Significance
The pseudokinase integrin-linked kinase (ILK) is a central component of focal adhesions, cytoplasmic multiprotein complexes that integrate and transduce biochemical and mechanical signals from the extracellular environment into the cell and vice versa. However, the precise molecular functions, particularly the mechanosensory properties of ILK and the significance of retained adenosine triphosphate (ATP) binding, are still unclear. Combining molecular-dynamics simulations with cell biology, we establish a role for ATP binding to pseudokinases. We find that ATP promotes the structural stability of ILK, allosterically influences the interaction between ILK and its binding partner parvin at adhesions, and enhances the mechanoresistance of this complex. On the cellular level, ATP binding facilitates efficient traction force buildup, focal adhesion stabilization, and efficient cell migration.
Keywords: focal adhesion, integrin-linked kinase, molecular dynamics, traction force microscopy
Abstract
Focal adhesions link the actomyosin cytoskeleton to the extracellular matrix regulating cell adhesion, shape, and migration. Adhesions are dynamically assembled and disassembled in response to extrinsic and intrinsic forces, but how the essential adhesion component integrin-linked kinase (ILK) dynamically responds to mechanical force and what role adenosine triphosphate (ATP) bound to this pseudokinase plays remain elusive. Here, we apply force–probe molecular-dynamics simulations of human ILK:α-parvin coupled to traction force microscopy to explore ILK mechanotransducing functions. We identify two key salt-bridge–forming arginines within the allosteric, ATP-dependent force-propagation network of ILK. Disrupting this network by mutation impedes parvin binding, focal adhesion stabilization, force generation, and thus migration. Under tension, ATP shifts the balance from rupture of the complex to protein unfolding, indicating that ATP increases the force threshold required for focal adhesion disassembly. Our study proposes a role of ATP as an obligatory binding partner for structural and mechanical integrity of the pseudokinase ILK, ensuring efficient cellular force generation and migration.
Cells sense and respond to a broad variety of biochemical and mechanical signals from the neighboring cells and the surrounding microenvironment, including the extracellular matrix (ECM). Produced and remodeled by the cells, the ECM serves as a physical scaffold for tissues, but also actively guides tissue development and homeostasis by regulating a broad range of cellular processes, such as adhesion, migration, growth, and differentiation (1, 2). ECM assembly and bidirectional cell–ECM signaling is mediated by the integrin family of cellular surface receptors. Integrin–ECM binding leads to recruitment of filamentous (F-)actin to allow force generation through the contractile actomyosin cytoskeleton. As integrins lack enzymatic activity and do not bind actin directly, their function depends on establishing and maintaining large, multiprotein complexes of actin-binding and -regulatory proteins, the focal adhesions (FAs) (3). FAs are crucial for the precise spatiotemporal coordination of integrin signaling. In addition to sensing ECM composition, integrins and FAs sense mechanical cues and transduce them into biochemical signals through a process termed mechanotransduction (4). These mechanical cues include ECM rigidity, tension, and shear. A key event of mechanosensing is modulation of actomyosin contractility and FA dynamics, determined by the balance of protein association and dissociation. According to current models, collectively termed the molecular clutch model, exposing cells to large, rapidly applied forces or plating cells on stiff substrates leads to large FAs with slower exchange rates of adapter molecules and longer FA lifetimes (5). Thus, precise coordination of the stability of FAs and their actomyosin linkage is required for effective generation of traction stresses during mechanosensing, as well as during directed cell migration (4, 6).
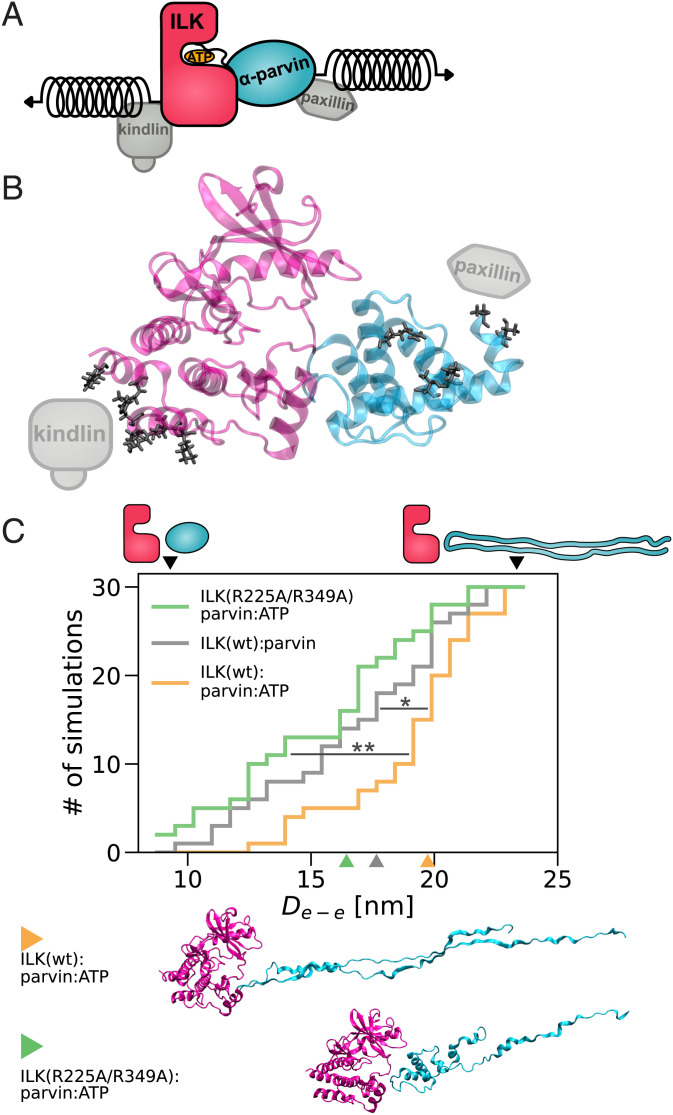
A central regulator of FA dynamics and stability downstream of β 1 integrins is integrin-linked kinase (ILK), one of the few essential and evolutionarily conserved components of FAs (7, 8). ILK is the central component of a tripartite IPP complex comprising ILK, PINCH (particularly interesting new cysteine–histidine rich protein), and α-parvin (9). ILK consists of two distinct domains (Fig. 1 A and B): an N-terminal ankyrin-repeat domain, associated with PINCH (10), and a C-terminal atypical kinase domain, which binds to the calponin-homology (CH2) domain of α-parvin (11). The current notion is that the IPP serves as a signal-processing platform by recruiting a variety of proteins. For example, α-parvin directly interacts with paxillin (12–14), PINCH influences receptor tyrosine kinases (15), and both parvin and PINCH bind F-actin (16, 17), connecting the IPP to the cytoskeleton. Although ILK was believed to directly associate with β-integrin tails (18), more recent studies show that ILK rather indirectly contacts integrins by binding to kindlin-2 (19, 20). As kindlin-2 directly interacts with β-integrin (21–25) and binds PIP2 via its PH domain (26, 27), this interaction recruits the IPP to the cell membrane at sites of FAs. While ILK contains a kinase domain with a typical kinase fold, the catalytic function of ILK has been heavily debated (8, 9, 11, 28–31), and ILK is now widely regarded as a pseudokinase. Its obligate interaction partner parvin binds to the putative substrate entry site within the kinase-like domain, and disruption of ILK:parvin binding reduces the localization of ILK to FAs (11). Interestingly, ILK retained its ability to bind adenosine triphosphate (ATP) in the nucleotide-binding cleft in an unusual binding mode (11), and recent work has identified a role for ATP binding in actin stress fiber formation and adhesion morphology (16). However, the molecular mechanism by which ATP impacts ILK functions remains fairly elusive.
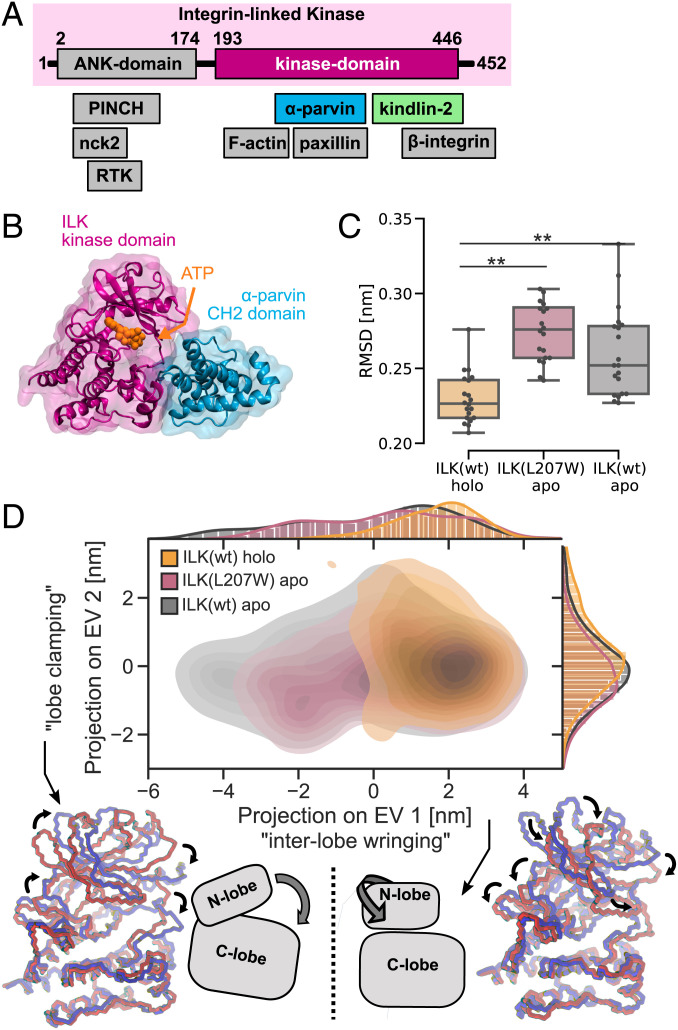
Fig. 1.
Destabilization of ILK pseudokinase without ATP. (A) Schematic overview of ILK and its associated proteins. (B) The ILK:parvin complex rendered from PDB ID code 3KMW (11). (C) Trajectory median backbone RMSD of ILK(WT) holo and apo and apo ILK(L207W). n = 20 trajectories. **P = 0.001, one-way ANOVA/Tukey honestly significant difference (HSD). (D) PCA of holo and apo ILK(WT) and ILK(L207W). Structures extracted from MD simulations are projected onto PC axes for the first and second PC. Extreme conformations (red and blue) of apo ILK along PC1 and PC2 are overlaid based on a least-squared fit to the C-lobe of the pseudokinase domain, and large-scale motions are indicated by arrows. A schematic of the described motions is presented (see also Movies S1 and S2).
The importance of the IPP in regulating the integrin–actin linkage is apparent in deletion studies in mice and cells: ILK depletion results in embryonic lethality caused by failure in epiblast polarization and severe defects in F-actin organization at adhesion sites (32). Furthermore, tissue-specific deletion of ILK leads to heart disease (33), while ILK is also associated with cancer progression (34) and might play a role in aging (35). At the cellular level, ILK deficiency leads to aberrant remodeling of the actin and microtubule cytoskeletons and decreased force generation, which comprises FA formation, cell migration, and ECM remodeling (9, 32, 36–38).
While ILK is clearly indispensable for the integrin–actin connection and efficient force generation through integrins, the precise molecular mechanism of force-induced IPP signaling and the role of ATP therein are unknown. One critical determinant of FA kinetics and thus mechanosensing is thought to be force-sensitive changes in protein conformations. These changes depend on the strength and duration of force application: Forces must be transmitted for a period sufficient to induce conformational changes, but large forces also trigger bond breakage or protein dissociation, which will terminate force transmission. Thus, a constant competition between conformational change and bond breakage likely exists at adhesions under high traction stresses (4, 6). Molecular examples include focal adhesion kinase (FAK) and talin: The competition between force-induced unfolding and dissociation from binding partners depends on parameters such as loading rate and direction, and defines their mechanosensing roles (39–44).
Regulated by the activity of the chaperone Hsp90, the stability of ILK is critical for the ability of cells to generate traction forces (45). This suggests that force-induced unfolding of ILK could play a role for FA signal transduction by promoting disassembly of force-bearing adhesions (45). To address the mechanosensitivity of ILK and the molecular mechanisms of FA regulation, we employ extensive molecular dynamics (MD) simulations of ILK in equilibrium conditions and under mechanical load coupled to biochemical and cellular experiments. Our studies provide evidence for an allosteric strengthening effect of ATP on the ILK:parvin binding and a role for ATP on the cellular force propagation toward α-parvin. Furthermore, previously unrecognized ILK salt-bridge residues are identified to be critical for ILK:parvin interaction and force transmission. We propose that ILK retained ATP as an obligatory binding partner for structural integrity and efficient force transmission of FAs.
Results
ATP Structurally Stabilizes the ILK Pseudokinase Domain.
To explore the effect of ATP on the ILK pseudokinase dynamics, we performed microsecond-scale, all-atom MD simulations with explicit water of wild-type (WT) ILK in the holo state (with ATP; Protein Data Bank [PDB] ID code 3KMW) and apo state (without ATP; PDB ID code 3KMU), as well as of the ILK(L207W) mutation in the pseudokinase domain that was shown to sterically occlude ATP binding to ILK without affecting the structural integrity of the protein [PDB ID code 6MIB (16)]. For each, we simulated the ILK pseudokinase domain in complex with the α-parvin CH2 domain to mimic the cellular conditions as closely as possible since parvin is the obligate binding partner of ILK. We found that the overall flexibility of the ILK pseudokinase domain is significantly increased in the apo state, as indicated by a higher RMSD from the respective starting structures observed without ATP and similarly in the ILK(L207W) mutation (Fig. 1C). Thus, ATP binding leads to a decrease in the internal kinase dynamics.
To further characterize the dynamic effect of ATP, we examined the differences in the large-scale coordinated motions using principal component analysis (PCA; Fig. 1D). We found that in the apo states of both ILK(WT) and ILK(L207W), the pseudokinase explores more of the available conformational space, especially along the direction of the most prominent motion (PC1). This motion describes an “interlobe wringing” motion, where the N-lobe of the kinase fold twists around the nucleotide binding pocket on top of the C-lobe (Fig. 1D and Movie S1). Conversely, PC2 describes a “clamping” motion of both lobes (Fig. 1D and Movie S2). Hence, the lack of ATP promotes the above-mentioned interlobe wringing motion, which contributes to the higher internal flexibility of apo ILK.
Loss of ILK:ATP Binding Destabilizes FAs in Particular on Rigid Substrates.
As the simulations showed that ATP is required to stabilize the pseudokinase domain, we sought to functionally test the consequences of this instability in the absence of ATP. To analyze the role of ATP binding on the ILK:α-parvin interaction, we reconstituted ILK-deficient murine fibroblasts (32) with either ILK(WT)–GFP or ILK(L207W)–GFP and performed coimmunoprecipitation assays. Western blot analyses of the immunoprecipitates showed that the L207W mutation had no substantial effect on steady-state α-parvin binding (Fig. 2 A and B), as also reported before (16). This was consistent with immunofluorescence analyses of α-parvin and ILK localization, which showed a comparable pattern of FA staining of both ILK and parvin in cells expressing ILK(WT) or the ILK(L207W) mutant (Fig. 2C and SI Appendix, Fig. S1A).
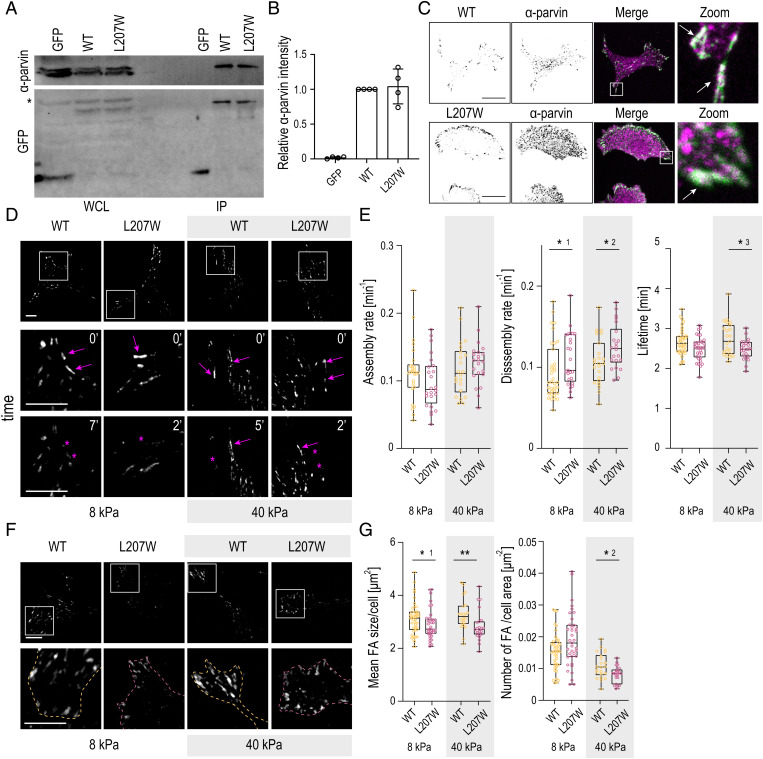
Fig. 2.
Loss of ILK ATP binding destabilizes FAs, in particular on rigid substrates. (A) Representative Western blot of GFP pull-down experiments from ILK -∕- fibroblasts expressing GFP, ILK(WT)–GFP, or ILK(L207W)–GFP. α-parvin is coprecipitated at similar levels in ILK(WT) and L207W cells. The asterisk marks unspecific antibody binding. IP, immunoprecipitation; WCL, whole-cell lysate. (B) Quantification of α-parvin to GFP ratio from immunoprecipitation experiments (mean ± SD, n = 4 independent experiments). (C) Representative immunofluorescence stainings of α-parvin and GFP. α-parvin localizes at FAs both in ILK(WT)–GFP and ILK(L207W)–GFP cells. C, Right shows zoom-ins of the areas indicated by the white rectangles. Arrows indicate colocalization of α-parvin (magenta) with ILK(WT)–GFP and ILK(L207W)–GFP (green). (D) Representative snapshots of Movies S3 and S4 of ILK(WT)–GFP and ILK(L207W)–GFP cells plated on 8-kPa and 40-kPa substrates and imaged over time to quantify adhesion dynamics. D, Bottom shows zoom-ins of the areas indicated by the white squares. Arrows indicate adhesion growth, while asterisks mark adhesion disassembly. (E) Quantification of the assembly, disassembly, and lifetime of adhesions in ILK(WT)–GFP and ILK(L207W)–GFP cells plated on 8-kPa and 40-kPa substrates (n > 22 cells per condition pooled across four independent experiments). *1P = 0.0469; *2P = 0.0437; *3P = 0.0479 (Mann–Whitney). (F) Representative images of adhesions in ILK(WT)–GFP and ILK(L207W)–GFP cells on 8-kPa and 40-kPa substrates. F, Lower shows zoom-ins of the areas indicated by the white squares. Dashed lines indicate cell edges defined by cytoplasmic fluorescence. (G) Quantification of the mean FA size per cell and number of adhesions per cell area in ILK(WT) and ILK(L207W)–GFP cells cultured on 8-kPa and 40-kPa substrates from live-imaging FA area threshold set to 20 pixels. (n > 22 cells per condition pooled across four independent experiments.) *1P = 0.0396; *2P = 0.0258; **P = 0.0022 (Mann–Whitney). (Scale bars: 20 µm.).
To understand the effect of this mutation on adhesion stability in conditions of low and high traction forces, we performed time-lapse imaging of FA dynamics in ILK(WT)–GFP and ILK(L207W)–GFP cells plated on polyacrylamide (PAA) gels with either low (8 kPa) or high (40 kPa) stiffness. Quantification of adhesion dynamics showed no significant differences in the assembly rates of FAs in WT and the L207W mutant, regardless of substrate stiffness. However, the L207W mutant showed faster disassembly rates as well as decreased adhesion lifetimes on both soft and stiff substrates. Interestingly, this difference was more pronounced on the stiff substrates (Fig. 2 D and E and Movies S3 and S4). Consistent with destabilization of adhesions, further analysis of FA morphology revealed an overall lower number of FAs per cell area in L207W cells compared to WT (Fig. 2 F and G), as also reported earlier (16). In addition, FAs were slightly smaller in size. Again, these differences were more pronounced on stiff substrates (Fig. 2 F and G). Decreased FA area was confirmed by paxillin staining on cells plated on crossbow micropatterns to remove cell shape heterogeneity (SI Appendix, Fig. S1B) (46). Collectively, and consistent with our MD simulations, these results indicate that although the inability of the ILK pseudocatalytic domain to bind ATP does not substantially impair the biochemical interaction with α-parvin, it destabilizes FAs, in particular, under high traction stresses that occur on rigid substrates.
ATP Allosterically Influences Parvin-Binding Salt Bridges.
The pronounced change in the internal collective kinase dynamics and the effects on FA stability suggested that ATP not only affects residues in its immediate binding pocket, but should also exhibit long-range effects within the kinase-like domain. We thus next asked which residues and interactions are specifically altered by ATP on a molecular level by calculating the change in time-averaged punctual stresses on each residue upon ATP binding (SI Appendix, Fig. S2) using Force Distribution Analysis [FDA (47); Materials and Methods]. We find notable differences in punctual stress across the residues of ILK, both within the ATP-binding pocket and also in regions farther away from the binding pocket. In contrast, the per-residue rms fluctuation showed only minor differences across a majority of ILK residues between the holo and apo state (SI Appendix, Fig. S3), highlighting the sensitivity of internal force calculations to reveal long-distance allostery.
To further determine the ATP-dependent allosteric pathways of force propagation, we calculated the residue-based, pairwise forces from FDA in the holo complex and determined the differences to the apo structure (SI Appendix, Table S1). Fig. 3A shows the connected networks of residue pairs that exhibit significant changes in pairwise forces larger than a given threshold. As already indicated by the punctual stress, many residues composing the ATP-binding pocket adapt upon ATP binding (Fig. 3A, orange arrow), and ATP also strongly affects residues outside of its binding pocket. Changes in force-distribution patterns extend into the kinase N- and C-lobe and strikingly even into parvin. This suggests that ATP has an allosteric effect on the obligatory binding partner of ILK. Particularly important for this intermolecular allostery were two arginine residues: R225 and R349 at the ILK:parvin interface (marked with purple arrows in Fig. 3A). R225 is located on a loop of the N-lobe, and R349 belongs to the activation segment of conventional kinases. Structural inspection of these arginines revealed that they form previously unrecognized salt bridges with E332 and D336 in parvin, and R349 additionally interacts with L333 of parvin.
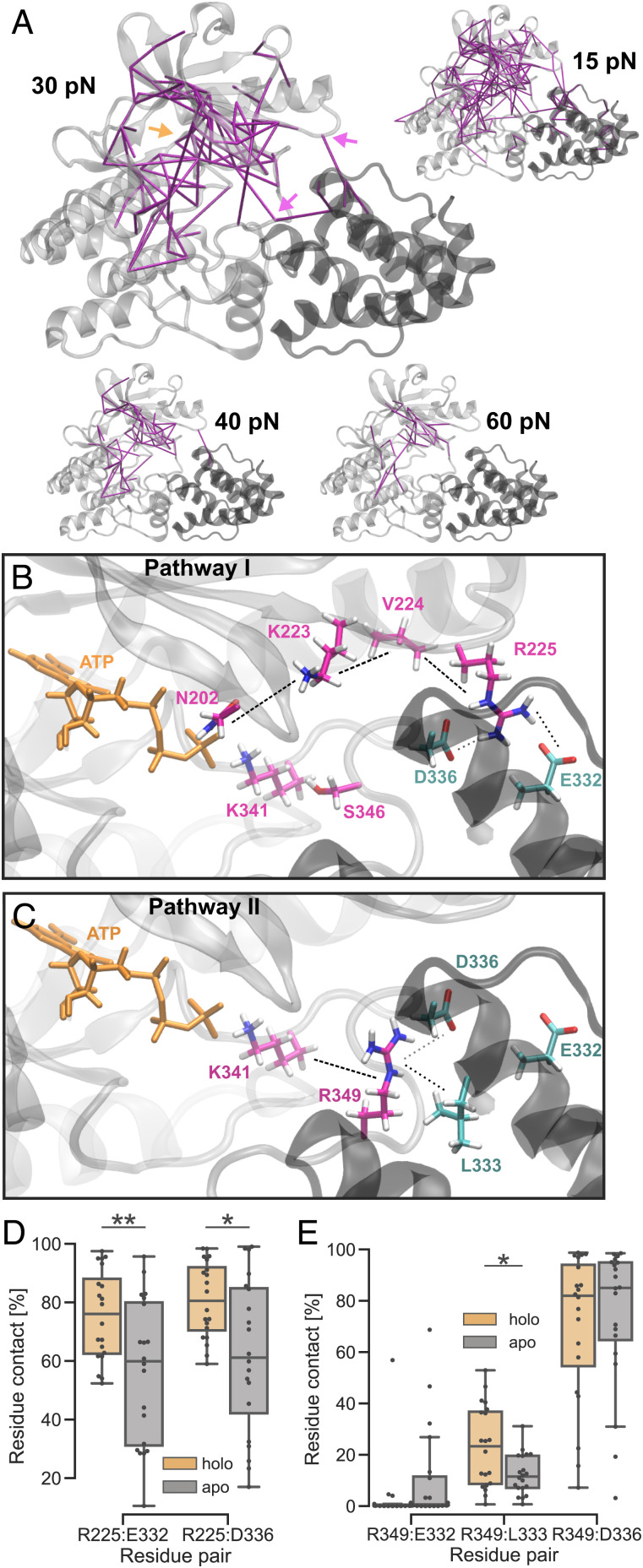
Fig. 3.
ATP allosterically influences parvin-binding salt bridges. (A) Residue-based, pairwise forces compared between ILK holo and apo with different force thresholds (purple lines). Approximate positions of the ATP-binding pocket (orange arrow) and salt-bridge–forming arginines R225 and R349 (purple arrows) are shown. (B and C) Proposed force transduction pathways from ATP to salt-bridge–forming arginines R225 and R349, respectively, and into parvin and as calculated by FDA (for force values, see SI Appendix, Fig. S4). (D) Trajectory-averaged residue contact probability (threshold 0.35 nm) between ILK R225 and parvin depending on the presence of ATP (n = 20 independent trajectories). **P = 0.0096; *P = 0.013 (Mann–Whitney). (E) Residue contact probability between ILK R349 and parvin as in D (n = 20 independent trajectories. *P = 0.017 (Mann–Whitney).
Using the calculated forces from FDA, we determined the shortest pathways of significantly changed pairwise forces from ATP toward the salt-bridge–forming residues (Fig. 3 C and D; and for detailed force values, see SI Appendix, Fig. S4). R225 is found to be allosterically influenced by ATP over a residue cascade involving N202, K223, and V224 (Fig. 3C and SI Appendix, Fig. S4A). FDA also provided hints toward a pathway involving K341 and S346, however, without complete statistical significance (SI Appendix, Fig. S4A). The pathway of internal force changes from ATP to R349 proceeds over the ATP-coordinating K341, corresponding to glycine from the crucial DFG motif of conventional kinases (Fig. 3D and SI Appendix, Fig. S4B) (11).
To further confirm the importance of the two salt-bridge–forming arginines, we performed MD simulations of holo ILK(WT) without parvin. Previous investigations of the ILK:parvin binding interface primarily focused on the parvin-binding helix (αG-helix) containing M402 and K403, that mutated to alanine lead to a loss of ILK:parvin binding (11). In the absence of parvin, this already-described parvin-binding helix shows a higher flexibility in our simulations (SI Appendix, Fig. S5). Additionally, the two salt-bridge–forming arginines R225 and R349 also increase in flexibility, indicating that one of their main functions is to bind parvin. To assess the ability of the arginines to form salt bridges with parvin, we calculated the residue contact probability (percentage of total simulation time where two residues are in contact; Fig. 3 D and E). The average occupancy of, on average, 80% for the R225–E332, R225–D336, and R349–D336 salt bridges corroborates that those are indeed functional and important parvin-binding residues. The average R349–L333 contact percentage is around 25%, rendering this residue pair less important for parvin binding. We confirmed the FDA results on the ATP-dependent ILK:parvin interactions by comparing the residue contact probability across the interface between the ATP-bound state and the apo state (Fig. 3 D and E). We found that the absence of ATP significantly lowered the occupancy of the R225–E332 and of the R225–D336 salt bridges, which suggests an allosteric destabilization of the N-lobe:parvin contacts (Fig. 3D). Likewise, the contact probability of R349–L333 is also decreased in the absence of ATP; however, the two salt bridges involving R349 (R349–E332 and R349–D336) did not show statistically significant changes upon ATP removal (Fig. 3D). Thus, the lack of ATP does not fully abolish the contacts between ILK and parvin, but appears to weaken the interface. Overall, our data suggested that the functional parvin-binding interface extends toward the ILK activation loop and the N-lobe involving R349 and R225, respectively, and those interactions between ILK and parvin putatively contribute to the stability of the complex.
Point Mutations in the Salt-Bridge–Coordinating Residues Destabilize α-Parvin Binding and FAs.
To understand the functional role of the two salt-bridge residues R255 and R349 that we identified by the simulations to mediate the allosteric effect of ATP on ILK:parvin binding, we generated single R255A and R349A, as well as a R255A/R349A double mutant, reconstituted ILK-∕- cells with these proteins, and performed coimmunoprecipitation assays with α-parvin. Both R255A and R349A single mutants showed reduced α-parvin binding compared to ILK(WT), whereas strikingly, R255A/R349A showed strong reduction of parvin binding (Fig. 4 A and B). Immunofluorescence analyses further revealed that although the R255A/R349A double mutant was able to localize to FAs, α-parvin failed to efficiently do so and, instead, displayed a punctate cytoplasmic staining pattern (Fig. 4C and SI Appendix, Fig. S6A).
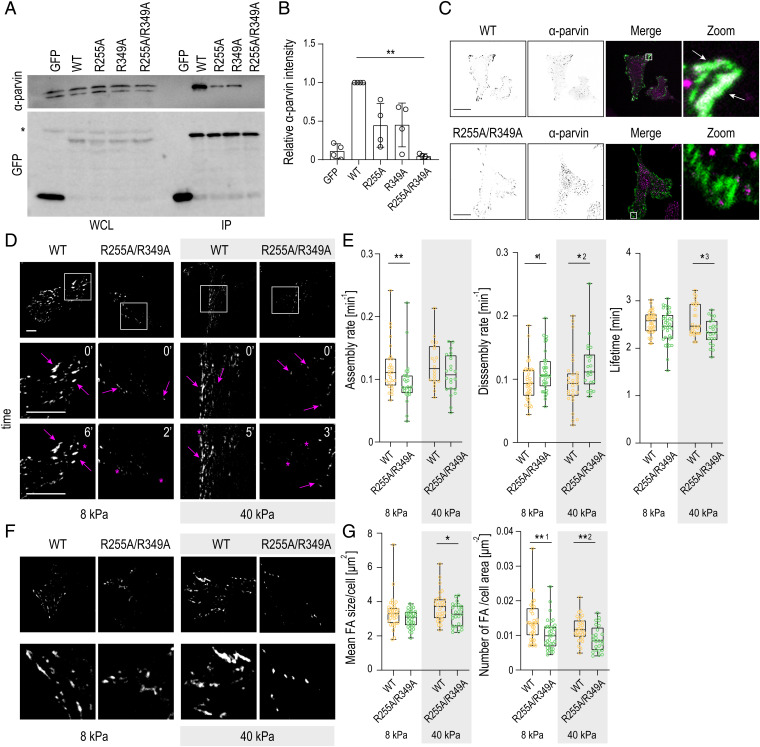
Fig. 4.
Point mutations in the predicted salt-bridge–coordinating residues destabilize α-parvin binding and FAs. (A) Representative Western blot of GFP pull-down experiments from ILK -∕- fibroblasts expressing GFP, ILK(WT)–GFP, ILK(R225A)–GFP, ILK(R349A)–GFP, and ILK(R225A/R349A)–GFP mutants. R225A–GFP and R349A–GFP show reduced α-parvin binding and ILK-(R225A/R349A) complete loss of interaction. The asterisk marks unspecific antibody binding. IP, immunoprecipitation; WCL, whole-cell lysate. (B) Quantification of α-parvin to GFP ratio from immunoprecipitation experiments (mean ± SD, n = 4 independent experiments). **P = 0.0035 (Friedman/Dunn). (C) Representative immunofluorescence images of α-parvin and ILK. α-parvin colocalizes at FAs with ILK(WT)–GFP cells (arrows), whereas no obvious localization of α-parvin to adhesions is seen in ILK-(R225A/R349A)–GFP cells. C, Right shows zoom-ins of the areas indicated by the white rectangles. (D) Representative snapshots of Movies S5 and S6 of ILK(WT)–GFP and ILK(R225A/R349A)–GFP cells plated on 8-kPa and 40-kPa substrates and imaged over time to assess adhesion dynamics. D, Bottom shows enlargements of the areas indicated by the white squares. Arrows indicate adhesion growth, while asterisks mark adhesion disassembly. (E) Quantification of adhesion assembly, disassembly, and lifetime in ILK(WT)–GFP and ILK(R225A/R349A)–GFP cells on 8-kPa and 40-kPa substrates (n > 20 cells per condition pooled across four independent experiments). *1P = 0.0394; *2P = 0.0210; *3P = 0.0113; **P = 0.0084 (Mann–Whitney). (F) Representative images of FAs in ILK(WT)–GFP and ILK(R225A/R349A)–GFP cells on 8-kPa and 40-kPa substrates. F, Bottom shows zoom-ins of the areas indicated by the white squares. Dashed lines indicate cell edges defined by cytoplasmic fluorescence. (G) Quantification of the mean FA size and adhesion number in ILK(WT)–GFP and ILK(R225A/R349A)–GFP cells on 8-kPa and 40-kPa substrates. FA area threshold was set to 20 pixels. (n > 25 cells per condition pooled across four independent experiments.) *P = 0.0454; **1P = 0.0016; *2P = 0.0054 (Mann–Whitney). (Scale bars: 20 µm.).
To understand the consequences of the salt-bridge mutations under high and low traction forces, we performed quantitative analysis of the adhesion dynamics in ILK(WT)–GFP and ILK(R255A/R349A)–GFP cells plated on soft (8 kPa) or stiff (40 kPa) substrates. In contrast to the ATP-binding mutant, ILK(R255A/R349A)–GFP cells showed decreased FA assembly rates on both soft and stiff substrates. In addition, and resembling the ATP-binding mutant, increased disassembly rates and decreased adhesion lifetimes were observed, in particular, on stiff substrates (Fig. 4 D and E and Movies S5 and S6). Consistently, morphological analyses of FAs revealed a significantly smaller size and a lower number of adhesions per cell area in ILK(R255A/R349A) cells compared to WT, with a more pronounced phenotype on 40-kPa stiff gels. Decreased FA area was confirmed by paxillin staining on cells plated on crossbow micropatterns (SI Appendix, Fig. S6B). Collectively, these results indicate that the disruption of ATP-induced allostery, which abolishes α-parvin binding, leads to slower adhesion assembly, as well as destabilization of FAs, in particular, on stiff substrates.
ATP Increases the Mechanical Stability of the ILK:Parvin Complex.
Having observed destabilization of FAs by disruption of ATP binding, in particular on stiff substrates with high traction forces, we asked how ATP affects ILK stability under mechanical load using force–probe MD simulations (Fig. 5A). Forces are transmitted from the ECM/integrins to the IPP and the actomyosin network. The interaction between ILK and integrins occurs via kindlin-2. Although a crystal structure of the ILK:kindlin-2 complex is not available, K423 and I427 of ILK have been experimentally validated as residues interacting with kindlin-2 (20). We determined a larger patch between ILK and kindlin-2, which includes these two residues using homology modeling and guided molecular docking (SI Appendix, Fig. S7). We defined those residues of ILK that were reproducibly identified to interact with kindlin-2 across the docked ensemble as the kindlin-2 binding patch and as the most probable force-bearing residues on the ILK surface toward the kindlin/integrin anchorage (Fig. 5B). On the other side of the IPP force-propagation pathway, α-parvin binds actin with its N terminus over the recently discovered WASP–homology2 domain (16), while the N-terminal part of the α-parvin CH2 domain is in contact with the LD1 domain of paxillin (13, 14). Therefore, the mechanical force transmitted from actin onto the N-terminal part of the CH2 domain would distribute over the residues that are encompassed by paxillin. Those residues were regarded as the pulling patch on parvin (Fig. 5B)
Fig. 5.
ATP stabilizes ILK:parvin under mechanical tension. (A) Stretching force applied to ILK and parvin in the form of virtual springs at indicated positions. (B) Contact residues (black) between ILK (pink) and a kindlin-2 model (gray) defined as the ILK pulling patch. Contact residues (black) between parvin (cyan) and paxillin–LD1 (gray) defined as the parvin pulling patch. (C) Cumulative number of complex dissociation events as a function of distance between the pulled residue patches until dissociation (De-e), which is defined by an interface area below 0.6 nm2 [n = 30 independent trajectories, 10 per velocity, ILK holo vs. ILK apo: *P = 0.028, ILK(WT) vs. ILK(R225A/R349A); **P = 0.001, one-way ANOVA/Tukey HSD]. Schemes and black arrowheads visualize the highest and lowest De-e before dissociation. Colored arrowheads and corresponding snapshots indicate the median structure of each condition at the time of dissociation; snapshots of 15th and 85th percentile per condition are shown in SI Appendix, Fig. S8B.
In force–probe MD simulations, we subjected the two sets of force-bearing residues to harmonic pulling potentials moving away from one another with constant pulling velocities of 1 m/s to 0.01 m/s (Fig. 5 A and C and Movies S7–S9). Mechanical force induced both ILK:parvin dissociation, as measured by the changes in interface area, and ILK or parvin unfolding, reflected by an increase in the end-to-end distance of the complex prior to dissociation. This general unfolding and dissociation mechanism was similar across different pulling velocities (SI Appendix, Fig. S8A). To quantify and compare the force-induced dynamics, we defined complex dissociation to occur at the time at which the interface area drops below 0.6 nm2. Dissociation was observed at various unfolding stages, after only minor parts of the protein complex had straightened and unfolded (end-to-end distance 9 nm) all the way to incidences at which major parts of parvin were already unfolded (end-to-end distance 28 nm). Interestingly, the mechanical stability of the ILK:parvin interface relative to the individual proteins decreases upon depletion of ATP: Across pulling velocities, the ATP-bound ILK:parvin complex dissociates at higher extensions—i.e., only after more extensive parvin unfolding—than the apo state. The preference of dissociation over unfolding seen for the WT apo complex was also observed in the ATP-bound double–salt-bridge mutant and was indistinguishable from the WT apo complex. These findings indicate a weaker ILK:parvin interface in the absence of ATP, which corroborates the notion of ATP as an allosteric regulator of ILK:parvin (Fig. 3) and suggests a mechanistic explanation of the observed destabilization of FAs on stiff substrates (Fig. 4).
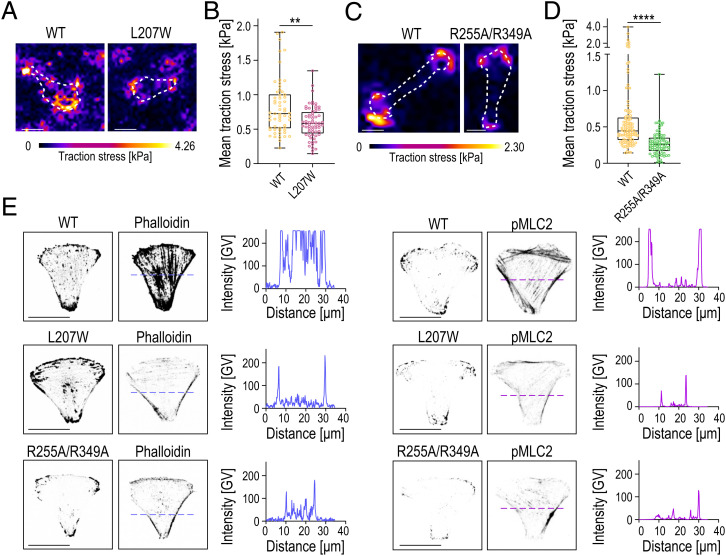
Loss of ATP and α-Parvin Binding Prevents Traction Force Generation and Actomyosin Contractility.
As the simulations indicated that ATP stabilizes the ILK:parvin complex under mechanical load, we predicted that destabilization of the complex would prevent buildup of traction stresses within FAs. To test this, we performed traction force microscopy. Interestingly, both L207W and R255A/R349A showed lower mean traction stresses compared to WT cells (Fig. 6 A–D). To assess whether the reduced traction stresses are associated with impaired actomyosin contractility, we analyzed F-actin and myosin II activity by phalloidin and phospho-Myosin Light Chain 2 (pMLC2) stainings. To this end, we plated cells on crossbow-shaped micropatterns to polarize the cells and to accurately quantify changes in myosin II activity across the nonadhesive edges (46). WT cells featured thick and mature ventral stress fibers and high pMLC2 (Fig. 6E). Quantification of the immunostainings showed substantial reduction in actin stress fibers and myosin II activity for both the L207W and the R255A/R349A mutants. Collectively, these results show that ATP binding and its allosteric salt-bridge interactions are required for the generation of traction stresses and actomyosin contractility.
Fig. 6.
Impaired ATP and α-parvin binding prevent traction force generation and actomyosin contractility. (A) Representative images and traction force heat maps of ILK(WT)–GFP and ILK(L207W)–GFP cells. (B) Quantification of mean traction stresses of ILK(WT) and ILK(L207W) cells (n > 59 cells per condition pooled across four independent experiments). **P = 0.0047 (Kolmogorov–Smirnov). (C) Representative images and traction force heat maps of ILK(WT)–GFP and ILK(R255A/R349A)–GFP cells plated on 8-kPa PAA gels. (D) Quantification of mean traction stresses of ILK(WT)–GFP and ILK(R255A/R349A)–GFP cells (n > 78 cells per condition pooled across four independent experiments). ****P = 0.0001 (Kolmogorov–Smirnov). (E) Representative images of ILK(WT)–GFP, ILK(L207W)–GFP, and ILK(R255A/R349A)–GFP cells stained with phalloidin and pMLC2 on crossbow micropatterns and corresponding line-scan quantifications of phalloidin (in blue) and pMLC2 (magenta) fluorescence intensity. GV, gray values. (Scale bars: 20 µm.).
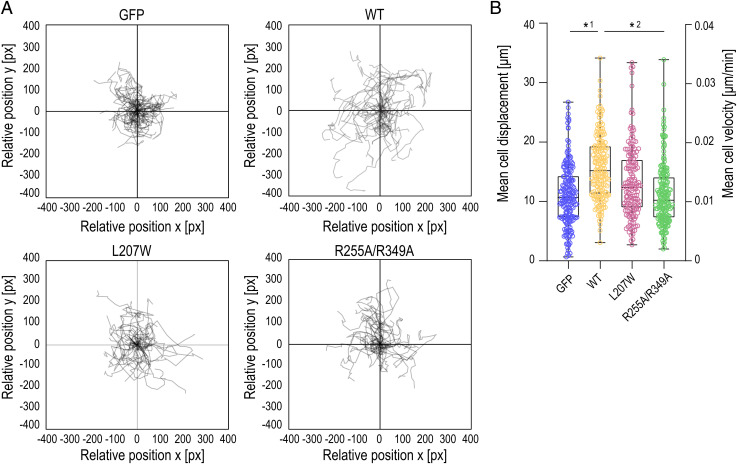
Loss of ATP and α-Parvin Binding Impairs Cell Migration.
We predicted that the inability of the ILK mutants to generate traction stresses, which stabilize FAs and promote actomyosin contractility, would impair cell migration that relies on these processes (48). To test this, we performed cell migration assays and analyzed migration trajectories of single cells using live imaging. As expected, the ILK -∕- cells showed reduced migration compared to cells where ILK expression was restored by expressing ILK(WT)–GFP (Fig. 7 and Movies S10 and S11). Interestingly, ILK(R255A/R349A)–GFP cells showed a migration defect with migration rates lower than ILK(WT) cells and comparable to ILK-∕- cells (Fig. 7 and Movies S12 and S13). Collectively, these data indicate that destabilization of the ILK:parvin interface prevents buildup of traction forces and actomyosin contractility and thus impairs the migratory capacity of cells.
Fig. 7.
Point mutation in the ATP pseudocatalytic domain and point mutations in the salt-bridge–coordinating residues lead to migration defects. (A) Representative trajectory plots showing ILK -∕- GFP, ILK(WT)–GFP, ILK(L207W)–GFP, and ILK(R255A/R349A)–GFP cell trajectories during 12-h acquisition. n > 30 cell tracks per condition were set to a common origin (intersection of x and y axes) (SI Appendix, Movies S10–S13). (B) Quantitative analysis of average distance (µm, left y axis) and velocity (µm/min, right y axis) of ILK -∕- GFP, ILK(WT)–GFP, ILK(L207W)–GFP, and ILK(R255A/R349A)–GFP cells. (n = 3 independent experiments with >35 cells per condition per experiment.) *1P = 0.0114; *2P = 0.0269 (Friedman/Dunn).
Discussion
In this study, we employ a combined computational and experimental approach to understand how the pseudokinase ILK integrates and transduces biochemical and mechanical signals. Despite being a pseudokinase, ILK still binds ATP, and we asked if and how ATP plays a role in ILK molecular interactions, function, and mechanotransduction. While pseudokinases regulate diverse cellular processes, despite lacking the ability to phosphorylate substrates, a significant portion of them bind ATP with largely unknown functions. For other pseudokinases, such as STRADα, ATP was shown to conformationally alter the pseudokinase to maintain a closed conformation for efficient target binding and downstream signaling (49). For ILK, first hints toward a function of ATP were provided by the observation that F-actin bundling is sensitized by ATP bound to ILK (16). However, the exact molecular mechanism of ATP function, in particular, in the context of mechanical force, has remained elusive. We here put forward a multifaceted role of ATP for ILK function. We find ATP to enhance the structural integrity of ILK, to allosterically impact the ILK:parvin interaction, and to enhance the resistance of the IPP complex to force.
In previous studies, purified ILK was shown to form highly insoluble aggregates in the absence of α-parvin (11), indicating that it likely coevolved with α-parvin as an obligatory binding partner. The inherent instability of ILK is further underlined by the requirement for the heat shock protein Hsp90 for a stable ILK:parvin interaction and force generation on the ECM (45). Thus, since ATP shows only little effect on the overall ILK structure (11), our data indicate that ATP instead alters the dynamics and stability of ILK and the ILK:parvin complex, proposing ATP as a secondary binding partner for structural integrity.
ATP-binding-deficient ILK(L207W) was described as a mutant that sterically occludes the ATP-binding site without affecting the structural integrity (16). The overall structural integrity is indeed maintained, as shown by the ability of this mutant to still bind parvin (ref. 16 and this study). However, on an atomistic level, the kinase-like domain is more flexible without ATP and displays a more pronounced interlobe wringing motion comparable to apo ILK(WT). On the cellular level, loss of ATP in the ILK pseudokinase leads to destabilization of FAs, resulting in decreased FA number. This is especially pronounced on stiff substrates, where high traction forces are generated.
Our simulation data further provide evidence for internal force-propagation pathways from ATP that allosterically influence previously unacknowledged parvin-binding salt bridges (R225 and R349). Indeed, we show that those residues confer parvin binding, herein cooperating with the previously analyzed M402/K403 in the C-lobe of ILK (11). Interestingly, while failing to recruit parvin to FAs due to impaired binding, the double–salt-bridge mutant (R349A/R225A) itself still localizes correctly to FAs, contrasting the M402A/K403A mutant. In this way, the double–salt-bridge mutant phenotypically resembles the ILK mutant deficient in ATP binding. In contrast to the L207W mutant, another ATP-binding-defective mutant (N200A/K341A) failed to localize correctly to FAs (11). Our FDA indicates that the reason for this might be the importance of K341 for the allosteric signal transduction to the parvin-binding salt bridges. Our data thus put forward a model that features two distinct parvin-binding pathways within ILK: one ATP-dependent pathway across the kinase N-lobe and activation segment, which governs the two salt-bridge–forming arginines, and the previously established C-lobe pathway, which is ATP-independent. Intriguingly, only the ATP-independent mode is required for localizing ILK into FAs, whereas ATP-dependent binding is required for adhesion stability.
Beyond the stabilizing effect of ATP on the pseudokinase in equilibrium conditions, we also observed that ATP conveys mechanical stabilization of the ILK:parvin complex under force. The molecular basis for this mechanical stabilization in our simulations is based on the assumption that forces in FAs are transmitted from integrins via kindlin to the IPP, ultimately reaching F-actin, and vice versa. This is reflected in the choice of residue patches for the force–probe MD. Using a combined molecular-modeling and docking approach for ILK:kindlin, we identify kindlin-binding residues and are able to robustly simulate the resultant pulling direction. A more comprehensive view onto mechanotransduction across the kindlin–ILK axis, potentially also involving force propagation via the membrane as for FAK (39, 50), can only be obtained once an experimental kindlin:ILK structure becomes available. Another plausible mechanotransduction pathway involves the N-terminal ankyrin-repeat domains of ILK (10). Given the critical role of the pulling direction on protein mechanical response (51), this additional force-application route might further alter the competition between unfolding and rupture of the IPP complex and the role of ATP therein.
As evidenced by the indistinguishable mechanical instability of apo ILK and the salt-bridge mutant that still binds ATP, we hypothesize that the stabilizing effect of ATP on ILK:parvin under mechanical force is at least partly mediated by the allosterically influenced salt bridges. Under the assumption that an intact ILK:parvin complex is a prerequisite for cellular force generation, we therefore predicted that the double–salt-bridge mutant is impaired in establishing cellular forces due to its apparently weaker interface under mechanical load. Experimental quantification of cell–ECM traction stresses showed that, indeed, disruption of both ATP and parvin binding in ILK leads to reduced force generation and cell migration, supporting the notion that stability of ILK:parvin interaction determines the ability of FAs to bear loads. Thus, we propose that ILK functions as a pseudomechanokinase, where ATP binding serves a mechanosensory role. We generally observe a slightly milder phenotype for the ATP-binding mutant compared to the double–salt-bridge mutant, which might be explained by fact that parvin is not recruited to FAs in the double–salt-bridge mutant. Since the IPP was shown to form in cells in suspension, parvin binding likely precedes recruitment of IPP to FAs (52). Therefore, our results of reduced FA assembly rates in the double–salt-bridge mutant that strongly disrupts parvin binding might be a hint toward a function of parvin in the IPP-driven FA assembly. In contrast, FA disassembly, especially on stiff substrates, is more severely perturbed in both mutants, which might indicate that both parvin and ATP regulate IPP stability under mechanical force. Hence, ATP might be regarded as one puzzle piece that keeps the ILK:parvin complex intact to precisely regulate the point of FA disruption, determined by the amount of force buildup, which ultimately leads to FA disassembly for efficient rear-end retraction.
In summary, this study sheds light onto the stabilizing function of ATP on the ILK pseduokinase leading to efficient traction force buildup, FA stabilization, and efficient migration. Apart from further advancing the knowledge on ILK-mediated integrin–actin communication and the dynamics of FAs, our study contributes to the understanding of pseudokinase evolution and function. Pseudokinases have emerged as useful models to study the noncatalytic properties of the kinase fold in isolation (53, 54). With several pseudokinases currently under investigation as therapeutic targets (55, 56), further understanding the function of ILK pseudokinase might culminate in therapeutic implementations to specifically target ILK not as a kinase, but a mechanotransducer. Mechanotransducing (pseudo)kinases are uniquely suited to integrate mechanical into other signaling pathways, and more members of this intriguing class of proteins likely remain to be uncovered.
Materials and Methods
A summary of materials and methods is outlined below. Detailed methods can be found in SI Appendix.
MD Simulations.
MD simulations were performed by using GROMACS 2018.1 (57), the Amber99sb*-ILDNP force-field (58, 59), the TIP3-water model (60), and ATP parameters (61). ILK crystal structures (11, 16) were solvated with 3 nm between periodic images, including sodium and chloride ions corresponding to 100 mM. Energy minimization was followed by 500 ps each in the number, volume, and temperature canonical ensemble and the isothermal–isobaric (NPT) ensemble with harmonic constraints on all protein atoms with a force constant of 1,000 kJ mol−1 nm−1. Systems were simulated in NPT without constraints on heavy atoms at 300 K for 20 individual production runs each for 500 ns. The first 20 ns were neglected, leaving a total simulation time analyzed of 9.6 µs per condition. To identify the major correlated structural motions, we performed PCA. All trajectories were projected onto the eigenvectors generated by the apo state. To calculate the changes in internal forces between the ILK holo and apo states, we used FDA (47). Statistically significant (Mann–Whitney test, P < 0.05) interactions are shown as graphs of at least three residues with force differences above a threshold.
Guided Molecular Docking.
To determine a patch of residues on ILK that is most likely in contact with kindlin-2, we employed a combined molecular modeling and guided docking approach. All available crystal structures of kindlin-2 (23, 26, 27, 62, 63) were structurally aligned by using UCSF-Chimera (64), according to an underlying sequence alignment with the human kindlin-2 generated with T-coffee (65). The full human kindlin-2 structures were generated by a homology modeling using MODELER (66) based on the positions of the placed fragments. Guided molecular docking was performed with Haddock2.2 (67). The experimentally validated ILK:kindlin-2 interaction residues [ILK: K423 and I427 (20); kindlin-2: L353, E354, L357, and E358 (62)] were chosen as the actively participating residues.
Force–Probe MD Simulations.
The ILK:kindlin-2 interaction residues from our guided docking within a cutoff of 0.35 nm with the highest occupancy across all docking poses (P419, H420, K423, I427, K435, M441, and K448) were regarded as the patch of residues for force–probe MD. The residues on parvin that were interacting with paxillin (13, 14) within a threshold of 0.35 nm (A249, T252, V264, T267, and R369) were regarded as the patch of residues on parvin for pulling. Those residue patches were subjected to harmonic spring potentials with a force constant of 50 kJ mol−1 nm−1 moving in opposite directions with a constant velocity (v = 1 m s−1, 0.1 m/s and 0.01 m s−1) for 10 simulations per velocity and condition. The interface area between ILK and parvin was calculated by the solvent-accessible surface area, and a complex dissociation event was defined if the interface area was below 0.6 nm2.
Plasmid Constructs, Transfection, and Cell Culture.
Described point mutations were generated by performing site-directed mutagenesis in mouse ILK complementary DNA cloned into pEGFP-N1 (Clontech). ILK -∕- mouse fibroblasts were obtained as described (45) and transiently transfected with Lipofectamine 3000 reagent (Invitrogen). GFP immunoprecipitation was performed by using the Miltenyi Biotec MultiMACS GFP Isolation Kit (Miltenyi Biotec catalog no. 130-091-125) and analyzed by Western blotting.
Substrate Engineering, Live Imaging, and Traction Force Microscopy.
PAA gels and traction force microscopy were performed essentially as described (68–70). Micropatterned adhesive surfaces were generated by using the PRIMO optical module (Alveole). Cells were live-imaged by using a Zeiss Axiovert inverted microscope coupled to a CSUX1 spinning-disk device (Yokogawa) with a 488-nm laser and scientific complementary metal–oxide–semiconductor camera (Hamamatsu). Images were acquired and analyzed by using the Focal Adhesion Analysis Server (71) for FA dynamics or the Fiji plugin Trackmate (72) for migration.
Immunofluorescence Stainings and Confocal Microscopy.
Cells were fixed in 4% paraformaldehyde and stained with antibodies, as described in detail in SI Appendix. Images were collected by laser-scanning confocal microscopy (SP8X; Leica) with Leica Application Suite software (LAS X version 2.0.0.14332a) and quantified by using Fiji (73).
Statistical Analysis.
Statistical analyses were performed by using GraphPad Prism (version 8) and Python. Statistical significance was determined by the specific tests indicated in the corresponding figure legends. All experiments presented in the manuscript were repeated in at least three independent experiments/biological replicates. No datapoints were excluded.
Supplementary Material
Acknowledgments
We acknowledge Anu M. Luoto for technical assistance. This work was supported by the Klaus Tschira Foundation; the state of Baden-Württemberg through bwHPC; the German Research Foundation through Grant INST 35/1134-1 FUGG (to F.G.); the Helsinki Institute of Life Science; the Sigrid Juselius Foundation; and Academy of Finland Grant 317597 (to S.A.W.). Molecular graphics and analyses were partially performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California San Francisco, with support from NIH Grant P41-GM103311.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
Plasmids data have been deposited in Addgene (accession nos. 176896 [mILK-EGFP], 176897 [mILK-R255A-EGFP], 176898 [mILK-R349A-EGFP], 176899 [mILK-R225A/R349A-EGFP], and 176900 [mILK-L207W-EGFP]). All study data are included in the article and/or supporting information.
References
- 1.Hynes R. O., Integrins: Bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Legate K. R., Wickström S. A., Fässler R., Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23, 397–418 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Schiller H. B., Friedel C. C., Boulegue C., Fässler R., Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12, 259–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman B. D., Grashoff C., Schwartz M. A., Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316–323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elosegui-Artola A., Trepat X., Roca-Cusachs P., Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356–367 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kechagia J. Z., Ivaska J., Roca-Cusachs P., Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20, 457–473 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Wu C., The PINCH-ILK-parvin complexes: Assembly, functions and regulation. Biochim. Biophys. Acta 1692, 55–62 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Legate K. R., Montañez E., Kudlacek O., Fässler R., ILK, PINCH and parvin: The tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 7, 20–31 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Wickström S. A., Lange A., Montanez E., Fässler R., The ILK/PINCH/parvin complex: The kinase is dead, long live the pseudokinase! EMBO J. 29, 281–291 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiswell B. P., Zhang R., Murphy J. W., Boggon T. J., Calderwood D. A., The structural basis of integrin-linked kinase-PINCH interactions. Proc. Natl. Acad. Sci. U.S.A. 105, 20677–20682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda K., Gupta S., Chen K., Wu C., Qin J., The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol. Cell 36, 819–830 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolopoulos S. N., Turner C. E., Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J. Biol. Chem. 276, 23499–23505 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Lorenz S., et al., Structural analysis of the interactions between paxillin LD motifs and alpha-parvin. Structure 16, 1521–1531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., et al., The structure of alpha-parvin CH2-paxillin LD1 complex reveals a novel modular recognition for focal adhesion assembly. J. Biol. Chem. 283, 21113–21119 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velyvis A., et al., Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling. Nat. Struct. Biol. 10, 558–564 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Vaynberg J., et al., Non-catalytic signaling by pseudokinase ILK for regulating cell adhesion. Nat. Commun. 9, 4465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H., et al., Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics. eLife 10, e64395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannigan G. E., et al., Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature 379, 91–96 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Montanez E., et al., Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 22, 1325–1330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadry Y. A., Huet-Calderwood C., Simon B., Calderwood D. A., Kindlin-2 interacts with a highly conserved surface of ILK to regulate focal adhesion localization and cell spreading. J. Cell Sci. 131, jcs221184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calderwood D. A., Campbell I. D., Critchley D. R., Talins and kindlins: Partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harburger D. S., Bouaouina M., Calderwood D. A., Kindlin-1 and -2 directly bind the C-terminal region of beta integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 284, 11485–11497 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., et al., Structural basis of kindlin-mediated integrin recognition and activation. Proc. Natl. Acad. Sci. U.S.A. 114, 9349–9354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qadota H., Moerman D. G., Benian G. M., A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (Kindlin) to integrin adhesion sites. J. Biol. Chem. 287, 28537–28551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahed Z., Haydari Z., Rathish A., Mofrad M. R. K., Kindlin is mechanosensitive: Force-induced conformational switch mediates cross-talk among integrins. Biophys. J. 116, 1011–1024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J., et al., Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation. J. Biol. Chem. 286, 43334–43342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Zhu Y., Ye S., Zhang R., Crystal structure of kindlin-2 PH domain reveals a conformational transition for its membrane anchoring and regulation of integrin activation. Protein Cell 3, 434–440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannigan G., Troussard A. A., Dedhar S., Integrin-linked kinase: A cancer therapeutic target unique among its ILK. Nat. Rev. Cancer 5, 51–63 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Fukuda K., Knight J. D. R., Piszczek G., Kothary R., Qin J., Biochemical, proteomic, structural, and thermodynamic characterizations of integrin-linked kinase (ILK): Cross-validation of the pseudokinase. J. Biol. Chem. 286, 21886–21895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maydan M., et al., Integrin-linked kinase is a functional Mn2+-dependent protein kinase that regulates glycogen synthase kinase-3β (GSK-3beta) phosphorylation. PLoS One 5, e12356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hannigan G. E., McDonald P. C., Walsh M. P., Dedhar S., Integrin-linked kinase: Not so ‘pseudo’ after all. Oncogene 30, 4375–4385 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Sakai T., et al., Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 17, 926–940 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannigan G. E., Coles J. G., Dedhar S., Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ. Res. 100, 1408–1414 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Zheng C. C., et al., Significance of integrin-linked kinase (ILK) in tumorigenesis and its potential implication as a biomarker and therapeutic target for human cancer. Am. J. Cancer Res. 9, 186–197 (2019). [PMC free article] [PubMed] [Google Scholar]
- 35.Olmos G., López-Ongil S., Ruiz Torres M. P., Integrin-linked kinase: A new actor in the ageing process? Exp. Gerontol. 100, 87–90 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Friedrich E. B., et al., Integrin-linked kinase regulates endothelial cell survival and vascular development. Mol. Cell. Biol. 24, 8134–8144 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuda T., Chen K., Shi X., Wu C., PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J. Biol. Chem. 278, 51324–51333 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Stanchi F., et al., Molecular dissection of the ILK-PINCH-parvin triad reveals a fundamental role for the ILK kinase domain in the late stages of focal-adhesion maturation. J. Cell Sci. 122, 1800–1811 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Zhou J., et al., Mechanism of focal adhesion kinase mechanosensing. PLOS Comput. Biol. 11, e1004593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tapia-Rojo R., Alonso-Caballero A., Fernandez J. M., Direct observation of a coil-to-helix contraction triggered by vinculin binding to talin. Sci. Adv. 6, eaaz4707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goult B. T., Yan J., Schwartz M. A., Talin as a mechanosensitive signaling hub. J. Cell Biol. 217, 3776–3784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mofrad M. R. K., Golji J., Abdul Rahim N. A., Kamm R. D., Force-induced unfolding of the focal adhesion targeting domain and the influence of paxillin binding. Mech. Chem. Biosyst. 1, 253–265 (2004). [PubMed] [Google Scholar]
- 43.Rahikainen R., Öhman T., Turkki P., Varjosalo M., Hytönen V. P., Talin-mediated force transmission and talin rod domain unfolding independently regulate adhesion signaling. J. Cell Sci. 132, jcs226514 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Bauer M. S., et al., Structural and mechanistic insights into mechanoactivation of focal adhesion kinase. Proc. Natl. Acad. Sci. U.S.A. 116, 6766–6774 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radovanac K., et al., Stabilization of integrin-linked kinase by the Hsp90-CHIP axis impacts cellular force generation, migration and the fibrotic response. EMBO J. 32, 1409–1424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Théry M., et al., Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. U.S.A. 103, 19771–19776 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costescu B. I., Gräter F., Time-resolved force distribution analysis. BMC Biophys. 6, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ridley A. J., et al., Cell migration: Integrating signals from front to back. Science 302, 1704–1709 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Zeqiraj E., et al., ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 7, e1000126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goñi G. M., et al., Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc. Natl. Acad. Sci. U.S.A. 111, E3177–E3186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carrion-Vazquez M., et al., The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10, 738–743 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., et al., Assembly of the PINCH-ILK-CH-ILKBP complex precedes and is essential for localization of each component to cell-matrix adhesion sites. J. Cell Sci. 115, 4777–4786 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Jacobsen A. V., Murphy J. M., The secret life of kinases: Insights into non-catalytic signalling functions from pseudokinases. Biochem. Soc. Trans. 45, 665–681 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Kung J. E., Jura N., Structural basis for the non-catalytic functions of protein kinases. Structure 24, 7–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byrne D. P., Foulkes D. M., Eyers P. A., Pseudokinases: Update on their functions and evaluation as new drug targets. Future Med. Chem. 9, 245–265 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Kung J. E., Jura N., Prospects for pharmacological targeting of pseudokinases. Nat. Rev. Drug Discov. 18, 501–526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abraham M. J., et al., Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 58.Best R. B., Hummer G., Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Phys. Chem. B 113, 9004–9015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aliev A. E., et al., Motional timescale predictions by molecular dynamics simulations: Case study using proline and hydroxyproline sidechain dynamics. Proteins 82, 195–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 61.Meagher K. L., Redman L. T., Carlson H. A., Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 24, 1016–1025 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Fukuda K., et al., Molecular basis of kindlin-2 binding to integrin-linked kinase pseudokinase for regulating cell adhesion. J. Biol. Chem. 289, 28363–28375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perera H. D., et al., Membrane binding of the N-terminal ubiquitin-like domain of kindlin-2 is crucial for its regulation of integrin activation. Structure 19, 1664–1671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen E. F., et al., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Di Tommaso P., et al., T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webb B., Sali A., Comparative protein structure modeling using modeller. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Zundert G. C. P., et al., The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Pelham R. J. Jr., Yl. Wang, Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. U.S.A. 94, 13661–13665 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dembo M., Wang Y. L., Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76, 2307–2316 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tseng Q., et al., A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab Chip 11, 2231–2240 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Berginski M. E., Gomez S. M., The Focal Adhesion Analysis Server: A web tool for analyzing focal adhesion dynamics. F1000 Res. 2, 68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tinevez J. Y., et al., TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids data have been deposited in Addgene (accession nos. 176896 [mILK-EGFP], 176897 [mILK-R255A-EGFP], 176898 [mILK-R349A-EGFP], 176899 [mILK-R225A/R349A-EGFP], and 176900 [mILK-L207W-EGFP]). All study data are included in the article and/or supporting information.