Abstract
Voltage-activated potassium (Kv) channels open upon membrane depolarization and proceed to spontaneously inactivate. Inactivation controls neuronal firing rates and serves as a form of short-term memory and is implicated in various human neurological disorders. Here, we use high-resolution cryo–electron microscopy and computer simulations to determine one of the molecular mechanisms underlying this physiologically crucial process. Structures of the activated Shaker Kv channel and of its W434F mutant in lipid bilayers demonstrate that C-type inactivation entails the dilation of the ion selectivity filter and the repositioning of neighboring residues known to be functionally critical. Microsecond-scale molecular dynamics trajectories confirm that these changes inhibit rapid ion permeation through the channel. This long-sought breakthrough establishes how eukaryotic K+ channels self-regulate their functional state through the plasticity of their selectivity filters.
Structures of the Shaker Kv channel reveal the mechanism of slow C-type inactivation involves dilation of the selectivity filter.
INTRODUCTION
Voltage-activated potassium (Kv) channels open and close in response to changes in membrane voltage, serving critical functions in electrical signaling in neurons and muscle, neurotransmitter and hormone secretion, cell proliferation and migration, and ion homeostasis (1, 2). Functional studies of Shaker Kv channels have been foundational for establishing key mechanistic principles such as voltage sensing, pore opening, and inactivation in eukaryotes (1, 3–5). The structure of the related Kv1.2/2.1 paddle chimera has provided a valuable framework to begin to understand these processes (6, 7), but it has been challenging to capture the protein in distinct conformations, thereby limiting mechanistic insights (8, 9). In addition, the phenotypes of well-characterized mutants in the Shaker Kv channel do not always translate to the Kv1.2 channel (10), leaving key questions unanswered. One of these outstanding questions is what the structural mechanism underlies the so-called C-type inactivation of Shaker Kv channels (11), a slow inactivation process that regulates the firing of action potentials in neurons and cardiac muscle by diminishing the availability of these channels to conduct ions. This intriguing process has been implicated in a range of neurological and psychiatric disorders (1, 12–15) and occurs in other Kv channels too. For example, the human ether-a-go-go–related Kv channel also enters a C-type inactivated state rapidly after opening, which translates into unique inwardly rectifying properties that are critical for shaping the cardiac action potential and preventing arrhythmias (16–19). C-type inactivation has also been proposed to serve important roles in Ca2+-activated K+ channels and K2P channels (20–22).
It has long been hypothesized that C-type inactivation in Shaker Kv channels involves a conformational change in the external pore, as it is influenced by mutations in this region and by ion and blocker occupancy of the selectivity filter (11, 14, 15, 23–35). X-ray structures of mutants stabilizing open and inactivated states of KcsA, a prokaryotic K+ selective channel that consists of a pore domain homologous to that found in Kv channels, support the prevailing view that slow inactivation in K+ channels is caused by a collapse of the ion selectivity filter (36–38). However, the extent to which slow inactivation in KcsA informs on the mechanism of C-type inactivation in Kv channels remains an open question (15, 39, 40). In the present study, we report the first-known structure of the Shaker Kv channel and that of a well-characterized mutant that markedly accelerates C-type inactivation (24, 30, 41, 42). Our findings reveal that C-type inactivation in Shaker involves an unanticipated conformational rearrangement of the external pore that leads to displacement of the P-loop and dilation of the ion selectivity filter.
RESULTS
Structure of the Shaker Kv channel in lipid nanodiscs
The Shaker Kv channel containing an mVenus-tag along with a deletion of residues 6 to 46 to remove fast inactivation (Shaker-IR) (43, 44) was expressed in mammalian cells, purified and reconstituted into lipid nanodiscs using MSP1E3D1 (fig. S1) (8), and its structure was solved using cryo–electron microscopy (cryo-EM; figs. S2 and S3 and table S1). The structural construct is identical to that used in extensive functional studies from many laboratories (6, 11, 23–26, 28–30, 32, 35, 41, 43–47). We determined the structure of the transmembrane (TM) regions of Shaker-IR to 3 Å resolution after focused refinement; the map was of high quality throughout, with clearly resolved densities for most side chains, making de novo atomic model building straightforward. Only two external loops within the voltage-sensing domains were unresolved (residues H254 to T276 and V330 to M356).
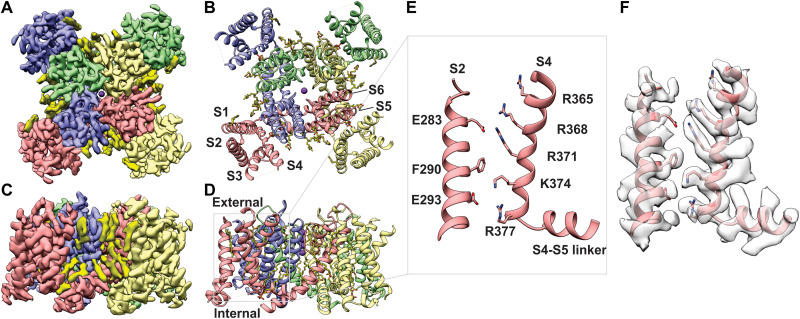
The fold of the tetrameric Shaker-IR Kv channel is remarkably similar to that of the Kv1.2/2.1 paddle chimera channel [Cα root mean square deviation (RMSD), 1.23 Å; TM score, 0.99; figs. S4 and S5] (7). Each subunit in Shaker-IR is composed of six TM helices (S1-S6), with the S1-S4 helices from each subunit forming individual voltage-sensing domains and the tetrameric arrangement of S5-S6 helices forming the central pore domain (Fig. 1, A and B). As in the Kv1.2/2.1 channel structure, Shaker-IR displays a domain-swapped architecture with the S1-S4 voltage-sensing domains positioned near the S5-S6 pore-forming helices of the adjacent subunit (Fig. 1, A and B). The cryo-EM map for Shaker-IR reveals clear densities corresponding to phospholipids that appear to be bound to the protein within both the external and internal leaflets of the lipid nanodisc (Fig. 1, C and D); many of these densities are similar to those seen in the Kv1.2/2.1 paddle chimera solved in mixed detergent/lipid micelles (7).
Fig. 1. Structure of the Shaker-IR Kv channel in lipid nanodisc.
(A and B) View of Shaker Kv channel model (A) and EM map (B) from an extracellular perspective. (C and D) TM side-view of Shaker-IR Kv channel model (C) and EM map (D). Each subunit is shown in different colors and EM densities that appear to be lipid molecules are colored in bright yellow. (E and F) Close-up view of a voltage-sensing domain. The S2 and S4 helices within the voltage-sensing domain highlighted with a gray box in (D) is shown without (E) and with (F) EM density.
The internal structure of the S1-S4 voltage-sensing domain of Shaker-IR is also similar to that in the Kv1.2/2.1 chimera, and in both cases, basic residues in S4 located near the middle of the TM region are stabilized by an occluded cation binding site or charge-transfer center that is composed of a highly conserved Phe residue in the middle of the S2 helix (F290) and acidic residues nearby in S2 and S3 (E293 and D316; Fig. 1, E and F) (48). Within the conserved core region of the S4 helix, four Arg residues (R362, R365, R368, and R371) are positioned external to F290 in S2, while one Lys (K374) is positioned near F290 and the innermost Arg (R377) and Lys (K380) are positioned internal to F290 (Fig. 1, E and F). The fact that the four basic residues in S4 known to carry most of the gating charge (49, 50) are accessible to the external solution suggests that our structure captures the voltage-sensing domains in the activated state, which, for this channel, is predominant at 0 mV. Accordingly, the intracellular S6 gate region (51) is open (Fig. 2, C to E), further indicating that the structure represents an activated/open state.
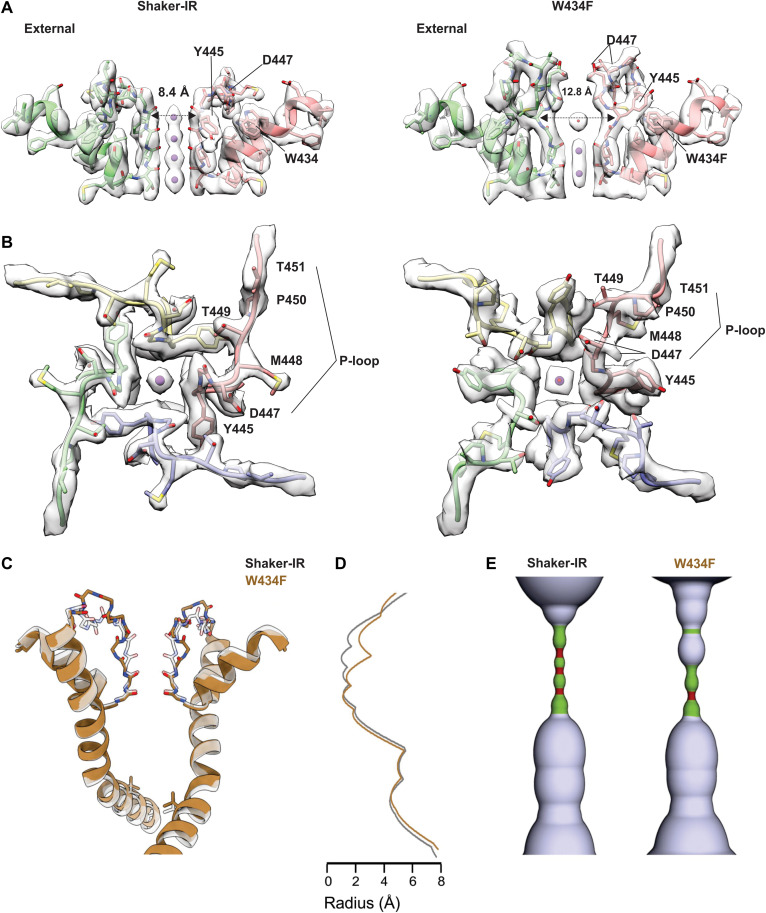
Fig. 2. Structure of the external pore of wild-type Shaker-IR and the W434F mutant.
(A) Side view of the selectivity filter of Shaker-IR (left) and W434F (right). The distance between Y445 CA is shown to highlight the dilation of the selectivity filter in W434F. (B) View of the pore domain of Shaker-IR (left) and W434F (right) viewed from an extracellular perspective. Residues experiencing a large movement are labeled to highlight structural reorientation of the outer pore domain in W434F. (C) Superposition of pore-lining regions for Shaker-IR and the W434F mutant. V478 is shown to mark the position of the internal S6 gate that prevents ion permeation across the internal pore when the Shaker Kv channel closes (51). (D) Plot of pore radius for Shaker-IR and the W434F mutant. (E) Hole diagrams illustrating the ion permeation pathways for Shaker-IR and W434F. Radii ≤1 Å are shown in red; radii >1 Å and ≤ 2 Å are shown in green, and radii larger than 2 Å are shown in light blue. The plot in (B) and the diagram in (C) are aligned with the protein model in (A).
Within the external pore, in the ion selectivity filter, backbone carbonyl oxygens line the ion permeation pathway and are positioned similarly to other K+ channel structures thought to represent a conducting state (Fig. 2A) (52). Strong EM densities can be clearly discerned at four positions within the filter and along its central axis (Fig. 2A); these densities are consistent with those revealed in previous structural studies (52) and appear to reflect a series of ion binding sites within the filter. In summary, there is every indication that the structure of Shaker-IR that we have resolved represents an activated, open, and conducting conformation.
Structure of the W434F mutant Shaker Kv channel in lipid nanodiscs
The W434F mutant of Shaker-IR has been extensively characterized and shown to be effectively nonconducting because the mutation accelerates entry of channels into the C-type inactivated state upon membrane depolarization (24, 30, 41, 42). We introduced the W434F mutation into our structural construct, expressed and purified the protein, and reconstituted it into lipid nanodiscs using the same approach as for Shaker-IR (fig. S1). We solved the structure of the W434F mutant with an overall resolution of 2.9 Å within the TM region; as for Shaker-IR, the cryo-EM map is of excellent quality throughout, enabling us to model the complete sequence of the TM region with the only exception of two external loops in the voltage sensors (figs. S3 and S6 and table S1). Structural alignment of the S1-S6 regions of the W434F mutant and Shaker-IR reveals that their backbone folds are very similar (fig. S7), with low Cα RMSD (1.07 Å) and comparably high TM score (0.9). The structures of the S1-S4 voltage-sensing domains in the two proteins are essentially indistinguishable, and in both instances, the internal pores are clearly open (Fig. 2, C to E, and fig. S7).
In contrast, a pronounced conformational change occurs within the external pore (Fig. 2, A and B, and movie S1), a region where both cryo-EM maps have the highest resolution (figs. S2 and S6). A key interaction between D447 in the P-loop and W434 in the pore helix, observed in Shaker-IR and most other K+ channels (7, 41, 52), is broken in W434F, resulting in a displacement of the P-loop toward the external solution by about 5 Å (Figs. 2, A and B, and 3A and fig. S7B). At the external end of the ion selectivity filter, the side chain of Y445 undergoes a marked reorientation in the mutant, breaking off its hydrogen bond contact with T439 in the adjacent subunit and rotating 90° to reposition behind the filter of the same subunit (Figs. 2, A and B, and 3A and fig. S7B). M448 and T449, two critical positions for C-type inactivation (25, 28, 32), also move large distances, with the Cα of M448 moving about 5 Å from an exposed position in Shaker-IR to a buried site near to W435 in the rapidly inactivating mutant (Fig. 3A and fig. S7B). In contrast, both T449 and P450 flip from being partially buried in Shaker-IR to completely exposed to solvent in W434F (Figs. 2, A and B, and 3A and fig. S7, B and C). The cryo-EM density map for the mutant indicates two distinct rotamers of the D447 side chain, one of which positions the acidic residue near T449 (Figs. 2B and 3A and fig. S7C). It is apparent, therefore, that there is an overall reorganization of side-chain interactions behind the filter in the inactivated state, with critical residues within the P-loop between Y445 and P450 undergoing substantial displacements. This reorganization translates into a marked expansion at the external portion of the ion selectivity filter, by about 4 Å (Fig. 2, A to E, and movie S1). Ion densities therein are perturbed, while those at the internal end of the filter are seemingly unaltered (Fig. 2A and fig. S8). Inspection of maps and half maps with either C4 or C1 symmetry imposed shows that the density within the dilated portion of the filter is relatively weak, suggestive of occupancy by water molecules or possibly K+ ions (Fig. 2A and fig. S8).
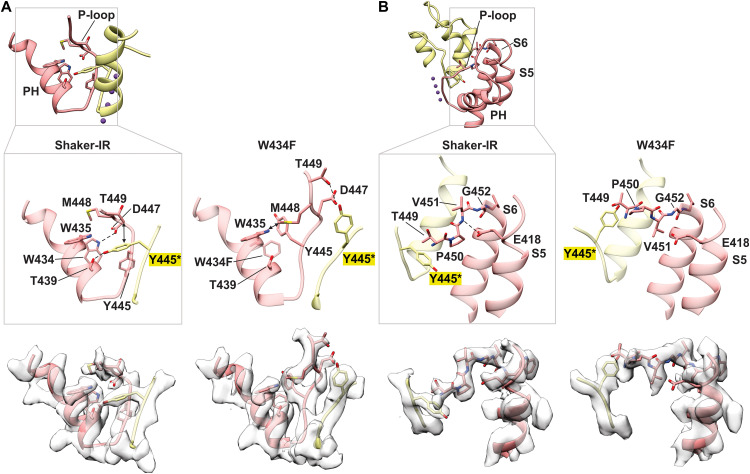
Fig. 3. Conformational changes reposition residues that are critical for C-type inactivation in Shaker Kv channels.
(A) Residue interactions within the pore helix (PH) and P-loop for Shaker-IR and the W434F mutant. Inset above provides the orientation of PH and P-loop for two subunits. Enlarged models are shown without or with cryo-EM density (below). D447 forms a hydrogen bond with W434 (OE − NE, 3.3 Å) in Shaker-IR but not in W434F. CG of T449 is 3.6 Å from Y445 in the adjacent subunit in Shaker-IR but 7.5 Å in W434F given movement of both residues. T439 forms a hydrogen bond with Y445 (2.4 Å) in the adjacent subunit in Shaker-IR, which is broken in W434F given the large movement of Y445. In W434F, OG of T449 is 3.2 Å from the OE of D447, and the side chain of M448 is 3.5 Å from W435. (B) Residue interactions between E418 in S5 with residues in the P-loop for Shaker-IR and the W434F mutant. The inset above provides the orientation of S5, PH, P-loop, and S6 for two subunits. Enlarged models are shown without or with cryo-EM density (below). E418 at the top of S5 forms a hydrogen bond with the backbone N of V451 in the P-loop (3.1 Å) in Shaker-IR but not in W434F (4.3 Å). The pronounced flipping of the side chain of V451 in the W434F mutant directs its side chain toward E418.
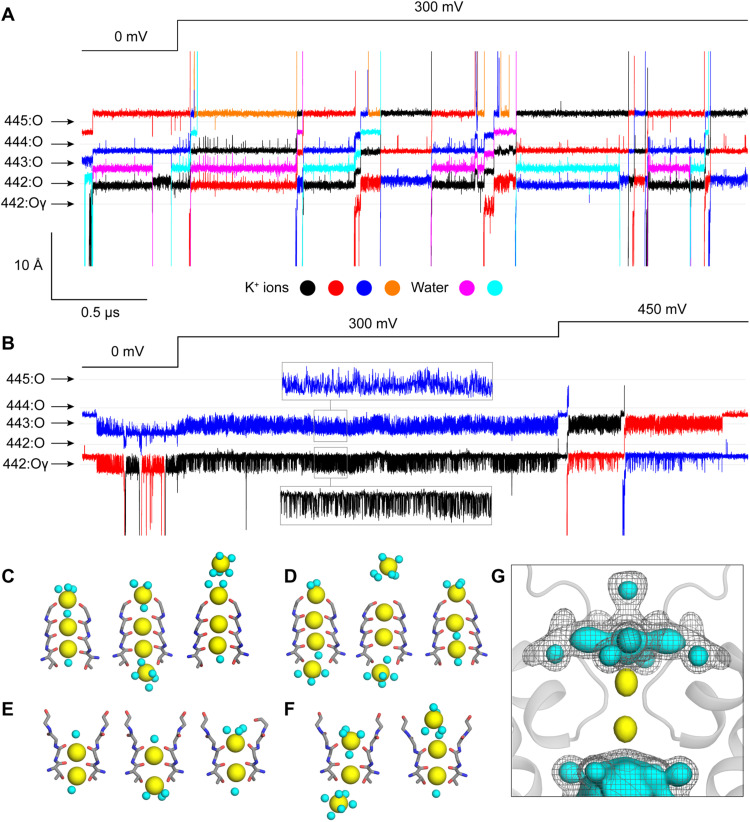
It seems apparent that the balance of ion-protein and ion-solvent interactions that controls K+ permeation in the activated state is somehow perturbed by the dilation of the external portion of the filter (Fig. 2, C to E) and the reconfiguration of key side chains such as D447. However, whether conduction is indeed impaired in W434F is not self-evident from the structure alone. To evaluate this question, we calculated a series of molecular dynamics (MD) trajectories specifically designed to evaluate the ion conducting properties of the pore domain in each of the two structures determined experimentally (Materials and Methods and fig. S9). For Shaker-IR, we calculated a trajectory of 3.5 μs that included a TM voltage jump from 0 to 300 mV at t = 0.5 μs. In symmetric 300 mM KCl, we observed a total of 12 complete permeation events after the voltage jump (Fig. 4, A, C, and D). In all cases, the K+ ions traversed the filter in the outward direction and in a stepwise manner, transiently interacting with each of the carbonyl groups along the way (movie S2). One occupancy state however emerges as the most populated at 300 mV (and 300 mM KCl), featuring three K+ ions: At the external end of the filter, one K+ is, in part, coordinated by the carbonyls of Y445 and, in part, hydrated; a second ion concurrently interacts with the carbonyls of G444 and V443, and the third is approximately at the same level as the T442 carbonyls (Fig. 4, C and D). As has been noted for other channels, K+ permeation proceeds through a knock-on mechanism (52–54); we observe that this mechanism is, at times, mediated by water molecules permeating in between adjacent K+ ions, while at other times, the K+ ions interact directly (Fig. 4, A and C, and movie S2). Note that the single-channel conductance inferred from this trajectory is approximately 3.5 pS or 10-fold slower than the value experimentally measured in symmetric 300 mM KCl (55). Given the myriad approximations and simplifications adopted in MD simulations, this discrepancy appears reasonable and consistent with existing literature (56). Errors in the order of 1 kcal/mol in the rate-limiting barriers for conduction are most certainly expected and would be sufficient to explain the observed discrepancy. Thus, while additional simulations will be no doubt of interest from both mechanistic and methodological standpoints, we posit that the existing data reaffirm the notion that the structure of Shaker-IR reported here captures the conductive form of the channel.
Fig. 4. Altered ion interactions with the filter during C-type inactivation in Shaker Kv channels.
(A) MD simulation of K+ permeation across the pore domain of Shaker-IR. The figure shows the time series of the position of K+ ions as they traverse or transiently enter the selectivity filter; the time series of water molecules traversing the filter are also shown. The time resolution is 120 ps. The relative position of coordinating backbone/side-chain oxygens in residues 442 to 445 is indicated. Note that no voltage was applied during the first 0.5 μs of the trajectory; a jump to 300 mV was introduced at this point and sustained for 3 μs thereafter. (B) Same as (C), for W434F mutant. Note that, in this case, a second voltage jump from 300 to 450 mV was introduced at 2.5 μs and sustained for 1 μs thereafter. (C and D) Snapshots of two K+ permeation events during the simulation of Shaker-IR. K+ ions and water molecules are shown as yellow and cyan spheres, respectively. The elapsed times between snapshots are approximately 3 and 4 ns, respectively. (E) Representative configurations of the selectivity filter in the simulation of the W434F mutant at 300 mV. Both ions fluctuate but fail to permeate in 2 μs of trajectory under this condition. (F) Snapshots of a K+ permeation event during the simulation of W434F after the jump to 450 mV. The elapsed time between snapshots is approximately 8 ns. (G) Calculated isodensity maps for K+ (yellow surface) and water (cyan surface, gray mesh) near and within the selectivity filter of W434F based on the analysis of 2 μs of trajectory under 300 mV. For water, two isovalues are used to illustrate the emergence of well-defined, fourfold symmetric structures resulting from interactions with ions and protein as well as confinement.
The simulation calculated for the W434F mutant, which was designed identically to that carried out for Shaker-IR and is equivalent in total length, shows radically different results. We did not observe any permeation events before or after the voltage jump from 0 to 300 mV for a total of 2.5 μs of simulation (Fig. 4B and movie S3). Instead, two K+ ions persistently reside in the filter, coordinated by the carbonyls of V443 and the side chain and backbone of T442 (Fig. 4E), while multiple water molecules occupy the pocket made available near G444 and Y445 upon dilation of the filter (Fig. 4G). The occupancy of water molecules within the external end of the filter in the simulations suggests that the weak density observed within the cryo-EM maps in this region (Fig. 2A and fig. S8) is more likely to be water than a K+ ion. These observations notwithstanding, we reasoned that because the selectivity filter of W434F is dilated, and not constricted, conduction ought to be feasible without a structural change provided a sufficiently strong driving force. A second voltage jump from 300 to 450 mV during the last 1 μs of the trajectory showed two full permeation events (Fig. 4, B and F). The permeation mechanism of W434F differs from that observed for Shaker-IR, however, not only in the number of K+ ions involved but also in that K+ ions do not dwell at the dilation site near G444 and Y445 before reaching the external solution (Fig. 4B and movie S3). Analogous voltage jumps from 300 to 600 mV and from 300 to 900 mV led to a larger number of K+ ion permeation events, but nevertheless, the preferred species at this distortion was again water. From these data, we can extract two tentative conclusions: First, the reconfiguration of the external pore observed in the W434F mutant severely impairs the conducting properties of the selectivity filter in comparison to Shaker-IR at the same conditions; second, this reconfiguration impairs conduction not by sterically blocking the ion pathway but rather by imposing a mechanism of K+ permeation that entails a substantially larger energetic barrier as a result of the filter dilation. Whether K+ ions can traverse this barrier at all in experimentally accessible conditions remains, however, an open question. It will be of great interest to further dissect and quantify these phenomena in future studies.
DISCUSSION
Our objective was to solve structures of the Shaker Kv channel to understand its mechanism of C-type inactivation, which is likely shared by many other eukaryotic channels. The structure of Shaker-IR provides an important framework for future studies aimed at exploring voltage-dependent gating mechanisms and for reexamining a large body of functional studies to gain new mechanistic insights (1, 4, 5). The value of structures of the same protein used in extensive functional studies is exemplified here with the W434F mutation, which renders the channel effectively nonconducting by greatly promoting C-type inactivation (24, 30, 42).
Is the conformational rearrangement and dilation of the selectivity filter that we observe in W434F representative of the structural change occurring during C-type inactivation in wild-type Shaker Kv channels? Although additional studies are warranted to explore this question further and C-type inactivation might involve multiple distinct conformational changes in the outer pore, the structural changes that we observe in W434F are remarkably consistent with many functional studies exploring the mechanism of C-type inactivation in the Shaker Kv channel. The hydrogen bond between D447 and W434 that breaks off in the structure of W434F (Fig. 3A) has been shown to stabilize the conducting state of the Shaker Kv channel (41), with mutation of either residue markedly promoting C-type inactivation (24, 30, 32, 41, 57). T449 is a particularly critical residue for C-type inactivation in the Shaker Kv channel because polar substitutions speed C-type inactivation and hydrophobic substitutions slow inactivation (25). Hydrophobic substitutions at T449 so markedly slow C-type inactivation that they can rescue normal ion conduction in W434F (29). The structure of Shaker-IR reveals that T449 is partially buried, as it is positioned within 4 Å of the aromatic ring of Y445 in the adjacent subunit, whereas it becomes completely solvent exposed in the C-type inactive state (Fig. 3A). Hydrophobic substitution at 449 would thus be expected to stabilize the conducting conformation by strengthening hydrophobic interactions between 449 and Y445. The large displacement of T449 in W434F places this side chain within 3.2 Å of D447 for one of the resolved rotamers for the D447 side chain (Fig. 3A), which explains why Cys substitutions at position 449 generate a strong metal binding site that forms at much faster rates when Shaker Kv channels are inactivated (26). Although these metal bridges had been envisioned to form between T449C residues in opposing subunits across the pore axis, thereby supporting the theory that C-type inactivation entails the collapse of the filter, our structures reveal an alternate interpretation, namely, the metal bridges form between T449C and D447 within the same subunit in the C-type inactivated state. External Ba2+ ions have been shown to inhibit the Shaker Kv channel at a site that is sensitive to substitutions at both T449 and D447 (57), which can be readily understood given the intimate relationship between these residues seen in the W434F structure. The increased solvent exposure of T449 and P450 observed in the W434F structure is also consistent with the enhanced rate of modification of introduced Cys residues at these positions by thiol-reactive compounds when Shaker C-type inactivates (28). The intersubunit hydrogen bond between T439 and Y445 observed in Shaker-IR, which is necessarily broken with the large movement of Y445 that occurs in the structure of W434F (Fig. 3A), has also been shown to stabilize the open state of Shaker Kv channels, with mutation of either residue markedly promoting C-type inactivation (41). Last, the structural changes that we observe can explain a series of important observations around E418 within the external end of the S5 helix near the perimeter of the pore domain (Fig. 3B). Introduction of fluorophores at position S424 in the Shaker Kv channel, which, in the structure of Shaker-IR, is nearby to E418, gives rise to fluorescence changes that closely track C-type inactivation (58). Mutations of E418 markedly speed C-type inactivation in Shaker Kv channels (59, 60), and substitutions with Cys at E418 and V451 within the P-loop lead to disulfide bond formation that stabilizes the C-type inactivated state (60). In the structure of Shaker-IR, E418 hydrogen bonds with the backbone amide of V451 in the P-loop and this interaction is lost as the P-loop is displaced in W434F (Fig. 3B). Notably, displacement of the P-loop in the W434F mutant is accompanied by flipping of the side chain of V451 toward E418 (Fig. 3B). Although side-chain density is missing for E418 in the W434F mutant, the EM density for the external end of the S5 helix is similar in Shaker-IR and W434F (Fig. 3B), consistent with E418C and V451C being optimally positioned to form a disulfide bond that stabilizes the C-type inactivated state.
It is important to clarify that our structures are not at all inconsistent with the observation that externally applied tetraethyl ammonium or K+ ions can interfere with C-type inactivation in Shaker Kv channels. While this observation had been rationalized using a foot-in-the-door analogy, in support of the notion that the filter collapses upon inactivation (26, 28), a mechanism whereby binding of these ions within the selectivity filter hinders inactivation by precluding its dilation is equally plausible (15). All that is required to explain this particular experimental observation is for the conducting filter to interact more favorably with external ions than the inactive state; the structure of the W434F mutant of the Shaker Kv channel is clearly consistent with this premise, as it lacks the external ion-binding sites seen in the conducting state.
The structural changes that we observe in the ion selectivity filter also provide important insight into how inactivation of the selectivity filter might be coupled with other conformational mechanisms elsewhere in the channel. For example, in KcsA, opening of the internal pore is thought to trigger collapse of the selectivity filter because a bulky Phe in the pore-lining M2 helix interacts with a conserved Thr at the base of the ion selectivity filter (fig. S10D) (38, 61). Our structures suggest that this coupling mechanism is unlikely to apply for the Shaker Kv channel, as the base of the ion selectivity filter is unchanged when comparing Shaker-IR and the W434F mutant (Fig. 3 and fig. S10C). The reason for this difference might be that the residue equivalent to the bulky Phe in the S6 helix of Shaker is I470 (fig. S10C), or it might simply be that the conformational changes during inactivation in KcsA are unrelated to those occurring in Shaker (fig. S10). C-type inactivation in Shaker is thought to be coupled to the conformation of the voltage-sensing domains (58, 60, 62). In the future, it will be interesting to further explore coupling between voltage sensor activation, internal pore opening, and C-type inactivation by solving structures of the Shaker Kv channel with voltage sensors in resting states and the pore in a closed state.
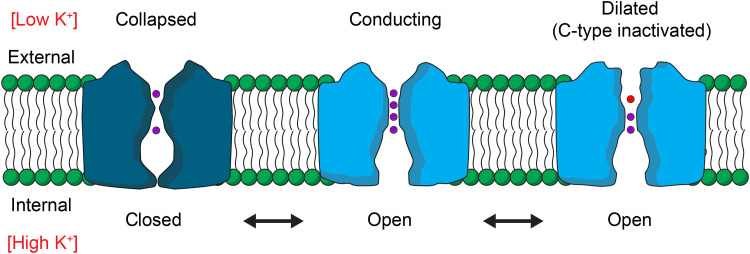
A structural mechanism for C-type inactivation through filter dilation raises the interesting question about what conditions might promote a more constricted or collapsed conformation, similar to that observed in the KcsA channel in the presence of low K+ concentrations (52). If a K+-dependent collapse of the filter also occurs in Kv channels, then it would likely represent a conformation adopted each time the internal gate of a K+ channel closes under physiological ionic conditions because, in that state, the filter will ultimately equilibrate with the external solution containing low [K+] (Fig. 5). Collapse of the filter would assure that ion permeation is minimized when the internal gate closes in the absence of an activating stimulus (46). It will be interesting to determine whether the ion selectivity filters in Kv channels collapse in low K+ because it would indicate that the filter adopts at least three distinct conformations during the gating cycle of a Kv channel. It will also be exciting to determine whether other K+ channels use filter dilation to regulate ion permeation. Although structural changes related to those that we see here have thus far not been resolved within the filter of K2P channels, for example, closure of those channels has been proposed to involve rearrangement of residues in the P-loop and dilation of the filter and has been shown to deplete ion occupancy in the external half of the filter while maintaining those within the inner half (63), suggesting that the mechanisms of C-type inactivation in Shaker and filter gating in K2P channels may be related.
Fig. 5. Gating cycle of the pore domain of a Kv channel.
Cartoons illustrating dynamic changes in structure of the ion conduction pore during the gating cycle of a Kv channel. With voltage sensor activation and opening of the internal S6 gate, the filter will equilibrate with the high potassium concentration inside the cell and adopt a conducting conformation (middle). With prolonged activation, the filter C-type inactivates and dilates to diminish ion permeation (right). Four K+ ion binding sites (purple) within the filter can be occupied in the conducting state, whereas in the C-type inactivated state, ions primarily occupy only the internal half of the filter and water molecules (red) likely occupy the dilated external part of the filter. At negative membrane voltages where the voltage-sensing domains are resting and the internal S6 gate is closed (left), the filter would equilibrate with the external solution containing a low potassium concentration and potentially collapse, as observed in structures of the KcsA potassium channel (52).
MATERIALS AND METHODS
Shaker Kv channel expression using baculovirus and mammalian expression system
To produce the Shaker Kv channel for cryo-EM, wild-type and W434F channels were cloned into the pEG vector in which enhanced green fluorescent protein was substituted with mVenus (64) and expressed in tsA201 cells using the previously published baculovirus-mammalian expression system with a few minor modifications (65). Briefly, P1 virus was generated by transfecting Sf9 cells (~2.5 million cells on a T25 flask with a vent cap) with 50 to 100 ng of fresh bacmid using Cellfectin. After 4 to 5 days of incubation in a humidified 28°C incubator, the cell culture medium was collected by centrifugation (3000g × 10 min), supplemented with 2% fetal bovine serum (FBS), and filtered through a 0.45-μm filter to harvest the P1 virus. To amplify the P1 virus, ~500 ml of Sf9 cell cultures at a density of ~1.5 million cells/ml was infected with 1 to 200 μl of the virus and incubated in a 28°C shaking incubator for 3 days. The cell culture medium was then collected by centrifugation (5000g × 20 min), supplemented with 2% FBS, and filter through a 0.45-μm filter to harvest P2 virus. The volume of P1 virus used for the amplification was determined by carrying out a small-scale amplification screening in which ~10 ml of Sf9 cell cultures at the same density were infected with different volumes of P1 virus and harvested after 3 days to transduce tsA201 cells and compare the expression level of Shaker Kv channels using mVenus fluorescence intensity. The P2 virus was protected from light using aluminum foil and stored at 4°C until use. To express the Shaker channels, tsA201 cells at ~1.5 million cells/ml in Freestyle medium with 2% FBS were transduced with 10% (v/v) P2 virus and incubated at a 37°C CO2 incubator. To boost the protein expression, sodium butyrate (2 M stock in H2O) was added to 10 mM at ~16 hours of posttransduction. The culture was continued at the 37°C CO2 incubator for another 24 hours, and the cells were harvested by centrifugation (5000g × 20 min) and frozen at −80°C until use.
Shaker Kv channel purification
Before extraction of the Shaker channels from tsA201 cells, membrane fractionation was carried out using a hypotonic solution and ultracentrifugation. The cells were first resuspended in a hypotonic solution [20 mM tris (pH 7.5) and 10 mM KCl] with protease inhibitors (pepstatin, aprotinin, leupeptin, benzamidine, trypsin inhibitor, and phenylmethylsulfonyl fluoride) using a Dounce homogenizer, incubated at 4°C for ~30 min, and centrifuged at 1000g for 10 min to remove cell debris. The supernatant was ultracentrifuged for 1 hour (45,000 rpm; Beckman Ti45 rotor), and collected membranes were stored at −80°C until use. To purify Shaker Kv channels, the fractionated membranes were resuspended in an extraction buffer [50 mM tris (pH 7.5), 150 mM KCl, 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 50 mM n-Dodecyl-B-D-Maltoside (DDM), and 5 mM Cholesteryl Hemisuccinate (CHS) with the protease inhibitor mixture used above] and extracted for 1 hour at 4°C. The solution was clarified by centrifugation (12,000g × 10 min) and incubated with CoTALON resins at 4°C for 1 hour, at which point the mixture was transferred to an empty disposable column (Econo-Pac; Bio-Rad). The resin was washed with 10-column volume of Buffer A [50 mM tris (pH 7.5), 150 mM KCl, 1 mM DDM, 0.1 mM CHS, and porcine brain total lipid extract (0.1 mg/ml)] with 10 mM imidazole, and bound proteins were eluted with Buffer A with 250 mM imidazole. The eluate was concentrated using Amicon Ultra (100 kDa) to ~350 to 450 μl and loaded onto a Superose 6 (10 mm by 300 mm) gel filtration column and separated with Buffer A. All purification steps described above were carried out at 4°C or on ice.
Lipid nanodisc reconstitution of the Shaker Kv channel
Lipid nanodisc reconstitution was performed following the previously published methods with minor modifications (8). On the day of nanodisc reconstitution, the Shaker Kv channel purified by Superose 6 in detergent was concentrated to ~1 to 3 mg/ml and incubated with histidine-tagged MSP1E3D1 and 3:1:1 mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine for 30 min at room temperature. The mixture was transferred to a tube with Bio-Beads (~30- to 50-fold of detergent; w/w) and incubated at room temperature for ~3 hours in the presence of Tobacco Etch Virus (TEV) protease (prepared in-house) and 2 mM TCEP to remove N-terminal fusion protein including poly-histidine and mVenus tag. The reconstituted protein was loaded onto Superose 6 column (10 mm by 300 mm) and separated using 20 mM tris and 150 mM KCl buffer at 4°C. The success of nanodisc reconstitution was confirmed by collecting separated fractions and running SDS–polyacrylamide gel electrophoresis to verify the presence of Shaker Kv and MSP1E3D1 bands at a similar ratio. Typically, optimal reconstitution required the incubation of 1:10:200 or 1:10:400 molar ratio of tetrameric Shaker Kv, MSP1E3D1, and the lipid mixture.
Cryo-EM sample preparation and data acquisition
Concentrated samples of Shaker-IR or the W434F mutant in nanodiscs (3 μl) were applied to glow-discharged Quantifoil grids (R 1.2/1.3 Cu 300 mesh). The grids were blotted for 2.5 s, with blot force of 4 and 100% humidity, at 16°C using an FEI Vitrobot Mark IV (Thermo Fisher Scientific), followed by plunging into liquid ethane cooled by liquid nitrogen. Images were acquired using an FEI Titan Krios equipped with a Gatan LS image energy filter (slit width, 20 eV) operating at 300 kV. A Gatan K3 Summit direct electron detector was used to record movies in superresolution mode with a nominal magnification of ×105,000, resulting in a calibrated pixel size of 0.43 Å per pixel. The typical defocus values ranged from −0.5 to −1.5 μm. Exposures of 1.6 s were dose-fractionated into 32 frames, resulting in a total dose of 52 e− Å−2. Images were recorded using the automated acquisition program SerialEM (66).
Image processing
All processing was completed in RELION (67) and cryoSPARC (68). The beam-induced image motion between frames of each dose-fractionated micrograph was corrected and binned by 2 using MotionCor2 (69) and contrast transfer function (CTF) estimation was performed using CTFFIND4 (70). Micrographs were selected, and those with outliers in defocus value and astigmatism, as well as low resolution (>5 Å) reported by CTFFIND4 were removed. The initial set of particles from 300 micrographs were picked using Gautomatch (www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/#gauto) and followed by reference-free two-dimensional (2D) classification in RELION. The good classes were then used as template to pick particles from all selected micrographs using Gautomatch. Particles [4,934,881 (Shaker-IR) and 7,446,104 (W434F)] were picked and extracted with 2× downscaling (pixel size, 1.72 Å). Several rounds of reference-free 2D classification were performed to remove ice spot, contaminants, and bad particles, yielding 2,246,076 (wild-type) and 2,347,164 (W434F) particles, respectively. The good particles were 3D classified using reference generated by 3D initial model in C1 symmetry. Good classes were then selected and followed by further 2D classification and 3D classification. The classes that show good features within the TM domains were merged. After removing duplicate particles, the selected particles were reextracted using box size of 296 pixels without binning (pixel size, 0.86 Å). To get good reconstruction within the TM domain, the reextracted particles were further 3D classified with a TM domain mask in C4 symmetry, followed by 3D auto-refine and CTF refinement. After that, Bayesian polishing was performed, and bad particles were removed from polishing particles using 2D classification. The selected polishing particles were subjected to 3D auto-refine in RELION and then nonuniform refinement in cryoSPARC. The final reconstruction was reported at 3.0 Å for Shaker-IR and 2.9 Å for W434F. To confirm whether ion density in the selectivity filter is real, the reference was shifted by 10 pixels in the X direction using relion_image_handler so that the selectivity filter is not in the center axis. C1 symmetry was applied for 3D auto-refine. The ion density without postprocessing is shown in fig. S8.
Model building and structure refinement
Model building was first carried out by manually fitting the TM domain of Kv1.2/2.1 paddle chimera channel (Protein Data Bank, 6EBM) into the EM density map using UCSF Chimera (71). The model was then manually built in Coot (72) and refined using real_space refinement in PHENIX (73) with secondary structure and geometry restraints. The final model was evaluated by comprehensive validation in PHENIX. Structural figures were generated using PyMOL (https://pymol.org/2/support.html) and UCSF Chimera.
Structural alignments
Alignments of single subunits (fig. S7) were performed using align in PyMOL. Alignments of tetramers (figs. S4 and S10) were performed using Fr-TM-Align (74). Alignments between KcsA and Shaker (fig. S10) were performed using the pore domain (S5-S6 helices) of all four subunits. Alignments between the Kv 1.2/2.1 paddle chimera and Shaker (fig. S4) were performed using the TM domain (S1-S6 helices) of all four subunits. Pore radii were calculated using the HOLE program (75).
MD simulations of Shaker-IR and the W434F mutant
The simulations of Shaker-IR and the W434F mutant used the cryo-EM structures reported in this study as the starting configuration; in both cases, the simulations examined a construct encompassing residues 382 to 485 (from S4-S5 linker helix to S6 helix), with neutral N and C termini and all ionizable side chains in their default protonation state at pH 7 (the net charge of these constructs is thus zero). For Shaker-IR, two K+ ions were initially positioned in the selectivity filter, one coordinated by residues 442 and 443 and another by residues 444 and 445; a third ion was positioned below the side chain of 442, and two water molecules were modeled between the three ions. For the W434F mutant, K+ ions were initially positioned between residues 443 and 444 and by residue 442, with a water molecule in between. (Note that neither configuration was ultimately the most populated in the resulting trajectories.) Dowser (76) was used to model water molecules buried within the protein structures. To complete the initial setup of the experimental cryo-EM structures, both constructs (including ions and water molecules) were briefly energy-minimized using CHARMM (77) and the CHARMM36m force field (78–80); specifically, the minimization consisted of 250 steps using the steepest-descent algorithm, followed by 250 steps using the conjugate-gradient algorithm. Both Shaker-IR and W434F were simulated in a POPC lipid bilayer flanked by a 300 mM KCl solution. To generate a molecular model of this membrane/solvent environment, we created a coarse-grained (CG) POPC-lipid bilayer in 300 mM KCl in an orthorhombic box of ~90 Å by 90 Å by 100 Å using insane.py (81). To equilibrate this CG system, we carried out a 20-μs MD simulation using GROMACS 2018.8 (82) and the Martini2.2 force field (83–86) at constant temperature (303 K) and constant semi-isotropic pressure (1 atm) and with periodic boundary conditions. The integration time step was 20 fs. To embed Shaker-IR in this environment, its atomic structure was first coarse-grained using martinize.py (83, 85) and overlaid onto the membrane/solvent system in a configuration that seemed plausible, removing all overlapping lipid/water molecules. To optimize the protein/lipid/solvent interfaces in the resulting model, we carried out a 10-μs MD simulation of the complete system using GROMACS 2018.8 (82) and Martini2.2 (83–86) at constant temperature (303 K) and constant semi-isotropic pressure (1 atm) and with periodic boundary conditions and an integration time step of 20 fs. Having ascertained the equilibration of the membrane structure and of the position of the protein in the lipid bilayer, the final snapshot of the CG trajectory was transformed into an all-atom representation based on the CHARMM36m force field (78–80). To do so, lipid and solvent molecules were back-mapped (87) onto all-atom models, and the CG version of Shaker-IR was replaced with the energy-minimized all-atom construct described above after optimally superposing the Cα trace of the latter onto that of the former. The resulting all-atom molecular system includes 201 POPC lipids, 14,376 water molecules, and 79 K+ and 79 Cl− (300 mM KCl) in an orthorhombic box of ca. 90 Å by 90 Å by 100 Å.
To further optimize this all-atom model, the simulation system was first energy-minimized for 5000 steps using with NAMD 2.13 (88, 89) and the conjugate-gradient algorithm. We then carried out a series of MD simulations wherein structural restraints are applied to the protein and ions/water in the selectivity filter progressively weakened over the course of ~150 ns. The scheme followed in this multistep equilibration is summarized in table S2. These simulations were carried out using NAMD 2.13 (88, 89) and CHARMM36m (78–80) at constant temperature (298 K) and constant semi-isotropic pressure (1 atm) with periodic boundary conditions and an integration time step of 2 fs. Electrostatic interactions were calculated using PME, with a real-space cutoff value of 12 Å; van der Waals interactions were also cut off at 12 Å, with a smooth switching function taking effect at 10 Å. The all-atom simulation system for the W434F mutant was constructed by replacing the structure of Shaker-IR with the structure of the W434F mutant at the end of step 6 in the equilibration protocol summarized in table S2. The resulting all-atom molecular system includes 201 POPC lipids, 14,375 water molecules, and 79 K+ and 79 Cl− (300 mM KCl) in an orthorhombic box of ca. 90 Å by 90 Å by 100 Å. The complete minimization/equilibration protocol (steps 1 through 9) was then repeated for the W434F mutant. No external electric field was applied during either the equilibration of Shaker-IR or the W434F mutant, so the TM voltage in both cases was zero. The dimensions of both simulation systems at the end of the equilibration protocol are ca. 89 Å by 89 Å by 92 Å. Last, to quantify the ion-conducting properties of Shaker-IR and the W434F mutant, a 3.5-μs MD trajectory was calculated for each construct using an Anton 2 supercomputer (90) and the CHARMM36m force field (78–80). The starting configuration for each of these simulations was the final configuration after step 9 in the corresponding equilibration. These simulations were also carried out at constant temperature (298 K) and semi-isotropic pressure (1 atm), set with the Nose-Hoover thermostat (91, 92) and the Martyna-Tobias-Klein barostat (93), respectively, and with periodic boundary conditions and an integration time step of 2.5 fs. Electrostatic interactions were calculated using the Gaussian split Ewald method (94); van der Waals interactions were cut off at 10 Å. To ensure that the simulations evaluate the properties of the functional states captured by the cryo-EM data, we precluded structural distortions that might occur in the microsecond time scale by supplementing the force field with a series of potential energy terms that bias but do not confine the dynamics of all Φ, Ψ, and χ1 dihedral angles toward the values observed in the experimental structures. The functional form of each of these terms is
which is identical to that used in previous studies based on microsecond simulations calculated on Anton (95, 96), but we used a weaker bias k equal to 0.6 kcal/mol. Accordingly, the conformational ensembles in our simulations of Shaker-IR and W434F are quite diverse; for example, typical RMSDs of the protein backbone from the cryo-EM structures range from 1 to 1.4 Å (fig. S9). No external electric field was applied during the first 0.5 μs of either of MD trajectories. At 0.5 μs, a voltage jump to 300 mV was introduced in the system, for both Shaker-IR and the W434F, through the application of an outwardly directed electric field of 0.075 kcal mol−1 Å−1 e−1 perpendicular to the membrane plane (1 kcal mol−1 Å−1 e−1 = 43.4 mV/Å). For a W434F, a second voltage jump to 450 mV was introduced at 2.5 μs through the application of an outwardly directed electric field of 0.1125 kcal mol−1 Å−1 e−1.
Acknowledgments
We thank A. Jara-Oseguera, M. Holmgren, M. Mayer, and members of the Swartz laboratory for helpful discussion and H. Wang of the NIH Multi-Institute Cryo-EM Facility (MICEF) for assistance in acquiring cryo-EM data. This work used the NIH MICEF and computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov), and the Pittsburgh Supercomputing Center provided access to an Anton2 computer donated by D. E. Shaw Research through NIH grant R01-GM1169161.
Funding: This research was supported by the Intramural Research Programs of the National Institute of Neurological Disorders and Stroke, NIH, Bethesda, MD to K.J.S. (NS002945) and the National Heart Blood and Lung Institute, NIH, Bethesda, MD to J.J. (HL006238) and J.D.F.-G. (HL006203).
Author contributions: Conceptualization: C.B., X.-F.T., and K.J.S. Methodology: X.-F.T., C.B., R.S., A.I.F.-M., K.H., T.-H.C., J.J., J.D.F.-G., and K.J.S. Investigation: X.-F.T., C.B., R.S., A.I.F.-M., T.-H.C., J.D.F.-G. Visualization: X.-F.T., C.B., R.S., A.I.F.-M., K.H., J.D.F.-G., and K.J.S. Funding acquisition: J.J., J.D.F.-G., and K.J.S. Project administration: C.B., J.D.F.-G., and K.J.S. Supervision: J.D.F.-G. and K.J.S. Writing (original draft): X.-F.T., C.B., R.S., A.I.F.-M., K.H., T.-H.C., J.J., J.D.F.-G., and K.J.S. Writing (review and editing): X.-F.T., C.B., R.S., A.I.F.-M., K.H., T.-H.C., J.J., J.D.F.-G., and K.J.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Maps of wild-type Shaker-IR and the W434F mutant have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-25147 and EMD-25152, respectively. Models of wild-type Shaker-IR and the W434F mutant have been deposited in the Protein Data Bank with accession codes 7SIP and 7SJ1, respectively.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.Yellen G., The voltage-gated potassium channels and their relatives. Nature 419, 35–42 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Jan L. Y., Jan Y. N., Voltage-gated potassium channels and the diversity of electrical signalling. J. Physiol. 590, 2591–2599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yellen G., The moving parts of voltage-gated ion channels. Q. Rev. Biophys. 31, 239–295 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Swartz K. J., Sensing voltage across lipid membranes. Nature 456, 891–897 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bezanilla F., How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9, 323–332 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Alabi A. A., Bahamonde M. I., Jung H. J., Kim J. I., Swartz K. J., Portability of paddle motif function and pharmacology in voltage sensors. Nature 450, 370–375 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long S. B., Tao X., Campbell E. B., MacKinnon R., Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Matthies D., Bae C., Toombes G. E., Fox T., Bartesaghi A., Subramaniam S., Swartz K. J., Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pau V., Zhou Y., Ramu Y., Xu Y., Lu Z., Crystal structure of an inactivated mutant mammalian voltage-gated K(+) channel. Nat. Struct. Mol. Biol. 24, 857–865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suarez-Delgado E., Rangel-Sandin T. G., Ishida I. G., Rangel-Yescas G. E., Rosenbaum T., Islas L. D., KV1.2 channels inactivate through a mechanism similar to C-type inactivation. J. Gen. Physiol. 152, e201912499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshi T., Zagotta W. N., Aldrich R. W., Two types of inactivation in Shaker K+ channels: Effects of alterations in the carboxy-terminal region. Neuron 7, 547–556 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Adelman J. P., Bond C. T., Pessia M., Maylie J., Episodic ataxia results from voltage-dependent potassium channels with altered functions. Neuron 15, 1449–1454 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Hubner C. A., Jentsch T. J., Ion channel diseases. Hum. Mol. Genet. 11, 2435–2445 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Kurata H. T., Fedida D., A structural interpretation of voltage-gated potassium channel inactivation. Prog. Biophys. Mol. Biol. 92, 185–208 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Hoshi T., Armstrong C. M., C-type inactivation of voltage-gated K+ channels: Pore constriction or dilation? J. Gen. Physiol. 141, 151–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith P. L., Baukrowitz T., Yellen G., The inward rectification mechanism of the HERG cardiac potassium channel. Nature 379, 833–836 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Keating M. T., Sanguinetti M. C., Molecular and cellular mechanisms of cardiac arrhythmias. Cell 104, 569–580 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Nerbonne J. M., Kass R. S., Molecular physiology of cardiac repolarization. Physiol. Rev. 85, 1205–1253 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Sanguinetti M. C., Tristani-Firouzi M., hERG potassium channels and cardiac arrhythmia. Nature 440, 463–469 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Yan J., Li Q., Aldrich R. W., Closed state-coupled C-type inactivation in BK channels. Proc. Natl. Acad. Sci. U.S.A. 113, 6991–6996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagriantsev S. N., Peyronnet R., Clark K. A., Honore E., Minor D. L. Jr., Multiple modalities converge on a common gate to control K2P channel function. EMBO J. 30, 3594–3606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zilberberg N., Ilan N., Goldstein S. A., KCNKO: Opening and closing the 2-P-domain potassium leak channel entails "C-type" gating of the outer pore. Neuron 32, 635–648 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Choi K. L., Aldrich R. W., Yellen G., Tetraethylammonium blockade distinguishes two inactivation mechanisms in voltage-activated K+ channels. Proc. Natl. Acad. Sci. U.S.A. 88, 5092–5095 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perozo E., MacKinnon R., Bezanilla F., Stefani E., Gating currents from a nonconducting mutant reveal open-closed conformations in Shaker K+ channels. Neuron 11, 353–358 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Barneo J., Hoshi T., Heinemann S. H., Aldrich R. W., Effects of external cations and mutations in the pore region on C-type inactivation of Shaker potassium channels. Receptors Channels 1, 61–71 (1993). [PubMed] [Google Scholar]
- 26.Yellen G., Sodickson D., Chen T. Y., Jurman M. E., An engineered cysteine in the external mouth of a K+ channel allows inactivation to be modulated by metal binding. Biophys. J. 66, 1068–1075 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baukrowitz T., Yellen G., Modulation of K+ current by frequency and external [K+]: A tale of two inactivation mechanisms. Neuron 15, 951–960 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., Jurman M. E., Yellen G., Dynamic rearrangement of the outer mouth of a K+ channel during gating. Neuron 16, 859–867 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., Yan Y., Sigworth F. J., The Shaker mutation T449V rescues ionic currents of W434F K+ channels. Biophys. J. 82, 234e (2002). [Google Scholar]
- 30.Yang Y., Yan Y., Sigworth F. J., How does the W434F mutation block current in Shaker potassium channels? J. Gen. Physiol. 109, 779–789 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starkus J. G., Kuschel L., Rayner M. D., Heinemann S. H., Ion conduction through C-type inactivated Shaker channels. J. Gen. Physiol. 110, 539–550 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina A., Castellano A. G., Lopez-Barneo J., Pore mutations in Shaker K+ channels distinguish between the sites of tetraethylammonium blockade and C-type inactivation. J. Physiol. 499 ( Pt. 2), 361–367 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogielska E. M., Aldrich R. W., A mutation in S6 of Shaker potassium channels decreases the K+ affinity of an ion binding site revealing ion-ion interactions in the pore. J. Gen. Physiol. 112, 243–257 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogielska E. M., Aldrich R. W., Functional consequences of a decreased potassium affinity in a potassium channel pore. Ion interactions and c-type inactivation. J. Gen. Physiol. 113, 347–358 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armstrong C. M., Hoshi T., K+ channel gating: C-type inactivation is enhanced by calcium or lanthanum outside. J. Gen. Physiol. 144, 221–230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuello L. G., Cortes D. M., Perozo E., The gating cycle of a K+ channel at atomic resolution. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuello L. G., Jogini V., Cortes D. M., Perozo E., Structural mechanism of C-type inactivation in K+ channels. Nature 466, 203–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuello L. G., Jogini V., Cortes D. M., Pan A. C., Gagnon D. G., Dalmas O., Cordero-Morales J. F., Chakrapani S., Roux B., Perozo E., Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature 466, 272–275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordero-Morales J. F., Jogini V., Chakrapani S., Perozo E., A multipoint hydrogen-bond network underlying KcsA C-type inactivation. Biophys. J. 100, 2387–2393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Shen R., Rohaim A., Mendoza Uriarte R., Fajer M., Perozo E., Roux B., Computational study of non-conductive selectivity filter conformations and C-type inactivation in a voltage-dependent potassium channel. J. Gen. Physiol. 153, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pless S. A., Galpin J. D., Niciforovic A. P., Kurata H. T., Ahern C. A., Hydrogen bonds as molecular timers for slow inactivation in voltage-gated potassium channels. eLife 2, e01289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bezanilla F., Gating currents. J. Gen. Physiol. 150, 911–932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zagotta W. N., Hoshi T., Aldrich R. W., Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science 250, 568–571 (1990). [DOI] [PubMed] [Google Scholar]
- 44.Hoshi T., Zagotta W. N., Aldrich R. W., Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science 250, 533–538 (1990). [DOI] [PubMed] [Google Scholar]
- 45.Demo S. D., Yellen G., The inactivation gate of the Shaker K+ channel behaves like an open- channel blocker. Neuron 7, 743–753 (1991). [DOI] [PubMed] [Google Scholar]
- 46.Soler-Llavina G. J., Holmgren M., Swartz K. J., Defining the conductance of the closed state in a voltage-gated K+ channel. Neuron 38, 61–67 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Soler-Llavina G. J., Chang T. H., Swartz K. J., Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron 52, 623–634 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Tao X., Lee A., Limapichat W., Dougherty D. A., MacKinnon R., A gating charge transfer center in voltage sensors. Science 328, 67–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aggarwal S. K., MacKinnon R., Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron 16, 1169–1177 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Seoh S. A., Sigg D., Papazian D. M., Bezanilla F., Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron 16, 1159–1167 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Holmgren M., Jurman M. E., Yellen G., Gated access to the pore of a voltage-dependent K+ channel. Neuron 19, 175–184 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y., Morais-Cabral J. H., Kaufman A., MacKinnon R., Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Morais-Cabral J. H., Zhou Y., MacKinnon R., Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature 414, 37–42 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Mironenko A., Zachariae U., de Groot B. L., Kopec W., The persistent question of potassium channel permeation mechanisms. J. Mol. Biol. 433, 167002 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Heginbotham L., MacKinnon R., Conduction properties of the cloned Shaker K+ channel. Biophys. J. 65, 2089–2096 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jensen M. O., Jogini V., Eastwood M. P., Shaw D. E., Atomic-level simulation of current-voltage relationships in single-file ion channels. J. Gen. Physiol. 141, 619–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hurst R. S., Toro L., Stefani E., Molecular determinants of external barium block in Shaker potassium channels. FEBS Lett. 388, 59–65 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Loots E., Isacoff E. Y., Protein rearrangements underlying slow inactivation of the Shaker K+ channel. J. Gen. Physiol. 112, 377–389 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ortega-Saenz P., Pardal R., Castellano A., Lopez-Barneo J., Collapse of conductance is prevented by a glutamate residue conserved in voltage-dependent K+ channels. J. Gen. Physiol. 116, 181–190 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larsson H. P., Elinder F., A conserved glutamate is important for slow inactivation in K+ channels. Neuron 27, 573–583 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Labro A. J., Cortes D. M., Tilegenova C., Cuello L. G., Inverted allosteric coupling between activation and inactivation gates in K+ channels. Proc. Natl. Acad. Sci. U.S.A. 115, 5426–5431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olcese R., Latorre R., Toro L., Bezanilla F., Stefani E., Correlation between charge movement and ionic current during slow inactivation in Shaker K+ channels. J. Gen. Physiol. 110, 579–589 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lolicato M., Natale A. M., Abderemane-Ali F., Crottes D., Capponi S., Duman R., Wagner A., Rosenberg J. M., Grabe M., Minor D. L. Jr., K2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions. Sci. Adv. 6, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rana M. S., Wang X., Banerjee A., An improved strategy for fluorescent tagging of membrane proteins for overexpression and purification in mammalian cells. Biochemistry 57, 6741–6751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goehring A., Lee C. H., Wang K. H., Michel J. C., Claxton D. P., Baconguis I., Althoff T., Fischer S., Garcia K. C., Gouaux E., Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pandit S. B., Skolnick J., Fr-TM-align: A new protein structural alignment method based on fragment alignments and the TM-score. BMC Bioinformatics 9, 531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 76.Zhang L., Hermans J., Hydrophilicity of cavities in proteins. Proteins 24, 433–438 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Brooks B. R., Brooks III C. L., Mackerell Jr. A. D., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M., CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B. L., Grubmuller H., MacKerell A. D. Jr., CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., Mackerell A. D. Jr., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klauda J. B., Venable R. M., Freites J. A., O’Connor J. W., Tobias D. J., Mondragon-Ramirez C., Vorobyov I., MacKerell A. D. Jr., Pastor R. W., Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wassenaar T. A., Ingolfsson H. I., Bockmann R. A., Tieleman D. P., Marrink S. J., Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 11, 2144–2155 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Pronk S., Pall S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M. R., Smith J. C., Kasson P. M., van der Spoel D., Hess B., Lindahl E., GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Jong D. H., Singh G., Bennett W. F., Arnarez C., Wassenaar T. A., Schafer L. V., Periole X., Tieleman D. P., Marrink S. J., Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 9, 687–697 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Marrink S. J., de Vries A. H., Mark A. E., Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B 108, 750–760 (2004). [Google Scholar]
- 85.Monticelli L., Kandasamy S. K., Periole X., Larson R. G., Tieleman D. P., Marrink S. J., The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comput. 4, 819–834 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., de Vries A. H., The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Wassenaar T. A., Pluhackova K., Bockmann R. A., Marrink S. J., Tieleman D. P., Going backward: A flexible geometric approach to reverse transformation from coarse grained to atomistic models. J. Chem. Theory Comput. 10, 676–690 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Phillips J. C., Hardy D. J., Maia J. D. C., Stone J. E., Ribeiro J. V., Bernardi R. C., Buch R., Fiorin G., Henin J., Jiang W., McGreevy R., Melo M. C. R., Radak B. K., Skeel R. D., Singharoy A., Wang Y., Roux B., Aksimentiev A., Luthey-Schulten Z., Kale L. V., Schulten K., Chipot C., Tajkhorshid E., Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fiorin G., Klein M. L., Henin J., Using collective variables to drive molecular dynamics simulations. Mol. Phys. 111, 3345–3362 (2013). [Google Scholar]
- 90.D. E. Shaw, J. P. Grossman, J. A. Bank, B. Batson, J. A. Butts, J. C. Chao, M. M. Deneroff, R. O. Dror, A. Even, C. H. Fenton, A. Forte, J. Gagliardo, G. Gill, B. Greskamp, C. R. Ho, D. J. Ierardi, L. Iserovich, J. S. Kuskin, R. H. Larson, T. Layman, L.-S. Lee, A. K. Lerer, C. Li, D. Killebrew, K. M. Mackenzie, S. Y.-H. Mok, M. A. Moraes, R. Mueller, L. J. Nociolo, J. L. Peticolas, T. Quan, D. Ramot, J. K. Salmon, D. P. Scarpazza, U. B. Schafer, N. Siddique, C. W. Snyder, J. Spengler, P. T. P. Tang, M. Theobald, H. Toma, B. Towles, B. Vitale, S. C. Wang, C. Young, paper presented at the SC14: International Conference for High Performance Computing, Networking, Storage and Analysis, New Orleans, LA, USA, 16 to 21 November 2014. [Google Scholar]
- 91.Nosé S., A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268 (1984). [Google Scholar]
- 92.Hoover W. G., Canonical dynamics—Equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 93.Martyna G. J., Tobias D. J., Klein M. L., Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994). [Google Scholar]
- 94.Shan Y., Klepeis J. L., Eastwood M. P., Dror R. O., Shaw D. E., Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J. Chem. Phys. 122, 054101 (2005). [DOI] [PubMed] [Google Scholar]
- 95.Pan A. C., Jacobson D., Yatsenko K., Sritharan D., Weinreich T. M., Shaw D. E., Atomic-level characterization of protein-protein association. Proc. Natl. Acad. Sci. U.S.A. 116, 4244–4249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jensen M. O., Jogini V., Borhani D. W., Leffler A. E., Dror R. O., Shaw D. E., Mechanism of voltage gating in potassium channels. Science 336, 229–233 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 and S2
Movies S1 to S3