MCB.01019-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis N. (2020). Transforming Growth Factor-β in Tissue Fibrosis. J. Exp. Med. 217 (3), e20190103. 10.1084/jem.20190103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Yu H. M., Chiu S. Y., Mirando A. J., Maruyama E. O., Cheng J. G., et al. (2014). Disruption of SUMO-specific Protease 2 Induces Mitochondria Mediated Neurodegeneration. Plos Genet. 10 (10), e1004579. 10.1371/journal.pgen.1004579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R., Wang X. N., Guo C. H., Li Y., Ding C. Y., Li Y. J., et al. (2021). Wuzhi Capsule Increased Systemic Exposure to Methotrexate by Inhibiting the Expression of OAT1/3 and P-Gp. Ann. Transl Med. 9 (10), 845. 10.21037/atm-21-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Wei Q., Su Y., Dong Z. (2015). SUMOylation Occurs in Acute Kidney Injury and Plays a Cytoprotective Role. Biochim. Biophys. Acta 1852 (3), 482–489. 10.1016/j.bbadis.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Wilkinson K. A., Evans A. J., Rubin P. P., Henley J. M. (2017). SENP3-mediated deSUMOylation of Drp1 Facilitates Interaction with Mff to Promote Cell Death. Sci. Rep. 7, 43811. 10.1038/srep43811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadanny A., Efrati S. (2020). The Hyperoxic-Hypoxic Paradox. Biomolecules 10 (6), 958. 10.3390/biom10060958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Liang Y., Dong J., Zhou L., Gao C., Jiang C., et al. (2017). SUMO E3 Ligase PIASy Mediates High Glucose-Induced Activation of NF-Κb Inflammatory Signaling in Rat Mesangial Cells. Mediators Inflamm. 2017, 1685194. 10.1155/2017/1685194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Xu L., Zhou X., Gao C., Yang M., Chen G., et al. (2013). High Glucose Induces Activation of NF-Κb Inflammatory Signaling through IκBα Sumoylation in Rat Mesangial Cells. Biochem. Biophys. Res. Commun. 438 (3), 568–574. 10.1016/j.bbrc.2013.07.065 [DOI] [PubMed] [Google Scholar]
- Imoto S., Ohbayashi N., Ikeda O., Kamitani S., Muromoto R., Sekine Y., et al. (2008). Sumoylation of Smad3 Stimulates its Nuclear export during PIASy-Mediated Suppression of TGF-Beta Signaling. Biochem. Biophys. Res. Commun. 370 (2), 359–365. 10.1016/j.bbrc.2008.03.116 [DOI] [PubMed] [Google Scholar]
- Ishiuchi N., Nakashima A., Doi S., Yoshida K., Maeda S., Kanai R., et al. (2020). Hypoxia-preconditioned Mesenchymal Stem Cells Prevent Renal Fibrosis and Inflammation in Ischemia-Reperfusion Rats. Stem Cell Res. Ther. 11 (1), 130. 10.1186/s13287-020-01642-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. W., Zhang Z. Z., He P. P., Jiang L. P., Chen J. Z., Zhang X. T., et al. (2021). Emerging Roles of Growth Differentiation Factor-15 in Brain Disorders (Review). Exp. Ther. Med. 22 (5), 1270. 10.3892/etm.2021.10705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. Y., Wei X. X., Zhi X. L., Wang X. H., Meng D. (2021). Drp1-dependent Mitochondrial Fission in Cardiovascular Disease. Acta Pharmacol. Sin. 42 (5), 655–664. 10.1038/s41401-020-00518-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- K S. T., Joshi G., Arya P., Mahajan V., Chaturvedi A., Mishra R. K. (2021). SUMO and SUMOylation Pathway at the Forefront of Host Immune Response. Front Cell Dev Biol 9, 681057. 10.3389/fcell.2021.681057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. S., Saunier E. F., Akhurst R. J., Derynck R. (2008). The Type I TGF-Beta Receptor Is Covalently Modified and Regulated by Sumoylation. Nat. Cell Biol. 10 (6), 654–664. 10.1038/ncb1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkula A., Ojala V. K., Mendez L. M., Sistonen L., Elenius K., Sundvall M. (2021). Therapeutic Potential of Targeting the SUMO Pathway in Cancer. Cancers (Basel) 13 (17), 4402. 10.3390/cancers13174402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz K., Piller T., Müller S. (2018). SUMO-specific Proteases and Isopeptidases of the SENP Family at a Glance. J. Cell Sci. 131 (6), jcs211904. 10.1242/jcs.211904 [DOI] [PubMed] [Google Scholar]
- Lao Y., Yang K., Wang Z., Sun X., Zou Q., Yu X., et al. (2018). DeSUMOylation of MKK7 Kinase by the SUMO2/3 Protease SENP3 Potentiates Lipopolysaccharide-Induced Inflammatory Signaling in Macrophages. J. Biol. Chem. 293 (11), 3965–3980. 10.1074/jbc.M117.816769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J., Kim H. J., Choi M. S., Chang J. E. (2021). Crosstalk between Autophagy and Inflammatory Processes in Cancer. Life (Basel) 11 (9), 903. 10.3390/life11090903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. S., Chang C., Liu D., Derynck R. (2003). Sumoylation of Smad4, the Common Smad Mediator of Transforming Growth Factor-Beta Family Signaling. J. Biol. Chem. 278 (30), 27853–27863. 10.1074/jbc.M301755200 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Kim P. H., Oh S. M., Park J. H., Yoo Y. C., Lee J., et al. (2014). SUMO Proteins Are Not Involved in TGF-Β1-Induced, Smad3/4-Mediated Germline α Transcription, but PIASy Suppresses it in CH12F3-2A B Cells. Immune Netw. 14 (6), 321–327. 10.4110/in.2014.14.6.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li A., Li M., Liu Y., Zhao W., Gao D. (2018). Ginkgolic Acid Exerts an Anti-inflammatory Effect in Human Umbilical Vein Endothelial Cells Induced by Ox-LDL. Pharmazie 73 (7), 408–412. 10.1681/ph.2018.8397 [DOI] [PubMed] [Google Scholar]
- Li O., Ma Q., Li F., Cai G. Y., Chen X. M., Hong Q. (2019). Progress of Small Ubiquitin-Related Modifiers in Kidney Diseases. Chin. Med. J. (Engl) 132 (4), 466–473. 10.1097/CM9.0000000000000094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. S., Sun Q., Hua M. R., Suo P., Chen J. R., Yu X. Y., et al. (2021). Targeting the Wnt/β-Catenin Signaling Pathway as a Potential Therapeutic Strategy in Renal Tubulointerstitial Fibrosis. Front. Pharmacol. 12, 719880. 10.3389/fphar.2021.719880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Liu H., Zhu J., Song N., Lu Z., Fang Y., et al. (2021). Inhibition of p53/miR-34a/SIRT1 axis Ameliorates Podocyte Injury in Diabetic Nephropathy. Biochem. Biophys. Res. Commun. 559, 48–55. 10.1016/j.bbrc.2021.04.025 [DOI] [PubMed] [Google Scholar]
- Liang Y. C., Lee C. C., Yao Y. L., Lai C. C., Schmitz M. L., Yang W. M. (2016). SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Sci. Rep. 6, 26509. 10.1038/srep26509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Crawford D. R., Lin S., Hwang J., Sebuyira A., Meng R., et al. (2011). Inducible COX-2-dependent Apoptosis in Human Ovarian Cancer Cells. Carcinogenesis 32 (1), 19–26. 10.1093/carcin/bgq212 [DOI] [PubMed] [Google Scholar]
- Liu J., Li Y., Bian X., Xue N., Yu J., Dai S., et al. (2021). Astragaloside IV Alleviates Heart Failure by Regulating SUMO-specific Protease 1. Exp. Ther. Med. 22 (4), 1076. 10.3892/etm.2021.10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Wang X., Li D., Xu D., Li D., Lv Z., et al. (2020). Ginkgolic Acid, a SUMO-1 Inhibitor, Inhibits the Progression of Oral Squamous Cell Carcinoma by Alleviating SUMOylation of SMAD4. Mol. Ther. Oncolytics 16, 86–99. 10.1016/j.omto.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen W., Wang Q., Li L., Wang C. (2013). Negative Regulation of TLR Inflammatory Signaling by the SUMO-Deconjugating Enzyme SENP6. Plos Pathog. 9 (6), e1003480. 10.1371/journal.ppat.1003480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Wang G., He D., Liu F. (2004). Repression of Smad4 Transcriptional Activity by SUMO Modification. Biochem. J. 379 (Pt 1), 23–29. 10.1042/BJ20031867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhou Q., Shi Y., Liu J., Zhong F., Hao X., et al. (2013). SUMOylation of PPARγ by Rosiglitazone Prevents LPS-Induced NCoR Degradation Mediating Down Regulation of Chemokines Expression in Renal Proximal Tubular Cells. PLoS One 8 (11), e79815. 10.1371/journal.pone.0079815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Wu F., Xue E., Huang L., Yan P., Pan X., et al. (2019). Hypoxia Preconditioning Promotes Bone Marrow Mesenchymal Stem Cells Survival by Inducing HIF-1α in Injured Neuronal Cells Derived Exosomes Culture System. Cell Death Dis 10 (2), 134. 10.1038/s41419-019-1410-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., North B. J., Shu J. (2021). Regulation of Topoisomerase II Stability and Activity by Ubiquitination and SUMOylation: Clinical Implications for Cancer Chemotherapy. Mol. Biol. Rep. 48 (9), 6589–6601. 10.1007/s11033-021-06665-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. M., Tang P. M., Li J., Lan H. Y. (2015). TGF-β/Smad Signaling in Renal Fibrosis. Front. Physiol. 6, 82. 10.3389/fphys.2015.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Dasso M. (2007). Modification in Reverse: the SUMO Proteases. Trends Biochem. Sci. 32 (6), 286–295. 10.1016/j.tibs.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Nowakowski P., Markiewicz-Żukowska R., Bielecka J., Mielcarek K., Grabia M., Socha K. (2021). Treasures from the forest: Evaluation of Mushroom Extracts as Anti-cancer Agents. Biomed. Pharmacother. 143, 112106. 10.1016/j.biopha.2021.112106 [DOI] [PubMed] [Google Scholar]
- Ohshima T., Shimotohno K. (2003). Transforming Growth Factor-Beta-Mediated Signaling via the P38 MAP Kinase Pathway Activates Smad-dependent Transcription through SUMO-1 Modification of Smad4. J. Biol. Chem. 278 (51), 50833–50842. 10.1074/jbc.M307533200 [DOI] [PubMed] [Google Scholar]
- Pengrattanachot N., Cherngwelling R., Jaikumkao K., Pongchaidecha A., Thongnak L., Swe M. T., et al. (2020). Atorvastatin Attenuates Obese-Induced Kidney Injury and Impaired Renal Organic Anion Transporter 3 Function through Inhibition of Oxidative Stress and Inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 1866 (6), 165741. 10.1016/j.bbadis.2020.165741 [DOI] [PubMed] [Google Scholar]
- Saitoh H., Hinchey J. (2000). Functional Heterogeneity of Small Ubiquitin-Related Protein Modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275 (9), 6252–6258. 10.1074/jbc.275.9.6252 [DOI] [PubMed] [Google Scholar]
- Sapir A. (2020). Not So Slim Anymore-Evidence for the Role of SUMO in the Regulation of Lipid Metabolism. Biomolecules 10 (8), 1154–6258. 10.3390/biom10081154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R., Zhang F. P., Tian F., Anders Friberg P., Wang X., Sjöland H., et al. (2004). Increase of SUMO-1 Expression in Response to Hypoxia: Direct Interaction with HIF-1alpha in Adult Mouse Brain and Heart In Vivo . FEBS Lett. 569 (1-3), 293–300. 10.1016/j.febslet.2004.05.079 [DOI] [PubMed] [Google Scholar]
- Shimizu Y., Lambert J. P., Nicholson C. K., Kim J. J., Wolfson D. W., Cho H. C., et al. (2016). DJ-1 Protects the Heart against Ischemia-Reperfusion Injury by Regulating Mitochondrial Fission. J. Mol. Cell. Cardiol. 97, 56–66. 10.1016/j.yjmcc.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Gao C., Xie L., Fan Y., Shen Y., Huang Q., et al. (2021). Astragaloside II Ameliorated Podocyte Injury and Mitochondrial Dysfunction in Streptozotocin-Induced Diabetic Rats. Front. Pharmacol. 12, 638422. 10.3389/fphar.2021.638422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Qing W., Qi R., Zou M., Gong L., Liu Y., et al. (2018). Inhibition of Sumoylation Alleviates Oxidative Stress-Induced Retinal Pigment Epithelial Cell Senescence and Represses Proinflammatory Gene Expression. Curr. Mol. Med. 18 (9), 575–583. 10.2174/1566524019666190107154250 [DOI] [PubMed] [Google Scholar]
- Taghvaei S., Sabouni F., Minuchehr Z., Taghvaei A. (2021). Identification of Novel Anti-cancer Agents, Applying In Silico Method for SENP1 Protease Inhibition. J. Biomol. Struct. Dyn. 3, 1–15. 10.1080/07391102.2021.1880480 [DOI] [PubMed] [Google Scholar]
- Thoms H. C., Stark L. A. (2021). The NF-Κb Nucleolar Stress Response Pathway. Biomedicines 9 (9), 1082. 10.3390/biomedicines9091082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossidou I., Himmelseher E., Teng B., Haller H., Schiffer M. (2014). SUMOylation Determines Turnover and Localization of Nephrin at the Plasma Membrane. Kidney Int. 86 (6), 1161–1173. 10.1038/ki.2014.198 [DOI] [PubMed] [Google Scholar]
- Tossidou I., Niedenthal R., Klaus M., Teng B., Worthmann K., King B. L., et al. (2012). CD2AP Regulates SUMOylation of CIN85 in Podocytes. Mol. Cell. Biol. 32 (6), 1068–1079. 10.1128/MCB.06106-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossidou I., Teng B., Drobot L., Meyer-Schwesinger C., Worthmann K., Haller H., et al. (2010). CIN85/RukL Is a Novel Binding Partner of Nephrin and Podocin and Mediates Slit Diaphragm Turnover in Podocytes. J. Biol. Chem. 285 (33), 25285–25295. 10.1074/jbc.M109.087239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utispan K., Koontongkaew S. (2021). Mucin 1 Regulates the Hypoxia Response in Head and Neck Cancer Cells. J. Pharmacol. Sci. 147 (4), 331–339. 10.1016/j.jphs.2021.08.007 [DOI] [PubMed] [Google Scholar]
- Wagner T., Kiweler N., Wolff K., Knauer S. K., Brandl A., Hemmerich P., et al. (2015). Sumoylation of HDAC2 Promotes NF-κb-dependent Gene Expression. Oncotarget 6 (9), 7123–7135. 10.18632/oncotarget.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakashima T., Tanaka T., Fukui K., Komoda Y., Shinozaki Y., Kobayashi H., et al. (2020). JTZ-951, an HIF Prolyl Hydroxylase Inhibitor, Suppresses Renal Interstitial Fibroblast Transformation and Expression of Fibrosis-Related Factors. Am. J. Physiol. Ren. Physiol 318 (1), F14–F24. 10.1152/ajprenal.00323.2019 [DOI] [PubMed] [Google Scholar]
- Wang B., Zhang C., Ma X., Yu T., Liu X., Hu C., et al. (2021). Astragaloside IV Improves Angiogenesis under Hypoxic Conditions by Enhancing Hypoxia-inducible F-actor-1α SUMOylation. Mol. Med. Rep. 23 (4), 244. 10.3892/mmr.2021.11883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., You G. (2019). The SUMO-specific Protease Senp2 Regulates SUMOylation, Expression and Function of Human Organic Anion Transporter 3. Biochim. Biophys. Acta Biomembr 1861 (7), 1293–1301. 10.1016/j.bbamem.2019.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang J., You G. (2019). Activation of Protein Kinase A Stimulates SUMOylation, Expression, and Transport Activity of Organic Anion Transporter 3. AAPS J. 21 (2), 30. 10.1208/s12248-019-0303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang Z., Fang A., Wu K., Chen X., Wang G., et al. (2020). Resveratrol Attenuates Inflammatory Bowel Disease in Mice by Regulating SUMO1. Biol. Pharm. Bull. 43 (3), 450–457. 10.1248/bpb.b19-00786 [DOI] [PubMed] [Google Scholar]
- Wang L., Wansleeben C., Zhao S., Miao P., Paschen W., Yang W. (2014a). SUMO2 Is Essential while SUMO3 Is Dispensable for Mouse Embryonic Development. EMBO Rep. 15 (8), 878–885. 10.15252/embr.201438534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhang T., Fang M., Shen N., Wang D., Teng J., et al. (2015). Podocytes Protect Glomerular Endothelial Cells from Hypoxic Injury via deSUMOylation of HIF-1α Signaling. Int. J. Biochem. Cell Biol 58, 17–27. 10.1016/j.biocel.2014.10.030 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhu J., Fang M., Zhang T., Xie H., Wang N., et al. (2014b). Inhibition of P53 deSUMOylation Exacerbates Puromycin Aminonucleoside-Induced Apoptosis in Podocytes. Int. J. Mol. Sci. 15 (11), 21314–21330. 10.3390/ijms151121314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang K., Wang R., Liu X. (2018). SUMOylation Regulates TGF-β1/Smad4 Signalling In-Resistant Glioma Cells. Anticancer Drugs 29 (2), 136–144. 10.1097/CAD.0000000000000578 [DOI] [PubMed] [Google Scholar]
- Wang B. S., Ma X. F., Zhang C. Y., Li Y. X., Liu X. Z., Hu C. Q. (2021). Astragaloside IV Improves Angiogenesis and Promotes Wound Healing in Diabetic Rats via the Activation of the SUMOylation Pathway. Biomed. Environ. Sci. 34 (2), 124–129. 10.3967/bes2021.018 [DOI] [PubMed] [Google Scholar]
- Wang Z., Jiang L., Wang J., Chai Z., Xiong W. (2021). Morphine Promotes Angiogenesis by Activating PI3K/Akt/HIF-1α Pathway and Upregulating VEGF in Hepatocellular Carcinoma. J. Gastrointest. Oncol. 12 (4), 1761–1772. 10.21037/jgo-20-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl F. A., Ambrosini S., Mohammed S. A., Kraler S., Lüscher T. F., Costantino S., et al. (2021). Inflammation in Metabolic Cardiomyopathy. Front. Cardiovasc. Med. 8, 742178. 10.3389/fcvm.2021.742178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K. A., Henley J. M. (2010). Mechanisms, Regulation and Consequences of Protein SUMOylation. Biochem. J. 428 (2), 133–145. 10.1042/BJ20100158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Li O., Zhu F., Wang X., Chen P., Cai G., et al. (2021). Krϋppel-like Factor 15 Suppresses Renal Glomerular Mesangial Cell Proliferation via Enhancing P53 SUMO1 Conjugation. J. Cell. Mol. Med. 25 (12), 5691–5706. 10.1111/jcmm.16583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B., Liu X., Yang J., Cheng J., Gu J., Xue S. (2018). PIAS1 Protects against Myocardial Ischemia-Reperfusion Injury by Stimulating PPARγ SUMOylation. BMC Cell Biol 19 (1), 24. 10.1186/s12860-018-0176-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Yu J., Ge S., Huang J., Fan X. (2020). SUMOylation Homeostasis in Tumorigenesis. Cancer Lett. 469, 301–309. 10.1016/j.canlet.2019.11.004 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zuo Y., Zhang H., Kang X., Yue F., Yi Z., et al. (2010). Induction of SENP1 in Endothelial Cells Contributes to Hypoxia-Driven VEGF Expression and Angiogenesis. J. Biol. Chem. 285 (47), 36682–36688. 10.1074/jbc.M110.164236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Li D. X., Cao B. Q., Liu S. J., Xu D. H., Zhu X. Y., et al. (2021). Exercise Training Ameliorates Early Diabetic Kidney Injury by Regulating the H 2 S/SIRT1/p53 Pathway. FASEB j. 35 (9), e21823. 10.1096/fj.202100219R [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhang Y., Sun S. (2019). Deciphering the SUMO Code in the Kidney. J. Cell. Mol. Med. 23 (2), 711–719. 10.1111/jcmm.14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E. T. (2009). SUMOylation and De-SUMOylation: Wrestling with Life's Processes. J. Biol. Chem. 284 (13), 8223–8227. 10.1074/jbc.R800050200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y. K., Wu W. F., Huang X. R., Li H. D., Ren Y. P., Zeng J. C., et al. (2021). Deletion of Smad3 Protects against C-Reactive Protein-Induced Renal Fibrosis and Inflammation in Obstructive Nephropathy. Int. J. Biol. Sci. 17 (14), 3911–3922. 10.7150/ijbs.62929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M., Liu W., Hu Y., Fu N. (2020). Sumoylation in Liver Disease. Clin. Chim. Acta 510, 347–353. 10.1016/j.cca.2020.07.044 [DOI] [PubMed] [Google Scholar]
- Zhang F. P., Mikkonen L., Toppari J., Palvimo J. J., Thesleff I., Jänne O. A. (2008). Sumo-1 Function Is Dispensable in normal Mouse Development. Mol. Cell. Biol. 28 (17), 5381–5390. 10.1128/MCB.00651-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wu C., Gao L., Du G., Qin X. (2020). Astragaloside IV Derived from Astragalus Membranaceus: A Research Review on the Pharmacological Effects. Adv. Pharmacol. 87, 89–112. 10.1016/bs.apha.2019.08.002 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Zhang K., Li Z., Zhang H., Fu F., Fu J., et al. (2021). High Migration and Invasion Ability of PGCCs and Their Daughter Cells Associated with the Nuclear Localization of S100A10 Modified by SUMOylation. Front. Cell Dev Biol 9, 696871. 10.3389/fcell.2021.696871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Zhang X., Rong J. (2021a). SUMOylation as a Therapeutic Target for Myocardial Infarction. Front. Cardiovasc. Med. 8, 701583. 10.3389/fcvm.2021.701583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Zhao J., Zhang X., Fan N., Rong J. (2021b). Upregulation of Small Ubiquitin-like Modifier 2 and Protein SUMOylation as a Cardioprotective Mechanism against Myocardial Ischemia-Reperfusion Injury. Front. Pharmacol. 12, 731980. 10.3389/fphar.2021.731980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Zhou M., Wang Z., Fu Y., Jia M., Wang X., et al. (2019). PGRN Acts as a Novel Regulator of Mitochondrial Homeostasis by Facilitating Mitophagy and Mitochondrial Biogenesis to Prevent Podocyte Injury in Diabetic Nephropathy. Cell Death Dis 10 (7), 524. 10.1038/s41419-019-1754-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Gao C., Huang W., Yang M., Chen G., Jiang L., et al. (2014). High Glucose Induces Sumoylation of Smad4 via SUMO2/3 in Mesangial Cells. Biomed. Res. Int. 2014, 782625. 10.1155/2014/782625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Liang W., Chen Z., Hu J., Feng J., Cao Y., et al. (2021). Mitoquinone Protects Podocytes from Angiotensin II-Induced Mitochondrial Dysfunction and Injury via the Keap1-Nrf2 Signaling Pathway. Oxid. Med. Cell. Longev. 2021, 1394486. 10.1155/2021/1394486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Wang J., Gao J., Han H., Fang Y. (2018). Phosphoproteomic Analysis of the Antitumor Effects of Ginsenoside Rg3 in Human Breast Cancer Cells. Oncol. Lett. 15 (3), 2889–2898. 10.3892/ol.2017.7654 [DOI] [PMC free article] [PubMed] [Google Scholar]