Summary
Background
Tigecycline is one of the few last-resort antibiotics for the treatment of carbapenem-resistant Enterobacteriaceae infection, the incidence of which has been rapidly increasing. However, the emergence and spread of tigecycline resistance genes tet(X) (including tet(X3) and tet(X4)) has largely compromised the efficient usage of tetracyclines in the clinical settings.
Methods
The synergistic effect was determined by a checkerboard minimum inhibitory concentration (MIC) assay, a time-killing assay and scanning electron microscopy (SEM) analysis. In-depth mechanisms were defined using an enzyme inhibition assay, western blotting, RT-PCR analysis, molecular dynamics (MD) simulations, biolayer interferometry (BLI) assay and metabolomics analysis.
Findings
Herein, our work identified a natural compound, plumbagin, as an effective broad-spectrum inhibitor of Tet(X) (also known as monooxygenase) by simultaneously inhibiting the activity and the production of Tet(X3)/Tet(X4). Plumbagin in combination with tetracyclines showed a synergistic bactericidal effect against Tet(X3)/Tet(X4)-producing bacteria. Mechanistic studies revealed that direct engagement of plumbagin with the catalytic pocket of Tet(X3)/Tet(X4) induced an alternation in its secondary structure to inhibit the activity of these monooxygenases. As a consequence, monotherapy or combination therapy with plumbagin increases the oxidative stress and metabolism in bacteria. Moreover, in a mouse systemic infection model of tet(X4)-positive E. coli, the combination of plumbagin and methacycline exhibited remarkable treatment benefits, as shown by a reduced bacterial load and the alleviation of pathological injury.
Interpretation
Plumbagin, as an inhibitor of Tet(X3)/Tet(X4), represents a promising lead drug, as well as an adjunct with tetracyclines to treat bacterial infections, especially for extensively drug-resistant bacteria harbouring Tet(X3)/Tet(X4).
Funding
The National Natural Science Foundation of China.
Keywords: Plumbagin, Tet(X), Tetracycline, Inhibitor, Monooxygenase
Research in context.
Evidence before this study
Antibiotic resistance is an increasing threat to public health and food security. With the occurrence and high prevalence of the tigecycline-resistance genes tet(X3) and tet(X4), the clinical efficacy of the last-resort antibiotic tigecycline is being challenged. Thus, it is imperative to develop novel antibiotics or anti-infective strategies to meet this serious clinical threat. Alarmingly, there have been few reports on alternative antibiotics or tetracyclines potentiators targeting the activity of Tet(X3)/Tet(X4).
Added value of this study
To potentiate the therapeutic effect of antibiotics or to prevent the development of antibiotic resistance, we focused on exploring the effective antibiotic combinations and tested the activity of several natural compounds with tetracyclines against tet(X3)/tet(X4)-positive bacteria. Plumbagin, served as a potential anti-cancer compound, has been extensively researched, however, its potency in the treatment of pathogenic bacteria has not been explored and utilized. In this study, we found that plumbagin, as an inhibitor of Tet(X3)/Tet(X4), potently potentiated the antibacterial activity of tetracyclines in vitro or in vivo. In-depth mechanism exploration indicated that plumbagin directly engaged with the catalytic pocket of Tet(X3)/Tet(X4), thereby altering their conformation and inhibiting the catalytic activity of Tet(X3)/Tet(X4). Further analysis revealed that plumbagin interferes with redox processes and metabolism needed for the bacterial survival, which offers an alternative target for fighting bacterial resistance. The discovery of plumbagin as a novel and safe tetracyclines adjuvant provides a promising therapeutic strategy for infections caused by tetracycline-resistant bacteria.
Implications of all the available evidence
The current study found a novel and promising tetracyclines adjuvant, plumbagin, to combat the multidrug-resistant Gram-negative bacteria infections, which is of highly potential practical value for clinical therapeutics.
Alt-text: Unlabelled box
Introduction
Therapeutic options to address infection caused by extensively drug-resistant (XDR) pathogenic bacteria in human clinical therapy and livestock breeding have become formidable challenges due to the emergence and rapid spread of carbapenem-resistant Enterobacteriaceae (CRE).1 Accordingly, tigecycline has been recognized as the last-resort antibiotic for the clinical treatment of some bacterial infections, especially for XDR Gram-negative bacteria. However, following the widespread use and abuse of antibiotics, obvious resistance to tetracyclines has been reported worldwide with worrying implications for public and food safety.2,3
A recent study by He et al. reported two plasmid-mediated and widely existing tigecycline resistance genes, namely, tet(X3) and tet(X4), which encode the drug resistance enzymes Tet(X3) and Tet(X4), respectively, that seriously impair the clinical efficacy of tigecyclines.4 The occurrence of plasmid-carried tigecycline resistance genes tet(X3) and tet(X4) in no less than 16 bacterial species, including Enterobacteriaceae and A. baumannii, would foreseeably indicate a major threat to the effective use of tigecycline as the last line of defence and severely threaten public health. Therefore, a potent agent to combat the looming tigecycline resistance is urgently needed in the clinic, especially for Enterobacteriaceae and A. baumannii. As reported in previous works,5 the antibiotic adjuvant strategy is a potential and cost-effective approach to boost antibiotic effectiveness. Thus, the identification of inhibitors by targeting Tet(X3)/Tet(X4) could effectively address such crisis.
The enzymes Tet(X3)/Tet(X4) have been reported as NADPH-requiring monooxygenases responsible for the inactivation of tetracycline antibiotics, and which could be the result of reductive electron transfer in the bacterial oxidation and reduction process, or hydroxylation reactions.4,6 Further, various small-molecule compounds have been identified as effective inhibitors to intervene in bacterial oxidation and reduction process, which reduce respiratory chain electron transport and ATP generation or enhance ROS generation.7,8 Hence, the identification of Tet(X3)/Tet(X4) inhibitors is a promising strategy for both the recovery of bacterial sensitivity to tetracycline antibiotics and the inhibition of bacterial viability.
Here, plumbagin, as a naturally occurring naphthoquinone isolated from plants, was identified as a potential Tet(X3)/Tet(X4) inhibitor that displays synergistic activity with tetracyclines against tet(X3)/tet(X4)-positive bacteria. Naphthoquinones have been well characterized to possess various pharmacological activities,9, 10, 11 including antiallergic, antiviral, anti-inflammatory, and antibacterial activity. Importantly, naphthoquinones are widely used as redox cycling agents owing to their redox activity in both hosts and pathogens. Although various pharmacological activities of plumbagin have been reported, its potential application in the treatment of drug-resistant pathogens has not been explored. This work successfully identified plumbagin as a novel tetracycline antibiotics adjuvant, thereby highlighting the potential for fighting resistant bacterial infection with natural compounds by reversing the resistance to antibiotics via multiple mechanisms of action. Accordingly, the discovery of plumbagin as a novel and safe tetracycline adjuvant provided a promising therapeutic strategy for treating tetracycline-resistant pathogen infections.
Methods
Bacteria and reagents
The bacterial strains used in this study included tet(X3)/tet(X4)-positive E. coli, tet(X4)-positive K. pneumoniae and tet(X3)-positive A. baumannii, as described in Supplementary Table 1.4 The tested Tet(X)-positive isolates of chicken or human origin are listed in Supplementary Table 2. Unless otherwise noted, all of the strains were grown in LB broth (BIOFOUNT). The plumbagin (dissolved in dimethyl sulfoxide) was obtained from Desite Biotechnology Co., Ltd. Antibiotics were purchased from the China Institute of Veterinary Drug Control, CSNpharm, Inc., MedChem Express, Inc. and Dalian Meilun Biotechnology Co., Ltd. The Hydrogen Peroxide Assay Kit (S0038), Reactive Oxygen Species (ROS) Assay Kit (S0033S), Total Superoxide Dismutase (SOD) Assay Kit (S0101S), ATP Assay Kit (S0026) and BCA Protein Assay Kit (P0012S) were obtained from Beyotime Biotechnology Co., Ltd., China. The coenzyme I NAD(H) Content Assay Kit (BC0310) were purchased from Solarbio (Beijing).
Cell lines and cell culture
Human embryonic kidney cells (HEK-293) (RRID: CVCL_0045) and lung carcinoma cells (A549) (RRID: CVCL_0023) were cultured in DMEM high glucose medium supplemented with 10% heat-inactivated fetal bovine serum (Biological Industries, BI), penicillin (100 units/mL), and streptomycin (100 μg/mL). Peritoneal macrophages were isolated and harvested as described in a previous report,12 and cultured in RPMI-1640 medium containing 10% heat-inactivated fetal bovine serum. Cells were cultured at 37 °C in a humidified incubator with 5% CO2 atmosphere. Cell lines in current study were obtained from the American Type Culture Collection (ATCC), authenticated by Short tandem repeat (STR) profiling and were not contaminated by bacteria and mycoplasma.
Construction, expression and purification of Tet(X3)/Tet(X4) and their mutants
The tet(X3) and tet(X4) genes were obtained from A. baumannii 34AB and E. coli 47EC, respectively, and cloned into the expression vector pGEX-4T-1 followed by digestion with the endonucleases BamHI and XhoI. The expression vectors for the Tet(X3)/Tet(X4) mutants were constructed using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) based on the E. coli BL21(DE3)-pGEX-4T-1-Tet(X3)/Tet(X4) construct and the gene-specific primers were listed in Supplementary Table 3. The purification of Tet(X3)/Tet(X4) and their mutants were performed using the prokaryotic expression system.
Enzyme inhibition assays
The absorbance at 400 nm was determined to examine the activity of Tet(X3)/Tet(X4) as previously described,6,13 and which require NADPH, Mg2+ and O2 for activity. The purified proteins of Tet(X3)/Tet(X4) were incubated with NADPH in the presence of Mg2+ and O2, and different concentrations of potential lead candidates for 20 min at 37°C. Then, each sample was mixed with an equal volume of tetracycline and the enzyme activity of Tet(X3)/Tet(X4) was monitored by the changes of the absorbance at OD400 nm for 1 h at 37°C. Ultimately, the inhibitory percentage of compounds on the catalytic activity of Tet(X3)/Tet(X4) was determined by comparing the changes in absorbance values of each samples with the untreated control, which was expressed as Inhibition (%) = 1 − (∆OD400 samples/∆OD400 untreated control) × 100%, ∆OD400 means the changes in absorbance values at 400 nm for 1 h.
Antimicrobial susceptibility testing
MICs of candidate natural compounds and antibiotics for the tested strains were determined by the standard broth microdilution method based on the Clinical and Laboratory Standards Institute (CLSI) guideline.14 Concisely, candidate natural compounds or antibiotics were diluted using 2-fold serial dilutions in a sterile 96-well microtiter plate with LB broth, and then 100 μL of the bacterial suspensions (1 × 106 CFUs/mL) were added to each well. After 16–24 h of coincubation at 37 °C, the lowest concentrations of the compounds with no visible bacterial growth were monitored as the MICs of natural compounds or antibiotics. The checkerboard microdilution method was further applied to evaluate the synergies between natural compounds and tetracyclines against Tet(X)-positive strains or Tet(X)-negative strains. Briefly, the synergies between antibiotics and compounds were determined by checkerboard assays with a 2-fold serial dilution of compounds (8 × 12 matrix) mixed with a bacterial suspension (5 × 105 CFUs/mL). The MIC values of each combination were monitored as described above, and then the fractional inhibitory concentration index (FICI) was calculated according to the following formula15: FIC index = (MIC of compounds in combination/MIC of compound alone) + (MIC of antibiotics in combination/MIC of antibiotics alone). The synergy effect was defined as an FIC index of ≤ 0.5.
Growth curves
For the growth curve assay, the tested strains were cultured in LB broth to obtain a starting OD600 nm = 0.3. Then, plumbagin (from 4 μg/mL to 32 μg/mL) was added to the bacterial culture medium and coincubated at 37°C with shaking at 200 rpm. The absorbance of each sample at 600 nm was monitored by a UV spectrophotometer every 30 min.
Time-killing assays
The potential bactericidal effect of plumbagin in combination with tetracyclines was evaluated via time-killing assays. Plumbagin, tetracyclines or both in combination were cultured with the bacterial suspensions (5 × 105 CFUs/mL) in 200 μL of LB broth. At the indicated time points, cocultured supernatants from different treatments were serially diluted 10-fold and then spotted onto LB agar. To assess the bactericidal activity, the number of bacterial colonies was counted and calculated. Additionally, various contractions of bacterial suspensions (from 1 × 105 CFUs/mL to 1 × 108 CFUs/mL) were used for determination of bactericidal effect by spot assay with the indicated treatment.
Live/Dead bacteria staining
Overnight bacterial cultures of E. coli DH5α+pAM401-tet(X3)/tet(X4) were diluted in LB broth and then incubated with plumbagin (32 μg/mL), tetracycline (32 μg/mL) or their combination for 8 h at 37°C. Then, the bacteria were collected, centrifuged, resuspended in PBS to obtain an OD600 nm of 0.5 and stained using the LIVE/DEAD BacLight Bacterial Viability Kit (Invitrogen) under the guidance of the manufacturer's instructions. Green fluorescently labeled bacteria were live, and red fluorescently labeled bacteria were dead.
Scanning electron microscope (SEM) analysis
Overnight E. coli DH5α+pAM401-tet(X3)/tet(X4) treated with plumbagin (32 μg/mL), tetracycline (32 μg/mL) or their combination were washed, centrifuged and resuspended in PBS to obtain an OD600 nm of 0.5. The samples preincubated with polylysine were washed with fresh PBS and fixed in glutaraldehyde overnight at 4°C. Then, scanning electron microscopy (SEM) was performed to observe the morphological changes of the bacteria.
Safety evaluation
The cell viability of A549 cells, HEK-293 cells, and peritoneal macrophages co-incubated with plumbagin for 5 h was determined using a cytotoxicity detection kit (LDH; Roche). Then, the absorbance of cell culture supernatant at 492 nm was detected, and the corresponding cytotoxicity was calculated.
In addition, rabbit red blood cells (RBCs) were treated with different concentrations of plumbagin for 1 h. The haemolytic activity of plumbagin was evaluated by measuring the absorbance values of the supernatant at OD570 nm.
Western blot assays
After treatment with various concentrations of plumbagin (0, 8, 32 μg/mL) for 4 h or 8 h, equal amounts of bacterial cultures were centrifuged, collected, mixed with 5 × loading buffer, and then boiled at 100°C for 10 min. Each sample was separated by SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. Then, the membrane was blocked with 5% skim milk for 2 h at room temperature and further incubated with primary antibodies against Tet(X3)/Tet(X4) (prepared and stored in our laboratory) and HRP-conjugated goat anti-mouse secondary antibodies (1:3000; RRID: AB_2722565; Cat No. SA00001-1, Proteintech). Finally, the membrane was visualized with an enhanced chemiluminescence substrate.
Determination of DNA and protein expression
Overnight cultures of E. coli DH5α+pAM401-tet(X3)/tet(X4) were incubated with plumbagin (0, 8, 32 μg/mL) for 6 h and then centrifuged and resuspended to obtain an OD600 nm of 0.8. PCR amplification was performed in a thermal cycler (Bio-Rad, T100TM). PCR products were separated by electrophoresis on a 1.5% agarose gel and visualized.
E. coli BL21(DE3)-pGEX-4T-1-Tet(X3)/Tet(X4) was cultured until the OD600 nm = 0.6. Isopropyl-β-D-thiogalactopyranoside (IPTG, 1 mM) and different concentrations of plumbagin (0, 8, 32 μg/mL) were added to the bacterial culture medium and cocultured for 4 h at 37°C with shaking at 200 rpm. Then, the OD600 nm of the bacterial suspension was standardized to 0.8 and collected and boiled with or without 5 × loading buffer at 100°C for 10 min for SDS-PAGE. The proteins from each sample were stained with Coomassie brilliant blue.
Molecular dynamics (MD) simulations
The PDB code for the initial 3D structure of Tet(X3)/Tet(X4) was 4A6N, and the standard docking procedures of plumbagin and Tet(X3)/Tet(X4) were performed using AutoDock Vina.16 All of the simulations and analyses of the trajectories were performed with Gromacs 5.1 software17 using the Amber99sb force field and TIP3P water model. After the simulation, the binding free energy between the protein and the ligand was analysed using the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method, as described in previous reports.18 Analysis of the trajectories were performed by using the VMD, PyMOL, and Gromacs analysis tools.
Fluorescence quenching analysis
The binding constants (KA) for plumbagin with the binding sites on WT-Tet(X3), M215A-Tet(X3), L222A-Tet(X3), F224A-Tet(X3), or WT-Tet(X4), L219A-Tet(X4), I234A-Tet(X4), F221A-Tet(X4) were determined using a fluorescence-quenching method, at the excitation wavelength of 280 nm and emission wavelength of 345 nm, following previously described methods.19
Biolayer interferometry (BLI)
Tet(X3)/Tet(X4) (GST-tagged proteins) were respectively captured on the surface of anti-GST antibody coated BLI sensors, followed by determination of detectable binding with different concentrations of plumbagin. The BLI assay was conducted on an Octet K2 (FortéBio) instrument and performed following a test cycle, which was pre-wetted in PBS for 900 s, and coupled GST-tagged Tet(X3)/Tet(X4) before adding to the indicated concentrations of compounds and the protein alone coupled to anti-GST antibody coated BLI sensors were used as a reference control, respectively. And the BLI results were analysed using FortéBio Data Analysis 11.0 software.20,21
Circular dichroism (CD) spectra analysis
The circular dichroism (CD) spectra of Tet(X3)/Tet(X4) with or without plumbagin were analyzed by a CD spectrophotometer (MOS-500; Bio-Logic) with the scanning wavelengths ranged from 190 to 250 nm as described in our previous study.22 And the BeStSel web server was used to analyze the secondary structure of the protein.23
Metabolomics analysis
Overnight cultures of E. coli DH5α+pAM401-tet(X4) were diluted 1/100 into fresh LB broth and coincubated with or without plumbagin (32 μg/mL) at 37°C for 8 h. Then, after washing with fresh PBS, the bacteria were centrifuged at 12,000 rpm for 10 min at 4°C and resuspended in sterile PBS. Metabolomics detection could be performed through following process. First of all, metabolite features were detected and screened. In addition, internal standard normalization method was employed for the data analysis. The resulted three-dimensional data including the peak number, sample name and normalized peak area were fed into R package metaX for principal component analysis (PCA) and orthogonal partial least square-discriminate analysis (OPLS-DA). Additionally, the enrichment analysis of the metabolic pathways between the two groups were analysed by KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.kegg.jp) and MetaboAnalyst (http://www.metaboanalyst.ca/).
NAD+/NADH level detection
Overnight cultures of E. coli DH5α+pAM401-tet(X4) were treated with plumbagin, tetracycline or the combinations at 37°C for 5 h, and bacterial cultures were collected, centrifuged and resuspended. The NAD+/NADH ratio regulated by the TCA cycle in E. coli DH5α+pAM401-tet(X4) was determined by using a coenzyme I NAD(H) Content Assay Kit, following the manufacturer's instructions.
Detection of ROS, H2O2 and SOD activity
The levels of ROS in E. coli DH5α+pAM401-tet(X4) treated with plumbagin, tetracycline or the combinations were measured with 2′,7′- dichlorofluorescein diacetate (DCFH-DA, 10 μM) following the manufacturer's instructions. After incubation and washing with sterile PBS buffer three times, the fluorescence intensity was detected by a spectrofluorimeter at 488 nm and 525 nm. Furthermore, the H2O2 and SOD activities in E. coli DH5α+pAM401-tet(X4) treated with plumbagin, tetracycline or in combination were measured with the Hydrogen Peroxide Assay Kit and the Total Superoxide Dismutase Assay Kit, respectively.
Intracellular ATP determination
Overnight cultures of E. coli DH5α+pAM401-tet(X4) were co-incubated with the indicated concentrations of plumbagin or tetracycline at 37°C for 5 h, and bacterial cultures were collected, centrifuged and resuspended. Then, following the manufacturer's protocol, the intracellular ATP levels were determined and calculated based on the standard curve.
Inner membrane (IM) permeability determination
The bacteria were probed with 10 nM Propidium Iodide (PI), with the indicated concentrations of drugs, and the fluorescence intensity was measured at the excitation wavelength of 535 nm and emission wavelength of 615 nm.
Reverse transcriptase PCR (RT-PCR) analysis
RT-PCR assays were used to examine the influence of plumbagin on the transcription of tet(X3)/tet(X4). In brief, overnight cultures of E. coli DH5α+pAM401-tet(X3)/tet(X4) were diluted 1:100 in LB broth and incubated with the indicated concentrations of plumbagin at 37°C for 8 h. Then, total RNA was extracted and prepared as previously described.24 The RNA was reverse-transcribed to cDNA using NovoScript® Plus All-in-one 1st Strand cDNA Synthesis SuperMix (E047; Novoprotein, China) according to the manufacturer's instructions. The levels of tet(X3), tet(X4), WrbA and ydhR in each sample were determined by quantitative real-time polymerase chain reaction (qRT-PCR) with NovoScript®SYBR qPCR SuperMix Plus (E096; Novoprotein, China) and the gene-specific primers were listed in Supplementary Table 4. Finally, the gene transcription levels were determined using the 2−ΔΔCt method. In addition, the E. coli 16S ribosomal RNA gene was used as an internal control to quantify the transcription levels of the samples.
Mice infection assays
Female BALB/c mice (6-8 weeks old) were obtained from Liaoning Changsheng Technology Industrial Co., Ltd. The mice were acclimated for 5 days in a specific pathogen-free (SPF) facility and used for experiments with enough food and water in a comfortable environment (24 ± 2°C, 55 ± 10% humidity).
In light of the in vivo pharmacokinetic profile and in vivo efficacy of plumbagin or methacycline in previous reports,25, 26, 27, 28 we developed the following combination therapy regimen including dosing interval and dosage. The mice were intraperitoneally infected with 100 μL of E. coli DH5α+pAM401-tet(X4) bacterial suspension (1 × 109 CFUs/mL) and assigned randomly to four groups (solvent control, methacycline alone, plumbagin alone, and methacycline + plumbagin). And mice were respectively treated with a single dose of methacycline (10 mg/kg), or plumbagin (5 mg/kg) via oral administration, a combination of methacycline and plumbagin or control solvent every 12 h. The sample size for each group, as indicated in the following figure legend, has high reliability and statistical significance of data management.
All the mice were euthanized at 48 h post infection, and the targeted organs (livers, spleens and kidneys) were collected and homogenized for calculation of the bacterial load via serial 10-fold dilution and plated onto LB agar plate medium at 37°C overnight. Then, the livers and spleens were fixed with 10% formaldehyde for the histological assessment and TNF-α, IL-6 and IFN-γ in the livers were assayed using ELISA kits (BioLegend) following the manufacturer's instructions.
Toxicity in vivo
Plumbagin was resuspended with 0.5% CMC (Carboxymethylcellulose sodium) and sonicated. Female BALB/c mice were fasting 2 h and divided randomly into three groups (n = 6 for each group). For the subacute toxicity test, plumbagin (0, 5, 25 mg/kg body weight per groups) was administered orally intermittently every 12 h within 48 hours, as described in mice infection assays. And general behavior of mice in each group was observed continuously for up to 7 days for any sign of toxicity, such as changes in body weight. After 7 days, mice were euthanized and the pathological changes of the different organs (livers, spleens and kidneys) were collected and observed.
Statistical analyses
Statistical analysis was performed using GraphPad Prism 8.0 and SPSS software. All data are presented as the mean ± SD, and for comparison of two groups, significance was determined using Student's t-test. All P values less than 0.05 were considered statistically significant and were indicated in the figure legends. *, P<0.05; **, P<0.01.
Ethics
All animal experiments were approved by the Animal Welfare and Research Ethics Committee at Jilin University (approval no. ALKT202012006) and were conducted following the guidelines of this committee. All the participants signed written informed consents.
Role of the funding source
The funders had no role in the study design, data collection, analysis, interpretation, or in the writing of the paper. The authors had full access to all the data in the study and accept responsibility to submit for publication.
Results
Plumbagin restored the antibacterial activity of tetracyclines by inhibiting the activity of Tet(X3)/Tet(X4) in vitro
To explore the potential antibacterial synerists against Tet(X3)/Tet(X4)-positive bacteria, we therefore tested the Tet(X) enzyme inhibitory activity of over 200 native compounds including flavones, pentacyclic triterpenoids, quinonoids and terpenoids, etc. Then checkerboard microdilution analysis were further performed for confirming the synergistic effects with tetracycline against Tet(X3)/Tet(X4)-positive E. coli. Ultimately, plumbagin was identified as an effective broad-spectrum inhibitor of Tet(X) with the antibacterial synergistic activity plus tetracyclines.
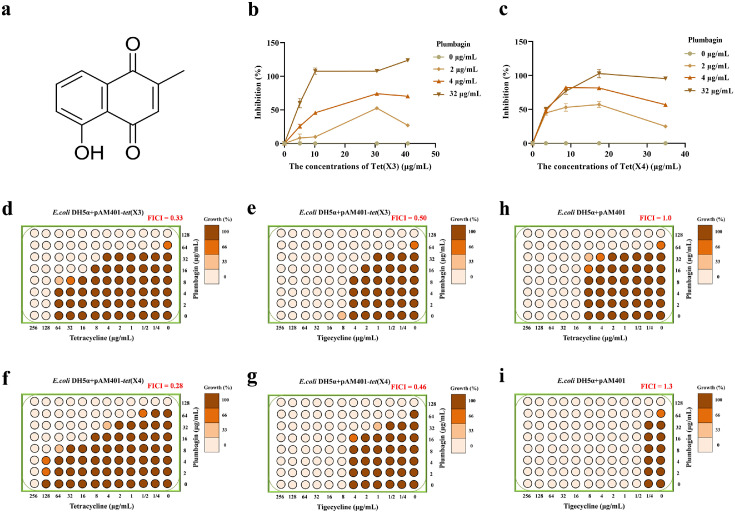
Similar to Tet(X), Tet(X3)/Tet(X4) are tetracycline-inactivating monooxygenase with the activity of catalysing the oxygenation of tetracyclines to induce a change in the optical absorption value at 400 nm.6,13 Thus, this change in absorption values were determined to identify inhibition of Tet(X3)/Tet(X4) for inhibitor screening. As shown in Figure 1b,c, the catalytic activities of Tet(X3)/Tet(X4) were observably inhibited by plumbagin (Figure 1a) in a concentration dependent manner, respectively. In addition, using the checkerboard method, a synergistic effect between plumbagin and tigecycline or tetracycline occurred for E. coli DH5α+pAM401-tet(X3) (FICItetracycline = 0.33, FICItigecycline = 0.50) (Figure 1d,e) and E. coli DH5α+pAM401-tet(X4) (FICItetracycline = 0.28, FICItigecycline = 0.46) (Figure 1f,g), whereas no synergies were observed for E. coli DH5α+pAM401 without Tet(X3)/Tet(X4) (FICI ≥ 1.0) (Figure 1h,i). Consistent with these results, plumbagin in combination with other tetracyclines, including methacycline, doxycycline, eravacycline and omadacycline, also exhibited a significant synergistic effect (Supplementary Fig. 1a,b). Thus, plumbagin treatment effectively restores the sensitivity of tetracyclines against Tet(X3)/Tet(X4)-producing bacteria by targeting Tet(X3)/Tet(X4).
Figure 1.
Plumbagin restored the antibacterial activity of tetracyclines. (a) Chemical structure of plumbagin. The catalytic activities of Tet(X3) (b) and Tet(X4) (c) with the indicated concentrations of plumbagin were detected using enzyme inhibition assays by measuring the absorbance at 400 nm. Checkerboard broth microdilution assays were performed to evaluate the synergistic activity between tigecycline/tetracycline and plumbagin against E. coli DH5α+pAM401-tet(X3) (d and e), E. coli DH5α+pAM401-tet(X4) (f and g), or negative-E. coli DH5α+pAM401 (h and i). Data are representative of three independent experiments.
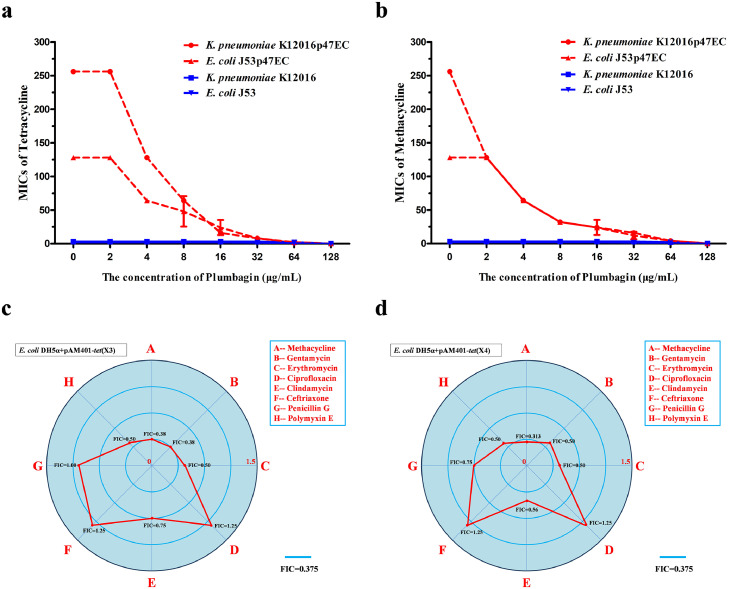
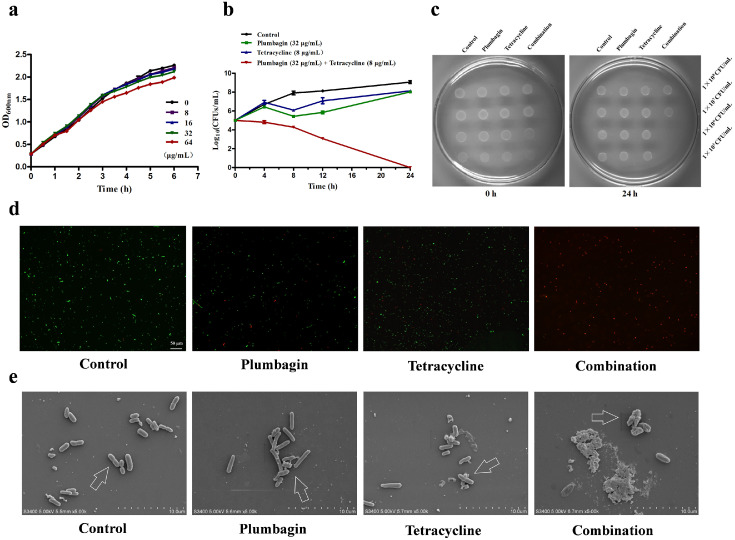
The clinical isolates K. pneumoniae K12016 and E. coli J53 received the plasmid p47EC encoding Tet(X4) and became resistant to tetracyclines as described previously4 (Figure 2a,b). As expected, plumbagin significantly enhanced the antibacterial effect of tetracycline and methacycline against K. pneumoniae K12016p47EC and E. coli J53p47EC in a dose-dependent manner (Figure 2a,b). In addition, plumbagin showed synergies with multiple tetracycline antibiotics, including tetracycline, methacycline and tigecycline, against the clinical isolates E. coli 47EC and A. baumannii 34AB carrying Tet(X3)/Tet(X4) (Supplementary Fig. 2). Notably, such synergies were also observed in tet(X4)-positive E. coli and tet(X6)-positive Proteus cibarius isolates of chicken origin, and tet(X5)-positive A. baumannii from human (Supplementary Table 2). In addition, plumbagin, in combination with the nontetracycline antibiotics gentamycin, erythromycin, clindamycin or polymyxin E, had synergistic or additive effects on E. coli DH5α+pAM401-tet(X3) (Figure 2c) and E. coli DH5α+pAM401-tet(X4) (Figure 2d) but not ciprofloxacin, ceftriaxone or penicillin (Figure 2c,d), suggesting that plumbagin is an anticipated and broad-spectrum synergist. E. coli DH5α+pAM401-tet(X4) was further used to determine whether plumbagin combined with tetracycline had an efficient bactericidal effect in vitro. As shown in Figure 3a, plumbagin, at concentrations no greater than 64 µg/mL, had no visible influence on the growth of E. coli DH5α+pAM401-tet(X4). However, plumbagin in combination with tetracycline could completely kill all tested bacteria in 24 h compared with both monotherapies without bactericidal effects (Figure 3b,c). Moreover, plumbagin at the concentrations required for the synergies exhibited no significant antibacterial effect against the other three tet(X3)/tet(X4)-positive stains (Supplementary Fig. 3a–c). Remarkably, the combination of plumbagin and different tetracycline antibiotics (such as tetracycline, methacycline and tigecycline) exhibited efficient bactericidal effects against tet(X4)-positive E. coli and tet(X3)-positive A. baumannii, as shown by the potent elimination of bacteria by 24 h post inoculation for the tested strains (Supplementary Fig. 3d–h).
Figure 2.
The antibacterial activity of tetracyclines and other antibiotics combined with plumbagin against tet(X3)/tet(X4)-positive strains. Microdilution checkerboard analysis showed that plumbagin restored the susceptibility of tet(X4)-carrying E. coli J53p47EC and K. pneumoniae K12016p47EC to tetracycline (a) and methacycline (b), in comparison to the tet(X4)-negative strains E. coli J53 and K. pneumoniae K12016. (c and d) FIC indices of the combination of plumbagin (32 μg/mL) and antibiotics of different types against tet(X3)/tet(X4)-positive strains. Synergy was defined as an FIC index of ≤ 0.5.
Figure 3.
Plumbagin in combination with tetracyclines effectively killed bacteria by damaging their surface structures without affecting bacterial growth. (a) Growth curves for E. coli DH5α+pAM401-tet(X4) cultured with various concentrations of plumbagin (0–64 µg/mL). (b) Time-killing assays of resistant E. coli DH5α+pAM401-tet(X4) by a combination of tetracycline and plumbagin. (c) LB agar plate images of the spot assays to analyse the effect of plumbagin in combination with tetracycline on the inhibition of E. coli DH5α+pAM401-tet(X4). The spotted cultures were continuously diluted and cultured for 24 h. Fluorescence labeling analysis (scale bar = 50 μm) (d) and SEM analysis (scale bar = 10 μm) (e) of E. coli DH5α+pAM401-tet(X4) with the indicated treatment were shown. Control, bacteria without any treatment; Plumbagin, bacteria treated with 32 μg/mL plumbagin; Tetracycline, bacteria treated with 32 μg/mL tetracycline; Combination, bacteria treated with 32 μg/mL plumbagin and 32 μg/mL tetracycline. The experiments were repeated three times independently, and only representative data are displayed.
In agreement with these results, most bacteria lived with a normal cell morphology in the samples with monotherapy or without treatment (Figure 3d,e); however, the combination therapy led to visible shrinkage of the cell wall and increased cell permeability and death of E. coli DH5α+pAM401-tet(X4), as evidenced by noticeably more red-dyed bacteria (dead bacteria) by the LIVE/DEAD BacLight Bacterial Viability Kit (Figure 3d) and visibly destroyed bacteria under SEM (Figure 3e). Likewise, the notable increase of dead bacteria and the cell membrane breakage, obvious vesicular and irregular processes on the surface of E. coli DH5α+pAM401-tet(X3) were also observed by the combination treatment (Supplementary Fig. 4a,b). Taken together, our results established that plumbagin, at concentrations that did not affect the bacterial viability, could effectively restore the bactericidal effect of tetracyclines by inhibiting the activity of Tet(X3)/Tet(X4) in vitro.
Furthermore, we evaluated the safety of plumbagin on A549 cells, HEK-293 cells, peritoneal macrophages and red blood cells (RBCs). Plumbagin, a naturally-derived phytochemical, showed no cytotoxicity to A549 cells, HEK-293 cells, or peritoneal macrophages at low concentrations of no more than 16 μg/mL, however, which presented a slight cytotoxicity at high concentrations at 5 h (IC50>32 μg/mL) (Supplementary Fig. 5a–c). And haemolysis activity examinations indicated that plumbagin had no poisonous effect on RBCs at the concentrations required for the synergism with tetracyclines (Supplementary Fig. 5d). These findings illustrated that plumbagin is a relatively safe compound to restore tetracyclines activity.
Identification of the mechanism of the interaction between plumbagin and Tet(X3)/Tet(X4)
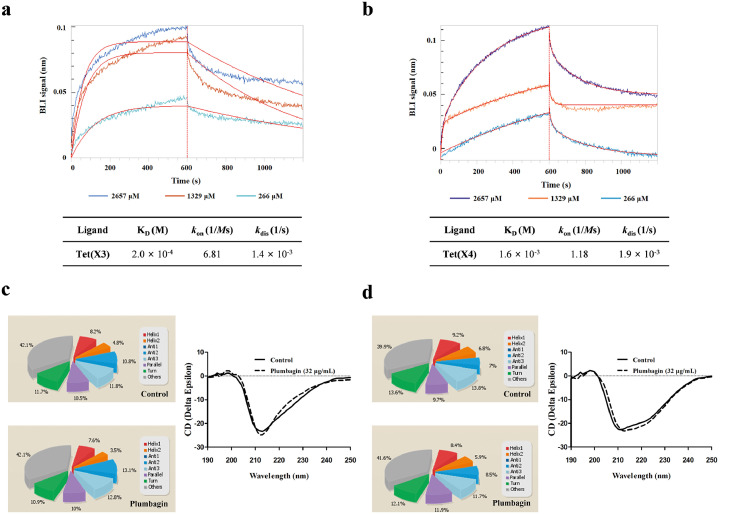
Biolayer interferometry (BLI), a convictive technique for measuring interactions between plumbagin and Tet(X3)/Tet(X4) in real time, was performed and demonstrated that a detectable affinity of Tet(X3)/Tet(X4) (Figure 4a,b) by plumbagin was observed, with an affinity constant KD of 2.0 × 10−4 M or 1.6 × 10−3 M, respectively. To examine the effect of plumbagin on the catalytic activity of Tet(X4) against the tetracyclines, tetracycline alone or the combination was severally captured on BLI sensors and the binding forces in different treatment groups were compared. As shown in Supplementary Fig. 6a, a clearly negative signal was observed on the interactions of tetracycline with Tet(X4) which indicated a detectable degradation or denaturation of ligand. Notably, incubation with different concentrations of plumbagin reversed the observed change in this interaction (Supplementary Fig. 6b).
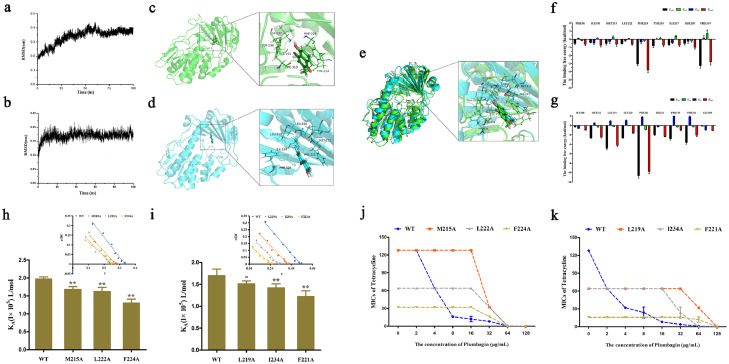
Figure 4.
Identified of plumbagin directly interact with Tet(X3)/Tet(X4) protein. (a,b) BLI sensors monitored the real-time association and dissociation kinetics of Tet(X3)/Tet(X4) (captured on the surface of anti-GST antibody coated BLI sensors) with different concentrations of plumbagin and the binding parameters of Tet(X3)/Tet(X4) with plumbagin were shown in the table, respectively. And circular dichroism (CD) spectra of Tet(X3) (c) and Tet(X4) (d) were obtained to further analyze the effect of plumbagin on the secondary structure of the protein. The wavelength of CD spectroscopy was set as 190-250 nm.
Moreover, CD spectroscopy analysis revealed that plumbagin treatment induced visible alterations in the secondary structures of both Tet(X3) and Tet(X4), as shown by reduced α-helix1, α-helix2 and turn conformations and increased anti-2 conformations (Figure 4c,d). These findings suggested plumbagin bound to Tet(X3)/Tet(X4), accompanied by the alternation of second structure.
To further examine these findings, molecular docking and molecular modeling were used to explore the binding model between plumbagin and Tet(X3)/Tet(X4). After 100 ns of simulation under equilibrium (Figure 5a,b), the binding 3D modes of Tet(X3)/Tet(X4) with plumbagin were obtained (Figure 5c,d). As shown in Figure 5c,d, plumbagin could bind to the catalytic pocket of Tet(X3)/Tet(X4) via hydrophobic interactions. Consistent with the similar three-dimensional structures, especially for the catalytic pocket shared by Tet(X), an extremely similar binding mode and sites were observed in the merger of 3D structures of Tet(X3)/plumbagin and Tet(X4)/plumbagin (Figure 5e), labeled in green and blue color respectively, suggesting that plumbagin-mediated inhibition of Tet(X3)/Tet(X4) by the similar mechanism of action. To verify the above theoretical results, the total binding free energy for the Tet(X3)/Tet(X4)-plumbagin complex and their detailed energy contributions were calculated by the MM-PBSA approach. The energy decomposition analysis indicated that the side chains of Tet(X3) (PHE56, ILE191, MET215, LEU222, PHE224, TYR234, ILE237, SER238 and PHE319) (Figure 5f) and Tet(X4) (ILE188, MET212, LEU219, LEU220, PHE221, ILE234, PRO315, PHE316 and GLU369) (Figure 5g) formed strong interactions with plumbagin (ΔEtotal of ≤ -0.5 kcal/mol), with van der Waals forces contributing the highest proportion to such an interaction, especially for resides PHE224 and PHE319 of Tet(X3) and LEU219 and PHE221 of Tet(X4) (Figure 5f,g).
Figure 5.
Determination of the binding model of plumbagin with Tet(X3)/Tet(X4). RMSD values of the Tet(X3)-plumbagin (a) and Tet(X4)-plumbagin (b) complex during the simulation. Three-dimensional (3D) structure determination of Tet(X3) (green color) (c) or Tet(X4) (blue color) (d) with plumbagin by molecular modeling analysis. (e) Overlapping 3D structures of Tet(X3)/plumbagin and Tet(X4)/plumbagin with similar binding sites. Decomposition of the binding energy on a per-residue basis in the binding sites of Tet(X3) (f) or Tet(X4) (g) with plumbagin. The binding constants (KA) of plumbagin with Tet(X3) and its mutants (h), Tet(X4) and its mutants (i). Further, the synergistic activity of plumbagin with tetracyclines for the bacteria harbouring Tet(X3) mutants (Tet(X3)-M215A, Tet(X3)-L222A and Tet(X3)-F224A) (j) and Tet(X4) mutants (Tet(X4)-L219A, Tet(X4)-I234A and Tet(X4)-F221A) (k) were significantly decreased compared with the bacteria carrying WT-Tet(X).
Additionally, to further illustrate the detailed action of plumbagin on Tet(X3)/Tet(X4), mutation experiments and fluorescence quenching were further performed. As shown in Figure 5h,i, the binding constants (KA) of all of the mutants were significantly lower than that of WT-Tet(X3)/Tet(X4) based on fluorescence spectroscopy quenching analysis. Furthermore, no synergistic effects were observed for the combination of tetracycline with plumbagin at concentrations of 32 μg/mL or lower against bacteria harbouring the mutant Tet(X3) (Figure 5j) and Tet(X4) (Figure 5k). Additionally, the bacteria carrying the mutants were more sensitive to tetracycline than those with the WT-Tet(X) (Figure 5j,k). These observations indicated that the residues MET215, LEU222 and PHE224 in Tet(X3) and LEU219, ILE234 and PHE221 in Tet(X4) are required for the modification of tetracyclines and the engagement of plumbagin with Tet(X3)/Tet(X4).
Taken together, our results established that direct engagement of plumbagin with the Tet(X3)/Tet(X4) catalytic pocket altered the conformation and then inhibited the catalytic activity of Tet(X3)/Tet(X4).
Plumbagin inhibited the expression of monooxygenases
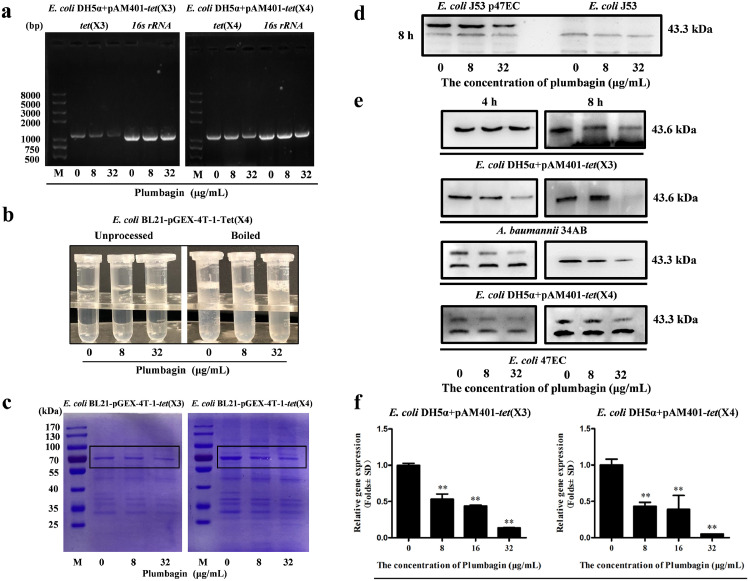
Tet(X3)/Tet(X4), also known as monooxygenases, evolved tetracycline resistance enzymes in most Gram-negative bacteria, as described in our previous report.4 As shown in Figure 6a, the DNA synthesis levels of tet(X3) and tet(X4) were visibly decreased following plumbagin treatment. And the production of Tet(X3)/Tet(X4) in E. coli BL21-pGEX-4T-1-Tet(X3)/Tet(X4) induced by IPTG was dose-dependently inhibited after the addition of plumbagin (Figure 6b,c). Consistent with these results, western blot analysis further revealed that plumbagin-mediated inhibition of Tet(X4) production was observed in tet(X4)-positive E. coli J53p47EC but not E. coli J53 without tet(X4) (Figure 6d). Meanwhile, plumbagin treatment resulted in a visible inhibition of Tet(X3)/Tet(X4) production in the gene engineering strains (E. coli DH5α+pAM401-tet(X3) and E. coli DH5α+pAM401-tet(X4)) and clinically isolated strains (tet(X3)-producing strain A. baumannii 34AB and tet(X4)-producing strain E. coli 47EC) (Figure 6e). Collectively, plumbagin addition showed different behaviors in different classes of bacteria, which may attribute to the different synergistic effects. Such a decrease was further detected in the RT-PCR analysis for the gene transcription of tet(X3) and tet(X4), the tetracycline-inactivating monooxygenase (Figure 6f). Moreover, to investigate whether plumbagin affects the transcription of other oxidoreductases, the gene transcription of WrbA (an FMN-dependent NAD(P)H: quinone oxidoreductase that has been implicated in oxidative defense)29 and ydhR (a monooxygenase in bacteria)30 were both observably inhibited by plumbagin treatment Supplementary Fig. 7a,b). Thus, our results indicated that plumbagin represents an effective Tet(X3)/Tet(X4) inhibitor that simultaneously neutralizes Tet(X3)/Tet(X4) activity and inhibits Tet(X3)/Tet(X4) expression.
Figure 6.
Plumbagin inhibited the transcription and expression of resistance genes tet(X3)/tet(X4). (a) Plumbagin suppressed the DNA synthesis of tet(X3)/tet(X4) in E. coli. The tet(X3)/tet(X4) gene in E. coli treated with the indicated concentrations of plumbagin was amplified by PCR analysis, and then the desired products were determined using DNA gel electrophoresis. And 16s rRNA was used as an internal control. (b) The expression of Tet(X4) in E. coli BL21-pGEX-4T-1-Tet(X4) was decreased by plumbagin with boiling. (c) Plumbagin addition decreased the expression of Tet(X3)/Tet(X4) in E. coli BL21-pGEX-4T-1-Tet(X3) or E. coli BL21-pGEX-4T-1-Tet(X4) induced by IPTG. Samples were loaded on SDS-PAGE gels and analysed by staining with Coomassie brilliant blue. (d) Tet(X4) production in E. coli J53p47EC with tet(X4) and E. coli J53 without tet(X4) following coculture with the indicated concentrations of plumbagin (0, 8 and 32 μg/mL) was determined by western blotting assays. (e) Tet(X3)/Tet(X4) production in tet(X3)/tet(X4)-positive engineering strains and clinical strains cocultured with the indicated treatment were determined by western blotting assays. (f) Dose-dependent inhibition of the transcription of the genes tet(X3)/tet(X4)-encoding monooxygenases by plumbagin was determined by RT-PCR analysis. *p < 0.05, **p < 0.01 (Student's t test). All data are representative of three independent experiments and are presented as the mean ± SD.
Plumbagin interfered with redox processes and metabolism in bacteria
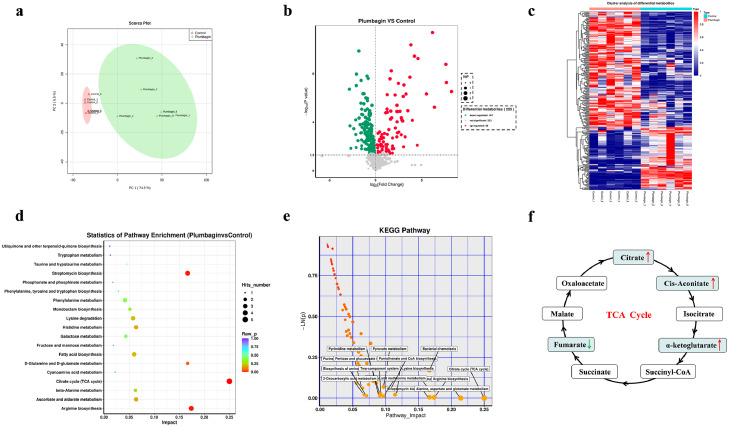
The inhibition of monooxygenases, such as Tet(X3) and Tet(X4), by plumbagin prompted us to further examine whether plumbagin treatment could interfere with redox processes and metabolism in bacteria carrying Tet(X3)/Tet(X4). As expected, the results of the metabonomics analysis, including principal component analysis, volcano plot of the univariate analysis, hierarchical cluster analysis of differential metabolites and metabolic pathway analysis, under negative ion mode (NEG) in E. coli DH5α+pAM401-tet(X4), revealed that 167 metabolites and 88 metabolites were downregulated and upregulated following plumbagin treatment, respectively (Figure 7a–c). KEGG enrichment analysis further showed that the differentially expressed genes (DEGs) were involved in streptomycin biosynthesis, arginine biosynthesis and bacterial metabolism-related pathways, especially for the tricarboxylic acid (TCA) cycle (Figure 7d,e). Further, intracellular metabolites analysis indicated that the abundance of citrate, cis-aconitate and α-ketoglutarate in the TCA cycle were increased significantly upon plumbagin treatment (Figure 7f).
Figure 7.
Metabolomic analysis (NEG mode) of E. coli DH5α+pAM401-tet(X4) treated with plumbagin. Principal component analysis (PCA) (a) and volcano plots (b) were used to identify differential metabolites between the plumbagin-treatment group and the control group. Significantly upregulated and downregulated metabolites are shown in red and green, respectively. Non-significantly different metabolites are shown in grey. (c) Cluster analysis in NEG mode was carried out based on the heatmap, and the x- and y-axes represent the different experimental groups and the different metabolites, respectively. (d and e) KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis of metabolic pathways. The most significantly enriched pathways are shown. (f) Analysis of the abundance of metabolites in TCA cycle. The decreasing and increasing abundances of metabolites were highlighted in green and red arrows, respectively. Data are presented as the means of six biological replicates.
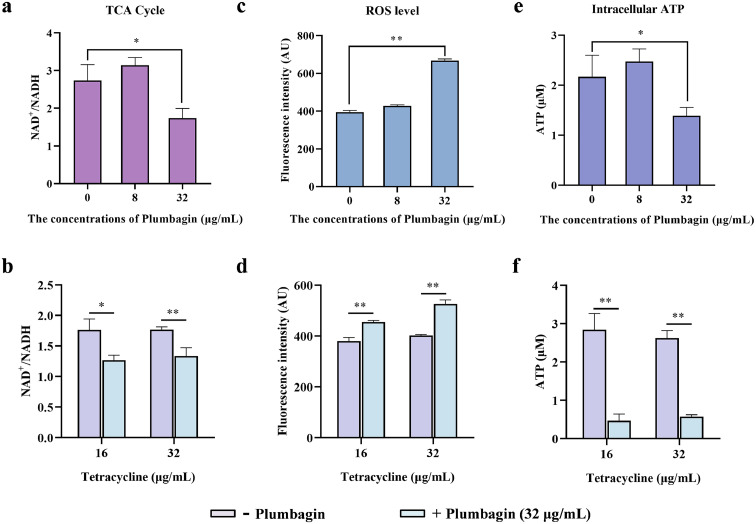
We therefore tested whether plumbagin alone, or in combination with tetracycline could accelerate the TCA cycle, and the NAD+/NADH ratio regulated by the TCA cycle in E. coli DH5α+pAM401-tet(X4) was determined. Plumbagin (32 μg/mL) treatment significantly decreased the NAD+/NADH ratio, suggesting an enhancement of the activities of TCA cycle rate (Figure 8a,b). The acceleration of TCA cycle promoted the generation and accumulation of reactive oxygen species (ROS), which can aggravate membrane damage and mediate oxidative damage involved in the bacteria. In this regard, the level of ROS in the energy metabolism pathway could be notably upregulated by the addition of plumbagin (Figure 8c,d). For determination of the ROS mediated by plumbagin redox-cycling, pyruvate as a potent H2O2 scavenger was exogenously added. And pyruvate addition significantly reduced the ROS generation (Supplementary Fig. 8a), and inhibited the upregulation of ROS by plumbagin (Supplementary Fig. 8b). Furthermore, with co-incubation with pyruvate, the synergistic effects between plumbagin and tigecycline or tetracycline against E. coli DH5α+pAM401-tet(X4) were greatly weakened (Supplementary Fig. 8c,d), which demonstrated the pivotal role of ROS generation by plumbagin or combination treatment in restoring the susceptibility of Tet(X)-positive bacteria to tetracyclines. Moreover, the monotherapy with plumbagin or combination therapy with plumbagin and tetracycline resulted in a significantly increased levels of H2O2 (Supplementary Fig. 9a) and an observably decreased of SOD activity (Supplementary Fig. 9b) in bacteria, which indicated that plumbagin, or combined with tetracycline, led to oxidative stress damage to the targeted bacteria. Consistent with a redox imbalance, the notable increase in ROS production often accompanied by a corresponding a reduction in intracellular ATP levels, as shown in Figure 8e,f, plumbagin treatment prominently inhibited the intracellular ATP generation. Meanwhile, we applied a fluorescent probe propidium iodide (PI) to further evaluate the effect of plumbagin on bacterial inner membrane (IM) and membrane permeability. Consistent with these findings, a significant increase of fluorescence was observed when co-incubated with plumbagin (Supplementary Fig. 9c,d).
Figure 8.
Synergistic mechanisms of plumbagin in combination with tetracycline. An accelerated TCA cycle was observed after the treatment with plumbagin (a) or the combination of plumbagin and tetracycline (b). (c and d) Plumbagin supplementation dramatically promoted the generation of ROS, whether it alone or combined with tetracycline. And the level of intracellular ATP in E. coli DH5α+pAM401-tet(X4) was further measured after coculture with plumbagin alone (e) or in combination with tetracycline (f). All of the data are representative of three independent experiments and presented as the mean ± SD. *p < 0.05, **p < 0.01 (Student's t test).
Collectively, these results suggested that inhibition of Tet(X3)/Tet(X4) by plumbagin interferes with monooxygenase activity and alters the metabolism of bacteria, with an attendant increase in the IM permeability, which contributes to the synergistic effect of plumbagin with the above antibiotics and provides a new perspective to combat resistant bacterial infection.
Plumbagin in combination with tetracyclines synergistically protected mice from systemic infection
For the toxicity test, compared with the control group, gross pathological inspection of various organs and body weight in mice receiving the indicated dose levels of plumbagin showed no significant changes (Supplementary Fig. 10a,b). And no death was observed up to 7 days for the mice received five times the effective dose plumbagin (25 mg/kg body weight) (Supplementary Table 5), further suggesting that plumbagin, at 5 mg/kg body weight, displayed no toxicity to mice.
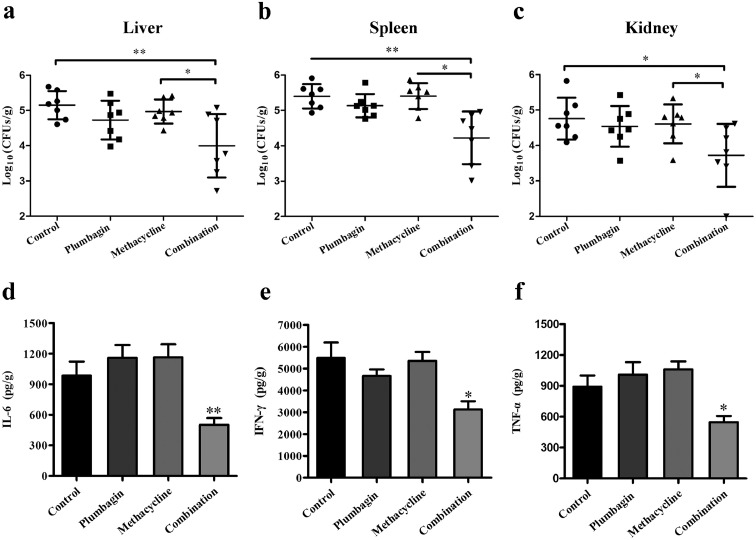
The synergy of plumbagin combined with methacycline was further illustrated by experimental therapeutics in the context of a mouse model of systemic infection in vivo. As shown in Figure 9a–c, the combination therapy significantly reduced the bacterial load in the livers, spleens and kidneys compared with the non-treatment mice and infected mice treated with methacycline or plumbagin. Notably, in comparison with the infected mice without treatment, monotherapy with plumbagin also decreased the bacterial load in different organs, even lower than those in mice which received monotherapy with methacycline, suggesting that plumbagin possesses the potent anti-infection activity. Additionally, infection with E. coli successfully induced a remarkable inflammatory response in mice. Neither methacycline nor plumbagin treatment notably reduced inflammation. However, the combination treatment of plumbagin and methacycline significantly decreased the levels of the typical inflammatory-related factors IL-6 (Figure 9d), IFN-γ (Figure 9e) and TNF-α (Figure 9f) in the livers. In line with these in vivo results, the infected mice that received a combination of plumbagin and methacycline showed alleviated histopathological injury to both the liver and spleen (Supplementary Fig. 10c). Taken together, our results indicated that plumbagin combined with methacycline may represent a novel therapeutic strategy against infection by drug-resistant bacteria carrying Tet(X3)/Tet(X4).
Figure 9.
Combined therapy of plumbagin with tetracyclines had a significant synergistic effect against tet(X4)-positive E. coli in a mouse systemic infection model. Mice were infected with E. coli DH5α+pAM401-tet(X4) and then treated with plumbagin, methacycline or plumbagin in combination with methacycline. The infected mice treated with control solvent served as controls. The bacterial burden in the livers (a), spleens (b), and kidneys (c) was calculated by plating (n=7 per group). The production of the inflammatory mediators interleukin IL-6 (d), IFN-γ (e) and tumour necrosis factor (TNF)-α (f) in the liver was detected using enzyme-linked immunosorbent assays (ELISA). All data are presented as the mean ± SD. *p < 0.05, **p < 0.01 (Student's t test).
Discussion
The occurrence and spread of plasmid-carried tigecycline resistance genes tet(X3) and tet(X4) severely compromise the efficiency of tetracyclines as the last resort for bacterial infection.4 According to our previous work,31,32 the identification of an effective inhibitor of Tet(X3)/Tet(X4) or other Tet(X) variants from natural compounds is a promising strategy to tackle this urgent challenge. Herein, we successfully identified plumbagin as an effective agent that directly inhibited the catalytic activity of Tet(X3)/Tet(X4) against tetracycline via an enzyme inhibition assay. As a consequence, the antibacterial activity of tetracycline was significantly restored from resistance (MIC > 64 μg/mL) to sensitivity (MIC < 8 μg/mL) by plumbagin treatment with a synergistic effect. Notably, plumbagin resulted in a 32-fold reduction in the MIC of tetracycline (MIC ≥ 128 μg/mL) for all tested bacteria in this study compared with tigecycline (MIC ≥ 8 μg/mL) with a 4-fold reduction (FIC ≤ 0.5), suggesting that plumbagin has a better synergistic effect against the higher tetracycline-resistant bacteria. Additionally, the structures of Tet(X) variants such as Tet(X3), Tet(X4), Tet(X5) and Tet(X6) share high homology; therefore, the synergistic effect of plumbagin and tetracyclines was also observed in the vast majority of Tet(X)-positive clinical isolates (Supplementary Table 2), which indicated that plumbagin is a broad-spectrum inhibitor of Tet(X). Interestingly, plumbagin treatment also restored the antibacterial activity of other antibiotics against Tet(X3)/Tet(X4)-positive bacteria, such as gentamycin, erythromycin, clindamycin and polymyxin. These results further suggested that the inhibition of oxidoreductases, such as monooxygenases Tet(X3)/Tet(X4), WrbA and ydhR by plumbagin potentiates its effect as an effective sensitizing agent for various antibiotics.
The mechanism research results revealed that a similar engagement of plumbagin with Tet(X3) and Tet(X4) by directly binding to the catalytic pocket, led to a significant alteration of the secondary structure and subsequent an inhibition of Tet(X3)/Tet(X4) activity. Meanwhile, plumbagin effectively inhibited the expression of flavin-dependent monooxygenases, including Tet(X3) and Tet(X4), at the transcriptional level. Due to the ubiquitous effect of monooxygenases on cell metabolism, plumbagin treatment significantly intervened with the metabolites of targeted bacteria, suggesting that plumbagin, as an agent, possesses a potential antibacterial activity. Although no ideal therapeutic performance was observed for plumbagin monotherapy, this treatment also exhibited an anti-infection effect against Tet(X4)-positive bacteria in a mouse model of systemic infection. Additionally, recent reports found that plumbagin treatment showed an antibacterial effect against methicillin-resistant Staphylococcus aureus (MRSA), with an MIC range of 4–8 μg/mL.33 Therefore, plumbagin could be further developed as an agent to treat drug-resistant pathogen infections, especially for MRSA, by structural modification and optimization.
The potential toxicity of plumbagin in vivo should be of great concern for better therapeutic efficacy and future drugs development.28 In this work, in consideration of virulence and specificity of the pathogens, Escherichia coli DH5α+pAM401-tet(X4) was used to establish the systemic infection model. Encouragingly, reduction of bacterial load with the treatment of plumbagin alone (5 mg/kg) or in combination with methacycline was observed, in the absence of significant toxicity in the subacute toxicity testing. Consistent with previous reports, plumbagin at the maximal tolerated oral dose of 100 mg/kg body weight exhibited no acute toxicity. And for the subacute toxicity testing, no death of mice were observed following daily oral doses of 25 mg/kg body weight plumbagin for 14 days.34 Although this dose successfully avoided toxicity in mice, a higher dose may be required for acute systemic infection. In contrast, 5 mg/kg or higher concentrations of plumbagin treatment did not achieve an ideal performance without significantly decreasing the survival rate of the infected mice due to the toxicity of plumbagin. Overall, further research is needed to explore the use of plumbagin for the treatment of localized infections such as mastitis in cows or develop a novel dosage form to improve its efficiency and depress the toxicity for further clinical application.35
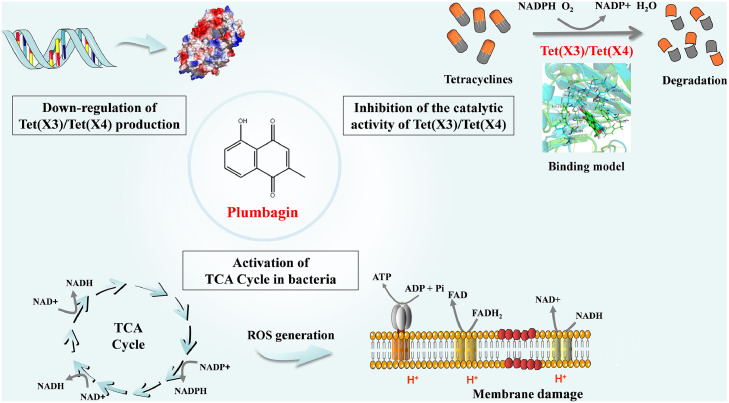
In summary, our findings first indicated that plumbagin restores the susceptibility of Tet(X)-positive bacteria to tetracyclines by the disturbance of membrane integrity and direct engagement with the catalytic pocket of Tet(X3)/Tet(X4), thereby inhibiting the activity and the production of Tet(X3)/Tet(X4). Meanwhile, plumbagin addition perturbed the redox processes and metabolism, and promoted the oxidative damage (Figure 10). Our results provide a promising therapeutic strategy with a novel natural compound plumbagin, with unique pharmacological effects, for the treatment of tigecycline-resistant bacteria infection by targeting Tet(X3)/Tet(X4).
Figure 10.
Scheme summarizing the synergistic mechanisms of plumbagin and tetracyclines. Plumbagin restored the susceptibility of Tet(X3)/Tet(X4)-positive bacteria to tetracyclines via different mechanisms.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgments
Contributors
YW, JFW, LX and YLZ designed this project. LX, YLZ, SN, ZYL, YNZ, DJL, YNY and XDN performed experiments; HHF and XMD analysed the data and prepared figures; YW, JFW, LX and YLZ drafted this manuscript. JFW, HHF and XMD accessed and verified the underlying data. All authors have read, reviewed and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant 81861138046, 32172912 and 31972742).
Data sharing statement
The data are available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103943.
Contributor Information
Yang Wang, Email: wangyang@cau.edu.cn.
Jianfeng Wang, Email: wjf927@jlu.edu.cn.
Appendix. Supplementary materials
References
- 1.Potter R.F., D'Souza A.W., Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Update. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yelin I., Kishony R. Antibiotic resistance. Cell. 2018;172(5):1136. doi: 10.1016/j.cell.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 3.Chopra I., Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol R. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He T., Wang R., Liu D.J., et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4(9):1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Jia Y.Q., Yang K.N., et al. Melatonin overcomes MCR-mediated colistin resistance in gram-negative pathogens. Theranostics. 2020;10(23):10697–10711. doi: 10.7150/thno.45951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W.R., Moore I.F., Koteva K.P., Bareich D.C., Hughes D.W., Wright GD. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem. 2004;279(50):52346–52352. doi: 10.1074/jbc.M409573200. [DOI] [PubMed] [Google Scholar]
- 7.Balcaen L.I.L., De Samber B., De Wolf K., Cuyckens F., Vanhaecke F. Hyphenation of reverse-phase HPLC and ICP-MS for metabolite profiling-application to a novel antituberculosis compound as a case study. Anal Bioanal Chem. 2007;389(3):777–786. doi: 10.1007/s00216-007-1303-2. [DOI] [PubMed] [Google Scholar]
- 8.Haagsma A.C., Podasca I., Koul A., et al. Probing the interaction of the diarylquinoline TMC207 with Its target mycobacterial ATP synthase. PLoS One. 2011;6(8):e23575. doi: 10.1371/journal.pone.0023575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lien J.C., Huang L.J., Wang J.P., Teng C.M., Lee K.H., Kuo SC. Synthesis and antiplatelet, antiinflammatory and antiallergic activities of 2,3-disubstituted 1,4-naphthoquinones. Chem Pharm Bull. 1996;44(6):1181–1187. doi: 10.1248/cpb.44.1181. [DOI] [PubMed] [Google Scholar]
- 10.Padhye S., Dandawate P., Yusufi M., Ahmad A., Sarkar F.H. Perspectives on medicinal properties of plumbagin and its analogs. Med Res Rev. 2012;32(6):1131–1158. doi: 10.1002/med.20235. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki K., Abe H., Yoshizaki F. In vitro antifungal activity of naphthoquinone derivatives. Biol Pharm Bull. 2002;25(5):669–670. doi: 10.1248/bpb.25.669. [DOI] [PubMed] [Google Scholar]
- 12.Kim S.J., Shin H.J., Lee B.J., et al. The antiinflammatory mechanism of Igongsan in mouse peritoneal macrophages via suppression of NF-kappa B/Caspase-1 activation. Phytother Res. 2014;28(5):736–744. doi: 10.1002/ptr.5058. [DOI] [PubMed] [Google Scholar]
- 13.Moore I.F., Hughes D.W., Wright GD. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry-Us. 2005;44(35):11829–11835. doi: 10.1021/bi0506066. [DOI] [PubMed] [Google Scholar]
- 14.CLSI . 30th ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2020. Performance Standards for Antimicrobial Susceptibility Testing. CLSI supplement M100. [Google Scholar]
- 15.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemoth. 2003;52(1):1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 16.Trott O., Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 18.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Dis. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X.Y., Yang Y.N., Gao Y.W., Niu XD. Discovery of the novel inhibitor against New Delhi metallo-beta-lactamase based on virtual screening and molecular modelling. Int J Mol Sci. 2020;21(10):3567. doi: 10.3390/ijms21103567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah N.B., Duncan TM. Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. Jove J Vis Exp. 2014;(84):e51383. doi: 10.3791/51383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen RL. Strategies using bio-layer interferometry biosensor technology for vaccine research and development. Biosensors-Basel. 2017;7(4):49. doi: 10.3390/bios7040049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y.L., Guo Y., Wen Z.M., et al. Isoalantolactone enhances the antimicrobial activity of penicillin G against staphylococcus aureus by inactivating beta-lactamase during protein translation. Pathogens. 2020;9(3):161. doi: 10.3390/pathogens9030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Micsonai A., Wien F., Kernya L., et al. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci USA. 2015;112(24):E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rio D.C., Ares M., Hannon G.J., Nilsen TW. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harb Protoc. 2010;2010(6) doi: 10.1101/pdb.prot5439. pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 25.Agwuh K.N., MacGowan A. Pharmacokinetics and pharmacodynamics of the tetracyclines including glycylcyclines. J Antimicrob Chemoth. 2006;58(2):256–265. doi: 10.1093/jac/dkl224. [DOI] [PubMed] [Google Scholar]
- 26.Sumsakul W., Plengsuriyakarn T., Na-Bangchang K. Pharmacokinetics, toxicity, and cytochrome P450 modulatory activity of plumbagin. BMC Pharmacol Toxicol. 2016;17(1):50. doi: 10.1186/s40360-016-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh Y.J., Lin L.C., Tsai TH. Measurement and pharmacokinetic study of plumbagin in a conscious freely moving rat using liquid chromatography/tandem mass spectrometry. J Chromatogr B. 2006;844(1):1–5. doi: 10.1016/j.jchromb.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Tripathi S.K., Panda M., Biswal BK. Emerging role of plumbagin: cytotoxic potential and pharmaceutical relevance towards cancer therapy. Food Chem Toxicol. 2019;125:566–582. doi: 10.1016/j.fct.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 29.Kishko I., Harish B., Zayats V., et al. Biphasic kinetic behavior of E. coli WrbA, an FMN-dependent NAD(P)H: quinone oxidoreductase. PLoS One. 2012;7(8):e43902. doi: 10.1371/journal.pone.0043902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revington M., Semesi A., Yee A., Shaw GS. Solution structure of the Escherichia coli protein ydhR: a putative mono-oxygenase. Protein Sci. 2005;14(12):3115–3120. doi: 10.1110/ps.051809305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S., Zhang J., Zhou Y.L., et al. Pterostilbene restores carbapenem susceptibility in New Delhi metallo-beta-lactamase-producing isolates by inhibiting the activity of New Delhi metallo-beta-lactamases. Br J Pharmacol. 2019;176(23):4548–4557. doi: 10.1111/bph.14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y.L., Wang J.F., Guo Y., et al. Discovery of a potential MCR-1 inhibitor that reverses polymyxin activity against clinical mcr-1-positive Enterobacteriaceae. J Infect. 2019;78(5):364–372. doi: 10.1016/j.jinf.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Periasamy H., Iswarya S., Pavithra N., Senthilnathan S., Gnanamani A. In vitro antibacterial activity of plumbagin isolated from Plumbago zeylanica L. against methicillin-resistant Staphylococcus aureus. Lett Appl Microbiol. 2019;69(1):41–49. doi: 10.1111/lam.13160. [DOI] [PubMed] [Google Scholar]
- 34.Sumsakul W., Plengsuriyakarn T., Chaijaroenkul W., Viyanant V., Karbwang J., Na-Bangchang K. Antimalarial activity of plumbagin in vitro and in animal models. BMC Complement Altern Med Ther. 2014;14:15. doi: 10.1186/1472-6882-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kromker V., Leimbach S. Mastitis treatment-reduction in antibiotic usage in dairy cows. Reprod Domest Anim. 2017;52:21–29. doi: 10.1111/rda.13032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.