Abstract
Nonactin is the parent compound of a group of ionophore antibiotics, known as the macrotetrolides, produced by Streptomyces griseus subsp. griseus ETH A7796. Nonactin is a significant compound because of its inhibitory effects on the P170 glycoprotein-mediated efflux of chemotherapeutic agents in multiple-drug-resistant cancer cells. Nonactin is also significant in that it is a highly atypical polyketide. Very little is presently known about the genes of the nonactin biosynthesis cluster. In this paper we describe our efforts to establish a connection between the product of a gene from the nonactin biosynthesis cluster and a known biochemical transformation in nonactin biosynthesis. Nonactate synthase is the enzyme which catalyzes the formation of nonactic acid from an acyclic precursor in nonactin biosynthesis. We have synthesized the substrate for this enzyme and have detected the in vitro cyclization activity of the substrate in cell-free preparations of S. griseus subsp. griseus ETH A7796. Previous studies by R. Plater and J. A. Robinson (Gene 112:117–122, 1992) had suggested, based on sequence homology, that the product of a partial open reading frame found close to the tetranactin resistance gene of S. griseus could be the nonactate synthase. We have therefore cloned, sequenced, and heterologously expressed this full gene (nonS), and we have shown that the gene product, NonS, does indeed catalyze the formation of the furan ring of nonactic acid as hypothesized.
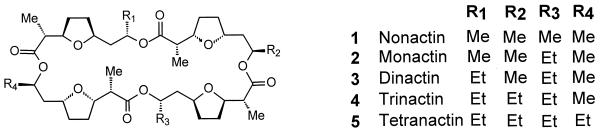
Nonactin (Fig. 1, compound 1) is the parent compound of a group of ionophore antibiotics, known as the macrotetrolides, produced by Streptomyces griseus subsp. griseus ETH A7796 (8, 16, 18, 19, 24, 29, 30, 33, 34). Nonactin has been shown to possess antitumor activity both against mammalian cell lines in vitro and against Crocker sarcoma 180 in studies with mice (8). Nonactin was recently shown to be a novel inhibitor of the 170-kDa P-glycoprotein-mediated efflux of 4-O′-tetrahydropyranyldoxorubicin in multidrug-resistant erythroleukemia K562 cells at subtoxic concentrations (10). The natural macrotetrolide homologues show a wide range of potency. For example, the MIC of nonactin against Staphylococcus aureus and Mycobacterium bovis is more than an order of magnitude greater than that of dinactin, a difference which is paralleled by the changes in the stability constants of their Na+ and K+ complexes (27, 29).
FIG. 1.
Structures of the naturally occurring macrotetrolides.
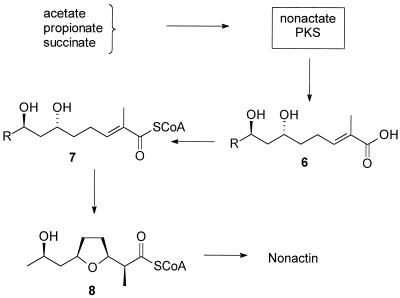
Initial biosynthesis studies using 14C-labeled compounds suggested that nonactin is made from acetate, propionate, and succinate and that nonactic acid and homononactic acid are present in the culture broth (36, 37, 46, 47). The early work was confirmed and extended by Robinson and coworkers, who used extensive feeding studies employing stable isotopes and radioisotopes (3–5, 14, 15, 44–46). Robinson et al. postulated a pathway for nonactin biosynthesis that was based on polyketide biosynthesis (Fig. 2) (3–6, 14, 15, 44, 45) and in the process demonstrated the highly atypical nature of nonactin biosynthesis. One of the unusual features of the nonactin biosynthesis pathway is that it produces both enantiomers of the precursor nonactic acid. Both enantiomers of nonactic acid are subsequently stereospecifically assembled into the final product. This feature raises the important hypothesis that the enzymes which catalyze reactions late in the biosynthesis of nonactic acid cannot discriminate, and indeed have evolved not to discriminate, between enantiomers of their substrates. At this point, however, an alternative hypothesis cannot be ruled out. There may exist a pair of enzymes for each reaction in the late stages of nonactin biosynthesis. Each enzyme would then stereospecifically act upon its appropriate substrate enantiomer. To answer these important questions, we decided to clone the entire nonactin biosynthesis gene cluster and to establish the nature of the chemical reactions catalyzed by each gene product of the cluster. This paper describes our research efforts with the first target enzyme, nonactate synthase.
FIG. 2.
Overview of the current hypothesis for nonactin biosynthesis. PKS, polyketide synthase.
It was known that a late, acyclic intermediate, such as compound 6 (Fig. 2), when activated as an N-caprylcysteamine thioester, was efficiently incorporated into nonactin when added to fermentative cultures of S. griseus (45). Furthermore, each enantiomer of the activated compound 6 was stereospecifically incorporated into the appropriate enantiomer of the monomer units of nonactin. These feeding study data lead Spavold and Robinson to conclude that there was an enzyme activity present in S. griseus which was capable of catalyzing the cyclization reaction to form the furan ring of nonactic acid (45). Here we refer to this enzyme as nonactate synthase.
Plater and Robinson isolated and sequenced a 3.3-kb fragment of DNA from S. griseus subsp. griseus ETH A7796 which conferred tetranactin resistance (nonR) on S. lividans TK24 (38). Analysis of the DNA sequence revealed three complete open reading frames (ORFs) and an incomplete ORF. Of great significance to our studies was the observation that the deduced product of the incomplete orfX (in this work called nonS) showed 27.9% amino acid sequence identity with the C-terminal end of the rat mitochondrial enoyl coenzyme A (enoyl-CoA) hydratase (35). The chemical reactions catalyzed by the enoyl-CoA hydratase family of enzymes and the hypothesized activity of nonactate synthase are very similar. Each reaction involves the addition of an oxygen nucleophile, in a Michael fashion, across the α,β-unsaturated system of the appropriate substrate. Due to the similarity of these reactions, Plater and Robinson (38) postulated that nonS may encode the nonactate synthase, that is, the enzyme catalyzing the formation of the furan ring of nonactic acid.
We set out to confirm the hypothesis of Plater and Robinson by cloning nonS in its entirety. We sought to synthesize putative substrates for the enzyme and to demonstrate in vitro conversion of these substrates by cell-free systems of the nonactin-producing organism. Final confirmation of the hypothesis would come from the heterologous overexpression of the gene product, NonS, and the demonstration that it indeed could catalyze the conversion of our synthesized substrates into nonactic acid precursors.
This paper describes our successful cloning, analysis, and overexpression of the active nonactate synthase, confirming that nonS does indeed encode the nonactate synthase.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. griseus subsp. griseus ETH A7796 (DSM 40695) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. Streptomyces lividans TK24 was obtained from D. A. Hopwood (John Innes Institute, Norwich, England). S. griseus was grown on 2× TSB medium (26) or on R2YE solid medium (26). Recombinant S. lividans TK24 strains were grown in liquid YEME medium (26) containing 1 μg of neomycin · ml−1 and were maintained on plates of R2YE solid medium containing neomycin at a concentration of 10 μg · ml−1. Escherichia coli DH5α (25) was used to propagate plasmids and maintain the partial genomic library of S. griseus subsp. griseus A7796 chromosomal DNA. LB medium (42) was used to grow E. coli; plasmids were introduced into E. coli by transformation by standard procedures (42). Ampicillin was added at a concentration of 100 μg · ml−1 to cultures of E. coli harboring plasmids.
Procedures for protoplast formation, transformation, and regeneration of protoplasts for S. lividans were carried out as outlined by Hopwood et al. (26). Plasmids in Streptomyces were prepared by methods described by Carter and Milton (13). Restriction mapping and other routine molecular methods used in this work are described by Sambrook et al. (42). Plasmids used and constructed in this work are given in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| pUC19 | 2.69 kbp; Ampr; E. coli cloning vector | 50 |
| pANT841 | 2.75 kbp; pUC19 containing a polylinker with additional cloning sites optimized for cloning DNA with high G+C content; blue-white selectable; Ampr | 17 |
| pANT797 | 8.43 kbp; 1.337-kbp NruI fragment and T4 DNA polymerase-blunted KpnI fragment into PvuII-digested pANT795; E. coli-Streptomyces shuttle vector; expression of genes cloned into polylinker is driven by SnpR-activated snpA promoter; Neor | 41 |
| pANT1400 | 9.05 kbp; 6.3-kbp BamHI fragment from S. griseus subsp. griseus ETH A7796 chromosome into BglII site of pANT841 (contains nonS, nonR, orfB, partial orfC, and region upstream of nonS) | This work |
| pANT1401 | 6.29 kbp; 3.55-kbp SstI fragment from pANT1400 into pANT841 (contains region upstream of nonS) | This work |
| pANT1402 | 5.22 kbp; 2.47-kbp SstI fragment from pANT1400 into pANT841 (contains complete nonS, nonR, and orfB) | This work |
| pANT1403 | 10.93 kbp; 2.50-kbp EcoRI-HindIII fragment from pANT1402 into pANT797 | This work |
Neor, neomycin resistance; Ampr, ampicillin resistance; SnpR, small neutral protease regulatory protein; snpA, promoter from Streptomyces sp. strain C5 for the small neutral protease.
Molecular cloning and sequence analysis of the nonS gene.
Chromosomal DNA from S. griseus was isolated by the procedure of Pospiech and Neumann (39) and digested at 37°C with restriction endonuclease BamHI, HincII, or SstI (U.S. Biochemicals, Cleveland, Ohio) according to the manufacturer’s directions. The digests were fractionated by agarose gel electrophoresis and then transferred to BA-85 nitrocellulose filters (Schleicher and Schuell, Inc., Keene, N.H.). The blots were hybridized with the 32P-end-labeled oligonucleotide 5′-GGAGGATTTCGACCGCGAACTGGCCGATCTG-3′, whose sequence was identical to that of part of the partial nonS previously identified by Plater and Robinson (38). Hybridization and washing were performed as described by Rajgarhia and Strohl (41). BamHI-digested DNA fragments (6.0 to 8.0 kb) from S. griseus subsp. griseus, which contained the largest fragment that hybridized to the labeled oligonucleotide, were isolated from agarose gel pieces and used to construct a partial, pUC19-based (50) genomic library in E. coli. The library was screened by using a labeled 48-mer oligonucleotide (5′-GGATGGCCGCGTTCACGGAGAAGCGGCCGCCCCGCTTCACCGGCGCCT-3′). DNA from colonies that hybridized to the probe was isolated, and Southern analysis was performed again to confirm the positive clones. A 6.3-kb BamHI fragment was cloned and subsequently sequenced with the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit and analyzed on an ABI model 310 automated sequencer (Perkin-Elmer/Applied Biosystems).
Analysis of sequence data.
The entire nonS ORF was determined by using FRAME (9) and CODON PREFERENCE (49) algorithms with IBM-PC programs, and the sequences were compared with those in the databases by using BLAST (2) and PSI-BLAST (2). Amino acid alignments were obtained by using CLUSTAL X (48).
Synthesis procedures.
Solvents were obtained from Fisher Scientific. All other chemicals, unless noted otherwise, were obtained from the Aldrich Chemical Company (Milwaukee, Wis.). The solvents CH2Cl2 (over CaH2), diethyl ether (over Na/K-benzophenone), and tetrahydrofuran (THF) (over Na/K-benzophenone) were dried and distilled prior to use. MgSO4 refers exclusively to anhydrous MgSO4. The term Hexanes refers to the commercially available solvent, which is a mixture containing predominantly n-hexane but also substantial amounts of the isomers of hexane. Solutions were concentrated by evaporation in vacuo. All synthesis procedures, unless noted otherwise, were carried out under a slight positive pressure of dry argon gas. Column chromatography was performed with Merck Silica Gel 60. Nuclear magnetic resonance spectra (1H and 13C) were acquired at either 400, 270, or 250 MHz, were referenced to the residual solvent, and are reported as chemical shift (δ, in parts per million), intensity, splitting pattern, and coupling constant (J, in hertz).
(5R*,S*)-Hydroxyocta-1,6-diene (compound 10).
Dibromoethane (3.1 ml, 36.5 mmol) was added dropwise via syringe to a stirred suspension of magnesium powder (3.54 g, 146 mmol) in dry THF (200 ml) and then warmed gently. The suspension was stirred until gas evolution had ceased, and it was then left to cool to room temperature. 4-Bromobut-1-ene (9.84 g, 72.9 mmol) was added in small portions via syringe to the constantly stirred suspension. The reaction mixture became moderately warm and maintained a gentle reflux. The mixture was stirred for 30 min. Freshly distilled crotonaldehyde (6.65 ml, 80.2 mmol) in dry THF (10 ml) was added dropwise. After 2 h, the reaction was carefully quenched by the addition of water (10 ml) followed by saturated, aqueous NaCl solution (brine) (250 ml). The organic phase was recovered. The aqueous phase was extracted three times with diethyl ether (150 ml). The combined organic phases were dried over MgSO4, filtered, and concentrated to give a pale yellow liquid. The product was purified by chromatography on silica gel, eluting with 25% ethyl acetate (EtOAc)–hexanes to give a colorless oil (8.21 g, 90.2%). b p 52 to 54°C, 0.5 torr; δH (270 MHz, CDCl3) 1.55 (2H, mult), 1.62 (3H, d, J = 6.7 Hz), 1.78 (1H, br-s, OH), 2.10 (2H, mult), 4.00 (1H, q, J = 6.8 Hz), 4.90 (1H, mult), 5.00 (1H, mult), 5.40 (1H, mult), 5.60 (1H, mult), and 5.80 (1H, mult); δc 17.7, 29.9, 38.6, 72.2, 114.8, 128.9, 134.4, and 138.6. High-resolution mass spectrometry (HRMS): found, 126.105163; calculated for C8H14O, 126.104465.
(5R*,7S*)-5,7-Dihydroxyoct-1-ene (compound 12).
The olefin compound 10 (6.35 g, 50.3 mmol), (+)-diethyl tartrate (0.78 g, 3.8 mmol), (−)-diethyl tartrate (0.78 g, 3.8 mmol), and powdered 13×(2μ) sieves (1.9 g) were added to dry CH2Cl2 (200 ml). The stirred suspension was cooled to −20°C. Titanium(IV) isopropoxide (1.5 ml, 5.0 mmol) was added dropwise via syringe, and the suspension was stirred for 40 min. Freshly dried and titrated cumene hydroperoxide (13.5 ml, 75 mmol) was added dropwise via syringe, and the stirred solution was allowed to warm to room temperature overnight. The suspension was poured into a solution of FeSO4 · 7H2O (33 g, 120 mmol) and tartaric acid (11 g, 60 mmol) in water (100 ml) and stirred for 30 min. The organic phase was recovered. The aqueous phase was extracted twice with CH2Cl2 (100 ml). The combined organic phases were dried (MgSO4), filtered, and concentrated. The oil obtained was fractionated on silica gel, eluting with 15% EtOAc–hexanes, to give the semipure epoxide compound 11.
The epoxide (compound 11) was dissolved in dry THF (125 ml) and cooled to 0°C. RedAl (Aldrich) (31.3 ml, 101 mmol) was added dropwise via syringe, and the solution was stirred overnight at 0°C. The reaction was quenched by the careful, dropwise addition of a aqueous solution of tartaric acid (5%, wt/vol) and sodium tartrate (5%, wt/vol) (200 ml). The cloudy white suspension was stirred vigorously at room temperature for 30 min and then diluted with water (100 ml) and EtOAc (150 ml). The organic phase was recovered. The aqueous phase was extracted twice with EtOAc (100 ml). The combined organic phases were dried (MgSO4), filtered, and concentrated to give a colorless oil which was evacuated at 0.2 torr for 40 min at room temperature to remove MeOCH2CH2OH. The oil was fractionated on silica gel, eluting with 50% EtOAc–hexanes, to give the pure diol compound 12 (4.79 g, 66.0% from the olefin compound 10). δH (270 MHz, CDCl3) 1.03 (3H, d, J = 6.7 Hz), 1.20 (4H, mult), 2.00 (2H, mult), 3.65 (1H, mult), 3.86 (1H, mult), 4.82 (1H, d, J = 11.3 Hz), 4.90 (1H, d, J = 20 Hz), and 5.72 (1H, mult); δc (67.9 MHz, CDCl3) 24.3, 31.0, 38.3, 47.1, 65.5, 68.8, 114.9, and 139.7.
(5R*,7S*)-5,7-bis-(tert-butylsilyloxy)oct-1-ene (compound 13).
The diol compound 12 (0.47 g, 3.3 mmol), tert-butyldimethylsilylchloride (2.46 g, 16.3 mmol), and N,N-dimethylaminopyridine (0.04 g, 0.33 mmol) were dissolved in dry pyridine (1.6 ml, 19.5 mmol). Sufficient dry dimethylformamide (2 ml) was added to allow the thick suspension to be stirred efficiently. After being stirred at room temperature for 48 h, the white suspension was poured into saturated aqueous CuSO4 solution (100 ml) and diluted with CH2Cl2 (50 ml). The organic phase was recovered. The aqueous phase was extracted with CH2Cl2 (50 ml). The combined organic phases were dried (MgSO4), filtered, and concentrated to give a colorless oil. The product was obtained by chromatography on silica gel, eluting with 4% EtOAc–hexanes, as a colorless oil (1.18 g, 96.9%). δH (270 MHz, CDCl3) 0.05 (12H, s), 0.87 (18H, s), 1.15 (3H, d, J = 7 Hz), 1.55 (4H, mult), 2.07 (2H, mult), 3.77 (1H, mult), 3.90 (1H, sext, J = 5.7 Hz), 4.92 (1H, d, J = 10.3 Hz), 5.0 (1H, d, J = 18.8 Hz), and 5.30 (1H, mult); δc (67.9 MHz, CDCl3), −4.2, −3.9, −3.8. −3.7, −2.7, 18.3, 24.8, 25.9, 26.2, 29.5, 37.4, 48.2, 66.6, 70.0, 114.5, and 139.1.
Ethyl (6S*,8R*)-6,8-bis(tert-butyldimethylsilyloxy)-2-methylnon-2E-enoate (compound 14).
Ozone gas was bubbled through a solution of the olefin compound 13 (4.67 g, 12.5 mmol) in dry CH2Cl2 (50 ml) which was stirred vigorously at −78°C. Excess ozone, seen as a bright blue color, was observed after 6 to 7 min. Argon gas was bubbled through the stirred solution until the blue color had gone. A solution of triphenylphosphine (3.44 g, 13.1 mmol) in CH2Cl2 (10 ml) was added to the stirred solution, still at −78°C. After 30 min, the reaction mixture was allowed to warm to room temperature and then stirred overnight. The mixture was concentrated in vacuo to a volume of approximately 25 ml. CH2Cl2 (15 ml) was introduced, and then the ylide Ph3P⩵CMeCO2Et (9.06 g, 25 mmol) was added. The reaction mixture was stirred at room temperature overnight. The mixture was concentrated and fractionated directly on silica gel, eluting with 2.5% EtOAc–hexanes, to give the product as a colorless oil (4.06 g, 71.6%). δH (270 MHz, CDCl3) 0.00 (12H, s), 0.80 (18H, s), 1.07 (3H, d, J = 5.6 Hz), 1.20 (3H, t, J = 7 Hz), 1.50 (4H, mult), 1.75 (3H, s), 2.15 (2H, br q), 3.70 (1H, pent, J = 5.6 Hz), 3.80 (1H, sext, J = 5.6 Hz), 4.10 (2H, q, J = 5.6 Hz), and 6.70 (1H, t, J = 7 Hz); δc (67.9 MHz, CDCl3) −4.4, −4.1, −4.0, −3.7, 12.4, 14.4, 18.2, 24.5, 24.7, 25.9, 26.1, 36.8, 48.0, 60.4, 66.5, 69.9, 128.1, 142.1, and 168.2 HRMS: found, 459.338592; calculated for C24H51O2Si2, 459.332592.
(6S*,8R*)-6,8-Bis(tert-butyldimethylsilyloxy)-2-methylnon-2E-enoic acid (compound 15).
Aqueous LiOH solution (1 ml, 2.5 mmol) was added to a vigorously stirred solution of the ester compound 14 (229 mg, 0.5 mmol) in a mixture of THF (3 ml) and methanol (2 ml). After 24 h at room temperature, the reaction mixture was diluted with water (50 ml) and acidified to pH 2.5 with 2 N HCl solution. The product was extracted twice with EtOAc (50 ml). The organic extracts were combined, dried (MgSO4), filtered, and concentrated. The product was obtained by preparative thin-layer chromatography on silica gel, eluting with 20% EtOAc–hexanes, as a colorless oil (211 mg, 98.0%). δH (270 MHz, CDCl3) 0.00 (12H, s), 0.90 (18H, s), 1.10 (3H, d, J = 7 Hz), 1.60 (4H, mult), 1.90 (3H, s), 2.20 (2H, mult), 3.80 (1H, mult), 3.90 (3H, mult), 6.90 (1H, t, J = 7 Hz), and 11.20 (1H, br s); δc (67.9 MHz, CDCl3) −4.2, −4.1, −3.96, −3.93, −3.7, 12.1, 18.3, 24.7, 26.1, 36.7, 48.1, 66.6, 70.0, 127.4, 145.0, and 173.7.
(6S*,8R*)-6,8-Bis(tert-butyldimethylsilyloxy)-2-methylnon-2E-enoate, N-octylcysteamine thioester (compound 17).
Dicyclohexylcarbodiimide (105 mg, 0.51 mmol) in dry THF (0.75 ml) was added to a stirred solution of the acid compound 15 (211 mg, 0.49 mmol) in dry THF (4 ml). The mixture was stirred for 10 min at room temperature. N-Caprylcysteamine (199 mg, 0.98 mmol), as a suspension in dry THF (2 ml), was added via syringe. After 24 h, the reaction mixture was diluted with aqueous, saturated NaHCO3 solution and extracted with CH2Cl2. The organic extracts were combined, dried (MgSO4), filtered, and concentrated. The product was obtained by chromatography on silica gel, eluting with 20% EtOAc–hexanes, as a colorless oil (159 mg, 52.7%). δH (270 MHz, CDCl3) 0.00 (12H, s), 0.82 (18H, s), 1.10 (3H, d, J = 6.5 Hz), 1.3 (10H, br s), 1.55 (8H, mult), 1.85 (3H, s), 2.10 (2H, t, J = 6.8 Hz), 2.22 (2H, mult), 3.05 (2H, t, J = 6.9 Hz), 3.40 (2H, q, J = 6.9 Hz), 3.77 (1H, p, J = 6.9 Hz), 3.85 (1H, sext, J = 6.7 Hz), 5.85 (1H, br t), and 6.75 (1H, t, J = 5.6 Hz); δc (67.9 MHz, CDCl3) −4.2, −3.9 (2C), −3.7, 12.6, 14.1, 18.3, 22.8, 24.7, 24.8, 25.9, 26.1, 28.7, 29.2, 31.9, 36.7, 37.0, 39.9, 48.1, 66.6, 70.0, 136.1, 141.9, 173.4, and 194.0. HRMS: found, 615.413666; calculated for C10H18O4, 615.417288.
(6S*,8R*)-6,8-Dihydroxy-2-methylnon-2E-enoate, N-octylcysteamine thioester (compound 18).
Acetic acid (3.0 ml) was added to a stirred suspension of the thioester compound 17 (91.0 mg, 0.15 mmol) in THF (1.0 ml) and water (1.0 ml). The solution was stirred at 23°C for 24 h. The volatile solvents were removed by evaporation in vacuo. The remaining mixture was diluted with water and extracted with EtOAc. The extracts were combined, dried (MgSO4), filtered, and concentrated. Chromatography on silica gel, eluting with AcOH-EtOAc (1:99), afforded the product compound 18 as a colorless oil (31.9 mg, 54.9%). δH (250 MHz, CDCl3) 0.85 (3H, t, J = 6.6 Hz), 1.30 (11H, mult and obscured d), 1.60 (4H, mult), 1.85 (3H, s), 2.15 (2H, t, J = 8.2 Hz), 2.32 (2H, mult), 3.05 (2H, t, J = 6.4 Hz), 3.45 (2H, q, J = 7.5 Hz), 3.9 (1H, mult), 4.17 (1H, mult), 6.12 (1H, br t), and 6.77 (1H, t, J = 8.3 Hz); δc (62.9 MHz, CDCl3) 12.6, 14.1, 22.7, 23.8, 25.4, 25.8, 28.7, 29.1, 29.4, 31.6, 36.2, 36.7, 39.9, 44.7, 65.5, 68.6, 136.3, 141.5, 173.8, and 194.1. HRMS: found, 387.243958; calculated for C20H37O4NS, 387.244330.
(6S*,8R*)-6,8-Dihydroxy-2-methylnon-2E-enoic acid (compound 16).
Aqueous 2 N HCl (1.0 ml) was added to a stirred solution of compound 15 (147 mg, 0.34 mmol) in THF (4.0 ml). The reaction mixture was stirred at 23°C for 30 min and then concentrated in vacuo. The mixture was diluted with water (5 ml) and extracted twice with EtOAc (10 ml). The organic extracts were combined, dried (MgSO4), filtered, and concentrated. The product was obtained after preparative thin-layer chromatography on silica gel, eluting with 1% AcOH in EtOAc, as a colorless oil (45 mg, 65.5%) δH (400 MHz, CDCl3) 1.12 (3H, d, J = 6.1 Hz), 1.55 (4H, mult), 1.81 (3H, s), 2.22 (2H, mult), 3.78 (1H, p, J = 5.7 Hz), 3.86 (1H, sext, J = 6.2 Hz), and 6.88 (1H, t, J = 6.5 Hz); δc (100.6 MHz, CDCl3) 12.1, 18.2, 27.7, 35.2, 47.9, 66.5, 69.8, 127.3, 145.1, and 173.9. HRMS: found, 202.115616; calculated for C10H18O4, 202.120509.
Enzyme assay procedures.
Cells from either a 96-h fermentative culture of S. griseus subsp. griseus ETH A7796 or a 48-h culture of S. lividans TK24 harboring plasmids were collected by centrifugation (10,000 × g, 30 min) and washed once with potassium phosphate buffer (50 mM, pH 7.0). The cells were resuspended in a minimal amount of potassium phosphate buffer (50 mM, pH 7.0) and lysed by two passages through a French pressure cell. The broken-cell suspensions were clarified by centrifugation (10,000 × g, 30 min) to produce the cell-free samples for enzyme assay. The protein concentrations in the samples were determined by the dye-binding assay of Bradford (12).
An enzyme assay consisted of a mixture of the substrate (25 μl, 64 mM in acetonitrile), cell-free preparation (100 μl), potassium phosphate buffer (2.7 ml, 50 mM, pH 7.0), and bovine serum albumin (2.7 mg). The reaction was initiated by addition of the cell-free preparation. UV spectra in the range of 230 to 450 nm were recorded every 5 min over a 2-h period. The rate of the reaction was calculated by using the absorbance change at 277 nm when the thioester compound 18 was employed as the substrate (ɛ277 was estimated to be 3,630 M−1 · cm−1).
HPLC analysis.
Some of the enzyme assay incubations (see above) were left for 12 h and then extracted twice with EtOAc (2 ml). The organic extracts were combined, dried (MgSO4), filtered, concentrated, and then resuspended in CH2Cl2. High-pressure liquid chromatography (HPLC) analysis was carried out on a Spherisorb silica column (150 by 4.6 mm; Aldrich). Detection of the substrate and product was achieved by monitoring the absorbance of the eluate at 230 nm. Isocratic elution at 1 ml · min−1 with 10% EtOH in hexanes afforded the optimal resolution (rt of compound 18 = 8.6 min; rt of N-caprylcysteamine thioester of nonactic acid = 6.3 min). Confirmation of the eluate identity was achieved by obtaining mass spectra for small collected samples of the eluate.
Nucleotide sequence accession number.
The nucleotide sequences described in this work have been submitted to the National Center for Biotechnology Information under accession no. AF074603.
RESULTS AND DISCUSSION
Synthesis of the substrate.
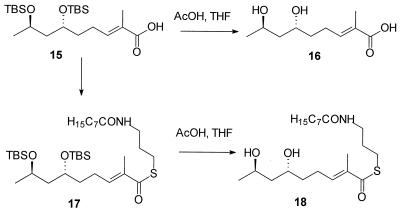
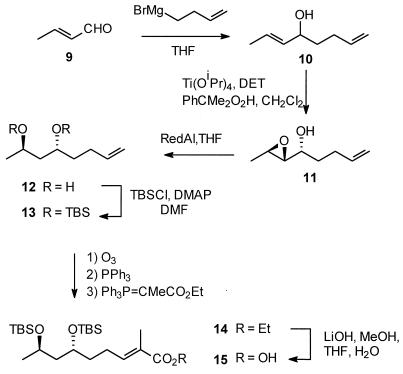
At the outset of this work, it was not known if the substrate for the nonactate synthase was the free acid compound 6 or the CoA thioester activated form of the substrate, compound 7 (Fig. 2). It was known, however, that the CoA analog compound 18 (see Fig. 4), when administered to a fermentative culture of S. griseus, was efficiently and stereospecifically incorporated into nonactic acid and, therefore, nonactin. Given that a search for in vitro nonactate synthase activity would require both compounds 6 and 18, we chose to base our synthetic strategy on the earlier work of Spavold and Robinson (44, 45). Both the free acid and the thioester analog, therefore, are available by divergent synthesis from a common, advanced intermediate (Fig. 3 and 4). Furthermore, we chose to synthesize a racemic mixture of the substrates, leaving questions of the stereospecificity of the process until the time when these issues can be appropriately addressed with a purified, homogeneous nonactate synthase.
FIG. 4.
Divergent synthesis of substrates from intermediate 15.
FIG. 3.
Early synthesis steps to generate intermediate 15 for later divergent synthesis of each substrate. TBSCl, tert-butyldimethylsilylchloride.
The racemic allylic alcohol compound 10 (Fig. 3) was formed via Grignard addition to crotonaldehyde; no 1,4-addition product was observed. Diastereoselective epoxidation of the allylic alcohol was achieved with titanium(IV) isopropoxide and a racemic mixture of diethyl tartrate. In this reaction no Payne rearrangement of the product was observed, in contrast to our earlier observations in the synthesis of similar terminal alkyne intermediates (20). RedAl reduction of the epoxide compound 11 was initially troublesome. Rapid consumption of the starting material was observed at 0°C. On isolation, the product of the reaction was found to be from a Payne rearrangement. On longer exposure (ca. 12 to 18 h) to RedAl, the first intermediate undergoes the expected reductive opening of the epoxide to form the 1,3-diol compound 12. Fortunately, the stereochemistry obtained in the product is the same when either the original epoxide or its rearrangement product is reductively opened. Protection of the diol proved uneventful and efficient when conducted at a sufficiently high concentration. A significant departure from earlier studies was undertaken in the use of a “one-pot” ozonolysis-Wittig tandem reaction with the readily obtained ylide Ph3P⩵CMeCO2Et. Once purified, the product showed no traces of contamination by any cis-olefin. The first putative substrate, compound 16 (Fig. 4), was then obtained by base-catalyzed hydrolysis of the ester and acid-catalyzed desilylation.
Base-catalyzed hydrolysis of the ester compound 14 was followed by coupling of N-caprylcysteamine to the acid compound 15, and then specific acid-catalyzed desilylation afforded the second putative substrate, compound 18 (Fig. 4).
These synthesis procedures gave the acid compound 16 and the thioester compound 18 in 28.5 and 14.5% overall yields, respectively, from the commercially available starting material, 2-bromobut-1-ene.
Cloning of the entire nonS gene from S. griseus subsp. griseus ETH A7796.
We were considerably helped in our cloning approaches by Plater and Robinson’s earlier isolation and analysis of the macrotetrolide resistance gene (38). A 32P-end-labeled oligonucleotide, identical in sequence to the C-terminus-encoding end of nonS, was used to probe, via Southern blotting, S. griseus genomic DNA. The largest hybridizing BamHI-digested fragments, of approximately 7 kb in size, were isolated from the gel and used to construct a pUC19-based partial genomic library in E. coli. Initial attempts to screen the library with the original 31-mer oligonucleotide were unsuccessful. The library was screened again with a longer 48-mer oligonucleotide identical to an alternative region of the known sequence of nonS. DNA isolated from colonies which hybridized to the latter probe was isolated and sequenced to confirm positive clones. These procedures yielded pANT1400, which contained 6.3 kb of S. griseus genomic DNA, a fragment almost certainly large enough to contain the entire nonS gene, based on the sizes of known enoyl-CoA hydratase enzymes.
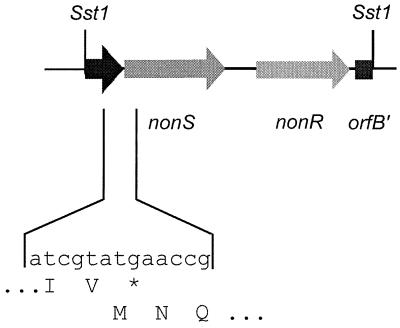
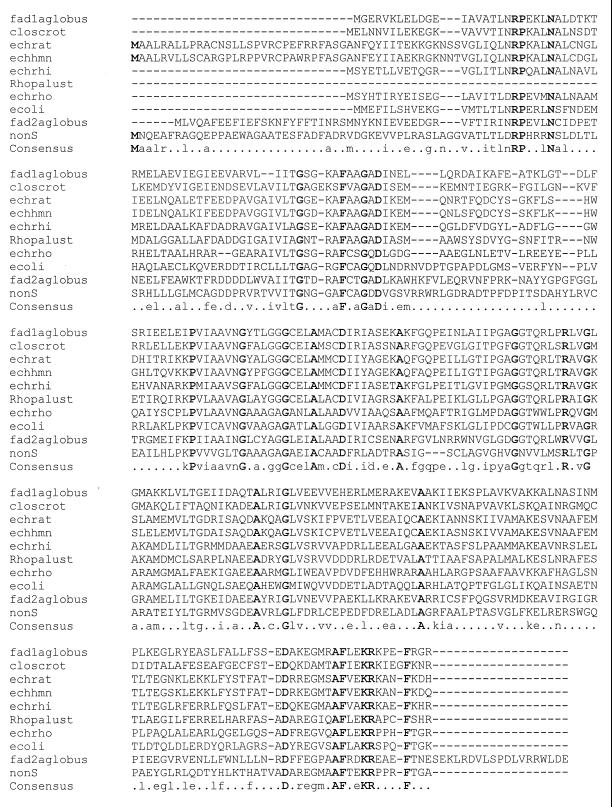
After extensive restriction mapping, an approximately 2.47-kb SstI-SstI fragment was subcloned to give pANT1402. Analysis of the sequence (Fig. 5 and 6) revealed the complete nonS ORF with an ATG start codon and TGA stop codon. The ORF encodes a protein of 297 amino acids with a predicted molecular mass of 31,670 Da. The sequence showed great similarity to a family of soluble enoyl-CoA hydratase enzymes (Fig. 7) when compared to known sequences in databases. Plater and Robinson had earlier noted the homology of the then-available C-terminal end of the predicted nonS product to the enoyl-CoA hydratase from the rat (38). Our sequencing data considerably reinforce this proposal, given the number of amino acid identities across the entire family (1, 7, 11, 21, 28, 31, 32, 51) and the consistency of the overall sizes of the enzymes. Furthermore, nonS appears to be translationally coupled to an unidentified ORF downstream.
FIG. 5.
Map of the 2.47-kbp SstI-SstI fragment cloned in pANT1402. nonR, tetranactin resistance gene.
FIG. 6.
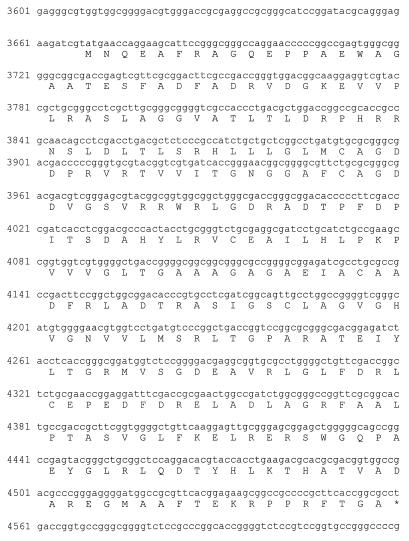
Annotated DNA sequence of the gene nonS and its product. The numbers refer to the nucleotide numbers of the deposited sequence (accession no. AF074603)
FIG. 7.
Amino acid sequence comparison of a number of related enoyl-CoA hydratase enzymes. fad1aglobus, enoyl-CoA hydratase from Archaeoglobus fulgidus (31); closcrot, crotonase from Clostridium acetobutylicum (11); echrat, enoyl-CoA hydratase from Rattus norvegicus (35); echhmn, short-chain enoyl-CoA hydratase from Homo sapiens (28); echrhi, enoyl-CoA hydratase from Rhizobium meliloti (32); Rhopalust, cyclohex-1-ene-1-carboxyl-CoA hydratase from Rhodopseudomonas palustris (21); echrho, probable enoyl-CoA hydratase homolog from Rhodobacter capsulatus (7); ecoli, probable enoyl-CoA hydratase from E. coli (1); fad2aglobus, enoyl-CoA hydratase from A. fulgidus (31); nonS, nonactate synthase (this work).
The 2.47-kb SstI-SstI fragment from pANT1402 was subcloned into the streptomycete expression vector pANT797 (41) to form pANT1403. In this construct, expression of nonS is driven by the SnpR-activated snpA promoter. The plasmid pANT1403 was introduced into S. lividans TK24 by protoplast fusion to allow for heterologous expression of NonS.
Enoyl hydratase assays.
In vitro assays for the conversion of substrate were based on established assays for enoyl-CoA hydratase enzymes (22, 23, 43). The Michael addition across the α,β-unsaturated acid or thioester leads to a loss of conjugation and a substantial decrease in UV absorbance. Initially we sought the nonactate synthase activity in the parent strain S. griseus subsp. griseus ETH A7796. Even at high protein concentrations, no significant change in the UV absorption of the assay solutions was observed when the free acid compound 16 was used as a substrate. When the thioester compound 18 was used, however, a decrease in absorption was observed, with a maximal change at 277 nm, the absorbance maximum of the α,β-unsaturated thioester. No change in absorbance was seen when either the protein or the substrate was omitted from the assay. A sample was left to incubate for a 12-h period and then extracted. The presence of the product in the extract was observed in HPLC analysis of the extract. The authentic product was obtained from earlier synthetic work (40). A rate for the reaction was measured and corresponded to a specific activity of 3.0 nmol · min · mg−1, which is a reasonable value in the context of the levels of expression of secondary metabolic enzymes and the fact that the N-caprylcysteamine thioester is a substrate analog of the native CoA thioester. We have therefore demonstrated the in vitro activity of nonactate synthase in the nonactin-producing species, S. griseus.
Further assays were carried out in a similar fashion with cell extracts obtained from S. lividans TK24(pANT1403). These assays resulted in nonactate synthase activity of 2.3 nmol · min · mg−1. The product identity was again confirmed by HPLC analysis. As a control, a cell extract of S. lividans(pANT797) (vector-only control) was examined. No nonactate synthase activity was observed. We conclude, therefore, that the product of nonS is capable of catalyzing the formation of the furan ring of nonactic acid from its acyclic precursor. The gene product NonS is the nonactate synthase.
Implications for nonactin biosynthesis.
We have demonstrated nonactate synthase activity in cell extracts of the nonactin-producing organism, S. griseus. We have further demonstrated, by heterologous expression of an active protein in S. lividans TK24, that the product of nonS is the nonactate synthase. The amino acid sequence of the nonactate synthase places it in a family of soluble enoyl-CoA hydratase enzymes. These data, together with the observation that the reaction occurs only with the thioester analog and not with the free acid, suggest that in vivo the true substrate is an activated thioester.
ACKNOWLEDGMENTS
We gratefully thank Stephen C. Bergmeier for many helpful discussions. We especially appreciate the support and technical assistance of Don Ordaz and the Ohio State University Fermentation Facility. The Campus Chemical Instrumentation Center is acknowledged for providing mass spectrometric analyses.
This work was supported by the College of Pharmacy, The Ohio State University, and by grant CA77347 from the National Cancer Institute.
REFERENCES
- 1.Aiba H, Baba T, Fujita K, Hayashi K, Inada T, Isono K, Itoh T, Kasai H, Kashimoto K, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Makade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Sivasundaram S, Tagami H, Takeda J, Takemoto K, Takeuchi Y, Wada C, Yamamoto Y, Horiuchi T. A 570-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 28.0-40.1 min region on the linkage map. DNA Res. 1996;3:363–377. doi: 10.1093/dnares/3.6.363. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashworth D M, Robinson J A. Biosynthesis of the macrotetrolide antibiotics: an investigation using carbon-13 and oxygen-18 labelled acetate and propionate. Chem Commun (J Chem Soc Sect D) 1983;1983:1327–1329. [Google Scholar]
- 4.Ashworth D M, Clark C A, Robinson J A. On the biosynthetic origins of the hydrogen atoms in the macrotetrolide antibiotics and their mode of assembly catalysed by a nonactin polyketide synthase. J Chem Soc Perkin Trans I. 1989;1989:1461–1467. [Google Scholar]
- 5.Ashworth D M, Robinson J A, Turner D L. Biosynthesis of nonactin from acetate, propionate, and succinate; the assignment of its carbon-13 N.M.R. spectrum by two-dimensional correlation spectroscopy. Chem Commun (J Chem Soc Sect D) 1982;1982:491–493. [Google Scholar]
- 6.Ashworth D M, Robinson J A, Turner D L. Biosynthesis of the macrotetrolide antibiotics; the incorporation of carbon-13 and oxygen-18 labelled acetate, propionate, and succinate. J Chem Soc Perkin Trans I. 1988;1988:1719–1727. [Google Scholar]
- 7.Beckman D L, Kranz R G. A bacterial homolog to the mitochondrial enoyl-CoA hydratase. Gene. 1991;107:171–172. doi: 10.1016/0378-1119(91)90313-z. [DOI] [PubMed] [Google Scholar]
- 8.Bennett R E, Brindle S A, Giuffre N A, Jackson P W, Kowald J, Pansy F E, Perlman D, Trejo W H. Production of a novel cytotoxic agent, SQ 15,859, by Streptomyces chrysomallus. 1962. pp. 169–172. . Antimicrob. Agents Chemother. 1961. [Google Scholar]
- 9.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 10.Borrel M N, Pereira E, Fiallo M, Garnier-Suillerot A. Mobile ionophores are a novel class of P-glycoprotein inhibitors. The effects of ionophores on 4′-O-tetrahydropyranyl-adriamycin incorporation in K562 drug-resistant cells. Eur J Biochem. 1994;223:125–133. doi: 10.1111/j.1432-1033.1994.tb18973.x. [DOI] [PubMed] [Google Scholar]
- 11.Boynton Z L, Bennet G N, Rudolph F B. Cloning, sequencing, and expression of clustered genes encoding β-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 13.Carter M J, Milton L D. An inexpensive and simple method for DNA purification on silica particles. Nucleic Acids Res. 1993;21:1044. doi: 10.1093/nar/21.4.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark C A. The biosynthesis of nonactin. Ph.D. thesis. Southampton, United Kingdom: University of Southampton; 1982. [Google Scholar]
- 15.Clark C A, Robinson J A. Biosynthesis of nonactin. The role of acetoacetyl-CoA in the formation of nonactic acid. Chem Commun (J Chem Soc Sect D) 1985;1985:1568–1569. [Google Scholar]
- 16.Corbaz R, Ettinger L, Gäumann E, Keller-Schlierlein W, Kradolfer F, Kyburz E, Neipp L, Prelog V, Zähner H. Stoffwechselprodukte von Actinomyceten. Nonactin Helv Chim Acta. 1955;38:1445. [Google Scholar]
- 17.DeSanti, C. L., J. S. Lampel, K. A. Lampel, and W. R. Strohl. Unpublished data.
- 18.Dobler M. Crystal structure of nonactin. Helv Chim Acta. 1972;55:1371–1384. doi: 10.1002/hlca.19720550504. [DOI] [PubMed] [Google Scholar]
- 19.Dutcher J D. Isolation and characterization of a cytotoxic agent, SQ 15,859, from Streptomyces chrysomallus. 1962. pp. 173–177. . Antimicrob. Agents Chemother. 1961. [Google Scholar]
- 20.Earle M J, Priestley N D. Synthesis and evaluation of a designed inhibitor for nonactin biosynthesis in S. griseus ETH A7796. Bioorg Med Chem Lett. 1997;7:2187–2192. [Google Scholar]
- 21.Egland P G, Pelletier D A, Dispensa M, Gibson J, Harwood C S. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita Y, Shimakata T, Kusaka T. Purification of two forms of enoyl-CoA hydratase. J Biochem. 1980;88:1045–1050. doi: 10.1093/oxfordjournals.jbchem.a133055. [DOI] [PubMed] [Google Scholar]
- 23.Furuta S, Miyazawa S, Osumi T, Hashimoto T, Ui N. Properties of mitochondrial and peroxisomal enoyl-CoA hydratases from rat liver. J Biochem. 1980;88:1059–1070. doi: 10.1093/oxfordjournals.jbchem.a133057. [DOI] [PubMed] [Google Scholar]
- 24.Gerlach H, Hutter R, Keller-Schlierlein W, Seibl J, Zähner H. Metabolic products of microorganisms. LVIII. New macrotetrolides from actinomycetes. Helv Chim Acta. 1967;50:1782–1793. doi: 10.1002/hlca.19670500711. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 26.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 27.Izatt R M, Bradshaw J S, Nielsen S A, Lamb J D, Christensen J J, Sen D. Thermodynamic and kinetic data for cation-macrocycle interaction. Chem Rev. 1985;85:271–339. [Google Scholar]
- 28.Kanazawa M, Ohtake A, Abe H, Yamamoto S, Satoh Y, Takayanagi M, Niimi H, Mori M, Hashimoto T. Molecular cloning and sequence analysis of the cDNA for human mitochondrial short-chain enoyl-CoA hydratase. Enzyme Protein. 1993;47:9–13. doi: 10.1159/000468650. [DOI] [PubMed] [Google Scholar]
- 29.Keller-Schlierlein W, Gerlach H. Macrotetrolides. Fortschr Chem Org Naturstoffe. 1968;26:161–189. [PubMed] [Google Scholar]
- 30.Kilbourne R T, Dunitz J D, Pioda L A R, Simon W. Structure of the K+ complex with nonactin, a macrotetrolide antibiotic possessing highly specific K+ transport properties J. Mol Biol. 1967;30:559–563. doi: 10.1016/0022-2836(67)90370-1. [DOI] [PubMed] [Google Scholar]
- 31.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus flugidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 32.Margolin W, Bramhill D, Long S R. The dnaA gene of Rhizobium meliloti lies within an unusual gene arrangement. J Bacteriol. 1995;177:2892–2900. doi: 10.1128/jb.177.10.2892-2900.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menshikov G P, Rubinsthein M M. Isolation of new antibiotic longisporin and a study of its chemical nature. J Gen Chem USSR. 1956;26:2267. [Google Scholar]
- 34.Meyers E, Pansy F E, Perlman D, Smith D A, Weisenborn F L. The in vitro activity of nonactin and its homologs: monactin, dinactin, and trinactin. J Antibiot. 1965;18:128. [PubMed] [Google Scholar]
- 35.Minami-Ishii N, Taketani S, Osumi T, Hashimoto T. Molecular cloning and sequence analysis of the cDNA for rat mitochondrial enoyl-CoA hydratase. Eur J Biochem. 1989;185:73–78. doi: 10.1111/j.1432-1033.1989.tb15083.x. [DOI] [PubMed] [Google Scholar]
- 36.Pape H. Metabolic products of microorganisms. 109. Biosynthesis of macrotetrolides. II. Biosynthesis of homononactinic acid. Arch Mikrobiol. 1972;85:233–238. [PubMed] [Google Scholar]
- 37.Pape H. Metabolic products of microorganisms. 97. Biosynthesis of macrotetrolides. I. Precursors of the carbon skeleton of nonactin. Arch Mikrobiol. 1972;82:254–264. [PubMed] [Google Scholar]
- 38.Plater R, Robinson J A. Cloning and sequence of a gene encoding macrotetrolide antibiotic resistance from Streptomyces griseus. Gene. 1992;112:117–122. doi: 10.1016/0378-1119(92)90312-d. [DOI] [PubMed] [Google Scholar]
- 39.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 40.Priestley N D. Studies on enzymes involved in the primary and secondary metabolism of antibiotic producing Streptomyces. Ph.D. thesis. Southampton, United Kingdom: University of Southampton; 1991. [Google Scholar]
- 41.Rajgarhia V R, Strohl W R. Minimal Streptomyces sp. strain C5 daunorubicin polyketide synthase genes required for aklanonic acid biosynthesis. J Bacteriol. 1997;179:2690–2696. doi: 10.1128/jb.179.8.2690-2696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Shimakata T, Fujita Y, Kusaka T. Involvement of one of two enoyl-CoA hydratases and enoyl-CoA reductase in the acetyl-CoA-dependent elongation of medium chain fatty acids by Mycobacterium smegmatis. J Biochem. 1980;88:1051–1058. doi: 10.1093/oxfordjournals.jbchem.a133056. [DOI] [PubMed] [Google Scholar]
- 44.Spavold Z M. Studies in antibiotic production. Ph.D. thesis. Southampton, United Kingdom: University of Southampton; 1987. [Google Scholar]
- 45.Spavold Z M, Robinson J A. Nonactin biosynthesis: On the role of (6R,8R)- and (6S,8S)-2-methyl-6,8-dihydroxynon-2E-enoic acids in the formation of nonactic acid. Chem Commun (J Chem Soc Sect D) 1988;1988:4–6. [Google Scholar]
- 46.Stahl P, Pape H. Metabolic products of microorganisms. 110. Biosynthesis of macrotetrolides. III. Isolation of free nonactinic acids and their function as precursors of macrotetrolides. Arch Mikrobiol. 1972;85:239–248. [PubMed] [Google Scholar]
- 47.Stahl P O. Untersuchungen zur Biosynthese der Makrotetrolide bei Streptomyces griseus. Ph.D. thesis. Tübingen, Germany: Eberhard-Karls-Universität Tübingen; 1975. [Google Scholar]
- 48.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright F, Bibb M. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 50.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 51.Zeelen J P, Hiltunen J K, Wierenga R K. Crystallization experiments with 2-enoyl-CoA hydratase, using an automated ‘fast-screening’ crystallization protocol. Acta Crystallogr Sect D. 1994;50:443–447. doi: 10.1107/S0907444994001277. [DOI] [PubMed] [Google Scholar]