Summary
Innate-like T cells, including invariant natural killer T (iNKT) cells, mucosal-associated invariant T (MAIT) cells and γδ T cells, are present in various barrier tissues, including the lung, where they carry out protective responses during infections. Here, we investigate their roles during pulmonary pneumococcal infection. Following infection, innate-like T cells rapidly increase in lung tissue, in part through recruitment, but TCR activation and cytokine production occur mostly in IL-17-producing NKT17 and γδ T cells. NKT17 cells are preferentially located within lung tissue prior to infection, as are CD103+ dendritic cells (cDC1), which are important both for antigen presentation to NKT17 cells and γδ T cell activation. Whereas IL-17-producing γδ T cells are numerous, GM-CSF is exclusive to NKT17 cells and is required for optimal protection. These studies demonstrate how particular cellular interactions and responses of functional subsets of innate-like T cells contribute to protection from pathogenic lung infection.
Keywords: T cell, innate, Streptococcus pneumoniae, lung infection, dendritic cell, natural killer T cell, γδ T cell
Introduction
The lung is a barrier tissue and contains populations of innate lymphoid cells (ILC) and innate-like T cells. Innate-like T cells carry out rapid activation and cytokine secretion, thereby bridging the innate and adaptive immune responses. They have a number of distinct properties, including the ability to respond to TCR stimulation or to cytokines in the absence of TCR signaling (Gutierrez-Arcelus et al., 2019). These lymphocyte types include invariant natural killer (iNKT) cells, γδ T cells and mucosal-associated invariant T (MAIT) cells. iNKT cells respond to glycolipid antigens presented by CD1d, a non-polymorphic MHC class I antigen presenting molecule. A number of bacteria have been shown to produce glycolipids recognized by iNKT cells, including pathogens such as Borrelia burgdorferi, group B Streptococcus, and Streptococcus pneumoniae (Chang et al., 2011; Kinjo et al., 2011; Kinjo et al., 2006; Mattner et al., 2005). γδ T cells are not exclusively specific for microbial antigens, although the most abundant population of γδ T cells in human blood recognizes pyrophosphate-containing microbial metabolites (Harly et al., 2012; Ribot et al., 2021; Sandstrom et al., 2014). MAIT cells recognize microbial riboflavin metabolites presented by MR1, a highly conserved and non-polymorphic MHC class I antigen presenting molecule. Many bacteria produce antigens that stimulate MAIT cells, including pathogens such as Mycobacterium tuberculosis, Francisella tularensis and S. pneumoniae (Hartmann et al., 2018; Le Bourhis et al., 2010; Meierovics et al., 2013). Several types of antigen presenting cells (APC) express CD1d and MR1, but considering iNKT cells, conventional dendritic cells type 1 (cDC1), marked by expression of CD8α or CD103, have most often been implicated in antigen presentation (Arora et al., 2014), although depending on the form of the lipid antigen, other APC types may be important (Barral et al., 2008; Kawasaki et al., 2013).
Innate-like T cells differentiate into functional subsets in the thymus. For example, thymic iNKT cells differentiate into NKT1, NKT2 and NKT17 cells (Lee et al., 2013; Watarai et al., 2012), analogous in their capacity to produce cytokines to Th1, Th2 and Th17 cells, respectively. Functional subsets of γδ T cells and MAIT cells that produce IFN-γ or IL-17 also are present in the thymus (Godfrey et al., 2015; Ribot et al., 2021). These populations likewise are found in peripheral tissues, including the lung, where they reside long-term (Khairallah et al., 2018; Murray et al., 2021; Salou et al., 2019; Thomas et al., 2011).
S. pneumoniae is a leading cause of pneumonia and meningitis in both children and the elderly (O’Brien et al., 2009; Wahl et al., 2018; Wroe et al., 2012). iNKT cell antigen-dependent responses to this microbe have been well characterized (Brigl et al., 2011; Girardi et al., 2011; Kinjo et al., 2011), and iNKT cells are essential for protection of mice from S. pneumoniae (Brigl et al., 2011; Kawakami et al., 2003; Kinjo et al., 2011). γδ T cells also have been shown to be important for protection from S. pneumoniae, with a role in the early recruitment of neutrophils (Hassane et al., 2017; Nakasone et al., 2007). Therefore, S. pneumoniae provides an example in which two types of innate-like T lymphocytes play non-redundant roles in protection from lung infection.
Although iNKT cells and γδ T cells are required for host defense, there is relatively little information on the mechanisms of activation, cell-cell contacts and immune responses underlying the role of innate-like T cells in host protection. Here, we addressed several issues pertaining to protective innate-like T cell responses to lung infection with S. pneumoniae. First, we sought to identify the underlying mechanisms leading to the rapid activation of protective iNKT and γδ T cell responses during S. pneumoniae infection, including the role of TCR activation of these cells. Second, we sought to identify a nonredundant function(s) of iNKT cells that would distinguish them from the more numerous γδ T cells that produce similar cytokines. Finally, we identified features of APC subsets important for activating iNKT cells and γδ T cells. Our data show how specialized responses of two populations of innate-like T cells, partially dependent on cDC1 activation and antigen presentation, provide host protection.
Results
Circulatory and tissue location of innate-like lung T cells
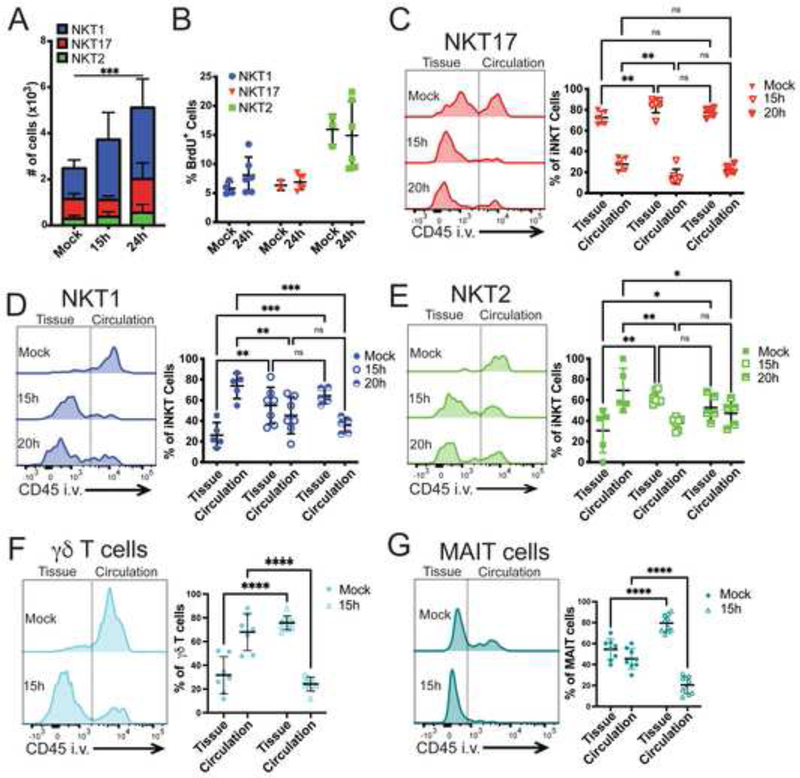
We analyzed the extent to which individual iNKT cell subsets, γδ T cells and MAIT cells were located in circulation, before and early after infection with S. pneumoniae. We utilized the S. pneumoniae strain URF918, a clinical isolate of the invasive serotype 3, because as shown previously, when C57BL/6J mice were infected with this strain, bacterial clearance and survival were highly dependent on the activation of iNKT cells (Brigl et al., 2011; Kawakami et al., 2003; Kinjo et al., 2011). Neutrophil recruitment was reported to be decreased in the absence of iNKT cells (Kawakami et al., 2003; Nakamatsu et al., 2007), which we confirmed (Supplementary Figure 1A). iNKT cell subsets were enumerated by flow cytometry using αGalCer-loaded CD1d tetramers together with iNKT cell subset-specific cell surface proteins (Supplementary Figure 1B) that have been shown to provide highly enriched populations (Engel et al., 2016; Murray et al., 2021; Zhao et al., 2018). At steady-state, the three prevalent iNKT cell subsets in the lungs were NKT1, NKT17, and NKT2 cells, in order of decreasing prevalence (Figure 1A). The number of lung iNKT cells significantly increased by 15 hours and further at 24 hours after infection, with the largest increase in NKT1 cells (Figure 1A). This was primarily due to recruitment, rather than cell proliferation, as shown by comparable BrdU incorporation in iNKT cells from mice analyzed at 24 hours after infection (Figure 1B). Despite increased BrdU incorporation, NKT2 cells remained a relatively minor subset after infection.
Figure 1: Innate-like T cells are recruited into the lung tissue after S. pneumoniae infection.
A) iNKT cell subset numbers in the lung before and after infection. N=7 mice per group, 2 independent experiments combined. Statistical significance was assessed via 2-way ANOVA, with Tukey’s multiple comparisons test. Source of variation: Interaction (*); Time (***), depicted in figure; Subset (****). NKT1 cells significantly expand at 15h (**) and 24h (****), Tukey’s multiple comparisons test. B) Percent of BrdU+ iNKT cell subsets in lung 24h after infection. N=6 mice per group, combined data from 2 independent experiments. C-G) Representative histograms and quantification of sub-tissue location of lung innate-like T cells, as determined by anti-CD45 antibody intravenous injection prior to tissue collection. Line separating tissue and circulation based on cell types from mice not injected with anti-CD45. Localization of NKT17 (C), NKT1 (D), and NKT2 (E); N=5–7 mice per group, combined data from 3 independent experiments. Statistical significance of NKT subset localization in Mock assessed via 2-way ANOVA, with Šídák’s multiple comparisons test. E &F) Sub-tissue location of lung γδ T cells (F) and MAIT cells (G); combination of 2 independent experiments, N=7–10 mice per group. Statistical significance assessed via 2-way ANOVA, with Šídák’s multiple comparisons test. See also Figure S1.
We analyzed the location of the iNKT cell subsets by intravenously injecting anti-CD45 antibody 3 minutes before tissue collection. This labels CD45+ cells located in circulation, and leaves cells in the interstitial tissue and alveoli unlabeled (Barletta et al., 2012; Reutershan et al., 2005). In agreement with a previous report (Salou et al., 2019), NKT17 cells had a significant presence in the lung, an average of more than 70% in the tissue, and this tendency was maintained or increased after infection (Figure 1C). In contrast, prior to infection, the majority of NKT1 and NKT2 cells were in the lung vasculature or circulation (Figure 1D, E). Within 15 hours of infection, however, a higher proportion of NKT1 and NKT2 cells were no longer in blood vessels (Figure 1D, E). Similarly, the majority of the lung γδ T cells were also in circulation but were recruited to the tissue by 15 hours (Figure 1F). Although MAIT cells were predominately located within the tissue at steady-state, the percentage of tissue MAIT cells also increased following infection (Figure 1G).
Differential activation of subsets
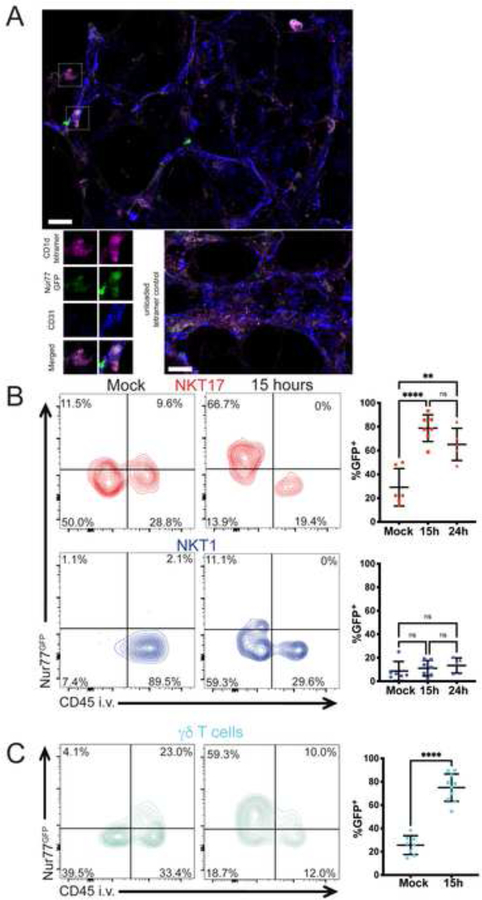
We determined if subsets of lung iNKT cells were activated and produced their signature cytokines. Previous research showed that iNKT cells produce cytokines as early as 12 hours after infection (Holzapfel et al., 2014; Kinjo et al., 2011), suggesting TCR activation occurs rapidly following infection in response to bacterial antigens that have been well characterized (Kinjo et al., 2011; Girardi et al., 2011). To measure T cell antigen receptor (TCR)-mediated activation, we immunized Nur77GFP mice, which express GFP under the control of the Nr4a1 (Nur77) promoter (Moran et al., 2011). Reporter expression has been shown to be a faithful readout of TCR activation in T lymphocytes, including iNKT cells (Holzapfel et al., 2014; Moran et al., 2011). We examined GFP expression in reporter mice by immunofluorescence staining of whole-mount vibratome sections of lung tissue from mice infected 14 hours earlier (Figure 2A, Supplementary Video 1). Although iNKT cells were infrequent, and therefore not present in every field, some cells stained brightly with the CD1d tetramer. Only duller staining was observed in sections stained with an unloaded CD1d tetramer. Many of the CD1d-tetramer+ cells also were positive for the GFP reporter, suggesting that some iNKT cells received a TCR signal following infection. Blood and lymphatic vessels express CD31 and iNKT cell staining in relation to CD31 staining suggests that most iNKT cells were outside of vessels following infection.
Figure 2: NKT17 and γδ T cells receive TCR signals after infection.
A) Immunofluorescence staining of lung tissue in Nur77GFP reporter mice 14 hours post-infection. CD31 (blue): blood vessels; CD1d-αGalCer-loaded tetramer (purple): iNKT cells; GFP (Green) indicating TCR reporter expression. Top: boxes indicate iNKT cells. Lower left: magnified view of the cells above, including one that is GFP+ and one that is not. Lower right: unloaded CD1d tetramer staining. Scale bar: 20 μm. B) Left: representative flow cytometry plot of sub-tissue location of TCR-activated (Nur77GFP+) lung NKT17 and NKT1 cells at 15 hours after infection. Right: Quantification of GFP+ cells of panel A at 15 and 24 hours after infection. N=5–7 mice per group, 3 independent experiments. **p=.0014, ****p<0.0001 (One-way ANOVA with Tukey’s multiple comparisons). C) Left: representative flow cytometry plot of sub-tissue location of TCR-activated (Nur77GFP+) lung γδ T cells at 15 hours after infection. Right: Quantification of GFP+ cells at 15 hours after infection. N= 8–10 mice per group, combined data from 3 independent experiments. ****p<0.0001 (unpaired t test). See also Figure S1.
To achieve quantitative data, we used flow cytometry and focused on the more abundant lung subsets, NKT1 and NKT17 cells. There was a significant GFP signal in unstimulated NKT17 cells, consistent with previous data indicating that a minority of lung iNKT cells showed signs of constitutive activation (Murray et al., 2021). By 15 hours, the majority of NKT17 cells were GFP+, and these reporter positive cells were inaccessible to the CD45 antibody and therefore were outside of circulation (Figure 2B). Very few NKT1 cells were GFP+ and the mean fluorescence intensity of the few GFP+ cells was lower, although these TCR-activated NKT1 cells were also unlabeled by anti-CD45 (Figure 2B). Therefore, NKT17 cells were the predominant iNKT cell subset receiving a TCR signal at early times after infection, and TCR responses correlated with their extravascular location. An increased percentage of γδ T cells also became GFP+ after infection, although like NKT17 cells, they also showed evidence for constitutive activation (Figure 2C). Additionally, MAIT cells received a TCR signal, but a considerably lower percentage of MAIT cells became GFP+ compared to NKT17 cells and γδ T cells (Supplementary Figure 1C).
Diverse S. pneumoniae strains have iNKT cell antigens
The URF918 strain of S. pneumoniae is a clinical isolate that is highly virulent in mice (Kinjo et al., 2011), and therefore we determined if other strains have antigens that activate iNKT cells. Production of antigens was analyzed in an APC-free, hybridoma stimulation assay. In this assay, total bacterial sonicates were loaded onto CD1d coated plates and TCR-mediated activation was measured with IL-2 secretion. Antigenic activity was observed in the D39 strain (serotype 2), a laboratory standard, as well as in some strains from a group of clinical isolates of serotype 19A, including those that caused invasive infections (Supplementary Figure 2A, B, Supplementary Table 1). Therefore, these data suggest that diverse clinical isolates of S. pneumoniae are likely to have antigens that activate iNKT cells. There was variability in antigenic content, however, as we observed previously for MAIT cell antigens in these strains (Hartmann et al., 2018).
Role of cytokines in protection
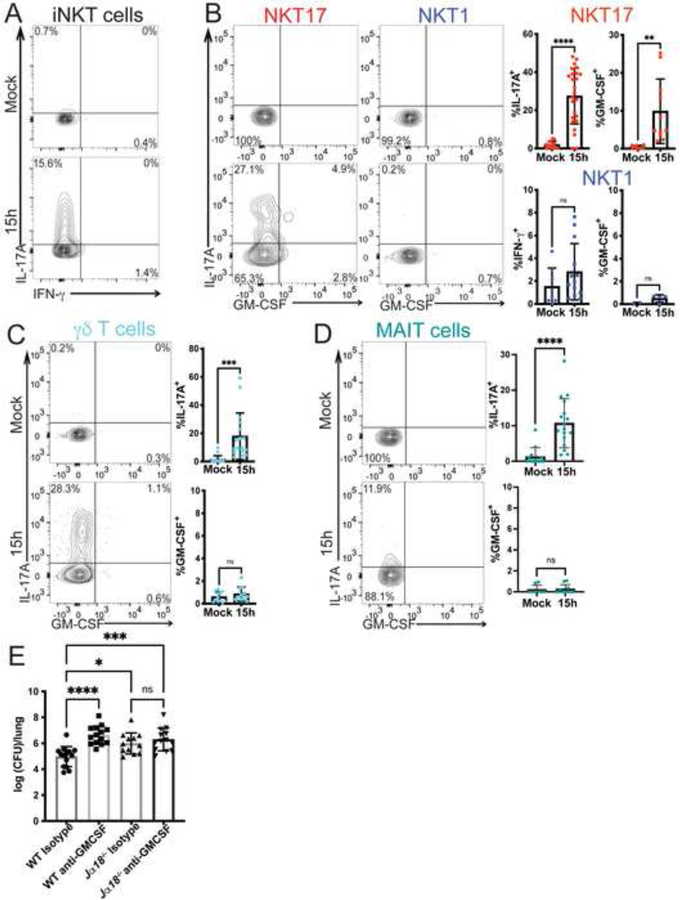
We assessed cytokine production by innate-like T cells following S. pneumoniae infection by intracellular cytokine staining. Cells were analyzed ex vivo after a brief culture, without further TCR re-stimulation. At 15 hours post-infection, a substantial fraction of the total population of iNKT cells produced IL-17A while a relatively small but variable percentage produced IFN-γ (Figure 3A, Supplementary Figure 3A). Considering the NKT17 subset, it expressed both IL-17A and GM-CSF (Figure 3B), including double (IL-17+, GM-CSF+) and single producers of these cytokines. NKT1 cells did not produce GM-CSF or IL-17A (Figure 3B). As previously reported, we confirmed that both IL-17A and GM-CSF were required for protection from S. pneumoniae infection (Brown et al., 2017; Ivanov et al., 2012; Zhang et al., 2009), with mice deficient in either of these cytokines displaying an increase in bacterial burden in the lung (Supplementary Figure 3B).
Figure 3. Innate-like T cells produce cytokines following infection.
A) Representative plot of cytokine production by lung iNKT cells at 15 hours after infection with URF918. B) Left: Representative plot of cytokine production by NKT17 and NKT1 cells 15 hours after infection with URF918. Right: quantification of cytokine production by lung NKT1 cells and NKT17 cells. IL-17A, IFN-γ: N=8–25 mice per group, 5 independent experiments. GM-CSF: N=9–11 mice per group, representative data of 4 independent experiments; *p=.0038, ****p<0.0001 (unpaired t test). C & D) Cytokine production by γδ T cells (C) and MAIT cells (D) 15 hours post infection with URF918. Representative flow cytometry plots on left, quantitation on right. C) N=11–17 mice per group, combined from 4 independent experiments, *** p = 0.0004, unpaired t test. D) IL-17A: N=13–17 mice per group, combined from 4 independent experiments; GM-CSF: N=6–10 mice per group, 2 independent experiments, **** p<0.0001, unpaired t test. E) Bacterial burdens in lung (16 hpi) of C57BL/6J mice and Jα18−/− mice treated with an isotype control (Rat IgG2a) or anti-GM-CSF antibody and infected with URF918. Combined from 3 independent experiments, N=12–14 mice per group; one-way ANOVA with Tukey’s multiple comparisons test, **** P<0.0001, * P = 0.0112, **** P = 0.0004. See also Figure S3.
γδ T cells and MAIT cells also rapidly increased IL-17A synthesis following infection, but neither cell type produced GM-CSF in response to S. pneumoniae infection (Figure 3C, 3D) (Hassane et al., 2020; Ivanov et al., 2014). MAIT cells were relatively infrequent, and analysis of Mr1−/− mice indicated they were not essential for protection (Supplementary Figure 3C). IL-17A-producing γδ T cells were more numerous after S. pneumoniae infection than iNKT cells (approximately 15,000 γδ T cells compared to less than 1,000 NKT17 cells following infection), suggesting that IL-17A production alone by iNKT cells would not account for their requirement for host protection.
Considering the strong effect on susceptibility of GM-CSF deficiency (Supplementary Figure 3B), the importance of neutrophil recruitment and activity for defense from S. pneumoniae (Garvy and Harmsen, 1996), and the early production of GM-CSF uniquely by iNKT cells, these data suggest that production of GM-CSF may be one factor responsible for the non-redundant function of iNKT cells. To determine if iNKT cell GM-CSF was required, we treated C57BL/6J mice and Jα18-deficient mice, lacking iNKT cells, with an anti-GM-CSF antibody prior to infection. If GM-CSF was required for iNKT cell-mediated protection, then blockade of GM-CSF in the absence of iNKT cells should have no effect on bacterial burden. Indeed, we found that C57BL/6J mice treated with an anti-GM-CSF antibody had an increased bacterial burden following infection compared to isotype-treated C57BL/6J control mice (Figure 3E). Jα18-deficient mice treated with the anti-GM-CSF or isotype control antibody, however, had similar bacterial burdens (Figure 3E), suggesting that iNKT cell production of GM-CSF contributed to protection from infection.
Differential responses of lung DC subtypes to infection
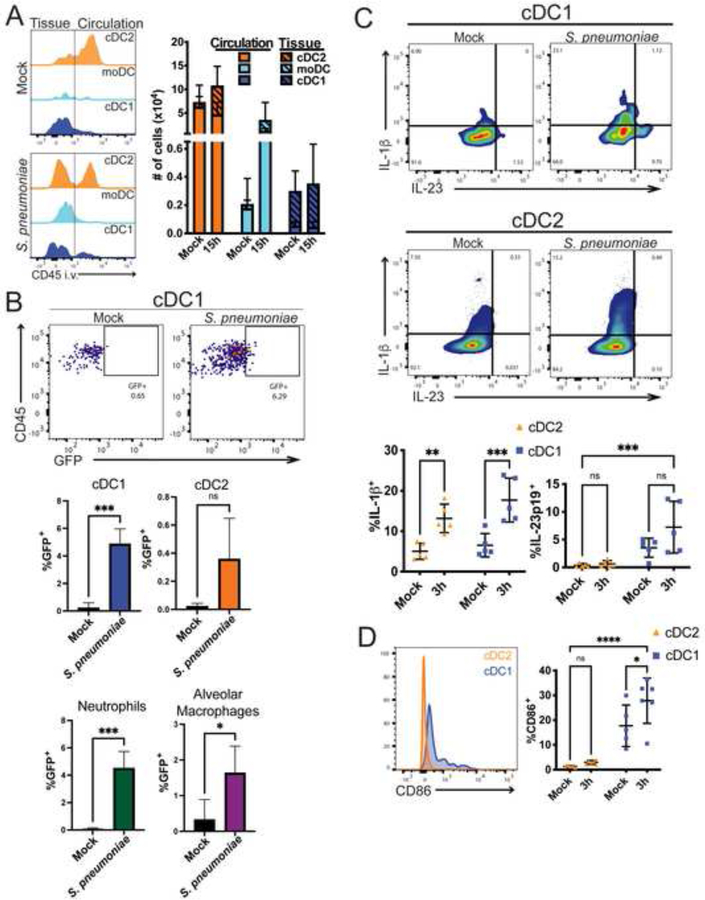
The lung contains several types of APCs that could participate in iNKT cell activation during infection. As shown in Figure 4A, prior to infection, cDC2 cells, gated as in Supplementary Figure 4A, were in both the lung tissue and circulation, while the less numerous CD103+ or cDC1 cells were almost exclusively outside of circulation and presumably within the tissue. By 15 hours after infection, there was a significant increase in cDC2 cells in both the tissue and circulation. There also was an influx of CD64+ monocyte-derived dendritic cells (moDCs) into the lung tissue, which were rare in uninfected mice. All three DC subsets and alveolar macrophages expressed surface CD1d and thus could have the capacity to present antigen to iNKT cells (Supplementary Figure 4B).
Figure 4: DC responses in the lung following infection.
A) Left: representative histograms of sub-tissue location of dendritic cell subtypes at 0 and 15 hours after infection. Line depicting tissue vs. circulatory localization drawn based on each cell type from mice not injected with anti-CD45. Right: Quantification of cells in tissue (hashed bars) and circulation (open bars) at 0 and 15 hours after infection. N=11–15 mice per group, 4 independent experiments combined. B) Mice were infected with GFP-expressing URF918 and euthanized 2 hpi. Bacterial uptake by lung APCs was assessed via flow cytometry. Top: Representative flow cytometry of cDC1. Bottom: Quantification. N=3–6 mice per group, combined from 3 independent experiments; statistical significance assessed via unpaired t test. C) Representative flow cytometry plot of cytokine production by lung cDC1 and cDC2 cells 3 hpi with URF918. Quantification below. Statistical significance was assessed via 2-way ANOVA, with Šídák’s multiple comparisons test. D. CD86 expression by cDC1 and cDC2 cells 3 hpi with URF918. Quantification on right. D & E. N = 5 mice per group, 2 independent experiments. Statistical significance was assessed via 2-way ANOVA, with Šídák’s multiple comparisons test. See also Figure S4.
To identify APCs that could be important for defense from S. pneumoniae, we tested the capacity of lung myeloid cell types to take up bacteria. Mice were infected with GFP-expressing S. pneumoniae and 2 hours post-infection, bacterial uptake was assessed by immunofluorescence and flow cytometry. For immunofluorescence analysis, we analyzed Langerin-YFP mice in which cDC1 cells express YFP (Ghigo et al., 2013; Zahner et al., 2011). In tissue sections of infected Langerin-YFP mice, some S. pneumoniae puncta (white pseudo color) were co-localized with cDC1 cells (purple pseudo color) (Supplementary Video 2). Considering S. pneumoniae is an extracellular bacterium, the bacteria and GFP signal might be degraded rapidly on uptake. This might account for the observation that more of the signal is from the surface rather than in the DC. By flow cytometry, we found a significant increase in GFP+ cDC1 cells (Figure 4B), as well as GFP+ neutrophils and alveolar macrophages. These data suggest that several myeloid cell types rapidly take up or bind to bacteria following infection.
iNKT cell production of IL-17A may be enhanced by either IL-23 or IL-1β, alone or in combination (Doisne et al., 2011; Rachitskaya et al., 2008; St Leger et al., 2018). Further, IL-23 has been shown to stimulate GM-CSF production by CD4+ Th17 cells, although it is not known if this is true for iNKT cells (Codarri et al., 2011; El-Behi et al., 2011; Rothchild et al., 2014). We therefore analyzed cytokines and cell surface molecules expressed by cDC1 and cDC2 that might be important for amplifying iNKT cell protective responses (Figure 4C). Pro-IL-1β production was increased in both cDC2 and cDC1 cells at 3 hours after infection (Figure 4C). IL-23 has been shown to be important for defense against S. pneumoniae (Kim et al., 2013), and IL-23p19 was produced mainly by cDC1 cells. These cells also expressed much higher levels of the costimulatory molecule CD86 (Figure 4D). Due to their localization outside the vasculature, CD1d expression, bacterial uptake, cytokine production and co-stimulatory molecule expression, these data suggest that cDC1 may be an important subset required to stimulate the protective anti-microbial response in the lung.
cDC1 activate lung T cells
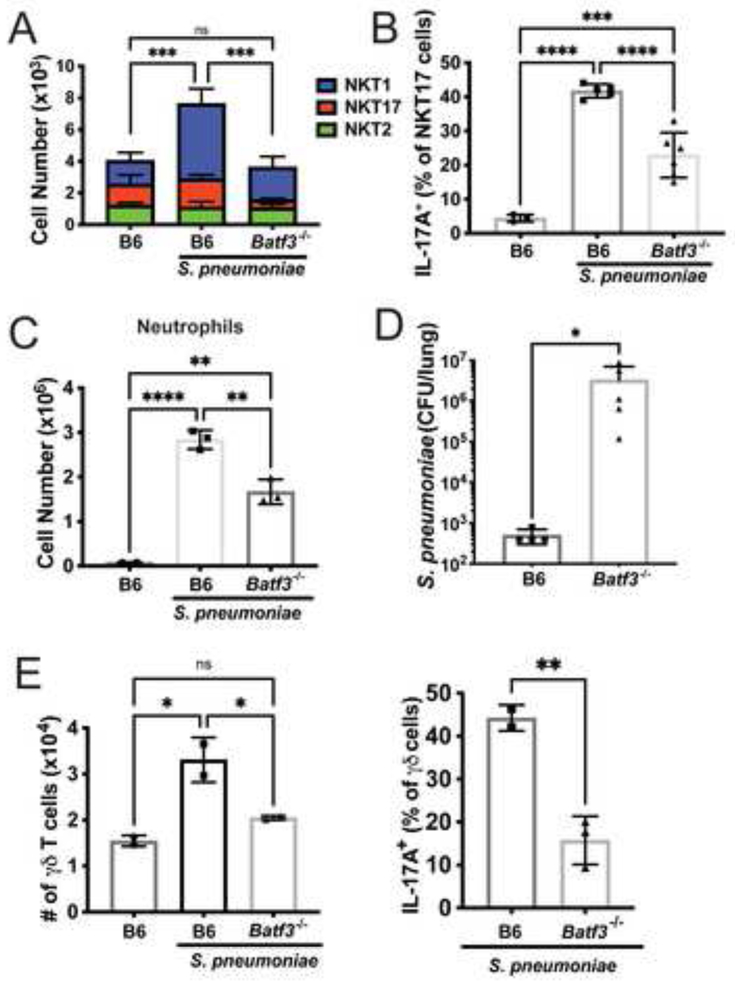
The bacterial load was increased in S. pneumoniae-infected Cd1d1f/f x CD11c-Cre+ mice that lack CD1d in all dendritic cells (Supplementary Figure 4C), suggesting the importance of DCs in controlling infection. To address the hypothesis that cDC1 are most important, we infected Batf3−/− mice, which lack these cells due to deletion of an essential transcription factor (Grajales-Reyes et al., 2015; Hildner et al., 2008; Seillet et al., 2013). When Batf3−/− mice were infected with S. pneumoniae, iNKT cells did not expand, and there was a 50% reduction in the frequency of IL-17A producing NKT17 cells compared to C57BL/6J mice (Figure 5A, B), similar to Cd1d1f/f x CD11c-Cre+ mice (Supplementary Figure 4C). This directly correlated with a reduction in neutrophil recruitment at 15 hours (Figure 5C), and an increase in bacterial burdens after 2 days (Figure 5D), altogether suggesting cDC1 cells are important for iNKT cell activation and protection from S. pneumoniae. Analysis of Batf3−/− mice revealed that expansion of γδ T cells, as well as cytokine production by these cells, was also dependent on cDC1 cells (Figure 5E).
Figure 5: Batf3−/− mice have reduced innate-like T cell responses.
A) iNKT cell numbers at 15 hours after infection in C57BL/6J and Batf3−/− mice. N=6 per group. ***p<0.001, One-way ANOVA, with Tukey’s multiple comparisons. B) IL-17A production by NKT17 cells at 15 hours after infection. N=6 per group, ***p=.0004, ****p<0.0001 (One-way ANOVA with Tukey’s multiple comparisons). C) Lung neutrophil numbers at 15 hours after infection. N=6 per group. ****p<0.0001 (One-way ANOVA with Tukey’s multiple comparisons). D) Bacterial loads at 2 days. N=5 per group, *p=0.0159 (Mann-Whitney Test). E) γδ T cells numbers (left) and IL-17A production (right) at 15 hours after infection in C57BL/6J and Batf3−/− mice. N=2–3 mice per group, 1 experiment. Left: One-way ANOVA with Tukey’s multiple comparisons; Right: ** p= 0.0078, unpaired t test.
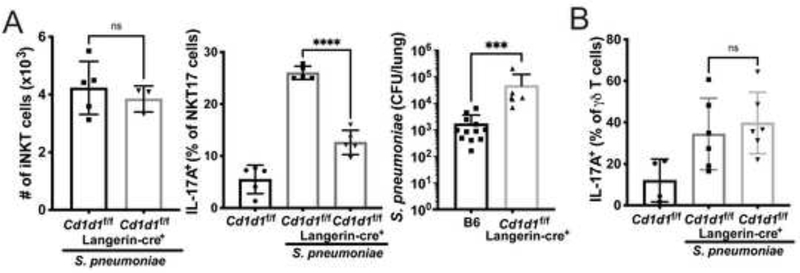
To determine if protection was dependent on direct stimulation of iNKT cells and/or γδ T cells by antigens presented by CD1d, we utilized mice with conditional deletion of Cd1d1 on cDC1 cells (Langerin-Cre+) (Zahner et al., 2011). The specificity of the deletion on populations of lung myeloid cells is shown in Supplementary Figure 4B. Deletion of Cd1d1 had no effect on iNKT cell recruitment (Figure 6A, left), suggesting that innate immune activation of cDC1 and other cell types likely leads to the synthesis of chemokines that recruit iNKT cells in the absence of antigen presentation. Loss of CD1d expression by cDC1 significantly reduced iNKT cell-derived IL-17A (Figure 6A, center), however, and led to increased bacterial loads at 2 days (Figure 6A, right). Therefore, cDC1 cells not only participated in recruiting iNKT cells in a CD1d-independent fashion, but they activated these lymphocytes through CD1d antigen presentation. In contrast, IL-17A production by γδ T cells was not reduced in mice lacking CD1d specifically on cDC1 cells (Figure 6B), and therefore TCR activation of these cells must have depended on different antigens that were not dependent on CD1d.
Figure 6: Dendritic cells deficient in CD1d have reduced iNKT cell responses.
A) LangerincreCd1d1f/f mice following infection with URF918. Left: total iNKT cell numbers at 15 hours post-infection. N=5 per group. Middle: IL-17A production by NKT17 cells at 15 hours after infection. N=5 per group. ****p<0.0001 (One-way ANOVA with Tukey’s Multiple Comparisons). Right: Bacterial loads 2 days after infection. ***p<0.0001 (Mann-Whitney Test). B) IL-17A production by γδ T cells in Cd1d1f/f × Langerin-Cre+ mice following URF918 infection (One-way ANOVA with Tukey’s Multiple Comparisons). See also Figure S4.
Discussion
Here we have identified the cellular interactions and the means by which innate-like T cells participate in protection from lung infection with an important pathogen, S. pneumoniae. Although γδ T cells are important, here we highlight the non-redundant role of GM-CSF production by NKT17 cells that are stimulated by antigens presented by cDC1. Lung infection of mice with strain URF918 S. pneumoniae has provided a striking example in which the response of iNKT cells has proven to be absolutely essential for controlling CFU, beginning as early as 24 hours (Brigl et al., 2011; Kawakami et al., 2003; Kinjo et al., 2011). In this case, the protective role of iNKT cells is elicited very rapidly, with kinetics similar to features of the innate immune response. In contrast, although S. pneumoniae has an antigen that activates MAIT cells (Hartmann et al., 2018), and they produced cytokines rapidly after infection, we did not find an essential role for MAIT cells, perhaps due to their lower cell numbers in the lung. γδ T cells also are essential for host defense (Hassane et al., 2017; Nakasone et al., 2007), however, and they also are activated within 16h to produce cytokines such as IL-17 (Hassane et al., 2020). Previous research showed IL-17 could be detrimental or protective, depending on the infecting S. pneumoniae strain (Ritchie et al., 2018). Regardless, the early production of IL-17 by γδ T cells is correlated with the recruitment of neutrophils, a critical feature of the protective response (Hassane et al., 2017; Hassane et al., 2020). The requirement for both innate-like T cell types raises the issue as to how the contributions of iNKT cells and γδ T cells differ.
Consistent with a requirement for early responses, iNKT and γδ T cells are present in the lung and in a partially activated state (Murray et al., 2021). Furthermore, the number of both cell types rapidly increased in the lung after infection. Although the majority of both iNKT and γδ T cells were in blood vessels prior to infection, they were found predominantly outside circulation, and likely in the tissue parenchyma, by 15 hours. For iNKT cells, however, there is evidence for a TCR signal even earlier, by 5 hours (Holzapfel et al., 2014), and intracellular cytokine could be detected directly ex vivo by 12 hours (Holzapfel et al., 2014; Kinjo et al., 2011). Therefore, despite their rapid recruitment from circulation, some of the very earliest and perhaps critical responses by iNKT cells may be carried out by those iNKT cells already residing outside the vasculature. Lung blood vessel localization prior to infection, however, was not an impediment for rapid TCR activation of lung γδ T cells. Furthermore, there was evidence for constitutive activation of some γδ T cells, perhaps by products derived from the commensal microbiota.
Following S. pneumoniae infection, γδ T cells produce IFN-γ and IL-17A (Hassane et al., 2020), and as noted, their role in protection has been linked to IL-17A production (Hassane et al., 2017; Hassane et al., 2020). Like γδ T cells, iNKT cells have been shown to participate in the protective response to a variety of infections, but in the majority of cases, protection was related to IFN-γ secretion by these cells (Kinjo et al., 2013). Even in other contexts, such as amelioration of arthritis, NKT1 cells were shown to be the effective population, which may in part reflect the prevalence of these cells in C57BL/6J mice (Zhao et al., 2018). Here our evidence suggests that the NKT17 cells are uniquely important for protection. The population of iNKT cells prior to infection within lung tissue were predominantly NKT17 cells, in agreement with earlier reports (Lee et al., 2015; Salou et al., 2019). Therefore, tissue residence and early immune responses by NKT17 cells were correlated. Lung NKT1 cells are not exhausted, however, because after priming with αGalCer, they produced IFN-γ which protected from infection with S. pneumoniae along with IL-17A (Ivanov et al., 2012). Furthermore, in several earlier studies (Brigl et al., 2011; Holzapfel et al., 2014; Ivanov et al., 2012; Kinjo et al., 2011; Nakamatsu et al., 2007) there was evidence for IFN-γ production by iNKT cells after S. pneumoniae infection, although the percentage of cytokine-producing cells varied. We do not discount the importance of NKT1 cells and IFN-γ in our mice. The influx of NKT1 cells recruited to the lung and NKT1 responses later than 24h could have protective roles.
Considering that the capacity for IL-17A synthesis is found in several types of innate or innate-like cells, including ILC3 (Cua and Tato, 2010; Rosine and Miceli-Richard, 2020) as well as T cells, there could be functional redundancy for these cell types. In agreement with a previous publication (Hassane et al., 2020), we found that lung γδ T cells were more numerous than iNKT cells, and a higher percentage of them produced IL-17A compared to iNKT cells after S. pneumoniae infection. Therefore, it seemed unlikely that the nonredundant function of iNKT cells was due solely to IL-17A secretion, although differences in neighboring cells, combined with differences in other signals, could provide for a divergence in the effects of iNKT and γδ T cell IL-17A. Some NKT17 cells in fact produce other cytokines including GM-CSF and IL-22 (Coquet et al., 2008; Doisne et al., 2011; Paget et al., 2012). The importance of GM-CSF synthesis by iNKT cells in controlling bacterial infections was identified following Mycobacterium tuberculosis infection, although production was not attributed to a particular iNKT cell subset (Rothchild et al., 2014). The requirement for GM-CSF in the response to S. pneumoniae was confirmed here, in agreement with previous data indicating the importance of GM-CSF (Brown et al., 2017; Steinwede et al., 2011). Among T cells, early GM-CSF production was specific to NKT17 cells. Although there are other potential sources of GM-CSF (Yamamoto et al., 2014), antibody blocking of GM-CSF was effective in reducing lung S. pneumoniae CFU only in wild type mice, but not in Traj18−/− mice, supporting a role for GM-CSF from NKT17 cells.
We determined that cDC1 cells were one important APC-type for activating innate-like T cells and optimal protection. Mice lacking this subset due to deletion of Batf3 had a reduced number of lung iNKT and γδ T cells after infection and a decreased percentage of these cells producing IL-17A, with a concomitant reduction in neutrophil recruitment and increased lung CFU. In a model in which αGalCer was administered prior to S. pneumoniae infection to elicit protective responses, lung cDC1 cells were implicated (Ivanov et al., 2012). Additionally, deletion of the gene encoding CD1d in cDC1 cells did not affect iNKT cell recruitment but led to a reduction in iNKT cell activation to produce IL-17A and an increased bacterial load. By contrast, γδ T cell cytokine production was not decreased, indicating these cells are dependent on the cDC1 cell type, likely for TCR activation, and perhaps the cytokines they produce, but they are not dependent on their CD1d-mediated presentation of either foreign antigen or perhaps self-ligand. The specificity of the responding γδ T cells and whether there is a requirement for antigen presentation remains unknown, although S. pneumoniae has a well-characterized foreign antigen for iNKT cells (Kinjo et al., 2011). Other myeloid populations also express CD1d and/or produce activating cytokines such as IL-1β, and therefore one or more of these populations could be responsible for the remaining iNKT cell and γδ T cell activation. This could occur either through the production of cytokines or other proteins, or in the case of iNKT cells, because of CD1d antigen presentation.
In summary, this study identified previously unknown mechanisms by which innate-like T cells, in part due to interactions with cDC1 cells, protect mice from lung infection with S. pneumoniae. For iNKT cells, TCR-mediated GM-CSF production by NKT17 cells, which are pre-located largely within lung tissue, was important. After infection, γδ T cells rapidly left circulation and produced cytokines including IL-17A, and while their activation depended on cDC1, it did not depend on their expression of CD1d. Therefore, the signals inducing γδ TCR activation remain unknown. Importantly, all of these T cell and DC populations are relatively infrequent in the lung, emphasizing the importance of rare and specialized cell types in protection from S. pneumoniae and potentially other infections. MAIT cells are more abundant in humans than in mice. Intriguingly, MAIT cell numbers increased in nasal biopsies from human subjects challenged with S. pneumoniae (Jochems et al., 2019), and IL-17A-producing MAIT cells were increased in children with community acquired pneumonia (Lu et al., 2020). Therefore, it is possible that similar mechanisms dependent on innate-like T cells apply to human lung bacterial infections, although with the more prevalent MAIT cells providing immune support.
Limitations of the Study
A limitation of this study is that although all the serotypes of S. pneumoniae that have been tested have an antigen(s) that activates iNKT cells, we have only analyzed a single invasive strain in vivo. It is possible that the requirement in vivo for a response by iNKT cells for host defense may be restricted to those strains that are more virulent (Barthelemy et al., 2016; Ivanov et al., 2012; Kawakami et al., 2003; Kinjo et al., 2011). Furthermore, we have only tested a single strain of mice, C57BL/6J, although in FVB strain mice we showed that optimal antigen presentation and activation of iNKT cells was correlated with survival from infection (Chandra et al., 2018). Additionally, it is possible that other cytokines and chemokines that we have not tested contribute to the non-redundant function of iNKT cells.
STAR Methods
Resource availability
Lead contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Mitch Kronenberg (mitch@lji.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Animals
Mice were bred and housed under specific pathogen-free conditions in the vivarium of the La Jolla Institute for Immunology (La Jolla, CA). Age and gender matched, male and female mice ages 8–16 weeks were used in all experiments. Inbred C57BL/6J, Batf3−/−, GM-CSF−/− (Csf2−/−) and Itgax-cre (Cd11c-cre) mouse strains were purchased from Jackson Laboratory (Bar Harbor, ME). Cd1d1f/f mice (Birkholz et al., 2015), Traj18−/− mice (Chandra et al., 2015), and Langerin-Cre knockin mice (Zahner et al., 2011) were generated as described previously. Nur77GFP mice were a gift from Kristin A. Hogquist (University of Minnesota, Minneapolis, Minnesota, USA) (Moran et al., 2011). All procedures were carried out under the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC) and approved by the La Jolla Institute for Immunology Institutional Animal Care and Use Committee (IACUC).
Streptococcus pneumoniae strains
S. pneumoniae serotype 3 strain URF918 is a clinical isolate originally from Japan (Kawakami et al., 2003). GFP-Streptococcus pneumoniae was generated in URF918 as previously described, using the construct graciously provided by Jan-Willem Veening (Kjos et al., 2015). S. pneumoniae serotype 19A isolates, used for in vitro iNKT cell activation experiments, originated from the sites listed in Supplementary Table 1.
Method details
Streptococcus pneumoniae infection
For mouse infections, URF918 was cultured in Todd-Hewitt broth (BD Biosciences) plus yeast extract at 37°C in an incubator at 5% CO2, collected at a mid-log phase and washed twice in PBS. For pulmonary infection, mice were anesthetized with isoflurane and elevated on a board. Mice were inoculated with S. pneumoniae (1–3×106 colony-forming units in a volume of 50 μl per mouse) by insertion of a pipet tip into the trachea. For calculation of total lung bacterial burden, tissues were collected at day 2 after infection and were homogenized in PBS to assess bacterial burden. Homogenates were inoculated at different dilutions in a volume of 50 μl on 5% sheep blood agar plates (Hardy Diagnostics, Santa Maria, CA) and cultured for 18 h, followed by counting of colonies. Note that the CFU in WT mice varied in different experiments and comparisons are therefore generally only valid within an experiment. For blocking experiments, mice were treated with 10 μg purified NA/LE Rat IgG2a isotype control antibody (BD Biosciences, San Diego, CA) or Ultra-LEAF purified anti-mouse GM-CSF antibody (BioLegend, San Diego, CA), as previously described (Brown et al., 2017). Lungs were harvested 16 hours post-infection. For bacterial uptake, mice were infected with 107 GFP- S. pneumoniae as described above and lungs were harvested 2 hpi and processed as described for flow cytometry or immunofluorescence.
Processing of lung tissue for flow cytometry
Mouse lungs were removed and rinsed with RPMI+10% FBS. The lungs were placed in a GentleMacs C tube (Miltenyi Biotec, Bergisch Gladbach, Germany) with 2 mL STEMCELL Spleen Dissociation Medium (STEMCELL Technologies, Vancouver, BC, Canada), and homogenized using the Miltenyi GentleMacs dissociator for 30 minutes. The cell suspensions were filtered with a 70 μm filter and washed with RPMI+10% FBS. For intracellular cytokine experiments, the cell suspensions were cultured with Golgi-Stop and Golgi-Plug (BD Biosciences) in 2 mL RPMI+10% FBS for 2 hours. Cell suspensions were washed and stained for flow cytometry.
Discrimination of tissue and circulating cells
Mice were anesthetized with isofluorane and injected retro-orbitally with 3 μg of Alexafluor700-labeled anti-CD45 antibody (30-F11), as described previously (Barletta et al., 2012; Reutershan et al., 2005). After 3 minutes, the lungs were removed for processing.
BrdU incorporation assay
At the time of infection, mice were injected intraperitoneally with BrdU. Lung tissues were collected at 24 hours and stained for surface markers and BrdU according to the manufacturer’s protocol (BD Biosciences).
Flow cytometry
For staining of cell surface molecules, cells were suspended in staining buffer (PBS, 2% BSA) and stained with fluorochrome-conjugated antibodies at a concentration of 1:200 for 20 min in a total volume of 50 μl. FcγR-blocking Ab anti-CD16/32 (2.4G2) was added to prevent nonspecific binding. Cells were fixed with 2% formaldehyde for 30 minutes on ice. For intracellular cytokine staining, cells were permeabilized with diluted ThermoFisher 10x permeabilization buffer for 5 minutes and stained overnight at 4° C in 1x permeabilization buffer. Cells were washed thoroughly, analyzed in an LSR II flow cytometer or Fortessa (BD Biosciences), and data were processed with Flow Jo software (Tree Star, Ashland, OR).
iNKT cells were stained using tetramers of CD1d loaded with αGalCer (BV421, in house preparation), live/dead yellow (ThermoFisher Scientific), anti-TCRβ-APC-eF780 (H57–597, ThermoFisher Scientific), anti-CD8α-BV605 (53–6.7, BioLegend) and anti-CD19-BV605 (1D3, BD Biosciences), anti-CD4-AF700 (GK1.5, BioLegend), anti-ICOS-Pe Cy7 (C398.4A, BioLegend), anti-CD49a-BV711 (HA31/8, BD Biosciences). For intracellular cytokine staining the following antibodies were used: anti-GM-CSF-PE (MP1–22E9, BioLegend); anti-IL-17A (TC11–18H10.1, BioLegend); anti-IFN-γ (XMG1.2, BioLegend). iNKT cell subsets were gated as follows (Supplementary Figure 1B): live lymphocytes, singlets, CD8−CD19−, CD45+, Tetramer+TCRβ+ iNKT cells and separated into NKT1, NKT2, and NKT17 cell subsets based on the following expression profiles: NKT1: CD49a+ICOS−; NKT2: CD49−ICOS+CD4+; NKT17: CD49a−ICOS+CD4−.
Staining of antigen presenting cells used the following reagents, gating strategy shown in Supplementary Figure 4A: live/dead yellow (ThermoFisher Scientific), anti-CD45-BV786 (30.F11, BD Biosciences); anti-siglecF-BV421 (E50–2440, BD Biosciences); anti-Ly6G-PE (1A8, BioLegend); anti-CD11b-PerCP-Cy5.5 (M1/70, BD Biosciences); anti-CD11c-APC (N418, eBioscience); anti-CD103-BV711 (M290, BD Biosciences); anti-CD24-FITC (M1/69, BD Biosciences); anti-CD64-PE Cy7 (X54–5/7.1, BioLegend); anti-CD86 (GL-1, BD Biosciences). For intracellular cytokine staining, anti-IL-23p19-AF488 (fc23cpg, Invitrogen); anti-IL-1β-APC (NJTEN3, eBioscience). Following selection for single, live, CD45+ cells, antigen presenting cells were gated as follows (Supplementary Figure 4A): cDC1: Ly6G−SiglecF−CD11c+CD103+; cDC2: Ly6G−SiglecF−CD11c+CD103−CD11b+CD64−CD24+; moDC: Ly6G−SiglecF−CD11c+CD103−CD11b+CD64+CD24+; alveolar macrophages: SiglecF+Ly6G−CD11c+; neutrophils: Ly6G+CD11b+.
MAIT cells were stained using 5-OP-RU-MR1-Tetramer or 6-FP-MR1-Tetramer (NIH Tetramer Core). Single cell suspensions were stained with tetramers for 40 min at room temperature. Cells were washed twice with staining buffer then incubated with antibodies for further surface staining. γδ T cells and MAIT cells were gated as follows: live lymphocytes, singlets, CD11b−CD19−CD45+γδTCR+ (anti-TCR γδ-FITC, clone GL3; γδ T cells); live lymphocytes, singlets, CD11b−CD19−CD45+γδ TCR−TCRβ+5-OPRU Tetramer+ (MAIT cells). Cytokine staining/antibodies described above.
Confocal microscopy
For bacterial uptake, Langerin-YFP+ mice were infected with 107 GFP- S. pneumoniae as described above and euthanized 2 hpi. Lungs were inflated with low-melting point agarose and processed into 300 μm sections with a Vibratome. Sections were individually placed in a multi-well plate, washed, and fixed with paraformaldehyde, washed, and embedded in Prolong Glass anitfade (ThermoFisher Scientific) under #1.5 coverslip.
Nur77GFP mice were inoculated with S. pneumoniae as described above and euthanized after 14h. Lungs were inflated with low-melting point agarose and processed into 300 μm sections with a Vibratome. Sections were individually placed in a multi-well plate, blocked with FcγR-blocking Ab (2.4G2) in PBS for 1 hour at RT, and then stained overnight at 4° C on a rocker in a 500 μl volume. PE- or AF647-labeled CD1d tetramer loaded with αGalCer (NIH tetramer core) was used at 1:200 dilution, and anti-CD31-BV421 antibody (clone 390, BioLegend) was added at 1:100 dilution. After washing, sections were fixed with paraformaldehyde, washed and embedded in Prolong Glass anitfade (ThermoFisher Scientific) under #1.5 coverslip.
Slides were imaged with ZEISS LSM780 confocal microscope using 40x/1.3NA EC Plan-Neofluar oil objective. Fluorescence of BV421, GFP, and PE was excited with 405 nm, 488 nm, and 561 nm laser lines, and emitted signals were collected on spectrally tuned bialkali PMT and GaAsP detectors. Single-stained controls were used to ensure no bleed-through between channels. Unloaded tetramers were used as a specificity control. Pixel size was set to 115 nm and Z-stacks were acquired with a 3 μm step size using tile scan function. Average field of view was 370×370×70 μm, and 3 regions were acquired per sample. Images were loaded into Imaris (9.7.2, Oxford Instruments) and 3D views were used to examine the location of cells positive for GFP and tetramer staining in respect to CD31-labeled vasculature. Channel brightness was adjusted to improve contrast in the same manner across all images. Orthogonal top views were created using Snapshot function in Imaris and figures were arranged in Photoshop.
in vitro iNKT cell activation and ELISA
Preparation of bacterial lysates and cell-free antigen-presentation assays have been described previously (Kinjo et al., 2011). Briefly, bacterial sonicates were incubated for 24 hours in microwells coated with soluble mouse CD1d. After washing, 1 × 105 mouse DN3A4–1.2 Vα14i NKT cell hybridoma cells, also known as 1.2 cells (Burdin et al., 2000) were added to the wells for 20 to 24 h. Mouse IL-2 was measured in the supernatants by enzyme-linked immunosorbent assay (BD Biosciences).
Quantification and statistical analysis
Graphs and statistical analyses were generated with Prism 7 and 9 software (Graphpad, 2016). Statistical details for each experiment can be found in the figure legends. Individual data points represent individual mice. The vertical bars in the graph represent standard deviation of the mean.
Supplementary Material
Supplementary Video 1. Nur77GFP-expressing iNKT cells during Streptococcus pneumoniae infection
Supplementary Video 2. Uptake of Streptococcus pneumoniae-GFP by Langerin-YFP cells in lung tissue
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-eF780 anti-TCRβ | ThermoFisher Scientific | H57-597, Cat# 47-5961-80, RRID:AB_1272209 |
| BV605 anti-CD8α | BioLegend | 53-6.7, Cat# 100743, RRID:AB_2561352 |
| BV605 anti-CD19 | BD Biosciences | 1D3, Cat# 563148, RRID:AB_2732057 |
| AF700 anti-CD4 | BioLegend | GK1.5, Cat# 100429, RRID:AB_493698 |
| PE Cy7 anti-ICOS | BioLegend | C398.4A, Cat# 313519, RRID:AB_10641839 |
| BV711 anti-CD49a | BD Biosciences | HA31/8, Cat# 564863, RRID:AB_2738987 |
| PE anti-GM-CSF | BioLegend | MP1-22E9, Cat# 505405, RRID:AB_315381 |
| AF700 anti-IL-17A | BioLegend | TC11-18H10.1, Cat# 506914, RRID:AB_536016 |
| PerCP-Cy5.5 anti-IFNγ | BioLegend | XMG1.2, Cat# 505821, RRID:AB_961361 |
| BV786 anti-CD45 | BD Biosciences | 30-F11, Cat# 564225, RRID:AB_2716861 |
| BV421 anti-Siglec-F | BD Biosciences | E50-2440, Cat# 562681, RRID:AB_2722581 |
| PE anti-Ly6G | BioLegend | 1A8, Cat# 127608, RRID:AB_1186099 |
| PerCP-Cy5.5 anti-CD11b | BD Biosciences | M1/70, Cat# 561114, RRID:AB_2033995 |
| APC anti-CD11c | ThermoFisher Scientific | N418, Cat# 17-0114-81, RRID:AB_469345 |
| BV711 anti-CD103 | BD Biosciences | M290, Cat# 564320, RRID:AB_2738743 |
| FITC anti-CD24 | BD Biosciences | M1/69, Cat# 553261, RRID:AB_394740 |
| PE Cy7 anti-CD64 | BioLegend | X54-5/7.1, Cat# 139314, RRID:AB_2563904 |
| PE anti-CD86 | BD Biosciences | GL-1, Cat# 553692, RRID:AB_394994 |
| AF488 anti-IL-23p19 | ThermoFisher Scientific | fc23cpg, Cat# 53-7023-82, RRID:AB_2574435 |
| APC anti-IL-1β | ThermoFisher Scientific | NJTEN3, Cat# 17-7114-80, RRID:AB_10670739 |
| FITC anti-TCRγδ | BioLegend | GL3, Cat# 118105, RRID:AB_313829 |
| anti-mouse CD16/CD32 | In house purification | 2.4G2 |
| BV421 anti-CD31 | BioLegend | 390, Cat# 102423, RRID:AB_2562186 |
| AF700 anti-CD45 | BioLegend | 30-F11, Cat# 103127, RRID:AB_493714 |
| Purified NA/LE Rat IgG2a | BD Biosciences | R35–95, Cat# 554687, RRID:AB_479678 |
| Ultra-LEAF purified anti-GM-CSF | BioLegend | MP1-22E9 |
| Purified Rat Anti-Mouse IL-2 | BD Biosciences | Cat# 554424, RRID:AB_395383 |
| Biotin Rat Anti-Mouse IL-2 | BD Biosciences | Cat# 554426, RRID:AB_395384 |
| Bacterial and Virus Strains | ||
| GFP- Streptococcus pneumoniae URF918 | Jan-Willem Veening (Kjos et al., 2015) | |
| S. pneumoniae serotype 19A isolates | Fadie T. Coleman, Joseph P. Mizgerd | |
| S. pneumoniae serotype 3 strain URF918 | Japan (Kawakami et al., 2003) | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| GolgiStop | BD Biosciences | Cat# 554724, RRID:AB_2869012 |
| Bromodeoxyuridine | BD Biosciences | Cat# 550891, RRID:AB_2868906 |
| Live dead yellow | ThermoFisher Scientific | Cat# L34968 |
| Ghost Dye™ UV 450 Viability Dye | Tonbo Biosciences | 13-0868-T500 |
| eBioscience™ Permeabilization Buffer (10X) | ThermoFisher Scientific | Cat# 00-8333-56 |
| mouse 5-OP-RU-MR1 tetramer | NIH Tetramer Core | |
| mouse 6-FP-MR1 tetramer | NIH Tetramer Core | |
| α-GalCer-CD1d tetramer | in-house preparation | |
| Soluble mouse CD1d | in-house preparation | |
| Peroxidase conjugated Streptavidin | Jackson Immuno Research | 016-030-084 |
| Critical Commercial Assays | ||
| mouse IL-2 ELISA | ||
| Experimental Models: Cell Lines | ||
| DN3A4-1.2 Va14i NKT cell hybridoma cell line | M. Bix, University of California, San Fransisco | |
| Experimental Models: Organisms/Strains | ||
| B6.129S(C)-Batf3tm1Kmm/J mice | The Jackson Laboratory | Cat# JAX:013755, RRID:IMSR_JAX:013755 |
| B6.129S-Csf2tm1Mlg/J mice | The Jackson Laboratory | Cat# JAX:026812, RRID:IMSR_JAX:026812 |
| B6.Cg-Tg(Itgax-cre)1-1Reiz/J mice | The Jackson Laboratory | Cat# JAX:008068, RRID:IMSR_JAX:008068 |
| Cd1d1f/f mice | Birkholz et al., 2015 | |
| Traj18−/− mice | Chandra et al., 2015 | |
| Langerin-Cre knockin mice | Zahner et al., 2011 | |
| Nur77GFP mice | Kristin A. Hogquist, University of Minnesota, Minneapolis, Minnesota, USA (Moran et al., 2011) | |
| C57BL/6J mice | The Jackson Laboratory | Cat# JAX:000664, RRID:IMSR_JAX:000664 |
| Software and Algorithms | ||
| FlowJo software | Tree Star, Ashland, OR | |
| Prism 7 and 9 software | Graphpad Software, San Diego, CA | |
Acknowledgements
We thank the Microscopy and Histology Core and Flow Cytometry Core at the La Jolla Institute for Immunology for technical assistance with these studies. We also thank the NIH Tetramer Core, Jan-Willem Veening and Kristin A. Hogquist for reagents. Supported by the American Lung Association Senior Research Training Fellowship RT-412662 to C.M.C; US National Institutes of Health R01 AI71922, AI105215, AI137230 to M.K.; T32 AI125179 to M.P.M.; R35 HL135756 and R01 AI115053 to J.P.M.; the German Research Foundation SFB1292, CL 419/2–2, CL 419/4–1, and the Research Center for Immunotherapy (FZI) Mainz to B.E.C.; Shared Instrumentation Grant (SIG) Program S10 OD018499 to the Flow Cytometry Core Facility at the La Jolla Institute for Immunology; S10 RR027366 for a FACSAria II cell sorter to Dr. Michael Croft; Tullie and Rickey Families SPARK Awards for Innovations in Immunology to C.M.C.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Arora P, Baena A, Yu KO, Saini NK, Kharkwal SS, Goldberg MF, Kunnath-Velayudhan S, Carreno LJ, Venkataswamy MM, Kim J, et al. (2014). A single subset of dendritic cells controls the cytokine bias of natural killer T cell responses to diverse glycolipid antigens. Immunity 40, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta KE, Cagnina RE, Wallace KL, Ramos SI, Mehrad B, and Linden J (2012). Leukocyte compartments in the mouse lung: distinguishing between marginated, interstitial, and alveolar cells in response to injury. J Immunol Methods 375, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral P, Eckl-Dorna J, Harwood NE, De Santo C, Salio M, Illarionov P, Besra GS, Cerundolo V, and Batista FD (2008). B cell receptor-mediated uptake of CD1d-restricted antigen augments antibody responses by recruiting invariant NKT cell help in vivo. Proc Natl Acad Sci U S A 105, 8345–8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelemy A, Ivanov S, Hassane M, Fontaine J, Heurtault B, Frisch B, Faveeuw C, Paget C, and Trottein F (2016). Exogenous Activation of Invariant Natural Killer T Cells by alpha-Galactosylceramide Reduces Pneumococcal Outgrowth and Dissemination Postinfluenza. mBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkholz AM, Girardi E, Wingender G, Khurana A, Wang J, Zhao M, Zahner S, Illarionov PA, Wen X, Li M, et al. (2015). A Novel Glycolipid Antigen for NKT Cells That Preferentially Induces IFN-gamma Production. J Immunol 195, 924–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, Cohen NR, Hsu FF, Besra GS, and Brenner MB (2011). Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med 208, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Sequeira RP, and Clarke TB (2017). The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun 8, 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdin N, Brossay L, Degano M, Iijima H, Gui M, Wilson IA, and Kronenberg M (2000). Structural requirements for antigen presentation by mouse CD1. Proc Natl Acad Sci U S A 97, 10156–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Gray J, Kiosses WB, Khurana A, Hitomi K, Crosby CM, Chawla A, Fu Z, Zhao M, Veerapen N, et al. (2018). Mrp1 is involved in lipid presentation and iNKT cell activation by Streptococcus pneumoniae. Nat Commun 9, 4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Zhao M, Budelsky A, de Mingo Pulido A, Day J, Fu Z, Siegel L, Smith D, and Kronenberg M (2015). A new mouse strain for the analysis of invariant NKT cell function. Nat Immunol 16, 799–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, Savage PB, Endo S, Yamamura T, Maaskant J, et al. (2011). Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest 121, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, and Becher B (2011). RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol 12, 560–567. [DOI] [PubMed] [Google Scholar]
- Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, Smyth MJ, and Godfrey DI (2008). Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci U S A 105, 11287–11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua DJ, and Tato CM (2010). Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol 10, 479–489. [DOI] [PubMed] [Google Scholar]
- Doisne JM, Soulard V, Becourt C, Amniai L, Henrot P, Havenar-Daughton C, Blanchet C, Zitvogel L, Ryffel B, Cavaillon JM, et al. (2011). Cutting edge: crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1.1-invariant NKT cells to bacteria. J Immunol 186, 662–666. [DOI] [PubMed] [Google Scholar]
- El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, and Rostami A (2011). The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol 12, 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, and Kronenberg M (2016). Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nature Immunology 17, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvy BA, and Harmsen AG (1996). The importance of neutrophils in resistance to pneumococcal pneumonia in adult and neonatal mice. Inflammation 20, 499–512. [DOI] [PubMed] [Google Scholar]
- Ghigo C, Mondor I, Jorquera A, Nowak J, Wienert S, Zahner SP, Clausen BE, Luche H, Malissen B, Klauschen F, et al. (2013). Multicolor fate mapping of Langerhans cell homeostasis. J Exp Med 210, 1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, Yu E, Li Y, Tarumoto N, Pei B, Wang J, Illarionov P, Kinjo Y, Kronenberg M, and Zajonc DM (2011). Unique Interplay between Sugar and Lipid in Determining the Antigenic Potency of Bacterial Antigens for NKT Cells. PLoS Biology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, and Moody DB (2015). The burgeoning family of unconventional T cells. Nat Immunol 16, 1114–1123. [DOI] [PubMed] [Google Scholar]
- Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W, Kretzer NM, Briseno CG, Durai V, Bagadia P, et al. (2015). Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol 16, 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Teslovich N, Mola AR, Polidoro RB, Nathan A, Kim H, Hannes S, Slowikowski K, Watts GFM, Korsunsky I, et al. (2019). Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat Commun 10, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harly C, Guillaume Y, Nedellec S, Peigne CM, Monkkonen H, Monkkonen J, Li J, Kuball J, Adams EJ, Netzer S, et al. (2012). Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human gammadelta T-cell subset. Blood 120, 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann N, McMurtrey C, Sorensen ML, Huber ME, Kurapova R, Coleman FT, Mizgerd JP, Hildebrand W, Kronenberg M, Lewinsohn DM, et al. (2018). Riboflavin Metabolism Variation among Clinical Isolates of Streptococcus pneumoniae Results in Differential Activation of Mucosal-associated Invariant T Cells. Am J Respir Cell Mol Biol 58, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassane M, Demon D, Soulard D, Fontaine J, Keller LE, Patin EC, Porte R, Prinz I, Ryffel B, Kadioglu A, et al. (2017). Neutrophilic NLRP3 inflammasome-dependent IL-1beta secretion regulates the gammadeltaT17 cell response in respiratory bacterial infections. Mucosal Immunol 10, 1056–1068. [DOI] [PubMed] [Google Scholar]
- Hassane M, Jouan Y, Creusat F, Soulard D, Boisseau C, Gonzalez L, Patin EC, Heuze-Vourc’h N, Sirard JC, Faveeuw C, et al. (2020). Interleukin-7 protects against bacterial respiratory infection by promoting IL-17A-producing innate T-cell response. Mucosal Immunol 13, 128–139. [DOI] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. (2008). Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel KL, Tyznik AJ, Kronenberg M, and Hogquist KA (2014). Antigen-dependent versus -independent activation of invariant NKT cells during infection. J Immunol 192, 5490–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Fontaine J, Paget C, Macho Fernandez E, Van Maele L, Renneson J, Maillet I, Wolf NM, Rial A, Leger H, et al. (2012). Key role for respiratory CD103(+) dendritic cells, IFN-gamma, and IL-17 in protection against Streptococcus pneumoniae infection in response to alpha-galactosylceramide. J Infect Dis 206, 723–734. [DOI] [PubMed] [Google Scholar]
- Ivanov S, Paget C, and Trottein F (2014). Role of non-conventional T lymphocytes in respiratory infections: the case of the pneumococcus. PLoS Pathog 10, e1004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochems SP, de Ruiter K, Solorzano C, Voskamp A, Mitsi E, Nikolaou E, Carniel BF, Pojar S, German EL, Reine J, et al. (2019). Innate and adaptive nasal mucosal immune responses following experimental human pneumococcal colonization. J Clin Invest 129, 4523–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Yamamoto N, Kinjo Y, Miyagi K, Nakasone C, Uezu K, Kinjo T, Nakayama T, Taniguchi M, and Saito A (2003). Critical role of Valpha14+ natural killer T cells in the innate phase of host protection against Streptococcus pneumoniae infection. Eur J Immunol 33, 3322–3330. [DOI] [PubMed] [Google Scholar]
- Kawasaki N, Vela JL, Nycholat CM, Rademacher C, Khurana A, van Rooijen N, Crocker PR, Kronenberg M, and Paulson JC (2013). Targeted delivery of lipid antigen to macrophages via the CD169/sialoadhesin endocytic pathway induces robust invariant natural killer T cell activation. Proc Natl Acad Sci U S A 110, 7826–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairallah C, Chu TH, and Sheridan BS (2018). Tissue Adaptations of Memory and Tissue-Resident Gamma Delta T Cells. Front Immunol 9, 2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Lee S, Berg RE, Simecka JW, and Jones HP (2013). Interleukin-23 (IL-23) deficiency disrupts Th17 and Th1-related defenses against Streptococcus pneumoniae infection. Cytokine 64, 375–381. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Illarionov P, Vela J, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, et al. (2011). Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nature Immunology 12, 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, Kitano N, and Kronenberg M (2013). The role of invariant natural killer T cells in microbial immunity. J Infect Chemother 19, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, et al. (2006). Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol 7, 978–986. [DOI] [PubMed] [Google Scholar]
- Kjos M, Aprianto R, Fernandes VE, Andrew PW, van Strijp JA, Nijland R, and Veening JW (2015). Bright fluorescent Streptococcus pneumoniae for live-cell imaging of host-pathogen interactions. J Bacteriol 197, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, Levy E, Dusseaux M, Meyssonnier V, Premel V, et al. (2010). Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol 11, 701–708. [DOI] [PubMed] [Google Scholar]
- Lee Y, Wang H, Starrett GJ, Phuong V, Jameson SC, and Hogquist KA (2015). Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity 43, 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Holzapfel KL, Zhu J, Jameson SC, and Hogquist KA (2013). Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 14, 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Liu M, Wang J, Fan H, Yang D, Zhang L, Gu X, Nie J, Chen Z, Corbett AJ, et al. (2020). IL-17 production by tissue-resident MAIT cells is locally induced in children with pneumonia. Mucosal Immunol 13, 824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J, Debord KL, Ismail N, Goff RD, Cantu C 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. (2005). Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434, 525–529. [DOI] [PubMed] [Google Scholar]
- Meierovics A, Yankelevich WJ, and Cowley SC (2013). MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci U S A 110, E3119–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, and Hogquist KA (2011). T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 208, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MP, Engel I, Seumois G, Herrera-De la Mata S, Rosales SL, Sethi A, Logandha Ramamoorthy Premlal A, Seo GY, Greenbaum J, Vijayanand P, et al. (2021). Transcriptome and chromatin landscape of iNKT cells are shaped by subset differentiation and antigen exposure. Nat Commun 12, 1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamatsu M, Yamamoto N, Hatta M, Nakasone C, Kinjo T, Miyagi K, Uezu K, Nakamura K, Nakayama T, Taniguchi M, et al. (2007). Role of interferon-gamma in Valpha14+ natural killer T cell-mediated host defense against Streptococcus pneumoniae infection in murine lungs. Microbes Infect 9, 364–374. [DOI] [PubMed] [Google Scholar]
- Nakasone C, Yamamoto N, Nakamatsu M, Kinjo T, Miyagi K, Uezu K, Nakamura K, Higa F, Ishikawa H, O’Brien R L, et al. (2007). Accumulation of gamma/delta T cells in the lungs and their roles in neutrophil-mediated host defense against pneumococcal infection. Microbes Infect 9, 251–258. [DOI] [PubMed] [Google Scholar]
- O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. [DOI] [PubMed] [Google Scholar]
- Paget C, Ivanov S, Fontaine J, Renneson J, Blanc F, Pichavant M, Dumoutier L, Ryffel B, Renauld JC, Gosset P, et al. (2012). Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: potential role in protection against lung epithelial damages. J Biol Chem 287, 8816–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachitskaya AV, Hansen AM, Horai R, Li Z, Villasmil R, Luger D, Nussenblatt RB, and Caspi RR (2008). Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J Immunol 180, 5167–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reutershan J, Basit A, Galkina EV, and Ley K (2005). Sequential recruitment of neutrophils into lung and bronchoalveolar lavage fluid in LPS-induced acute lung injury. Am J Physiol Lung Cell Mol Physiol 289, L807–815. [DOI] [PubMed] [Google Scholar]
- Ribot JC, Lopes N, and Silva-Santos B (2021). gammadelta T cells in tissue physiology and surveillance. Nat Rev Immunol 21, 221–232. [DOI] [PubMed] [Google Scholar]
- Ritchie ND, Ritchie R, Bayes HK, Mitchell TJ, and Evans TJ (2018). IL-17 can be protective or deleterious in murine pneumococcal pneumonia. PLoS Pathog 14, e1007099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosine N, and Miceli-Richard C (2020). Innate Cells: The Alternative Source of IL-17 in Axial and Peripheral Spondyloarthritis? Front Immunol 11, 553742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothchild AC, Jayaraman P, Nunes-Alves C, and Behar SM (2014). iNKT cell production of GM-CSF controls Mycobacterium tuberculosis. PLoS Pathog 10, e1003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, Richer W, Goubet AG, Daviaud C, Menger L, et al. (2019). A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med 216, 133–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom A, Peigne CM, Leger A, Crooks JE, Konczak F, Gesnel MC, Breathnach R, Bonneville M, Scotet E, and Adams EJ (2014). The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vgamma9Vdelta2 T cells. Immunity 40, 490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Jackson JT, Markey KA, Brady HJ, Hill GR, Macdonald KP, Nutt SL, and Belz GT (2013). CD8alpha+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood 121, 1574–1583. [DOI] [PubMed] [Google Scholar]
- St Leger AJ, Hansen AM, Karauzum H, Horai R, Yu CR, Laurence A, Mayer-Barber KD, Silver P, Villasmil R, Egwuagu C, et al. (2018). STAT-3-independent production of IL-17 by mouse innate-like alphabeta T cells controls ocular infection. J Exp Med 215, 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwede K, Tempelhof O, Bolte K, Maus R, Bohling J, Ueberberg B, Langer F, Christman JW, Paton JC, Ask K, et al. (2011). Local delivery of GM-CSF protects mice from lethal pneumococcal pneumonia. J Immunol 187, 5346–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SY, Scanlon ST, Griewank KG, Constantinides MG, Savage AK, Barr KA, Meng F, Luster AD, and Bendelac A (2011). PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. J Exp Med 208, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, Luksic I, Nair H, McAllister DA, Campbell H, et al. (2018). Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Health 6, e744–e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H, et al. (2012). Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol 10, e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroe PC, Finkelstein JA, Ray GT, Linder JA, Johnson KM, Rifas-Shiman S, Moore MR, and Huang SS (2012). Aging population and future burden of pneumococcal pneumonia in the United States. J Infect Dis 205, 1589–1592. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ahyi AN, Pepper-Cunningham ZA, Ferrari JD, Wilson AA, Jones MR, Quinton LJ, and Mizgerd JP (2014). Roles of lung epithelium in neutrophil recruitment during pneumococcal pneumonia. Am J Respir Cell Mol Biol 50, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner SP, Kel JM, Martina CA, Brouwers-Haspels I, van Roon MA, and Clausen BE (2011). Conditional deletion of TGF-betaR1 using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity. J Immunol 187, 5069–5076. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Clarke TB, and Weiser JN (2009). Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest 119, 1899–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Svensson MND, Venken K, Chawla A, Liang S, Engel I, Mydel P, Day J, Elewaut D, Bottini N, et al. (2018). Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat Commun 9, 2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. Nur77GFP-expressing iNKT cells during Streptococcus pneumoniae infection
Supplementary Video 2. Uptake of Streptococcus pneumoniae-GFP by Langerin-YFP cells in lung tissue
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.